Abstract
MSH4 encodes a MutS protein that plays a specialized role in meiosis. In eukaryotic species, such as budding yeast, mice, Caenorhabditis elegans, and Arabidopsis, msh4 mutants display meiotic defects with a reduced number of chiasmata. Here, we characterized rice MSH4 by map-based cloning. In Osmsh4 mutants, the chiasma frequency was dramatically decreased to ∼10% of the wild type, but the synaptonemal complex was normally installed. The double mutant analysis showed that in the Osmsh4 Osmsh5 mutant, the reduction of chiasmata was greater than other zmm mutants. This was consistent with the absence of localization for OsZIP4 and OsMER3 in Osmsh4 and suggests an earlier role for OsMSH4 and OsMSH5 than other ZMM proteins where they may be required to stabilize progenitor Holliday junctions. Using yeast two-hybrid and pull-down assays, we verified the direct physical association between OsMSH4 and OsMSH5 and OsMSH5 and HEI10 in plants for the first time. The MSH4–MSH5 heterodimer has been demonstrated in mammals to stabilize the formation of progenitor and double Holliday junctions that may be resolved as crossovers (COs). We propose that OsMSH4 interacts with OsMSH5 to promote formation of the majority of COs in rice.
Keywords: OsMSH4, crossover, meiosis, rice
MEIOSIS is a special form of cell division that generates haploid gametes for sexual propagation. Meiocytes undergo an intricate and elaborate process: one round of chromosome replication is followed by two rounds of cell division, the unique meiosis I and the mitosis-like meiosis II, generating four haploid gametes. The first division segregates homologous chromosomes, whereas the second one splits sister chromosomes, thereby halving the chromosome content from diploid to haploid. The first division has been recognized as the crucial stage of meiosis, in which prophase I is particularly important. Prophase I can be divided into five dissimilar but inseparable stages according to their different chromosome structures and behaviors: leptotene, zygotene, pachytene, diplotene, and diakinesis, during which homologous chromosomes pair, synapse, and recombine (Zickler and Kleckner 1999; Li and Ma 2006).
Pairing, synapsis, and recombination are well linked in a carefully coordinated mechanism. Homologs search out and pair with each other, facilitating the initiation of recombination. Meanwhile, early stages of recombination are also used to promote global pairing and synapsis of homologs. Mature synaptonemal complexes (SCs) form as the homologous chromosomes approach each other at a distance of 100 nm and are disassembled during diplotene (Sym and Roeder 1994). After the disassembly of SCs, homolog pairs are held together only by crossovers (COs) to ensure their alignment on the metaphase I plate. COs also create physical connections between homologous chromosomes to guarantee their normal segregation to the opposite poles. In consequence, the absence of COs causes random segregation of homologs at anaphase I, leading to the formation of aneuploid dyads and a reduction of bivalents, followed by the segregation of sister chromosomes in anaphase II to produce aneuploid tetrads at the end of meiosis (Ross-Macdonald and Roeder 1994; de Vries et al. 1999; Lipkin et al. 2002; Chen et al. 2005). COs are believed to correspond to homologous recombination sites, visualized as chiasmata under the microscope.
Homologous recombination begins with DNA double-strand breaks (DSBs) catalyzed by SPO11 protein (Keeney et al. 1997). DSBs are further processed by MRX complex (Mre11–Rad50–Xrs2) and two DNA strand-exchange proteins DMC1/RAD51 to initiate the strand exchange and produce double Holliday junctions (dHjs) (Baumann et al. 1996; Li et al. 1997; Schwacha and Kleckner 1997; Puizina et al. 2004; Osman et al. 2011). Reciprocal exchanges between homologous chromosomes occur to generate COs or noncrossovers (NCOs) (Allers and Lichten 2001; Borner et al. 2004; Lynn et al. 2007).
The ZMM proteins (Zip1–4, Mer3, Msh4, Msh5, and Spo16), initially characterized in Saccharomyces cerevisiae and found conserved in almost all eukaryotes, have been shown to play critical roles in processing of a subset of recombination interactions at sites destined to become COs (Borner et al. 2004; Lynn et al. 2007; Youds and Boulton 2011). Accordingly, elimination of the majority of COs was detected in zmm mutants of many species, including budding yeast (Ross-Macdonald and Roeder 1994; Tsubouchi et al. 2006), mammals (Mercier et al. 2001; Nakagawa et al. 2001; Novak et al. 2001; Neyton et al. 2004), and plants (Higgins et al. 2004; Wang et al. 2009; Shen et al. 2012).
MutS family proteins together with MutH and MutL function in repair of DNA mismatches in bacteria. In eukaryotic species, there are six MutS proteins (Msh1–6). Among them, Msh2, Msh3, and Msh6 have been shown to take part in DNA mismatch repair (MMR) (Borts et al. 2000; Kolas and Cohen 2004), whereas only Msh4 and Msh5 are revealed to specifically get involved in the meiotic recombination process but not in MMR (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Zalevsky et al. 1999; Kneitz et al. 2000; Novak et al. 2001; Higgins et al. 2004; Hoffmann and Borts 2004; Kolas and Cohen 2004; Neyton et al. 2004; Her et al. 2007). In S. cerevisiae, mutations in Msh4 and Msh5 lead to the disruption of CO formation and the nondisjunction of homologous chromosomes (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). In Caenorhabditis elegans, MSH4 plays a decisive role in CO formation, as research has shown that the mutation in him-14, the ortholog of budding yeast Msh4, completely eliminates the COs (Zalevsky et al. 1999). In mouse strains lacking MSH4 or MSH5, COs are entirely eliminated and homologous chromosomes fail to align properly, resulting in both male and female sterility (de Vries et al. 1999; Edelmann et al. 1999; Kneitz et al. 2000). In Arabidopsis, chiasma frequency is greatly reduced in Atmsh4 and Atmsh5 in comparison to the wild type (Higgins et al. 2004, 2008). MSH4 and MSH5 can form a heterodimer (Bocher et al. 1999; Snowden et al. 2004), which binds to single Holliday junctions or three-armed progenitor Holliday junctions as a sliding clamp embracing the homologous chromosomes and promoting the formation of COs (Borner et al. 2004; Snowden et al. 2004).
Rice is a popular plant model for molecular biological studies and one of the world’s most important food crops. With the complete sequencing of the rice genome, there have been great achievements in functional genomic studies. Using genetic analysis combined with cytology, many genes related to rice meiosis have been discovered and their functions further characterized (Luo et al. 2014). In this study, we identified the rice homolog of MSH4 by map-based cloning, investigated its function in CO formation, and demonstrated its relationship with other ZMM proteins. Our genetic and cytological data suggested that the loss of OsMSH4 function resulted in a more severe diminution in CO formation than other zmm mutants. We also validated that OsMSH4 in combination with OsMSH5 might promote formation of the majority of crossover. In addition, we demonstrated an interaction between OsMSH5 and HEI10, providing further evidence for the intimate relationships among ZMM proteins.
Materials and Methods
Plant materials
Osmsh4-1 and Osmsh4-4 were identified from an indica rice variety, Zhongxian 3037, induced by 60Co-γ ray radiation. Osmsh4-2 and Osmsh4-3 were TOS17 insertion lines provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences (https://tos.nias.affrc.go.jp/). The Osmsh5 allele employed in this study is Osmsh5-1 (Luo et al. 2013). Zhongxian 3037 was used as the wild type in the related experiments. Paddy fields were applied to cultivate rice materials required for both gene mapping and functional characterization.
Molecular cloning of OsMSH4
STS markers were designed based on the sequence differences between the indica rice variety 9311 and the japonica rice variety Nipponbare on the basis of data published on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) (Supporting Information, Table S1).
The reverse transcription was performed as previously described (Shen et al. 2012). The BD SMART RACE cDNA Amplification kit (Clontech) was used to perform the following amplification according to the manufacturer’s instructions. Two gene-specific primers, 5RACER1 (5′-CTGATAAATGTTGCGAAG-3′) and 5RACER2 (5′-CTAGTTGAGTCAATGTTCA-3′) were combined with the general adaptor primers supplied by the kit. For 3′-RACE, two rounds of PCR were performed with the same 3′-adaptor primer (5′-CTGATCTAGAGGTACCGGATCC-3′) and two OsMSH4 gene-specific primers, 3RACEF1 (5′-GTTGCTCGTAGATCTTTCTGT-3′) and 3RACEF2 (5′-ACTTCACGGATCACAGAACA-3′). The products of 3′-RACE-PCR and 5′-RACE-PCR were cloned and sequenced.
Construction of RNAi cassette and rice transformation
A 380-bp fragment was amplified and cloned into pMD18T vector (Takara). A single clone, proven by sequencing to be an accurate copy with no mutations, was used to generate a 380-bp fragment and further cloned into the BglII/XhoI and BamHI/SalI sites of the pUCRNAi vector. The resulting stem-loop fragment was then cloned into the binary vector pCAMBIA2300-actin. Finally, the RNAi construct was transformed into Agrobacterium tumefacien strains for further plant transformation.
Yeast two-hybrid assay
The yeast two-hybrid assays were conducted using the Matchmaker Gold Yeast Two-Hybrid system with ZMM proteins, including OsMSH4, OsMSH5, and HEI10. ORFs of the related genes were amplified using the 2× Power Taq PCR Mastermix I (BioTeKe) and cloned into pGADT7 and pGBKT7 to generate AD and BD recombinants. Those plasmids were cotransformed into Y2H Gold strain in an AD–BD-coupled manner.
α-Galactosidase activity was qualitatively monitored using the blue color reporter assay on quadruple dropout (QDO) selection medium with aureobasidin A and the chromagenic substrate X-α-Gal (SD −Leu −Trp −Ade −His +aureobasidin A +X-α-Gal). Positive clones turned blue in the presence of X-α-Gal, while negative clones were even unable to grow. The yeast strains transformed with both vectors were also plated on double dropout (DDO) selecting medium (SD −Leu −Trp) to confirm the successful transformation.
Pull-down assay
OsMSH4 and HEI10 coding sequences were separately cloned into pMAL-c5x (NEB), while the OsMSH5 coding sequence was cloned into pGEX4T-2 (Amersham). All these recombinant vectors and empty vectors were transformed into Escherichia coli BL21 (DE3) individually and 0.2 mM isopropyl β-D-thiogalactoside (IPTG) was added to induce the expression of those proteins. The cell lysate containing 25–50 mg GST-fused OsMSH5 or GST tag were incubated with 200 μl 50% glutathione sepharose 4B beads (GE Healthcare) at 4° for 2 hr under gentle agitation. After that, the coated GST-fused protein beads were washed three times in 1× phosphate-buffer saline (PBS) solution. Then the cell lysates containing 25–50 mg MBP-fused proteins or MBP tag were added into the washed GST-fused protein beads. After 1 hr of incubation at 4° under gentle agitation, the bound protein–bead complexes were sedimented by centrifugation and washed five times in 1× PBS solution. Then, the beads were resuspended in 50 μl SDS-PAGE loading buffer and heated for 10 min in the boiled water. Protein samples were resolved on 10% SDS-PAGE gels for further blotting analysis using an anti-MBP antibody.
Meiotic chromosome preparation
Young panicles from both wild-type and Osmsh4 plants were collected and fixed in Carnoy’s solution (ethanol:glacial acetic = 3:1). Anthers at meiotic stages were quickly squashed in an acetocarmine solution onto glass slides and then frozen in liquid nitrogen. After lifting and removing coverslips, slides were dehydrated using an ethanol gradient (70, 90, and 100%). Chromosome spreads were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in an antifade solution (Vector Laboratories, Burlingame, CA). Images were captured under a Zeiss A2 fluorescence microscope with a micro-charge-coupled device camera.
Immunofluorescence
Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature. Anthers in the meiotic stage were squashed with a dissecting needle in 1× PBS solution and covered with a coverslip. After prying up coverslips, slides were dehydrated using an ethanol gradient (70, 90, and 100%). Slides harboring anthers were incubated in a humid chamber at 37° for ∼4 hr with different combinations of antibodies, all diluted 1:500 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent). After three rounds of washing in 1× PBS, Texas Red-conjugated goat antirabbit antibody and fluorescein isothiocyanate-conjugated sheep antimouse antibody (1:1000) were added to the slides.
Results
Isolation of a sterile mutant
We isolated a mutant from the self-fertilization line of an indica rice variety, Zhongxian 3037, induced by 60Co-γ ray radiation. The mutant plant showed normal vegetative growth and development (Figure S1A), but was almost sterile (Figure S1B). Its pollen grains were empty and shrunken and could not be stained by I2-KI (Figure S1, C and D). Furthermore, we checked the viability of female gametes by pollinating its flowers with the wild-type pollens. There was no seed produced, indicating that the female gametes were also affected by this mutation.
We also verified that the sterile phenotype of Osmsh4-1 was controlled by a single recessive gene, via calculating the segregation ratio of the progeny delivered from a self-fertilized heterozygous plant. There were 73 fertile plants and 25 sterile plants, which is consistent with a 3:1 segregation (χ2 = 0.22, P > 0.05).
Identification of the OsMSH4 gene
To identify the gene responsible for the sterile phenotype, a large F2 mapping population was constructed by crossing the heterozygous plants with a japonica rice variety Zhonghua11. The F2 and F3 sterile segregates, amounting to 732 individual plants in total, were collected for further map-based cloning. The gene was initially mapped to the long arm of chromosome 7 and finally confined to a 110-kb region.
Within this region, a candidate gene (Os07g0486000) was identified and the protein was depicted as a MutS family protein resembling MSH4 in other organisms such as Arabidopsis (75% identity/87% positive), Brachypodium distachyo (91% identity/95% positive), mouse (35% identity/58% positive), and S. cerevisiae (29% identity/50% positive). In addition, this mutant was found to have similar defects with msh4 mutants in other species, including reduced fertility on a macroscopic scale and desynapsis on a microscopic scale, which increased the likelihood that OsMSH4 was the candidate gene. We therefore amplified and sequenced the candidate gene. A point mutation, the transition of G to A, was detected in the seventh exon, creating a new stop codon and causing premature termination of protein translation.
To confirm the mutant phenotypes were caused by the loss of function of OsMSH4, an RNA interference transformation vector was constructed and transformed into the wild-type rice to silence its expression. From the transgenic plants, sterile plants were selected for further analysis. RT-PCR analysis demonstrated that the expression of OsMSH4 in the sterile RNAi plants was markedly reduced compared to the wild type (Figure S3). Moreover, chromosome behavior in the meiocytes of sterile plants resembled that in the Osmsh4 mutants (Figure S4D). We therefore drew the conclusion that the knockdown of OsMSH4 led to the sterile phenotype.
In addition, three more Osmsh4 mutants were identified (Figure S2A). Two of them are Tos17-insertion mutants provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences; the other one was identified using the same mapping strategy with Osmsh4-1 and this allele had a T-to-C mutation in the splicing site of intron 18. As Osmsh4-1 is a deletion line creating a new stop codon, we selected it for further cytological investigation.
Characterization of the OsMSH4 gene
We performed 5′-RACE and 3′-RACE to find the 5′-UTR and 3′-UTR of OsMSH4, respectively. Meanwhile, primers were designed in the 5′-UTR and 3′-UTR to amplify the open reading frame (ORF). The full length of OsMSH4 cDNA is 2853 bp with an ORF of 2397 bp, which gives rise to 798 amino acids. The whole length of this gene is 9995 bp, comprising 24 exons and 23 introns (Figure S2A). OsMSH4 not only displays notable homology with MSH4 in Arabidopsis but also shows a close relationship to MSH4 in mammals and budding yeast (Figure S2B).
The expression patterns of OsMSH4 in different organs were determined by RT-PCR analysis. OsMSH4 transcripts were distributed among all organs; however, the expression in panicles was much higher than in other organs (Figure S3). In addition, the expression of OsMSH4 was shown to be dramatically reduced in both Osmsh4-1 and OsMSH4RNAi plants (Figure S3).
Meiotic defects occur in Osmsh4 mutants
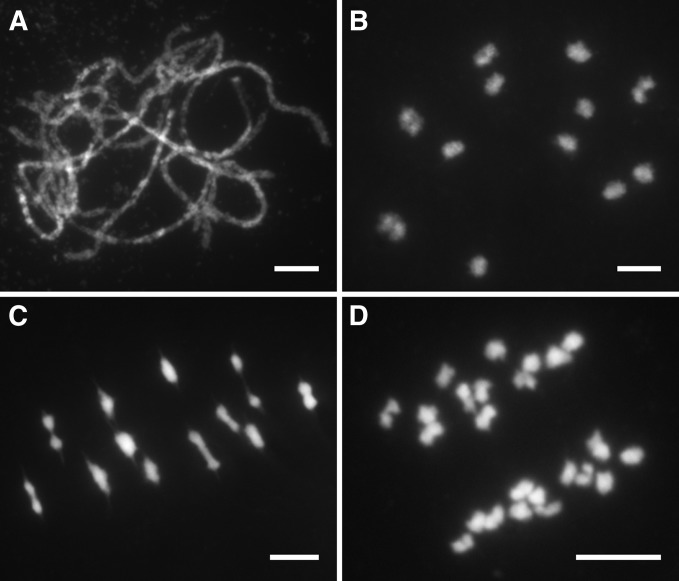
DAPI-stained meiotic chromosomes of pollen mother cells (PMCs) at different stages were investigated in both wild type and Osmsh4-1. In wild type, meiotic chromosomes were first seen as thin and tangled threads at leptotene and started to pair and synapse at zygotene. At pachytene, SCs were well established between homologous chromosomes (Figure 1A). Paired chromosomes were condensed into small dots and 12 rod-like or ring-like bivalents were distinctly visible at diakinesis (Figure 1B). All the bivalents lined up on the equatorial plate at metaphase I (Figure 1C), followed by the separation of homologous chromosomes at anaphase I, which then migrated into two opposite poles (Figure 1D). The second meiotic division resembled mitosis, and during this process, sister chromatids of each chromosome separated from each other, generating four sets of 12 chromosomes in a tetrad at the end of meiosis.
Figure 1.
Meiotic chromosome behaviors of PMCs in the wild type. (A) Pachytene. (B) Diakinesis. (C) Metaphase I. (D) Anaphase I. Chromosomes were stained with DAPI. Bars, 5 μm.
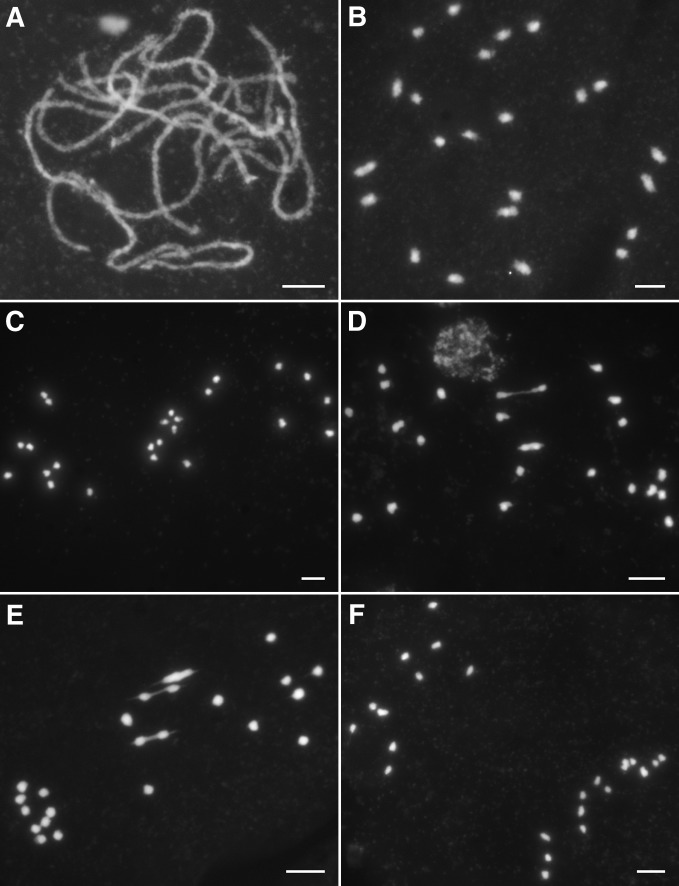
Cytological investigation on the DAPI-stained meiotic chromosomes revealed a defective meiosis in Osmsh4-1 meiocytes. From leptotene to pachytene, all the homologous chromosomes paired and synapsed (Figure 2A), showing the same chromosomes behave as the wild type. However, divergences from the wild type became apparent at diakinesis with the occurrence of many univalents (Figure 2B). At metaphase I, only 1.65 bivalents per PMC on average situated on the equatorial plate (n = 204), while the univalents dispersed within the whole nucleus (Figure 2, C–E). From anaphase I to telophase I, due to the random segregation of the univalents, chromosomes were separated unevenly into two daughter cells (Figure 2F). As a result, microspores with different numbers of chromosomes formed at the end of meiosis II. We also investigated chromosome behaviors in Osmsh4-2, Osmsh4-3, Osmsh4-4, and OsMSH4RNAi plant meiocytes, which were very similar to those in Osmsh4-1 (Figure S4, A–D).
Figure 2.
Meiotic chromosomes in Osmsh4-1 meiocytes. (A) Pachytene. (B) Diakinesis. (C) Metaphase I with no bivalents. (D) Metaphase I with two bivalents. (E) Metaphase I with three bivalents. (F) Anaphase I. Bars, 5 μm.
The chiasma frequency is dramatically reduced in Osmsh4
We quantified the number of chiasmata both in Osmsh4-1 and the wild type by evaluating the shape of bivalents at metaphase I, using the criteria described in a previous study (Sanchez Moran et al. 2001). Among the 45 PMCs surveyed in the wild type, all the meiocytes have 12 bivalents at metaphase I, with a mean of 20.56 ± 1.49 (from 18 to 24) chiasmata per cell (Figure S5A). However, only 1.65 ± 1.03 bivalents with 1.71 ± 1.25 chiasmata per cell were observed among 204 PMCs in Osmsh4-1 (Figure S5B). We also generated a double mutant between Osmsh4 and Osmsh5 and analyzed its chiasma number. The mean chiasma number per cell was 1.76 ± 1.20 (n = 115) in Osmsh4 Osmsh5 (Figure S4F). Compared with 1.71 ± 1.25 in Osmsh4-1 and 2.10 ± 0.91 in Osmsh5 (Luo et al. 2013), there was no significant difference detected (t317 = 0.45, P > 0.5), suggesting OsMSH4 and OsMSH5 probably work in the same pathway (Table 1).
Table 1. Chiasma frequency in different genotypes.
| Genotype | Chiasma/cell | Reference |
|---|---|---|
| Wild type | 20.56 ± 1.49 (n = 47)a | This study |
| Osmsh4-1 | 1.71 ± 1.25 (n = 204) | This study |
| Osmsh5-1 | 2.10 ± 0.91 (n = 97) | Luo et al. (2013) |
| Oszip4 | 6.05 ± 1.97 (n = 164) | Shen et al. (2012) |
| Osmer3 | 5.59 ± 2.07 (n = 64) | Shen et al. (2012) |
| hei10 | 6.50 ± 2.10 (n = 130) | Wang et al. (2012) |
| Osmsh4 Osmsh5 | 1.76 ± 1.20 (n = 115) | This study |
Mean ± SD n, number of pollen mother cells observed.
Complete synapsis is achieved in Osmsh4
As one of the most important events in prophase I, successful synapsis necessitates teamwork by numerous genes, otherwise normal synapsis will fail. In Arabidopsis, unsynapsed axes are detected in the absence of AtMSH4 (Higgins et al. 2004). In mice, far more severe defects were detected in msh4, as homologous chromosomes even failed to align properly (Kneitz et al. 2000).
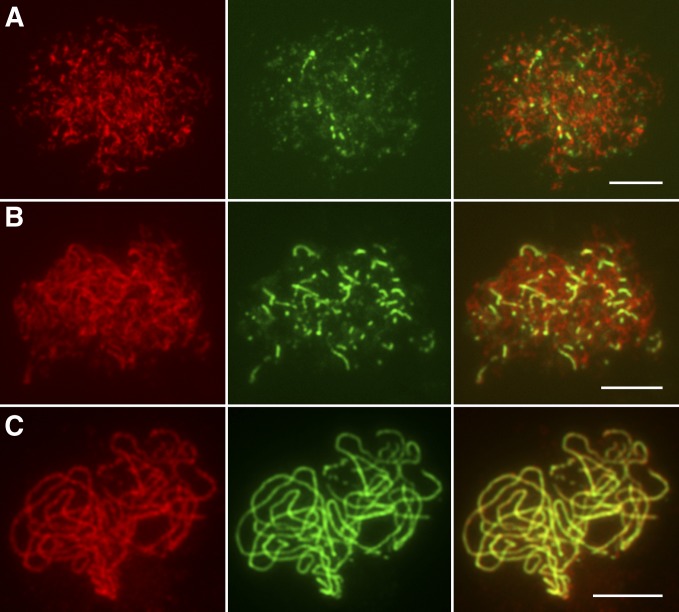
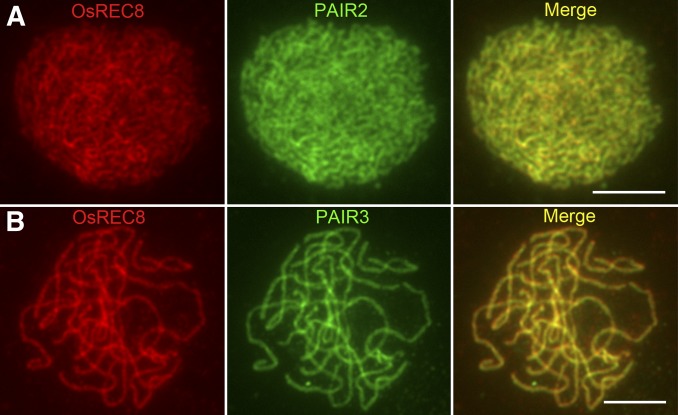
Dual immunodetection was carried out in Osmsh4-1 meiocytes with antibodies related to either lateral or central elements of SCs to explore the role of OsMSH4 in synapsis. OsREC8, the rice homolog of yeast SCC1/SYN1, is a member of the meiotic cohesion complex. The temporal and spatial distribution of OsREC8 protein covers the whole of prophase I in rice meiosis, making it a perfect marker for monitoring SCs (Shao et al. 2011). ZEP1, a transverse filament protein that constitutes the central element of SCs, localizes on chromosomes as punctate foci at leptotene and aligns along the whole length of chromosomes at pachytene when synapsis is complete (Figure S6, A–C). Hundreds of Osmsh4-1 PMCs were used for dual immunodetection experiments, in which the ZEP1 signals duplicated those in the wild type. Punctate ZEP1 signals were observed at early leptotene (Figure 3A), and these gradually elongated and formed short linear signals from late leptotene to zygotene (Figure 3B). Well-formed continuous linear signals along the whole pachytene chromosomes were also detected (Figure 3C), suggesting that synapsis was fully achieved in Osmsh4-1. PAIR2, an axel element of SCs in rice (Nonomura et al. 2006), persists on meiotic chromosomes in wild-type PMCs until the end of zygotene (Figure S7A). PAIR3 also works as a meiotic axial element in rice (Wang et al. 2011), with a very similar localization pattern to OsREC8 (Figure S7B). We found PAIR2 and PAIR3 both loaded onto chromosomes normally in Osmsh4-1 meiocytes (Figure 4, A and B).
Figure 3.
Immunolocalization of OsREC8 and ZEP1 in Osmsh4-1. (A) The localization of ZEP1 at leptotene. (B) The localization of ZEP1 at zygotene. (C) The localization of ZEP1 at pachytene. OsREC8 signals (red) were used to indicate chromosomes. Bars, 5 μm.
Figure 4.
Immunolocalization of OsREC8, PAIR2, and PAIR3 in Osmsh4-1. (A) The localization of PAIR2 at zygotene. (B) The localization of PAIR3 at pachytene. OsREC8 signals were used to indicate chromosomes. Bars, 5 μm.
OsMSH4 acts in conjunction with OsMSH5
It has been shown in other organisms that MSH4 and MSH5 directly interact and drive CO formation as a heterodimer (Bocher et al. 1999; Borner et al. 2004; Snowden et al. 2004; Her et al. 2007; Higgins et al. 2008). However, no evidence has revealed this direct connection in plants.
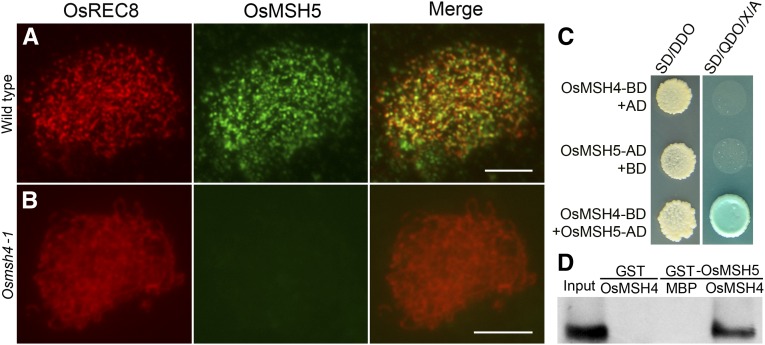
We conducted the yeast two-hybrid assay on OsMSH5 and OsMSH4. ORFs of OsMSH5 and OsMSH4 were separately cloned into the related yeast AD vector pGADT7 and the BD vector pGBKT7. These two recombinant vectors were cotransformed into the Y2H Gold yeast strain to trigger their coexpression. We observed bright spots on the double dropout medium, suggesting that the coexpression occurred, and the specific blue-colored spots on the selecting medium represented the positive clones (Figure 5C). Furthermore, we also conducted pull-down assays in vitro to confirm this interaction, using GST-fusion proteins and MBP-fusion proteins expressed in E. coli. In this assay, GST–OsMSH5, but not GST alone, showed the specific affinity to MBP–OsMSH4 (Figure 5D), further confirming the results from Y2H assays. Thus, the direct interaction between OsMSH4 and OsMSH5 was verified in rice.
Figure 5.
OsMSH4 is essential for the loading of OsMSH5 onto chromosomes. (A) Dual immunolocalization of OsMSH5 in wild type. (B) Dual immunolocalization of OsMSH5 in Osmsh4-1. Bars, 5 μm. (C) The interaction between OsMSH4 and OsMSH5 in yeast two-hybrid assays. SD/DDO: SD −Leu −Trp was used to test the cotranformation efficiency. The interaction was tested by the growth of yeast on selection medium SD/QDO/X/A (SD −Ade −His −Leu −Trp +X +A). X: X-α-Gal. A, aureobasidin A; SD medium, minimal, synthetically defined medium for yeast; BD, bait vector; AD, prey vector. (D) The interaction between OsMSH4 and OsMSH5 in GST pull-down assays. Purified GST–OsMSH5 protein was used as positive control (input).The result of MBP–OsMSH4 incubated with GST tag was used as the negative control. Glutathione Sepharose beads coated with GST-OsMSH5 were used to capture either MBP tag or MBP–OsMSH4. The specific affinity of GST–OsMSH5 to MBP–OsMSH4 was shown.
The localization of OsMSH5 was investigated in the Osmsh4-1 mutant. In the PMCs observed, in comparison to the dot-like OsMSH5 signals observed in the wild type (Figure 5A), no OsMSH5 signals were visible on chromosomes of Osmsh4-1 (Figure 5B), suggesting that the loading of OsMSH5 depends on OsMSH4.
The loading of OsMER3 and OsZIP4 depends on OsMSH4
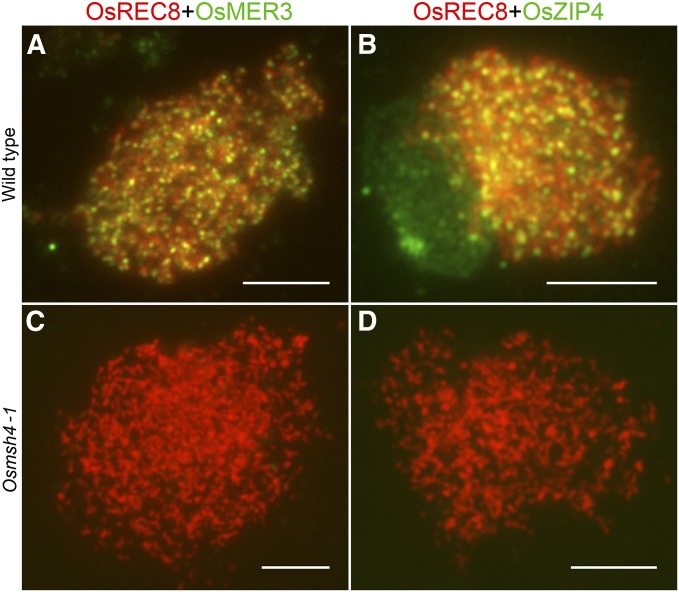
In rice, the two zmm mutants, Osmer3 and Oszip4, show the similar meiotic defects with decreased numbers of chiasmata (Wang et al. 2009; Shen et al. 2012). To show the relationship of OsMSH4 with OsMER3 and OsZIP4, we performed immunodetection of OsMER3 and OsZIP4 in both wild type and Osmsh4-1. In contrast to the dot-like signals observed in the wild type at zygotene (Figure 6, A and B), there was no OsMER3 and OsZIP4 foci in Osmsh4-1 (Figure 6, C and D), implying that the loading of OsMER3 and OsZIP4 depends on OsMSH4.
Figure 6.
Immunodetection of OsMER3 and OsZIP4 in wild type and Osmsh4-1. (A) Staining of OsMER3 at zygotene in the wild type. (B) Staining of OsZIP4 at zygotene in the wild type. (C) Staining of OsMER3 at zygotene in Osmsh4-1. (D) Staining of OsZIP4 at zygotene in Osmsh4-1. Bars, 5 μm.
The formation of normal HEI10 prominent foci at late pachytene requires OsMSH4
HEI10 is the rice homolog of yeast Zip3 and its loss-of-function mutant showed markedly reduced number of chiasmata as seen in other zmm mutants. Immunostaining tests were carried out to investigate its relationship with OsMSH4.
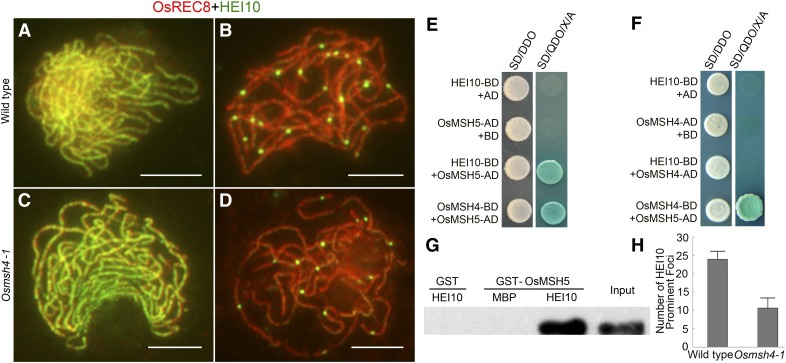
In wild-type meiocytes, HEI10 was first detected at leptotene as punctate foci, and linear signals extended to the entire chromosomes from zygotene to early pachytene (Figure 7A). At late pachytene, some bright foci appeared (Figure 7B), which were proposed to indicate COs (Wang et al. 2012).
Figure 7.
OsMSH4 affects the formation of HEI10 prominent foci. (A and C) Staining for HEI10 at zygotene in wild type and Osmsh4-1. (B and D) Staining for HEI10 at late pachytene in wild type and Osmsh4-1. Bars, 5 μm. (E) HEI10 interacted with OsMSH5 in yeast two-hybrid assays. The interactions were verified by the growth of yeast on selection medium SD/QDO/X/A. The blue dots from coexpression of OsMSH4 and OsMSH5 were used as positive control. (F) HEI10 did not interact with OsMSH4 in yeast two-hybrid assays. (G) OsMSH5 bound to HEI10 in GST pull-down assays. Purified GST–OsMSH5 protein was used as positive control (input). The result of MBP–HEI10 incubated with GST tag was used as the negative control. Glutathione Sepharose beads coated with GST-OsMSH5 were used to capture either MBP tag or MBP–HEI10. GST–OsMSH5 specifically bound to HEI10. (H) Statistical contrast of HEI10 prominent foci between wild type and Osmsh4-1.
The immunostaining pattern of HEI10 in Osmsh4-1 differed from that of OsMER3 and OsZIP4. In Osmsh4-1, the loading of HEI10 onto chromosomes was indistinguishable from that of wild type from leptotene to early pachytene when the small dot signals gradually developed into linear signals (Figure 7C). However, at late pachytene, divergence was apparent as the prominent foci did not develop as normal (Figure 7D). We surveyed 20 PMCs from Osmsh4-1 in late pachytene and found 10.65 ± 2.80 prominent foci in comparison to 24.05 ± 2.17 in wild-type meiocytes (n = 22) (Figure 7H). It seems that HEI10 was loaded normally onto chromosomes until early pachytene in Osmsh4-1; however, the formation of prominent foci at late pachytene was severely disrupted.
In addition, a further yeast two-hybrid assay confirmed the association between OsMSH5 and HEI10. OsMSH5–pGADT7 vector was cotransformed with HEI10–pGBKT7, and their interaction was confirmed by the presence of blue-colored spots on the selecting medium (Figure 7E). In the GST pull-down assays, GST–OsMSH5 could bind to MBP–HEI10 (Figure 7G), providing additional evidence for their direct interaction. However, we did not detect the interaction between OsMSH4 and HEI10 (Figure 7F). Thus, we propose that OsMSH5 may serve as a bridge between OsMSH4 and HEI10.
Discussion
OsMSH4 plays a pivotal role during CO formation
Recombination nodules are large protein complexes that associate with SCs (Stack et al. 1993). Early nodules (ENs) and late nodules (LNs) are the two types of recombination nodule involved in SC and CO formation, in which ENs have been presumed to correspond to the initial recombination sites and correlate with the initiation of SC formation (Stack et al. 1993), while LNs appear to be associated with COs that are assumed to mature into chiasmata. In budding yeast, Zip3 proteins are regarded as LNs and are considered to mark the final sites of COs (Agarwal and Roeder 2000; Fung et al. 2004; Tsubouchi et al. 2008). However, Zip3 also interacts with components of ENs, such as Rad51 and Rad57, providing a potential link between ENs and LNs (Agarwal and Roeder 2000). Zip3 presents on meiotic chromosomes as discrete foci and localizes in CO-designated sites (Agarwal and Roeder 2000). However, ZHP-3, the C. elegans ortholog of budding yeast Zip3, shows highly dynamic localization (Bhalla et al. 2008); RNF212, a protein in mammals with homology to Zip3 and ZHP-3, also dynamically localizes onto chromosomes (Kong et al. 2008; Chowdhury et al. 2009; Reynolds et al. 2013). In rice, HEI10 is the first protein confirmed to mark COs. It also displays dynamic positioning, with dot signals turning to linear signals from leptotene to early pachytene and restricted prominent foci observed from late pachytene to diakinesis (Wang et al. 2012). The early staining of HEI10 may represent early recombination, while the restricted foci seen during late pachytene may represent COs.
All ZMM foci are regarded as LNs in budding yeast and are considered to mark the final sites of COs (Agarwal and Roeder 2000; Fung et al. 2004; Tsubouchi et al. 2008). All MSH4-marked interactions may progress into COs (Lynn et al. 2007). However, in mouse and Arabidopsis, MSH4 foci occur in substantially greater numbers than the final COs, suggesting that MSH4 may participate in the processing of intermediates that do not generate COs but assist in CO formation (Higgins et al. 2004; de Boer et al. 2006). In Arabidopsis, most MSH4 foci colocalize with RAD51 (Higgins et al. 2004), and in mammals MSH4 and RAD51/DMC1 interact directly (Neyton et al. 2004), consistent with a direct role of MSH4 in recombination. Notably, in mammals, MSH4 is also found to act in association with the CO-specific markers MLH3 and MLH1 (Santucci-Darmanin et al. 2000, 2002; Marcon and Moens 2003), indicating its presence at CO-designated sites and its significance in CO formation.
In Osmsh4, HEI10 displayed the same localization pattern with the wild type until early pachytene, indicating the normal processing of early recombination. However, the large bright dots of HEI10 at late pachytene were reduced dramatically compared to wild type. The specific localization pattern of HEI10 suggested a probable defect in the final aggregation onto CO-destined sites, conforming to the working model in which MSH4 forms a heterodimer with MSH5 and plays a role in late recombination processes. The heterodimers bind to the progenitors of dHjs to ensure their stable formation during zygotene to early pachytene and promote the final resolution of dHjs to mature COs at late pachytene, suggesting a role in late recombination (Borner et al. 2004; Page and Hawley 2004; Snowden et al. 2004). Thus, the absence of either protein will prevent the formation of the heterodimer, which in turn may cause instability of dHj formation and interfere with their development into COs; proteins associated with COs may then be unable to find specific sites to localize to. Since OsMSH5 was unable to localize onto chromosomes in the absence of OsMSH4, the formation of OsMSH4–OsMSH5 heterodimer was completely disrupted. Thus, the sites designated as COs could not form as usual, leading to the disrupted localization pattern of prominent HEI10 foci in Osmsh4. In general, OsMSH4 appears to mainly function in late recombination and CO formation but not the early processes, as the SCs were well formed and early localization of HEI10 was not affected in the mutants. Together, these data provided vital clues concerning the partnership between OsMSH5 and OsMSH4 in processing recombination intermediates during the formation of COs.
Except for the results from immunostaining, our studies also revealed a reduction in the chiasma frequency in Osmsh4. In Osmsh4-1, the chiasma number was reduced to no more than 10% of the wild type. Besides, the number of chiasmata per cell in Osmsh5 was also decreased to 10% of the wild type (Luo et al. 2013). The reduction in Osmsh4 and Osmsh5 was much greater than the other zmm mutants in rice, such as Oszip4 and Osmer3, which affected the formation of 70–80% of COs, leaving ∼30% chiasmata in the mutants (Wang et al. 2009, 2012; Shen et al. 2012). Considering these results, we suggest that OsMSH4 in association with OsMSH5 plays a far more crucial role in the formation of COs than other ZMM proteins.
The relationship of OsMSH4 with other ZMM proteins
In budding yeast, the ZMM protein groups are involved in many meiotic events. Zip2–4 proteins are implicated in ubiquitinylation and/or SUMOylation processes and are likely to work cooperatively in the modification of protein interactions (Perry et al. 2005; Cheng et al. 2006). Furthermore, Mer3 and Msh4/Msh5 probably mediate local recombination interactions in the early and late stages, respectively (Hollingsworth et al. 1995; Novak et al. 2001; Nakagawa and Kolodner 2002). During pachytene, these proteins extensively colocalize, as shown by the colocalization of Zip2–Zip3, Zip4–Zip2, and Zip2–Msh4 (Agarwal and Roeder 2000; Novak et al. 2001; Tsubouchi et al. 2006). In Arabidopsis, Atmsh4 and Atmsh5 mutants show great reduction in CO formation and these proteins colocalize with each other in wild-type PMCs (Higgins et al. 2008). Moreover, HEI10 in Arabidopsis colocalizes with AtMLH1 from pachytene to diakinesis (Chelysheva et al. 2012). In rice, colocalization between OsMER3 and early HEI10 signals has also been shown, suggesting their cooperation in processing recombination intermediates in the early stages of meiotic prophase I (Wang et al. 2012). In addition to the known classical ZMM genes, other genes with ZMM-related phenotypes have also been discovered in recent years. PARTING DANCERS (PTD), a novel gene, was found to be epistatic to ZMMs in Arabidopsis, and mutation in this gene causes a reduction in COs (Wijeratne et al. 2006). Moreover, recent studies have revealed that PTD interacts with SHOC1, the Arabidopsis ortholog to Zip2, demonstrating its close relationship with classical ZMM proteins (Macaisne et al. 2008, 2011).
OsMSH4 plays a pivotal role in the localization of other ZMM proteins. In Osmsh4, chiasma frequency was shown to exhibit a catastrophic reduction, and the localization of OsMER3 and OsZIP4 onto chromosomes was eliminated. These findings led us to propose that OsMSH4 functions upstream of both OsMER3 and OsZIP4. The absence of OsMSH4 also affected the formation of prominent HEI10 foci; in Osmsh5, the formation of prominent HEI10 foci was also disrupted (Luo et al. 2013), implying that as a heterodimer, the two function in the same way. In the yeast two-hybrid assays and pull-down tests, we showed evidence of multiple interactions among different ZMM proteins. In addition to confirming the OsMSH4–OsMSH5 interaction, we concurrently detected a physical association between OsMSH5 and HEI10. So far, no direct evidence has been presented concerning the relationships between ZMM proteins in plants, even though there have been many claims that they are functionally related, based solely on their colocalization in cytological experiments. Therefore, these findings may offer great help in drawing the interaction network of ZMM proteins. Furthermore, the interaction between OsMSH5 and HEI10 suggested an indirect interaction between OsMSH4 and HEI10 with OsMSH5 serving as a bridge, providing indirect evidence for the indispensable role of OsMSH4 in the localization of HEI10.
Taken together, our data suggest that OsMSH4 functions in conjunction with OsMSH5 to regulate the formation of the majority of crossovers in rice and these two proteins function upstream of other ZMMs. We can also deduce that the ZMM complex in rice may not only be a functional complex but also a real physical one held together by direct or indirect associations at specific times, if only transiently.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Sciences and Technology of China (2012AA10A301 and 2011CB944602), and the National Natural Science Foundation of China (31360260, 31230038, and U1302261).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.168732/-/DC1.
Communicating editor: A. Houben
Literature Cited
- Agarwal S., Roeder G. S., 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Baumann P., Benson F. E., West S. C., 1996. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87: 757–766. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Wynne D. J., Jantsch V., Dernburg A. F., 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocher T., Barusevicius A., Snowden T., Rasio D., Guerrette S., et al. , 1999. hMSH5: a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Res. 59: 816–822. [PubMed] [Google Scholar]
- Borner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Chambers S. R., Abdullah M. F., 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Chambon A., Gendrot G., Pereira L., et al. , 2012. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang W., Timofejeva L., Gerardin Y., Ma H., 2005. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J. 43: 321–334. [DOI] [PubMed] [Google Scholar]
- Cheng C. H., Lo Y. H., Liang S. S., Ti S. C., Lin F. M., et al. , 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Bois P. R., Feingold E., Sherman S. L., Cheung V. G., 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5: e1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E., Stam P., Dietrich A. J., Pastink A., Heyting C., 2006. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA 103: 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S. S., Baart E. B., Dekker M., Siezen A., de Rooij D. G., et al. , 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., et al. , 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127. [DOI] [PubMed] [Google Scholar]
- Fung J. C., Rockmill B., Odell M., Roeder G. S., 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Her C., Zhao N. X., Wu X. L., Tompkins J. D., 2007. MutS homologues hMSH4 and hMSH5: diverse functional implications in humans. Front. Biosci. 12: 905–911. [DOI] [PubMed] [Google Scholar]
- Higgins J. D., Armstrong S. J., Franklin F. C., Jones G. H., 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. D., Vignard J., Mercier R., Pugh A. G., Franklin F. C. H., et al. , 2008. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55: 28–39. [DOI] [PubMed] [Google Scholar]
- Hoffmann E. R., Borts R. H., 2004. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 107: 232–248. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C., 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kneitz B., Cohen P. E., Avdievich E., Zhu L., Kane M. F., et al. , 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Cohen P. E., 2004. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107: 216–231. [DOI] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Stefansson H., Masson G., Helgason A., et al. , 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319: 1398–1401. [DOI] [PubMed] [Google Scholar]
- Li W., Ma H., 2006. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 16: 402–412. [DOI] [PubMed] [Google Scholar]
- Li Z., Golub E. I., Gupta R., Radding C. M., 1997. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl. Acad. Sci. USA 94: 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin S. M., Moens P. B., Wang V., Lenzi M., Shanmugarajah D., et al. , 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31: 385–390. [DOI] [PubMed] [Google Scholar]
- Luo Q., Tang D., Wang M., Luo W., Zhang L., et al. , 2013. The role of OsMSH5 in crossover formation during rice meiosis. Mol. Plant 6: 729–742. [DOI] [PubMed] [Google Scholar]
- Luo Q., Li Y., Shen Y., Cheng Z., 2014. Ten years of gene discovery for meiotic event control in rice. J. Genet. Genomics 41: 125–137. [DOI] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Borner G. V., 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Macaisne N., Novatchkova M., Peirera L., Vezon D., Jolivet S., et al. , 2008. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 18: 1432–1437. [DOI] [PubMed] [Google Scholar]
- Macaisne N., Vignard J., Mercier R., 2011. SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J. Cell Sci. 124: 2687–2691. [DOI] [PubMed] [Google Scholar]
- Marcon E., Moens P., 2003. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Vezon D., Bullier E., Motamayor J. C., Sellier A., et al. , 2001. SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15: 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kolodner R. D., 2002. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol. Cell. Biol. 22: 3281–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Flores-Rozas H., Kolodner R. D., 2001. The MER3 helicase involved in meiotic crossing over is stimulated by single-stranded DNA-binding proteins and unwinds DNA in the 3′ to 5′ direction. J. Biol. Chem. 276: 31487–31493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyton S., Lespinasse F., Moens P. B., Paul R., Gaudray P., et al. , 2004. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol. Hum. Reprod. 10: 917–924. [DOI] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Eiguchi M., Suzuki T., Kurata N., 2006. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119: 217–225. [DOI] [PubMed] [Google Scholar]
- Novak J. E., Ross-Macdonald P. B., Roeder G. S., 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Higgins J. D., Sanchez-Moran E., Armstrong S. J., Franklin F. C., 2011. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190: 523–544. [DOI] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558. [DOI] [PubMed] [Google Scholar]
- Perry J., Kleckner N., Borner G. V., 2005. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. USA 102: 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J., Siroky J., Mokros P., Schweizer D., Riha K., 2004. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., et al. , 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., Roeder G. S., 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Sanchez Moran E., Armstrong S. J., Santos J. L., Franklin F. C., Jones G. H., 2001. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9: 121–128. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Walpita D., Lespinasse F., Desnuelle C., Ashley T., et al. , 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14: 1539–1547. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Neyton S., Lespinasse F., Saunieres A., Gaudray P., et al. , 2002. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL. homolog in mammalian meiotic recombination. Hum. Mol. Genet. 11: 1697–1706. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shao T., Tang D., Wang K., Wang M., Che L., et al. , 2011. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Tang D., Wang K., Wang M., Huang J., et al. , 2012. The role of ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 125(Pt 11): 2581–2591. [DOI] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Stack, S., J. Sherman, L. Anderson, and L. Herickhoff, 1993 Meiotic nodules in vascular plants, pp. 301–311 in Chromosomes Today. Springer-Verlag, New York. [Google Scholar]
- Sym M., Roeder G. S., 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Zhao H., Roeder G. S., 2006. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev. Cell 10: 809–819. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Macqueen A. J., Roeder G. S., 2008. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 22: 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Qin B., et al. , 2011. PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Miao C., et al. , 2012. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. J., Tang D., Wang M., Lu J. F., Yu H. X., et al. , 2009. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122: 2055–2063. [DOI] [PubMed] [Google Scholar]
- Wijeratne A. J., Chen C., Zhang W., Timofejeva L., Ma H., 2006. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol. Biol. Cell 17: 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds J. L., Boulton S. J., 2011. The choice in meiosis: defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 124: 501–513. [DOI] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.