Abstract
Cancer is driven by the acquisition of somatic DNA lesions. Distinguishing the early driver mutations from subsequent passenger mutations is key to molecular sub-typing of cancers, understanding cancer progression, and the discovery of novel biomarkers. The advances of genomics technologies (whole-genome exome, and transcript sequencing, collectively referred to as NGS(Next Gengeration Sequencing)) have fueled recent studies on somatic mutation discovery. However, the vision is challenged by the complexity, redundancy, and errors in genomic data, and the difficulty of investigating the proteome translated portion of aberrant genes using only genomic approaches. Combination of proteomic and genomic technologies are increasingly being employed.
Various strategies have been employed to allow the usage of large scale NGS data for conventional MS/MS searches. This paper provides a discussion of applying different strategies relating to large database search, and FDR(False Discovery Rate) based error control, and their implication to cancer proteogenomics. Moreover, it extends and develops the idea of a unified genomic variant database that can be searched by any mass spectrometry sample. A total of 879 BAM files downloaded from TCGA repository were used to create a 4.34 GB unified FASTA database which contained 2, 787, 062 novel splice junctions, 38, 464 deletions, 1, 105 insertions, and 182, 302 substitutions. Proteomic data from a single ovarian carcinoma sample (439, 858 spectra) was searched against the database. By applying the most conservative FDR measure, we have identified 524 novel peptides and 65, 578 known peptides at 1% FDR threshold. The novel peptides include interesting examples of doubly mutated peptides, frame-shifts, and non-sample-recruited mutations, which emphasize the strength of our approach.
Introduction
Cancer is driven by the acquisition of somatic DNA lesions. Understanding of the progression of the lesions, distinguishing the early driver mutations from subsequent passenger mutations, deciphering the role of somatic mutations in regulating protein expression are all under active investigation. The availability of genomics technologies (mainly whole-genome and exome sequencing, and transcript sampling via RNA-seq, collectively referred to as NGS) have fueled recent studies on these topics1,2. It is very likely that the some of the discovered mutations will aid in molecular sub-typing of cancers, and act as diagnostic and prognostic bio-markers.
A challenge to this vision comes from the complexity, redundancy, and errors in genomic data, and the difficulty of investigating the proteome translated portion of aberrant genes using only genomic approaches. In comparative studies, while protein and RNA expression matched for the most abundant molecules, the correlation for lower abundance molecules was much worse (~ 0.4).3 Others found that as many as 20% of transcripts do not have a matching protein identification, often due to a different frame of translation.4 The high variability between protein and genomic expression in these studies suggests that a combination of proteomic and genomic technologies are the best bet for identifying coding variants and their use as biological markers of cancer, and such searches are increasingly employed5-7. Moreover, one cannot rely on comparison of RNA and protein data from the same sample.
The problem of searching all protein samples and all RNA samples becomes a significant challenge for proteogenomics, especially for bottom up mass spectrometric protocols, where a short peptide spectrum is matched against theoretical databases of spectra derived from genomic sequences. The chance of a false identification grows with increasing database sizes. A typical RNA-seq alignment file is around 10 GB, and is different for each sample. The TCGA resources8 alone lists around 5Tb of RNA-seq data for Ovarian Carcinoma. In order to utilize large-scale NGS data in proteomics search, efficient methods for managing the large data-size are essential. This paper provides an efficient method to search the large search space of NSG data and discussion of applying more accurate FDR based error control strategies, and their implication to cancer proteogenomics.
Large Database Search
As our goal is to discover aberrant peptides in cancer, including fusion genes, splicing variants, and possibly even novel expressed genes, we cannot rely on the human proteome, hence large databases. First, a six-frame translation of the human genome is already ~ 6Gb, but that pales in comparison to the available transcript data that encodes many of the variants. For example, the TCGA resources8 lists around 5Tb of RNA-seq data for a single tumor type (Ovarian Carcinoma).
Some approaches have been suggested to handle the big-data overload. These include, sample specific search to reduce the search space by generating a curated individual database for RNA-seq obtained from each sample5-7, and direct translation of the outputs from available genomics assembly tools.9-13 Alternatively, our method favors a graph structured accumulative approach14 that combines multiple sample NGS data into a unified database. A graph based approach enables us to efficiently encode cumulative/large information from multiple RNA-seq datasets into a compact unified database. Moreover, unified database approach also enables us to maintain a single FDR threshold throughout the entire analysis. Finally, our approach also enables peptide identifications with combinatorial multiple splice junctions or variants. Based on our proposed method, we released a JAVA and python based tool called SpliceDB (https://bixlab.ucsd.edu/display/CCMSwebsite) which generates FASTA formatted splice graph database from multiple RNA-seq alignments.
False discovery rate based error control strategies
One of the challenges with proteogenomic studies is the aggressive and variable choices of False Discovery Rates (FDR) strategies, all designed to maximize the discovery of aberrant peptides. In most conventional proteomic studies, a global peptide level FDR with 1% FDR cut-off is used. However it has been shown from other studies that FDR threshold can be biased in a larger database search space such as PTM and SNP5 tolerant searches. In this study, in order to discuss the effect of applying different FDR strategies, we performed a benchmark study under identical condition applying three different FDR based peptide error control strategies which are Combined FDR (Supplemental Figure S.1(a)), Separate FDR (Supplemental Figure S.1(b)), and Two-stage FDR(Supplemental Figure S.1(c)). While more sophisticated FDR approaches can be further applied in combination of our FDR strategies, here we calculated standard global FDR threshold in order to mainly focus on the effect of separate, multi-stage calculation strategies. As shown in our result, in applying conservative FDR error control, our results are robust to the choice of FDR.
Calling peptides versus events
An important part of proteogenomics search for discovering aberrant events is that we are looking for events (alternative splicing, gene-fusions, etc.), not peptides. The SpliceDB tool described here can be used in stand-alone fashion just for FASTA database creation, but also can be paired with our integrative proteogenomics pipeline Enosi.15-18 To focus on the effect of choosing different strategies in ‘peptide identifications results’, we will not describe cancer specific event calling here, but will present some results describing events we could identify in a proteogenomic search of a single primary ovarian carcinoma sample.
To summarize, the manuscript makes the following contributions. First, we extend the SpliceDB database construction to scale to human cancer data-sets, and include all different types of variation. We build and present a generic ovarian cancer database that can be searched with any proteomic data-sets. We utilized a total of 879 BAM files downloaded from TCGA1,2 repository and created total 4.34 GB (103× compression) of unified FASTA database which contained 2, 787, 062 novel splice junctions, 38, 464 deletions, 1105 insertions, and 182, 302 substitutions.
Next, we systematically test the impact of applying different strategies regarding to database construction and FDR based error control on the identification of aberrant peptides in cancer. Total 439, 858 spectra collected from a single ovarian cancer sample were searched against the both the created FASTA database as well as a sample specific database. By applying most conservative FDR measure, we could identified 524 novel peptides and 65, 578 known peptides at 1% FDR threshold. Moreover, selected detailed examples of doubly mutated peptide and different-sample-recruited mutation identifications were shown to emphasize the strength of our method, and the large number of identifications from a single sample underscore the value of proteogenomic searches in identifying aberrant peptides in cancer.
Method
Following our previous study,14 we extended the splice graph database construction method to encode a more extended list of genomic variants. Splice graph14 is a data structure which represents exons as nodes, and splice junctions as edges. The graph is constructed from the junction information extracted from the RNA-seq alignments and all types of mutations reported from VCF (Variant Call Format) files. For variant calling from RNA-seq alignments (in BAM format), we used GATK12,13 tool with parameters ‘—stand_call_conf 30.0 —stand_emit_conf 10.0’. Detailed descriptions on initial RNA-seq information handling, graph algorithms, and FASTA conversion algorithms, are described in our previous study.14 The graph construction is done in an accumulative fashion, and the last FASTA conversion step must be performed each time when additional information is incorporated into the graph. Our graph construction approach also conserves the property of compactness14,19 and completeness14,19 of the original search space (state of proof shown in our previous study14). In this study, we introduce a concept of variant graph which enables additional nodes and edges representing arbitrary length deletions, insertions, and substitutions.
Database creation from RNA-seq data
RNA-seq data is downloaded from TCGA data repository8 in BAM formatted files. Total size of the downloaded BAM files are shown in Figure 1. Our first step in database creation is to extract useful information from RNA-seq alignment/coordinate files (BAM/SAM,20 GFF, BED) and variant calls (VCF). Details of BAM file processing can be found in our previous study.14
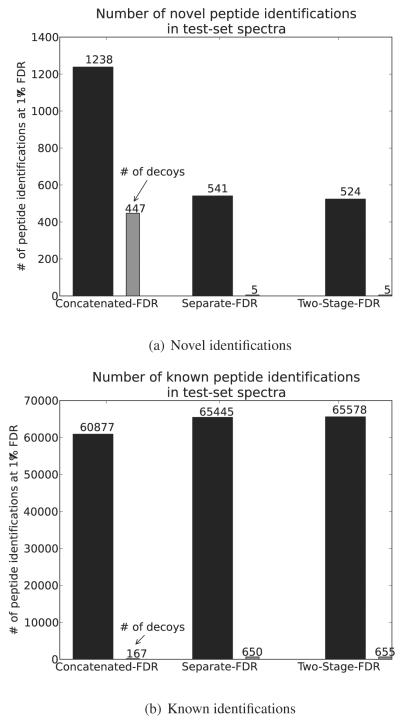
Figure 1.
Number of peptide identifications in 439,858 spectra collected from a single sample (sample id: TCGA-24-1467) using different FDR based error control strategies.
Storing genomic information for post-processing usage
RNA-seq level information (sample ID, read counts, junctions, variants, and so on) is not only used in database creation but also heavily utilized in further proteogenomics analysis. In order to efficiently maintain and retrieve various types of RNA-seq level information, we implemented a multiple depth hash table structure which enables fast access to the source information.
As described in Supplemental Figure S.2, SpliceDB21 extracts information from input files (BAM/SAM, GFF, BED, VCF) and generates a hash table using three key-data pairs. Three key values used in this hashing stage are, (1) category of a variant call (splice, deletion, insertion, and substitution), (2) chromosome name, and (3) beginning coordinate of a junction/variant. For example, in case when VCF file calls an ‘AT’ insertion in chromosome 1 at 30000th base pair, an entry containing RNA-seq level information (sample ID, read counts, junctions, variants, and so on.) is created using three key pairs of (insertion, chr1, 30000). Information maintained in the hash table is written to an intermediate file (.spl) for future usages such as cumulative database concatenation and validation of proteogenomic discoveries.18
Variant graph construction
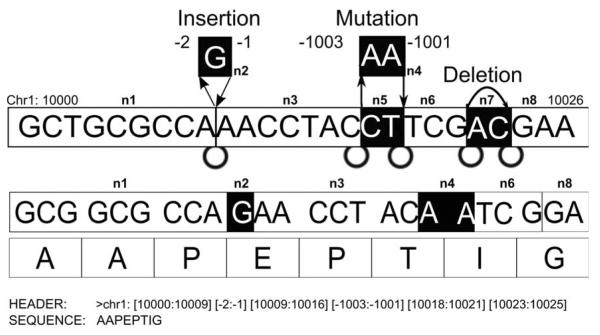
Our next step in database creation is to construct a graph data structure using information collected from the previous stage. The underlying method in graph construction and FASTA conversion for variant peptide database is shared from our previous study.14 In this study, we extended our method to a population based study where individual genomes differ from the standard genomic reference due to the presence of mutations. These mutations may be a germ-line, somatic mutations, or even polymorphic, i.e. Somatic mutations can be distinguished from germ-line mutations by comparison with DNA from the same individual. Since protein level data used in this study does not include non-tumor samples, we treated both types of mutations equally during the MS/MS search but only differentiate it in the post-processing stage while retrieving the originated genomic level information. In order to encode all types of variants into the graph structure, we added additional types of nodes and edges. While deletions can be expressed similar to splice junctions within the graph, insertions and substitutions cannot be incorporated using the same concept. Since insertions and substitutions cannot share the coordinate system of the reference DNA, we introduced insertion and substitution nodes having artificial coordinates which can be inserted to the existing graph. As described in Figure 6, negative numbers in different ranges are used to distinguish between inserted and substituted nodes.
Figure 6.
Insertions and substitutions are represented as additional node and edges having negative coordinate values. Deletions are represented same as splice junctions with actual DNA coordinates.
The variant graph can be written in FASTA format by applying the conversion strategy introduced in our previous study.14 In the FASTA conversion stage, the coordinate information of each entry is written in the FASTA header in order to reconstruct the original genomic coordinates of identified peptides.
Restoring genomic information
In proteogenomic analysis, genomic information of identified peptides such as original coordinates, and RNA-seq meta-data must be restored after the MS/MS search. We restore this information by using FASTA file headers and intermediate (.spl) files created during the database creation process. First, original coordinates of identified peptide sequence are calculated according to the corresponding FASTA header entry.
After having the peptide identifications, we can reconstruct the original coordinate of each peptide. For example, if we have a variant graph shown in the Figure 6, the graph can traverse the path ‘n1-n2-n3-n4-n6-n8’, and its corresponding nucleotide and amino acid sequence will be ‘GCTGCGCCAGAACCTACAATCGGA’, and ‘AAPEPTIG’. Next let’s assume that we have an identified peptide ‘PEPTI’, then we can find the coordinate of the peptide from its FASTA header. In this example, ‘PEPTI’ begins at the third amino acid of ‘AAPEPTIG’, so the beginning coordinate of the peptide will be ‘10006’ and the ending coordinate will be located after traversing 15 nucleotide starting from ‘10006’. In this case, ‘chr1: [10006:10009] [−2:−1] [10009:10016] [−1003:−1001] [10018:10020]’ is the actual coordinate of ‘PEPTI’. Moreover, these restored genomic coordinates are next used in retrieving the hashed RNA-seq level data. In the above example, a set of hash keys indicating the first insertion and the second substitution will be generated as (insertion, chr1, 10009) and (substitution, chr1, 10016).
Database Search Details
MS/MS data used in this study was generated from PNNL (Pacific Northwest National Laboratory) as part of the CPTAC22 (Clinical Proteomic Tumor Analysis Consortium) project. From the total MS/MS data generated by our collaborators, in this study we used a single sample. Additionally, iTRAQ quantification information is not utilized in this study since the goal of this study is focused on aberrant peptide identification. The 439, 858 spectra acquired from a single ovarian cancer sample (sample id: TCGA-24-1467, see Methods) were used in this study, and searched against all proteogenomic databases (Table 1). We used MSGF+23 for MS/MS database search with following parameters: parent mass tolerance 20ppm, semi-tryptic, Fixed Carbamidomethyl C, optional Oxidized methionine, and fixed iTRAQ related modifications. Known protein database was downloaded from Ensembl24,25(version GRCh37.70) which contained 104, 785 sequences. We attempt to use this comparably richer set of known protein database in order to be more conservative in our novel sequence calling. By categorizing any previously known genomic variants included in the Ensembl known protein database as ‘known peptide sequences’, we tempt to focus more on identifying possible ‘cancer related’ mutations. The reversed decoy database of the same size was created for each database and also searched for all databases to apply the target-decoy approach. Using 100 CPU nodes of the CCMS cluster server in parallel, the total search took 28.63 wall clock hours. For each spectrum, we selected PSMs with the lowest SpecProb reported by MSGFDB across all database search results (known proteins, 6-frame, and proteogenomics FASTA).
Table 1.
Statistics of created cancer databases
| Single OV sample | OV(PNNL samples) | OV(splice only) | BRCA(splice only) | |
|---|---|---|---|---|
|
| ||||
| # samples | 1 | 67 | 228 | 484 |
| BAM size | 7.2 GB | 750 GB | 2.0 TB | 3.2 TB |
| FASTA size | 187 MB | 607 MB | 395 MB | 814 MB |
| Novel splice | 168,289 | 321,587 | 498,233 | 646,629 |
| Deletions | 3150 | 38,464 | - | - |
| Insertions | 62 | 1,105 | - | - |
| substitutions | 7109 | 182,302 | - | - |
FDR based error control strategies
In this study, we applied three different FDR based error control strategies approaches for testing their effect on novel peptide identifications. In order to design accurate benchmark comparisons and to highlight the effect of combined, separation, and multi-stage FDR strategies, we calculated the global level FDR without applying further more sophisticated FDR calculations. (for example, further sub-classifying PSMs into different charge states, or utilizing peptide length and modification rates)
Indeed, the challenges with current proteogenomic studies is the aggressive and variable choice of False Discovery Rates (FDR) based error control strategies, all designed to maximize the discovery of aberrant peptides. In most conventional proteomic studies, a global peptide level FDR calculation with 1% FDR cut-off is used. However it has been shown from other studies that FDR based error control can be biased in a larger database search space such as PTM and SNP5 tolerant searches. Here, we discuss based on a single conservative choice of FDR based error control strategies, how different ways to execute a search (Supplemental Figure S.1) may lead to different results.
Combined FDR (Supplemental Figure S.1(a))
(1) Merge MS/MS search results from every target and decoy database according to the best scored PSM per each spectra. (2) Calculate FDR using the combined PSM result.
This FDR is identical to the conventional peptide level FDR based error control used in most proteomics studies. The term ‘combined’ refers to combining reference protein databases and translated RNA databases into a single database, to be used for proteogenomic searches.
Separate FDR (Supplemental Figure S.1(b))
(1) Merge MS/MS search results from every target and decoy database according to the best scored PSM per each spectra. (2) Iterate all merged PSM and check the origin of the matched database entry. (3) If a PSM is matched to the known protein and it’s decoy database, put the corresponding PSM to the known sequence PSM set. (4) If a PSM is matched to any proteogenomic database and their decoy, put this into the novel sequence PSM set. (5) Calculate separate FDR in each known and novel PSM set.
Following the FDR approach suggested in the study of Jing et al. (2011),5 the search uses a combined database to score and rank peptides, but separates known and novel PSMs prior to FDR calculation. This results in conservative novel peptide identifications.5 The separation step is done strictly by iterating over every PSM to extract peptides that have string matches within the known and known-decoy database. Note that this procedure is different from simply launching the MS/MS search using only the proteogenomics database (excluding the known protein database) and calculating conventional FDR. Since most proteogenomics databases partially overlap with the known protein database, the proteogenomic database search might contain known peptide hits.
Two-stage FDR (Supplemental Figure S.1(c))
1. Search only known protein database and it’s decoy. 2. Calculate FDR in the known database only PSM result. 3. Search all proteogenomic databases and calculate FDR using only the spectra that are not identified through the previous known protein database search.
The two stage FDR is very similar to separate FDR but differ in some aspect. For example, it is possible that a search algorithm may assign higher score to a spectrum in a novel peptide sequence compared to a similar(homologous) known peptide sequence. This can happen in the case when the spectrum contains high noise or missing peaks. This suggested multi-stage process guarantees that every known peptide that can be identified by conventional MS/MS search is not misinterpreted as novel peptide (this is important especially in the case of SNV mutated peptides due to the sequence similarity).
Sample preparation and LC-MS/MS analysis
TCGA ovarian tumor tissues were cryo-pulverized and homogenized in lysis buffer (8M urea, 100mM NH4HCO3, pH 7.8, 0.1%NP−40 , 0.5% sodium deoxycholate, and protease inhibitors), after which the extracted proteins were reduced, alkylated and tryptically digested (Promega, Madison, WI) overnight. The resulting tryptic peptides were then cleaned up using strong cation exchange SPE and reversed phase C18 SPE columns (Supelco, Bellefonte, PA), dried and labeled with 4-plex iTRAQ reagents according to the manufacturer’s instructions (AB Sciex, Foster City, CA). The 4-plex iTRAQ labeled sample was separated on a XBridge C18 column (Waters, Milford, MA) using a LC gradient starting with a linear increase of solvent A (10mM triethylammonium bicarbonate, pH 7.5) to 10% B(10mM triethylammonium bicarbonate, pH 7.5, 90% acetonitrile) in 6 min, then 86 min to 30%B, 10 min to 42.5%B, 5 min to 55%B and another 5 min to 100%B. The flow rate was 0.5mL/min. A total of 96 fractions were collected and concatenated into 24 fractions by combining 4 fractions that are 24 fractions apart. The concatenated fractions were dried down and re-suspended in 0.1% trifluoroacetic acid to a peptide concentration of 0.15μg/μL for LC-MS/MS analysis.
The LC system was custom built using Agilent 1200 nanoflow pumps (Agilent Technologies, Santa Clara, CA). A 35cm × 360μmo.d. × 75μm i.dreversed-phase column wasslurry packed with 3μm Jupiter C18 (Phenomenex, Torrence, CA). Mobile phase flow rate was 300nL/min and consisted of 0.1% formic acid in water (A) and 0.1% formic acid acetonitrile (B) with a gradient profile as follows (min:%B); 0:5, 1:10, 85:28, 93:60, 98:75, 100:75. MS analysis was performed using a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) outfitted with a custom electrospray ionization interface. The ion transfer tube temperature and spray voltage were 300 °C and 1.8 kV, respectively. Orbitrap spectra (AGC 3×106) were collected from 300 – 1800m/z at a resolution of 30K followed by data-dependent HCD MS/MS (centroid mode, at a resolution of 7500, collision energy 45%, activation time 0.1ms, AGC 5×104) of the ten most abundant ions using an isolation width of 2.5 Da. Charge state screening was enabled to reject unassigned and singly charged ions. A dynamic exclusion time of 30 sec was used to discriminate against previously selected ions (within 0.55 Da to 2.55 Da).
Results
Database statistics
RNA-seq data used in this study was downloaded from TCGA1,2 repository.
The statistics of RNA-seq data-set and corresponding constructed FASTA databases are illustrated in Table 1. We used ovarian and breast cancer sample data with a total of 6.27 TB of RNA-seq alignments in 879 BAM formatted files. In order to predict small nucleotidevariants expressed the protein samples, we only used the subset of 67 files among 879 files that TCGA-sample-id that match with the PNNL selected samples for CPTAC22 study. Thus, a totalof 879 files are used in junction prediction, and 67 BAM files matching samples selected by PNNL for the ovarian cancer CPTAC22 study. These 67 selected BAM files were plugged into the GATK12,13 tool for variant calling analysis. Using our SpliceDB workflow, a total of 879 BAM files were used in creating the splice graph, and separately, 67 VCF formatted GATK12,13 output files were used in the variant graph construction. Our final FASTA database size was 4.34 GB in total, and contained 1, 466, 449 novel junctions (which includes 1, 180, 071 canonical GT-AG splice junction sites, and 24, 433 small deletions less than 10bp), 38, 464 deletions, 1, 105 insertions, and 182, 302 substitutions. The database will be made available for the usage researchers working on cancer proteogenomics in a way that agrees with the TCGA1,2 data usage guide lines.
Moreover, for the comparison experiment performed in the following section, we additionally created a genomic database from a single RNA-seq sample (sample id: TCGA-24-1467). From this sample, using our SpliceDB workflow applying same parameters, we created a 187 MB splice graph and variant graph database in FASTA format. This single sample variant graph encoded 168, 289 novel splice junctions (which includes 161, 935 canonical GT-AG splice junction sites, and 3, 322 small introns less than 10bp), 62 insertions, 3, 150 deletions, and 7, 109 substitutions.
Comparison between different FDR based error control strategies
In order to test the effect of different FDR calculation strategies in proteogenomic searches, we applied three different FDR approaches to our PSM results. Figure 1 shows the number of known and novel peptide identifications using different target-decoy based FDR strategies. The diagram showing the descriptions of each FDR strategy is shown in Supplemental Figure S.1.
With combined FDR, we identified 60, 877 known peptides and 1238 novel peptides. In contrast, the two stage FDR resulted 65, 578 known and 524 novel peptide identifications. However, in combined FDR, we note that the number of peptides hitting the decoy database under a certain FDR threshold is very different in novel database versus known database. After applying combined FDR approach, we explicitly separated the PSMs from known, known decoy, novel, novel decoy, database and calculated the FDR in both novel and known peptide hits. As shown in Figure 1, we get 36% FDR in novel peptides, and 0.03% FDR in known peptides while combined FDR was calculated as 1%. This indicates that combining the two PSM distributions raises the FDR cut-off for known peptides (lower identifications) and lowers it for novel peptides.
We choose a two-stage FDR approach in recognition of the differences in database sizes for the two searches. While results from the separate FDR is very similar to the two-stage FDR, two-stage FDR shows more conservative threshold on ‘novel identifications’.
Comparison between single-sample-matched and unified database search
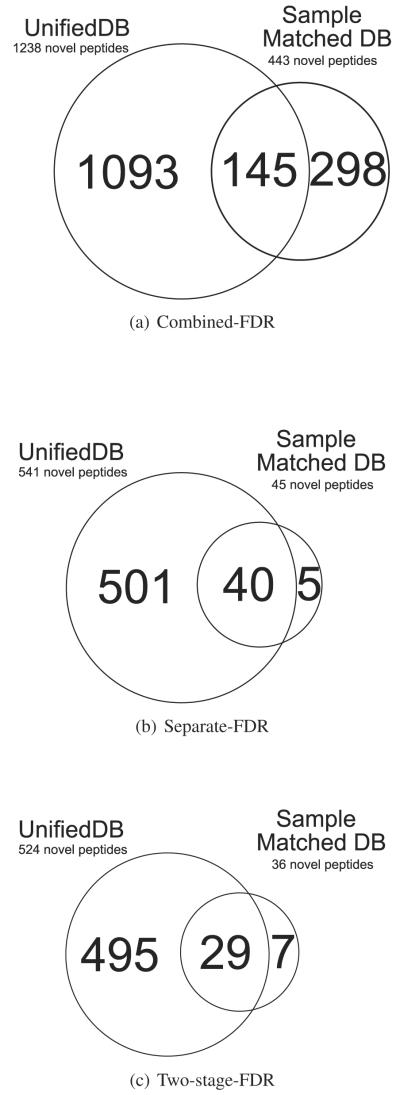
In order to explore the trade-off between creating a single sample targeted database versus multiple sample unified database, we performed a computational experiment using 439,858 spectra collected from an identical sample (sample id: TCGA-24-1467). Figure 2 shows the comparison of MS/MS search results between the single sample database (187 MB in FASTA) and all the sample unified database (total 4.34 GB in FASTA). As shown in Figure 2, the unified database shows a higher number of novel peptide identifications for every FDR estimation strategy, even with much a larger (×20) search space. Moreover, we observed that the overlapping portion of peptide identifications between the unified and sample matched database increases while applying more accurate novel peptide FDR calculations.
Figure 2.
Overlap between novel identifications from unified and single sample database.
MS-MS search results
The MS/MS search identified 524 novel peptides and 65, 578 known peptides at 1% FDR threshold. (using two-stage FDR strategy S.1(c)) By applying our integrative proteogenomics pipeline,14,18 470 novel findings were called (Table 2) from 524 identified novel peptides. In assigning proteogenomic events, we removed all peptides that can be mapped to more than 3 genomic locations, and multiply located peptides are used only as supportive evidences of uniquely located peptides to prevent overestimation of our findings.15-18 Details in handling multiply located peptides and event level error control will be discussed in our further study. Peptides identified as ‘novel sequence’ from our pipeline carry mutations which are not part of the wild-type proteome. We analyzed the novel peptide events, and selected examples that showcase the strength of our method. The other results can be found in Supplementary data ‘Supp_1_novel_ identifications.xlsx’. (list of known peptide identifications can be found in ‘Supp_2_known_ identifications.xlsx’)
Table 2.
List of novel findings (alternative splice junctions indicate novel junctions that shares identical splice site with a RefSeq gene in one side.)
| Type of novel findings | # of novel findings |
|---|---|
|
| |
| Substitution | 236 |
| Deletion | 5 |
| Novel splice junctions | 90 |
| Alternative splice junctions | 74 |
| Novel gene | 49 |
| TranslatedUTR | 2 |
| Exon boundary | 4 |
| Novel exon | 6 |
| Reverse strand | 4 |
Examples of identified mutated peptides
As mentioned, a key advantage of our method lies in the capability of identifying combinatorial multiple variants and the possibility of utilizing large scale information from multiple sample data. Note that the CPTAC22 project which provided the proteomic data, did not include matched tumor-normal controls which could help in identifying somatic versus genetic variants. It is possible to mine the ‘genomic’ data to distinguish between genetic and somatic variants in post-processing analysis that can be performed after the peptide identification. However, in this study we didn’t perform in depth diseases related analysis in order to strictly focus on providing the solid peptide identification results along with benchmark reports while applying different approaches.
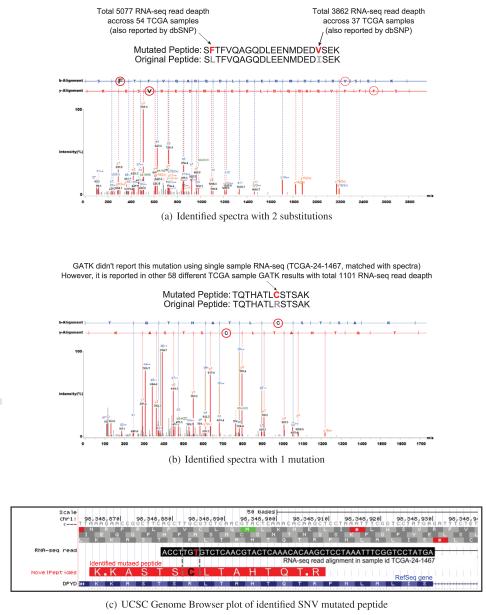
Our novel peptides include 13 multiply mutated peptide identifications. Figure 3(a) shows a selected example with two substitutions within a single peptide (‘S(F)TFVQAGQDLEENMDED(V)SEK’, spectra count:2). Both substitutions are supported by significant read-depth across multiple samples. Note that both substitutions are reported by dbSNP26 which also supports the validity of our finding.
Figure 3.
Alignment of identified spectra of mutated peptides.
The next example in Figure 3(b) is a case where we identified a SNV mutated peptide (‘TQTHATL(C)STSAK’, spectra count:2) using distinct sample RNA evidence (selected out of a total of 285 similar substitution events). Interestingly, this mutation was not found within the GATK12,13 variant call result the matched sample RNA-seq but heavily reported by 58 different samples. In order to explore the possibility that genomics alignment or variant calling tools might have filtered out this mutation, we went back and examined the original BAM file of this particular sample. As shown in Figure 3(c), we found a RNA-seq read alignment in this region that carries this exact mutation (note that this RNA-seq read also spans known splice junctions of RefSeq gene DPDY). This indicates that GATK filtered out this mutation while processing the single sample RNA-seq file due to the presence of splice junction, low quality score, or insufficient read depth.
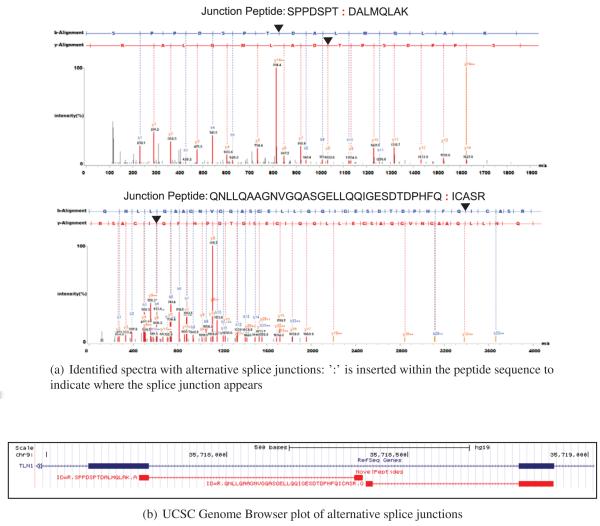
Junction peptides are particularly difficult because the span on one end is often too small to make a definitive call. In Figure 4, we show an example of multiple junction peptides that confirm a single alternative splice junction event. Two identified peptides ‘SPPDSPT:DALMQLAK’ (spectral count:3) and ‘QNLLQAAGNVGQASGELLQQIGESDTDPHFQ:ICASR’ (spectral count:1) both indicate alternative splice junctions which share one junction each with the refseq gene ‘TLN1’. Exon in the middle of each peptide also shares the same translation frame indicating a possible novel exon region. Moreover junctions in both peptides had strong RNA-seq level coverage evidence of 22,559 read depth in ‘SPPDSPT:DALMQLAK’ and 17,749 read depth in ‘QNLLQAAGNVGQASGELLQQIGESDTDPHFQ:ICASR’, across multiple samples.
Figure 4.
Alignment of identified spectra of novel junction peptides.
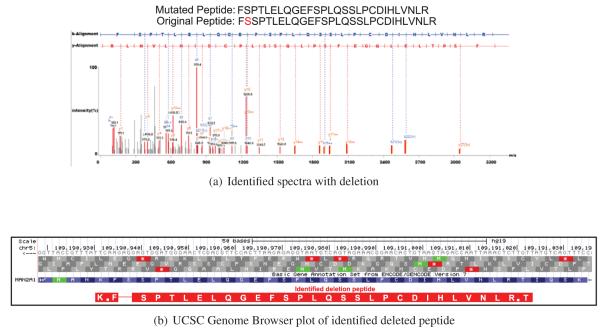
Figure 5(a) is an example of deletion peptide identification (selected from a total of 3 such cases). In this peptide identification, amino acid ’S’ was deleted from the original peptide sequence of ’F(S)SPTLELQGEFSPLQSSLPCDIHLVNLR’ (spectra count:1). This deletion site was expressed across 16 different RNA-seq samples. Together, these examples illustrate the power of proteogenomics searches in confirming translation of DNA lesions.
Figure 5.
Alignment of identified spectra of mutated peptides.
Novel peptides identified from outside of general protein coding regions
The bulk of our novel peptides are mutations on known proteins (275). One of the suggested strategies to reduce false identifications is to limit identification of novel peptides to genes where unmodified peptides have already been discovered. To test the usefulness of a more general method, we investigated peptides not from known gene regions. We do see 60 peptides within immunoglobulin regions (Supplemental Figure S.3) which (because of their high variability) are detected by our pipeline as various types of novel sequences such as alternative/novel splice junctions, fusion genes, and substitutions. A detailed description of proteogenomic event handling and observations from global multiple protein sample results are beyond the scope of this study and will be addressed in our future work. We also find peptides in annotated pseudo-genes (Supplemental Figure S.4), and yet another novel peptide in an unannotated region (Supplemental Figure S.5) which had previously been marked as a gene by a computational tool. Our pipeline automatically identifies many peptides outside of general protein coding regions. However, more detailed study will be necessary to investigate the underlying biology of these regions.
Discussion
Few approaches have been suggested in the community to search large scale genomic data using conventional MS/MS search algorithms. Here, we briefly revisit other approaches and compare with our suggested method. First, a sample specific search can be done to reduce the genomic data size and MS/MS search space. To apply this, one can generate a curated individual database for RNA-seq obtained from each sample5-7, and search it against proteomic data from the same sample. This targeted database approach has advantages in increasing the peptide identifications by assigning lower FDR threshold due to reduced database search space. However, as shown in Table 2, we claim that multiple sample driven database can improve the search results by incorporating shared information from many samples. Moreover, it requires coordination between genomic and proteomic laboratories to ensure that both sets of data are created for the same individual. Finally, each sample will use different FDR error control strategy which results in having multiple FDR thresholds throughout the whole process.
A second alternative approach is to assemble the transcript and DNA evidence into compact isoforms and variant calls using available tools,9-13 and then search translated versions of the isoforms and variants with proteomic data. The advantage of this approach is that it doesn’t require a genomic reference27 and is very easy to implement. However, the assembly of transcripts from multiple samples is unsolved, and the confidence with which we can assemble and compact all RNA data is limited. There is no study to date which has integrated a large multi-sample RNA-seq data into a single compacted transcript database, and used it for proteomic searches.
In comparison, our method implements an approach that merges and compress large-scale RNA-seq data (all) into a single database by applying graph-based algorithm. We achieve the large scale incorporation of RNA-seq data (from multiple samples) with no loss of information,14 while maintaining the reasonable database size for conventional MS/MS search engines. This unified database approach also enables us to maintain a single and more conservative FDR threshold throughout the entire analysis. Moreover, a graph based approach enables us to efficiently encode cumulative/large information from multiple RNA-seq data-sets into a compact unified database. Our approach also enables peptide identifications with combinatorial multiple splice junctions or variants. Based on our proposed method, we released a JAVA and python based tool called SpliceDB21 which generates FASTA formatted splice graph database from multiple RNA-seq alignments.
FDR based error control strategies in large database search
Due to the larger database search space, major criticisms of large-scale proteogenomics studies have been focused on the possibilities of false positive novel peptide identifications included in the result, which naturally emphasizes the need of stringent FDR estimation strategy.28 We agree that different approaches can dramatically change the number of novel peptide identifications. In this study we have shown that combined FDR strategy might boost the total novel identification results by introducing a global bias towards high-scored known peptide PSMs. Even the separate FDR approach still bears the possibility of identifying homologous peptides as novel sequences. Therefore, without introducing any unverified-novel FDR calculations, we applied a simple multi-stage approach where it can remove the FDR calculation bias in combined approach and additionally remove ambiguities in high scored false matches. As expected, two-stage FDR approach shows most conservative FDR measure (Figure 1) in novel identifications compared to other approaches. Therefore, we claim that two-stage FDR approach is most suitable for large database searches.
Further study
In this study, we have shown that multiple RNA-seq data-sets can be efficiently incorporated and utilized by the unified MS/MS search database. However, due to the limited MS/MS data access (obtained from a single sample), further integrative analysis is not shown in this study. Note that the results shown in this study are based on peptide level identifications without introducing advanced proteogenomic events. Detailed strategies of proteogenomic events to handle multiple genomic locations properly and advanced event level analysis will be shown in our future work.
Interestingly, novel sequence identifications shown in this study already includes results from highly variable regions such as Immunoglobulin genes and other novel sequence identifications (such as translated pseudo genes, transcript genes, novel gene areas, and so on) where known peptide identifications may not exist near by. Approaches on properly handling these identification in proteogenomic event level will be discussed in our future work.
While our approach focus mostly on ‘novel sequence identifications’ and follow conventional/standard approaches in ‘known sequence identifications’, other proteogenomic approaches5-7,29 focus heavily on improving ‘known sequence identifications’. Since we used relatively richer set of known proetins (Ensembl), we reason that by utilizing genomic level information to create a targeted and curated known protein database following other studies5-7,29 will improve our known identification results.
In conclusion, we reason that applying different philosophies in proteomic database creation will show various trade-offs. As we can see in the test experiment of this study (Figure 2), we claim that incorporation of large transcriptome data can increase the chance of novel peptide identification.
Spectra used in this study can be found at ftp://MSV000078631@massive.ucsd.edu.
Supplementary Material
Acknowledgement
V. B. and S. W. were supported by a grant from the NIH (P41-RR024851). S. C. was supported in part by the NSF IGERT Plant Systems Biology training grant # DGE-0504645. This work was supported by grant U24-CA-160019 (to R.D.S. and K.R.N.) from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The authors acknowledge partial support for proteomics analyses from P41GM103493 (to R.D.S.). The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory (EMSL), a U.S. Department of Energy (DOE) national scientific user facility located at PNNL in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830. N. S. is supported by an DOE early career grant (to S.H.P.).
References
- 1.Koboldt DC, Fulton RS, McLellan MD, Schmidt H. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. e. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell D, Berchuck A, Birrer M, Chien J, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponnala L, Wang Y, Sun Q, van Wijk KJ. Correlation of mRNA and protein abundance in the developing maize leaf. Plant J. 2014 doi: 10.1111/tpj.12482. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Su Z, Ma ZQ, Slebos RJ, et al. A bioinformatics workflow for variant peptide detection in shotgun proteomics. Mol. Cell Proteomics. 2011;10:M110.006536. doi: 10.1074/mcp.M110.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Duncan DT, Zhang B. CanProVar: a human cancer proteome variation database. Hum. Mutat. 2010;31:219–228. doi: 10.1002/humu.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Slebos RJ, Wang D, Halvey PJ, et al. Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 2012;11:1009–1017. doi: 10.1021/pr200766z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genomics Hub Repository. UC Santa Cruz: https://cghub.ucsc.edu.
- 9.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D, Pertea G, Trapnell C, Pimentel H, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, Banks E, Poplin R, Garimella KV, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo S, Cha SW, Merrihew G, He Y, et al. Proteogenomic database construction driven from large scale RNA-seq data. J. Proteome Res. 2014;13:21–28. doi: 10.1021/pr400294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner S, Shen Z, Ng J, Florea L, et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellana NE, Payne SH, Shen Z, Stanke M, et al. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 2010;73:2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellana NE, Shen Z, He Y, Walley JW, et al. An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol. Cell Proteomics. 2014;13:157–167. doi: 10.1074/mcp.M113.031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards NJ. Novel peptide identification from tandem mass spectra using ESTs and sequence database compression. Mol. Syst. Biol. 2007;3:102. doi: 10.1038/msb4100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Splice DB. UC San Diego, CCMS: http://bix.ucsd.edu/tmp/SpliceDB/SpliceDB.zip.
- 22.Clinical Proteomic Tumor Analysis Consortium Clinical Proteomic Tumor Analysis Consortium. http://proteomics.cancer.gov.
- 23.Kim S, Mischerikow N, Bandeira N, Navarro JD, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P, Ahmed I, Amode MR, Barrell D, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ensembl.
- 26.Sherry ST, Ward MH, Kholodov M, Baker J, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans VC, Barker G, Heesom KJ, Fan J, et al. De novo derivation of proteomes from transcriptomes for transcript and protein identification. Nat. Methods. 2012;9:1207–1211. doi: 10.1038/nmeth.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venter E, Smith RD, Payne SH. Proteogenomic analysis of bacteria and archaea: a 46 organism case study. PLoS ONE. 2011;6:e27587. doi: 10.1371/journal.pone.0027587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Zhang B. customProDB: an R package to generate customized protein databases from RNA-Seq data for proteomics search. Bioinformatics. 2013;29:3235–3237. doi: 10.1093/bioinformatics/btt543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.