Background: TIP60 can be regulated by autoacetylation and deacetylated by SIRT1.
Results: Novel lysine residues of TIP60 autoacetylation were identified, and TIP60 can be functionally regulated by HDAC3 through deacetylation.
Conclusion: HDAC3 promotes TIP60 ubiquitination and cytoplasmic localization and protects cells from apoptosis after DNA damage.
Significance: Our findings provide a better understanding of TIP60 regulatory mechanisms and its cellular functions.
Keywords: Apoptosis, DNA Damage, Histone Acetylase, Histone Deacetylase 3 (HDAC3), Ubiquitylation (Ubiquitination)
Abstract
The key member of the MOZ (monocyticleukaemia zinc finger protein), Ybf2/Sas3, Sas2, and TIP60 acetyltransferases family, Tat-interactive protein, 60 kD (TIP60), tightly modulates a wide array of cellular processes, including chromatin remodeling, gene transcription, apoptosis, DNA repair, and cell cycle arrest. The function of TIP60 can be regulated by SIRT1 through deacetylation. Here we found that TIP60 can also be functionally regulated by HDAC3. We identified six lysine residues as its autoacetylation sites. Mutagenesis of these lysines to arginines completely abolishes the autoacetylation of TIP60. Overexpression of HDAC3 increases TIP60 ubiquitination levels. However, unlike SIRT1, HDAC3 increased the half-life of TIP60. Further study found that HDAC3 colocalized with TIP60 both in the nucleus and the cytoplasm, which could be the reason why HDAC3 can stabilize TIP60. The deacetylation of TIP60 by both SIRT1 and HDAC3 reduces apoptosis induced by DNA damage. Knockdown of HDAC3 in cells increased TIP60 acetylation levels and increased apoptosis after DNA damage. Together, our findings provide a better understanding of TIP60 regulation mechanisms, which is a significant basis for further studies of its cellular functions.
Introduction
TIP60 was first identified as a cofactor of the HIV Tat protein and is a key member of the MYST7 (termed for the founding members MOZ, Ybf2/Sas3, Sas2, and TIP60) histone acetyltransferase (HAT) family (1, 2). MYST HATs are characteristic of a highly conserved MYST domain that is responsible for the acetyl-CoA binding and acetylation activity. TIP60 is involved in the regulation of diverse cellular activities, including chromatin remodeling, gene transcription, DNA damage responses, and tumorigenesis (3, 4). The acetylation of core histones by HATs is essential in the loosening of nucleosomes and the activation of gene transcription. The core histones H2AX, H3, and H4 are known substrates of TIP60 acetyltransferase activity (5–7). TIP60 also serves as a cofactor of several transcription factors, such as human immunodeficiency virus, type 1 Tat (1), β-amyloid precursor protein (8), STAT3 (9), and CCAAT/enhancer-binding protein α (10), to modulate gene transcription.
The close association of TIP60 with DNA damage response has been studied extensively (4, 11, 12). DNA damage response is involved in the activation of diverse cellular signaling pathways leading to cell cycle arrest, DNA repair, and apoptosis. TIP60 acts at multiple levels in DNA damage response through interaction with ATM (Ataxia telangiectasia mutated), p53, and p21, and so forth. Ectopic expression of mutated TIP60 lacking HAT activity results in the deficiency of cell apoptosis and DNA double strand break repair (3). TIP60 is part of an evolutionarily conserved nuclear multimeric protein complex, NuA4, that possesses histone deacetylase, DNA helicase, ATPase, and structural DNA binding activity (4). This complex is recruited by many transcriptional factors to their target promoters, such as the critical mediator of DNA damage response p53. TIP60 acetylates p53 at lysine 120 within the DNA-binding domain, followed by the p53-dependent transcriptional activation of proapoptotic genes, which is a key event in determining cell fate under DNA damage (13). Mutant p53 (K120R) lacking acetylation by TIP60 totally abolishes cell apoptosis without affecting cell cycle arrest, implicating that TIP60-mediated acetylation of p53 Lys-120 is involved in the choice between cell cycle arrest and apoptosis. Upon DNA double strand breakage, the activation of the protein kinase ATM is essential for DNA repair and requires complicated posttranslational modifications, including phosphorylation and acetylation. Acetylation of ATM Lys-3016 by TIP60 is indispensable for the activation of the kinase activity of ATM in response to DNA damage (14, 15). Interestingly, a recent study reported that TIP60 is also required for p21-dependent DNA damage-induced cell cycle arrest (16). Lysines 161 and 163 of p21 have been identified as novel targets of TIP60-mediated acetylation. Depletion of TIP60 leads to the degradation of unacetylated p21 protein and the disruption of G1 arrest, whereas p21 2KQ (a mimetic of acetylated p21 at Lys-161 and Lys-163) significantly augments the induction of cell cycle arrest compared with that of cells expressing wild-type p21. Because of the increasing involvement of TIP60 in more and more biological processes, clarification of the regulation models of TIP60 will provide a better understanding of its cellular functions. Here we found that TIP60 is autoacetylated at multiple lysine sites within the N terminus and that the autoacetylation is negatively regulated by SIRT1 and HDAC3 deacetylase activity. Moreover, the autoacetylation is necessary for its nuclear localization and maximal HAT activity.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HEK293, H1299, and U2OS cells were obtained from the ATCC. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Anti-acetylated lysine polyclonal antibody and anti-HDAC3 polyclonal antibody were purchased from Cell Signaling Technology. Anti-p53 monoclonal antibody (DO-1), anti-SIRT1 monoclonal antibody, and anti-TIP60 monoclonal antibody were purchased from Santa Cruz Biotechnology. Anti-FLAG monoclonal antibody, cycloheximide, etoposide, trichostatin A (TSA), and nicotinamide were purchased from Sigma-Aldrich. Anti-acetyl lysine-agarose was purchased from Immunechem. Transfection of plasmids in HEK293 cells was performed by using the calcium phosphate precipitation method.
In Vitro Acetylation Assay
GST-p53CT, GST, and GST-TIP60 proteins were expressed and purified from BL21 bacteria. A 20-μl reaction contained 10 μl of reaction buffer (50 mm Tris (pH 7.9), 10% glycerol, 1 mm DTT, and 10 mm sodium butyrate), 1 μl of [14C]acetyl-CoA (55 mCi/mmol, Amersham Biosciences), 1 μg of GST or GST fusion protein, 0.1 μg of p300HAT, and double-distilled H2O as needed. The reaction mixture was then incubated at 30 °C for 2 h, followed by SDS-PAGE and radioautography. To show the amount of proteins used in the in vitro acetylation assay, another SDS-PAGE gel was run in parallel using the same amount and loading sequence of GST or GST fusion proteins, followed by gel code blue staining (Thermo Scientific).
Acetylation/Deacetylation Assay in cells
HEK293 cells were seeded in a 10-cm Petri dish at the density of 2 × 106 cells/dish the day before transfection. FLAG-tagged plasmids, as indicated, were transfected into HEK293 cells by using the calcium phosphate method. At 24 h, for the acetylation assay, cells were incubated with 1 μm of TSA and 5 mm nicotinamide for an additional 6 h before harvest. For the deacetylation assay, cells were not treated with any drugs before harvest. After collection of cells by centrifugation, whole cell lysates were prepared in FLAG lysis buffer (50 mm Tris-HCl (pH 7.8), 137 mm NaCl, 1 mm NaF, 1 mm NaVO3, 1% Triton X-100, 0.2% sarkosyl, 1 mm DTT, and 10% glycerol) containing fresh protease inhibitors, 10 μm TSA, and 5 mm nicotinamide. Cell extracts were then incubated with anti-FLAG M2 beads (Sigma-Aldrich) at 4 °C overnight. After washing the beads five times with BC100 buffer (50 mm Tris-HCl (pH 7.8), 100 mm NaCl, 0.2% Triton X-100, and 10% glycerol), FLAG peptide was added, and the beads were incubated for an additional 2 h to elute the bound proteins. Immunoprecipitated proteins were subjected to SDS-PAGE and analyzed by Western blotting with different antibodies as indicated.
RNA Extraction and Real-time PCR
Total RNA was extracted by using the total RNA kit I (Omega). The first-strand cDNA was synthesized with a Moloney murine leukemia virus first strand cDNA synthesis kit (Omega). Real-time PCR analysis was performed with an ABI7500 (Applied Biosystems) using the Super-Real PreMix Plus (SYBR Green) kit. β-Actin (forward primer, 5′TCATGTTTGAGACCTTCAA; reverse primer, 5′GTCTTTGCGGATGTCCACG) was used as the endogenous control of TIP60 (forward primer, 5′CGTAAGAACAAGAGTTATTCCCAG; reverse primer, 5′GTCTTCCGTTGATTCTTTCTCC). All experiments were performed in triplicate. The relative expression was determined using the ΔΔCT method.
Immunofluorescence Assay
H1299 cells were seeded onto sterile coverslips in a 6-well plate at 30–40% confluence. The next day, transfection with plasmids as indicated was performed by using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. 24 h post-transfection, cells were washed with PBS, fixed in 4% paraformaldehyde for 20 min, and permeabilized with 0.2% Triton X-100 for 5 min. After blocking for 30 min with blocking buffer (1% BSA in PBS buffer (pH 7.4), cells were incubated with antibodies against HA, FLAG, HDAC3, and SIRT1 for 1 h, followed by fluorophore-conjugated secondary antibody incubation for an additional hour. Finally, coverslips were sealed with nail polish onto glass slides and then subjected to fluorescence microscopy.
Apoptosis Assay
U2OS cells were transfected with plasmids or siRNA as indicated for 24 h and then treated with 0.1% dimethyl sulfoxide or 20 μm etoposide for an additional 24 h. After harvesting cells by digestion with 0.05% trypsin/EDTA solution (Invitrogen), an apoptosis assay was performed with the annexin V/FITC apoptosis detection kit I according to the instructions of the manufacturer (BD Biosciences). The FACS data were analyzed by Flowjo software.
RESULTS
Identification of Autoacetylation Lysine Residues of TIP60
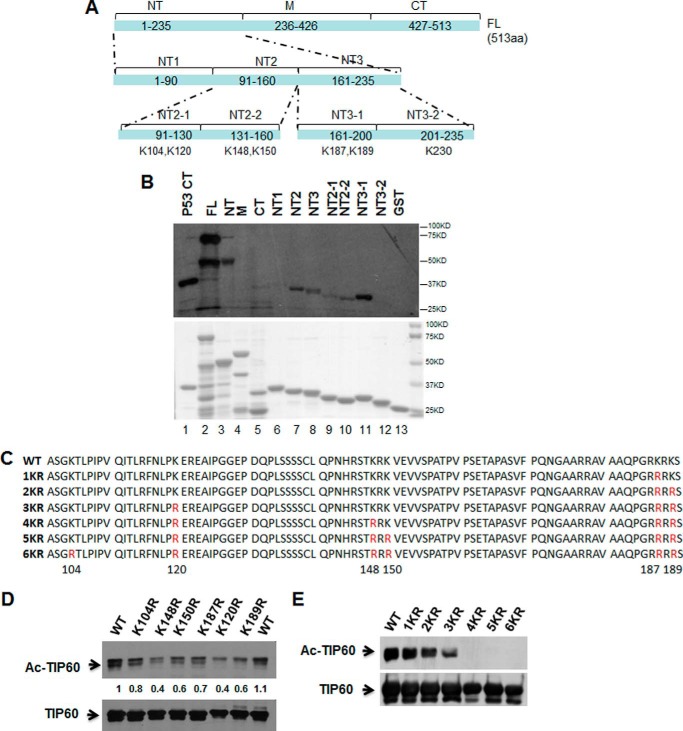
To identify TIP60 autoacetylation sites, we purposely separated the TIP60 protein into three fractions on the basis of the functional domains reported previously: NT, including the chrome domain; M, including the zinc finger and the MYST domain; and CT, including a short nuclear receptor interaction box (Fig. 1A) (7). Diverse GST-TIP60 constructs, as indicated in Fig. 1A, were established and expressed in Escherichia coli. After purification using GST affinity resins, 1.0 g of each GST fusion protein was employed to the in vitro acetylation assay (Fig. 1B, top panel), whereas GST-p53CT served as the positive control and GST as the negative control. TIP60 NT, but not M and CT, clearly showed the strong acetylation signal (Fig. 1B, third through fifth lanes), indicating that the major acetylation sites are located at the N terminus. We further separated the NT fraction into three pieces: NT1, NT2, and NT3. The same acetylation assay found that NT2 and NT3 gave strong acetylation signals (Fig. 1B, sixth through eighth lanes). There are seven lysine residues in NT2 and NT3 (Fig. 1A). Accordingly, NT2 and NT3 were separated further into NT2-1, NT2-2, NT3-1, and NT3-2. We detected acetylation signals in NT2-1, NT2-2, and NT3-1 but not in NT3-2, which contains Lys-230 (Fig. 1B, ninth through twelfth lanes). Therefore, Lys-230 was excluded from the acetylation sites, and we finally identified six putative lysine residues of TIP60 autoacetylation (Lys-104, Lys-120, Lys-148, Lys-150, Lys-187, and Lys-189) through this in vitro acetylation assay.
FIGURE 1.
Identification of the acetylation sites of TIP60. A, schematic of various truncated TIP60 proteins fused with GST and putative acetylated sites of TIP60 protein. aa, amino acids; FL, full-length. B, various GST-TIP60 fusion proteins were subjected to an in vitro acetylation assay. Top panel, representative radioautography data. Bottom panel, gel code blue staining. C, schematic representation of various point mutant constructs of TIP60. D and E, HEK293 cells were transfected with wild-type or mutant FLAG-tagged TIP60 constructs for 24 h and then incubated with 1 μm TSA and 5 mm nicotinamide for an additional 6 h. The acetylation levels of TIP60 (Ac-TIP60) and total TIP60 protein were detected after anti-FLAG immunoprecipitation. The polyclonal antibody against acetylated lysine (Cell Signaling Technology, catalog no. 9441) was employed in these experiments.
To further confirm our results via in vivo acetylation assay, we established a number of TIP60 constructs with point mutations in which lysines were mutated to arginines (Fig. 1C). HEK293 cells were transfected with wild-type or mutant FLAG-TIP60 constructs, as indicated, followed by IP and Western blot analyses. Compared with the WT, the single mutations of all six lysines resulted in reduced acetylation levels (Fig. 1D). Further analysis with multiple lysine mutations showed that 1KR, 2KR, and 3KR gradually reduced the autoacetylation of TIP60, whereas 4KR, 5KR, and 6KR totally abolished autoacetylation (Fig. 1E). These results are in agreement with the in vitro acetylation assay, suggesting that TIP60 autoacetylation is located at multiple lysine residues within the N terminus. Interestingly, Lys-104 and Lys-120 lie in the alternative exon (amino acids 96–147) of TIP60 and TIP60β (17), implying that these two sites may possess distinct functions from other autoacetylation lysine residues.
TIP60 Interacts with and Is Deacetylated by HDAC3
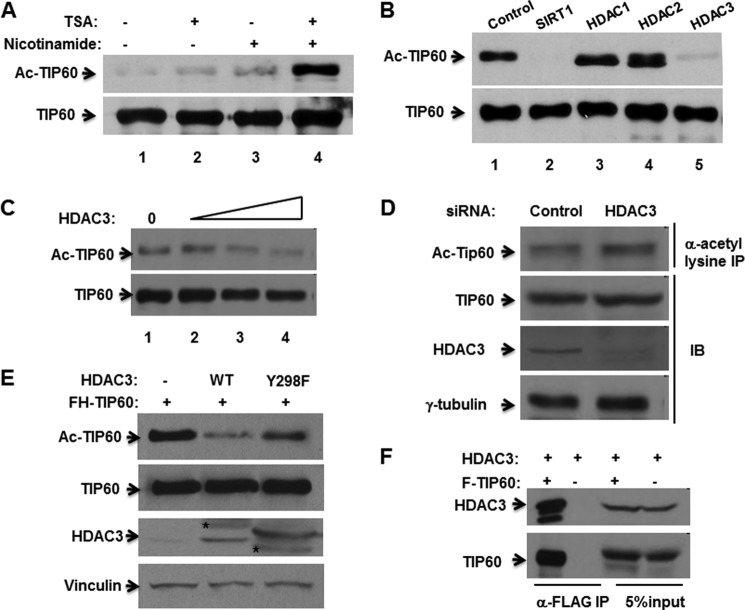
It has been reported that TIP60 can be regulated by SIRT1 through deacetylation (18–20). We further investigated this regulation. TSA and nicotinamide are the specific inhibitors of the class I/II HDACs and sirtuins (class III HDACs), respectively. Cells were treated with TSA and nicotinamide individually or simultaneously. As expected, nicotinamide treatment increased TIP60 acetylation levels. However, TSA treatment also increased TIP60 acetylation levels. Both TSA and nicotinamide treatment yielded the highest level of acetylation of TIP60 (Fig. 2A). These results indicate that, besides SIRT1, other HDACs also play a role in mediating TIP60 deacetylation. To identify which HDACs plays a role in TIP60 deacetylation, we performed a deacetylation assay with SIRT1, HDAC1, HDAC2, and HDAC3. As shown in Fig. 2B, SIRT1 can clearly deacetylate TIP60 (lane 2), and HDAC3 can also deacetylate TIP60 (lane 5), whereas HDAC1 and HDAC2 cannot deacetylate TIP60 (lanes 3 and 4). We further performed a TIP60 deacetylation assay with HDAC3 and found that TIP60 was deacetylated by HDAC3 in a dose-dependent manner (Fig. 2C). HDAC3 was down-regulated in cells, as expected, and TIP60 acetylation levels were increased (Fig. 2D). We also investigated the effect of catalytically dead HDAC3 on TIP60 acetylation. It has been reported that HDAC3-Y298F renders the in vitro-translated HDAC3 proteins completely inactive, as measured by HDAC assay (21). As shown in Fig. 2E, HDAC3 wild-type significantly suppressed TIP60 acetylation, whereas HDAC3-Y298F only exhibited modest suppression, even with much higher protein expression than HDAC3 wild-type. Interaction between TIP60 and HDAC3 was examined by co-IP assay. As shown in Fig. 2F, HDAC3 can clearly be coimmunoprecipitated by TIP60. Overall, these results demonstrated that TIP60 can interact with and be deacetylated by HDAC3.
FIGURE 2.
TIP60 interacts with and is deacetylated by HDAC3. A, HEK293 cells were transfected with pTOPO-FLAG-TIP60 for 24 h and then incubated with or without 1 μm TSA and/or 5 mm nicotinamide, as indicated, for an additional 6 h. An in vivo acetylation assay (Ac-TIP60) and Western blot analysis were then performed. B, HEK293 cells were transfected with FLAG-TIP60 alone or with deacetylases as indicated. Deacetylation assays were then performed. C, HEK293 cells were transfected with FLAG-TIP60 alone or with increasing amounts of HDAC3 constructs, followed by a deacetylation assay. D, H1299 cells were transfected with luciferase siRNA or HDAC3 siRNA using Lipofectamine 2000 reagent. Whole cell lysates were subjected to anti-acetyl lysine IP. The immunoprecipitates and whole cell lysates were then analyzed by Western blotting with the indicated antibodies. IB, immunoblot. E, HEK293 cells were transfected with FH-TIP60 alone or with HDAC3 or F-HDAC3-Y298F, followed by a deacetylation assay. The asterisks indicate nonspecific bands. F, HEK293 cells were transfected with the indicated plasmids. Whole cell lysates were subjected to M2 bead IP, and the resulting immunoprecipitates and 5% input were analyzed by antibodies against HDAC3 and TIP60.
HDAC3 Affects TIP60 Stability
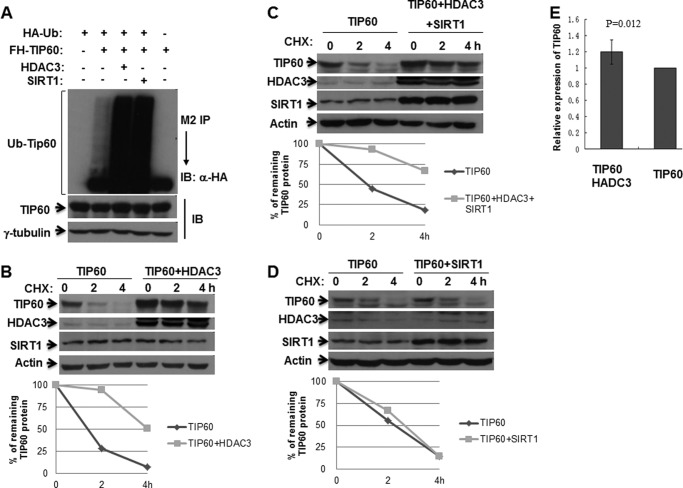
To investigate the functional consequence of HDAC3 deacetylation of TIP60, we first examined its effect on TIP60 stability. The ubiquitination assay was performed for TIP60 with HDAC3 or SIRT1. Similar to SIRT1, HDAC3 can also increase the ubiquitination level of TIP60 (Fig. 3A). However, when we examined the effect of HDAC3 on the half-life of TIP60, we found that, unlike SIRT1, which did not change the TIP60 half-life when cotransfected into cells (Fig. 3D), HDAC3 increased the TIP60 half-life (Fig. 3B). This TIP60 half-life increase is due to protein stabilization because the TIP60 mRNA levels are almost same in TIP60 alone or TIP60 cotransfected with HDAC3 (Fig. 3E). When TIP60 was cotransfected with both SIRT1 and HDAC3, the TIP60 half-life was increased (Fig. 3C), indicating that HDAC3 overcame the SIRT1 effect. These results demonstrate that HDAC3 can, surprisingly, increase TIP60 ubiquitination levels. Unlike SIRT1, HDAC3 increased the TIP60 half-life, and this stabilization has dominant effects over SIRT1.
FIGURE 3.
HDAC3 promotes the ubiquitination and protein stability of TIP60. A, HEK293 cells were transfected with the indicated plasmids. Whole cell lysates were prepared and subjected to M2 bead IP. The ubiquitination (Ub) level of TIP60 was measured by Western blot analysis with the indicated antibodies. IB, immunoblot. B–D, HEK293 cells were transfected with TIP60 alone and TIP60 plus HDAC3 (B), TIP60 plus HDAC3 and SIRT1 (C), or TIP60 plus SIRT1 (D) for 24 h. Cells were treated with 100 μg/ml cycloheximide (CHX) for the indicated time before harvest. TIP60 expression levels were determined by Western blot analysis. The graphs represent the values obtained after densitometry analysis by ImageJ software. The percentage of remaining protein after cycloheximide addition is plotted. E, HEK293 cells were transfected with the indicated plasmids. TIP60 mRNA expression was determined by quantitative real-time PCR.
HDAC3 Affects TIP60 Cellular Localization
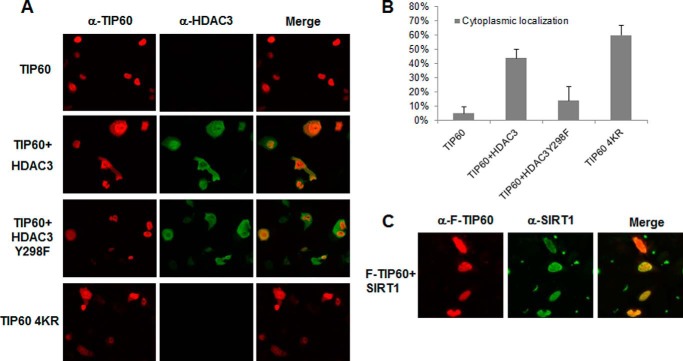
To further investigate the regulation of HDAC3 on TIP60, we examined the possibility of HDAC3 affecting TIP60 cellular localization. It is believed that TIP60 proteins normally localize in the nucleus and translocate to the perinuclear compartment or cytoplasm because of certain stimulations (17, 22–24). To elucidate the possibility of HDAC3 affecting the localization of TIP60, we transfected H1299 cells with TIP60 alone, with HDAC3, with HDAC3-Y298F, or with SIRT1, followed by an immunofluorescent assay. As shown in Fig. 4A, first row, it is clear that wild-type TIP60 exclusively localizes in the nucleus. However, when TIP60 was cotransfected with HDAC3, TIP60 was detected in both the nucleus and cytoplasm in more than 40% of cells (Fig. 4A, second row, and Fig. 4B). As expected, HDAC3-Y298F, which exhibited a limited capability for TIP60 deacetylation, was unable to induce TIP60 cytoplasmic localization compared with HDAC3 wild-type (Fig. 4A, third row, and Fig. 4B, 14% versus 44%). When TIP60 was cotransfected with SIRT1, TIP60 remained in the nucleus (Fig. 4C). We also transfected the TIP60 acetylation mutant TIP60–4KR, which could not be acetylated (Fig. 1E), in the same immunofluorescent assay and found that it was located in both the nucleus and the cytoplasm in 60% of cells (Fig. 4A, fourth row, and Fig. 4B). These results demonstrate that HDAC3 regulates TIP60 cellular localization by deacetylation, whereas SIRT1 does not have this effect.
FIGURE 4.
HDAC3 affects the cellular localization of TIP60. A and B, H1299 cells were transfected with FH-TIP60, FH-TIP60 plus HDAC3, FH-TIP60 plus HDAC3-Y298F, or FH-TIP60 4KR for 24 h. An immunofluorescence assay was performed by using primary antibodies against HA and HDAC3 (see details under “Experimental Procedures”). The bar chart is representative of the percentage of cells with TIP60 cytoplasmic localization (B). C, H1299 cells were transfected with FH-TIP60 plus SIRT1 for 24 h. An immunofluorescence assay was performed using primary antibodies against FLAG and SIRT1 (see details under “Experimental Procedures”).
HDAC3 Mediates TIP60 Deacetylation Resistant to Apoptosis Induced by DNA Damage
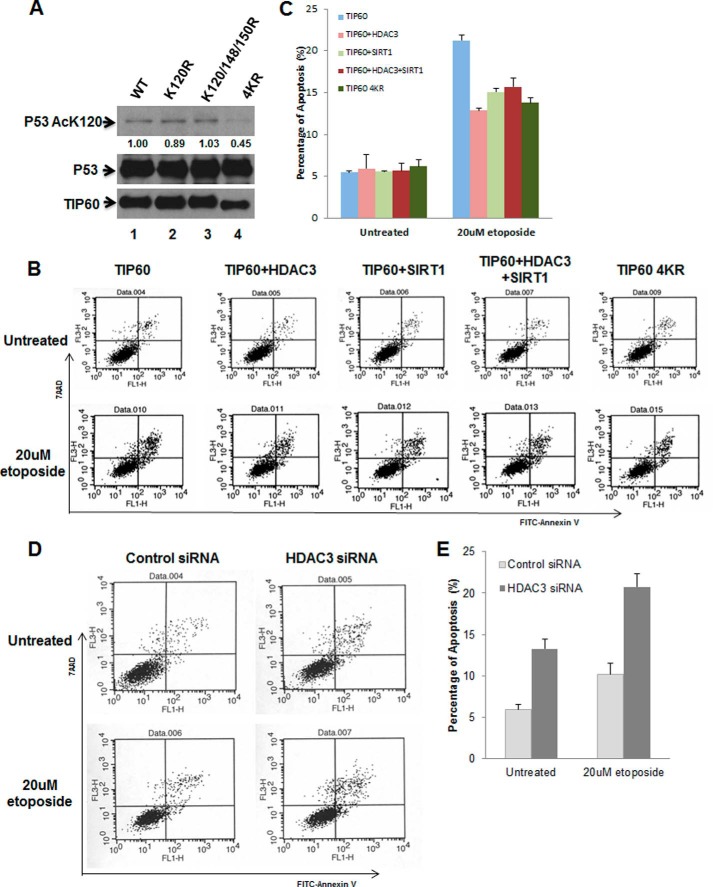
As the most famous target of TIP60, p53 was employed to clarify the role of autoacetylation in TIP60 HAT activity. HEK293 cells were transfected with a combination of p53 and wild-type or mutant TIP60 constructs. 24 h post-transfection, whole cell lysates were prepared and subjected to anti-p53 IP and Western blot analysis. As shown in Fig. 5A, TIP60 4KR partially decreased the acetylation levels of p53 lysine 120 (top panel, lane 4). The immunoprecipitates were also analyzed with antibodies against TIP60 and p53 proteins. Compared with the wild type, TIP60 4KR had a comparable capacity of binding to p53 protein (Fig. 5A, bottom panel, lane 1 versus lane 4). Taken together, the impairment of p53 Lys-120 acetylation levels was attributed to the reduction of TIP60 HAT activity and not the binding ability. Our findings suggest that autoacetylation is necessary for maximal HAT activity of TIP60 proteins.
FIGURE 5.
HDAC3-mediated TIP60 deacetylation promotes cell survival upon DNA damage. A, HEK293 cells were transfected with p53 plus wild-type or mutant TIP60 plasmids as indicated. An immunoprecipitation assay was performed using anti-p53 antibody, and acetylated p53 Lys-120, total p53, and total TIP60 were determined by Western blot analysis. B and D, U2OS cells were transfected with the indicated plasmids or siRNA for 24 h, followed by etoposide treatment for an additional 24 h, and then stained by annexin V and PI (propidium iodide) to analyze apoptosis by FACS (see details under “Experimental Procedures”). C and E, the bar charts were plotted with the percentage of annexin V-positive cells in B and D. Data are representative of three independent experiments.
We further investigated the effect of TIP60 deacetylation in apoptosis induced by DNA damage. TIP60 was transfected alone or cotransfected with HDAC3, SIRT1, or HDAC3 + SIRT1 into U2OS cells, and an apoptosis assay was performed. As shown in Fig. 5, B and C, TIP60 deacetylation has no effect on apoptosis without DNA damage. However, cotransfection of HDAC3, SIRT1, or both showed resistance to apoptosis after DNA damage treatment. Similar results were obtained when the TIP60 acetylation mutant 4KR was transfected into cells. We further down-regulated HDAC3 in cells by RNAi and found that cells showed more apoptotic behavior under normal conditions and with DNA damage treatment (Fig. 5, D and E). Together, our results demonstrate that TIP60 autoacetylation regulates its HAT activity on p53 K120 acetylation and that TIP60 deacetylation increases its resistance to apoptosis induced by DNA damage.
DISCUSSION
TIP60 is a multifunctional protein involved in various cellular processes ranging from chromatin remodeling to gene transcription, apoptosis, DNA repair, and cell cycle arrest. The function of TIP60 can be regulated by autoacetylation and SIRT1-mediated deacetylation. Here we found that TIP60 can also be functionally regulated by HDAC3 through deacetylation. Overexpression of HDAC3 increased TIP60 ubiquitination levels. However, unlike SIRT1, HDAC3 increased the TIP60 half-life. Further research found that HDAC3 colocalized with TIP60 both in the nucleus and the cytoplasm, which could be the reason why HDAC3 can stabilize TIP60. Deacetylation of TIP60 by both SIRT1 and HDAC3 reduced apoptosis induced by DNA damage. The knockdown of HDAC3 in cells increased the TIP60 acetylation level and increased apoptosis after DNA damage. Together, our findings provide a better understanding of TIP60 regulatory mechanisms, which is a significant basis for further studies of its cellular functions.
TIP60 autoacetylation was first observed by Tang et al. (13) when they performed an acetylation assay to identify the acetylated lysine of p53 by TIP60. More recently, Yang and colleagues performed an LC-MS/MS analysis and suggested that TIP60 autoacetylation occurs at seven lysine residues (Lys-76, Lys-80, Lys-104, Lys-150, Lys-187, Lys-327, and Lys-383) (32). It is very interesting that there is an overlap (Lys-104, Lys-150, and Lys-187) but not complete agreement in the identified lysines between Yang and colleagues and our studies. The employment of different experimental methods is most likely the reason. In their study, Lys-327 is confirmed within the conserved MYST domain, which possesses HAT activity, and the mutation of Lys-327 to arginine led to the loss of both autoacetylation activity and cognate HAT activity. Because of the localization specificity of Lys-327, it is hard to conclude that the loss of autoacetylation upon K327R mutation is due to Lys-327, which is the key lysine of autoacetylation, but not due to the impairment of HAT activity. Moreover, our study demonstrated that TIP60 6KR, without any disturbance of the MYST domain, can totally abolish autoacetylation. Both Yang and colleagues and our group are in agreement that autoacetylation plays a substantial role on the TIP60 enzymatic activity and that its maximal HAT activity requires autoacetylation. Conversely, overexpression of SIRT1 leads to the abolishment of TIP60 autoacetylation and suppression of TIP60-mediated acetylation of H2AX (18).
On the basis of sequence conservation within the HAT domain, histone acetyltransferases can be separated into at least four families, including Gcn5/p300/CBP associated factor (pCAF), P300/CREB-binding protein (CBP), Rtt109, and MYST proteins. The majority of HATs have been reported to have autoacetylation, such as p300 (25, 26), Rtt109 (27), pCAF (28), human males absent on the first (hMOF) (29, 30), yEsa1 (29), and TIP60. Autoacetylation is considered to be an important regulatory model for HATs. pCAF autoacetylation is required for its nuclear localization. Deacetylated pCAF by HDACs or mutagenesis accumulates in the cytoplasm (28). The autoacetylation of p300 is primarily an intramolecular event and triggers the specific structural changes of its HAT domain, followed by the up-regulation of HAT activity, transactivation activity, and complex formation (25, 31). The effect of autoacetylation on HAT activity was investigated most. Although the specific lysines of autoacetylation and mechanisms of autoacetylation on enzymatic activity are distinct in each case, autoacetylation acting an important role in regulating HATs functions was demonstrated. It seems that autoacetylation is a common regulation pattern of enzymatic activity for histone acetyltransferases.
Previous studies regarding the TIP60 deacetylation have focused on the SIRT1 protein (18, 19). TIP60 autoacetylation has been reported to cause the dissociation of the TIP60 oligomer and enhance its interaction with substrates. SIRT1 abolished TIP60 autoacetylation through its deacetylase activity and, consequently, reduced TIP60 HAT activity and facilitated proteasome-dependent TIP60 degradation. Our study found, for the first time, that HDAC3 also mediates TIP60 deacetylation, opening another regulating mechanism for TIP60. Although both SIRT1 and HDAC3 can deacetylate TIP60 and increase its ubiquitination levels, the outcome is different. SIRT1 reduced the TIP60 half-life (Fig. 3D) (20), whereas HDAC3 increased the TIP60 half-life (Fig. 3B), and this stabilization is dominant over the effects of SIRT1 because cotransfection of both HDAC3 and SIRT1 with TIP60 resulted in an increase in the TIP60 half-life (Fig. 3C). The mechanism of this stabilization is still not clear. However, we found that cotransfection of HDAC3 and TIP60 changed TIP60 cellular localization from the nucleus to both the nucleus and the cytoplasm (Fig. 4A, center column), whereas cotransfection of SIRT1 and TIP60 does not have this effect (Fig. 4C). This localization change could be one of the reasons for TIP60 stabilization mediated by HDAC3. The other possibility could be the tight interaction between HDAC3 and TIP60, preventing TIP60 from moving to the proteasome for degradation. Nevertheless, the precise mechanism for this regulation needs further investigation.
The acetylation mutant TIP60–4KR led to the cytoplasmic translocation of TIP60 proteins (Fig. 4A, fourth row), suggesting that autoacetylation is involved in the regulation of TIP60 cellular localization. This situation is similar to that of another histone acetyltransferase, pCAF. pCAF(352–832)L606A has limited HAT activity, is unable to be autoacetylated, and is distributed in both the nucleus and the cytoplasm, whereas pCAF(352–832) is localized only in the nucleus (28).
Regulation of apoptosis is one of the most important functions of TIP60. TIP60 acetylates p53 at lysine 120, followed by the p53-dependent transcriptional activation of proapoptotic genes, which is a key event in determining cell fate in response to DNA damage (13). Mutant p53 (K120R) lacking acetylation by TIP60 totally abolishes cell apoptosis without affecting cell cycle arrest, suggesting that TIP60-mediated acetylation of p53 Lys-120 is involved in the choice between cell cycle arrest and apoptosis. Because autoacetylation mutant TIP60–4KR reduced its ability to acetylate p53 Lys-120 (Fig. 5A), it should be resistant to p53-dependent apoptosis induced by DNA damage. Indeed, overexpression of TIP60–4KR in U2OS cells did show reduced apoptosis after DNA damage (Fig. 5, B and C). Similar results were obtained in the cotransfection of TIP60 with HDAC3, SIRT1, or both (Fig. 5, B and C), indicating that deacetylation of TIP60 reduced its ability to acetylate p53 at Lys-120, therefore reducing its ability to induce apoptosis in response to DNA damage. Conversely, when HDAC3 was down-regulated in cells which increased TIP60 acetylation, cells became more vulnerable to apoptosis in response to DNA damage (Fig. 5, D and E).
Our findings regarding HDAC3 deacetylating TIP60 and regulating its function provide a better understanding of TIP60 regulatory mechanisms, which is a significant basis for further studies of its cellular functions. Several questions still remain to be answered. How does HDAC3-mediated deacetylation affect TIP60 enzymatic activity, protein stability, and cellular functions of TIP60? What is the correlation and differentiation of the SIRT1- and HDAC3-mediated regulation of TIP60? It will be interesting and noteworthy for further studies to answer these questions.
Acknowledgments
We thank Dr. Mitchell A. Lazar for the HDAC3-Y298F constructs. We thank Dr. Si-Qing Zhang for technical support and suggestions. We also thank Wei Fan and Kai Li for technical support and John Luo for proofreading.
Footnotes
- MYST
- MOZ, Ybf2/Sas3, Sas2, TIP60
- HAT
- histone acetyltransferase
- MOZ
- monocyticleukaemia zinc finger protein
- CREB
- cAMP-response element-binding protein
- TSA
- trichostatin A
- IP
- immunoprecipitation
- ATM
- Ataxia telangiectasia mutated.
REFERENCES
- 1. Kamine J., Elangovan B., Subramanian T., Coleman D., Chinnadurai G. (1996) Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology 216, 357–366 [DOI] [PubMed] [Google Scholar]
- 2. Utley R. T., Côté J. (2003) The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 274, 203–236 [DOI] [PubMed] [Google Scholar]
- 3. Ikura T., Ogryzko V. V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 [DOI] [PubMed] [Google Scholar]
- 4. Squatrito M., Gorrini C., Amati B. (2006) Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16, 433–442 [DOI] [PubMed] [Google Scholar]
- 5. Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., Kimura H., Ikura M., Nishikubo S., Ito T., Muto A., Miyagawa K., Takeda S., Fishel R., Igarashi K., Kamiya K. (2007) DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell Biol. 27, 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murr R., Loizou J. I., Yang Y. G., Cuenin C., Li H., Wang Z. Q., Herceg Z. (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91–99 [DOI] [PubMed] [Google Scholar]
- 7. Sapountzi V., Logan I. R., Robson C. N. (2006) Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38, 1496–1509 [DOI] [PubMed] [Google Scholar]
- 8. Cao X., Südhof T. C. (2001) A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 [DOI] [PubMed] [Google Scholar]
- 9. Xiao H., Chung J., Kao H. Y., Yang Y. C. (2003) Tip60 is a co-repressor for STAT3. J. Biol. Chem. 278, 11197–11204 [DOI] [PubMed] [Google Scholar]
- 10. Bararia D., Trivedi A. K., Zada A. A., Greif P. A., Mulaw M. A., Christopeit M., Hiddemann W., Bohlander S. K., Behre G. (2008) Proteomic identification of the MYST domain histone acetyltransferase TIP60 (HTATIP) as a co-activator of the myeloid transcription factor C/EBPα. Leukemia 22, 800–807 [DOI] [PubMed] [Google Scholar]
- 11. Sun Y., Jiang X., Price B. D. (2010) Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischle W. (2009) Tip60-ing the balance in DSB repair. Nat. Cell Biol. 11, 1279–1281 [DOI] [PubMed] [Google Scholar]
- 13. Tang Y., Luo J., Zhang W., Gu W. (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 [DOI] [PubMed] [Google Scholar]
- 14. Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Y., Xu Y., Roy K., Price B. D. (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell Biol. 27, 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee M. S., Seo J., Choi D. Y., Lee E. W., Ko A., Ha N. C., Yoon J. B., Lee H. W., Kim K. P., Song J. (2013) Stabilization of p21 (Cip1/WAF1) following Tip60-dependent acetylation is required for p21-mediated DNA damage response. Cell Death Differ. 20, 620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee H. J., Chun M., Kandror K. V. (2001) Tip60 and HDAC7 interact with the endothelin receptor a and may be involved in downstream signaling. J. Biol. Chem. 276, 16597–16600 [DOI] [PubMed] [Google Scholar]
- 18. Yamagata K., Kitabayashi I. (2009) Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem. Biophys. Res. Commun. 390, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 19. Wang J., Chen J. (2010) SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J. Biol. Chem. 285, 11458–11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng L., Ling H., Yuan Z., Fang B., Bloom G., Fukasawa K., Koomen J., Chen J., Lane W. S., Seto E. (2012) SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol. Cell Biol. 32, 2823–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Z., Feng D., Fang B., Mullican S. E., You S. H., Lim H. W., Everett L. J., Nabel C. S., Li Y., Selvakumaran V., Won K. J., Lazar M. A. (2013) Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheridan A. M., Force T., Yoon H. J., O'Leary E., Choukroun G., Taheri M. R., Bonventre J. V. (2001) PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A(2), induces apoptosis, and potentiates prostaglandin production. Mol. Cell Biol. 21, 4470–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sliva D., Zhu Y. X., Tsai S., Kamine J., Yang Y. C. (1999) Tip60 interacts with human interleukin-9 receptor α-chain. Biochem. Biophys. Res. Commun. 263, 149–155 [DOI] [PubMed] [Google Scholar]
- 24. Yamamoto T., Horikoshi M. (1997) Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 272, 30595–30598 [DOI] [PubMed] [Google Scholar]
- 25. Stiehl D. P., Fath D. M., Liang D., Jiang Y., Sang N. (2007) Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation. Cancer Res. 67, 2256–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thompson P. R., Wang D., Wang L., Fulco M., Pediconi N., Zhang D., An W., Ge Q., Roeder R. G., Wong J., Levrero M., Sartorelli V., Cotter R. J., Cole P. A. (2004) Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11, 308–315 [DOI] [PubMed] [Google Scholar]
- 27. Albaugh B. N., Arnold K. M., Lee S., Denu J. M. (2011) Autoacetylation of the histone acetyltransferase Rtt109. J. Biol. Chem. 286, 24694–24701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blanco-García N., Asensio-Juan E., de la Cruz X., Martínez-Balbás M. A. (2009) Autoacetylation regulates P/CAF nuclear localization. J. Biol. Chem. 284, 1343–1352 [DOI] [PubMed] [Google Scholar]
- 29. Yuan H., Rossetto D., Mellert H., Dang W., Srinivasan M., Johnson J., Hodawadekar S., Ding E. C., Speicher K., Abshiru N., Perry R., Wu J., Yang C., Zheng Y. G., Speicher D. W., Thibault P., Verreault A., Johnson F. B., Berger S. L., Sternglanz R., McMahon S. B., Côté J., Marmorstein R. (2012) MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 31, 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang C., Wu J., Sinha S. H., Neveu J. M., Zheng Y. G. (2012) Autoacetylation of the MYST lysine acetyltransferase MOF protein. J. Biol. Chem. 287, 34917–34926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arif M., Kumar G. V., Narayana C., Kundu T. K. (2007) Autoacetylation induced specific structural changes in histone acetyltransferase domain of p300: probed by surface enhanced Raman spectroscopy. J. Phys. Chem. B 111, 11877–11879 [DOI] [PubMed] [Google Scholar]
- 32. Yang C., Wu J., Zheng Y. G. (2012) Function of the active site lysine autoacetylation in TIP60 catalysis. Plos One. 7, e32886. [DOI] [PMC free article] [PubMed] [Google Scholar]