Abstract
Chagas disease is a tropical neglected disease endemic in Latin America caused by the protozoan Trypanosoma cruzi. The parasite has four major life stages: epimastigote, metacyclic trypomastigote, bloodstream trypomastigote, and amastigote. The differentiation from infective trypomastigotes into replicative amastigotes, called amastigogenesis, takes place in vivo inside mammalian host cells after a period of incubation in an acidic phagolysosome. This differentiation process can be mimicked in vitro by incubating tissue-culture-derived trypomastigotes in acidic DMEM. Here we used this well-established differentiation protocol to perform a comprehensive quantitative proteomic and phosphoproteomic analysis of T. cruzi amastigogenesis. Samples from fully differentiated forms and two biologically relevant intermediate time points were Lys-C/trypsin digested, iTRAQ-labeled, and multiplexed. Subsequently, phosphopeptides were enriched using a TiO2 matrix. Non-phosphorylated peptides were fractionated via hydrophilic interaction liquid chromatography prior to LC-MS/MS analysis. LC-MS/MS and bioinformatics procedures were used for protein and phosphopeptide quantitation, identification, and phosphorylation site assignment. We were able to identify regulated proteins and pathways involved in coordinating amastigogenesis. We also observed that a significant proportion of the regulated proteins were membrane proteins. Modulated phosphorylation events coordinated by protein kinases and phosphatases that are part of the signaling cascade induced by incubation in acidic medium were also evinced. To our knowledge, this work is the most comprehensive quantitative proteomics study of T. cruzi amastigogenesis, and these data will serve as a trustworthy basis for future studies, and possibly for new potential drug targets.
More than 100 years after its initial description, Chagas disease remains a neglected tropical disease affecting almost 8 million people worldwide (1). The illness is endemic in Latin America, but an increasing number of cases are being reported in non-endemic regions, including the United States and Europe (1–5). The infection is caused by the protozoan Trypanosoma cruzi, a single-cell parasite that undergoes differentiation events throughout its life cycle to adapt to a variety of environments inside mammalian hosts and triatomine bug vectors. The parasite has four major life stages: epimastigote and metacyclic trypomastigote in the insect vector and bloodstream trypomastigote and amastigote in mammalian hosts (6, 7). So far, there is no vaccine or effective chemotherapeutic treatment; the drugs commonly used to treat human infection, nifurtimox and benznidazol, are highly toxic and do not prevent the progression of the chronic form of the illness (8).
Among the differentiation processes that T. cruzi undergoes during its lifecycle, metacyclogenesis (differentiation from epimastigotes to metacyclic trypomastigotes) and amastigogenesis (from trypomastigotes to amastigotes) have been more deeply studied (9). Metacyclogenesis occurs naturally inside the triatomine vector's gut, probably in response to environmental changes such as a decline in available nutrients. The molecular mechanisms involved in triggering this process remain unknown. However, metacyclogenesis can be mimicked in vitro using well-defined media that stimulate the differentiation process by simulating the vector's gut physicochemical conditions (10). This process results in important changes that have been studied intensively through proteomic approaches (11, 12).
Parasite differentiation, as well as host-cell infection, involves the intracellular release of Ca2+ in the coordination of molecular events involved in those processes. However, the precise molecular mechanisms responsible for controlling Ca2+ release are not fully known. Two pathways related to this event have already been identified: (i) the engagement of gp82 with an unknown ligand, which triggers a signaling cascade in metacyclic trypomastigotes that involves the phosphorylation of PKC and a 175-kDa protein (13); and (ii) the release of Ca2+ reservoirs from the endoplasmatic reticulum mediated by inositol-3-phosphate (14).
Amastigogenesis occurs when metacyclic trypomastigotes turn into amastigotes inside mammalian nucleated host cells (first amastigogenesis) or after infection by tissue-derived bloodstream trypomastigotes, when the so-called second amastigogenesis is observed, also inside host cells (15). This process, which is not completely understood at the molecular level, takes place preferably inside host cells, although extracellular amastigotes may be observed during the infection's acute phase (16).
The study of T. cruzi differentiation strongly relies on in vitro assays. For instance, Contreras and collaborators developed a protocol in which metacyclic trypomastigotes can be differentiated to amastigotes by means of incubation in MEMTAU media, a combination of MEM and TAU3AAA medium, enriched with FBS at 37 °C (17). In fact, the production of axenic amastigotes by means of incubation of tissue-culture-derived trypomastigotes in acidic media has been widely employed since the first report by Kanbara and collaborators (18). Another study, carried out by Tomlinson and collaborators, demonstrated that axenic amastigotes not only resemble intracellular amastigotes morphologically, but also share several biochemical features, stage-specific surface antigens, and replicative capability, an attribute essential for fulfilling the life cycle (19). This study also showed that the acidic incubation accelerates the morphological differentiation, and also seems to be a key factor in triggering DNA replication. Overall, the axenic differentiation systems yield relatively large quantities of parasites, free of host-cell contaminations, and allow the collection of samples at defined time points during the differentiation.
In order to unravel the mechanisms that mediate T. cruzi differentiation, it is essential to identify the molecules responsible for the intracellular signaling mechanisms that trigger the process in response to environmental changes (20). For instance, little is known about how cAMP molecules are involved in amastigogenesis; however, several studies have reported the involvement of this ubiquitous intracellular second messenger in the control of metacyclic trypomastigote differentiation in vitro (21). It has been demonstrated that intracellular cAMP levels are higher in metacyclic trypomastigotes than in epimastigotes (22), and also that the addition of exogenous cAMP or its analogs accelerates metacyclogenesis in vitro (23). The elevated cAMP levels strongly suggest a role for cAMP in the coordination of metacyclic T. cruzi differentiation.
In trypanosomatids and most eukaryotes, cAMP is synthesized by adenylate cyclases and degraded by phosphodiesterases (24). Numerous physiological processes are regulated through cAMP-dependent protein kinases (PKAs)1 that are activated upon cAMP binding to the PKA regulatory subunit and release of its active catalytic subunit (25).
Grellier and collaborators have shown that tissue-culture-derived trypomastigotes that are exposed to phosphatase inhibitors, such as calyculin A, start to differentiate into rounded, extracellular amastigote-like cells even at a physiological pH (26). Furthermore, okadaic acid, another phosphatase inhibitor, represses axenic acidic-pH-induced amastigogenesis, strengthening the hypothesis that phosphorylation/dephosphorylation events are implicated in T. cruzi amastigogenesis (27).
Approximately 2% of the T. cruzi genome encodes protein kinases, suggesting that these enzymes play a major regulatory role in the parasite (28). A comparative survey of the kinomes (kinase repertoire) from trypanosomatids showed that Trypanosoma brucei, T. cruzi, and Leishmania major have 176, 190, and 199 protein kinases, respectively, with ∼12% of them being unique from trypanosomatids (28, 29). In fact, phosphorylation is among the most widespread post-translational modifications in nature. For example, it is estimated that over 30% of the protein repertoire in a mammalian cell is phosphorylated at some point during expression (30). This post-translational modification also regulates a number of biological processes involving signal transduction (31), recognition and molecular interactions, and other cellular events. With the current interest in protein kinases as molecular targets for treating a variety of diseases (32, 33), the possibility that kinase inhibitors could represent new anti-parasitic agents is being explored (34). Hence, studies regarding the dynamics of protein phosphorylation during cell differentiation in human-hosted T. cruzi life stages become particularly relevant.
This work aimed at performing the first large-scale quantitative proteome and phosphoproteome analysis of T. cruzi amastigogenesis. For that, the differentiation of cell-culture-derived trypomastigotes to axenic amastigotes was induced in vitro, and parasite protein samples from initial, intermediate, and final points of the differentiation process were subjected to iTRAQ labeling and mass spectrometric analysis.
MATERIALS AND METHODS
FBS was purchased from Sorali Biotecnologia (Campo Grande, Brazil). GeLoader tips were from Eppendorf (Hamburg, Germany). Modified trypsin was obtained from Promega (Madison, WI). Lysyl endopeptidase (Lys-C) was purchased from Wako Pure Chemical Industries (Osaka, Japan). A TSKGel Amide-80 2-mm, 3-μm particle size HILIC column was obtained from Tosoh Bioscience (Stuttgart, Germany). PorosOligoR3 reversed phase material was acquired from PerSeptive Biosystems (Framingham, MA). StageTips were obtained from Thermo Scientific (Odense, Denmark). Titanium dioxide TitanSphere beads were from GL Sciences Inc. (Tokyo, Japan). iTRAQ reagents were from Applied Biosystems (Foster City, CA). Complete cocktails of protease and phosphatase inhibitors (Complete Mini and PhosStop) were from Roche (Meylan, France). DMEM and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO). All chemicals and solvents used were HPLC grade or higher, and all sample preparation procedures were performed in low-binding polypropylene microtubes from Sorenson Bioscience (Salt Lake City, UT).
Trypomastigote in Vitro Culture and Acidic pH–induced Axenic Amastigogenesis Assay
HeLa cell culture was maintained in DMEM, pH 7.5, supplemented with 5% FBS and 100 μg/ml gentamicin at 37 °C under a 5% CO2 atmosphere. Tissue-culture-derived trypomastigotes (Y strain) were collected from infected HeLa cell culture monolayers (35) during the second day of outbreak, which was the most abundant. The parasite population, which consisted of over 98% trypomastigotes, was washed three times with DMEM without FBS prior to differentiation induction. Axenic amastigotes were generated by incubating trypomastigotes in DMEM, pH 5.0, without FBS for 9 h at 37 °C under a 5% CO2 atmosphere as previously described (19, 36). Besides the trypomastigote and amastigote samples (corresponding to 0 and 9 h of induction, respectively), intermediary samples were collected at 30 min and 2 h of induction. All samples were prepared in biological triplicate.
Sample Preparation for LC-MS/MS
Each sample containing the parasite life forms was lysed in 100 μl of 8 m urea containing a complete mixture of protease and phosphatase inhibitors through three rapid freeze-thaw cycles. After centrifugation at 14,000g at 4 °C for 15 min, the supernatants were transferred to new tubes, diluted four times with ice-cold ethanol, and vortexed. The same volume of ice-cold acetone was added to the tubes, which were then vortexed vigorously and incubated overnight at −20 °C. After incubation, the material was centrifuged at 20,000g at 4 °C for 15 min, and the supernatant was discarded. The pellet was washed three times with ice-cold 40% ethanol/40% acetone solution. Finally, the sample was resuspended in 8 m urea in 20 mm triethylammonium bicarbonate, reduced with 20 mm DTT at room temperature for 60 min, alkylated with 40 mm iodoacetamide at room temperature in the dark for at least 60 min, and digested overnight at room temperature with Lys-C (0.05 U). After digestion, the solution was diluted to a final urea concentration of 1 m, 1 μg of modified trypsin was added (1:100 (w/w) trypsin:substrate ratio), and the samples were incubated for 3 h at room temperature. The peptide samples were acidified with TFA (0.1% (v/v) final concentration) and desalted on homemade microcolumns of Poros Oligo R3 packed resin (1 cm long) in p200 tips (adapted from Ref. 37). Prior to lyophilization, the peptide concentration was determined by means of amino acid analysis using a Biochrom 30 amino acid analyzer (Biochrom, Cambridge, UK) according to the manufacturer's protocol.
iTRAQ Labeling
Three biological replicates of each sample were labeled with iTRAQ according to manufacturer's specifications, with minor modifications. Briefly, 100 μg of dried, desalted digest from each sample was individually resuspended in 30 μl of 300 mm triethylammonium bicarbonate and added to an iTRAQ label vial already resuspended with 70 μl of ethanol. The vials were briefly mixed and incubated at room temperature for 1 to 2 h, and then all four labeled samples were mixed in equal proportion (trypomastigotes were labeled with 114; 30 min of induction with 115; 2 h of induction with 116; and axenic amastigotes with 117).
HILIC Fractionation
Prior to LC-MS/MS analysis, around 20 μg of each multiplexed iTRAQ-labeled sample replicate was separated into 10 chromatographic fractions. For that, dried samples were resuspended in solvent B (90% acetonitrile, 0.1% TFA) and applied onto a TSKGel Amide 80 HILIC HPLC column (length, 15 cm; diameter, 2 mm; particle size, 3 μm) as described elsewhere (38, 39). Peptides were eluted at 6 μl/min by decreasing solvent B in a linear manner (100% to 60% solvent B) over 26 min and collected every 2 min. The fractions were combined based on their intensities detected at a 216-nm wavelength to an average of 1200 au. All eluted fractions prior to and after the elution of the 10 most intense fractions were added to the first and last sample fractions, respectively, to avoid missing any peptide.
Phosphopeptide Enrichment
Phosphopeptides were enriched in batch via TiO2 affinity as described elsewhere (40), with minor modifications. Briefly, iTRAQ-labeled multiplexed samples (around 380 μg and 200 μg for the third replicate) were re-resuspended in 1 m glycolic acid in 80% acetonitrile/5% TFA (v/v), and 0.6 mg of TiO2 beads were added per 100 μg of peptide sample before incubation under vigorous shaking for 10 to 15 min. Beads were spun down, and the supernatant was transferred to new microtubes. Addition of TiO2 beads to the supernatants (using 0.3 mg TiO2 per 100 μg of peptide) was repeated two more times. The TiO2 beads from the three rounds of enrichment were combined and washed first with 80% acetonitrile/1% TFA (v/v) and then with 10% acetonitrile/0.1% TFA (v/v) to remove non-phosphorylated peptides bound to TiO2 in a HILIC way. Phosphopeptides were then eluted with ammonia solution (0.28%), pH 11, and lyophilized.
LC-MS/MS
Samples (10 HILIC fractions and 1 phosphopeptide-enriched fraction) were analyzed in biological triplicate, with two technical repetitions each, using an EASY-nano LC system (Proxeon Biosystems, Odense, Denmark) coupled online with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA). Each fraction was loaded onto an 18-cm fused silica emitter (75-μm inner diameter) packed in-house with reverse phase capillary column ResiproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH, Ammerbuch, Germany) and eluted using a gradient from 100% solvent A (0.1% formic acid) to 26% solvent B (0.1% formic acid, 95% acetonitrile) for 77 min, 26% to 100% solvent B for 5 min, and 100% solvent B for 8 min (a total of 90 min at 200 nL/min) for HILIC fractions and 100% solvent A to 26% solvent B for 180 min, 26% to 100% solvent B for 5 min, and 100% solvent B for 8 min (a total of 193 min at 200 nL/min) for phosphopeptide-enriched fraction. After each run, the column was washed with 90% solvent B and re-equilibrated with solvent A. Mass spectra were acquired in positive ion mode applying data-dependent automatic survey MS scan and tandem mass spectra (MS/MS) acquisition modes. Each MS scan in the Orbitrap analyzer (mass range = m/z 350–1800, resolution = 100,000) was followed by MS/MS of the seven most intense ions in the LTQ. Fragmentation in the LTQ was performed by high-energy collision-activated dissociation, and selected sequenced ions were dynamically excluded for 30 s. Raw data were viewed in Xcalibur v.2.1 (Thermo Scientific), and data processing was performed using Proteome Discoverer v.1.3 beta (Thermo Scientific). The Raw files were submitted to a database search using Proteome Discoverer with an in-house Mascot v.2.3 algorithm against the T. cruzi database, downloaded (in early 2012) using the Database on Demand tool (41) containing the 12,574 proteins of the parasite found in UniProt/Swiss-Prot and UniProt/TrEMBL. Contaminant proteins (several types of human keratins, BSA, and porcine trypsin) were also added to the database, and all contaminant proteins identified were manually removed from the result lists. The searches were performed with the following parameters: MS accuracy, 10 ppm; MS/MS accuracy, 0.05 Da; two missed cleavage sites allowed; carbamidomethylation of cysteine as a fixed modification; and oxidation of methionine, N-terminal iTRAQ tagging, phosphorylation of S, T, and Y residues, and protein N-terminal acetylation as variable modifications. The numbers of proteins, protein groups, and peptides were filtered for false discovery rates less than 1% and only peptides with rank 1. A minimum of two peptides per protein was accepted for identification using Proteome Discoverer. The identification lists from technical repetitions were merged, and repeated protein groups were removed. Phosphopeptide identifications with phosphorylation site probability scores less than 75% were removed, as suggested by the software developer (42).
Data Analysis and Statistics
ProteinCenter software (Thermo Scientific) was used to interpret the results at the protein level (e.g. statistical GO classification and number of transmembrane domains). More detailed annotations of the identified proteins and enzyme activity prediction and classification (Enzyme Commission numbers) were acquired using Blast2GO software (43) with the default parameters, and predictions of kinase recognition sites on protein sequences were performed using iGPS software (44) with a low threshold, Homo sapiens as the organism, and “all protein kinases” prediction. Sequence motif enrichment analyses of phosphorylation sites were performed using the MotifX algorithm (45) with windows of 13, 15, and 17 amino acids. The same T. cruzi database employed previously for protein identification was used as a background file.
Statistical analysis was performed with R software. The data consisted of three replicates containing normal fractions and fractions enriched for phosphopeptides. The intensity values of each fraction and each iTRAQ label were log-transformed and median-normalized. Multiple measurements of the same peptide were merged using the RRollup function of the DanteR package (46); we allowed only one peptide measurement and chose the “mean” instead of the “median” option. The values of non-phosphorylated peptides were converted into protein quantitations (Rrollup, “mean”), and we modified the original function to require a minimum of three peptides per protein. Statistically significant regulations require more sophisticated tools than just application of the standard t test. Limma (47) and rank products (48) provide sufficient power to deal with low replicate numbers and additional missing values (49). We therefore carried out both statistical tests on all phosphopeptides and protein ratios against label 114 and corrected them for multiple testing (50). All phosphopeptides/proteins with smaller q-values (from both tests) below 0.05 (5% false discovery rate) were considered regulated.
For the cluster analysis, we calculated the mean over all three replicated values for each condition. Phosphopeptides and proteins were merged into one dataset. Fuzzy c-means clustering (51, 52) was applied after we determined the value of the fuzzifier and obtained the number of clusters according to Schwämmle and Jensen (53).
Data Availability
Mass spectrometer output files (Raw data), peptide and protein identification files (MGF files), the reference protein sequence database, and annotated spectra of bona fide phosphopeptide identifications have been deposited in a public repository, the PeptideAtlas database (www.peptideatlas.org/PASS/PASS00434), under the dataset tag Tc Amastigogenesis and database identifier PASS00434.
RESULTS AND DISCUSSION
Protein and Phosphopeptide Identification and Relative Expression Levels
In this large-scale proteome analysis of T. cruzi amastigogenesis, parasite samples were collected in four defined induction periods (Fig. 1): (i) trypomastigotes (0 min); (ii) 30 min after induction, where we expected to observe the first changes in relevant biological processes; (iii) 2 h after induction, corresponding to the average time that parasites remain inside acidic phagolysosomes in vivo (54); and (iv) axenic amastigotes (9 h of induction). Relative quantitation was obtained through iTRAQ labeling, and samples were analyzed in biological triplicate, with each replicate analyzed twice to increase the number of protein groups identified. High-confidence peptide and phosphopeptide identification lists, with their respective iTRAQ reporter-ion intensities, were analyzed with R software to identify significant changes in protein and phosphorylation levels during the differentiation.
Fig. 1.
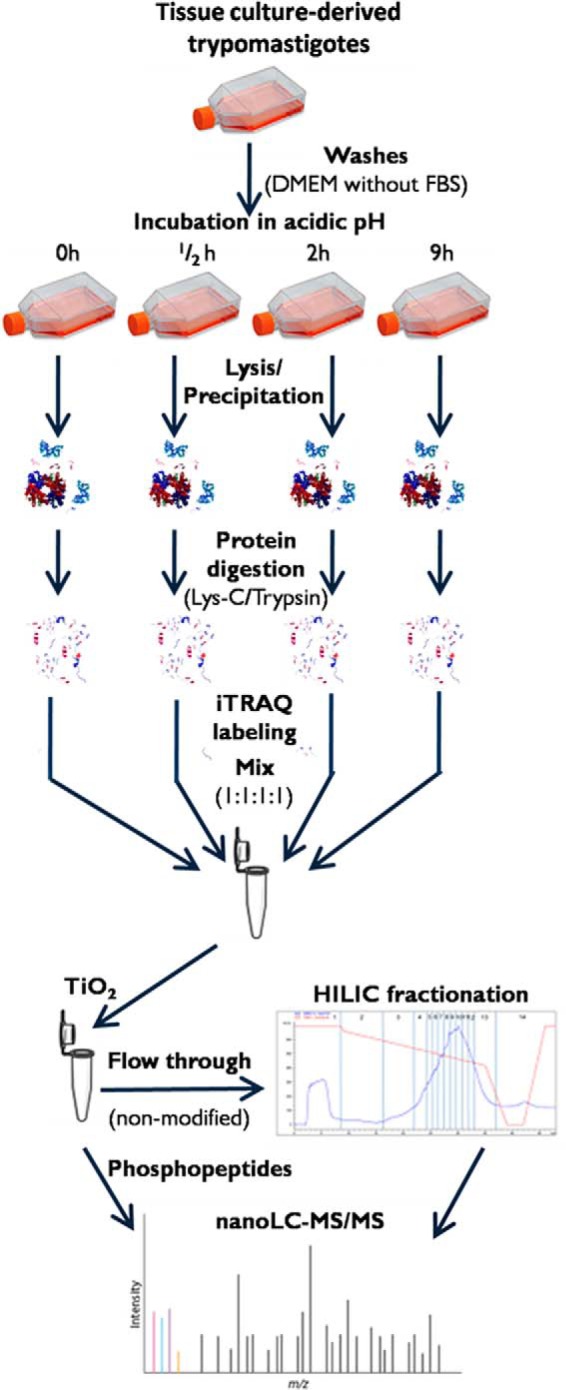
Experimental setup. Tissue-culture-derived trypomastigotes were harvested and washed before incubation in DMEM at pH 5.0 for 0 h, 0.5 h, 2 h, and 9 h. After incubation, the parasites were lysed in 8 m urea. Following protein digestion, peptides were labeled with iTRAQ and combined in 1:1:1:1 ratio. Phosphopeptides were enriched via TiO2 chromatography, and non-modified peptides (flow-through and washes) were pre-fractionated offline on a TSKGel Amide 80 HILIC HPLC column and subjected to LC-MS/MS analysis.
Mass spectrometric analysis of the iTRAQ-labeled peptide samples provided a total of 1339 protein identifications (supplemental Table S1). It was also observed that 551 of them presented statistically significant expression regulation throughout amastigogenesis (supplemental Table S2). Concerning protein phosphorylation, a total of 381 phosphopeptides (belonging to 225 proteins) were identified (supplemental Table S3), of which 183 phosphopeptides (132 proteins) had regulated phosphorylation patterns during amastigogenesis (supplemental Table S4). We observed that all the phosphopeptide regulations were independent of the corresponding protein expression profile.
The Principal Component Analysis plot showed that the variability between replicates at each time point was less than that between different time points, which means relatively good reproducibility was achieved in each condition (supplemental Fig. S1).
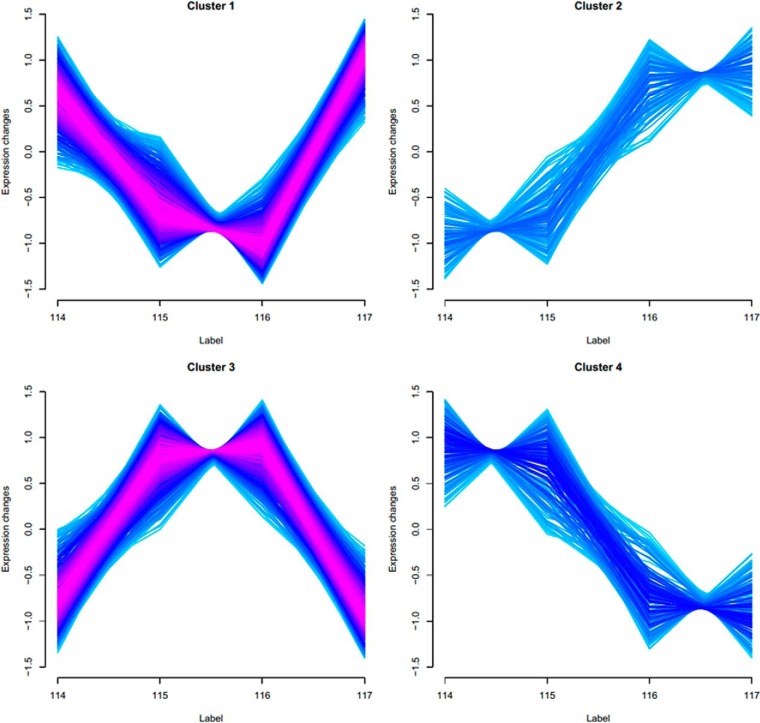
Two validation indices (minimal centroid distance (53) and Xie–Beni index (55)) showed that regulated proteins and phosphopeptides could be grouped in only four clusters (supplemental Fig. S1 and supplemental Table S5). The resulting expression profiles potentially correlate to in vivo events the parasite undergoes inside host cells. As Fig. 2 shows, most differences in abundance were found at intermediate times of the process without large differences between trypomastigote and axenic amastigote life stages (clusters 1 and 3). The other major changes occurred after 2 h of acidic pH incubation, in a situation where the expression in the first two time points (0 and 30 min) was significantly different from that in the last two time points (2 h and 9 h) (clusters 2 and 4). The latter protein expression changes were in agreement with the average induction period of 2 h inside phagolysosomes in vivo (54). Therefore, the results strongly suggest that the distribution of protein expression profiles encompasses protein regulation and phosphorylation events relevant to the major molecular processes involved in T. cruzi amastigogenesis. It was also observed that most of the protein differential expression profiles were grouped in clusters 1 and 3, with 183 and 219 members, respectively, whereas cluster 4 possessed 72 members and cluster 2 had 77 members (supplemental Table S2).
Fig. 2.
Expression profile of regulated proteins and phosphopeptides during axenic amastigogenesis. Regulated proteins and phosphopeptides were grouped in only four clusters: trypomastigotes (114), 30 min of induction (115), 2 h of induction (116), and axenic amastigotes (117).
Profiling Proteome Changes during Amastigogenesis
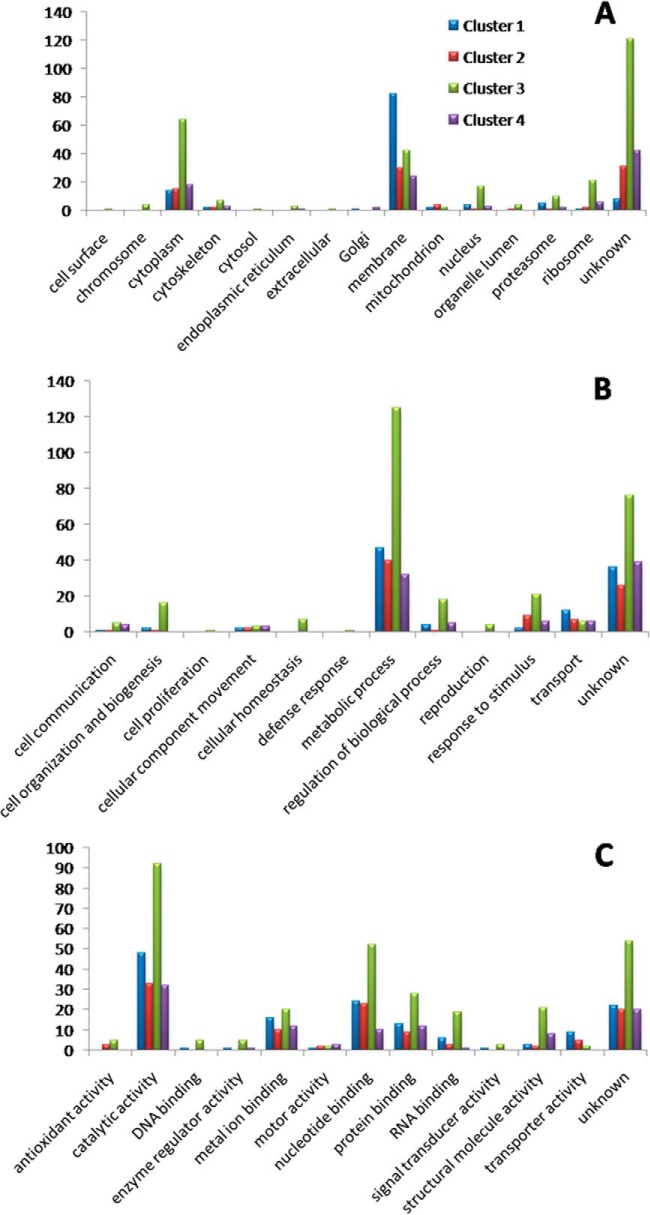
Fig. 3A shows the distribution of GO slim cellular component terms; clusters 1 and 3 covered the greatest diversity of terms. Most of the regulated proteins were annotated under “membrane,” and this term was the most frequent in all clusters. In fact, around 40% of regulated proteins (212 proteins) possessed at least one predicted transmembrane domain (supplemental Table S2). The comparison of the percentage of membrane proteins between all identified proteins and all regulated proteins showed no significant change when we used Fisher's exact text in ProteinCenter software. This indicates that membrane proteins were not preferentially regulated throughout amastigogenesis. Furthermore, the data show that our sample preparation procedures were efficient in recovering membrane proteins, which are prone to be lost in proteomic sample preparations and/or not be well detected in whole cell analysis because of their very low relative abundance in the total cell extract.
Fig. 3.
GO slim terms of proteins with regulated expression levels during axenic amastigogenesis. GO slim cellular component (A), biological activity (B), and molecular function (C). Y-axis represents the number of each GO slim term.
Recently, two comprehensive proteomic surveys of T. cruzi plasma membrane proteins have been reported (56, 57). We observed that most of the 212 proteins regulated during amastigogenesis with predicted transmembrane domains were also detected in the reported plasma membrane subproteome of trypomastigotes and axenic amastigotes. The majority of them (143 proteins) were shared by both life forms; 5 and 34 were reported exclusively on the surface of trypomastigotes and axenic amastigotes, respectively, and only 30 were not detected in the cell surface subproteome.
Proteins involved in metabolic processes were also the most frequent in all clusters concerning biological process (Fig. 3B), as well as proteins annotated with “catalytic activity,” regarding molecular function (Fig. 3C). This indicates pronounced functional and metabolic reorganization throughout the differentiation process, besides the already known ultrastructural rearrangement, which is a consequence of pH induction.
Enzymes
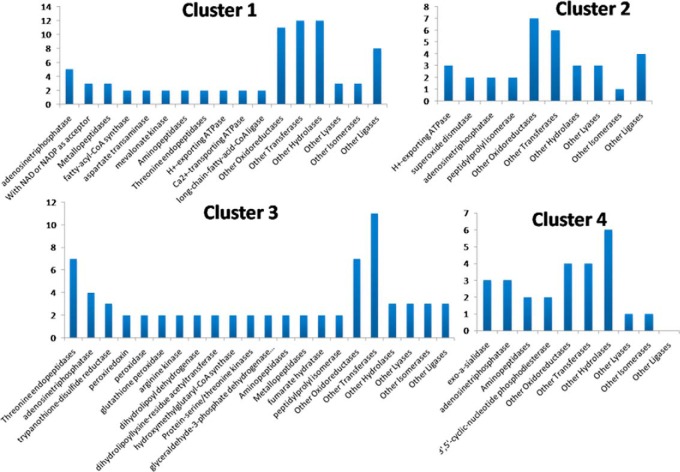
As mentioned before, in all clusters, proteins annotated with “catalytic activity” were the most represented among the molecular function terms (Fig. 3C). Fig. 4 depicts the most represented enzyme activities predicted for proteins in each cluster. Adenosinetriphosphatases (EC:3.6.1.3) had representatives in all clusters. Among them, putative dynein heavy chains reduced expression levels at intermediate points of differentiation (clusters 1 and 4), as did putative proteasome regulatory ATPase subunit 1 (gi 407861152), although subunits 2 and 3 (gi 407849457 and gi 407844541) were overexpressed at intermediate times. These proteasome subunits are annotated under “reproduction.” Also grouped in cluster 3 was a putative transitional endoplasmic reticulum ATPase (gi 322820539), a membrane protein necessary for Golgi stack fragmentation during mitosis and reassembly afterward. This protein is also involved in the formation of the transitional endoplasmic reticulum, the ubiquitin conjugation pathway, and the response to DNA damage (58–61). Furthermore, it demonstrates a qualification for replication. After 2 h of induction, only two ATPases related to protein degradation were overexpressed: the chaperone ClpB (caseinolytic peptidase B), and the putative proteasome activator protein PA26 (gi 407848921). In T. brucei, PA26 was proposed to fulfill regulatory functions in the control of proteasomal degradation of peptides generated by unidentified protein degradation machinery (62).
Fig. 4.
Most represented enzyme activities predicted for proteins in each cluster of regulated proteins during axenic amastigogenesis. Y-axis represents the number of enzymes predicted with each activity.
Membrane proteins with proton-exporting ATPase activity (EC:3.6.3.6) were grouped in clusters 1 and 2. Among them, those underexpressed at intermediate time points (cluster 1) were the SERCA-type calcium-ATPase (gi 4165126) and putative calcium-translocating P-type ATPase (gi 322827561), which also perform Ca2+-transporting activities (EC:3.6.3.8). It is known that the increase in cytosolic Ca2+ concentrations is a key event in T. cruzi metacyclogenesis (63). Reported experiments using fura 2–loaded T. cruzi trypomastigotes upon myoblast cell infection showed that the increase in parasite intracellular Ca2+ concentration occurs after association with the host cell, and after 1 to 2 h of contact, highly fluorescent parasites could be detected inside host cells, as their Ca2+ content was three to five times greater than in extracellular non-associated parasites (64). We might suppose that Ca2+ release is an early event occurring even before 30 min. Also, the acidic pH would repress the expression of the Ca2+ transporters, within at least 2 h, to maintain the intracellular concentration of this second messenger and thereby coordinate processes relevant to differentiation.
Noticeably, T. cruzi expressed a different set of protein kinases at each time point during amastigogenesis. Enzymes with Ser/Thr kinase activity (EC:2.7.11.0) were grouped only in clusters 1 and 3. In cluster 3, NIMA-related kinase (gi 322819192) and protein kinase A catalytic subunit isoform 1 (gi 407847507) are, respectively, known for regulating crucial processes such as the cell cycle (65) and being part of signaling pathways that regulate metabolic processes and differentiation. Several reviews have mentioned the action of PKAs in trypanosomatids (66–68). It has already been reported that T. cruzi PKA regulatory subunits interact with P-type ATPases and also act together with other parasite membrane proteins such as TS family members (69, 70). In cluster 1, the putative MAPK (gi 407831435) was identified. Proteins from the MAPK family are known for regulating cellular proliferation, gene expression, differentiation, mitosis, apoptosis, and other functions (71).
Two transmembrane cyclic-nucleotide phosphodiesterases (EC:3.1.4.17) (gi 55741314 and gi 37543960) were detected only in cluster 4, meaning that their expression was down-regulated during amastigogenesis. These enzymes regulate the intracellular levels of cAMP and cGMP, intracellular second messengers of pathways whose importance to T. cruzi cell proliferation is already known (68, 72). The decrease in their expression after the period during which the parasite remains inside phagolysosomes in vivo reinforces the hypothesis that key processes of induction/regulation of amastigogenesis are achieved within these 2 h. After that point, the process may be irreversibly triggered, with the remaining time possibly necessary for ultrastructural adjustments. Besides that, in T. cruzi cAMP phosphodiesterases have been described as associated with flagellum and seem to be implicated in osmoregulation. Members of this class of enzymes, together with others involved in cAMP signaling pathways, are compartmentalized, and consequently there are microenvironments with distinct levels of this second messenger (73–76).
Also underexpressed after 2 h (cluster 4) were surface glycoproteins such as 85-kDa surface antigen (gi 544414), surface glycoprotein Tc-85/16 (gi 37778153), and trans-sialidase (gi 642913), all of them presenting predicted exo-α-sialidase activities (EC:3.2.1.18). These proteins are involved in host-cell invasion processes and also facilitate escape from the parasitophorous vacuole (77). Therefore, it was not surprising to find a higher content of those proteins in trypomastigotes (0 h) and a decrease in their expression after 2 h. However, we also identified another member of this class of enzyme, the putative trans-sialidase (gi 322826620), with the opposite regulation profile, which possibly indicates another role for this last protein.
Protein Kinases, Phosphatases, and Other Proteins Related to Signal Transduction and Regulation of Metabolic Processes
Among the proteins with regulated expression profiles during amastigogenesis, 19 were annotated with biological functions related to signal transduction, and 31 were involved with the regulation of biological processes, comprising 44 distinct protein kinases, protein phosphatases, and other proteins related to signal transduction and regulation of biological processes (Table I).
Table I. Protein kinases, protein phosphatases, and other proteins related to signal transduction and regulation of biological processes with regulated expression during amastigogenesis.
| I.D. (NCBI) | Cluster | Description (UniProt) | Description (Blast2GO) |
|---|---|---|---|
| gi 407850988 | 1 | Hypothetical protein TCSYLVIO_003802 | Suppressive immunomodulating |
| gi 322820826 | 1 | Isoleucine-tRNA ligase | Isoleucyl-tRNA synthetase |
| gi 322819420 | 1 | Mitochondrial RNA binding protein 1, putative | Mitochondrial RNA binding protein 1 |
| gi 322816735 | 1 | Mitochondrial RNA-binding protein 2, putative | Mitochondrial RNA binding protein 2 |
| gi 407831435 | 1 | Mitogen-activated protein kinase, putative | Mitogen-activated protein |
| gi 407852188 | 1 | Phosphatase 2C, putative | Phosphatase 2C |
| gi 322828909 | 1 | Phosphatase-like protein, putative | Phosphatase-like protein |
| gi 407861152 | 1 | Proteasome regulatory ATPase subunit 1, putative | Proteasome regulatory ATPase subunit 1 |
| gi 407835032 | 1 | Protein kinase, putative, mitogen-activated protein kinase, putative | Mitogen-activated protein kinase |
| gi 322818689 | 1 | Putative uncharacterized protein | ADP-ribosylation factor GTPase activating |
| gi 407843924 | 1 | Serine/threonine protein phosphatase 2A regulatory subunit, putative | Serine threonine protein phosphatase 2A regulatory subunit |
| gi 407847529 | 2 | Poly(A)-binding protein, putative, polyadenylate-binding protein, putative | Poly(A)-binding protein |
| gi 407847514 | 2 | rab7 GTP binding protein, putative | rab7 GTP binding protein |
| gi 407859812 | 3 | 14–3-3 protein, putative | 14–3-3 protein |
| gi 70867395 | 3 | ADP-ribosylation factor 1, putative | ADP-ribosylation factor |
| gi 70883358 | 3 | ADP-ribosylation factor 3, putative | ADP ribosylation factor 3 |
| gi 322823357 | 3 | ADP-ribosylation factor-like protein, putative | ADP-ribosylation factor-like protein |
| gi 6166121 | 3 | Dihydrolipoyl dehydrogenase | Dihydrolipoamide dehydrogenase |
| gi 407848174 | |||
| gi 70886939 | 3 | Eukaryotic initiation factor 5a, putative | Eukaryotic initiation factor 5a |
| gi 34922663 | 3 | Kinetoplastid membrane protein 11 | Kinetoplastid membrane protein-11 |
| gi 322815184 | 3 | Mitochondrial RNA binding protein, putative | RNA binding protein rbp16 |
| gi 322819192 | 3 | NIMA-related kinase, putative | Folate biopterin transporter |
| gi 407849690 | 3 | Protein disulfide isomerase, putative | Protein disulfide isomerase |
| gi 407853650 | 3 | Protein disulfide isomerase, putative | Protein disulfide |
| gi 407847507 | 3 | Protein kinase A catalytic subunit isoform 1, putative | Protein kinase A catalytic subunit isoform 1 |
| gi 28195111 | 3 | Protein kinase A regulatory subunit | Protein kinase A regulatory subunit |
| gi 407850919 | 3 | Protein phosphatase 2C, putative | Protein phosphatase 2C-like protein |
| gi 322815489 | 3 | Protein phosphatase, putative | Protein phosphatase |
| gi 322822336 | 3 | Putative uncharacterized protein | faz1_tryb2 ame: full = flagellar attachment zone protein 1 |
| gi 322820941 | 3 | Putative uncharacterized protein | GTPase activating protein |
| gi 322818216 | 3 | Putative uncharacterized protein | Activator of 90 KDa heat shock protein ATPase-like protein 1 |
| gi 407853753 | 3 | Small GTP-binding protein Rab11, putative, Rab11 GTPase, putative | Small GTP-binding protein |
| gi 407835694 | |||
| gi 322826564 | 3 | Thimet oligopeptidase, putative | Thimet oligopeptidase |
| gi 124109207 | 3 | Thiol transferase Tc52 | Thiol transferase Tc52 |
| gi 136620 | 3 | Trypanothione reductase | Trypanothione reductase |
| gi 19171158 | 3 | Tryparedoxin | Tryparedoxin |
| gi 55741314 | 4 | Cyclic nucleotide phosphodiesterase | cAMP specific |
| gi 37543960 | |||
| gi 407846684 | 4 | GTP-binding nuclear protein rtb2, putative | GTP-binding nuclear protein |
| gi 407850451 | 4 | Hypothetical protein TCSYLVIO_004099 | ras-related protein rab-2a |
| gi 9967363 | 4 | Proteasome regulatory non-ATPase subunit 1–1 | Proteasome regulatory non-ATPase subunit |
| gi 70887024 | - | 60S ribosomal protein L23, putative | 60S ribosomal protein L23 |
In cluster 3 (overexpression at intermediate times of differentiation), we found monomeric GTP-binding proteins such as ADP-ribosylation factors 1 and 3 (gi 70867395 and gi 70883358); putative ADP-ribosylation factor-like (gi 322823357), which regulates vesicle formation, intracellular vesicle traffic, actin remodeling, and phospholipase D activation (78–83); and rab 11 proteins (gi 407853753 and gi 407835694), which also have a role in intracellular vesicle traffic, particularly in endosome recycling (84, 85). In contrast, putative rab 7 (gi 407847514), commonly associated with late endosomes (86), was overexpressed only after 2 h of induction (cluster 2), and a hypothetical protein (gi 407850451) described by Blast2GO software as “ras-related protein rab-2a” whose localization was reported in the endoplasmic reticulum and Golgi apparatus (87, 88) was underexpressed after 2 h (cluster 4). Therefore, our results indicate an increase in vesicular traffic probably due to drastic morphological modification. In other organisms, Rab proteins and their effectors coordinate consecutive stages of transport, such as vesicle formation, vesicle and organelle motility, and tethering of vesicles to their target compartments (89).
Another regulatory protein overexpressed at intermediate time points was the 14–3-3 protein (gi 407859812), which has been shown to interact with a variety of signaling proteins with diverse functions, including protein kinases, phosphatases, and membrane receptors. This means it can act in a variety of vital regulatory processes such as mitogenic signal transduction, apoptosis, and cell-cycle control (90). In addition, in T. brucei, this protein has a central role in motility, cytokinesis, and the cell cycle (91), but to our knowledge so far there has been no study regarding the 14-3-3 protein family in T. cruzi.
The cAMP signaling pathway is among the major process coordinating pathways in T. cruzi (68, 92); therefore, it was not surprising to find MAPK and PKA catalytic subunits with regulated expression levels during amastigogenesis. The fact that these proteins showed opposite expression patterns (clusters 1 and 3, respectively) is novel and relevant information. It was also observed that two membrane cAMP phosphodiesterases that convert cAMP in AMP presented expression patterns distinct from MAPK and PKA. However, no adenylate cyclase was identified with regulated expression.
PKA is a well-known signal transduction mediator in several biological systems, including T. cruzi metacyclogenesis (92). This cAMP-dependent Ser/Thr kinase family is activated upon an increase in cAMP concentration (due to activation of adenylate cyclases and/or inhibition of cAMP phosphodiesterases). Both catalytic and regulatory subunits of TcPKA are localized in the flagellum region and in trypomastigote plasma membrane (70, 93), and the reported interaction with P-type ATPases seems to suggest a role for these ATPases in TcPKA anchoring to the parasite plasma membrane (94). Bao and collaborators published a screening of TcPKA-interacting proteins (69). That study, using a two-hybrid system, listed 38 candidates. Eighteen of them were hypothetical proteins, and eight were proteins presumably relevant in parasite growth regulation, adaptations, and differentiation, including a MAPK and a cAMP-specific phosphodiesterase. MAPKs are also known mediators of signal transduction in higher eukaryotes and are activated, directly or not, by a wide range of stimuli and in many ways, including through Tyr-kinase receptors, G protein–coupled receptors, and PKCs (71). In T. brucei, the deletion of TbMAPK5, a homolog of MAPK, resulted in diminished parasitemia and premature differentiation (95). Also, the deletion of TbMAPK2 affected differentiation (96), and it was reported that TbMAPK4 confers resistance to temperature stress (97). Recently, the TcMAPK2 was also characterized and, although it had high homology with ERK2 from lower eukaryotes, presented significant differences from mammalian homologs (98). In trypomastigotes, these MAPK2s were located mostly along the flagellum, whereas in intracellular amastigotes they were concentrated in the plasma membrane. The phosphorylated (active) form of TcMAPK2 was reported to be more abundant in trypomastigotes than in amastigotes (98). Its interaction with PKA and cAMP-phosphodiesterases indicates cross-talk between different signaling pathways and suggests that TcMAPK plays an important role in regulating proliferation and differentiation. Previous reports on interactions between cAMP and MAPK pathways in T. cruzi, added to our findings of distinct differential protein abundances of key enzymes involved in those pathways during amastigogenesis, indicate the relevance of such pathways in coordinating molecular events during parasite differentiation. Thus, further characterization of such pathways will provide relevant data on T. cruzi biology and lead to potential chemotherapeutic targets.
Protein phosphatases catalyze the dephosphorylation of kinase substrates, including PKA and MAPK, and the use of phosphatase inhibitors has revealed the relevance of such enzymes in T. cruzi amastigogenesis (26, 27). Calyculin A, a potent inhibitor of PP1A and PP2A, induced the transformation of T. cruzi trypomastigotes into amastigote-like forms at pH 7.5 (26). Furthermore, when trypomastigotes were treated with okadaic acid, a potent PP2A inhibitor, during the axenic transformation, after 2 h proteins remained phosphorylated, and this cellular process was blocked (27). In fact, PP2A of T. cruzi has a critical role in the transformation of trypomastigotes into axenic amastigotes related to microtubule stability, probably because after dephosphorylation the flagellar proteins are degraded and the parasite finally reaches the typical round form (27). In our study, we did not detect significant differential expression of the PP2A catalytic subunit, but its putative regulatory subunit (gi 407843924) was underexpressed at intermediate time points in differentiation (cluster 1).
Phosphatase-like (gi 32282890) and a putative PP2C (gi 407852188) were also found in cluster 1. Another putative PP2C (gi 407850919) and a putative protein phosphatase (gi 322815489) presented the opposite expression profile and were placed in cluster 3. Overall the results suggest that serine/threonine phosphatases are involved in phosphorylation and dephosphorylation processes occurring during amastigogenesis.
The Phosphoproteome during Amastigogenesis
Our analysis of phosphorylation levels employing iTRAQ labeling and TiO2-affinity enrichment revealed that unlike in the total proteome, where most of the regulations were observed at intermediate time points (clusters 1 and 3), most of the regulated phosphorylation events were grouped in clusters 2 and 4. In all, 69 phosphopeptides from 49 proteins were grouped in cluster 2, and 61 phosphopeptides from 41 proteins were grouped in cluster 4. Cluster 1 contained 30 phosphopeptides from 27 different proteins, and cluster 3 had 23 phosphopeptides from 15 proteins. Bona fide phosphosite assignments from phosphopeptides with differential abundance in amastigogenesis showed that 165 phosphopeptides presented only one phosphorylated residue, whereas 18 were multi-phosphorylated. All together, we detected, with high confidence, 119 pS, 21 pT, and 1 pY in regulated phosphopeptides in 157, 27, and 1 protein, respectively.
The majority of the significant changes in phosphorylation levels occurred after 2 h of incubation in acidic DMEM (cluster 2 and 4). This indicates that in vivo the control of protein kinase and phosphatase activities induced by acidic medium might happen while the parasite is inside the phagolysosome.
A total of 46 proteins with regulated phosphorylation levels were annotated in ProteinCenter with the term “membrane” and/or presented at least one predicted transmembrane domain and up to nine as in the putative dynein heavy chain (gi 407853700). Also remarkable was the amount of those proteins described as hypothetical, unknown, or uncharacterized that, in total, reached 77 proteins with regulated phosphorylation levels.
As already mentioned, protein kinases regulate several cellular processes and are involved in the most well-known metabolic pathways, especially in signal transduction pathways. Therefore, their activity must be sharply regulated. There are several reviews regarding regulation mechanisms of protein kinases (99–101). One of these mechanisms involves the phosphorylation of residues in the activation loop or in sites in the C or N termini of the protein kinases. We also point out that phosphorylation of residues may play a role in activation or inhibition, depending on the residue and protein.
We detected a total of 10 protein kinases and one phosphatase with a differential phosphorylation pattern in at least one residue during amastigogenesis, and many of these proteins are potentially associated with membranes by at least one transmembrane domain (Table II). Indeed, the phosphorylation occurred in amino acids inside or immediately prior to (three or four residues) the activation loop of three protein kinases including a PKA catalytic subunit.
Table II. Kinases and phosphatase peptides differentially phosphorylated during amastigogenesis.
| I.D. (NCBI) | Cluster | Phosphopeptides | Description (UniProt) | Domain containing the phosphopeptide |
|---|---|---|---|---|
| gi 70885555 | 1 | PEEFDpSQEVDFTK | S-phase kinase-associated protein, putative | – |
| gi 407855987 | 1 | NTIALWEAQApSEGADEERGFGEGPVWVPK | Regulatory subunit of protein kinase A-like protein, putative | – |
| gi 322828093 | 2 | NYMNpSPTQPPVDAAR, GVAEPDpTPR | Serine/threonine-protein phosphatase | – |
| gi 407862997 | 2 | pSLAGSIDYQAPEVLK | Protein kinase+A1:F14, putative | Activation loop/catalytic region |
| gi 10119900 | 2 | GGMpTSHAAVVAR, SGAAApSMPGMMDTVLNLGMNK | Pyruvate phosphate dikinase 2 | PEP-utilizing enzyme, mobile domain; PEP/pyruvate binding domain |
| gi 322818077 | 2 | YFDRYPEpSPRHPLPPLTAK, pTFTLCGTPEYLAPEVIQSR | Protein kinase A catalytic subunit, putative | – |
| gi 407832869 | 3 | AVATPPHpSQQQQQLTAEAIGEATDGR | Serine/threonine protein kinase, putative | – |
| gi 407832551 | 4 | DAASpSDAGDDVTPAK | Regulatory subunit of protein kinase A-like protein, putative | – |
| gi 407846847 | 4 | KLSPSEPNVApYICSR | Glycogen synthase kinase-3 α, putative | Activation loop |
| gi 407847507 | 4 | pTFTLCGTPEYLAPEVIQSK | Protein kinase A catalytic subunit isoform 1, putative | Activation loop/catalytic subunit/regulatory subunit interface |
| gi 407831435 | 4 | AATAALApSAENVLGADHMLR | Mitogen-activated protein kinase, putative | – |
Catalytic subunits of PKA and putative PKA-like presented differential phosphorylation patterns in Ser and Thr residues grouped in clusters 2 and 4, respectively, whereas their regulatory subunits were grouped in clusters 1 and 4. A putative MAPK also presented differential phosphorylation in cluster 4, but this post-translational modification was located in a residue that was not part of a known domain. Also grouped in cluster 4 was the only regulated phospho-Tyr detected on an activation loop that belongs to putative glycogen synthase kinase-3 α (gi 407846847). This protein kinase was already identified and acts on over 40 different proteins in several pathways, influencing cell structure, growth, motility, and apoptosis (102). Although T. cruzi Tyr-specific protein kinases are still unknown, phosphorylation of Tyr-residues in T. cruzi and T. brucei (103–105) has been reported. Compared with other eukaryotic genomes, T. cruzi has a lower proportion of Tyr-phosphatases, a higher number of Ser/Thr-phosphatases, and atypical phosphatases of which the double-specific are the majority, accounting for more than one-third of these enzymes (106). It is also worth mentioning that so far no Tyr-specific kinase has been described in T. cruzi (105).
Only one protein phosphatase presented differentially phosphorylated residues. It was a phospho-Ser and a phospho-Thr in a putative Ser/Thr-phosphatase (gi 322828093) whose profile was grouped in cluster 2.
Prediction of Kinases Responsible for Regulated Phosphorylation Events and Enriched Phosphorylation Motif Analysis
It is accepted that short linear motifs around the phosphorylation sites provide primary specificity for kinase-substrate recognition (107–110). We used iGPS software (44) to predict the potential phosphorylation sites in the sequences from the proteins that presented regulated phosphorylation levels, as well as the protein kinases responsible for these events. In addition, we manually correlated the predicted phosphopeptides to our experimental data (supplemental Table S6). Only 19 identified phosphosites (18 proteins) could be correlated to at least one protein kinase group through the prediction algorithm. This is probably due to the fact that the prediction is based on human kinases and the evolutionary distance between organisms might hamper any assignment, or because the current knowledge of kinase recognition motifs is not complete. Indeed, it has already been reported that many trypanosomatid eukaryotic protein kinases show no strong similarity to any of the recognized eukaryotic protein kinase families, suggesting that these enzymes differ in structure and function from mammalian eukaryotic protein kinases (29).
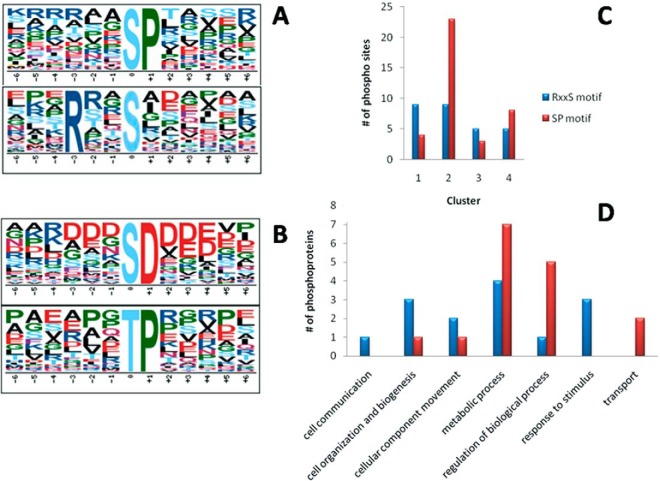
Consequently we decided to search for enriched phosphorylation motifs in both total and regulated phosphopeptides identified. Using windows of 13, 15, and 17 amino acids around the phosphorylation site did not change the outcome. Also, motif enrichment analysis against human and yeast (instead of T. cruzi) databases did not provide different results (data not shown). Our analysis showed that two phosphorylation motifs could be enriched out of regulated phosphorylation sites during amastigogenesis (Fig. 5A), and using all bona fide phosphorylation sites detected further evinced an additional two motifs (Fig. 5B). Also, no phosphoprotein contained phosphorylation events in both enriched motifs. These motifs can be used to direct future studies regarding kinases relevant to amastigogenesis.
Fig. 5.
Consensus motif enrichment analysis. Overrepresented phosphorylation motifs among regulated phosphorylation sites relative to total T. cruzi database (A). Overrepresented phosphorylation motifs among all detected phosphorylation sites relative to total T. cruzi database (A and B). Cluster-wise distribution of regulated motifs (C). GO biological process categorization of annotated phosphoproteins containing regulated phosphorylation events within enriched motifs (D).
The abundance of the regulated phosphorylation sites within the RxxS motif was somewhat homogeneous throughout the four clusters, whereas the SP motif was remarkably more prominent in clusters 2 and 4 (Fig. 5C). This correlates the SP motif regulation to the events deriving from 2 h of acidic incubation. Furthermore, the biological nature of the SP and RxxS regulatory events was investigated through GO biological process annotation of phosphoproteins containing those motifs (Fig. 5D). The SP motif was shown to be more related to intracellular events such as transport, regulation of biological processes, and mainly metabolic processes, reinforcing the relevance of this incubation period to amastigogenesis. The RxxS motif was correlated to events like response to stimuli, communication, cell organization, and biogenesis. Indeed, all those processes are highly relevant to the differentiation process, and their cluster-wise distribution and annotation indicate differences in the pathways coordinating each.
CONCLUSIONS
This comprehensive proteome and phosphoproteome analysis of acidic-pH-induced amastigogenesis showed that differential phosphorylation events, as well as regulated protein expression levels, could be grouped in four clusters. Their profile consisted of up- and down-regulations at intermediate times without major differences in fully differentiated life forms, where most proteins were grouped. In addition, up- and down-regulations were also prominent after 2 h of induction, where most phosphopeptides were gathered.
Even without membrane enrichment procedures, we observed that a significant proportion of all regulated proteins consisted of membrane proteins with at least one transmembrane domain. Most of the regulated proteins were enzymes involved in metabolic processes. Another line of evidence that supports this conclusion is the overexpression of reproduction-related proteasome subunits and putative transitional endoplasmic reticulum ATPase.
Curiously, we observed that Ca2+ transporters were down-regulated during amastigogenesis, although it is known that an increase in cytosolic Ca2+ concentration is a key event in parasite differentiation. We assume that the release of Ca2+ through these transporters is a very early event occurring even before 30 min of induction and that by that time these proteins are already being degraded. Even so, Ca2+-dependent proteins, such as calmodulin, were overexpressed at the intermediate time points.
The cAMP signaling pathway has been reported to be involved in T. cruzi metacyclogenesis. Here, we observed indications that this pathway also is involved in amastigogenesis. Protein Ser/Thr-kinases, including PKA, were overexpressed at intermediate times, whereas MAPK, another relevant kinase known to communicate with PKA, showed the opposite profile. Moreover, we could also detect differential phosphorylation events on both MAPK and PKA during amastigogenesis. Also, two cAMP phosphodiesterases were differentially expressed, but no adenylate cyclase enzyme was identified with regulated expression.
Phosphatases play a crucial role in amastigogenesis, and we observed down-regulation of regulatory subunits of PP2A and other protein phosphatases post-induction, reinforcing the idea that phosphatases are involved in phosphorylation/dephosphorylation events that coordinate transformation.
We could not correlate most of the regulated phosphorylations to any human protein kinase prediction, which indicates that T. cruzi protein kinases recognize motifs different from those found in humans. This further justifies targeting parasite kinases for chemotherapeutic intervention because of their potential specificity, and we have provided some phosphorylation motifs that could be used to guide future studies on T. cruzi kinases.
Overall, this work provides a comprehensive understanding of the molecular basis of T. cruzi amastigogenesis by showing several potential proteins and pathways directly related to pH-induced differentiation and consequent phosphorylation events that regulate or might regulate biochemical processes.
The regulated enzymes, pathways, and phosphorylation events shown here are good potential candidates for chemotherapeutic intervention, because they are key participants in the differentiation process of human-dwelling T. cruzi. Several protein inhibitors could be assigned and the parasite's response to them could be reported in future studies.
Supplementary Material
Footnotes
Author contributions: R.M.Q., S.C., P.R., and C.A.R. designed research; R.M.Q. and S.C.M. performed research; S.C., B.D.L., P.R., and C.A.R. contributed new reagents or analytic tools; R.M.Q., S.C., V.S., P.R., and C.A.R. analyzed data; R.M.Q., S.C., S.C.M., V.S., B.D.L., P.R., and C.A.R. wrote the paper.
* This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq Grant No. 563998/2010-5 and Ph.D. fellowship to R.M.L.Q.), FAPEG (Fundação de Amparo a Pesquisa do Estado de Goiás), CAPES (Programa Nacional de Incentivo a Pesquisa em Parasitologia Basica Grant No. 23038.005298/2011-83), and FINEP (Financiadora de Estudos e Projetos).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PK
- protein kinase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- GO
- gene ontology
- HILIC
- hydrophilic interaction liquid chromatography
- MAPK
- mitogen-activated protein kinase
- PCA
- principal component analysis.
REFERENCES
- 1. Rassi A., Jr., Rassi A., Marin-Neto J. A. (2010) Chagas disease. Lancet 375, 1388–1402 [DOI] [PubMed] [Google Scholar]
- 2. Albajar-Vinas P., Jannin J. (2011) The hidden Chagas disease burden in Europe. Euro Surveillance 16, 2–4 [DOI] [PubMed] [Google Scholar]
- 3. Roca C., Pinazo M. J., Lopez-Chejade P., Bayo J., Posada E., Lopez-Solana J., Gallego M., Portus M., Gascon J. (2011) Chagas disease among the Latin American adult population attending in a primary care center in Barcelona, Spain. PLoS Neglect. Trop. Dis. 5, e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coura J. R., Vinas P. A. (2010) Chagas disease: a new worldwide challenge. Nature 465, S6–S7 [DOI] [PubMed] [Google Scholar]
- 5. Bern C., Montgomery S. P. (2009) An estimate of the burden of Chagas disease in the United States. Clin. Infect. Dis. 49, e52–e54 [DOI] [PubMed] [Google Scholar]
- 6. de Souza W. (1984) Cell biology of Trypanosoma cruzi. Int. Rev. Cytol. 86, 197–283 [DOI] [PubMed] [Google Scholar]
- 7. Coura J. R., Dias J. C. (2009) Epidemiology, control and surveillance of Chagas disease: 100 years after its discovery. Mem. Inst. Oswaldo Cruz 104 Suppl 1, 31–40 [DOI] [PubMed] [Google Scholar]
- 8. Urbina J., Docampo R. (2003) Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 19, 495–501 [DOI] [PubMed] [Google Scholar]
- 9. de Souza W., de Carvalho T. M., Barrias E. S. (2010) Review on Trypanosoma cruzi: host cell interaction. Int. J. Cell Biology. 2010, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Contreras V. T., Salles J. M., Thomas N., Morel C. M., Goldenberg S. (1985) In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 16, 315–327 [DOI] [PubMed] [Google Scholar]
- 11. Parodi-Talice A., Monteiro-Goes V., Arrambide N., Avila A., Duran R., Correa A., Dallagiovanna B., Cayota A., Krieger M., Goldenberg S., Robello C. (2007) Proteomic analysis of metacyclic trypomastigotes undergoing Trypanosoma cruzi metacyclogenesis. J. Mass Spectrom. 42, 1422–1432 [DOI] [PubMed] [Google Scholar]
- 12. de Godoy L. M., Marchini F. K., Pavoni D. P., Rampazzo Rde C., Probst C. M., Goldenberg S., Krieger M. A. (2012) Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics 12, 2694–2703 [DOI] [PubMed] [Google Scholar]
- 13. Favoreto S., Jr., Dorta M. L., Yoshida N. (1998) Trypanosoma cruzi 175-kDa protein tyrosine phosphorylation is associated with host cell invasion. Exp. Parasitol. 89, 188–194 [DOI] [PubMed] [Google Scholar]
- 14. Yoshida N., Favoreto S., Jr., Ferreira A. T., Manque P. M. (2000) Signal transduction induced in Trypanosoma cruzi metacyclic trypomastigotes during the invasion of mammalian cells. Braz. J. Med. Biol. Res. 33, 269–278 [DOI] [PubMed] [Google Scholar]
- 15. Vanhamme L., Pays E. (1995) Control of gene expression in trypanosomes. Microbiol. Rev. 59, 223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brack C. (1968) [Electron microscopic studies on the life cycle of Trypanosoma cruzi with special reference to developmental forms in the vector rhodnius prolixus]. Acta Trop. 25, 289–356 [PubMed] [Google Scholar]
- 17. Contreras V. T., Navarro M. C., De Lima A. R., Arteaga R., Duran F., Askue J., Franco Y. (2002) Production of amastigotes from metacyclic trypomastigotes of Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 97, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 18. Kanbara H., Uemura H., Nakazawa S., Nussenzweig V. (1990) Effect of low pH on transformation of Trypanosoma cruzi trypomastigote to amastigote. Jpn. J. Parasitol. 39, 226–228 [Google Scholar]
- 19. Tomlinson S., Vandekerckhove F., Frevert U., Nussenzweig V. (1995) The induction of Trypanosoma cruzi trypomastigote to amastigote transformation by low pH. Parasitology 110 (Pt 5), 547–554 [DOI] [PubMed] [Google Scholar]
- 20. Rangel-Aldao R., Allende O., Cayama E. (1985) A unique type of cyclic AMP-binding protein of Trypanosoma cruzi. Mol. Biochem. Parasitol. 14, 75–81 [DOI] [PubMed] [Google Scholar]
- 21. Heath S., Hieny S., Sher A. (1990) A cyclic AMP inducible gene expressed during the development of infective stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 43, 133–141 [DOI] [PubMed] [Google Scholar]
- 22. Rangel-Aldao R., Allende O., Triana F., Piras R., Henriquez D., Piras M. (1987) Possible role of cAMP in the differentiation of Trypanosoma cruzi. Mol. Biochem. Parasitol. 22, 39–43 [DOI] [PubMed] [Google Scholar]
- 23. Gonzales-Perdomo M., Romero P., Goldenberg S. (1988) Cyclic AMP and adenylate cyclase activators stimulate Trypanosoma cruzi differentiation. Exp. Parasitol. 66, 205–212 [DOI] [PubMed] [Google Scholar]
- 24. Laxman S., Beavo J. A. (2007) Cyclic nucleotide signaling mechanisms in trypanosomes: possible targets for therapeutic agents. Mol. Interventions 7, 203–215 [DOI] [PubMed] [Google Scholar]
- 25. Shalaby T., Liniger M., Seebeck T. (2001) The regulatory subunit of a cGMP-regulated protein kinase A of Trypanosoma brucei. Eur. J. Biochem. 268, 6197–6206 [DOI] [PubMed] [Google Scholar]
- 26. Grellier P., Blum J., Santana J., Bylen E., Mouray E., Sinou V., Teixeira A. R., Schrevel J. (1999) Involvement of calyculin A-sensitive phosphatase(s) in the differentiation of Trypanosoma cruzi trypomastigotes to amastigotes. Mol. Biochem. Parasitol. 98, 239–252 [DOI] [PubMed] [Google Scholar]
- 27. Gonzalez J., Cornejo A., Santos M. R., Cordero E. M., Gutierrez B., Porcile P., Mortara R. A., Sagua H., Da Silveira J. F., Araya J. E. (2003) A novel protein phosphatase 2A (PP2A) is involved in the transformation of human protozoan parasite Trypanosoma cruzi. Biochem. J. 374, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parsons M., Worthey E. A., Ward P. N., Mottram J. C. (2005) Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naula C., Parsons M., Mottram J. C. (2005) Protein kinases as drug targets in trypanosomes and Leishmania. Biochim. Biophys. Acta 1754, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hubbard M. J., Cohen P. (1993) On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18, 172–177 [DOI] [PubMed] [Google Scholar]
- 31. Graves J. D., Krebs E. G. (1999) Protein phosphorylation and signal transduction. Pharmacol. Ther. 82, 111–121 [DOI] [PubMed] [Google Scholar]
- 32. Force T., Kuida K., Namchuk M., Parang K., Kyriakis J. M. (2004) Inhibitors of protein kinase signaling pathways: emerging therapies for cardiovascular disease. Circulation 109, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 33. Arslan M. A., Kutuk O., Basaga H. (2006) Protein kinases as drug targets in cancer. Curr. Cancer Drug Targets 6, 623–634 [DOI] [PubMed] [Google Scholar]
- 34. Doerig C. (2004) Protein kinases as targets for anti-parasitic chemotherapy. Biochim. Biophys. Acta 1697, 155–168 [DOI] [PubMed] [Google Scholar]
- 35. Andrews N. W., Colli W. (1982) Adhesion and interiorization of Trypanosoma cruzi in mammalian cells. J. Protozool. 29, 264–269 [DOI] [PubMed] [Google Scholar]
- 36. Hernandez-Osorio L. A., Marquez-Duenas C., Florencio-Martinez L. E., Ballesteros-Rodea G., Martinez-Calvillo S., Manning-Cela R. G. (2010) Improved method for in vitro secondary amastigogenesis of Trypanosoma cruzi: morphometrical and molecular analysis of intermediate developmental forms. J. Biomed. Biotechnol. 2010, 283842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116 [DOI] [PubMed] [Google Scholar]
- 38. McNulty D. E., Annan R. S. (2008) Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics 7, 971–980 [DOI] [PubMed] [Google Scholar]
- 39. Palmisano G., Lendal S. E., Engholm-Keller K., Leth-Larsen R., Parker B. L., Larsen M. R. (2010) Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat. Protoc. 5, 1974–1982 [DOI] [PubMed] [Google Scholar]
- 40. Jensen S. S., Larsen M. R. (2007) Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645 [DOI] [PubMed] [Google Scholar]
- 41. Reisinger F., Martens L. (2009) Database on Demand—an online tool for the custom generation of FASTA-formatted sequence databases. Proteomics 9, 4421–4424 [DOI] [PubMed] [Google Scholar]
- 42. Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 43. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 44. Song C., Ye M., Liu Z., Cheng H., Jiang X., Han G., Songyang Z., Tan Y., Wang H., Ren J., Xue Y., Zou H. (2012) Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell. Proteomics 11, 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 46. Taverner T., Karpievitch Y. V., Polpitiya A. D., Brown J. N., Dabney A. R., Anderson G. A., Smith R. D. (2012) DanteR: an extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 28, 2404–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smyth G. K. (2005) Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer, New York: Editors: Robert Gentleman, Vince Carey, Wolfgang Huber, Rafael Irizarry, Sandrine Dudoit, 397–420 [Google Scholar]
- 48. Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 [DOI] [PubMed] [Google Scholar]
- 49. Schwammle V., Leon I. R., Jensen O. N. (2013) Assessment and improvement of statistical tools for comparative proteomics analysis of sparse data sets with few experimental replicates. J. Proteome Res. 12, 3874–3883 [DOI] [PubMed] [Google Scholar]
- 50. Storey J. D. (2002) A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498 [Google Scholar]
- 51. Futschik M. E., Carlisle B. (2005) Noise-robust soft clustering of gene expression time-course data. J. Bioinformat. Comput. Biol. 3, 965–988 [DOI] [PubMed] [Google Scholar]
- 52. Bezdek J. C. (1973) Cluster validity with fuzzy sets. J. Cybernetics 3, 58–73 [Google Scholar]
- 53. Schwammle V., Jensen O. N. (2010) A simple and fast method to determine the parameters for fuzzy c-means cluster analysis. Bioinformatics 26, 2841–2848 [DOI] [PubMed] [Google Scholar]
- 54. Burleigh B. A., Andrews N. W. (1995) The mechanisms of Trypanosoma cruzi invasion of mammalian cells. Ann. Rev. Microbiol. 49, 175–200 [DOI] [PubMed] [Google Scholar]
- 55. Xie X. L., Beni G. (1991) A validity measure for fuzzy clustering. IEEE Trans. Pattern Anal. Mach. Intell. 13, 841–847 [Google Scholar]
- 56. Queiroz R. M., Charneau S., Bastos I. M., Santana J. M., Sousa M. V., Roepstorff P., Ricart C. A. (2014) Cell surface proteome analysis of human-hosted Trypanosoma cruzi life stages. J. Proteome Res. 13, 3530–3541 [DOI] [PubMed] [Google Scholar]
- 57. Queiroz R. M., Charneau S., Motta F. N., Santana J. M., Roepstorff P., Ricart C. A. (2013) Comprehensive proteomic analysis of Trypanosoma cruzi epimastigote cell surface proteins by two complementary methods. J. Proteome Res. 12, 3255–3263 [DOI] [PubMed] [Google Scholar]
- 58. Ishigaki S., Hishikawa N., Niwa J., Iemura S., Natsume T., Hori S., Kakizuka A., Tanaka K., Sobue G. (2004) Physical and functional interaction between Dorfin and Valosin-containing protein that are colocalized in ubiquitylated inclusions in neurodegenerative disorders. J. Biol. Chem. 279, 51376–51385 [DOI] [PubMed] [Google Scholar]
- 59. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 60. Acs K., Luijsterburg M. S., Ackermann L., Salomons F. A., Hoppe T., Dantuma N. P. (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 61. Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hubscher U., Meyer H., Ramadan K. (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 13, 1376–1382 [DOI] [PubMed] [Google Scholar]
- 62. Yao Y., Huang L., Krutchinsky A., Wong M. L., Standing K. G., Burlingame A. L., Wang C. C. (1999) Structural and functional characterizations of the proteasome-activating protein PA26 from Trypanosoma brucei. J. Biol. Chem. 274, 33921–33930 [DOI] [PubMed] [Google Scholar]
- 63. Lammel E. M., Barbieri M. A., Wilkowsky S. E., Bertini F., Isola E. L. (1996) Trypanosoma cruzi: involvement of intracellular calcium in multiplication and differentiation. Exp. Parasitol. 83, 240–249 [DOI] [PubMed] [Google Scholar]
- 64. Moreno S. N., Silva J., Vercesi A. E., Docampo R. (1994) Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J. Exp. Med. 180, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O'Regan L., Blot J., Fry A. M. (2007) Mitotic regulation by NIMA-related kinases. Cell Div. 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Naula C., Seebeck T. (2000) Cyclic AMP signaling in trypanosomatids. Parasitol. Today 16, 35–38 [DOI] [PubMed] [Google Scholar]
- 67. Seebeck T., Gong K., Kunz S., Schaub R., Shalaby T., Zoraghi R. (2001) cAMP signalling in Trypanosoma brucei. Int. J. Parasitol. 31, 491–498 [DOI] [PubMed] [Google Scholar]
- 68. Huang H. (2011) Signal transduction in Trypanosoma cruzi. Adv. Parasitol. 75, 325–344 [DOI] [PubMed] [Google Scholar]
- 69. Bao Y., Weiss L. M., Braunstein V. L., Huang H. (2008) Role of protein kinase A in Trypanosoma cruzi. Infect. Immun. 76, 4757–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bao Y., Weiss L. M., Ma Y. F., Kahn S., Huang H. (2010) Protein kinase A catalytic subunit interacts and phosphorylates members of trans-sialidase super-family in Trypanosoma cruzi. Microbes Infect. 12, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocrine Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 72. Santos D. O., Oliveira M. M. (1988) Effect of cAMP on macromolecule synthesis in the pathogenic protozoa Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 83, 287–292 [DOI] [PubMed] [Google Scholar]
- 73. Kunz S., Oberholzer M., Seebeck T. (2005) A FYVE-containing unusual cyclic nucleotide phosphodiesterase from Trypanosoma cruzi. FEBS J. 272, 6412–6422 [DOI] [PubMed] [Google Scholar]
- 74. Alonso G. D., Schoijet A. C., Torres H. N., Flawia M. M. (2007) TcrPDEA1, a cAMP-specific phosphodiesterase with atypical pharmacological properties from Trypanosoma cruzi. Mol. Biochem. Parasitol. 152, 72–79 [DOI] [PubMed] [Google Scholar]
- 75. Alonso G. D., Schoijet A. C., Torres H. N., Flawia M. M. (2006) TcPDE4, a novel membrane-associated cAMP-specific phosphodiesterase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 145, 40–49 [DOI] [PubMed] [Google Scholar]
- 76. Schoijet A. C., Miranda K., Medeiros L. C., de Souza W., Flawia M. M., Torres H. N., Pignataro O. P., Docampo R., Alonso G. D. (2011) Defining the role of a FYVE domain in the localization and activity of a cAMP phosphodiesterase implicated in osmoregulation in Trypanosoma cruzi. Mol. Microbiol. 79, 50–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Andrews N. W. (1994) From lysosomes into the cytosol: the intracellular pathway of Trypanosoma cruzi. Braz. J. Med. Biol. Res. 27, 471–475 [PubMed] [Google Scholar]
- 78. Boman A. L., Kahn R. A. (1995) Arf proteins: the membrane traffic police? Trends Biochem. Sci. 20, 147–150 [DOI] [PubMed] [Google Scholar]
- 79. Moss J., Vaughan M. (1995) Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J. Biol. Chem. 270, 12327–12330 [DOI] [PubMed] [Google Scholar]
- 80. Park J. B., Kim J. H., Kim Y., Ha S. H., Yoo J. S., Du G., Frohman M. A., Suh P. G., Ryu S. H. (2000) Cardiac phospholipase D2 localizes to sarcolemmal membranes and is inhibited by alpha-actinin in an ADP-ribosylation factor-reversible manner. J. Biol. Chem. 275, 21295–21301 [DOI] [PubMed] [Google Scholar]
- 81. Pasqualato S., Renault L., Cherfils J. (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Rep. 3, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nie Z., Hirsch D. S., Randazzo P. A. (2003) Arf and its many interactors. Curr. Opin. Cell Biol. 15, 396–404 [DOI] [PubMed] [Google Scholar]
- 83. Kawasaki M., Nakayama K., Wakatsuki S. (2005) Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr. Opin. Struct. Biol. 15, 681–689 [DOI] [PubMed] [Google Scholar]
- 84. Stenmark H., Olkkonen V. M. (2001) The Rab GTPase family. Genome Biol. 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Grosshans B. L., Ortiz D., Novick P. (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Soldati T., Rancano C., Geissler H., Pfeffer S. R. (1995) Rab7 and Rab9 are recruited onto late endosomes by biochemically distinguishable processes. J. Biol. Chem. 270, 25541–25548 [DOI] [PubMed] [Google Scholar]
- 87. Short B., Preisinger C., Korner R., Kopajtich R., Byron O., Barr F. A. (2001) A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 89. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 90. Fu H., Subramanian R. R., Masters S. C. (2000) 14–3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 [DOI] [PubMed] [Google Scholar]
- 91. Inoue M., Nakamura Y., Yasuda K., Yasaka N., Hara T., Schnaufer A., Stuart K., Fukuma T. (2005) The 14–3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J. Biol. Chem. 280, 14085–14096 [DOI] [PubMed] [Google Scholar]
- 92. Flawia M. M., Tellez-Inon M. T., Torres H. N. (1997) Signal transduction mechanisms in Trypanosoma cruzi. Parasitol. Today 13, 30–33 [DOI] [PubMed] [Google Scholar]
- 93. Huang H., Weiss L. M., Nagajyothi F., Tanowitz H. B., Wittner M., Orr G. A., Bao Y. (2006) Molecular cloning and characterization of the protein kinase A regulatory subunit of Trypanosoma cruzi. Mol. Biochem. Parasitol. 149, 242–245 [DOI] [PubMed] [Google Scholar]
- 94. Bao Y., Weiss L. M., Hashimoto M., Nara T., Huang H. (2009) Protein kinase A regulatory subunit interacts with P-Type ATPases in Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 80, 941–943 [PubMed] [Google Scholar]
- 95. Domenicali Pfister D., Burkard G., Morand S., Renggli C. K., Roditi I., Vassella E. (2006) A mitogen-activated protein kinase controls differentiation of bloodstream forms of Trypanosoma brucei. Eukaryot. Cell 5, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Muller I. B., Domenicali-Pfister D., Roditi I., Vassella E. (2002) Stage-specific requirement of a mitogen-activated protein kinase by Trypanosoma brucei. Mol. Biol. Cell 13, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Guttinger A., Schwab C., Morand S., Roditi I., Vassella E. (2007) A mitogen-activated protein kinase of Trypanosoma brucei confers resistance to temperature stress. Mol. Biochem. Parasitol. 153, 203–206 [DOI] [PubMed] [Google Scholar]
- 98. Bao Y., Weiss L. M., Ma Y. F., Lisanti M. P., Tanowitz H. B., Das B. C., Zheng R., Huang H. (2010) Molecular cloning and characterization of mitogen-activated protein kinase 2 in Trypanosoma cruzi. Cell Cycle 9, 2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Johnson L. N., Noble M. E., Owen D. J. (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158 [DOI] [PubMed] [Google Scholar]
- 100. Hubbard S. R. (2002) Protein tyrosine kinases: autoregulation and small-molecule inhibition. Curr. Opin. Struct. Biol. 12, 735–741 [DOI] [PubMed] [Google Scholar]
- 101. Hubbard S. R. (1999) Src autoinhibition: let us count the ways. Nat. Struct. Biol. 6, 711–714 [DOI] [PubMed] [Google Scholar]
- 102. Jope R. S., Johnson G. V. (2004) The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 103. Parsons M., Valentine M., Deans J., Schieven G. L., Ledbetter J. A. (1991) Distinct patterns of tyrosine phosphorylation during the life cycle of Trypanosoma brucei. Mol. Biochem. Parasitol. 45, 241–248 [DOI] [PubMed] [Google Scholar]
- 104. Nett I. R., Davidson L., Lamont D., Ferguson M. A. (2009) Identification and specific localization of tyrosine-phosphorylated proteins in Trypanosoma brucei. Eukaryot. Cell 8, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marchini F. K., de Godoy L. M., Rampazzo R. C., Pavoni D. P., Probst C. M., Gnad F., Mann M., Krieger M. A. (2011) Profiling the Trypanosoma cruzi phosphoproteome. PLoS One 6, e25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brenchley R., Tariq H., McElhinney H., Szoor B., Huxley-Jones J., Stevens R., Matthews K., Tabernero L. (2007) The TriTryp phosphatome: analysis of the protein phosphatase catalytic domains. BMC Genomics 8, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ubersax J. A., Ferrell J. E., Jr. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 108. Kobe B., Kampmann T., Forwood J. K., Listwan P., Brinkworth R. I. (2005) Substrate specificity of protein kinases and computational prediction of substrates. Biochim. Biophys. Acta 1754, 200–209 [DOI] [PubMed] [Google Scholar]
- 109. Kreegipuu A., Blom N., Brunak S., Jarv J. (1998) Statistical analysis of protein kinase specificity determinants. FEBS Lett. 430, 45–50 [DOI] [PubMed] [Google Scholar]
- 110. Songyang Z., Lu K. P., Kwon Y. T., Tsai L. H., Filhol O., Cochet C., Brickey D. A., Soderling T. R., Bartleson C., Graves D. J., DeMaggio A. J., Hoekstra M. F., Blenis J., Hunter T., Cantley L. C. (1996) A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16, 6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometer output files (Raw data), peptide and protein identification files (MGF files), the reference protein sequence database, and annotated spectra of bona fide phosphopeptide identifications have been deposited in a public repository, the PeptideAtlas database (www.peptideatlas.org/PASS/PASS00434), under the dataset tag Tc Amastigogenesis and database identifier PASS00434.