Abstract
The cytoplasm of the eukaryotic cell is subdivided into distinct functional domains by the presence of a variety of membrane-bound organelles. The remaining aqueous space may be further partitioned by the regulated assembly of discrete ribonucleoprotein (RNP) complexes that contain particular proteins and messenger RNAs. These RNP granules are conserved structures whose importance is highlighted by studies linking them to human disorders like amyotrophic lateral sclerosis. However, relatively little is known about the diversity, composition, and physiological roles of these cytoplasmic structures. To begin to address these issues, we examined the cytoplasmic granules formed by a key set of signaling molecules, the protein kinases of the budding yeast Saccharomyces cerevisiae. Interestingly, a significant fraction of these proteins, almost 20%, was recruited to cytoplasmic foci specifically as cells entered into the G0-like quiescent state, stationary phase. Colocalization studies demonstrated that these foci corresponded to eight different granules, including four that had not been reported previously. All of these granules were found to rapidly disassemble upon the resumption of growth, and the presence of each was correlated with cell viability in the quiescent cultures. Finally, this work also identified new constituents of known RNP granules, including the well-characterized processing body and stress granule. The composition of these latter structures is therefore more varied than previously thought and could be an indicator of additional biological activities being associated with these complexes. Altogether, these observations indicate that quiescent yeast cells contain multiple distinct cytoplasmic granules that may make important contributions to their long-term survival.
Keywords: P-bodies, stress granules, stationary phase, ribonucleoprotein (RNP) granules, protein kinases
THE cytoplasm of eukaryotic cells is subdivided into discrete functional units by the presence of a plethora of membrane-bound organelles. This compartmentalization serves multiple purposes and is essential for the normal functioning of the cell. For example, proteins involved in related processes can be concentrated within a limited space so as to increase overall reaction efficiencies. Interestingly, recent studies have suggested that additional levels of compartmentalization exist within eukaryotic cells. In particular, specific sets of proteins and messenger RNAs (mRNAs) have been found to colocalize to discrete sites in the cytoplasm in response to a variety of external and internal cues (Anderson and Kedersha 2009; Thomas et al. 2011). This coalescence results in the formation of distinct ribonucleoprotein (RNP) granules that differ in composition from the surrounding environment. However, in contrast to the more traditional organelles, these RNP structures lack a limiting membrane, and granule constituents have been found to rapidly exchange with their respective cytoplasmic pools (Anderson and Kedersha 2009). Biophysical and microscopy studies have further suggested that these granules behave like discrete liquid droplets within the cytoplasm and that they may form as a result of a phase transition process (Brangwynne et al. 2009; Weber and Brangwynne 2012). In all, these attributes could provide the cell with the means for a more dynamic and reversible segregation of cytoplasmic components.
Processing bodies (P-bodies) and stress granules are two of the best-characterized cytoplasmic RNP granules in eukaryotic cells. These structures have been conserved through evolution and contain nontranslating mRNAs and specific sets of proteins (Anderson and Kedersha 2009; Balagopal and Parker 2009). Both types of granules are transient in nature and are induced by a variety of overlapping stress conditions. Stress granules contain several translation factors and are thought to be sites of storage for mRNAs that will be translated following the removal of the inducing stress (Kedersha and Anderson 2002; Yamasaki and Anderson 2008). In contrast, P-bodies contain proteins that are involved in mRNA processing, including the Dcp1/Dcp2 decapping enzyme and the Xrn1 exonuclease (Bashkirov et al. 1997; Van Dijk et al. 2002; Sheth and Parker 2003; Cougot et al. 2004; Eulalio et al. 2007b). These observations led to the initial suggestion that P-bodies represent cytoplasmic sites of mRNA decay. However, more recent work has demonstrated that mRNA turnover can occur normally in cells that lack P-body foci (Stoecklin et al. 2006; Decker et al. 2007; Eulalio et al. 2007a). As a result, the precise role of P-body granules in mRNA processing remains unclear. In mammalian cells, P-bodies have also been implicated in the microRNA-mediated inhibition of translation and in the replicative life cycle of several viruses (Liu et al. 2005; Bhattacharyya et al. 2006; Beckham and Parker 2008; Reineke and Lloyd 2013). Therefore, P-bodies, and perhaps other RNP granules, may be associated with different biological activities depending upon the particular proteins present in these structures. Defining the constituents of RNP granules will thus be essential for a complete description of their roles in eukaryotic cells.
RNP granules can also be induced by specific developmental and cell growth transitions. For example, polar granules in Drosophila melanogaster are produced in the oocyte and serve to specify the germ-cell lineage in the developing embryo (Leatherman and Jongens 2003; Tadros and Lipshitz 2005; Thomson et al. 2008). These granules contain specific maternal transcripts that are translated after fertilization. RNP granules also appear to be induced when eukaryotic cells stop dividing and become quiescent (An et al. 2008; Narayanaswamy et al. 2009; Noree et al. 2010). This latter induction has been most thoroughly documented in Saccharomyces cerevisiae where several observations indicate that such granules are prevalent in stationary-phase cells. First, both P-bodies and stress granules are efficiently induced during the entry into this quiescent phase, and the former appear to be required for the long-term survival of these nondividing cells (Ramachandran et al. 2011; Shah et al. 2013). In addition, proteins associated with the actin cytoskeleton and the proteasome are found in actin bodies and proteasome storage granules, respectively, in stationary-phase cells (Sagot et al. 2006; Laporte et al. 2008). These structures may not contain an RNA component and have been proposed to act as sites of storage during this period of quiescence. Finally, high-throughput microscopy studies have identified a number of cytoplasmic proteins that localize to discrete foci upon stationary-phase entry (Narayanaswamy et al. 2009). However, it was not determined whether these latter proteins were recruited to known granules or if the identified foci represent novel cytoplasmic structures. Nevertheless, the data altogether indicate that the cessation of cell growth is associated with a significant redistribution of cytoplasmic protein.
In this study, we examined the stationary-phase localization of a defined set of proteins, the protein kinases of the budding yeast S. cerevisiae. The primary goal of this work was to gain insight into the variety and composition of the cytoplasmic granules present in quiescent cells. Using a microscopy-based analysis, we found that multiple, distinct cytoplasmic granules were present in stationary-phase cells and identified novel components of both P-bodies and stress granules. Almost one-fifth of the protein kinases examined were found to form cytoplasmic foci specifically in growth-arrested cells. Approximately one-half of these foci were coincident with P-bodies and/or stress granules with the remainder identifying several discrete cytoplasmic granules that had not been reported previously. All of these cytoplasmic structures were observed to disassemble rapidly upon the resumption of growth, and the presence of each was correlated with cell viability in stationary-phase cultures. In all, the results suggested that signaling proteins have a significant presence in RNP (and perhaps protein-only) granules and are consistent with recent observations indicating that these cytoplasmic structures might be hubs of signaling activity (Kedersha et al. 2013). The data also suggest that the protein composition of known RNP foci, like the P-body and stress granule, is more varied than initially thought and could be an indicator of additional biological activities being associated with these structures. As a result, it will be important to define the composition of the different granules that coexist in the resting cell and to ascertain the biological roles of each of these structures.
Materials and Methods
Yeast strains and growth conditions
Standard Escherichia coli and yeast growth conditions and media were used throughout this work. The yeast-rich growth medium, YPAD, and the minimal YM and SC media have been described (Kaiser et al. 1994; Chang et al. 2004; Ramachandran and Herman 2010). For the microscopy analysis, yeast cultures were started in rich medium at an initial OD600 of ∼0.5 and incubated at 30° with agitation for the indicated number of days. The GFP-tagged kinase strains were obtained from the Yeast GFP Fusion Localization collection (Invitrogen). The S. cerevisiae proteome contains ∼120 protein kinases, and we examined the 95 strains that were available in this collection. Strains carrying GFP-tagged versions of Tor1 and Tor2 were generously provided by Ted Powers. The carbon starvation experiments were performed in SC- or YPA-based media lacking glucose for strains with and without plasmids, respectively. For the colocalization experiments, cells were typically transformed with a plasmid expressing an mCherry (mCh)-tagged reporter, like Edc3 for P-bodies or Pbp1 for stress granules. These strains were then grown in an SC-based minimal medium and examined after either 3 or 4 days of growth at 30°. Strains carrying the MET3-RAS2val19 allele were grown in medium containing 500 μM methionine to repress expression from the MET3 promoter (Howard et al. 2002). Expression from this promoter was induced by transferring cells to a medium that lacked methionine. A PCR-based strategy was used to delete the PAT1 locus in the GFP-tagged reporter strains (Baudin et al. 1993). The presence of the null allele was confirmed initially with a PCR analysis and subsequently with a growth assay that assesses the chronological life span (CLS) of the candidates. Strains that lack the PAT1 locus have been shown to exhibit a significantly decreased CLS (Ramachandran et al. 2011).
Plasmid construction
The plasmids used in this study are listed in Supporting Information, Table S1. The MET3-RAS2val19 plasmids, pPHY795 and pPHY796, have been described (Howard et al. 2001; Ramachandran and Herman 2010). The plasmid expressing Edc3-mCherry (pRP1574) was kindly provided by Roy Parker. To construct the Cdc28-GFP plasmid (pPHY3754), the CDC28-GFP locus was PCR-amplified from genomic DNA derived from the GFP-tagged yeast strain and cloned into the EcoRI and SpeI sites of pRS413. The amplified fragment contained the CDC28 promoter, the fusion open reading frames, and the ADH1 terminator. The Cdc28-mCherry plasmid pPHY3765 was made by replacing the GFP locus with that of mCherry in the above Cdc28-GFP plasmid. The additional mCherry plasmids were made in a similar fashion, starting with the appropriate GFP-tagged yeast strain. These constructs included fusions with the Pat1, Pbp1, Pab1, Fpk1, Kss1, Kin28, Abp1, and Scl1 proteins.
Fluorescence microscopy
Cells expressing the appropriate fusion construct(s) were grown as indicated and then collected by centrifugation, resuspended in the appropriate medium, and spotted onto microscope slides as described (Budovskaya et al. 2005; Yeh et al. 2010). The cells were then imaged with a ×100/1.45 numerical aperture Plan-Apo objective lens on a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY) equipped with an Andor Zyla digital camera and the appropriate Nikon HC filter sets. For the quantitation, the data in all cases represent the average of two or more experiments where at least 100 cells were examined in each experiment. The percentage of colocalization for any two reporters was calculated by scoring the proportion of coincident foci in those cells that possessed at least one of each type of focus. The final merged images were created with the Image J software package.
Protein analysis
Protein samples for Western blotting were prepared with a glass-bead lysis protocol, separated on 7.5–10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes as described (Budovskaya et al. 2002; Deminoff et al. 2006). The membranes were then probed with the appropriate primary and secondary antibodies. The Supersignal chemiluminescent substrate (Pierce) was subsequently used to detect the reactive bands.
Low-complexity domain analysis
Sequence regions of low complexity were identified using the SEG program with default parameter settings from the National Center for Biotechnology Information (NCBI) website. Sequences of this type tend to be unstructured and are often referred to as intrinsically disordered domains (IDDs) (Dunker et al. 2002). The SEG score for each protein kinase was determined by counting the number of amino acids in the longest continuous region of low complexity as predicted by the SEG program. For each of the kinases, the percentage of cells with foci on day 7 was plotted against the SEG score and the relevant R2 and P-values were calculated.
Propidium iodide staining of yeast cells
Yeast strains were grown to the indicated times and then stained as described with propidium iodide (PI) to identify the dead cells in the culture (Ocampo and Barrientos 2011). PI is a charged molecule that is unable to cross the plasma membrane of living cells.
Results
Protein kinase localization to cytoplasmic foci during stationary-phase entry
We examined the intracellular localization of 97 protein kinases fused at their C terminus to the reporter GFP at three different times: in log-phase cultures and after 1 or 7 days of growth in the rich medium YPAD. In this medium, S. cerevisiae cells grow initially by the fermentation of glucose even in the presence of oxygen. Cells growing exponentially in this manner are referred to as “log phase” in this study. The glucose is depleted within 24 hr of culture, and the cells then switch to a respiratory mode of growth (Werner-Washburne et al. 1993; Herman 2002). This switch is known as the diauxic shift, and the day 1 samples correspond to a time shortly after this change. The cells then grow by respiration for an additional 4 or 5 days on the ethanol that was produced during the period of fermentation. Finally, the cells enter stationary phase and at day 7 are in this quiescent state. Previous work demonstrated that P-bodies are present by day 1 under these growth conditions but that stress granules do not appear until cells begin to enter the stationary phase (Shah et al. 2013). Both types of foci then persist throughout this period of quiescence.
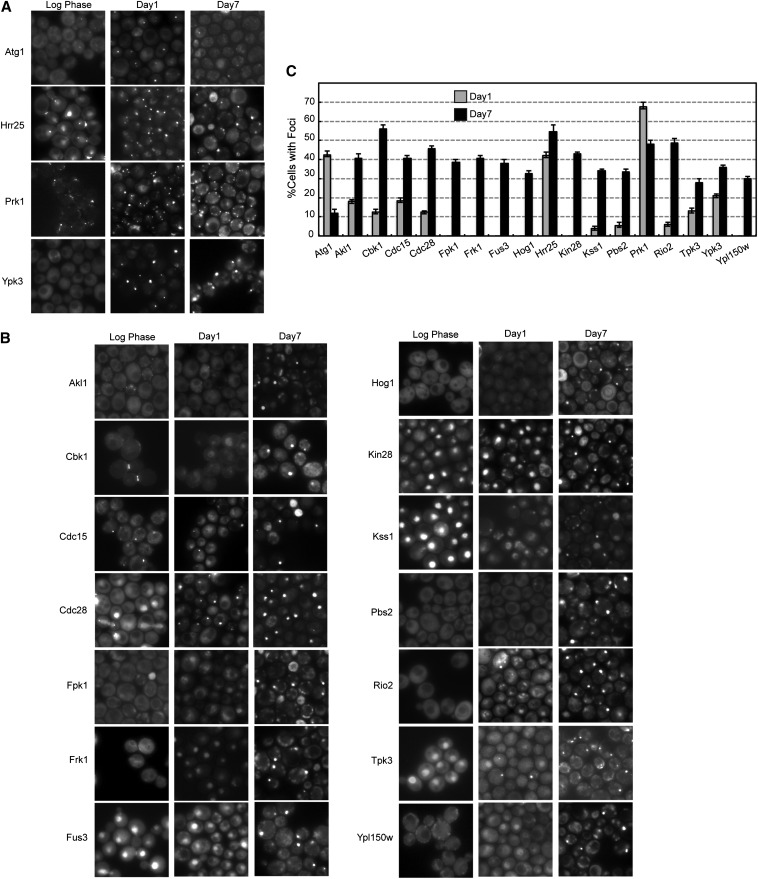
This microscopy analysis identified 17 protein kinases that were present in cytoplasmic foci in >30% of the cells at either day 1 or 7 of growth (Figure 1). These enzymes were generally not associated with cytoplasmic granules in log-phase cells. Three of these kinases, Atg1, Hrr25, and Prk1, were recruited to foci in >40% of the population at day 1 (Figure 1, A and C). The other 14 enzymes in this group generally had significantly more punctae at day 7 (Figure 1, B and C). The remaining protein kinases either were not found in foci at these growth times or were present to lesser degrees (Table S2 and Figure S1). However, in all cases, the fraction of cells with a focus appeared to reach a maximum of ∼50% of the culture (Figure 1C). This was also true for all of the examined reporters for both P-body and stress granule foci, including Edc3 and Pbp1 (Table S2). Although potentially interesting, it is not yet clear why only half of the cells in these stationary-phase cultures appear to be able to form cytoplasmic granules. One interesting possibility is suggested by previous work indicating that only ∼50% of the cells in stationary-phase cultures exhibit the ability to remain viable for long periods of time (Allen et al. 2006; Aragon et al. 2008). These cells have been termed the Q, or quiescent, fraction of this population and may correspond to those cells that have a focus in our studies here.
Figure 1.
Protein kinase localization to cytoplasmic foci in stationary-phase yeast cells. (A and B) Cells expressing the indicated protein kinase-GFP fusions were examined by fluorescence microscopy during log phase and after 1 or 7 days of growth in rich medium. (C) Quantitation of the microscopy data shown in A and B. The fraction of cells with a visible focus is shown for each protein kinase at days 1 and 7. The data represent the average of at least two independent experiments where n = 100 in each case.
The appearance and number of protein kinase foci per cell varied, but most enzymes were detected in one to two punctae per cell (Figure 1). The most notable exception was Prk1, which was observed in numerous small foci in each cell at day 7 (Figure 1A). As with both P-bodies and stress granules, the kinase foci were found to persist throughout stationary phase; the cultures here were examined for up to 28 days (Figure S2). The 17 candidates identified are involved in a broad spectrum of cellular processes and belong to several protein kinase families (Table 1). An examination of the associated Gene Ontology terms did not reveal any obvious enrichment for a particular type of function. The remainder of this report is focused on these 17 enzymes. Tpk3, a catalytic subunit of the cAMP-dependent protein kinase, was also included in the subsequent analysis because it had been shown previously to be present in P-body foci (Tudisca et al. 2010).
Table 1. Gene Ontology terms associated with foci-forming protein kinases.
| Kinase | Gene Ontology Terms |
|---|---|
| Akl1 | Endocytosis, actin cytoskeleton organization |
| Atg1 | Autophagy |
| Cbk1 | Cell polarity, cell separation, cell-wall synthesis |
| Cdc15 | Exit from mitosis, cytokinesis, meiotic spindle disassembly |
| Cdc28 | Cell-cycle regulation |
| Fpk1 | Phospholipid translocation, response to pheromone |
| Frk1 | Unknown |
| Fus3 | Cell mating, invasive growth |
| Hog1 | Response to osmotic stress |
| Hrr25 | Ribosome maturation, DNA repair, vesicle transport |
| Kin28 | mRNA transcription and processing, subunit of TFIIH |
| Kss1 | Pheromone response, filamentous growth |
| Pbs2 | Response to osmotic stress |
| Prk1 | Actin cytoskeleton organization, endocytosis |
| Rio2 | Ribosome maturation |
| Tpk3 | Catalytic subunit of PKA, cellular growth |
| Ypk3 | Unknown |
| Ypl150w | Unknown |
Several observations indicated that the fusion proteins being studied retained the normal functions of the given enzymes. First, the localization patterns observed in log-phase cells were consistent with those reported previously. For example, the nuclear proteins Cdc28, Fus3, Kin28, and Kss1 were all found in the nucleus of dividing cells in this study (Figure 1B). However, the timing of the relocation to cytoplasmic foci varied for each enzyme. Whereas Kin28 and Fus3 were still found in the nucleus at day 1, Kss1 and Cdc28 had left this compartment by this time. All of these kinases were largely absent from the nucleus by day 7. Second, we found that those strains with GFP fused to essential proteins did not exhibit any significant growth defects (Figure S3); these data are consistent with the fusion proteins having sufficient levels of the essential activities associated with these enzymes. Where possible, nonessential kinases, like Hog1 and Atg1, were examined with assays specific to these enzymes (Figure S3). Third, the endpoints of the fusion constructs were verified with a PCR analysis, and the size of the encoded GFP proteins was confirmed by gel electrophoresis (data not shown). Finally, it is important to note that the presence of any protein in a focus was not specific to the GFP reporter and its tendency to form dimers (Tsien 1998). In all cases tested, similar results were obtained with other reporters, most notably mCh, that exist as monomers in the cell (see below). Altogether, these studies demonstrated that a significant fraction of yeast protein kinases was recruited to cytoplasmic foci in stationary-phase cells.
Protein kinase granule formation was reversible
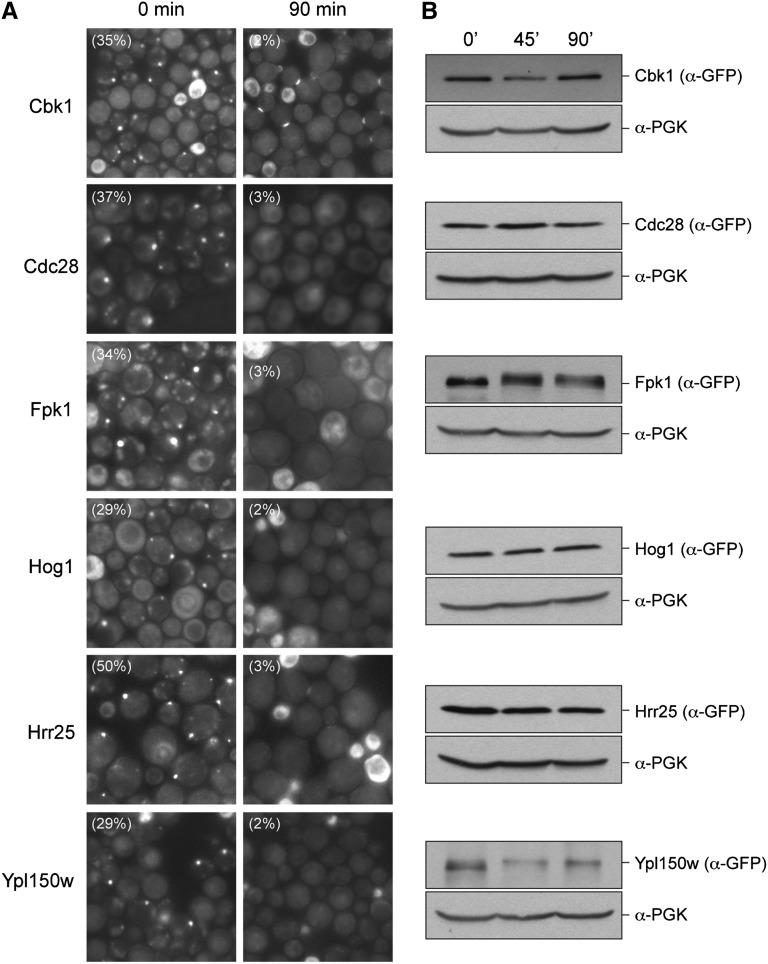
The presence of a visible focus could be the result of the localized aggregation of misfolded forms of the protein kinase under study. The formation of such structures is very often an irreversible event. An example of this type of compartment is the so-called IPOD, or “Insoluble Protein Deposit,” which is thought to contain misfolded protein that forms amyloid and other types of aggregate structures (Kaganovich et al. 2008; Chen et al. 2011). To test whether any kinase focus might correspond to such a compartment, we asked whether the observed foci would disassemble upon the reinitiation of growth. For these experiments, cells expressing the protein kinases were grown in minimal medium for several days to allow foci formation and then transferred to fresh medium for 90 min. We found that the foci in all 18 strains were largely dispersed after this exposure to fresh medium; in each case, the number of cells with punctae was reduced by at least 90%. The data for six representative kinases are shown in Figure 2A (see also Figure S4). At the time of examination, most of the cells in these cultures had not yet completed a round of cell division. This disassembly was not significantly affected by the presence of pharmacological inhibitors of either translation elongation (cycloheximide) or proteasomal degradation (MG132) (data not shown). Finally, a Western blot analysis indicated that the levels of the protein kinase fusions were similar before and after a 90-min incubation with fresh medium in the presence of cycloheximide (Figure 2B). These latter results collectively argue against a model whereby the protein kinases are degraded in response to the addition of the fresh medium. Instead, the data suggest that the foci are rapidly disassembled upon the resumption of growth and that they may represent physiologically relevant assemblages of these proteins.
Figure 2.
The protein kinase foci disassemble upon the reinitiation of growth. (A) The protein kinase foci disassemble upon the addition of fresh medium to growth-arrested cells. Cells expressing the indicated GFP fusion proteins were grown for several days in minimal medium to induce protein kinase foci and then transferred to fresh medium for 90 min. The cells were examined by fluorescence microscopy before (0 min) and after (90 min) the incubation in fresh medium. (B) The protein kinase-GFP fusion proteins were not degraded upon the resumption of growth. The relative level of each of the fusion proteins was assessed by Western blotting with cell extracts prepared before (0 min) and after 45 or 90 min of incubation with fresh medium containing the translation elongation inhibitor, cycloheximide (35 μg/ml). The levels of phosphoglycerate kinase (PGK) served as a loading control for this experiment.
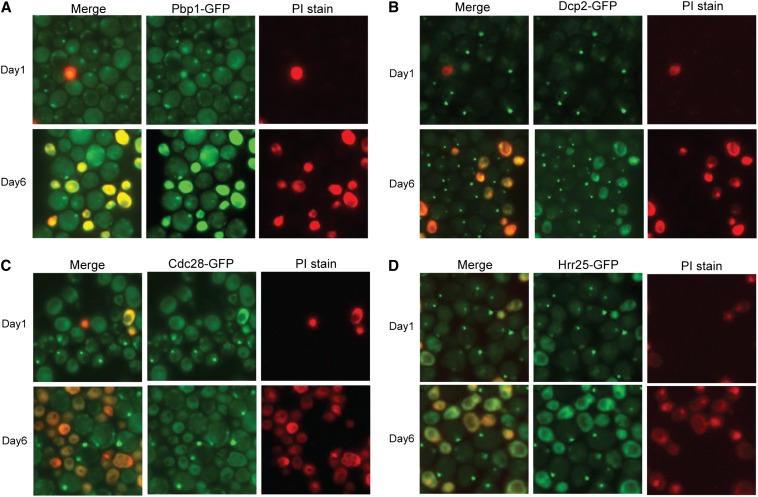
We also found that the presence of a protein kinase focus was correlated with cell viability in the stationary-phase cultures. For this analysis, we used PI to identify the dead or dying yeast cells in the culture (Ocampo and Barrientos 2011). PI is a charged molecule that is not efficiently taken up by living cells. PI staining has been used to assess the length of time that stationary-phase cells remain viable, the time period referred to as the CLS (Kennedy 2008). Mutants defective for P-body formation, like pat1Δ, have been found to exhibit a diminished CLS, suggesting that P-body foci are important for the long-term survival of stationary-phase cells (Ramachandran et al. 2011; Shah et al. 2013). Here, we found that cells that possessed a protein kinase focus were generally unstained by PI, suggesting that these cells were viable. The results for two representative protein kinases, Hrr25 and Cdc28, are shown in Figure 3. A similar correlation with viability was observed for both P-body and stress granule foci (Figure 3). However, it is important to emphasize that additional studies are needed before we can say whether any of the kinase foci identified here contribute in a meaningful way to cell viability during stationary phase.
Figure 3.
The presence of a Cdc28 or Hrr25 protein kinase focus was correlated with cell viability. Cells containing the indicated GFP fusion proteins were grown for 1 or 6 days in SC minimal medium and then stained with PI as described in Materials and Methods. PI preferentially stains dying and dead cells in the culture. Pbp1 is a reporter for stress granules and Dcp2 for P-bodies. The proteins examined were as follows: (A) Pbp1, (B) Dcp2, (C) Cdc28, and (D) Hrr25.
Localization to cytoplasmic granules did not require the presence of an intrinsically disordered domain
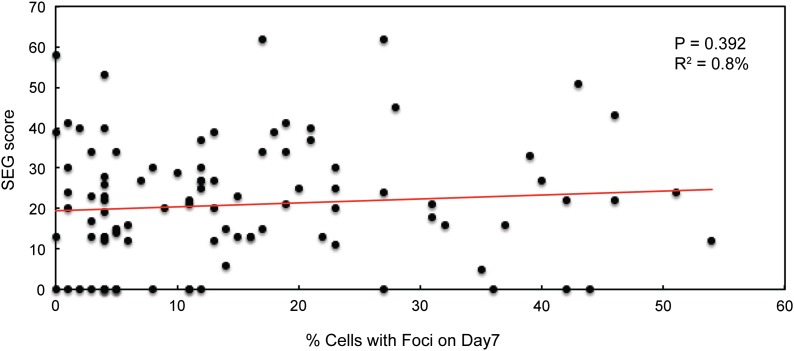
Recent studies have found that many proteins in RNP granules have unstructured domains or IDDs (Gilks et al. 2004; Decker et al. 2007; Reijns et al. 2008; Kato et al. 2012). This work led to the suggestion that the presence of such a region was an important determinant of protein localization to RNP structures (Jonas and Izaurralde 2013; Malinovska et al. 2013). Therefore, we tested whether there was a correlation between the presence (and size) of an IDD and the localization to cytoplasmic foci. Potential IDDs in the set of protein kinases were identified with the SEG algorithm available at the NCBI website (Wootton and Federhen 1996). Using the highest scoring domain detected in each protein, we found no significant correlation between the SEG value and the frequency of cytoplasmic foci at either day 1 or 7 of growth (Figure 4). Similar observations were made when a second algorithm, known as PONDR, was used to predict the presence of IDDs in the protein kinases (Romero et al. 2001). Finally, we observed no correlation between foci formation and the relative abundance of the protein kinases; the abundance values were obtained from the Saccharomyces Genome Database website. The protein levels of a set of kinases encompassing a wide range of predicted expression levels were confirmed by Western blotting (data not shown). In all, these data suggested that protein features, other than the presence of an IDD, were often important for the observed localization to cytoplasmic foci.
Figure 4.
The frequency of protein kinase foci did not correlate with the presence of an intrinsically disordered domain. The graph plots the SEG score obtained from the NCBI website against the fraction of cells with foci at day 7 of growth in rich medium for all 97 protein kinases examined. A least-squares linear regression analysis was performed with the data (Minitab 16, Minitab Inc.), and no significant linear relationship was found.
Protein kinase foci defined eight different stationary-phase granules
The protein kinases recruited to cytoplasmic foci could be associated with either known RNP granules or novel cytoplasmic structures. To distinguish between these possibilities, we performed a series of colocalization experiments with mCh-tagged reporters for a variety of known granules. These experiments identified nine different localization patterns that corresponded to as many as eight distinct cytoplasmic granules (Table 2). Four of these latter structures had not been described previously. The majority of these colocalization studies were performed in minimal medium to select for the presence of the plasmid-borne mCh constructs. Although the relative fraction of cells with foci was generally similar to that observed in rich medium, the kinetics of foci appearance was accelerated. The following sections summarize the localization patterns observed with the different protein kinases.
Table 2. Localization patterns observed with foci-forming protein kinases.
| Class | Colocalization pattern | Protein kinases |
|---|---|---|
| I | With P-bodies | Cdc15, Hrr25, Tpk3, Ypk3 |
| II | With stress granules | Rio2 |
| III | With P-bodies and stress granules | Cbk1, Cdc28, Fus3, Pbs2 |
| IV | With actin bodies | Prk1 |
| V | With phagophore assembly site | Atg1 |
| VI | Novel-1 | Akl1, Frk1, Kin28 |
| VII | Novel-2 | Fpk1, Kss1 |
| VIII | Novel-3 | Hog1 |
| IX | Novel-4 | Ypl150w |
Protein kinases associated primarily with P-bodies:
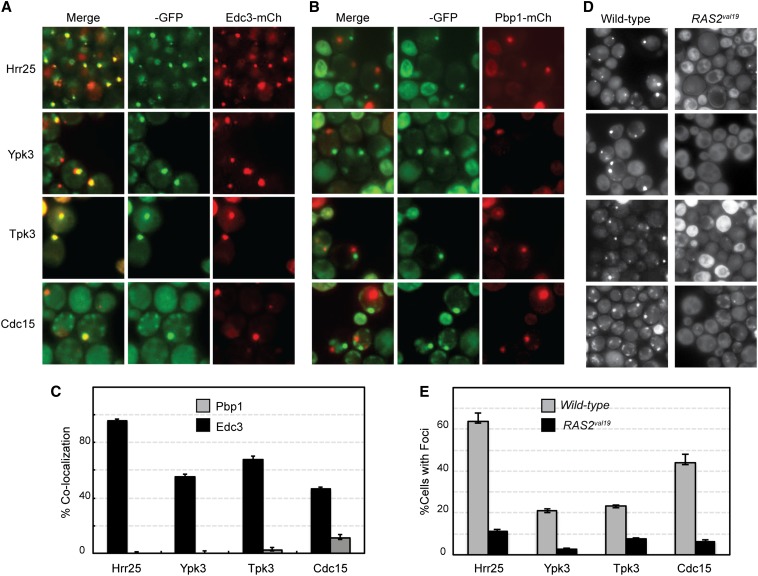
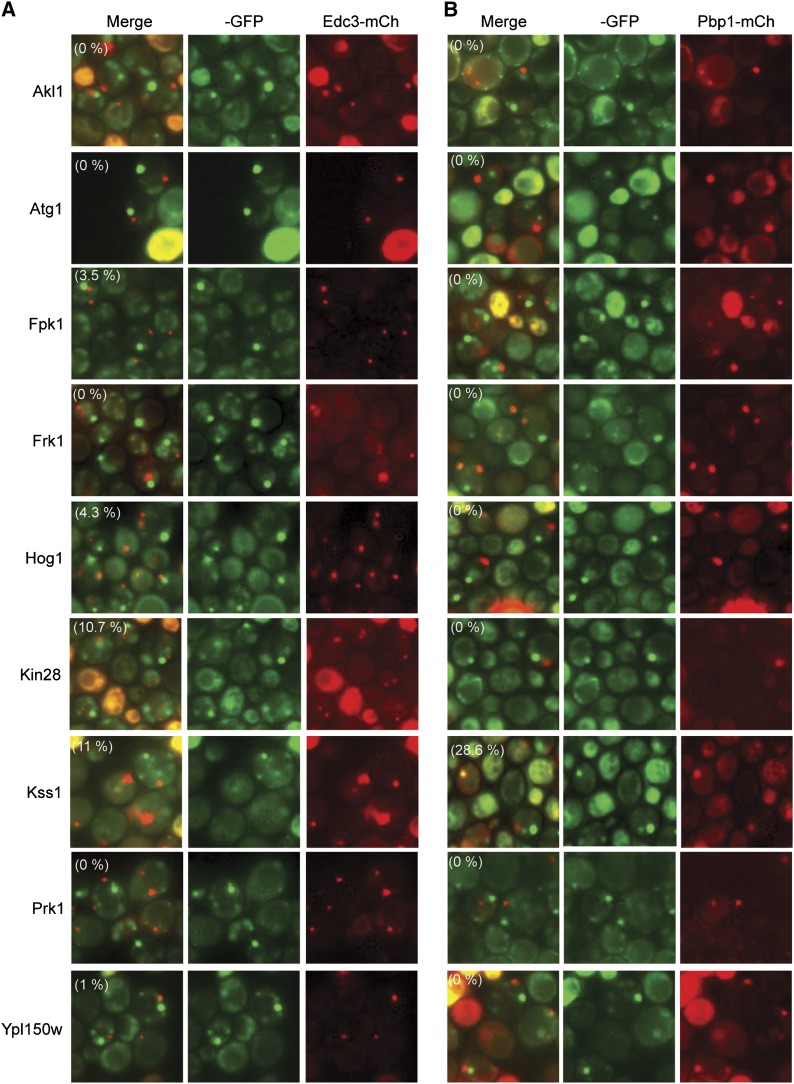
Four protein kinases, Cdc15, Hrr25, Tpk3, and Ypk3, showed significant colocalization with multiple P-body markers (Figure 5 and Figure S5, A and B). This result was expected for Tpk3 as this PKA subunit had been found previously to associate with P-body foci (Tudisca et al. 2010). In contrast, this localization had not been observed for any of the other three enzymes. Hrr25 is in the casein kinase I family of protein kinases and is most similar to the mammalian δ/ε enzymes (Demaggio et al. 1992; Fish et al. 1995). Cdc15 is involved in the mitotic exit network, and Ypk3 is an AGC family kinase that is distantly related to the mammalian serum- and glucocorticoid-inducible kinase SGK (Casamayor et al. 1999; Noton and Diffley 2000). The degree of colocalization observed here with the P-body marker Edc3 varied from >95% with Hrr25 to just under 50% for Cdc15 (Figure 5C). In contrast, no significant colocalization was detected with the stress granule marker Pbp1 for Hrr25, Tpk3, and Ypk3; ∼10% of the Cdc15 foci were found to colocalize with this stress granule reporter (Figure 5C). Previous work had indicated that P-body numbers are diminished in cells that lack Pat1, a critical scaffolding protein for P-body formation, or have elevated levels of PKA signaling (Ramachandran et al. 2011; Shah et al. 2013). PKA has been shown to inhibit P-body assembly in S. cerevisiae via the direct phosphorylation of Pat1 (Ramachandran et al. 2011). Consistent with the colocalization studies, we found that the relative number of foci for all four of these kinases was significantly diminished in cells that lacked Pat1 or contained the constitutively active RAS2val19 allele (Figure 5, D and E; Figure S5, C and D). The presence of the latter dominant allele results in elevated PKA levels and thus in decreased numbers of P-body foci. In all, the data indicated that these four protein kinases were recruited preferentially to P-body foci in quiescent cells.
Figure 5.
Four protein kinases were associated preferentially with P-bodies in stationary-phase cells. (A and B) The colocalization of the four GFP-tagged protein kinases, Hrr25, Ypk3, Tpk3, and Cdc15, with a reporter for P-bodies (Edc3-mCh) and stress granules (Pbp1-mCh). (C) Quantitation of the colocalization data. (D and E) The presence of the hyperactive RAS2val19 allele resulted in diminished numbers of protein kinase foci. Note that this analysis was performed in minimal medium to maintain selection for the RAS2val19 plasmid. The values shown in each graph represent the average of at least two independent experiments (n = 100).
Rio2 was recruited preferentially to stress granule foci:
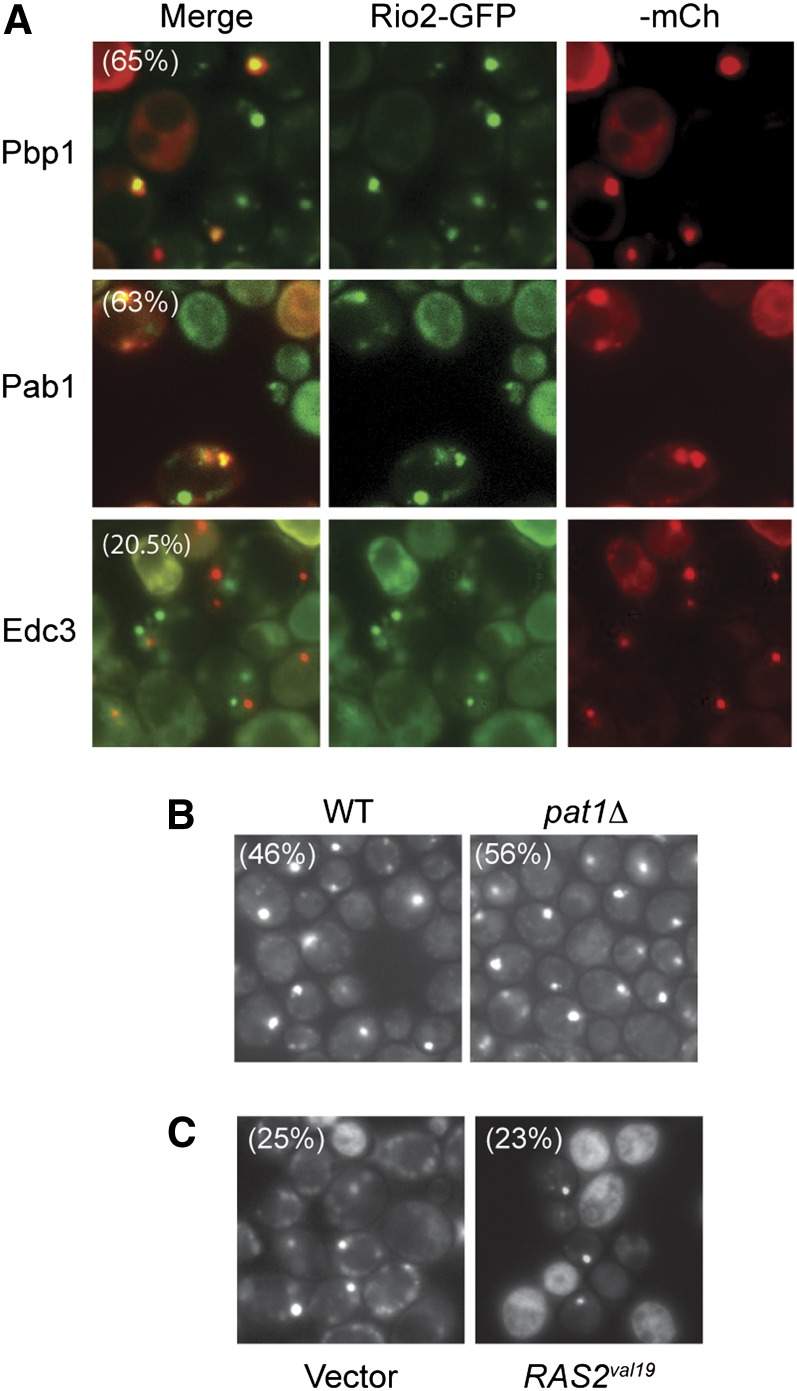
In contrast to the above kinases, Rio2 was found to colocalize with two mCh-tagged constituents of stress granules, Pbp1 and Pab1 (Figure 6A). Significantly less colocalization was observed with the P-body reporter Edc3. Consistent with these data, the number of Rio2-GFP foci was not dramatically affected by alterations that decrease P-body numbers (Figure 6, B and C). Therefore, Rio2 appeared to associate preferentially with stress granules in stationary-phase cells. Rio2 has been shown to have a conserved role in the regulation of protein translation by promoting the maturation of the 40S small ribosomal subunit (Laronde-Leblanc and Wlodawer 2005). Thus, Rio2 could be another translation factor recruited to stress granules perhaps to facilitate the downregulation of protein synthesis that occurs during this period of growth arrest.
Figure 6.
The Rio2 protein kinase was preferentially associated with stress granules in stationary-phase cells. (A) The colocalization of a Rio2-GFP fusion protein with reporters for either stress granules (Pbp1- and Pab1-mCh) or P-bodies (Edc3-mCh). The extent of co-localization is indicated by the percentage values in the merged panels for Pbp1 and Edc3. (B and C) The frequency of Rio2 foci was not affected by either the loss of the PAT1 locus (B) or the presence of the hyperactive RAS2val19 allele (C). The fraction of cells with Rio2-GFP foci in each experiment is indicated by the percentage values shown. The experiments shown in C were performed in minimal medium to maintain selection for the RAS2val19 plasmid.
Protein kinases present in both P-bodies and stress granules:
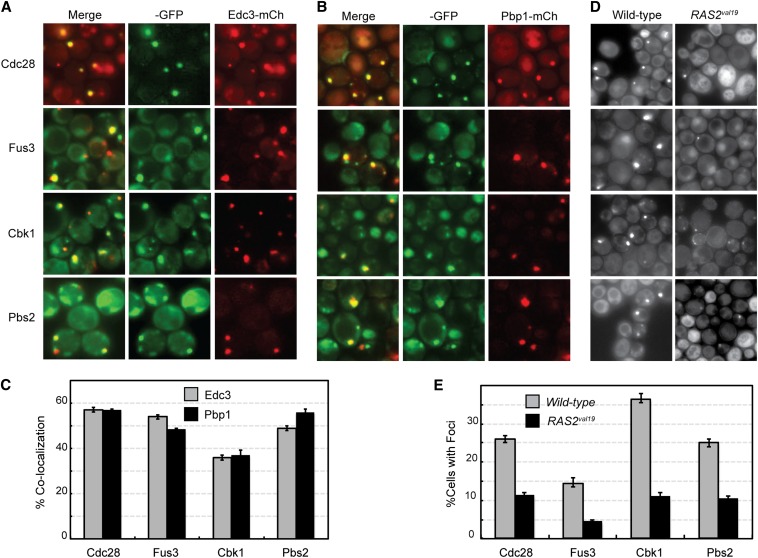
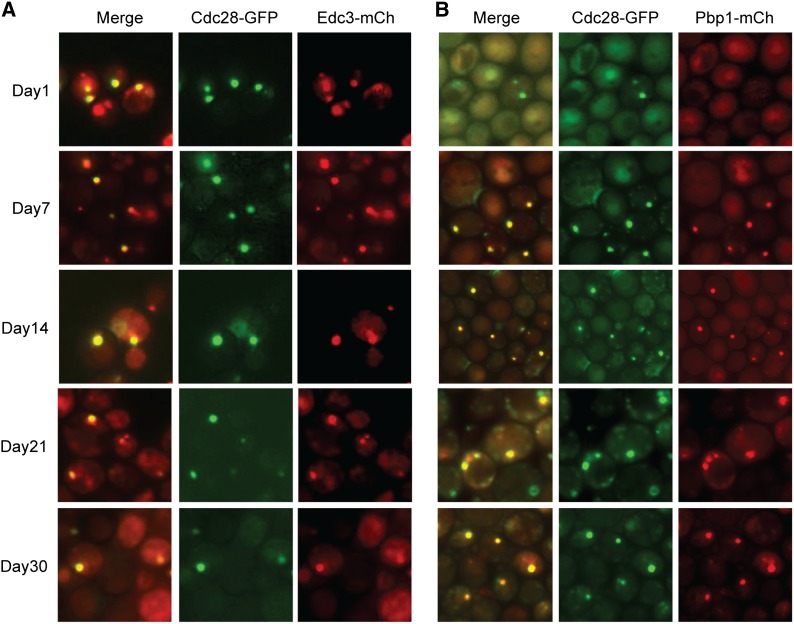
Although P-bodies and stress granules are distinct structures in stationary-phase cells (Liu et al. 2012; Shah et al. 2013), we found that four protein kinases were associated with both types of granule (Figure 7). These four enzymes, Cbk1, Cdc28, Fus3, and Pbs2, exhibited similar levels of colocalization with both P-body and stress granule reporters (Figure 7, A–C; Figure S6). Cdc28 is the major cyclin-dependent kinase involved in cell-cycle regulation in this budding yeast (Reed and Wittenberg 1990; Mendenhall and Hodge 1998). Although this enzyme has been studied extensively, its appearance in such cytoplasmic foci has not been reported previously. Cbk1 is an Ndr/LATS family kinase involved in the regulation of cell morphogenesis, and Fus3 and Pbs2 are members of two distinct MAP kinase cascades (Brewster et al. 1993; Bardwell et al. 1996; Bidlingmaier et al. 2001). Fus3 is a terminal MAP kinase in a cascade involved in cell mating, and Pbs2 is the MAP kinase kinase in a cascade important for the response to osmotic stress. The degree of colocalization observed with both types of granule was ∼50% for Cdc28, Fus3, and Pbs2 (Figure 7C). Consistent with the observed association with P-bodies, we found that the numbers of foci for each of these kinases was diminished by about one-half in cells that lacked Pat1 or contained the RAS2val19 allele (Figure 7, D and E; Figure S6, D and E). To test whether these associations for Cdc28 persisted throughout stationary phase, the chromosomal CDC28 locus was fused to GFP in strains with an integrated copy of either Edc3- or Pbp1-mCh. These cells were then grown in rich medium and imaged at the indicated times. These experiments clearly showed that Cdc28 was present in both P-bodies and stress granules at all times examined and indicated that these associations were not specific to a particular growth medium (Figure 8). It is important to reiterate that very little colocalization is observed between P-body and stress granule proteins in stationary-phase cells. For example, we have found <5% colocalization between the Edc3 and Pbp1 reporters at this time. As a result, the presence of the above four kinases in both types of granule was unexpected, and determining the underlying reasons for this shared localization could provide important insights into the biological functions of these physically distinct RNP structures.
Figure 7.
Protein kinases that were associated with both P-bodies and stress granules in stationary-phase cells. The colocalization of the four GFP-tagged protein kinases, Cdc28, Fus3, Cbk1, and Pbs2, with a reporter for either P-bodies (A, Edc3-mCh) or stress granules (B, Pbp1-mCh). (C) Quantitation of the colocalization data shown in A and B. (D) The frequency of foci formation was diminished by the presence of the hyperactive RAS2val19 allele. (E) Quantitation of the RAS2val19 effects on protein kinase foci formation.
Figure 8.
The colocalization of Cdc28 with either a P-body (A, Edc3-mCh) or a stress granule (B, Pbp1-mCh) reporter persisted throughout the stationary phase of growth. Representative images at the indicated days of growth in a rich medium are shown.
Prk1 was associated with actin bodies:
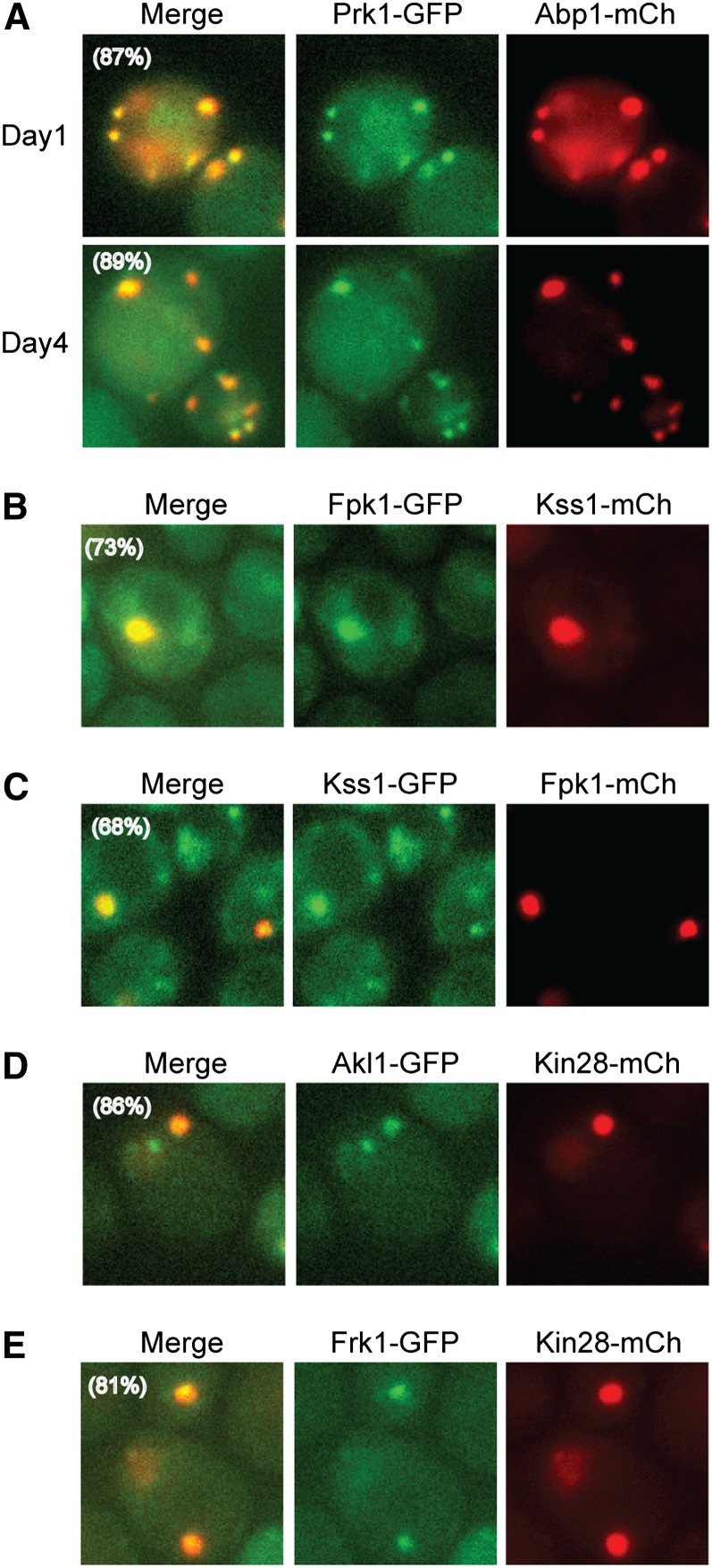
The remaining nine protein kinases did not colocalize significantly with reporters of either P-bodies or stress granules (Figure 9). In addition, when tested, the loss of Pat1 or the presence of RAS2val19 did not have a significant effect on the number of foci observed with these protein kinases (Figure S7; data not shown). Since Prk1 is known to associate with actin in dividing cells (Cope et al. 1999), we tested whether this enzyme might be present in actin bodies in stationary phase. Consistent with this possibility, Prk1 showed a striking colocalization with the actin-binding protein Abp1, which is known to be present in actin bodies (Figure 10A) (Sagot et al. 2006). A second foci-forming kinase, Akl1, is related in sequence to Prk1 and is also associated with the actin cytoskeleton in dividing cells (Smythe and Ayscough 2003). However, Akl1 was not found to colocalize with Abp1 or Prk1 and thus appears to be in a distinct structure in stationary-phase cells (see below). This latter result was not altogether unexpected as the Akl1- and Prk1-containing foci had a markedly different appearance (see Figure 1). Therefore, the Prk1 protein kinase appears to be associated specifically with the actin body structures that form upon S. cerevisiae growth arrest. It will be important to test whether this enzyme is required for either the assembly or disassembly of these actin-based storage granules.
Figure 9.
Foci-forming protein kinases that did not exhibit a significant degree of colocalization with either a P-body (A, Edc3-mCh) or a stress granule (B, Pbp1-mCh) reporter. Representative images for strains expressing the indicated protein kinase-GFP fusions are shown. The extent of colocalization for each protein kinase is indicated by the percentage value in parentheses in the merged image.
Figure 10.
Colocalization studies with protein kinases that were not found in either P-bodies or stress granules defined a number of novel cytoplasmic granules. Cells expressing the indicated reporter proteins were examined by fluorescence microscopy (n = 200) after the indicated days of growth in minimal medium. Representative images for the following protein combinations are shown: (A) Prk1 and Abp1; (B and C) Fpk1 and Kss1; (D) Akl1 and Kin28; and (E) Frk1 and Kin28.
Atg1 was the only protein kinase present at the autophagy-related phagophore assembly site:
The Atg1 protein kinase is a key regulator of autophagy that is recruited to a conserved cytoplasmic structure, the phagophore assembly site (PAS), under conditions that induce this degradative pathway (Nakatogawa et al. 2009). The PAS is the nucleation site from which the autophagosome, the primary transport intermediate in this pathway, emerges. In S. cerevisiae, there is typically one PAS punctae per cell that contains a number of different Atg proteins, including Atg1 (Suzuki and Ohsumi 2007; Xie and Klionsky 2007). Previous work demonstrated that autophagy is induced upon glucose deprivation and that Atg1 is present at the PAS under these conditions (Takeshige et al. 1992; Stephan et al. 2009; Stephan et al. 2010). Here, we found that Atg1 was present in cytoplasmic foci by the first day of culture but that there was no significant colocalization between these foci and either P-body or stress granule reporters nor any of the other foci-forming kinases (see Figure 9). Thus, Atg1 was the only protein kinase in this study that was associated with the PAS. In addition, the data here failed to identify a direct connection between this autophagy-related structure and either P-bodies or stress granules. This observation is interesting in light of recent observations suggesting that stress granules may be targeted by a specific autophagy-based turnover pathway (Buchan et al. 2013). However, the details of this “granulophagy” are still unclear, and our failure to see any colocalization between Atg1 and stress granules could be due simply to the fact that this degradative process is not operational under the growth conditions examined here. Finally, it is interesting to note that the fraction of cells with an Atg1 focus peaked at day 1 and that after this point the number of cells with a PAS steadily declined (Figure 1). Since the presence of this structure can serve as an indicator of an active autophagy pathway, the data are consistent with autophagy activity being highest in cells proceeding through the diauxic shift. Cells at this time are likely to be carrying out an extensive remodeling of cytoplasmic constituents to change from a fermentative to a respiratory mode of growth (Gray et al. 2004). The corollary from these observations is that autophagy activity might be lower in quiescent cells and therefore perhaps not as important then for cell survival.
Protein kinases localizing to novel cytoplasmic granules:
The remaining seven protein kinases appear to be present in cytoplasmic foci that are not coincident with P-bodies, stress granules, actin bodies, or proteasomal storage granules; the Scl1 protein was used as the reporter for the latter structures (Laporte et al. 2008). To assess whether these enzymes were in separate or shared granules, we performed further colocalization experiments with mCh-tagged derivatives of Fpk1, Kss1, and Kin28. These experiments indicated that the remaining kinases could be separated into the following groups: (1) Kin28, Akl1, and Frk1; (2) Kss1 and Fpk1; (3) Hog1; and (4) Ypl150w (Figure 10 and Table 2). Since we were unable to generate a stable mCherry fusion with either Hog1 or Ypl150w, the assignment of these two kinases into separate groups is still tentative. For now, they have been placed into separate classes based on subtle differences in foci morphology. It is not yet clear why the enzymes in the first two groups were found in the same structures nor what the physiological roles of each granule might be in the stationary-phase cell. For example, in the first grouping, Kin28 is a subunit of the TFIIH general transcription factor, Akl1 is associated with the actin cytoskeleton, and the function of Frk1 is unknown (Feaver et al. 1994; Smythe and Ayscough 2003). Nonetheless, this work highlights the fact that there are multiple distinct RNP (or protein-only) granules that coexist in quiescent cells and suggests that there are likely to be additional structures that have yet to be identified. How the formation of each of these structures is regulated and how the structures are able to maintain their distinct identities within the same cytoplasmic space are important questions that remain to be answered.
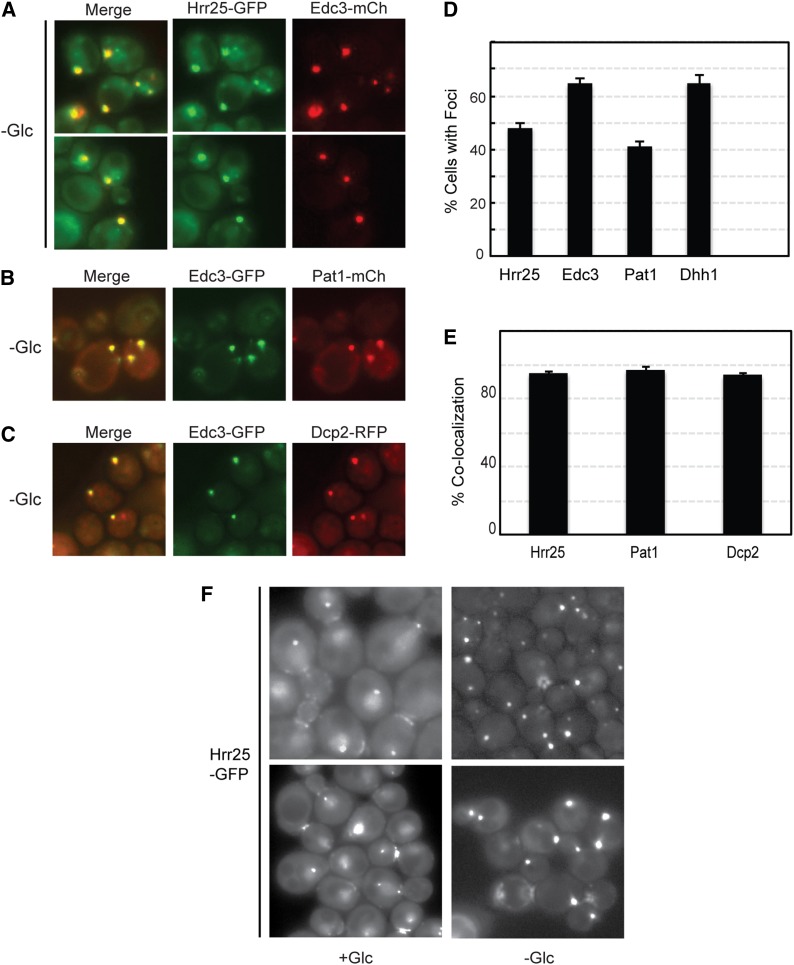
Hrr25 rapidly localizes to P-body foci upon an acute glucose deprivation
The Hrr25 protein was analyzed further as this kinase exhibited the highest level of colocalization with P-body reporters. In yeast, P-bodies are most commonly induced by an acute starvation for glucose, and we tested here whether Hrr25 was recruited to these RNP structures under these conditions (Nissan and Parker 2008). For these experiments, log-phase cells were transferred to a medium that lacks glucose for 15 min and were examined by fluorescence microscopy. In dividing cells, Hrr25 was located in nuclei at the spindle pole bodies (SPBs) and at the bud neck as reported previously (Lusk et al. 2007). However, after glucose starvation, this protein kinase was also observed in cytoplasmic foci that were coincident with P-bodies (Figure 11A). These latter foci were not associated with the nuclear compartment and thus were distinct from the SPB; the nucleus was labeled with a histone H2B-RFP reporter in these experiments (Figure S8). The level of colocalization between Hrr25 and the different P-body markers was as high as that observed between the P-body reporters themselves (Figure 11, B–E). Therefore, Hrr25 was by this measure a bona fide constituent of P-body granules. Interestingly, the glucose starvation also resulted in a rapid decrease in the level of Hrr25 at its normal locales in dividing cells (Figure 11F). For example, almost 70% of the cells in log phase exhibited strong Hrr25 staining in the nucleus and/or at the bud neck. This number dropped to ∼10% after 15 min of glucose starvation. These data are thus consistent with Hrr25 performing a specific function in P-body granules and/or in the relocalization, resulting in diminished levels of Hrr25 activity at its usual sites of action. The generation of a Hrr25 variant that retains its essential activities during mitotic growth but fails to be localized to P-bodies would help us to discern between these two possibilities.
Figure 11.
Hrr25 was efficiently recruited to P-body foci after a brief period of glucose starvation. (A) Yeast cells expressing the Hrr25-GFP fusion protein were grown to log phase and then transferred for 15 min to a medium that lacked glucose. Two representative images of the colocalization observed between Hrr25 and the Edc3-mCh reporter are shown. (B and C) Fluorescence microscopy images showing the colocalization observed between Edc3-GFP and either Pat1-mCh (B) or Dcp2-RFP (C) after a 15-min starvation for glucose. (D) Hrr25 formed cytoplasmic foci at a frequency similar to that observed for known P-body reporters. The foci were induced by a 15-min starvation for glucose as described in A. The graph values represent the quantitation of the microscopy data shown in A–C and are the average of at least two independent experiments (n = 100). (E) Hrr25 displayed a high level of colocalization with the P-body reporter Edc3. Quantitation of the colocalization data presented in A–C; the bars represent the degree of colocalization observed between Edc3 and the indicated proteins. (F) The intracellular localization of Hrr25 was altered by an acute starvation for glucose. Cells expressing a Hrr25-GFP fusion protein were examined by fluorescence microscopy before (+Glc) and after a 15-min starvation for glucose (−Glc). Two representative images are shown for each.
This rapid recruitment to cytoplasmic foci upon glucose deprivation was not observed with any of the other 17 protein kinases examined, including those that were found at P-bodies in stationary-phase cells (data not shown). Of the latter, only Tpk3 showed any significant relocalization to cytoplasmic foci after a 15-min glucose starvation; however, in this case, typically <5% of the cells contained a Tpk3 focus. These studies therefore suggest that distinct mechanisms govern the recruitment of different enzymes to foci and emphasize the potential importance of Hrr25 activity for P-body-related functions.
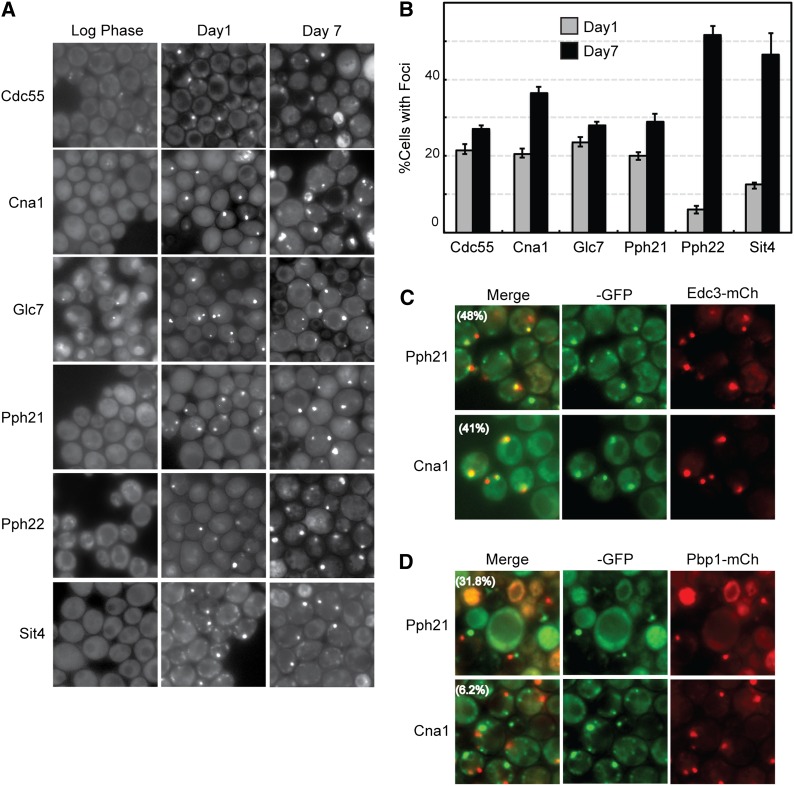
Protein phosphatases also relocate to cytoplasmic foci during stationary-phase entry
To ensure that our observations with protein kinases were not specific to this class of signaling molecule, we also examined the subcellular localization of a set of S. cerevisiae protein phosphatases. Thirty of these enzymes were available in the collection of GFP fusions, and the intracellular location of each was assessed at the same three points of growth. This analysis revealed that six of these phosphatases were in cytoplasmic foci in >25% of the cells at day 7 (Figure 12, A and B). The fraction of phosphatases found in foci was very similar to that observed for the set of protein kinases tested and with other studies of protein localization in stationary-phase cells. As with the protein kinases, no correlation was found between the presence of an IDD and the frequency with which a given phosphatase was associated with a cytoplasmic focus (data not shown). Finally, particular protein phosphatases were also found to be localized to specific RNP granules. For example, Cna1, the catalytic subunit of calcineurin (Garrett-Engele et al. 1995), was associated preferentially with P-body foci (Figure 12, C and D). This association appears to be evolutionarily conserved as this same protein has been found in P-bodies in the human fungal pathogen Cryptococcus neoformans (Kozubowski et al. 2011). In contrast, Pph21, a catalytic subunit of the protein phosphatase 2A (Sneddon et al. 1990), was found to colocalize with both P-body and stress granule reporters (Figure 12, C and D). In all, these data were consistent with those presented above for protein kinases and support the contention that signaling proteins are sorted to a variety of cytoplasmic granules in stationary-phase cells.
Figure 12.
Particular S. cerevisiae protein phosphatases are also recruited to cytoplasmic foci in stationary-phase cells. (A) Cells expressing the indicated protein phosphatase-GFP fusions were examined by fluorescence microscopy during log phase and after 1 or 7 days of growth in rich medium. (B) Quantitation of the microscopy data shown in A. (C and D) The colocalization of the protein phosphatase subunits Pph21 and Cna1 with a reporter for P-bodies (C, Edc3-mCh) or stress granules (D, Pbp1-mCh). The extent of colocalization is indicated by the percentage value in parentheses in the merged images.
Discussion
In this report, we examined protein localization in yeast cells that were entering the quiescent, G0-like, stationary phase of growth. The impetus for these studies were observations indicating that particular proteins relocate to discrete cytoplasmic foci during this period of quiescence. Our goal was to examine this phenomenon further to gain insight into the nature of these cytoplasmic granules and whether their formation was a general and physiologically relevant phenomenon. We focused here on a particular set of key signaling molecules, the protein kinases of the budding yeast S. cerevisiae. These proteins were examined by microscopy at different stages of growth, and a significant fraction, almost one-fifth of those tested, were found to be recruited to distinct cytoplasmic foci in >30% of the cells in stationary-phase cultures. These cytoplasmic structures rapidly disassembled upon the resumption of growth, and their presence was correlated with cell viability. In all, the data here were consistent with these cytoplasmic granules being physiologically relevant structures that could play a role in the long-term survival of quiescent cells.
This study extended our understanding of this protein reassortment in stationary phase in two important ways. First, it identified new constituents of two well-characterized RNP granules, the P-body and stress granule. This observation is significant as the biological activities associated with a particular RNP granule are likely determined by the proteins resident within. For example, P-bodies in certain virus-infected cells have been found to contain viral proteins and to be required for efficient virus genome replication (Reineke and Lloyd 2013). Determining the protein constituents could therefore provide insight into the physiological role of a particular granule. In this study, we found that four protein kinases were recruited preferentially to P-bodies and that another was associated primarily with stress granules. Four additional protein kinases were found to be present in both P-bodies and stress granules. Only one of these enzymes, the Tpk3 subunit of PKA, was known previously to be associated with these RNP granules (Tudisca et al. 2010). The presence of these enzymes suggests that they might target specific proteins within these granules, and it will be important to determine whether they influence either the assembly or disassembly of these different structures. Finally, given that this study examined only a fraction of the proteome, it is very likely that there are many additional proteins in these RNP granules that remain to be identified. Characterizing these components will be essential for a full description of the activities associated with these cytoplasmic structures.
The second and perhaps more significant finding is the recognition that a variety of distinct RNP (and perhaps protein-only) granules coexist within the cytoplasm of a quiescent cell. This point was demonstrated here by a series of colocalization studies that defined as many as eight different cytoplasmic foci, including four that had not been identified previously. These observations suggest that specific mechanisms are in place in stationary-phase cells to ensure that these proteins are delivered to the appropriate sites in the cytoplasm. The foci-forming proteins were not simply aggregating at a number of different but nonspecific sites in the cytoplasm nor were all of them recruited to a solitary location for misfolded protein. Instead, the data suggest that the cytoplasm of a quiescent cell is subdivided into a rather large number of different functional domains. Although the global significance of this partitioning is not yet clear, it is important to note that the presence of at least one of these structures, the P-body, is necessary for the long-term survival of stationary-phase cells (Ramachandran et al. 2011). It seems very likely that at least some of the other granules present at this time will be important as well for this survival. The identification of mutants defective for the formation of a particular granule would allow for a direct test of this possibility.
Despite the insights gained here, many important questions remain about this redistribution of protein that appears to occur in stationary-phase cells. For example, it is not known what specific events trigger the formation of the different foci and what signaling pathways ultimately regulate granule assembly and dissolution. Although PKA signaling has been shown to have a key role in regulating P-body formation (Ramachandran et al. 2011; Shah et al. 2013), this pathway does not seem to influence any of the other cytoplasmic foci identified here. Certainly a key question is why the proteins examined here localize to granules specifically in stationary-phase cells. This localization for the most part did not occur in response to an acute, short-term starvation for glucose. This is especially interesting for those proteins that are found in P-bodies and stress granules, two RNP structures that are known to form under this starvation condition. The ultimate trigger could be related to the decrease in overall energy charge associated with a stationary-phase arrest or to a change in the levels of a specific metabolite. It will be equally important to determine why only particular proteins are recruited to these different cytoplasmic sites. This specificity is consistent with this recruitment being a regulated event and with these structures being physiologically relevant in the resting cell. There are clearly a number of possible reasons for the presence in a focus including that these polypeptides are being stored to be available when growth resumes. The existence of significant stores of key essential proteins could facilitate a timely re-entry into the cell cycle. Alternatively, these proteins could be recruited to specifically target, and thus regulate, other granule constituents or to effectively lower their concentration at their normal cellular sites of action. In the latter case, the recruitment to the granule would result in the downregulation of any normal activities associated with that protein. Understanding the precise contributions of each of these different granules is therefore likely to reveal new insights into the biology of the quiescent cell and the ways in which order can be established within the cytoplasmic milieu.
Supplementary Material
Acknowledgments
We thank Anita Hopper, James Hopper, Roy Parker, Ted Powers, and Claudio de Virgilio for strains and plasmids used in this study; Katie Gressel, Sanandan Malhotra, and Benita Wu for technical assistance; Qian Shi for assistance with statistical analyses; Jian-Qiu Wu for the use of his fluorescence microscope in the early stages of this work; and members of the Herman lab for many thoughtful discussions and valuable comments on the manuscript. This work was supported by research grants GM101191 and GM65227 from the National Institutes of Health (to P.K.H.) and by a graduate student fellowship from the Pelotonia Fellowship Program (to K.H.S.)
Footnotes
Communicating editor: J. Heitman
Literature Cited
- Allen C., Buttner S., Aragon A. D., Thomas J. A., Meirelles O., et al. , 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Kumar R., Sheets E. D., Benkovic S. J., 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320: 103–106. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10: 430–436. [DOI] [PubMed] [Google Scholar]
- Aragon A. D., Rodriguez A. L., Meirelles O., Roy S., Davidson G. S., et al. , 2008. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell 19: 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V., Parker R., 2009. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L., Cook J. G., Chang E. C., Cairns B. R., Thorner J., 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16: 3637–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V. I., Scherthan H., Solinger J. A., Buerstedde J. M., Heyer W. D., 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C. J., Parker R., 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3: 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W., 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71: 513–521. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S., Weiss E. L., Seidel C., Drubin D. G., Snyder M., 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., et al. , 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C., 1993. An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763. [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Kolaitis R. M., Taylor J. P., Parker R., 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Hama H., DeWald D. B., Herman P. K., 2002. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 277: 287–294. [DOI] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K., 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102: 13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R., 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9: 186–197. [DOI] [PubMed] [Google Scholar]
- Chang Y. W., Howard S. C., Herman P. K., 2004. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15: 107–116. [DOI] [PubMed] [Google Scholar]
- Chen B., Retzlaff M., Roos T., Frydman J., 2011. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3: a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope M. J., Yang S., Shang C., Drubin D. G., 1999. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 144: 1203–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B., 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R., 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaggio A. J., Lindberg R. A., Hunter T., Hoekstra M. F., 1992. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc. Natl. Acad. Sci. USA 89: 7008–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff S. J., Howard S. C., Hester A., Warner S., Herman P. K., 2006. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics 173: 1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradovic Z., 2002. Intrinsic disorder and protein function. Biochemistry 41: 6573–6582. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E., 2007a P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8: 9–22. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E., 2007b P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27: 3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W. J., Svejstrup J. Q., Henry N. L., Kornberg R. D., 1994. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Fish K. J., Cegielska A., Getman M. E., Landes G. M., Virshup D. M., 1995. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J. Biol. Chem. 270: 14875–14883. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele P., Moilanen B., Cyert M. S., 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15: 4103–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., et al. , 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15: 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., et al. , 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68: 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P. K., 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5: 602–607. [DOI] [PubMed] [Google Scholar]
- Howard S. C., Chang Y. W., Budovskaya Y. V., Herman P. K., 2001. The Ras/PKA signaling pathway of Saccharomyces cerevisiae exhibits a functional interaction with the Sin4p complex of the RNA polymerase II holoenzyme. Genetics 159: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S. C., Budovskaya Y. V., Chang Y. W., Herman P. K., 2002. The C-terminal domain of the largest subunit of RNA polymerase II is required for stationary phase entry and functionally interacts with the Ras/PKA signaling pathway. J. Biol. Chem. 277: 19488–19497. [DOI] [PubMed] [Google Scholar]
- Jonas S., Izaurralde E., 2013. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27: 2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J., 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kato M., Han T. W., Xie S., Shi K., Du X., et al. , 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P., 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., Anderson P., 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B. K., 2008. The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J. Intern. Med. 263: 142–152. [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Aboobakar E. F., Cardenas M. E., Heitman J., 2011. Calcineurin colocalizes with P-bodies and stress granules during thermal stress in Cryptococcus neoformans. Eukaryot. Cell 10: 1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B., Sagot I., 2008. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N., Wlodawer A., 2005. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim. Biophys. Acta 1754: 14–24. [DOI] [PubMed] [Google Scholar]
- Leatherman J. L., Jongens T. A., 2003. Transcriptional silencing and translational control: key features of early germline development. BioEssays 25: 326–335. [DOI] [PubMed] [Google Scholar]
- Liu I. C., Chiu S. W., Lee H. Y., Leu J. Y., 2012. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell 23: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R., 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Waller D. D., Makhnevych T., Dienemann A., Whiteway M., et al. , 2007. Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic 8: 647–660. [DOI] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Alberti S., 2013. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim. Biophys. Acta 1834: 918–931. [DOI] [PubMed] [Google Scholar]
- Mendenhall M. D., Hodge A. E., 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y., 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10: 458–467. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O’Connell J. D., et al. , 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106: 10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T., Parker R., 2008. Analyzing P-bodies in Saccharomyces cerevisiae. Methods Enzymol. 448: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C., Sato B. K., Broyer R. M., Wilhelm J. E., 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton E., Diffley J. F., 2000. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell 5: 85–95. [DOI] [PubMed] [Google Scholar]
- Ocampo A., Barrientos A., 2011. Quick and reliable assessment of chronological life span in yeast cell populations by flow cytometry. Mech. Ageing Dev. 132: 315–323. [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Herman P. K., 2010. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 187: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K., 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Wittenberg C., 1990. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 5697–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M. A., Alexander R. D., Spiller M. P., Beggs J. D., 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121: 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Lloyd R. E., 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., et al. , 2001. Sequence complexity of disordered protein. Proteins 42: 38–48. [DOI] [PubMed] [Google Scholar]
- Sagot I., Pinson B., Salin B., Daignan-Fornier B., 2006. Actin bodies in yeast quiescent cells: An immediately available actin reserve? Mol. Biol. Cell 17: 4645–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K. H., Zhang B., Ramachandran V., Herman P. K., 2013. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193: 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R., 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E., Ayscough K. R., 2003. The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. EMBO Rep. 4: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon A. A., Cohen P. T., Stark M. J., 1990. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 9: 4339–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K., 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 106: 17049–17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K., 2010. The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 6: 294–295. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P., 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7: 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y., 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581: 2156–2161. [DOI] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D., 2005. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232: 593–608. [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y., 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119: 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. G., Loschi M., Desbats M. A., Boccaccio G. L., 2011. RNA granules: the good, the bad and the ugly. Cell. Signal. 23: 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T., Liu N., Arkov A., Lehmann R., Lasko P., 2008. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 125: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y., 1998. The green fluorescent protein. Annu. Rev. Biochem. 67: 509–544. [DOI] [PubMed] [Google Scholar]
- Tudisca V., Recouvreux V., Moreno S., Boy-Marcotte E., Jacquet M., et al. , 2010. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur. J. Cell Biol. 89: 339–348. [DOI] [PubMed] [Google Scholar]
- van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., et al. , 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. C., Brangwynne C. P., 2012. Getting RNA and protein in phase. Cell 149: 1188–1191. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M., Braun E., Johnston G. C., Singer R. A., 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57: 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J. C., Federhen S., 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266: 554–571. [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J., 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Anderson P., 2008. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. Y., Wrasman K., Herman P. K., 2010. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.