Abstract
Protein–protein interactions (PPIs) are fundamental to the structure and function of protein complexes. Resolving the physical contacts between proteins as they occur in cells is critical to uncovering the molecular details underlying various cellular activities. To advance the study of PPIs in living cells, we have developed a new in vivo cross-linking mass spectrometry platform that couples a novel membrane-permeable, enrichable, and MS-cleavable cross-linker with multistage tandem mass spectrometry. This strategy permits the effective capture, enrichment, and identification of in vivo cross-linked products from mammalian cells and thus enables the determination of protein interaction interfaces. The utility of the developed method has been demonstrated by profiling PPIs in mammalian cells at the proteome scale and the targeted protein complex level. Our work represents a general approach for studying in vivo PPIs and provides a solid foundation for future studies toward the complete mapping of PPI networks in living systems.
Protein–protein interactions (PPIs)1 play a key role in defining protein functions in biological systems. Aberrant PPIs can have drastic effects on biochemical activities essential to cell homeostasis, growth, and proliferation, and thereby lead to various human diseases (1). Consequently, PPI interfaces have been recognized as a new paradigm for drug development. Therefore, mapping PPIs and their interaction interfaces in living cells is critical not only for a comprehensive understanding of protein function and regulation, but also for describing the molecular mechanisms underlying human pathologies and identifying potential targets for better therapeutics.
Several strategies exist for identifying and mapping PPIs, including yeast two-hybrid, protein microarray, and affinity purification mass spectrometry (AP-MS) (2–5). Thanks to new developments in sample preparation strategies, mass spectrometry technologies, and bioinformatics tools, AP-MS has become a powerful and preferred method for studying PPIs at the systems level (6–9). Unlike other approaches, AP-MS experiments allow the capture of protein interactions directly from their natural cellular environment, thus better retaining native protein structures and biologically relevant interactions. In addition, a broader scope of PPI networks can be obtained with greater sensitivity, accuracy, versatility, and speed. Despite the success of this very promising technique, AP-MS experiments can lead to the loss of weak/transient interactions and/or the reorganization of protein interactions during biochemical manipulation under native purification conditions. To circumvent these problems, in vivo chemical cross-linking has been successfully employed to stabilize protein interactions in native cells or tissues prior to cell lysis (10–16). The resulting covalent bonds formed between interacting partners allow affinity purification under stringent and fully denaturing conditions, consequently reducing nonspecific background while preserving stable and weak/transient interactions (12–16). Subsequent mass spectrometric analysis can reveal not only the identities of interacting proteins, but also cross-linked amino acid residues. The latter provides direct molecular evidence describing the physical contacts between and within proteins (17). This information can be used for computational modeling to establish structural topologies of proteins and protein complexes (17–22), as well as for generating experimentally derived protein interaction network topology maps (23, 24). Thus, cross-linking mass spectrometry (XL-MS) strategies represent a powerful and emergent technology that possesses unparalleled capabilities for studying PPIs.
Despite their great potential, current XL-MS studies that have aimed to identify cross-linked peptides have been mostly limited to in vitro cross-linking experiments, with few successfully identifying protein interaction interfaces in living cells (24, 25). This is largely because XL-MS studies remain challenging due to the inherent difficulty in the effective MS detection and accurate identification of cross-linked peptides, as well as in unambiguous assignment of cross-linked residues. In general, cross-linked products are heterogeneous and low in abundance relative to non-cross-linked products. In addition, their MS fragmentation is too complex to be interpreted using conventional database searching tools (17, 26). It is noted that almost all of the current in vivo PPI studies utilize formaldehyde cross-linking because of its membrane permeability and fast kinetics (10–16). However, in comparison to the most commonly used amine reactive NHS ester cross-linkers, identification of formaldehyde cross-linked peptides is even more challenging because of its promiscuous nonspecific reactivity and extremely short spacer length (27). Therefore, further developments in reagents and methods are urgently needed to enable simple MS detection and effective identification of in vivo cross-linked products, and thus allow the mapping of authentic protein contact sites as established in cells, especially for protein complexes.
Various efforts have been made to address the limitations of XL-MS studies, resulting in new developments in bioinformatics tools for improved data interpretation (28–32) and new designs of cross-linking reagents for enhanced MS analysis of cross-linked peptides (24, 33–39). Among these approaches, the development of new cross-linking reagents holds great promise for mapping PPIs on the systems level. One class of cross-linking reagents containing an enrichment handle have been shown to allow selective isolation of cross-linked products from complex mixtures, boosting their detectability by MS (33–35, 40–42). A second class of cross-linkers containing MS-cleavable bonds have proven to be effective in facilitating the unambiguous identification of cross-linked peptides (36–39, 43, 44), as the resulting cross-linked products can be identified based on their characteristic and simplified fragmentation behavior during MS analysis. Therefore, an ideal cross-linking reagent would possess the combined features of both classes of cross-linkers. To advance the study of in vivo PPIs, we have developed a new XL-MS platform based on a novel membrane-permeable, enrichable, and MS-cleavable cross-linker, Azide-A-DSBSO (azide-tagged, acid-cleavable disuccinimidyl bis-sulfoxide), and multistage tandem mass spectrometry (MSn). This new XL-MS strategy has been successfully employed to map in vivo PPIs from mammalian cells at both the proteome scale and the targeted protein complex level.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Bovine cytochrome C (>95% purity) was purchased from Sigma Aldrich (St. Louis, MO). Amicon Ultra 100-kDa, 30-kDa, and 10-kDa NMWL centrifugal filters were purchased from EMD Millipore (Billerica, MA). LaminA/C antibody was purchased from Cell Signaling Technology, Inc. (Danvers, MA). Calnexin and GAPDH antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). Streptavidin agarose resin, high-capacity streptavidin agarose resin, HRP-conjugated streptavidin, and Super Signal West Pico chemiluminescent substrate were purchased from Thermo Scientific (Rockford, IL). Sequencing-grade trypsin was purchased from Promega Corp. (Madison, WI). Endoproteinase Lys-C was purchased from WAKO Chemicals (Osaka, Japan). TPCK-treated trypsin was purchased from Worthington Biochemical Corp (Lakewood, NJ). All other general chemicals for buffers and culture media were purchased from Fisher Scientific or VWR International (Radnor, PA).
Synthesis of Azide-A-DSBSO and BARAC
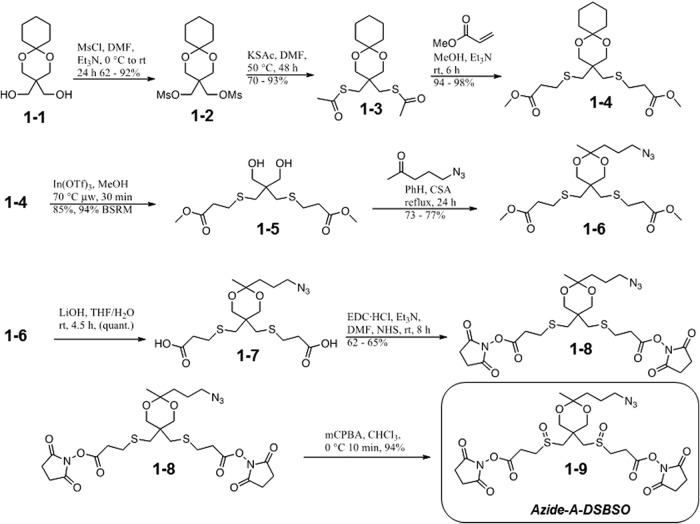
The synthesis and characterization of the Azide-A-DSBSO cross-linker are described in Ref. 55. The simplified scheme is depicted in Fig. 1. BARAC reagent was synthesized as described elsewhere (45).
Fig. 1.
Schematic synthesis scheme for Azide-A-DSBSO.
SDS-PAGE and Immunoblotting Analysis
Protein samples were separated via SDS-PAGE and either stained using Coomassie Blue or transferred to a PVDF membrane and analyzed via immunoblotting. Biotin-conjugated proteins and HB-tagged proteins were detected by streptavidin-HRP conjugate. Cross-linked and non-cross-linked Rpn11-HB and HB-Rpt6 were also detected with streptavidin-HRP conjugate. Lamin A/C, calnexin, and GAPDH were detected using specific primary antibodies and either rabbit or mouse secondary HRP-conjugated antibody. Biotin-conjugated peptides were blotted onto nitrocellulose membrane and detected with streptavidin-HRP conjugate.
In Vitro Cross-linking, Biotin Conjugation, and Enrichment of Azide-A-DSBSO Cross-linked Cytochrome C
Azide-A-DSBSO cross-linking of bovine cytochrome C was similar to that described elsewhere (35). The reaction was quenched with 500 mm NH4HCO3, and samples were ultracentrifuged on 10-kDa NMWL Amicon Ultra centrifugal filters to remove excess cross-linker. Various amounts of BARAC were then reacted with the cross-linked cytochrome C in either phosphate or 8 m urea lysis buffer with agitation overnight. The reaction efficiency for each condition was evaluated via immunoblotting. Following conjugation, excess BARAC was removed by ultracentrifugation and washed with 25 mm NH4HCO3. Biotin-conjugated cytochrome C was purified through binding to streptavidin beads (15).
In Vivo Azide-A-DSBSO Cross-linking of HEK 293 Cells
HEK 293 cells were grown on DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were grown to 80% confluence, trypsinized, washed with PBS, and cross-linked with 2 mm Azide-A-DSBSO in PBS for 1 h with rotation at 37 °C. Following quenching of cross-linking reactions by the addition of 125 mm glycine, cells were pelleted and stored at −80 °C after removal of the supernatant. Frozen cell pellets were lysed in 8 m urea lysis buffer and clarified via centrifugation (15).
Biotin Conjugation and Enrichment Strategy for Azide-A-DSBSO Cross linked Proteins
Azide-A-DSBSO cross-linked 293 cell lysate was reacted with varying concentrations of BARAC with agitation overnight. The resulting biotin-conjugated lysates were analyzed via SDS-PAGE and immunoblot analysis to determine the conjugation efficiency. Bound proteins were reduced with 2 mm tris(2-carboxyethyl)phosphine for 30 min at room temperature and alkylated using 50 mm chloroacetamide in the dark at room temperature for 30 min prior to overnight digestion with 2% Lys-C (w/w) at 37 °C and subsequent overnight digestion with 2% trypsin TPCK (w/w) at 37 °C. The Lys-C/trypsin combination is preferred for proteins purified under fully denaturing conditions to achieve optimal digestion efficiency. Non-cross-linked peptides were extracted and analyzed directly via LC-MS/MS, whereas streptavidin-bound peptides were first acid-cleaved from beads with 20% formic acid, 20% acetonitrile overnight before LC-MSn analysis.
Biotin Conjugation and Enrichment Strategy for Azide-A-DSBSO Cross-linked Peptides
In vivo cross-linked proteins in 293 cell lysates were concentrated using 100-kDa NMWL Amicon Ultra centrifugal filters, and the resulting filtrates were then passed through 30-kDa NMWL Amicon Ultra centrifugal filters. Proteins remaining on both membranes were reduced with 2 mm tris(2-carboxyethyl)phosphine for 30 min at room temperature and then alkylated with 50 mm chloroacetamide at room temperature in the dark for 30 min prior to a 3% Lys-C (w/w) overnight digestion at 37 °C and subsequent 3% trypsin TPCK (w/w) overnight digestion at 37 °C. Digests were collected via centrifugation and reacted with 100 μm BARAC at room temperature with agitation overnight. The biotin-conjugated peptides were then enriched using high-capacity streptavidin agarose resin. Bound peptides were acid-cleaved and then submitted for LC-MSn analysis.
Affinity Purification of in Vivo Azide-A-DSBSO Cross-linked Proteasome Complexes and Subsequent Enrichment of Cross-linked Peptides
Stable 293 cell lines expressing an HB tagged proteasome subunit (Rpn11 or Rpt6) were grown to confluence in DMEM containing 10% FBS and 1% Pen/strep. The cells were washed with PBS and cross-linked and quenched as described above. The cells were lysed in 8 m urea denaturing lysis buffer (15). The cleared lysates were subjected to HB-tag-based tandem affinity purification, which involved binding to Ni2+-Sepharose beads followed by binding to streptavidin resins (15). Proteins bound on beads were reduced, alkylated, and then incubated in 250 μm BARAC with rotation at room temperature overnight in 8 m urea buffer. After extensive washing, bound proteins were digested by Lys-C and trypsin (15). The peptides freed into solution during digestion were subjected to further enrichment through binding to Neutravidin resin for 1 h at room temperature, and cross-linked peptides were acid-eluted as described. The enriched cross-linked peptides were then subjected to LC-MSn analysis.
Analysis of Cross-linked Peptides by LC-MSn
Most of the enriched cross-linked peptides were analyzed via LC-MSn using an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA) coupled on-line with either an Eksigent NanoLC system (Dublin, CA) or an EASY-nLC-1000 (Thermo Scientific, San Jose, CA). A few cross-linked samples from intact cells were analyzed using an Orbitrap Elite mass spectrometer (courtesy of Thermo Scientific Demo Lab, San Jose, CA) coupled on-line with an EASY-nLC 1000 (Thermo Scientific). LC-MSn data acquisition and analysis were as described (20). Only ions with charge of 3+ or more in the MS1 scan were selected for MS2 analysis.
Identification of Cross-linked Peptides via Database Searching
Because of the similarity between DSBSO and DSSO, the general data analysis workflow for the identification of DSBSO interlinked peptides via LC-MSn is the same as the analysis of DSSO cross-linked peptides (20, 36). Using the Batch-Tag software within a developmental version of Protein Prospector (v5.10.10, University of California San Francisco), MS2 and MS3 spectra were searched against a decoy database consisting of a normal Swiss-Prot database concatenated with its randomized version (SwissProt.2013.3.1.random.concat with a total of 454,402 protein entries). Homo sapiens was set as the species (20,501 entries) for analyzing data from human cells. The mass tolerances for parent ions and fragment ions were set as ±20 ppm and 0.6 Da, respectively. For Lys-C/trypsin digests, trypsin was set as the enzyme with a maximum of three missed cleavages allowed. Cysteine carbamidomethylation was set as a constant modification. Protein N-terminal acetylation, asparagine deamidation, N-terminal conversion of glutamine to pyroglutamic acid, and methionine oxidation were selected as variable modifications. Similar to DSSO cross-linked peptides, DSBSO cross-linked peptides display unique and characteristic MS2 fragmentation patterns corresponding to their cross-linking types. Therefore, three additional defined modifications on uncleaved lysines and free protein N termini were chosen: alkene (C3H2O, + 54 Da), sulfenic acid (C3H4O2S, + 254 Da), and unsaturated thiol (C3H2SO, + 236 Da). These are modifications resulting from collision-induced dissociation cleavage of the DSBSO cross-linked peptides. Proteins were identified with expectation values ≤ 0.01 and a minimum of two unique peptides (15).
The in-house program Link-Hunter is a revised version of the previously written Link-Finder program, designed to automatically validate and summarize cross-linked peptide sequences based on MSn data and database searching results as previously described (20, 36). In addition to checking MS2 spectra for predicted patterns, Link-Hunter automatically correlates sequence data from MS3 to MS2 and MS1 parent masses and reports identified interlinked peptides with two associated sequences.
Interaction Network Mapping and Functional Enrichment Analysis
PPI network graphs resulting from cross-links identified in this work were generated manually in Excel from the final list of identified cross-linked proteins. For comparison with known interactions, the final list of cross-linked interacting proteins was fed into an analysis pipeline that automatically extracts physical (but not genetic) interactions from BioGRID, MINT, and IntAct using their Web services (15). Both PPI networks were imported into and visualized by Cytoscape v2.8.3. Functional enrichment was performed using the DAVID Bioinformatics Resources (v.6.7) Functional Annotation Tool (46). Gene I.D.s were submitted, and the Functional Enrichment Chart with enriched Gene Ontology annotations (cellular compartment and biological processes) was downloaded. Only high-confidence functional annotations (false discovery rate < 0.01) were reported.
Mapping Cross-links to 2.5-Å Nucleosome Crystal Structure
The 2.5-Å nucleosome crystal structure (PDB 3AV1) was downloaded from the Protein Data Bank, and cross-linked residues were mapped and visualized using PyMOL. Distances were measured from the amino group of lysine side chains, also using PyMOL.
RESULTS
Developing a New in Vivo XL-MS Platform for Mapping PPIs in Living Cells
In order to establish a robust in vivo XL-MS workflow, we have designed and synthesized a novel cross-linking reagent, Azide-A-DSBSO (Fig. 1). This multifunctional cross-linker combines the unique features of both enrichable (i.e. azide disuccinimidyl glutarate) (35) and MS-cleavable (i.e. DSSO) cross-linkers previously developed in our lab (36). Azide-A-DSBSO is membrane permeable and has a spacer length of ∼14 Å. In addition, it carries a bio-orthogonal azide tag that functions as an enrichment handle permitting selective isolation of cross-linked proteins and peptides through azide-based conjugation chemistry and subsequent affinity purification (35). The incorporation of an acid-cleavable site adjacent to the azide tag facilitates the purification and recovery of cross-linked peptides. Moreover, the integration of two symmetric sulfoxide groups in the spacer region of Azide-A-DSBSO results in robust MS-cleavable bonds that enable fast and unambiguous identification of cross-linked peptides via MSn analysis (20, 36). Together, these features make Azide-A-DSBSO an ideal reagent for studying PPIs, especially from living cells.
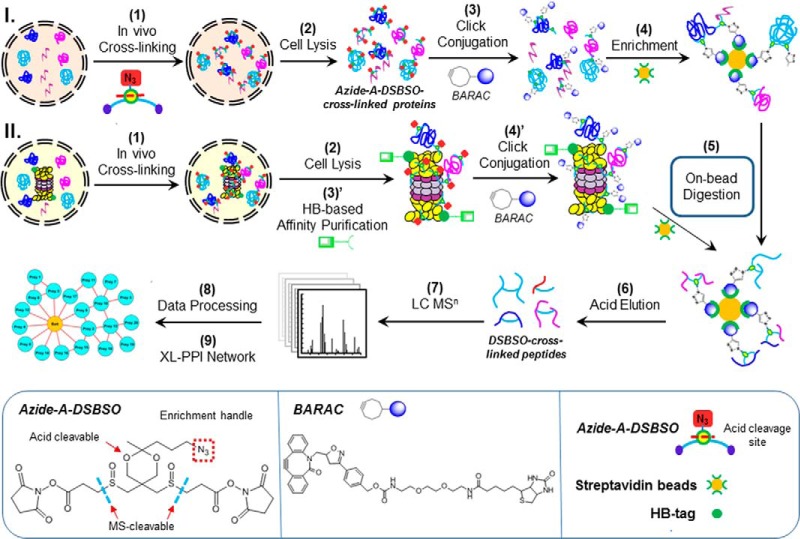
There are nine steps in the general Azide-A-DSBSO-based XL-MS workflow for mapping in vivo PPIs in mammalian cells illustrated in Fig. 2. As shown, in vivo Azide-A-DSBSO cross-linking is first carried out in intact human cells (e.g. HEK 293) (step 1), which are then lysed under fully denaturing conditions (e.g. 8 m urea) to effectively solubilize cross-linked proteins (step 2). To map PPIs on the global scale (path I), the Azide-A-DSBSO cross-linked proteins in cell lysates are conjugated with a biotin-tagged strained alkyne (i.e. BARAC) through copper-free click chemistry (step 3) (47). The resulting biotinylated cross-linked proteins are then enriched via binding to streptavidin resin (step 4). After removal of the non-cross-linked proteins, bound proteins are directly digested on beads (step 5). The biotin-tagged cross-linked peptides are separated from non-cross-linked peptides, as only cross-linked peptides remain bound to streptavidin beads while other peptides are released to the supernatant during digestion. The bound cross-linked peptides are eluted from streptavidin beads by acid cleavage and become the acid-cleaved products of Azide-A-DSBSO peptides, that is, DSBSO cross-linked peptides (step 6) for subsequent LC-MSn analysis (step 7). The presence of an acid cleavage site in Azide-A-DSBSO not only improves enrichment selectivity, but also facilitates subsequent MS analysis by serving to remove the conjugated enrichment handle to yield a smaller mass tag (∼308 Da) on cross-linked peptides. The analysis of LC-MSn data to identify cross-linked peptides (step 8) is similar to that described elsewhere (20, 36). Finally, the identified interlinked peptides can be used to generate an experimentally derived in vivo cross-linked protein–protein interaction network (step 9).
Fig. 2.
The Azide-A-DSBSO-based in vivo XL-MS platform for mapping PPIs in living cells. Path I: at the proteome scale. Path II: at the targeted protein complex level. The inset displays structures of Azide-A-DSBSO and BARAC.
In addition to mapping PPIs in cells at the proteome scale, the same strategy can be modified to study in vivo PPIs of protein complexes (Fig. 2, path II). In this workflow, HB-tag-based tandem affinity purification under fully denaturing conditions is implemented to enable the effective purification of in vivo cross-linked protein complexes as previously reported (12–15, 48). This step is crucial for enhancing the sensitivity and selectivity of subsequent analyses of the selected protein complexes. As shown in Fig. 2, after in vivo cross-linking of 293 cells stably expressing an HB-tagged proteasome subunit (e.g. Rpn11-HB), affinity purification of cross-linked HB-tagged protein complexes is carried out through binding first to Ni2+-Sepharose resins, and then to streptavidin beads (step 3′). The proteins bound to streptavidin beads are directly conjugated with BARAC (step 4′), on-bead digested, and eluted and analyzed via LC-MSn similar to the corresponding steps in path I (steps 5–9).
Selective Enrichment of Azide-A-DSBSO Cross-linked Peptides
One of the key steps in the Azide-A-DSBSO cross-linking strategy is the selective enrichment of cross-linked products. This is achieved by incorporating an affinity tag (e.g. biotin tag) to Azide-A-DSBSO cross-linked products through azide-based conjugation chemistry. Although both copper-catalyzed click chemistry and Staudinger ligation were effective, they have proven to be technically challenging (35). In order to develop a more robust methodology, we adopted a copper-free click chemistry reaction using a biotin-tagged strained alkyne BARAC (47). The tests using Azide-A-DSBSO cross-linked standard protein cytochrome C showed that copper-free conjugation was efficient in both phosphate buffer and buffers containing 8 m urea (supplemental Fig. S1), well suited to our goal of capturing protein interactions in living cells using denaturing buffer. Our results suggest that relative to other azide-based conjugation chemistry methods (35), copper-free chemical conjugation is simpler, more efficient, easier in terms of sample handling, and less labor intensive.
Identification of Azide-A-DSBSO Cross-linked Peptides via LC-MSn
As described above, LC-MSn analysis was performed on the acid-cleaved products of Azide-A-DSBSO cross-linked peptides (i.e. DSBSO cross-linked peptides) (Fig. 3). DSBSO is symmetric and contains two sulfoxide groups that result in four C–S bonds. However, only the two outer C–S bonds proximal to the cross-linked lysines can be cleaved during collision-induced dissociation; the two inner C–S bonds cannot undergo fragmentation because of the lack of β hydrogens (Fig. 3). Given that the same types of MS-cleavable C–S bonds are present in both DSBSO and DSSO, the identification of DSBSO cross-linked peptides by MSn should be as robust as that of DSSO cross-linked peptides (36). This is exemplified by a representative MSn analysis of a DSBSO interlinked peptide (α-β) from in vivo cross-linked 293 cells (Fig. 3). As shown, the cleavage of either of the two MS-cleavable C–S bonds during MS2 analysis leads to the physical separation of the two DSBSO cross-linked peptide constituents, α and β, yielding two characteristic fragment ion pairs (i.e. αA/βT and αT/βA) (Fig. 3A). These MS2 fragment ions are composed of single peptide chains with defined mass modifications (alkene (A) and thiol (T) remnants of DSBSO), which are then subjected to MS3 sequencing for easy identification by conventional database searching tools (Fig. 3B) (36). In addition to MS2 and MS3 data, the MS1 parent ion information is used to further confirm the identities of cross-linked peptides by matching their measured peptide masses to the theoretical masses of predicted cross-linked peptides (Fig. 3C). In this representative example, integration of the MSn data identified the peptide unambiguously as FANYIDK120VR cross-linked to QK139QASHAQLGDAYDQEIR, describing a new interprotein interaction between vimentin and neurofilament medium polypeptide protein in human 293 cells.
Fig. 3.
MSn analysis of a representative DSBSO interlinked peptide (α-β) from in vivo cross-linked human 293 cells. A, top: graphic illustration of characteristic fragmentation of a DSBSO interlinked peptide (α-β) during collision-induced dissociation (CID) in MS2. Bottom: MS1 and MS2 spectra of the representative cross-linked peptide (m/z 869.15524+). In the MS2 spectrum, two pairs of peptide fragments (i.e. αA/βT (m/z 590.312+/1139.502+) and αT/βA (681.312+/1048.002+)) were detected. αA, βA, and KA are alkene modified species (+54 Da); αT and βT are unsaturated thiol modified species (+236 Da). B, respective MS3 spectra of αA (m/z 590.312+) and βA (1048.002+) fragments detected in MS2. The detection of a series of y and b ions unambiguously identified their sequences as FANYIDKAVR and QKAQASHAQLGDAYDQEIR, respectively. C, integration of MSn data identified this peptide as a DSBSO interlinked peptide (i.e. [FANYIDK120VR] cross-linked to [QK139QASHAQLGDAYDQEIR]).
Profiling PPIs from Intact HEK 293 Cells
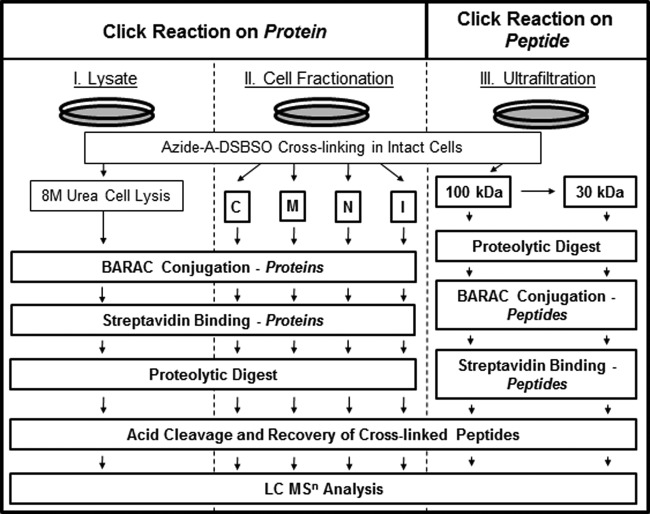
In order to maximize the information acquired on protein interaction interfaces from in vivo cross-linked HEK 293 cells, we explored three different sample preparation strategies, paths I, II, and III (Fig. 4). Path I describes the direct analysis of cross-linked cells as illustrated in Fig. 2. As shown in supplemental Fig. S2, protein cross-linking, BARAC conjugation, and purification of cross-linked products were as effective for in vivo cross-linked cells as for standard proteins. Path II incorporates a subcellular fractionation step before BARAC conjugation, thus dividing cell lysates into four fractions: cytosolic, membrane, nuclear, and insoluble fractions. Immunoblotting analysis revealed that such subcellular fractionation was still possible under our experimental conditions, although there was a marked decrease in the amount of proteins in the cytosolic fraction (supplemental Fig. S3). Nonetheless, BARAC reactions and subsequent purification were also effective for each subcellular fraction (supplemental Fig. S3). The third sample preparation strategy (path III) involved sequential filtration of in vivo cross-linked cell lysates through 100-kDa and then 30-kDa cutoff centrifugal filters to remove small and non-cross-linked components (Fig. 4). The two-step filtration was used to recover a wider range of cross-linked proteins larger than 30 kDa and to improve the dynamic range of subsequent MSn analysis. Proteins remaining on the two different membranes were digested, and the resulting peptides were collected for BARAC conjugation and streptavidin enrichment of cross-linked peptides. As shown in supplemental Fig. S3E, biotin-conjugation and subsequent enrichment were efficient for peptide digests as well, further suggesting the robustness of copper-free click chemistry using BARAC.
Fig. 4.
Three sample preparation strategies for analyzing in vivo Azide-A-DSBSO cross-linked human 293 cells. Biotin conjugation with BARAC on the protein level (Paths I and II) and on the peptide level (Path III) for subsequent enrichment. Path I, BARAC conjugation after cell lysis; Path II, BARAC conjugation after cell fractionation (C = cytosolic; M = membrane; N = nuclear; I = insoluble fractions); Path III, BARAC conjugation and enrichment of cross-linked peptides after 100-kDa and 30-kDa ultrafiltration and digestion.
From all of the samples prepared, we identified a total of 938 proteins in our analyses (supplemental Table S1), 584 of which were identified with at least one cross-linker modified peptide, amounting to 4812 redundant Azide-A-DSBSO labeled peptides, including dead-end, intralinked, and interlinked peptides. Functional annotation of the 584 proteins revealed that they are localized in various cellular compartments and involved in diverse biological processes (supplemental Table S2), demonstrating that Azide-A-DSBSO is well suited for capturing PPIs in cells.
Protein–Protein Interaction Network Mapping
Because of their unique capability for describing PPI interfaces, only interlinked peptides are reported here (supplemental Table S3). In this work, 240 unique interlinked peptides were identified, including 136 intrasubunit and 104 intersubunit interlinks. Using this data, we established an in vivo PPI network map with 85 protein–protein interactions between 54 proteins (supplemental Fig. S4). In comparison to existing PPI databases, 50 novel intra- and intersubunit interactions were identified with direct physical evidence at specific amino acid residues. Among them, an interesting one is between two intermediate filament proteins, NFM and VIME, an interaction confirmed by seven unique interlinked peptides representing seven unique K–K linkages (supplemental Table S3). The identification of interlinked peptides between various domains of NFM and VIME suggests that extensive interaction interfaces exist between these two proteins and potentially implicate VIME involvement in the polymerization or regulation of neurofilament proteins in HEK 293 cells. It has been reported that VIME co-localizes with neurofilament proteins dynamically during neuronal differentiation, and its co-purification with NFH and NFL has been observed (49). Given the close relationship among the three neurofilament subunits NFL, NFM, and NFH, our findings corroborate well with the known function of vimentin in the development of neurofilaments. In general, identifying intermediate filament protein interactions from the native cellular environment is a major challenge, and therefore the ability to capture and directly identify not only which intermediate filament proteins interact, but at which residues, represents a major step forward in this area of research.
It is noted that the most abundant interactions identified in this work resulted from histones and structural proteins (supplemental Table S3), most likely attributed to their abundance as previously reported (24). In total, we identified 118 unique cross-linked peptides among the four (H2A, H2B, H3.2, and H4) core histones, with 47 from H2A-H2B, 13 from H2A-H3.2, 10 from H2B-H4, 8 from H2B-H3.2, 4 from H3.2-H4, and 1 from H2A–H4 pairs. Additionally, we identified 35 unique intraprotein interlinked peptides, with 20 from H2B, 11 from H3.2, 3 from H4, and 1 from H2A (supplemental Table S3). Based on the known nucleosome crystal structures (PDB 3AV1) (supplemental Fig. S5), the distances between the identified cross-linked lysines are <26 Å, which is consistent with other cross-linking studies (23, 36), suggesting that Azide-A-DSBSO has an ideal spacer length for studying protein structures. Collectively, these results have demonstrated the feasibility of the Azide-A-DSBSO-based XL-MS strategy for mapping PPI network topologies from intact cells.
Mapping in Vivo Subunit Interactions of Proteasome Complexes
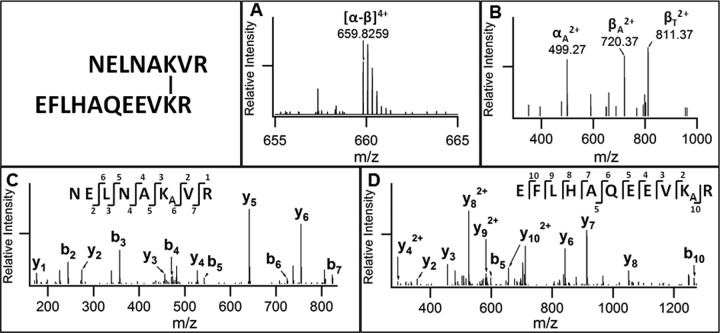
In order to establish an in vivo XL-MS workflow for protein complexes (Fig. 2), we next employed the Azide-A-DSBSO cross-linking strategy to study interactions of proteasome complexes. To capture and isolate proteasome interacting proteins, we used 293 cell lines stably expressing an HB-tagged proteasome subunit (i.e. Rpn11-HB or HB-Rpt6). We first optimized in vivo cross-linking of HB-tagged proteasome complexes. As shown in supplemental Fig. S6, in vivo Azide-A-DSBSO cross-linking of human proteasome complexes and their subsequent HB-tag-based purification were effective. The BARAC conjugation and enrichment of cross-linked peptides was carried out after purification of in vivo cross-linked proteasome complexes as illustrated in Fig. 2 (path II). Fig. 5 illustrates a representative MSn analysis of an in vivo intersubunit interlinked peptide (m/z 659.82594+) from human proteasome complexes. MS2 analysis of this peptide yielded three detectable fragments, αA (m/z 499.272+), βA (m/z 720.372+), and βT (m/z 811.372+), displaying characteristic fragmentation of DSBSO interlinked peptides. Together with MS3 sequencing of αA (m/z 499.272+) and βT (m/z 811.372+) fragments, MSn analysis identified this cross-linked peptide as NELNAK55VR interlinked to EFLHAQEEVK80R unambiguously, which represents an intersubunit interaction between proteasome subunits Rpt6:K55 and Rpt3:K80. In total, MS3 sequencing identified 119 non-redundant cross-linker modified peptides, 54 of which represent 27 unique interlinked peptides (supplemental Tables S4 and S5). Among them, 22 unique interlinked peptides resulted from inter- or intrasubunit cross-links between known subunits of the proteasome complex, including 8 unique intersubunit (i.e. α3-Rpt6, Rpt2-Rpt6, Rpt3-Rpt6, Rpt4-Rpt6, Rpn5-Rpn6, Rpn11-Rpt6, Rpn5-Rpn9, Rpn2-Rpt6) and 11 unique intrasubunit interactions (supplemental Tables S4 and S5). In addition to interactions between proteasome subunits, we identified one interprotein interlink between HSPA1A and HSP8A and three intraprotein interlinked peptides from three known proteasome interacting proteins, HSPA1A, EEF2, and RPS15. In summary, our results demonstrate that our new XL-MS workflow can be generalized to determine protein interaction interfaces of protein complexes in cells.
Fig. 5.
MSn analysis of a representative DSBSO interlinked peptide of in vivo cross-linked proteasome subunits. A, MS1 spectrum of a representative DSBSO interlinked peptide [α-β] (m/z 659.82594+). B, MS2 spectrum of [α-β] (m/z 659.82594+) in which interlinked peptide [α-β] was separated into fragment ions αA (m/z 499.272+), βA (m/z 720.372+), and βT (m/z 811.372+). MS3 spectra of (C) αA and (D) βT fragment ions identified them as peptide NELNAKAVR of proteasome subunit Rpt6 and peptide EFLHAQEEVKTR of proteasome subunit Rpt3, respectively. Integration of MSn data identified this peptide as an intersubunit interlink between Rpt3 and Rpt6. KA, alkene modified lysine; KT, unsaturated thiol modified lysine.
DISCUSSION
Here we have presented a new and general XL-MS workflow based on Azide-A-DSBSO for studying PPIs in living cells. This new XL-MS workflow differs from existing approaches by its collective abilities allowing (i) effective protein cross-linking in vivo to capture authentic protein interactions, (ii) selective enrichment of cross-linked proteins and peptides to improve their detection, (iii) simplified and unambiguous identification of cross-linked peptides by MSn, and (iv) direct coupling with affinity purification of in vivo cross-linked protein complexes to study their interactions. In comparison to existing reagents for in vivo studies (24), the integration of several unique features (i.e. small size, proper spacer length, bio-orthogonal affinity handle, robust MS-cleavable bonds, and acid cleavage site) makes Azide-A-DSBSO a much more attractive reagent for defining protein–protein interactions in cells.
Apart from mapping PPIs at the proteome level, we have successfully coupled the Azide-A-DSBSO-based XL-MS strategy with HB-tag-based affinity purification to delineate the in vivo subunit connectivity of human proteasome complexes for the first time. This work expands the utility of previously developed cross-linking methodologies such as the QTAX (quantitative analysis of tandem affinity purified in vivo cross-linked (x) protein complexes) strategy in studying in vivo interaction networks of protein complexes beyond the identification of interacting partners (12, 13). Interestingly, seven out of eight identified pair-wise interactions corroborated well with previous in vitro XL-MS studies of yeast proteasome complexes (19, 20, 36), suggesting that interaction similarity exists between orthologs as well as between in vivo and in vitro proteasome structures. In this work, we further confirmed the close association between Rpt3 and Rpt6 through the identification of two interlinked peptides at their N-terminal (Rpt3:K80-Rpt6:K55) and central (Rpt3:K238-Rpt6:K222) regions. In addition, the identified contacts between the N termini of Rpt6 and Rpn11, as well as Rpn2, correlate with the electron microscopy structures of yeast proteasomes in which the N-terminal sequences of Rpt3 and Rpt6 form a coil structure for Rpn2 and the lid subcomplex to attach to the base (50, 51). Moreover, a novel interaction between α3 and Rpt6 identified here implies the intimate relationship of Rpt6 and the 20S α ring. It is worth noting that the identification of these in vivo proteasome subunit contacts was possible only when HB-based affinity purification was incorporated into the workflow, indicating the necessity of targeted analysis for profiling PPIs of protein complexes in cells. Importantly, our results have proven the feasibility of the Azide-A-DSBSO based XL-MS strategy for such targeted analysis, demonstrating a unique capability that current strategies do not possess. Although other proteasome components were captured and identified from affinity purified Azide-A-DSBSO cross-linked Rpn11-HB or HB-Rpt6 containing proteasome complexes (data not shown), it appears that direct interactions of protein baits are enriched, as eight of the interactions identified were directly with Rpt6. Although additional baits would be needed to generate a more comprehensive in vivo subunit topology map of the proteasome complex, this would be advantageous when only the direct binding partner needs to be identified. Collectively, this work represents a significant step toward a full understanding of the in vivo PPIs of protein complexes.
In summary, we have successfully developed a new, versatile, and general XL-MS workflow for mapping PPIs at both the proteome scale and the targeted protein complex level, representing a technological advancement in defining protein interactions in living systems. In comparison to previous AP-MS and quantitative tandem affinity purification studies relying on multiple reciprocal purifications and/or existing PPI databases for interaction validation and the construction of in silico interaction network maps (2, 13, 14, 52), our new in vivo XL-MS strategy allows the identification of direct protein interaction contacts for generating interaction networks experimentally. In addition, this information can be used for determining protein structural topologies in future studies. In combination with stable isotope labeling (53) and cross-linking chemistry targeting other residues such as acidic residues (54), new reagents can be further developed to describe PPI dynamics in cells. The potential of this technology is enormous, and with improvements in instrumentation and sample preparation, a vast variety of unexplored biological applications can be envisioned.
Supplementary Material
Acknowledgments
We thank Drs. A. L. Burlingame and Robert Chalkley for supporting the use of Protein Prospector and David Vong for help in data sorting.
Footnotes
Author contributions: R.M.K., X.W., and L.H. designed research; R.M.K., X.W., and A.B. performed research; R.M.K., X.W., A.B., C.Y., W.K., Y.Y., E.J.N., T.S., J.D., A.K., S.G., D.V., S.D.R., and L.H. contributed to developing new reagents and analytical tools, optimizing protocols, and/or analyzing samples; R.M.K., X.W., C.Y., and L.H. analyzed data; R.M.K., X.W., and L.H. wrote the paper; A.B. and S.D.R. designed chemical synthesis; C.Y. contributed to figures, tables, and text; S.D.R. supervised chemical synthesis and edited the paper.
* This work was supported by National Institutes of Health Grant Nos. R01GM074830 and R21CA161807 to L.H. and R01GM106003 to L.H. and S.R.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PPI
- protein–protein interaction
- XL-MS
- cross-linking mass spectrometry
- AP-MS
- affinity purification mass spectrometry
- Azide-A-DSBSO
- azide-tagged, acid-cleavable disuccinimidyl bis-sulfoxide
- BARAC
- biarylazacycloctynone
- HB
- His-Bio
- DSSO
- disuccinimidyl sulfoxide
- MS
- mass spectrometry
- MS2
- tandem mass spectrometry
- MSn
- multi-stage tandem mass spectrometry
- LC
- liquid chromatography
- TPCK
- L-1-tosylamido-2-phenylethyl chloromethyl ketone.
REFERENCES
- 1. Ryan D. P., Matthews J. M. (2005) Protein-protein interactions in human disease. Curr. Opin. Struct. Biol. 15, 441–446 [DOI] [PubMed] [Google Scholar]
- 2. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 3. Kocher T., Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 4. Guan H., Kiss-Toth E. (2008) Advanced technologies for studies on protein interactomes. Adv. Biochem. Eng. Biotechnol. 110, 1–24 [DOI] [PubMed] [Google Scholar]
- 5. Ryan C. J., Cimermancic P., Szpiech Z. A., Sali A., Hernandez R. D., Krogan N. J. (2013) High-resolution network biology: connecting sequence with function. Nat. Rev. Genet. 14, 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng Y., Zhang C., Croucher D. R., Soliman M. A., St-Denis N., Pasculescu A., Taylor L., Tate S. A., Hardy W. R., Colwill K., Dai A. Y., Bagshaw R., Dennis J. W., Gingras A. C., Daly R. J., Pawson T. (2013) Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., Pawson T., Bonner R., Tate S., Gingras A. C. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 9. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasilescu J., Guo X., Kast J. (2004) Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 11. Schmitt-Ulms G., Hansen K., Liu J., Cowdrey C., Yang J., DeArmond S. J., Cohen F. E., Prusiner S. B., Baldwin M. A. (2004) Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat. Biotechnol. 22, 724–731 [DOI] [PubMed] [Google Scholar]
- 12. Guerrero C., Tagwerker C., Kaiser P., Huang L. (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 13. Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaake R. M., Milenkovic T., Przulj N., Kaiser P., Huang L. (2010) Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J. Proteome Res. 9, 2016–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fang L., Kaake R. M., Patel V. R., Yang Y., Baldi P., Huang L. (2012) Mapping the protein interaction network of the human COP9 signalosome complex using a label-free QTAX strategy. Mol. Cell. Proteomics 11, 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tardiff D. F., Abruzzi K. C., Rosbash M. (2007) Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl. Acad. Sci. U.S.A. 104, 19948–19953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greber B. J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J. P., Leibundgut M., Aebersold R., Ban N. (2014) Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 [DOI] [PubMed] [Google Scholar]
- 19. Lasker K., Forster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kao A., Randall A., Yang Y., Patel V. R., Kandur W., Guan S., Rychnovsky S. D., Baldi P., Huang L. (2012) Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. Mol. Cell. Proteomics 11, 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leitner A., Joachimiak L. A., Bracher A., Monkemeyer L., Walzthoeni T., Chen B., Pechmann S., Holmes S., Cong Y., Ma B., Ludtke S., Chiu W., Hartl F. U., Aebersold R., Frydman J. (2012) The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmstrom L., Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 [DOI] [PubMed] [Google Scholar]
- 24. Chavez J. D., Weisbrod C. R., Zheng C., Eng J. K., Bruce J. E. (2013) Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteomics 12, 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang H., Tang X., Munske G. R., Zakharova N., Yang L., Zheng C., Wolff M. A., Tolic N., Anderson G. A., Shi L., Marshall M. J., Fredrickson J. K., Bruce J. E. (2008) In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical cross-linkers. J. Proteome Res. 7, 1712–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 27. Sutherland B. W., Toews J., Kast J. (2008) Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 43, 699–715 [DOI] [PubMed] [Google Scholar]
- 28. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Panchaud A., Singh P., Shaffer S. A., Goodlett D. R. (2010) xComb: a cross-linked peptide database approach to protein-protein interaction analysis. J. Proteome Res. 9, 2508–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Forster F., Beck M., Aebersold R. (2012) False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9, 901–903 [DOI] [PubMed] [Google Scholar]
- 31. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 32. Trnka M. J., Baker P. R., Robinson P. J., Burlingame A. L., Chalkley R. J. (2014) Matching cross-linked peptide spectra: only as good as the worse identification. Mol. Cell. Proteomics 13, 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chu F., Mahrus S., Craik C. S., Burlingame A. L. (2006) Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 128, 10362–10363 [DOI] [PubMed] [Google Scholar]
- 34. Chowdhury S. M., Du X., Tolic N., Wu S., Moore R. J., Mayer M. U., Smith R. D., Adkins J. N. (2009) Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 5524–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vellucci D., Kao A., Kaake R. M., Rychnovsky S. D., Huang L. (2010) Selective enrichment and identification of azide-tagged cross-linked peptides using chemical ligation and mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 1432–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kao A., Chiu C. L., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., Huang L. (2011) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu Y., Tanasova M., Borhan B., Reid G. E. (2008) Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal. Chem. 80, 9279–9287 [DOI] [PubMed] [Google Scholar]
- 38. Petrotchenko E., Borchers C. (2010) ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muller M. Q., Dreiocker F., Ihling C. H., Schafer M., Sinz A. (2010) Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 [DOI] [PubMed] [Google Scholar]
- 40. Tang X., Munske G. R., Siems W. F., Bruce J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 41. Kasper P. T., Back J. W., Vitale M., Hartog A. F., Roseboom W., de Koning L. J., van Maarseveen J. H., Muijsers A. O., de Koster C. G., de Jong L. (2007) An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. Chembiochem 8, 1281–1292 [DOI] [PubMed] [Google Scholar]
- 42. Nessen M. A., Kramer G., Back J., Baskin J. M., Smeenk L. E., de Koning L. J., van Maarseveen J. H., de Jong L., Bertozzi C. R., Hiemstra H., de Koster C. G. (2009) Selective enrichment of azide-containing peptides from complex mixtures. J. Proteome Res. 8, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petrotchenko E. V., Xiao K., Cable J., Chen Y., Dokholyan N. V., Borchers C. H. (2009) BiPS, a photocleavable, isotopically coded, fluorescent cross-linker for structural proteomics. Mol. Cell. Proteomics 8, 273–286 [DOI] [PubMed] [Google Scholar]
- 44. Luo J., Fishburn J., Hahn S., Ranish J. (2012) An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol. Cell. Proteomics 11, M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jewett J. C., Sletten E. M., Bertozzi C. R. (2010) Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 47. Gordon C. G., Mackey J. L., Jewett J. C., Sletten E. M., Houk K. N., Bertozzi C. R. (2012) Reactivity of biarylazacyclooctynones in copper-free click chemistry. J. Am. Chem. Soc. 134, 9199–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 49. Yabe J. T., Chan W. K., Wang F. S., Pimenta A., Ortiz D. D., Shea T. B. (2003) Regulation of the transition from vimentin to neurofilaments during neuronal differentiation. Cell Motil. Cytoskeleton 56, 193–205 [DOI] [PubMed] [Google Scholar]
- 50. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., Sakata E., Nickell S., Plitzko J. M., Villa E., Baumeister W., Forster F. (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 109, 14870–14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gavin A. C., Maeda K., Kuhner S. (2011) Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 [DOI] [PubMed] [Google Scholar]
- 53. Yu C., Kandur W., Kao A., Rychnovsky S., Huang L. (2014) Developing new isotope-coded mass spectrometry-cleavable cross-linkers for elucidating protein structures. Anal. Chem. 86, 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leitner A., Joachimiak L. A., Unverdorben P., Walzthoeni T., Frydman J., Forster F., Aebersold R. (2014) Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9455–9460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burke A. M. (2011) Ph.D. thesis, Reagents for in vivo Protein Cross-Linking and Automated Analysis of Protein-Protein Interactions with Tandem Mass Spectrometry University of California, Irvine [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.