Abstract
The oocytes of most sexually reproducing animals arrest in meiotic prophase I. Oocyte growth, which occurs during this period of arrest, enables oocytes to acquire the cytoplasmic components needed to produce healthy progeny and to gain competence to complete meiosis. In the nematode Caenorhabditis elegans, the major sperm protein hormone promotes meiotic resumption (also called meiotic maturation) and the cytoplasmic flows that drive oocyte growth. Prior work established that two related TIS11 zinc-finger RNA-binding proteins, OMA-1 and OMA-2, are redundantly required for normal oocyte growth and meiotic maturation. We affinity purified OMA-1 and identified associated mRNAs and proteins using genome-wide expression data and mass spectrometry, respectively. As a class, mRNAs enriched in OMA-1 ribonucleoprotein particles (OMA RNPs) have reproductive functions. Several of these mRNAs were tested and found to be targets of OMA-1/2-mediated translational repression, dependent on sequences in their 3′-untranslated regions (3′-UTRs). Consistent with a major role for OMA-1 and OMA-2 in regulating translation, OMA-1-associated proteins include translational repressors and activators, and some of these proteins bind directly to OMA-1 in yeast two-hybrid assays, including OMA-2. We show that the highly conserved TRIM-NHL protein LIN-41 is an OMA-1-associated protein, which also represses the translation of several OMA-1/2 target mRNAs. In the accompanying article in this issue, we show that LIN-41 prevents meiotic maturation and promotes oocyte growth in opposition to OMA-1/2. Taken together, these data support a model in which the conserved regulators of mRNA translation LIN-41 and OMA-1/2 coordinately control oocyte growth and the proper spatial and temporal execution of the meiotic maturation decision.
Keywords: oocyte meiotic maturation, oocyte growth, translational regulation, RNA-binding proteins, ribonucleoprotein particle purification
MEIOSIS ensures that the embryo inherits a proper genome (reviewed by Page and Hawley 2003; Bhalla and Dernburg 2008), whereas inheritance of the oocyte cytoplasm and its cellular organelles enables that genome to function (reviewed by Houston 2013). The oocytes of most sexually reproducing animals arrest in the diplotene or diakinesis stage of meiotic prophase I (reviewed by Masui and Clarke 1979; Downs 2010; Kim et al. 2013). Oocyte growth, which occurs during this period of arrest, enables oocytes to acquire the cytoplasmic components needed to produce healthy progeny and to gain competence to complete meiosis. Oocyte meiotic arrest is an ancient reproductive strategy and many of its molecular underpinnings are deeply conserved in evolution. Meiotic resumption (also called meiotic maturation) involves the transition to metaphase I (M phase), which is triggered by maturation-promoting factor (MPF) (Masui and Markert 1971; reviewed by Masui 2001). MPF consists of the Cdk1 catalytic subunit and the cyclin B regulatory subunit (Dunphy et al. 1988; Gautier et al. 1988, 1990; Lohka et al. 1988; reviewed by Nurse 1990). Species-specific hormonal signals and soma–germline interactions regulate oocyte meiotic maturation. A failure of oocytes to undergo meiotic maturation results in infertility, whereas improper execution of the meiotic divisions causes aneuploidy (reviewed by Nagaoka et al. 2012). The timing of meiotic maturation also is crucial. If oocytes undergo meiotic maturation prior to completing the growth process, their capacity to produce healthy offspring is diminished.
Active MPF phosphorylates substrates that function in the cellular processes of meiotic maturation including nuclear envelope breakdown, chromosome condensation, and meiotic spindle assembly. By contrast, the regulation of cytoplasmic events of oocyte meiotic maturation, which include rearrangement of the cytoskeleton, redistribution of cellular organelles, and post-translational modifications, are comparatively less well understood (reviewed by Li and Albertini 2013; Mao et al. 2014). Because the full-grown oocytes of most animals are transcriptionally quiescent, translational regulation is a major control point (reviewed by Kong and Lasko 2012).
Most animal oocytes store mRNAs that are translated upon meiotic resumption or after fertilization. Translation of key regulators promotes meiotic progression in response to hormonal stimulation (Sagata et al. 1988; Ferby et al. 1999; Lenormand et al. 1999; Hochegger et al. 2001; Haccard and Jessus 2006; Chen et al. 2011). Other classes of maternal mRNAs remain repressed until the oocyte-to-embryo transition (reviewed by Li et al. 2010; Robertson and Lin 2013). Studies in several systems provide a paradigm for the translational regulation of oogenesis and meiotic maturation (Kadyk and Kimble 1998; Brent et al. 2000; Wang et al. 2002; Barnard et al. 2004; Benoit et al. 2008; Cui et al. 2008, 2013). Repressed mRNAs possess short poly(A) tails and bind proteins that exclude translation initiation factors. In Xenopus, progesterone triggers Cdk1-dependent phosphorylation of the cytoplasmic polyadenylation element binding protein, which activates the GLD-2 cytoplasmic poly(A) polymerase to promote translation (see Ivshina et al. 2014 for a review). These studies highlight the importance of translational regulation in oogenesis, and they suggest these mechanisms might drive the oogenic program through the coordinate control of key reproductive mRNAs. A challenge is to identify the battery of regulated mRNAs, discern their roles in promoting and integrating oocyte growth and meiotic progression, and elucidate their regulatory modes.
In this and the accompanying article in this issue (Spike et al. 2014), we address how conserved regulators of mRNA translation coordinately control oocyte growth and the proper spatial and temporal execution of the meiotic maturation decision in the nematode Caenorhabditis elegans. Sexual development of C. elegans depends on the ratio of X chromosomes to autosomes—diploid animals with two X chromosomes are hermaphrodites, whereas those with a single X are males (Brenner 1974; Madl and Herman 1979; Farboud et al. 2013). The self-fertile hermaphrodite (Figure 1) produces sperm before switching to oogenesis as an adult (sperm-to-oocyte switch). Feminizing mutations block sperm production, resulting in self-sterility (Ellis and Schedl 2007). In the absence of sperm, oocytes arrest in diakinesis (McCarter et al. 1999). After insemination, meiotic maturation and fertilization occur in an assembly-line fashion, though meiotic maturation rates decline when sperm becomes limiting. Oocytes develop in close association with the gonadal sheath cells, follicle-like cells that regulate meiotic maturation (Greenstein et al. 1994; McCarter et al. 1997; Miller et al. 2003; Govindan et al. 2006, 2009; Kim et al. 2012; Starich et al. 2014). C. elegans sperm utilize the major sperm protein (MSP) as an unconventionally secreted hormone to promote meiotic maturation (Miller et al. 2001; Kosinski et al. 2005). The sheath cells function as the main MSP sensors. Protein kinase A (PKA) signaling in the sheath cells is required for all MSP responses in the germ line (Govindan et al. 2006, 2009; Kim et al. 2012), which include activation of MPK-1 mitogen-activated protein kinase (Miller et al. 2001; Lee et al. 2007; Arur et al. 2009), rearrangement of the oocyte microtubule cytoskeleton (Harris et al. 2006), localization of the AIR-2 Aurora B kinases to oocyte chromatin (Schumacher et al. 1998; Govindan et al. 2009), reorganization of oocyte RNPs (Schisa et al. 2001; Jud et al. 2008), and the stimulation of the actomyosin-dependent cytoplasmic flows that drive oocyte growth (Wolke et al. 2007; Govindan et al. 2009; Nadarajan et al. 2009; Figure 1). In turn, the sheath cells form gap junctions with oocytes (Hall et al. 1999; Starich et al. 2014). Sheath–oocyte gap junctions function as negative regulators of the MSP response (Govindan et al. 2006, 2009; Whitten and Miller 2007; Starich et al. 2014) and are needed for the oocyte growth-promoting cytoplasmic flows to cease in the absence of MSP (Nadarajan et al. 2009).
Figure 1.
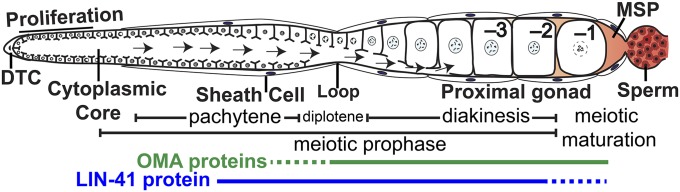
Adult hermaphrodite gonad arm: DTC, distal tip cell; –1 to –3, proximal oocytes; arrows, cytoplasmic flow for oocyte growth. The –1 oocyte undergoes meiotic maturation in response to MSP secreted from sperm in a process that requires the redundant function of OMA-1 and OMA-2 (OMA proteins). The expression patterns of the OMA proteins (Detwiler et al. 2001; Lee and Schedl 2004) and LIN-41 (Spike et al. 2014) are indicated.
The function of cytoplasmic RNPs appears to represent a key downstream target of MSP signaling. In the absence of MSP, large RNP foci condense in oocytes, and MSP signaling results in dynamic RNP decondensation (Schisa et al. 2001; Jud et al. 2008; Hubstenberger, et al. 2013). Genetic analysis revealed the SACY-1 DEAD-box RNA helicase as a strong negative regulator of meiotic maturation (Kim et al. 2012), which functions in oocytes downstream of somatic PKA signaling but upstream of the TIS11 zinc-finger RNA-binding proteins OMA-1 and OMA-2 (hereafter referred together as the OMA proteins). The OMA proteins are redundantly required for oocyte meiotic maturation (Detwiler et al. 2001). In oma double mutants, multiple readouts of MSP signaling are defective (Detwiler et al. 2001) and oocytes grow abnormally large because they receive sustained low rates of MSP-dependent cytoplasmic flows (Detwiler et al. 2001; Govindan et al. 2009). Consistent with an essential role in transducing the MSP signal, the OMA proteins function upstream of the conserved cell cycle regulator CDK-1 (Detwiler et al. 2001). The OMA proteins localize to the cytoplasm, bind the 3′-untranslated regions (3′-UTRs) of nos-2, zif-1, glp-1, and mom-2 mRNAs, and repress their translation (Detwiler et al. 2001; Jadhav et al. 2008; Guven-Ozkan et al. 2010; Kaymak and Ryder 2013; Oldenbroek et al. 2013). Repression of zif-1 and mom-2 in oocytes also requires the eIF4E-binding protein IFET-1 (Guven-Ozkan et al. 2010; Oldenbroek et al. 2013). nos-2, zif-1, glp-1, and mom-2 are not required for meiotic maturation, yet their regulation suggests a general function for OMA proteins in controlling translation in oocytes.
Here we purify OMA-1 ribonucleoprotein particles (OMA RNPs) and define many of their mRNA and protein components. As a class, mRNAs enriched in OMA RNPs have reproductive functions. Several mRNAs enriched in OMA RNPs were tested and found to be targets of OMA-mediated translational repression, dependent on sequences in their 3′-UTRs. Consistent with a major role in regulating translation, OMA RNP protein components include translational repressors and activators. Cardinal among OMA RNP components is the highly conserved TRIM-NHL RNA-binding protein LIN-41, which also represses several OMA target mRNAs. In the accompanying article (Spike et al. 2014), we show that LIN-41 and the OMA proteins exhibit an antagonistic relationship—LIN-41 inhibits M-phase entry and oocyte cellularization, whereas the OMA proteins promote these events. Taken together, these studies reveal the OMA RNP as a master regulator of the oogenic program that coordinates and controls oocyte growth and meiotic maturation.
Materials and Methods
Strains
The genotypes of strains used in this study are reported in Supporting Information, Table S1. The following mutations were used: LGI: fog-1(q253ts), unc-13(e1091), lin-41(n2914), lin-41(ma104), lin-41(tn1487ts), fog-3(q470), spe-9(hc88ts); LGIII: cdc-25.3(ok358), unc-119(ed3); LGIV: oma-1(zu405te33); LGV: acy-4(ok1806), oma-2(te51), rnp-1(ok1549), fog-2(q71), and fog-2(oz40). The following rearrangements were used: hT2[bli-4(e937) let-?(q782) qIs48] (I; III) and nT1[qIs51] (IV; V). The following transgene insertions were used: teIs1[pRL475 oma-1p::oma-1::gfp, pDPMM016 unc-119(+)], tnIs17[pCS410 oma-1p::oma-1::s::tev::gfp, pDPMM0016B unc-119(+)], teIs114[pRL2701 pie-1p::gfp::h2b::zif-1 3′UTR, unc-119(+)], tnIs36[pCS450 pie-1p::gfp::h2b::cdc-25.3 3′UTR, unc-119(+)], tnIs48[pCS450 pie-1p::gfp::h2b::cdc-25.3 3′UTR, unc-119(+)], tnIs53[pCS456 pie-1p::gfp::h2b::rnf-5 3′UTR, unc-119(+)], tnIs54[pCS456 pie-1p::gfp::h2b::rnf-5 3′UTR, unc-119(+)], tnIs57[pCS458 pie-1p::gfp::h2b::rnp-1 3′UTR, unc-119(+)], tnIs64[pCS464 pie-1p::gfp::h2b::fce-1 3′UTR, unc-119(+)], tnIs77[pCS466 pie-1p::gfp::h2b::pqn-70 3′UTR, unc-119(+)], tnIs80[pCS468 pie-1p::gfp::h2b::wdr-23 3′UTR, unc-119(+)], tnIs87[pDC5 pie-1p::gfp::h2b::rom-1 3′UTR, unc-119(+)], tnIs93[pDC22(pie-1p::gfp::h2b::gap-2 3′UTR, unc-119(+)], and tnIs95[pDC18 pie-1p::gfp::h2b::fbf-2 3′UTR, unc-119(+)].
OMA-1 immunopurifications
fog-1(ts); oma-1; tn1s17, and spe-9(ts); oma-1; tnIs17 embryos were hatched at 25° in the absence of food. Animals were collected for lysate preparation as young adults, ∼48 hr after being placed on food at 25°. Animals were raised on peptone-enriched plates seeded with the bacterial strain NA22. Lysate preparation, OMA-1::S::TEV::GFP immunopurification and tobacco etch virus (TEV) protease digestion were performed as described for RT-qPCR of OMA-1 target mRNAs (Oldenbroek et al. 2013). OMA-1 has two CCCH zinc fingers, and the buffers used minimize Zn2+ chelation. Negative controls used the anti-GFP immunopurification antibody with lysates prepared from either fog-1(ts); oma-1 or spe-9(ts); oma-1 animals, which lack the OMA-1::S::TEV::GFP fusion protein. Digestion with 5 μg/ml RNase A (Sigma) was performed for 15 min at room temperature in immunopurification wash buffer. RNase A was not added to the buffer in negative controls.
Microarrays and RNAseq
The RNase inhibitor RNAsin (Promega) was added to OMA-1 immunopurification lysates and buffers to inhibit RNA degradation. RNAs were isolated from 50 μl input lysate or five 1 ml OMA-1 immunopurifications using Trizol (Invitrogen). RNAs were further purified and concentrated using RNAqueous-Micro columns (Ambion) and eluted in a 20 μl final volume.
To prepare samples for microarray analysis, the MessageAmp III RNA Amplification kit (Ambion) was used to linearly amplify 500–600 ng input RNA or 3–5 μl immunopurified RNA (IP RNA) and fragment 20 μg of amplified RNA (aRNA). Input RNA and aRNA samples were examined using a Bioanalyzer RNA Nano chip (Agilent). RNA integrity number (RIN) scores were RIN > 9.5 for all input RNA samples, and the IP aRNA and input aRNA samples had similar profiles. aRNA was hybridized to C. elegans GeneChip arrays (Affymetrix). Three biological replicates comparing IP RNA to input RNA were performed for OMA-1::S purifications from each strain. For RNA sequencing, a TruSeq RNA library (Illumina) was prepared from 5 μl of IP RNA with no poly(A) mRNA purification and sequenced using a HiSeq2000 instrument (Illumina) and standard protocols. The IP RNA sample chosen for sequencing derived from a lysate made from fog-1(ts); oma-1; tn1s17 animals and had been analyzed on arrays. This sample was chosen because qRT-PCR suggested it contained the most IP RNA. Microarray detection steps subsequent to RNA amplification and all RNA sequencing steps were performed at the University of Minnesota Genomics Center.
Data analysis to identify RNAs enriched in IP RNA relative to input RNA and compare the IP RNA samples from strains of different genotypes was performed using Genespring GX12 (Agilent Technologies). Data were summarized using MAS5 and baseline transformed to the median of all samples. For each probe set, flags were required to be called present in at least five of the six samples in each experiment. Significance analysis utilized either a paired or unpaired T-test (purifications were paired with their cognate input sample) and a Benjamini–Hochberg correction for multiple testing [P(corr) < 0.05]. Probe sets significantly increased at least twofold in IP RNA relative to input RNA in both experiments are considered OMA-1-associated, as described in the text. A concordant list of 1383 probe sets was identified from the same data using Robust Multi-array Average summarization with quantile normalization (1108 probe sets overlap with the MAS5 list), indicating that most OMA-1-associated probe sets are identified independent of the summarization and normalization method (C. Spike, unpublished results). OMA-1 purifications were highly correlated with each other (rs ≥ 0.97) but more weakly correlated with same-genotype input lysate samples (rs = 0.50–0.59), consistent with the observation that the populations of mRNAs in these samples are quite distinct.
Sequencing data from 98 million 50-bp paired-end reads were mapped to the C. elegans genome (WS220/ce10) using TopHat v1.4.1 (Trapnell et al. 2009) and a University of California, Santa Cruz Illumina iGenome reference annotation file (ce10) to facilitate alignment. A total of 41 million reads from the OMA-1 purification mapped to regions containing rRNA genes and were discarded. A total of 54 million uniquely mapped reads were used to estimate the fragments per kilobase of transcript per million mapped reads (FPKM) values of 24,244 defined transcripts. FPKM values corrected for fragment bias were estimated using Cufflinks v1.3.0 (Trapnell et al. 2010; Roberts et al. 2011); estimates were not quartile normalized. Datasets were integrated using DAVID tools (http://david.abcc.ncifcrf.gov) to cross-reference Affymetrix probe set identifiers and the 1250 germline intrinsic and 1652 fem-1-enriched genes from Reinke et al. (2004) with National Center for Biotechnology Information (NCBI) reference sequences in the Cufflinks output. Microarray and RNAseq data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession no. GSE54518 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54518).
The 3′-UTR reporter constructs
Several criteria, including the predicted function of the encoded protein, influenced the choice of OMA-1-associated mRNAs for 3′-UTR-based reporter analysis. However, all selected mRNAs were strongly enriched in OMA-1 purifications (fourfold or more; File S1) with some evidence of expression in the germ line or early embryos. The mRNAs initially selected were chosen in part because they appear to be abundant in OMA-1 purifications (e.g., cdc-25.3, rnp-1, rnf-5, pqn-70, fce-1, and wdr-23). Less abundant mRNAs encoding proteins with interesting functions were subsequently chosen for analysis (e.g., gap-2 and rom-1).
The 3′-UTR reporter constructs were generated by recombining the following entry clones with the destination vector pCG150 (Merritt et al. 2008) using the Multisite Gateway system (Life Technologies). Entry clones pCG142 and pCM1.35 supplied the pie-1 promoter and GFP::histone H2B coding sequences, respectively (Merritt et al. 2008). Gene-specific 3′-UTR sequences were amplified from C. elegans fosmid library clones (Source BioScience) and recombined with the Gateway donor vector pDONR P2R-P3 to make 3′-UTR entry clones. Each 3′-UTR entry clone includes the longest 3′-UTR sequence identified for that particular gene by Jan et al. (2011) and at least 290 bp of downstream sequence; the sequence of each clone is available upon request. Transgenes were inserted into unc-119(ed3) animals using the Biolistic PDS-1000/He particle delivery system (Bio-Rad) with tungsten or gold beads and published protocols (Praitis et al. 2001; Merritt et al. 2010). Experiments with strains containing 3′-UTR reporter constructs were generally performed at 25° to prevent transgenes from silencing.
Microscopy and image quantification
DIC and fluorescent images were acquired on a Zeiss motorized Axioplan 2 microscope with a 63X Plan-Apochromat (numerical aperture 1.4) objective lens using a AxioCam MRm camera and AxioVision software (Zeiss). Images used to compare levels of GFP expression from 3′-UTR reporter transgenes, either visually or graphically, were taken with identical exposure settings, unless noted otherwise. Nuclear GFP accumulation was quantified using AxioVision software and background corrected relative to oocyte cytoplasm.
In situ hybridization
In situ hybridizations with dissected C. elegans gonads were performed as described (Voronina et al. 2012) with the following modifications. Methanol fixation and 4% paraformaldehyde postfixation steps and washes were as described (Voronina et al. 2012). Fixed samples were then incubated with a freshly mixed solution of 0.1% NaBH4 in PBS for 5 min on ice to reduce autofluorescence, washed a minimum of three times with PBT, twice with 2× SSC, and hybridized with custom Stellaris Quasar 570 dye-labeled oligonucleotide probes (Biosearch Technologies) as described (Raj et al. 2008). Gonads were washed once with PBT and three times with PBS before mounting in Vectashield containing DAPI (Vector Laboratories). Imaging used the Axioplan 2 microscope described above with an apotome adaptor (Zeiss).
Immunofluorescence
Dissected gonads were fixed in 3% paraformaldehyde, as described (Rose et al. 1997). Primary antibodies were a mixture of two purified mouse monoclonal anti-MSP antibodies (Kosinski et al. 2005, each at 1:300) and rabbit anti-RME-2 antibody (Grant and Hirsh 1999; kindly provided by B. Grant, Rutgers University, 1:50). Secondary antibodies were Alexa 488-conjugated goat anti-rabbit (Life Technologies, 1:500), and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch, 1:500).
Protein gels, western blots, and mass spectrometry
Proteins were separated using NuPage 4–12% Bis-Tris gels (Invitrogen) and visualized using SYPRO Ruby protein gel stain (Invitrogen) or by western blotting. Primary antibodies used to detect proteins on western blots include rabbit anti-OMA-1 (Detwiler et al. 2001, 1:50), mouse anti-GFP (Clontech; 1:30,000), goat anti-S-tag (Abcam; 1:30,000), chicken anti-CAR-1 (Boag et al. 2005; kindly provided by K. Blackwell, Joslin Diabetes Center; 1:5,000), and a rabbit antibody raised against the amino terminus of CGH-1 (I. Yamamoto and D. Greenstein, unpublished results; 1:30,000). Secondary antibodies used for western blots were peroxidase-conjugated goat anti-mouse (Pierce), donkey anti-rabbit (Pierce), donkey anti-goat (Abcam), and donkey anti-chicken (Jackson ImmunoResearch) antibodies diluted 1:30,000. Blots stained with the anti-S-tag antibody were blocked with 1.5% purified BSA (Sigma); other blots were blocked with 5% nonfat dried milk.
Five 1 ml OMA-1 immunopurifications were combined for mass spectrometry. Proteins were precipitated with 10% trichloroacetic acid, briefly separated on a 12% NuPage gel, stained with colloidal Coomassie (Invitrogen), and lanes were subdivided into gel slices. Proteins close in size to TEV protease (∼25–30 kD) may not have been identified, as this gel slice was discarded. Mass spectrometry was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School) using an LTQ ion-trap mass spectrometer (Thermo Electron). Proteins identified as possible contaminants in control purifications included the following: (1) proteins identified in an RNase-treated mock OMA-1 purification using anti-GFP antibody and lysate from fog-1(ts); oma-1 animals (i.e., proteins that bind to the antibodies or beads); (2) proteins eluted from this mock OMA-1 purification by RNase treatment (i.e., abundant, possibly RNA-binding proteins); and (3) proteins identified in one of two immunopurifications using anti-MSP antibody and lysate from young adult hermaphrodites (i.e., abundant proteins; I. Yamamoto and D. Greenstein, unpublished data). The identification and removal of abundant proteins as possible contaminants was deemed necessary because only a small number of C. elegans proteins were identified in the RNase-treated negative control (10 proteins were identified by two or more peptides). All proteins repeatedly identified in OMA-1 purifications, including possible contaminants, are shown in File S2.
Yeast two-hybrid analyses
Yeast two-hybrid experiments were performed using the GAL4-based transcription system. The following bait vector constructs were generated in pRL865, which is a derivative of pDEST32 (Invitrogen): ZYG-11 (pRL1973), OMA-1 (pRL1485), and OMA-1 E141K (pRL2277). pGBKT7-derived bait vector constructs: OMA-1N(1-117) (pRL575). The following prey vector constructs were generated in pRL864, which is a pDEST22 (Invitrogen) derivative: PQN-59 (pRL1909) and ZYG-11 (pRL1972). The following prey constructs were generated in pRL1058, which is a pACT2 (Clontech) derivative: TAF-4 (pRL1368), SPN-4 (pRL2063), MEX-3 (pRL2027), GLD-1 (pRL2022), C27B7.2 (pRL976), DH11.5 (isoform e; pRL938), and OMA-2N (pRL2428). The bait and prey vector controls used were pGBKT7 and pACT2, respectively. Plasmids derived from pACT2 and pGBKT7 are high-copy number and those derived from pDEST22 and pDEST32 are low-copy number. Low-copy number plasmids were used when high-copy number plasmids were toxic or self-activating. Yeast strains AH109 (Clontech) and Mav203 (Invitrogen) were used as indicated.
RNA interference
Gene-specific RNA interference (RNAi) was performed by feeding C. elegans with dsRNA-expressing Escherichia coli (Timmons and Fire 1998) using the RNAi culture media described by Govindan et al. (2006). Most RNAi clones were obtained from a C. elegans RNAi feeding library (Source BioScience). RNAi clones for several genes were constructed de novo (gld-3, mex-1, ifet-1, spn-4, ccf-1, pqn-59, ife-3, sqd-1, hrp-2, H27M09.1, gcn-1, and daf-21); their sequences are available upon request. Exposure to dsRNA-expressing E. coli was initiated: (1) at the first larval stage in experiments examining translational derepression of 3′-UTR reporter constructs; (2) at the third larval stage in screens for suppressors of oma-1; oma-2, enhancers of oma-1 and oma-2, and proteins that repress translation of the zif-1 3′-UTR reporter construct; and (3) at the fourth larval stage in the cdc-25.2(RNAi) experiment. Animals were examined and imaged as young adults, ∼24 hr after the mid-L4 stage. The synthetic lethal phenotype exhibited by oma-2; puf-5(RNAi) embryos was identified both as described in the text and in independent experiments (Y. Nishi and R. Lin, unpublished results). The confirmatory experiment described in the text used sdz-18(RNAi) as a negative control and RNAi as in Oldenbroek et al. (2013); essentially identical results were obtained using the RNAi protocols described above and an empty vector negative control.
Results
OMA-1 is a component of oocyte RNPs
C. elegans oocytes contain several different, and likely overlapping, types of RNPs. When sperm are absent, oocyte RNPs containing CAR-1, CGH-1, and several other RNA-binding proteins enlarge and become cortically localized, a process that is inhibited by the MSP signaling pathway (Jud et al. 2008; Noble et al. 2008). Similarly, GFP-tagged OMA-1, which is diffusely localized in the presence of sperm (Figure 2, E and F), is found in large cortical foci in the absence of sperm (Figure 2, G and H; Figure S1, D, F, N, and P). Large foci of OMA-1 are also found when MSP signaling is compromised in acy-4 mutants (Figure 2, I and J), but are absent from spe-9(ts) animals (Figure S1, G and H) that have fertilization-incompetent sperm able to stimulate meiotic maturation (Singson et al. 1998). In addition, OMA-1 foci in females are disrupted by RNAi-mediated knockdown of genes encoding the RNA-binding proteins PUF-5 and CAR-1 (Figure S1, Q and R; C. Spike, unpublished results), as described for CAR-1 and CGH-1-containing RNPs in female oocytes (Noble et al. 2008; Hubstenberger et al. 2013). These observations suggest that OMA-1 is a component of oocyte RNPs, at least in the absence of sperm.
Figure 2.
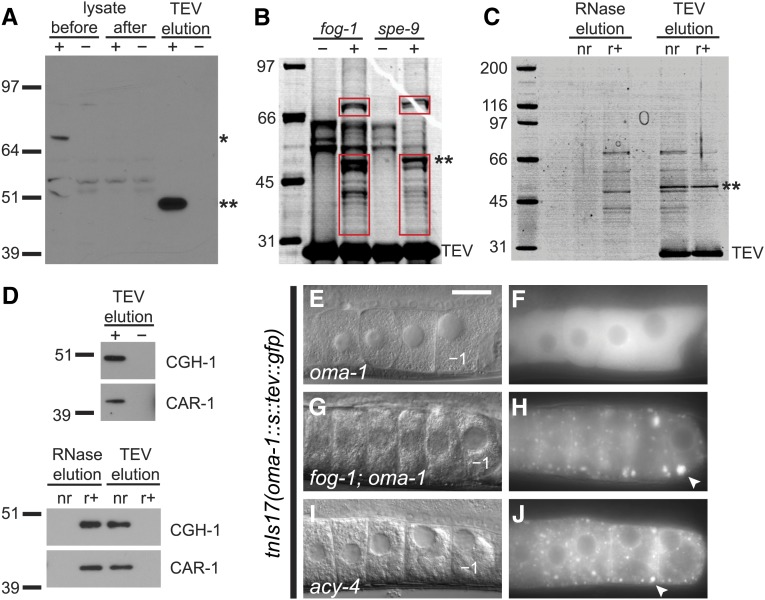
OMA-1 is a component of riboncleoprotein particles (RNPs). (A) OMA-1::S::TEV::GFP (asterisk) is depleted after incubation with matrix-coupled anti-GFP antibodies (compare lanes 1 and 3). OMA-1::S (double asterisk in A–C) is subsequently eluted from the affinity matrix by digestion with TEV protease. A total of 0.25% of each lysate and 1% of each TEV-eluted sample were loaded. Purifications and protease cleavage were monitored by western blotting using either anti-OMA-1 (shown), anti-S-tag, or anti-GFP antibodies. Here, and in subsequent panels, samples marked with a plus (+) sign were prepared from lysates containing OMA-1::S::TEV::GFP. Samples marked with a minus (−) sign are negative controls prepared from lysates lacking OMA-1::S::TEV::GFP. All purifications were performed in an oma-1(zu405te33) genetic background and were from fog-1(ts) females, unless otherwise specified. (B) Abundant proteins that copurify with OMA-1::S from fog-1 and spe-9 extracts were visualized by staining a polyacrylamide gel with SYPRO-Ruby (red boxes). Proteins in negative controls [minus (−) sign] are similar in size to human keratins (Moll et al. 2008), common contaminants of protein purifications. (C and D) Many proteins require RNA for their association with OMA-1. (C) Treatment with RNase A, prior to incubation with TEV protease (RNase elution, r+), causes proteins to elute from the immunoaffinity matrix. Proteins are not eluted by a mock RNase treatment (RNase elution, nr). Comparatively few proteins copurify with OMA-1::S after RNase A treatment (TEV elution, r+). Proteins were visualized using SYPRO-Ruby. (D) Western blots show that CGH-1 and CAR-1 copurify with OMA-1::S in an RNA-dependent manner. (E–J) OMA-1 reorganizes into large RNPs (arrowheads) when the sperm-dependent signal promoting meiotic maturation is absent or not transmitted to oocytes. Oocytes expressing the rescuing OMA-1::S::TEV::GFP fusion protein show a diffuse pattern of GFP localization (E and F), similar to spe-9(ts) animals raised at 25° (Figure S1). If sperm are absent, as in fog-1(ts) animals raised at 25°, OMA-1::S::TEV::GFP reorganizes into large foci (G and H). Similar foci form in the presence of sperm when the MSP-dependent signaling pathway active in sheath cells is disrupted, as in acy-4(ok1806) mutants (I and J). Explicit genotypes are specified in the legend to Figure S1. Bar, 20 μm.
OMA-1 and OMA-2 are oocyte-specific CCCH zinc finger RNA-binding proteins (Detwiler et al. 2001). The OMA proteins repress the translation of a few known target mRNAs in oocytes (Jadhav et al. 2008; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013; Kaymak and Ryder 2013). After fertilization, OMA-1 and OMA-2 become phosphorylated and interfere with transcription by preventing TAF-4, a subunit of the TFIID RNA polymerase II transcriptional complex, from entering the nucleus (Guven-Ozkan et al. 2008). We isolated the mRNAs and proteins that associate with OMA-1 in oocytes using immunoaffinity purification (Figure S2). To avoid isolating OMA-1 complexes from embryos, protein extracts were made from sterile adults. These animals make normal oocytes but either lack sperm [fog-1(ts)] or have sperm that are unable to fertilize oocytes [spe-9(ts)] when grown at 25°. To facilitate purification, OMA-1 was tagged at the C terminus using a reversed version of the tag described by Cheeseman et al. (2004), which includes an S-tag, TEV protease cleavage site, and GFP. OMA-1::S::TEV::GFP is able to restore fertility to oma-1; oma-2 mutant animals, indicating that it functions properly in vivo. We conducted purifications in the oma-1(zu405te33) protein null mutant background (Detwiler et al. 2001) to avoid competition with the endogenous protein. Immunoaffinity purification was performed using anti-GFP antibodies followed by cleavage with TEV protease to elute OMA-1 complexes (Figure 2A; Figure S2) and reduce nonspecific background (Gerber et al. 2004). An S-tag/S-protein purification step would introduce RNase activity (Raines et al. 2000) and was not a part of our purification scheme; instead the S-tag was used to detect OMA-1::S after cleavage with TEV protease. Total protein stains indicate that numerous proteins copurify with OMA-1 (Figure 2B). Furthermore, the banding patterns appear similar in independent purifications (Figure 2, B and C; C. Spike, unpublished results), suggesting reproducible purification of the same collection of proteins. Furthermore, most proteins that copurify with OMA-1 are eluted from the affinity matrix by RNase treatment prior to cleavage with TEV protease (Figure 2C), indicating that their interactions with OMA-1 either require or are stabilized by RNA.
Identification of OMA-1-associated mRNAs
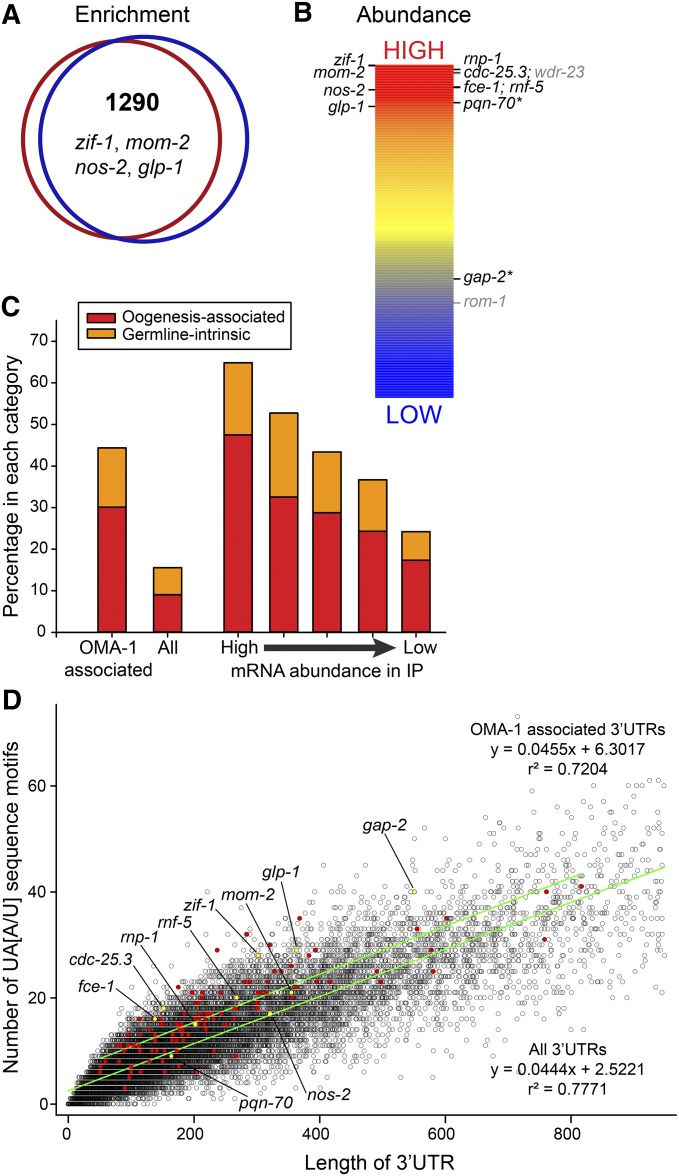
Because OMA-1/2 are essential for meiotic maturation, and function downstream of the MSP signaling pathway (Detwiler et al. 2001; Govindan et al. 2006, 2009; Kim et al. 2012), we hypothesized that OMA-1 might regulate, and therefore associate with, different mRNAs in the presence and absence of sperm-dependent MSP signaling. To test this hypothesis, we used Affymetrix microarrays to compare the mRNAs that copurify with OMA-1 in the presence and absence of sperm-dependent signaling [i.e., spe-9(ts) and fog-1(ts) strains, respectively]. Contrary to our expectation, essentially the same mRNAs were identified as significantly enriched in OMA-1 purifications relative to total lysate mRNA (input mRNA) in three biological replicates for both sets of OMA-1 purifications (Figure 3A; ∼1079 mRNAs corresponding to 1290 probe sets enriched at least twofold). Furthermore, when we directly compared the OMA-1 purifications from spe-9(ts) and fog-1(ts) animals, only two mRNAs were significantly enriched only in purifications from either strain (File S1), and neither mRNA was significantly enriched in OMA-1 purifications compared to input mRNA. Since several of the enriched mRNAs are in vivo targets of OMA-dependent translational repression (see below), these observations suggest that OMA-1 stably associates with the same mRNA targets in the presence and absence of sperm-dependent MSP signaling. We consider mRNAs identified as OMA-1-associated in both sets of purifications to be the best candidates for targets of OMA-dependent regulation in vivo (Figure 3A; File S1). After removing duplicates, this list of ∼1079 genes includes all previously identified mRNA targets of OMA-dependent translational repression in oocytes: zif-1, mom-2, nos-2, and glp-1 (Jadhav et al. 2008; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013; Kaymak and Ryder 2013). mei-1 mRNA was not identified (File S1), likely because this mRNA is a target of OMA-1-mediated repression in embryos, but not oocytes (Li et al. 2009), which was the basis for our purification.
Figure 3.
Identification and analysis of mRNAs that copurify with OMA-1. (A) GeneChip microarrays identified OMA-1-associated mRNAs (1290 probe sets) that are enriched at least twofold in OMA-1 purifications relative to input samples [P(corr) ≤ 0.05]. Similar sets of mRNAs were identified in OMA-1 purifications from fog-1(ts) (red circle) and spe-9(ts) (blue circle) lysates, including known targets of OMA-dependent translational repression (zif-1, mom-2, nos-2, and glp-1). Note that most mRNAs that are significantly enriched in a single experiment are enriched less than twofold, or are enriched but fail data-quality metrics, in the other experiment (File S1). (B) The relative abundances of candidate target mRNAs in a representative OMA-1 purification were determined by high-throughput sequencing. All candidate mRNA targets with FPKM values >1 are shown; colors indicate FPKM values (red > 569, yellow = 127, blue < 18.5). Genes whose 3′-UTRs have been tested for their ability to confer OMA-dependent translational repression in oocytes are shown; those with positive results are in black. Two targets of OMA-dependent translational repression have multiple mRNA isoforms with FPKM values >1 (asterisks); only the most abundant isoform is indicated. (C) A significant percentage of OMA-1 target genes are highly expressed during oogenesis (oogenesis-associated) or identified as germline-intrinsic (P = 1.4 × 10–21). Enrichment of both gene classes is most dramatic among OMA-1 targets of high abundance in the immunopurified samples (P = 2.3 × 10–95). The FPKM value of the most abundant transcript of each gene was used to estimate target abundance. Significance was determined using a hypergeometric probability test. (D) OMA-1 target mRNAs tend to have more OMA-1-binding motifs in their 3′-UTRs than other mRNAs with similarly sized 3′-UTRs. The numbers of UA[A/U] motifs found in a genome-wide collection of C. elegans UTRs (Jan et al. 2011) are plotted relative to 3′-UTR length (black circles). All 3′-UTRs <950 nucleotides were plotted (98.2% of identified UTRs). Red- and yellow-filled circles correspond to likely and known targets of OMA-dependent translational repression, respectively (n = 120). Linear regression of these datasets generated lines with similar slopes but significantly different y-intercepts (P < 0.0001), suggesting that there are additional UA[A/U] motifs in the 3′-UTRs of likely OMA-1 target mRNAs.
We noticed that zif-1, mom-2, nos-2, and glp-1 appear to be more abundant in OMA-1 purifications than many other OMA-1-associated mRNAs (Figure S3). Because the relative levels of different mRNAs in the same sample cannot be reliably measured using microarrays (Draghici et al. 2006), we used Illumina high-throughput sequencing to identify and quantify the mRNAs present in a representative OMA-1 purification. The mRNA sample selected was also analyzed in the microarray experiments and found to be similar to other OMA-1 purifications (Spearman rank-order correlations rs ≥ 0.967). Only 42% of the mapped reads from this sample correspond to rRNA, which represents a significant depletion considering that no rRNA depletion or mRNA enrichment steps other than OMA-1 purification were performed (see Materials and Methods), and total RNA is >80% rRNA. We eliminated rRNA sequences prior to determining transcript abundance in the immunopurified samples, which used the FPKM measurement developed by Trapnell et al. (2010). Indeed, zif-1, mom-2, nos-2, and glp-1 are more abundant than most other OMA-1-associated mRNAs (Figure 3B) and among the 200 most abundant mRNAs in the immunopurified sample analyzed (File S1). We verified that this analysis provides an accurate estimate of the relative levels of OMA-1-associated mRNAs across all samples by comparing to quantitative RT-PCR data. In the sequenced sample, zif-1 has a 5.1-fold higher FPKM value than nos-2, and zif-1 was consistently 4–7-fold more abundant than nos-2 by qRT-PCR in independent OMA-1 purifications (Table S2).
We next assessed whether OMA-1-associated mRNAs are present in oocytes; such mRNAs could be in vivo components of OMA-1 RNPs. Genes that are highly expressed during oogenesis or identified as germline intrinsic by Reinke et al. (2004) are likely expressed in oocytes and are significantly enriched among genes encoding OMA-1-associated mRNAs (Figure 3C; P = 1.4 × 10−21). Enrichment of these gene categories was even more significant among genes encoding OMA-1-associated mRNAs of high abundance in the immunopurifications (Figure 3C; P = 2.3 × 10−95). OMA-associated mRNAs that are abundant in OMA-1 purifications are therefore the most likely to be present in oocytes and in vivo targets of OMA-dependent translational repression. We examined the biological processes associated with these high-confidence mRNA targets of OMA-1 using DAVID tools (http://david.abcc.ncifcrf.gov) to identify enriched Gene Ontology (GO) terms (Huang et al. 2009a,b). Enriched GO terms and GO term clusters related to reproduction, embryonic development, and germline sex determination were identified (see File S1 for complete lists), consistent with identified functions of OMA-1/2 in vivo (see Discussion).
OMA-1/2 regulate the translation of OMA-1-associated mRNAs
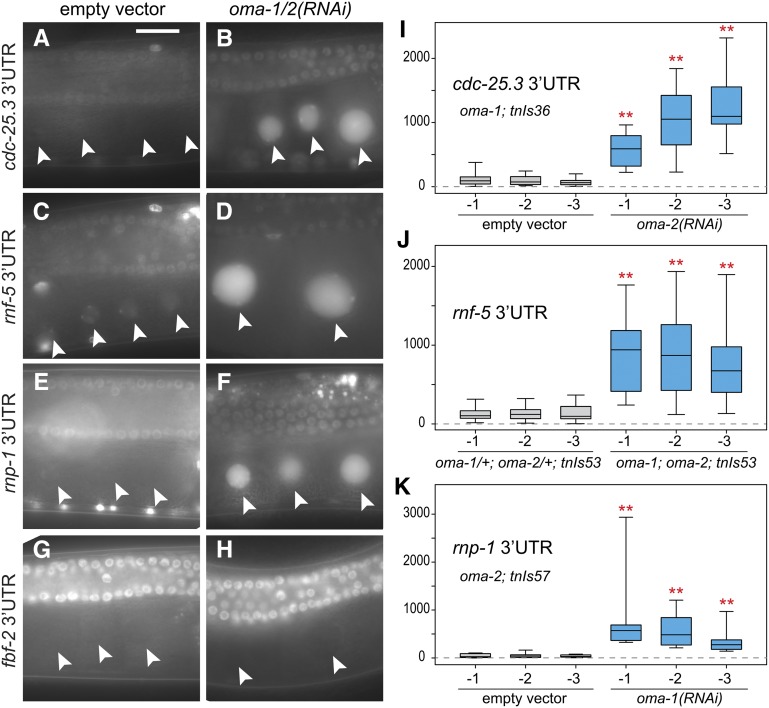
Sequences in the 3′-UTRs of mRNAs are important for regulating protein accumulation in the oogenic germ line (Merritt et al. 2008) and mediate the OMA-dependent translational repression of zif-1, mom-2, nos-2, and glp-1 in oocytes (Jadhav et al. 2008; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013; Kaymak and Ryder 2013). To examine the regulation of OMA-1-associated mRNAs in vivo, we generated animals expressing reporter transgenes containing the 3′-UTRs of eight different candidate targets (Figure 3B) and fbf-2, a germline-expressed mRNA that is not OMA-1-associated (File S1). Each transgene uses the pie-1 promoter to express a GFP::histone 2B (H2B)-coding mRNA in the germ line, but has a distinct 3′-UTR sequence. Based on genome-wide expression profiles, all of the OMA-1-associated mRNAs we tested for 3′-UTR-dependent regulation are plausibly present in oocytes and maternally provided to embryos (Baugh et al. 2003; Reinke et al. 2004; Gerstein et al. 2010). As expected, GFP expression from the fbf-2 reporter transgene was unaffected by oma-1/2(RNAi) (Figure 4, G and H). However, six of the eight transgenes containing the 3′-UTRs of OMA-1-associated mRNAs had higher levels of GFP expression in oma-1/2-depleted oocytes compared to control oocytes (Figure 4, A–F; Figure S4, A–F). Increased GFP expression requires the simultaneous depletion of oma-1 and oma-2; GFP levels appear unchanged after oma-1(RNAi) if oma-2 is undepleted and the converse is also true (D. Coetzee and C. Spike, unpublished results). Interestingly, reporter constructs containing the cdc-25.3, rnf-5, and rnp-1 3′-UTRs were more strongly derepressed after oma-1/2(RNAi) than the other reporters (fce-1, pqn-70, and gap-2 3′-UTRs). We quantified these changes after crossing 3′-UTR transgenes into oma-1(zu405te33) or oma-2(te51) loss-of-function mutants, because single-gene RNAi tends to be more effective (Gönczy et al. 2000). Oocytes expressing reporter constructs containing the cdc-25.3, rnf-5, and rnp-1 3′-UTRs had 5- to 18-fold more GFP in Oma oocytes compared to controls, and these increases were highly significant (Figure 4, I–K). Oma oocytes expressing reporter constructs containing the fce-1 and pqn-70 3′-UTRs also had significant increases in GFP expression, but were more modestly affected with only two- to threefold more GFP compared to controls (Figure S4, K and L). Distinct spatial–temporal patterns of translation were noted for the different 3′-UTR reporter constructs regulated by OMA-1 and OMA-2. Reporters containing the cdc-25.3 and rnp-1 3′-UTRs are repressed in oocytes, but expressed in fairly young embryos (Figure S5). Similar patterns of regulation have been noted for other targets of OMA-dependent translational repression in oocytes (Evans et al. 1994; Subramaniam and Seydoux 1999; Guven-Ozkan et al. 2010, Oldenbroek et al. 2012, 2013), suggesting that the OMA proteins function—at least in part—to repress maternally provided mRNAs that are translated during embryogenesis.
Figure 4.
The 3′-UTRs of mRNAs that copurify with OMA-1 can mediate OMA-dependent translational repression in oocytes. GFP::histone 2B (H2B) expression from a 3′-UTR reporter transgene from either cdc-25.3 (A, B, and I), rnf-5 (C, D, and J), or rnp-1 (E, F, and K) is strongly increased in oocytes after reducing oma-1/2 function. GFP::H2B expression from a reporter transgene containing the fbf-2 3′-UTR (G and H), which is not OMA-1-associated, is unaffected. The images shown here (and in Figure S4, Figure S5, Figure S6, and Figure S7) often include germline pachytene nuclei, which are smaller than and dorsal to the oocyte nuclei (arrowheads). oma-1/2 function was compromised using oma-1/2(RNAi) (B, D, F, and H), by combining RNAi with a loss-of-function mutation in oma-1 or oma-2 (I and K) or by crossing the transgene into oma-1(te33zu405); oma-2(te51) double mutants (J). Background-corrected nuclear GFP intensity (in arbitrary fluorescence units, plotted on the y-axis) was measured for the three oocytes closest to the spermatheca (x-axis, oocytes –1 to –3) and is significantly increased in oma-1/2 oocytes in every position compared to controls (two asterisks, P < 0.001 using a Mann–Whitney U-test). Box plots represent the data from 10 (I and K) or 28–29 (J) oocytes in each position. Box plot whiskers indicate the minimum and maximum intensity values. Bar, 20 μm.
Oma oocytes do not undergo meiotic maturation and remain in the gonad indefinitely. We examined 3′-UTR reporters regulated by OMA-1/2 in fog-2(oz40) female and gsa-1(RNAi)-treated oocytes, which mature infrequently because MSP-dependent signaling is absent (fog-2) or has been compromised [gsa-1(RNAi); Govindan et al. 2006, 2009]. The amount of GFP::H2B expressed in these oocytes was not dramatically different from the wild type (Figure S6), suggesting that reduced rates of oocyte maturation per se do not cause or substantially contribute to increased GFP expression in Oma oocytes. Furthermore, these results suggest that the translational repression of OMA targets occurs even in the absence of MSP-dependent signaling. Indeed, oma-1/2 depletion in fog-2 females increases GFP expression from the OMA-target 3′-UTR transgenes tested (cdc-25.3, rnp-1, and rnf-5; Figure S7). Thus, OMA-1 and OMA-2 are required for repression in the absence of MSP-dependent signaling, which is consistent with the result that substantially the same mRNAs copurify with OMA-1 irrespective of the presence of the MSP signal. Collectively, these results are consistent with the idea that OMA-1 and OMA-2 repress the translation of many OMA-1-associated mRNAs in oocytes, independent of the MSP signaling pathway and suggest that OMA-dependent translational repression can be relatively weak or strong, depending on the 3′-UTR of the mRNA target. Further these results indicate that the change in OMA-1 RNP localization in the absence of MSP-dependent signaling is independent of its translational repression activity.
OMA-1-binding motifs in the 3′-UTRs of OMA-1-associated mRNAs
OMA-1 binds with high affinity to RNAs that contain multiple copies of a short UA[A/U] consensus sequence in vitro (Kaymak and Ryder 2013). However, OMA-1-binding motifs are extremely common in C. elegans 3′-UTRs (>99% have at least one OMA-1-binding motif; Figure 3D), which tend to be AU-rich (Mangone et al. 2010; Jan et al. 2011). Thus, we examined whether OMA-1-binding motifs are unusually prevalent in the 3′-UTRs of OMA-1-associated mRNAs. For this analysis, we used 120 OMA-1-associated mRNAs that are relatively abundant in OMA-1 purifications or established targets of OMA-1-dependent translational repression in oocytes (Figure 3B) and chose the 3′-UTR identified by Jan et al. (2011) that most closely matches our RNAseq data (File S1). Such OMA-1-associated 3′-UTRs tend to be longer than typical C. elegans 3′-UTRs (median length of 205 instead of 130 nucleotides), consistent with the idea that they contain important regulatory sequences. Although there was a poor correlation between the absolute number of OMA-1-binding motifs in each 3′-UTR and mRNA abundance after OMA-1 purification (R2 = 0.09), OMA-1-binding motifs tend to be slightly enriched in OMA-1-associated 3′-UTRs relative to similarly sized C. elegans 3′-UTRs (Figure 3D), suggesting that a subset of OMA-1-binding motifs might be important for OMA-1 binding. We examined whether a longer consensus sequence containing UA[A/U] motifs was present in this collection of OMA-1-associated 3′-UTRs, but were unable to identify one (using MEME; Bailey et al. 2009). Together, these results suggest that at least some OMA-1-binding motifs may be important for OMA-1 binding, but also imply that additional determinants of OMA-1-binding specificity likely exist, as postulated by Kaymak and Ryder (2013). Our finding that the set of OMA-1-associated proteins includes many RNA-binding proteins (see below) is consistent with this view.
Involvement of OMA-1 target mRNAs in oogenesis
The set of OMA-associated mRNAs and proteins (see below) will provide insight into the role of OMA-1 and OMA-2 in the regulation of oocyte meiotic maturation and the oocyte-to-embryo transition, a point illustrated in the companion article (Spike et al. 2014). We initially focused on cdc-25.3 and rnp-1 because their 3′-UTRs appear to mediate strong translational repression by OMA-1 and OMA-2 and both genes are highly conserved (Shaye and Greenwald 2011). cdc-25.3 encodes one of four members of the Cdc25 family of phosphatases that activate meiotic maturation by removing inhibitory phosphorylations of CDK at residues Thr14 and Tyr15 (Kumagai and Dunphy 1991), catalyzed by the Wee1 or Myt1 kinases (Kornbluth et al. 1994; Mueller et al. 1995). The identification of a CDK activator as a target of OMA-1 and OMA-2-mediated translational repression was counterintuitive because oocytes in oma-1; oma-2 double mutants fail to undergo meiotic maturation. Nonetheless, we tested whether the regulation or function of cdc-25.3 might be critical for oogenesis, a possibility that later proved correct, though not in the ways we originally imagined (Spike et al. 2014). We first examined the phenotype of an existing deletion in cdc-25.3 to determine whether its function is important for oocyte or embryo development. cdc-25.3(ok358) animals are viable and fertile, but exhibit partially penetrant larval arrest, as described in our companion article (Spike et al. 2014). Additional analysis suggests that cdc-25.3 functions during embryogenesis but is redundant with cdc-25.2, a different cdc25-family gene important during oogenesis (Kim et al. 2010a); weak cdc-25.2(RNAi) causes highly penetrant embryonic lethality in cdc-25.3(ok358) animals (100%; n = 298), but not in the wild type (3%; n = 349). Potentially, overexpression of CDC-25.3 in oma-1; oma-2 double mutants might interfere with CDK-1 activation. We tested this possibility genetically by constructing cdc-25.3(ok358); oma-1(zu405te33); oma-2(te51) triple mutants, which were found to exhibit the Oma phenotype and were completely sterile.
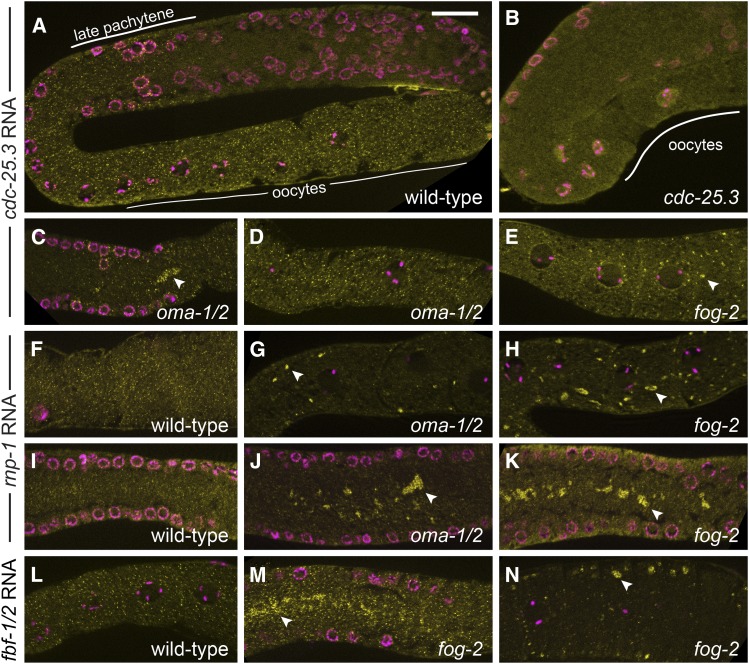
Although OMA-1 and OMA-2 negatively regulate the translation of 3′-UTR reporter constructs, their target mRNAs might be destabilized in their absence. We tested this possibility using fluorescent in situ hybridization (FISH) to examine the spatial–temporal pattern of endogenous cdc-25.3 and rnp-1 mRNA accumulation in the germ line. Both OMA-1-associated mRNAs are present in oocytes (Figure 5, A and F). cdc-25.3 mRNA is first detected in late pachytene (Figure 5A), while rnp-1 mRNA is detected throughout the germ line (Figure 5, F and I; D. Coetzee, unpublished results). cdc-25.3 and rnp-1 mRNAs are also clearly present in oma-1; oma-2 mutant germ lines and oocytes (Figure 5, C, D, G, and J). Finally, cdc-25.3 mRNA was not detected in the oocytes of the cdc-25.3(ok358) deletion mutant (Figure 5B), confirming the specificity of this probe and procedure. A similar experiment was not feasible for rnp-1 due to germline abnormalities in the deletion mutant (see below), but this probe is predicted to be highly specific (D. Coetzee, unpublished results). We conclude that OMA-1-associated mRNAs are not degraded in the absence of OMA-1 and OMA-2. We cannot exclude the possibility that transcript abundances are modestly altered, however, because older Oma oocytes are somewhat difficult to image after FISH and the localization patterns of both mRNAs change when meiotic maturation is inhibited. rnp-1 mRNAs coalesce, or aggregate, into higher order structures in oma-1; oma-2 mutant and fog-2 female germ lines, both in the rachis (Figure 5, J and K) and in oocytes (Figure 5, G and H), similar to what has been described for total RNA and a few specific mRNAs in female germ lines (Schisa et al. 2001; Jud et al. 2008; Noble et al. 2008). Some aggregation of the cdc-25.3 mRNA was also observed (Figure 5, C and E), but its localization more closely resembles the wild type, particularly in oma-1; oma-2 mutant oocytes (Figure 5D). The highly similar fbf-1 and fbf-2 mRNAs (Zhang et al. 1997), which are not OMA-1-associated (File S1), also aggregate in oma-1; oma-2 mutant and fog-2 female germ lines (Figure 5, L–N; D. Coetzee, unpublished results), although not as dramatically as rnp-1 mRNA.
Figure 5.
cdc-25.3 and rnp-1 mRNAs are present in wild-type and oma-1/2 oocytes. Fluorescent in situ hybridization (FISH) was used to examine cdc-25.3 (A–E), rnp-1 (F–K) and fbf-1/2 (L–N) mRNA expression and localization in wild-type (A, F, I, and L), oma-1(zu405te33); oma-2(te51) (C, D, G, and J), and fog-2(oz40) (E, H, K, M, and N) germ lines. mRNAs are visualized as small yellow puncta; DAPI-stained DNA is magenta. cdc-25.3 mRNAs are absent from the distal germ line, present during late pachytene, diplotene, and diakinesis (A), and absent from cdc-25.3(ok358) mutants (B). cdc-25.3 mRNA expression in oma-1/2 and fog-2 germ cells is similar to wild type, although there is some aggregation of individual mRNAs in late pachytene in oma-1/2 animals (C) and in older fog-2 oocytes (E) and possibly somewhat fewer puncta in older oma-1/2 oocytes (not shown). rnp-1 and fbf mRNAs were present throughout the germ line, including pachytene-stage germ cells (I) and oocytes (F and L). Both mRNAs aggregate in the rachis (J, K, and M) and in oocytes (G, H, and N) of oma-1/2 and fog-2 animals compared to the wild type. Aggregation of the rnp-1 mRNA was more dramatic than the aggregation of either cdc-25.3 or fbf mRNAs and was most distinct in fog-2 animals. Bar, 20 μm.
While cdc-25.3 is dispensable for oogenesis, rnp-1 appears to be required. Adult rnp-1(ok1549) hermaphrodites are completely sterile with small germ lines that fail to transition from spermatogenesis to oogenesis (Figure 6B). Genetic analysis has identified many of the key genes that control sex determination in C. elegans (reviewed by Ellis and Schedl 2007; Kimble and Crittenden 2007). fog-3 is one of the most downstream genes in the germline sex-determination pathway and is required for spermatogenesis (Chen et al. 2000). Furthermore, the fog-3 gene is epistatic to rnp-1 with respect to germline sex determination. fog-3; rnp-1 double mutants fail to make sperm, or express MSP, but some animals make tiny underdeveloped oocytes (6 of 13; Figure 6C). Thus, rnp-1 functions upstream of fog-3 in the germline sex determination pathway and promotes both oogenesis and normal oocyte development. Since rnp-1 mRNA is present throughout the germ line (Figure 5, F and I; D. Coetzee, unpublished results), but the OMA proteins are first expressed in late pachytene (Detwiler et al. 2001; Figure 1), RNP-1 is likely expressed earlier in germline development.
Figure 6.
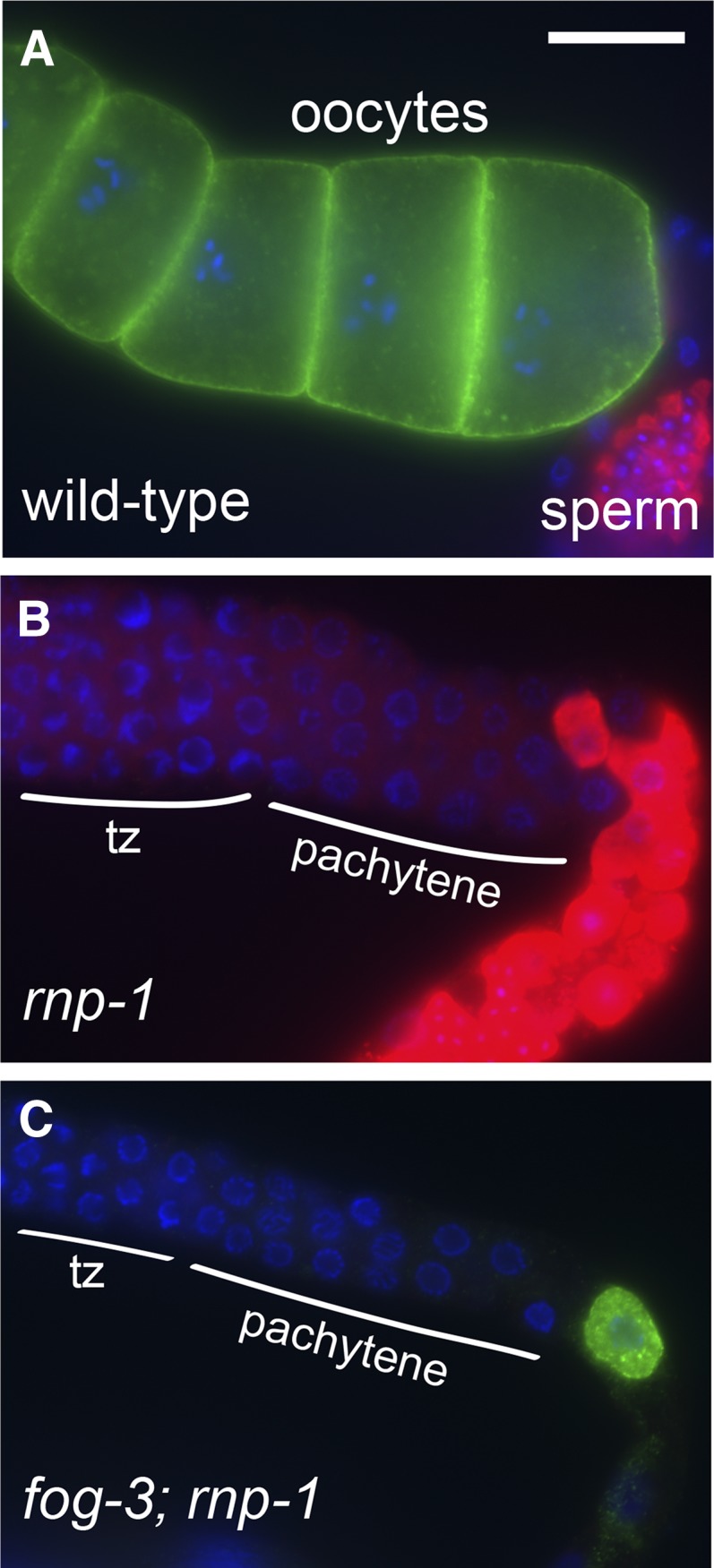
rnp-1 promotes the switch from spermatogenesis to oogenesis and is required for normal oocyte development. (A) Wild-type animals make both sperm (red, anti-MSP) and oocytes (green, anti-RME-2). (B) rnp-1(ok1549) animals have small germ lines with sperm (red) but no oocytes. Nuclei transitioning from mitosis to meiosis (tz) and in the pachytene stage of meiotic prophase are indicated in B and C. (C) fog-3(q470) unc-13(e1091); rnp-1(ok1549) animals lack sperm but can make tiny underdeveloped oocytes (green). Six of 13 germ lines examined expressed RME-2 and none expressed MSP. Bar, 20 μm.
Identification of OMA-1-associated proteins
We next investigated the proteins that copurify with OMA-1. OMA-1 RNPs were isolated in the presence and absence of MSP-dependent signaling and copurifying proteins were identified using mass spectrometry. OMA-1 was well covered (≥59%) and identified by a large number of peptides in both purifications (≥60 peptides; File S2). Most of the other proteins identified were also present in both purifications (Figure S8), including the abundantly represented proteins CGH-1 and CAR-1 (File S2). Both RNA-binding proteins are components of germline RNPs (Boag et al. 2005) and copurify with OMA-1 in the presence and absence of sperm (Figure 2D, File S2). Interestingly, western blots consistently detect CGH-1 and CAR-1 in OMA-1 purifications from fog-1(ts) extracts (n ≥ 3), but do not detect either protein in purifications from spe-9(ts) extracts (n = 3), suggesting that CGH-1 and CAR-1 are more abundant in purifications from fog-1(ts) females, though mass spectrometry clearly shows they are present in purifications from spe-9(ts) extracts (File S2). Finally, the interaction of CGH-1 and CAR-1 with OMA-1 is RNA dependent (Figure 2D), as observed for many other proteins that copurify with OMA-1 RNPs (Figure 2C; File S2).
Mass spectrometry is extremely sensitive, and >250 different proteins were identified by at least two peptides in both OMA-1 RNP purifications (Figure S8, File S2). Many of these proteins, like CGH-1 and CAR-1, have RNA-related functions and could be important components of OMA-1 RNPs in vivo. Other copurifying proteins likely represent abundant contaminants (e.g., UNC-54/myosin and VIT-1/vitellogenin; see Materials and Methods). We focused on the subset of proteins that copurify with OMA-1 from fog-1(ts) females after RNase treatment based on the expectation that close associations with OMA-1 (be they direct or indirect) might be at least partially resistant to RNase treatment. Many proteins, including some with RNA-related functions (e.g., CGH-1, EDC-3, and CEY-4), were depleted from OMA-1 RNPs by RNase treatment, leaving a much smaller pool of candidates (133 different proteins; Figure S8). Importantly, the eIF4E-binding protein IFET-1 continued to copurify with OMA-1 in the presence of RNase A (File S2). Prior work established that IFET-1 interacts with OMA-1 in vitro and represses the translation of OMA target 3′-UTR reporters in vivo (Li et al. 2009; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013). Next, proteins identified in negative controls or as abundant contaminants were excluded from consideration, leaving a smaller list of OMA-1-associated proteins (51 different proteins; Figure S8). This step eliminated CAR-1, but again retained IFET-1 (File S2). It is difficult to eliminate all contaminants identified by mass spectrometry using a limited number of negative controls (Mellacheruvu et al. 2013), so we examined the biological functions of the remaining proteins in more detail (Table 1, File S2).
Table 1. Proteins that copurify with OMA-1 and either interact with OMA-1 or regulate mRNA translation, cytoplasmic polyadenylation, or deadenylation.
| Protein | Coverage (%)a | Known physical interaction or phenotypic similarity |
|---|---|---|
| PQN-59 | 9 | Interacts with OMA-1 (Figure 7) |
| mRNA translation | ||
| GLD-1 | 24 | Interacts with OMA-1 (Figure 7) |
| LIN-41 | 19 | Represses OMA targets (Figure 8) |
| MEX-3 | 47 | Interacts with OMA-1 (Figure 7) |
| OMA-2 | 21 | Interacts with OMA-1 (Figure 7) |
| MEX-1 | 26 | None |
| PUF-5 | 10 | Enhances oma-2(te51) and represses glp-1 translation in oocytes (this work; Lublin and Evans 2007) |
| SPN-4 | 6 | Interacts with OMA-1 (Figure 7) |
| IFG-1 | 10 | None |
| IFE-3 | 14 | Interacts with IFET-1 (Li et al. 2009) |
| IFET-1 | 6 | Interacts with OMA-1 and represses OMA targets in oocytes (Li et al. 2009; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013) |
| POS-1 | 21 | Interacts with SPN-4 (Ogura et al. 2003) and is expressed in oocytes (T. Guven-Ozkan and R. Lin, unpublished results)b |
| Cytoplasmic polyadenylation | ||
| GLD-3 | 21 | None |
| GLD-2 | 16 | None |
| Deadenylation | ||
| LET-711 | 4 | None |
| CCF-1 | 18 | None |
| NTL-9 | 11 | None |
Peptide coverage in a single gel slice assessed by mass spectrometry in an OMA-1 purification after RNase treatment. OMA-1 was purified from a fog-1(ts) female lysate.
POS-1 expression was detected in oocytes by antibody staining (using antibodies described in Tabara et al. 1999), albeit at a level lower than in embryos (T. Guven-Ozkan and R. Lin, unpublished results).
Many OMA-1-associated proteins have RNA-related functions (27 of 51; 53%), including mRNA translation, cytoplasmic polyadenylation and deadenylation (Table 1, File S2), and most of these proteins function or are found in C. elegans oocytes (see below; reviewed in Nousch and Eckmann 2013). OMA-2 is one of these proteins, suggesting that OMA-1 and OMA-2 are closely associated in oocytes. OMA-1-associated proteins such as IFET-1, PUF-5, MEX-1, MEX-3, POS-1, and SPN-4 (Table 1) are present in oocytes and regulate the translation of OMA-1-associated mRNAs, either in oocytes (IFET-1 and PUF-5) or in early embryos (Ogura et al. 2003; Lublin and Evans 2007; Jadhav et al. 2008; Pagano et al. 2009; Guven-Ozkan et al. 2010; Oldenbroek et al. 2012, 2013). IFET-1 interacts with multiple eIF4E isoforms (Li et al. 2009), including the OMA-1-associated isoform IFE-3 (14% coverage, Table 1), which is one of the germline-enriched eIF4E proteins in C. elegans (Amiri et al. 2001). eIF4E interacts with eIF4G/IFG-1 (10% coverage, Table 1), which is important for oocyte development in C. elegans (Contreras et al. 2008); these proteins form complexes that both inhibit and activate translation (Hentze 1997; Rajyaguru et al. 2012). OMA-1-associated proteins important for cytoplasmic polyadenylation include GLD-2 (16% coverage; Table 1) and GLD-3 (21% coverage; Table 1), which are present in oocytes and required for normal oocyte development (Kadyk and Kimble 1998; Kim et al. 2010b). GLD-2 is the catalytic poly(A)polymerase subunit (Wang et al. 2002), and many of the GLD-2-associated transcripts identified by Kim et al. (2010b) are also OMA-1-associated (52%; 284/544). Finally, OMA-1-associated proteins important for deadenylation include CCF-1 and LET-711/NTL-1, the two Ccr4–Not complex subunits shown to be crucial for fertility and normal oocyte development (Nousch et al. 2013). Interestingly, components of the Ccr4–Not complex were found to interact with the mammalian TIS11 zinc-finger protein tristetraprolin (reviewed by Brooks and Blackshear 2013). Thus, many of the proteins identified as OMA-1-associated are important components of oocyte or germline RNPs and possibly relevant to OMA-1 function in vivo.
OMA-1-associated proteins interact with OMA-1
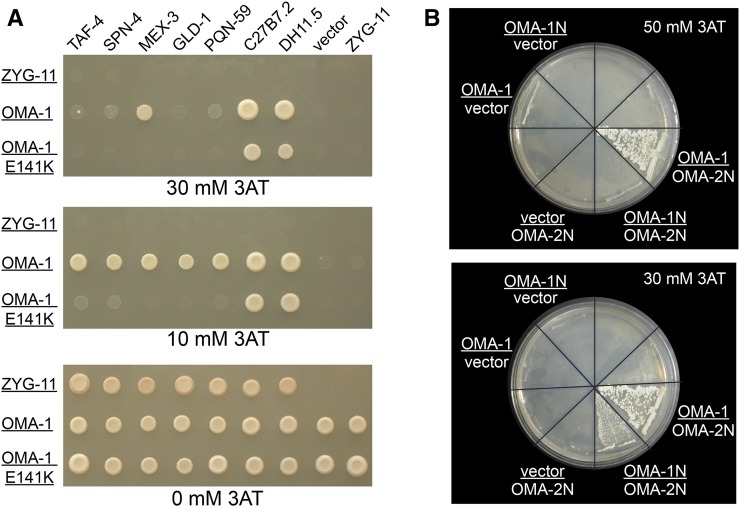
Yeast two-hybrid assays suggest that OMA-1 physically interacts with several of the proteins identified by mass spectrometry (Table 1; Figure 7, A and B). Importantly, these interactions were identified independent from, and blind to, the mass spectrometry results and represent an orthogonal dataset. Instead, candidate proteins were identified in a yeast two-hybrid screen (PQN-59) or tested for a physical interaction with OMA-1 based on in vivo biological function (SPN-4, MEX-3, GLD-1, and OMA-2). The interactions of SPN-4, MEX-3, and GLD-1 with OMA-1 map to a region containing the two OMA-1 zinc fingers (amino acids 111–188; Y. Nishi and R. Lin, unpublished results), and are abolished by the E141K zinc finger mutation (Figure 7A). However, the amino terminus of OMA-1 and OMA-2 mediates the yeast two-hybrid interaction between these two proteins (amino acids 1–117 and 1–111, respectively; Figure 7B). Each interaction is of weak-to-moderate strength, but stronger than negative controls and comparable to the positive control TAF-4 (Guven-Ozkan et al. 2008; Figure 7, A and B). Combined with previous data indicating that IFET-1 interacts with OMA-1 (Li et al. 2009), these results indicate that OMA-1 physically interacts with some of the proteins that regulate the translation of OMA target mRNAs. Notably, our OMA-1 RNP purifications identified all the previously known and newly identified OMA-1-interacting proteins save for three exceptions: TAF-4, C27B7.2, and DH11.5. The TAF-4–OMA-1/2 interaction requires MBK-2 phosphorylation of OMA-1/2, which occurs in the embryo (Guven-Ozkan et al. 2008), and thus we might not expect its representation in purifications from sterile adult hermaphrodites and females. Less is known about C27B7.2 and DH11.5, including their expression patterns, so their absence in our purifications is not easily interpreted.
Figure 7.
Several proteins that copurify with OMA-1 interact with OMA-1 in yeast two-hybrid assays. (A) MEX-3, SPN-4, GLD-1, and PQN-59 interact with OMA-1 in GAL4-based yeast two-hybrid assays on 10 mM 3-amino-1, 2,4-triazole (3AT). Bait vectors are underlined; ZYG-11 bait and prey vectors are negative controls. TAF-4, C27B7.2, and DH11.5 were identified in a GAL4-based yeast two-hybrid screen for proteins that interact with the OMA-1 N terminus (Guven-Ozkan et al. 2008; R. Lin, unpublished results) and are positive controls. PQN-59 was independently identified as an OMA-1-interacting protein using an SRS-based yeast two-hybrid screen (R. Lin, unpublished results). MEX-3, SPN-4, and GLD-1 were tested as candidate OMA-1-interacting proteins based on their expression patterns and biological functions. OMA-1 E141K is a point mutation that affects the first OMA-1 zinc finger; this residue is critical for OMA-1 function in vivo (Detwiler et al. 2001). MEX-3, SPN-4, and GLD-1 all interact with a region of OMA-1 containing the CCCH zinc fingers in similar assays (Y. Nishi and R. Lin, unpublished results). (B) OMA-1 and OMA-2 interact in a yeast two-hybrid assay. The interaction of the N-terminal domains of OMA-1 and OMA-2 is weaker than the interaction of full-length OMA-1 with the OMA-2 N terminus. Full-length OMA-1 and OMA-2 also interact, but less reproducibly (T. Guven-Ozkan and R. Lin, unpublished results). Yeast strains used were AH109 (A) and Mav203 (B).
puf-5(RNAi) exhibits synthetic lethality with oma-2
The set of proteins identified in our immunopurifications comprise most or all independently defined OMA-1-interacting proteins. Thus, we expect that this set will prove informative for understanding the biological functions of OMA-1/2. To begin to assess the relevance of this set, we conducted a screen for suppressors and enhancers of oma-1 and oma-2. We used gene-specific RNAi to reduce the function of most OMA-1-associated proteins (File S2). These experiments focused on OMA-1-associated proteins with RNA-related functions, but included many other proteins present in OMA-1 purifications, including CGH-1 and CAR-1. RNAi was performed in a variety of genetic backgrounds, including the wild type, oma-1(zu405te33); oma-2(te51) double mutants, and oma-1(zu405te33) and oma-2(te51) single mutants. No suppressors of the oma-1; oma-2 oocyte meiotic maturation defect were identified. This analysis uncovered a synthetic lethal interaction between puf-5(RNAi) and oma-2(te51). puf-5(RNAi) causes low-penetrance embryonic lethality in the wild type and oma-1(zu405te33) mutants (9%; n ≥ 172) but high-penetrance embryonic lethality in oma-2(te51) mutants (97%; n = 223). oma-2(te51) mutants do not exhibit appreciable embryonic lethality in control RNAi experiments (2%; n = 85). This result suggests that PUF-5 and OMA-2 function redundantly and, somewhat unexpectedly, that OMA-1 and OMA-2 are not completely redundant either during oogenesis or early embryogenesis. Furthermore, it confirms that PUF-5 is likely relevant to OMA function in vivo, as surmised from the observation that each of these proteins represses the translation of glp-1 mRNA in oocytes (Lublin and Evans 2007; Kaymak and Ryder 2013).
LIN-41 represses the translation of OMA-1/2 targets in oocytes
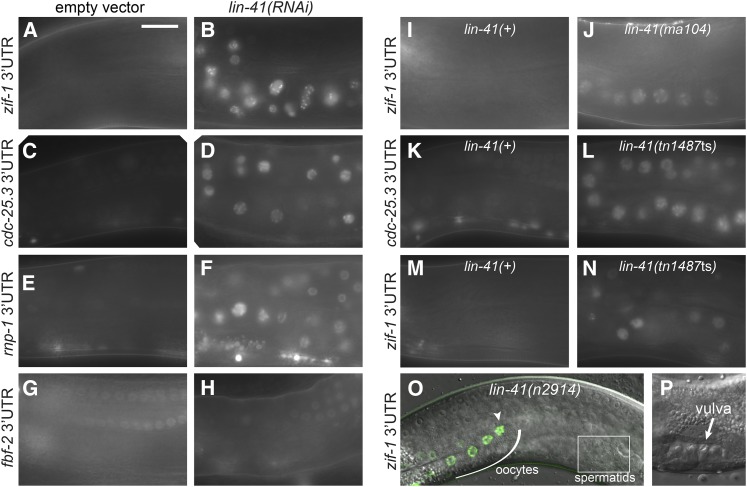
In a second approach for identifying proteins relevant to OMA-1/2 function, we conducted an RNAi screen for regulators of 3′-UTR-dependent OMA-1/2-mediated translational repression. Animals expressing the zif-1 3′-UTR reporter were used in our initial RNAi screen because this reporter is strongly repressed in wild-type oocytes (Figure 8, A, I, and M; Guven-Ozkan et al. 2010). Interestingly, IFET-1 and LIN-41 were the only OMA-1-associated proteins we identified that strongly repress zif-1 translation in oocytes. IFET-1 was previously shown to repress OMA-1/2 targets in oocytes (Li et al. 2009; Guven-Ozkan et al. 2010; Oldenbroek et al. 2013). LIN-41 is a conserved member of the TRIM-NHL family of proteins and has been proposed to repress mRNA translation in C. elegans as well as other organisms (Slack et al. 2000; Loedige et al. 2013; Worringer et al. 2014). GFP expression from the zif-1 3′-UTR reporter was increased after lin-41(RNAi) near the loop region (Figure 8, A and B); these germ cells are young abnormal oocytes and are described in detail in our companion article (Spike et al. 2014). We examined other 3′-UTR reporters strongly repressed by OMA-1/2 to see if they are also repressed by LIN-41 in oocytes. Indeed, GFP expression from the cdc-25.3 and rnp-1 3′-UTR reporter contructs is dramatically increased after lin-41(RNAi) (Figure 8, C–F). lin-41(RNAi) may modestly increase GFP expression from the rnf-5 3′-UTR construct, but this transgene was much less strongly regulated (C. Spike, unpublished results). We also examined GFP expression from a subset of OMA target 3′-UTR constructs in the oocytes of strong loss-of-function alleles of lin-41, including the temperature-sensitive sterile lin-41(tn1487ts) allele (Spike et al. 2014). As expected, GFP expression from the cdc-25.3 and zif-1 3′-UTR constructs is increased in lin-41(tn1487ts) oocytes at the restrictive temperature (25°) relative to controls. GFP expression from the cdc-25.3 3′-UTR construct was robust and pervasive (Figure 8, K and L), while expression from the zif-1 3′-UTR construct was relatively faint and predominantly visible in young oocytes near the loop (Figure 8, M and N). A similar pattern was observed when the zif-1 3′-UTR construct was crossed into the strong loss-of-function lin-41(n2914) allele, suggesting that, in these mutants, the zif-1 3′-UTR construct is translated during early oogenesis. We confirmed that GFP is expressed from the zif-1 3′-UTR construct in lin-41(n2914) oocytes by examining young animals that are just beginning to produce oocytes. In such animals, GFP expression is observed in most, if not all, oocytes and appears to correlate temporally with oocyte formation (Figure 8, O and P).
Figure 8.
LIN-41 represses the translation of OMA targets in oocytes. (A–H) lin-41(RNAi) (B, D, F, and H) strongly enhances GFP::H2B expression from reporter transgenes containing the zif-1 (A and B), cdc-25.3 (C and D), or rnp-1 (E and F) 3′-UTRs in oocytes. There was no apparent increase in GFP::H2B expression from the reporter transgene containing the control fbf-2 3′-UTR (G and H). (I and J) GFP::H2B expression from the zif-1 reporter transgene can be seen in the oocytes of fertile lin-41(ma104) animals (67%; n = 54), but not wild-type controls (0%; n > 36). (K–N) GFP::H2B expression from the cdc-25.3 (K and L) and zif-1 (M and N) reporter transgenes is increased in the oocytes of lin-41(tn1487ts) mutants raised at 25° relative to controls. (O and P) Expression from the zif-1 reporter transgene (green) begins as lin-41(n2914) germ cells start to develop into oocytes (O); the most proximal oocyte is GFP positive (arrowhead). The germ line of this lin-41(n2914) animal was imaged just prior to vulval eversion (P), which would normally occur around the time of the L4-to-adult molt (Sharma-Kishore et al. 1999). Identical exposure times were used to collect the paired images with the exception that the control in A was 40% overexposed compared to B. Bar, 20 μm.
Strong losses of lin-41 function [as in lin-41(n2914), lin-41(tn1487ts), or lin-41(RNAi)] cause severe defects in oocyte growth and meiotic progression (Spike et al. 2014). However, GFP expression from OMA-1/2 target 3′-UTR constructs does not appear to be a secondary consequence of these abnormalities. GFP expression from the fbf-2 3′-UTR construct is not increased or altered after lin-41(RNAi) (Figure 8, G and H), indicating that lin-41 oocytes are still capable of repressing the translation of some mRNAs. Furthermore, GFP is visibly expressed from the zif-1 3′-UTR construct in lin-41(ma104) oocytes, but not wild-type oocytes (Figure 8, I and J). lin-41(ma104) is a hypomorphic allele that is viable and fertile (Slack et al. 2000). The oocytes produced by homozygous lin-41(ma104) animals tend to be small but appear relatively normal and do not exhibit defects in meiotic prophase progression (Figure 8J, C. Spike, unpublished results). After outcrossing, homozygous lin-41(ma104) animals exhibited only low levels of embryonic lethality (2%; n = 1765) and a brood size of 181 progeny (n = 12) at 20°. This is ∼56% of the wild-type brood size (∼320 progeny), consistent with the observation that oocyte development is only modestly impaired in lin-41(ma104) animals. Together, these results indicate that LIN-41 represses the translation of several OMA targets in oocytes and is likely relevant to OMA function in vivo as we show in the accompanying article (Spike et al. 2014).
Discussion
In this and the accompanying article in this issue (Spike et al. 2014), we integrated biochemical and genomic approaches with genetic analyses to address the requirement of the OMA proteins in the regulation of oocyte growth and meiotic maturation in C. elegans. Prior genetic analysis established that OMA-1 and OMA-2 are redundantly required for oocyte meiotic maturation (Detwiler et al. 2001). In oma-1; oma-2 double mutants, multiple germline readouts of MSP signaling are defective: MPK-1 mitogen-activated protein kinase activation is not sustained, reorganization of the cortical microtubule cytoskeleton does not occur, the AIR-2 Aurora B kinase fails to localize to oocyte chromatin, and nuclear envelope breakdown does not occur properly (Detwiler et al. 2001; Harris et al. 2006). While depletion of the inhibitory WEE-1.3 kinase by RNA interference in oma-1oma-2 double mutants can drive oocytes into M phase, fertility is not restored. Instead, oocytes mature in ectopic positions, and are not properly ovulated and fertilized (Detwiler et al. 2001; Burrows et al. 2006). A second defect observed in oma-1oma-2 double mutants is that oocytes grow abnormally large only in the presence of sperm (Detwiler et al. 2001). Actomyosin-dependent cytoplasmic streaming from the core cytoplasm of the gonad drives oocyte growth and requires the continued presence of sperm (Wolke et al. 2007). MSP meiotic maturation signaling in sheath cells is sufficient to drive cytoplasmic streaming, and gap junction communication between sheath cells and oocytes is required for cytoplasmic streaming to cease when sperm are absent (Govindan et al. 2009; Starich et al. 2014). Oocytes in oma-1oma-2 double mutant hermaphrodites appear to grow abnormally large because they receive low rates of cytoplasmic flow for a longer period of time (Govindan et al. 2009). Thus, the OMA proteins appear to promote and coordinate cytoplasmic and nuclear events of oocyte meiotic maturation.
The OMA proteins also function to coordinate oocyte meiotic maturation with events needed for the oocyte-to-embryo transition. Upon oocyte meiotic maturation, the dual-specificity tyrosine-phosphorylation-regulated protein kinase MBK-2 becomes fully active during meiosis II and phosphorylates the OMA proteins (Pellettieri et al. 2003; Nishi and Lin 2005; Shirayama et al. 2006; Stitzel et al. 2006; Cheng et al. 2009). Phosphorylation of OMA-1 on threonine-residue 239 facilitates its interaction with TAF-4 (Guven-Ozkan et al. 2008), a subunit of TFIID required for RNA polymerase II-mediated transcription (Walker et al. 2004). The OMA–TAF-4 interaction sequesters TAF-4 in the cytoplasm to prevent RNA polymerase II transcription in the early germline blastomeres P0 and P1 (Guven-Ozkan et al. 2008). The maintenance of transcriptional repression in the C. elegans germline lineage is critical for germline development (Mello et al. 1996; Seydoux et al. 1996; Seydoux and Dunn 1997; Schaner et al. 2003). This transcriptional repression function of the OMA proteins is likely not relevant for the regulation of meiotic maturation because this activity is genetically separable from the meiotic maturation requirement and only manifests upon phosphorylation by MBK-2 (Guven-Ozkan et al. 2008), which is dependent on meiotic maturation (Stitzel et al. 2006; Cheng et al. 2009). Likewise, the role of the OMA proteins in regulating the translation of mei-1/katanin, appears restricted to the embryo (Li et al. 2009). mei-1/katanin is needed for the meiotic divisions of the oocyte but must be degraded for the mitotic divisions of the embryonic blastomeres to occur properly (Mains et al. 1990; Clandinin and Mains 1993; Clark-Maguire and Mains 1994a,b; Dow and Mains 1998; Srayko et al. 2000; Pellettieri et al. 2003; Pintard et al. 2003).
To address the role of the OMA proteins in oogenesis, we affinity purified OMA-1-containing RNPs and characterized their mRNA and protein components. Purifications were conducted using sterile strains that produce oocytes but not embryos so as to focus on the role of OMA-1 during oogenesis. We conducted purifications in the presence and absence of sperm because oocyte meiotic maturation is dependent on MSP signaling (McCarter et al. 1999; Miller et al. 2001; Kosinski et al. 2005). It was also of interest to conduct purifications in the absence of sperm because under these conditions RNP foci dramatically condense in oocytes to form large cortically localized aggregates of high stability (Schisa et al. 2001; Jud et al. 2008; Hubstenberger et al. 2013). Surprisingly, our analysis revealed that a set of ∼1079 mRNAs is significantly and reproducibly enriched in OMA RNPs irrespective of the presence of sperm. This group of mRNAs contains all mRNA targets of OMA-mediated translational repression identified in prior work (zif-1, mom-2, nos-2, and glp-1; Jadhav et al. 2008; Guven-Ozkan et al. 2010; Kaymak and Ryder 2013; Oldenbroek et al. 2013). As a class, mRNAs enriched in OMA RNPs have reproductive functions. Our analysis of eight new mRNA targets reveals that the OMA proteins mediate 3′-UTR-dependent translational repression of six. The OMA proteins mediate translational repression of the mRNA targets tested (cdc-25.3, rnp-1, and rnf-5) in both the presence and absence of sperm. As shown in the accompanying article, translational regulation of the CDK-1 activator CDC-25.3 is relevant to the regulation of oocyte meiotic maturation (Spike et al. 2014). Among the mRNA targets tested, the level of OMA-mediated translational repression varies considerably depending on the sequence of the 3′-UTR. This observed variation in the strength of 3′-UTR-dependent translational repression might be biologically relevant for ensuring that oocytes contain the proper levels of proteins commensurate with their roles in oogenesis. Consistent with this view, genetic analysis of rnp-1 indicates that it is required both for the hermaphrodite sperm-to-oocyte switch and for normal oocyte development.
Our study is consistent with prior work that shows the OMA proteins contribute to translational repression in oocytes (Jadhav et al. 2008; Guven-Ozkan et al. 2010; Kaymak and Ryder 2013; Oldenbroek et al. 2013). However, we cannot exclude the possibility that OMA RNPs might function to activate the translation of some mRNA targets. In our 3′-UTR reporter analysis, we focused on a limited number of mRNA targets that were stably associated with OMA-1; most were also relatively abundant in the OMA-1 immunopurification. A more comprehensive examination will be needed to test whether OMA RNPs can also activate the translation of a subset of OMA-1 target mRNAs. Consistent with this possibility, we observed a significant overlap between GLD-2-associated (Kim et al. 2010b) and OMA-1-associated mRNAs. GLD-2 is thought to be a translational activator (reviewed in Ivshina et al. 2014) and it has been reported to stabilize its mRNA targets (Kim et al. 2010b). Taken together, our analysis is consistent with the idea that the OMA proteins promote oogenesis in part through the translational regulation of a battery of mRNA targets that mediate myriad biochemical and reproductive functions.
To complement the analysis of OMA target mRNAs, we also analyzed OMA-1-associated proteins using mass spectrometry. We focused our analysis on proteins that copurify with OMA-1 in the presence of RNase A, based on the prediction that this set might include proteins with the tightest association with OMA-1 or its associated factors. In addition, the RNase A treatment is expected to reduce the identification of biologically irrelevant interactions that might occur between proteins in the lysate and mRNAs in OMA RNPs. Since RNPs are heterogeneous and dynamic and the oogenic germ line contains cells in the continuum of stages of meiotic prophase, our purifications likely report the average composition of a variety of distinct OMA-1-containing RNPs. Nonetheless, this set provides a useful starting point for unraveling the mechanism by which the OMA proteins regulate oocyte growth and meiotic maturation.
We took two approaches to validate this set of OMA-1-associated proteins. In the first approach, we took advantage of yeast two-hybrid screens for OMA-1-interacting proteins. In the second approach, we identified proteins required for the translational repression of zif-1, an OMA target mRNA. Interactions identified using the yeast two-hybrid system must occur in the absence of biologically relevant target mRNAs and their 3′-UTR sequences; however, we cannot exclude the possibility that nonspecific mRNA might mediate some of the protein–protein interactions in the heterologous system. A critical experimental design element of the two-hybrid analyses was that they were conducted independently from and blind to the affinity purifications. Strikingly, our purifications identified all oocyte proteins known to interact with OMA-1, including PQN-59, GLD-1, MEX-3, OMA-2, SPN-4, and IFET-1. Importantly, many of these proteins have established roles in translational control. For example, IFET-1 was proposed to have a wide-ranging role in translational repression during germline development (Sengupta et al. 2013). Indeed, IFET-1 was one of the two OMA-1-associated proteins identified as being required for translational repression of the zif-1 3′-UTR reporter. The other protein was LIN-41, and in the accompanying article we demonstrate an in vivo requirement for LIN-41 in the control of oocyte growth and meiotic maturation (Spike et al. 2014).
The set of OMA-1-associated proteins contains translational repressors (RNA-binding proteins and components of the Ccr4–Not deadenylase complex) and translational activators (GLD-2 and GLD-3), consistent with the idea that the OMA proteins play a major role in translational regulation during oogenesis. OMA-1-associated proteins include several well-documented RNA-binding proteins shown to mediate 3′-UTR-dependent translational repression in the context of germline development, including GLD-1 (Jones and Schedl 1995; Jan et al. 1999; Lee and Schedl 2001; Schumacher et al. 2005; Jungkamp et al. 2011; Wright et al. 2011; Doh et al. 2013) and MEX-3 (Jadhav et al. 2008; Pagano et al. 2009; Mainpal et al. 2011). The TRIM-NHL protein LIN-41 might also bind RNA because the NHL-repeat domain of the Drosophila Brain Tumor protein was recently shown to bind RNA via the positively charged surface of its six-bladed β propeller NHL-repeat domain (Edwards et al. 2003; Loedige et al. 2014). OMA-1, and presumably OMA-2, binds strongly to RNA containing multiple copies of UA[A/U] sequences in vitro (Kaymak and Ryder 2013). However, OMA-1 binds RNA with ∼50-fold lower affinity than its mammalian paralog tristetraprolin, which binds AU-rich elements (Brewer et al. 2004; reviewed by Brooks and Blackshear 2013). While we detected a modest enrichment of OMA-1-binding motifs in OMA-1-associated 3′-UTRs, these sequences do not appear to be sufficient to guarantee association with or translational repression by OMA-1. Interactions with other RNA-binding proteins, which we detected in our affinity purifications and by two-hybrid assays, might confer greater specificity in vivo. Different mRNA targets might recruit distinct constellations of RNA regulators to generate their appropriate gene-specific expression patterns. In fact, combinatorial interactions between multiple translational regulators were recently shown to restrict the expression of the OMA target mom-2/wnt to embryonic blastomeres that transmit the Wnt signal (Oldenbroek et al. 2013).
Our analysis of OMA RNPs prompts two related questions: Can defects in translational repression alone explain the oma-1; oma-2 mutant phenotype and are all OMA target mRNAs repressed? In one model, a global defect in 3′-UTR-mediated translational repression would cause the missexpression of a cohort of proteins in mutant oocytes, thereby interfering with their capacity to respond to the MSP meiotic maturation signal. While this model cannot be excluded, several observations suggest this might not be the case. In the absence of sperm, oma-1; oma-2 mutant oocytes appear superficially normal, and double mutant oocytes retain the capacity to activate CDK-1 (Detwiler et al. 2001). If a large-scale abnormality in translational repression prevented meiotic maturation, one might imagine that such a block might be recapitulated by other mutations that perturb gene expression in oocytes or other insults to oocyte physiology. Extensive genetic analyses highlight the relative uniqueness of the oma-1; oma-2 double mutant phenotype. Besides removing sperm from the gonad due to germ line feminization (McCarter et al. 1999), the only single gene mutations that block oocyte meiotic maturation interfere with PKA activation in gonadal sheath cells. This is observed in acy-4 (encodes adenylate cyclase 4) null mutants and in genetically mosaic animals whose gonadal sheath cells contain strong loss-of-function mutations in kin-1 (encodes the catalytic subunit of PKA) and gsa-1 (encodes the stimulatory G protein Gαs; Govindan et al. 2009; Kim et al. 2012). Our analysis of the OMA-1-associated protein LIN-41 provides the best argument that a defect in translational repression might be an insufficient explanation for the oma-1; oma-2 mutant phenotype (this work; Spike et al. 2014). Our data here show that LIN-41 represses the translation of OMA targets in oocytes, including cdc-25.3. In the accompanying article in this issue, we report a comprehensive analysis of the essential roles of lin-41 in oogenesis (Spike et al. 2014). LIN-41 and the OMA proteins exhibit an antagonistic relationship—LIN-41 inhibits M-phase entry and oocyte cellularization, whereas the OMA proteins promote these events. Taken together, these studies suggest the OMA RNP is a key regulator of the oogenic program that coordinates and controls oocyte growth and meiotic maturation.
Supplementary Material
Acknowledgments
We are grateful to Keith Blackwell, Barth Grant, Alexandre Paix, and Geraldine Seydoux for providing strains, reagents, or protocols. We thank Mary Kroetz and Tim Schedl for their helpful suggestions during the course of this work. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by grant P40OD010440 from the National Institutes of Health (NIH) Office of Research Infrastructure Programs. Next generation DNA sequencing and microarray analyses were carried out at the University of Minnesota Genomics Center with the assistance of Aaron Becker, Nicole Peterson, and Kenny Beckman and with instrumentation supported by grant 1S10RR026342-01 from the NIH National Center for Research Resources. Genome-wide analysis of OMA RNP components was carried out in part using software and hardware provided by the University of Minnesota Supercomputing Institute. Mass spectrometry was carried out at the Taplin Mass Spectrometry Facility with the assistance of Ross Tomaino. This work was supported by NIH grants GM57173 and GM65115 to D.G. and HD37933 and GM84198 to R.L. M.O. was supported by an NIH genetics training grant (5T32GM083831).
Note added in proof: See Spike et al. 2014 (pp. 1535–1558) in this issue for a related work.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.168823/-/DC1.
Communicating editor: M. V. Sundaram
Literature Cited
- Amiri A., Keiper B. D., Kawasaki I., Fan Y., Kohara Y., et al. , 2001. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128: 3899–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., Ohmachi M., Naya S., Hayes M., Miranda A., et al. , 2009. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl. Acad. Sci. USA 106: 4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D. C., Ryan K., Manley J. L., Richter J. D., 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651. [DOI] [PubMed] [Google Scholar]
- Baugh L. R., Hill A. A., Slonim D. K., Brown E. L., Hunter C. P., 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900. [DOI] [PubMed] [Google Scholar]
- Benoit P., Papin C., C., J. E. Kwak, M. Wickens, and M. Simonelig, 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F., 2008. Prelude to a division. Annu. Rev. Cell Dev. Biol. 24: 397–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag P. R., Nakamura A., Blackwell T. K., 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132: 4975–4986. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent A. E., MacQueen A., Hazelrigg T., 2000. The Drosophila wispy gene is required for RNA localization and other microtubule-based events of meiosis and early embryogenesis. Genetics 154: 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. Y., Malicka J., Blackshear P. J., Wilson G. M., 2004. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 279: 27870–27877. [DOI] [PubMed] [Google Scholar]
- Brooks S. A., Blackshear P. J., 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows A. E., Sceurman B. K., Kosinski M. E., C. T. Richie, P. L. Sadler et al, 2006. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development 133: 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., et al. , 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18: 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Singal A., Kimble J., Ellis R. E., 2000. A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev. Biol. 217: 77–90. [DOI] [PubMed] [Google Scholar]
- Chen J., Melton C., Suh N., Oh J. S., Horner K., et al. , 2011. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Nat. Cell Biol. 15: 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Klancer R., Singson A., Seydoux G., 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139: 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T. R., Mains P. E., 1993. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics 134: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P. E., 1994a Localization of the mei-1 gene product of Caenorhabditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P. E., 1994b mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136: 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras V., Richardson M. A., Hao E., Kieper B. D., 2008. Depletion of the cap-associated isoform of translation factor eIF4G induces apoptosis in C. elegans. Cell Death Differ. 15: 1232–1242. [DOI] [PubMed] [Google Scholar]
- Cui J., Sackton K. L., Horner V. L., Kumar K. E., Wolfner M., 2008. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics 178: 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Sartain C. V., Pleiss J. A., Wolfner M. F., 2013. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 383: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. [DOI] [PubMed] [Google Scholar]
- Doh J. H., Jung Y., Reinke V., Lee M. H., 2013. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm 2: e26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow M. R., Mains P. E., 1998. Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics 150: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs S. M., 2010. Regulation of the G2/M transition in rodent oocytes. Mol. Reprod. Dev. 77: 566–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S., Khatri P., Eklund A. C., Szallasi Z., 2006. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 22: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy W. G., Brizuela L., Beach D., Newport J., 1988. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 54: 423–431. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T. A., Wilkinson B. D., Wharton R. P., Aggarwal A. K., 2003. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 17: 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R. and T. Schedl, 2007 Sex determination in the germ line (March 5, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.82.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T. C., Crittenden S. L., Kodoyianni V., Kimble J., 1994. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell 77: 183–194. [DOI] [PubMed] [Google Scholar]
- Farboud B., Nix P., Jow M. M., Gladden J. M., Meyer B. J., 2013. Molecular antagonism between X-chromosome and autosome signals determines nematode sex. Genes Dev. 27: 1159–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I., Blazquez M., Palmer A., Eritja R., Nebreda A. R., 1999. A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 13: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Norbury C., Lohka M., Nurse P., Maller J., 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54: 433–439. [DOI] [PubMed] [Google Scholar]
- Gautier J., Minshull J., Lohka M., Glotzer M., Hunt T., et al. , 1990. Cyclin is a component of maturation-promoting factor from Xenopus. Cell 60: 487–494. [DOI] [PubMed] [Google Scholar]
- Gerber A. P., Herschlag D., Brown P. O., 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Echeverri C., Oegema K., Coulson A., Jones S. J., et al. , 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., Greenstein D., 2006. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., Hird S., Plasterk R. H., Andachi Y., Kohara Y., et al. , 1994. Targeted mutations in the Caenorhabditis elegans POU homeo box gene ceh-18 cause defects in oocyte cell cycle arrest, gonad migration, and epidermal differentiation. Genes Dev. 8: 1935–1948. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R., 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Robertson S. M., Nishi Y., Lin R., 2010. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development 137: 3373–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccard O., Jessus C., 2006. Redundant pathways for Cdc2 activation in Xenopus oocyte: either cyclin B or Mos synthesis. EMBO Rep. 7: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. [DOI] [PubMed] [Google Scholar]
- Harris J. E., Govindan J. A., Yamamoto I., Schwartz J., Kaverina I., et al. , 2006. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev. Biol. 299: 105–121. [DOI] [PubMed] [Google Scholar]
- Hentze M. W., 1997. eIF4G: A multipurpose ribosome adapter? Science 275: 500–501. [DOI] [PubMed] [Google Scholar]
- Hochegger H., Klotzbücher A., Kirk J., Howell M., le Guellec K., et al. , 2001. New B-type cyclin synthesis is required between meiosis I and II during Xenopus oocyte maturation. Development 128: 3795–3807. [DOI] [PubMed] [Google Scholar]
- Houston D. W., 2013. Regulation of cell polarity and RNA localization in vertebrate oocytes. Int. Rev. Cell Mol. Biol. 306: 127–185. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A., 2009a Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang, W., B. T. Sherman, X. Zheng, J. Yang, T. Imamichi et al., 2009b Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics Chapter 13: Unit 13.11. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A., Noble S. L., Cameron C., Evans T. C., 2013. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev. Cell 27: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M., Lasko P., Richter J. D., 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 30: 393–415. [DOI] [PubMed] [Google Scholar]
- Jadhav S., Rana M., Subramaniam K., 2008. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135: 1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P., 2011. Formation, regulation, and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., Goodwin E. B., 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18: 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., Schedl T., 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9: 1491–1504. [DOI] [PubMed] [Google Scholar]
- Jud M. C., Czerwinski M. J., Wood M. P., Young R. A., Gallo C. M., et al. , 2008. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 318: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkamp A. C., Stoeckius M., Mecenas D., Grün D., Mastrobuoni G., et al. , 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kaymak E., Ryder S. P., 2013. RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J. Biol. Chem. 288: 30463–30472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kawasaki I., Shim Y. H., 2010a cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. J. Cell Sci. 123: 993–1000. [DOI] [PubMed] [Google Scholar]
- Kim K. W., Wilson T. L., Kimble J., 2010b GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 107: 17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Govindan J. A., Tu Z. J., Greenstein D., 2012. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics 192: 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433. [DOI] [PubMed] [Google Scholar]
- Kong J., Lasko P., 2012. Translational control in cellular and developmental processes. Nat. Rev. Genet. 13: 383–394. [DOI] [PubMed] [Google Scholar]
- Kornbluth S., Sebastian B., Hunter T., Newport J., 1994. Membrane localization of the kinase which phosphorylates p34cdc2 on threonine 14. Mol. Biol. Cell 5: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G., 1991. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell 64: 903–914. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. H. and T. Schedl, 2006 RNA-binding proteins (April 18, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.79.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007. Multiple functions and dynamic activation of MPK-1 ERK signaling in C. elegans germline development. Genetics 177: 2039–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand J. L., Dellinger R. W., Knudsen K. E., Subramani S., Donoghue D. J., 1999. Speedy: a novel cell cycle regulator of the G2/M transition. EMBO J. 18: 1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zheng P., Dean J., 2010. Maternal control of early mouse development. Development 137: 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Albertini D. F., 2013. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 14: 141–152. [DOI] [PubMed] [Google Scholar]
- Li W., DeBella L. R., Guven-Ozkan T., Lin R., Rose L. S., 2009. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J. Cell Biol. 187: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Gaidatzis D., Sack R., Meister G., Filipowicz W., 2013. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Stotz M., Qamar S., Kramer K., Hennig J., et al. , 2014. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka M. J., Hayes M. K., Maller J. L., 1988. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc. Natl. Acad. Sci. USA 85: 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin A. L., Evans T. C., 2007. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev. Biol. 303: 635–649. [DOI] [PubMed] [Google Scholar]
- Madl J. E., Herman R. K., 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainpal R., Priti A., Subramaniam K., 2011. PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 360: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains P. E., Kemphues K. J., Sprunger S. A., Sulston I. A., Wood W. B., 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 126: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L. H. Lou, Y. Lou, N. Wang, and F. Jin, 2014. Behavior of cytoplasmic organelles and cytoskeleton during oocyte maturation. Reprod. Biomed. Online 28: 284–299. [DOI] [PubMed] [Google Scholar]
- Masui Y., 2001. From oocyte maturation to the in vitro cell cycle: the history of discoveries of maturation-promoting factor (MPF) and cytostatic factor (CSF). Differentiation 69: 1–17. [DOI] [PubMed] [Google Scholar]
- Masui Y., Markert C. L., 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 177: 129–145. [DOI] [PubMed] [Google Scholar]
- Masui Y., Clarke H. J., 1979. Oocyte maturation. Int. Rev. Cytol. 57: 185–282. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1997. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181: 121–143. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., et al. , 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10: 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Schubert C., Draper B., Zhang W., Lobel R., et al. , 1996. The PIE-1 protein and germline specification in C. elegans embryos. Nature 382: 710–712. [DOI] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, C., C. M. Gallo, D. Rasoloson, and G. Seydoux, 2010 Transgenic solutions for the germline (February 8, 2010), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.148.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Miller M. A., Ruest P. J., Kosinski M., Hanks S. K., Greenstein D., 2003. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 17: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Moll R., Divo M., Langbein L., 2008. The human keratins: biology and pathology. Histochem. Cell Biol. 129: 705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. R., Coleman T. R., Kumagai A., Dunphy W. G., 1995. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science 270: 86–90. [DOI] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S. I., Hassold T. J., Hunt P. A., 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y., Lin R., 2005. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev. Biol. 288: 139–149. [DOI] [PubMed] [Google Scholar]
- Noble S. L., Allen B. L., Goh L. K., Nordick K., Evans T. C., 2008. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182: 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Eckmann C. R., 2013. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757: 205–247. [DOI] [PubMed] [Google Scholar]
- Nousch M., Techritz N., Hampel D., Millonigg S., Eckmann C. R., 2013. The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci. 126: 4274–4285. [DOI] [PubMed] [Google Scholar]
- Nurse P., 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Ogura K., Kishimoto N., Mitani S., Gengyo-Ando K., Kohara Y., 2003. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 130: 2495–2503. [DOI] [PubMed] [Google Scholar]
- Oldenbroek M., Robertson S. M., Guven-Ozkan T., Gore S., Nishi Y., et al. , 2012. Multiple RNA-binding proteins function combinatorially to control the soma-restricted expression pattern of the E3 ligase subunit ZIF-1. Dev. Biol. 363: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenbroek M., Robertson S. M., Guven-Ozkan T., Spike C., Greenstein D., et al. , 2013. Regulation of maternal Wnt mRNA translation in C. elegans embryos. Development 140: 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano J. M., Farley B. M., Essien K. I., Ryder S. P., 2009. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. USA 106: 20252–20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G., 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell 5: 451–462. [DOI] [PubMed] [Google Scholar]
- Pintard L., Kurz T., Glaser S., Willis J. H., Peter M., et al. , 2003. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13: 911–921. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines R. T., McCormick M., Van Oosbree T. R., Mierendorf R. C., 2000. The S.Tag fusion system for protein purification. Methods Enzymol. 326: 362–376. [DOI] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5: 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P., She M., Parker R., 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45: 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J. L., Pachter L., 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S., Lin R., 2013. The oocyte-to-embryo transition. Adv. Exp. Med. Biol. 757: 351–372. [DOI] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T., et al. , 1997. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192: 59–77. [DOI] [PubMed] [Google Scholar]
- Sagata N., Oskarsson M., Copeland T., Brumbaugh J., Vande Woude G. F., 1988. Function of c-mos proto-oncogene product in meiotic maturation in Xenopus oocytes. Nature 335: 519–525. [DOI] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D., Kelly W. G., 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. [DOI] [PubMed] [Google Scholar]
- Schumacher J. M., Golden A., Donovan P. J., 1998. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M. S., Low W. Y., Patterson J. R., Kim H. M., Traven A., et al. , 2013. ifet-1 is a broad-scale translational repressor required for normal P granule formation in C. elegans. J. Cell Sci. 126: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Mello C. C., Pettitt J., Wood W. B., Priess J. R., et al. , 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382: 713–716. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Dunn M. A., 1997. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124: 2191–2201. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R., White J. G., Southgate E., Podbilewicz B., 1999. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development 126: 691–699. [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Soto M. C., Ishidate T., Kim S., Nakamura K., et al. , 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16: 47–55. [DOI] [PubMed] [Google Scholar]
- Singson A., Mercer K. B., L’Hernault S. W., 1998. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 93: 71–79. [DOI] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D., et al. , 2014. The TRIM- NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of C. elegans oocytes. Genetics 198: 1535–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Buster D. W., Bazirgan O. A., McNally F. J., Mains P. E., 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14: 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis. elegans. Genetics 198: 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Pellettieri J., Seydoux G., 2006. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 16: 56–62. [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G., 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871. [DOI] [PubMed] [Google Scholar]
- Tabara H., Hill R. J., Mello C. C., Priess J. R., Kohara Y., 1999. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126: 1–11. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E., Paix A., Seydoux G., 2012. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139: 3732–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Shi Y., Blackwell T. K., 2004. An extensive requirement for transcription factor IID-specific TAF-1 in Caenorhabditis elegans embryonic transcription. J. Biol. Chem. 279: 15339–15347. [DOI] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Whitten S. J., Miller M. A., 2007. The role of gap junctions in Caenorhabditis elegans oocyte maturation and fertilization. Dev. Biol. 301: 432–446. [DOI] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. [DOI] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., et al. , 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E., et al. , 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 30: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.