Background: MyoGEF is implicated in regulating cytokinesis and breast cancer cell invasion.
Results: Binding of the MyoGEF carboxyl-terminal region to its DH domain suppresses breast cancer cell invasion and interferes with cytokinesis.
Conclusion: Intramolecular interactions of MyoGEF act as an autoinhibitory mechanism to regulate MyoGEF functions.
Significance: Autoinhibitory intramolecular interactions of MyoGEF serve as a control point to regulate cytokinesis and breast cancer cell invasion.
Keywords: Cell Migration, Cytokinesis, Guanine Nucleotide Exchange Factor (GEF), Myosin, Rho (Rho GTPase), MyoGEF
Abstract
We have reported previously that nonmuscle myosin II-interacting guanine nucleotide exchange factor (MyoGEF) plays an important role in the regulation of cell migration and cytokinesis. Like many other guanine nucleotide exchange factors (GEFs), MyoGEF contains a Dbl homology (DH) domain and a pleckstrin homology domain. In this study, we provide evidence demonstrating that intramolecular interactions between the DH domain (residues 162–351) and the carboxyl-terminal region (501–790) of MyoGEF can inhibit MyoGEF functions. In vitro and in vivo pulldown assays showed that the carboxyl-terminal region (residues 501–790) of MyoGEF could interact with the DH domain but not with the pleckstrin homology domain. Expression of a MyoGEF carboxyl-terminal fragment (residues 501–790) decreased RhoA activation and suppressed actin filament formation in MDA-MB-231 breast cancer cells. Additionally, Matrigel invasion assays showed that exogenous expression of the MyoGEF carboxyl-terminal region decreased the invasion activity of MDA-MB-231 cells. Moreover, coimmunoprecipitation assays showed that phosphorylation of the MyoGEF carboxyl-terminal region by aurora B kinase interfered with the intramolecular interactions of MyoGEF. Furthermore, expression of the MyoGEF carboxyl-terminal region interfered with RhoA localization during cytokinesis and led to an increase in multinucleation. Together, our findings suggest that binding of the carboxyl-terminal region of MyoGEF to its DH domain acts as an autoinhibitory mechanism for the regulation of MyoGEF activation.
Introduction
There are approximately 80 guanine nucleotide exchange factors (GEFs)2 in humans. They belong to a family of signaling proteins that play a critical role in the regulation of a variety of physiological processes, such as cell migration, cell cycle progression, membrane trafficking, angiogenesis, morphogenesis, apoptosis, and gene expression (1–3). GEFs generally execute their biological functions through activation of overlapping downstream Rho GTPase proteins such as RhoA, Rac1, and Cdc42 (2, 4). Rho GTPase proteins exist in active, GTP-bound forms or in inactive, GDP-bound forms. GEFs activate Rho GTPase proteins by enhancing the exchange of bound GDP for GTP (5). Dysfunctional regulation of Rho GTPase signaling has been associated with numerous human diseases and disorders (6). Both clinical and animal studies have demonstrated that Rho GTPase signaling plays an important role in promoting tumor progression and metastasis (7).
It is generally thought that the invasion of tumor cells into the surrounding tissues is a prerequisite for tumor metastasis (8). Therefore, the roles of Rho GTPase proteins in the regulation of cell migration and invasion have been investigated intensively (9). Activation of Rho GTPase proteins leads to the remodeling of cytoskeleton organization that plays a central role in the regulation of cell migration and invasion (10). Accordingly, it has been demonstrated that RhoA and RhoC are key promoters of breast cancer invasion and metastasis (11, 12). Consistent with the fact that numerous RhoGEFs exist in humans (1), multiple RhoGEFs, such as leukemia-associated RhoGEF (LARG), PDZ-RhoGEF, and myosin II-interacting GEF (MyoGEF), are implicated in promoting breast cancer cell invasion through activation of RhoA and/or RhoC (13–15).
Rho GTPase signaling also plays a central role in the regulation of cytokinesis. The localization and activation of the Rho GTPase protein RhoA at the cleavage furrow are essential for the assembly of the actomyosin contractile ring during cytokinesis (16–18). RhoA can activate Rho kinase, which, in turn, inhibits myosin phosphatase and directly phosphorylates myosin light chain, hence increasing myosin contractile activity (19, 20). It is generally thought that Ect2, a GEF for RhoA, is a key activator of RhoA at the cleavage furrow during cytokinesis (18, 21–23). Nonetheless, several other RhoGEFs, such as GEF-H1, LARG, and MyoGEF, have also been implicated in regulating cytokinesis (24–26). Furthermore, a line of evidence has demonstrated that two mitotic kinases, aurora B and polo-like kinase (Plk1), play a central role in mitotic progression and cytokinesis (27, 28). Aurora B and Plk1 can activate Ect2 and MyoGEF during cytokinesis and localize both proteins to the central spindle (29–32).
GEFs invariably contain at least a Dbl homology (DH) domain and a pleckstrin homology (PH) domain (1). The DH domain is the catalytic unit of the GEFs. It can catalyze the dissociation of GDP from the inactive, GDP-bound forms of Rho GTPase proteins, thus stimulating the exchange of GDP for GTP and promoting the formation of the active, GTP-bound forms of GTPase proteins (33, 34). The PH domain is adjacent and carboxyl-terminal to the DH domain. The tandem DH and PH domains often form a functional unit to catalyze the guanine nucleotide exchange activity (34–36). In addition, the PH domain can bind to phospholipids and participate in protein-protein interactions, thus targeting GEFs to proper subcellular locations such as the cell membrane and cytoskeleton (34). Activation of GEFs can be achieved through various modes of regulation, such as protein-protein interactions, associations with the membrane and cytoskeleton, and relief of autoinhibitory intramolecular interactions (1, 34, 37–39). It has been demonstrated that intramolecular interactions between the DH domain and the amino- or carboxyl-terminal regions of GEFs can inhibit the catalytic activity of the DH domain (40–42). Various molecular events, such as protein-protein interactions and posttranslational modifications, can disrupt such intramolecular interactions and result in the activation of GEFs (40, 41). Intermolecular interactions have also been implicated in the regulation of GEFs. However, unlike intramolecular interactions, intermolecular interactions of GEFs can be either activatory or inhibitory. For instance, oligomerization of Dbl through the DH domain is required for the oncogenic activity of Dbl (43). On the other hand, hetero- and homo-oligomerization of PDZ-RhoGEF, LARG, and p115RhoGEF through their carboxyl-terminal regions lead to the inhibition of the respective GEFs (44, 45).
We have reported previously that MyoGEF plays an important role in the regulation of cytokinesis and breast cancer cell invasion (15, 24, 31, 32, 46, 47). MyoGEF is highly expressed in invasive breast cancer cells and tissues (15). Depletion of MyoGEF decreases RhoA/RhoC activation and suppresses breast cancer cell invasion (15). We also found that phosphorylation of MyoGEF at Thr-544 by aurora B creates a docking site for Plk1, thus leading to the binding of Plk1 to MyoGEF (31). In turn, Plk1 phosphorylates MyoGEF at Thr-574 (32). Such sequential phosphorylation of MyoGEF by aurora B and Plk1 localizes MyoGEF to the central spindle and leads to MyoGEF activation during cytokinesis. In this study, we provide evidence demonstrating that intramolecular interactions between the DH domain and the carboxyl-terminal region of MyoGEF could serve as an autoinhibitory mechanism for the regulation of MyoGEF functions.
EXPERIMENTAL PROCEDURES
Cell Culture
MDA-MB-231, U2OS, and HeLa cells were purchased from the ATCC. MDA-MB-231 cells were grown in Leibovitz's L-15 medium supplemented with 10% fetal bovine serum. U2OS and HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum. Lipofectamine was used for transfection with plasmids according to the instructions of the manufacturer (Invitrogen). For immunofluorescence analysis, the transfected cells were trypsinized 24 h after transfection and grown on fibronectin-coated coverglasses for an additional 3 h.
Plasmids
Plasmids encoding Myc- and GFP-tagged full-length MyoGEF were generated as described previously (15, 24). Gateway destination vectors (pDEST27, pCS 3MT DEST, pCS Cherry DEST, pDest-mCherry-N1, pCS EGFP DEST, and pDest-eGFP-N1) were used to generate mammalian expression constructs for GST-, Myc-, mCherry-, and GFP-tagged MyoGEF fragments according to the instructions of the manufacturer (Invitrogen). The MyoGEF T544D and T544E mutants were generated by site-directed mutagenesis according to the instructions of the manufacturer (Agilent Technologies) and confirmed by DNA sequencing (Eurofins MWG Operon). pDEST27 was purchased from Invitrogen. pcDNA3-EGFP-RhoA (plasmid 12965) (48), pCS 3MT DEST (plasmid 13070), pCS Cherry DEST (plasmid 13075), pDest-mCherry-N1 (plasmid 31907) (49), pCS EGFP DEST (plasmid 13071), and pDest-eGFP-N1 (plasmid 31796) (49) were purchased from Addgene.
Expression and Purification of Recombinant Polypeptides
A bacterial expression system was used to express GST-and His-tagged recombinant polypeptides. BL21 bacterial cells expressing GST-tagged recombinant polypeptides were resuspended in PBS and homogenized by sonication. Triton X-100 was added to a final concentration of 1%. The bacterial lysates were then incubated by shaking at 23 °C for 1 h. A glutathione-conjugated agarose (Sigma) column was used to purify GST-tagged recombinant polypeptides. The proteins were eluted with 100 mm Tris-HCl (pH 8.0) and 10 mm glutathione and dialyzed against 50 mm Tris-HCl (pH 7.5) and 50 mm NaCl. BL21 bacterial cells expressing His-tagged recombinant polypeptides were resuspended in 50 mm sodium phosphate (pH 8.0) and 300 mm NaCl and homogenized by sonication. A nickel-nitrilotriacetic acid-agarose (Qiagen) column was used to purify His-tagged recombinant polypeptides. The proteins were eluted with 50 mm sodium phosphate (pH 8.0), 300 mm NaCl, and 250 mm imidazole and dialyzed against 50 mm Tris-HCl (pH 7.5) and 50 mm NaCl.
GST Pulldown Assays
GST pulldown assays were done as described previously (47, 50). Briefly, the immobilized GST-MyoGEF polypeptides were incubated with in vitro-translated Myc-MyoGEF fragments or transfected cell lysates overnight at 4 °C. After washing four times with binding buffer (50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 0.05% Triton-X-100, 10% glycerol, 0.2 mm EDTA, and 1 mm DTT), the beads were resuspended in SDS loading buffer to elute the bound proteins. In vitro-translated Myc-MyoGEF fragments were synthesized using the TNT SP6 quick-coupled transcription/translation system (Promega, Madison, WI) according to the instructions of the manufacturer.
Coimmunoprecipitation Assays
5 μg of purified His- or GST-MyoGEF fragments was mixed with the in vitro-translated MyoGEF fragments in BC100 buffer (20 mm HEPES (pH 7.9), 100 mm NaCl, 10% glycerol, 0.01% Nonidet P-40, and 1 mm DTT). The mixtures were precleared with protein A/G-agarose beads. The precleared mixtures were incubated with agarose-conjugated anti-Myc antibody overnight at 4 °C. After three washes with BC100 buffer, the bound proteins were eluted with SDS loading buffer. To examine the effect of aurora B- and Plk1-mediated phosphorylation on intramolecular interactions of MyoGEF, 5 μg of purified GST-tagged MyoGEF polypeptides was incubated with buffer alone or with 1 μg of recombinant His-aurora B or His-Plk1 (Invitrogen) in kinase buffer (5 mm MOPS (pH 7.2), 2.5 mm β-glycerophosphate, 1 mm EGTA, 4 mm MgCl2, 0.05 mm DTT, and 250 μm ATP). The reaction mixtures (50 μl) were incubated at 30 °C for 30 min. The treated GST-MyoGEF polypeptides were then used for coimmunoprecipitation assays.
Immunoblot Analysis
Cell lysates or immunoprecipitates were separated on 10% or 4–12% SDS-PAGE gels (Bio-Rad), transferred to an Immobilon-P transfer membrane (Millipore), blocked in 5% nonfat milk, and incubated with primary antibodies as indicated. The following primary antibodies were used: mouse anti-Myc (1:1000, 9E10, catalog no. sc-40, Santa Cruz Biotechnology), rabbit anti-Ect2 (1:1000, Santa Cruz Biotechnology), mouse anti-GST (1:250, Bio-Rad), and rabbit anti-MyoGEF (1:250) (24). After three washes, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5000, Santa Cruz Biotechnology) for 1 h at 23 °C and visualized by SuperSignal West Pico Luminol/enhancer solution (Pierce). A ScanLater kit was used to process some of the immunoblots according to the instructions of the manufacturer (Molecular Devices, LLC).
Immunofluorescence Analysis
Cells grown on coverslips were fixed with 4% paraformaldehyde for 12 min at 23 °C. After blocking with 1% bovine serum albumin for 1 h at 23 °C, the fixed cells were incubated with primary antibodies as indicated for 3 h at 23 °C or overnight at 4 °C, followed by incubation with secondary antibodies for 40 min at 23 °C. For RhoA staining, cells were fixed with 10% TCA for 15 min on ice. The primary antibodies used for immunofluorescence analysis were mouse anti-Myc (1:1000, 9E10, catalog no. sc-40, Santa Cruz Biotechnology), mouse anti-RhoA (1:100, catalog no. sc-418, Santa Cruz Biotechnology), and rabbit anti-MyoGEF (1:100) (24). Alexa Fluor 594 goat anti-mouse IgG (1:500), Alexa Fluor 488 goat anti-mouse IgG (1:500), and Alexa Fluor 350 goat anti-mouse IgG (1:250) were purchased from Invitrogen. Nuclei were visualized by DAPI (Sigma). The coverslips were mounted using a Prolong antifade kit (Invitrogen). Images were collected using the Nikon TiE Perfect Focus digital fluorescence imaging system (Morrell Instrument Co., Inc.) with an Andor Zyla sCMOS 2560 × 2160 camera. The images were processed by deconvolution.
Quantification of Actin Filaments
To assess the effect of the MyoGEF carboxyl-terminal region on actin filament formation, MDA-MB-231 cells transfected with the mCherry-MyoGEF-501–790 plasmid were stained with phalloidin, and the fluorescence intensities in transfected cells were quantitated using NIH ImageJ software (51). Briefly, the freehand selection tool was used to mark the cells of interest in the same field. Regions next to cells were also selected and served as a background. The corrected total cell fluorescence was calculated on the basis of the integrated density from transfected and surrounding untransfected cells as well as background readings: corrected total cell fluorescence = integrated density − (area of selected cell × mean fluorescence of background readings). A more than 2-fold decrease in the corrected total cell fluorescence in transfected cells compared with that in surrounding untransfected cells was used as the threshold for an indicator of decreased actin filament formation.
To assess the effect of the MyoGEF carboxyl-terminal region on actin filament formation induced by the MyoGEF amino-terminal region, we used NIH ImageJ software to count the actin filaments in transfected cells. Briefly, the line plot profile tool was used to examine the elongated actin stress fibers. Each peak in the line plot profile image represents a stress fiber. It should be noted that the lines were drawn perpendicular to the parallel actin bundles. Also, the areas containing large actin aggregates were avoided.
Rhotekin Pulldown Assay
The Rhotekin pulldown assay was carried out as described previously (24, 52).
Matrigel Invasion Assays
Transfected MDA-MB-231 cells were trypsinized, and ∼1 × 105 cells (in Leibovitz's L-15 medium containing 3% of BSA) were seeded on the upper wells of Biocoat Matrigel chambers (Corning). The lower wells were filled with Leibovitz's L-15 medium containing 10% FBS. The transfected cells then underwent chemoattraction across the Matrigel and filter (pore size, 8 μm) to the lower surface of the transwells for 22 h. The nonmigrating cells on the upper chambers were removed by a cotton swab. A fluorescence microscope was used to image the migrated cells on the lower surface of the membrane at five different and random fields at ×10 magnification. Data were collected from three independent experiments, each done in triplicate. Migrated cells were counted and mean differences (± S.E.) between groups were analyzed using Student's t test.
RESULTS
The Carboxyl-terminal Region of MyoGEF Binds to the DH Domain
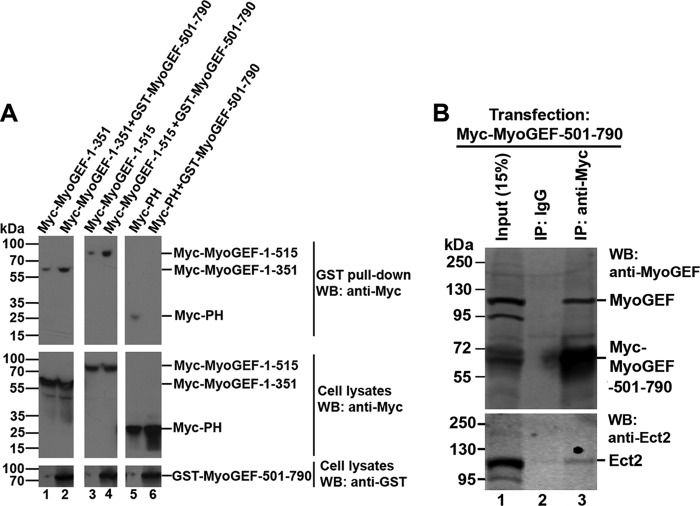
It has been shown that intramolecular interactions between the DH domains and the amino- or carboxyl-terminal regions of GEFs can act as an autoinhibitory mechanism to regulate the activation of numerous GEFs (1, 34). Like many other GEFs, MyoGEF contains a DH domain and a PH domain at the amino-terminal region of the molecule (Fig. 1A). Therefore, we asked whether an autoinhibitory intramolecular interaction exists for the regulation of MyoGEF activation. To this end, we used in vitro pulldown assays to examine the interaction between the amino- and carboxyl-terminal regions of MyoGEF. GST pulldown assays showed that a GST-MyoGEF carboxyl-terminal fragment (GST-MyoGEF-501–790) coprecipitated with the in vitro translated Myc-tagged amino-terminal fragment (Myc-MyoGEF-1–515) (Fig. 1C; compare lane 2 with lane 3). These results suggested that the carboxyl-terminal region of MyoGEF could bind to the amino-terminal region of the molecule. As shown in Fig. 1A, the amino-terminal region (residues 1–515) of MyoGEF contains a DH domain (residues 162–351) and a PH domain (residues 363–515). We then asked whether the carboxyl-terminal region of MyoGEF could bind to its DH and/or PH domains. Immunoprecipitation assays showed that Myc-DH (but not Myc-PH) coprecipitated with His-MyoGEF-501–790 (Fig. 1D, compare lane 1 with lane 2 in the top panel), suggesting that the MyoGEF carboxyl-terminal region could bind to the DH domain. To confirm the in vivo interaction between the DH domain and the carboxyl-terminal region of MyoGEF, HeLa cells transfected with plasmids encoding GST-MyoGEF-501–790 and a Myc-tagged amino-terminal fragment (Myc-MyoGEF-1–351, Myc-MyoGEF-1–515, or Myc-PH) were subjected to GST pulldown assays. A plasmid encoding GST-MyoGEF-501–790 was cotransfected into HeLa cells with a plasmid encoding Myc-MyoGEF-1–351, Myc-MyoGEF-1–515, or Myc-MyoGEF-PH. As shown in Fig. 2A, GST-MyoGEF-501–790 coprecipitated with Myc-MyoGEF-1–351 (containing the DH domain) and Myc-MyoGEF-1–515 (containing both the DH and PH domains). However, Myc-PH did not coprecipitate with GST-MyoGEF-501–790. Therefore, our results suggested that the carboxyl-terminal region of MyoGEF could bind to the DH domain (but not the PH domain) in transfected cells.
FIGURE 1.
In vitro interactions between the amino- and carboxyl-terminal regions of MyoGEF. A, schematic of MyoGEF polypeptides that were used for the GST pulldown assays in C and the immunoprecipitation assays in D. The numbers indicate the amino acid residues. B, Coomassie Blue staining image of GST-MyoGEF-501–790 used in the GST pulldown assays in C. C, GST pulldown assays. Agarose beads associated with GST-MyoGEF-501–790 were used to pull down the in vitro-translated Myc-MyoGEF-1–515. D, coimmunoprecipitation of His-MyoGEF-501–790 and the in vitro-translated Myc-DH or Myc-PH. Purified His-MyoGEF-501–790 was incubated with the in vitro-translated Myc-DH or Myc-PH and then subjected to coimmunoprecipitation assays with an antibody specific for Myc, followed by immunoblotting with antibodies specific for MyoGEF and Myc to detect His-MyoGEF-501–790 (top panel) and Myc-DH/Myc-PH (bottom panel), respectively. IP, immunoprecipitation; WB, Western blot; IgG-HC, immunoglobulin heavy chain; IgG-LC, immunoglobulin light chain.
FIGURE 2.
Intramolecular interactions of MyoGEF. A, interactions between amino- and carboxyl-terminal regions of MyoGEF in transfected cells. Plasmids encoding Myc-MyoGEF fragments (1–351, 1–515, or PH) were cotransfected into HeLa cells with empty vectors (lanes 1, 3, and 5) or with a plasmid encoding GST-MyoGEF-501–790 (lanes 2, 4, and 6). The transfected cells were subjected to GST pulldown assays, followed by immunoblot analysis with anti-Myc antibody to detect Myc-tagged MyoGEF fragments. B, the MyoGEF carboxyl-terminal region binds to endogenous MyoGEF but not to Ect2. MDA-MB-231 cells transfected with a plasmid encoding Myc-MyoGEF-501–790 were subjected to immunoprecipitation (IP) assays with anti-Myc antibody, followed by immunoblot analyses with an antibody specific for MyoGEF (compare lane 2 with lane 3 in the top panel) or Ect2 (compare lane 2 with lane 3 in the bottom panel). WB, Western blot.
To determine the specificity of the interaction between the MyoGEF carboxyl-terminal region and the DH domain, we examined whether the MyoGEF carboxyl-terminal region could bind to other RhoGEFs such as Ect2 (21). MDA-MB-231 cells transfected with a plasmid encoding Myc-MyoGEF-501–790 were subjected to coimmunoprecipitation assays with anti-Myc antibody. As shown in Fig. 2B, endogenous MyoGEF coprecipitated with Myc-MyoGEF-501–790 (Fig. 2B, top panel). However, endogenous Ect2 (a closely related RhoGEF) did not coprecipitate with Myc-MyoGEF-501–790 (Fig. 2B, bottom panel). These results suggested that the MyoGEF carboxyl-terminal region could specifically bind to its DH domain.
Exogenous Expression of the MyoGEF Carboxyl-terminal Region Suppresses Actin Filament Formation
GST pulldown and immunoprecipitation assays showed that the carboxyl-terminal region of MyoGEF could bind to the DH domain (Figs. 1 and 2). These findings led us to speculate that the intramolecular interaction between the carboxyl-terminal region and the DH domain could suppress the activation of MyoGEF. We have shown previously that MyoGEF is highly expressed in MDA-MB-231 breast cancer cells and that it can promote the cell invasion activity through activation of RhoA and RhoC (15). Furthermore, exogenous expression of MyoGEF induces actin bundling (15). Therefore, we asked whether exogenous expression of the MyoGEF carboxyl-terminal region in MDA-MB-231 cells interfered with actin filament formation. MDA-MB-231 breast cancer cells were transfected with a plasmid encoding mCherry-MyoGEF-501–790. 24 h after transfection, the transfected cells were trypsinized and cultured on fibronectin-coated coverglasses for 3 h. The transfected cell were then fixed with paraformaldehyde and stained for actin filaments. As shown in Fig. 3B, a–c, exogenous expression of mCherry-MyoGEF-501–790 in MDA-MB-231 cells decreased the formation of actin filaments (green, n = 82/97 cells). These results suggested that the carboxyl-terminal region of MyoGEF (residues 501–790) could inhibit MyoGEF activation through interactions with the DH domain.
FIGURE 3.

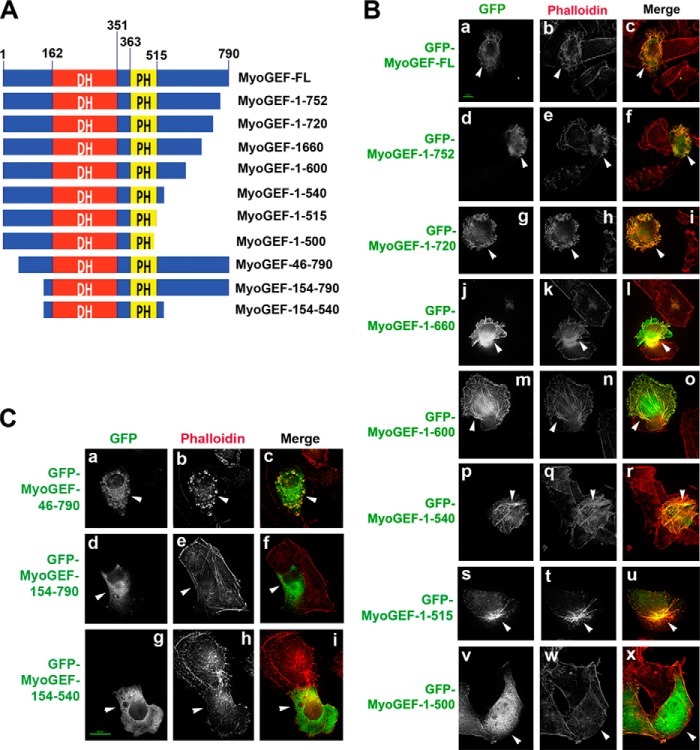
Effect of MyoGEF carboxyl-terminal regions on actin filament formation. A, schematic of truncated MyoGEF fragments that were used to transfect MDA-MB-231 cells in B. The numbers indicate the amino acid residues. B, MDA-MB-231 cells transfected with plasmids encoding various truncated MyoGEF fragments (red or green as indicated) were subjected to immunofluorescence staining for actin filaments (red or green as indicated; b, e, h, k, n, and q). Arrowheads indicate cells expressing truncated MyoGEF fragments. Scale bar = 20 μm.
Our previous findings have shown that the carboxyl-terminal end of MyoGEF contains a PDZ-binding motif that can interact with the PDZ domain-containing protein GIPC1 (46). Therefore, it is possible that exogenous expression of MyoGEF-501–790, which contains the carboxyl-terminal PDZ-binding motif, could interfere with actin organization by disrupting the GIPC1-MyoGEF interaction. To test this possibility, we examined whether exogenous expression of a carboxyl-terminal fragment lacking the carboxyl-terminal PDZ-binding motif (MyoGEF-501–780-ΔSEV) interfered with actin filament formation. We have found previously that MyoGEF polypeptides that lack the carboxyl-terminal three residues (SEV) do not bind to GIPC1 (46). As shown in Fig. 3B, d–f, exogenous expression of MyoGEF-501–790-ΔSEV also decreased actin filament formation (n = 59/101 cells), suggesting that exogenous expression of MyoGEF carboxyl-terminal fragments could interfere with actin filament formation without disrupting GIPC1-MyoGEF interactions. However, we do not yet know whether binding of GIPC1 to the carboxyl-terminal end of MyoGEF has an impact on the intramolecular interactions of MyoGEF. It is of note that the impact of MyoGEF-501–790 on actin filament formation (n = 82/97 cells, 84%) was much greater than that of MyoGEF-501–790-ΔSEV (n = 59/101 cells, 58%).
Next we asked which regions of the MyoGEF carboxyl-terminal half were involved in disrupting actin filament formation in transfected MDA-MB-231 cells. Plasmids encoding various truncated versions of the MyoGEF carboxyl-terminal half were transfected into MDA-MB-231 cells. The transfected cells were then fixed and stained for actin filaments. The following fragments were used: 526–660 (containing the aurora B site Thr-544 and the Plk1 site Thr-57, Fig. 3B, g–i), 361–515 (containing the PH domain, Fig. 3B, j–l), 681–790 (containing the carboxyl-terminal PDZ-binding motif, Fig. 3B, m–o), and 515–660 (containing the aurora B site Thr-544 and the Plk1 site Thr-574 as well as 15 residues from the PH domain, Fig. 3B, p–r). Our results showed that exogenous expression of these truncated versions of MyoGEF carboxyl-terminal half did not affect actin filament formation in transfected MDA-MB-231 cells (Fig. 3B, compare b and e with h, k, n, and q). These findings suggested that the autoinhibitory intramolecular interaction required the whole carboxyl-terminal region of MyoGEF (residues 501–790).
Both the DH and PH Domains Are Required for MyoGEF to Induce Actin Bundling
Our results showed that the carboxyl-terminal region of MyoGEF could bind to its DH domain (Figs. 1 and 2) and that exogenous expression of the MyoGEF carboxyl-terminal fragment 501–790 inhibited actin filament formation (Fig. 3B, b and e). Therefore, we asked whether carboxyl-terminal truncation of MyoGEF could affect the ability of MyoGEF to induce actin bundling. Plasmids encoding GFP-tagged full-length (MyoGEF-FL) and truncated versions of MyoGEF were transfected into MDA-MB-231 cells (Fig. 4A). 24 h after transfection, the transfected cells were fixed with paraformaldehyde and stained for actin filaments (red). Our results showed that exogenous expression of MyoGEF-FL induced actin bundling compared with untransfected cells (Fig. 4B, a–c, arrowheads). In addition, GFP-MyoGEF-FL also localized to the newly induced actin bundles (Fig. 4B, a–c, arrowheads). The following carboxyl-terminally truncated versions of MyoGEF also induced actin bundling: MyoGEF-1–752 (Fig. 4B, d–f, arrowheads), MyoGEF-1–720 (Fig. 4B, g–i, arrowheads), MyoGEF-1–660 (Fig. 4B, j–l, arrowheads), MyoGEF-1–600 (Fig. 4B, m–o, arrowheads), MyoGEF-1–540 (Fig. 4B, p–r, arrowheads), and MyoGEF-1–515 (Fig. 4B, s–u, arrowheads). However, MyoGEF-1–500 was not able to induce actin bundling and did not colocalize with actin filaments (Fig. 4B, v–x, arrowheads). These findings suggested that amino acid residues 501–515 were required for MyoGEF to induce actin bundling and to localize to actin bundles. It is of note that the carboxyl-terminally truncated fragments MyoGEF-1–600, MyoGEF-1–540, and MyoGEF-1–515 could induce stress fiber formation and colocalize with the induced stress fibers (Fig. 4B, n, q, and t, arrowheads). In contrast, MyoGEF-FL and the truncated fragments MyoGEF-1–752, MyoGEF-1–720, and MyoGEF-1–660 did not induce the formation of stress fibers (Fig. 4B, b, e, h, and k, arrowheads; compare b, e, h, and k with n, q, and t). Instead, MyoGEF-FL and these fragments (residues 1–752, 1–720, and 1–660) induced actin bundles that were associated with membrane ruffling (Fig. 4B, b, e, h, and k, arrowheads). It would be interesting to know how carboxyl-terminal residues 600–790 suppress the ability of MyoGEF to induce stress fiber formation.
FIGURE 4.
The ability of the tandem DH-PH domain to induce actin bundling. A, schematic of truncated MyoGEF fragments that were used to transfect MDA-MB-231 cells in B and C. The numbers indicate the amino acid residues. B, MDA-MB-231 cells transfected with plasmids encoding carboxyl-terminally truncated GFP-MyoGEF fragments (green) were subjected to immunofluorescence staining for actin filaments (red; b, e, h, k, n, q, t, and w). C, MDA-MB-231 cells transfected with plasmids encoding amino-terminally truncated GFP-MyoGEF fragments (green) were subjected to immunofluorescence staining for actin filaments (red; b, e, and h). The arrowheads in B and C indicate cells expressing the truncated MyoGEF fragments. Scale bar = 20 μm.
We next examined the ability of amino-terminally truncated MyoGEF fragments to induce actin bundling in transfected MDA-MB-231 cells. The following MyoGEF fragments were examined (Fig. 4A): MyoGEF-46–790 (lacking the amino-terminal 45 amino acid residues), MyoGEF-154–790 (lacking the amino-terminal 153 amino acid residues), and MyoGEF-154–540 (containing the tandem DH-PH domain only). Our results showed that MyoGEF-46–790 could induce actin bundling and localize to the actin bundles (Fig. 4C, a–c, arrowheads). However, neither MyoGEF-154–790 nor MyoGEF-154–540 could induce actin bundling (Fig. 4C, d–i). These results suggested that the tandem DH-PH domain (residues 154–515) alone was not sufficient to induce actin bundling in transfected cells. Currently, we do not know why the presence of the amino-terminal region (residues 46–153) was required for the DH-PH domain of MyoGEF to induce actin bundling and to localize to actin bundles. Although the PH domain is often required for the DH domain to function in numerous GEFs (35, 36), it has also been shown that the PH domain can bind to the DH domain and inhibit the DH domain (53). It is unlikely that the PH domain of MyoGEF could inhibit the function of the DH domain because exogenous expression of the MyoGEF fragment 1–354 (containing the amino-terminal region and the DH domain) did not induce actin bundling in transfected cells (data not shown). Also, exogenous expression of a MyoGEF fragment (residues 361–515, containing the PH domain) did not affect actin filament formation (Fig. 3B, j–l).
The MyoGEF Carboxyl-terminal Fragment Can Inhibit MyoGEF-induced Actin Bundling
Our results showed that exogenous expression of MyoGEF-501–790 could inhibit actin filament formation in transfected MDA-MB-231 cells (Fig. 3B, a–c, arrowheads). Furthermore, pulldown assays showed that the MyoGEF carboxyl-terminal region could bind to the DH domain of MyoGEF (Figs. 1D and 2B). Fig. 4B also shows that exogenous expression of MyoGEF-1–540 could induce actin stress fiber formation. Therefore, we asked whether exogenous expression of MyoGEF-501–790 could inhibit stress fiber formation induced by MyoGEF-1–540. MDA-MB-231 cells were transfected with plasmids encoding GFP-MyoGEF-1–540 and Myc-MyoGEF-501–790. The transfected cells were stained for Myc-MyoGEF-501–790 (blue) and actin filaments (red). As shown in Fig. 5, in the absence of Myc-MyoGEF-501–790, exogenous expression of GFP-MyoGEF-1–540 could induce stress fiber formation (Fig. 5A, a, b, and d, arrows). However, the presence of Myc-MyoGEF-501–790 (blue) markedly decreased stress fiber formation induced by GFP-MyoGEF-1–540 (Fig. 5A, e–h, arrowheads; compare e–h with a–d). Quantification of actin filament formation also confirmed that expression of Myc-MyoGEF-501–790 could significantly decrease actin bundling induced by GFP-MyoGEF-1–540 (Fig. 5, B–D). These results suggested that MyoGEF carboxyl-terminal region could bind to the DH domain and suppress MyoGEF-induced actin filament formation.
FIGURE 5.
Effect of the MyoGEF carboxyl-terminal region on actin bundling induced by the MyoGEF amino-terminal region. A, MDA-MB-231 cells transfected with a plasmid encoding GFP-MyoGEF-1–540 (a–d) or with plasmids encoding GFP-MyoGEF-1–540 and Myc-MyoGEF-501–790 (e–h) were subjected to immunofluorescence staining for actin filaments (red) and Myc-MyoGEF-501–790 (blue). White arrows indicate cells expressing GFP-MyoGEF-1–540 alone (a, b, and d). White arrowheads indicate cells expressing both GFP-MyoGEF-1–540 and Myc-MyoGEF-501–790 (e–h). Red arrows in b and f indicate the positions at which immunofluorescence intensities were measured as shown in B and C. Scale bar = 20 μm. B, representative line plot profile of the transfected cell in A, b. C, representative line plot profile of the transfected cell in A, f. Only the fluorescence peaks with a gray value of larger than 50 were counted. Each peak in B and C represents a stress fiber. D, quantification of actin filaments in cells transfected with a plasmid encoding GFP-MyoGEF-1–540 (n = 20 cells) or with plasmids encoding GFP-MyoGEF-1–540 and Myc-MyoGEF-501–790 (n = 19 cells). **, p < 0.01.
Exogenous Expression of the MyoGEF Carboxyl-terminal Region Inhibits the Invasion Activity of MDA-MB-231 Breast Cancer Cells
We have found previously that MyoGEF can promote the invasion activity of MDA-MB-231 breast cancer cells through activation of RhoA and RhoC (15). In this study, we further showed that the MyoGEF carboxyl-terminal region could bind to the DH domain and inhibit MyoGEF-induced actin bundling (Figs. 1, 2, and 5). Our results also showed that exogenous expression of a carboxyl-terminally truncated MyoGEF fragment (MyoGEF-1–540) induced stress fiber formation (Figs. 4B and 5). Therefore, we asked whether the intramolecular interaction between the carboxyl-terminal region of MyoGEF and its DH domain played a role in the regulation of cell invasion activity. MDA-MB-231 cells were transfected with either the mCherry empty vector or a plasmid encoding mCherry-MyoGEF-501–790. Matrix gel invasion assays were used to examine the invasion activity of the transfected cells. The transfected cells were allowed to invade Matrigel for 22 h. A fluorescence microscope was used to examine the fluorescence-positive cells that successfully migrated to the underside of the Matrigels. As shown in Fig. 6, exogenous expression of mCherry-MyoGEF-501–790 significantly decreased the invasion activity of MDA-MB-231 cells (Fig. 6, A and B). Our results suggested that the carboxyl-terminal region of MyoGEF could bind to the DH domain of MyoGEF and inhibit MyoGEF activation, thus decreasing cell invasion activity.
FIGURE 6.
Exogenous expression of a MyoGEF carboxyl-terminal fragment decreases cell invasion activity and RhoA activation in MDA-MB-231 cells. A, MDA-MB-231 cells transfected with the mCherry-vector (a) or a plasmid encoding mCherry-MyoGEF-501–790 (b) were subjected to Matrigel invasion assays. A fluorescence microscope was used to image the transfected cells that migrated to the underside of the membrane at ×10 magnification. It is of note that, on the basis of the percentage of fluorescence-positive cells, the transfection efficiency was similar for the mCherry vector and mCherry-MyoGEF-501–790 plasmid. Scale bar = 200 μm. B, quantitative results of the Matrigel invasion assays in A. **, p < 0.01. C, a representative Rhotekin pulldown assay. A plasmid encoding GFP-RhoA was cotransfected into MDA-MB-231 cells with an empty vector (lane 1) or a plasmid encoding mCherry-MyoGEF-501–790 (lane 2). The transfected cells were subjected to Rhotekin pulldown assays as described previously (24). D, quantitation of the results of Rhotekin pulldown assays from three independent experiments. *, p < 0.05.
We have found previously that MDA-MB-231 breast cancer cells highly express MyoGEF and that RNAi-mediated depletion of MyoGEF in MDA-MB-231 cells decreased the levels of activated RhoA and RhoC (15). We next asked whether exogenous expression of the MyoGEF carboxyl-terminal region (mCherry-MyoGEF-501–790) could suppress RhoA activation in MDA-MB-231 cells. A plasmid encoding GFP-RhoA was cotransfected into MDA-MB-231 cells with the mCherry empty vector or a plasmid encoding mCherry-MyoGEF-501–790. The transfected cells were subjected to Rhotekin pulldown assays. As shown in Fig. 6C, exogenous expression of mCherry-MyoGEF-501–790 decreased the levels of activated RhoA in transfected MDA-MB-231 cells (Fig. 6C, top panel, compare lane 1 with lane 2). Quantification of Rhotekin pulldown assays also confirmed that exogenous expression of mCherry-MyoGEF-501–790 significantly inhibited RhoA activation (Fig. 6D). This finding suggested that the MyoGEF carboxyl-terminal region could bind to MyoGEF and inhibit its ability to activate RhoA.
Exogenous Expression of the MyoGEF Carboxyl-terminal Region Leads to Cytokinesis Defects
We have reported previously that phosphorylation of MyoGEF by aurora B and Plk1 can activate MyoGEF during cytokinesis (31, 32). Phosphorylation of MyoGEF at Thr-544 creates a docking site for Plk1 to bind MyoGEF (31). In turn, Plk1 can phosphorylate MyoGEF at Thr-574 (32). Therefore, we asked whether phosphorylation of MyoGEF by aurora B could inhibit the intramolecular interaction between the amino- and carboxyl-terminal regions of MyoGEF. Purified His-aurora B (Invitrogen) was used to phosphorylate GST-MyoGEF-501–790 in vitro. The phosphorylated GST-MyoGEF-501–790 was then incubated with the in vitro-translated Myc-MyoGEF-1–500, followed by immunoprecipitation assays with an antibody specific for Myc. Fig. 7A shows that Myc-MyoGEF-1–500 coprecipitated with GST-MyoGEF-501–790. In contrast, Myc-MyoGEF-1–500 did not coprecipitate with the GST-MyoGEF-501–790 that was phosphorylated by aurora B (Fig. 7A, top row, compare lane 2 with lane 4), suggesting that phosphorylation of MyoGEF at Thr-544 by aurora B could inhibit the intramolecular interactions of MyoGEF. Next, we asked whether phosphomimetic mutation at Thr-544 interfered with the intramolecular interactions of MyoGEF. As shown in Fig. 7B, the phosphomimetic mutant mCherry-MyoGEF-501–790-T544E did not coprecipitate with Myc-MyoGEF-1–515, further confirming that phosphorylation of MyoGEF at Thr-544 by aurora B inhibits the intramolecular interactions of MyoGEF. It is of note that wild-type MyoGEF-501–790 could bind to MyoGEF-1–515 (Figs. 1 and 2).
FIGURE 7.
Effect of the MyoGEF carboxyl-terminal region on cytokinesis. A, effect of aurora B-mediated phosphorylation on the intramolecular interactions of MyoGEF. Purified GST-MyoGEF-501–790 treated with buffer (lane 4) or aurora B (lane 2) was mixed with the in vitro-translated Myc-1–500 and then subjected to immunoprecipitation (IP) assays with anti-Myc antibody, followed by immunoblotting with antibodies specific for MyoGEF and Myc to detect GST-MyoGEF-501–790 (top row) and Myc-MyoGEF-1–500 (bottom row), respectively. B, effect of the T544E phosphomimetic mutation on the intramolecular interactions of MyoGEF. The in vitro-translated mCherry-MyoGEF-501–790-T544E was mixed with the in vitro-translated Myc-MyoGEF-1–515 and then subjected to immunoprecipitation assays with anti-Myc antibody, followed by immunoblot analyses with antibodies specific for MyoGEF and Myc to detect mCherry-MyoGEF-501–790-T544E (top panel) and Myc-MyoGEF-1–515 (bottom panel), respectively. Note that the anti-MyoGEF antibody does not recognize the amino-terminal fragment Myc-MyoGEF-1–515. WB, Western blot. C, MDA-MB-231 cells transfected with a plasmid encoding mCherry vector (a–c), mCherry-MyoGEF-501–790-WT (d–f), or mCherry-MyoGEF-501–790-T544D were subjected to immunofluorescence staining for nuclei (DAPI). Scale bar = 20 μm. D, multinucleated U2OS cells were identified and counted following transfection with the indicated plasmids. **, p < 0.01.
We next asked whether exogenous expression of the MyoGEF carboxyl-terminal region in U2OS cells interfered with cytokinesis. U2OS cells were used for this experiment because this cell line expresses endogenous MyoGEF and because the background levels of multinucleation in U2OS cells are much lower than those in MDA-MB-231 cells (data not shown). U2OS cells were transfected with the mCherry empty vector or a plasmid encoding mCherry-MyoGEF-501–790. Forty-eight hours after transfection, the cells were stained for nuclei. As shown in Fig. 7, C and D, we observed multinucleation in U2OS cells expressing mCherry-MyoGEF-501–790 (Fig. 7C, d–f, arrowheads) but not in cells expressing mCherry alone (Fig. 7C, compare a–c with d–f). Fig. 7A shows that in vitro phosphorylation of the MyoGEF carboxyl-terminal region by aurora B could inhibit the intramolecular interaction between the amino- and carboxyl-terminal regions of MyoGEF. Therefore, we also asked whether exogenous expression of the phosphomimetic MyoGEF carboxyl-terminal fragment (MyoGEF-501–790-T544D) had an impact on cytokinesis. We found that exogenous expression of MyoGEF-501–790-T544D did not cause multinucleation in U2OS cells (Fig. 7C, g–i), suggesting that phosphorylation of MyoGEF at Thr-544 by aurora B could relieve the autoinhibitory intramolecular interactions of MyoGEF.
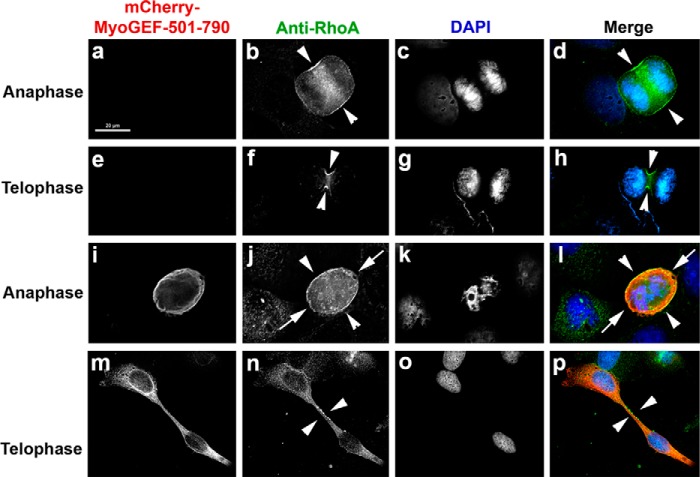
It is well known that localization and activation of RhoA at the cleavage furrow are essential for the completion of cytokinesis (16–18). We next asked whether exogenous expression of the MyoGEF carboxyl-terminal fragment MyoGEF-501–790 affected RhoA activation and localization at the cleavage furrow. Immunofluorescence staining of TCA-fixed cells with anti-RhoA antibody has been used to monitor the localization of activated RhoA at the cleavage furrow (54, 55). The cortical staining of RhoA is generally considered as evidence for the activated forms of the protein. U2OS cells transfected with a plasmid encoding mCherry-MyoGEF-501–790 were fixed with TCA and then subjected to immunofluorescence analysis with antibodies specific for RhoA and MyoGEF. The MyoGEF antibody is a peptide antibody that can specifically recognize the carboxyl-terminal end of MyoGEF (24). Therefore, this MyoGEF peptide antibody was used to detect the expression of mCherry-MyoGEF-501–790 in TCA-fixed U2OS cells. In cells that did not express mCherry-MyoGEF-501–790, RhoA concentrated at the cleavage furrow (Fig. 8, b, d, f, and h, arrowheads). In contrast, RhoA was diffusely localized to the cleavage furrow (Fig. 8, j and l, arrowheads) and the pole regions (Fig. 8, j and l, arrows) during anaphase in cells expressing mCherry-MyoGEF-501–790 (Fig. 8, compare b and d with j and l). Furthermore, we found that RhoA did not concentrate to the midbody at late stages of cytokinesis (Fig. 8, n and p, arrowheads, compare f and h with n and p). Our results suggested that exogenous expression of the MyoGEF carboxyl-terminal region interfered with the localization of RhoA at the cleavage furrow. These observations are consistent with our previous findings that RNAi-mediated depletion of MyoGEF can disrupt RhoA localization at the cleavage furrow (47).
FIGURE 8.
Effect of the MyoGEF carboxyl-terminal terminal region on RhoA localization during cytokinesis. U2OS cells transfected with a plasmid encoding mCherry-MyoGEF-501–790 were fixed with 10% TCA and subjected to immunofluorescence analysis with antibodies specific for MyoGEF (red) and RhoA (green). The arrowheads in b, d, f, h, j, l, n, and p point to the cleavage furrow. The arrows in j and l point to the pole regions. Scale bar = 20 μm.
The T544D Phosphomimetic Carboxyl-terminal Fragment Does Not Affect Actin Filament Formation
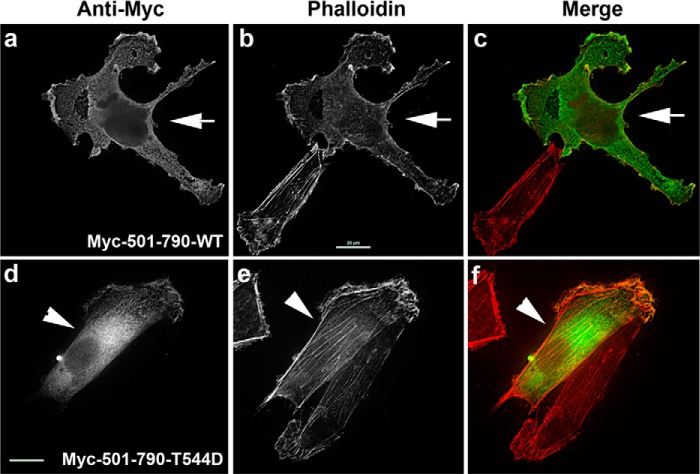
Our results showed that expression of the phosphomimetic mutant MyoGEF-501–790-T544D did not cause multinucleation in transfected U2OS cells (Fig. 7C). This finding was also in agreement with our observations that phosphorylation of the MyoGEF carboxyl-terminal region by aurora B could suppress the intramolecular interactions of MyoGEF (Fig. 7, A and B). We next asked whether phosphomimetic mutation of the MyoGEF carboxyl-terminal region interfered with actin filament formation in interphase cells. We found that exogenous expression of the phosphomimetic mutant MyoGEF-501–790-T544D did not affect actin filament formation (Fig. 9; compare a–c with d–f), further confirming that phosphorylation of MyoGEF at Thr-544 by aurora B could inhibit the intramolecular interactions of MyoGEF. It has been shown that aurora B is also implicated in the regulation of cell spreading and cell migration (56, 57). However, treatment of transfected MDA-MB-231 cells with an inhibitor specific for aurora B did not interfere with actin bundling induced by exogenously expressed MyoGEF (data not shown), suggesting that, during interphase, phosphorylation of MyoGEF by aurora B was not likely to be a major pathway to activate MyoGEF.
FIGURE 9.
Effect of the T544D phosphomimetic mutation on actin filament formation. MDA-MB-231 cells transfected with a plasmid encoding either a wild-type MyoGEF carboxyl-terminal fragment (Myc-501–790-WT, a–c) or a phosphomimetic MyoGEF carboxyl-terminal fragment (Myc-MyoGEF-501–790-T544D, d–f) were subjected to immunofluorescence staining for actin filaments (red) and Myc-tagged polypeptides (green). Arrows indicate cells expressing Myc-MyoGEF-501–790-WT. Arrowheads indicate cells expressing Myc-MyoGEF-501–790-T544D. Scale bar = 20 μm.
MyoGEF Forms Homodimers/Oligomers
Our results demonstrated that the MyoGEF carboxyl-terminal region could bind to its DH domain and suppress MyoGEF activation. It is conceivable that the interaction between the DH domain and the carboxyl-terminal region of MyoGEF can occur either intramolecularly or intermolecularly. Therefore, we asked whether MyoGEF could form homodimers/oligomers. As shown in Fig. 10A, in vitro pulldown assays demonstrated that GFP-MyoGEF could coprecipitate with Myc-MyoGEF. Furthermore, coimmunoprecipitation assays also showed that endogenous MyoGEF coprecipitated with Myc-MyoGEF from transfected MDA-MB-231 cells (Fig. 10B). These results suggested that MyoGEF could form homodimers/oligomers. Next we asked whether the interaction between the MyoGEF carboxyl-terminal region and the DH domain was required for MyoGEF oligomerization. In vitro pulldown assays demonstrated that aurora B-mediated phosphorylation of the MyoGEF carboxyl-terminal region and the T544E phosphomimetic mutation interfered with the interaction between the MyoGEF carboxyl-terminal region and the DH domain (Fig. 7, A and B). Therefore, we asked whether the T544E phosphomimetic mutation interfered with the formation of MyoGEF dimers/oligomers. Fig. 10C shows that the T544E phosphomimetic mutation did not interfere with the intermolecular interactions of MyoGEF. Therefore, our findings indicated that MyoGEF could form homodimers/oligomers independently of the interaction between the MyoGEF carboxyl-terminal region and the DH domain. However, it is still not clear which domains mediate MyoGEF dimerization/oligomerization.
FIGURE 10.
Dimerization and/or oligomerization of MyoGEF. A, intermolecular interactions of MyoGEF in vitro. The in vitro-translated GFP-MyoGEF and Myc-MyoGEF were mixed and subjected to immunoprecipitation (IP) assays with anti-Myc antibody, followed by immunoblot analysis with anti-MyoGEF antibody (compare lane 2 with lane 3). WB, Western blot. B, intermolecular interactions of MyoGEF in transfected cells. MDA-MB-231 cells transfected with a plasmid encoding Myc-MyoGEF were subjected to immunoprecipitation assays with anti-Myc antibody, followed by immunoblot analysis with anti-MyoGEF antibody (compare lane 2 with lane 3). C, effect of the T544E phosphomimetic mutation on MyoGEF dimerization/oligomerization. The in vitro-translated GFP-MyoGEF-T544E and Myc-MyoGEF-T544E were mixed and subjected to immunoprecipitation assays with anti-Myc antibody, followed by immunoblot analysis with anti-MyoGEF antibody (compare lane 2 with lane 3). D, schematic showing that intramolecular interactions of MyoGEF can be disrupted by phosphorylation and protein-protein interactions. The question mark indicates that binding of CSPP to phospho-Thr-574 remains to be confirmed. NMII, nonmuscle myosin II; AURKB, aurora kinase B; Plk1, polo-like kinase 1; pT544, phosphothreonine 544; pT574, phosphothreonine 574; GIPC1, GAIP interacting protein, C terminus 1.
DISCUSSION
In this study, we demonstrated that the intramolecular interaction between the DH domain and the carboxyl-terminal region of MyoGEF serves as an autoinhibitory mechanism to regulate MyoGEF activation. In vitro and in vivo pulldown assays show that the carboxyl-terminal region of MyoGEF could bind to its DH domain. We also demonstrated that exogenous expression of the MyoGEF carboxyl-terminal region interferes with actin filament formation, inhibits breast cancer cell invasion activity, and leads to the formation of multinucleated cells. Our findings suggest that the autoinhibitory intramolecular interactions of MyoGEF serve as a control point to regulate cytokinesis and breast cancer cell invasion.
Autoinhibitory Regulation of MyoGEF Functions
Our previous findings have shown that MyoGEF is implicated in the regulation of cytokinesis and breast cancer cell invasion (15, 24). In this study, we found that the intramolecular interaction between the DH domain and the carboxyl-terminal region of MyoGEF could inhibit MyoGEF functions. Our findings demonstrate that exogenous expression of the MyoGEF carboxyl-terminal region interferes with MyoGEF functions, leading to cytokinesis defects and decreasing breast cancer cell invasion. It has been shown that the carboxyl-terminal region of MyoGEF can interact with centrosome/spindle-associated protein (CSPP), ezrin, and GIPC1 (46, 47, 58). Aurora B and Plk1 can phosphorylate the carboxyl-terminal region of MyoGEF (31, 32). Therefore, exogenous expression of the MyoGEF carboxyl-terminal region in transfected cells could potentially cause cell phenotypes through interference with protein-protein interactions and phosphorylation involving the carboxyl-terminal region of MyoGEF. However, exogenous expression of the T544D phosphomimetic MyoGEF carboxyl-terminal fragment did not interfere with actin filament formation in interphase cells (Fig. 9) and did not cause multinucleation (Fig. 7C). Furthermore, phosphorylation of the MyoGEF carboxyl-terminal region by aurora B decreased the intramolecular interactions of MyoGEF. Moreover, the MyoGEF carboxyl-terminal region did not bind to a closely related RhoGEF Ect2 (Fig. 2B). Therefore, the dominant negative effects of the MyoGEF carboxyl-terminal region are likely dependent on its interaction with the DH domain, leading to the inhibition of MyoGEF functions.
Intramolecular interactions have been shown to serve as an autoinhibitory mechanism for the regulation of GEFs (34). It is conceivable that domains or regions participating in an intramolecular interaction are also used for intermolecular interactions. Our results demonstrate that the MyoGEF carboxyl-terminal region can bind to its DH domain and suppress MyoGEF activation, raising the possibility that intermolecular interactions of MyoGEF can occur between the carboxyl-terminal region and the DH domain. However, we found that phosphomimetic mutation at Thr-544 interfered with the interaction between the carboxyl-terminal region and the DH domain (Fig. 7B). In contrast, phosphomimetic mutation at Thr-544 did not disrupt the formation of MyoGEF dimers/oligomers (Fig. 10C). Therefore, our findings suggest that the interaction between the MyoGEF carboxyl-terminal region and its DH domain can occur intramolecularly. However, we cannot completely rule out the possibility that the interaction between the carboxyl-terminal region and the DH domain of MyoGEF also occurs intermolecularly.
Our results show that the carboxyl-terminal region of MyoGEF could bind to its DH domain, thus interfering with MyoGEF activation (Figs. 3 and 5). However, exogenous expression of full-length MyoGEF also induced actin bundling in transfected cells compared with that of carboxyl-terminally truncated MyoGEF fragments (Fig. 4B), suggesting that an unknown mechanism exists to activate full-length MyoGEF in the transfected MDA-MB-231 cells (see below).
Regulation of Breast Cancer Cell Invasion by MyoGEF
We have reported previously that MyoGEF can activate RhoA/RhoC in MDA-MB-231 breast cancer cells (15). Furthermore, MyoGEF-induced actin bundling can be inhibited by a dominant negative mutant of RhoA but not by a dominant negative mutant of Rac1 (15). Therefore, our findings suggest that RhoA and RhoC are implicated in MyoGEF-induced actin bundling. Cell migration involves lamellipodium formation and membrane ruffles at the front as well as retraction of the rear part of migrating cells (59). Actin polymerization at the front of migrating cells is critical for lamellipodium formation and membrane ruffles, whereas myosin-based contractility is required for retraction of the rear part of migrating cells (59–62). It is generally thought that Rac1 is restricted to the front of migrating cells and is responsible for lamellipodium formation and membrane ruffles, whereas RhoA is localized to the rear of migrating cells and is responsible for tail retraction (62–65). This appears to be at odds with our observations that exogenous expression of MyoGEF resulted in the formation of actin bundles that are often associated with membrane ruffles (Fig. 4B), which are a typical function of Rac1 (66). However, another line of research also demonstrates that RhoA/RhoC can be activated in the front of migrating cells (67, 68), consistent with findings that RhoA/RhoC can induce membrane ruffles (69–71). In support of this concept, a study has shown that S100A4 and Rhotekin (a downstream effector of RhoA) can form a complex with the activated forms of RhoA, thus converting an activated RhoA from a stress fiber inducer to a membrane ruffle inducer (72).
It has been shown that the PH domain of GEFs can associate with actin filaments and phospholipids, thus targeting GEFs to the actin cytoskeleton and/or the cell membrane (1, 34). We have found previously that the PH domain of MyoGEF can interact with nonmuscle myosin II (15, 24). The PDZ domain of GIPC1 can bind to the carboxyl-terminal PDZ-binding motif of MyoGEF, and the GIPC1-MyoGEF interaction is implicated in promoting breast cancer cell invasion (46). The ezrin-radixin-moesin proteins can link the plasma membrane to actin filaments and play a critical role in organizing the cell cortex and promoting breast tumor metastasis (73, 74). It has been shown that ezrin can bind to the carboxyl-terminal region (residues 579–790) of MyoGEF/PLEKHG6 (58). In addition, ezrin is localized to membrane ruffles and required for membrane ruffling (75–77). In this study, we found that full-length MyoGEF could induce and localize to membrane ruffle-associated actin bundles (Fig. 4B). In contrast, carboxyl-terminally truncated MyoGEF with deletion of 190–250 amino acid residues from the carboxyl-terminal end of MyoGEF predominantly localized to stress fibers (Fig. 4B). Therefore, we propose that binding of proteins such as ezrin and GIPC1 to the carboxyl-terminal region of MyoGEF may not only interfere with the autoinhibitory intramolecular interactions, leading to the activation of MyoGEF, but also recruit MyoGEF to the cell membrane, where MyoGEF activates RhoA and induces actin bundling and membrane ruffling, thus promoting cell invasion (Fig. 10D). Accordingly, carboxyl-terminally truncated MyoGEF mutants lacking the binding sites for ezrin and GIPC1 predominantly localize to the stress fiber (Fig. 4B). It would be interesting to know whether binding of ezrin or GIPC1 to the carboxyl-terminal region of MyoGEF has an impact on the intramolecular interactions of MyoGEF.
Regulation of Cytokinesis by MyoGEF
We have reported previously that MyoGEF is implicated in promoting cytokinesis (24, 47). Aurora B and Plk1 are two mitotic kinases that play critical roles in the regulation of cytokinesis (27, 28). Aurora B and Plk1 can promote MyoGEF activation and localization at the central spindle during cytokinesis by phosphorylating MyoGEF at Thr-544 and Thr-574, respectively (31, 32). Our findings in this study show that phosphorylation of the MyoGEF carboxyl-terminal region by aurora B relieves the intramolecular interaction between the DH domain and the carboxyl-terminal region of MyoGEF (Figs. 7 and 10D). These findings are consistent with our previous observations that aurora B-mediated phosphorylation of MyoGEF at Thr-544 leads to the activation of MyoGEF during cytokinesis. Our previous studies have also demonstrated that phosphorylation of MyoGEF at Thr-544 creates a Plk1 docking site and promotes the binding of Plk1 to MyoGEF (31). In turn, Plk1 phosphorylates MyoGEF at Thr-574, leading to the activation and localization of MyoGEF at the central spindle during cytokinesis (32). However, it is still not clear how Plk1-mediated phosphorylation of MyoGEF at Thr-574 is implicated in the regulation of MyoGEF localization and activation during cytokinesis. One possibility is that Plk1-mediated phosphorylation of MyoGEF promotes the binding of signaling molecules such as CSPP to the carboxyl-terminal region of MyoGEF, thus suppressing the intramolecular interactions of MyoGEF (Fig. 10D). In support of this speculation, we have found previously that CSPP can bind to the carboxyl-terminal region of MyoGEF in a phosphorylation-dependent manner and that the CSPP-MyoGEF interaction plays a role in promoting MyoGEF localization to the central spindle (47). In addition, phosphorylation of the MyoGEF carboxyl-terminal region by Plk1 promotes the binding of CSPP to MyoGEF.3 In summary, we propose that phosphorylation and protein-protein interactions involving the carboxyl-terminal regions of MyoGEF decrease the autoinhibitory intramolecular interactions of MyoGEF, thus leading to the activation and localization of MyoGEF at the central spindle during cytokinesis (Fig. 10D). It would be interesting to know whether binding of CSPP to the carboxyl-terminal region of MyoGEF has an impact on the intramolecular interactions of MyoGEF.
Acknowledgments
We thank Dr. Robert S. Adelstein and Dr. Mary Anne Conti for critical reading and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R15GM097702 (to Q. W.).
D. Wu, M. Jiao, S. Zu, C. C. Sollecito, K. Jimenez-Cowell, A. J. Mold, R. M. Kennedy, and Q. Wei, unpublished data.
- GEF
- guanine nucleotide exchange factor
- LARG
- leukemia-associated RhoGEF
- DH
- Dbl homology
- PH
- pleckstrin homology
- FL
- full-length
- CSPP
- centrosome/spindle-associated protein.
REFERENCES
- 1. Schmidt A., Hall A. (2002) Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 2. Rossman K. L., Der C. J., Sondek J. (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 3. Jaffe A. B., Hall A. (2005) Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 4. Heasman S. J., Ridley A. J. (2008) Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 5. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 6. Hall A. (2012) Rho family GTPases. Biochem. Soc. Trans. 40, 1378–1382 [DOI] [PubMed] [Google Scholar]
- 7. Jaffe A. B., Hall A. (2002) Rho GTPases in transformation and metastasis. Adv. Cancer Res. 84, 57–80 [DOI] [PubMed] [Google Scholar]
- 8. Yamaguchi H., Wyckoff J., Condeelis J. (2005) Cell migration in tumors. Curr. Opin. Cell Biol. 17, 559–564 [DOI] [PubMed] [Google Scholar]
- 9. Ridley A. J. (2001) Rho GTPases and cell migration. J. Cell Sci. 114, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 10. Price L. S., Collard J. G. (2001) Regulation of the cytoskeleton by Rho-family GTPases: implications for tumour cell invasion. Semin. Cancer Biol. 11, 167–173 [DOI] [PubMed] [Google Scholar]
- 11. Hakem A., Sanchez-Sweatman O., You-Ten A., Duncan G., Wakeham A., Khokha R., Mak T. W. (2005) RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 19, 1974–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Golen K. L., Wu Z. F., Qiao X. T., Bao L. W., Merajver S. D. (2000) RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 60, 5832–5838 [PubMed] [Google Scholar]
- 13. Aittaleb M., Boguth C. A., Tesmer J. J. (2010) Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 77, 111–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Worzfeld T., Swiercz J. M., Looso M., Straub B. K., Sivaraj K. K., Offermanns S. (2012) ErbB-2 signals through Plexin-B1 to promote breast cancer metastasis. J. Clin. Invest. 122, 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu D., Asiedu M., Wei Q. (2009) Myosin-interacting guanine exchange factor (MyoGEF) regulates the invasion activity of MDA-MB-231 breast cancer cells through activation of RhoA and RhoC. Oncogene 28, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glotzer M. (2005) The molecular requirements for cytokinesis. Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 17. Kosako H., Yoshida T., Matsumura F., Ishizaki T., Narumiya S., Inagaki M. (2000) Rho-kinase/ROCK is involved in cytokinesis through the phosphorylation of myosin light chain and not ezrin/radixin/moesin proteins at the cleavage furrow. Oncogene 19, 6059–6064 [DOI] [PubMed] [Google Scholar]
- 18. Kamijo K., Ohara N., Abe M., Uchimura T., Hosoya H., Lee J. S., Miki T. (2006) Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol. Biol. Cell 17, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K. (1996) Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 [DOI] [PubMed] [Google Scholar]
- 20. Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., Kaibuchi K. (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271, 20246–20249 [DOI] [PubMed] [Google Scholar]
- 21. Yüce O., Piekny A., Glotzer M. (2005) An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishimura Y., Yonemura S. (2006) Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 119, 104–114 [DOI] [PubMed] [Google Scholar]
- 23. Cook D. R., Solski P. A., Bultman S. J., Kauselmann G., Schoor M., Kuehn R., Friedman L. S., Cowley D. O., Van Dyke T., Yeh J. J., Johnson L., Der C. J. (2011) The ect2 rho guanine nucleotide exchange factor is essential for early mouse development and normal cell cytokinesis and migration. Genes Cancer 2, 932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu D., Asiedu M., Adelstein R. S., Wei Q. (2006) A novel guanine nucleotide exchange factor MyoGEF is required for cytokinesis. Cell Cycle 5, 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Birkenfeld J., Nalbant P., Bohl B. P., Pertz O., Hahn K. M., Bokoch G. M. (2007) GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev. Cell 12, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martz M. K., Grabocka E., Beeharry N., Yen T. J., Wedegaertner P. B. (2013) Leukemia-associated RhoGEF (LARG) is a novel RhoGEF in cytokinesis and required for the proper completion of abscission. Mol. Biol. Cell 24, 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barr F. A., Silljé H. H., Nigg E. A. (2004) Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 28. Carmena M., Earnshaw W. C. (2003) The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 29. Petronczki M., Glotzer M., Kraut N., Peters J. M. (2007) Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell 12, 713–725 [DOI] [PubMed] [Google Scholar]
- 30. Wolfe B. A., Takaki T., Petronczki M., Glotzer M. (2009) Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu D., Asiedu M., Matsumura F., Wei Q. (2014) Phosphorylation of myosin II-interacting guanine nucleotide exchange factor (MyoGEF) at threonine 544 by Aurora B kinase promotes the binding of Polo-like kinase 1 to MyoGEF. J. Biol. Chem. 289, 7142–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asiedu M., Wu D., Matsumura F., Wei Q. (2008) Phosphorylation of MyoGEF on Thr-574 by Plk1 promotes MyoGEF localization to the central spindle. J. Biol. Chem. 283, 28392–28400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cherfils J., Chardin P. (1999) GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24, 306–311 [DOI] [PubMed] [Google Scholar]
- 34. Zheng Y. (2001) Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 35. Aittaleb M., Gao G., Evelyn C. R., Neubig R. R., Tesmer J. J. (2009) A conserved hydrophobic surface of the LARG pleckstrin homology domain is critical for RhoA activation in cells. Cell Signal. 21, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chhatriwala M. K., Betts L., Worthylake D. K., Sondek J. (2007) The DH and PH domains of Trio coordinately engage Rho GTPases for their efficient activation. J. Mol. Biol. 368, 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Z., Guo L., Sprang S. R., Sternweis P. C. (2011) Modulation of a GEF switch: autoinhibition of the intrinsic guanine nucleotide exchange activity of p115-RhoGEF. Protein Sci. 20, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bi F., Debreceni B., Zhu K., Salani B., Eva A., Zheng Y. (2001) Autoinhibition mechanism of proto-Dbl. Mol. Cell. Biol. 21, 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu B., Martins I. R., Li P., Amarasinghe G. K., Umetani J., Fernandez-Zapico M. E., Billadeau D. D., Machius M., Tomchick D. R., Rosen M. K. (2010) Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li P., Martins I. R., Amarasinghe G. K., Rosen M. K. (2008) Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat. Struct. Mol. Biol. 15, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yohe M. E., Rossman K., Sondek J. (2008) Role of the C-terminal SH3 domain and N-terminal tyrosine phosphorylation in regulation of Tim and related Dbl-family proteins. Biochemistry 47, 6827–6839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahmad K. F., Lim W. A. (2010) The minimal autoinhibited unit of the guanine nucleotide exchange factor intersectin. PLoS ONE 5, e11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu K., Debreceni B., Bi F., Zheng Y. (2001) Oligomerization of DH domain is essential for Dbl-induced transformation. Mol. Cell. Biol. 21, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chikumi H., Barac A., Behbahani B., Gao Y., Teramoto H., Zheng Y., Gutkind J. S. (2004) Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene 23, 233–240 [DOI] [PubMed] [Google Scholar]
- 45. Eisenhaure T. M., Francis S. A., Willison L. D., Coughlin S. R., Lerner D. J. (2003) The Rho guanine nucleotide exchange factor Lsc homo-oligomerizes and is negatively regulated through domains in its carboxyl terminus that are absent in novel splenic isoforms. J. Biol. Chem. 278, 30975–30984 [DOI] [PubMed] [Google Scholar]
- 46. Wu D., Haruta A., Wei Q. (2010) GIPC1 interacts with MyoGEF and promotes MDA-MB-231 breast cancer cell invasion. J. Biol. Chem. 285, 28643–28650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Asiedu M., Wu D., Matsumura F., Wei Q. (2009) Centrosome/spindle pole-associated protein regulates cytokinesis via promoting the recruitment of MyoGEF to the central spindle. Mol. Biol. Cell 20, 1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Subauste M. C., Von Herrath M., Benard V., Chamberlain C. E., Chuang T. H., Chu K., Bokoch G. M., Hahn K. M. (2000) Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 275, 9725–9733 [DOI] [PubMed] [Google Scholar]
- 49. Hong T. T., Smyth J. W., Gao D., Chu K. Y., Vogan J. M., Fong T. S., Jensen B. C., Colecraft H. M., Shaw R. M. (2010) BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLoS Biol. 8, e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pal D., Wu D., Haruta A., Matsumura F., Wei Q. (2010) Role of a novel coiled-coil domain-containing protein CCDC69 in regulating central spindle assembly. Cell Cycle 9, 4117–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu B. P., Burridge K. (2000) Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol. Cell. Biol. 20, 7160–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das B., Shu X., Day G. J., Han J., Krishna U. M., Falck J. R., Broek D. (2000) Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275, 15074–15081 [DOI] [PubMed] [Google Scholar]
- 54. Charras G. T., Hu C. K., Coughlin M., Mitchison T. J. (2006) Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yonemura S., Hirao-Minakuchi K., Nishimura Y. (2004) Rho localization in cells and tissues. Exp. Cell Res. 295, 300–314 [DOI] [PubMed] [Google Scholar]
- 56. Floyd S., Whiffin N., Gavilan M. P., Kutscheidt S., De Luca M., Marcozzi C., Min M., Watkins J., Chung K., Fackler O. T., Lindon C. (2013) Spatiotemporal organization of Aurora-B by APC/CCdh1 after mitosis coordinates cell spreading through FHOD1. J. Cell Sci. 126, 2845–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu X. P., Liu Z. L., Peng A. F., Zhou Y. F., Long X. H., Luo Q. F., Huang S. H., Shu Y. (2014) Inhibition of Aurora-B suppresses osteosarcoma cell migration and invasion. Exp. Ther. Med. 7, 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. D'Angelo R., Aresta S., Blangy A., Del Maestro L., Louvard D., Arpin M. (2007) Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol. Biol. Cell 18, 4780–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lauffenburger D. A., Horwitz A. F. (1996) Cell migration: a physically integrated molecular process. Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 60. Webb D. J., Parsons J. T., Horwitz A. F. (2002) Adhesion assembly, disassembly and turnover in migrating cells: over and over and over again. Nat. Cell Biol. 4, E97–E100 [DOI] [PubMed] [Google Scholar]
- 61. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Cell migration: integrating signals from front to back. Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 62. Raftopoulou M., Hall A. (2004) Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 63. Kraynov V. S., Chamberlain C., Bokoch G. M., Schwartz M. A., Slabaugh S., Hahn K. M. (2000) Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 [DOI] [PubMed] [Google Scholar]
- 64. Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K. M. (2004) Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 [DOI] [PubMed] [Google Scholar]
- 65. Sastry S. K., Rajfur Z., Liu B. P., Cote J. F., Tremblay M. L., Burridge K. (2006) PTP-PEST couples membrane protrusion and tail retraction via VAV2 and p190RhoGAP. J. Biol. Chem. 281, 11627–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 67. Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 68. Kurokawa K., Matsuda M. (2005) Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol. Biol. Cell 16, 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kawano Y., Fukata Y., Oshiro N., Amano M., Nakamura T., Ito M., Matsumura F., Inagaki M., Kaibuchi K. (1999) Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147, 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bravo-Cordero J. J., Sharma V. P., Roh-Johnson M., Chen X., Eddy R., Condeelis J., Hodgson L. (2013) Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J. Cell Sci. 126, 3356–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. O'Connor K. L., Nguyen B. K., Mercurio A. M. (2000) RhoA function in lamellae formation and migration is regulated by the α6β4 integrin and cAMP metabolism. J. Cell Biol. 148, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen M., Bresnick A. R., O'Connor K. L. (2013) Coupling S100A4 to Rhotekin alters Rho signaling output in breast cancer cells. Oncogene 32, 3754–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fehon R. G., McClatchey A. I., Bretscher A. (2010) Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Elliott B. E., Meens J. A., SenGupta S. K., Louvard D., Arpin M. (2005) The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 7, R365–R373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Borm B., Requardt R. P., Herzog V., Kirfel G. (2005) Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp. Cell Res. 302, 83–95 [DOI] [PubMed] [Google Scholar]
- 76. Lamb R. F., Ozanne B. W., Roy C., McGarry L., Stipp C., Mangeat P., Jay D. G. (1997) Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr. Biol. 7, 682–688 [DOI] [PubMed] [Google Scholar]
- 77. Menager C., Vassy J., Doliger C., Legrand Y., Karniguian A. (1999) Subcellular localization of RhoA and ezrin at membrane ruffles of human endothelial cells: differential role of collagen and fibronectin. Exp. Cell Res. 249, 221–230 [DOI] [PubMed] [Google Scholar]