Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) modification of proteins has been shown to be involved in many different cellular processes, such as cell cycle control, nutrient sensing, signal transduction, stress response and transcriptional regulation. Cells have developed complex regulatory systems in order to regulate gene expression appropriately in response to environmental and intracellular cues. Control of eukaryotic gene transcription often involves post-translational modification of a multitude of proteins including transcription factors, basal transcription machinery, and chromatin remodeling complexes to modulate their functions in a variety of manners. In this review we describe the emerging functional roles for and techniques to detect and modulate the O-GlcNAc modification and illustrate that the O-GlcNAc modification is intricately involved in at least seven different general mechanisms for the control of gene transcription.
Keywords: O-GlcNAc, transcriptional regulation, post-translational modification, review
INTRODUCTION
Cells have developed a highly regulated system to respond to environmental and intracellular signals to specifically and coordinately express gene products [1, 2]. Surprisingly, the number of protein-coding genes in a genome does not reflect organism complexity, thus it has been hypothesized that increased complexity in gene regulation leads to increased organism complexity [3]. Indeed, the regulation of eukaryotic gene transcription involves a multitude of proteins including transcription factors, basal transcription machinery, and chromatin remodeling complexes [4]. An additional layer of complexity results from a wide variety of post-translational modifications on regulatory proteins [5]. Herein, we describe the emerging role of the O-GlcNAc post-translational modification of nuclear/cytosolic proteins in the regulation of transcription.
In the 1980’s, Hart and coworkers reported a nucleocytoplasmic, post-translational sugar modification on serine and/or threonine residues of polypeptides, O-GlcNAc [6–8]. All metazoans currently studied contain the O-GlcNAc modification on proteins involved in many different cell processes, such as cell cycle control [9–11], nutrient sensing [12], signal transduction [13–16], stress response [17, 18], and transcriptional regulation (the focus of this review) [19–23]. Furthermore, O-GlcNAc transferase (OGT) [24–26], the enzyme required for O-GlcNAc addition, is required for mouse embryonic stem cell viability, emphasizing the importance of this modification [27]. O-GlcNAc is more akin to phosphorylation than complex glycosylation in that it is not elongated, its cycling enzymes, OGT and O-GlcNAcase (OGA) [28, 29], are nucleocytoplasmic, it is dynamic and inducible, and it can regulate intracellular protein activity, localization, stability, and molecular interactions. O-GlcNAc is often found on the same residues as known phosphorylation sites, suggesting reciprocity between the modifications in some cases, Fig. (1) [13, 30, 31]. However, unlike phosphorylation, which is modulated by hundreds of kinases and phosphatases, the cycling of the O-GlcNAc modification is accomplished by the gene products of single genes for OGT and OGA in most metazoans.
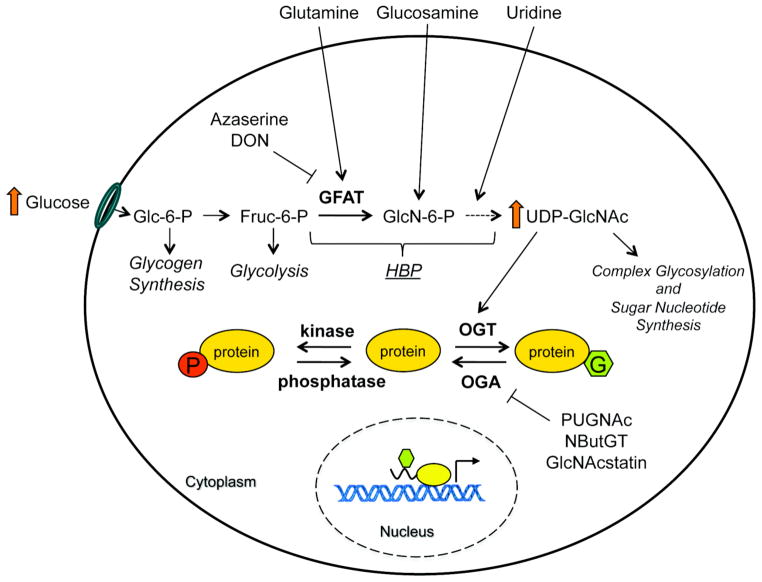
Fig. 1. Modulation of cellular O-GlcNAc levels using HBP flux and specific enzyme inhibitors.
The end product of the HBP, UDP-GlcNAc, is sensitive to changes in nutrient levels. Glucosamine enters the HBP downstream of the rate-limiting enzyme GFAT to elevate UDP-GlcNAc levels. The use of the amidotransferase inhibitors azaserine or DON decreases UDP-GlcNAc levels. Proteins can be reciprocally modified by glycosylation and phosphorylation. However, unlike phosphorylation, which is regulated by hundreds of kinases and phosphatases, O-GlcNAc modification is cycled by the result of gene products from only two genes, ogt and oga. OGT transfers the GlcNAc onto serine and threonine residues of nuclear and cytosolic proteins and is responsive to changes in UDP-GlcNAc concentrations. Global O-GlcNAc levels can also be raised by the use of OGA inhibitors PUGNAc, NButGT and GlcNAc-statin. Enzymes are depicted in bold and biological pathways are in italics.
O-GlcNAc modification of transcription regulatory proteins could fine tune their regulation in response to nutrient levels in the cell because the synthesis of its sugar donor, UDP-GlcNAc, via the hexosamine biosynthetic pathway (HBP), responds to amino acid, fatty acid, nucleotide and glucose metabolism [12, 21]. There are several ways to modulate O-GlcNAc levels on proteins (for review see [32]) Fig. (1). OGT is responsive to physiological levels of UDP-GlcNAc, so increased HBP flux by hyperglycemia or by the addition of glucosamine results in globally elevated levels of O-GlcNAc modification [33]. Decreased O-GlcNAc levels can be achieved by blocking glutamine-fructose-6-phosphate transaminase (GFAT), the rate limiting enzyme of the HBP, using the pharmacological inhibitors azaserine or 6-diazo-5-oxo-L-norleucine (DON) or by decreasing glucose levels. However, the alteration of HBP flux may lead to off-target effects as azaserine and DON are general amidotransferase inhibitors. A more specific way to alter global O-GlcNAc levels is by the use of pharmacological OGA inhibitors such as the widely used O-(2-acetamido-2-deoxy-D-glucopyrano-sylidene)amino-N-phenylcarbamate (PUGNAc) [34], or the more specific inhibitors 1,2-dideoxy-2′-propyl-α-D-gluco-pyranoso-[2,1-D]-Δ2′-thiazoline (NButGT) [35] and GlcNAcstatin [36, 37]. Several OGT inhibitors have also been recently characterized [38] although their specificity and in vivo utility has not been adequately explored. Alternatively, O-GlcNAc steady state levels can be modulated genetically by over expression or knockdown of OGT and/or OGA.
O-GlcNAc DETECTION AND SITE MAPPING
Over the last 20 years more than 400 proteins have been shown to be modified by O-GlcNAc using a variety of detection methods [21, 39–42]. Interestingly, most RNA Polymerase II transcription factors are glycosylated; many of which respond to nutrient abundance [19, 43]. There are several methods to identify O-GlcNAc modification of proteins [44] and the relevant methods will be briefly discussed here. The first step in identifying O-GlcNAc modified proteins generally involves modification-specific enrichment. Detection or enrichment of O-GlcNAc modified proteins can be achieved using O-GlcNAc specific antibodies, such as RL2 [45, 46] and CTD110.6 [47], and by lectin-blotting or chromatography using succinylated Wheat Germ Agglutinin, a terminal GlcNAc-binding lectin. The presence of O-GlcNAc on proteins can also be determined by labeling with radiolabeled galactose using purified β-1,4-galactosyltransferase (GalT), a galactosyltransferase that transfers galactose onto terminal GlcNAc moieties [6]. Click-iT™ chemistry available from Invitrogen offers two different approaches for in vitro and in vivo labeling of O-GlcNAc residues. In vitro labeling takes advantage of a mutant form of GalT that transfers ketone-modified galactose onto the GlcNAc residues of proteins [48]. The ketone group introduces a chemically reactive group that can be tagged with biotin and then enriched with streptavidin [48]. Using an in vivo approach, introduction of N-azidoacetylglucosamine (GlcNAz) into the cells allows this azidosugar to be converted via the salvage pathway to UDP-GlcNAz and transferred onto proteins by OGT [49]. The azido group of GlcNAz acts as a bio-orthogonal handle for enrichment by the addition of functional groups using the Staudinger ligation [50]. However, there are limitations to using the in vivo approach, since it requires the UDP-GlcNAz to compete with the existing UDP-GlcNAc in the cell. These O-GlcNAc enrichment techniques can be combined with mass spectrometry to identify the actual residues of modification [54–56]. Proteomic efforts in this area have identified hundreds of modified polypeptides with proteins involved in transcriptional regulation being a major class [45, 48, 54–56]; however, only about 75 proteins have had their sites of modification mapped. The modification is extremely labile, small, uncharged, and usually substoichiometric [32, 51] making detection difficult using standard mass spectrometry techniques.
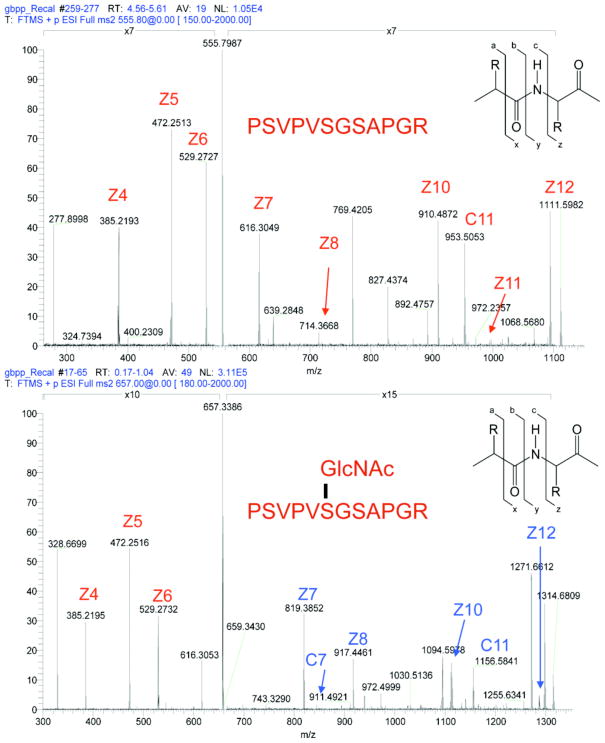
Recently, several methods have been developed to make O-GlcNAc site-mapping by mass spectrometry (MS) feasible in biologically relevant tissues. O-GlcNAc enrichment techniques can be combined with mass spectrometry to identify the actual residues of modification [41, 52]. Collision-induced dissociation (CID) mass spectrometry tends to cleave PTMs, so a non-labile tag added to the site of O-GlcNAc modification facilitates identification. Site-mapping studies using β-elimination followed by Michael addition with dithiothreitol attach a non-labile tag to the site of O-GlcNAc modification so it can be identified by CID MS [42]. In addition, enrichment of O-GlcNAc containing peptides by chemoenzymatic labeling assists in detection [39, 48, 49, 51]. An advantage of these methods is that more O-GlcNAc peptides, which are generally substoichiometric in a total peptide pool, can be detected leading to a more prolific site mapping experiment. The development of electron transfer dissociation fragmentation and related dissociation techniques that often retain CID-labile PTMs have allowed for the identification of O-GlcNAc modified fragments directly [53, 54]. In Fig. (2), we show an example of an electron dissociation technique (electron capture dissociation) for definitively mapping a site of O-GlcNAc on UL32, a synthetic glycosylated peptide, to one particular residue on a peptide containing three potential sites of attachment. Unlike CID, the fragmented peptides containing the modified amino acid retain the mass of the sugar. Electron dissociation techniques are an emerging technology for O-GlcNAc site-mapping that show great promise [51, 54–57].
Fig. 2. Site-mapping of O-GlcNAc sites is facilitated by electron dissociation techniques.
UL32, a synthetic O-GlcNAc modified protein, is efficiently fragmented and the site of modification (from three possible sites) is easily assigned via electron capture dissociation. When comparing the spectra from unglycosylated (top) and glycosylated peptide (bottom), singly charged fragments retaining the O-GlcNAc modified serine (shown in BLUE) show an increase in mass to charge of 203 daltons, the weight of a single GlcNAc residue.
O-GlcNAc REGULATION OF EUKARYOTIC GENE EXPRESSION
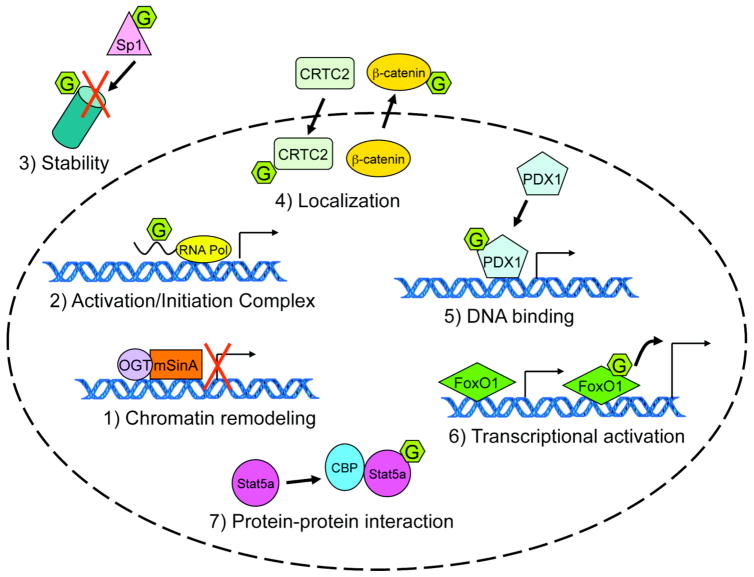
Specialized transcription factor regulation occurs through the actions of multiple post-translational modifications (PTMs) (reviewed in [5, 58]) such as phosphorylation [59, 60], SUMOylation [61], acetylation [62], and the focus of this review, O-GlcNAc modification. Transcriptional control can occur via at least seven different general mechanisms, Fig. (3), and examples of O-GlcNAc modification participation in each of these regulatory steps are explored below.
Fig. 3. Transcriptional regulation by O-GlcNAc can occur via seven different mechanisms.
The O-GlcNAc modification has been demonstrated to regulate transcription by modulating proteins involved in chromatin remodeling and transcriptional initiation, as well as protein-protein associations, localization, stability, DNA binding, and transactivation capacity of individual transcription factors.
Chromatin Remodeling
Chromatin not only provides compact packaging for DNA, it also regulates transcription. For transcription to occur, nucleosomes, the histone proteins/DNA subunits of chromatin, must be positioned to allow transcriptional machinery to access both the promoter and upstream regulatory elements and to allow transcriptional elongation [63]. Access to DNA is regulated by chromatin remodeling enzymes, which recognize PTM’s on histones [63, 64]. Acetylation, the most well studied histone PTM, is added by histone acetyltransferases and removed by histone deacetylases (HDACs) [63, 64]. Transcriptional regulation is associated with altered histone acetylation and movement, restructuring, and ejection of nucleosomes [63]. Methylation of certain histone lysines by histone methyltransferases also plays a role in both gene silencing and activation [65]. The actual chromatin remodeling enzymes are thought to be regulated by PTM’s such as phosphorylation and acetylation [63]. Several studies have found glycosylation affects the regulation of chromatin remodeling [66, 67].
The first evidence for O-GlcNAc’s role in transcriptional regulation was the observation that Drosophila melanogaster polytene chromosomes contain more O-GlcNAc modified proteins at the transcriptionally repressed condensed regions than at the active puff regions of the chromatin [68]. Further studies implicated OGT in transcriptional repression through the identification of an interaction between mSin3a and OGT [66]. mSin3a is a corepression scaffolding protein that forms a multi-protein complex with HDAC and can be recruited by transcription factors to modify histones and repress transcription [69]. Several transcription factors involved in cell survival and apoptosis, such as p53, an O-GlcNAc modified protein [70, 71], recruit mSin3a [72]. The paired amphipathic helix domain 4 of mSin3A was shown to bind to the tetracopeptide repeat (TPR) domain of OGT, suggesting a mechanism where mSin3a recruits OGT for gene silencing [66]. Although both the TPR and catalytic domain of OGT promote transcriptional repression, catalytically active OGT is required for full transcriptional repression [66]. The other proteins in the repression complex, mSin3A and HDAC1, were also found to be O-GlcNAc modified [66] and, although the functional significance is still to be elucidated, may explain why the catalytic activity of OGT is necessary. In agreement with the data seen in Drosophila melanogaster polytene chromosomes, a chromatin immunoprecipitation assay showed an increase in both O-GlcNAc modified proteins and mSin3a presence on the promoters of silenced genes [66]. In another study, OGT was found to interact with both mSin3A and Sp3 and was associated with the prevention of transcriptional repression of angiopoietin-2 during hyperglycemic conditions [73]. However, it is unclear whether the association of mSin3A with Sp3 or the direct O-GlcNAc modification of Sp3 was responsible for the transcriptional activation of angiopoietin-2 [73].
A landmark study recently identified a key role for O-GlcNAc in modulating the activity of MLL5, a histone lysine methyltransferase [67]. MLL5 was found to co-activate RARα (retinoic acid receptor α) induction of promyelocyte-like differentiation into granulocyte-like HL60 cells. OGT forms a complex with MLL5. Elevation of O-GlcNAc levels in undifferentiated HL60 cells increase retinoic acid (RA) stimulated differentiation. Upon RA stimulation, RARα activates the expression of C/EBPε, a major differentiation facilitating transcription factor. Expression of a T440A, the major site of O-GlcNAc modification, mutant of MLL5 failed to activate C/EBPε expression and enhancement of the RA effect on differentiation. Further experiments established that OGT is necessary for MLL5 methylation of H3K4, which allows the transcriptional activation of pro-differentiation genes [67]. Thus, this manuscript clearly illustrates a causal relationship between O-GlcNAc modification of a protein and its enzymatic activity, which is directly involved in chromatin remodeling.
Transcriptional Initiation and Elongation
O-GlcNAc modification has also been implicated in regulating transcriptional initiation via RNA Polymerase II (RNAP II). Transcriptional initiation is achieved in part by several general transcription factors that recruit hypophosphorylated RNAP II to the core promoter and form a preinitiation complex [74]. RNAP II has a carboxyl terminal domain (CTD) that consists of several tandem consensus sequence repeats that are modified by phosphate and O-GlcNAc [47, 74]. The phosphorylation of the CTD is involved in promoter clearance, passage through promoter proximal pause sites, stabilization of the elongation complex, and recruitment of mRNA processing machinery [2]. The CTD exists in two states with regards to its phosphorylation status; IIO is the phosphorylated form and is found predominantly in the elongation complex, while IIA is the unphosphorylated form generally found in the initiation complex [74].
When purified fractions of RNAP II were labeled with GalT, it was shown that only the IIA form, the unphosphorylated form, of CTD was modified with O-GlcNAc [75]. In an additional study, OGT failed to label a CTD consensus sequence that had been phosphorylated in vitro by CTD kinase, and CTD kinase would not label a CTD consensus sequence that had been synthetically glycosylated on the Thr 4 of each repeat, suggesting mutual exclusivity between the modifications [47]. This yin-yang relationship between phosphorylation and O-GlcNAc on the CTD suggests that the O-GlcNAc modification may prevent elongation from occurring by blocking phosphorylation or may help to recycle RNAP II after elongation has occurred to allow the complex to reattach to the promoter [47]. Further in vivo investigation is needed to clarify the function of glycosylation on the CTD of RNAP II; however, the suggestion that glycosylation regulates transcription initiation is not unprecedented.
Degradation
The proper maintenance of transcription factor levels in cells is often accomplished by degradation via the ubiquitin-proteasome system [76]. Degradation is achieved by two steps: first, ubiquitin is added by an E3 ubiquitin ligase to lysine residues on proteins targeted for destruction, and second, the polyubiquitinylated proteins are degraded by the 26S proteasome [77]. The 26S proteasome is comprised of two major subcomplexes: two 19S regulatory particle caps and the 20S catalytic core [77]. The 20S core catalyzes the proteolysis of protein substrates. The 19S particle caps contain six ATPases that work to recognize and unfold substrates for entry into the 20S core [77, 78]. Glycosylation and phosphorylation have been suggested to regulate both the activity of the proteasome and the targeting of proteins to the proteasome [23, 79].
The most well-studied O-GlcNAc modified transcription factor is Sp1, a ubiquitous transcription factor for TATA-less genes. Sp1 target genes are involved in many different processes including metabolism, cell proliferation and oncogenesis [80]. In 1988, Jackson and Tjian determined that Sp1 is O-GlcNAc modified [81]. Since then, glycosylation has been described to affect Sp1 function by modulating its stability, protein-protein interactions, DNA binding, and localization [23, 81]. An initial study found that glucose starvation plus adenylate cyclase activation in normal rat kidney cells resulted in decreased Sp1 protein levels and Sp1 hypoglycosylation [82]. The authors suggested that hypoglycosylation of Sp1 promotes degradation through a proteasome-like mechanism [82]. However, it was subsequently shown that the degree of Sp1 glycosylation was independent of its degradation, and instead it was discovered that OGT inhibits and OGA activates the ATPase activity of the 19S regulatory particle caps of the proteasome [79]. OGT catalytic activity is necessary for this inhibition of the proteasome [79]. O-GlcNAc modification of Rpt2, one of the six ATPases present in the 19S cap, blocks the ATPase activity that provides the energy for hydrophobic proteins to unfold and be translocated inside the catalytic core of the proteasome for degradation [79]. Subsequently, in the 26S proteasome of Drosophila melanogaster, five out of nineteen regulatory subunits of the 19S cap and nine out of fourteen subunits of the 20S catalytic core were shown to be O-GlcNAc modified by immunoblotting with monoclonal antibodies and wheat germ agglutinin [83]. O-GlcNAc modification of the proteasome may function to regulate protein degradation in response to nutrient availability, which could potentially regulate transcription by altering transcription factor steady-state levels of transcription factors, such as in the case of Sp1.
Besides its global effect on proteasome function, O-GlcNAc modification is also associated with altered stability of individual transcription factors such as c-Myc, estrogen receptor β (ER-β), and p53. These transcription factors have been shown to be regulated by the ubiquitin proteasome pathway via phosphorylation [71, 84, 85]. A reciprocal relationship between phosphorylation and O-GlcNAc modification is observed for both c-Myc and ER-β [84–87].
c-Myc, a proto-oncogene, was one of the earliest proteins to be site-mapped for O-GlcNAc modification. c-Myc is O-glycosylated on Thr 58 in the N-terminal transcriptional activation domain region [87, 88]. Thr 58 is in the major region of mutation seen in Burkitt’s lymphomas [89], and phosphorylation at this site leads to c-Myc polyubiquitinylation and degradation [90]. T58A mutants have increased stability, suggesting that glycosylation via blocking of phosphorylation on this residue may result in increased stability, although the specific mechanism is not known [85]. c-Myc is targeted by several signaling pathways and regulates a plethora of target genes involved in cell proliferation, differentiation, and apoptosis [91]. Thus, PTM’s on c-Myc including phosphorylation and glycosylation appear to influence the specificity and stability of c-Myc [90].
ER-β, an ER-α homologue, is important in many processes such as growth and development, response to stress, and control of energy balance [92, 93]. Phosphorylation of ER-α by GSK-3 (glycogen synthase kinase-3) promotes its stability and full transcriptional activation, and this regulation of ER-α has emerged as an important theme in estrogen signaling [94, 95]. Although this theme is not as well-studied for ER-β, phosphorylation of the ER-β AF-1 domain has been shown to affect its proteasome-dependent degradation [96]. Glycosylation may also play a role in regulating ER-β stability. Ser 16 of ER-β is reciprocally glycosylated and phosphorylated [86]. S16A and S16E mutants were generated to mimic no modification and constitutive phosphorylation, respectively. The S16A mutant had a longer half-life (15–16 hours) and the S16E mutant had a shorter half-life (4–5 hours) than the wild type ER-β (7–8 hours), which suggests that glycosylation may promote ER-β stability by blocking phosphorylation and subsequent targeting for degradation [84].
p53 is a tumor suppressor gene required for cell cycle arrest and apoptosis. Normally, cellular p53 levels, which are highly regulated, are kept very low via degradation by the ubiquitin-dependent proteosome system [97]. Factors such as DNA damage or the activation of oncogenes induce increased p53 stability and activation [97]. p53 is found to be mutated and dysfunctional in many human cancers [97]. An early study determined p53 is O-GlcNAc modified and the presence of the modification was suggested to increase p53’s ability to bind DNA [70]. A later study determined a role for O-GlcNAc modification in p53 stability [71]. p53 is O-GlcNAc modified on Ser 149, which is located on the DNA binding domain. Mutation of Ser 149 to alanine increases Thr 155 phosphorylation. Since elevated Thr 155 phosphorylation is associated with increased degradation of p53, Ser 149 glycosylation has been hypothesized to play an important role in p53 stabilization [71].
Localization
Several papers have been published showing a functional relationship between O-GlcNAc modification and nuclear or cytoplasmic localization [98–101]. Transcription factors must localize to the nucleus to activate transcription, so sequestering latent transcription factors to the cytoplasm provides an additional mechanism of transcriptional regulation. In response to signals, latent cytoplasmic transcription factors are activated by several mechanisms, many of which depend on phosphorylation or other PTM’s, such as glycosylation [1].
The transducer of regulated cyclic adenosine 3′-5′ monophosphate response element (CREB) protein (CRTC2) associates with CREB to regulate gluconeogenic genes, including glucose-6-phosphatase (G6Pase), in response to insulin and glucagon [102]. Gluconeogenic genes fail to be inactivated during chronic hyperglycemic conditions, leading to gluconeogenesis during energy prevalent conditions. CRTC2 associates with CREB to bind the cAMP response element on the G6Pase promoter. When insulin is present, SIK2 (salt-induced kinase 2) is activated by Akt and phosphorylates Ser 171 of CRTC2, which allows it to be sequestered in the cytoplasm by 14-3-3 proteins and targeted for degradation [103]. Glucagon signaling prevents SIK2 from phosphorylating CRTC2 [104]. The dephosphorylated form of CRTC2 is no longer sequestered in the cytosol by 14-3-3 proteins and is free to translocate to the nucleus and activate transcription of target genes. CRTC2 is reciprocally modified by O-GlcNAc and phosphate on Ser 171 and Ser 70, suggesting alternative roles for the modifications. Hyperglycemia or elevating O-GlcNAc levels via genetic or pharmacological methods decreases CRTC2 phosphorylation and increases its O-GlcNAc modification, nuclear localization, and G6Pase promoter activation [98]. Mutation of these sites to aspartate, which simulates phosphorylation, prevents hyperglycemic stimulation of G6Pase promoter activation. Overexpression of OGA in the liver of diabetic db/db mice restores their gluconeogenic profiles to nearly normal levels, suggesting that elevated O-GlcNAc levels contribute to the nuclear localization of CRTC2 and the subsequent deregulation of gluconeogenesis during hyperglycemic conditions [98].
O-GlcNAc modification appears to be required for the nuclear localization of NeuroD1 (neurogenic differentiation 1). NeuroD1 is required for the terminal differentiation of neurons and for the development and insulin production of pancreatic β-cells [105]. Hyperglycemia results in increased phosphorylation of NeuroD1 on Ser 274, nuclear translocation, and increased NeuroD1 binding to the insulin promoter. Mutation to S274A results in the cytoplasmic accumulation of NeuroD1 even in hyperglycemic conditions [99]. Elevation of global O-GlcNAc levels using PUGNAc increased NeuroD1 nuclear localization, binding to the insulin promoter, and insulin expression even in normoglycemic conditions, suggesting that phosphorylation and O-GlcNAc modification are acting cooperatively. This result may be due to a similar increase in NeuroD1 glycosylation in both hyperglycemic and PUGNAc-treated conditions. OGT was found to associate with NeuroD1 in hyperglycemic conditions and OGA was found to associate in normoglycemic conditions [100]. Identifying the NeuroD1 glycosylation sites would help to distinguish whether the effect on localization and subsequent insulin transcriptional activation results from the specific glycosylation of NeuroD1, the interplay between glycosylation and phosphorylation, or from the alteration of global O-GlcNAc levels [100].
β-catenin glycosylation has been shown to regulate its cellular localization [101]. β-catenin plays two major roles in the cell: first, it associates with E-cadherin to form cellular adhesions, and secondly, it is the major downstream signaling molecule for the canonical arm of the Wnt signaling pathway. Wnt signaling pathways are involved in cell growth, movement, and cell survival and are associated with several types of cancer [106]. GSK-3 phosphorylation of β-catenin on its N-terminus targets it for ubiquitination and degradation. Wnt-activated signaling regulates β-catenin by inactivating GSK-3, allowing for the accumulation of β-catenin and its translocation to the nucleus. Here it can activate transcription of target genes by activating TCF (T-cell factor) and recruiting chromatin remodeling proteins [106]. β-catenin has been shown to be O-GlcNAc modified [107]. PUGNAc treatment of several cancer cell lines resulted in the redistribution of glycosylated β-catenin from the nucleus to the cytoplasm without affecting total protein levels [101]. The increase in cytoplasmic localization was associated with decreased expression of two downstream targets genes, cyclin D and vascular endothelial growth factor A, and decreased promoter activation [101]. More work is needed to determine how the glycosylation of β-catenin influences its interaction with many regulatory binding partners, such as GSK-3 and TCF, and in turn the role of O-GlcNAc in regulating its degradation and transcriptional activation [101].
DNA Binding and Transcriptional Activation
All classical transcription factors share two features: a DNA binding domain for binding to a specific sequence of DNA and a transactivation domain for response to regulatory factors. Sequence-specific transcription factors recruit coactivators to initiate transcription. These coactivators include chromatin remodeling enzymes that are needed to allow the basal transcription machinery to access the DNA and form the pre-initiation complex with the help of additional regulatory proteins [4]. PTM’s, such as glycosylation, can affect the ability of transcription factors to bind DNA and activate transcription [5].
The transcription factors PDX-1 (pancreatic/duodenal homeobox-1) protein, NeuroD1, and V-maf musculoaponeurotic fibrosarcoma oncogene homologue A co-regulate insulin transcription. The exact mechanisms of regulation are not clear, which is probably due to the number and complexity of post-translational modifications and cofactor interactions. PDX-1 is necessary for pancreatic development, and it activates several β-cell specific genes, such as insulin [105]. In response to changing glucose concentrations, PDX-1 recruits chromatin remodeling enzymes and other cofactors and regulates transcriptional elongation. PDX-1 phosphorylation is associated with its translocation to the nucleoplasm and its transactivation potential [105]. PDX-1 is also O-GlcNAc modified on at least two sites [108]. Hyperglycemia or PUGNAc treatment of MIN6 mouse insulinoma cells increases global O-GlcNAc protein levels, enhances PDX-1 binding to the insulin promoter, and is associated with an increase in insulin secretion [108]. The addition of azaserine, which inhibits GFAT and results in lower UDP-GlcNAc levels, decreases global O-GlcNAc levels and glucose-stimulated insulin secretion [108]. Treatment with siRNA against OGT also results in decreased glucose-stimulated insulin secretion, suggesting that the O-GlcNAc modification modulates insulin secretion, perhaps by activating PDX-1 binding to the insulin promoter [108, 109]. O-GlcNAc seems to be extensively involved in β-cell transcription factor regulation and may play an important role in controlling gene expression in response to glucose levels.
Like CRTC2, the forkhead transcription factor family, of which FoxO1 is a member, plays a major role in regulating energy homeostasis [110]. In the liver, FoxO1 and its coactivator, peroxisome proliferator activated receptor γ coactivator 1α (PGC1α), participate in the regulation of gluconeogenesis by activating the expression of G6Pase and phosphoenolpyruvate carboxykinase [111, 112]. Insulin signaling induces Akt to phosphorylate FoxO1 on residues Thr 24, Ser 256, and Ser 319, which results in FoxO1 cytoplasmic localization [113]. FoxO1 is subject to many PTM’s, including glycosylation [114]. Increasing global O-GlcNAc levels by hyperglycemia, PUGNAc, or overexpression of OGT in HEK293 or rat hepatoma cells increases FoxO1 activation of a G6Pase promoter reporter construct [115, 116]. A triple alanine mutant of the Akt phosphorylation sites on FoxO1 is still able to be glycosylated, suggesting that the FoxO1 O-GlcNAc sites are not directly reciprocal with the Akt phosphorylation sites [115]. Consistent with this result, O-GlcNAc modification does not seem to be required for FoxO1 translocation to the nucleus [115, 116]. FoxO1 has been shown to be O-GlcNAc modified on the following residues: Ser 550, Thr 648, Ser 654, and either Thr 317 or Ser 318 [115]. These sites were mutated to alanine and tested for activation of the G6Pase promoter. Only the T317A mutant had a small decrease in promoter activation under hyperglycemic conditions [115]. A follow-up study found that PGC1α interacts with OGT and enhances both OGT interaction and modification of FoxO1 [117]. Coexpression of PGC1α and FoxO1 in HEK293 cells cooperatively increases promoter activation in response to hyperglycemia [117].
Protein/Protein Interactions
Modification of proteins by O-GlcNAc has been shown to modulate protein-protein interactions that regulate nuclear localization [98, 118], stability [71], chromatin remodeling [66, 67], and transcriptional activation [101, 119, 120].
O-GlcNAc modification of Sp1 and β-catenin has been shown to decrease transcriptional activity possibly through inhibition of binding to co-activators [101, 120]. In addition, O-GlcNAc modification of a small peptide segment of Sp1 has been shown in vitro to prevent binding to the general transcription factor TAF110 (TATA-binding-protein-associated factor) [121].
Glycosylation of STAT5a (signal transducer and activator of transcription 5a) was found to be important for its interaction with CREB-binding protein (CBP) [119]. STAT proteins are activated by tyrosine phosphorylation in response to various cytokines and growth factors [122]. They initiate downstream transcriptional activation by dimerizing, translocating to the nucleus and activating transcription partly through the binding to co-activator molecules, such as CBP, that have histone acetyltransferase activity [123]. Mass spectrometry analysis and mutational studies of STAT5a showed that Thr 92 and potentially Thr 97 are O-GlcNAc modified [119]. The mutant T92A prevented STAT5a interaction with CBP and transactivation without affecting DNA binding [119].
NFκB (Nuclear factor κB) signaling has been implicated in a wide range of cellular processes, such as cell immune response, survival, differentiation, and proliferation. In the canonical NFκB signaling pathway, NFκB is normally bound to IκB and sequestered in the cytoplasm [124]. Phosphorylation of IκB by IκB kinase (IKK) leads to IκB degradation via the ubiquitin-proteasome pathway and this allows NFκB to translocate to the nucleus where it can activate transcription [124]. PTM of NFκB subunits can alter transcriptional activation by affecting interactions with transcriptional coactivators and corepressors. NFκB is activated by many pathways, so differential PTMs may specify the particular targets of NFκB. IKK is also regulated by PTMs [124].
Manipulation of the HBP in mesangial cells showed that hyperglycemia increases glycosylation of the p65 subunit of NFκB and promoter activation of a target gene, VCAM-1 (vascular cell adhesion molecule 1) [125]. Hyperglycemia or OGT overexpression decreased the association of the p65 subunit of NFκB with IκB and increased NFκB nuclear localization. Overexpression of OGA in rat vascular smooth muscle cells resulted in lower global O-GlcNAc levels and the reversal of NFκB activation by hyperglycemia. OGT overexpression resulted in the same effects as NFκB activation by hyperglycemia. Mutation of an NFκB O-GlcNAc modification site, Thr 352, to an alanine was found to abrogate promoter activation, DNA binding affinity, association with IκB, nuclear localization, and the expression of VCAM-1 induced by PUGNAc or OGT overexpression [118]. The primary effect of NFκB O-GlcNAc modification may be to prevent p65/IκB interaction, which would lead to nuclear localization and downstream target activation however, more investigation is needed to target the exact mechanism.
A recent paper tied p53 repression of NFκB activation to the O-GlcNAc modification of IKKβ [126]. p53 inactivation leads to an increase in glycolysis through enhanced NFκB activation and results in a positive feedback loop where glycolysis further activates NFκB signaling [127]. The authors proposed that O-GlcNAc modification of IKKβ could be acting as a glucose-sensor to potentiate the feedback loop. In a hepatic cancer cell line, hyperglycemia enhanced IKKβ O-GlcNAc modification and TNFα (tumor necrosis factor α) -stimulated NFκB promoter activation and prolonged NFκB DNA binding and IKKβ activity. Since phosphorylation of IKKβ at Ser 733 is known to inhibit its activation [128], O-GlcNAc modification of Ser 733 is suggested to prevent phosphorylation-stimulated inactivation leading to an increased in activation of NFκB in transformed cells [126]. These studies establish a clear role for O-GlcNAc in the activation of NFκB.
OGT/OGA Targeting to Substrates – A Special Case of Protein/Protein Interactions
O-GlcNAc modification regulates the function of many target proteins, so aberrant modification by OGT needs to be avoided for proper cellular function. However, the mechanism by which OGT selects its targets is not currently known. No consensus sequence for O-GlcNAc attachment has been found, so it has been proposed that interaction with OGT’s TPR domain may determine which proteins it modifies [26, 129, 130]. OGT may also use adaptor proteins that help to modulate its specificity and increase the complexity of its regulation [31, 131]. Cheung et al. used a yeast two-hybrid screen to identify proteins that interact with OGT from a human fetal brain cDNA library [131]. Two of the twenty-seven putative OGT-interacting proteins identified, MYPT1 (myosin phosphatase target subunit 1) and CARM1 (coactivating arginine methyltransferase), were shown to interact with OGT and be O-GlcNAc modified by independent methods [131]. Knockdown of MYPT1 using siRNA in Neuro-2a cells reduced the O-GlcNAc levels of several proteins, suggesting that MYPT1 might target OGT to substrates in vivo [131]. CARM1 is a histone methyltransferase and functions as part of the p160 coactivator complex, which contributes to chromatin remodeling and transcriptional activation [132]. CARM1 may help to target OGT to substrates that are involved in transcriptional activation [131]. Trak1 (also known as OIP106) was identified by another yeast two-hybrid screen of OGT interacting proteins [130]. Trak1 associates with RNAP II, so it has been proposed that Trak1 targets OGT to the transcriptional machinery [130, 133]. Finally, as mentioned above, PGC-1α may act as an adaptor protein for OGT recruitment to FoxO1 [117].
Although little is known about targeting of OGT to its substrates, even less is known about the regulation of OGA [134]. In some cases, OGT and OGA are found in the same complex [135]. As described above, NeuroD1 can associate with either OGT or OGA depending on glucose concentration [100]. The identification of more OGA-interacting proteins might provide insight into the mechanism of deglycosylation. Using a similar strategy as the OGT experiments, we used a yeast two-hybrid assay obtained from Proquest to identify human OGA binding partners using a cDNA library from human skeletal muscle. Proteins not in frame, proteins identified only once, and proteins known to commonly give false positives were removed from the results. A total of ten proteins were identified by this screen as shown in Table (1). Several of these proteins, including Fragile X mental retardation-related protein 1 (FXR1), Interferon-related developmental regulator 1 (IFRD1), and TANK-Binding Kinase 1 (TBK1)-binding protein 1 (TBKBP1), are relevant to eukaryotic gene expression.
The leading cause of inherited mental retardation is Fragile X syndrome, which is caused by the reduction in an RNA binding protein, Fragile X Mental Retardation protein (FMRP) [136]. FMRP binds polyribosomes and suppresses translation [137]. FMRP has two homologs, FXR1 and FXR2, which share about 60% sequence homology to FMRP and have been shown to repress TNF translation [138]. Several other RNA-binding proteins, including Ewing-sarcoma RNA-binding protein, eukaryotic initiation factor 4A1, elongation factor 1, and the small and large ribosomal subunits, have been shown to be O-GlcNAc modified, suggesting a possible functional role for O-GlcNAc in post-transcriptional regulation as well [17, 21, 42].
IFRD1 has been shown to play a role in development by induction of differentiation by repression of a specific set of genes through interactions with the co-repressor complex mSin3B/HDAC1 [139, 140]. IFRD1 is implicated in the prevention of Sp1 binding to a common DNA element in IFRD1 regulated genes. It has also been implicated in recruiting HDAC to β-catenin in order to repress its transcriptional activity on downstream targets, such as osteopontin [141, 142]. Since IFRD1 interacts with already known O-GlcNAc targets, it will be interesting to see if the interaction with OGA is required to modify these targets for their function or for interaction with IFRD1.
TBKBP1 was found to interact with TBK1 and inducible IκB kinase (IKKi), which are members of the IKK family that regulate interferon regulatory factor (IRF) [143]. IRF and NFκB coordinate to regulate innate antiviral immunity [144]. TBK1 and IKKi phosphorylate and activate IRF in response to TLR3 (Toll-like receptor 3) activation. Like NFκB, upon activation, IRF dimerizes and translocates to the nucleus to initiate transcriptional activation. TBKBP1, which is also named Similar to NAP1 TBK1 adaptor (SINTBAD), along with two other cofactors, TANK and NAP1, are needed for full activation of IRF3 in response to the Sendai virus [143]. These cofactors might serve as a link between downstream signaling from TLR3 and activation of TBK1 and IKKi [143]. Since OGA interacts with TBKBP1 and the O-GlcNAc modification is intricately involved in NFκB signaling that is similar to the IRF pathway, it is plausible that the IRF pathway is also regulated by O-GlcNAc modification. Future work will need to establish the relevance of this hypothesis.
SUMMARY
The O-GlcNAc modification of nuclear and cytoplasmic proteins plays a variety of roles in transcription factor regulation including recruiting chromatin remodeling factors, affecting protein stability, changing nuclear localization, and altering DNA binding and transcriptional activation. O-GlcNAc modification can either exert its effects directly on the modified transcription factor or indirectly by altering protein-protein interactions with other modified co-factors. It is becoming increasingly clear that transcription factors do not function in a solely “on” or “off” state but are subject to a number of modifications, such as O-GlcNAc, that fine-tune their regulation [145]. This is advantageous to the cell because transcription factors must interpret a wide range of signals, including nutrient/metabolic signals, and specifically respond to regulate a subset of target genes.
A key feature of the O-GlcNAc modification is that the levels of its sugar donor, UDP-GlcNAc, are directly responsive to the changes in cellular glucose flux. A nutrient sensing ability is valuable for the cell because it prevents it from being a slave to its extracellular environment [12]. Because altering glucose flux readily modulates global protein O-GlcNAc levels and not just the O-GlcNAc modification on specific proteins, many O-GlcNAc studies to date are correlative. Specific mechanistic and functional studies that show O-GlcNAc modification is indispensable for protein function are beginning to appear in the literature, primarily in relationship to transcriptional control (illustrated above). Advances in the O-GlcNAc site-mapping technology along with the initial experiments for understanding targeting mechanisms for OGT and OGA substrate recognition and the highlighted recent “smoking gun” experiments should facilitate increased interest in understanding functional mechanisms for O-GlcNAc on a wider range of proteins in an increasing number of systems.
Acknowledgments
Thanks to the members of the Wells group for the critical reading of this manuscript. This work was supported in part by a grant from NIH/NIDDK (1RO1DK075069 to LW). CFT is an American Heart Association predoctoral fellow (Southeast affiliate, 0715377B). LW is a Georgia Cancer Coalition Distinguished Scholar.
ABBREVIATIONS
- CBP
CREB-binding protein
- CID
Collision-induced dissociation
- CREB
Cyclic adenosine 3′-5′ monophosphate (cAMP) response element
- CRTC2
Cyclic adenosine 3′-5′ monophosphate response element (CREB) protein 2
- CTD
Carboxyl terminal domain
- ER-β
Estrogen receptor β
- FMRP
Fragile X Mental Retardation protein
- FoxO1
Forkhead box other-1
- G6Pase
Glucose-6-phosphatase
- GalT
β-1,4-galactosyltransferase
- GFAT
Glutamine-fructose-6-phosphate transaminase
- GSK-3
Glycogen Synthase Kinase-3
- HBP
Hexosamine biosynthetic pathway
- HDACs
Histone deacetylases
- IFRD1
Interferon-related developmental regulator 1
- IKK
IκB kinase
- IRF
interferon regulatory factor
- NeuroD1
Neurogenic differentiation 1
- NFκB
Nuclear factor κB
- OGA
O-GlcNAcase, neutral β-N-acetlyglucosaminidase (HexC)
- O-GlcNAc
O-linked β-N-acetylglucosamine
- OGT
O-GlcNAc transferase
- PDX-1
Pancreatic/duodenal homeobox-1 protein
- PGC1α
Peroxisome proliferator activated receptor γ co-activator 1α
- PTMs
Post-translational modifications
- PUGNAc
O-(2-acetamido-2-deoxy-D-glucopyrano-sylidene)amino-N-phenylcarbamate
- RA
Retinoic acid
- TBK1
TANK-Binding Kinase 1
- TBKBP1
TANK-Binding Kinase 1 (TBK1)-binding protein 1
- TPR
Tetratricopeptide repeat
References
- 1.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;1;295:813–8. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–51. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 3.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khidekel N, Hsieh-Wilson LC. A ‘molecular switchboard’--covalent modifications to proteins and their impact on transcription. Org Biomol Chem. 2004;2:1–7. doi: 10.1039/b312466e. [DOI] [PubMed] [Google Scholar]
- 6.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–17. [PubMed] [Google Scholar]
- 7.Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 1986;261:8049–57. [PubMed] [Google Scholar]
- 8.Holt GD, Haltiwanger RS, Torres CR, Hart GW. Erythrocytes contain cytoplasmic glycoproteins. O-linked GlcNAc on Band 4. 1. J Biol Chem. 1987;262:14847–50. [PubMed] [Google Scholar]
- 9.Dehennaut V, Lefebvre T, Sellier C, et al. O-linked N-acetylglucosaminyltransferase inhibition prevents G2/M transition in Xenopus laevis oocytes. J Biol Chem. 2007;282:12527–36. doi: 10.1074/jbc.M700444200. [DOI] [PubMed] [Google Scholar]
- 10.Dehennaut V, Hanoulle X, Bodart JF, et al. Microinjection of recombinant O-GlcNAc transferase potentiates Xenopus oocytes M-phase entry. Biochem Biophys Res Commun. 2008;369:539–46. doi: 10.1016/j.bbrc.2008.02.063. [DOI] [PubMed] [Google Scholar]
- 11.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–56. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 12.Wells L, Vosseller K, Hart GW. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol Life Sci. 2003;60:222–8. doi: 10.1007/s000180300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–8. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 14.Gandy JC, Rountree AE, Bijur GN. Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1. FEBS Lett. 2006;580:3051–8. doi: 10.1016/j.febslet.2006.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci USA. 2002;99:5313–8. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Ongusaha PP, Miles PD, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–9. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 17.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–31. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Comer FI, Hart GW. O-GlcNAc and the control of gene expression. Biochim Biophys Acta. 1999;1473:161–71. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 20.Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3:766–72. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 21.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 22.Vosseller K, Sakabe K, Wells L, Hart GW. Diverse regulation of protein function by O-GlcNAc: a nuclear and cytoplasmic carbohydrate post-translational modification. Curr Opin Chem Biol. 2002;6:851–7. doi: 10.1016/s1367-5931(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 23.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine: peptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 1990;265:2563–8. [PubMed] [Google Scholar]
- 25.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–15. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 26.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–8. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 27.Shafi R, Iyer SP, Ellies LG, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–9. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–45. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 29.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269:19321–30. [PubMed] [Google Scholar]
- 30.Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 2000;275:29179–82. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- 31.Wells L, Kreppel LK, Comer FI, Wadzinski BE, Hart GW. O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J Biol Chem. 2004;279:38466–70. doi: 10.1074/jbc.M406481200. [DOI] [PubMed] [Google Scholar]
- 32.Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295:E17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haltiwanger RS, Blomberg MA, Hart GW. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine: polypeptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 1992;267:9005–13. [PubMed] [Google Scholar]
- 34.Haltiwanger RS, Grove K, Philipsberg GA. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins using the peptide O-GlcNAc-beta-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-D-glucopyranosylidene) amino-N-phenylcarbamate. J Biol Chem. 1998;273:3611–7. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- 35.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 2005;280:25313–22. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 36.Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, van Aalten DM. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 2006;128:16484–5. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorfmueller HC, Borodkin VS, Schimpl M, van Aalten DM. GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem J. 2009;420:221–7. doi: 10.1042/BJ20090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross BJ, Swoboda JG, Walker S. A strategy to discover inhibitors of O-linked glycosylation. J Am Chem Soc. 2008;130:440–1. doi: 10.1021/ja078125s. [DOI] [PubMed] [Google Scholar]
- 39.Vosseller K, Trinidad JC, Chalkley RJ, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–34. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci USA. 2004;101:13132–7. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khidekel N, Ficarro SB, Clark PM, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–48. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 42.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Issad T, Kuo M. O-GlcNAc modification of transcription factors, glucose sensing and glucotoxicity. Trends Endocrinol Metab. 2008;19:380–9. doi: 10.1016/j.tem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Zachara NE. Detecting the “O-GlcNAcome”; detection, purification, and analysis of O-GlcNAc modified proteins. Methods Mol Biol. 2009;534:251–79. doi: 10.1007/978-1-59745-022-5_19. [DOI] [PubMed] [Google Scholar]
- 45.Hanover JA, Cohen CK, Willingham MC, Park MK. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem. 1987;262:9887–94. [PubMed] [Google Scholar]
- 46.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–56. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–52. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- 48.Khidekel N, Arndt S, Lamarre-Vincent N, et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–3. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 49.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci USA. 2003;100:9116–21. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–10. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 51.Whelan SA, Hart GW. Identification of O-GlcNAc sites on proteins. Methods Enzymol. 2006;415:113–33. doi: 10.1016/S0076-6879(06)15008-9. [DOI] [PubMed] [Google Scholar]
- 52.Nandi A, Sprung R, Barma DK, Zhao Y, Kim SC, Falck JR. Global identification of O-GlcNAc-modified proteins. Anal Chem. 2006;78:452–8. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 53.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–6. [Google Scholar]
- 54.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikesh LM, Ueberheide B, Chi A, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–22. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8:4466–83. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 57.Viner RI, Zhang T, Second T, Zabrouskov V. Quantification of post-translationally modified peptides of bovine alpha-crystallin using tandem mass tags and electron transfer dissociation. J Proteomics. 2009 Feb 24; doi: 10.1016/j.jprot.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 58.O’Malley BW, Qin J, Lanz RB. Cracking the coregulator codes. Curr Opin Cell Biol. 2008;20:310–5. doi: 10.1016/j.ceb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitmarsh AJ, Davis RJ. Regulation of transcription factor function by phosphorylation. Cell Mol Life Sci. 2000;57:1172–83. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–27. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 61.Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–92. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 63.Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 64.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 66.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;12;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 67.Fujiki R, Chikanishi T, Hashiba W, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–9. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 68.Kelly WG, Hart GW. Glycosylation of chromosomal proteins: localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;21;57:243–51. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- 69.Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–95. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–30. [PubMed] [Google Scholar]
- 71.Yang WH, Kim JE, Nam HW, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–83. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 72.Murphy M, Ahn J, Walker KK, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao D, Taguchi T, Matsumura T, et al. High glucose increases angiopoietin-2 transcription in microvascular endothelial cells through methylglyoxal modification of mSin3A. J Biol Chem. 2007;282:31038–45. doi: 10.1074/jbc.M704703200. [DOI] [PubMed] [Google Scholar]
- 74.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–7. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 75.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–24. [PubMed] [Google Scholar]
- 76.Desterro JM, Rodriguez MS, Hay RT. Regulation of transcription factors by protein degradation. Cell Mol Life Sci. 2000;57:1207–19. doi: 10.1007/PL00000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 79.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–25. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 80.Wierstra I. Sp1: Emerging roles--Beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 81.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–33. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 82.Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–8. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sumegi M, Hunyadi-Gulyas E, Medzihradszky KF, Udvardy A. 26S proteasome subunits are O-linked N-acetylglucosamine-modified in Drosophila melanogaster. Biochem Biophys Res Commun. 2003;312:1284–9. doi: 10.1016/j.bbrc.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 84.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–5. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 85.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–20. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- 87.Chou TY, Dang CV, Hart GW. Glycosylation of the c-Myc transactivation domain. Proc Natl Acad Sci USA. 1995;92:4417–21. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–5. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 89.Yustein JT, Dang CV. Biology and treatment of Burkitt’s lymphoma. Curr Opin Hematol. 2007;14:375–81. doi: 10.1097/MOH.0b013e3281bccdee. [DOI] [PubMed] [Google Scholar]
- 90.Vervoorts J, Luscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281:34725–9. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 91.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 92.Kuiper GG, Gustafsson JA. The novel estrogen receptor-beta subtype: potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett. 1997;410:87–90. doi: 10.1016/s0014-5793(97)00413-4. [DOI] [PubMed] [Google Scholar]
- 93.Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen receptor beta: an overview and update. Nucl Recept Signal. 2008;6:e003. doi: 10.1621/nrs.06003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grisouard J, Medunjanin S, Hermani A, Shukla A, Mayer D. Glycogen synthase kinase-3 protects estrogen receptor alpha from proteasomal degradation and is required for full transcriptional activity of the receptor. Mol Endocrinol. 2007;21:2427–39. doi: 10.1210/me.2007-0129. [DOI] [PubMed] [Google Scholar]
- 95.Reid G, Hubner MR, Metivier R, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 96.Picard N, Charbonneau C, Sanchez M, et al. Phosphorylation of activation function-1 regulates proteasome-dependent nuclear mobility and E6-associated protein ubiquitin ligase recruitment to the estrogen receptor beta. Mol Endocrinol. 2008;22:317–30. doi: 10.1210/me.2007-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 98.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–5. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 99.Petersen HV, Jensen JN, Stein R, Serup P. Glucose induced MAPK signalling influences NeuroD1-mediated activation and nuclear localization. FEBS Lett. 2002;528:241–5. doi: 10.1016/s0014-5793(02)03318-5. [DOI] [PubMed] [Google Scholar]
- 100.Andrali SS, Qian Q, Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J Biol Chem. 2007;282:15589–96. doi: 10.1074/jbc.M701762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sayat R, Leber B, Grubac V, Wiltshire L, Persad S. O-GlcNAc-glycosylation of beta-catenin regulates its nuclear localization and transcriptional activity. Exp Cell Res. 2008;314:2774–87. doi: 10.1016/j.yexcr.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 102.Koo SH, Flechner L, Qi L, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–11. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 103.Dentin R, Liu Y, Koo SH, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–9. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 104.Screaton RA, Conkright MD, Katoh Y, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 105.Andrali SS, Sampley ML, Vanderford NL, Ozcan S. Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem J. 2008;415:1–10. doi: 10.1042/BJ20081029. [DOI] [PubMed] [Google Scholar]
- 106.Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- 107.Zhu W, Leber B, Andrews DW. Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. EMBO J. 2001;20:5999–6007. doi: 10.1093/emboj/20.21.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao Y, Miyazaki J, Hart GW. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch Biochem Biophys. 2003;415:155–63. doi: 10.1016/s0003-9861(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 109.Akimoto Y, Hart GW, Wells L, et al. Elevation of the post-translational modification of proteins by O-linked N-acetylglucosamine leads to deterioration of the glucose-stimulated insulin secretion in the pancreas of diabetic Goto-Kakizaki rats. Glycobiology. 2007;17:127–40. doi: 10.1093/glycob/cwl067. [DOI] [PubMed] [Google Scholar]
- 110.Zhang W, Patil S, Chauhan B, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–17. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 111.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Puigserver P, Rhee J, Donovan J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–5. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 113.Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–9. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 114.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–36. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 115.Housley MP, Rodgers JT, Udeshi ND, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–92. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008;582:829–34. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 117.Housley MP, Udeshi ND, Rodgers JT, et al. A PGC-1{alpha}-O-GlcNAc Transferase Complex Regulates FoxO Transcription Factor Activity in Response to Glucose. J Biol Chem. 2009;284:5148–57. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang WH, Park SY, Nam HW, et al. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci USA. 2008;105:17345–50. doi: 10.1073/pnas.0806198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–72. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 120.Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci USA. 2001;5;98:6611–6. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roos MD, Su K, Baker JR, Kudlow JE. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol Cell Biol. 1997;17:6472–80. doi: 10.1128/mcb.17.11.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 123.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;30;279:703–7. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 124.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 125.James LR, Tang D, Ingram A, et al. Flux through the hexosamine pathway is a determinant of nuclear factor kappaB- dependent promoter activation. Diabetes. 2002;51:1146–56. doi: 10.2337/diabetes.51.4.1146. [DOI] [PubMed] [Google Scholar]
- 126.Kawauchi K, Araki K, Tobiume K, Tanaka N. Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification. Proc Natl Acad Sci USA. 2009;106:3431–6. doi: 10.1073/pnas.0813210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–8. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 128.Schomer-Miller B, Higashimoto T, Lee YK, Zandi E. Regulation of IkappaB kinase (IKK) complex by IKKgamma-dependent phosphorylation of the T-loop and C terminus of IKKbeta. J Biol Chem. 2006;281:15268–76. doi: 10.1074/jbc.M513793200. [DOI] [PubMed] [Google Scholar]
- 129.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–22. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 130.Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2003;278:5399–409. doi: 10.1074/jbc.M209384200. [DOI] [PubMed] [Google Scholar]
- 131.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283:33935–41. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–45. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- 133.Iyer SP, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278:24608–16. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 134.Hurtado-Guerrero R, Dorfmueller HC, van Aalten DM. Molecular mechanisms of O-GlcNAcylation. Curr Opin Struct Biol. 2008;18:551–7. doi: 10.1016/j.sbi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 135.Whisenhunt TR, Yang X, Bowe DB, Paterson AJ, Van Tine BA, Kudlow JE. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology. 2006;16:551–63. doi: 10.1093/glycob/cwj096. [DOI] [PubMed] [Google Scholar]
- 136.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 137.Garber K, Smith KT, Reines D, Warren ST. Transcription, translation and fragile X syndrome. Curr Opin Genet Dev. 2006;16:270–5. doi: 10.1016/j.gde.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 138.Garnon J, Lachance C, Di Marco S, et al. Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J Biol Chem. 2005;280:5750–63. doi: 10.1074/jbc.M401988200. [DOI] [PubMed] [Google Scholar]
- 139.Vietor I, Huber LA. Role of TIS7 family of transcriptional regulators in differentiation and regeneration. Differentiation. 2007;75:891–7. doi: 10.1111/j.1432-0436.2007.00205.x. [DOI] [PubMed] [Google Scholar]
- 140.Vietor I, Vadivelu SK, Wick N, et al. TIS7 interacts with the mammalian SIN3 histone deacetylase complex in epithelial cells. EMBO J. 2002;21:4621–31. doi: 10.1093/emboj/cdf461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vietor I, Kurzbauer R, Brosch G, Huber LA. TIS7 regulation of the beta-catenin/Tcf-4 target gene osteopontin (OPN) is histone deacetylase-dependent. J Biol Chem. 2005;280:39795–801. doi: 10.1074/jbc.M509836200. [DOI] [PubMed] [Google Scholar]
- 142.Wick N, Schleiffer A, Huber LA, Vietor I. Inhibitory effect of TIS7 on Sp1-C/EBPalpha transcription factor module activity. J Mol Biol. 2004;336:589–95. doi: 10.1016/j.jmb.2003.11.060. [DOI] [PubMed] [Google Scholar]
- 143.Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 2007;26:3180–90. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene. 2006;25:6844–67. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–22. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]