Abstract
Staphylococcus aureus is an important cause of infections and HIV-infected individuals are frequently susceptible to this pathogen. The aim of this study was to perform a systematic review to identify both the risk factors associated with colonization/infection by methicillin-resistant S. aureus in HIV patients and the methods used for characterization of isolates. An electronic search of articles published between January 2001 and December 2013 was first conducted. Among 116 studies categorized as being at a quality level of A, B or C, only 9 studies were considered to have high methodological quality (level A). The majority of these studies were retrospective (4/9 studies). The risk factors associated with colonization/infection by S. aureus were use of antimicrobials (4/9 studies), previous hospitalization (4/9 studies) and low CD4+ T lymphocyte counts (<200 cells/μl) (3/9 studies). Culture in mannitol salt agar (3/9 studies) and the latex agglutination test (5/9 studies) were the main methods used for bacterial phenotypic identification. Genotypic profiles were accessed by pulsed-field gel electrophoresis (6/9 studies) and USA300 was the most prevalent lineage (5/9 studies). Most isolates were resistant to erythromycin (3/9 studies) and susceptible to vancomycin (4/9 studies). Ultimately, use of antimicrobials and previous hospitalization were the main risk factors for colonization/infection by methicillin-resistant S. aureus in HIV-infected individuals. However, the numbers of evaluated patients, the exclusion and inclusion criteria and the characterization of the S. aureus isolates were not uniform, which made it difficult to establish the characteristics associated with HIV patients who are colonized/infected by S. aureus.
Keywords: S. aureus, HIV, Infection and Colonization
INTRODUCTION
HIV/AIDS is characterized as a serious public health problem and is considered as a major challenge worldwide 1. According to the World Health Organization 2, approximately 33.3 million people were infected with HIV/AIDS in 2009 and it is estimated that a total of 2.6 million individuals are infected annually. In 2010, these figures already exceeded 34 million infected individuals, including adults and children.
An infected person may not show any clinical symptoms that are suggestive of the infection for years. However, continued viral replication and the consequent decline in CD4+ T cells may lead to the onset of clinical manifestations. In addition, upon acquiring immunodeficiency, the infected person becomes susceptible to the development of other infections. In fact, bacterial infections are responsible for most of the events that affect HIV-infected patients 3. Although the use of antiretroviral therapy has improved this scenario, a few pathogens can cause clinical complications, such as Staphylococcus aureus. This pathogen, which has been identified as being responsible for morbidity and mortality among HIV patients, produces important virulence factors and frequently acquires resistance to different antibiotics 4,5.
Methicillin-resistant S. aureus (MRSA) expresses an additional penicillin-binding protein, PBP2a, which is encoded by the mecA gene. This gene is located in a mobile genetic element referred to as staphylococcal chromosomal cassette mec (SCCmec) 6. SCCmec types II and III are commonly associated with hospital-acquired isolates, whereas community-acquired isolates normally carry type IV or V 7,8.
Decreased host immunity among HIV-infected patients places them at increased risk for several infections, including those caused by S. aureus and MRSA. Certain authors have evaluated the risk factors related to infection and/or colonization of HIV-infected individuals with S. aureus and have shown the importance of various behavioral and clinical aspects 9-11. However, only few studies have used microbiological methods to characterize isolates to validate of their data 9,10. Thus, this systematic review aimed to determine both the risk factors associated with colonization/infection by MRSA in HIV-infected individuals and the methods used for characterization of isolates.
MATERIALS AND METHODS
The present study may be characterized as a descriptive, exploratory and systematic literature review. This review was conducted by two examiners, who performed a non-biased analysis of the studies selected based on the inclusion and exclusion criteria described below.
The search covered articles published between January 2001 and December 2013 in five online health sciences databases: PubMed, the Scientific Electronic Library Online (SciELO), the Latin American and Caribbean Health Sciences (LILACS) database, Ovid, Google Scholar. The search was conducted using six keywords: “MRSA, methicillin-resistant Staphylococcus aureus”, “HIV”, “Staphylococcus”, “recurrent infections”, “molecular typing” and “epidemiology”. These keywords were selected as being reflective of the topic of interest, as analyzed previously using the Medline MeSH.
Articles were selected from each database based on the following inclusion criteria: a title and abstract compatible with the given subject; published within the last 13 years (from January 1, 2001 to December 31, 2013); published in Portuguese, English or Spanish; and based on original research. Articles were excluded when the main subject was not compatible with the aim of the present study (such as phytochemistry, biology of lymphocytes and kidney disease, among other topics); when they were published more than 13 years ago; when they dealt with S. aureus-related infections in bone or gynecological areas or with endocarditis, which have characteristics that distinguish them from other infections; when their links for online access had no data available for consultation; and when they were case reports, literature reviews, book chapters, theses or letters to the editor.
Papers that fulfilled the selection criteria were analyzed for their methodological quality using a quality assessment questionnaire based on criteria established by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISM) statement. The scores for determining the presence of features regarded as important in the study are shown in Table 1. In this classification, each item presented by a study corresponds to a score of one. The studies that received scores between 8 and 10 were classified as level A, which indicates high methodological quality and studies that had scores of 5 to 7 points were classified as level B, implying intermediate methodological quality. Meanwhile, scores of less than or equal to 4 points were included in level C, indicating low methodological quality. Only studies classified as level A were included in the current review.
Table 1.
Quality evaluation criteria and their scores.*
| Number | Quality Assessment | Yes | No |
| 1 | Representative sample of the target population | 1 | 0 |
| 2 | Definition of the type of study | 1 | 0 |
| 3 | Definition of the inclusion and exclusion criteria applied to the sample | 1 | 0 |
| 4 | Presence of a control group | 1 | 0 |
| 5 | Description of risk factors for colonization and/or infection | 1 | 0 |
| 6 | Phenotypic analysis: Gram staining and catalase and coagulase tests | 1 | 0 |
| 7 | Molecular analysis: SCCmec (typing), PFGE and MLST | 1 | 0 |
| 8 | Antimicrobial susceptibility analysis: MIC and disk diffusion tests | 1 | 0 |
| 9 | Statistical analysis | 1 | 0 |
| 10 | Discussion of the limitations of the study | 1 | 0 |
Based on criteria established by the PRISM statement. SCCmec: staphylococcal cassette chromosome mec; PFGE: pulsed-field gel electrophoresis; MLST: multilocus sequence typing; MIC: minimum inhibitory concentration.
RESULTS
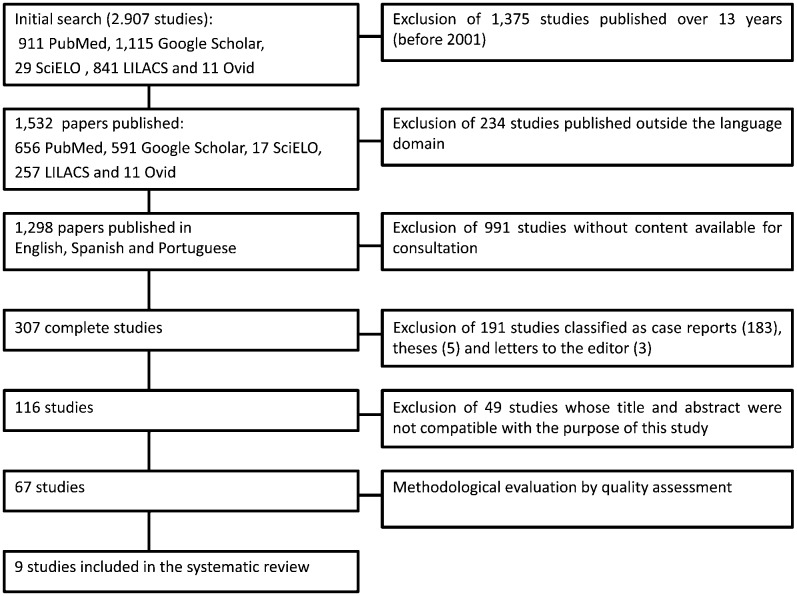
Figure 1 outlines the steps for the selection of the studies. The number of articles found in and selected from the databases is listed in Table 2. An initial electronic search of the health sciences databases using the keywords uncovered a total of 911, 29, 841, 11 and 1,115 articles from PubMed, SciELO, LILACS, Ovid and Google Scholar, respectively. After applying the inclusion and exclusion criteria, 67 studies were selected and analyzed for quality. The quality assessment revealed 9 papers in category A, which were included in the present systematic review (Table 3). Of these papers, eight received 8 points and only one received 9 points. The data from the selected studies are listed in Table 3. Among the most relevant data, the size of the target population was considered to be very discrepant, ranging from 107 to 4,674 patients.
Figure 1.
- Outline of the steps for the selection of the studies.
Table 2.
Databases and search results.
| Reference source | Keywords | Total articles | Articles found after applying the inclusion and exclusion criteria | Articles selected based on quality assessment |
| PubMed | HIVc; Staphylococcus; MRSAd | 911 | 23 | 9 |
| SciELOa | HIV; Staphylococcus; MRSA | 29 | 12 | 0 |
| LILACSb | HIV; Staphylococcus; MRSA | 841 | 17 | 0 |
| Ovid | HIV; Staphylococcus; HIV; MRSA | 11 | 5 | 0 |
| Google Scholar | MRSA; HIV; recurrent infections; molecular typing; epidemiology | 1.115 | 10 | 8 |
Scientific Electronic Library Online;
Latin American and Caribbean Health Sciences;
human immunodeficiency virus;
methicillin-resistant Staphylococcus aureus.
Table 3.
Characteristics of the nine selected studies involving HIV patients with colonization and/or infection by Staphylococcus aureus.
| Type of study (country; authors) | Category (score) | Sample size | Clinical source(s) | Factors associated with colonization and/or infection* by MRSA | Phenotypic analysis of isolates | Molecular analysis and virulence/resistance genes investigated | Susceptibility test(s) | Antibiotic resistance rates for S. aureus isolates |
| Case-control(Italy; Tumbarello et al., 2002) | A (8) | 4,674 HIV-infected patients/129 S. aureus/41 MRSA/88 MSSA | Blood | Nosocomial episodes, previous antibiotic therapy and/or bacterial infections | API® test ID 32 STAPH | ND | MIC | CLI 39%; MCL 57%; MET 32%; PEN 73%; QUI 49%; TMP/SXT 70%; VAN 0% |
| Case-control (USA; Hidron et al., 2005) | A (9) | 726 patients/81 HIV-infected patients (14 MRSA)/645 HIV-negative (39 MRSA) | Anterior nares | No antimicrobial use in the past three months before the current admission (only for HIV-infected patients) | SMA; latex agglutination test | PFGE; SCCmec typing; PVL | Disk diffusion; OAS | CLI 32%; DOX 10%; GEN 23%; MCL 92.5%; OXA 100%; QUI 83%; RIF 20%; TMP/SXT 21%; VAN 0% (data from MRSA isolates, independent of patient classification) |
| Retrospective case-control (Italy; Drapeau et al., 2007) | A (8) | 3,000 patients/28 MRSA | Blood; skin and soft tissue; sputum, bronchoalveolar lavage and pleural fluids | T CD4+ T lymphocyte counts <200 cells/μl; previous hospitalization; an invasive procedure in the previous year | ND | ND | Automated system | ND |
| Retrospective (USA; Burkey et al., 2008) | A (8) | 4,607 patients/216 S. aureus/94 MRSA | Blood | Intravenous drug use; CD4+ T lymphocyte counts <200 cells/μl; end-stage renal disease | ND | ND | OAS | ND |
| Prospective (USA; Shet et al., 2009) | A (8) | 107 patients/41 S. aureus/21 MRSA | Anterior nares | Previous antibiotic therapy | SMA; latex agglutination test | PFGE; SCCmec typing; PVL, ACME, spa, mecA, and mupA genes | MIC | CLI 43%; MCL 100%; MUP 39%; QUI 95%; TET 24%; TMP/SXT 5% |
| Experimental, randomized controlled trial, prospective (USA; Gordon et al., 2010) | A (8) | 191 S. aureus/27% MRSA | Anterior nares | Relapse of drug or alcohol use within the previous month; previous antibiotic therapy | SMA; latex agglutination test | PFGE | MIC; disk diffusion | CLI 83%; FOX 20%; GEN 9%; MCL 95%; OXA 27%; PEN 99%; QUI 45%; RIF 2%; TET 16%; TMP/SXT 89%; VAN 0% |
| Retrospective et al., 2010) | A (8) | 900 patients/72 MRSA | Skin or soft tissue and normally sterile sites | Previous antibiotic therapy and/or hospitalization; CD4+ T lymphocyte counts <200 cells/μl; drug use; hepatitis B co-infection; OI in the previous year; alternative housing**; incarceration; alcohol abuse | ND | PFGE | ND | TMP/STX 2% |
| Observational (USA; Popovich et al., 2013) | A (8) | 745 patients/374 HIV-infected (74 MRSA)/371 HIV-negative (41 MRSA) | Nares; throat; bilateral axillae; bilateral inguinal regions; peri-rectal area and a chronic wound | Incarceration; male gender | ChromID MRSA; latex agglutination | PFGE | Disk diffusion | ND |
| Cohort (USA; Farley et al., 2013) | A (8) | 498 patients/68 HIV-infected (10 MRSA)/430 HIV-negative (20 MRSA) | Anterior nares; axilla and wound | History of or current abscess; isolation outside the hospital unit; HIV infection (data from all patients, independent of whether they were infected with HIV) | Gram stain; catalase; latex agglutination test | tufB, nuc, mecA, mupA, ileS-2 genes, PVL, TSST-1; PFGE; MLST; an automated platform for pathogen identification and strain typing | CHROMagar MRSA; automated system | TMP/STX 0%; MUP 7.7% (data from MRSA isolates, independent of patient classification) |
Factors for which odds ratios and 95% confidence intervals were calculated. **Includes patients residing in personal-care homes, nursing homes, long-term care facilities, and correctional facilities as well as those who were homeless. MRSA: methicillin-resistant S. aureus; HIV: human immunodeficiency virus; OI: opportunistic infection; SMA: culture in mannitol salt agar; PFGE: pulsed-field gel electrophoresis; PCR: polymerase chain reaction; PVL: Panton-Valentine leukocidin; TSST1: toxic shock toxin 1; ACME: arginine catabolic mobile element; MIC: minimum inhibitory concentration; CLI: clindamycin; DOX: doxycycline; FOX: cefoxitin; GEN: gentamicin; MCL: macrolides; MET: methicillin; MUP: mupirocin; OXA: oxacillin; PEN: penicillin; QUI: quinolones; RIF: rifampin; TET: tetracycline; TMP/STX: trimethoprim/sulfamethoxazole; VAN: vancomycin; ND: not determined; OAS: oxacillin agar screening.
The study designs were mostly retrospective (4/9 studies) and the most common risk factors associated with colonization/infection were previous use of antimicrobials (4/9 studies), previous hospitalization (4/9 studies) and the CD4+ T lymphocyte count (typically <200 cells/μl) (3/9 studies) (Table 3).
The main methodologies used for S. aureus phenotypic identification were culture in salt mannitol agar (3/9 studies) and the latex agglutination test (5/9 studies), which was used for the detection of the clumping factor or bound coagulase. Pulsed-field gel electrophoresis (PFGE) was the main molecular analytical method used to define the search community/hospital clones (6/9 studies) and the USA300 lineage was considered to be the most prevalent in the majority of the articles that assessed the clonality of isolates. The exception was a study performed by Gordon et al. (2010) from New York, USA, which found a larger number of isolates related to the USA500 lineage, a hospital clone.
Susceptibility, as determined by the disk diffusion test, was evaluated in only 3 of the 9 studies. However, 5 studies determined the minimum inhibitory concentration (MIC) or the susceptibility of isolates using an automated technique.
MRSA isolates were detected and analyzed by the authors of 6 of the 9 selected papers, whereas the other 3 studies were retrospective, evaluating data from laboratory and hospital databases. The studies that evaluated antimicrobial resistance described erythromycin-resistant S. aureus (3/9 studies), vancomycin-susceptible S. aureus (4/9 studies) and/or rifampicin-susceptible S. aureus (3/9 studies). Two studies also performed SCCmec typing and found that type IV was a common or unique cassette among MRSA isolates.
DISCUSSION
The use of HAART has increased the life expectancy of HIV-infected patients, whether pediatric or adult, via the reduction of opportunistic infections, with a consequent improvement in quality of life 13. In this context, it remains unclear whether these patients have an increased risk of these microbial infections due to a higher prevalence of colonization by pathogens or due to other factors. Therefore, knowledge of the risk factors that may favor colonization is critical to develop strategies to prevent colonization and infection in patients and their contacts 4,9,14,15. In the current systematic review, both the risk factors associated with colonization/infection by MRSA in HIV-infected individuals and the methods for characterization of this pathogen were evaluated.
Among the 9 studies that qualified for this review, 2 were conducted in Europe and 7 were conducted in North America. In 4 of these studies, use of antibiotics was detected as an important risk factor for S. aureus infections in HIV-positive individuals 9,. In addition, most of these isolates were MRSA, suggesting that the use of antibiotics can lead to the selection of resistant microorganisms. However, there was no consensus among the authors about previous exposure to antibiotics during sampling 9,. Thus, although it has been well established that prior antimicrobial therapy is a risk factor for MRSA colonization/infection, it is difficult to determine exactly what duration of antibiotic use may be considered as a risk factor.
Previous hospitalization 15,18,19 and invasive medical procedures 19 were also described as risk factors for the acquisition of MRSA by HIV patients. In particular, a multivariate analysis showed that HIV patients hospitalized for more than five days had a 14% increased risk of acquiring MRSA 19. Other factors, such as diabetes 20, previous infections or co-infections (including skin and soft tissue infections) 15,16,18, co-morbidities 20 and a history of colonization and infection by MRSA 15, were also reported as risk factors for acquiring this pathogen.
Certain behavioral risk factors have also been associated with colonization by MRSA, including imprisonment 18, alcohol abuse 18, heterosexual behavior 20, homelessness 18 and illicit drug use 17,18. Although excluded based on its quality criteria, a study by Mimiaga et al. reported that crystal methamphetamine users are more likely to have multiple partners and to engage in risky sexual intercourse 21. Similarly, strong associations between HIV infection, use of injectable drugs and the development of bacteremia caused by MRSA have been shown 20.
The samples selected for these types of studies should be clearly representative of the target population of HIV-infected individuals. However, in the present review, the majority of the studies selected their samples from a single institution, without sample calculation, which may be related to the convenience of sample collection or to a low number of cases. The resultant variation made the data analyses challenging. Moreover, certain studies used only one sample (one clinical specimen per patient), whereas others used more than one sample per patient 9,11,15-17,20,22,, making the analysis difficult. Additionally, there were differences in the type of design used among the studies analyzed.
In the present review, nearly half of the articles (3/9 studies) used culture in selective media (mannitol salt agar), followed by the latex agglutination test, to identify S. aureus isolates. Although latex tests are simple and rapid, they do not present 100% sensitivity 24 because other species, such as S. lugdunensis, that are positive for clumping factor may be misidentified as S. aureus using this test 25. In contrast, the coagulase tube test, which is a more reliable method for identifying S. aureus, was not performed in any of the studies evaluated in this review 24.
The disk diffusion test, which was used in only three studies 15,17,26, is the simplest and least expensive assay and presents high sensitivity and specificity for detecting MRSA isolates 28. However, MIC determination and oxacillin agar screening (OAS) were also used to determine the methicillin resistance in several studies 9,15-17,. Although these procedures are recommended by CLSI (2009) 12, MIC determination is usually more laborious than other techniques and OAS presents low sensitivity and may be unable to detect SCCmec IV isolates 28. It is important to mention that SCCmec IV is traditionally found in community-acquired MRSA isolates and in specific risk groups, such as children, athletes, members of the military and men who have sex with men 29. However, in this review, only two studies performed SCCmec typing and they found that type IV was a common 15 or unique cassette 9 among the analyzed MRSA isolates.
Six studies employed PFGE to verify the clonality of the MRSA isolates obtained from their samples. The USA300 clone was the most common lineage, found in four studies 9,18,26,27, whereas Hidron et al. detected isolates of the USA300 (30.8%), USA100 (40.4%) and USA500 (19.2%) lineages 15. The USA500 clone was also detected among 15% of MRSA isolates in the study conducted by Gordon et al. 17. The USA100 and USA500 isolates, which carry SCCmec II and I, respectively, were the most common lineages recovered from hospital-acquired infections in the USA and Europe, respectively, in the past decade. However, these lineages are currently being replaced by other lineages that were once limited to the community, such as USA300/SCCmec IV 30. In the USA, where all studies that applied the PFGE technique were conducted, the USA300 clone is considered to be the predominant cause of community-acquired S. aureus infections and colonization 30,31, confirming the findings demonstrating that the USA300 lineage is more frequent among HIV patients. In contrast, in other geographical regions, such as Brazil, Australia and eastern Asia, USA300 is rarely found 32. In these localities, other lineages are spreading throughout the community 30,33. Thus, more studies examining the prevalence of MRSA isolates from HIV patients around the world should be performed to clarify the molecular epidemiology of MRSA isolates recovered from patients in other countries.
The findings of the nine articles selected in this systematic review indicate that the main risk factors for colonization/infection by MRSA in HIV-infected individuals are use of antimicrobials and previous hospitalization. However, the numbers of evaluated patients, the exclusion and inclusion criteria and the characterization of S. aureus isolates were not uniform, limiting identification of the characteristics associated with HIV patients who are colonized/infected by S. aureus. Infections caused by MRSA isolates are an important issue in the clinical management of HIV patients, so more studies evaluating risk factors and the best techniques to identify and characterize isolates from these patients are necessary.
ACKNOWLEDGMENTS
This study was supported in part by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Foundation, the Ministry of Education of Brazil and Brazilian governmental institutions.
Footnotes
No potential conflict of interest was reported.
REFERENCES
- 1.UNAIDS. Report on the global AIDS epidemic 2013. (2013). Available at: http://www.unaids.org/en/HIV_data/2006GlobalReport/default.asp. Accessed on 10 June 2013.
- 2.World Health Organization. Antiretroviral therapy guidelines, 2010. (2012). Available at: http://whqlibdoc.who.int/publications/2010/9789241599764_eng.pdf. Accessed on 16 May 2013.
- 3.Madhivanan P, Mothi Sn, Kumarasamy N, Yepthomi T, Venkatesan C, Lambert JS, et al. Clinical manifestations of HIV infected children. Indian J Pediatr. 2003;70(8):615–20. doi: 10.1007/BF02724249. [DOI] [PubMed] [Google Scholar]
- 4.Weinke T, Schiller R, Fehrenbach FJ, Pohle HD. Association between Staphylococcus aureus nasopharyngeal colonization and septicemia in patients infected with the human immunodeficiency virus. Eur J Clin Microbiol Infect Dis. 1992;11(11):985–9. doi: 10.1007/BF01967787. [DOI] [PubMed] [Google Scholar]
- 5.Senthilkumar A, Kumar S, Sheagren JN. Increased incidence of Staphylococcus aureus bacteremia in hospitalized patients with acquired immunodeficiency syndrome. Clin Infect Dis. 2001;33(8):1412–6. doi: 10.1086/322656. [DOI] [PubMed] [Google Scholar]
- 6.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin-resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549–55. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conceição T, Tavares A, Miragaia M, Hyde K, Aires-de-Sousa M, de Lencastre H. Prevalence and clonality of methicillin-resistant Staphylococcus aureus (MRSA) in the Atlantic Azores islands: predominance of SCCmec types IV, V and VI. Eur J Clin Microbiol Infect Dis. 2010;29(5):543–50. doi: 10.1007/s10096-010-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66(5):1061–9. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- 9.Shet A, Mathema B, Mediavilla JR, Kishii K, Mehandru S, Jeane-Pierre P, et al. Colonization and subsequent skin and soft tissue infection due to Methicillin-Resistant Staphylococcus aureus in a cohort of otherwise healthy adults infected with HIV type I. J Infect Dis. 2009;200(1):88–93. doi: 10.1086/599315. [DOI] [PubMed] [Google Scholar]
- 10.Furuno JP, Johnson JK, Schweizer ML, Uche A, Stine OC, Shurland SM, et al. Community-associated methicillin-resistant Staphylococcus aureus bacteremia and endocarditis among HIV patients: a cohort study. BMC Infect Dis. 2011;11:298. doi: 10.1186/1471-2334-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szumowski JD, Wener KM, Gold HS, Wong M, Venkataraman L, Runde CA, et al. Methicillin-resistant Staphylococcus aureus colonization, behavioral risk factors, and skin and soft-tissue infection at an ambulatory clinic serving a large population of HIV-infected men who have sex with men. Clin Infect Dis. 2009;49(1):118–21. doi: 10.1086/599608. [DOI] [PubMed] [Google Scholar]
- 12.Clinical Laboratory Standards Institute. CLSI document M100-S19. Wayne, PA: 2009. Performance standards for antimicrobial susceptibility testing; Nineteenth informational supplement. [Google Scholar]
- 13.Ferreira Dde C, Passos MR, Rubini Nde P, Knupp RR, Curvelo JA, dos Reis HL, et al. Validation study of a scale of life quality evaluation in a group of pediatric patients infected by HIV. Cien Saude Colet. 2011;16(5):2643–52. doi: 10.1590/s1413-81232011000500034. [DOI] [PubMed] [Google Scholar]
- 14.Holbrook KA, Klein RS, Hartel D, Elliott DA, Barsky TB, Rothschild LH, et al. Staphylococcus aureus nasal colonization in HIV-seropositive and HIV-seronegative drug users. J Acquir Immune Defic Syndr Hum Retrovir. 1997;16(4):301–6. doi: 10.1097/00042560-199712010-00012. [DOI] [PubMed] [Google Scholar]
- 15.Hidron AI, Kourbatova EV, Halvosa JS, Terrell BJ, McDougal LK, Tenover FC, et al. Risk factors for colonization with Methicillin-Resistant Staphylococcus aureus (MRSA) in patients admitted to an urban hospital: emergence of community-associated MRSA nasal carriage. Clin Infect Dis. 2005;41(2):159–66. doi: 10.1086/430910. [DOI] [PubMed] [Google Scholar]
- 16.Tumbarello M, Donati KG, Tacconelli E, Citton R, Spanu T, Leone F, et al. Risk factors and predictors of mortality of methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia in HIV-infected patients. J Antimicrob Chemother. 2002;50(3):375–82. doi: 10.1093/jac/dkf126. [DOI] [PubMed] [Google Scholar]
- 17.Gordon RJ, Chez N, Jia H, Zeller B, Sobieszczyk M, Brennan C, et al. The NOSE Study (Nasal Ointment for Staphylococcus aureus Eradication): A Randomized Controlled Trial of Monthly Mupirocin in HIV-Infected Individuals. J Acquir Immune Defic Syndr. 2010;55(4):466–72. doi: 10.1097/QAI.0b013e3181ec2a68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsetty SK, Stuart LL, Blake RT, Parsons CH, Salgado CD. Risks for methicillin-resistant Staphylococcus aureus colonization or infection among patients with HIV infection. HIV Med. 2010;11(6):389–94. doi: 10.1111/j.1468-1293.2009.00802.x. [DOI] [PubMed] [Google Scholar]
- 19.Drapeau CMJ, Angeletti C, Festa A, Petrosillo N. Role of previous hospitalization in clinically-significant MRSA infection among HIV-infected inpatients: results of a case-control study. BMC Infect Dis. 2007;7:36. doi: 10.1186/1471-2334-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkey MD, Wilson LE, Moore RD, Lucas GM, Francis J, Gebo KA. The incidence of and risk factors for MRSA bacteremia in an HIV infected cohort in the HAART era. HIV Med. 2008;9(10):858–62. doi: 10.1111/j.1468-1293.2008.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimiaga MJ, Fair AD, Mayer KH, Koenen K, Gortmaker S, Tetu AM, et al. Experiences and sexual behaviors of HIV-infected MSM who acquired HIV in the context of crystal methamphetamine use. AIDS Educ Prev. 2008;20(1):30–41. doi: 10.1521/aeap.2008.20.1.30. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan A, Seifried S, Zhu L, Bitar W, Srivastava DK, Shenep JL, et al. Short communication: methicillin-resistant Staphylococcus aureus infections in children and young adults infected with HIV. AIDS Res Hum Retroviruses. 2009;25(12):1219–24. doi: 10.1089/aid.2009.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schechter-Perkins EM, Mitchell PM, Murray KA, Rubin-Smith JE, Weir S, Gupta K. Prevalence and predictors of nasal and extra nasal staphylococcal colonization in patients presenting to the emergency department. Ann Emerg Med. 2011;57(5):492–9. doi: 10.1016/j.annemergmed.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 24.Andriesse GI, Elberts S, Vrolijk A, Verhulst C, Kluytmans JA. Evaluation of a fourth-generation latex agglutination test for the identification of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2011;30(2):259–64. doi: 10.1007/s10096-010-1080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira EM, Teixeira CA, Alvarenga AL, Schuenck RP, Giambiagi-Demarval M, Holandino C, et al. A Brazilian lineage of Staphylococcus lugdunensis presenting rough colony morphology may adhere to and invade lung epithelial cells. J Med Microbiol. 2012;61(Pt 4):463–9. doi: 10.1099/jmm.0.033001-0. [DOI] [PubMed] [Google Scholar]
- 26.Popovich KJ, Hota B, Aroutcheva A, Kurien L, Patel J, Lyles-Banks R, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization burden in HIV-infected patients. Clin Infect Dis. 2013;56(8):1067–74. doi: 10.1093/cid/cit010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farley JE, Ross T, Krall J, Hayat M, Caston-Gaa A, Perl T, et al. Prevalence, risk factors, and molecular epidemiology of methicillin-resistant Staphylococcus aureus nasal and axillary colonization among psychiatric patients on admission to an academic medical center. Am J Infect Control. 2013;41(3):199–203. doi: 10.1016/j.ajic.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 28.Matos PD, Schuenck RP, Cavalcante FS, Caboclo RM, Santos KR, et al. Accuracy of phenotypic methicillin susceptibility methods in the detection of Staphylococcus aureus isolates carrying different SCCmec types. Mem Inst Oswaldo Cruz. 2010;105(7):931–4. doi: 10.1590/s0074-02762010000700017. [DOI] [PubMed] [Google Scholar]
- 29.Cooke FJ, Brown NM. Community-associated methicillin-resistant Staphylococcus aureus infections. Br Med Bull. 2010;94:215–27. doi: 10.1093/bmb/ldq010. [DOI] [PubMed] [Google Scholar]
- 30.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(4):e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freitas EA, Harris RM, Blake RK, Salgado CD. Prevalence of USA300 strain type of methicillin-resistant Staphylococcus aureus among patients with nasal colonization identified with active surveillance. Infect Control Hosp Epidemiol. 2010;31(5):469–75. doi: 10.1086/651672. [DOI] [PubMed] [Google Scholar]
- 32.Schuenck RP, Cavalcante FS, Emery E, Giambiagi-de Marval M, dos Santos KR. Staphylococcus aureus isolates belonging to different multilocus sequence types present specific virulence gene profiles. FEMS Immunol Med Microbiol. 2012;65(3):501–4. doi: 10.1111/j.1574-695X.2012.00958.x. [DOI] [PubMed] [Google Scholar]
- 33.Scribel LV, Silva-Carvalho MC, Souza RR, Superti SV, Kvitko CH, Figueiredo AM, et al. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus carrying SCCmecIV in a university hospital in Porto Alegre, Brazil. Diagn. Microbiol. Infect Dis. 2009;65(4):457–61. doi: 10.1016/j.diagmicrobio.2009.08.012. [DOI] [PubMed] [Google Scholar]