Nitric oxide regulation of stem cell decisions in Arabidopsis roots is related to auxin biosynthesis, transport, and signaling.
Abstract
Nitric oxide (NO) is a unique reactive nitrogen molecule with an array of signaling functions that modulates plant developmental processes and stress responses. To explore the mechanisms by which NO modulates root development, we used a pharmacological approach and NO-deficient mutants to unravel the role of NO in establishing auxin distribution patterns necessary for stem cell niche homeostasis. Using the NO synthase inhibitor and Arabidopsis (Arabidopsis thaliana) NO biosynthesis mutants (nitric oxide-associated1 [noa1], nitrate reductase1 [nia1] and nia2, and nia1 nia2 noa1), we show that depletion of NO in noa1 reduces primary root elongation and increases flavonol accumulation consistent with elevated reactive oxygen species levels. The elevated flavonols are required for the growth effect, because the transparent testa4 mutation reverses the noa1 mutant root elongation phenotype. In addition, noa1 and nia1 nia2 noa1 NO-deficient mutant roots display small root meristems with abnormal divisions. Concomitantly, auxin biosynthesis, transport, and signaling are perturbed. We further show that NO accumulates in cortex/endodermis stem cells and their precursor cells. In endodermal and cortical cells, the noa1 mutant acts synergistically to the effect of the wuschel-related homeobox5 mutation on the proximal meristem, suggesting that NO could play an important role in regulating stem cell decisions, which has been reported in animals.
Root growth enables plants to explore their substrate and extract nutrients and water from larger areas and greater depths while also providing anchorage to the substrate. To properly regulate primary root elongation, multiple intrinsic and extrinsic signals must be integrated. Plant hormones are important long-distance signals, and auxin in particular regulates many aspects of root development. Auxin regulates primary root growth in three important ways: by positioning the stem cell niche, affecting divisions in a transit-amplifying zone known as the meristem, and increasing cell volume in the elongation zone (for review, see Bennett and Scheres, 2010). These processes are dependent on the establishment of auxin gradients. Models generated on the basis of available information predict a maximum auxin concentration in the quiescent center (QC) and a steep auxin gradient in the proximal meristem, which decreases with distance to the QC (Grieneisen et al., 2007; Kramer et al., 2008; Laskowski et al., 2008).
Interestingly, reactive oxygen species (ROS) and reactive nitrogen species (RNS) have been found to play an important role in the response to intrinsic and extrinsic growth-modulating signals. Research over the last decades has shown several crucial roles of nitric oxide (NO; an RNS) during plant development. NO is involved in the promotion of seed germination and alleviation of seed dormancy, the shaping of root architecture, the repression of floral transition, and the regulation of mitochondrial respiratory complexes, stomatal closure, fruit maturation, senescence, and iron metabolism (for review, see Freschi, 2013). Synergistic effects of auxin and NO have been observed during the regulation of a series of plant responses (for review, see Freschi, 2013). In virtually all of these cases, NO was identified to function downstream of auxins, apparently through linear signaling pathways (Pagnussat et al., 2003, 2004). Increased NO production has frequently been observed after exogenous auxin application (Pagnussat et al., 2002; Correa-Aragunde et al., 2004; Hu et al., 2005; Lombardo et al., 2006) or in auxin-overproducing mutants (Chen et al., 2010), and it is especially evident in plant tissues or cells undergoing auxin-dependent physiological responses. Other than these impacts of auxin on NO production, recent studies have shown that NO might also modulate auxin metabolism, transport, and signaling. For example, NO has been shown to elevate root indole-3-acetic acid (IAA) levels in cadmium-treated alfalfa (Medicago truncatula) seedlings by reducing its degradation by IAA oxidase activity, thereby positively impacting auxin equilibrium and ameliorating cadmium toxicity (Xu et al., 2010). In addition, pharmacological treatments and NO-overproducing mutants indicated that, at high concentrations, NO inhibits rootward auxin transport in Arabidopsis (Arabidopsis thaliana) roots by reducing the abundance of the auxin efflux protein PIN-FORMED1 (PIN1) through a proteasome-independent posttranscriptional mechanism (Fernández-Marcos et al., 2011). Finally, a direct influence of NO on auxin perception and signal transduction has also been suggested based on the recent demonstration that the auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) undergoes S-nitrosylation at two particular Cys residues (Terrile et al., 2012). This S-nitrosylation of TIR1 seems to promote its interaction with auxin/IAA proteins, which are transcriptional repressors of genes associated with auxin responses.
Genetic tools are powerful in the understanding of NO action in key physiological processes. Plants defective in genes encoding nitrate reductase (NIA1 and NIA2) and nitric oxide-associated1 (AtNOA1) exhibit reduced NO levels (Lozano-Juste and León, 2010a, 2010b). Here, we show that nia1 nia2 noa1 mutant roots display small root meristems with abnormal divisions. In addition, the reduced primary root elongation phenotype of noa1 mutant is correlated with an increase of flavonol and ROS accumulation. Furthermore, by introducing different auxin reporters into plants with mutations in NOA1, NIA1, and NIA2, we gain deeper insight into NO and auxin interactions that regulate primary root growth. Thus, the abnormal phenotypes present in these NO mutants are related to perturbations in auxin biosynthesis, transport, and/or signaling. Previous studies using animal cell models showed that manipulation of the NO pathway contributes to stem cell fate determination. Treatment with a combination of NO donors enhanced the differentiation of stem cells (Mujoo et al., 2011). Because auxin plays a main role in QC/initial/columella cell organization (Kramer et al., 2008; for review, see Bennett and Scheres, 2010) and NO accumulates in cortex/endodermis stem cells in Arabidopsis root tips (Fernández-Marcos et al., 2011), we hypothesized that NO, acting downstream of auxin, might act redundantly with regulators of stem cell niche patterning. The phenotypic, genetic, and molecular characterization of the noa1, nia1 nia2, and nia1 nia2 noa1 mutant root tips illustrates how the interplay between auxin and NO shapes stem cell homeostasis.
RESULTS
Depletion of NO, Either Genetically or by a Synthesis Inhibitor, Reduces Primary Root Growth in Arabidopsis
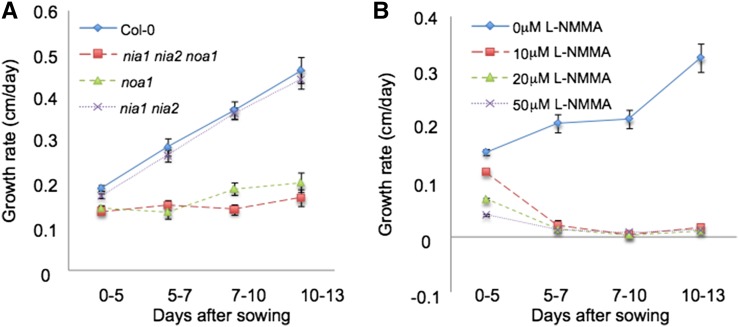
The accumulation of NO (an RNS) increases in response to different stress situations and during developmental processes (Delledonne et al., 2001; Hu et al., 2005). Previous reports have determined that this molecule is also detected in normal/basal growth conditions, accumulating in the root tip and the beginning of the elongation/differentiation zone (Illés et al., 2006; Fernández-Marcos et al., 2011, 2012). Triple nia1 nia2 noa1 mutants are impaired in nitrate reductase and AtNOA1-mediated NO biosynthetic pathways. NO content in roots of nia1 nia2 and noa1-2 plants is lower than in wild-type plants and below the detection limit in nia1 nia2 noa1 plants (Lozano-Juste and León, 2010a, 2010b; Supplemental Fig. S1). In addition to reduced size and seed germination potential (Lozano-Juste and León, 2010a, 2010b), NO-deficient triple mutant seedlings display significantly reduced root growth (47% decrease after 10 d; P < 0.0001) compared with wild-type controls under the same growth conditions (Fig. 1A). Although the double nia1 nia2 mutation has a marginal effect in root growth inhibition (6% decrease; P = 0.1), noa1-2 mutation has a highly significant effect (41% decrease after 10 d; P < 0.0001), indicating that the mutation in NOA1 plays a major role in primary root growth inhibition phenotype of the triple mutant (Fig. 1A; Supplemental Fig. S1A).
Figure 1.
NO is necessary for normal primary root elongation. A, Growth rate (from 0–13 d after sowing) of primary root of Col-0, noa1, nia1 nia2, and nia1 nia2 noa1 seedlings grown vertically under standard conditions. Error bars represent se; n > 40. B, Growth rate (from 0–13 d after sowing) of primary root of Col-0 seedlings grown vertically in the presence of 0, 10, 20, and 50 µm L-NMMA (+dimethyl sulfoxide). Error bars represent se; n > 40.
To investigate whether the root growth phenotypes of noa1-2 and nia1 nia2 noa1 were linked to the impairment in NO biosynthesis, we supplemented the growth medium with NO synthase inhibitor NG-monomethyl-L-arginine (L-NMMA), a mammalian NO synthase inhibitor able to also block NO synthase activity in plants (Foissner et al., 2000; Supplemental Fig. S1B). Exogenous L-NMMA decreased germination in wild-type seeds (60% decrease 3 d after sowing nonstratified seeds upon treatment with 50 µm L-NMMA; P < 0.0001; Supplemental Fig. S1C) without affecting seed germination potential (Supplemental Fig. S1D). Interestingly, treatments with L-NMMA produced roots that experienced limited growth (similar to that of the NO-deficient mutants), resulting in primary roots of less than 0.7 cm in length from day 5 onward (22% decrease after 5 d and 80% decrease after 10 d of treatment with 10 µm L-NMMA; P < 0.0001; Fig. 1B; Supplemental Figs. S1E and S2A). To evaluate the potential toxicity of L-NMMA in these conditions, Columbia-0 (Col-0) seedlings were grown in the presence of L-NMMA (10 and 20 µm) for 5 d and then transferred during 2, 5, or 7 d to plates containing Murashige and Skoog (MS) media. When grown for 5 d on 10 or 20 µm L-NMMA, Arabidopsis seedlings can recover root growth (Supplemental Fig. S2B), suggesting that L-NMMA is not toxic to plant roots but inhibits root elongation. Furthermore, confocal analysis of root apices shows fewer and enlarged cells in 5-d-old seedlings treated with L-NMMA compared with nontreated seedlings. Seven days after transferring to plates containing MS media, no evident morphological differences with nontreated seedlings were detected in root apices (Supplemental Fig. S2, C and D). Thus, the depletion of NO, either genetically or by a synthesis inhibitor, reduces primary root growth in Arabidopsis.
NO Is Required for the Maintenance of Primary Root Meristem Organization
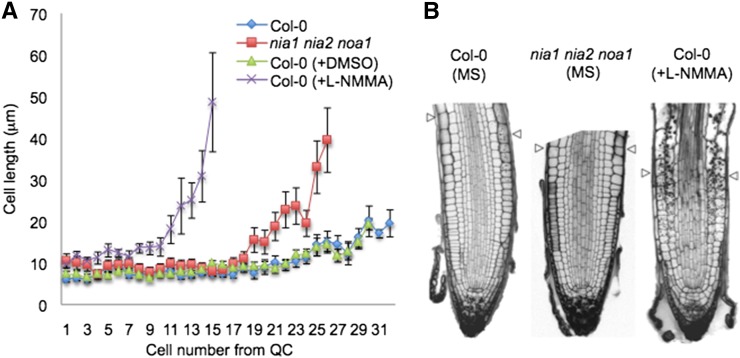
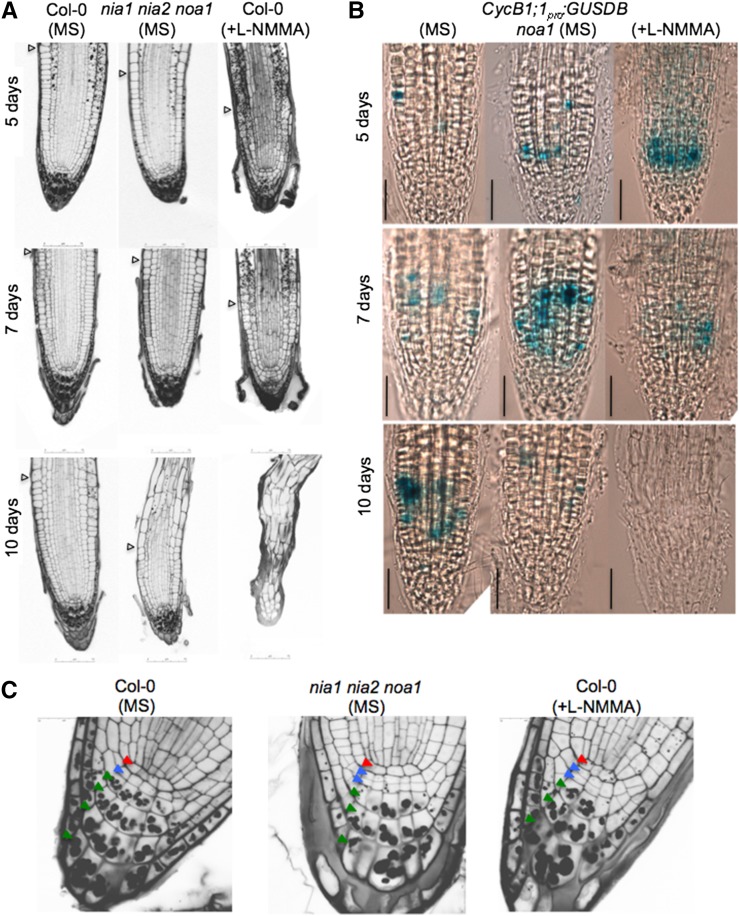
To gain deeper insight into the mechanism by which NO regulates root meristem organization, root tips of 5-, 7-, and 10-d-old nia1 nia2 noa1 triple mutants were stained with propidium iodide (PI) and analyzed compared with wild-type controls (Figs. 2 and 3A). We measured the size and number of root cells in a cortical cell file, determining the length of cells from the initials adjacent to the QC to the rapid elongation/differentiation zone. A significant decrease in the size of nia1 nia2 noa1 primary root meristems can be observed within the division zone (Fig. 2). At earlier time points (5 and 7 d), we detected normal or even enhanced mitotic cyclin B1 (CYCB1) GUS reporter expression in L-NMMA-treated wild-type and noa1 mutant seedlings (Fig. 3B). However, reduced expression of CycB1;1pro:GUSDB cell division reporter was observed in roots of 10-d-old noa1 seedlings compared with the wild-type controls. Exogenous L-NMMA also reduced the expression of CycB1;1pro:GUSDB. In addition, in wild-type untreated 5-d-old seedlings, most of the roots (75%; n = 20) have only one layer of columella stem cells, which was shown by the absence of starch-containing amyloplasts, whereas 5-d-old nia1 nia2 noa1 has two layers of undifferentiated columella stem cells (60%; n = 10; Fig. 3C, blue arrows). The disorganization of the primary root meristem is even more evident in L-NMMA-treated plants (Fig. 3A). Germination of seedlings on medium supplemented with L-NMMA displayed reduced rates of columella stem cell differentiation (Fig. 3C). Therefore, compared with wild-type seedlings, 5-d-old treated seedlings have two layers of undifferentiated columella stem cells (64.3%; n = 14; Fig. 3C, blue arrows). Furthermore, a significant decrease in the size of the primary root meristem can be observed 7 d after treatment (Figs. 2 and 3A). The lack of cells expressing CycB1;1pro:GUSDB after 10 d in the noa1 mutant and upon treatment with L-NMMA is consistent with growth arrest (Fig. 3B).
Figure 2.
NO is necessary for normal meristem organization. A, Average cell sizes in the cortical cell layer (cells 1–31 from the QC) of 5-d-old nia1 nia2 noa1 mutant and wild-type (Col-0) roots grown in the presence (+L-NMMA) or absence (Col-0 and Col-0 + dimethyl sulfoxide [DMSO]) of 20 μm L-NMMA. B, Meristematic regions of 7-d-old nia1 nia2 noa1 mutant and wild-type (Col-0) roots (grown in the presence or absence of 20 μm L-NMMA). Arrowheads indicate the length of the meristem from QC to the beginning of the elongation/differentiation zone.
Figure 3.
Meristem organization requires L-NMMA-dependent NO synthesis. A, Meristematic regions of 5-, 7-, and 10-d-old nia1 nia2 noa1 mutant and wild-type (Col-0) roots (grown in the presence or absence of 20 μm L-NMMA). Arrowheads indicate the end of the meristem and the beginning of the elongation/differentiation zone. B, CycB1;1pro:GUSDB expression in 5-, 7-, and 10-d-old noa1 mutant and wild-type (Col-0) seedlings grown in the presence or absence of 20 μm L-NMMA. Bars = 50 µm. C, QC/columella stem cells (CSC) disorganization and starch accumulation in 5-d-old nia1 nia2 noa1 mutant and wild-type (Col-0) seedlings grown in the presence or absence of 20 µm L-NMMA using modified pseudo-Schiff PI stain. Red, blue, and green arrowheads indicate QC, CSC, and columella cells, respectively.
Elevated Flavonol Accumulation in noa1 Contributes to the Root Growth Phenotype
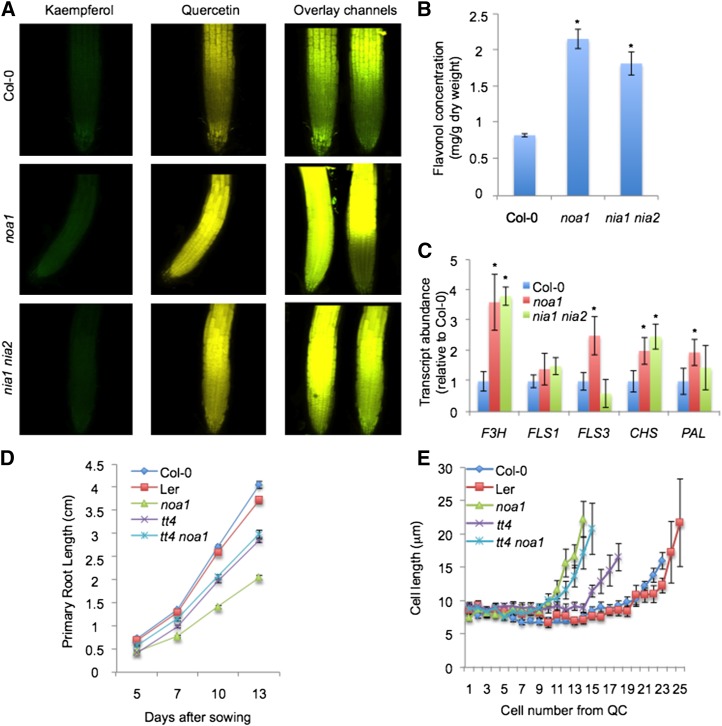
Because a metabolic interaction between flavonoids, NO, and ROS has been previously reported (Rice-Evans et al., 1997; Pollastri and Tattini, 2011; Tossi et al., 2012) and because flavonol accumulation affects root growth and development (Brown et al., 2001; Buer and Muday, 2004; Lewis et al., 2011; Grunewald et al., 2012), we tested whether reduced primary root growth in NO-deficient mutants was accompanied by a differential accumulation of flavonols and ROS. We detected flavonols by using the fluorescent indicator diphenylboric acid-2-aminoethyl ester (DPBA) in roots of the wild type (Col-0) and noa1 and nia1 nia2 mutants. In addition, we have confirmed ROS levels by 2',7'-dichlorofluorescein (H2DCF) in roots of the noa1, nia1 nia2, and nia1 nia2 noa1 mutant backgrounds, revealing enhanced ROS accumulation. Similarly, L-NMMA treatment increases ROS levels in root cells (Supplemental Fig. S3A). Our results indicate that flavonol and ROS levels increase in the region of the root in which developmental transitions are occurring. Specifically, the flavonol quercetin (gold fluorescence) shows elevated accumulation in roots of noa1 and nia1 nia2 mutants, whereas kaempferol levels (yellow-green fluorescence) are not significantly altered in these genetic backgrounds (Fig. 4A). To quantify the accumulation of flavonols, a quantitative analysis of specific flavonols glycosides was performed using HPLC-diode array detector (DAD)-mass spectrometry analysis on extracts from 8-d-old seedlings of the wild type (Col-0) and noa1 and nia1 nia2 mutants (Supplemental Tables S1–S3). Eight major flavonols were detected in the samples analyzed that were tentatively identified based on their chromatographic and spectroscopic behavior in the HPLC-DAD-mass spectrometry analyses (retention time, absorption spectrum, and molecular ion and mass spectrometry fragmentation pattern in tandem mass spectrometry and triple mass spectrometry; Supplemental Table S3). Flavonols identified in 8-d-old Arabidopsis seedlings were quercetin, kaempferol, and isorhamnetin glycosides, the most abundant which were kaempferol 3-O-rhamnosyl 7-O-glucoside and kaempferol 3-O-rahmnoside-7-O-rhamnoside (Supplemental Tables S1 and S2). Consequently, flavonol accumulation is significantly enhanced in noa1 (61% increase; P < 0.05) and nia1 nia2 (54% increase; P < 0.05) mutants (Fig. 4B; Supplemental Table S2). To elucidate whether transcripts of the genes encoding enzymes in the flavonol biosynthetic pathway were consistently altered in NO-deficient mutants, we measured the expression levels of FLAVONOL SYNTHASE1 (FLS1), FLS3, FLAVANONE 3β-HYDROXYLASE (TRANSPARENT TESTA6; F3H), CHALCONE SYNTHASE (CHS), and PHE AMMONIA-LYASE (PAL) genes in roots of the wild type (Col-0) and noa1 and nia1 nia2 mutants (Supplemental Fig. S3B; Supplemental Table S4). Our results indicate that F3H shows a strong induction in roots of noa1 and nia1 nia2 mutants, whereas FLS1, FLS3, CHS, and PAL show only a moderate induction in roots of nia1 nia2 and noa1 (Fig. 4C). To test whether differential accumulation of flavonoids could be contributing to the reduction of primary root growth in NO-deficient mutants, the transparent testa4 (tt4-1) mutant that lacks all flavonoids (Saslowsky et al., 2000) was crossed with the noa1 mutant (Supplemental Figs. S3B and S4A). Interestingly, the double mutant tt4 noa1 exhibited significantly elevated root growth (31% increase after 13 d; P < 0.005) compared with the noa1 mutant under the same growth conditions, indicating that flavonol accumulation plays a major role in the primary root growth inhibition phenotype of noa1 (Fig. 4D), although slight differences were associated with number, cell length (Fig. 4E; Supplemental Fig. S3C), and ROS homeostasis (Supplemental Fig. S3D).
Figure 4.
NO-deficient mutants are altered in the pattern of flavonol accumulation. A, Accumulation of flavonols (kaempferol and quercetin) by DPBA staining in roots of the wild type (Col-0) and noa1 and nia1 nia2 mutants. B, Quantification of flavonols by HPLC-DAD-mass spectrometry analysis in 8-d-old seedlings of the wild type (Col-0) and noa1 and nia1 nia2 mutants. Flavonol concentration is expressed as milligrams per gram of dry weight. Values represent the average of three independent experiments. *, Statistically significant difference from the wild type (P < 0.05). C, F3H, FLS1, FLS3, CHS, and PAL transcription levels in roots of the wild type (Col-0) and noa1 and nia1 nia2 mutants. *, Statistically significant difference from the wild type (P < 0.05). D, Time course (from 5–13 d after sowing) of longitudinal primary root growth of Col-0, Ler, noa1, tt4, and tt4 noa1 seedlings grown vertically under standard conditions. Error bars represent se; n > 40. E, Average cell sizes in the cortical cell layer (cells 1–25 from the QC) of 5-d-old noa1, tt4, and tt4 noa1 mutants and wild-type (Col-0 and Ler) roots.
NO Alters Auxin Sensitivity
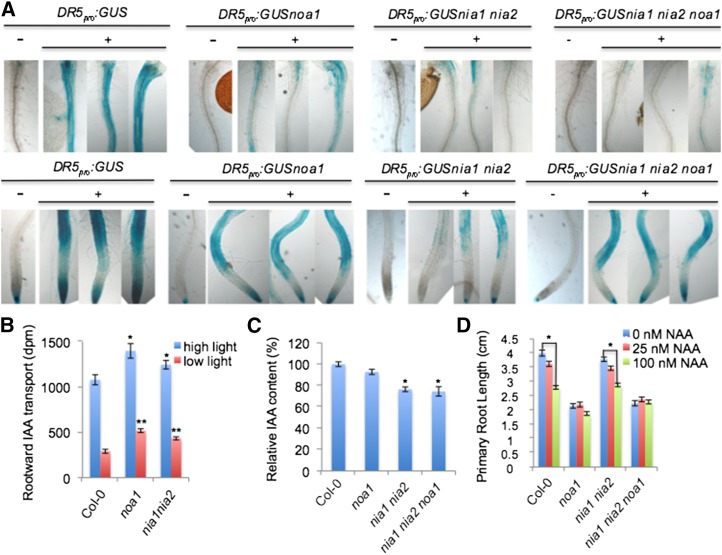
Because auxin plays a main role in primary root meristem organization, we decided to analyze whether the abnormal phenotypes present in NO-deficient mutants and upon treatment of wild-type plants with L-NMMA were related to modifications in the distribution of auxin signaling. To test whether auxin-induced gene expression is altered in noa1, nia1 nia2, and nia1 nia2 noa1 mutant backgrounds, the auxin response DR5pro:GUS reporter line was crossed with mutant plants and characterized genetically (Supplemental Fig. S4, B and C). Interestingly, DR5pro:GUS expression was reduced in roots (both in the root-to-shoot junction zone and root tips) of 3-, 5-, and 7-d-old NO-deficient seedlings compared with wild-type controls (Supplemental Fig. S5A). Similarly, exogenous L-NMMA also reduced expression of this auxin reporter (Supplemental Fig. S5, B and C), although this defect was largely recovered after transferring to MS media discarded L-NMMA toxicity (Supplemental Fig. S5C). In agreement with these genetic findings, the induction of DR5pro:GUS expression after auxin treatment in noa1, nia1 nia2, and nia1 nia2 noa1 mutant plants was reduced compared with control seedlings (Fig. 5A). We further tested whether the reduction in DR5-driven GUS expression in roots of 7-d-old NO-deficient seedlings was linked to an alteration in auxin transport and/or levels. Unexpectedly, we found that the tritium-labeled IAA rootward or acropetal transport in noa1 and nia1 nia2 mutants was increased 1.8-fold (P < 0.0001) and 1.5-fold (P = 0.005), respectively, relative to Col-0 plants (Fig. 5B). We asked whether PIN1:GFP fluorescence was altered in the noa1 mutant, consistent with an earlier reported action of elevated NO levels on this protein (Fernández-Marcos et al., 2011), but found no effect on localization or levels of this reporter. Only a slight effect on the transcripts encoding the IAA efflux proteins PIN1, PIN3, and PIN7 was observed (Supplemental Fig. S6). We quantified absolute IAA content in 7-d-old wild-type plants, noa1 single mutants, nia1 nia2 double mutants, and nia1 nia2 noa1 triple mutants. Although the noa1 mutation had no significant effect on IAA levels (7% decrease; P = 0.7), the nia1 nia2 mutations reduced IAA levels (24% decrease; P < 0.05; Fig. 5C). A similar reduction in IAA levels was observed between wild-type controls and nia1 nia2 noa1 seedlings (26% decrease; P = 0.05), indicating that the reduction in free IAA levels of nia1 nia2 may require more 1-naphthaleneacetic acid (NAA) to achieve the threshold of DR5 activation, with no difference in sensitivity (Fig. 5C). Along with the detected differences in auxin levels, we examined whether the reduced DR5pro:GUS expression in NO-deficient plants was possibly caused by impaired auxin response. To this purpose, we tested root growth inhibition by applying auxin exogenously (25 and 100 nm NAA). The primary root elongation of wild-type seedlings was inhibited by 100 nm auxin (30% decrease after 13 d of treatment; P < 0.0001; Fig. 5D), whereas primary root elongation of noa1 and nia1 nia2 seedlings was less inhibited (12% decrease in noa1; P < 0.05; 23% decrease in nia1 nia2; P < 0.0001). Interestingly, nia1 nia2 noa1 triple mutant seedlings did not show any significant reduction (3% decrease after 13 d of treatment; P = 0.6; Fig. 5D), pointing to a reduced sensitivity to exogenous auxin. Together, these results show that NO is required in maintaining auxin sensitivity.
Figure 5.
Auxin content, transport, and signaling are all additively impacted by mutations in NOA1, NIA1, and NIA2. A, The GUS-stained DR5pro:GUS-expressing wild type (Col-0) and noa1, nia1 nia2, and nia1 nia2 noa1 mutants grown vertically under standard conditions for 5 d and then transferred to control media (−) or media containing 100 nm NAA for 12 h (+). B, Acropetal auxin transport measured in roots of the 7-d-old wild type (Col-0) and 7-d-old noa1 and nia1 nia2 mutants grown at two fluence rates/light levels: 100 µE (high) and 20 µE (low). Error bars represent se; n > 15. *, Statistically significant difference from the wild type (P < 0.05). **, Statistically significant difference from the wild type (P < 0.01). C, Relative IAA content in the 7-d-old wild type (Col-0) and 7-d-old noa1, nia1 nia2, and nia1 nia2 noa1 mutants. Error bars represent se; n > 6. *, Statistically significant difference from the wild type (P < 0.05). D, Dose-response analysis of the effect of NAA on primary root length of the 13-d-old wild type (Col-0) and 13-d-old noa1, nia1 nia2, and nia1 nia2 noa1 mutants. Error bars represent se; n > 40. *, Significant differences compared with the untreated control (P < 0.05).
NO Affects Stem Cell Activity
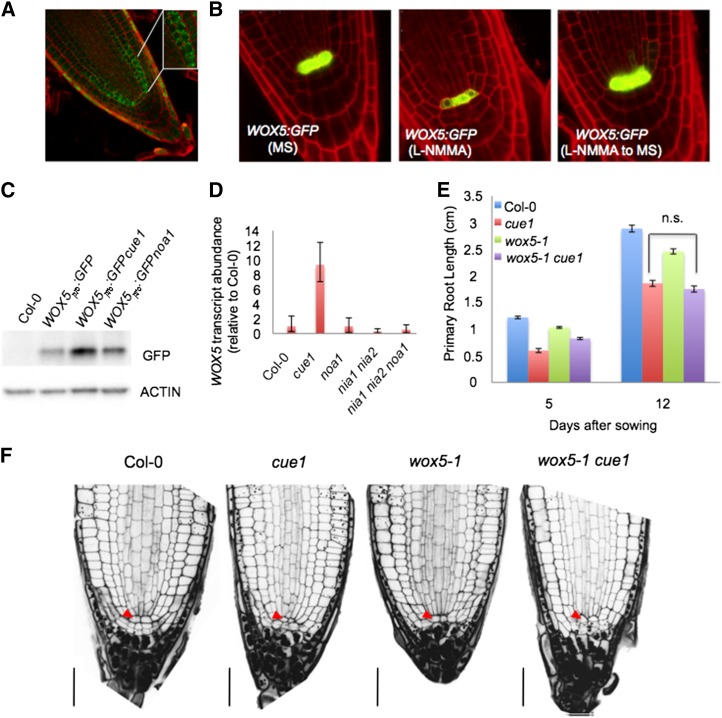
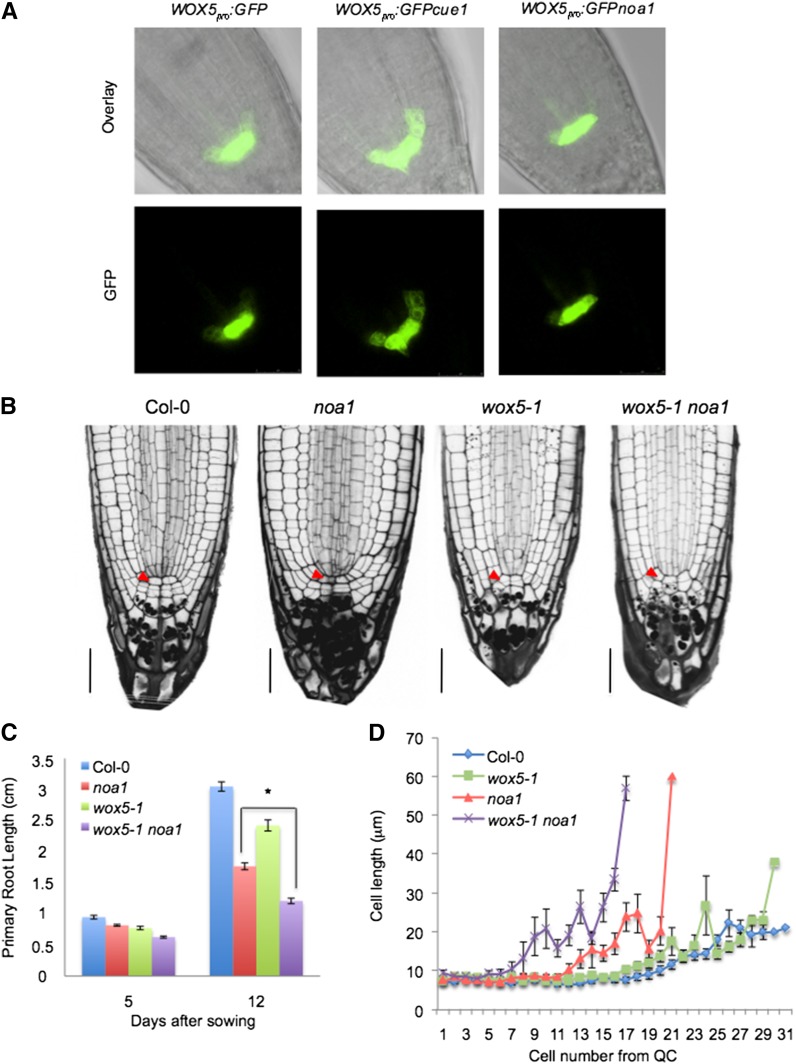
In seedlings of the triple mutant nia1 nia2 noa1 and wild-type seedlings upon treatment with L-NMMA, the cells in the QC have an abnormal shape and are enlarged compared with the wild type (Figs. 2B and 3, A and C). Notably, adjacent stem cells are even larger, suggesting that they have undergone differentiation. Because auxin is a key regulator of stem cell activity and NO alters auxin response (Fig. 5A; Fernández-Marcos et al., 2011), we hypothesized that NO could be directly affecting stem cell activity. Interestingly, careful examination by 4,5-diaminofluorescein diacetate (DAF-2DA) analysis of 5-d-old wild-type roots reveals that NO accumulates in cortex/endodermis stem cells. Additionally, the NO molecule is also detected in the immediate progeny, which generates endodermal and cortical tissues (Fig. 6A), where the signal appears to be predominantly extranuclear in most cells (Fig. 6A, inset). Farther up the immediate progeny, the signal becomes gradually weaker. WUSCHEL-related homeobox5 (WOX5) activity is required to noncell autonomously maintain the stem cells as undifferentiated, and its gene expression in the QC is dependent on auxin (Ding and Friml, 2010). Consistent with a lack of morphologically identifiable QC cells, the expression of WOX5 was considerably reduced in transgenic plants expressing functional WOX5pro:GFP subjected to L-NMMA treatment (Fig. 6B), whereas noa1 (Figs. 6, C and D and 7A), nia1 nia2, and nia1 nia2 noa1 mutations had almost no effect on WOX5 levels (Fig. 6D). Interestingly, treatments with the NO donor S-nitroso-N-acetyl-d,l-penicillamine to plants subjected to L-NMMA were able to revert the low levels of WOX5 expression to normal, suggesting that this effect was caused by a reduction in the NO content (Supplemental Fig. S7). Thus, WOX5 expression was significantly increased in chlorophyll a/b binding protein underexpressed1 (cue1), an NO-overaccumulator mutant (Figs. 6D and 7A). To find out whether different NO levels could be altering WOX5-related phenotypes, wox5-1 mutation was introduced in noa1 and cue1 genetic backgrounds (Supplemental Fig. S8). Thus, whereas the inhibition of primary root length in the double mutant wox5-1 cue1 was similar to cue1 (Fig. 6, E and F), the inhibition of primary root length in the double mutant wox5-1 noa1 (61% decrease after 12 d; P < 0.0001) was more pronounced than in noa1 (42% decrease after 12 d; P < 0.0001) and wox5-1 (20% decrease after 12 d; P < 0.0001) single mutants, showing that genetic association of wox5-1 and noa1 mutants had an additive effect on root growth (Fig. 7, B and C). Because WOX5 is only required for columella stem cell maintenance, we investigated whether the effect of the wox5-1 mutation on the proximal meristem might be masked by NOA1 function. Microscopic analysis revealed that, in noa1 single mutant, the wox5-1 mutation promotes a significant decrease in size of primary root meristem as assayed by cell number of division zone (Fig. 7D). At the same time, the inflection point on the cell-length curve marking the transition to the rapid elongation zone occurs around cell numbers 8 to 10 in the wox5-1 noa1 double mutant. These results further support a synergistic interaction of the noa1 and wox5-1 mutations on the proximal meristem, suggesting an important role for NO in the regulation of stem cell decisions.
Figure 6.
Genetic analysis between NO altered mutant backgrounds and wox5. A, Detection of endogenous NO production using DAF-2DA in 5-d-old wild-type (Col-0) roots. The RNS-dependent biosensor fluoresces strongly in cortex/endodermis initials (inset) and the immediate progeny generating endodermis and cortex tissues in meristem cells. B, WOX5 marker analysis in wild-type (Col-0) roots grown in the absence or presence of 20 µm L-NMMA and then transferred to MS medium. C, Immunoblot analysis with anti-GFP antiserum of in vivo levels of GFP protein in root extracts of WOX5pro:GFP, WOX5pro:GFPcue1, and WOX5pro:GFPnoa1 5-d-old seedlings. Actin protein levels also were determined as a loading control. D, WOX5 transcription levels in roots of the wild type (Col-0) and cue1, noa1, nia1 nia2, and nia1 nia2 noa1 mutants. E, Primary root lengths in the 5- and 12-d-old wild types (Col-0) and 5- and 12-d-old cue1, wox5-1, and wox5-1 cue1 mutants. Error bars represent se; n > 40. n.s., No statistically significant difference. F, Root sections of 5-d-old wild-type (Col-0), cue1, wox5-1, and wox5-1 cue1 seedlings. Red arrowheads indicate QC. Bars = 30 µm.
Figure 7.
NO effect in the stem cell homeostasis through WOX5. A, WOX5 marker analysis in the wild type (Col-0) and cue1 and noa1 mutants. B, Root sections of the 5-d-old wild type (Col-0) and 5-d-old noa1, wox5-1, and wox5-1 noa1 seedlings. Red arrowheads indicate QC. Bars = 30 µm. C, Primary root length in 5- and 12-d-old wild-type (Col-0) seedlings and noa1, wox5-1, and wox5-1 noa1 mutants. Error bars represent se; n > 40. *, Statistically significant difference (P < 0.05). D, Average cell sizes in the cortical cell layer (cells 1–31 from QC) of 7-d-old wox5-1, noa1, and wox5-1 noa1 mutant and wild-type (Col-0) roots grown vertically under standard conditions.
DISCUSSION
Plants with mutations in the genes encoding proteins that synthesize or metabolize NO have linked NO to plant development (nia1 nia2 noa1 [Lozano-Juste and León, 2010a, 2010b], NO overproducer1 [nox1]/chlorophyll a/b binding protein underexpressed1 [cue1; He et al., 2004], S-Nitrosoglutathione reductase [gsnor; Kwon et al., 2012], and Arabidopsis nonsymbiotic hemoglobin1 [AHb1; Perazzolli et al., 2004]). Triple mutant nia1 nia2 noa1 is impaired in nitrate reductase- and AtNOA1-mediated NO biosynthetic pathways. nia1 nia2 noa1 plants displaying reduced root length (Fig. 1) reverted after NO donor application (Lozano-Juste and León, 2010a, 2010b). To investigate whether the root growth phenotype of nia1 nia2 noa1 was directly linked to the impairment in NO biosynthesis in the noa1, nia1, or nia2 mutations, we examined the wild-type response to L-NMMA, which is a useful tool for testing the biological role of NO in plants (Foissner et al., 2000). Treatment with L-NMMA did not affect seed germination potential (Supplemental Fig. S1) but produced roots that experienced limited growth (Fig. 1) and were unable to generate and maintain the root meristematic tissue (Fig. 3A). However, treated plants were able to recover after transferred to normal media, suggesting lack of toxicity (Supplemental Fig. S2). Interestingly, L-NMMA treatment induced ROS accumulation (Supplemental Fig. S3) and mimicked the root phenotype of Arabidopsis glutathione-deficient mutant root meristemless1 (rml1; Cheng et al., 1995), resulting in primary roots of limited growth. Taken together, our results suggest that depletion of NO could alter intracellular redox status in a way similar to that of the rml1 mutant or growing plants in the presence of buthionine sulphoximine (BSO), a specific inhibitor of γ-glutamylcysteine synthetase (Koprivova et al., 2010). Several lines of experimental evidence indicate that NO acts as a potent antioxidant in response to methylviologen damage (Beligni et al., 2002) as well as in aleurone cells (Beligni and Lamattina, 1999; Beligni et al., 2002). Furthermore, NOA1 protein attenuates oxidative stress and ROS (Ahlfors et al., 2009).
Results presented in this work further show that plants impaired in certain aspects of NO production (i.e. noa1 and nia1 nia2 mutants) also increased accumulation of flavonols (Fig. 4; Supplemental Fig. S2; Supplemental Tables S1 and S2) and ROS levels (Supplemental Fig. S3). We, therefore, propose that flavonoids are specifically synthesized in noa1 and nia1 nia2 mutants to rescue the inactivation of NOA1 and NIA1/NIA2. This increase may function to buffer the ROS accumulation produced in NO-deficient mutants, probably preserving cell redox homeostasis. Interestingly, our data also show that the reduced root length of noa1 plants is caused by flavonoid accumulation rather than NO homeostasis Thus, the combined noa1 mutant with mutations affected in flavonoid biosynthesis genes (like tt4) support a role of flavonoids in this NO-deficient mutant phenotype (Fig. 4; Supplemental Fig. S3). Flavonoids act as ROS-scavenging compounds (Rice-Evans et al., 1997; Pollastri and Tattini, 2011), and their accumulation affects root growth and development (Brown et al., 2001; Buer and Muday, 2004; Lewis et al., 2011; Grunewald et al., 2012), being generally considered a hallmark of plant stress. We note that accumulation of flavonoids in NADPH-dependent thioredoxin reductase (ntra ntrb) mutants has been shown, leading to tolerance against UV light (Bashandy et al., 2009).
Plants with mutations in genes encoding proteins that modify ROS accumulation (Dunand et al., 2007) or distribution (Tsukagoshi et al., 2010) display altered root growth. According to Bashandy et al., 2010, NADPH-dependent thioredoxin reductase and glutathione biosynthesis mutations reduce root meristem size. Similarly, double mutant RADICAL-INDUCED CELL DEATH (RCD) and SIMILAR TO RADICAL-INDUCED CELL DEATH ONE1 (SRO1; rcd1 sro1) accumulates ROS, and rcd1 sro1 roots display short root apical meristems (Teotia and Lamb, 2011). Our observations indicate that the reduction of primary root growth in nia1 nia2 noa1 or plants grown in the presence of L-NMMA is linked to a reduction in both the number of cells and the length of the primary root meristem (Figs. 2 and 3A). Thus, analysis regarding the expression of the mitotic marker CYCB1;1 reveals that cell division ceased 10 d after germination in the presence of L-NMMA and in noa1 genetic background compared with the wild type (Fig. 3B). We, therefore, hypothesize that L-NMMA-sensitive generation of NO is required for normal cell proliferation at the root tip.
Previous reports in alfalfa cell culture indicated that application of L-NMMA inhibited the activation of the plant cell cycle (Otvös et al., 2005). Similarly, phenotypic analysis of rml mutants indicated that RML is specifically involved in activating the cell division cycle in root apical cells (Cheng et al., 1995). More recently, Shen et al. (2013) showed that NO deficiency caused by loss of NOA1 impairs cellular development, such as the duration of the mitotic phase and the timing of the transition to endocycles in noa1 mutant leaves, which can be reverted by constitutive expression of the cell-cycle gene CYCD3;1. Because NO seems to accumulate at the immediate progeny of the ground tissue stem cells (cortex/endodermis initial cells; Fig. 6A), where NOA1 and NIA2 are highly expressed (Sozzani et al., 2010), we tentatively suggest a specific link between NO and the divisions of endodermis, a rate-limiting tissue for root growth (Ubeda-Tomás et al., 2008), based on that NOA/NIA-dependent, L-NMMA-sensitive NO synthesis is required for growth-limiting stem cell division in Arabidopsis roots.
Experimental evidence indicates that exposure of Arabidopsis to BSO leads to disappearance of QC cell markers (Koprivova et al., 2010). Similarly, QC identity is compromised in rcd1 sro1 roots (Teotia et al., 2010). Consistent with a lack of morphologically identifiable QC cells, the expression of the QC marker WOX5 was considerably reduced in transgenic plants expressing functional WOX5pro:GFP subjected to L-NMMA treatment (Fig. 6B), although this defect was largely recovered after transferring to normal media and NO donor application (Supplemental Fig. S7). Whereas expression analysis revealed that noa1, nia1 nia2, and nia1 nia2 noa1 mutations had almost no effect on WOX5 levels (Figs. 6, C and D and 7A), increased NO content in the cue1 mutant slightly expanded the WOX5pro:GFP domain compared with noa1 and the wild type (Fig. 7A). Although WOX5 is mainly expressed in QC cells, it does not seem to be a major factor in QC specification. WOX5 is required to noncell autonomously maintain the columella stem cells (distal stem cells) as undifferentiated. Therefore, in wox5-1 roots, the cells at the stem cell position but not the QC accumulate starch granules. In contrast, the proximal meristem appears normal, because the wox5-1 mutation might be masked by redundant function of SHORTROOT/SCARECROW, PLETHORA1, and PLETHORA2 activity (Sarkar et al., 2007). In animals, there are several studies indicating that NO plays an important role in regulating stem cell decisions, contributing to cell fate determination of stem cells (Mujoo et al., 2011). Because NO presents a maxima at cortex/endodermis stem cells, we hypothesized that NO might act redundantly with WOX5 function to induce differentiation of stem cells into cortex and endodermis cells. Interestingly, association of wox5-1 and noa1 mutations had an additive effect on root growth (Fig. 7, B–D). Thus, careful microscopic analysis revealed that, in the noa1 single mutant, the wox5-1 mutation promotes a significant decrease in size of primary root meristem as assayed by cell number of division zone (Fig. 7, B and D). This result confirms that the effect of wox5-1 mutation on the proximal meristem might be masked by the redundant functions reported by Sarkar et al. (2007), including NOA1 activity, and contributes to our understanding of stem cell niche homeostasis by the interplay between auxin and NO on root growth.
Our results also implicate NO in auxin response. Analysis of the DR5pro:GUS expression in noa1, nia1 nia2, and nia1 nia2 noa1 mutants showed that DR5pro:GUS expression was reduced in roots of all different genotypes, indicating that auxin response is reduced in NO-deficient mutants (Fig. 5A; Supplemental Fig. S5A). Furthermore, the inhibitory effects of auxin on root growth were partially suppressed in NO-deficient plants (Fig. 5D).
By using ROS-inducer compounds, Blomster et al. (2011) and Cheng et al. (2011) reported that ROS treatment caused auxin signaling to be transiently suppressed. Similarly, analysis of the DR5 expression in rcd1 sro1 plants (Teotia and Lamb, 2011) or Arabidopsis MONOTHIOL GLUTAREDOXIN (Cheng et al., 2011) revealed that auxin response maxima are impaired to some extent while accumulating higher levels of ROS. Consistent with this evidence, Peer et al. (2013) suggested a model where ROS accumulation reduces auxin response by conjugation or catabolism of auxin (IAA) to an irreversible and inactive catabolic product, which would be unable to induce DR5 expression.
Finally, compared with controls, NO-deficient plants exhibited reduced DR5 induction in response to exogenously applied auxin (Fig. 5A). We further show that auxin levels are perturbed in NO-deficient mutants, suggesting that NO alters auxin metabolism (Fig. 5C). Although the reduction in free IAA levels of nia1 nia2 may indicate that more NAA is required to achieve the threshold of DR5 activation with no difference in sensitivity, noa1 mutation seems to impair auxin sensitivity by specifically altering auxin response. These results are consistent with the results for the ntra ntrb cadmium sensitive2 mutant, which shows that auxin concentration was almost 3-fold lower in triple mutant seedlings (Bashandy et al., 2010). Interestingly, although BSO treatment resulted in loss of the auxin carrier PIN1 (Koprivova et al., 2010), rootward auxin transport is enhanced in mutants with lower levels of NO, such as noa1 and nia1 nia2, in a mechanism not mediated by changes in PIN1 protein levels. Oppositely, high levels of endogenous NO reduce rootward auxin transport (Fernández-Marcos et al., 2011), supporting the hypothesis that altered NO levels cause an important effect in auxin transport capacity. Based on our previous report (Fernández-Marcos et al., 2011) and this study, the alteration in the homeostasis of NO seems to be detrimental for root growth, with very similar developmental consequences in both high- and low-NO concentrations.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and ecotype Landsberg erecta of Arabidopsis (Ler) were obtained from the Nottingham Arabidopsis Stock Centre. Loss-of-function noa1 (Guo et al., 2003), nia1 nia2 (Wilkinson and Crawford, 1993), nia1 nia2 noa1 (Lozano-Juste and Leon, 2010a, 2010b), cue1 (He et al., 2004), and wox5-1 (Sarkar et al., 2007) in the Col-0 background were previously described. Loss-of-function tt4-1 (Koornneef, 1990) in the Ler background was also previously described. CycB1;1pro:GUSDB (Colón-Carmona et al., 1999), DR5pro:GUS/GFP (Ulmasov et al., 1997), WOX5pro:GFP (Sarkar et al., 2007), and PIN1pro:GFP-PIN1 (Heisler et al., 2005) were obtained from the Arabidopsis Biological Resource Center. Genetic crosses were performed using noa1, nia1 nia2, nia1 nia2 noa1, and cue1 mutants as donors in all the cases and the corresponding acceptor lines. For in vitro culture, Arabidopsis seeds were surface sterilized in 75% (v/v) sodium hypochlorite and 0.01% (v/v) Triton X-100 for 5 min and washed three times in sterile water before sowing. Seeds were then stratified for 3 d at 4°C. For all analyses, plants were grown vertically under a 16-h-light/8-h-dark photoperiod at 22°C on one-half-strength MS root medium (Duchefa) supplemented with 7.5 g L−1 Suc and 15 g L−1 agar. Values were analyzed with the two-tailed Student’s t test.
To assess the effects of L-NMMA on root growth and seed germination, Col-0, CycB1;1pro:GUSDB, and DR5pro:GUS/GFP seeds were sown on the modified MS medium with or without 10, 20, or 50 µm L-NMMA (Sigma).
DR5pro:GUS, DR5pro:GUSnoa1, DR5pro:GUSnia1 nia2, and DR5pro:GUSnia1 nia2 noa1 lines were grown or transferred onto media supplemented with 100 nm NAA (Sigma). GUS staining was compared after 12 h.
Flavonoid Extraction
Twenty milligrams of freeze-dried Arabidopsis seedlings were macerated in 70% (v/v) MeOH containing 0.5% (v/v) trifluoracetic acid; the mixture was vortexed, sonicated for 10 min, and then centrifuged at 4,000g. The supernatant was collected, and the residue was further submitted two times to the same process. The extracts were combined and concentrated under vacuum until the methanol was removed, and the resultant aqueous phase was washed with n-hexane to eliminate chlorophyll. The extract without chlorophyll was deposited onto a C18 Sep-Pak Cartridge (Waters) for its purification. The cartridge had been previously activated with methanol followed by water. More polar substances (sugars and acids) were removed from the cartridge with ultrapure water; then, flavonoids were eluted with acetonitrile:H2O (80:20) containing 0.5% (v/v) trifluoracetic acid. The flavonoid fraction was vacuum concentrated and dissolved in aqueous 2.5% (v/v) acetic acid for HPLC-DAD-electrospray ionization (ESI)/mass spectrometry analysis. For each sample, three independent extracts from different treatments were prepared, purified, and analyzed.
HPLC-DAD-ESI/Mass Spectrometry Analysis of Flavonoids
The flavonoid extracts were analyzed using a Hewlett-Packard 1100 Series Liquid Chromatograph (Agilent Technologies). Separation was achieved on an AQUA (Phenomenex) reverse-phase C18 column (5 µm, 150- × 4.6-mm i.d.) thermostatted at 35°C. The HPLC conditions described by Di Paola-Naranjo et al. (2004) were used with minor modifications. The solvents used were 2.5% (v/v) acetic acid in water (solvent A) and 100% HPLC-grade acetonitrile (solvent B). The gradient used was isocratic 10% solvent B for 5 min, 10% to 13% solvent B over 15 min, and 15% to 35% over 25 min at a flow rate of 0.5 mL min−1. Double online detection was carried out in a DAD using 360 as the preferred wavelength and a mass spectrometer connected to the HPLC system by the DAD cell outlet.
Mass spectrometry analyses were performed using a Finnigan LCQ MS Detector (Thermoquest) equipped with an atmospheric pressure ionization source using an ESI interface. Both the sheath gas and the auxiliary gas were nitrogen at flow rates of 1.2 and 6 L min−1, respectively. The capillary and source voltages were 4 V and 4.5 kV, respectively, and the capillary temperature was 195°C. Spectra were obtained in negative ion mode, and mass spectrometry was programmed to carry out a series of three consecutive scans: a full mass from between mass-to-charge ratio (m/z) 150 and m/z 1,500, a zoom scan of the most abundant ion in a ±5-mass units range, and a mass spectrometry-mass spectrometry scan of the most abundant ion in the full mass using a normalized energy of collision of 45%. Flavonols were identified based on their chromatographic behavior, and UV and mass spectra were compared with our library and literature data. Pseudomolecular and tandem mass spectrometry product ions of the detected compounds are shown in Supplemental Table S3.
Flavonoid Quantification
Flavonol quantification was performed from the areas of the peaks recorded at 360 nm by comparison with external calibration curves obtained for the corresponding aglycones (i.e. quercetin, kaempferol, and isorhamnetin purchased from Extrasynthese). Each sample was analyzed in triplicate using three independent extracts, and the results were expressed in milligrams per gram of dry weight (aglycone equivalents). Total flavonol content was obtained by summing up the concentrations of the individual compounds.
DPBA Staining of Arabidopsis Seedlings and Quantification
Individuals were submerged in an aqueous solution containing 0.01% (v/v) Triton X-100 and 2.52 mg mL−1 DPBA for 7 min followed by a distilled water wash for 7 min. Roots were mounted in distilled water and imaged on a Zeiss 710 Laser-Scanning Confocal Microscope (Carl Zeiss Microimaging) with excitation at 458 nm. DPBA fluoresces with distinct spectral properties in complex with kaempferol and quercetin (Neu, 1956; Sheahan and Rechnitz, 1992; Peer et al., 2001), with quercetin DPBA having a longer wavelength fluorescence emission than kaempferol DPBA. We optimized laser-scanning confocal microscope settings to spectrally separate fluorescence of kaempferol DPBA and quercetin DPBA (which reflect the fluorescence of the glycosylated forms of kaempferol and quercetin, respectively) by capturing a window of the emission spectrum for each compound where they do not overlap (Lewis et al., 2011).
Estimation of Protein Levels by Immunoblots
A total of 30 to 50 mg of root tissue was ground in liquid nitrogen and resuspended in 1 mL of 50 mm Tris-HCl (pH 7.5) supplemented with 75 mm NaCl, 15 mm EGTA, 15 mm MgCl2, 1 mm dithiothreitol, 0.1% (v/v) Tween 20, 1 mm NaF, 0.2 m NaV, 2 mm Na-pyro-P, 60 mm β-glycerol-P, and 1× proteases inhibitor mix (Roche), and it was incubated on ice for 10 min followed by centrifugation for 10 min at 13,000g at 4°C. Protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad) based on the method by Bradford (1976); 10 µg of total protein was loaded per well in SDS-acrylamide/bisacrylamide gel electrophoresis using Tris-Gly-SDS buffer. Proteins were electrophoretically transferred to an Inmobilon-P polyvinylidene difluoride membrane (Millipore) using the Trans-Blot Cell (Bio-Rad). Membranes were blocked in phosphate-buffered saline-Tween containing 2% (w/v) ECL Advance Blocking Agent (Amersham) and probed with antibodies diluted in blocking buffer. Anti-GFP (Clontech), anti-PIN1 (provided by Klaus Palme, University of Freiburg, Germany), anti-Actin clone 10-B3 Purified Mouse Immunoglobulin (Sigma), and ECL-Peroxidase-Labeled Anti-Mouse (Amersham) antibodies were used in the western-blot analyses. Detection was performed using the ECL Advance Western Blotting Detection Kit (Amersham), and the chemiluminescence was detected using an Intelligent Dark-Box II, LAS-1000 Scanning System (Fujifilm).
Transcript Quantification
For quantification of transcripts, plants were grown vertically, and roots were harvested upon removal of hypocotyls and shoot with a razor blade. RNA was isolated with Tri-Reagent (Ambion), and complementary DNA was synthesized using the Ambion Retroscript Kit. Relative quantification using real-time PCR was performed using ACTIN8 for normalization (Fernández-Marcos et al., 2011). Primer sequences for quantification of F3H, FLS1, FLS3, CHS, PAL, PIN1, PIN3, PIN7, and WOX5 transcripts and the analysis of mutants are listed in Supplemental Table S3.
Determination of IAA in Plant Tissue
For each sample, 150 mg of 7-d-old seedlings was pooled. Plant material was grounded in liquid nitrogen and resuspended in methanol to a concentration of 0.15 g fresh weight mL−1. Each sample was spiked with 30 pmol of [2H2]-IAA (internal standard; C/D/N Isotopes) and incubated under slight agitation at 70°C over 1 h. After centrifugation (13.2 rpm at 4°C for 20 min), the supernatant was transferred into fresh microreaction tubes and dried under vacuum for subsequent gas chromatography-tandem mass spectrometry analysis, which was conducted with minor variation as previously described by Pollmann et al. (2009). In brief, the dried residues were dissolved in 30 µL of methanol, to which 200 µL of diethyl ether was added followed by a 20-min ultrasonic treatment (BRANSONIC 5510 DTH; Branson Ultrasonics). The solution was centrifuged (13.2 rpm at 4°C for 10 min), and the particle-free supernatant was transferred into a fresh tube. Next, the sample was applied to custom-made microscale aminopropyl solid phase extraction cartridges. The cartridge was washed two times with 250 µL of CHCl3:2-propanol (2:1, v/v), and the IAA-containing fraction was eluted with 400 µL of acidified diethyl ether (2% acetic acid, v/v). The eluates were taken to dryness, dissolved in 50 µL of diethyl ether, and transferred into 0.8-mL autosampler vials (Supelco; Chromacol). The eluates were dried under a gentle stream of nitrogen and then dissolved in 20 µL of N,O-Bis(trimethylsilyl)trifluoroacetamide (Supelco). Trimethylsilylization was carried out at 70°C over 30 min. To analyze auxin contents in the samples, 1 µL of each was injected splitless by a CombiPAL Autoinjector (CTC Analytics) into a Bruker Scion-455 Gas Chromatograph (BRUKER Daltonics) equipped with a 30-m × 0.25-mm i.d. fused silica capillary column with a chemical bond 0.25-µm ZB35 stationary phase (Phenomenex). Helium at a flow rate of 1 mL min−1 served as the mobile phase. A pressure pulse of 40 pounds per square inch over 1 min was used to force the transfer of compounds from the injector into the column. The injector temperature was 250°C, and the column temperature was held at 50°C for 1 min and then increased by 20°C per minute to 250°C and held for another 4 min at 250°C. The column effluent was introduced into the ion source of a Bruker Scion-TQ Triple Quadrupole Mass Spectrometer. The mass spectrometer was used in ESI-multiple reaction monitoring mode. The transfer line temperature was set to 250°C, and the ion source temperature was set to 200°C. Ions were generated with −70 eV at a filament emission current of 80 µA. The dwell time was 100 ms, and the reactions m/z 247 to m/z 130 (endogenous IAA) and m/z 249 to m/z 132 ([2H2]-IAA; internal standard) were recorded. Argon set at 2.0 mTorr was used as the collision gas. The amount of the endogenous compound was calculated from the signal ratio of the unlabeled over the stable isotope-containing mass fragment observed in the parallel measurements.
Measurement of Rootward Auxin Transport in Arabidopsis Roots
This method was previously described in Lewis and Muday (2009) and Fernández-Marcos et al. (2011) and used the application of 100 nm tritium-labeled IAA (American Radiolabeled Chemicals) to the root-shoot junction. After 8 h, the radioactivity accumulating in a 5-mm section at the root apex was determined. To accurately compare noa1 and nia1 nia2 with the wild type, seedlings were grown in 20 and 100 µmol m−2 s−2 cool white light. Under reduced light intensity, the roots grew more similarly, allowing us to measure transport over equivalent distances from site of application to root tips.
GUS Staining
GUS staining of CycB1;1pro:GUSDB, CycB1;1pro:GUSDBnoa1, DR5pro:GUS, DR5pro:GUSnoa1, DR5pro:GUSnia1 nia2, and DR5pro:GUSnia1 nia2 noa1 lines was performed as previously described (Fernández-Marcos et al., 2011). Staining was examined after a 2-h incubation at 37°C.
GFP, DAF-2DA, and DCF Imaging
Root apical meristem morphology and pericycle cell sizes were examined as described in Fernández-Marcos et al., 2011. NO accumulation in the root tip was detected using DAF-2DA (Sigma) as described in Fernández-Marcos et al. (2011), and ROS levels were detected using H2DCF. For live imaging of GFP, DAF-2DA, and H2DCF, cell walls were counterstained with 4 mg mL−1 PI (Sigma) in water, and GFP, DAF-2DA, H2DCF, and PI fluorescence was examined with a Leica SP 2 Confocal Microscope. For fluorescence detection, the excitation source was an argon ion laser at 488 nm, and detection filters were between 515 and 530 nm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genetic and pharmacological effects of NO depletion on primary root growth.
Supplemental Figure S2. Effect of L-NMMA on primary root growth.
Supplemental Figure S3. Contribution of flavonol and ROS accumulation to the NO-deficient mutant phenotype.
Supplemental Figure S4. Genetic characterization of noa1-based double mutants and reporter lines under NO-deficient mutant backgrounds.
Supplemental Figure S5. Effect of NO depletion on auxin signaling in the primary root.
Supplemental Figure S6. Effect of NO-deficient mutants on PIN1 transcript and protein accumulation.
Supplemental Figure S7. Recovery of WOX5 marker expression in the presence of L-NMMA and S-nitroso-N-acetyl-d,l-penicillamine.
Supplemental Figure S8. Genetic characterization of wox5-based double mutants and reporter lines.
Supplemental Table S1. Concentration of individual flavonols (milligrams per gram of dry weight) in Col-0 wild-type seedlings and NO-deficient mutants nia1 nia2 and noa1.
Supplemental Table S2. Total flavonol concentration (milligrams per gram of dry weight) in Col-0 wild-type seedlings and NO-deficient mutants nia1 nia2 and noa1.
Supplemental Table S3. Pseudomolecular and tandem mass spectrometry product ions of the analyzed flavonols.
Supplemental Table S4. List of primers used in the quantitative real-time PCR analysis and mutant genetic characterization.
Supplementary Material
Acknowledgments
We thank Roberto Solano, Crisanto Gutierrez, and Dolores Rodriguez for critical reading of the article and stimulating discussions and Centro de Investigación del Cáncer-Universidad de Salamanca for excellent technical fluorescence microscopy assistance.
Glossary
- BSO
buthionine sulphoximine
- Col-0
Columbia-0
- DAD
diode array detector
- DPBA
diphenylboric acid-2-aminoethyl ester
- ESI
electrospray ionization
- H2DCF
2',7'-dichlorofluorescein
- IAA
indole-3-acetic acid
- Ler
ecotype Landsberg erecta of Arabidopsis
- L-NMMA
nitric oxide synthase inhibitor
- MS
Murashige and Skoog
- m/z
mass-to-charge ratio
- NAA
naphthylacetic acid
- NO
nitric oxide
- PI
propidium iodide
- QC
quiescent center
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
Footnotes
This work was supported by the Ministerio de Educación y Ciencia Spain (grant nos. EcoSeed-311840 ERC.KBBE.2012.1.1–01, BIO2011–26940, and CSD2007–00057 [TRANSPLANTA]), the Junta de Castilla y León (grant no. SA239U13 to O.L.), the U.S. National Science Foundation (grant no. IOS–0820717), and the National Science Foundation Major Research Instrumentation Program (grant nos. MRI–0722926 to G.K.M. and DBI–1039755 to G.K.M.) for purchase of the confocal microscope.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahlfors R, Brosché M, Kollist H, Kangasjärvi J (2009) Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J 58: 1–12 [DOI] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Taconnat L, Renou JP, Meyer Y, Reichheld JP (2009) Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol Plant 2: 249–258 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L (1999) Nitric oxide protects against cellular damage produced by methylviologen herbicides in potato plants. Nitric Oxide 3: 199–208 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B (2010) Root development-two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Blomster T, Salojärvi J, Sipari N, Brosché M, Ahlfors R, Keinänen M, Overmyer K, Kangasjärvi J (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol 157: 1866–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Seeley KA, Sung ZR (1995) RML1 and RML2, Arabidopsis genes required for cell proliferation at the root tip. Plant Physiol 107: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Liu X, Wu Q, Thompson SM, Lin J, Chang J, Whitham SA, Park S, Cohen JD, et al. (2011) Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J Biol Chem 286: 20398–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L (2004) Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola-Naranjo RD, Sánchez-Sánchez J, González-Paramás AM, Rivas-Gonzalo JC (2004) Liquid chromatographic-mass spectrometric analysis of anthocyanin composition of dark blue bee pollen from Echium plantagineum. J Chromatogr A 1054: 205–210 [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J (2010) Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O (2011) Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc Natl Acad Sci USA 108: 18506–18511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lorenzo O (2012) Nitric oxide: an emerging regulator of cell elongation during primary root growth. Plant Signal Behav 7: 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner I, Wendehenne D, Langebartels C, Durner J (2000) In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J 23: 817–824 [DOI] [PubMed] [Google Scholar]
- Freschi L. (2013) Nitric oxide and phytohormone interactions: current status and perspectives. Front Plant Sci 4: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hu X, Neill SJ, Tang Z, Cai W (2005) Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol 137: 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illés P, Schlicht M, Pavlovkin J, Lichtscheidl I, Baluška F, Ovečka M (2006) Aluminium toxicity in plants: internalization of aluminium into cells of the transition zone in Arabidopsis root apices related to changes in plasma membrane potential, endosomal behaviour, and nitric oxide production. J Exp Bot 57: 4201–4213 [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1990) Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf Serv 28: 1–4 [Google Scholar]
- Koprivova A, Mugford ST, Kopriva S (2010) Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Draye X, Bennett MJ (2008) Modelling root growth and development. SEB Exp Biol Ser 61: 195–211 [PubMed] [Google Scholar]
- Kwon E, Feechan A, Yun BW, Hwang BH, Pallas JA, Kang JG, Loake GJ (2012) AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236: 887–900 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BS, Muday GK (2011) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol 156: 144–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MC, Graziano M, Polacco JC, Lamattina L (2006) Nitric oxide functions as a positive regulator of root hair development. Plant Signal Behav 1: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J (2010a) Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol 152: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J (2010b) Nitric oxide modulates sensitivity to ABA. Plant Signal Behav 5: 314–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujoo K, Krumenacker JS, Murad F (2011) Nitric oxide-cyclic GMP signaling in stem cell differentiation. Free Radic Biol Med 51: 2150–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu R. (1956) Aromatische borsäuren als bathochrome reagentien für flavone. J Anal Chem 151: 328–332 [Google Scholar]
- Otvös K, Pasternak TP, Miskolczi P, Domoki M, Dorjgotov D, Szucs A, Bottka S, Dudits D, Fehér A (2005) Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J 43: 849–860 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lamattina L (2003) Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol 132: 1241–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lombardo MC, Lamattina L (2004) Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol 135: 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L (2002) Nitric oxide is required for root organogenesis. Plant Physiol 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Cheng Y, Murphy AS (2013) Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64: 2629–2639 [DOI] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M (2004) Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri S, Tattini M (2011) Flavonols: old compounds for old roles. Ann Bot (Lond) 108: 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Düchting P, Weiler EW (2009) Tryptophan-dependent indole-3-acetic acid biosynthesis by ‘IAA-synthase’ proceeds via indole-3-acetamide. Phytochemistry 70: 523–531 [DOI] [PubMed] [Google Scholar]
- Rice-Evans CA, Miller NJ, Paganga G (1997) Antioxidant properties of phenolic compounds. Trends Plant Sci 2: 152–159 [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Dana CD, Winkel-Shirley B (2000) An allelic series for the chalcone synthase locus in Arabidopsis. Gene 255: 127–138 [DOI] [PubMed] [Google Scholar]
- Sheahan JJ, Rechnitz GA (1992) Flavonoid-specific staining of Arabidopsis thaliana. Biotechniques 13: 880–883 [PubMed] [Google Scholar]
- Shen Q, Wang YT, Tian H, Guo FQ (2013) Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol Plant 6: 1214–1225 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia S, Lamb RS (2011) RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J Exp Bot 62: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia S, Muthuswamy S, Lamb RS (2010) Radical-induced cell death1 and similar to RCD one1 and the stress-induced morphogenetic response. Plant Signal Behav 5: 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrile MC, París R, Calderón-Villalobos LI, Iglesias MJ, Lamattina L, Estelle M, Casalongué CA (2012) Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J 70: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi V, Lombardo C, Cassia R, Lamattina L (2012) Nitric oxide and flavonoids are systemically induced by UV-B in maize leaves. Plant Sci 193-194: 103–109 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, Hedden P, Bhalerao R, Bennett MJ (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10: 625–628 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1993) Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet 239: 289–297 [DOI] [PubMed] [Google Scholar]
- Xu J, Wang W, Yin H, Liu X, Sun H, Mi Q (2010) Exogenous nitric oxide improves antioxidative capacity and reduces auxin degradation in roots of Medicago truncatula seedlings under cadmium stress. Plant Soil 326: 321–330 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.