Abstract
Chromosome 8q24 has emerged as an important region for genetic susceptibility to various cancers, but little is known about the contribution of DNA methylation at 8q24. To evaluate variability in DNA methylation levels at 8q24 and the relationship with cancer susceptibility single nucleotide polymorphisms (SNPs) in this region, we quantified DNA methylation levels in peripheral blood at 145 CpG sites nearby 8q24 cancer susceptibility SNPs or MYC using pyrosequencing among 80 Caucasian men in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. For the 60 CpG sites meeting quality control, which also demonstrated temporal stability over a 5-year period, we calculated pairwise Spearman correlations for DNA methylation levels at each CpG site with 42 8q24 cancer susceptibility SNPs. To account for multiple testing, we adjusted p-values into q-values reflecting the False Discovery Rate (FDR). In contrast to the MYC CpG sites, most sites nearby the SNPs demonstrated good reproducibility, high methylation levels, and moderate-high between-individual variation. We observed 10 statistically significant (FDR<0.05) CpG site–SNP correlations. These included correlations between an intergenic CpG site at Chr8:128393157 and the prostate cancer SNP rs16902094 (rho=−0.54; p-value=9.7×10−7; q-value=0.002), a PRNCR1 CpG site at Chr8:128167809 and the prostate cancer SNP rs1456315 (rho=0.52; p-value=1.4×10−6; q-value=0.002), and 2 POU5F1B CpG sites and several prostate/colorectal cancer SNPs (for Chr8:128498051 and rs6983267, rho=0.46; p-value=2.0×10−5; q-value=0.01). This is the first report of correlations between blood DNA methylation levels and cancer susceptibility SNPs at 8q24, suggesting that DNA methylation at this important susceptibility locus may contribute to cancer risk.
Keywords: cancer susceptibility, chromosome 8q24, DNA methylation, epigenetic variation, single nucleotide polymorphism
INTRODUCTION
Chromosome 8q24 has emerged as an important region for genetic susceptibility to a number of cancers, including prostate, breast, colorectal, bladder, and ovarian, as well as glioma, Hodgkin lymphoma (HL), and chronic lymphocytic leukemia (CLL) (1–8). This region is of particular importance for prostate cancer, as genome-wide association studies (GWAS) have identified multiple independent single nucleotide polymorphisms (SNPs) at 8q24 to be associated with prostate cancer risk (2, 9–14). Recently, known susceptibility SNPs at 8q24 were also shown to interact with pesticide use to increase prostate cancer risk in the Agricultural Health Study (15). However, the biological mechanism underlying these associations is unclear.
The 8q24 region has traditionally been described as a gene desert, with most of the susceptibility SNPs spanning an ~800kb region that lies more than 200kb upstream of the MYC oncogene. It has been proposed that epigenetic mechanisms might contribute to risk based on the identification of gene regulatory elements at 8q24 and evidence of long-range interactions with MYC for several susceptibility regions (16–18), although most studies directly evaluating the relationship between the susceptibility SNPs and MYC expression have been null (19). Other genes, specifically POU5F1B and PSCA, and non-coding RNAs (ncRNAs), including PRNCR1, CCAT2, and PVT1, have been identified in the region and might contribute to cancer susceptibility. Although POU5F1B was originally thought to be a pseudogene, recent evidence suggests it may encode a functional protein (20) and it was observed to be overexpressed in prostate cancer tissue (21). PSCA contains a bladder cancer susceptibility SNP and has been shown to be upregulated in bladder cancer and prostate cancer tissues (22, 23). PRNCR1 and CCAT2 have been shown to be overexpressed in prostate or colorectal cancer, respectively (24, 25), and expression of PVT1 has been associated with an upstream prostate cancer susceptibility SNP (26), suggesting a potential role in cancer risk.
Alteration in DNA methylation, a type of epigenetic change, is thought to be one mechanism by which genes and the environment may interact in the development of disease and may contribute to the observed associations for 8q24 genetic variants with cancer risk and their interactions with pesticides. Current evidence suggests that there is both an inherited and an environmental component to DNA methylation (27). Changes in DNA methylation could potentially affect cancer risk by altering the expression of genes or ncRNAs or by influencing genetic stability (28). A number of studies have demonstrated altered DNA methylation levels at CpG sites in gene promoter regions, repetitive DNA elements, and CpG islands (regions containing a higher density of CpG sites) in cancer tissue when compared with histologically normal tissue (27, 28), and there is growing evidence that DNA methylation in peripheral blood may also be associated with cancer (29–33). Evidence also indicates that DNA methylation may mediate the association between genetic variants and disease outcomes. Recently, DNA methylation was suggested to mediate the association between FTO gene variants and obesity (34), as well as the genetic susceptibility to rheumatoid arthritis (35). However, DNA methylation in peripheral blood at chromosome 8q24 has not been well characterized.
To evaluate variability in DNA methylation levels at 8q24 in peripheral blood and the relationship with cancer susceptibility SNPs in the region, we conducted a study of 80 non-Hispanic Caucasian male participants in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Specifically, we aimed to evaluate between-individual and temporal variation in DNA methylation at CpG sites at 8q24 to identify CpG sites with variability in the population and to investigate the stability of DNA methylation levels in this region over time. In addition, we aimed to investigate the underlying genetic structure in the region by evaluating correlations for DNA methylation levels at the CpG sites with each other and with the cancer susceptibility SNPs at 8q24. Correlations between DNA methylation levels at 8q24 in peripheral blood and SNPs in this region have not been previously studied. Such knowledge may provide additional insight into the genetic/epigenetic architecture of this complex cancer susceptibility region. As a secondary aim, we also aimed to evaluate associations between DNA methylation levels at 8q24 and established cancer risk factors [e.g., age, smoking, and body mass index (BMI)].
MATERIALS AND METHODS
Study population
The PLCO Cancer Screening Trial is a randomized trial of more than 150,000 men and women ages 55 to 74 who were enrolled from 10 centers across the United States between 1993 and 2001 to evaluate the impact of specific cancer screening regimens on the risk of mortality from prostate, lung, colorectal, and ovarian cancers (36). At baseline, study subjects provided written informed consent and completed a questionnaire regarding demographic and environmental/lifestyle characteristics (e.g., diet and smoking). Screening arm participants were also asked to provide blood samples at baseline and at yearly follow-up visits for 5 years (36). In the present study, we evaluated DNA methylation levels at chromosome 8q24 using DNA extracted from peripheral blood samples for 80 non-Hispanic Caucasian male PLCO participants in the screening arm without a previous diagnosis of cancer. To allow assessment of temporal variation in DNA methylation levels at 8q24, we selected paired blood samples from baseline (T0) and year 5 (T5) from a separate group of 80 non-Hispanic Caucasian men in PLCO (40 prostate cancer cases and 40 controls). We chose to use samples from T0 and T5, the latest available time point, to maximize the possibility of a change in DNA methylation levels over time. We also selected an independent group of 273 non-Hispanic Caucasian men in PLCO who had no previous history of cancer at enrollment in the PLCO Trial and had genotyping data for the SNPs of interest to serve as a replication set. All subjects selected for these studies were included in a previous genome-wide association study for prostate cancer (14). All participants provided informed consent to participate in genetic studies of cancer and other diseases, and the study was approved by the institutional review boards at the ten centers and the National Cancer Institute.
DNA methylation assay design
We developed targeted assays to quantify DNA methylation at specific CpG sites at 8q24 using pyrosequencing on bisulfite-modified DNA, which is a highly sensitive method to detect differences in DNA methylation between individuals. We chose to develop targeted assays instead of using DNA microarray technology, such as the Illumina Infinium HumanMethylation450 BeadChip (Illumina HM450), because the existing microarrays have limited coverage of the 8q24 region. In addition, CpG sites on the microarrays were not necessarily chosen based on between-individual variation, and in this study, we aimed to identify specific CpG sites with substantial between-individual variation in DNA methylation levels.
Since the 8q24 region is large and because previous research has suggested a greater role of cis than trans regulation of DNA methylation levels by genetic variants (37, 38), we targeted assay development within 50kb of 10 different prostate cancer susceptibility SNPs at 8q24: rs4242382, rs1447295, rs6983267, rs16901979, rs10086908, rs620861, rs1016343, rs1456315, rs13252298, and rs7837328. These SNPs were selected on the basis of an association with prostate cancer in populations of European ancestry, with preference given to SNPs that also demonstrated interactions with pesticide exposures with respect to prostate cancer risk in a report from the Agricultural Health Study (15). However, several of the SNP regions of interest were not pursued because there were few CpG sites or because of the highly repetitive nature of the sequences. We avoided assay development in highly repetitive regions as the assay reproducibility was expected to be low. We focused on SNPs that were associated with prostate cancer risk in the assay design because the 8q24 region appears to be particularly important for prostate cancer, but susceptibility SNPs for other cancers were often located nearby as well. Given the suspected importance of the MYC oncogene at 8q24, we also developed assays in or near MYC, including promoter regions, as well as intron, exon, and 3′ regions. Assay design for all assays was conducted using the forward strand.
In total, we analyzed DNA methylation levels at 145 specific CpG sites at 8q24. Although we did not run the Illumina HM450 microarray in our study, we compared the list of CpG sites that we analyzed to the sites included in this microarray. Of the 145 CpG sites evaluated in our study, 19 were included in the Illumina HM450 microarray, and 18 of the 19 were in the MYC gene or MYC promoter regions.
Pyrosequencing assays
DNA was extracted from all samples using the Qiagen QIAsymphony method. For quality control and to allow assessment of assay reproducibility, 15% duplicates were randomly selected from the study participants (for total of 12 duplicate pairs) and randomly interspersed among the samples, blinded to the analysis laboratory. DNA samples for the study participants were shipped overnight on 96-well plates to EpigenDx, Inc. on dry ice and subsequently stored at −20°C. The DNA was bisulfite-converted using a Zymo Research EZ DNA Methylation kit, which results in conversion of unmethylated cytosines to uracil, whereas the methylated cytosines are not converted. The DNA was then PCR-amplified within the chromosome 8q24 regions of interest, resulting in incorporation of cytosine (C) for the methylated cytosines and thymine (T) for the unmethylated cytosines, using 45 cycles per PCR. Four artificial control samples were included on each plate [one negative control sample (no DNA added), as well as three positive control samples with known global DNA methylation levels: low (0%), partial (50%), and highly methylated (100%)]. Sequencing was performed using the Pyrosequencing PSQ96 HS System (Pyrosequencing Qiagen). The methylation status at each CpG site was analyzed as an artificial C/T SNP using QCpG software (Pyrosequencing Qiagen), and the percent of methylation was calculated for each CpG site as methylated cytosine divided by the sum of methylated and unmethylated cytosines.
ENCODE data
We obtained ENCODE data annotations [ChIP-seq, which assesses transcription factor binding sites (TFBS), DNase I hypersensitivity, and histone methylation] for regions of interest at 8q24 in histologically normal tissues (PrEC, ColonOC) from prostate or colon cancer patients and cancer cell lines (Caco-2, HCT-116, LNCaP) of prostate or colon origin from the hg19 build UCSC ENCODE file browser (http://genome.ucsc.edu/cgi-bin/hgFileSearch). CpG site coordinates were converted from hg18 to hg19 using the liftOver tool and intersections were performed with intersectBed (BEDTools package), allowing a gap of 400bp. We determined average alignability for CpGs by taking the average alignability score (from the 36mer CRG Alignability track in the UCSC Genome Browser, http://genome.ucsc.edu) for the bases in a 200bp window (100bp upstream and 100bp downstream) surrounding each CpG site. We identified peaks corresponding to putative TFBS, DNase I hypersensitivity sites, or histone methylation marks based on a p-value<0.05. We translated p-values into q-values, which reflected the False Discovery Rate (FDR) using the Benjamini and Hochberg method (39), accounting for the number of peaks for each mark within a given cell line.
Genotyped SNPs
We considered 42 cancer susceptibility SNPs at 8q24 in our study (Supplementary Table S1). These SNPs were selected based on an association with any cancer in a genome-wide association study with a p-value<10−6. We also included one SNP (rs7837328) that was not associated with prostate cancer at the genome-wide significance level in GWAS, but that previously was shown to interact with pesticide exposure to increase prostate cancer risk (p-interaction<0.05) in the Agricultural Health Study (15). Genotyping was previously conducted using the Omni2.5 platform. Of the 42 SNPs of interest, 18 were not directly genotyped in our study participants, and we used data that were imputed for these specific SNPs using 1,000 Genomes Project data release version 3 and IMPUTE2 (40).
Statistical analysis
Based on the 12 pairs of duplicate samples in our study, we calculated the coefficient of variation (CV) and Intraclass Correlation Coefficient (ICC) for each CpG site separately to assess assay reproducibility. For the 60 CpG sites with a CV<25, we calculated correlations for DNA methylation levels at each site with each other and with the 42 cancer susceptibility SNPs at 8q24 by calculating pairwise CpG site – CpG site and CpG site – SNP Spearman correlations (rho). We used ordinal variables for the SNPs, coded according to the number of variant alleles (0 corresponding to homozygous wild-type, 1 to heterozygous, and 2 to homozygous variant). We squared the rho values for the CpG site – CpG site correlations to obtain r2 values. For each measured correlation, we translated the p-value into a q-value, which reflected the FDR using the Benjamini and Hochberg method (39), accounting for the number of comparisons. An FDR threshold of 0.05 was used for defining significant associations.
To evaluate within-individual temporal variation in DNA methylation levels at 8q24, we conducted paired t-tests comparing the mean DNA methylation level between the T0 and T5 visits for each of the 60 CpG sites separately using paired blood samples for the 40 prostate cancer cases and 40 controls combined. We also explored the pattern among cases and controls separately.
For our secondary aim of evaluating the association between DNA methylation levels at 8q24 and established cancer risk factors, we calculated pairwise Spearman correlations for DNA methylation levels at each of the 60 CpG sites and the following factors: age at blood draw, smoking (separately for smoking status and pack-years), and BMI. Age, BMI, and pack-years of smoking were maintained as continuous variables and smoking status was treated as an ordinal variable (0 for never smoker, 1 for former smoker, and 2 for current smoker).
RESULTS
We quantified DNA methylation levels at a total of 145 specific CpG sites, including 46 sites nearby cancer susceptibility SNPs at 8q24 and 99 sites in or near MYC (Table 1). Most of the CpG sites evaluated upstream of MYC were located in intergenic regions; however, several of the assays conducted were located in PRNCR1 or POU5F1B (Figure 1).
Table 1.
DNA methylation levelsa and QC results by assay
| Chr8q24 Assay ID | Coordinatesb | Assay location | Closest SNPc | Cancer associated with SNP | Distance to SNP (bp)d | # CpGs meet QC/analyzede | Avg CV all CpGs | Avg mean all CpGs | Avg CV QC subsetf | Avg ICC QC subsetf | Avg Mean QC subsetf | Avg Range QC subsetf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADS2775-FS1 | Chr8:128081544- 128081546 | Intergenic | rs10086908 | Prostate | 425 | 2/2 | 2.6 | 92.8 | 2.6 | 0.41 | 92.8 | 19.2 |
| ADS2775-FS2 | Chr8:128081584- 128081614 | Intergenic | rs10086908 | Prostate | 465 | 5/5 | 2.1 | 92.6 | 2.1 | 0.70 | 92.6 | 13.9 |
| ADS2774-FS1 | Chr8:128167771- 128167788 | PRNCR1 | rs7841060 | Prostate | 2112 | 3/3 | 1.9 | 94.1 | 1.9 | 0.64 | 94.1 | 14.5 |
| ADS2774-FS2 | Chr8:128167809- 128167834 | PRNCR1 | rs7841060 | Prostate | 2150 | 4/4 | 6.7 | 73.9 | 6.7 | 0.65 | 73.9 | 29.6 |
| ADS2771-FS1 | Chr8:128393038 | Intergenic | rs445114 | Prostate | 675 | 1/1 | 3.2 | 51.3 | 3.2 | 0.68 | 51.3 | 15.6 |
| ADS2771-FS2re | Chr8:128393157- 128393166 | Intergenic | rs445114 | Prostate | 794 | 3/3 | 2.8 | 85.8 | 2.8 | 0.81 | 85.8 | 20.9 |
| ADS2772-FS1 | Chr8:128398828 | Intergenic | rs620861 | Prostate | −6027 | 1/1 | 0.9 | 76.3 | 0.9 | 0.61 | 76.3 | 5.0 |
| ADS2772-FS2 | Chr8:128398847- 128398868 | Intergenic | rs620861 | Prostate | −5987 | 2/2 | 0.6 | 78.4 | 0.6 | 0.77 | 78.4 | 3.8 |
| ADS2772-FS3 | Chr8:128398897- 128398908 | Intergenic | rs620861 | Prostate | −5947 | 2/2 | 1.6 | 78.8 | 1.6 | 0.55 | 78.8 | 13.0 |
| ADS3548-FS1 | Chr8:128465744 | Intergenic | rs1562430 | Breast | 5150 | 1/1 | 1.9 | 84.3 | 1.9 | 0.86 | 84.3 | 19.7 |
| ADS3548-FS2 | Chr8:128465833- 128465853 | Intergenic | rs1562430 | Breast | 5239 | 2/2 | 2.0 | 89.1 | 2.0 | 0.82 | 89.1 | 15.9 |
| ADS3549-FS1 | Chr8:128498051 | POU5F1B Exon 2 | rs7014346 | Colorectal | 4077 | 1/1 | 0.8 | 90.9 | 0.8 | 0.78 | 90.9 | 16.3 |
| ADS3549-FS2 | Chr8:128498079- 128498113 | POU5F1B Exon 2 | rs7014346 | Colorectal | 4105 | 3/3 | 2.7 | 89.2 | 2.7 | 0.62 | 89.2 | 14.0 |
| ADS3549-FS3 | Chr8:128498131- 128498173 | POU5F1B Exon 2 | rs7014346 | Colorectal | 4157 | 5/5 | 2.2 | 69.0 | 2.2 | 0.66 | 69.0 | 12.3 |
| ADS2778-FS1 | Chr8:128513861- 128513898 | Intergenic | rs7000448 | Prostate | 3509 | 3/5 | 2.4 | 53.7 | 2.4 | 0.79 | 53.6 | 11.7 |
| ADS2778-FS2 | Chr8:128513944- 128513966 | Intergenic | rs7000448 | Prostate | 3592 | 2/2 | 2.4 | 34.6 | 2.4 | 0.89 | 34.6 | 14.8 |
| ADS3546-FS | Chr8:128569063- 128569088 | Intergenic | rs1447295 | Prostate | 3785 | 4/4 | 2.5 | 47.0 | 2.5 | 0.46 | 47.0 | 6.2 |
| 8q24 overall | Chr8:128081544- 128569088 | -- | -- | -- | -- | 44/46 | 2.5 | 74.3 | 2.5 | 0.67 | 75.2 | 14.7 |
| MYC Assay ID | ||||||||||||
| ADS2415-FS1re | Chr8:128815467-128815522 | Promoter | rs9642880 | Bladder | 28217 | 0/6 | 93.5 | 0.8 | -- | -- | -- | -- |
| ADS2415-FS2 | Chr8:128815565- 128815613 | Promoter | rs9642880 | Bladder | 28315 | 1/5 | 43.6 | 1.4 | 11.7 | 0.53 | 1.8 | 2.6 |
| ADS2415-FS3 | Chr8:128815638- 128815661 | Promoter | rs9642880 | Bladder | 28388 | 0/4 | 54.0 | 1.7 | -- | -- | -- | -- |
| ADS1967-FS2 | Chr8:128816421- 128816434 | Promoter | rs9642880 | Bladder | 29171 | 0/4 | 77.1 | 1.2 | -- | -- | -- | -- |
| ADS1967-FS3 | Chr8:128816501- 128816530 | Promoter | rs9642880 | Bladder | 29251 | 2/4 | 62.0 | 3.1 | 14.7 | 0.49 | 5.5 | 5.8 |
| ADS1966-FS | Chr8:128816695- 128816746 | Promoter | rs9642880 | Bladder | 29445 | 2/6 | 49.9 | 1.7 | 19.2 | 0.79 | 2.0 | 4.3 |
| ADS2554-FS1 | Chr8:128817270- 128817289 | Promoter | rs9642880 | Bladder | 30020 | 0/4 | 117.7 | 0.5 | -- | -- | -- | -- |
| ADS2554-FS2 | Chr8:128817329- 128817396 | Promoter | rs9642880 | Bladder | 30079 | 0/4 | 99.8 | 1.1 | -- | -- | -- | -- |
| ADS2554-FS3 | Chr8:128817416- 128817566 | Promoter & 5′UTR | rs9642880 | Bladder | 30166 | 0/15 | 126.5 | 1.2 | -- | -- | -- | -- |
| ADS3575-FS1 | Chr8:128818454- 128818537 | Intron 1 | rs9642880 | Bladder | 31204 | 0/9 | 98.6 | 0.8 | -- | -- | -- | -- |
| ADS3575-FS2 | Chr8:128818552- 128818607 | Intron 1 | rs9642880 | Bladder | 31302 | 0/11 | 115.6 | 0.7 | -- | -- | -- | -- |
| ADS3574-FS1 | Chr8:128820041- 128820053 | Exon 2 | rs9642880 | Bladder | 32791 | 0/4 | 86.2 | 0.9 | -- | -- | -- | -- |
| ADS3574-FS2 | Chr8:128820098- 128820122 | Exon 2 | rs9642880 | Bladder | 32848 | 1/5 | 37.2 | 1.2 | 22.7 | 0.79 | 1.4 | 2.0 |
| ADS3574-FS3 | Chr8:128820154- 128820197 | Exon 2 | rs9642880 | Bladder | 32904 | 0/8 | 93.0 | 1.0 | -- | -- | -- | -- |
| ADS3573-FS | Chr8:128822327- 128822382 | Exon 3 | rs9642880 | Bladder | 35077 | 5/5 | 3.2 | 73.2 | 3.2 | 0.71 | 73.8 | 14.0 |
| ADS3572-FS1 | Chr8:128823273- 128823334 | 3′ | rs9642880 | Bladder | 36023 | 3/3 | 6.2 | 36.0 | 6.2 | 0.81 | 36.2 | 26.4 |
| ADS3572-FS2 | Chr8:128823437- 128823493 | 3′ | rs9642880 | Bladder | 36187 | 2/2 | 6.6 | 42.3 | 6.6 | 0.77 | 42.4 | 33.3 |
| MYC overall | Chr8:128815467- 128823493 | -- | -- | -- | -- | 16/99 | 80.9 | 6.7 | 9.4 | 0.71 | 36.3 | 15.0 |
Abbreviations: bp, base pairs; CV, coefficient of variation; ICC, Intraclass Correlation Coefficient; SNP, single nucleotide polymorphism; UTR, Untranslated Region
Percent DNA methylation
NCBI36/hg18 coordinates encompassing the CpG sites analyzed
Closest cancer susceptibility SNP at 8q24
Negative value indicates the CpG sites are upstream of the SNP; positive value indicates the CpG sites are downstream
CpG sites with CV<25 were designated as meeting QC
Among the subset of CpG sites meeting QC
Figure 1. Landscape of DNA methylation assays, cancer susceptibility loci, non-coding RNAs, and protein-coding genes within 800kb region of 8q24.21.
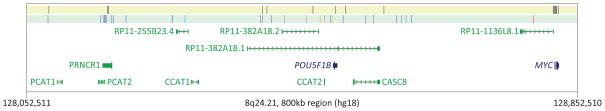
Black vertical lines within the top yellow strip depict the locations of the DNA methylation assays and the color vertical lines within the second strip depict the locations of the cancer susceptibility loci (blue, prostate cancer; sky blue, chronic lymphocytic leukemia; pink, breast cancer; orange, colorectal cancer; red, bladder cancer) with p-values<10−6 according to published genome-wide association studies. One locus (rs6983267) 3′ to RP11-382A18.2, which is both a prostate and colorectal cancer susceptibility locus, is colored half-blue, half-orange. Non-coding RNAs are colored green and protein coding genes are colored dark blue. PVT1 is located 119.5kb telomeric and PSCA 14.9Mb telomeric to the region plotted here. Other cancer susceptibility loci (Hodgkin lymphoma, breast cancer, ovarian cancer, glioma, bladder cancer) have been reported at least 290kb telomeric to the region plotted here.
When we calculated CVs using the duplicate samples in our study, we found that, in contrast to the MYC CpG sites, most of the CpG sites nearby the cancer susceptibility SNPs at 8q24 demonstrated good reproducibility (CV<25) (Table 1). Of the 99 MYC CpG sites evaluated, only 16 sites (16%) displayed a CV<25, whereas of the 46 CpG sites evaluated nearby the susceptibility SNPs, 44 sites (96%) displayed a CV<25. The poor reproducibility for most of the MYC sites appeared to result from the relatively low DNA methylation levels observed for these sites. The average mean level was 6.7% across the MYC CpG sites evaluated, in contrast to 74.3% across the CpG sites evaluated nearby the susceptibility SNPs (Table 1). Considering the 60 CpG sites with good reproducibility (Supplementary Table S2), the CpG sites nearby the susceptibility SNPs tended to demonstrate moderate to high between-individual variation in our population, with an average range by assay from 3.8 to 29.6 percentage points and an average ICC by assay from 0.41 to 0.89 (Table 1). The MYC sites tended to demonstrate lower between-individual variation, except for the exon 3 and 3′ regions evaluated, which showed an average range by assay from 14.0 to 33.3 percentage points and an average ICC from 0.71 to 0.81 (Table 1).
For the 60 CpG sites meeting our QC threshold, we also explored the extent to which DNA methylation levels at the CpG sites were correlated with each other, as well as with the known cancer susceptibility SNPs at 8q24, by calculating pairwise correlations. The strongest correlations for DNA methylation levels with each other (r2>0.36, i.e. rho>0.6) occurred for CpG site pairs located within/nearby POU5F1B or MYC (highest r2 in each of the gene regions=0.55, with p-values of 7.1×10−15 and 2.6×10−14, respectively) (Figure 2). For these highly correlated CpG site pairs, the average distance apart was 56bp, and the associations remained statistically significant after adjustment for multiple comparisons (FDR<0.05) (data not shown).
Figure 2.
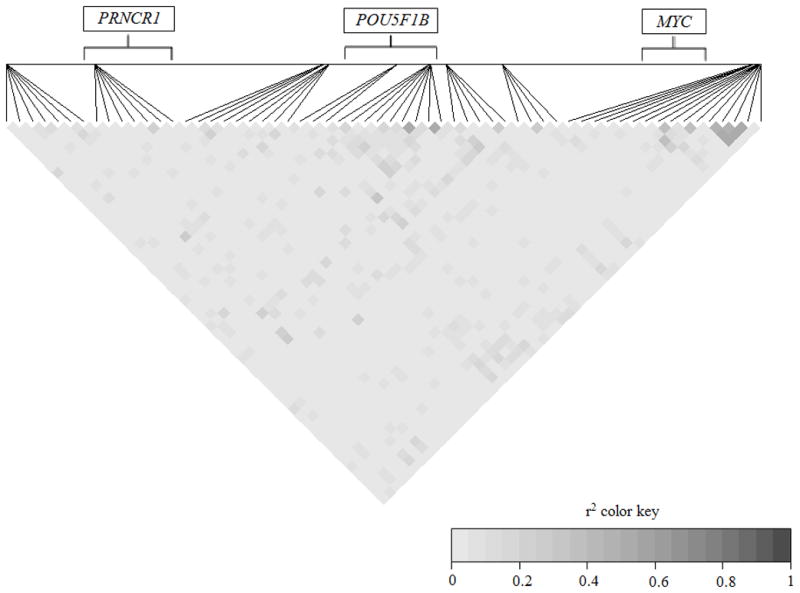
Pairwise CpG site – CpG site correlations for the 60 CpG sites with a CV<25, spanning a 741.9kb region.
We also observed significant correlations between DNA methylation levels and several cancer susceptibility SNPs at 8q24 (CpG site – SNP correlations). Of the 41 CpG site – SNP correlations with a p-value<0.01 (Table 2), 10 correlations met an FDR<0.05. Boxplots displaying the distribution of DNA methylation levels by genotype for the top CpG site – SNP correlations are shown in Supplementary Figure S1. The most significant association was observed for the intergenic CpG site at Chr8:128393157 and the prostate cancer susceptibility SNP rs16902094 (rho=−0.54; p-value=9.7×10−7; q-value=0.002) (Table 2). Other associations that met an FDR<0.05 included a CpG site at Chr8:128167809 in PRNCR1 and 2 nearby correlated (r2=0.80) prostate cancer susceptibility SNPs, rs1456315 and rs13254738 (for rs1456315, rho=0.52; p-value=1.4×10−6; q-value=0.002). The CpG site at Chr8:128167809 was also significantly correlated with the CLL susceptibility SNP rs2466032 (rho=−0.42, p-value=1.2×10−4; q-value=0.04). In addition, associations between 2 moderately correlated CpG sites at Chr8:128498051 and Chr8:128498134 in POU5F1B (r2=0.09) and several correlated prostate/colorectal cancer susceptibility SNPs (r2=0.51–1.0) met an FDR<0.05 (for Chr8:128498051 and rs6983267, rho=0.46; p-value=2.0×10−5; q-value=0.01) (Table 2). All of the correlations that met an FDR<0.05 were also significantly correlated with a p-value<0.05 in a replication set of 273 non-Hispanic Caucasian men (Table 3).
Table 2.
CpG site – SNP correlations with a p-value<0.01, grouped by CpG site and assaya
| CpG site coordinateb | Assay | Location | SNPc | Rho | p-value | q-valued | Distance SNP to CpG site (bp)e | Cancer associated with SNP |
|---|---|---|---|---|---|---|---|---|
| Chr8:128393157 | ADS2771-FS2re | Intergenic | rs16902094 | −0.54 | 9.7×10−7 | 0.002 | 3629 | Prostate |
| Chr8:128393157 | ADS2771-FS2re | Intergenic | rs13281615 | 0.40 | 4.9×10−4 | 0.10 | −31643 | Breast |
| Chr8:128393157 | ADS2771-FS2re | Intergenic | rs445114 | 0.33 | 4.6×10−3 | 0.40 | 794 | Prostate |
| Chr8:128393157 | ADS2771-FS2re | Intergenic | rs620861 | 0.33 | 4.6×10−3 | 0.40 | −11698 | Prostate |
| Chr8:128393162 | ADS2771-FS2re | Intergenic | rs16902094 | −0.42 | 2.5×10−4 | 0.06 | 3634 | Prostate |
| Chr8:128393166 | ADS2771-FS2re | Intergenic | rs16902094 | −0.42 | 3.0×10−4 | 0.06 | 3638 | Prostate |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs1456315 | 0.52 | 1.4×10−6 | 0.002 | −5310 | Prostate |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs2466032 | −0.42 | 1.2×10−4 | 0.04 | −111193 | CLL |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs13254738 | 0.41 | 1.7×10−4 | 0.04 | −5716 | Prostate |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs13252298 | −0.33 | 2.6×10−3 | 0.31 | 3471 | Prostate |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs7837688 | −0.32 | 3.8×10−3 | 0.38 | −440733 | Prostate |
| Chr8:128167777 | ADS2774-FS1 | PRNCR1 | rs6470494 | 0.29 | 8.4×10−3 | 0.57 | 10691 | Prostate |
| Chr8:128167834 | ADS2774-FS2 | PRNCR1 | rs10505477 | 0.30 | 7.6×10−3 | 0.56 | −308791 | Colorectal |
| Chr8:128167834 | ADS2774-FS2 | PRNCR1 | rs6983267 | 0.30 | 7.6×10−3 | 0.56 | −314653 | Prostate and colorectal |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs6983267 | 0.46 | 2.0×10−5 | 0.01 | 15564 | Prostate and colorectal |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs10505477 | 0.46 | 2.0×10−5 | 0.01 | 21426 | Colorectal |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs7014346 | −0.46 | 2.0×10−5 | 0.01 | 4077 | Colorectal |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs7837328 | −0.44 | 6.0×10−5 | 0.02 | 5742 | Prostate |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs1447293 | −0.29 | 9.1×10−3 | 0.59 | −43451 | Prostate |
| Chr8:128498079 | ADS3549-FS2 | POU5F1B | rs10505477 | 0.34 | 2.3×10−3 | 0.29 | 21454 | Colorectal |
| Chr8:128498079 | ADS3549-FS2 | POU5F1B | rs6983267 | 0.34 | 2.3×10−3 | 0.29 | 15592 | Prostate and colorectal |
| Chr8:128498079 | ADS3549-FS2 | POU5F1B | rs7837328 | −0.30 | 8.1×10−3 | 0.57 | 5770 | Prostate |
| Chr8:128498113 | ADS3549-FS2 | POU5F1B | rs10505477 | 0.33 | 3.1×10−3 | 0.32 | 21488 | Colorectal |
| Chr8:128498113 | ADS3549-FS2 | POU5F1B | rs6983267 | 0.33 | 3.1×10−3 | 0.32 | 15626 | Prostate and colorectal |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs7837328 | −0.44 | 4.8×10−5 | 0.02 | 5825 | Prostate |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs7014346 | −0.42 | 1.3×10−4 | 0.04 | 4160 | Colorectal |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs10505477 | 0.35 | 1.6×10−3 | 0.23 | 21509 | Colorectal |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs6983267 | 0.35 | 1.6×10−3 | 0.23 | 15647 | Prostate and colorectal |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs2294008 | −0.33 | 3.1×10−3 | 0.32 | −15260799 | Bladder |
| Chr8:128498149 | ADS3549-FS3 | POU5F1B | rs7014346 | −0.36 | 1.1×10−3 | 0.19 | 4175 | Colorectal |
| Chr8:128498149 | ADS3549-FS3 | POU5F1B | rs7837328 | −0.36 | 1.1×10−3 | 0.19 | 5840 | Prostate |
| Chr8:128081544 | ADS2775-FS1 | Intergenic | rs6470494 | −0.30 | 7.0×10−3 | 0.55 | −75542 | Prostate |
| Chr8:128081544 | ADS2775-FS1 | Intergenic | rs13252298 | 0.30 | 8.4×10−3 | 0.57 | −82794 | Prostate |
| Chr8:128081544 | ADS2775-FS1 | Intergenic | rs2608053 | −0.29 | 9.6×10−3 | 0.60 | −1063470 | HL |
| Chr8:128081544 | ADS2775-FS1 | Intergenic | rs1456315 | −0.29 | 9.8×10−3 | 0.60 | −91575 | Prostate |
| Chr8:128081593 | ADS2775-FS2 | Intergenic | rs10088218 | −0.32 | 4.3×10−3 | 0.40 | −1531538 | Ovarian |
| Chr8:128081600 | ADS2775-FS2 | Intergenic | rs9642880 | −0.29 | 9.1×10−3 | 0.59 | −705650 | Bladder |
| Chr8:128569077 | ADS3546-FS | Intergenic | rs16902094 | −0.32 | 4.3×10−3 | 0.40 | 179549 | Prostate |
| Chr8:128569088 | ADS3546-FS | Intergenic | rs7017300 | −0.31 | 6.0×10−3 | 0.49 | −25362 | Prostate |
| Chr8:128816702 | ADS1966-FS | MYC | rs7837328 | 0.35 | 1.5×10−3 | 0.23 | 324393 | Prostate |
| Chr8:128816702 | ADS1966-FS | MYC | rs7014346 | 0.31 | 5.1×10−3 | 0.43 | 322728 | Colorectal |
Abbreviations: bp, base pairs; CLL, chronic lymphocytic leukemia; HL, Hodgkin’s lymphoma; SNP, single nucleotide polymorphism
Bolding denotes False Discovery Rate<0.05
NCBI36/hg18 coordinate
Cancer susceptibility SNP at 8q24
False Discovery Rate after adjustment for multiple comparisons using the Benjamini Hochberg method
Negative value indicates the CpG site is upstream of the associated SNP; positive value indicates the CpG site is downstream
Table 3.
Replication study results for the 10 CpG site – SNP correlations meeting an FDR<0.05
| CpG site coordinatea | Assay | Location | SNPb | Rho | p-value |
|---|---|---|---|---|---|
| Chr8:128393157 | ADS2771-FS2re | Intergenic | rs16902094 | −0.46 | 8.5×10−16 |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs1456315 | 0.34 | 6.4×10−9 |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs2466032 | −0.15 | 0.02 |
| Chr8:128167809 | ADS2774-FS2 | PRNCR1 | rs13254738 | 0.29 | 1.5×10−6 |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs6983267 | 0.36 | 1.8×10−9 |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs10505477 | 0.38 | 2.0×10−10 |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs7014346 | −0.41 | 2.0×10−12 |
| Chr8:128498051 | ADS3549-FS1 | POU5F1B | rs7837328 | −0.39 | 3.9×10−11 |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs7837328 | −0.21 | 4.5×10−4 |
| Chr8:128498134 | ADS3549-FS3 | POU5F1B | rs7014346 | −0.25 | 3.6×10−5 |
Abbreviations: FDR, False Discovery Rate; SNP, single nucleotide polymorphism
NCBI36/hg18 coordinate
Cancer susceptibility SNP at 8q24
We followed up on the 4 CpG sites that were significantly associated with 8q24 cancer susceptibility SNPs after adjustment for multiple comparisons in our study by searching for TFBS, DNase hypersensitivity sites, and histone methylation marks within 400bp of the CpG sites in histologically normal tissues from prostate or colon cancer patients and cancer cell lines of prostate or colon origin in ENCODE. Although these 4 regions were relatively repetitive, resulting in poor alignability (average alignability for each region: 46%, 3%, 36%, and 37%, respectively), which likely reduced the ENCODE data that were available (typically ~100% alignability is required for confident peak calls), we identified several marks with a p-value<0.05 nearby the CpG sites of interest. In the colorectal cancer cell line Caco-2 (colorectal adenocarcinoma), we identified a DNase I hypersensitivity site about 340bp away (p-value=1.6×10−3; q-value=2.3×10−3) from the POU5F1B CpG site at Chr8:128498051. In addition, in the colorectal cancer cell line HCT-116 (colorectal carcinoma), we identified a TFBS for CTCF approximately 254bp or 171bp from the 2 POU5F1B CpG sites at Chr8:128498051 and Chr8:128498134, respectively (p-value=0.04; q-value=0.04). We did not observe significant peaks (p<0.05) for any of the marks evaluated in the available histologically normal tissue from prostate or colon cancer patients in ENCODE.
When we evaluated temporal variation in DNA methylation levels for the 60 CpG sites meeting QC criteria, we identified 3 CpG sites (Chr8:128498097, Chr8:128393166, and Chr8:128465853) that demonstrated significant changes in DNA methylation levels during the approximately 5-year time period examined (p-values=0.01, 0.02, and 0.04, respectively). However, none of these associations remained significant after adjustment for multiple comparisons with an FDR<0.05 (data not shown).
Further evaluating DNA methylation levels at the 60 CpG sites with established cancer risk factors, we observed a number of significant associations with age, BMI, smoking status, and pack-years of smoking, although none remained significant after adjustment for multiple comparisons with an FDR<0.05 (data not shown). Notably, the intergenic CpG site at Chr8:128393157, which was significantly associated with a prostate cancer susceptibility SNP at 8q24 in our study, was associated with BMI (p=0.003). In addition, the CpG site at Chr8:128167809 in PRNCR1, which also demonstrated significant associations with 8q24 cancer susceptibility SNPs in our study, was associated with smoking status (p=0.04) and pack-years of smoking (p=0.05).
DISCUSSION
Our study is the first to investigate patterns of DNA methylation in peripheral blood at chromosome 8q24 and the relationship with cancer susceptibility SNPs at 8q24. In this population of non-Hispanic Caucasian males, the CpG sites located nearby the cancer susceptibility SNPs at 8q24 tended to show high levels of DNA methylation, moderate to high between-individual variation, and good reproducibility. In contrast, the CpG sites in or near MYC tended to display lower levels of DNA methylation, lower between-individual variation, and poor reproducibility, although the CpG sites in the MYC exon 3 and 3′ regions performed well and also demonstrated moderate to high between-individual variation. DNA methylation levels at the CpG sites evaluated tended to be stable over time in the relatively short 5-year period examined in our study. There was some evidence of associations between a number of CpG sites and established risk factors for cancer (age, smoking, and BMI); however, these associations did not remain significant after adjustment for multiple comparisons. Given the relatively small sample size in our study, these associations may warrant reevaluation in larger populations. We identified correlations for DNA methylation levels with each other within/nearby POU5F1B and MYC, as well as correlations between DNA methylation levels and cancer susceptibility SNPs at 8q24. Of the CpG site – SNP correlations, 10 remained significant after adjustment for multiple comparisons, encompassing a CpG site at Chr8:128393157 in an intergenic region, a CpG site located at Chr8:128167809 in PRNCR1, and two CpG sites located in POU5F1B at Chr8:128498051 and Chr8:128498134. All of the top 10 CpG site – SNP correlations were also significant in an independent population of non-Hispanic Caucasian males.
Our finding of correlations for DNA methylation levels at CpG sites with each other within or nearby POU5F1B and MYC, while potentially due to chance, suggests a coordination of DNA methylation levels within these gene regions. Although the origin of these patterns is unclear (e.g. role of inheritance/genetic factors vs. environment), our findings are consistent with a previous study that observed correlations for DNA methylation levels at CpG sites located near one another, particularly those within distances up to 1–2kb apart (41).
Our finding of correlations between DNA methylation levels at specific CpG sites and cancer susceptibility SNPs at 8q24 is also intriguing. Although it is possible that these findings may be due to chance, we adjusted for multiple comparisons in our study and 10 of the findings remained statistically significant. Moreover, these correlations replicated in an independent population of non-Hispanic Caucasian men. Our results may reflect a shared inheritance or perhaps a differential propensity conferred by the SNPs to epigenetic alteration (42). A number of studies have identified loci containing genetic variants that are associated with DNA methylation levels, termed methylation quantitative trait loci (meQTLs) (37, 38, 42–44). Similar to what has been observed for other regions, our strongest CpG site – SNP correlations were generally observed between CpG sites and SNPs located in relatively close proximity, with an average distance for our significant findings of 18,264bp. No recombination hotspots were observed between these highly correlated CpG sites and SNPs. Some of the meQTLs in the prior studies were also associated with gene expression (i.e. expression quantitative trait loci, eQTLs), and associations were observed between DNA methylation levels and gene expression as well (37, 38). Although we did not have gene expression data to explore this question further, it is possible that the SNP-associated variation in DNA methylation observed in our study could in turn affect expression of genes or ncRNAs in the region and thereby contribute to cancer risk.
The most significant association between the CpG sites and cancer susceptibility SNPs evaluated in our study was observed for the intergenic CpG site at Chr8:128393157 and rs16902094, which has been associated with prostate cancer risk in GWAS (12). Based on the Repeat masker track of the UCSC Genome Browser (http://genome.ucsc.edu/), this CpG site is located in a L1HS long interspersed nuclear element 1 (LINE-1) repetitive element. As we observed reduced DNA methylation associated with an increasing number of risk alleles (which was the variant allele) for rs16902094, it is possible that altered DNA methylation in this region could in turn affect genomic stability and thereby contribute to cancer risk. We also observed significant associations between a CpG site at Chr8:128167809 in PRNCR1 and 2 prostate cancer susceptibility SNPs, rs1456315 and rs13254738, which were highly correlated with each other and therefore not independent of one another. Rs1456315, also located in PRNCR1, and rs13254738, located nearby, have been associated with prostate cancer risk in several populations of different ancestry, although the associations were stronger among populations of African or Asian as opposed to European ancestry (13, 45, 46). PRNCR1 has been shown to be upregulated in prostate cancer cells and prostate intraepithelial neoplasia and may contribute to prostate carcinogenesis by affecting androgen receptor activity (24). The CpG site at Chr8:128167809 was also significantly associated with the CLL susceptibility SNP rs2466032, suggesting that DNA methylation in this important cancer susceptibility region may play a role in other cancers as well as prostate.
We also observed significant associations between 2 CpG sites at Chr8:128498051 and Chr8:128498134 in POU5F1B and several SNPs that have been associated with prostate and/or colorectal cancer (8, 15, 47, 48). These 2 CpG sites were moderately correlated with each other (r2=0.09) and the SNPs were moderately to highly correlated with each other (rho=0.51–1.0), suggesting that these findings are not independent of one another. Although rs7837328 was not associated with prostate cancer risk at the strict genome-wide significance level in GWAS, it was associated with prostate cancer risk in the Agricultural Health Study, and the association was shown to be increased among participants with increasing exposure to specific pesticides (15). Our finding of an association between this SNP and DNA methylation levels at 8q24 suggests a potential role of DNA methylation in these findings. Notably, we also observed associations between DNA methylation levels in POU5F1B and rs6983267, which has been associated with prostate and colorectal cancers in GWAS (14, 48) and has also previously demonstrated long-range interactions with MYC in colon cancer cells (49), lending some plausibility for an epigenetic mechanism contributing to risk for SNPs in this region. We also identified ENCODE marks nearby both POU5F1B CpG sites, which included a DNAse I hypersensitivity site and a TFBS for CTCF, in colon cancer cell lines.
Our study was limited by the fact that we did not sequence the entire 8q24 region and therefore we were not able capture all CpG sites. In addition, we did not evaluate CpG sites in some regions of interest because the sequences were highly repetitive, which we expected would result in poor assay reproducibility. As a further limitation, we did not have gene expression data, which would have allowed study of whether variation in DNA methylation at these sites was associated with gene expression. We also acknowledge that the relationships we observed between DNA methylation levels in peripheral blood and cancer susceptibility SNPs at 8q24 may differ in different tissues; however, we did not have available data to address this question in our study.
Our study also has several strengths. We quantified DNA methylation levels using pyrosequencing, which is considered one of the most sensitive approaches available to detect differences in DNA methylation between individuals. In addition, by conducting our study within a population-based study, we were able to quantify normal variation in DNA methylation levels at 8q24. Moreover, the availability of serial blood samples and information on a variety of environmental exposures allowed for study of temporal variation and the relationship between DNA methylation and important cancer risk factors. A further strength was our ability to evaluate both genetic and epigenetic data together as this allowed for more comprehensive study of the underlying genetic structure in this complex region, providing some clues about the biological mechanism that might underlie the associations for 8q24 SNPs with the risk of various cancers. We were also able to demonstrate that our most significant CpG site – SNP correlations replicated in an independent population.
In conclusion, among non-Hispanic Caucasian men in PLCO, we identified a number of specific CpG sites at 8q24 that demonstrated good reproducibility, temporal stability, and moderate to high between-individual variation and thus are well-suited for evaluation in relation to cancer risk in future studies. These sites are largely located nearby cancer susceptibility SNPs at 8q24 as opposed to in or near MYC. In addition, our study is the first to report correlations between DNA methylation levels at specific CpG sites in peripheral blood and cancer susceptibility SNPs at 8q24, suggesting that there may also be a role of DNA methylation at this important cancer susceptibility locus in cancer risk.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute and the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention at the National Cancer Institute, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr. Thomas Riley, Mr. Craig Williams, Mr. Michael Furr, and Dr. William Wheeler at Information Management Services, Inc., and Ms. Barbara O’Brien and staff at Westat, Inc. for their contributions to the PLCO Cancer Screening Trial. We also thank the PLCO study participants for their contributions to making this study possible.
Footnotes
Potential Conflicts of Interest: L. Yan and A. Meyer are employed by EpigenDx, Inc., and L. Yan is the major stockholder of EpigenDx. The other authors do not have any potential conflicts of interest to disclose.
References
- 1.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24. 1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–6. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 3.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24. 21 and 10p14 (GATA3) Nat Genet. 2010;42:1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. 361e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multistage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 9.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 10.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 15.Koutros S, Beane Freeman LE, Berndt SI, Andreotti G, Lubin JH, Sandler DP, et al. Pesticide use modifies the association between genetic variants on chromosome 8q24 and prostate cancer. Cancer Res. 2010;70:9224–33. doi: 10.1158/0008-5472.CAN-10-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107:9742–6. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–5. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–74. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panagopoulos I, Moller E, Collin A, Mertens F. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol Rep. 2008;20:1029–33. [PubMed] [Google Scholar]
- 21.Kastler S, Honold L, Luedeke M, Kuefer R, Moller P, Hoegel J, et al. POU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinoma. Prostate. 2010;70:666–74. doi: 10.1002/pros.21100. [DOI] [PubMed] [Google Scholar]
- 22.Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012;109:4974–9. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohaar I, Porter-Gill P, Lenz P, Fu YP, Mumy A, Tang W, et al. Genetic variant as a selection marker for anti-prostate stem cell antigen immunotherapy of bladder cancer. J Natl Cancer Inst. 2013;105:69–73. doi: 10.1093/jnci/djs458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–52. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 25.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer KB, Maia AT, O’Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7:e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 29.Brennan K, Garcia-Closas M, Orr N, Fletcher O, Jones M, Ashworth A, et al. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res. 2012;72:2304–13. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 30.Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, et al. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev. 2012;21:1293–302. doi: 10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, et al. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study. Epigenetics. 2012;7:291–9. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. J Clin Oncol. 2011;29:1133–9. doi: 10.1200/JCO.2010.31.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almen MS, Jacobsson JA, Moschonis G, Benedict C, Chrousos GP, Fredriksson R, et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–7. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes RB, Sigurdson A, Moore L, Peters U, Huang WY, Pinsky P, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005;592:147–54. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 40.Chen J, Zhang JG, Li J, Pei YF, Deng HW. On combining reference data to improve imputation accuracy. PLoS One. 2013;8:e55600. doi: 10.1371/journal.pone.0055600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore LE, Nickerson ML, Brennan P, Toro JR, Jaeger E, Rinsky J, et al. Von Hippel-Lindau (VHL) inactivation in sporadic clear cell renal cancer: associations with germline VHL polymorphisms and etiologic risk factors. PLoS Genet. 2011;7:e1002312. doi: 10.1371/journal.pgen.1002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res. 2009;2:862–7. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- 45.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13. 4. Nat Genet. 2012;44:1231–5. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24. 21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 49.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–20. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


