Abstract
Runx2 is the master switch controlling osteoblast differentiation and formation of the mineralized skeleton. The post-translational modification of Runx2 by phosphorylation, ubiquitinylation, and acetylation modulates its activity, stability, and interactions with transcriptional co-regulators and chromatin remodeling proteins downstream of osteogenic signals. Characterization of Runx2 by electron transfer dissociation tandem mass spectrometry revealed sites of O-linked N-acetylglucosamine (O-GlcNAc) modification, a nutrient-responsive post-translational modification that modulates the action of numerous transcriptional effectors. O-GlcNAc modification occurs in close proximity to phosphorylated residues and novel sites of arginine methylation within regions known to regulate Runx2 transactivation. An interaction between Runx2 and the O-GlcNAcylated, O-GlcNAc transferase enzyme was also detected. Pharmacological inhibition of O-GlcNAcase (OGA), the enzyme responsible for the removal of O-GlcNAc from Ser/Thr residues, enhanced basal (39.9%) and BMP2/7-induced (43.3%) Runx2 transcriptional activity in MC3T3-E1 pre-osteoblasts. In bone marrow-derived mesenchymal stem cells differentiated for 6 days in osteogenic media, inhibition of OGA resulted in elevated expression (24.3%) and activity (65.8%) of alkaline phosphatase (ALP) an early marker of bone formation and a transcriptional target of Runx2. Osteogenic differentiation of bone marrow-derived mesenchymal stem cells in the presence of BMP2/7 for 8 days culminated in decreased OGA activity (39.0%) and an increase in the abundance of O-GlcNAcylated Runx2, as compared with unstimulated cells. Furthermore, BMP2/7-induced ALP activity was enhanced by 35.6% in bone marrow-derived mesenchymal stem cells differentiated in the presence of the OGA inhibitor, demonstrating that direct or BMP2/7-induced inhibition of OGA is associated with increased ALP activity. Altogether, these findings link O-GlcNAc cycling to the Runx2-dependent regulation of the early ALP marker under osteoblast differentiation conditions.
Runx2 [also known as: core binding factor α 1 (CBFA1); acute myeloid leukemia transcription factor 3 (AML3); polyoma enhancer-binding protein 2α (PEPB2α)] is a member of the runt-domain gene family of DNA binding proteins (Runx1, Runx2, and Runx3), which control the expression of numerous genes involved in cell growth, proliferation, and determination of cell lineage (1). Aberrant expression and activity of Runx proteins are implicated in leukemia, metastatic breast cancer, and defects in skeletal development and bone remodeling (2–4). Runx2 is expressed during early embryonic development in mesenchymal and cartilage condensations of the developing bone anlagen, and its transcription, activity, and stability are tightly regulated (5–7). Considered the master regulator of osteoblast differentiation, Runx2 also contributes to chondrocyte maturation (8, 9) and, through these cell-types, controls intramembranous and endochondral ossification culminating in the formation of the mineralized skeleton (5, 6, 10). A gain-of-function mutation of Runx2, resulting from duplication of exons 3 to 5, is linked to dysplastic long bone formation, enlarged clavicles, and thickening of the cranial vault (11). Conversely, the rare autosomal dominant disorder, cleidocranial dysplasia, has been traced to multiple loss-of-function mutations within the Cbfa1 gene, and is characterized by defective formation or absence of the clavicle, enlarged fontanelles, and dental abnormalities (9, 12, 13). Mice nullizygous for Runx2 develop a cartilaginous skeletal framework that fails to undergo mineralization caused by the absence of osteoblasts, and these mice die at birth because of respiratory failure (5, 10).
Runx2 is post-translationally modified (PTM)1 downstream of a diverse set of signaling pathways whose coordinated action controls osteoblast differentiation and bone development. The expression of genes associated with matrix formation, remodeling, and mineralization, such as alkaline phosphatase (ALP), osteocalcin (OCN), osteopontin, bone sialoprotein, and matrix metalloprotease 13 is governed by Runx2 [reviewed in (14)]. Runx2 activation, interactions with transcriptional and epigenetic co-regulators, and PTM status are modulated by endocrine, autocrine, and paracrine signals such as estrogen, vitamin D3, parathyroid hormone (PTH), TGF-β and bone morphogenetic proteins (BMPs), fibroblast growth factor (FGF), Wnt ligands, and insulin-like growth factor 1 (IGF-1) [reviewed in (1, 15, 16)]. Phosphorylation of Runx2 by ERK/MAPK (downstream of IGF-1, BMP2 and 7, and FGF2) (17–19), GSK3β (downstream of Wnt/β-catenin) (20), and PKA (downstream of PTH) (21) modulates transcriptional activity and the specificity of target gene expression. Runx2 activity is also regulated through interactions with lysine acetyltransferases (e.g. p300, CBP) and class I/II histone deacetylases (HDACs 3, 4, 5, and 6) (22–25), and acetylation of Runx2 in response to BMP2-SMAD-p300, FGF-ERK, or HDAC inhibition enhances Runx2 activity and markers of osteoblast differentiation (22, 26).
Previous studies have linked enhanced expression of differentiation markers in chondrocytes and pre-osteoblasts with elevated modification of proteins by O-linked N-acetylglucosamine (O-GlcNAc), and suggested O-GlcNAcylation of Runx2 contributes to osteoblast differentiation (27, 28). Cycling of O-GlcNAc modification on protein Ser/Thr residues is essential to mammalian tissue specification, cell survival, and embryonic development (29–32), and there are numerous examples of O-GlcNAc modification impinging on phosphorylated-mediated signal transduction (33–36). The entirety of cellular O-GlcNAc cycling is controlled by the actions of the highly conserved enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). OGT catalyzes the transfer of GlcNAc to target proteins utilizing the donor substrate UDP-GlcNAc, a product of the hexosamine biosynthetic pathway that requires glucose, glutamine, acetyl CoA, and ATP (37). Inheritance of a null OGT allele is embryonically lethal at an early stage (31), and the elucidation of mutations within the OGT coding region of Drosophila super sex combs, which display aberrant spatial tissue patterning and increased lethality, led to the discovery that OGT is a member of the multiprotein polycomb group repressor (PcG) complex that regulates transcription of homeotic genes during embryonic development (38, 39). O-GlcNAc levels fluctuate in a tissue-specific manner during embryogenesis (40), embryonic stem cell differentiation (41–43), and during differentiation of mesenchymal stem cell-derived osteoblast (murine and human), adipocyte, myoblast, and chondrocyte lineages (27, 28, 44–46). In embryonic stem cells, OGT localizes to the promoters of multiple transcription factors critical to osteoblast differentiation including Runx1, Runx2, Osterix, and Twist2 (47). Although we have detected O-GlcNAcylation of proteins integral to the BMP2-TAK1-MAPK signaling axis in pre-osteoblasts (44), the regulation and contribution of O-GlcNAc cycling enzymes during osteoblast differentiation and bone formation is virtually unexplored.
The aim of the present study was to characterize sites of Runx2 O-GlcNAc modification and evaluate the regulation of O-GlcNAc cycling by BMP2/7 in osteogenic, murine bone marrow mesenchymal stem cells (BMMSCs). Human Runx2 (type II; MASNS…), expressed in HEK293 cells, was characterized by tandem mass spectrometry (MS/MS). O-GlcNAc modification was detected within the N-terminal activation domain and the C-terminal proline/serine/threonine-rich (PST) region that governs Runx2 activity and interaction with transcriptional co-regulators. A chemoenzymatic labeling strategy was employed to verify O-GlcNAc modification of Runx2 in osteogenic BMMSCs. Differentiation in the presence of BMP2/7 was associated with a decrease in OGA enzyme activity, as well as an elevation in global protein O-GlcNAcylation. Pharmacological inhibition of OGA enhanced basal and BMP2/7-stimulated Runx2 transactivation and ALP activity. These data demonstrate for the first time O-GlcNAc modification of the critical osteogenic transcription factor, Runx2, at specific residues, and link O-GlcNAc cycling to the regulation of the early osteoblast marker and Runx2 target, ALP, during early osteoblast differentiation.
EXPERIMENTAL PROCEDURES
Materials
Cell line origins were as follows: mesenchymal stem cells derived from bone marrow (BMMSCs) of C57BL/6 mice were obtained from Invitrogen (Carlsbad, CA); immortalized HEK293A cells are from Qbiogene (Carlsbad, CA); C57BL/6 calvarial MC3T3E1 pre-osteoblasts (subclone E4) were from ATCC (Manassas, VA). Minimum essential medium alpha (αMEM) was obtained from Cellgro-Mediatech (Manassas, VA). Fetal bovine serum (FBS), mesenchymal stem cell (MSC)-verified FBS, and modified improved essential medium (IMEM) were from Invitrogen (Grand Island, NY). Penicillin/streptomycin and penicillin G/streptomycin/amphotericin antibiotic/antimycotic solutions were from HyClone (Logan, UT). Human, N-terminal 3XFLAG-tagged runt-related transcription factor 2 (Runx2) (transcript variant 1; GenBank accession #: NM 001024630) under the control of the CMV promoter is from GeneCopoeia Inc. (Rockville, MD). Antibody providers were as follows: the pan-specific, monoclonal O-GlcNAc antibody raised against YSPTS(O-GlcNAC)PSK (CTD110.6) was from ThermoScientific (Rockford, IL); GAPDH (H-12) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); Runx2 (D1H7) was from Cell Signaling (Danvers, MA); OGT antibody was from Sigma-Aldrich (St. Louis, MO); MGEA5 (OGA) antibody was from ProteinTech Group Inc. (Chicago, IL); anti-DYKDDDDK (FLAG) epitope tag was from Fisher Scientific (Pittsburgh, PA); Secondary, dye-conjugated goat anti-rabbit IRDye 680RD and goat anti-mouse 800CW IgG antibodies were from Li-Cor (Lincoln, NE). Immunoblots employing the CTD 110.6 O-GlcNAc antibody were incubated with the secondary Alexa Fluor goat anti-mouse 680 IgM antibody (Invitrogen). Thiamet G (48) was from Cayman Chemical (Ann Arbor, MI) and recombinant human bone morphogenetic protein 2/7 heterodimer was from R&D Systems Inc. (Minneapolis, MN).
Cell Culture
HEK293 cells were maintained in IMEM with 10% heat inactivated FBS, 1% antibiotic/antimycotic. For production of 3XFLAG-tagged Runx2, cells were transfected with 4 μg plasmid encoding the N-terminal 3XFLAG-tagged human Runx2 cDNA in the presence of PolyFect transfection reagent (Qiagen). Cells were maintained in growth medium for 24 h after which they were washed twice with unamended IMEM and incubated for an additional 18 h under serum-fasting conditions (1% heat-inactivated FBS) in the presence of Thiamet G (20 μm). Alternatively, HEK293 cells were infected with the AdBioR2 adenoviral vector (2 × 106 ifu/ml), which encodes the Type II (P1) isotype of murine Runx2 containing a minimal biotinylation sequence tag for 24 h (kindly provided by Renny Franceschi, University of Michigan) (17, 49).
BMMSCs and MC3T3E1 cells were maintained in αMEM with 10% MSC-FBS, 1% penicillin/streptomycin. To induce osteogenesis, BMMSCs were seeded at 1 × 104 cells per cm2 in growth medium and allowed to attach for 18 h. Cells were then provided with medium containing ascorbic acid (50 μg/ml) and β-glycerol phosphate (5 mm) (osteogenic medium). This time point was defined as Day 0. Additional parameters for differentiation of BMMSCs are described in the appropriate subsections for specific experiments. All cells were incubated at 37 °C under 5% CO2 conditions.
Luciferase Assays
MC3T3E1 cells were plated at a density of 2 × 104 cells/cm2 and transfected with the indicated plasmids in the absence of antibiotics using Lipofectamine 2000 (Invitrogen). Each well was transfected with 2.5 μg of the 6OSE2-luciferase reporter gene [contains six copies of the Runx2-specific cis-regulatory element (OSE2) (50)] and 0.025 μg of the pGL4-hRluc reporter (Promega, Madison, WI) in which Renilla reformis luciferase cDNA expression is driven by the constitutive SV40 promoter. After 6 h, cells were washed twice and incubated for an additional 48 h in growth medium containing 5% serum and amended with Thiamet G, BMP2/7, or their respective vehicles. Luciferase activity was assayed using the Dual-Luciferase Assay System (Promega) with a FLUOstar OPTIMA plate reader (BMG Labtech, Ortenberg, Germany).
LC-MS/MS Analysis of Runx2
HEK293 cells expressing 3XFLAG-tagged Runx2 were sonicated in 50 mm Tris HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 amended with protease/phosphatase inhibitors (1:100 HALT inhibitor mixture; Pierce, Rockford, IL), and an OGA inhibitor, Thiamet G (20 μm). Insoluble material was removed from the supernatant by centrifugation and resuspended in 50 mm Tris HCl (pH 7.4), 150 mm NaCl, and 1 mm MgCl2 with 250 U (1% v/v) benzonase nuclease (EMD Chemicals, Inc., San Diego, CA) for 1 h (4 °C) to release any additional DNA-bound transcription factor. After spinning at 8000 × g the benzonase-digested fraction was pooled with the previously collected supernatant.
Prior to the immunoprecipitation of Runx2, total protein was diluted to 2 mg/ml with 50 mm Tris HCl (pH 7.4), 150 mm NaCl and precleared with Protein A (ProtA) agarose beads (EMD Chemicals, Inc.) to remove nonspecific interactors. Beads were collected by centrifugation and the supernatant was incubated with Anti-FLAG M2-agarose affinity beads (Sigma) for 18 h at (4 °C). Beads were washed extensively with 50 mm Tris HCl (pH 7.4), 150 mm NaCl, and immunoprecipitates were eluted 2–3× with an equal bead volume of 2 × XT sample buffer (BioRad), 10% β-mercaptoethanol at 100 °C for 5 min. Eluates were pooled and stored at −80 °C until further analysis.
Immunoprecipitated protein was resolved on a 4–12% gradient Criterion XT gel (BioRad) and zinc stained (E-Zinc reversible stain kit; Pierce) to visualize Runx2 (521 aa; 56.6 kDa). The band corresponding to Runx2 [61.1 kDa including 3XFLAG-tag/linker (∼4.5 kDa)] was excised and de-stained using E-zinc eraser solution (Pierce). Gel pieces were washed twice for 10 min with ammonium bicarbonate (100 mm), dehydrated with acetonitrile, and dried by vacuum centrifugation. To reduce cysteine residues, gel pieces were then incubated with dithiothreitol (5 mg/ml in ammonium bicarbonate for 30 min) prior to alkylating in iodoacetamide (15 mg/ml in ammonium bicarbonate) in the dark for 30 min. Gel pieces were again washed with ammonium bicarbonate, dehydrated, and dried under vacuum prior to digestion with trypsin (Promega) at 37 °C for 18 h. To extract peptides, gel pieces were washed twice with 50% acetonitrile, 5% formic acid, and then twice with 85% acetonitrile and 5% formic acid. Peptides were dried under vacuum and reconstituted in 0.1% TFA.
For separation by C18 reversed phase nano-LC, peptides were reconstituted in solvent A (2% acetonitrile and 0.2% formic acid) and loaded on a 300 μm i.d. × 5 mm trap column [C18 PepMap 100, 5 μm, 100 Å (Thermo Scientific)] and washed for 10 min in solvent A. Peptides were then separated on a 75 μm i.d. x 15 cm analytical column [C18 YMC ODS-AQ 120 Å S5 (Waters Corporation) packed-in house] at 200 nl/min with a gradient of 3–50%B in 180 min (B = 98% acetonitrile, 2% H20, and 0.2% formic acid) using an Ultimate 3000 nanoflow system (Dionex). The top ten most abundant ions in the full mass spectrum survey scan (m/z 400–1700; resolution 60,000) were fragmented by higher energy collisional dissociation (HCD) (35% normalized collision energy) on an Orbitrap Elite (ThermoScientific). The presence of diagnostic HexNAc oxonium ions (m/z 138.06, 186.08, and 204.09) within the top 30 product ions in the HCD scan triggered the acquisition of an electron transfer dissociation (ETD) fragmentation spectrum of the precursor ions in the Velos ion trap. Charge-state dependent reaction time for ETD fragmentation was enabled. Because of the labile nature of the O-linkage by collisional dissociation, supplemental activation was not utilized. Dynamic exclusion was enabled with a repeat count of 3, a repeat duration of 30 s, and exclusion duration of 180 s. A lock-mass (m/z 445.120024) was utilized for recalibration (51).
Data Analysis
Tandem mass spectra were searched against the human Runx2 sequence (UniProtKB accession number: Q13950) using Proteome Discoverer v. 1.4.0.288 or a human UniProtKB protein database using Maxquant 1.4.1.2. Within both searches trypsin was selected as the proteolytic enzyme, allowing for a maximum of two missed cleavages, and spectra were searched allowing for the following variable modifications: phosphorylation (STY; +79.97 Da), HexNAc (STY; +203.08 Da); monomethylation (KR; +14.02); dimethylation (KR; +28.03); trimethylation (K; +42.05); and oxidation (M; +15.995). Cysteine carbamidomethylation was allowed as a static modification. Also, an additional allowance for the neutral loss of GlcNAc from precursor and fragment ions was enabled. Within Proteome Discoverer dual search nodes (Mascot; SEQUEST HT) were enabled. HCD and ETD spectra were searched with precursor ion mass tolerances set at 20 ppm and fragment ion mass tolerances at 0.1 Da and 0.8 Da, respectively. Within Maxquant, spectra were searched using the Andromeda algorithm (52) with precursor ion mass tolerances set at 20 ppm for the first search, 5 ppm for the second search, and fragment ion mass tolerances at 0.1 Da and 0.8 Da, respectively. A decoy database (reward) was employed to filter peptide identifications to an FDR <0.01. After the main database search, unmatched tandem mass spectra were re-searched for unanticipated modifications using the dependent peptide feature. This search reveals unanticipated mass shifts that may be of interest or chemical artifacts that arise during sample preparation. The dependent peptide search utilizes a database constrained to the peptides positively identified in the first search. These results were filtered by an FDR <0.1 and a positional probability of >0.99. With the exception of the O-GlcNAc modified peptides, the positional probability of the assigned sites of modification of all modified peptides reported was >0.99. The search results are provided in supplemental Tables. The sites of modification on reported peptides were manually confirmed by comparing experimental sequence ions to theoretical fragment ions generated by Protein Prospector 2 v. 5.10.8.
Immunoblotting and qPCR
For analysis of total cell lysate protein, monolayers were washed twice with ice-cold PBS and lysed in 15 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 amended with protease, phosphatase, and OGA inhibitors (Thiamet G). Insoluble material was pelleted at 8000 × g and soluble protein saved for immunoblot analysis. Alternatively crude cytosolic and nuclear fractions were collected using NE-PER extraction reagents (ThermoScientific) according to manufacturer's instructions. Protein concentration was determined by Bradford assay (ThermoScientific) and total protein (25 μg) resolved by SDS-PAGE. After transfer to a nitrocellulose membrane immunoblots were incubated with protein- or modification-specific antibodies prior to detection with dye- conjugated secondary antibodies (1:20,000) on an Odyssey Infrared imaging system (Li-Cor). Image analysis was performed with Image Studio Software v.2.1.10 for quantitation of antibody signal by densitometry.
For mRNA analysis, total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen) and RNA concentration determined by UV absorbance at 260 nm [NanoDrop 2000 UV-Vis spectrophotometer (ThermoScientific)]. Messenger RNA (1 μg) was reverse transcribed with iScript cDNA Synthesis (BioRad) reagents and cDNA amplified in the presence of SYBR Select Master Mix (Applied Biosystems, Foster City, CA) and sequence specific primers on an StepOnePlus real-time PCR system (Applied Biosystems). Primers used were as follows for the following mouse (Mus musculus) gene products: ALP (liver/bone/kidney), forward 5′-ATCGGAACAACCTGACTGACCCTT-3′, reverse 5′-ACCCTCATGATGTCCGTGGTCAAT-3′; peroxisome proliferator-activated receptor (PPAR) γ, forward 5′-ACATAAAGTCCTTCCCGCTGACCA-3′, reverse 5′-AAATTCGGATGGCCACCTCTTTGC-3′; and GAPDH, forward 5′-CAGCAAGGACACTGAGCAAG-3′, reverse 5′-GGTCTGGGATGGAAATTGTG-3′. Amplification parameters were as described previously (44) and products were quantified with StepOneTM system software v2.0 (Applied Biosystems).
Chemoenzymatic Labeling of O-GlcNAc Modified Nuclear Lysate Proteins
Chemoenzymatic labeling of proteins containing terminal O-GlcNAc residues with a 5122 Da polyethylene glycol mass tag (PEG, 98% pure; Creative PEGWorks; Winston-Salem, NC) was used to visualize glycosylation of Runx2 by immunoblotting (53). First, azide-labeled galactose (GalNAz) is enzymatically incorporated onto terminal GlcNAc residues utilizing a mutant galactosyltransferase (Y298L β-1,4-GalT1) enzyme. O-GlcNAc modified protein from HEK293 cellular lysates or BMMSC nuclear lysates was labeled in this manner in the presence of the GalT1 enzyme (4.8% v/v) and UDP-GalNAz using the Click-iT O-GlcNAc Enzymatic Labeling System (Invitrogen). Next, the alkyne-labeled PEG tag was reacted with GalNAz in the presence of a copper [Cu(I)] catalyst using the Click-iT Protein Analysis Detection kit (Invitrogen) according to the manufacturer's instructions with the exception that the alkyne-labeled biotin was replaced with alkyne-labeled PEG at 6 mm final reaction concentration (53). PEG-labeled protein was resolved by SDS-PAGE and immunoblotted with a Runx2-specific antibody. To estimate the stoichiometry of Runx2 O-GlcNAc modification in BMMSC nuclear lysates we performed densitometry. The MW values of unmodified Runx2 and the three most abundant PEG-labeled populations (e.g. PEG1, 2, and 3) were quantified relative to a protein standard (Precision Plus Protein standard; Bio-Rad). Average MW values for each band were calculated from at least three independent replications. To estimate the number of O-GlcNAc modifications present on Runx2 under Thiamet G conditions, the unmodified Runx2 MW value was subtracted from the MW value of the PEG-labeled population, and this value was then divided by the MW value of the PEG mass tag.
ALP Activity
ALP activity was assayed in BMMSC total cell lysates using the Lab Assay ALP kit (Wako Pure Chemical Industries, Osaka, Japan) according to the manufacturer's instructions. For these assays, BMMSCs were cultured in 5% FBS under osteogenic conditions for 24 h (Day 1) before supplementing the medium with Thiamet G (20 μm) or its DMSO vehicle (0.025% v/v final concentration). Cells were cultured for an additional 6 days (7 days total culture) in the absence or presence of Thiamet G. The medium was replaced on Day 4. For ALP activity assays, BMMSCs were lysed in 10 mm Tris-HCl (pH 8.0), 1 mm MgCl2, 0.5% TX-100 (54) amended with complete-mini EDTA-free protease inhibitor mixture (Sigma-Aldrich) and OGA inhibitors [PUGNAc (50 μm) or Thiamet G (20 μm)]. Crude lysate protein (20–40 μg) was incubated with p-nitrophenylphosphate for 15 min at 37 °C (pH 9.8). Sample absorbance was quantified at 405 nm on a FLUOstar OPTIMA plate reader. ALP activity (U/μl) was calculated for blank (buffer only) subtracted values relative to a p-nitrophenol standard curve, were U is defined as 1 nmol p-nitrophenol released per minute. Enzyme activity was normalized by protein concentration (μg/μl) determined by Bradford assay at 600 nm. For each experimental replicate, three monolayers were assayed in duplicate. Alternatively, cell monolayers were stained to visualize ALP activity. For this, cells were washed twice with PBS, fixed for 10 min in 4% (v/v) paraformaldehyde in PBS with 0.3% (v/v) Triton X-100, and then washed three more times with PBS before incubating with Permanent FastRed Quanto stain (ThermoScientific) according to manufacturer's instructions. Plates were imaged and image quality (brightness, contrast, red channel saturation) was adjusted equally across represented images with Adobe Photoshop 7.0.1.
BMMSC Proliferation
To measure the effect of OGA inhibition on BMMSC proliferation, cultures were incubated with 5-bromo-2′-deoxyuridine (BrdU) for 24 h prior to detection with the BrdU Cell Proliferation Assay kit (Cell signaling Technology; Danvers, MA). BrdU incorporation was quantified at 450 nm.
OGA Activity
OGA activity was analyzed in BMMSC total cell lysates by measuring the colorimetric conversion of p-nitrophenyl N-acetyl-β-d-glucosaminide (p-NPAGA) to p-nitrophenol in a molar excess of N-acetylgalactosamine (55). BMMSCs were cultured in osteogenic medium for 24 h and the next day (Day 1) cells were supplemented with BMP2/7 (50 ng/ml) or vehicle (4 mm HCl and 0.1% BSA in PBS; brought to 0.5% v/v final concentration in culture media). Cells were cultured for 8 days. The osteogenic medium was changed on Days 3 and 6 at which time cells were resupplied with BMP2/7 or its vehicle. BMMSCs were lysed in 15 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, and 0.5% Triton X-100 in the absence of Thiamet G. Aliquots were reserved from each treatment replicate for subsequent quantitation of OGA protein levels by immunoblot. These aliquots were amended with Thiamet G (20 μm) at the time of harvest to inhibit residual OGA activity and stored at −80 °C for subsequent analysis. For analysis of OGA activity, crude lysate protein (0.2–0.5 mg) in 100 μl lysis buffer was diluted 1:1 in reaction buffer (50 mm sodium cacodylate, pH 6.4 and 0.3% BSA) in the presence of 2 mm p-NPAGA and 50 mm N-acetylgalactosamine. All treatments assayed for OGA activity in the absence or presence of Thiamet G, to control for background signal.
Absorbance was measured at 405 nm at 25, 35, and 65 min. The concentration (c) of p-nitrophenol (εmm = 17.7) released by OGA was calculated using Beer's law (A405 = εLc). OGA activity (μmol/mg/min) was determined from background subtracted-values, and averaged across the three time points for each treatment replicate. To control for potential variations in OGA protein expression, reserved lysate protein was immunoblotted and OGA protein expression was quantified by densitometry. Relative OGA expression for each treatment replicate was determined by normalizing against GAPDH signal intensity. OGA activity was then normalized by OGA protein expression. Three monolayers were assayed per independent replicate.
Statistics
Data are represented as mean values ± S.E. Statistical significance was assessed using Student's pairwise t test (α = 0.05).
RESULTS
Pharmacological Inhibition of OGA Enhances Runx2 Activity in MC3T3E1 Pre-osteoblasts
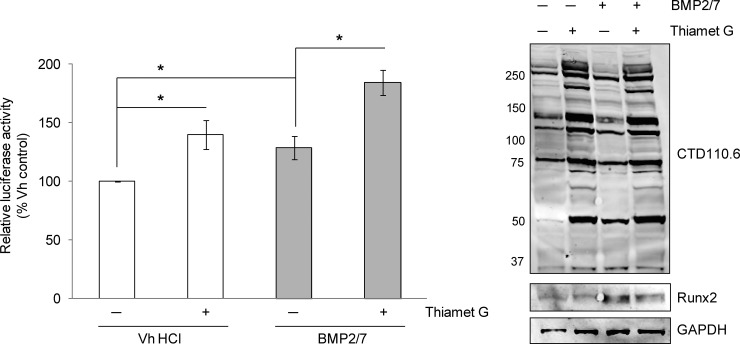
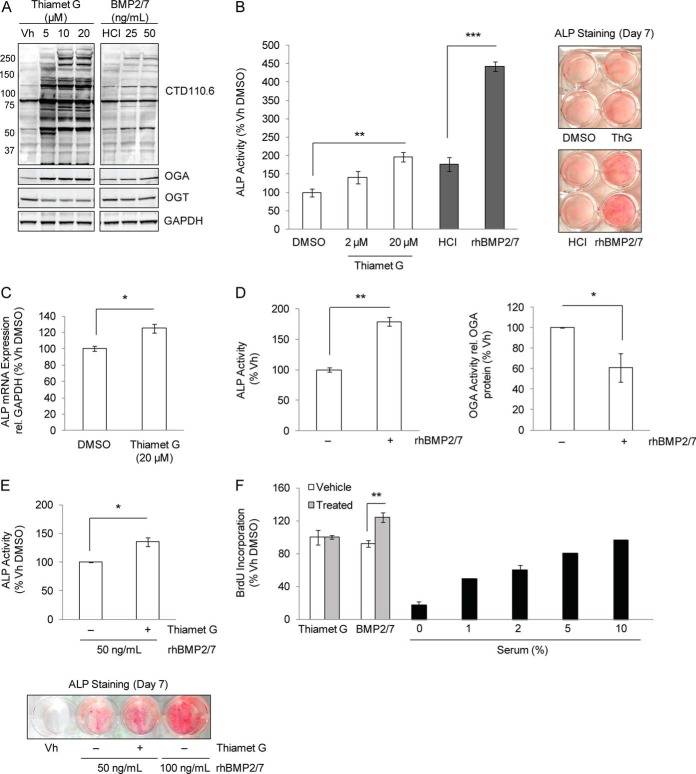
It was previously reported that inhibition of hexosaminidases with O-(2-Acetamido-2-deoxy-d-glucopyranosylidenamino) N-phenylcarbamate (PUGNAc) in MC3T3E1 pre-osteoblasts amplified the effects of PTH and forskolin on Runx2 transcriptional activity and osteocalcin expression (28). To confirm that OGA inhibition, and the concomitant rise in protein O-GlcNAc modification, augments Runx2-dependent transactivation, the activity of the 6OSE2-luciferase reporter gene was measured in MC3T3E1 pre-osteoblasts cultured in the presence or absence of BMP2/7 and a selective OGA inhibitor, Thiamet G. BMP2/7 enhanced Runx2 activity after 48 h as expected and inhibition of OGA elevated Runx2 activity to a similar extent (Fig. 1). In the presence of both BMP2/7 and Thiamet G, Runx2 activity was further enhanced. Protein lysates probed for O-GlcNAc modification and Runx2 by immunoblotting confirmed the expected rise in total protein O-GlcNAc modification and demonstrated that the increase in Runx2 activity was independent of changes in Runx2 protein expression. Altogether these results support previous findings by Kim et al. that Runx2 activity is enhanced through global elevation of O-GlcNAc modification.
Fig. 1.
Runx2 transcriptional activity is enhanced by inhibition of OGA. MC3T3E1 cells were transfected with the 6OSE2-luc reporter and the pGL4-hRLuc plasmid and incubated for 48 h in the absence or presence of both Thiamet G (20 μm) and BMP2/7 (50 ng/ml). Firefly luciferase activities (6OSE2-luc) were normalized to corresponding Renilla luciferase activities to control for transfection efficiency. Relative luciferase activity was averaged across three independent replications and is expressed as a percentage of the vehicle control mean. On the right, lysate protein (20 μg) from each treatment was immunoblotted for O-GlcNAc modification (CTD110.6), Runx2 and GAPDH as a loading control. Error bars are ± S.E.: p < 0.05 (*).
Runx2 is O-GlcNAc Modified Proximal to Phosphorylation at Ser28
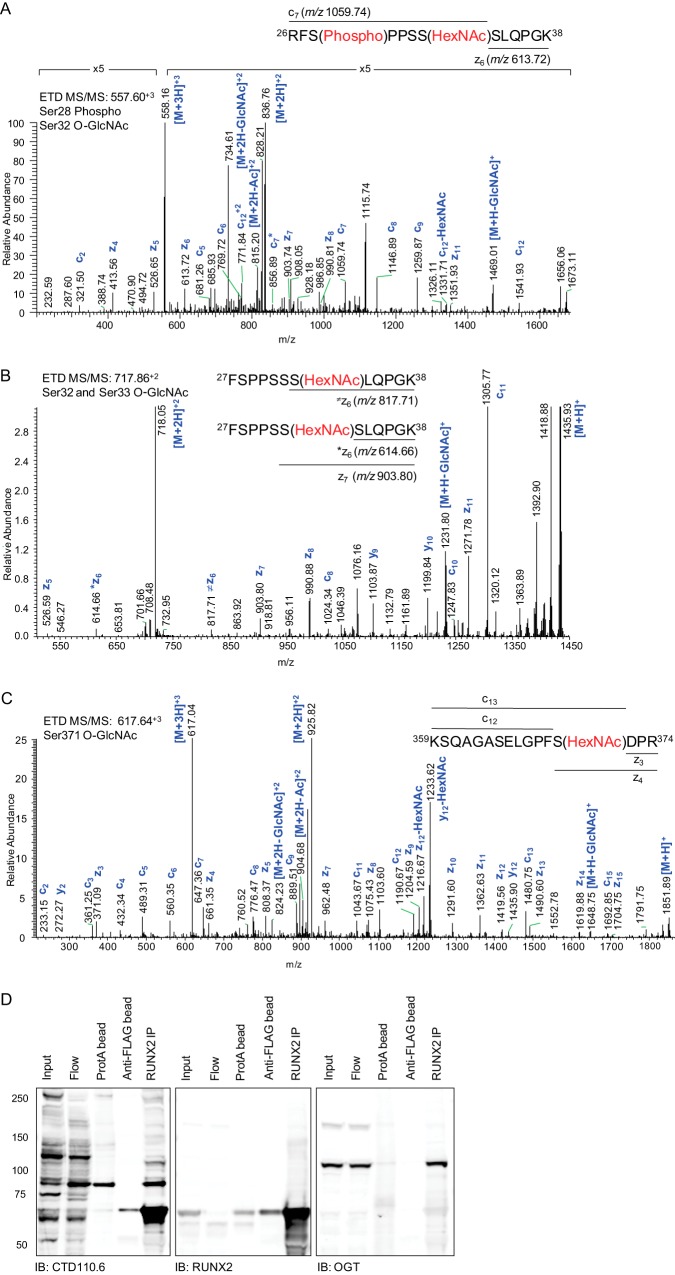
To identify sites of O-GlcNAc modification, Runx2 was characterized by tandem mass spectrometry. To facilitate mass spectrometric analysis, FLAG-tagged Runx2 was expressed in HEK293 cells in the presence of Thiamet G. The protein was affinity purified with anti-FLAG beads, gel purified, trypsin digested, and the peptides were analyzed by tandem mass spectrometry (MS/MS) using higher energy collisional dissociation (HCD) and electron transfer dissociation (ETD). The presence of diagnostic N-acetylhexosamine (HexNAc) product ions in the HCD MS/MS spectrum triggered the acquisition of ETD fragmentation spectra, enabling unambiguous assignment of O-GlcNAc modification sites (56). Seventy-eight percent of the Runx2 sequence was observed (supplemental Table 1). One large peptide (residues 392–468) containing the His/Thr/Tyr motif required for BMP2-induced osteoblastogenesis (57) was not detected. The peptide 26RFSPPSSSLQPGK38 was simultaneously phosphorylated at Ser28 and O-GlcNAc modified. The ETD spectrum (Fig. 2A) is consistent with fragmentation of the phosphorylated peptide that is mono-glycosylated at Ser32 (c7 and z6 ions at m/z 1059.74 and 613.72, respectively), whereas the ETD fragmentation pattern of the unphosphorylated, fully tryptic 27FSPPSSSLQPGK38 peptide is consistent with co-elution and cofragmentation of the mono-O-GlcNAcylated peptide, modified at Ser32 and Ser33 (Fig. 2B and Table I). The peptide, residues 26–38 phosphorylated at Ser28, was also observed without O-GlcNAc modification. Ser28 is phosphorylated downstream of ERK/MAPK, PI3K/Akt, and PTH/PKA pathways (17, 21, 58, 59), and phosphorylation at this site has been implicated in PTH-induced expression of MMP13 (21) as well as invasive properties of metastatic breast cancer cells (58).
Fig. 2.
Runx2 is O-GlcNAc modified and co-immunoprecipitates with O-GlcNAc transferase. 3XFLAG-Runx2 was expressed in HEK293 cells for 24 h before treating cells with Thiamet G (20 μm) for 18 h. A, The ETD MS/MS fragmentation spectrum of residues 26–38 (557.60+3 m/z) modified by phosphorylation at Ser28 and HexNAc at Ser32. B, The ETD MS/MS fragmentation spectrum for residues 27–38 (717.86+2 m/z). The fragmentation pattern observed in the ETD MS/MS is consistent with co-elution of two mono-glycosylated peptides that are O-GlcNAc modified at Ser32 (*z6 and z7 ions at m/z 614.66 and 903.80 respectively) or Ser33 (⊠ z6 at m/z 817.71). C, The ETD MS/MS fragmentation spectrum for residues 359–374 (617.64+3 m/z). For (A-C) the corresponding HCD MS/MS spectra (supplemental Data) are annotated according to the fragmentation pattern of the unmodified peptide because of neutral loss of the HexNAc moiety prior to peptide backbone dissociation. C, Aliquots from all steps of the immunoprecipitation (IP) procedure [total cell lysate (input); post-IP supernatant (Flow); elimination of nonspecifically binding proteins (ProtA); post-IP anti-FLAG beads; elution of Runx2 (Runx2 IP)] were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (bottom).
Table I. Identification of O-GlcNAc modification, phosphorylation, and methylation sites on human Runx2.
| Peptide | Residues | Modification | Site | Observed (m/z) | ΔM [ppm] | Charge | MS/MS Mode | Previously described? |
|---|---|---|---|---|---|---|---|---|
| RFSPPSSSLQPGK | 26–38 | Phospho | 28 | 489.90 | 0.36 | +3 | HCD, ETD | Yes (17, 21, 58, 59, 88) |
| RFSPPSSSLQPGK | 26–38 | Phospho/HexNAc | p28/g32 | 557.60 | 0.41 | +3 | HCD, ETD | No |
| FSPPSSSLQPGK | 27–38 | HexNAc | 32 | 717.86 | 1.52 | +2 | HCD, ETD | No |
| FSPPSSSLQPGK | 27–38 | HexNAc | 33 | 717.86 | 1.52 | +2 | HCD, ETD | No |
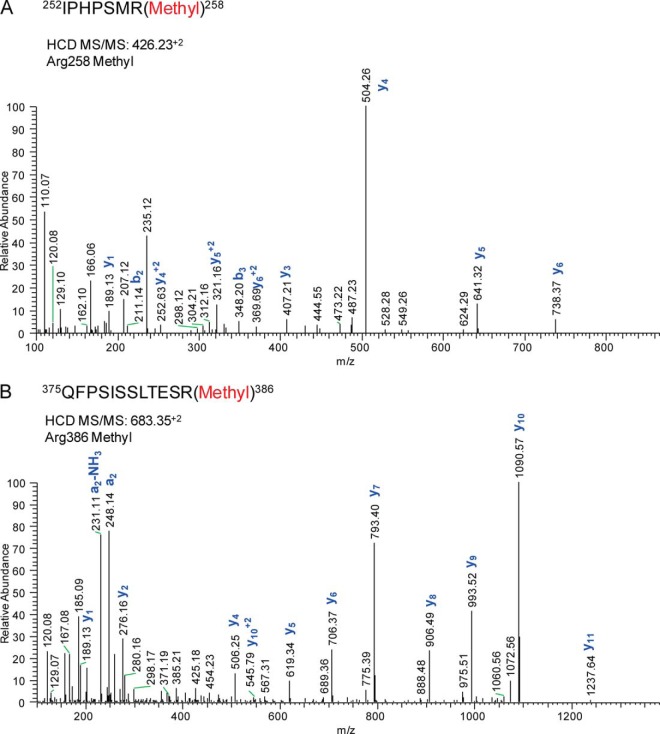
| IPHPSMR | 252–258 | Methyl | 258 | 426.23 | −0.56* | +2 | HCD | No |
| VGVPPQNPRPSLNSAPSPFNPQGQSQITDPR | 259–289 | Methyl | 267 | 1099.89 | −5.63* | +3 | HCD | No |
| VGVPPQNPRPSLNSAPSPFNPQGQSQITDPR | 259–289 | Dimethyl | 267 | 1104.90 | −6.72* | +3 | HCD | Yes (62) |
| RISDDDTATSDFC[Cam] LWPSTLSK | 338–358 | Phospho | 340 | 1248.55 | −0.99 | +2 | HCD | Yes (17, 21) |
| KSQAGASELGPFSDPR | 359–374 | HexNAc | 371 | 617.64 | 0.01 | +3 | HCD, ETD | No |
| QFPSISSLTESR | 375–386 | Methyl | 386 | 683.35 | −1.48 | +2 | HCD | No |
| QFPSISSLTESRFSNPR | 375–391 | Methyl | 386 | 984.01 | 0.15 | +2 | HCD | No |
| QFPSISSLTESRFSNPR | 375–391 | Dimethyl | 386 | 991.52 | 1.57 | +2 | HCD | No |
Bold residues denote localization of PTMs either by HCD or ETD MS/MS. [Cam] = carbamidomethyl. Peptide precursor masses were recalibrated in MaxQuant. Mass errors of peptides only detected by Proteome Discoverer are indicated with an asterisk.
Runx2 O-GlcNAc Modification at Ser371
O-GlcNAc modification was also identified within the C-terminal peptide, 359KSQAGASELGPFSDPR374, at Ser371 (Fig. 2C and Table I). Residing within the PST-rich domain, this residue and those immediately surrounding it are conserved among human Runx1, Runx2, and Runx3. Phosphorylation of the homologous residue in Runx3 (Ser243) was previously detected in a global phosphoproteomic analysis of Hela cells (60), and may be a site of reciprocal or mutually exclusive modification.
All of the O-GlcNAcylated peptides observed exhibited the expected neutral loss of the GlcNAc monosaccharide from precursor peptide and fragment ions upon higher energy collisional dissociation, as well as generation of three or more diagnostic HexNAc fragment ions at m/z 126, 138, 144, 168, 186, or 204 (supplemental Data). The ETD fragmentation yielded primarily c, z, and y ions, partial neutral loss of HexNAc from the precursor ions, and, in some instances, neutral loss of the HexNAc acetyl group. In addition to the complex fragmentation pattern of O-GlcNAc modified peptides generated by MS/MS, identification of O-GlcNAc modification is also complicated by the inability of MS/MS to distinguish N-acetylglucosamine (GlcNAc) from N-acetygalactosamine (GalNAc), which are isobaric and exhibit the same fragmentation pattern. Thus the 203.2 Da mass increase occurring at Ser, Thr, and Tyr residues is annotated as HexNAc. Proteins containing a single O-linked GalNAc monosaccharide are typically associated with the endoplasmic reticulum, plasma membrane, or are extracellularly localized (61).
O-GlcNAc Transferase (OGT) Co-immunoprecipitates with Runx2
To further confirm O-GlcNAc modification of Runx2, immunoprecipitated 3XFLAG-tagged Runx2 was immunoblotted with the monoclonal pan-specific anti-O-GlcNAc antibody (CTD110.6) (Fig. 2D). In addition to a strong signal corresponding to O-GlcNAc modified Runx2, the presence of a prominent band at 110 kDa prompted us to probe for OGT, which is known to be O-GlcNAcylated and interact with chromatin remodeling proteins and transcription factors. The presence of endogenous OGT in Runx2 immunoprecipitates, but not in the Protein A-agarose control, suggests a specific and detectable interaction between the O-GlcNAc cycling enzyme and Runx2.
Runx2 is Mono- and Di-methylated at Multiple Arginine Residues
In addition to O-GlcNAc modification, multiple sites of methylation were observed by MS/MS (Table I, supplemental Tables). Arginine residues can be monomethylated, symmetrically dimethylated, or asymmetrically dimethylated, and we observed mono- (Me1) and dimethylation (Me2) of the 259VGVPPQNPRPSLNSAPSPFNPQGQSQITDPR289 peptide at Arg267 (supplemental Data). This site of modification was recently observed in a large-scale screen of asymmetrically dimethylated proteins that were immunoaffinity enriched from murine embryonic tissue (62). Novel sites of methylation were also identified at Arg258 (Me1) (Fig. 3A) and Arg386 (Me1 and Me2) (Fig. 3B and supplemental Data). Arg386 occurs in close proximity to Ser388, which in murine type II Runx2 is targeted by the MAP3K mixed-lineage kinase 3 (MLK3), an upstream regulator of ERK1/2- and p38MAPK-mediated Runx2 activation (59). This raises the possibility of functional regulation of Runx2 phosphorylation by proximal methylation, which has been described on other transcription factors such as FoxO1 (63, 64).
Fig. 3.
Runx2 is mono- and di-methylated at multiple arginines. The HCD MS/MS fragmentation spectra for residues A, 252–258 (426.23+2 m/z) and B, 375–386 (683.35+2) revealed mono-methylation of arginines 258 and 356. Annotated spectra of dimethylated peptides are provided in the supplemental Data.
Runx2 is O-GlcNAc Modified in BMMSCs
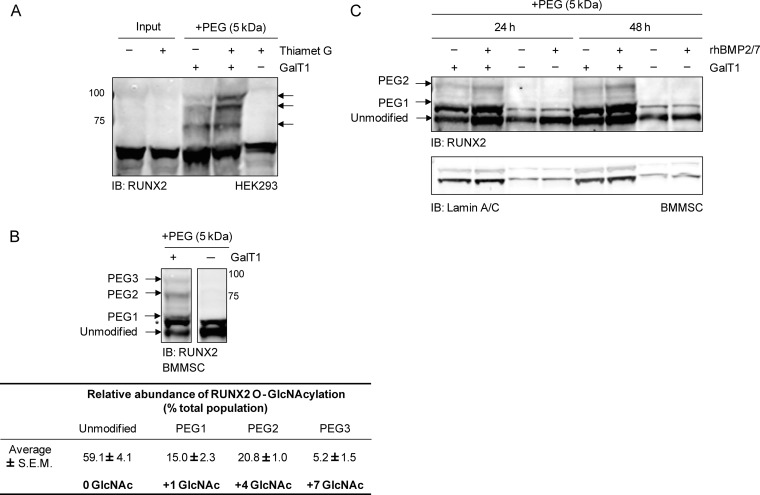
Initial attempts to confirm O-GlcNAcylation of Runx2 immunoprecipitated from MC3T3E1 pre-osteoblasts were inconclusive because of cross-reactivity of the CTD110.6 O-GlcNAc antibody with an immunogen of similar molecular weight in IgG control immunoprecipitates (data not shown). As an alternative approach, O-GlcNAcylated Runx2 was chemoenzymatically labeled (53). Briefly, a mutant galactosyl transferase (Y298L β-1,4-GalT) catalyzes the transfer of azido-modified UDP-GalNAc (GalNAz) to terminal GlcNAc moieties. Subsequently, a 5 kDa polyethylene glycol (PEG) alkyne tag is reacted with the azido-labeled sugar through azide-alkyne Huisgen cycloaddition to slow the electrophoretic mobility of O-GlcNAc modified proteins by SDS-PAGE. This strategy was initially applied to lysates from Thiamet G-stimulated (18 h) HEK293 cells transduced with an adenoviral construct encoding murine type II Runx2, which contains a minimal biotinylation sequence tag (17). Labeled O-GlcNAcylated proteins were separated by SDS-PAGE and probed for Runx2 by Western blot. Multiple antibody reactive bands were detected above the unmodified Runx2 band (Fig. 4A). The intensities of the two highest MW bands were increased by OGA inhibition consistent with increased O-GlcNAc modification.
Fig. 4.
Endogenously expressed Runx2 is O-GlcNAc modified in BMMSCs. Total or nuclear protein was chemoenzymatically labeled with 5 kDa PEG (+PEG), resolved by SDS-PAGE, and immunoblotted with the Runx2 antibody. As a negative control, reactions were performed in the absence of the GalT1 enzyme (-GalT1). A, HEK293 cells were transduced with the AdBioR2 adenovirus encoding murine type II Runx2 for 24 h before treating cells with Thiamet G (20 μm) or its DMSO carrier with 1% serum (18 h). Total lysate protein (150 μg) was then chemoenzymatically labeled, black arrows indicate mass-shifted, O-GlcNAc modified Runx2. B, BMMSCs pretreated with BMP2/7 for 48 h under osteogenic conditions were incubated with Thiamet G with 1% serum (18 h). Nuclear protein (75 μg) was chemoenzymatically labeled and the Runx2 protein detected by immunoblotting. The shift in MW of the most prominent bands were calculated to determine the number of GlcNAc moieties on the protein. The relative abundance of O-GlcNAc modified Runx2 was estimated by densitometry. C, BMMSCs were serum-starved (18 h) before stimulation with BMP2/7 (50 ng/ml) or its vehicle for 24 or 48 h under osteogenic conditions. Nuclear protein was chemoenzymatically labeled in the presence or absence of GalT1 (+GalT1, 150 μg protein; -GalT1, 50 μg protein). The asterisk denotes an unidentified protein that reacts with the Runx2 antibody but remains unchanged with chemoenzymatic labeling. Lamin A/C served as a loading control.
Next, this strategy was applied to nuclear protein isolated from bone marrow mesenchymal stem cells (BMMSCs) that had been cultured under osteogenic conditions in the presence of BMP2/7 for 48 h, and then treated with Thiamet G for 18 h. Three major mass-shifted Runx2 bands could be visualized after chemoenzymatic labeling and immunoblotting (Fig. 4B). The average MW of these bands (± S.E.) was 58.5 ± 0.4 (unmodified Runx2), 63.9 ± 0.2 kDa (PEG1), 78.5 ± 1.7 kDa (PEG2), and 93.3 ± 1.9 (PEG3), which correspond to 0, 1, 4, and 7 sites of O-GlcNAc modification, respectively. Two minor mass-shifted bands corresponding to Runx2 proteoforms labeled at two or three O-GlcNAc sites were also consistently observed between PEG1 and PEG2 populations but these faint bands were below the limit of quantitation. Densitometry was employed to estimate the fraction of nuclear localized Runx2 that is O-GlcNAcylated (Fig. 4B). The PEG1 band (1 O-GlcNAc) represented ∼15% of the total population, whereas PEG2 (4 O-GlcNAc) and PEG3 (7 O-GlcNAc) bands made up ∼21 and 5% of the total population, respectively. The Runx2 antibody also detected a band of unknown identity above unmodified Runx2, which may represent a more heavily modified (e.g. phosphorylated) form of Runx2. This band (Fig. 4B*) was unaffected by the chemoenzymatic reaction. Under conditions of Thiamet G-treatment an estimated 40% of the nuclear Runx2 in BMMSCs was O-GlcNAc modified.
BMPs are known to stimulate osteoblast differentiation by increasing Runx2 phosphorylation and transactivation (17, 65). To test whether acute BMP2/7-stimulation alone affected the relative abundance of Runx2 O-GlcNAc modification, BMMSCs were serum-starved for 18 h and cultured under osteogenic conditions for 24 or 48 h in the absence or presence of BMP2/7. Nuclear lysates were chemoenzymatically labeled with the 5 kDa PEG mass tag. Although treatment with BMP2/7 increased Runx2 protein levels and facilitated detection of the chemoenzymatically labeled protein, we did not observe a change in the extent of Runx2 O-GlcNAcylation (Fig. 4C).
Inhibition of OGA Increases ALP Expression and Activity in BMMSCs
To examine the impact of OGA inhibition on osteoblast differentiation at an early stage, we measured the expression and activity of the Runx2-transcriptional target, ALP, in BMMSCs induced toward the osteoblast lineage. Cells were cultured for 6 days in medium containing 5% serum, ascorbic acid, β-glycerol phosphate (66–69), and Thiamet G or its vehicle. Differentiation toward the osteoblast lineage was confirmed by measuring ALP and PPARg transcript expression, markers of osteoblast and adipocyte differentiation, respectively (supplemental Fig. S1). As a positive control, BMMSCs were also differentiated in the presence of BMP2/7. Chronic pharmacological inhibition of OGA increased global protein O-GlcNAc levels and induced an expected compensatory increase in OGA transcript and protein expression (27) (Fig. 5A, supplemental Fig. S2). Furthermore, treatment with Thiamet G increased ALP activity in a dose-dependent manner (Fig. 5B) and increased ALP transcript expression (Fig. 5C). On average, treatment with 20 μm Thiamet G enhanced ALP activity by 65.8% and ALP transcript expression by 24.3% compared with vehicle treatments.
Fig. 5.
OGA mediates induction of the early osteogenic marker ALP. BMMSCs were cultured for 6 days (Days 1–7) in osteogenic medium containing 5% serum and Thiamet G (0–20 μm), BMP2/7 (50 ng/ml), or their respective DMSO and HCl vehicles (Vh). A, Changes in total protein O-GlcNAc modification, OGT, and OGA protein levels in response to OGA inhibition or BMP2/7 treatment were probed by immunoblotting. B, The effect of inhibition of OGA on ALP activity was measured in BMMSC lysates and fixed monolayers. OGA enzyme activity (Units/μg total protein) is expressed as a percentage of the control (left panel). ALP staining of BMMSC monolayers treated with Thiamet G (20 μm) or BMP2/7 (50 ng/ml) (right panel). C, Inhibition of OGA increases ALP mRNA transcript expression. Values are expressed as a percentage of control. D, Chronic BMP2/7 treatment decreases OGA activity. BMMSCs were cultured in osteogenic medium containing 10% serum and BMP2/7 (50 ng/ml) or its vehicle. The enzyme activities of ALP and OGA were measured on Day 7 and Day 8, respectively. OGA activity was normalized by OGA expression as determined by densitometric analysis of immunoblots (supplemental Fig. S3) and is expressed as a percentage of the control. E, OGA inhibition enhances BMP2/7-stimulated ALP activity as measured in lysates or monolayers. BMMSCs were cultured for 6 days with BMP2/7 and Thiamet G (20 μm) or its DMSO vehicle. ALP activity was determined on Day 7 by enzyme assay (top panel) or by staining (lower panel). F, OGA inhibition does not impact the rate of cellular proliferation. The incorporation of BrdU was measured at A450 on Day 6 of differentiation of BMMSCs treated with Thiamet G or BMP2/7. BMP2/7- and serum-treatments were employed as positive and negative controls for proliferation, respectively. Error bars are ± S.E.: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
Immunoblot analysis also revealed a modest increase in global O-GlcNAc modification in BMP2/7-treated BMMSCs compared with vehicle controls (Fig. 5A), suggesting that BMP2/7 may affect the activity of O-GlcNAc cycling enzymes. To test this, we measured the expression and activity of OGA in BMMSCs after 8 days of culture in osteogenic media. Although OGA and OGT protein expression remained unchanged (supplemental Fig. S3), OGA enzyme activity was significantly decreased by 39.0% (Fig. 5D). To test whether inhibition of OGA activity enhances BMP2/7-induced effects on ALP activity, BMMSCs were cultured under 5% serum osteogenic conditions for 6 days with 50 ng/ml BMP2/7 in the presence or absence of Thiamet G. Compared with treatments lacking the OGA inhibitor, addition of Thiamet G significantly enhanced the effect of BMP2/7 on the induction of ALP activity by 35.6% (p < 0.05) (Fig. 5E). In summary, these data suggest that induction of the early osteogenic marker ALP is associated with down-regulation of OGA enzyme activity that can be induced by BMP2/7-treatment.
BMMSCs remain proliferative in culture for several days past the point of visible confluence, creating a dense monolayer resulting from cell packing. To investigate whether OGA inhibition affected BMMSC proliferation, the incorporation of the BrdU pyrimidine analog into the DNA of actively proliferating cells was measured. BMMSCs were cultured in osteogenic medium containing 5% serum and amended with Thiamet G (20 μm), BMP2/7 (50 ng/ml), or their respective vehicles. Although BMP2/7 increased proliferation as expected, Thiamet G-treatment did not affect BMMSC proliferation at Days 2, 3, 4, or 6 (Day 6 time point shown in Fig. 5F), similar to a previous report in lung and colon cancer cell lines (70).
DISCUSSION
The PTM of Runx2 allows for the discrimination of complex signals that influence co-regulator interactions, localization, and protein stability during osteoblast differentiation (15, 16). We report for the first time novel sites of O-GlcNAc modification and methylation on human type II Runx2. A summary of known and novel PTMs (this study), as well as regions of protein and DNA interactions on Runx2, is provided in Fig. 6. O-GlcNAc modification of endogenously expressed Runx2 was confirmed in BMMSCs by chemoenzymatic labeling, and using this method it was estimated by densitometry that 40% of the nuclear localized Runx2 pool was O-GlcNAcylated in response to OGA inhibition. Additionally, the abundance of O-GlcNAcylated Runx2 was enhanced with BMP2/7 treatment, and inhibition of OGA augmented the activity of a Runx2-specific luciferase reporter and the ALP marker under both basal and BMP2/7-stimulated conditions. This suggests that O-GlcNAc modification of Runx2 increases its transcriptional activity, consistent with a previous report (28). To date, we have observed three sites of O-GlcNAc modification on Runx2 at S32, S33, and S371 that reside within known transactivation domains and that are proximal to regulatory sites of phosphorylation. Ongoing studies are aimed at the mass spectrometric characterization of Runx2 isolated from osteogenic cell lines to facilitate functional assessment of site-specific Runx2 O-GlcNAc modification on transcriptional activation and protein–protein interactions.
Fig. 6.
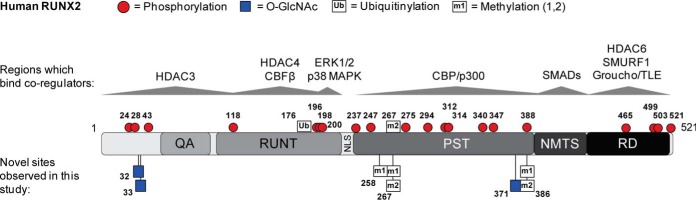
Post-translational Modifications and Protein Interactions of RUNX2. Cartoon of human type II RUNX2 with previously described sites of PTM (top) and novel sites of PTM identified in this study (below) indicated. Interactions with select transcriptional co-regulators are indicated by gray arrows. QA: glutamine-alanine rich domain; RUNT: DNA-binding domain; NLS: nuclear localization sequence; PST: proline/serine/threonine-rich domain; NMTS: nuclear matrix targeting sequence. A summary of known modifications on RUNX2 with references can be found at www.phosphosite.org.
Chronic treatment of BMMSCs with Thiamet G enhanced the activity of the Runx2-target ALP, an early marker of differentiation, in both unstimulated and BMP2/7-induced cells after a week of culture. The timing associated with observed effects of OGA inhibition on ALP expression are in-line with studies in chondrocytes that showed increased ALP transcript levels after 9 days of differentiation in the presence of Thiamet G (27). Similarly, in human MSCs, altered regulation of ALP activity by alanine-substitution at MAPK-regulated Runx2 phosphorylation sites were observed after 6 days (71). Of note, Runx2 transcriptional activity was enhanced by Thiamet G treatment after 48 h in MC3T3E1 pre-osteoblasts, a cell stage approximated in BMMSCs after 6 days of culture under osteogenic conditions. In addition to Runx2, Thiamet G may also induce changes in ALP expression through regulation of Msx2 or Dlx5 which also target the promoter of the ALP gene (ALPL) (72). Future studies utilizing site-directed mutagenesis will more directly address the role of O-GlcNAc modified residues of Runx2 on the regulation of ALP expression.
This study revealed the presence of a potentially novel mechanism for BMP2/7 regulation of ALP expression. The observation that BMP2/7 stimulation decreases OGA activity suggests that regulation of O-GlcNAc modification may contribute, in part, to the pro-osteogenic effects elicited by select BMP growth factors. Previous studies demonstrate interactions between OGT and ERK/p38MAPK pathway components (40, 73, 74), and the close proximity of ERK and p38MAPK-regulated sites of phosphorylation at Ser24, Ser28, and Ser31 (17, 59) to O-GlcNAcylation at Ser32 and Ser33, as well as the simultaneous occurrence of O-GlcNAcylation and phosphorylation of peptide residues 26–38, raises the possibility of cooperativity among these modifications. ERK1/2 and p38MAPK-dependent pathways are integral to proper bone formation and osteoblast function, and are stimulated by BMPs, FGFs, and insulin/IGF-1, as well as loading-induced biophysical signals that result in activating phosphorylation of Runx2 and increased bone formation (75, 76). The critical role of the TGF-β activated kinase 1 (TAK1)-MAPK kinase 3/6-p38MAPK-Runx2 axis was revealed in TAK1-deficient mice that exhibited impaired Runx2 phosphorylation, activation, and a skeletal phenotype similar that of Runx2-haploinsufficient mice (71). Sequential alanine-substitution at sites homologous to Ser31, Ser275, and Ser312 of the human Type II isoform additively attenuated BMP2/7-induced activation of the OCN promoter by type I RUNX2, and the triple mutant failed to drive osteoblast differentiation of human mesenchymal stem cells (71). Also, simultaneous alanine-substitution of Ser28, Ser301, and Ser319 (numbering for murine Runx2 type II, Ser294 and Ser312 human Runx2) attenuated the stimulatory effects of mixed lineage kinase 3 (MLK3), a MAP3K that activates both p38MAPK and ERK1/2, on Runx2 transcriptional activity (59). Very little is known regarding the upstream regulation of OGA, and to our knowledge BMP2/7-mediated inhibition of OGA activity has not been described.
Novel sites of arginine methylation (Arg258, Arg267, and Arg386) were observed within the PST-domain close to the runt-DNA binding domain and the nuclear matrix targeting sequence (NTMS). Previous studies support a role for protein methylation in the modulation of osteogenesis and bone maintenance. S-adenosylmethionine (SAM), the universal donor for the methylation of target residues (77), is depleted with low folic acid intake, a condition associated with increased fracture risk in the elderly (78–80). In chondrocytes the functional interaction between the H3K9 histone methyltransferase ESET (i.e. SETDB1), Runx2, and HDAC4 represses Runx2 responsive genetic programs that regulate terminal chondrocyte differentiation during endochondral ossification, and mesenchymal ESET-deficient mice display premature chondrocyte hypertrophy, lower levels of trabecular bone formation, and defective expression of mineralization markers in long bones (81). Although it has yet to be determined whether direct methylation of Runx2 affects its function, inhibition of SAM methyltransferases in C2C12 stromal cells attenuated BMP2-induced activation of Runx2 (78). This effect was not observed in response to a specific reduction in DNA methylation, suggesting that methylation cycling at the protein level is necessary for Runx2 transcriptional activation. We observed methylation at Arg386 that occurs within close proximity to phosphorylation at Ser388 (59). Recent studies demonstrate mutually exclusive functions for protein arginine methyltransferase (PRMT)-mediated methylation of FOXO1 at Arg248 and Arg250, which antagonizes its AKT-mediated phosphorylation at Ser253 and nuclear export (63, 64). It is therefore possible that sites of methylation and phosphorylation may interact to influence Runx2 function.
Differential PTM of Runx2, its interacting co-regulators, and associated histones provides a complex and dynamic mechanism for the regulation of bone remodeling in response to hormones, nutrients, stress, and mechanical load, and the effects of PTM cross-talk on the modulation of Runx2 function are only beginning to be clarified. Such examples include the partial dependence of FGF2-induced Runx2 acetylation on its phosphorylation at the MEK1/2-regulated murine Ser301 site, a process that regulates Runx2 ubiquitinylation and stability (82). Also, phosphorylation modulates Runx2 interactions with p300, CBP, and PCAF histone acetyltransferases and co-repressors (HDAC4) (71, 83, 84), as well as the prolyl cis-trans isomerase (PIN1)-mediated stabilization of Runx2 (85). Within the Runx2 protein proximal sites of O-GlcNAc modification and phosphorylation may be mutually exclusive (33, 86), or may yield distinct regulatory functions as suggested by a study demonstrating that only 8% of O-GlcNAcylation sites, (of over 1750 identified in murine synaptosomes), occurred at phosphorylation sites (61). Additionally, the observed interaction of Runx2 with OGT is consistent with this protein's ability to scaffold chromatin remodeling factors and co-regulators such as HDACs 3–6, p300/CBP, retinoblastoma protein (pRb), SMADs, SMURF1, TLE/Groucho, mSin3a, STAT1, glucocorticoid receptor, etc. [reviewed in (16, 87)]. The availability of modification-specific Runx2 antibodies will greatly facilitate elucidation of its PTM regulation and the co-occurrence of modified Runx2 forms on activated promoters of genes that drive osteogenesis.
In summary, we report O-GlcNAc modification and methylation of the critical transcription factor Runx2. We also present findings that suggest a role for OGA in the modulation of Runx2-dependent pathways that influence the production of the ALP matrix maturation marker during early phases of osteoblast differentiation. Our findings link the activity of O-GlcNAc cycling enzymes to the regulation of osteoblast function, and have implications for pathological bone loss observed under conditions of dysregulated nutrient metabolism such as type I and type II diabetes and age-related osteopenia and osteoporosis. It remains to be determined whether O-GlcNAc modification of Runx2 increases expression of gene targets such as ALP through direct effects on DNA-binding and transactivation, or through modulation of functional complex formation, which allows discrimination of complex upstream signals governing Runx2-dependent transcriptional programs.
Supplementary Material
Acknowledgments
We thank Susana Comte-Walters, Mary Berkaw, Dr. Courtney Haycraft, and Dr. Neil Olszewski for technical assistance. We thank Jennifer R. Bethard and the Mass Spectrometry Facility, a University Sponsored Research Resource.
Footnotes
Author contributions: A.K.N. and L.E.B. designed research; A.K.N. performed research; A.K.N. and L.E.B. analyzed data; A.K.N. and L.E.B. wrote the paper.
* This work was supported by National Institutes of Health Grants R01 DE020925 and T32 DE017551 05 from the NIDCR, and S10 D010731–01 from the NCRR.
 This article contains supplemental Figs. S1 to S3, Tables, and Data.
This article contains supplemental Figs. S1 to S3, Tables, and Data.
1 The abbreviations used are:
- PTM
- post-translational modification
- ALP
- alkaline phosphatase
- BMMSC
- bone-marrow derived mesenchymal stem cell
- BMP2/7
- recombinant human bone morphogenetic protein 2/7 heterodimer
- ETD
- electron transfer dissociation
- C18 nLC-MS/MS
- C18 reversed phase nano-liquid chromatography tandem mass spectrometry
- GalNAz
- azido-labeled N-acetyl-galactosamine
- GalT1
- galactosyltransferase
- HCD
- higher energy collisional dissociation
- HexNAc
- N-acetylhexosamine
- MMP13
- matrix metalloproteinase 13
- MW
- molecular weight
- OCN
- osteocalcin
- OGA
- O-GlcNAcase
- O-GlcNAc
- β-O-linked N-Acetylglucosamine
- OGT
- O-GlcNAc transferase
- PEG
- polyethylene glycol
- PST
- proline/serine/threonine rich region
- PTH
- parathyroid hormone
- Runx2
- runt-related transcription factor 2.
REFERENCES
- 1. Martin J. W., Zielenska M., Stein G. S., van Wijnen A. J., Squire J. A. (2011) The role of RUNX2 in osteosarcoma oncogenesis. Sarcoma 2011, 282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tandon M., Chen Z., Pratap J. (2014) Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast Cancer Res. 16, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soung do Y., Talebian L., Matheny C. J., Guzzo R., Speck M. E., Lieberman J. R., Speck N. A., Drissi H. (2012) Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J. Bone Miner. Res. 27, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim E. J., Cho S. W., Shin J. O., Lee M. J., Kim K. S., Jung H. S. (2013) Ihh and Runx2/Runx3 signaling interact to coordinate early chondrogenesis: a mouse model. PloS One 8, e55296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 6. Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 7. Lengner C. J., Drissi H., Choi J. Y., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2002) Activation of the bone-related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech. Dev. 114, 167–170 [DOI] [PubMed] [Google Scholar]
- 8. Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., Komori T. (1999) Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 214, 279–290 [DOI] [PubMed] [Google Scholar]
- 9. Kim I. S., Otto F., Zabel B., Mundlos S. (1999) Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80, 159–170 [DOI] [PubMed] [Google Scholar]
- 10. Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 11. Moffatt P., Ben Amor M., Glorieux F. H., Roschger P., Klaushofer K., Schwartzentruber J. A., Paterson A. D., Hu P., Marshall C., Consortium F. C., Fahiminiya S., Majewski J., Beaulieu C. L., Boycott K. M., Rauch F. (2013) Metaphyseal dysplasia with maxillary hypoplasia and brachydactyly is caused by a duplication in RUNX2. Am. J. Hum. Genet. 92, 252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffroy V., Ducy P., Karsenty G. (1997) Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 16, 307–310 [DOI] [PubMed] [Google Scholar]
- 13. Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 14. Komori T. (2010) Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 339, 189–195 [DOI] [PubMed] [Google Scholar]
- 15. Jonason J. H., Xiao G., Zhang M., Xing L., Chen D. (2009) Post-translational Regulation of Runx2 in Bone and Cartilage. J. Dent. Res. 88, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lian J. B., Stein G. S., Javed A., van Wijnen A. J., Stein J. L., Montecino M., Hassan M. Q., Gaur T., Lengner C. J., Young D. W. (2006) Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 7, 1–16 [DOI] [PubMed] [Google Scholar]
- 17. Ge C., Xiao G., Jiang D., Yang Q., Hatch N. E., Roca H., Franceschi R. T. (2009) Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J. Biol. Chem. 284, 32533–32543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao G., Jiang D., Gopalakrishnan R., Franceschi R. T. (2002) Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J. Biol. Chem. 277, 36181–36187 [DOI] [PubMed] [Google Scholar]
- 19. Qiao M., Shapiro P., Kumar R., Passaniti A. (2004) Insulin-like growth factor-1 regulates endogenous RUNX2 activity in endothelial cells through a phosphatidylinositol 3-kinase/ERK-dependent and Akt-independent signaling pathway. J. Biol. Chem. 279, 42709–42718 [DOI] [PubMed] [Google Scholar]
- 20. Kugimiya F., Kawaguchi H., Ohba S., Kawamura N., Hirata M., Chikuda H., Azuma Y., Woodgett J. R., Nakamura K., Chung U. I. (2007) GSK-3beta controls osteogenesis through regulating Runx2 activity. PloS One 2, e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Selvamurugan N., Shimizu E., Lee M., Liu T., Li H., Partridge N. C. (2009) Identification and characterization of Runx2 phosphorylation sites involved in matrix metalloproteinase-13 promoter activation. FEBS Lett. 583, 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 23. Westendorf J. J., Zaidi S. K., Cascino J. E., Kahler R., van Wijnen A. J., Lian J. B., Yoshida M., Stein G. S., Li X. (2002) Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 22, 7982–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 25. Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 26. Deplus R., Delatte B., Schwinn M. K., Defrance M., Mendez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andres-Bergos J., Tardio L., Larranaga-Vera A., Gomez R., Herrero-Beaumont G., Largo R. (2012) The increase in o-linked N-acetylglucosamine protein modification stimulates chondrogenic differentiation both in vitro and in vivo. J. Biol. Chem. 287, 33615–33628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S. H., Kim Y. H., Song M., An S. H., Byun H. Y., Heo K., Lim S., Oh Y. S., Ryu S. H., Suh P. G. (2007) O-GlcNAc modification modulates the expression of osteocalcin via OSE2 and Runx2. Biochem. Biophys. Res. Commun. 362, 325–329 [DOI] [PubMed] [Google Scholar]
- 29. Heckel D., Comtesse N., Brass N., Blin N., Zang K. D., Meese E. (1998) Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum. Mol. Genet. 7, 1859–1872 [DOI] [PubMed] [Google Scholar]
- 30. Lubas W. A., Frank D. W., Krause M., Hanover J. A. (1997) O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 [DOI] [PubMed] [Google Scholar]
- 31. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Donnell N., Zachara N. E., Hart G. W., Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z., Gucek M., Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Griffith L. S., Schmitz B. (1999) O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur. J. Biochem. 262, 824–831 [DOI] [PubMed] [Google Scholar]
- 36. Wang Z., Pandey A., Hart G. W. (2007) Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell. Proteomics 6, 1365–1379 [DOI] [PubMed] [Google Scholar]
- 37. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gambetta M. C., Oktaba K., Muller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
- 39. Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., Honda B. M. (2009) Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dehennaut V., Slomianny M. C., Page A., Vercoutter-Edouart A. S., Jessus C., Michalski J. C., Vilain J. P., Bodart J. F., Lefebvre T. (2008) Identification of structural and functional O-linked N-acetylglucosamine-bearing proteins in Xenopus laevis oocyte. Mol. Cell. Proteomics 7, 2229–2245 [DOI] [PubMed] [Google Scholar]
- 41. Kim H. S., Park S. Y., Choi Y. R., Kang J. G., Joo H. J., Moon W. K., Cho J. W. (2009) Excessive O-GlcNAcylation of proteins suppresses spontaneous cardiogenesis in ES cells. FEBS Lett. 583, 2474–2478 [DOI] [PubMed] [Google Scholar]
- 42. Maury J. J., Chan K. K., Zheng L., Bardor M., Choo A. B. (2013) Excess of O-linked N-acetylglucosamine modifies human pluripotent stem cell differentiation. Stem Cell Res. 11, 926–937 [DOI] [PubMed] [Google Scholar]
- 43. Jang H., Kim T. W., Yoon S., Choi S. Y., Kang T. W., Kim S. Y., Kwon Y. W., Cho E. J., Youn H. D. (2012) O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 44. Nagel A. K., Schilling M., Comte-Walters S., Berkaw M. N., Ball L. E. (2013) Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation. Mol. Cell. Proteomics 12, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ogawa M., Mizofuchi H., Kobayashi Y., Tsuzuki G., Yamamoto M., Wada S., Kamemura K. (2012) Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 1820, 24–32 [DOI] [PubMed] [Google Scholar]
- 46. Hsieh T. J., Lin T., Hsieh P. C., Liao M. C., Shin S. J. (2012) Suppression of Glutamine:fructose-6-phosphate amidotransferase-1 inhibits adipogenesis in 3T3-L1 adipocytes. J. Cell. Physiol. 227, 108–115 [DOI] [PubMed] [Google Scholar]
- 47. Chen Q., Chen Y., Bian C., Fujiki R., Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 49. de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 100, 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ducy P., Karsenty G. (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cox J., Michalski A., Mann M. (2011) Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 22, 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 53. Rexach J. E., Rogers C. J., Yu S. H., Tao J., Sun Y. E., Hsieh-Wilson L. C. (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ogata N., Chikazu D., Kubota N., Terauchi Y., Tobe K., Azuma Y., Ohta T., Kadowaki T., Nakamura K., Kawaguchi H. (2000) Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J. Clin. Invest. 105, 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robinson K. A., Ball L. E., Buse M. G. (2007) Reduction of O-GlcNAc protein modification does not prevent insulin resistance in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 292, E884–E890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao P., Viner R., Teo C. F., Boons G. J., Horn D., Wells L. (2011) Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Proteome Res. 10, 4088–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Javed A., Afzal F., Bae J. S., Gutierrez S., Zaidi K., Pratap J., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs 189, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pande S., Browne G., Padmanabhan S., Zaidi S. K., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2013) Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J. Cell. Physiol. 228, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zou W., Greenblatt M. B., Shim J. H., Kant S., Zhai B., Lotinun S., Brady N., Hu D. Z., Gygi S. P., Baron R., Davis R. J., Jones D., Glimcher L. H. (2011) MLK3 regulates bone development downstream of the faciogenital dysplasia protein FGD1 in mice. J. Clin. Invest. 121, 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cantin G. T., Yi W., Lu B., Park S. K., Xu T., Lee J. D., Yates J. R., 3rd (2008) Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 7, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 61. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., Burlingame A. L. (2012) Global Identification and Characterization of Both O-GlcNAcylation and Phosphorylation at the Murine Synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Choi D., Oh K. J., Han H. S., Yoon Y. S., Jung C. Y., Kim S. T., Koo S. H. (2012) Protein arginine methyltransferase 1 regulates hepatic glucose production in a FoxO1-dependent manner. Hepatology 56, 1546–1556 [DOI] [PubMed] [Google Scholar]
- 64. Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., Fukamizu A. (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 65. Ge C., Yang Q., Zhao G., Yu H., Kirkwood K. L., Franceschi R. T. (2012) Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J. Bone Miner. Res. 27, 538–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu W., Rawlins B. A., Boachie-Adjei O., Myers E. R., Arimizu J., Choi E., Lieberman J. R., Crystal R. G., Hidaka C. (2004) Combined bone morphogenetic protein-2 and -7 gene transfer enhances osteoblastic differentiation and spine fusion in a rodent model. J. Bone Miner. Res. 19, 2021–2032 [DOI] [PubMed] [Google Scholar]
- 67. Franceschi R. T., Iyer B. S. (1992) Relationship between collagen synthesis and expression of the osteoblast phenotype in MC3T3-E1 cells. J. Bone Miner. Res. 7, 235–246 [DOI] [PubMed] [Google Scholar]
- 68. Franceschi R. T., Iyer B. S., Cui Y. (1994) Effects of ascorbic acid on collagen matrix formation and osteoblast differentiation in murine MC3T3-E1 cells. J. Bone Miner. Res. 9, 843–854 [DOI] [PubMed] [Google Scholar]
- 69. Quarles L. D., Yohay D. A., Lever L. W., Caton R., Wenstrup R. J. (1992) Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J. Bone Miner. Res. 7, 683–692 [DOI] [PubMed] [Google Scholar]
- 70. Mi W., Gu Y., Han C., Liu H., Fan Q., Zhang X., Cong Q., Yu W. (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 1812, 514–519 [DOI] [PubMed] [Google Scholar]
- 71. Greenblatt M. B., Shim J. H., Zou W., Sitara D., Schweitzer M., Hu D., Lotinun S., Sano Y., Baron R., Park J. M., Arthur S., Xie M., Schneider M. D., Zhai B., Gygi S., Davis R., Glimcher L. H. (2010) The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J. Clin. Invest. 120, 2457–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim Y. J., Lee M. H., Wozney J. M., Cho J. Y., Ryoo H. M. (2004) Bone morphogenetic protein-2-induced alkaline phosphatase expression is stimulated by Dlx5 and repressed by Msx2. J. Biol. Chem. 279, 50773–50780 [DOI] [PubMed] [Google Scholar]
- 73. Cheung W. D., Hart G. W. (2008) AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 283, 13009–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goldberg H., Whiteside C., Fantus I. G. (2011) O-linked beta-N-acetylglucosamine supports p38 MAPK activation by high glucose in glomerular mesangial cells. Am. J. Physiol. Endocrinol. Metab. 301, E713–E726 [DOI] [PubMed] [Google Scholar]
- 75. Boutahar N., Guignandon A., Vico L., Lafage-Proust M. H. (2004) Mechanical strain on osteoblasts activates autophosphorylation of focal adhesion kinase and proline-rich tyrosine kinase 2 tyrosine sites involved in ERK activation. J. Biol. Chem. 279, 30588–30599 [DOI] [PubMed] [Google Scholar]
- 76. You J., Reilly G. C., Zhen X., Yellowley C. E., Chen Q., Donahue H. J., Jacobs C. R. (2001) Osteopontin gene regulation by oscillatory fluid flow via intracellular calcium mobilization and activation of mitogen-activated protein kinase in MC3T3-E1 osteoblasts. J. Biol. Chem. 276, 13365–13371 [DOI] [PubMed] [Google Scholar]
- 77. Wang Y. C., Peterson S. E., Loring J. F. (2014) Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24, 143–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vaes B. L., Lute C., van der Woning S. P., Piek E., Vermeer J., Blom H. J., Mathers J. C., Muller M., de Groot L. C., Steegenga W. T. (2010) Inhibition of methylation decreases osteoblast differentiation via a non-DNA-dependent methylation mechanism. Bone 46, 514–523 [DOI] [PubMed] [Google Scholar]
- 79. Imbard A., Benoist J. F., Blom H. J. (2013) Neural tube defects, folic acid and methylation. Int. J. Environ. Res. Public Health 10, 4352–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dhonukshe-Rutten R. A., Pluijm S. M., de Groot L. C., Lips P., Smit J. H., van Staveren W. A. (2005) Homocysteine and vitamin B12 status relate to bone turnover markers, broadband ultrasound attenuation, and fractures in healthy elderly people. J, Bone Miner. Res. 20, 921–929 [DOI] [PubMed] [Google Scholar]
- 81. Yang L., Lawson K. A., Teteak C. J., Zou J., Hacquebord J., Patterson D., Ghatan A. C., Mei Q., Zielinska-Kwiatkowska A., Bain S. D., Fernandes R. J., Chansky H. A. (2013) ESET histone methyltransferase is essential to hypertrophic differentiation of growth plate chondrocytes and formation of epiphyseal plates. Dev. Biol. 380, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park O. J., Kim H. J., Woo K. M., Baek J. H., Ryoo H. M. (2010) FGF2-activated ERK mitogen-activated protein kinase enhances Runx2 acetylation and stabilization. J. Biol. Chem. 285, 3568–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sierra J., Villagra A., Paredes R., Cruzat F., Gutierrez S., Javed A., Arriagada G., Olate J., Imschenetzky M., Van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L., Montecino M. (2003) Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 23, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee M., Partridge N. C. (2010) Parathyroid hormone activation of matrix metalloproteinase-13 transcription requires the histone acetyltransferase activity of p300 and PCAF and p300-dependent acetylation of PCAF. J. Biol. Chem, 285, 38014–38022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoon W. J., Islam R., Cho Y. D., Woo K. M., Baek J. H., Uchida T., Komori T., van Wijnen A., Stein J. L., Lian J. B., Stein G. S., Choi J. Y., Bae S. C., Ryoo H. M. (2013) Pin1-mediated Runx2 modification is critical for skeletal development. J. Cell. Physiol. 228, 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schroeder T. M., Jensen E. D., Westendorf J. J. (2005) Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res. C Embryo Today 75, 213–225 [DOI] [PubMed] [Google Scholar]
- 88. Wee H. J., Huang G., Shigesada K., Ito Y. (2002) Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. EMBO Rep. 3, 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.