Abstract
The Chinese hamster ovary cell (CHO) is the major host cell factory for recombinant production of biological therapeutics primarily because of its “human-like” glycosylation features. CHO is used for production of several O-glycoprotein therapeutics including erythropoietin, coagulation factors, and chimeric receptor IgG1-Fc-fusion proteins, however, some O-glycoproteins are not produced efficiently in CHO. We have previously shown that the capacity for O-glycosylation of proteins can be one limiting parameter for production of active proteins in CHO. Although the capacity of CHO for biosynthesis of glycan structures (glycostructures) on glycoproteins are well established, our knowledge of the capacity of CHO cells for attaching GalNAc-type O-glycans to proteins (glycosites) is minimal. This type of O-glycosylation is one of the most abundant forms of glycosylation, and it is differentially regulated in cells by expression of a subset of homologous polypeptide GalNAc-transferases. Here, we have genetically engineered CHO cells to produce homogeneous truncated O-glycans, so-called SimpleCells, which enabled lectin enrichment of O-glycoproteins and characterization of the O-glycoproteome. We identified 738 O-glycoproteins (1548 O-glycosites) in cell lysates and secretomes providing the first comprehensive insight into the O-glycosylation capacity of CHO (http://glycomics.ku.dk/o-glycoproteome_db/).
The Chinese hamster ovary (CHO)1 cell is the major host cell used for production of recombinant therapeutic biologics (1). A number of subclones including ones with mutations in the dihydrofolate reductase and glutamine synthase (GS) genes as well as ones adapted to suspension culture in defined media have been developed and provided important improvements to the original CHO-K1 line. The genomes of the original CHO-K1 cell line and several subclones used in the industry today as well as the original Chinese hamster were recently sequenced (2, 3). The transcriptome of CHO-K1 was also characterized, and wider applications of Omics strategies to analyze and potentially design improved CHO clones for specific purposes are now possible (4, 5).
One of the major reasons for using CHO for production of human therapeutic glycoproteins is its general glycosylation capacity, which is largely compatible with and nonimmunogenic in man (1, 6). The glycosylation capacity of CHO cells with respect to N-glycan and O-glycan (GalNAc-type) structures is well understood from analysis of the glycosylation of recombinant expressed glycoproteins (7), but the capacity for this type of O-glycosylation with respect to sites and types of O-glycoproteins is largely unexplored. GalNAc-type O-glycosylation is controlled by a large family of up to 20 polypeptide GalNAc-transferases (GalNAc-Ts), which initiate O-glycosylation by catalyzing transfer of GalNAc to selected Ser and Thr (and potentially Tyr) residues in proteins (8). Each of these isoenzymes is differentially expressed in cells and tissues, and they have distinct albeit partly overlapping peptide acceptor substrate specificities. Our ability to predict O-glycosylation capacity of cells as well as overall O-glycosylation of proteins and particular sites of O-glycosylation is limited. An improved algorithm for overall prediction of O-glycosylation has recently been introduced (9), but this does not take into account the contribution of individual GalNAc-T isoforms and hence cannot be used to predict the O-glycosylation capacity of particular cells. One recent study identified several O-glycoproteins shed from CHO cells using metabolic labeling with UDP-GalNAz (10), but the identified O-glycoproteins were not characterized further in terms of O-glycan structures and sites. The O-glycoproteome of CHO cells is therefore virtually unexplored.
CHO-K1 was recently reported to express a limited subset of the GalNAc-T isoforms (2), and it is therefore expected that CHO can only support O-glycosylation of a fraction of human O-glycoproteins. Because only few O-glycoproteins have so far been expressed in CHO and analyzed in detail, our knowledge of potential problems with expression and O-glycosylation of proteins in CHO is quite limited. We have previously demonstrated that expression of fibroblast growth factor 23 (FGF23) in wild-type CHO is problematic, because FGF23 is cleaved at a proprotein convertase (PC) cleavage site (RHTR179⇓) and inactivated. CHO cells do not express the GalNAc-T3 isoform required to O-glycosylate T178 in the PC site, but if FGF23 is co-expressed with GalNAc-T3 in CHO cells the uncleaved active form of FGF23 is efficiently secreted (11). FGF23 is an important regulator of serum phosphate homeostasis and a potential therapeutic target. The repertoire of GalNAc-Ts in host cells can lead to surprises as was demonstrated when IL-17A was expressed in HEK293. Natural IL-17A is not O-glycosylated but when expressed in HEK293 the recombinant protein was found to carry one O-glycan (12). It is therefore clear that it is important to define the capacity for O-glycosylation of recombinant expression host cells and possibly modify this to meet specific requirements for O-glycoproteins.
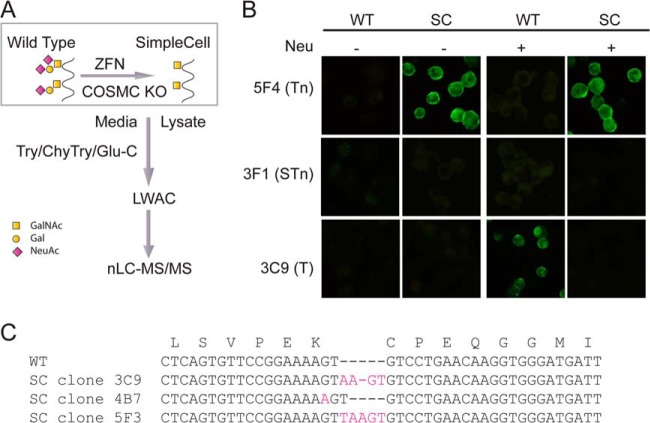
We previously developed the so-called SimpleCell strategy enabling proteome-wide discovery of O-glycoproteins and sites of O-glycan attachments (13). This strategy relies on stable genetic blockage of the O-glycan elongation pathway in cell lines leading to expression of the O-glycoproteome with homogeneous truncated O-glycans, which enables simple enrichment of all O-glycoproteins or O-glycopeptides and sensitive sequencing and identification by mass spectrometry (Fig. 1A) (14). Here, we applied this strategy to CHO cells by knocking out the Cosmc gene with a Zinc-finger nuclease (ZFN). Cosmc is a private ER chaperone required for the O-glycan core 1 synthase, C1Gal-T1, that catalyzes the second step in O-glycan elongation adding β3Gal to the initial GalNAc residues (Galβ1–3GalNAcα1-O-Ser/Thr) attached to the protein backbone (15). Thus, loss of Cosmc leads to abrogation of O-glycan elongation and synthesis of O-glycoproteins with only the initial GalNAc residue (GalNAcα1-O-Ser/Thr). Using CHO SimpleCells we characterized the O-glycoproteome in total lysates as well as the secretome and identified a total of 738 O-glycoproteins and 1548 O-glycosites. Partial analysis of the O-glycoproteome of CHO wildtype (WT) cells using a different lectin capture strategy further increased the total number of identified O-glycoproteins (824) and glycosites (1727). Analysis of this data set confirm that CHO cells have limited capacity for O-GalNAc glycosylation, and this opens for engineering strategies to produce CHO cells with improved properties.
Fig. 1.
The SimpleCell O-GalNAc glycoproteome strategy. A, ZFN targeting of the core-1 synthesis step by knockout of Cosmc simplifies all O-glycosylation to GalNAc (Tn), which allows isolation of GalNAc-glycopeptides released in total proteolytic digests of cells by VVA lectin chromatography. nLC-MS/MS coupled with ETD is used to sequence glycopeptides and determine sites of O-glycosylation. Symbols for monosaccharides GalNAc, Gal, and sialic acid are indicated. B, Fluorescence micrographs showing immunocytochemical staining of the SimpleCell lines 3C9 and corresponding wild-type cell line (Tn: monoclonal antibody 5F4; STn: monoclonal antibody 3F1; T: monoclonal antibody 3C9; plus or minus neuraminidase (Neu). C, Sequences of ZFN target site in the Cosmc gene, highlighting ZFN-introduced mutations in CHO cells. Only one allele was detected in CHO SimpleCell. WT, wild type; SC, SimpleCell.
MATERIALS AND METHODS
Generation of O-GalNAc CHO SimpleCells
A ZFN targeting construct for Cosmc was custom produced (Sigma-Aldrich, St. Louis, MO) targeted toward the Cosmc sequence 5′-GCCTTCTCAGTGTTCCGGAaaagtgTCCTGAACAAGGTGGGAT-3′ (FokI nuclease cutting site is indicated in lowercase). CHO-GS (glutamine synthetase deficient) (Sigma-Aldrich) cells, which is used as CHO WT cell in this study, were maintained as suspension cultures in EX-CELL CHO CD Fusion serum-free media supplemented with 4 mm l-glutamine. All culture media, supplements, and other reagents used were obtained from Sigma-Aldrich unless otherwise specified. Cells were seeded at 0.5 × 106 cells/ml in T25 flasks (NUNC, Roskilde, Denmark) 1 day prior to transfection. Transfections were conducted with 2 × 106 cells and 2 μg endotoxin free plasmid DNA of each Z by electroporation using Amaxa kit V and program U24 with Amaxa Nucleofector 2B (Lonza, Switzerland). Electroporated cells were subsequently placed in 3 ml growth media and 5 days later plated out as single cells in round bottom 96-well plates. Single cell sorted knockout clones were identified by immunocytochemistry using monoclonal antibody (MAb) 5F4, which detects the truncated Tn O-glycan (16). Selected clones were verified by PCR followed by Sanger sequencing to define the nature of the induced mutations in the Cosmc gene. Mutants were further characterized in detail by immunocytochemistry using MAbs to other O-glycan structures STn (3F1) and T (3C9) with and without neuraminidase pretreatment as described previously (16).
Sample Preparation
CHO SimpleCells: Two T175 flasks with SimpleCells (0.5 × 106 cell/ml in 200 ml) cultured for 48–72 h were harvested and cells were washed in ice-cold PBS. Total cell lysates (TCL) were prepared by resuspending the cell pellet in 1 ml 50 mm ammonium bicarbonate, 0.1% RapiGest followed by sonication (13). For preparation of secretomes conditioned media was dialyzed and glycoproteins first enriched by capture on a short (300 μl) VVA agarose column that selectively binds αGalNAc (17). Glycoproteins were eluted by heating (4 × 90 °C 10 min) with 0.05% RapiGest. Cell lysates and enriched glycoproteins from media were reduced by 5 mm dithiothreitol (30 min, 60 °C) and alkylated by 10 mm iodoacetamide (30 min, room temperature (RT)). Each sample was then digested with trypsin, purified by C18 solid phase extraction and diluted in binding buffer and subjected to VVA LWAC as previously described (13, 14). In order to increase the depth of analysis sets of cell lysate and enriched media were prepared by the same way using chymotrypsin and Glu-C instead of a trypsin as an alternative digestion strategy. Thus, six GalNAc-glycopeptide containing samples were prepared for analysis.
CHO WT cells: A similar enrichment strategy was applied to CHO WT cells relying on enrichment by PNA lectin binding selectively to Galβ1–3GalNAc after neuraminidase as described previously (13). Total cell lysates were prepared as described above and treated with 100 units neuraminidase (Clostridium perfringens) (New England Biolabs, Ipswich, MA) (37 °C, 2 h). Conditioned medium was treated with 0.1 U/ml neuraminidase (Clostridium perfringens) (Sigma) (37 °C, 2 h), and glycoproteins with Galβ1–3GalNAc O-glycans enriched on a short PNA agarose column (800 μl). Cell lysates and enriched glycoproteins from media were digested with trypsin and subjected to PNA LWAC as previously described (13). After the enrichment of T-glycopeptides by PNA LWAC, the flow through (containing a mixture of peptides and potential Tn (GalNAcα)-glycopeptides) was further re-enriched by VAA LWAC.
Mass Spectrometry
All samples were further fractionated by isoelectric focusing to reduce sample complexity (17), desalted by Stage Tips (Empore disk-C18, 3 m), and dissolved in 0.1% formic acid. Samples were analyzed on an EASY-nLC 1000 UHPLC (Thermo Scientific) interfaced via nanoSpray Flex ion source to an LTQ-Orbitrap Velos Pro spectrometer (Thermo Scientific). A precursor MS1 scan (m/z 350–1700) of intact peptides was acquired in the Orbitrap at a nominal resolution setting of 30,000, followed by Orbitrap HCD-MS2 and ETD-MS2 (m/z of 100–2000) of the five most abundant multiply charged precursors in the MS1 spectrum; a minimum MS1 signal threshold of 50,000 was used for triggering data-dependent fragmentation events; MS2 spectra were acquired at a resolution of 7500 for HCD MS2 and 15,000 for ETD MS2. Activation times were 30 and 200 ms for HCD and ETD fragmentation, respectively; isolation width was four mass units, and usually one microscan was collected for each spectrum. Automatic gain control targets were 1,000,000 ions for Orbitrap MS1 and 100,000 for MS2 scans, and the automatic gain control for fluoranthene ion used for ETD was 300,000. Supplemental activation (20%) of the charge-reduced species was used in the ETD analysis to improve fragmentation. Dynamic exclusion for 60 s was used to prevent repeated analysis of the same components. Polysiloxane ions at m/z 445.12003 were used as a lock mass in all runs.
Data Analysis
Data processing was performed using Proteome Discoverer 1.4 software (Thermo Scientific) as previously described with small changes (9). Sequest HT node was used instead of Sequest. Because of the high speed of Sequest HT data processing, all spectra were initially searched with the full cleavage specificity, filtered according to the confidence level (medium, low, and unassigned) and further searched with the semispecific enzymatic cleavage. In all cases the precursor mass tolerance was set to 6 ppm and fragment ion mass tolerance to 50 mmu. Carbamidomethylation on cysteine residues was used as a fixed modification. Methionine oxidation and HexNAc and HexHexNAc attachment to serine, threonine, and tyrosine were used as variable modifications for ETD MS2. All HCD MS2 were preprocessed as described (9) and searched under the same conditions mentioned above using only methionine oxidation as variable modification. All spectra were searched against a concatenated forward and reverse CHO-specific database (UniProt, October 2012, containing 24,382 canonical entries) using a target false discovery rate (FDR) of 1%. FDR was calculated using target decoy PSM validator node, a part of the Proteome Discoverer workflow. The resulting list was filtered to include only peptides with glycosylation as a modification. This resulted in a final glycoprotein list identified by at least one unique glycopeptide. Only ETD MS2 data were used for unambiguous site assignment. Single peptide and PMS identifications are compiled as indexed reference spectra for each sample (supplemental Spectra). Indexed reference spectra are stored as zip folder containing index.xlsx (for navigation) and data folder containing all supportive spectra with assignment. Upon removal of data duplicates the best score glycopeptides are listed as Excel table for each individual sample with the related information such as: accession number, protein name, range, score, etc. (supplemental Tables S2–S11). These tables represent the detailed information of the summary table (supplemental Table S1).
Expression of Recombinant Human EPO in CHO WT/SC
An expression construct containing the entire coding sequence of human erythropoietin (EPO) in pcDNA3.1/myc-His (C-terminal tags) was synthesized (Genewiz, Carol Stream, IL). CHO WT and SC cells were transfected by nucleofection and stable transfectants selected in 0.4 mg/ml Zeocin (Invitrogen, Carlsbad, CA) and screened by immunocytochemistry using anti-myc MAb (9E10) and by direct enzyme-linked immunosorbent assays (ELISA) using monoclonal anti-His antibody (C-Term)-HRP antibody (Invitrogen). Stable clones were selected after two rounds of limited dilution. His-tagged EPO was purified by nickel affinity purification (Qiagen, Valencia, CA). Media was mixed 3:1 (v/v) in 4× binding buffer (200 mm sodium phosphate, pH 8.0, 1.2 m NaCl) and applied to prepacked nickel-nitrilotriacetic acid (Ni-NTA) affinity resin column (Qiagen), pre-equilibrated in binding buffer (50 mm sodium phosphate, pH 8.0, 300 mm NaCl). The column was washed with binding buffer and bound protein eluted with 300 mm imidazole in binding buffer. EPO containing fractions were identified by SDS-PAGE and further purified by reverse-phase HPLC (1100 Hewlett Packard system) with a Jupiter 5 μ C4 300A column (Phenomenex, Torrence, CA), using 0.1% trifluoroacetic acid (TFA) and a gradient of 10–100% acetonitrile. For mass spectrometry analysis 1∼5 μg purified EPO was reduced (5 mm DTT, 60 °C, 30 min) and alkylated (10 mm iodoacetamide, RT, 30 min), followed by digestion with trypsin (100 ng, Roche) and purified by Stage Tip columns (Empore disk-C18, 3 m). Mass spectrometry analysis was performed as previously described (18).
RESULTS
Development of CHO SimpleCells
The SimpleCell strategy is depicted in Fig. 1. The key feature is stable gene inactivation of Cosmc whereby the common elongation pathway of O-glycosylation is abrogated and O-glycan structures are simplified to the most immature GalNAcα1-O-Ser/Thr (Tn) structure. Cosmc knockout cells were easily identified by immunocytochemistry with MAb 5F4 detecting the de novo induction of the truncated Tn-structure. MAb 5F4 did not stain CHO WT cells but stained cloned CHO SimpleCells homogeneously (Fig. 1B). Importantly, CHO SC does not produce sialyl-Tn (STn) structures as demonstrated by immunocytochemistry using MAb 3F1 recognizing STn. In agreement with previous studies (16), CHO WT cells produce sialyl-T O-glycans as demonstrated by positive MAb 3C9 staining only after neuraminidase treatment (Fig. 1B).
Knockout of Cosmc was verified by target specific PCR followed by Sanger sequencing of TOPO cloned products (Fig. 1C). The results indicate that CHO cells only have one copy of the Cosmc gene, and the introduced mutations in the selected clones gave rise to small insertions, that is, +4bp, +1bp, and +5bp in the CHO SC clones designated 3C9, 4B7, and 5F3, respectively. The mutations are all out-of-frame resulting in disruption of the Cosmc protein. All SC clones were fully viable and no gross variations in growth and morphology were observed. Furthermore, in a preliminary analysis of the transcriptome of four of the CHO cell lines (CHO SC clones 3C9 and 4B7, WT cell, and the original CHO-K1 clone) by RNAseq, we did not observe changes in expression of any of the GALNTs (not shown).
Mapping of O-Glycosites in CHO SimpleCells
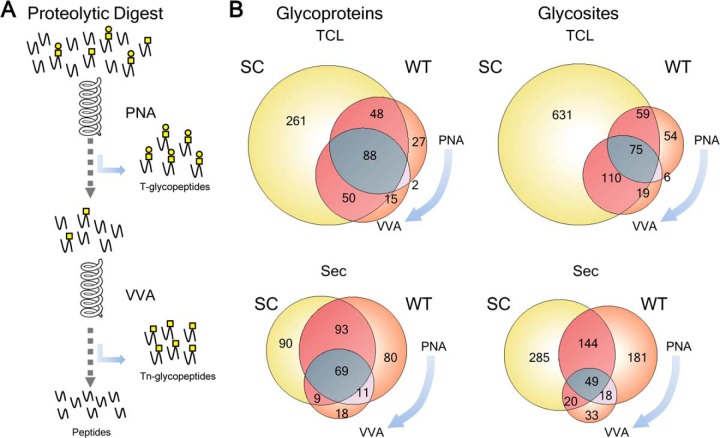
CHO SC produce homogenous truncated O-glycan structures with only the αGalNAc monosaccharide attached to the protein backbone, and this greatly simplify isolation of O-glycoproteins and glycopeptides by VVA lectin chromatography as well as spectral interpretation and data processing for sequencing of O-glycopeptides and identification of O-glycosites (Fig. 1A) (9, 13). CHO SC cells produce only Tn structures without further capping by sialic acid, as the responsible gene ST6GalNAc-I is not expressed in CHO cells (Fig. 1B) (16). However, CHO WT cells exclusively produce core1 mono- and disialylated structures (7). We used two lectins to compare T capture by PNA lectin in WT cells (after pretreatment with neuraminidase) and Tn capture by VVA in SC cells, respectively. We analyzed both total cell lysates (TCL) and secretomes (Sec) using three proteases for digestion (trypsin, chymotrypsin, and Glu-C) and orthogonal fractionation by IEF prior to nLC-MS analysis (17). Fractions were analyzed by nLC-MS with data-dependent acquisition protocols including HCD-MS2 and ETD-MS2 from the same precursors and identified glycoproteins, glycopeptides, and glycosites are listed in Supplemental Table 1. Analysis of TCLs yielded more identified O-glycoproteins and glycosites than secretomes, but analysis of secretomes led to identification of a new subset of O-glycoproteins (Fig. 2). Capture of secreted O-glycoproteins in conditioned media utilizing a short VVA lectin chromatography step for enrichment before digestion was originally developed to capture endogenously secreted proteins from SimpleCell lines growing in serum containing medium to avoid bovine O-glycoproteins (19). As CHO GS grow in serum-free media the short VVA column mainly served to enrich for Tn glycoproteins and reduce the sample volume. The use of chymotrypsin and Glu-C in addition to the golden-standard, trypsin, substantially increased the identified O-glycoproteins and O-glycosites although trypsin produced the largest data set (Supplemental Fig. S1). In total 738 O-glycoproteins and 1548 unambiguously assigned O-glycosites were identified in SC (Fig. 2) (http://glycomics.ku.dk/o-glycoproteome_db).
Fig. 2.
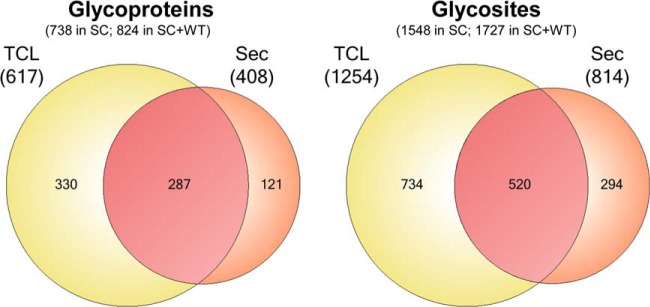
Summary of O-glycoproteins and O-glycosites identified in CHO SC and WT cells. Comparative analysis of glycoproteins and glycosites determined in CHO SC lysate and secretome from a cumulative data of three different proteolytic digests: trypsin, chymotrypsin, and GluC (see supplemental Fig. S1). Contribution of the tryptic digest of CHO WT data (Fig. 3) to the entire compendium is shown as cumulative numbers in parenthesis as “SC+WT”.
Mapping of O-Glycosites in CHO WT Cells
CHO WT cells produce T (Galβ3GalNAcα) based O-glycans with sialic acid capping (Fig. 1B). Thus, removal of sialic acids by neuraminidase treatment produces homogeneous O-glycoproteins with T O-glycan structures that enable capture by the lectin PNA. PNA lectin chromatography is less efficient compared with VVA (14), but still applicable to lectin chromatography enrichment. In order to compare efficiency of this strategy we analyzed and compared trypsin digests of both a total cell lysate and secretome of CHO WT cells (Fig. 3). The strategy resulted in identification of substantially lower numbers of O-glycoproteins and glycosites in total cell lysates with 230 O-glycoproteins compared with 447 identified from CHO SC trypsin digests (overlapping 186) (Fig. 3B), and 323 O-glycosites compared with 875 from CHO SC trypsin digests (Fig. 3B). Somewhat surprising, the same analysis of secretomes yielded a slightly different picture where the identified O-glycoproteins (total 280) overlapped less well (overlap 171) with those identified from CHO SC (total 261) (Fig. 3B). Although these results clearly show that the SimpleCell strategy is more sensitive for discovery of O-glycoproteins, we were puzzled by the relative poor overlap with the secretomes from SC and WT cells, and therefore tested if a subset of O-glycoproteins with truncated Tn O-glycans were secreted from WT cells. We applied the pass through of the trypsin digests of WT lysates and secretomes from the PNA LWAC step to VVA LWAC (Fig. 3A). This resulted in identification of a subset of the same proteins identified from the PNA LWAC in addition to O-glycoproteins only identified by the CHO SC strategy. Interestingly, the latter fractions of O-glycoproteins found in CHO SC were substantially larger for lysates than for secretomes, which would be consistent with the notion that analysis of lysates would enable identification of early precursor stages of O-glycoproteins that eventually maturate with sialylated T-glycans. In agreement with this analysis the secretome should have more completely glycosylated O-glycoproteins. This supports the hypothesis that the SimpleCell strategy is more sensitive. Nevertheless, the contribution of this data to the CHO SC O-glycoproteome compendium increased the final numbers to 824 O-glycoproteins and 1727 O-glycosites.
Fig. 3.
Analysis of the CHO WT O-glycoproteome. A, Depiction of glycopeptides enrichment strategy (T- and Tn-epitopes) based on a sequential PNA and VVA LWAC. B, Comparative analysis of a tryptic digest subset of O-glycoproteins and O-glycosites identified in CHO SC (left) and a tryptic digest of CHO WT (right). O-glycoproteins and glycosites discovered in a cell lysate and a secretome in CHO WT are after PNA and subsequent VVA LWAC enrichment.
A First Generation GalNAc-Type O-Glycoproteome of CHO
We previously presented a first draft of the human O-glycoproteome developed from 12 human cancer SimpleCells (9). This O-glycoproteome was assembled with less sensitive methods compared with the current process because we mainly processed trypsin digests and analysis was done on an older generation of the OrbiTrap instrument (OrbiTrap XL). Nevertheless it is valuable to compare the O-glycoproteomes of human and CHO cells (Fig. 4), and to do this we first assembled the CHO O-glycoproteins for which it was possible to establish unambiguous orthologous human proteins (713/824). Interestingly, out of these 713 CHO O-glycoproteins only 258 overlapped with 926 human previously identified O-glycoproteins (9) (unpublished). Given that the human O-glycoproteome is based on >12 cell lines with different organ origins it seems likely that the human O-glycoproteome would be larger and that CHO would represent a subset, but the CHO O-glycoproteome included a larger fraction of proteins not found previously than the fraction that overlapped. At first sight this may suggest that there is a fundamental difference in the O-glycoproteomes of CHO and man. Two main factors could explain such a fundamental difference: (1) differences in expression and secretion of proteins; and (2) differences in expression of GalNAc-Ts and hence O-glycosylation of proteins. However, technical differences are likely also at play in that CHO was thoroughly mapped using three proteases and the analysis was performed on the OrbiTrap Velos Pro rather than the XL as used for human cell lines (9). More studies are clearly needed to fully address these concerns. Nevertheless, the finding that the overlap of the CHO and human O-glycoproteomes is limited and the larger number of identified human and CHO O-glycoproteins being nonoverlapping (Fig. 4), does appear to be in agreement with the finding that CHO cells only express a limited subset of GalNAc-Ts.
Fig. 4.
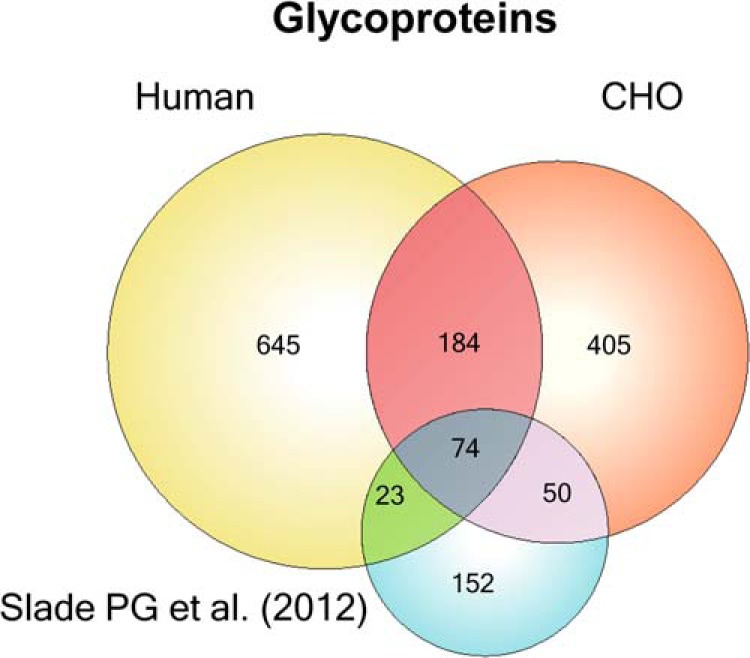
Comparative analysis of CHO and human O-glycoproteomes. O-GalNAc proteins previously discovered in Human SimpleCell lines (9, 17), CHO glycoproteins discovered by GalNAz click chemistry (10), and CHO glycoproteins from the current manuscript. In order to obtain ortholog mappings from CHO to human, InParanoid 4.1 was run on the CHO proteome as obtained from UniProt, as well as the human reference proteome. Proteins from the Slade et al. data set were first mapped to the CHO proteome (yielding 286 protein identifiers) using InParanoid and then mapped to the human proteome using the previous mapping. It should be noted that the orthology mappings obtained may not necessarily comprehensive because of the draft nature of the CHO genome.
A recent study used the metabolic labeling strategy with UDP-GalNAz developed by the Bertozzi group (20) to probe the O-glycoproteome of the secretome of CHO DG44 and S cells (10). The study demonstrated that endogenous CHO O-glycoproteins could be identified by this strategy and 352 putative O-glycoproteins were identified in CHO DG44 and CHO S cells, although actual sites of glycosylation were not determined (10). We compared this data set with our data set from CHO SimpleCells as well as the larger human O-glycoproteome established by multiple human SimpleCells (Fig. 4). We only analyzed CHO proteins with identifiable human orthologs, which was 299 out of the 352 putative O-glycoproteins (10). Of these 299 proteins, 124 overlapped with the O-glycoproteins identified in the present study. When including our human O-glycoproteome data set an additional 23 were overlapping (Fig. 4). Of the remaining 152 nonoverlapping proteins most of these (127) are predicted to be glycosylated using the NetOGlyc4.0 predictor (9).
We have recently identified a significant number of Tyr O-GalNAc glycosylation sites in the human O-glycoproteome (9). Here, we also found 29 unambiguously assigned Tyr O-glycosites in CHO SC, and a further 14 Tyr glycosites in the secretome of CHO WT. Interestingly, we identified essentially equal number of Tyr glycosites in lysate and secretome of CHO SC, but in CHO WT we only found these sites in the secretome.
Recently, the most comprehensive proteome analysis of CHO was performed identifying over 6000 proteins, where almost 1300 were identified as N-glycoproteins (4). Comparison of the O-glycoproteome identified here with this proteome showed overlap for about half of the O-glycoproteins (Fig. 5), and as many as 375 (out of 803) of the O-glycoproteins were not identified in the proteome analysis. Comparison of the O-glycoproteome with the N-glycoproteome established by Baycin-Hizal and colleagues (4) demonstrated that 217 proteins are both N- and O-glycosylated (not shown). We also compared our O-glycoproteome with results from a recent study of the secretome of CHO searching for autocrine growth factors using serum-free media (21). The identified proteins from this data set show relatively low overlap between both the complete proteome analysis and our data set. That the bulk of sites from both our data set and the autocrine growth factor data set were not identified in the complete proteomic study points toward the importance of a variety of enrichment strategies to obtain more comprehensive proteome sets.
Fig. 5.
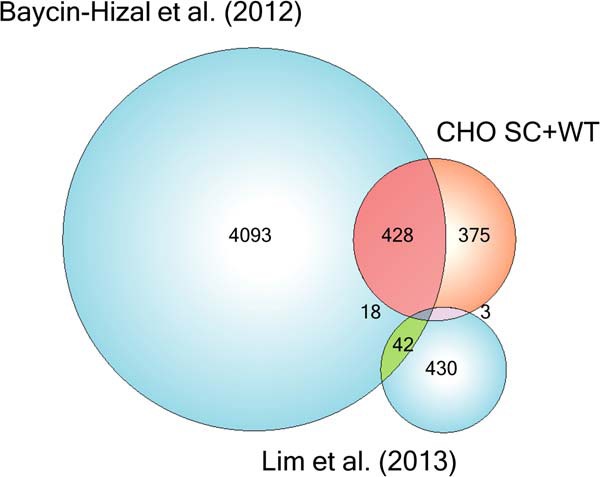
Comparison of analyses of the CHO proteome and glycoproteome. The sets of proteins identified in the current manuscript, a complete proteomic study (4), a study of secreted autocrine growth factors (21), and their respective overlaps.
Cellular and Functional Classification of CHO O-Glycoproteins
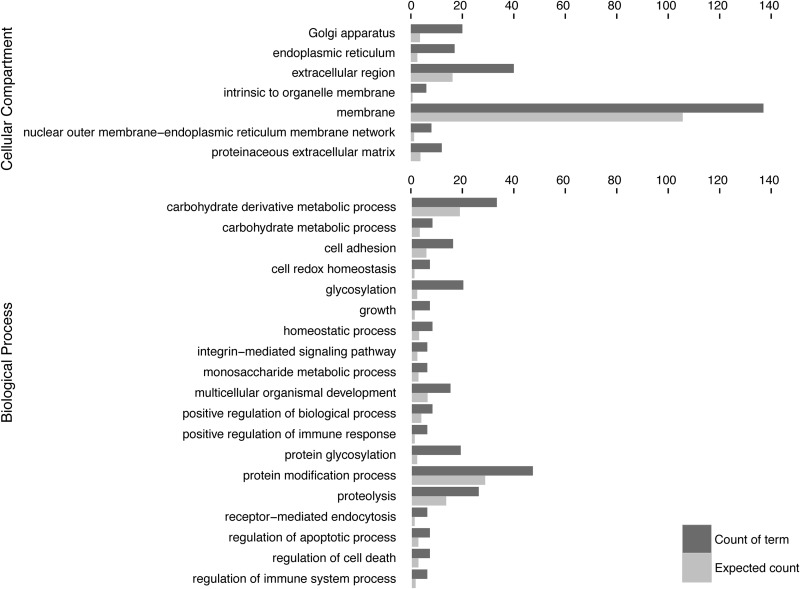
Cellular component gene ontology analysis (GO) (22) of the identified CHO O-glycoproteins showed overrepresentation of ER/Golgi and extracellular compartment terms in agreement with our previous studies (Fig. 6) (9). This was further in agreement with biological process analysis where overrepresentation of glycosylation and protein modification was found.
Fig. 6.
Cellular component gene ontology analysis. Gene Ontology enrichment for the WT and SC CHO glycoproteomes were calculated for biological process and cellular component. The cellular component enrichment clearly illustrates a preference for extracellular and membrane-associated localizations, whereas the biological process enrichment illustrates terms commonly found associated with O-glycoproteins. The GO enrichment analysis was performed in R using the GOStats package and GO annotations from UniProt-GOA.
GlycoDomainViewer Analysis of the CHO O-Glycoproteome
We recently used the GlycoDomainViewer to perform an analysis of both the global and local features of the human O-glycoproteome characterized to date (23), and we used the same strategy to examine features of the CHO O-glycoproteome. The analyses were performed on the subset of 1632 unambiguously defined O-glycosites that had sufficient domain annotation information (Conserved Domain Database (CDD)). The global site distribution on proteins is similar to earlier studies of the human O-glycoproteome (9), as the bulk of the O-glycosites (45%) in the CHO O-glycoproteome was found as single isolated sites on proteins, with another 20% of the proteins having two identified sites, and 10% having three identified sites. Proteins that undergo O-GalNAc glycosylation should enter ER and the secretory pathway through a signal peptide and enrichment of proteins with signal peptides in the O-glycoproteome is therefore expected. Curiously SignalP predicted that only 25% of the CHO O-glycoproteins identified have a signal sequence. This is somewhat in contrary to our previous studies where 68% of the identified human O-glycoproteins were predicted to have a signal sequence by SignalP. Manually inspecting the list of CHO O-glycoproteins, however, it is evident that the majority of the proteins identified have N-terminal hydrophobic sequences most likely constituting signal peptides and hence enter the secretory pathway.
Although the lectin enrichment strategy utilized is highly selective for O-GalNAc glycopeptides, we still find a substantial fraction of peptides in the eluate and presumably by abundance also a minor fraction O-GlcNAc glycopeptides derived from cytosolic O-glycoproteins (24). Because we are unable to distinguish O-GalNAc from O-GlcNAc by the utilized mass spectrometry strategy, a minor amount of O-GlcNAc glycopeptides may contaminate the data set. We have previously shown that this is a very minor problem in our studies of the human O-GalNAc glycoproteome (9), and in the present data set we identified 10 proteins, representing <2% of the VVA enriched glycoproteome, which are most likely modified by O-GlcNAc (supplemental Table S1).
Analysis of the human O-glycoproteome for local site context features previously revealed that most of the O-glycosites are positioned in unstructured regions on proteins, specifically on linker regions and stem regions of transmembrane proteins (type I and II), whereas just under 25% of the O-glycosites were in structured regions (9, 23). When the same analysis was performed on the CHO O-glycoproteome, it was similarly seen that most of the unambiguously identified O-glycosites were found in unstructured regions. Stem regions of transmembrane proteins often carry O-glycosylation, and this may be associated with protection and perhaps modulation of ectodomain shedding (25). In stem regions of type II transmembrane proteins we found 51 O-glycosites, and these proteins were characterized by having ectodomains with predicted glycosyltransferase function (GT-A folds, GT29 domains) or TNF superfamily domains. We found 162 O-glycosites in stem regions of type I transmembrane proteins and the ectodomains of these frequently carried Erv1/ALR family, TNFR, and HA binding link domains. Only 12% of the O-glycosites (208 of 1632 unambiguously defined sites that could be analyzed) were observed in conserved domain folds. A number of conserved domains with glycosites were found in multiple proteins, and the most frequently observed domains include a macrophage colony stimulating factor fold, various IG domain folds, and a thioredoxin fold.
Linker regions between conserved domains account for a large proportion of the protein context classifications of the O-glycoproteome (40%, 646 sites). Linker regions are defined as short flexible regions placed in-between conserved folds and they presumably function to extend the distance between domains. These linker regions carrying O-glycosites were found most frequently associated with domains such as fibronectin, laminin G or EGF domains, SEA domains, and Ig domains.
Recombinant Expression of EPO in CHO
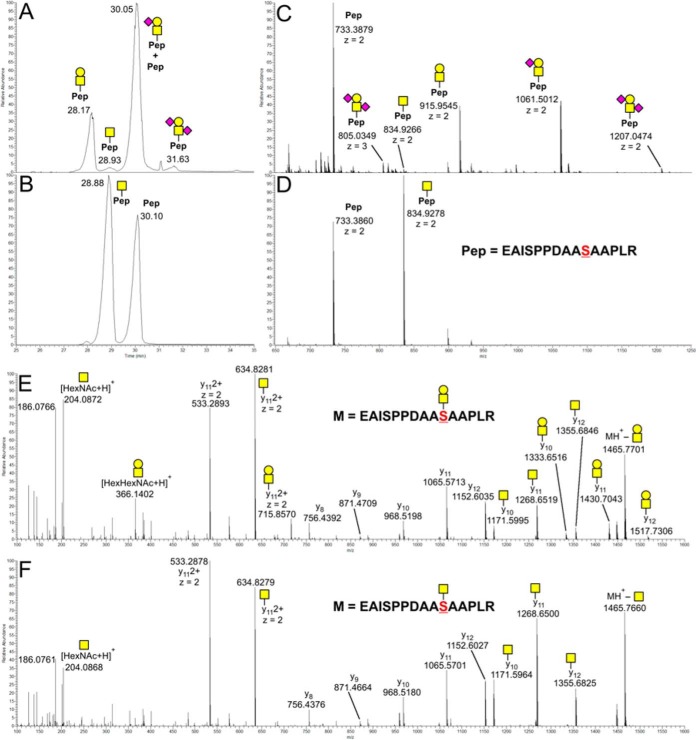
We assessed O-glycosylation capacity of CHO WT and SC by expressing human EPO. EPO was selected as reporter protein because of its well-characterized single O-glycosylation site (Ser126), which is known to have partial occupancy and carry mono and disialyl-T O-glycan structures (26). EPO also has three N-glycosylation sites (Asn24, Asn38, and Asn83) that are occupied with heterogeneous tri- and tetraantennary N-glycans (26). EPO from WT cells was found to have partial O-glycan occupancy (∼50%) with a sialylated core1 structure (Fig. 7). In contrast, EPO expressed in CHO SC carried only the truncated Tn structure. This demonstrates that CHO SC can be used for production of O-glycoproteins with truncated Tn O-glycans.
Fig. 7.
nLC-MS Analysis of a tryptic digest from human EPO recombinantly expressed in CHO. O-glycosylation capacity of CHO WT and SC is shown for the recombinantly expressed human EPO using a tryptic glycopeptide (144)EAISPPDAASAAPLR(158) with the well-characterized O-glycosylation site (Ser126) as an example. A, XIC of the LC range from 25–35 min of a tryptic digest from human EPO expressed in CHO WT. The corresponding mass spectrum acquired in the same range is shown on panel C. B, XIC of the LC range from 25–35 min of a tryptic digest from human EPO expressed in CHO SC. The corresponding mass spectrum acquired in the same range is shown on panel D. E, ESI-Orbitrap-HCD-MS2 spectrum of the precursor ions at m/z 915.9545, z = 2+ (panel C). F, ESI-Orbitrap-HCD-MS2 spectrum of the precursor ions at m/z 834.9278, z = 2+ (panel D).
DISCUSSION
Here, we developed CHO SimpleCells and provided the first in-depth characterization of the CHO O-glycoproteome with defined O-glycosylation sites. The O-glycoproteome was found to constitute a subfraction of the human O-glycoproteome identified from 12 different human cancer cell lines, which is in agreement with expression data on polypeptide GalNAc-T isoforms in CHO cells suggesting that CHO indeed has limited capacity for O-glycosylation. This finding has implications for use of CHO for production of O-glycoproteins and paves the way for design of engineered CHO cells with improved properties. We further demonstrate that CHO SimpleCells are useful for recombinant production of O-glycoproteins with homogeneous GalNAcα1-O-Ser/Thr O-glycans.
The CHO cell is the major production platform for recombinant therapeutics and most of these are glycoproteins (27). The major focus on glycosylation capacity in CHO cells has been devoted to the N-glycosylation pathway primarily with respect to sialylation and core fucosylation as these parameters serve important functions for circulatory half-life of therapeutics and effector functions of IgGs, respectively (28, 29). Little attention has been drawn to O-glycosylation in CHO and only a few therapeutic proteins produced in CHO have been characterized with respect to their O-glycans. EPO is the best characterized example and the first to demonstrate that CHO produce simple sialylated T O-glycans without branching (7). The TNFα receptor IgG1-Fc-fusion protein (Etanercept, Enbrel) is perhaps the therapeutic with the highest number of O-glycans (up to 12) identified to date and considerable efforts have been applied to identify and characterize these glycosites and the O-glycans structures (30). Receptor fusion proteins designed like Etanercept will likely often have clustered O-glycans in the linker region between the extracellular receptor part and the Fc domain, because stem regions of cell membrane receptors often have O-glycosylation in this region to help extend and protrude the receptor binding domain and prevent proteolytic release of the ectodomain. O-glycosylation on therapeutic fusion proteins may thus be very important for stability, and it will be desirable to ensure that as many O-glycosites as possible or at least those used in the natural protein in vivo are utilized when expressed recombinantly in CHO. The SimpleCell approach enables sensitive identification of O-glycoproteins with glycosites utilized in cells from total cell extracts and media using shotgun mass spectrometry approaches, whereas determination of the stoichiometry of occupancy at individual glycosites can be performed with recombinantly expressed and purified O-glycoproteins as exemplified here with EPO.
The GalNAc-type O-glycoproteome has for long remained elusive because of analytical obstacles and lack of reliable predictors. Several different O-glycoproteomics strategies have been developed in the last decade including different enrichment techniques (31, 32), metabolic incorporation of GalNAz enabling tagging and isolation of O-glycoproteins (20), and the SimpleCell strategy (13). The latter has so far produced the deepest coverage/largest data set with human cancer SimpleCells (9), and applied to CHO we obtained the largest number of O-glycoproteins and O-glycosites identified in a single cell so far. This was partly because of the use of three enzymes for proteolysis (17), and partly through use of the OrbiTrap Velos Pro with ETD.
In principle any mammalian cell will have similar capacity for attaching N-glycans to N-glycosylation sites encoded in the protein sequence by the oligosaccharyltransferase complex (33). O-glycosylation is in contrast highly differentially regulated in cells through the repertoire of polypeptide GalNAc-Ts (8) and the identified O-glycoproteome may be used to estimate the CHO cells' capacity for O-glycosylation. The first view of the CHO O-glycoproteome and comparison with our extended human O-glycoproteome data suggests that CHO has limited capacity for O-glycosylation, which is also supported by recent whole transcriptome analysis where only a small subset of GalNAc-Ts were found to be expressed (2).
Slade and colleagues (10) used the GalNAz metabolic labeling of O-glycoproteins and identified secreted putative O-glycoproteins from CHO-S and DG44 cells, and this set showed some overlap with the O-glycoproteins identified here (Fig. 4). Both the SimpleCell and metabolic labeling strategies rely on an enrichment step to isolate endogenously produced and secreted glycoproteins, and the lower coverage provided by the latter strategy is likely partly because of lower incorporation of GalNAz compared with GalNAc. Furthermore, care should be taken to interpret all GalNAz identified glycoproteins as O-GalNAc type glycosylated as UDP-GalNAz may be converted to UDP-GlcNAz in cells by the C4-Glc/GlcNAc epimerase and incorporated into O-GlcNAc glycoproteins (34). Baycin-Hizal and colleagues (4) also determined the largest N-glycoproteome of CHO with over 1200 N-glycoproteins using hydrazide-beads enrichment strategy of N-glycopeptides. It is important to note that this study used “normal” H16O water during PNGase F release of N-glycans but even the replacement of “normal” water by H18O water during enzymatic digestion to circumvent natural deamidation of Asn, may not completely abolish this issue (35).
The CHO cell has a long history with respect to mutants with altered glycosylation (36). A large number of lectin resistant mutants have been developed in the past and the enzymatic and in some cases the genetic deficiencies underlying the altered glycosylation capacity have been mapped through elegant work. Given the accomplishment of whole genome sequencing of several CHO lines and the original Chinese hamster (2, 3), CHO has entered a new era with respect to genetic engineering to improve desirable features. The first successful design engineering of glycosylation in CHO was performed by two rounds of highly laborious and time consuming homologous recombinations to knockout FUT8 (37), but with the emerging precise gene editing technologies the efforts required have been drastically reduced as demonstrated by fast knock out of FUT8 with ZFN-mediated gene targeting (38). More recently, MGAT1 was knocked out in CHO GS cells to design a CHO platform for production of N-glycoproteins with homogeneous high mannose N-glycosylation, which is desirable for crystallization studies (39). The emergence of the precise gene editing technologies is expected to have major impact on custom design of glycosylation capacities of CHO and other cells (40).
Here, we demonstrated that trimming the entire elongation pathway of O-glycosylation in CHO cells is possible by targeting the Cosmc gene, which results in a cell factory enabling production of O-glycoproteins with truncated homogenous Tn (GalNAcα) O-glycans. This is important not only for the O-glycoproteomics studies performed here but also for production of O-glycoproteins with enhanced immunogenicity. The Tn glycoform is one of the most broadly expressed cancer glycoforms, natural IgM anti-Tn antibodies in man account for the polyagglutinability phenomenon, and spontaneous Tn O-glycopeptide specific IgG antibodies are found in cancer patients (41–43). Moreover, such Tn O-glycopeptide specific antibodies can have true cancer-specific reactivity (16), and they may be elicited in cancer patients in part by recognition of Tn O-glycoproteins by the MGL lectin receptor of dendritic cells (44). CHO SimpleCells may thus be valuable for production of O-glycoproteins for use in producing cancer vaccines (41).
Another interesting application for CHO SimpleCells would be in the site-directed O-glycopegylation strategy designed to enhance half-life of therapeutic proteins. We previously developed an in vitro O-glycosylation strategy to introduce GalNAc residues at specific sites in E. coli produced proteins (e.g. CSF, INFα2b, and GM-CSF), which could subsequently be used for transfer of sialic acid with a PEG molecule using a sialyltransferase and CMP-sialic acid with a linear methoxypolyethylene glycol linked to the 5′-amino nitrogen of the sialic acid residue (CMP-SiaPEG-20K) (45). The limitations of this strategy were requirement for expression of the protein to be pegylated in E. coli with potential problems with refolding and activity of protein and the subsequent in vitro enzymatic incorporation of GalNAc residues. CHO SimpleCells can in principle now be used to express folded O-glycoproteins with appropriate GalNAc residues for direct enzymatic pegylation.
In summary, the CHO SimpleCell system established in this report has enabled the first comprehensive insight into the O-glycoproteome and the O-glycosylation capacity of the CHO cell. The CHO SimpleCell serves as a discovery platform for further engineering of CHO to improve O-glycosylation capacity, and it may in itself have interesting applications as a host cell for production of novel therapeutics.
Supplementary Material
Footnotes
Author contributions: S.Y.V. and H.C. designed research; Z.Y., A.H., C.S., K.T.S., S.B.L., and S.Y.V. performed research; Y.N., H.J.J., C.S., K.T.S., M.A.S., N.R.S., K.J.K., and E.P.B. contributed new reagents or analytic tools; Z.Y., A.H., H.J.J., S.B.L., S.Y.V., and H.C. analyzed data; Z.Y., A.H., S.Y.V., and H.C. wrote the paper.
* This work was supported by The Novo Nordisk Foundation, The Mizutani Foundation, and The Danish National Research Foundation (DNRF107).
 This article contains supplemental Fig. S1, supplemental spectra, and Tables S1 to S11.
This article contains supplemental Fig. S1, supplemental spectra, and Tables S1 to S11.
1 The abbreviations used are:
- CHO
- Chinese hamster ovary
- GS
- glutamine synthase
- MAb
- monoclonal antibody
- WT
- wild type
- EPO
- erythropoietin
- GalNAc
- N-acetylgalactosamine
- VVA
- Vicia Villosa Agglutinin
- PNA
- Peanut Agglutinin
- LWAC
- lectin weak affinity chromatography
- HCD
- high energy collision dissociation
- ETD
- electron-transfer dissociation
- PSM
- peptide-spectrum matches
- SC
- Simple Cell.
REFERENCES
- 1. Walsh G. (2010) Biopharmaceutical benchmarks 2010. Nat. Biotechnol. 28, 917–924 [DOI] [PubMed] [Google Scholar]
- 2. Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., Andersen M. R., Neff N., Passarelli B., Koh W., Fan H. C., Wang J., Gui Y., Lee K. H., Betenbaugh M. J., Quake S. R., Famili I., Palsson B. O., Wang J. (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis N. E., Liu X., Li Y., Nagarajan H., Yerganian G., O'Brien E., Bordbar A., Roth A. M., Rosenbloom J., Bian C., Xie M., Chen W., Li N., Baycin-Hizal D., Latif H., Forster J., Betenbaugh M. J., Famili I., Xu X., Wang J., Palsson B. O. (2013) Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat. Biotechnol. 31, 759–765 [DOI] [PubMed] [Google Scholar]
- 4. Baycin-Hizal D., Tabb D. L., Chaerkady R., Chen L., Lewis N. E., Nagarajan H., Sarkaria V., Kumar A., Wolozny D., Colao J., Jacobson E., Tian Y., O'Meally R. N., Krag S. S., Cole R. N., Palsson B. O., Zhang H., Betenbaugh M. (2012) Proteomic analysis of Chinese hamster ovary cells. J. Proteome Res. 11, 5265–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker J., Timmermann C., Rupp O., Albaum S. P., Brinkrolf K., Goesmann A., Puhler A., Tauch A., Noll T. (2014) Transcriptome analyses of CHO cells with the next-generation microarray CHO41K: Development and validation by analysing the influence of the growth stimulating substance IGF-1 substitute LongR. J. Biotechnol. 178C, 23–31 [DOI] [PubMed] [Google Scholar]
- 6. Kramer O., Klausing S., Noll T. (2010) Methods in mammalian cell line engineering: from random mutagenesis to sequence-specific approaches. Appl. Microbiol. Biotechnol. 88, 425–436 [DOI] [PubMed] [Google Scholar]
- 7. Sasaki H., Bothner B., Dell A., Fukuda M. (1987) Carbohydrate structure of erythropoietin expressed in Chinese hamster ovary cells by a human erythropoietin cDNA. J. Biol. Chem. 262, 12059–12076 [PubMed] [Google Scholar]
- 8. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Paul Bennett E., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slade P. G., Hajivandi M., Bartel C. M., Gorfien S. F. (2012) Identifying the CHO secretome using mucin-type O-linked glycosylation and click-chemistry. J. Proteome Res. 11, 6175–6186 [DOI] [PubMed] [Google Scholar]
- 11. Kato K., Jeanneau C., Tarp M. A., Benet-Pages A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 12. Geoghegan K. F., Song X., Hoth L. R., Feng X., Shanker S., Quazi A., Luxenberg D. P., Wright J. F., Griffor M. C. (2013) Unexpected mucin-type O-glycosylation and host-specific N-glycosylation of human recombinant interleukin-17A expressed in a human kidney cell line. Protein Expres. Purif. 87, 27–34 [DOI] [PubMed] [Google Scholar]
- 13. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 14. Steentoft C., Bennet E. P., Clausen H. (2013) Glycoengineering of human cell lines using zinc finger nuclease gene targeting-SimpleCells with homogeneous GalNAc O-glycosylation allow isolation of the O-glycoproteome by one-step lectin affinity chromatography. Methods Mol. Biol. 1022, 387–402 [DOI] [PubMed] [Google Scholar]
- 15. Ju T., Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc. Natl. Acad. Sci. U. S. A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarp M. A., Sorensen A. L., Mandel U., Paulsen H., Burchell J., Taylor-Papadimitriou J., Clausen H. (2007) Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17, 197–209 [DOI] [PubMed] [Google Scholar]
- 17. Vakhrushev S. Y., Steentoft C., Vester-Christensen M. B., Bennett E. P., Clausen H., Levery S. B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vester-Christensen M. B., Halim A., Joshi H. J., Steentoft C., Bennett E. P., Levery S. B., Vakhrushev S. Y., Clausen H. (2013) Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 110, 21018–21023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U. S. A. 109, 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hang H. C., Yu C., Kato D. L., Bertozzi C. R. (2003) A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. U. S. A. 100, 14846–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim U. M., Yap M. G., Lim Y. P., Goh L. T., Ng S. K. (2013) Identification of autocrine growth factors secreted by CHO cells for applications in single-cell cloning media. J. Proteome Res. 12, 3496–3510 [DOI] [PubMed] [Google Scholar]
- 22. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 23. Joshi H. J., Steentoft C., Schjoldager T., Vakhrushev S. Y., Wandall H., Clausen H. (2014) Protein O-GalNAc glycosylation – the most complex and differentially regulated PTM. Glycosciences: Biology and Medicine. Eds N. Taniguchi, in press [Google Scholar]
- 24. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U. S. A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schjoldager K. T., Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing - deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 [DOI] [PubMed] [Google Scholar]
- 26. Sasaki H., Ochi N., Dell A., Fukuda M. (1988) Site-specific glycosylation of human recombinant erythropoietin: analysis of glycopeptides or peptides at each glycosylation site by fast atom bombardment mass spectrometry. Biochemistry 27, 8618–8626 [DOI] [PubMed] [Google Scholar]
- 27. Walsh G. (2010) Post-translational modifications of protein biopharmaceuticals. Drug Disc. Today 15, 773–780 [DOI] [PubMed] [Google Scholar]
- 28. Fukuda M. N., Sasaki H., Lopez L., Fukuda M. (1989) Survival of recombinant erythropoietin in the circulation: the role of carbohydrates. Blood 73, 84–89 [PubMed] [Google Scholar]
- 29. Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., Shitara K. (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 [DOI] [PubMed] [Google Scholar]
- 30. Houel S., Hilliard M., Yu Y. Q., McLoughlin N., Martin S. M., Rudd P. M., Williams J. P., Chen W. (2014) N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal. Chem. 86, 576–584 [DOI] [PubMed] [Google Scholar]
- 31. Nilsson J., Ruetschi U., Halim A., Hesse C., Carlsohn E., Brinkmalm G., Larson G. (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 [DOI] [PubMed] [Google Scholar]
- 32. Darula Z., Sherman J., Medzihradszky K. F. (2012) How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol. Cell. Proteomics 11, 111:016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kornfeld R., Kornfeld S. (1985) Assembly of asparagine-linked oligosaccharides. Ann. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 34. Boyce M., Carrico I. S., Ganguli A. S., Yu S. H., Hangauer M. J., Hubbard S. C., Kohler J. J., Bertozzi C. R. (2011) Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. U. S. A. 108, 3141–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., Larsen M. R. (2012) Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 36. Patnaik S. K., Stanley P. (2006) Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 416, 159–182 [DOI] [PubMed] [Google Scholar]
- 37. Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R., Wakitani M., Niwa R., Sakurada M., Uchida K., Shitara K., Satoh M. (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 87, 614–622 [DOI] [PubMed] [Google Scholar]
- 38. Malphettes L., Freyvert Y., Chang J., Liu P. Q., Chan E., Miller J. C., Zhou Z., Nguyen T., Tsai C., Snowden A. W., Collingwood T. N., Gregory P. D., Cost G. J. (2010) Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol. Bioeng. 106, 774–783 [DOI] [PubMed] [Google Scholar]
- 39. Sealover N. R., Davis A. M., Brooks J. K., George H. J., Kayser K. J., Lin N. (2013) Engineering Chinese hamster ovary (CHO) cells for producing recombinant proteins with simple glycoforms by zinc-finger nuclease (ZFN)-mediated gene knockout of mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (Mgat1). J. Biotechnol. 167, 24–32 [DOI] [PubMed] [Google Scholar]
- 40. Steentoft C., Bennett E. P., Schjoldager K. T., Vakhrushev S. Y., Wandall H. H., Clausen H. (2014) Precision genome editing: A small revolution for glycobiology. Glycobiology 24, 663–680 [DOI] [PubMed] [Google Scholar]
- 41. Tarp M. A., Clausen H. (2008) Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 1780, 546–563 [DOI] [PubMed] [Google Scholar]
- 42. Wandall H. H., Blixt O., Tarp M. A., Pedersen J. W., Bennett E. P., Mandel U., Ragupathi G., Livingston P. O., Hollingsworth M. A., Taylor-Papadimitriou J., Burchell J., Clausen H. (2010) Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 70, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Springer G. F. (1984) T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 [DOI] [PubMed] [Google Scholar]
- 44. Napoletano C., Zizzari I. G., Rughetti A., Rahimi H., Irimura T., Clausen H., Wandall H. H., Belleudi F., Bellati F., Pierelli L., Frati L., Nuti M. (2012) Targeting of macrophage galactose-type C-type lectin (MGL) induces DC signaling and activation. Eur. J. Immunol. 42, 936–945 [DOI] [PubMed] [Google Scholar]
- 45. DeFrees S., Wang Z. G., Xing R., Scott A. E., Wang J., Zopf D., Gouty D. L., Sjoberg E. R., Panneerselvam K., Brinkman-Van der Linden E. C., Bayer R. J., Tarp M. A., Clausen H. (2006) GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology 16, 833–843 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.