Abstract
Intervertebral disc (IVD) homeostasis is mediated through a combination of micro-environmental and biomechanical factors, all of which are subject to genetic influences. The aim of this study is to develop and characterize a genetically tractable, ex vivo organ culture model that can be used to further elucidate mechanisms of intervertebral disc disease. Specifically, we demonstrate that IVD disc explants (1) maintain their native phenotype in prolonged culture, (2) are responsive to exogenous stimuli, and (3) that relevant homeostatic regulatory mechanisms can be modulated through ex-vivo genetic recombination. We present a novel technique for isolation of murine IVD explants with demonstration of explant viability (CMFDA/propidium iodide staining), disc anatomy (H&E), maintenance of extracellular matrix (ECM) (Alcian Blue staining), and native expression profile (qRT-PCR) as well as ex vivo genetic recombination (mT/mG reporter mice; AdCre) following 14 days of culture in DMEM media containing 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. IVD explants maintained their micro-anatomic integrity, ECM proteoglycan content, viability, and gene expression profile consistent with a homeostatic drive in culture. Treatment of genetically engineered explants with cre-expressing adenovirus efficaciously induced ex vivo genetic recombination in a variety of genetically engineered mouse models. Exogenous administration of IL-1ß and TGF-ß3 resulted in predicted catabolic and anabolic responses, respectively. Genetic recombination of TGFBR1fl/fl explants resulted in constitutively active TGF-ß signaling that matched that of exogenously administered TGF-ß3. Our results illustrate the utility of the murine intervertebral disc explant to investigate mechanisms of intervertebral disc degeneration.
Introduction
Low back pain (LBP) is a significant healthcare concern worldwide [1] and is a leading reason for seeking medical attention with the lifetime prevalence approaching 84% [2]. Degeneration of the intervertebral disc (IVD) is a major contributing factor to LBP. The presence of individuals over the age of 50 with evidence of IVD degeneration (IVDD) approaches 90%, correlating with the lifetime prevalence of LBP [3].
The IVD is the cartilaginous joint between two vertebral bodies. It is comprised of an outer ring of collagen lamellae, the annulus fibrosus (AF), an inner gelatinous nucleus pulposus (NP), and two cartilaginous end plates (CEP) lying adjacent to the superior and inferior vertebral bodies. The AF is comprised primarily of type I collagen, while the NP is comprised of type II collagen and proteoglycans, principally aggrecan [4].
Due to the complex structure and distinct characteristics of the IVD tissues, challenges in translating in vitro assays to overall organ function persist. Monolayer culture expansion of both AF and NP tissue results in down regulation of collagen and proteoglycan production compared to freshly isolated tissue. However, phenotypic genetic expression can be restored with sophisticated micro-mass culture techniques [5], [6]. While this has been instrumental in deciphering the specific cellular characteristics of a particular IVD tissue, mechanisms of IVD organ homeostasis remains elusive. Certainly, dysfunction of a particular IVD constituent will affect the function of another under in vivo conditions [7], [8].
To assess IVD organ function, ex vivo whole IVD organ culture models have been developed. These explant models provide a useful tool to study the IVD. Insofar, rabbit, rat, sheep, bovine, and human explants have been investigated [9], [10], [11], [12], [13], [14], [15], [16], [17]. Functional assays utilizing these models have provided insight into cellular response and resultant anatomic changes in conditions simulating degeneration in vivo [18] and have shed light on the etiology of IVDD [19]. While these models are useful constructs to study the IVD, inherent difficulty lies in deciphering genetic mechanisms of IVD degeneration. Classic twin studies have estimated the heritability index of IVDD to be 74% in the lumbar spine [20]. Indeed, over 20 genetic polymorphisms have been identified in association with IVDD, reviewed in [21]. These data underscore the need to account for both genetic and micro-environmental factors when studying the IVD. Therefore, in comparison to other IVD organ culture models, an advantageous novel IVD organ culture model would require the potential for genetic manipulation and ability to evaluate functional organ response. Herein, we have developed a genetically tractable murine IVD explant culture. What follows is a characterization of the murine IVD as an organ culture model, verification of ex vivo genetic tractability, and examples of utility in functional assays. We also provide a demonstration of coupling genetic engineered models with functional IVD organ response, ex vivo.
Methods
Mice
Van Andel Institute Institutional Animal Care and Use Committee approved this study. Experiments were carried out according to institutional animal care and use committee protocols. Wild-type mice from c57bl/6, FVB, and mixed background were utilized. For experiments utilizing transgenic mice, mT/mG, TGFßR1fl/fl, and NF1fl/fl were used. mT/mG reporter mice fluoresce red at baseline; however, when exposed to cre-recombinase, they express green fluorescence protein and are useful to visually track cellular genetic recombination events [22]. TGFßR1fl/fl mice constitutively activate TGFß receptor 1 when exposed to cre-recombinase [23] while NF1fl/fl mice under the same conditions exhibit loss of neurofibromatosis1 function [24] and have demonstrable effects on the IVD [25].
Explant harvest
Mice were euthanized with carbon dioxide asphyxiation and washed with ethanol. The skin was dissected free from the anterior abdomen and thorax and subsequently discarded. The peritoneal sac was removed en bloc. The contents of the thoracic cavity were removed and the ribs transected transversely at their midpoint for increased exposure. With the aid of a dissecting scope (Leica) the prevertebral tissue and psoas muscles were removed, exposing the anterior spine (figure 1a). The anterior longitudinal ligament was removed with dissecting scissors and an 11 blade scalpel. The junction of the bony endplate (BEP) and CEP was identified. An 11 blade scalpel was used to transect the superior CEP from the BEP (figure 1b). The inferior vertebral body was transected through cortical bone in the upper third region. The IVD and remaining inferior vertebral body was removed from the spine. A scalpel was used to remove the remaining bony tissue from the IVD. The IVD was inspected with the dissection microscope for gross anatomic integrity. Slight anterior to posterior compression was applied with forceps; if any extravasation of NP tissue occurred during compression, the IVD was discarded. IVDs that passed compressive inspections were immediately placed in ice cold PBS. Individual discs were then placed in Dulbecco's Modified Eagle Medium containing 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. Media was changed every other day; explants were cultured for up to 14 days.
Figure 1. Method of IVD organ harvest for ex vivo culture.
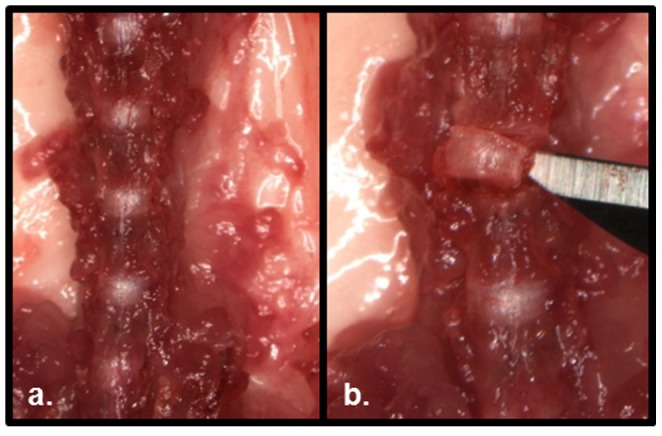
(a) Anterior spine is exposed after removal of peritoneal sac and paraspinal musculature. (b) Excision of a lumbar IVD after the superior and inferior extents were freed with sharp dissection at 4× magnification.
Histology
For histological analysis, explant IVDs were fixed with 10% neutral buffered formalin for 24 hours, embedded in paraffin blocks, and 5 µm sections were prepared. Slides were de-paraffinized in Citrisolv (Fisher; Pittsburgh, PA), rehydrated through ethanol series and stained with hematoxylin and eosin or alcian blue.
Cell viability
Explant IVDs were incubated with CellTracker Green CMFDA (5-Chloromethylfluorescein Diacetate) (Invitrogen) and propidium iodide (Sigma-Aldrich) according to manufacturer's protocol. Explants were rotated for 10 minutes at 4 degrees Celsius. Explant IVDs were then incubated at 37 degrees Celsius and 5% CO2 for 1 hour. 1/2 of explants were incubated in the presence of 70% methanol, serving as a positive control for cellular death. After fixing overnight at 4 degrees Celsius in 10% neutral buffered formalin, explants were dehydrated by immersion in 15% and 30% sucrose solutions for 24 hours at 4 degrees Celsius. Explants were imbedded in O.C.T. compound (Tissue-Tek) and frozen over dry ice. IVDs were kept at −80 degrees Celsius until sectioning. Sections were cut in 5 µm sections and imaged on a digital inverted fluorescent microscope (EVOSfl, fisher scientific).
Allele-specific PCR
Genomic DNA was harvested from tissues using DirectPCR reagent (Viagen) according to the manufacturer's instructions. PCR reactions were performed with Syzygy MeanGreen Taq Master Mix (Syzygy Biotech) in 20 µl reactions with 1 µl crude DNA lysate. Primers designed to detect the mutated NF1flx allele (flanked by loxp sites, 350 base pair amplicon) and cre-recombined NF1 allele (280 base pair amplicon) and cycling conditions were used exactly as previously described [24].
Quantitative real-time PCR
Total RNA was extracted from explant IVDs using TRIzol reagent (Invitrogen). Synthesis of cDNA was performed using 1 µg of RNA according to the High Capacity cDNA Reverse Transcription Kit (Invitrogen). Primers (table 1) were obtained from Integrated DNA technologies. Quantitative real-time PCR reactions were performed using SYBR, Select Mastermix (Applied Biosystems) in 10 ul reactions. PCR was performed according to manufacturer's instructions using a StepOnePlus cycler (Applied Biosystems; Carlsbad, CA). Relative gene expression was normalized to GAPDH, measured by quantitative real-time PCR, and expressed using the ΔΔCT method. The paired Student's t test was utilized to identify statistically significant differences in gene expression; significance was set to a p value <0.05.
Table 1. Murine primers for genes analyzed.
| Primer | Forward Sequence | Reverse Sequence |
| GAPDH | 5′-AACTTTGGCATTGTGGAAGG-3′ | 5′-GGATGCAGGGATGATGTTCT-3′ |
| ACAN | 5′-ACCCGGTACCCTACAGAGAC-3′ | 5′-GTCCACCCCTCCTCACATTG-3′ |
| ADAMTS4 | 5′-GAGTCCCATTTCCCGCAGAA-3′ | 5′-ATAACCGTCAGCAGGTAGCG-3′ |
| COL1 | 5′-CTGACGCATGGCCAAGAAGA-3′ | 5′-TCTCACCATTGGGGACCCTT-3′ |
| COL2 | 5′-CATCTTGCCGCATCTGTGTG-3′ | 5′-TGCCCCTTTGGCCCTAATTT-3′ |
| TIMP1 | 5′-GATACCATGATGGCCCCCTTT-3′ | 5′-CGCTGGTATAAGGTGGTCTCG-3′ |
| TIMP2 | 5′-CAGGTACCAGATGGGCTGTG-3′ | 5′-TGGTGCCCATTGATGCTCTT-3′ |
Explant treatments
Explants were harvested and cultured in accordance with the aforementioned conditions. After 48 hours of culture, explants were treated with IL-1ß at 10 ng/mL for 12 or 24 hours to simulate degenerative conditions. For cultures simulating anabolic conditions, TGF-ß3 at 10 ng/mL for 24 hours was utilized.
For genetic recombination experiments in mT/mG [22], NF1fl/fl mice, and TGFßR1fl/fl explants were harvested. Following 24 hours of serum and antibiotic free culture, explants were treated with either cre-expressing adenovirus (5×107 pfu/mL) or a non-cre-expressing adenovirus control (5×107 pfu/mL). Serum was replaced to the culture conditions after 24 hours. At 48 hours, the explants were washed and cultured according to the aforementioned protocol. Explants were harvested from culture at 10 days and used for experimentation.
Western blot analysis
Explants were then washed in ice cold PBS and sharply morsilized with a 15 blade scalpel in the presence of lysis buffer (RIPA lysis buffer) supplemented with complete Protease Inhibitor Cocktail (Roche). Protein was measured using the BCA assay (Pierce), and 20 µg of explant lysate was run on a 10% SDS/polyacrylamide gel. The proteins were transferred onto a nitrocellulose membrane, and membranes were probed overnight at 4 degrees Celsius with primary antibody. P44/P42(MAPK) 1∶500 (cell signaling), p-P44/P42 (activated MAPK) 1∶500 (Cell Signaling), primary antibodies were utilized. Membranes were then probed with horseradish-peroxidase-conjugated secondary antibody for 1.5 hours at room temperature before detection using an enhanced chemiluminescence (ECL) detection system (Pierce).
Confocal microscopy
Explants of mT/mG mice were treated with cre-expressing adenovirus or non-cre-expressing adenovirus, as described above. Following culture for 10 days, explants were then fixed overnight at 4 degrees Celsius in 10% neutral buffered formalin, dehydrated by immersion in 15% and 30% sucrose solutions for 24 hours at 4 degrees Celsius, imbedded in O.C.T. compound (Tissue-Tek) and frozen over dry ice. IVDs were kept at −80 degrees Celsius until sectioning. Sections were cut in 5 µm sections. Slides were dried and mounted in Vectashield (Vector Labs). All images were obtained using a 20× objective on an A1plus confocal system (Nikon) with GFP and Tomato fluorescence imaged in series. Maximum intensity projection-based Z-stack image rendering was performed in NIS-Elements software (Nikon).
Results
Explants retain micro-anatomic integrity and cellular viability in ex vivo culture
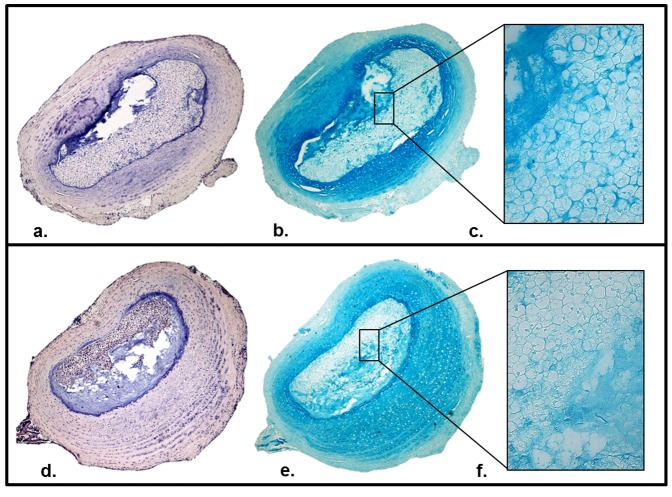
Explants were harvested and cultured for 14 days. Histological comparison of freshly harvested and cultured explants reveals a retained integrity of micro-anatomic features (figure 2). The AF and NP were clearly demarcated. The collagen lamellar bundles of the AF were preserved in culture. The NP stained positively for proteoglycan. After 14 days of culture, the cellular architecture of the NP demonstrated characteristic vacuolated cells when examined under 40× magnification.
Figure 2. Histological analysis of explant micro-anatomic features.
IVDs are shown in axial slices. (top pane) Freshly harvested murine IVDs. (bottom pane) Murine IVDs after 14 days of ex vivo culture. (a,d) Hematoxylin and eosin (b,e) Alcian blue. (c,f) 40× magnification of alcian blue stained NP.
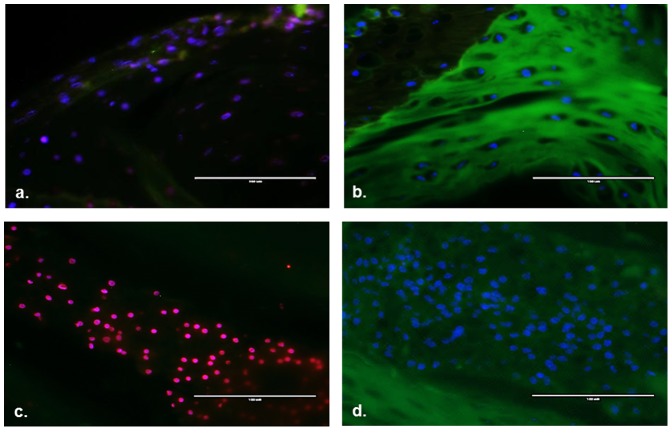
Explants incubated with CMFDA, a cell permeable chemical that live cells will convert to a cell impermeable green fluorescent compound [26], and PI, a DNA binding red fluorescent dye impermeable to living cells [27], were imaged on a digital inverted fluorescent microscope. IVDs treated with 70% methanol demonstrated strong PI uptake and no CMFDA signal, indicative of widespread cellular death (figure 3). Explants incubated at 14 days did not demonstrate appreciable PI uptake in either the NP or AF. CMFDA uptake was noted in both the AF and NP of cultured explants.
Figure 3. Cell viability in 14 day explant organ culture.
Sections were stained with propidium iodide (red), Cell tracker green (green), and DAPI nuclear co-stain. Annulus fibrosus tissue section after treatment with 70% methanol (positive control) for 24 hours (a) and after 14 days of ex vivo organ culture (b). Nucleus pulposus tissue section after treatment with 70% methanol for 24 hours (c) and after 14 days of ex-vivo organ culture (d). Purple staining is indicative of PI and DAPI counterstaining.
Explants demonstrate characteristic gene expression patterns in ex vivo culture
We chose to compare gene expression of COL1, the principal extracellular matrix protein of the AF, as well as ACAN and COL2, the principal extracellular matrix proteins of the NP, between 14 d explants and freshly harvested IVDs. No statistically significant difference in gene expression was identified in COL1 (fold change: 0.7; 0.5–1.0; p = 0.3), COL2 (fold change: 1.6; 0.7–2.3; p = 0.5), and ACAN (fold change: 0.8; 0.7–0.8; p = 0.7) (figure 4a). Next, we assessed the gene expression of the protease ADAMTS4 and the protease inhibitors: TIMP1 and TIMP2 (figure 4b). We found a statistically significant increase in both the tissue protease ADAMTS4 (fold change: 4.9; 3.9–5.2; p = 0.007) and the protease inhibitor TIMP1 (fold change: 8.1; 5.7–11.8; p = 0.01), indicating that homeostatic mechanisms remain intact at day 14 in the cultured explants.
Figure 4. Real-time PCR analysis of 14 day murine IVD explants compared to freshly harvested IVD explants.
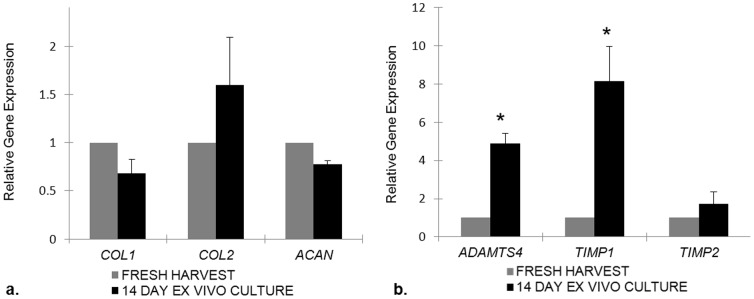
Genes related to structural extracellular matrix production (a) and extracellular matrix remodeling genes (b) were analyzed. After 14 days in culture, murine IVD explants do not change the gene expression of the principal structural genes of the IVD, while ADAMTS4 and TIMP1 are significantly upregulated. Analysis was normalized to GAPDH. Results are presented relative to freshly harvested murine IVDs where the expression value was set to 1. * denotes statistical significance (p<0.05).
IVD genetic recombination is successful in ex vivo culture conditions
Wild type and mT/mG reporter explants were treated with cre-expressing adenovirus and control adenovirus. Both untreated and adenovirus controls failed to demonstrate GFP fluorescence conversion, while explants treated with cre-expressing adenovirus demonstrated evidence of genetic recombination ex vivo with GFP signal demonstrated in both the AF and NP (figure 5).
Figure 5. Demonstration of ex vivo murine IVD genetic recombination.
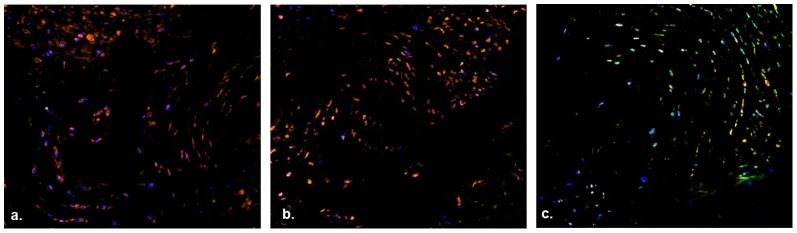
mT/mG mice IVDs were cultured for 10 days in the presence of (a) standard culture conditions, (b) control adenovirus, or (c) cre-recombinase expressing adenovirus. Images are 20× magnification Slides are counterstained with DAPI (blue). Red fluorescence represents no genetic recombination, green fluorescence represents successful genetic recombination, and yellow fluorescence represents an ongoing genetic recombination event.
To further assess the ability of the murine explants to undergo genetic ex vivo recombination, NF1fl/fl IVD explants were cultured with cre-expressing adenovirus. Individual IVDs were subjected to allele specific PCR after culture. All individual IVDs cultured with cre-expressing adenovirus demonstrated evidence of genetic recombination (figure S1).
Explant Response to IL-1ß
IL-1ß is a pro-catabolic factor in IVDD [28], [29] and has been shown to stimulate IVDD both in vitro and in vivo experiments [30], [31], [32]. Upon challenge with IL-1ß (10 ng/ml), wild-type IVD explants demonstrate a significant down regulation of COL2 (fold change: 0.07; 0.05–0.08; p = 7.4×10−11) and ACAN (fold change: 0.3; 0.2–0.5; p = 3.0×10−5) while ADAMTS4 (fold change: 3.1; 2.5–4.2; p = 0.009) gene expression was upregulated (figure 6a). Exogenous IL-1ß administration induced sustained MAPK phosphorylation at 14 days (figure 6b) [33].
Figure 6. IVD explant response to IL-1ß.
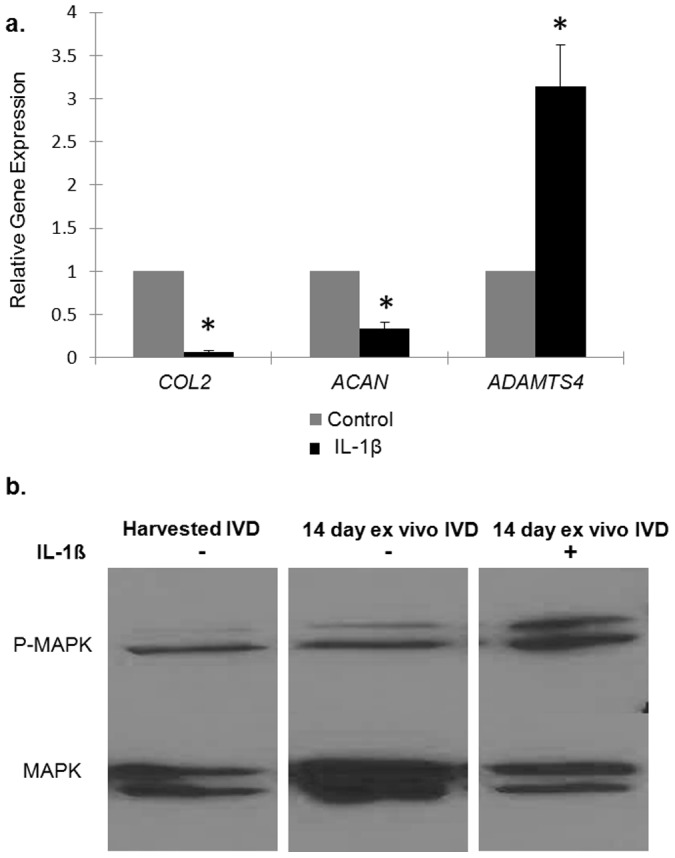
(a) When treated with IL-1ß at 10 ng/mL for 12 hours, IVD explants demonstrate significant decreases in gene expression of COL2 (p = 7.4×10−11) and ACAN (p = 3.0×10−5); ADAMTS4 (p = 0.009) gene expression is increased. Values are normalized to GAPDH. Results are presented relative to IVDs cultured without IL-1ß where the expression value was set to 1 (b) Murine IVD explants demonstrate increased MAPK phosphorylation when treated with IL-1ß at 10 ng/mL for 24 hours. * denotes statistical significance (p<0.05).
Explant Response to TGF-ß
We chose to investigate TGF-ß for its stimulatory and protective effects on the IVD [34], [35], [36], [37]. Explants treated with exogenous TGF-ß3 (10 ng/mL) demonstrated statistically significant gene expression upregulation of ACAN (fold change: 2.0; 1.8–2.5; p = 0.005) and TIMP1 (fold change: 3.1; 2.7–3.4; p = 0.007), implicating an anabolic and protective effect within the cultured IVD explant (figure 7a). COL2 gene expression demonstrated a more variable response to TGF-ß3 but trended towards upregulation (fold change 4.7; 2.1–7.4; p = 0.06).
Figure 7. IVD explant response to TGF-ß.
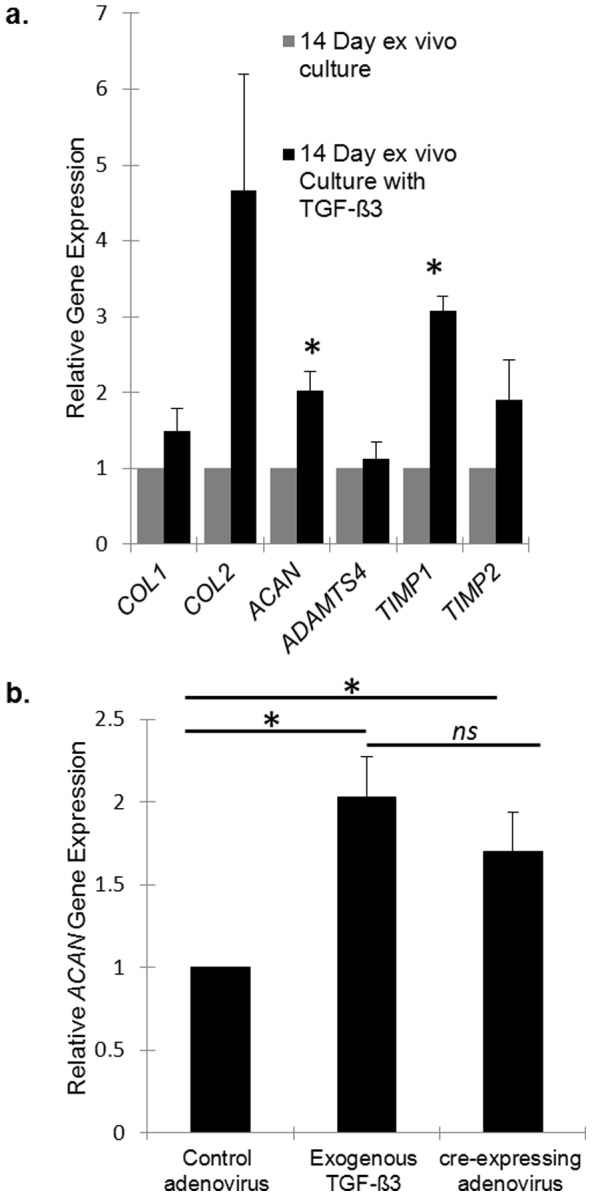
(a) Real-time PCR analysis of murine IVD explant gene expression response to stimulation with exogenously administered TGF-ß3 (10 ng/mL for 24 hours) demonstrates significant increase in the gene expressions of ACAN and TIMP1. Values are normalized to GAPDH. Results are presented relative to IVDs cultured without TGF-ß3 where the expression value was set to 1. Values are normalized to GAPDH. (b) Murine IVDs from TGFßR1fl/fl demonstrate significant upregulation in ACAN gene expression when cultured with cre-expressing adenovirus. Exogenous TGF-ß3 at 10 ng/mL for 24 hours was cultured with wild-type murine IVDs. Results are presented relative to IVDs cultured with control adenovirus where the expression value was set to 1. Values are normalized to GAPDH. * denotes statistical significance (p<0.05). ns denotes not statistically significant (p>0.05).
To evaluate whether IVD explants could be used to couple genetic recombination into a functional response, TGF-ßR1fl/fl mice were utilized. Tissues in TGF-ßR1fl/fl mice demonstrate constitutive activation of the TGF-ß1 receptor following recombination [23]. Aggrecan gene expression was significantly upregulated in treated TGF-ßR1fl/fl explants (fold change: 1.7; 1.0–2.2; p = 0.01) compared to controls (figure 7b). No difference in ACAN expression was observed between explants treated with exogenous TGF-ß3 and the recombined TGF-ßR1fl/fl explants (p = 0.4).
Discussion
Herein, we describe a novel technique for utilizing murine IVD explants as a functional unit to study IVD homeostasis and degeneration. We demonstrate that the IVD can be harvested and maintained as an organ unit with its native phenotype intact. We also demonstrate a high degree of genetic tractability in the model as we successfully recombined explants harvested from mice bearing conditional mutations (TGFßR1fl/fl, mT/mG; and NF1fl/fl). Given the recent advances in genetically-engineered mouse models [38], [39], the impact of genetic factors on IVDD progression can now be studied in greater detail.
We also demonstrate that ex vivo murine explants maintain their ability to respond to both catabolic and anabolic signals based on expression profiling of key structural and extracellular matrix genes. While there is upregulation of the aggrecanase ADAMTS4 over time in culture, there appears to be a compensatory increase in the metalloproteinase inhibitor TIMP1, indicating that IVD explants retain a homeostatic drive under ex vivo conditions. In human tissue, aggrecanase expression is upregulated in degenerative IVDs [40]. The murine IVD may be a useful model to help elucidate the mechanistic role aggrecanases contribute to IVDD as they are candidate drug targets in other models of arthritis [41], [42]. It is worth noting that polymorphisms in the aggrecan gene are associated with the development of IVDD [43] and herniated discs are common in aggrecan(+/−) mice [44].
Additionally, we induced a degenerative state under ex vivo culture conditions using exogenous IL-1ß, which is known to be upregulated in degenerative human IVDs [29], [45]. Our findings parallel those of others insofar as murine explants demonstrate downregulated aggrecan gene expression, upregulated ADAMTS4 gene expression, and increased activated MAPK protein expression in response to IL-1ß stimulation [32]. Genetically engineered murine IVD explants may be a useful adjunct to characterize how the organ as a whole responds to inflammatory signals.
It is important to recognize current IVD explant models [9], [10], [11], [12], [13], [14], [15], [16], [17]. Because a strong genetic influence in IVDD exists [20], [21], [46], an ex vivo model of IVDD that allows efficient manipulation of both genetic and micro-environmental parameters will help broaden our understanding of targetable mechanisms underlying IVDD. As proof of concept, we assessed the murine IVD explant response to TGF-ß signaling. Exogenous TGF-ß3 treatment in rat explants resulted in upregulated expression of aggrecan and decreased degradation products [10]. Additionally, endogenous TGF-ß expression in disc tissue was shown to be protective by suppressing an inflammatory state [34] and upregulating connective tissue growth factor [35]. Our results with exogenous TGF-ß3 treatment confirm upregulated ACAN and TIMP1 gene expression, consistent with previous studies in whole IVD explants [10]. Furthermore, by utilizing the TGFßR1fl/fl genetic construct, we also demonstrated significant upregulation of ACAN gene expression as a direct result of constitutive TGF-ßR1 activation. These data confirm that by activating TGF-ß signaling at the receptor-level, either through direct administration of ligand or genetic recombination, we were able to increase gene expression of the principal NP proteoglycan, aggrecan.
It is important to recognize the limitations of our model and how other models may be better suited for certain investigations. While we can manipulate micro-environmental and genetic factors to recapitulate disc degeneration over 14 days, IVDD in humans is a chronic process spanning many years. Therefore, observations gleaned with the murine IVD are limited in this regard and would certainly benefit from human corollary data. Additionally, as the size and geometry of larger animal explant models more closely resemble the human IVD, biomechanical testing may be better suited for these models [11], [16]. Experimental surgical interventions are well studied in larger animal models and would be difficult to perform in the murine spine. Reproducible IVDD with needle puncture of the AF is well characterized in the rabbit spine [47] and in vivo biophysical parameter measurements, such as intradiscal pressure, have been measured in the ovine spine [48].
Conclusions
The murine IVD explant is a useful tool to study the IVD. Under standard culture conditions the microanatomy of the IVD remains intact, the cells are viable, and the genetic expression pattern is consistent with an intact homeostatic drive. The murine IVD explant can be manipulated to induce a degenerative state or protective state. It can also be readily modified to reflect the diverse genetic factors underlying IVDD. We expect that this model will facilitate a more complete understanding of the complex mechanisms underlying IVDD.
Supporting Information
(a) IVDs from wild-type (WT) and NF1fl/fl mice were genotyped with allele specific PCR. (b) IVDs from wild-type (control) and NF1fl/fl were cultured with cre-expressing adenovirus. Allele specific PCR demonstrates successful ex vivo genetic recombination in all IVDs tested.
(TIF)
Acknowledgments
Special thanks to Dr. Xiaohong Li for the generous gift of TGFßR1fl/fl mice.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information Files.
Funding Statement
This work was supported by the Jay and Betty Van Andel Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoy D, Bain C, Williams G, March L, Brooks P, et al. (2012) A systematic review of the global prevalence of low back pain. Arthritis and rheumatism 64:2028–2037. [DOI] [PubMed] [Google Scholar]
- 2. Cassidy JD, Carroll LJ, Cote P (1998) The Saskatchewan health and back pain survey. The prevalence of low back pain and related disability in Saskatchewan adults. Spine 23:1860–1866 discussion 1867. [DOI] [PubMed] [Google Scholar]
- 3. Cheung KM, Karppinen J, Chan D, Ho DW, Song YQ, et al. (2009) Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty-three individuals. Spine 34:934–940. [DOI] [PubMed] [Google Scholar]
- 4. Choi KS, Cohn MJ, Harfe BD (2008) Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Developmental dynamics: an official publication of the American Association of Anatomists 237:3953–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith LJ, Chiaro JA, Nerurkar NL, Cortes DH, Horava SD, et al. (2011) Nucleus pulposus cells synthesize a functional extracellular matrix and respond to inflammatory cytokine challenge following long-term agarose culture. European cells & materials 22:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, et al. (2012) Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nature communications 3:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dahia CL, Mahoney E, Wylie C (2012) Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PloS one 7:e35944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang M, Tang D, Shu B, Wang B, Jin H, et al. (2012) Conditional activation of beta-catenin signaling in mice leads to severe defects in intervertebral disc tissue. Arthritis and rheumatism 64:2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haschtmann D, Stoyanov JV, Ettinger L, Nolte LP, Ferguson SJ (2006) Establishment of a novel intervertebral disc/endplate culture model: analysis of an ex vivo in vitro whole-organ rabbit culture system. Spine 31:2918–2925. [DOI] [PubMed] [Google Scholar]
- 10. Risbud MV, Di Martino A, Guttapalli A, Seghatoleslami R, Denaro V, et al. (2006) Toward an optimum system for intervertebral disc organ culture: TGF-beta 3 enhances nucleus pulposus and anulus fibrosus survival and function through modulation of TGF-beta-R expression and ERK signaling. Spine 31:884–890. [DOI] [PubMed] [Google Scholar]
- 11. Chiba K, Andersson GB, Masuda K, Momohara S, Williams JM, et al. (1998) A new culture system to study the metabolism of the intervertebral disc in vitro. Spine 23:1821–1827 discussion 1828. [DOI] [PubMed] [Google Scholar]
- 12. Seol D, Choe H, Ramakrishnan PS, Jang K, Kurriger GL, et al. (2013) Organ culture stability of the intervertebral disc: rat versus rabbit. Journal of orthopaedic research: official publication of the Orthopaedic Research Society 31:838–846. [DOI] [PubMed] [Google Scholar]
- 13. Chan SC, Gantenbein-Ritter B (2012) Preparation of intact bovine tail intervertebral discs for organ culture. Journal of visualized experiments: JoVE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts S, Menage J, Sivan S, Urban JP (2008) Bovine explant model of degeneration of the intervertebral disc. BMC musculoskeletal disorders 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korecki CL, MacLean JJ, Iatridis JC (2007) Characterization of an in vitro intervertebral disc organ culture system. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society 16:1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gantenbein B, Grunhagen T, Lee CR, van Donkelaar CC, Alini M, et al. (2006) An in vitro organ culturing system for intervertebral disc explants with vertebral endplates: a feasibility study with ovine caudal discs. Spine 31:2665–2673. [DOI] [PubMed] [Google Scholar]
- 17. Parolin M, Gawri R, Mwale F, Steffen T, Roughley P, et al. (2010) Development of a whole disc organ culture system to study human intervertebral disc. Evidence-based spine-care journal 1:67–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ponnappan RK, Markova DZ, Antonio PJ, Murray HB, Vaccaro AR, et al. (2011) An organ culture system to model early degenerative changes of the intervertebral disc. Arthritis research & therapy 13:R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markova DZ, Kepler CK, Addya S, Murray HB, Vaccaro AR, et al. (2013) An organ culture system to model early degenerative changes of the intervertebral disc II: profiling global gene expression changes. Arthritis research & therapy 15:R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sambrook PN, MacGregor AJ, Spector TD (1999) Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis and rheumatism 42:366–372. [DOI] [PubMed] [Google Scholar]
- 21. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, et al. (2013) Genetic polymorphisms associated with intervertebral disc degeneration. The spine journal: official journal of the North American Spine Society 13:299–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double-fluorescent Cre reporter mouse. Genesis 45:593–605. [DOI] [PubMed] [Google Scholar]
- 23. Sonnylal S, Denton CP, Zheng B, Keene DR, He R, et al. (2007) Postnatal induction of transforming growth factor beta signaling in fibroblasts of mice recapitulates clinical, histologic, and biochemical features of scleroderma. Arthritis and rheumatism 56:334–344. [DOI] [PubMed] [Google Scholar]
- 24. Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, et al. (2001) Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes & development 15:859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang W, Nyman JS, Ono K, Stevenson DA, Yang X, et al. (2011) Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Human molecular genetics 20:3910–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J (2009) Microfluidic control of cell pairing and fusion. Nature methods 6:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muders MH, Zhang H, Wang E, Tindall DJ, Datta K (2009) Vascular endothelial growth factor-C protects prostate cancer cells from oxidative stress by the activation of mammalian target of rapamycin complex-2 and AKT-1. Cancer research 69:6042–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le Maitre CL, Hoyland JA, Freemont AJ (2007) Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis research & therapy 9:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kepler CK, Markova DZ, Dibra F, Yadla S, Vaccaro AR, et al. (2013) Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL-1beta in painful human intervertebral discs. Spine 38:873–880. [DOI] [PubMed] [Google Scholar]
- 30. Wang J, Tian Y, Phillips KL, Chiverton N, Haddock G, et al. (2013) Tumor necrosis factor alpha- and interleukin-1beta-dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis and rheumatism 65:832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phillips KL, Jordan-Mahy N, Nicklin MJ, Le Maitre CL (2013) Interleukin-1 receptor antagonist deficient mice provide insights into pathogenesis of human intervertebral disc degeneration. Annals of the rheumatic diseases 72:1860–1867. [DOI] [PubMed] [Google Scholar]
- 32. Tian Y, Yuan W, Fujita N, Wang J, Wang H, et al. (2013) Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-kappaB. The American journal of pathology 182:2310–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wuertz K, Vo N, Kletsas D, Boos N (2012) Inflammatory and catabolic signalling in intervertebral discs: the roles of NF-kappaB and MAP kinases. European cells & materials 23:103–119 discussion 119-120. [DOI] [PubMed] [Google Scholar]
- 34. Zhu Y, Ohba T, Ando T, Fujita K, Koyama K, et al. (2013) Endogenous TGF-beta activity limits TSLP expression in the intervertebral disc tissue by suppressing NF-kappaB activation. Journal of orthopaedic research: official publication of the Orthopaedic Research Society 31:1144–1149. [DOI] [PubMed] [Google Scholar]
- 35. Tran CM, Markova D, Smith HE, Susarla B, Ponnappan RK, et al. (2010) Regulation of CCN2/connective tissue growth factor expression in the nucleus pulposus of the intervertebral disc: role of Smad and activator protein 1 signaling. Arthritis and rheumatism 62:1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin H, Shen J, Wang B, Wang M, Shu B, et al. (2011) TGF-beta signaling plays an essential role in the growth and maintenance of intervertebral disc tissue. FEBS letters 585:1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baffi MO, Slattery E, Sohn P, Moses HL, Chytil A, et al. (2004) Conditional deletion of the TGF-beta type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Developmental biology 276:124–142. [DOI] [PubMed] [Google Scholar]
- 38. Inui M, Miyado M, Igarashi M, Tamano M, Kubo A, et al. (2014) Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Scientific reports 4:5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharpless NE, Depinho RA (2006) The mighty mouse: genetically engineered mouse models in cancer drug development. Nature reviews Drug discovery 5:741–754. [DOI] [PubMed] [Google Scholar]
- 40. Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, et al. (2013) Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. The spine journal: official journal of the North American Spine Society 13:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fosang AJ, Little CB (2008) Drug insight: aggrecanases as therapeutic targets for osteoarthritis. Nature clinical practice Rheumatology 4:420–427. [DOI] [PubMed] [Google Scholar]
- 42. Huang K, Zhang C, Zhang XW, Bao JP, Wu LD (2011) Effect of dehydroepiandrosterone on aggrecanase expression in articular cartilage in a rabbit model of osteoarthritis. Molecular biology reports 38:3569–3572. [DOI] [PubMed] [Google Scholar]
- 43. Eser B, Cora T, Eser O, Kalkan E, Haktanir A, et al. (2010) Association of the polymorphisms of vitamin D receptor and aggrecan genes with degenerative disc disease. Genetic testing and molecular biomarkers 14:313–317. [DOI] [PubMed] [Google Scholar]
- 44. Watanabe H, Nakata K, Kimata K, Nakanishi I, Yamada Y (1997) Dwarfism and age-associated spinal degeneration of heterozygote cmd mice defective in aggrecan. Proceedings of the National Academy of Sciences of the United States of America 94:6943–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Risbud MV, Shapiro IM (2014) Role of cytokines in intervertebral disc degeneration: pain and disc content. Nature reviews Rheumatology 10:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Battie MC, Videman T, Parent E (2004) Lumbar disc degeneration: epidemiology and genetic influences. Spine 29:2679–2690. [DOI] [PubMed] [Google Scholar]
- 47. Masuda K, Aota Y, Muehleman C, Imai Y, Okuma M, et al. (2005) A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine 30:5–14. [DOI] [PubMed] [Google Scholar]
- 48. Reitmaier S, Schmidt H, Ihler R, Kocak T, Graf N, et al. (2013) Preliminary investigations on intradiscal pressures during daily activities: an in vivo study using the merino sheep. PloS one 8:e69610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) IVDs from wild-type (WT) and NF1fl/fl mice were genotyped with allele specific PCR. (b) IVDs from wild-type (control) and NF1fl/fl were cultured with cre-expressing adenovirus. Allele specific PCR demonstrates successful ex vivo genetic recombination in all IVDs tested.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information Files.