Background: Caprin-2 is a newly identified regulator in canonical Wnt signaling.
Results: Mutants targeting trimer contacts of Caprin-2 CRD rather than calcium-binding sites affect the function of Caprin-2 in canonical Wnt signaling.
Conclusion: Caprin-2 CRD forms a flexible homotrimer mediated by calcium, and this trimeric assembly is required for the function of Caprin-2.
Significance: This work facilitates our understanding of how Caprin-2 functions in canonical Wnt signaling.
Keywords: Calcium-binding Protein, Crystal Structure, Mutagenesis, Wnt Pathway, Wnt Signaling, C1q, Caprin-2
Abstract
Previously, we have identified Caprin-2 as a new regulator in canonical Wnt signaling through a mechanism of facilitating LRP5/6 phosphorylation; moreover, we found that its C-terminal C1q-related domain (Cap2_CRD) is required for this process. Here, we determined the crystal structures of Cap2_CRD from human and zebrafish, which both associate as a homotrimer with calcium located at the symmetric center. Surprisingly, the calcium binding-deficient mutant exists as a more stable trimer than its wild-type counterpart. Further studies showed that this Caprin-2 mutant disabled in binding calcium maintains the activity of promoting LRP5/6 phosphorylation, whereas the mutations disrupting Cap2_CRD homotrimer did impair such activity. Together, our findings suggested that the C-terminal CRD domain of Caprin-2 forms a flexible homotrimer mediated by calcium and that such trimeric assembly is required for Caprin-2 to regulate canonical Wnt signaling.
Introduction
Caprin-1 and Caprin-2 were initially identified as cytoplasmic activation/proliferation-associated proteins, and share two homologous regions (HR1 and HR2)4 (1). Compared with Caprin-1, Caprin-2 contains an extra C1q-related domain (Cap2_CRD) at its C terminus. C1q domain was first identified as the recognition subunit of the C1 complex complement and was later identified in other proteins with a variety of functions (2, 3). C1q family proteins share a homologous three-dimensional folding and a trimeric assembly with the TNF family despite low sequence identity. The trimeric assembly is essential for the function of TNF family proteins (4, 5), although it remains unclear whether this is also the case for the C1q family. Interestingly, most C1q family proteins contain one or multiple calcium ions at their trimetric centers, whereas their structurally homologous TNF family proteins usually do not bind any metals. It is proposed that calcium may contribute to the stabilization of C1q family proteins as well as their trimeric assemblies (6, 7). Calcium may also function in mediating and regulating the interactions between C1q family proteins and their ligands (8–10). However, experimental evidence supporting these proposals is very limited, and the significance of calcium for the trimerization and function of C1q proteins remains to be determined.
So far, most well characterized C1q family proteins are extracellular ones, whereas knowledge regarding the structures and functions of intracellular ones remains scant. Caprin-2 is an intracellular protein that was identified as a new regulator of canonical Wnt signaling (11). In canonical Wnt signaling, upon Wnt stimulation, LRP5/6 is aggregated and forms a large ensemble with Frizzled, Dvl, Axin, and GSK3 (also known as LRP5/6 signalosomes) (12), which triggers LRP5/6 phosphorylation and initiates the signal transduction. Previously, we found that Caprin-2 binds to LRP5/6 and facilitates its phosphorylation, thus playing a positive role in canonical Wnt signaling (11). Further studies have revealed that Caprin-2 is associated with several components in LRP5/6 signalosomes, such as Axin and Dvl (data not shown), implying that Caprin-2 may serve as a scaffold protein in LRP5/6 signalosomes. Importantly, our previous data suggested that the C-terminal C1q-related domain (CRD) is required for the function of Caprin-2 in canonical Wnt signaling (11): 1) only Caprin-2 with an extra CRD domain, but not Caprin-1, can co-immunoprecipitate LRP5; 2) the HR2 and CRD region of Caprin2 is required to interact with LRP5; and 3) the functional effect of Caprin-2 knockdown can only be rescued by full-length Caprin-2 but not the truncation mutant missing CRD domain.
To get a better understanding of the role of the CRD domain in the regulation of Caprin-2 of LRP5/6 phosphorylation, we determined the crystal structures of Cap2_CRD from human and zebrafish. In both cases, CRD domain associates as a homotrimer with a typical jellyroll topology. Interestingly, human Cap2_CRD (hCap2_CRD) binds two calcium ions mediated by residues Asp1078 and Glu1084 and zebrafish (zCap2_CRD) binds one through Glu871. We also determined the crystal structure of the hCap2_CRD double mutant D1078A/E1084A (Cap2_DAEA), which is disabled in binding calcium. Surprisingly, Cap2_DAEA retains the trimeric assembly and exists as an even more stable trimer in solution than its wild-type counterpart. In line with it, the full-length Caprin-2 DAEA double mutant maintains the activity in promoting LRP5/6 phosphorylation. However, the mutants targeting Cap2_CRD trimeric interface show reduced activity toward promoting LRP6 phosphorylation, although their bindings with LRP6 are not significantly affected. Collectively, our findings indicated that hCap2_CRD exists as a flexible trimer mediated by calcium and that its trimerization is essential for the function of Caprin-2 in promoting LRP5/6 phosphorylation.
EXPERIMENTAL PROCEDURES
Cloning of Constructs, Protein Expression, and Purification
zCap2_CRD (amino acids Ala784–Asp914 of zebrafish Caprin-2) was amplified by PCR. After being cleaved by NcoI and XhoI, the PCR product was inserted into PET-28a (Novagen) cut with the same two restriction enzymes. After being verified by DNA sequencing, the plasmid was transformed into BL21-CodonPlus-RIL (Agilent Technologies), which provides extra copies of argU, ileY, and leuW tRNA genes. Protein expression was induced at 16 °C for 22 h or 37 °C for 3 h after A600 arrived ∼0.6. The pellet was dissolved in buffer A containing 20 mm Hepes, 500 mm NaCl, 1 mm DTT, 20 mm imidazole. After sonication, the lysate was centrifuged, and the supernatant was applied to a nickel affinity column (GE Healthcare). The target protein eluted from the nickel column was concentrated and applied to Superdex 75 10/300 GL gel filtration column (GE Healthcare) using buffer A but with a lower NaCl concentration of 150 mm plus 0.1% CHAPS. The protein after Superdex 75 was concentrated into ∼7.4 mg/ml and used for crystallization. hCap2_CRD (amino acids Arg997–Asp1127 of human Caprin-2) and its double mutant D1078A/E1084A (Cap2_DAEA) were expressed and purified in a way similar to that of zCap2_CRD and were concentrated into ∼3.3 and ∼8.6 mg/ml, respectively, for crystallization. The HA-tagged and GFP-tagged full-length human Caprin-2 constructs were conducted as described before (11)
Mutagenesis
Mutants I1048R, D1078A/E1084A, I1091S, W1114S, and Y1122S were introduced into hCap2-CRD using a method similar to that described in the Stratagene QuikChange kit manual, among which I1048R, D1078A/E1084A, and I1091S were also introduced into the full-length HA-hCaprin2. All the mutants were subsequently confirmed by DNA sequencing.
Crystallization and Structure Determination
zCap2_CRD crystals were obtained under the condition of 4.0 m sodium nitrate, 0.1 m Bis-Tris propane, pH 7.0. Crystals were flash-cooled at liquid nitrogen in mother liquid containing 25% glycerol. Diffraction data were collected at Shanghai Synchrotron Radiation Facility Beamline BL17u and processed with the HKL2000 software (13). zCap2_CRD crystals belonged to space group I23 with cell parameters a = b = c = 94.985 Å. The crystal structure was determined by molecular replacement using Phaser as part of the CCP4 suite (14–16) and Protein coordinates of Collagen Viii Nc1 Domain monomer structure (Protein Data Bank code 1O91) was used as the search model (17). Refinement steps were firstly performed using REFMAC (18) with data up to 1.5 Å, and model corrections were carried out using Coot (19). Next, anisotropic refinement was conducted using SHELX package by randomly selecting 5% of the observed reflections as cross-validation (20). Using data in resolution range 10–1.5 Å, the model was first refined to convergence with an R factor of 20.57% and a free R factor of 23.65%. Then thermal factor refinement was added, and water molecules were also added gradually, accompanied with the gradual extension of resolution limit up to 1.05 Å. Alternative conformations were modeled for eight residues: Pro803, Thr811, Met816, Val833, Ile835, Ile838, Ser874, and Lys902 using 2Fo − Fc, as well as clues in the diagnostic table output by SHELXL. Riding hydrogen atoms based on stereo-chemical requirements were added at the last refinement step.
hCap2_CRD WT and hCap2_DAEA double mutant crystals were grown under the condition of 15–20% PEG 8000, 0.2 m calcium acetate, 0.1 m carcodylate, pH 6.6, and 100 mm sodium citrate, pH 5.6, 12% 2-propanol, 11% PEG 4000, respectively. hCap2_CRD WT crystals belonged to space group P1 with cell parameters a = 48.353 Å; b = 48.342 Å; c = 80.884 Å; α = 89.509°; β = 87.241°; and γ = 71.743°, whereas hCap2_DAEA double mutant crystals belonged to space group R3 with cell parameters a = b = 77.498 Å; c = 53.447 Å; α = β = 90.000°; and γ = 120.000°. Both structures were determined and refined in a similar way as that of zCap2_CRD with zCap2_CRD as a search model except without thermal factor refinement in SHELX.
Buried surface areas were calculated with PDBePISA. The final Rwork/Rfree factors were 0.152/0.161, 0.204/0.249, and 0.186/0.217, respectively, for zCap2_CRD, hCap2_CRD, and hCap2_DAEA. The quality of these models was examined with the program MolProbity (21). The data collection and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| hCap2_CRD | zCap2_CRD | hCap2_CRD_EADA | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | I23 | R3 |
| Unit cell parameters (Å) | |||
| a, b, c (Å) | 48.35, 48.34, 80.88 | 94.99, 94.99, 94.99 | 77.50, 77.50, 53.45 |
| α, β, γ (°) | 89.51, 87.24, 71.74 | 90, 90, 90 | 90, 90, 120 |
| Resolution range (Å) | 50.0–1.95 (1.98–1.95)a | 50–1.05 (1.09–1.05) | 50.0–1.49 (1.54–1.49) |
| Total reflections | 223,387 | 826,345 | 181,859 |
| Unique reflections | 21,926 | 66,313 | 19,550 |
| Completeness (%) | 90 (96.7) | 95.2 (100) | 99.3 (100) |
| Redundancy | 1.9 (2.0) | 3.9 (4.1) | 5.3 (5.2) |
| Rmerge | 0.024 (0.251) | 0.092 (0.445) | 0.054 (0.207) |
| <I/σI> | 24.22 (2.52) | 17.86 (3.26) | 29.27 (12.31) |
| Refinement | |||
| Rwork/Rfree | 0.204/0.249 | 0.152/0.161 | 0.186/0.217 |
| No. atoms | |||
| Protein | 6276 | 1065 | 1070 |
| Ligand | 4 | 1 | 64 |
| Water | 139 | 130 | 82 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.005 | 0.007 |
| Bond angles (°) | 1.29 | 1.13 | 1.19 |
| Ramachandran | |||
| Favored (%) | 94 | 97 | 96 |
| Outliers (%) | 0.52 | 0 | 0 |
| Average B factors (Å2) | 36.2 | 11.7 | 22.6 |
a The values in parentheses are for the highest resolution shell.
Light Scattering Experiments
WT and mutant hCap2_CRD proteins (∼1 mg/ml) were filtered firstly through 0.45-μm cellulose acetate membranes before used for light scattering studies on DynaPro NanoStar (WYATT Techology). Each sample was measured 50 times with an acquisition time of 10 s at 5 °C. The data were then analyzed with the DYNAMICS software package. The dispersity of the solution was assessed, and the average of the hydrodynamic radius (RH) was calculated.
Native Gel
6% native gel was prepared in a similar way to that of the conventional SDS-PAGE gel, except removing SDS from both running buffer and sample loading buffer. The running buffer was prechilled on ice. Then the gel was put in an ice bucket under a constant current of 20 mA.
Cross-linking Analysis
Cross-linking analysis using BS3 (Thermo Scientific) was carried out according to the product manual. Briefly, the proteins (∼1 mg/ml) of wild-type and mutant hCap2_CRDs were mixed with BS3 (1 mm) and incubated at room temperature for 30 min. The cross-linking reaction was quenched by 20 mm Tris buffer, pH 7.5.
Cell Culture and Transfection
HEK-293T cells were propagated in DMEM (Invitrogen) plus 10% FBS (Invitrogen). Cells were seeded in plates 24 h before transfection. Plasmids were transfected using Lipo2000 (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation and Western Bolt Analysis
After transfection, the cells were harvested and lysed in protein lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% (v/v) Triton X-100, 5 mm EDTA, and proteinase inhibitors) and centrifuged at 16,000 × g for 15 min at 4 °C. The lysates were incubated with primary antibody for 1 h at 4 °C. Protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc.) was added and incubated at 4 °C for 3 h. Samples were washed three times, eluted by SDS loading buffer, separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with antibodies. The results were visualized using ImageQuantTM LAS 4000 biomolecular imager (GE Healthcare). Anti-GFP (Roche), anti-HA (Covance), anti-Flag (Cell Signaling), and anti-Actin (Santa Cruz) antibodies were used in this work. Anti-LRP6 and anti-phospho-LRP6 antibodies were obtained from Cell Signaling Technology.
RESULTS
Crystal Structures of Cap2_CRD
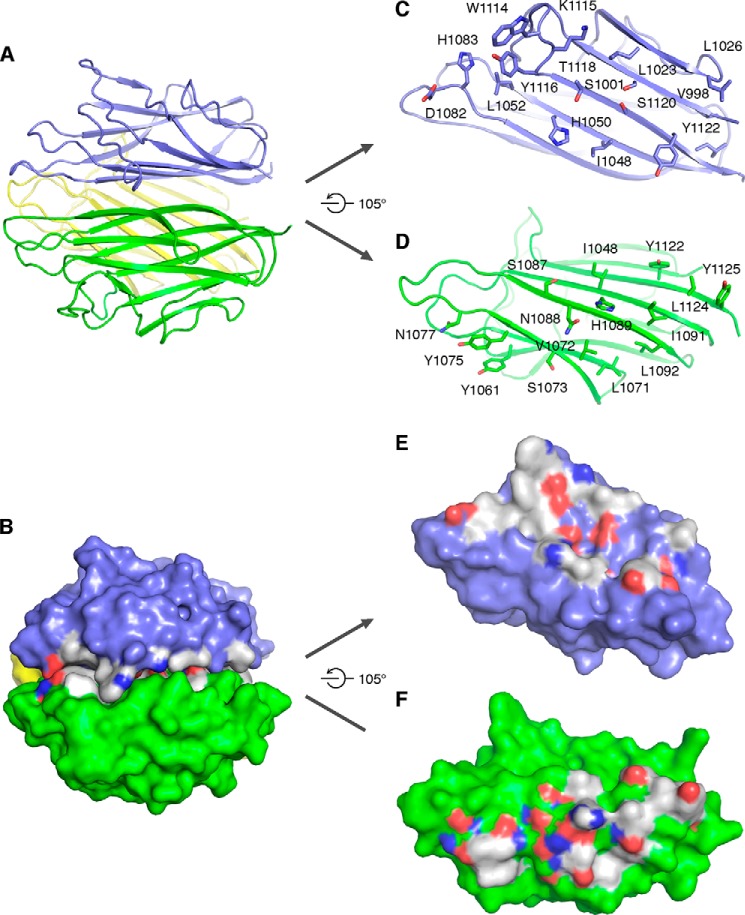
As introduced previously, Caprin-2 is composed of three domains: HR1, HR2, and CRD (Fig. 1A). HR1 and HR2 have been implicated in a function related to RNA binding (22–24). In addition, HR1 plays a role in mediating oligomerization of the Caprin proteins (1). Compared with HR1 and HR2, the function of CRD domain—present only in Caprin-2 but not in Caprin-1—remains largely unknown. In human, there are four isoforms of Caprin-2, and the longest one contains 1127 amino acids, whereas in zebrafish, only one isoform of Caprin-2 exists, encompassing 914 amino acids. These two proteins share ∼36% sequence identity over the full length but ∼85% over their CRD domains, indicating a possibly significant role of this highly conserved domain in the function of Caprin-2. To better understand the function of Caprin-2, we determined the crystal structures of the CRD domain of human and zebrafish Caprin-2. zCap2_CRD contained one molecule in each asymmetric unit and formed a crystallographic 3-fold axis related trimer, whereas hCap2_CRD contained six molecules in each asymmetric unit, forming two noncrystallographic symmetry related homotrimers (Fig. 1, B–D). The crystal structures of zCap2_CRD and hCap2_CRD are highly similar with an r.m.s. deviation of 0.3656 Å for the main chain atoms. For brevity, we hereafter use hCap2_CRD for discussion unless noted otherwise. hCap2_CRD exhibited a classic 10-stranded jelly roll folding topology similar to that of Collagen Viii Nc1 (r.m.s. deviation of 1.3471 Å; Protein Data Bank code 1O91) (17), although the sequence homology between them is low (35.4%).
FIGURE 1.
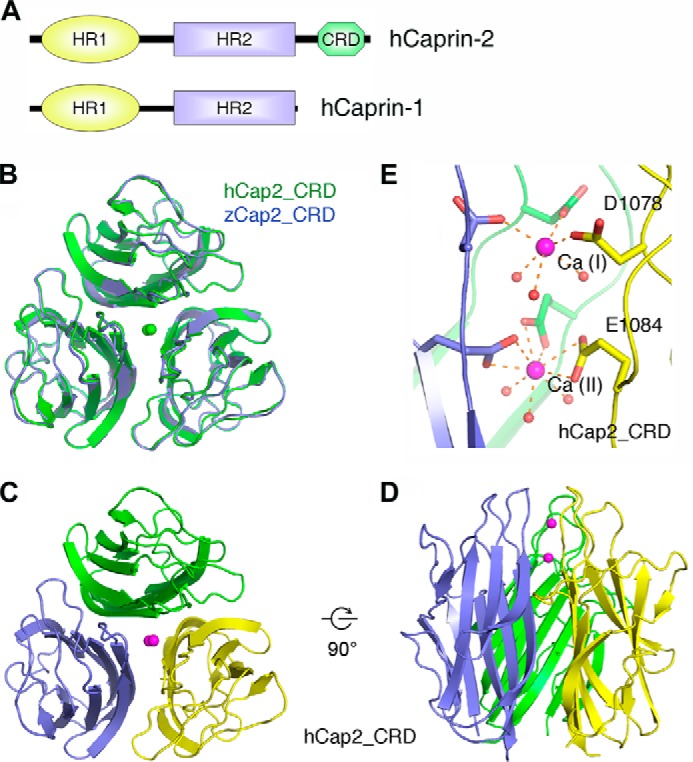
Structures of hCap2_CRD and zCap2_CRD. A, schematic representation of domain organization of Caprin family proteins. B, hCap2_CRD (green) and zCap2_CRD (blue) superimposed well, with an r.m.s. deviation of 0.3656 Å over 130 pairs of Cα atoms. C and D, hCap2_CRD exhibited a classic 10-strand jelly roll folding topology and a trimeric assembly characteristic for both C1q family and TNF family proteins. E, there are two calcium ions, designated as Ca(I) and Ca(II), aligned along the 3-fold axis of hCap2_CRD trimer. Ca(I) was coordinated by Asp1078 in a unidentate way and three water molecules, whereas Ca(II) interacted with Asp1084 in a bidentate way and three water molecules. The Ca2+-oxygen distances spanned a range from 2.29 to 2.81 Å with the average distance being 2.49 Å.
The homotrimerization of Cap2_CRD buried a total surface of ≈5600 Å2 (∼40% of total accessible molecular surface) (Fig. 2, A and B). The homotrimeric interaction of Cap2_CRD is largely mediated by hydrophobic packing. Specifically, residues Ile1048, Tyr1061, Leu1071, Val1072, Tyr1075, His1089, Ile1091, Leu1092, Tyr1122, Leu1124, and Tyr1125 from one subunit interacted with Leu1023, Leu1026, Ile1048, His1050, Leu1052, His1083, Trp1114, Lys1115, Tyr1116, and Tyr1122 from the other one through hydrophobic interaction (Fig. 2, C–F). Meanwhile, side chains of residues Ser1073, Tyr1075, Asn1077, Ser1087, Asn1088, His1089, Tyr1122, and Tyr1125 and main chains of residues Ser1073 and Leu1071 from one subunit formed hydrogen bond or ion pair with side chains of Ser1001, His1050, Asp1082, Lys1115, Thr1118, Ser1120, and Tyr1122 and main chains of Val998, Trp1114, Lys1115, and Leu1123 from the other subunit (Fig. 2, C–F). Interestingly, the molecular surface of the Cap2_CRD homotrimer exhibits patches of residues that may participate in recognizing ligands or binding partners.
FIGURE 2.
Details of the interface of hCap2_CRD trimer. A, C, and D, cartoon views of hCap2_CRD trimer interface. The trimer interface of hCap2_CRD is mediated by both hydrophobic and hydrophilic interactions. B, E, and F, surface view of hCap2_CRD trimeric assembly with a total buried surface of ≈5600 Å2. Surfaces were colored by electrostatic potential.
Two metal ions were found to align along the 3-fold symmetry axis, which were coordinated by the carboxylate groups of residues Glu1084 and Asp1089 from all three subunits, as well as by six water molecules (Fig. 1E). Because calcium is the only metal reported so far for C1q family proteins, we therefore assigned these two metal ions as calcium. The identity of the calcium was subsequently confirmed by structural refinement and chemical analyses. Ca(I) was coordinated by Asp1078 in a unidentate way and three water molecules, producing a coordination number of 6, whereas Ca(II) interacted with Asp1084 in a bidentate way as well as three water molecules, resulting in a coordination number of 9. The Ca2+-oxygen distances spanned a range from 2.29 to 2.81 Å with the average distance being 2.49 Å. Moreover, the average B factor of Ca2+ matched that of the solvent atoms (8.8 Å2 versus 8.7 Å2 for zCap2_CRD and 35.7 Å2 versus 35.5 Å2 for hCap2_CRD), and difference maps for calcium ions were featureless. Ca(II) bound by Glu1084 in hCap2_CRD was also observed in zCap2_CRD through Glu871, whereas Ca(I) bound by Asp1078 in hCap2_CRD was absent in zCap2_CRD.
hCap2_DAEA Disabled in Binding Calcium Retains a Trimeric Assembly
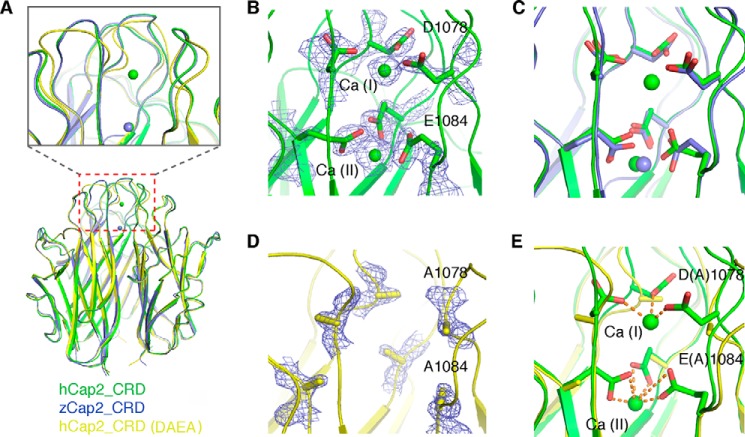
Because calcium is believed to be important for the stabilization of C1q family trimer (6, 7), we generated a calcium binding-deficient double mutant, D1078A/E1084A, to disrupt the trimer of hCap2_CRD, and next we determined its crystal structure. However, unexpectedly, hCap2_DAEA exhibited a trimeric assembly that is almost identical to that of the wild-type hCap2_CRD (r.m.s. deviation of 0.415 Å for the main chain atoms) (Fig. 3A). The 2Fo − Fc map contoured at 1.5 verified the replacement of residues Asp1078 and Glu1084 by two alanines and also clearly demonstrated the absence of densities corresponding to the two calcium ions observed in wild-type hCap2_CRD (Fig. 3, B and D). Despite overall similarity, local conformational changes around residue Asp1078 and Glu1084 were observed in hCap2_DAEA, compared with its wild-type counterpart (Fig. 3, A, C, and E), whereas the three-dimensional conformation of these areas are rather conserved between human and zebrafish Cap2_CRD, despite their relatively low sequence homology (Fig. 3C). These results indicated that replacement of Asp1078 and Glu1084 with alanine has an impact mainly restricted to the regions close to Asp1078 and Glu1084 and that hCap2_DAEA without binding calcium maintains the ability to form a trimer.
FIGURE 3.
Cap2_DAEA mutant retains a trimeric assembly in crystal structure. A, Cap2_DAEA mutant surprisingly exists as a trimer. Shown is a side view of the superimposed structures of WT hCap_CRD (green), zCap2_CRD (blue), and DAEA mutant (yellow). B and D, zoomed in view of the calcium-binding region of hCap2_CRD and Cap2_DAEA, respectively. The 2Fo − Fc density map is contoured at the 1.5 σ level. C and E, the superimposed structures of zCap2_CRD and Cap2_DAEA with that of wild-type hCap2_CRD.
Calcium-mediated Wild-type hCap2_CRD Trimer Is an Unstable One
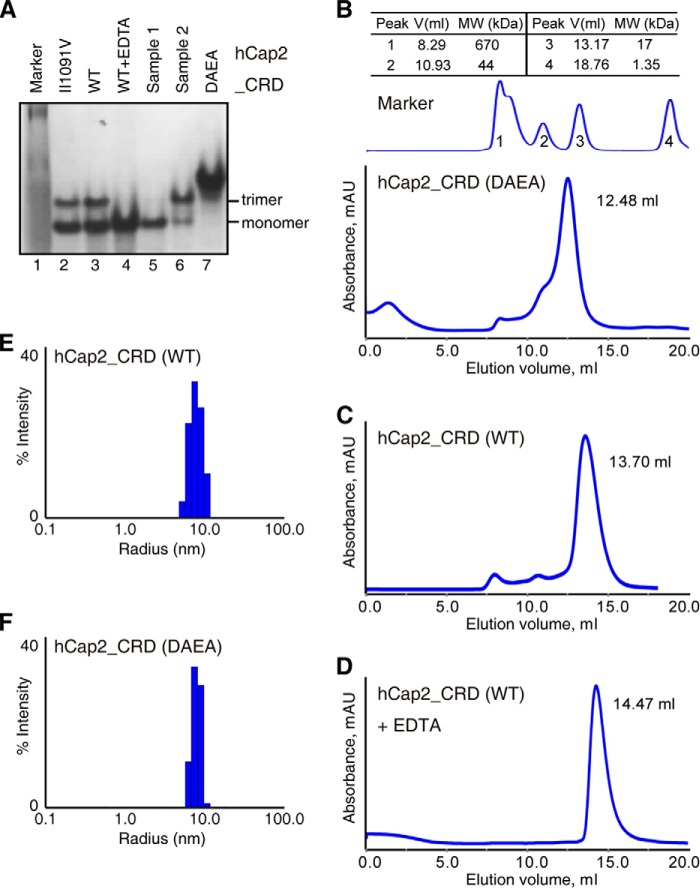
To explore the potential role of calcium for Cap2_CRD, we examined the oligomeric state of wild-type Cap2_CRD with the presence or absence of EDTA. EDTA forms complex with metals and thus could deplete Cap2_CRD of calcium. First, we observed on native gel that wild-type Cap2_CRD, when untreated with EDTA, showed two bands (Fig. 4A) that were very likely the trimetric and monomeric band. By contrast, hCap2_DAEA only displayed one band. Notably, wild-type Cap2_CRD, when treated with 1 mm EDTA, showed only the monomeric band, which could be shifted substantially to the trimeric one when 5-fold more calcium was added.
FIGURE 4.
Wild-type hCap2_CRD trimer mediated by calcium is unstable. A, native gel analysis showed that wild-type Cap2_CRD exists as a mixture of trimer and monomer, which could be shifted substantially to monomer by EDTA treatment or trimer by calcium. Sample 1 is obtained by loading EDTA-treated wild-type hCap2_CRD on Superdex 75 in a buffer containing 0.5 mm EDTA. Sample 2 is obtained from adding a final concentration of 5 mm CaCl2 to sample 1. B–D, DAEA mutant, wild-type Cap2_CRD and the latter treated with EDTA were eluted from gel filtration column as trimer, a mixture of trimer and monomer, and monomer, respectively. However, their apparent molecular masses (25, 14, and 9 kDa) calculated from gel filtration standard are obviously lower than expected for unknown reasons. E and F, dynamic light scattering analysis showed that wild-type hCap2_CRD and hCap2_DAEA both exhibit a single peak, and they have a similar average hydrodynamic radius with an estimated molecular mass (MW) of 60 kDa.
To investigate the oligomeric state of wild-type hCap2_CRD and the DAEA mutant in solution, we next checked their apparent molecular masses on gel filtration column Superdex 75 10/300 GL (GE Healthcare). hCap2_DAEA was eluted as a single peak with an apparent molecular mass of ∼25 kDa calculated from gel filtration standard (Bio-Rad) (Fig. 4B), whereas the elution peak of wild-type hCap2_CRD lagged significantly behind that of the DAEA mutant with an apparent molecular mass of ∼14 kDa (Fig. 4C). We next subjected wild-type Cap2_CRD treated with excessive EDTA to Superdex 75 column pre-equilibrated with buffer containing 1 mm EDAT. EDTA-treated wild-type Cap2_CRD was eluted with an apparent molecular mass of ∼9 kDa (Fig. 4D). This indicated that wild-type Cap2_CRD exists most likely as a mixture of trimer and monomer on gel filtration, whereas hCap2_DAEA tends to form stable trimer. For unknown reasons, these MWs calculated from gel filtration strand are obviously lower than the theoretical molecular masses (∼48 kDa and ∼16 kDa for trimetric and monomeric states, respectively). Because C1q family proteins normally exist as trimers, we speculated that the monomeric form of wild-type hCap2_CRD observed in the experiments of gel filtration and native gel was probably due to partial dissociation of hCap2_CRD trimer caused by these conditions. To address this concern, we turned to dynamic light scattering, a more gentle and noninvasive technique, to further examine the oligomeric state of hCap2_CRD in solution. As shown in Fig. 4 (E and F), the results of DSL showed that the average hydrodynamic radius of wild-type hCap2_CRD is similar to that of the DAEA mutant, both of which show an apparent molecular mass of ∼60 kDa, indicating that wild-type hCap2_CRD exists as a trimer in solution. However, we observed a polydispersity value for wild-type hCap2_CRD (25.6%) on dynamic light scattering higher than that of its DAEA mutant (17.2%), supporting the possibility that wild-type hCap2_CRD trimer is indeed less stable and thus appeared to be less monodisperse than the DAEA mutant. Overall, these results suggest that calcium is essential for the stabilization of Cap2_CRD trimer; however, the calcium-mediated wild-type Cap2_CRD trimer is an unstable one compared with its DAEA mutant.
Residues at Trimer Interface but Not Calcium Triggers the trimerization of Cap2_CRD
To further explore the role of calcium during trimerization of Cap2_CRD, we next carried out cross-linking analyses using BS3 (Thermo Scientific). BS3 is a regent that is capable of capturing a transient complex by connecting the interacting partners covalently. Only if the trimerization could be initiated in a similar way, the trimeric state would be captured and secured by BS3 in a similar manner regardless of its subsequent stability. That is, both the stable and the transient trimer, after BS3 treatment, would display similar trimeric bands on SDS-PAGE gel. To investigate the role of calcium in the formation of Cap2_CRD trimer, we treated Cap2_CRD with EDTA or calcium and then subjected these samples to cross-linking analysis using BS3. As shown in Fig. 5A, all of these samples, regardless of whether they contained calcium or not, displayed similar trimeric bands, indicating that calcium is not required for the initial formation of Cap2_CRD trimer.
FIGURE 5.
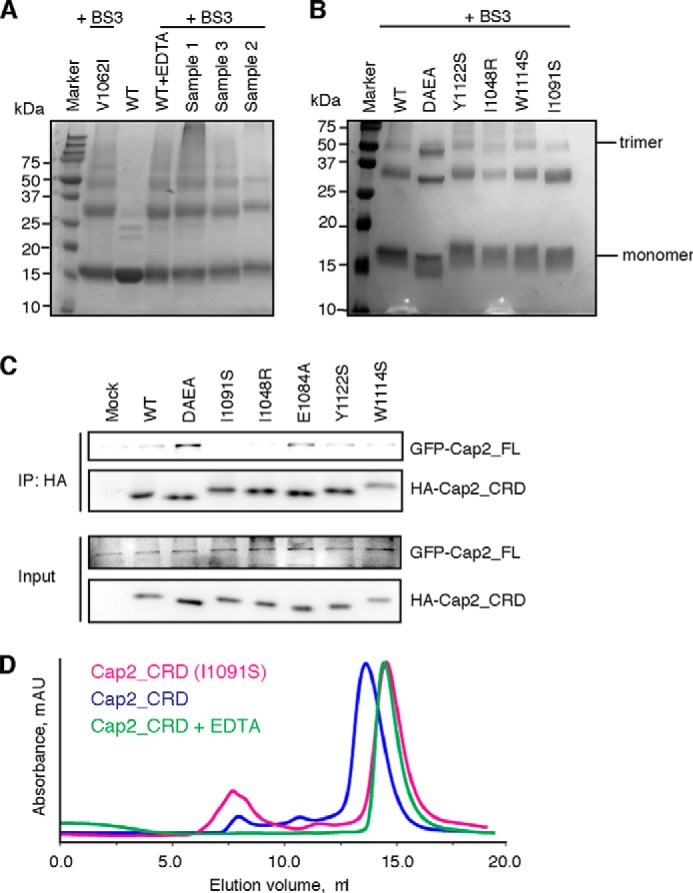
Mutations targeting trimeric interface disrupt Cap2_CRD trimer. A, BS3 cross-linking analysis showed that the trimetric bands are similar for all hCap2_CRD samples regardless of whether they contain calcium or not. Sample 1 is obtained from adding EDTA to WT Cap2_CRD. Sample 2 is obtained by further purification of sample 1 on Superdex 75 in a buffer containing 0.5 mm EDTA. Sample 3 is obtained from adding a final concentration of 5 mm CaCl2 to sample 2. V1062I is a nonsense point mutant also as a positive control. B, BS3 cross-linking analysis for mutants targeting the trimeric interface. C, HEK293T cells were co-transfected with GFP-tagged full-length Caprin-2 and HA-tagged Cap2_CRD wild type or the mutants as indicated. The lysates were then immunoprecipitated (IP) using anti-HA and then immunoblotted with anti-GFP. Compared with wild-type Cap2_CRD, I1048R and I1091S showed decreased binding affinities with the full-length Caprin-2, whereas DAEA and E1084A showed an increased binding with the full-length Caprin-2. D, the elution peak of I1091S lagged behind that of its wild-type counterpart and showed a similar position with the EDTA-treated wild-type Cap2_CRD.
Next, to disrupt Cap2_CRD trimerization, we generated a group of mutants targeting Cap2_CRD trimeric interface (I1048R, I1091S, W1114S, and Y1122S). We purified these mutants and subjected them to BS3 cross-linking. Among these mutants, the trimeric bands for I1048R and I1091S decreased markedly compared with that of other mutants, indicating that I1048 and I1091 may play an important role in the initial formation of Cap2_CRD trimer, and thus single mutation of them impairs significantly the ability of Cap2_CRD to form a trimer (Fig. 5B).
To further confirm the disruption of Cap2_CRD trimerization by I1048R or I1091S, we performed co-immunoprecipitation assay and co-transfected HEK293T cells with the GFP-tagged full-length Caprin-2 and HA-tagged Cap2_CRD wild type or the related mutants. The lysates were immunoprecipitated using anti-HA and then immunoblotted with anti-GFP. As shown in Fig. 5C, compared with wild-type Cap2_CRD, I1048R, and I1091S showed decreased binding with the full-length Caprin-2, supporting our notion that I1048R and I1091S leads to disruption of hCap2_CRD homotrimerization. Meanwhile, we observed an increased binding of DAEA or E1084A with the full-length Caprin-2, which is also consistent with our finding that replacement of Glu1084 with alanine could increase the stability of Cap2_CRD trimer. Finally, we did a large scale purification for the mutant I1091S and checked its oligomeric state on Superdex 75 column. As shown in Fig. 5D, the elution peak of I1091S lagged behind that of its wild-type counterpart and showed a similar elution profile with that of Cap2_CRD treated with EDTA, further supporting the possibility that a single mutation like I1091S could disrupt Cap2_CRD trimer as did the EDTA, although through different mechanisms; I1091S did it through disrupting trimer contacts and EDTA through removing calcium from Cap2_CRD. Together, these results indicated that trimer contacts rather than calcium are required for mediating the initial trimeric assembly of Cap2_CRD, and mutations targeting the trimeric interface, such as I1048R and I1091S, could effectively disrupt the Cap2_CRD trimer.
Trimerization Is Essential for Caprin-2 to Facilitate LRP6 Phosphorylation
To investigate the role of calcium as well as trimerization for the function of Caprin-2 in canonical Wnt signaling, we then transfected HEK293T cells with HA-tagged full-length Caprin-2 harboring the mutation of I1048R, I1091S, or DAEA. As described before, DAEA is a double mutant disabled in binding calcium. I1048R and I1091S are two single mutants targeting and disrupting Cap2_CRD trimeric interface. As shown in Fig. 6A, compared with wild-type Caprin-2, DAEA double mutant maintained the activity in terms of facilitating Wnt3a-induced LRP6 phosphorylation; by contrast, the two single mutants, I1048R and I1091S, displayed a remarkable decrease of such activity.
FIGURE 6.
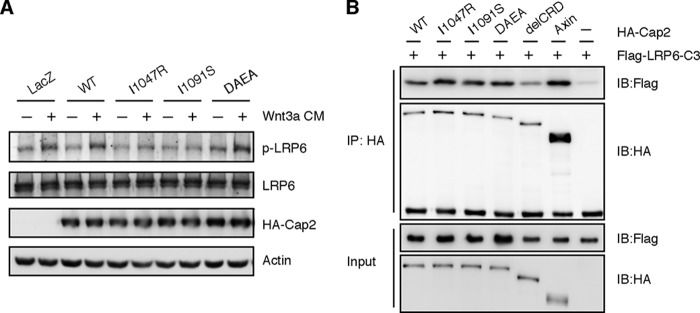
Trimerization of Cap2_CRD is essential for the function of Caprin-2. A, HEK293T cells were transfected with HA-tagged full-length Caprin-2 harboring the mutation of I1048R, I1091S, or D1078A/E1084A, and the cells were treated with control or Wnt3a CM for half an hour before lysis. Compared with WT Caprin-2, hCap2_DAEA maintained the activity in facilitating Wnt3a-induced LRP6 phosphorylation. By contrast, I1048R and I1091S displayed a remarkable decrease of activity. B, HEK293T cells were transfected with LRP6-C3 (intracellular fragment of LRP6) together with Capin-2 wild type or the above mutants. Axin, a known LRP5/6-binding protein, was used as a positive control. The interaction of I1048R or I1091S with LRP6-C3 showed no obvious decrease compared with the wild type. IP, immunoprecipitation; IB, immunoblot.
Our previous data indicated that both the C-terminal CRD and the middle HR2 region of Caprin-2 are involved in mediating Caprin2-LRP5/6 association, but CRD plays a major role in mediating Caprin2-LRP5/6 interaction (11). To investigate whether trimerization of Cap2_CRD is required for its interaction with LRP5/6, the HEK293T cells were transfected with LRP6-C3 (intracellular fragment of LRP6) together with Capin-2 wild type or the above mutants. Axin, a known LRP-binding protein, was used as a positive control. As shown in Fig. 6B, consistent with our previous result, Caprin-2 truncation without CRD exhibited markedly decreased binding with LRP6-C3. However, the interaction of I1048R or I1091S with LRP6-C3 showed no obvious decrease compared with their wild-type counterpart. These observations indicated that trimerization of Cap2_CRD is not necessary for Caprin-2 to interact with LRP6. Taken together, these results indicated that trimerization of the CRD domain is crucial for Caprin-2 to prompt LRP5/6 phosphorylation rather than interact with LRP5/6.
DISCUSSION
Most C1q family proteins identified so far are extracellular membrane-associated receptors. They play important roles in cell development, apoptosis, and immunological processes. Compared with extracellular ones, few intracellular C1q family proteins have been characterized. Caprin-2, as an intracellular protein, comprising a C1q domain, provides an opportunity to extend our knowledge about C1q family proteins. Our previous data indicated that the C-terminal C1q-related domain is required for the function of Carpin-2 in the canonical Wnt pathway (11). In the present work, we conducted crystallographic and biochemical studies on this domain of Caprin-2. To our knowledge, Cap_CRD is the first crystal structure of an intramolecular C1q protein, and our findings present direct evidence for the involvement of an intracellular C1q-related protein in regulating cell signaling.
According to our results, the homotrimerization of Cap2_CRD is most likely triggered by hydrophobic and hydrophilic interactions among residues on the homotrimeric interface in a manner independent of calcium. However, upon homotrimerization, the negatively charged residue (Glu1084 in hCap2_CRD) from three protomers is brought closer, resulting in electrostatic repulsion and/or steric hindrance. Such homotrimer is hence unstable and tends to dissociate as in the case of EDTA treatment. However, binding of calcium by these three glutamine residues balances the charge and stabilizes their conformations, thus leading to a relatively stable trimer. This interpretation is further supported by the fact that substitution of the bulky polar residue Glu1084 by alanine significantly stabilized the homotrimer as shown in the case of the double mutant hCap2_DAEA.
C1q and TNF family proteins are thought to derived from a common ancestor. However, calcium binding is an inherent property for C1q family proteins. TNF family proteins usually contain no metals, with few exceptions (25). According to our results, the conserved polar residue corresponding to Glu1084 of hCap2_CRD is responsible, at least partially, for the intrinsic calcium binding nature of C1q family proteins. When this calcium ion was replaced by alanine in hCap2_DAEA mutant, local structural rearrangement occurs around the calcium-binding sites (Fig. 3, A, C, and E), leading to a more stable trimer. Thus, we speculate that the TNF domain may generally form a more stable trimer than the C1q domain. In fact, for most members of C1q family, the C1q domain is preceded by a collagen-like region, which may help to stabilize the C1q homotrimer such as in the case of mouse adiponectin (26). The C1q domain of mouse adiponectin, when expressed alone, exists as both monomer and trimer but forms trimers and hexamers when the collagen-like region is included for expression (26). Such collagen-like region is not present in Cap2_CRD. The amount of buried molecular surface upon Cap2_CRD homotrimerization is comparable with that of mouse adiponectin globular domain (5600 Å2 versus 5150 Å2) and is significantly smaller than that of the collagen X NC1 (7610 Å2), which maintains a stable homotrimer even under very harsh conditions such as 2 m urea containing 2% SDS (6, 7). The lack of an N-terminal collagen-like region and a relatively small trimeric interface suggest that instability is probably an intrinsic nature of Caprin-2 homo-oligomer that could be exploited for a regulatory function. This assumption is reminiscent of human GITRL, a member of TNF family, which also modulates its function through changing its oligomeric states via its C-terminal tail (27).
In addition to LRP5 cytoplasmic domain (11), we found that Cap2_CRD can also bind several other components in the LRP5/6 signalosomes, such as Axin and Dvl (data not shown), indicating that Cap2_CRD may act as a scaffold protein to recruit a diversity of partners. In fact, C1q family proteins are well appreciated for their activities in binding amazingly diverse ligands. Based on mutagenesis analyses, as well as the available three-dimensional structures of C1q protein-ligand complex, some ligands appear to bind to the surface of single subunit of C1q homotrimer (10, 28–31) rather than to a cleft formed by two neighboring subunits as observed in TNF family (28, 32). Consistent with this ligand-binding model, we found that mutants of Cap2_CRD disrupting its homotrimerization showed no apparent decrease in binding with LRP6, indicating that Cap2_CRD may also interact with LRP5/6 through the exposed molecular surface of protomer, instead of the homotrimeric interface. However, these mutants exhibited a dramatic decrease in the activity of promoting LRP6 phosphorylation, indicating that homotrimerization is required for Caprin-2 to regulate Wnt signaling. Moreover, the oligomerization state of Caprin-2 could probably be regulated by Wnt stimulation, thereby facilitating LRP5/6 function dynamically. Indeed, we observe a shift of Caprin-2 wild type, but not that of the I1091S mutant, to higher molecular mass fractions on gel filtration column Superose 6 in response to Wnt stimulation, and importantly this change of Caprin-2 is concordant with a shift of LRP6 to the same fractions (data not shown). Given that LRP5/6 aggregation is essential for LRP5/6 phosphorylation (12), it is conceivable that Caprin-2 may act as a scaffold protein mediating LRP5/6 aggregation at least partially through a homointeraction of its CRD domain. As such, monomeric Cap2_CRD maintains the ability to interact with LRP5/6, but it fails to facilitate the aggregation and phosphorylation of LRP5/6. In the future, structural information of Cap2_CRD in complex with LRP5/6 and other Wnt signaling components will help to clearly address these questions. In summary, our studies here not only provide structural insights into the function of Caprin-2 in canonical Wnt signaling but may also facilitate understanding about the function of intracellular C1q proteins as well as the role of calcium for C1q family proteins.
Acknowledgments
We thank the staff at Beamline BL17U of the Shanghai Synchrotron Radiation Facility for help of data collection.
This work was supported by Ministry of Science and Technology of China Grants 2010CB912100 (to L. L.) and 2011CD966300 (to X. S.) and National Natural Science Foundation of China Grants 31230044, 30930052, and 91213304 (to L. L.) and 31100532 (to X. S.).
The atomic coordinates and structure factors (codes 4OUS, 4OUL, and 4OUM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HR
- homologous region
- CRD
- C1q-related domain
- r.m.s.
- root mean square
- BS3
- bis[sulfosuccinimidyl] suberate.
REFERENCES
- 1. Grill B., Wilson G. M., Zhang K. X., Wang B., Doyonnas R., Quadroni M., Schrader J. W. (2004) Activation/division of lymphocytes results in increased levels of cytoplasmic activation/proliferation-associated protein-1: prototype of a new family of proteins. J. Immunol. 172, 2389–2400 [DOI] [PubMed] [Google Scholar]
- 2. Carland T. M., Gerwick L. (2010) The C1q domain containing proteins: where do they come from and what do they do? Dev. Comp. Immunol. 34, 785–790 [DOI] [PubMed] [Google Scholar]
- 3. Kishore U., Reid K. B. (2000) C1q: structure, function, and receptors. Immunopharmacology 49, 159–170 [DOI] [PubMed] [Google Scholar]
- 4. Smith R. A., Baglioni C. (1987) The active form of tumor necrosis factor is a trimer. J. Biol. Chem. 262, 6951–6954 [PubMed] [Google Scholar]
- 5. He M. M., Smith A. S., Oslob J. D., Flanagan W. M., Braisted A. C., Whitty A., Cancilla M. T., Wang J., Lugovskoy A. A., Yoburn J. C., Fung A. D., Farrington G., Eldredge J. K., Day E. S., Cruz L. A., Cachero T. G., Miller S. K., Friedman J. E., Choong I. C., Cunningham B. C. (2005) Small-molecule inhibition of TNF-α. Science 310, 1022–1025 [DOI] [PubMed] [Google Scholar]
- 6. Shapiro L., Scherer P. E. (1998) The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 [DOI] [PubMed] [Google Scholar]
- 7. Bogin O., Kvansakul M., Rom E., Singer J., Yayon A., Hohenester E. (2002) Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure 10, 165–173 [DOI] [PubMed] [Google Scholar]
- 8. Roumenina L. T., Kantardjiev A. A., Atanasov B. P., Waters P., Gadjeva M., Reid K. B., Mantovani A., Kishore U., Kojouharova M. S. (2005) Role of Ca2+ in the electrostatic stability and the functional activity of the globular domain of human C1q. Biochemistry 44, 14097–14109 [DOI] [PubMed] [Google Scholar]
- 9. Ghai R., Waters P., Roumenina L. T., Gadjeva M., Kojouharova M. S., Reid K. B., Sim R. B., Kishore U. (2007) C1q and its growing family. Immunobiology 212, 253–266 [DOI] [PubMed] [Google Scholar]
- 10. Gaboriaud C., Juanhuix J., Gruez A., Lacroix M., Darnault C., Pignol D., Verger D., Fontecilla-Camps J. C., Arlaud G. J. (2003) The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 278, 46974–46982 [DOI] [PubMed] [Google Scholar]
- 11. Ding Y., Xi Y., Chen T., Wang J. Y., Tao D. L., Wu Z. L., Li Y. P., Li C., Zeng R., Li L. (2008) Caprin-2 enhances canonical Wnt signaling through regulating LRP5/6 phosphorylation. J. Cell Biol. 182, 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. (2007) Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 [DOI] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 16. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 17. Kvansakul M., Bogin O., Hohenester E., Yayon A. (2003) Crystal structure of the collagen α1(VIII) NC1 trimer. Matrix Biol. 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 18. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 21. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Solomon S., Xu Y., Wang B., David M. D., Schubert P., Kennedy D., Schrader J. W. (2007) Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27, 2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiina N., Tokunaga M. (2010) RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J. Biol. Chem. 285, 24260–24269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shiina N., Shinkura K., Tokunaga M. (2005) A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 25, 4420–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oren D. A., Li Y., Volovik Y., Morris T. S., Dharia C., Das K., Galperina O., Gentz R., Arnold E. (2002) Structural basis of BLyS receptor recognition. Nat. Struct. Biol. 9, 288–292 [DOI] [PubMed] [Google Scholar]
- 26. Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., Eto K., Akanuma Y., Froguel P., Foufelle F., Ferre P., Carling D., Kimura S., Nagai R., Kahn B. B., Kadowaki T. (2002) Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 8, 1288–1295 [DOI] [PubMed] [Google Scholar]
- 27. Zhou Z., Song X., Berezov A., Zhang G., Li Y., Zhang H., Murali R., Li B., Greene M. I. (2008) Human glucocorticoid-induced TNF receptor ligand regulates its signaling activity through multiple oligomerization states. Proc. Natl. Acad. Sci. U.S.A. 105, 5465–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukai Y., Nakamura T., Yoshikawa M., Yoshioka Y., Tsunoda S., Nakagawa S., Yamagata Y., Tsutsumi Y. (2010) Solution of the structure of the TNF-TNFR2 complex. Sci. Signal. 3, ra83. [DOI] [PubMed] [Google Scholar]
- 29. Garlatti V., Chouquet A., Lunardi T., Vivès R., Païdassi H., Lortat-Jacob H., Thielens N. M., Arlaud G. J., Gaboriaud C. (2010) Cutting edge: C1q binds deoxyribose and heparan sulfate through neighboring sites of its recognition domain. J. Immunol. 185, 808–812 [DOI] [PubMed] [Google Scholar]
- 30. Païdassi H., Tacnet-Delorme P., Garlatti V., Darnault C., Ghebrehiwet B., Gaboriaud C., Arlaud G. J., Frachet P. (2008) C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kojouharova M. S., Gadjeva M. G., Tsacheva I. G., Zlatarova A., Roumenina L. T., Tchorbadjieva M. I., Atanasov B. P., Waters P., Urban B. C., Sim R. B., Reid K. B., Kishore U. (2004) Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q-IgG interaction. J. Immunol. 172, 4351–4358 [DOI] [PubMed] [Google Scholar]
- 32. Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H. J., Broger C., Loetscher H., Lesslauer W. (1993) Crystal structure of the soluble human 55 kd TNF receptor-human TNF β complex: implications for TNF receptor activation. Cell 73, 431–445 [DOI] [PubMed] [Google Scholar]