Abstract
Background
Two closely related species of mutans streptococci, namely Streptococcus mutans and Streptococcus sobrinus, are associated with dental caries in humans. Their acidogenic and aciduric capacity is directly associated with the cariogenic potential of these bacteria. To survive acidic and temporarily harsh conditions in the human oral cavity with hundreds of other microbial co-colonizers as competitors, both species have developed numerous mechanisms for adaptation.
Objectives
The recently published novel genome information for both species is used to elucidate genetic similarities but especially differences and to discuss the impact on cariogenicity of the corresponding phenotypic properties including adhesion, carbohydrate uptake and fermentation, acid tolerance, signaling by two component systems, competence, and oxidative stress resistance.
Conclusions
S. sobrinus can down-regulate the SpaA-mediated adherence to the pellicle. It has a smaller number of two-component signaling systems and bacteriocin-related genes than S. mutans, but all or even more immunity proteins. It lacks the central competence genes comC, comS, and comR. There are more genes coding for glucosyltransferases and a novel energy production pathway formed by lactate oxidase, which is not found in S. mutans. Both species show considerable differences in the regulation of fructan catabolism. However, both S. mutans and S. sobrinus share most of these traits and should therefore be considered as equally virulent with regard to dental caries.
Keywords: Mutans streptococci, comparative genomics, adhesion, sugar metabolism, two-component-systems, competence, bacteriocins, cariogenicity
Dental caries is a complex disease that results from interactions of acidogenic/aciduric bacteria colonizing the tooth surface and the oral environment. Although other species, especially lactobacilli, bifidobacteria, and less investigated aciduric species such as Atopobium sp. or Slackia exigua, are also involved, the mutans streptococci (MS) group, in humans represented by Streptococcus mutans (serotypes c, e, f and k) and Streptococcus sobrinus (serotypes d and g) are still considered major etiological agents of dental decay. Over time, their attributed role changed from more or less true pathogens [specific plaque hypothesis (1)] to up-regulators of a sugar-triggered vicious cariogenic circle under destabilization of the homeostasis [extended caries ecological hypothesis (2, 3)] and the discussion is not finished, as outlined by Rosier et al. recently (4).
However, it is generally accepted that S. mutans can be more frequently isolated from carious lesions than S. sobrinus and is by far the most relevant cariogenic Streptococcus species, although we should not rule out the acidogenic properties of several non-MS (5–7). A number of epidemiological and in-vitro studies suggested that S. sobrinus – under circumstances yet to discover – may be even more cariogenic than S. mutans (8–11). In addition, clinical studies have suggested that pre-school and 15-year-old school children harboring both S. mutans and S. sobrinus had a higher incidence of dental caries than those with S. mutans alone (12, 13).
The virulence of MS is directly related to properties that enable these organisms to colonize and thrive on the tooth surfaces during acidic conditions. These properties include the production and regulation of adhesion proteins, glucosyltransferases (GTFs), and extracellular polysaccharides such as glucans that allow the bacteria to firmly adhere to the tooth surface in a biofilm. However, both species follow different strategies for adherence: S. mutans mainly using pellicle directed and specific surface antigens, S. sobrinus mainly using glucans and, as a consequence, both are found on different surfaces (S. sobrinus more buccal, S. mutans more occlusal). Comparing the recently explored genetic inventory of both species, this review will discuss consequences of specific genotypes for cariogenic phenotypes for each species. We focus on genes responsible for adhesion, metabolism of sugars (obtained from salivary glycoproteins and from the host diet to generate lactic acid, leading to acidogenicity), acid tolerance (ability to tolerate abundant amounts of lactic acid, leading to aciduricity), signaling, competence, bacteriocins and related immunity proteins, and oxidative stress resistance. The genetic inventory of S. mutans strains UA159, NN2025, 5DC8, AC4446, KK21, KK23, ATCC25175, and NCTC11060 as well as S. sobrinus DSM 20742 and TCI-107 was compared using the OrthoMCL software. Additional strains and data were included found in the NCBI database (http://blast.ncbi.nlm.nih.gov/) and the Human Oral Microbiome Database (HOMD at http://www.homd.org), respectively. Please refer to references 31 and 41 for details of our methods.
Adhesion
In several clinical studies, the role of S. sobrinus in relationship to caries is suggested to be additive to that of S. mutans. The latter is strongly associated with caries, but in situations of more severe caries, it is frequently found together with S. sobrinus (14–17). The acquisition of these bacteria by the susceptible host is a crucial step. The ‘infection dose’(with respect of the ecological plaque hypothesis, we might call it ‘stress dose’) may play an important role, since more transmission, or higher bacterial counts from the source of transmission, will result in a higher chance for successful acquisition (18, 19). But when the bacterium cannot adhere in the oral cavity, the acquisition will never be successful. Therefore, adherence to epitopes in the oral cavity is a crucial step for streptococci to become resident.
In S. mutans, cell surface antigen I/II (AgI/II), the glucan-binding region of the GTF enzymes and additional glucan-binding proteins (Gbp) have been implicated in its initial and specific adherence to saliva-coated (acquired enamel pellicle) tooth surfaces. These contact regions induce immune responses in mice effective in protection against colonization of S. mutans (20) and have been suggested as anti-caries vaccination agents. Cell wall proteins from the AgI/II family (synonyms SAI/II, PAc, P1, or SpaP; encoded by spaP; binding to salivary agglutinin glycoproteins, extracellular matrix molecules, and ligands of other oral bacteria) are not exclusively found in S. mutans but homologs have been reported in a variety of oral streptococci including S. intermedius, S. gordonii, S. pyogenes, and even non-oral S. agalactiae (21, 22). The S. sobrinus variant has been described as SpaA (or PAg), but with structural differences and less adhesive potential (23) possibly due to down regulation by Par, see Table 1. The AgI/II family is highly conserved throughout different streptococcal species and seems to be associated with the M-protein in other streptococci. It has been found that monoclonal antibodies against S. sobrinus SpaA are able to change the adhesion of this bacterium on hydroxyapatite disks in a triple species biofilm, suggesting a role of SpaA in the interspecies adherence in biofilms (24). In a review on streptococcal adherence factors, it has been reported that SpaA interacts with multiple host and microbial factors, from which binding to the salivary glycoprotein gp340 is most important (25). By our whole genome sequencing approach, we found another ‘adhesive protein’ encoding gene (D823_10858) in S. sobrinus DSM 20742. It seems to be the adherence component of an ATP binding cassette (ABC)-type Zn2 + and Mn2 + transporter system with homologs in many streptococci, including S. mutans, and related to the pneumococcal surface antigen PsaA (26).
Table 1.
Comparing proteins and corresponding genes involved in adherence on pellicle coated tooth surfaces between S. mutans [eight strains according to (31)] and S. sobrinus DSM 20742 and TCI-107
| S. mutans | S. sobrinus | |||
|---|---|---|---|---|
|
|
||||
| Class | Name | Function | Eight strainsa | DSM 20742, TCI-107 |
| Surface adhesins | AgI/II, Spa, PA | Specific adherence to acquired enamel pellicle | SMU.610, spaP, pac | D823_07515, spaA, pag |
| Par | Negative regulator of surface antigen | Absent? | D823_01230 or D823_08637, par | |
| Unnamed | Surface adhesin, part of ABC ion transporter | SMU.1302 | D823_10858 | |
| Glucosyltransferases | Gtf-I | Glucosyltransferase-I (insoluble) | SMU.1004, gtfB | D823_05448, gtfI |
| Gtf-SI | Glucosyltransferase-SI | SMU.1005, gtfC | D823_05918, gtfSI | |
| Gtf-S | Glucosyltransferase-S (soluble) | SMU.910, gtfD | D823_03428, gtfS1 | |
| Gtf-S | Glucosyltransferase-S (soluble) | Absent | D823_01485, gtfS2 | |
| Gtf-T | Glucosyltransferase-T (soluble) | Absent | D823_07585 or D823_10815, gtfTb | |
| Gtf-U | Glucosyltransferase-U (soluble) | Absent | D823_07585 or D823_10815, gtfUb | |
| Gtf | Glucosyltransferase | Absent | D823_03433, gtf | |
| Glucan-binding proteins | GbpA | Glucan-binding protein A | SMU.2112, gbpA | D823_05458 and D823_05463 |
| GbpB | Glucan-binding protein B | SMU.22, gbpB | D823_01475 | |
| GbpC | Glucan-binding protein C | SMU.1396, gbpC | D823_02626 and D823_02641 | |
| GbpD | Glucan-binding protein D and lipase | SMU.772, gbpD | D823 00935 | |
All genes shown are conserved for at least seven out of eight strains and the UA159 gene variant is shown as representative.
The exact assignment between D823_07585, D823_10815 and gtfT, gtfU was not possible.
Summarizing the literature, it seems that initial attachment of S. sobrinus to the pellicle is minimal and in a less specific manner (25, 27) but, once attached, it can, in the presence of sucrose, accumulate by massive glucan formation (7, 28). Any GTFs (and especially those secreted from S. mutans) present in the pellicle, promote the initial attachment of S. sobrinus (28, 29), explaining why S. sobrinus is rarely found without S. mutans. Different GTFs of different classes are combined to synthesize glucans. The enzyme called GTF-S synthesizes a soluble α-(1-6)-branched dextran, the enzyme called GTF-I, synthesizes an insoluble α-(1-3) rich D-mutan (7, 30). A third class (GTF-SI) does exist producing a semi-soluble glucan with mixed α-(1-6)-α-(1-3) linkages. By phenotype, S. mutans appears to form primarily GTF-S, whereas S. sobrinus has both GTF-S and GTF-I activities [see (7) for review]. In animal models, S. sobrinus GTF activity at slow growth rates consisted mainly of GTF-S activity (30) but at higher growth rates, such as it might occur in plaque during exposure to dietary sucrose, the proportions of GTF-I increased, resulting in more insoluble dextran (30). If this finding is extrapolated to humans, then frequent sucrose pulses allow S. sobrinus to accumulate on smooth surfaces via mutan production and this contributes to the increase in smooth-surface decay we indeed see in S. sobrinus positive subjects.
Comparing both species on a whole genome level (Table 1) reveals new details. S. sobrinus DSM 20742 has seven instead of three genes encoding for GTFs, one which is insoluble (GTF-I), one intermediate (GTF-SI), two ‘regular’ soluble (GTF-S1 and -S2), two which are α-(1-6)-branched glucan synthases [GTF-T or U respectively, see (32)], and one not further specifiable (‘GTF’). These multiple glucans apparently provide a potential by which S. sobrinus extends its niche from the retentive fissure site to the non-retentive smooth surfaces. It appears that GTF and glucans may play a minor role in fissure decay and perhaps no role at all where S. mutans is concerned. This also may explain why vaccines directed against the GTF of S. sobrinus (but not against the GTF of S. mutans) are mainly protective on smooth surfaces in animal models [reviewed by Loesche (7)].
S. mutans also synthesizes four Gbps: GbpA, GbpB, GbpC, and GbpD. The loss of any of the Gbps has an impact on adhesion and biofilm formation including dextran-dependent aggregation, dextranase inhibition, plaque cohesion, and perhaps cell wall synthesis (33). A whole genome comparison reveals that S. sobrinus has a similar repertoire in terms of Gbps (GbpA–D) but with two copies for GbpA and C encoding genes.
Uptake and metabolism of carbohydrates
Even more important than adherence is the successful growth of oral streptococci in their ecological niche. Where some years ago it was thought that pathogenic species were actively involved in acidogenicity of the ecosystem, nowadays we have more evidence that carbohydrate uptake and the resulting pH effects are, among other, the driving etiological factors, responsible for destabilization of oral biofilm homeostasis (3). MS get selected as they can compete with other species easily because of their capability to ferment many kinds of carbohydrates very efficiently. But interspecies competition is also found within MS. For instance, S. mutans out-competes S. sobrinus in the presence of the amino acid sugar N-acetylglucosamine (GlcNAc) together with glucose. It is suggested that GlcNAc inhibited growth of S. sobrinus in media containing both glucose and GlcNAc, by competing with glucose for the glucose phosphotransferase, depleting intracellular levels of phosphoenolpyruvate, or possessing lower levels of N-acetyl-glucosamine-6-phosphate deacetylase and/or glucosamine-6-phosphate deaminase activity (34). However, we found the deacetylase and deaminase genes in both species so that the difference might be due to transport systems [phosphotransferase system (PTS), see below] and/or promoter activity.
For S. mutans, the accumulation of genes related to carbohydrate uptake and metabolism was an essential evolutionary advancement contributing to the survival in the oral cavity and to the success as a caries ‘pathogen’ or – more correct – trigger. The transport of various oligosaccharides, including melibiose, raffinose, stachyose, and maltodextrans, is primarily conducted by the activity of ABC transporters, which include the multiple sugar metabolism (msm) and malXFGK transport systems. The predominant route for uptake of mono- and disaccharides is the phosphoenolpyruvate-sugar PTS, for review see (35). So far, we did not find essential differences in the genetic configuration related to sugar uptake between S. mutans and S. sobrinus here, as homologs for, e.g. msmG (D823_01675 in S. sobrinus), msmK (D823_07028), malX (WP_019787850.1), ptsH (D823_05408), or ptsI (WP_019775468) were found; however, this needs further in-depth investigation.
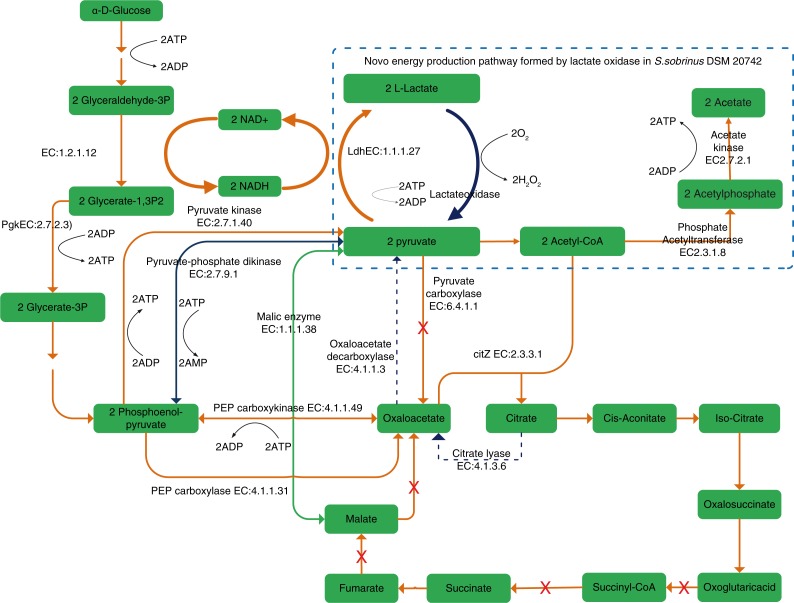
Due to their key roles in carbohydrates metabolism and energy production, glycolysis/gluconeogenesis, TCA cycle and pyruvate metabolism pathways are generally considered to be highly conserved among oral bacteria. Interestingly, between other MS and S. sobrinus, differences in the central carbon metabolic pathways were found by our group (31) and shown in Fig. 1. Facultative anaerobes such as lactic acid bacteria including Streptococcus lack cytochrome oxidases of a respiratory chain and ATP required for survival and growth is generated by substrate level phosphorylation in the glycolysis pathway almost exclusively (36). Interestingly, two L-lactate oxidases (with similarity between 65 and 73% to lactate oxidases of lactobacilli) are found to be conserved in S. sobrinus (so far confirmed for strains AC153, DSM 20742, TCI-107) but are absent in all S. mutans strains. These two enzymes catalyze the reaction of L-Lactate+O2→Pyruvate+H2O2 and/or D-Lactate+O2→Pyruvate+H2O2. Indeed, three strains of S. sobrinus have been shown to produce hydrogen peroxide in vitro (37). It has been reported that in S. pneumoniae concerted action of lactate oxidase and pyruvate oxidase forms a novel energy-generation pathway by converting lactate acid to acetic acid under aerobic growth conditions (38). Because there is no pyruvate oxidase identified in S. sobrinus DSM 20742, the function of the lactate oxidases in S. sobrinus DSM 20742 should be different to that of S. pneumoniae. By a close examination we hypothesize that lactate oxidase, together with pyruvate dehydrogenase, phosphate acetyl transferase and acetate kinase, could form a novel energy production pathway to convert lactate acid to acetate and simultaneously produce one additional ATP, as depicted. By doing so, the lactate oxidases of S. sobrinus DSM 20742 could also play a role in consuming lactate to regulate pH, which would be an advantage for S. sobrinus in resistance to acid stress. In addition, this pathway could replenish Acetyl-CoA, an important intermediate for the biosynthesis of fatty acids and amino acids. Furthermore, lactate oxidase and lactate dehydrogenase could form a local NAD+ regeneration system, which would be certainly advantageous to S. sobrinus DSM 20742 under aerobic growth conditions. Favored by possessing the lactate oxidases, S. sobrinus has the potential ability of producing H2O2 to kill not only competitors (oxygen sensitive S. mutans, oral anaerobes) but also macrophages (39), and defend its ecological niche. In contrast to the unique harboring of lactate oxidases in S. sobrinus DSM 20742, citrate lyase (EC 4.1.3.6), which catalyzes the cleavage of citrate into oxaloacetate and acetate, and oxaloacetate decarboxylase (EC 4.1.1.3), catalyzing the irreversible decarboxylation of oxaloacetate to pyruvate and CO2, are not found in S. sobrinus DSM 20742, as shown in Fig. 1 by the blue dotted lines. The absence of citrate lyase and oxaloacetate decarboxylase implies that S. sobrinus DSM 20742 might lack the ability in anaerobic utilization of citrate as a substrate. The disadvantages of S. sobrinus DSM 20742 in citrate utilization could be offset by the novel energy production pathway from lactate to acetate, as proposed above.
Fig. 1.
Central metabolism pathways of mutans streptococci. The orange lines represent enzyme reactions conserved across the mutans streptococci strains compared in our recent study (31), whereas the blue lines represent enzyme reactions specifically present (solid line) or absent (dashed line) in S. sobrinus DSM 20742. Red crosses: the corresponding enzymes were not present in any strain investigated.
Acid tolerance and two-component signal systems
Bacterial transduction two component systems (TCSs) play important roles by enabling cells to detect and respond to diverse changes/stresses in the environment with the pH to be one of the most important. A bacterial TCS comprises in general a trans-membrane sensor histidine kinase (HK) and a corresponding cytoplasmic response regulator (RR) encoded by genes located adjacently within the same operon, although stand-alone genes (‘orphans’) coding for HKs or RRs have also been reported (40). In a recent study, our group investigated differences in TCSs among MS including several S. mutans strains and S. sobrinus DSM 20742 (41). Totally, 18 TCS clusters comprising HK-RR pairs were identified. In Table 2, similarities and differences for both species in TCSs conserved for S. mutans are summarized. S. sobrinus DSM 20742 demonstrated deficits in the signal transduction systems related to acid tolerance and fructan catabolism (TCS-3, consisting of CovS/CovR). CovS/CovR is involved in the acid tolerance of S. mutans (42) and has also been reported to play a role in counteracting oxidative stress and reducing susceptibility to phagocytic killing (43). Therefore, the absence of TCS-3 can be interpreted as a selective disadvantage for S. sobrinus which might at least partially explain its lower prevalence and abundance in the oral cavity in general and in caries in particular.
Table 2.
Comparing two component systems between S. mutans [eight strains according to (31)] and S. sobrinus DSM 20742
| S. mutans | S. sobrinus | |||
|---|---|---|---|---|
|
|
||||
| TCS cluster | TCS protein | Function | 8 strainsa | DSM 20742 |
| TCS-1 | HK-VicK RR-VicR | Biofilm development, competence development, oxidative stress tolerance, acid tolerance, autolysin production, glucan metabolism, fructan metabolism | SMU.1516 SMU.1517 | D823_04656 D823_04651 |
| TCS-2 | HK-CiaH RR-CiaR | Sucrose-dependent biofilm formation, competence development, multiple stress response, bacteriocin production | SMU.1128 SMU.1129 | D823_05868 D823_05873 |
| TCS-3 | HK-CovS RR-CovR | Acid tolerance, hydrogen peroxide resistance, murine macrophage killing | SMU.1145c SMU.1146c | Absent Absent |
| TCS-4 | HK-KinF RR-LlrF | Acid tolerance, pp(G)pp metabolism, control of alarmone synthesis | SMU.928SMU.927 | D823_08322D823_08327 |
| TCS-5 | HK-ScnK RR-ScnR | Bacteriocin production | SMU.1814SMU.1815 | AbsentAbsent |
| TCS-6 | HK-SpaK RR-SpaR | Bacteriocin production, self-protection against anti-microbial peptides | SMU.660SMU.659 | D823_02456D823_02461 |
| TCS-7 | HK-PhoR RR-YcbL | Unknown | SMU.1037cSMU.1038c | AbsentAbsent |
| TCS-8 | HK-KinG RR-LlrG | Bacteriocin resistance, substrate transport in cell envelope stress | SMU.1009SMU.1008 | D823_04566D823_04561 |
| TCS-9 | HK-LevS RR-LevR | Biofilm formation, acid tolerance, fructan metabolism | SMU.1965cSMU.1964c | AbsentAbsent |
| TCS-10 | HK-LytS RR-LytT | Biofilm formation, oxidative stress tolerance, autolysis, fructan metabolism, cell wall metabolism | SMU.577 SMU.576 | D823_00965 D823_00970 |
| TCS-11 | HK-LiaS RR-LiaR | Biofilm formation, acid tolerance, cell envelope stress response, bacteriocin production & resistance, sucrose-dependent adherence | SMU.486 SMU.487 | D823_03016 D823_03011 |
| TCS-12 | HK-HK11 RR-RR11 | Unknown | SMU.1548cSMU.1547c | D823_06808D823_06803 |
| TCS-13 | HK-ComD RR-ComE | Biofilm formation, quorum sensing, competence development, bacteriocin production | SMU.1916 SMU.1917 | D823_05333 D823_05328 |
Only those conserved in S. mutans are discussed for both species. For more information see ref. (41).
All genes shown are conserved for at least seven out of eight strains and the UA159 gene is shown as representative.
TCS-7 (PhoR/YcbL) was shared by the eight S. mutans strains but was absent in S. sobrinus. PhoR is known for sensing environmental phosphate – which can be a limiting factor – in other species (44), but the clear function of TCS-7 in S. mutans is still unknown.
TCS-9 (LevRS), which affects the acid tolerance response, is also absent in S. sobrinus DSM 20742. In S. mutans UA159, the levRS gene cluster is flanked by levQ and levT, which code for two carbohydrate-binding proteins. These four genes together constitute a four-component signal transduction system levQRST controlling the transcription of the fructan hydrolase gene (fruA) and a four-gene cluster levDEFG, which encodes a fructose/mannose sugar-PTS located immediately downstream of levQRST (45). S. sobrinus was also found to lack the levQ, levT and levDEFG genes. Taking together, these findings indicate dramatic differences in the regulation of fructan catabolism and the acid tolerance response of S. sobrinus DSM 20742 in comparison to the S. mutans strains. However, other S. sobrinus genes (D823_00365, D823_00410) not shared with S. mutans and related to acid tolerance were found in strain DSM 20742.
Finally, TCS-5 (ScnKR) was found to be absent in some S. mutans strains and in S. sobrinus DSM 20742. In S. pyogenes ScnKR is essential for the production of a bacteriocin (SAFF22). In our recent study (41), we therefore inferred that TCS-5 might be involved in the regulation of mutacin production but not in acid tolerance.
Development of competence
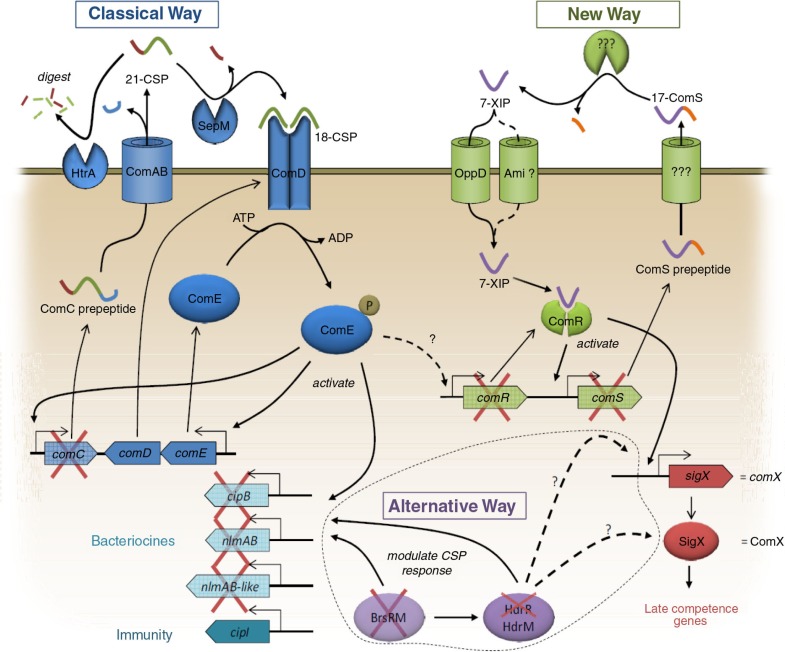
Competence development is a complex process involving sophisticated regulatory networks that trigger the capacity of bacterial cells to take up exogenous DNA from the environment. This phenomenon is frequently encountered in bacteria of the oral cavity, e.g. S. mutans (46). In S. mutans, ComX (or SigX), an alternative sigma factor, drives the transcription of the so-called ‘late-competence genes’ required for genetic transformation. ComX activity is induced by the inputs from two types of signaling pathways, namely the competence-stimulating peptide (CSP)-dependent competence regulation system (‘Classical way’, Fig. 2, left side and Table 3) and the XIP-dependent competence regulation system (‘New way’, Fig. 2, right side, Table 3). ComX and the ‘late-competence genes’ regulated by ComX are highly conserved even between the species, indicating that all MS might have the principal ability to be induced to genetic competence. On the other hand, the upstream signaling pathways, described in-depth below, regulating the activity of ComX show large differences.
Fig. 2.
Key differences in competence-related genes between S. mutans and S. sobrinus. Those which are missing in S. sobrinus are crossed off. Adapted from references (47, 48).
Table 3.
Comparing competence development-related systems between S. mutans UA159 [and orthologs of seven additional strains according to (31)] and S. sobrinus DSM 20742
| S. mutans | S. sobrinus | |||
|---|---|---|---|---|
| Group | Name | Function | Eight strainsa | DSM 20742 |
| Classical way | ComC | Competence stimulating peptide, precursor | SMU.1915 | Absent |
| ComA/NlmT | Competence factor and non-lantibiotic mutacin transporter ATP-binding/permease protein | SMU.286 SMU.1881c | D823_05343 D823_01400 | |
| ComB/NlmE | Accessory factor for NlmT | SMU.287 | D823_05923 | |
| SepM | Cell surface-associated protease cleavage CSP | SMU.518 | D823_08607 | |
| ComD | Histidine kinase | SMU.1916 | D823_05333 | |
| ComE | Response regulator | SMU.1917 | D823_05328 D823_7992b | |
| HtrA | Serine protease | SMU.2164 | D823_03191 | |
| New way | ComS | comX-inducing peptide (XIP) precursor | NC_004350.2 | Absent |
| (62613-62666)a | ||||
| ComR | ComS receptor | SMU.61 | Absent | |
| Alternative way | HdrM | High-density responsive membrane protein | SMU.1855 | D823_08222 |
| HdrR | High-density responsive regulator | SMU.1854 | Absent | |
| BrsM | SMU.2081 | Absent | ||
| BrsR | SMU.2080 | Absent | ||
| OppD | Oligopeptide ABC transporter | SMU.258 | D823_04322 | |
| Late competence | ComX (SigX) | Competence-specific sigma factor | SMU.1997 | D823_08887 |
| ComEA | Competence protein | SMU.625 | D823_08107 | |
| ComEC | Competence protein; possible integral membrane protein | SMU.626 | D823_08117 | |
| CoiA | Competence protein CoiA | SMU.644 | D823_01025 | |
| EndA | Competence-associated membrane nuclease (DNA-entry nuclease) | SMU.1523 | D823_09687 | |
| ComG | Competence protein G | SMU.1981c | D823_01170 | |
| ComYD | Competence protein ComYD | SMU.1983 | D823_01160 | |
| ComYC | Competence protein ComYC, | SMU.1984 | D823_01155 | |
| Possible competence-induced protein | SMU.2075c | D823_03558 | ||
| CinA | Competence damage-inducible protein A | SMU.2086 | D823_03593 | |
| ComYB | Competence protein; general (type II) secretory pathway protein | SMU.1985 | D823_01150 | |
| ComYA | Late competence protein; type II secretion system protein | SMU.1987 | D823_01145 | |
| ComFC | Late competence protein required for DNA uptake | SMU.499 | D823_02981 | |
| ComFA | Late competence protein F | SMU.498 | D823_02986 | |
| CinA | Competence damage-inducible protein A | SMU.2086 | D823_03593 | |
All genes shown are conserved for at least seven out of eight strains and the UA159 gene variant is shown as representative.
The gene D823_7992 is very similar to D823_5328 but found distantly on a different contig.
CSP-dependent competence regulation
In S. mutans, the prepeptide of CSP is encoded by comC. Whereas comC is present in all S. mutans strains investigated so far, it is absent in the two annotated strains S. sobrinus DSM 20742 and TCI-107. Apart from this synthase, all genes required for CSP-dependent signaling that are found in S. mutans are also present in S. sobrinus. The membrane bound ABC-transporter (ComAB), the two-component signal transduction system ComDE, the extracellular protease SepM, which is involved in the processing of 21-CSP to the mature 18-CSP, as well as the HtrA protease, which is thought to degrade extracellular CSP (49), were all identified in S. sobrinus. But as the central comC homologue is missing in S. sobrinus, this species cannot develop competence through this signaling pathway, but what about alternatives?
XIP-dependent competence regulation
A new peptide regulatory system (ComSR) that is independent of CSP and directly activates ComX has been identified by Mashburn-Warren et al. (47). The novel autoinducer XIP (sigX inducing peptide) is synthesized as a prepeptide by the synthase ComS. The membrane protein that processes and exports the 17-mer ComS precursor to the active 7-mer pheromone XIP is unknown. Extracellular XIP is internalized through the peptide transporter OppD. Internalized XIP binds to the transcriptional regulator ComR, which is thereby dimerized and activated. The expression of the synthase ComS, as well as the expression of the alternative sigma-factor ComX, resulting in transcription of the complete transformasome, is controlled by ComR. Deletion of the comR or comS gene completely abolished the competence in S. mutans (47). Thus, ComR is the central regulator for competence in S. mutans and is also required for CSP induced competence development. In our previous study (31), the ComSR regulatory system was identified in all of the S. mutans strains, but not in S. sobrinus DSM 20742. Accordingly, despite the presence of comX and the ‘late-competence genes’, we were not able to obtain the genetic competence state of S. sobrinus DSM 20742 experimentally. Interestingly, S. sobrinus DSM 20742 does harbor the OppD peptide transporter for import of extracellular XIP. Thus, all the genes for both signaling pathways for competence are present, with the exception of the respective autoinducer synthases comC and comS, and the essential transcriptional regulator comR.
Autoinducer-independent competence regulation
Under conditions of biofilm growth the HdrMR system, a novel two-gene regulatory system, has been shown to contribute to competence development through the activation of ComX (50, 51). Microarray analysis revealed that both regulators, ComE and HdrR, activate a large set of genes (50, 51). Recently, Xie et al. (52) identified another regulatory system in S. mutans, designated BrsRM, that regulates bacteriocin-related genes but also affects the HdrRM system. In our recent study, HdrR and the complete BrsRM system were found absent in S. sobrinus.
Distribution of bacteriocin-related proteins
Bacteriocins are proteinaceous antimicrobials produced by bacteria to kill or inhibit the growth of similar or closely related bacterial strains. Bacteriocins produced by MS are named ‘mutacins’. As dental plaque, the dominating niche of MS, is a multispecies biofilm community that harbors many microorganisms, mutans group strains have developed a variety of mutacins to inhibit the growth of competitors, such as mitis group streptococci (53–55). In our previous study (31), information about known mutacins as well as mutacin-immunity proteins was collected from the NCBI (http://www.ncbi.nlm.nih.gov) and Oralgen (http://www.oralgen.lanl.gov/) databases. The collected protein sequences were used to blast against the proteomes of eight S. mutans strains and S. sobrinus DSM 20742 to see whether or not these known mutacins and corresponding immunity proteins exist in both species in a similar or different pattern. Distributions of identified mutacins and mutacin-immunity proteins are summarized in Table 4. Diversity of Streptococcus bacteriocins has been reported previously (56, 57). An interesting new result is that, in contrast to S. mutans strains, S. sobrinus DSM 20742 does not possess any genes coding for mutacin or mutacin-like proteins.
Table 4.
Comparing bacteriocin and corresponding immunity proteins between S. mutans [eight strains according to (31)] and S. sobrinus DSM 20742
| S. mutans | S. sobrinus | |
|---|---|---|
|
|
||
| Mutacin/immunity protein | Eight strains | DSM 20742 |
| Lantibiotic mutacins | ||
| Mutacin-Smb | Rare | Absent |
| Mutacin-I | Rare | Absent |
| Mutacin-II | Rare | Absent |
| Mutacin-III | Rare | Absent |
| Mutacin-K8 | Rare | Absent |
| Non-lantibiotic bacteriocins | ||
| Mutacin-IV (NlmA) | Frequent | Absent |
| Mutacin-IV (NlmB) | Frequent | Absent |
| Mutacin-IV like (SMU.283) | Conserved | Absent |
| Immunity protein of Mutacin-IV | Highly conserved | Present |
| Mutacin-V (CipB) | Frequent | Absent |
| CipI, immunity protein of CipB | Very frequent | Absent |
| Homolog of CipI | Frequent | Present |
| SMU.423 (possible bacteriocin) | Conserved | Present |
| NlmT/ComA | Conserved | Present |
| ATP-binding protein of NlmTE | Frequent | Present |
| NlmE/ComB (accessory factor for NlmT) | Highly conserved | Present |
Mutacin-Smb has been identified in S. mutans and S. ratti previously (58, 59). Mutacin-K8 is an ortholog of the bacteriocin Streptococcin A-FF22 identified in group-A streptococci (60), and its production system has also been previously identified in the S. mutans strains K8 (61), KK23, and NN20125 (31). Possibly caused by transposase activity, the complete mutacin-K8 production system can be disrupted leaving partial orthologs which we found in S. mutans AC4446, UA159, 5DC8, and KK21 (31). Lantibiotic mutacins, mutacin-I (62), mutacin-II (63) and mutacin-III (64) are found in only a few strains of S. mutans so far. Mutacin-IV, non-lantibiotic bacteriocins coded by nlmA/B, was discovered first in S. mutans UA140 to be active against the mitis group streptococci (65). We found nlmA/B in most S. mutans strains but not in S. sobrinus DSM 20742, the latter even negative for mutacin-IV-like protein encoding genes. Interestingly, the immunity protein for mutacin-IV (SMU.152) was identified in all mutacin-IV-negative strains including S. sobrinus, consistent with the fact that no inhibition phenomenon has been observed yet among different MS strains. Mutacin-V, another non-lantibiotic peptide coded by cipB, was frequently found in S. mutans strains (exceptions are, however, reference strains ATCC 15175 and NCTC 11060) but not in S. sobrinus DSM 20742. There are two homologs of mutacin-V immunity protein in S. mutans UA159, namely the product of SMU.1913 and CipI (SMU.925) (66, 67); the latter is supposed to be the key factor of immunity. All the S. mutans strains investigated by our group together with S. sobrinus DSM 20742 possess at least one orthologous gene encoding one of the two mutacin-V immunity proteins so that they might not be inhibited by mutacin-V producing strains. Furthermore, a possible non-lantibiotic bacteriocin peptidegene (SMU.423) was found to be conserved in S. mutans and present in S. sobrinus DSM 20742. In addition, putative ComAB (NlmTE), which has been proved to be the transporter complex of mutacin IV or – more likely – for multi-type non-lantibiotic bacteriocins in S. mutans (31, 68), are identified in all S. mutans strains and S. sobrinus. It may very be possible that this bacteriocin is similar to the mutacin isolated from S. sobrinus strain MT6223 that proved to be competitive with S. mutans in rat models (69).
To summarize, S. sobrinus DSM 20742 (and confirmed in TCI-107) does not possess any genes coding for mutacin or mutacin-like proteins including Mutacin-Smb, Mutacin-K8, Mutacin-I-III, Mutacin-IV (NlmA and B), and Mutacin-V, although bacteriocin-like proteins from S. sobrinus have been found for individual strains.
However, searching among the bacteriocin/immunity genes discovered exclusively in S. sobrinus (and not in S. mutans), two immunity protein encoding genes but no additional bacteriocin-gene were identified (function in brackets): D823_05508 (immunity protein PlnI-like) and D823_03977 (putative bacteriocin immunity protein).
The accumulation and conservation of mutacin immunity proteins apparently play an important role for the survival of all MS strains and species in a bacteriocin-rich environment.
Oxidative stress defense systems
For protection against reactive oxygen species (such as O2 −, H2O2, HO·) or adaptation to oxidative stresses, aerobes and facultative anaerobes have evolved efficient defense systems, comprising an array of antioxidant enzymes such as catalase, superoxide dismutase (SOD), alkylhydroperoxide reductase (AhpCF), Dps-like peroxide resistance protein (Dpr), thioredoxin reductase, and glutathione reductase, which have been identified in many bacterial species. By searching for known antioxidant systems in the genomes of sequenced mutans streptococcal strains, we obtained an overview of putative oxidative defense systems (31) summarized in Table 5. Catalase, which catalyzes the decomposition of hydrogen peroxide, was never found in any of the mutans streptococcal strains but other classes of oxygen tolerance-related proteins do exist.
Table 5.
Comparing proteins involved in oxygen tolerance between S. mutans [eight strains according to (31)] and S. sobrinus DSM 20742
| S. mutans | S. sobrinus | |||
|---|---|---|---|---|
|
|
||||
| Class | Name | Function | Eight strainsa | DSM 20742 |
| SOD | Sod | Superoxide dismutase | SMU.629 | D823_08152 |
| AhpF/AhpC system | AhpC | Alkyl hydroperoxide reductase, subunit C | SMU.764 | Absent |
| AhpF (Nox1) | Alkyl hydroperoxide reductase, subunit F | SMU.765 | Absent | |
| Dpr | Dpr | Peroxide resistance protein/iron binding protein | SMU.540 | D823_02352 |
| Thioredoxin system | TrxB | Thioredoxin reductase (NADPH) | SMU.463 | D823_01947 |
| TrxB | Thioredoxin reductase | SMU.869 | D823_01550 | |
| TrxA | Thioredoxin | SMU.1869 | D823_06913 | |
| TrxH | Thioredoxin family protein | SMU.1971c | D823_08552 | |
| Thioredoxin family protein | SMU.1169c | Absent | ||
| Tpx | Thiol peroxidase | SMU.924 | D823_07595 | |
| Glutaredoxin system | GshAB | Glutathione biosynthesis bifunctional protein | SMU.267c | D823_06703 |
| GshR | Glutathione reductase | SMU.838 | D823_04976 | |
| GshR | Glutathione reductase | SMU.140 | Absent | |
| NrdH | Glutaredoxin | SMU.669c | D823 05398 | |
All genes shown are conserved for at least seven out of eight strains and the UA159 gene variant is shown as representative.
First, SOD, which catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide, was found in all strains of our study conserved including S. sobrinus DSM 20742 and it is present in S. sobrinus TCI-107 in two copies.
Next, it has been reported that both the bi-component peroxidase system AhpF/AhpC (catalyzing the NADH-dependent reduction of organic hydroperoxides and/or H2O2 to their respective alcohol) and Dpr (a ferritin-like iron-binding protein) confer tolerance to oxidative stress in S. mutans (70). AhpF/AhpC-genes were present in all S. mutans strains, but were absent in S. sobrinus DSM 20742 (31) indicating that these genes do not form an essential peroxide tolerance system for MS. Indeed, Higuchi et al. (36) found that a double ahpF/ahpC mutant still showed the same level of peroxide tolerance as the defect could be complemented by the S. mutans-dpr gene. Dpr homologs were found in all S. mutans strains and in S. sobrinus underlining the essential function in oxygen tolerance.
Thioredoxins are a class of small redox mediator proteins known to be present in all organisms. They are involved in many important biological processes, including redox signaling. The flavor enzyme thioredoxin reductase keeps thioredoxins in the reduced state in a NADPH-dependent reaction (71). They act as electron donors to many proteins including thiol peroxidases (72). Thioredoxin, thioredoxin reductase and thiol peroxidase, the components of the thioredoxin system, were identified in all S. mutans strains and in S. sobrinus DSM 20742. Thioredoxin family proteins (SMU.1971c and SMU.1169c) are found to be present in nearly all strains, except for S. sobrinus DSM 20742, which lacks any ortholog of SMU1169c (31).
Finally, glutaredoxins share many functions of thioredoxins. But they are oxidized by their corresponding substrates and reduced by glutathione (GSH) (73). The resulting oxidized glutathione (GSSG) is regenerated by glutathione reductase. Together, these components comprise the glutathione system (74). Several S. mutans strains possess two glutathione reductase orthologs (SMU.140 and SMU.838). In contrast, S. sobrinus DSM 20742 possesses an ortholog for SMU.838 but not for SMU.140, possibly leading to a reduced potential for re-generation of GSH from GSSG and weakening its oxidative resistance.
Link between acidogenicity, aciduricity, biofilm formation, mutacin production, competence and – ultimately – cariogenicity
The main virulence traits of S. mutans – adherence, acidogenicity, aciduricity, biofilm formation and mutacin production – as well as its ability to incorporate foreign DNA into its genome (genetic competence) are controlled or modulated by quorum sensing and thus depending on its own cell number but maybe also on cell numbers of cohabitants. For S. mutans, it has recently been shown that the complete quorum sensing system is induced by co-culture with the human pathogenic fungus Candida albicans (75). Additionally, both strains grow better together than as a monoculture, suggesting that this synergism may lead to enhanced cariogenicity. This could recently be confirmed in an animal model (76). Interestingly, one main cariogenic trait of S. mutans, the synthesis of extracellular glucans and fructans, was strongly inhibited in co-culture (75). Similar studies on S. sobrinus are missing but – without competence so far investigated – such a co-stimulation is not expected.
Clearly the influence of inter-species communication for caries development warrants further studies.
In summary, this work compares two main cariogenic MS on a whole genome level. Although more aciduric and acidogenic, by many other ecologically important features, S. sobrinus seems to be weaker in its cariogenic potential. As rapid adherence to the pellicle coated tooth surface is crucial for colonization and expressing cariogenic potential, the down-regulation of surface antigen SpaA by the negative regulator Par, which is only found in S. sobrinus but not in S. mutans, could be essential. Furthermore, the lack of genetic competence of S. sobrinus limits its evolutionary potential. However, we have to keep in mind that this analysis is based on about 8–20 annotated S. mutans strains but on only two S. sobrinus strains. In total we found about 470 genes in S. sobrinus DSM 20742 – about half of them hypothetical proteins with no allocated or known function so far – with no orthologs in S. mutans. Thus, S. sobrinus possess much more potential yet to be discovered. Finally, this comparison of genetic inventory (genotypes) might help to describe or predict phenotypes but there are more factors, especially in a very complex habitat like the human oral cavity, which certainly influence the true cariogenic potential of those organisms.
Acknowledgements
This work is based on two former publications (Song et al. BMC Genomics 2012 and 2013) and we wish to thank the co-authors Padhmanand Sudhakar, Wei Wang, Anke Rheinberg (former Brock), Jibin Sun, and Michael Reck for their contributions.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors. This work was funded as part of the BioInSys project No. 0315411D by the German Ministry of Education and Research (BMBF, call MedSys). We thank all collaborative partners here.
References
- 1.Loesche WJ. Chemotherapy of dental plaque infections. Oral Sci Rev. 1976;9:65–107. [PubMed] [Google Scholar]
- 2.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–71. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi N, Nyvad B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008;42:409–18. doi: 10.1159/000159604. [DOI] [PubMed] [Google Scholar]
- 4.Rosier BT, De Jager M, Zaura E, Krom BP. Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Front Cell Infect Microbiol. 2014;4:92. doi: 10.3389/fcimb.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Soet JJ, Nyvad B, Kilian M. Strain-related acid production by oral streptococci. Caries Res. 2000;34:486–90. doi: 10.1159/000016628. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson P, Gandour IA, Olsson B, Rickardsson B, Abbas K. High prevalence of mutans streptococci in a population with extremely low prevalence of dental caries. Oral Microbiol Immunol. 1987;2:121–4. doi: 10.1111/j.1399-302x.1987.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 7.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Soet JJ, Toors FA, de Graaff J. Acidogenesis by oral streptococci at different pH values. Caries Res. 1989;23:14–17. doi: 10.1159/000261148. [DOI] [PubMed] [Google Scholar]
- 9.de Soet JJ, van Loveren C, Lammens AJ, Pavicic MJ, Homburg CH, ten Cate JM, et al. Differences in cariogenicity between fresh isolates of Streptococcus sobrinus and Streptococcus mutans . Caries Res. 1991;25:116–22. doi: 10.1159/000261353. [DOI] [PubMed] [Google Scholar]
- 10.Okada M, Kawamura M, Oda Y, Yasuda R, Kojima T, Kurihara H. Caries prevalence associated with Streptococcus mutans and Streptococcus sobrinus in Japanese schoolchildren. Int J Paediatr Dent. 2012;22:342–8. doi: 10.1111/j.1365-263X.2011.01203.x. [DOI] [PubMed] [Google Scholar]
- 11.Okada M, Taniguchi Y, Hayashi F, Doi T, Suzuki J, Sugai M, et al. Late established mutans streptococci in children over 3 years old. Int J Dent. 2010;2010:732468. doi: 10.1155/2010/732468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada M, Soda Y, Hayashi F, Doi T, Suzuki J, Miura K, et al. Longitudinal study of dental caries incidence associated with Streptococcus mutans and Streptococcus sobrinus in pre-school children. J Med Microbiol. 2005;54:661–5. doi: 10.1099/jmm.0.46069-0. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Acedo M, Montiel-Company JM, Dasi-Fernandez F, Almerich-Silla JM. Streptococcus mutans and Streptococcus sobrinus detection by polymerase chain reaction and their relation to dental caries in 12 and 15 year-old schoolchildren in Valencia (Spain) Med Oral Patol Oral Cir Bucal. 2013;18:839–e45. doi: 10.4317/medoral.18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holbrook WP, Magnusdottir MO. Studies on strains of Streptococcus mutans isolated from caries-active and caries-free individuals in Iceland. J Oral Microbiol. 2012;4:10611. doi: 10.3402/jom.v4i0.10611. http://dx.doi.org/10.3402/jom.v6.2658410611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindquist B, Emilson CG. Interactions between and within Streptococcus mutans and Streptococcus sobrinus isolated from humans harboring both species. Scand J Dent Res. 1991;99:498–504. doi: 10.1111/j.1600-0722.1991.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 16.Lindquist B, Emilson CG. Dental location of Streptococcus mutans and Streptococcus sobrinus in humans harboring both species. Caries Res. 1991;25:146–52. doi: 10.1159/000261358. [DOI] [PubMed] [Google Scholar]
- 17.Lindquist B, Emilson CG. Colonization of Streptococcus mutans and Streptococcus sobrinus genotypes and caries development in children to mothers harboring both species. Caries Res. 2004;38:95–103. doi: 10.1159/000075932. [DOI] [PubMed] [Google Scholar]
- 18.Caufield PW, Cutter GR, Dasanayake AP. Initial acquisition of mutans streptococci by infants: evidence for a discrete window of infectivity. J Dent Res. 1993;72:37–45. doi: 10.1177/00220345930720010501. [DOI] [PubMed] [Google Scholar]
- 19.Straetemans MM, van Loveren C, de Soet JJ, de Graaff J, ten Cate JM. Colonization with mutans streptococci and lactobacilli and the caries experience of children after the age of five. J Dent Res. 1998;77:1851–5. doi: 10.1177/00220345980770101301. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, Jespersgaard C, Lamberty-Mallory L, Katz J, Huang Y, Hajishengallis G, et al. Enhanced immunogenicity of a genetic chimeric protein consisting of two virulence antigens of Streptococcus mutans and protection against infection. Infect Immun. 2002;70:6779–87. doi: 10.1128/IAI.70.12.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubovics NS, Stromberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol. 2005;55:1591–605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Green NM, Sitkiewicz I, Lefebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus . Infect Immun. 2006;74:4200–13. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demuth DR, Davis CA, Corner AM, Lamont RJ, Leboy PS, Malamud D. Cloning and expression of a Streptococcus sanguis surface antigen that interacts with a human salivary agglutinin. Infect Immun. 1988;56:2484–90. doi: 10.1128/iai.56.9.2484-2490.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Raamsdonk M, de Soet JJ, Jones CL, de Graaff J. Effect of antibodies on the chain length and growth of Streptococcus sobrinus . Caries Res. 1997;31:35–40. doi: 10.1159/000262371. [DOI] [PubMed] [Google Scholar]
- 25.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–50. doi: 10.1128/MMBR.00014-09. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MC, Pilling PA, Epa VC, Berry AM, Ogunniyi AD, Paton JC. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998;6:1553–61. doi: 10.1016/s0969-2126(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 27.Staat RH, Peyton JC. Adherence of oral streptococci: evidence for nonspecific adsorption to saliva-coated hydroxylapatite surfaces. Infect Immun. 1984;44:653–9. doi: 10.1128/iai.44.3.653-659.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons RJ, Cohen L, Hay DI. Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect Immun. 1986;52:555–61. doi: 10.1128/iai.52.2.555-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolla G, Ciardi JE, Schultz SA. Adsorption of glucosyltransferase to saliva coated hydroxyapatite. Possible mechanism for sucrose dependent bacterial colonization of teeth. Scand J Dent Res. 1983;91:112–17. doi: 10.1111/j.1600-0722.1983.tb00786.x. [DOI] [PubMed] [Google Scholar]
- 30.Walker GJ, Brown RA, Taylor C. Activity of Streptococcus mutans alpha-D-glucosyltransferases released under various growth conditions. J Dent Res. 1984;63:397–400. doi: 10.1177/00220345840630030801. [DOI] [PubMed] [Google Scholar]
- 31.Song L, Wang W, Conrads G, Rheinberg A, Sztajer H, Reck M, et al. Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics. 2013;14:430. doi: 10.1186/1471-2164-14-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanbu A, Hayakawa M, Takada K, Shinozaki N, Abiko Y, Fukushima K. Production, characterization, and application of monoclonal antibodies which distinguish four glucosyltransferases from Streptococcus sobrinus . FEMS Immunol Med Microbiol. 2000;27:9–15. doi: 10.1111/j.1574-695X.2000.tb01405.x. [DOI] [PubMed] [Google Scholar]
- 33.Banas JA, Vickerman MM. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- 34.Homer KA, Patel R, Beighton D. Effects of N-acetylglucosamine on carbohydrate fermentation by Streptococcus mutans NCTC 10449 and Streptococcus sobrinus SL-1. Infect Immun. 1993;61:295–302. doi: 10.1128/iai.61.1.295-302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moye ZD, Zeng L, Burne RA. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl Environ Microbiol. 2014;80:972–85. doi: 10.1128/AEM.03579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higuchi M, Yamamoto Y, Kamio Y. Molecular biology of oxygen tolerance in lactic acid bacteria: functions of NADH oxidases and Dpr in oxidative stress. J Biosci Bioeng. 2000;90:484–93. [PubMed] [Google Scholar]
- 37.Garcia-Mendoza A, Liebana J, Castillo AM, de la Higuera A, Piedrola G. Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol. 1993;39:434–9. doi: 10.1099/00222615-39-6-434. [DOI] [PubMed] [Google Scholar]
- 38.Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, et al. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol. 2008;190:3572–9. doi: 10.1128/JB.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okahashi N, Nakata M, Sumitomo T, Terao Y, Kawabata S. Hydrogen peroxide produced by oral streptococci induces macrophage cell death. PLoS One. 2013;8:e62563. doi: 10.1371/journal.pone.0062563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, et al. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics. 2012;13:128. doi: 10.1186/1471-2164-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levesque CM, Mair RW, Perry JA, Lau PC, Li YH, Cvitkovitch DG. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett Appl Microbiol. 2007;45:398–404. doi: 10.1111/j.1472-765X.2007.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen PM, Chen HC, Ho CT, Jung CJ, Lien HT, Chen JY, et al. The two-component system ScnRK of Streptococcus mutans affects hydrogen peroxide resistance and murine macrophage killing. Microbes Infect. 2008;10:293–301. doi: 10.1016/j.micinf.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Gardner SG, Johns KD, Tanner R, McCleary WR. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol. 2014;196:1741–52. doi: 10.1128/JB.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng L, Wen ZT, Burne RA. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans . Mol Microbiol. 2006;62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
- 46.Tanzer JM, Livingston J, Thompson AM. The microbiology of primary dental caries in humans. J Dent Educ. 2001;65:1028–37. [PubMed] [Google Scholar]
- 47.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.365-2958.010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Federle MJ, Morrison DA. One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol Microbiol. 2012;86:241–5. doi: 10.1111/mmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol. 2012;86:258–72. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okinaga T, Niu G, Xie Z, Qi F, Merritt J. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J Bacteriol. 2010;192:1844–52. doi: 10.128/JB.01667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okinaga T, Xie Z, Niu G, Qi F, Merritt J. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans . Mol Oral Microbiol. 2010;25:165–77. doi: 10.1111/j.2041-1014.2010.00574.x. [DOI] [PubMed] [Google Scholar]
- 52.Xie Z, Okinaga T, Niu G, Qi F, Merritt J. Identification of a novel bacteriocin regulatory system in Streptococcus mutans . Mol Microbiol. 2010;78:1431–47. doi: 10.111/j.365-2958.010.07417.x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alaluusua S, Takei T, Ooshima T, Hamada S. Mutacin activity of strains isolated from children with varying levels of mutans streptococci and caries. Arch Oral Biol. 1991;36:251–5. doi: 10.1016/0003-9969(91)90094-b. [DOI] [PubMed] [Google Scholar]
- 54.Baba T, Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. doi: 10.1016/S0966-842X(97)01196-7. [DOI] [PubMed] [Google Scholar]
- 55.Hossain MS, Biswas I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol. 2011;77:2428–34. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bekal-Si Ali S, Hurtubise Y, Lavoie MC, LaPointe G. Diversity of Streptococcus mutans bacteriocins as confirmed by DNA analysis using specific molecular probes. Gene. 2002;283:125–31. doi: 10.1016/s0378-1119(01)00875-7. [DOI] [PubMed] [Google Scholar]
- 57.Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus . J Bacteriol. 2007;189:1189–98. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob Agents Chemother. 2005;49:541–8. doi: 10.1128/AAC.49.2.541-548.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyink O, Balakrishnan M, Tagg JR. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol Lett. 2005;252:235–41. doi: 10.1016/j.femsle.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Johnson DW, Tagg JR, Wannamaker LW. Production of a bacteriocine-like substance by group-A streptococci of M-type 4 and T-pattern 4. J Med Microbiol. 1979;12:413–27. doi: 10.1099/00222615-12-4-413. [DOI] [PubMed] [Google Scholar]
- 61.Robson CL, Wescombe PA, Klesse NA, Tagg JR. Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology. 2007;153:1631–41. doi: 10.1099/mic.0.2006/003756-0. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, et al. Genes involved in the repression of mutacin I production in Streptococcus mutans . Microbiology. 2009;155:551–6. doi: 10.1099/mic.0.021303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen P, Qi F, Novak J, Caufield PW. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl Environ Microbiol. 1999;65:1356–60. doi: 10.1128/aem.65.3.1356-1360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi F, Chen P, Caufield PW. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl Environ Microbiol. 1999;65:3880–7. doi: 10.1128/aem.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009;72:905–17. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–9. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hale JD, Heng NC, Jack RW, Tagg JR. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans . J Bacteriol. 2005;187:5036–9. doi: 10.1128/JB.187.14.5036-5039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loyola-Rodriguez JP, Morisaki I, Kitamura K, Hamada S. Purification and properties of extracellular mutacin, a bacteriocin from Streptococcus sobrinus . J Gen Microbiol. 1992;138:269–74. doi: 10.1099/00221287-138-2-269. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. Role of the dpr product in oxygen tolerance in Streptococcus mutans . J Bacteriol. 2000;182:3740–7. doi: 10.1128/jb.182.13.3740-3747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346:1–8. [PMC free article] [PubMed] [Google Scholar]
- 72.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–9. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 73.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–6. [PubMed] [Google Scholar]
- 74.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 75.Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans . ISME J. 2014;8:2256–71. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–81. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]