Abstract
Accurate identification of mycetoma causative agent is a priority for treatment. However, current identification tools are far from being satisfactory for both reliable diagnosis and epidemiological investigations. A rapid, simple, and highly efficient molecular based method for identification of agents of black grain eumycetoma is introduced, aiming to improve diagnostic in endemic areas. Rolling Circle Amplification (RCA) uses species-specific padlock probes and isothermal DNA amplification. The tests were based on ITS sequences and developed for Falciformispora senegalensis, F. tompkinsii, Madurella fahalii, M. mycetomatis, M. pseudomycetomatis, M. tropicana, Medicopsis romeroi, and Trematosphaeria grisea. With the isothermal RCA assay, 62 isolates were successfully identified with 100% specificity and no cross reactivity or false results. The main advantage of this technique is the low-cost, high specificity, and simplicity. In addition, it is highly reproducible and can be performed within a single day.
Author Summary
Treatment of eumycetoma largely depends on the causative pathogen. Identification of mycetoma agent with phenotypic features is too limited, and physiological and biochemical techniques are laborious, time-consuming and nonspecific, whereas the currently available molecular methods based on DNA sequencing are specific but extremely expensive. We describe rolling circle amplification method for identification of black grain eumycetoma using species-specific padlock probes. Eight probes were designed and successfully used for species identification and the results were easily visualized in 1% agarose gel. RCA provides a simple, reproducible, and cost-effective method for rapid identification of mycetoma agent that can be used in low-resource clinical settings.
Introduction
Black grain eumycetoma represents the most common fungal mycetoma worldwide. This chronic, erosive infection of subcutaneous tissues particularly affects the lower extremities and leads to severe disability [1]. The disease is considered a major health problem in tropical areas and is prevalent among people of low socio-economic status [2].
Mycetoma presents as a subcutaneous mass with multiple sinuses that discharge pus, serous fluid and grains, i.e. the characteristic compact grains of the causative agent formed inside the lesion [3].
A wide range of microorganisms has been reported to cause mycetoma. For treatment, not only differentiation between (fungal) eumycetoma and (bacterial) actinomycetoma is important, but also the identity of the causative agent, since species differ in their response to antimicrobial drugs [4]. In endemic countries, clinical diagnosis may be the only diagnostic method. A fully developed mycetoma lesion is easily identified clinically, whereas in early stages with the absence of grains, the infection may be confused with phaeomycosis or soft tissue tumors [1]. In such cases fine needle aspiration cytology or deep surgical biopsy for histological examination are useful [1], [5]. Some fungal and bacterial grains have a characteristic histological appearance which helps in provisional identification, but recognition of the causative species remains impossible [6]. Isolation of the pathogen from discharged grains or from biopsies allows identification of agents that sporulate, but most of the species lack phenotypic characteristics [3]. Molecular techniques have been introduced to facilitate the identification of nondescript organisms [7], [8], [9], but are of high cost and time-consuming. Thus, there is a need for a fast, simple and reliable method for identification.
Rolling circle amplification (RCA) is a powerful diagnostic method based on detection of specific nucleic-acid sequences and enzymatic amplification of circularized oligonucleotide probes under isothermal conditions [10]. The probes are linear oligonucleotides that contain two target-complementary sequences at their ends joined by linkers [11]. The ends of the probe hybridize to the complementary target in juxtaposition and then ligate which allows the circularization of the probe [11]. The circular structured molecule then amplifies with DNA polymerase that has strand displacement and progressive DNA synthesis activity resulting in series of repeats of the original circular template [11], [12]. The technique has been proven to be rapid, specific and low-cost for molecular identification of viruses, bacteria, and fungi [13], [14], [15], [16]. It has been applied for identification of a rare black grain mycetoma species Exophiala jeanselmei [17]. In addition, RCA has been used successfully for identification of white grain mycetoma species Scedosporium boydii [18]. The aim of the present study is to develop RCA-based diagnostics for the most common agents of black-grain eumycetoma.
Materials and Methods
Strains analyzed
The study included 62 isolates belonging to eight species causing black grain mycetoma: Madurella mycetomatis (n = 32), M. fahalii (n = 1), M. pseudomycetomatis (n = 3), M. tropicana (n = 2), Trematosphaeria grisea (n = 10), Falciformispora senegalensis (n = 6), F. tompkinsii (n = 2), and Medicopsis romeroi (n = 6). Strains were obtained from the reference collections of CBS-KNAW Fungal Biodiversity Centre (Utrecht, The Netherlands) and the Mycetoma Research Centre (MRC, Khartoum, Sudan) and are listed with metadata in Table 1. Type strains of all tested species were included. All strains were identified down to species level by sequencing of the rDNA ITS region [19], [20].
Table 1. Isolation source, origin, of strains analyzed.
| Name | No. | Source | Origin | |
| 1. | Falciformispora senegalensis | CBS 196.79 T | Mycetoma | Senegal |
| 2. | Falciformispora senegalensis | CBS 197.79 | Human | Senegal |
| 3. | Falciformispora senegalensis | CBS 198.79 | Mycetoma | Senegal |
| 4. | Falciformispora senegalensis | CBS 199.79 | Human | Senegal |
| 5. | Falciformispora senegalensis | CBS 132257 | Mycetoma | Sudan |
| 6. | Falciformispora senegalensis | CBS 132272 | Mycetoma | Sudan |
| 7. | Falciformispora tompkinsii | CBS 200.79 | Man | Senegal |
| 8. | Falciformispora tompkinsii | CBS 201.79 | Man | Senegal |
| 9. | Medicopsis romeroi | CBS 252.60 T | Mycetoma | Venezuela |
| 10. | Medicopsis romeroi | CBS 132878 | Mycetoma | India |
| 11. | Medicopsis romeroi | CBS 122784 | Plant | |
| 12. | Medicopsis romeroi | CBS 123975 | Phaeohyphomycosis | India |
| 13. | Medicopsis romeroi | CBS 128765 | Subcutaneous cyst | Kuwait |
| 14. | Medicopsis romeroi | CBS 135987 | onychomycosis | Netherlands |
| 15. | Trematosphaeria grisea | CWZ 29591 | ||
| 16. | Trematosphaeria grisea | CBS 332.50 T | Mycetoma | Chili |
| 17. | Trematosphaeria grisea | CBS 246.66 | Submandibular abscess | India |
| 18. | Trematosphaeria grisea | CBS 120271 | Tap water | The Netherlands |
| 19. | Trematosphaeria grisea | CBS 135982 | Pastry gel | The Netherlands |
| 20. | Trematosphaeria grisea | CBS 135984 | Water | The Netherlands |
| 21. | Trematosphaeria grisea | CBS 136543 | Water | The Netherlands |
| 22. | Trematosphaeria grisea | CBS 135985 | Water | The Netherlands |
| 23. | Trematosphaeria grisea | CBS 135983 | Water | The Netherlands |
| 24. | Trematosphaeria grisea | CBS 136537 | Water | The Netherlands |
| 25. | Madurella mycetomatis | CBS 132258 (Mm10) | Mycetoma | Sudan |
| 26. | Madurella mycetomatis | CBS 132259 (Mm13) | Mycetoma | Sudan |
| 27. | Madurella mycetomatis | CBS 132260 (Mm14) | Mycetoma | Sudan |
| 28. | Madurella mycetomatis | CBS 132261 (Mm16) | Mycetoma | Sudan |
| 29. | Madurella mycetomatis | CBS 132262 (Mm18) | Mycetoma | Sudan |
| 30. | Madurella mycetomatis | CBS 132263 (Mm22) | Mycetoma | Sudan |
| 31. | Madurella mycetomatis | CBS 132265 (Mm28) | Mycetoma | Sudan |
| 32. | Madurella mycetomatis | CBS 132266 (Mm29) | Mycetoma | Sudan |
| 33. | Madurella mycetomatis | CBS 132267 (Mm30) | Mycetoma | Sudan |
| 34. | Madurella mycetomatis | CBS 132269 (Mm33) | Mycetoma | Sudan |
| 35. | Madurella mycetomatis | CBS 132270 (Mm36) | Mycetoma | Sudan |
| 36. | Madurella mycetomatis | CBS 132273 (Mm44) | Mycetoma | Sudan |
| 37. | Madurella mycetomatis | CBS 132274 (Mm45) | Mycetoma | Sudan |
| 38. | Madurella mycetomatis | CBS 132285 (Mm46) | Mycetoma | Sudan |
| 39. | Madurella mycetomatis | CBS 132276 (Mm49) | Mycetoma | Sudan |
| 40. | Madurella mycetomatis | CBS 109814 (Mm54) | Mycetoma | Sudan |
| 41. | Madurella mycetomatis | CBS 131320 (Mm55) | Mycetoma | Sudan |
| 42. | Madurella mycetomatis | CBS 132280 (Mm58) | Mycetoma | Sudan |
| 43. | Madurella mycetomatis | CBS 132281 (Mm63) | Mycetoma | Sudan |
| 44. | Madurella mycetomatis | CBS 132282 (Mm64) | Mycetoma | Sudan |
| 45. | Madurella mycetomatis | CBS 132283 (Mm68) | Mycetoma | Sudan |
| 46. | Madurella mycetomatis | CBS 132284 (Mm71) | Mycetoma | Sudan |
| 47. | Madurella mycetomatis | CBS 132285 (Mm72) | Mycetoma | Sudan |
| 48. | Madurella mycetomatis | CBS 132286 (Mm73) | Mycetoma | Sudan |
| 49. | Madurella mycetomatis | CBS 132287 (Mm78) | Mycetoma | Sudan |
| 50. | Madurella mycetomatis | CBS 132288 (Mm83) | Mycetoma | Sudan |
| 51. | Madurella mycetomatis | CBS 109801T | Mycetoma | Sudan |
| 52. | Madurella mycetomatis | CBS 110087 | Mycetoma | Sudan |
| 53. | Madurella mycetomatis | CBS 110359 | Mycetoma | Mali |
| 54. | Madurella mycetomatis | CBS 110356 | Mycetoma | Mali |
| 55. | Madurella mycetomatis | CBS 132419 | Mycetoma | India |
| 56. | Madurella mycetomatis | CBS 132589 | Mycetoma | India |
| 57. | Madurella tropicana | CBS 201.38 T | Mycetoma | Indonesia |
| 58. | Madurella tropicana | CBS 331.50 | ||
| 59. | Madurella pseudomycetomatis | CBS 129177 T | Mycetoma | China |
| 60. | Madurella pseudomycetomatis | CBS 216.29 | Mycetoma | |
| 61. | Madurella pseudomycetomatis | CBS 248.48 | Mycetoma | New Mexico |
| 62. | Madurella fahalii | CBS129176 T | Mycetoma | Sudan |
(CBS Centraalbureau voor Schimmelcultures; Between brakets Erasmus collection number for strains from Sudan; Type strains marked withT)
DNA extraction and target amplification
DNA was extracted using cetyltrimethyl ammonium bromide (CTAB) method as described by Möller et al. [21]. Amplification of the ITS region was performed using primers V9G and LS266 [22] in a 25 µL reaction mixture containing: 10 ng of template DNA, 0.1 mM each dNTP, 0.6 U Taq polymerase (GC Biotech, Alphen aan den Rijn, The Netherlands), 1 µL of each primer (10 pmol) and 2.5 µL reaction buffer (0.1 M Tris-HCl, 0.5 M KCl, 25 mM MgCl2, 0.1% gelatin, 1% Triton X-100). PCR reactions consisted of a 5 min predenaturation step at 95°C, followed by 30 cycles of 95°C for 30 s, 52°C for 30 s and 72°C for 1 min, with final post elongation step at 72°C for 7 min. PCR products were detected by electrophoresis using 1% agarose gels.
Padlock probe design
Sequences of the ITS region were used to design 8 probes specific for each species used in this study. Two alignments were generated since the analyzed species were known to belong to two different fungal orders [19], [20]. ITS derived from Madurella (Sordariales) were aligned with 200 isolates of Chaetomiaceae including Chaetomium, Thielavia, and Achaetomium. For the remaining species (Pleosporales) an alignment was constructed to include representative isolates of the family Trematosphaeriaceae and of coelomycetes in the suborder Pleosporineae. Sequences were aligned using BioNumerics v4.61 (Applied Maths, Sint-Martens-Latem, Belgium). Probes were designed with minimum secondary structure and were checked using PrimerSelect (DNASTAR Lasergene, WI, U.S.A.). To insure specificity of the probes, target-specific sites of each padlock probe was submitted to BLAST in NCBI sequence database for homologous sequences. The melting temperature of the 5′ end of the probe binding arm was designed to be>63°C while for the 3′ end binding arm it was designed to be at least 15°C below the annealing temperature. The probes were phosphorylated at the 5′ end and are listed in Table 2. Probe linkers were taken from Zhou et al. [23].
Table 2. Oligonucleotide padlock probes and probe-specific primers used for species identification with RCA.
| Species name | Probe and primer name | Sequences |
| M. tropicana | MTROP | 5′pGAGAGCAAACAGGGTGTTGTATAgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaCAGAGAGGCCATA-3′ |
| M. pseudomycetomatis | MPSEU | 5′pGGAGCAACAGGGTGTTGTATAATgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaAGAGAGGCCATAC -3′ |
| M. fahalii | MFAH | 5′pTGATACTACTACGCTCGGAGTGACgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaCCCTGAGCGAGG -3′ |
| T. grisea | TGRIS | 5′pACCCGTAGGTCCTCCCAAAAGCGgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaTGGACGCCAGTCC-3′ |
| F. senegalensis | FSEN | 5′pACATAGACAAGGGTGTTGCCGGCgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaCAACGTACGGTAC-3′ |
| F. tompkinsii | FTOM | 5′pTCTTCCCAAAGTGCGCAAAGTGCgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaCTATGCCACCAAG-3′ |
| M. romeroi | MRO | 5′pAAGGCGAGTCCACGCACTCTGGgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtctaCTGCCAATGACTTT -3′ |
| M. mycetomatis | MYC | 5′pACTACACTACCGGGAGGCCCgatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctac CGCGCAGACACGATAgtctaAGGGGGCCGAGGGAC-3′ |
| RCA1 | 5′-ATGGGCACCGAAGAAGCA-3′ | |
| RCA2 | 5′-CGCGCAGACACGATA-3′ |
5′p- indicate phosphorylation of 5′ end, probes binding arms are underlined, the arms joined with non specific region lower case and RCA1 and RCA2 primer binding regions are bolded.
Probe ligation
Padlock probe ligation was performed in a mixture consisting of 1 µl purified ITS amplicons, 2 U pfu DNA ligase (Epicentre Biotechnologies, Madison, WI, U.S.A.), and 0.1 µmol/l padlock probe in buffer (20 mM Tris-HCl pH 7.5, 10 mM MgCl2, 20 mM KCl, 0.1% Igepal, 0.01 mM rATP, 1 mM DTT), with a total reaction volume of 10 µl. Ligation conditions were: 5 min denaturation at 94°C, followed by 7 cycles of 94°C for 30 sec, 63°C for 4 min, and final cooling at 10°C.
Exonucleolysis
Prior to RCA amplification reaction and in order to reduce the ligation-independent amplification, ligation products were treated by addition of 10 U exonucleases I and 10 U exonucleases III (New England Biolabs, Hitchin, U.K.) with a final volume of 20 µl. The mixture was then incubated for 30 min at 37°C, followed by 3 min at 94°C to deactivate the endonuclease enzymes.
Rolling circle amplification (RCA)
RCA amplification reaction was performed in a 50 µl mixture containing; 2 µl ligation product, 8 U Bst DNA polymerase (New England Biolabs), 10 pmol of each RCA primer (Table 2), and 400 µM dNTP mix. The mixture was incubated at 65°C for 60 min and cooled at 10°C. Electrophoresis on a 1% agarose gel was used to visualize RCA products. A positive reaction is indicated by the presence of ladder-like pattern. The reaction was also visualized by adding 1.0 µl of a 10-fold diluted SYBR Green I (Cambrex BioScience, Workingham, U.K.) to 10 µl of the amplification product. Accumulated double stranded DNA was detected with UV transilluminator (Vilber Lourmat, Marne-la-Vallée, France).
Determination of analytical specificity and sensitivity
The specificity of the 8 RCA probes was tested using strains of black-grain mycetoma causative species listed in table 1. Analytical sensitivity was determined using 10-fold serial dilution of M. mycetomatis (CBS 109801) and M. fahalii (CBS 129176) DNA and the test was performed as mentioned above. In addition, RCA was performed directly using DNA samples without amplification of the target gene. To evaluate the detection limit from direct DNA samples two-fold serial dilutions of target DNA were tested.
The sensitivity of the RCA probes was also determined by 10-fold serial dilution of MYC and MFAH probes tested with amplified ITS of M. mycetomatis and M. fahalii respectively.
Results
RCA was used to identify 62 strains belonging to eight species causing human eumycetoma. Since black grain eumycetoma species are known to be phylogenetically distant, it is easy to find unique sites for their identification. The ribosomal ITS region was sufficient for identification of all species and showed no intraspecific variability within a set of 100 M. mycetomatis strains in our collection. For M. mycetomatis, M. tropicana, M. pseudomycetomatis, and F. senegalensis the ITS1 region was selected for probe design, while for M. fahalii, T. grisea, F. tompkinsii and M. romeroi the ITS 2 region was found to be more suitable.
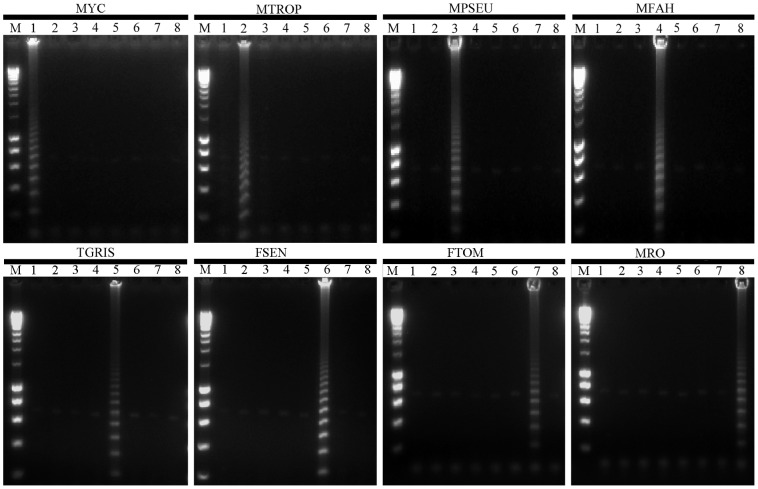
RCA results for the tested strains were easily visualized in 1% agarose gel. Positive reactions demonstrated ladder like patterns while negative reactions resulted in a clear background (Fig. 1). With SYBR green, positive results showed green fluorescence when exposed to UV light, while negatives did not. When exonucleolysis was performed some inhibition was observed with low RCA positive signals on gel or with fluorescence. Faint non-specific bands were observed when this step was omitted. RCA reactions were performed successfully without digestion with exonucleases, as the non-specific bands did not interfere with RCA results.
Figure 1. Specificity of rolling circle amplification probes.
Agarose gel electrophoresis analysis of rolling circle amplification products. Positives probe signal seen as band pattern was only present with matched template–probe mixtures. Probe names are indicated on the top of the gel. Lanes; 1 M. mycetomatis CBS 109801T, 2 M. tropicana CBS 201.38T, 3 M. pseudomycetomatis CBS 129177T, 4 M. fahalii CBS 129176T, 5 T. grisea CBS 332.50T, 6 F. senegalensis CBS 196.79T, 7 F. tompkinsii CBS 200.70, 8 M. romeroi CBS 252.60T, M DNA ladder.
All M. mycetomatis strains were correctly identified with RCA, irrespective of their geographical origin (Sudan, India, Mali) (Fig. 2). For the other agents, each individual species-specific probes yielded positive results with their corresponding species and with 100% agreement with ITS sequencing (Fig. 2, Table 3). No cross reactivity or false positive and negative results were observed. The sensitivity of RCA when using amplified product of the target gene was less than 32×10−3 ng of DNA. A higher concentration of 100 ng is needed when the test is carried out directly from the DNA samples without amplification of the ITS. The probes were very sensitive and a concentration of 6.6×10−5 ng was successfully ligated and then amplified with RCA.
Figure 2. Madurella mycetomatis identification by RCA.
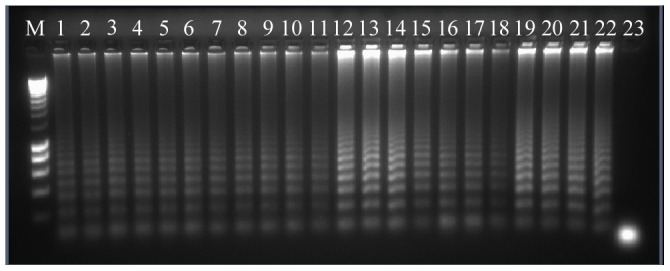
Gel representation of rolling circle amplification reaction using Madurella mycetomatis probe (MYC) for strains recovered from mycetoma patient of origin: lane 1–18 Sudan; lane 19, 20 Mali; lane 21, 22 India; lane 23 negative control water; lane M ladder.
Table 3. Rolling circle amplification results of analysed strain.
| Strains | RCA result for species-specific padlock probe | |||||||
| MYC | MTROP | MPSEU | MFAH | TGRIS | FSEN | FTOM | MRO | |
| M. mycetomatis (32) | + | - | - | - | - | - | - | - |
| M. tropicana (2) | - | + | - | - | - | - | - | - |
| M. pseudomycetomatis (3) | - | - | + | - | - | - | - | - |
| M. fahalii(1) | - | - | - | + | - | - | - | - |
| T. grisea (10) | - | - | - | - | + | - | - | - |
| F. senegalensis(6) | - | - | - | - | - | + | - | - |
| F. tompkinsii (2) | - | - | - | - | - | - | + | - |
| M. romeroi (6) | - | - | - | - | - | - | - | + |
Positive results (+), negative results (-).
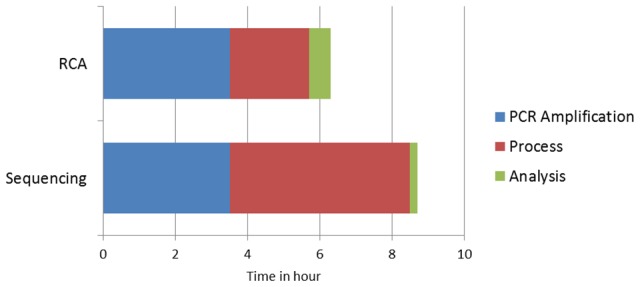
The turnaround time required for conducting the entire experiment including PCR amplification of target DNA, RCA processing and analysis was found to be 6 hours. DNA sequencing of the ITS region took more than 8 hours to be performed (Fig. 3).
Figure 3. Identification time of species using rolling circle amplification (RCA) and sequencing of ITS.
Discussion
Mycetoma is a unique tropical disease, endemic in many tropical and subtropical regions that has been recently added to the WHO list of neglected tropical diseases. [24]. It is mainly prevalent in what is known as “mycetoma belt” which includes Mexico, Senegal, Sudan, India and other countries between tropic of cancer [1]. In 2014, a mycetoma consortium of scientists and physicians published research gaps on mycetoma which need to be addressed in the coming years [2]. One of the research priorities identified was the need to develop a reliable and cost-effective method for species identification to improve diagnosis [2].
Mycetoma agents have been extensively studied in recent years [8], [9], [20]. The large phylogenetic distance between a number of these agents provides the possibility to use a moderately variable marker like rDNA ITS for species identity. Ahmed et al. [25] developed PCR-restriction fragment length polymorphism (RFLP) for identification of M. mycetomatis targeting the ITS region. However, with the description of the molecular siblings M. fahalii, M. pseudomycetomatis, and M. tropicana [26] the method might be insufficiently accurate. Moreover, there is a need for identification these siblings species; Madurella grisea appeared to be distantly related and was re-named as T. grisea [20].
In the present study we developed a simple, fast and highly specific molecular method for the identification of agents of black grain mycetoma. In this method, the ITS region is easily amplified using one set of primers, which simplifies the use. In a second, isothermal amplification reaction padlock probes are used to identify the species by RCA. The only equipment necessary is a thermocycler for the PCR reaction and a water bath or heating block for the RCA reaction. This relative simplicity enhances possible use in routine laboratories in endemic areas. Due to its robustness, high potential, and reproducibility, RCA is increasingly used as a diagnostic tool in pathogenic fungi, e.g. agents of chromoblastomycosis, dermatophytes, Aspergillus, Candida, and Talaromyces marneffei [16], [23], [27], [28]. The method does not require DNA sequencing and is therefore considered as a rapid and cost-effective. Applications are being expanded to nano- and biotechnology [29].
In the present study eight species-specific probes were designed and used for identification of 62 isolates. For the RCA reaction species probe hybridization to the 3′ and 5′ ends of target DNA and joining of adjacent ends by DNA ligase when both show perfect complementarity. The ligation appears to be highly specific and thus the method can detect single nucleotide polymorphism [30]. The amplification reaction is driven by an isothermal DNA polymerase to amplify the circularized probes with high efficiency and an estimated capacity to synsthesize more than 70,000 bp per hour [31]. RCA products can be detected with different methods including gel electrophoresis, radiolabeling, UV absorbance, fluorescence, and single molecule detection [32]. It was known that the positive signals can be detected within 15 min after starting the RCA reaction by real time PCR [23]. In the present study the RCA positive signal was easily visualized using both gel electrophoresis and fluorescent dye. The duration of our RCA protocol was 2 h, but additional time is required for DNA extraction and ITS amplification. Compared to the DNA sequencing the turnaround time for RCA is 2 hours less than sequencing and this even more if there is no in-house sequencer available.
Our results with eight padlock probes showed that RCA accurately identified all species with no cross reactivity (Fig. 1). It may be concluded that RCA is extremely useful for specific identification of agents of mycetoma. Performance and rapid turnaround time features make the RCA suitable for quick and reliable diagnosis, which is an enormous improvement compared to the current phenotypic identification of mostly non-sporulating cultures. Future application of RCA could be the detection of agents DNA directly from clinical samples without requirement of culturing.
Supporting Information
STARD flowchart for RCA.
(PDF)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Fahal AH (2006) Mycetoma, Clinicopathological Monograph. Khartoum: Khartoum University Press.
- 2. van de Sande WW, Maghoub el S, Fahal AH, Goodfellow M, Welsh O, et al. (2014) The mycetoma knowledge gap: identification of research priorities. PLoS Negl Trop Dis. 8: e2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon-Chung KJ, Bennet JE (1992) Medical Mycology. Philadelphia: Lea and Febiger.
- 4. Fahal AH (2010) Management of mycetoma. Expert Rev Dermatol 5: 87–93. [Google Scholar]
- 5. Yousif BM, Fahal AH, Shakir MY (2010) A new technique for the diagnosis of mycetoma using fixed blocks of aspirated material. Trans R Soc Trop Med Hyg 104: 6–9. [DOI] [PubMed] [Google Scholar]
- 6. Alam K, Maheshwari V, Bhargava S, Jain A, Fatima U, et al. (2009) Histological diagnosis of madura foot (mycetoma): a must for definitive treatment. J Glob Infect Dis 1: 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Hoog G S, Buiting A, Tan C S, Stroebel A B, Ketterings C, et al. (1993) Diagnostic problem with imported cases of mycetoma in The Netherland. Mycoses 36: 81–87. [DOI] [PubMed] [Google Scholar]
- 8. Desnos-Ollivier M, Bretagne S, Dromer F, Lortholary O, Dannaoui E (2006) Molecular identification of black-grain mycetoma agents. J Clin Microbiol 44: 3517–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ahmed AO, Desplaces N, Leonard P, Goldstein F, De Hoog S, Verbrugh H, et al. (2003) Molecular detection and identification of agents of eumycetoma: detailed report of two cases. J Clin Microbiol. 41: 5813–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert W, Dressler D (1968) DNA Replication: The Rolling Circle Model. Cold Spring Harb Symp Quant Biol 33: 473–484. [DOI] [PubMed] [Google Scholar]
- 11. Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, et al. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265: 2085–2088. [DOI] [PubMed] [Google Scholar]
- 12. Inoue J, Shigemori Y, Mikawa T (2006) Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res 34: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang B, Potter SJ, Lin Y, Cunningham AL, Dwyer DE, et al. (2005) Rapid and sensitive detection of severe acute respiratory syndrome Coronavirus by rolling circle amplification. J Clin Microbiol 43: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macera L, Cortey M, Maggi F, Segalés J, Kekarainen T (2011) A novel rolling circle amplification assay to detect members of the family Anelloviridae in pigs and humans.Virus Res. 160: 424–427. [DOI] [PubMed] [Google Scholar]
- 15. Chen X, Wang B, Yang W, Kong F, Li C, et al. (2014) Rolling circle amplification for direct detection of rpoB gene mutations in Mycobacterium tuberculosis isolates from clinical specimens. J Clin Microbiol 52: 1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Najafzadeh MJ, Sun J, Vicente VA, de Hoog GS (2011) Rapid identification of fungal pathogens by rolling circle amplification using Fonsecaea as a model. Mycoses 54: e577–e582. [DOI] [PubMed] [Google Scholar]
- 17. Najafzadeh MJ, Dolatabadi S, Saradeghi Keisari M, Naseri A, Feng P, et al. (2013) Detection and identification of opportunistic Exophiala species using the rolling circle amplification of ribosomal internal transcribed spacers. J Microbiol Methods 94: 338–342. [DOI] [PubMed] [Google Scholar]
- 18. Lackner M, Najafzadeh MJ, Sun J, Lu Q, Hoog GS (2012) Rapid identification of Pseudallescheria and Scedosporium strains by using rolling circle amplification. Appl Environ Microbiol 78: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Hoog GS, Ahmed SA, Najafzadeh MJ, Sutton DA, Keisari MS, et al. (2013) Phylogenetic Findings Suggest Possible New Habitat and Routes of Infection of Human Eumyctoma. PLoS Negl Trop Dis 7: e2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmed SA, van de Sande WWJ, Stevens DA, Fahal A, van Diepeningen A, et al. (2014) Revision of agents of black-grain eumycetoma in the order Pleosporales Persoonia. 33: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Möller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20: 6115–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerrits van den Ende AHG, De Hoog GS (1999) Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana . Stud Mycol 43: 152–162. [Google Scholar]
- 23. Zhou X, Kong F, Sorrell TC, Wang H, Duan Y, et al. (2008) Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rollingcircle amplification. J Clin Microbiol 46: 2423–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO (2013) The 17 neglected tropical diseases. Geneva: World Health Organization. Availabe: http://www.who.int/neglected_diseases/diseases/en/. Accessed 10 Nov 2014.
- 25. Ahmed AO, Mukhtar MM, Kools-Sijmons M, Fahal AH, de Hoog S, et al. (1999) Development of a species-specific PCR-restriction fragment length polymorphism analysis procedure for identification of Madurella mycetomatis . J Clin Microbiol 37: 3175–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Hoog GS, van Diepeningen AD, Mahgoub el S, van de Sande WW (2012) New species of Madurella, causative agents of black-grain mycetoma. J Clin Microbiol 50: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamzehei H, Yazdanparast SA, Davoudi MM, Khodavaisy S, Golehkheyli M, et al. (2013) Use of rolling circle amplification to rapidly identify species of Cladophialophora potentially causing human infection. Mycopathologia 175: 431–438. [DOI] [PubMed] [Google Scholar]
- 28. Kong F, Tong Z, Chen X, Sorrell T, Wang B, et al. (2008) Rapid identification and differentiation of Trichophyton species, based on sequence polymorphisms of the ribosomal internal transcribed spacer regions, by rolling-circle amplification. J Clin Microbiol 46: 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ali MM, Li F, Zhang Z, Zhang K, Kang DK, et al. (2014) Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev 43: 3324–3341. [DOI] [PubMed] [Google Scholar]
- 30. Nilsson M, Krejci K, Koch J, Kwiatkowski M, Gustavsson P, et al. (1997) Padlock probes reveal single-nucleotide differences, parent of origin and in situ distribution of centromeric sequences in human chromosomes 13 and 21. Nat Genet 16: 252–255. [DOI] [PubMed] [Google Scholar]
- 31. Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, et al. (1989) Highly efficient DNA synthesis by the phage phi Ф 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem 264: 8935–8940. [PubMed] [Google Scholar]
- 32. Banér J, Nilsson M, Mendel-Hartvig M, Landegren U (1998) Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res 26: 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STARD flowchart for RCA.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.