Abstract
We report the first systems biology investigation of regulators controlling arterial plaque macrophage transcriptional changes in response to lipid lowering in vivo in two distinct mouse models of atherosclerosis regression. Transcriptome measurements from plaque macrophages from the Reversa mouse were integrated with measurements from an aortic transplant-based mouse model of plaque regression. Functional relevance of the genes detected as differentially expressed in plaque macrophages in response to lipid lowering in vivo was assessed through analysis of gene functional annotations, overlap with in vitro foam cell studies, and overlap of associated eQTLs with human atherosclerosis/CAD risk SNPs. To identify transcription factors that control plaque macrophage responses to lipid lowering in vivo, we used an integrative strategy – leveraging macrophage epigenomic measurements – to detect enrichment of transcription factor binding sites upstream of genes that are differentially expressed in plaque macrophages during regression. The integrated analysis uncovered eight transcription factor binding site elements that were statistically overrepresented within the 5′ regulatory regions of genes that were upregulated in plaque macrophages in the Reversa model under maximal regression conditions and within the 5′ regulatory regions of genes that were upregulated in the aortic transplant model during regression. Of these, the TCF/LEF binding site was present in promoters of upregulated genes related to cell motility, suggesting that the canonical Wnt signaling pathway may be activated in plaque macrophages during regression. We validated this network-based prediction by demonstrating that β-catenin expression is higher in regressing (vs. control group) plaques in both regression models, and we further demonstrated that stimulation of canonical Wnt signaling increases macrophage migration in vitro. These results suggest involvement of canonical Wnt signaling in macrophage emigration from the plaque during lipid lowering-induced regression, and they illustrate the discovery potential of an epigenome-guided, systems approach to understanding atherosclerosis regression.
Author Summary
Atherosclerosis, a progressive accumulation of lipid-rich plaque within arteries, is an inflammatory disease in which the response of macrophages (a key cell type of the innate immune system) to plasma lipoproteins plays a central role. In humans, the goal of significantly reducing already-established plaque through drug treatments, including statins, remains elusive. In mice, atherosclerosis can be reversed by experimental manipulations that lower circulating lipid levels. A common feature of many regression models is that macrophages transition to a less inflammatory state and emigrate from the plaque. While the molecular regulators that control these responses are largely unknown, we hypothesized that by integrating global measurements of macrophage gene expression in regressing plaques with measurements of the macrophage chromatin landscape, we could identify key molecules that control macrophage responses to the lowering of circulating lipid levels. Our systems biology analysis of plaque macrophages yielded a network in which the Wnt signaling pathway emerged as a candidate upstream regulator. Wnt signaling is known to affect both inflammation and the ability of macrophages to migrate from one location to another, and our targeted validation studies provide evidence that Wnt signaling is increased in plaque macrophages during regression. Our findings both demonstrate the power of a systems approach to uncover candidate regulators of regression and to identify a potential new therapeutic target.
Introduction
Underlying many cardiovascular diseases is atherosclerosis, an arterial accumulation of lipid-laden macrophages (foam cells) with attendant chronic inflammation [1]. Atherosclerosis is endemic in adults [2] and can progress over decades before presenting with overt symptoms [3]. Atherosclerosis is correlated with risk factors such as hyperlipidemia, especially high plasma levels of low-density lipoprotein (LDL) cholesterol [4], [5]. There is significant interest in finding new therapies to promote the regression of atherosclerotic plaques [6]–[8], with particular emphasis on the plaque macrophage as a potential target because of its roles in lipoprotein uptake and vascular inflammation [9]–[12], plaque destabilization [13], [14], and plaque remodeling in response to dietary changes [15].
In mice, plaque regression has been achieved through expression [16]–[18] or inactivation [19], [20] of lipid metabolism-modifying genes; aortic tissue grafting [21]–[23]; and treatment with small-molecule therapeutics [24]–[26] or biologics [27], [28], frequently in conjunction with a diet alteration. We have been studying the role of macrophages during plaque regression in two mouse models of regression, the Reversa mouse [19], [20], [29], [30] and an aortic arch transplant model [21], [22], [31]. Reversa mice are Ldlr −/− Apob 100/100 [32], [33] and they possess a loxP-flanked allele of the gene encoding microsomal triglyceride transfer protein (Mttp) and a drug-inducible Cre recombinase (Cre) transgene [19], [34]. In the Reversa mouse, hyperlipidemia can be reversed by the transient induction of Cre expression in the liver, which causes recombinant knockout of Mttp, and thus significantly lowers the plasma levels of very low density lipoprotein (VLDL) and LDL [19]. In prior work we have shown that inactivation of Mttp in the Reversa mouse, when combined with a switch from Western diet to chow, leads to plaque regression at the aortic root [20]. In the aortic transplant model, an atherosclerotic aortic arch from an Apoe −/− mouse [35] is grafted in place of a segment of the abdominal aorta [31] of either a wild-type (WT) or an Apoe −/− mouse. Grafts within WT recipients exhibit plaque regression, while plaques within Apoe −/ recipients continue to progress [22]. A common feature of the Reversa and transplant regression models is the substantial reduction in plaque macrophages during regression [20], [23], [29], [30], [36]. It is at present unknown whether there are molecular pathways that may initiate the depletion of plaque macrophages that are shared across regression models. Identifying such core pathways could yield new therapeutic approaches to stimulate regression and/or beneficial remodeling of plaque.
Recently, we studied transcriptome differences between macrophages in regressing versus progressing plaques in the aortic transplant model [36]. We observed patterns of differential expression that suggest that macrophages in regressing plaques up-regulate genes associated with cell motility, down-regulate genes associated with cell adhesion, and up-regulate genes associated with an anti-inflammatory, “M2 macrophage” [37] phenotype. Although our analysis pointed to several candidate molecular regulators, it is unknown whether there are transcription factors that are common to the transplant and Reversa regression models that act as master controllers for the responses of plaque macrophages to lipid lowering.
In the current study we carried out a systems investigation of the Reversa model, with three goals: (i) to identify pathways and gene functions that are associated with plaque macrophage responses to lipid lowering in vivo; (ii) to understand the connections between gene sets identified in our study and gene sets from previous studies of macrophage foam cell formation, aortic wall changes during plaque progression and regression, and atherosclerosis risk alleles in humans; and (iii) to identify transcriptional regulators that are associated with macrophage responses to lipid lowering in vivo. We therefore carried out microarray transcriptome profiling of plaque macrophages isolated from the aortic root of Reversa mice undergoing lipid lowering in vivo and analyzed differentially expressed genes using multiple bioinformatics approaches. Gene functional enrichment analysis (i) detected over-representation of cytoskeletal-binding and Rho GTPase genes among genes that are upregulated in response to lipid lowering, pointing to cytoskeletal reorganization during regression. Comparative analysis (ii) of gene sets from our study with those from previous studies of in vitro models of macrophage foam cell formation revealed a statistically significant overlap. Additionally, we found a significant overlap between human monocyte expression quantitative trait loci (eQTLs) for human orthologs of genes that are differentially expressed during plaque regression, and genetic loci that are associated with risk of atherosclerosis or coronary artery disease. Promoters of genes identified in our study were analyzed for transcription factor binding site over-representation (iii) using a novel chromatin-guided method, REMINISCE (Figure S1 and Methods). REMINISCE leverages macrophage epigenomic and chromatin measurements, such as histone acetylation [38], [39] and DNase I hypersensitive sites [40], in the analysis of the 5′ regulatory regions of differentially expressed genes, including enhancers as well as promoters. In both the Reversa and aortic transplant regression models, we found that the consensus binding site sequence for the T-cell specific, HMG-box factors (TCF) and lymphoid enhancer factors (LEF), together known as the TCF/LEF family of transcription factors, was overrepresented within the 5′ regulatory regions of genes that are upregulated in plaque macrophages during regression.
TCF/LEF transcription factors are activated by nuclear β-catenin (CTNNB1) that accumulates in response to activation of the canonical Wnt signaling pathway [41], [42]. The Wnt pathway controls multiple functions in development, tissue organization, cell proliferation, cell migration [42], and inflammation [43]. In macrophages, canonical Wnt signaling is thought to promote cell motility through a β-catenin-dependent mechanism [44]. In this study we show, for the first time, that activation of canonical Wnt signaling, defined by up-regulation of β-catenin, occurs within macrophage-rich plaque in two mechanistically distinct lipid-lowering models of plaque regression, the Reversa and aortic transplant models.
Results
Identification of pathways and gene functions that are associated with plaque macrophage responses to lipid lowering in vivo
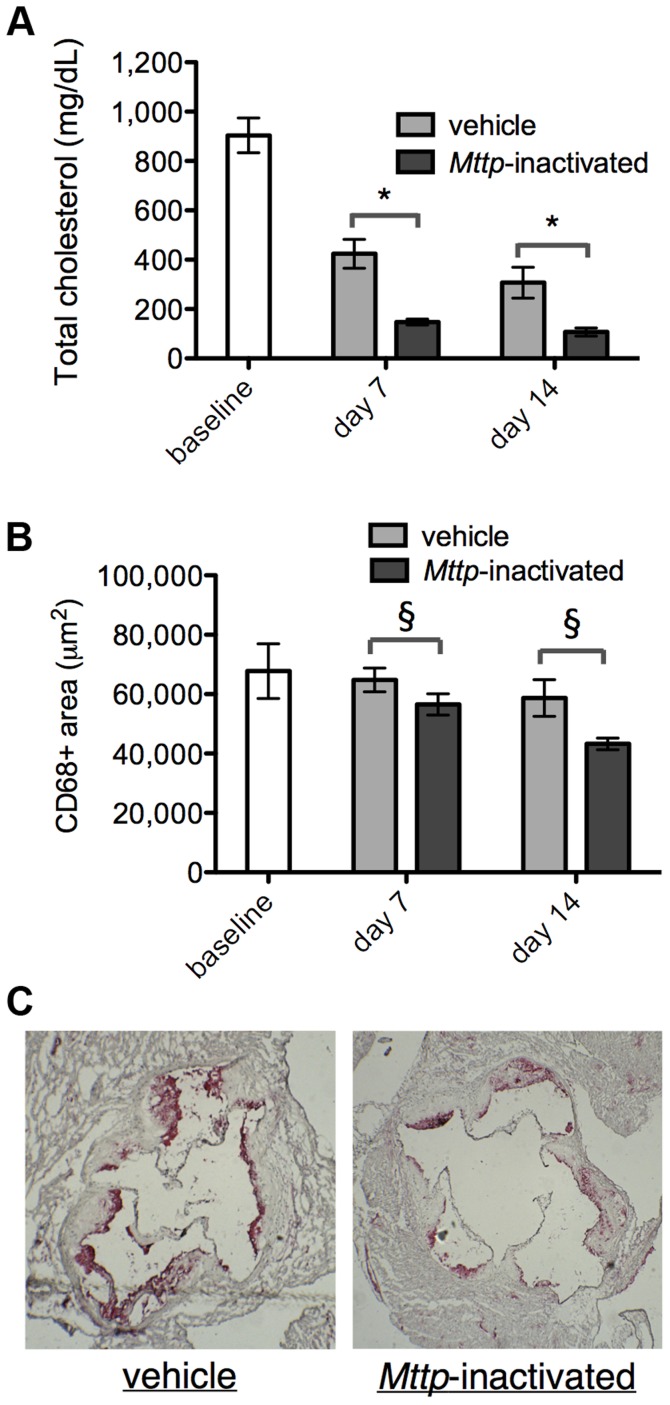
Reversa mice were maintained on Western diet for 16 weeks, which allowed for the development of hypercholesterolemia (903±71 mg/dL total cholesterol [mean ± standard error, SE]) and substantial, macrophage-rich plaques at the aortic root, as determined by immunostaining with the macrophage marker Cluster of Differentiation 68 (CD68). At 16 weeks, a group of animals were sacrificed to measure the baseline plasma total cholesterol levels and CD68+ areas within plaque within aortic root sections. The remaining animals were switched to chow diet and divided into two groups, an experimental group that received four injections of polyinosinic:polycytidylic acid (poly I:C) to induce Mx1:Cre in order to inactivate Mttpfl/fl, and a control group that was injected with vehicle (saline) on the same schedule. On day seven or 14 after the final injection, animals were sacrificed and plasma cholesterol and plaque CD68+ areas were measured. By day 14, cholesterol levels had decreased by 88% in the Mttp-inactivated animals vs. baseline (Figure 1A and Table 1, P<0.001) and plaque CD68+ area had decreased by 36% in the Mttp-inactivated animals vs. baseline (Figure 1B and Table 2, P<0.05). Additionally, plaque CD68+ area was reduced in Mttp-inactivated vs. vehicle-treated animals at seven and 14 days (P<0.05); at day 14, CD68+ area was 26% smaller in the Mttp-inactivated animals to the vehicle-treated animals (Figure 1C). These results indicated that there is plaque regression that is attributable to inactivation of Mttp. In contrast, comparisons of the saline-treated sample groups to baseline showed no statistically significant differences in CD68+ area that could be attributed to diet switch alone.
Figure 1. Reversal of hyperlipidemia in Reversa mice reduces plaque macrophage content.
(A) Plasma cholesterol levels were reduced upon switching to chow diet and upon inactivation of Mttp. Bars are mean ± SE. Cholesterol differences between vehicle and Mttp-inactivated are: at day seven, 65% reduction; at day 14, 65% reduction. The cholesterol reduction in Mttp-inactivated vs. vehicle, at days seven and 14, is significant with P<10−4 [(*) two-way analysis of variance (ANOVA)]. For all bars, N = 5 except for baseline, where N = 7. (B) Mttp inactivation contributes to a reduction in macrophage content of the plaque. Bars are mean ± SE. CD68+ area differences between vehicle and Mttp-inactivated are: at day seven, 13%; at day 14, 26%. The CD68+ area reduction in Mttp-inactivated vs. vehicle groups, at both days seven and 14, is significant with P<0.05 (§, two-way ANOVA). For all bars, N = 7 except day seven, where N = 9. The difference in CD68 areas between day 14 (or day seven) saline-treated groups and baseline were not statistically significant. (C) Mttp inactivation reduces CD68 cellular content within aortic root plaque. Micrographs are representative aortic root sections from day 14 animals (from vehicle and Mttp-inactivated groups, as indicated) that have been counterstained with hematoxylin (blue) and immunostained for CD68 (red).
Table 1. Mttp inactivation lowers plasma cholesterol in the treatment vs. control groups.
| Group | Number of animals | Plasma total cholesterol level (mg/dL) |
| baseline | 7 | 903±71 |
| vehicle, day 7 | 5 | 424±59 |
| Mttp-inactivated, day 7 | 5 | 148±12 |
| vehicle, day 14 | 5 | 307±62 |
| Mttp-inactivated, day 14 | 5 | 107±16 |
Reversa mice were fed a Western diet for 16 weeks. One group of animals was sacrificed at 16 weeks as the baseline group. The rest of the mice were switched to chow diet and assigned to two groups, vehicle and Mttp-inactivated; animals within these two groups received injections of saline or poly I:C, respectively (see Methods). Animals were sacrificed on day seven or day 14 after the injections, and plasma total cholesterol levels were measured (see Methods). Measurements are summarized as mean ± SE.
Table 2. In vivo lipid lowering leads to reduced aortic root CD68+ area, independent of diet switch.
| Group | Number of animals | Area of CD68+ cells within aortic root sections (µm2) |
| baseline | 7 | 67,800±9,200 |
| vehicle, day 7 | 9 | 64,800±4,000 |
| Mttp-inactivated, day 7 | 9 | 56,500±3,600 |
| vehicle, day 14 | 7 | 58,700±6,200 |
| Mttp-inactivated, day 14 | 7 | 43,300±2,000 |
Reversa mice were fed a Western diet for 16 weeks. One group of animals was sacrificed at 16 weeks as the baseline group. The rest of the mice were switched to chow diet and assigned to two groups, vehicle and Mttp-inactivated; animals within these two groups received injections of saline or poly I:C, respectively (see Methods). Animals were sacrificed on day seven or day 14 after the injections, and aortic roots were sectioned, immunostained for CD68, and the CD68+ areas were measured (see Methods). Measurements are summarized as mean ± SE.
In order to focus on the molecular signaling events that initiate the reduction in plaque macrophages, we selected an early time point (day seven) when there was a statistically significant difference in the CD68+ area in the treatment group vs. vehicle (Figure 1, B and C). CD68+ plaque macrophages were isolated from aortic root sections from Mttp-inactivated Reversa, vehicle-treated Reversa, and poly I:C-injected Ldlr −/− animals (Table S1) by laser capture microdissection (LCM) [45], [46], a procedure that we have previously demonstrated specifically enriches for the mRNAs of macrophage-related genes by more than 30-fold over levels seen in aortic homogenates [45]. Global transcriptome profiles were then obtained from the LCM-derived samples using high-density oligonucleotide microarray hybridization. A total of 213 genes were detected as differentially expressed in plaque macrophages from Mttp-inactivated vs. vehicle-treated animals (93 had higher expression levels in Mttp-inactivated than in control, and 120 had lower levels; Table S2). In order to control for any effect of the poly I:C injections on the macrophage transcriptional profile we compared the transcriptomes of plaque CD68+ cells from saline-treated Reversa mice with those from poly I:C-treated Ldlr −/− mice. We found that only one of the 213 lipid-responsive genes, Trnt1, was detected as differentially expressed in the Reversa saline vs. Ldlr −/− poly I:C comparison, and this gene was excluded from further bioinformatic analysis. To further substantiate that the 213 genes in plaque CD68+ cells are responding to lipid lowering rather than directly to poly I:C, the list of 213 genes was compared with a list of genes that are differentially expressed (vs. vehicle) in bone marrow-derived macrophages stimulated in vitro with poly I:C (Table S3) to determine whether the degree of overlap in the two gene lists is greater than would be expected by chance. The analysis revealed that there is not more overlap with in vitro poly I:C responses than would be expected by chance (P>0.1, Fisher's Exact Test).
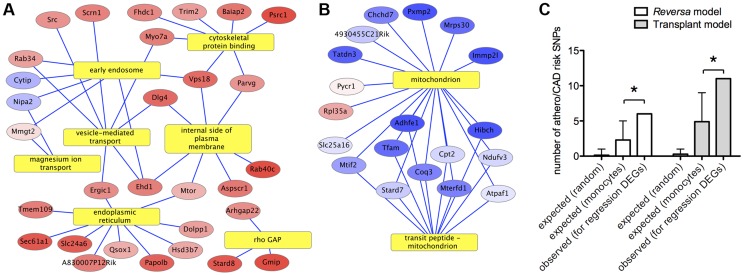
To gain insight into the biological processes that occur during lipid lowering in vivo, we performed gene functional annotation enrichment analysis. The most significantly overrepresented processes associated with the upregulated genes are related to cell motility, cytoskeletal binding, and Rho GTPases (Figure 2A and Table S4). For the genes that were downregulated in macrophages from lipid-lowered vs. control animals, the gene annotation term that was the most significantly overrepresented was mitochondrion (Figure 2B and Table S4). In order to determine whether lipid lowering in vivo might have increased plaque macrophage apoptosis, multiple gene functional enrichment analysis tools were applied, and in each case, no finding of a statistical enrichment for apoptosis-related gene annotations was detected (see Methods).
Figure 2. Functional bioinformatic analysis of genes that respond to lipid lowering in vivo indicates connections with cell motility and risk of human CAD/atherosclerosis.
Yellow rectangles represent gene annotation categories. Ellipses represent genes that are differentially expressed between Mttp-inactivated and vehicle-treated sample groups in plaque macrophages in the Reversa mouse model of plaque regression (red = up in Mttp-inactivated, blue = down). Genes are labeled by official gene symbol. Color saturation indicates the magnitude of the fold-change (see Table S2). Edges connect genes to the statistically enriched annotation categories with which they are associated (see Methods; enrichment P values are given in Table S4). (A) A statistically significant cluster of upregulated genes is associated (by gene annotations) with the cytoskeleton. (B) A statistically significant cluster of downregulated genes is associated with the mitochondrion. (C) Human monocyte expression QTLs for genes that are differentially expressed in plaque CD68+ cells in regression are enriched for SNPs associated with risk of atherosclerosis or CAD, versus the number of atherosclerosis/CAD SNPs for monocyte eQTLs overall. The bars marked “expected” show the range of numbers of SNPs associated with risk of atherosclerosis or CAD that would be expected to occur by chance within the eQTLs of the set of 213 lipid lowering-responsive genes, based on the overall percentage of SNPs that are associated with risk of atherosclerosis/CAD (“random”) and based on the percentage of SNPs within all monocyte eQTLs that are associated with atherosclerosis/CAD (“monocytes”). Error bars indicate >90% confidence intervals for the number of atherosclerosis/CAD SNPs that would be expected by chance (see Methods). (*) P<0.05, Fisher's Exact Test (contingency table data are in Table S6).
Contextualization of lipid lowering-responsive genes with genes identified in previous studies and atherosclerosis risk alleles in humans
We hypothesized that among the plaque macrophage genes that are responsive to lipid lowering in vivo, genes that had been previously reported to be differentially expressed in response to lipid loading in vitro would be statistically overrepresented. We therefore combined differentially expressed gene lists from 21 datasets from 15 studies in the atherosclerosis and macrophage foam cell literature (Table S5), and scored genes based on the number of studies in which they were detected as differentially expressed during foam cell formation or disease progression (similar to the approach that was used in a recent meta-analysis to identify obesity-related genes [47]). We found that 19 out of the 213 differentially expressed genes in our study were also detected among the high-scoring genes in the in vitro lipid-loaded macrophage meta-analysis gene set, corresponding to a 2.9-fold enrichment (P<0.0001) vs. for all mouse genes.
Because in vivo lipid lowering is associated with plaque regression in the Reversa and aortic transplant models (Figure 1 and [19], [20], [23], [36]), we conjectured that the gene expression changes in plaque CD68+ cells in response to lipid lowering would be associated with atheroprotective effects in humans. Because such effects might be subtle for any individual gene, and because “master regulators” of the transcriptional response are often not transcriptionally regulated themselves, we tested our hypothesis by computationally searching for connections between human atherosclerosis/CAD risk-associated single nucleotide polymorphisms (SNPs) and monocyte eQTLs for orthologs of genes that are differentially expressed in plaque CD68+ cells in response to lipid lowering (as had been previously applied to a transcriptome study of whole aortic arches [30]). We assumed that if there are no atheroprotective effects associated with the observed gene expression changes in CD68+ cells, then the fraction of atherosclerosis/CAD risk SNPs within monocyte eQTLs for the 213 lipid lowering-responsive genes should be the same as the fraction of atherosclerosis/CAD SNPs within all monocyte eQTLs. Using a list of human eQTLs from two population genetic studies of peripheral blood monocyte gene expression [48], [49], we identified SNPs associated with mRNA levels (eSNPs) of genes whose mouse orthologs are differentially expressed in CD68+ cells during regression in the two mouse models. We expanded the regression-gene-set-associated eSNPs to include all validated SNPs in linkage disequilibrium (using haplotype information from the 1,000 Genomes Project [50]), obtaining a set of regression-associated monocyte eQTLs. We then assessed the frequency of atherosclerosis/CAD risk SNPs (starting with a list of 761 risk SNPs obtained from the NCBI PheGenI database [51]) within these regression-associated monocyte eQTLs and compared these frequencies to the frequency of atherosclerosis/CAD risk SNPs within monocyte eQTLs in general. We found that the regression-associated eQTLs are substantially more enriched for atherosclerosis/CAD risk SNPs than monocyte eQTLs in general (Figure 2C and Table S6), in both regression models (P<0.05 for each model); overall, a total of 17 atherosclerosis/CAD risk-associated SNPs were found within regression-associated eQTLs (Table S7). Further investigation revealed that one of the Reversa regression-associated eQTLs encompasses rs599839, a SNP in the 1p13.3 locus that has been found to be associated with LDL cholesterol (LDL-C) levels and with risk of CAD and MI [52]–[55], as well as with hepatic mRNA expression of the intracellular lipoprotein sorting receptor Sortilin 1 (SORT1) [56], [57]. In human monocytes [48] and in liver cells [56], variant rs599839 is also an eSNP for mRNA expression of the gene Proline/serine-rich coiled coil 1 (PSRC1), which is one of most strongly upregulated genes in CD68+ cells in Mttp-inactivated vs. vehicle-treated Reversa mice (Table S2).
Identification and targeted validation of transcriptional regulators that are associated with macrophage responses to lipid lowering in vivo
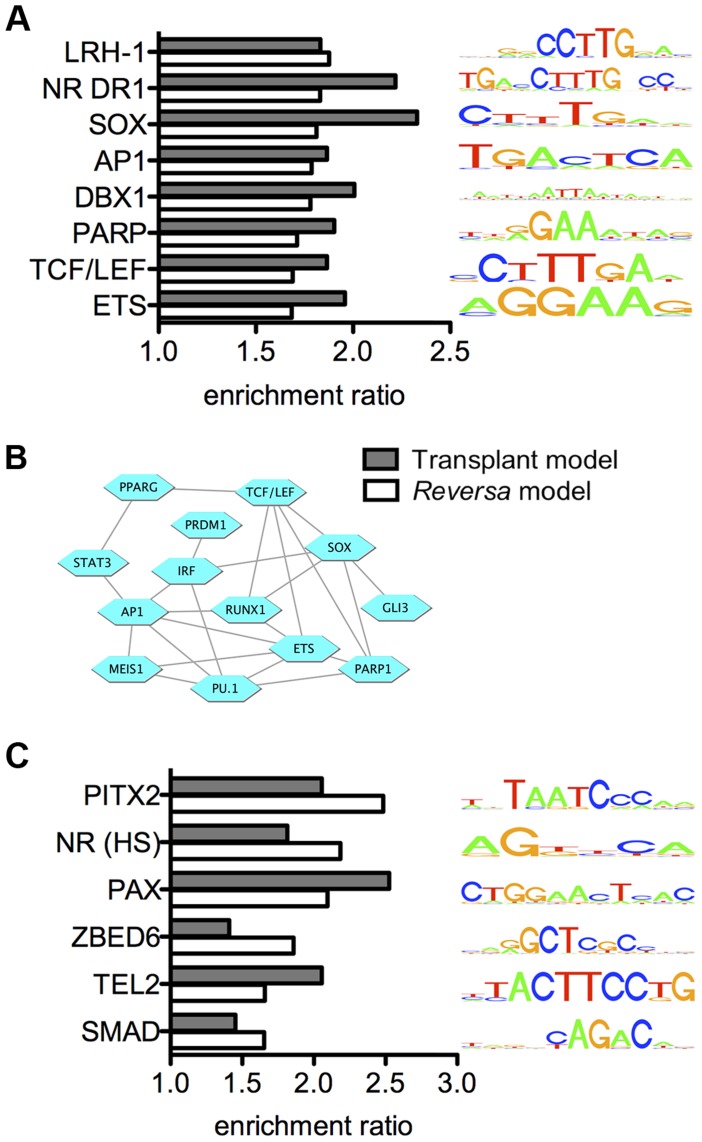
We applied our novel promoter scanning analysis method, REMINISCE (Figure S1), to identify candidate transcription factors that are associated with the transcriptional responses of plaque macrophages to lipid lowering in vivo. REMINISCE combines 17 different types of macrophage chromatin and epigenomic measurements (see Table S8) that were obtained from NCBI GEO and from the Mouse ENCODE project [58] in a machine-learning algorithm (using genome regions that coincide with transcription factor binding sites that are obtained from ChIP-seq as a training dataset of regulatory regions) to map macrophage cis-regulatory regions, and then scans genomic sequences within these regions to identify statistically overrepresented transcription factor binding site sequence patterns (see Methods). In a direct comparison of REMINISCE with non-epigenome-guided motif-scanning of all noncoding sequence within ±1 kbp or ±5 kbp of the transcription start site for gene sets derived from macrophage transcriptional responses to the atherogenic lipoprotein oxidized LDL, REMINISCE detected substantially more binding site motifs above the significance threshold (Figure S2). Applying REMINISCE to the 213 lipid lowering-responsive genes from the Reversa model, we identified 15 transcription factor binding site sequence motifs that are statistically overrepresented within the 5′ regulatory regions of upregulated genes (Figure S3A), and 14 motifs that were associated with the downregulated genes (Figure S3B). Because any experimental model is subject to model-specific artifacts, we also analyzed our previously published data on the transcriptional response in CD68+ cells isolated from regressing plaques in the aortic transplant model using the REMINISCE method. We found that of the 15 motifs that were overrepresented among upregulated genes in the Reversa model, eight were also overrepresented among upregulated genes in the aortic transplant model (Figure 3A), including the motifs for PPARγ (nuclear receptor direct repeat 1, or NR DR1), the TCF/LEF family of transcription factors, and activating protein 1 (AP-1). The transcription factors represented by the 15 motifs that were associated with upregulated genes in the Reversa model are highly interconnected in the human protein-protein interactome [59] (clustering coefficient of 0.31, vs. 0.14 for the whole interactome [60]) (Figure 3B). Similarly to the case with transcription factors for upregulated genes, of the 14 motifs that were overrepresented among downregulated genes in the Reversa model, six were also detected in the aortic transplant model (Figure 3C and Table S9).
Figure 3. Integrated analysis reveals candidate regulators of the macrophage transcriptional response to lipid lowering in vivo.
(A) Transcription factor binding site motifs whose matches are statistically overrepresented within 5′ regulatory regions of genes that are upregulated in plaque macrophages in response to lipid lowering in vivo. Bars indicate the ratio of the number matches of the indicated motifs per kbp, within an upregulated gene set, to the number of motif matches per kbp for randomly selected sets of genes expressed in CD68+ cells. Bar color indicates the regression model: white, Reversa model; gray, aortic transplant model. Only motifs for which the ratio exceeds 1.5 (with a statistical significance of P<0.01) in both models are shown (complete list in Table S9). Motifs are labeled by the transcription factor family name with which they are associated (motif IDs are provided in Table S9). Each motif is represented by a four-color motif logo. (B) The transcription factors that are associated with upregulated genes are highly interconnected at the level of protein-protein interaction network. Blue hexagons represent transcription factors or transcription factor families, and edges denote protein-protein interactions between them. (C) Transcription factor binding site motifs whose matches are statistically overrepresented within 5′ regions of genes that are downregulated in plaque macrophages in response to lipid lowering in vivo. “NR (HS)”, nuclear receptor half-site.
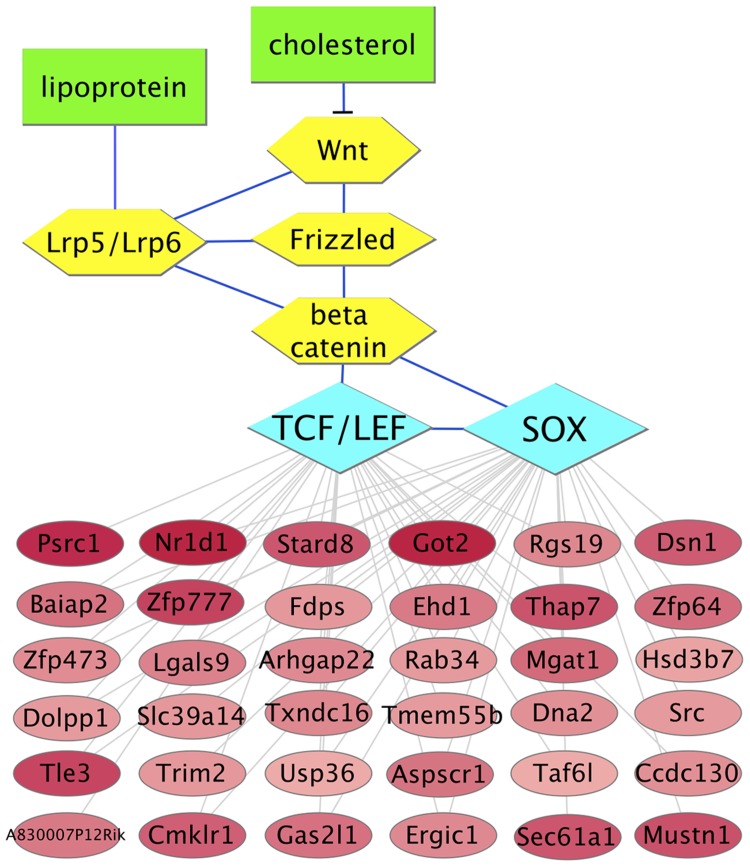
Under the hypothesis that the transcription factors that control macrophage responses to lipid lowering in vivo would have binding site sequences within the 5′ regulatory regions of a high proportion of differentially expressed genes, we ranked the motifs from Figure 3 by the fraction of the differentially expressed genes that possess a match for each motif (Table S10). Notably, for the upregulated genes, the motif whose matches were present within the largest percentage of genes, 33%, was the CTTTGA motif that is recognized by TCF/LEF and SOX transcription factor families. The finding of enrichment of TCF/LEF binding site sequences within 5′ regulatory regions of genes that are upregulated in the Reversa mouse was confirmed by additional computational analysis using high-precision motifs based on three-dimensional structural modeling of protein-DNA interactions [61] (Table S9). Because the reduced macrophage content of the plaque in the transplant regression model has been associated with macrophage emigration from the plaque, we also ranked transcription factor binding site motifs based on the proportion of cytoskeletal-binding and RhoGAP domain-containing genes (Table S4) whose 5′ regulatory regions contain matches for each motif. We found that the motif with the highest percentage, 50%, was the CTTTGA motif (corresponding to TCF/LEF and SOX) (Table S10). Next, we used the global mammalian protein interaction network to examine which signaling pathways could connect between lipoproteins or cholesterol and these transcription factors. We found that the Wnt signaling pathway [62], [63], through β-catenin and through TCF/LEF and SOX family members [64], satisfies these criteria (Figure 4).
Figure 4. Predicted molecular interaction network for upregulated genes that possess CTTTGA elements connects cholesterol to motility.
Red ellipses represent genes whose expression levels are elevated in plaque macrophages from Mttp-inactivated vs. vehicle-treated animals (darker color indicates a greater differential expression ratio; see Table S2). Yellow hexagons and blue edges represent the upstream signaling pathway based on the literature (the blunt-ended edge indicates inhibition of signaling), with green rectangles representing lipids (cholesterol: membrane cholesterol; lipoprotein: apoB-containing lipoprotein). Blue diamonds represent transcription factor binding site motifs. Gray edges connect the transcription factors to the downstream genes that possess corresponding binding site sequence matches.
Gene regulation likely occurs at multiple levels in plaque macrophages. Therefore, we scanned the 3′ untranslated regions (UTRs) of the two sets of differentially expressed genes in plaque CD68+ cells in the Reversa model for microRNA target sequences. We identified eighteen microRNAs whose target sequences were overrepresented within the 3′ UTRs of the upregulated and downregulated genes (six for upregulated genes, and twelve for downregulated) (Figure S4, Table S11). Under the hypothesis that some of these microRNAs might work in concert in response to lipid lowering in vivo, we analyzed molecular pathways for enrichment of the number of pathway-associated genes that possess target sequences for any of the microRNAs in Figure S4A, using a microRNA pathway analysis tool [65]. For the upregulated genes, the two pathways with the strongest enrichment were axon guidance (P<10−4) and focal adhesion (P<0.001); also enriched was the Wnt signaling pathway (P<0.05) (Table S12).
β-catenin-dependent activation of TCF/LEF is the final step in the canonical Wnt signaling pathway. To investigate the possibility of canonical Wnt pathway activation during plaque regression, we measured the relative mRNA level of β-catenin (Ctnnb1) in CD68+ cells isolated from aortic grafts from WT and Apoe −/− recipient mice in the aortic transplant regression model. By quantitative PCR (qPCR), we found that the relative level of Ctnnb1 mRNA is approximately 2.2-fold higher in CD68+ cells in regressing plaques than in progressing plaques (Figure 5A) (P<0.05). Additionally, we measured mRNA levels of two known Wnt target genes, Lrp6 (for which loss-of-function has been associated with premature CAD [66]) and Gja1 [67], both of which were indicated by microarray profiling to be upregulated in regressing vs. progressing plaque CD68+ cells in the aortic transplant model [36]. By qPCR, both Lrp6 and Gja1 were found to be upregulated in CD68+ cells in regressing vs. progressing plaques (Figure S5). Given a previous report that β-catenin deficiency impairs macrophage migration [44] and in light of evidence of plaque macrophage emigration during regression in both the Reversa [20] and aortic transplant [22] models, we investigated whether canonical Wnt pathway activation stimulates macrophage migration in vitro. In a scratch-wound cell migration assay [68], [69], primary murine macrophages treated with the canonical Wnt pathway agonist Wnt3a scored more than two-fold higher than vehicle-treated macrophages (Figure S6).
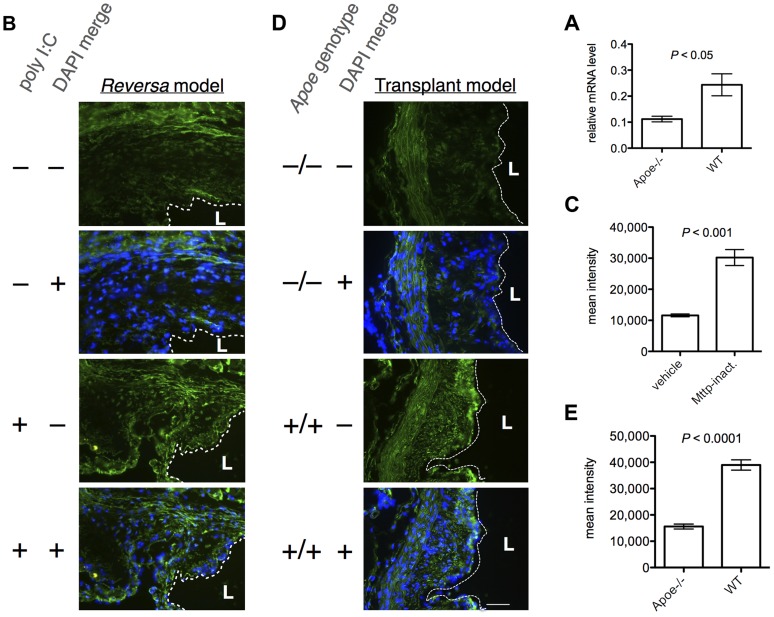
Figure 5. β-catenin expression is increased in regressing vs. progressing plaques, in both models of plaque regression.
(A) The relative Ctnnb1 (β-catenin mRNA) levels, as measured by qPCR, are higher in CD68+ cells in regressing plaques than in progressing plaques, in the aortic transplant model (day five post-transplant). Bars, mean ± SE (N = 6 animals per sample group). (B, C) Plaques in Mttp-inactivated Reversa mice have higher levels of β-catenin than plaques in vehicle-treated animals. (B) Representative fluorescence micrographs from an immunohistochemical analysis of β-catenin expression at the aortic root in the Reversa mouse at day seven post-injection to inactivate Mttp (N = 3 animals per group, six sections analyzed per animal). Green, β-catenin immunofluorescence; blue, diamidino-2-phenylindole (DAPI) fluorescence; dashed white line divides lumen (“L”) from intima; scale bar, 50 µm. The green signal in the upper part of the top panel is autofluorescence from the elastic lamina. (C) Plaque β-catenin expression for each treatment group was quantified by morphometric image analysis. Bars, mean ± SE (N = 4 per treatment group); “Mttp-inact.”, Mttp-inactivated. (D, E) Plaques in WT recipient animals have higher levels of β-catenin than do plaques in Apoe−/− recipients, in the transplant regression model. (D) Representative fluorescence micrographs from an immunohistochemical analysis of β-catenin expression in the aortic transplant model (N = 3 animals in each genotype group, six sections per graft), at day five post-transplant. Color channels and annotations are as for (B). (E) Plaque β-catenin expression for each genotype group, by morphometric analysis. Bars, mean ± SE (N = 4 per genotype group). P values in (A,C,E) are from unpaired, two-tailed, Student's t-tests.
To confirm the prediction that Wnt signaling is activated in plaque macrophages during regression in vivo, we analyzed the expression of β-catenin in plaque within aortic root sections from Reversa mice at day seven following Mttp-inactivation or vehicle treatment, by fluorescence immunohistochemistry. Consistent with the systems biological analyses, the level of β-catenin was significantly greater (approximately 2.6-fold, P<0.001) in the plaques isolated from the Mttp-inactivated vs. vehicle-treated animals (Figure 5B, 5C). Because the TCF/LEF binding site was detected as being overrepresented among genes that are upregulated during plaque regression in the transplant regression model as well, we also analyzed β-catenin expression in plaques in the aortic arch graft of WT and Apoe −/− recipient animals. As with the Reversa model, we found that β-catenin is expressed at an approximately 2.5-fold higher level (P<0.0001) in regressing vs. progressing plaques (Figure 5D, Figure 5E, Figure S7), with increased abundance in the nuclear area (Figure S8).
Discussion
It is clear that atherosclerosis regression involves the interactions of many cellular pathways and multiple cell types and plaque components. To address this complexity, a systems biology approach employing large-scale molecular profiling is needed to identify core, model-independent mechanisms [8], [10], [70], [71]. To our knowledge, the present work, along with complementary measurements obtained from the aortic transplant regression model [36], represent the first reports of transcriptome measurements specifically of CD68+ macrophages within regressing plaques. In a previous transcriptome study [29] of changes in vessel wall tissue after lipid lowering in vivo in the Reversa mouse, a modest number of genes were detected as differentially expressed in the aortic wall (37 genes in [29], and 42 genes for arteries containing early lesions in [30]). Notably, none of these was detected as differentially expressed in our study of responses of plaque macrophages to lipid lowering, pointing to the need to separately measure the gene expression changes of constituent cell types of the plaque [71], as we have done for macrophages in the present work. While the 213 genes we identified as differentially expressed in plaque CD68+ cells in the Reversa mouse in response to lipid lowering are likely only a subset of the transcriptional changes in plaque macrophages over various time points, the enrichment of atherosclerosis/CAD risk alleles within human monocyte expression QTLs for orthologs of these genes (Figure 2C) provides intriguing support for their collective function in macrophages in the context of atherosclerosis.
Given the central role of macrophages in vascular inflammation [72] and in plaque destabilization in humans [73]–[75], our finding that Mttp-inactivation in the Reversa mouse reduces the macrophage content of the plaque (Figure 1 and [20]) points to the potential for beneficial remodeling. However, it also raises the question of how much of this reduction is driven by three processes (all of which have been reported in different models of regression): emigration from the plaque [22], deceased infiltration of monocytes [17], and increased macrophage cell death [25]. To the extent that accumulation of apoptotic macrophages within shoulder regions of advanced plaque is considered a marker of vulnerable plaques [76], the balance of these mechanisms is potentially clinically significant.
In the aortic transplant regression model, plaque macrophages in WT recipients rapidly emigrate from the graft to regional lymph nodes [22], while remaining macrophages do not exhibit abnormal levels of apoptosis [22]. Measurements of macrophage transcriptional changes during plaque regression versus progression in the aortic transplant model are consistent with an induced emigration hypothesis; macrophages in regressing (vs. progressing) plaques had lower expression levels of genes associated with adhesion and higher expression levels of genes associated with cell motility [36]. In regards to the emigration hypothesis in the Reversa model (which is supported by macrophage trafficking studies in vivo [20]), the plaque macrophage transcriptome measurements in this work provide several key insights. First, among genes that are upregulated in macrophages from Mttp-inactivated versus control animals, genes encoding cytoskeletal-binding proteins and Rho GTPases are overrepresented, consistent with increased motility. Second, among genes that are differentially expressed, no overrepresentation of apoptosis-related genes was found at the time point that we examined. A third insight, involving the Wnt signaling pathway, comes from the transcription factor analysis as described below.
In this study, we used an integrative computational approach, REMINISCE, to identify transcription factors that are likely to mediate macrophage responses during plaque regression. A novel aspect of the approach is that it incorporates macrophage epigenome measurements to guide the search for transcription factor binding sites (resulting in improved sensitivity; Figure S2) in the context of an enrichment analysis. In the analysis, we made use of transcriptome measurements from both the aortic transplant and Reversa regression models as well as macrophage-specific chromatin measurements to detect overrepresentation of known transcription factor binding site sequences upstream of genes that are differentially expressed during plaque regression. To focus on transcription factors that may have a model-independent role in macrophage responses to lipid lowering, we selected factors whose binding site sequences were overrepresented in both regression models. The significant overlap between the sets of transcription factors that were identified from the two models was unexpected due to the distinct differences in genetic backgrounds, plaque locations, and regression time-scales of the models. Of these factors, the association of PPARγ with genes that are upregulated during plaque regression is consistent with one of the principal findings from a previous transcriptome profiling study of whole aortic arches in the Reversa mouse with early lesions [30]. While our previous work investigating the effect of treatment of Reversa mice with a PPARγ agonist did not observe a significant effect of the agonist on the abundance of macrophages in the plaque [20], PPARγ is certainly worthy of future studies to clarify its precise function in plaque regression (especially given conflicting findings in different models and disease stages [20], [77]–[79]).
Among the other transcription factors whose binding site motifs were found to be associated with genes that are upregulated in CD68+ cells in regressing plaques in both regression models, the TCF/LEF and SOX transcription factor families (whose binding site sequences were associated with genes that are upregulated in response to lipid lowering) were particularly notable because of their connection to Wnt signaling, and thus, cell motility. Additionally, pathway enrichment analysis based on genes with target sequences for the microRNAs that were associated with upregulation in plaque CD68+ cells identified axon guidance and Wnt signaling as enriched pathways. Based on the transcription factor binding site analysis, we hypothesized that canonical Wnt signaling may be increased in plaque macrophages during regression. This hypothesis was confirmed by multiple observations including Ctnnb1 mRNA measurements in CD68+ cells, mRNA measurements of Wnt pathway target genes, and immunohistochemical analysis demonstrating increased β-catenin in regressing plaques in the Reversa and aortic transplant mouse models. Although the molecular mechanisms that cause increased Wnt signaling in plaque in response to lipid lowering are not known, and, of course, in spite of our associative findings, it is possible that plaque regression is mediated by Wnt-independent pathways, nonetheless, the observation of increased Wnt signaling in response to lipid lowering is consistent with previous reports that, in cultured cells, depletion of cholesterol increases canonical Wnt signaling [63].
In both the Reversa and aortic transplant models of regression, lipid lowering in vivo leads to decreased macrophage content in plaque (Figure 1B, [20], [23], [28]–[30], [36]), and trafficking studies in both of these models suggest that lipid lowering triggers macrophage emigration from the plaque [20], [22]. Our own functional studies of macrophage responses to Wnt3a stimulation (Figure S6) as well as previous studies of the role of β-catenin in macrophage migration [44], [80], are consistent with the model that increased canonical Wnt signaling – resulting in increased β-catenin levels – stimulates macrophage motility. A transitioning of macrophages from a sessile to a motile state would be expected to facilitate macrophage egress from the plaque [81].
This work is the first to our knowledge to propose that canonical Wnt signaling is involved in atherosclerosis regression in response to lipid lowering in vivo, and adds to several lines of evidence that support that alterations in Wnt signaling play a role in the progression of atherosclerosis. LRP6 genetic variants that impair Wnt/β-catenin signaling have been reported that are associated with increased risk of carotid atherosclerosis [82] and early CAD [66] in humans, suggesting that decreased canonical Wnt/β-catenin signaling may be pro-atherogenic; conversely, by this reasoning, increased Wnt signaling would be expected to have protective, pro-regressive effects. Consistent with this model, the canonical Wnt signaling molecule Wnt3a has been demonstrated to mediate anti-inflammatory effects via β-catenin-dependent suppression of the expression of proinflammatory cytokines [83]. Canonical Wnt signaling is also thought to play an important role in maintaining vascular cellular homeostasis, for example, in orchestrating lineage commitment within pericytes in the vessel wall [84]. Furthermore, Wnt signaling has been shown to potently inhibit lipid accumulation in cultured cells and to regulate plasma cholesterol levels in vivo [85]–[87], as well as (noted before) to promote macrophage migration ([44] and Figure S6).
While the present work provides evidence implicating β-catenin-interacting transcription factors in atherosclerosis regression, it is highly probable that other transcription factors are involved, given the complexity of macrophage transcriptional regulation in response to changing lipoprotein levels [70]. Of particular interest for future studies are the transcription factors that have overrepresented binding site sequences within the upregulated gene group, and that are connected to the TCF/LEF family members in the protein interaction network (Figure 3B). It should also be noted that a potential limitation of the analytical approach used in this work is the partial reliance on proximity to key factors in the global protein interaction network, in prioritizing transcriptional regulators for targeted validation; the use of such a proximity criterion could introduce bias against transcription factors that are less well-studied.
While this work used curated chromatin measurements from primary macrophages to guide the bioinformatic search for transcription factor binding sites, the numbers of cultured cells that are required for a ChIP-seq assay is rapidly being reduced; continued assay improvements may make possible genome-wide location analysis of chromatin factors within specific cellular constituents of plaque, which would likely enable more precise mapping of regulatory elements that mediate transcriptional changes during plaque regression. Equally important, comprehensive datasets of genetic variants and their associations with cardiovascular phenotypes (which may achieve significance only in a meta-analysis), such as those provided by the CARDIoGRAMplusC4D and dbGAP databases, will undoubtedly be beneficial for contextualizing the functions of these regulatory elements in atherosclerosis. Another enhancement in future studies would be to use lineage-tracing techniques in vivo to definitively characterize the extent of cellular phenotypic conversion in the Reversa model at the aortic root. Our previous work, however, has demonstrated that CD68-guided LCM of aortic root plaque yields a highly macrophage-enriched cell population [45]. Furthermore, no transcriptional changes were observed during plaque regression that would suggest a change in the proportion of macrophages in the cells captured by CD68-guided LCM in regressing vs. progressing plaques.
Beyond the enrichment of TCF/LEF transcription factor binding sites, the finding of enrichment of binding sites for the oxidative stress-responsive transcription factor nuclear respiratory factor 2 (NRF-2) within the regulatory regions of the downregulated gene group (Figure S3) is interesting in light of the observed down-regulation of mitochondrial transcription factor A (Tfam, a regulator of mitochondrial biogenesis and a known NRF target gene [88]). Downregulation of NRF-2 activity might reflect decreased oxidative stress in plaque macrophages due to the substantially reduced plasma lipoprotein levels [89].
Another intriguing finding from our work is the higher level of expression of Psrc1 in plaque macrophages in the Reversa regression model. Psrc1 mRNA has been previously been reported to be elevated in macrophage-rich regions of human atheroma [90]. The function of PSRC1 in macrophages is not known, but in other cell types it stabilizes microtubules [91], [92] and functions in cell migration [92]. Given our findings of elevated β-catenin in regressing plaques, it is interesting that a study of PSRC1 function found that PSRC1 stimulates the β-catenin pathway [93]. While strong evidence indicates that the effect of SNP rs599839 on LDL-C is mediated by hepatic expression of Sortilin 1 (SORT1) [56], the link between rs599839 and monocyte expression of PSRC1 seems worthy of further study in the context of plaque regression.
The multi-model analyses of plaque regression we have carried out is likely to ultimately shed light on the process of plaque stabilization in humans, given that atherosclerosis in genetically modified mice recapitulates many features of clinical disease. Large clinical trials have shown that in humans treated with lipid-lowering drugs, plaque area, as assessed by intravascular ultrasound (IVUS), changes very little [94], [95], while atherothrombotic event rates are dramatically decreased [96], [97], presumably due to stabilization resulting from changes in plaque macrophages not detected by IVUS. Indeed, even in mouse models, we have found that plaque size measurements do not always reflect changes in plaque macrophage content because of increased matrix proteins (e.g., [98]). Demonstrating similar phenomena in human plaques awaits improvements in non-invasive imaging. Overall, in human plaques, a drastic reduction in the abundance of macrophages within the plaque would be presumed to be associated with protective remodeling of the atheroma, and given the similarities in the disease process between mice and people, some of the factors and pathways we have identified are likely to be clinically relevant (e.g., Wnt signaling [66]).
In conclusion, this work demonstrates the value of a genomic, systems biology approach for elucidating the cellular pathways and transcriptional regulatory mechanisms that are dysregulated or altered by perturbations in vivo. Although the present study is focused on macrophage responses to lipid lowering in vivo, our approach of integrating LCM-derived transcriptome measurements with epigenomic and chromatin structural information from primary cell models could be applied to other constituents of the plaque (e.g., smooth muscle cells and endothelial cells) and then ultimately combined in a comprehensive regulatory network. More broadly, with today's panoply of publicly available epigenomic and chromatin structural measurements from a wide array of mouse and human cell types and tissues (e.g., through the ENCODE project), we anticipate that integrative approaches such as the one used in this work could be useful in the regulatory analysis of transcriptome measurements from unique, highly specific cell populations in other in vivo contexts.
Materials and Methods
Animal work
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health), and in accordance with animal use protocols that were approved by the NYU Langone Medical Center Institutional Animal Care and Use Committee (PHS Animal Welfare Assurance Number A3435-01) or the Institute for Systems Biology Institutional Animal Care and Use Committee (PHS Animal Welfare Assurance Number A4355-1). All tissue harvest (except mouse peritoneal macrophage harvest) and all survival surgeries were carried out under ketamine/xylazine-induced anesthesia, and for survival surgeries, animals were carefully monitored post-surgery for signs of pain or distress. For peritoneal macrophage harvest, mice were euthanized by CO2 asphyxiation following standard operating procedures established by the Institute for Systems Biology IACUC.
The development of the Reversa mouse (JAX Stock Number 004192, Ldlr −/− Apob 100/100 Mttp fl/fl Mx1:Cre +/+ animals on mixed background, in which injection of poly I:C causes silencing of hepatic Mttp expression) has been previously described [19]. In this work, for experimental studies of the Reversa model of plaque regression, four-week-old Reversa mice were weaned onto a Western diet (Catalog no. 100244, 40% kcal from milk fat; Dyets Inc., Bethlehem, PA) for 16 weeks (baseline time point) to establish aortic plaque and then switched to a chow diet and injected intraperitoneally every other day, for a total of four injections, with either 500 µg poly I:C (Sigma) or vehicle (saline). Blood samples were obtained from the retro-orbital plexus at baseline, seven days after the last poly I:C injection, and 14 days post-injection. Animals were sacrificed for arterial tissue collection at either baseline, seven days after the last poly I:C injection, or 14 days post-injection, as indicated in the Results section.
The aortic transplant model of plaque regression has been previously described [21], [23], [31]. In this work, for studies of plaque regression in the aortic transplant model, C57BL/6–Apoe −/− mice [35] were maintained on Western diet for 16 weeks and then the aortic arches were harvested and grafted into the abdominal aortas of Apoe −/− and WT mice. The grafts were harvested at three or five days post-surgery, as indicated in the Results section.
For all experimental data reported in this work, animal cohort sizes are as reported in the corresponding figure captions, table captions, or main article text, except for the microarray transcriptome profiling of plaque macrophages in the Reversa model, for which the cohort sizes are described in Table S1.
Cell culture
All cell culture was performed at 37°C and in the presence of 5% CO2.
Bone marrow-derived macrophages (BMDM) were prepared as follows: L929-conditioned medium was prepared by growing L929 cells to confluency in Dulbecco's Modified Eagle Medium (DMEM)+10% (v/v) fetal bovine serum (FBS)+100 I.U./mL penicillin and 100 µg/mL streptomycin (hereafter, 1% penicillin/streptomycin). The media was then replaced with DMEM containing 2% FBS+1% penicillin/streptomycin. Conditioned media was collected every three days, filtered through a 0.22 µm filter and fresh media containing 2% FBS was added. Bone marrow cells were isolated from the femur and tibia of 8–12 week old mice. After red blood cell lysis (RLB, Sigma), cells were plated in Petri dishes in L929-conditioned medium and incubated for seven days. Cells were then serum-starved in 0.2% FBS overnight and treated with 400 ng/mL Wnt3a (R&D Systems, Minneapolis, MN) for 24 h. RNA was isolated from cells using TRIzol (Life Technologies). cDNA synthesis was performed using Verso cDNA kit (Thermo Scientific) and qPCR analysis was performed as described below (see section qPCR).
Peritoneal macrophages were prepared as follows: In each of three separate replicate experiments, resident peritoneal macrophages were isolated from 2–4 female C57BL/6J mice (ages 8–12 weeks) using intraperitoneal lavage as described previously [70]. Peritoneal exudate cells were pooled, resuspended in peritoneal macrophage medium (Roswell Park Memorial Institute 1640 medium plus L-glutamine plus 10% FBS plus 1% penicillin/streptomycin) and then plated into two tissue culture wells at approximately 106 cells/well. After two hours, wells were triple-washed with warm PBS to select for adherent cells. Cells were incubated for 24 h in the presence of 25 µg/mL (by protein content) CuSO4-oxidized human LDL (oxLDL, Intracel, Frederick, MD) or vehicle.
Scratch-wound macrophage migration assay
BMDM (prepared from N = 3 mice) were differentiated in six well plates and grown to confluency. Cells were serum-starved in 0.2% FBS overnight and a single scratch wound was made with a plastic p200 tip. Cells were washed with starvation media to remove non-adherent cells and incubated with or without Wnt3a (400 ng/mL) for 24 h. The cells were then fixed in 3.7% formaldehyde, and wound area visualized. The percentages of area covered in 24 h were quantified using NIH ImageJ software. All conditions were done in quadruplicates.
Blood and tissue analysis
Plasma cholesterol levels were determined by a colorimetric enzymatic assay (Wako Diagnostics, Richmond, VA). Mice were sacrificed at the time points indicated below (see Results) and hearts (for aortic root analysis) or aortic arch grafts were harvested and frozen in Optimum Cutting Temperature (OCT) compound and serial-sectioned at a thickness of 6 µm onto positively charged glass slides (Superfrost Plus, Fisher Scientific, Pittsburgh, PA).
For CD68 immunostaining and laser capture microdissection, slides were hematoxylin-stained, cleared with xylenes, air-dried, and foam cells were identified by light microscopy. Every seventh section was immunostained with a CD68-specific antibody (AbD Serotec, MCA 1957) as previously described [23] and used as a guide slide for laser-capture microdissection for the next six serial sections. Sections were imaged at 40× on a Leica DM4000B microscope. In addition, slides that were used for morphometric analysis were counterstained with eosin and the CD68-immunostained areas were quantified by computer-aided morphometric analysis of digitized images (Image-Pro Plus software v5, Media Cybernetics, Silver Spring, MD). For isolation of CD68+ cells from the plaques, laser-capture microdissection (LCM) was performed with the PixCell II instrument (Arcturus Bioscience) as previously described [45], [46]. All LCM procedures were performed under RNase-free conditions. Student's t-test was used for comparisons of CD68+ pixel areas and cholesterol levels for each sample group vs. baseline. A two-way ANOVA was used to test for differences in CD68+ pixel areas and cholesterol levels, between Mttp-inactivated and vehicle-treated sample groups at the two time points (seven and 14 days post-injection). In the linear model for the ANOVA, the post-treatment time point (day seven or day 14) was taken as the first factor, and the treatment (poly I:C or saline) was taken as the second factor.
β-catenin immunohistochemistry (IHC) was carried out using frozen tissue sections from aortic grafts from eight animals (N = 4 animals per genotype group). Sections were fixed in acetone for 10 min, blocked with DAKO blocking buffer (DAKO, Cat. # X0909) and incubated with β-catenin rabbit polyclonal antibody (Santa Cruz, Cat, # sc7199, 1∶200) overnight at 4°C. As a negative control, sections from Apoe −/− recipient animals were assayed without the primary antibody. Sections were then incubated with FITC-conjugated secondary antibody for one hour, washed and mounted onto slides using VECTASHIELD mounting medium with diamidino-2-phenylindole (DAPI) (Vector Labs, Burlingame, CA). Images at 63× magnification were obtained using a Leica SP5 confocal microscope. Quantification of the mean fluorescent pixel intensity was performed on immunofluorescent images using Image-Pro Plus software v5. Six sections per animal were analyzed. An unpaired, two-tailed Student's t-test was used to test for differences in the numbers of pixels that were positive for β-catenin fluorescence, between the sample groups.
Microarrays
For LCM-derived samples, RNA was isolated from LCM caps using the PicoPure RNA Isolation Kit (Life Technologies) following the manufacturer's instructions. Total RNA was quantified using the NanoDrop spectrophotometer and assayed for quality using the Agilent Bioanalyzer 2100. Single-stranded cDNA was produced in microgram quantities from the RNA template using the Ovation Pico WTA System v2 (NuGEN), with the QIAquick PCR Purification Kit (QIAGEN) used for cDNA purification. The cDNA was analyzed on the Agilent 2100 Bioanalyzer and then fragmented and biotin-labeled using the Encore Biotin Module (NuGEN).
For peritoneal macrophage-derived samples, total RNA was isolated using TRIzol and RNA quality was analyzed with an Agilent 2100 Bioanalyzer. The three independent experiments thus yielded six RNA samples. Sample mRNA was amplified and labeled with the Affymetrix One-Cycle Eukaryotic Target Labeling Assay protocol and reagents. Labeled cDNA was hybridized to the Affymetrix GeneChip Mouse Exon 1.0 ST array using standard protocols and reagents from Affymetrix.
Fragmented, biotin-labeled cDNA was hybridized to the Affymetrix Mouse Exon Array 1.0 ST GeneChip and the GeneChip was washed and stained using the protocol and reagents in the Affymetrix GeneChip Hybridization, Wash, and Stain Kit. The stained GeneChips were imaged using the Affymetrix GeneChip Scanner 3000 and probe intensities were quantify from the scanned images using the Gene Chip Operating Software from Affymetrix. The transcriptome profiling data for this study are available online in the NCBI Gene Expression Omnibus (GEO) database, under accession numbers GSE52482 and GSE58913.
qPCR
LCM-derived samples
Real time quantitative PCR was performed using Taqman probes (Life Technologies) and using the Applied Biosystems 7300 Real time PCR system (Life Technologies). The input for each assay was 50 ng of cDNA amplified from LCM-isolated RNA using the Ovation Pico WTA system V2 (NuGEN). Taqman Universal Master Mix II (Life Technologies) was used for all reactions. Ct values were extracted from the fluorescence-versus-cycle measurements using the instrument software (Applied Biosystems). Relative mRNA levels were quantified from the Ct values using the exp(ΔΔCt) method [99], using Gapdh as the endogenous normalizer. The specific Taqman assays that were used for the study are listed in Table S13.
Computational meta-analysis
Affymetrix microarray data from the Cho et al. study [100] and the Hägg et al. study [101] (see Table S5 for references) were obtained from NCBI GEO and processed using the Robust Multichip Average method [102] in the software package Bioconductor, to obtain background-adjusted, quantile-normalized, and probeset-summarized log2 intensities. Human gene probesets were mapped to mouse orthologs using the Bioconductor tools for querying NCBI HomoloGene, and differential expression testing was carried out using a two-way ANOVA with a Benjamini-Hochberg [103] false discovery rate cutoff of 0.15. From all other studies, lists of differentially expressed genes were obtained from data tables from the publications referenced in Table S5, and mapped to mouse orthologs using HomoloGene. For each mouse gene in the Entrez Gene database, the number of independent analyses in which the gene was reported as differentially expressed during foam cell formation or atherosclerosis progression (among the studies in Table S5) was tabulated and used as the gene's meta-analysis score. Genes with a score greater than or equal to two (see Table S2, column L) were counted as high-scoring in the meta-analysis, for statistical enrichment test (see Results). Statistical enrichment of the set of genes with meta-analysis scores of two or greater, within the set of 213 genes that are differentially expressed in plaque CD68+ cells, was tested using Fisher's Exact Test, with the larger sample set consisting of the set of 21,594 mouse genes for which a meta-analysis score was tabulated.
Microarray data analysis
For microarray transcriptome profiling of plaque macrophages in the Reversa model, GeneChip scan images were processed into probe intensities using the Affymetrix GeneChip Operating Software system and then background-adjusted, quantile-normalized, and probeset-summarized using the Robust Multichip Average feature of Affymetrix Power Tools, with transcript-level probeset definitions from the Version 15 of the CustomCDF Project [104] (Ensembl Transcript-derived probeset definitions). Probe intensities were separately processed into exon-level normalized probeset intensities using the Robust Multichip Average Feature of Affymetrix Power Tools (version 1.10.2), and (separately) using exon-level probeset definitions from the CustomCDF project (Ensembl Exon) and from the Affymetrix Mouse Exon Array 1.0 ST NetAffx Annotations version 29. Exon-level detect-above-background calls were made using the two separate sets of (CustomCDF and Affymetrix) exon-level probesets using the Detection Above Background algorithm in Affymetrix Power Tools. For each exon-level probeset, detect-above-background P values were combined for all samples within a sample group using the geometric mean, and then across sample groups by taking the minimum P value, to obtain a single “present” P value for each exon-level probeset. For each Ensembl Transcript corresponding to one of the transcript-level probesets for which at least 75% of the constituent exon-level probesets was interrogated on the array, the transcript was considered to be above background in the sample if at least half of its constituent exons (that were interrogated with probes on the array) were detected above background with a “present” P value of less than or equal to 0.01 [105], [106]. For all probesets that passed the filter, log2 probeset intensities across all 24 Reversa samples (see Table S1) were analyzed for differential expression between the Mttp-inactivated and control sample groups, as described below.
For comparative transcriptome profiling of plaque macrophages in the transplant regression model, probe intensity values were obtained from a two-color 3′ microarray dataset of CD68+ cells from aortic arch grafts harvested from Apoe −/− and WT host animals three days post-surgery (N = 14 for Apoe −/−, N = 18 for WT). This previously published dataset was obtained from the lab of one of the authors (EAF) [36], and was based on the Rosetta/Merck Mouse 23.6K 3.0 A1 microarray (custom-synthesized by Agilent; design files are available at GEO accession number GPL9733). Probe intensities in each of the two color channels (corresponding to sample-derived RNA and a universal mouse RNA reagent (Stratagene) that was used as an internal control) were combined to quantify absolute expression (geometric mean) and normalized expression (ratio of the macrophage sample-derived intensity to the internal control-derived intensity) for each of the samples. Probes were filtered for minimum absolute log2 intensity (greater than or equal to −5) to remove probable below-background probe intensities. For all probes that passed the filter, log2 expression ratios across all 32 samples were analyzed for differential expression between the WT and Apoe −/− sample groups, as described below.
Testing for differential expression was performed using a two-factor model (sex and treatment for the Reversa study, sex and genotype for the transplant study) with empirical Bayes variance estimates, using the Limma package in Bioconductor [107], with a proportion factor of 0.03. Transcripts were selected as differentially expressed between the sample groups if the treatment factor P value was less than 0.01. For each unique gene among the set of differentially expressed transcripts, a representative transcript was selected by selecting (from among the differentially expressed transcripts that are associated with the gene in the Ensembl database) the transcript with the largest number of exons represented on the microarray. Enrichment tests for gene annotation terms were carried out separately for the sets of upregulated and downregulated genes in the Reversa transcriptome dataset, using the gene annotation analysis tool DAVID [108] with a P value cutoff of 0.05. Gene sets were analyzed with three different functional annotation enrichment tools to determine whether gene annotations related to apoptosis or cell death are overrepresented: DAVID, Gene Set Enrichment Analysis [109], and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Microarray analysis of poly I:C-treated macrophages was carried out using the raw microarray intensity data from a previous study [110] in which primary murine bone marrow-derived macrophages (BMDM) were treated in vitro for two hours with 6 µg/mL poly I:C and then profiled using the Affymetrix Mouse Exon Array 1.0 ST GeneChip. Six CEL files (three from each of the two sample groups, untreated and poly I:C-treated) were obtained from NCBI GEO (accession number GSE7052) and processed as described above (save for the proportion factor used for the empirical Bayes step in LIMMA being 0.1). The statistical cutoff used for differential expression tests was P<0.01. The statistical test for overlap between the poly I:C-responsive genes in BMDM and the genes that are transcriptionally responsive to lipid lowering in vivo was carried out using Fisher's Exact Test (two-sided).
Microarray transcriptome profiling probe-level measurements from mouse peritoneal macrophages were quantile-normalized without background adjustment and then summarized for probeset intensities using the Affymetrix PLIER algorithm [111] and for detection above background using the Affymetrix DABG algorithm [112] using probe-to-probeset mappings from the CustomCDF project [104] version 15, based on the mouse transcripts and mouse exons lists from the Ensembl database release 65. Transcripts were marked as “present” only if at least 2/3 of the constituent exons had a DABG P value of less than 0.01 [105]. Differential expression testing for each Ensembl transcript was carried out using a t-test with empirical Bayes variance estimates, using the Limma package [107] in Bioconductor with 0.1 as the prior expected proportion of differentially expressed genes. P values for differential expression between the sample groups (vehicle and oxLDL) were converted to q-values using the Bioconductor package qvalue [113]. Transcripts were filtered with a false discovery rate cutoff of 0.05 and a minimum absolute fold-change cutoff of 2.0, and filtered to include only transcripts that could be mapped to the RefSeq NM identifier. Transcripts were mapped start site coordinates on the mm9 genome assembly, and noncoding sequences within ±5 kbp of the transcription start sites were retrieved, using the BioMart tool in Ensembl release 65.
Transcription factor binding site motif analysis
In this section, we describe the computational method REMINISCE (Regulatory Element Machine-learning to Infer Networks Instructing Specific Cellular Expression; see Figure S1) that was used to identify transcription factor binding site motifs whose matches are overrepresented within 5′ regulatory regions of differentially expressed genes. Using the BioMart interface to the Ensembl database (release 65), transcription start site locations (on the NCBI M37 mouse genome assembly) were obtained for the sets of differentially expressed transcripts that were detected by microarray in CD68+ cells in plaques in the Reversa or transplant regression models. Noncoding DNA sequences (defined by excluding locations marked as protein coding within at least one exon within the Ensembl database) within ±5 kbp of a transcription start site were analyzed using a Random Forest classifier [114] to identify probable cis-regulatory regions, with a score assigned at 100 bp intervals based on the voting fraction of trees in the classifier. The features used for the classification (Table S8) include genome-wide measurements (binarized peak locations) of histone acetylation in bone marrow-derived macrophages obtained by ChIP-seq [39], “valley scores” in histone acetylation ChIP-seq signal [39], DNase I hypersensitive sites in bone marrow-derived macrophages [39] detected by short-read sequencing, vertebrate PhastCons interspecies sequence conservation scores from the University of California Santa Cruz Genome Bioinformatics database [115], GC content, computational predictions of CpG island locations [116], and basepair distances to the nearest binarized feature locations of each type. The Random Forest classifier for predicting cis-regulatory regions was trained using compendium of transcription factor binding site locations obtained by combining ChIP-seq datasets targeting 19 transcriptional regulators in murine macrophages, that were obtained from the literature (Table S8). Noncoding sequence within regions predicted to have a cis-regulatory function by the Random Forest model (voting fraction greater than 0.5) was scanned for transcription factor binding site motif matches using a combined set of vertebrate binding site position weight matrices from the TRANSFAC Professional 2010 [117] and JASPAR 2010 [118] databases. The scanning was performed using the software tool Clover [119] (Feb. 19, 2010 release) on noncoding sequences within cis-regulatory regions (after masking low-complexity repeats using NCBI DustMasker) proximal to transcription start sites of differentially expressed transcripts as well as proximal to transcription start sites of a set of 1,000 randomly selected genes that were detected above background in plaque macrophages by microarray hybridization (see Microarray Analysis). For the Clover scanning, a pseudocount value of 0.3 [120] was used. Matches for binding site sequence patterns within the promoter sequences for upregulated or downregulated genes were assessed for statistical overrepresentation using the frequencies compiled on the promoter regions for the 1,000 randomly selected macrophage-expressed genes, using the binomial test (P value threshold of 0.01). Subsequent to the motif analysis using TRANSFAC 2010, 5′ cis-regulatory regions for the upregulated Reversa gene set were re-analyzed using a substantially larger set (numbering 56) of TCF/LEF transcription factor family motifs from an updated motif database (TRANSFAC Professional 2013.4), using the same methods.
Transcription factor network analysis
Official gene symbols for genes encoding protein components of transcription factors that were implicated in the transcription factor binding site enrichment analysis (see Figure 3A) were uploaded to the GeneMANIA network analysis web application [59]. The interaction network for the transcription factors was assembled using protein-protein interactions (direct physical interactions only), and visualized using Cytoscape [121].
MicroRNA target sequence enrichment analysis
MicroRNA enrichment analysis was carried out using the CORNA software algorithm [122] using conserved microRNA target predictions from miRBase (v5) and using Ensembl BioMart [123] (release 70) for gene identifier translation. Statistical enrichment was assessed using Fisher's Exact Test (P value threshold of 0.01). Pathway enrichment analysis of genes with target sequences for the microRNAs shown in Figure S4 was carried out using the analysis program DIANA-miRPath [65], with target sequences predictions from the DIANA-microT-CDS v.5.0 algorithm [124].
Human SNP analysis of plaque regression gene sets
A set of 239,165 eSNP-to-gene associations (that collectively connect 157,668 eSNPs to monocyte expression levels of 13,989 genes) was compiled by merging (eSNP,gene) pair lists from two human population genetic studies of monocyte gene expression regulation [48], [49] comprised of samples from 1,490 and 283 individuals, respectively. From these 239,165 pairs, eSNPs that were associated with human genes whose mouse orthologs (obtained via successive mapping using the HomoloGene version 68 and the INPARANOID [125] version 8.0 databases) were detected as differentially expressed in plaque CD68+ cells in the Reversa model of plaque regression (for Reversa, the 213 genes in Table S2) or in the aortic transplant model of plaque regression (from Table S2 of [36], the 549 genes with absolute geometric-mean expression ratio >1.25 between the WT and Apoe −/− recipient sample groups) were then obtained, resulting in lists of 2,380 (Reversa) and 5,024 (transplant) plaque regression-associated eSNPs, respectively. The three sets of eSNPs – Reversa eSNPs, transplant eSNPs, and the full 157,668 “monocyte eSNPs” – were expanded to include proxy SNPs that are both in linkage disequilibrium (LD) with an eSNP (R>0.9) and within 250 kbp of the eSNP, yielding LD-expanded sets of 6,214, 13,248, and 363,283 SNPs, respectively. The expansion to proxy SNPs was carried out using LD maps from the 1,000 Genomes Project (CEU population group) and using the SNP analysis software SNAP [126]. A set of 761 SNPs that are associated with risk of atherosclerosis or CAD was obtained by searching the NCBI Phenotype-Genotype Integrator database [51] for the MeSH terms “atherosclerosis”, “coronary artery disease”, and “carotid stenosis”. Overlaps were computed for the set of 761 atherosclerosis/CAD SNPs with four different sets of SNPs: the 6,214 LD-expanded eSNPs from the Reversa model (overlap = 6), the 13,248 LD-expanded eSNPs from the aortic transplant model (overlap = 11), the 363,283 LD-expanded monocyte eSNPs (overlap = 134), and the set of all 44,278,189 validated SNPs in dbSNP build 138 (overlap = 761). From these overlaps, two 2×2 contingency tables were constructed to test for association between atherosclerosis/CAD SNPs and regression gene-associated eSNPs in the Reversa and aortic transplant models, respectively, using the LD-expanded set of monocyte eSNPs as a background set. Additionally, two 2×2 contingency tables were constructed to test for association between atherosclerosis/CAD SNPs and regression gene-associated eSNPs in the Reversa and aortic transplant models, respectively, using the set of all validated SNPs as a background set. P values and odds ratios (Figure 2C and Table S6) were obtained for these four contingency tables using Fisher's Exact Test. Confidence intervals (shown in Figure 2C) on the expected overlap of atherosclerosis/CAD SNPs and regression-associated eSNPs were obtained using the quantile function of the hypergeometric distribution.
Analysis software
Computational analyses were carried using scripts in the R statistical computing environment, also using the Bioconductor set of packages for R), and in the Perl programming language. Bar graphs were prepared using Prism (GraphPad Software, La Jolla, CA). All custom software scripts used in the computational analysis are available from the authors upon request. Statistical tests were carried out using R and GraphPad Prism.
Supporting Information
Diagram of REMINISCE method for detecting enrichments of transcription factor binding sites for specific transcription factors, within predicted regulatory regions. Cell type-specific epigenomic and chromatin information (A) are combined with cell type-generic genomic information (B) within an ensemble decision tree classifier, Random Forest (C). The classifier is trained to use the input features (A,B) to predict the locations of cis-regulatory regions, identified by combining transcription factor location datasets (D) from the specific cell type (macrophages). The classifier predicts cis-regulatory regions (E) upstream of genes that are differentially expressed in plaque macrophages in response to lipid lowering in vivo, and these regions are scanned to identify matches for transcription factor binding site motifs from a precompiled library of vertebrate motifs (F). For each transcription factor, frequencies per bp of predicted cis-regulatory region sequence for its binding sites are tabulated and tested for enrichment (G) above a background frequency from cis-regulatory regions for a list of genes that are expressed above background level in murine macrophages. See Methods section for details.
(TIFF)
Sensitivity for in silico detection of enrichments of transcription factor binding sites is compared between the epigenome-guided method, REMINISCE, and approaches in which all noncoding sequence within ±1 kbp or ±5 kbp of transcription start sites are analyzed. The three analysis methods were applied to a transcriptome profiling study of macrophage foam cells in which mouse resident peritoneal macrophages (N = 3 independent replicate experiments; see Methods) were cultured with oxLDL or vehicle for 24 h. High-confidence sets of differentially expressed genes were identified (false discovery rate cutoff of 0.05 and minimum absolute fold-change of 2.0) as upregulated (94 genes) or downregulated (342 genes), and a background set of 2,000 genes that were randomly selected from the set of genes that were detected above background in at least one sample group (vehicle or oxLDL). The 5′ regulatory regions for the genes were scanned for matches to transcription factor binding site motifs using the epigenome-guided method, REMINISCE, as described in the Methods section (“REMINISCE method”, white bars) and by analyzing all noncoding sequence within ±1 kbp of transcription start sites (“1 kbp classic”, gray bars) or within ±5 kbp of transcription start sites (“5 kbp classic”, charcoal bars). For all three methods, the number of matches for a binding site motif in the biological gene sets (genes up- or downregulated by oxLDL) per kbp of sequence analyzed, was compared to the number that would be expected by chance for genes that are expressed above background in resident peritoneal macrophages. Bar group labels represent transcription factor binding site motifs. Bar lengths represent the ratio of the number of motif matches (per kbp of sequence analyzed) for the indicated oxLDL-response gene set (left, upregulated in oxLDL; right, downregulated in oxLDL) to the number of motif matches (per kbp of sequence analyzed) for the background set of genes. For each bar group (motif), a missing bar of a particular shade indicates that the motif's binding sites were not detected as enriched (P<0.01) using that method. The results indicate that the REMINISCE method detected substantially more motifs, and at higher enrichment ratios, than the two methods based on analysis of all noncoding sequence.
(TIFF)
Motif scanning enrichment analysis for promoters of genes that are differentially expressed in plaque macrophages during plaque regression, in the Reversa and/or in the transplant regression models. Bars represent the ratio of the number of transcription factor (TF) binding site motif matches (per kbp of cis-regulatory sequence analyzed) for the indicated differentially expressed gene set, to the number of binding site motif matches per kbp for randomly generated sets of genes that are expressed above background in plaque CD68+ cells. Bar shading indicates the regression model from which the gene set was derived (white, Reversa model, gray, transplant regression model). A missing gray bar indicates that binding sites for the indicated TF binding site motif were not detected as significantly higher than the number expected by chance, for the transplant regression model. TF motifs shown on this figure were selected based on a significance threshold (see Methods) for enrichment for gene sets derived from the Reversa mouse, and thus, every TF shown here has a white bar. (A) TF binding site motifs for genes that are upregulated in CD68+ cells from regressing animals vs. control animals. (B) TF binding site motifs for genes that are downregulated in CD68+ cells from regressing animals vs. control animals.
(TIFF)
microRNA target sequence enrichment analysis identifies many microRNAs whose target sequences are overrepresented within the 3′ UTRs of genes that are differentially expressed in CD68+ cells in plaques in Mttp-inactivated vs. vehicle-treated Reversa mice. (A) Enrichment analysis for genes that are upregulated in Mttp-inactivated vs. vehicle-treated macrophages. Bars indicate the ratio of the number of target sequence matches within the gene set, vs. the number expected by chance for randomly selected sets of genes (see Methods). (B) Enrichment analysis for genes that are downregulated in Mttp-inactivated vs. vehicle-treated macrophages.
(TIFF)
Upregulation of Wnt pathway target genes in CD68+ cells in regressing vs. progressing plaques. qPCR-measured relative mRNA levels for Wnt signaling pathway target genes Lrp6 (A) and Gja1 (B) in CD68+ cells in aortic grafts in Apoe −/− and WT recipient animals (N = 6 per genotype group) in the aortic transplant regression model on day five post-transplant show increased expression during plaque regression vs. progression. Bars, mean ± SE. P values reported are for an unpaired, two-tailed Student's t-test.
(TIFF)
Canonical Wnt pathway activation stimulates macrophage migration in vitro. (A) Micrographs of a scratch-wound migration assay for primary murine macrophages incubated for 24 h in medium alone or medium plus Wnt3a (400 ng/mL). Dotted black lines demarcate the scratch region. (B) Quantification of percentage of scratch area recovered after 24 h incubation. Bars, mean ± SE (N = 4 replicates per sample group). (‡) P<0.01 (unpaired, two-tailed Student's t-test vs. control).
(TIFF)
β-catenin immunofluorescence has minimal background signal in the arterial intima of aortic graft sections from the aortic transplant model of plaque regression. Representative images of the negative control (primary antibody excluded) are shown, with and without DAPI merge. The elastic lamina is identified by its autofluorescence. Green, β-catenin immunofluorescence; blue, DAPI fluorescence. Scale bar, 100 µm.
(TIFF)
β-catenin immunofluorescence shows increased abundance in the nuclear area, in WT vs. Apoe−/− recipient mice in the aortic transplant regression model. (A) Images are confocal fluorescence micrographs from representative sections from a total of six grafts (N = 3 for each recipient animal genotype; six sections per graft). Green, β-catenin immunofluorescence; blue, DAPI fluorescence; dashed white line divides lumen from intima; scale bars, 20 µm. White arrows indicate areas of overlap of β-catenin and DAPI fluorescence signals in the regressing plaque section (bottom image). (B) Zoomed images of regions that are indicated with yellow dotted lines in the merged images from progressing and regressing plaques (A, second and fourth images from the top).
(TIFF)
List of sample groups for the transcriptome profiling of plaque macrophages from the Reversa plaque regression model. Columns are (A) genotype; (B) sex; (C) treatment; (D) time point (from last injection of poly I:C for the poly I:C treatment group); (E) number of animals in the sample group.
(XLSX)
Genes that were detected as differentially expressed in plaque macrophages from Mttp-inactivated vs. vehicle-treated animals, in the Reversa model. Columns are: (A) Ensembl Gene ID; (B) Chromosome; (C) Strand; (D) Transcript start coordinate; (E) Transcript end coordinate; (F) Official gene symbol; (G) Gene description; (H) RefSeq ID (if applicable); (I) Entrez Gene ID(s); (J) P value for the treatment factor (poly I:C vs. saline) in the statistical analysis; (K) signed ratio of the average expression levels between the sample groups (Mttp-inactivated and vehicle-treated groups); (L) meta-analysis score (see section Meta Analysis, in Methods); an empty cell indicates a lack of gene symbol mapping from the meta-analysis).
(XLSX)
Genes that are differentially expressed by poly I:C treatment in bone marrow-derived macrophages. Columns are: (A) Ensembl Transcript ID; (B) Chromosome; (C) Strand; (D) Transcript start coordinate; (E) Transcript end coordinate; (F) Official gene symbol; (G) Gene description; (H) RefSeq ID (if applicable); (I) Entrez Gene ID(s); (J) Entrez Gene ID(s); (K) signed ratio of the average expression levels between the sample groups (poly I:C and unstimulated, respectively).
(XLSX)
Gene functional enrichment analysis results for genes that are differentially expressed in plaque CD68+ cells in Mttp-inactivated vs. vehicle-treated Reversa mice. Columns are: (A) Type of gene function annotation; (B) Gene function term; (C) Count of genes with this term; (D) Percent of genes with this term; (E) P value for enrichment (Fisher's Exact Test); (F) Ensembl Gene IDs for genes with this term.
(XLSX)
Studies from the literature that were used in the meta-analysis of gene expression changes during foam cell formation and atherosclerosis. Columns are (A) Authors; (B) Journal; (C) Bibliographic reference; (D) Organism; (E) Cell/tissue type; (F) treatment, and (G) study type.
(XLSX)
Numbers of risk SNPs for atherosclerosis or CAD that coincide with monocyte expression QTLs and with regression-associated monocyte expression QTLs. (see Figure 2C and Methods). Columns are as follows: (A) Row labels; (B) Starting total SNP counts; (C) Number of SNPs after expanding to proxy SNPs within an LD block; (D) Number that overlap with known risk SNPs for atherosclerosis or CAD; (E) Frequency of atherosclerosis/CAD SNPs within each row's LD-expanded SNP set; (F) P value for enrichment of atherosclerosis/CAD SNPs (Fisher's Exact Test); (G) Odds ratio from Fisher's Exact Test.
(XLSX)
Risk SNPs for atherosclerosis or CAD that coincide with regression-associated monocyte expression QTLs. Columns are as follows: (A) RefSNP ID; (B) Trait (MeSH term); (C) Context (location relative to nearest gene; “nearGene-3” indicates that the SNP is located downstream of the 3′ end of a gene).
(XLSX)
Sources of epigenomic and transcription factor binding site ChIP-seq data used for features and training (respectively) for the computational model for mapping cis-regulatory regions Columns are as follows: (A) Transcriptional regulator or chromatin mark; (B) Proposed function; (C) Source; (D) Accession number of the ChIP-seq data set (where applicable).
(XLSX)
Statistically overrepresented motifs within up- or down-regulated genes in the Reversa model. Results for the sets of upregulated and downregulated genes are presented in separate blocks of rows, each labeled “for upregulated genes” and “for downregulated genes” respectively. The results of a motif scan of the upregulated genes using TCF/LEF family motifs from TRANSFAC 2013 are reported in a third block of rows, marked “TCF/LEF Scan of upregulated genes with TRANSFAC 2013”. Columns are as follows: (A) Motif ID; (B) Motif description (note: “HS” indicates a half-site for a bipartite binding site sequence); (C) Consensus sequence pattern; (D) Binding site match frequency enrichment ratio in the differentially expressed gene set vs. the frequency expected by chance among macrophage-expressed genes; (E) P value for the binomial test on the frequency of matches; binding site frequency enrichment ratio in differentially expressed genes in the transplant regression model.
(XLSX)
Numbers of genes with transcription factor binding site sequence matches of the indicated types, within their promoter regions. Results for the sets of upregulated and downregulated genes are presented in separate sections labeled “for upregulated genes” and “for downregulated genes” respectively. Columns are: (A) motif identifier; (B) motif description; (C) number of genes possessing a match for the motif within their promoter regions; (D) fraction of genes; (E) number of cytoskeleton-related genes (based on GO annotation) possessing a match for the motif within their promoter regions.
(XLSX)
Results of microRNA target sequence enrichment analysis for sets of genes that are differentially expressed in plaque CD68+ cells from Mttp-inactivated vs. vehicle-treated Reversa mice. Columns are as follows: (A) MicroRNA name; (B) Total number of target sequences among all mouse genes; (C) Expected number of target sequences for the indicated gene set (upregulated or downregulated genes); (D) Observed number of target sequences for the indicated gene set; (E) P value from the hypergeometric test; (F) ratio of observed to expected count.
(XLSX)
Results of pathway enrichment analysis of genes possessing target sequences for microRNAs listed in Figure 4A, derived from transcriptome profiling of plaque CD68+ cells from Reversa mice in plaque regression. Columns are as follows: (A) Pathway/function name; (B) Pathway ID; (C) Number of genes in the pathway that have a target sequence for one of the microRNAs in Figure S4A; (D) P value for enrichment test; (E) Official gene symbols; (F) Ensembl Gene IDs.
(XLSX)
qPCR primers that were used in this study. Columns are: (A) Official gene symbol; (B) Taqman assay ID.
(XLSX)
Acknowledgments
The authors thank the Array Core staff at the Institute for Systems Biology (Seattle, Washington, USA) for technical assistance, and Holly Arnold, Jichen Yang, and Mudra Choudhury for a critical reading of the manuscript.
Funding Statement
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01HL084312 (to EAF and ESG), K25HL098807 (to SAR), T32HL098129 (to YV), T32HL098129 (to PM), F30AG029748 (to JEF), R01AI032972 (to AA), R01AI025032 (to AA), and U19AI100627 (to AA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alwan A, Armstrong T, Bettcher D, Boerma T, Branca F, et al. (2011) Global Atlas on Cardiovascular Diseases Prevention and Control. Mendes S, Puska P, Norrving B, editors Geneva: World Health Organization. [Google Scholar]
- 2. Oliva A, Flores J, Merigioli S, LeDuc L, Benito B, et al. (2011) Autopsy investigation and Bayesian approach to coronary artery disease in victims of motor-vehicle accidents. Atherosclerosis 218: 28–32 10.1016/j.atherosclerosis.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 3. McGill HC, McMahan CA, Gidding SS (2008) Preventing heart disease in the 21st century: implications of the Pathobiological Determinants of Atherosclerosis in Youth (PDAY) study. Circulation 117: 1216–1227 10.1161/CIRCULATIONAHA.107.717033 [DOI] [PubMed] [Google Scholar]
- 4. Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, et al. (1998) Prediction of coronary heart disease using risk factor categories. Circulation 97: 1837–1847 10.1161/01.CIR.97.18.1837 [DOI] [PubMed] [Google Scholar]
- 5. Reardon MF, Nestel PJ, Craig IH, Harper RW (1985) Lipoprotein predictors of the severity of coronary artery disease in men and women. Circulation 71: 881–888 10.1161/01.CIR.71.5.881 [DOI] [PubMed] [Google Scholar]
- 6. Williams KJ, Feig JE, Fisher EA (2008) Rapid regression of atherosclerosis: insights from the clinical and experimental literature. Nat Clin Pract Cardiovasc Med 5: 91–102 10.1038/ncpcardio1086 [DOI] [PubMed] [Google Scholar]
- 7. Francis AA, Pierce GN (2011) An integrated approach for the mechanisms responsible for atherosclerotic plaque regression. Exp Clin Cardiol 16: 77–86. [PMC free article] [PubMed] [Google Scholar]
- 8. Hewing B, Fisher EA (2012) Preclinical mouse models and methods for the discovery of the causes and treatments of atherosclerosis. Expert Opin Drug Discov 7: 207–216 10.1517/17460441.2012.660143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansson GK, Hermansson A (2011) The immune system in atherosclerosis. Nat Immunol 12: 204–212 10.1038/ni.2001 [DOI] [PubMed] [Google Scholar]
- 10. Moore KJ, Tabas I (2011) Macrophages in the pathogenesis of atherosclerosis. Cell 145: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li AC, Glass CK (2002) The macrophage foam cell as a target for therapeutic intervention. Nat Med 8: 1235–1242 10.1038/nm1102-1235 [DOI] [PubMed] [Google Scholar]
- 12. Ross R (1999) Atherosclerosis–an inflammatory disease. N Engl J Med 340: 115–126 10.1056/NEJM199901143400207 [DOI] [PubMed] [Google Scholar]
- 13. Shah PK, Falk E, Badimon JJ, Fernandez-Ortiz A, Mailhac A, et al. (1995) Human monocyte-derived macrophages induce collagen breakdown in fibrous caps of atherosclerotic plaques. Potential role of matrix-degrading metalloproteinases and implications for plaque rupture. Circulation 92: 1565–1569. [PubMed] [Google Scholar]
- 14. Libby P, Geng YJ, Aikawa M, Schoenbeck U, Mach F, et al. (1996) Macrophages and atherosclerotic plaque stability. Curr Opin Lipidol 7: 330–335. [DOI] [PubMed] [Google Scholar]
- 15. Daoud AS, Fritz KE, Jarmolych J, Frank AS (1985) Role of macrophages in regression of atherosclerosis. Ann N Y Acad Sci 454: 101–114. [DOI] [PubMed] [Google Scholar]
- 16. Tangirala RK, Tsukamoto K, Chun SH, Usher D, Pure E, et al. (1999) Regression of atherosclerosis induced by liver-directed gene transfer of apolipoprotein A-I in mice. Circulation 100: 1816–1822 10.1161/01.CIR.100.17.1816 [DOI] [PubMed] [Google Scholar]
- 17. Potteaux S, Gautier EL, Hutchison SB, van Rooijen N, Rader DJ, et al. (2011) Suppressed monocyte recruitment drives macrophage removal from atherosclerotic plaques of Apoe −/− mice during disease regression. J Clin Invest 121: 2025–2036 10.1172/JCI43802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raffai RL, Weisgraber KH (2002) Hypomorphic apolipoprotein E mice: a new model of conditional gene repair to examine apolipoprotein E-mediated metabolism. J Biochem 277: 11064–11068 10.1074/jbc.M111222200 [DOI] [PubMed] [Google Scholar]
- 19. Lieu HD, Withycombe SK, Walker Q, Rong JX, Walzem RL, et al. (2003) Eliminating atherogenesis in mice by switching off hepatic lipoprotein secretion. Circulation 107: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 20. Feig JE, Parathath S, Rong JX, Mick SL, Vengrenyuk Y, et al. (2011) Reversal of hyperlipidemia with a genetic switch favorably affects the content and inflammatory state of macrophages in atherosclerotic plaques. Circulation 123: 989–998 10.1161/CIRCULATIONAHA.110.984146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trogan E, Fayad ZA, Itskovich VV, Aguinaldo J-GS, Mani V, et al. (2004) Serial studies of mouse atherosclerosis by in vivo magnetic resonance imaging detect lesion regression after correction of dyslipidemia. Arterioscler Thromb Vasc Biol 24: 1714–1719 10.1161/01.ATV.0000139313.69015.1c [DOI] [PubMed] [Google Scholar]
- 22. Llodra J, Angeli V, Liu J, Trogan E, Fisher EA, et al. (2004) Emigration of monocyte-derived cells from atherosclerotic lesions characterizes regressive, but not progressive, plaques. Proc Natl Acad Sci USA 101: 11779–11784 10.1073/pnas.0403259101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trogan E, Feig JE, Dogan S, Rothblat GH, Angeli V, et al. (2006) Gene expression changes in foam cells and the role of chemokine receptor CCR7 during atherosclerosis regression in ApoE-deficient mice. Proc Natl Acad Sci U S A 103: 3781–3786 10.1073/pnas.0511043103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feig JE, Shang Y, Rotllan N, Vengrenyuk Y, Wu C, et al. (2011) Statins promote the regression of atherosclerosis via activation of the CCR7-dependent emigration pathway in macrophages. PLoS ONE 6: e28534 10.1371/journal.pone.0028534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toomey S, Harhen B, Roche HM, Fitzgerald D, Belton O (2006) Profound resolution of early atherosclerosis with conjugated linoleic acid. Atherosclerosis 187: 40–49 10.1016/j.atherosclerosis.2005.08.024 [DOI] [PubMed] [Google Scholar]
- 26. Hewing B, Parathath S, Mai CK, Fiel MI, Guo L, et al. (2013) Rapid regression of atherosclerosis with MTP inhibitor treatment. Atherosclerosis 227: 125–129 10.1016/j.atherosclerosis.2012.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah PK, Yano J, Reyes O, Chyu KY, Kaul S, et al. (2001) High-dose recombinant apolipoprotein A-IMilano mobilizes tissue cholesterol and rapidly reduces plaque lipid and macrophage content in apolipoprotein E-deficient mice. Potential implications for acute plaque stabilization. Circulation 103: 3047–3050 10.1161/hc2501.092494 [DOI] [PubMed] [Google Scholar]
- 28. Feig JE, Rong JX, Shamir R, Sanson M, Vengrenyuk Y, et al. (2011) HDL promotes rapid atherosclerosis regression in mice and alters inflammatory properties of plaque monocyte-derived cells. Proceedings of the National Academy of Sciences 108: 7166–7171 10.1073/pnas.1016086108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skogsberg J, Lundstrom J, Kovacs A, Nilsson R, Noori P, et al. (2008) Transcriptional profiling uncovers a network of cholesterol-responsive atherosclerosis target genes. PLoS Genet 4: e1000036 10.1371/journal.pgen.1000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Björkegren JLM, Hägg S, Talukdar HA, Foroughi Asl H, Jain RK, et al. (2014) Plasma cholesterol–induced lesion networks activated before regression of early, mature, and advanced atherosclerosis. PLoS Genet 10: e1004201 10.1371/journal.pgen.1004201.s019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chereshnev I, Trogan E, Omerhodzic S, Itskovich V, Aguinaldo J-G, et al. (2003) Mouse model of heterotopic aortic arch transplantation. J Surg Res 111: 171–176 10.1016/S0022-4804(03)00039-8 [DOI] [PubMed] [Google Scholar]
- 32. Véniant MM, Zlot CH, Walzem RL, Pierotti V, Driscoll R, et al. (1998) Lipoprotein clearance mechanisms in LDL receptor-deficient “Apo-B48-only” and “Apo-B100-only” mice. J Clin Invest 102: 1559–1568 10.1172/JCI4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Véniant MM, Sullivan MA, Kim SK, Ambroziak P, Chu A, et al. (2000) Defining the atherogenicity of large and small lipoproteins containing apolipoprotein B100. J Clin Invest 106: 1501–1510 10.1172/JCI10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raabe M, Véniant MM, Sullivan MA, Zlot CH, Bjorkegren J, et al. (1999) Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Invest 103: 1287–1298 10.1172/JCI6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plump AS, Smith JD, Hayek T, Aalto-Setälä K, Walsh A, et al. (1992) Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71: 343–353. [DOI] [PubMed] [Google Scholar]
- 36. Feig JE, Vengrenyuk Y, Reiser V, Wu C, Statnikov A, et al. (2012) Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS ONE 7: e39790 10.1371/journal.pone.0039790.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martinez FO, Gordon S, Locati M, Mantovani A (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177: 7303–7311. [DOI] [PubMed] [Google Scholar]
- 38. Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, et al. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro . EMBO J 15: 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 39. Ramsey SA, Knijnenburg TA, Kennedy KA, Zak DE, Gilchrist M, et al. (2010) Genome-wide histone acetylation data improve prediction of mammalian transcription factor binding sites. Bioinformatics 26: 2071–2075 10.1093/bioinformatics/btq405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, et al. (2011) Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767 10.1101/gr.121541.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 10.1146/annurev.cellbio.20.010403.113126 [DOI] [PubMed] [Google Scholar]
- 42. Mosimann C, Hausmann G, Basler K (2009) Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10: 276–286 10.1038/nrm2654 [DOI] [PubMed] [Google Scholar]
- 43. Pereira CP, Bachli EB, Schoedon G (2009) The Wnt pathway: a macrophage effector molecule that triggers inflammation. Curr Atheroscler Rep 11: 236–242. [DOI] [PubMed] [Google Scholar]
- 44. Amini-Nik S, Cambridge E, Yu W, Guo A, Whetstone H, et al. (2014) β-Catenin-regulated myeloid cell adhesion and migration determine wound healing. J Clin Invest 124: 2599–2610 10.1172/JCI62059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trogan E, Choudhury RP, Dansky HM, Rong JX, Breslow JL, et al. (2002) Laser capture microdissection analysis of gene expression in macrophages from atherosclerotic lesions of apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A 99: 2234–2239 10.1073/pnas.042683999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trogan E, Fisher EA (2005) Laser capture microdissection for analysis of macrophage gene expression from atherosclerotic lesions. Methods Mol Biol 293: 221–231. [DOI] [PubMed] [Google Scholar]
- 47. English SB, Butte AJ (2007) Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics 23: 2910–2917 10.1093/bioinformatics/btm483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, et al. (2010) Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5: e10693 10.1371/journal.pone.0010693.t007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, et al. (2012) Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet 44: 502–510 10.1038/ng.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Genomes Project Consortium (2012) Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, et al. (2014) Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet 22: 144–147 10.1038/ejhg.2013.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kleber ME, Renner W, Grammer TB, Linsel-Nitschke P, Boehm BO, et al. (2010) Association of the single nucleotide polymorphism rs599839 in the vicinity of the sortilin 1 gene with LDL and triglyceride metabolism, coronary heart disease and myocardial infarction. The Ludwigshafen Risk and Cardiovascular Health Study. Atherosclerosis 209: 492–497 10.1016/j.atherosclerosis.2009.09.068 [DOI] [PubMed] [Google Scholar]
- 53. Zhou L, Ding H, Zhang X, He M, Huang S, et al. (2011) Genetic variants at newly identified lipid loci are associated with coronary heart disease in a Chinese Han population. PLoS ONE 6: e27481 10.1371/journal.pone.0027481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. (2007) Genomewide association analysis of coronary artery disease. N Engl J Med 357: 443–453 10.1056/NEJMoa072366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coronary Artery Disease Consortium (2009) Samani NJ, Deloukas P, Erdmann J, Hengstenberg C, et al. (2009) Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscler Thromb Vasc Biol 29: 774–780 10.1161/ATVBAHA.108.181388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, et al. (2010) From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466: 714–719 10.1038/nature09266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, et al. (2010) Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab 12: 213–223 10.1016/j.cmet.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 58. Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, et al. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q (2008) GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 9 Suppl 1: S4 10.1186/gb-2008-9-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stumpf M, Balding DJ, Girolami M (2011) Handbook of Statistical Systems Biology. First ed. West Sussex: John W. Wiley & Sons. [Google Scholar]
- 61. Gabdoulline R, Eckweiler D, Kel A, Stegmaier P (2012) 3DTF: a web server for predicting transcription factor PWMs using 3D structure-based energy calculations. Nucleic Acids Research 40: W180–W185 10.1093/nar/gks551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsaousi A, Mill C, George SJ (2011) The Wnt pathways in vascular disease: lessons from vascular development. Curr Opin Lipidol 22: 350–357 10.1097/MOL.0b013e32834aa701 [DOI] [PubMed] [Google Scholar]
- 63. Mermelstein CS, Portilho DM, Mendes FA, Costa ML, Abreu JG (2007) Wnt/beta-catenin pathway activation and myogenic differentiation are induced by cholesterol depletion. Differentiation 75: 184–192 10.1111/j.1432-0436.2006.00129.x [DOI] [PubMed] [Google Scholar]
- 64. Kormish JD, Sinner D, Zorn AM (2010) Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev Dyn 239: 56–68 10.1002/dvdy.22046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, et al. (2012) DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Research 40: W498–W504 10.1093/nar/gks494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mani A, Radhakrishnan J, Wang H, Mani A, Mani M-A, et al. (2007) LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 315: 1278–1282 10.1126/science.1136370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van der Heyden MA, Rook MB, Hermans MM, Rijksen G, Boonstra J, et al. (1998) Identification of connexin43 as a functional target for Wnt signalling. J Cell Sci 111 (Pt 12) 1741–1749. [DOI] [PubMed] [Google Scholar]
- 68. Todaro GJ, Lazar GK, Green H (1965) The initiation of cell division in a contact-inhibited mammalian cell line. J Cell Physiol 66: 325–333. [DOI] [PubMed] [Google Scholar]
- 69. Koth LL, Cambier CJ, Ellwanger A, Solon M, Hou L, et al. (2010) DAP12 is required for macrophage recruitment to the lung in response to cigarette smoke and chemotaxis toward CCL2. J Immunol 184: 6522–6528 10.4049/jimmunol.0901171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gold ES, Ramsey SA, Sartain MJ, Selinummi J, Podolsky I, et al. (2012) ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J Exp Med 209: 807–817 10.1084/jem.20111202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ramsey SA, Gold ES, Aderem A (2010) A systems biology approach to understanding atherosclerosis. EMBO Mol Med 2: 79–89 10.1002/emmm.201000063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brasier AR (2010) The nuclear factor-kappaB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc Res 86: 211–218 10.1093/cvr/cvq076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mallat Z, Besnard S, Duriez M, Deleuze V, Emmanuel F, et al. (1999) Protective role of interleukin-10 in atherosclerosis. Circ Res 85: e17–e24. [DOI] [PubMed] [Google Scholar]
- 74. Virmani R, Burke AP, Farb A, Kolodgie FD (2002) Pathology of the unstable plaque. Prog Cardiovasc Dis 44: 349–356. [DOI] [PubMed] [Google Scholar]
- 75. Kolodgie FD, Narula J, Burke AP, Haider N, Farb A, et al. (2000) Localization of apoptotic macrophages at the site of plaque rupture in sudden coronary death. Am J Pathol 157: 1259–1268 10.1016/S0002-9440(10)64641-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Laufer EM, Winkens MHM, Narula J, Hofstra L (2009) Molecular imaging of macrophage cell death for the assessment of plaque vulnerability. Arterioscler Thromb Vasc Biol 29: 1031–1038 10.1161/ATVBAHA.108.165522 [DOI] [PubMed] [Google Scholar]
- 77. Li AC, Binder CJ, Gutierrez A, Brown KK, Plotkin CR, et al. (2004) Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J Clin Invest 114: 1564–1576 10.1172/JCI18730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thorp E, Kuriakose G, Shah YM, Gonzalez FJ, Tabas I (2007) Pioglitazone Increases Macrophage apoptosis and plaque necrosis in advanced atherosclerotic lesions of nondiabetic low-density lipoprotein receptor null mice. Circulation 116: 2182–2190 10.1161/CIRCULATIONAHA.107.698852 [DOI] [PubMed] [Google Scholar]
- 79. Nakaya H, Summers BD, Nicholson AC, Gotto AM, Hajjar DP, et al. (2009) Atherosclerosis in LDLR-knockout mice is inhibited, but not reversed, by the PPARgamma ligand pioglitazone. Am J Pathol 174: 2007–2014 10.2353/ajpath.2009.080611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gong K, Zhou F, Huang H, Gong Y, Zhang L (2012) Suppression of GSK3β by ERK mediates lipopolysaccharide induced cell migration in macrophage through β-catenin signaling. Protein Cell 3: 762–768 10.1007/s13238-012-2058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Gils JM, Derby MC, Fernandes LR, Ramkhelawon B, Ray TD, et al. (2012) The neuroimmune guidance cue netrin-1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat Immunol 13: 136–143 10.1038/ni.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sarzani R, Salvi F, Bordicchia M, Guerra F, Battistoni I, et al. (2011) Carotid artery atherosclerosis in hypertensive patients with a functional LDL receptor-related protein 6 gene variant. Nutr Metab Cardiovasc Dis 21: 150–156 10.1016/j.numecd.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 83. Neumann J, Schaale K, Farhat K, Endermann T, Ulmer AJ, et al. (2010) Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming Mycobacterium tuberculosis-infected macrophages. FASEB J 24: 4599–4612 10.1096/fj.10-160994 [DOI] [PubMed] [Google Scholar]
- 84. Marinou K, Christodoulides C, Antoniades C, Koutsilieris M (2012) Wnt signaling in cardiovascular physiology. Trends Endocrinol Metab 23: 628–636 10.1016/j.tem.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 85. Terrand J, Bruban V, Zhou L, Gong W, Asmar El Z, et al. (2009) LRP1 controls intracellular cholesterol storage and fatty acid synthesis through modulation of Wnt signaling. J Biochem 284: 381–388 10.1074/jbc.M806538200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Go G-W, Srivastava R, Hernandez-Ono A, Gang G, Smith SB, et al. (2014) The combined hyperlipidemia caused by impaired Wnt-LRP6 signaling is reversed by Wnt3a rescue. Cell Metab 19: 209–220 10.1016/j.cmet.2013.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fujino T, Asaba H, Kang M-J, Ikeda Y, Sone H, et al. (2003) Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA 100: 229–234 10.1073/pnas.0133792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hock MB, Kralli A (2009) Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol 71: 177–203 10.1146/annurev.physiol.010908.163119 [DOI] [PubMed] [Google Scholar]
- 89. Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, et al. (2004) Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res 94: 609–616 10.1161/01.RES.0000119171.44657.45 [DOI] [PubMed] [Google Scholar]
- 90. Puig O, Yuan J, Stepaniants S, Zieba R, Zycband E, et al. (2011) A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ Cardiovasc Genet 4: 595–604 10.1161/CIRCGENETICS.111.960773 [DOI] [PubMed] [Google Scholar]
- 91. Hsieh P-C, Chiang M-L, Chang J-C, Yan Y-T, Wang F-F, et al. (2012) DDA3 stabilizes microtubules and suppresses neurite formation. J Cell Sci 125: 3402–3411 10.1242/jcs.099150 [DOI] [PubMed] [Google Scholar]
- 92. Zhang L, Shao H, Zhu T, Xia P, Wang Z, et al. (2013) DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration. Sci Rep 3: 1681 10.1038/srep01681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hsieh P-C, Chang J-C, Sun W-T, Hsieh S-C, Wang M-C, et al. (2007) p53 downstream target DDA3 is a novel microtubule-associated protein that interacts with end-binding protein EB3 and activates beta-catenin pathway. Oncogene 26: 4928–4940 10.1038/sj.onc.1210304 [DOI] [PubMed] [Google Scholar]
- 94. Nissen SE, Tuzcu EM, Schoenhagen P, Brown BG, Ganz P, et al. (2004) Effect of intensive compared with moderate lipid-lowering therapy on progression of coronary atherosclerosis: a randomized controlled trial. JAMA 291: 1071–1080 10.1001/jama.291.9.1071 [DOI] [PubMed] [Google Scholar]
- 95. Zhao XQ, Brown BG, Hillger L, Sacco D, Bisson B, et al. (1993) Effects of intensive lipid-lowering therapy on the coronary arteries of asymptomatic subjects with elevated apolipoprotein B. Circulation 88: 2744–2753 10.1161/01.CIR.88.6.2744 [DOI] [PubMed] [Google Scholar]
- 96. Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, et al. (2008) Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 359: 2195–2207 10.1056/NEJMoa0807646 [DOI] [PubMed] [Google Scholar]
- 97. Nissen SE, Nicholls SJ, Sipahi I, Libby P, Raichlen JS, et al. (2006) Effect of very high-intensity statin therapy on regression of coronary atherosclerosis: the ASTEROID trial. JAMA 295: 1556–1565 10.1001/jama.295.13.jpc60002 [DOI] [PubMed] [Google Scholar]
- 98. Parathath S, Grauer L, Huang L-S, Sanson M, Distel E, et al. (2011) Diabetes adversely affects macrophages during atherosclerotic plaque regression in mice. Diabetes 60: 1759–1769 10.2337/db10-0778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cho HJ, Shashkin P, Gleissner CA, Dunson D, Jain N, et al. (2007) Induction of dendritic cell-like phenotype in macrophages during foam cell formation. Physiol Genomics 29: 149–160 10.1152/physiolgenomics.00051.2006 [DOI] [PubMed] [Google Scholar]
- 101. Hägg DA, Jernas M, Wiklund O, Thelle DS, Fagerberg B, et al. (2008) Expression profiling of macrophages from subjects with atherosclerosis to identify novel susceptibility genes. Int J Mol Med 21: 697–704. [PubMed] [Google Scholar]
- 102.Bolstad BM (2004) Low-level analysis of high-density oligonucleotide array data: background, normalization, and summarization. Ph.D. Thesis, Berkeley: University of California, Berkeley. [Google Scholar]
- 103. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300. [Google Scholar]
- 104. Dai M, Wang P, Boyd AD, Kostov G, Athey B, et al. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Affymetrix (2006) Identifying and validating alternative splicing events. Affymetrix Technical Note [Google Scholar]
- 106. Okoniewski MJ, Miller CJ (2008) Comprehensive analysis of Affymetrix exon arrays using BioConductor. PLoS Comput Biol 4: e6 10.1371/journal.pcbi.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 108. Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, et al. (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- 109. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, et al. (2012) A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 10.1038/nature11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Affymetrix (2005) Guide to probe logarithmic intensity error (PLIER) estimation. Affymetrix Technical Note [Google Scholar]
- 112.Affymetrix (2005) Exon array background correction. First ed. Affymetrix Whitepaper. [Google Scholar]
- 113. Dabney A, Storey JD (2003) QVALUE: the manual version 1.0. Computer Software User Guide, University of Washington, Department of Biostatistics [Google Scholar]
- 114. Breiman L (2001) Random forests. Machine learning 45: 5–32. [Google Scholar]
- 115. Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, et al. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP (2010) Redefining CpG islands using hidden Markov models. Biostatistics 11: 499–514 10.1093/biostatistics/kxq005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wingender E, Chen X, Hehl R, Karas H, Liebich I, et al. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, et al. (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–D110 10.1093/nar/gkp950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Frith MC, Fu Y, Yu L, Chen J-F, Hansen U, et al. (2004) Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32: 1372–1381 10.1093/nar/gkh299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nishida K, Frith MC, Nakai K (2009) Pseudocounts for transcription factor binding sites. Nucleic Acids Research 37: 939–944 10.1093/nar/gkn1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wu X, Watson M (2009) CORNA: testing gene lists for regulation by microRNAs. Bioinformatics 25: 832–833 10.1093/bioinformatics/btp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Smedley D, Haider S, Ballester B, Holland R, London D, et al. (2009) BioMart–biological queries made easy. BMC Genomics 10: 22 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG (2012) Functional microRNA targets in protein coding sequences. Bioinformatics 28: 771–776 10.1093/bioinformatics/bts043 [DOI] [PubMed] [Google Scholar]
- 125. Remm M, Storm CE, Sonnhammer EL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 10.1006/jmbi.2000.5197 [DOI] [PubMed] [Google Scholar]
- 126. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939 10.1093/bioinformatics/btn564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagram of REMINISCE method for detecting enrichments of transcription factor binding sites for specific transcription factors, within predicted regulatory regions. Cell type-specific epigenomic and chromatin information (A) are combined with cell type-generic genomic information (B) within an ensemble decision tree classifier, Random Forest (C). The classifier is trained to use the input features (A,B) to predict the locations of cis-regulatory regions, identified by combining transcription factor location datasets (D) from the specific cell type (macrophages). The classifier predicts cis-regulatory regions (E) upstream of genes that are differentially expressed in plaque macrophages in response to lipid lowering in vivo, and these regions are scanned to identify matches for transcription factor binding site motifs from a precompiled library of vertebrate motifs (F). For each transcription factor, frequencies per bp of predicted cis-regulatory region sequence for its binding sites are tabulated and tested for enrichment (G) above a background frequency from cis-regulatory regions for a list of genes that are expressed above background level in murine macrophages. See Methods section for details.
(TIFF)
Sensitivity for in silico detection of enrichments of transcription factor binding sites is compared between the epigenome-guided method, REMINISCE, and approaches in which all noncoding sequence within ±1 kbp or ±5 kbp of transcription start sites are analyzed. The three analysis methods were applied to a transcriptome profiling study of macrophage foam cells in which mouse resident peritoneal macrophages (N = 3 independent replicate experiments; see Methods) were cultured with oxLDL or vehicle for 24 h. High-confidence sets of differentially expressed genes were identified (false discovery rate cutoff of 0.05 and minimum absolute fold-change of 2.0) as upregulated (94 genes) or downregulated (342 genes), and a background set of 2,000 genes that were randomly selected from the set of genes that were detected above background in at least one sample group (vehicle or oxLDL). The 5′ regulatory regions for the genes were scanned for matches to transcription factor binding site motifs using the epigenome-guided method, REMINISCE, as described in the Methods section (“REMINISCE method”, white bars) and by analyzing all noncoding sequence within ±1 kbp of transcription start sites (“1 kbp classic”, gray bars) or within ±5 kbp of transcription start sites (“5 kbp classic”, charcoal bars). For all three methods, the number of matches for a binding site motif in the biological gene sets (genes up- or downregulated by oxLDL) per kbp of sequence analyzed, was compared to the number that would be expected by chance for genes that are expressed above background in resident peritoneal macrophages. Bar group labels represent transcription factor binding site motifs. Bar lengths represent the ratio of the number of motif matches (per kbp of sequence analyzed) for the indicated oxLDL-response gene set (left, upregulated in oxLDL; right, downregulated in oxLDL) to the number of motif matches (per kbp of sequence analyzed) for the background set of genes. For each bar group (motif), a missing bar of a particular shade indicates that the motif's binding sites were not detected as enriched (P<0.01) using that method. The results indicate that the REMINISCE method detected substantially more motifs, and at higher enrichment ratios, than the two methods based on analysis of all noncoding sequence.
(TIFF)
Motif scanning enrichment analysis for promoters of genes that are differentially expressed in plaque macrophages during plaque regression, in the Reversa and/or in the transplant regression models. Bars represent the ratio of the number of transcription factor (TF) binding site motif matches (per kbp of cis-regulatory sequence analyzed) for the indicated differentially expressed gene set, to the number of binding site motif matches per kbp for randomly generated sets of genes that are expressed above background in plaque CD68+ cells. Bar shading indicates the regression model from which the gene set was derived (white, Reversa model, gray, transplant regression model). A missing gray bar indicates that binding sites for the indicated TF binding site motif were not detected as significantly higher than the number expected by chance, for the transplant regression model. TF motifs shown on this figure were selected based on a significance threshold (see Methods) for enrichment for gene sets derived from the Reversa mouse, and thus, every TF shown here has a white bar. (A) TF binding site motifs for genes that are upregulated in CD68+ cells from regressing animals vs. control animals. (B) TF binding site motifs for genes that are downregulated in CD68+ cells from regressing animals vs. control animals.
(TIFF)
microRNA target sequence enrichment analysis identifies many microRNAs whose target sequences are overrepresented within the 3′ UTRs of genes that are differentially expressed in CD68+ cells in plaques in Mttp-inactivated vs. vehicle-treated Reversa mice. (A) Enrichment analysis for genes that are upregulated in Mttp-inactivated vs. vehicle-treated macrophages. Bars indicate the ratio of the number of target sequence matches within the gene set, vs. the number expected by chance for randomly selected sets of genes (see Methods). (B) Enrichment analysis for genes that are downregulated in Mttp-inactivated vs. vehicle-treated macrophages.
(TIFF)
Upregulation of Wnt pathway target genes in CD68+ cells in regressing vs. progressing plaques. qPCR-measured relative mRNA levels for Wnt signaling pathway target genes Lrp6 (A) and Gja1 (B) in CD68+ cells in aortic grafts in Apoe −/− and WT recipient animals (N = 6 per genotype group) in the aortic transplant regression model on day five post-transplant show increased expression during plaque regression vs. progression. Bars, mean ± SE. P values reported are for an unpaired, two-tailed Student's t-test.
(TIFF)
Canonical Wnt pathway activation stimulates macrophage migration in vitro. (A) Micrographs of a scratch-wound migration assay for primary murine macrophages incubated for 24 h in medium alone or medium plus Wnt3a (400 ng/mL). Dotted black lines demarcate the scratch region. (B) Quantification of percentage of scratch area recovered after 24 h incubation. Bars, mean ± SE (N = 4 replicates per sample group). (‡) P<0.01 (unpaired, two-tailed Student's t-test vs. control).
(TIFF)
β-catenin immunofluorescence has minimal background signal in the arterial intima of aortic graft sections from the aortic transplant model of plaque regression. Representative images of the negative control (primary antibody excluded) are shown, with and without DAPI merge. The elastic lamina is identified by its autofluorescence. Green, β-catenin immunofluorescence; blue, DAPI fluorescence. Scale bar, 100 µm.
(TIFF)
β-catenin immunofluorescence shows increased abundance in the nuclear area, in WT vs. Apoe−/− recipient mice in the aortic transplant regression model. (A) Images are confocal fluorescence micrographs from representative sections from a total of six grafts (N = 3 for each recipient animal genotype; six sections per graft). Green, β-catenin immunofluorescence; blue, DAPI fluorescence; dashed white line divides lumen from intima; scale bars, 20 µm. White arrows indicate areas of overlap of β-catenin and DAPI fluorescence signals in the regressing plaque section (bottom image). (B) Zoomed images of regions that are indicated with yellow dotted lines in the merged images from progressing and regressing plaques (A, second and fourth images from the top).
(TIFF)
List of sample groups for the transcriptome profiling of plaque macrophages from the Reversa plaque regression model. Columns are (A) genotype; (B) sex; (C) treatment; (D) time point (from last injection of poly I:C for the poly I:C treatment group); (E) number of animals in the sample group.
(XLSX)
Genes that were detected as differentially expressed in plaque macrophages from Mttp-inactivated vs. vehicle-treated animals, in the Reversa model. Columns are: (A) Ensembl Gene ID; (B) Chromosome; (C) Strand; (D) Transcript start coordinate; (E) Transcript end coordinate; (F) Official gene symbol; (G) Gene description; (H) RefSeq ID (if applicable); (I) Entrez Gene ID(s); (J) P value for the treatment factor (poly I:C vs. saline) in the statistical analysis; (K) signed ratio of the average expression levels between the sample groups (Mttp-inactivated and vehicle-treated groups); (L) meta-analysis score (see section Meta Analysis, in Methods); an empty cell indicates a lack of gene symbol mapping from the meta-analysis).
(XLSX)
Genes that are differentially expressed by poly I:C treatment in bone marrow-derived macrophages. Columns are: (A) Ensembl Transcript ID; (B) Chromosome; (C) Strand; (D) Transcript start coordinate; (E) Transcript end coordinate; (F) Official gene symbol; (G) Gene description; (H) RefSeq ID (if applicable); (I) Entrez Gene ID(s); (J) Entrez Gene ID(s); (K) signed ratio of the average expression levels between the sample groups (poly I:C and unstimulated, respectively).
(XLSX)
Gene functional enrichment analysis results for genes that are differentially expressed in plaque CD68+ cells in Mttp-inactivated vs. vehicle-treated Reversa mice. Columns are: (A) Type of gene function annotation; (B) Gene function term; (C) Count of genes with this term; (D) Percent of genes with this term; (E) P value for enrichment (Fisher's Exact Test); (F) Ensembl Gene IDs for genes with this term.
(XLSX)
Studies from the literature that were used in the meta-analysis of gene expression changes during foam cell formation and atherosclerosis. Columns are (A) Authors; (B) Journal; (C) Bibliographic reference; (D) Organism; (E) Cell/tissue type; (F) treatment, and (G) study type.
(XLSX)
Numbers of risk SNPs for atherosclerosis or CAD that coincide with monocyte expression QTLs and with regression-associated monocyte expression QTLs. (see Figure 2C and Methods). Columns are as follows: (A) Row labels; (B) Starting total SNP counts; (C) Number of SNPs after expanding to proxy SNPs within an LD block; (D) Number that overlap with known risk SNPs for atherosclerosis or CAD; (E) Frequency of atherosclerosis/CAD SNPs within each row's LD-expanded SNP set; (F) P value for enrichment of atherosclerosis/CAD SNPs (Fisher's Exact Test); (G) Odds ratio from Fisher's Exact Test.
(XLSX)
Risk SNPs for atherosclerosis or CAD that coincide with regression-associated monocyte expression QTLs. Columns are as follows: (A) RefSNP ID; (B) Trait (MeSH term); (C) Context (location relative to nearest gene; “nearGene-3” indicates that the SNP is located downstream of the 3′ end of a gene).
(XLSX)
Sources of epigenomic and transcription factor binding site ChIP-seq data used for features and training (respectively) for the computational model for mapping cis-regulatory regions Columns are as follows: (A) Transcriptional regulator or chromatin mark; (B) Proposed function; (C) Source; (D) Accession number of the ChIP-seq data set (where applicable).
(XLSX)
Statistically overrepresented motifs within up- or down-regulated genes in the Reversa model. Results for the sets of upregulated and downregulated genes are presented in separate blocks of rows, each labeled “for upregulated genes” and “for downregulated genes” respectively. The results of a motif scan of the upregulated genes using TCF/LEF family motifs from TRANSFAC 2013 are reported in a third block of rows, marked “TCF/LEF Scan of upregulated genes with TRANSFAC 2013”. Columns are as follows: (A) Motif ID; (B) Motif description (note: “HS” indicates a half-site for a bipartite binding site sequence); (C) Consensus sequence pattern; (D) Binding site match frequency enrichment ratio in the differentially expressed gene set vs. the frequency expected by chance among macrophage-expressed genes; (E) P value for the binomial test on the frequency of matches; binding site frequency enrichment ratio in differentially expressed genes in the transplant regression model.
(XLSX)
Numbers of genes with transcription factor binding site sequence matches of the indicated types, within their promoter regions. Results for the sets of upregulated and downregulated genes are presented in separate sections labeled “for upregulated genes” and “for downregulated genes” respectively. Columns are: (A) motif identifier; (B) motif description; (C) number of genes possessing a match for the motif within their promoter regions; (D) fraction of genes; (E) number of cytoskeleton-related genes (based on GO annotation) possessing a match for the motif within their promoter regions.
(XLSX)
Results of microRNA target sequence enrichment analysis for sets of genes that are differentially expressed in plaque CD68+ cells from Mttp-inactivated vs. vehicle-treated Reversa mice. Columns are as follows: (A) MicroRNA name; (B) Total number of target sequences among all mouse genes; (C) Expected number of target sequences for the indicated gene set (upregulated or downregulated genes); (D) Observed number of target sequences for the indicated gene set; (E) P value from the hypergeometric test; (F) ratio of observed to expected count.
(XLSX)
Results of pathway enrichment analysis of genes possessing target sequences for microRNAs listed in Figure 4A, derived from transcriptome profiling of plaque CD68+ cells from Reversa mice in plaque regression. Columns are as follows: (A) Pathway/function name; (B) Pathway ID; (C) Number of genes in the pathway that have a target sequence for one of the microRNAs in Figure S4A; (D) P value for enrichment test; (E) Official gene symbols; (F) Ensembl Gene IDs.
(XLSX)
qPCR primers that were used in this study. Columns are: (A) Official gene symbol; (B) Taqman assay ID.
(XLSX)