Abstract
The lubricative, heavily glycosylated mucin-like synovial glycoprotein lubricin has previously been observed to contain glycosylation changes related to rheumatoid and osteoarthritis. Thus, a site-specific investigation of the glycosylation of lubricin was undertaken, in order to further understand the pathological mechanisms involved in these diseases. Lubricin contains an serine/threonine/proline (STP)-rich domain composed of imperfect tandem repeats (EPAPTTPK), the target for O-glycosylation. In this study, using a liquid chromatography–tandem mass spectrometry approach, employing both collision-induced and electron-transfer dissociation fragmentation methods, we identified 185 O-glycopeptides within the STP-rich domain of human synovial lubricin. This showed that adjacent threonine residues within the central STP-rich region could be simultaneously and/or individually glycosylated. In addition to core 1 structures responsible for biolubrication, core 2 O-glycopeptides were also identified, indicating that lubricin glycosylation may have other roles. Investigation of the expression of polypeptide N-acetylgalactosaminyltransferase genes was carried out using cultured primary fibroblast-like synoviocytes, a cell type that expresses lubricin in vivo. This analysis showed high mRNA expression levels of the less understood polypeptide N-acetylgalactosaminyltransferase 15 and 5 in addition to the ubiquitously expressed polypeptide N-acetylgalactosaminyltransferase 1 and 2 genes. This suggests that there is a unique combination of transferase genes important for the O-glycosylation of lubricin. The site-specific glycopeptide analysis covered 82% of the protein sequence and showed that lubricin glycosylation displays both micro- and macroheterogeneity. The density of glycosylation was shown to be high: 168 sites of O-glycosylation, predominately sialylated, were identified. These glycosylation sites were focused in the central STP-rich region, giving the domain a negative charge. The more positively charged lysine and arginine residues in the N and C termini suggest that synovial lubricin exists as an amphoteric molecule. The identification of these unique properties of lubricin may provide insight into the important low-friction lubricating functions of lubricin during natural joint movement.
Human diarthrodial joints are surrounded by synovial fluid (SF),1 a dense extracellular matrix fluid composed of proteins, glycoproteins, hyaluronic acid, proteoglycans, and phospholipids (1). During movement, the cartilage surfaces of the articulating joints slide over each other with an extremely low coefficient of friction that ranges from 0.0005 to 0.04 (2) and handle pressures up to ∼200 atm (3). In a healthy state, the joint surface and SF constitute a system of reduced friction that results in lifelong lubrication and wear resistance, primarily due to biolubricating molecules such as hyaluronic acid and lubricin (4). Human synovial lubricin is encoded by the proteoglycan 4 (Prg4) gene (5, 6) and is synthesized by fibroblast-like synoviocytes (FLSs) and superficial zone chondrocytes. Its 1404-amino-acid sequence contains a central mucin-like domain consisting of 59 imperfectly repeated sequences of EPAPTTPK. The O-glycosylation (in particular core 1 and sialylated core 1) of lubricin is suggested to be responsible for its lubricating properties (7), as the removal of these residues results in the loss of boundary lubrication. The molecule has also been suggested to play a key role in protecting the cartilage surface from excessive adsorption of proteins and cells (8).
Arthritis results in the loss of this joint surface, leading to severe pain and a restricted range of motion. The two most common arthritic diseases, osteoarthritis (OA) and rheumatoid arthritis (RA), have different mechanisms of degradation. RA is an autoimmune systemic high inflammatory disease that increases the friction between articulating cartilage surfaces, resulting in degradation of the joint (9), whereas OA is a result of mechanical stress (10). Degeneration of the cartilage can be detected from proteoglycan fragments in the SF (11, 12). Because of the limited efficacy of available treatments, particularly for OA, understanding the biological factors related to arthritis is essential.
The joints of arthritis patients, both RA and OA, have shown a down-regulation of expression and changes in glycosylation of lubricin (13). Studies using OA animal models suggest that there is a relationship between pathogenesis and the down-regulation of lubricin (9, 14, 15). This decrease in lubricin expression exacerbates the disease by accelerating the joint destruction, suggesting that certain characteristics of lubricin may be indicators of disease progression in RA and OA. Given the critical nature of lubricin glycosylation, we initiated a site-specific glycopeptide characterization of the lubricin mucin-like domain using liquid chromatography–tandem mass spectrometry with both collision-induced and electron-transfer dissociation fragmentation methods (LC-CID/ETD-MS2) after tryptic digestion of both intact and partly de-glycosylated lubricin.
Collision-induced dissociation–tandem mass spectrometry (CID-MSn) of O-linked (and N-linked) glycopeptides is capable of generating sequence information both for the attached glycan (in MS2) and for the de-glycosylated peptide (in MS3), but it lacks the site-specific information of the modified amino acids (16). This is due to extensive glycosidic bond cleavage of the precursor ion in MS2 producing B/C and Y/Z ions (Domon and Costello carbohydrate fragmentation nomenclature (17)). In addition, the identification of the modified amino acids is even more difficult for peptides containing several Ser/Thr residues because of the lack of a consensus sequence for mucin-type O-glycosylation. Electron-capture dissociation and ETD are fragmentation techniques used for the site-specific characterization of protein post-translational modifications including phosphorylation (18) and glycosylation (19). Both techniques induce cleavage of the N-Cα bonds of the peptide backbone, producing c- and z-type fragment ions, while leaving the post-translational modification unaffected.
In order to understand the biosynthesis of O-linked glycoproteins, one needs to link site localization of glycosylation to the expression of enzymes responsible for GalNAc-type (or mucin-type) O-glycosylation. This is necessary because the prediction of the site of GalNAc-type O-glycosylation is difficult. One reason for this is the large, redundant UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase (ppGalNAc T) gene family containing 20 gene-encoded isoenzymes, all possessing unique and/or overlapping substrate specificities (20, 21). These ppGalNAc Ts transfer GalNAc from the sugar nucleotide donor UDP-GalNAc to the hydroxyl groups of Ser and Thr residues in the proteins traversing the Golgi/endoplasmic reticulum. Altered protein O-glycosylation, suggested to be due to changes in the expression of distinct ppGalNAc Ts, has been reported in various disease states, including ulcerative colitis and cancer (21, 22). Thus, the connection of site-specific O-glycosylation with the responsible ppGalNAc Ts is important for understanding the functions of lubricin, as site-specific O-glycosylation has been shown to regulate the functions of proteins (23, 24) and may be involved in the pathological transformation of the joint in arthritis diseases.
Although the type of glycosylation present on lubricin has been investigated previously, the site-specific glycopeptide characterization, including the analysis of the glycan types at these locations, was investigated for the first time in this study. In order to understand the nature of glycoproteins, it is essential to not only define the protein component or the glycan characteristics, but also understand how these two essential components interact. The macro- (different site occupation) and micro-heterogeneity (different glycan structure at each site) provided a heterogeneous mixture of lubricin O-linked glycopeptides that might help to explain the extraordinary properties of lubricin and how it can function as a lubricating agent in a demanding environment.
EXPERIMENTAL PROCEDURES
Human Tissues and Cells
Synovial tissue specimens were obtained from patients with RA and OA during joint replacement surgery at Sahlgrenska University Hospital (Gothenburg, Sweden). Primary FLS cultures were established using collagenase/dispase and used in passage 5.
Isolation of Acidic Glycoproteins from SF
SF samples from RA and OA patients (n = 5) were collected during therapeutic joint aspiration at the Rheumatology Clinic of Sahlgrenska University Hospital. All patients gave informed consent, and the hospital's ethics committee approved the procedure. The arthritis patients fulfilled the American College of Rheumatology 1987 revised criteria for RA (25). The samples were clarified by centrifugation at 10,000 × g for 10 min and stored at −80 °C before use. The acidic proteins, including lubricin, were purified from RA and OA samples separately as previously described (26, 27). Protein concentration was determined with a bicinchoninic acid protein assay kit using bovine serum albumin as the standard.
Enzymatic Digestion of Lubricin
Enriched lubricin fractions (30 μg) from RA and OA patients were reduced (20 mm DTT, 70 °C for 1 h) and alkylated (50 mm iodoacetamide for 30 min at room temperature in the dark) separately. The DTT (VWR, Radnor, PA) and iodoacetamide (Sigma-Aldrich, St. Louis, MO) were removed using a spin filter with a 30-kDa cutoff (Merck Millipore, Billerica, MA). The samples were de-sialylated via incubation with 5 mU of sialidase A (Prozyme Inc., Hayward, CA) at 37 °C for 16 h. Partially de-glycosylated lubricin was generated after de-sialylation via incubation with 5 mU of O-glycanase (Prozyme) specific for removal of core 1 type O-glycan (Galβ1–3GalNAcα1-O-Ser/Thr) from glycoproteins and glycopeptides at 37 °C for 4 h in 50 mm sodium phosphate buffer, pH 6.0.
In order to investigate the accessibility of the heavily O-glycosylated mucin-like domain of lubricin to proteolytic enzymes, the enriched sample (in solution) was digested with trypsin (Promega, Madison, WI) prior to sialidase A and O-glycanase treatment. In brief, the reduced and alkylated samples were buffer exchanged with 50 mm ammonium bicarbonate (Sigma-Aldrich) and incubated with trypsin (1:30 enzyme to protein) at 37 °C for 16 h. The reaction was quenched by heating the sample in NuPAGE lithium dodecyl sulfate sample buffer, pH 8.4 (Life Technologies, Carlsbad, CA), at 95 °C for 10 min prior to loading onto a 3–8% Tris acetate NuPAGE gel (Life Technologies).
One-dimensional Isoelectric Focusing and Transfer of Proteins from IPG to Membrane
The concentration of the sample before and after de-sialylation was adjusted (0.05 to 0.1 mg/ml) by dilution with 7 m urea, 2 m thiourea (Sigma-Aldrich), and 2% CHAPS (MCLAB, San Francisco, CA). The sample was reduced with 5 mm tributylphosphine solution in 2-propanol (Sigma-Aldrich) for 2 h and alkylated with 15 mm iodoacetamide for 1 h at room temperature. Bromphenol blue tracking dye solution (5 μl of 0.2 mg/ml solution; Sigma-Aldrich) was added prior to the samples' addition to the rehydration/equilibration tray. An IPG strip of pH 3–10 (Bio-Rad, Hercules, CA) was placed with the gel facing the sample. The strip was covered with paraffin oil (to prevent evaporation), and the gel was rehydrated for 16 h at room temperature. The proteins in the rehydrated IPG strip were focused using a Protean IEF Cell (Bio-Rad). A linear gradient in voltage was set as follows: 100 V in 5 min, then a gradient up to 10,000 V in 8 h. Once 10,000 V was reached, the focusing was continued for an additional 8 h.
The proteins from the IPG gel were transferred to PVDF membrane via passive diffusion as previously described (28, 29), with minor changes. Briefly, the IPG strips (gel facing upward), after being washed with water and soaked in 50 mm Tris buffer (Sigma-Aldrich) for 10 min, were placed above two filter papers soaked in the same 50 mm Tris buffer. The PVDF membrane (Immobilon P, Millipore), after being soaked in methanol for 2 min and 50 mm Tris buffer for 10 min, was placed above the IPG gel, which was covered with two additional Tris-wetted filter papers. The sandwich was covered in plastic foil and compressed by a weight of about 4 kg (to ensure good contact), and the proteins were blotted for 24 h at room temperature. After transfer, the membrane was washed with water for subsequent immunodetection as described in the section “SDS-PAGE Gel Separation, Western Blotting, and Staining.”
RNA Extraction, Reverse Transcription, and Real-time Quantitative PCR
RNA from primary human FLSs of RA (n = 2) and OA (n = 2) patients was extracted separately using an RNeasy Mini Kit (Qiagen, Valencia, CA), and the concentration was determined with a NanoDrop (Thermo Scientific, Wilmington, DE) at 260 and 280 nm. Taq-man-based real-time PCR was performed on cDNA derived from RNA using High Capacity cDNA Reverse Transcription Kits as recommended by the supplier (Applied Biosystems, CA). Profiling of all 20 GALNTs was performed as described previously using β-actin as an internal normalization standard (21).
SDS-PAGE Gel Separation, Western Blotting, and Staining
The samples before and after enzymatic treatment (trypsin digestion and partial de-glycosylation) were separated using 3–8% Tris acetate NuPAGE gels (Invitrogen). The separated proteins were transferred to PVDF membrane using a semi-dry blotter (Bio-Rad) and probed with lubricin-specific antibody (mouse anti-lubricin mAb13, Pfizer Research, Cambridge, MA) and carbohydrate-specific biotinylated lectins including peanut agglutinin (PNA) (Vector Laboratories, Burlingame, CA) and wheat germ agglutinin (WGA) (Vector Laboratories) as previously described (12). For peptide and glycopeptide identification, the SDS-NuPAGE gels were stained with Coomassie Brilliant Blue (Thermo Scientific, Waltham, MA) for 30 min and de-stained with 5% aqueous acetic acid.
Cotton Wool Glycopeptide Enrichment
The major Coomassie Blue–stained protein bands (3–8% Tris acetate NuPAGE gel) before and after partial de-glycosylation (sialidase and O-glycanase treatment) were excised and subjected to in-gel trypsin digestion (30) for subsequent LC-MS2 analysis. The generated glycopeptides were also enriched offline using cotton wool hydrophilic interaction liquid chromatography (HILIC) solid phase extraction microtips packed in house (31). In brief, a microtip (10 μl) packed with cotton wool was washed five times with LC-MS water and conditioned seven times with 83% acetonitrile (water and acetonitrile were from Merck Millipore). The samples, solubilized in 83% acetonitrile, were applied by pipetting up and down at least 25 times. Peptides were eluted with 83% acetonitrile containing 0.1% TFA (Sigma-Aldrich), and glycopeptides were eluted with water. The eluted fractions were concentrated in a vacuum centrifuge for subsequent LC-MS2 analysis.
LC-MS2 Analysis of Lubricin
C18 pre- and analytical columns packed in-house (3-μm particles; Dalco Chromtech, Stockholm, Sweden) were used, with inner diameters of 75 μm and lengths of 4 cm and 20 cm, respectively. Mobile phases consisted of aqueous 0.2% formic acid (Sigma-Aldrich) for solvent A and 0.2% formic acid in 80% acetonitrile for solvent B. A linear gradient was set as follows: 0% B for 5 min, then a gradient up to 35% B in 70 min and to 80% B in 5 min. A 20-min wash at 80% B was used to keep the column sensitive and prevent carryover, and a 25-min equilibration with 100% A completed the gradient. The column was attached to an Agilent 1100 series HPLC (Agilent Technologies, Santa Clara, CA). An LTQ-Orbitrap XL (Thermo Scientific) was used in positive ion mode for MS and MS2 analysis. The spray voltage was set to 2 kV, and the ion transfer tube was set at 200 °C. The full scans were acquired in a Fourier transform MS mass analyzer that covered an m/z range of 400–2000. The MS2 analysis was performed under data-dependent mode to fragment the top five precursors using either CID or ETD. For CID, a normalized collision energy of −35 eV, an isolation width of m/z 1.0, an activation Q value of 0.250, and a time of 30 ms were used. In the case of ETD, an isolation width of m/z 2.0 and activation times of 100 and 150 ms were used.
Data Analysis
The raw files containing centroid MS2 spectra were searched against UniProt (AC# Q92954, released February 19, 2014), NCBI (released August 13, 2013; 251,429 human entries), and Swiss-Prot (version 2013 05; 20,257 human entries) human protein databases using the in-house version of the Mascot software (v.2.2.04, Matrix Science Inc., Boston, MA). The raw file was also converted to mzXML and mgf formats by Software from Seattle Proteome Center (ReAdW 4.3.1), and searches were carried out using the online GPM (32) and Byonic software (Protein Matrics, San Carlos, CA) (33). The search parameters for GPM software were set as follows: peptide tolerance, 4 ppm; MS2 tolerance, 0.5 Da; enzyme, trypsin; one missed cleavage allowed; fixed carbamidomethyl modification of cysteines; and variable modifications of HexNAc (203.0794 Da) of Ser. For Mascot and Byonic software, variable modifications of HexNAc (203.0794), HexHexNAc (365.1323), Hex2HexNAc2 (730.2644), and NeuAcHexHexNAc (656.2278) of Ser and Thr were also included in the search parameters used for GPM. For positive protein identification, the minimum criteria were three unique peptides with scores above the significance threshold (p < 0.05). In the case of glycopeptides, the criteria were MS2 spectra that sequenced 75% of the peptide including the identification of the modified Ser/Thr residue (glycan location) for positive identification. However, the majority of the glycopeptide identifications were based on manual interpretation because of the lack of sufficient glycopeptide information obtained using the software. The software-identified glycopeptides were all manually validated.
The interpretation (manual and software annotation) of the CID-MS2 glycopeptide spectra generated glycan sequence information. The presence of oxonium ions (m/z 204 (HexNAc), 292 (NeuAc), 366 (HexHexNAc), etc.) in the CID-MS2 spectra was used to validate glycopeptides identified by the software. In order to obtain peptide information, the human synovial lubricin (UniProt Q92954) was theoretically trypsin digested using from the Swiss Institute of Bioinformatics. This allowed the comparison of the m/z of the de-glycosylated peptide in MS2 spectra with the theoretically generated peptide list for peptide identification. The LC-MS2 analyses for RA and OA SF lubricin were carried out separately, and the final data were combined, as there was no major difference observed in the glycopeptide analyses of RA and OA SF lubricin.
pI Modeling of Lubricin
The isoelectric point (pI) dependence for sialic acid of full length and STP-rich regions of lubricin was simulated as described by Henriksson et al. (34, 35). For pI simulation, the amino acid composition, pKa values for the amino acid side chains and for the N- (pKa = 8) and C-terminal (pKa = 3.1) groups, and presence of charged sialic acid groups were taken into account. The pKa values used were Lys 10, Arg 12, His 6, Glu and Asp 4.1, Tyr 10.4, and sialic acid 2.6 (36).
RESULTS
Accessibility and Characterization of the Glycosylated Region of Lubricin
Human synovial lubricin was purified from SF of RA and OA patient samples (n = 5). After purification, lubricin was detected as a major band in both RA and OA samples through the use of lubricin-specific antibody with an apparent molecular mass of >200 kDa after SDS-PAGE (Fig. 1A, lane 1). The antibody also detected an additional, faint high-mass band that was due to lubricin complexes (37). All bands were previously confirmed to contain lubricin when subjected to in-gel trypsin digestion and subsequent LC-MS2 analysis (38).
Fig. 1.
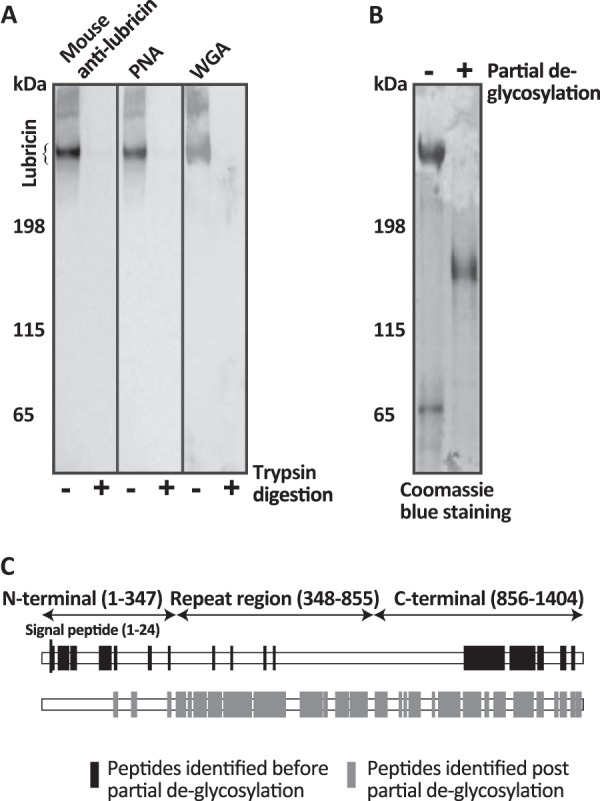
Accessibility and characterization of the glycosylated region of lubricin. A, the enriched synovial lubricin samples, before and after trypsin digestion, were separated on a 3–8% Tris acetate gel, blotted onto PVDF membrane, and then probed with lubricin-specific antibody (mouse anti-lubricin) and carbohydrate-specific biotinylated lectins PNA, specific for core 1 (Galβ1–3GalNAc) O-glycan, and WGA, specific for sialic acid and terminal GlcNAc. B, SDS-PAGE (3–8% Tris acetate gel) of the acidic glycoprotein fractions of the SF before (−) and after (+) partial de-glycosylation stained with Coomassie Brilliant Blue. C, SF lubricin was in-solution digested, and non-modified peptides were identified via mass spectrometry for protein coverage determination (black). The low protein coverage (in particular the mucin domain) suggests that the mucin domain is extensively glycosylated. Some of the core 1 structures were removed by partial de-glycosylation, and the previously glycosylated peptides were identified for protein coverage (gray). The results suggest that lubricin contains an extended STP-rich region relative to the mucin domain previously defined by UniProt.
In order to indicate the localization of the glycosylated mucin-like domain within lubricin, the samples (reduced and alkylated) were subjected to in-solution trypsin digestion and separated on SDS-PAGE gels prior to subsequent Western blotting using lubricin-specific antibody and biotinylated lectins (PNA, Galβ1–3GalNAcα1-O-Ser/Thr and WGA, sialic acid, and terminal GlcNAc). The effectiveness of the trypsin digestion was shown by the fact that the generated peptides were too small to be detected on the gel (Fig. 1). This showed that both the less glycosylated N- and C-terminals and the mucin-like domain were accessible for digestion (Fig. 1A, PNA and WGA). The lubricin mucin domain is different from traditional indigestible mucin domains, allowing Lys residues (trypsin cleavage site) in the imperfect repeat EPAPTTPK to be protease accessible. This also suggested that the glycans of lubricin were smaller and/or less frequent than other mucins, allowing the trypsin site to be accessible despite the surrounding glycosylation. The positive lectin (PNA and WGA) binding of the reduced and alkylated but not trypsin digested samples suggested that SF lubricin predominantly contained short core 1 and sialylated core 1 structures (Fig. 1A). This was further verified by partial de-glycosylation using sialidase and O-glycanase to remove sialylated and unsialylated core 1 structures. This treatment resulted in a substantial decrease in size (Fig. 1B), with an apparent mass of >155 kDa, close to the predicted size of apolubricin (151 kDa).
The dominating lubricin band seen in SDS-PAGE was subjected to in-gel trypsin digestion, and unmodified lubricin peptides were identified via LC-MS2. The identified peptides were predominately from the N- and C-terminal regions. Even though the mucin-like domain was indicated to be less extensively glycosylated, only a few non-modified peptides from the mucin domain could be identified (Fig. 1C, black). In a stretch of 507 amino acids in the central region (aa 348–855) there were only two (one unique) peptides (KPAPTTPK) (3% coverage) identified. After partial de-glycosylation, a total of 99 (13 unique) unmodified peptides from this lubricin mucin-like domain were identified via LC-MS2 (Fig. 1C, gray), providing a coverage of 84% of the mucin-like domain (aa 348–855) rich in Thr (29.5%), Pro (30.5%), and, to a lesser extent, Ser (2.4%). These data suggested that this entire region was highly glycosylated with small glycans, as even though tryptic peptides could be created, unmodified peptides could not be identified. The current domain model of lubricin consists of less glycosylated N and C terminals separated by a glycosylated mucin-like domain (aa 348–855) region of a tandemly repeated amino acid sequence. However, the data shown here indicate that lubricin consists of an extended glycosylated STP-rich region (aa 232–1056) (Fig. 4A) larger than the mucin-like domain previously defined by UniProt. The molecular mass of glycosylated lubricin is estimated to be ∼350 kDa, and that of apomucin to be >151 kDa, indicating glycosylation constitutes 57% of the total protein mass. Given that the estimated average mass of an oligosaccharide on lubricin is 600 to 1000 Da (38), it is expected that lubricin holds 200 to 300 oligosaccharide chains.
Fig. 4.
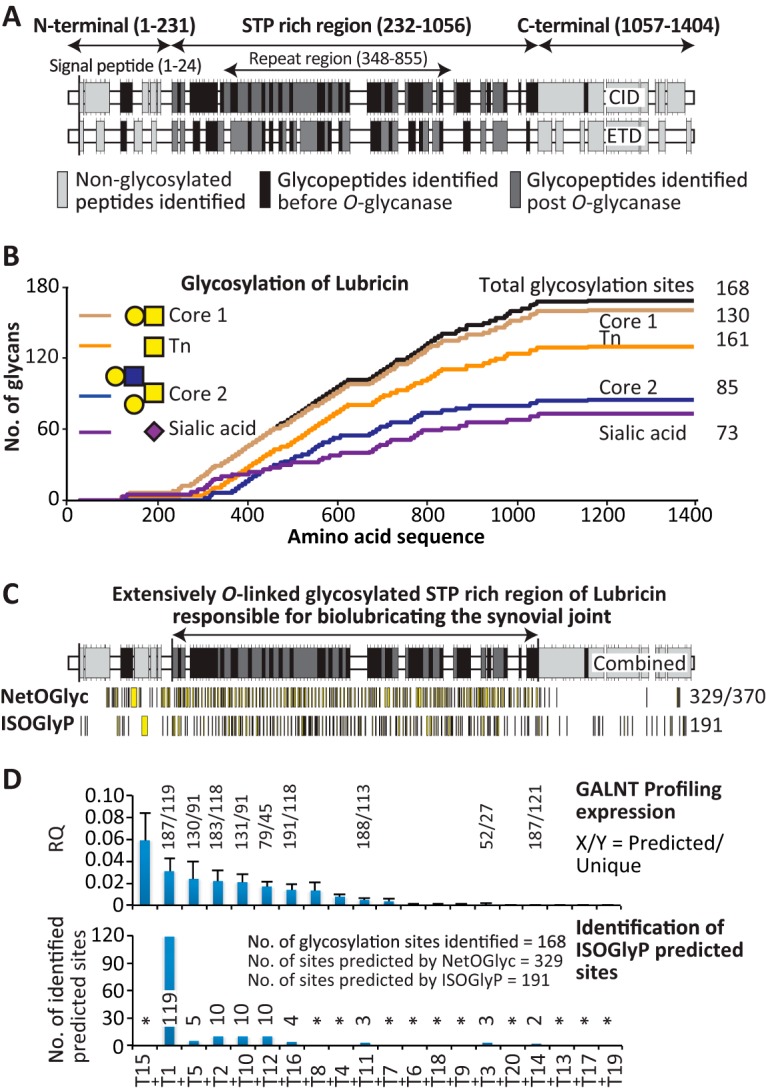
The O-glycosylation map of lubricin. A, the glycosylated peptides identified in the STP-rich region and non-glycosylated peptides (light gray) in the end domains of the lubricin sequence before (black) and after (dark gray) partial de-glycosylation using CID and ETD fragmentation. B, graphic representation of all of the identified glycans and their positions in the protein sequence. The figure includes the identified glycopeptide containing GalNAcα1- (dark orange, 130), core 1 Galβ1–3GalNAcα1- (chocolate, 161), core 2 Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1- (blue, 85), and sialylated core 1 and 2 (pink, 73) glycans with their positions in the protein sequence. The graph also includes the total number of identified glycosylation sites (168 sites) in the protein sequence (black). The total number of sites is based on the identified peptides and the fact that two threonines in the imperfect tandem repeat variants (e.g. EPAPTTPK, SAPTTPK, EPAPTTTK) can be glycosylated. It indicates a high number of glycosylations in the STP-rich region, whereas the N- and C-terminal regions are scarcely glycosylated. C, the combined CID and ETD identified glycopeptides in the STP-rich region (black and dark gray) and non-glycosylated peptides (light gray) in the end domains of lubricin before (black) and after partial de-glycosylation (dark gray). The software used (NetOGlyc4.0 and ISOGlyP) predicted sites of lubricin O-glycosylated Ser/Thr in the protein sequence (yellow) and the total number of potential Ser/Thr sites (370). The results indicate that the gene-based ISOGlyP software predicted a similar number of glycosylation sites (191 sites) as identified by the data presented in this report (168 sites). D, the GALNT profiling expression analysis of the 20 known human GALNT genes (top) in primary FLSs and the ISOGlyP-predicted sites by the available GALNT genes identified by the data presented in this report (bottom). The genes were arranged in descending order of their expression. RQ, relative quantification. Asterisk denotes genes not included in the ISOGlyP software.
Identification of Lubricin Mucin Glycopeptides Using CID and ETD
In order to analyze the STP-rich region and identify the number of glycosylation sites on synovial lubricin, we adopted a combined approach using both CID- and ETD-MS2 to identify the types of glycans attached, as well as their position. Tryptic glycopeptides of both RA and OA samples were generated both before and after partial de-glycosylation (Fig. 1B) for subsequent mass spectrometric analysis. The LC-CID/ETD-MS2 approach successfully identified 185 O-glycosylated peptides. They are presented, together with the identified O-linked glycans, glycan attachment sites, and annotation method (software or manual) of each individual glycopeptide, in Table I (and in supplemental Table S1). Predominantly core 1 and monosialylated core 1 (NeuAcα2–3Galβ1–3GalNAcα1-) O-linked glycopeptides were identified. A small proportion of disialylated core 1 (NeuAcα2–3Galβ1–3(NeuAcα2–6)GalNAcα1-) peptides were also detected. The peptides (KPAPTTPK) identified as non-glycosylated in the previously defined mucin-like domain and in the STP-rich region (VLAKPTPK and KPAPTTPK) were also shown to be glycosylated. The glycosylation of the threonine in the KPAPTTPK repeat was also shown, as indicated by the data presented in this report (supplemental Table S1). In addition to core 1, a small proportion of core 2 (Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1-) and monosialylated core 2 (e.g. NeuAcα2–3(Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1-)) glycopeptides were also identified. These findings are consistent with the O-linked glycans identified in our and others' previous studies (7, 26, 38).
Table I. A list of the CID- and ETD-identified glycopeptides with their peptide sequence, types of glycans, position in the peptide sequence, and annotation method used.
| Lubricin peptide | Glycan compositiona | Assignmentb | Annotation | Lubricin peptide | Glycan composition | Assignment | Annotationc |
|---|---|---|---|---|---|---|---|
| K.STTK.R | 2, 3 | CID | Manual | K.KAPPSGASQTIK.S | 3, 4 | Both | Both |
| K.STTKR.S | 3 | CID | Manual | K.APPPSGASQTIK.S | 3, 4 | Both | Manual |
| R.SPKPPNKK.K | 3 | CID | Manual | K.VTTPDTSTTQHNK.V | 3 | Both | Manual |
| K.VSTSPK.I | 1, 3 | CID | Both | K.ITTAKPINPR.P | 3 | ETD | Both |
| K.ETTVETK.E | 3 | CID | Manual | K.ETSLTVNKETTVETK.E | 4 | ETD | Both |
| K.ETTTTNK.Q | 3, 4 | CID | Both | K.ETTVETKETTTTNK.Q | 3 | ETD | Both |
| K.TTSAK.E | 3, 6 | CID | Manual | K.ETTTTNKQTSTDGK.E | 3 | CID | Manual |
| K.DLAPTSK.V | 1, 2, 3, 5, 8, 10, 11 | CID | Manual | K.TTSAKETQSIEK.T | 1, 3, 4 | Both | Both |
| K.VLAKPTPK.A | 1, 3, 4 | Both | Both | K.ETQSIEKTSAK.D | 3, 4 | Both | Both |
| K.GPALTTPK.E | 1, 3, 4 | CID | Manual | K.TSAKDLAPTSK.V | 1, 3, 4, 5, 10 | Both | Both |
| K.EPTPTTPK.E | 1, 3 | CID | Manual | K.EPTPTTPKEPASTTPK.E | 3 | CID | Manual |
| K.EPASTTPK.E | 1, 3 | CID | Manual | K.EPTPTTIKSAPTTPK.E | 3, 4 | CID | Manual |
| K.EPTPTTIK.S | 1, 3, 8 | CID | Manual | K.SAPTTPKEPAPTTTK.S | 1, 3 | ETD | Both |
| K.EPAPTAPK.K | 3, 8, 10 | CID | Manual | K.EPAPTTTKEPAPTTPK.E | 3 | Both | Both |
| K.EPSPTTPK.E | 1, 3 | CID | Manual | K.EPAPTTTKEPAPTTTK.S | 3 | Both | Manual |
| K.SAPTTTK.E | 1 | CID | Manual | K.SAPTTPKEPAPTTPK.K | 1, 3 | ETD | Both |
| K.EPSPTTTK.E | 6, 8 | ETD | Both | K.EPAPTTPKEPTPTTPK.E | 3 | CID | Manual |
| K.ETAPTTPKK.L | 3 | Both | Both | K.EPTPTTPKEPAPTTK.E | 8 | CID | Manual |
| K.KLTPTTPEK.L | 1, 3 | Both | Manual | K.EPAPTTPKEPAPTAPK.K | 3 | Both | Both |
| K.LTPTTPEK.L | 3 | Both | Both | K.EPAPTTTKEPSPTTPK.E | 3 | CID | Manual |
| K.AAAPNTPK.E | 3, 4, 8, 10 | CID | Manual | K.SAPTTTKEPAPTTTK.S | 1, 3 | ETD | Both |
| K.GTAPTTLK.E | 3, 4, 8, 10 | CID | Manual | K.SAPTTPKEPSPTTTK.E | 1, 6 | ETD | Both |
| K.ELAPTTTK.E | 1, 3 | Both | Manual | a.EPAPTTPKETAPTTPK.a | 3 | ETD | Both |
| K.GTAPTTPK.E | 1, 3 | CID | Manual | K.GTAPTTLKEPAPTTPK.K | 3 | ETD | Both |
| K.GPTST TSDK.P | 1, 3 | ETD | Both | K.EPAPTTPKKPAPK.E | 3 | ETD | Manual |
| K.EPTTIHK.S | 8, 10 | CID | Manual | K.GPTSTTSDKPAPTTPK.E | 1 | ETD | Both |
| K.ALENSPK.E | 3, 4 | CID | Manual | K.ETAPTTPKEPAPTTPK.K | 3 | ETD | Both |
| K.EPGVPTTK.T | 1, 3, 4, 10 | Both | Manual | K.SPDESTPELSAEPTPK.A | 3, 4 | CID | Manual |
| K.ETATTTEK.T | 1, 3 | CID | Manual | K.ALENSPKEPGVPTTK.T | 1, 3 | Both | Manual |
| K.TTTLAPK.V | 1, 2, 3 | CID | Manual | R.TTPETTTAAPK.M | 3 | Both | Manual |
| K.VTTTK.K | 3 | CID | Manual | K.ITTLKTTTLAPK.V | 1, 3 | ETD | Manual |
| R.ATNSK.A | 3 | CID | Both | K.ITATTTQVTSTTTQDTTPFK.I | 1, 3 | ETD | Both |
| K.KPTSTK.K | 1, 3, 4 | CID | Manual | K.KPTSTKKPK.T | 1, 3, 4, 5, 8, 10 | CID | Manual |
| K.TMPR.V | 3 | CID | Manual | R.KPKTTPTPR.K | 1, 3, 8 | CID | Both |
| K.TTPTPR.K | 1, 3, 4 | CID | Manual | R.NGTLVAFR.G | 1, 6, 10 | CID | Manual |
| aa.EPAPTTPK.aa | 1, 3, 4, 8, 10 | CID | Manual | K.ETAPTTPK.K | 1, 3 | Both | Manual |
| aa.EPAPTTTK.aa | 1, 3, 8 | CID | Manual | K.EPAPTTTKSAPTTPK.E | 3 | CID | Manual |
| K.KPAPTTPK.E | 1, 3 | CID | Both | K.SAPTTPK.E | 1, 3, 8 | CID | Manual |
Notes: The line under an amino acid in a peptide sequence represents the position of glycosylation. The data showed glycosylation of threonine in the EPAPTTPK and KPAPTTPK repeats and two of the three sites in SAPTTPK and EPAPTTTK.
a 1, GalNAcα1-; 2, NeuAcα2–6GalNAcα1-; 3, Galβ1–3GalNAcα1-; 4, NeuAcα2–3Galβ1–3GalNAcα1-/NeuAcα2–6(Galβ1–3)GalNAcα1-; 5, NeuAcα2–3Galβ1–3(NeuAα2–6)GalNAcα1-; 6, GlcNAc-GalNAcα1-; 7, NeuAcα2–6(GlcNAcβ1–3)GalNAcα1-; 8, Galβ1–3(GlcNAcβ1–6) GalNAcα1-; 9, NeuAcα2–3Galβ1–3(GlcNAcβ1–6)GalNAcα1-; 10, Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1-; 11, NeuAcα2–3Galβ1–3 (Galβ1–4GlcNAcβ1–6)GalNAcα1-. Sequence assumed based on identified sequences on lubricin (38).
b Assignment based on MS/MS using CID, ETD, or both types of fragmentation.
c Glycopeptide MS/MS spectra identified by means of manual annotation or both manual and software-assisted annotation.
The CID-MS2 approach effectively identified the nature of the glycans attached to lubricin. CID-MS2 spectra of four different O-linked isoforms of the same amino acid sequence (EPAPTTPK) located in the STP-rich region are presented in Figs. 2A–2D. The spectrum of the [M+2H]2+ ions at m/z 603.3 (Galβ1–3GalNAcα1-O-[EPAPTTPK]) resulted in fragmentation of the glycan component into y-type ions (Domon and Costello nomenclature) and b-type ions corresponding to [HexNAc+H]+ and [HexHexNAc+H]+ oxonium ions at m/z 204 and 366, respectively (Fig. 2A). This corresponded to a glycopeptide with a core 1 glycan at one of the Thr residues. The spectrum showed the neutral loss of Hex residue (m/z 1043.6), which was followed by a loss of HexNAc residue (m/z 840.3), establishing the glycan sequence as Hex-HexNAc. However, the CID-MS2 spectrum did not show which of the Thr residues was glycosylated. The sialylated version of this glycopeptide was also identified (Fig. 2B). The presence of an oxonium ion at m/z 292 (sialic acid, NeuAc) in the CID-MS2 spectrum of the [M+2H]2+ ions at m/z 748.8 (NeuAc-Hex-HexNAc-O-[EPAPTTPK]) showed that this glycopeptide can also be sialylated (Fig. 2B). The loss of a NeuAc residue (m/z 1205.3) followed by the loss of Hex (m/z 1043.3) and finally the loss of HexNAc (m/z 840.5) indicated a NeuAc-Hex-HexNAc- structure attached to a Thr residue in the peptide sequence (Fig. 2B). This, together with previous O-glycan analysis, suggested that the attached structure was NeuAcα2–3Galβ1–3GalNAcα1-O-Thr.
Fig. 2.
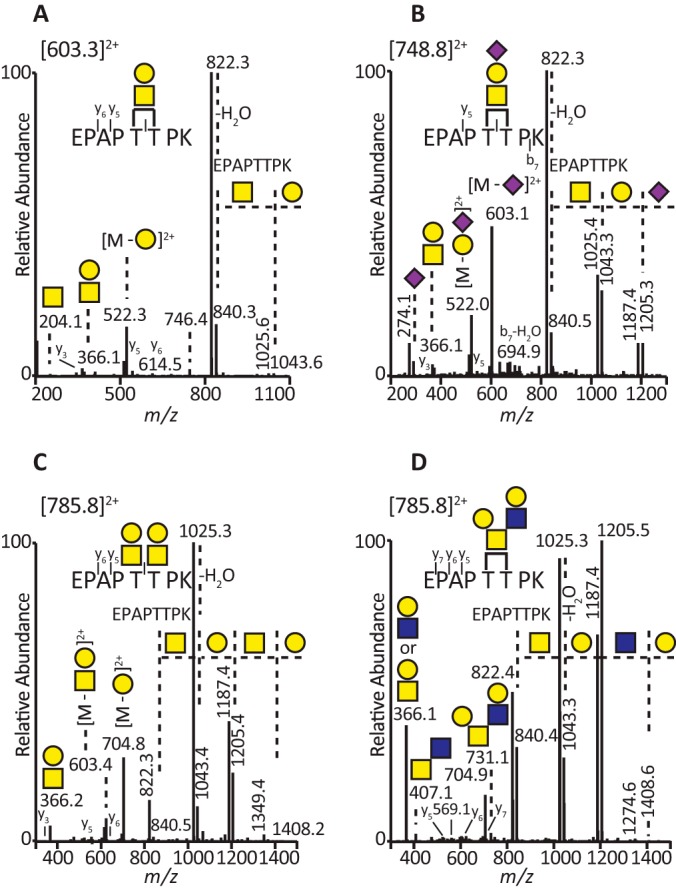
Identification of lubricin mucin glycopeptides using CID. The figure shows the different glycoforms of the same tryptic peptide within the STP-rich region. A, CID-MS2 spectrum of the tandem repeat (EPAPTTPK) containing core 1 glycan (Galβ1–3GalNAcα1-) of the [M+2H]2+ ions at m/z 603.3. B, CID-MS2 spectrum of the same tandem repeat containing sialyl T antigen (NeuAcα2–3Galβ1–3GalNAcα1-) of the [M+2H]2+ ions at m/z 748.8. C, CID-MS2 spectrum of the same tandem repeat (EPAPTTPK) containing two separate core 1 glycans (Galβ1–3GalNAcα1-) on two adjacent threonines of the [M+2H]2+ ions at m/z 785.8. D, CID-MS2 spectrum of the same tandem repeat containing core 2 glycan Galβ1–3(Galβ1–4GlcNAcβ1–6)GalNAcα1- of the [M+2H]2+ ions at m/z 785.8. This shows that the same peptide (A, B, and D) can be differently glycosylated, indicating the complexity associated with lubricin O-glycosylation, and that CID was able to provide information about the nature of glycans and peptides. In the figure, yellow squares and circles represent N-acetylgalactosamine and galactose, respectively, and blue squares and pink diamonds represent N-acetylglucosamine and N-acetylneuramininc acid (sialic acid), respectively.
The CID-MS2 spectra allowed the identification of isomeric glycopeptides, showing differences in the number of glycosylation sites and glycan sequences. The presence of diagnostic ions at m/z 407 [HexNAc2+H]+ and 569 [Hex(HexNAc)HexNAc+H]+ (Fig. 2D) were used to differentiate a single substituted core 2 O-glycan from a doubly substituted core 1 O-glycan on both Thr residues in EPAPTTPK repeats. These results indicated that there were heavily glycosylated regions of the STP-rich region, such as the doubly glycosylated EPAPTTPK repeat shown in Fig. 2C. The STP region also displayed more complex O-glycosylation such as the core 2 structure shown in Fig. 2D.
ETD-MS2 analysis was used for the identification of glycosylation sites within the STP-rich region, particularly for the identification of non-consensus repeats. Generally, the ETD-MS2 highly charged glycopeptide precursor ions fragmented efficiently, allowing the site of glycosylation to be further narrowed down, in most cases to single amino acid residues (Table I and supplemental Table S1). All ETD-MS2 spectra were manually annotated for verification of the location of uniquely modified Ser/Thr residues in order to remove all possible ambiguity.
The ETD-MS2 spectrum of the [M+3H]3+ ions at m/z 673.3 glycopeptide (K972ITTLKTTTLAPK985V) allowed the identification of the glycan-modified Thr residues within the peptide sequence. The ETD-MS2 spectrum, [M+3H]3+ ions at m/z 673.3 (Fig. 3A), displayed the z10+1-, z9+1-, and c7-ions at m/z 1788.6, 1687.8, and 1506.6, respectively, indicating that Thr974, Thr975, and Thr980 were unmodified. However, the c6-ion at m/z 1040.4 was observed with the addition of a Hex-HexNAc residue (365 Da), indicating that the Thr979 was modified with an unsialylated core 1 structure. The c5-ion at m/z 574.4 showed that the adjacent Thr978 was modified with a second Hex-HexNAc residue (Fig. 3A). The z7-ion at m/z 1444.7 denoted that the two core 1 glycans (Hex-HexNAc) were still intact and attached to the peptide. However, the z5+1-ion at m/z 513.2 confirmed that both Thr978 and Thr979 were core 1 modified, as it was the loss of two threonines and two Hex-HexNAc- units. Lubricin displayed macroheterogeneity as shown by the identification of an isomeric glycopeptide (Fig. 3B). The ETD-MS2 spectrum of the [M+3H]3+ ions at m/z 673.3 indicated the same peptide sequence (K972ITTLKTTTLAPK985V) with two core 1 glycans on separate threonine residues (Fig. 3B). The c7-ion at m/z 1141.6 indicated the loss of the Hex-HexNAc Thr980. The c6-ion at m/z 1040.6 was the loss of unmodified Thr979. The Thr978 was modified with the second Hex-HexNAc residue identified by the c5-ion at m/z 574.5, the loss of Thr978 with a HexHexNAc. The c5-ion at m/z 574.5, a peptide with two core 1 glycans, indicated that Thr974 and Thr975 were unmodified. The ETD-MS2 spectra of the isomeric [M+3H]3+ ion at m/z 673.3 (Figs. 3A and 3B) revealed the site occupancy within the glycopeptide K972ITTLKTTTLAPK985V of SF lubricin.
Fig. 3.
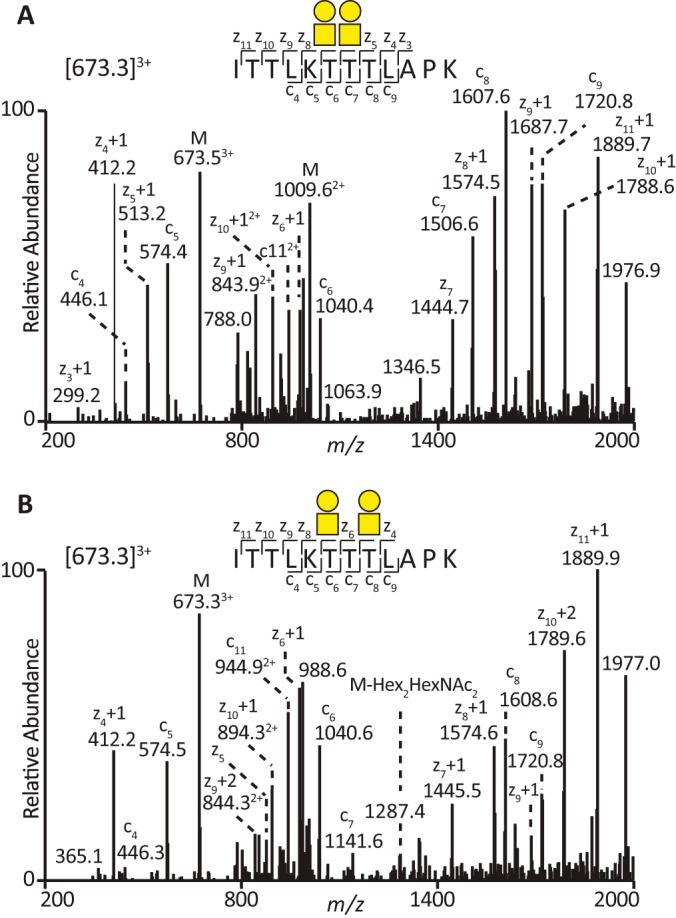
Identification of glycosylation sites using electron transfer dissociation (ETD). A, ETD-MS2 spectrum of the [M+3H]3+ ions at m/z 673.3 indicates a tryptic peptide (ITTLKTTTLAPK) with two separate core 1 glycans (Galβ1–3GalNAcα1-) on two adjacent threonine residues. B, ETD-MS2 spectrum of the [M+3H]3+ ions at m/z 673.3 reveals the same tryptic peptide (ITTLKTTTLAPK) with two separate core 1 glycans (Galβ1–3GalNAcα1-) on two separate threonine residues, indicating that lubricin glycosylation displays macroheterogeneity within the same peptide sequence. M denotes the glycopeptide parent mass.
Overall, the dual fragmentation approach identified 185 lubricin glycopeptides, primarily from the STP-rich region. This allowed us to characterize 168 glycosylation sites, predominantly in the STP-rich region (aa 232–1056), covering 71% of the Ser/Thr in this STP-rich region. This, together with the identified non-glycosylated Ser/Thr (mainly in the N and C termini), covered 72% of the Ser/Thr (266 out of 370 Ser/Thr were identified) in the entire protein sequence. The Ser/Thr coverage provided one of the most extensive O-glycosylation maps of a mucin-type protein (supplemental Fig. S2 and supplemental Table S1). The identified glycosylated and non-glycosylated Ser/Thr (both in the N and C termini and in the STP-rich region) are shown in supplemental Fig. S2. In addition to the Ser/Thr coverage, the mass spectrometric approach covered 82% of the entire protein sequence, and the coverages for the N terminus (aa 1–231), STP-rich region (aa 232–1056), and C terminus (aa 1057–1404) were 79%, 80%, and 85%, respectively.
The O-glycosylation Map of Lubricin
The identified O-glycopeptides, glycan composition, fragmentation technique, annotation technique (software/manual), and glycosylation sites are listed in Table I (and in supplemental Table S1). Regions of glycosylation sites identified via CID and ETD both before and after partial de-glycosylation are shown in Fig. 4A. The identified O-glycopeptides characterized 168 glycosylation sites. This indicated that 63% of the identified Ser/Thr residues (266 Ser/Thr, both glycosylated and non-glycosylated, were identified) in lubricin were O-glycan modified (Fig. 4B) with a bias toward Thr glycosylation due to the high Thr content (supplemental Fig. S2). An extended STP-rich region was also apparent spanning amino acids 232–1056, larger than the previously defined mucin-like domain suggested in UniProt. The entire lubricin molecule has in total 370 potential O-glycosylation sites. Of these, 35% of Ser/Thr (130 glycans) were HexNAc (GalNAc) modified with a distribution throughout the extensively glycosylated STP-rich region (Fig. 4B). Core 1 modified (43%, 161 glycans) and larger core 2 modified (23%, 85 glycans) glycopeptides were also identified (Fig. 4B). The high-abundant core 1 modified Ser/Thr were uniformly distributed throughout the STP-rich region, whereas the low-abundant core 2 modified residues were limited to the previously defined mucin-like domain (aa 348–856) (Fig. 4B). This was likely because the accumulative nature of glycopeptides from the repeat region made them easier to detect, and it might not necessarily be a reflection of core 2 enrichment in the repeat area. Outside the STP region, only a single GalNAc-modified Thr (1159NGTLVAFR1166) was identified. This residue, in the hemopexin 1 domain (1148–1191), was also shown to be glycosylated with core 2 structures (Figs. 4A and 4B; supplemental Fig. S1; supplemental Table S2).
The identification of core 2 glycopeptides confirmed the presence of core 2 structures on lubricin. The identification of core 2 together with single HexNAc (GalNAc)-modified glycopeptides suggested that lubricin glycosylation might also have other roles in addition to lubrication. The majority of the GalNAc extended into either core 1 or core 2 sialylated structures (73 glycans; Fig. 4B). Both mono- and disialylated core 1 and core 2 modified Ser/Thr were identified (Table I and supplemental Table S1), but monosialylation was more prevalent, which is consistent with previously identified synovial lubricin O-glycans (38).
Comparison of the Lubricin O-glycomap with Predicted O-glycosylation Sites
The glycosylations identified here were compared with currently available O-glycosylation prediction tools. The glycosylation prediction tool NetOGlyc4.0 (39) is based on in vivo identified O-glycosylation sites and predicted almost double the number of O-glycosylation sites identified here (almost 90% of all Ser/Thr residues) (Fig. 4C). In addition to the STP-rich region, most of the Ser/Thr residues in the N-terminal region were predicted to be O-glycosylated (Fig. 4C). An individual ppGalNAc T in vitro enzymatic specificity–based prediction tool, ISOGlyP (40), predicted that 51% (191) of the sites were glycosylated (Figs. 4C and 4D) utilizing all ppGalNAc T specificities available in the tool. This prediction is closer to the 168 detected in this study (Fig. 4B). ISOGlyP predicted that a high proportion of the Thr in the STP-rich region would be glycosylated, as was also shown by the MS analysis (Table I, supplemental Fig. S2, supplemental Table S1). However, no single ppGalNAc T of those included in the tool was able to glycosylate all the sites identified via MS. The ubiquitously expressed GALNT1 and -2 were suggested to glycosylate only 76% of the total sites found via MS (Fig. 4D). In total, 166 out of 191 (87%) sites predicted by the nine available genes in the software were identified through MS analysis (Fig. 4D). This suggested that a GALNT not included in ISOGlyP might be responsible for at least some of the glycosylation on synovial lubricin (Fig. 4D and supplemental Table S2).
Investigating the Expression of Glycosyltransferase Genes and Glycosylation
An alternate method for understanding glycosylation is to investigate the Golgi apparatus glycosyltransferases responsible for glycosylating lubricin. Because the ISOGlyP results suggested less common transferases were necessary for lubricin glycosylation, the expression of ppGalNAc Ts from human primary FLSs isolated from RA and OA patients was investigated. These types of cell lines are known to produce lubricin (1). The relative quantifications of all transcripts were normalized against β-actin expression. The average (n = 4, except for GALNT8, where n = 3) expression of the GALNT genes is arranged in descending order of expression in Fig. 4D. High mRNA expression was observed for GALNT1, -2, -5, and 15, and lower expression was noted for the GALNT8, -10, -12, and 16 genes. The high expression of GALNT1 and -2 was in agreement with the suggestion that these two genes are ubiquitously expressed. In contrast, GALNT5 and GALNT15 have been shown to display restricted expression profiles, suggesting these isoforms serve unique functions in the tissue where they are expressed (21). The high expression of GALNT5 in FLSs indicated a potential role of this gene, and its relevance is increasing, as the expression of this gene has also been shown in chondrocytes (neXtProt). The GALNT5 gene was also able to correctly predict 54% of the sites identified via MS (Fig. 4D), which also indicates potential involvement of this gene in lubricin glycosylation. The data showed that the highest expression was of the GALNT15 gene in the FLS cultures. The specificity of this enzyme toward mucin-type domains is not currently understood (41), making its further investigation essential, especially as the gene has been shown to be one of the most expressed genes in chondrocytes and bone (21, 42).
The Implications of the Identification of the Site-specific Glycosylation of Lubricin and Its Role in Lubrication
Sialylated and sulfated glycans will alter the charge of heavily glycosylated proteins. Apomucins are usually neutral or acidic, secreted with a predicted pI of 2 to 4.7 (43, 44). The predicted pI of apolubricin is exceptional in this respect in that it can be as high as 9.8, but the protein can end up acidic after glycosylation. With a detailed glycosylation map, the dependence of glycosylation and the amount of sialylation for the charge and pI of the lubricin can be modeled (Fig. 4B). Given that the majority of glycans of lubricin are mono- rather than disialylated (Table I and supplemental Table S1), an upper limit of 168 possible sialic acid residues was suggested. The positive charge buffering capacity of lubricin required ∼60 sialic acids to give the STP-rich region of lubricin a negative charge at the physiological pH (7.2–7.4) of SF (43). An additional 10 sialic acids were required to render the whole lubricin negatively charged (Fig. 5A). Beyond 80 sialic acids, lubricin and its STP-rich region both were negatively charged and capable of maintaining the negative charge during pH shifts of SF and/or limited chemical/enzymatical agents that partially lowered the sialic acid content of lubricin. This is likely the number of sialic acid residues required in order for lubricin to sustain its function on the cartilage surface. We carried out isoelectric focusing before and after de-sialylation in order to better understand the contribution of sialic acid to the physical properties of lubricin. The pI of lubricin before de-sialylation ranged from 4 to 7.5 in a chaotropic environment (Fig. 5B), whereas after de-sialylation the pI of lubricin was ∼7.5. This suggests that the removal of sialic acids changed the molecule from highly acidic to basic and that in addition to the N and C termini, the mucin domain also became positively charged because of the presence of abundant Lys residues and the loss of sialic acid.
Fig. 5.
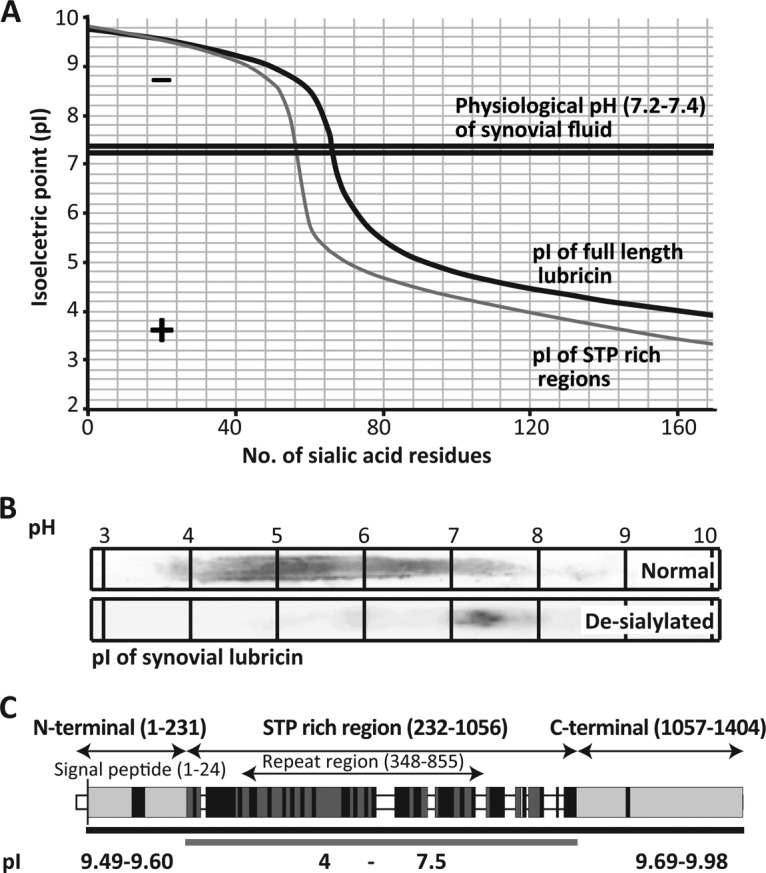
The implication of the type and distribution of O-glycosylation on the charge and role of lubricin as a lubricating agent. A, the predicted isoelectric point (pI) of full-length (black) and STP-rich regions (dark gray) for varying numbers of sialic acids identified, indicating that the pI decreases as the number of sialic acids increases. B, enriched acidic SF fractions, before and after sialidase treatment, were separated on pH 3–10 IPG gels and blotted to PVDF membrane. Western blot analysis using lubricin-specific antibody (mAb13) indicated a pH range of synovial lubricin before sialidase treatment (pH 4–7.5) and a drastic increase in pH (pH 7.5) after sialidase treatment. C, the pI of the N- (9.45–9.6) and C-terminal regions (9.69–9.98) (light gray), where very few glycosylation sites were identified, and the extended STP-rich region, which contained the majority of the protein glycosylation (4–7.5).
This analysis showed that lubricin is an amphoteric, mucin-like molecule with a negatively charged central domain that can become highly hydrated due to its glycosylation and is flanked by positively charged unglycosylated regions (pI 9.49–9.98) (Fig. 5C). The substantial change in the pI and the drastic alteration of the charge of lubricin with around 60 to 70 sialic acids indicates that there is a critical point where the number of glycosylation sites (controlled by the ppGalNAc Ts) and the amount of sialic acid (controlled by sialyltransferases) will significantly alter the properties of lubricin. This shows that pathological alteration of the glycosylation of lubricin may contribute to an altered lubricating surface of articular joints.
Overall, in this study we used a combined CID/ETD-MS2 fragmentation approach to successfully characterize the heavily glycosylated STP-rich region of lubricin and identify an unprecedented 168 glycosylation sites on a single protein. This approach allowed the identification of not only the site of glycosylation, but also its nature, providing a new understanding of the nature of this unique zwitterionic protein. The use of prediction software uncovered the potential importance of novel transferases, which was confirmed by GALNT expression showing that the less understood GALNT5 and -15 were highly expressed in FLSs.
DISCUSSION
Decreased expression and increased degradation of lubricin have been suggested in the joints of RA and OA patients, making the changing characteristics of lubricin a potential indicator of arthritic disease progression. In addition to boundary lubrication, suggested to be established by core 1 O-glycosylation, the multiple protein domains of lubricin may serve other biological functions, such as the protection of chondrocytes and signaling (45). The method adopted in this study allowed us to investigate the specific location of glycans on lubricin. The combination of CID and ETD methods not only enabled evaluation of the protein component and the location of the glycosylation, but also provided details of the attached glycan. Although this was a very effective approach, manual interpretation of CID/ETD-MS2 data was essential because of the lack of universal software in the field of glycoproteomics. The detailed analysis allowed further understanding of the zwitterionic nature of the protein. The inclusion of molecular biology to evaluate the expression of important glycosyltransferases of this highly specialized tissue showed that it has a very different profile from other tissues of the body.
Western blot analysis revealed that unlike that of traditional mucous-forming mucin (46), the mucin-like domain of lubricin could be completely digested by trypsin (Figs. 1A and 1C). Extensive degradation of lubricin by papain and Pronase and partial degradation by pepsin have also been reported previously (47). In addition to these, neutrophil elastase (a serine protease) and cathepsin B (a cysteine protease) have also been shown to degrade lubricin in vitro (13, 48). Given that lubricin was found to have an abundance of closely located occupied glycosylation sites, this suggests that it was the smaller size, rather than a smaller number of glycans, that made lubricin more enzyme accessible than other heavily glycosylated proteins such as mucins.
The glycans identified included the previously reported released O-linked glycans of lubricin (7, 26, 38). However, the confirmation of core 2 O-linked glycans, identified as core 2 glycopeptides from the previously defined mucin-like domain (aa 348–855), and the site-specific glycopeptide characterization of lubricin (in particular the STP-rich region) are shown for the first time in this report. Core 2 structures are the oligosaccharide precursors of inflammatory epitopes such as sialyl Lewis x and sulfated sialylated type 2 structures (49). These types of structures on lubricin have previously been indicated to influence joint inflammation (38). Core 2 structures can also have other functions—for example, cell surface glycans reduce cell–cell interaction (50) and can even be used as cell surface markers to distinguish effector and memory CD8+ T cells (51).
CID-MS2 fragmentation of the O-linked glycopeptides produced sequence information for different glycoforms of the same tryptic peptide (EPAPTTPK) (Figs. 2A–2D). This showed that lubricin glycosylation displayed both macro- (two separate core 1–like glycans) and site-specific micro-heterogeneity (different glycans at a single amino acid position) (Figs. 2C and 2D). However, because CID-MS2 resulted in extensive glycosidic fragment ions, it was not always possible to identify the site-specific location of the glycan in peptides with more than one Thr or Ser (Figs. 2A, 2B, and 2D). To overcome this, ETD was used as an additional technique for the site-specific identification of glycans because it induces peptide backbone cleavage, leaving the glycan unaffected. An additional complication of ETD fragmentation in this study was the abundance of the small repeat (EPAPTTPK), as its low mass reduces the higher charge state advantages of ETD. Therefore, it was the novel combined use of CID and ETD that allowed the site-specific glycan localization and glycan determination of this difficult protein. The site-specific glycopeptide analysis (Fig. 4B) redefined the mucin-like domain to an extended STP-rich region (aa 232–1056). This was due to the identification of extensive O-linked glycopeptides (e.g. peptide K972ITTLKTTTLAPK985V found outside the repeat domain showing four out of five glycosylated sites) in the vicinity of the tandem repeat region of the previously defined mucin repeat domain suggested by UniProt (Figs. 4A and 4B).
In contrast to N-linked glycosylation, the identification of O-glycan attachment sites is made more difficult by the lack of a consensus sequence and the heterogeneity associated with extensive O-glycosylation. The recent increase in O-glycan data has allowed the development of prediction tools including NetOGlyc4.0 and ISOGlyP. It was obvious for lubricin glycosylation that without knowledge about the types of transferases present, the specificity of software such as NetOGlyc4.0 (39), based on neural network predictions of mucin type O-glycosylation sites from all 20 GalNAc Ts, will have some limitations (Fig. 4C). In contrast, software such as ISOGlyP (40), based on individual glycosyltransferase prediction specificity, is likely to be more successful (Figs. 4C and 4D). ISOGlyP predicted 191 O-glycosylation sites, more similar to the data presented in this report (168 Ser/Thr O-linked glycosylation sites). Interestingly, the MS data identified a GalNAc and core 2 modified Thr (1159NGTLVAFR1166) in the hemopexin 1 domain of the C-terminal region (Figs. 4A and 4B; Table I; supplemental Fig. S1), which was not predicted to be glycosylated by either of the software programs (supplemental Table S2). This might indicate a potential regulatory role associated with a particular ppGalNAc T, as the C-terminal recombinant construct of lubricin has been shown to be involved in binding to the cartilage surface (43).
The GALNT profiling expression analysis using primary FLSs showed high expression of the ubiquitous GALNT1 and -2 genes. In addition, high expression levels of GALNT5 and, particularly, GALNT15 were also shown. GALNT5 has been shown to exhibit a restricted expression pattern (21), including expression in chondrocytes (neXtProt). GALNT15 has been suggested to have a broader expression pattern; its dominant expression in the FLSs indicated a particular role in the synovial tissue. The identification of GALNT15 as the 17th most abundant enzyme in chondrocytes (42) indicated that this less studied enzyme could be particularly important for the glycosylation of synovial lubricin.
The site-specific glycopeptide analysis showed that majority of lubricin O-glycans were composed of core 1 structures with terminal galactose (Table I and supplemental Table S1). Terminal galactose is a ligand for galectins, known to increase expression during RA (52) and suggested, along with fibrinogen, to play a pro-inflammatory role by regulating neutrophil activation and degranulation (53). The high proportion of sialylated core 1 glycopeptides identified has biosynthetic importance, as the sialic-acid-terminated glycan end cannot be extended any further by glycosyltransferases in the Golgi/endoplasmic reticulum (54). This might also explain the low proportion of core 2 structures identified as core 2 glycopeptides, which could be a consequence of low core 2 GlcNAc transferase activity or high sialyltransferase activity, or both. The high proportion of sialylated core 1 glycans on lubricin reduces the possibility of the formation of larger, potentially immunologically reactive glycans, restricting lubricin to short, negatively charged glycans.
The terminal domains of lubricin have a large number of positively charged arginine and lysine residues, whereas the STP-rich region is negatively charged because of the attached sialic acid, making lubricin an amphoteric polyelectrolyte. Lubricin is suggested to be a good lubricant for negatively charged surfaces such as the surface of the outermost layers (lamina splendens) of the articular cartilage. This is mainly due to an increase in repellent charge forces between the negatively charged STP-rich region and the negatively charged components of the outermost layers of the cartilage such as hyaluronic acid, lipids, and proteoglycans (55). The pI of synovial lubricin ranged from 4 to 7.5 as measured by isoelectric focusing (Figs. 5B and 5C). De-sialylation increased the pI substantially to close to 7.5 (Fig. 5B). The lower pI relative to the theoretical calculation for apolubricin (pI 9.8) is likely due to the influence of the pKa of individual amino acid residues by the chaotropic reagants and the remaining sulfated residues on the lubricin oligosaccharides (38). Although lubrication might not be totally dependent on sialic acid, it might be enhanced through an increase in repellent charge forces due to an increase in negative charges around the STP-rich region domain (55).
The terminal somatomedin B-like and hemopexin-like domains have been shown to promote integrin-mediated attachment of cells to the extracellular matrix (6, 8). It has also been reported that lubricin lacking these end domains binds only weakly to the cartilage surface (56). This weak binding is suggested to result in inefficient lubrication (43). Therefore, it can be speculated that for efficient lubricin function, both positively charged end domains and the negatively charged STP-rich region are essential.
Concluding Remarks
The mass spectrometric site-specific glycopeptide characterization performed in this study mapped the glycosylation profile of lubricin within the STP-rich region and indicates that lubricin glycosylation displays both micro- and macroheterogeneity. The presence of two adjacent simultaneously glycosylated Thr residues in the consensus repeat unit EPAPTTPK indicated that there are regions within the lubricin domain that are highly glycosylated. The data presented here redefine an extended STP-rich region relative to the mucin domain previously defined by UniProt. Screening of ppGalNAc Ts from primary FLSs showed high expression of the less understood GALNT15 and GALNT5 genes, indicating that lubricin glycosylation is unique. Overall, this study showed that heavy glycosylation, particularly sialylation, is essential for creating the amphoteric nature of lubricin, a property that may facilitate its efficient biolubrication function.
Supplementary Material
Acknowledgments
We are grateful to Marshall Bern from Protein Matrics Inc. for providing help in using the Byonic software for glycopeptide data analysis. We thank C. R. Flannery (Pfizer Research, Cambridge, MA) for providing lubricin-specific antibody (mAb 13).
Footnotes
Author contributions: L.A., S.A.F., and N.G.K. designed research; L.A., S.A.F., C.J., and E.P.B. performed research; C.J., A.H.E., and N.G.K. contributed new reagents or analytic tools; L.A., S.A.F., E.P.B., and N.G.K. analyzed data; L.A. and N.G.K. wrote the paper; A.H.E. provided rheumatoid and osteoarthritis samples.
* This work was supported by the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), the King Gustav V Memorial Foundation, and Petrus and Augusta Hedlund's foundation. The LTQ-Orbitrap mass spectrometer was obtained with a grant from the Knut and Alice Wallenberg Foundation (KAW2007.0118). The HPLC instrument was obtained with a gift from the Ingabritt and Arne Lundbergs Research Foundation.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SF
- synovial fluid
- PNA
- peanut agglutinin
- WGA
- wheat germ agglutinin
- CID
- collision-induced dissociation
- ETD
- electron transfer dissociation
- RA
- rheumatoid arthritis
- OA
- osteoarthritis
- FLS
- fibroblast-like synoviocyte
- GALNT
- polypeptide N-acetylgalactosaminyltransferase gene
- IPG
- immobilized pH gradient gel
- ppGalNAc T
- polypeptide GalNAc-transferase
- aa
- amino acids.
REFERENCES
- 1. Jay G. D., Tantravahi U., Britt D. E., Barrach H. J., Cha C. J. (2001) Homology of lubricin and superficial zone protein (SZP): products of megakaryocyte stimulating factor (MSF) gene expression by human synovial fibroblasts and articular chondrocytes localized to chromosome 1q25. J. Orthop. Res 19, 677–687 [DOI] [PubMed] [Google Scholar]
- 2. Jay G. D., Fleming B. C., Watkins B. A., McHugh K. A., Anderson S. C., Zhang L. X., Teeple E., Waller K. A., Elsaid K. A. (2010) Prevention of cartilage degeneration and restoration of chondroprotection by lubricin tribosupplementation in the rat following anterior cruciate ligament transection. Arthritis Rheum. 62, 2382–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morrell K. C., Hodge W. A., Krebs D. E., Mann R. W. (2005) Corroboration of in vivo cartilage pressures with implications for synovial joint tribology and osteoarthritis causation. Proc. Natl. Acad. Sci. U.S.A. 102, 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greene G. W., Banquy X., Lee D. W., Lowrey D. D., Yu J., Israelachvili J. N. (2011) Adaptive mechanically controlled lubrication mechanism found in articular joints. Proc. Natl. Acad. Sci. U.S.A. 108, 5255–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swann D. A., Silver F. H., Slayter H. S., Stafford W., Shore E. (1985) The molecular structure and lubricating activity of lubricin isolated from bovine and human synovial fluids. Biochem. J. 225, 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rhee D. K., Marcelino J., Baker M., Gong Y., Smits P., Lefebvre V., Jay G. D., Stewart M., Wang H., Warman M. L., Carpten J. D. (2005) The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J. Clin. Invest. 115, 622–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jay G. D., Harris D. A., Cha C. J. (2001) Boundary lubrication by lubricin is mediated by O-linked beta(1–3)Gal-GalNAc oligosaccharides. Glycoconj. J. 18, 807–815 [DOI] [PubMed] [Google Scholar]
- 8. Schaefer D. B., Wendt D., Moretti M., Jakob M., Jay G. D., Heberer M., Martin I. (2004) Lubricin reduces cartilage–cartilage integration. Biorheology 41, 503–508 [PubMed] [Google Scholar]
- 9. Ungethuem U., Haeupl T., Witt H., Koczan D., Krenn V., Huber H., von Helversen T. M., Drungowski M., Seyfert C., Zacher J., Pruss A., Neidel J., Lehrach H., Thiesen H. J., Ruiz P., Blass S. (2010) Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol. Genomics 42A, 267–282 [DOI] [PubMed] [Google Scholar]
- 10. Lorenz H., Richter W. (2006) Osteoarthritis: cellular and molecular changes in degenerating cartilage. Prog. Histochem. Cytochem. 40, 135–163 [DOI] [PubMed] [Google Scholar]
- 11. Goggs R., Carter S. D., Schulze-Tanzil G., Shakibaei M., Mobasheri A. (2003) Apoptosis and the loss of chondrocyte survival signals contribute to articular cartilage degradation in osteoarthritis. Vet. J. 166, 140–158 [DOI] [PubMed] [Google Scholar]
- 12. Ali L., Jin C., Karlsson N. G. (2012) Glycoproteomics of Lubricin-Implication of Important Biological Glyco- and Peptide-Epitopes in Synovial Fluid, Rheumatoid Arthritis—Etiology, Consequences and Co-Morbidities (Lemmey A., Ed), pp. 131–150, Intech, New York, NY [Google Scholar]
- 13. Elsaid K. A., Fleming B. C., Oksendahl H. L., Machan J. T., Fadale P. D., Hulstyn M. J., Shalvoy R., Jay G. D. (2008) Decreased lubricin concentrations and markers of joint inflammation in the synovial fluid of patients with anterior cruciate ligament injury. Arthritis Rheum. 58, 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Young A. A., McLennan S., Smith M. M., Smith S. M., Cake M. A., Read R. A., Melrose J., Sonnabend D. H., Flannery C. R., Little C. B. (2006) Proteoglycan 4 downregulation in a sheep meniscectomy model of early osteoarthritis. Arthritis Res. Ther. 8, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elsaid K. A., Jay G. D., Chichester C. O. (2007) Reduced expression and proteolytic susceptibility of lubricin/superficial zone protein may explain early elevation in the coefficient of friction in the joints of rats with antigen-induced arthritis. Arthritis Rheum. 56, 108–116 [DOI] [PubMed] [Google Scholar]
- 16. Halim A., Nilsson J., Ruetschi U., Hesse C., Larson G. (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell. Proteomics 11, M111.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Domon B., Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 [Google Scholar]
- 18. Lu H., Zong C., Wang Y., Young G. W., Deng N., Souda P., Li X., Whitelegge J., Drews O., Yang P. Y., Ping P. (2008) Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol. Cell. Proteomics 7, 2073–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanisch F. G. (2012) O-glycoproteomics: site-specific O-glycoprotein analysis by CID/ETD electrospray ionization tandem mass spectrometry and top-down glycoprotein sequencing by in-source decay MALDI mass spectrometry. Methods Mol. Biol. 842, 179–189 [DOI] [PubMed] [Google Scholar]
- 20. Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., Kato K., Irimura T., Suryanarayanan G., Hollingsworth M. A., Clausen H. (2007) The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology 17, 374–387 [DOI] [PubMed] [Google Scholar]
- 21. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato K., Jeanneau C., Tarp M. A., Benet-Pages A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 23. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., Clausen H. (2010) O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 285, 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arnett F. C., Edworthy S. M., Bloch D. A., McShane D. J., Fries J. F., Cooper N. S., Healey L. A., Kaplan S. R., Liang M. H., Luthra H. S., Medsger T. A., Mitchell D. M., Neustadt D. H., Pinals R. S., Schaller J. G., Sharp J. T., Wilder R. L., Hunder G. G. (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31, 315–324 [DOI] [PubMed] [Google Scholar]
- 26. Estrella R. P., Whitelock J. M., Packer N. H., Karlsson N. G. (2010) The glycosylation of human synovial lubricin: implications for its role in inflammation. Biochem. J. 429, 359–367 [DOI] [PubMed] [Google Scholar]
- 27. Flowers S. A., Ali L., Lane C. S., Olin M., Karlsson N. G. (2013) Selected reaction monitoring to differentiate and relatively quantitate isomers of sulfated and unsulfated core 1 O-glycans from salivary MUC7 protein in rheumatoid arthritis. Mol. Cell. Proteomics 12, 921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Towbin H., Ozbey O., Zingel O. (2001) An immunoblotting method for high-resolution isoelectric focusing of protein isoforms on immobilized pH gradients. Electrophoresis 22, 1887–1893 [DOI] [PubMed] [Google Scholar]
- 29. Wopereis S., Grunewald S., Morava E., Penzien J. M., Briones P., Garcia-Silva M. T., Demacker P. N., Huijben K. M., Wevers R. A. (2003) Apolipoprotein C-III isofocusing in the diagnosis of genetic defects in O-glycan biosynthesis. Clin. Chem. 49, 1839–1845 [DOI] [PubMed] [Google Scholar]
- 30. Kuster B., Wheeler S. F., Hunter A. P., Dwek R. A., Harvey D. J. (1997) Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal. Biochem. 250, 82–101 [DOI] [PubMed] [Google Scholar]
- 31. Selman M. H., Hemayatkar M., Deelder A. M., Wuhrer M. (2011) Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83, 2492–2499 [DOI] [PubMed] [Google Scholar]
- 32. Zhang C. C., Rogalski J. C., Evans D. M., Klockenbusch C., Beavis R. C., Kast J. (2011) In silico protein interaction analysis using the global proteome machine database. J. Proteome Res. 10, 656–668 [DOI] [PubMed] [Google Scholar]
- 33. McClintock C. S., Parks J. M., Bern M., Ghattyvenkatakrishna P. K., Hettich R. L. (2013) Comparative informatics analysis to evaluate site-specific protein oxidation in multidimensional LC-MS/MS data. J. Proteome Res. 12, 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henriksson G., Englund A. K., Johansson G., Lundahl P. (1995) Calculation of the isoelectric points of native proteins with spreading of pKa values. Electrophoresis 16, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 35. Henriksson H., Stahlberg J., Isaksson R., Pettersson G. (1996) The active sites of cellulases are involved in chiral recognition: a comparison of cellobiohydrolase 1 and endoglucanase 1. FEBS Lett. 390, 339–344 [DOI] [PubMed] [Google Scholar]
- 36. Arnberg N., Kidd A. H., Edlund K., Nilsson J., Pring-Akerblom P., Wadell G. (2002) Adenovirus type 37 binds to cell surface sialic acid through a charge-dependent interaction. Virology 302, 33–43 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt T. A., Plaas A. H., Sandy J. D. (2009) Disulfide-bonded multimers of proteoglycan 4 PRG4 are present in normal synovial fluids. Biochim. Biophys. Acta 1790, 375–384 [DOI] [PubMed] [Google Scholar]
- 38. Jin C., Ekwall A. K., Bylund J., Bjorkman L., Estrella R. P., Whitelock J. M., Eisler T., Bokarewa M., Karlsson N. G. (2012) Human synovial lubricin expresses sialyl Lewis x determinant and has L-selectin ligand activity. J. Biol. Chem. 287, 35922–35933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerken T. A., Jamison O., Perrine C. L., Collette J. C., Moinova H., Ravi L., Markowitz S. D., Shen W., Patel H., Tabak L. A. (2011) Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng L., Tachibana K., Iwasaki H., Kameyama A., Zhang Y., Kubota T., Hiruma T., Tachibana K., Kudo T., Guo J. M., Narimatsu H. (2004) Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 566, 17–24 [DOI] [PubMed] [Google Scholar]
- 42. Kumar S., Badger A. M., Lee J. C., Gowen M., Lark M. W., Connor J. R., Dodds R. A., Halsey W., Van Horn M., Mao J., Sathe G., Mui P., Agarwal P. (2001) Identification and initial characterization of 5000 expressed sequenced tags (ESTs) each from adult human normal and osteoarthritic cartilage cDNA libraries. Osteoarthr. Cartil. 9, 641–653 [DOI] [PubMed] [Google Scholar]
- 43. Lee S., Muller M., Rezwan K., Spencer N. D. (2005) Porcine gastric mucin (PGM) at the water/poly(dimethylsiloxane) (PDMS) interface: influence of pH and ionic strength on its conformation, adsorption, and aqueous lubrication properties. Langmuir 21, 8344–8353 [DOI] [PubMed] [Google Scholar]
- 44. Bushnak I. A., Labeed F. H., Sear R. P., Keddie J. L. (2010) Adhesion of microorganisms to bovine submaxillary mucin coatings: effect of coating deposition conditions. Biofouling 26, 387–397 [DOI] [PubMed] [Google Scholar]
- 45. Waller K. A., Zhang L. X., Elsaid K. A., Fleming B. C., Warman M. L., Jay G. D. (2013) Role of lubricin and boundary lubrication in the prevention of chondrocyte apoptosis. Proc. Natl. Acad. Sci. U.S.A. 110, 5852–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao R., Wang T. T., DeMaria G., Sheehan J. K., Kesimer M. (2012) Mapping the protein domain structures of the respiratory mucins: a mucin proteome coverage study. J. Proteome Res. 11, 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flannery C. R., Hughes C. E., Schumacher B. L., Tudor D., Aydelotte M. B., Kuettner K. E., Caterson B. (1999) Articular cartilage superficial zone protein (SZP) is homologous to megakaryocyte stimulating factor precursor and is a multifunctional proteoglycan with potential growth-promoting, cytoprotective, and lubricating properties in cartilage metabolism. Biochem. Biophys. Res. Commun. 254, 535–541 [DOI] [PubMed] [Google Scholar]
- 48. Elsaid K. A., Jay G. D., Warman M. L., Rhee D. K., Chichester C. O. (2005) Association of articular cartilage degradation and loss of boundary-lubricating ability of synovial fluid following injury and inflammatory arthritis. Arthritis Rheum. 52, 1746–1755 [DOI] [PubMed] [Google Scholar]
- 49. Tsuboi S., Fukuda M. (2001) Roles of O-linked oligosaccharides in immune responses. Bioessays 23, 46–53 [DOI] [PubMed] [Google Scholar]
- 50. Kenny D. T., Skoog E. C., Linden S. K., Struwe W. B., Rudd P. M., Karlsson N. G. (2012) Presence of terminal N-acetylgalactosaminebeta1–4N-acetylglucosamine residues on O-linked oligosaccharides from gastric MUC5AC: involvement in Helicobacter pylori colonization? Glycobiology 22, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 51. Harrington L. E., Galvan M., Baum L. G., Altman J. D., Ahmed R. (2000) Differentiating between memory and effector CD8 T cells by altered expression of cell surface O-glycans. J. Exp. Med. 191, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ohshima S., Kuchen S., Seemayer C. A., Kyburz D., Hirt A., Klinzing S., Michel B. A., Gay R. E., Liu F. T., Gay S., Neidhart M. (2003) Galectin 3 and its binding protein in rheumatoid arthritis. Arthritis Rheum. 48, 2788–2795 [DOI] [PubMed] [Google Scholar]
- 53. Fernandez G. C., Ilarregui J. M., Rubel C. J., Toscano M. A., Gomez S. A., Beigier Bompadre M., Isturiz M. A., Rabinovich G. A., Palermo M. S. (2005) Galectin-3 and soluble fibrinogen act in concert to modulate neutrophil activation and survival: involvement of alternative MAPK pathways. Glycobiology 15, 519–527 [DOI] [PubMed] [Google Scholar]
- 54. Brockhausen I. (1999) Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta 1473, 67–95 [DOI] [PubMed] [Google Scholar]
- 55. Zappone B., Ruths M., Greene G. W., Jay G. D., Israelachvili J. N. (2007) Adsorption, lubrication, and wear of lubricin on model surfaces: polymer brush-like behavior of a glycoprotein. Biophys. J. 92, 1693–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swann D. A., Hendren R. B., Radin E. L., Sotman S. L., Duda E. A. (1981) The lubricating activity of synovial fluid glycoproteins. Arthritis Rheum. 24, 22–30 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


