Abstract
Previously, we identified the stress-induced chaperone, Hsp27, as highly overexpressed in castration-resistant prostate cancer and developed an Hsp27 inhibitor (OGX-427) currently tested in phase I/II clinical trials as a chemosensitizing agent in different cancers. To better understand the Hsp27 poorly-defined cytoprotective functions in cancers and increase the OGX-427 pharmacological safety, we established the Hsp27-protein interaction network using a yeast two-hybrid approach and identified 226 interaction partners. As an example, we showed that targeting Hsp27 interaction with TCTP, a partner protein identified in our screen increases therapy sensitivity, opening a new promising field of research for therapeutic approaches that could decrease or abolish toxicity for normal cells. Results of an in-depth bioinformatics network analysis allying the Hsp27 interaction map into the human interactome underlined the multifunctional character of this protein. We identified interactions of Hsp27 with proteins involved in eight well known functions previously related to Hsp27 and uncovered 17 potential new ones, such as DNA repair and RNA splicing. Validation of Hsp27 involvement in both processes in human prostate cancer cells supports our system biology-predicted functions and provides new insights into Hsp27 roles in cancer cells.
Heat Shock Proteins (Hsps)1 are molecular chaperones dedicated to protein homeostasis maintenance by associating with key regulatory proteins such as transcriptional factors, protein kinases, and hormone receptors. Hsps expression is induced in response to a wide variety of stresses and provides strong cytoprotective effects from otherwise lethal conditions (1). They are often abnormally overexpressed in cancer cells, provoking cancer cell survival and therapy-resistance (2, 3). We previously identified Hsp27 (or HSPB1, member of the HSPB family (4)) as a highly overexpressed protein in castration-resistant prostate cancer (CRPC) (5, 6). Hsp27 knockdown using antisense oligonucleotides (ASO) and small interference RNA (siRNA) increases apoptotic rates and enhances hormone- and chemo-therapy activity in CRPC (5–7). We consequently developed and patented a second generation ASO targeting Hsp27 (Patent PCT no 10/605, 498, 2005) that has been licensed (OGX-427). Phase I/II clinical trials are ongoing in prostate, breast, lung, ovarian, and bladder cancers (http://www.oncogenex.ca/) (8). These trials show promising results calling for further definition of the mechanisms leading to Hsp27 cytoprotection. Understanding Hsp27 mechanisms of action in cancer cells will allow the improvement of OGX-427 pharmacological safety and the development of new therapeutic targets and treatment strategies that would have no toxicity for normal tissues.
The best defined role of Hsp27 is its ATP-independent molecular chaperone activity that aids in proper folding and stabilization of non-native proteins (9). Beyond this chaperone activity, evidence show the involvement of Hsp27 in cell cycle progression (10), presentation of oxidized proteins to the proteasome (11), apoptosis inhibition (12), regulation of translation initiation (13), and actin cytoskeleton organization (14). The diversity of Hsp27 cellular functions is the result of specific interactions with partner proteins. For instance, Hsp27 inhibits components of the mitochondrial cell death pathway through interaction with cytochrome-c and caspase-3 (15). Interaction with P53 implicates Hsp27 in cell cycle regulation (10). Identifying Hsp27-interaction partners and functional modules on a large-scale should shed a new light on the cellular processes by which Hsp27 exerts its cytoprotective effects on normal and cancer cells. Indeed, interaction networks are increasingly used to decipher the molecular bases of protein cellular functions because proteins involved in the same molecular complex, pathway, or biological process tend to interact which each other, thereby forming “functional modules” (16).
To decipher Hsp27 interactions and cellular functions, we undertook an interactome mapping strategy followed by a network analysis. We identified 226 Hsp27-interacting partners. Among them, we showed that targeting Hsp27 interaction with TCTP, a partner protein identified in our screen increases therapy sensitivity, opening a new promising field of research for therapeutic approaches that could decrease or abolish toxicity for normal cells. We further predicted 17 newly described Hsp27 cellular functions based on its presence in functional modules such as DNA repair as well as RNA splicing, two processes for which we uncovered and experimentally validated the involvement Hsp27 in CRPC cells, supporting our system biology predicted functions, and providing new insights into Hsp27 roles in cancer cells.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid SRS Yeast Medium and Screening
We used the SRS Y2H CytoTrap (Stratagene, Agilent Technologies, Inc. Massy, France) to screen Hsp27 partner proteins using two human cDNA libraries from testis and cervix cancer HeLa, respectively (Stratagene, Agilent Technologies, Inc., catalog numbers 975205 and 975212 respectively), as previously described (17). The transcriptome repertoire of each library is composed of ∼500,000 transcripts; we screened 5.106 transformants from each library, thus covering each transcriptome 10 times, reaching 95% saturation of each screen. This percentage was calculated by counting the number of already identified interactors over the total number of interactors by screen.
Cell Culture
The CRPC cell line PC-3 and castration sensitive LNCaP were purchased from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco's Modified Eagle's Medium (PC-3) and RPMI 1640 (Roswell Park Memorial Institute) (LNCaP) (Invitrogen, Cergy Pontoise, France), supplemented with 10% fetal bovine serum (FBS).
Interactome Validation by Co-IP and Western blot
Co-IP experiments were carried out to validate Y2H interactions. Cleared lysates from PC-3 cells with adjusted protein concentration (Pierce BCA Protein assay kit, Fisher Scientific, France) were used for co-IP with 8 μg/ml rabbit anti-Hsp27 antibody (Stressgen Bioreagents, Enzo Life Sciences, Villeurbanne, France), rabbit anti-spindlin-1 (SPIN1) (Abnova Le Perray En Yvelines, France), rabbit anti-ENPP2 (Abcam, France), rabbit anti-Ku80 (Cell signaling, Ozyme Montigny-le-Bretonneux, France). Immune complexes were precleared with trueblot anti-rabbit Immunoglobulin (Ig) IP beads (eBiosciences, Paris, France) for 1h at +4 °C, then spinned. The supernatant was incubated with the corresponding antibody or rabbit IgG (as internal control) for 2.5h or O/N at +4 °C. Immune complexes were precipitated after 1h incubation with 30 μl of trueblot anti-rabbit Ig IP beads (eBiosciences, Paris, France). After three washes in cold lysis buffer, the complexes were resuspended in protein sample buffer (Bio-Rad) and boiled for 5min before Western blot was performed as described previously (6) with 1:5000 rabbit anti-Hsp27 polyclonal antibody (Stressgen), 1:50 rabbit anti-ENPP2 polyclonal antibody (Abcam), 1:1000 rabbit anti-FTH1 polyclonal antibody (Cell signaling, Ozyme, France), 1:500 rabbit anti-HUWE1 polyclonal antibody (Abnova, Tebu-bio, France), 1:2000 rabbit anti-SF3A3 polyclonal antibody (Abcam), 1:250 mouse anti-RAB43 monoclonal antibody (Abcam), 1:1000 rabbit anti-DFF45/DFF35 polyclonal antibody (Cell signaling, Ozyme), 1:1000 rabbit anti-PAF1 polyclonal antibody (Abnova), 1:1000 rabbit anti-calreticulin polyclonal antibody (Cell signaling, Ozyme), 1:1000 rabbit anti-IGBP1 polyclonal antibody (Abcam), 1:200 mouse anti-PGM2 monoclonal antibody (Abnova), 1:200 mouse anti-FKBP4 monoclonal antibody (Abnova), 1:500 rabbit anti-GSTO1 polyclonal antibody (Abnova), 1:200 mouse anti-EIF4G2 monoclonal antibody (Abnova), 1:200 rabbit anti-SPIN1 polyclonal antibody (Abnova), 1:1000 rabbit anti-EIF4A polyclonal antibody (Cell signaling, Ozyme), 1:1000 rabbit anti-Ku80 polyclonal antibody (Cell signaling, Ozyme), 1:1000 mouse anti-DNAPKs monoclonal antibody (Cell signaling), and 1:2000 rabbit anti-TCTP polyclonal antibody (Abcam).
Hsp27 Interaction Partners Functional Annotation and Enrichment
The DAVID suite (18) was used to search for Gene Ontology (GO) Biological Process annotations and enrichments. Q-values are hypergeometric p values corrected for multiple testing according to the Benjamini & Hochberg procedure. The significant GO Biological Process terms (q-value<0.02) enriched among the Hsp27 interaction partners were represented using SimCT (19), a web-based tool that provides a simplified subgraph of the ontology, facilitating the interpretation of functional annotations.
Integrated Hsp27 Interactome and Functional Module Identification
A PPI network containing 80,877 binary interactions between 10,282 proteins was built by fusing the 226 interactions we identified in our screen and the protein–protein interaction dataset assembled by Bossi and Lehner (20). The obtained network was partitioned into 751 overlapping modules using the OCG algorithm (21). Among those, 54 modules contained the Hsp27 protein. The biological processes in which the modules are involved were determined using the following: for each module, significant enrichments in GO Biological Process terms were searched among the annotations of their constituent proteins using the DAVID suite (18). Terms with q-values <1E−6 were selected to annotated the modules. When numerous terms were significantly enriched, the complexity was reduced using SimCT (19) in order to choose the more representative terms. When none of the GO terms reached the q-value threshold of 10−6, a majority rule was used to annotate the module. All networks were visualized using Cytoscape (22).
Hsp27 siRNA Transfection
Cells were seeded in a 100 mm dish and were transfected 24h later with 20 nm CTL-siRNA or Hsp27-siRNA, following a pre-incubation for 10–20 min with 20 μl of Lipofectamine® RNAiMAX reagent (Invitrogen, Cergy Pontoise, France) in serum-free OPTI-MEM (Invitrogen, Cergy Pontoise, France). The sequence of Hsp27-siRNA corresponds to human Hsp27 (5′-GCUGCAAAA UCCGAUGAGACdTdT-3′; Dharmacon, Lafayette, CO). The CTL-siRNA duplex (5′-AUC AAACUGUUGUCAGCGCUGdTdT-3′) was used as control. Nontransfected cells were simply treated with Lipofectamine® and OPTI-MEM. The medium was replaced 24 h later by fresh medium and cells were incubated for 72 h.
Hsp27 Deletion Mutants Transfection
Poly-histidine-tagged Hsp27 wild type (WT) and three deletion mutants: of the N-terminal domain of Hsp27 with (N2) or without (N1) the entire α-crystallin domain and the C-terminal domain mutant comprised of a truncated α-crystallin domain (C1), in pcDNA4 containing His-tag epitope at N-terminal of the inserted fragment (23) were kindly provided by Pr O'Brien (Ottawa University, Ontario, Canada). LNCaP cells were transfected with 10 μg WT or deletion mutants using Fugene reagent (Roche Diagnostics, GmbH Mannheim, Germany) according to the manufacturer's instructions. Cleared lysates were obtained 48 h post-transfection according to (6) were used for immunoprecipitation experiments and in vitro survival assays as previously described (24).
DNA DSB Experiments
ZeocinTM (a generous gift from Dr. Mauro Modesti, Invitrogen) is a copper-containing glycopeptide antibiotic (bleomycin family) able to cleave DNA. Cells were treated with 100 μg/ml ZeocinTM for 2 h at 37 °C. After treatment, cells were collected for immunofluorescence, RNA/DNA isolation and nuclear/cytoplasmic protein extraction, or washed with fresh medium and returned in the incubator for 1 h to allow DNA repair, then collected for the following experiments.
Western blot
Proteins were extracted using nuclear extract kit (Active Motif, La Hulpe, Belgium) according to the manufacturer's guidelines. Western blot was carried out using extracts with adjusted protein concentration (Pierce BCA Protein assay kit, Fisher Scientific, France) with 1:5000 rabbit anti-Hsp27 polyclonal antibody (Stressgen), 1:1000 rabbit anti-γ-H2AX (Abcam), 1:1000 rabbit anti-H2AX (Abcam, Paris, France), and 1:3000 mouse anti-vinculin (Sigma Aldrich, France) used as internal loading control.
Immunofluorescence
Immunofluorescence analysis was performed as described previously (25) on PC-3 cells with 1:2500 primary mouse monoclonal anti-γ-H2AX antibody (Upstate, Millipore SAS, France) and secondary fluorescent goat anti-mouse Alexa Fluor 488 antibody (Invitrogen). Images were captured using a Zeiss 510 META fluorescence confocal microscope plan 40X/1.4 (Le Pecq, France).
NHEJ Assay
The NHEJ assay was conducted according to previous work with slight modifications (26). Briefly, 20 μg of nuclear extracts prepared from PC-3 (72 h after transfection, as described above), were mixed with 250 ng of StuI digested pDEST26 plasmid DNA in NHEJ buffer (200 mm Hepes-KOH, pH7.5, 800 mm KCl, and 100 mm MgCl2), 1 mm ATP, and 1 mm DTT. The 20 μl reactions were incubated at 37 °C for 2 h followed by protease (10 mg/ml), 0.5% SDS, 0.5 M EDTA treatment at 37 °C for 30min. The reactions were separated on a 0.8% agarose gel and stained with SYBR green (Invitrogen). The stained gels were scanned, and the percentage of end joining was calculated by dividing the sum of (ligated) dimer by the sum of (unligated) monomer and dimer.
Exonic Expression Profiling
We hybridized two independent biological replicates for each experimental condition (CTL-siRNA and Hsp27-siRNA PC-3 cells) on Affymetrix Human Exon 1.0 ST splice arrays. Total RNA was extracted using the all-in-one Miniprep kit (Bio Basic Canada, Inc.) and RNA integrity assessed using the 2100 Agilent bioanalyser (Agilent, Palo Alto, CA). Preparation of cDNA from 200 ng total RNA, hybridizations, washes, and detection was performed as recommended (www.affymetrix.com). Scanning was performed with Affymetrix GeneArray scanner 3000 7G, and quantification with Affymetrix® GeneChip® Command Console® Software (AGCC). Hybridization images were inspected for artifacts. Expression data were analyzed in R software using aroma.affymetrix and associated packages (http://www.cran.r-project.org). Analysis was performed at the core level using the Ensembl definition of transcripts. Probes mapping was performed with the customized array annotation file from the Aroma Project (i.e. “HuEx-1_0-st-v2, U-Ensembl49, G-Affy.cdf,” http://www.aroma-project.org), and retained 326,983 probe sets corresponding to 22,035 defined genes. These probe sets were then background-adjusted and quantile-normalized with Robust Multichip Analysis (RMA). Then, a filtering process retained the 241,259 probe sets (20,337 genes) with expression level superior to 100 units in at least one of four samples. The alternative splicing level of each probe set in a sample was defined by its FIRMA score (27), computed as its expression level relative to that of corresponding gene. To identify probe sets and genes with differential splicing between the two experimental conditions, we applied supervised analyses (moderated t-statistics) to both expression and FIRMA levels of each probe set using linear models with empirical Bayes statistic included in the limma R packages (28). Probe sets were considered significantly differential if they showed a fold change of at least two and in both analytic levels a p value of at least 5% with a 25% false discovery rate (FDR). The list of corresponding genes was then confronted to the ASAPII database (www.bioinformatics.ucla.edu/ASAP2) and Cancer Gene Census and analyzed using the DAVID suite (18). Hierarchical clustering was performed using Cluster program and displayed using Treeview (29).
RT-PCR Validation of Alternative Splicing
Total RNA was extracted from PC-3 cells using RNeasy kit (Qiagen, Qiagen, Courtaboeuf, France) and mRNA was reverse transcribed to cDNA using SuperScript II Reverse Transcriptase and oligo-dT primers (Invitrogen) prior to PCR amplification using GoTaq Hot Start Polymerase (Promega, Charbonnières-les-Bains, France) and specific primers designed against SLC45A3 exons 2 and 3 (Fig. 5C) (forward sequence: 5′-TTTGGCCTGGAGGTGTGTTT-3′ and reverse sequence: 5′-TCCCGGAAGAGGTCAGAGAG-3′) and against exons 2 and 4 (forward sequence: 5′-TTTGGCCTGGAGGTGTGTTT-3′ and reverse sequence: 5′-ACTGCTCGAGTGCCGAATCG-3′).
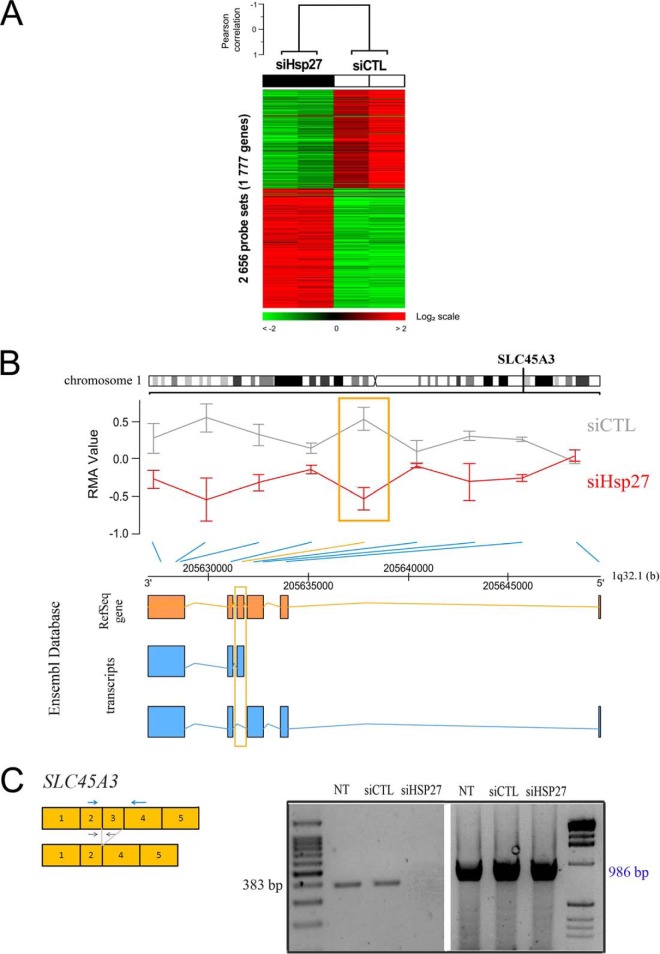
Fig. 5.
Hsp27 regulates the differential splicing of 1777 genes. A, Hierarchical clustering of four samples and 2656 probe sets (grouped by gene) identified by supervised analysis with significant differential splicing between the two experimental conditions (siCTL and siHsp27). Each row of the data matrix represents a probe set and each column a sample. Expression levels are depicted according to the color scale shown at the bottom. Red and green indicate expression levels respectively above and below the median. The magnitude of deviation from the median is represented by the color saturation. The dendrogram (above matrixes) represents the overall similarity of samples in expression profiles: two groups of samples are evidenced and correspond to the two different experimental conditions. Clustering parameters are data median-centering on probe sets, Pearson correlation as similarity metrics and centroid linkage. B, Plot of the SLC45A3 gene localized on the long arm of chromosome 1. Exon-level expression values (RMA normalized) represented as red and gray lines correspond to average expression level of siHsp27 and siCTL respectively and error bars represent standard deviation. The significant probe set is enlightened by an orange box. Link between each probe set and their respective position on the 1q32.1 is shown by an oblique blue line; or orange for the significant probe set. Under the chromosome position axis are described the exons from RefSeq Gene and transcripts as reported on the Ensembl database (Orange and Blue box respectively). The exon showing significant variation is enlightened by an orange frame. C, Schematic representation of amplifying forward and reverse primers design in the exon 2/exon 3 junction (black arrows) and exon2/exon 4 (blue arrows) and RT-PCR validation of exon 3 differential splicing of SLC45A3 in nontransfected cells, siCTL and siHsp27 transfected PC3 cells as well as presence of alternatively spliced variant. The PCR product sizes are 383 bp and 986 bp respectively.
Cell Survival Assay
The in vitro effects of Zeocin™ treatment (in the presence or absence of Hsp27) on cell survival were assessed using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) mitogenic assay as previously described (30). Briefly, transiently transfected PC-3 cells with Hsp27-siRNA or CTL-siRNA, as described above, for 72 h, were treated with 20, 50, and 100 μg/ml of ZeocinTM for 2 h. Medium in each well was then refreshed and MTT assay was performed in triplicate after 72 h. LNCaP cells transiently transfected with Hsp27 truncated mutants (see above) were treated with 1 nm Docetaxel, 48h after transfection, in serum-free media. Cell viability was assayed the day after using Crystal violet dye as previously described (30).
Statistical Analysis for Survival Assays
Results from survival assay were expressed as mean ± S.E. Statistical analysis was performed using one-way analysis of variance followed by Fisher's protected least significant difference test (Statview 512, Brain Power Inc., Calabases, CA). *p ≤ 0.05 was considered significant, with **p ≤ 0.01 and ***p ≤ 0.001.
RESULTS
Hsp27 Interaction Map Using Two-hybrid Identifies 226 Interacting Proteins
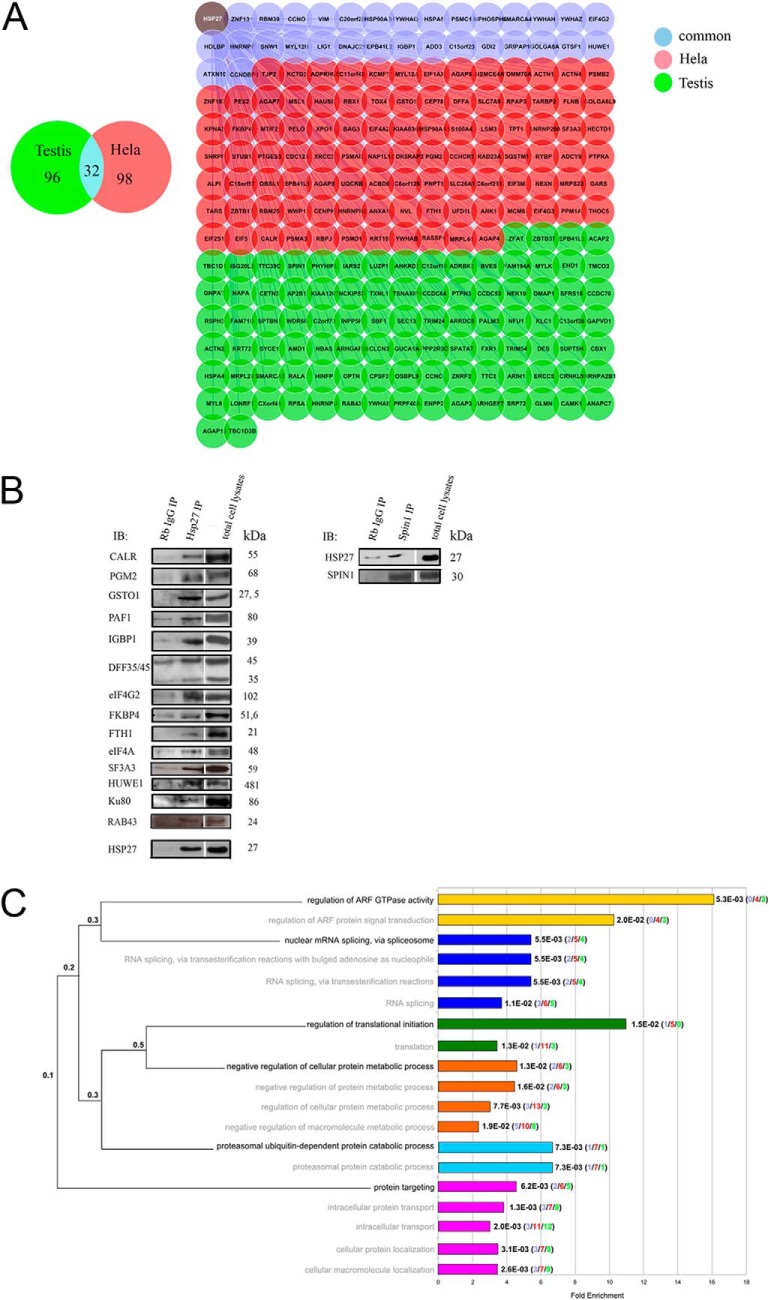
To understand how Hsp27 exerts its cytoprotective effects, we established the interaction map of Hsp27. We used the yeast two-hybrid (Y2H) technique Sos Recruitment System (SRS) to identify Hsp27 interactions in the yeast cytoplasm (17). Y2H screens were performed on two human cDNA libraries from different tissues (HeLa cervix cancer cells and normal testis cells). To avoid sampling effect and to reach saturation (31), ten screens per library were performed (Experimental Procedures) allowing the identification of 226 proteins among which only one (PPM1A, (32)) had already been identified as an Hsp27 interacting protein. Of these 226 interactors, 98 and 96 were identified from HeLa and testis cDNA libraries respectively, and 32 from both (Fig. 1A, supplemental Table S1).
Fig. 1.
Large-scale Y2H screen deciphers Hsp27 PPI. A, The 226 Hsp27-interacting proteins identified in the Y2H screen of HeLa cDNA library (red nodes), testis cDNA library (green nodes), or in common in both libraries (blue nodes). B, Validation of 6% of the Hsp27 interactors by Co-IP from PC-3 cells total protein extracts. Rb (Rabbit), IgG (Immunoglobulin G), IP (Immunoprecipitation), IB (Immuno-blot). C, GO Biological Process enrichments (Benjamini-corrected p values (q-value) < 0.02) of the Hsp27 interaction partners. Barplots on the right indicate the fold enrichment of each annotation, and the exact q-values. Each term corresponds to a colored bar and related terms share the same color. In brackets are represented the numbers of interactors identified from the testis (green), HeLa (red), or both (blue) libraries. The dendrogram on the left depicts the hierarchical organization of the significantly over-represented terms in the ontology. The most precise terms are written in black and the less specialized terms in gray, with the associated term precision at the branching points.
We subsequently tested 24 interactions (10.6%) by co-immunoprecipitation (co-IP) in human PC-3 prostate cancer cells (Experimental Procedures, Supplemental Table S1). Sixty percent were scored positive (Fig. 1B and (30)). Importantly, these validations in mammalian cells, in which translation and post-translational modifications are physiological, suggest that the interactions observed in yeast were not caused by Hsp27 recognition of misfolded proteins. All PPI reported here have been submitted to the International Molecular Exchange (IMEx) Consortium (33) through IntAct and assigned the identifier IM-20864.
Binary Analysis of Hsp27 Interactions Gives a Global View of Hsp27 Functions
According to their Gene Ontology (GO) annotations, the Hsp27 interactors were enriched in proteins involved in the regulation of ARF GTPase activity (Benjamini-corrected hypergeometric p-val = q-val = 5.3E-3), the regulation of translation initiation (q-val = 1.5E-2), the negative regulation of cellular protein metabolic process (q-val = 1.3E-3), and the proteasomal ubiquitin-dependent protein catabolic process (q-val = 7.3E-3) (Fig. 1C), all being cellular functions in which Hsp27 has already been implicated (Table I). This validated the quality of the Y2H interactome and further identified the molecular actors at work with Hsp27 in these processes. For instance, whereas only one Hsp27 interaction partner involved in translation regulation had been described previously (eIF4G1, (34)), we discovered seven additional interactions of Hsp27 with translation initiation factors (eIF1AX, eIF2S1, eIF3M, eIF4A2, eIF4G2, eIF4G3, and eIF5), and one ribosomal protein (RPSA), thereby reinforcing the relationship between Hsp27 and the translation initiation complex. Similarly, we identified five different subunits of the proteasome as interacting with Hsp27 (PSMD1, A3, A6, B2, and C1) suggesting a molecular scenario for Hsp27 enhancement of pro-apoptotic proteins degradation through the proteasomal pathway in cancer cells (11). Importantly, our screen directly associated Hsp27 with two novel cellular processes, namely nuclear mRNA splicing via spliceosome (q-val = 5.5E-3) through interactions with six ribonucleoproteins (SNRNP200, SNRPF, HNRNPA2B1, HNRPF, HNRNPH2, and HNRNPU) and two splicing factors (SF3A3 and SFRS18), and protein targeting (q-val = 6.2E-3) via interactions with exportin 1 (XPO1) and importin alpha 4 (KPNA3) among others (Fig. 1C). Overall, the identification of Hsp27 interactors informed on the molecular entities acting with the chaperone and provided a global view of its functions. It is worth noting that all these functions were significantly enriched in interactors identified from the HeLa cancer cells library (red color in brackets) compared with interactors identified from the normal testis cells library (green color in brackets). Our data suggest that these functions could be specifically related to cancer progression.
Table I. Biological processes associated with Hsp27 identified after Hsp27 interactome integration to the human interactome (M = module).
Targeting Hsp27-partner Protein Interaction may Represent an Interesting Alternative Strategy to Increase Therapy-sensitivity in Cancer Cells
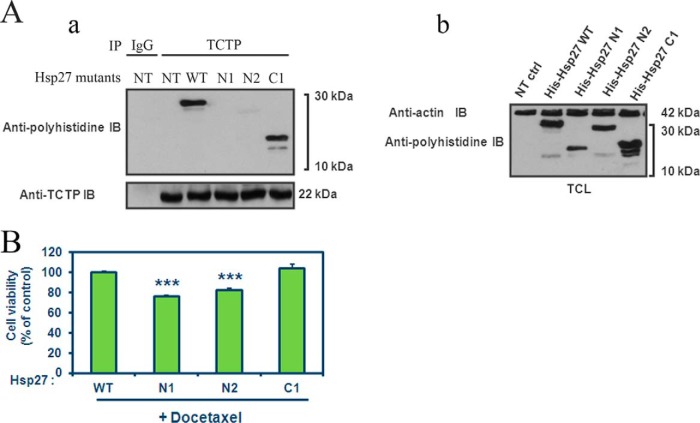
Among the 226 Hsp27 partner proteins identified, we chose to further study TCTP (translationally controlled tumor protein). Our aim was to confirm that our interactome identification from the HeLa library screen is related to cancer cell functions and may allow the definition of specific anti-cancer therapeutic strategies, which would have no or little effect on normal cells. We previously demonstrated that Hsp27 protects TCTP from its proteasomal degradation leading to Hsp27 cytoprotection in CRPC and that TCTP was scarcely expressed in normal cells (30). We started by studying the disruption of the Hsp27-TCTP interaction. To determine the relevance of this interaction in Hsp27 cytoprotective effects in PC cells, LNCaP cells were transfected either with Hsp27 WT (wild-type), with N-terminal deletion mutants N1 and N2 or the C-terminal domain mutant C1 (23). All constructions contained a 6-his (6-polyhistidine) tag, fused to the Hsp27 protein, allowing the recognition of the endogenous from the transfected Hsp27 mutant proteins (Fig. 2A, b). These deletion mutants allowed us to map the C-terminal region of Hsp27 as necessary for TCTP interaction, as we observed a strong interaction between TCTP and Hsp27 WT and C1 but not with N1 or N2 (Fig. 2A, a). Interestingly, only the N1 and N2 mutants, unable to bind TCTP, were able to sensitize LNCaP cells to docetaxel after androgen withdrawal (Fig. 2B). Therefore, molecules targeting Hsp27-TCTP interaction might be promising therapeutic drugs. Moreover, this example reinforces our confidence in our screening results and strongly suggests that Hsp27 interactions and associated functions identified from the cancer cDNA library HeLa, seem to be related to cancer progression.
Fig. 2.
The loss of Hsp27-TCTP interaction increases cell treatment-sensitivity. A, (a) LNCaP cells, transfected or not (NT) with Hsp27 wild-type (WT) or deletion mutants (N1, N2, or C1), cell lysates have been used to immunoprecipitate (IP) TCTP or rabbit anti-immunoglobulin (IgG). The membrane has been then immunoblotted (IB) with anti-polyhistidine and anti-TCTP antibodies. (b) Total cell lysates (TCL) immunoblot was used as a control of transfection and represent proteins from LNCaP cells, extracted from cultured cells and blotted with anti-polyhistidine or anti-actin (loading control) antibodies. B, LNCaP cell viability was determined using crystal violet dye. Cells were transiently transfected with Hsp27 WT and truncated mutants (N1, N2, and C1) during 48h and then have been treated with 1 nm docetaxel in serum-free media (mimics androgen withdrawal in vitro). Error bars represent the S.E., ***p ≤ 0.001.
Merging our Hsp27 Interaction Map with the Human Interactome Enhances the Functional Landscape of Hsp27 and Reveals 17 New Cellular Functions
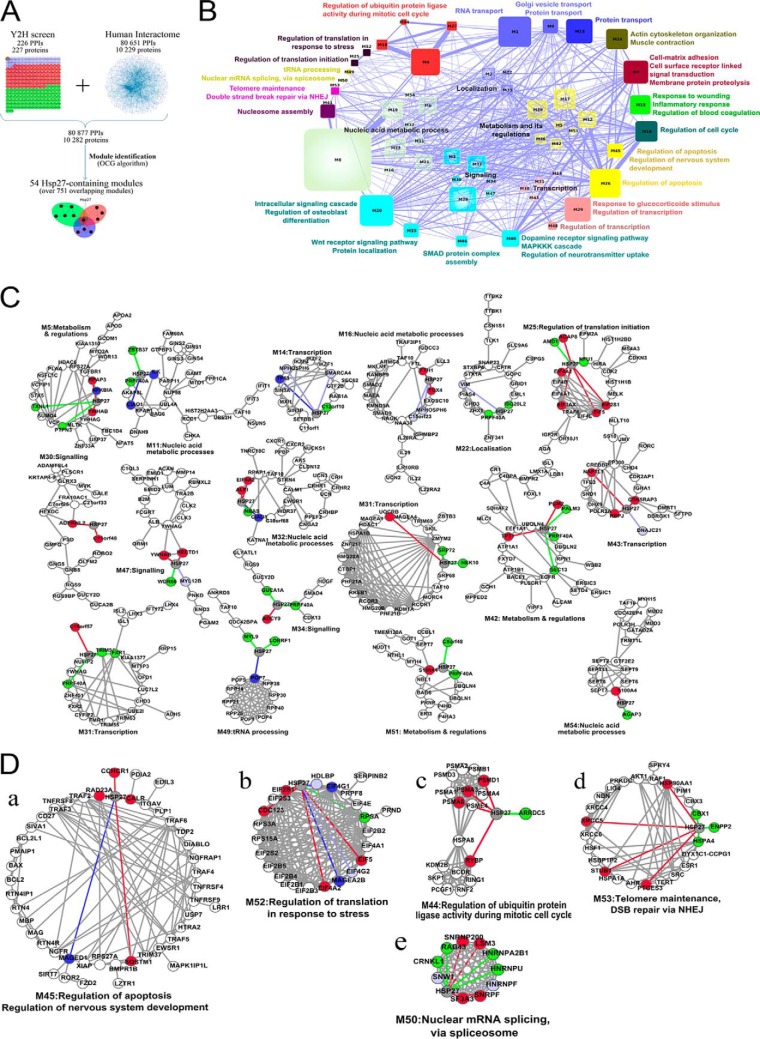
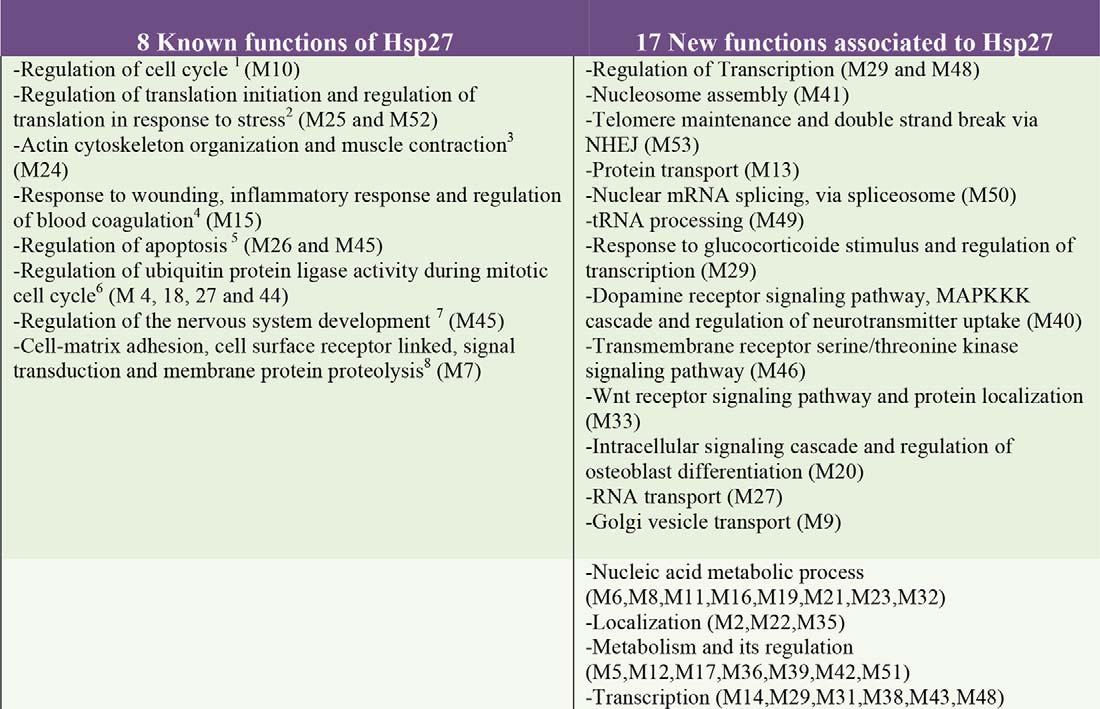
The analysis of the binary interactions issued from the Y2H screens was a starting point toward the elucidation of Hsp27 cellular processes. However, this strictly local approach did not consider the extended functional information contained in the whole human protein-protein interaction (PPI) network. PPI networks are modular and composed of groups of highly connected proteins involved in the same cellular function (16). These modules correspond to the functional units of the network and permit function prediction when containing uncharacterized proteins (35, 36). We merged our Hsp27 interaction map and the human interactome consisting of 80,651 interactions between 10,229 proteins (20). This raised the total number of Hsp27 interactors to 254 by complementing our Y2H screen results with 28 supplementary interactions detected in different experimental conditions (supplemental Table S2). We identified the functional modules composing the resulting PPI network using the OCG (Overlapping Cluster Generator) algorithm (21) (Fig. 3A). Among the 751 overlapping modules identified, 54 contained Hsp27 and a total of 206 out of its 254 interactors (supplemental Table S3, supplemental Figs. S1–S54). These 54 modules form a comprehensive map of Hsp27 cellular functions (Fig. 3B). Indeed, according to the GO terms annotating their constituent proteins (Experimental Procedures), these modules range from biological processes already reported to be associated with Hsp27 to completely novel ones, thereby confirming and complementing (see thereafter) both previous knowledge and our Y2H screens. Thirteen modules are involved in eight cellular processes already associated with Hsp27 (Table I), such as regulation of the cell cycle (M10, Fig. 3C, supplemental Table S3), regulation of translation initiation and regulation of translation in response to stress (M25 and M52), actin cytoskeleton organization and muscle contraction (M24), response to wounding, inflammatory response and regulation of blood coagulation (M15), regulation of apoptosis (M26 and M45), regulation of ubiquitin protein ligase activity during mitotic cell cycle (M4, M18, M27, and M44), regulation of the nervous system development (M45) and cell-matrix adhesion, cell surface receptor linked, signal transduction, and membrane protein proteolysis (M7). Interestingly, our Hsp27 module map predicts a role for Hsp27 in 17 additional cellular functions in which the protein has never been implicated before (Table I, supplemental Table S3): regulation of transcription (M29, M48), nucleosome assembly (M41), telomere maintenance and double strand break via NHEJ (M53), protein transport (M13), nuclear mRNA splicing via spliceosome (M50), tRNA processing (M49), response to glucocorticoide stimulus and regulation of transcription (M29), dopamine receptor signaling pathway, MAPKKK cascade and regulation of neurotransmitter uptake (M40), transmembrane receptor serine/threonine kinase signaling pathway (M46), WNT receptor signaling pathway and protein localization (M33), intracellular signaling cascade and regulation of osteoblast differentiation (M20), RNA transport (M27), and Golgi vesicle transport (M9), nucleic acid metabolic process (M6, M8, M11, M16, M19, M21, M23, and M32), localization (M2, M22, and M35), metabolism and its regulation (M5, M12, M17, M36, M39, M42, and M51), and transcription (M14, M29, M31, M38, M43, and M48). The module map, hence, remarkably extends the functional landscape of Hsp27 to a number of functions that were neither known beforehand nor revealed by the study of the binary interaction map alone.
Fig. 3.
Hsp27 functional module analysis. A, Flowchart of the network approach. The 226 interactions identified in our Y2H screen were added to the human interactome (20). The resulting network was partitioned in 751 overlapping modules with the OCG algorithm (21). B, The 54 functional modules containing Hsp27 are represented as a network where the nodes correspond to the functional modules and the edges to the shared proteins between modules. Node size and edge width are proportional to the number of proteins they represent. Modules are colored and grouped according to their GO Biological Process annotations. C, Examples of 17 functional modules. Colored nodes are direct Hsp27 interactors. Green nodes represent interactors identified from testis library, red from the HeLa one, light blue from both, dark blue nodes are Hsp27 partners present in the human interactome dataset. The same color code is used for edges representing interactions identified in the Y2H screen. For sake of clarity, only the 17 smallest Hsp27-containing modules are represented. D, Details of the five investigated functional modules.
A Detailed Analysis of Functional Modules Reveals New Pieces in the Puzzle of Hsp27 Roles
Insights for Hsp27 Role in Apoptosis
The functional module annotated regulation of apoptosis (q-val<3.53E-17, Module 45, Fig. 3D, a) highlights a well-described function of Hsp27 (1, 3). The anti-apoptotic properties of Hsp27 have been widely described and it is clear that Hsp27 interferes at different levels of apoptotic pathways in cancer cells through interactions with cytochrome c, AKT, caspase 3, and DAXX (37). This functional module contains 47 proteins, among which 24 are known to be involved in apoptosis, such as BCL2, BAX, and TNF-receptor associated factors 2–6. Hsp27 interacts with five partners (CCHCR1, CALR, SQSTM1, RAD23A, and MAGED1), which are directly or indirectly involved in apoptotic pathways. Our data provided new elements for the role of Hsp27 in apoptosis. Hsp27 interaction with RAD23A could indirectly interfere with DIABLO, a pro-apoptotic protein antagonizing IAP proteins (inhibitors of apoptosis) (38), and involved in the mitochondrial apoptotic pathway. In addition, Hsp27 could interfere with pathways dependent on XIAP, TRAF4 and 6 (TNF receptor-associated factor 4) (39) through its direct interactions with MAGED1, RAD23A, CALR, and SQSTM1. CALR has been recently shown to be a major mutated gene in myeloproliferative neoplasms (40). In accordance with previous work (37), our results strongly reinforce the fact that Hsp27 uses different strategies to negatively modulate apoptotic signaling pathways.
A Reinforced Relationship Between Hsp27 and the Translation Initiation Complex
The participation of Hsp27 to the functional module annotated as regulation of translation in response to stress (Module 52, Fig. 3D, b, q-val = 10−19) is because of its direct interactions with eIF4G1, MAGEA2B, eIF3S1, eIF4A2, CDC123, eIF5, and RPSA. The involvement of Hsp27 in regulating the translation initiation process has already been described in particular via its interaction with eIF4E (24, 25) and eIF4G1 (34). We complemented this relationship with six novel interactions of Hsp27 with key factors of the translation initiation steps. This mechanism involves interactions of certain protein factors with mRNA, and the small ribosomal subunit (40S subunit), thus holding the mRNA in place (41). We identified an interaction of Hsp27 with RPSA, a 40S ribosomal-associated protein component. Hsp27 interacts with eIF3S1, a component of the eIF3 complex that binds to the 40S ribosomal subunit and plays a role in keeping the large ribosomal subunit from prematurely binding to the 40S subunit (41). eIF3 also interacts with the eIF4F complex consisting of eIF4E, eIF4A, and eIF4G (42), all three interacting with Hsp27. Moreover, we showed that Hsp27 interacts with eIF5, a translation initiation factor allowing the release of the eIF2 complex, and with CDC123, a regulator of eIF2 (43). Our data, hence allowed to considerably strengthen the link between the translation initiation machinery and Hsp27.
Regulation of Ubiquitin Protein Ligase Activity During Mitotic Cell Cycle by Hsp27
We found that Hsp27 participates in a module dedicated to the regulation of ubiquitin protein ligase activity during mitotic cell cycle (q-val = 4.73E-12, Module 44, Fig. 3D, c). Hsp27 has a high ubiquitin-binding ability and promotes the ubiquitination of a number of cellular proteins through mechanisms remaining elusive. We found that Hsp27 interacts with RYBP, an ubiquitin-binding protein interacting with the ubiquitin E3 ligase RING1. This ligase recognizes proteins to be ubiquinated and targets their degradation toward the proteasome (44), an essential mechanism inducing cell-cycle arrest after stress. A role of Hsp27 in protein degradation was previously suggested from its colocalization with the 26S proteasome a multicatalytic proteinase complex (11). The interaction of Hsp27 with the 26S proteasome is required to activate the proteasome. The three interactions of Hsp27 with different subunits of the 26S proteasome (PSMD1, A3, and A6) that we have discovered not only confirmed the role of Hsp27 in the ubiquitin/proteasome degradation pathway but also identified the potential molecular actors at work that could in part explain cancer progression.
Undisclosed Hsp27 Cancer-related Functions: Involvement in DNA Repair and mRNA Splicing Revealed by the Module Map
Given its overexpression in cancer cells, understanding Hsp27 cytoprotective functions through the identification of interactions that are specific to cancer cells may provide targets for drug development (45, 46). Among the novel Hsp27 functions predicted by the module map (Fig. 3D, d and e, Table I), double-strand breaks repair via NHEJ and telomere maintenance and nuclear mRNA splicing, via spliceosome were particularly relevant for cancer progression, therefore calling for experimental investigations.
Hsp27 Negatively Regulates DNA Repair of Double-strand Breaks in Cancer Cells
We found that Hsp27 belongs to a functional module implicated in double-strand break repair via NHEJ and telomere maintenance (q-val = 7.44E−9) through its interactions with 1) XRCC5/KU80 (Fig. 3D, d), an ATP-dependent DNA helicase II involved in double-strand break (DSBs) repair mediated by nonhomologous end-joining (NHEJ) (47), 2) Hsp90AA1, an inducible form of Hsp90 and its cochaperone PTGES3 (or p23) both interacting with TERT (Fig. 3D, d) for the assembly of active telomerase (48), and 3) STUB1, known to negatively regulate telomerase activity (49). The cytoprotective effect of Hsp27 in cancer cells could therefore be related to a possible function of the protein in DNA repair function.
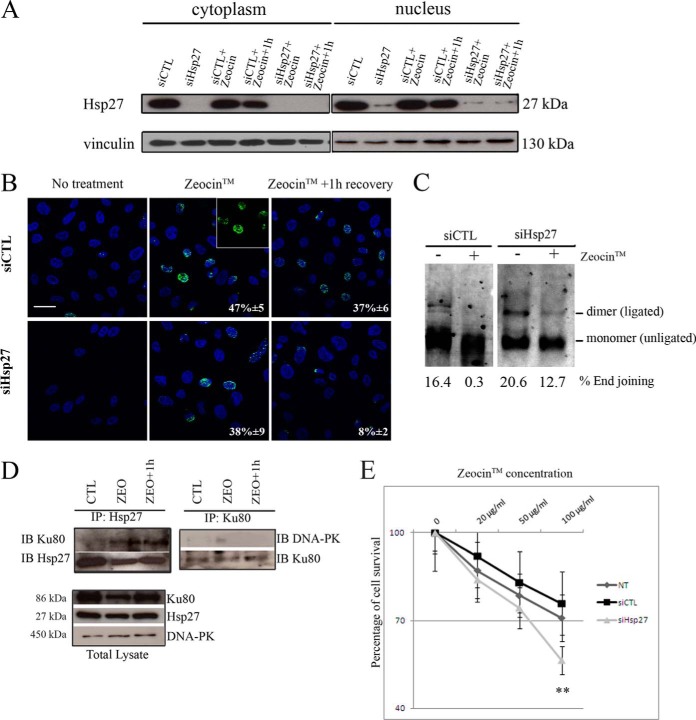
DSBs are lethal DNA damages that can occur in response to ionizing radiation or chemotherapeutic agents (50). They are usually repaired by two pathways, homologous recombination (HR) and NHEJ that, when deregulated, lead to repair defaults contributing to chromosome translocations, genomic instability, and ultimately to cancer (51). DSBs can be followed through the formation of nuclear foci corresponding to the accumulation of phosphorylated histone H2AX (γ-H2AX) (52). There is a close correlation between nuclear γ-H2AX foci and DSBs and between the rate of foci loss and DSBs repair, providing a sensitive and reliable method to monitor DSBs repair. To confirm the implication of Hsp27 in DSBs repair through NHEJ, we monitored DNA repair through the accumulation of γ-H2AX in CRPC cells. Hsp27-overexpressing PC-3 cells were transfected with Hsp27-siRNA and treated with ZeocinTM, a radiomimetic cationic antibiotic, causing DNA damage by cleaving both strands of the DNA molecule (53). After transfection with Hsp27-siRNA, a drastic decrease in Hsp27 protein level was observed in both cytoplasm and nucleus compartments (Fig. 4A). γ-H2AX stained nuclei were significantly less abundant in the absence of Hsp27 (38%±9) than in controls (CTL-siRNA, 47%±5) (Fig. 4B). After one hour recovery of ZeocinTM treatment nuclei stained with γ-H2AX foci were more abundant in control cells (37%±6) than in Hsp27-depleted cells (8%±2) (Fig. 4B). DSBs thus might be more efficiently repaired in the absence of Hsp27 thereby suggesting that cancer cells overexpressing Hsp27 could present defects in DSBs reparation. In addition, in vitro plasmid-based NHEJ assays performed in CTL-siRNA and Hsp27-siRNA transfected cells showed reduced end joining after ZeocinTM treatment in control cells, indicating that the Hsp27-related interference with NHEJ is DNA damage-dependent (Fig. 4C). Co-immunoprecipitation experiments were then preformed after treatment with ZeocinTM treatment and 1h recovery after treatment in order to evaluate Hsp27 interaction with Ku80 (Fig. 4D). We observed that Hsp27 interacts with Ku80 upon ZeocinTM treatment and recovery for repair (Fig. 4D, left panel). In the same conditions, Ku80 did not interact with DNA-PK (Fig. 4D, right panel). To correlate the effect of Hsp27 expression on DNA repair defect and cancer cells survival, we performed survival assays of PC-3 cells transfected with CTL-siRNA or Hsp27-siRNA and then treated with increasing concentrations of ZeocinTM. As shown in Fig. 4E, the survival of the Hsp27-depleted cells was decreased as compared with control conditions, further supporting the link between Hsp27 overexpression, lack of repair ability and abnormal survival in cancer cells. Altogether, our results functionally validated the involvement of Hsp27 in DNA repair predicted by the module map, by showing its implication in DSB repair negative regulation. Our data suggest that Hsp27 interacts with Ku80 preventing the later to bind DNA DSBs and consequently to interact with DNA-PK, leading to DSBs accumulation and conferring survival advantage for cancer cells.
Fig. 4.
Hsp27 negatively regulates DNA repair of DSBs induced by Zeocin™. A, Western-blot analysis of Hsp27 expression in cytoplasm and nucleus protein extracts of PC-3 cells transfected with CTL-siRNA (siCTL) and Hsp27-siRNA (siHsp27) then treated with Zeocin™ and allowed to recover for 1h. Vinculin was used as internal loading control. B, Representative Immunofluorescence of PC-3 cells transfected with siCTL and siHSP27 showing the γ-H2AX-stained nuclei recognized by specific antibody. The presence of cells in the observation field is indicated by DAPI-stained nuclei. Bar, 50 μm. C, NHEJ assay was conducted using nuclear extracts from Hsp27 expressing (siCTL) and nonexpressing (siHSP27) PC-3 cells. The percentage of end joining (calculated by the sum of (ligated) dimer and multimer divided by the sum of (unligated) monomer, dimer and multimer) is shown in each lane. D, Co-IP of Hsp27 and Ku80 in PC-3 cells E, Cell viability (expressed in percentage) analysis of PC-3 72h after treatment with Zeocin™ at 20, 50, and 100 μg/ml using MTT test. The experiment was repeated in triplicate. ** differs between PC-3 cells transfected with siCTL and siHSP27 (p ≤ 0.01).
Hsp27 Regulates mRNA Alternative Splicing in Cancer Cells
We found that Hsp27 belongs to a functional module implicated in nuclear mRNA splicing, via spliceosome (q-val = 9.98E-13) (Fig. 3B, supplemental Table S3) through nine interactions identified in our Y2H screen (Fig. 3D, e), including three splicing factors (SF3A3, CRNKL1, and SNW1), three components of the spliceosome (SNRPF, LSM3, and SNRNP200), and three splicing regulators (HNRNPF, HNRNPU, and HNRNPA2B1). Splicing events are a fundamental mechanism controlling gene expression and their alteration plays a role in human disease and cancer (54). The spliceosome complex is characterized by a dynamic composition and conformation. It results from the ordered interactions of small nuclear ribonucleoprotein (snRNPs) and numerous splicing factors (55). Some reports have suggested an indirect role of Hsp27 in mRNA splicing (56–58), but a direct association of Hsp27 with spliceosome function has never been described. To study the involvement of Hsp27 in splicing, we performed a whole-genome exonic expression profiling of PC-3 cells treated by Hsp27-siRNA or CTL-siRNA. Splicing events can be monitored using Affymetrix exon arrays, enabling two complementary levels of analysis: gene expression and alternative splicing of hundreds of thousands of known and predicted exons. The splicing level (FIRMA, Finding Isoforms Using Robust Multichip Analysis) of each probe set was defined in each sample. A supervised analysis identified 2656 probe sets corresponding to 1777 genes (9% of the 22,035 tested genes) that were differentially spliced between Hsp27-siRNA and control conditions, including 1461 up-regulated probe sets (55%) and 1195 down-regulated (45%) probe sets after Hsp27 silencing (Experimental Procedures, Fig. 5A, and supplemental Table S4). Noticeably, 153 of these 1777 genes were reported as alternatively spliced genes in the ASAPII database containing extensive referenced alternative spliced genes and their spliced variants (supplemental Table S4) (59), among which 16 genes displayed an alternative splicing associated to cancer (Table II).
Table II. Genes alternatively spliced in absence of Hsp27 and previously reported as alternatively spliced in cancers.
A comparison with the Cancer Gene Census (60) showed that 51 out of the 1777 genes have mutations causally implicated in cancers (supplemental Table S5). Among them, solute carrier family 45, member 3 (SLC45A3) has been largely described as an androgen-driven prostate specific gene. It can form transcript chimeras when fused with gene transcripts encoding ETS-family oncogenic transcription factors such as ELK4 (61, 62). Although we did not detect such chimeric transcripts in PC-3 cells, SLC45A3 was differentially spliced at the level of exon 3 and overall underexpressed in the absence of Hsp27 as compared with control (Fig. 5B and supplemental Table S4). We validated the differential splicing of exon 3 in nontransfected, control and Hsp27-siRNA-treated PC-3 cells by RT-PCR (Fig. 5C) by showing the presence of exon 3 in Hsp27-expressing cells (Fig. 5C, lanes NT for nontransfected, siCTL left panel), and its absence in cells devoid of Hsp27 (lane siHsp27, left panel). We have also validated the presence of the alternatively spliced variant in siHsp27 cells using primers against exon 2 and exon 4 (Fig. 5C, right panel). Altogether, by demonstrating a differential splicing program of 9% of the tested genes in the presence or absence of Hsp27, we functionally validated the involvement of Hsp27 in mRNA alternative splicing in CRPC cells predicted by the module map.
DISCUSSION
Interactome and Module Maps Revealed Hsp27 Functions
We combined experimental and bioinformatics approaches to identify Hsp27 interaction partners, and to map and validate its participation in functional modules. Several lines of evidence showed the specificity of the 226 Hsp27-interactions detected from the large-scale Y2H screen, despite Hsp27 propensity to recognize misfolded proteins (63). First, 60% out of the 24 interactions re-tested by co-IP in human CRPC scored positive, a higher proportion than most of the previously published validations (64–66). Second, the set of Hsp27 interactors was significantly enriched in actors of certain cellular processes (Fig. 1C). This underlined the overall specificity of the interactions detected by the screen because no such statistical over-representation is expected from a (random) list of misfolded proteins. Furthermore, whereas some of these functions acted as internal controls since corresponding to well-known Hsp27 processes (67, 68), we were able to experimentally validate the novel implication of Hsp27 in other functions. Altogether, this emphasizes our confidence in the screening results.
Beyond the functional insights brought by the establishment of the Hsp27 interaction map, we proposed an Hsp27-module map resulting from a global human interactome network analysis. This led to a comprehensive picture of Hsp27 involvement at the crossroad of multiple functions (Fig. 3B and Table I). Again, some of these Hsp27 functions being previously documented, the rationale of our approach was validated. Moreover, whereas certain functional modules recapitulated and confirmed the results of the binary interaction map analysis (such as Module 52 dedicated to the regulation of translation in response to stress, and Module 44, annotated to the regulation of ubiquitin protein ligase activity during mitotic cell cycle (Fig 3D, b, c), some others revealed known Hsp27 functions that were not unveiled by the binary interaction map alone (such as Module 45 involved in regulation of apoptosis and regulation of nervous system development) (Fig. 3D, a). Thus, the involvement of Hsp27 in DNA repair via NHEJ, later demonstrated experimentally in this work, was predicted by the module map whereas blurred and not apparent in the Hsp27 binary interaction map. This definitely illustrates the predictive power of the module approach that amplified the functional signal by taking into account neighboring interactions in the PPI network.
We chose to focus on functional modules closely related to Hsp27 functions in cancer survival (regulation of apoptosis, telomere maintenance and double strand break via NHEJ, regulation of ubiquitin protein ligase activity during mitotic cell cycle, nuclear mRNA splicing via spliceosome) (Figs. 3D) composed predominantly of PPI we identified from screening the cancer HeLa cDNA library. The mechanisms previously demonstrated to account for the Hsp27 cytoprotective effects were: ATP-independent chaperone activity (folding and aggregation modulation of the denatured proteins) (63), interference with effectors of the apoptotic machinery (3), and stabilization or proteasomal degradation of selected proteins (11, 25, 30, 69). Our study revealed 17 exciting new aspects of Hsp27 involvement in essential metabolic and cellular processes such as: telomere maintenance and double strand break via NHEJ, nuclear mRNA splicing via the spliceosome, Golgi vesicles, protein and RNA transport, and regulation of transcription. We provided experimental evidence for the involvement of Hsp27 in DNA repair and mRNA splicing thereby confirming the predictions made from the functional module analysis.
The Role of Hsp27 in DNA Repair and mRNA Splicing Uncovers Hsp27 Functions in Cancer Progression
The integrated bioinformatics analysis of our Y2H screen with an existing collection of human PPI revealed a functional module where Hsp27 interacted with Ku80 (Figs. 1A, 1B and 3D, d). Along with the subunit Ku70, the Ku heterodimer mediates NHEJ in which DSBs bind to one another. Normal cells undergo growth arrest to repair DSBs (70). Conversely, malignant cells (such as PC-3 cells) often present aberrant cell cycle checkpoints control and have a decreased capacity of DSBs repair (71). Cells with deficiencies in DSBs repair are prone to translocations, gene amplifications, and deletions leading to chromosomal alterations and cancer predisposition (51). We validated the involvement of Hsp27 in DNA repair of DSBs induced by Zeocin™ in PC-3 cells (Fig. 4B and 4C) and demonstrated that control cells cumulate unrepaired DSBs and survive better than cells lacking Hsp27 (Fig. 4E). This is in agreement with the fact that nonrepaired or mis-repaired DSBs and aberrant NHEJ activity are highly linked to increased tumor progression and genetic instability (72). Moreover, our results in PC-3 cells were consistent with previous data showing that defective NHEJ DNA repair is determinant for prostate cancer progression (71). Our results strongly support that Hsp27, when overexpressed in cancer cells, negatively regulates NHEJ DNA repair process through its interaction with Ku80. Because we observed no modification in Ku80 protein expression upon Hsp27 silencing by specific siRNA (data not shown), we exclude a proteasomal degradation mechanism. When overexpressed, Hsp27 interacts with Ku80 after DSB formation (Fig. 4D), which may be responsible for preventing Ku80 interaction with DNA-PK (Fig. 4D), member of the repair complex that binds DNA DSBs through Ku heterodimer interaction (50), therefore contributing to less repair of DSB (Fig. 4B and 4C). It is noteworthy that Hsp27, which also interacts with proteins involved in telomere maintenance (Ku80, STUB1, HSP90, and Fig. 3D, d), can be important for maintaining genomic stability. Accordingly, our results suggest that overexpressed Hsp27 in CRPC could contribute to genomic instability and malignant survival cells by titrating out Ku80, therefore preventing it to act on DSBs and dysfunctional telomeres. In accordance with our results, it was recently published that the inhibition of Hsp27 enhances radio-sensitivity in head-and-neck cancer through modulating DNA repair (73).
mRNA alternative splicing is a gene-expression regulation strategy used by cells. In response to stresses that can affect the execution of normal metabolic processes (including DNA replication, transcription, and translation), mRNA splicing is blocked as a survival defense mechanism leading to regulation of this process (74). Few reports indicate an indirect role of induced Hsps in splicing regulation (75). Hsp27 regulates SRp38, a serine/arginine-rich protein involved in splicing, by enhancing its phosphorylation (56). Moreover, Hsp27 involvement in splicing complex assembly regulation was implied by the indirect interactions of Hsp27 with Ddx20 and SMN through Hsp22 (57). Up to date, no report accounts for a direct participation of Hsp27 in the spliceosome function. Our discovery of direct interactions of Hsp27 with splicing factors, regulators and spliceosome ribonucleoproteins (Fig. 3C, d) strongly suggests a regulatory role for Hsp27 in this process. By providing experimental evidence for the alternative splicing of 1777 genes in PC-3 cells in the absence of Hsp27, the involvement of Hsp27 in differential splicing events is heavily supported (Fig. 5A, supplemental Table S4). Interestingly, the functional annotations of the 1777 genes refer to cellular processes related to malignant survival and tumor progression such as cell adhesion, motility and invasion, proliferation, and differentiation (data not shown) (54). These results are in accordance with the accumulating evidence indicating that alternative splicing contributes to the heterogeneity of prostate cancer (76) but further analysis is necessary to confirm the expression of alternative transcripts and proteins corresponding to the reported 1777 genes in prostate cancer samples. Alternative splicing is known to be regulated at two major levels: the spliceosome assembly and expression and post-translational modifications of the splicing factors, which impact on their intracellular localization, PPI/protein-RNA interactions, and intrinsic activity (54). Hsp27 does not have an effect on the expression or intracellular localization of some of its interacting proteins (data not shown). It may be hypothesized that Hsp27 regulates alternative splicing through interaction and stabilization of splicing factors affecting spliceosome assembly and therefore activity. Our results are in accordance with the accumulating evidence indicating that alternative splicing contributes to the heterogeneity of prostate cancer (76). Further analysis is necessary to confirm the expression of alternative transcripts and proteins corresponding to the reported 1777 genes in prostate cancer samples and to better understand how Hsp27 regulates alternative splicing through its interactions and downstream effectors.
From Interactors to Drug Targets
By means of its overexpression and cytoprotective role in cancer cells, Hsp27 represents a promising therapeutic target for various types of cancers (8). Understanding Hsp27 cytoprotective functions through the identification of its interactions specific to cancer cells, represents a great step forward to identifying potent sets of targets for drug development (46, 77). Mapping of the Hsp90 protein interaction network previously led to a remarkable progress in the development of Hsp90 inhibitors nowadays in advanced cancer clinical trials (78). Given the extended and elevated number of Hsp27 client proteins, more directed means of disrupting the Hsp27 network could be considered. We previously provided a rationale to this concept by showing that the Hsp27-interacting protein TCTP (Fig. 1A and Supplemental Table S2) is involved in Hsp27 cytoprotection in CRPC (30). We have shown here that Hsp27 stabilized TCTP by inhibiting its stress-induced ubiquitination and proteasomal degradation. We therefore provided proof of principle that TCTP is an inhibitor of apoptosis in human CRPC, plus a rational target for therapy of CRPC that could avoid undesirable toxicity in normal tissues through TCTP inhibition (ASO) (30). In the present study, we have provided evidence of the potential therapeutic interest in targeting the Hsp27-TCTP interaction as we demonstrated increased treatment-sensitivity of cancer cells after interruption of this interaction (Fig. 2B). These results open a promising field of developing compounds efficiently disrupting the Hsp27-TCTP interaction. The same strategy has been previously described for sepherdin and AICAR, two novel inhibitors of the Hsp90/survivin interaction (79). Selective targeting of essential survival processes of malignant cells may be useful in future therapeutics development. For instance, because of the involvement of telomerase activity and lengthening of telomeres in tumor cell progression, these mechanisms have emerged as promising targets. In addition, the spliceosome machinery has been proposed as a novel target for anticancer drug development (80). Components of the DSBs repair pathways such as Ku80 are also targets for anticancer drugs' design (81). By demonstrating the involvement of Hsp27 in these functions, we propose that targeting specific interactions or proteins within these pathways in cancer cells could potentiate the anti-tumor effects of Hsp27 inhibition.
Altogether, our present work provides an in-depth picture of Hsp27 PPI networks as well as new insights into Hsp27 multifunctionality, considerably contributing in both basic cell function and medical therapeutics. Precisely, we provide an enriched understanding of Hsp27 mechanisms of cytoprotection contributing to cancer progression and open a new promising field of research for multitarget therapeutic approaches.
Supplementary Material
Acknowledgments
We thank Christophe de la Roche Sainte André for his valuable advice on this work. We are particularly grateful to Mauro Modesti for his expertise in DSB experiments and for kindly providing us with Zeocin TM and to Andreas Zanzoni for carefully reading the manuscript. We also thank José Adélaïde for his technical advice.
Footnotes
Author contributions: M.K., C.B., and P.R. designed research; M.K., C.A., V.B., A.B., O.G., and P.F. performed research; N.J.D., O.G., and P.F. contributed new reagents or analytic tools; M.K., A.B., F.B., and C.B. analyzed data; M.K., A.B., C.B., and P.R. wrote the paper; M.K. read, approved and revised the final manuscript; C.A., V.B., N.J.D., O.G., P.F., C.G., and F.B. read and approved the final manuscript; A.B. critically revised the manuscript; D.B., C.B., and P.R. participated in the coordination and revision.
* This work was supported by grants from: French Cancer Institute (InCa, PAIR prostate program #R10111AA), ITMO Cancer (BioSys call, #A12171AS), Institut National de la Santé et de la Recherche Médicale (Inserm), Association pour la Recherche sur le Cancer (ARC, MK), Association pour la Recherche sur les Tumeurs de la Prostate (ARTP, CA), French Ministère de l'Enseignement Supérieur et de la Recherche (MRT, VB), Agence Nationale pour la Recherche (ANR, Emergence Program #ANR-11-EMMA-0022), Aix-Marseille University and competitivity pole Eurobiomed. Authors declare to have no financial, personal, or professional competing interest.
 This article contains supplemental Figs. S1 to S54 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S54 and Tables S1 to S5.
1 The abbreviations used are:
- Hsp
- Heat shock protein
- TCTP
- translationally controlled tumor protein
- CRPC
- castration-resistant prostate cancer
- ASO
- antisense oligonucleotides
- siRNA
- small interference RNA
- co-IP
- co-immunoprecipitation
- GO
- Gene Ontology
- OCG
- Overlapping Cluster Generator
- DSBs
- double-strand breaks
- NHEJ
- nonhomologous end-joining
- HR
- homologous recombination
- γ-H2AX
- phosphorylated histone H2AX.
REFERENCES
- 1. Didelot C., Schmitt E., Brunet M., Maingret L., Parcellier A., Garrido C. (2006) Heat shock proteins: endogenous modulators of apoptotic cell death. Handb. Exp. Pharmacol. 172, 171–198 [DOI] [PubMed] [Google Scholar]
- 2. Parcellier A., Schmitt E., Brunet M., Hammann A., Solary E., Garrido C. (2005) Small heat shock proteins HSP27 and alphaB-crystallin: cytoprotective and oncogenic functions. Antioxid. Redox Signal 7, 404–413 [DOI] [PubMed] [Google Scholar]
- 3. Garrido C., Brunet M., Didelot C., Zermati Y., Schmitt E., Kroemer G. (2006) Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle 5, 2592–2601 [DOI] [PubMed] [Google Scholar]
- 4. Kampinga H. H., Hageman J., Vos M. J., Kubota H., Tanguay R. M., Bruford E. A., Cheetham M. E., Chen B., Hightower L. E. (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperon. 14, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rocchi P., Beraldi E., Ettinger S., Fazli L., Vessella R. L., Nelson C., Gleave M. (2005) Increased Hsp27 after androgen ablation facilitates androgen-independent progression in prostate cancer via signal transducers and activators of transcription 3-mediated suppression of apoptosis. Cancer Res. 65, 11083–11093 [DOI] [PubMed] [Google Scholar]
- 6. Rocchi P., So A., Kojima S., Signaevsky M., Beraldi E., Fazli L., Hurtado-Coll A., Yamanaka K., Gleave M. (2004) Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 64, 6595–6602 [DOI] [PubMed] [Google Scholar]
- 7. Rocchi P., Jugpal P., So A., Sinneman S., Ettinger S., Fazli L., Nelson C., Gleave M. (2006) Small interference RNA targeting heat-shock protein 27 inhibits the growth of prostatic cell lines and induces apoptosis via caspase-3 activation in vitro. BJU Int. 98, 1082–1089 [DOI] [PubMed] [Google Scholar]
- 8. Hotte S.J., Yu E.Y., Hirte H.W., Higano C.S., Gleave M., Chi K.N. (2009) OGX-427, a 20 methoxyethyl antisense oligonucleotide (ASO), against Hsp27: Results of a first-in-human trial. J Clin Oncol. ASCO Annual Meeting Proceedings 2010; 28(15) suppl, 2010: 3077 [Google Scholar]
- 9. Ehrnsperger M., Graber S., Gaestel M., Buchner J. (1997) Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Venkatakrishnan C. D., Dunsmore K., Wong H., Roy S., Sen C. K., Wani A., Zweier J. L., Ilangovan G. (2008) HSP27 regulates p53 transcriptional activity in doxorubicin-treated fibroblasts and cardiac H9c2 cells: p21 upregulation and G2/M phase cell cycle arrest. Am. J. Physiol. Heart Circ. Physiol. 294, H1736–1744 [DOI] [PubMed] [Google Scholar]
- 11. Parcellier A., Brunet M., Schmitt E., Col E., Didelot C., Hammann A., Nakayama K., Nakayama K. I., Khochbin S., Solary E., Garrido C. (2006) HSP27 favors ubiquitination and proteasomal degradation of p27Kip1 and helps S-phase re-entry in stressed cells. FASEB J. 20, 1179–1181 [DOI] [PubMed] [Google Scholar]
- 12. Concannon C. G., Gorman A. M., Samali A. (2003) On the role of Hsp27 in regulating apoptosis. Apoptosis 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 13. Doerwald L., van Genesen S. T., Onnekink C., Marin-Vinader L., de Lange F., de Jong W. W., Lubsen N. H. (2006) The effect of alphaB-crystallin and Hsp27 on the availability of translation initiation factors in heat-shocked cells. Cell Mol. Life Sci. 63, 735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mounier N., Arrigo A. P. (2002) Actin cytoskeleton and small heat shock proteins: how do they interact? Cell Stress Chaperon. 7, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pandey P., Farber R., Nakazawa A., Kumar S., Bharti A., Nalin C., Weichselbaum R., Kufe D., Kharbanda S. (2000) Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene 19, 1975–1981 [DOI] [PubMed] [Google Scholar]
- 16. Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. (1999) From molecular to modular cell biology. Nature 402, C47–52 [DOI] [PubMed] [Google Scholar]
- 17. Thalappilly S., Suliman M., Gayet O., Soubeyran P., Hermant A., Lecine P., Iovanna J. L., Dusetti N. J. (2008) Identification of multi-SH3 domain-containing protein interactome in pancreatic cancer: a yeast two-hybrid approach. Proteomics 8, 3071–3081 [DOI] [PubMed] [Google Scholar]
- 18. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 19. Herrmann C., Berard S., Tichit L. (2009) SimCT: a generic tool to visualize ontology-based relationships for biological objects. Bioinformatics 25, 3197–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bossi A., Lehner B. (2009) Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker E., Robisson B., Chapple C. E., Guenoche A., Brun C. (2012) Multifunctional proteins revealed by overlapping clustering in protein interaction network. Bioinformatics 28, 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Madhoun A. S., Chen Y. X., Haidari L., Rayner K., Gerthoffer W., McBride H., O'Brien E. R. (2007) The interaction and cellular localization of HSP27 and ERbeta are modulated by 17beta-estradiol and HSP27 phosphorylation. Mol. Cell. Endocrinol. 270, 33–42 [DOI] [PubMed] [Google Scholar]
- 24. Baylot V., Andrieu C., Katsogiannou M., Taieb D., Garcia S., Giusiano S., Acunzo J., Iovanna J., Gleave M., Garrido C., Rocchi P. (2011) OGX-427 inhibits tumor progression and enhances gemcitabine chemotherapy in pancreatic cancer. Cell Death Dis. 2, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrieu C., Taieb D., Baylot V., Ettinger S., Soubeyran P., De-Thonel A., Nelson C., Garrido C., So A., Fazli L., Bladou F., Gleave M., Iovanna J. L., Rocchi P. (2010) Heat shock protein 27 confers resistance to androgen ablation and chemotherapy in prostate cancer cells through eIF4E. Oncogene 29, 1883–1896 [DOI] [PubMed] [Google Scholar]
- 26. Yin H., Glass J. (2006) In prostate cancer cells the interaction of C/EBPalpha with Ku70, Ku80, and poly(ADP-ribose) polymerase-1 increases sensitivity to DNA damage. J. Biol. Chem. 281, 11496–11505 [DOI] [PubMed] [Google Scholar]
- 27. Purdom E., Simpson K. M., Robinson M. D., Conboy J. G., Lapuk A. V., Speed T. P. (2008) FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics 24, 1707–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1544–6115 [DOI] [PubMed] [Google Scholar]
- 29. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baylot V., Katsogiannou M., Andrieu C., Taieb D., Acunzo J., Giusiano S., Fazli L., Gleave M., Garrido C., Rocchi P. (2012) Targeting TCTP as a new therapeutic strategy in castration-resistant prostate cancer. Mol. Ther. 20, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venkatesan K., Rual J. F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K. I., Yildirim M. A., Simonis N., Heinzmann K., Gebreab F., Sahalie J. M., Cevik S., Simon C., de Smet A. S., Dann E., Smolyar A., Vinayagam A., Yu H., Szeto D., Borick H., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Cusick M. E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabasi A. L., Vidal M. (2009) An empirical framework for binary interactome mapping. Nat. Methods 6, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W., Chen H., Jin C., Wei J., Zhang W., Yang Y., Liu Q., Zhou Y., Zhang C., Wu Z., Xu W., Zhang Y., Liu T., Yu D., Chen L., Zhu D., Zhong X., Kang L., Gan X., Yu X., Ma Q., Yan J., Zhou L., Liu Z., Zhu Y., Zhou T., He F., Yang X. (2011) Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 7, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F. S., Cesareni G., Chatr-aryamontri A., Chautard E., Chen C., Dumousseau M., Goll J., Hancock R. E., Hannick L. I., Jurisica I., Khadake J., Lynn D. J., Mahadevan U., Perfetto L., Raghunath A., Ricard-Blum S., Roechert B., Salwinski L., Stumpflen V., Tyers M., Uetz P., Xenarios I., Hermjakob H. (2012) Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods 9, 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cuesta R., Laroia G., Schneider R. J. (2000) Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 14, 1460–1470 [PMC free article] [PubMed] [Google Scholar]
- 35. Brun C., Chevenet F., Martin D., Wojcik J., Guenoche A., Jacq B. (2003) Functional classification of proteins for the prediction of cellular function from a protein-protein interaction network. Genome Biol. 5, R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharan R., Ulitsky I., Shamir R. (2007) Network-based prediction of protein function. Mol. Syst. Biol. 3, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paul C., Simon S., Gibert B., Virot S., Manero F., Arrigo A. P. (2010) Dynamic processes that reflect anti-apoptotic strategies set up by HspB1 (Hsp27). Exp. Cell Res. 316, 1535–1552 [DOI] [PubMed] [Google Scholar]
- 38. Smolewski P., Robak T. (2011) Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Curr. Mol. Med. 11, 633–649 [DOI] [PubMed] [Google Scholar]
- 39. Eckelman B. P., Salvesen G. S., Scott F. L. (2006) Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7, 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thorsten G. H., Harutyunyan A. S., Nivarthi H., Rumi E., Jelena D., Milosevic N., Them C. C., Berg T., Gisslinger B., Pietra D., Chen D., Vladimer G. I., Bagienski K., Milanesi C., Carola Casetti I., Sant'Antonio E., Ferretti V., Elena C., Schischlik F., Cleary C., Six M., Schalling M., Schönegger A., Bock C., Malcovati L., Pascutto C., Superti-Furga G., Cazzola M., Kralovics R. (2013) LBA-1 Frequent Mutations in the Calreticulin Gene CALR in Myeloproliferative Neoplasms. ASH Annual Meeting 55th edition, ASH Annual Meeting [Google Scholar]
- 41. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun C., Todorovic A., Querol-Audi J., Bai Y., Villa N., Snyder M., Ashchyan J., Lewis C. S., Hartland A., Gradia S., Fraser C. S., Doudna J. A., Nogales E., Cate J. H. (2011) Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl. Acad. Sci. U.S.A. 108, 20473–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perzlmaier A. F., Richter F., Seufert W. (2013) Translation initiation requires cell division cycle 123 (Cdc123) to facilitate biogenesis of the eukaryotic initiation factor 2 (eIF2). J. Biol. Chem. 288, 21537–21546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Metzger M. B., Hristova V. A., Weissman A. M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Farkas I. J., Korcsmaros T., Kovacs I. A., Mihalik A., Palotai R., Simko G. I., Szalay K. Z., Szalay-Beko M., Vellai T., Wang S., Csermely P. (2011) Network-based tools for the identification of novel drug targets. Sci. Signal 4, pt3. Available at: (http://stke.sciencemag.org/content/4/173/pt3.long) [DOI] [PubMed] [Google Scholar]
- 46. Vidal M., Cusick M. E., Barabasi A. L. (2011) Interactome networks and human disease. Cell 144, 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malyarchuk S., Castore R., Harrison L. (2008) DNA repair of clustered lesions in mammalian cells: involvement of nonhomologous end-joining. Nucleic Acids Res. 36, 4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Toogun O. A., Dezwaan D. C., Freeman B. C. (2008) The hsp90 molecular chaperone modulates multiple telomerase activities. Mol. Cell. Biol. 28, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee J. H., Khadka P., Baek S. H., Chung I. K. (2010) CHIP promotes human telomerase reverse transcriptase degradation and negatively regulates telomerase activity. J. Biol. Chem. 285, 42033–42045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lees-Miller S. P., Meek K. (2003) Repair of DNA double strand breaks by nonhomologous end joining. Biochimie 85, 1161–1173 [DOI] [PubMed] [Google Scholar]
- 51. Khanna K. K., Jackson S. P. (2001) DNA double-strand breaks: signaling, repair, and the cancer connection. Nat. Genet. 27, 247–254 [DOI] [PubMed] [Google Scholar]
- 52. Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ehrenfeld G. M., Shipley J. B., Heimbrook D. C., Sugiyama H., Long E. C., van Boom J. H., van der Marel G. A., Oppenheimer N. J., Hecht S. M. (1987) Copper-dependent cleavage of DNA by bleomycin. Biochemistry 26, 931–942 [DOI] [PubMed] [Google Scholar]
- 54. Germann S., Gratadou L., Dutertre M., Auboeuf D. (2012) Splicing programs and cancer. J. Nucleic Acids 2012, Art. No. 269570 Volume 2012, Article ID 269570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matlin A. J., Moore M. J. (2007) Spliceosome assembly and composition. Adv. Exp. Med. Biol. 623, 14–35 [DOI] [PubMed] [Google Scholar]
- 56. Marin-Vinader L., Shin C., Onnekink C., Manley J. L., Lubsen N. H. (2006) Hsp27 enhances recovery of splicing as well as rephosphorylation of SRp38 after heat shock. Mol. Biol. Cell 17, 886–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun X., Fontaine J. M., Hoppe A. D., Carra S., DeGuzman C., Martin J. L., Simon S., Vicart P., Welsh M. J., Landry J., Benndorf R. (2010) Abnormal interaction of motor neuropathy-associated mutant HspB8 (Hsp22) forms with the RNA helicase Ddx20 (gemin3). Cell Stress Chaperones 15, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 59. Xing Y., Lee C. (2006) Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 7, 499–509 [DOI] [PubMed] [Google Scholar]
- 60. Futreal P. A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M. R. (2004) A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar-Sinha C., Kalyana-Sundaram S., Chinnaiyan A. M. (2012) SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing. Cancer Discov. 2, 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Perner S., Rupp N. J., Braun M., Rubin M. A., Moch H., Dietel M., Wernert N., Jung K., Stephan C., Kristiansen G. (2013) Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int. J. Cancer 132, 807–812 [DOI] [PubMed] [Google Scholar]
- 63. Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 64. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., Vidal M. (2009) An experimentally derived confidence score for binary protein–protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boxem M., Maliga Z., Klitgord N., Li N., Lemmens I., Mana M., de Lichtervelde L., Mul J. D., van de Peut D., Devos M., Simonis N., Yildirim M. A., Cokol M., Kao H. L., de Smet A. S., Wang H., Schlaitz A. L., Hao T., Milstein S., Fan C., Tipsword M., Drew K., Galli M., Rhrissorrakrai K., Drechsel D., Koller D., Roth F. P., Iakoucheva L. M., Dunker A. K., Bonneau R., Gunsalus K. C., Hill D. E., Piano F., Tavernier J., van den Heuvel S., Hyman A. A., Vidal M. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pilot-Storck F., Chopin E., Rual J. F., Baudot A., Dobrokhotov P., Robinson-Rechavi M., Brun C., Cusick M. E., Hill D. E., Schaeffer L., Vidal M., Goillot E. (2010) Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol. Cell. Proteomics 9, 1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kammoun M., Picard B., Henry-Berger J., Cassar-Malek I. (2013) A network-based approach for predicting Hsp27 knock-out targets in mouse skeletal muscles. Comput. Struct. Biotechnol. J. 6, e201303008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arrigo A. P. (2007) The cellular “networking” of mammalian Hsp27 and its functions in the control of protein folding, redox state, and apoptosis. Adv. Exp. Med. Biol. 594, 14–26 [DOI] [PubMed] [Google Scholar]
- 69. Baylot V., Katsogiannou M., Andrieu C., Taieb D., Acunzo J., Giusiano S., Fazli L., Gleave M., Garrido C., Rocchi P. (2012) Targeting TCTP as a New Therapeutic Strategy in Castration-resistant Prostate Cancer. Mol. Ther. 20, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ohnishi T., Mori E., Takahashi A. (2009) DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat. Res. 669, 8–12 [DOI] [PubMed] [Google Scholar]
- 71. Fan R., Kumaravel T. S., Jalali F., Marrano P., Squire J. A., Bristow R. G. (2004) Defective DNA strand break repair after DNA damage in prostate cancer cells: implications for genetic instability and prostate cancer progression. Cancer Res. 64, 8526–8533 [DOI] [PubMed] [Google Scholar]
- 72. Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B. S. (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14, 611–623 [DOI] [PubMed] [Google Scholar]
- 73. Guttmann D. M., Hart L., Du K., Seletsky A., Koumenis C. (2013) Inhibition of Hsp27 radiosensitizes head-and-neck cancer by modulating deoxyribonucleic acid repair. Int. J. Radiat. Oncol. Biol. Phys. 87, 168–175 [DOI] [PubMed] [Google Scholar]
- 74. Biamonti G., Caceres J. F. (2009) Cellular stress and RNA splicing. Trends Biochem. Sci. 34, 146–153 [DOI] [PubMed] [Google Scholar]
- 75. Corell R. A., Gross R. H. (1992) Splicing thermotolerance maintains Pre-mRNA transcripts in the splicing pathway during severe heat shock. Exp. Cell Res. 202, 233–242 [DOI] [PubMed] [Google Scholar]
- 76. Sette C. (2013) Alternative splicing programs in prostate cancer. Int. J. Cell Biol. 2013, Art. No. 458727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khan S. H., Ahmad F., Ahmad N., Flynn D. C., Kumar R. (2011) Protein–protein interactions: principles, techniques, and their potential role in new drug development. J. Biomol. Struct. Dyn. 28, 929–938 [DOI] [PubMed] [Google Scholar]
- 78. Pearl L. H., Prodromou C., Workman P. (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 410, 439–453 [DOI] [PubMed] [Google Scholar]
- 79. Meli M., Pennati M., Curto M., Daidone M. G., Plescia J., Toba S., Altieri D. C., Zaffaroni N., Colombo G. (2006) Small-molecule targeting of heat shock protein 90 chaperone function: rational identification of a new anticancer lead. J. Med. Chem. 49, 7721–7730 [DOI] [PubMed] [Google Scholar]
- 80. van Alphen R. J., Wiemer E. A., Burger H., Eskens F. A. (2009) The spliceosome as target for anticancer treatment. Br. J. Cancer 100, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bolderson E., Richard D. J., Zhou B. B., Khanna K. K. (2009) Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin. Cancer Res. 15, 6314–6320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.