Background: RNF4 is a transcription activator, yet the mechanism is unknown.
Results: RNF4 mediates ubiquitination and turnover of MeCP2 and thus derepresses transcription from DNA methylation.
Conclusion: RNF4 regulates transcription by creating a permissive epigenetic environment.
Significance: This study lays the basis for better understanding the role of RNF4 in epigenetics and transcription.
Keywords: DNA Methylation, Protein Turnover, RING Finger Protein 4 (RNF4), Transcription, Ubiquitylation (Ubiquitination), Methyl CpG-binding Protein 2 (MeCP2)
Abstract
RNF4 is an E3 ubiquitin ligase originally identified as a transcription co-activator. The mechanism by which RNF4 promotes transcription remains unclear. In this study, I found that RNF4 antagonizes transcriptional repression mediated by DNA methylation. RNF4 does not promote DNA demethylation, but mediates the ubiquitination of MeCP2, a methyl-CpG-binding domain (MBD) protein. Removal of MeCP2 from gene promoters activates transcription. This study thus not only uncovers how RNF4 functions as a transcription activator, but also reveals the mechanism by which MeCP2 protein stability is regulated.
Introduction
Epigenetic modifications play important roles in regulating gene expression. Epigenetic modifications occur on both histone and DNA. For example, methylation on histone H3 lysines 4 and 36 and acetylation on H3 lysines 9 and 14 correspond to active transcription, whereas methylation on H3 lysines 9 and 27 usually correlates to transcriptional repression (1). Besides histones, DNA can be methylated on the 5 position of cytosine, and methylation of promoter and enhancer DNA represses transcription (2). It is well established that transcriptional repression by DNA methylation is achieved through the recruitment of methyl cytosine-binding domains (MBD)2 family proteins, which in turn recruit repressor complexes such as histone deacetylase complexes (3, 4).
Great progress has been made in understanding the mechanisms of DNA demethylation during the past several years. It is now clear that Ten-eleven translocation (TET) family proteins mediate the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (5, 6), which is either passively diluted during DNA replication (7) or further oxidized by TET proteins to 5-formylcytosine and 5-carboxylcytosine (5caC) (8). Finally, 5-formylcytosine and 5caC are removed by thymine-DNA glycosylase (TDG) and replaced with unmethylated cytosine through base excision repair (9). Active DNA demethylation plays key roles in regulating gene expression in diverse biological processes (10).
RNF4 was initially identified as a transcriptional cofactor (11). It is an interesting protein as it contains a SUMO-interacting motif (SIM) and a RING finger domain that functions as an E3 ubiquitin ligase, thus linking sumoylation to ubiquitination (12). The biological function of RNF4 is broad as it targets many proteins for degradation, such as promyelocytic leukemia protein (PML), CENP-I, hypoxia-inducible factor (HIF), PARP-1, and MDC1 (13). Interestingly, a study revealed that RNF4 modulates DNA demethylation by activating the base excision repair machinery (14), although the precise mechanism remains unclear. Taking advantage of an in vitro methylated GFP reporter, I find that RNF4 only activates the reporter when it is methylated. Rather than promoting DNA demethylation, I found that RNF4 mediates the ubiquitination and turnover of MeCP2, an MBD protein that represses transcription.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
293T cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected with Lipofectamine 2000 for DNA plasmids and with RNAiMAX for siRNA (Life Technologies) following the manufacturer's protocol. Cycloheximide treatment was performed as described (15).
Antibodies and Reagents
GFP, MeCP2, and Myc antibodies were from Santa Cruz Biotechnology; RNF4, tubulin, ubiquitin, and FLAG (M2) antibodies were from Sigma; and 5caC antibody was described previously (8). RNF4 WT, ΔSIM, and CS plasmids were gifts from Dr. Zhenkun Lou (Mayo Clinic). TET2, TDG, and AID plasmids were described previously (16). pEGFP-N1 plasmid was from Clontech. To generate in vitro methylated GFP reporter, pEGFP-N1 plasmid was treated with CpG methyltransferase M.SssI (New England Biolabs) following the manufacturer's protocol and then purified with Qiagen miniprep kit. siRNAs against RNF4 and MeCP2 were from GE Dharmacon.
Glycosylase Assay
Myc-RNF4 and FLAG-TDG proteins were purified from 293T cells with Myc (MBL International) and M2-agarose (Sigma), respectively. Glycosylase assay was described previously (17). Briefly, 10 μl of reaction mixture contained 20 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 0.1 mg/ml BSA, 0.1 pmol of 32P-labeled 5caC DNA oligonucleotide, 4 pmol of purified TDG, and 0–16 pmol of RNF4. After incubation at 37 °C for 30 min, reaction was terminated by 1 μl of 1 m NaOH, incubated at 97 °C for 3 min, and then neutralized with 1 μl of 1 m acetic acid. Samples were analyzed with 15% denaturing gel and exposed against autoradiographic film (Kodak).
Bisulfite Sequencing
48 h after transfection of the GFP reporter, total DNA was extracted by the DNeasy kit (Qiagen). Bisulfite conversion was performed with the EpiTech bisulfite kit (Qiagen). The CMV promoter of GFP reporter was amplified with primers 5′-ataacccatatataaaattc-3′ and 5′-cgaaattagttttgtttatatag-3′ and ligated to pCR2.1-TOPO vector (Life Technologies) for DNA sequencing. The result of bisulfite conversion was analyzed with the BiQ analyzer.
ChIP-qPCR and RT-qPCR
Chromatin Immunoprecipitation was performed with the Imprint ChIP kit (Sigma) following the manufacturer's protocol. Total RNA extraction was performed with the RNeasy mini kit (Qiagen), and cDNA library was reversely transcribed via the ImProm-II reverse transcription system (Promega). qPCR reactions in SsoFast EvaGreen supermixes (Bio-Rad) were run and analyzed by the CFX384 real time PCR system (Bio-Rad).
In Vitro Ubiquitination
Myc-RNF4 and FLAG-MeCP2 proteins were overexpressed and purified from 293T cells. In vitro ubiquitination assay was performed with a kit from Enzo Life Sciences following the manufacturer's protocol.
Western Blotting
Cells were lysed with NETN buffer (0.5% Nonidet P-40, 150 mm NaCl, 50 mm Tris, and 1 mm EDTA) at 4 °C. To extract MeCP2, buffer containing 500 mm NaCl was used. Cell debris was removed by centrifugation, and 30–50 μg of supernatant was analyzed by SDS-PAGE and blotted with respective antibodies.
Immunofluorescence Microscopy
Cells grown in 4-well chamber slides (Lab-Tek) were first fixed with 4% paraformaldehyde for 20 min and then permeabilized with 0.5% Triton X-100 and blocked with buffer containing 5% donkey serum. Cells were sequentially incubated with primary antibodies for 2 h at room temperature and sequentially incubated with secondary antibodies for 30 min at room temperature. Slides were visualized under fluorescence microscopy (Zeiss), and images were processed by Photoshop.
RESULTS
RNF4 Activates Genes That Are Repressed by DNA Methylation
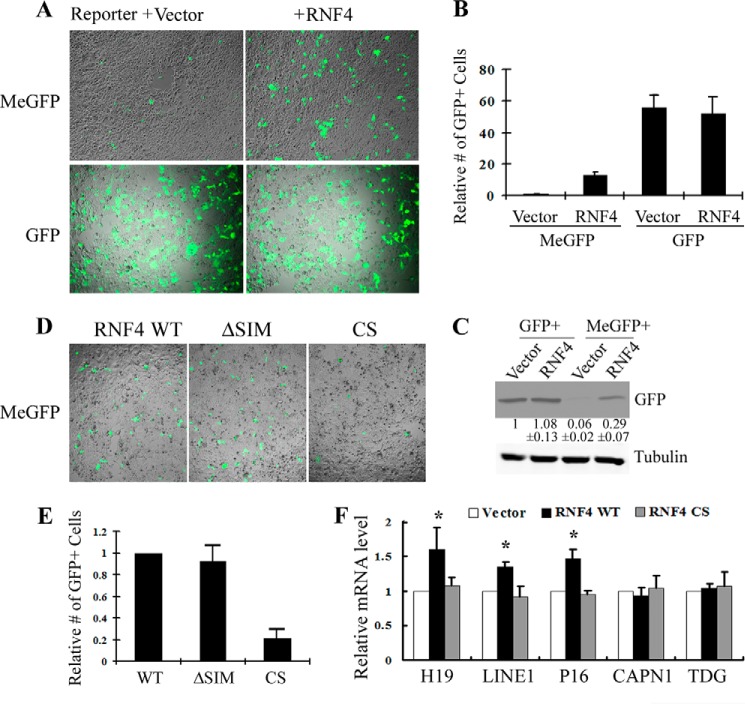
RNF4 was originally identified as a transcriptional co-activator for steroid receptors. However, the molecular mechanism is unclear. To address this question, I took advantage of a GFP reporter that is in vitro methylated by M.SssI (MeGFP), which almost completely shut down GFP expression (Fig. 1A). Interestingly, co-transfection of RNF4 significantly activates GFP expression, increasing the number of GFP-positive cells by about 15-fold (Fig. 1, A and B), which was confirmed by Western blot using a GFP antibody (Fig. 1C). Importantly, the effect of RNF4 is specific to methylated GFP vector as it does not affect transcription when the GFP reporter is not methylated (Fig. 1, A–C). RNF4 is a small protein with only two defined domains, an N-terminal SIM and a C-terminal Ring finger domain, which functions as an E3 ubiquitin ligase. To understand how RNF4 regulates gene expression, I tested loss of function mutations to each of the two domains and analyzed their effects on MeGFP expression. As shown in Fig. 1D, whereas deletion of the SIM domain (ΔSIM) does not modify the effect of RNF4 on the MeGFP reporter, Ring domain mutant (CS) fails to turn on MeGFP (Fig. 1, D and E). To rule out the possibility of an in vitro artifact, I tested whether RNF4 activates endogenous transcriptions that are repressed by DNA methylation. Indeed, I found that RNF4 significantly derepresses the expression of an imprinted gene H19, as well as P16 and LINE1 known to be regulated by DNA methylation, but not genes that are constitutively expressed such as CAPN1 (Calpain1) and TDG. Again, this activity is abolished by RNF4 CS mutant (Fig. 1F). Taken together, these data suggest that RNF4 derepresses gene expression caused by DNA methylation, and the Ring finger domain is important for this activity.
FIGURE 1.
RNF4 promotes transcription of methylated genes. A, GFP vector was in vitro methylated by M.SssI and then co-transfected in 293T cells with either vector control or RNF4. Upper panels, MeGFP expresses at a very low level, but is significantly activated by RNF4. Lower panels, RNF4 does not affect the expression of unmethylated GFP. B, quantification of GFP-positive cells in A. The number of vector control is normalized to 1. Quantification of three independent experiments ± S.D. is shown. C, representative Western blot demonstrates that RNF4 significantly enhances expression from methylated, but not unmethylated, GFP vector. Densitometry quantification of GFP signal relative to tubulin loading control is provided. Values represent the mean of three independent experiments ± S.D. D, MeGFP vector was co-transfected in 293T cells with RNF4 WT, SIM, and CS mutants, and RNF4 CS failed to activate GFP expression. E, the number of GFP-positive cells per field in D was quantified, and the value of RNF4 WT was normalized to 1. Results from three independent experiments ± S.D. are shown. F, RNF4 WT, but not RNF4 CS mutant, enhances transcription of endogenous genes that are suppressed by DNA methylation, including H19, LINE1, and P16, but not active genes such as CAPN1 and TDG. RT-qPCR results from three independent experiments ± S.D. are shown, and the value of vector control is normalized to 1 (*, p < 0.05).
RNF4 Does Not Promote DNA Demethylation
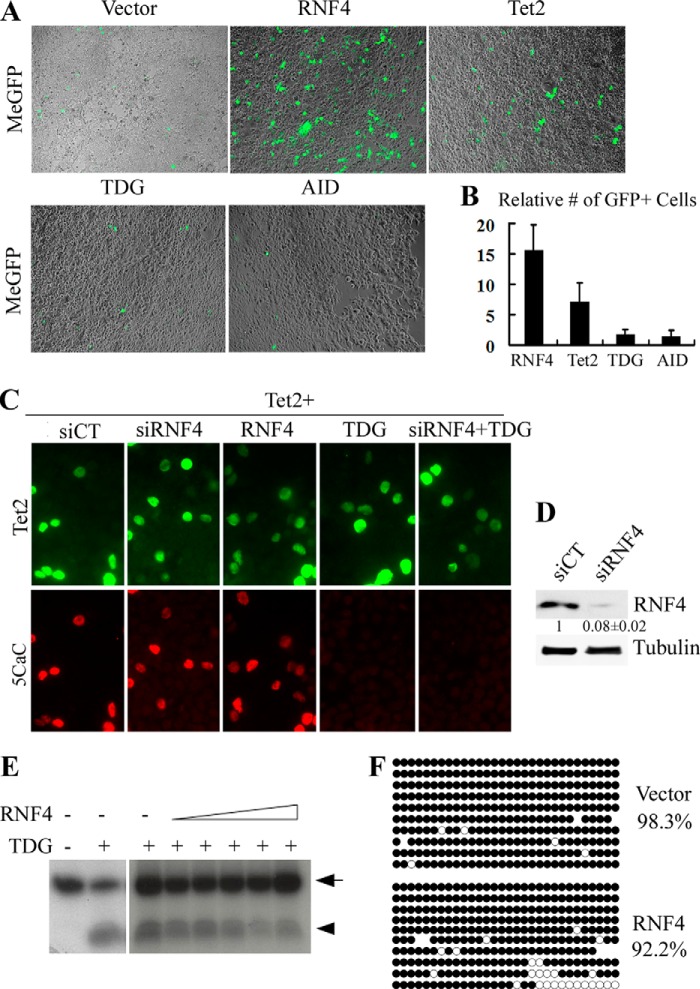
The roles of TET and TDG during DNA demethylation are well established, whereas the involvement of other factors such as AID has been implicated (18). I hypothesize that RNF4 activates MeGFP through DNA demethylation by enhancing the activity of TET, TDG, or AID. To test this possibility, I co-transfected MeGFP with vectors encoding those factors and monitored GFP expression. Surprisingly, although Tet2 activates GFP expression, it is significantly less effective than RNF4 (Fig. 2, A and B). Moreover, I failed to detect any effects of TDG or AID. These data argue against the notion that RNF4 activates gene expression through DNA demethylation.
FIGURE 2.
RNF4 does not induce active DNA demethylation. A, MeGFP vector was co-transfected in 293T cells with RNF4 or factors involved in active DNA demethylation, including Tet2, TDG, and AID. Representative images show the expression of GFP. B, the number of GFP-positive cells per field in A was quantified, and the value of vector control was set as 1. Results from three independent experiments ± S.D. are shown. C, RNF4 does not affect 5caC level. Immunostaining with 5caC antibody detects accumulation of 5caC in Tet2-expressing 293T cells. 5caC level is not affected by either knockdown or overexpression of RNF4. In contrast, expression of TDG completely removes 5caC, and the activity of TDG is not impaired by RNF4 knockdown. siCT, scrambled siRNA control. D, RNF4 knockdown is confirmed by Western blot analysis. Densitometry quantification of RNF4 signal relative to tubulin is provided. E, purified RNF4 and TDG proteins were incubated with 5caC-containing oligonucleotide (arrow) for in vitro glycosylase assay. The lower bands (arrowhead) represent oligonucleotide nicked by TDG, which is not affected by the addition of RNF4. F, MeGFP co-expressed with empty vector or RNF4 was extracted from cells for bisulfite sequencing. The CMV promoter remains predominantly methylated (filled circles) in both control and RNF4 cells.
To further test whether RNF4 regulates active DNA demethylation, I monitored the level of 5caC, a key intermediate product. As shown in Fig. 2C, 5caC is readily detectable in cells transfected with Tet2. However, neither knockdown nor overexpression of RNF4 affects the level of 5caC (Fig. 2C, second and third panel). As a control, expression of TDG completely removes 5caC, and the activity of TDG is not affected by RNF4 knockdown (Fig. 2C, fourth and fifth panel). Thus, although RNF4 is reported to enhance the activity of TDG toward G/T mismatch (14), it does not affect the activity of TDG toward 5caC. To confirm this result, I purified RNF4 and TDG proteins for an in vitro glycosylase assay. As shown in Fig. 2E, TDG but not RNF4 nicks the 5caC probe, which leads to a fragmented DNA product, and the addition of RNF4 does not enhance the glycosylase activity of TDG toward 5caC.
Finally, I analyzed the methylation status of the GFP reporter. MeGFP vector was extracted from cells with or without co-transfection of RNF4, and promoter methylation was determined by bisulfite sequencing. As expected, MeGFP is predominantly methylated (98.3%) in control sample (Fig. 2F), consistent with silenced GFP expression. Surprisingly, despite derepression of transcription, MeGFP from RNF4-expressing cells remains to be highly methylated (92.2%). Taken together, I conclude that RNF4 activates transcription independent of active DNA demethylation.
RNF4 Mediates the Turnover of MeCP2
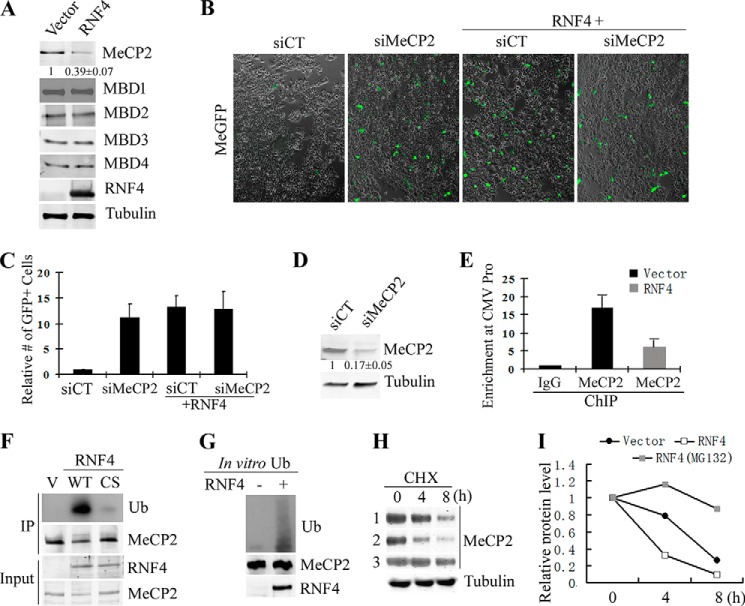
Methylated DNA is known to repress transcription by recruiting the MBD family protein co-repressor complexes. To determine whether RNF4 functions through this mechanism, I asked whether RNF4 may negatively regulate MBD proteins. To this end, I co-transfected MBD1–4 as well as MeCP2 with empty vector or RNF4 and then analyzed protein levels by Western blotting. Interestingly, the level of MeCP2, but not other MBD proteins, is significantly reduced when RNF4 is co-transfected (Fig. 3A). To determine whether the effect of RNF4 on MeCP2 is linked to activation of MeGFP reporter, I treated cells with MeCP2 siRNA prior to transfection of MeGFP, and indeed GFP expression is significantly elevated in MeCP2 knockdown cells when compared with control (Fig. 3, B and C). The knockdown of MeCP2 is confirmed by Western blot (Fig. 3D). There is no synergic effect between RNF4 and knockdown of MeCP2 (Fig. 3, B and C), suggesting that MeCP2 functions downstream of RNF4. Moreover, ChIP-qPCR analysis revealed binding of MeCP2 to the methylated CMV promoter of MeGFP reporter, and the binding is reduced by expression of RNF4 (Fig. 3E).
FIGURE 3.
RNF4 ubiquitinates and turns over MeCP2. A, Western blot analysis indicates that the level of MeCP2, but not other MBD proteins, is reduced by co-expression of RNF4. Values represent the mean of three independent experiments ± S.D. B and C, representative images (B) and quantification (C) of the expression of MeGFP reporter in scrambled siRNA control and MeCP2 knockdown cells. GFP expression is derepressed by MeCP2 knockdown and is not further activated by RNF4. The number of GFP-positive cells per field was quantified, and the value of control was set as 1. Results from three independent experiments ± S.D. are shown. siCT, scrambled siRNA control. D, Western blot confirms the knockdown of MeCP2. Values represent the mean of three independent experiments ± S.D. E, ChIP-qPCR reveals that MeCP2 binds to the promoter of the methylated GFP reporter, and MeCP2 binding is significantly reduced by RNF4 expression. Results from three independent experiments ± S.D. are shown. F, RNF4 ubiquitinates MeCP2. MeCP2 was immunoprecipitated (IP) from cells expressing RNF4 WT or CS mutant, and ubiquitination (Ub) level was analyzed by Western blotting. RNF4 WT, but not the CS mutant, significantly promotes MeCP2 ubiquitination. V means empty vector control. G, RNF4 directly ubiquitinates MeCP2. Purified RNF4 and MeCP2 proteins were subjected to in vitro ubiquitination assay. H, Western blot of MeCP2 levels in vector control (lane 1) and RNF4 (lanes 2 and 3) transfected cells. Cells were treated with cycloheximide (CHX) and collected for analysis at different time points. Samples of lane 3 were co-treated with MG132 to block proteasome activity. I, densitometry quantification for panel H shows that RNF4 significantly accelerates the turnover of MeCP2, which is blocked by inhibition of proteasome.
MeCP2 is among the best studied epigenetic factors. However, the mechanisms that regulate MeCP2 protein turnover remain unknown. Because RNF4 is an E3 ubiquitin ligase, I tested whether RNF4 mediates ubiquitination of MeCP2. MeCP2 was immunoprecipitated from cells expressing the wild type or the Ring domain mutant (CS) RNF4, and the ubiquitination level was determined by blotting for ubiquitin. As shown in Fig. 3F, RNF4 significantly enhances MeCP2 ubiquitination when compared with vector control and RNF4 CS mutant. Moreover, in vitro assay with purified proteins suggests that RNF4 can directly ubiquitinate MeCP2 (Fig. 3G). Next, I tested whether RNF4 promotes MeCP2 turnover. The half-life of MeCP2 is determined by blocking protein synthesis with cycloheximide. As shown in Fig. 3, H and I, MeCP2 is a dynamic protein with a half-life of 6.5 h (Fig. 3H, lane 1), which is significantly reduced by the presence of RNF4 (Fig. 3H, lane 2), and co-treatment of the proteasome inhibitor MG132 significantly blocks MeCP2 turnover (Fig. 3H, lane 3). Taken together, I conclude that RNF4 promotes ubiquitination and turnover of MeCP2 and thus derepresses gene expression.
DISCUSSION
In this study, I provide evidence that RNF4 activates genes that are repressed by DNA methylation. RNF4 does not regulate DNA methylation per se, but mediates the ubiquitination and turnover of a MBD protein MeCP2, which in turn affects gene expression.
RNF4 was initially identified as a transcription co-activator for androgen receptor (11). It was later found to possess E3 ubiquitin ligase activity (19), but no transcription regulators have been identified as RNF4 substrates. Thus, for a long time, people have believed that RNF4 plays a structural role. A more recent study found that RNF4 interacts with TDG and enhances the glycosylase activity of TDG (14), but the role of RNF4 as an E3 ligase was not addressed. This study demonstrates that the E3 ubiquitin ligase activity of RNF4 is critical for transcriptional activation, and further identified MeCP2 as a novel RNF4 substrate. Because the reporter assay revealed that the effect of RNF4 on transcription depends on DNA methylation, the scope of this investigation has been limited to a few MBD proteins (Fig. 3A). In addition to MeCP2, I believe that RNF4 can target other proteins for ubiquitination-dependent turnover to create a permissive transcriptional environment. This study lays the basis for better understanding the role of RNF4 in epigenetics and transcription regulation in the future.
In addition to revealing how RNF4 contributes to transcriptional regulation, I also provide the first evidence that MeCP2 is subjected to regulation of protein turnover. MeCP2 is one of the best studied epigenetic factors, yet surprisingly little is known about the regulation of MeCP2 protein stability. I found that RNF4 promotes MeCP2 ubiquitination and turnover, which leads to many interesting questions. For example, which residues on MeCP2 are ubiquitinated? How many ubiquitin molecules are added to MeCP2? Does ubiquitination directly target MeCP2 for degradation, or does it lead to MeCP2 mislocalization, which in turn results in degradation? Importantly, imbalance of MeCP2 protein level causes neurodevelopmental disorders (20, 21), and it will be interesting to test whether RNF4 has a role in the context of these diseases.
Acknowledgment
I thank Dr. Yi Zhang for support and critical reading of this manuscript.
This article was selected as a Paper of the Week.
- MBD
- methyl cytosine-binding domains
- TET
- Ten-eleven translocation
- 5caC
- 5-carboxylcytosine
- TDG
- thymine-DNA glycosylase
- SIM
- SUMO-interacting motif
- SUMO
- small ubiquitin-like modifier
- qPCR
- quantitative PCR
- MeCP2
- methyl CpG-binding protein 2
- MeGFP
- methylated GFP.
REFERENCES
- 1. Zhou V. W., Goren A., Bernstein B. E. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 [DOI] [PubMed] [Google Scholar]
- 2. Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 [DOI] [PubMed] [Google Scholar]
- 3. Fatemi M., Wade P. A. (2006) MBD family proteins: reading the epigenetic code. J. Cell Sci. 119, 3033–3037 [DOI] [PubMed] [Google Scholar]
- 4. Jones P. L., Veenstra G. J., Wade P. A., Vermaak D., Kass S. U., Landsberger N., Strouboulis J., Wolffe A. P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 [DOI] [PubMed] [Google Scholar]
- 5. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoue A., Zhang Y. (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu H., Zhang Y. (2011) Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moilanen A. M., Poukka H., Karvonen U., Häkli M., Jänne O. A., Palvimo J. J. (1998) Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 18, 5128–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 13. Sriramachandran A. M., Dohmen R. J. (2014) SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75–85 [DOI] [PubMed] [Google Scholar]
- 14. Hu X. V., Rodrigues T. M., Tao H., Baker R. K., Miraglia L., Orth A. P., Lyons G. E., Schultz P. G., Wu X. (2010) Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc. Natl. Acad. Sci. U.S.A. 107, 15087–15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y., Zhang Y. (2014) Regulation of TET protein stability by calpains. Cell Rep. 6, 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nabel C. S., Jia H., Ye Y., Shen L., Goldschmidt H. L., Stivers J. T., Zhang Y., Kohli R. M. (2012) AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L. K., Bartolomei M. S., Rambow F., Bassi M. R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A. J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J. U., Su Y., Zhong C., Ming G. L., Song H. (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Häkli M., Lorick K. L., Weissman A. M., Jänne O. A., Palvimo J. J. (2004) Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett. 560, 56–62 [DOI] [PubMed] [Google Scholar]
- 20. Collins A. L., Levenson J. M., Vilaythong A. P., Richman R., Armstrong D. L., Noebels J. L., David Sweatt J., Zoghbi H. Y. (2004) Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 13, 2679–2689 [DOI] [PubMed] [Google Scholar]
- 21. Samaco R. C., Fryer J. D., Ren J., Fyffe S., Chao H. T., Sun Y., Greer J. J., Zoghbi H. Y., Neul J. L. (2008) A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum. Mol. Genet. 17, 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]