Abstract
Investigation of the response of coral microbial communities to seasonal ecological environment at the microscale will advance our understanding of the relationship between coral-associated bacteria community and coral health. In this study, we examined bacteria community composition from mucus, tissue and skeleton of Porites lutea and surrounding seawater every three months for 1 year on Luhuitou fringing reef. The bacterial communities were analyzed using pyrosequencing of the V1-V2 region of the 16S rRNA gene, which demonstrated diverse bacterial consortium profiles in corals. The bacterial communities in all three coral compartments studied were significantly different from the surrounding seawater. Moreover, they had a much more dynamic seasonal response compared to the seawater communities. The bacterial communities in all three coral compartments collected in each seasonal sample tended to cluster together. Analysis of the relationship between bacterial assemblages and the environmental parameters showed that the bacterial community correlated to dissolved oxygen and rainfall significantly at our study site. This study highlights a dynamic relationship between the high complexity of coral associated bacterial community and seasonally varying ecosystem parameters.
Coral reefs are the largest structures made by living creatures. They contain an enormous diversity of organisms, and have been referred to as the rainforests of the ocean1. The coral holobiont contains symbiotic dinoflagellates as well as diverse fungi, bacteria, archaea and viruses. The microniches within the coral holobiont including the surface mucus layer, tissue and calcium carbonate skeleton, provide habitats for diverse microbial life. The importance of coral associated microbes is being recognized due to the findings that microbes play an active role in maintaining coral nutrition and health2.
High coral bacterial species diversity has been emphasized in numerous studies, and site-specificity and/or coral host species-specificity has been demonstrated3,4,5,6,7. Seasonal factors have also been reported to cause shifts in coral-associated bacterial diversity8,9,10. Hong and colleagues8 suggested a multifactor interaction model with variables that influence coral microbe associations that includes: environmental conditions, coral species, colony physiology, and temporal factors. Findings show high complexity of coral associated bacterial assemblages8,9,11 and no conclusive diversity pattern of coral-associated bacteria has been reported to date.
The spatial scale of variations in bacterial community composition is important in considering coral bacteria interactions and their relationship to environmental stresses. Heterogeneity in bacterial diversity has been reported not only in different geographic locations and coral species3, but also within individual coral colonies12. Moreover, spatial structure of the bacterial assemblages has even been described from a single coral colony3,13. Corals' biological structures provide different microhabitats, which will likely have unique physiochemical environments that may influence the bacterial community structure2. However, most studies investigate the diversity of coral-associated bacteria without considering this factor. A focus on bacterial assemblages associated with corals that simultaneously addresses temporal and spatial structure will significantly enhance our understanding of the dynamic of coral-associated bacterial community.
The dynamic population of coral-associated bacteria is considered to be a mechanism that allows for the rapid adaptation of corals in a changing environment14. Garren et al.15 further emphasized that the resilience capability of microbial communities within a single coral colony fundamentally contribute to the resilience and health of reef ecosystems. We now know some spatial or seasonal biodiversity transitions profiles of the coral-associated microbes3,8,9,16. Thinking about the microbial community in the context of the temporal transition and spatial partition will contribute to illuminating the dynamics of microbial community compositions within the coral holobiont. Our previous research demonstrated that relatively small common bacterial communities are in corals distributed in the South China Sea17. We speculated that these bacterial associations may be structured by multiple factors at different scales and that corals may associate with microbes in terms of similar function, rather than identical species17.
As a massive reef-building coral, Porites lutea is certainly important in reef systems. In this study, we comprehensively investigated the bacterial communities associated with the dominant coral species P. lutea on Luhuitou fringing reef over 1 year in consideration both of time and spatial structures. This allowed us to determine the variations in coral associated bacterial diversity in different holobiont compartments response to the seasonal environmental changes. We tested the hypotheses that: 1) seasonal transitions would cause shift in bacterial community associated with the coral P. lutea distributed in the South China Sea, and 2) the seasonal variations might happen in three compartments including the coral mucus, tissue and skeleton. This study provides the first comparison of microbial communities associated with P. lutea over a seasonal time scale, and will aid in advancing our understanding of the response of coral holobiont to changing environment and developing effective coral reef management strategies without overlooking the microbial ecology of the microscale ecosystems.
Results
Coral-associated bacterial diversity
Quality filtering recovered a total of 164,352 reads from pyrosequencing of the mixture of PCR amplicons of the pooled DNA, with sequence lengths ranging from 277 to 369 bp after primer and barcode removal. The number of OTUs and diversity estimates at a 3% dissimilarity level are listed in Table 1. The rarefaction curves (with a 3% cutoff value) of the samples showed that the sample size (read number) was generally sufficient (Fig. S1).
Table 1. Numbers of sequences and OTUs (97%) and diversity estimates of Porites lutea coral-associated bacteria. M represents mucus; T represents tissue; S represents skeleton; SW represents seawater.
| Index | Feb-M | Feb-S | Feb-SW | May-M | May-T | May-S | May-SW | Aug-M | Aug-T | Aug -S | Aug -SW | Nov-M | Nov-T | Nov-S | Nov-SW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Seq. | 6,359 | 14,774 | 21,368 | 10,330 | 7,042 | 8,279 | 8,957 | 8,463 | 10,823 | 9,896 | 9,629 | 12,235 | 13,733 | 15,418 | 7,046 |
| Good's coverage | 0.81 | 0.92 | 0.96 | 0.99 | 0.93 | 0.88 | 0.97 | 0.95 | 0.98 | 0.99 | 0.97 | 0.89 | 0.93 | 0.97 | 0.95 |
| OTUs | 1823 | 2265 | 1226 | 114 | 792 | 1571 | 547 | 617 | 353 | 197 | 570 | 2002 | 1397 | 875 | 569 |
| invsimpson | 33.18 | 64.55 | 13.63 | 1.16 | 22.28 | 14.16 | 9.85 | 5.20 | 2.89 | 3.48 | 5.46 | 52.41 | 11.75 | 7.21 | 8.71 |
| Chao | 4781.26 | 4066.54 | 3551.50 | 321.77 | 1786.17 | 3699.46 | 1204.94 | 1682.68 | 842.73 | 682.65 | 1340.80 | 4907.46 | 3558.44 | 1542.52 | 1538.85 |
| ace | 8672.32 | 5526.81 | 5652.58 | 539.77 | 2936.58 | 5912.46 | 1738.26 | 3295.64 | 1568.87 | 1020.61 | 1982.04 | 9312.61 | 6023.57 | 2053.99 | 3095.73 |
| Shannon | 5.79 | 5.70 | 3.85 | 0.44 | 4.24 | 5.00 | 3.54 | 2.99 | 1.93 | 1.92 | 3.04 | 5.32 | 4.04 | 3.29 | 3.47 |
The highest number of OTUs was found in the P. lutea skeleton collected in February and the lowest in mucus collected in May (Table 1). The Shannon indices were similar in mucus and skeleton (5.79 and 5.70) collected in February, as well as in three parts of corals (mucus 5.32, tissue 4.04 and skeleton 3.29) collected in November. In contrast to this relatively high diversity, the Shannon indices in corals collected in August were 2.99 in mucus, 1.93 in tissue and 1.92 in skeleton. The Shannon indices were 4.24 and 5.00 in tissue and skeleton respectively collected in May; however it was only 0.44 in mucus, which also possessed the lowest OTUs number. The value of the Shannon index in sea water ranged from 3.04 to 3.85.
Bacterial community compostition
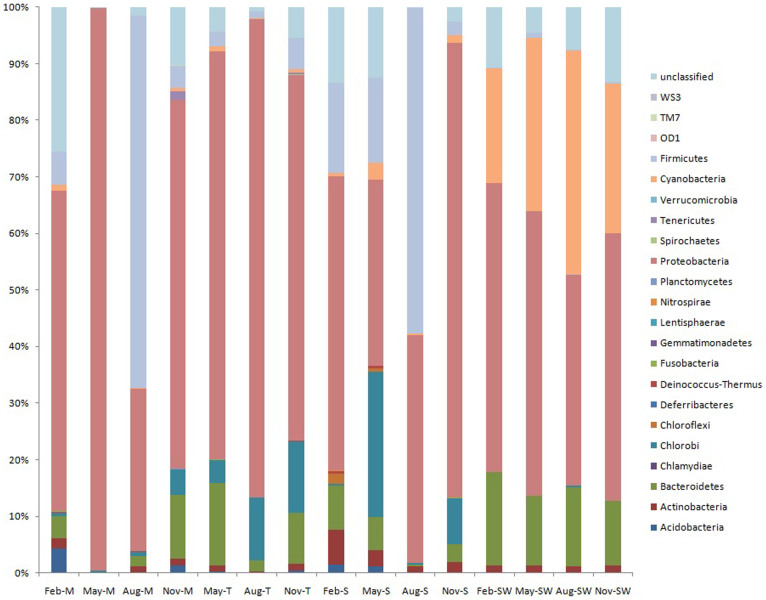
Eighteen formally described phyla and three candidate phyla were recovered from the coral-associated bacterial communities, while 12 formally described phyla and 1 candidate phylum were recovered from the seawater bacterial communities (Fig. 1).
Figure 1. Bacterial composition profiles.
Taxonomic classification of bacterial reads into phylum using the classify.seqs command in Mothur program. The RDP reference files and a bootstrap confidence level of 80% were applied for classification. M represents mucus; T represents tissue; S represents skeleton; SW represents seawater.
Proteobacteria dominated pyrosequencing libraries from both corals (28.7–99.4%) and seawater samples (37.2–51.1%). Alphaproteobacteria were ubiquitous and dominant in corals and seawater collected in different months except in coral mucus collected in May (Fig. S2 and S3). Most coral bacterial communities contained more abundant Betaproteobacteria than seawater bacterial communities, except mucus collected in May and skeleton collected in August (Fig. S2 and S3). Gammaproteobacteria were predominant in coral libraries, especially in coral mucus collected in May, in which most of the reads (8075 reads, account 78.2% of reads in the mucus library) were affiliated with the genus Vibrio (Table S1). Pseudomonadaceae contributed to the large amount of Gammaproteobacteria in tissues collected in August. In addition, a much larger number of Gammaproteobacteria was observed in seawater in May compared with other months. The spike of Gammaproteobacteria (13.6%) resulted from an increasing presence of Alteromonas in seawater in May (Table S2). A higher number of unclassified proteobacteria were found in the seawater in comparison to corals (Fig. S2).
Firmicutes were more predominant in coral mucus (65.7%) and skeleton (57.5%) collected in August compared to other months (<15.9%); they were rare (0.1–0.8%) in the seawater libraries. Actinobacteria group was ubiquitous, dominant and relatively stable in corals and seawater, except in coral mucus in May (Fig. 1 and Fig. S2). More sequences affiliated with Bacteroidetes were detected in the corals collected in February, May and November (3.1–14.6%) and in seawater libraries (11.3–16.5%) in contrast to the August coral libraries (0.3–2.0%). Chlorobi, primarily Prosthecochloris, were abundant (4.0–25.6%) in the coral samples collected in May, August and November, but they were rare in February and nearly absent in seawater (Fig. 1 and S3, Table S1). Cyanobacteria were dominant in all four seawater libraries and were rare in the coral libraries (Fig. 1).
Comparison of coral-associated bacterial communities
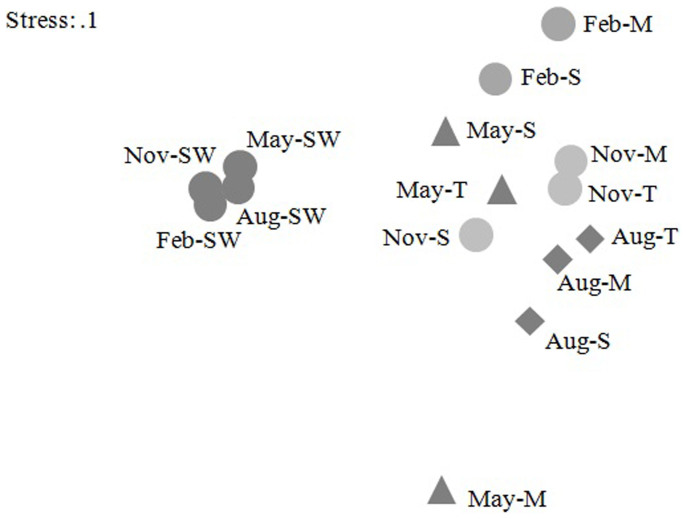
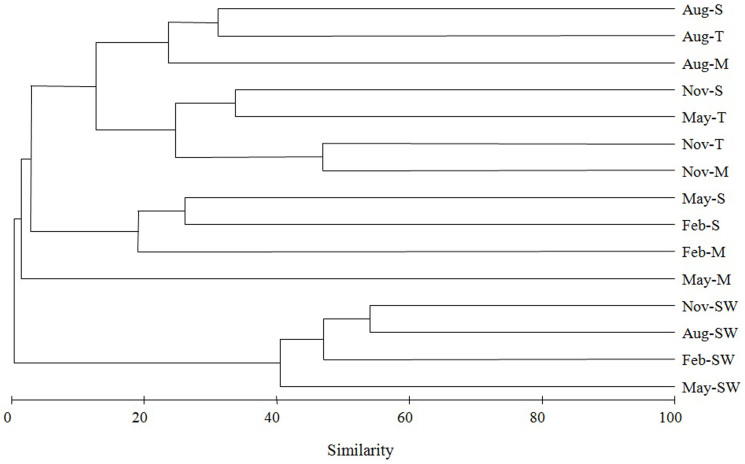
Based on sequence similarity, a total of 10,118 OTUs were classified by running cluster command with the average neighbor algorithm in Mothur. The nMDS matrix was generated from the OTU percentages in each sample and was computed to compare the similarity of the bacterial communities among different samples (Fig. 2). nMDS ordination revealed distinct microbial communities associated with corals and in seawater (Fig. 2). One-way analysis showed that the R value was 0.78 and P value was 0.002. The bacterial communities associated with coral compartments from the same month generally clustered together (Fig. 3) and this cluster was well separated from each other (two-way analysis R = 0.391, P = 0.009), except the mucus, tissue and skeleton samples collected in May.
Figure 2. Non-metric multidimensional scaling (nMDS) plot of bacterial community structure from each sample.
In this map, each dot represents the bacterial population of each sample. Letters M, T, S and SW respectively represent mucus, tissue, skeleton and seawater. The bacterial communities associated with corals and in seawater were distinct. Moreover, the bacterial compositions of the coral compartments from the same month tended to clustered together, except the mucus, tissue and skeleton samples collected in May.
Figure 3. Hierarchical cluster analysis of bacterial communities associated with different coral and surrounding seawater samples.
Clustering was based on Bray-Curtis similarity estimated from the OTU matrix by using the complete linkage method. M represents mucus; T represents tissue; S represents skeleton; SW represents seawater.
In detailed compositions, the dominant bacterial groups were diverse in different compartments of P. lutea collected in different months (Fig. S2, S3, S4 and S5). Such as in the mucus layer, Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria and Gammaproteobacteria were dominant (>5%) in February, while the Vibrionaceae was dominant in May (Fig. S5). A shift towards Alphaproteobacteria, Gammaproteobacteria and Bacilli was observed in August mucus (Fig. S2). At the final sampling time (November), more diverse groups including Sphingobacteria, Flavobacteria, Chlorobia, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria were dominant (>5%, Fig. S2). In tissue, Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria were constant major groups in May, August and November, while Flavobacteria and Chlorobia were alternatively presented in May and August and appeared together in November (Fig. S2). In skeleton, Actinobacteria, Sphingobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Bacilli and Clostridia were predominant in February samples. Chlorobia, Alphaproteobacteria, Gammaproteobacteria, Bacilli and Clostridia were dominant groups in May. Alphaproteobacteria, Gammaproteobacteria and Bacilli were predominant in August. Chlorobia, Alphaproteobacteria and Gammaproteobacteria were dominant in November (Fig. S2).
Analysis of the relationships between bacterial community composition and environment
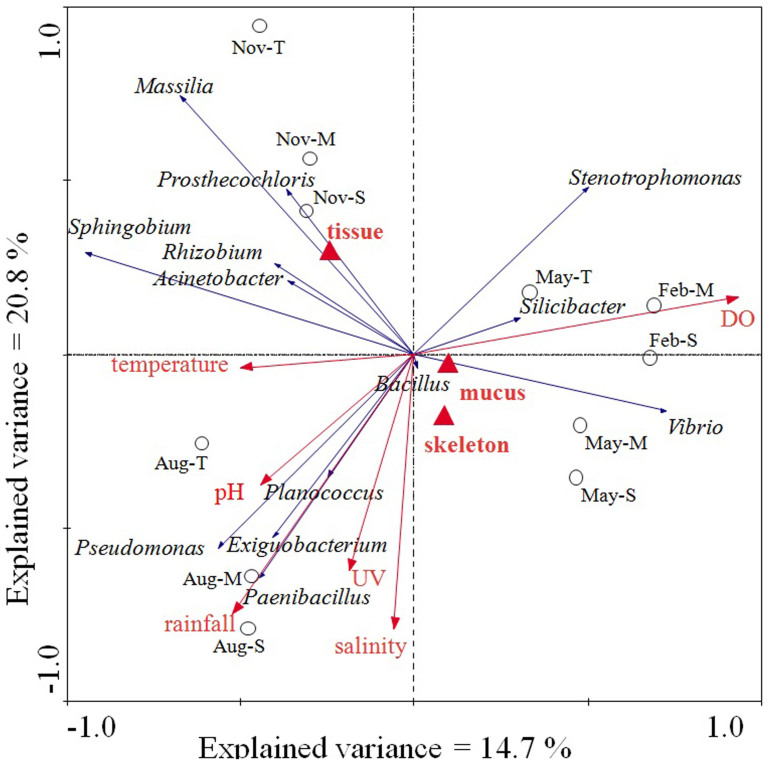
To discern the possible relationship between microbial community composition and environmental parameters, redundancy analysis (RDA) was performed. The results indicated that 60.9% of the total variance in the coral-associated bacterial composition could be explained by the environmental parameters and the coral fraction types, while 20.8% and 14.7% of the variance were explained by the first and second axes, respectively (Fig. 4 and Table S3). Both the first and second axes showed good correlation with environmental data (r = 0.998 and 0.894) (Table S3). The first and second axes clearly distinguished the bacterial assemblages of corals collected in different months (Fig. 4). This result was consistent with the nMDS ordination analysis. Of all the environmental factors analyzed, dissolved oxygen (DO, P = 0.013; 999 Monte Carlo permutations) and rainfall (P = 0.037; 999 Monte Carlo permutations) contributed significantly to the coral bacterium-environment relationship. A triplot illustrating the relationship between major bacterial genera and these environmental parameters showed that Planococcus, Pseudomonas, Exiguobacterium and Paenibacillus were highly positively correlated with rainfall and seawater pH value (Fig. 4). In contrast, Silicibacter, Stenotrophomonas and Vibrio were negatively correlated with rainfall and seawater pH value, but positively correlated with DO (Fig. 4).
Figure 4. RDA ordination triplot showing the relationship among environmental variables, coral samples, and bacterial genus compositions.
Automatic forward selection with Monte Carlo permutation tests was applied to build the parsimonious model, which included temperature, ultraviolet radiation (UV), dissolved oxygen (DO), pH value of seawater (pH), salinity, rainfall and nominal variables including coral compartments mucus, tissue and skeleton, explaining the variance in the bacterial communities. Correlations between environmental variables and the first two RDA axes are represented by the lengths and angles of the arrows (environmental-factor vectors). Only abundant bacteria genera (>5%) were showed in the triplot.
Specific taxa associated with each of the three coral compartments
Using INDVAL, the strict specialists, which tend to be present only in coral mucus, tissue or skeleton, were identified. Table 2 shows the result containing 37 OTUs that seemed to be strict specialists (IndVal>0.6, P<0.05) which associated with P. lutea, representing 0.5% of the total OTU data set. There were 7 strict specialists OTUs in coral mucus, 24 in tissue and 6 in skeleton.
Table 2. Specialists OTUs were identified with INDVAL. For each of the specialists OTUs, we indicate the Indicator Value (IndVal), the statistical significance of the association (p-value), the number of sequences corresponding to the specialist OTU (size) and the lowest taxonomic rank assigned with a confidence greater than 80%.
| OTUs | cluster | IndVal | p value | Size | Phylogenetic affiliation |
|---|---|---|---|---|---|
| Otu00158 | mucus | 0.75 | 0.037 | 82 | Unclassified bacteria |
| Otu00771 | mucus | 0.75 | 0.031 | 10 | Sphingobacteriales |
| Otu00943 | mucus | 0.75 | 0.031 | 8 | Gammaproteobacteria |
| Otu01141 | mucus | 0.75 | 0.036 | 6 | Gammaproteobacteria |
| Otu00519 | mucus | 0.6964 | 0.041 | 17 | Actinobacteria |
| Otu00581 | mucus | 0.675 | 0.042 | 14 | Gammaproteobacteria |
| Otu00693 | mucus | 0.6429 | 0.04 | 11 | Desulfobulbaceae |
| Otu00527 | tissue | 1 | 0.002 | 16 | Muricauda |
| Otu00844 | tissue | 1 | 0.002 | 9 | Hymenobacter |
| Otu00037 | tissue | 0.9108 | 0.005 | 593 | Oxalobacteraceae |
| Otu00008 | tissue | 0.9039 | 0.005 | 5989 | Massilia |
| Otu00345 | tissue | 0.9032 | 0.018 | 27 | Anaplasmataceae |
| Otu00223 | tissue | 0.8929 | 0.021 | 49 | Unclassified bacteria |
| Otu00695 | tissue | 0.8 | 0.014 | 11 | Prosthecochloris |
| Otu00247 | tissue | 0.7391 | 0.015 | 43 | Comamonadaceae |
| Otu00850 | tissue | 0.7273 | 0.025 | 9 | Acinetobacter |
| Otu00267 | tissue | 0.7123 | 0.038 | 37 | Flavobacterium |
| Otu00029 | tissue | 0.6987 | 0.036 | 753 | Chryseobacterium |
| Otu00238 | tissue | 0.6897 | 0.021 | 45 | Unclassified bacteria |
| Otu00637 | tissue | 0.6667 | 0.021 | 13 | Unclassified bacteria |
| Otu01048 | tissue | 0.6667 | 0.03 | 7 | Clostridium_XI |
| Otu01185 | tissue | 0.6667 | 0.026 | 6 | Fusobacterium |
| Otu01488 | tissue | 0.6667 | 0.034 | 5 | Planococcaceae |
| Otu01738 | tissue | 0.6667 | 0.023 | 4 | Unclassified bacteria |
| Otu01751 | tissue | 0.6667 | 0.018 | 4 | Alphaproteobacteria |
| Otu01767 | tissue | 0.6667 | 0.036 | 4 | Unclassified bacteria |
| Otu01782 | tissue | 0.6667 | 0.026 | 4 | Pseudomonas |
| Otu01986 | tissue | 0.6667 | 0.037 | 3 | Acinetobacter |
| Otu02107 | tissue | 0.6667 | 0.02 | 3 | Alphaproteobacteria |
| Otu03434 | tissue | 0.6667 | 0.028 | 2 | Proteobacteria |
| Otu00387 | tissue | 0.625 | 0.036 | 23 | Acinetobacter |
| Otu00099 | skeleton | 0.9191 | 0.03 | 185 | Staphylococcus |
| Otu00290 | skeleton | 0.7071 | 0.025 | 34 | Gammaproteobacteria |
| Otu00360 | skeleton | 0.7 | 0.045 | 25 | Anoxybacillus |
| Otu00258 | skeleton | 0.6522 | 0.042 | 39 | Clostridiales |
| Otu00879 | skeleton | 0.6429 | 0.044 | 9 | Finegoldia |
| Otu00243 | skeleton | 0.6188 | 0.047 | 44 | Friedmanniella |
Discussion
Seasonal influences on the P. lutea–associated bacterial community
Seasonal factors influencing coral-associated bacteria communities have been reported for several coral species distributed throughout different regions9,10,18. This temporal variation was also present within the bacterial communities associated with other invertebrate19, but not in sponges20. In this study, we suggest that bacterial populations in the coral P. lutea on Luhuitou fringing reef showed variation with seasons (Fig. 2 and 3).
Similar to previous observations on the coral Acropora in the Great Barrier Reef4 and Isopora palifera at Ken-Ting9, Alphaproteobacteria and Gammaproteobacteria were the two constant predominant groups in all three compartments of P. lutea. However, when analyzed based on lower taxonomic levels, the compositions of these two constant major groups in three coral compartments in different time were distinct (Table S1 and Fig. S3, S4 and S5). Such as Rhodobacteraceae was the most abundant alphaproteobacterial group in both of mucus and skeleton in February and in coral tissue and skeleton collected in May, while Sphingobium was most abundant in all of mucus, tissue and skeleton in August and November. In addition, Rhizobium and unclassified Rhiziobales were the following abundant groups in the coral skeleton collected in November. Mutualistic benefits of coral-associated bacterial communities have been suggested3,21,22,23, it makes sense that different bacterial combinations fit the requirements of their host exposed to dynamic environmental factors.
Consistent with previous observations, distinct partitioning has been observed in the composition of bacterial communities inhabiting the coral and overlying seawater9,17,24. Furthermore, the bacterial communities were much more stable in surrounding seawater in contrast to bacteria associated with corals on Luhuitou fringing reef. Flavobacteria, Alphaproteobacteria and Cyanobacteria were dominant in seawater in all four sampling months. Even at the lower taxonomic level, the major groups, such as Flavobacteriaceae, Ruegeria, unclassified Rhodobacteraceae and GpIIa, showed stability (Table S2). nMDS and ANOSIM analysis further determined the similar composition of bacterial communities in seawater at different times. The result that coral-associated bacteria community was more susceptible compared with water column-associated bacteria suggested that the relationship between corals and their associated microbes might more directly linked to coral holobiont health. However, recent reports showed that the mucus community from Brazilian coral species was more stable and resistant to seasonal variations compared to the water and sediment communities7. Whether the contradictions are due to environmental conditions in different coral reef ecosystems, or due to differing coral species, might be resolved when we investigate a larger number of coral species distributed in various geographic positions.
Bacterial communities correlated to the P. lutea compartments
Investigation of the spatial organization of bacterial communities within the coral holobiont is crucial for understanding the relationship between coral and bacterial assemblages. We investigated the spatial structures of bacterial communities in different microhabitats of coral including the mucus layer, tissue and skeleton at every three months throughout a year. It should be noticed that the composition of bacteria showed dramatically low similarities (< 30%) among different compartments of coral sampled at the same time (Fig. 3). This is similar to the results presented by Sweet and colleagues13 that bacterial communities within separated coral compartments were significantly different, with average similarity between each other ranging from 24%–46%.
In this study, the bacterial communities of all three coral fractions, mucus, tissue and skeleton, tended to cluster together according to the sampling time (Fig. 2 and 3). In the previously published research, the bacterial communities in all Acropora eurystoma fractions were influenced by the pH and coral fractions were distributed within the clusters divided according to the pH level to which the coral was exposed25. These results suggested that the bacterial assemblages in the microhabitats of corals including the surface mucus layer, tissue and the skeletal matrix, may synchronously respond to environmental shift.
Specialists associated with coral compartments
INDVAL analysis showed that several species (OTUs) were associated with a specific compartment. Bacteria inhabiting a specific compartment may reflect the properties of contemporaneous microhabits provided by the coral as well as the coral physiological and adaptive requirements. In this study, we noticed that OTUs belonging to Prosthecochloris and Clostridium strongly associated with coral tissue, both of which were previously reported potential nitrogen fixers26,27. It seems that these potential nitrogen fixers specifically live in the tissue of P. lutea. Several bacterial groups such as Acinetobacter28 and Pseudomonadaceae29, considered to be related to coral bleaching and/or disease, were representative specialists in P. lutea tissue. In addition, Vibrio species have been previously reported as an implication for bleaching of some coral species30,31,32,33, the coral samples collected in May, in which mucus bacterial community was dominated by Vibrio, did not show any visible signs of disease or bleaching. These bacteria may form a natural part of the healthy coral microbiota34, though when the balance of bacterial consortium is disrupted, they may become enriched and switch on the virulence factors35,36,37. Whether these small specialist assemblages play role in restoring the bacterial community balance as a functional conserved community38,39 remains to be investigated.
Relationship of environmental factors and the P. lutea bacterial community
Six environmental factors (Table S4) and three separate coral compartments were included in the analysis with the bacterial groups. Dissolved oxygen (DO) and rainfall appeared as the most influential environmental parameters out of all those measured that significantly contributed to the variation in bacterial community of P. lutea on Luhuitou fringing reef. Chen et al9 suggested that the rainfall was a factor with greatest effect on the bacterial community of I. palifera at Ken-Ting, and the changes in the pattern of Bacilli were associated with rainfall. The significant influence of rainfall observed in this study further supports the previous viewpoint. In our study, the Bacilli were the most abundant group in mucus and skeleton during August; the highest rainfall occurred from June to August (Fig. S6). The predominant Bacilli, Exiguobacterium and Planococcus in mucus and Paenibacillus in the skeleton had a high positive correlation to rainfall in RDA analysis (Fig. 4). This supported the speculation that these bacteria might be derived from terrestrial soil and might occur transiently in coral8,9. Moreover, Exiguobacterium and Paenibacillus are facultative anaerobes, and the oxygen depleted microhabitats within the coral holobiont may favor their existence.
In this study, 60.9% of variance in P. lutea–associated bacterial community composition could be explained both by the measured environmental parameters and the three coral compartments. It implies there are other undetected drivers of the shift of bacterial assemblages. We noticed that Alteromonas was dominant in seawater during May, but it was rarely observed in the other three months. At this sampling time, Vibrio was the absolute dominant group in coral mucus (Table S2). This synchronous phenomenon might reflect the potential correlations between certain bacterial groups in water column and corals. Comprehensively investigating the ecological network between the coral associated microbiota and water column-derived microbes may shed light on this hypothesis. Differential bacterial communities detected from separate coral compartments have been seen in this study as well as previous studies3,13,34, reflecting the significant influence of coral microhabitats. These findings highlight the internal drivers of the structure of coral associated bacterial communities3,13,34. Therefore, including the physiochemical properties of coral microhabitats into the environmental parameters might better explain the variance in the coral-associated bacterial communities.
In conclusion, our results suggest that bacterial populations in all three compartments of the coral P. lutea showed seasonal variations. Dissolved oxygen and rainfall were the most influential environmental factors, out of all those measured, that significantly contributed to the variation in bacterial community of P. lutea on Luhuitou fringing reef. Results of analysis of relationship between bacterial assemblages and the environment showed that the P. lutea–associated bacterial community composition variance could not be completely explained by both the environmental parameters measured and the coral structure compartments, which suggested that differences in microbial populations could be a result of factors other than those included in our analysis. Such factors could include the interactions with water column bacteria communities and the properties of physiochemical environments of coral microhabitats. The probability of bacterial adaptation to a specific compartment was extremely small; however, the study of the ecological role of the small number of specialists is highly desirable. In order to illuminate the relationship between coral and the associated microbes and further understand the roles coral-associated microbes have in maintaining coral holobiont health, investigation of coral-associated microbial consortium at different time scales, their restoring capability and mechanism, and measurement of the microenvironmental conditions of coral compartments is definitely needed in future studies.
Methods
Sample collection
The Luhuitou fringing reef located in Sanya, southern Hainan Island, is approximately 3500 m long and 250–500 m wide and consists of approximately 70% of the coral species so far reported for Hainan Island and its surrounding islands40. Three healthy P. lutea coral colonies and seawater samples were collected in February, May, August and November in 2012 at a depth of 3–5 m from the Luhuitou fringing reef (109°28′E, 18°13′N). Coral fragments (approximately 10 × 10 cm) were collected from the side of the colonies using a punch core and hammer.
Coral samples were thoroughly washed with sterile sea water and then soaked in 300 ml PBS buffer for 30 min. The PBS solution, which contained mucus dispersed in it was then centrifuged for 10 min at 15000 g at 4°C, discarded the supernatant and stored the pellets at −80°C for later analysis. The coral fragments were then washed several times with sterile seawater and the tissue was then sprayed from the coral by using a sprayer with TE buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA (pH 8.0)]. The skeleton was thoroughly cleaned by airbrush for removing the tissue residues, followed by several washes with sterile seawater, only then the skeleton was kept. Overlying seawater (1 L) adjacent to the coral colonies was also collected, filtered through 0.22 μm filters (Millipore). Coral fractions and filters were stored at −80°C until DNA extraction.
DNA extraction, PCR amplification and pyrosequencing
The coral tissue and skeleton samples were homogenized separately in liquid nitrogen with a mortar and pestle. The powdered samples were added to the PowerBead Tubes provided in the PowerSoil DNA Isolation Kit (MoBio, Solana Beach, CA, USA). The 0.22 μm polycarbonate filter membranes with adsorbed microbial cells from seawater were cut into pieces before added to the PowerBead Tubes. The mucus pellet samples mentioned above were transferred to the PowerBead Tubes as well. Total DNA was extracted using the PowerSoil DNA Isolation Kit according to the manufacturer's instructions. Bacterial V1-V2 hypervariable regions of the 16S ribosomal RNA gene were amplified using the bacterial forward primer 27F41, which includes the primer A adaptor and a unique 10 bp barcode on the 5′ end (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNAGAGTTTGATCCTGGCTCA-3′), and the reverse primer 357R42 with the primer B adaptor on the 5′ end (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG CTGCTGCCTYCCGTA-3′). PCR amplifications were performed in a Mastercycler pro (Eppendorf, Hamburg, Germany) in a final volume of 50 μl, containing 4 μl of 2.5 mM deoxynucleotide triphosphate mixture (TaKaRa), 2 μl of 10 μM each primer, 5 μl (10–20 ng) template DNA and 2.5 units Ex Taq DNA polymerase (TaKaRa, with its recommended reaction buffer). The PCR conditions were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 60°C −0.5°C/cycle for 30 s, 72°C for 30 s; followed by 72°C for 10 min. Each genomic DNA sample was amplified in triplicate PCR reactions. The same multiplex identifier used for the same compartment (i.e. mucus, tissue or skeleton) samples from three individual colonies collected in the same month. Amplicons with the same barcode were pooled and purified using the E.Z.N.A.® Gel Extraction Kit (Omega Bio-Tek). The quality of the purified PCR products was assessed using a Nanodrop spectrophotometer (Thermo Scientific, Vantaa, Finland). Pooled 200 ng of the purified tagged amplicons from each sample with multiplex identifier were pyrosequenced on the Roche 454 Genome Sequencer FLX System.
Diversity indices and taxonomic identification of pyrosequencing reads
The pyrosequencing data were deposited in the NCBI Sequence Read Archive (SRA) database under the accession number SRR1263017. The samples were demultiplexed. Reads that did not meet the following criteria were removed: homopolymers less than 6 bp, application of a quality window of 50 bp with an average flowgram score > 20, sequence length > 150 bp and no ambiguous bases in the sequence. Chimera detection and removal were performed on the remaining reads by running chimera.uchime packaged in Mothur v.1.30.243. Qualified reads were identified using the classify.seqs command using the RDP reference files with a bootstrap confidence level of 80%, and those classify as mitochondria or chloroplast were considered contaminants and removed from further analysis. Sequences were clustered into operational taxonomic units (OTUs) with a 97% threshold using the cluster command. The taxonomy information for each of our OTUs were analysed using the classify.otu command together with the taxonomy file generated from the sequences classify step. Species richness and diversity estimates were performed using Mothur43. To standardize all datasets, the smallest number 6,359 of sequences was randomly selected from each sample 1,000 times.
Comparison of microbial communities and their relationship with the environment
The relationships among bacterial assemblages of coral and seawater samples were analyzed by non-Metric Multidimensional scaling (nMDS) ordination44. The Bray-Curtis distance matrix was estimated from the OTU matrix, and then, the nMDS profile was generated by the PRIMER 5 software (PRIMER-E, Lutton, Ivybridge, UK). Differences in bacterial communities between categories were tested with an analysis of similarities (ANOSIM), with 10,000 replicates44.
The correlations between bacterial community composition and environmental variables were analyzed by ordination methods using the software package CANOCO 4.5 for Windows45. The numbers of reads assigned to different genera in each library were converted into percentages used as the species input. Environmental data included temperature, ultraviolet radiation (UV), dissolved oxygen, pH, salinity and rainfall (Table S4) were standardized and in combination with nominal variables including coral compartments mucus, tissue and skeleton served as the environmental input. Detrended correspondence analysis (DCA) with detrending by segments was first applied to determine the lengths of gradient of axes. Exploratory DCA analyses of the species data set indicated that the gradient length of the first axis in standard deviation units below 3 units. Therefore, the redundancy analysis (RDA) was applied to determine the relationship of environmental parameters and bacterial community compositions. The significance of the relation between explanatory variables and community composition was tested using Monte Carlo permutation tests (999 unrestricted permutations, P < 0.05).
Identification of strict habitat specialists
The INDVAL46 (INDicator VALues) analysis was performed for detecting the indicator species individually in P. lutea mucus, tissue and skeleton based on OTU fidelity and relative abundance. Good indicator species can be considered as strict habitat specialist, which should be found mostly in samples from a single compartment47. The smallest number 6,359 of sequences was randomly subsampled and the OTU6359 table was generated. Analyses were run using the data set OTU6359 (7395 OTUs) and the package labdsv (http://ecology.msu.montana.edu/labdsv/R/) within the R environment. Only OTUs with significant (p value<0.05) INDVAL values that were>0.5 were considered as significant associations with coral mucus, tissue and skeleton46.
Author Contributions
J.L., S.Z. and L-J.L. conceived the works. J.L. and Q.C. performed the experiments. J-D.D. and J.Y. contributed the materials. J.L. wrote the manuscript. All authors reviewed the final manuscript.
Supplementary Material
Supplementary information
Acknowledgments
Tropical Marine Biological Research Station in Hainan is gratefully acknowledged for collecting coral samples. Authors gratefully acknowledge the use of the HPCC at the South China Sea Institute of Oceanology, Chinese Academy of Sciences. We thank Prof. Farooq Azam for helpful discussions. Thank you to Mr. Ryan Guillemette for insightful edits. This research was supported by National Natural Science Foundation of China (No. 41106139, 41230962) and Pearl River Nova Program of Guangzhou (No. 2014J2200075), Administration of Ocean and Fisheries of Guangdong Province (No. GD2012-D01-002) and the Knowledge Innovation Program of the Chinese Academy of Sciences (No. SQ201301).
References
- Rosenberg E., Koren O., Reshef L., Efrony R. & Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- Bourne D. G. & Webster N. S. [Coral reef bacterial communities.] [Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E. & Thompson, F. (eds.)] [163–187] (Springer-Verlag, Heidelberg, 2013). [Google Scholar]
- Rohwer F., Seguritan V., Azam F. & Knowlton N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10 (2002). [Google Scholar]
- Littman R. A., Willis B. L., Pfeffer C. & Bourne D. G. Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163 (2009). [DOI] [PubMed] [Google Scholar]
- Lee O. O. et al. Spatial and Species Variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Appl. Environ. Microbiol. 78, 7173–7184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K. M., Moss A. G., Chadwick N. E. & Liles M. R. Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos C., Torres T. T. & Ottoboni L. M. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci. Rep. 3, 1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M. J., Yu Y. T., Chen C. A., Chiang P. W. & Tang S. L. Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 75, 7797–7806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. P., Tseng C. H., Chen C. A. & Tang S. L. The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5, 728–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceh J., van Keulen M. & Bourne D. G. Coral-associated bacterial communities on Ningaloo Reef, Western Australia. FEMS Microbiol. Ecol. 75, 134–144 (2011). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lanetty M., Granados-Cifuentes C., Barberan A., Bellantuono A. J. & Bastidas C. Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol. Ecol. 22, 4349–4362 (2013). [DOI] [PubMed] [Google Scholar]
- Daniels C. A. et al. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Prog. Ser. 426, 29–40 (2011). [Google Scholar]
- Sweet M. J., Croquer A. & Bythell J. C. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52 (2011). [Google Scholar]
- Reshef L., Koren O., Loya Y., Zilber-Rosenberg I. & Rosenberg E. The Coral Probiotic Hypothesis. Environ. Microbiol. 8, 2068–2073 (2006). [DOI] [PubMed] [Google Scholar]
- Garren M., Raymundo L., Guest J., Harvell C. D. & Azam F. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS ONE 4, e7319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O. & Rosenberg E. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. App. Env. Microbiol. 72, 5254–5259 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Highly heterogeneous bacterial communities associated with the South China Sea reef corals Porites lutea, Galaxea fascicularis and Acropora millepora. PLoS ONE 8, e71301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes N. E. et al. The Montastraea faveolata microbiome: ecological and temporal influences on a Caribbean reef-building coral in decline. Environ. Microbiol. 15, 2082–2094 (2013). [DOI] [PubMed] [Google Scholar]
- Daniels C. & Breitbart M. Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovata. FEMS Microbiol. Ecol. 82, 90–101 (2012). [DOI] [PubMed] [Google Scholar]
- Erwin P. M., Pita L., López-Legentil S. & Turon X. Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M. P., Mazel C. H., Gorbunov M. Y. & Falkowski P. G. Discovery of symbiotic nitrogen-fixing Cyanobacteria in corals. Science 305, 997–1000 (2004). [DOI] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H. & Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719 (2007). [DOI] [PubMed] [Google Scholar]
- Kimes N. E., Van Nostrand J. D., Weil E., Zhou J. Z. & Morris P. J. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12, 541–556 (2010). [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J., Zerkle A. L., Bonheyo G. T. & Fouke B. W. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68, 2214–2228 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D. et al. The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Toth J. & Kasap M. Nitrogen-fixation genes and nitrogenase activity in Clostridium acetobutylicum and Clostridium beijerinckii. J. Ind. Microbiol. Biotechnol. 27, 281–286 (2001). [DOI] [PubMed] [Google Scholar]
- Overmann J. [The family Chlorobiaceae.] [Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K-H. & Stackebrandt, E. (eds.)] [359–378] (Springer-Verlag, Heidelberg, 2006). [Google Scholar]
- Koren O. & Rosenberg E. Bacteria associated with the bleached and cave coral Oculina patagonica. Microb. Ecol. 55, 523–529 (2008). [DOI] [PubMed] [Google Scholar]
- Roder C. et al. Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J. 8, 31–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A., Loya Y., Fine M. & Rosenberg E. Bacterial infection and coral bleaching. Nature 380, 396 (1996). [Google Scholar]
- Kushmaro A., Rosenberg E., Fine M. & Loya Y. Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Ecol. Prog. Ser. 147, 159–165 (1997). [Google Scholar]
- Ben-Haim Y. & Rosenberg E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar. Biol. 141, 47–55 (2002). [Google Scholar]
- Ben-Haim Y., Zicherman-Keren M. & Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G. & Munn C. B. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7, 1162–1174 (2005). [DOI] [PubMed] [Google Scholar]
- Mao-Jones J., Ritchie K. B., Jones L. E. & Ellner S. P. How microbial community composition regulates coral disease development. PLoS Biol. 30, e1000345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman R., Willis B. L. & Bourne D. G. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660 (2011). [DOI] [PubMed] [Google Scholar]
- Kims et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14 (2006). [Google Scholar]
- Rypien K. L., Ward J. W. & Azam F. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12, 28–39 (2010). [DOI] [PubMed] [Google Scholar]
- Liu Z. F. et al. [Sedimentology.] [Wang, P. X. & Li, Q. Y. (eds.)] [229–236] (Springer Science+Bussiness Media B. V., Dordrecht, Heidelberg, London, New York, 2009). [Google Scholar]
- Hongoh Y., Ohkuma M. & Kudo T. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isopteran; Rhinotermitidae). FEMS Microbiol. Ecol. 44, 231–242 (2003). [DOI] [PubMed] [Google Scholar]
- Liu Z., Lozupone C., Hamady M., Bushman F. D. & Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35, e120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143 (1993). [Google Scholar]
- ter Braak C. J. F. & Šmilauer P. CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5). (Microcomputer Power, Ithaca, NY, 2002). [Google Scholar]
- Dufrêne M. & Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366 (1997). [Google Scholar]
- Legendre P. & Legendre L. [Numerical ecology. 2nd English edition.] (Elsevier, Amsterdam, 1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information