Abstract
Background
With limited funding and biological specimen availability, choosing an optimal sampling design to maximize power for detecting gene-by-environment (G-E) interactions is critical. Enriched sampling is often used to select subjects with rare exposures for genotyping to enhance power for tests of G-E effects. However, exposure misclassification (MC) combined with biased sampling can affect characteristics of tests for G-E interaction and joint tests for marginal association and G-E interaction. Here, we characterize the impact of exposure-biased sampling under conditions of perfect exposure information and exposure MC on properties of three methods for conducting these tests.
Methods
We assess the power, Type I error, bias, and mean squared error of case-only, case-control, and empirical Bayes methods for testing G-E interaction and a joint marginal G (or E) effect and G-E interaction across three biased sampling schemes. Properties are evaluated via simulation. We also consider the role of gene-environment independence.
Results
With perfect exposure information, enriched sampling schemes enhance power as compared to random selection of subjects irrespective of exposure prevalence but yield bias in estimation of the G-E interaction and marginal E parameters. G-E independence affects the relative properties of the interaction detection methods, with the case-only approach suffering most severely. Exposure MC distorts the relative performance of sampling designs when compared to the case of perfect exposure information.
Conclusions
Those conducting G-E studies should be aware of exposure MC properties and the prevalence of exposure when choosing an ideal sampling scheme and method for detecting G-E interactions and joint effects.
Keywords: sampling design, gene-environment interaction, interaction, genetic epidemiology, case-control, exposure misclassification
BACKGROUND
Elucidating the role of interactions between genes and the environment in complex disease etiology has become a recent focus of many epidemiologic investigations [1, 2]. This expansion upon traditional genetic association studies that examine the relation between genetic variants and disease status is driven both by biological interest in the interplay between these genetic and environmental risk factors and by the potential increase in power that interactions may offer for detecting the existence of any genetic effect [3, 4]. Further, joint tests for marginal environmental and gene-environment (G×E) interaction effects may be helpful for identifying genetic subgroups among which the effects of environmental exposures are enhanced. Several approaches for estimating/testing for multiplicative G×E interaction have been proposed in the literature; the case-only, case-control, and empirical Bayes methods have been widely used in the post genome-wide association study (GWAS) era [5–8]. The case-only and empirical Bayes approaches capitalize on gene-environment independence to improve power to detect gene-environment interaction, with the empirical Bayes approach achieving this goal in a more robust, data-adaptive manner.
Despite the potential for G×E interaction studies to unravel clues about the multi-factorial etiology of a complex disease, a significant obstacle in conducting a successful study lies in achieving enough power to detect potentially modest effect sizes. Thus, during the design stage, practical considerations including the optimization of constrained financial and biologica l sample resources remain a strategic challenge. While a power-maximizing sampling scheme for selecting GWAS subjects from specific exposure risk groups has been described for testing marginal association of the genes with disease, this approach is valid only when a lack of strong G×E interaction can be assumed [9]. When the goal of the study is actually to detect an interaction, some have proposed that leveraging environmental exposure information during sample selection for genotyping provides power advantages [10]. Exposure-enriched sampling designs may increase power by allowing for the oversampling of subjects from relatively rare exposure categories. However, the benefits of exposure-biased sampling designs for G×E interaction detection have not been rigorously evaluated. Further, it is known that non-differential misclassification (MC) of environmental exposure data can lead to biased estimates of interaction effects and can affect the power and type I error properties of statistical tests for interaction [11–14]. In addition, because differences in exposure recall between cases and controls are frequently noted, differential MC of exposure by disease status poses more severe of a concern for bias in estimation and inflated type 1 error as well as reduced power for statistical testing [13, 15, 16]. It remains unclear how either form of MC impacts the relative performance of different exposure enrichment designs implemented during the sampling phase, as the sampling strategy is now based on a contaminated exposure instead of a perfect exposure.
This report presents the results from a simulation study that examines the statistical advantages and disadvantages of various exposure-enriched sampling schemes for detecting G×E interactions and joint tests of both marginal and interaction effects. In addition, we aim to describe how exposure misclassification impacts the choice of such designs. Our focus here is to examine the influence that study design and sample selection decisions have on power/type 1 error for testing and efficiency/bias in estimation, with the goal of informing future analytic approaches.
It is true that the literature on two-phase or biased sampling design is now rich and methods for appropriately handling such data for inference about effects of the exposure are readily available (particularly in the absence of MC) [17–19]. For instance, a simple offset term using known sampling fractions for cases and controls can be derived to separate the effect of exposure under a biased sampling scheme from the actual effect of exposure under a logistic regression for case-control studies. The various estimators considered in this paper can likewise be modified to appropriately account for the biased sampling design. Thus, a relevant analytic issue here would be to obtain an optimal design using the corrected estimators/tests that fully account for the biased sampling scheme. However, we do not adopt that strategy and simply illustrate the consequences of what is typically done in testing for genetic effects in large GWA studies where the design is optimized to enhance power for discovery (rightfully so, as the interest lies in detection of susceptibility loci and not estimating the true effect size very precisely) and standard analytic tools without finer adjustments for biased sampling are used. The effect of MC on such a design and analysis strategy is not present in the literature. Two sources of potential bias, exposure-enriched sampling and misclassification of exposure, are explored simultaneously in this study, adding new knowledge to the existing literature.
METHODS
Simulation Design: Generation of the 2×4 table
We consider the analysis of simulated data generated from an unmatched case-control study with a binary genetic factor (G), assuming a dominant genetic model, and a binary environmental variable (E). G takes a value of 1 for a genetically susceptible subject and 0 for a subject without a copy of the risk allele. For power comparisons, binary G is a reasonable assumption to consider because dichotomization maintains the relative performance of the methods and sampling designs expected from the more general co-dominant or additive scenario. In terms of exposure, E = 1 signifies exposed, and E = 0 designates unexposed subjects. Similarly, D represents disease status, where D = 1 indicates affected, and D = 0 denotes unaffected. Table 1 describes the distribution of cell counts in the underlying case-control study base. Using the notations in Table 1, the vectors of cell frequencies for cases and controls are realizations from two independent multinomial distributions, r1 ~ Multinomial (n1, p11, p12, p13, p14) and r0 ~ Multinomial (n0, p01, p02, p03, p04), respectively. OR11 = (p01 p14)/(p04p11) denotes the odds ratio for individuals with G = 1 and E = 1 in relation to those baseline subjects with G = 0 and E = 0. OR01 = (p01p13)/(p03p11) signifies the G-D odds ratio among the unexposed, capturing the main effect of G, and OR10 = (p01 p12)/(p02p11) represents the E-D association among the genetically non-susceptible, capturing the main effect of E. The multiplicative interaction parameter under study is represented by OR11/OR10OR01. We generate the control frequency vector first using given values of the prevalence of G, prevalence of E and odds ratio of gene-environment association in controls, namely ORge. Using the control probability vector and given values of the main effect and interaction parameters, the case frequency vector is then generated [7].
Table 1.
Data generated from an unmatched case-control study with binary genetic and environmental factors.
| G=0 | G=1 | ||||
|---|---|---|---|---|---|
| E=0 | E=1 | E=0 | E=1 | ||
| D=0 | r01 | r02 | r03 | r04 | n0 |
| D=1 | r11 | r12 | r13 | r14 | n1 |
Tests for G×E interaction
The three tests for G×E interaction employed here have been thoroughly described in the literature. In brief, we utilize the case-control (CC) approach leveraging simple logistic regression, maximum likelihood estimation, and a standard Wald test for the null hypothesis of βg×e = 0. In addition, we conduct a case-only (CO) analysis using a retrospective likelihood-based test that requires an assumption of gene-environment (G-E) independence. Finally, we employ the Empirical Bayes (EB) method which provides a trade-off between bias and efficiency of the previous two approaches (CC and CO) using a shrinkage estimator with adaptive weights attached to the CC and CO estimators. The adaptive weights for the EB estimator are identical to the one proposed in [7], with more weight towards CC if gene-environment independence is violated. Thus, the EB estimator is less susceptible to bias and Type 1 error than a CO estimator if there is gene-environment dependence.
Joint test for marginal genetic (or environmental) association and G ×E interaction
We employ the recently published approach that combines the use of two models (1 and 2 below) to test the marginal effect of G (or E) and the G×E interaction [3]. The key differences between this method and a two degrees of freedom (df) likelihood ratio test for H0 : βg = βg×e = 0 for G (or H0 : βe = βg×e = 0 for E) as proposed in [4] are that this approach tests for the marginal association of G (or E), as in (1), rather than the main effect in a full G×E model (2), and for the interaction component it takes advantage of possible G-E independence when testing for G×E interaction through extension to the CO and EB approaches [3]. Let us consider the two models that yield the components of the joint tests that are considered in [3]:
| (1) |
| (2) |
The composite hypothesis being tested for G is H0 : αg= 0 and H0 : βg×e = 0; and the composite hypothesis for E is H0 : αe = 0 and H0 : βg×e = 0. Now, βg×e can be estimated using any of the three methods described above (CC, CO, or EB). The Wald test χ2 statistics for the marginal association tests in (1) and interaction tests in (2) are independent [3]. Thus, adding the two test statistics yields a 2 df χ2 test statistic of one of the following 3 forms (depending on which test is used for the interaction part: CC, CO or EB) :
We will study effect of exposure-biased designs in conjunction with potential misclassification on these three analytical strategies. Recall that CC does not use G-E independence, CO assumes it to be true, and EB adaptively uses it without making a blanket assumption like CO.
Exposure-enriched sampling designs
Following generation of data for the underlying case-control study base, we consider three alternative sub-sampling schemes with differential enrichment for exposed subjects. As the genotyping budget is often limited, we assume a hypothetical scenario that resources are available to obtain genetic data on only half of the cases and controls in the original study base.
-
-
Scheme 1: select 50% of cases and 50% of controls from the full 2×4 table, preferentially including all exposed subjects (both cases and controls) and a random sample of unexposed subjects to reach the genotyping capacity. Since same rate of exposure enrichment is assumed for cases and controls, this design will not generate significant bias for exposure effect estimates, in spite of exposure enrichment.
-
-
Scheme 2: genotype 50% of cases and 50% of controls, including all exposed cases, a fixed percentage of exposed controls (50%, 70%, or 90%), and a random sample of unexposed individuals to reach the genotyping capacity. Note that differential sampling rates across cases and controls will generate bias in the estimate of the exposure main effect as well as G ×E interaction term.
-
-
Scheme 3: randomly sample 50% of cases and 50% of controls for genotyping irrespective of their exposure status. This scheme represents the traditional sampling design with no exposure enrichment but may lack power for testing G×E interaction.
Simulation settings
The base case simulates 5000 cases and 5000 controls as the underlying case-control study base, from which a subsample of 2500 cases and 2500 controls are drawn for genotyping and inclusion in the interaction study. We assume that the prevalence of exposure (P(E)) is 0.1 (relatively rare) and that the risk allele frequency for G (a single marker) is relatively common at 0.2. The true interaction odds ratio parameter (ORg×e) is varied from the null value of 1.0 up to 1.8. Further, for this base case we assume that gene-environment independence holds and that perfect exposure information is available. The main effect of G (OR01 = ORg = exp(βg)) and the main effect of E (OR10 = ORe = exp(βe)) are both set to 1. We examine the relative performance of the G×E and joint effect detection methods under each sampling design with respect to power (the probability of rejecting the null hypothesis), type I error, mean squared error (MSE), and bias in parameter estimation. Biases are calculated with respect to deviation from the true marginal effects αg, αe, and the interaction effect βg×e. The reported results are based on 5000 simulated datasets. Subsequently, the following sensitivity analyses are conducted for the perfect exposure measurement scenario by deviating from the base case. We vary the sample size of the underlying original study base down to 2000 cases and 2000 controls, the prevalence of exposure up to 0.3, ORg up to 1.3, and ORe up to 1.5. Also, we examine the impact of gene-environment independence violations by varying the gene-environment association parameter, ORge, from 1.0 down to 0.8 and up to 1.1 (Supplementary Material).
Exposure misclassification (MC)
While we first test the three sampling schemes under the assumption of perfect exposure information, we then examine the impact of exposure MC on the performance of these designs. When MC is considered, the underlying study sample is generated in exactly the same way as described above and includes 5000 cases and 5000 controls. The true exposure (E) is generated first, followed by the contaminated exposure (E) based on the following probabilities associated with exposure measurement:
P (E=1 | E=1) = sensitivity (SE)
P (E=1 | E=0) = 1-specificity (SP)
Note that this generates non-differential misclassification because we do not consider disease status in the misclassification model. Then, with a sub-sampling rate of 50% (i.e. 50% of cases and 50% of controls are selected for each sampling scheme), the final number of genotyped samples is 2500 cases and 2500 controls. Nine combinations of SE and SP parameters are assessed, with individual values ranging from 0.6–1.0. Because disease diagnosis may increase sensitivity and reduce specificity of recalling exposure history in cases as compared to controls, we also consider a situation of differential misclassification, where SE=1.0 and SP=0.6 among cases and SE=0.8 and SP=0.8 among controls [16]. Subsequently, the enriched sampling designs based on the observed misclassified exposures are employed, followed by interaction and joint effect testing as before.
RESULTS
Exposure-biased sampling with perfect exposure information
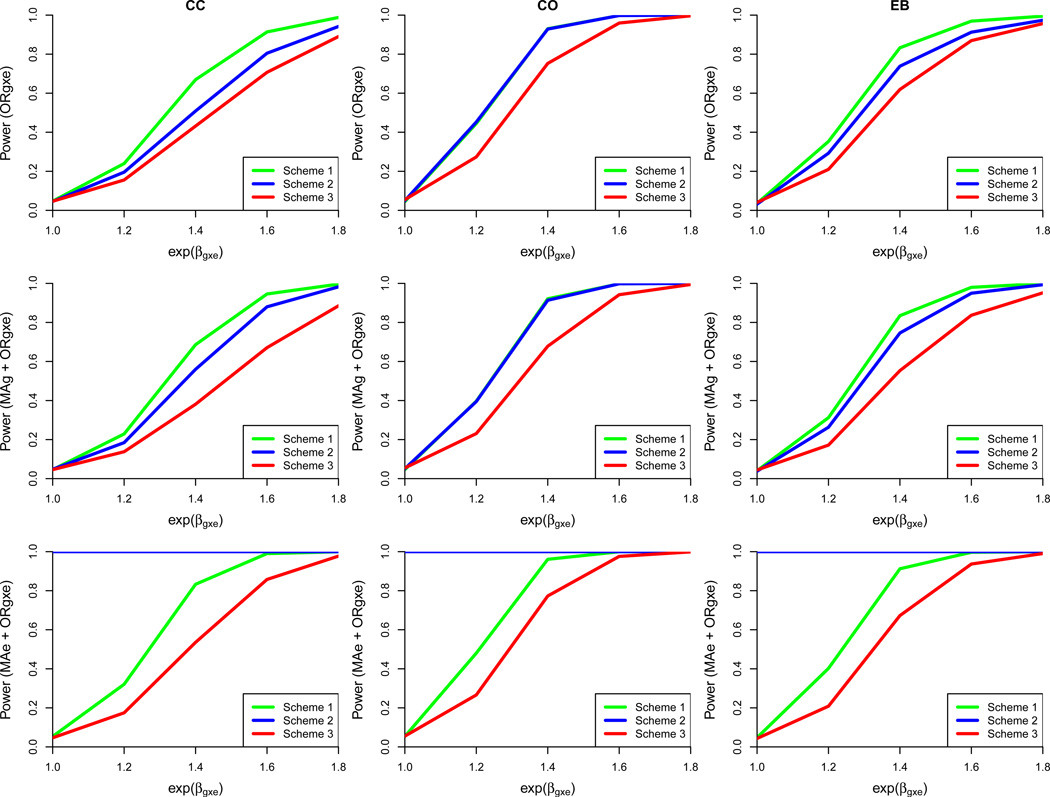
Overall, the base case with 2500 cases and 2500 controls sampled from among 5000 cases and 5000 controls, an exposure prevalence of 10%, and G-E independence demonstrates that exposure-enriched sampling increases power but yields bias in estimation of the interaction OR and the marginal effects of G and E (Figure 1; Supplementary Table 1) under the alternative hypothesis. Below, results for each of the proposed sampling schemes are presented individually, followed by a direct comparison of the designs.
Figure 1.
Power comparison of three exposure-biased sampling designs under the base case with perfect exposure measurement. Each row represents one of three tests: G×E interaction (ORg×e), joint marginal G and G ×E interaction (MAg + ORg×e), or joint marginal E and G×E interaction (MAe + ORg×e). Each column designates one of three different approaches for ORg×e (CC = case-control, CO = case-only, EB = Empirical Bayes). Based on 5000 simulated datasets, 2500 cases and 2500 controls, genotype information on 1 marker, ORg = ORe = 1, an exposed control subsampling rate of 0.5 for Scheme 2, and gene-environment independence. Scheme 1: Genotype 50% of cases and 50% of controls including all E = 1 and a random sample of E = 0. Scheme 2: Genotype 50% of cases and 50% of controls including all exposed cases, 50% of exposed controls, and a random sample of E = 0. Scheme 3: Randomly sample 50% of cases and 50% of controls for genotyping, irrespective of exposure status.
Scheme 1
Scheme1, with all exposed cases and controls genotyped as well as a random sample of unexposed subjects, performs quite well with respect to power across the CC, CO, and EB methods when testing only for the interaction as well as joint tests (Figure 1). Over 80% power can be achieved by using one of the three interaction detection methods described for the ORg×e, joint MAg + ORg×e, and joint MAe + ORg×e tests once the true ORg×e reaches 1.4. Type I error is tightly controlled across all tests. A small amount of bias is present for estimating the marginal effects of G and E, but the estimate of ORg×e remains relatively unbiased (Supplementary Table 1).
Scheme 2
In Scheme 2, 50% of 5000 cases and 50% of 5000 controls are genotyped according to the following distribution: all exposed cases, a predefined percentage of exposed controls, and a random sample of all unexposed subjects to fill the remaining samples. Scheme 2 performs increasingly similar to Scheme 1 as the proportion of exposed control samples selected for genotyping increases. Thus, only the lowest proportion of 0.5 that was considered is discussed below. When the sampling proportion of exposed controls is set to 0.5, power for detecting the interaction OR and joint effect of G across the range of ORg×e values considered is adequate, with the CO method yielding the greatest power at the lowest effect size among the three approaches (Figure 1). This scheme is always able to detect a marginal effect of E due to the sampling design, even under the null hypothesis of H0 : αe = 0 and H0 : βg×e = 0. Type I error for the ORg×e and joint effect of G tests are well-controlled under this scheme, hovering around the 0.05 level. Bias in ORg×e measurement is essentially nonexistent, but bias in MAg measurement is present at a modest level, and the downward bias for MAe is quite substantial (Supplementary Table 1).
Scheme 3
Scheme 3 randomly samples 50% of cases and 50% of controls, irrespective of exposure status. This scheme yields the lowest power for the interaction and joint tests as compared to the other two designs, not achieving 80% power by at least one of the CC, CO, or EB methods until the true interaction OR reaches 1.6 (Figure 1). Again, the case-only approach is the most powerful method employed in all three ORg×e and joint tests. Type I error is steadfast near 0.05 across all tests and detection methods. This scenario remains unbiased for detecting the interaction effect and the marginal effects of G and E as expected (Supplementary Table 1).
Sampling Design Comparison
In the settings for this scenario with gene-environment independence being true, case-only analysis is unbiased under random sampling of cases and perfect exposure data. Thus, any source of bias for this G×E detection method can be directly attributed to the exposure-enriched sampling scheme. Scheme 1 performs consistently the best with respect to power across all 3 methods for the ORg×e and joint G tests (Figure 1). Also, Scheme 1 yields low bias in estimating ORg×e because the sampling fraction is the same for cases and controls. However, it does yield slightly greater bias in estimation of the MAg, and MAe parameters as compared to Scheme 3, which employs completely random sampling. MSE is low for ORg×e, MAg, and MAe and comparable across all 3 sampling designs (Supplementary Table 1). While Scheme 2 appears to yield the greatest power for detecting the joint effect of E, this result is potentially driven by Type I error. Across all scenarios in the base case, the CO approach is the most powerful for detecting ORg×e. When the main effect of G is increased to 1.3, power for detecting the interaction OR and the joint effect of E increases marginally across all designs. However, as expected, power for detecting the joint effect of MAg plus the interaction ORg×e increases dramatically. Similarly, when the main effect of E (ORe) is increased to 1.5, power for detecting ORg×e and the joint effect of G experience minimal boosts across all schemes but the detection rates of E effect increase substantially.
Alternative settings were explored to examine the impact of different parameter values on power and bias as compared to the base case. If the prevalence of exposure is increased to 30%, there is very little separation between the three sampling schemes with respect to power or bias for the interaction or joint tests (Supplementary Table 2). The CO approach remains the most powerful when the G-E independence assumption still holds, and the relative performance of the 3 sampling schemes remains the same. When the sample size is decreased to 2000 cases and 2000 controls (prior to genotyping 50% of the subjects), there is an overall depression of power across all 3 methods, and the differential across sampling schemes is exacerbated (Supplementary Table 3). Gene-environment dependence yields greater bias in estimation of ORg×e for CO and EB approaches but does not affect the relative performance of the 3 exposure-enriched sampling designs with respect to power (Supplementary Tables 4 and 5). The CO approach suffers the most, but this phenomenon under violation of gene-environment independence has been well-characterized in the literature.
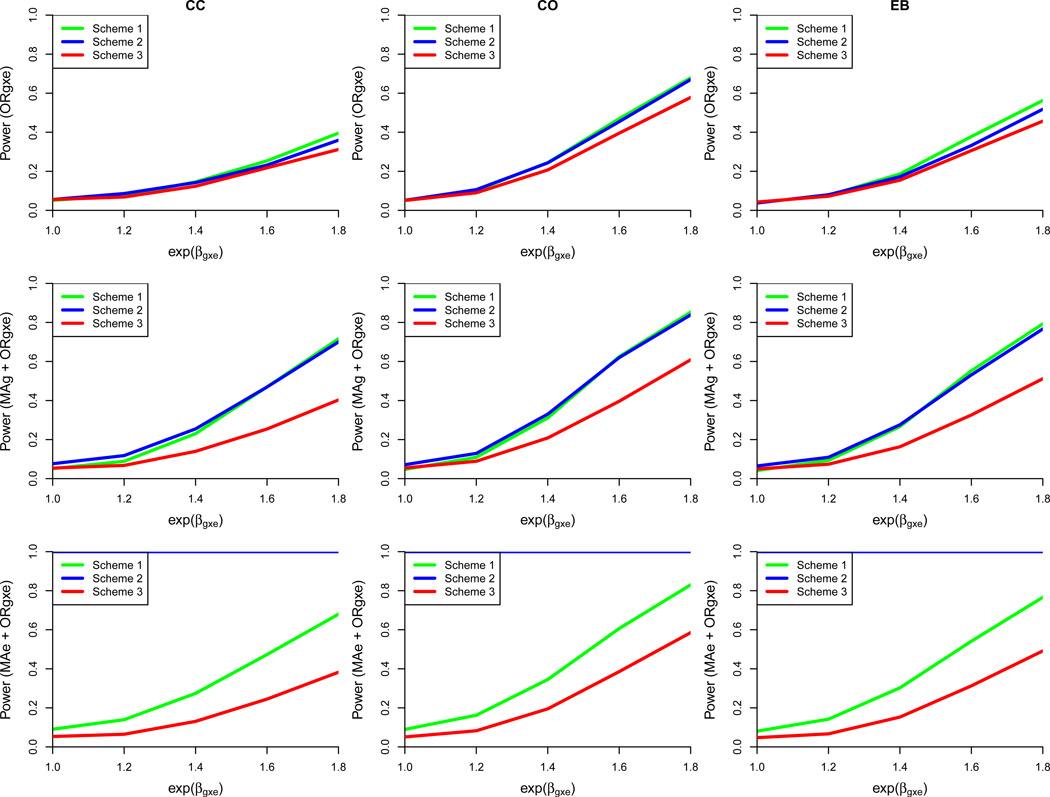
Exposure-biased sampling based on misclassified exposure information
The same exposure-biased sampling schemes were reexamined under the scenario of imperfect exposure information across a range of SE and SP values. Here, the base case includes 5000 cases and 5000 controls with prevalence of exposure set to 10%, ORg = ORe = 1, and scheme 2 sub-sampling rate of exposed control subjects set to 50%. We present results for non-differential misclassification in detail, followed by a brief commentary on the relative performance of sampling designs when exposure misclassification is differential by disease status.
Scheme 1
This design with the strongest exposure-enriched sampling experiences a severe depression in power for detecting G×E interaction across the CC, CO, and EB methods, especially for the anticipated modest effect sizes (Supplementary Table 6). As expected, at each combination of SE and SP, power increases as the true ORg×e increases (Supplementary Table 7). The CO approach is the most powerful at all combinations of SE and SP, but it achieves only 68% power with SE and SP of 0.8 when the true interaction OR is 1.8. For this same effect size and SE-SP configuration, CC only has power of 40%, and it is only when SE ≥ 0.6 and SP = 1 that CC achieves power greater than 90%. The EB approach yields intermediate power results that reflect its hybrid nature between the CO and CC methods. A decrease in SP depresses power to a greater extent than an identical decrease in SE. Power for the joint test of a marginal effect of G (or E) and the G×E interaction on disease status is also negatively impacted by MC but is affected to a slightly lesser extent than the interaction-only test. Type I error is tightly controlled for all 3 methods of detecting G×E interaction across the range of sensitivities, specificities, and true ORg×e values. This is to be expected as SP and SE do not make an impact under the null hypothesis under this misclassification scheme.
Supplementary Table 7 also demonstrates that the reduction of SE and SP leads to increasingly biased estimates of ORg×e and the marginal effect of E (MAe; the marginal effect of G, MAg, is minimally biased). Type I error for both joint tests is 0.05 or below across all SE, SP, and true ORg×e ranges.
Scheme 2
Scheme 2 also experiences a decrease in power for detecting the stand-alone G×E interaction and the joint effect of G plus G×E interaction in the presence of exposure MC (Supplementary Table 8). For the interaction effect, the CO analysis still performs best in the base case, and the CC and EB approaches are influenced to a greater extent by the MC. Power for detecting the joint effect of G and G ×E is less affected by a decrease in SE than SP. Type I error is reasonably controlled for the ORg×e test even at low SP and SE combinations but becomes slightly inflated for the joint test for G across all 3 methods. The joint test for E in this design is dominated by sampling bias reflected in type I error. The reduction of SE and SP leads to more positively biased estimates of ORg×e and extreme negatively biased estimates of MAe; again, the estimate of MAg is mostly unaffected.
Scheme 3
This design which is blind to exposure status also suffers in terms of power when SP and SE of exposure measurement drop below one (Supplementary Table 9). The test for interaction is severely impacted, to a greater extent than either of the joint tests for a given combination of SE and SP. The power achieved for detecting a true interaction parameter of 1.8 is a maximum of 0.58 when SE = SP = 0.8 and all three of the CC, CO, and EB approaches are considered (Supplementary Table 9). This scheme yields very tightly controlled type 1 error for the interaction and joint G (or E) tests across all 3 methods for detecting ORg×e. Bias in ORg×e estimation increases rapidly as SP of exposure measurement drops. There is no bias in MAg estimation, but there exists modest bias in MAe, particularly as SE and SP approach 0.6 and the true interaction parameter nears 2.0.
Sampling Design Comparison
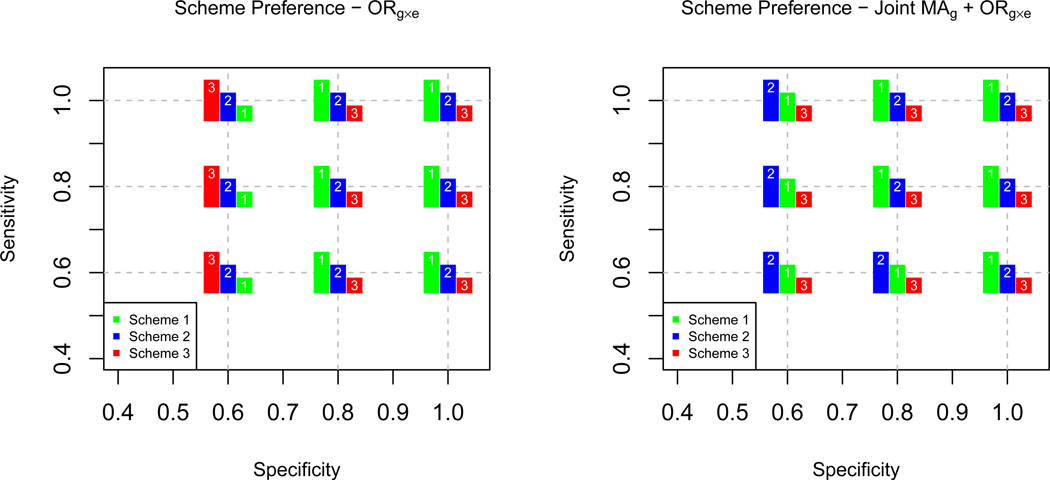
As SP and SE decrease, power decreases across all three tests and methods for ORg×e detection for all 3 sampling designs (Supplementary Tables 7–9). Figure 2 visually summarizes the power comparison across sampling designs when SP and SE are both set to 0.8 (Supplementary Table 6). The apparent benefit of Scheme 1 that was demonstrated in the presence of perfect exposure information does not carry over when exposure is misclassified. There is no uniformly most powerful design across the range of true interaction parameter values for the interaction-only test (Figure 3). For detecting ORg×e, Scheme 1 overtakes Scheme 2 as the most powerful around a true OR of 1.4. Scheme 2 performs marginally better than the other two designs across the range of true values below 1.4 down to 1.0. For the joint effect of G, Scheme 2 outperforms Scheme 1 with respect to power at more modest true ORg×e levels. Interestingly, for all 3 scenarios, the joint test for G is not always more powerful than the marginal test for G. Among the values examined, the joint test for G is more powerful than the marginal test for G only when SP ≥ 0.8 and SE ≥ 0.6 (Schemes 1 and 2) and when SP ≥ 0.6 and SE ≥ 0.8 (Scheme 3). While Scheme 2 also appears to yield the greatest power for detecting the joint effect of E, this result is likely driven by Type I error. Ignoring Scheme 2, Scheme 1 achieves higher power than Scheme 3 regardless of the ORg×e detection method chosen. Across all scenarios in the base case, the CO approach has the potential to achieve the most power for detecting ORg×e and the joint effect of G (or E).
Figure 2.
Power comparison of three exposure-biased sampling designs under the base case with exposure misclassification (sensitivity = 0.8; specificity = 0.8). Each row represents one of three tests: G ×E interaction (ORg×e), joint marginal G and G×E interaction (MAg + ORg×e), or joint marginal E and G ×E interaction (MAe + ORg×e). Each column designates one of three different approaches for estimating the G ×E interaction parameter (CC = case-control, CO = case-only, EB = Empirical Bayes). Based on 5000 simulated datasets, 2500 cases and 2500 controls, genotype information on 1 marker, ORg = ORe = 1, an exposed control subsampling rate of 0.5 for Scheme 2, and gene-environment independence. Scheme 1: Genotype 50% of cases and 50% of controls including all E = 1 and a random sample of E = 0. Scheme 2: Genotype 50% of cases and 50% of controls including all exposed cases, 50% of exposed controls, and a random sample of E = 0. Scheme 3: Randomly sample 50% of cases and 50% of controls for genotyping, irrespective of exposure status.
Figure 3.
Sampling scheme preference with respect to power maximization across different specificity (SP) and sensitivity (SE) combinations. The Empirical Bayes method was used to estimate ORg×e. Based on 5000 simulated datasets, 2500 cases and 2500 controls, genotype information on 1 marker, gene-environment independence, ORg = ORe = 1.0, and an exposed control subsampling rate of 0.5 for Scheme 2. Scheme 1: Genotype 50% of cases and 50% of controls including all E = 1 and a random sample of E = 0. Scheme 2: Genotype 50% of cases and 50% of controls including all exposed cases, 50% of exposed controls, and a random sample of E = 0. Scheme 3: Randomly sample 50% of cases and 50% of controls for genotyping, irrespective of exposure status.
Again focusing on the scenario where SE = SP = 0.8, type I error is tightly controlled for all tests when employing Schemes 1 or 3, with slight inflation for Scheme 1’s joint test for E (Supplementary Table 6). Type I error is marginally inflated for the joint G test and drastically inflated for the joint E test when Scheme 2 sampling is conducted (Supplementary Table 6). Decreasing SP introduces a large bias in ORg×e estimation across all three schemes (Supplementary Tables 7–9). However, the bias in MAe is more substantial for Scheme 2 than for either or Schemes 1 or 3, which demonstrated relatively comparable bias in estimation of this parameter. There is minimal bias in MAg estimation in Schemes 1 and 2 and none for Scheme 3. Like the scenario of perfect exposure information, MSE for MAg and MAe estimation is minimal across all 3 sampling designs. However, MSE for ORg×e is quite severely penalized as SP and SE drop. The impact of gene-environment independence violations on the above observations can be visualized in Supplementary Figures 1 (ORge=0.8) and 2 (ORge=1.1).
Sampling Design Comparison with Differential Misclassification of Exposure (data not shown)
Similar to the scenario of non-differential misclassification with SE = 0.8 and SP = 0.8 in both cases and controls, the base case with differential misclassification (SEcase = 1.0; SPcase = 0.6; SEcontrol = SPcontrol = 0.8) shows a loss of Scheme 1’s power advantage. Scheme 3 now performs best with respect to power for detecting the interaction-only effect. For the joint effect of G, Schemes 1 and 2 achieve comparable power across the range of ORg×e parameters considered. Type I error remains tightly controlled for all sampling designs across all tests except for the joint test for E. Bias in ORg×e estimation is slightly more severe than in the case of non-differential misclassification, but the levels of bias are still comparable across sampling designs. The bias in MAe is substantially greater for differential as compared to non-differential MC, but Scheme 2 remains the sampling design with the most bias. If gene-environment independence is violated (ORge = 0.8) in addition to differential MC, the relative performance of sampling schemes remains the same as when ORge = 1.0.
DISCUSSION
This simulation study characterizes the benefits and limitations of various exposure-enriched sampling designs for detecting G×E interactions or joint genetic (or environmental) effects in unmatched case-control studies. It also considers biased sampling in the context of exposure misclassification. In the absence of exposure misclassification, a design that samples all exposed individuals (Scheme 1) is an optimal approach with respect to power maximization while yielding tolerable levels of bias in estimation for both the interaction parameter and marginal G (or E) effects. Further, type I error is controlled under this enriched sampling scheme. In addition to the test for pure G×E interaction which has been studied in the past, we characterize the impact of different exposure-biased sampling designs on simultaneous testing when both a marginal genetic (or environmental) effect and gene-environment interaction are of interest. These joint tests are particularly useful in that they may increase power for detecting the presence of a genetic (or environmental) effect in the presence of G ×E interaction. With perfect exposure information, an exposure-enriched sampling design further enhances the power for detecting the joint effect of G in presence of non-null interaction.
It is important to recognize that these advantages of exposure-enriched sampling and the comparison between sampling designs do not hold uniformly under all conditions that vary from the base case. First, the power advantages are accentuated when the exposure is relatively rare. When the environmental factor is more common, exposure-biased sampling designs become less distinguishable from random sampling. Further, exposure misclassification is a common problem plaguing epidemiologic studies. Thus, we sought to address this practical limitation by examining the impact of exposure MC on the relative performance of the three sampling designs. Although Scheme 1 is ideal under perfect exposure measurement conditions, in the situation of imperfect information, we demonstrate that no single exposure-biased sampling scheme is consistently the optimal design. The preferred approach in terms of power depends on the strength of the anticipated interaction OR, which method of ORg×e detection is used (CC, CO, or EB), whether interest is in the interaction itself or in boosting power to detect any genetic effect that exists (an emphasis on the joint test for G and G×E interaction), and whether misclassification is differential or non-differential.
Although power is a key concern for G ×E interaction studies, bias in parameter estimation is another important element when evaluating these statistical methods. Previous work has shown that differential exposure MC by disease status does not lead to bias in ORg×e estimation when a multiplicative interaction truly exists if exposure SE and SP measures are non-differential by genotype and G-E independence is present among controls [13]. We show here that bias in estimating the ORg×e parameter does not exist in the presence of exposure-enriched sampling when exposure information is perfect. However, in the situation of exposure MC (differential and non-differential), bias in ORg×e estimation appears across all sampling designs as the true interaction OR increases. With respect to marginal effect estimation, there is substantial bias for MAe without exposure misclassification for the design with unequal proportions of exposed cases and controls (Scheme 2) and for all 3 designs when exposure MC occurs. Thus, investigators conducting G ×E interaction studies who are considering an exposure-enriched sampling scheme should be cognizant of this potential increase in bias when exposure is difficult to measure accurately. MC is a well-acknowledged problem that is not often considered seriously in the design stage.
This study has several limitations that could be addressed in future work. First, this analysis focuses on a single genetic marker. Often, G×E interaction studies are conducted in the context of post-GWAS analyses, where a multitude of candidate markers are of interest. As multiple testing rapidly increases, the power simulated in this study becomes severely penalized. However, even for a single marker, it would be interesting to consider if other genetic models or rarer genetic variation make any impact on the relative performance of different sampling schemes. In addition, we do not consider here the misclassification of G in addition to imperfect exposure information. This misclassification is possible given that genotyping arrays have associated error rates, and other studies have considered the impact of such errors [20, 21]. Further studies may investigate in greater depth the influence of both G and E MC on the relative performance of different sampling designs. Also, we consider only a binary environmental exposure, but many such exposures are ordinal or continuous in practice. Furthermore, our focus here is on case-control studies, but it would be useful to examine the impact of exposure-biased sampling on other types of study designs.
Although we demonstrate that exposure MC distorts the benefit of using an exposure-biased design, the investigator is typically unaware of the extent to which an epidemiologically-assessed environmental exposure is misclassified. This makes it difficult to differentiate if Scheme 1 will have clear advantages. One solution may be to estimate the SE and SP of exposure measurement using a validation sub-study [22, 23]. However, even if no validation sample exists to estimate SE and SP, we have demonstrated that knowledge of a range for the SP and SE parameters can assist in choosing the optimal sampling design for that range. When considering the maximization of power, Scheme 2 has benefits over Scheme 1 under the scenario of modest non-differential exposure MC when anticipated effect sizes are low. On the other hand, Scheme 3 remains consistently unbiased but can severely lack power with perfect exposure. However, contrary to expectations, Scheme 3 has the power advantage when exposure MC is differential by disease status. In addition, some have proposed that correction for the effect of exposure MC is possible [23–25]. Some have presented solutions to handle measurement errors in environmental factors under genome-wide G×E interaction scans. Lobach et al [26] proposed a pseudo- likelihood model in which the environmental covariate is non-parametrically modeled and its dependence on disease-status is also considered. Alternatively, Lobach et al [27] have illustrated a semi-parametric Bayesian approach where the potentially skewed inferences on the risk parameters due to measurement errors are handled through the symmetric normality prior on the risk parameter. While not a replacement for better exposure measurement, it would be worthwhile to learn if these correct ion methods restore the advantages of exposure-biased sampling that are visible with perfect exposure information.
Overall, results from this study provide rigorous support for a method of prioritizing subjects for genotyping. Overall, exposure enrichment appears to be a beneficial strategy for maximizing available resources for G×E interaction detection under the scenario of perfect exposure information. Genetic epidemiology studies are expensive to design and implement, so this strategy that boosts power on a fixed budget is quite advantageous. However, those conducting G×E studies should be careful to consider exposure measurement MC properties and the prevalence of exposure when choosing an ideal sampling scheme and method for G×E interaction detection. Finally, it is important to recognize that no matter how powerful the test, any G×E interactions that are identified require validation and replication, which has proven to be challenging due to differences in the distribution of both genetic and environmental exposure across populations.
Supplementary Material
Acknowledgments
SOURCES OF FINANCIAL SUPPORT
Support for this study was provided by National Science Foundation DMS 1007494, National Institutes of Health ES 20811, National Institutes of Health CA 156608, and National Institutes of Health CA 148107. Funding for SLS was provided by the National Human Genome Research Institute at the National Institutes of Health (T32 HG00040), the National Institute of Environmental Health Sciences at the National Institutes of Health (T32 ES013678), and a fellowship from the University of Michigan Rackham Graduate School. Funding for PB and BM was partially provided by the University of Michigan Cancer Center Support Grant NIH P30 CA 046592.
NOTATION
- CC
case-control
- CO
case-only
- EB
empirical Bayes
- G
genetic variant
- E
environmental exposure
- D
disease/outcome status
- αg
marginal log-odds ratio associated with the genetic factor
- MAg
marginal genetic association
- αe
marginal log-odds ratio associated with the environmental factor
- MAe
marginal environmental association
- ORge
odds ratio for the association between the genetic and environmental variables in controls
- βg
main effect log-odds ratio associated with the genetic factor
- ORg
exp(βg)
- βe
main effect log-odds ratio associated with the environmental factor
- ORe
exp(βe)
- βg×e
gene by environment interaction log-odds ratio
- ORg×e
exp(βg×e)
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Hunter DJ. Gene-environment interactions in human diseases. Nature reviews Genetics. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 2.Thomas D. Gene-environment-wide association studies: emerging approaches. Nature reviews Genetics. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai JY, Logsdon BA, Huang Y, et al. Simultaneously testing for marginal genetic association and gene-environment interaction. American journal of epidemiology. 2012;176:164–173. doi: 10.1093/aje/kwr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Human heredity. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 5.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Statistics in medicine. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee N, Carroll RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92:399–418. [Google Scholar]
- 7.Mukherjee B, Chatterjee N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade-off between bias and efficiency. Biometrics. 2008;64:685–694. doi: 10.1111/j.1541-0420.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee B, Ahn J, Gruber SB, Chatterjee N. Testing gene-environment interaction in large-scale case-control association studies: possible choices and comparisons. American journal of epidemiology. 2012;175:177–190. doi: 10.1093/aje/kwr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oexle K, Meitinger T. Sampling GWAS subjects from risk populations. Genetic epidemiology. 2011;35:148–153. doi: 10.1002/gepi.20562. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Kang G, Vanderweele T, Zhang C, Mukherjee B. Efficient designs of gene-environment interaction studies: implications of Hardy-Weinberg equilibrium and gene-environment independence. Statistics in medicine. 2012;31:2516–2530. doi: 10.1002/sim.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Closas M, Rothman N, Lubin J. Misclassification in case-control studies of gene-environment interactions: assessment of bias and sample size. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8:1043–1050. [PubMed] [Google Scholar]
- 12.Rothman N, Garcia-Closas M, Stewart WT, Lubin J. The impact of misclassification in case-control studies of gene-environment interactions. IARC scientific publications. 1999:89–96. [PubMed] [Google Scholar]
- 13.Garcia-Closas M, Thompson WD, Robins JM. Differential misclassification and the assessment of gene-environment interactions in case-control studies. American journal of epidemiology. 1998;147:426–433. doi: 10.1093/oxfordjournals.aje.a009467. [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom S, Yen YC, Spiegelman D, Kraft P. The impact of gene-environment dependence and misclassification in genetic association studies incorporating gene-environment interactions. Human heredity. 2009;68:171–181. doi: 10.1159/000224637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll RJ, Gail MH, Lubin JH. Case-control studies with errors in covariates. Journal of the American Statistical Association. 1993;88:185–199. [Google Scholar]
- 16.Rothman KJ, Greenland S, Lash TL. Section II: Study Design and Conduct. Modern Epidemiology. (Third Edition ed) 2008;111:38. [Google Scholar]
- 17.Breslow NE, Chatterjee N. Design and analysis of two-phase studies with binary outcome applied to Wilms tumour prognosis. Journal of the Royal Statistical Society: Series C (Applied Statistics) 1999;48(4):457–168. [Google Scholar]
- 18.Lee AJ, Scott AJ, Wild CJ. Efficient estimation in multi-phase case-control studies. Biometrika. 2010;97(2):361–374. [Google Scholar]
- 19.Lumley T. Package Survey, Version 3.2.4. 2011. R for analyzing data from complex surveys. [Google Scholar]
- 20.Cheng KF. Analysis of case-only studies accounting for genotyping error. Annals of human genetics. 2007;71:238–248. doi: 10.1111/j.1469-1809.2006.00314.x. [DOI] [PubMed] [Google Scholar]
- 21.Wong MY, Day NE, Luan JA, Wareham NJ. Estimation of magnitude in gene-environment interactions in the presence of measurement error. Statistics in medicine. 2004;23:987–998. doi: 10.1002/sim.1662. [DOI] [PubMed] [Google Scholar]
- 22.Greenland S. Statistical uncertainty due to misclassification: implications for validation substudies. Journal of clinical epidemiology. 1988;41:1167–1174. doi: 10.1016/0895-4356(88)90020-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Mukherjee B, Ghosh M, Gruber S, Moreno V. Accounting for error due to misclassification of exposures in case-control studies of gene-environment interaction. Statistics in medicine. 2008;27:2756–2583. doi: 10.1002/sim.3044. [DOI] [PubMed] [Google Scholar]
- 24.Rice K. Full- likelihood approaches to misclassification of a binary exposure in matched case-control studies. Statistics in medicine. 2003;22:3177–3194. doi: 10.1002/sim.1546. [DOI] [PubMed] [Google Scholar]
- 25.Spiegelman DRB, Logan R. Estimation and inference for logistic regression with covariate misclassification and measurement error, in main study/validation study designs. Journal of the American Statistical Association. 2000;95:51–61. [Google Scholar]
- 26.Lobach I, Fan R, Carroll RJ. Genotype-based association mapping of complex diseases: gene-environment interactions with multiple genetic markers and measurement error in environmental exposures. Genetic epidemiology. 2010;34:792–802. doi: 10.1002/gepi.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobach I, Mallick B, Carroll RJ. Semiparametric Bayesian analysis of gene-environment interactions with error in measurement of environmental covariates and missing genetic data. Statistics and its interface. 2011;4:305–316. doi: 10.4310/sii.2011.v4.n3.a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.