Abstract
The C. elegans pharyngeal neuron M4 is a multi-functional cell that acts as a cholinergic motor neuron to stimulate peristaltic pharyngeal muscle contraction and as a neuroendocrine cell secreting neuropeptides and growth factors to affect other cells both inside and outside the pharynx. The conserved transcription factors ZAG-1 and CEH-28 are co-expressed in M4 through most of development, and here we examine how these factors contribute to M4 differentiation. We find ZAG-1 functions upstream of CEH-28 in a branched pathway to activate expression of different sets of M4 differentiation markers. CEH-28 activates expression of the growth factor genes dbl-1 and egl-17, and the neuropeptide genes flp-5 and flp-2, while ZAG-1 activates expression of the serotonin receptor ser-7, as well as expression of ceh-28 and its downstream targets. Other markers of M4 differentiation are expressed normally in both zag-1 and ceh-28 mutants, including the neuropeptide gene flp-21 and the acetylcholine biosynthetic gene unc-17. Unlike ceh-28 mutants, zag-1 mutants completely lack peristaltic muscle contractions resulting from broader defects in M4 differentiation. Despite these defects, neither ZAG-1 nor CEH-28 are terminal selectors of the M4 phenotype, and we suggest they function in a hierarchy to regulate different aspects of M4 differentiation.
Introduction
Determining the mechanisms controlling motor neuron differentiation is essential to understanding nervous system development and to ultimately design cell-based therapies for human motor neuron diseases [reviewed in [1]]. However, the complexity of most nervous systems make it difficult to characterize these mechanisms for individual cell types.
The C. elegans pharynx is emerging as an exceptionally simple model to examine neuronal differentiation and function [2]. The pharynx is a rhythmically contracting neuromuscular pump located at the anterior of the digestive system, and it transports food through a central lumen into the intestine. The pharynx contains 20 neurons of 14 different types that make up a small nervous system separate from the somatic nervous system, and 20 muscle cells that contract during feeding [3]. These muscles exhibit two distinct types of contractions, called pumps and peristalses [4]. Pumping is a simultaneous contraction of the muscles in the anterior and very posterior regions of the pharynx, and these contractions concentrate food in the anterior pharyngeal lumen. In contrast, peristalsis is a wave-like contraction of a single muscle cell type that makes up a narrow region in the center of the pharynx called the isthmus, and this peristalsis carries a bolus of food through the isthmus lumen toward the intestine. Pumping occurs frequently, approximately 100–200 times per minute, while peristalses are relatively infrequent, occurring after every 4th to 40th pump. Our current challenge is understanding the mechanisms that produce the diverse neuron types that control pharyngeal contractions.
The pharyngeal M4 neuron is a multi-functional cell that both controls muscle contraction and secretes signaling molecules. M4 is a cholinergic motor neuron that stimulates isthmus muscle peristalsis, and in its absence the pharyngeal lumen becomes stuffed with food and the animals starve [5], [6]. Recently M4 has also been shown to have neurosecretory functions. M4 secretes the FMRFamide-like peptide neurotransmitter FLP-21 and the insulin-like growth factor INS-10, which function under hypoxic conditions to systemically modulate gustatory behavior and anterior touch neuron sensitivity, respectively [7], [8]. M4 also secretes the TGF-ß-family growth factor DBL-1 to affect the morphology of the nearby pharyngeal gland cells [9]. A number of additional neuropeptide and growth factor genes are also expressed in M4 [10], [11], and M4 can be considered part of a primitive neuroendocrine system [7], [9]. We are interested in how M4 differentiation is controlled to produce this complex, multifunctional phenotype.
The NK-2 family homeodomain transcription factor CEH-28 plays a key role in regulating synapse formation and gene expression in M4. ceh-28 mutants exhibit abnormal and mispositioned synapses in M4 and a highly penetrant stuffed pharynx phenotype [12]. In contrast to animals that lack M4 and do not peristalse, ceh-28 mutants can hyperstimulate isthmus muscle peristalses, and we believe this defect leads to inefficient feeding [5], [12]. ceh-28 mutants fail to express the dbl-1 gene in M4, and this loss of TGF-ß signaling leads to defects in morphology of the nearby g1 gland cells [9]. However other differentiation markers such as the serotonin receptor gene ser-7b and the vesicular ACh transporter gene unc-17 are expressed normally in the M4 cell of ceh-28 mutants [12]. Thus, other factors also contribute to M4 differentiation.
We are also interested in the role the conserved zinc-finger/homeodomain transcription factor ZAG-1 plays in M4. ZAG-1 is the sole C. elegans member of the ZEB-family of transcription factors, which in humans are mutated in Mowat-Wilson Syndrome and overexpressed in some metastatic cancers [reviewed in [13]]. C. elegans zag-1 is widely expressed in the nervous system, including in M4, as well as in embryonic pharyngeal muscles [14], [15]. zag-1 null mutants exhibit larval lethality and an inability to feed, and this feeding defect could result from defects in M4 or pharyngeal muscle development [15].
Here we explore the role of CEH-28 and ZAG-1 in regulating gene expression in M4, and we find that these factors function in a hierarchical pathway to progressively regulate distinct aspects of M4 differentiation. In addition to activating dbl-1, CEH-28 activates expression of the FGF gene egl-17 and the FMRFamide peptide genes flp-5 and flp-2. In contrast, ZAG-1 functions upstream and activates expression of ceh-28 and its downstream targets, but it also is necessary for expression of ser-7b, which is expressed independently of CEH-28 [12]. Other genes are expressed normally in M4 in both ceh-28 and zag-1 mutants, indicating neither of these factors is a terminal selector of M4 fate [16]. This understanding of how these conserved factors function in M4 may guide work developing therapies by manipulating mammalian ZAG-1 and CEH-28 orthologs to produce specific neuronal differentiation patterns.
Results
CEH-28 activates egl-17, flp-5, and flp-2 expression in M4
CEH-28 is an NK-2 family homeodomain transcription factor that is expressed exclusively in the M4 pharyngeal neuron from mid-embryogenesis through adulthood, and it regulates M4 synapse assembly and signaling [9], [12]. The only previously known transcriptional target of CEH-28 is dbl-1, which encodes a TGF-ß family growth factor secreted from M4 to affect the nearby g1 pharyngeal gland cells [9]. We sought to identify additional targets by comparing expression of gfp reporters regulated by the egl-17, flp-5, flp-2 and flp-21 promoters in wild-type animals and ceh-28 mutants (Figure 1A). These reporters are expressed in M4 [10], [11], and some contain potential CEH-28 binding sites, suggesting they may be direct targets of CEH-28 regulation.
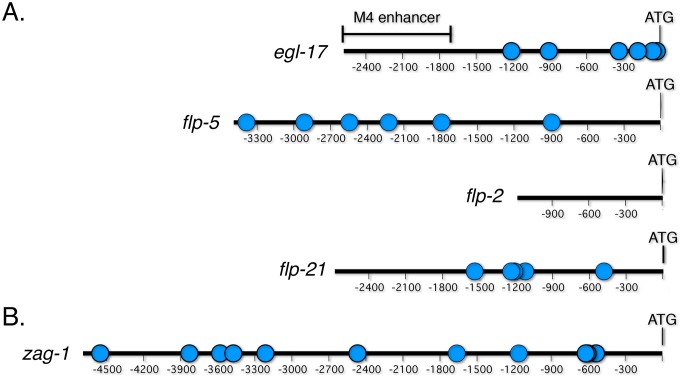
Figure 1. Promoters of potential CEH-28 target genes.
Schematic diagrams of promoter fragments in gfp fusions used in this study with potential CEH-28 binding sites indicated (blue dots). The translational start site (ATG) is numbered as bp 1. (A) egl-17 contains an M4 specific enhancer (bar). Potential CEH-28 binding sites are located in egl-17 at −1212, −906, −334, −179, −59, and −24; in flp-5 at −3387, −2914, −2546, −2225, −1793, and −892; in flp-21 at −1536, −1238, −1212, −1123, and −480. (B) Schematic diagram of the zag-1 promoter sufficient for zag-1 expression in M4 and other neurons [15]. Our studies used fosmid WRM063aA08 containing a gfp translational fusion [45], which is expressed in similar pattern. zag-1 contains potential CEH-28 binding sites at −4552, −3830, −3581, −3474, −3214, −2468, −1664, −1162, −619, −604, and −536.
egl-17 encodes a fibroblast growth factor (FGF) expressed in M4 and the vulva [10], and we found that CEH-28 activates egl-17 expression specifically in M4. egl-17::gfp expression was completely lost in M4 in ceh-28 mutants, while expression in the vulva was unaffected (Figure 2A–C; Table 1). In the dbl-1 promoter, separable sequences mediate expression in M4 and other neurons, and CEH-28 directly targets an M4-specific enhancer in this promoter [9]. Previous studies suggest the egl-17 promoter has a similar organization [17]. This work identified a region from −2589 to −1756 bp upstream of the translational start site necessary for egl-17::gfp expression in M4, but it had no role in vulval cell expression. We asked if this fragment was sufficient to enhance expression of the basal pes-10 promoter fused to gfp (Δpes-10::gfp), which is sensitive to linked enhancers [18]. We found transgenic animals bearing this reporter expressed GFP exclusively in M4 (Figure 1A; Figure 2D). While this enhancer does not contain any recognizable CEH-28 binding sites, its activity was lost in ceh-28 mutants, indicating that it functions downstream of CEH-28 (Figure 2E; Table 1). We suggest either that enhancer is directly activated by CEH-28 through non-consensus binding sites, or it is activated indirectly by another CEH-28 dependent factor.
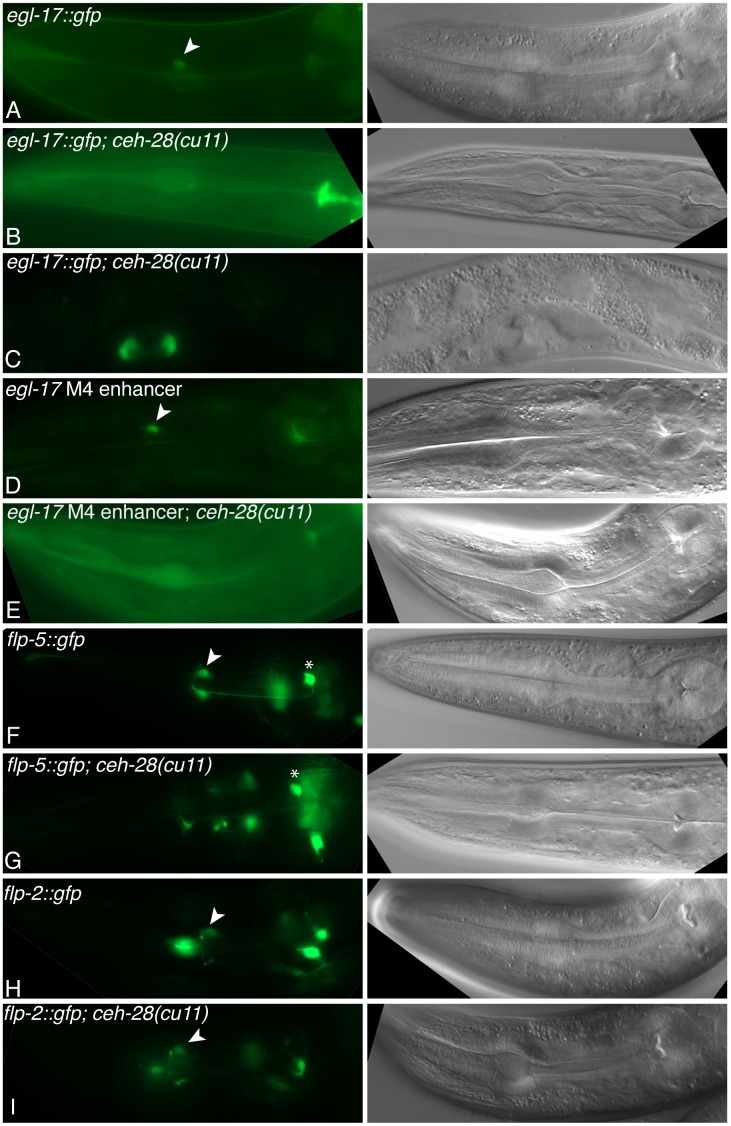
Figure 2. Expression of M4 differentiation markers in ceh-28(cu11) mutants.
Fluorescence (left) and DIC (right) micrographs of L4 to adult animals of the indicated genotypes bearing egl-17::gfp ayIs4 (A–C), the egl-17 M4 enhancer::Δpes-10::gfp cuEx793 (D,E), the flp-5::gfp ynIs49 (F,G), or the flp-2::gfp ynIs57 (H,I). (A,B,D–I) Expression in the pharynx with M4 (arrowhead) or I4 (asterisk, F and G) indicated. (C) egl-17::gfp expression in the vulva, which is unaffected in ceh-28 mutants.
Table 1. Frequency of animals expressing GFP in M4 in wild-type and ceh-28 mutants.
| Reporter | Percent animals expressing GFP inM4 in wild type (n)a | Percent animals expressing GFPin M4 in ceh-28(cu11) (n)a , b |
| ayIs4[egl-17::gfp] | 100 (35) | 0 (40)** |
| egl-17 M4 enhancer::gfp | 80 (30) | 0 (30)** |
| ynIs49[flp-5::gfp] | 100 (30) | 0 (37)** |
| ynIs57[flp-2::gfp] | 100 (30) | 80 (45)* |
| ynIs80[flp-21::gfp] | 100 (32) | 100 (35) |
| wgIs83[zag-1::gfp] | 100 (40) | 66 (45)** |
Transgenic adults were scored for GFP expression in M4.
Statistically significant difference between ceh-28(cu11) and wild type. (*p<0.01; **p<0.0001). Calculated using the two-tailed, Fisher’s exact test.
flp-2, flp-5 and flp-21 encode FMRFamide-like neuropeptides expressed in M4 and other neurons, and we found that CEH-28 activates flp-5 and flp-2 expression in M4, but flp-21 is expressed independently of CEH-28. Expression of flp-5::gfp was eliminated in ceh-28 mutant M4 cells, while the frequency of flp-2::gfp expression was modestly but significantly reduced (Figure 2F–I, Table 1). In both cases expression was unaffected in other neurons. In contrast, flp-21::gfp expression was unaffected in M4 and other neurons in ceh-28 mutants (Table 1).
These results expand our understanding of gene regulation in M4, and together with our previous work, identify dbl-1, egl-17, and flp-5 as downstream targets of CEH-28 [9], [12]. CEH-28 contributes to flp-2 expression, but other factors must also activate flp-2 in M4. In contrast ser-7b, unc-17, and flp-21 are expressed in M4 independently of CEH-28 [12].
ZAG-1 is essential for isthmus peristalsis
ZAG-1 is a ZEB-family C2H2 zinc-finger/homeodomain factor that regulates neuron pathfinding and differentiation in C. elegans [14], [15]. It is believed to be expressed in M4 and many other neurons, and in some pharyngeal muscles during embryogenesis. zag-1(hd16) null mutants arrest after hatching and exhibit a stuffed pharynx phenotype [15]. Because this phenotype can result from M4 defects, we characterized pharyngeal muscle contractions and M4 function in zag-1(hd16) mutants.
We found zag-1(hd16) mutants completely lack isthmus peristalses. These mutants pump, although at a slower rate than wild-type L1s (Table 2; Movie S1 and S2). However, while wild-type L1s peristalse approximately after every 9th pump, zag-1(hd16) mutants never exhibited a peristalsis (Table 2). Both of these phenotypes are observed in animals lacking M4 [5], [19], suggesting motor neuron function of M4 is defective in zag-1 mutants.
Table 2. Summary of feeding behavior in wild-type and zag-1 mutants.
| Genotype | Pump Rate (pumps/min) | Duration of Procorpus contractions (ms) | Duration of Posterior Bulb Contractions (ms) | Duration of Isthmus peristalsis (ms) | % Pumps followed by Isthmus Peristalsis |
| N2a | 116±2 | 159±1 | 173±2 | n.d. | 11% |
| zag-1(hd16) b | 58±21 | 118±3 | 96±6 | n.d. | 0% |
| N2+ serotoninc | 177±39 | n.d. | n.d. | n.d. | 30% |
| zag-1(hd16)+ serotonind | 158±41 | n.d. | n.d. | n.d. | 0% |
| N2+ arecolinee | 50±16 | n.d. | n.d. | 216±22 | 100% |
| zag-1(hd16) + arecolinef | 34±4 | n.d. | n.d. | 128±22 | 100% |
4 N2 L1s were recorded for 35–40 s and a total of 213 pumps were analyzed.
4 zag-1(hd16) L1s were recorded for 35–60 s and a total of 203 pumps were analyzed.
4 N2 L1s were treated with 20 mM serotonin and recorded for 15–20 s each and a total of 193 pumps were analyzed.
5 zag-1(hd16) L1s were treated 20 mM serotonin and recorded for 12–25 s each and a total of 222 pumps were analyzed.
4 N2 L1s were treated with 5 mM arecoline and recorded for 28–50 s each and a total of 115 pumps were analyzed.
6 zag-1(hd16) L1s were treated with 5 mM arecoline and recorded for 45–60 s each and a total of 166 pumps were analyzed.
To determine if the lack of peristalses in zag-1(hd16) mutants results from defects in M4 or the pharyngeal muscles, we examined pharyngeal muscle contractions in animals treated with compounds that stimulate either of these cell types. Serotonin stimulates the MC and M4 neurons, and this leads to increased pumping and peristalsis, respectively [20]. Wild-type L1s treated with serotonin exhibited a moderate increase in the pump rate and frequency of peristalsis compared to untreated animals (Table 2; Movie S3). In comparison, zag-1(hd16) mutants treated with serotonin exhibited a strong increase in the pump rate compared to untreated animals, but they still failed to peristalse (Table 2; Movie S4). Arecoline directly stimulates acetylcholine receptors in the isthmus muscles [12], [19], and we found that arecoline treatment stimulated very frequent peristalses in both wild-type L1s and zag-1(hd16) mutants (Table 2; Movies S5 and S6). Together these results demonstrate that the isthmus muscle of zag-1(hd16) mutants can produce a peristaltic contraction, but the M4 cell in these animals cannot stimulate this contraction. While arecoline treated zag-1 mutants did peristalse, these contractions were shorter than those in wild-type animals, suggesting the functional M4 in wild-type animals still affects peristalsis under these conditions (Table 2).
ZAG-1 regulates ceh-28 and other markers of M4 differentiation
zag-1 mutants exhibit differentiation defects in several neurons outside of the pharynx [14], [15], and we were interested in asking if M4 differentiation is similarly affected in these mutants. To examine gfp reporter gene expression, zag-1(hd16)/+ hermaphrodites that were heterozygous for these chromosomally integrated reporters were generated, and we compared reporter expression in progeny zag-1(hd16) homozygotes and their viable wild-type or heterozygous zag-1(hd16)/+ siblings at the L1 stage.
We first examined expression of ceh-28::gfp and reporters for the CEH-28 targets dbl-1, egl-17, flp-5 and flp-2. Both the frequency and intensity of ceh-28::gfp expression was reduced in zag-1(hd16) homozygotes (Figure 3A; Table 3). Likewise, we also observed loss or reduced expression of the CEH-28 targets, strongly suggesting that expression of endogenous ceh-28 is reduced in zag-1(hd16) homozygotes (Figure 3B–E; Table 3). The only CEH-28 target that retained some expression in zag-1(hd16) mutants was flp-2::gfp, which was also only partially affected in ceh-28 mutants (Table 1).
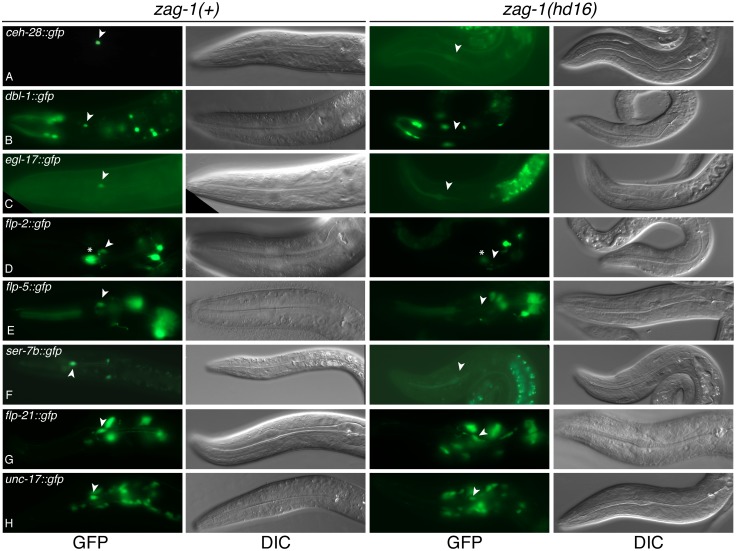
Figure 3. Expression of M4 differentiation markers in zag-1(hd16) mutants.
Fluorescence (left) and DIC (right) micrographs of transgenic L1-L2 animals bearing ceh-28::gfp (A), dbl-1::gfp (B), egl-17::gfp (C), flp-2::gfp (D), flp-5::gfp (E), ser-7b::gfp (F), flp-21::gfp (G), unc-17::gfp (H) in a zag-1(+) or zag-1(hd16) mutants. The position of M4 is marked (arrowheads). Asterisk indicates flp-2::gfp expression in the MC neuron (D).
Table 3. Expression of M4 differentiation markers in zag-1(+) and zag-1(hd16) mutants.
| Transgene | % animals expressing GFP in M4 in zag-1(+) (n)a | % animals expressing GFP in M4 in zag-1(hd16) (n)b , c |
| nIs177[ceh-28::gfp] d | 69 (54) | 22 (58)** |
| ctIs43[dbl-1::gfp] d | 74 (43) | 0 (32)** |
| ayIs4[egl-17::gfp] d | 74 (34) | 0 (36)** |
| ynIs49[flp-5::gfp] d | 63 (40) | 0 (40)** |
| ynIs57[flp-2::gfp] d | 67 (30) | 38 (45)* |
| cuEx469[ser-7b::gfp] e | 63 (30) | 0 (30)** |
| ynIs80[flp-21::gfp] d | 74 (34) | 68 (25) |
| mdIs18[unc-17::gfp] d | 65 (40) | 73 (30) |
GFP expression in the phenotypically wild-type +/+ or zag-1(hd16)/+ progeny of zag-1(hd16)/+ hermaphrodites, which we refer to as zag-1(+).
GFP expression in the zag-1(hd16) homozygous progeny of zag-1(hd16)/+ hermaphrodites.
Statistically significant difference between zag-1(hd16) and zag-1(+). (*p<0.02; **p<0.0001). Calculated using the two-tailed, Fisher’s exact test.
Chromosomally integrated transgene is expected to be present in 75% of the progeny of transgenic hermaphrodites.
cuEx469 is an extrachromosomal transgene that is unlinked to any of the chromosomes.
We next examined reporters for the ser-7b, flp-21, and unc-17 genes that are expressed in M4 independently of CEH-28 (Table 1) [12]. While flp-21::gfp and unc-17::gfp were expressed normally in zag-1(hd16) mutants, expression of ser-7b::gfp was completely lost (Figure 3F–H; Table 3). This loss is consistent with our observation that zag-1(hd16) mutants do not peristalse when treated with serotonin, and it suggests that ZAG-1 is essential for endogenous ser-7b expression in M4.
CEH-28 activates zag-1 expression in a positive feedback loop
While ZAG-1 functions upstream to activate expression of ceh-28, we observed that the zag-1 promoter also contains several potential CEH-28 binding sites (Figure 1B). To test if CEH-28 also regulates zag-1 expression, we examined expression of a zag-1::gfp reporter and found that it is indeed expressed in M4 as previously suggested (Figure 4A) [15]. The frequency of zag-1::gfp expression was moderately but significantly reduced in ceh-28(cu11) mutants compared to wild type (Figure 4B; Table 1), indicating CEH-28 functions in a positive feedback loop to activate zag-1 expression, perhaps to maintain stable expression of both zag-1 and ceh-28 after initial activation.
Figure 4. Expression of zag-1::gfp in wild-type and ceh-28 mutants.
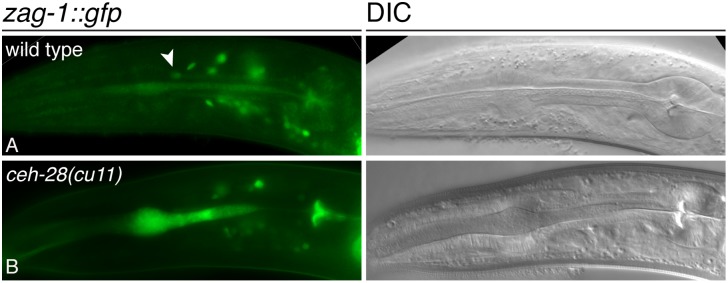
Fluorescence (left) and DIC (right) micrographs of zag-1::gfp wgIs83 in wild-type (A) and. ceh-28(cu11) mutant (B) adults. M4 is indicated (arrowhead) in (A). The anterior pharyngeal lumen is stuffed with bacteria in (B), and this results in non-specific autofluorescence.
Discussion
Here we show that the transcription factors CEH-28 and ZAG-1 function in a hierarchy to regulate multiple aspects of M4 differentiation (Figure 5). We previously showed that ceh-28 mutants fail to express the TGF-ß family gene dbl-1 in M4 [9], and we now find these mutants lack or exhibit reduced expression of a subset of additional M4 differentiation markers, including egl-17::gfp, flp-5::gfp and flp-2::gfp. We also find that ZAG-1 functions upstream of CEH-28 and plays a broader role in M4 differentiation. zag-1 mutants exhibit strongly reduced frequency and intensity of expression of a ceh-28::gfp marker, and reduced or eliminated expression of markers regulated by CEH-28. We hypothesize that ZAG-1 is required for strong expression of the endogenous ceh-28 gene in M4, and the reduced ceh-28 expression in zag-1 mutants leads to loss of expression of the CEH-28 downstream targets dbl-1, egl-17 and flp-5. Notably, zag-1 mutants also lack expression of a ser-7b::gfp, which is expressed normally in ceh-28 mutants, demonstrating ZAG-1 functions upstream of CEH-28 and regulates the ser-7 promoter independently of CEH-28 [12]. zag-1::gfp expression is also reduced in ceh-28 mutants, indicating CEH-28 contributes to zag-1 expression through a positive feedback loop. While flp-2::gfp expression is reduced in both zag-1 and ceh-28 mutants, it is difficult to know if flp-2 is directly downstream of either of these genes, or whether both function in parallel to activate flp-2 (Figure 5). Because of positive feedback between ceh-28 and zag-1, mutations affecting in either of these alter expression of the other gene, which in turn could flp-2 expression. Finally, the M4 differentiation markers unc-17::gfp and flp-21::gfp are expressed normally in both zag-1 and ceh-28 mutants, indicating other factors promote aspects of M4 neuronal differentiation independently of ZAG-1 and CEH-28. We suggest an additional factor(s) (‘X’ in Figure 5) activates expression of these genes, as well as zag-1 in M4.
Figure 5. zag-1 and ceh-28 function in a hierarchy in the M4 neuron.
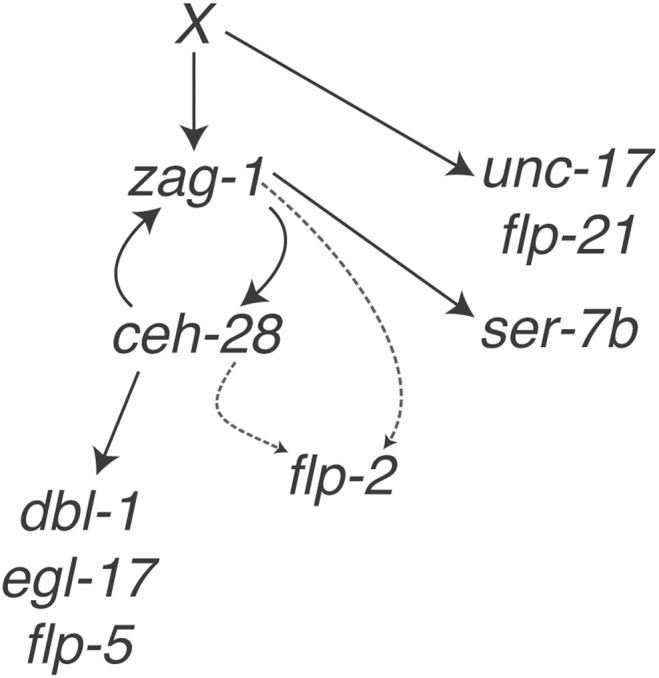
Model indicating gene regulatory interactions in M4 discussed in the text. CEH-28 is necessary to activate expression of dbl-1, egl-17, and flp-5. CEH-28 also activates zag-1 gene expression in a positive feedback loop. Either CEH-28, ZAG-1 or both activate flp-2 expression (dashed arrows). ZAG-1 functions upstream to activate ceh-28 and ser-7b expression, while another factor(s) activate zag-1, unc-17, and flp-21 (indicated as X).
We further show that defects in M4 differentiation in zag-1 mutants result in an absence of peristaltic contractions in the pharyngeal isthmus muscles. This phenotype results from defects in the M4 neuron rather than in the muscle, because stimulating the isthmus muscles with arecoline stimulates isthmus peristalsis in zag-1 mutants, whereas stimulating M4 with serotonin has no effect in these animals. This severe peristalsis defect likely contributes to the L1 arrest phenotype of zag-1(hd16) as previously suggested [15].
CEH-28 regulates M4 signaling by activating growth factors and neuropeptides
CEH-28 is not generally required for M4 neuronal differentiation. However, ceh-28 mutants are defective in expressing dbl-1, egl-17, flp-5, and flp-2 [9], [12], indicating that CEH-28 regulates multiple neurosecretory functions of M4. Like dbl-1, the egl-17, flp-5 and flp-2 genes are also expressed in cells other than M4, and this expression is unaffected in ceh-28 mutants. Both dbl-1 and egl-17 contain M4 specific enhancers that are separable from sequences controlling expression in other cells [9], [17], and this modular organization may be a common feature of promoters active in M4. Activity of the dbl-1 enhancer depends on CEH-28 binding sites [9], but, while the egl-17 promoter region contains potential CEH-28 sites, none of these are located in the M4 enhancer (Figure 1). We speculate that this enhancer is activated by CEH-28 through non-consensus sites or through other CEH-28-dependent factors. Additional studies are necessary to determine the functional significance of potential binding sites in other promoters regulated by CEH-28.
DBL-1 secreted from M4 affects the morphology of the nearby pharyngeal g1 gland cells [9], but the functions of the newly identified CEH-28 targets in M4 are unknown. EGL-17 has no known role in the pharynx, while exogenous FLP-5 and FLP-2 neuropeptides can excite pumping in pharyngeal explants [21]. None of the mutants egl-17(n1377), flp-5(gk3123) or flp-2(gk1039) exhibit a stuffed pharynx phenotype similar to that of ceh-28 mutants, suggesting these secreted proteins are not necessary for normal feeding (data not shown), and we believe other CEH-28 targets are important for M4 synapse assembly and motor neuron function. Alternatively, the functions of these genes are redundant with each other or with other signaling pathways, as has been observed for cholinergic and neuropeptide control of egg laying [22].
ZAG-1 plays a crucial role in regulating M4 differentiation
ZAG-1 is an ortholog of the vertebrate ZEB family transcription factors and Drosophila Zfh1 [14], [15]. In vertebrates these proteins regulate epithelial to mesenchymal transitions during development and in cancer metastasis, and control differentiation of particular neuronal types [13], [23]. Mutations affecting human ZEB proteins have been implicated in Mowat Wilson syndrome and corneal dystrophies [24]–[27]. In C. elegans and Drosophila, ZEB family proteins function in axonal path finding, neuronal differentiation, and neuronal cell fate [14], [15], [28], [29].
Our results indicate ZAG-1 is a major regulator of M4 differentiation. M4 is present and partially differentiated in zag-1 mutants, but these mutants lack expression of several markers of M4 differentiation. Moreover zag-1 mutants exhibit a complete loss of peristaltic contraction of the isthmus muscles. This contractile defect results from defects in M4 rather than the pharyngeal muscles themselves, because stimulation of the muscles with exogenous arecoline restores peristalses, while stimulation of M4 with serotonin has no effect. In wild-type animals the ability of serotonin to stimulate pharyngeal pumping and peristalses is mediated by the SER-7 receptor in the MC and M4 motor neurons, respectively [20], and the failure of exogenous serotonin to simulate peristalsis in zag-1 mutants is consistent with the loss of expression of the endogenous ser-7 gene in M4 in these animals.
ZEB family proteins most often function as transcriptional repressors, but they can also activate transcription [reviewed in [30]]. Mammalian ZEB1 activates transcription of the ovalbumin gene in response to estrogen signaling [31], as well as the MMP-1 and CDK-4 genes [32], [33]. Likewise, Drosophila Zfh1 can repress expression of mef2 during muscle development [34], while it activates expression of FMRFa gene in neurons [35]. This ability of ZEB family factors to function either as activators and repressors may result from cell type specific cofactors or post-translational modifications [36]–[38] or different DNA binding activities mediated through the multiple binding domains in these proteins [39].
Like its vertebrate and Drosophila orthologs, C. elegans ZAG-1 also functions as both a repressor and an activator. ZAG-1 negatively regulates its own expression and expression of unc-25, which is required for GABA synthesis [14], [15]. Our results now suggest ZAG-1 can also function as a transcriptional activator of the ser-7b and ceh-28 promoters (Figure 5). Current whole animal ChIP-seq analyses performed by the modENCODE consortium have not revealed significant ZAG-1 binding within these promoters [40], so we do not know if this regulation is direct, but binding might be undetectable if it only occurs in M4 or a small number of other cells.
Recently, ZEB2 was found to repress Nkx-2.1 expression in the developing mouse cerebral cortex, and loss of this regulation may contribute to Mowat Wilson syndrome [23], [41], [42]. While this regulation is opposite to what we have observed between ZAG-1 and ceh-28, it suggests ZEB-family factors may be common regulators of NK-2 family homeobox genes.
A hierarchy of transcription factors control M4 differentiation
In both invertebrates and vertebrates, ‘terminal selector’ transcription factors have been shown to be key activators of batteries of genes involved in terminal differentiation of specific neuronal types [reviewed in [16]]. Such terminal selector genes are expressed in these specific neurons throughout development, and, after initial activation, they maintain their own expression through positive autoregulation. While mutants defective for terminal selector genes express markers of pan-neural differentiation, they fail to express markers of neuron type-specific differentiation.
While both ceh-28 and zag-1 are expressed in M4 throughout development, neither appears to have other characteristics of a terminal selector for the M4 phenotype. CEH-28 does not maintain its own expression in M4, and ceh-28 mutants strongly express a ceh-28::gfp reporter in M4 throughout their life-cycle [12]. In comparison, ZAG-1 does partially activate its own expression indirectly in M4 via CEH-28 in a positive feedback loop (Figure 5), but it represses its own promoter in many neurons [14], [15]. More importantly, neither of these factors appears necessary for expression of batteries of genes for a specific aspect of M4 differentiation. For example, the flp-5, flp-2, and flp-21 genes encoding FMRFamide-family neuropeptides all are regulated differently in M4. Instead, our observations indicate ZAG-1 and CEH-28 function in a branched, hierarchical network to regulate M4 gene expression (Figure 5). Other genes are regulated independently of both CEH-28 and ZAG-1, and additional transcription factors must function upstream in this hierarchy. zag-1 and ceh-28 could themselves be activated by a terminal selector of M4 differentiation. Alternatively, different aspects of M4 differentiation might be independently regulated without a terminal selector transcription factor, perhaps resulting from the multifunctional nature of M4. More comprehensive analyses of M4 gene expression and the promoters of M4 expressed genes will distinguish between these possibilities.
Materials and Methods
Nematode handling, transformation and strains
C. elegans strains were grown under standard conditions [43]. Germline transformations were performed using pRF4 (100 ng/µl) and indicated gfp reporters (15 ng/µl) [44].
The following strains were used in this study: NH2466 ayIs4[egl-17::gfp dpy-20(+)]; dpy-20(e1282ts) [10], OK0978 ayIs4; ceh-28(cu11), OK0975 cuEx793 [egl-17 M4 enhancer], OK0976 ceh-28(cu11); cuEx793, NY2049 ynIs49[flp-5::gfp] [11], OK0979 ynIs49; ceh-28(cu11), NY2057 nyIs47; him-5(e1490) [11], OK0980 ynIs57; ceh-28(cu11), NY2080 ynIs80[flp-21::gfp] [11], OK1013 ynIs80; ceh-28(cu11), OP83 unc-119(ed3); wgIs83[zag-1::TY1::gfp::3xFLAG + unc-119(+)] [45], OK0974 unc-119(ed3); ceh-28(cu11); wgIs83, VH514 zag-1(hd16)/unc-17(ed113) dpy-14(e184) [15] MT15672 nIs177[ceh-28::4xNLS::gfp] [46], BW1946 ctIs43[Pdbl-1::gfp]; unc-42(e270) [47], OK516 cuEx469[ser-7b::gfp] [12], RM2258 pha-1(e2132ts); mdIs18[unc-17::gfp] [48].
To visualize the expression of gfp reporters in zag-1(hd16) homozygotes, transheterozygous zag-1(hd16)/+; gfp/+ were generated by crossing gfp/+ males with zag-1(hd16)/unc-17(ed113) dpy-14(e184) hermaphrodites and picking GFP expressing F1 cross progeny to individual plates. F1 animals were allowed to produce progeny for 2 days (25°C), and zag-1(hd16)/+; gfp/+ plates segregating zag-1(hd16) homozygotes in the F2 were identified. GFP expression was examined in F2 zag-1(hd16) homozygous progeny, recognized by their Unc, Coiler phenotype, and their +/+ or zag-1(hd16)/+ siblings.
General methods for nucleic acid manipulations and plasmid construction
Standard methods were used to manipulate all DNAs [49], and plasmids sequences are available from the authors. The egl-17 M4 enhancer containing bp 18,928–19,857 of cosmid F38G1 (accession # FO080171) was amplified from N2 genomic DNA and inserted into HindIII-SalI digested Δpes-10::gfp to generate pOK293.01.
Identification of candidate CEH-28 binding sites potential targets
Candidate CEH-28 binding sites were identified as described previously [9] by scanning the promoter sequences using the WormBase function Annotate Sequence Motif using the GBrowse plugin MotifFinder (www.wormbase.org;gmod.org/wiki/MotifFinder.pm) with the JASPAR position-frequency-matrix MA0264.1 (jaspar.cgb.ki.se) for the closely related homeodomain factor CEH-22 at a threshold of 0.82 [50].
Analysis of feeding behavior and drug studies
To analyze pharyngeal muscle contractions, wild-type or zag-1(hd16) embryos were hatched in the absence of food on unseeded NGM plates. L1 larvae from each genotype were suspended in 5 µl of M9 buffer containing OP50 and imaged on a 2% agarose pad under a coverslip. To stimulate M4 with serotonin, L1 animals obtained from a mixed stage population were placed on an unseeded NGM plate for 20 min and subsequently soaked 20 min in 20 mM serotonin on a 2% agarose pad before imaging. Pharyngeal muscles were stimulated with 5 mM arecoline as previously described [12].
Individual N2 or zag-1(hd16) animals that pumped were recorded at 25 frames/sec for 2 min using a Zeiss AxioImager microscope with an MRm camera and ZEN Software. For each genotype or drug treatment the feeding behavior was analyzed in at least 4 animals. Video frames and QuickTime movies of feeding behavior were exported and processed using ImageJ (developed at the US NIH and available at http://rsb.info.nih.gov/nih-image/) and quantifications were performed using Microsoft Excel.
Supporting Information
Pumping and peristalsis in a wild-type L1 larva. Five pumps of a wild-type L1 animal played at 1/5th speed (5 frames/sec). A pump occurs when the muscles in the anterior and posterior pharynx synchronously contract to open the pharyngeal lumen. A wave-like, peristaltic contraction is observed in the isthmus only after the third pump.
(MOV)
zag-1 mutants completely lack isthmus peristalsis. Seven pumps of a zag-1(hd16) mutant animal played at 1/5th speed (5 frames/sec). Note that the animal pumps somewhat more slowly than a wild-type animal, and that peristaltic contraction in the isthmus was never observed.
(MOV)
Pumping and peristalsis in serotonin treated wild-type L1 larva. Three pumps of a wild-type L1 treated with 20 mM serotonin played at 1/5th speed (5 frames/sec). A peristaltic contraction was observed only after the second pump.
(MOV)
Feeding behavior of serotonin treated zag-1(hd16 ) mutants. Seven pumps of a zag-1(hd16) mutant L1 larva treated with 20 mM serotonin played at 1/5th speed (5 frames/sec). Note that the animal pumps normally, however a peristaltic contraction in the isthmus.
(MOV)
Wild-type L1 larva treated with acetylcholine receptor agonist arecoline. Four pumps of the wild-type L1 treated with 5 mM arecoline played at 1/5th speed (5 frames/sec). Note that every pump is followed by a prolonged peristaltic contraction in which a larger region of the isthmus lumen is open at any given time.
(MOV)
zag-1(hd16) mutant L1 larva treated with acetylcholine receptor agonist arecoline. Two pumps of a zag-1(hd16) mutant L1 treated with 5 mM arecoline played at 1/5th speed (5 frames/sec). Both the pumps are followed by a strong peristaltic contraction.
(MOV)
Acknowledgments
The authors are indebted to Harald Hutter, Chris Li, Takashi Hirose, Robert Horvitz, Yo Suzuki, Jim Rand, Michael Stern, Yang Dai and Janet Richmond for plasmids, strains and advice, and to Paul Huber, Alena Kozlova and anonymous reviewers for critical reading of this manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by a grant from the National Institutes of Health grant R01 GM82865 (http://www.nih.gov/), the University of Illinois at Chicago's College of Liberal Arts and Sciences and the Research Open Access Publishing Fund (http://www.uic.edu/), and the State of Illinois (https://www.illinois.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Winner B, Marchetto MC, Winkler J, Gage FH (2014) Human-induced pluripotent stem cells pave the road for a better understanding of motor neuron disease. Hum Mol Genet. [DOI] [PubMed]
- 2. Franks CJ, Holden-Dye L, Bull K, Luedtke S, Walker RJ (2006) Anatomy, physiology and pharmacology of Caenorhabditis elegans pharynx: a model to define gene function in a simple neural system. Invert Neurosci 6:105–122. [DOI] [PubMed] [Google Scholar]
- 3. Albertson DG, Thomson JN (1976) The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 275:299–325. [DOI] [PubMed] [Google Scholar]
- 4. Avery L, Shtonda BB (2003) Food transport in the C. elegans pharynx. J Exp Biol 206:2441–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avery L, Horvitz HR (1987) A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duerr JS, Han HP, Fields SD, Rand JB (2008) Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol 506:398–408. [DOI] [PubMed] [Google Scholar]
- 7. Chen X, Chalfie M (2014) Modulation of C. elegans Touch Sensitivity Is Integrated at Multiple Levels. J Neurosci 34:6522–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pocock R, Hobert O (2010) Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci 13:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramakrishnan K, Ray P, Okkema PG (2014) CEH-28 activates dbl-1 expression and TGF-beta signaling in the C. elegans M4 neuron. Dev Biol 390:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burdine RD, Branda CS, Stern MJ (1998) EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125:1083–1093. [DOI] [PubMed] [Google Scholar]
- 11. Kim K, Li C (2004) Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol 475:540–550. [DOI] [PubMed] [Google Scholar]
- 12. Ray P, Schnabel R, Okkema PG (2008) Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Dev Neurobiol 68:421–433. [DOI] [PubMed] [Google Scholar]
- 13. Vandewalle C, Van Roy F, Berx G (2009) The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci 66:773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark SG, Chiu C (2003) C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130:3781–3794. [DOI] [PubMed] [Google Scholar]
- 15. Wacker I, Schwarz V, Hedgecock EM, Hutter H (2003) zag-1, a Zn-finger homeodomain transcription factor controlling neuronal differentiation and axon outgrowth in C. elegans. Development 130:3795–3805. [DOI] [PubMed] [Google Scholar]
- 16. Hobert O (2011) Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol 27:681–696. [DOI] [PubMed] [Google Scholar]
- 17. Cui M, Han M (2003) Cis regulatory requirements for vulval cell-specific expression of the Caenorhabditis elegans fibroblast growth factor gene egl-17. Dev Biol 257:104–116. [DOI] [PubMed] [Google Scholar]
- 18. Thatcher JD, Fernandez AP, Beaster-Jones L, Haun C, Okkema PG (2001) The Caenorhabditis elegans peb-1 gene encodes a novel DNA-binding protein involved in morphogenesis of the pharynx, vulva, and hindgut. Dev Biol 229:480–493. [DOI] [PubMed] [Google Scholar]
- 19. Raizen DM, Lee RY, Avery L (1995) Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141:1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song BM, Avery L (2012) Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J Neurosci 32:1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papaioannou S, Marsden D, Franks CJ, Walker RJ, Holden-Dye L (2005) Role of a FMRFamide-like family of neuropeptides in the pharyngeal nervous system of Caenorhabditis elegans. J Neurobiol 65:304–319. [DOI] [PubMed] [Google Scholar]
- 22. Ringstad N, Horvitz HR (2008) FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci 11:1168–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, et al. (2013) Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron 77:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cacheux V, Dastot-Le Moal F, Kaariainen H, Bondurand N, Rintala R, et al. (2001) Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet 10:1503–1510. [DOI] [PubMed] [Google Scholar]
- 25. Krafchak CM, Pawar H, Moroi SE, Sugar A, Lichter PR, et al. (2005) Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet 77:694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riazuddin SA, Zaghloul NA, Al-Saif A, Davey L, Diplas BH, et al. (2010) Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet 86:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, et al. (2001) Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat Genet 27:369–370. [DOI] [PubMed] [Google Scholar]
- 28. Layden MJ, Odden JP, Schmid A, Garces A, Thor S, et al. (2006) Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Dev Biol 291:253–263. [DOI] [PubMed] [Google Scholar]
- 29. Smith CJ, O'Brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, et al. (2013) Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79:266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G (2012) Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci 69:2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dillner NB, Sanders MM (2004) Transcriptional activation by the zinc-finger homeodomain protein delta EF1 in estrogen signaling cascades. DNA Cell Biol 23:25–34. [DOI] [PubMed] [Google Scholar]
- 32. Hu F, Wang C, Du J, Sun W, Yan J, et al. (2010) DeltaEF1 promotes breast cancer cell proliferation through down-regulating p21 expression. Biochim Biophys Acta 1802:301–312. [DOI] [PubMed] [Google Scholar]
- 33. Hu F, Wang C, Guo S, Sun W, Mi D, et al. (2011) deltaEF1 promotes osteolytic metastasis of MDA-MB-231 breast cancer cells by regulating MMP-1 expression. Biochim Biophys Acta 1809:200–210. [DOI] [PubMed] [Google Scholar]
- 34. Postigo AA, Dean DC (1999) ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci U S A 96:6683–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vogler G, Urban J (2008) The transcription factor Zfh1 is involved in the regulation of neuropeptide expression and growth of larval neuromuscular junctions in Drosophila melanogaster. Dev Biol 319:78–85. [DOI] [PubMed] [Google Scholar]
- 36. Costantino ME, Stearman RP, Smith GE, Darling DS (2002) Cell-specific phosphorylation of Zfhep transcription factor. Biochem Biophys Res Commun 296:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Long J, Zuo D, Park M (2005) Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem 280:35477–35489. [DOI] [PubMed] [Google Scholar]
- 38. Watanabe Y, Kawakami K, Hirayama Y, Nagano K (1993) Transcription factors positively and negatively regulating the Na,K-ATPase alpha 1 subunit gene. J Biochem 114:849–855. [DOI] [PubMed] [Google Scholar]
- 39. Ikeda K, Kawakami K (1995) DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem 233:73–82. [DOI] [PubMed] [Google Scholar]
- 40. Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330:1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomassy GS, Lodato S, Arlotta P (2013) A sip of GABA for the cerebral cortex. Neuron 77:1–3. [DOI] [PubMed] [Google Scholar]
- 42. van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, et al. (2013) Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron 77:70–82. [DOI] [PubMed] [Google Scholar]
- 43.Lewis JA, Fleming JT (1995) Basic Culture Methods. Methods in Cell Biology-Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press. pp. 4–30. [Google Scholar]
- 44.Mello C, Fire A (1995) DNA Transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press. pp. 451–482. [Google Scholar]
- 45. Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, et al. (2012) A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150:855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hirose T, Galvin BD, Horvitz HR (2010) Six and Eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. Proc Natl Acad Sci U S A 107:15479–15484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, et al. (1999) A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126:241–250. [DOI] [PubMed] [Google Scholar]
- 48. Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB (1993) The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261:617–619. [DOI] [PubMed] [Google Scholar]
- 49.Ausubel FM (1990) Current protocols in molecular biology. New York: Greene Pub. Associates and Wiley-Interscience: J. Wiley. 3 v. (loose-leaf) p.
- 50. Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW 3rd, et al. (2006) Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24:1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pumping and peristalsis in a wild-type L1 larva. Five pumps of a wild-type L1 animal played at 1/5th speed (5 frames/sec). A pump occurs when the muscles in the anterior and posterior pharynx synchronously contract to open the pharyngeal lumen. A wave-like, peristaltic contraction is observed in the isthmus only after the third pump.
(MOV)
zag-1 mutants completely lack isthmus peristalsis. Seven pumps of a zag-1(hd16) mutant animal played at 1/5th speed (5 frames/sec). Note that the animal pumps somewhat more slowly than a wild-type animal, and that peristaltic contraction in the isthmus was never observed.
(MOV)
Pumping and peristalsis in serotonin treated wild-type L1 larva. Three pumps of a wild-type L1 treated with 20 mM serotonin played at 1/5th speed (5 frames/sec). A peristaltic contraction was observed only after the second pump.
(MOV)
Feeding behavior of serotonin treated zag-1(hd16 ) mutants. Seven pumps of a zag-1(hd16) mutant L1 larva treated with 20 mM serotonin played at 1/5th speed (5 frames/sec). Note that the animal pumps normally, however a peristaltic contraction in the isthmus.
(MOV)
Wild-type L1 larva treated with acetylcholine receptor agonist arecoline. Four pumps of the wild-type L1 treated with 5 mM arecoline played at 1/5th speed (5 frames/sec). Note that every pump is followed by a prolonged peristaltic contraction in which a larger region of the isthmus lumen is open at any given time.
(MOV)
zag-1(hd16) mutant L1 larva treated with acetylcholine receptor agonist arecoline. Two pumps of a zag-1(hd16) mutant L1 treated with 5 mM arecoline played at 1/5th speed (5 frames/sec). Both the pumps are followed by a strong peristaltic contraction.
(MOV)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.