Background: A genetic variant within RNF207 is associated with a prolonged QT interval. QT interval prolongation may lead to arrhythmia, a risk factor for sudden cardiac death.
Results: RNF207 regulates action potential duration and the repolarizing channel HERG.
Conclusion: RNF207 is an important regulator of cardiac excitation.
Significance: Understanding the composition and dynamics of membrane complexes is important in unraveling the mechanism of arrhythmias.
Keywords: Endoplasmic Reticulum (ER), HERG, Protein Complex, SNP, Zebrafish, HSP70, RNF207, Action Potential Duration, Conduction Velocity
Abstract
Two recent studies (Newton-Cheh, C. et al. (2009) Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. 41, 399–406 and Pfeufer, A. et al. (2009) Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat. Genet. 41, 407–414) identified an association, with genome-wide significance, between a single nucleotide polymorphism within the gene encoding RING finger protein 207 (RNF207) and the QT interval. We sought to determine the role of RNF207 in cardiac electrophysiology. Morpholino knockdown of RNF207 in zebrafish embryos resulted in action potential duration prolongation, occasionally a 2:1 atrioventricular block, and slowing of conduction velocity. Conversely, neonatal rabbit cardiomyocytes infected with RNF207-expressing adenovirus exhibited shortened action potential duration. Using transfections of U-2 OS and HEK293 cells, Western blot analysis and immunocytochemistry data demonstrate that RNF207 and the human ether-a-go-go-related gene (HERG) potassium channel interact and colocalize. Furthermore, RNF207 overexpression significantly elevated total and membrane HERG protein and HERG-encoded current density by ∼30–50%, which was dependent on the intact N-terminal RING domain of RNF207. Finally, coexpression of RNF207 and HSP70 increased HERG expression compared with HSP70 alone. This effect was dependent on the C terminus of RNF207. Taken together, the evidence is strong that RNF207 is an important regulator of action potential duration, likely via effects on HERG trafficking and localization in a heat shock protein-dependent manner.
Introduction
A prolonged QT interval increases the likelihood for ventricular arrhythmias and sudden cardiac death in the general population (1). Although 13 genes, mainly encoding ion channels, have been identified as causing Mendelian forms of long QT syndrome (2–4), mutations in these genes are rare and can, therefore, account for only a small percentage of the overall population risk for sudden cardiac death (5). Because approximately one-third of QT interval variability is heritable (6–8), a number of genome-wide association studies have been performed to identify genes underlying this variation (9–14). For example, two recent genome-wide association studies (10, 11) identified five novel loci associated with the QT interval. These polymorphisms may cumulatively contribute to QT interval variation and, hence, to the net risk for cardiac arrhythmias (5). Importantly, gene variants affecting QT interval duration may also have a detrimental impact through effects to decrease the “repolarization reserve” (15) in the context of sensitivity to drugs, sex hormones, or other environmental factors that prolong the QT interval. A comprehensive understanding of the genetic basis of repolarization will require knowledge of all the relevant genes, their mechanisms of action, and their interactions with each other or with environmental stressors.
One SNP associated with prolongation of the QT interval (approximately 1.7 (10, 11) or 2.9 ms (16)) has been mapped within the gene encoding RNF207 (10, 11). The SNP rs846111 actually lies within the open reading frame of the RNF207 gene and not in its 3′ untranslated region as reported (10, 11). Interestingly, RNF207 contains a RING domain (17), suggesting that the protein may be a ubiquitin ligase. On the basis of these findings and our experience in studying the trafficking of the voltage-gated potassium channels Kv1.5, HERG, and KvLQT1 (18–21), we hypothesized that RNF207 is a candidate for regulation of the QT interval and thus cardiac repolarization. We further hypothesized that RNF207 potentially acts through effects on ion channel synthesis, trafficking, and/or recycling or degradation through proteasomes or lysosomes (22). Therefore, we set out to investigate the possible role of RNF207 in action potential duration (APD)3 regulation in zebrafish and neonatal rabbit cardiomyocytes (NRbCM) and to identify the associated ion channels responsible for these effects. Here we present data that support a role for RNF207 in the regulation of cardiac repolarization, which appears to be highly conserved throughout vertebrate evolution from bony fish to mammals.
EXPERIMENTAL PROCEDURES
DNA
Human RNF207 was cloned from human heart RNA (Agilent Technologies). The HA-tagged open reading frame of RNF207 was cloned into a modified form of pENTR 1A Dual (Invitrogen), which contains a downstream internal ribosomal entry site linked to GFP. This bicistronic expression cassette was then transferred to the vector pAd/CMV/V5-DEST (Invitrogen) using the Gateway cloning system (Invitrogen). HEK293 cells (Invitrogen) were transfected with PacI-digested pAd/CMV/V5-DEST-RNF207-GFP, and adenoviral stocks were prepared according to the instructions of the manufacturer. As a control, we used the “empty” vector pAd/CMV/V5-DEST-GFP that allows the expression of GFP. HA-or FLAG-tagged RNF207 open reading frames were cloned into pcDNA3 (Invitrogen). The deletions of the RING (ΔRING), B-box 1 (ΔB-box), and B-box C-terminal (ΔBBC) domains; the C-terminal region (ΔC); and the SNP-containing RNF207 (G603A) were generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Agilent Technologies). Rabbit RNF207 was cloned from total RNA isolated from wild-type New Zealand White rabbit hearts. To obtain the full sequence of the rabbit RNF207 mRNA, 5′ and 3′ rapid amplification of cDNA ends was performed as described previously (23). The rabbit RNF207 mRNA sequence was deposited in the GenBank database (GenBank accession number JQ754141). pcDNA3-HERG and pcDNA3-KvLQT1 express the human HERG and KvLQT1 channels (20). phERG-mEGFP, described previously (21), encodes an N-terminal fusion of HERG and monomeric enhanced GFP (mEGFP), which contains an A206K mutation resulting in mEGFP. The biophysical properties of phERG-mEGFP resemble that of wild-type HERG (21). The following plasmids were purchased from Addgene: pcDNA5/FRT/TO HIS HSPA1A expressing His-tagged HSP70 (catalog no. 19537) (24), pcDNA5/FRT/TO HIS HSPA8 expressing His-tagged HSC70 (catalog no. 19541) (24), and pcDNA5/FRT/TO HIS DNAJA1 expressing His-tagged DJA1 (catalog no. 19545) (24). The plasmid pEGFP was obtained from Clontech.
Transfections
Human osteosarcoma U-2 OS cells (ATCC) were cultured at 37 °C with 5% CO2 in McCoy's 5A medium (Invitrogen) supplemented with 10% FBS. HEK293 cells (ATCC) and rat H9c2 cardiomyoblasts (ATCC) were cultured in DMEM (Invitrogen). All cell lines were split at ∼50% confluency and only used at low passage numbers (<10). Transient transfections into U-2 OS, HEK293, and H9c2 cells were performed using Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. In initial experiments, we varied the ratio of RNF207 to HERG expression plasmids (1.5:1, 3:1, and 6:1 molar ratios). We noticed a small but significant increase in HERG expression (approximately 20%) at a 3:1 molar ratio in HEK293 cells but chose a 6:1 molar ratio in all subsequent transfections because we generally observed a more pronounced HERG increase (approximately 50%, data not shown). Therefore, expression plasmids for RNF207, its deletions, or HSP70 (HSC70, DJA1) were used at a 6-fold molar excess compared with the HERG (KvLQT1) expression plasmid for all transient transfections. For example, 400 ng of DNA (4 ng of GFP expression plasmid, 32 ng of HERG expression plasmid, 182 ng of RNF207 expression plasmid, and 182 ng of pcDNA3) per well of a 12-well plate were used for transfections.
Preparation of Neonatal Rabbit Ventricular Cardiomyocytes
Cardiomyocytes were isolated from 3- to 5-day-old New Zealand White rabbits in accordance with Institutional Animal Care and Use Committee-approved protocols as described previously (21).
Preparation of Adult Rabbit Left Ventricular Cardiomyocytes
Cardiomyocytes were isolated from the hearts of 3- to 4-month-old New Zealand White rabbits (both sexes) with standard enzymatic techniques in accordance with Institutional Animal Care and Use Committee-approved protocols as published elsewhere (25). The filtered cells were maintained in 45 mm KCl, 65 mm K-glutamate, 3 mm MgSO4, 15 mm KH2PO4, 16 mm taurine, 10 mm HEPES, 0.5 mm EDTA, and 10 mm glucose (pH 7.3) for 1 h. In five subsequent steps, the Ca2+ concentration was increased to 1.8 mm. Cells were centrifuged, resuspended in medium 199 supplemented with 10% FBS and antibiotics, plated on laminin-coated coverglasses, and allowed to attach. After 2 h, the medium was replaced, and adenovirus (multiplicity of infection of 100) was added to the cells. Cells were maintained at 37 °C with 5% CO2, and, 40 h later, cells were used for immunofluorescence or biochemistry.
Immunoblot Analysis
Coimmunoprecipitations (co-IPs) and immunoblot analyses were carried out as in previous studies (20).
Immunofluorescence
Immunofluorescence was carried out as reported previously (21).
Aquaculture
Experiments were performed on zebrafish (Danio rerio) from WT WIK breeding colonies in accordance with Institutional Animal Care and Use Committee-approved protocols. The care and breeding of zebrafish were performed as described previously at 28.5 °C with standard medium (E3) (26).
Morpholino Knockdown
Morpholinos were designed to target the start site and the splice acceptor of exon 3 of RNF207. The specific morpholino sequences were as follows: 5′-TCGTATTGCTCTTGGCACAAGTGGC-3′ (RNF207ATG) and 5′-TTGCATCCACATCCTAGAGAGATTA-3′ (RNF207Ex3SA).
Morpholinos (Gene Tools LLC) were resuspended in 1× Danieu's solution, and 125–500 μm (2-nl drop size, 1.9–7.6 ng) of morpholino was injected into fertilized eggs from TuAB fish at the single cell stage. The effectiveness of splice targeting morpholinos was assessed by quantitative RT-PCR (data not shown). As an additional measure of morpholino specificity, we tested all morpholinos used in the setting of p53 knockdown to verify that the phenotypes seen in wild-type embryos were not secondary to nonspecific activation of p53 (27).
Optical Mapping: Zebrafish
Hearts were isolated from embryos 48 and 72 h post-fertilization (hpf) and stained with the transmembrane potential-sensitive dye di-8-ANEPPS (Invitrogen) for measurement of action potential propagation. Resulting fluorescence intensities were recorded with a high-speed charge-coupled device (CCD) camera (RedShirtImaging), and images were analyzed using software implemented in MATLAB (26). For all comparisons, regions of interest were determined and compared between morphant embryos and controls from at least two separate mate pairings using two-sided Student's t test.
Digital Imaging: Neonatal Rabbit Ventricular Cardiomyocytes
Transduced NRbCMs were stimulated using a field stimulation of 20 V/cm (MyoPacer EP cell stimulator, IonOptix LLC) and 1.0-Hz cycle length with two platinum stimulation electrodes, placed on the temperature-regulated chamber (35 °C) of a Nikon Ti microscope (Nikon Instruments, ×20 magnification), and perfused at 0.2 ml/min using a syringe pump (Aladdin-100, World Precision Instruments). Action potentials were recorded in RH237 (Invitrogen, λexcitation = 520, λemission > 610 nm) loaded cardiomyocytes using an Evolve 128 EM-CCD instrument (Photometrics, 2 × 2 binning resulting in a pixel resolution of 2.4 × 2.4 μm at 979 f/s).
Electrophysiological Recording
U-2 OS and HEK293 cells were transfected with GFP, HERG, and RNF207 expression plasmids (or pcDNA3 as control) 48 h before recording. Whole cell patch clamp recordings of HERG currents were performed with an Axopatch-200B amplifier (Axon Instruments) at room temperature (21–23 °C). Data were filtered at 1 kHz and analyzed using Origin (Origin Lab). The pipette solution contained 50 mm KCl, 65 mm K-glutamate, 5 mm MgCl2, 5 mm EGTA, 10 mm HEPES, 5 mm glucose, 5 mm K2-ATP, and 0.25 mm Na2-GTP (pH 7.2). The pipette resistance was 2–4 MΩ. The bath solution contained 140 mm NaCl, 5.4 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 0.33 mm NaH2PO4, 7.5 mm glucose, and 5 mm HEPES (pH 7.4). Two-way analysis of variance followed by Tukey's post hoc test was used to test for differences in the I-V relationship for normalized peak tail currents.
RESULTS
Cloning of Cardiac RNF207
We cloned RNF207 from human and rabbit hearts (Fig. 1). The overall structure of RNF207 is reminiscent of tripartite motif-containing (TRIM) proteins (28–30). These proteins are characterized by a zinc-binding RING domain, one or two zinc-binding B-box domains, and a coiled coil domain in the amino-terminal region. Most TRIM proteins also have ubiquitin ligase activity. Although TRIM proteins with one B-box always have a type 2 B-box (histidine is the second zinc-coordinating residue), the single B-box in RNF207 is of type 1 (cysteine is the second zinc-coordinating residue, Fig. 1). The SNP reported to be associated with the QT interval lies in the C-terminal non-homologous region of human RNF207, resulting in a glycine to alanine change (G603A). Interestingly, zebrafish have two RNF207 genes: rnf207b, which encodes a 634-amino acid protein highly homologous to human, rabbit, and mouse RNF207 (Fig. 1), and rnf207a, which encodes a 199-amino acid protein with similarity to the C-terminal homologous region of RNF207 molecules from other species (Fig. 1).
FIGURE 1.
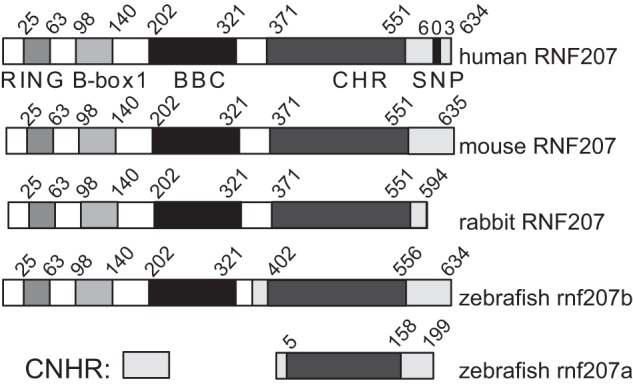
Structure of RNF207. Structural comparison of cardiac RNF207 from human, mouse, and rabbit with zebrafish RNF207. The conserved RING (C-x2-C-x11-C-x2-H-x2-C-x2-C-x11-C-x2-C), B-box 1 (C-x2-C-x14-C-x2-C-x4-C-x2-C-x4-H-x7-H) and B-box C-terminal (BBC) domains as well as a C-terminal homologous region (CHR) that contains no known protein domain, are depicted. No sequence homology is found in the C-terminal non-homologous region (CNHR). Also shown is the SNP (G603A) in human RNF207, which is associated with QT interval prolongation (10, 11, 16).
Action Potential Prolongation and Conduction Velocity Slowing in Zebrafish RNF207 Morphants
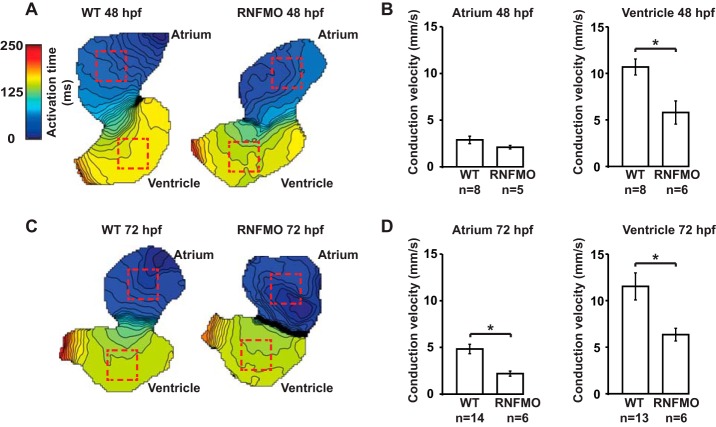
To explore the effects of RNF207 on repolarization in vivo, we designed two morpholinos to the zebrafish ortholog rnf207b (Fig. 1). One targets the ATG of the dominant isoform, and a second targets the splice acceptor site of exon 3 (shared by all isoforms of the transcript). In initial dose-ranging studies, we did not see any evidence of systemic toxicity but did note abnormalities of contractility and looping in the developing zebrafish heart at higher doses of morpholino (data not shown). Electrophysiology studies of the atrial and ventricular myocardium revealed a significant prolongation of APD (Fig. 2, A–D) 48 h hpf (mean ventricular APD80 ± S.E.: morphants, 283 ± 10 ms versus WT 247 ± 9 ms, p < 0.05; atrium, morphants 136 ± 5 ms versus WT 120 ± 4 ms, p < 0.05). Knockdown of RNF207 caused significant slowing of conduction in the ventricle but not in the atrium at 48 hpf (Fig. 3, A and B) and in both chambers at 72 hpf (Fig. 3, C and D). The reduced wavefront propagation velocity is depicted by the crowding of the isochrones in Fig. 3, A and C, with local conduction velocity estimates shown in Fig. 3, B and D. By 120 hpf, when the morpholino effect on morphology is no longer evident, there is persistent APD prolongation that, when pronounced, can lead to 2:1 atrioventricular block in the morphant embryos (data not shown). These data suggest that the emergence of mature physiologic repolarization at around 43 hpf in the zebrafish (13, 31), which is associated with the appearance of IKr at the membrane (32, 33), does not occur with knockdown of RNF207. Because a 2:1 atrioventricular block is observed with ZERG (zebrafish ether-a-go-go related gene) mutant or morphant zebrafish larvae and with an IKr-blocking drug in WT zebrafish larvae (34, 35), we hypothesize that the main effect of RNF207 knockdown is through inhibiting ZERG transport to the membrane.
FIGURE 2.
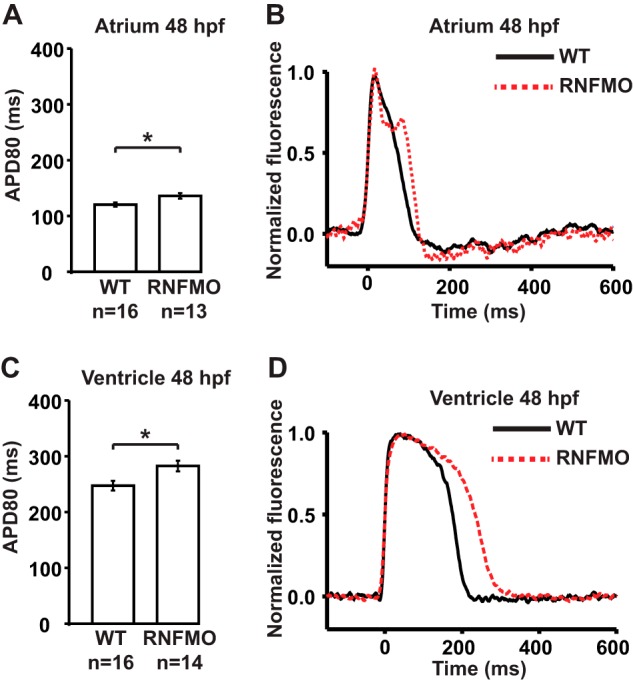
Action potential prolongation in zebrafish RNF207 morphants. Hearts were stained with di-8-ANEPPS. They were transferred into a perfusion chamber with integrated field stimulation electrodes that contained 15 μm blebbistatin to prevent motion artifacts. The chamber was mounted on an inverted microscope (Nikon TE-2000U). Samples were illuminated (525/50 nm excitation filter), and fluorescence images were acquired (685/80 nm emission filter) at 2 kHz by a charge-coupled device camera. Action potentials were acquired during field stimulation at 80 bpm, and action potential durations (APD80) were measured from the fluorescence data using established algorithms in MATLAB. A, mean atrial APD80 from WT and RNF207 morphants' (RNFMO) hearts measured in the atrial regions of interest (ROIs) shown in Fig. 3. B, representative action potential traces from single sites in the atrium of WT (black) and RNFMO hearts (red). C, mean ventricular APD80 from WT and RNFMO hearts measured in the ventricular ROIs shown in Fig. 3. D, representative ventricular action potential traces from single sites in the ventricle of WT (black) and RNFMO hearts (red). Data are mean values ± S.E. *, p < 0.01.
FIGURE 3.
Conduction velocity slowing in zebrafish RNF207 morphants. Hearts were stained with di-8-ANEPPS and motion-arrested. Conduction velocity was estimated in unpaced hearts during sinus rhythm before APD80 was measured during field pacing using established algorithms in MATLAB. A, isochronal maps of WT (left panel) and RNF207 morphant (right panel) hearts at 48 hpf. Each line represents the action potential wavefront position at 5-ms intervals. The color code depicts the timing of electrical activation (blue areas activated before red areas). Red squares represent 35 × 35 μm ROIs located in the middle of the atrial and ventricular chambers. APD80s and conduction velocities were averaged in the ROIs. B, mean estimated atrial (left panel) and ventricular (right panel) conduction velocities for 48 hpf WT and RNFMO hearts. C, isochronal maps of WT (left panel) and RNFMO (right panel) hearts at 72 hpf with ROIs. D, mean estimated atrial (left panel) and ventricular (right panel) conduction velocities for WT and RNFMO hearts. The data shown are mean values ± S.E. *, p < 0.01.
Action Potential Shortening of Overexpressed RNF207 in Neonatal Rabbit Ventricular Cardiomyocytes
Because IKr is a major repolarizing current in NRbCMs (21), we next wanted to test the effect of overexpression of wild-type RNF207 (containing the major allele) and its variant-containing version (RNF207-SNP, containing the minor allele) in these cells. For this purpose, adenovirus-expressing HA-tagged RNF207, the variant-containing RNF207, or GFP (multiplicity of infection of 1) was incubated with NRbCMs for 48 h. Digital imaging of transduced cells demonstrated that RNF207 overexpression significantly shortened APD by ∼15% compared with the control (control, 495 ± 61 ms; RNF207, 418 ± 39 ms) (Fig. 4, A and B), implying a possible role for RNF207 in cardiac repolarization and putatively reflecting additional rabbit ERG (RERG) polypeptides in the membrane. Indeed, we observed a trend toward a 30% increase in RERG protein (135- and 155-kDa bands) upon RNF207 overexpression that did not reach statistical significance, possibly because of variability in RERG expression in these cells in cultures (p < 0.1, data not shown). The observed APD shortening was also seen with the variant form of RNF207 (RNF207-SNP, 409 ± 81 ms) (Fig. 4, A and B). Western blotting (Fig. 4C) and immunofluorescence experiments (Fig. 4D) did not reveal any differences in the expression or localization of RNF207 and RNF207-SNP. These data confirm that the minor allelic variant (G603A) behaves like the wild-type major allele RNF207 in this system and do not support any direct effect of the polymorphism on the function of RNF207. Furthermore, we did not observe any differences in the stability of wild-type and SNP-containing RNF207 polypeptides in vitro (data not shown).
FIGURE 4.
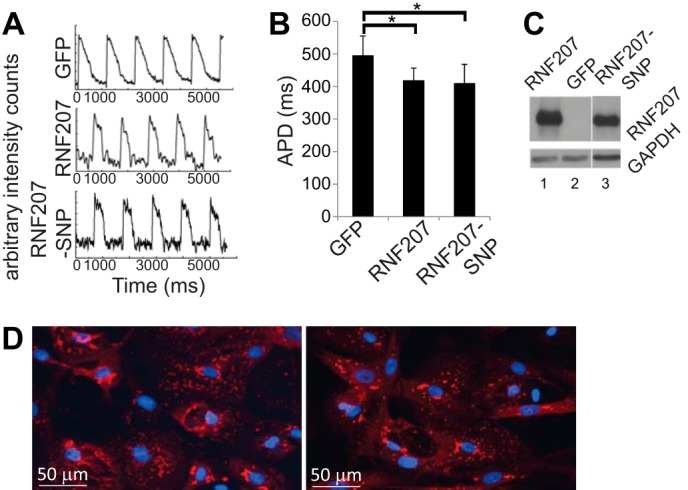
Action potential shortening by overexpressed RNF207 in neonatal rabbit cardiomyocytes. Cells were infected with adenovirus containing Ad-GFP, Ad-RNF207, and Ad-RNF207-SNP. A, APD signals. B, respective mean ± S.D. APDs from two independent experiments (n = 14, 14, and 13 for GFP, RNF207, and RNF207-SNP, respectively). *, p < 0.01 for GFP versus RNF207 and GFP versus RNF207-SNP. C, Western blot data from cell extracts to measure expression of RNF207 and RNF207-SNP. D, immunostaining of NRbCMs overexpressing HA-tagged RNF207 (left panel) or RNF207-SNP (right panel) using HA antiserum and DAPI for nucleus staining.
RNF207 Affects HERG Expression and Function in Vitro
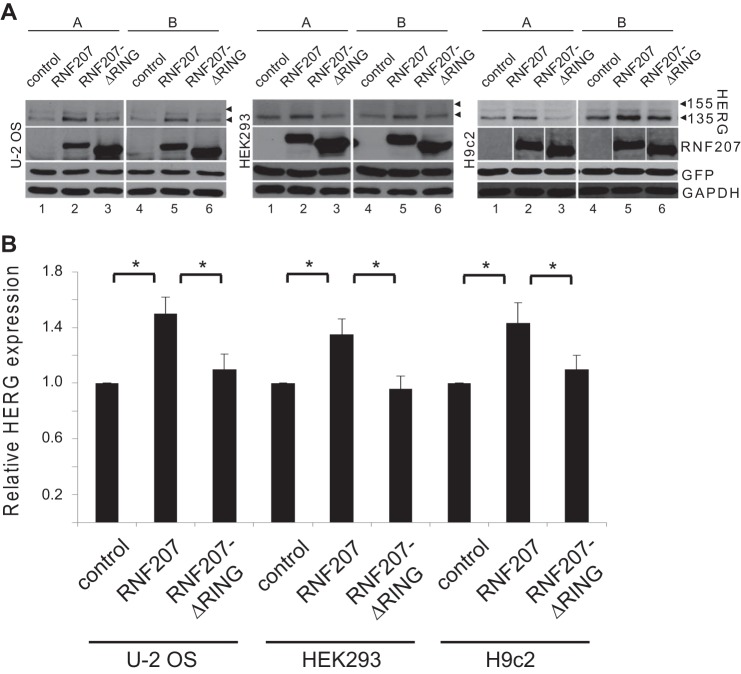
Our data obtained from zebrafish and NRbCMs imply a possible role for RNF207 in the regulation of HERG expression. Therefore, we directly tested the effect of RNF207 on HERG expression in vitro. Transiently cotransfected RNF207 significantly increased total HERG protein levels in U-2 OS, HEK293, and rat cardiomyoblast H9c2 cells by ∼30–50% (Fig. 5, A and B). These effects of RNF207 on HERG are dependent on a functional RING domain because overexpression of RNF207 lacking its RING domain (Fig. 5, A and B, RNF207-ΔRING) or with mutation of the first zinc-coordinating cysteine of the RING domain to arginine (RNF207-C25R, data not shown) had no significant effect on HERG protein levels. Further, biotinylation of surface protein from HEK293 cells indicated that RNF207 expression led to an approximately 30% increase in HERG protein on the membrane (Fig. 6, A and B). The specificity of RNF207 for HERG was further supported by the finding that levels of cotransfected KvLQT1 were not affected by wild-type RNF207 (Fig. 7, A and B). Coimmunoprecipitation experiments in U-2 OS and HEK293 cells (Fig. 8, A and B) demonstrated an interaction between RNF207 and the core-glycosylated form of HERG (135 kDa) but not the fully glycosylated form of HERG (155 kDa), which is mainly found on the membrane. This interaction implies that RNF207 is probably located in the ER and/or possibly cis-Golgi apparatus. Immunofluorescence data further support these findings by showing colocalization of overexpressed HA-tagged RNF207 and HERG-mEGFP in structures surrounding the nuclei of U-2 OS cells (Fig. 9A). We next wanted to localize endogenous HERG and RNF207 in adult rabbit cardiomyocytes. However, because of the lack of specificity of commercially available antisera, we had to overexpress HA-tagged RNF207 and FLAG-tagged HERG in these cells. We noticed colocalization of both proteins in perinuclear areas as well as in other parts of the cell (Fig. 9B).
FIGURE 5.
Effect of RNF207 on total HERG levels. A, HERG expression in U-2 OS, HEK293, and H9c2 cells cotransfected with expression plasmids for HERG, GFP (lanes 1–6), pcDNA3 (control, lanes 1 and 4), and expression plasmids for RNF207 (lanes 2 and 5) or RNF207-ΔRING (lanes 3 and 6). For each condition, depicted immunoblots represent two (A and B) of three technical replicates from one of three independent experiments. B, respective relative expression levels (mean ± S.D.) of HERG normalized to GAPDH expression from three independent experiments performed in triplicate. *, p < 0.05.
FIGURE 6.
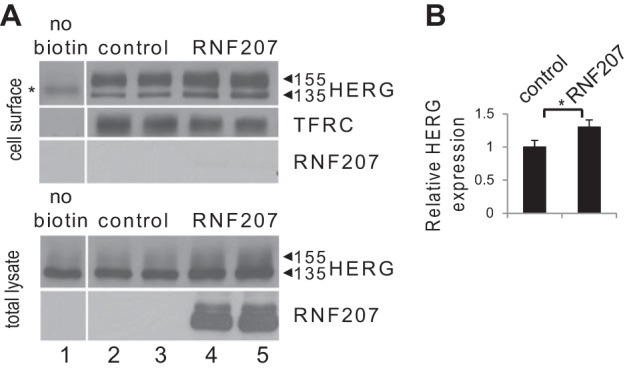
Effect of RNF207 on surface HERG levels. A, top panel, representative immunoblot of cell-surface HERG and transferrin receptor (TFRC). HEK293 cells were transfected with HERG (lanes 1–5), pcDNA3 as a control (lanes 1–3), and RNF207 plasmids (lanes 4 and 5). Cell surface proteins were biotinylated using sulfo-NHS-SS-biotin, purified with neutravidin beads from total cell lysates, subjected to SDS-PAGE, and blotted onto a nitrocellulose membrane. HERG and transferrin receptor levels were measured using specific antibodies. Cytosolic HA-tagged RNF207 was not detected (lanes 4 and 5), confirming that biotinylation was specific for cell membrane proteins. No cell surface HERG (the asterisk indicates a nonspecific band) or transferrin receptor was detected from HEK293 cells that were not incubated with biotin (lane 1). Bottom panel, representative immunoblot of whole-cell lysates from transfected HEK293 cells following biotinylation. HERG and HA-tagged RNF207 protein were detected using specific antibodies. B, respective relative cell surface expression levels (mean ± S.D.) of HERG normalized to transferrin receptor expression from two independent experiments performed in duplicate. *, p < 0.05.
FIGURE 7.
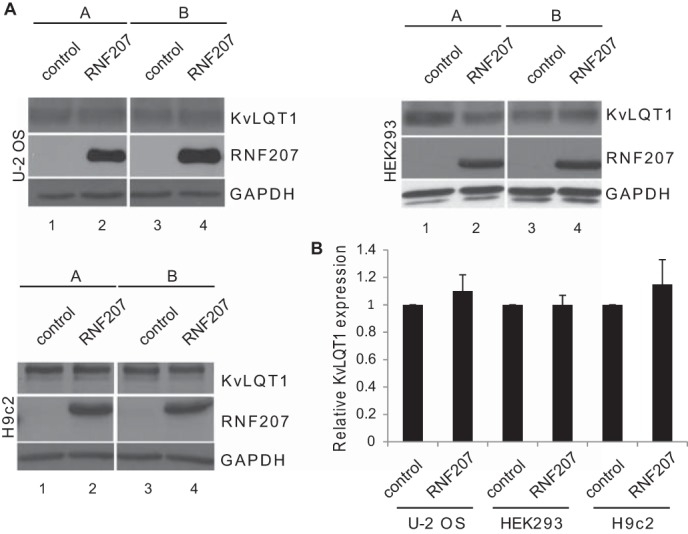
Effect of RNF207 on total KvLQT1 levels. A, KvLQT1 expression in U-2 OS (top left panel), HEK293 (top right panel), and H9c2 cells (bottom left panel) cotransfected with expression plasmids for KvLQT1 (lanes 1–4), pcDNA3 (control, lanes 1 and 3), and expression plasmid for RNF207 (lanes 2 and 4). For each condition, the depicted immunoblots represent two (A and B) of three technical replicates from one of three independent experiments. B, respective relative expression levels (mean ± S.D.) of KvLQT1 normalized to GAPDH expression from three independent experiments performed in triplicates. *, p < 0.05.
FIGURE 8.
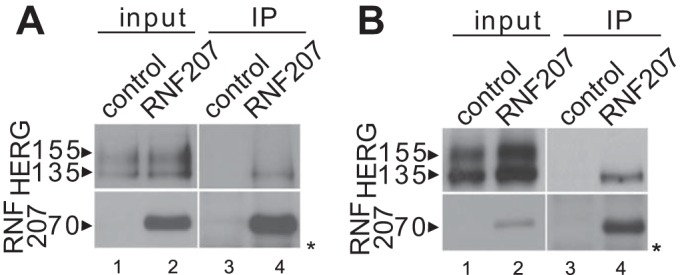
Interaction of RNF207 and HERG. A, U-2 OS cells were transfected with HERG and FLAG-tagged RNF207 expression plasmids (1:6 molar ratio) or pcDNA3 as a control. IP was performed on U-2 OS cell extracts with anti-FLAG antiserum to pull down RNF207-interacting proteins. The immunoblot against HERG protein reveals an interaction between pulled-down RNF207 and core-glycosylated, 135-kDa HERG. Input samples were probed against FLAG to detect RNF207 and HERG (the asterisk indicates nonspecific bands). B, Western blot analysis of a similar IP experiment using HEK293 cells. HEK293 cells were transfected with HERG and FLAG-tagged RNF207 expression plasmids (1:6 molar ratio) or pcDNA3 as a control.
FIGURE 9.
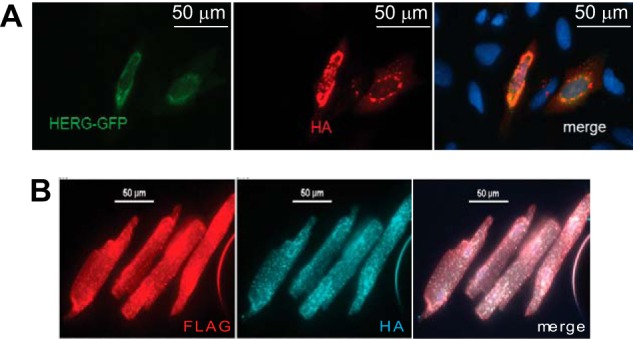
Colocalization of RNF207 and HERG. A, colocalization of mEGFP-tagged HERG and HA-tagged RNF207 in U-2 OS cells cotransfected with expression plasmids for HERG-mEGFP and HA-RNF207 were fixed, immunostained with antibodies against HA, and analyzed under a microscope. Nuclei were stained with DAPI (blue). B, colocalization of FLAG-tagged HERG and HA-tagged RNF207 in adult rabbit cardiomyocytes transduced with adenoviral expression vectors for FLAG-HERG and HA-RNF207. Cells were fixed, immunostained with antibodies against FLAG and HA, and analyzed under a microscope.
We next tested whether RNF207 affects HERG function by measuring IKr. To this end, we cotransfected U-2 OS cells with expression plasmids for GFP, HERG, and RNF207 (or control plasmid) and studied IKr in these cells. RNF207-overexpressing U-2 OS cells also exhibited a significantly higher HERG tail current density than control cells (e.g. peak tail current at 40 mV, 40.4 ± 5.2 pA/picofarad for RNF207 and 29.3 ± 4.4 pA/picofarad for control; Fig. 10, A and B), which is consistent with an approximate 40% increase in HERG expression on the membrane, as determined by surface biotinylation experiments (Fig. 10, C and D). To estimate the V1/2 of activation for control and RNF207-coexpressing cells, we averaged Boltzmann fits to the I-V data generated. The results show no significant difference in V1/2 of activation (control, V1/2 = −12.0 ± 1.5 mV (n = 19); RNF207, V1/2 = −10.2 ± 1.4 mV (n = 23); p = 0.41). Similarly, no differences in slopes were detected (control, slope factor = 7.8 ± 0.2 mV (n = 19); RNF207, slope factor = 7.4 ± 0.2 mV (n = 23); p = 0.12). These data indicate that voltage-dependent activation is not significantly affected by RNF207, suggesting that the observed increase in membrane expression level is the underlying reason for the increase in current density. We also cotransfected U-2 OS cells with expression plasmids for GFP, HERG, and the RNF207-ΔRING mutant (or control plasmid) and studied IKr in these cells. Cotransfected U-2 OS cells showed unaltered HERG tail currents compared with control cells (87 ± 5% of control cells (n = 24) for the RNF207-ΔRING mutant (n = 23), p > 0.05 (data not shown)). This finding underscores the importance of a functional RING domain for an RNF207-dependent effect on IKr. Not surprisingly, experiments in transiently cotransfected HEK293 cells showed a similar increase in HERG current in RNF207-overexpressing cells compared with control cells (data not shown). We also noticed that the steady-state levels of RNF207 transcripts in cultured cardiomyocytes and the cell lines used in these experiments were very low (barely detectable), therefore preventing us from knocking down endogenous RNF207 (data not shown).
FIGURE 10.
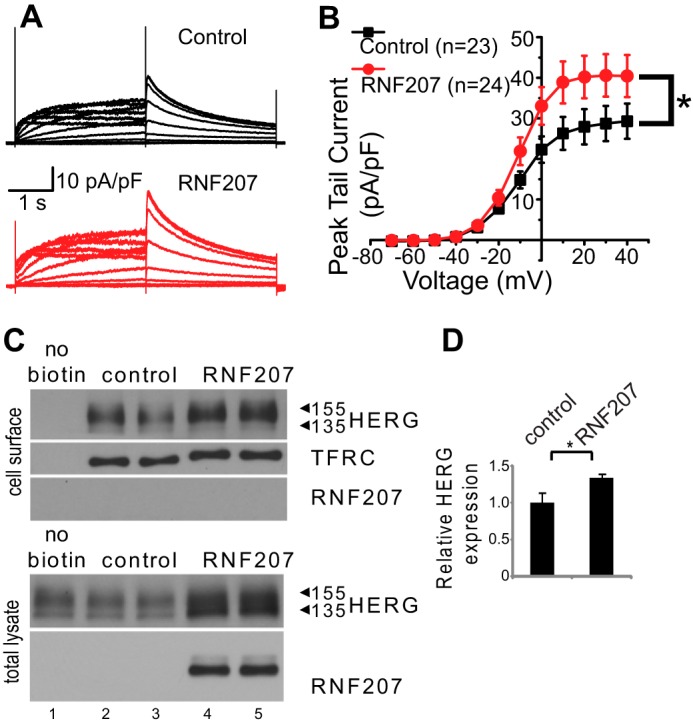
Effect of RNF207 on HERG currents. U-2 OS cells were transfected with GFP, HERG, and RNF207 expression plasmids or control. A, representative original recordings of HERG currents from control (black) and RNF207-expressing cells (red). The membrane potential was held at −80 mV. Voltage steps between −70 and 40 mV were used to activate HERG channels before returning to −60 mV to record tail currents. B, peak tail current amplitudes at −60 mV were normalized to cell capacitance as a measurement of current density. Average results were plotted as a function of preceding depolarizing potentials (mean ± S.E.). The IKr peak tail current density was increased significantly by RNF207 expression. *, p < 0.05 by two-way analysis of variance. C, top panel, representative immunoblot of cell surface HERG and transferrin receptor (TFRC). U-2 OS cells were transfected with HERG (lanes 1–5), pcDNA3 as a control (lanes 1–3), and RNF207 plasmids (lanes 4 and 5). Bottom panel, representative immunoblot of whole-cell lysates from transfected U-2 OS cells following biotinylation. D, respective relative cell surface expression levels (mean ± S.D.) of HERG normalized to transferrin receptor expression from two independent experiments performed in duplicate. *, p < 0.05.
RNF207 and HSP70 Additively Increase HERG Expression in Vitro
Because several proteins are involved in the quality control, folding, and export of HERG, including HSP90, FKBP38, HSP70, HSC70, and HSP40 chaperones and CHIP (36, 37), we looked into the possible role of RNF207 in these steps. We performed co-IP (Fig. 11A) and observed strong interactions between RNF207 and the HSP40 type 1 chaperone DJA1, HSP70, and HSC70. The interaction between RNF207 and HSP70 is dependent on the C terminus of RNF207 (Fig. 11B, lanes 9 and 10) because we detected almost no appreciable interaction between the heat shock protein and RNF207-ΔC, which lacks the C-terminal region (Fig. 1). In contrast, the deletion of RING, B-box1, or BBC domains of RNF207 did not abolish the interaction with HSP70 (Fig. 11B, lanes 3–8). We next tested whether the C-terminal deletion of RNF207 affected HERG function by measuring IKr. We cotransfected HEK293 cells with expression plasmids for GFP, HERG, and RNF207-ΔC (or a control plasmid) and measured IKr. RNF207-ΔC-overexpressing cells exhibited a significantly lower HERG tail current density than control cells (e.g. peak tail current at 40 mV, 17.5 ± 1.3 pA/picofarad for RNF207-ΔC and 28.4 ± 1.9 pA/picofarad for the control; Fig. 11, C and D; p < 0.05), implying that the C-terminal deletion of RNF207 acts as a dominant negative mutant. We also estimated the V1/2 of activation and slope factors for control and RNF207-ΔC-coexpressing cells but noticed no significant differences (data not shown).
FIGURE 11.
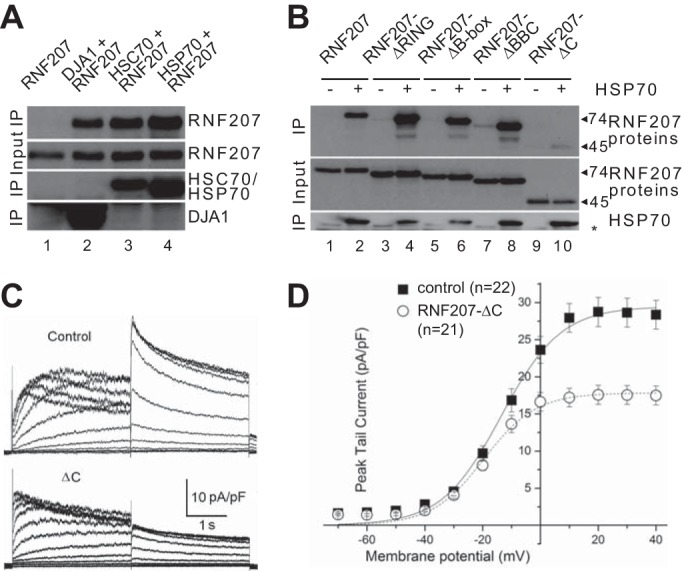
Dominant negative effect of RNF207-ΔC on HERG currents. A, HEK293 cells were transfected with expression plasmids for RNF207 and His-tagged DJA1, HSC70, HSP70, or control. Coimmunoprecipitation was performed with anti-His antiserum. The immunoblot against RNF207 reveals (top panel) an interaction between RNF207 and the chaperones (lanes 2–4). B, anti-His co-IP between RNF207, its deletions, and His-tagged HSP70. C, HEK293 cells were transfected with GFP, HERG, and RNF207-ΔC expression plasmids or control. Shown are representative original recordings of HERG currents from control and RNF207-ΔC-expressing cells. The membrane potential was held at −80 mV. Voltage steps between −70 and 40 mV were used to activate HERG channels before returning to −60 mV to record tail currents. D, peak tail current amplitudes at −60 mV were normalized to cell capacitance as a measurement of current density. Average results were plotted as a function of preceding depolarizing potentials (mean ± S.E.). The IKr peak tail current density was decreased significantly by RNF207-ΔC expression. *, p < 0.05.
The aforementioned RNF207/HSP70 interaction (Fig. 11, A and B) may explain the additive effect of these proteins to increase the expression of HERG in cotransfected HEK293 cells (RNF207 versus control, +33%; HSP70 versus control, +89%; RNF207 + HSP70 versus control, +131%; p < 0.05 each; Fig. 12, A and B). Similar total HERG expression levels were observed in H9c2 cardiomyoblasts (RNF207 versus control, +45%; HSP70 versus control, +81%; RNF207+HSP70 versus control, +154%; p < 0.05 each; Fig. 12, C and D). Furthermore, in each case, RNF207-ΔC expression resulted in down-regulation of total HERG protein compared with controls in HEK293 and H9c2 cells (RNF207-ΔC versus control, −27% and −18%; RNF207-ΔC + HSP70 versus HSP70, −25% and −34%; p < 0.05 each; Fig. 12, A–D), which corroborates the dominant negative effect of the C-terminal deletion of RNF207 on IKr.
FIGURE 12.
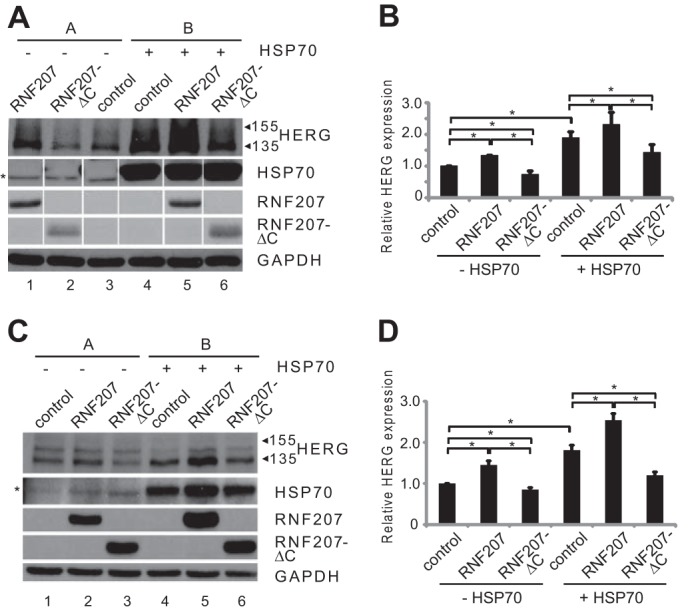
Additive increase of HERG expression by RNF207 and HSP70. A, representative Western blot analysis with protein extracts from HEK293 cells that were cotransfected with pcDNA3 (lanes 1–4) and HERG (lanes 1–6), RNF207 (lanes 1 and 5), RNF207-ΔC (lanes 2 and 6), or HSP70 (lanes 4–6) plasmids. The asterisk indicates nonspecific bands. For each condition, the depicted immunoblots represent two (A and B) of three technical replicates from one of three independent experiments. B, respective normalized HERG expression levels (mean ± S.D.) from three independent experiments performed in triplicates. C, representative Western blot analysis with protein extracts from H9c2 cells that were cotransfected with pcDNA3 (lanes 1–4) and expression vectors for HERG (lanes 1–6), RNF207 (lanes 2 and 5), RNF207-ΔC (lanes 3 and 6), or HSP70 (lanes 4–6) (the asterisk indicates nonspecific bands). D, respective normalized HERG expression levels (mean ± S.D.) from three independent experiments performed in triplicate.
DISCUSSION
Our data provide the first mechanistic insight for the observed association between RNF207 and QT interval variation reported in the aforementioned genome-wide association studies (10, 11, 16). Not surprisingly, RNF207 appears to regulate expression of the HERG potassium channel, a major player in cardiac repolarization. On the basis of the reported effects of RNF207 on APD in vivo (Fig. 2, A–D) and in vitro (Fig. 4, A and B), we propose to add genetic variants of RNF207 to the growing list of genetic risk factors for sporadic or drug-induced arrhythmias. Identification and expansion of molecular risk factors could help to identify “vulnerable” patients and aid in the development of new strategies targeting individuals with a prolonged QT interval.
The Ensembl gene tree (38) for RNF207 was used to identify orthologs in five vertebrate classes (data not shown): mammals, birds (e.g. chicken), reptiles (anole lizard), amphibian (Xenopus), and bony fish (zebrafish). Although we were not able to identify any putative RNF207 orthologs in jawless and cartilaginous fish using BLAST searches in GenBank (data not shown), we currently cannot completely rule out the existence of RNF207 orthologs in these lower vertebrate classes. The high degree of sequence conservation among known RNF207 orthologs (Fig. 1 and data not shown) implies a conservation of function during vertebrate evolution from bony fish to mammals. Here we provide strong evidence for a role of RNF207 in the regulation of APD both in a teleost, i.e. zebrafish (Fig. 2), and in cardiomyocytes derived from neonatal rabbit hearts (Fig. 4, A and B). Therefore, these data, for the first time, provide a potential mechanism for the observed association between RNF207 and QT interval variation originally suggested by the two human genetic studies (10, 11). Because we occasionally observed a 2:1 atrioventricular block in RNF207 zebrafish morphants, reminiscent of the phenotype of zebrafish ZERG mutants or morphants or those treated with an IKr-blocking drug (34, 35), we focused on the possible effect of RNF207 on the ERG potassium channel. Our biochemical data indeed indicate that RNF207 is a regulator of HERG trafficking in vitro (Fig. 5).
We also performed a thorough analysis of microarray data available from the NCBI public functional genomics data repository (GEO, data not shown, (39)). Interestingly, RNF207 levels were significantly down-regulated in hearts of mice with isoproterenol-induced cardiomyopathy (GEO accession nos. GDS3596 and GDS3684 (40, 41)), which serves as a model for human heart failure. The down-regulation of RNF207 could be recapitulated in vitro when rat ventricular CMs were treated with either the pyridine activator of myocyte hypertrophy or phenylephrine for 48 h (GEO accession no. GDS902 (42)). We currently speculate that loss of function of RNF207 in heart failure could contribute to a reduced repolarization reserve (reduction of IKr) and, thereby, an arrhythmogenic phenotype (early and/or delayed afterdepolarizations).
Interestingly, we also noticed a significant slowing of conduction velocity in the heart of embryonic zebrafish RNF207 morphants (Fig. 3). Conduction velocity depends on electrical coupling, excitability, and tissue architecture (43). Therefore, possible downstream targets of RNF207 could well be the gap junction forming connexin 43 or the sodium channel SCN5A, both of which are crucial to cardiac excitability. However, it remains to be shown whether RNF207-dependent mechanisms for SCN5A or connexin 43 forward trafficking exist and whether they are similar to what we report for HERG. Because of this profound effect of the RNF207 knockdown on conduction velocity in zebrafish, we currently speculate that genetic variants of RNF207 may be associated with the QRS interval, which reflects depolarization and conduction time. Although there has been a recent genome-wide association meta-analysis (44) identifying 22 loci associated with QRS duration (p < 5 × 10−8), RNF207 variants may have been overlooked because of the statistical threshold required in such studies. It will be interesting to see whether any of the common RNF207 variants are actually associated with variations in the QRS interval by revisiting the data from this meta-analysis.
Because the aforementioned genome-wide association studies (10, 11, 16) linked the QT interval to a SNP found within the open reading frame of RNF207, we decided to characterize the coding SNP that causes the amino acid substitution G603A first. Therefore, we looked for any differences between RNF207-SNP and wild-type RNF207 with respect to function (Fig. 4, A and B) or effects on HERG expression and stability. We first used the Sorting Tolerant from Intolerant (SIFT) algorithm (45) (on the basis of evolutionary conservation) to predict whether the coding SNP would have any impact on the function of RNF207. A SIFT analysis predicted that the G603A substitution is tolerated (data not shown). A similar result was obtained using the Protein Variation Effect Analyzer (PROVEAN) tool (46) (data not shown). These predictions were supported by our experimental findings, which did not identify any significant differences between variant and wild-type RNF207 function. As the SNP is found in the C-terminal non-homologous region of RNF207 and because the glycine-alanine mutation is highly conservative because of similar physiochemical properties of both amino acids, these findings are not surprising. It remains possible that the coding SNP may actually affect pretranslational steps at the DNA or RNA level, which could not be detected in our experimental system, but, more likely, that other genetic variants that are in linkage disequilibrium with the coding SNP are the actual cause for a putative loss-of-function phenotype associated with prolongation of the QT interval (10, 11, 16, 47, 48).
After demonstrating that the function of wild-type RNF207 is similar to the SNP variant G603A, we next investigated domains or motifs in RNF207 that are critical for the reported effects on HERG expression. The presence of a RING domain suggests that RNF207 may be a RING domain ubiquitin ligase (17). We demonstrated a requirement for the RING domain for the effects of RNF207 on total HERG expression (Fig. 5) and IKr (see above) in vitro and also observed that the intact RING domain is crucial for RNF207 (auto-) ubiquitination in U-2 OS cells (data not shown). To study any possible ubiquitination activity conferred by RNF207, we first tried to identify RNF207-interacting ubiquitin-conjugating enzymes (49). These interacting ubiquitin-conjugating enzymes generally confer the type of ubiquitin linkage on the target molecule, which, in turn, determines its cellular fate and may contribute to ubiquitin ligase substrate specificity (49). Using a yeast two-hybrid screen with 30 ubiquitin-conjugating enzymes (50), we were not able to identify an interaction between RNF207 and a bona fide ubiquitin-conjugating enzyme (data not shown). These data imply that RNF207, in common with other RING proteins such as BARD1 and MDMX (51, 52), has no intrinsic ubiquitin ligase activity. However, such proteins are able to oligomerize with other RING domain proteins (e.g. BRCA1 and MDM2) and, on formation of a heterodimer, dramatically increase the E3 activity of their binding partners (17, 51, 52). We propose that RNF207 may serve a similar function, resulting in increased ubiquitination of RNF207 target molecules, such as HERG, or proteins involved in HERG maturation at the ER. Importantly, coimmunoprecipitation experiments ruled out any RNF207-dependent changes in the ubiquitination of total HERG protein in vitro (data not shown).
It is currently believed that large complexes consisting of the key chaperones HSC70/HSP70 and HSP90, assisted by cochaperones such as HDJ-1/2 (HSP40), HOP, calnexin, calreticulin, and FKBP38 (36, 37, 53–55), are involved in proper HERG folding, assembly, and ER export as well as in the retention of trafficking-deficient or misfolded HERG in the ER. The RNF207-dependent HERG expression (Fig. 5), as well as RNF207 interaction (Fig. 8) and colocalization (Fig. 9) with core-glycosylated HERG, when combined with our observation of interaction between RNF207 and HSP70 (Figs. 11 and 12), suggest that RNF207 may interact with cytosolic chaperones such as HSP70 and HSP40 on the ER to assist synthesis, folding, or ER export of HERG (Fig. 13). We currently speculate that RNF207 may be involved in one of these chaperone-mediated steps, e.g. that it may act in a cochaperone-like manner and increase either nucleotide exchange activity on HSP70 (56) or diminish ATPase activity on HSP90 (57) to enhance respective chaperone activities. Alternatively, one could envisage that RNF207 negatively interferes with the function or stability (e.g. through ubiquitination) of the ubiquitin ligase CHIP (58). CHIP is responsible for the ubiquitination of misfolded chaperone substrates such as HERG (59) or CFTR (60), which targets them for proteasomal degradation. Because CHIP competes with HOP for binding to HSC70/HSP70 and HSP90 and because the outcome of this competition decides whether an HSC70/HSP70-HSP90 client protein will be degraded or exported from the ER (61), a possible RNF207-dependent titration of CHIP would allow more HERG protein to exit the ER. We also hypothesize that the aforementioned dominant-negative RNF207-ΔC may repress HERG synthesis/folding/ER exit and probably acts as a “sponge” to absorb/quench the endogenous protein(s) required to assist RNF207-dependent HERG maturation.
FIGURE 13.
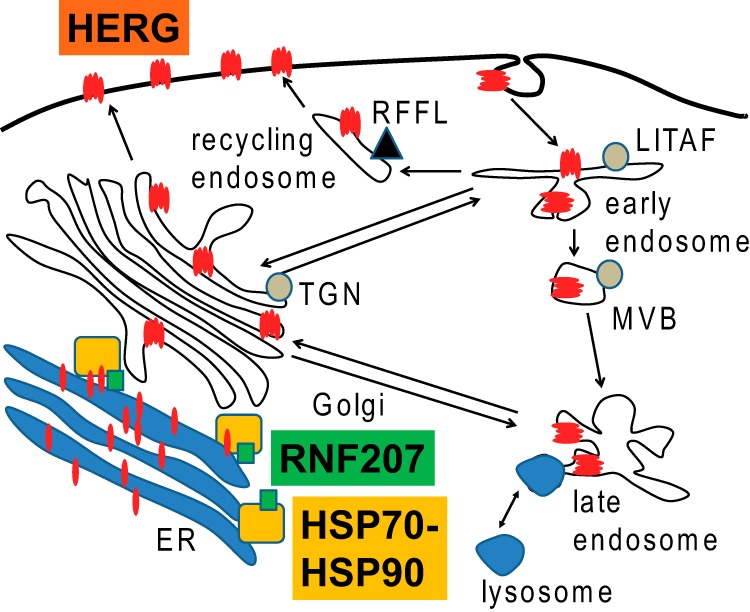
Model of RNF207-assisted HERG maturation in the heart. RNF207 binds HERG, HSP70, and other chaperones and, thereby, possibly assists in HERG synthesis, folding, or ER exit. Also depicted are RFFL (62, 63) and LITAF (64, 65), two genes with nearby loci associated with prolonged QT (10, 11, 16). MVB, multivesicular body; TGN, trans-Golgi network.
Importantly, there is increasing evidence that the stoichiometry of macromolecular ion channel complexes is precisely orchestrated through extensive coregulation of synthesis, trafficking, and degradation of the participating proteins, possibly in conjunction with specific lipids. Our observation of conduction slowing in the zebrafish suggests that RNF207 may contribute to the broader coregulation of other ion channel proteins beyond the IKr complex. Future studies will explore the full repertoire of RNF207-interacting proteins.
In summary, our data suggest that RNF207 regulation of HERG trafficking to the membrane is a fundamental mechanism underlying the physiological effects of RNF207 on the APD in both zebrafish and humans. These findings emphasize the importance of a complete understanding of the composition and dynamics of macromolecular membrane complexes in unraveling the mechanisms of common arrhythmias.
Acknowledgments
We thank Dr. Harm Kampinga (University of Groningen, University Medical Center Groningen, Department of Cell Biology, Groningen, The Netherlands) for the HSP70, HSC70, and DJA1 plasmids; Dr. Rachel Klevit (School of Medicine, University of Washington, Seattle, WA) for the ubiquitin-conjugating enzyme yeast expression library, and Dr. Richard Clements (Rhode Island Hospital, Cardiovascular Research Center, Providence, RI) for help with statistics.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s)JQ754141.
- APD
- action potential duration
- NRbCM
- neonatal rabbit ventricular cardiomyocyte
- mEGFP
- monomeric enhanced GFP
- IP
- immunoprecipitation
- hpf
- hours post-fertilization
- OS
- osteosarcoma
- ER
- endoplasmic reticulum
- RNFMO
- RNF207 morphants
- ROI
- region of interest.
REFERENCES
- 1. Tomaselli G. F., Beuckelmann D. J., Calkins H. G., Berger R. D., Kessler P. D., Lawrence J. H., Kass D., Feldman A. M., Marban E. (1994) Sudden cardiac death in heart failure: the role of abnormal repolarization. Circulation 90, 2534–2539 [DOI] [PubMed] [Google Scholar]
- 2. Lu J. T., Kass R. S. (2010) Recent progress in congenital long QT syndrome. Curr. Opin. Cardiol. 25, 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang Y., Yang Y., Liang B., Liu J., Li J., Grunnet M., Olesen S. P., Rasmussen H. B., Ellinor P. T., Gao L., Lin X., Li L., Wang L., Xiao J., Liu Y., Zhang S., Liang D., Peng L., Jespersen T., Chen Y. H. (2010) Identification of a Kir3.4 mutation in congenital long QT syndrome. Am. J. Hum. Genet. 86, 872–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amin A. S., Pinto Y. M., Wilde A. A. (2013) Long QT syndrome: beyond the causal mutation. J. Physiol. 591, 4125–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newton-Cheh C., Shah R. (2007) Genetic determinants of QT interval variation and sudden cardiac death. Curr. Opin. Genet. Dev. 17, 213–221 [DOI] [PubMed] [Google Scholar]
- 6. Hong Y., Rautaharju P. M., Hopkins P. N., Arnett D. K., Djoussé L., Pankow J. S., Sholinsky P., Rao D. C., Province M. A. (2001) Familial aggregation of QT-interval variability in a general population: results from the NHLBI Family Heart Study. Clin. Genet. 59, 171–177 [DOI] [PubMed] [Google Scholar]
- 7. Newton-Cheh C., Larson M. G., Corey D. C., Benjamin E. J., Herbert A. G., Levy D., D'Agostino R. B., O'Donnell C. J. (2005) QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: the Framingham Heart Study. Heart Rhythm 2, 277–284 [DOI] [PubMed] [Google Scholar]
- 8. Akylbekova E. L., Crow R. S., Johnson W. D., Buxbaum S. G., Njemanze S., Fox E., Sarpong D. F., Taylor H. A., Newton-Cheh C. (2009) Clinical correlates and heritability of QT interval duration in blacks: the Jackson Heart Study. Circ. Arrhythm. Electrophysiol. 2, 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marjamaa A., Newton-Cheh C., Porthan K., Reunanen A., Lahermo P., Väänänen H., Jula A., Karanko H., Swan H., Toivonen L., Nieminen M. S., Viitasalo M., Peltonen L., Oikarinen L., Palotie A., Kontula K., Salomaa V. (2009) Common candidate gene variants are associated with QT interval duration in the general population. J. Intern. Med. 265, 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfeufer A., Sanna S., Arking D. E., Müller M., Gateva V., Fuchsberger C., Ehret G. B., Orrú M., Pattaro C., Köttgen A., Perz S., Usala G., Barbalic M., Li M., Pütz B., Scuteri A., Prineas R. J., Sinner M. F., Gieger C., Najjar S. S., Kao W. H., Mühleisen T. W., Dei M., Happle C., Möhlenkamp S., Crisponi L., Erbel R., Jöckel K. H., Naitza S., Steinbeck G., Marroni F., Hicks A. A., Lakatta E., Müller-Myhsok B., Pramstaller P. P., Wichmann H. E., Schlessinger D., Boerwinkle E., Meitinger T., Uda M., Coresh J., Kääb S., Abecasis G. R., Chakravarti A. (2009) Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat. Genet. 41, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newton-Cheh C., Eijgelsheim M., Rice K. M., de Bakker P. I., Yin X., Estrada K., Bis J. C., Marciante K., Rivadeneira F., Noseworthy P. A., Sotoodehnia N., Smith N. L., Rotter J. I., Kors J. A., Witteman J. C., Hofman A., Heckbert S. R., O'Donnell C. J., Uitterlinden A. G., Psaty B. M., Lumley T., Larson M. G., Stricker B. H. (2009) Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. 41, 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Volpi S., Heaton C., Mack K., Hamilton J. B., Lannan R., Wolfgang C. D., Licamele L., Polymeropoulos M. H., Lavedan C. (2009) Whole genome association study identifies polymorphisms associated with QT prolongation during iloperidone treatment of schizophrenia. Mol. Psychiatry 14, 1024–1031 [DOI] [PubMed] [Google Scholar]
- 13. Milan D. J., Kim A. M., Winterfield J. R., Jones I. L., Pfeufer A., Sanna S., Arking D. E., Amsterdam A. H., Sabeh K. M., Mably J. D., Rosenbaum D. S., Peterson R. T., Chakravarti A., Kääb S., Roden D. M., MacRae C. A. (2009) Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation 120, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aberg K., Adkins D. E., Liu Y., McClay J. L., Bukszár J., Jia P., Zhao Z., Perkins D., Stroup T. S., Lieberman J. A., Sullivan P. F., van den Oord E. J. (2012) Genome-wide association study of antipsychotic-induced QTc interval prolongation. Pharmacogenomics J. 12, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roden D. (1998) Taking the “idio” out of “idiosyncratic”: predicting torsades de pointes. Pacing Clin. Electrophysiol. 21, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 16. Noseworthy P. A., Havulinna A. S., Porthan K., Lahtinen A. M., Jula A., Karhunen P. J., Perola M., Oikarinen L., Kontula K. K., Salomaa V., Newton-Cheh C. (2011) Common genetic variants, QT interval, and sudden cardiac death in a Finnish population-based study. Circ. Cardiovasc. Genet. 4, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 18. Folco E. J., Liu G. X., Koren G. (2004) Caveolin-3 and SAP97 form a scaffolding protein complex that regulates the voltage-gated potassium channel Kv1.5. Am. J. Physiol. Heart Circ. Physiol. 287, H681–H690 [DOI] [PubMed] [Google Scholar]
- 19. Jindal H. K., Folco E. J., Liu G. X., Koren G. (2008) Posttranslational modification of voltage-dependent potassium channel Kv1.5: COOH-terminal palmitoylation modulates its biological properties. Am. J. Physiol. Heart Circ. Physiol. 294, H2012–H2021 [DOI] [PubMed] [Google Scholar]
- 20. Ren X. Q., Liu G. X., Organ-Darling L. E., Zheng R., Roder K., Jindal H. K., Centracchio J., McDonald T. V., Koren G. (2010) Pore mutants of HERG and KvLQT1 downregulate the reciprocal currents in stable cell lines. Am. J. Physiol. Heart Circ. Physiol. 299, H1525–H1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Organ-Darling L. E., Vernon A. N., Giovanniello J. R., Lu Y., Moshal K., Roder K., Li W., Koren G. (2013) Interactions between hERG and KCNQ1 α-subunits are mediated by their COOH termini and modulated by cAMP. Am. J. Physiol. Heart Circ. Physiol. 304, H589–H599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ciechanover A. (2006) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Exp. Biol. Med. 231, 1197–1211 [DOI] [PubMed] [Google Scholar]
- 23. Roder K., Koren G. (2006) The K+ channel gene, Kcnb1: genomic structure and characterization of its 5′-regulatory region as part of an overlapping gene group. Biol. Chem. 387, 1237–1246 [DOI] [PubMed] [Google Scholar]
- 24. Hageman J., Kampinga H. H. (2009) Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 14, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brunner M., Peng X., Liu G. X., Ren X. Q., Ziv O., Choi B. R., Mathur R., Hajjiri M., Odening K. E., Steinberg E., Folco E. J., Pringa E., Centracchio J., Macharzina R. R., Donahay T., Schofield L., Rana N., Kirk M., Mitchell G. F., Poppas A., Zehender M., Koren G. (2008) Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J. Clin. Invest. 118, 2246–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panáková D., Werdich A. A., Macrae C. A. (2010) Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel. Nature 466, 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marín I. (2012) Origin and diversification of TRIM ubiquitin ligases. PLoS ONE 7, e50030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Napolitano L. M., Meroni G. (2012) TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 64, 64–71 [DOI] [PubMed] [Google Scholar]
- 30. Ozato K., Shin D. M., Chang T. H., Morse H. C. (2008) TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Milan D. J., Giokas A. C., Serluca F. C., Peterson R. T., MacRae C. A. (2006) Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133, 1125–1132 [DOI] [PubMed] [Google Scholar]
- 32. Werdich A. A., Brzezinski A., Jeyaraj D., Khaled Sabeh M., Ficker E., Wan X., McDermott B. M., Jr., Macrae C. A., Rosenbaum D. S. (2012) The zebrafish as a novel animal model to study the molecular mechanisms of mechano-electrical feedback in the heart. Prog. Biophys. Mol. Biol. 110, 154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leong I. U., Skinner J. R., Shelling A. N., Love D. R. (2010) Identification and expression analysis of kcnh2 genes in the zebrafish. Biochem. Biophys. Res. Commun. 396, 817–824 [DOI] [PubMed] [Google Scholar]
- 34. Langheinrich U., Vacun G., Wagner T. (2003) Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol. Appl. Pharmacol. 193, 370–382 [DOI] [PubMed] [Google Scholar]
- 35. Arnaout R., Ferrer T., Huisken J., Spitzer K., Stainier D. Y., Tristani-Firouzi M., Chi N. C. (2007) Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 104, 11316–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker V. E., Atanasiu R., Lam H., Shrier A. (2007) Co-chaperone FKBP38 promotes HERG trafficking. J. Biol. Chem. 282, 23509–23516 [DOI] [PubMed] [Google Scholar]
- 37. Walker V. E., Wong M. J., Atanasiu R., Hantouche C., Young J. C., Shrier A. (2010) Hsp40 chaperones promote degradation of the HERG potassium channel. J. Biol. Chem. 285, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hubbard T. J., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Fitzgerald S., Fernandez-Banet J., Graf S., Haider S., Hammond M., Herrero J., Holland R., Howe K., Johnson N., Kahari A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Melsopp C., Megy K., Meidl P., Ouverdin B., Parker A., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Severin J., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wood M., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Flicek P., Kasprzyk A., Proctor G., Searle S., Smith J., Ureta-Vidal A., Birney E. (2007) Ensembl 2007. Nucleic Acids Res. 35, D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galindo C. L., Skinner M. A., Errami M., Olson L. D., Watson D. A., Li J., McCormick J. F., McIver L. J., Kumar N. M., Pham T. Q., Garner H. R. (2009) Transcriptional profile of isoproterenol-induced cardiomyopathy and comparison to exercise-induced cardiac hypertrophy and human cardiac failure. BMC Physiol. 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong J., Chang C., Agrawal R., Walton G. B., Chen C., Murthy A., Patterson A. J. (2010) Gene expression profiling: classification of mice with left ventricle systolic dysfunction using microarray analysis. Crit. Care Med. 38, 25–31 [DOI] [PubMed] [Google Scholar]
- 42. Bush E. W., Hood D. B., Papst P. J., Chapo J. A., Minobe W., Bristow M. R., Olson E. N., McKinsey T. A. (2006) Canonical transient receptor potential channels promote cardiomyocyte hypertrophy through activation of calcineurin signaling. J. Biol. Chem. 281, 33487–33496 [DOI] [PubMed] [Google Scholar]
- 43. van Rijen H. V., de Bakker J. M., van Veen T. A. (2005) Hypoxia, electrical uncoupling, and conduction slowing: role of conduction reserve. Cardiovasc. Res. 66, 9–11 [DOI] [PubMed] [Google Scholar]
- 44. Sotoodehnia N., Isaacs A., de Bakker P. I., Dörr M., Newton-Cheh C., Nolte I. M., van der Harst P., Müller M., Eijgelsheim M., Alonso A., Hicks A. A., Padmanabhan S., Hayward C., Smith A. V., Polasek O., Giovannone S., Fu J., Magnani J. W., Marciante K. D., Pfeufer A., Gharib S. A., Teumer A., Li M., Bis J. C., Rivadeneira F., Aspelund T., Köttgen A., Johnson T., Rice K., Sie M. P., Wang Y. A., Klopp N., Fuchsberger C., Wild S. H., Mateo Leach I., Estrada K., Völker U., Wright A. F., Asselbergs F. W., Qu J., Chakravarti A., Sinner M. F., Kors J. A., Petersmann A., Harris T. B., Soliman E. Z., Munroe P. B., Psaty B. M., Oostra B. A., Cupples L. A., Perz S., de Boer R. A., Uitterlinden A. G., Völzke H., Spector T. D., Liu F. Y., Boerwinkle E., Dominiczak A. F., Rotter J. I., van Herpen G., Levy D., Wichmann H. E., van Gilst W. H., Witteman J. C., Kroemer H. K., Kao W. H., Heckbert S. R., Meitinger T., Hofman A., Campbell H., Folsom A. R., van Veldhuisen D. J., Schwienbacher C., O'Donnell C. J., Volpato C. B., Caulfield M. J., Connell J. M., Launer L., Lu X., Franke L., Fehrmann R. S., te Meerman G., Groen H. J., Weersma R. K., van den Berg L. H., Wijmenga C., Ophoff R. A., Navis G., Rudan I., Snieder H., Wilson J. F., Pramstaller P. P., Siscovick D. S., Wang T. J., Gudnason V., van Duijn C. M., Felix S. B., Fishman G. I., Jamshidi Y., Stricker B. H., Samani N. J., Kääb S., Arking D. E. (2010) Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar P., Henikoff S., Ng P. C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 46. Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P. (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dudbridge F., Gusnanto A. (2008) Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barrett J. C., Cardon L. R. (2006) Evaluating coverage of genome-wide association studies. Nat. Genet. 38, 659–662 [DOI] [PubMed] [Google Scholar]
- 49. Wenzel D. M., Stoll K. E., Klevit R. E. (2011) E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Christensen D. E., Brzovic P.S., Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 51. Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 52. Badciong J. C., Haas A. L. (2002) MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J. Biol. Chem. 277, 49668–49675 [DOI] [PubMed] [Google Scholar]
- 53. Ficker E., Dennis A. T., Wang L., Brown A. M. (2003) Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ. Res. 92, e87–e100 [DOI] [PubMed] [Google Scholar]
- 54. Li P., Ninomiya H., Kurata Y., Kato M., Miake J., Yamamoto Y., Igawa O., Nakai A., Higaki K., Toyoda F., Wu J., Horie M., Matsuura H., Yoshida A., Shirayoshi Y., Hiraoka M., Hisatome I. (2011) Reciprocal control of hERG stability by Hsp70 and Hsc70 with implication for restoration of LQT2 mutant stability. Circ. Res. 108, 458–468 [DOI] [PubMed] [Google Scholar]
- 55. Wang Y., Huang X., Zhou J., Yang X., Li D., Mao H., Sun H. H., Liu N., Lian J. (2012) Trafficking-deficient G572R-hERG and E637K-hERG activate stress and clearance pathways in endoplasmic reticulum. PLoS ONE 7, e29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sadis S., Hightower L. E. (1992) Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry 31, 9406–9412 [DOI] [PubMed] [Google Scholar]
- 57. Li J., Soroka J., Buchner J. (2012) The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823, 624–635 [DOI] [PubMed] [Google Scholar]
- 58. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Iwai C., Li P., Kurata Y., Hoshikawa Y., Morikawa K., Maharani N., Higaki K., Sasano T., Notsu T., Ishido Y., Miake J., Yamamoto Y., Shirayoshi Y., Ninomiya H., Nakai A., Murata S., Yoshida A., Yamamoto K., Hiraoka M., Hisatome I. (2013) Hsp90 prevents interaction between CHIP and HERG proteins to facilitate maturation of wild-type and mutant HERG proteins. Cardiovasc. Res. 100, 520–528 [DOI] [PubMed] [Google Scholar]
- 60. Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 61. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gopalakrishnan K., Morgan E. E., Yerga-Woolwine S., Farms P., Kumarasamy S., Kalinoski A., Liu X., Wu J., Liu L., Joe B. (2011) Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension 57, 764–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gopalakrishnan K., Kumarasamy S., Yan Y., Liu J., Kalinoski A., Kothandapani A., Farms P., Joe B. (2012) Increased expression of Rififylin in a < 330 kb congenic strain is linked to impaired endosomal recycling in proximal tubules. Front Genet. 3, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang Y., Bennett C. (2007) Litaf/Simple protein is increased in intestinal tissues from patients with CD and UC, but is unlikely to function as a transcription factor. Inflamm. Bowel. Dis. 13, 120–121 [DOI] [PubMed] [Google Scholar]
- 65. Shirk A. J., Anderson S. K., Hashemi S. H., Chance P. F., Bennett C. L. (2005) SIMPLE interacts with NEDD4 and TSG101: evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J. Neurosci. Res. 82, 43–50 [DOI] [PubMed] [Google Scholar]