Background: Capsule polymerases of Neisseria meningitidis serogroups W and Y comprise hexosyl- and sialyltransferase activity.
Results: Hexosyltransferase activity is encoded by the predicted N-terminal GT-B fold. Sialyltransferase activity requires 168 additional amino acids upstream of the predicted C-terminal GT-B fold.
Conclusion: The sialyltransferase domains of NmW/Y define a new glycosyltransferase (CAZy) family.
Significance: The new CAZy family comprises sequences from distantly related species.
Keywords: Bioinformatics, Enzyme Catalysis, Glycosyltransferase, Phylogenetics, Polysaccharide, Fluorescence-based Testing of Glycosyltransferases, Neisseria meningitidis Serogroup W and Y, Capsule Polymerases, Hexosyltransferases, Sialyltransferases
Abstract
Crucial virulence determinants of disease causing Neisseria meningitidis species are their extracellular polysaccharide capsules. In the serogroups W and Y, these are heteropolymers of the repeating units (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n in NmW and (→6)-α-d-Glc-(1→4)-α-Neu5Ac-(2→)n in NmY. The capsule polymerases, SiaDW and SiaDY, which synthesize these highly unusual polymers, are composed of two predicted GT-B fold domains separated by a large stretch of amino acids (aa 399–762). We recently showed that residues critical to the hexosyl- and sialyltransferase activity are found in the predicted N-terminal (aa 1–398) and C-terminal (aa 763–1037) GT-B fold domains, respectively. Here we use a mutational approach and synthetic fluorescent substrates to define the boundaries of the hexosyl- and sialyltransferase domains. Our results reveal that the active sialyltransferase domain extends well beyond the predicted C-terminal GT-B domain and defines a new glycosyltransferase family, GT97, in CAZy (Carbohydrate-Active enZYmes Database).
Introduction
Bacterial capsules are a protective layer of extracellular polysaccharides that are firmly attached to the cell surface (1). Capsules both enhance bacterial survival in the environment by providing a highly hydrated physical barrier surrounding the cell (2) and contribute to symbiotic and pathogenic interactions with their host (1, 3). Capsules are divided into four groups based on characteristics of the polysaccharide and export machinery (4, 5). Group II capsules are characterized by polysaccharides with a high charge density and export via an ATP-binding cassette transporter-dependent system (6). Neisseria meningitidis (also referred to as meningococcus), a group of strictly human pathogens, produce group II capsules and are divided into 12 serogroups based on the chemical properties of the capsular polysaccharides (CPSs)4 (7). Six of these serogroups (A, B, C, W, X, and Y) are important pathogens (8) (the serogroup nomenclature is simplified according to Harrison et al. (7)). In serogroups B, C, W, and Y, the negative charge of the CPS results from the building block sialic acid (Sia or Neu5Ac), a nonasugar carrying a carboxylate function in position 1 (9). Whereas CPSs in NmB and NmC are homopolymers with Neu5Ac in α2,8- and α2,9-glycosidic linkage, respectively, the CPSs of NmW and NmY represent heteropolymers of the repeating units (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n in NmW and (→6)-α-d-Glc-(1→4)-α-Neu5Ac-(2→)n in NmY (10). These latter structures are highly unusual because sialic acid occurs as an internal sugar. As a rule, sialic acid is a terminal (non-reducing end) sugar and, if internal, is conjugated with other sialic acid residues (11).
We recently succeeded with the molecular cloning and functional characterization of the capsule polymerases of NmW (SiaDW) and NmY (SiaDY) and demonstrated bifunctionality for both enzymes (12). The large proteins with molecular masses of 120 kDa are 98% identical at the amino acid level and were predicted to comprise two glycosyltransferase (GT) domains of the GT-B fold (13). Bioinformatics and rational mutagenesis showed that the N-terminal domains encompass hexosyltransferase (HexTF) activity and that the C-terminal domains encode the sialyltransferase (SiaTF) activity (12). Point mutations introduced to destroy one catalytic function fully inactivated the capsule polymerases SiaDW and SiaDY. However, if the single mutant enzymes were combined in the same reaction tube, 70% of wild-type activity was restored. Beyond the demonstration that the two functional domains are capable of complementing each other in trans, these data indicated that chain elongation needs the successive activity of HexTF and SiaTF domains (12). Additional proof for the independence of the HexTF and SiaTF domains was obtained by saturation transfer difference NMR, showing the simultaneous binding of both sugar-nucleotides (CMP-Neu5Ac and UDP-Gal) and the priming oligosaccharide acceptor (12).
The N-terminal HexTF domains of SiaDW and SiaDY are classified in CAZy family GT4 (14). On the other hand, due to the absence of similarity to known sialyltransferase families and the lack of defined boundaries for the SiaTF domains in SiaDW and SiaDY, these domains could not be assigned to a CAZy family.
Here we aimed to determine whether the putative SiaTF domains of SiaDW and SiaDY could be physically separated from the respective HexTF domains and to delineate the boundaries of the SiaTF and HexTF domains. Crucial toward this goal was the availability of a sensitive assay system that would enable the unequivocal detection of single sugar transfers. To this end, we synthesized fluorescently labeled acceptor substrates with which HexTF and SiaTF could be specifically primed. High performance liquid chromatography separation and fluorescence detection (HPLC-FD) of the labeled reaction products provided the single-product resolution necessary to delineate the two glycosyltransferase activities. We show that the N-terminal GT-B folds contain all of the information for HexTF activity, whereas the predicted C-terminal GT-B domains were insufficient to generate active SiaTFs. Using a mutational approach, the SiaTFs were shown to require an additional 168 amino acids immediately upstream of the predicted GT-B domains. Bioinformatics analyses demonstrate that the newly identified SiaTF domains are the first characterized members of a new CAZy family, GT97.
EXPERIMENTAL PROCEDURES
Generation of Truncation Mutants and Expression of Recombinant Proteins
To separate HexTF and SiaTF domains, truncated variants of SiaDW and SiaDY (Fig. 4) were designed with care not to destroy secondary structure elements as predicted by the Phyre2 server (13). These constructs were generated by PCR using pHC4 (SiaDW) and pHC5 (SiaDY) (15) as templates. Hot Start Phusion-DNA-Polymerase (Thermo Scientific/Fermentas) was used in these experiments, and PCR conditions were as follows: 1 cycle of 98 °C/120 s; 30 cycles of 98 °C/15 s, 65 °C/30 s, 72 °C/30 s; and 1 cycle of 72 °C/300 s. Used primers, containing NdeI or XhoI sites, together with the obtained truncation variants are listed in Table 1. PCR products after digestion with NdeI and XhoI (New England Biolabs, Inc.) were purified and ligated into the respective sites of the expression vector pET22-b (Novagen). After transformation into Escherichia coli XL-1 Blue (Stratagene), transformed colonies were selected on ampicillin, and constructs were controlled by restriction analysis and sequencing. Expressed proteins carried a C-terminal His6 epitope. If recombinant proteins were purified, the protocol described by Romanow et al. (12) was used.
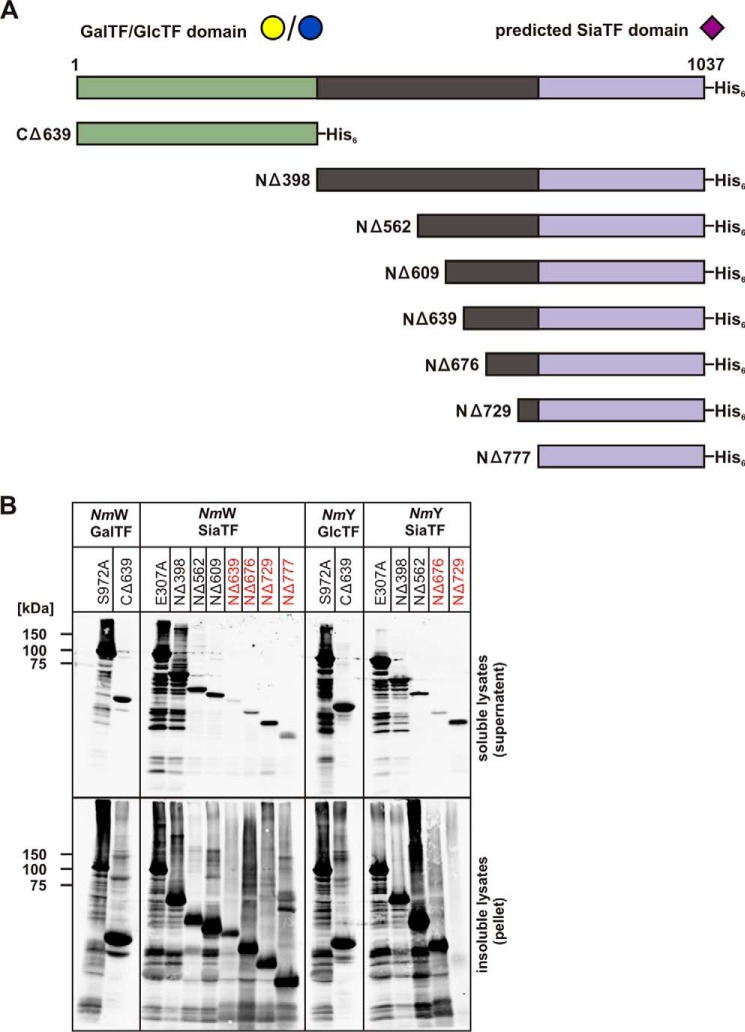
FIGURE 4.
The capsule polymerases from NmW and NmY and deletion mutants made thereof. A, schematic illustration of the full-length capsule polymerases NmW/NmY. The GT-B folds predicted to comprise HexTF and SiaTF are highlighted in green and purple, respectively (13). The linker region connecting the GT-B folded domains is shown in black. Mutants made in the course of this project were named according to the introduced deletion. All proteins were expressed with an N-terminal His6 tag. B, Western blot to monitor protein expression. Constructs as indicated were expressed in BL21(DE3), transformed cells were lysed, and lysates were separated into soluble and insoluble fractions. After electrophoretic separation on 10% SDS-PAGE and Western blotting, proteins were displayed with an anti-penta-His antibody. Protein variants that were inactive in the in vitro test system are marked in red.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strains/Plasmids/Primers | Description/Sequence | Source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | B; F−ompT hsdSB(rB−mB−) gal dcm (DE3) | Novagen |
| XL1-Blue | RecA1 ebdA1 gyrA96 thi-1 hsdR17 supE44 | Stratagene |
| Plasmids | ||
| pET-22b(+) | Novagen | |
| Primer pairs used for cloning | ||
| KS422/KS273 | 5′-GCATCTCATATGGCTGTTATTATATTTGTTAACG-3′ | |
| WT NmW | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS422/KS273 | 5′-GCATCTCATATGGCTGTTATTATATTTGTTAACG-3′ | |
| WT NmY | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| Primer pairs used for the introduction of mutation in SiaDW | ||
| KS350/KS351 | 5′-CTGATCATGACATCAGAAAGTGCGGGATTTCCATATATATTTATG-3′ | |
| E307A | 5′-CATAAATATATATGGAAATCCCGCACTTTCTGATGTCATGATCAG-3′ | |
| KS370/KS371 | 5′-ATCTCGCGTTGCTGTAGGTGTTTATGCAACTAGCTTATTTG-3′ | |
| S972A | 5′-CAAATAAGCTAGTTGCATAAACACCTACAGCAACGCGAGAT-3′ | |
| Primer pairs used for the introduction of mutation in SiaDY | ||
| AR11/AR12 | 5′-ATACAGATATCCTAATCATGACATCTCAAAGCGCAGGCTTTGGTTATATAT-3′ | |
| E307A | 5′-atatataaccaaagcctgcgctttgagatgtcatgattaggatatctgtat-3′ | |
| KS370/KS371 | 5′-ATCTCGCGTTGCTGTAGGTGTTTATGCAACTAGCTTATTTG-3′ | |
| S972A | 5′-CAAATAAGCTAGTTGCATAAACACCTACAGCAACGCGAGAT-3′ | |
| Primer pairs used for the generation of SiaDW truncations | ||
| KS422/KS421 | 5′-GCATCTCATATGGCTGTTATTATATTTGTTAACG-3′ | |
| CΔ639 | 5′-CCGCTCGAGGCTGCGCGGAAGAATAGTG-3′ | |
| AR2/KS273 | 5′-GCATCTCATATGTTTAATAACGTATCATTATCGTC-3′ | |
| NΔ398 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| AR1/KS273 | 5′-GCATCTCATATGACTGATGATAATTTAATACCTAT-3′ | |
| NΔ562 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| AR9/AR273 | 5′-GCATCTCATATGAAATATTCTTATAAATATATCTA-3′ | |
| NΔ609 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| AR10/KS273 | 5′-GCATCTCATATGTCTTGGGAACTTATTCGTGCCTC-3′ | |
| NΔ639 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS433/KS273 | 5′-GCATCTCATATGGGTAAGCGTTCGATGGATG-3′ | |
| N-Δ676 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS434/KS273 | 5′-GCATCTCATATGTCACTGAAAAGTAATGTAGTTG-3′ | |
| NΔ729 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS435/KS273 | 5′-GCATCTCATATGAATATCGAAGCATTTCTAAAACC-3′ | |
| NΔ777 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| Primer pairs used for the generation of SiaDY truncations | ||
| KS422/KS421 | 5′-GCATCTCATATGGCTGTTATTATATTTGTTAACG-3′ | |
| CΔ639 | 5′-CCGCTCGAGGCTGCGCGGAAGAATAGTG-3′ | |
| AR2/KS273 | 5′-GCATCTCATATGTTTAATAACGTATCATTATCGTC-3′ | |
| NΔ398 | 5-´CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| AR1/KS273 | 5′-GCATCTCATATGACTGATGATAATTTAATACCTAT-3′ | |
| NΔ562 | 5′-CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS433/KS273 | 5′- GCATCTCATATGGGTAAGCGTTCGATGGATG-3′ | |
| NΔ676 | 5-´CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
| KS434/KS273 | 5′-GCATCTCATATGTCACTGAAAAGTAATGTAGTTG-3′ | |
| NΔ729 | 5-´CCGCTCGAGTTTTTCTTGGCCAAAAAACTG-3′ | |
Synthesis and Purification of Fluorescently Labeled Acceptors to Prime SiaDW and SiaDY Reactions
Because activity testing in the classical radioactive incorporation assay depends on polymer formation (short oligosaccharides are mobile in the paper chromatography step (16)), the reliable testing of monofunctional mutants required a test system in which single sugar transfers could be clearly observed. We exploited the capability of SiaDW and SiaDY to extend sialic acid derivatives carrying the fluorescent label 2′-(4-methylumbelliferyl) (4-MU) at the reducing end. The recombinant C-terminally His6-tagged monofunctional full-length enzymes NmW-(S972A)-His6 and NmY-(S972A)-His6 were used as HexTFs, and NmW-(E307A)-His6 and NmY-(E307A)-His6 were used as SiaTFs. 4-MU-Sia-Gal/Glc were obtained by mixing 4 mm 4-MU-Sia (Sigma-Aldrich or Iris Biotech GmbH) with 4 mm donor sugar UDP-Gal/UDP-Glc (Sigma) and 20 μg ml−1 of the respective HexTF (NmW-(S972A)-His6 or NmY-(S972A)-His6) in reaction buffer (20 mm Tris/HCl, pH 8.0, 20 mm MgCl2, and 2 mm DTT). After 24 h at 25 °C, enzymes were removed by ultrafiltration (Amicon®Ultra 10 molecular weight cut-off, Millipore), and filtrates containing the reaction products (4-MU-Sia-Gal or 4-MU-Sia-Glc) were lyophilized (Alpha 1-2 LD Plus, Martin Christ). After dissolution in water, samples were desalted on P2-gel filtration columns (Bio-Rad). Reaction products were identified by high performance liquid chromatography using an UFLC-RX system (Shimadzu) coupled to a fluorescence detector (RF-10A XL). Samples were excited at 315 nm and monitored at 375 nm. Although, under the conditions used, this reaction did not give product yields of >90% (see Fig. 2), further product purification was omitted because the acceptor quality increased significantly from 4-MU-Sia to 4-MU-Sia-Hex, making 4-MU-Sia an irrelevant contaminant. The obtained 4-MU-Sia-Gal/Glc were then the starting material for Sia transfer to obtain 4-MU-Sia-Gal/Glc-Sia. This reaction was carried out with either NmW-(E307A)-His6 or NmY-(E307A)-His6 in the presence of 2 mm CMP-Neu5Ac (Nacalai Tesque). Reaction conditions were identical to those described in the first reaction step. However, due to the significantly improved acceptor, the sialylation in this step was complete after 1 h of incubation. After the removal of enzymes and desalting, obtained compounds were used in iterative rounds to synthesize primers of the needed size.
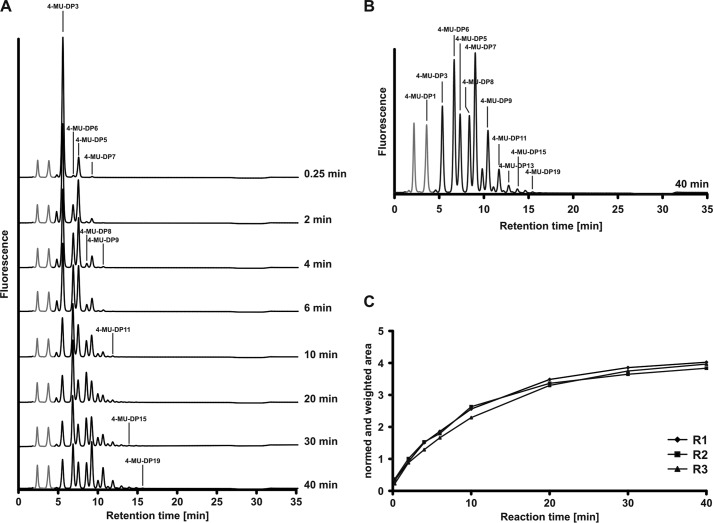
FIGURE 2.
Time lapse recording of the SiaDW reaction. A, SiaDW was primed with 1 mm 4-MU-Sia-Gal-Sia (4-MU-DP3) in the presence of 2 mm UDP-Gal and 2 mm CMP-Sia. At the indicated time points, product profiles were recorded by HPLC-FD. Gray peaks, 4-MU and 4-MU-DP1, which were left in the priming 4-MU-DP3 pool. Due to the steep increase in acceptor quality from DP2 to DP3, both peaks remained unchanged over the reaction course. B, enlarged display of the HPLC-FD profile obtained at 40 min and labeling of individual DPs. C, reaction progress curves were generated by quantification of each chromatogram and plotting these values against the reaction time point. Three independent experiments (R1–R3) document the high reproducibility of the reaction.
Enzyme Testing
Optimal buffer conditions (pH and metal ion concentration) for SiaDW/Y activity were established using a radioactive incorporation assay (12). Identical to other neisserial capsule polymerases, SiaDW/Y activity was optimal at slightly basic pH. Conditions identified with the wild type enzymes were maintained for all assays in this study. For activity testing with fluorescent compounds, reactions were carried out in a total volume of 25 μl. Mixtures contained 50 mm Tris/HCl, pH 8.0, 20 mm MgCl2, 2 mm DTT, 1 mm acceptor (4-MU-Sia-Gal/Glc, 4-MU-Sia-Gal/Glc-Sia, or 4-MU-Sia-Gal/Glc-Sia-Gal/Glc) plus 2 mm of the nucleotide sugar (UDP-Gal/Glc (Sigma) and/or 2 mm CMP-Neu5Ac (Nacalai Tesque)), depending on the tested enzyme. Reactions were started by the addition of 20 μg ml−1 purified enzyme or, if the soluble fraction of bacterial lysates was used as the enzyme source, with 72–100 μg ml−1 total protein. After appropriate incubation times, reactions were stopped by shock freezing in liquid nitrogen.
Synthesized oligo- and polymers were analyzed and quantified via the fluorescent tag using an ultrafast HPLC system (UFLC-RX, Shimadzu) with coupled FD (detector RF-10A XL). The separation of 4-MU-labeled products was achieved with an anion exchange column (CarboPac® PA-100 column, Dionex). Before loading onto the column, samples were diluted 500-fold in water. The buffers A (20 mm NaNO3) and B (1 m NaNO3) were used to establish a curved elution gradient, reaching 21.65% buffer B over 35 min at 0.6 ml min−1 and a column temperature of 50 °C. The curved portion of the gradient is described by the following formula, in which the index −1.425 describes the slope (LS Solution, Shimadzu) (17–19).
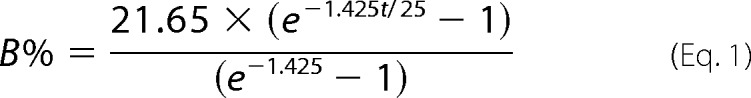 |
Elution profiles were monitored via fluorescence emission at 375 nm with 315 nm as the excitation wavelength. Under these conditions, the separation of polysaccharides up to a degree of polymerization (DP) of 21 was easily achieved (see “Results”).
The HPLC profiles can be quantified to determine reaction progress. Peak areas represent the relative amount of each DP in the product profile and were calculated by integration of HPLC chromatograms with the LC Solution software (Shimadzu). Normalizing peak areas to the total area under the curve, weighting each peak according to the number of transfers required to form this product, and summing over all peaks gives the “normalized and weighted area” according to the formula,
 |
where A is the peak area, and n is the number of transfers required to form this product. The normalized and weighted area is directly proportional to the total number of sugar transfers at that reaction time point.
SDS-PAGE and Immunoblotting
SDS-PAGE was performed under reducing conditions using 2.5% (v/v) β-mercaptoethanol. Western blot analysis was done on a PVDF membrane (Millipore). For detection of the hexahistidine (His6) tag, penta-His antibody (Qiagen) was used as primary antibody at a concentration of 1 μg ml−1 and subsequently detected with 0.05 μg ml−1 IgG1 anti-mouse IR Dye800 antibody (Odyssey Infrared Imaging®). Protein bands were visualized and quantified with the infrared fluorescence detection system (LI-COR®) according to the manufacturer's instructions.
Bioinformatics
The most similar homologs of the identified SiaTF domain were found by iterative PSI-BLAST searches of the non-redundant protein sequences database using the PSI-BLAST server (20). In each round, sequences with identity of >20% and coverage of >70% were included in the following iteration. After four iterations, the collected sequences were filtered for an E-value of better than 1e−50 and for the presence of conserved motifs (see “Results”). These sequences were aligned using the MAFFT server (G-INS-i strategy; BLOSUM30 scoring matrix; unalignlevel: 0.4) (21). Poorly fitting sequences were removed with MaxAlign (22) and by visual inspection of alignments and preliminary trees. The maximum likelihood tree was calculated from 359 ungapped positions using the PhyML software with bootstrapping (23). The tree was visualized with Archaeopterix (24).
RESULTS
Synthesis of Fluorescently Labeled Oligosaccharide Substrates
With the intention to delineate HexTF and SiaTF activities and functionally express the individual enzyme domains, it was first necessary to have defined acceptors to prime each of these activities. As detailed under “Experimental Procedures,” this was achieved using the previously described active site mutants bearing only the HexTF or SiaTF activity (12). The commercially available, but poorly used, fluorescent substrate 4-MU-Sia was first extended by a single hexose using either NmW-(S972A)-His6 or NmY-(S972A)-His6 to give 4-MU-Sia-Gal or 4-MU-Sia-Glc, respectively (henceforth referred to as 4-MU-Sia-Hex or 4-MU-DP2). The 4-MU-DP2 was then extended by the respective SiaTF (NmW-(E307A)-His6 or NmY-(E307A)-His6) to give 4-MU-Sia-Hex-Sia (4-MU-DP3), which proved to be an efficient acceptor for the HexTFs. Structures of the new acceptors (as an example shown for oligosaccharides with Gal as hexose) are given in Fig. 1.
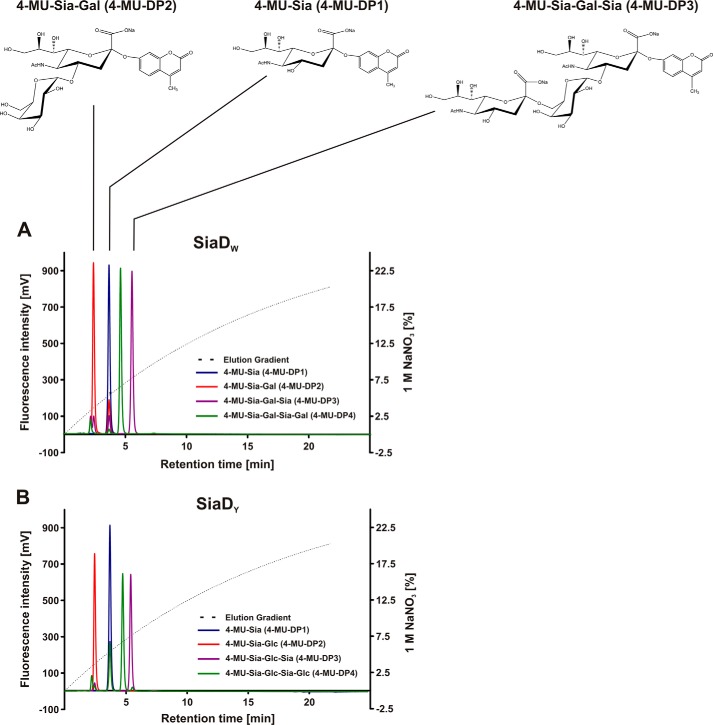
FIGURE 1.
Fluorescently labeled primers for the separate testing of hexosyl- and sialyltransferase activity and chromatographic behavior in anion exchange chromatography. Starting from 4-MU-Sia, the monofunctional active site mutants of SiaDW/SiaDY were used in iterative steps to generate labeled acceptors of defined length. Reaction samples were analyzed on a CarboPac® PA-100 column, and products eluted with a curved NaNO3 gradient as shown. A, structures of the labeled SiaDW glycans (top) and their running behavior on anion exchange chromatography (bottom) are shown. Note that the retention time increases with the addition of a Sia residue and decreases with the addition of Hex, leading to the elution order DP2, DP1, DP4, and then DP3. B, the elution behavior of labeled SiaDY glycans is similar to that of SiaDW. Of note, the chromatograms show that the conversion of 4-MU-DP1 to 4-MU-DP2 was incomplete under the conditions used, whereas all other reactions proceeded to completion.
Anion exchange separation of 4-MU-Sia and the elongated products 4-MU-DP2 to 4-MU-DP4 was achieved by HPLC with a CarboPac PA-100 column and NaNO3 gradient. Product profiles for the NmW and NmY substrates show baseline resolution of all compounds (Fig. 1, A and B). Although the elution behavior of each product is dominated by the total charge, charge density also plays a role. This interplay is manifested as large increases in retention time upon the addition of a negatively charged sialic acid and a smaller reduction in retention time upon the addition of a neutral hexose. The subtle effect of different Gal and Glc stereochemistries on the conformation of NmW and NmY oligosaccharides also results in small differences in the observed retention times. Although transfer of the first hexose onto 4-MU-DP1 remained incomplete under the conditions used, subsequent transfer reactions proceeded to completion. Notably, the wild-type enzymes were unable to initiate polymerization on 4-MU-Gal and 4-MU-Glc. The finding suggests that the SiaTF domain requires at least a disaccharide to prime activity or, alternatively, recognition of the acceptor may require the presence of at least one sialic acid residue.
The Fluorescent Substrates Enable Sensitive and Quantitative Detection of Capsule Polymerase Activity
To observe the initial steps in chain elongation, the wild-type SiaDW/Y were primed with 1 mm 4-MU-DP3 in the presence of a 4-fold excess of donor sugar (2 mm CMP-Sia and 2 mm UDP-Gal/Glc). Reactions were sampled over 40 min, and products were examined by HPLC-FD (Fig. 2). Shown are representative results obtained with SiaDW. Under these conditions, we observed the successive elongation of 4-MU-DP3 up to a maximum of DP19 (DP16 in the case of SiaDY).
The new assay system enabled a precise quantification of SiaDW/Y activity. The normalized and weighted area (see “Experimental Procedures”) is directly proportional to the total number of sugars transferred at a given reaction time point. Progress curves are generated by plotting these values against reaction time (Fig. 2C). The congruence of reaction progress curves obtained in three independent experiments demonstrates the reliability of the assay system.
Observation of Single Sugar Transfers Confirms Monofunctionality of Active Site Mutants
We previously generated active site mutants that disabled either the SiaTF or HexTF function of SiaDW (12). However, monofunctionality was tested in a complementation assay using paper chromatography to detect incorporation of radiolabeled sugars into long polysaccharides. In order to unequivocally observe the transfer of single sugar residues, we decided to test the single point mutant enzymes with the new fluorescent acceptor substrates. In separate reactions, NmW-(S972A)-His6 and NmW-(E307A)-His6 were incubated with both donor sugars and primed with either 4-MU-DP2 or 4-MU-DP3 (Fig. 3). As expected, each enzyme transferred a single sugar residue onto the appropriate acceptor, clearly demonstrating the monofunctionality of these enzymes. These monofunctional enzymes served as positive controls in the subsequent experiments to delineate the HexTF and SiaTF domains.
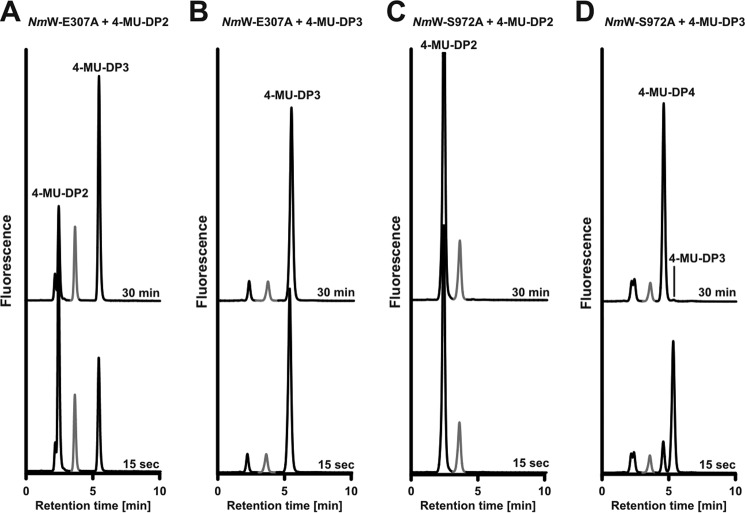
FIGURE 3.
Use of fluorescent primers to monitor the reaction products of single point mutant enzymes. Reaction profiles of the monofunctional mutants NmW-(E307A)-His6 and NmW-(S972A)-His6 were recorded in the presence of both donor sugars using the fluorescent primers as indicated. Reaction samples were taken after 15 s and 30 min. Peaks marked in gray represent 4-MU-DP1 and are unmodified in all reactions. A, NmW-(E307A)-His6 with 4-MU-DP2. The only reaction product formed was 4-MU-DP3. B, NmW-(E307A)-His6 primed with 4-MU-DP3. No product was formed. C, NmW-(S972A)-His6 primed with 4-MU-DP2. No product was formed. D, NmW-(S972A)-His6 primed with 4-MU-DP3. The only product formed was 4-MU-DP4.
Truncation Mutagenesis to Separate HexTF and SiaTF in SiaDW and SiaDY
A schematic representation of the linear sequence of SiaDW/Y (Fig. 4A) shows the previously predicted HexTF and SiaTF domains and a stretch of 365 amino acids linking these domains (12). In bioinformatics analyses, neither sequence homologies nor structural folds could be identified in the central 365 amino acids. Therefore, we initially hypothesized that this sequence is not part of the catalytic domains but is necessary for tertiary organization of the two GT-B domains. To test this hypothesis, constructs harboring the predicted catalytic domains CΔ639 and NΔ777 (see Fig. 4A) were expressed and tested with the new fluorescent acceptors.
The predicted HexTF domain displayed similar activity to the respective active site mutants (Fig. 5A; see Fig. 3D for comparison), confirming that the predicted N-terminal GT-B fold (NmW-CΔ639-His6) comprises the active domain.
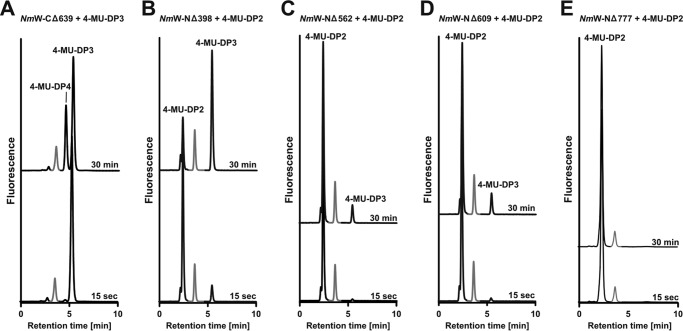
FIGURE 5.
Activity testing of truncation mutants. Truncation mutant comprising the HexTF (A) and the SiaTF with variant N-terminal extensions (B–E) were assayed in the presence of both donor sugars and a suited fluorescently labeled acceptor as indicated. Reaction samples were taken after 15 s and 30 min. Peaks marked in gray represent 4-MU-DP1. These peaks are unchanged over the reaction course. A, NmW-(CΔ639)-His6 with 4-MU-DP3 catalyzes the transfer of UDP-Gal by creating 4-MU-DP4. B, NmW-(NΔ398)-His6 primed with 4-MU-DP2 catalyzes the transfer of CMP-Sia by producing 4-MU-DP3. C, NmW-(NΔ562)-His6 primed with 4-MU-DP2 catalyzes the transfer of CMP-Sia by creating 4-MU-DP3. D, NmW-(NΔ609)-His6 primed with 4-MU-DP2 catalyzes the transfer of CMP-Sia by producing 4-MU-DP3. E, NmW-(NΔ777)-His6 primed with 4-MU-DP2 does not catalyze the transfer of CMP-Sia.
In contrast, no activity could be detected with the predicted SiaTF domain (NΔ777; Fig. 5E), suggesting that the linker region, or part thereof, might be necessary to form the active SiaTF domain. Thus, the NΔ398 truncation, comprising the SiaTF domain and the entire linker region (see Fig. 4A) was generated and tested. Indeed, this construct showed SiaTF activity comparable with the positive control (compare Figs. 5B and 3A), confirming that the SiaTF domain extends beyond the predicted GT-B fold. Further truncation constructs were tested to more precisely define the boundary of the SiaTF domain. We found that NΔ562 and NΔ609 (Fig. 5, C and D) retained SiaTF activity, but further truncation completely inactivated the enzyme. The results described for the HexTF and SiaTF domains of SiaDW were similarly obtained for the two domains in SiaDY. The expression of SiaDY truncation mutants is displayed in Fig. 4B. Activity data are not shown.
Taken together, the truncation studies defined the boundaries of the SiaTF domains of these capsule polymerases and demonstrated that the domains can be separated and expressed as active transferases. As originally predicted, the N-terminal HexTF domain comprises a classic GT-B fold belonging to CAZy family GT4 (25). In contrast, the C-terminal SiaTF domain extends beyond the predicted GT-B fold and includes ∼170 additional amino acids of the linker region. The sequence stretch connecting the two GT-B folds is thus not a linking element but part of the functional SiaTF domains in SiaDW/Y.
The Sialyltransferase Domains in SiaDW/Y Define a New Glycosyltransferase Family
BLAST searches carried out with the newly identified SiaTF domain (SiaDW aa 563–1037) failed to reveal similarity to any of the known SiaTF families classified in the CAZy database (14). Further, searching the Pfam database (26) revealed no matches to HMM profiles (27, 28) constructed with the known SiaTF families. However, iterative PSI-BLAST searches of the non-redundant protein sequence database (29) revealed similarity to a number of uncharacterized protein sequences. The identified sequences were filtered for e-value (better than 1e−50) and for the presence of conserved motifs that have previously been identified in SiaDW (12). The presented data support the inclusion of these proteins as a new family of glycosyltransferases, GT97, in the CAZy database.
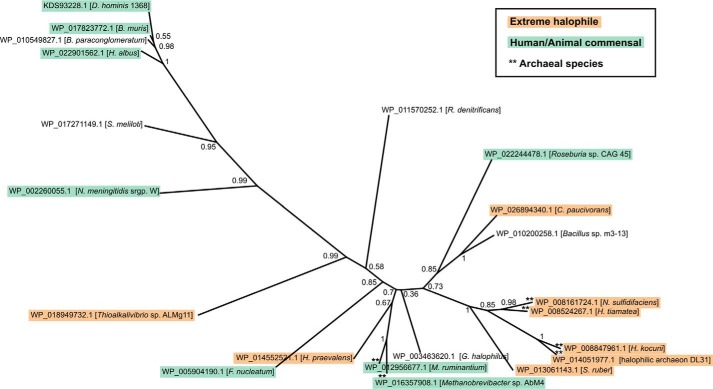
Members of the new family are only found in a handful of taxonomically scattered species of bacteria and archaea. No homologues were found in eukaryotes. Notably, the identified sequences are primarily found in two types of organisms with very distinct habitats: (i) commensal bacteria inhabiting humans and animals, including a number of opportunistic pathogens, as well as (ii) extremely halophilic bacteria and archaea isolated from saturated brines, salt lakes, and ponds (Fig. 6). Because archaea are not known to incorporate sialic acid into their glycans but have been shown to use homologous biosynthetic pathways for the incorporation of the related nonulosonic acids (NulOs), legionaminic acid and pseudaminic acid (30, 31), it is possible that GT97 glycosyltransferases exhibit specificity for these CMP-activated sugars. Supporting the putative function of GT97 family members as NulO transferases, the genome sequences of all of the identified organisms were found to harbor homologs of other nonulosonic acid biosynthesis genes, specifically NulO synthases (32, 30) and CMP-NulO synthetase (33, 30). Although the role of sialylated structures in commensal organisms and opportunistic pathogens is generally understood to be the avoidance of the immune system by molecular mimicry of host structures (34), the functional significance of GT97 family proteins in extreme halophiles is less clear. The phylogenetic tree for the new family revealed clustering that is largely at odds with the true phylogenetic relationships among these organisms (Fig. 6), suggesting that convergent evolution or horizontal gene transfer may have played a role in evolution of the GT97 family.
FIGURE 6.
Phylogenetic tree of family GT97. Phylogenetic analysis was carried out using the PhyML software with bootstrapping (23) using the alignment shown in Fig. 7 as input. Protein accession numbers and species names are indicated, and bootstrapping values are given at the nodes. GT97 family members were only found in the bacterial and archaeal domains of life, and the majority of these species are either human or animal commensals (including several opportunistic pathogens) or extreme halophiles, as indicated.
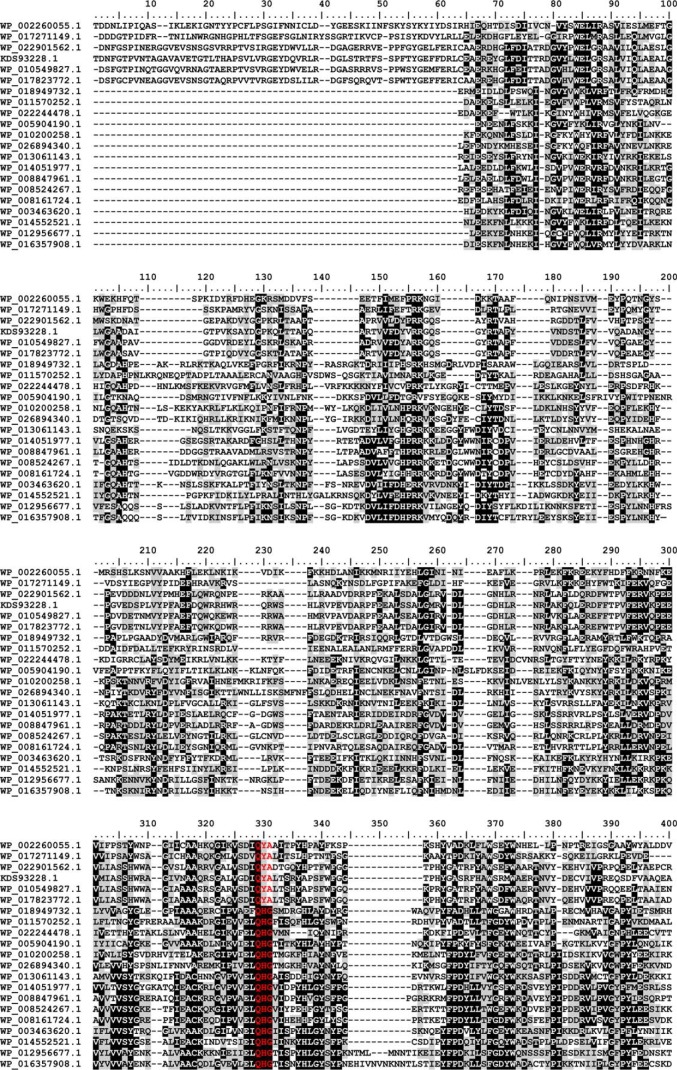
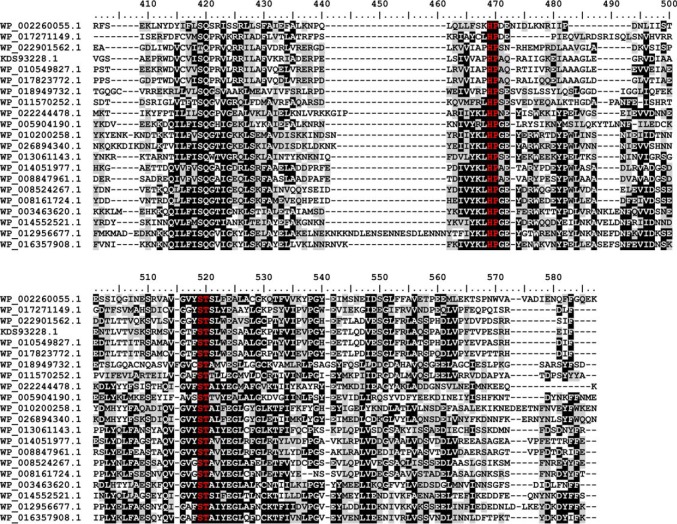
An alignment of the new family (Fig. 7) reveals slight differences from the known bacterial sialyl motifs (indicated in the alignment in red letters). The (D/E)(D/E)G motif (35), which is part of the catalytic center in bacterial sialyltransferases of CAZy families GT38, GT52, and GT80, was found to be replaced by QYA or QHG in the new family. Two other bacterial sialyl motifs, the HP motif (35) and the S(S/T) motif (36), which are involved in binding the nucleotide sugar (35), are also highly conserved in GT97. However, the S(S/T) motif is strictly ST in the GT97 family. The functional importance of the HP and the ST motifs has previously been demonstrated for SiaDW/Y (12). To interrogate the importance of the QYA motif, the point mutant SiaDW-Y836F was prepared and completely abolished activity (data not shown).
FIGURE 7.
Multiple sequence alignment of family GT97. Protein sequences were aligned with the MAFFT server (G-INS-I strategy; BLOSUM20 scoring matrix; unalignlevel: 0.4) (21) and visualized using BioEdit (42). High conservation is indicated by white letters on a black background, less conserved regions have black letters on a gray background, and the sialyl-motifs are highlighted in red. Accession numbers correspond to SiaDW and uncharacterized proteins from the following organisms: WP_002260055.1 (aa 563–1037/N. meningitidis serogroup W), WP_017271149.1 (aa 545–999/Sinorhizobium meliloti), WP_022901562.1 (aa 653–1117/Humibacter albus), KDS93228.1 (aa 678–1144/Dermabacter hominis 1368), WP_010549827.1 (aa 651–1116/Brachybacterium paraconglomeratum), WP_017823772.1 (aa 664–1129/Brachybacterium muris), WP_018949732.1 (aa 18–458/Thioalkalivibrio sp. ALMg11), WP_011570252.1 (aa 101–539/Roseobacter denitrificans), WP_022244478.1 (aa 8–439/Roseburia sp. CAG:45), WP_005904190.1 (aa 21–464/Fusobacterium nucleatum), WP_010200258.1 (aa 9–467/Bacillus sp. m3-13), WP_026894340.1 (aa 8–467/Clostridiisalibacter paucivorans), WP_013061143.1 (aa 8–446/S. ruber), WP_014051977.1 (aa 44–483/halophilic archaeon DL31), WP_008847961.1 (aa 33–474/H. kocurii), WP_008524267.1 (aa 8–446/H. tiamatea), WP_008161724.1 (aa 9–445/N. sulfidifaciens), WP_003463620.1 (aa 15–456/Gracilibacillus halophilus), WP_014552521.1 (aa 15–457/Halanaerobium praevalens), WP_012956677.1 (aa 15–497/Methanobrevibacter ruminantium), WP_016357908.1 (aa 13–470/Methanobrevibacter sp. AbM4).
Taken together, the work presented in the current study (i) defines the minimum boundaries of the SiaTF domains of SiaDW/Y, (ii) shows that these domains are functional in isolation, and (iii) allows us to define a novel glycosyltransferase family, GT97, which is present in commensal bacteria of humans and animals and in extremely halophilic archaea and bacteria.
DISCUSSION
With the recent demonstration that the capsule polymerases of the N. meningitidis serogroups W and Y (SiaDW and SiaDY) are chimeric proteins comprising family GT4 HexTFs at their N terminus and a SiaTF at their C terminus (12), the question arose of whether the two enzymatic functions depend on their common presence in one polypeptide chain or if the functional subunits can be expressed separately. Here we demonstrate that the latter is the case, and we report a truncation study that delineates the boundary of the functionally active C-terminal SiaTF domain. This functional domain is ∼170 residues longer than the previously predicted GT-B fold domain (12). However, from our results, it is not clear whether the additional region (amino acids 610–777) serves a chaperone function ensuring correct folding of the SiaTF domain or if it directly participates in catalysis.
This study sees SiaDW/Y classified into two CAZy families, GT4 based on the HexTF domain and GT97 based on the newly delineated SiaTF domain. Several other bifunctional GTs also belong to different CAZy families, including the Pasteurella multocida heparosan synthases (GT2 and GT45) (37) and the bifunctional human glycosyltransferase LARGE (GT8 and GT49) (38).
Successful delineation of the two catalytic units required a novel assay system for the detection of single sugar transfers. To this end, we exploited two monofunctional active site mutants, capable of transferring either a single hexose or sialic acid residue, for the controlled synthesis of fluorescently labeled acceptor substrates. The new substrates (4-MU-Sia-Hex for SiaTFs and 4-MU-Sia-Hex-Sia for HexTFs), combined with HPLC-FD analysis of reaction products, provided a highly sensitive and robust assay of these enzymatic activities. The stability and specificity of the new substrates for the target activity enabled activity measurements directly from bacterial lysates, which accelerates mutant testing enormously. In addition, we also demonstrate that the new assay system can be used to quantitatively monitor the product distribution and time course of the polymerization reaction.
The results of the current study reveal some details about the acceptor specificity of the HexTF and SiaTF domains of SiaDW/Y. For the HexTF domain, 4-MU-Sia proved to be a very poor acceptor compared with 4-MU-Sia-Hex-Sia; however, under the reaction conditions used, up to 90% of 4-MU-Sia could be converted to 4-MU-Sia-Hex. In contrast, neither the wild-type nor monofunctional SiaTFs used 4-MU-Gal/Glc as an acceptor, but these enzymes were highly efficient with 4-MU-Sia-Hex. These results indicate that a minimum of one Sia residue is necessary for recognition by the acceptor binding sites of both the HexTF and SiaTF domain. Further, the addition of the hydrophobic group MU improved acceptor quality because free Sia was not a substrate for the enzymes (12). These findings may reflect the nature of the in vivo priming acceptor. Willis and Whitfield (2) recently demonstrated for a number of Gram-negative bacteria, including N. meningitidis serogroup B, that the capsular polysaccharide is anchored via the reducing end to a unique glycolipid consisting of a lysophosphatidylglycerol moiety β-linked to oligo-3-deoxy-d-manno-oct-2-ulosonic acid. The observed acceptor specificities of SiaDW/Y agree well with the proposition that this universal anchor structure also primes in vivo capsule synthesis in NmW and NmY. This would explain both the requirement for a Sia residue, because this is a structural analog of 3-deoxy-d-manno-oct-2-ulosonic acid, and the improvement of acceptor quality by the addition of a hydrophobic group that may occupy the lipid binding site.
Using the newly defined SiaTF domain to search protein databases revealed a new family of glycosyltransferases that is classified as GT97 in the CAZy database. The phylogenetic tree of GT97 sequences showed anomalous clustering, with high similarity occurring between sequences from distantly related species. This may suggest that horizontal gene transfer or convergent evolution has played a role in the history of the GT97 family. A case in point is the grouping of the Bacteroidetes Salinibacter ruber protein (WP_013061143) with those from haloarchaea Natronorubrum sulfidifaciens, Halorhabdus tiamatea, and Halorubrum kocurii. Indeed, the genome sequence of the extremely halophilic bacterium S. ruber revealed that considerable amounts of genetic material have been shared with its haloarchaea coinhabitants in concentrated sea water brines (39). These findings are in agreement with the well studied phylogeny of NulO biosynthetic pathways, which include pathways for sialic acid, pseudaminic acid, and legionaminic acid (30, 31). These pathways are also taxonomically scattered among bacteria and archaea and show complex phylogenetic relationships suggestive of horizontal gene transfer and convergent evolution (30, 31, 40). The presence of GT97 sequences in archaea raises the question of whether this family exhibits specificity for different CMP-activated donor sugars in the different domains of life. Archaea are not known to incorporate sialic acid into their glycans but rather pseudaminic acid and legionaminic acid, which share related biosynthetic pathways (30). Thus, it is possible, if not likely, that some of the identified GT97 proteins are actually pseudaminic acid or legionaminic acid transferases.
A notable finding is the presence of GT97 proteins in several extreme halophiles. Interestingly, in this respect, adaptation to hypersaline environments has been shown to involve acidification of the proteome, the replacement of neutral amino acids with acidic residues to maintain hydration (41). Thus, one may speculate that nonulosonic acid biosynthesis pathways confer a selective advantage by similarly increasing the acidity of the glycome. However, further work is needed to confirm the function of other members of the GT97 family and to interrogate the possibility of glycome acidification in extreme halophiles.
This work arose as a new research aspect in the frame of Deutsche Forschungsgemeinschaft (DFG) Research Unit 548. This work was supported in part through DFG Grant Ge801/10-1 and in part through impact-derived resources of the Institute for Cellular Chemistry.
- CPS
- capsular polysaccharide
- CAZy
- Carbohydrate-Active enZYmes Database
- GT
- glycosyltransferase
- HexTF
- hexosyltransferase
- 4-MU
- 2′-(4-methylumbelliferyl)
- Sia
- sialic acid
- SiaTF
- sialyltransferase
- FD
- fluorescence detection
- DP
- degree of polymerization
- aa
- amino acids
- NulO
- nonulosonic acid.
REFERENCES
- 1. Willis L. M., Whitfield C. (2013) Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44 [DOI] [PubMed] [Google Scholar]
- 2. Willis L. M., Whitfield C. (2013) KpsC and KpsS are retaining 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) transferases involved in synthesis of bacterial capsules. Proc. Natl. Acad. Sci. U.S.A. 110, 20753–20758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stephens D. S., Greenwood B., Brandtzaeg P. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 [DOI] [PubMed] [Google Scholar]
- 4. Whitfield C., Roberts I. S. (1999) Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 5. Whitfield C. (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 [DOI] [PubMed] [Google Scholar]
- 6. Roberts I. S. (1996) The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50, 285–315 [DOI] [PubMed] [Google Scholar]
- 7. Harrison O. B., Claus H., Jiang Y., Bennett J. S., Bratcher H. B., Jolley K. A., Corton C., Care R., Poolman J. T., Zollinger W. D., Frasch C. E., Stephens D. S., Feavers I., Frosch M., Parkhill J., Vogel U., Quail M. A., Bentley S. D., Maiden M. C. (2013) Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 19, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosenstein N. E., Perkins B. A., Stephens D. S., Popovic T., Hughes J. M. (2001) Meningococcal disease. N. Engl. J. Med. 344, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 9. Schauer R., de Freese A., Gollub M., Iwersen M., Kelm S., Reuter G., Schlenzka W., Vandamme-Feldhaus V., Shaw L. (1997) Functional and biosynthetic aspects of sialic acid diversity. Indian J. Biochem. Biophys. 34, 131–141 [PubMed] [Google Scholar]
- 10. Bhattacharjee A. K., Jennings H. J., Kenny C. P., Martin A., Smith I. C. (1976) Structural determination of the polysaccharide antigens of Neisseria meningitidis serogroups Y, W-135, and BO1. Can. J. Biochem. 54, 1–8 [DOI] [PubMed] [Google Scholar]
- 11. Angata T., Varki A. (2002) Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 12. Romanow A., Haselhorst T., Stummeyer K., Claus H., Bethe A., Mühlenhoff M., Vogel U., von Itzstein M., Gerardy-Schahn R. (2013) Biochemical and biophysical characterization of the sialyl-/hexosyltransferase synthesizing the meningococcal serogroup W135 heteropolysaccharide capsule. J. Biol. Chem. 288, 11718–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 14. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Claus H., Vogel U., Mühlenhoff M., Gerardy-Schahn R., Frosch M. (1997) Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol. Gen. Genet. 257, 28–34 [DOI] [PubMed] [Google Scholar]
- 16. Weisgerber C., Troy F. A. (1990) Biosynthesis of the polysialic acid capsule in Escherichia coli K1. The endogenous acceptor of polysialic acid is a membrane protein of 20 kDa. J. Biol. Chem. 265, 1578–1587 [PubMed] [Google Scholar]
- 17. Inoue S., Lin S. L., Lee Y. C., Inoue Y. (2001) An ultrasensitive chemical method for polysialic acid analysis. Glycobiology 11, 759–767 [DOI] [PubMed] [Google Scholar]
- 18. Inoue S., Inoue Y. (2001) A challenge to the ultrasensitive chemical method for the analysis of oligo- and polysialic acids at a nanogram level of colominic acid and a milligram level of brain tissues. Biochimie 83, 605–613 [DOI] [PubMed] [Google Scholar]
- 19. Keys T. G., Freiberger F., Ehrit J., Krueger J., Eggers K., Buettner F. F., Gerardy-Schahn R. (2012) A universal fluorescent acceptor for high-performance liquid chromatography analysis of pro- and eukaryotic polysialyltransferases. Anal. Biochem. 427, 107–115 [DOI] [PubMed] [Google Scholar]
- 20. Schäffer A. A., Aravind L., Madden T. L., Shavirin S., Spouge J. L., Wolf Y. I., Koonin E. V., Altschul S. F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29, 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katoh K., Kuma K., Toh H., Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gouveia-Oliveira R., Sackett P. W., Pedersen A. G. (2007) MaxAlign: maximizing usable data in an alignment. BMC. Bioinformatics 8, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 24. Kuraku S., Zmasek C. M., Nishimura O., Katoh K. (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Claus H., Stummeyer K., Batzilla J., Mühlenhoff M., Vogel U. (2009) Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol. Microbiol. 71, 960–971 [DOI] [PubMed] [Google Scholar]
- 26. Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finn R. D., Clements J., Eddy S. R. (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eddy S. R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis A. L., Desa N., Hansen E. E., Knirel Y. A., Gordon J. I., Gagneux P., Nizet V., Varki A. (2009) Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U.S.A. 106, 13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kandiba L., Eichler J. (2013) Analysis of putative nonulosonic acid biosynthesis pathways in Archaea reveals a complex evolutionary history. FEMS Microbiol. Lett. 345, 110–120 [DOI] [PubMed] [Google Scholar]
- 32. Hao J., Balagurumoorthy P., Sarilla S., Sundaramoorthy M. (2005) Cloning, expression, and characterization of sialic acid synthases. Biochem. Biophys. Res. Commun. 338, 1507–1514 [DOI] [PubMed] [Google Scholar]
- 33. Gilbert M., Watson D. C., Wakarchuck W. W. (1997) Purification and characterization of the recombinant CMP-sialic acid synthetase from Neisseria meningitidis. Biotechnol. Lett. 19, 417–420 [Google Scholar]
- 34. Severi E., Hood D. W., Thomas G. H. (2007) Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 35. Freiberger F., Claus H., Günzel A., Oltmann-Norden I., Vionnet J., Mühlenhoff M., Vogel U., Vann W. F., Gerardy-Schahn R., Stummeyer K. (2007) Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol. Microbiol. 65, 1258–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamamoto T., Hamada Y., Ichikawa M., Kajiwara H., Mine T., Tsukamoto H., Takakura Y. (2007) A β-galactoside α2,6-sialyltransferase produced by a marine bacterium, Photobacterium leiognathi JT-SHIZ-145, is active at pH 8. Glycobiology 17, 1167–1174 [DOI] [PubMed] [Google Scholar]
- 37. Kane T. A., White C. L., DeAngelis P. L. (2006) Functional characterization of PmHS1, a Pasteurella multocida heparosan synthase. J. Biol. Chem. 281, 33192–33197 [DOI] [PubMed] [Google Scholar]
- 38. Inamori K., Yoshida-Moriguchi T., Hara Y., Anderson M. E., Yu L., Campbell K. P. (2012) Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science 335, 93–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mongodin E. F., Nelson K. E., Daugherty S., Deboy R. T., Wister J., Khouri H., Weidman J., Walsh D. A., Papke R. T., Sanchez Perez G., Sharma A. K., Nesbø C. L., MacLeod D., Bapteste E., Doolittle W. F., Charlebois R. L., Legault B., Rodriguez-Valera F. (2005) The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 18147–18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bravo I. G., García-Vallvé S., Romeu A., Reglero A. (2004) Prokaryotic origin of cytidylyltransferases and α-ketoacid synthases. Trends Microbiol. 12, 120–128 [DOI] [PubMed] [Google Scholar]
- 41. Deole R., Challacombe J., Raiford D. W., Hoff W. D. (2013) An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content. J. Biol. Chem. 288, 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hall T. A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]