Abstract
In many animals, oocytes enter meiosis early in their development but arrest in meiotic prophase I. Oocyte growth, which occurs during this arrest period, enables the acquisition of meiotic competence and the capacity to produce healthy progeny. Meiotic resumption, or meiotic maturation, involves the transition to metaphase I (M phase) and is regulated by intercellular signaling and cyclin-dependent kinase activation. Premature meiotic maturation would be predicted to diminish fertility as the timing of this event, which normally occurs after oocyte growth is complete, is crucial. In the accompanying article in this issue, we identify the highly conserved TRIM-NHL protein LIN-41 as a translational repressor that copurifies with OMA-1 and OMA-2, RNA-binding proteins redundantly required for normal oocyte growth and meiotic maturation. In this article, we show that LIN-41 enables the production of high-quality oocytes and plays an essential role in controlling and coordinating oocyte growth and meiotic maturation. lin-41 null mutants display a striking defect that is specific to oogenesis: pachytene-stage cells cellularize prematurely and fail to progress to diplotene. Instead, these cells activate CDK-1, enter M phase, assemble spindles, and attempt to segregate chromosomes. Translational derepression of the CDK-1 activator CDC-25.3 appears to contribute to premature M-phase entry in lin-41 mutant oocytes. Genetic and phenotypic analyses indicate that LIN-41 and OMA-1/2 exhibit an antagonistic relationship, and we suggest that translational regulation by these proteins could be important for controlling and coordinating oocyte growth and meiotic maturation.
Keywords: oocyte meiotic maturation, oocyte growth, translational regulation, RNA-binding proteins, TRIM-NHL protein
THE program of oocyte development depends on two key subroutines: one governing oocyte differentiation and a second controlling progression through the meiotic pathway of development (reviewed by Kim et al. 2013). A characteristic aspect of oocyte differentiation is growth: oocytes are often among the largest cells in an organism. The process of oocyte growth ensures the maternal inheritance of cellular organelles and factors essential for embryonic development, including ribosomes, ER, Golgi, mitochondria, cytoplasmic determinants, maternal messenger RNAs (mRNAs), and gene products essential for “housekeeping” functions, among many others. A second defining feature of animal oogenesis is meiotic arrest: developing oocytes arrest in meiotic prophase I, and oocytes typically grow during this period of arrest (reviewed by Masui and Clarke 1979; Downs 2010; Kim et al. 2013). Oocytes resume meiosis in the process of meiotic maturation, which is often regulated by hormonal signaling and soma–germline interactions. Meiotic maturation involves the transition to metaphase I (M phase), and its hallmarks are nuclear envelope breakdown, cortical cytoskeletal rearrangement, and meiotic spindle assembly. Activation of the Cdk1/cyclin B kinase (maturation-promoting factor) is sufficient to drive prophase-arrested oocytes into M phase (reviewed by Nurse 1990; Masui 2001). Meiotic resumption must be tightly regulated to ensure that only fully grown oocytes of the highest quality mature. The mechanisms that control and coordinate oocyte growth and meiotic maturation are incompletely understood.
Caenorhabditis elegans provides an attractive system for studies of oogenesis because the adult hermaphrodite gonad is spatially organized (Supporting Information, Figure S1). The differentiating progeny of germline stem cells enter meiosis distally and form oocytes as they progress proximally. As in other organisms, intercellular signaling plays a major role in organizing the development and function of the gonad (reviewed by Lesch and Page 2012; Hansen and Schedl 2013; Kim et al. 2013). The oocyte in the most proximal position (–1 oocyte) undergoes meiotic maturation in response to the major sperm protein (MSP) hormone (McCarter et al. 1999; Miller et al. 2001). The presence of sperm in the gonad also stimulates oocyte formation (Ward and Carrel 1979). MSP signaling promotes the actomyosin-dependent cytoplasmic flows that drive oocyte growth (Wolke et al. 2007; Nadarajan et al. 2009). This mode of regulation ensures that meiotic maturation and ovulation occur in assembly-line fashion when sperm are available for fertilization. Extracellular MSP exhibits a graded distribution in the proximal gonad arm (Kosinski et al. 2005), but this seems an insufficient explanation for the spatial restriction of meiotic maturation because more distally located oocytes (i.e., the oocytes in the –2 and –3 positions) often exhibit molecular evidence of MSP signal reception. For example, these distal oocytes often display mitogen-dependent protein kinase (MAPK) activation, the localization of the Aurora B AIR-2 kinase to chromatin, and rearrangement of the microtubule cytoskeleton (Miller et al. 2001; Harris et al. 2006; Lee et al. 2007; Govindan et al. 2009). The mechanisms that might restrict meiotic maturation to the –1 oocyte are poorly understood but are thought to involve a combination of intercellular signaling and germline intrinsic mechanisms (see McCarter et al. 1999; Harris et al. 2006; Kim et al. 2013 for a discussion).
Studies of C. elegans gld-1, which encodes a STAR-family RNA-binding protein, provide a precedent for how germline-intrinsic mechanisms might coordinate oocyte differentiation and meiotic progression (Francis et al. 1995a,b; Jones and Schedl 1995; Jones et al. 1996). In gld-1 null mutants, oogenesis is abolished: germ cells that adopt a female sexual fate do not exhibit any evidence of oocyte differentiation (Francis et al. 1995a,b; Jones et al. 1996). Instead, gld-1 mutant female germ cells exit from the pachytene stage of meiotic prophase and reenter mitosis, resulting in the formation of a germline tumor (Francis et al. 1995a,b). GLD-1 functions in part to promote oocyte differentiation by repressing the translation of mRNA targets (Lee and Schedl 2001, 2004; Schumacher et al. 2005; Biedermann et al. 2009; Wright et al. 2011). In this work, we investigate the mechanisms that coordinately regulate oocyte growth and spatially restrict meiotic maturation in C. elegans. In the accompanying article in this issue (Spike et al. 2014), we affinity-purified ribonucleoprotein particles (RNPs) containing the TIS11 zinc-finger RNA-binding proteins OMA-1 and OMA-2 (referred to as the OMA proteins), which are redundantly required for meiotic maturation (Detwiler et al. 2001). The TRIM-NHL protein LIN-41 was identified as a component of OMA-1-containing ribonucleoprotein particles (OMA RNPs), and it participates in 3′ UTR-mediated translational repression (Spike et al. 2014). Here we reveal LIN-41 as an essential regulator of the oocyte fate that promotes oocyte growth, inhibits M-phase entry, and maintains oocyte quality. Taken together with the results of the accompanying article in this issue (Spike et al. 2014), these studies reveal OMA RNPs as key regulators of oogenesis coordinately required for the control of oocyte growth and the proper spatial and temporal execution of the meiotic maturation decision.
Materials and Methods
Strains
The genotypes of strains used in this study are reported in Table S1. The following mutations were used: LGI—gld-2(q497), unc-13(e1091), rrf-1(pk1417), lin-41(n2914), lin-41(tn1505), lin-41(tn1487ts), lin-41(tn1487tstn1515), lin-41(tn1487tstn1516), lin-41(tn1487tstn1536), lin-41(tn1487tstn1539), lin-41(tn1541[gfp::tev::s::lin-41]), and lin-11(n566); LGII—daz-1(tj3), unc-4(e120), and lin-29(n333); LGIII—cdc-25.3(ok358), glp-1(bn18ts), emb-30(tn377ts), ced-7(n1892), unc-119(ed3), and unc-64(e246); LGIV—unc-24(e1172), oma-1(zu405te33), mbk-2(pk1427), and ced-3(n717); LGV—oma-2(te51) and fog-2(oz40); LGX—let-7(n2853ts). The following rearrangements were used: hT2[bli-4(e937) let-?(q782) qIs48] (I;III), mIn1[dpy-10(e128) mIs14] II, and nT1[qIs51] (IV; V). The following complex-array transgenes and transgene insertions were used: tnEx196[lin-41(fosmid)::gfp, myo-2p::TdTomato], tnEx198[lin-41(fosmid), myo-2p::TdTomato], isIs18[pSL445 pie-1p::gfp::sas-6 (Leidel et al. 2005), unc-119(+)], itIs37[pie-1p::mCherry::H2B::pie-1 3′ UTR, unc-119(+)] (McNally et al. 2006), and otIs45[unc-119p::gfp] V (Altun-Gultekin et al. 2001).
Antibody preparation and purification
lin-41 cDNA sequences were cloned into the Escherichia coli expression vector pMal-c2 to create an inducible fusion protein wherein maltose-binding protein (MBP) was fused to amino acids 203–420 of LIN-41 (MBP::LIN-41). MBP::LIN-41 was column- and gel-purified and used to immunize both guinea pigs and rabbits. Immunizations and sera collection were performed using standard protocols (Cocalico Biologicals, Reamstown, PA). Guinea pig antibodies (GP49, GP50) were affinity-purified on an MBP::LIN-41-coupled column after antibodies recognizing MBP were removed. Rabbit antibodies (R214) were similarly affinity-purified after antibodies recognizing MBP, E. coli proteins, and C. elegans proteins in a lysate made from sterile glp-1(ts) adults were removed. All purified antibodies recognize LIN-41 on Western blots and exhibit similar patterns by immunofluorescence in adult hermaphrodite germ lines.
Immunofluorescence, fluorescent labeling, and microscopy
Dissected gonads stained with the following antibodies were fixed in 3% paraformaldehyde as described (Rose et al. 1997) with some modifications. Gonads stained with tetramethylrhodamine-labeled phalloidin (Molecular Probes, 0.165 μM) were treated post-fix with acetone (2 min) instead of methanol. Gonads stained with rat anti-tubulin (YL1/2; 1:100; Accurate Chemical) often used a modified fixation buffer composed of 75 mM PIPES (pH 6.9) to help preserve microtubules. Gonads stained with goat anti-SYP-1 (1:4000; kindly provided by Abby Dernburg, University of California-Berkeley) and rabbit anti-SYP-2 (1:500; also provided by Abby Dernburg) were fixed with 1% paraformaldehyde for 10 min. Other primary antibodies were a mixture of two purified mouse monoclonal anti-MSP antibodies (each at 1:300) (Kosinski et al. 2005), rabbit anti-RME-2 antibody (1:50; kindly provided by B. Grant, Rutgers University) (Grant and Hirsh 1999), rabbit anti-phospho-histone H3 (Ser10; 1:400; Millipore), guinea pig anti-LMN-1 (1:800; kindly provided by J. Liu, Cornell University) (Liu et al. 2000), mouse anti-MAPKYT (1:400; Sigma), and guinea pig anti-LIN-41 (1:100; this work). Secondary antibodies were Alexa 488-conjugated donkey anti-rabbit (1:500; Jackson ImmunoResearch), Alexa 488-conjugated goat anti-rabbit (1:500; Life Technologies), Cy3-conjugated donkey anti-goat (1:500; Jackson ImmunoResearch), Cy3-conjugated goat anti-mouse (1:500; Jackson ImmunoResearch), Cy3-conjugated goat anti-rat (1:500; Jackson ImmunoResearch), Cy3-conjugated goat anti-rabbit (1:500; Jackson ImmunoResearch), and DyLight 488-conjugated donkey anti-guinea pig (1:500; Jackson ImmunoResearch). 4′,6-diamidino-2-phenylindole (DAPI) was used to detect DNA. Acridine orange staining was performed as described (Gartner et al. 2004) using 0.5 ml of a 20 μg/ml acridine orange solution (Fisher Scientific). DIC and fluorescent images were acquired on a Zeiss motorized Axioplan 2 microscope with either a 40× Plan-Neofluar (numerical aperture 1.3) or a 63× Plan-Apochromat (numerical aperture 1.4) objective lens using a AxioCam MRm camera and AxioVision software (Zeiss). Imaging of gonads stained with SYP-1, SYP-2, and phalloidin used an apotome adaptor (Zeiss), as did a few other experiments, as indicated in the figure legends.
Dissected gonads stained with rabbit anti-GLD-1 (1:100; kindly provided by T. Schedl, Washington University School of Medicine) (Jones et al. 1996), mouse anti-CYE-1 (1:20; kindly provided by E. Kipreos, University of Georgia) (Brodigan et al. 2003), rat anti-REC-8 (1:200; kindly provided by J. Loidl, University of Vienna) (Pasierbek et al. 2001), and rabbit anti-HIM-3 (1:500; kindly provided by M. Zetka, McGill University, Montreal) (Zetka et al. 1999) were fixed and stained as described (Jones et al. 1996; Hansen et al. 2004a,b). Fluorescent images were captured using a Zeiss Imager Z.1 microscope with an AxioCam MRm camera (Zeiss).
Western blots
Proteins were separated using NuPage 4-12% Bis–Tris gels (Invitrogen) and visualized after western blotting. Primary antibodies used to detect proteins were guinea pig anti-LIN-41 (1:2000; this work) and mouse monoclonal anti-actin C4 (1:80,000; MP Biomedicals). Secondary antibodies used for western blots were peroxidase-conjugated donkey anti-guinea pig (Jackson ImmunoResearch) and goat anti-mouse (Pierce) antibodies diluted 1:60,000.
Genetic screen for new alleles of lin-41
L4-stage let-7(n2853ts) animals were mutagenized with EMS at 15° and shifted to 25° as adults. An estimated 60,000–120,000 F1 progeny were screened for dominant suppressors of let-7 larval lethality. Suppressor mutations with recessive phenotypes (e.g., sterile or dumpy) that could be balanced by the hT2 (I;III) rearrangement, and suppressor mutations with no phenotype, were tested for their ability to complement lin-41(n2914). Suppressors of lin-41(tn1487ts) sterility were selected in the F2 generation following EMS mutagenesis. Four intragenic revertants were isolated from ∼300,000 EMS-mutagenized haploid genomes.
GFP-tagging LIN-41 in genomic and fosmid contexts
To fuse GFP::TEV::S (Cheeseman and Desai 2005) at the N terminus of endogenous LIN-41, we targeted a specific site near the initiator methionine for Cas9 scission [protospacer-associated motif (PAM) site at position 9341872 in the genome] with a single-guide RNA (sgRNA). Primers U6prom EcoRI F/lin-41gRNAR and lin-41gRNAF/U6prom HindIII R were used to generate overlapping PCR products that could be amplified and used to replace the EcoRI/HindIII insert in pU6::unc-119sgRNA to generate pU6::lin-41sgRNA1 following Friedland et al. (2013). To generate a repair template (pCE2-2), a 5′-lin-41 homology arm was generated by PCR using primers GA-B52 and GA-1R and a lin-41 fosmid (WRM064dG06) as template. To generate the 3′-lin-41 homology arm, nested PCR was performed using primers lin-41-1F and lin-41-1R, followed by GA-B3 and GA-3F. The gfp::tev::s insert was generated by PCR using primers GA-2F and GA-2R with pIC26 as the template. A four-piece Gibson assembly (New England BioLabs) was used to stitch together the 5′-lin-41 homology arm, the gfp::tev::s insert, the 3′-lin-41 homology arm, and the pBluescript KS– vector, digested with SacI and KpnI. Site-directed mutagenesis was performed using a Q5 site-directed mutagenesis kit (New England BioLabs) and primers C13-PAMF and C13-PAMR to alter the PAM site from TTGG to CTTG without changing the coding sequence of LIN-41. For transformation, the pCE2-2 repair template (100 ng/μl), the lin-41 sgRNA1 construct (100 ng/μl), the Peft-3::Cas9-SV40 NLS::tbb-2 3′ UTR (150 ng/μl), and a myo-2p::TdTomato marker (4 ng/μl) were microinjected into 58 wild-type adult hermaphrodites. Two independent targeted lines were isolated from screening the progeny of 452 F1 transgenic animals using a 40× Plan-Neofluar (numerical aperture 1.3) objective lens on a Zeiss Axioskop microscope. The alleles lin-41(tn1541[gfp::tev::s::lin-41]) and lin-41(tn1542[gfp::tev::s::lin-41]) were validated by PCR and sequencing and outcrossed three times against the wild type. The brood size of lin-41(tn1541[gfp::tev::s::lin-41]) was measured and found to be 319 ± 28 (n = 30). Primer sequences used were the following: lin-41gRNAF—5′-GACCATCGTGCCATGCTCATGTTTTAGAGCTAGAAATAGCAAGTTA-3′; lin-41gRNAR—5′-ATGAGCATGGCACGATGGTCAAACATTTAGATTTGCAATTCAATTATATAG-3′; GA-B52—5′-CTATAGGGCGAATTGGAGCTCGTGTGTAGAGAGTGGGAACGAC-3′; GA-1R—5′-CTGCAGCCCGGGGGATCCCATTTCACTTTTTCCAAGTCTGAAAAG-3′; lin-41-1F—5′-CGGGAATGCGACGTTGGAAACG-3′; lin-41-1R—5′-ACTAAATTGGCCCTCCGACT-3′; GA-B3—5′-AGGGAACAAAAGCTGGGTACCGTCGTGTTGAGACGCAAAAAGC-3′; GA-3F—5′-CATGGACAGCGGAGGTGGAGGTATGGCGACCATCGTGCCATGC-3′; GA-2F—5′-CTTTTCAGACTTGGAAAAAGTGAAATGGGATCCCCCGGGCTGCAG-3′; GA-2R—5′-GCATGGCACGATGGTCGCCATACCTCCACCTCCGCTGTCCATG-3′; C13-PAMF—5′-GCCATGCTCACTTGAGAAAGAAGAAG-3′; and C13-PAMR—5′-ACGATGGTCGCCATACCT-3′; other primer sequences are available on request.
Transgenic animals expressing lin-41::gfp were generated using recombineering (Warming et al. 2005; Tursun et al. 2009) and microinjection (Stinchcomb et al. 1985). To create the C-terminal LIN-41::GFP fusion, the fosmid WRM064dG06 was used. lin-41::gfp or unmodified fosmid (1 μg/ml) were injected into lin-41(n2914)/unc-13(e1091) lin-11(n566) animals along with ScaI-digested N2 genomic DNA (100 μg/ml) and myo-2p::TdTomato (2 μg/ml), a co-injection marker. lin-41(n2914) sterility was rescued by the unmodified fosmid but not rescued by lin-41(fosmid)::gfp.
RNA interference and 5-ethynyl-2′-deoxyuridine incorporation
Gene-specific RNA interference (RNAi) was performed by feeding C. elegans with double-stranded RNA (dsRNA)-expressing E. coli (Timmons and Fire 1998) at 22° using the RNAi culture media described by Govindan et al. (2006). The identity of RNAi clones was verified by DNA sequencing. Exposure to dsRNA-expressing E. coli was initiated at the beginning of the first larval stage in lin-41(RNAi) experiments and during the fourth larval stage in cdk-1(RNAi), wee-1.3(RNAi), and plk-1(RNAi) experiments. Adult rrf-1(pk1417) animals (24 hr past mid-L4 at 22°) that had been exposed to lin-41(RNAi) or control bacteria containing a nontargeting RNAi vector were individually picked, washed, and transferred to 5-ethynyl-2′-deoxyuridine- (EdU)-labeled MG1693 bacteria for 4 hr, followed by immediate gonad dissection and fixation. Incorporated EdU was detected after gonad dissection using an Alexa 488 Click-iT EdU detection kit (Invitrogen). MG1693 bacteria that had incorporated EdU were prepared as described (Ito and McGhee 1987; Fox et al. 2011).
Results
lin-41 promotes oocyte growth and meiotic progression
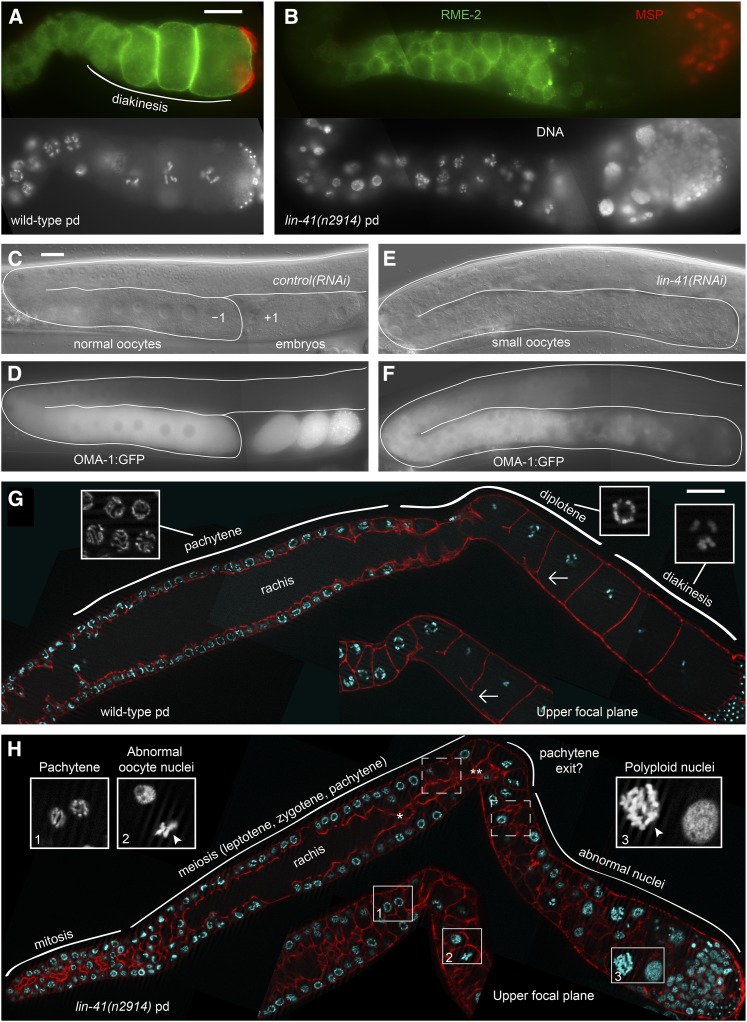
Phenotypic analyses of a strong loss-of-function lin-41 allele previously demonstrated that lin-41(n2914) adults are sterile: they lack normal oocytes but make apparently normal sperm (Slack et al. 2000). LIN-41 protein was also isolated as a component of OMA RNPs (Spike et al. 2014), and the OMA proteins are expressed in oocytes (Detwiler et al. 2001). Together, these observations suggest that LIN-41 might be required in oocytes for normal oogenesis. To determine the basis for the lin-41 sterile phenotype, we examined strong loss-of-function and reduction-of-function alleles. Strong loss-of-function lin-41 alleles confer somatic heterochronic defects, including molting defects that make the animals sickly and complicate analysis of the germ line. Since passage through the dauer stage of development suppresses several heterochronic mutations (Liu and Ambros 1991; Abrahante et al. 2003), we analyzed germline phenotypes in strong loss-of-function lin-41 mutant animals that had developed through the alternative dauer larval stage. Post-dauer-recovered lin-41 mutant animals have some somatic defects, including a short and fat (Dumpy) body shape, but are much healthier than lin-41 animals that do not pass through the dauer stage. lin-41 post-dauer animals are also completely sterile with large germ lines that are easier to analyze. Essentially all of the lin-41 germline phenotypes that we describe are also observed in non-postdauer animals [e.g., lin-41(RNAi) and lin-41(tn1487ts) at 25°]; none appear to depend on life history. A more limited analysis of non-postdauer lin-41(n2914) adults involving an assessment of the oocyte fate and DAPI staining of dissected gonads (n = 9) (Figure 1, A and B) is consistent with this conclusion.
Figure 1.
Sterile lin-41 animals produce sperm and small, abnormal oocytes. (A and B) Immunofluorescence staining identifies sperm (red, MSP) and oocytes (green, RME-2) in wild-type (A) and lin-41(n2914) mutant germ lines (B). (C–F) OMA-1::GFP is present in both control (C and D) and lin-41(RNAi) oocytes (E and F), but the most proximal cells lose expression of OMA-1::GFP and no embryos form. (G and H) Phalloidin stain of the actin cytoskeleton (red) of wild-type (G) and lin-41(n2914) (H) post-dauer (pd) adult gonads reveals that the rachis narrows (single asterisk in H) and appears to terminate (double asterisk in H) around the time when lin-41 germ cells exit from pachytene. The last oocyte connected to the rachis in the wild type is indicated by an arrow (G). All insets are magnified two-fold relative to the main images (G and H). Insets in G show examples of pachytene, diplotene, and diakinesis nuclei. Insets in H show pachytene-stage nuclei (1), abnormal nuclei immediately after exit from pachytene (2), and more proximal nuclei (3). Abnormal nuclei have condensed (arrowheads) or decondensed chromosomes. Bar, 20 μm.
We examined lin-41(n2914) mutants and lin-41(RNAi) adults and noted that germ cells in the proximal arms of these animals contain a rough-appearing cytoplasm similar to the cytoplasm that characterizes wild-type oocytes and early embryos; however, the oocytes appeared small and were arranged in a disorganized fashion throughout the proximal arm (Figure 1, C and E; Figure S2). We examined the expression of MSP, a sperm-specific protein important for sperm motility and meiotic maturation signaling (Italiano et al. 1996; Miller et al. 2001; Kosinski et al. 2005), and RME-2, an oocyte-specific yolk receptor protein (Grant and Hirsh 1999), and confirmed that lin-41 gonads contain both sperm and small disorganized oocytes (Figure 1, A and B). These observations suggest that lin-41(n2914) animals are sterile because they make small, abnormal oocytes. Small, disorganized oocytes expressing OMA-1::GFP are also observed in sterile lin-41(RNAi) animals (Figure 1, C–F). Interestingly, both experiments suggest that lin-41 oocytes become progressively abnormal as they age and move proximally toward the spermatheca. Oocyte marker expression (e.g., RME-2 and OMA-1::GFP) is reduced or absent in the oldest oocytes, and nuclei in this region are often polyploid (Figure 1, B, F, and H). Consistent with the idea that these cells lose the oocyte fate, we observed that a minority of the most proximal cells expressed unc-119p::gfp, a neuronal marker (Maduro and Pilgrim 1995). On the first 2 days of adulthood, 65 of 96 gonad arms of post-dauer lin-41(n2914) animals contained unc-119p::gfp-positive cells adjacent to the spermatheca (average 2.2, range 0–14). The number of cells expressing unc-119p::gfp was sensitive to growth conditions (i.e., growth medium and bacterial food source) (D. Greenstein, unpublished results and as described below).
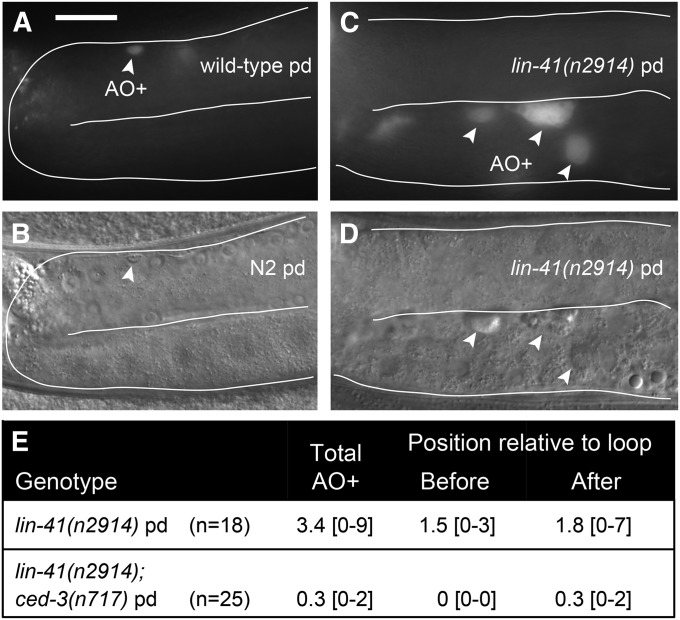
To further confirm that lin-41 germ cells are oogenic, we examined whether lin-41(n2914) germ cells are able to undergo programmed cell death (apoptosis). Germline programmed cell death is restricted to oogenic germ lines (Gumienny et al. 1999) and occurs in late pachytene, just before the gonad turns ventrally and oocytes begin to grow and transition from pachytene into diplotene (Figure 2, A and B). lin-41(n2914) germ lines usually contain several dying cells; however, they are often observed proximally and are larger than those in the wild type (Figure 2, C–E). These observations suggest that lin-41 germ cell death may be abnormal and not restricted to late pachytene. However, because most lin-41 germ cell death requires ced-3 (Figure 2E), which encodes the caspase required for programmed germ cell death (Gumienny et al. 1999), these results are also consistent with the interpretation that lin-41 germ cells can undergo programmed cell death and are oogenic.
Figure 2.
Abnormal germ cells in lin-41 null mutants retain the capacity to undergo programmed cell death and are thus oogenic. (A–D) Acridine orange (AO) accumulates in lin-41(n2914) oocytes (arrowheads in C and D) that are much larger than the late-pachytene-stage cells that undergo programmed cell death in wild type (arrowhead in A and B). (E) Average number of acridine-orange-stained cells (AO+) in animals of the indicated genotypes with the range of observed values in brackets. Bar, 20 μm.
In addition to being small and disorganized, lin-41 oocytes have highly abnormal nuclei, even in the early stages of oogenesis (Figure 1H, inset 2). lin-41(n2914) germ cell nuclei appear normal until late pachytene, but thereafter contain highly condensed or completely decondensed chromosomes (Figure 1H) that generally do not resemble diplotene or diakinesis-stage chromosomes. Wild-type oocytes make the transition from pachytene to diplotene and diakinesis in the context of oocyte growth and cellularization (Figure 1G). When we examined the rachis or cytoplasmic core of the syncytial gonad, a structure essential for normal oocyte growth (Wolke et al. 2007), we found that it narrows and appears to terminate prematurely in lin-41(n2914) mutants (Figure 1H, n = 26). Interestingly, lin-41 oocyte nuclei exit pachytene at approximately the same time, or shortly after, the apparent end of the rachis. Together, these observations suggest that lin-41 oocytes prematurely cellularize and exit meiotic prophase during late pachytene or during the pachytene-to-diplotene transition.
LIN-41 is expressed and functions in the oogenic germ line independently of the heterochronic gene pathway
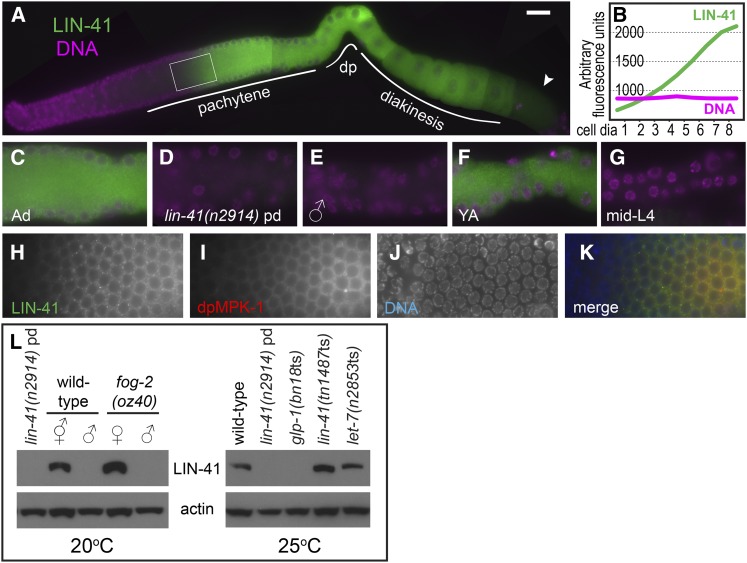
LIN-41-specific antibodies were generated to examine LIN-41 expression and localization in the germ line (Figure 3; Figure S3; Figure S4). LIN-41 is strongly expressed in adult hermaphrodite germ cells beginning in mid-pachytene (Figure 3A). LIN-41 protein levels rise dramatically at this time, increasing several-fold over a distance corresponding to just a few cell diameters (Figure 3B). We explored the timing of this rise and found that it consistently occurs just before the peak of MPK-1 MAPK activation in mid-pachytene (Figure 3, H–K, n = 14), which may promote the progression of oogenic germ cells from early to late pachytene (Lee et al. 2007). LIN-41 is also abundantly expressed in diplotene- and diakinesis-stage oocytes, but is generally reduced in abundance in the –1 oocyte as it undergoes meiotic maturation (14/17 oocytes identified), suggesting that LIN-41 may be eliminated at this time. Consistent with this interpretation, an N-terminal GFP::LIN-41 fusion protein, generated by CRISPR-Cas9 genome editing (Dickinson et al. 2013; Friedland et al. 2013; Tzur et al. 2013) of the endogenous locus, is expressed in oocytes, disappears during meiotic maturation, and is absent from early embryos (Figure 4). A similar observation was made for a nonrescuing C-terminal LIN-41::GFP fusion protein (Figure S5). Most LIN-41 is diffusely cytoplasmic, with a small amount of punctate LIN-41 that is most evident during pachytene (Figure 3, C and H). As expected, no strong staining is apparent in lin-41(n2914) hermaphrodite germ lines (Figure 3D; Figure S3C). LIN-41 was also not detected in the germ lines of males or mid-L4 stage larvae (Figure 3, E and G; Figure S3, D and F), both of which have late-pachytene-stage germ cells undergoing spermatogenesis (Ellis and Schedl 2007; Ellis and Stanfield 2014), suggesting that pachytene-stage LIN-41 expression is restricted to germ cells committed to oogenesis. The primary, and possibly exclusive, site of LIN-41 expression in adults appears to be the oogenic germ line. LIN-41 is detectable by western blot in oogenic adult hermaphrodites and fog-2(oz40) females, but is not detected in wild-type and fog-2 spermatogenic adult males or sterile glp-1(bn18ts) adults (Figure 3L; Figure S4), which lack most germ cells (Austin and Kimble 1987). LIN-41 was not detected in lin-41(n2914) (Figure 2L; Figure S4) or lin-41(tn1505) adults by Western blot (C. Spike, unpublished results), again confirming the specificity of the LIN-41 antibodies. We conclude that LIN-41 expression in the C. elegans germ line is strongly associated, both developmentally and spatially, with the oogenic fate.
Figure 3.
LIN-41 is expressed during oogenesis. (A and B) Immunofluorescence staining of wild-type hermaphrodites shows that LIN-41 expression (green) begins in mid-pachytene and continues throughout oogenesis. Expression begins abruptly and is reduced in oocytes undergoing meiotic maturation (arrowhead). The signal in the area enclosed by the rectangle is quantitated in B. (C–G) LIN-41 expression in late pachytene in an adult hermaphrodite (C), lin-41(n2914) post-dauer hermaphrodite (D), adult male (E), young adult hermaphrodite (F), and mid-L4-stage hermaphrodite (G); images in (C–E) and (F and G) are from the same experiments respectively, and have identical exposure times. Figure S3 shows the entire germ line of each genotype and stage. (H–K) LIN-41 expression begins just before MPK-1 MAP kinase activation (red) peaks in pachytene. Bar, 20 μm (A) and 10 μm (C–K). (L) Western blots examining LIN-41 expression in adult worms of the indicated genotypes; 40–50 worms were used in each lane. The slight mobility shift of LIN-41 in let-7(ts) was not reproducible. Figure S4 contains uncropped images of both LIN-41 blots and shows the positions of size standards.
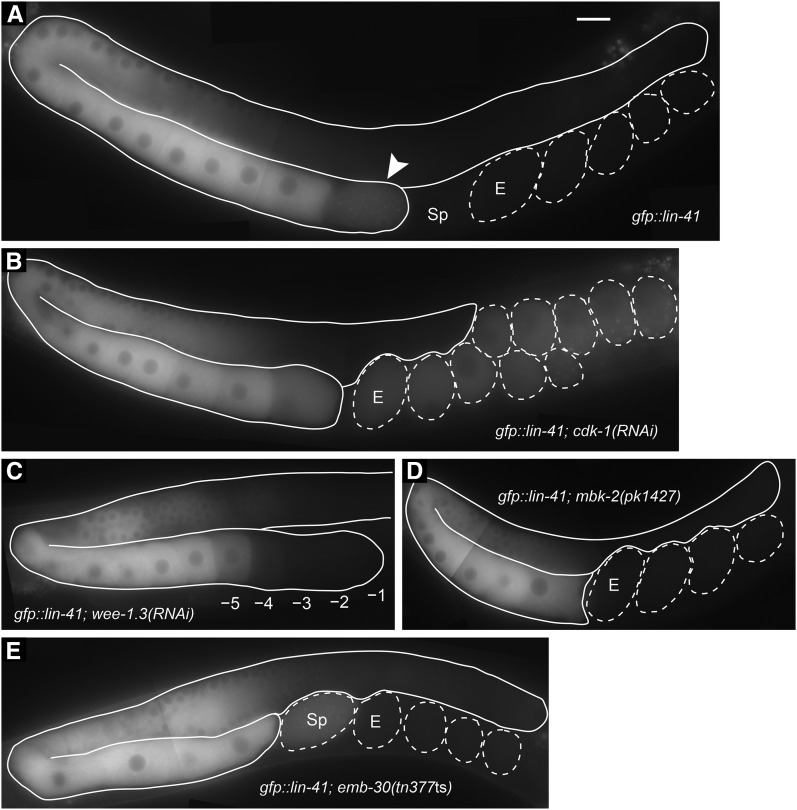
Figure 4.
GFP::LIN-41 begins to be eliminated from oocytes during meiotic maturation. (A–E) A functional GFP::LIN-41 fusion protein was generated by CRISPR-Cas9 genome editing of the lin-41 locus and used to examine the localization of the protein in otherwise wild-type hermaphrodites (A) after RNAi of cdk-1 (B) or wee-1.3 (C) in mbk-2(pk1427) mutants (D) that lack the function of the MBK-2 minibrain-related DYRK-family kinase (Pellettieri et al. 2003) or in emb-30(tn377ts) mutants at 25° (E) that contain a mutation in the APC4 subunit of the anaphase-promoting complex (Furuta et al. 2000). The elimination of GFP::LIN-41 depends on CDK-1 (B). A mildly affected wee-1.3(RNAi) gonad arm shows the elimination of GFP::LIN-41 from the –1 to –4 oocytes, with the GFP signal intensity appearing lower in the –5 oocyte (C). DIC microscopy showed that all oocytes in the gonad arm had intact nuclear envelopes. The arrowhead indicates an oocyte undergoing meiotic maturation that contains a reduced level of GFP::LIN-41 (A). Sp, spermatheca; E, first embryo in the uterus. Bar, 20 μm. Genotypes also included itIs37[pie-1p::mCherry::H2B::pie-1 3′UTR, unc-119(+)] (B and C) and unc-24(e1172) (D).
To determine if lin-41 functions in the germ line or somatic gonad to exert its role in oocyte development, we performed lin-41(RNAi) on rrf-1(pk1417) mutant worms, which are sensitive to RNAi in germ cells, but resistant to RNAi in many somatic cells (Sijen et al. 2001; Kumsta and Hansen 2012). Consistent with the interpretation that lin-41 functions in the germ line for fertility, rrf-1; lin-41(RNAi) adults are sterile with small, disorganized oocytes (Figure S2, C and D), and they display the meiotic progression defect described above for lin-41(n2914) mutant oocytes, but lack somatic lin-41 phenotypes. These phenotypes include the short and fat (Dumpy) body phenotype, which is characteristic of most lin-41 loss-of-function mutants (Slack et al. 2000) and is phenocopied by lin-41(RNAi) in wild-type hermaphrodites (C. Spike, unpublished results).
lin-41 and other members of the heterochronic gene pathway regulate the timing of cell division and terminal differentiation of lateral hypodermal seam cells during larval development (Reinhart et al. 2000; Slack et al. 2000; reviewed by Rougvie and Moss 2013). In this pathway, lin-41 represses the precocious accumulation of LIN-29 protein, and lin-29 is epistatic to a fertile allele of lin-41 with respect to seam cell development (Slack et al. 2000). However, lin-29 is not epistatic to the sterile lin-41(n2914) allele with respect to fertility (Figure S2, A and B), indicating that LIN-29 is not a key target of lin-41 repression in the adult germ line. Likewise, the let-7 microRNA is upstream of lin-41 in the heterochronic pathway and represses LIN-41 translation in hypodermal cells through sequences in the lin-41 3′ UTR, a relationship that is conserved in other systems (Reinhart et al. 2000; Worringer et al. 2014; reviewed by Ecsedi and Grosshans 2013). We examined LIN-41 protein accumulation in let-7(ts) adults raised at the restrictive temperature, but did not see an increase in LIN-41 protein accumulation in whole animals by western blot or in the adult germ line by immunofluorescence (Figure 3L; C. Spike, unpublished data). Although the mature let-7 microRNA is present in adults, it may be confined to somatic cells (Lau et al. 2001) and may not regulate LIN-41 protein accumulation in the germ line. We conclude that LIN-41 is expressed and functions in the oogenic germ line in a manner that is independent of the heterochronic gene pathway.
lin-41 is not required for spermatogenesis
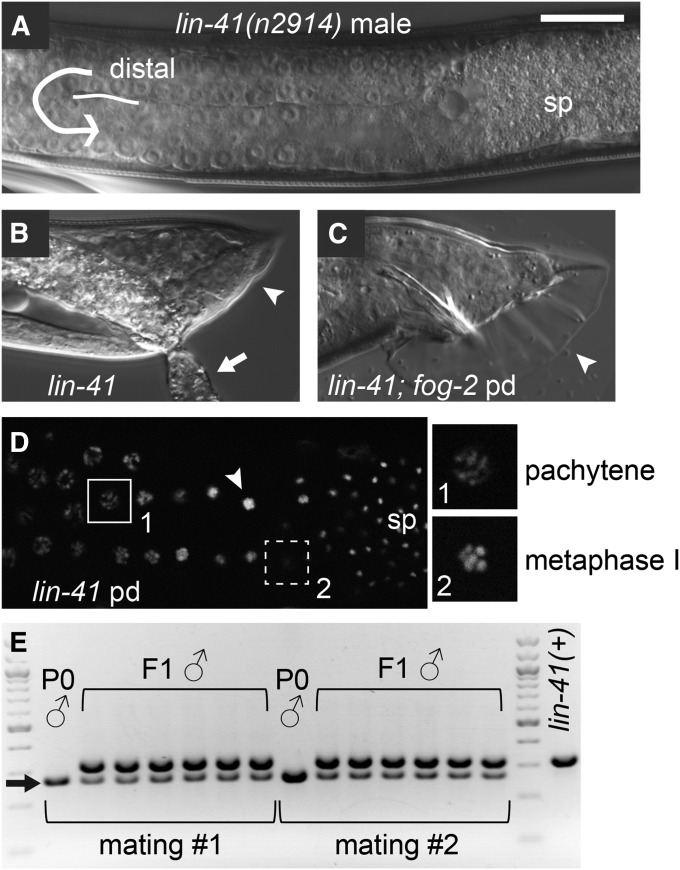
The absence of LIN-41 from spermatogenic germ cells (Figure 3, E, G, and L) and the presence of morphologically normal sperm in lin-41(n2914) adult hermaphrodites (Slack et al. 2000) (Figure 1B) suggests that spermatogenesis is likely normal in lin-41(n2914) mutants. We therefore examined lin-41(n2914) males raised under normal growth conditions; these animals are very small as adults, approximately the size of normal L3-stage males, and have abnormal tails (Figure 5, A and B). Similar male tail abnormalities were previously described for the hypomorphic lin-41(ma104) allele (Del Rio-Albrechtsen et al. 2006). Despite these somatic abnormalities, the lin-41(n2914) adult male germ line appears to be fully developed and produces abundant spermatids (Figure 5A). lin-41 males recovered from starved plates with many dauers also have normal-appearing germ lines (Figure 5D) and are suppressed for the small body size and tail defects (Figure 5C). We tested whether lin-41 post-dauer animals produce viable sperm using a mating test; 3 of 30 lin-41(n2914) post-dauer males sired cross-progeny (Figure 5E). Thus, although lin-41(n2914) post-dauer males have a low mating efficiency, likely due to incomplete suppression of lin-41 male somatic defects (C. Spike, unpublished results), they produce fertilization-competent sperm.
Figure 5.
Spermatogenesis appears normal in lin-41 males. (A and B) lin-41(n2914) males produce spermatids (sp), but have abnormal tails (B, arrowhead). These animals are small and sickly; the arrow points to intestinal material that has leaked through the rectum. (C and D) lin-41(n2914) post-dauer males appear more normal with respect to tail morphology (C, arrowhead) and do not exhibit defects in meiotic progression during spermatogenesis (D). Insets in D are magnified two-fold relative to the main image and show pachytene (1) and metaphase I images (2); nuclei with male-specific karyosome (arrowhead) and spermatid (sp) morphologies are indicated. Images in D were taken using an apotome adaptor. (E) lin-41(n2914); fog-2 post-dauer males (P0s) are able to sire progeny (F1s) when mated to unc-64 hermaphrodites. Male parents and progeny were genotyped by PCR. lin-41(n2914) is a small deletion that results in a smaller PCR product (arrow) than lin-41(+). non-Unc F1 hermaphrodites also segregated Lin-41 animals among their progeny. The fog-2(oz40) marker was used for convenience; fog-2 is dispensable for male development (Schedl and Kimble 1988). Bar, 20 μm (A–D).
LIN-41 NHL-repeat domain is critical for fertility and oocyte quality
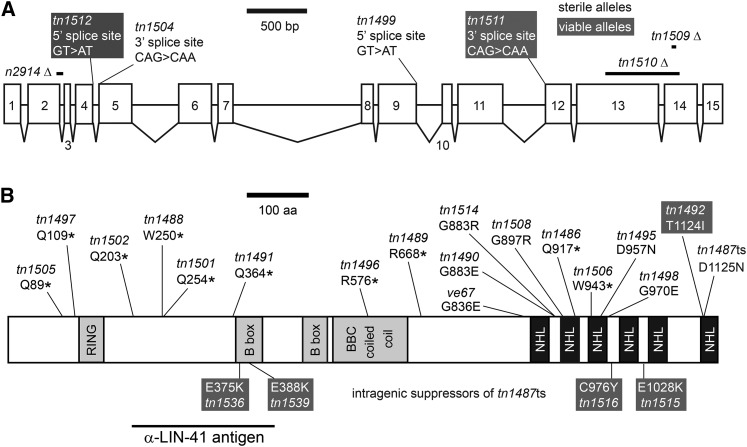
Studies focused on the heterochronic pathway demonstrated that lin-41 is a dominant suppressor of let-7(ts) lethality and identified two sterile alleles of lin-41, including lin-41(n2914), an early out-of-frame deletion, and lin-41(mg187), a DNA rearrangement allele that is no longer extant (Slack et al. 2000). To identify additional sterile alleles of lin-41, we screened for new dominant let-7(ts) suppressors and identified lin-41 alleles based on chromosomal linkage and complementation tests with lin-41(n2914). Sequence changes within the lin-41 gene were identified for 25 of 29 lin-41 alleles, and most of these alleles cause sterility (Figure 6). Importantly, 8 of the sterile lin-41 alleles are missense mutations in conserved residues within the LIN-41 NHL-repeat domain (ve67, tn1490, tn1514, tn1508, tn1495, tn1498/tn1500, tn1487ts). lin-41(tn1487ts) is described in more detail below. One of the three viable lin-41 alleles (tn1492), for which we isolated and identified a molecular lesion, has a missense mutation in the NHL-repeat domain (Figure 6), similar to the viable alleles described by Slack et al. (2000) who first identified this region as critical for LIN-41 function. Based on the new sterile alleles, we conclude that the LIN-41 NHL-repeat domain is critical for fertility as well as seam cell development. This domain is likely to be a sequence-specific RNA-binding domain important for regulating mRNA translation (Loedige et al. 2013, 2014). Indeed, LIN-41 appears to regulate translation in C. elegans oocytes: it copurifies with OMA-1, a known translational regulator, and represses the translation of several targets of oma-1 and oma-2, as we show in the accompanying article in this issue (Spike et al. 2014).
Figure 6.
New lin-41 alleles indicate that the NHL-repeat domain is critical for fertility. (A) A drawing depicting the exon–intron structure of lin-41 (C12C8.3a in WormBase WS242). Deletions predicted to cause frameshifts and point mutations that disrupt splice sites are illustrated above the lin-41 gene. (B) A drawing depicting the LIN-41 protein, predicted domains, and the region recognized by anti-LIN-41 antibodies. Premature stop codons (*) and amino acid substitutions that alter the size or composition of the LIN-41 protein are shown. We identified many new alleles that cause sterility (black text), including amino acid substitutions in the C-terminal NHL-repeat domain (region with black boxes), and a few alleles that are fertile or viable (white text, shaded background). Intragenic suppressor mutations located in the NHL-repeat domain (tn1515, tn1516) restore fertility more effectively than those in the B-box zinc finger (tn1536, tn1539). lin-41(n2914) was previously described (Slack et al. 2000). lin-41(ve67) was identified and provided by J. Tennessen and A. Rougvie. We also identified three viable and fertile alleles (tn1485, tn1493, and tn1503) and one sterile allele (tn1494) for which we did not find a molecular lesion in exons, introns, or 3′ UTR sequences (C. Eichten, C. Spike, and D. Greenstein, unpublished results).
Our screen permitted the isolation of temperature-sensitive alleles, and we identified one lin-41 allele with temperature-sensitive sterility. lin-41(tn1487ts) animals are 100% fertile at 15° but almost completely sterile at 25° (Table 1). At the intermediate temperature of 22°, lin-41(tn1487ts) animals are mostly fertile but display reduced fecundity and embryonic viability, resulting in dramatically reduced brood sizes. These defects are likely caused by poor-quality oocytes. lin-41(tn1487ts) oocytes at 22° are often small and sometimes disorganized, but can make a relatively normal transition from pachytene into diakinesis, unlike lin-41(n2914) oocytes (compare Figure 7, D–I; Figure 8, I–L; and Figure S3G). Furthermore, the progeny of lin-41(tn1487ts) XX hermaphrodites raised at 22° are more frequently XO males, suggesting that there are elevated levels of chromosome nondisjunction during oocyte meiosis at this temperature (Table 1). Male frequency is ∼10- to 20-fold higher than in the wild type (Table 1) (Hodgkin et al. 1979; Rose and Baillie 1979). Increased numbers of males are also observed among the progeny of lin-41 alleles isolated as intragenic suppressors of lin-41(tn1487ts) sterility at 25° (Table 1). Males are most evident among the progeny of two relatively weak suppressors of tn1487ts with amino acid changes in the first B-box zinc-finger domain (tn1487tstn1536 and tn1487tstn1539), but are also notable among the progeny of two stronger suppressors with amino acid changes in the NHL-repeat domain (tn1487tstn1515 and tn1487tstn1516). It is presently unknown how these second-site mutations in the lin-41 gene suppress the temperature-sensitive sterility of tn1487ts. Temperature-sensitive alleles can cause protein destabilization and degradation at restrictive temperatures, but lin-41(tn1487ts) does not reduce LIN-41 protein accumulation in adult hermaphrodites raised at 25° (Figure 3L). Furthermore, LIN-41 expression and localization is relatively normal in lin-41(tn1487ts) germ lines at both 22° and 25°, although there appears to be a gradual, rather than dramatic, increase in LIN-41 protein levels during pachytene at both temperatures (Figure S3G; C. Spike, unpublished results). It is possible, therefore, that tn1487ts and its intragenic suppressor mutations identify LIN-41 residues with critical roles in regulating LIN-41 RNA binding or mRNA translation activities. Consistent with this possibility, the lin-41(tn1487ts) mutation in the NHL-repeat domain derepresses translation of several 3′ UTR reporter constructs, including the OMA mRNA targets, zif-1 and cdc-25.3 (Spike et al. 2014).
Table 1. Reduction of lin-41 function reduces oocyte quality and increases meiotic chromosome nondisjunction.
| Genotype | Temperature | % Fertilea | Brood sizeb | % Male |
|---|---|---|---|---|
| WTc | 25° | 100 (n = 258) | 201.5 ± 61.8 (n = 30) | 0.1 (n = 6044) |
| lin-41(tn1487 ts) | 15° | 100 (n = 224) | 103.8 ± 37.8d (n = 64) | 0.3 (n = 6642) |
| lin-41(tn1487 ts) | 22° | 83.7 (n = 86) | 3.9 ± 3.8e (n = 86) | 2.4 (n = 338) |
| lin-41(tn1487 ts)c | 25° | 0.3 (n = 310) | 0 ± 0.1 (n = 168) | ND |
| lin-41(tn1487 tstn1515)c | 25° | 98.1 (n = 472) | 66 ± 36 (n = 31) | 1.6 (n = 435) |
| lin-41(tn1487 tstn1516)c | 25° | 99.7 (n = 324) | 108 ± 36 (n = 25) | 2.5 (n = 1586) |
| lin-41(tn1487 tstn1536)c | 25° | 33.9 (n = 230) | 5.4 ± 6.3f (n = 83) | 5.1 (n = 455) |
| lin-41(tn1487 tstn1539)c | 25° | 86.2 (n = 232) | 14.0 ± 10.9g (n = 82) | 5.7 (n = 1152) |
Fertile animals produced at least one viable offspring.
Number of progeny hatching per adult.
Newly fertilized embryos were collected at 15° and shifted to 25°. The fertility, brood size, incidence of male progeny, and embryonic lethality were measured by examination of the F1 progeny.
lin-41(tn1487 ts) adults lay 139.7 eggs per adult (n = 32), with 75.9% hatching at 15°.
lin-41(tn1487 ts) adults lay 9.8 eggs per adult (n = 29), with 39.1% hatching at 22°.
lin-41(tn1487 tstn1536) adults lay 29.1 eggs per adult (n = 39), with 20.1% hatching at 25°.
lin-41(tn1487 tstn1539) adults lay 57.3 eggs per adult (n = 40), with 27.3% hatching at 25°.
Figure 7.
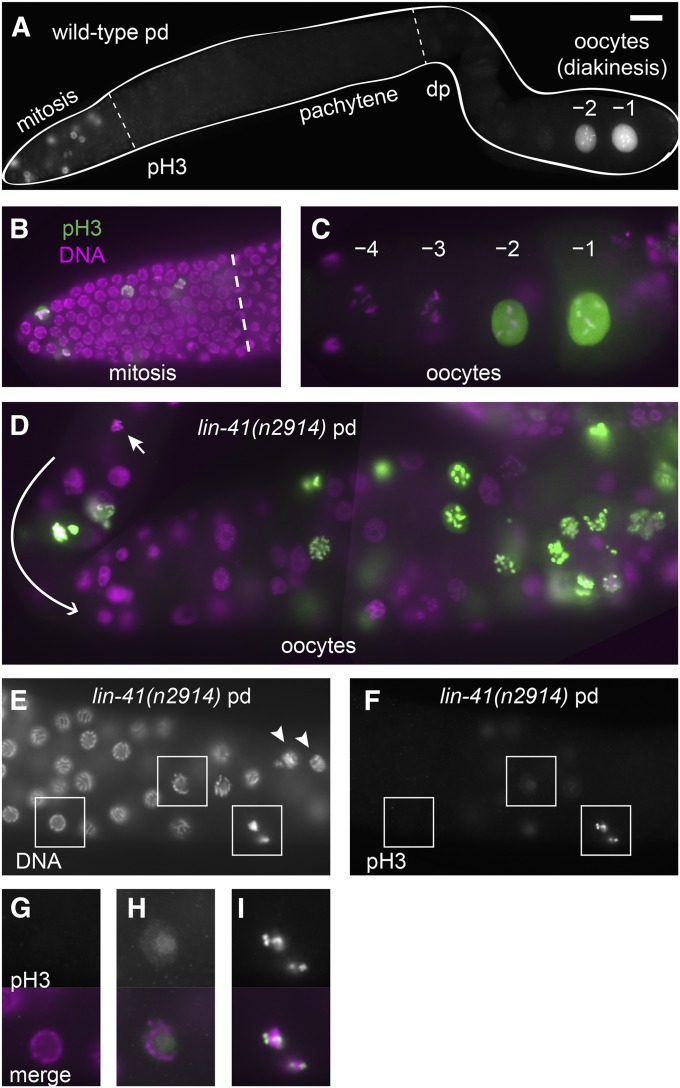
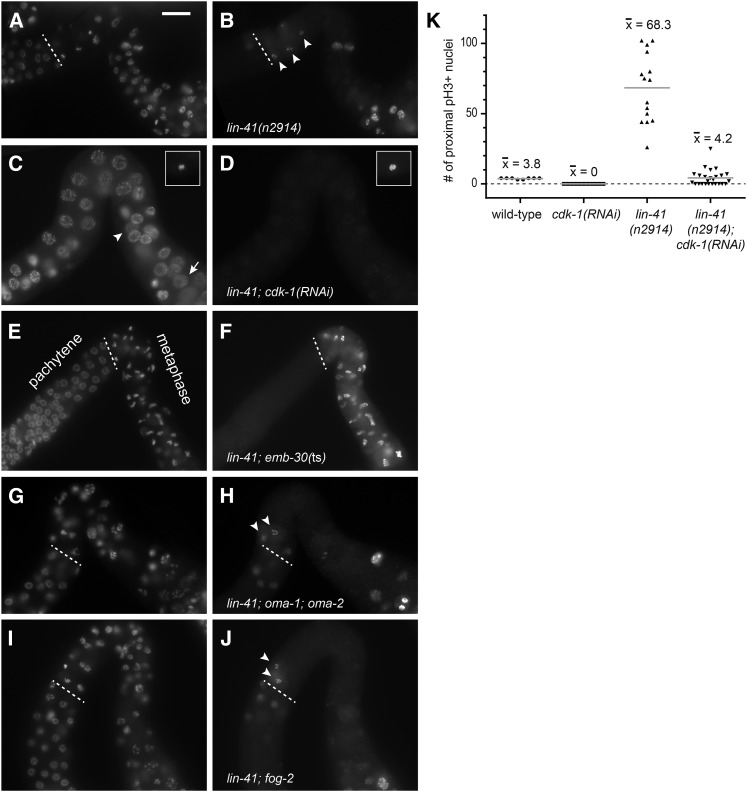
Distinct patterns of phospho-histone H3 accumulation in the wild-type and lin-41 mutants. (A–C) In wild-type animals, antibodies detecting phosphorylated histone H3 (pH3) strongly stain some of the nuclei in the distal mitotic region and also stain the nuclei of the most proximal oocytes (–1 and –2), but do not stain pachytene or diplotene (dp) nuclei (A). The overlap of pH3 (green) with a nuclear DNA stain (DAPI, magenta) is shown at higher magnification in the mitotic region (B) and in the last few oocytes (C). (D–I) In lin-41(n2914) animals, many oocyte nuclei accumulate pH3, including nuclei near the loop (D, curved arrow; the earliest nucleus accumulating pH3 on chromosomes is indicated by a short arrow). The first pH3-positive chromosomes are found just after lin-41(n2914) germ cells exit from pachytene (E and F). Arrowheads in E indicate nuclei with decondensed chromosomes, which are located proximal to the first pH3-positive nuclei. Nuclei in boxes are magnified two-fold in G–I and include a typical pachytene-stage nucleus with no pH3 accumulation (G), a later pachytene-stage nucleus with faint nuclear pH3 (H), and condensed pH3-positive chromatin immediately after pachytene (I). Bar, 20 μm (A); 10 μm (B–F); and 5 μm (G–I).
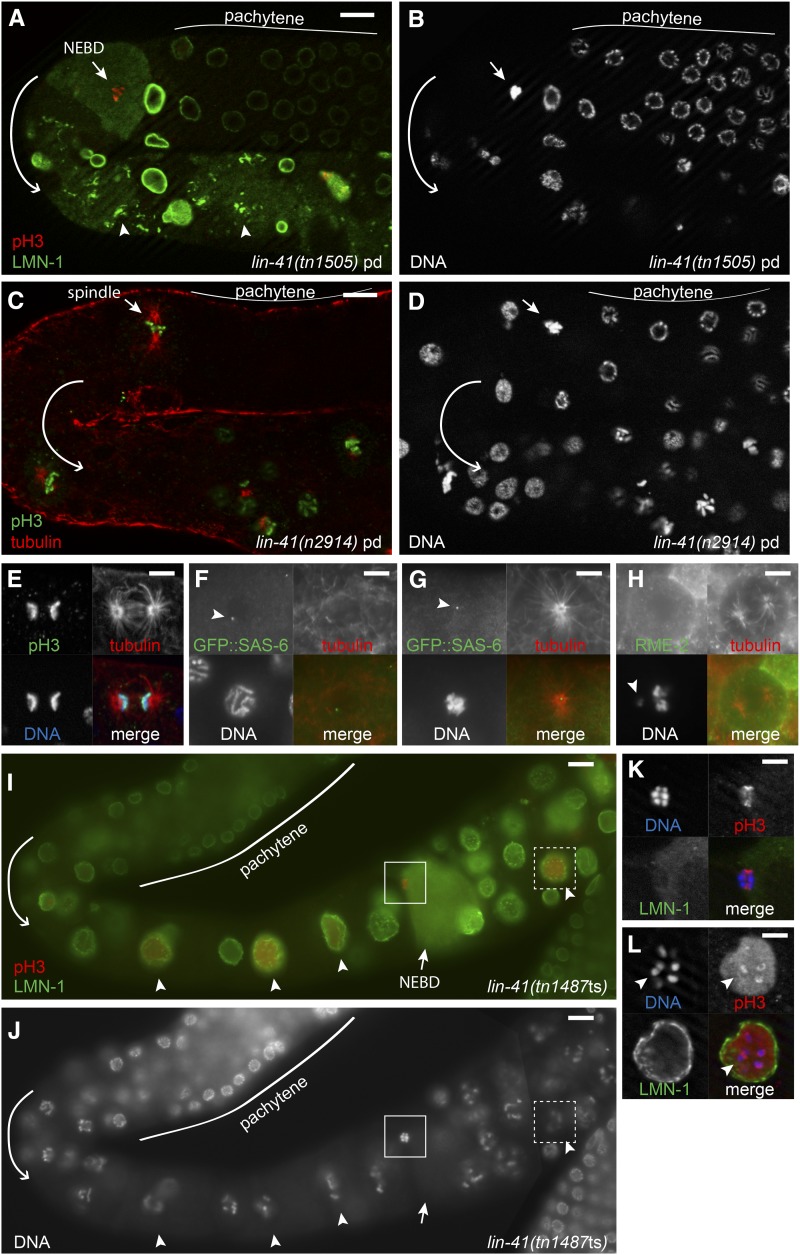
Figure 8.
lin-41 germ cells enter M phase prematurely. (A and B) Staining with anti-LMN-1 (A, green) reveals a lin-41(tn1505) oocyte that has undergone nuclear envelope breakdown (short arrow) immediately after pachytene. The oocyte has condensed pH3-positive (A, red) chromosomes (B) and is in M phase. Abnormal aggregates of LMN-1 that are not associated with the nuclear envelope are evident proximally (A, arrowheads). (C–E) Microtubules (C and E, red) form spindles immediately after pachytene in lin-41(n2914). Condensed pH3-positive (C and E, green) chromosomes (D and E) are in metaphase-like (C, short arrow) and late-anaphase-like (E) configurations. (F and G) Centrioles (GFP::SAS-6, green) are present after lin-41(RNAi) in late-pachytene-stage nuclei (F) and at the center of spindle poles (tubulin, red) immediately after pachytene (G). (H) Spindle (tubulin, red) in a lin-41(n2914) RME-2-expressing oocyte (green); the arrowhead indicates a chromosome that has not aligned with the others. (I–L) A mildly affected lin-41(tn1487ts) germ line at 25°; several pH3-positive (I, red) oocyte nuclei are in diakinesis (arrowheads) and one has undergone NEBD (short arrow) and is in M phase. Insets are shown in greater detail in K and L. Chromosomes in the M-phase oocyte (K) have congressed, but otherwise resemble the separated bivalents typical of diakinesis-stage oocytes (L, arrowhead). Images in A–E, K, and L were taken using an apotome adaptor. Bars, 5 μm (E–H, K, and L) and 10 μm (A–D, I, and J).
lin-41 prevents oocytes from entering M phase
Wild-type C. elegans oocytes exit meiotic prophase just prior to ovulation and fertilization at the proximal end of the germ line. We examined events associated with exit from meiotic prophase and entry into meiotic M phase in lin-41(n2914) mutant oocytes and found that many of these events occur prematurely (Figure 7; Figure 8; Figure S6). One of these events is the phosphorylation of histone H3 (Ser10), which occurs just prior to and during M-phase entry (Hsu et al. 2000). In wild-type hermaphrodites, chromosome-associated phosphorylated histone H3 (pH3) is strongly detected in nuclei in the distal mitotic region (proliferation zone) and in the most developed diakinesis-stage oocytes; there is no pH3 accumulation at earlier stages of oogenesis (Figure 7, A–C). By contrast, lin-41(null) oocytes prematurely accumulate phosphorylated histone H3 in early oogenesis. Faint pH3 accumulation is often observed in the last few pachytene-stage nuclei, with bright chromosome-associated pH3 appearing on condensed chromosomes immediately after pachytene (Figure 7, E–I). However, only some lin-41(n2914) oocyte nuclei accumulate pH3; it is typically absent from nuclei with decondensed chromosomes (Figure 7, D–F). This pattern suggests that lin-41 oocytes enter M phase immediately after pachytene and likely cycle out of and back into M phase, perhaps multiple times.
Nuclear envelope breakdown (NEBD) is one of the defining characteristics of M-phase entry. In wild-type animals, this event occurs exclusively in the most developed oocyte a few minutes before ovulation (McCarter et al. 1999). We monitored the integrity of the nuclear envelope in oocytes using antibodies that recognize the nuclear lamina (Liu et al. 2000) and observed that lin-41 mutant oocytes undergo NEBD immediately after pachytene (Figure 8, A and B; Figure S6). We consider oocytes with condensed, pH3-positive chromosomes that have undergone NEBD to be in M phase. Such oocytes are observed immediately after pachytene in a majority of lin-41(null) gonads [lin-41(n2914) and lin-41(tn1505)] and in the gonads of some lin-41(tn1487ts) animals raised at 25° (Table 2). Premature M-phase entry is also observed after lin-41(RNAi) in both the wild-type and rrf-1(pk1417) mutants (C. Spike, unpublished results).
Table 2. Oocytes prematurely enter M phase in lin-41 mutants.
| Genotype | Temperature | No. of proximal M-phase nucleia | % Fixed gonads with M-phase nucleia |
|
|---|---|---|---|---|
| Immediately after pachytene (%) | Proximally (%) | |||
| lin-41(n2914) | 20° | 14.2 ± 9.7 (n = 13) | 63 (n = 16) | 100 (n = 16) |
| lin-41(tn1505) | 20° | ND | 90 (n = 21) | 100 (n = 21) |
| lin-41(tn1487 ts)b | 25°c | 1.5 ± 1.4 (n = 20) | 25 (n = 20) | 85 (n = 20) |
| lin-41(tn1487 ts)b | 22°c | 1.2 ± 1.7 (n = 22) | 0 (n = 22) | 54 (n = 22) |
| lin-41(tn1487 ts)b | 15° | 0.2 ± 0.4 (n = 14) | 0 (n = 14) | 21 (n = 14) |
| lin-41(tn1487 ts)b | 22°d | 0.6 ± 0.6 (n = 24) | 0 (n = 24) | 54 (n = 24) |
| lin-41(tn1487 ts); oma-1(zu405te33); oma-2(te51)b | 22°d | 0 ± 0 (n = 27) | 0 (n = 27) | 0 (n = 27) |
| lin-41(tn1487 ts); fog-2(oz40)e | 22°f | 0.5 ± 0.8 (n = 28) | 0 (n = 28) | 32 (n = 28) |
Nuclei that have undergone NEBD and are positive for phospho-histone H3 (Ser10).
lin-41(tn1487 ts) homozygous parent.
Animals were upshifted from 15° as embryos.
Animals were upshifted from 15° as L4 larvae.
lin-41(tn1487 ts) heterozygous parent.
Animals were upshifted from 15° as L2–L4 larvae.
M-phase entry of lin-41 mutant oocytes immediately after pachytene requires cdk-1, as does expression of the unc-119p::gfp reporter:
We confirmed that lin-41 mutant oocytes prematurely enter M phase by disrupting conserved cell-cycle genes. Entry into M phase requires the cyclin-dependent kinase CDC2/CDK-1 (Boxem et al. 1999; Chase et al. 2000; Burrows et al. 2006), and cdk-1(RNAi) prevents the premature accumulation of pH3 in lin-41 mutant oocytes, as well as chromosome condensation and NEBD (Figure 9, C, D, and K). Because cdk-1 is required for postembryonic cell divisions, including those of germline stem cells (Boxem et al. 1999), the cdk-1(RNAi) treatment must be initiated late in development—in the L4 larval stage. The few proximal pH3-positive nuclei observed in lin-41; cdk-1(RNAi) germ lines (Figure 9K) were located near the spermatheca, and they likely correspond to the earliest oocytes formed after the initiation of the RNAi treatment.
Figure 9.
CDK-1 promotes M-phase entry in lin-41 oocytes. (A and B) lin-41 nuclei enter M phase (arrowheads) immediately after pachytene (dashed line); this does not occur in lin-41; cdk-1(RNAi) animals (C and D). Instead, lin-41; cdk-1(RNAi) oocyte nuclei enlarge, but remain in pachytene (arrowhead) until the DNA completely decondenses (arrow). Insets in C and D show an oocyte in M phase in a lin-41 animal treated with control RNAi; this treatment did not suppress the lin-41 phenotype (see K in this figure and Figure S7C). (E and F) Oocytes exit from pachytene and arrest with condensed pH3-positive (F) chromosomes (E) in lin-41; emb-30(ts) animals upshifted to 25°. Although cdk-1 is essential, oma-1 and oma-2, which redundantly promote oocyte meiotic maturation, are not needed for lin-41 oocyte nuclei to enter M phase after pachytene (G and H), and neither is the presence of sperm required (I and J). DNA (A, C, E, G, and I) and pH3 accumulation (B, D, F, H, and J) are shown; the nuclear lamina was also examined to identify oocytes in M phase (not shown). Bar, 20 μm. (K) Post-dauer L4-stage hermaphrodites were fed control or cdk-1 dsRNA-expressing bacteria for 2 days at 22°, and the number of post-pachytene nuclei positive for phospho-histone H3 were counted. Wild-type animals treated with cdk-1(RNAi) made oocytes that failed to accumulate pH3, as previously described (Boxem et al. 1999).
We noted that post-pachytene germ cells in lin-41(n2914); cdk-1(RNAi) adults appeared to retain the oocyte fate, they possessed a rough-appearing cytoplasm and a cuboidal shape, although they were smaller than normal oocytes (Figure S7E). We therefore asked whether the capacity of proximal lin-41(n2914) mutant cells to express a somatic marker (e.g., unc-119p::gfp) depends on the premature activation of cdk-1. We compared post-dauer lin-41(n2914); otIs45[unc-119p::gfp] adults fed control or cdk-1 dsRNA-expressing bacteria. In the control RNAi treatment, all lin-41(n2914) otIs45[unc-119p::gfp] adult gonad arms contained unc-119p::gfp-positive cells extending to the loop region, and in many gonad arms the unc-119p::gfp-positive cells extended distally to the loop (Figure S7). By contrast, in the cdk-1(RNAi) treatment, far fewer cells expressed unc-119p::gfp, and the expressing cells rarely extended to the loop region (Figure S7). The few cells observed to express unc-119p::gfp were located adjacent to the spermatheca (Figure S7) in a similar position to that observed for residual pH3-positive cells following cdk-1(RNAi) (Figure 9). Based on this result, we conclude that the loss of the oocyte fate and the expression of a somatic marker are likely a secondary consequence of the premature activation of CDK-1 at the end of pachytene in lin-41 null mutant oogenic cells.
Most or all developing oocytes enter M phase immediately after pachytene in lin-41 mutants:
Progression from metaphase to anaphase requires EMB-30/APC4, a subunit of the anaphase-promoting complex (APC) (Furuta et al. 2000). lin-41(n2914) mutant oocytes with compromised emb-30 activity appear to be trapped in metaphase with condensed pH3-positive chromosomes and a disassembled nuclear lamina (Figure 9, E and F). This M-phase block reveals that most or all developing oocytes appear to enter M phase immediately after pachytene (Figure 9, E and F).
lin-41 mutant oocytes assemble spindles and attempt to segregate chromosomes immediately after pachytene:
lin-41 oocyte nuclei appear to transition from pachytene into M phase without going through diplotene (Figure 1H; Figure 7, E and F; Figure 8, A–D; Figure 9, A and B). In C. elegans oocytes, centriole elimination occurs during the diplotene stage of meiotic prophase I (Mikeladze-Dvali et al. 2012), resulting in meiotic spindles that are both acentriolar and anastral (Albertson and Thomson 1993). However, the spindles that form in lin-41 mutant oocytes have obvious astral microtubules and therefore resemble mitotic, rather than meiotic, spindles (Figure 8, C–E), suggesting that centriole elimination has not occurred. We confirmed that lin-41 mutant M-phase oocytes contain centrioles by examining the localization of the centriolar protein GFP::SAS-6 (Leidel et al. 2005) after lin-41(RNAi). All spindle poles identified after lin-41(RNAi) contained GFP::SAS-6 at their core (Figure 8G; n = 38), including 20 observed immediately after pachytene. We conclude that centriole elimination does not occur prior to M-phase entry in lin-41 oocytes, consistent with the observation that lin-41 oocyte nuclei do not progress through diplotene.
lin-41 mutant oocytes retain a meiotic cell fate as they prematurely enter M phase:
We considered the possibility that lin-41 mutant oocytes enter mitosis after exiting meiotic prophase, similar to gld-1(null) oogenic germ cells (Francis et al. 1995a,b). In gld-1(null) mutant hermaphrodites, however, there are no cytological signs of oogenesis. Instead, the proximal gonad contains many small, undifferentiated but proliferative germ cell nuclei (Francis et al. 1995b; Jones et al. 1996). In gld-1(null) mutants, germ cell exit from meiotic prophase likely occurs at an early, programmed cell death-resistant stage of pachytene (Gumienny et al. 1999). Because GLD-1 is abundant throughout pachytene, diminishing only as oocytes transition into diplotene (Jones et al. 1996; Lee and Schedl 2001), we hypothesized that premature elimination of GLD-1 might cause, or contribute to, the lin-41 mutant phenotype. GLD-1 is abundant throughout pachytene in lin-41(n2914) mutants, however, diminishing only as lin-41 oocytes transition from pachytene into M phase and with a similar spatial-temporal pattern as in the wild type (Figure S8, A and B).
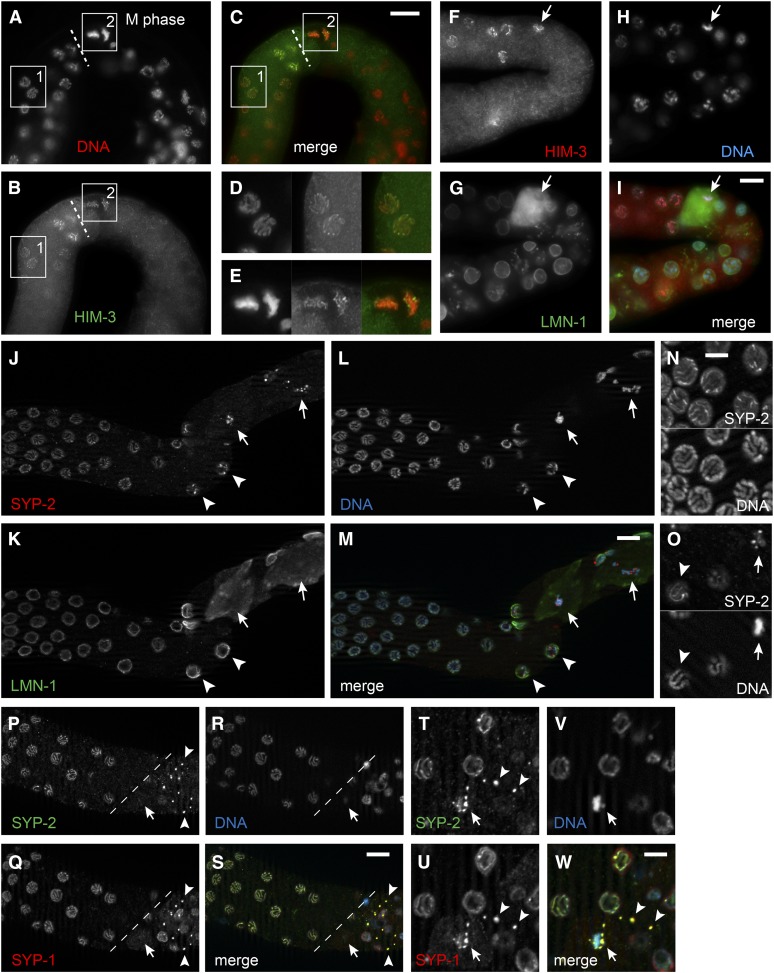
Proliferative germ cell nuclei can be distinguished from those in meiotic prophase by staining with REC-8- and HIM-3-specific antibodies, respectively, under the appropriate fixation conditions (Hansen et al. 2004a). We examined lin-41 germ lines for mitotic and meiotic germ cells using this assay. REC-8 staining was present in distal proliferative nuclei, disappearing from germ cells as they transition to meiosis, as in the wild type (Figure S8, C and D). Meiotic nuclei were HIM-3-positive and REC-8-negative, including likely M-phase nuclei identified immediately after pachytene based on their position and morphology (Figure 10, A–E); 27 of 39 were HIM-3-positive; all 39 were REC-8-negative). When we stained lin-41(n2914) germ lines with HIM-3 and lamin, we observed that nuclei having undergone NEBD within a few cell diameters of the end of pachytene were HIM-3-positive (Figure 10, F–I). Since HIM-3 is a meiosis-specific chromosome protein that localizes to the axial cores of synapsed homologous chromosomes during pachytene (Zetka et al. 1999), these results indicate that lin-41 mutant oogenic cells possess a meiotic fate at the time of their initial M-phase entry. Because there is no true diplotene stage in lin-41 null mutants, the oocytes that enter M phase are likely derived from pachytene.
Figure 10.
Analysis of axial and central components of the synaptonemal complex suggests that lin-41(n2914) oocytes enter M phase immediately after pachytene. (A–E) HIM-3 (B, green) localizes to chromosomes (A, red) both before (inset 1 and D) and immediately after (inset 2 and E) lin-41(n2914) oocytes exit from pachytene (dashed line). HIM-3-positive nuclei after pachytene exit have condensed chromosomes in metaphase-like configurations, suggesting that they are in M phase. Bar, 20 μm (A–C) or 10 μm (D and E). (F–I) A similar lin-41(n2914) oocyte (arrow) with condensed HIM-3-positive chromosomes (red and blue in the merge, respectively, and in F and H) has a disassembled nuclear lamina (G, green), confirming that it has undergone nuclear envelope breakdown. Bar, 10 μm. (J–M) SYP-2 (J, red) localizes to foci on or near chromosomes (L, blue) in young lin-41(n2914) oocytes that have undergone nuclear envelope breakdown (arrows), as evidenced by a disassembled nuclear lamina (K, green). Nearby nuclei with intact nuclear laminae (arrowheads) have an asymmetric pattern of SYP-2 localization along their chromosomes. Bar, 10 μm. (N and O) Wild-type nuclei just before diplotene (N) and a lin-41(n2914) pachytene-stage nucleus just prior to M phase (O, arrowhead). These nuclei have asymmetric SYP-2 localization patterns along their chromosomes. A nearby lin-41(n2914) oocyte in M phase (O, arrow) displays SYP-2 foci that might represent a disassembling synaptonemal complex. Bar, 5 μm (P–S) Remnants of the synaptonemal complex (arrowheads) that contain both SYP-1 (Q, red) and SYP-2 (P, green) persist after lin-41(n2914) oocytes exit from pachytene (dashed line). The location of the M-phase oocyte shown in T–W is indicated with an arrow. The nucleus of this oocyte is in a slightly different focal plane. Bar, 10 μm. (T–W) SYP-1 (U, red) and SYP-2 (T, green) appear to localize both on and near the condensed chromosomes (V, blue) of the M-phase oocyte (arrow). Two cytoplasmic remnants of the synaptonemal complex that appear to be in other cells are indicated by arrowheads. Bar, 5 μm.
To further examine the meiotic status of oogenic cells upon M-phase entry, we stained for lamin and SYP-2, which is a structural protein of the central region of the synaptonemal complex (Colaiácovo et al. 2003). We observed that lin-41(n2914) mutant pachytene-stage nuclei exhibit a normal SYP-2-staining pattern, suggesting that homologs are fully synapsed (Figure 10, J–W). Meiotic nuclei appear to progress to the late pachytene stage (Figure 10, N and O) when synaptonemal complex proteins begin to exhibit uneven localization along the bivalent (Nabeshima et al. 2005). In M-phase nuclei undergoing or having completed NEBD, SYP-2 localizes to discrete foci that are sometimes no longer associated with chromosomes (Figure 10, J–W). SYP-1, which is also a structural protein of the central region of the synaptonemal complex (MacQueen et al. 2002), colocalizes with SYP-2 at these foci in M-phase lin-41 mutant oogenic nuclei, suggesting that they might represent remnants of a disassembling synaptonemal complex (Figure 10, P–W). These results suggest that homologous chromosomes are synapsed as they begin to condense and prematurely enter M phase in lin-41 oocyte nuclei. Taken together with the analysis presented above, these observations suggest that late-pachytene-stage, or possibly early-diplotene-stage nuclei, are driven into M phase prematurely in lin-41 mutants via CDK-1 activation. Consistent with this interpretation, pachytene-stage nuclei often persist proximally to the loop following cdk-1(RNAi) in lin-41(n2914) mutants, although the chromosomes ultimately decondense and the nuclei enlarge (Figure 9C).
Nuclei near the spermatheca in lin-41(n2914) mutants were often HIM-3-negative and REC-8-positive (Figure S8, C and D), suggesting that mutant oocytes might acquire proliferative characteristics as they move even further proximally, subsequent to their initial M-phase entry immediately after pachytene. We examined whether proximal lin-41(RNAi) oocyte nuclei enter S phase by assessing their ability to incorporate the nucleoside analog EdU. DNA replication is completed during the very early stages of meiosis, so normal oocyte nuclei do not incorporate EdU after a short pulse of exposure (4 hr); only mitotic and premeiotic cells in the distal arm incorporate EdU as they pass through S phase (Figure S9, A and C). We anticipated that lin-41 nuclei in the proximal gonad arm would incorporate EdU, particularly since some of these nuclei become polyploid (Figure 1, B and H). Contrary to our expectation, when we examined the germ lines of rrf-1; lin-41(RNAi) animals, which incorporate EdU in distal nuclei (Figure S9B), we did not observe strong EdU incorporation in nuclei in the proximal arm either near the loop or more proximally near the spermatheca (Figure S9D) (C. Spike, unpublished results). However, upon overexposure, some weak EdU incorporation was noted in oocyte nuclei shortly after the end of pachytene (Figure S9D).
We think that the relatively weak and patchy EdU incorporation observed in lin-41 nuclei immediately after pachytene likely reflects repair DNA synthesis. DNA damage might occur when late-pachytene-stage chromosomes are driven into M phase prematurely. The polyploidy that we observed proximally in lin-41 mutant oocytes (Figure 1, B and H) might arise from the fusion of proximal nuclei undergoing repeated rounds of M-phase entry and involve the collapse of cellular partitions between cells. Alternatively, uptake or detection of the EdU label might be compromised in this abnormal proximal cell population. Thus, we are unable to exclude the possibility that some proximal lin-41 nuclei replicate their DNA after exiting from M phase. Nonetheless, the absence of an apparent S phase immediately after premature M-phase entry of lin-41 mutant oocytes is consistent with the interpretation that they retain a meiotic fate at the point of their initial M-phase entry.
In summary, we find that nuclei in null and strong loss-of-function lin-41 animals exit meiotic prophase and enter M phase near the end of pachytene, possibly at the pachytene–diplotene transition. M-phase entry occurs in cells specified to become oocytes (Figure 8H) with synapsed homologous chromosomes (Figure 10). Chromosome separation or segregation occurs at least occasionally (Figure 8E) and is mediated by a centriole-containing spindle (Figure 8G). The LIN-41 expression pattern suggests that lin-41 could be required to prevent M-phase entry in all developing oocytes, which would normally grow, eliminate centrioles, and organize meiotic chromosomes into bivalents before undergoing meiotic maturation. We were able to test this hypothesis using the temperature-sensitive lin-41(tn1487ts) mutant. lin-41(tn1487ts) mutants exhibit lower rates of M-phase entry at the end of pachytene, relative to lin-41(null) alleles (Table 2), which permits some (at 25°) or most (at 22°) lin-41(tn1487ts) mutant oocytes to progress through diplotene and into diakinesis. We examined lin-41(tn1487ts) oocytes at 22° and 25° and identified oocytes with bivalent chromosomes that had entered M phase prematurely (Figure 8, I–K). Our phenotypic analysis of lin-41(tn1487ts) is therefore consistent with the idea that lin-41 prevents meiotic M-phase entry during late, as well as early, stages of oogenesis.
Premature M-phase entry of lin-41 mutant oogenic nuclei is inhibited when germ cells arrest in pachytene
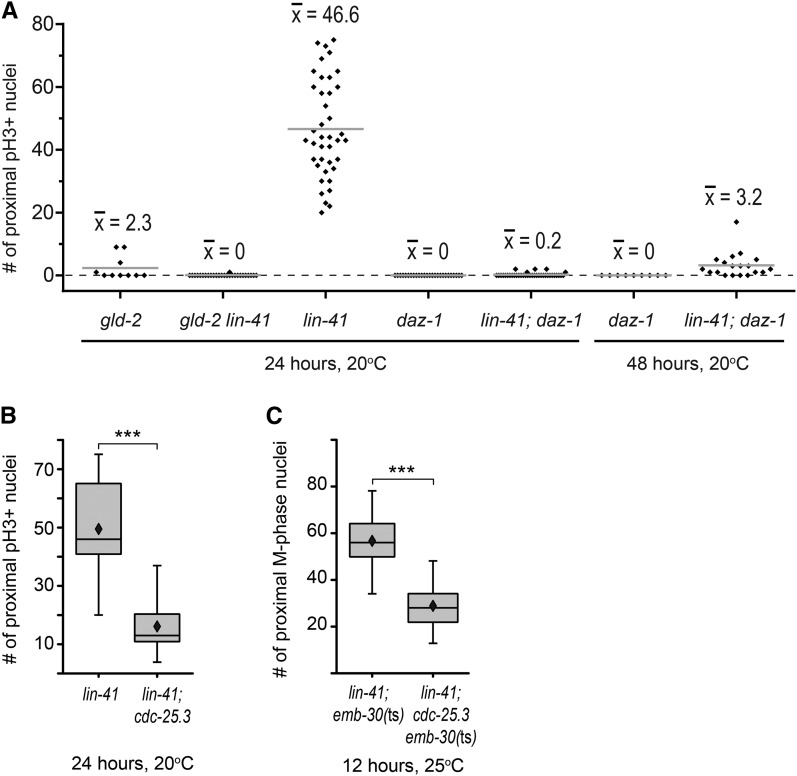
Oocyte nuclei in strong loss-of-function lin-41 animals appear to enter M phase near the very end of pachytene. Thus, causing oogenic germ cells to arrest in pachytene might reduce or eliminate M-phase entry in lin-41 mutants, depending on the efficiency of the imposed block to pachytene progression. We tested this hypothesis using a strong loss-of-function mutation in the daz-1 gene. In daz-1(tj3) mutants, oogenic cells arrest during pachytene and do not progress into diplotene and diakinesis or accumulate phosphorylated histone H3 (Karashima et al. 2000; Maruyama et al. 2005). However, smaller germ cell nuclei were occasionally observed interspersed with arrested pachytene-stage oogenic nuclei (Karashima et al. 2000). We verified these observations and determined that daz-1 is largely, but not entirely, epistatic to lin-41. Most lin-41(n2914); daz-1(tj3) oogenic germ cells remain in pachytene, with only a small number of nuclei exhibiting condensed chromosomes that accumulate phosphorylated histone H3 (Figure 11A). Such nuclei resemble those found in lin-41 mutants and are often observed near pH3-negative nuclei exhibiting decondensed chromosome morphologies (Figure S10, G–I). Although the majority of nuclei remain arrested in pachytene in lin-41; daz-1 double mutants, the number of gonads with pH3-positive nuclei, as well as the typical number of pH3-positive nuclei in each gonad, increases as the animals age (Figure 11A). One possibility is that a small fraction of daz-1 mutant pachytene-stage nuclei, although not able to progress to diplotene, can nonetheless progress to the point in pachytene at which lin-41 is required to prevent premature M-phase entry. Multiple nuclear divisions could contribute to the increase in numbers of pH3-positive nuclei over time. Alternatively, our observations could in part reflect a slight temporal delay in the switch from spermatogenesis to oogenesis, since younger daz-1 mutant adults sometimes appear engaged in spermatogenic meiotic divisions (Figure S10, A–C); however, such nuclei were excluded in Figure 11A through the simultaneous assessment of sexual fate (Figure S10). Yet, our results might be explained if daz-1 mutant pachytene-stage germ cells are sometimes confused as to their sexual fate, on occasion only committing to the female fate late in pachytene. Indeed, daz-1 also functions in the hermaphrodite sperm-to-oocyte switch (Otori et al. 2006). In any event, the number of lin-41; daz-1 nuclei that exit from pachytene is small enough for us to conclude that most lin-41; daz-1 pachytene-stage nuclei must be unable to enter M phase either because they have not developed to the very late stage of pachytene where lin-41 nuclei enter M phase or because they are abnormal in some other way.
Figure 11.
Genes that regulate meiotic progression or the cell cycle promote M phase in lin-41 oocytes. (A) The number of postmitotic nuclei with chromosomally associated phospho-histone H3 (pH3+) in the germ lines of animals 24 or 48 hr past the mid-L4 stage. Individual germline counts (black diamonds) and average numbers (shaded bars) are shown for each genotype. (B and C) cdc-25.3 stimulates M-phase entry in lin-41(n2914) hermaphrodites. lin-41(n2914); cdc-25.3(ok358) germ lines (n = 30) have a significantly reduced number of proximal pH3-positive oocyte nuclei immediately after pachytene compared to equivalently staged lin-41 controls (n = 27) (B). Similar results are observed when the anaphase-promoting complex is disabled in emb-30(ts) animals, so that lin-41 (n = 25) and lin-41; cdc-25.3 (n = 43) nuclei are trapped in M phase after exiting from pachytene (C). Box plot whiskers and diamonds indicate minimum/maximum values and averages, respectively, and asterisks indicate significance (P < 0.001) in B and C. All animals in C were also homozygous for the linked ced-7(n1892) mutation.
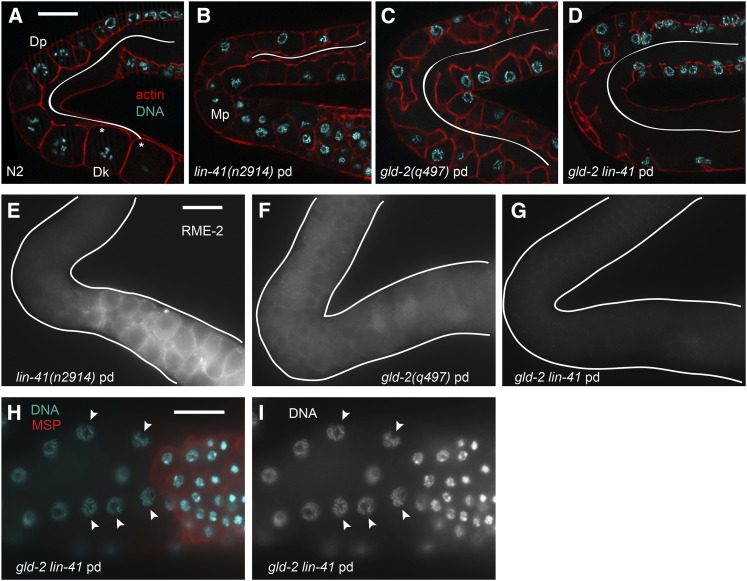
Pachytene arrest may also prevent lin-41-dependent M-phase entry in gld-2(q497) lin-41(n2914) germ lines. We constructed this double-mutant combination because the GLD-2 cytoplasmic poly(A) polymerase (Wang et al. 2002) and LIN-41 copurify with OMA RNPs and function at similar stages of oogenesis, suggesting that they might regulate overlapping sets of oocyte mRNAs (Spike et al. 2014). Indeed, in the accompanying article in this issue (Spike et al. 2014), we observed an overlap between the mRNA components of OMA RNPs and those copurifying with GLD-2 (Kim et al. 2010b). Like lin-41 mutants, strong loss-of-function gld-2 mutants make oocytes that reach the pachytene stage of meiotic prophase, but then become abnormal and fail to enter diplotene or diakinesis (Kadyk and Kimble 1998). Unlike lin-41 mutants, however, gld-2 oocytes do not enter M phase immediately after pachytene. Instead, a small number of older gld-2 oocytes with abnormal pH3-positive nuclei are sometimes present (Figure 11A); a few of these nuclei undergo NEBD and appear to be in M phase (C. Spike, unpublished results). gld-2lin-41 mutant oocytes do not enter M phase prematurely (Figure 11A), but instead they appear to arrest in pachytene (Figure 12, A–D, H, and I), a phenotype not observed in either single mutant (Figure 1H) (Kadyk and Kimble 1998). gld-2lin-41 double mutants therefore appear to exhibit a synthetic phenotype that we examined in more detail. Oocyte cellularization occurs prematurely in lin-41 mutants (Figure 1H; Figure 12B) but is spatially delayed in gld-2 mutants relative to the wild type (Figure 12, A and C). Cellularization is even more abnormal in gld-2lin-41 mutants, however, and completely fails in many animals (Figure 12D). Likewise, expression of the oocyte marker RME-2, which appears to be reduced and spatially altered in gld-2 mutants, is further reduced or absent from gld-2lin-41 mutants (Figure 12, E–G). Expression of MSP is unaltered, however, suggesting that gld-2lin-41 germ cells are not sexually transformed (Figure 12, H and I). We conclude that multiple events that normally occur around the time when germ cells transition from pachytene into diplotene fail in gld-2lin-41 mutants, consistent with the interpretation that meiotic germ cells arrest in pachytene in these animals.
Figure 12.
gld-2 lin-41 oogenic germ cells arrest in pachytene. (A–D) Oocyte cellularization in gld-2 lin-41 and control day-1 adults at 20°; Z-series images of the rhodamine-phalloidin-labeled actin cytoskeleton (red) were used to determine the approximate extent of the rachis (white line) in animals of each genotype. Wild-type oocytes cellularize after transitioning from diplotene (Dp) into diakinesis (Dk), immediately after the loop (A). Asterisks in A indicate the last two detectable connections between oocytes and the rachis in the wild type; both are in a different focal plane. lin-41 oocytes cellularize prematurely (B), near the transition from pachytene into M phase (Mp). The rachis remains open longer in gld-2 and gld-2 lin-41 animals (C and D), with cellularization failing completely in some gld-2 lin-41 animals; 59% of gld-2 lin-41 germ lines had regions where oocyte cellularization appeared to have failed (n = 22) compared to 0% of gld-2 and lin-41 germ lines (n = 20 and n = 32, respectively). (E–G) RME-2 expression is reduced in gld-2 lin-41 animals compared to either single mutant. (H and I) The nonspermatogenic germ cells of gld-2 lin-41 animals have synapsed chromosomes and appear to arrest in pachytene (arrowheads). Spermatogenic cells are MSP-positive (red), and nonspermatogenic cells are MSP-negative. Bars, 20 μm.
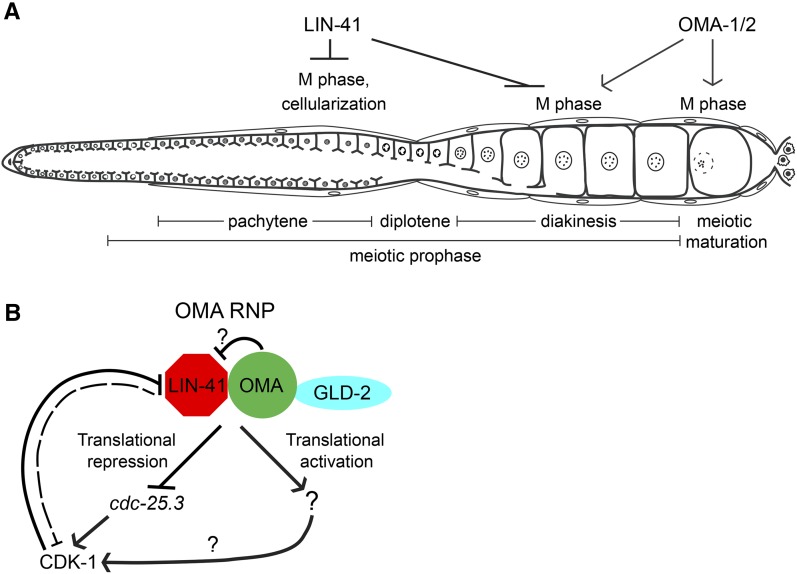
LIN-41 is antagonized by OMA-1/2 and functions upstream of CDC-25.3
In wild-type oocytes, exit from meiotic prophase is stimulated by MSP-dependent signaling and requires the functionally redundant CCCH zinc-finger proteins OMA-1 and OMA-2. oma-1/2 function upstream of wee-1.3, a conserved kinase known to inhibit CDK-1 activation and repress premature oocyte meiotic maturation and M phase (Detwiler et al. 2001; Burrows et al. 2006). RNAi depletion of CDK-1 prevents lin-41(n2914) oocytes from entering M phase after pachytene (Figure 9, C and D), but this phenotype is not suppressed when sperm or OMA-1/2 is absent (Figure 9, G–J), indicating that these factors are either upstream of, or irrelevant to, lin-41 function at this early stage of oogenesis. We also examined more developed lin-41(tn1487ts) oocytes, which are able to progress into diakinesis before entering M phase (Figure 8, I–L). M-phase entry in lin-41(tn1487ts) oocytes at 22° requires OMA-1/2 but not the presence of sperm (Table 2). lin-41(tn1487ts); oma-1; oma-2 oocytes are also notably larger than lin-41(tn1487ts) oocytes (C. Spike, unpublished results), as might be expected since OMA-1/2 inhibits oocyte growth (Detwiler et al. 2001; Govindan et al. 2009). Thus, LIN-41 and OMA-1/2 exhibit an antagonistic relationship: LIN-41 inhibits M-phase entry and oocyte cellularization whereas OMA-1/2 promotes these events. Furthermore, our observations suggest that lin-41 is epistatic to oma-1/2 for its function in preventing the premature M-phase entry of pachytene-stage oogenic cells, whereas oma-1/2 appears to be required for the entry of proximal oocytes into M phase in lin-41(tn1487ts) mutants.
LIN-41 was identified as a component of OMA RNPs, and these two proteins appear to repress the translation of some of the same target mRNAs during oogenesis (Spike et al. 2014). One of these target mRNAs encodes CDC-25.3, a CDC-25-family phosphatase predicted to activate CDK-1 by removing inhibitory phosphorylations. Other CDC-25-family proteins are known to play essential roles in germline development in C. elegans (Ashcroft et al. 1999; Ashcroft and Golden 2002; Clucas et al. 2002; Kim et al. 2009; Kim et al. 2010a; Yoon et al. 2012), but CDC-25.3 appears to be more important for larval development. Animals homozygous for the cdc-25.3(ok358) deletion are fertile and produce an approximately normal number of embryos (312 ± 39, n = 5). Most of these embryos hatch (92%, n = 1544), but only 32% develop into L4-stage larvae or adults in a timely fashion (within 2.5 days after egg laying at 20°; n = 828), and most appear to arrest as young larvae (Figure S11, A and B). Although the cdc-25.3 mRNA is present during oogenesis (Spike et al. 2014), cdc-25.3(ok358) germ lines are indistinguishable from the wild type (Figure S11, C–E), consistent with the idea that CDC-25.3 does not normally play a significant role in germline development. Because CDC-25.3 likely activates CDK-1, which is required for lin-41-dependent M-phase entry, we used a genetic epistasis test to examine whether ectopic expression of CDC-25.3 might contribute to premature M-phase entry in lin-41(n2914) oocytes. lin-41; cdc-25.3 oocytes clearly enter M phase after pachytene, but there is a significant reduction in the number of oocyte nuclei accumulating phospho-histone H3 in lin-41; cdc-25.3 animals compared to lin-41 alone (Figure 11B) , suggesting that CDC-25.3 might help to stimulate, but is not sufficient to trigger, M-phase entry in lin-41(n2914) mutants. Because the oocytes of lin-41 animals appear to cycle in and out of M phase (Figure 7D), we trapped them in metaphase using emb-30(ts). Animals were upshifted to the restrictive temperature prior to oogenesis to ensure that oocytes would be trapped in M phase immediately after exiting from pachytene. Fewer oocytes in M phase were observed in lin-41; cdc-25.3emb-30 triple-mutant animals compared to lin-41; emb-30 controls (Figure 11C), consistent with the idea that ectopic CDC-25.3 stimulates lin-41 oocytes to enter M phase immediately after pachytene.
CDK-1 is required for the elimination of GFP::LIN-41
The preceding analysis established that lin-41 inhibits M-phase entry of developing oocytes both at the late pachytene stage and at diakinesis. Our results suggest that LIN-41 regulates M-phase entry in part through the translational repression of the CDK-1 inhibitor, CDC-25.3. For the most proximal oocyte to mature in the wild type, there needs to be a mechanism that antagonizes LIN-41 function. Genetic epistasis analysis suggests that OMA-1/2 may be part of this mechanism because oocytes do not undergo meiotic maturation in lin-41(tn1487ts); oma-1(zu405te33); oma-2(te51) triple mutants (Table 2). We were therefore intrigued by the observation that LIN-41 and GFP::LIN-41 begin to be eliminated from oocytes as they undergo meiotic maturation (Figure 3; Figure 4). To ask whether the elimination of GFP::LIN-41 requires CDK-1 function, we fed lin-41(tn1541[gfp::tev::s::lin-41]) L4-stage hermaphrodites with cdk-1 dsRNA-expressing bacteria. GFP::LIN-41 persisted in all fertilized one-cell arrested embryos accumulating in the uterus following cdk-1(RNAi) (Figure 4B; n = 50 day-1 and day-2 adults), but did not persist after control (n = 37 animals) or plk-1(RNAi) (n = 32 animals), which also causes a one-cell arrest (Chase et al. 2000). We next tested whether CDK-1 activation is sufficient to cause GFP::LIN-41 elimination by feeding lin-41(tn1541[gfp::tev::s::lin-41]) L4-stage hermaphrodites with wee-1.3 dsRNA-expressing bacteria. Indeed, GFP::LIN-41 disappeared prematurely from most of the wee-1.3(RNAi) adult gonad arms examined (69/73 day-2 adults). In the more severely affected wee-1.3(RNAi) animals, the proximal gonad arm filled with small abnormal oocytes resulting from premature meiotic maturation as previously reported (Burrows et al. 2006), and GFP::LIN-41 was typically absent from the entire proximal arm. In animals more mildly affected by the wee-1.3(RNAi) treatment, normal-appearing oocytes formed in the proximal arm, but GFP::LIN-41 was eliminated prematurely from oocytes that had not undergone NEBD (Figure 4C). By contrast, the disappearance of GFP::LIN-41 during meiotic maturation required neither the MBK-2 M-phase kinase nor the activity of the anaphase-promoting complex, as revealed by analysis of mbk-2(pk1427) (n = 65 gonad arms examined) and emb-30(tn377ts) mutants (n = 40 gonad arms examined), respectively (Figure 4, D and E). Since OMA-1/2 is required for CDK-1 activation (Detwiler et al. 2001), the antagonism between the OMA proteins and LIN-41, and the regulation of LIN-41 by CDK-1, might be components of the mechanism that spatially restricts meiotic maturation to the most proximal oocyte.
Discussion
Here we present genetic and phenotypic analyses of the essential roles of the TRIM-NHL protein LIN-41 during oogenesis in C. elegans. Our analysis reveals LIN-41 as an inhibitor of oocyte M-phase entry, a promoter of oocyte growth, and a maintainer of oocyte quality. In this capacity, LIN-41 appears to function in opposition to the OMA RNA-binding proteins with which it associates in an oogenic translational regulatory RNP complex (Spike et al. 2014). By contrast, lin-41 is not required for spermatogenesis or male fertility. In lin-41 null mutants, pachytene-stage oogenic cells fail to progress to diplotene, the stage in which centrioles are eliminated for acentriolar meiotic spindle assembly (Mikeladze-Dvali et al. 2012). Instead, developing oocytes in lin-41 null mutants exhibit a remarkable phenotype: they activate CDK-1, enter M phase, assemble a spindle, and attempt to segregate chromosomes.
Several observations suggest the lin-41 null mutant phenotype is not simply a “return to mitosis” phenotype as observed in gld-1 mutant pachytene-stage oogenic cells (Francis et al. 1995a,b) or in puf-8 mutant primary spermatocytes (Subramaniam and Seydoux 2003), both of which ultimately form germline tumors of mitotically cycling undifferentiated germ cells. First, examination of HIM-3, a meiosis-specific protein that localizes to the axial cores of synapsed homologous chromosomes during pachytene, and SYP-1 and SYP-2, proteins of the central region of the synaptonemal complex, confirms that lin-41 null mutant oogenic cells proceed directly from late pachytene into M phase. Second, instead of producing undifferentiated germline tumors, pachytene-stage cells that enter M phase appear oogenic and begin to express the oocyte yolk receptor RME-2 (and OMA-1::GFP), continuing to do so for a period of time. In the most proximal region of the gonad, many cells lose expression of oocyte markers (RME-2 and OMA-1::GFP); however, this correlates with their increasing state of disorganization and polyploidy and their acquisition of the capacity to express a somatic marker (unc-119p::gfp).
Third, we used EdU labeling to test whether the abnormal oogenic nuclei in lin-41 null mutants enter S phase after pachytene, as would be expected from a “return to mitosis” phenotype. While our conditions detected S phase in the distal stem cell population in lin-41 null mutants, we found scant evidence that oogenic cells replicate their genome after pachytene. Rather, we observed relatively weak and patchy EdU incorporation immediately after pachytene, possibly reflecting repair DNA synthesis. It would not be hard to believe that DNA damage might ensue when chromosomes are driven into M phase from late pachytene or early diplotene. Indeed, we observed that components of the central region of the synaptonemal complex (SYP-1 and SYP-2) relocalized to foci and disassembled as oocyte nuclei entered M phase. This abrupt disassembly of the synaptonemal complex might represent an aberrant process because cytoplasmic remnants of the synaptonemal complex persist, which is not observed in the wild type. Finally, in lin-41 null mutants, we observed large apoptotic cell corpses in the proximal gonad arm that are engulfed by sheath cells, as revealed by acridine orange staining. Importantly, both developmental and DNA-damage-induced apoptosis of germ cells are restricted to oogenesis (Gumienny et al. 1999; Gartner et al. 2000; Jaramillo-Lambert et al. 2010). By this criteria also, the proximal germ cells in lin-41 mutants appear to have adopted the oocyte fate; however, they are defective in completing the oogenic program. In any case, the final fate of the abnormal cells in the most proximal region likely reflects the downstream consequences of premature M-phase entry because cdk-1(RNAi) reduces their capacity to express a somatic marker (Figure S7).
Instead of a “return to mitosis,” the lin-41 null mutant phenotype appears to be a consequence of the premature activation of CDK-1 in oogenic cells in late pachytene or early diplotene. Consistent with this view, a null mutation in cdc-25.3, which encodes a Cdc25-family phosphatase predicted to activate CDK-1, reduces the number of pachytene-stage cells that enter M phase in lin-41 null mutants. In the accompanying article in this issue, we show that the cdc-25.3 mRNA, like LIN-41, is an OMA RNP component (Spike et al. 2014). The germline expression of OMA-1/2 and LIN-41 are defining features of the oocyte fate, and these proteins mediate 3′ UTR-dependent translational repression of cdc-25.3. The simplest interpretation is that translational derepression of cdc-25.3 contributes to premature M-phase entry of lin-41 mutant oocytes early in their development.
While this work was under review, Tocchini et al. (2014) reported the isolation of two strong loss-of-function lin-41 alleles in a genetic screen for mutants that express a reporter of zygotic gene transcription in the germ line. The activation of zygotic transcription normally occurs after the oocyte-to-embryo transition, which is dependent on oocyte meiotic maturation (reviewed by Robertson and Lin 2013). Our data suggest that a major aspect of the oocyte meiotic maturation program, M-phase entry resulting from CDK-1 activation, occurs prematurely in lin-41 mutants and that the abnormal end-stage phenotype of mutant oocytes is likely a consequence of this defect. Tocchini et al. (2014) proposed that LIN-41 has a specific function as a regulator of pluripotency in the germ line. The finding that OMA RNPs contain regulators of embryonic cell-fate determination (e.g., MEX-1, MEX-3, POS-1, and SPN-4) in addition to LIN-41 (Spike et al. 2014) might be relevant to the potential involvement of LIN-41 in the regulation of pluripotency (Tocchini et al. 2014). However, it is worth noting that, in C. elegans, pachytene-stage germ cells appear to be the most transcriptionally active with RNA polymerase II activity declining as oocytes transition from diplotene to diakinesis (Starck 1977; Gibert et al. 1984; Schisa et al. 2001; Kelly et al. 2002; Walker et al. 2007). Since lin-41 null mutant oocytes do not as a rule progress to diplotene or diakinesis, they might not be predicted to downregulate RNA polymerase II transcriptional activity after pachytene. Thus, premature M-phase entry might have multiple consequences that contribute to the terminal fate of pachytene-stage oogenic cells in lin-41 null mutants.
Analysis of lin-41 mutant germ lines also suggests a major role in promoting oocyte growth. Developing oocytes are connected to the cytoplasmic core of the gonad by “ring channels” (Wolke et al. 2007) through which they receive proteins, RNAs, metabolites, and organelles via actomyosin-dependent cytoplasmic streaming (Hirsh et al. 1976; Maddox et al. 2005; Wolke et al. 2007; Govindan et al. 2009; Nadarajan et al. 2009; Amini et al. 2014). Analysis of the syncytial organization of adult lin-41 null mutant gonads indicates that the cytoplasmic core of the gonad narrows and terminates near the loop region, coincident with the premature cellularization of developing oocytes. Both the premature cellularization and M-phase entry of pachytene-stage oogenic cells in lin-41 null mutants require the function of the GLD-2 cytoplasmic poly(A) polymerase, which is also a component of OMA RNPs (Spike et al. 2014); lin-41gld-2 double mutants exhibit a synthetic phenotype in which developing oocytes fail to cellularize but arrest in pachytene.
Analysis of lin-41 reduction-of-function mutants also supports a role in facilitating the production of high-quality oocytes that can complete embryogenesis. A high incidence of embryonic lethality (60–80%) and X-chromosome nondisjunction (∼10- to 20-fold higher than in the wild type) is observed in lin-41(tn1487ts) mutants at a semipermissive temperature (22°) and in several of its intragenic revertants at 25°. Thus, LIN-41 is not simply an arbiter of the meiotic maturation decision: its proper function is needed to produce high-quality oocytes. The LIN-41 expression pattern in the germ line is consistent with this role. LIN-41 is present in the oogenic germ line from the mid-pachytene stage, but begins to be eliminated during meiotic maturation and is absent from embryos. One possibility is that the high levels of embryonic lethality and meiotic chromosome nondisjunction in lin-41 reduction-of-function mutants are a secondary consequence of subtle defects in the oocyte growth process. Alternatively, defects in translational regulation might disrupt the protein composition of lin-41 mutant oocytes.
LIN-41 is best known for its role in the developmental timing pathway in somatic cells where it is a target of the let-7 microRNA (Reinhart et al. 2000; Slack et al. 2000; Vella et al. 2004; Bagga et al. 2005; Del Rio-Albrechtsen et al. 2006; Harris and Horvitz 2011; Vladia et al. 2012; Zou et al. 2013). LIN-41 is highly conserved, as is its regulation by the let-7 microRNA (Lancman et al. 2005; Schulman et al. 2005; Maller Schulman et al. 2008; Worringer et al. 2014). Our data indicate that reduced let-7 function does not affect LIN-41 levels in the germ line, which is consistent with observations suggesting that the let-7 microRNA might not be expressed in the germ line (Lau et al. 2001). Rather, the expression of LIN-41 in the germ line is associated with the oocyte fate.
The prevailing view is that LIN-41 proteins function as regulators of post-transcriptional gene expression. This model stems from the observation that LIN-41 prevents the accumulation of the LIN-29 protein in the L3 larval stage (Slack et al. 2000), whereas lin-29 mRNA expression is first observed in the L2 stage (Rougvie and Ambros 1995; Bettinger et al. 1996). LIN-41 was proposed to function in one of two ways: either as an RNA-binding protein that might repress the translation of LIN-29 or as a RING-domain-containing E3 ubiquitin ligase that might regulate LIN-29 stability (Slack et al. 2000). Several reports are consistent with the idea that LIN-41 proteins can function as E3 ubiquitin ligases (Rybak et al. 2009; Loedige et al. 2013); however, the RING domain needed for ubiquitin ligase activity appears dispensable for mRNA repression (Loedige et al. 2013; Worringer et al. 2014). We did not isolate mutations in the RING domain; however, a definitive interpretation is not possible because of its small target size. Tocchini et al. (2014) used site-directed mutagenesis to alter five key cysteine residues that contribute to the coordination of zinc molecules necessary for maintaining the structure and function of the RING domain. Because this mutant form of LIN-41 appears fully functional, LIN-41 E3 ubiquitin ligase activity appears to be dispensable for oocyte and somatic development.
Consistent with a role in mRNA regulation, the Drosophila NHL-repeat domain protein Brat was recently shown to bind the hunchback mRNA and mediate translational repression (Loedige et al. 2014). By artificially tethering LIN-41 to reporter mRNA constructs via bacteriophage λ N protein sequences, a role for mammalian LIN-41 as a repressor of mRNA function was demonstrated (Loedige et al. 2013). Recent results show that human LIN-41 promotes reprogramming of induced pluripotent stem cells in a mechanism that may involve translational repression of the prodifferentiation transcription factor EGR1 (Worringer et al. 2014). In this and in the accompanying article in this issue (Spike et al. 2014), we isolated LIN-41 as a protein component of a translational regulatory RNP complex critical for oogenesis. Our mutational analysis of lin-41 underscores the importance of the NHL-repeat domain: missense mutations in conserved residues cause sterility. We further showed a role for LIN-41 in regulating oocyte M-phase entry in part through the 3′ UTR-mediated translational repression of cdc-25.3. These results suggest that LIN-41 functions in the germ line as a translational regulator to control oocyte growth and meiotic maturation.
This genetic study represents an extension of biochemical and genomic analyses undertaken to understand how the OMA-1/2 RNA-binding proteins regulate oocyte growth and meiotic maturation (Spike et al. 2014). LIN-41 was identified as a protein component of OMA RNPs and as one of two proteins shown to repress the translation of tested OMA target mRNAs. Phenotypic analysis shows that the LIN-41 and the OMA proteins exhibit an antagonistic relationship: LIN-41 inhibits M-phase entry and oocyte cellularization whereas the OMA proteins promote these events (Figure 13). Genetic analysis reveals a complex epistatic relationship between lin-41 and oma-1/2. Analysis of the triple mutant using null alleles demonstrates that lin-41 is epistatic: pachytene-stage oogenic cells prematurely enter M phase irrespective of oma-1/2 activity. The translational derepression of cdc-25.3 contributes to this phenotype in part. We were able to explore the genetic relationship between lin-41 and oma-1/2 in proximal diakinesis-stage oocytes using the lin-41(tn1487ts) mutation at a semipermissive temperature (22°). Under these conditions, many lin-41(tn1487ts) gonads exhibit evidence of premature M-phase entry (meiotic maturation) of proximal oocytes irrespective of the presence of sperm, further demonstrating that lin-41 is a negative regulator of oocyte meiotic maturation. Importantly, oocytes in lin-41(tn1487ts); oma-1; oma-2 triple mutants do not enter M phase. This result confirms the redundant requirement of oma-1 and oma-2 for oocyte meiotic maturation and suggests that lin-41 may not be needed at this stage, with the caveat that this analysis depends on the use of a non-null lin-41 allele. This interpretation is consistent with our observation that LIN-41 protein levels decline rapidly upon the onset of oocyte meiotic maturation and that the elimination of LIN-41 from oocytes undergoing meiotic maturation requires CDK-1 activity. These observations raise the possibility that LIN-41 must be inactivated and degraded for meiotic maturation to occur properly. Taken together, these data support a model in which LIN-41 and the OMA proteins coordinately control oocyte growth and the proper spatial and temporal execution of the meiotic maturation decision (Figure 13). One possibility is that the OMA proteins function to inhibit LIN-41, thereby promoting CDK-1 activation and presumably LIN-41 degradation. Alternatively, the OMA proteins might antagonize LIN-41 through the activation of CDK-1.
Figure 13.
LIN-41 and OMA-1/2 control oocyte growth and spatially regulate M-phase entry during oogenesis. (A) LIN-41 promotes oocyte growth and inhibits M-phase entry, whereas OMA-1/2 inhibits oocyte growth and promotes M-phase entry. (B) LIN-41 inhibits M-phase entry in part through the 3′ UTR-mediated translational repression of cdc-25.3, which encodes an activator of CDK-1. In turn, the activation of CDK-1 during meiotic maturation promotes the elimination of LIN-41. We speculate that the OMA proteins antagonize LIN-41 function either through GLD-2-dependent translational activation of mRNA targets or directly through some other means.
Since the oma-1/2 and lin-41 mutant phenotypes are polar opposites, it is intriguing that their proteins are found in an RNP and repress the translation of shared mRNA targets, including cdc-25.3. While the failure to repress cdc-25.3 translation contributes to the lin-41 null mutant phenotype, translational derepression of cdc-25.3 in oma-1; oma-2 double mutants is likely phenotypically irrelevant because these oocytes fail to enter M phase. OMA RNPs contain translational activators (e.g., GLD-2), and gld-2 and lin-41 interact genetically. Thus, OMA-LIN-41 RNPs might toggle crucial mRNA targets between activation and repression, as dictated by upstream signaling; however, such mRNAs have not been identified (Figure 13). A second possibility is that LIN-41 might function in part as a component of an OMA-independent RNP complex and that such complexes might be dynamic and highly regulated during oogenesis. In this regard, it is notable that LIN-41 expression commences in mid-pachytene, whereas the onset of OMA-1 expression occurs in late pachytene just prior to the loop (Detwiler et al. 2001; Lee and Schedl 2004). A third possibility is that a subset of LIN-41 or OMA-1/2 activities might occur independently of translational regulation. A goal for future work will be to explain the in vivo genetic behavior of lin-41, oma-1, and oma-2 in the regulation of oocyte growth and meiotic maturation at a biochemical level.
Supplementary Material
Acknowledgments
We thank Ann Rougvie for the key suggestion of examining germline development in lin-41 post-dauer animals; Nanette Pazdernik and Tim Schedl for advice on EdU labeling; Abby Dernburg, Monica Colaiácovo, Pierre Gönczy, Barth Grant, Jun Kelly Liu, Josef Loidl, Edward Kipreos, Ann Rougvie, Tim Schedl, Anne Villeneuve, and Monique Zetka for providing strains or reagents; and Seongseop Kim, Tim Schedl, Todd Starich, and Ann Rougvie for their help and suggestions during the course of this work. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by grant P40OD010440 from the National Institutes of Health (NIH) Office of Research Infrastructure Programs. This work was supported by NIH grants GM57173 and GM65115 (to D.G.) and Canadian Institutes of Health Research grant MOP-97815 (to D.H.).
Note added in proof: See Spike et al. 2014 (pp. 1513–1533) in this issue for a related work.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.168831/-/DC1.
Communicating editor: M. V. Sundaram
Literature Cited
- Abrahante J. E., Daul A. L., Li M., Volk M. L., Tennessen J. M., et al. , 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y., et al. , 2001. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128: 1951–1969. [DOI] [PubMed] [Google Scholar]
- Amini R., Goupil E., Labella S., Zetka M., Maddox A. S., et al. , 2014. C. elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J. Cell Biol. 206: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft N., Golden A., 2002. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Ashcroft N. R., Srayko M., Kosinski M. E., Mains P. E., Golden A., 1999. RNA-mediated interference of a cdc25 homolog in Caenorhabditis elegans results in defects in embryonic cortical membrane, meiosis, and mitosis. Dev. Biol. 206: 15–32. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., et al. , 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Bettinger J. C., Lee K., Rougvie A. E., 1996. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122: 2517–2527. [DOI] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. [DOI] [PubMed] [Google Scholar]
- Boxem M., Srinivasan D. G., van den Heuvel S., 1999. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126: 2227–2239. [DOI] [PubMed] [Google Scholar]
- Brodigan T. M., Liu J., Park M., Kipreos E. T., Krause M., 2003. Cyclin E expression during development in Caenorhabditis elegans. Dev. Biol. 254: 102–115. [DOI] [PubMed] [Google Scholar]
- Burrows A. E., Sceurman B. K., Kosinski M. E., Richie C. T., Sadler P. L., et al. , 2006. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development 133: 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D., Serafinas C., Ashcroft N., Kosinski M., Longo D., et al. , 2000. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26: 26–41. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A., 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005: pl1. [DOI] [PubMed] [Google Scholar]
- Clucas C., Cabello J., Büssing I., Schnabel R., Johnstone I. L., 2002. Oncogenic potential of a C. elegans cdc25 gene is demonstrated by a gain-of-function allele. EMBO J. 21: 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Del Rio-Albrechtsen T., Kiontke K., Chiou S. Y., Fitch D. H., 2006. Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev. Biol. 297: 74–86. [DOI] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs S. M., 2010. Regulation of the G2/M transition in rodent oocytes. Mol. Reprod. Dev. 77: 566–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M., Grosshans H., 2013. LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev. 27: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R., Schedl T., 2007. Sex determination in the germ line (March 5, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.82.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., Stanfield G. M., 2014. The regulation of spermatogenesis and sperm function in nematodes. Semin. Cell Dev. Biol. 29C: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Vought V. E., Hanazawa M., Lee M. H., Maine E. M., et al. , 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T., 1995a Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T., 1995b gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T., Tuck S., Kirchner J., Koch B., Auty R., et al. , 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Gartner A., MacQueen A. J., Villeneuve A., 2004. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol. Biol. 280: 257–274. [DOI] [PubMed] [Google Scholar]
- Gibert M. A., Starck J., Beguet B., 1984. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50: 77–85. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., Greenstein D., 2006. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., Hengartner M. O., 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Hubbard E. J., Schedl T., 2004a Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268: 342–357. [DOI] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004b Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. [DOI] [PubMed] [Google Scholar]
- Harris D. T., Horvitz H. R., 2011. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138: 4051–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. E., Govindan J. A., Yamamoto I., Schwartz J., Kaverina I., et al. , 2006. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev. Biol. 299: 105–121. [DOI] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., et al. , 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291. [DOI] [PubMed] [Google Scholar]
- Italiano J. E., Roberts T. M., Stewart M., Fontana C. A., 1996. Reconstitution in vitro of the motile apparatus from the amoeboid sperm of Ascaris shows that filament assembly and bundling move membranes. Cell 84: 105–114. [DOI] [PubMed] [Google Scholar]
- Ito K., McGhee J. D., 1987. Parental DNA strands segregate randomly during embryonic development of Caenorhabditis elegans. Cell 48: 329–336. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., Engebrecht J., 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., Schedl T., 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affects a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9: 1491–1504. [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T., 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180: 165–183. [DOI] [PubMed] [Google Scholar]
- Kadyk L. C., Kimble J., 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Karashima T., Sugimoto A., Yamamoto M., 2000. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127: 1069–1079. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee A. R., Kawasaki I., Strome S., Shim Y. H., 2009. A mutation of cdc-25.1 causes defects in germ cells but not in somatic tissues in C. elegans. Mol. Cells 28: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kawasaki I., Shim Y. H., 2010a. cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. J. Cell Sci. 123: 993–1000. [DOI] [PubMed] [Google Scholar]
- Kim K. W., Wilson T. L., Kimble J., 2010b. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 107: 17445–17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. [DOI] [PubMed] [Google Scholar]
- Kumsta C., Hansen M., 2012. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS ONE 7: e35428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancman J. J., Caruccio N. C., Harfe B. D., Pasquinelli A. E., Schageman J. J., et al. , 2005. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev. Dyn. 234: 948–960. [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P., 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P., 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Lesch B. J., Page D. C., 2012. Genetics of germ cell development. Nat. Rev. Genet. 13: 781–794. [DOI] [PubMed] [Google Scholar]
- Liu J., Rolef Ben-Shahar T., Riemer D., Treinin M., Spann P., et al. , 2000. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11: 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. C., Ambros V., 1991. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature 350: 162–165. [DOI] [PubMed] [Google Scholar]
- Loedige I., Gaidatzis D., Sack R., Meister G., Filipowicz W., 2013. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Stotz M., Qamar S., Kramer K., Hennig J., et al. , 2014. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiácovo M. P., McDonald K., Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A. S., Habermann B., Desai A., Oegema K., 2005. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 132: 2837–2848. [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D., 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman B. R., Liang X., Stahlhut C., DelConte C., Stefani G., et al. , 2008. The let-7 microRNA target gene, Mlin41/Trim71, is required for mouse embryonic survival and neural tube closure. Cell Cycle 7: 3935–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R., Endo S., Sugimoto A., Yamamoto M., 2005. Caenorhabditis elegans DAZ-1 is expressed in proliferating germ cells and directs proper nuclear organization and cytoplasmic core formation during oogenesis. Dev. Biol. 277: 142–154. [DOI] [PubMed] [Google Scholar]
- Masui Y., 2001. From oocyte maturation to the in vitro cell cycle: the history of discoveries of maturation-promoting factor (MPF) and cytostatic factor (CSF). Differentiation 69: 1–17. [DOI] [PubMed] [Google Scholar]
- Masui Y., Clarke H. J., 1979. Oocyte maturation. Int. Rev. Cytol. 57: 185–282. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J., 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., von Tobel L., Strnad P., Knott G., Leonhardt H., et al. , 2012. Analysis of centriole elimination during C. elegans oogenesis. Development 139: 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Colaiácovo M. P., 2005. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168: 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Otori M., Karashima T., Yamamoto M., 2006. The Caenorhabditis elegans homologue of deleted in azoospermia is involved in the sperm/oocyte switch. Mol. Biol. Cell 17: 3147–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G., 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell 5: 451–462. [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Robertson S., Lin R., 2013. The oocyte-to-embryo transition. Adv. Exp. Med. Biol. 757: 351–372. [DOI] [PubMed] [Google Scholar]
- Rose A. M., Baillie D. L., 1979. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92: 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T., et al. , 1997. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192: 59–77. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Ambros V., 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121: 2491–2500. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Moss E. G., 2013. Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105: 153–180. [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E. A., et al. , 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11: 1411–1420. [DOI] [PubMed] [Google Scholar]
- Schedl T., Kimble J., 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Schulman B. R., Esquela-Kerscher A., Slack F. J., 2005. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 234: 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., et al. , 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., et al. , 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M., et al. , 2014. Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198: 1513–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J., 1977. Radioautographic study of RNA synthesis in Caenorhabditis elegans (Bergerac variety) oogenesis. Biol. Cell. 30: 181–182. [Google Scholar]
- Stinchcomb D. T., Shaw J. E., Carr S. H., Hirsh D., 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5: 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G., 2003. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 13: 134–139. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tocchini C., Keusch J. J., Miller S. B., Finger S., Gut H., et al. , 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10: e1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., Hobert O., 2009. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4(3): e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M. C., Choi E. Y., Lin S. Y., Reinert K., Slack F. J., 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 18: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladia B., Kemper K., Alaimo J., Heine C., Moss E. G., 2012. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8: e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Boag P. R., Blackwell T. K., 2007. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev. Biol. 304: 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J., 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Ward S., Carrel J. S., 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73: 304–321. [DOI] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. [DOI] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., et al. , 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. E., Gaidatzis D., Senften M., Farley B. M., Westhof E., et al. , 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 30: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Kawasaki I., Shim Y. H., 2012. CDC-25.1 controls the rate of germline mitotic cell cycle by counteracting WEE-1.3 and by positively regulating CDK-1 in Caenorhabditis elegans. Cell Cycle 11: 1354–1363. [DOI] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., Müller F., 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Zinovyeva A., Ambros V., Chuang C. F., et al. , 2013. Developmental decline in neuronal regeneration by the progressive change in two intrinsic timers. Science 340: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.