Background: Proteins involved in expression of histone genes co-localize to discrete nuclear structures called histone locus bodies (HLBs).
Results: Two main components of HLBs directly interact with each other through their C-terminal regions.
Conclusion: Formation of HLBs depends on a subset of critical protein-protein interactions.
Significance: Our work may help to understand the mechanisms that integrate transcription of histone genes with maturation of the nascent histone transcripts.
Keywords: Gene Expression, Histone, Nuclear Organization, Protein-Protein Interaction, RNA Processing, Histone Locus Bodies, 3' End Processing, NPAT, FLASH, YARP, mute
Abstract
Nuclear protein, ataxia-telangiectasia locus (NPAT) and FLICE-associated huge protein (FLASH) are two major components of discrete nuclear structures called histone locus bodies (HLBs). NPAT is a key co-activator of histone gene transcription, whereas FLASH through its N-terminal region functions in 3′ end processing of histone primary transcripts. The C-terminal region of FLASH contains a highly conserved domain that is also present at the end of Yin Yang 1-associated protein-related protein (YARP) and its Drosophila homologue, Mute, previously shown to localize to HLBs in Drosophila cells. Here, we show that the C-terminal domain of human FLASH and YARP interacts with the C-terminal region of NPAT and that this interaction is essential and sufficient to drive FLASH and YARP to HLBs in HeLa cells. Strikingly, only the last 16 amino acids of NPAT are sufficient for the interaction. We also show that the C-terminal domain of Mute interacts with a short region at the end of the Drosophila NPAT orthologue, multi sex combs (Mxc). Altogether, our data indicate that the conserved C-terminal domain shared by FLASH, YARP, and Mute recognizes the C-terminal sequence of NPAT orthologues, thus acting as a signal targeting proteins to HLBs. Finally, we demonstrate that the C-terminal domain of human FLASH can be directly joined with its N-terminal region through alternative splicing. The resulting 190-amino acid MiniFLASH, despite lacking 90% of full-length FLASH, contains all regions necessary for 3′ end processing of histone pre-mRNA in vitro and accumulates in HLBs.
Introduction
Transcription of animal replication-dependent histone genes gives rise to intronless mRNA precursors (pre-mRNAs) that undergo a specialized 3′ end processing reaction to form mature histone mRNAs (1–3). In this reaction, histone pre-mRNAs are cleaved downstream of a highly conserved stem-loop structure, and the upstream cleavage product is not polyadenylated. Cleavage of histone pre-mRNAs requires the stem-loop binding protein (SLBP),2 which tightly interacts with the upstream stem-loop (4) and the U7 snRNP, which binds to the histone downstream element (HDE) located 3′ to the cleavage site (2). SLBP remains bound to the stem-loop following processing and assists mature histone mRNA on its way to the cytoplasm, where it stimulates translation of histone mRNAs into histone proteins (3).
The U7 snRNP consists of a short ∼60-nucleotide U7 snRNA and a unique Sm ring characterized by the presence of Lsm10 and Lsm11 proteins that replace the related SmD1 and SmD2 proteins found in the same position in the spliceosomal snRNPs (5, 6). The interaction between the U7 snRNP and histone pre-mRNA is mediated by the 5′ end of the U7 snRNA, which base pairs with the HDE. Following processing, the downstream cleavage product is degraded, liberating the U7 snRNP from the HDE for another round of processing (7, 8). The N-terminal region of Lsm11 interacts with the N-terminal region of FLICE-associated huge protein (FLASH), a protein of 220 kDa (9), and these two fragments form a platform that recruits the histone pre-mRNA cleavage complex (HCC) composed of several proteins involved in cleavage/polyadenylation, including the endonuclease CPSF73, its homologue CPSF100, and the scaffolding protein symplekin (10, 11).
In all eukaryotes, expression of replication-dependent histone genes is restricted to S phase and tightly coordinated with DNA replication (12–15). In mammalian cells, the cell cycle regulation of histone gene expression involves a 3–5-fold increase in the rate of transcription as cells enter S phase, but it is primarily achieved at the post-transcriptional level (3). Up-regulation of histone gene transcription during S phase occurs via promoter-specific factors (16–19) and critically depends on Nuclear protein, ataxia-telangiectasia locus (NPAT), which functions as a transcriptional co-activator (20–24). NPAT is phosphorylated by CDK2/cyclin E at the G1-S phase transition at multiple sites within the C-terminal half, and these modifications are essential for enhanced transcription of histone genes (20, 22). The post-transcriptional regulation involves activation of the U7-dependent processing of histone pre-mRNAs in S phase and rapid degradation of mature histone mRNAs upon completion of DNA replication (3). This regulation is largely achieved by the cell cycle-dependent synthesis of SLBP, which reaches the highest level in S phase and is degraded by the proteasome pathway in early G2 phase (25).
Proteins involved in biogenesis of histone mRNAs, including NPAT and components of the U7 snRNP, are concentrated in histone locus bodies (HLBs) that assemble at the histone gene clusters. HLBs have been identified in a broad range of animals, including Drosophila (26–29), zebrafish (30), Xenopus (31), and mammals (20, 22, 32–34), suggesting that they play an important role in executing proper expression of histone genes. Primary diploid human cells in S phase contain four HLBs that assemble on the two major histone gene clusters on chromosomes 1 and 6 (20), (22, 33). The numbers of HLBs in transformed cells, including HeLa cells, is typically higher than four due to aneuploidy of these cells (35, 36).
HLBs are believed to coordinate expression of the five types of histone genes and increase production of mature histone mRNAs by creating high local concentrations of the limiting factors. How HLBs (and other nuclear bodies) are formed remains a matter of debate, and models, including both highly hierarchical and random events, have been proposed (29, 37–41). In Drosophila, the initial step in HLB formation involves the recognition of chromatin at a specific site in the histone gene loci by a yet unidentified mechanism and subsequent recruitment of multi sex combs (Mxc), the Drosophila orthologue of NPAT, and FLASH, followed by the recruitment of the U7 snRNP and other pre-mRNA processing factors (42). At least some of these factors are recruited to HLBs prior to expressing histone genes, i.e. in a manner independent of the presence of the histone pre-mRNAs (27, 29, 42–44), indicating that the initial phase of HLB assembly is primarily driven by protein-protein interactions. Protein-RNA interactions that emerge following the initiation of transcription on histone genes likely contribute to formation of the fully developed and readily detectable HLBs (45). Recognizing the main players in forming this complex network of interactions is important for our understanding of both the biogenesis and function of HLBs.
Here, we report the identification of a conserved interaction of human NPAT with a C-terminal domain shared by FLASH and an unrelated protein known as Yin Yang 1-associated protein-related protein (YARP) or Gon4l, a transcriptional repressor important in early development. YARP is a homologue of Drosophila Mute that localizes to HLBs in Drosophila cells (28, 29) and likely acts there as a repressor of histone gene transcription (28). We show that YARP localizes to HLBs in HeLa cells and that the NPAT-interacting domain shared by FLASH and YARP serves as an HLB localization signal. Overall, our studies suggest that the direct interaction of NPAT with FLASH and YARP plays an essential role in the biogenesis of HLBs in animal cells and may provide an important bridge that integrates various nuclear events in histone gene expression.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid System
The C-terminal region of FLASH (amino acids 1880–1982) was cloned into the pGBKT7 vector (Clontech) and used to screen a normalized Mate and PlateTM library from HeLa S3 (Clontech, catalogue no. 630479), as suggested by the manufacturer. Human MiniFLASH was isolated by screening a normalized Universal Human Mate and PlateTM library (Clontech, catalogue no. 630480) against full-length Lsm11 cloned in pGBKT7 (9). Yeast diploid cells expressing proteins potentially interacting with the bait proteins were initially selected on plates lacking histidine and containing 3.5 or 5 mm 3-aminotriazole (3-AT) and subsequently tested on plates containing up to 100 mm 3-AT.
Antibodies
The following antibodies were used in this study: mouse monoclonal anti-HA 16B12 (Covance), mouse monoclonal anti-Myc 9E10 (Santa Cruz Biotechnology), rabbit anti-CPSF73 A301–090A (Bethyl Laboratories), rabbit anti-SLBP (46), rabbit anti-FLASH (9), sheep anti-mouse IgG NA931V (GE Healthcare), donkey anti-rabbit IgG Na934V (GE Healthcare), goat anti-rabbit IgG labeled with green fluorescent Alexa Fluor 488 (Invitrogen), goat anti-rabbit IgG labeled with red fluorescent Alexa Fluor 594 (Invitrogen), goat anti-mouse IgG labeled with red fluorescent Alexa Fluor 594 (Invitrogen), and goat anti-mouse IgG labeled with green fluorescent Alexa Fluor 488 (Invitrogen).
Protein Expression and Purification
Various fragments of FLASH, YARP, NPAT, Drosophila FLASH, Mute, and Mxc, each N-terminally fused to glutathione S-transferase (GST), were expressed in bacteria from the pET-42a vector and purified on nickel beads (Qiagen) via the His tag present on each fusion protein. For 35S labeling, each protein was cloned in a vector containing the SP6 RNA promoter and expressed in rabbit reticulocytes using the Transcription and Translation-coupled kit (TnT, Promega), according to the manufacturer's protocol.
Analysis of Protein-Protein Interactions by the GST Pulldown Assay
Various combinations of two different proteins, one tagged with GST (∼5 μg) and the other labeled with 35S (2.5–20 μl of the TnT reaction, depending on the efficiency of labeling) were mixed in 100 μl of binding buffer (100 mm NaCl, 20 mm Tris, pH 7.5, 10% glycerol, 0.1% Nonidet P-40) and incubated on ice for 30 min. The volume was subsequently increased to 600 μl with the binding buffer, and each sample was rotated with 30 μl of glutathione-agarose beads (Sigma) at 4 °C for 1 h. The beads were rinsed several times with the binding buffer, rotated with the same buffer for an additional 30 min, transferred to a new tube, and resuspended in an SDS sample buffer, and bound proteins were analyzed on an SDS-polyacrylamide gel. Each gel was initially stained with Coomassie Blue to monitor the amount of the GST-tagged proteins and subsequently dried and used for autoradiography to detect 35S-labeled proteins. The following peptides were used in competition experiments: Mock1, CLDLYEEILTEEGTAKEA; Mock2, CMLATGGFLQGDEADSY; and Mock3, CQDFYRIRDLLYEQYAIV.
Co-precipitation of Proteins Transiently Expressed in HeLa Cells
cDNA inserts encoding either human SLBP alone or human SLBP extended at the C terminus with the last 103 amino acids of human FLASH (amino acids 1880–1982) were cloned into pcDNA3/HA downstream from the region encoding three hemagglutinin (HA) tags. The same C-terminal fragment of human FLASH as well as a longer fragment (amino acids 1808–1982) containing a bipartite nuclear localization sequence were also cloned into the pEGFP-C3 vector (Clontech) to express GFP fusion proteins. The C-terminal regions of human NPAT (amino acids 1297–1427) and Drosophila Mxc (amino acids 1669–1837) were cloned into the pcDNA3/Myc vector immediately downstream of a single Myc epitope. Approximately 106 HeLa cells grown in monolayers were co-transfected in the presence of Lipofectamine 2000 (Invitrogen) with various combinations of constructs expressing the HA- and Myc-tagged proteins and 24 h later collected and lysed in Nonidet P-40 buffer (0.5% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8, 10 mm sodium azide, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride). The cell lysates were supplemented with EDTA to a final concentration of 10 mm and incubated 30 min on ice with 50 ng of the stem-loop RNA containing biotin at the 5′ end (Biot-SL) or with appropriate antibodies. Bound proteins were collected on streptavidin-Sepharose beads (RNA-mediated purification) or protein A-agarose beads (immunoprecipitation), separated in an SDS-polyacrylamide gel, and analyzed by Western blotting, using anti-HA and anti-Myc antibodies.
Immunofluorescence
Control HeLa cells or HeLa cells transiently expressing proteins with various tags were fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100. The intracellular localization of HA and Myc fusion proteins was analyzed by confocal microscopy using appropriate anti-tag primary antibodies and secondary antibodies. The localization of GFP fusion proteins was analyzed by directly detecting the GFP signal. Histone locus bodies were detected with the monoclonal DH3 anti-NPAT antibody (kindly provided by E. Harlow) (22) or a rabbit polyclonal antibody against N-terminal FLASH (αFLASH/N) (9).
RESULTS
C-terminal Domains of FLASH and YARP Interact with the C Terminus of NPAT
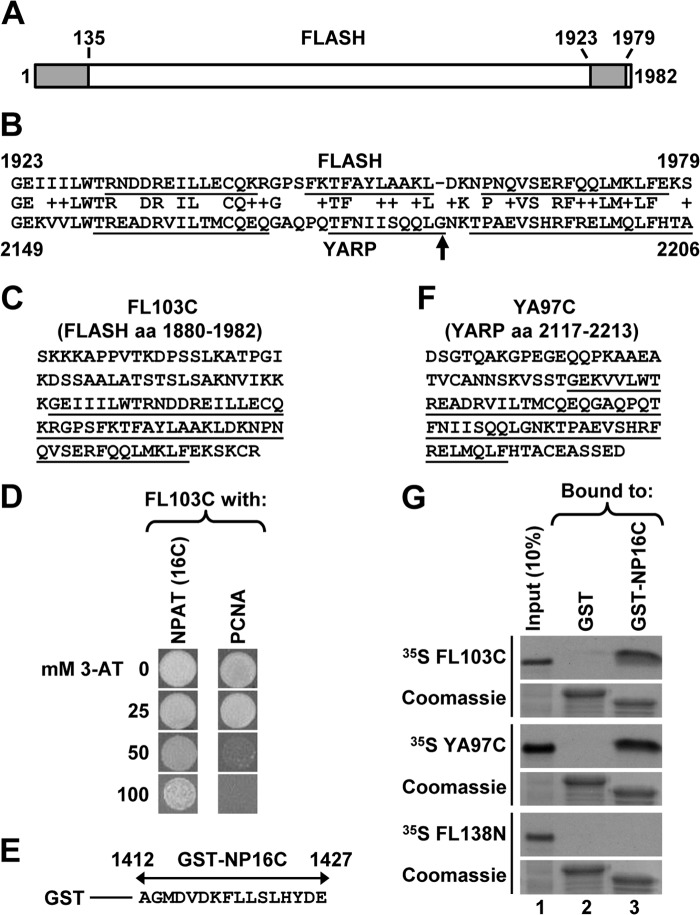
Human FLASH is a 220-kDa protein consisting of 1982 amino acids (Fig. 1A). Its N-terminal region of ∼135 amino acids is essential for 3′ end processing of histone pre-mRNAs in vitro by forming a platform with Lsm11 that recruits the HCC to the U7 snRNP and hence to histone pre-mRNA (11). This is the most highly conserved region of FLASH, sharing a weak but recognizable homology with its Drosophila orthologue (9, 47). The C-terminal region of human FLASH contains a domain of about 55 amino acids (Fig. 1A) that is highly conserved in all vertebrates but is missing in Drosophila FLASH. Strikingly, a similar domain is present at the end of an unrelated vertebrate protein known as YARP or Gon4l (Fig. 1B) (48, 49). As part of our interest in understanding the functional organization of FLASH, we investigated the role of its C terminus and potential links between FLASH and YARP.
FIGURE 1.
C-terminal regions of FLASH and YARP interact with NPAT. A, diagram of human FLASH consisting of 1982 amino acids. Positions of the conserved N- and C-terminal domains are indicated with gray boxes. B, protein BLAST alignment of the C-terminal domains of FLASH (top, amino acids 1923–1979) and YARP (bottom, amino acids 2149–2206). Amino acid substitutions that are unlikely to change the overall protein structure, as determined by the BLOSUM62 scoring matrix, are marked by “+”. α-Helices, as predicted by NMR structural studies (see “Discussion” for details), are underlined. The highly conserved glycine present in vertebrate YARP is indicated with an arrow. C, sequence of the last 103 amino acids of human FLASH. The C-terminal domain shared with YARP is underlined. D, yeast two-hybrid screen identifies NPAT and PCNA as potential binding partners of the FLASH C-terminal region. Growth of yeast cells expressing the C-terminal region of FLASH and either the last 16 amino acids of NPAT (left vertical row) or the C-terminal half of PCNA (right vertical row) in the presence of increasing concentration of 3-AT. E, sequence of the last 16 amino acids of human NPAT fused to the N-terminal GST (GST-NP16C). F, sequence of the last 97 amino acids of human YARP. The C-terminal domain shared with FLASH is underlined. G, GST pulldown assay to analyze in vitro interaction of 35S-labeled C-terminal regions of FLASH (FL103C) or YARP (YA97C) with the last 16 amino acids of NPAT fused to GST (GST-NP16C). 35S-Labeled N-terminal region of FLASH (FL138N, amino acids 1–138) and GST alone were used as negative controls. The amount of each GST protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (bottom panels). Note that GST alone migrates higher than GST-NP16C due to containing random amino acids at the C terminus from translating the multiple cloning site present in the pET-42a vector.
To identify binding partners of the highly conserved C-terminal domain common to FLASH and YARP, we cloned the last 103 amino acids of FLASH (Fig. 1C) in the yeast pGBKT7 bait vector (Clontech) and screened this fragment against a two-hybrid library from HeLa cells (Clontech). Of the more than 10 million diploids screened, seven colonies contained the same insert encoding the last 16 amino acids of NPAT, a known co-activator of histone gene transcription and a resident of HLBs (20–22, 34). The next most frequently identified positive clone (four independent isolates) contained the C-terminal half of proliferating cell nuclear antigen (PCNA), a component of the DNA replication machinery. Judging from the growth rate in the presence of increasing concentrations of 3-AT, the C-terminal region of FLASH interacts stronger with NPAT than with PCNA (Fig. 1D).
Because NPAT and FLASH reside in HLBs, their interaction was likely biologically relevant and raised the possibility that YARP may also interact with NPAT to control histone gene expression in HLBs. We used a GST pulldown assay to determine whether the C-terminal regions of FLASH and YARP directly interact in vitro with the C terminus of NPAT. We cloned the last 16 amino acids of NPAT downstream of GST (Fig. 1E), creating the GST-NP16C fusion protein. The C-terminal regions of FLASH (FL103C, amino acids 1880–1982) and YARP (YA97C, amino acids 2117–2213) were expressed and labeled with [35S]methionine in rabbit reticulocyte lysate using the TnT kit (Promega). In the pulldown assay, more than 20% of 35S-labeled FL103C used in the experiment bound to the GST-NP16C fusion protein (Fig. 1G, top two panels, lane 3). The corresponding domain of YARP also bound to GST-NP16C, although the interaction was less efficient, with on average 15% input recovered from glutathione beads (Fig. 1G, two middle panels, lane 3). In contrast, the N-terminal domain of FLASH (GST-FL138N, amino acids 1–138) does not detectably interact with GST-NP16C (Fig. 1G, two bottom panels, lane 3).
To confirm the interaction of the C-terminal NPAT with the C-terminal regions of FLASH or YARP, we carried out a reciprocal experiment. The C-terminal amino acids of FLASH(1880–1982) or YARP(2117–2213) were cloned downstream of an N-terminal GST tag and expressed in bacteria (GST-FL103C and GST-YA97C, respectively). The C-terminal region of NPAT encompassing the last 131 amino acids (Fig. 2A), NP131C, was labeled with 35S by in vitro translation. About 20% of the input NP131C was consistently bound by GST-FL103C or GST-YA97C (Fig. 2B, top panel, lanes 3 and 4, respectively). To determine whether the interaction involves only the last 16 amino acids of NPAT, we deleted these amino acids from the NP131C protein, creating NPΔ16C. This deletion reduced the interaction with GST-FL103C and abolished the interaction with GST-YA97C (Fig. 2B, middle panel, lanes 3 and 4, respectively). Deleting an additional nine amino acids (NPΔ25C) eliminated the interaction with GST-FL103C (Fig. 2B, bottom panel, lane 3).
FIGURE 2.
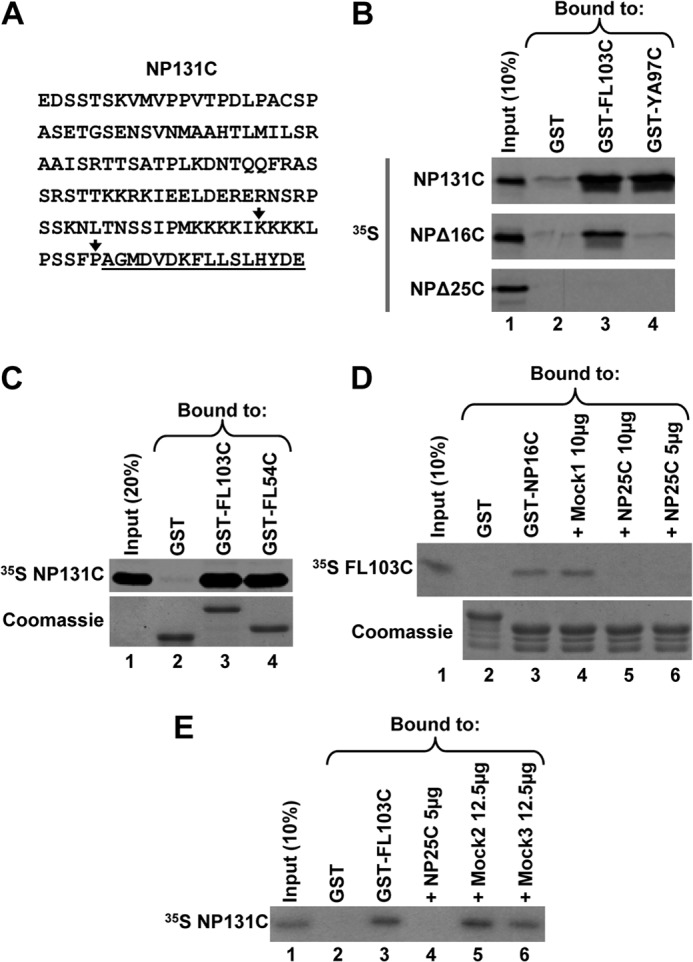
C-terminal domain shared by FLASH and YARP is sufficient for the interaction with NPAT. A, sequence of the last 131 amino acids of human NPAT (NP131C). The 16-amino acid extreme C-terminal region cloned in the yeast two-hybrid screen and capable of interacting with the C-terminal regions of FLASH and YARP is underlined. The last amino acids of the truncated NPΔ16C and NPΔ25C proteins are indicated with vertical arrows. B, GST pulldown assay to analyze in vitro interaction of 35S-labeled NP131C or its truncated versions (NPΔ16C and NPΔ25C) with the C-terminal regions of FLASH (GST-FL103C) or YARP (GST-YA97C), each fused to N-terminally positioned GST. GST alone (lane 2) was used as a negative control. The amount of each GST protein purified on glutathione beads was monitored by staining gels with Coomassie Blue and was equal in each lane (data not shown). C, GST pulldown assay to analyze the interaction of 35S-labeled NP131C with GST-FL103C or its shorter version GST-FL54C containing only the 54-amino acid region of homology with YARP (underlined in Fig. 1C). The amount of each GST protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (bottom panel). D, GST pulldown assay to analyze competition between GST-NP16C (lane 3) and Mock1 peptide (lane 4) or NP25C peptide (lanes 5 and 6) for binding 35S-labeled FL103C. GST alone (lane 2) was used as negative control. The amounts of the GST-NP16C and GST proteins purified on glutathione beads were monitored by staining the gel with Coomassie Blue (bottom panel). E, GST pulldown assay to analyze competition between 35S-labeled NP131C and NP25C peptide (lane 4) or two Mock peptides (lanes 5 and 6) for binding GST-FL103C. GST alone (lane 2) was used as negative control. The amount of each protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (data not shown).
Previous studies by others demonstrated that the endogenous FLASH and NPAT co-immunoprecipitated but failed to determine whether they interact directly or exist in a common complex containing additional proteins (34, 50). We now conclude that FLASH and NPAT directly contact each other via their C-terminal regions.
We created GST-FL54C containing only the 54-amino acid domain of FLASH that is shared with YARP (underlined in Fig. 1C). NP131C interacted with GST-FL54C as efficiently as with the longer C-terminal FLASH fragment consisting of 103 amino acids (Fig. 2C, lanes 3 and 4). We synthesized a peptide containing the last 25 amino acids of NPAT (NP25C) and tested its ability to compete with GST-NP16C in binding 35S-labeled FL103C. The NP25C peptide eliminated the interaction of GST-NP16C with FL103C (Fig. 2D, lanes 5 and 6), whereas a nonspecific peptide (Mock1) had no effect (Fig. 2D, lane 4). The NP25C peptide, but not two other nonspecific peptides, Mock2 and Mock3, also blocked the interaction in a reciprocal assay where the last 131 amino acids of NPAT were labeled with 35S, and the FLASH C terminus was fused to GST (Fig. 2E, compare lane 4 with lanes 5 and 6). Altogether, these results demonstrate that the 25-amino acid C-terminal region of NPAT alone is capable of strongly interacting with the C-terminal domain of FLASH.
C-terminal Domains of FLASH and NPAT Interact When Transiently Expressed in HeLa Cells
To determine whether the C-terminal regions of FLASH and NPAT interact when expressed in human cells, we constructed the HA-SLBP/F clone encoding human SLBP extended with a 3×HA tag at the N terminus and with the last 103 amino acids of FLASH at the C terminus (Fig. 3A). We also constructed a clone that expresses the last 131 amino acids of human NPAT (amino acids 1297–1427) as a fusion with an N-terminal Myc epitope (Myc-NP131C). As negative controls, we used the HA-SLBP fusion protein that lacks the C-terminal FLASH extension and the Myc-Mxc169C fusion protein containing the last 169 amino acids of Mxc (amino acids 1669–1837). Mxc, the Drosophila orthologue of NPAT (Fig. 3A), localizes to HLBs in Drosophila (29) but shares no recognizable homology with the C terminus of human NPAT. Each HA-tagged version of SLBP was co-expressed in HeLa cells with either Myc-NP131C or Myc-Mxc169C. Approximately 24 h following transfection, HeLa cells were collected and lysed, and SLBP from each lysate was affinity-purified on streptavidin-agarose beads using a short RNA containing the stem-loop structure from histone mRNA and biotin on the 5′ end (Fig. 3B, Biot-SL). This method of affinity-purifying SLBP was used by us previously (51) and is efficient and specific.
FIGURE 3.
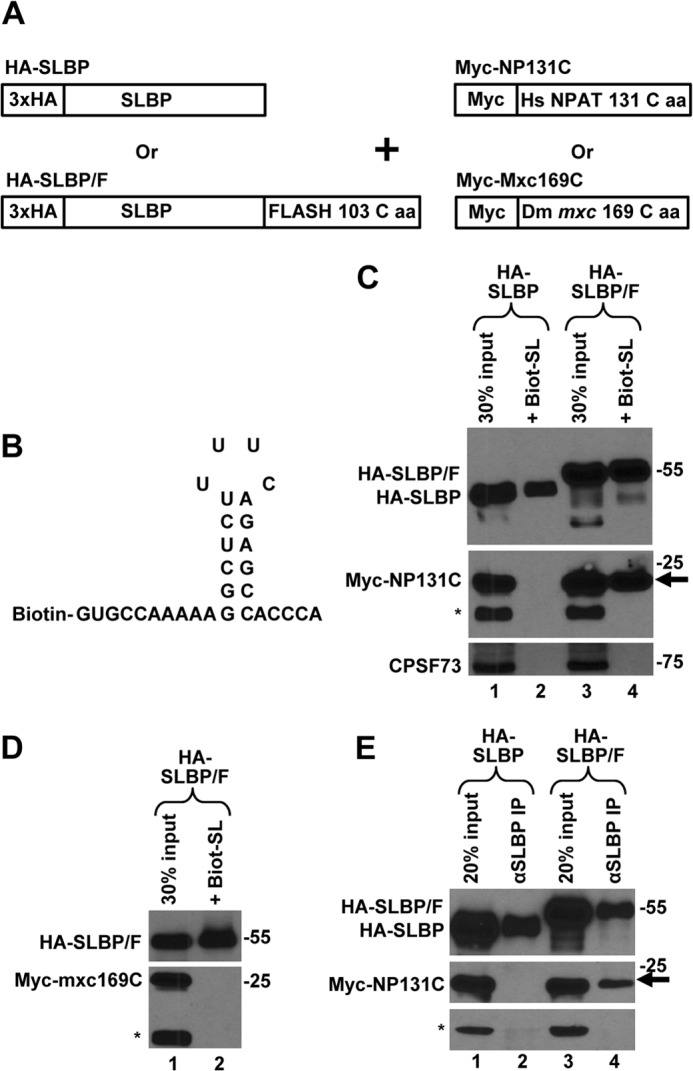
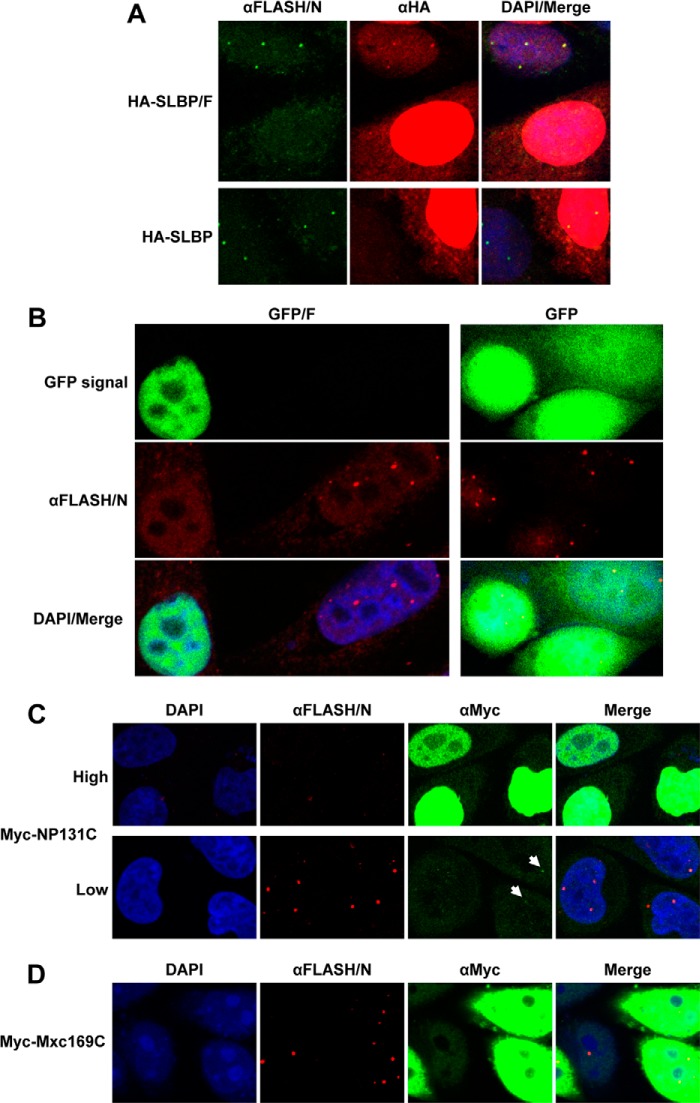
C-terminal regions of FLASH and NPAT interact in HeLa cells. A, schematic representation of clones transiently expressed in HeLa cells. B, nucleotide sequence of the Biot-SL RNA used to purify SLBP. C, Myc-NP131C was co-expressed in HeLa cells with either HA-SLBP (lanes 1 and 2) or HA-SLBP/F (lanes 3 and 4). SLBP was affinity-purified by the Biot-SL RNA from whole cell lysates prepared from transfected HeLa cells and the precipitated material tested for the presence of the transiently expressed HA- and Myc-tagged proteins (lanes 2 and 4). Lanes 1 and 3 contain 30% of the material used for purification. Myc-NP131C co-purified by virtue of interacting with HA-SLBP/F is indicated with an arrow. A protein cross-reacting with the anti-Myc antibody (indicated with an asterisk) served together with CPSF73 as a control to measure the extent of nonspecific background present in the purified material. D, Myc-Mxc169C and HA-SLBP/F were co-expressed in HeLa cells and the interaction between these two proteins tested as described in C. A protein cross-reacting with the anti-Myc antibody is indicated with an asterisk. E, Myc-NP131C was co-expressed in HeLa cells with either HA-SLBP (lanes 1 and 2) or HA-SLBP/F (lanes 3 and 4). SLBP was precipitated by an anti-SLBP antibody (αSLBP) and the precipitated material tested for the presence of the transiently expressed HA- and Myc-tagged proteins (lanes 2 and 4). A protein cross-reacting with the anti-Myc antibody is indicated with an asterisk, and the co-precipitated Myc-NP131C is indicated with an arrow.
The Biot-SL RNA purifies HA-SLBP and HA-SLBP/F with the efficiency varying between experiments from 10 to 40% of the input (Figs. 3C and 6A). The precipitate also contains the endogenous SLBP, which does not react with the anti-HA antibody and can only be detected by an anti-SLBP antibody (data not shown). We used anti-Myc antibodies and Western blotting to analyze the material purified by the stem-loop RNA. Importantly, HA-SLBP/F but not HA-SLBP co-purified with Myc-NP131C (Fig. 3C, arrow, compare lanes 2 and 4), demonstrating that the C-terminal regions of FLASH and NPAT can form a complex when present at physiologically relevant concentrations. As expected, the material did not contain detectable amounts of CPSF73 and other proteins of the whole cell lysate, confirming that the RNA-mediated purification is very selective. The specificity of the method was additionally confirmed by expressing an HA-tagged protein that lacks SLBP. This protein fails to interact with the Biot-SL RNA (see Fig. 8F, lane 4).
FIGURE 6.
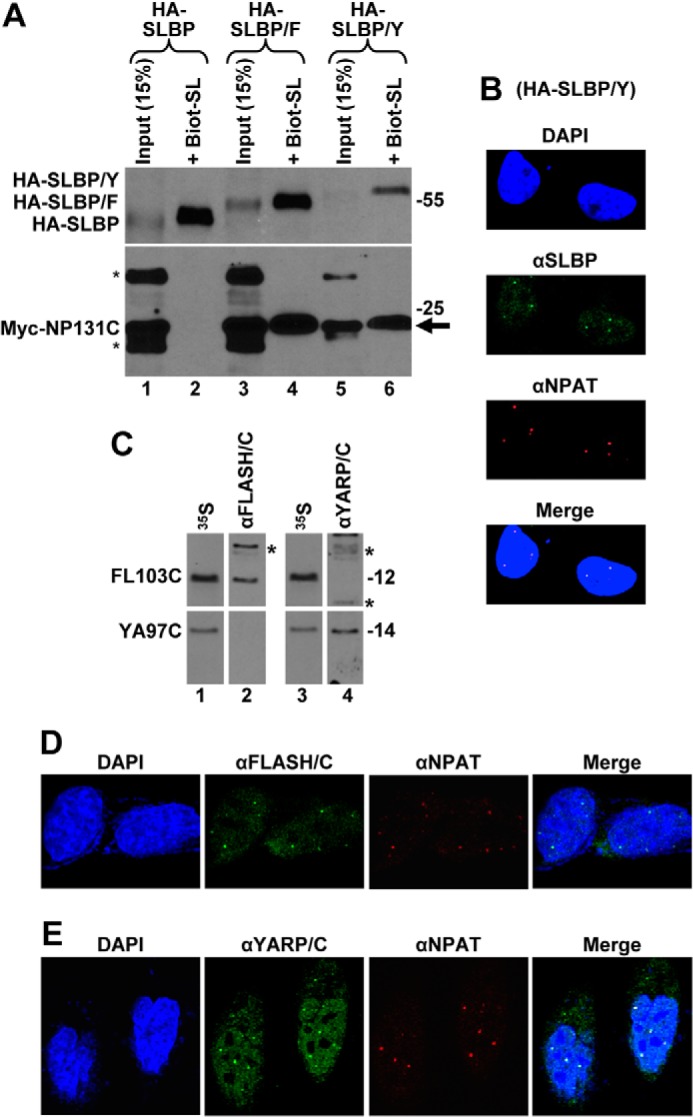
YARP is a component of HLBs in HeLa cells. A, Myc-NP131C was co-expressed in HeLa cells with either HA-SLBP (lanes 1 and 2), HA-SLBP/F (lanes 3 and 4), or HA-SLBP/Y (lanes 5 and 6). SLBP was affinity-purified by the Biot-SL RNA from whole cell lysates prepared from transfected HeLa cells, and the precipitated material was tested for the presence of the transiently expressed HA- and Myc-tagged proteins (lanes 2, 4, and 6). Lanes 1, 3, and 5 contain 15% of the material used for purification. Co-purified Myc-NP131C is indicated with an arrow, and proteins cross-reacting with the anti-Myc antibody are indicated with asterisks. B, fraction of SLBP in HeLa cells transiently expressing HA-SLBP/Y localizes to HLBs. SLBP was detected by an anti-SLBP antibody and stained green. HLBs were detected using the DH3 mouse monoclonal antibody (αNPAT) and stained red. C, rabbit antibodies targeted to the C-terminal regions of FLASH or YARP are free of cross-reacting activities. FL103C and YA97C were expressed in rabbit reticulocyte lysate and labeled by incorporating [35S]methionine. Duplicate samples of each labeled protein were separated in an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, detected by autoradiography (lanes 1 and 3), and subsequently immunoblotted with the cross-adsorbed αFLASH/C (lane 2) or αYARP/C (lane 4) antibodies. Proteins of the rabbit reticulocyte lysate that cross-react with each antibody are indicated with asterisks. D and E, endogenous FLASH (D) and YARP (E) were detected in HeLa cells by αFLASH/C and αYARP/C antibodies, respectively, and stained green. HLBs were detected by an antibody against NPAT and stained red. Nuclei were visualized by staining with DAPI.
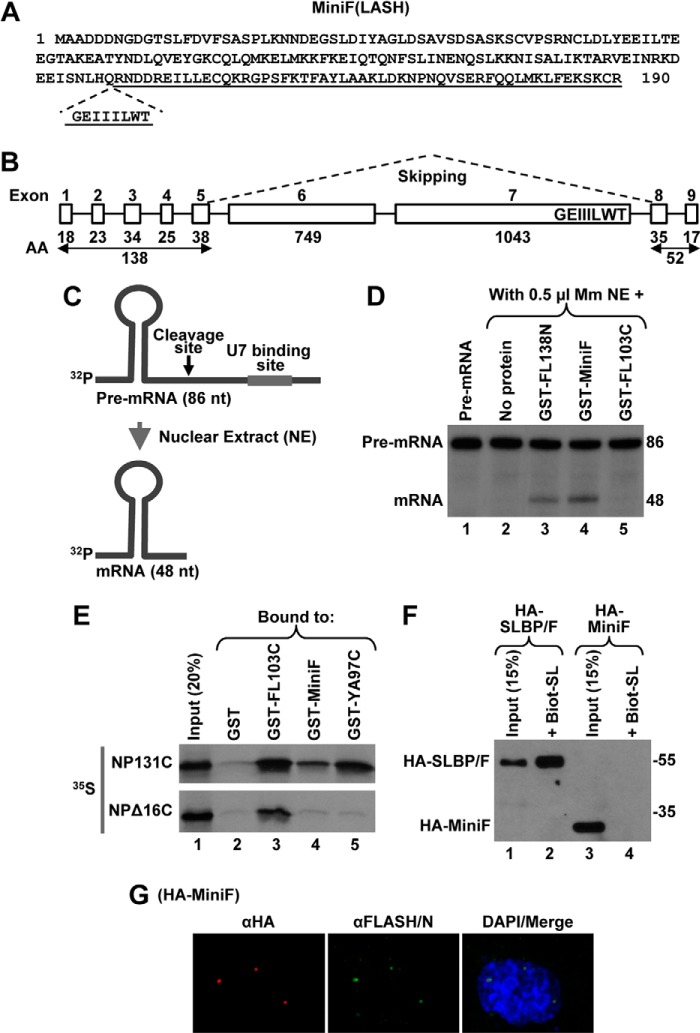
FIGURE 8.
Identification of human MiniFLASH (MiniF). A, amino acid sequence of 190-amino acid human MiniF. The incomplete C-terminal domain is underlined, and the missing GEIIILWT sequence is shown below. B, exon/intron organization of the human FLASH gene. The number of amino acids (AA) encoded by individual exons 1–9 of FLASH are shown at the bottom. The MiniF mRNA results from skipping the two large exons 6 and 7, as depicted by dashed lines, and in-frame linking of the N- and C-terminal exons. The highly conserved GEIIILWT sequence of the C-terminal domain of FLASH that is encoded by the end of exon 7 and hence missing in MiniF is indicated. C, diagram of the histone pre-mRNA substrate (86 nucleotides (nt)) and the product of in vitro 3′ end processing (48 nucleotides) carried out in a nuclear extract (NE). The HDE that is located downstream of the cleavage site (arrow) and serves as binding site for U7 snRNP is indicated with a thick line. D, 3′ end processing of the 86-nucleotide pre-mRNA substrate in 0.5 μl of mouse myeloma (Mm) nuclear extract alone (lane 2) or in the presence of 100 ng of indicated GST fusion FLASH proteins (lanes 3–5). The input substrate is shown in lane 1. E, GST-pulldown assay to analyze the interaction between MiniF and the C-terminal region of NPAT. Bacterially expressed proteins indicated at the top of lanes 2–5 were incubated with 35S-labeled NP131C or NPΔ16C, and the amount of each radioactive protein absorbed on glutathione beads was determined by autoradiography. As determined by Coomassie Blue staining, the amount of GST proteins in each lane was comparable (data not shown). Lane 1 contains 20% of each radioactive protein used in the experiment. F, transient expression of HA-MiniF in HeLa cells was compared with expression of HA-SLBP/F (lanes 3 and 1, respectively). As expected, only HA-SLBP/F can be purified by binding to the Biot-SL (lane 2). G, immunofluorescence of HeLa cells transiently expressing HA-MiniF. HA-MiniF was detected by a mouse anti-HA antibody and stained red. HLBs were detected by an antibody against the N-terminal FLASH and stained green. Nuclei were visualized by staining with DAPI.
We also co-expressed HA-SLBP/F with Myc-Mxc169C, containing the C-terminal 169 amino acids of Drosophila Mxc. Whole cell lysate was prepared from HeLa cells co-expressing HA-SLBP/F and Myc-Mxc169C, and the fusion SLBP protein was purified by the Biot-SL RNA. As determined by Western blotting with anti-HA and anti-Myc antibodies, the material collected on streptavidin beads contained HA-SLBP/F but lacked any detectable Myc-Mxc169C (Fig. 3D, lane 2), demonstrating that these two proteins do not form a stable complex.
Finally, we tested the interaction between HA-SLBP/F and Myc-NP131C using immunoprecipitation. Lysates from HeLa cells co-expressing Myc-NP131C with either HA-SLBP or HA-SLBP/F were incubated with an anti-SLBP antibody, and the precipitated material was analyzed by Western blotting using anti-HA and anti-Myc antibodies. Again, HA-SLBP/F but not HA-SLBP co-purified with Myc-NP131C (Fig. 3E, lanes 4 and 2, respectively).
C-terminal Domain of FLASH Targets Heterologous Proteins to HLBs
SLBP is a 3′ end processing factor that tightly binds the highly conserved stem-loop structure in histone pre-mRNA located upstream of the cleavage site and remains associated with the mature histone mRNA during translation (3). Immunostaining experiments demonstrate that endogenous SLBP, although present in the cytoplasm, is predominantly nuclear and displays no detectable enrichment in HLBs (52). This is in a sharp contrast to the U7 snRNP, including its core component Lsm11, which is concentrated in HLBs in both mammalian and Drosophila cells (27, 32).
To investigate the biological role of the interaction between the C-terminal regions of FLASH and NPAT, we tested whether the SLBP fused with the last 103 amino acids of FLASH is concentrated in HLBs. We transiently expressed HA-SLBP and HA-SLBP/F in HeLa cells and used fluorescent confocal microscopy to determine their intracellular localization 24 h following transfection. To ensure expression at a physiological level, HeLa cells were transfected with small amounts of plasmid DNA encoding HA-SLBP or HA-SLBP/F. The steady-state levels of these proteins were comparable with the level of endogenous SLBP (Fig. 4A, lanes 2 and 3).
FIGURE 4.
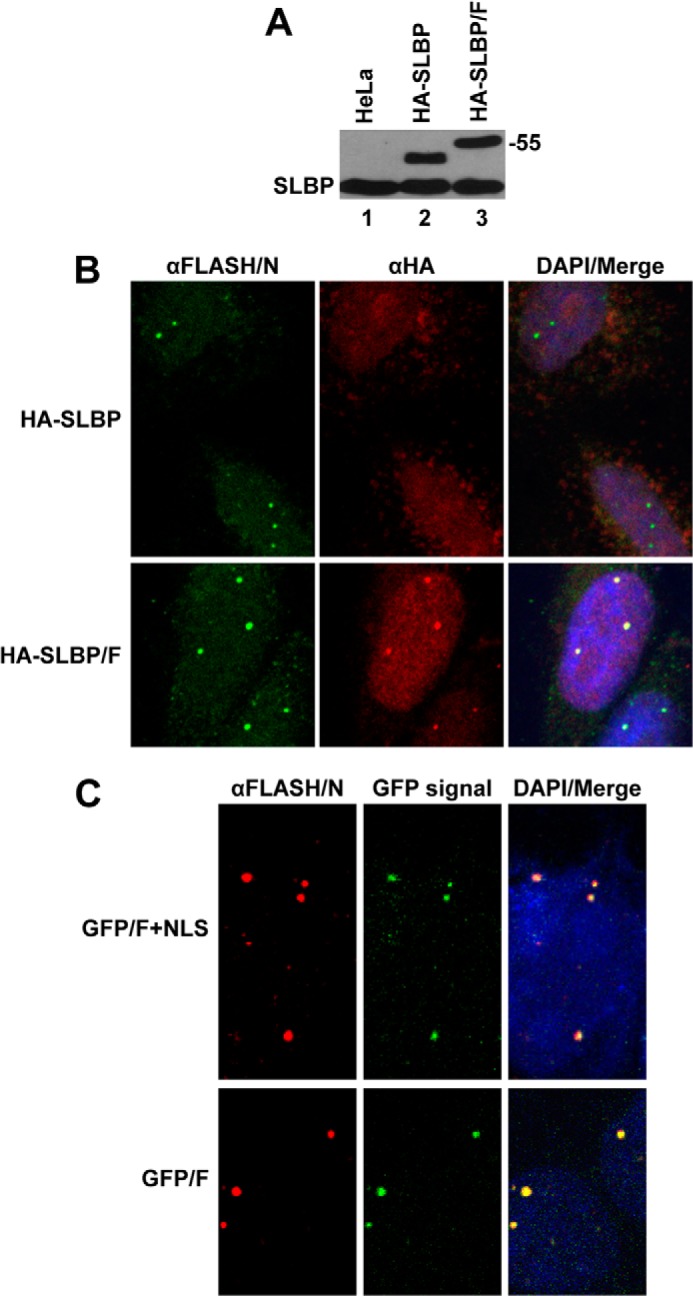
C-terminal region of FLASH targets SLBP and GFP to HLBs in HeLa cells. A, HA-tagged versions of SLBP (HA-SLBP) or SLBP extended at the C terminus by the last 103 amino acids of FLASH (HA-SLBP/F) were transiently expressed in HeLa cells and their levels compared relative to the endogenous SLBP using an anti-SLBP antibody (αSLBP). B, HeLa cells transiently expressing HA-SLBP (top panels) and HA-SLBP/F (bottom panels) were assayed by immunofluorescence. HLBs were detected by the αFLASH/N antibody and stained green. The intracellular localization of the HA-SLBP and HA-SLBP/F fusion proteins was detected using a mouse anti-HA antibody (αHA) and stained red. Nuclei were visualized with DAPI. HA-SLBP/F, but not HA-SLBP, in addition to showing a diffuse nuclear localization is enriched in a number of distinct foci that co-localize with the HLBs detected by the αFLASH/N antibody. C, immunofluorescence of HeLa cells transiently expressing GFP/F+NLS (top panels) and GFP/F (bottom panels). HLBs were detected using a rabbit antibody against N-terminal FLASH (αFLASH/N) and stained red. Nuclei were visualized with DAPI.
HeLa HLBs were stained with an antibody against the N-terminal portion of FLASH (αFLASH/N), and the intracellular localization of the HA-tagged SLBP proteins expressed from transfected plasmids was analyzed by an anti-HA antibody (αHA). HA-SLBP, which differs from the endogenous SLBP only by the presence of the HA tag, was localized predominantly to the nucleoplasm, showing no detectable foci that co-localized with HLBs (Fig. 4B, top panels). In contrast, the HA-SLBP/F fusion protein, in addition to displaying a uniform distribution, was concentrated in distinct nuclear structures, and this punctate pattern of staining was coincident with HLBs detected by the αFLASH/N antibody (Fig. 4B, bottom panels).
To further confirm this result and to exclude the possibility that the localization of HA-SLBP/F to HLBs was partly facilitated by SLBP, we appended the same fragment of FLASH (amino acids 1880–1982) to the C terminus of green fluorescent protein (GFP), thus creating the GFP/F fusion protein. We also constructed a second clone, GFP/F+NLS, in which GFP was followed by the last 175 amino acids of human FLASH (amino acids 1808–1982). This longer fragment contains a motif located between amino acids 1834 and 1851 that conforms to the bipartite nuclear localization sequence (NLS) and could potentially facilitate targeting the fusion protein to the nucleus. We transiently expressed the two fusion proteins in HeLa cells and tested their localization 24 h post-transfection by monitoring the GFP signal. Both GFP/F+NLS and GFP/F when expressed at a very low level (see below) were concentrated in HLBs detected by the αFLASH/N (Fig. 4C). We conclude that the C-terminal domain of FLASH that interacts with NPAT functions as a signal for localizing proteins to HLBs.
Overexpression of the C-terminal Regions of FLASH or NPAT Results in a Diffused Nuclear Distribution of Endogenous FLASH
When we analyzed HeLa cells transfected with the HA-SLBP/F clone, we observed that cells highly expressing this fusion protein lacked the normal punctate pattern detectable by the αFLASH/N (Fig. 5A, top panels). In contrast, cells overexpressing HA-SLBP contain readily detectable FLASH bodies (Fig. 5A, bottom panels). This observation suggested that an excess of the C-terminal FLASH saturates the C-terminal region of endogenous NPAT, resulting in the failure of endogenous FLASH to localize to HLBs.
FIGURE 5.
Effects of overexpressing the C-terminal regions of FLASH or NPAT on HLB formation. A, overexpression of HA-SLBP/F (top panels) but not HA-SLBP (bottom panels) by individual HeLa cells prevents localization of endogenous FLASH to HLBs. Localization of FLASH was monitored by a rabbit antibody against N-terminal FLASH (αFLASH/N) and stained green. HA fusion proteins were detected using a mouse anti-HA antibody (αHA) and stained red. For comparison, each field contains one cell with a low or undetectable level of expression of HA-SLBP/F or HA-SLBP and one cell overexpressing each protein. Nuclei were visualized with DAPI. B, overexpression of the GFP/F fusion protein (left panels) but not GFP lacking the C-terminal region of FLASH (right panels) results in mislocalization of endogenous FLASH. FLASH was detected by the αFLASH/N antibody and stained red. Two HeLa cells, one lacking and one overexpressing GFP/F, are shown on the left for comparison. Nuclei were visualized with DAPI. C, mislocalization of endogenous FLASH by overexpressing Myc-NP131C. Three cells with high expression (top panels) and three cells with low expression (bottom panels) of Myc-NP131C are shown for comparison. All cells come from the same field. Myc fusion protein was detected using a mouse anti-Myc antibody (αMyc) and stained green. FLASH was detected by the αFLASH/N antibody and stained red. Weakly stained foci of Myc-NP131C that co-localize with FLASH bodies (bottom) are indicated with arrows. Nuclei were visualized with DAPI. D, overexpression of Myc-Mxc169C has no effect on formation of FLASH bodies. Cells were stained as described in C.
We further tested this effect by overexpressing GFP/F. Also in this case, cells highly expressing the GFP fusion protein lacked foci enriched in endogenous FLASH (Fig. 5B, left panels). In contrast, cells that were not transfected or expressed low levels of this protein displayed a typical pattern of HLBs detectable by the αFLASH/N antibody. No effect on formation of HLBs was observed in cells highly expressing GFP alone (Fig. 5B, right panels). We also investigated the effect of overexpressing the C-terminal region of NPAT fused to Myc (Myc-NP131C). HeLa cells were transfected with high doses of DNA encoding this protein and analyzed for the presence of FLASH bodies using the αFLASH/N primary antibody and an anti-rabbit secondary antibody conjugated to red Alexa Fluor 594. As determined by an anti-Myc primary antibody and an anti-mouse secondary antibody conjugated to green fluorescent Alexa Fluor 488, overexpressed Myc-NP131C resulted in a strong and uniform green staining in the nucleus (Fig. 5C, top panels). Importantly, all cells with the high level of expression lacked any detectable red-stained FLASH bodies. In contrast, cells overexpressing Myc-Mxc169C that does not form a stable complex with human FLASH contained readily detectable FLASH bodies (Fig. 5D). Altogether, our results are consistent with the model that high expression of the C-terminal regions of FLASH or NPAT in HeLa cells saturates the limiting amounts of endogenous NPAT and FLASH, respectively, thus preventing a direct interaction between these two proteins and hence proper localization of FLASH to HLBs. Interestingly, Myc-NP131C expressed at a very low level does not affect FLASH localization but, in addition to showing a faint uniform distribution, is weakly enriched in HLBs (Fig. 5C, bottom panels, arrows).
Intracellular Localization of YARP
The presence of the FLASH-like domain at the C terminus of YARP and its ability to interact with NPAT in vitro suggested that this protein may also be enriched in mammalian HLBs. This possibility was strongly supported by the fact that YARP shares a significant homology with Mute, a Drosophila protein that localizes to HLBs in Drosophila cultured cells and embryos (28, 29). We first tested whether YARP and NPAT can form a complex when both proteins are expressed at physiologically relevant concentrations in human cells. An HA-tagged version of SLBP extended at the C terminus with amino acids 2117–2213 of YARP (HA-SLBP/Y) was transiently co-expressed with Myc-NP131C in HeLa cells and purified from whole cell lysate 24 h post-transfection by the Biot-SL RNA. The material collected on streptavidin beads in the presence of this RNA was highly enriched in HA-SLBP/Y and contained Myc-NP131C (Fig. 6A, lane 6, arrow), suggesting that these two proteins interact in HeLa cells. As controls, we again tested the interaction of Myc-NP131C with the two other SLBP fusion proteins, HA-SLBP and HA-SLBP/F. Consistent with the data presented above, only HA-SLBP/F co-purified with Myc-NP131C (Fig. 6A, lane 4). No background amounts of Myc-NP131C were detected in the material purified from cells expressing HA-SLBP (Fig. 6A, lane 2), confirming that the interaction requires the C-terminal region of either FLASH or YARP.
We transfected HeLa cells with low doses of DNA encoding HA-SLBP/Y and monitored the localization of the endogenous SLBP and transiently expressed HA-SLBP/Y by an anti-SLBP antibody. The antibody, in addition to detecting a uniform nucleoplasmic distribution characteristic of the endogenous SLBP, stained a number of much brighter nuclear foci that were coincident with NPAT (Fig. 6B). This result suggests that the C-terminal portion of YARP functions in the same manner as the homologous C-terminal portion of FLASH and, when appended to SLBP, promotes its enrichment in HLBs.
To investigate whether endogenous YARP localizes to HLBs, we generated an anti-YARP antibody using GST-YA97C as an immunogen. We also generated an antibody against GST-FL103C. Because the C-terminal regions of YARP and FLASH in addition to containing unique sequences share significant similarity, we used a two-step purification method to deplete each serum of cross-reactive activities. We preincubated the anti-YARP serum with an excess of GST-FL103C and the anti-FLASH serum with an excess of GST-YA97C and used the resulting supernatants to affinity-purify specific anti-YARP/C (αYARP/C) and anti-FLASH/C (αFLASH/C) antibodies with the proteins used for immunization. The lack of cross-reactivity following this procedure was confirmed by Western blotting (Fig. 6C). The C-terminal FLASH and YARP fragments were expressed and 35S-labeled in vitro, separated in an SDS-/polyacrylamide gel, and transferred to nitrocellulose membrane. Bands corresponding to each protein were identified by autoradiography and subsequently tested for their reactivity with the αYARP/C or αFLASH/C antibodies. The 35S-labeled C-terminal FLASH reacted only with the αFLASH/C antibody (Fig. 6C, top, compare lanes 2 and 4), and the C-terminal YARP was specifically recognized by the αYARP/C antibody (Fig. 6C, bottom, compare lanes 2 and 4), demonstrating that each antibody was effectively depleted of the cross-reacting activities.
We analyzed the intracellular localization of endogenous YARP and FLASH in HeLa cells using the cross-adsorbed and affinity-purified αYARP/C or αFLASH/C primary antibodies and a goat anti-rabbit secondary antibody conjugated to green fluorescent Alexa Fluor 488. HLBs were detected in the same cells using a mouse monoclonal antibody against NPAT (22) and stained red with a secondary anti-mouse antibody conjugated to Alexa Fluor 594. The αFLASH/C antibody detected a number of foci in HeLa nuclei that were coincident with the NPAT-stained HLBs (Fig. 6D). Importantly, the αYARP/C antibody also stained a number of foci that co-localized with NPAT (Fig. 6E). Based on these results, we conclude that YARP is a newly identified component of HLBs in mammalian cells, and its localization is likely mediated by the C-terminal domain that interacts with NPAT. In addition to detecting HLBs, the αYARP/C antibody stained the nucleoplasm, excluding the nucleolus. Thus, YARP, although present in HLBs, may also bind to other regions of chromatin, consistent with its proposed role in controlling expression of multiple genes (see “Discussion”).
Mute, the Drosophila Homologue of YARP, Interacts with the C Terminus of Mxc
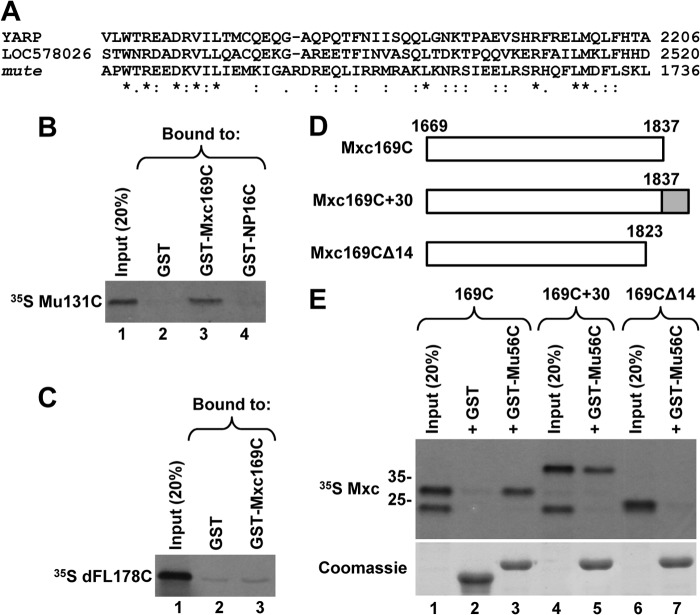
YARP throughout large parts of its length shares homology with Drosophila Mute, a component of the HLBs in Drosophila cells (28, 29), arguing that these two proteins are functionally related. The homology, although very limited, also extends to the C-terminal regions (Fig. 7A) (28). In contrast, no obvious sequence similarity can be observed between the C-terminal regions of NPAT and Mxc (data not shown). We tested the possibility that the C-terminal region of Mute directly interacts with the C-terminal region of Mxc, thus providing a plausible mechanism of how Mute is targeted to HLBs in Drosophila cells. We labeled a fragment of Mute encompassing the last 131 amino acids (Mu131C) with 35S and tested binding of this fragment to the last 169 amino acids of Mxc fused to GST (GST-Mxc169C). GST alone or fused to the last 16 amino acids of human NPAT served as negative controls. GST-Mxc169C reproducibly interacted with Mu131C, and on average precipitated nearly 20% of the input to glutathione beads (Fig. 7B, lane 3), whereas the two negative controls showed a background interaction (Fig. 7B, lanes 2 and 4). We also tested GST-Mxc169C and 35S-labeled dFL178C containing the last 178 amino acids of Drosophila FLASH. Both this pair (Fig. 7C, lane 3) and a reciprocal pair where the same C-terminal region of Drosophila FLASH was fused to GST and the last 169 amino acids of Mxc was labeled with 35S (data no shown) did not interact in vitro.
FIGURE 7.
Mute, the Drosophila homologue of YARP, interacts with Mxc. A, Clustal 2.1 multiple sequence alignment of the C-terminal regions of human YARP (2240 amino acids), Strongylocentrotus purpuratus protein LOC578026 (2553 amino acids), and Drosophila melanogaster Mute (1739 amino acids). Identical residues are denoted with an asterisk; conserved (strongly similar properties) and semi-conserved (weakly similar properties) residues are indicated with a colon and a period, respectively. B and C, GST pulldown assay to analyze in vitro interaction of 35S-labeled C-terminal regions of Mute (Mu131C, B) or Drosophila FLASH (dFL178C, C) with the last 169 amino acids of Mxc fused to GST (GST-Mxc169C). GST alone and GST fused to the last 16 amino acids of human NPAT (GST-NP16) were used as negative controls. The amount of each GST protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (data not shown). D and E, 35S-labeled Mxc169C and its two mutant versions, as depicted in D, were tested for interaction with the last 56 amino acids of Mute fused to GST (GST-Mu56C) (top panel in E). GST was used as a negative control. The amount of each GST protein purified on glutathione beads was monitored by staining the gel with Coomassie Blue (bottom panel in E). Note that inputs for Mxc169C and Mxc169C+30 (lanes 1 and 4) contain an additional 35S-labeled protein migrating below the 25-kDa size marker. This protein is likely generated as a result of premature termination of in vitro translation and it does not interact with GST-Mu56C.
We next expressed the last 56 amino acids of Mute (amino acids 1684–1739) as a fusion with GST (GST-Mu56C) and labeled Mxc169C and its two mutants with 35S (Fig. 7D). The last 56 amino acids of Mute share a limited similarity with the C-terminal domain of YARP (Fig. 7A). GST-Mu56C and Mxc169C clearly interact in vitro (Fig. 7E, lane 3), demonstrating that interaction is mediated through the homology domain in Mute. To map more precisely the region of Mxc that is required for the interaction, we deleted 14 amino acids from Mxc169C (Fig. 7D). The deletion mutant Mxc169CΔ14 was unable to interact with GST-Mu56C (Fig. 7E, lane 7). Thus, the extreme end of Mxc is important for the interaction, resembling the situation in human NPAT. To determine whether this location at the very C terminus is important for the interaction, we deleted the stop codon, hence extending Mxc at the C terminus with an additional 30 amino acids (Mxc169C+30). This modification had no effect on the interaction (Fig. 7E, lane 5), arguing that the region of NPAT/Mxc interacting with the C-terminal domain of FLASH/YARP/Mute can be located more internally.
MiniFLASH, a Short Form of FLASH, Is Localized to HLBs
The N-terminal region of FLASH (amino acids 1–138) interacts with Lsm11 (9, 53), and together these two proteins form a platform that recruits the HCC to the U7 snRNP (11). This short fragment of FLASH stimulates the activity of nuclear extracts in 3′ end processing of histone pre-mRNAs (9, 53), indicating that it mediates all essential processing functions of FLASH in nuclear extracts and that the remaining 95% of the protein is dispensable in vitro. Here, we have determined that the C-terminal domain of FLASH (amino acids 1923–1982) by itself can target FLASH to HLBs. Thus, out of nearly 2000 amino acids of FLASH, fewer than 200 residues derived from the opposing N- and C-terminal ends may be sufficient to support full processing activity of this protein in vivo.
As part of our continued effort to identify proteins interacting with Lsm11, we screened several independent cDNA libraries using the yeast two-hybrid system. In most screens, we consistently isolated clones encoding various N-terminal portions of FLASH that included the Lsm11- and HCC-binding sites (9). Strikingly, a universal cDNA library obtained from a collection of adult human tissues representing a broad range of expressed genes in both male and female donors yielded a FLASH variant in which the N- and C-terminal regions were directly linked, and nearly 1800 amino acids of the central part of the protein were missing. Of the 190-amino acid total length of this variant, the first 138 residues and the last 52 residues originated from the N and C terminus of full-length FLASH, respectively (Fig. 8A). We refer to this protein as MiniFLASH (MiniF). MiniF is encoded by an alternatively spliced form of FLASH mRNA that lacks two internal exons, consisting of over 2000 and 3000 nucleotides (Fig. 8B). These exons are among the largest internal exons in the genome, vastly exceeding the average length of 150 nucleotides (54, 55).
We tested whether MiniF is capable of supporting in vitro processing of a 5′-labeled histone pre-mRNA derived from the mouse histone H2a-614 gene (9, 53). This 86-nucleotide substrate contains all necessary processing sequences and upon cleavage is converted to a 48-nucleotide mature product ending in the stem-loop (Fig. 8C). A highly diluted mouse nuclear extract (equivalent of 0.5 μl of undiluted extract) is virtually inactive in processing due to limiting amounts of FLASH (Fig. 8D, lane 2). Consistent with our previous reports (9, 53), the GST-tagged N-terminal region of FLASH (GST-FL138N) stimulates 3′ end processing of the H2a pre-mRNA in this extract, increasing processing efficiency from a nearly undetectable level to 10% (Fig. 8D, lane 3). As expected, the C-terminal region of FLASH encompassing amino acids 1880–1982 (GST-FL103C) had no effect on processing (Fig. 8D, lane 5). The GST-MiniF fusion protein was functional (Fig. 8D, lane 4), indicating that 52 C-terminal amino acids of FLASH do not interfere with the ability of the N-terminal region to stimulate in vitro processing.
The last 52 amino acids of MiniF correspond to a truncated NPAT-binding domain that lacks the conserved GEIIILWT sequence at the N terminus (Fig. 8, A and B). We tested whether this incomplete C-terminal domain is sufficient to interact with the C-terminal NPAT and can support proper localization of MiniF to HLBs in vivo. In the GST pulldown assay, MiniF when compared with FL103C showed reduced ability to interact with the C terminus of NPAT (Fig. 8E, top panel, lanes 3 and 4), suggesting that the missing 8-amino acid region may be important for the optimal interaction. Shortening the C-terminal region of NPAT by removing the last 16 amino acids eliminated the interaction with MiniF (Fig. 8E, bottom panel, lane 4). Consistent with the data shown in Fig. 2, the interaction of NPΔ16C with GST-FL103C and GST-YA97C was significantly reduced and eliminated, respectively (Fig. 8E, bottom panel, lanes 3 and 5). Altogether, these data indicate that the incomplete C-terminal FLASH domain in MiniF retains partial ability to interact with NPAT.
To determine whether the incomplete C-terminal domain is sufficient to deliver MiniF to HLBs, we transiently expressed a 3×HA-tagged version of MiniF in HeLa cells and monitored its intracellular localization by immunofluorescence. MiniF expressed at the level comparable with HA-SLBP/F and migrated at the expected size of 30 kDa (Fig. 8F, lane 3). HLBs were detected by the αFLASH/N (which also detects MiniF) and stained green, whereas MiniF was detected by an anti-HA antibody and stained red (Fig. 8G). These experiments showed that the HA-tagged MiniF is concentrated in HLBs.
DISCUSSION
Human FLASH, a protein of nearly 2000 amino acids, is essential for U7-dependent 3′ end processing of histone pre-mRNAs and resides in HLBs (9, 33, 34, 56). Its N-terminal region of about 135 amino acids interacts with Lsm11, a component of the U7 snRNP, and together these two proteins recruit a subset of polyadenylation factors, including the endonuclease, to histone pre-mRNA for 3′ end cleavage (9, 53, 57). A short motif located in the center of FLASH mediates its interaction with Ars2, a stable component of the Cap binding complex (50). The structural and functional organization of FLASH outside these regions has not been characterized. The 55-amino acid domain located near the C terminus is among the most highly conserved regions in vertebrate FLASH. Strikingly, a similar sequence with about 50% identical residues is also present at the end of an unrelated protein known as YARP or Gon4-like (Gon4l). Studies on YARP suggested that it functions as a transcriptional regulator involved in controlling critical developmental decisions but provided no information on the role of its C-terminal domain (49, 58). YARP is a homologue of Drosophila Mute, a protein of unknown function shown to localize to HLBs in Drosophila cells and embryos (28).
C-terminal Domains of FLASH and YARP Interact with the Extreme C Terminus of NPAT
Using yeast two-hybrid screens, we identified NPAT as a factor that binds the C-terminal domains of FLASH and YARP. This finding was validated by in vitro GST pulldown assays and studies in HeLa cells. An in vitro interaction can also be detected between the C-terminal regions of Mute (28) and Mxc (29), the recently identified orthologues of vertebrate YARP and NPAT, respectively.
The structures of the C-terminal domains of FLASH (www.ncbi.nlm.nih.gov) and YARP have been determined by structural genomic projects, and both proteins adopt the same overall fold, consisting of three α-helices (underlined in Fig. 1B). Various homology searches, including SMART (Simple Modular Architecture Research Tool), predict that the same fold is likely adopted by the C-terminal domain of Mute (29). This fold structurally resembles the SANT and Myb domains and is referred to as the SANT/Myb-like domain. The Myb domain functions as a DNA recognition module in Myb-related transcriptional activators (59), whereas the SANT domain is an essential element of many chromatin-remodeling proteins (60). In contrast to the Myb domain, the SANT domain is unable to bind DNA (61) and instead it may function as a module that interacts with histone tails (60). Our studies demonstrated that the related C-terminal SANT/Myb-like domain in FLASH and YARP functions in vertebrates as an NPAT-interacting module. Interestingly, compared with FLASH orthologues, YARP orthologues in vertebrates contain an insertion of a single amino acid, glycine, that is located at the C-terminal end of helix 2 (Fig. 1B). The absolute conservation of this amino acid in YARP suggests that it is functionally important and may provide a critical feature that discriminates the YARP domain from the C-terminal domain of FLASH.
The region of NPAT that binds the C-terminal domains of FLASH and YARP was mapped to the last 25–30 amino acids, with as few as the last 16 C-terminal NPAT residues being sufficient to mediate a strong interaction in vitro. This short peripheral region of NPAT is among the most highly conserved regions of NPAT in vertebrates, although it does not resemble any known domain. Removing the last 14 amino acids from Mxc abolishes the interaction with the SANT/Myb-like domain of Mute, indicating that the interacting domain in Mxc is also located at the very end of the protein. Strikingly, NPAT and Mxc display no recognizable homology within this region.
Drosophila FLASH is similar to vertebrate FLASH only within the N-terminal region, which interacts with Lsm11 and polyadenylation factors (9, 11). The C-terminal region of Drosophila FLASH lacks a recognizable SANT/Myb-like domain and does not interact with the last 169 amino acids of Mxc in vitro. Intriguingly, the localization of FLASH to HLBs in Drosophila also requires the C-terminal regions of FLASH and Mxc (29, 47, 62). It is possible that these two proteins can only interact in the presence of a third component and/or a post-translational modification, which are missing in the in vitro pulldown experiment. Clearly, the interaction between the C-terminal regions of FLASH/YARP/Mute and NPAT/Mxc, whether direct or mediated by other proteins, is conserved in all animals.
SANT/Myb-like Domain of FLASH and YARP Functions as an HLB Localization Signal
What is the role of the direct interaction between the SANT/Myb-like domain of FLASH and the C terminus of NPAT? It has been previously shown that down-regulation of FLASH by RNAi in human cells results in a gradual redistribution of NPAT to the nucleoplasm (34, 50). Thus, NPAT on its own is not able to form HLBs in human cells. The formation of HLBs can be restored by ectopic expression of full-length FLASH but not by a FLASH mutant that lacks the C-terminal region (50). This result is consistent with our findings and emphasizes the importance of the FLASH C terminus for HLB formation.
We showed that the C terminus of FLASH fused to SLBP and GFP localizes these proteins to HLBs. Moreover, overexpression of the C-terminal regions of either FLASH or NPAT in HeLa cells results in mislocalization of endogenous FLASH, which becomes dispersed in the nucleus rather than concentrated in HLBs. This effect likely occurs due to titrating limiting amounts of endogenous NPAT and FLASH, respectively, with the excess of the interacting C-terminal fragments. The C-terminal fragment of NPAT consisting of only 131 amino acids, when expressed in HeLa cells at a very low level, forms faint foci that co-localize with the FLASH bodies. It is likely that this short fragment of NPAT is recruited to HLBs through a direct interaction with the C terminus of the endogenous FLASH. This possibility is consistent with the notion that the proper localization of these two proteins to HLBs is interdependent. We speculate that the interacting C-terminal domains of NPAT and FLASH form a platform that either directly or indirectly recognizes a specific DNA sequence or chromatin structure at the histone gene loci, thus providing the initial nucleation step in assembling HLBs in mammalian cells.
A number of observations indicate that during Drosophila embryogenesis components of the U7 snRNP are recruited to HLBs sequentially rather than together as a complete particle, with FLASH and Mxc associating first, followed by the core U7 snRNP (U7 snRNA plus the Sm ring that contains Lsm10 and Lsm11) (29). Drosophila FLASH and Lsm11 form a tight complex in vitro (9, 53), and it is unknown how these two proteins are temporarily kept apart in the nucleus and whether their association is regulated by cell cycle signals, including phosphorylation mediated by the CDK2-cyclin E complex. Surprisingly, an Lsm11 mutant unable to interact with FLASH is still enriched in HLBs (47), suggesting that at least in Drosophila the recruitment of the core U7 snRNP to HLBs is independent of FLASH and must be mediated by other yet unknown interactions. Collectively, these data suggest that the interaction between FLASH and Lsm11 and the assembly of the composite U7 snRNP containing the CPSF73 endonuclease (10, 11) is a tightly regulated step, which likely occurs late in the HLB biogenesis in conjunction with the activation of histone gene transcription, with phosphorylation of Mxc by CDK2/cyclin E serving as a triggering signal. This mechanism likely ensures the high efficiency of the U7-dependent cleavage and prevents generation of incorrectly processed and possibly harmful polyadenylated histone mRNAs (63). It is unclear whether in mammalian cells FLASH also interacts with NPAT as an independent entity, hence initiating the process of organizing HLBs, and the rest of the U7 snRNP is recruited later in HLB biogenesis. Immunofluorescence studies in synchronized human embryonal stem cells and somatic diploid WI-38 fibroblasts indicate that NPAT and the U7 snRNP, as judged by staining of Lsm10, co-localize early in the G1 phase, as soon as HLBs are detected (33). However, detailed studies are required to determine whether FLASH and the core U7 snRNP join the HLBs together or as two separate entities to form a complex at the G1/S phase transition, concomitantly with the activation of histone gene transcription.
Despite the universal nature of the interaction between the SANT/Myb-like domain and the C-terminal region of NPAT, in vivo studies suggest that both this interaction and formation of HLBs are not essential for cell cycle progression and production of histone mRNAs in cultured cells (50). In Drosophila, the SANT/Myb-like domain at the end of Mute is dispensable for the normal development of flies (28) and a C-terminal mutation of Mxc that removes the last 193 amino acids, while preventing formation of HLBs, only causes female sterility (29, 62). Furthermore, our recent results indicate that the inability to assemble HLBs due to the C-terminal deletion of FLASH does not result in any major defects in adult flies.3 Thus, although the assembly of HLBs is likely to increase the efficiency and fidelity of histone mRNA biosynthesis, the presence of these nuclear structures is not absolutely essential for viability of cells and organisms (29, 62).
YARP and Drosophila Mute Are Likely Functionally Related
Mute is a resident of HLBs in Drosophila cells (28, 29), and Mute mutations result in an increased accumulation of histone mRNAs in fly embryos, suggesting that one of the functions of Mute is to repress transcription of histone genes (28). Consistent with the presence of the NPAT-interacting domain, we demonstrate that YARP localizes to HLBs in HeLa cells, supporting the notion that the two proteins are functionally related. Indeed, a number of observations suggest that YARP may function in the same fashion as Mute by negatively regulating transcription of histone genes. It has been shown, for example, that YARP forms a complex consisting of the multifunctional transcriptional factor Ying Yang 1 (YY1), the transcriptional co-repressor Sin3a, and histone deacetylase 1 (HDAC1) and that this complex functions as a transcriptional repressor that commits B cell progenitors to B lymphopoiesis (58). Interestingly, virtually all vertebrate replication-dependent histone genes contain elements that bind YY1 (64–66), providing a plausible mechanism for recruiting YARP and the repressive complex to negatively regulate transcription of histone genes in vertebrates.
Both YARP and Mute in addition to regulating histone genes may affect expression of multiple genes, including those essential for development (28, 49, 67). Deficiency of Mute was shown to affect Drosophila muscle development, hence Mute is also referred to as “Muscle wasted” (28). Some of these functions may be executed by various splice variants and paralogues of YARP and Mute. The human genome contains a gene encoding a YARP homologue, YY1-associated protein (YY1AP) that arose as a result of partial duplication of the YARP gene during evolution of anthropoids (48, 68). YY1AP corresponds to the central part of YARP (amino acids 601–1338), and the two regions share nearly 100% sequence identity, consistent with the evolutionarily recent duplication event. YY1AP also binds YY1 (69) but lacks the SANT/Myb-like domain and is therefore unlikely to localize to HLBs.
MiniF, a Splicing Variant of FLASH
FLASH is encoded by several exons with the potential to generate multiple splice variants. Intriguingly, in a cDNA library constructed on mRNA isolated from a mixture of human tissues, we identified a number of independent clones encoding a short form of FLASH that we refer to as MiniFLASH (MiniF). MiniF consists of only 190 amino acids and results from skipping two extremely large internal exons and an in-frame fusion of exons encoding the N- and C-terminal regions. The first 138 amino acids of MiniF derive from the N terminus of FLASH and contain all elements necessary for 3′ end processing of histone pre-mRNAs in vitro (9, 11, 53). The remaining 52 amino acids of MiniF correspond to the C terminus of FLASH and encompass almost the entire SANT/Myb-like domain. This incomplete domain retains some ability to interact with NPAT and is sufficient to localize MiniF to HLBs, the site of histone pre-mRNA processing in vivo.
The two exons skipped in MiniF are among the longest internal exons found in the mammalian genome. It is unknown whether their skipping is an aberrant process or a result of a highly regulated alternative splicing specific for some tissues or developmental stages. The central region of mammalian FLASH deleted from MiniF consists of nearly 1800 amino acids and plays a largely unknown role. Parts of this region may control apoptosis (70) and transcription of selected genes (71), two processes previously linked to FLASH. A short 13-amino acid motif located between amino acids 931 and 943 that is missing in MiniF interacts with Ars2 (50). Ars2 exists in a tight complex with Cap-binding proteins of 20 and 80 kDa, and its interaction with FLASH may coordinate transcription of histone genes with maturation of histone mRNA at the 5′ and 3′ ends. Indeed, a mutant FLASH lacking the Ars2-interacting motif is inefficient in supporting proliferation and cell cycle progression of human pharyngeal carcinoma-derived KB cells (50), suggesting that MiniFLASH, although efficiently localized to the HLB and active in histone pre-mRNA processing in vitro, may not effectively promote histone mRNA biosynthesis in vivo. Clearly, the absence of the Ars2-interacting motif and other functional domains may also liberate MiniFLASH from tight regulatory constraints, potentially resulting in important consequences for cell metabolism and phenotype.
By searching human EST databases, we found two cDNA clones encoding MiniFLASH (accessions numbers BC029386 and CN356213). BC029386 was isolated from kidney cells, whereas CN356213 originated from a mixture of H1, H7, and H9 embryonic stem cells. Inspection of high throughput sequencing data from the ENCODE project as well as other publicly available data also revealed a small percentage of the alternatively spliced form encoding MiniFLASH in other cell types and organs. We used antibodies directed against the two terminal regions of FLASH for Western blotting and multiple pairs of primers for RT-PCR, but we failed to detect expression of MiniFLASH in H1 and H9 cells. Further studies are required to determine whether this splice variant is expressed at a very low level and/or only under certain physiological conditions.
Acknowledgments
We thank B. Xu for constructing SLBP expression plasmids, K. Krajewski (High-Throughput Peptide Synthesis Facility) for the 25-amino acid NPAT peptide, and D. Tatomer, K. Curry, and T. Perdue (Biology Microscopy Facility) for help with the immunofluorescence microscopy. We also thank E. Harlow for anti-NPAT antibody, R. Dumitru (Human Pluripotent Stem Cell Core Facility) for embryonal stem cells, D. Davis and J. Purvis for H9-specific cDNA, and G. Stein, J. Stein, and P. Ghule for stimulating discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM29832.
D. C. Tatomer, I. Sabath, Z. Dominski, W. F. Marzluff, and R. J. Duronio, submitted for publication.
- SLBP
- stem-loop binding protein
- HLB
- histone locus body
- NPAT
- nuclear protein, ataxia-telangiectasia locus
- FLASH
- Flice-associated huge protein
- YARP
- Yin Yang 1-associated protein-related protein
- PCNA
- proliferating cell nuclear antigen
- 3-AT
- 3-aminotriazole
- HDE
- histone downstream element
- HCC
- histone pre-mRNA cleavage complex
- NLS
- nuclear localization sequence.
REFERENCES
- 1. Dominski Z., Marzluff W. F. (1999) Formation of the 3′ end of histone mRNA. Gene 239, 1–14 [DOI] [PubMed] [Google Scholar]
- 2. Dominski Z., Marzluff W. F. (2007) Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396, 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marzluff W. F., Wagner E. J., Duronio R. J. (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan D., Marzluff W. F., Dominski Z., Tong L. (2013) Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 339, 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pillai R. S., Grimmler M., Meister G., Will C. L., Lührmann R., Fischer U., Schümperli D. (2003) Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17, 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schümperli D., Pillai R. S. (2004) The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 61, 2560–2570 [DOI] [PubMed] [Google Scholar]
- 7. Walther T. N., Wittop Koning T. H., Schümperli D., Müller B. (1998) A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4, 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang X. C., Sullivan K. D., Marzluff W. F., Dominski Z. (2009) Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 29, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X. C., Burch B. D., Yan Y., Marzluff W. F., Dominski Z. (2009) FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell 36, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabath I., Skrajna A., Yang X. C., Dadlez M., Marzluff W. F., Dominski Z. (2013) 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 19, 1726–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X. C., Sabath I., Dębski J., Kaus-Drobek M., Dadlez M., Marzluff W. F., Dominski Z. (2013) A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 33, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osley M. A. (1991) The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60, 827–861 [DOI] [PubMed] [Google Scholar]
- 13. Marzluff W. F., Duronio R. J. (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14, 692–699 [DOI] [PubMed] [Google Scholar]
- 14. Heintz N. (1991) The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta 1088, 327–339 [DOI] [PubMed] [Google Scholar]
- 15. Stein G. S., Stein J. L., van Wijnen A. J., Lian J. B. (1994) Histone gene transcription: A model for responsiveness to an integrated series of regulatory signals mediating cell cycle control and proliferation/differentiation interrelationships. J. Cell. Biochem. 54, 393–404 [DOI] [PubMed] [Google Scholar]
- 16. Mitra P., Xie R. L., Medina R., Hovhannisyan H., Zaidi S. K., Wei Y., Harper J. W., Stein J. L., van Wijnen A. J., Stein G. S. (2003) Identification of HiNF-P, a key activator of cell cycle-controlled histone H4 genes at the onset of S phase. Mol. Cell. Biol. 23, 8110–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fletcher C., Heintz N., Roeder R. G. (1987) Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51, 773–781 [DOI] [PubMed] [Google Scholar]
- 18. Gallinari P., La Bella F., Heintz N. (1989) Characterization and purification of H1TF2, a novel CCAAT-binding protein that interacts with a histone H1 subtype-specific consensus element. Mol. Cell. Biol. 9, 1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng L., Roeder R. G., Luo Y. (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- 20. Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., Adams P. D., Wang J., Qin J., Chow L. T., Harper J. W. (2000) Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14, 2298–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye X., Wei Y., Nalepa G., Harper J. W. (2003) The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 23, 8586–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., Fletcher J. A., Harlow E. (2000) NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14, 2283–2297 [PMC free article] [PubMed] [Google Scholar]
- 23. Miele A., Braastad C. D., Holmes W. F., Mitra P., Medina R., Xie R., Zaidi S. K., Ye X., Wei Y., Harper J. W., van Wijnen A. J., Stein J. L., Stein G. S. (2005) HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 25, 6140–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeRan M., Pulvino M., Greene E., Su C., Zhao J. (2008) Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 28, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng L., Dominski Z., Yang X. C., Elms P., Raska C. S., Borchers C. H., Marzluff W. F. (2003) Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol. Cell. Biol. 23, 1590–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J. L., Buszczak M., Gall J. G. (2006) Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 14, 465–475 [DOI] [PubMed] [Google Scholar]
- 27. Liu J. L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J. G. (2006) The Drosophila melanogaster Cajal body. J. Cell Biol. 172, 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bulchand S., Menon S. D., George S. E., Chia W. (2010) Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci. 123, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 29. White A. E., Burch B. D., Yang X. C., Gasdaska P. Y., Dominski Z., Marzluff W. F., Duronio R. J. (2011) Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193, 677–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strzelecka M., Oates A. C., Neugebauer K. M. (2010) Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 1, 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nizami Z. F., Deryusheva S., Gall J. G. (2010) Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb. Symp. Quant. Biol. 75, 313–320 [DOI] [PubMed] [Google Scholar]
- 32. Pillai R. S., Will C. L., Lührmann R., Schümperli D., Müller B. (2001) Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14-kDa Sm D1-like protein. EMBO J. 20, 5470–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghule P. N., Dominski Z., Yang X. C., Marzluff W. F., Becker K. A., Harper J. W., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2008) Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barcaroli D., Bongiorno-Borbone L., Terrinoni A., Hofmann T. G., Rossi M., Knight R. A., Matera A. G., Melino G., De Laurenzi V. (2006) FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. U.S.A. 103, 14808–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bongiorno-Borbone L., De Cola A., Vernole P., Finos L., Barcaroli D., Knight R. A., Melino G., De Laurenzi V. (2008) FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 7, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 36. Ghule P. N., Dominski Z., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2009) The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J. Cell. Physiol. 220, 129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Handwerger K. E., Gall J. G. (2006) Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16, 19–26 [DOI] [PubMed] [Google Scholar]
- 38. Matera A. G., Izaguire-Sierra M., Praveen K., Rajendra T. K. (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell 17, 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dundr M., Misteli T. (2010) Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nizami Z., Deryusheva S., Gall J. G. (2010) The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2, a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Machyna M., Heyn P., Neugebauer K. M. (2013) Cajal bodies: where form meets function. Wiley Interdiscip. Rev. RNA 4, 17–34 [DOI] [PubMed] [Google Scholar]
- 42. Salzler H. R., Tatomer D. C., Malek P. Y., McDaniel S. L., Orlando A. N., Marzluff W. F., Duronio R. J. (2013) A sequence in the Drosophila H3-H4 promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev. Cell 24, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abbott J., Marzluff W. F., Gall J. G. (1999) The stem loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol. Biol. Cell 10, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu C.-H., Gall J. G. (1993) U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. U.S.A. 90, 6257–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shevtsov S. P., Dundr M. (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 [DOI] [PubMed] [Google Scholar]
- 46. Wang Z.-F., Whitfield M. L., Ingledue T. C., 3rd, Dominski Z., Marzluff W. F. (1996) The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10, 3028–3040 [DOI] [PubMed] [Google Scholar]
- 47. Burch B. D., Godfrey A. C., Gasdaska P. Y., Salzler H. R., Duronio R. J., Marzluff W. F., Dominski Z. (2011) Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17, 1132–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohtomo T., Horii T., Nomizu M., Suga T., Yamada J. (2007) Molecular cloning of a structural homolog of YY1AP, a coactivator of the multifunctional transcription factor YY1. Amino Acids 33, 645–652 [DOI] [PubMed] [Google Scholar]
- 49. Lu P., Hankel I. L., Knisz J., Marquardt A., Chiang M. Y., Grosse J., Constien R., Meyer T., Schroeder A., Zeitlmann L., Al-Alem U., Friedman A. D., Elliott E. I., Meyerholz D. K., Waldschmidt T. J., Rothman P. B., Colgan J. D. (2010) The Justy mutation identifies Gon4-like as a gene that is essential for B lymphopoiesis. J. Exp. Med. 207, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kiriyama M., Kobayashi Y., Saito M., Ishikawa F., Yonehara S. (2009) Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 29, 4729–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dominski Z., Yang X. C., Kaygun H., Dadlez M., Marzluff W. F. (2003) A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12, 295–305 [DOI] [PubMed] [Google Scholar]
- 52. Erkmann J. A., Wagner E. J., Dong J., Zhang Y., Kutay U., Marzluff W. F. (2005) Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol. Biol. Cell 16, 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang X. C., Xu B., Sabath I., Kunduru L., Burch B. D., Marzluff W. F., Dominski Z. (2011) FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 31, 1492–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hawkins J. D. (1988) A survey on intron and exon lengths. Nucleic Acids Res. 16, 9893–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bolisetty M. T., Beemon K. L. (2012) Splicing of internal large exons is defined by novel cis-acting sequence elements. Nucleic Acids Res. 40, 9244–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barcaroli D., Dinsdale D., Neale M. H., Bongiorno-Borbone L., Ranalli M., Munarriz E., Sayan A. E., McWilliam J. M., Smith T. M., Fava E., Knight R. A., Melino G., De Laurenzi V. (2006) FLASH is an essential component of Cajal bodies. Proc. Natl. Acad. Sci. U.S.A. 103, 14802–14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dominski Z., Carpousis A. J., Clouet-d'Orval B. (2013) Emergence of the β-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 1829, 532–551 [DOI] [PubMed] [Google Scholar]
- 58. Lu P., Hankel I. L., Hostager B. S., Swartzendruber J. A., Friedman A. D., Brenton J. L., Rothman P. B., Colgan J. D. (2011) The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem. 286, 18311–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., Sarai A., Ishii S., Nishimura Y. (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648 [DOI] [PubMed] [Google Scholar]
- 60. Boyer L. A., Latek R. R., Peterson C. L. (2004) The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163 [DOI] [PubMed] [Google Scholar]
- 61. Horton J. R., Elgar S. J., Khan S. I., Zhang X., Wade P. A., Cheng X. (2007) Structure of the SANT domain from the Xenopus chromatin remodeling factor ISWI. Proteins 67, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rajendra T. K., Praveen K., Matera A. G. (2010) Genetic analysis of nuclear bodies: from nondeterministic chaos to deterministic order. Cold Spring Harb. Symp. Quant. Biol. 75, 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wagner E. J., Burch B. D., Godfrey A. C., Salzler H. R., Duronio R. J., Marzluff W. F. (2007) A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell 28, 692–699 [DOI] [PubMed] [Google Scholar]
- 64. Eliassen K. A., Baldwin A., Sikorski E. M., Hurt M. M. (1998) Role for a YY1 binding site in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18, 7106–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Palko L., Bass H. W., Beyrouthy M. J., Hurt M. M. (2004) The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117, 465–476 [DOI] [PubMed] [Google Scholar]
- 66. Last T. J., van Wijnen A. J., Birnbaum M. J., Stein G. S., Stein J. L. (1999) Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J. Cell. Biochem. 72, 507–516 [PubMed] [Google Scholar]
- 67. Liu Y., Du L., Osato M., Teo E. H., Qian F., Jin H., Zhen F., Xu J., Guo L., Huang H., Chen J., Geisler R., Jiang Y. J., Peng J., Wen Z. (2007) The zebrafish udu gene encodes a novel nuclear factor and is essential for primitive erythroid cell development. Blood 110, 99–106 [DOI] [PubMed] [Google Scholar]
- 68. Kuryshev V. Y., Vorobyov E., Zink D., Schmitz J., Rozhdestvensky T. S., Münstermann E., Ernst U., Wellenreuther R., Moosmayer P., Bechtel S., Schupp I., Horst J., Korn B., Poustka A., Wiemann S. (2006) An anthropoid-specific segmental duplication on human chromosome 1q22. Genomics 88, 143–151 [DOI] [PubMed] [Google Scholar]
- 69. Wang C. Y., Liang Y. J., Lin Y. S., Shih H. M., Jou Y. S., Yu W. C. (2004) YY1AP, a novel co-activator of YY1. J. Biol. Chem. 279, 17750–17755 [DOI] [PubMed] [Google Scholar]
- 70. Imai Y., Kimura T., Murakami A., Yajima N., Sakamaki K., Yonehara S. (1999) The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature 398, 777–785 [DOI] [PubMed] [Google Scholar]
- 71. Alm-Kristiansen A. H., Saether T., Matre V., Gilfillan S., Dahle O., Gabrielsen O. S. (2008) FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene 27, 4644–4656 [DOI] [PubMed] [Google Scholar]