Background: Many functions are subjected to regulation by protein-protein interactions. An example is the SERCA1a-SLN8 complex.
Results: Rabbit and pig SLN have two types of fatty acid anchors (palmitic and oleic acid) attached to an intramembranous cysteine residue.
Conclusion: A role and evolutionary significance of these S-acylations are suggested.
Significance: First demonstration of SLN S-acylation and of the intramembranous S-oleoylation of a membrane protein.
Keywords: Calcium ATPase, Mass Spectrometry (MS), Membrane Protein, Protein Acylation, Protein Palmitoylation, Sarcoplasmic Reticulum (SR), Protein Oleoylation, Sarcolipin
Abstract
Sarcolipin (SLN) is a regulatory peptide present in sarcoplasmic reticulum (SR) from skeletal muscle of animals. We find that native rabbit SLN is modified by a fatty acid anchor on Cys-9 with a palmitic acid in about 60% and, surprisingly, an oleic acid in the remaining 40%. SLN used for co-crystallization with SERCA1a (Winther, A. M., Bublitz, M., Karlsen, J. L., Moller, J. V., Hansen, J. B., Nissen, P., and Buch-Pedersen, M. J. (2013) Nature 495, 265–2691; Ref. 1) is also palmitoylated/oleoylated, but is not visible in crystal structures, probably due to disorder. Treatment with 1 m hydroxylamine for 1 h removes the fatty acids from a majority of the SLN pool. This treatment did not modify the SERCA1a affinity for Ca2+ but increased the Ca2+-dependent ATPase activity of SR membranes indicating that the S-acylation of SLN or of other proteins is required for this effect on SERCA1a. Pig SLN is also fully palmitoylated/oleoylated on its Cys-9 residue, but in a reverse ratio of about 40/60. An alignment of 67 SLN sequences from the protein databases shows that 19 of them contain a cysteine and the rest a phenylalanine at position 9. Based on a cladogram, we postulate that the mutation from phenylalanine to cysteine in some species is the result of an evolutionary convergence. We suggest that, besides phosphorylation, S-acylation/deacylation also regulates SLN activity.
Introduction
Sarcolipin (SLN)2 from sarcoplasmic reticulum (2) is an important protein in the muscle contraction/relaxation process. The activity of SLN is affected by phosphorylation (3, 4). Despite its small size (31 residues, Mr = 3773 Da; Ref. 5), a number of enzymological studies have underlined its regulatory role in connection with the sarcoendoplasmic reticulum calcium ATPase (SERCA1a) activity, such as Ca2+-dependent ATPase activity, Ca2+ affinity to SERCA1a, and Ca2+ transport (4, 6). Recently, Sahoo et al. (7) have shown that SLN primarily lowers the Vmax of SERCA-mediated Ca2+ uptake, but not pump affinity for Ca2+, in agreement with the finding that the Ca2+-dependent ATPase activity at saturating Ca2+ concentration in sealed or unsealed membrane preparations is unaffected by the presence of SLN (8, 9). SLN binds to SERCA1a, as proposed initially by co-purification (2), and later by co-immunoprecipitation (10), and shown more precisely by crosslinking experiments (7, 11). The exact binding site of SLN was recently revealed to be in a groove close to helices M2, M6, and M9 of SERCA1a in two similar three-dimensional structures of the SLN-SERCA1a complex (1, 12). SLN is mainly present as a single transmembrane α-helix, as previously demonstrated by NMR studies of the synthetic peptide in detergent (13–15), but some studies point out its binding to SERCA1a in association with phospholamban, forming a ternary complex (4, 6, 10, 11). However, in rabbits, it is not clear whether this ternary complex is frequent or not. Indeed phospholamban, a peptide homologous to SLN but with a cytoplasmic extension, is mainly present in cardiac muscle while SLN is predominantly expressed in skeletal muscle (7, 16, 17). The ternary association also depends on the animal species, being possible in atria of smaller rodents but not in rabbits or pigs (16).
Only few studies have addressed the stoichiometry of SLN/SERCA1a. Some indicate an approximate 1/1 ratio (see e.g. Refs. 2, 4), which may however vary depending on the species or tissue (16–18). Besides its regulatory role, complemented by site-directed mutagenesis experiments (see e.g. Refs. 4, 6, 7, 10, 11), SLN was also recently implicated as a regulator of muscle-based thermogenesis, based on a mouse knock-out model (19).
The possible link of SLN to thermogenesis is thought to depend on uncoupling of SERCA-mediated ATP hydrolysis from Ca2+ transport (i.e. by the formation of a futile cycle; Refs. 7–9, 20), but also the role of the ryanodine receptor 1 Ca2+ channel must be considered in thermogenesis (19, 21).
In the present report, we demonstrate that rabbit and pig SLNs are fully palmitoylated or oleoylated at a membrane-embedded Cys-9 residue. Oleoylation of a cysteine residue appears to be a novel post-translational modification for a membrane protein. Hydroxylamine treatment of sarcoplasmic reticulum (SR), which leads to partial removal of these fatty acids, increases Ca2+-dependent ATPase activity. We discuss our observations with respect to recently determined crystal structures of the SERCA1a-SLN complex and analyze phylogenetic data on SLN.
EXPERIMENTAL PROCEDURES
Chemicals
Octaethylene glycol mono-n-dodecyl ether (C12E8) was purchased from Nikkol Chemical (Tokyo, Japan), and n-dodecyl-β-d-maltopyranoside (DDM) from Affymetrix (Anatrace detergents and lipids, United Kingdom). TSK3000SW silica gel column was purchased from Tosoh Biosciences (Tessenderlo, Belgium). Products for SDS-PAGE and gel filtration standards are from Bio-Rad. Most of the other chemical products were purchased from Sigma-Aldrich; see below for possible exceptions.
Membrane Preparations
Ca2+-transporting sarcoplasmic reticulum (SR) vesicles were isolated from rabbit skeletal muscle according to published procedures (22, 23). From these preparations, purified Ca2+-ATPase membranes were obtained by extraction of extrinsic proteins with a low concentration of deoxycholate (DOC-extracted SR; Refs. 24). Ca2+-transporting sarcoplasmic reticulum (SR) vesicles were also isolated from pig and rat back skeletal muscle according to published procedures (22), except that the final supernatant after removal of large aggregates was carefully recovered, transferred into ultracentrifugation tubes in the presence of 2 m KCl and 100 mm MgATP, and centrifuged for 90 min at 60,000 × gav (45Ti Beckman Coulter rotor) to achieve extrinsic proteins removal and SR vesicles recovery. As for rabbit SR vesicles, the final pellet containing pig or rat SR vesicles was resuspended in 0.3 m sucrose for long-time storage at −80 °C.
Solubilization and Hydroxylamine Treatment
SR vesicles were suspended at a concentration of 4 mg protein·ml−1 in the appropriate buffer (see figure legends for details, buffer A: 50 mm MOPS-Tris, pH 7.5, 25 mm NaCl, 1 mm MgCl2, and 1 mm CaCl2; buffer B: 50 mm MOPS-Tris, pH7.5, 25 mm NaCl, 50 mm MgCl2) supplemented with 40 mg·ml−1 DDM or C12E8 for solubilization (detergent:protein ratio 10:1 w/w; Ref. 25), and incubated for 10 min at 20 °C. Volume of the sample here is defined as “one volume.” Depalmitoylation can be achieved by addition of one volume of a freshly prepared 2 m hydroxylamine solution (buffered at pH 7.5 with saturated Tris) and incubation for 1 h at 20 °C. For untreated “control” sample, one volume of initial buffer (or 2 m Tris-Cl at pH 7.5) was added instead of hydroxylamine. Hydroxylamine treatments were performed under various conditions: (i) on the detergent-solubilized and SEC-purified enzyme: the ATPase was first solubilized, injected on the column equilibrated with a buffer containing 50 mm MOPS-Tris pH7.5, 100 mm NaCl, 0.4 mg·ml−1 DDM, selected elution fractions were treated with hydroxylamine and finally submitted to a desalting step to decrease the amount of NaCl and hydroxylamine, which may prevent efficient analysis of the samples by MALDI-TOF mass spectrometry; (ii) in treating the native or solubilized ATPase before SEC purification and to purify the complex on a column initially equilibrated with the above buffer but with only 25 mm NaCl. This allowed us to omit the additional desalting step which was time consuming and deleterious for some samples.
Delipidation and Elution of the SERCA1a-SLN Complex in the Presence of Detergent
Solubilized material was recovered after centrifugation for 10 min at 50,000 × g on a TLA100.3 rotor (Beckman coulter TL100 ultracentrifuge) and injected on a detergent equilibrated size exclusion column (TSK3000SW silica gel column, 50 mm MOPS-Tris at pH 7, 25 mm NaCl, and 0.4 mg·ml−1 DDM unless otherwise stated in the figure legends), essentially as previously described (26). Two 1-ml fractions were collected; one corresponding to the major peak known to contain SERCA1a (Ve ∼6.5–7.5 ml), and a second fraction corresponding to the end of the previous elution peak (Ve ∼8.0–9.0 ml), containing mainly mixed micelles and most of the SR endogenous lipids (27). Selected fractions were used for SDS-PAGE and MALDI-TOF analysis (see below and figure legends). For SDS-PAGE, two methods were used to visualize proteins after electrophoresis. The first technique consists of a classical Coomassie Blue staining (40% methanol (v/v), 10% acetic acid (v/v), and 0.1% Coomassie Blue R250 (w/v)). The second technique involves a light-induced modification of the tryptophanyl residues by haloalkane compounds (Mini-Protean TGX Stain-free Precast 4–20% gels from Bio-Rad) (28, 29). Whereas tryptophan has a fluorescence emission spectrum in the UV domain (about 310–350 nm, depending on its environment), the covalent indole derivative obtained after the reaction is now visible at a wavelength of about 410–420 nm. Assuming that the environment of each tryptophan in the SDS-containing gel is the same, the resulting in-gel fluorescence intensity can be used to estimate protein stoichiometries if the number of tryptophans in the sequences is known (29). Gels can be stained with Coomassie Blue in a subsequent step, following the above described procedure. Note that before assessing the stoichiometry of SERCA1a and SLN, which contain thirteen and only one tryptophan, respectively, this stain-free technique was validated on another complex purified in the laboratory.3
Analysis by MALDI-TOF Mass Spectrometry
1 μl of sample was mixed with 3 μl of saturated solution of sinapinic acid in 30% acetonitrile, 0.3% trifluoroacetic acid. 1 μl of the mixture was loaded into a MALDI-TOF spectrometer (Perseptive Biosystems, Voyager STR-DE) equipped with a nitrogen laser (337 nm). Spectra were obtained in linear mode using delayed extraction. Two ranges were analyzed: 2000–6000 Da for SLN alone (accelerating voltage of 20 kV) and 3000–140,000 Da for SLN and ATPase (accelerating voltage of 25 kV). The external standards used for calibration in the range 2000–6000 Da were Escherichia coli thioredoxin and insulin.
Carbamidomethylation of Samples
SEC-purified samples (2 μl at about 1 mg·ml−1), treated or not with NH2OH were incubated with 50 mm iodoacetamide in 50 mm ammonium bicarbonate, for 15 min in the dark at room temperature. The reaction was stopped by addition of the matrix (sinapinic acid in 30% acetonitrile, 0.3% trifluoroacetic acid) and analyzed by MALDI-TOF as above.
N-terminal Sequencing
The following method was previously described (30): following SDS-PAGE separation of proteolytic peptides (performed on DOC-extracted SR vesicles) on Tricine gels (31) and transfer of the proteins onto a PVDF membrane, the bands corresponding to the various peptides were cut from the stained membranes and analyzed by Edman degradation. The sequence corresponding to the first 8 residues of SLN was found in the 6 kDa region.
Steady-state ATPase Activity Measurements
These measurements were performed using a classical enzyme-coupled assay, by measuring the rate of NADH oxidation (32, 33). The initial buffer for the assay was 50 mm Tes-NaOH pH 7.5, 1 mm MgCl2, 100 mm KCl, 5 mm MgATP, 1 mm phosphoenolpyruvate, 0.1 mg·ml−1 lactate dehydrogenase, 0.05 mg·ml−1 pyruvate kinase, and 0.2–0.3 mm NADH (to reach about 1.5–1.7 OD at 340 nm). In some cases, 1 mg·ml−1 C12E8 or 2 μg·ml−1 of the calcium ionophore A23187 (unless otherwise stated in figure legends) was added, depending of the conditions of the experiment (see text and figures legends for details). The initial concentration of free Ca2+ was about 50 μm. Low concentrations of free Ca2+ were obtained by buffering with EGTA, using the following apparent dissociation constant for Ca2+ at pH 7: Kd Ca-EGTA ∼0.04 μm. Free Ca2+ concentrations were also calculated with the maxchelator program (Ref. 34; maxchelator.stanford.edu). Values of specific activities indicated are the mean of two or three assays and error bars show the distribution of the values. When error bars are not visible, it means that they were smaller than the symbols.
RESULTS
Rabbit SLN Is Palmitoylated/Oleoylated on Cysteine 9
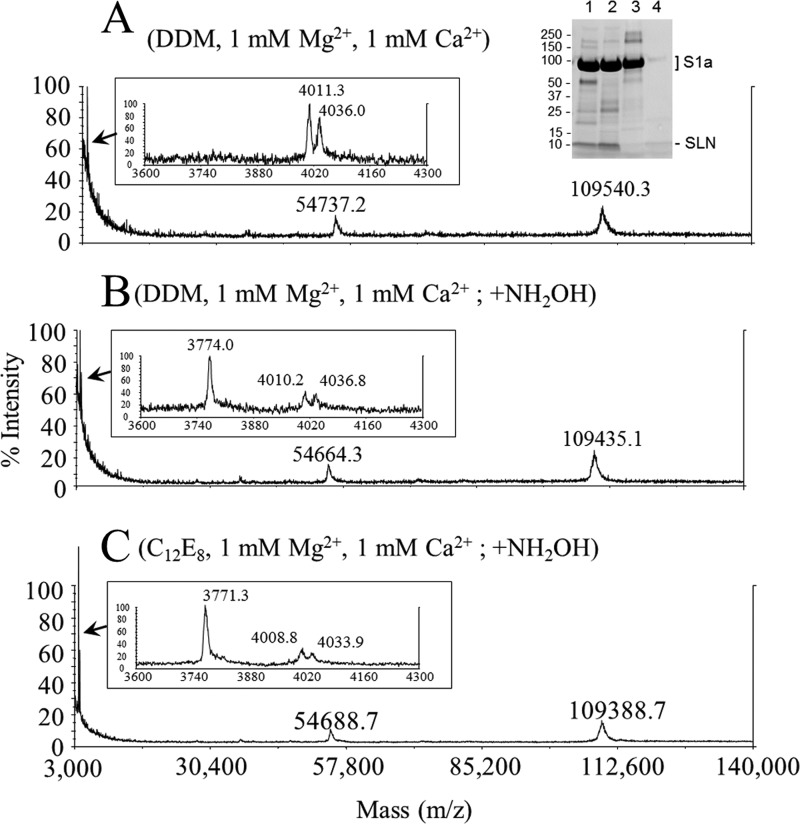
MALDI-TOF mass determination of proteins in SR vesicles followed an established procedure (26). In short, SR vesicles were solubilized under different conditions and loaded on a detergent-containing SEC column (Fig. 1). Besides a small peak of aggregated SERCA1a in the void volume, solubilized Ca2+-ATPase elutes in a single peak as monomeric Ca2+-ATPase (25). A mixed micelle peak containing most of the lipids elutes after the main peak as smaller sized particles. Removal of endogenous lipids, use of low salt concentration and tight control of detergent concentration in the column were found to be the key points for obtaining mass spectrometry spectra by MALDI-TOF of hydrophobic peptides and large membrane proteins, such as SERCA1a. The spectrum obtained for the monomeric Ca2+-ATPase peak (Fig. 2A) revealed essentially the mass of intact Ca2+-ATPase, which, deduced from the amino acid sequence, is 109,532. The mass obtained by MALDI-TOF was 109,493.9 ± 93.1 Da (average of ten independent experiments, taking into account the detection of the monoionized species at about m/z ∼109,500 together with its twice ionized equivalent at about m/z ∼54,750). Two peptides of m/z ∼4,011 and 4,036 were also present in the main SERCA1a peak (Fig. 2A, left inset). We postulated that these correspond to SLN (Mr = 3774 Da, mass of the protonated form ((M + H)+), which interacts with the Ca2+-ATPase (see references in the introduction), but with some post-translational modifications, which would lead to their increased mass. Cysteine 9, the only cysteine in the rabbit SLN sequence (inset of Fig. 4), was an obvious choice to look for as the site of this potential modification. To test this hypothesis, we treated the sample with 1 m hydroxylamine for 1 h (35), a classical treatment for removal of cysteine-bound fatty acids; this was performed in a number of conditions, either before (not shown) or after (Fig. 1, B and C) detergent solubilization, followed by gel filtration and mass spectrometry (Fig. 2, B and C), with essentially the same results. After hydroxylamine treatments we found that the mass corresponding to a large part of the peptides was reduced to that of unmodified SLN. Furthermore, the mass differences that we found before and after the hydroxylamine treatment, i.e. between 4011/4036 and 3774 are 238 Da and 264 Da, (average of 10 experiments), matching a linked palmitic acid, C16:0, and an octadecenoic acid, C18:1, most probably the cis-9-octadecenoic acid derivative also commonly referred to as oleic acid.
FIGURE 1.
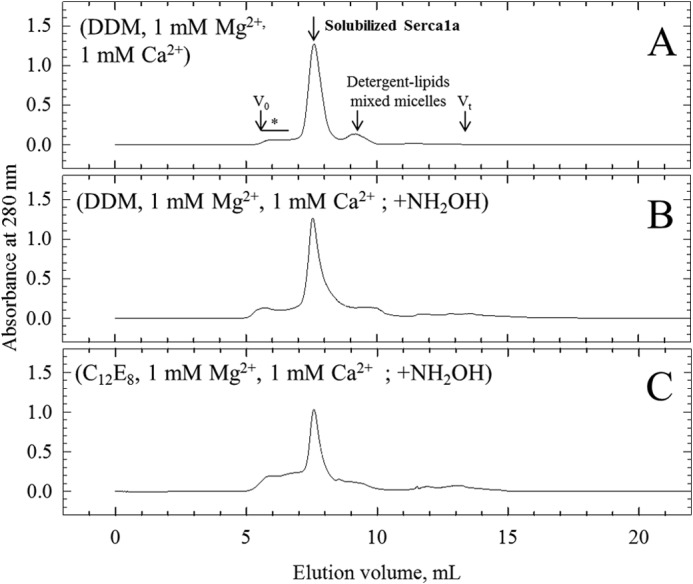
Delipidation and purification of a SERCA1a-SLN complex by size exclusion chromatography. The TSK 3000SW silica gel column was pre-equilibrated and eluted in 50 mm MOPS-Tris, pH 7.5, 25 mm NaCl, 0.4 mg·ml−1 DDM, supplemented with 1 mm MgCl2 and 1 mm CaCl2, at room temperature (20–23 °C). Conditions followed for sample preparations before injection on the column are indicated in parentheses for each panel with full description under “Experimental Procedures.” DDM or C12E8 was used at 40 mg·ml−1 and NH2OH was used at 1 m. Arrows indicate peaks corresponding to monomeric solubilized ATPase and to detergent-lipid mixed micelles. Asterisk indicates the region of the chromatogram where multimeric forms and aggregates are eluted. Excluded volume (Vo) and total volume (Vt) of the column are also indicated.
FIGURE 2.
MALDI-TOF analysis of purified fractions. Fractions corresponding to the elution peak of solubilized SERCA1a (see Fig. 1) were collected, and an aliquot of each was submitted to MS analysis. Spectra were acquired either in a large range of molecular masses (m/z from 3000 to 140,000 Da) or on a smaller range, from 2000–6000 Da (zoom in the 3600–4300 Da region in the left insets). Conditions for sample preparations, before (A) or after (B, C) hydroxylamine treatment are indicated on each spectrum. The precision on the mass determination is quite high considering the large range studied and the resulting difficulty to achieve an accurate calibration in these particular acquisition conditions: the experimental mass for SERCA1a is about 109,493.9 ± 93.1 Da for an expected mass of 109,532 Da, and the experimental mass for SLN is about 3772.0 ± 1.1 Da for an expected mass of 3774 Da (NB: in this figure and in all the other figures we report the masses of the protonated forms (M + H)+). Right inset of panel A: SR vesicles (lane 1), DOC-extracted SR vesicles (lane 2), SEC purified SERCA1a/SLN complex (lane 3), and the mixed micelles peak (lane 4) (Ve = 6.5–7.8 ml and Ve = 8.6–9.6 ml, respectively) were loaded on a TGX Stain-Free Precast 4–20% SDS-PAGE for in-gel fluorescence analysis (Bio-Rad). A band corresponding to SLN was visualized at an apparent molecular weight of 8–10 kDa as expected (see “Results”). In this experiment about 20–25 μg of protein has been applied in each lane. Under these conditions the Ca2+-ATPase band is oversaturated but SLN can easily be spotted. For quantitative comparison, dilutions of the same samples were loaded too (not shown). A profile analysis was done to estimate the intensity of fluorescence of SERCA1a and SLN, which respectively contain 13 and only one tryptophan residue. Treatment by hydroxylamine had no effect on the SERCA1a/SLN ratio (not shown).
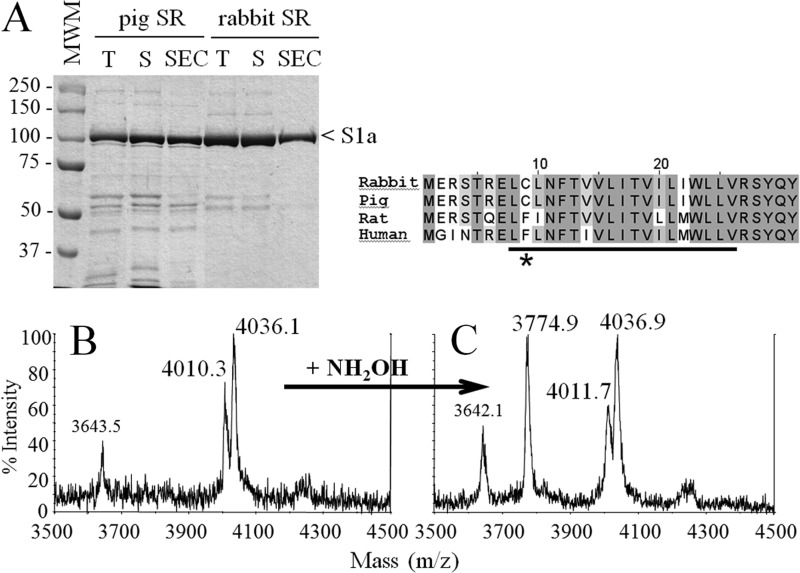
FIGURE 4.
Analysis of pig SR. Purification of the putative pig SERCA1a-SLN complex was achieved as for the rabbit SR (see procedure for Fig. 1). A, Coomassie Blue stained SDS-PAGE of the purified samples. T corresponds to the total fraction before solubilization by DDM, S corresponds to the supernatant, which contains solubilized proteins after centrifugation, SEC corresponds to the SEC-purified fraction that contains the purified SERCA1a-SLN complex. S1a indicates SERCA1a. In pig SR, the minor band running immediately below the Ca2+-ATPase is phosphorylase, not present in the rabbit sample because the latter were fasting 1 day before sacrifice (54). The molecular weight marker (MWM) used was Prestained Plus Precision All Blue Standard from Bio-Rad. Samples were loaded on a 8% Laemmli type Tris-glycine SDS-PAGE. B and C, MS analysis of the SEC-purified DDM-solubilized pig SR without or with treatment with NH2OH (panels B and C, respectively). Spectra were acquired either on a large range of masses as in Fig. 2 (i.e. on a m/z range going from 3000 to 140,000 Da) which allows simultaneous detection of SLN and of SERCA1a (data not shown) or on a smaller range, from 2000–6000 Da (only windows going from 3500 to 4500 Da are shown here). Panel B corresponds to the untreated and panel C to the hydroxylamine-treated sample. The peak at 3643 Da is likely to be an impurity, in line with the less pure pig SR. Right inset, sequence alignment of rabbit, pig, rat, and human sarcolipin. Conserved amino acids are highlighted in gray. The asterisk indicates cysteine or phenylalanine at position 9. The transmembrane region expected to match with the hydrophobic part of the lipid bilayer is underlined (see also Fig. 6B).
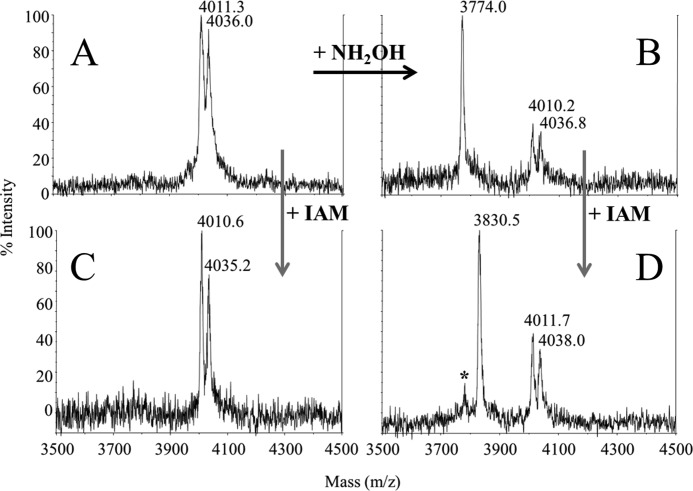
Judging from the ratio of the various peaks in Fig. 2, B and C, the yields of depalmitoylation and deoleoylation were very high, reaching values of about 80% both after solubilization in DDM or C12E8 and low Mg2+ concentration. It was however less (about 30%) when hydroxylamine treatment was performed directly on the membrane or in DDM or C12E8 and high Mg2+ concentration (data not shown). No or very little unmodified SLN was detected in untreated samples (Fig. 2, panel A) or in a MALDI-TOF analysis of the detergent-lipid mixed micelles peak, which also contained SLN (not shown). This suggests that almost all the SLN present in the membrane, whether associated to SERCA1a or not, is modified, and that the fatty acids additions are not restricted to those in association with SERCA1a. The overall ratio between SLN and SERCA1a cannot be inferred from the peak sizes in MALDI-TOF experiments, but a rough estimate can be made from SDS-PAGE after modification of tryptophan by haloalkanes, or by Coomassie Blue staining (right inset of Fig. 2A). The comparison of the amount of SLN present before and after gel filtration indicates that a significant amount (about 70–80%) of the SLN is removed from the SERCA1a peak and is present in the mixed micelles peak (right inset of Fig. 2A). To further assess that cysteine 9 is the residue bearing the post-translational modification, we treated the proteins with iodoacetamide before or after hydroxylamine treatment (Fig. 3). Iodoacetamide only affected the peak corresponding to the deacylated SLN, with an expected increase of 57 Da corresponding to a carbamidomethylation of the free sulfhydryl group.
FIGURE 3.
Hydroxylamine and iodoacetamide treatment to unambiguously identify Cys-9 as the modified residue. Samples from the experiments depicted in Fig. 2, A and B were treated with iodoacetamide to unambiguously identify the palmitoylated/oleylated residue (panels A and B, respectively). As described above, 2 peaks were detected in the untreated sample (panel A; m/z = 4011.3 and 4036.0 Da). Treatment with hydroxylamine (+ NH2OH, black arrow) resulted in the apparition of a unique peak at m/z = 3774.0 Da suggesting that post-translational modifications are present on a cysteine residue. Differences in masses of the peaks before and after treatment correspond to the mass of a palmitic acid and an oleic acid (Δm/z = 236.2 and 262.8, respectively). Iodoacetamide was added on the two samples (untreated and hydroxylamine treated, +IAM, gray arrows). As depicted in panels C and D, iodoacetamide only affected the peak corresponding to the deacylated SLN whose m/z increased from 3774.0 to 3830.5 Da: the difference should correspond to the acetamide addition to the SH side-chain of the cysteine. IAM reaction was probably not total, as suggested by the presence of some residual unmodified SLN (*).
Crystallization Conditions and Sample Preparation Are Not Responsible for the Absence of Visible Palmitate/Oleoate on the Recent SLN/SERCA1a Complex Structure
Because no cysteine modification was observed and modeled in the recent sarcolipin/SERCA1a crystal structures (1, 12) we wondered if the crystallization conditions could have promoted the loss of the fatty acids. Indeed palmitoylations are reversible reactions, and the enzymes necessary for palmitate addition or removal are present in the endoplasmic reticulum (for review, see Refs. 36, 37). Alternatively, the crystallization conditions may have favored the segregation of non-acylated SLN. Therefore, to approach the crystallization conditions in our experiments, we increased the Mg2+ concentration to 50 mm in the absence of Ca2+, during both the solubilization and the gel filtration steps, and we also changed the detergent DDM for C12E8. Apart from the formation of a larger amount of aggregates in the C12E8/50 mm Mg2+ condition, the MALDI-TOF results showed also full palmitoylation/oleoylation of the purified SLN (data not shown). This demonstrates that the fatty acids remain attached to SLN even in conditions close to the preparation of samples for crystallogenesis. We then wondered if the presence of fatty acid was linked to individual differences between rabbits used for the different SR preparation, due for instance to a difference in the feeding protocols or due to a slightly different preparation method of SR. The initial experiments (Figs. 1 to 3) were performed from rabbits raised in France. Using SR and DOC-extracted SR vesicles prepared in Denmark for the crystallization trials we obtained the same results as before, including about the same palmitate/oleate ratio (data not shown). Thus, we conclude that our initial observations were not linked to specific individuals or preparation procedures. Furthermore we conclude that SLN is fully palmitoylated/oleoylated prior to crystallization. We noted also that the Ca2+-ATPase was not very stable under such crystallization conditions while it is stable in the other conditions, even after hydroxylamine treatment (data not shown). The absence of the modification in the two recent crystal structures is likely the consequence of a large flexibility of the fatty chains in the crystal, preventing their observation in the crystal structure (see “Discussion”).
Pig SLN Is Also Palmitoylated/Oleoylated
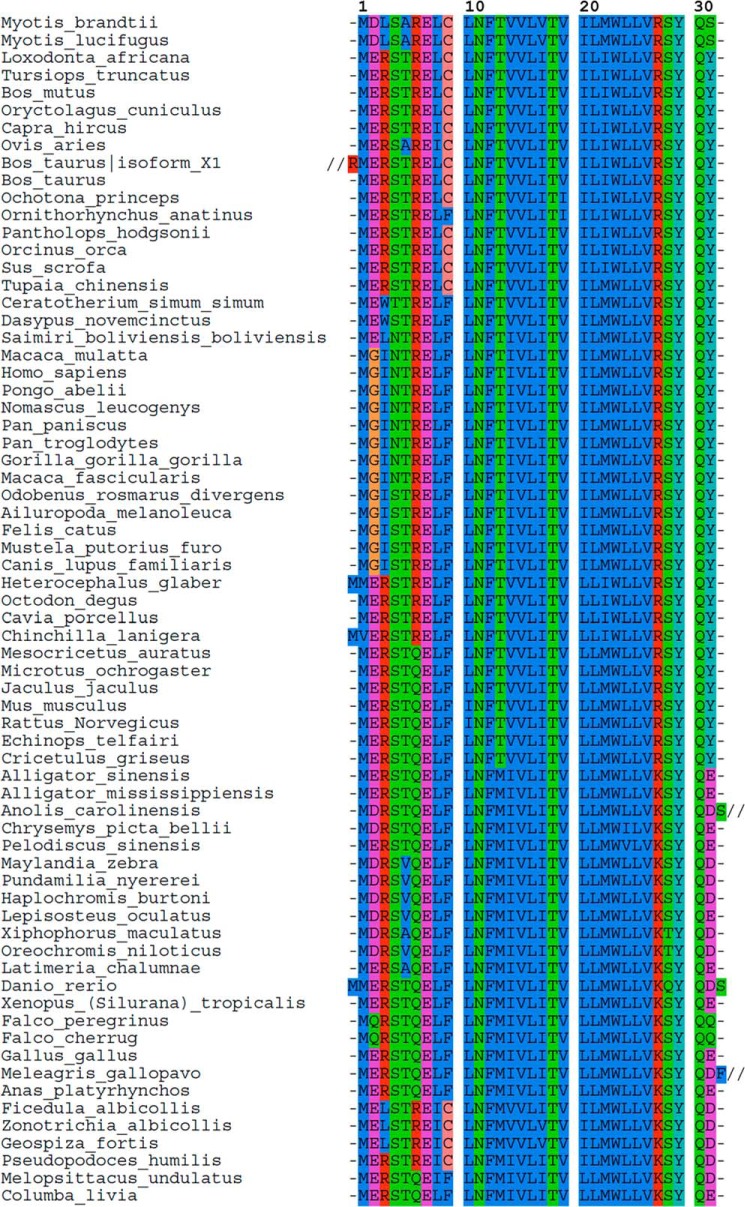
Interestingly, many mammals have a phenylalanine instead of a cysteine in position 9 (see below for discussion on this subject). Considering the available sequences in protein sequence databases (Uniprot KB and NCBI; see Fig. 7 legend for the detailed procedure), we chose pig as an alternative species bearing a cysteine to run similar tests. Pig SLN has the same sequence as the rabbit (right inset of Fig. 4). The SR preparation led to SDS-PAGE (Fig. 4A) and gel filtration profiles (not shown) that reveal a less pure preparation: a number of additional bands were present (compare pig or rabbit SR in Fig. 4A). Nevertheless, after gel filtration the SLN associated to the pig Ca2+-ATPase was clearly detected with the same two acylations, essentially also fully modified, with the noticeable feature that the oleoyl peak was larger than the palmitoyl peak in this case, representing about 60% of the peaks (Fig. 4B). As for the rabbit samples, hydroxylamine treatment led also to partial depalmitoylation/deoleoylation of pig SLN (Fig. 4C).
FIGURE 7.
Alignment of available sarcolipin peptide sequences. The human SLN sequence (Uniprot no. O00631) was used as a template for a BLASTp search in the non-redundant protein sequence database (55). The PAM-30 algorithm was used as it is recommended for analysis of short peptide sequences. Following this strategy, 110 sequences were retrieved from the resulting taxonomy report, covering 67 species. However, when several sequences were available in the database for a given species (e.g. 13 sequences for human), all the available sequences were exactly the same, except for the cow and the ferret. In these two cases, a second SLN isoform was predicted from whole genome sequencing. Those sequences are identical in the membrane part but have putative additional residues in the N and/or C termini. The alignment presented here was done with the Seaview program (56, 57). Three sequences were shortened to optimize the figure presented here: Bos taurus isoform_X1, residues M1SSLFASQSEKALPRAQKSRQPGRQEGVGAQVGEREEKPIAAQCVLDPIQEVRTSRRSLVCPQKFAFSSHQATL75; Anolis carolinensis, residues R33KEARRCGEWGAGFLMPPMMQGNLVAE59; Meleagris gallopavo, residues L33FAKDTLEERWLSVFTGFCHQPFEV57.
Deacylation Increases the Ca2+-ATPase Activity
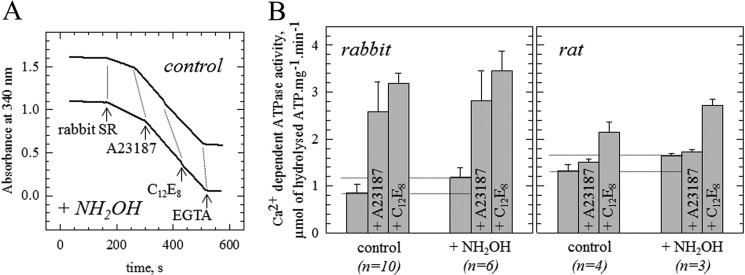
Finally, we checked if the presence of palmitate/oleate affected the Ca2+-dependent ATPase activity of rabbit, pig, and rat SERCA1a, (the sarcolipin of the latter cannot be acylated as it contains a Phe instead of a Cys residue at position 9, see inset of Fig. 4). We found that NH2OH treatment has a small effect on the activity of the detergent-solubilized enzyme, and on the activity of SR vesicles treated by the ionophore A23187 for the three mammals, whereas the affinity of SERCA1a for Ca2+ was unaltered (data not shown; tested only for the rabbit). In contrast, the Ca2+-ATPase activity was increased in the intact SR vesicles, by about 30% for the rabbit and 20% for the rat (Fig. 5, data not shown for pig SR, which, in a single experiment, was also increased).
FIGURE 5.
Hydroxylamine treatment of SR membranes results in an increase of calcium ATPase activity. A, ATPase activity. Rabbit SR membranes were initially suspended at 4 mg·ml−1 in the assay buffer and stored on ice. When employed, an equal volume of a freshly prepared 2 m hydroxylamine solution (NH2OH, pH adjusted at 7.5 with saturated Tris) was added prior to incubation at 20 °C for 1 h. As a control, an equal volume of buffer was added before incubation at 20 °C for 1 h. ATPase activity was estimated by an enzyme-coupled assay as described under “Experimental Procedures.” The reaction was started by addition of 5 μg·ml−1 ATPase (SR) to record the basal activity. It was followed by successive additions of 5 μg·ml−1 of the A23187 ionophore (A23187) and 1 mg·ml−1 C12E8 to reach maximum activity (in absence of any accumulation of calcium). The Ca2+-independent activity was estimated after addition of 1 mm EGTA. Note that this Ca2+-independent activity was systematically lower after treatment with hydroxylamine, suggesting that it had inactivated some contaminants. For presentation purpose, one of the traces was manually lowered by −0.5 units of absorbance. The initial “true” OD was almost the same for all the assays. When the SR membranes were treated with 1 m hydroxylamine, the final concentration of hydroxylamine in the cuvette was 5 mm. For comparison, 5 mm hydroxylamine was added to the cuvette prior to addition of the untreated SR. Note that we observed that 5 mm hydroxylamine slightly decreases the rate of regeneration of ATP (data not shown). B, effect of hydroxylamine on ATPase activity in rabbit and rat SR. Experiments depicted in panel A for rabbit SR were repeated 10 times for control samples and 6 times for hydroxylamine-treated samples, and are summarized in the left panel. The same series of experiments was done with rat SR and is presented in the right panel (n = 4 for control and n = 3 for hydroxylamine pre-treated membranes).
DISCUSSION
There is an expanding list of mammalian proteins shown to be modified by S-palmitoylation (38), some of them being integral membrane proteins (39). Recent interesting additions in the context of skeletal muscle is a report describing palmitoylation of the rabbit ryanodine receptor RyR1, the α-subunit of the L-type calcium channel CaV1.1 and SERCA1a (40). For some species, we can now add SLN (see below). Furthermore, we report the presence of a rare modification, namely oleoylation in addition to palmitoylation of SLN. While oleoylation has been observed on the N-terminal methionine and on a lysine of the aquaporin AQP0 (41), this is to the best of our knowledge the first demonstration of oleoylation on a cysteine of a membrane protein. In addition, in the case of the AQP0, the two palmitoylated/oleoylated amino acid side chains are localized at the membrane interface. Surprisingly, in the case of SLN, the cysteine 9 side chain is about 8–10 Å below the membrane interface and neighboring Trp-931 from SERCA1a, as can be judged from the recent crystal structures, suggesting that the added acyl chain is diving deep into the membrane (Fig. 6A). This is also further demonstrated from a molecular dynamics simulation of SERCA1a in a lipid environment (42) as shown in Fig. 6B. At present we cannot provide any explanation for the puzzling observation that about half of SLN carries a palmitate and the other half an oleate. One could assume that oleoylation could replace palmitoylation of a membrane protein with no particular functional consequences. Concerning the proposed extensive palmitoylation of SERCA1a reported by Chaube et al. (40), we cannot confirm it since the molecular mass that we obtained for this enzyme by MALDI-TOF (Fig. 2) corresponds exactly to that of the unmodified enzyme (except for the already described N-terminal acylation (43)). Furthermore, in previous works we identified peptides 1–119 or 1–181 (26, 44) and 351–505 (45) as non-modified, which excludes the cysteine residues at position 12, 364 and 471 from the list given by (40). Also nearly full DTNB modifications of all 19 reduced cysteines of SERCA1a have been described in the past (46). Nevertheless, we cannot completely exclude a partial modification of one or two cysteines. Pig and rat SERCA1a appear also unmodified (data not shown). N-terminal sequencing experiments of rabbit SR in the low molecular mass region of Tricine-PAGE gels (about 6000–10,000 Da), revealed the presence of a peptide with the following sequence: MERSTREL, corresponding to SLN. Interestingly, after the first 8 amino acids, the sequencing process stopped, which gave a hint of a post-translational modification of the cysteine at position 9.4 MacLennan et al. were the first to purify SLN and found 143–198 nmol of bound fatty acid per mg of protein, which corresponds to 0.6–0.8 molecules of fatty acid per peptide, but later the idea of a modified SLN was abandoned, because Wawrzynow et al. reported that, by fast atom bombardment mass spectrometry, the intact mass of SLN was found (5). However, they performed the purification and analysis under somewhat severe conditions and the final yield of SLN was only 1–2%, which leads to the possibility that only a minor subfraction was studied in their mass spectrometry experiments. In our case, we obtain about 100% modification with a ratio palmitate/oleate of 60/40 in rabbit and an opposite ratio in pig. It is interesting to note that there are programs available which, based on sequence data, predict the probability of palmitoylation: thus with the CSS-Palm 3.0 program we obtained for Cluster C a score of 3.686 with a cutoff of 0.951, i.e. SLN has, by this criterium, a high probability to possess a palmitoylation site (47).
FIGURE 6.
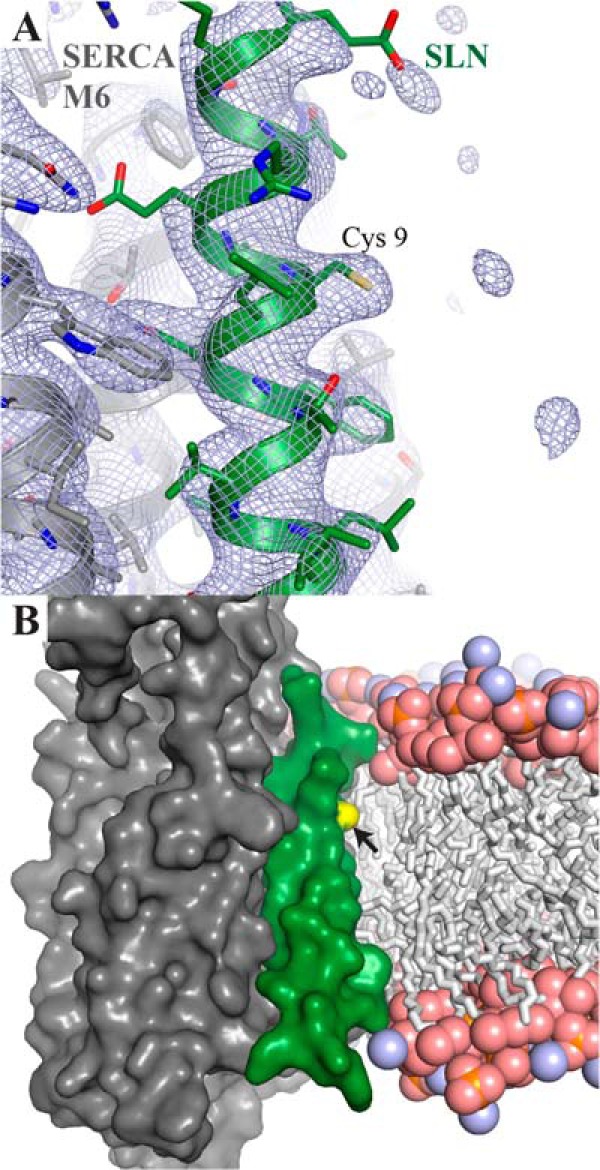
Zoom-in on the sarcolipin Cys 9 side chain in the SERCA-SLN complex structure (PDB ID 4H1W). A, sarcolipin and SERCA1a are depicted with a green and a gray cartoon backbone, respectively, with the Cys-9 side chain in stick representation. The blue mesh represents the final refined 2Fo-Fc electron density map, contoured at 1.0 σ within a radius of 10 Å around SLN. The Cys-9 side chain is well defined in the density, but there is no indication for an attached acyl moiety. This does, however, not rule out the presence of a modification, since local structural disorder leads to a loss of diffraction signal, making highly mobile elements invisible in electron density maps. B, SERCA1a-SLN complex in a surface representation, colored as in panel A. Cys-9 (arrowhead) is highlighted in yellow. The position of the lipid bilayer is indicated by a ball-and-stick representation of a POPC bilayer (carbon, oxygen, nitrogen and phosphorus colored in gray, light red, light blue, and orange, respectively), derived from molecular dynamics simulations of SERCA in a lipid environment (42).
Co-crystallization of SERCA1a with SLN was only obtained recently (1, 12), thus SLN is only found in two of the 53 structures deposited in the Protein Data Bank, even though SLN has probably always been present in the samples as also confirmed here. However, despite a reasonable resolution (3.0–3.1 Å) for the two SLN complex structures, no indications were observed of the acylations that we describe here. A careful re-examination of the two crystal structures shows electron density for the α and β carbon from the cysteine side chain whereas the final sulfhydryl group becomes invisible (see Fig. 6A). This is typical for local disorder and suggests a high flexibility of the linkage between the cysteine β–carbon and its modified sulfhydryl group. A possibility for the rare occurrence of well-diffracting SERCA1a-SLN complex crystals, is that there could be flexibility of the binding interfaces of both partners. Finally, micro-heterogeneity due to two different fatty acids, i.e. the simultaneous presence of both a palmitate and oleoate chain in the crystal, as well as a SLN/SERCA1a ratio, which is perhaps different from 1 (mol/mol) could influence order in the crystal. Although some reports point to a 1:1 ratio in rabbit skeletal muscles (4) others have measured rather different ratios in other muscle tissues or in different species (16, 17). For instance, according to these authors, rodents have very little sarcolipin in skeletal muscles, a fact that we could verify since we could not detect rat SLN associated to SERCA1a in mass spectrometry experiments (the main peak observed was rat SERCA1a with a mass of 109,518 Da and no peak in the region 3000–4000 Da, data not shown). Vangheluwe et al. pointed out that a nearly 3-fold excess of SERCA over SLN is present in rabbit extensor digitorum longus (16). In our experiments we cannot be very confident about the amount of SLN associated to SERCA1a, because quantification is difficult given the size differences between the two proteins. The use of tryptophan modification by haloalkane after SDS-PAGE (right inset of Fig. 2A) led us to a rough estimate of only 1 molecule SLN for 10 molecules of SERCA1a after gel filtration but close to 1/3 before gel filtration i.e. in the crystallization conditions (1): this was measured based on the ratio of tryptophan between the two proteins, which is supposed to be a more reliable parameter than the Coomassie Blue staining to reach quantitative estimate of proteins (28, 29). Our attempts to suppress SLN binding by performing SERCA1a solubilization under different conditions (high lipid content, or additional presence of high Ca2+ and AMPPCP to stabilize a Ca2·E1·AMPPCP intermediate, or presence of beryllium fluoride or thapsigargin to stabilize E2P- or E2-like forms) were unsuccessful, as judged by the Coomassie Blue staining of Tricine gels (data not shown). This is somewhat surprising since the binding interface is state-specific (1) but not so surprising if one takes into consideration the crosslinking results of (7) showing that SLN can bind to various kinetic states of SERCA pump. We also noted that the palmitate/oleate ratio was not modified either under these various conditions.
What is then the role of palmitoylation/oleoylation? Charollais and van der Goot have reviewed a number of processes that are regulated by palmitoylation such as membrane protein folding, conformation of transmembrane domains (e.g. tilting of α-helices), trafficking, targeting, and association with specific membrane domains, the interplay with other post-translational modifications and interactions with other proteins (39, 48). Palmitoylation is reversible and the involved enzymes present in various membranes including the sarco/endoplasmic reticulum (36, 37). Thus, acylation/deacylation (when Cys-9 is present) is a regulatory modification of SLN, similar to SLN phosphorylation (3, 4) but this remains mainly an open question for the future. Phospholemman, a transmembrane peptide member of the FXYD family (49) and somewhat analogous to SLN, inhibits the Na+,K+-ATPase in a palmitoylation-dependent fashion (35). In our hands, depalmitoylation through hydroxylamine treatment performed on SR membranes led to an increase of the Ca2+-ATPase activity in SR vesicles of 30% for rabbit, slightly more than for the rat (20%, Fig. 5). Given the fact that rat SLN cannot be acylated, and is present in only low amounts in noncardiac muscles in this species (Ref. 16, 17 and present study), the reason for this effect in rat SR should be different from the SLN-induced uncoupling of Ca2+ transport that has been suggested for rabbit SR membrane under different contexts (7–9, 20, 35). As to a possible involvement of the ryanodine receptor, which is also Cys-palmitoylated, it should be noted that according to Chaube et al. (40) the effect of depalmitoylation of this protein by bromopalmitate addition is to reduce, rather than to increase Ca2+ efflux from SR, and so hydroxylamination will be expected to be counterproductive to an increased ATPase activity. We thus have to conclude that the matter cannot be definitely settled at this time, but may indicate hydroxylamine-sensitive modification of SLN and so far unidentified component(s) of SR. It should also be considered that the SLN/SERCA1a molar ratio is less than 1 in rabbit (Fig. 2A, right inset), that the hydroxylamine treatment does not lead to full depalmitoylation of SLN (Fig. 2), and that other hydroxylamine-sensitive components of the SR membrane may affect the ATPase activity.
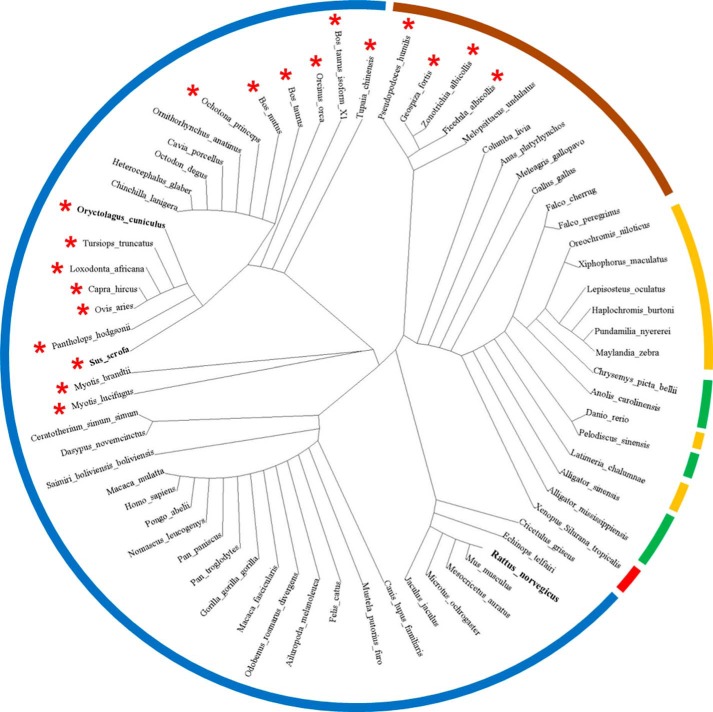
SLN is generally well conserved along the transmembrane helix sequence (Fig. 7). The cysteine at position 9 is present in e.g. rabbit and pig SLN but replaced by a phenylalanine in most species (19 and 48 out of 67 species total, respectively, see Fig. 7). As depicted in the cladogram (Fig. 8), most of the cysteine-containing sequences from our set of data are found in mammals. Among rather closely related species, such as in bats and primates, one would expect the same amino acid, but cysteine is present in the former and phenylalanine is present in the latter, which suggests a rather recent mutation. Interestingly, our analysis showed that four bird species also express a cysteine-containing SLN. The most parcimonious interpretation is that all these sequences are orthologs that have evolved from a common single ancestor (50). We did not find any organism harboring two sequences of SLN paralogs, one with a cysteine and the other with a phenylalanine. Under that assumption, we can deduce from the distribution of sequences containing a cysteine in the cladogram that the mutation from phenylalanine to cysteine is the result of an evolutionary convergence: distant or close species living in different environment may sometimes explore the same set of mutations which may or may not have an impact on the function of a given protein. The hypothesis of a convergent evolution is reinforced by the observation that the cysteine was coded in the four birds by the codon TGC, while it was TGT in the mammals. Interestingly, in a general analysis of many membrane proteins, Mokrab et al. (51) have noted that, specifically for transmembrane α-helices as it is the case here, cysteine and phenylalanine are often substituted by isoleucine, leucine, or valine, but also that phenylalanine is never substituted by a cysteine and that the opposite substitution is also very rare. This general analysis argues against Cys/Phe substitutions at random and further hints at an important role for that exact variation observed at position 9 in SLN. High conservation of SLN transmembrane region (starting at position 8, see Fig. 7), suggests a very high purifying selection as tested by the Ka/Ks criteria (the value obtained is well below 1 as expected: average Ka/Ks ± S.E. is 0.17 ± 0.07 n = 130). Furthermore, it is generally observed that α-helices of membrane proteins facing the lipid environment have a higher mutation rate than e.g. those facing the inside of the protein (51–53). By this criterion, the complete conservation of the SLN hydrophobic α-helix, is intriguing, and may reflect that it may also have other interaction partners in the membrane besides SERCA1a, e.g. the ryanodine receptor, or it could be exploring an oligomeric state like phospholamban when not bound to SERCA1a.
FIGURE 8.
Cladogram representation of available SLN sequences. Sequences used as template for the cladogram construction are presented in Fig. 7. Sequence alignment and tree construction were done with the PhyML program starting from the 67 unique sequences (57). Prior to tree construction, sequence alignment was analyzed with the Gblock program to eliminate gaps and generate clearly defined flanking regions. The dendroscope program (58, 59) was used to enhance the visualization of the generated tree presented here. The color code is the following: amphibians in red, reptiles in green, fishes in brown, birds in orange, and mammals in blue. Cysteine-containing sequences are indicated with a red asterisk. Rabbit, pig, and rat SLN are bolded.
Acknowledgments
We thank Raphaël Guérois (iBiTec-S, UMR 8221, CEA-Saclay, France) for stimulating discussions on some evolutionary considerations. We thank Morten Buch-Perdersen and Anne-Marie Winther (Pumpkin, Aarhus, Denmark) for interesting discussion on SLN structure, Nadège Jamin and Véronica Beswick (Membrane Protein Laboratory, iBitec-S, UMR 8221, CEA-Saclay, France) for discussion on this work, and Zexian Liu (University of Science & Technology of China, Hefei, Anhui, China) for his help on the CSS-Palm program for predicting potential palmitoylation. We thank Maria Muusgard (Structural Bioinformatics and Computational Biochemistry Unit, Dept. of Biochemistry, University of Oxford, UK) for providing an atomic model of the POPC bilayer around the Ca2+-ATPase, derived from MD simulation experiments.
This work was supported by grants from the Agence Nationale de la Recherche (to M. l. M. and C. M.) and the Domaines d'Intérêt Majeur Maladies Infectieuses Région Ile de France (to M. l. M. and C. M.).
Azouaoui, H., Montigny, C., Ash, M. R., Fijalkowski, F., Jacquot, A., Grønberg, C., López-Marqués, R. L., Palmgren, M. G., Garrigos, M., le Maire, M., Decottignies, P., Gourdon, P., Nissen, P., Champeil, P., and Lenoir, G., in press.
M. le Maire and L. Denoroy, unpublished observation.
- SLN
- sarcolipin
- SR
- sarcoendoplasmic reticulum
- SERCA1a
- sarcoendoplasmic reticulum Ca2+-ATPase isoform 1a
- DDM
- n-dodecyl-β-D-maltopyranoside
- C12E8
- octaethylene glycol monododecyl ether
- DOC
- deoxycholate
- SEC
- size exclusion chromatography
- MS
- mass spectrometry
- MALDI-TOF
- matrix-assisted laser desorption ionization-time of flight
- IAM
- iodoacetamide
- PVDF
- polyvinylidene fluoride.
REFERENCES
- 1. Winther A. M., Bublitz M., Karlsen J. L., Møller J. V., Hansen J. B., Nissen P., Buch-Pedersen M. J. (2013) The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495, 265–269 [DOI] [PubMed] [Google Scholar]
- 2. MacLennan D., Yip C., Iles G., Seeman P. (1973) Isolation of sarcoplasmic reticulum proteins. Cold Spring Harb. Symp. Quant. Biol. 37, 469–477 [Google Scholar]
- 3. Bhupathy P., Babu G. J., Ito M., Periasamy M. (2009) Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. J. Mol. Cell Cardiol. 47, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Odermatt A., Becker S., Khanna V. K., Kurzydlowski K., Leisner E., Pette D., MacLennan D. H. (1998) Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 273, 12360–12369 [DOI] [PubMed] [Google Scholar]
- 5. Wawrzynow A., Theibert J. L., Murphy C., Jona I., Martonosi A., Collins J. H. (1992) Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch. Biochem. Biophys. 298, 620–623 [DOI] [PubMed] [Google Scholar]
- 6. Gorski P. A., Glaves J. P., Vangheluwe P., Young H. S. (2013) Sarco(endo)plasmic reticulum calcium ATPase (SERCA) inhibition by sarcolipin is encoded in its luminal tail. J. Biol. Chem. 288, 8456–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sahoo S. K., Shaikh S. A., Sopariwala D. H., Bal N. C., Periasamy M. (2013) Sarcolipin protein interaction with sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) is distinct from phospholamban protein, and only sarcolipin can promote uncoupling of the SERCA pump. J. Biol. Chem. 288, 6881–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith W. S., Broadbridge R., East J. M., Lee A. G. (2002) Sarcolipin uncouples hydrolysis of ATP from accumulation of Ca2+ by the Ca2+-ATPase of skeletal-muscle sarcoplasmic reticulum. Biochem. J. 361, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mall S., Broadbridge R., Harrison S. L., Gore M. G., Lee A. G., East J. M. (2006) The presence of sarcolipin results in increased heat production by Ca(2+)-ATPase. J. Biol. Chem. 281, 36597–36602 [DOI] [PubMed] [Google Scholar]
- 10. Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., MacLennan D. H. (2003) Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. U.S.A. 100, 5040–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morita T., Hussain D., Asahi M., Tsuda T., Kurzydlowski K., Toyoshima C., Maclennan D. H. (2008) Interaction sites among phospholamban, sarcolipin, and the sarco(endo)plasmic reticulum Ca(2+)-ATPase. Biochem. Biophys. Res. Commun. 369, 188–194 [DOI] [PubMed] [Google Scholar]
- 12. Toyoshima C., Iwasawa S., Ogawa H., Hirata A., Tsueda J., Inesi G. (2013) Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 495, 260–264 [DOI] [PubMed] [Google Scholar]
- 13. Mascioni A., Karim C., Barany G., Thomas D. D., Veglia G. (2002) Structure and orientation of sarcolipin in lipid environments. Biochemistry 41, 475–482 [DOI] [PubMed] [Google Scholar]
- 14. Buffy J. J., Traaseth N. J., Mascioni A., Gor'kov P. L., Chekmenev E. Y., Brey W. W., Veglia G. (2006) Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry 45, 10939–10946 [DOI] [PubMed] [Google Scholar]
- 15. Buffy J. J., Buck-Koehntop B. A., Porcelli F., Traaseth N. J., Thomas D. D., Veglia G. (2006) Defining the intramembrane binding mechanism of sarcolipin to calcium ATPase using solution NMR spectroscopy. J. Mol. Biol. 358, 420–429 [DOI] [PubMed] [Google Scholar]
- 16. Vangheluwe P., Schuermans M., Zádor E., Waelkens E., Raeymaekers L., Wuytack F. (2005) Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem. J. 389, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Babu G. J., Bhupathy P., Carnes C. A., Billman G. E., Periasamy M. (2007) Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J. Mol. Cell Cardiol. 43, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Odermatt A., Taschner P. E., Scherer S. W., Beatty B., Khanna V. K., Cornblath D. R., Chaudhry V., Yee W. C., Schrank B., Karpati G., Breuning M. H., Knoers N., MacLennan D. H. (1997) Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics 45, 541–553 [DOI] [PubMed] [Google Scholar]
- 19. Bal N. C., Maurya S. K., Sopariwala D. H., Sahoo S. K., Gupta S. C., Shaikh S. A., Pant M., Rowland L. A., Bombardier E., Goonasekera S. A., Tupling A. R., Molkentin J. D., Periasamy M. (2012) Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat. Med. 18, 1575–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee A. G. (2002) A calcium pump made visible. Curr. Opin. Struct. Biol. 12, 547–554 [DOI] [PubMed] [Google Scholar]
- 21. Gillard E. F., Otsu K., Fujii J., Khanna V. K., de Leon S., Derdemezi J., Britt B. A., Duff C. L., Worton R. G., MacLennan D. H. (1991) A substitution of cysteine for arginine 614 in the ryanodine receptor is potentially causative of human malignant hyperthermia. Genomics 11, 751–755 [DOI] [PubMed] [Google Scholar]
- 22. De Meis L., Hasselbach W. (1971) Acetyl phosphate as substrate for Ca2+ uptake in skeletal muscle microsomes. Inhibition by alkali ions. J. Biol. Chem. 246, 4759–4763 [PubMed] [Google Scholar]
- 23. Champeil P., Guillain F., Vénien C., Gingold M. P. (1985) Interaction of magnesium and inorganic phosphate with calcium-deprived sarcoplasmic reticulum adenosine triphosphatase as reflected by organic solvent induced perturbation. Biochemistry 24, 69–81 [DOI] [PubMed] [Google Scholar]
- 24. Meissner G., Fleischer S. (1973) Ca2+ uptake in reconstituted sarcoplasmic reticulum vesicles. Biochem. Biophys. Res. Commun. 52, 913–920 [DOI] [PubMed] [Google Scholar]
- 25. le Maire M., Arnou B., Olesen C., Georgin D., Ebel C., Møller J. V. (2008) Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat. Protoc. 3, 1782–1795 [DOI] [PubMed] [Google Scholar]
- 26. Lenoir G., Picard M., Gauron C., Montigny C., Le Maréchal P., Falson P., Le Maire M., Møller J. V., Champeil P. (2004) Functional properties of sarcoplasmic reticulum Ca(2+)-ATPase after proteolytic cleavage at Leu119-Lys120, close to the A-domain. J. Biol. Chem. 279, 9156–9166 [DOI] [PubMed] [Google Scholar]
- 27. Møller J. V., le Maire M. (1993) Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J. Biol. Chem. 268, 18659–18672 [PubMed] [Google Scholar]
- 28. Kazmin D., Edwards R. A., Turner R. J., Larson E., Starkey J. (2002) Visualization of proteins in acrylamide gels using ultraviolet illumination. Anal. Biochem. 301, 91–96 [DOI] [PubMed] [Google Scholar]
- 29. Ladner C. L., Yang J., Turner R. J., Edwards R. A. (2004) Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal. Biochem. 326, 13–20 [DOI] [PubMed] [Google Scholar]
- 30. Juul B., Turc H., Durand M. L., Gomez de Gracia A., Denoroy L., Møller J. V., Champeil P., le Maire M. (1995) Do transmembrane segments in proteolyzed sarcoplasmic reticulum Ca(2+)-ATPase retain their functional Ca2+ binding properties after removal of cytoplasmic fragments by proteinase K? J. Biol. Chem. 270, 20123–20134 [DOI] [PubMed] [Google Scholar]
- 31. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 32. Møller J. V., Lind K. E., Andersen J. P. (1980) Enzyme kinetics and substrate stabilization of detergent-solubilized and membraneous (Ca2+ + Mg2+)-activated ATPase from sarcoplasmic reticulum. Effect of protein-protein interactions. J. Biol. Chem. 255, 1912–1920 [PubMed] [Google Scholar]
- 33. Montigny C., Arnou B., Marchal E., Champeil P. (2008) Use of glycerol-containing media to study the intrinsic fluorescence properties of detergent-solubilized native or expressed SERCA1a. Biochemistry 47, 12159–12174 [DOI] [PubMed] [Google Scholar]
- 34. Bers D. M., Patton C. W., Nuccitelli R. (2010) A practical guide to the preparation of Ca(2+) buffers. Methods Cell Biol. 99, 1–26 [DOI] [PubMed] [Google Scholar]
- 35. Tulloch L. B., Howie J., Wypijewski K. J., Wilson C. R., Bernard W. G., Shattock M. J., Fuller W. (2011) The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. J. Biol. Chem. 286, 36020–36031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aicart-Ramos C., Valero R. A., Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808, 2981–2994 [DOI] [PubMed] [Google Scholar]
- 37. Nadolski M. J., Linder M. E. (2007) Protein lipidation. Febs J. 274, 5202–5210 [DOI] [PubMed] [Google Scholar]
- 38. Resh M. D. (2013) Covalent lipid modifications of proteins. Curr. Biol. 23, R431–R435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charollais J., Van Der Goot F. G. (2009) Palmitoylation of membrane proteins (Review). Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 40. Chaube R., Hess D. T., Wang Y. J., Plummer B., Sun Q. A., Laurita K., Stamler J. S. (2014) Regulation of the Skeletal Muscle Ryanodine Receptor/Ca2+-release Channel RyR1 by S-Palmitoylation. J. Biol. Chem. 289, 8612–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schey K. L., Gutierrez D. B., Wang Z., Wei J., Grey A. C. (2010) Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry 49, 9858–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Musgaard M., Thøgersen L., Schiøtt B., Tajkhorshid E. (2012) Tracing cytoplasmic Ca(2+) ion and water access points in the Ca(2+)-ATPase. Biophys. J. 102, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tong S. W. (1977) The acetylated NH2-terminus of CaATPase from rabbit skeletal muscle sarcoplasmic reticulum: a commom NH2-terminal acetylated methionyl sequence. Biochem. Biophys. Res. Commun. 74, 1242–1248 [DOI] [PubMed] [Google Scholar]
- 44. Montigny C., Jaxel C., Shainskaya A., Vinh J., Labas V., Møller J. V., Karlish S. J., le Maire M. (2004) Fe2+ -catalyzed oxidative cleavages of Ca2+ -ATPase reveal novel features of its pumping mechanism. J. Biol. Chem. 279, 43971–43981 [DOI] [PubMed] [Google Scholar]
- 45. Champeil P., Menguy T., Soulié S., Juul B., de Gracia A. G., Rusconi F., Falson P., Denoroy L., Henao F., le Maire M., Moller J. V. (1998) Characterization of a protease-resistant domain of the cytosolic portion of sarcoplasmic reticulum Ca2+-ATPase. Nucleotide- and metal-binding sites. J. Biol. Chem. 273, 6619–6631 [DOI] [PubMed] [Google Scholar]
- 46. Andersen J. P., Moller J. V. (1977) Reaction of sarcoplasmic reticulum Ca2+-ATPase in different functional states with 5,5′-dithiobis(2-nitrobenzoate). Biochim. Biophys. Acta 485, 188–202 [DOI] [PubMed] [Google Scholar]
- 47. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blaskovic S., Blanc M., van der Goot F. G. (2013) What does S-palmitoylation do to membrane proteins? Febs J. 280, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 49. Sweadner K. J., Rael E. (2000) The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68, 41–56 [DOI] [PubMed] [Google Scholar]
- 50. Koonin E. V. (2005) Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338 [DOI] [PubMed] [Google Scholar]
- 51. Mokrab Y., Stevens T. J., Mizuguchi K. (2010) A structural dissection of amino acid substitutions in helical transmembrane proteins. Proteins 78, 2895–2907 [DOI] [PubMed] [Google Scholar]
- 52. Stevens T. J., Arkin I. T. (2001) Substitution rates in alpha-helical transmembrane proteins. Protein Sci. 10, 2507–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Donnelly D., Overington J. P., Ruffle S. V., Nugent J. H., Blundell T. L. (1993) Modeling α-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 2, 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Picard M., Toyoshima C., Champeil P. (2006) Effects of inhibitors on luminal opening of Ca2+ binding sites in an E2P-like complex of sarcoplasmic reticulum Ca2+-ATPase with Be2+-fluoride. J. Biol. Chem. 281, 3360–3369 [DOI] [PubMed] [Google Scholar]
- 55. Altschul S., Madden T., Schäffer A., Zhang J., Zhang Z., Miller W., Lipman D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Galtier N., Gouy M., Gautier C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 [DOI] [PubMed] [Google Scholar]
- 57. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 58. Huson D. H., Richter D., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huson D. H., Scornavacca C. (2012) Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biology 61, 1061–1067 [DOI] [PubMed] [Google Scholar]