Background: Protein partnerships regulate specific functions of cells. BCOR protein-protein interactions are critical to aspects of B cell biology.
Results: IRF8 pairs with BCOR in the nucleus to enhance its transcriptional repressive activity.
Conclusion: Partnering of IRF8 with BCOR defines a novel mechanism for controlling B cell gene expression.
Significance: Understanding gene regulation in specific cell types requires identification of protein complexes that modulate expression.
Keywords: Gene Expression, Immunology, Lymphocyte, Protein Complex, Protein-Protein Interaction, Transcription Repressor
Abstract
B cell lymphoma 6 (BCL6) corepressor (BCOR) was discovered as a BCL6-interacting corepressor, but little is known about its other biological activities in normal B cell development and function. Previously, we found that interferon regulatory factor 8 (IRF8), also known as interferon consensus sequence-binding protein, directly targets a large number of genes in germinal center B cells including BCL6. In this study, we screened potential binding partners of IRF8 using a retrovirus-based protein complementation assay screen in a mouse pre-B cell line. We found that IRF8 interacts directly with BCOR and that the α-helical region of IRF8 and the BCL6 binding domain of BCOR are required for this interaction. In addition, IRF8 protein interacts directly with BCL6. Using an siRNA-mediated IRF8 knockdown mouse B cell lymphoma cell line, we showed that IRF8 represses Bcor and enhances Bcl6 transcription. Taken together, these data suggest that a complex comprising BCOR-BCL6-IRF8 modulates BCL6-associated transcriptional regulation of germinal center B cell function.
Introduction
Interferon regulatory factor 8 (IRF8)5 is a member of the IRF family of transcription factors that was first identified in dissections of cellular responses to type I interferon (IFN I) (1). Since then, IRF family members have been shown to be involved in a wide range of biologic responses including regulation of hematopoietic differentiation, responses to pathogen-derived signals, control of the cell cycle, and apoptosis (2–4). IRF8, expressed primarily in cells of the immune system, is perhaps best known for its roles in directing the development and function of myeloid and dendritic cells (5–7). The target genes and transcriptional programs that mediate the effects of IRF8 in these cell types are becoming increasingly well understood. IRF8 binds to sites in its transcriptional targets as a heterodimer paired with other members of the IRF family, members of the E26 transformation-specific (ETS) family, or other partners to activate or repress gene expression (8–11).
IRF8 is also expressed in cells of the T and B lymphocyte lineages. Analyses of B lineage cells showed that IRF8 contributes to a series of transcriptional networks affecting the earliest stages of lineage commitment and specification in the bone marrow (12), rearrangements at the immunoglobulin (Ig) κ locus (13), and distribution of mature cells into splenic B cell subcompartments (14). IRF8 was shown to exert its influences on the germinal center (GC) reaction, class switch recombination, and plasma cell development through partnering with PU.1 and IRF4 to promote the activation of Aicda and Bcl6, genes central to the maintenance of the B cell program, while repressing genes such as Prdm1 that promote terminal differentiation (15, 16).
Previous studies demonstrated that IRF8 is involved in the regulation of Bcl6 expression in GC B cells (15). BCL6 is a transcriptional repressor with critical roles in several immunological processes including B and T cell functions, especially GC development and generation. BCL6 is highly expressed in B cells undergoing affinity maturation in GC, and its expression is down-regulated upon selection for apoptosis or differentiation (17, 18). The critical function of BCL6 in GC biology is associated with the BCL6 BTB/POZ domain physically interacting with the corepressor proteins BCOR (19), NCoR, SMRT (20), Mi-2/NuRD (21), and histone deacetylase complexes to mediate its potent transrepressor activity.
To determine whether there are other partners for IRF8 that might contribute to this complex and late developmental transcriptional program of B cells, we adopted the technique of enhanced retroviral mutagen protein complementation assay (22). We identified 32 potential interaction partners that included BCOR, a transcriptional corepressor that specifically inhibits gene expression when recruited to promoter regions by BCL6 (19). Aside from the established importance of BCOR as a BCL6-interacting corepressor, there have been few studies about the role of BCOR in GC B cell development and function. Here we show that BCOR interacts directly with IRF8 and that the BCOR-IRF8 complex enhances transcriptional repression by BCL6.
EXPERIMENTAL PROCEDURES
Cell Culture and Stimulation
HEK293 cells were maintained at 37 °C with 5% CO2 in DMEM (Quality Biological Inc.) supplemented with 10% FBS, penicillin, and streptomycin. NFS202, 18-81, 18-81 Tet-On, WEHI231, and MPC11 cells (all from our laboratory) and OCI-Ly1 (originally provided by Dr. Riccardo Dalla-Favera, Columbia University) were cultured with RPMI 1640 complete medium supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm non-essential amino acids, 50 μm β-mercaptoethanol, 1 mm sodium pyruvate, and HEPES.
Plasmids and Transfection
Plasmids for the retrovirus-based protein complementation assay (RePCA) screen were from Odyssey Thera, Inc. (San Ramon, CA). Green fluorescent protein (GFP)-tagged full-length and truncated forms (1–390, 356, Del-N, and Del-C) of Irf8 plasmid were described previously and were kindly provided by Dr. Keiko Ozato (National Institute of Child Health and Human Development, National Institutes of Health). The full-length ORFs of Bcor (GenBankTM accession number NM_029510.3) and ankyrin repeat (ANK) domain- or BCL6 binding domain (BBD)-deleted forms of Bcor were cloned in pcDNA4-myc and pTOPO-V5-His (Invitrogen) vectors, respectively.
For Lipofectamine LTX (Invitrogen) cotransfection, 5 × 105 HEK293 cells were plated into a 60-mm dish with 2 ml of medium. Each 1.5 μg of DNA was mixed with 2.5 μl of Plus reagent in 500 μl of serum-free medium for 5 min. Then 7.5 μl of Lipofectamine LTX was added, incubated for 20 min at room temperature, and loaded onto the cells. Cells were harvested 48 h after transfection.
RePCA
The RePCA screens were performed as described previously (22) with some modifications. Briefly, the mouse Irf8 gene was cloned by RT-PCR and stably transfected to 18-81 Tet-on cells. Retrovirus-infected cells were selected with 0.5 μg/ml puromycin. After induction of GFP by doxycycline, fluorescent cells were sorted by flow cytometry using an Aria-Green sorter (BD Biosciences). To identify target genes, cDNA was synthesized from expanded clones, and PCR-amplified with a specific intensely fluorescent protein (IFP) C-terminal portion primer and T7 primer, and PCR products were sequenced.
Immunostaining
Cells were fixed for 20 min in 4% paraformaldehyde and rinsed three times in PBT (0.1% Tween 20 in PBS). Samples were incubated with antibody specific for BCOR (Abcam) at 37 °C for 2 h followed by incubation with anti-rabbit IgG Alexa Fluor 546-conjugated secondary antibody (Invitrogen) for 1 h at room temperature (1:500). For confocal microscopic analyses, fixed and stained cells were mounted with Vectashield mounting medium with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Images were taken using a Zeiss LSM 710 laser-scanning microscope (Carl Zeiss) equipped with a Plan-Apochromat 63×/1.4 oil immersion objective at ambient temperature and processed using ZEN 2009 software (Carl Zeiss).
RNA Preparation and Quantitative Real Time PCR (qRT-PCR)
Total RNA was extracted from cultured cells using an RNeasy Miniprep kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized with the Superscript III first strand synthesis system (Invitrogen) using 1 μg of total RNA in a 20-μl reaction volume, one-tenth of which was used as a template for qRT-PCR in the ABI PRISM 7900 HT Fast Real-Time PCR system (Applied Biosystems) using the TaqMan probe according to the manufacturer's instructions. Each PCR procedure included a non-template negative control reaction. The Hprt level was used as the internal control, and ΔC(t) values were calculated according to the 2−ΔΔC(t) method (23). -Fold change was determined by normalizing the ΔC(t) values for each gene. Each sample subjected to qRT-PCR was analyzed in triplicate.
Luciferase Reporter Assays
For promoter repression assays, HEK293 cells were transiently transfected with 0.25 μg of either pGL3-containing 5× BCL6 or a control promoter reporter construct, 0.1 μg of human Bcor (pcDNA4-Bcor-Myc) or pcDNA4-Irf8, and 0.05 μg of Bcl6 expression plasmid made up to a total 0.35 μg using empty vector. After 30 h, cells were extracted in 110 μl of passive lysis buffer and analyzed in a MicroBeta Luminescence counter (PerkinElmer Life Sciences) using a luciferase reporter assay kit (Promega). All firefly luciferase activities were normalized to the expressed Renilla luciferase activity transfected with pAc-LacZ vector (50 ng). Student's t test was performed to evaluate statistical significance.
Coimmunoprecipitation and Immunoblotting
Cells were lysed in IP Lysis Buffer (Pierce) with protease inhibitor mixtures (Roche Applied Science). Cell lysates were precleared with Dynabeads protein G (Cell Signaling Technology) for 0.5 h at room temperature and then incubated with 2.5 μg of antibody of interest-conjugated Dynabeads protein G for 16 h at 4 °C. The immune complexes were precipitated, washed, and eluted in Laemmli sample buffer with reducing reagent. For immunoblotting, cell lysates were boiled in Laemmli sample buffer with reducing reagent and separated through 4–12% NuPAGE Bis-Tris gradient gels (Invitrogen). Following transfer, the membrane was incubated with various antibodies as described; anti-IRF8, -BCOR, -BCL6, -MYC, -V5, and -FLAG antibodies were used at a 1:1000–2000 dilution. Goat anti-rabbit or mouse IgG secondary antibodies (Pierce; 1:5000) were subsequently incubated for 1 h at room temperature. The proteins were visualized using the SuperSignal® West Dura Substrate kit (Pierce) according to the manufacturer's instructions.
In Vitro Pulldown Assay
Biotinylated BCOR protein and IRF8 proteins were synthesized in vitro using the TnT coupled transcription/translation system (Promega) according to the manufacturer's instructions. Synthesized protein efficiency was checked with SDS-PAGE, and mixtures of both proteins were incubated on ice for 1 h. Dynabeads streptavidin (Invitrogen) was used for precipitation. Precipitated proteins were subjected to 4–12% NuPAGE Bis-Tris gradient gels (Invitrogen) followed by immunoblotting.
Chromatin Immunoprecipitation (ChIP) and ReChIP Assay
For ChIP assays, cells were fixed with 1% formaldehyde for 15 min, quenched by adding 0.125 m glycine, and washed twice with PBS. Fixed samples were homogenized with a Dounce homogenizer in swelling buffer (0.1% Nonidet P-40, 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride), and the pellet was suspended in sonication buffer (50 mm HEPES, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5 mm phenylmethylsulfonyl fluoride supplemented with a protease inhibitor mixture). Sonication efficiency was checked on 2% agarose gels to obtain 100–400 bp of sheared chromatin. Anti-rabbit BCOR antibody (Abcam)-conjugated protein A-agarose was used for precipitation with sheared chromatin. The immune complexes were washed with LiCl buffer (100 mm Tris-HCl, pH 8.0, 500 mm LiCl, 0.1% Nonidet P-40, 0.1% deoxycholic acid) and then eluted by elution buffer (1% SDS, 0.1 m NaHCO3). Eluted proteins were diluted and subjected to the ReChIP procedure with anti-goat IRF8 antibody (Santa Cruz Biotechnology). A goat/rabbit serum was used as a control. DNA was recovered by phenol/chloroform extraction and ethanol precipitation, and DNA diluted 1:10 was used for PCR.
RESULTS
Mouse BCOR Binds Directly with Mouse IRF8
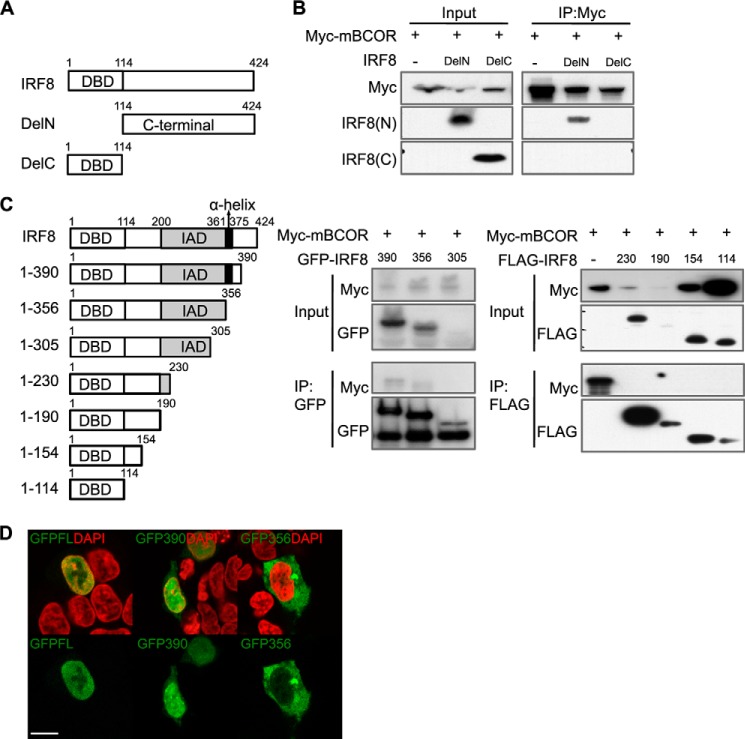
To identify potential partner proteins that influence the biological function of IRF8 in B lineage cells, we screened proteins interacting directly with IRF8 using a retrovirus-based protein complementation assay (22). As bait, we used intensely fluorescent protein N-terminal portion-conjugated full-length mouse IRF8. The bait vector was co-transfected with an IFP C-terminal portion-conjugated cDNA library target vector into the 18-81 mouse pre-B cell line, and cotransfected cells were incubated for 24–48 h. GFP-positive cells (Fig. 1) were purified by FACS to develop sequence information on targets.
FIGURE 1.
Screen for interacting partners of IRF8. 18-81 pre-B cells were transfected to express IFP N-terminal portion-IRF8. The cell population was infected with RePCA viruses to create endogenous protein fusions with IFP C-terminal portion. Fusion of IFP C-terminal portion in-frame to a protein results in restoring fluorescence. Green fluorescence-expressing cells were sort-purified by FACS and cloned, and target genes were identified by RT-PCR. BF, bright field; GREEN, green-filtered images.
From sequencing analyses of interacting target clones, a series of potential interaction partners of mIRF8 was identified (Table 1). Importantly, SFPI1 (PU.1) was identified as a binding partner of IRF8 by RePCA, providing a proof of principle because both PU.1 and IRF8 are well known heterodimeric transcription factors sharing IRF binding motifs on target DNA (9). In particular, the identified potential binding partners, SFPI1, TAF8, GFI1B, NRID2, POLK (DNA polymerase κ), THRB (thyroid hormone receptor β), and BCOR, generally bind to promoter or enhancer DNA sequences to regulate transcriptional activity. Among the binding partners, BCOR was hit three times, implying that BCOR was highly disposed to physically bind or interact with IRF8.
TABLE 1.
Interaction partners of IRF8 identified in RePCA screen
| Gene symbol | Times identified |
|---|---|
| Sfpi1 | 1 |
| Sh3bgrl2 | 4 |
| Csf1 | 1 |
| Thrb | >1 |
| Polk | 1 |
| Fbxl18 | >1 |
| Mef2d | 1 |
| Gfi1b | 1 |
| Nr1d2 | >1 |
| Bcor | 3 |
| Blnk | 1 |
| Ccnd | 1 |
| Sost | >1 |
| Taf8 | 1 |
| Prtn3 | 1 |
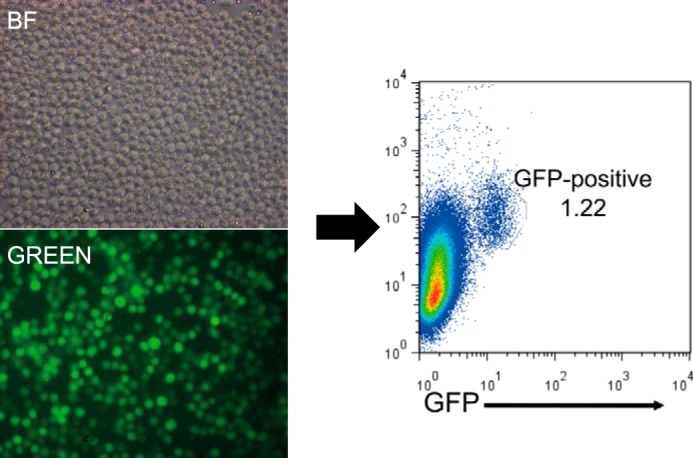
To confirm the physical interactions between IRF8 and BCOR, we performed a pulldown study using Myc-tagged mouse BCOR (mBCOR) or human BCOR protein. For this, HEK293 cells were co-transfected with a full-length mouse IRF8 expression vector and a Myc-tagged mBCOR construct. mBCOR was pulled down by anti-Myc antibody 48 h after transfection. BCOR was not detected in IRF8 singly transfected control pulldown products, but IRF8 was clearly detected by pulling down Myc-tagged mBCOR/BCOR (Fig. 2A). To determine whether IRF8 and BCOR can interact directly in the absence of other proteins, we translated biotin-BCOR and IRF8 in vitro. Biotin-BCOR alone or mixed with IRF8 was pulled down with streptavidin-coated beads. The results (Fig. 2C) showed that IRF8 forms a complex with BCOR directly in vitro. We also tested for IRF8-BCOR interactions by immunostaining for endogenous BCOR in HEK293 cells transiently expressing GFP-IRF8. Transfected GFP-IRF8 overlapped completely with DAPI-stained nuclei, indicating a dominant location for IRF8 in the nucleus. The pattern for BCOR staining also overlapped with that of GFP-IRF8 (Fig. 2B). These results indicate that interactions between BCOR and IRF8 are likely to occur mainly in the nucleus.
FIGURE 2.
BCOR interacts with IRF8. A, HEK293 cells expressing IRF8 and Myc-tagged human BCOR or mBCOR were lysed, pulled down with anti-Myc antibody, and subsequently blotted for BCOR and IRF8. IRF8 was co-immunoprecipitated with Myc-BCOR/mBCOR. B, images of immunostaining with anti-BCOR (red) antibody in transiently GFP-IRF8 (green)-expressing HEK293 cells. Nuclei were counterstained with DAPI (blue). BCOR colocalized with GFP-IRF8 in the nucleus. Scale bar, 100 μm. C, a mixture of in vitro translated biotin-BCOR and IRF8 proteins or biotin-BCOR alone was pulled down with Dynabeads streptavidin and blotted for BCOR and IRF8. In vitro synthesized IRF8 co-immunoprecipitated with biotin-BCOR. IP, immunoprecipitation.
The C-terminal Region of IRF8 Contains an α-Helical Region That Is Required for Interaction with BCOR as Well as Localization to the Nucleus
IRF8 comprises two major functional domains, an N-terminal DNA-binding region and a C-terminal IRF association domain involved in protein-to-protein interactions with other IRF family members as well as additional partner proteins (5). To determine which domain of IRF8 is responsible for binding to BCOR, deletion constructs lacking either the N- or C-terminal domains of IRF8 (DelN and DelC, respectively) were prepared (Fig. 3A) and co-transfected with Myc-tagged BCOR into HEK293 cells. Expression levels of Myc-tagged BCOR were comparable in lines co-transfected with IRF8 DelN or DelC. Western blotting using anti-IRF8 antibody identified IRF8 DelN in pulldowns utilizing anti-Myc antibodies. However, IRF8 DelC was not detected in Western blot analyses of cells co-transfected with Myc-BCOR (Fig. 3B). This result clearly showed that the C-terminal domain of IRF8 is required for interactions with BCOR.
FIGURE 3.
BCOR interaction with the C terminus of IRF8 and nuclear localization. A, diagram of N and C termini-deleted IRF8 constructs. DBD, DNA binding domain. B, HEK293 cells expressing N (DelN) and C termini deleted (DelC) IRF8s and Myc-tagged full-length BCOR were lysed, pulled down with anti-Myc antibody, and subsequently blotted for BCOR and both the N-terminal and C-terminal IRF8s. The C terminus-deleted IRF8 failed to co-immunoprecipitate with Myc-mBCOR. IRF8(N), antibody raised against the N-terminal region of IRF8; IRF8(C), antibody raised against the C-terminal region of IRF8. C, diagram of a series of C termini-truncated GFP/FLAG-IRF8 constructs. DBD, DNA binding domain (white box); IAD, IRF association domain (gray box); α-helix, α-helix conserved region (black box). HEK293 cells expressing a series of C termini-truncated GFP/FLAG-IRF8 and Myc-tagged full-length BCOR were lysed, pulled down with anti-GFP or anti-FLAG antibody, and subsequently blotted for GFP, FLAG, and Myc. BCOR was co-precipitated with 390 IRF8 that only contains the α-helix region. D, images of C termini-truncated GFP-IRF8 (green)-transfected HEK293 cells. Nuclei were counterstained with DAPI (red). GFPFL, GFP-full-length IRF8 (amino acids 1–424); GFP390, GFP-IRF8 (amino acids 1–390); GFP356, GFP-IRF8 (amino acids 1–356). Scale bar, 100 μm. IP, immunoprecipitation.
To better understand the nature of the IRF8 C-terminal binding domain responsible for the interaction with BCOR, we generated a series of C-terminal truncation mutants of IRF8 (Fig. 3C) for co-immunoprecipitation analyses. The 390 construct co-precipitated with Myc-tagged BCOR, but the 356 construct did not (Fig. 3C). In addition, we expressed each of the GFP-IRF8 C-terminal deletion mutants in mammalian cells to determine the subcellular localization of IRF8-BCOR interactions by confocal microscopy. The GFP-tagged α-helix-deleted form of IRF8 (amino acids 1–356) was found to accumulate in the cytoplasm, whereas the full-length and 390 proteins localized to the nucleus (Fig. 3D). These results indicate that the α-helix domain in the C terminus of IRF8 mediates interactions with BCOR much as it does for other IRF8 partner proteins. It also affects the nuclear localization of IRF8 protein and thus the ability of IRF8 to form a complex with BCOR in the nucleus.
The N-terminal Region of BCOR Contains a BBD That Is Required for Interaction with IRF8
The structure of BCOR includes a BBD and an ANK domain as shown in Fig. 4. The BCOR BBD in residues 498–514 associates with the BCL6 POZ/BTB domain to form dimers, and the BTB/BBD domain determines the functional activity of BCL6 as a transcriptional repressor (24, 25). Thus, we tested V5-tagged ANK domain- or BBD-deleted BCOR in pulldown assays to determine the region required for binding with IRF8. IRF8 was co-precipitated only with the ANK domain-deleted BCOR (Fig. 4). This result shows that the region of BCOR containing the BBD is important for binding with IRF8 directly as well as with BCL6.
FIGURE 4.
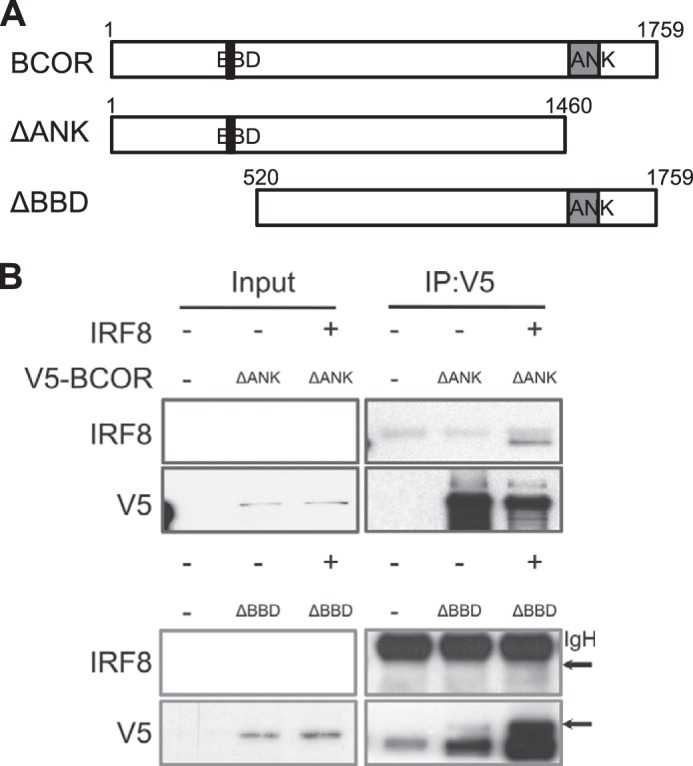
IRF8 interacts with the BCL6 binding domain of BCOR. A, diagram of ANK domain- and BBD-deleted BCOR constructs. ANK, gray box; BBD, black box. B, HEK293 cells expressing V5-tagged ANK domain- or BBD-deleted BCORs and full-length IRF8 were lysed, pulled down with anti-V5 antibody, and subsequently blotted for V5 and IRF8. The BCL6 binding domain-deleted BCOR failed to co-immunoprecipitate with IRF8. The arrow indicates IRF8 protein. IgH, immunoglobulin heavy chain; IP, immunoprecipitation.
The BCOR-IRF8 Complex Enhances Gene Repression by BCL6, and BCL6 Interacts Directly with IRF8
BCOR interacts with the POZ domain of BCL6 and acts as transcriptional corepressor to amplify transcriptional repression by BCL6 (19). To determine whether the interaction of IRF8 with BCOR enhances its corepressor activity, we performed luciferase assays in HEK293 cells using a 5× BCL6 binding site construct. In cells co-transfected with BCOR and BCL6, the transcriptional activity of the reporter construct was 10-fold lower than in cells transfected with BCL6 alone (Fig. 5A). Reporter activity in cells co-transfected with BCL6 and IRF8 was 4-fold lower than that in cells transfected with BCL6 alone. Remarkably, when cells were co-transfected with the combination of IRF8, BCOR, and BCL6 expression vectors, luciferase activity was 2-fold lower than in cells co-transfected with the BCOR and BCL6 expression constructs and 20-fold lower than in cells transfected with BCL6 alone. These results indicate that IRF8 augments the corepressor activity of BCOR. These findings prompted us to ask whether IRF8 might be interacting directly with BCL6 or only indirectly through binding to BCOR. We generated FLAG-tagged BCL6 recombinant protein that was co-transfected with IRF8 into HEK293 cells. The exogenous IRF8 protein was co-precipitated with anti-FLAG antibody. When we increased the amount of IRF8, the amount of precipitated FLAG-BCL6 also increased in a dose-dependent manner (Fig. 5B). We also detected endogenous protein-protein interaction of IRF8 and BCL6 in the 18-81 pre-B cell line using immune complexes formed using an anti-IRF8 antibody (Fig. 5C). The endogenous BCL6 protein level was comparable for the 18-81 and MPC11 cell lines, but BCL6 was not precipitated with anti-IRF8 antibody in MPC11 cells that expressed no or very little IRF8 protein. This result strongly suggests that IRF8 interacts directly with BCL6 in B lineage cells.
FIGURE 5.
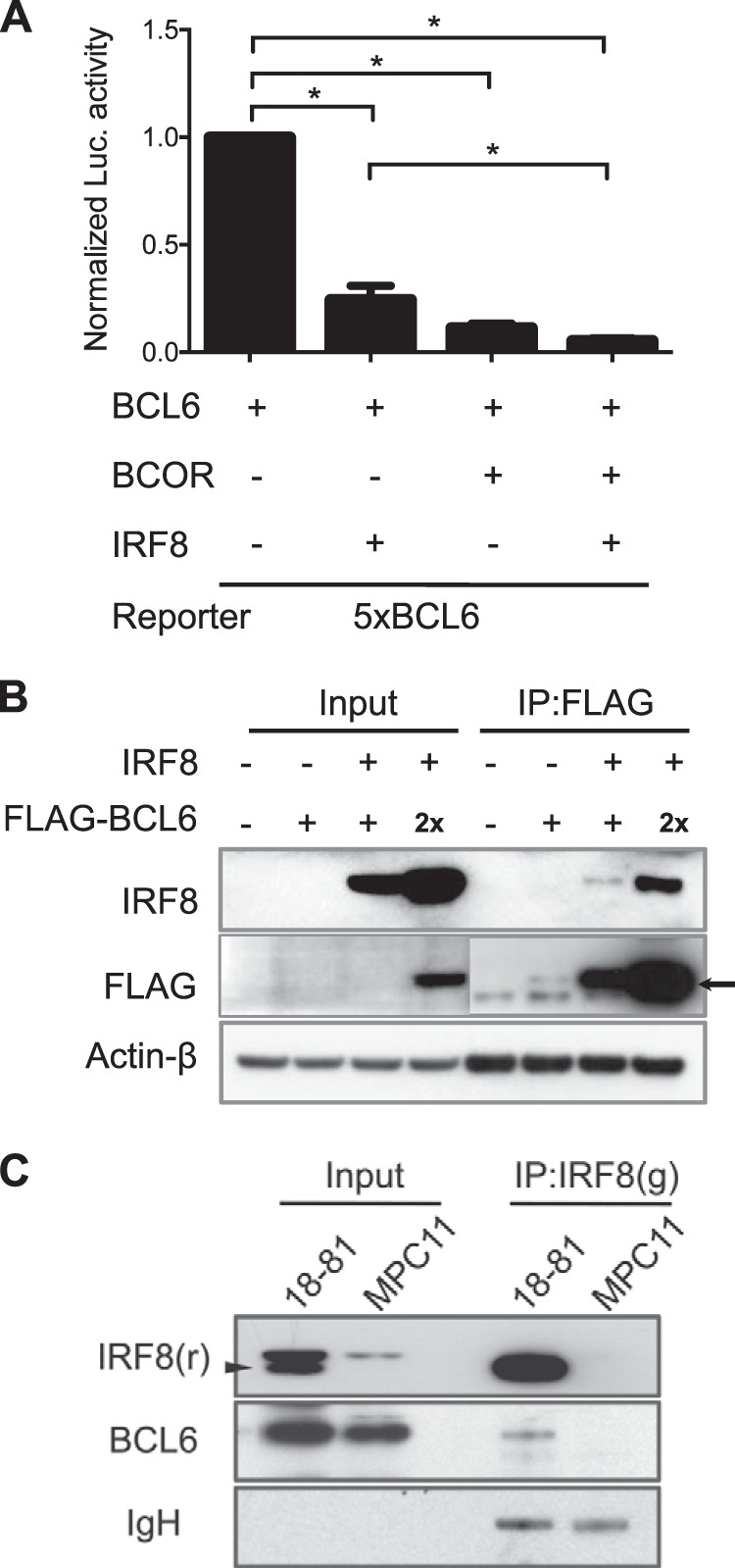
Synergistic repression of BCL6 activity by BCOR and IRF8. A, HEK293 cells containing each expression plasmid (BCL6, BCOR, and IRF8) and BCL6-responsive luciferase were lysed, and a luciferase assay was performed. Normalized luciferase (Luc) activity is the ratio of firefly luciferase activity to the expressed Renilla luciferase activity. Data shown are means ± S.E. (error bars). Student's t test was performed to evaluate statistical significance. *, p < 0.05. B, HEK293 cells expressing IRF8 and FLAG-tagged BLC6 were lysed, pulled down with anti-FLAG antibody, and subsequently blotted for FLAG and IRF8. The arrow indicates FLAG-BCL6 protein. IRF8 co-immunoprecipitated with FLAG-tagged BCL6. C, 18-81 and MPC11 cell lysates were precipitated with anti-IRF8 antibody and blotted for BCL6 and IRF8. Endogenous BCL6 was co-immunoprecipitated with IRF8 only in 18-81 cells. Immunoglobulin heavy chain (IgH) is shown as a loading control. The arrowhead indicates IRF8 protein. g, goat; r, rabbit; IP, immunoprecipitation.
To determine whether the proposed nuclear interactions of IRF8 and BCOR are physiologically relevant, we examined the relationships of IRF8 and BCOR DNA targets by comparing ChIP-chip (16) and ChIP-sequencing data (26), respectively, generated with the OCI-Ly1 human B lymphoma cell line (Fig. 6). Data generated by the two different techniques do not allow for a full comparison as ChIP-chip assessed binding only at sites 3.5 kb upstream to 0.75 kb downstream of transcriptional start sites (16). To lessen the impact of this issue, we chose to identify common targets for IRF8 and BCOR by examining their binding peaks, the threshold being a distance between the IRF8 peak and the BCOR peak of less than 1 kb. This analysis identified 1816 putative common target sites (Fig. 6A). Selected targets were validated by the ChIP-ReChIP assay using antibodies to IRF8 and BCOR (Fig. 6B), which showed that IRF8 and BCOR proteins occupy the same DNA regulatory elements in OCI-Ly1 cells. A functional categorization of other targets is listed in Table 2.
FIGURE 6.
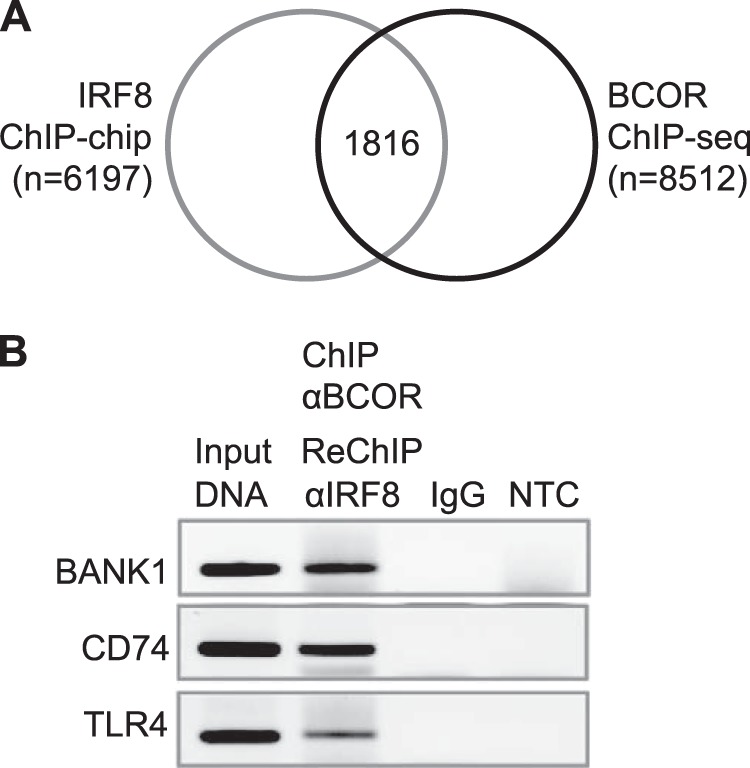
Identification of common targets of IRF8 and BCOR in the human OCI-Ly1 B cell line. A, Venn diagram showing IRF8 and BCOR targets in OCI-Ly1 human diffuse large B cell lymphoma cells analyzed by ChIP-chip (gray line) and ChIP-sequencing (ChIP-seq) (black line), respectively. One thousand eight hundred sixteen genes were identified as putative common IRF8 and BCOR targets. B, co-occupancy by BCOR and IRF8 in BANK1, CD74, and TLR4 fragments. Formaldehyde-fixed chromatin from OCI-Ly1 cells was precipitated by anti-BCOR and subjected to ReChIP by anti-IRF8 antibody. DNA was recovered and amplified with primer sets for each target gene. NTC, non-template control.
TABLE 2.
Functional annotation of common targets of IRF8 and BCOR in human GC B cell line OCI-Ly1
| Function annotation | Common targets of IRF8 and BCOR |
|---|---|
| Quantity of lymphocytes | ARID5B, BANK1, CD74, CIITA, DDX58, ETV6, ISG15, LSP1, MS4A1, TLR4 |
| Proliferation of lymphocytes | BANK1, CD37, CD74, CIITA, ISG15, MS4A1, TLR4 |
| Differentiation of lymphocytes | CD74, CIITA, DDX58, ETV6, STAT2, TLR4 |
| Differentiation of B lymphocytes | CD74, DDX58, ETV6, TLR4 |
BCOR and IRF8 Are Co-expressed in Various B Cell Subsets
Bcor is expressed widely in most tissues of the mouse (27) including all B lineage subpopulations from prepro-B cells to all mature subsets in peripheral lymphoid compartments (ImmGen). Irf8 is expressed primarily in hematopoietic cells including all stages of B cell differentiation up to terminally differentiated plasma cells (12). To determine whether IRF8 might affect expression of BCOR and vice versa, we studied transcript levels for Irf8 and Bcor in sublines of the mouse NFS202 B cell lymphoma of GC origin that express an IRF8-suppressive or a scrambled siRNA (15). Bcor transcript levels were 3–4.5-fold higher in the knockdown cell line than in the scrambled siRNA transfectants, whereas Bcl6 transcript levels were dramatically reduced more than 90% in the Irf8 knockdown cells (Fig. 7A). Similarly, Bcor transcript levels were 6-fold higher in the immature B cell line WEHI231, whereas Irf8 transcript levels were reduced 2.8-fold in the pre-B cell line 18-81 (Fig. 7B). These results demonstrated that IRF8 normally represses Bcor transcription and indicated that these proteins are involved in an interdependent transcriptional network as well as interacting physically.
FIGURE 7.
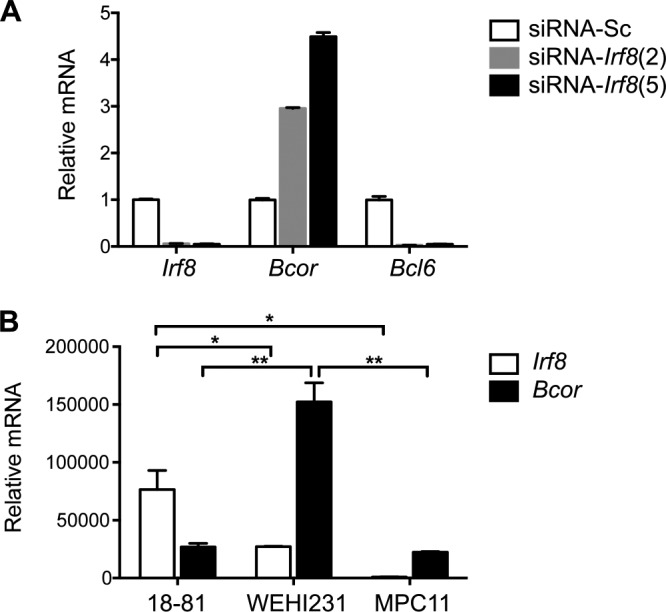
Effect of IRF8 on expression of BCOR in mouse B cells. A, Expression of Bcor, Irf8, and Bcl6 in NFS-202 cells with stably transfected pSR-mIRF8 (clones 2 and 5) were analyzed by qRT-PCR. Sc, scrambled siRNA control. Results are normalized to Hprt and indicate -fold change relative to those of the scrambled siRNA control. B, expression of Bcor and Irf8 in 18-81, WEHI231, and MPC11 cells was analyzed by qRT-PCR. Data shown are means ± S.E. (error bars). Student's t test was performed to evaluate statistical significance. *, p < 0.05; **, p < 0.01.
DISCUSSION
IRF8 is expressed throughout B cell differentiation and was shown previously to play important roles in B cell lineage specification and commitment (12), anergy (28), rearrangement at the Ig κ locus (13), and distribution of B cells to peripheral lymphoid compartments (29). IRF8 is highly expressed in mouse and human GCs (15, 30) and shares a large number of common target genes in GC B cells, about half of which are also targeted by PU.1 (16). IRF8 has also been implicated in the development of B cell-driven systemic autoimmune diseases in humans and mice (31, 32). Finally, several recent studies have identified roles for IRF8 in the pathogenesis of human diffuse large B cell lymphoma (33, 34) and probably mouse diffuse large B cell lymphoma as well (15).
The studies presented here shed new light on a complex regulatory network that involves contributions of IRF8, BCOR, BCL6, and PU.1 to B cell biology. Previous studies had documented important protein-protein interactions between BCL6 and BCOR (19), BCL6 and PU.1 (35), and PU.1 and IRF8 (36). The relationship between IRF8 and BCOR as a candidate partner protein was first suggested by the results of the RePCA analysis that identified 15 potential partners of IRF8, only one of which, SFPI1 (PU.1), had been identified previously. An analysis of the interactions between IRF8 and BCOR demonstrated a requirement for the C-terminal region of IRF8, which contains an α-helix protein-protein interaction domain, and the N-terminal region of BCOR, which contains the sequences required for BCL6 binding. Accumulation in the cytoplasm of IRF8 mutant protein lacking the α-helix region clearly supports previous observations of IRF8 function and interaction with partner proteins in the nucleus (37, 38).
A possible relationship among IRF8, PU.1, and BCOR was of interest in view of the demonstration that PU.1 is reported to direct BCL6 to its target sites (35). In this regard, it is also noteworthy that the ability of IRF8 to bind DNA on its own is quite weak and that tight binding to target sequences requires interaction with another transcription factor such as PU.1. It thus appears that PU.1 recruits IRF8 to DNA regions containing both ETS and IRF binding elements presented in an overlapping manner. Therefore IRF8 might compete with BCL6 or BCOR for physical interaction with PU.1 and subsequent protein partner recruitment for functional activities because BCL6 can physically interact with IRF8 both in vitro and in vivo as shown here (Fig. 5, B and C).
The functional consequences of this complex formation are indicated by the fact that the presence of IRF8 enhances the repressive activity of BCOR on BCL6 but also that IRF8 has a significant effect on the repressive activity of BCL6 in the absence of BCOR. It is well established that the BCOR complex itself is structurally complex, comprising seven or more interacting proteins and BCL6 with two other corepressors, NCoR and SMRT, associated with large transcriptional regulatory complexes (20, 39). Although different combinations of protein partners can determine their functional activity, these BCOR complexes occupy the BCL6 promoter region for BCL6 autoregulation as well as other target genes in B cells (40) and constrain immune responses (41). Also, the interplay between BCL6 and BCOR for interaction with IRF8 might modulate the activity of IRF8 as an activator or repressor in the germinal center reaction.
Additional considerations regarding the composition of the complex come from our studies showing that IRF8 represses Bcor transcription while promoting transcription of Bcl6. These and other aspects of our studies must be viewed with the understanding that although the eventual goal has been to define these interactions as they occur in GCs the vast majority of the data have been developed in vitro. Identifying interacting components of IRF8-BCOR complexes and developing new approaches for examining the GC reaction will clearly be important to future studies.
Acknowledgments
We thank Dr. Vivian Bardwell (University of Minnesota) for providing BCL6 reporter construct and discussions. We thank Dr. Keiko Ozato (National Institute of Child Health and Human Development, National Institutes of Health) for providing various truncated forms and GFP-tagged IRF8 plasmids.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NIAID.
- IRF
- interferon regulatory factor
- BCOR
- BCL6 corepressor
- BLC6
- B cell lymphoma 6
- GC
- germinal center
- RePCA
- retrovirus-based protein complementation assay
- BBD
- BCL6 binding domain
- IFP
- intensely fluorescent protein
- ANK
- ankyrin repeat
- qRT-PCR
- quantitative real time PCR
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- mBCOR
- mouse BCOR
- NCoR
- nuclear receptor corepressor
- SMRT
- silencing mediator of retinoid and thyroid hormone receptor
- BTB
- BR-C, ttk and bab
- POZ
- pox virus and zinc finger.
REFERENCES
- 1. Driggers P. H., Ennist D. L., Gleason S. L., Mak W. H., Marks M. S., Levi B. Z., Flanagan J. R., Appella E., Ozato K. (1990) An interferon γ-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. U.S.A. 87, 3743–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K. P., Gabriele L., Waring J. F., Bachmann M. F., Zinkernagel R. M., Morse H. C., 3rd, Ozato K., Horak I. (1996) Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 [DOI] [PubMed] [Google Scholar]
- 3. Gobin S. J., Biesta P., Van den Elsen P. J. (2003) Regulation of human β2-microglobulin transactivation in hematopoietic cells. Blood 101, 3058–3064 [DOI] [PubMed] [Google Scholar]
- 4. Yang D., Thangaraju M., Browning D. D., Dong Z., Korchin B., Lev D. C., Ganapathy V., Liu K. (2007) IFN regulatory factor 8 mediates apoptosis in nonhemopoietic tumor cells via regulation of Fas expression. J. Immunol. 179, 4775–4782 [DOI] [PubMed] [Google Scholar]
- 5. Tamura T., Nagamura-Inoue T., Shmeltzer Z., Kuwata T., Ozato K. (2000) ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13, 155–165 [DOI] [PubMed] [Google Scholar]
- 6. Tsujimura H., Nagamura-Inoue T., Tamura T., Ozato K. (2002) IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J. Immunol. 169, 1261–1269 [DOI] [PubMed] [Google Scholar]
- 7. Schiavoni G., Mattei F., Sestili P., Borghi P., Venditti M., Morse H. C., 3rd, Belardelli F., Gabriele L. (2002) ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bovolenta C., Driggers P. H., Marks M. S., Medin J. A., Politis A. D., Vogel S. N., Levy D. E., Sakaguchi K., Appella E., Coligan J. E., Ozato K. (1994) Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc. Natl. Acad. Sci. U.S.A. 91, 5046–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brass A. L., Kehrli E., Eisenbeis C. F., Storb U., Singh H. (1996) Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10, 2335–2347 [DOI] [PubMed] [Google Scholar]
- 10. Kuwata T., Gongora C., Kanno Y., Sakaguchi K., Tamura T., Kanno T., Basrur V., Martinez R., Appella E., Golub T., Ozato K. (2002) γ interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell. Biol. 22, 7439–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu C., Rao K., Xiong H., Gagnidze K., Li F., Horvath C., Plevy S. (2003) Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J. Biol. Chem. 278, 39372–39382 [DOI] [PubMed] [Google Scholar]
- 12. Wang H., Lee C. H., Qi C., Tailor P., Feng J., Abbasi S., Atsumi T., Morse H. C., 3rd (2008) IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood 112, 4028–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma S., Turetsky A., Trinh L., Lu R. (2006) IFN regulatory factor 4 and 8 promote Ig light chain κ locus activation in pre-B cell development. J. Immunol. 177, 7898–7904 [DOI] [PubMed] [Google Scholar]
- 14. Feng J., Wang H., Shin D. M., Masiuk M., Qi C. F., Morse H. C., 3rd (2011) IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol. 186, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee C. H., Melchers M., Wang H., Torrey T. A., Slota R., Qi C. F., Kim J. Y., Lugar P., Kong H. J., Farrington L., van der Zouwen B., Zhou J. X., Lougaris V., Lipsky P. E., Grammer A. C., Morse H. C., 3rd (2006) Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin D. M., Lee C. H., Morse H. C., 3rd (2011) IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS One 6, e27384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cattoretti G., Shaknovich R., Smith P. M., Jäck H. M., Murty V. V., Alobeid B. (2006) Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J. Immunol. 177, 6930–6939 [DOI] [PubMed] [Google Scholar]
- 18. Cattoretti G., Chang C. C., Cechova K., Zhang J., Ye B. H., Falini B., Louie D. C., Offit K., Chaganti R. S., Dalla-Favera R. (1995) BCL-6 protein is expressed in germinal-center B cells. Blood 86, 45–53 [PubMed] [Google Scholar]
- 19. Huynh K. D., Fischle W., Verdin E., Bardwell V. J. (2000) BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14, 1810–1823 [PMC free article] [PubMed] [Google Scholar]
- 20. Huynh K. D., Bardwell V. J. (1998) The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17, 2473–2484 [DOI] [PubMed] [Google Scholar]
- 21. Fujita N., Jaye D. L., Geigerman C., Akyildiz A., Mooney M. R., Boss J. M., Wade P. A. (2004) MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119, 75–86 [DOI] [PubMed] [Google Scholar]
- 22. Ding Z., Liang J., Lu Y., Yu Q., Songyang Z., Lin S. Y., Mills G. B. (2006) A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc. Natl. Acad. Sci. U.S.A. 103, 15014–15019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Ghetu A. F., Corcoran C. M., Cerchietti L., Bardwell V. J., Melnick A., Privé G. G. (2008) Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell 29, 384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatzi K., Melnick A. (2014) Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 20, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatzi K., Jiang Y., Huang C., Garrett-Bakelman F., Gearhart M. D., Giannopoulou E. G., Zumbo P., Kirouac K., Bhaskara S., Polo J. M., Kormaksson M., MacKerell A. D., Jr., Xue F., Mason C. E., Hiebert S. W., Prive G. G., Cerchietti L., Bardwell V. J., Elemento O., Melnick A. (2013) A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 4, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wamstad J. A., Bardwell V. J. (2007) Characterization of Bcor expression in mouse development. Gene Expr. Patterns 7, 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pathak S., Ma S., Shukla V., Lu R. (2013) A role for IRF8 in B cell anergy. J. Immunol. 191, 6222–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang H., Morse H. C., 3rd (2009) IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 43, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez A., Pittaluga S., Rudelius M., Davies-Hill T., Sebasigari D., Fountaine T. J., Hewitt S., Jaffe E. S., Raffeld M. (2008) Expression of the interferon regulatory factor 8/ICSBP-1 in human reactive lymphoid tissues and B-cell lymphomas: a novel germinal center marker. Am. J. Surg. Pathol. 32, 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chrabot B. S., Kariuki S. N., Zervou M. I., Feng X., Arrington J., Jolly M., Boumpas D. T., Reder A. T., Goulielmos G. N., Niewold T. B. (2013) Genetic variation near IRF8 is associated with serologic and cytokine profiles in systemic lupus erythematosus and multiple sclerosis. Genes Immun. 14, 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baccala R., Gonzalez-Quintial R., Blasius A. L., Rimann I., Ozato K., Kono D. H., Beutler B., Theofilopoulos A. N. (2013) Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc. Natl. Acad. Sci. U.S.A. 110, 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bouamar H., Abbas S., Lin A. P., Wang L., Jiang D., Holder K. N., Kinney M. C., Hunicke-Smith S., Aguiar R. C. (2013) A capture-sequencing strategy identifies IRF8, EBF1, and APRIL as novel IGH fusion partners in B-cell lymphoma. Blood 122, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J., Grubor V., Love C. L., Banerjee A., Richards K. L., Mieczkowski P. A., Dunphy C., Choi W., Au W. Y., Srivastava G., Lugar P. L., Rizzieri D. A., Lagoo A. S., Bernal-Mizrachi L., Mann K. P., Flowers C., Naresh K., Evens A., Gordon L. I., Czader M., Gill J. I., Hsi E. D., Liu Q., Fan A., Walsh K., Jima D., Smith L. L., Johnson A. J., Byrd J. C., Luftig M. A., Ni T., Zhu J., Chadburn A., Levy S., Dunson D., Dave S. S. (2013) Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110, 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei F., Zaprazna K., Wang J., Atchison M. L. (2009) PU.1 can recruit BCL6 to DNA to repress gene expression in germinal center B cells. Mol. Cell. Biol. 29, 4612–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakano N., Nishiyama C., Masuoka N., Nishiyama M., Yamane H., Okumura K., Ogawa H. (2005) Analysis of PU.1/ICSBP (IRF-8) complex formation with various PU.1 mutants: molecular cloning of rat Icsbp (Irf-8) cDNA. Immunogenetics 56, 871–877 [DOI] [PubMed] [Google Scholar]
- 37. Dror N., Rave-Harel N., Burchert A., Azriel A., Tamura T., Tailor P., Neubauer A., Ozato K., Levi B. Z. (2007) Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J. Biol. Chem. 282, 5633–5640 [DOI] [PubMed] [Google Scholar]
- 38. Tsujimura H., Tamura T., Gongora C., Aliberti J., Reis e Sousa C., Sher A., Ozato K. (2003) ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood 101, 961–969 [DOI] [PubMed] [Google Scholar]
- 39. Dhordain P., Albagli O., Lin R. J., Ansieau S., Quief S., Leutz A., Kerckaert J. P., Evans R. M., Leprince D. (1997) Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. U.S.A. 94, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gearhart M. D., Corcoran C. M., Wamstad J. A., Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barish G. D., Yu R. T., Karunasiri M. S., Becerra D., Kim J., Tseng T. W., Tai L. J., Leblanc M., Diehl C., Cerchietti L., Miller Y. I., Witztum J. L., Melnick A. M., Dent A. L., Tangirala R. K., Evans R. M. (2012) The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 15, 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]