Background: Sequence and structure comparisons suggested that the heme-binding protein HbpS from Streptomyces reticuli might bind cobalamin.
Results: HbpS binds aquo-cobalamin and the responsible histidine was identified.
Conclusion: The calculated Kd of 34 μm suggests that HbpS might bind cobalamin in both bacterial cultures and in the Streptomyces natural environment the soil.
Significance: The results suggest an evolutionary path between tetrapyrrole binding roles in the HbpS-like protein family.
Keywords: Bioinformatics, Ligand-binding Protein, Metalloprotein, Microbiology, Spectroscopy
Abstract
The extracellular protein HbpS from Streptomyces reticuli interacts with iron ions and heme. It also acts in concert with the two-component sensing system SenS-SenR in response to oxidative stress. Sequence comparisons suggested that the protein may bind a cobalamin. UV-visible spectroscopy confirmed binding (Kd = 34 μm) to aquo-cobalamin (H2OCbl+) but not to other cobalamins. Competition experiments with the H2OCbl+-coordinating ligand CN− and comparison of mutants identified a histidine residue (His-156) that coordinates the cobalt ion of H2OCbl+ and substitutes for water. HbpS·Cobalamin lacks the Asp-X-His-X-X-Gly motif seen in some cobalamin binding enzymes. Preliminary tests showed that a related HbpS protein from a different species also binds H2OCbl+. Furthermore, analyses of HbpS-heme binding kinetics are consistent with the role of HbpS as a heme-sensor and suggested a role in heme transport. Given the high occurrence of HbpS-like sequences among Gram-positive and Gram-negative bacteria, our findings suggest a great functional versatility among these proteins.
Introduction
Vitamin B12 and its derivatives are corrinoid macrocycles (Fig. 1) usually referred to as cobalamins (Cbl)2 and corrinoids (1, 2). Vitamin B12 is popularly known as an essential part of the human diet, but these corrinoids are also essential for some bacteria (3, 4). They promote growth in some algae as part of a symbiotic relationship with bacteria (5), and it has even been stated that corrinoids in soil act as growth factors in some plants (6). Some bacteria must take cobalamins from the environment, but others such as Propionibacterium, Pseudomonas, and Streptomyces and some archaea such as Halobacterium, Methanobacterium, and Methanosarcina synthesize cobalamins in considerable amounts. Two synthetic routes (aerobic and anaerobic) have been documented, and both are quite complex, involving ∼30 genes (7–9).
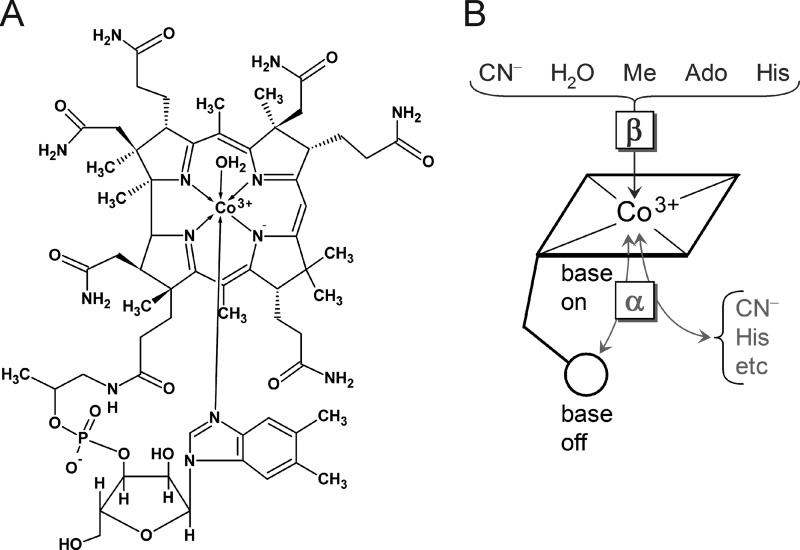
FIGURE 1.
Structure of aquo-cobalamin and base-on/base-off conformations. A, aquo-cobalamin in the base-on conformation in which the 5′,6′-dimethyl-benzimidazole base is coordinated to the Co3+ ion at the α-site. B, the base-on and base-off conformations. The variable interacting ligands as well as the α- and β-sites are indicated. Ado, AdoCbl; Me, MeCbl.
Because some bacteria, algae, and plants benefit from Cbl in the environment and because the synthesis occurs in a number of bacteria (10), there is a remarkable traffic of corrinoids in soils involving diffusion and transport proteins, both intra- and extracellular. The relationship between the soil-dwelling streptomycetes and plants and insects has been described as symbiotic (11, 12), but it is really part of a larger ecosystem. The bacteria form mycelia that penetrate the insoluble remains of fungi, plants, and other organisms. Secreted hydrolytic enzymes break larger insoluble molecules into smaller species that can be taken up (13). In addition, there is traffic in secreted secondary metabolites including antibiotics that modulate competition and cooperation between species.
There are different forms of Cbl in nature such as 5′-deoxyadenosylcobalamin (coenzyme B12/AdoCbl), methylcobalamin (MeCbl), and aquo-cobalamin, (vitamin B12a/H2OCbl+). Cyanocobalamin, known as vitamin B12 (CNCbl), is the main industrially produced Cbl. The formal oxidation state of the cobalt ion in AdoCbl, MeCbl, H2OCbl+, and CNCbl is +3 (14). Fig. 1 shows some other relevant features of cobalamins. Aside from the corrin ring with the central cobalt, there is a nucleotide loop whose 5′,6′-dimethyl-benzimidazole base coordinates the metal at the lower axial position (α-site). Other active or inactive groups such as methyl, adenosyl, cyanide, hydroxyl, and histidine may be coordinated to cobalt at the upper axial position (β-site) (Fig. 1) (1, 15). The variety of active groups and the different oxidation states of the cobalt (+1, +2, and +3) allow the cofactor to participate in many different kinds of biochemistry (16). One can also see that the cobalamins are large ligands with many potential hydrophobic and polar interactions that can lead to association constants as high as 1015 m−1 (15).
There is a wide variety of proteins that interact with cobalamins. Usually one associates cobalamins with their role as cofactors for mutases, dehydratases, deaminases, ribonucleotide reductases, methyl transferases, methionine synthases, and methylmalonyl-CoA-mutases (2). In streptomycetes there are Cbl-dependent enzymes that catalyze a set of modifications to peptides or polyketides or other chemical backbones during the biosynthesis of antibiotics. For instance, the methylation of the antibiotics clorobiocin and fosfomycin is a cobalamin-dependent reaction (17, 18). Looking further afield, it has been reported that Cbl interacts with a riboswitch to regulate the expression of the ribonucleotide reductase nrdABS operon in Streptomyces coelicolor A3(2) (19). Croft et al. (5) suggest that cobalamin transport mechanisms have evolved several times, just within algae. There are probably more roles waiting to be found. Obviously, there is a wealth of proteins that bind cobalamins via very different modes.
Some of these binding mechanisms involve contacts with the upper or lower face shown in Fig. 1. In the “base-off” mode an imidazole group from a histidine residue displaces the 5′,6′-dimethyl-benzimidazole ligand from the α-position, and the protein sequence usually has an Asp-X-His-X-X-Gly motif (16, 20, 21). In the “base-on” binding mode the nucleotide base remains coordinated to the cobalt, and the sequence motif is absent. This has been seen in both enzymes (22) and transport proteins (23, 24). In the Cbl-transporter transcobalamin, the upper axial ligand of H2OCbl+ is replaced by a histidine residue of the protein (23, 24).
Our previous work has focused on the extracellular protein HbpS from the soil bacterium Streptomyces reticuli and its binding of heme. This multifunctional protein sequesters large quantities of ferrous iron ions that might protect S. reticuli from the effects of peroxide- and iron-based oxidative stress (25). HbpS is also an unusual heme-binding protein in which a threonine residue (Thr-113) apparently binds to the tetrapyrrole macrocycle (26, 27). In vitro and in vivo studies have also shown that HbpS can degrade the heme group. This activity may be responsible for HbpS-mediated protection against toxic concentrations of heme (28). Furthermore, HbpS acts as an accessory module of the two-component system SenS-SenR from S. reticuli (29, 30). In this system extracellular HbpS interacts with the membrane-embedded sensor kinase SenS. Under conditions of oxidative stress this leads to the autophosphorylation of SenS that, in turn, phosphorylates the transcriptional response regulator SenR. This activates the transcription of anti-oxidative genes (30, 31).
HbpS is a homo-octamer in both the crystal structure (Protein Data Bank acquisition codes 3FPV and 3FPW) and solution with a molecular mass of 8 × 15.5 kDa. This oligomerization is essential for the interaction with iron ions as well as with the sensor kinase SenS but not for the interaction with heme (25, 27, 28, 32). Sequence comparisons using the S. reticuli HbpS protein showed a large number of related proteins from both Gram-positive and Gram-negative bacteria, including species from the genera of Streptomyces, Arthrobacter, Rhodococcus, Nocardia, Leifsonia, Vibrio, Klebsiella, Pseudomonas, and Agrobacterium. Some of the hbpS-like genes are situated within operons encoding proteins that either degrade aromatic compounds or are involved in the metabolism of propane-1,2-diol or glycerol (26, 30). However, the exact role of these HbpS-like proteins is poorly understood. HbpS and all HbpS-like sequences have also been labeled as DUF336 domains with the vague annotation of “cofactor binding” (25, 26).
In this work we describe sequence comparisons which suggest that HbpS may also interact with cobalamin. Binding studies using different Cbl compounds and UV-visible spectroscopy showed that HbpS specifically binds aquo-cobalamin. Binding kinetics were characterized spectrophotometrically. Mutant versions of HbpS were used to identify the amino acid in HbpS that coordinates the cobalt ion of aquo-cobalamin. An HbpS-related protein also bound aquo-cobalamin, suggesting this feature may be common among HbpS-like proteins. Rate constants of heme binding by HbpS were also determined. The role of HbpS as a sensor or transporter of both heme and cobalamin is discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Media, and Culture Conditions
Streptomyces venezuelae ATCC 10712 was cultivated in complete (R2) liquid medium as previously described (33). Escherichia coli strains BL21(DE3)pLysS and DH5α were cultivated in LB medium. The plasmid vector pETM11 as well as the plasmid constructs pETHbpS, pETHbpS-H28A, pETHbpS-H51A, pETHbpS-H156A, pETHbpS-T113A, and pETHbpS-T113H (27, 28, 32) were used.
Isolation and Cleavage of DNA, Ligation, and Agarose gel Electrophoresis
Chromosomal DNA of S. venezuelae was isolated after growth in a sucrose-containing R2 medium for 2 days (33). Plasmids were isolated from E. coli using a mini plasmid kit (Qiagen) and cleaved with various restriction enzymes according to the suppliers (New England BioLabs; Thermo Scientific) instructions. Ligation was performed with T4 ligase. Gel electrophoresis was carried out in 0.8–2% agarose gels using Tris borate EDTA buffer. Plasmids were used to transform E. coli DH5α by electroporation or E. coli BL21 (DE3)pLys with the CaCl2 method (34).
Chemicals and Enzymes
Chemicals for SDS- and native-PAGE were obtained from ROTH. Cobalamin compounds (AdoCbl, MeCbl, CNCbl, and H2OCbl+), hemin, apomyoglobin, and tris(2-carboxyethyl)phosphine were purchased from Sigma. Molecular weight markers for DNA and protein, restriction enzymes, T4 ligase, and DNA polymerase for PCR were obtained from Thermo Scientific or New England Biolabs.
Cloning of a hbpS-like Gene from S. venezuelae
The coding region (without the codons encoding for the signal peptide) of the hbpS-homologous gene from S. venezuelae (SVEN_5961), referred to here as hbpSv, was amplified by PCR using the isolated chromosomal DNA as template and the primers PForVen *5′-GATGCCATGGCCACCGGCGAGAACACGG-3′ (containing an NcoI restriction site, underlined)) and PRevVen (5′-GATGAAGCTTCTACTTGCCGAGGGAGG-3′ (containing a HindIII restriction site)). The PCR product was digested with NcoI and HindIII, ligated with NcoI/HindIII-cleaved pETM11, and subsequently used to transform E. coli DH5α. The correctness of the hbpSv gene and its in-frame fusion with the His-tag codons were confirmed by sequencing the resulting plasmid, pETHbpSv.
Site-directed Mutagenesis
A single PCR reaction was used to replace the codon encoding Lys-161 with a histidine in the hbpSv gene on the plasmid pETHbpSv. The oligonucleotide PforVSal (5′-CGTCGTCGACCGCAACGGCAACACGATC-3′ (containing a SalI restriction site, underlined)) was used as a forward primer. The reverse primer PRevVKH (5′-GCGCAAGCTTCTAGTGGCCGAGGGAGGCGACG (containing a HindIII restriction site (underlined) and the reverse complement codon for His, in italics)) was used to generate HbpSv-K161H. After PCRs, the amplicons were restricted with SalI and HindIII and then ligated with the longer SalI-HindIII fragment of pETHbpSv. The ligation products were used to transform E. coli DH5α. Each of the plasmid constructs was then analyzed with restriction enzymes and sequencing. The resulting correct plasmid was named pETHbpSv-K161H. To overproduce the corresponding proteins, this plasmid as well as pETHbpSv, pETHbpS, pETHbpS-H28A, pETHbpS-H51A, pETHbpS-H156A, pETHbpS-T113A, and pETHbpS-T113H were used to transform E. coli BL21 (DE3)pLysS as reported by Zou et al. (32).
Production and Purification of Holo and Apoproteins
Protein was produced and purified as described by Zou et al. (32) with slight modifications. The synthesis of the His-tag fusion proteins in the respective E. coli BL21 (DE3) pLysS transformant was induced at A600 of 0.5 by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside to the culture medium. Cells were grown for 4 h at 37 °C, harvested, washed with a chilled solution W (100 mm Tris/HCl, 150 mm NaCl, pH 8.0), and disrupted by ultrasonication (Branson sonifier, 5 × 10 s, with 10-s intervals) in the presence of 1 μg/ml DNase I. Cell debris were centrifuged at 30,000 × g at 4 °C. The supernatant containing soluble proteins was subsequently used for protein purification.
To isolate the holoprotein, the supernatant was incubated with 0.5 mg/ml concentrations of each cobalamin compound either in the dark or at ambient light for 2 h at room temperature, and then Ni2+-NTA-agarose beads were added to the solution. His-tag proteins were eluted by adding 250 mm imidazole in solution W. To analyze the protein eluates by UV-visible spectroscopy, imidazole was removed by dialysis using solution W in the presence of 5 mm tris(2-carboxyethyl)phosphine. In parallel, a His-tag tobacco etch virus protease was isolated by Ni2+-NTA affinity chromatography (32) and used to cleave the His tag from the fusion protein. Further Ni2+-NTA affinity and anion exchange chromatography on a DEAE-Sepharose column were used to obtain the pure holoprotein. The apoprotein was obtained in a similar way but without the initial incubation with cobalamin. The homogeneity of the His-tag free proteins was analyzed by SDS- and native PAGE as well as by mass spectrometry. Protein concentration was calculated using the Bradford method (35). To analyze the interaction of the apoprotein with the cobalamin, HbpS apoprotein (2–20 μm) was incubated with 10 μm cobalamin in 20 mm Tris/HCl, pH 7.0, for 2 h at 25 °C.
Cobalamin Binding Assays
Cbl binding was monitored by absorption spectroscopy in the range of 250–700 nm using dual-beam Specord 205 UV-visible (Analytik Jena) or Varian Cary 50 (Varian) spectrophotometers. Experiments were performed in triplicate.
Kinetics of Cobalamin Binding
The binding kinetics were measured at a fixed concentration of H2OCbl+ (19 μm) mixed with varying concentrations of HbpS (15–280 μm) in 20 mm Tris, pH 7.5, at room temperature. The optical changes after mixing were monitored by measuring a difference of absorbance at 358 and 352 nm. The recorded curves were used to calculate the rate constants of ligand binding and dissociation. The obtained fitting parameters are presented as the best estimate ± S.E.
Analysis of Cbl-binding Proteins by Native PAGE
Proteins solutions were loaded on to native PAA gels (10%). After electrophoresis the gel was immediately scanned and subsequently incubated with the protein-staining solution PageBlue (Thermo Scientific).
Heme Binding Assays
Hemin (Fe3+ form of heme) at fixed concentration (5 μm) was incubated with increasing concentrations of the apoprotein (0–15 μm at 0.2 μm increments up to 5 μm and in 1 μm increments from 5 to 15 μm) in 20 mm Tris/HCl, pH 7.5, at 30 °C for 2 h. The absence of heme in the apoprotein was confirmed by UV-visible spectroscopy. Hemin was dissolved in 100 mm NaOH, and its concentration was determined using ϵ385 = 58.4 mm−1cm−1 (36). Fresh dilutions were always made using 10 mm NaOH. Heme binding was monitored spectrophotometrically, and experiments were performed in triplicate. Measurements were performed using a reference cuvette containing 5 μm hemin. Kd was calculated using Equation 1 based on the difference spectrum at 411 nm as HbpS apoprotein was added to hemin.
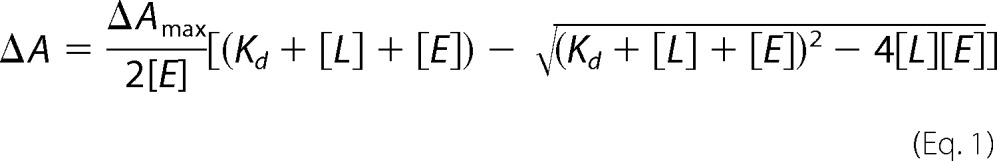 |
Here ΔA is the observed change in absorbance, ΔAmax is the maximum of absorbance, [E] is the concentration of HbpS, and [L] the concentration of hemin.
Dissociation of Heme from HbpS
The rate constants of heme dissociation from heme-binding proteins can be determined using apomyoglobin as a heme scavenger (37, 38). Time-dependent heme transfer was followed by UV-visible spectroscopy. HbpS holoproteins were obtained by incubation of the apoprotein (5 μm) with heme (20 μm) in 20 mm Tris/HCl, pH 7.5, at 25 °C for 4 h. Free heme was removed using CentriPure MINI Spin Desalt Z-50 columns (purchased from SERVA) with the separation performed twice. 4 μm holoprotein samples were then incubated with apomyoglobin (4 μm) for 20 min at 25 °C. Absorbance at 408 nm was recorded at intervals of 5 s. The dissociation rate constant (k−) was calculated by fitting the change in absorbance at 408 nm to a single exponential decay (38) using the GraphPad Prism software.
Sequence Searches, Alignments, and Tree Construction
Homologues of HbpS were collected with a simple blast search in the non-redundant sequence database (Oct 2013) up to a maximum e-value of 10−6 (39). The full sequences were retrieved from the same database and realigned with MAFFT (40) using the most accurate mode and up to 100 iterations. A maximum likelihood tree was built with RAxML (41) using the gamma model for rate heterogeneity, the alpha parameter empirically derived from the data, the BLOSUM62 matrix for substitutions, and 1000 bootstraps. The most likely tree (not a consensus) was drawn with dendroscope (42). Sequence alignments from selected HbpS-like proteins were generated using Clustal Omega (43).
RESULTS
HbpS as a Putative Cobalamin-binding Protein
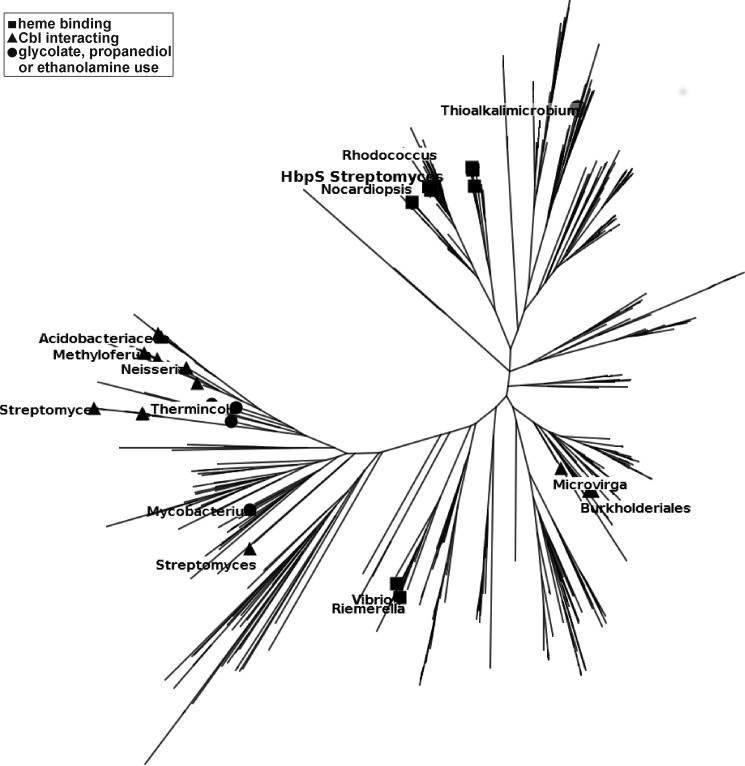
Fig. 2 shows a maximum likelihood tree for 619 close sequence homologues of HbpS. The set only includes sequences related to HbpS with an e-value ≤10−6, and even the most distant protein has a >35% sequence identity with HbpS. There are no large gaps in the alignments, and there is no evidence of saturation, so it is likely that there are no gross errors. At the same time we know that the set of sequences does not reflect nature. It is just the sequences in the data bank that happen to be most closely related to HbpS. One-third of the sequences can be accounted for by four genera with 11% of the sequences from Streptomyces and 9% from Thioalkalivibrio. 6% come from each of Acinetobacter and Pseudomonas. Only a few branches have been labeled with their species for orientation in the diagram.
FIGURE 2.
Unrooted maximum likelihood tree for proteins related to HbpS. ■, sequences annotated as heme binding; ▴, sequences labeled as cobalamin-interacting; ●, proteins annotated as involved in glycolate, propanediol, or ethanolamine use.
This is clearly a gene or protein tree and not a species tree. The species S. reticuli may not be a close relative of Rhodococcus, but their HbpS-like proteins are very similar. There are examples of other Streptomyces species on the left of the tree whose proteins have been labeled as involved in cobalamin binding and that are closer to Neisseria and Thermincola potens than to S. reticuli. It is also interesting that closely related HbpS-like proteins span a range of bacteria from Gram-positive Saccharopolyspora and Rhodococcus to Gram-negative types such as Thiomonas and Riemerella.
The real interest does not lie in species phylogeny but in the functional annotations. Unfortunately, most sequences come from genome sequencing and have no annotation. The few with a clear function have been marked with squares (heme binding), triangles (cobalamin interacting), and circles (involved in glycolate, propanediol, or ethanolamine use) according to keyword matching (Fig. 2). Heme-binding proteins might be closely related to those labeled as Cbl-interacting, but proteins involved in glycolate/propanediol or ethanolamine chemistry are also closely related. This is the crux of this part of the work. On available data, one could not reasonably say there is any functional partitioning over the tree. It could be that heme binders bind cobalamins or vice versa and there is some gradual variation and overlap of function. This calculation leads to the clear question. Does HbpS, a protein known in the literature as a heme binder, bind a cobalamin?
HbpS Binds Aquo-cobalamin
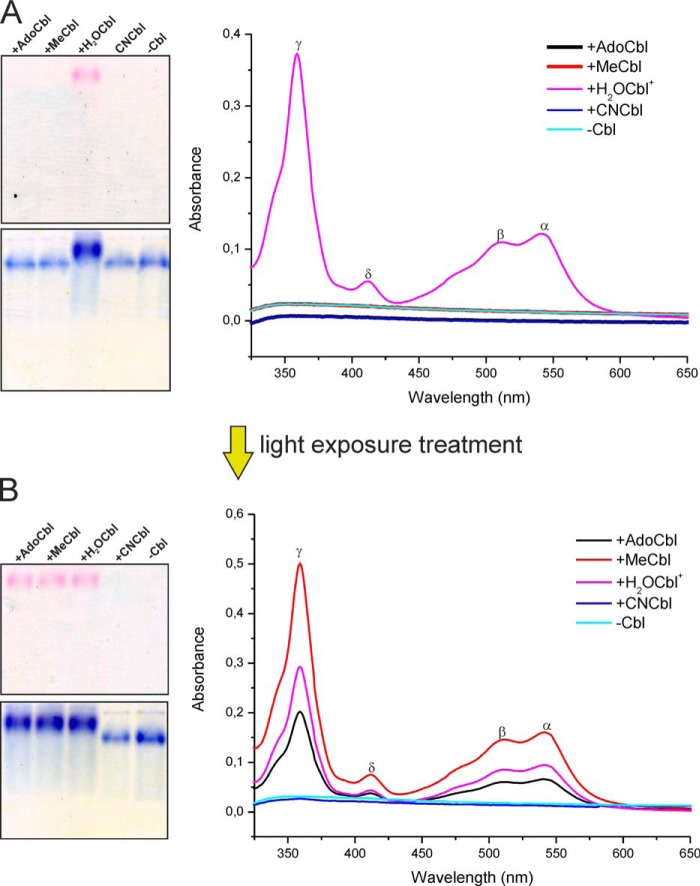
To check for cobalamin binding, the protein extracts containing recombinant HbpS were mixed with an excess of different cobalamins (AdoCbl, CNCbl, MeCbl, and H2OCbl+) in the dark at room temperature for 2 h. HbpS was then isolated by Ni2+-NTA affinity chromatography as described under experimental procedures. The protein eluate obtained after the incubation with H2OCbl+ (aquo-cobalamin) was pink-colored, indicating the presence of Cbl (not shown). Other protein-ligand combinations gave colorless products, suggesting an absence of Cbl. Aliquots of protein eluates containing His-tagged and imidazole-free HbpS were loaded onto a native PAA gel (Fig. 3A, left) and analyzed by UV-visible spectroscopy (Fig. 3A, right). Of the four cobalamins tested, only H2OCbl+ bound to HbpS, because the latter migrated as a pink band (Fig. 3A, left, top). Staining of the proteins with PageBlue on the same gel showed that the lane with HbpS and H2OCbl+ migrated slowest, consistent with a higher molecular weight of the octameric protein-ligand complex (Fig. 3A, left, bottom), presumably saturated with eight molecules of Cbl. UV-visible spectroscopy showed a distinctive cobalamin spectrum only for the HbpS preincubated with H2OCbl+ (Fig. 3A, right). Interestingly, the absorbance peaks of the HbpS-bound cobalamin were red-shifted compared with those of free H2OCbl+ under the same experimental conditions (black spectrum on Fig. 4B, left). This observation suggests substitution of the original Co3+-coordinated water by another ligand, which was confirmed by the results discussed below.
FIGURE 3.
HbpS interactions with H2OCbl+ on native gel electrophoresis and UV-visible spectroscopy in the dark (A) and after exposure to ambient light (B). Gels (containing 10 μg of each protein) are shown immediately after running (top) and after PageBlue staining (bottom). Labels show the different cobalamin compounds and the absorbance maxima of the HbpS-H2OCbl+ complexes (γ = 358 nm, δ = 420 nm, β = 514 nm, and α = 539 nm).
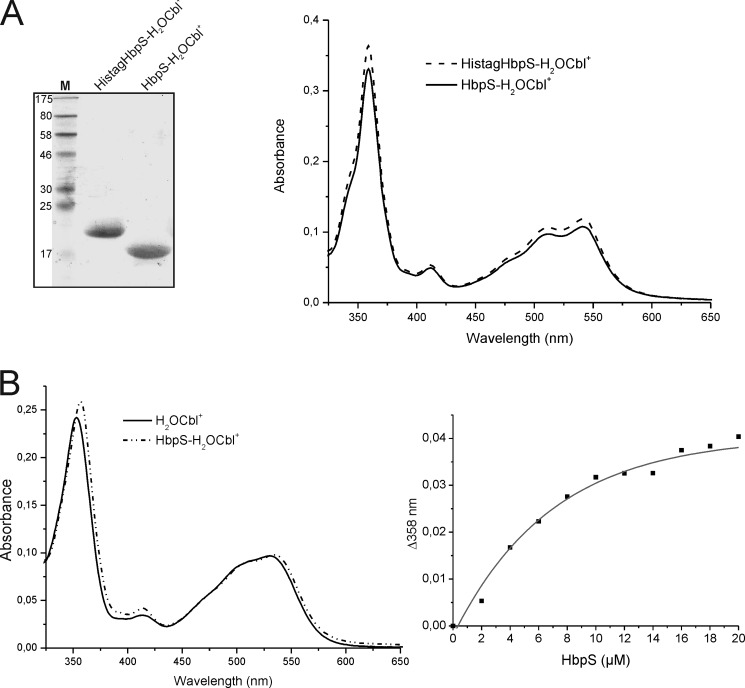
FIGURE 4.
Comparison of Cbl binding between His-tagged and His tag-free HbpS proteins and titration experiments. A, isolated His-tagged HbpS with bound H2OCbl+ (HistagHbpS-H2OCbl+) were treated with a tobacco etch virus protease. After subsequent chromatography, His tag-free HbpS with bound H2OCbl+ was obtained (HbpS-H2OCbl+). 10 μg of each protein was analyzed either SDS-PAGE (left) or UV-visible spectroscopy (right). The molecular mass (in kDa) of protein markers (lane M) is indicated (left). B, binding of H2OCbl+ by the HbpS apoprotein was monitored. Aquo-cobalamin (10 μm) was incubated with increasing concentrations (2–20 μm, with 2 μm increments) of the HbpS apoprotein for 2 h at 25 °C. UV-visible spectra of H2OCbl+ alone (black spectrum) and bound to 14 μm HbpS (dot-dashed spectrum) are shown (left). The difference absorbance at 358 nm (Δ358) was plotted against HbpS concentrations (right).
To better characterize interactions with different Cbl derivatives, an additional experiment was conducted. Samples of the same protein extract were incubated with different cobalamins (AdoCbl, MeCbl, CNCbl, and H2OCbl+) but now under exposure to ambient laboratory light. The protein fractions were separated from free cobalamins by Ni2+-NTA affinity chromatography. This gave pink protein eluates for the samples incubated with AdoCbl, MeCbl, and H2OCbl+ but not for CNCbl (not shown). Native PAGE (Fig. 3B, left) and UV-visible spectroscopy (Fig. 3B, right) confirmed that the colored protein eluates contained the bound Cbl. The original ligands MeCbl and AdoCbl (incubated in the presence of HbpS at ambient light) showed the same spectral patterns as for H2OCbl+ bound to HbpS (γ = 358 nm, δ = 420 nm, β = 514 nm, and α = 539 nm) (Fig. 3, A and B, right). Such a result is consistent with the expected photolysis of these cofactors in an oxygenated solution yielding H2OCbl+ (1). All spectra resembled the one observed upon formation of the complex between H2OCbl+ and transcobalamin in which Co3+-coordinated water is substituted by a His residue of the binding protein (24, 44). We can conclude that HbpS interacts exclusively with H2OCbl+ whether it is added directly or formed upon illumination of MeCbl and AdoCbl samples. This explains why HbpS did not bind either MeCbl or AdoCbl in the dark. The Co3+ ion of MeCbl and AdoCbl was protected from coordination with the external ligands by the respective β-groups, tightly associated with the metal ion (2, 16). The same is true for CNCbl, which has a higher photostability of its carbon-cobalt bond.
It seems rather improbable that H2OCbl+ binds to the His tag of the recombinant HbpS protein because this interaction would have hampered protein purification on the Ni2+-NTA column. This was checked experimentally. We removed the His tag using a tobacco etch virus protease and isolated HbpS in its native form (Fig. 4A, left) by adsorption of the His tag on a Ni2+-NTA affinity column followed by gel filtration and anion exchange chromatography. The His tag-free HbpS solution (in 20 mm Tris/HCl, pH 7.0) retained its pink color after purification and displayed the UV-visible spectrum typical of the HbpS-Cbl complex (Fig. 4A, right).
The interaction of the HbpS apoprotein with H2OCbl+ was also monitored in a binding experiment, in which H2OCbl+ (10 μm; black spectrum on Fig. 4B, left) was mixed with increasing concentrations (2–20 μm) of the apoprotein and UV-visible spectra of samples were recorded after 2 h of incubation. The presence of the apoprotein caused a shift of the H2OCbl+ absorbance maxima. For example, the main γ peak shifted from 353 to 358 nm (Fig. 4B, left), indicating formation of the HbpS-Cbl complex in which water is substituted by a ligand with higher electron-donating properties (1). Moreover, the absorbance at 358 nm increased with increasing concentrations of the protein (Fig. 4B, right).
Identification of the Interacting Ligand
H2OCbl+ has a tendency to bind electron-donating ligands such as CN−, SO32−, N3−, NO2−, imidazole, and other N-heterocycles (1). Coordination of a His residue can be reversed by adding another ligand with higher affinity for the β-site of Cbl, e.g. CN− or N3− (44). To check for a similar interaction (histidine-Cbl) in HbpS, we used a competition experiment with potassium cyanide (KCN). The CN− ion forms a very strong coordination bond with Cbl (1) and can displace the Cbl-interacting histidine (44). This reaction is expected to give CNCbl that will dissociate from HbpS unless other binding mechanisms are involved. Two parallel samples of the HbpS apoprotein (each 20 μm) were incubated with H2OCbl+ (80 μm) for 2 h. 1 mm KCN was then added to one sample, and incubation was continued for 16 h. Proteins were subsequently subjected to either native PAGE or gel filtration chromatography.
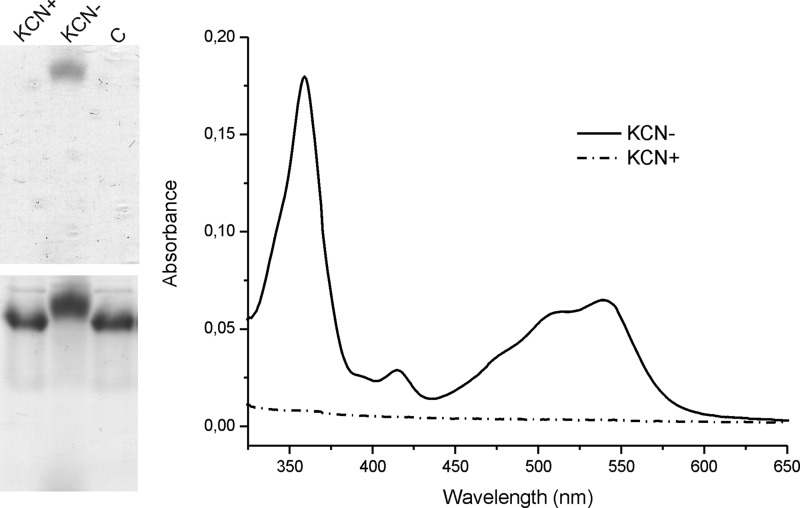
After native PAGE, a pink protein band (seen as black protein) was observed only in the sample without KCN treatment (Fig. 5, left, top). Protein staining by PageBlue also showed that the untreated sample migrated faster on the native gel than the KCN-treated sample. This suggests that KCN disrupts the interaction of HbpS with Cbl (Fig. 5, left, bottom). The apoprotein was also loaded onto the native gel and migrated in the same way as the HbpS-Cbl sample treated with KCN (Fig. 5, left, bottom, lane C). These observations were corroborated by UV-visible spectroscopy. Before measurements, the samples of HbpS + H2OCbl+ with/without KCN were subjected to gel filtration to remove free low molecular weight ligands. The absorbance spectra showed no Cbl in the KCN-treated protein fraction (Fig. 5, right). This suggests that an amino acid residue in HbpS (most likely His) coordinates to the cobalt ion of Cbl if it is not protected by the strongly associated β-ligands (e.g. Ado-, Me-, or CN-group). No other HbpS-Cbl interactions (insensitive to the presence of CN−) were detected.
FIGURE 5.
Disruption of HbpS with H2OCbl+ interactions by potassium cyanide. Two parallel samples of the HbpS apoprotein (20 μm) were incubated with H2OCbl+ (80 mm) for 2 h at 25 °C. KCN (1 mm) was added to one sample, and incubation was continued overnight. The mixtures (containing 10 μg of each protein) were either loaded onto a native PAA gel (left) or analyzed by UV-visible spectroscopy after gel filtration (right). The native gel was scanned after electrophoresis (left, top) and subsequently stained with PageBlue (left, bottom). Treatment of samples with (KCN+) or without (KCN−) potassium cyanide is indicated. The apoprotein HbpS without a previous incubation with H2OCbl+ was used as a control (left, lane C).
Binding Kinetics
The spectral shift of the major γ-peak of cobalamin was used to follow HbpS and H2OCbl+ interactions. We used a constant concentration of H2OCbl+ (19 μm) with varying concentrations of HbpS (15–280 μm) to maintain the same scale of absorbance at 352 and 358 nm associated with Cbl. With comparable concentrations of the two reactants (HbpS and H2OCbl+) the time equation of the binding reaction A + B ↔ AB can be expressed as follows (45).
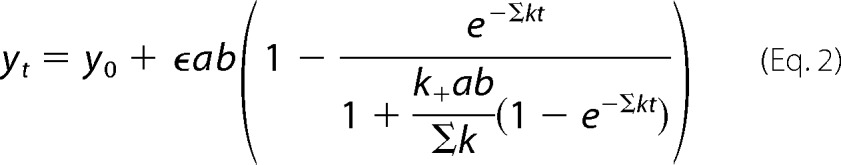 |
where
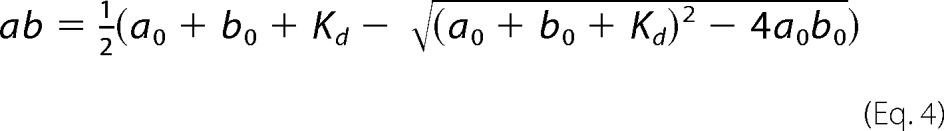 |
Here yt is the measured absorbance at time t, y0 is the initial absorbance at zero time, ϵ is the molar amplitude of response upon formation of AB (ϵ units of μm−1cm−1), ab is the equilibrium concentration of the complex AB, a0 and b0 are the initial concentrations of the two reactants A and B (e.g. Cbl and HbpS); k+ and k− are the rate constants of binding and dissociation, and Kd is the dissociation constant of the complex AB (Kd = k−/k+). The rate constants (k+ and k−) and the initial absorbance y0 were the fitting parameters, given known values for a0, b0, and ϵ.
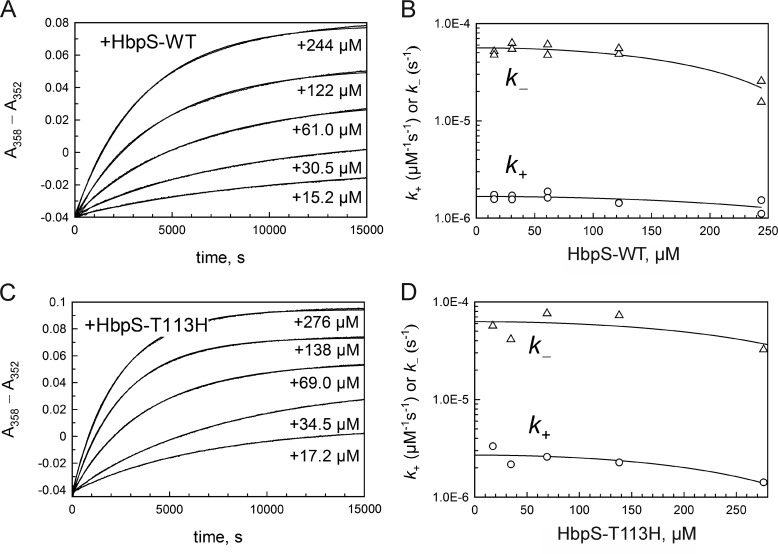
Fig. 6A shows the time-dependent kinetics of interaction between H2OCbl+ and HbpS (wild type (WT)) fit to Equation 2. The maximal amplitude of optical changes ΔY at an infinite concentration of HbpS monomers was estimated by extrapolation (0.122 and 0.125 in two different experiments). This ΔY allowed the estimation of ϵ = ΔY/a0 (where a0 is the fixed concentration of H2OCbl+). The value of ϵ was substituted into Equation 2. The calculated values of k+ and k− decreased at high protein concentrations (Fig. 6B). More or less proportional decrease of both rate constants was apparently caused by high viscosity of the medium (protein concentration up to 4 mg/ml) and/or weak unspecific protein-protein interactions shielding the Cbl binding site. In this case, both attachment and detachment of the ligand are slower. The predicted values of rate constants in a relatively diluted protein solution are k+ = 1.67 ± 0.13 m−1s−1 and k− = (5.62 ± 0.52) × 10−5 s−1 as judged from an empirical polynomial fitting equation. Their ratio gives Kd = 34 ± 4.2 μm.
FIGURE 6.
Kinetics of interaction between HbpS (wild type and mutant T113H) and H2OCbl+. A, binding reaction of HbpS-WT. The reactants H2OCbl+ (a0 = 19.0 μm) and HbpS (monomer concentrations of b0 = 15 - 244 μm) were mixed, and absorbance changes (A358 − A352) were traced over time. Curves were approximated by Equation 2 to calculate k+ and k− (ϵ = 0.00658 μm−1cm−1). B, rate constants versus protein concentration. The values of k+ and k− from panel A (and the data of a parallel experiment, not shown) were plotted versus the concentration of HbpS-WT (monomers). The charts were approximated by the empirical functions k+ = 1.67·10−6 − 6.32·10−12x2 and k− = 5.62·10−5 − 5.78·10−10x2. The first parameter of each function predicts the rate constant at a low protein concentration: k+ = 1.67 ± 0.14 m−1s−1, and k− = (5.62 ± 0.52)·10−5 s−1. C, binding reaction of HbpS-T113H. The reactants H2OCbl+ (a0 = 19.0 μm) and HbpS (b0 = 17 - 276 μm) were mixed. and the time course of absorbance (A358 − A352) was recorded. Curves were approximated by Equation 2 to calculate k+ and k− (ϵ = 0.00779 μm−1cm−1). D, rate constants versus protein concentration. The values of k+ and k− from panel C were plotted versus the concentration of HbpS-T113H mutant (monomers). The charts were approximated by the empirical functions k+ = 2.77·10−6 − 1.72·10−11x2 and k− = 6.27·10−5 − 3.34·10−10x2. The first parameter of each function predicts the rate constant at a low protein concentration: k+ = 2.77 ± 0.61 m−1s−1, and k− = (6.27 ± 0.23)·10−5 s−1.
HbpS Uses His-156 to Interact with the Cobalt Ion of Cbl
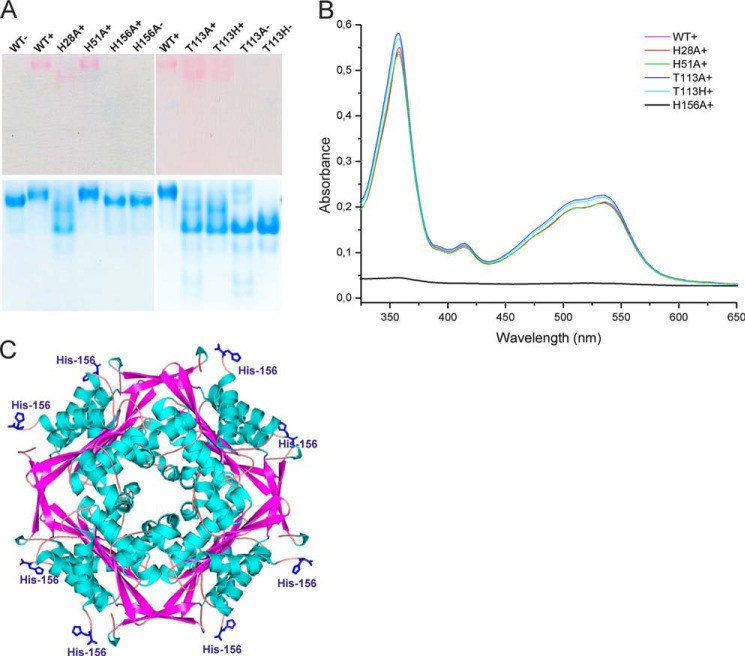
HbpS contains three histidine residues (His-28, His-51, and His-156) per chain. His-28 is important for the stability of the HbpS octamer, but the other two His residues are not (28). His to Ala mutants (27, 28) were used to prepare the three corresponding His tag-free proteins for comparison with the wild type protein. Each protein (20 μm) was incubated with H2OCbl+ (80 μm) and analyzed by native PAGE. Only the HbpS-H156A mutant did not migrate as a pink protein band (Fig. 7A, lane H156+), indicating the absence of bound Cbl in this sample. PageBlue staining of the proteins on the same native PAA gel showed that the migration behavior of this mutant is identical to the wild type (Fig. 7A, lane WT−) and the mutant HbpS-H156A (Fig. 7A, lane H156−) not exposed to H2OCbl+. In parallel, unbound H2OCbl+ was separated from the protein solutions by gel filtration, and then identical concentrations of proteins were analyzed by UV-visible spectroscopy. In contrast to the HbpS-H156A sample, the wild type as well as HbpS-H28A and HbpS-H51A samples displayed the characteristic protein-Cbl spectrum (Fig. 7B). Clearly His-156 is essential for Cbl binding. Noteworthy, His-156 is exposed on the surface of the HbpS octamer (Fig. 7C).
FIGURE 7.
HbpS uses His-156 to bind H2OCbl+. 20 μm concentrations of either HbpS wild type apoprotein or its mutant versions were incubated with 80 μm H2OCbl+ (WT+, H28A+, H51A+, H156A+, T113A+, and T113H+). As a control, four samples were incubated in buffer lacking H2OCbl+ (WT−, H156−, T113A−, and T113H−). The mixtures (containing 10 μg of each protein) were either loaded onto a native PAA gel (A) or analyzed by UV-visible spectroscopy after gel filtration (B). The native gel was scanned after electrophoresis (A, top) and subsequently stained with PageBlue (A, bottom). C, the exposed His-156 (in blue) on the surface of the HbpS octamer (PDB code 3FPV) is shown.
Binding of Aquo-cobalamin to a HbpS-like Protein
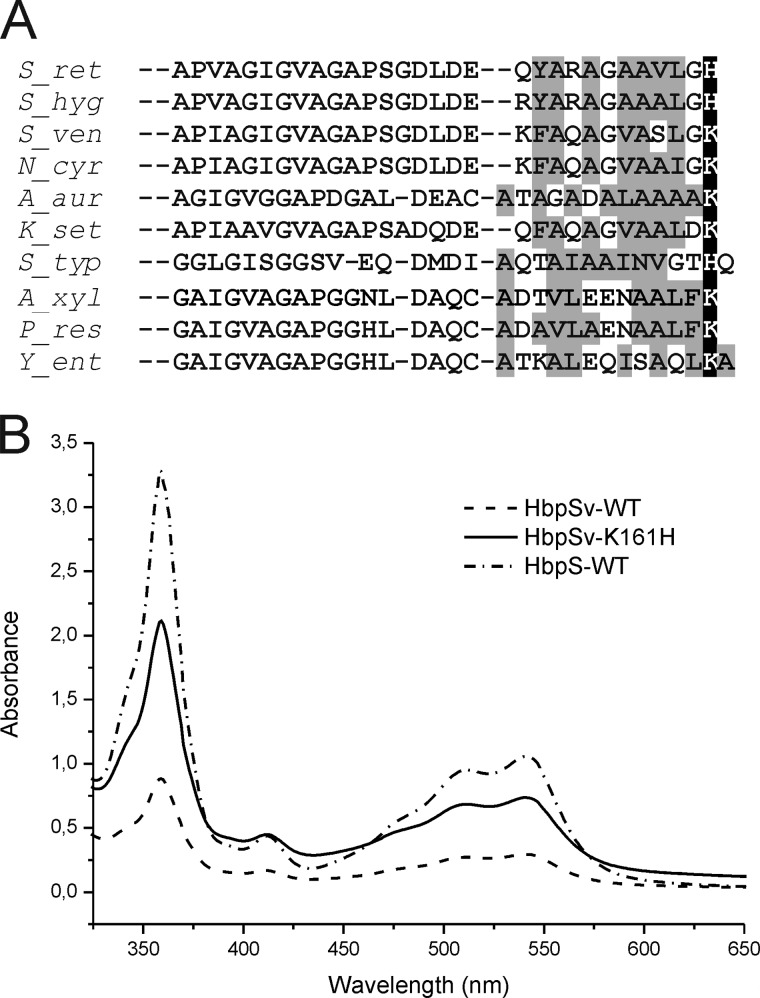
The C-terminal regions of several HbpS-like proteins display a marked predominance of hydrophobic residues, in particular in positions 10 to 12 (Fig. 8A). Together with His-156 in HbpS, these hydrophobic residues could conceivable contribute to aquo-Cbl binding. Interestingly, many homologues have a lysine at the analogous C-terminal position (Fig. 8A). To see if these proteins lose their Cbl binding properties, an HbpS-like protein with a C-terminal lysine (residue 161) was cloned from S. venezuelae and is referred to as HbpSv. For comparison, an HbpSv mutant with histidine at the C terminus (HbpSv-K161H) was also prepared after cloning from E. coli transformants. Protein extracts containing either HbpSv-WT or HbpSv-K161H or HbpS-WT were incubated with H2OCbl+. After Ni2+-NTA chromatography and dialysis, the protein eluates were analyzed by UV-visible spectroscopy. All three proteins clearly showed H2OCbl+ binding, which was strongest with HbpS-WT and weakest with HbpSv-WT (Fig. 8B). Apparently, a lysine can substitute for a histidine to some extent. This is not surprising, because terminal amino groups can weakly interact with H2OCbl+, which was used for preparation of affinity materials with bound Cbl (46, 47). Replacing Lys-161 by His in HbpSv noticeably increased the binding (Fig. 8B).
FIGURE 8.
Alignment of C-terminal amino acids of HbpS-like proteins and cobalamin binding by HbpSv. A, HbpS-like proteins from Streptomyces hygroscopicus (S_hyg; GI: 451797635), S. venezuelae (S_ven; GI: 408681679), Nocardia asteroides (N_ast; GI: 517878279), Arthrobacter aurescens TC1 (A_aur; GI: 119961831), Kitasatospora setae (K_set; GI: 357389326), S. typhimurium (S_typ; GI: 5069458), Achromobacter xylosoxidans (A_xyl; GI: 566051808), Pseudomonas resinovorans (P_res; GI: 512376536), and Yersinia enterocolitica (Y_ent; GI: 595644304) were compared with HbpS from S. reticuli (S_ret; GI:5834772). The S. typhimurium sequence is the C-terminal part of the PduO (PduOC) protein. In contrast to PduOC all listed HbpS-like proteins show >35% amino acid identity to HbpS. PduOC was included as many of the HbpS-like proteins are annotated as PduO-like proteins. Hydrophobic amino acids are marked with a gray background. His-156 in HbpS as well as His and Lys at the corresponding position in the other proteins are marked with a black background and written in white. Sequences were aligned with Clustal Omega. B, protein extracts containing either HbpSv wild type (HbpSv-WT) or HbpSv with substituted Lys-161 by His (HbpSv-K161H) or HbpS wild type (HbpS-WT) were incubated with H2OCbl+. HbpS and HbpSv proteins were then isolated and subsequently analyzed by UV-visible spectroscopy.
Heme Affinity of HbpS
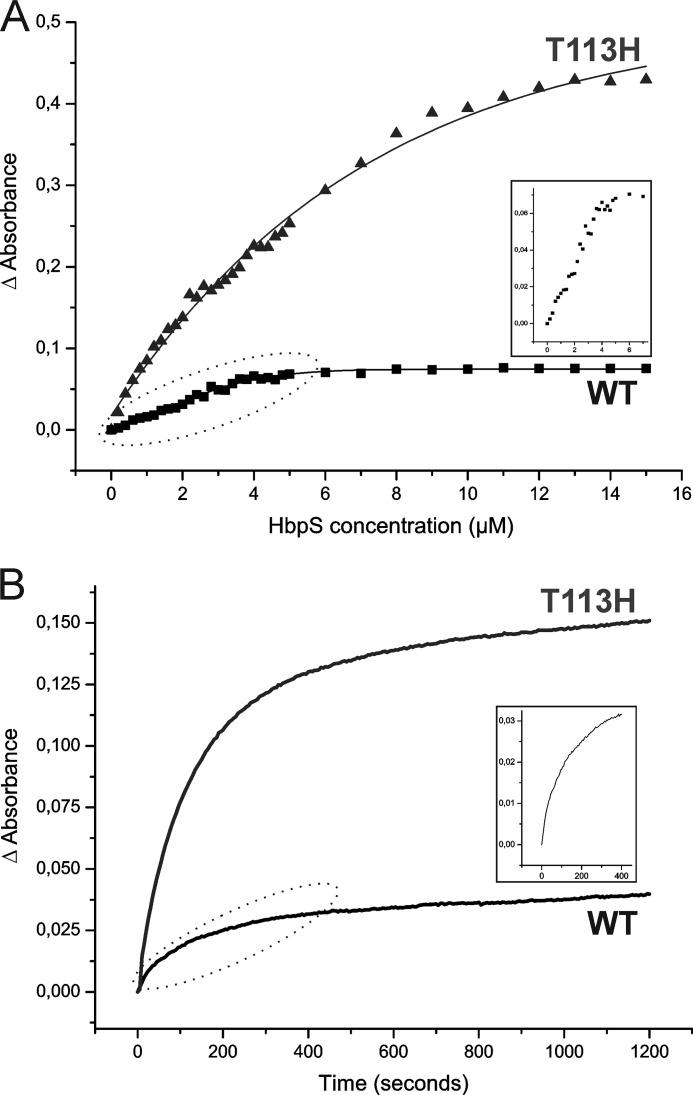
This work focuses on cobalamin binding, but HbpS has been regarded as a heme binder, and it is impossible to avoid some comparisons. We, therefore, first analyzed heme binding kinetics of HbpS. Titration measurements (Fig. 9A) using a fixed concentration of hemin (5 μm) and an increasing concentration of HbpS protein (0–15 μm at 0.2 μm increments up to 5 μm and in 1 μm increments from 5 to 15 μm) led to a calculated Kd of 1.0 ± 0.3 μm for wild type HbpS. The titration curve in Fig. 9A (WT) indicates that the binding of the wild type HbpS to hemin is nearly stoichiometric; thus, the calculated Kd is the maximal level for HbpS-heme binding and should be considered as ≤1 μm. We also used HbpS-T113H mutant for comparison as it was previously shown that Thr-113 is involved in heme binding. Mutagenesis of Thr-113 to alanine abolishes heme binding, but mutation to a histidine yields a protein that has an apparently higher heme binding activity than the wild type (26). The calculated Kd for this mutant is 1.1 ± 0.3 μm.
FIGURE 9.
Titration assays and heme transfer to apomyoglobin. A, increasing concentrations (0–15 μm at 0.2 μm increments up to 5 μm and in 1 μm increments from 5 to 15 μm) of either the wild type or the T113H mutant protein were incubated with a fixed concentration of hemin (5 μm) at 30 °C for 2 h in 20 mm Tris/HCl, pH 7.5, and subjected to UV-visible spectroscopy. Measurements were performed using a reference cuvette containing 5 μm hemin. The plot shows the difference absorbance (Δ Absorbance) at 411 nm versus protein concentration. The inset shows Δ Absorbance values in the wild type sample before a saturation was achieved (dotted ellipse) in an enlarged scale. Kd was calculated using the Equation 1 in “Experimental Procedures.” B, the time course of heme transfer from holo-HbpS (4 μm) to apomyoglobin (4 μm) was measured for 20 min at 5-s intervals. The inset shows the heme transfer in the wild type sample during the first 6 min (dotted ellipse) in an enlarged scale. The dissociation rate (k−) was calculated by fitting the change in absorbance at 408 nm to a single exponential decay. k− was calculated using the GraphPad Prism software.
We also measured the dissociation rate constant using apomyoglobin as a heme scavenger. The heme transfer from holo-HbpS to apomyoglobin was followed by UV-visible spectroscopy, and the time course was fit to a single exponential (Fig. 9B). For the wild type and HbpS-T113H, we found k− of (4.0 ± 0.08) × 10−3 s−1 and (5.6 ± 0.08) × 10−3 s−1, respectively. Using the equilibrium constants Kd given above, one can calculate an associate rate constant k+ = (4.0 ± 0.38) × 103 m−1 s−1 for the wild type and (5.1 ± 0.38) × 103 m−1 s−1 for T113H. The differences in binding kinetics between both proteins should not be over-interpreted. HbpS-T113H has the same migration behavior on a native gel as the HbpS-H28A (Fig. 7A) mutant, which has been shown to be monomeric in solution (28). We also know that in the crystal structure Thr-113 has zero solvent accessibility. Mutating Thr-113 changes the monomer/octamer equilibrium, which must have an effect in accessibility of heme binding sites and consequently on the binding kinetics.
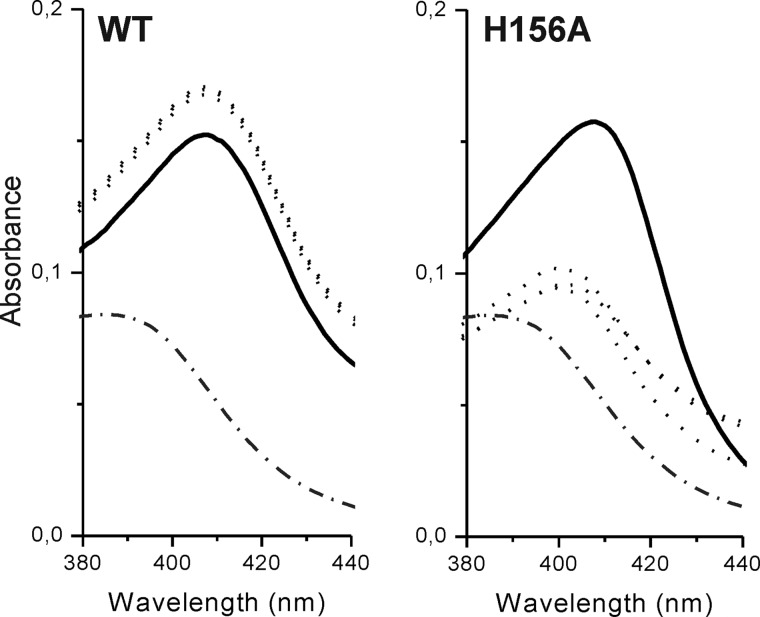
Given that HbpS-H156A lacks Cbl binding activity, we checked whether its heme binding activity is also lost. Our previous report showed that this mutant apparently binds heme as strongly as the wild type protein. In that work heme binding was measured after 16 h of incubation (26). In this work binding was measured after 1, 2, 4, and 16 h. The spectrum of free hemin with an absorbance maximum at 385 nm was used as a reference (Fig. 10, both panels; dot-dashed line). The wild type protein shows a comparable heme binding after just 1 h (Fig. 10, left; dotted spectra). The intensity of the Soret peak (411 nm) slightly decreased after 16 h of incubation (Fig. 10, left; black spectrum). In contrast, the HbpS-H156A shows a little heme binding after 1, 2, or 4 h (Fig. 10, right; dotted spectra) as the absorbance maximum of hemin in each sample was shifted from 385 nm to 399 nm. This relatively short shift likely resulted from a nonspecific binding to the protein. After 16 h the absorbance maximum was shifted to 411 nm (Fig. 10, right; black spectrum), with a comparable intensity as recorded for the wild type. The observed late heme binding by HbpS-H156A as well as by the wild type is probably due to nonspecific binding to the protein, which is likely partially denatured. The spectrum may also include signals from hemin dimers, that can form in aqueous solutions (48, 49). These data indicate that His-156 is involved in heme binding.
FIGURE 10.
Heme binding by HbpS-H156A. 20 μm HbpS proteins (either wild type or H156A mutant) were incubated with 10 μm hemin for either 1 or 2 or 4 or 16 h in 20 mm Tris/HCl, pH 7.5, at 30 °C. In parallel, a control sample containing only 10 μm hemin was prepared. Heme binding was analyzed by UV-visible spectroscopy using a reference cuvette containing the control sample. Spectra in the region between 380 and 440 nm are shown. Spectra recorded after 1, 2, or 4 h of incubation are shown as dotted lines and the spectrum after 16 h is shown as a solid line. The spectrum of free hemin (2 μm) is shown as a dot-dashed line.
We also tested whether the Cbl binding activity of the Thr-113 mutants is affected. After incubation of the mutant proteins (T113H and T113A) with aquo-cobalamin followed by native PAGE (Fig. 7A) and UV-visible spectroscopy (Fig. 7B), almost identical Cbl binding activity was observed compared with the wild type HbpS. Additionally, a control experiment was conducted to calculate Cbl binding kinetics of HbpS-T113H as described for the wild type protein. Fig. 6, C and D, shows the spectra for HbpS-T113H. The reaction was characterized by a somewhat higher maximal optical response (ΔY = 0.148) and a higher binding rate constant k+ = 2.77 ± 0.61 m−1s−1. On the other hand, the dissociation constant did not significantly change k− = (6.27 ± 0.23) × 10−5 s−1. This provided a little better affinity for H2OCbl+ equal to Kd = 23 ± 5 μm. The data indicate that Thr-113 and His-156 play a role in heme binding, but Thr-113 is not involved in Cbl binding.
DISCUSSION
The S. reticuli protein HbpS clearly binds aquo-cobalamin but not other common cobalamin compounds, including MeCbl, AdoCbl, and CNCbl. Although there is no crystal structure, there is strong evidence as to the type of binding between HbpS and aquo-cobalamin. First, the HbpS-cobalamin spectrum resembles that of transcobalamin/aquo-cobalamin and that of the His-Cbl coordination complex (44). Next, the binding is competitively disrupted by CN− ions, which bind at the β-site of Cbl. Finally, HbpS does not have the Asp-X-His-X-X-Gly motif typical of base-off interactions (20, 21). Taken together, it seems very likely that Cbl binds via the base-on mode, which is typical of Cbl-transporting proteins (23, 24).
From the site-directed mutagenesis, one knows that His-156 is essential for binding aquo-cobalamin, and in the octamer crystal structure His-156 is relatively accessible (Fig. 7C). There are also several hydrophobic residues in this region that may well be involved in binding the large aromatic system of a cobalamin. It is also interesting to compare kinetics in the protein with those of free His in solution. The binding rate constant of HbpS (k+ = 1.76 m−1 s−1) is of the same order of magnitude as k+ = 0.92 m−1 s−1 of free His (44), although ionic strength in the two experiments was somewhat different. The dissociation rates, however, differ much more (6.0 × 10−5 s−1 for HbpS and 2.2 × 10−4 s−1 for free His). If the rate of collisions is the same in both cases, it means that the cobalamin-protein interaction is stabilized by additional contacts.
H2OCbl+ affinity of native HbpS was ∼7-fold higher than free His, and the affinity of the T113H mutant was still higher, accompanied by larger spectral changes. At the same time, these Kd values were still orders of magnitude lower than three Cbl-transporting proteins in humans (around 10−14 m) (50). This means that HbpS is not a remarkably strong cobalamin binder (Kd = 34 μm), but it guarantees some interaction between Cbl and HbpS in soil (the natural environment of streptomycetes) where the concentration of extracellular Cbl was from 0.2 to 10 μm (51). In bacterial cultures, values of 1–200 μm have been recorded (52). Furthermore, the slow speed of Cbl binding to HbpS will have little impact on the potential process of Cbl internalization, as bacterial populations in soils exist over long periods of time. The importance can be illustrated with an example. The mixture of 10 μm Cbl + 10 μm HbpS will give a concentration of the complex of 1.9 μm (19% of Cbl bound). For 5 μm Cbl + 10 μm HbpS it will be 1.04 μm (21% of Cbl bound). This is not strong binding, but if the complex HbpS-Cbl is cleared with a reasonable speed and HbpS concentration is larger than that of Cbl, nearly all extracellular Cbl will be internalized. Dissociation of Cbl from HbpS within a cell does not present a problem because Cbl is reduced to 2+ (or/and 1+) form, whereupon the β ligand immediately dissociates. Unfortunately, there are no data for the extracellular concentration of HbpS, which one would need to assess the importance of the interactions under real conditions. At the moment we would simply state that HbpS is not a strong Cbl-binding protein, but it might have a function in bacteria as a transporter of aquo-cobalamin and aquo-corrinoids.
HpbS-heme binding has a low k+ and a relative high Kd value when compared with other proteins involved in heme transport (Table 1). At the same time, the dissociation rate constant k− (4 × 10−3 s−1) is within the range of some heme transporters such as HasA, Shp, Rv0203, and PhuS (Table 1). From this point of view, HbpS is not a tight heme binder, but it might be involved in the transport of heme. Unfortunately, one does not have dissociation constants for other extracellular heme binders from streptomycetes or other soil bacteria that would be necessary to better assess the likelihood of a transporting role. At the same time, the Kd value of heme binding by HbpS is within the range (∼10−6 m) reported for some heme-sensing proteins such as AppA and PpsR (Table 1). Such a Kd value in heme-sensing proteins might be an indication of a flexible heme-binding pocket that is required during sensing of heme. From structural comparisons (27), one knows that HbpS is similar to the heme binding domains within DosS and DosT from Mycobacterium tuberculosis. These are two-component membrane-bound kinases that are involved in heme sensing (61, 62). Analogously, HbpS acts as an accessory module that regulates the activity of the membrane-bound sensor kinase, SenS, in a heme-dependent manner (63). In this context, the heme binding of HbpS is exactly what one would expect.
TABLE 1.
Kinetic and equilibrium parameters of heme binding to some hemeproteins
ND, not determined.
| Protein | k+ | k− | Kd | Function | Reference |
|---|---|---|---|---|---|
| m−1 s−1 | s−1 | m | |||
| Myoglobin | 7 × 107 | 8.4 × 10−7 | 1.3 × 10−14 | O2 transport | (53) |
| HasA | 1.6 × 107 | 3 × 10−4 | 2 × 10−11 | Heme aqcuisition in Pseudomonas aeruginosa | (54, 55) |
| Shp | 1.6 × 106 | 3 × 10−4 | 2 × 10−10 | Heme transfer in Streptococcus pyogenes | (56) |
| Rv0203 | 1.3 × 108 | 8.2 × 10−2 | 6 × 10−10 | Heme uptake in M. tuberculosis | (57) |
| PhuS | 1.8 × 105 | 3.6 × 10−2 | 2 × 10−7 | Heme aqcuisition in P. aeruginosa | (38) |
| Rbt5 | ND | ND | 5.4 × 10−6 | Heme transport in Candida albicans | (58) |
| AppA | ND | ND | 1.3 × 10−6 | Heme sensing in Rhodobacter sphaeroides | (59) |
| PpsR | ND | ND | 1.9 × 10−6 | Heme sensing in R. sphaeroides | (60) |
| HbpS | 4 × 103 | 4 × 10−3 | ≤1 × 10−6 | Sensor protein from S. reticuli | this work |
| HbpS-T113H | 5 × 103 | 5.6 × 10−3 | 1.1 × 10−6 | Mutant version of HbpS | this work |
Because cobalamin binding is an order of magnitude weaker than heme binding, the equilibrium distributions of protein-ligand complexes will be sensitive to the concentrations of the interacting species (ignoring potential synergetic or antagonistic interactions between the two ligands). There will certainly be circumstances where the concentration of cobalamins in the soil is higher than that of heme. One should also note that heme and various cobalamins are not the only pyrrole-based secondary metabolites produced by streptomycetes. There are also tri-pyrroles such as the antibiotics prodigiosin and undecylprodigiosin, bi-pyrroles such as staurosporine and rebeccamycin, and mono-pyrroles such as clorobiocin (64). Binding to these compounds should be measured because some cross-reactivity seems to be inevitable.
Secondary metabolites have been shown to act as signals that interact with sensory proteins in signaling pathways (65). hbpS as well as different hbpS-like genes are clustered with two-component system genes (30), leading to the assumption that the interaction between an HbpS-like protein with the respective metabolite might trigger a signal cascade. Considering the traffic in pyrrole-based metabolites, HbpS and related proteins could be a part of signaling networks that remain to be explored.
Sequence comparisons (Fig. 2) show that HbpS is on the edge of the family of proteins, sometimes labeled as PduO proteins (66). The N-terminal domain of PduO has an ATP:cob(I)alamin adenosyltransferase activity, whereas the C-terminal part consists of the DUF336 domain. HbpS-like proteins are only annotated as DUF336. The exact role of the C-terminal domain of PduO is currently unknown. Interestingly, the C-terminal domain of PduO from Salmonella typhimurium contains a histidine residue at the same position of the Cbl-coordinating His-156 in HbpS (Fig. 8A). Searches for structural similarities show that the structure of HbpS (PDB code 3FPV) is easily superimposed on the OrfY protein from Klebsiella pneumoniae (PDB code 2A2L) (Fig. 11). OrfY and the C-terminal domain of PduO show 36% amino acid identity. The role of OrfY is currently unknown, but the location on the Klebsiella genome is interesting. The orfY gene lies within the dha regulon that also includes genes for the enzymes glycerol dehydratase, 1,3-propanediol oxidoreductase, glycerol-dehydrogenase, and dihydroxyacetone kinase. These are key enzymes in the anaerobic metabolism of glycerol (67). Glycerol dehydratase as well as the isofunctional enzyme diol dehydratase is an adenosylcobalamin-dependent enzyme. The diol dehydratase gene is located within the pdu operon that encodes among others the adenosyltransferase PduO. The metabolism of glycerol and 1,2-diols is a multistep process also comprising the reduction of aquo-cob(III)alamin to cob(I)alamin. The adenosyl transferase subsequently catalyzes the transfer of ATP to cob(I)alamin resulting in adenosyl-cobalamin (68). Aquo-cobalamin regularly appears under accidental termination of catalytic cycles and is quite vulnerable to degradation because of its “freely accessible” β-site (1). We can conjecture that the HbpS-like protein OrfY as well as the C-terminal part of PduO might be involved in the protective binding and/or transport of this cobalamin species.
FIGURE 11.
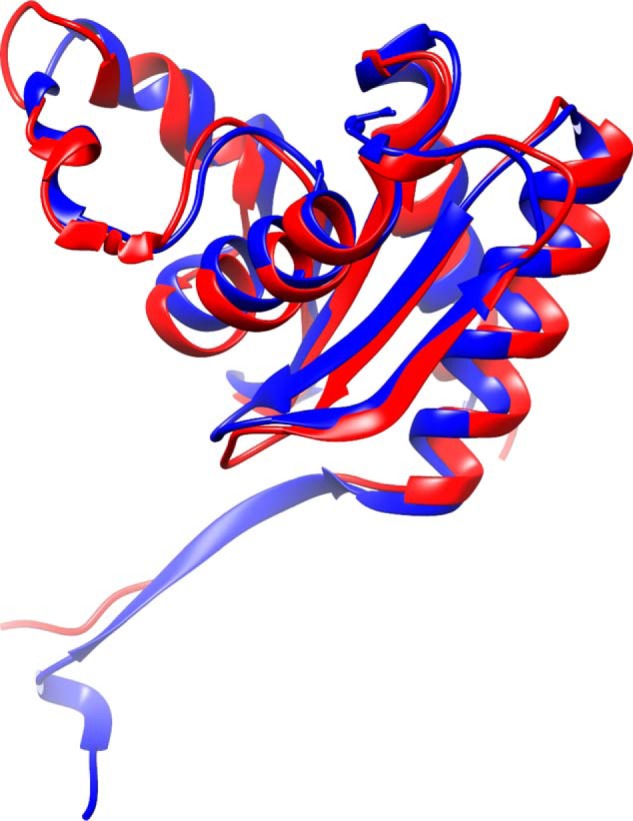
Structural alignment of one chain of HbpS from S. reticuli (blue; PDB code 3FPV) superimposed to 2.5 Å over 125 residues to OrfY from Klebsiella pneumoniae (red; PDB code 2A2L) calculated with SALAMI (69).
The interaction of HbpS with both heme and aquo-cobalamin is now clear. Fig. 2 shows HbpS and a set of closely related proteins, labeled as heme binders, which also showed cobalamin binding features. Are all HbpS-like proteins heme and/or cobalamin binders? This question will not be answered quickly. We showed that the related protein HbpSv also bound aquo-cobalamin. From the structures in the databases, one knows that HpbS and OrfY are closely related. The questions are, whether heme and/or cobalamin binding is metabolically important and, perhaps, which proteins have been erroneously annotated. Ultimately, the answers will be of interest for the evolution of protein function. Hopefully, one will be able to map the function onto phylogeny and be able to see an evolutionary path for the change of one type of porphyrin binding to another.
Acknowledgment
We are very grateful to Prof. Dr. H. Schrempf for continuous support (from the Applied Genetics of the Microorganisms, University of Osnabrueck).
This work was supported by Deutsche Forschungsgemeinschaft Grants OR 224/2-1 and OR 224/4-1.
- Cbl
- cobalamin
- PAA
- polyacrylamide
- AdoCbl
- 5′-deoxyadenosylcobalamin
- MeCbl
- methylcobalamin
- H2OCbl+
- aquo-cobalamin or vitamin B12a
- CNCbl
- cyanocobalamin
- PDB
- Protein Data Bank
- Ni2+-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Pratt J. M. (1972) Inorganic Chemistry of Vitamin B12. Academic Press, Inc. London [Google Scholar]
- 2. Kräutler B. (2005) Vitamin B12: chemistry and biochemistry. Biochem. Soc. Trans. 33, 806–810 [DOI] [PubMed] [Google Scholar]
- 3. Lochhead A. G., Thexton R. H. (1951) Vitamin B12 as a growth factor for soil bacteria. Nature 167, 1034. [DOI] [PubMed] [Google Scholar]
- 4. Lochhead A. G., Burton M. O. (1956) Soil as a habitat of vitamin-requiring bacteria. Nature 178, 144–145 [Google Scholar]
- 5. Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005) Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 [DOI] [PubMed] [Google Scholar]
- 6. Mozafar A., Oertli J. J. (1992) Uptake of a microbially-produced vitamin (B12) by soybean roots. Plant Soil 139, 23–30 [Google Scholar]
- 7. Friedmann H. C., Cagen L. M. (1970) Microbial biosynthesis of B12-like compounds. Annu. Rev. Microbiol. 24, 159–208 [DOI] [PubMed] [Google Scholar]
- 8. Roth J. R., Lawrence J. G., Rubenfield M., Kieffer-Higgins S., Church G. M. (1993) Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175, 3303–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roth J. R., Lawrence J. G., Bobik T. A. (1996) Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 [DOI] [PubMed] [Google Scholar]
- 10. Warren M. J., Raux E., Schubert H. L., Escalante-Semerena J. C. (2002) The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19, 390–412 [DOI] [PubMed] [Google Scholar]
- 11. Meschke H., Schrempf H. (2010) Streptomyces lividans inhibits the proliferation of the fungus Verticillium dahliae on seeds and roots of Arabidopsis thaliana. Microb. Biotechnol. 3, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Book A. J., Lewin G. R., McDonald B. R., Takasuka T. E., Doering D. T., Adams A. S., Blodgett J. A., Clardy J., Raffa K. F., Fox B. G., Currie C. R. (2014) Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically-linked capacity for the degradation of lignocellulose. Appl. Environ. Microbiol. 80, 4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chater K. F., Biró S., Lee K. J., Palmer T., Schrempf H. (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34, 171–198 [DOI] [PubMed] [Google Scholar]
- 14. Gruber K., Puffer B., Kräutler B. (2011) Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem. Soc. Rev. 40, 4346–4363 [DOI] [PubMed] [Google Scholar]
- 15. Fedosov S. N., Fedosova N. U., Kräutler B., Nexø E., Petersen T. E. (2007) Mechanisms of discrimination between cobalamins and their natural analogues during their binding to the specific B12-transporting proteins. Biochemistry 46, 6446–6458 [DOI] [PubMed] [Google Scholar]
- 16. Kräutler B. (2006) Cobalt: B12 enzymes and coenzymes. Encyclopedia of Inorganic Chemistry [Google Scholar]
- 17. Flinspach K., Westrich L., Kaysser L., Siebenberg S., Gomez-Escribano J. P., Bibb M., Gust B., Heide L. (2010) Heterologous expression of the biosynthetic gene clusters of coumermycin A(1), clorobiocin, and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93, 823–832 [DOI] [PubMed] [Google Scholar]
- 18. Allen K. D., Wang S. C. (2014) Initial characterization of Fom3 from Streptomyces wedmorensis: the methyltransferase in fosfomycin biosynthesis. Arch. Biochem. Biophys. 543, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borovok I., Gorovitz B., Schreiber R., Aharonowitz Y., Cohen G. (2006) Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J. Bacteriol. 188, 2512–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffmann B., Konrat R., Bothe H., Buckel W., Kräutler B. (1999) Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium. Eur. J. Biochem. 263, 178–188 [DOI] [PubMed] [Google Scholar]
- 21. Sjuts H., Dunstan M. S., Fisher K., Leys D. (2013) Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2. Acta Crystallogr. D Biol. Crystallogr. 69, 1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shibata N., Masuda J., Tobimatsu T., Toraya T., Suto K., Morimoto Y., Yasuoka N. (1999) A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: x-ray structure of diol dehydratase. Structure 7, 997–1008 [DOI] [PubMed] [Google Scholar]
- 23. Wuerges J., Garau G., Geremia S., Fedosov S. N., Petersen T. E., Randaccio L. (2006) Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc. Natl. Acad. Sci. U.S.A. 103, 4386–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wuerges J., Geremia S., Fedosov S. N., Randaccio L. (2007) Vitamin B12 transport proteins: crystallographic analysis of beta-axial ligand substitutions in cobalamin bound to transcobalamin. IUBMB Life 59, 722–729 [DOI] [PubMed] [Google Scholar]
- 25. Wedderhoff I., Kursula I., Groves M. R., Ortiz de Orué Lucana D. (2013) Iron binding at specific sites within the octameric HbpS protects streptomycetes from iron-mediated oxidative stress. PLoS One 8, e71579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ortiz de Orué Lucana D., Schaa T., Schrempf H. (2004) The novel extracellular Streptomyces reticuli heme-binding protein HbpS influences the production of the catalase-peroxidase CpeB. Microbiology 150, 2575–2585 [DOI] [PubMed] [Google Scholar]
- 27. Torda A. E., Groves M. R., Wedderhoff I., Ortiz de Orué Lucana D. (2013) Elucidation of heme-binding sites in the actinobacterial protein HbpS. FEMS Microbiol. Lett. 342, 106–112 [DOI] [PubMed] [Google Scholar]
- 28. Ortiz de Orué Lucana D., Bogel G., Zou P., Groves M. R. (2009) The oligomeric assembly of the novel heme-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase. J. Mol. Biol. 386, 1108–1122 [DOI] [PubMed] [Google Scholar]
- 29. Ortiz de Orué Lucana D., Roscher M., Honigmann A., Schwarz J. (2010) Iron-mediated oxidation induces conformational changes within the redox-sensing protein HbpS. J. Biol. Chem. 285, 28086–28096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ortiz de Orué Lucana D., Groves M. R. (2009) The three-component signalling system HbpS-SenS-SenR as an example of a redox sensing pathway in bacteria. Amino Acids 37, 479–486 [DOI] [PubMed] [Google Scholar]
- 31. Siedenburg G., Groves M. R., Ortiz de Orué Lucana D. (2012) Novel redox-sensing modules: accessory proteins- and nucleic acids-mediated signaling. Antioxid. Redox. Signal. 16, 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zou P., Groves M. R., Viale-Bouroncle S. D., Ortiz de Orué Lucana D. (2008) Cristallization and preliminary characterization of a novel heme-binding protein of Streptomyces reticuli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 386–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hopwood D. A., Bibb M. J., Chater K. F., Kieser T., Bruton C. J., Kieser H. M., Lydiate D. J., Smith C. P., Ward J. M., Schrempf H. (1985) Genetic Manipulation of Streptomyces: A Laboratory Manual, John Innes Foundation, Norwich, UK [Google Scholar]
- 34. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2 Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 35. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 36. Dawson J. H., Andersson L. A., Sono M. (1983) The diverse spectroscopic properties of ferrous cytochrome P-450-CAM ligand complexes. J. Biol. Chem. 258, 13637–13645 [PubMed] [Google Scholar]
- 37. Hargrove M. S., Singleton E. W., Quillin M. L., Ortiz L. A., Phillips G. N., Jr., Olson J. S., Mathews A. J. (1994) His64(E7) → Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J. Biol. Chem. 269, 4207–4214 [DOI] [PubMed] [Google Scholar]
- 38. Bhakta M. N., Wilks A. (2006) The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the delta-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry 45, 11642–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katoh K., Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huson D. H., Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 [DOI] [PubMed] [Google Scholar]
- 43. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fedosov S. N., Fedosova N. U., Nexø E., Petersen T. E. (2000) Conformational changes of transcobalamin induced by aquocobalamin binding: mechanism of substitution of the cobalt-coordinated group in the bound ligand. J. Biol. Chem. 275, 11791–11798 [DOI] [PubMed] [Google Scholar]
- 45. Fedosov S. N., Fedosova N. U., Berglund L., Moestrup S. K., Nexø E., Petersen T. E. (2005) Composite organization of the cobalamin binding and cubilin recognition sites of intrinsic factor. Biochemistry 44, 3604–3614 [DOI] [PubMed] [Google Scholar]
- 46. Nexo E. (1975) A new principle in biospecific affinity chromatography used for purification of cobalamin-binding proteins. Biochim. Biophys. Acta 379, 189–192 [PubMed] [Google Scholar]
- 47. Fedosov S. N., Laursen N. B., Nexø E., Moestrup S. K., Petersen T. E., Jensen E. Ø., Berglund L. (2003) Human intrinsic factor expressed in the plant Arabidopsis thaliana. Eur. J. Biochem. 270, 3362–3367 [DOI] [PubMed] [Google Scholar]
- 48. de Villiers K. A., Kaschula C. H., Egan T. J., Marques H. M. (2007) Speciation and structure of ferriprotoporphyrin IX in aqueous solution: spectroscopic and diffusion measurements demonstrate dimerization, but not mu-oxo dimer formation. J. Biol. Inorg. Chem. 12, 101–117 [DOI] [PubMed] [Google Scholar]
- 49. Asher C., de Villiers K. A., Egan T. J. (2009) Speciation of ferriprotoporphyrin IX in aqueous and mixed aqueous solution is controlled by solvent identity, pH, and salt concentration. Inorg. Chem. 48, 7994–8003 [DOI] [PubMed] [Google Scholar]
- 50. Fedosov S. N., Berglund L., Nexo E., Petersen T. E. (1999) Comparative analysis of cobalamin binding kinetics and ligand protection for intrinsic factor, transcobalamin and haptocorrin. J. Biol. Chem. 274, 26015–26020 [DOI] [PubMed] [Google Scholar]
- 51. Mozafar A. (1994) Enrichment of some B-vitamins in plants with application of organic fertilizers. Plant Soil 167, 305–311 [Google Scholar]
- 52. Martens J. H., Barg H., Warren M. J., Jahn D. (2002) Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58, 275–285 [DOI] [PubMed] [Google Scholar]
- 53. Hargrove M. S., Barrick D., Olson J. S. (1996) The association rate constant for heme binding to globin is independent of protein structure. Biochemistry 35, 11293–11299 [DOI] [PubMed] [Google Scholar]
- 54. Yukl E. T., Jepkorir G., Alontaga A. Y., Pautsch L., Rodriguez J. C., Rivera M., Moënne-Loccoz P. (2010) Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 49, 6646–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deniau C., Gilli R., Izadi-Pruneyre N., Létoffé S., Delepierre M., Wandersman C., Briand C., Lecroisey A. (2003) Thermodynamics of heme binding to the HasA(SM) hemophore: effect of mutations at three key residues for heme uptake. Biochemistry 42, 10627–10633 [DOI] [PubMed] [Google Scholar]
- 56. Nygaard T. K., Blouin G. C., Liu M., Fukumura M., Olson J. S., Fabian M., Dooley D. M., Lei B. (2006) The mechanism of direct heme transfer from the streptococcal cell surface protein Shp to HtsA of the HtsABC transporter. J. Biol. Chem. 281, 20761–20771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Owens C. P., Du J., Dawson J. H., Goulding C. W. (2012) Characterization of heme ligation properties of Rv0203, a secreted heme binding protein involved in Mycobacterium tuberculosis heme uptake. Biochemistry 51, 1518–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kuznets G., Vigonsky E., Weissman Z., Lalli D., Gildor T., Kauffman S. J., Turano P., Becker J., Lewinson O., Kornitzer D. (2014) A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog. 10, e1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yin L., Dragnea V., Feldman G., Hammad L. A., Karty J. A., Dann C. E., 3rd, Bauer C. E. (2013) Redox and light control the heme-sensing activity of AppA. MBio. 4, e00563–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yin L., Dragnea V., Bauer C. E. (2012) PpsR, a regulator of heme and bacteriochlorophyll biosynthesis, is a heme-sensing protein. J. Biol. Chem. 287, 13850–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cho H. Y., Cho H. J., Kim Y. M., Oh J. I., Kang B. S. (2009) Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 284, 13057–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Podust L. M., Ioanoviciu A., Ortiz de Montellano P. R. (2008) 2.3 Å x-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry 47, 12523–12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bogel G., Schrempf H., Ortiz de Orué Lucana D. (2009) The heme-binding protein HbpS regulates the activity of the Streptomyces reticuli iron-sensing histidine kinase SenS in a redox-dependent manner. Amino Acids 37, 681–691 [DOI] [PubMed] [Google Scholar]
- 64. Walsh C. T., Garneau-Tsodikova S., Howard-Jones A. R. (2006) Biological formation of pyrroles: nature's logic and enzymatic machinery. Nat. Prod. Rep. 23, 517–531 [DOI] [PubMed] [Google Scholar]
- 65. Romero D., Traxler M. F., López D., Kolter R. (2011) Antibiotics as signal molecules. Chem. Rev. 111, 5492–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johnson C. L., Pechonick E., Park S. D., Havemann G. D., Leal N. A., Bobik T. A. (2001) Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:Cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183, 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun J., van den Heuvel J., Soucaille P., Qu Y., Zeng A. P. (2003) Comparative genomic analysis of dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol. Prog. 19, 263–272 [DOI] [PubMed] [Google Scholar]
- 68. Parsons J. B., Lawrence A. D., McLean K. J., Munro A. W., Rigby S. E., Warren M. J. (2010) Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One 5, e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Margraf T., Schenk G., Torda A. E. (2009) The SALAMI protein structure server. Nucleic Acids Res. 37, W480–W484 [DOI] [PMC free article] [PubMed] [Google Scholar]