SUMMARY
Background
Depolymerization of actin filaments is vital for the morphogenesis of dynamic cytoskeletal arrays and actin-dependent cell-motility. Cofilin is necessary for actin disassembly in cells, and it severs filaments most efficiently at low cofilin to actin ratios, whereas higher concentrations of cofilin suppress severing. However, the cofilin concentration in thymocytes is too high to allow the severing of single actin filaments.
Results
We observed that filaments sever efficiently in thymus cytosol. We identified Aip1 as a critical factor responsible for the severing and destabilization of actin filaments even in the presence of high amounts of cofilin. By FRET-based spectroscopy and single-filament imaging of actin, we show that besides driving the rapid severing of cofilin-actin filaments, Aip1 also augments the monomer dissociation rate at both the barbed and pointed ends of actin. Our results also demonstrate that Aip1 does not cap the barbed ends of actin filaments, as was previously thought.
Significance
Our results indicate that Aip1 is a cofilin-dependent actin depolymerization factor and not a barbed-end capping factor as was previously thought. Aip1 inverts the rules of cofilin-mediated actin disassembly such that increasing ratios of cofilin to actin now result in filament destabilization through faster severing and accelerated monomer loss from barbed and pointed ends. Aip1 therefore offers a potential control point for disassembly mechanisms in cells to switch from a regime of cofilin-saturation and stabilization to one that favors fast disassembly and destabilization.
INTRODUCTION
The highly dynamic nature of the actin cytoskeleton in cells allows for the rapid cytoskeletal reorganization that accompanies a variety of cellular processes such as cytokinesis, endocytosis, and cell motility. The cell has an array of factors that execute the disassembly of actin filaments at rates far greater than those achieved by actin alone in pure solution [1]. Cofilin is essential for cellular actin disassembly [2] and influences the morphogenesis of filamentous arrays in several ways [reviewed in [3]]. In vitro, cofilin severs actin filaments at low ratios of cofilin to actin [4], and this severing activity has been shown to be important for cells [5–7] as severing creates more filament ends that can grow or shrink, and thus increases filament dynamics.
Cofilin binds actin cooperatively [8, 9] and alters the configuration of actin protomers within the filament [10]. Contiguous binding sites occupied with cofilin are stable, but lateral interfaces between cofilin-occupied and unoccupied sites on actin are unstable and sever [11, 12]. Interestingly, therefore, cofilin severs actin filaments most efficiently when filaments are bound with an intermediate amount of cofilin. Severing is inefficient at higher ratios of cofilin to actin and filaments are stable in saturating concentrations of cofilin [4]. Nevertheless, actin filament severing activity detected in vitro with pure cofilin alone cannot account for the behavior of Listeria actin comet tails which disassemble faster with increasing concentrations of cofilin [13, 14]. Furthermore, the intrinsic cofilin severing and actin depolymerization rates do not account for the rapid actin turnover rates of yeast actin patches [15, 16]. Thus, cofilin-dependent auxiliary factors present in cytoplasm may be responsible for the destabilization of actin filaments even in the presence of saturating cofilin concentrations.
Thymus extract rapidly disassembles Listeria actin comet tails even though it is thought to contain high amounts of cofilin that should prevent actin disassembly [17]. Factors that compete with cofilin binding to F-actin such as myosin II can restore filament severing when saturating amounts of cofilin are present [18], by preventing cofilin binding and increasing the number of heterotypic junctions that promote severing on the filament. The presence of these factors in cytosol or on the actin comet tail could help account for this observation. However, fractionation and biochemical complementation identified Aip1, coronin and Cyclase Associated Protein (CAP) as the major auxiliary factors present in thymus cytosol that directly potentiate cofilin-mediated disassembly of Listeria actin comet tails [17, 19]. The mechanisms through which these factors promote cofilin mediated filament disassembly are poorly understood but are thought not to be limited to preventing the binding of cofilin. In fact, coronin increased the loading of cofilin on Listeria actin comet tails and stabilized them, whereas Aip1 was able to rapidly disassemble these stabilized filaments [17]. CAP could substitute for coronin but not Aip1 to augment comet tail disassembly [19] and previous work has indicated that Aip1 can exert its activity in the presence of higher cofilin to actin ratios [20]. This indicated to us that Aip1 was a likely candidate to explain the contradictory behaviors of pure cofilin and actin disassembly in complex cell extracts.
Aip1 facilitates cofilin-mediated actin disassembly in vitro [21–23] and mutations in Aip1 or perturbation of its function in cells lead to ectopic accumulation of F-actin [24, 25], defects in actin turnover dynamics [26], suppression of filament elongation [27], and depletion of the actin monomer pool that fuels actin assembly [28]. Thus, the in vitro and in vivo data are consistent with Aip1 playing a role in cytoskeletal organization by enhancing cofilin-mediated filament disassembly, but the underlying molecular mechanism is not yet known.
The apparent ability of Aip1 to cap filament barbed ends led to a popular model in which Aip1 facilitates cofilin-mediated disassembly by preventing the reannealing of severed filaments [21, 29, 30]. In this model, cofilin alone mediates severing while Aip1 simply blocks the back reaction and does not alter the mechanism of filament destabilization. Aip1 is therefore thought to control actin filament dynamics in a manner similar to capping protein (CapZ). CapZ is a well-characterized barbed end binding protein that caps barbed ends with nanomolar affinity thus preventing barbed end growth or shrinkage [31]. This model was supported by the fact that CapZ and Aip1 exhibit strong genetic interactions in yeast where null mutations in both CapZ and Aip1 elicit a more severe disruption in actin organization than a null mutation for either gene alone [27]. However, CapZ cannot substitute for Aip1 to disassemble Listeria actin comet tails or single filaments in the presence of cofilin and coronin [32].
In order to test our hypothesis that Aip1 is responsible for the disassembly of filaments in the presence of stabilizing concentrations of cofilin, and to reinvestigate the existing model of Aip1 function, we used single filament imaging of actin, and FRET-based bulk actin depolymerization assays.
RESULTS
Opposing behavior of recombinant cofilin and cofilin in thymus extract on depolymerization of single filaments
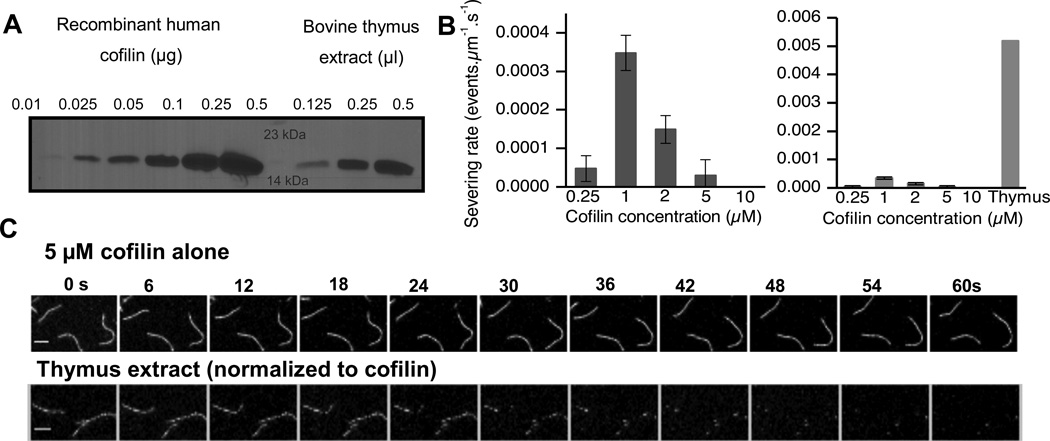
Previous work has shown that the amount of cofilin in thymocytes is high (up to the order of 20 µM) [17]. We confirmed this result by quantitative western blotting and found the amount of cofilin in thymus extract to be 21 ± 6 µM (Fig 1A). In our severing assays on single actin filaments, we found that recombinant human cofilin severed actin most efficiently at 1 µM (Fig. 1B), whereas severing was inhibited at higher cofilin concentrations consistent with previous results [4]. In order to test if cellular extract can sever single filaments, bovine thymus extract was diluted so that the final concentration of cofilin was 5 µM. At these concentrations, thymus extract is able to sever single actin filaments (Video S1) whereas an equimolar amount of recombinant cofilin was unable to sever pure actin filaments efficiently (Fig. 1B right panel, 1C). Actin filament severing rates were in fact ten times faster in the presence of thymus cytosol than the fastest rates detected with pure cofilin alone.
FIGURE 1.
Thymus extract is more efficient at depolymerizing single actin filaments than a normalized amount of recombinant cofilin. (A) Quantitative western blot showing standard amounts of recombinant cofilin (0.01–0.5 µg) (left lanes, increasing order) & fixed amounts of thymus extract. (B) Cofilin severs only across a narrow range of concentrations (left graph), with activity peaking at approximately 1 µM. Thymus extract is roughly 10-fold more effective at severing than the peak cofilin severing concentration (right graph) Error bars represent S.D. n= at least 2 movies. (C) Frames from time lapse movies showing that when normalized to a cofilin concentration of 5 µM, filaments are stable in the presence of pure cofilin (top panel), whereas they are disassembled within 60s with thymus extract (bottom panel). Scale bar= 2 µM.
Aip1 can depolymerize actin filaments even in the presence of saturating amounts of cofilin
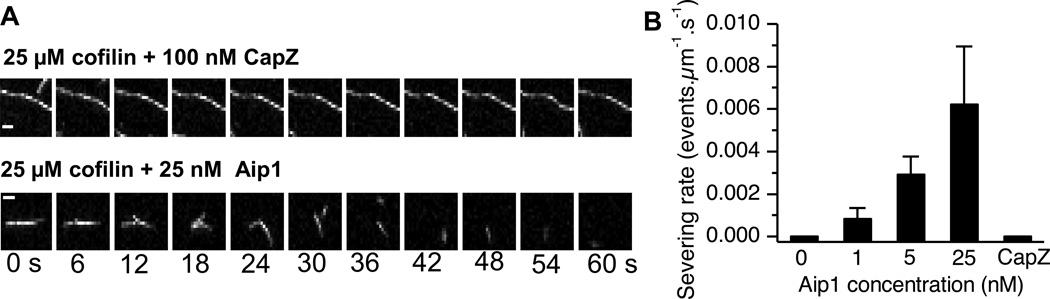
Previous work has shown that coronin increases cofilin loading on Listeria actin comet tails and stabilizes them. These filaments are destabilized by Aip1 [17]. Thus, Aip1 appeared to us as the most likely candidate for depolymerization of cofilin-saturated stabilized filaments and could possibly explain this behavior of extract. To test this, we imaged single filaments in the presence of saturating amounts of cofilin, in the presence or absence of Aip1. Actin filaments did not sever in the presence of 25 µM cofilin. As a control, we also tested severing in the presence of 25 µM cofilin and 100 nM capping protein (CapZ) (Fig. 2A, top panels, Video S2) and detected no severing events. However, filaments did sever in the presence of 25 nM Aip1 and 25 µM cofilin (Fig. 2A, bottom panels, Video S3). Severing rates increased from 0 events per micron per second in the presence of 25 µM cofilin to 0.006 events per micron per second in the presence of 25 nM Aip1 (Fig. 2B). This experiment provided us with the first evidence that Aip1 and capping protein do not appear to act by the same mechanism.
FIGURE 2.
Aip1 can sever filaments saturated by cofilin. (A) Frames from time lapse movies showing the dynamics of filaments in the presence of a saturating amount of cofilin+ CapZ (upper panels) or cofilin + Aip1 (lower panels). Filaments fragment in the presence of Aip1 only, not cofilin alone or cofilin + CapZ. (B) Quantitation of severing rates of filaments in the presence of saturating amounts of cofilin. No severing events were detected in the presence of 25 µM cofilin alone or cofilin + 100 nM CapZ. Severing rates increased with increasing concentrations of Aip1 (n=3 movies, error bars represent S.D.). Scale bar= 1 µM.
Aip1 does not displace cofilin to promote severing
Cofilin-mediated severing involves the destabilization of lateral interfaces between cofilin-bound and unbound sections of actin. Therefore, severing by Aip1 could operate by two mechanisms. Either Aip1 could be displacing cofilin from actin, thus creating additional unstable lateral interfaces and exploiting cofilin’s intrinsic ability to sever, or alternatively Aip1 could be potentiating severing by a different mechanism, but in co-operation with the bound cofilin.
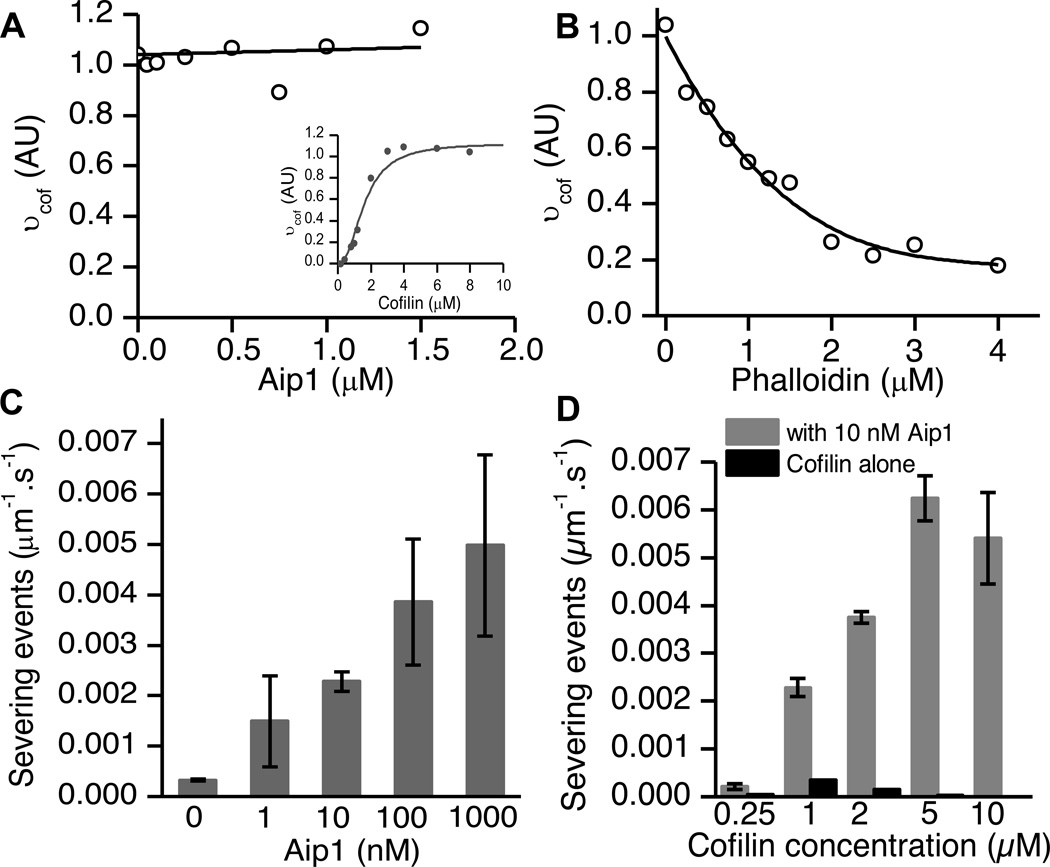
In order to test if Aip1 displaced bound cofilin from actin filaments, we validated that our recombinant cofilin binds co-operatively to actin as demonstrated previously [9] (Fig. 3A, inset). Next, we carried out competitive equilibrium binding assays on actin in the presence of a high initial occupancy of cofilin (3 µM, corresponding to binding density νcof > 0.9) and increasing concentrations of Aip1 (Fig. 3A). Aip1 did not affect pyrene fluorescence by itself (data not shown). As a control, we also carried out the experiment with a known competitive inhibitor of cofilin, phalloidin, as described previously (Fig. 3B) [18]. Concentrations of phalloidin as low as 0.25 µM displaced cofilin by ~20%. Increasing concentrations of phalloidin displaced cofilin from actin nearly completely. However, in the presence of Aip1 there appeared to be no displacement of cofilin at the concentrations at which we assayed its activity. There was little to no displacement even at 1:1 concentrations of Aip1: cofilin (Fig. 3A). Thus, Aip1 does not displace cofilin from actin.
Figure 3.
Aip1 does not displace cofilin from F-actin to sever (A) 2 µM fluorescent pyrene-actin was treated with 3 µM cofilin corresponding to vcof>0.9 and increasing concentrations of Aip1 at pH 6.8 to assay competitive binding by equilibrium fluorescence titration experiments. The graph was normalized to amount of polymer monitored by a FRET assay. Aip1 does not compete for binding by cofilin, as shown by a linear fit of the data. We carried out a binding assay to monitor cooperative binding of cofilin to fluorescently labelled pyrene actin in order to select the concentration used for graph (A) (inset). (B) Phalloidin, a known competitive inhibitor of actin, can displace cofilin from actin. (C) To validate our results from (A), we measured severing rates of actin in the presence of 1 µM cofilin that, in our single filament assays, showed maximal severing rates. Adding increasing amounts of Aip1 caused a consistent increase in severing rate. At equimolar cofilin to Aip1 ratios, the rate exceeded 10x that obtained by cofilin alone. (D) Aip1 boosts severing across a wide range of cofilin-actin ratios, even when it is present in sub-stoichiometric quantities (roughly 1000x less than the cofilin concentrations). Error bars represent S.D. and data from at least 2 movies was used to compute severing rates.
If Aip1 were to compete with cofilin for binding to F-actin, then we would predict that Aip1 should inhibit severing when cofilin is present at the optimal concentration where severing rates are highest. In our assay, filaments severed fastest in the presence of 1 µM cofilin corresponding to ν = 0.3. Using this concentration of cofilin, we observed that severing rates increased with increasing concentrations of Aip1 (Fig. 3C). This is the opposite of what is expected if Aip1 functions by displacing cofilin.
Aip1’s effect was seen even at extremely low ratios of Aip1: cofilin with a roughly four-fold increase at 10 nM Aip1. At equimolar concentrations of Aip1: cofilin, severing rates were increased over ten times the maximal severing rates achieved by any concentration of cofilin alone despite the fact that Aip1 does not displace cofilin at these concentrations (Fig. 3A).
The displacement hypothesis predicts that Aip1 will inhibit severing at cofilin binding densities of 0.5 or less, and only accelerate severing at concentrations corresponding to ν >0.5 [18]. Therefore, to further test whether Aip1 enhances severing by displacing cofilin, we compared severing rates across a range of cofilin concentrations in the presence or absence of Aip1. We observed that 10 nM Aip1 accelerated actin filament severing rates at all cofilin concentrations (Fig. 3D). This result is inconsistent with Aip1 displacing cofilin from F-actin.
Aip1 accelerates disassembly from the barbed and pointed ends of filaments
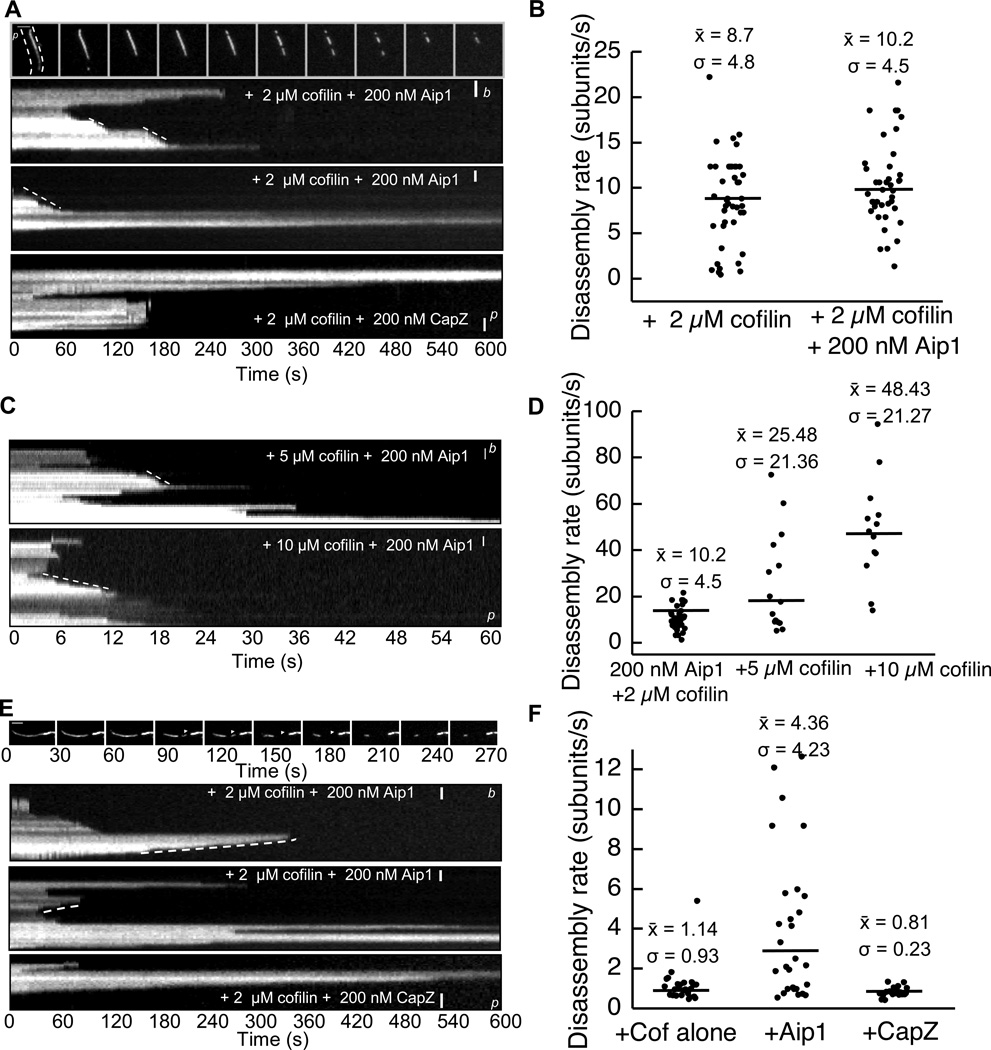
We observed that filaments treated with Aip1 in the presence of saturating concentrations of cofilin appeared to depolymerize rapidly suggesting that Aip1 also accelerates subunit loss from actin filament ends. In order to measure depolymerization rates from ends, filaments were immobilized to the coverslip with the actin bundling protein, filamin, unlike previous experiments where the filaments were free-floating and imaged in the presence of methylcellulose.
In the presence of 2 µM cofilin and 0.2 µM Aip1, filaments rapidly disassembled by shrinking from both barbed and pointed ends. Quantitation of depolymerization rates from barbed ends of polarity marked actin filaments revealed an average rate of 8 – 10 subunits per second in the presence of 2 µM cofilin with or without 0.2 µM Aip1 (Fig. 4A top panels, Fig. 4B, Videos S4,S5). Barbed ends of filaments in the presence of cofilin and CapZ, however, were stable, thus once again contradicting previous hypotheses that Aip1 acts in a manner similar to CapZ (Fig. 4A bottom panel).
FIGURE 4.
Aip1 increases rate of subunit dissociation from barbed and pointed ends. (A) Frames and resultant kymographs from a time lapse movie of polarity marked actin filaments [small letters b (barbed) and p (pointed) indicate orientation of ends] showing filaments shrinking from the barbed end in the presence of cofilin +/− Aip1, but not in the presence of CapZ. (B) Quantitation of filament depolymerization rates from (A). Filaments shrink at an average of 8-10 subunits in the presence or absence of Aip1. Black line represents mean of observations, n=at least 3 movies in each scenario, dots represent individual events. Mean, S.D. indicated. (C) Kymographs of polarity marked actin filaments disassembled in the presence of 5 and 10 µM cofilin and 200 nM Aip1. (D) Quantitation of rates from (C) for the barbed end shows barbed end dissociation rates roughly doubling (from 10 to 25 to 40) subunits per second for each 2x increase in cofilin concentration. Enhanced barbed end disassembly is cofilin-dependent. (E) Frames from a time lapse movie showing filaments shrinking from the pointed end after a severing event (white arrowhead) and resultant kymographs. These indicate that filaments depolymerize at increasing rates from the pointed end in the presence of 200 nM Aip1. (F) Quantitation of rates from (E) shows a roughly 4x increase in subunit dissociation rate from 1 to 5 subunits per second in the presence of Aip1, but not in the presence of cofilin or CapZ. Scale bar= 1 µm.
We also measured the effects of increasing amounts of cofilin on depolymerization rates. In the presence of 0.2 µM Aip1, barbed end depolymerization rates increased from an average of 10 subunits/second in the presence of 2 µM cofilin to 48 subunits/second in the presence of 10 µM cofilin (Fig. 4C, D). These results reveal a new function for Aip1 in accelerating cofilin-mediated barbed end depolymerization rates anywhere from 5–10 times those measured in the presence of cofilin alone. This is the opposite of what is expected of a high affinity barbed end capping factor and the opposite of what is observed in the presence of CapZ.
Aip1-disassembled cofilin-actin filaments also have a higher pointed end depolymerization rate. Pointed ends depolymerized at a rate of approximately 1 subunit per second in the presence of 2 µM cofilin with or without 0.2 µM CapZ. In contrast, pointed ends depolymerized approximately 4x faster on average in the presence of 2 µM cofilin and 0.2 µM Aip1 (Fig 4E,F).
We also observed a statistically significant increase in severing rates in the presence of Aip1 and cofilin, but not CapZ and cofilin (Figure S1). On average, we also noticed that filament severing rates were ~2-fold faster when the filaments were bound to filamin than when they were floating freely in solution (compare figure 1B to Figure S1B). The boost in severing rates could be the result of attachment of filaments along multiple points across their lengths, as this has been demonstrated to contribute to severing [33].
Aip1 and CapZ have differing effects on disassembly rate and critical concentration
There were multiple lines of evidence indicating that Aip1 was not acting as a barbed end capping factor and had additional biochemical roles distinct from CapZ. To further compare Aip1 and CapZ in actin disassembly, we sought to analyze changes in bulk actin polymer mass in the presence of high concentrations of cofilin.
Changes in pyrene fluorescence are a classic measure of the amount of actin polymer. However, pyrene-actin is quenched in the presence of cofilin and therefore cannot accurately report on polymer mass during cofilin-mediated disassembly [14]. We therefore used fluorescence energy transfer (FRET) to measure the decrease in polymer mass in the presence of the depolymerizers. FRET has been used successfully in the past to measure actin polymerization [34, 35]. We performed a number of controls to help validate the assay to measure F-actin in the presence of cofilin and those experiments are presented in figure S2.
We first examined the effect of cofilin alone on actin, using FRET. The addition of cofilin to 2 µM pre-polymerized F-actin at pH 7.8 caused an initial decrease in FRET, representing a decrease in polymer mass, in a dose dependent manner over 25 minutes (Fig. 5A). This is consistent with the fact that cofilin binds to the newly dissociated ADP actin monomers with an affinity of 150 nM and suppresses ADP to ATP exchange [36, 37].
FIGURE 5.
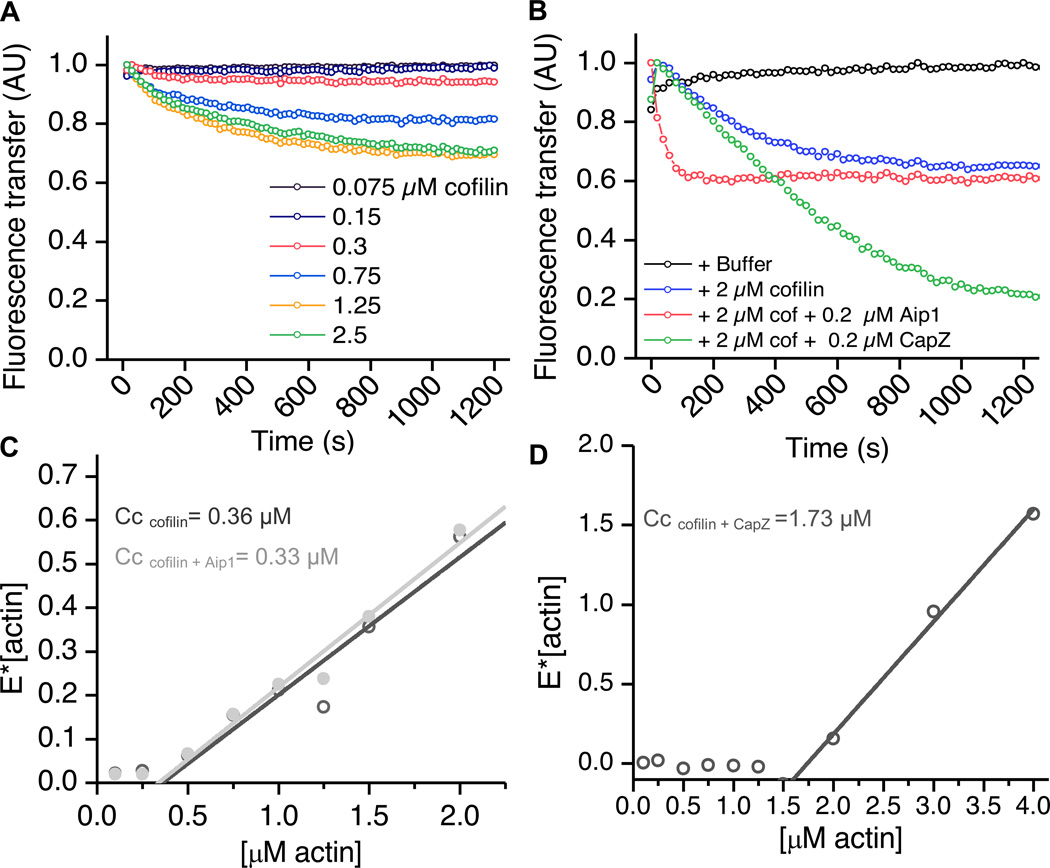
CapZ and Aip1 have differing effects on the rates and extents of cofilin-mediated disassembly. (A) Increasing concentrations of cofilin cause an initial disassembly of actin in a dose-dependent manner. 2 µM pre-polymerized F-actin was mixed with varying amounts of cofilin, and the fluorescence of Oregon green actin was measured over time. The decrease in FRET represents loss in polymer mass. (B) CapZ and Aip1 depolymerize actin at differing rates and to different extents. Pre-polymerized actin was treated with 2 µM cofilin +/− 0.2 µM Aip1 or CapZ. Actin depolymerizes initially in the presence of cofilin alone as described (blue line). In the presence of Aip1 (pink line), the reaction proceeds roughly 6x faster, but to the same extent as cofilin alone. 0.2 µM CapZ cause the reaction to proceed at roughly the same rate as cofilin alone but almost completely to monomer. (C, D) FRET assays to determine the critical concentration (Cc) of actin as a function of cofilin, CapZ or Aip1. Fluorescence intensity in the presence of polymerizing and non-polymerizing conditions was measured at various actin concentrations and the equation [E]= Emax* [(actin-Cc)/actin] was used to fit the data. The x-intercept represents the critical concentration. In (C), the dark grey line shows the condition actin+ cofilin, and in the presence of 1 µM cofilin alone, the critical concentration was only moderately raised (from 0.2 to 0.36). The addition of 0.1 µM Aip1 (light grey) did not significantly affect the critical concentration, however (D) the addition of 0.1 µM CapZ to cofilin, raised the critical concentration over ten-fold to 1.73 µM. Thus, Aip1 and CapZ have differing effects on actin critical concentration in the presence of cofilin. See also figure S3.
We used FRET to compare the disassembly characteristics of cofilin-actin in the presence of capping protein versus Aip1 (Fig. 5B). In the presence of cofilin and Aip1, actin polymer mass decayed roughly 6x faster but to the same extent as it did in the presence of cofilin alone. This indicated that Aip1 was accelerating the rate of actin depolymerization in conjunction with cofilin. However in the presence of cofilin and CapZ, actin polymer mass decayed more slowly than in the presence of Aip1, but it was converted nearly completely to monomer over a period of 25 minutes. Thus, CapZ appeared to affect the extent, but not the rate of actin depolymerization in the presence of cofilin, unlike Aip1.
Next, we tested the effects of Aip1 and CapZ on actin critical concentration (Cc) in the presence of cofilin by FRET. The concentration of actin at which we first begin to see polymerization signifies the critical concentration of actin. Using this approach, we estimate the Cc for pure actin to be 0.2 µM (Fig. S3), which is in agreement with previous measurements [38]. Addition of 1 µM cofilin increased the critical concentration to between 0.3 – 0.4 µM (Fig. 5C). We conclude that cofilin has only a modest effect on the critical concentration which is consistent with previous results [4]. We measured the Cc of actin in the presence of 1 µM cofilin and 0.1 µM Aip1 and found it to be nearly identical to that in the presence of cofilin alone (Cc = 0.36 µM in 1 µM cofilin alone versus Cc = 0.33 µM in the presence of 1 µM cofilin and 0.1 µM Aip1) (Fig. 5C). Similarly, treatment of actin with CapZ alone had only a modest effect on Cc (Fig. S3). However, the combination of 1 µM cofilin and 0.1 µM CapZ raised the critical concentration more than five-fold relative to that of pure actin to 1.7 µM (Fig. 5D) which is the pointed end critical concentration for ADP-actin [38]. This is consistent with the barbed ends being capped by CapZ and ADP-G-actin in a high affinity complex with cofilin. Therefore CapZ affects the critical concentration to a greater extent than Aip1, and the two proteins are not functionally redundant.
Aip1 does not cap filament barbed ends
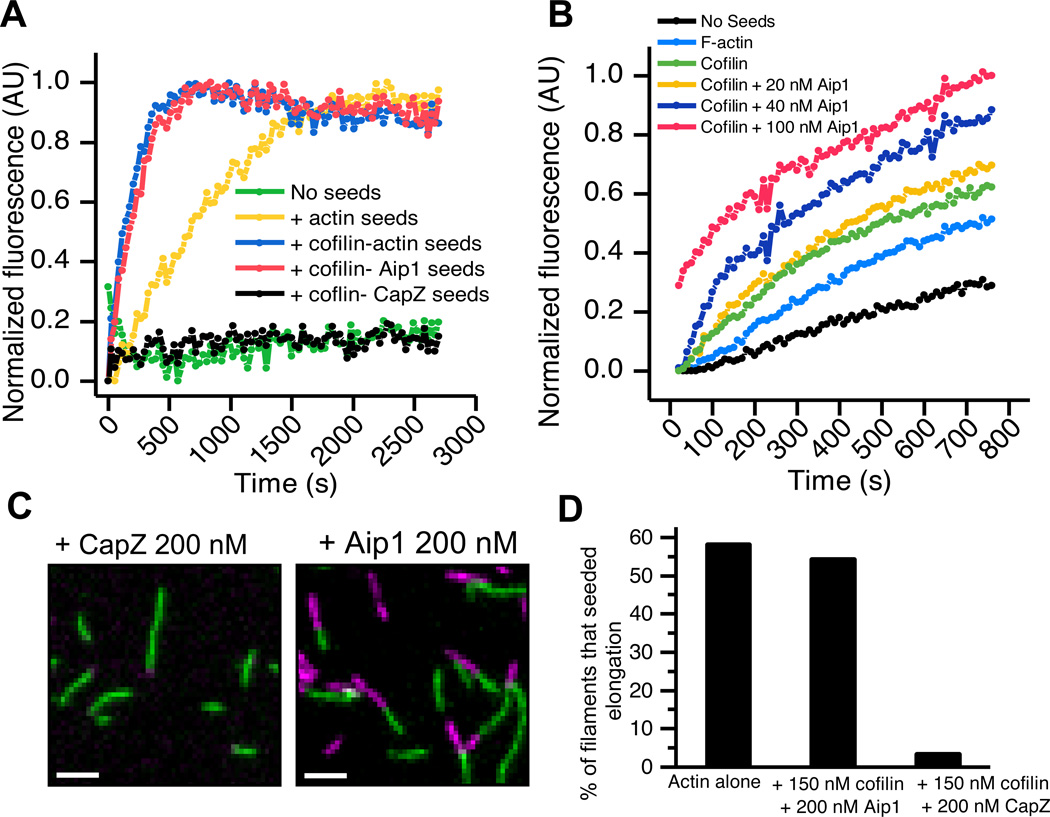
Differences in kinetics of cofilin-mediated depolymerization in the presence of Aip1 versus CapZ and cofilin as well as differences in the critical concentrations prompted us to directly reinvestigate whether or not Aip1 caps barbed ends. An established assay for barbed end capping is the inability of capped barbed ends to seed new actin polymerization. New actin monomer adds to the barbed ends of pre-existing short actin filament seeds thus shortening the lag phase of polymerization. Filaments with occluded barbed ends such as those bound by CapZ will not be able to reduce the lag phase of new actin assembly. When products of the disassembly reactions were used to seed new actin polymerization, F-actin seeds enhanced the initial rate of polymerization (Fig. 6A yellow line), and cofilin-actin filaments seeded polymerization even more efficiently due to a large number of free severed ends. As expected, CapZ-bound seeds were unable to enhance the rate of actin polymerization (black line). However, filaments depolymerized by cofilin and Aip1 seeded actin assembly as efficiently as the cofilin-actin seeding mixture (blue and pink lines). Thus, Aip1 does not form a high affinity cap on actin filaments. Additionally, the presence of increasing amounts of Aip1 in the presence of a fixed concentration of cofilin increased the number of pre-existing short filament seeds, consistent with Aip1’s ability to increase severing rates in the presence of cofilin as seen by light microscopy (Fig 6B).
FIGURE 6.
Aip1 does not cap filament ends. (A) Aip1 does not inhibit seeding of pyrene actin polymerization. Actin filaments were mixed with 2 uM cofilin +/− 0.2 uM Aip1 or CapZ for 15 minutes and then 0.25 µM total actin was used to seed polymerization of 2 µM pyrene actin. While the lag phase of unseeded actin is long (green line), actin seeds shorten the lag phase of polymerization (yellow line). Cofilin creates many severed ends that seed polymerization more efficiently whereas Aip1 does not inhibit this reaction (blue and pink lines). CapZ however inhibits the seeding reaction (black line). (B) Increasing amounts of Aip1 in the presence of cofilin produce more filament seeds for elongation. Filaments depolymerized in the presence of 20, 40 and 100 nM Aip1 (yellow, blue and pink lines respectively) and (a fixed amount) of cofilin show enhanced seeding as compared to cofilin alone seeds (green line). (C) Oregon Green 488 actin filaments elongate with Alexa 647 G-actin even in the continuous presence of 150 nM cofilin and 200 nM Aip1 (right panel), however, the presence of 200 nM CapZ inhibits this reaction (left panel). Scale bar= 2 µm. (D) Quantitation of the relative numbers of elongating ends in the presence of actin alone or actin and 150 nM Cofilin +/− 200 nM CapZ or Aip1.
We used single filament imaging to further test whether Aip1 mediates cofilin dependent barbed end capping. Fluorescently labeled Oregon green 488-actin filaments were polymerized on a bed of filamin in a perfusion chamber, and filaments were allowed to elongate by addition of Alexa 647 G-actin monomer, in the presence of 0.15 µM cofilin and 0.2 µM of either Aip1 or CapZ (Fig. 6C, 6D). Unlike CapZ, Aip1 did not inhibit elongation of preformed actin filaments at the level of single actin filaments. Therefore, Aip1 does not suppress barbed end growth even when it is continuously present at high concentrations along with cofilin.
DISCUSSION
We found that thymus extracts rapidly sever and disassemble single actin filaments despite having cofilin concentrations that are too high to sever actin filaments in pure solution. We demonstrated that fast actin disassembly in the presence of saturating cofilin can be attributed to, at least in part, Aip1 but not to Capping Protein and our analysis further revealed that Aip1 alters the characteristics of cofilin-mediated filament disassembly while capping protein does not.
Previous results proposed that Aip1 caps barbed ends with a high affinity and would therefore be functionally redundant with Capping Protein [26]. We found, however, that Aip1 does not prevent growth of filament ends, and filaments shrink at accelerated rates in the presence of Aip1, which is the opposite of what we would predict if the filaments were capped. In addition, Aip1 had little to no effect on the critical concentration while CapZ did. Our results are therefore inconsistent with Aip1 forming a stable cap that effectively terminates barbed end actin dyanmcis. Rather, our results are more consistent with models proposing that Aip1’s side binding and not its end-binding is more important for severing cofilin decorated filaments [22, 39] to create more filament ends that can grow or shrink. Interestingly though, small quantities of Aip1 show a strong effect on severing cofilin-actin filaments as observed by microscopy but a more modest effect on creating new barbed ends when assayed with bulk filament polymerization assays. Aip1 might possess the ability to create some unconventional ends that are transiently resistant to growth, as posited in previous work [32].
While Aip1 and CapZ are biochemically distinct, they show strong genetic interactions in yeast, playing critical roles in the assembly and morphogenesis of Arp2/3-derived actin arrays by maintaining a pool of actin subunits available for assembly [27]. CapZ can help maintain a pool of assembly competent actin by suppressing non-productive barbed end elongation and funneling actin monomer towards Arp2/3 nucleation sites [40]. Given our results, Aip1 might help maintain a pool of assembly-competent actin by triggering fast depolymerization of cofilin-F-actin. Thus, we propose that Aip1 and CapZ genetically complement one another through distinct mechanisms.
Cofilin binding to F-actin disrupts lateral interactions between actin subunits [41]. However, stretches of actin polymer saturated with cofilin are stable [4, 10] because cofilin has two actin binding sites allowing it to bridge two longitudinal subunits in the filament [8, 42, 43]. Severing therefore occurs at junctions between decorated and undecorated polymer [12] explaining why severing is maximal at intermediate levels of cofilin occupancy [4, 12, 18]. We found that Aip1 does not displace cofilin from F-actin and promotes actin filament disassembly at all cofilin occupancies. Our results imply that it is the sites of actin polymer occupied by cofilin themselves that are destabilized by Aip1 (Figure 7). This model is consistent with yeast two hybrid, binding, and modeling data supporting that Aip1 forms a ternary complex with F-actin and cofilin [22, 44, 45].
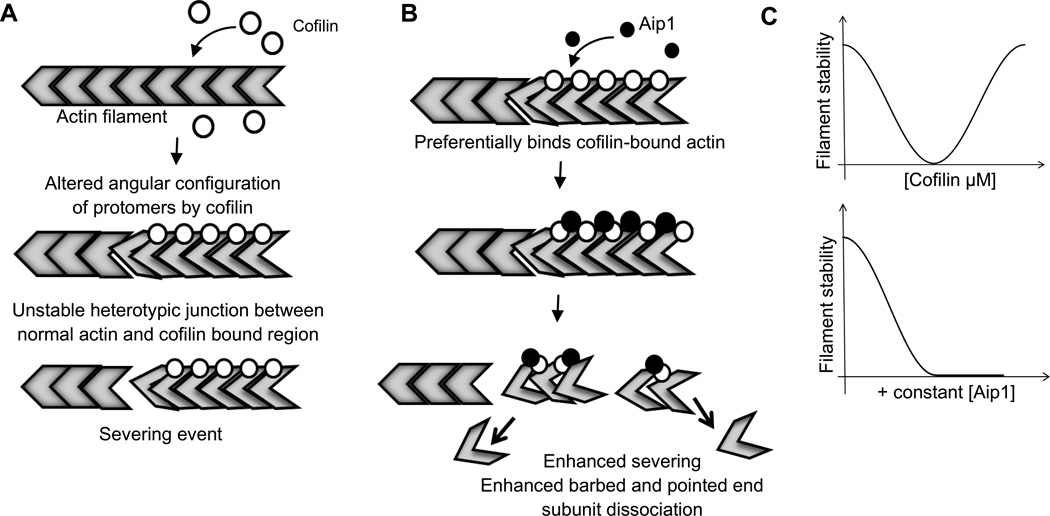
FIGURE 7.
Model representing mode of Aip1 action on cofilin-actin filaments. (A) Cofilin alone binds to the filament and alters the angular configuration of actin protomers within the polymer lattice. Severing is caused due to unstable heterotypic junctions between cofilin-bound and unbound regions on the actin filament. (B) Aip1 preferentially binds stretches of actin polymer occupied with cofilin leading to enhanced severing and faster disassembly from both barbed and pointed ends. (C) In the presence of Aip1, cofilin-saturated filaments are no longer stable and can be destabilized.
Mutagenesis and modeling studies on Aip1 demonstrate that Aip1 contacts cofilin while bridging two contiguous actin subunits in the filament [44]. Therefore, we can consider two alternative mechanisms through which Aip1 could promote cofilin mediated severing and subunit dissociation. In the first, Aip1 binding to cofilin occupied polymer might disrupt cofilin’s stabilizing interaction with the adjacent actin subunit to cause severing. Mutations in cofilin that compromise its F-actin specific binding interaction increase severing [46] and the Aip1 and cofilin binding sites on actin would appear to overlap [22, 45] making this an attractive model. However, studies with cofilin and Aip1 from C. elegans have shown that the ability of Aip1 to enhance actin disassembly requires cofilin’s F-actin binding site [47]. An alternative possibility then is that Aip1 further distorts actin structure in the presence of cofilin to promote severing and increase subunit dissociation rates from filament ends. It has been hypothesized that a slow isomerization step follows cofilin binding [48]. If this proposed conformational change were coupled to severing, we could speculate that Aip1 catalyzes the transition between the two states to destabilize the filament. Recent work on cofilin from Plasmodium falciparum shows that it contacts a novel binding site on F-actin to sever filaments without decorating the polymer as human cofilin does [49]. Thus, multiple cofilin binding modes might permit multiple modes of filament disassembly, and it is tempting to speculate that Aip1 alters filament structure or induces a conformational change in cofilin allowing mammalian cofilin to access the novel P. falciparum binding site to destabilize the polymer.
Our results demonstrating that the combination of Aip1 and cofilin accelerate actin subunit dissociation rates offers one possible mechanism for attaining faster depolymerization rates in vivo. Aip1 therefore offers a potential control point to switch cofilin action from one that favors actin assembly [4, 50] to one that favors fast depolymerization. Other factors in addition to Aip1 facilitate cofilin mediated actin disassembly. It will be important to re-examine each of these auxiliary factors to test if they, like Aip1, alter the mechanism of filament disassembly.
EXPERIMENTAL PROCEDURES
Full experimental procedures are available in the supplemental material.
Imaging of actin single filaments
Single actin filaments were either prepolymerized, flowed into perfusion chambers and imaged in solution (Figures 1, 2 and 3) or polymerized in the chamber and attached to coverslips via filamin (Figures 4 and 6). Relevant combinations depolymerizers were flowed into the perfusion chamber and filaments were imaged in a buffer containing oxygen-scavengers (1xPhotoBuffer). Severing events were enumerated as the number of visual breaks per second normalized to the amount of polymer measured in microns.
Depolymerization rates were calculated from kymographs generated using Fiji software.
Elongation of polymerized Oregon green actin filaments was carried out in the presence of 2 µM monomeric Alexa 647 actin, 150 nM cofilin and 200 nM of either CapZ or Aip1 for 60 seconds. This was compared to a control with actin alone.
Fluorescence equilibrium binding and competition assays
Quenching of pyrene fluorescence was used to quantify cofilin binding to F-actin in the presence or absence of phalloidin or Aip1 as described previously. [9,18]. FRET was used to normalize for the amount of polymer present at various concentrations of Aip1.
Measurement of FRET
12–15% labelled Oregon green actin and 35–40 % tetramethylrhodamine actin were premixed at 20 µM in G buffer (pH 7.4). 1xFBuffer was added to initiate polymerization. Spectroscopic monitoring of fluorescence quenching of OG488 (λEx= 490 nm, λEm = 530 nm) over time was used to report on assembly. For disassembly reactions, actin was prepolymerized and combinations of depolymerizers were added to final concentrations of 2 µM cofilin, 0.2 µM Aip1 or 0.2 µM CapZ. Fluorescence of Oregon Green actin was monitored as mentioned. Depolymerization leads to dequenching of OG488 fluorescence. Normalized readings were obtained by using Origin graphing software.
Measurement of critical concentration using FRET
F-actin was pre-polymerized at 10 µM and diluted to the desired concentrations in 1xF-buffer, in the presence of the indicated concentrations of the depolymerizers. Reactions were allowed to incubate overnight (12 hours) before reading. This was compared to equimolar actin concentrations where there was no FRET. Data was converted to E values as described previously by using the equation E=1-(FDA/FD) [51]. Readings were fit to the equation E*[actin]=Emax*[actin]-Emax*Cc as described previously [51].
Seeding reaction with actin pyrene
Actin filaments treated with or without cofilin and increasing concentrations of Aip1 for 10 mins (Fig 6A) and 2 mins (Fig 6B) were added to a solution of 2 µM pyrene labeled G-actin. The final concentration of F-actin seeds was 0.25 µM (Fig 6A) and 0.5 µM (Fig 6B) and the final cofilin concentration was 100 nM (Fig 6B). Final Aip1 concentrations are provided in the main text.
Supplementary Material
HIGHLIGHTS.
Aip1 destabilizes filaments in the presence of saturating concentrations of cofilin.
Aip1 potentiates cofilin-mediated severing and augments barbed and pointed end depolymerization rates.
Filaments severed by cofilin and Aip1 are not capped, thus contradicting previous models that Aip1 is redundant with capping protein.
Acknowledgements
Spectroscopic studies were carried out at the Roy J. Carver Metabolomics Center (UIUC). We thank Hui-Chia Yu and Kieran Normoyle for the purification of CapZ, ActA and filamin. We also thank the members of the Brieher Lab for their valuable input, and Karthik Murali for math-related discussions. This work was supported by National Institutes of Health grant R01-GM106106 to WMB.
Glossary
- Aip1
Actin Interacting Protein 1
- FRET
Förster Resonance Energy Transfer
- TMR
TetraMethylRhodamine
- Cc
Critical concentration
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 2.Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 3.Bravo-Cordero JJ, Magalhaes MaO, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono S. UNC-60B, an ADF/Cofilin Family Protein, Is Required for Proper Assembly of Actin into Myofibrils in Caenorhabditis elegans Body Wall Muscle. J. Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 1999;18:6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden SM, Miller PS, Brauweiler a, Bamburg JR. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- 9.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol. Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 10.McGough a, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De La Cruz EM. How cofilin severs an actin filament. Biophys. Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann A, Guérin C, Martiel J, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenblatt J, Agnew BJ, Abe H, Bamburg JR, Mitchison TJ. Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J. Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility [see comments] J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berro J, Sirotkin V, Pollard TD. Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol. Biol. Cell. 2010;21:2905–2915. doi: 10.1091/mbc.E10-06-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer–insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elam WA, Kang H, De La Cruz EM. Competitive displacement of cofilin can promote actin filament severing. Biochem. Biophys. Res. Commun. 2013;438:728–731. doi: 10.1016/j.bbrc.2013.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Normoyle KPM, Brieher WM. Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J. Biol. Chem. 2012;287:35722–35732. doi: 10.1074/jbc.M112.396051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohri K, Vorobiev S, Fedorov Aa, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- 21.Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- 22.Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 1999;112(Pt. 1):1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- 24.Ren N, Charlton J, Adler PN. The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics. 2007;176:2223–2234. doi: 10.1534/genetics.107.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato A, Kurita S, Hayashi A, Kaji N, Ohashi K, Mizuno K. Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells. Biochem. J. 2008;414:261–270. doi: 10.1042/BJ20071655. [DOI] [PubMed] [Google Scholar]
- 26.Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- 27.Michelot A, Grassart A, Okreglak V, Costanzo M, Boone C, Drubin DG. Article Actin Filament Elongation Is Controlled by Three Distinct Mechanisms. 2013:182–195. doi: 10.1016/j.devcel.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okreglak V, Drubin DG. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada K, Blanchoin L, Abe H, Chen H, Pollard TD, Bamburg JR. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- 30.Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wear MA, Yamashita A, Kim K, Maéda Y, Cooper JA. How capping protein binds the barbed end of the actin filament. Curr. Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- 32.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J. Mol. Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang YL, Taylor DL. Probing the dynamic equilibrium of actin polymerization by fluorescence energy transfer. Cell. 1981;27:429–436. doi: 10.1016/0092-8674(81)90384-6. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DL, Reidler J, Spudich JA, Stryer L. Detection of actin assembly by fluorescence energy transfer. J. Cell Biol. 1981;89:362–367. doi: 10.1083/jcb.89.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- 37.Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 1998;273:4647–4652. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 38.Howard J. Mechanics of Motor Proteins & the Cytoskeleton. 2001st ed. Sinaeur Associates; 2001. [Google Scholar]
- 39.Shi M, Xie Y, Zheng Y, Wang J, Su Y, Yang Q, Huang S. Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover. Plant J. 2012 doi: 10.1111/tpj.12065. [DOI] [PubMed] [Google Scholar]
- 40.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, Cao W, Suarez C, Martiel J-L, Blanchoin L, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lappalainen P, Fedorov EV, Fedorov a a, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pope BJ, Gonsior SM, Yeoh S, McGough A, Weeds AG. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 2000;298:649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- 44.Clark MG, Teply J, Haarer BK, Viggiano SC, Sept D, Amberg DC. A genetic dissection of Aip1p’s interactions leads to a model for Aip1p-cofilin cooperative activities. Mol. Biol. Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark MG, Amberg DC. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 2007;176:1527–1539. doi: 10.1534/genetics.107.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono S, McGough a, Pope BJ, Tolbert VT, Bui a, Pohl J, Benian GM, Gernert KM, Weeds aG. The C-terminal tail of UNC-60B (actin depolymerizing factor/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J. Biol. Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- 47.Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- 48.De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys. J. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong W, Webb AI, Olshina MA, Infusini G, Tan YH, Hanssen E, Catimel B, Suarez C, Condron M, Angrisano F, et al. A mechanism for actin filament severing by malaria parasite actin depolymerizing factor 1 via a low affinity binding interface. J. Biol. Chem. 2014;289:4043–4054. doi: 10.1074/jbc.M113.523365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 51.Bugyi B, Papp G, Hild G, Lõrinczy D, Nevalainen EM, Lappalainen P, Somogyi B, Nyitrai M. Formins regulate actin filament flexibility through long range allosteric interactions. J. Biol. Chem. 2006;281:10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.