Abstract
Octaprenyl pyrophosphate synthase (OPPs) catalyzes consecutive condensation reactions of one allylic substrate farnesyl pyrophosphate (FPP) and five homoallylic substrate isopentenyl pyrophosphate (IPP) molecules to form a C40 long-chain product OPP, which serves as a side chain of ubiquinone and menaquinone. OPPs belongs to the trans-prenyltransferase class of proteins. The structures of OPPs from Escherichia coli were solved in the apo-form as well as in complexes with IPP and a FPP thio-analog, FsPP, at resolutions of 2.2 to 2.6 Å, and revealed the detailed interactions between the ligands and enzyme. At the bottom of the active-site tunnel, M123 and M135 act in concert to form a wall which determines the final chain length. These results represent the first ligand-bound crystal structures of a long-chain trans-prenyltransferase and provide new information on the mechanisms of catalysis and product chain elongation.
Keywords: prenyltransferase, site-directed mutagenesis, product chain length, crystal structure
INTRODUCTION
Prenyltransferases transfer one or more molecules of isopentenyl pyrophosphate (IPP) to farnesyl pyrophosphate (FPP) for chain elongation to yield a diverse range of polyprenyl pyrophosphate products.1–4 Many natural products, including carotenoids, steroids, terpenoids and the lipid carriers that play important roles in cell wall biosynthesis and in cell signaling pathways are derived from these polyprenyl pyrophosphates.5,6 Prenyltransferases are classified into E- and Z-types, where E-type enzymes catalyze the formation of trans-double bond containing species while Z-type enzymes form cis-double bond containing species. We have determined the ligand-bound structures of cis-type undecaprenyl pyrophosphate synthase (UPPs) from E. coli, which reveals residue D26-mediated IPP binding and pyrophosphate dissociation from FPP to form the UPP product.7 Unlike cis-prenyltransferases, trans-prenyltransferases contain two conserved DDXXD motifs. To elucidate the trans-prenyltransferase mechanism, most recently, 27 structures of 19 trans-prenyltransferases were solved in apo forms or complexed with substrate analogs or products.8 However, the roles played by these two Asp-rich motifs are not fully understood yet.
Octaprenyl pyrophosphate synthase (OPPs), belonging to the E-type prenyltransferase family, catalyzes the sequential condensation reaction of FPP with five IPP molecules to generate trans C40-octaprenyl pyrophosphate (OPP).4 Although the ternary complex structure of the short-chain trans-prenyltransferases FPPs and GGPPs are available, no ligand-bound 3-D structure of long-chain trans-prenyltransferase has been reported, but only the apo-form structures of Thermotoga maritima OPPs (PDB code 1V4E) and Helicobacter pylori OPPs (PDB code 3TC1) are known.9–12 A homology model of the E. coli OPPs (EcOPPs) structure was built from the T. maritima OPPs (TmOPPs) structure using MODELLER, and structure-based mutagenesis studies of EcOPPs have been conducted to probe the possible substrate binding pattern and catalytic machinery.13,14 Site-directed mutagenesis results suggested that FPP is bound to the first DDXXD motif, and IPP was bound to several cationic residues (e.g. Arg, Lys, and His) via PPi, but not to the second DDXXD, consistent with the mechanism derived from the short-chain FPPs and GGPPs complexed structures.7,10 Nevertheless, structural analyses of substrate-bound OPPs complexes are required in order to more fully understand the catalytic and chain-length determining mechanisms.
Both trans- and cis-prenyltransferases contain an elongated tunnel-shaped crevice as the active site to accommodate their reactants and products, despite completely different structural folds.11,15 Large amino acids such as F132 of TmOPPs and L137 of EcUPPs were found to seal the bottom of the tunnels and determine the ultimate product chain lengths. Substitution of these large amino acids with the smaller Ala leads to formation of longer products due to removal of the blocking “floor”.11,16 In TmOPPs, sequential removal of F132, L128, I123, and D62 near the tunnel bottom resulted in a highly elongated (C95) product.17 The EcOPPs crystal structure as reported here clearly shows that two Met rather than Phe play a key role in determining product chain length and a “double-floor” hypothesis is proposed.
MATERIALS AND METHODS
Materials
Radiolabeled [14C]15IPP (55 mCi/mmol) was purchased from Amersham Pharmacia Biotech, and FPP was obtained from Sigma. Reverse-phase thin layer chromatography (TLC) plates were purchased from Merck. The PfuTurbo DNA polymerase was obtained from Life Technologies, Inc. The plasmid mini-prep kit and DNA gel extraction kit were purchased from Qiagen. The NiNTA resin was purchased from GE Healthcare. The protein expression kit including the pET46 Ek/LIC vector, E. coli DH5α and E. coli BL21trxB (DE3) competent cells were from Novagen. The QuikChange site-directed mutagenesis kit was obtained from Agilent Technologies, Inc. All commercial buffers and reagents were of the highest grade.
Protein expression, purification, crystallization and data collection
Protein expression and purification procedures were described previously.18 Briefly, the OPPs gene (Protein ID: NP_417654.1) from E. coli was cloned into pET46 Ek/LIC and then the plasmid transformed to E. coli BL21trxB (DE3). EcOPPs protein was induced with 0.8 mM isopropyl-thiogalactopyranoside (IPTG) at 37°C for 4 hours. The target protein was purified by using a Ni-NTA column and then a DEAE Sepharose Fast Flow column (GE Healthcare Life Sciences). The eluted EcOPPs was then dialyzed twice against 5 L buffer (25 mM Tris-HCl, pH 7.5, and 150 mM NaCl), and concentrated to 3 mg/mL for crystallization screening. All crystallization experiments were conducted at 22°C using the sitting-drop vapor-diffusion method. In general, 2 μL of EcOPPs-containing solution (25 mM Tris-HCl, 150 mM NaCl, pH 7.5; 3 mg/mL) was mixed with 2 μL of reservoir solution in 24-well Cryschem Plates (Hampton Research), and equilibrated against 300 μL of the reservoir solution. The optimized crystallization condition was 0.3 M magnesium chloride, 0.1 M Tris-HCl, pH 8.5, and 24% w/v polyethylene glycol 3350. Within 3–4 days, the crystals grew to about 0.1 mm × 0.1 mm × 1.5 mm. Prior to data collection, crystals were mounted in a cryoloop and soaked with cryoprotectant solution (0.3 M magnesium chloride, 0.1 M Tris-HCl, pH 8.5, 28% w/v polyethylene glycol 3350 and 4% v/v glycerol) for 3 s. The OPPs crystal in complex with IPP was obtained by soaking the apo-crystal with mother liquor containing 5 mM FsPP or IPP for 3 h. The X-ray diffraction datasets of OPPs apo-form and in complex with FsPP and IPP were collected to 2.2 Å, 2.6 Å and 2.45 Å resolution at beam line BL13B1 of the National Synchrotron Radiation Research Center (NSRRC, Hsinchu, Taiwan). Diffraction images were processed using the HKL-2000 program.19
Structure determination and refinement
Crystals of EcOPPs have been obtained before, but they were not suitable for structural analysis due to their low diffraction resolution (3.9 Å) and large unit cell dimensions.20 Crystals with better diffraction resolution (2.2 Å) were obtained by screening other crystallization conditions.21 As reported here, the crystal structure of EcOPPs apo-form was solved by molecular replacement (MR) method with Phaser program from the CCP4 suite using the hypothetical EcOPPs apo-form model, generated from the structure of Rhodobacter capsulatus nonaprenyl pyrophosphate synthase (RcNPPs; PDB code 3MZV; 45.2% sequence identity with EcOPPs apo-form) by SWISS-MODEL website, as a search model.8,22–25 Further refinement was carried out using the programs Coot and CNS.26,27 The complex crystals, including EcOPPs-FsPP and EcOPPs-IPP, were isomorphous to the EcOPPs apo-form crystal and were directly refined using the EcOPPs apo-form structure as a start.
There are two molecules in an asymmetric unit. The 2Fo−Fc difference Fourier map showed clear electron densities for most amino acid residues, but the loop regions from 152 to 157, 219 to 225 (A chain of the apo-structure), 92 to 102, 218 to 224 (B chain of the apo-structure), 152 to 157, 219 to 225 (A chain of the EcOPPs-FsPP structure), 91 to 102, 218 to 224 (B chain of the EcOPPs-FsPP structure), and 153 to 161, 219 to 226 (A chain of the EcOPPs-IPP structure), 92 to 102, 221 to 224 (B chain of the EcOPPs-IPP structure) lacked electron density, presumably due to disorder. Subsequent incorporation of FsPP, IPP and water molecules used a 1.0 σ map. All structural refinements were carried out with the programs Coot and CNS.26,27 These structures were validated by RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). Data collection and structure refinement statistics are summarized in Table I. All of the structure diagrams were drawn by using PyMol software (http://pymol.sourceforge.net/).
Table I.
Data collection and refinement statistics for EcOPPs crystals
| EcOPPs-apo | EcOPPs-FsPP | EcOPPs-IPP | |
|---|---|---|---|
|
| |||
| PDB code | 3WJK | 3WJN | 3WJO |
| Data co1lection | |||
| Space group | P21212 | P21212 | P21212 |
| Unit-cell | |||
| a [Å] | 116.9 | 116.7 | 117.5 |
| b [Å] | 128.4 | 128.2 | 130.9 |
| c [Å] | 46.4 | 46.5 | 46.4 |
| Resolution [Å] | 25–2.20 (2.28–2.20) | 25–2.60 (2.69–2.60) | 25–2.45 (2.54–2.45) |
| Unique reflections | 36335 (3529) | 22242 (2144) | 27265 (2658) |
| Redundancy | 3.9 (3.9) | 4.6 (4.8) | 6.3 (6.4) |
| Completeness [%] | 99.8 (99.4) | 99.8 (99.7) | 99.9 (100.0) |
| Average I/σ(I) | 27.5 (3.0) | 24.0 (3.6) | 28.5 (5.3) |
| Rmerge [%] | 5.8 (51.7) | 8.5 (45.4) | 6.2 (45.4) |
| Refinement | |||
| Rwork | 0.228 (0.315) | 0.216 (0.298) | 0.223 (0.267) |
| Rfree | 0.281 (0.335) | 0.265 (0.359) | 0.275 (0.308) |
| r.m.s.d. bonds [Å] | 0.007 | 0.007 | 0.007 |
| r.m.s.d. angles [º] | 1.3 | 1.2 | 1.3 |
| Ramachandran plot[%]) | |||
| most favored [%] | 95.1 | 96.3 | 95.5 |
| allowed [%] | 4.5 | 3.4 | 4.2 |
| disallowed [%] | 0.3 | 0.3 | 0.3 |
| no. of non-H atoms / average B [Å2] | 4965/45.94 | 4824/55.01 | 4993/49.95 |
Values in the parentheses are for the highest resolution shells.
Site-directed mutagenesis of OPPs
OPPs mutants were prepared by using a QuikChange site-directed mutagenesis kit in conjunction with the EcOPPs gene template cloned in the vector described previously.18,28 The mutagenic oligonucleotides were prepared by the Beijing Genomics Institution, Inc. (BGI) and were: 5′-CCCTGATCGAGTTTATCCACACGTACACTCTGCTACACGACGACG-3′ (A79Y); 5′-TACCCGCGCGTTCCAGATGGCGACCAGCCTCGGTTCACTCAA-3′ (M123A) and 5′-CTCAAAGTGCTGGAAGTCGCGTCAGAAGCCGTAAACGTCATCGC-3′ (M135A). The basic mutagenesis procedure utilizes a super-coiled double-stranded DNA (dsDNA) vector with the insert of interest and two synthetic oligonucleotide primers (forward and backward) containing the desired mutation. The procedure for protein purification followed the previously reported protocol.21
Product analysis for wild type and mutant EcOPPs
For product analysis, the reaction mixture contained 0.5 μM EcOPPs, 50 μM [14C]IPP, 5 μM FPP, 0.1% Triton X-100, 0.5 mM MgCl2, and 50 mM KCl in 100 mM Hepes buffer (pH 7.5) and was carried out for 12 h at 25°C. 10 mM EDTA was used to terminate the reactions. The radio-labeled polyprenyl pyrophosphate products were extracted with 1-butanol and were converted to prenyl alcohols by using acid pyrophosphatase, as described previously.11 The radio-labeled products were identified by autoradiography using a FUJIFILM BAS-1500 bioimaging analyzer, based on their reported Rf values.
RESULTS AND DISCUSSION
Comparison of amino acid sequences of trans-prenyltransferases
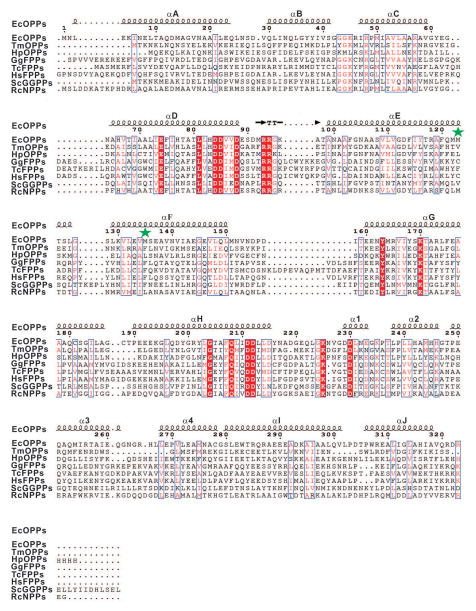
The amino acid sequences of several trans-prenyltransferases including C40-OPPs from E. coli (Gene Bank Accession Number: NP_417654.1), T. maritima (Gene Bank Accession Number: NP_229335.1) and H. pylori (Gene Bank Accession Number: 345110994); C15-FPPs from Gallus gallus (Gene Bank Accession Number: 157831115), Trypanosoma cruzi (Gene Bank Accession Number: 85543983) and Homo sapiens (Gene Bank Accession Number: 90109287); C20-GGPPs from Saccharomyces cerevisiae (Gene Bank Accession Number: 93279242) and C45-nonaprenyl pyrophosphate synthase (NPPs) from R. capsulatus (Gene Bank Accession Number: 297787795), are aligned with ClustalW2 and shown with ESPript, and the results obtained are shown in Fig. 1.8,11,12,15,29–34 These proteins all contain two DDXXD motifs which are considered crucial for FPP and IPP binding and catalytic activity. The amino acid located in the fifth position before the first DDXXD is an alanine inEcOPPs, TmOPPs, HpOPPs, and RcNPPs, and is known to be important in determining the chain length of the product, which will be discussed later.11 As shown below, the enzymatic product of the A79Y mutant of EcOPPs was greatly shortened, using FPP and IPP as substrates.35
Figure 1.
Amino acid alignment of EcOPPs, TmOPPs, H. pylori OPPs, G. gallus, T. cruzi, H. sapiens FPPs, S. cerevisiae GGPPs and R. capsulatus NPPs. Strictly conserved residues are highlighted by using red background and conservatively substituted residues are boxed. The secondary structural elements (helices-α, turns-T) of EcOPPs are shown above the aligned sequences. The M123 and M135 are indicated by green asterisks.
Overall structure of EcOPPs
In this study, we determined the crystal structures of EcOPPs in its apo form and in complex with either FsPP or IPP. We tried to solve the OPPs structure in complex with both FsPP and IPP together by soaking and co-crystallization, but no crystals survived under these conditions, despite some effort. The soaking experiments were possibly hampered by a dramatic conformational change upon simultaneous binding of FsPP and IPP, which would disrupt crystal packing. Co-crystallization was also unsuccessful, probably due to EcOPPs being highly active, the thiol-analog FsPP still reacting with IPP. Mg2+ is required for EcOPPs catalytic activity. However, unlike our previously solved S. cerevisiae GGPPs (ScGGPPs) complex structures, no Mg2+ was found to bind to FsPP in the active site, although a high concentration of magnesium chloride was present in the crystallization solution. EcOPPs is among the most active trans-prenyltransferases, and the Mg2+ binding region might be too flexible, or its formation too transient, to be seen in our X-ray structures.28
The overall structure of EcOPPs-FsPP is shown in Fig. 2A and B. The IPP molecule is taken from the EcOPPs-IPP structure. The overall structures of EcOPPs and TmOPPs are similar with a root mean square deviation (RMSD) of 1.91 Å for 180 matched pairs of Cα atoms (A chain). Compared with TmOPPs, EcOPPs has two more helices (J and α4), longer helices A and B, and a more outward helix F. The extra helices (J and α4) are distant from the active site and may play a structural role by providing EcOPPs higher flexibility. The EcOPPs structure contains 14 α helices, 10 of which (helices A to J) surround a large central cavity. An elongated tunnel-shaped cavity surrounded by five helices (helices C, D, E, F, and H) is, we propose, the active site. Two conserved DDXXD sequences are located on helices D and H near the opening cleft of the substrate-binding pocket, facing each other to create the FPP and IPP binding sites. The loops on the top of helices F-H are stabilized by hydrophobic interactions with several hydrophobic residues and are thus kept away from the substrate entrance, between helices D and F.
Figure 2.
Overall structure and the superimposition of FsPP and IPP from EcOPPs and ScGGPPs. (A) The side view and (B) the top view of EcOPPs are shown by using a cylinder diagram. Two identical subunits are associated into a dimer by forming a four layer helix bundle. The two identical subunits are shown in green and cyan, respectively. Two conserved DDXXD motifs are shown in red. The FsPP and IPP observed in OPPs complex structure are shown with stick, colored in blue and yellow, respectively. The M123 and M135 located at the bottom of the active-site tunnel are shown in sphere and colored in magenta. (C) A stereo view of two homologous proteins, EcOPPs (green) including FsPP (green) and IPP (lime) and ScGGPPs containing FsPP and IPP (all shown in yellow, PDB 2E8T), are superimposed by PyMol.
In the EcOPPs-IPP structure, one pyrophosphate in the FPP site and one IPP in the IPP site were observed. In the EcOPPs-FsPP structure, a FsPP was only seen in the FPP site. These two sites are located at the positions corresponding to those occupied by FsPP and IPP in the ScGGPPs ternary structure.10,11,15 Likewise, the DDXXD motifs of EcOPPs and ScGGPPs superimpose very well (Fig. 2C). The EcOPPs active site in apo-, FsPP- and IPP-bound structures all adopt more compact, closed structures than that seen in the ScGGPPs-FsPP-IPP structure (Fig. 2C). From the superimposition of the EcOPPs-FsPP, EcOPPs-IPP and ScGGPPs structures, it can be seen that the IPP from the EcOPPs structure occupies the same position as does the IPP in ScGGPPs, while the FsPP tail of EcOPPs points in a different direction. ScGGPPs catalyzes the condensation reaction of FPP and one molecule of IPP to produce the C20 product, GGPP. From previously solved structures, it can be seen that Y112 seals the FPP binding tunnel to prevent longer (e.g. C25 or C30) product formation.10 EcOPPs produces an even longer product (C40), so the FsPP tail observed in EcOPPs-FsPP extends further away from the active site towards the product tunnel (Fig. 2C). FsPP adopted a slightly different conformation upon binding to ScGGPPs than that seen in EcOPPs and the distance between the last prenyl planars from ScGGPPs and EcOPPs was 4.1 Å with 86.3° rotation (Figure S1). ScGGPPs catalyzes the condensation reaction by using IPP and FPP to produce GGPP (C20), which only needs to be blocked around the FPP binding site (no need to enter the product tunnel). In longer chain prenyltransferases (e.g. C30 to C50), however, the products need to extend into the elongated product tunnel for proper chain elongation.
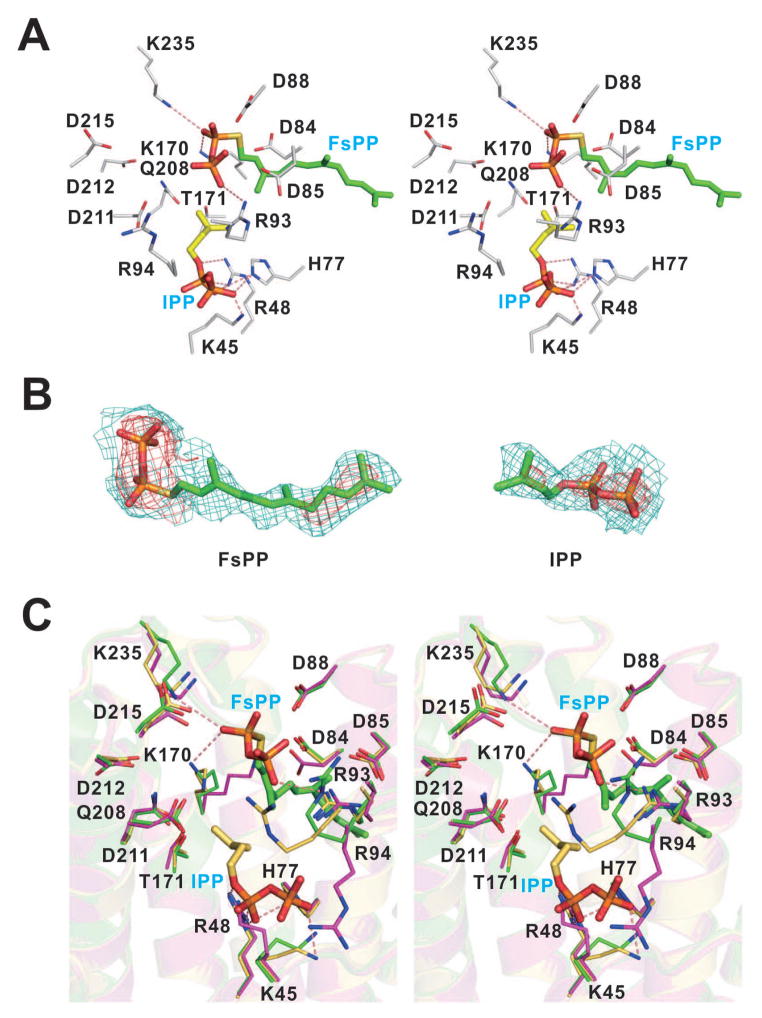
Details of FsPP and IPP binding, and conformational changes
The FsPP and IPP binding sites are shown in Fig. 3A. Electron densities are shown in Fig. 3B. Around the active site, some polar (mostly positively charged) residues including K45, R48, H77, R93, R94, K170, K235 were found close to FsPP and IPP (Fig. 3A). To gain insight into the EcOPPs catalytic mechanism, the EcOPPs-FsPP and EcOPPs-IPP structures were superimposed. The active site residues were taken from the EcOPPs-FsPP structure. When comparing the EcOPPs-apo, EcOPPs-FsPP and EcOPPs-IPP structures, it can be seen that all structures adopt a similar closed form and there are no significant conformational changes observed (RMSD < 0.4 Å). Nevertheless, the conformations of the side chains of K45, K170 and R94 are significantly altered upon ligand binding (Fig. 3C). The first DDXXD motif is responsible for FsPP binding. Based on previous studies, the pyrophosphate of FsPP should interact with three Mg2+ bound to the first DDXXD motif, but no Mg2+ was observed in our structures. Instead, there are multiple interactions between the pyrophosphate head of FsPP and R93, K170 and K235, and between the IPP head group and K45, R48 and H77. R93 and R94 may facilitate catalysis by stabilizing the leaving pyrophosphate group, and these residues are highly conserved among both short and long chain trans-prenyltransferases (e.g. R90 and R91 for TmOPPs; R112 and R113 for rat FPPs; R109 and R110 for yeast FPPs),36,37 K235 is in a flexible loop and may also interact with FPP and is also essential for catalysis, and is thought to trigger a closed conformation change on substrate entry.14
Figure 3.
Detailed interaction networks in the active site and conformational change upon substrate binding. (A) A stereo view of detailed active site interaction networks of the EcOPPs in complex with FsPP (green) and IPP (yellow). The FsPP and the secondary structures are from EcOPPs-FsPP and the IPP is adopted from EcOPPs-IPP structure. Residues K45, R48, H77, R93, R94, K170, T171, Q208 and K235 around the active site and two DDXXD motifs (D84, D85, D88, D211, D212 and D215) are shown. Hydrogen bonds with a distance below 3.5 Å are shown as red dashed lines. (B) The Fo-Fc omit maps of FsPP from the EcOPPs-FsPP structure and IPP from the EcOPPs-IPP structure are contoured at 1.0 σ (teal) and 3.0 σ level (red). (C) A stereo view of superimposition of active site residues from EcOPPs-apo, EcOPPs-FsPP and EcOPPs-IPP is shown. Detailed interaction networks of the apo-form, EcOPPs-FsPP and EcOPPs-IPP active site residues are shown in magenta, green and yellow, respectively. Some subtle conformational changes are observed for K45, K170 and R94 side chains. Hydrogen bonds with a distance below 3.5 Å are shown as red dashed lines. For clarity, only hydrogen bonds from EcOPPs-FsPP structure were shown for FsPP and hydrogen bonds from EcOPPs-IPP structure were shown for IPP.
Mechanism of product chain length determination for EcOPPs
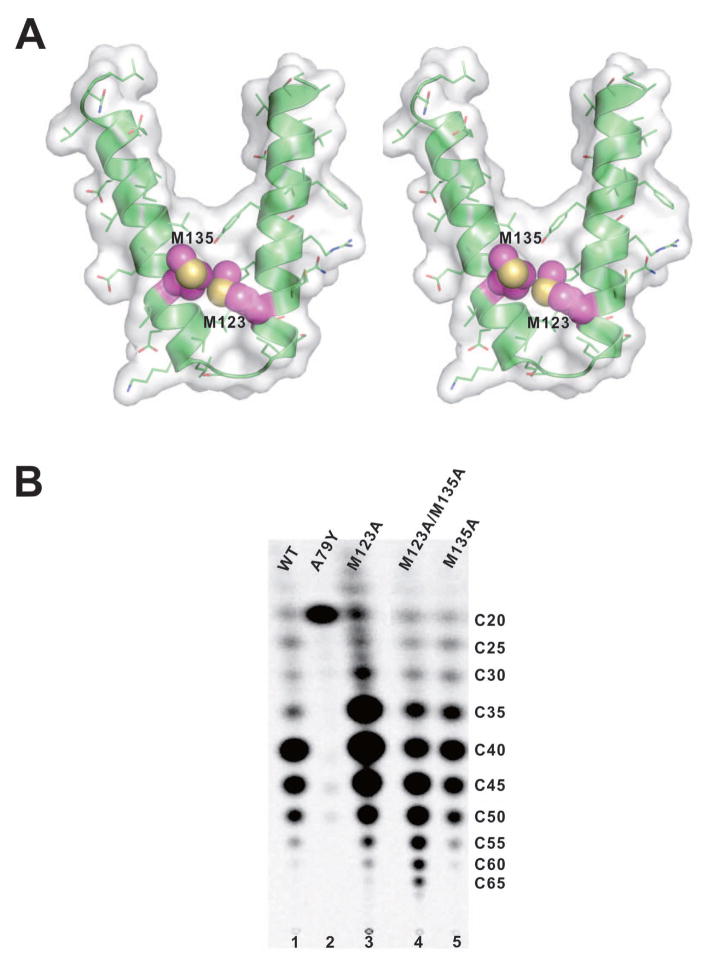
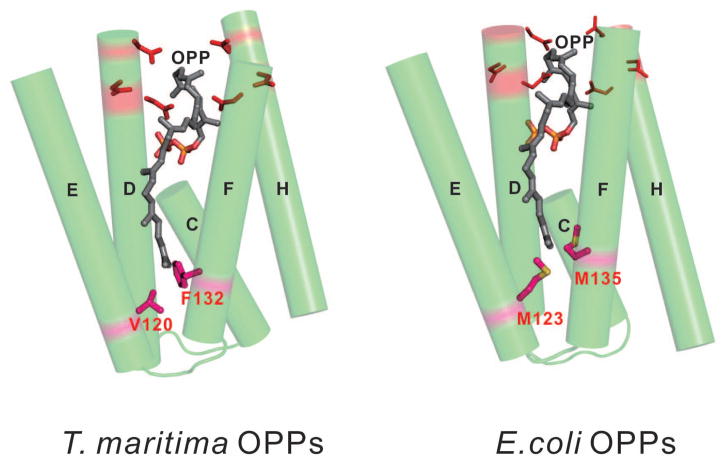
The EcOPPs structure is similar to that of TmOPPs at the bottom of the hydrophobic tunnel, because the shape of the tunnel of the EcOPPs (3.5–11.1 Å width and 22.7–25.9 Å length, Figure S2A) is similar to that of TmOPPs (3.8–10.6 Å width and 22.8–23.9 Å length, Figure S2B), but the proposed “floor” formation is different: one “single-floor” F132 is observed in TmOPPs, while two “double-floor” Met residues (M123 and M135) are found in EcOPPs (Figs. 4A and 5). Indeed, the sequence alignment (Fig. 1) shows that the amino acid corresponding to F132 in TmOPPs is M135 in EcOPPs. The other residue (M123) in EcOPPs corresponds to V120 in TmOPPs, which is also located at the bottom of the tunnel (Figs. 4A and 5), but is not involved in product chain length determination in the TmOPPs reaction.11 To determine whether either one or both of the Met residues are required for the correct product chain length in EcOPPs, we substituted either one or both Met residues with Ala and examined the products generated by the mutant enzymes. As shown in Fig. 4B, wild-type OPPs synthesized mainly a C40 product (lane 1), as did the mutants M123A (lane 3) and M135A (lane 5). Only when both M123 and M135 were replaced with Ala, the longer products (C55) were formed (lane 4). Therefore, we propose that the “double-floor” at the bottom of the active site of EcOPPs consists of two Met residues.
Figure 4.
The surface representation of the active site tunnels from the apo-structure and different chain length products synthesized by mutants. (A) A stereo view of the surface representation of the helices E and F for the product accommodating tunnel from EcOPPs-apo structure. The M123 (helix E) and M135 (helix F) are shown in sphere and colored in magenta. (B) Products synthesized by EcOPPs mutants were analyzed by TLC for WT, A79Y, M123A, M123A/M135A and M135A, respectively. The longest product (C55) was synthesized by M123A/M135A double mutant.
Figure 5.
The “single-floor” and “double-floor” mechanisms for determining product chain lengths of TmOPPs and EcOPPs. Five α-helices (C, D, E, F, and H) surrounding the active site of OPPs are shown. Two Met residues (M123 and M135) in EcOPPs and F132 in TmOPPs, which seal the bottom of the active site tunnels, are critical in determining the product chain lengths.
Trans-prenyltransferases generate products with correct chain lengths according to a molecular ruler mechanism, where one or two bulky amino acids occupy the bottom of each of the enzyme active sites to block extra chain elongation of the products, thereby determining the ultimate chain lengths. As revealed by the sequence alignment (Fig. 1) and their 3-D structures, F132, L127, F112/F113, Y107/H139 in TmOPPs, HpOPPs, GgFPPs, ScGGPPs suggest that the side chains of these residues are important for determining the ultimate length of the hydrocarbon chains, and site-directed mutagenesis studies have proven that substitution of these “floors” with small amino acids yields longer products.11,12,15,17,30,38,39 Therefore, the active site is “double-floored” by M123/M135 in EcOPPs but “single-floored” in TmOPPs and HpOPPs.
As shown in Fig. 4B (lane 2), substitution of the smaller A79 at the fifth position upstream of the first DDXXD with a larger Tyr led to a shorter chain length (C20) product, confirming a role for A79 in permitting chain elongation. A corresponding residue to A79 in EcOPPs is located in the fifth position upstream from the first DDXXD motif in TmOPPs (A76; Fig. 1). The A76Y mutant of TmOPPs also has altered the product from C40 to C20.11 The corresponding residue at this position in GgFPPs, TcFPPs, HsFPPs is a large Phe or His, where the amino acid near the DDXXD motif restricts the chain elongation of FPP. On the other hand, the mutant S71Y (the fourth amino acid prior to the first DDXXD motif of type-III ScGGPPs) created a blockage at the upper part of the active site crevice, and the mutant produced shorter C15 FPP as the final product.15
The stability of trans-prenyltransferases and product chain length determination might also be influenced by oligomer formation (as homomers or heteromers).35,40 For example, the heterodimeric EcOPPs of wild-type and A79Y produced a shorter (C30) product.35 Based upon an inspection of the current crystal structures, it is likely that the A79Y mutation (in helix D) might push helix E towards the wild-type monomer, blocking the product tunnel and turning out only the C30 product. In conclusion, the EcOPPs structures reported here provide interesting new information for the catalytic and product chain length determination mechanisms of a long chain trans-prenyltransferase, in which a “double-floor” structure is likely to block further chain elongation of the C40 product (Fig. 5).
Supplementary Material
Acknowledgments
The synchrotron data collection was conducted at beam line BL13B1 of NSRRC (National Synchrotron Radiation Research Center, Taiwan, R.O.C.) supported by the National Science Council (NSC) of Taiwan.
The abbreviations used are
- EDTA
ethylenediaminetetraacetic acid
- FPP
farnesyl pyrophosphate
- FPPs
farnesyl pyrophosphate synthase
- PPi
pyrophosphate
- FsPP
farnesyl-thiolo-pyrophosphate
- GGPPs
geranylgeranyl pyrophosphate synthase
- Hepes
4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid
- HexPPs
hexaprenyl pyrophosphate synthase
- IPP
isopentenyl pyrophosphate
- NPPs
nonaprenyl pyrophosphate synthase
- OPPs
octaprenyl pyrophosphate synthase
- Tris
tris (hydroxymethyl) aminomethane
- UPPs
undecaprenyl pyrophosphate synthase
Footnotes
This work was supported by grants from National Basic Research Program of China (2011CBA00805 and 2011CB710800), Tianjin Municipal Science and Technology Commission (12ZCZDSY12500), National ScienceCouncilof Taiwan (102-2113-M-001-005-MY3 to PHL) and in part by NIH grants GM065307 and CA158191. X.F. was supported by a Pre-doctoral Fellowship from the American Heart Association, Midwest Affiliate (13PRE14510056).
The atomic coordinates and structure factors for the E. coli OPPs in apo-form (code: 3WJK) and in complex with FsPP (code: 3WJN) and IPP (code: 3WJO) have been deposited in the RCSB Protein Data Bank.
References
- 1.Kellogg BA, Poulter CD. Chain elongation in the isoprenoid biosynthetic pathway. Curr Opin Chem Biol. 1997;1(4):570–578. doi: 10.1016/s1367-5931(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 2.Ogura K, Koyama T. Enzymatic Aspects of Isoprenoid Chain Elongation. Chem Rev. 1998;98(4):1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- 3.Soballe B, Poole RK. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology. 1999;145(Pt 8):1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 4.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. Eur J Biochem. 2002;269(14):3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 5.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 6.Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 7.Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH, Liang PH. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J Biol Chem. 2005;280(21):20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 8.Wallrapp FH, Pan JJ, Ramamoorthy G, Almonacid DE, Hillerich BS, Seidel R, Patskovsky Y, Babbitt PC, Almo SC, Jacobson MP, Poulter CD. Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc Natl Acad Sci U S A. 2013;110(13):E1196–1202. doi: 10.1073/pnas.1300632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J Biol Chem. 2004;279(10):8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 10.Guo RT, Cao R, Liang PH, Ko TP, Chang TH, Hudock MP, Jeng WY, Chen CK, Zhang Y, Song Y, Kuo CJ, Yin F, Oldfield E, Wang AH. Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases. Proc Natl Acad Sci U S A. 2007;104(24):10022–10027. doi: 10.1073/pnas.0702254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo RT, Kuo CJ, Chou CC, Ko TP, Shr HL, Liang PH, Wang AH. Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J Biol Chem. 2004;279(6):4903–4912. doi: 10.1074/jbc.M310161200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Zhang X, Zhang R, Wu C, Guo Y, Mao X, Guo G, Zhang Y, Wang DC, Li D, Zou Q. Modeling studies with Helicobacter pylori octaprenyl pyrophosphate synthase reveal the enzymatic mechanism of trans-prenyltransferases. Int J Biochem Cell Biol. 2012;44(12):2116–2123. doi: 10.1016/j.biocel.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 14.Chang KM, Chen SH, Kuo CJ, Chang CK, Guo RT, Yang JM, Liang PH. Roles of amino acids in the Escherichia coli octaprenyl diphosphate synthase active site probed by structure-guided site-directed mutagenesis. Biochemistry. 2012;51(16):3412–3419. doi: 10.1021/bi300069j. [DOI] [PubMed] [Google Scholar]
- 15.Chang TH, Guo RT, Ko TP, Wang AH, Liang PH. Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J Biol Chem. 2006;281(21):14991–15000. doi: 10.1074/jbc.M512886200. [DOI] [PubMed] [Google Scholar]
- 16.Ko TP, Chen YK, Robinson H, Tsai PC, Gao YG, Chen AP, Wang AH, Liang PH. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J Biol Chem. 2001;276(50):47474–47482. doi: 10.1074/jbc.M106747200. [DOI] [PubMed] [Google Scholar]
- 17.Guo RT, Kuo CJ, Ko TP, Chou CC, Liang PH, Wang AH. A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products. Biochemistry. 2004;43(24):7678–7686. doi: 10.1021/bi036336d. [DOI] [PubMed] [Google Scholar]
- 18.Kuo TH, Liang PH. Reaction kinetic pathway of the recombinant octaprenyl pyrophosphate synthase from Thermotoga maritima: how is it different from that of the mesophilic enzyme. Biochim Biophys Acta. 2002;1599(1–2):125–133. doi: 10.1016/s1570-9639(02)00410-7. [DOI] [PubMed] [Google Scholar]
- 19.Zbyszek O, Wladek M. Processing of X-ray Diffraction Data Collected in Oscillation Mode Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Guo RT, Ko TP, Chou CC, Shr HL, Chu HM, Tsai YH, Liang PH, Wang AH. Preliminary X-ray diffraction analysis of octaprenyl pyrophosphate synthase crystals from Thermotoga maritima and Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 12):2265–2268. doi: 10.1107/s0907444903018985. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Han X, Ko TP, Chen CC, Zhu Z, Hua E, Guo RT, Huang CH. Preliminary X-ray diffraction analysis of octaprenyl pyrophosphate synthase from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 3):328–331. doi: 10.1107/S1744309113003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 25.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Pan JJ, Kuo TH, Chen YK, Yang LW, Liang PH. Insight into the activation mechanism of Escherichia coli octaprenyl pyrophosphate synthase derived from pre-steady-state kinetic analysis. Biochim Biophys Acta. 2002;1594(1):64–73. doi: 10.1016/s0167-4838(01)00283-7. [DOI] [PubMed] [Google Scholar]
- 29.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 30.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Crystal structure of recombinant farnesyl diphosphate synthase at 2. 6-Å resolution. Biochemistry. 1994;33(36):10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 31.Gabelli SB, McLellan JS, Montalvetti A, Oldfield E, Docampo R, Amzel LM. Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: implications for drug design. Proteins. 2006;62(1):80–88. doi: 10.1002/prot.20754. [DOI] [PubMed] [Google Scholar]
- 32.Rondeau JM, Bitsch F, Bourgier E, Geiser M, Hemmig R, Kroemer M, Lehmann S, Ramage P, Rieffel S, Strauss A, Green JR, Jahnke W. Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs. ChemMedChem. 2006;1(2):267–273. doi: 10.1002/cmdc.200500059. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kainou T, Okada K, Suzuki K, Nakagawa T, Matsuda H, Kawamukai M. Dimer formation of octaprenyl-diphosphate synthase (IspB) is essential for chain length determination of ubiquinone. J Biol Chem. 2001;276(11):7876–7883. doi: 10.1074/jbc.M007472200. [DOI] [PubMed] [Google Scholar]
- 36.Joly A, Edwards PA. Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J Biol Chem. 1993;268(36):26983–26989. [PubMed] [Google Scholar]
- 37.Song L, Poulter CD. Yeast farnesyl-diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc Natl Acad Sci U S A. 1994;91(8):3044–3048. doi: 10.1073/pnas.91.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD. Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci U S A. 1996;93(26):15018–15023. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemmi H, Noike M, Nakayama T, Nishino T. An alternative mechanism of product chain-length determination in type III geranylgeranyl diphosphate synthase. Eur J Biochem. 2003;270(10):2186–2194. doi: 10.1046/j.1432-1033.2003.03583.x. [DOI] [PubMed] [Google Scholar]
- 40.Cui TZ, Kaino T, Kawamukai M. A subunit of decaprenyl diphosphate synthase stabilizes octaprenyl diphosphate synthase in Escherichia coli by forming a high-molecular weight complex. FEBS letters. 2010;584(4):652–656. doi: 10.1016/j.febslet.2009.12.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.