Abstract
Glucocorticoids (GCs) selectively trigger cell death in the multiple myeloma cell line MM1S which express NR3C1/Glucocorticoid Receptor (GR) protein, but fail to kill MM1R cells which lack GR protein. Given recent demonstrations of altered microRNA profiles in a diverse range of haematological malignancies and drug resistance, we characterized GC inducible mRNA and microRNA transcription profiles in GC sensitive MM1S as compared to GC resistant MM1R cells. Transcriptome analysis revealed that GCs regulate expression of multiple genes involved in cell cycle control, cell organization, cell death and immunological disease in MM1S cells, which remain unaffected in MM1R cells. With respect to microRNAs, mir-150-5p was identified as the most time persistent GC regulated microRNA, out of 5 QPCR validated microRNAs (mir-26b, mir-125a-5p, mir-146-5p, mir-150-5p, and mir-184), which are GC inducible in MM1S but not in MM1R cells. Functional studies further revealed that ectopic transfection of a synthetic mir-150-5p mimics GR dependent gene expression changes involved in cell death and cell proliferation pathways. Remarkably, despite the gene expression changes observed, overexpression of mir-150-5p in absence of GCs did not trigger significant cytotoxicity in MM1S or MM1R cells. This suggests the requirement of additional steps in GC induced cell death, which can not be mimicked by mir-150-5p overexpression alone. Interestingly, a combination of mir-150-5p transfection with low doses GC in MM1S cells was found to sensitize therapy response, whereas opposite effects could be observed with a mir-150-5p specific antagomir. Although mir-150-5p overexpression did not substantially change GR expression levels, it was found that mir-150-5p evokes GR specific effects through indirect mRNA regulation of GR interacting transcription factors and hormone receptors, GR chaperones, as well as various effectors of unfolded protein stress and chemokine signalling. Altogether GC-inducible mir-150-5p adds another level of regulation to GC specific therapeutic responses in multiple myeloma.
Introduction
Multiple myeloma (MM) is a B-cell neoplasm characterized by the accumulation of clonal malignant plasma cells in the bone marrow and often correlated with various cytogenetic abnormalities such as del(13), t(11∶14), non-hyperdiploidy, and del(17p) [1], [2]. The disease accounts for 10% of the haematological malignancies and approximately 1% of cancer-related deaths in Western countries [3]. Therapy against multiple myeloma consists of drugs which can decrease the clonal plasma cell population. Initial treatment towards the disease depends mainly on patients age and comorbidities. The ability of glucocorticoids (GCs) to efficiently kill lymphoid cells has led to their inclusion in essentially all chemotherapy protocols for lymphoid malignancies. For patients under the age of 65 high doses of chemotherapy of different combinations such as thalidomide–dexamethasone-bortezomib based regimens, and lenalidomide–dexamethasone followed by autologous haematopoietic stem cell transplantation has been a practice in the clinic in the recent years [4], [5], [6], [7], [8]. Despite the progress in therapy, MM remains largely incurable, due to low remission rates of conventional therapies resulting in short survival times (3–4 years) and the development of drug resistance. Numerous novel drug combinations are currently being tested to prevent resistance and improve GC efficacy in the therapy of lymphoid malignancies [9].
Glucocorticoids (GCs) are steroid hormones, which exert their pro- or anti-apoptotic actions via the glucocorticoid receptor (GR, NR3C1) and alterations in gene expression [10], [11]. The GRα isoform, which is the main mediator of GC effects, comprises three main functional domains: an N-terminal domain (NTD), a DNA binding domain (DBD) and a ligand binding domain (LBD) [12]. The inactive form of GRα primarily resides in the cytoplasm forming a chaperone complex with its binding partners such as heat shock proteins, FKBP51, FKBP52, CYP40 and other proteins [13]. Upon binding with the GC’s, the GR becomes activated and translocates to the nucleus. The activated GR has several different modes of action at the cellular level. The most prominent one is transactivation in which the activated GR binds to a glucocorticoid-response element (GRE) and induces gene expression. Further, GR can bind negative GRE sites (nGREs) or tether bound transcription factors (NFκB, AP1, P53 or STAT) and hence (trans)represses gene expression [14]. Although being known as a strong inducer of apoptosis in lymphoid cells for almost a century, the signaling pathways regulating the susceptibility or resistance of cancer cells to GCs are only partly revealed. Besides slow genomic GC actions depending on nuclear gene regulation, also rapid non-genomic effects of GCs in the cytoplasm modulate the apoptotic response [15], [16]. With respect to development of GC therapy resistance, various mechanisms have been proposed including GR degradation, Akt dependent GR phosphorylation, reduced histone deacetylase-2 (HDAC2) activity, excessive activation of the transcription factor NFkB or AP1, elevated macrophage migration inhibitory factor, oxidative stress and P-glycoprotein mediated regulation of drug efflux [17], [18], [19], [20]. A novel emerging hypothesis is that microRNAs modulate GC therapy response via (post-transcriptional) regulation of multiple mRNA targets involved in GR signaling related to cell proliferation and cell death [21], [22], [23], [24]. MiRNAs are short non-coding RNAs of approximately 22 nucleotides length which regulate expression of tens to hundreds of genes via mRNA degradation or translational repression, based on the sequence complementarity to the miRNA seed region (positions 2–8) at the 3′UTR of the mRNA [25]. Alternatively, miRNAs may indirectly reprogram transcriptional responses by changing mRNA levels of transcription factor levels [26]. As such, microRNAs may represent a possible feedback mechanism to fine-tune the magnitude of GC induced cell death via direct regulation of GR mRNA/protein levels or indirect regulation of GR interaction partners or downstream GC/GR responsive target mRNAs. Since miRNA dysregulation has a profound impact on cell differentiation, development, apoptosis and cell cycle progression, their expression profiles can be used as biomarkers for cancer type and grade (prognosis) and/or personalized therapy response [27]. To characterize the regulatory role of GC inducible microRNAs in multiple myeloma therapy response, we have compared mRNA and microRNA transcription profiles in GC sensitive MM1S cells and GC resistant MM1R cells upon exposure to GC. The latter cell model is frequently used for elucidation of regulatory mechanisms involved in GC therapy resistance in MM cells [28].
Materials and Methods
Cell Culture and cell viability
GC sensitive MM1S (CRL-2974) and GC resistant MM1R cell lines (CRL-2975) have been previously described [28] and were purchased from ATCC. Briefly, MM1S cells are a B-lymphoblast myeloma cell line derived from a 42 years old black female. The MM1R cell line is a steroid resistant subclone derived from MM1S cells, chronically exposed to dexamethasone (Dex). Cells were cultivated in RPMI-1640 Medium supplied with 10% Fetal Bovine Serum (E.U Approved; South American Origin) and 0.5% Penicillin/Streptomycin Solution (Invitrogen). The cell lines were additionally supplemented with 1% MEM Non-Essential Amino Acids and 1% Sodium Pyruvate (Invitrogen). Each cell line was maintained at 37°C in the atmosphere of 5% CO2 and 95% air, 95–98% humidity, until confluence. Cell viability was assessed by using colorimetric assay with 3-(4, 5-dimethylthiozol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma Aldrich, St. Louis, MO, United States of America) as previously described [29].
Antibodies and reagents
Water soluble dexamethasone was purchased from (Sigma Aldrich, St. Louis, MO, United States of America) and 10 mM stock solutions were prepared in sterile water. Anti-GRα (sc-8992) antibody, anti-PARP antibody (sc-7150) were purchased from Santa Cruz Biotechnology (Dallas, United States of America), and anti-α-tubulin (T5168) was purchased from Sigma Aldrich (St. Louis, MO, US).
RNA extraction and microarray processing
Total RNA from MM1S and MM1R cells, either Dex treated (1 µM) or transfected with mock or mir-150-5p mimetics, from three independent experiments was isolated using RNeasy Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol. Following extraction and concentration measurement (NanoDrop 1000, Thermo Scientific, Waltham, MA, USA) total RNA was quality controlled on a Bio-Rad Experion (Bio-Rad, Hercules, CA, USA). 500 ng of total RNA was amplified using the Illumina Total Prep RNA Amplification kit (Life Technologies, Carlsbad, CA, USA). Briefly, RNA was reverse transcribed using T7 oligo(dT) primers, after which biotinylated cRNA was synthesized through an in vitro transcription reaction. 750 ng of amplified cRNA was hybridized to a corresponding array of a HumanHT12 beadchip (Illumina, San Diego, CA, USA). Multiplicates were performed. The beadchip was incubated for 18 hours at 58°C in a hybridization oven under continuous rocking. After several consecutive washing steps (see manufacturer’s protocol), bead intensities were read on an Illumina Iscan.
Microarray data analysis
All the raw data intensities generated were read using the “beadarray” package (v2.8.1) in R [30]. Intensities were quantile normalized and differential gene expression between samples was estimated using “Limma” (v3.14.1). Resulting p-values were corrected for multiple hypothesis testing using the Benjamini-Hochberg method. Next to estimating gene expressions, euclidean distances between samples and genes were calculated and used as a distance metric in a hierarchical cluster analysis with MeV [31]. Pathway analysis was performed in the Ingenuity Pathway Knowledge Base (Ingenuity Systems, Redwood City, CA, USA) according to the instructions provided. A fold change cut-off of 2 as well as false discovery rate of 0.1% were set to identify genes whose expression was significantly differentially regulated. Raw array data were uploaded to the Gene Expression Omnibus (GEO) database and have accession number GSE59805.
cDNA conversion and QPCR analysis
Briefly 1 µg of RNA was converted into cDNA using Go Script reverse transcription system according to manufacturer’s instructions (Promega, Madison, WI, United States of America). Sybr green was purchased from (Bioline, London, UK). QPCR was performed using ABI7300 system (Applied biosystems, Life Technologies Corporation).
miRNA analysis
miRNA isolation was performed with a miRNeasy kit (Qiagen, Venlo, Netherlands) according to manufacturer instructions. Stem-loop RT-qPCR miRNA expression profiling of 760 microRNAs was done as previously described [32], [33]. Raw expression values were normalized using the global mean normalization strategy [32]. Validation of QPCR array results was done by reverse transcription of total RNA containing miRNA and miRNA detection by real-time PCR, using respectively miScript II RT Kit and miScript SYBR Green PCR Kit (Qiagen, Venlo, Netherlands). MicroRNA specific QPCR primer sets against human mir-146a (MS00003535), mir-491-5p (MS00031899), mir-146b-5p (MS00003542), mir-26b-5p (MS00003234), mir-150-5p (MS00003577), mir-330-3p (MS00031738), mir-195 (MS00003703), mir-184(MS00003640), mir-125a-5p (MS00003423) were purchased from Qiagen (Venlo, Netherlands). For microRNA target and pathway prediction, miRWalk software was applied [34]. Correlation of mRNA and microRNA transcription profiles of MM1S and MM1R cells either non-treated or treated with Dex, or following mock transfection or transfection with mir-150-5p mimetic was performed with IPA software.
In vitro transfection of MM cells with synthetic miR-150-5p mimetic and antagomir
Mir-150-5p mimetic and antagomir were purchased from Qiagen (Venlo, Netherlands). A total of 5×106 cells was electroporated with mock control or synthetic mir-150-5p at a final concentration of 150 nmol/L, with Nucleofector Kit V, using AMAXA Transfection System, program O23 from (LONZA, Basel, Switzerland) as described elsewhere [35].
Protein isolation
5×106 cells were seeded in 10 cm dishes and protein lysates of different experimental setups were prepared for Western analysis with 1XSDS Sample buffer containing 62.5 mM Tris-HCL pH-6.8, 2% SDS, 10% Glycerol, 0.5% Bromophenol Blue, DTT (5 mM).
Statistical Analysis
Statistics were performed using one way ANOVA, two way ANOVA followed by Dunnett’s and Bonferroni post-test and one-tailed paired student t-test using graph pad prism5 software.
Results
Dexamethasone decreases cell viability in a dose and time dependent manner in GC sensitive MM1S but not GC resistant MM1R cells
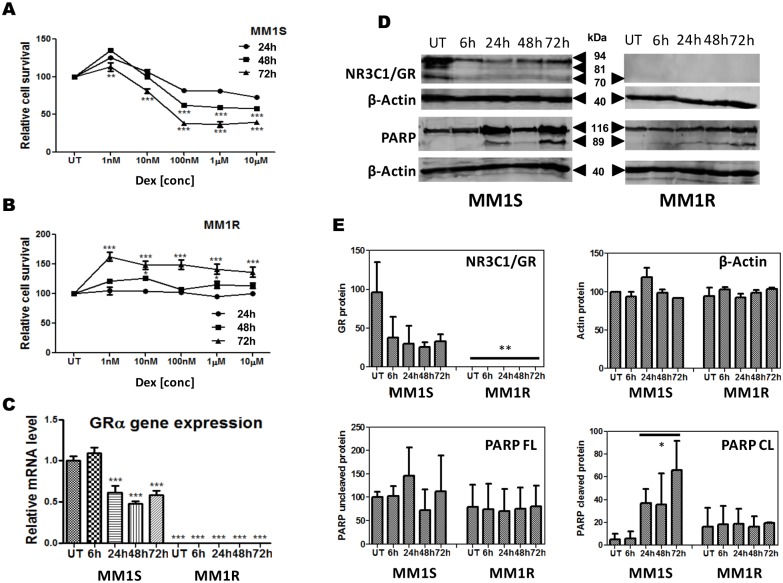
Before investigation of the regulatory role of GC inducible microRNA in NR3C1/GR responsive gene expression and cell death responses, we first characterized the GC therapy response in MM1S and MM1R cells. Briefly, MM1S and MM1R cells were left untreated or treated with 1 µM of the synthetic GC agonist dexamethasone (Dex) for respectively 6 h, 24 h, 48 h, 72 h. mRNA and proteins were isolated at each time point and analysed for corresponding NR3C1/GRα expression. Treating the cells with different concentrations of Dex for 24 h, 48 h and 72 h followed by measuring cell viability by a MTT colorimetric assay, revealed a dose and time dependent decrease in cell survival of MM1S cells upon GC exposure (Fig. 1A & 1B). In contrast Dex lacks the capacity to induce cytotoxicity in MM1R cells and rather triggers a weak growth stimulatory effect [28], [35], [36], [37]. In line with the different GC response in both cell lines, QPCR analysis demonstrates significant NR3C1/GRα mRNA transcription in MM1S, but near background levels of NR3C1/GRα mRNA in MM1R cells (Fig. 1C). Western analysis finally confirms significant levels of GRα protein expression in MM1S and undetectable levels of GRα protein in MM1R cells (Fig. 1D–E). As can be observed from Fig. 1D, MM1S cells express multiple translational isoforms of GRα [38], [39], of which expression decreases following GC treatment.
Figure 1. Dexamethasone decreases cell viability in a dose and time dependent manner in GC sensitive MM1S but not GC resistant MM1R cells.
(A–B) dose response curves of MM1S and MM1R cells treated with dexamethasone at different concentrations for 24, 48 and 72 h as determined by MTT colorimetric assay. Data represent (mean ± SEM) values of three independent experiments. (C) time dependent changes in GRα mRNA levels upon 1 µM dexamethasone treatment. Data represent (mean ± SEM) values of three independent experiments normalized to 28sRNA housekeeping gene and relative to MM1S untreated condition (S UT). Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05 and ns are not significantly different as determined by one-way ANOVA (Dunnett’s Post-test) (D) Time dependent changes in GRα protein levels and PARP cleavage in MM1S and MM1R cells following treatment of 1 µM Dex. (E) Densitometric analysis of western blot signals (n = 2) of protein levels of GR (92 kDa), β-Actin (40 kDa), full length PARP (116 kDa) and cleaved PARP (89 kDa) in MM1S and MM1R, following treatment of 1 µM Dex. Significant differences between western signals in MM1R and MM1S are indicated.
Despite the time dependent reduction of GR mRNA and protein levels upon 24–72 h GC treatment (Fig. 1C–1D), the cell death response in MM1S is not abrogated. Furthermore, GC induced cell death triggers time dependent cleavage of the apoptotic marker PARP in MM1S but not in MM1R cells (Fig. 1D–E). In contrast to GR levels observed in MM1S, expression levels of most GR isoforms in MM1R are below detection limit, although background levels of a 70 kDa variant can still be detected upon prolonged exposure (data not shown). Remarkably, although Dex completely lacks capacity to induce cytotoxicity in MM1R cells, a weak growth stimulatory effect can be observed upon prolonged exposure of GC in MM1R cells. This maybe attributed to residual activity of a possible anti-apoptotic translational isoform of GR [38].
Dexamethasone triggers GC specific transcriptional changes in cell cycle, DNA damage/repair, cell death and immunological pathways in MM1S but not MM1R
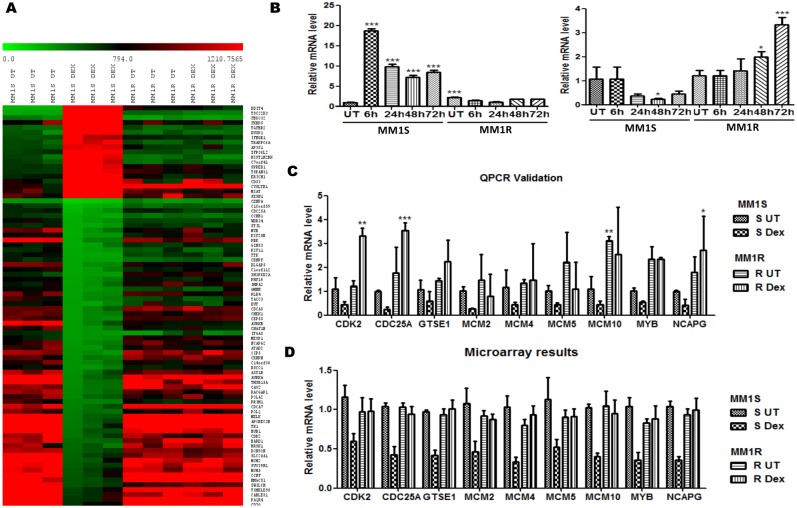
To evaluate specific gene expression changes linked to GC therapy response in multiple myeloma, mRNA was isolated from three independent experiments from MM1S and MM1R cells exposed for 72 h to the GC Dex (1 µM). After mRNA isolation, genome-wide transcription profiles were determined by Illumina array hybridisation. Results were analysed with the R-package “Limma” (v3.14.1) [40], [41], which resulted in the identification of 268 downregulated and 151 upregulated genes in MM1S, after filtering for a minimal two-fold transcriptional change in Dex treated samples versus control setup (absolute fold change >2) and adjusted P-value <0.05 (Gene Expression Omnibus (GEO) database GSE59805). In MM1R, no significant Dex responsive changes could be detected, in line with the GC resistant phenotype and the absence of GRα in MM1R cells (see Fig. 1). The heatmap (Fig. 2A) represents a graphical presentation of Dex specific transcriptional changes in MM1S and lack of Dex response in MM1R cells in three biological replicates, revealing highly reproducible and robust patterns of gene expression. By QPCR analysis, we confirmed GC dependent transactivation of GILZ (TSC22D3) target gene as well as GC specific (trans)repression of CDK2 in MM1S but not in MM1R, in line with the Illumina array data and previous reports [37], [42], [43], [44] (Fig. 2B). Remarkably, prolonged GC exposure (72 h) of MM1R cells reveals weakly increased levels of CDK2 according to QPCR analysis, which could not be detected by less sensitive microarray analysis. This effect may be attributed to residual activity of anti-apoptotic GR isoforms in MM1R, which may also contribute to weak growth stimulatory effects (Fig. 1A) upon prolonged exposure [38]. Integrated pathway analysis (IPA) further revealed that GCs trigger gene expression changes which promote cell death, decrease cell proliferation (cell cycle, DNA replication) and regulate immunological haematological pathways in MM1S but not MM1R (Table 1), in line with the reduced cell viability assay in MM1S (see Fig. 1). Next, we further validated microarray results of some additional target genes involved in cell cycle, DNA damage/repair and cell death. As can be observed from (Fig. 2C & 2D), the QPCR validation results correlate with the obtained microarray data. Furthermore, by analyzing patterns of coregulated genes, IPA software allows to identify activation or inhibition of possible upstream regulators (kinases, receptors, transcription factors) involved in gene expression changes [45]. The latter analysis predicts highly significant activation of Dex liganded NR3C1 and TP53 transcription factors, in regulating MM1S cell cycle arrest and/or apoptosis [46] (Table 2). Furthermore, predicted activation of IKK and cyclin dependent kinases CDKN2A, CDKN1A may be indicators of DNA damage/cell death and cell cycle events triggered by Dex [47], [48], [49], [50]. Finally, possible inhibition of anti-apoptotic prosurvival signals stimulated by TNF and AKT kinases may also contribute to Dex induced cell death in MM1S (Table 2).
Figure 2. Dexamethasone triggers GC specific transcriptional changes in cell cycle, DNA damage/repair, cell death and immunological pathways in MM1S but not MM1R.
(A) The heat map represents relative gene expression levels of MM1S and MM1R cells, left untreated (solvent) or treated with 1 µM Dex for 72 h. Gene expression changes >2-fold and with a p-value <0.05 are plotted on the heat map. (B) Left panel: Realtime quantitative PCR (QPCR) validation of Dex dependent activation of GILZ (TSC22D3) mRNA transcription Right panel: Realtime quantitative PCR (QPCR) validation of Dex dependent repression of CDK2 mRNA in MM1S but not in MM1R. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05 respectively) as compared to control setups, determined by one-way (Dunnett’s Post-test) (C–D) Realtime quantitative PCR (QPCR) validation and Illumina microarray observation of the top GC regulated genes involved cell cycle, DNA damage/repair and cell death in S = MM1S but not R = MM1R. All the QPCR validation studies have been normalized to the 28S RNA housekeeping gene and are represented relative to MM1S untreated condition (S UT). Bar graphs represent relative mRNA (mean ± SEM) levels of three independent experiments. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05 respectively) from control setups as determined by two-way ANOVA (Bonferroni post-tests).
Table 1. Top molecular and cellular biofunctions upon 1 µM Dex treatment in MM1S cells for 72 h.
| Cellular function | P-value | Predicted Activating state | Z-score |
| Cell Death and Survival | 1.03E-12 | Increased | 3.301 |
| Cellular Assembly and Organization,DNA Replication, Recombination, Repair | 1.98E-09 | Decreased | −2.711 |
| Cell Cycle | 9.63E-09 | Decreased | −2.043 |
| DNA Replication, Recombination, and Repair | 2.89E-07 | Decreased | −2.22 |
| Cellular Assembly and Organization,DNA Replication, Recombination, Repair | 9.74E-07 | Decreased | −2.138 |
| Cellular Development, CellularGrowth and Proliferation | 1.28E-06 | Decreased | −3.067 |
| Cellular Growth and Proliferation | 1.81E-06 | Decreased | −3.185 |
| Cell Cycle | 5.96E-06 | Decreased | −2.205 |
| Cell Cycle | 7.98E-06 | Decreased | −2.305 |
| Cancer, Hematological Disease,Immunological Disease | 1.04E-05 | ||
| Immunological Disease | 2.27E-05 |
Table 2. Top upstream regulators upon 1 µM Dex treatment in MM1S cells for 72 h.
| Upstreamregulator | Molecule type | P-value | Predicted activation state | Activation Z-score |
| TP53 | Transcription regulator | 1.53E-49 | Activated | 5.504 |
| CDKN1A | Kinase | 5.31E-47 | Activated | 4.322 |
| CDKN2A | Transcription Regulator | 1.80E-37 | Activated | 6.844 |
| Dexamethasone | Chemical drug | 1.14E-11 | Activated | 2.719 |
| NR3C1 | Ligand-dependent nuclear receptor | 4.58E-08 | Activated | 2.359 |
| I kappa b kinase | Complex | 4.09E-07 | Activated | 2.629 |
| PPARG | ligand-dependent nuclear receptor | 1.35E-05 | Inhibited | −2.804 |
MicroRNAs responsive to GC in MM1S cells, but unresponsive to GC in MM1R cells, are predicted to modulate cell cycle, cell growth and proliferation pathways
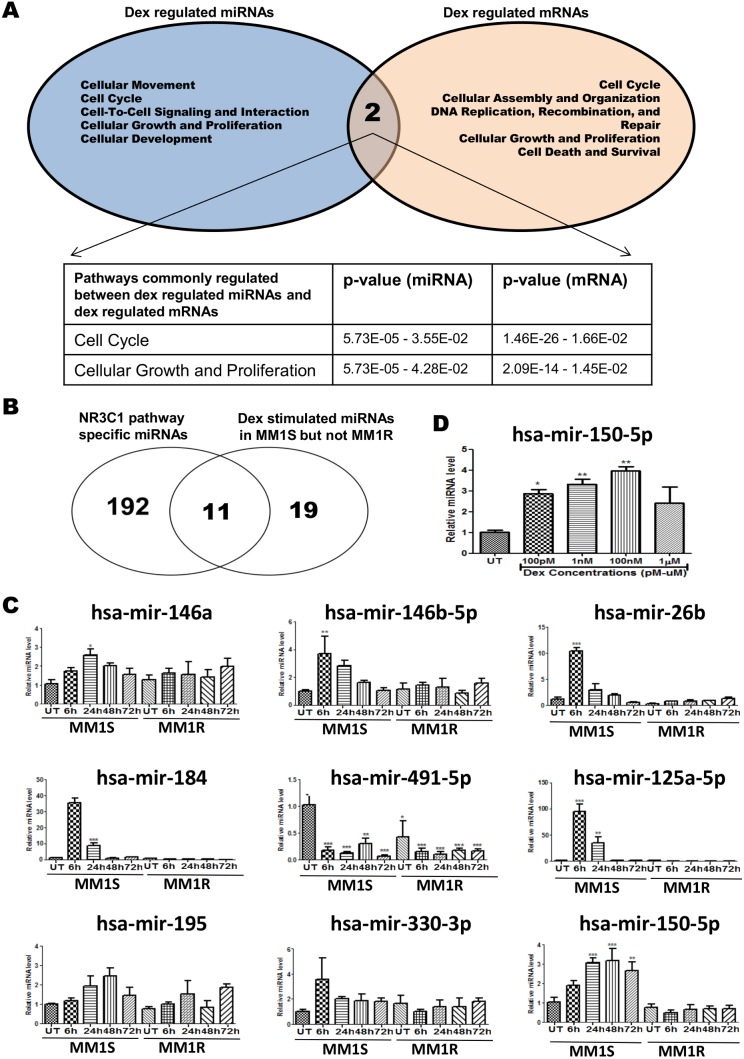
Total RNA was isolated from MM1S and MM1R cells treated with 1 µM Dex for 72 h, qPCR profiling of 760 miRNAs as previously described [32]. This allowed to identify 30 GC inducible and 14 GC repressed miRNAs (>2-fold) in MM1S, which do not change transcription in MM1R cells (summarized in Table 3). Next, IPA analysis was applied to cross compare microRNA and mRNA profiles in MM1S to predict target genes and top pathways which are affected by microRNA expression. Venn diagram analysis of the top 5 ranked molecular and cellular pathways enriched for genes differentially expressed in MM1S upon Dex exposure and predicted microRNA target genes responsive to Dex treatment reveals that GC responsive microRNAs may target mRNAs involved in cell cycle and cell proliferation pathways (Fig. 3A). For example, Dex inducible microRNAs mir-150-5p, mir-152, mir-146b-5p, mir-1290, mir-125a-5p, mir-1206 are predicted to modulate multiple target genes involved in cell cycle, cellular growth and proliferation pathways (as yellow marked in Table S1).
Table 3. Dex regulated miRNAs in MM1S but not MM1R upon 1 µM Dex treatment for 72 h.
| hsa-miR | Fold activation | Log2FC | hsa-miR | Fold repression | Log2FC |
| hsa-miR-146a | 2.10E+00 | 1.4 | hsa-miR-492 | 3.62E+08 | 0.00 |
| hsa-miR-152 | 2.17E+00 | 1.5 | hsa-miR-1275 | 4.13E+04 | 0.00 |
| hsa-miR-491-5p | 2.18E+00 | 1.5 | hsa-miR-766 | 3.14E+04 | 0.01 |
| hsa-miR-28-5p | 2.20E+00 | 1.5 | hsa-miR-412 | 6.25E+03 | 0.01 |
| hsa-miR-590-5p | 2.20E+00 | 1.5 | hsa-miR-636 | 3.81E+03 | 0.02 |
| hsa-miR-454# | 2.23E+00 | 1.5 | hsa-miR-181c | 3.34E+03 | 0.02 |
| hsa-miR-146b-5p | 2.24E+00 | 1.5 | rno-miR-29c# | 2.70E+03 | 0.02 |
| hsa-miR-627 | 2.43E+00 | 1.6 | hsa-miR-603 | 1.91E+03 | 0.02 |
| hsa-miR-26b | 2.58E+00 | 1.6 | hsa-miR-100# | 1.64E+03 | 0.02 |
| hsa-miR-95 | 2.69E+00 | 1.6 | hsa-miR-17# | 3.71E+02 | 0.05 |
| hsa-miR-324-3p | 3.16E+00 | 1.8 | hsa-miR-106b# | 1.03E+01 | 0.31 |
| hsa-miR-150-5p | 3.54E+00 | 1.9 | hsa-miR-27b | 5.46E+00 | 0.43 |
| hsa-miR-484 | 3.91E+00 | 2.0 | hsa-miR-92a-1# | 4.61E+00 | 0.47 |
| hsa-miR-1290 | 4.07E+00 | 2.0 | hsa-miR-301b | 2.11E+00 | 0.69 |
| hsa-miR-652 | 4.09E+00 | 2.0 | |||
| hsa-miR-330-3p | 4.19E+00 | 2.0 | |||
| hsa-miR-195 | 4.62E+00 | 2.1 | |||
| hsa-miR-184 | 8.25E+00 | 2.9 | |||
| hsa-miR-125a-5p | 2.49E+02 | 15.8 | |||
| hsa-miR-126 | 2.67E+02 | 16.4 | |||
| hsa-miR-515-3p | 3.28E+02 | 18.1 | |||
| hsa-miR-623 | 4.98E+02 | 22.3 | |||
| hsa-miR-1206 | 8.74E+02 | 29.6 | |||
| hsa-miR-802 | 1.01E+03 | 31.7 | |||
| hsa-miR-451 | 1.11E+03 | 33.4 | |||
| hsa-miR-892b | 4.12E+03 | 64.2 | |||
| hsa-miR-629 | 4.78E+03 | 69.1 | |||
| hsa-miR-518b | 7.14E+03 | 84.5 | |||
| hsa-miR-663B | 4.05E+04 | 201.3 | |||
| hsa-miR-34B | 5.69E+05 | 754.2 |
Figure 3. Experimental validation of GC inducible microRNAs, potentially involved in regulation of the NR3C1(GR) pathway in MM1S cells.
(A) Venn-diagram which summarizes the overlap between the top 5 predicted molecular and cellular functions generated by IPA from GC regulated miRNAs and GC regulated mRNAs (B) Venn-diagram which represents the crosscomparison of the list of GC responsive miRNAs identified in MM1S with the list of GR(NR3C1) pathway specific miRNAs according to MiRWalk prediction freeware. (C) Real-time QPCR validation of the transcription levels of 9 common miRNAs selected in Fig. 3B, in MM1S and MM1R cells treated for 24, 48 or 72 h with 1 µM Dex. (D) GC dose responsive regulation of mir-150-5p levels in MM1S cells treated for 72 h with GC. All the miRNA QPCR validation studies have been normalized to SNORD 95 and SNORD 96A housekeeping miRNAs according to manufacturer instructions and relative to MM1S untreated condition (S UT). Bar graphs represent relative miRNA (mean ± SEM) levels of three independent experiments as compared to control setups. Means with ***, **, * are significantly different to control setups (p<0.001, <0.01 or <0.05) as determined by one-way ANOVA (Dunnetts Post test).
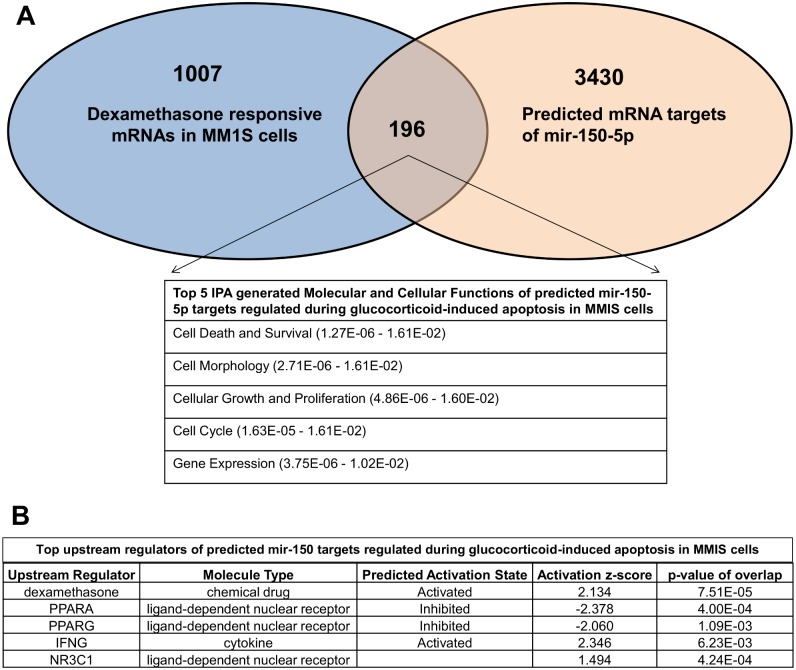
To reduce the number of Dex responsive microRNA for further validation and functional studies, we next cross compared the list of GC responsive miRNA identified in MM1S with a list of miRNAs predicted to regulate the GR(NR3C1) pathway (Biocarta) according to the MiRWalk database [34] (Fig. 3B). This generated a shortlist of 11 miRNAs, which were selected for further QPCR validation. Along the same line, IPA correlation of Dex repressed mRNAs and 30 Dex inducible miRNAs identified a similar GR specific role for mir-150-5p, mir-146b-5p and mir-125a-5p in the cell cycle and cellular growth and proliferation pathway (Table S1). Time dependent GC responsive changes in transcription levels of 11 selected microRNAs were evaluated by QPCR in MM1S and MM1R cells exposed for different time points (6 h, 24 h, 48 h and 72 h) to 1 µM of Dex. QPCR analysis could confirm time and GC specific induction of 5 hsa-microRNAs i.e. of mir-26b, mir-125a-5p, mir-146-5p, mir-150-5p, and mir-184 (Fig. 3C). Finally, hsa-mir-150-5p was selected for further functional studies because of its persistent GC responsive transcription between 6 and 72 h, and GC dose dependent regulation of mir-150-5p following 72 h treatment (Fig. 3D). Upon cross comparing 1203 Dex responsive target genes from MM1S cells with 1 µM Dex treatment for 72 h (gene selection was done based on p-value <0.05 and absolute fold change >1.5) and 3626 predicted hsa-mir-150-5p targets via the miRWalk database (prediction by at least 4 out of 10 available algorithms, i.e. DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22 and Targetscan), 196 common mRNA/gene targets (Fig. 4A) were identified in MM1S, which remain unaffected in MM1R cells (detailed list of genes can be found in Table S2). Upon employing IPA analysis, the 196 mRNA predicted GC responsive mir-150-5p target genes are enriched for cell death, cell morphology, cell cycle, cellular growth and proliferation pathways (Fig. 4A). Of special note, IPA upstream analysis predicts that mir-150-5p gene target genes are responsive for regulation by GC (Dex)/NR3C1, PPAR or STAT/IFNγ signaling (Fig. 4B).
Figure 4. Predicted role for mir-150-5p in regulation of cell death and survival, cell cycle, cellular growth and proliferation in MM1S cells.
(A) Venn-diagram which represents the crosscomparison of 3626 predicted hsa-mir-150-5p targets according to miRWalk software and 1203 dexamethasone responsive target genes in MM1S cells, which remain unaffected in MM1R cells (B) IPA upstream analysis of the predicted microRNA target mRNAs in MM1S cells.
Transfection of a mir-150-5p mimetic triggers GR specific gene expression in MM1S but not in MM1R cells, without modulating GR protein levels
To validate possible mir-150-5p effects on the predicted mRNA targets and/or cell cycle or cell proliferation pathways, we performed transfection experiments in MM1S and MM1R cells with a synthetic mir-150-5p mimetic in comparison to Dex treatment. Remarkably, although MM1S and MM1R show similar transcription levels of mir-150-5p upon mimetic transfection (Fig. 5A), we could only observe significant changes in mir-150-5p responsive mRNAs in MM1S but not in MM1R cells, upon analysing Illumina mRNA profiles. Interestingly, upon cross comparing Dex treated and mir-150-5p mimetic transfected MM1S/MM1R cells, IPA analysis revealed a remarkable overlap in mRNAs affected related to cell cycle-proliferation, DNA repair and cell death pathways in MM1S, as well as lack of effects for both setups in MM1R cells (Fig. 5B) (Table 4 and 5). To evaluate whether mir-150-5p may change GR/NR3C1 mRNA levels or protein stability, we performed QPCR and Western blot analysis following mir-150-5p transfection in MM1S cells. From Fig. 5C and D, it can be observed that mir-150-5p transfection does not change GR/NR3C1 mRNA or protein levels respectively. These results are in line with the in silico data, showing that none of the validated GC-inducible miRs are targeting GR-3′UTR mRNA (Fig. 5E and Table S3). Altogether, these results suggest that the mir-150-5p may mediate indirect GR like effects.
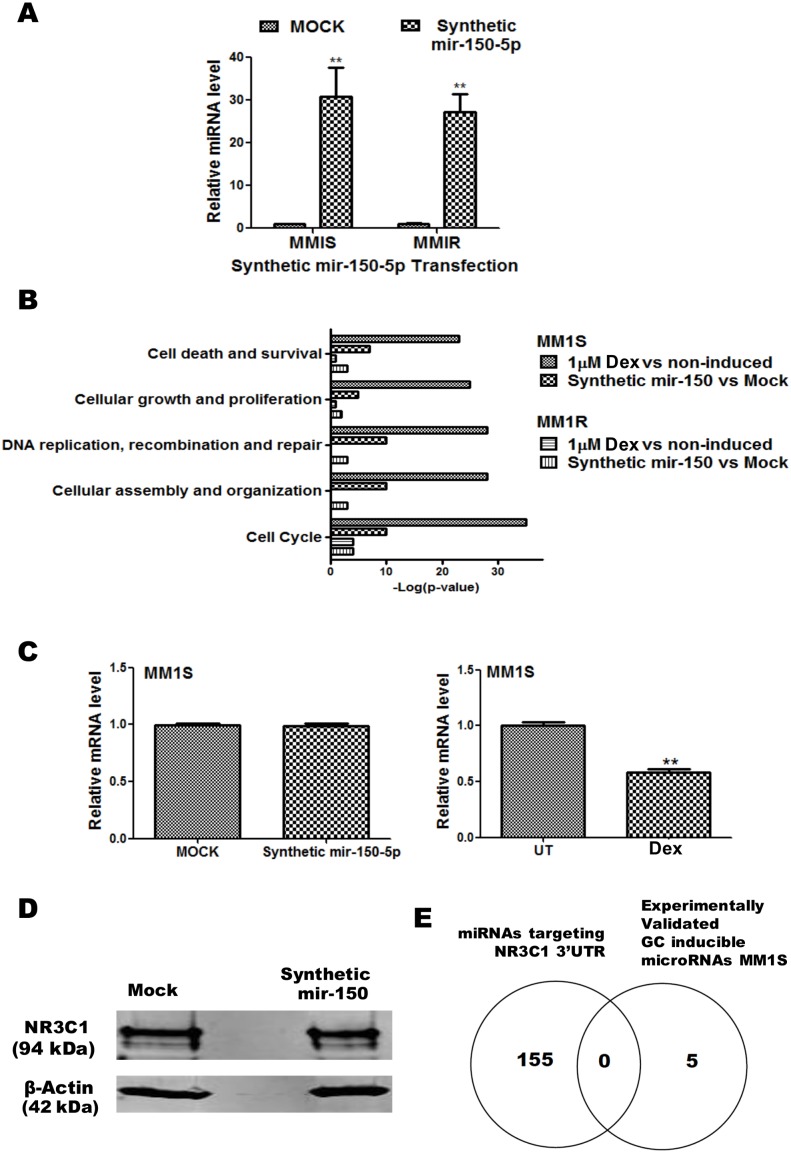
Figure 5. Mir-150-5p mimetic transfection triggers GR specific gene expression in MM1S but not in MM1R cells, without modulating GR protein levels.
(A) QPCR based quantification of synthetic mir-150-5p levels present 72 h after transfection of MM1S and MM1R cells. The miRNA expression levels have been normalized to SNORD 95 and SNORD 96A housekeeping miRNAs and are presented relative to MOCK control. Bar graphs represent relative miRNA (mean ± SEM) levels of three independent experiments. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05) from control setups according to two-way ANOVA (Bonferroni post-tests). (B) Comparison of top enriched pathways (IPA) of mRNA changes in MM1S cells treated for 72 h to 1 µM Dex versus changes in MM1S and MM1R cells after 72 h mock transfection of transfection with synthetic mir-150-5p. (C) NR3C1/GRα protein levels detected following 72 h transfection of a synthetic mir-150-5p mimetic in MM1S cells. (D) Venn-diagram which represents the overlap of experimentally validated GC inducible microRNAs in MM1S cells with the list of miRNAs predicted to target the 3′UTR of NR3C1. (E) QPCR analysis of NR3C1/GRα mRNA levels present in MM1S cells following 72 h transfection of a synthetic mir-150-5p mimetic (left panel) or following 72 h Dex (1 µM) treatment (right panel).
Table 4. Top molecular and cellular functions regulated upon synthetic mir-150-5p transfection in MM1S cells.
| Category | p-Value | Predicted Activation State | Activation z-score |
| Cell Cycle | 3.11E-06 | Decreased | –2.232 |
| Cellular Growth and Proliferation | 7.90E-05 | Decreased | –2.94 |
| Cellular Assembly and Organization | 2.45E-04 | Decreased | –2 |
| DNA Replication, Recombination,and Repair | 2.45E-04 | Decreased | –2 |
| Cellular Movement | 9.24E-03 | Decreased | –2.201 |
| Cell-To-Cell Signaling and Interaction | 9.24E-03 | Decreased | –2.201 |
| Immune Cell Trafficking | 9.24E-03 | Decreased | –2.201 |
| Inflammatory Response | 9.24E-03 | Decreased | –2.201 |
Table 5. Top upstream regulators in mir-150-5p transfected MM1S cells.
| UpstreamRegulator | FoldChange | MoleculeType | Predicted Activation State | Activation z-score | p-value ofoverlap |
| TP53 | 1.156 | transcription regulator | Activated | 2.056 | 9.11E-23 |
| CDKN1A | –1.86 | kinase | Activated | 2.433 | 1.91E-13 |
| IL6 | –1.027 | cytokine | Inhibited | –2.407 | 3.40E-13 |
| CDKN2A | 1.08 | transcription regulator | Activated | 2.656 | 1.44E-09 |
| dexamethasone | chemical drug | Activated | 2.668 | 3.35E-09 | |
| NR3C1 | –1.218 | ligand-dependent nuclear receptor | Activated | 2.007 | 1.07E-05 |
| PPARG | 1.04 | ligand-dependent nuclear receptor | Inhibited | –3.153 | 5.40E-05 |
| NFkB (complex) | complex | Inhibited | –2.669 | 3.71E-03 |
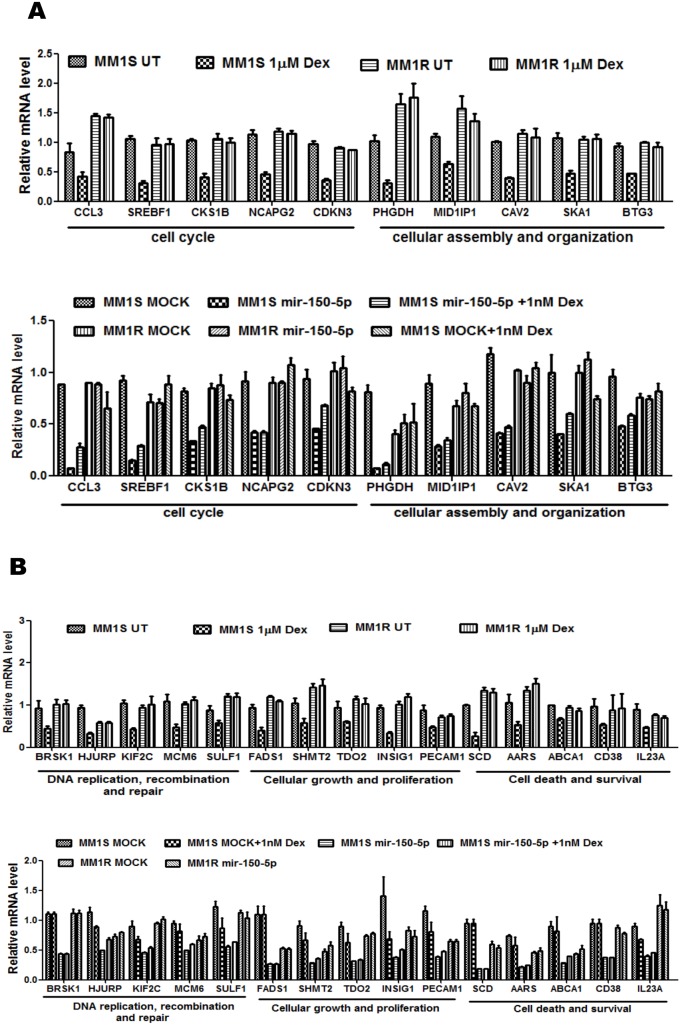
The genes involved in cell cycle-proliferation, DNA repair and cell death pathways which are regulated in Dex treated and mir-150-5p mimetic transfected MM1S/MM1R cells are indicated in Figure 6A and 6B or summarized in Table S4. To further characterize crosstalk between GC treatment and mir-150-5p transfection, we also performed microarray analysis of combination setups of mir-150-5p with 1 nM Dex, since 1 µM Dex treatment combinations did not yield array quality RNA because of excessive cytotoxicity. As can be noted from Fig. 6A and 6B, 1 nM Dex treatment triggers weaker gene expression changes as compared to 1 µM Dex treatment. Furthermore, since mir-150-5p levels upon transfection increase 30-fold (Fig. 6A) as compared to 3-fold induction upon Dex treatment (Fig. 3D), mir-150-5p mimetic overexpression triggers stronger gene expression changes than 1 nM Dex treatment. As such, combining mir-150-5p transfection with 1 nM Dex treatment did not further increase gene expression changes elicited by mir-150-p transfection alone (Fig. 7A & B). A further detailed comparison of GC and mir-150-5p responsive activation (i.e. GILZ and FKBP5) (Fig. 7A) or repression (IL23A, SKP2, BUB, SREBF1) (Fig. 7B) of typical GR target genes revealed similar effects in MM1S but lack of effects in MM1R. For example, transactivation of the FKBP5 and GILZ genes can be observed following mir-150-5p transfection and GC treatment in MM1S cells, although mir-150-5p effects on FKBP5 expression are stronger. Along the same line, repression of GR target genes IL23A, SKP2, BUB or SREBF1 can be observed upon Dex treatment as well following mir-150-5p transfection. Finally, besides the common regulated genes, additional GR target genes could be identified which were more selectively affected by mir-150-5p transfection (SCNN1G) or else GC treatment (GILZ, DUSP1, SGK1) indicating sophisticated fine tuning of GR regulation (Fig. 7C).
Figure 6. Mir-150-5p specific mRNA changes in MM1S are associated with pathways related to cell death and survival, cell cycle, cellular growth and proliferation (A–B).
mir-150-5p specific mRNA regulation of the genes involved in (i) cell cycle, (ii) cellular assembly and organization, (iii) DNA replication, recombination and repair, (iv) cell growth and proliferation, (v) cell death and survival. Illumina BeadChip Gene Expression Array results are presented as bar graphs, reflecting mean of gene expression fold change from three independent experiments of MM1S cells, treated for 72 h with 1 µM Dex vs control cells (upper panels A–B), or else of MM1S cells transfected for 72 h with synthetic mir-150-5p versus mock transfection, treated for 72 h with 1 nM Dex or a combination thereof versus control samples (lower panels A–B).
Figure 7. Mir-150-5p transfection mimics GR like gene activation and repression response in MM1S and lack of response in MM1R cells.
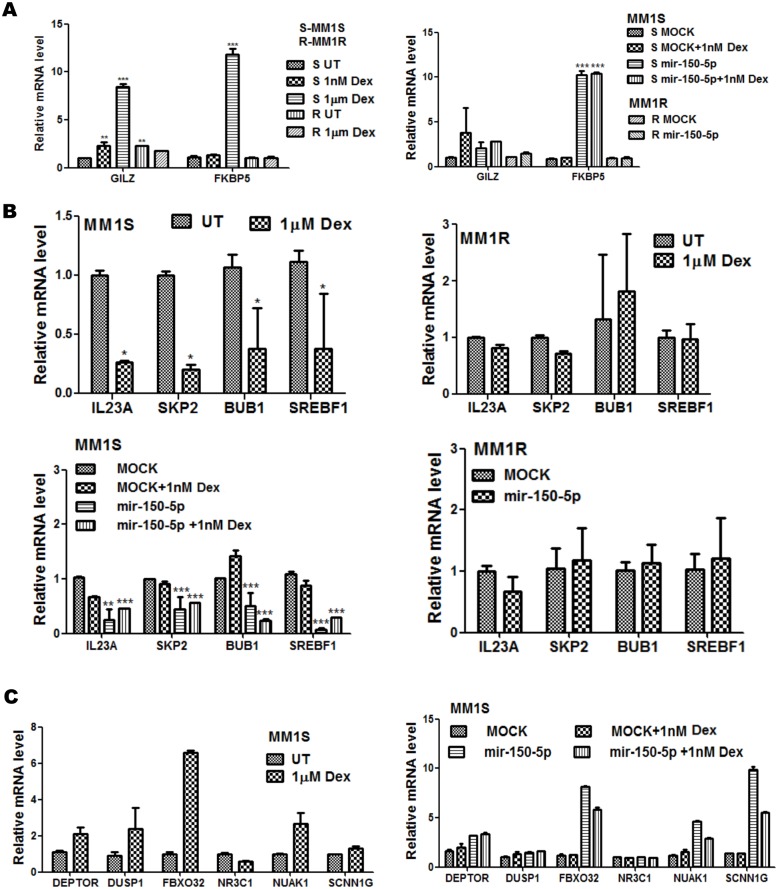
(A) Realtime quantitative PCR (QPCR) validation of GILZ (TSC22D3) and FKBP5 mRNA induction in MM1S and in MM1R cells, following 72 h treatment with 1 nM or 1 µM Dex (left panel), or following 72 h transfection with synthetic mir-150-5p, 72 h treatment with 1 nM Dex, or a combination thereof (right panel). Data represent (mean ± SEM) values of three independent experiments normalized to 28S RNA housekeeping gene and relative to the respective untreated/solvent (UT) or MOCK control setups. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05) from control setup as determined by two-way ANOVA (Bonferroni posttest). (B) Realtime quantitative PCR (QPCR) validation of MYB, IL23A, SKP2, BUB1, SREBF1 mRNA repression levels in MM1S or MM1R cells, following 72 h treatment with 1 nM or 1 µM Dex, or following 72 h transfection with synthetic mir-150-5p, 72 h treatment with 1 nM Dex, or a combination thereof. Data represent (mean ± SEM) values of three independent experiments normalized to 28S RNA housekeeping gene and relative to the respective untreated/solvent (UT) or MOCK control setups. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05) from control setup as determined by two-way ANOVA (Bonferroni posttests) (C) Illumina BeadChip Gene Expression Array results of selected genes are presented as bargraphs, reflecting mean of gene expression fold change from three independent experiments of MM1S cells, treated for 72 h with 1 µM Dex, or else of MM1S cells transfected for 72 h with synthetic mir-150-5p versus mock transfection, treated for 72 h with 1 nM Dex or a combination thereof versus control samples.
The GR-like gene response triggered by mir150-5p transfection is associated with complex regulation of transcription factors, hsp chaperones, kinases, cell cycle proteins, unfolded protein stress (UPR), AMPK/mTOR and chemokine specific signalling networks
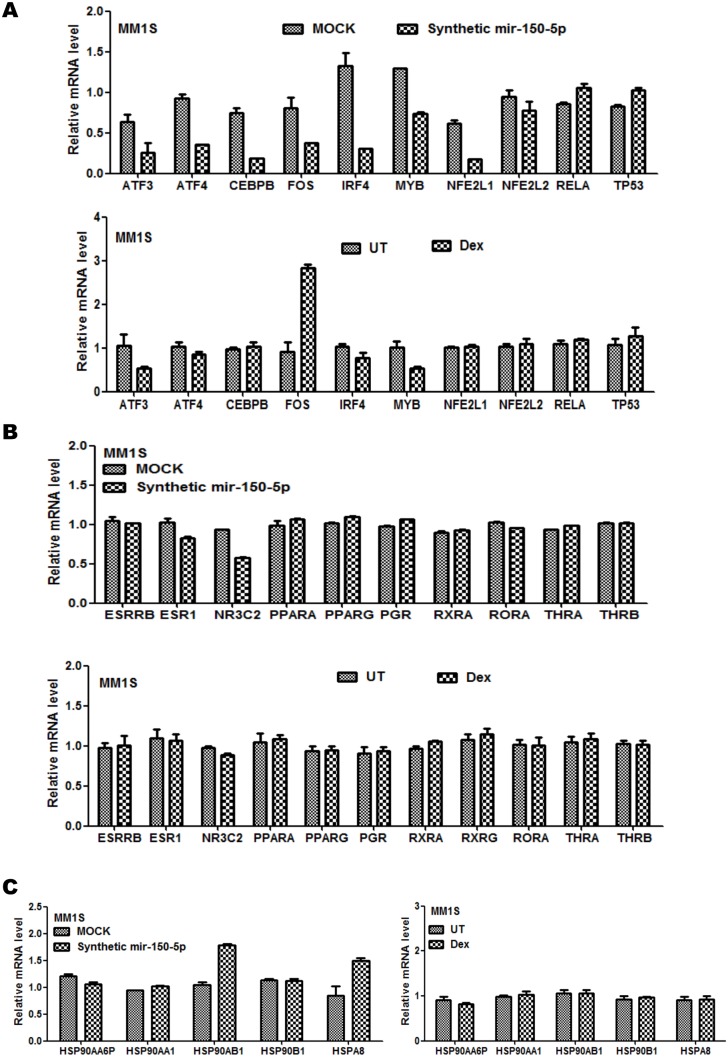
Due to lack of direct mir-150-5p effects on GR(NR3C1) mRNA and protein levels, we assumed that GR like downstream effects may be caused by mir-150-5p regulation of possible GR interaction partners (transcription factors, chaperones) or alternative nuclear hormone receptors (Fig. 8A–B). Although mir-150-5p and Dex setups both decrease MYB and ATF3 mRNA levels, opposite responses were observed for FOS transcription factors. Of particular interest, MYB is a key regulator of GR dependent gene expression in leukemia [51], [52], [53], [54], [55], [56], [57] (Fig. 8A).
Figure 8. Mir-150-5p transfection is associated with complex regulation of transcription factors, hormone receptors and chaperones.
Illumina BeadChip Gene Expression results of transcription factors (A), nuclear hormone receptors (B), and chaperones (C), are presented as bargraphs, reflecting mean of gene expression fold change from three independent experiments of MM1S, treated for 72 h with 1 µM Dex versus control setup (UT), or else of MM1S cells transfected for 72 h with synthetic mir-150-5p versus mock transfection.
With respect to hormone receptors, most hormone receptors remained constant upon mir-150-5p transfection (Fig. 8B), although weakly decreased levels of NR3C2 (MR) could be detected in the setup with mir-150-5p, whereas NR3C2 levels remain stable in presence of Dex. Interestingly, as a consequence of reciprocal crosstalk of NR3C2(MR) and NR3C1(GR) actions, suppressing MR function may indeed result in ligand independent stimulation of GR actions [58]. Next, we evaluated changes in transcription levels of hormone receptor chaperone molecules upon presence of mir-150-5p. Although most HSP mRNAs remain unaffected, significant changes in HSPA8 and HSP90AB1 levels could be detected following mir-150-5p transfection but not with Dex treatment, which may further contribute in fine tuning basal activity of nuclear hormone receptors (Fig. 8C) [59], [60], [61]. Furthermore mir-150-5p selectively decreases transcript levels of ATF4, JUN, NFE2L1(NRF1), CEBP/B and IRF4 but not of NFE2L2, TP53 and RelA (Fig. 8A).
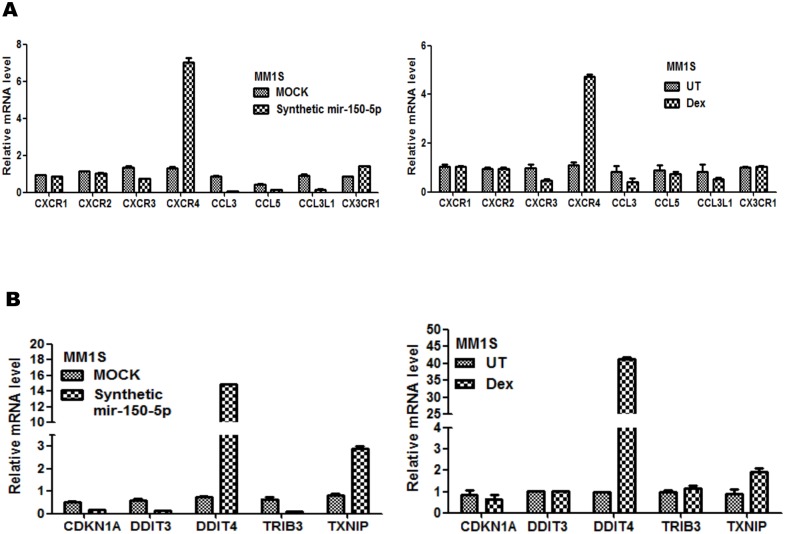
Finally, we also explored possible upstream effects of mir-150-5p on receptor kinase signalling pathways. Ligand independent hormone receptor (in)activation via kinases, chemokines and cytokines is well documented nowadays [62], [63], [64], [65], [66]. With respect to chemokine signalling, mir-150-5p transfection and GC treatment were found to increase mRNA levels of CXCR4 and CX3CR1 and decrease levels of CXCR3, CCL3, CCL5 whereas other members remain unaffected (Fig. 9A). Interestingly, various reports demonstrate a positive role for CXCR4/CXCL12 in hormone independent receptor activation [65], [66], [67], [68], [69]. Surprisingly, we observed a significant increase in CXCR4 levels, upon mir-150-5p transfection, in contrast to earlier reports demonstrating CXCR4 mRNA silencing of its 3′ UTR via mir-150-5p [68], [70]. Although various studies illustrate tumor promoting and prometastatic effects of CXCR4 [71], [72], its expression also plays a pivotal role in haematopoiesis [73] and has recently been reported as a good prognostic indicator in multiple myeloma [74]. Finally, we also identified remarkable effects on effectors in cell cycle (CDKN1A) and UPR/mTOR signalling pathways (DDIT3, DDIT4, TXNIP) and the Tribble family Ser/Thr pseudokinase (TRIB3) [75] which altogether may crosstalk with glucocorticoid receptor signalling and GC therapy response in MM cells [76], [77], [78] (Fig. 9B). Of special note, involvement of C/EBP and ATF4 (Fig. 8A) has been demonstrated in transcriptional regulation of UPR stress proteins DDIT3, DDIT4, TXNIP [79], [80], further corroborating our results.
Figure 9. Mir-150-5p transfection is associated with complex regulation of chemokine, cell cycle, UPR/mTOR signaling.
Illumina BeadChip Gene Expression results of chemokine receptors or ligands (A), or regulators of cell cycle, unfolded protein stress (UPR/mTOR signaling) (B), are presented as bargraphs, reflecting mean of gene expression fold change from three independent experiments of MM1S, treated for 72 h with 1 µM Dex versus control setup (UT), or else of MM1S cells transfected for 72 h with synthetic mir-150-5p versus mock transfection.
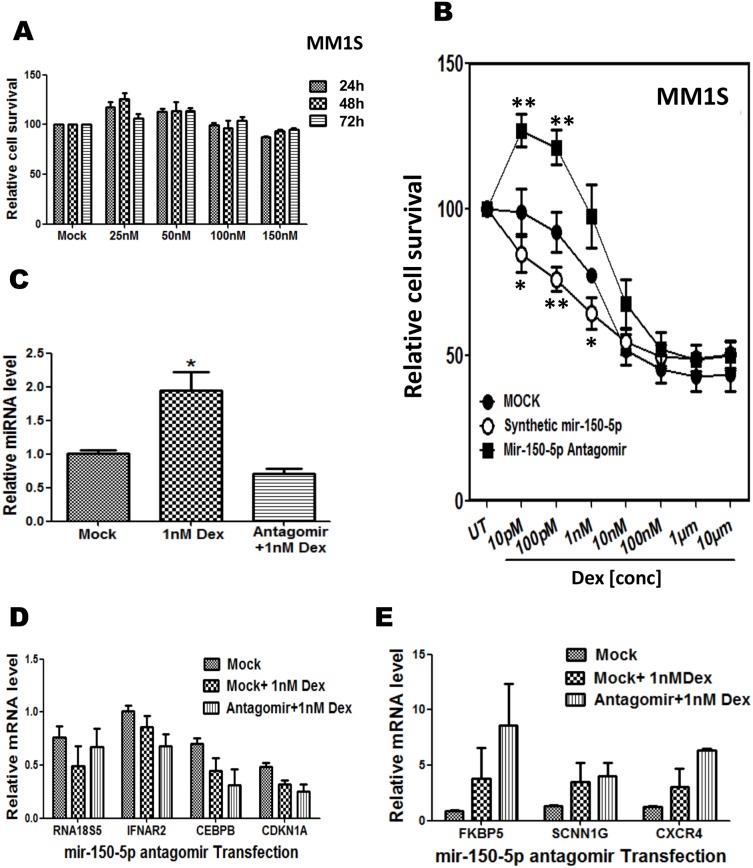
Weak sensitisation of low dose GC therapy response by mir-150-5p mimetic transfection can be reversed by transfection of a mir-150-5p-specific antagomir in MM1S cells
Since mir-150-5p modulates various genes related to cell cycle, cell proliferation and cell death pathways, we next wanted to measure cell death induced by ectopic mir-150-5p transfection. Surprisingly, no substantial induction of cell death could be observed by MTT-assay at 24, 48 and 72 h following transfection of MM1S cells by increasing amounts of mir150-5p (Fig. 10A). The latter suggests that mir-150-5p induced gene expression changes are not sufficient to trigger GC induced cell death and additional GC actions are required to kill the cells [15]. Next, we evaluated whether ectopic mir-150-5p transfection may sensitize for GC induced cell death. In this respect, mock and mir-150-5p transfected MM1S cells were exposed to various doses of Dex for 72 h and cell survival was again evaluated by a MTT-assay. From Fig. 10B, it can be observed that presence of mir-150-5p weakly increases GC induced cell death at nanomolar (nM) concentrations, although no significant effects could be discriminated at micromolar (µM) concentrations of Dex treatment. Moreover, upon GC treatment of MM1S cells transfected with a mir-150-5p specific antagomir, to reverse mir-150-5p effects, cell death could be reversed at low GC doses. This prompted us to evaluate corresponding changes in gene expression following GC treatment of MM1S cells transfected with mir-150-5p antagomir. Although we could confirm weak GC specific upregulation of mir150-5p and mild reduction of mir 150-5p levels upon GC treatment of MM1S cells transfected with mir-150-5p specific antagomir (Fig. 10C), array approaches seemed not sensitive enough to identify weak changes in gene repression or activation in GC treated cells in presence or absence of the antagomir (Fig. 10D–E and data not shown).
Figure 10. Weak sensitisation of low dose GC therapy response by mir-150-5p mimetic transfection can be reversed by transfection of a mir-150-5p-specific antagomir in MM1S cells.
(A) MTT based quantification of MM1S cell survival, following 24, 48, or 72 h transfection with increasing doses of synthetic mir-150-5p mimetic as compared to mock transfection control (B) MTT based quantification of MM1S cell survival, following 72 h treatment with increasing doses of Dex, in combination with either mock transfection, else transfection with a synthetic mir-150-5p mimetic or mir-150-5p specific antagomir (as indicated in figure). Data represented are relative cell survival levels (mean ± SEM) of three independent experiments. Means with ***, **, * are significantly different (p<0.001, <0.01 or <0.05) from control setup as determined by two-way ANOVA (Bonferroni posttests). (C) Realtime quantitative PCR (QPCR) quantification of mir-150-5p transcription levels following 72 h treatment with 1 nM Dex, either or not in combination with transfection of a mir-150-5p specific antagomir. (D–E) Illumina BeadChip Gene Expression results of a subset of genes are presented as bargraphs, reflecting the mean fold change from three independent experiments of MM1S cells treated for 72 h with 1 nM Dex vs. control setup, either or not in combination with transfection of a mir-150-5p specific antagomir.
Discussion
Decreased proliferation and induction of cell death by apoptosis are two major cellular events to reduce the growth of and to eliminate cancer cells. GCs are known to induce a cell cycle arrest preceding apoptosis. Treating the cancerous cells with higher doses of Dex can destabilize the glucocorticoid receptor (GR) which ultimately results in GC resistance [38], [81], [82], [83]. The characterization of miRNAs and their target mRNAs involved in regulation of the GC therapy response and/or resistance is an area of intense research and relatively little is known regarding GC specific miRNA regulatory networks. By combining genome-wide transcriptome analysis and Ingenuity Pathway Analysis, we envisioned to unravel the biological effects of GC responsive miRNAs and their putative mRNA targets in MM GC therapy. We identified 30 GC inducible and 14 GC repressed microRNAs which might modulate multiple aspects of GC therapy response in MM, including cell cycle, cell proliferation, DNA replication, gene expression and cell death pathways. Similarly, a study of GC responsive microRNAs in mucosal biopsies of eosinophilic esophagitis patients characterized 32 GC inducible and 4 GC repressed microRNAs, of which 7 GC inducible microRNAs are in common with our list, irrespective of the cellular context [84]. Next, we selected mir-150-5p as the most interesting GC inducible microRNA for further investigation because of its persistent upregulation by GC in MM1S cells and its possible tumor suppressor functions in (malignant) haematopoiesis [26], [54], [85], [86], [87]. According to IPA analysis, predicted mir-150-5p mRNA targets were related to cell cycle and cell proliferation responses preceding apoptosis. In line with IPA analysis, ectopic mir-150-5p expression in presence of low GC concentrations was found to weakly sensitize for GC induced cell death, whereas a mir-150-5p antagomir could reverse this effect. Remarkably, upon ectopic transfection of mir-150-5p or GC treatment in MM1S cells, various mRNAs were identified which respond similarly to increased mir-150-5p levels or GC treatment (i.e. MYB, IL23A, SKP2, BUB1, SREBP1, FKBP5). This suggests that mir-150-5p effects are strongly linked to GR function. Moreover, GR specific mir-150-5p effects are completely lost in MM1R cells and mir-150-5p can not restore GC sensitivity in MM1R cells lacking GR protein expression. Since mir-150-5p overexpression did not substantially change GR expression levels, we believe the GR like effects may be triggered via complex indirect regulation of GR interacting transcription factors (MYB, C/EBP, JUN, FOS) or hormone receptors (NR3C2/MR), GR chaperones (FKBP5, HSPA8, HSP90AB1), as well as various effectors of unfolded protein stress (ATF4, DDIT3, DDIT4, TXNIP) and chemokine specific signalling (CXCR3, CCL3, CCL5, CXCR4). Of special note, mir-150-5p selective decrease in NR3C2 (MR) levels may result in reciprocal stimulation of GR actions [58]. The GR dependent mir-150-5p effects also suggest that GR protein may contribute in mir-150-5p specific post-transcriptional regulation. For example, NR3C1 (GR) dependent post-transcriptional regulation of CCL2 and CCL7 but not of CCL5 mRNA has been demonstrated upon RNA binding of GR to a GU rich motif at the 5′UTR [88]. Alternatively, mir-150-5p may modulate GR/DNA binding, in analogy to the long noncoding RNA Gas5 [24].
Altogether, we identified various novel and previously published GC inducible microRNAs. Furthermore, multiple mir-150-5p responsive mRNAs are associated with GC induced cell proliferation and cell death therapy response in multiple myeloma. Functional cell survival studies in MM1S cells confirmed weak GC therapy sensitizing effects of mir-150-5p overexpression at low GC dose, whereas opposite effects could be observed in presence of a mir-150-5p specific antagomir. However, although QPCR assays revealed reversal of Dex induced mir-150-5p levels following antagomir transfection and attenuation of GC induced cell death, subsequent microarray analysis failed to identify significant mir-150-5p specific changes in GC regulated mRNA levels (Fig. 10D–E and data not shown). We believe that the array study may lack sensitivity and statistical power to identify significant mRNA changes when reversing a 2-fold time-dependent GC specific induction of mir-150p levels by its antagomir, whereas a 30-fold overexpression in mir150-5p levels following transfection may elicit more robust effects in gene expression arrays. In a similar study, failure of antagomirs to reverse mir-92 specific gene expression changes despite reduced levels of mir-92 in presence of its antagomir, was explained by possible redundancy in mir-92 regulated mRNA networks [89]. Along the same line, since we identified various GC inducible microRNAs related to the NR3C1 pathway, we can also not exclude redundant mir-150-5p regulated mRNA networks involved in NR3C1 regulation. For example, early GC inducible microRNAs, i.e. mir-26b, mir125-5p and mir184 could be responsible for subsequent increase of mir150-5p levels to fine tune GR activity (Fig. 3C). Recent reports also show coupling of mir150-5p to mir124- and mir155-networks [90], [91]. Alternatively, redundant mir-150-5p isomirs may escape control of the antagomir [91]. Future research will be required to further untangle specificity or redundancy of mir-150-5p-specific NR3C1 networks by complementary transcriptomic and proteomic approaches. Finally, recent reports demonstrating mir-150-5p secretion into microvesicles may allow development of a diagnostic biomarker to follow GC therapy response or else, combination of low GC doses with synthetic mir-150-5p vesicles may sensitize GC therapy responses in lymphoid cancers [92], [93].
Supporting Information
IPA prediction of microRNA targets based on correlation of transcriptional changes of specific microRNAs (up/down) with changes in mRNA levels (opposite direction down/up) and corresponding pathways in MM1S cells treated for 72 h with Dex.
(XLSX)
Crosscomparison of 1203 Dex responsive target genes from MM1S cells exposed for 72 h to 1 µM Dex (gene selection was done based on p-value <0.05 and absolute fold change >1.5) and 3626 predicted hsa-mir-150-5p targets via the miRWalk database (prediction by at least 4 out of 10 available algorithms, i.e. DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22 and Targetscan) 196 common mRNA/gene targets were identified in MM1S, which remain unaffected in MM1R cells.
(XLSX)
In silico prediction of microRNAs targeting the NR3C1 gene locus versus the NR3C1 3′ UTR.
(XLSX)
Detailed list of genes involved in cell cycle-proliferation, DNA repair and cell death pathways which are similarly regulated in Dex treated and mir-150-5p mimetic transfected MM1S cells, but which remain unaffected in MM1R cells (values represent (fold change versus control cells)).
(XLSX)
Acknowledgments
The authors thank all lab members for valuable scientific discussions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Raw gene expression array data were uploaded to the Gene Expression Omnibus (GEO) database and have accession number GSE59805.
Funding Statement
Research has been supported by the Strategic Basic Research (SBO) grant of the Agency for Innovation by Science and Technology (IWT, Belgium), the Multiple Myeloma Research Foundation (MMRF, US), FWO and NOI/DOCPRO (UA) research grants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, et al. (2004) Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res 64:1546–1558. [DOI] [PubMed] [Google Scholar]
- 2. Bommert K, Bargou RC, Stuhmer T (2006) Signalling and survival pathways in multiple myeloma. Eur J Cancer 42:1574–1580. [DOI] [PubMed] [Google Scholar]
- 3. Kastrinakis NG, Gorgoulis VG, Foukas PG, Dimopoulos MA, Kittas C (2000) Molecular aspects of multiple myeloma. Ann Oncol 11:1217–1228. [DOI] [PubMed] [Google Scholar]
- 4. Kyle RA, Rajkumar SV (2008) Multiple myeloma. Blood 111:2962–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niesvizky R, Jayabalan DS, Christos PJ, Furst JR, Naib T, et al. (2008) BiRD (Biaxin [clarithromycin]/Revlimid [lenalidomide]/dexamethasone) combination therapy results in high complete- and overall-response rates in treatment-naive symptomatic multiple myeloma. Blood 111:1101–1109. [DOI] [PubMed] [Google Scholar]
- 6. Gay F, Rajkumar SV, Coleman M, Kumar S, Mark T, et al. (2010) Clarithromycin (Biaxin)-lenalidomide-low-dose dexamethasone (BiRd) versus lenalidomide-low-dose dexamethasone (Rd) for newly diagnosed myeloma. Am J Hematol 85:664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rossi A, Mark T, Jayabalan D, Christos P, Zafar F, et al. (2013) BiRd (clarithromycin, lenalidomide, dexamethasone): an update on long-term lenalidomide therapy in previously untreated patients with multiple myeloma. Blood 121:1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korde N, Kristinsson SY, Landgren O (2011) Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM): novel biological insights and development of early treatment strategies. Blood 117:5573–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, et al. (2013) Direct Reversal of Glucocorticoid Resistance by AKT Inhibition in Acute Lymphoblastic Leukemia. Cancer Cell 24:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turner JD, Muller CP (2005) Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol 35:283–292. [DOI] [PubMed] [Google Scholar]
- 11. Schmidt S, Rainer J, Ploner C, Presul E, Riml S, et al. (2004) Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ 11 Suppl 1: S45–55. [DOI] [PubMed] [Google Scholar]
- 12. De Bosscher K (2010) Selective Glucocorticoid Receptor modulators. J Steroid Biochem Mol Biol 120:96–104. [DOI] [PubMed] [Google Scholar]
- 13. Kumar R, Thompson EB (2005) Gene regulation by the glucocorticoid receptor: structure: function relationship. J Steroid Biochem Mol Biol 94:383–394. [DOI] [PubMed] [Google Scholar]
- 14. De Bosscher K, Beck IM, Haegeman G (2010) Classic glucocorticoids versus non-steroidal glucocorticoid receptor modulators: survival of the fittest regulator of the immune system? Brain Behav Immun 24:1035–1042. [DOI] [PubMed] [Google Scholar]
- 15. Kfir-Erenfeld S, Yefenof E (2014) Non-genomic events determining the sensitivity of hemopoietic malignancies to glucocorticoid-induced apoptosis. Cancer Immunol Immunother 63:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kfir-Erenfeld S, Sionov RV, Spokoini R, Cohen O, Yefenof E (2010) Protein kinase networks regulating glucocorticoid-induced apoptosis of hematopoietic cancer cells: fundamental aspects and practical considerations. Leuk Lymphoma 51:1968–2005. [DOI] [PubMed] [Google Scholar]
- 17. De Iudicibus S, Franca R, Martelossi S, Ventura A, Decorti G (2011) Molecular mechanism of glucocorticoid resistance in inflammatory bowel disease. World J Gastroenterol 17:1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barnes PJ, Adcock IM (2009) Glucocorticoid resistance in inflammatory diseases. Lancet 373:1905–1917. [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Yao Y, Xiao F, Lan X, Yu C, et al. (2013) Administration of dexamethasone protects mice against ischemia/reperfusion induced renal injury by suppressing PI3K/AKT signaling. Int J Clin Exp Pathol 6:2366–2375. [PMC free article] [PubMed] [Google Scholar]
- 20. Schlossmacher G, Stevens A, White A (2011) Glucocorticoid receptor-mediated apoptosis: mechanisms of resistance in cancer cells. J Endocrinol 211:17–25. [DOI] [PubMed] [Google Scholar]
- 21. Li XJ, Luo XQ, Han BW, Duan FT, Wei PP, et al. (2013) MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong Y, Wu J, Zhao J, Wang H, Liu Y, et al. (2013) miR-29b and miR-29c Are Involved in Toll-Like Receptor Control of Glucocorticoid-Induced Apoptosis in Human Plasmacytoid Dendritic Cells. PLoS One 8:e69926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pore SK, Choudhary A, Rathore B, Ganguly A, Sujitha P, et al. (2013) Hsp90-targeted miRNA-liposomal formulation for systemic antitumor effect. Biomaterials 34:6804–6817. [DOI] [PubMed] [Google Scholar]
- 24. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673–676. [DOI] [PubMed] [Google Scholar]
- 26. Morris VA, Zhang A, Yang T, Stirewalt DL, Ramamurthy R, et al. (2013) MicroRNA-150 expression induces myeloid differentiation of human acute leukemia cells and normal hematopoietic progenitors. PLoS One 8:e75815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M (2012) microRNAs in cancer management. Lancet Oncol 13:e249–258. [DOI] [PubMed] [Google Scholar]
- 28. Greenstein S, Krett NL, Kurosawa Y, Ma C, Chauhan D, et al. (2003) Characterization of the MM.1 human multiple myeloma (MM) cell lines. Experimental Hematology 31:271–282. [DOI] [PubMed] [Google Scholar]
- 29. Suttana W, Mankhetkorn S, Poompimon W, Palagani A, Zhokhov S, et al. (2010) Differential chemosensitization of P-glycoprotein overexpressing K562/Adr cells by withaferin A and Siamois polyphenols. Mol Cancer 9:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23:2183–2184. [DOI] [PubMed] [Google Scholar]
- 31. Saeed AI, Sharov V, White J, Li J, Liang W, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 32. Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, et al. (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, et al. (2008) High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 36:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847. [DOI] [PubMed] [Google Scholar]
- 35. Tessel MA, Benham AL, Krett NL, Rosen ST, Gunaratne PH (2011) Role for microRNAs in regulating glucocorticoid response and resistance in multiple myeloma. Horm Cancer 2:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murray MY, Rushworth SA, Zaitseva L, Bowles KM, Macewan DJ (2013) Attenuation of dexamethasone-induced cell death in multiple myeloma is mediated by miR-125b expression. Cell Cycle 12:2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grugan KD, Ma C, Singhal S, Krett NL, Rosen ST (2008) Dual regulation of glucocorticoid-induced leucine zipper (GILZ) by the glucocorticoid receptor and the PI3-kinase/AKT pathways in multiple myeloma. J Steroid Biochem Mol Biol 110:244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gross KL, Oakley RH, Scoltock AB, Jewell CM, Cidlowski JA (2011) Glucocorticoid receptor alpha isoform-selective regulation of antiapoptotic genes in osteosarcoma cells: a new mechanism for glucocorticoid resistance. Mol Endocrinol 25:1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez-Vega B, Krett N, Rosen ST, Gandhi V (2006) Glucocorticoid receptor transcriptional isoforms and resistance in multiple myeloma cells. Mol Cancer Ther 5:3062–3070. [DOI] [PubMed] [Google Scholar]
- 40.Gentleman R (2005) Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science+Business Media. xix, 473 p. [Google Scholar]
- 41. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. [DOI] [PubMed] [Google Scholar]
- 42. Itani OA, Liu KZ, Cornish KL, Campbell JR, Thomas CP (2002) Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am J Physiol Endocrinol Metab 283:E971–979. [DOI] [PubMed] [Google Scholar]
- 43. Granes F, Roig MB, Brady HJ, Gil-Gomez G (2004) Cdk2 activation acts upstream of the mitochondrion during glucocorticoid induced thymocyte apoptosis. Eur J Immunol 34:2781–2790. [DOI] [PubMed] [Google Scholar]
- 44. Kullmann MK, Grubbauer C, Goetsch K, Jakel H, Podmirseg SR, et al. (2013) The p27-Skp2 axis mediates glucocorticoid-induced cell cycle arrest in T-lymphoma cells. Cell Cycle 12:2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kramer A, Green J, Pollard J Jr, Tugendreich S (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H, Qian W, Weng X, Wu Z, Zhuang Q, et al. (2012) Glucocorticoid receptor and sequential P53 activation by dexamethasone mediates apoptosis and cell cycle arrest of osteoblastic MC3T3-E1 cells. PLoS One 7:e37030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Obexer P, Hagenbuchner J, Rupp M, Salvador C, Holzner M, et al. (2009) p16INK4A sensitizes human leukemia cells to FAS- and glucocorticoid-induced apoptosis via induction of BBC3/Puma and repression of MCL1 and BCL2. J Biol Chem 284:30933–30940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baghdassarian N, Catallo R, Mahly MA, Ffrench P, Chizat F, et al. (1998) Glucocorticoids induce G1 as well as S-phase lengthening in normal human stimulated lymphocytes: differential effects on cell cycle regulatory proteins. Exp Cell Res 240:263–273. [DOI] [PubMed] [Google Scholar]
- 49. Nicholson L, Hall AG, Redfern CP, Irving J (2010) NFkappaB modulators in a model of glucocorticoid resistant, childhood acute lymphoblastic leukemia. Leuk Res 34:1366–1373. [DOI] [PubMed] [Google Scholar]
- 50. Bladh LG, Liden J, Pazirandeh A, Rafter I, Dahlman-Wright K, et al. (2005) Identification of target genes involved in the antiproliferative effect of glucocorticoids reveals a role for nuclear factor-(kappa)B repression. Mol Endocrinol 19:632–643. [DOI] [PubMed] [Google Scholar]
- 51. Bian Z, Li L, Cui J, Zhang H, Liu Y, et al. (2011) Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol 225:544–553. [DOI] [PubMed] [Google Scholar]
- 52. Chen S, Wang Z, Dai X, Pan J, Ge J, et al. (2013) Re-expression of microRNA-150 induces EBV-positive Burkitt lymphoma differentiation by modulating c-Myb in vitro. Cancer Sci 104:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang X, Huang H, Li Z, Li Y, Wang X, et al. (2012) Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell 22:524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adams BD, Guo S, Bai H, Guo Y, Megyola CM, et al. (2012) An in vivo functional screen uncovers miR-150-mediated regulation of hematopoietic injury response. Cell Rep 2:1048–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zheng Q, Zhou L, Mi QS (2012) MicroRNA miR-150 is involved in Valpha14 invariant NKT cell development and function. J Immunol 188:2118–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sarvaiya PJ, Schwartz JR, Geng CD, Vedeckis WV (2012) c-Myb interacts with the glucocorticoid receptor and regulates its level in pre-B-acute lymphoblastic leukemia cells. Mol Cell Endocrinol 361:124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Geng CD, Vedeckis WV (2011) A new, lineage specific, autoup-regulation mechanism for human glucocorticoid receptor gene expression in 697 pre-B-acute lymphoblastic leukemia cells. Mol Endocrinol 25:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hattori T, Murase T, Iwase E, Takahashi K, Ohtake M, et al. (2013) Glucocorticoid-induced hypertension and cardiac injury: effects of mineralocorticoid and glucocorticoid receptor antagonism. Nagoya J Med Sci 75:81–92. [PMC free article] [PubMed] [Google Scholar]
- 59. Jaaskelainen T, Makkonen H, Palvimo JJ (2011) Steroid up-regulation of FKBP51 and its role in hormone signaling. Curr Opin Pharmacol 11:326–331. [DOI] [PubMed] [Google Scholar]
- 60. Tatro ET, Everall IP, Kaul M, Achim CL (2009) Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res 1286:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Beck IM, Drebert ZJ, Hoya-Arias R, Bahar AA, Devos M, et al. (2013) Compound A, a selective glucocorticoid receptor modulator, enhances heat shock protein Hsp70 gene promoter activation. PLoS One 8:e69115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robertson S, Hapgood JP, Louw A (2013) Glucocorticoid receptor concentration and the ability to dimerize influence nuclear translocation and distribution. Steroids 78:182–194. [DOI] [PubMed] [Google Scholar]
- 63. Hu A, Josephson MB, Diener BL, Nino G, Xu S, et al. (2013) Pro-asthmatic cytokines regulate unliganded and ligand-dependent glucocorticoid receptor signaling in airway smooth muscle. PLoS One 8:e60452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eickelberg O, Roth M, Lorx R, Bruce V, Rudiger J, et al. (1999) Ligand-independent activation of the glucocorticoid receptor by beta2-adrenergic receptor agonists in primary human lung fibroblasts and vascular smooth muscle cells. J Biol Chem 274:1005–1010. [DOI] [PubMed] [Google Scholar]
- 65. Kasina S, Macoska JA (2012) The CXCL12/CXCR4 axis promotes ligand-independent activation of the androgen receptor. Mol Cell Endocrinol 351:249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sauve K, Lepage J, Sanchez M, Heveker N, Tremblay A (2009) Positive feedback activation of estrogen receptors by the CXCL12-CXCR4 pathway. Cancer Res 69:5793–5800. [DOI] [PubMed] [Google Scholar]
- 67. Boudot A, Kerdivel G, Habauzit D, Eeckhoute J, Le Dily F, et al. (2011) Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS One 6:e20898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rolland-Turner M, Goretti E, Bousquenaud M, Leonard F, Nicolas C, et al. (2013) Adenosine stimulates the migration of human endothelial progenitor cells. Role of CXCR4 and microRNA-150. PLoS One 8:e54135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rhodes LV, Short SP, Neel NF, Salvo VA, Zhu Y, et al. (2011) Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res 71:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tano N, Kim HW, Ashraf M (2011) microRNA-150 regulates mobilization and migration of bone marrow-derived mononuclear cells by targeting Cxcr4. PLoS One 6:e23114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, et al. (2013) Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther 6:1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Han TT, Fan L, Li JY, Xu W (2014) Role of chemokines and their receptors in chronic lymphocytic leukemia: Function in microenvironment and targeted therapy. Cancer Biol Ther 15:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR (1998) Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393:595–599. [DOI] [PubMed] [Google Scholar]
- 74. Bao L, Lai Y, Liu Y, Qin Y, Zhao X, et al. (2013) CXCR4 is a good survival prognostic indicator in multiple myeloma patients. Leuk Res 37:1083–1088. [DOI] [PubMed] [Google Scholar]
- 75. Liang KL, Rishi L, Keeshan K (2013) Tribbles in acute leukemia. Blood 121:4265–4270. [DOI] [PubMed] [Google Scholar]
- 76. Das I, Png CW, Oancea I, Hasnain SZ, Lourie R, et al. (2013) Glucocorticoids alleviate intestinal ER stress by enhancing protein folding and degradation of misfolded proteins. J Exp Med 210:1201–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ghavami S, Eshragi M, Ande SR, Chazin WJ, Klonisch T, et al. (2010) S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res 20:314–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li C, Chen H, Ding F, Zhang Y, Luo A, et al. (2009) A novel p53 target gene, S100A9, induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. Biochem J 422:363–372. [DOI] [PubMed] [Google Scholar]
- 79. Polman JA, Hunter RG, Speksnijder N, van den Oever JM, Korobko OB, et al. (2012) Glucocorticoids modulate the mTOR pathway in the hippocampus: differential effects depending on stress history. Endocrinology 153:4317–4327. [DOI] [PubMed] [Google Scholar]
- 80. Protiva P, Hopkins ME, Baggett S, Yang H, Lipkin M, et al. (2008) Growth inhibition of colon cancer cells by polyisoprenylated benzophenones is associated with induction of the endoplasmic reticulum response. Int J Cancer 123:687–694. [DOI] [PubMed] [Google Scholar]
- 81. Ramamoorthy S, Cidlowski JA (2013) Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocr Dev 24:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Barnes PJ (2013) Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol 131:636–645. [DOI] [PubMed] [Google Scholar]
- 83. Gross KL, Lu NZ, Cidlowski JA (2009) Molecular mechanisms regulating glucocorticoid sensitivity and resistance. Mol Cell Endocrinol 300:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, et al. (2012) MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS One 7:e40676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. He Y, Jiang X, Chen J (2013) The role of miR-150 in normal and malignant hematopoiesis. Oncogene. [DOI] [PubMed] [Google Scholar]
- 86. Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, et al. (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159. [DOI] [PubMed] [Google Scholar]
- 87. He Y, Jiang X, Chen J (2014) The role of miR-150 in normal and malignant hematopoiesis. Oncogene 33:3887–3893. [DOI] [PubMed] [Google Scholar]
- 88. Ishmael FT, Fang X, Houser KR, Pearce K, Abdelmohsen K, et al. (2011) The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J Immunol 186:1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sengul A, Santisuk R, Xing W, Kesavan C (2013) Systemic administration of an antagomir designed to inhibit miR-92, a regulator of angiogenesis, failed to modulate skeletal anabolic response to mechanical loading. Physiol Res 62:221–226. [DOI] [PubMed] [Google Scholar]
- 90. Manoharan P, Basford JE, Pilcher-Roberts R, Neumann J, Hui DY et al. (2014) Reduced miR-124a and miR-150 levels are associated with increased pro-inflammatory mediator expression in KLF2-deficient macrophages. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang M, Tan LP, Dijkstra MK, van Lom K, Robertus JL, et al. (2008) miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol 215:13–20. [DOI] [PubMed] [Google Scholar]
- 92. Li J, Zhang Y, Liu Y, Dai X, Li W, et al. (2013) Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem 288:23586–23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Almanza G, Anufreichik V, Rodvold JJ, Chiu KT, DeLaney A, et al. (2013) Synthesis and delivery of short, noncoding RNA by B lymphocytes. Proc Natl Acad Sci U S A 110:20182–20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IPA prediction of microRNA targets based on correlation of transcriptional changes of specific microRNAs (up/down) with changes in mRNA levels (opposite direction down/up) and corresponding pathways in MM1S cells treated for 72 h with Dex.
(XLSX)
Crosscomparison of 1203 Dex responsive target genes from MM1S cells exposed for 72 h to 1 µM Dex (gene selection was done based on p-value <0.05 and absolute fold change >1.5) and 3626 predicted hsa-mir-150-5p targets via the miRWalk database (prediction by at least 4 out of 10 available algorithms, i.e. DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22 and Targetscan) 196 common mRNA/gene targets were identified in MM1S, which remain unaffected in MM1R cells.
(XLSX)
In silico prediction of microRNAs targeting the NR3C1 gene locus versus the NR3C1 3′ UTR.
(XLSX)
Detailed list of genes involved in cell cycle-proliferation, DNA repair and cell death pathways which are similarly regulated in Dex treated and mir-150-5p mimetic transfected MM1S cells, but which remain unaffected in MM1R cells (values represent (fold change versus control cells)).
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Raw gene expression array data were uploaded to the Gene Expression Omnibus (GEO) database and have accession number GSE59805.