Abstract
Protein interaction domain (PID) linear peptide motif interactions direct diverse cellular processes in a specific and coordinated fashion. PID specificity, or the interaction selectivity derived from affinity preferences between possible PID-peptide pairs is the basis of this ability. Here, we develop an integrated experimental and computational cellulose peptide conjugate microarray (CPCMA) based approach for the high throughput analysis of PID specificity that provides unprecedented quantitative resolution and reproducibility. As a test system, we quantify the specificity preferences of four Src Homology 2 domains and 124 physiological phosphopeptides to produce a novel quantitative interactome. The quantitative data set covers a broad affinity range, is highly precise, and agrees well with orthogonal biophysical validation, in vivo interactions, and peptide library trained algorithm predictions. In contrast to preceding approaches, the CPCMAs proved capable of confidently assigning interactions into affinity categories, resolving the subtle affinity contributions of residue correlations, and yielded predictive peptide motif affinity matrices. Unique CPCMA enabled modes of systems level analysis reveal a physiological interactome with expected node degree value decreasing as a function of affinity, resulting in minimal high affinity binding overlap between domains; uncover that Src Homology 2 domains bind ligands with a similar average affinity yet strikingly different levels of promiscuity and binding dynamic range; and parse with unprecedented quantitative resolution contextual factors directing specificity. The CPCMA platform promises broad application within the fields of PID specificity, synthetic biology, specificity focused drug design, and network biology.
Protein interaction domains (PIDs)1 often compete for the same linear motif binding sites across a range of affinities, resulting in many potential interactions that may enable the rapid assembly and disassembly of signaling proteins in response to external and internal cues (1, 2). PID-peptide interactions have small binding interfaces, resulting in moderate affinity interactions mediated primarily by a few amino acid “hot-spots” within motifs specific for a particular PID family (3–5). The power of individual residues to direct interactions, the absence of structural constraint for linear motifs, and the modularity of PIDs has enabled the rapid evolution of these networks resulting in many large multimember PID families in higher eukaryotes (6–9). For these large families dedicated to the recognition of similar ligands, PID specificity—or the interaction selectivity derived from affinity preferences between possible PID-peptide pairs—underpins the effective conveyance of specific cell signals. High throughput interaction mapping efforts are used to decipher how this PID “specificity space” is populated, thereby providing insight into protein function and the principles of network architecture and evolution (10–15). The extent of binding overlap or interaction promiscuity within and between PID families for physiological ligands, the affinity range of overlapping interactions, and the biological relevancy of these interactions are important questions thus far poorly resolved by existing high throughput methods. Here, we develop and apply a quantitative high throughput method capable of addressing these questions.
Peptide arrays (16, 17), degenerate libraries (18, 19), and phage display (20) are the most frequently applied high throughput approaches for investigating PID specificity. Phage display and degenerate library approaches sample a large ligand space and can produce consensus selectivity motifs that represent the most preferred residues at every position panned. This selectivity data is used to predict interactions, often via position specific scoring matrices (PSSMs) (21–23). However, neither approach can explicitly measure nonbinding events and only large phage display data sets can resolve a limited subset of high-affinity contextual binding information (24). Nonbinding information and contextual interplay, that is, correlated contributions between ligand positions to binding affinity, play important roles in defining the specificity landscapes for multiple PID families (25–27). Having explicit nonbinding or low-affinity information available helps uncover contextual binding information, and improves the accuracy of interaction priority assignment between multiple competing PIDs. Correspondingly, the availability of nonbinding and contextual information improves interaction prediction performance (28, 29). Peptide arrays using physiological ligands do not have these limitations, yet may under-sample PID specificity space because of smaller library sizes. Newly emerging ultrahigh density peptide arrays avoid this particular limitation and are capable of sampling the entire proteome (30). However, a common limitation for all of these techniques is their dependence on nonquantitative interaction information.
A comprehensive understanding of PID specificity space requires the quantitative assessment of pairwise interactions across a broad dynamic range of affinities. Common low-throughput biophysical techniques used to measure protein-peptide interaction affinities require highly pure and often large amounts of interactants along with prior knowledge of their interaction. To facilitate discovery and lessen the stringency of the purity and/or quantity requirements of interacting molecules, multiple quantitative high throughput methodologies have been developed. Thus far, the protein microarray (PMA) (31) and high-throughput fluorescence polarization (HTFP) (11, 32) quantitative approaches have been used to examine PID specificity. Unfortunately, PMAs suffer from poor sensitivity, reproducibility, and measurement discrepancies (32, 33). The alternative HTFP assay is more sensitive than PMAs, but also has poor reproducibility and is biased toward high affinity interactions (32). Further, PMAs and HTFP have minimal KD sensitivities of 2 and 20 μm, respectively. This boundary limits their scope of application considering the importance of moderate affinity interactions for many PID families (34) and the general importance of these interactions in directing emergent phenomena such as ultrasensitivity and interaction gating (35, 36).
The Src Homology 2 (SH2) domain phosphotyrosine interactome is a system of prominent physiological importance that has been the subject of multiple preceding interaction mapping and platform development efforts (17, 19, 31, 32, 37, 38). Tyrosine phosphorylation and its recognition by SH2 domains is a uniquely metazoan adaptation (39), prominently involved in the transduction of growth signals (1, 2, 40). SH2 domain specificity has been leveraged in multiple contexts to rewire signaling pathways (15), and specificity dysregulation is associated with multiple diseases, including cancer (40–42). Despite the importance of directing specific signals, many SH2 domains share sequence preferences, resulting in substantial binding crossover over a broad dynamic range of affinities (27). Yet, the extent of this crossover and the peptide motif characteristics responsible for its direction are poorly resolved. In light of these biological attributes and the availability of preceding information to guide platform development and assess its performance, we chose a significantly novel set of 124 physiological phosphopeptides and four SH2 domains as a pilot interactome for our arrays.
Here we develop a cellulose peptide conjugate microarray (CPCMA) based approach that is the first broadly applicable protein interaction mapping approach capable of explicitly capturing binding and nonbinding information, resolving the contextual interplay between amino acids that contribute to binding, and providing a quantitative measure of interaction affinity. We optimize multiple production and incubation parameters for the arrays and develop an experimental and computational pipeline to control for systematic artifacts and robustly estimate affinities, ultimately producing a physiological interactome of 368 measurements between 92 peptides and four domains. In contrast to preceding platforms, we find our approach is highly sensitive, extremely precise, and consistent with multiple forms of orthogonal biophysical and in vivo validation. Leveraging unique modes of analysis enabled by the CPCMAs, we demonstrate that SH2 domains vary considerably in promiscuity and binding dynamic range in a manner aligned with biological function. Consistent with recent evidence that pathway crosstalk is subjected to negative selection (10–12, 43) we demonstrate for the first time that the degree distribution of the interactome is a function of affinity, and therefore little binding overlap is found among the highest affinity interactions, suggesting that such sites may be important points of in vivo coregulation. We highlight a few such interactions and corroborate others using available domain-site specific in vivo interaction information. Next, in order to provide insight into the specificity determinants responsible for this segregation we produce for the first time predictive domain specific binding motif affinity matrices. Lastly, we demonstrate the unique ability of the CPCMAs to resolve contextual contributions to binding, and highlight examples where contextual information contributes to binding specificity.
Quantitative analysis of systems-level PID properties is a necessary prerequisite for improved mathematical modeling of signal transduction systems, specificity focused drug design, and engineering advanced synthetic biology circuits. Thus, our quantitative CPCMA based approach should have broad application in future network biology and PID specificity efforts.
EXPERIMENTAL PROCEDURES
SH2 Domain Production and Purification
SH2 domain sequences were cloned into pGEX vectors as GST fusions as described (27) and expressed in E. coli BL21 cells using autoinduction recipes and protocols according to Studier et al. (44). Following overnight culturing, cells were pelleted and resuspended in 50 mm HEPES, 150 mm NaCl, 1 mm EDTA, 1 mm dithiotreitol, and 0.1% Triton X-100 pH 7.5 + protease inhibitor mixture (EMD). Unless stated otherwise, all purification steps were carried out at 4 °C. The suspension was sonicated, pelleted, and the supernatant was mixed with polyethyleneimine (PEI) to .1% (v/v) forming a white precipitate. This precipitate was pelleted in an ultracentrifuge, and the GST-SH2 domain containing supernatant was subjected to batch affinity chromatography using glutathione (GSH) functionalized resin (Fisher) for 2 h at RT or overnight. The proteins were eluted from the beads with 10 mm GSH in .1 m HEPES pH 8.0. Eluted protein was buffer exchanged using Nap10 (GE) columns for cation exchange chromatography. Proteins were further purified with an AKTA purifier system (GE) using a cation exchange resin SP (GE), CM (GE), or heparin (GE) to remove nucleic acid contamination. In isolated cases, proteins were subjected to gel filtration (GE) for additional cleanup. Following centrifugal concentration (Thermo), proteins were buffer exchanged into 50 mm HEPES, 100 mm NaCl, 1 mm EDTA, 1 mm DTT, 20% v/v glycerol pH 7.5 using Nap10 or PD MiniTrap (GE) columns, flash frozen, and stored at −80 °C until needed. Aliquots were subjected to SDS-PAGE with Coomassie Blue stain to verify purity. Concentration measurements were performed via BCA assay (Thermo) according to the manufacturer's instructions.
Soluble Peptide Production and Fluorescence Polarization
Peptides were synthesized using 9-Fluorenylmethoxycarbonyl (FMOC)-chemistry onto preloaded tenta-gel resins. Peptides were labeled N-terminally with Rhodamine B (Abbey Color, Fremont, CA) or 5–6 carboyfluorescein (Anaspec) and cleaved using trifluoroacetic acid. Peptides were purified and mass quantified using a LC/MS or MALDI mass analyzer (Agilent 2100). Peptides were lyophilized and stored under nitrogen at −20 °C until use. A subset of fluorescein labeled peptides was also purchased (Genscript, Piscataway, NJ). FP experiments were conducted using the Beacon 2000 (Invitrogen, Carlsbad, CA).
SPOT Synthesis
Membranes were synthesized using the MultiPep high throughput peptide synthesizer (Intavis, Cologne, Germany). All Fmoc-Amino Acids were purchased from Bachem, except Fmoc-pTyr (Anaspec). All solvents were purchased from Fisher unless otherwise indicated. Cellulose membranes were purchased from Intavis. The SPOT synthesis method was carried out on cellulose membranes using Fmoc chemistry as described in detail elsewhere (45). Each 11-mer peptide chain was synthesized with pTyr or Tyr in the 5th position relative to the N terminus. To prevent oxidation mediated artifacts, all cysteine residues were replaced with serine and all methionine residues were replaced with norleucine. All peptides were acetylated at the N terminus.
SC2 Synthesis
Following synthesis, each of the 384 peptide spots were manually removed from the membrane using a hole-puncher and placed into individual wells of a 96 well plate (Matrix Technologies, Toledo, OH). Each well was treated with 150 μl of a deprotection mixture consisting of 95% trifluoroacetic acid, 3% triisopropylsilane (TIPS), and 2% H2O for 1.5 h. Deprotection mixture is then aspirated from each well and replaced with 250 μl of trifluoroacetic acid (93.5%), Trifluoromethanesulfonic acid (TFMSA) (2.5%), H2O (2.5%), TIPS (1.5%), or one of the alternative dissolution cocktails (see Table I) for 17 h at RT with minor agitation. Subsequently each CPC is precipitated and washed three times with cold MTBE using the Precision 2000 pipetting robot (BioTek, Winooski, VT). The final wash supernatant is removed and each CPC is dissolved in 100 μl dry dimethyl sulfoxide with agitation. The dissolved conjugates are transferred in 20 μl aliquots to five 384 well plates (Genetix) by robotic or manual pipetting. The plates are then centrifuged and covered with metallic sealer to prevent evaporation (Dot Scientific). The plates are flush or flash frozen and stored at −20 in a desiccator until used for printing.
Table I. Assay sensitivity as a function of dissolving cocktail acidity.
| Dissolving cocktail | CBD sensitivity | Anti-pY sensitivity |
|---|---|---|
| TFA (92.5%), TFMSA (4%), H2O (1%), TIPS (2.5%)a | N.A. | N.A. |
| TFA (88.5%), TFMSA (4%), H2O (5%), TIPS (2.5%) - “High” | ++ | +++ |
| TFA (93.5%), TFMSA (2.5%), H2O (2.5%), TIPS (1.5%) - “Medium” | +++ | +++ |
| TFA (93.5%), TFMSA (1%), H2O (3%), TIPS (2.5%) - “Low” | + | + |
| TFA (82%), H2O (5%), TIPS (3%), DCM (10%)b | N.A. | N.A. |
(+++) superior, (++) good, (+) poor, “N.A.” not applicable.
a Precipitate not recoverable following acid mediated dissolution.
b Ineffective acid mediated dissolution.
Microarray Fabrication
Plates are thawed to RT in a desiccator before removing the seal. Five microliter aliquots were used for printing each batch of arrays. Each print batch was fabricated using the Omnigrid 100 (Digilab, Marlborough, MA) contact arrayer using a single SMP3 split pin (Arrayit, Sunnyvale, CA). Arrays were printed on amine, aldehyde, acrylic, epoxy, or nontreated slides. All slides were purchased from Genetix with the exception of the nontreated slides that were purchased from Fisher. Each unique CPC or CPC dilution combination was spotted four times per microarray for control experiments, 12 times per microarray for quantitative arrays and substrate comparisons (one/well), and 24 times per microarray for dissolution condition comparisons (two/well). After printing slides were stored in a desiccator at 4 °C until ready for use.
CPCMA Incubations and Analysis
All array incubations were performed using a custom designed ArraySlide and ArrayMix (Gel Company, San Francisco, CA) orbital shaker at 300 rpm according to the protocol outlined here. All steps were performed at RT unless otherwise indicated. All blocking and incubation steps were performed covered with seal plates to prevent evaporation. All wash steps were performed with 250 μl of solution unless otherwise indicated. Microarrays were swollen with two 10 min washes of 500 μl H2O followed by two 5 min washes with PBS, and two 5 min washes with PBS-T (.1% v/v). The arrays were then blocked for 1.5 h with 250 μl PBS-T/dry milk (2% w/v) and 1 mm dithiotreitol. The arrays were then washed three times with 250 μl PBS-T for 5 min. The following protocol was used for all CPCMAs. Primary analyte incubations were performed for 20 h at 4 °C with 100 μl PBS-T/dry milk (2% w/v), with (SH2 domains) or without (Anti-pY/CBD) 1 mm DTT. Anti-pY antibodies were arrayed at 2.5 nm and CBD-Protein L fusion (Fluka) at 45 nm. The arrays were then washed three times with PBS-T for 5 min. Secondary incubations were performed for 5 h with 100 μl PBS-T/dry milk (2% w/v). Anti-GST (Genscript monoclonal A00865) for SH2 domains and control experiments was arrayed at 7 nm, human IgG for CBD-L experiments was arrayed at 70 nm. After another round of PBS-T washes, tertiary incubations were performed for 2.5 h with 100 μl PBS-T/dry milk (2% w/v). AF633 conjugated anti-mouse and anti-human secondary detection antibodies (Invitrogen) were arrayed at 7 nm. The arrays were then washed thrice with PBS-T, twice with PBS, and twice with H2O. After the final wash with H2O slides were removed from the gaskets and washed in a bath of H2O covered in a glass tray (Ted Pella, Inc., Redding, CA) with slight agitation for 10 min. Slides were removed from their gaskets and dried with a stream of extra-dry Ar. Slides were placed covered in a desiccator at 4 °C until scanned. Slides were usually scanned within 1 week of arraying.
Array Scanning
All images were acquired on a Genepix 4000B scanner. Genepix Pro 6.1 software was used for feature identification and image processing. All images were acquired with maximal laser power at 5 μm resolution. Nonquantitative arrays were scanned at a single photomultiplier voltage such that saturated pixels were minimized. Spots with artifacts such as saturated pixels or bleed in from adjacent spots were omitted from further analysis. Average intensities were calculated for each feature on the microarray. For quantitative experiments, voltage levels were chosen such that the lowest setting captured no saturated pixels. Each array was scanned at a minimum of three different voltages, in steps of 100 V.
Data Analysis
Most data analysis steps were performed using the R statistical computing environment (http://www.R-project.org/). Some analysis was also performed using Originlab8.1 and Excel.
For CPCMAs used for substrate comparisons unique CPC/dilution factor replicates (n = 4) for each bioassay were averaged for each slide. For CPCMAs used for dissolution condition comparisons unique CPC/dissolution condition replicates (n = 24/slide) were averaged. For substrate and dissolution condition comparisons, each unique CPC condition was pooled into a single bioassay specific group for statistical comparisons. Group distributions were tested for normality. Normally distributed groups were compared by two sample t-tests and non-normally distributed groups were compared by Wilcoxon rank-sum test. Unless otherwise indicated, hypothesis testing was performed using a two-tailed t test. PCC comparisons were performed using Fisher's r-to-z transformations and a two tailed test of significance. Venn diagrams were produced using the R package “VennDiagram.” The network graphic was produced using cytoscape (46).
Receiver Operating Characteristic (ROC) Analysis
The array produces numerical estimates of affinity and not binary affinity class assignments. We tested the performance of our array derived affinity estimates as a ranking classifier, using our paired FP affinity estimates as the reference set. The KD cutoff corresponds to the reference set of FP values and is used to define the actual class set. For example, in the case of the 10 μm cutoff, if an interaction has an affinity greater than 10 μm as measured by FP it is assigned a 1, and if not a 0. To produce the ROC curve, the interactions with class affiliation are ordered by apparent KD, and the apparent KD (ranking classifier) values are then thresholded, or stepped sequentially throughout the data range, producing different true positive (tpr) and false positive rates (fpr) at each step. Where and such that tp and tn indicate those interactions correctly classified as above or below the threshold and fp and fn indicate those interactions incorrectly classified as above or below the threshold. ROC curves from single concentration data and SMM derived predictions were generated in an analogous fashion. For the 10 μm cutoff, the apparent KD value that produced the maximum accuracy (where ) was also calculated. ROC analyses and the associated plots were produced with the R package “ROCR.”
Literature Curation, SMALI, and Scansite Implementation
The literature was manually mined along with the databases Phosphosite (47), MINT (48), HPRD (49), DOMINO (50), and Pepcyber (51) for protein-protein, domain-protein, and domain-phosphopeptide in vivo interaction information (supplemental Table S2). SMALI (22) and Scansite (21) (low stringency) assigned a binding score to our 124 peptides in accordance with database instructions.
SMM and Logo Generation
The availability of high-quality binding affinity measurements for the SH2 domains offered us an opportunity to build accurate in silico models of the energy contributions from amino acid residues at different positions. We used the stabilized matrix method (SMM) algorithm to build such models (52). Briefly, SMM is a linear regression based method that generates a Position Specific Scoring Matrix (PSSM) given a set of peptides and their measured affinities. The algorithm optimizes a PSSM during “training” such that predicted affinities using the matrix are as close as possible to the measured ones as determined by a sum of squared errors. To predict a binding affinity of a peptide to an SH2 domain, corresponding entries in the domain's PSSM (i.e. residues/positions) are read off and summed. SMM is distinguished from simple applications of a linear regression technique by the following features: (1) trained models are adapted to noise in the affinity measurements, and (2) the algorithm can deal with bounded measurements. The SMM was applied to each domain as outlined previously (52) using the “reactive-quantitative” subset of peptides. Signals were converted into pKD scale, with higher numbers representing stronger affinities. Nonbinding interactions were assigned a value of 5, whereas binding interactions were assigned a value of 6. For each SH2 domain ten fivefold cross validations were performed. The scoring matrices shown in this study were averages of the 50 matrices. Matrices were converted to logos using the “Seq2logo” web tool (53).
RESULTS
CPCMA Production and Optimization
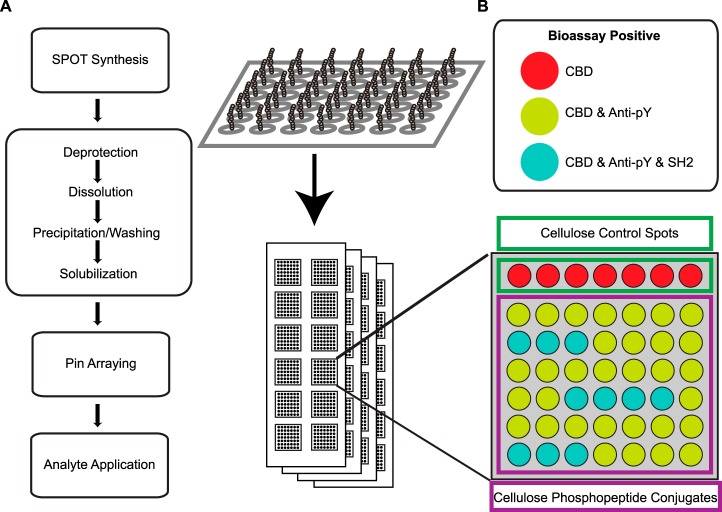
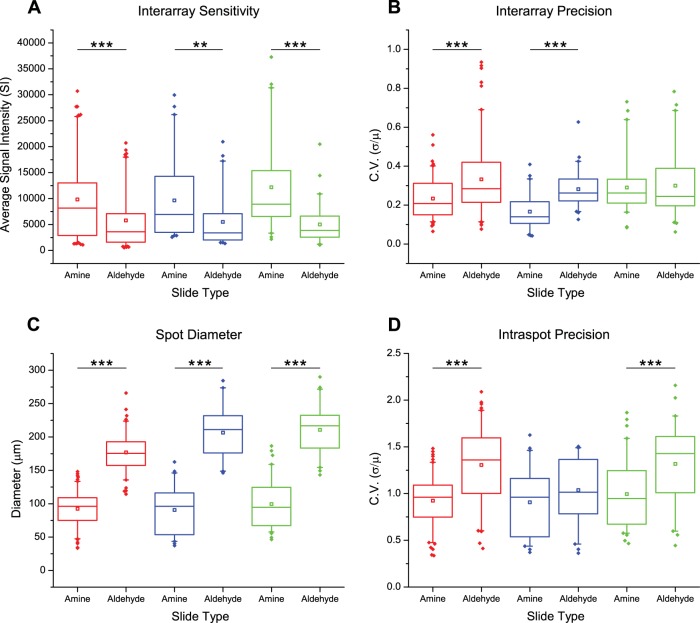
Frank and colleagues recently introduced the spotting compound conjugates (SC2) technique (54) to manufacture multiple peptide microarrays from a single SPOT synthesis cellulose membrane (16) (Fig. 1A). To optimize the CPCMA platform, we varied substrate type and cellulose polymer length and analyzed their effect on assay sensitivity and precision using a series of bioassays. A cellulose-binding domain (CBD) was used to report the density of the printed cellulose matrix, and both a phosphotyrosine-specific antibody (Anti-pY) and a representative SH2 domain from the protein VAV1 were used to report accessible phosphopeptides (Fig. 1B). Noting superior performance in preliminary tests, we examined assay sensitivity, precision, and throughput capacity (spot size) of amine and aldehyde coated surfaces. Using a panel of physicochemically diverse CPCs (supplemental Table S1), a maximum of 1024 measurements was made per bioassay. Amine functionalized slides displayed a clear advantage in sensitivity, precision, spot size, and spot uniformity compared with aldehyde slides (Fig. 2), and were therefore used for subsequent analyses.
Fig. 1.
Overview of CPCMA fabrication, layout, and bioassay interactions. A, 11-mer phosphopeptides are SPOT synthesized on a cellulose membrane via Fmoc SPPS. The CPC spots are manually removed and placed into microplates where they are simultaneously deprotected and acid solubilized. Following precipitation, washing and resolubilization (arrow), they are pin arrayed onto glass substrates with twelve addressable subarrays. Each subarray serves as an independent reaction vessel for each bioassay. B, Example subarray layout and interaction profile. Each subarray contains control spots consisting of cellulose alone (green square) and experimental CPC spots (purple square). The control spots can only be recognized by the CBD bioassay (red spots). All of the properly synthesized and displayed CPCs can be recognized by the CBD and Anti-pY antibody (yellow spots) whereas a subset of these can be recognized by the CBD, Anti-pY antibody, and SH2 domain (blue spots).
Fig. 2.
Amine functionalized substrates demonstrate superior characteristics. Two slides of each type were subjected to the three bioassays (red = CBD; blue = SH2 domain; green = Anti-pY antibody). A, Analysis of inter-array sensitivity indicates that amine coated slides are superior for all three bioassays. (CBD n = 109, SH2 n = 47, Anti-pY n = 49) B, Inter-array precision as measured by the coefficient of variation (C.V.) of signals across two arrays. CBD and the SH2 domain show superior precision for the amine coated slides, whereas Anti-pY shows comparable precision between the two substrates. (CBD n = 109, SH2 n = 47, Anti-pY n = 49) C, The average inter-array feature diameter is much smaller for amine slides, indicating that the amine substrate is capable of increased throughput. (CBD n = 95, SH2 n = 47, Anti-pY n = 51) D, The average intra-spot precision was measured by calculating the feature specific pixel C.V. The amine coated substrates produce more regular spots. (CBD n = 95, SH2 n = 47, and Anti-pY n = 52) *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001.
In SC2, the cellulose support is cleaved via acid mediated hydrolysis of glycosidic bonds, and the final cellulose polymer length depends on acid concentration (55). A longer cellulose chain may attach more robustly to the slide, thereby increasing CPC density and assay sensitivity, consistent with previous reports of increased sensitivity for protein and peptide microarrays employing three-dimensional polymer scaffolds for surface attachment (56, 57). However, this advantage must be balanced against the general insolubility of cellulose, which may lead to precipitation and printing anomalies. To further optimize the CPCMAs we performed a separate synthesis of supplemental Table S1 peptides, subjecting them to five different dissolution cocktails (Table I). Although we could not recover peptide from the highest and lowest acidity conditions, we identified a “medium” acidity condition with the highest CPCMA sensitivity (supplemental Fig. S1, Table I), which we used in subsequent assays.
CPCMA Affinity Estimation Pipeline
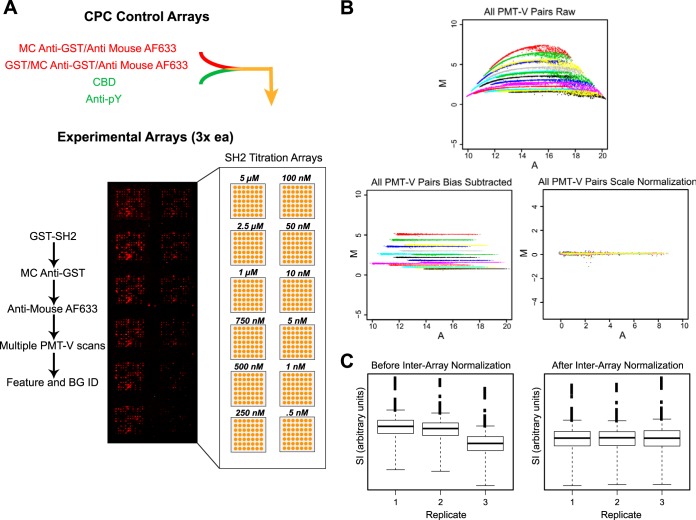
The experimental pipeline includes a series of control arrays followed by SH2 domain titration arrays (Fig. 3A) and a CBD bioassay-mediated print control assay (supplemental Fig. S2B). We measured affinities for 124 physiological phosphopeptides against four SH2 domains (supplemental Table S2). We independently processed eighteen of these peptides and nonadjacently arrayed them twice to assess the precision of replicate measurements (supplemental Fig. S2A). In total, we spotted 1728 features per array with 144 per subarray. A subset of the peptides are known to bind to specific SH2 domains with high affinity, and almost all phosphopeptides arrayed are phosphorylated in vivo (47). We comprehensively covered selected well characterized receptor tyrosine kinases (RTKs) such as the ERBB family, as well as some under characterized RTKs such as DDR1 and DDR2 that are known to be highly phosphorylated in cancer contexts (58). Well over one third (53 of 124) of our phosphopeptide set has never been investigated in high throughput before (supplemental Table S2). This design enabled comparisons with preceding platforms and orthogonal biophysical data while producing an interactome with considerable novelty. We selected SH2 domains from GRB2, NCK1, ABL1, and SHP2 (N-terminal domain) proteins. Each of these domains derive from proteins with different functions and have distinct selectivity preferences (supplemental Table S3), so overlap is thought to be relatively rare and for physiological peptides, potentially consequential in vivo. We produced each domain as GST fusions and applied each to three replicate arrays from 0.5 nm to 5 μm (Fig. 3A).
Fig. 3.
Quantitative CPCMA workflow with corrections for experimental bias. A, Experimental design. Two negative (red) and positive (green) interaction controls were performed on a pilot panel of CPCs. The intersection (yellow) of those CPCs that scored negative for detection antibody and GST tag binding (red) and positive for anti-pY and CBD (green) binding were printed for experimental assessment. One set of CBD print control arrays (not shown) and four sets of experimental SH2 domain titration arrays were performed as outlined. Each array in a set is scanned at multiple different photomultiplier voltages (PMT-V) such that the lowest applied voltage does not include any saturated features. B, Scanner bias correction and SI calibration. Colored M-A plots (where M = ln [SIPMT−Vhigh/SIPMT−Vlow] and A = ln [SIPMT−Vhigh*SIPMT−Vlow]) of all PMT-V feature pairs for a single experimental array are shown before (top) and after correction for voltage specific offset (bottom left) and gain (bottom right). The bias corrected SI measurements are used to estimate a single SI value for each feature on the array, simultaneously correcting for nonlinearity and saturated pixels. C, Inter-array correction. Representative box plots of the SI distributions between replicate arrays before and after inter-array normalization are shown. In this instance, replicate “1” and “2” were printed in succession, whereas “3” was printed six slides after replicate “2.”
We encountered and addressed three sources of systematic error in the CPCMAs: image capture gain and image saturation, printing efficiency between arrays, and printing efficiency across blocks within a single array. Because we use a wide SH2 concentration range, we cannot capture all features from all arrays without saturation with a single photomultiplier voltage setting. We therefore scanned each array multiple times, varying the voltage and ensuring that at least one scan had no saturated pixels. Bias-free measurements of the same array at different voltages would yield intensity values that differ only through instrument gain. Therefore, M-A scatterplots of one voltage versus a second voltage should produce uniform horizontal lines with homogeneous scatter. However, a clear curvilinear bias exists at low intensities, whereas at high intensities saturated pixels underestimate the signal (Fig. 3B, top). To correct for this bias and control for saturated pixels, we applied an affine model of scanner characteristics (see supplemental Experimental Procedures in supplemental material) (59). This model allows us to use the signals from each voltage as replicate measures of probe content, after bias subtraction and scale normalization (Fig. 3B, bottom panels). The median of these replicates yields a robust estimate of fluorophore quantity on a common linear scale.
Array production often suffers from a systematic, gradual loss of printed mass during a production run (60, 61). Our system revealed significant loss across three replicate arrays (Fig. 3C). Using the CBD to monitor printed mass, we observe a similar loss across print blocks within a single array (supplemental Fig. S2C). We therefore applied a correction using model-derived estimates for printing effects to achieve consistent signal distributions (see supplemental Experimental Procedures in supplemental material).
We assigned SH2-CPC interactions and subsequently the CPCs themselves into categories (supplemental Fig. S3A and S3B). Using this empirical approach to classification, a hierarchy emerged reflecting the information provided from each peptide. Approximately 74% of our peptide set bound to at least one domain. The great majority of these reactive peptides scored at least one quantitative reaction, yielding a rich interaction data set of 324 quantitative measurements and 44 binary measurements.
Validation of CPCMA Quantitative Performance
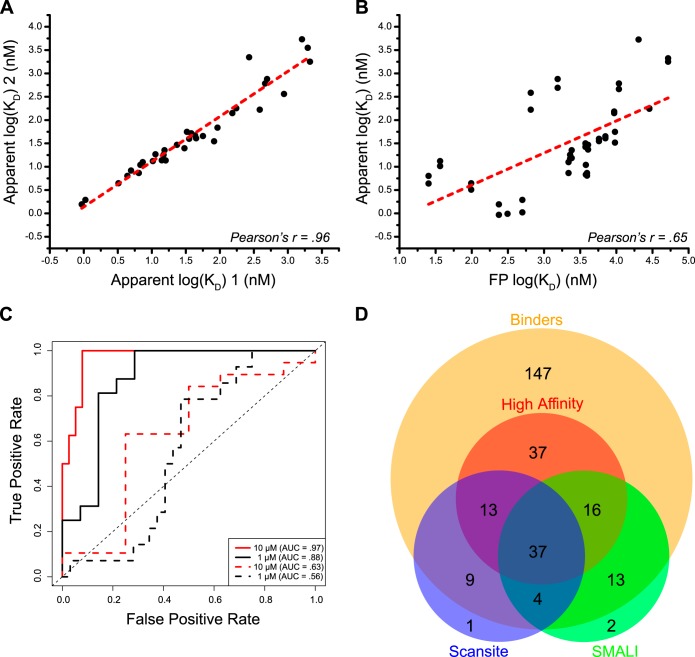
For quantitative binder interactions, we estimated the affinity using a simplified mass action model (see supplemental Experimental Procedures in supplemental material). The majority of binding isotherms saturated between 0.1–1 μm (supplemental Fig. S4A), yielding 211 pairwise interactions with reliable KD estimates. Included in this set are 34 replicate interactions drawn from 18 replicated and randomly positioned peptides on the arrays. The reproducibility of affinity estimates between the replicates was excellent, with a Pearson's correlation coefficient (PCC) approaching unity (PCC = 0.96, Fig. 4A). Moreover, the reproducibility of the KD estimates was superior to single concentration measurements, the usual measurement mode for high throughput assays (PCC = 0.87, p = 0.016, supplemental Fig. S4B).
Fig. 4.
CPCMA affinity measurements are reproducible, can be validated, and are useful for categorizing interactions. A, Scatterplot of affinity estimates derived from replicated nonadjacent peptide spots on the array (n = 34 pairs). Each replicate corresponds to a separate synthesis disc on the parent cellulose membrane and was subjected to a separate workup procedure. Dashed red-line represents the best fit unconstrained linear regression line. PCC value inset. B, Scatterplot of 44 SH2-peptide interaction affinities estimated by array, and in solution by fluorescence polarization (FP). Dashed red line represents the best-fit unconstrained linear regression line. Error bars represent S.E. and PCC values inset. C, ROC curves for array estimated KD values (solid lines) and 5 μm SI measurements (dashed lines) at 1 and 10 μm cutoffs. Area under the curve (AUC) values are provided in the inset legend. D, Dataset overlap with peptide library derived predictive algorithms. Four-way Venn diagram depicting overlap of array based measurements and interactions predicted from the Scansite (low stringency) and SMALI algorithms. Binders are the set of interactions measured above noise via array. The subset of high-affinity interactions are those with array logKD values < 1.47 nm.
To further validate the affinity measurements, we compared a subset of identical peptide-SH2 interactions using fluorescence polarization (FP). The FP assay does not suffer from interfacial assay artifacts and is often used as an orthogonal measurement technique (11, 27, 33). We examined 10 peptides and each SH2 domain across an affinity range spanning four orders of magnitude. These peptides were printed twice from separate syntheses, at nonadjacent positions on the array (supplemental Tables S2 and S4). Note that the array overestimates the absolute affinities because of an avidity effect imparted by the constitutively dimerized GST expression tag (62). Although the affinity measurements must be interpreted in a relative sense, the correlation of the CPCMA and FP estimates for this diverse panel of interactions is good (PCC = 0.65), as shown in Fig. 4B. Once again, single concentration measurements performed poorly (PCC = −0.28, supplemental Fig. S4C).
Because the CPCMAs outperformed single concentration estimates of binding, we assessed the classification performance of each type using receiver operating characteristic (ROC) analysis (63) (Fig. 4C). ROC curves were generated with the set of peptides having both array and FP derived affinity estimates (Fig. 4B) using the FP values as the reference set for classification. We chose cutoff values—that is KD values from the FP data set to define the classification boundary—in the low micromolar range because this is near our median affinity and because low and submicromolar interactions are common for well-characterized SH2 domains. At cutoff levels of 10 and 1 μm, apparent KD values classify more efficiently than single SI measurements (Fig. 4C). Apparent KD estimates at the 10 μm cutoff performed extremely well (AUC = 0.97), with maximal threshold accuracy achieved with Array log KD ∼ 1.74 nm. We chose this value to serve as a boundary between two affinity classes, “high” and “low” (corresponding to <10 μm and >10 μm, respectively). We further identified a third class of CPC-SH2 interactions, the “nonbinders.”
To further assess the appropriateness of the 10 μm boundary, and to enrich our data set with measured affinities and in vivo interaction information, we manually mined the literature and multiple databases (supplemental Table S2). All of the 34 biophysically quantified in vitro interactions and almost all of the in vivo domain-site specific interactions in our data set (41/43) we classified as binders. Moreover, 91% (31/34) of the in vitro interactions mapped to the correct affinity category, and 85% (35/41) of the in vivo interactions we identified as high-affinity, as expected for relevant interactions among numerous competitors.
Next we compared our interactions to those predicted by two algorithms trained on in vitro peptide libraries, Scansite (21) and SMALI (22) (Fig. 4D). Interactions that produce a probability value or score above a threshold are predicted to be high-affinity binders. Consistent with this, our interaction set almost completely encompasses the predictions from these two algorithms, the bulk of which mapping to the high-affinity class (∼70%, Fig. 4D).
To assess the performance of CPCMAs relative to the preceding quantitative PMA and HTFP platforms, we compared an overlapping set of 64 and 84 interactions, respectively (31, 32, 64). Our comparisons show that the CPCMAs are ∼fivefold more sensitive, have improved reproducibility, and more consistently align with SMALI and Scansite algorithmic predictions, known binding motif preferences, affinity measurements, and in vivo interactions (see Supplemental Results in supplemental material, supplemental Fig. S4D and S4E, supplemental Table S5).
Systems Level Analyses of the Quantitative SH2 Domain-phosphotyrosine Interactome
The CPCMA platform yielded a high quality quantitative interactome of 280 interactions between 92 peptides (89/92 of these peptides are physiological, 69/89 currently annotated as phosphorylated in vivo, with the remainder potentially phosphorylated) and four SH2 domains. A network graphic depicting interactions between SH2 domains and physiological peptides, along with interaction strength, phosphorylation data, and in vivo interaction metadata parsed into protein-protein, domain-protein, and domain-site categories is supplied as supplemental Fig. S5. Given that over one-third of our peptides have never been queried in high throughput before and preceding approaches suffered from low sensitivity and/or lack of quantification (see Supplemental Results), this interactome is considerably novel. Unlike preceding interactomes, here interactions can be prioritized based on affinity and (for quantitative peptides) nonbinding events, or negative information can also be incorporated into analyses of signaling systems. The phosphorylation data and in vivo interaction data can also be used to prioritize interactions for further investigations. For instance, an interaction between a domain and a site, which is validated in vivo, lends greater confidence that a high affinity interaction identified for another domain without in vivo information may be physiologically relevant, simply because this phosphorylation site has already mediated a functional SH2 domain interaction.
Peptides that bind to multiple domains are potential sites of coregulation, possibly serving as critical points for pathway cross-talk. The four SH2 domains examined here are from different functional subfamilies where binding motifs differ with little overlap (17, 19, 33, 37) and therefore high affinity binding overlap may be functionally relevant in vivo. Previous work in bacteria and yeast have shown that pathway cross-talk mediated by interactions between domains or kinases and short specificity determining peptide motifs may be subjected to negative evolutionary pressure (10, 43), resulting in highly specific domain-peptide interactions with little binding overlap between domain family members. In higher eukaryotes, such as humans, this may not be the case at the interaction site level given the role of tissue specific expression in mediating signaling specificity. In addition, because of the expansion in the size of protein domain families in higher eukaryotes, secondary contacts (interactions outside of the primary domain-ligand binding interface), and coincident detection (multiple domain-ligand interactions are needed for a functional binding interaction) may play a more prominent role in insulating signaling pathways compared with the primary interaction interfaces.
The degree of interaction overlap between SH2 domains has heretofore not been examined with this level of quantitative resolution. Therefore, it is not known if the degree distribution of the interaction network is a function of interaction affinity. If pathway cross-talk is subjected to negative selection in the SH2 domain phosphopeptide interactome at the level of the primary interaction interface, the relative number of promiscuous peptides should decrease as the affinity of the interactions (and presumably the physiological relevancy) examined increases. Consistent with this hypothesis, promiscuous peptides within our high affinity subset are rare (Fig. 5A). Considering only high-affinity interactions, 63 peptides bind to at least one SH2 domain, whereas only six peptides bind to three SH2 domains and no peptides bind to all four domains. Indeed, a comparison of the degree distributions between the entire quantitative data-set (n = 81 peptides and 267 interactions) and another for the high affinity subset (n = 63 peptides and 102 interactions) reveals that node degree is a function of interaction affinity (supplemental Fig. S6A and S6B, Fisher's Exact Test p = 1.0 × 10−7). This finding is consistent with the hypothesis that physiological domain-motif interactions evolved with signaling specificity as a constraint. To our knowledge, this is the first quantitative examination of this property for any domain-motif interactome in higher eukaryotes, and therefore may be representative of other families, such as the SH3, PDZ, Chromo, and Bromo domain families.
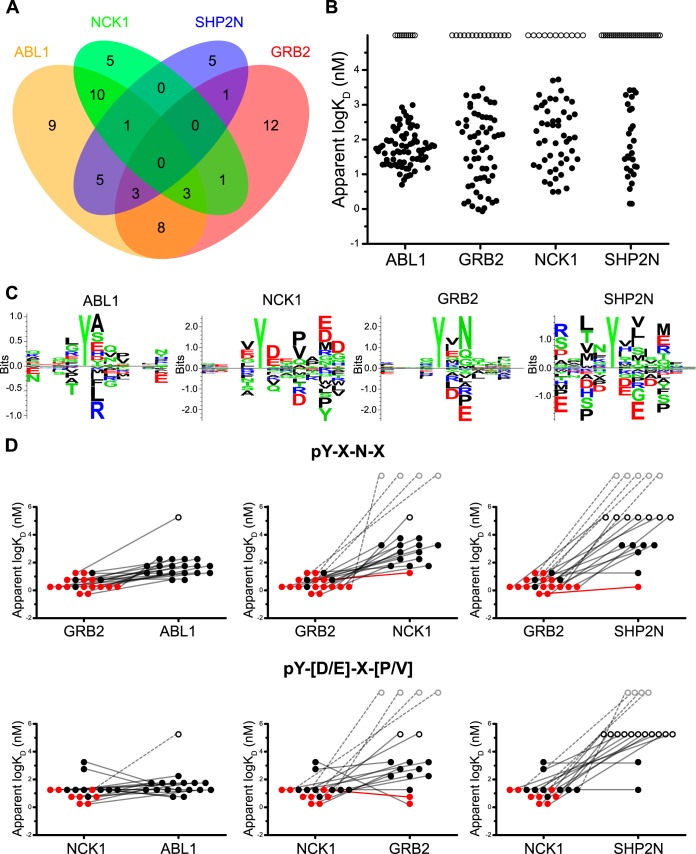
Fig. 5.
Multi-scale analyses of the binding profiles and overlap between the ABL1, GRB2, NCK1, and SHP2N SH2 domains. A, Four-way Venn diagram depicting “high affinity” SH2 domain-CPC binding overlap. B, Binding affinity distributions for each SH2 domain within the quantitative peptide class (n = 81). Interactions assigned an apparent KD are represented as closed circles, “binders” are represented as open circles and assigned a value of 5 for visualization purposes. C, SMM-derived interaction motifs for each SH2 domain. The size of each AA is proportional to its predicted global contribution to binding energy. The fifth position corresponds to the pY residue, which is constant in all experiments. D, GRB2 (top) and NCK1 (bottom) core-motif constrained dot-plots depicting the binding distributions and interaction crossover between domains. Dots represent core-motif conforming peptides. “Binders” are represented as in A, and “nonbinders” are represented as gray open circles and assigned a value above the ordinate for clarity. Black dots represent peptides without domain-site specific corroboration and red dots are in vivo corroborated domain-site specific interactions. Lines connect the same peptides between distributions. A dotted line indicates one interaction scored as a nonbinder. A solid red line denotes a known point of coregulation.
The binding mode of individual domains is best assessed quantitatively so that the distribution of binding affinities can be judged. The high sensitivity and quantitative accuracy provided by the CPCMAs enabled, for the first time a quantitative assessment of protein domain binding distributions (Fig 5B). Here, we tested the hypotheses that each domain has a similar central tendency and distribution shape. This novel data analysis mode revealed that SH2 domains do not differ greatly in their average affinity for physiological phosphopeptides (ANOVA p = 0.182, Kruskal-Wallis p = 0.362, supplemental Fig. S6C). However, there were differences in promiscuity (number of interactions/domain) and selectivity (dynamic range of interactions) between the domains. Q-Q plots of the binding distributions for the four domains reveal similar centers for all the distributions but a large difference in slope for the ABL1 SH2 domain compared with the others, indicating less spread (selectivity) for its interactions (supplemental Fig. S6C). Two-way Anderson-Darling tests with multiple testing correction also supports a difference between the distributions uniquely for ABL1 at the p = 0.05 significance level.
These systems level quantitative properties are likely to have important consequences in vivo. For ABL1, the high interaction promiscuity (large number of binders) and moderate selectivity (tight affinity distribution) is consistent with the role of the SH2 domain within the full length tyrosine kinase. There it enables the hyper-phosphorylation of diverse target proteins by tethering peptide ligands after initial phosphorylation, allowing processive phosphorylation of adjacent target sequences in cis (65). The binding profiles of the adapter proteins GRB2 and NCK1 are also consistent with their biological function, as they discriminate between potential ligands and also achieve high-affinity anchoring interactions to ensure effective signal transduction. Similarly, a highly-selective binding profile with high-affinity binding is consistent with the role of the N-terminal SH2 of the SHP2 tyrosine phosphatase. Here, SH2 ligands of SHP2 simultaneously release intra-molecular inhibition and anchor the phosphatase to sites of intended action (66).
Quantitative Affinity Matrix Construction and Interaction Prediction
Previous PSSMs provide insight into the sequence motifs critical for high-affinity binding (supplemental Table S3). However, because these matrices were trained by sequence frequency and not interaction affinities, they may be sensitive to the binding/nonbinding threshold while ignoring information from low affinity interactions. The combined high sensitivity and quantitative capacity of our approach enables for the first time the application of a quantitative binding motif summarization methodology to a HTP domain-peptide data set. Here, we have applied the stabilized matrix method (SMM) (52), which scores each amino acid at every position according to its approximated contribution to the binding energy to produce an affinity matrix.
All of the matrices produced here are consistent with previously reported motifs (Fig. 5C) while yielding further information. For instance, the core pY-X-N-X and pY-[D/E]-X-[V/P] motifs for GRB2 and NCK1, respectively, are apparent, whereas a preference for C-terminal acidic residues is seen here for NCK1. Likewise, SHP2 has an extended motif for hydrophobic residues at the −2 and the +1 through +4 positions. We verified the predictive capacity of our matrices using cross-validation. This analysis revealed excellent classification accuracy for the GRB2 and NCK1 motifs, good accuracy for SHP2, and poor accuracy for ABL1, likely because of the high promiscuity and narrow interaction affinity distribution for this particular domain (supplemental Fig. S7A–S7D). For all SH2 domains except ABL1, the predictive accuracy is improved when the classifier boundary is restricted to high-affinity interactions, which are likely most important for biological function (supplemental Fig. S7A–S7D). Overall, the performance of these motifs demonstrates the value of quantitative affinity estimates for ligand discrimination, and they serve as a useful guide for identifying residues impacting binding specificity (see below).
Motif Targeted Investigation of Contextual Affinity Modulation
Selectivity summarization matrices are a fundamentally useful tool for visualizing general sequence preferences and in this case predicting binding interactions. However, because of the independence assumption they cannot incorporate contextual interplay between AAs into predictions. Although the entire sequence always affects binding, context is most important for high-affinity interactions anchored by hot-spot residues, such as +2 Asn for GRB2. Neighboring residues modulate affinities, contributing to promiscuity and the assignment of binding priority across multiple domains (25, 27). We (27) and others (67) have shown that the GRB2 SH2 domain is sensitive to particular nonpermissive residues within the pY-X-N-X motif. To investigate if contextual interplay can be resolved by the CPCMAs, we parsed our 19 pY-X-N-X peptides into two groups depending on whether nonpermissive residues are in the motif. Indeed, we found a highly significant (p < 0.001) reduction in binding affinity for peptides containing nonpermissive residues (supplemental Fig. S7E).
Given that the CPCMA platform was able to resolve submotif differences in affinity, we investigated interaction overlap within a core-motif defined context, focusing on GRB2 and NCK1. The matrices for these domains display excellent predictive accuracies (AUC ∼ 0.9), agree with the literature, and contain known hot-spot residues. We show peptide affinities for pairs of SH2 domains in Fig. 5D. Consistent with the affinity defining nature of the core motifs, all known domain and site-specific in vivo interactions (red dots) and points of coregulation (multiple interacting domains, red lines) are within the high-affinity subset. Generally, high-affinity overlap is common with ABL1 because of its promiscuous nature, yet much less common for the other three domains.
Contextual information modulates interaction crossover for many peptides. For example, GRB2 and SHP2 both interact with IRS1 Y895 (68) in vivo, and are predicted here to interact with the Y39 peptide RVQIYHNPTAN within vasodilator-stimulated phosphoprotein (VASP). ABL1 also binds with high-affinity to both peptides, and is known to phosphorylate VASP Y39 in vivo (69). Although IRS1 Y895 contains no hot-spot residues for NCK1, VASP Y39 contains a +3P. However, this peptide lacks other permissive residues beyond the +3 position for NCK1, preventing crossover with this domain (Fig. 5C). Perhaps not surprisingly given the impact of contextual information, core-motif overlap does not guarantee binding overlap, nor does the absence of motif overlap prevent binding overlap. For example, of the two peptides that conform to both the GRB2 and NCK1 core-motifs, only one peptide binds with high affinity to both domains. The transmembrane adaptor protein non-T cell activation linker (NTAL) Y136 peptide DANSYENVLIS only binds strongly to GRB2. This selection may be accomplished by a combination of a −1 Ser and +4 Leu, which are detrimental for NCK1 (Fig. 5C). Similarly, ABL1 prefers a +2 Asn and +3 Pro, yet the HER4 site Y1162 PKQEYLNPVEE is this domain's lowest affinity pY-X-N-X peptide, perhaps because of a +1 Leu (Fig. 5C). Conversely, this peptide binds to NCK1 because of its acidic C terminus, despite not conforming to the NCK1 core-motif (Fig. 5C). This motif-focused analysis highlights the exquisite specificity in PID-peptide interactions, which can only be revealed through quantitative affinity measurements.
DISCUSSION
We find that the CPCMA based approach has greatly improved sensitivity, reproducibility, and quantitative resolution compared with earlier methods, allowing us to examine PID domain specificity space with confidence. The performance of the CPCMA platform was achieved through the careful application of a combination of critical experimental and computational processing decisions. New tools, such as the ArrayMix™ and ArraySlide™ enabled prolonged, reproducible, kinetically controlled incubations while novel applications of existing tools, such as the CBD for quality control enabled the application of systematic error corrections. Here we provide an approach that addresses issues related to the adequate display of peptide, optimal incubation conditions, image capture, correction of systematic error, estimation of affinity, and categorization of interactions. The procedure developed herein is therefore the most comprehensive approach for mapping PID specificity to date. At the most direct level, our approach enables a confident assessment of binding priority between competing PIDs for binding sites. This ability is critically important for the development of testable hypotheses examining time-dependent signal propagation and information integration at hub proteins (70), which are often aberrant processes in cancers (42, 71) and require the processing of high and low affinity interactions (34, 72–74). In addition, the CPCMA platform is capable of explicit measurement of nonbinding events, can resolve interplay between neighboring amino acids within a ligand, does not require the purification of both interacting components, and is economical to employ. Taken together, the CPCMA platform provides an appealing choice relative to existing alternatives for all manner of PID specificity investigations (supplemental Table S6). In support of this view, a number of new analysis modes for investigating PID specificity space were enabled by our approach.
Here we provided, to our knowledge for the first time, a quantitative investigation of domain specific binding distributions. These PID affinity signatures likely play an important role in shaping the systems level function for a given domain. For instance, a “promiscuous,” or highly connected domain may bind to many peptides but do so at low affinity, thereby producing interactions unlikely (in the absence of other in vivo mechanisms such as spatial clustering or interactions potentiated by other domains) to be competitive with other domains in vivo. However, this may not be the case if multiple interaction sites are available on the same protein (in cis), where avidity may enable successful competition with domains that have higher interaction affinities. We found the promiscuity and selectivity signatures of SH2 domain family members aligned with systems level protein function. For instance, the ABL1 SH2 domain demonstrated a promiscuous but nonselective distribution signature. A common moderate affinity for its many ligands ensures that ABL1 SH2 interactions are readily outcompeted by other phosphopeptide binding domains and may be an important component of the ABL kinase processive hyper-phosphorylation ability (65, 75). Together these properties may help confer ABL family kinases with the dynamism required to rapidly transition within and between its diverse sites of regulation (76). SH2 domain dependent processive multisite phosphorylation of diverse substrate proteins is also a hallmark of SRC family kinases (77), and it remains to be seen if this signature is common between these two families. More generally, how PID affinity signatures cluster within families, have changed during evolution, and can inform drug and biological circuit design are important future directions for exploration.
We demonstrated the ability of the CPCMA platform to resolve contextual contributions to binding affinity and produce predictive PID specific affinity matrices from a single quantitative data set. To our knowledge, this is the first demonstration of these abilities by a quantitative high-throughput platform and the first demonstration of the ability to resolve nonpermissive contributions to binding affinity in high-throughput. Previous work has incorporated quantitative high-throughput interaction information from multiple PIDs into unified interaction prediction algorithms (11, 28, 78). Although useful for predicting interactions for uncharacterized or under-characterized PID family members, such approaches are sensitive to the sequence similarity of test domains to the domains used in the training set (28). Going forward, we suggest that models trained incorporating the idiosyncratic contextual binding preferences of single domains are more likely to fully represent ligand binding preferences than those trained from entire PID families. Considering the significant impact of contextual binding information on PID specificity space (15), the combination of larger CPCMA derived interaction data sets with algorithms capable of incorporating residue correlations is therefore an important future application. For instance, when such models are combined with in vivo information more accurate network (re)constructions are possible (14, 17).
Using quantitative CPCMAs we recapture general themes derived from preceding PID specificity studies and demonstrate the ability to extend these concepts in new ways. PID-ligand interactions serve to transmit specific cell signals in a dynamic fashion and are therefore thought to possess modest interaction affinities (1). Consistent with previous reports to this effect (33, 34, 79) and recent findings that very high affinity PID-peptide interactions can impair cell signaling and viability (80, 81), we found our SH2 domain interactions span a moderate affinity range. Within a PID family, domain selectivity profiles cluster into separate binding motif classes (23, 33, 37, 82) that are aligned with sequence similarity and protein function (40, 83). Yet, likely as a consequence of selection against the emergence of signaling systems with excessive cross-reactivity (10, 43), PID family members have selectivity preferences that are evenly distributed across a continuum (11). This results in PID family specific interaction networks with perhaps surprisingly low levels of binding-site competition (12). Given the relatively evenly distributed selectivity profiles across family members and the moderate binding affinity norm, overlapping high-affinity interactions are thought to be uncommon, but potentially important sites of signaling cross-talk.
Here, we also observe low levels of high affinity interaction overlap across four SH2 domains with unique selectivity profiles. We demonstrate, for the first time that the degree distribution of a PID derived interactome is a function of affinity, with node degree decreasing with increasing affinity threshold. Moreover, consistent with the notion that CPCMA identified sites of high-affinity interaction overlap may represent rare sites of signaling coregulation, the vast majority of the known domain-site specific in vivo interactions map to our high-affinity subset, including some overlapping pairs known to perform a coregulatory function. For instance, we identified a high affinity overlap site between GRB2 and SHP2 at IRS1 Y895 that is known to function as a phenotypic switch regulating differentiation and proliferation in vascular smooth muscle cells. Here, SHP2 dephosphorylates this site upon stimulation with insulin-like growth factor-I, which serves as a switch to block GRB2/SOS mediated migration and proliferation while maintaining the PI3K/AKT signaling pathway, and thereby the differentiated cell state (68). SHP2 mediated dephosphorylation does not occur in dedifferentiated cells, resulting in cell proliferation and migration (68).
The ligand sequence determinants responsible for directing this high-affinity PID overlap have important implications within a systemic context, especially for ligands like phosphopeptides, which can serve as binding sites for kinases, phosphatases, and multiple PIDs. As demonstrated here using affinity matrices as a guide, core-motif defining hotspot residues are necessary prerequisites for high affinity interactions. However, consistent with our previous analysis of SH2 domain binding specificity (27), we find that high-affinity interaction overlap is not solely directed by core motif overlap, and subtle combinations of permissive and nonpermissive residues play important roles in defining specificity space. This concept seems to be a general one for PIDs (15), and highlights that selectivity space—as defined by preferred binding motifs where residues are assumed to contribute independently to binding affinity—does not predetermine specificity space, and many physiologically important interactions may be dictated by submotif contextual factors. Because of the low sensitivity, quantitative resolution, or inherent limitations of previous high throughput techniques, such important modulations have remained elusive. The quantitative CPCMA based approach developed here is well poised to help address the functional role each individual residue within a ligand plays in directing cellular signals.
Further application or in vivo investigation of findings derived from in vitro PID-peptide interaction mapping should be applied in a context specific fashion. Fundamentally, interacting proteins must be coexpressed and colocalized for an interaction to occur. Given this, although it is generally true that interactions between a given PID and its ligand are necessary for an interaction to occur in vivo, they are not always sufficient. There are multiple instances where secondary contacts (interactions outside the ligand motif and binding interface of the PID) are also necessary for a biological outcome (5, 15, 84). These “secondary” interactions can be mediated by alternative regions of the domain containing protein or of the domain itself. In addition, the functional significance of any given biophysical interaction identified in vitro may be contingent upon combinatorial interactions between a given PID and multiple ligands (35) or between multiple domain-ligand pairs (85). Because these and other mechanisms exist in vivo to potentiate the functional consequences of low-affinity interactions, it is unwise to ignore such interactions in follow-up investigations. However, given the concordance of in vivo corroborated interactions and decrease in ligand promiscuity within the “high affinity” subset of our results, it is likely that functional interactions within this subset rely less on these mechanisms than those within the “low affinity” subset, and therefore are the priority subset for downstream investigation.
Regardless of the affinity, many of these interactions take place only when a cell responds to an external stimulus. Therefore, time resolved quantitative in vivo analysis approaches that can resolve the state of protein complexes such as those based on affinity purification mass spectrometry (86–88) hold promise in the effort to tease apart the functional significance of putative PID-ligand interactions. In this way, in vitro interaction mapping and in vivo protein complex analysis provide highly complementary systems level approaches for understanding and manipulating cell signaling circuits.
The CPCMA based approach developed here can be readily applied to other PID ligand interaction systems. In addition to other natural systems, our approach is compatible with those incorporating unnatural amino acids, peptoids, and small molecules. Unlike other techniques, our approach does not require the maintenance of protein structure at an interface, the purification of each interacting component, sophisticated microfluidics, or imaging apparatus, is capable of multiplexing, and can use existing commercially-available equipment and open-source software suites. Taken together, the CPCMA platform should have broad applicability within the protein interaction network field to discover new interactions, new interaction features, and more effectively investigate emergent systems level properties of interactomes.
Supplementary Material
Acknowledgments
We thank Eric Yan and Steven Kron for their assistance synthesizing soluble peptides. We thank Greg Richardson and Joesph Camacho from Gelcompany for their work developing the ArrayMixTM and ArraySlideTM.
Footnotes
Author contributions: B.W.E., R.S.R., and P.D.N. designed research; B.W.E. performed research; B.W.E., Y.K., and B.P. contributed new reagents or analytic tools; B.W.E., Y.K., M.W., and B.P. analyzed data; B.W.E., R.S.R., and P.D.N. wrote the paper.
* This project was supported by funds from the National Science Foundation (NSF MCB-0819125). BWE was supported in part by NIH TG-GM07183. R.S.R. is supported by a grant from the National Institutes of Health (GM 078450).
 This article contains Supplemental Figs. S1 to S7 and Tables S1 to S6.
This article contains Supplemental Figs. S1 to S7 and Tables S1 to S6.
1 The abbreviations used are:
- PMT-V
- photomultiplier tube voltage
- CPC
- cellulose peptide conjugate
- CPCMA
- cellulose peptide conjugate microarray
- PMA
- protein microarray
- HTFP
- high throughput fluorescence polarization
- PID
- protein interaction domain
- SH2
- Src Homology 2
- RTK
- receptor tyrosine kinase
- pY/pTyr
- phosphotyrosine
- FP
- fluorescence polarization
- ROC
- receiver operator characteristic
- PCC
- pearson correlation coefficient
- SMM
- stabilized matrix method
- CV
- coefficient of variation
- GST
- glutathione s-transferase
- SI
- signal intensity
- PWM
- position weight matrix
- PSSM
- position specific scoring matrix.
REFERENCES
- 1. Pawson T., Nash P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 2. Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 3. Puntervoll P., Linding R., Gemund C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D. M., Ausiello G., Brannetti B., Costantini A., Ferre F., Maselli V., Via A., Cesareni G., Diella F., Superti-Furga G., Wyrwicz L., Ramu C., McGuigan C., Gudavalli R., Letunic I., Bork P., Rychlewski L., Kuster B., Helmer-Citterich M., Hunter W. N., Aasland R., Gibson T. J. (2003) ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. London N., Movshovitz-Attias D., Schueler-Furman O. (2010) The structural basis of peptide-protein binding strategies. Structure 18, 188–199 [DOI] [PubMed] [Google Scholar]
- 5. Stein A., Aloy P. (2008) Contextual specificity in peptide-mediated protein interactions. PloS One 3, e2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin J., Xie X., Chen C., Park J. G., Stark C., James D. A., Olhovsky M., Linding R., Mao Y., Pawson T. (2009) Eukaryotic protein domains as functional units of cellular evolution. Sci. Signal. 2, ra76. [DOI] [PubMed] [Google Scholar]
- 7. Bhattacharyya R. P., Remenyi A., Yeh B. J., Lim W. A. (2006) Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 [DOI] [PubMed] [Google Scholar]
- 8. Neduva V., Russell R. B. (2005) Linear motifs: evolutionary interaction switches. FEBS Lett. 579, 3342–3345 [DOI] [PubMed] [Google Scholar]
- 9. Mosca R., Pache R. A., Aloy P. (2012) The role of structural disorder in the rewiring of protein interactions through evolution. Mol. Cell. Proteomics 11, M111 014969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zarrinpar A., Park S. H., Lim W. A. (2003) Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 [DOI] [PubMed] [Google Scholar]
- 11. Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xin X., Gfeller D., Cheng J., Tonikian R., Sun L., Guo A., Lopez L., Pavlenco A., Akintobi A., Zhang Y., Rual J. F., Currell B., Seshagiri S., Hao T., Yang X., Shen Y. A., Salehi-Ashtiani K., Li J., Cheng A. T., Bouamalay D., Lugari A., Hill D. E., Grimes M. L., Drubin D. G., Grant B. D., Vidal M., Boone C., Sidhu S. S., Bader G. D. (2013) SH3 interactome conserves general function over specific form. Mol. Syst. Biol. 9, 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan C. S., Bodenmiller B., Pasculescu A., Jovanovic M., Hengartner M. O., Jorgensen C., Bader G. D., Aebersold R., Pawson T., Linding R. (2009) Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2, ra39. [DOI] [PubMed] [Google Scholar]
- 14. Miller M. L., Jensen L. J., Diella F., Jorgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., Sicheritz-Ponten T., Olhovsky M., Pasculescu A., Alexander J., Knapp S., Blom N., Bork P., Li S., Cesareni G., Pawson T., Turk B. E., Yaffe M. B., Brunak S., Linding R. (2008) Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu B. A., Engelmann B. W., Nash P. D. (2012) High-throughput analysis of peptide-binding modules. Proteomics 12, 1527–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frank R. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232 [Google Scholar]
- 17. Tinti M., Kiemer L., Costa S., Miller M. L., Sacco F., Olsen J. V., Carducci M., Paoluzi S., Langone F., Workman C. T., Blom N., Machida K., Thompson C. M., Schutkowski M., Brunak S., Mann M., Mayer B. J., Castagnoli L., Cesareni G. (2013) The SH2 Domain Interaction Landscape. Cell Reports 3, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodriguez M., Li S. S., Harper J. W., Songyang Z. (2004) An oriented peptide array library (OPAL) strategy to study protein-protein interactions. J. Biol. Chem. 279, 8802–8807 [DOI] [PubMed] [Google Scholar]
- 19. Zhou S., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J., Neel B. G., Birge R. B., Fajardo J. E., Chou M. M., Hanafusa H., Schaffhausen B., Cantley L. C. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 [DOI] [PubMed] [Google Scholar]
- 20. Tonikian R., Zhang Y., Boone C., Sidhu S. S. (2007) Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2, 1368–1386 [DOI] [PubMed] [Google Scholar]
- 21. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Wu C., Huang H., Zhang K., Gan J., Li S. S. (2008) Prediction of phosphotyrosine signaling networks using a scoring matrix-assisted ligand identification approach. Nucleic Acids Res. 36, 3263–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S. (2008) A specificity map for the PDZ domain family. PLoS Biol. 6, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gfeller D., Butty F., Wierzbicka M., Verschueren E., Vanhee P., Huang H., Ernst A., Dar N., Stagljar I., Serrano L., Sidhu S. S., Bader G. D., Kim P. M. (2011) The multiple-specificity landscape of modular peptide recognition domains. Mol. Syst. Biol. 7, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alexander J., Lim D., Joughin B. A., Hegemann B., Hutchins J. R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E. A., Fry A. M., Musacchio A., Stukenberg P. T., Mechtler K., Peters J. M., Smerdon S. J., Yaffe M. B. (2011) Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., Duan S., Wasney G. A., Li Z., Vedadi M., Schapira M., Min J., Arrowsmith C. H. (2011) Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu B. A., Jablonowski K., Shah E. E., Engelmann B. W., Jones R. B., Nash P. D. (2010) SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell. Proteomics 9, 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao X., Tan C. S., Voss C., Li S. S., Deng N., Bader G. D. (2011) A regression framework incorporating quantitative and negative interaction data improves quantitative prediction of PDZ domain-peptide interaction from primary sequence. Bioinformatics 27, 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kundu K., Costa F., Huber M., Reth M., Backofen R. (2013) Semi-supervised prediction of SH2-peptide interactions from imbalanced high-throughput data. PloS One 8, e62732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forsstrom B., Bislawska Axnas B., Stengele K. P., Buhler J., Albert T. J., Richmond T. A., Jingxin Hu F., Nilsson P., Hudson E. P., Rockberg J., Uhlen M. (2014) Proteome-wide epitope mapping of antibodies using ultra-dense peptide arrays. Mol. Cell. Proteomics 13, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones R. B., Gordus A., Krall J. A., MacBeath G. (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 32. Hause R. J., Jr., Leung K. K., Barkinge J. L., Ciaccio M. F., Chuu C. P., Jones R. B. (2012) Comprehensive binary interaction mapping of SH2 domains via fluorescence polarization reveals novel functional diversification of ErbB receptors. PloS One 7, e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu B. A., Engelmann B. W., Jablonowski K., Higginbotham K., Stergachis A. B., Nash P. D. (2012) SRC Homology 2 Domain Binding Sites in Insulin, IGF-1 and FGF receptor mediated signaling networks reveal an extensive potential interactome. Cell Commun. Signal.10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davey N. E., Van Roey K., Weatheritt R. J., Toedt G., Uyar B., Altenberg B., Budd A., Diella F., Dinkel H., Gibson T. J. (2012) Attributes of short linear motifs. Mol. Biosyst. 8, 268–281 [DOI] [PubMed] [Google Scholar]
- 35. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 36. Dueber J. E., Mirsky E. A., Lim W. A. (2007) Engineering synthetic signaling proteins with ultrasensitive input/output control. Nature Biotechnol. 25, 660–662 [DOI] [PubMed] [Google Scholar]
- 37. Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S., Li C., Roy P., Ho K., Songyang Z., Pawson T., Gao Y., Li S. S. (2008) Defining the specificity space of the human SRC homology 2 domain. Mol. Cell. Proteomics 7, 768–784 [DOI] [PubMed] [Google Scholar]
- 38. Songyang Z., Cantley L. C. (1995) SH2 domain specificity determination using oriented phosphopeptide library. Methods Enzymol. 254, 523–535 [DOI] [PubMed] [Google Scholar]
- 39. Liu B. A., Shah E., Jablonowski K., Stergachis A., Engelmann B., Nash P. D. (2011) The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes. Sci. Signal. 4, ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu B. A., Jablonowski K., Raina M., Arce M., Pawson T., Nash P. D. (2006) The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol. Cell 22, 851–868 [DOI] [PubMed] [Google Scholar]
- 41. Lappalainen I., Thusberg J., Shen B., Vihinen M. (2008) Genome wide analysis of pathogenic SH2 domain mutations. Proteins 72, 779–792 [DOI] [PubMed] [Google Scholar]
- 42. Reimand J., Bader G. D. (2013) Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Molecular Syst. Biol. 9, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Capra E. J., Perchuk B. S., Skerker J. M., Laub M. T. (2012) Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expres. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 45. Hilpert K., Winkler D. F., Hancock R. E. (2007) Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 [DOI] [PubMed] [Google Scholar]
- 46. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P.-L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined posttranslational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. (2002) MINT: a Molecular INTeraction database. FEBS Lett. 513, 135–140 [DOI] [PubMed] [Google Scholar]
- 49. Prasad T. S., Kandasamy K., Pandey A. (2009) Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol. Biol. 577, 67–79 [DOI] [PubMed] [Google Scholar]
- 50. Ceol A., Chatr-aryamontri A., Santonico E., Sacco R., Castagnoli L., Cesareni G. (2007) DOMINO: a database of domain-peptide interactions. Nucleic Acids Res. 35, D557–D560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gong W., Zhou D., Ren Y., Wang Y., Zuo Z., Shen Y., Xiao F., Zhu Q., Hong A., Zhou X., Gao X., Li T. (2008) PepCyber:P∼PEP: a database of human protein protein interactions mediated by phosphoprotein-binding domains. Nucleic Acids Res. 36, D679–D683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peters B., Sette A. (2005) Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics 6, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thomsen M. C., Nielsen M. (2012) Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dikmans A., Beutling U., Schmeisser E., Thiele S., Frank R. (2006) SC2: a novel process for manufacturing multipurpose high-density chemical microarrays. QSAR Comb. Sci. 25, 1069–1080 [Google Scholar]
- 55. Klemm D. (1998) Comprehensive cellulose chemistry, Wiley-VCH, Weinheim; New York [Google Scholar]
- 56. Mori T., Yamanouchi G., Han X., Inoue Y., Shigaki S., Yamaji T., Sonoda T., Yasui K., Hayashi H., Niidome T., Katayama Y. (2009) Signal-to-noise ratio improvement of peptide microarrays by using hyperbranched-polymer materials. J. Appl. Phys. 105:102020 [Google Scholar]
- 57. Zubtsov D. A., Savvateeva E. N., Rubina A. Y., Pan'kov S. V., Konovalova E. V., Moiseeva O. V., Chechetkin V. R., Zasedatelev A. S. (2007) Comparison of surface and hydrogel-based protein microchips. Anal. Biochem. 368, 205–213 [DOI] [PubMed] [Google Scholar]
- 58. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 59. Bengtsson H., Jonsson G., Vallon-Christersson J. (2004) Calibration and assessment of channel-specific biases in microarray data with extended dynamical range. BMC Bioinformatics 5, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smyth G. K., Speed T. (2003) Normalization of cDNA microarray data. Methods 31, 265–273 [DOI] [PubMed] [Google Scholar]
- 61. Quackenbush J. (2002) Microarray data normalization and transformation. Nat. Genet. 32, 496–501 [DOI] [PubMed] [Google Scholar]
- 62. Ladbury J. E., Lemmon M. A., Zhou M., Green J., Botfield M. C., Schlessinger J. (1995) Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. U. S. A. 92, 3199–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fawcett T. (2006) An introduction to ROC analysis. Pattern Recogn. Lett. 27, 861–874 [Google Scholar]
- 64. Kaushansky A., Gordus A., Chang B., Rush J., MacBeath G. (2008) A quantitative study of the recruitment potential of all intracellular tyrosine residues on EGFR, FGFR1, and IGF1R. Mol. Biosyst. 4, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mayer B. J., Hirai H., Sakai R. (1995) Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 5, 296–305 [DOI] [PubMed] [Google Scholar]
- 66. Sugimoto S., Wandless T. J., Shoelson S. E., Neel B. G., Walsh C. T. (1994) Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. J. Biol. Chem. 269, 13614–13622 [PubMed] [Google Scholar]
- 67. Cho S., Velikovsky C. A., Swaminathan C. P., Houtman J. C., Samelson L. E., Mariuzza R. A. (2004) Structural basis for differential recognition of tyrosine-phosphorylated sites in the linker for activation of T cells (LAT) by the adaptor Gads. EMBO J. 23, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hayashi K., Shibata K., Morita T., Iwasaki K., Watanabe M., Sobue K. (2004) Insulin receptor substrate-1/SHP-2 interaction, a phenotype-dependent switching machinery of insulin-like growth factor-I signaling in vascular smooth muscle cells. J. Biol. Chem. 279, 40807–40818 [DOI] [PubMed] [Google Scholar]
- 69. Maruoka M., Sato M., Yuan Y., Ichiba M., Fujii R., Ogawa T., Ishida-Kitagawa N., Takeya T., Watanabe N. (2012) Abl-1-bridged tyrosine phosphorylation of VASP by Abelson kinase impairs association of VASP to focal adhesions and regulates leukaemic cell adhesion. Biochem. J. 441, 889–899 [DOI] [PubMed] [Google Scholar]
- 70. Hlavacek W. S., Faeder J. R., Blinov M. L., Perelson A. S., Goldstein B. (2003) The complexity of complexes in signal transduction. Biotechnol. Bioeng. 84, 783–794 [DOI] [PubMed] [Google Scholar]
- 71. Creixell P., Schoof E. M., Erler J. T., Linding R. (2012) Navigating cancer network attractors for tumor-specific therapy. Nat. Biotechnol. 30, 842–848 [DOI] [PubMed] [Google Scholar]
- 72. Oh D., Ogiue-Ikeda M., Jadwin J. A., Machida K., Mayer B. J., Yu J. (2012) Fast rebinding increases dwell time of Src homology 2 (SH2)-containing proteins near the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 109, 14024–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jones R. B. (2013) Do low-affinity ErbB receptor protein interactions represent the base of a cell signaling iceberg? Exp. Rev. Proteomics 10, 115–118 [DOI] [PubMed] [Google Scholar]
- 74. Mayer B. J. (2012) Perspective: Dynamics of receptor tyrosine kinase signaling complexes. FEBS Lett. 586, 2575–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Duyster J., Baskaran R., Wang J. Y. J. (1995) Src homology 2 domain as a specificity determinant in the c-Abl-mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc. Natl. Acad. Sci. U. S. A. 92, 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Colicelli J. (2010) ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci. Signal. 3, re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patwardhan P., Miller W. T. (2007) Processive phosphorylation: mechanism and biological importance. Cell. Signal. 19, 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen J. R., Chang B. H., Allen J. E., Stiffler M. A., MacBeath G. (2008) Predicting PDZ domain-peptide interactions from primary sequences. Nat. Biotechnol. 26, 1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aasland R., Abrams C., Ampe C., Ball L. J., Bedford M. T., Cesareni G., Gimona M., Hurley J. H., Jarchau T., Lehto V. P., Lemmon M. A., Linding R., Mayer B. J., Nagai M., Sudol M., Walter U., Winder S. J. (2002) Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 513, 141–144 [DOI] [PubMed] [Google Scholar]
- 80. Kaneko T., Huang H., Cao X., Li X., Li C., Voss C., Sidhu S. S., Li S. S. (2012) Superbinder SH2 domains act as antagonists of cell signaling. Sci. Signal. 5, ra68. [DOI] [PubMed] [Google Scholar]
- 81. Zhang Y., Appleton B. A., Wiesmann C., Lau T., Costa M., Hannoush R. N., Sidhu S. S. (2009) Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5, 217–219 [DOI] [PubMed] [Google Scholar]
- 82. Carducci M., Perfetto L., Briganti L., Paoluzi S., Costa S., Zerweck J., Schutkowski M., Castagnoli L., Cesareni G. (2012) The protein interaction network mediated by human SH3 domains. Biotechnol. Adv. 30, 4–15 [DOI] [PubMed] [Google Scholar]
- 83. Teyra J., Sidhu S. S., Kim P. M. (2012) Elucidation of the binding preferences of peptide recognition modules: SH3 and PDZ domains. FEBS Lett. 586, 2631–2637 [DOI] [PubMed] [Google Scholar]
- 84. Bae J. H., Lew E. D., Yuzawa S., Tome F., Lax I., Schlessinger J. (2009) The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 138, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Scott J. D., Pawson T. (2009) Cell signaling in space and time: where proteins come together and when they're apart. Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bisson N., James D. A., Ivosev G., Tate S. A., Bonner R., Taylor L., Pawson T. (2011) Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 29, 653–658 [DOI] [PubMed] [Google Scholar]
- 87. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 88. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., Pawson T., Bonner R., Tate S., Gingras A. C. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.