Summary
Cellular microenvironments established by the spatial and temporal expression of specific signaling molecules are critical for both the maintenance and lineage-specific differentiation of progenitor cells. In Drosophila, a population of hematopoietic progenitors, or prohemocytes, within the larval lymph gland [1] gives rise to three mature cell types: plasmatocytes, lamellocytes, and crystal cells. Removal of the secreted signaling molecules Hedgehog [2] and PVF1 [3] from the Posterior Signaling Center (PSC)[2, 4, 5] which acts as a niche, leads to a loss of progenitors and complete differentiation of the lymph gland. Here, we characterize a novel population of signaling cells within the lymph gland, distinct from the PSC, that are required for lineage specific differentiation of crystal cells. We provide evidence that Yorkie[6] and Scalloped[7], the Drosophila homologues of YAP and TEAD, are required in Lineage Specifying Cells to regulate expression of Serrate, the Notch ligand responsible for the initiation of the crystal cell differentiation program[8] [5]. Genetic manipulation of yorkie and scalloped in the lymph gland specifically alters Serrate expression and crystal cell differentiation. Furthermore, Serrate expression in Lineage Specifying Cells is eliminated in the Lymph Gland upon the immune response induced by wasp parasitization to ensure the proper differentiation of lamellocytes at the expense of crystal cells. These findings expand the roles for Yorkie/Scalloped beyond growth to encompass specific cell fate determination in the context of blood development. Similar regulatory functions may extend to their homologues in vertebrate progenitor cell niches that are required for specifying cell fate.
Results and Discussion
Yorkie and Scalloped are required for crystal cell formation in the lymph gland
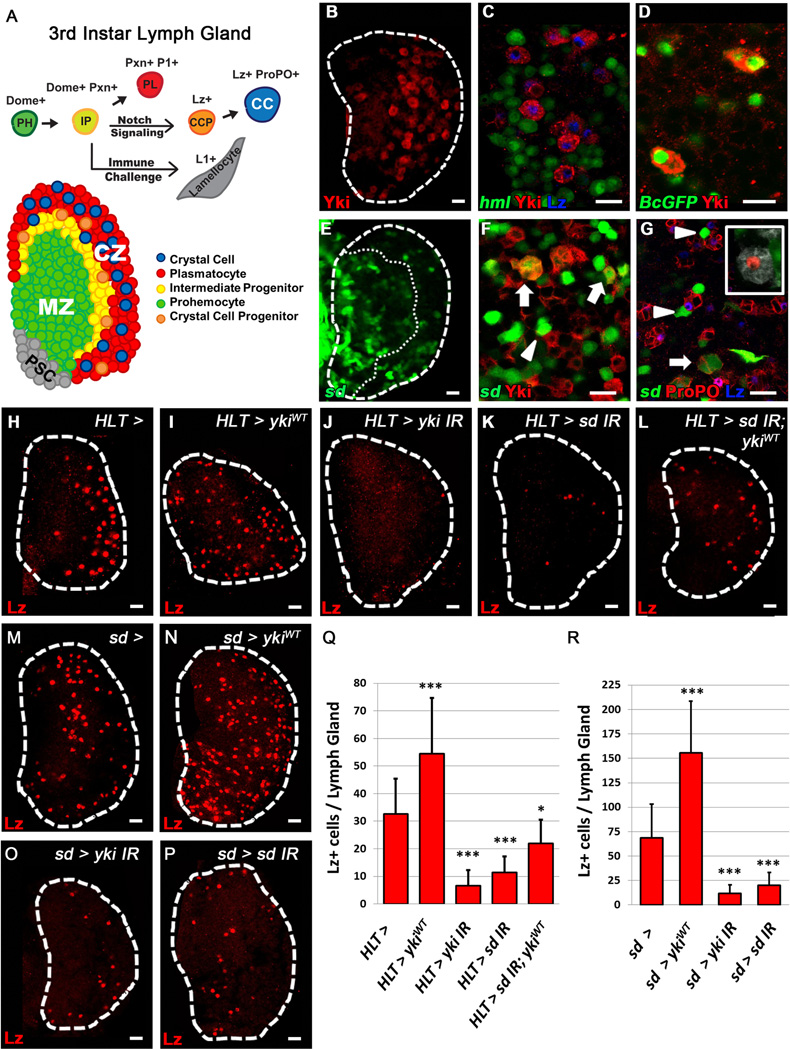
Differentiating hemocytes in the lymph gland (LG) are restricted to the periphery or Cortical Zone (CZ) of the organ (Fig. 1A). These hemocytes originate from a population of progenitors termed prohemocytes (PH) located in the Medullary Zone (MZ, Fig 1A) that are maintained by the PSC (Fig1A). PHs transition through an intermediate progenitor (IP) [9] state (Fig. 1A) where they express both progenitor (dome+) and early differentiation (Pxn+) markers [10]. These IPs will eventually fully mature into plasmatocytes (PL), crystal cells (CC) which are specified by Notch signaling [5, 8], or lamellocytes . CCs are marked by crystalline inclusions which contain Prophenoloxidase (ProPO) that is essential in the immune response [1]. These cells mature from newly specified CC progenitors (CCP), which express Lozenge (Lz) [5], the Drosophila homolog of Runx1, into functional ProPO+ cells.
Figure 1. Scalloped and Yorkie are required for proper crystal cell differentiation.
Crystal cell progenitors (CCP) are labeled with Lz (H–P, red). (A) Schematic of the 3rd Instar lymph gland and hemocyte differentiation. PSC in grey, prohemocytes (PH, green) of the MZ, intermediate progenitors (IP, yellow), plasmatocytes (PL, red) and crystal cells (CC, blue) in the CZ. (B) Yki (red) is expressed in scattered cells of the CZ in a 3rd instar lymph gland. (C) Yki (red) is observed in CCPs (Lz, blue) amongst differentiating hemocytes (hml, green) of the CZ. (D) Yki (red) is present in mature CCs labeled with Black cells-GFP (green). (E) sd (sd-gal4 > UAS-2xEGFP, green) is expressed in clusters of cells scattered throughout the lymph gland. CZ is demarcated by a dotted line. (F–G) sd (green) is present in a subset of Yki+ cells (F, arrows) and mature CCs (G, arrows) and is also seen adjacent to Yki+ cells (F, arrowhead) and CCs (G, arrowheads). (G, inset) Lineage traced (sd-gal4, UAS-GFP > UAS-FLP, A5C-FRT-STOP-FRT-LacZ) (red) mature CCs (ProPO, white) do not express sd (green). H–L For each panel, its corresponding pattern of HLT> GFP expression is demonstrated in Fig. S1F–J (H) WT lymph gland (I) Widespread over-expression of ykiWT in the lymph gland increases CCP numbers while (J) depletion of yki or (K) sd blocks CC formation. (L) sd knock-down blocks the increase of CCPs observed upon over-expression of ykiWT. (M) WT lymph gland. (N) Over-expression of ykiWT in sd expressing cells (sd-gal4 >) increases CCP numbers, while (O) depletion of yki or (P) sd strongly inhibits CC differentiation. (Q–R) Quantification of H–P (n=10). * p value <.05, *** p value < .001. Scale bar 10 μm. See also Fig. S1.
Scattered amongst differentiating cells, we observe a population of Yorkie (Yki) expressing cells (Fig. 1B–D). Similarly, Yki’s binding partner Scalloped (Sd) is expressed in clusters of cells found throughout the CZ (Fig. 1, E–G) where it is co-expressed with Yki (Fig. 1F, Arrows). In addition, Yki+ and sd+ cells are observed adjacent to each other (Fig. 1F, Arrowhead). Yki is also observed in 77% of Lz+ CCPs [5] (Fig. 1C) , but only in 8% of sd+ cells. Similarly, only a small percentage of Lz+ cells express sd (Fig. S1A, arrowheads). Yki is also present in Black cells-GFP+ cells (Fig. 1D), a marker of mature CCs (Fig S1B). A small number of sd+ cells are also ProPO+ ( Fig. 1G, Arrow) , while a subset of sd+ cells is observed adjacent to mature CCs, but do not express CC markers (Fig. 1G, Arrowheads). Furthermore, lineage tracing analysis with sd-gal4, UAS-GFP, identified ProPO+ traced cells which do not express GFP (Fig. 1G, inset), suggesting that sd is only transiently expressed in this population of CCs. Notch is also observed in a subset of sd+ cells (Fig. S1C, Arrow), but the majority of Notch+ cells do not co-express sd but are located adjacent to sd+ cells (Fig. S1C, Arrowhead). These observations demonstrate that Yki and Sd are present both in CCs and in neighboring populations.
We next generated sd and yki mutant clones to interrogate their function in the LG. While yki clones are extremely small or absent in the LG (data not shown), we do observe a very striking absence of mature ProPO+ CCs in sd loss of function mutant clones (Fig S1D–E), confirming a requirement for Sd in CC formation. To gain further insight into their role in CC differentiation, we manipulated yki and sd expression using the Hand Lineage Tracing (HLT) driver, which clonally expresses gal-4 throughout the LG (Fig. S1F–J correspond to Fig. 1H–L). We observe an increase of Lz+ CCPs (Fig. 1H–I, Q) upon LG specific over-expression of ykiWT. Conversely, depletion of yki (Fig. 1J, Q) or sd (Fig. 1K, Q). causes a decrease in Lz+ cells. Importantly, depletion of sd blocks the increase in CCPs observed upon ykiWT over-expression (Fig. 1L, Q), demonstrating that Sd is required for Yki’s function in CC differentiation. The extent of CC loss in this background is milder compared to sd depletion alone (Fig. 1Q), which could be explained bylow levels of remaining Sd interacting with an over-abundance of Yki.
Based on the pattern of expression (Fig. 1E–G) and the functional results upon sd depletion (Fig. 1K–L), we further investigated the relationship between Yki and Sd in the context of CC differentiation by manipulating yki and sd levels with sd-gal4. We observe a significant increase in CCP numbers (Fig. 1M–N, R) when ykiWT is over-expressed in sd+ cells. Similarly, depletion of yki in sd+ cells causes a dramatic loss of Lz+ cells (Fig. 1O, R) as does sd down-regulation (Fig. 1P, R). Importantly, manipulating levels of yki and sd with sd-gal4 or HLT drivers does not significantly alter differentiation of plasmatocytes (Fig. S1K–L). Taken together, these observations provide evidence of an integral role for both Yki and Sd specifically in CC differentiation.
While over-expression of sd using the CCP driver lz-gal4, increases CC numbers (Fig. S1M, N–O), over-expression of ykiWT does not affect CCs (Fig. S1M, P). We do observe a remarkable decrease in mature CCs when both sd and yki are depleted in CCPs (Fig. S1M, Q–R). In addition, we observe striking ectopic expression of Yki and Lz in early 2nd instar LGs upon over-expression of an activated form of Notch (Fig. S1S–T). Furthermore, while Notch mutant LGs do not express Yki(Fig. S1V–W), we do observe Yki expression in scattered cells of the CZ in lzR15 mutant LGs (Fig. S1U). These findings indicate that Yki is specifically upregulated by Notch signaling independent of Lz early in the CC differentiation program, and that Yki and Sd are required within CCPs to maintain normal CC numbers.
Yorkie and Scalloped promote Serrate expression in Lineage Specifying Cells
While over-expression of yki throughout the LG (Fig. 1I) or specifically in sd expressing cells (Fig. 1N) significantly increases CCP numbers, a similar increase in CCs is not observed when yki is over-expressed in CCPs that have already been specified (Fig. S1P). This discrepancy suggests that Yki can promote CC formation independent of any effects within already committed CCPs, perhaps due to limited availability of Sd in these cells. This finding, along with the observation that sd+ cells are frequently observed adjacent to CCs (Fig. 1G), suggested that there may be a non-cell autonomous role for Yki in CC differentiation, possibly through regulation of the Notch ligand Serrate.
Serrate (Ser) is highly expressed in the PSC (Fig. 2A, [5, 12]), however, Ser function in this compartment is not required for CC differentiation (Fig. S2A–C), [4]). Interestingly, both CCs [2, 12] and Ser+ cells [12] are still observed in LGs which lack the PSC, and Ser+ cells have also been observed outside of the PSC [5]. We confirmed the presence of Ser+ cells within the CZ of third instar LGs (Fig. 2A Arrowhead). Inhibition of Serrate in differentiating hemocytes of the CZ (Fig. S2 D–F) or MZ prohemocytes (Fig. S2G–I) does not affect CC differentiation. However, LG-wide inhibition of Serrate significantly decreases CC differentiation (Fig. S2 J–L) demonstrating that Serrate function is required in a subset of cells that are distinct from the PSC, hematopoietic progenitors, or differentiating hemocytes.
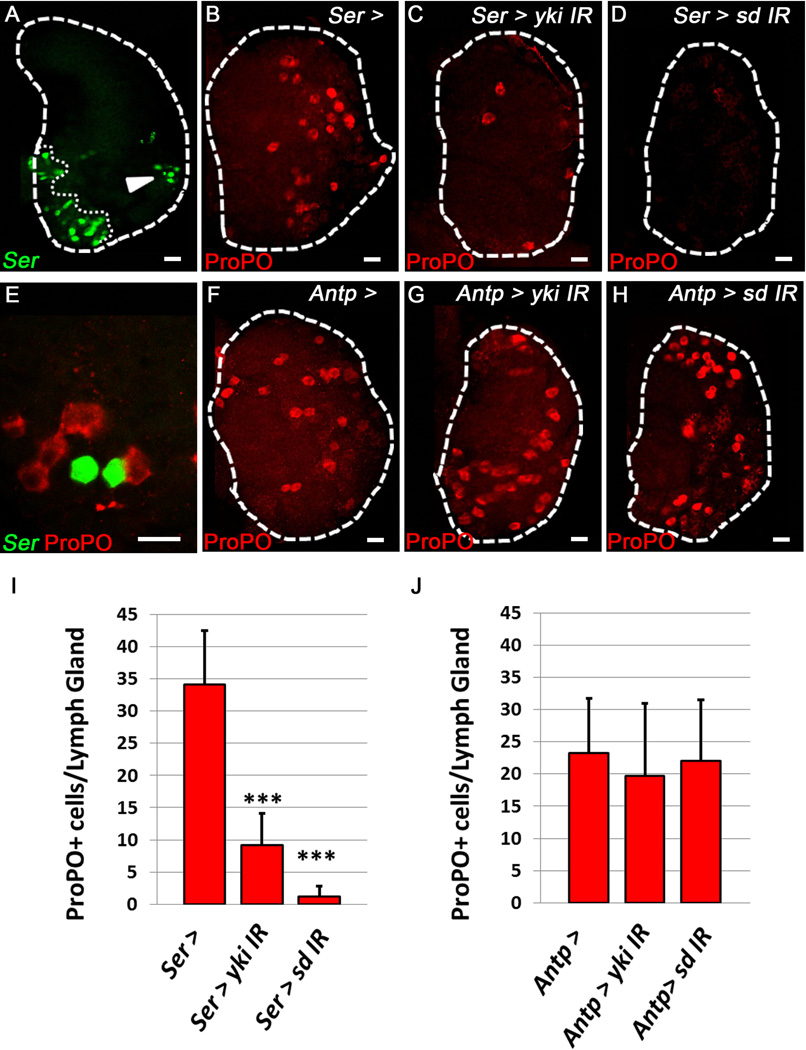
Figure 2. Yorkie and Scalloped are required specifically in Serrate-expressing cells for proper Crystal Cell differentiation.
Green labels Ser+ cells (Ser-gal4, UAS-GFP) (A, E) and red labels ProPO+ CCs. (A) Ser expressing cells (arrowhead) observed in the periphery of the Cortical Zone, distinct from the PSC (outlined by a dotted line). (B) WT (C) Knockdown of yki or (D) sd in Ser+ cells blocks CC formation. (E) Ser+ cells observed in direct contact with CCs in the Cortical Zone. (F) WT (G) Knockdown of yki or (H) sd in the PSC (Antp-gal4 >) has no effect on CC formation. (I) Quantification of yki and sd Knockdown in Ser+ cells. (J) Quantification of yki and sd Knockdown in the PSC. (n=10) *** indicates pValue <.001. Scale bar 10 μm. See also Fig. S2.
Having demonstrated that Yki and Sd can regulate CC numbers within the LG, we asked if they are specifically required in Ser+ cells for CC formation. Indeed, we observe a significant decrease in CC numbers upon depletion of yki or sd in these Ser+ cells (Fig. 2B–D, I) demonstrating a requirement for Yki and Sd in these signaling cells which are also observed adjacent to CCs (Fig. 2E). Depletion of yki or sd in the PSC using the Antp-gal4 driver does not affect CC differentiation (Fig. 2F–H, J). Therefore, Yki and Sd function is required specifically in Ser+ cells independent of the PSC for proper CC differentiation.
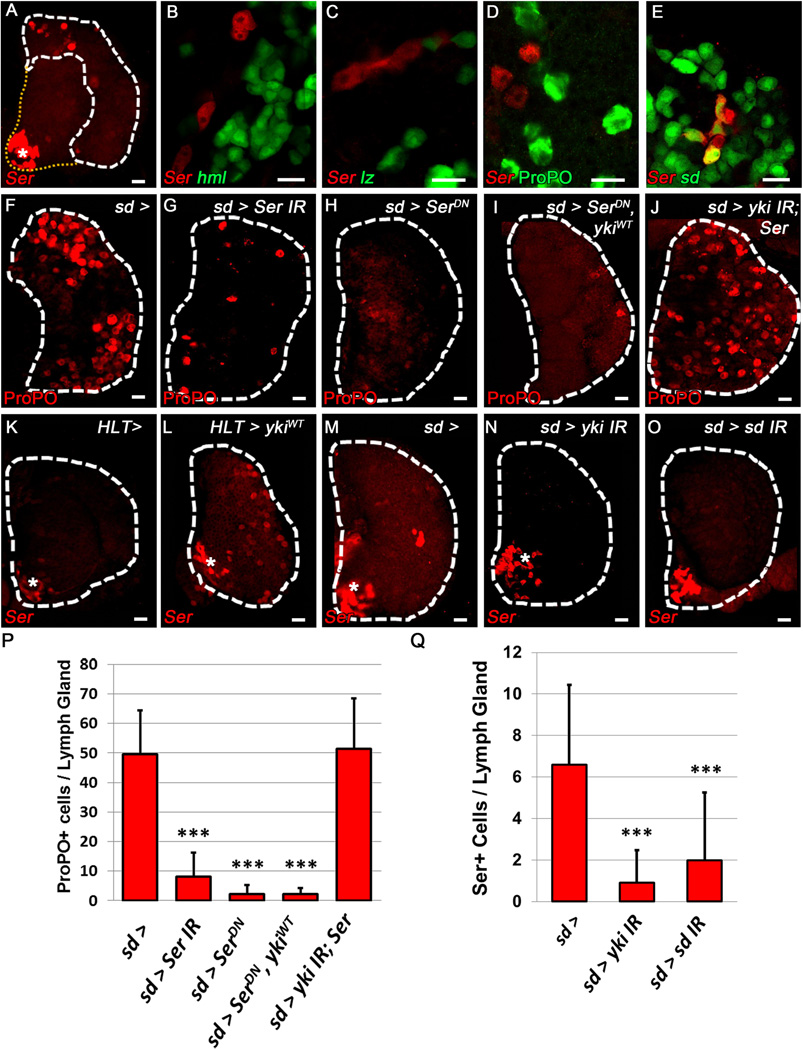
To gain further insight into the identity of Ser+ cells in the LG we performed a comprehensive analysis of hemocyte differentiation markers. Using a LacZ reporter of Ser expression, we confirmed that the population of Ser+ cells is located in the CZ (Fig. 3A). These cells do not express markers of differentiating hemocytes (Fig. 3B), but are observed in close proximity to both CCPs (Fig. 3C) and mature CCs (Fig. 3D). Furthermore, Ser+ cells in the CZ co-express sd (Fig 3E) and Yki (Fig. S3A–A”). It is important to reiterate that these Yki+ sd+ Ser+ cells do not express any other hemocyte markers (Fig. 3B–D), and are lineage traced from a sd+ cell (Fig. S3B–B”). We also observe a subset of Ser+ cells that arise from a dome+ precursor (Fig. S3C–C”, Arrowhead), but not all Ser+ cells originate from this population (Fig. S3C–C”, Arrow). Importantly, Ser expressing cells do not contribute to the CC lineage (Fig. S3D–D”). These data demonstrate that this unique population of Ser+ cells expresses both sd and Yki and represents a dedicated signaling cell that is distinct from other cell types in the LG.
Figure 3. Yorkie and Scalloped regulate Serrate-expressing lineage specifying cell (LSC) numbers.
Red labels Ser+ LSCs (SerLacZ, A–E, K–O) and CCs (F–J). (A) Ser expressing cells are located in the CZ, outlined by white hatch marks (asterisk denotes PSC) (B) Ser does not co-localize with the PL marker hml. (C–D) Ser+ cells are observed adjacent to lz+ CCPs (C, green) and ProPO+ mature CCs (D, green) but do not co-localize. (E) sd (green) is co-expressed with Ser (red) (F) WT lymph gland. (G) Depletion or (H) inhibition of Ser function in sd expressing cells (sd-gal4>) blocks CC differentiation. (I) CC differentiation is similarly blocked by inhibition of Ser after over-expression of ykiWT. (J) Loss of CCs observed upon yki depletion is rescued by over-expression of Ser. (K) Ser expression in larvae containing a single copy of SerLacZ is only observed in the PSC (asterisk). (L) Over-expression of ykiWT (HLT >)greatly increases Ser expression in lymph glands containing a single copy of SerLacZ. (M) WT lymph gland. (N) Depletion of yki or (O) sd in sd expressing cells (sd-gal4>) blocks Ser expression outside of the PSC (asterisk). (P–Q) Quantification of F–J and M–O (n=10). *** p value < .001. Scale bar 10 μm. See also Fig. S3.
Similar to the requirement of Yki and Sd in Ser+ cells, depletion or inhibition of Ser in sd+ cells is sufficient to block CC differentiation (Fig. 3F–H, P). This demonstrates that Ser is uniquely required in sd+ cells and no other LG cell populations (compare to Fig S2A–I) for CC differentiation. The Yki-mediated increase in CC numbers previously observed (Fig. 1N) is blocked by over-expression of SerDN (Fig. 3I, P), while over-expression of Ser rescues (Fig. 3J, P) the loss of CCs observed upon yki knockdown (Fig. 1O). In addition, over-expression of ykiWT in the LG increases Ser expression in the CZ (Fig. 3K–L). Similarly, down-regulation of yki or sd specifically in sd+ cells causes a significant decrease in the number of Ser+ signaling cells (Fig. 3M–O, Q) and a corresponding decrease in CCP numbers (Fig. 1O–P, R). However, over-expression of either ykiWT (Fig. S3E, G) or Ser (Fig. S3F, G) specifically in Ser+ cells, does not affect CC differentiation (compare to Fig. 2B), suggesting that changes in CC number upon ykiWT over-expression are due to an increase in the number of Ser+ cells (Fig. 3L). These results demonstrate that Yki and Sd have definitive roles in CC specification by regulating Ser expression in a distinct population of cells within the LG that we have termed Lineage Specifying Cells (LSCs).
Wasp parasitization triggers cell fate decisions required for the lymph gland immune response by altering Serrate expression
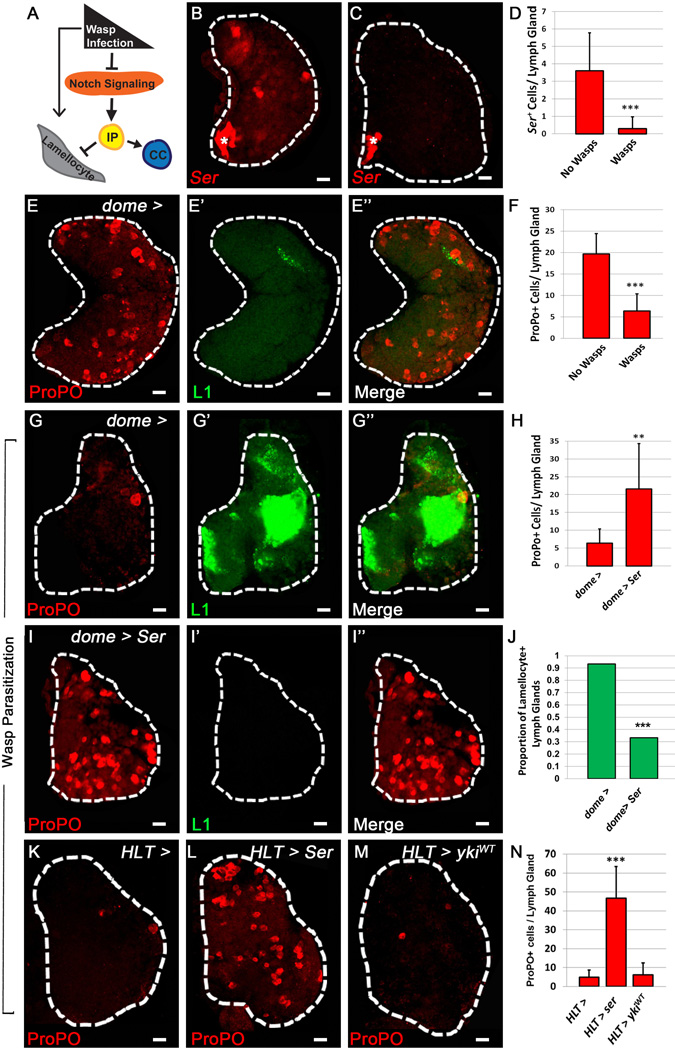
Larval parasitization by the wasp Leptopilina boulardi elicits a strong cellular immune response in the Drosophila LG REF 11 [13] characterized by lamellocyte differentiation (Fig. 4A) which is rarely observed in WT LGs. These large, flat cells defend the larva by engulfing invading pathogens or parasites, such as the L. boulardi eggs. Upon wasp parasitization there is a robust increase in lamellocyte differentiation along with a corresponding decrease in CC differentiation [9]. However, the mechanism by which this change in lineage fate decisions is regulated has not been definitively determined, although it has been recently shown that Notch signaling blocks lamellocyte formation (Fig. 4A) [14].
Figure 4. Serrate expression in LSCs is down-regulated in the lymph gland of immune challenged larvae.
(A) Schematic representation of the immune response generated upon wasp infection. Notch signaling, which promotes crystal cell (CC) differentiation in the lymph gland, is blocked by wasp parasitization, allowing intermediate progenitors (IP) to differentiate into lamellocytes [14]. Asterisk denotes PSC (B–C). LSCs (Ser) are red in B–C while CCs (ProPO) are red in E, E", G, G", I, I", K–M. Lamellocytes are labeled by green (L1). (B) Ser expression in WT. (C) Loss of LSC Ser expression upon wasp parasitization. (D) Quantification of Fig.4B–C, (n= 10) (E–E”) WT lymph gland contains CCs (E) and lacks lamellocytes (E'). (F) Quantification of Fig.4E, G (n=10). (G–G”) Wasp parasitization eliminates CCs (G) and promotes lamellocytes (G’). (H) Quantification of Fig.4G, I (n=10) (I–I”) Enforced expression of Ser upon wasp parasitization rescues CC loss (I), and inhibits lamellocyte formation (I'). (J) Quantification of Fig. 4G’, I’ (n= 10) (K) WT lymph gland. (L) Over-expression of Ser (HLT >) rescues CC numbers upon wasp parasitization, but (M) over-expression of ykiWT has no affect compared to WT. (N) Quantification of Fig.4 K–M. (n=10) ** pValue < .01, *** pValue < .001. Scale bar 10 μm. See also Fig. S4.
A possible explanation for the loss of CCs in the LG upon wasp parasitization could be that expression of Ser in LSCs is down-regulated under these conditions as a requirement for lamellocyte differentiation. Indeed, we observe a significant decrease in the numbers of Ser+ LSCs in parasitized larvae (Fig. 4B–C, D) associated with an up-regulation of lamellocytes (Fig. 4E', G') and a decrease in CCs (Fig. 4 E, F, G) [9]. To further verify that this down-regulation of Ser is required for a proper immune response, we ectopically expressed Ser in the LG and subjected these larvae to wasp parasitization. Unlike the WT parasitized control (Fig. 4G–G"), we observe a significant increase in CCs when Ser is over-expressed either in prohemocytes and IPs (Fig. 4H– I) or ubiquitously by HLT (Fig.4K– L, N). Most strikingly, there is a significant inhibition of lamellocyte differentiation upon enforced expression of Ser (Fig. 4G', I', J). These findings demonstrate that down-regulation of Ser is responsible for the decreased numbers of CCs observed in the LGs of wasp parasitized larvae and is essential for lamellocyte differentiation in the LG.
We next examined if the alterations in cell fate and observed changes in Ser expression upon immune challenge were due to changes in Yki and Sd function. Interestingly, both Yki and sd are strongly expressed in lamellocytes, while expression of Yki and sd in other cells of the LG is severely diminished upon wasp parasitization (Fig. S4 A–A", B–B"). Given the expression of sd in lamellocytes, we used sd-gal4 to interrogate the function of Yki and Sd in these cells. Down-regulating levels of yki or sd has no effect on lamellocyte differentiation (Fig. S4C–E) , and over-expression of ykiWT is not sufficient to rescue loss of CCs in immune challenged LGs (Fig. 4M, N). These findings indicate that wasp parasitization regulates Ser expression in LSCs to allow for lamellocyte differentiation at the expense of CCs, while emphasizing the dynamic role for LSCs in maintaining LG homeostasis under normal and stress conditions.
Our findings demonstrate a novel role for Yki and Sd in the Notch dependent lineage specification of CCs. While expression of the Yki and Sd homologues, YAP1/TAZ and TEAD, has been previously described in mammalian Hematopoietic Stem Cells (HSCs) [15, 16], no phenotypes have been observed upon manipulation of these factors in the HSC compartment [16]. Alternatively, we propose that a conserved role for YAP and TEAD signaling may reside in a non-cell autonomous manner originating from lineage specifying or niche cells, such as stromal cells of the bone marrow, thymic epithelium, and other sites of differentiation such as the liver and spleen. . Here, we have provided evidence for a novel regulatory role for Yki and Sd in promoting Ser expression in LSCs of the Drosophila LG while demonstrating LSC plasticity in immune challenged larvae. Parasitization by the wasp L. boulardi necessitates a lineage switch from CCs to lamellocytes, that is achieved by down-regulating Serrate, allowing a common pool of hematopoietic progenitors to differentiate into lamellocytes.
Recently, it was shown that YAP regulates expression of Jagged1, the mammalian homolog of Serrate, in hepatocytes [17, 18]. Hippo pathway signaling through YAP regulates liver cell fate decisions[18], while misregulation of YAP leads to increased Jagged1 expression in a TEAD-dependent manner, causing irregular activation of the Notch pathway and hepatocellular carcinoma [17]. In addition, a biphasic lineage specification mechanism involving Notch signaling is required in the specification of Megakaryocyte-Erythroid Progenitor into erythrocytes at the expense of megakaryocytes[19] under stress conditions. Given the presence of YAP and TEAD within mammalian hematopoietic compartments[15, 16, 20, 21], a similar requirement for these factors may be necessary for the regulation of Notch-dependent lineage specification.
We have described a similar role for Yki and Sd , in regulating Serrate+ LSCs in the Drosophila LG.. Our results demonstrate a mechanism where a small number of Ser expressing LSCs are tightly regulated by limiting availability of Yki and Sd, as perturbations to either of these factors alters CC differentiation. Furthermore, over-expression of Ser in the CZ increases CC numbers significantly (Ferguson and Martinez-Agosto, unpublished results), demonstrating the sensitivity to changes in Notch ligand availability in the LG [14]. Our finding that Ser expression in LSCs is specifically down-regulated upon wasp parasitization further demonstrates that these signaling cells are in fact dynamically regulated within the LG. In total, these mechanisms further expand our understanding of hematopoietic niches and the regulation of signaling molecules that characterize hematopoietic microenvironments.
Experimental Procedures (See also Text S1)
Genetic analysis
All crosses were reared at 29 degrees Celsius. Multiple yorkie (VDRC:104253 and NIG:4005R-1) and scalloped (VDRC: 101497 and NIG:8544R-3) RNAi constructs were tested and yielded similar phenotypes. In the case of HLT experiments, the Hand-gal, UAS-2xEGFP,UAS-FLP; A5C-FRT-STOP-FRT-GAL4 genetic background was used to generate lymph gland specific clones expressing the UAS construct of interest. For sd knockdown experiments, UAS-dicer2 was used in the background to enhance phenotypes. Inhibition of Serrate in hematopoietic progenitors of the Medullary Zone was achieved using the dome-gal4, UAS-mCD8::GFP; gal80ts stock, with larvae reared at 18 degrees Celsius. After 48hr, larvae were then shifted to 29 degrees Celsius. Notchts larvae were raised at 29 degerees Celsius. All crosses involving SerLacZ reporter analysis were performed using two copies of SerLacZ in the background, except for the experiment involving over-expression of UAS-ykiWT or its corresponding control.
Immunohistochemistry
Lymph glands (LGs) were dissected as previously described in 1xPBS and fixed in 3.7% paraformaldehyde for 20 minutes. LGs were then blocked for 30 minutes in 10% NGS in 0.4% TritonX/PBS (PBT). Antibodies were appropriately diluted in PBT and allowed to incubate with samples overnight at 4 degrees Celsius. LGs were washed 4× 15minutes in PBT and blocked again with 10% NGS in PBT for 30 minutes. Secondary antibodies were appropriately diluted in 10% NGS in PBT and allowed to incubate with samples overnight. LGs were then washed 4×15 minutes in PBT and mounted in Vectashield Mounting Medium. Samples were imaged using a Carl Zeiss LSM 310 Laser Scanning Confocal Microscope. A middle section of a Z-stack was used in every image.
Supplementary Material
Highlights.
-
-
Yki and Sd regulate crystal cell differentiation in the lymph gland
-
-
Ser expression in Lineage Specifying Cells depends on Yki and Sd
-
-
Loss of Ser expression induced by wasp infection inhibits crystal cell formation
Acknowledgments
We would like to thank the following laboratories for providing reagents: Kenneth Irvine, Utpal Banerjee, Mike Kanost, John Fessler, Nancy Fossett, Doujia Pan, Ben Ohlstein, Jin Jiang, Todd Schlenke, Istvan Ando, VDRC, DGRC, and NIG. A number of antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by the David Geffen School of Medicine at UCLA and the Ruth L. Kirchstein National Research Service Award GM007185.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: GBF and JAMA both developed concepts and approach, analyzed data, and wrote the manuscript. GBF performed all experiments.
References
- 1.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila Hematopoietic Lineage by Conserved Transcription Factors. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 2.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mondal Bama C, Mukherjee T, Mandal L, Evans Cory J, Sinenko Sergey A, Martinez-Agosto Julian A, Banerjee U. Interaction between Differentiating Cell- and Niche-Derived Signals in Hematopoietic Progenitor Maintenance. Cell. 2011;147:1589–1600. doi: 10.1016/j.cell.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- 5.Lebestky T, Jung S-H, Banerjee U. A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes & Development. 2003;17:348–353. doi: 10.1101/gad.1052803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo Signaling Pathway Coordinately Regulates Cell Proliferation and Apoptosis by Inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF Family Protein Scalloped Mediates Transcriptional Output of the Hippo Growth-Regulatory Pathway. Developmental Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Duvic B, Hoffmann JA, Meister M, Royet J. Notch Signaling Controls Lineage Specification during Drosophila Larval Hematopoiesis. Current Biology. 2002;12:1923–1927. doi: 10.1016/s0960-9822(02)01297-6. [DOI] [PubMed] [Google Scholar]
- 9.Krzemien J, Oyallon J, Crozatier M, Vincent A. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Developmental Biology. 2010;346:310–319. doi: 10.1016/j.ydbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Dragojlovic-Munther M, Martinez-Agosto JA. Multifaceted roles of PTEN and TSC orchestrate growth and differentiation of Drosophila blood progenitors. Development. 2012;139:3752–3763. doi: 10.1242/dev.074203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizki TM, Rizki RM. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Developmental & Comparative Immunology. 1992;16:103–110. doi: 10.1016/0145-305x(92)90011-z. [DOI] [PubMed] [Google Scholar]
- 12.Crozatier M, Ubeda J-M, Vincent A, Meister M. Cellular Immune Response to Parasitization in Drosophila Requires the EBF Orthologue Collier. PLoS Biol. 2004;2:e196. doi: 10.1371/journal.pbio.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorrentino RP, Carton Y, Govind S. Cellular Immune Response to Parasite Infection in the Drosophila Lymph Gland Is Developmentally Regulated. Developmental Biology. 2002;243:65–80. doi: 10.1006/dbio.2001.0542. [DOI] [PubMed] [Google Scholar]
- 14.Small C, Ramroop J, Otazo M, Huang LH, Saleque S, Govind S. An Unexpected Link Between Notch Signaling and ROS in Restricting the Differentiation of Hematopoietic Progenitors in Drosophila. Genetics. 2013 doi: 10.1534/genetics.113.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": Transcriptional Profiling of Embryonic and Adult Stem Cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 16.Jansson L, Larsson J. Normal Hematopoietic Stem Cell Function in Mice with Enforced Expression of the Hippo Signaling Effector YAP1. PLoS ONE. 2012;7:e32013. doi: 10.1371/journal.pone.0032013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, et al. Yes-Associated Protein Up-regulates Jagged-1 and Activates the NOTCH Pathway in Human Hepatocellular Carcinoma. Gastroenterology. 2013;144:1530–1542. e1512. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yimlamai D, Christodoulou C, Galli Giorgio G, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger Ben Z, Camargo Fernando D. Hippo Pathway Activity Influences Liver Cell Fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, van Handel B, Ibrahim S, Greve J, Mikkola H, et al. In Vivo Mapping of Notch Pathway Activity in Normal and Stress Hematopoiesis. Cell Stem Cell. 2013;13:190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing W, Kim J, Wergedal J, Chen S-T, Mohan S. Ephrin B1 Regulates Bone Marrow Stromal Cell Differentiation and Bone Formation by Influencing TAZ Transactivation via Complex Formation with NHERF1. Molecular and Cellular Biology. 2010;30:711–721. doi: 10.1128/MCB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo E, Basu-Roy U, Gunaratne Preethi H, Coarfa C, Lim D-S, Basilico C, Mansukhani A. SOX2 Regulates YAP1 to Maintain Stemness and Determine Cell Fate in the Osteo-Adipo Lineage. Cell Reports. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.