Abstract
The use of dense SNPs to predict the genetic value of an individual for a complex trait is often referred to as “genomic selection” in livestock and crops, but is also relevant to human genetics to predict, for example, complex genetic disease risk. The accuracy of prediction depends on the strength of linkage disequilibrium (LD) between SNPs and causal mutations. If sequence data were used instead of dense SNPs, accuracy should increase because causal mutations are present, but demographic history and long-term negative selection also influence accuracy. We therefore evaluated genomic prediction, using simulated sequence in two contrasting populations: one reducing from an ancestrally large effective population size (Ne) to a small one, with high LD common in domestic livestock, while the second had a large constant-sized Ne with low LD similar to that in some human or outbred plant populations. There were two scenarios in each population; causal variants were either neutral or under long-term negative selection. For large Ne, sequence data led to a 22% increase in accuracy relative to ∼600K SNP chip data with a Bayesian analysis and a more modest advantage with a BLUP analysis. This advantage increased when causal variants were influenced by negative selection, and accuracy persisted when 10 generations separated reference and validation populations. However, in the reducing Ne population, there was little advantage for sequence even with negative selection. This study demonstrates the joint influence of demography and selection on accuracy of prediction and improves our understanding of how best to exploit sequence for genomic prediction.
Keywords: genomic selection, whole-genome sequence, high-density SNP, GenPred, shared data resource
METHODOLOGY has been developed to predict genetic value for polygenic traits in livestock and crops by exploiting high-density genome-wide SNP genotypes that are fitted simultaneously in an analytical model (Meuwissen et al. 2001). The same methodology can be applied in human genetics, for example, to predict complex disease risk (reviewed in De los Campos et al. 2010). In livestock and plants this analytical approach is often referred to as “genomic selection” because the genomic predictions are used for selection decisions. First, a large “reference population” with genotypes and phenotypes is required to jointly estimate genome-wide SNP effects. Then the accuracy of prediction using the estimated SNP effects is reevaluated in an independent “validation population,” before the genomic prediction equation is routinely applied on individuals with genotypes but no phenotypes.
Genomic prediction (GP) methods generally use dense genome-wide SNP genotypes and therefore rely on exploiting linkage disequilibrium (LD) between these SNPs and unknown causative mutations or quantitative trait loci (QTL). The lower the LD is between the SNP and causal mutations, the lower the accuracy will be of GP. As the number of generations separating the reference and validation populations increases, the LD between SNPs and causative mutations is further eroded by recombination and therefore accuracy of GP will fall. The impact of recombination could be eliminated if the prediction was based on the causal mutations themselves. This would be possible if we had access to whole-genome sequence and this is increasingly likely as the cost of sequencing falls. Furthermore a number of species-specific databanks of whole-genome sequence are being generated and, using these as reference genomes, it is possible to impute full sequence for many thousands of individuals that have been genotyped with high-density SNP chips.
Several simulation studies have shown that there would be a significant advantage for genomic prediction using sequence compared to the equivalent of 30,000–60,000 genome-wide SNPs in an ∼30-M genome (Meuwissen and Goddard 2010; Clark et al. 2011; Druet et al. 2014). However, these studies did not compare use of sequence data with the higher-density commercial SNP arrays that are now commonly used for a number of species (for example, ≥600,000 SNPs in humans and cattle).
An additional argument for using sequence for GP is that it should be particularly advantageous when QTL have been under long-term negative selection (such as disease or fertility traits): causal variants are then more likely to be rare and therefore in low LD with SNPs on commercial chips that typically have minor allele frequency (MAF) > 0.1. A study by Druet et al. (2014) indirectly investigated this potential advantage of sequence by simulating genotype data in which QTL were represented only as rare variants. Given this approach, these authors conclude that sequence data could significantly improve the accuracy of GP compared to the equivalent of 50,000 SNPs genome-wide. However, simulation studies in which only rare variants are chosen to act as surrogate QTL may not provide an adequate model of loci under long-term negative selection. For example, changes in ancestral demography such as a recent bottleneck in effective population size (Ne) also exert a strong influence on the distribution of allele frequencies (e.g., Marth et al. 2004) and even mutations with a deleterious effect on fitness may drift to higher frequencies than would be expected in a population with no recent bottleneck. Also, patterns of LD surrounding loci that are under long-term negative selection may be quite different from those surrounding neutral loci due to “background selection” (Charlesworth et al. 1993).
In this study we investigate the potential advantages of sequence data for genomic prediction and demonstrate that this will jointly depend on the ancestral demography of a population, the presence or absence of long-term negative selection acting on QTL, and the method of analysis. We investigated two contrasting demographic scenarios in which QTL were represented by neutral loci or loci subjected to long-term negative selection. The first scenario was a large constant Ne at the upper limit for estimates of recent human and maize Ne (e.g., Vigouroux et al. 2002; Schaffner et al. 2005; McEvoy et al. 2011). The second scenario mimicked sequence data from a single cattle breed (MacLeod et al. 2013). We compared realized accuracies, using either sequence data or dense SNP panels chosen to reflect the high-density commercial SNP arrays currently applied in human and livestock genetics. The GP analytical methods used were genomic best linear unbiased prediction (“GBLUP”) (e.g., Habier et al. 2007; VanRaden 2008; Goddard 2009; Hayes et al. 2009) and a Bayesian method referred to as BayesR (Erbe et al. 2012), which is conceptually similar to BayesB (Meuwissen et al. 2001). We chose these two analytical methods because both are commonly applied in genomic prediction studies. A key difference between these two approaches is that GBLUP assumes a quasi-infinitesimal model while the Bayesian method assumes a large proportion of loci have no effect.
Materials and Methods
Simulated genotypes
Sequence data were forward-in-time simulated, using FREGENE software (Hoggart et al. 2007; Chadeau-Hyam et al. 2008) in a Wright–Fisher panmictic population. Two different populations were simulated: one with a constant effective population size (Ne) of 25,900 (“constant”) and the other with a decreasing population size (“bovine”). The full Ne parameters used for the bovine population were from a study that inferred demography in the Holstein cattle breed, using whole-genome sequence data (MacLeod et al. 2013, supplementary information, table S1). For both populations the mutation (µ) and recombination (r) rates were chosen as similar to recent mammalian estimates for these parameters (Kumar and Subramanian 2002; Arias et al. 2009; Roach et al. 2010; Campbell et al. 2012) and to generate realistic single-locus heterozygosity rates. Both µ and r were assumed constant across the genome, with r = 1 × 10−8 and µ = 9.4 × 10−9/bp per generation. The expected single-base pair heterozygosity was 9.7 × 10−4 in both the bovine and constant populations, similar to observed heterozygosity in Holstein dairy cattle (MacLeod et al. 2013) and some human populations (Venter et al. 2001; Voight et al. 2005).
As specified in FREGENE, the parameter scaling option was used to reduce the time taken for simulations (Hoggart et al. 2007; Chadeau-Hyam et al. 2008) where Ne > 2000. For the final output in the constant-sized population, the FREGENE “unscale” method was implemented to restore Ne to the actual size while rescaling all other rate parameters appropriately (Chadeau-Hyam et al. 2008). In the bovine population the Ne reduced to 90 individuals in the final three generations but we required a sample size of 5000 individuals. This was achieved by scaling up the Ne of 90 and the time period of three generations by a factor of 56, with the reciprocal scaling down of mutation rate, recombination rate, and selection coefficients.
The simulation of the large constant-sized population ran for 370,000 generations to ensure that it had reached a drift–recombination–mutation–selection (drm) equilibrium while in the bovine population, and only the most ancestral population reached drm equilibrium. The bovine and constant-sized populations were simulated both as neutral populations (Bov-Neut and Const-Neut) and as populations with long-term negative selection (Bov-Sel and Const-Sel). Twenty replicates were simulated for each of these four scenarios.
In both Bov-Sel and Const-Sel, long-term negative selection was simulated on a random selection of 0.1% of all new mutations with a constant selection coefficient (s) of −2 × 10−4 with additive effects (h = 0.5). Selection coefficients were constant over time but in FREGENE there is a user-specified probability that selection is switched off at any given site. We set this probability to 1/1,000,000 in the Bov-Sel population, which is equivalent to a mean of selected sites being switched off after 1,000,000 generations if not naturally lost or fixed. Similarly in the Const-Sel population the probability of selection being switched off was set to 1/750,000. This simply ensured that a drift–recombination–mutation–selection equilibrium was reached in the most ancestral large Ne of the bovine population and in the constant-sized population.
To make the study computationally feasible for genomic prediction using sequence data, we generated a genome size of 50 Mb, under the scaling argument demonstrated by Meuwissen and Goddard (2010) (that is, accuracy of GP is proportional to number of reference individuals per morgan length of the genome). At the end of the simulation, in each replicate of Const-Sel and Const-Neut, we used “SAMPLE” software (Chadeau-Hyam et al. 2008) to randomly sample haplotype pairs to generate sequence genotype data for 5000 individuals, and we henceforth refer to these individuals as “0_Gen”. Due to the low Ne in the bovine population, we first sampled 10,000 genotyped individuals for each replicate. Then, from these 10,000 individuals, we randomly selected 5000, having first discarded any one of a pair if they differed at <1500 genotypes of a random 10,000 SNP loci tested. We did this to ensure that there were no extremely close relatives (or near duplicates) in the final 5,000 0_Gen individuals. We then continued the simulations for a further 10 generations and then used SAMPLE to generate sequence genotypes for a further 2000 individuals and refer to this set of genotyped individuals as “10_Gen”. For each scenario we ran these 10 generations with Ne at the final full population size, that is, 5040 in the bovine population and 25,900 for the constant-sized population. Although an Ne of 5040 for the bovine populations represented an increase in the present-day bovine population size, this had only a very minimal impact on the observed LD pattern in the population because it spans a relatively short period of time. The scripts and parameter files used to generate simulated genotype data for each of the four scenarios are given in supporting information, File S1, File S2, File S3, and File S4.
High-density SNP genotypes (“HD SNPs”) were generated for all individuals by selecting 10,000 loci uniformly at random from the sequence variants to represent an HD SNP array (i.e., approximately one SNP/5 kbp). To mimic the ascertainment bias of commercial SNP arrays, HD SNPs were selected only if their MAF > 0.1. Additionally, for the Bov-Neut and Bov-Sel data, the same procedure was followed to generate medium-density SNP genotypes (“MD SNPs”) of 1000 loci.
Simulated phenotypes
A total of 50 additive QTL effects were simulated for all genotyped individuals in each of the Bov-Neut, Bov-Sel, Const-Neut, and Const-Sel replicated populations. In the Const-Neut and Bov-Neut populations, loci were randomly selected from polymorphic loci in the sequence data. In Const-Sel and Bov-Sel, 50 QTL loci were randomly chosen from segregating loci that had been subjected to long-term negative selection. In five of the Bov-Sel populations, there were just under 50 selected loci still segregating (49, 47, 46, 46, and 41) and therefore the shortfall was accommodated by randomly selecting QTL loci from neutral loci chosen with MAF < 0.1.
QTL allele substitution effects (αi) were sampled from a normal distribution (mean of zero) and additive genetic values for each individual, at each QTL (i = 1 to 50) were calculated as
and
where pi and qi are the major and minor allele frequencies for locus i (Falconer and Mackay 1996). Genetic values (GV) for each QTL were summed to provide a true additive genetic value (TGVj) for each of the genotyped individuals (j = 1 to 5000):
Phenotypes were generated by adding an appropriate residual term to the TGVj, drawn from a normal distribution, N(0, σ2e). The σ2e was chosen to generate a specified heritability (h2) by first estimating the variance of the TGVs (σ2TGV) in the simulated population and then
For each replicate we simulated a number of phenotypic data sets for a range of h2: 0.45, 0.15, 0.1, and 0.01. In Bov-Neut and Bov-Sel, for h2 = 0.1 we also generated phenotypic data sets with 15 QTL.
Accuracy of predicted genetic values
“Reference” individuals (T) with phenotypes and genotypes were used to estimate the prediction equations for SNP effects, where SNP genotypes were MD SNPs, HD SNPs, or full-genome sequence SNPs (SEQ). Realized accuracy of GP was tested in “validation” individuals (V) by assessing the correlation between their predicted genetic value (PGV) and the TGV. For each scenario, accuracy was calculated from the average of 20 replicated simulations. Both 0_Gen reference and validation groups were randomly chosen as nonoverlapping subsets (NT and NV) from the 5000 genotyped individuals in each replicated data set (NT = 3750 and NV = 1250 or NT = 2500 and NV = 2500). The 2000 individuals from 10_Gen were used as a second validation group for each replicate.
GBLUP analysis
Genomic prediction was implemented using best linear unbiased predictor (BLUP) methodology (Henderson 1984) but with a “genomic relationship matrix” (GRM) replacing the traditional pedigree relationship matrix. This approach is often referred to as “GBLUP” and has been shown to be equivalent to the original ridge regression BLUP approach implemented by Meuwissen et al. (2001) (e.g., Habier et al. 2007; Goddard 2009). The GBLUP analysis was implemented in ASReml software (Gilmour et al. 2005), using the model
where μ is the population mean, 1 is a vector of ones, Z is the incidence matrix for random individual effects, g is a vector of genetic values, and e is the vector of residuals. The g and e random effects are assumed to be normally distributed as N(0, Gσ2g) and N(0, Iσ2e), where G is the GRM derived using the approach of Yang et al. (2010). The G matrix was estimated from either the set of SNP markers (HD SNPs or MD SNPs) or the entire set of sequence SNPs, which also included the causal mutations (SEQ).
The variance components for random effects (σ2g and σ2e) were estimated first in ASReml and then used to estimate the genetic value ([]) for reference and validation individuals as
BayesR analysis
We implemented Bayesian genomic predictions, using the “BayesR” method detailed in Erbe et al. (2012). Briefly, this approach is similar to BayesB (Meuwissen et al. 2001) but SNP effects are assumed to come from one of four normal distributions, each with a mean of zero and variance
The models fitted to our data were as described in Erbe et al. (2012) except that we omitted a polygenic effect because individuals were randomly bred with no formal pedigree structure,
where μ is the mean, 1 is a vector of ones, W is the design matrix allocating records to SNP effects represented by the vector u, and e is the vector of random residuals. The dimensions of W were NT × Nm, where NT is the number of reference individuals and Nm is the number of marker genotypes. Each element of the W matrix was scaled by the allele frequency of SNP i, as for GBLUP: where xij is the genotype for SNP i in animal j (genotypes coded as 0 = 11, 1 = 12, and 2 = 22).
The true heritability of the trait was used to furnish an a priori estimate of the proportion of the total variance due to causal mutations (σ2g). We specified a total of 20,000 iterations with the first 10,000 discarded as burn-in, based on previous experience with the BayesR method and preliminary tests with our simulated data. For the MD and HD SNP data we ran four BayesR chains within each of the 20 replicated data sets, while for the SEQ data we ran two chains for each data set to moderate computational requirements. From the consistency of the results within and across replicates we were confident that this was adequate.
Deterministic prediction of accuracy
Deterministic equations have been developed to furnish an a priori estimate of the accuracy of GBLUP GP (“”) (Daetwyler et al. 2008, 2010; Goddard 2009; Goddard et al. 2011). Here we applied the approach of Goddard et al. (2011), where the predicted squared accuracy of GP is given as
and solving the above equation as a quadratic with the accuracy is
| (1) |
where θ = NTbh2/Me, NT is the number of reference individuals, b is a correction factor for the proportion of the QTL variance captured by the markers, and Me is an estimate of the average number of independently segregating chromosome segments genome-wide (Goddard 2009). (Goddard et al. 2011), where represents the pairwise measure of LD averaged across all pairs of loci on each chromosome. Me can be calculated analytically but requires the assumption of constant Ne so we estimated it empirically from the simulated sequence data with a random sample of 2500 SNPs in each of the 20 replicates. We determined that 2500 SNPs were an adequate sample size because we found a very similar result using 5000 SNPs in one replicate of the bovine and constant Ne population.
Results
The HD SNP density in this study is approximately equivalent to ∼600,000 genome-wide SNPs and MD SNPs to ∼60,000 SNPs in cattle and humans with their approximate genome size of 30 M (Venter et al. 2001; Bovine Genome Sequencing and Analysis Consortium et al. 2009). All results are expressed as an average across 20 replicated data sets. Allele frequency distributions and average LD in the bovine and constant Ne scenarios are presented first because these have a strong influence on the realized accuracy of GP.
Allele frequency distributions and LD
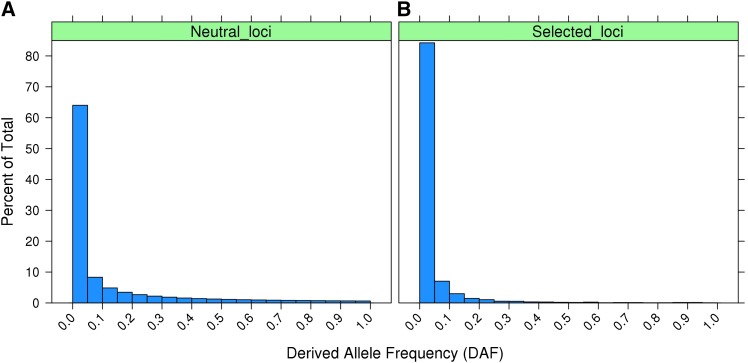
In the large constant-sized neutral population (Const-Neut) the distribution of derived allele frequency (DAF) among segregating loci (Figure 1A) followed the classic expectation for a neutral population (e.g., Marth et al. 2004). In Const-Neut 72% of segregating neutral loci had a DAF < 0.1 while 91% of the loci under long-term negative selection in Const-Sel had a DAF < 0.1 (Figure 1, A and B).
Figure 1.
(A and B) Histogram showing (A) the DAF distribution for segregating neutral loci in Const-Neut populations (20 replicates) and (B) the DAF distribution for loci subjected to negative selection that were still segregating in Const-Sel populations (20 replicates).
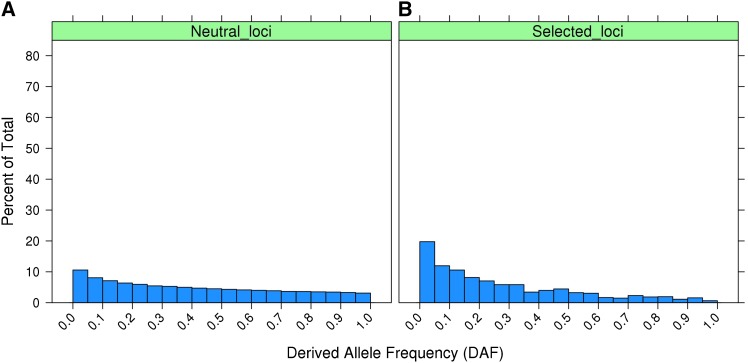
In contrast to Const-Neut, the DAF distribution for neutral loci in Bov-Neut is relatively flat (Figure 2A) because of the reduction in recent Ne. Although the bovine Ne in the most ancestral equilibrium phase was 62,000, the final DAF distribution was strongly influenced by random drift because the bovine demography underwent a sharp decline in Ne from ∼3000 generations ago to the present day. As a result, only 18% of loci have a DAF < 0.1 in Bov-Neut, and even for loci under long-term negative selection in Bov-Sel there were only 32% with DAF < 0.1 (Figure 2, A and B).
Figure 2.
(A and B) Histogram showing (A) the DAF distribution for segregating neutral loci in Bov-Neut populations (20 replicates) and (B) the DAF distribution for loci subjected to negative selection that were still segregating in Bov-Sel populations (20 replicates).
The pairwise nucleotide diversity or heterozygosity per base pair was expected to be 9.7 × 10−4 for all neutral populations. The Const-Neut populations matched this expectation, but Bov-Neut had a slightly lower heterozygosity than predicted (average of 8.9 × 10−4). This was probably a result of the scaling used for computational efficiency (see Materials and Methods): FREGENE simulations employ a two-allele finite site model and therefore repeat mutations at one site may have occurred more often than in an unscaled population. The heterozygosity in populations with long-term negative selection was only slightly lower than that of the neutral populations because only 0.1% of sites were subjected to selection. The average total numbers of segregating sites on the 50-Mb genome in 0_Gen populations were 404,589 in Const-Neut, 402,687 in Const-Sel, 142,973 in Bov-Neut, and 135,692 in Bov-Sel.
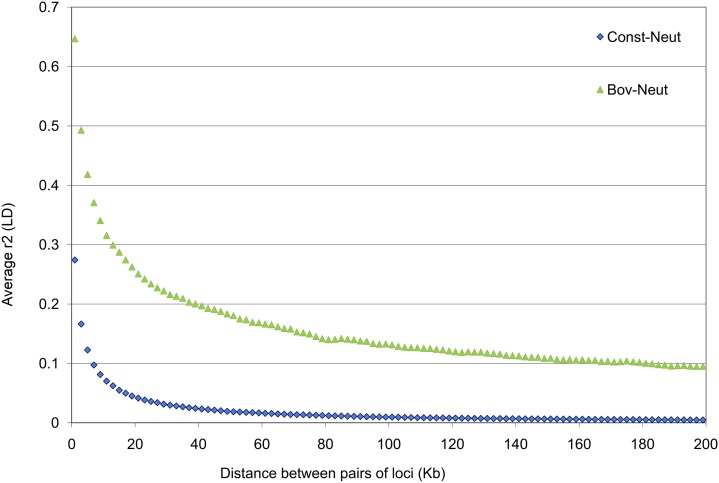
As expected, the average LD (pairwise r2) in Bov-Neut was much higher than in Const-Neut due to the recent population bottleneck in the bovine demography (Figure 3). We estimated Me (i.e., the number of effectively independent chromosome segments) directly from the average r2 in the sequence and found in Const-Neut Me = 1786, while in Bov-Neut Me = 59 on the 50-Mb genome.
Figure 3.
Observed LD (linkage disequilibrium—pairwise r2) averaged over bins of 2 kbp (Kb) for the Const-Neut (blue) and Bov-Neut (green) populations. The simulation recombination rate was set to 1 × 10−8; therefore 1 Mb ≡ 1 cM.
GBLUP accuracy: effects of long-term negative selection and demography
Table 1 compares the GBLUP realized accuracies for GP in the constant Ne and bovine populations for scenarios with either neutral or negatively selected causal mutations. The realized accuracy of GP is much higher in the bovine population than in the large constant-size population for a trait with the same heritability because of the higher LD in the bovine population. In the bovine scenarios the GBLUP accuracies were similar for SEQ and HD SNP, and in the constant Ne scenarios only a small improvement in GBLUP accuracy was observed with SEQ compared to HD SNPs.
Table 1. Realized and deterministic GBLUP accuracy with estimated heritabilities (h2) in bovine and constant Ne populations with either neutral QTL (Neut) or QTL under negative selection (Sel).
| Scenario | True h2 | Neut population estimated h2 (SE) | Sel population estimated h2 (SE) | Neut population realized accuracy (SE) | Sel population realized accuracy (SE) | Relative reduction in accuracy due to selection (%) | Deterministic prediction of accuracya |
|---|---|---|---|---|---|---|---|
| Constant SEQb | 0.45 | 0.459 (0.005) | 0.452 (0.001) | 0.595 (0.005) | 0.593 (0.005) | 0 | 0.662 |
| 0.15 | 0.161 (0.006) | 0.147 (0.006) | 0.427 (0.008) | 0.405 (0.007) | 5.1 | 0.420 | |
| Constant HD SNPb | 0.45 | 0.401 (0.006) | 0.371 (0.008) | 0.585 (0.006) | 0.562 (0.007) | 3.9 | 0.572 |
| 0.15 | 0.14 (0.005) | 0.120 (0.005) | 0.418 (0.009) | 0.378 (0.008) | 9.6 | 0.361 | |
| Bovine SEQb | 0.45 | 0.401 (0.005) | 0.413 (0.008) | 0.960 (0.001) | 0.961 (0.001) | 0 | 0.985 |
| 0.15 | 0.148 (0.005) | 0.145 (0.004) | 0.892 (0.003) | 0.888 (0.004) | 0.4 | 0.938 | |
| 0.01 | 0.010 (0.001) | 0.009 (0.001) | 0.591 (0.018) | 0.585 (0.013) | 1.0 | 0.624 | |
| Bovine HD SNPb | 0.45 | 0.440 (0.007) | 0.461 (0.010) | 0.961 (0.001) | 0.960 (0.001) | 0 | 0.983 |
| 0.15 | 0.154 (0.006) | 0.152 (0.005) | 0.894 (0.003) | 0.888 (0.004) | 0.7 | 0.935 | |
| 0.01 | 0.01 (0.001) | 0.009 (0.001) | 0.591 (0.018) | 0.586 (0.014) | 1.0 | 0.621 |
Standard errors across replicates are given in parentheses.
These accuracies are estimated using Equation 1 with our empirically estimated Me.
Accuracies are given for full sequence (SEQ) and high-density SNPs (HD SNPs) averaged across 20 replicates. The number of reference and validation individuals was 2500, except for the bovine scenario h2 = 0.01 where it was 3750.
When causal mutations were subject to long-term negative selection, we observed some reduction in the GBLUP realized accuracy in both Const-Sel and Bov-Sel compared to the neutral populations (Table 1). The lower the heritability was, the greater the difference between the accuracies of GP in selected and neutral populations. Notably, the difference between Const-Neut and Const-Sel GBLUP accuracies was more extreme with HD SNPs compared to SEQ. In contrast, the differences between GBLUP accuracies in Bov-Neut and Bov-Sel for a given heritability were the same regardless of using SEQ or HD SNPs.
The GBLUP heritability (h2) estimates were generally close to the true heritabilities (Table 1), but tended to be lower in Const-Sel relative to Const-Neut particularly when using HD SNPs. For example, in Const-Sel, when true h2 = 0.45, the h2 estimate using HD SNPs implies that ∼14% of the QTL variance was not captured by the SNPs. When the true h2 was 0.45, there was a trend for the SEQ data to estimate a downward bias in Bov-Neut, indicating that the sequence data introduce unwanted noise to the estimate of variance compared to HD SNPs. However, the regression of TGV on PGV was on average equal to one with both SEQ and HD SNPs, indicating that there was no obvious bias detected in the PGV (results not shown).
The deterministic predictions of accuracy using the Me estimated directly from the data are reasonably close to the realized GBLUP values, although they tend to slightly overestimate accuracy in the bovine population (Table 1).
BayesR accuracy: effects of long-term negative selection and demography
The BayesR method resulted in a very marked improvement in realized accuracies for GP, using SEQ compared to HD SNPs in in both Const-Neut and Const-Sel (Table 2). However, there was no improvement in BayesR accuracy for the bovine scenarios from using SEQ rather than HD SNPs (Table 2). In Table 2 the h2’s are lower for the bovine population simply to allow the bovine and constant Ne results to be compared at similar levels of GBLUP accuracy [remembering that accuracy is proportional to NTh2/Me (Equation 1)]. The realized BayesR accuracy in Const-Neut and Const-Sel was considerably higher than GBLUP accuracy, particularly with SEQ but also for HD SNPs, while in the bovine scenarios there was no difference between BayesR and GBLUP accuracies (Table 2). Although BayesR accuracy using HD SNPs was more variable than GBLUP accuracy across the constant Ne replicates, the within-replicate BayesR accuracy was always higher than GBLUP accuracy (results not shown).
Table 2. Realized accuracies for GP in Const-Neut, Const-Sel, Bov-Neut, and Bov-Sel, using BayesR methodology with sequence (SEQ) or high-density SNPs (HD SNPs).
| Population (size, T) | Accuracy BayesR with SEQ (SE) | Accuracy BayesR with HD SNPs (SE) | Increased accuracy BayesR vs. GBLUP with SEQ (%) | Increased accuracy BayesR vs. GBLUP with HD SNPs (%) |
|---|---|---|---|---|
| Const-Neut h2 = 0.45 (T = 2500) | 0.957 (0.002) | 0.779 (0.01) | 36.2 | 19.4 |
| Const-Sel h2 = 0.45 (T = 2500) | 0.952 (0.003) | 0.703 (0.015) | 35.9 | 14.1 |
| Const-Neut h2 = 0.15 (T = 2500) | 0.791 (0.012) | 0.614 (0.019) | 36.4 | 19.6 |
| Const-Sel h2 = 0.15 (T = 2500) | 0.832 (0.008) | 0.534 (0.025) | 42.7 | 15.6 |
| Bov-Neut h2 = 0.1 (T = 3750) | 0.895 (0.004) | 0.883 (0.004) | 0 | 0 |
| Bov-Sel h2 = 0.1 (T = 3750) | 0.896 (0.004) | 0.882 (0.004) | 0 | 0 |
| Bov-Neut h2 = 0.01 (T = 3750) | 0.587 (0.017) | 0.592 (0.017) | 0 | 0 |
| Bov-Sel h2 = 0.01 (T = 3750) | 0.579 (0.012) | 0.587 (0.013) | 0 | 0 |
Also shown is the absolute increase in accuracy from using BayesR rather than GBLUP. Standard errors of the accuracies across replicates are given in parentheses.
Persistency of accuracy across generations
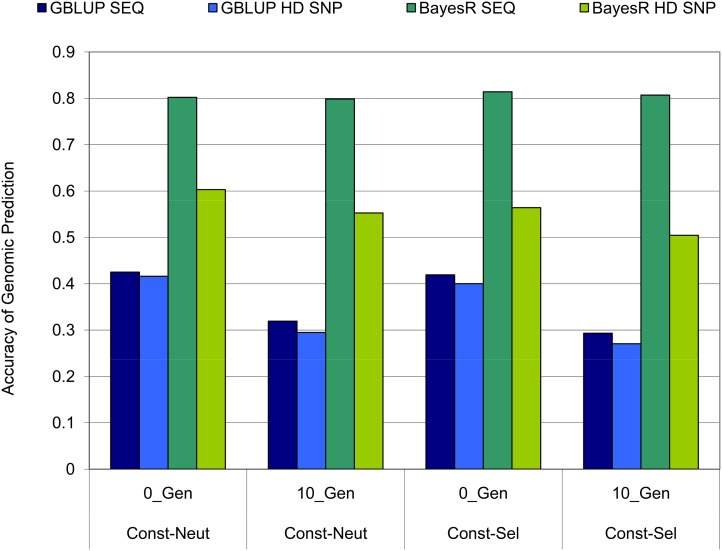
The persistency of accuracy was determined by comparing the accuracy of GP in validation individuals either from the same generation as the reference individuals (0_Gen) or separated by 10 generations (10_Gen). Figure 4 compares these accuracies in the constant Ne populations when h2 = 0.1 (3750 reference individuals). For GBLUP, there was ∼25% reduction in the accuracy of GP from 0_Gen to 10_Gen. This drop in the GBLUP accuracy was observed with both SEQ and HD and is slightly more pronounced in Const-Sel compared to Const-Neut. In contrast, the BayesR accuracy with SEQ for Const-Neut and Const-Sel showed only a marginal reduction from 0_Gen to 10_Gen. This high persistency of SEQ BayesR conferred the greatest advantage to Const-Sel where accuracy in 10_Gen was 37% higher with SEQ compared to HD SNPs. Although the BayesR accuracy using HD SNPs did fall in 10_Gen, it still remained considerably higher than GBLUP accuracy for both Const-Neut and Const-Sel. Similar results as shown in Figure 4 were obtained for Const-Neut and Const-Sel with trait h2 = 0.45 (results not shown).
Figure 4.
GBLUP and BayesR accuracies in the Const-Neut and Const-Sel populations when validation individuals were either from the same generation as reference individuals (0_Gen) or separated by 10 generations (10_Gen). The trait h2 = 0.1, number of reference individuals = 3750, and number of QTL = 50.
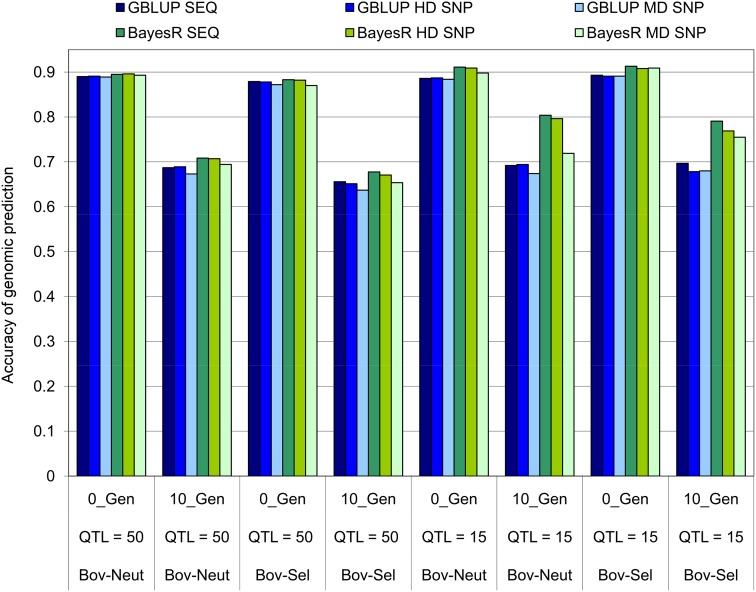
The same test of persistency was made in the bovine populations in simulations with either 50 or 15 QTL and a heritability of 0.1 (Figure 5). In addition to SEQ and HD SNPs, we tested the persistency of accuracy, using the MD SNP density (equivalent to a 60,000 SNP density in the bovine genome). In the bovine populations we simulated an additional scenario with 15 QTL because it is expected that Bayesian methods will perform better than GBLUP only if the number of QTL is considerably lower than Me (e.g., Daetwyler et al. 2010). As observed in the constant Ne, there was an ∼25% drop in GBLUP accuracy from 0_Gen to 10_Gen in all scenarios, and no very clear difference was observed between GBLUP SEQ, HD, and MD SNP accuracies (Figure 5). When the number of QTL = 50, the persistency was only slightly better for BayesR compared to GBLUP, with little difference between SEQ, HD, and MD SNPs. When the number of QTL = 15, the advantage in the persistency of BayesR accuracy compared to GBLUP is much clearer with both SEQ and HD SNPs: the reduction in accuracy was only ∼13% from 0_Gen to 10_Gen. However, even in Bov-Sel with QTL = 15, the BayesR accuracy in 10_Gen was only 2.8% higher with SEQ compared to HD SNPs.
Figure 5.
GBLUP and BayesR accuracies in the Bov-Neut and Bov-Sel populations when validation individuals were either from the same generation as reference individuals (0_Gen) or separated by 10 generations (10_Gen). The trait h2 = 0.1, number of reference individuals = 3750, and number of QTL = 50 or 15.
Discussion
This is a novel study evaluating the effect on GP accuracy of demography, long-term negative selection, use of SEQ compared to HD SNPs, the analytical method (Bayesian vs. GBLUP), and an interval of 10 generations between the reference and validation data. Below we discuss the effects and interactions of these variables, but first we consider how well our simulated data reflect real genomic data from contrasting populations such as domesticated livestock, humans, or outbred plant populations.
Model populations
We simulated two contrasting population scenarios to create LD, variant densities, and allele frequency spectra that reflect opposite ends of the range observed in real populations, because these parameters are critical in evaluating genomic prediction accuracy. Importantly, the simulation LD, nucleotide diversities, and allele frequency spectra are detailed in this article to allow further comparison as more real population sequence data become available.
Our bovine simulation models a single breed of domesticated livestock species, where effective population sizes have reduced dramatically since domestication and breed formation. The LD, nucleotide diversity, and allele frequency distribution in this bovine simulation were generally similar to those observed within a range of cattle breeds, in particular, Holstein dairy cattle (Bovine HapMap Consortium 2009; Villa-Angulo et al. 2009; MacLeod et al. 2013). A recent study of 234 whole-genome sequences from three dairy cattle breeds (Daetwyler et al. 2014) showed a higher proportion of rare variants (MAF < 0.05) compared to those in our simulation but this is expected because their allele frequencies were calculated across breeds. Additionally, there is some evidence that a considerable proportion of these rare variants may be sequencing errors even though relatively strict variant calling filters were used (O. Gonzalez-Recio and H. Daetwyler, personal communication). It is possible that the allele frequency spectrum of our simulated variants under selection is closer to the true distribution in error-free sequence than our neutral simulation. The total number of SNPs segregating in our bovine simulation is equivalent to ∼8 million in a 2800-Mb genome (2800/50 × 143,000). Although this is considerably lower than the ∼15 million variants reported by Daetwyler et al. (2014) for Holstein breed sequence data, their number includes ∼1.5 million indels (not included in our simulation) as well as homozygous nonreference alleles. Their estimate is also inflated by sequencing errors (possibly 1–1.5 million errors) and potentially has an upward bias from population substructure because they included black and white Holsteins from North America, Europe, and Australia as well as a subpopulation of red and white Holsteins. We chose a dairy cattle breed demography because a number of countries are already implementing genomic prediction in dairy cattle populations (Lund et al. 2011; VanRaden et al. 2011; Pryce and Daetwyler 2012). This is likely to reflect one of the more extreme livestock breeds in terms of the very sharp recent reduction in Ne, because the widespread use of artificial insemination enables popular bulls to sire tens of thousands of daughters.
We contrast the bovine demography with a population having a large constant Ne to study the effect of other variables in a simple demography with low LD. The low LD in the constant Ne scenario is characteristic of some outbred commercial plant populations such as maize (Rafalski and Morgante 2004) and pines (Neale and Savolainen 2004). While the average LD observed in Const-Neut is lower than that found in most human studies, it follows a similar pattern to that in African populations, which generally show the lowest levels of LD compared to a range of ethnic groups (McEvoy et al. 2011, supplementary figure 5 and figure 6B). Most human demography studies have found that there has been recent rapid expansion to the present-day Ne and, with the possible exception of African populations, this was preceded by a bottleneck 2000–4000 generations ago (e.g., Schaffner et al. 2005; Li and Durbin 2011).
In an expanding population, there would be a higher proportion of very rare recent mutations compared to those in a constant-sized population and this would exaggerate the differences observed in our constant-sized population. In a European and Asian human demography model (ancestrally large and then a bottleneck followed by rapid recent expansion) the past bottleneck would increase LD and average MAF but this would then be partly reversed by the rapid recent expansion in Ne (Simons et al. 2014). Therefore, although the nucleotide diversity and allele frequency distribution in Const-Neut are similar to those reported for humans (International HapMap Consortium 2007), it is possible that in some human populations there is a larger proportion of very rare recent variants compared to our constant-sized Ne. However, a recent study concluded that for many complex human diseases, rare alleles account for only a small proportion of the total genetic variation and therefore recent population growth will have little impact on these traits (Simons et al. 2014).
We therefore argue that our results give a reasonable indication of trends that would be expected in human and some outbred plant and livestock populations. Furthermore, our selection model is of interest because there is recent empirical evidence that genome-wide nonsynonymous mutations are more often at lower derived allele frequencies than neutral mutations, indicating that these variants often result in deleterious fitness effects (1000 Genomes Project Consortium 2012; Tennessen et al. 2012; Simons et al. 2014).
Genetic variance explained by SNPs and sequence variants
The genetic variance explained by the HD SNPs in the constant population is less than the total genetic variance (Table 1) presumably because the causal variants are often rare and hence in incomplete LD with the SNPs that have MAF > 0.1. This interpretation is supported by the fact that the variance explained is even lower in the Const-Sel population (where causal variants have even lower MAF) than the Const-Neut population and by the fact that the full genetic variance is explained by the sequence data that include the causal variants. In recent studies of human height and schizophrenia, there was an even higher proportion of the genetic variance missing than found here (Yang et al. 2010; Lee et al. 2012). If the “missing heritability” is due to low MAF variants and not to other factors such as overestimation of heritability using pedigree records, this suggests that causal variants for these traits may have lower MAF than those in our Const-Sel population.
The underestimate of the genetic variance in the bovine demography using sequence data in a trait with h2 = 0.45 is harder to explain. It may occur because the GRM calculated from the sequence variants assumes that low MAF variants each cause as much genetic variance as high MAF variants, whereas in the simulated data the high MAF variants cause more genetic variance than low MAF variants. We reanalyzed Bov-Neut data (h2 = 0.45), using an alternative method of constructing the GRM that allows higher MAF variants to cause more variance (VanRaden 2008, method 1). In this case the genetic variance was closer to the true variance for SEQ, but with a tendency to be overestimated in both SEQ and HD SNPs (estimated h2 of 0.464 and 0.470, respectively). The prediction accuracies did not change.
Demography, method of analysis, and genetic architecture of the trait
The higher GBLUP accuracy in the bovine compared to the constant Ne population is due to the recent sharp reduction in the bovine Ne, resulting in a higher level of LD and a lower number of effectively independent chromosome segments (Me). The GBLUP model estimates a SNP effect from a single normal distribution on each effectively independent chromosome segment. It is therefore an appropriate method when the effects of chromosome segments follow a normal distribution (such as the bovine demography) when the number of QTL is very similar to the number of effectively independent chromosome segments (QTL = 50 and Me = 59).
Previous studies have demonstrated that Bayesian methods similar to BayesR will show higher accuracies than GBLUP only when chromosome segment effects are not normally distributed (e.g., Daetwyler et al. 2010; Meuwissen and Goddard 2010; Clark et al. 2011). This can occur either because a large proportion of the effectively independent segments contain no QTL or the distribution of QTL effects is markedly nonnormal. BayesR, by using a mixture of four normal distributions, one of which has a zero variance, is able to more accurately estimate the SNP effects than GBLUP in the constant Ne population where many chromosome segments have no effect (Me = 1786). In this scenario the prior distribution assumed by BayesR is a closer approximation to the simulated distribution than the prior implied by GBLUP. This is the reason that BayesR has an advantage over GBLUP in the bovine demography when the number of QTL is only 15 (Figure 5).
In real single-breed cattle populations with SNP densities equivalent to our HD or MD SNPs there is often little difference between accuracies for GP with GBLUP or Bayesian methods (Habier et al. 2010; Erbe et al. 2012; Pryce et al. 2012; Su et al. 2012; Gao et al. 2013). The main exception has been for traits that are known to be affected by one or several genes of large effect and an unknown number of very much smaller effects, where a Bayesian approach was more accurate (Hayes et al. 2010). This implies that the GBLUP infinitesimal model generally provides a good approximation of real QTL effects for complex traits in a single cattle breed because of the high LD.
Clearly, the accuracy of the Bayesian analysis in our bovine population is quite sensitive to the density of QTL across the genome and the distribution of their effects. We chose a QTL density of 50 QTL per 50 Mb because this is equivalent to ∼3000 QTL effects on a 30-Mb genome and is of a similar order for recent estimates of the number of QTL affecting human height and complex diseases (Park et al. 2011; Stahl et al. 2012). Our scenario of 15 QTL is approximately equivalent to 900 QTL genome-wide, but if far fewer QTL explain most of the trait variance, then we would expect clear differences between the BayesR and GBLUP accuracies for a bovine population.
For this study we chose to simulate QTL effects from a single normal distribution although there is evidence that effects may follow a more leptokurtic distribution (e.g., Hayes and Goddard 2001; Flint and Mackay 2009; Park et al. 2011). Our approach avoids the issue of odd results from some simulations where one or a few very large QTL are segregating, particularly given our small genome. Previous studies have demonstrated that the use of a gamma (shape parameter <1) or double exponential distribution will generally result in higher Bayesian accuracies compared to normally distributed effects, but that GBLUP is little affected by the distributions used (Daetwyler et al. 2010; Clark et al. 2011). Thus, had we used for example a gamma distribution of QTL effects, some of the observed differences between GBLUP and BayesR might have been larger where the number of QTL was considerably lower than the Me, but overall our conclusions would not change. Clark et al. (2011) found a clearer advantage compared to our study for SEQ over MD SNPs with a Bayesian analysis of a bovine-like simulation and this is probably largely due to their QTL effects being simulated as a double exponential distribution.
Long-term negative selection model
Our study focused on modeling long-term negative selection because there has been considerable focus recently on the so-called missing heritability of polygenic traits (i.e., a proportion of the genetic variance not accounted for by dense SNP marker associations). This phenomenon could be due to a significant proportion of rare causal variants that are in very low LD with the generally common SNPs used on commercial SNP arrays (Yang et al. 2010). A high proportion of rare variants affecting a trait may be due to large or recently expanding Ne and/or long-term selection pressure, and evidence to date suggests that negative selection is far more common than positive or balancing selection (reviewed by Eyre-Walker and Keightley 2007; Boyko et al. 2008).
In Const-Sel the BayesR accuracy with HD SNPs was clearly reduced by negative selection because causal variants were mainly at very low frequency (Figure 1B), resulting in lower LD with HD SNPs. To maintain a minimum r2 of 0.5 between SNPs and causal variants there must be no more than 0.15 difference between their respective allele frequencies (Wray 2005). This difficulty was overcome by SEQ because the causal variants were present in the data. Even in Const-Neut the causal variants were at often at lower MAF than the HD SNPs due to the large Ne, and therefore there was also a large advantage for SEQ accuracies in Const-Neut. In a simulation study, Meuwissen and Goddard (2010) found that removing only causal mutations from sequence data reduced accuracy by 2.5–3.7% with a Bayesian analysis, confirming the importance of having causal variants (or markers in complete LD with them) included in the data.
Conversely, accuracy in Bov-Sel was less affected by long-term negative selection because the sharp reduction in the recent Ne allows many of the loci under the influence of selection to drift to relatively high frequencies, while others were purged more rapidly than in the large constant Ne. The allele frequency distribution of causal variants in Bov-Sel is therefore quite similar to that of the neutral SNPs and often one or more HD SNPs (with MAF > 0.1) are likely be in good LD with a causal variant. Therefore, even with BayesR, the high LD in the bovine population will still tend to spread the effect of the causal variant across a number of SNPs in very high LD with the causal variant. Druet et al. (2014) found a marked improvement in accuracy from use of sequence compared to MD SNPs even though they used a very similar bovine demography to ours. However, they simulated QTL only on rare variants as an indirect means of modeling long-term negative selection, while our results indicate that random drift is likely to prevent such an extreme distribution of low-frequency causal variants.
In a recent study of genomic prediction in Drosophila, Ober et al. (2012) found no improvement from using either a Bayesian or a GBLUP analysis with “sequence data” compared to much less dense markers. This seems surprising because the Ne of Drosophila is believed to be very large. However, the authors had first discarded all rare variants from their sequence because they had a very small reference population (<200). It is plausible that this approach discarded many rare causal variants because the traits analyzed were fitness traits and could explain why there was no improvement under their Bayesian analysis. However, as suggested by the authors, it could equally be a result of the highly polygenic nature of their traits saturating the effective number of chromosome segments so that the Bayesian analysis had no advantage over GBLUP (Ober et al. 2012).
The real world is more complex than our simple model of negative selection, so would this affect our conclusions? For example, domestic animals and plants have in recent time been subjected to positive selection. A recent study found no clear selection signatures related to complex economic traits in cattle (Kemper et al. 2014), and the authors postulate that this is in part due to the highly polygenic nature of these traits and pleiotropic effects with opposing fitness costs. The size and direction of our QTL effects were independent of the selection coefficient as for a pleiotropic model, which is also consistent with evidence from some human disease susceptibility loci (Park et al. 2011). Although the reality for many traits is likely to lie somewhere between this and complete dependence, even a moderate correlation between QTL effect and selection coefficient would result in little change in the contribution of rare alleles to the genetic variance (Simons et al. 2014). Therefore it is unlikely that accounting for recent positive selection or for a moderate correlation between s (the selection coefficient) and the QTL effect would change our conclusions, particularly in species such as cattle that have undergone a sharp reduction in recent Ne, which in itself causes a major reduction in the proportion of rare deleterious mutations segregating due to random drift.
The critical parameter in determining the strength of selection in any population is the absolute value of Nes, where s is the selection coefficient. When |Nes| < 1, the mutation will remain effectively neutral and conversely, if |Nes| > 100, the mutation is likely to be lost very rapidly. Therefore, our selection coefficient was chosen to be of similar magnitude to moderate fitness cost estimates in humans (reviewed by Eyre-Walker and Keightley 2007): |Nes| ≈ 5 in the constant-size population and |Nes| ≈ 12 in the most ancestral bovine population, lowering to near neutral in very recent time as Ne is reduced. A larger selection coefficient would therefore have accentuated the differences between Const-Sel and Const-Neut (and vice versa). In the bovine simulation we would expect a more subtle effect from a stronger s because of the moderating influence of random drift in the small recent Ne. We applied a constant selection coefficient, while the true distribution of fitness effects is believed to be closest to a gamma or mixture model (Boyko et al. 2008; Keightley and Halligan 2009). However, this is a reasonable simplification because modeling a mixture distribution is likely to give similar but more variable results where mutations with higher s would be more rapidly lost, while many variants with weaker s would be effectively neutral. It is interesting to note that the loci under negative selection in Const-Sel reflected a similar allele frequency distribution to that found in a study of human exome data, in which variants are more likely to be influenced by negative selection (Tennessen et al. 2012).
Accuracy 10 generations after the estimation of SNP effects
The accuracy of GP is often evaluated by a cross-validation design in which the data set is randomly divided into a reference set and a validation set. This is likely to result in a closer relationship between reference and validation populations than between the reference population and the population where the GP is to be applied in the real world, which may be separated in space or time from the reference population. We have attempted to model this by assessing the accuracy of GP 10 generations after the reference population.
GBLUP predicts the effect of each QTL by using a linear combination of many SNPs that are in LD with the QTL, spread over a wide genome region. Over 10 generations this LD is eroded by recombination, causing a marked decline in the accuracy of prediction. BayesR makes more use of causal variants or markers close to them and so the accuracy of prediction is eroded more slowly or not at all in the case of SEQ in the constant Ne population. In the bovine demography, the LD is so extensive that BayesR may still assign an effect to markers a long distance from the QTL and therefore recombination still reduces the accuracy after 10 generations. In fact, the best accuracy in the constant Ne population after 10 generations was higher than the best accuracy in the bovine population after 10 generations (a reversal of generation 0 results). This is presumably because the lower LD in the constant population helps BayesR to find the causal mutations in the sequence data and thus make predictions that are stable over generations. This is an important result because it demonstrates a potential economic advantage for BayesR, because phenotypes would need to be measured less often when prediction equations are more stable over time. We did observe an advantage for BayesR after 10 generations in the bovine scenario with 15 QTL even though there was little difference at 0 generations. This might explain why published studies with real cattle data find little difference between Bayesian and GBLUP accuracies because their reference and validation animals are generally more closely related than in our 10_Gen scenario.
Deterministic accuracy of GP
Past studies demonstrate that the deterministically derived accuracy of GP provides a reasonable match with observed GBLUP accuracies in simulated data when the assumption of a constant population size holds (Daetwyler et al. 2008, 2010; Goddard 2009; Goddard et al. 2011). However, with a more complex demography it is not clear what the appropriate Ne is to estimate Me. For example, when we used our true bovine present-day Ne of 90 to estimate Me for Equation 1, the deterministic prediction of accuracy was much higher than our realized accuracy because the expected r2 in a constant Ne of 90 would be much higher than for our more complex demography. Our empirically calculated Me resulted in good a priori estimates of the realized accuracy. We estimated that the equivalent constant-sized Ne that would give rise to this Me would be ≈330 (applying the analytical formula of Goddard et al. 2011). The deterministic formula provides a lower limit for the BayesR accuracy but the realized accuracy for BayesR will depend on the actual number of QTL and the distribution of their effects as well as the Me. This is hard to predict a priori without knowing the genetic architecture underlying a specific trait. A priori calculation of the expected accuracy of GP is useful in planning for effective use of resources: this is particularly important when considering use of sequence data, given the extra costs involved in generating, storing, and analyzing the data.
Current limitations for the application of sequence data
It is important to point out that current sequencing technology is not perfect and this represents a real limitation not considered in our simulation: for example, there are base-calling errors, and causal deletions/insertions may not be correctly mapped. These errors mean that some causal variants may still not be captured in sequence data and random errors will add noise. Furthermore, these errors will also reduce imputation accuracies and accurate imputation of rare variants is difficult until large banks of genome sequences are available for a range of populations. This will erode part of the predicted improvement from using sequence data, but this field is rapidly evolving with much emphasis on improving both sequencing quality and cost as well as imputation accuracy. A further important practical difficulty of implementing genomic prediction with whole-genome sequence is the enormous number of genotypes and training individuals that need to be processed; therefore it will be important to develop more computationally efficient approaches.
Conclusion
The accuracy of GP depends on two factors (Goddard 2009): (1) the proportion of the genetic variance explained by the SNP and (2) the accuracy with which the SNP effects are estimated.
The first factor depends on the MAF of QTL, the density of SNPs, and Me. If QTL have low MAF, SNPs have low density and Me is large, then the LD between SNPs and QTL is reduced and the SNPs fail to explain all the genetic variance. In human complex traits half the genetic variance is often missing using HD SNPs and this will severely limit the accuracy of GP based on SNPs with MAF > 0.1. Our simulation suggests that the missing genetic variance could be recovered by the use of sequence data, provided the current limitations of sequence data, imputation, and computational efficiency can be surmounted.
The second factor, the accuracy with which SNP effects are estimated, depends on an interaction between all the variables considered in this study. When the data are analyzed by GBLUP, the accuracy depends on NTh2/Me but is unaffected by the genetic architecture of the trait. Although GBLUP may have high accuracy in the generation used for reference, the accuracy drops sharply when tested 10 generations later because recombination breaks up the chromosome segments whose effects GBLUP estimated. BayesR has an advantage when the number of causal variants <<Me and when the distribution of QTL effects is far from normal. BayesR is then able to more accurately estimate effects than GBLUP, using HD SNP or sequence data, and accuracy persists better than under GBLUP when more generations separate the reference and validation populations.
Evidence from our study indicates that in populations with low LD and large Ne, such as some human and outbred plant populations, there is likely to be a significant advantage in using sequence data, but only with a Bayesian analysis. This advantage could be even greater if mutations affecting the complex trait have been under the influence of long-term negative selection. An example would be for risk prediction of complex diseases in humans, such as diabetes, where there is evidence of long-term negative selection operating on causal mutations (Park et al. 2010). Furthermore the persistency of accuracy with sequence even after 10 generations separated the validation and reference population indicates that the predictions of risk would be more robust across space and time than when using HD SNP data or analysis by GBLUP.
In domestic livestock, such as within a breed of cattle, where the Ne has been reducing from ancestrally large to very small, sequence data and Bayesian analysis have an advantage over HD SNPs and GBLUP only for some traits. However, the predictions are not robust and decay in accuracy with time. This could be overcome by continually retraining the prediction model but this may involve the expense of continually collecting more phenotypes. As an alternative, we suggest combining sequence data from two or more breeds of cattle to reduce long-range LD. This, together with a Bayesian analytical approach, should improve the robustness of the prediction. To compensate for reduced LD in the mixed-breed population, it will also be necessary to increase the size of the reference population.
Data availability
The scripts and FREGENE parameter files used to generate simulated sequence data for each of the four scenarios are given in File S1, File S2, File S3, and File S4. Due to the large size and number of data sets generated for this study, the data have not been deposited in a public repository, but the authors are willing to share data files on request.
Supplementary Material
Acknowledgments
We thank the reviewers for suggestions to improve this manuscript. I.M.M. acknowledges partial funding of this study from the Dairy Futures Cooperative Research Centre, Australia.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.168344/-/DC1.
Communicating editor: E. A. Stone
Literature Cited
- Arias J., Keehan M., Fisher P., Coppieters W., Spelman R., 2009. A high density linkage map of the bovine genome. BMC Genet. 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine Genome Sequencing and Analysis Consortium , C. G. Elsik, R. L. Tellam, and K. C. Worley, 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324: 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine HapMap Consortium , 2009. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. D., Chong J. X., Malig M., Ko A., Dumont B. L., et al. , 2012. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 44: 1277–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadeau-Hyam M., Hoggart C., O’Reilly P., Whittaker J., De Iorio M., et al. , 2008. Fregene: simulation of realistic sequence-level data in populations and ascertained samples. BMC Bioinformatics 9: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S., Hickey J., van der Werf J., 2011. Different models of genetic variation and their effect on genomic evaluation. Genet. Sel. Evol. 43: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Villanueva B., Woolliams J. A., 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3: e3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Capitan A., Pausch H., P. Slothard, R. van Binsbergenet al. , 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46: 858–865. [DOI] [PubMed] [Google Scholar]
- De Los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. [DOI] [PubMed] [Google Scholar]
- Druet T., Macleod I. M., Hayes B. J., 2014. Toward genomic prediction from whole-genome sequence data: impact of sequencing design on genotype imputation and accuracy of predictions. Heredity 112: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbe M., Hayes B. J., Matukumalli L. K., Goswami S., Bowman P. J., et al. , 2012. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 95: 4114–4129. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Addison-Wesley Longman, Harlow, Essex, UK. [Google Scholar]
- Flint J., Mackay T. F. C., 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Lund M. S., Zhang Y., Su G., 2013. Accuracy of genomic prediction using different models and response variables in the Nordic Red cattle population. J. Anim. Breed. Genet. 130: 333–340. [DOI] [PubMed] [Google Scholar]
- Gilmour, A. R., B. R. Cullis, B. J. Gogel, S. J. Welham, and R. Thompson, 2005 ASReml User Guide Release 2.0. VSN International, Hemel Hempstead, UK. [Google Scholar]
- Goddard M., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–257. [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., Meuwissen T. H. E., 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 128: 409–421. [DOI] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F.-R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B., Goddard M. E., 2001. The distribution of the effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 33: 209–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E., 2009. Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92: 433–443. [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6: e1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. R., 1984. Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, Ontario, Canada. [Google Scholar]
- Hoggart C. J., Chadeau-Hyam M., Clark T. G., Lampariello R., Whittaker J. C., et al. , 2007. Sequence-level population simulations over large genomic regions. Genetics 177: 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Hap-Map Consortium , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K., Saxton S., Bolormaa S., Hayes B. J., Goddard M. E., 2014. Selection for complex traits leaves little or no classic signatures of selection. BMC Genomics 15: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P., Halligan D., 2009. Analysis and implications of mutational variation. Genetica 136: 359–369. [DOI] [PubMed] [Google Scholar]
- Kumar S., Subramanian S., 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA 99: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., DeCandia T. R., Ripke S., Yang J., Sullivan P. F., et al. , 2012. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 44: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M., de Roos A., de Vries A., Druet T., Ducrocq V., et al. , 2011. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 43: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod I. M., Larkin D. M., Lewin H. A., Hayes B. J., Goddard M. E., 2013. Inferring demography from runs of homozygosity in whole-genome sequence, with correction for sequence errors. Mol. Biol. Evol. 30: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth G. T., Czabarka E., Murvai J., Sherry S. T., 2004. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics 166: 351–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy B. P., Powell J. E., Goddard M. E., Visscher P. M., 2011. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 21: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T., Goddard M., 2010. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale D. B., Savolainen O., 2004. Association genetics of complex traits in conifers. Trends Plant Sci. 9: 325–330. [DOI] [PubMed] [Google Scholar]
- Ober U., Ayroles J. F., Stone E. A., Richards S., Zhu D., et al. , 2012. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 8: e1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, 2012 An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed]
- Park J.-H., Wacholder S., Gail M. H., Peters U., Jacobs K. B., et al. , 2010. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-H., Gail M. H., Weinberg C. R., Carroll R. J., Chung C. C., et al. , 2011. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc. Natl. Acad. Sci. USA 108: 18026–18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J. E., Daetwyler H. D., 2012. Designing dairy cattle breeding schemes under genomic selection: a review of international research. Anim. Prod. Sci. 52: 107–114. [Google Scholar]
- Pryce J. E., Arias J., Bowman P. J., Davis S. R., Macdonald K. A., et al. , 2012. Accuracy of genomic predictions of residual feed intake and 250-day body weight in growing heifers using 625,000 single nucleotide polymorphism markers. J. Dairy Sci. 95: 2108–2119. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Morgante M., 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Roach J. C., Glusman G., Smit A. F. A., Huff C. D., Hubley R., et al. , 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328: 636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S. F., Foo C., Gabriel S., Reich D., Daly M. J., et al. , 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15: 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y. B., Turchin M. C., Pritchard J. K., Sella G., 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E. A., Wegmann D., Trynka G., Gutierrez-Achury J., Do R., et al. , 2012. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., R. F. Brøndum, P. Ma, B. Guldbrandtsen, G. P. Aamand et al, 2012. Comparison of genomic predictions using medium-density (∼54,000) and high-density (∼777,000) single nucleotide polymorphism marker panels in Nordic Holstein and Red Dairy Cattle populations. J. Dairy Sci. 95: 4657–4665. [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., W. Fu, E. E. Kennyet al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P., O’Connell J., Wiggans G., Weigel K., 2011. Genomic evaluations with many more genotypes. Genet. Sel. Evol. 43: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., et al. , 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y., Jaqueth J. S., Matsuoka Y., Smith O. S., Beavis W. D., et al. , 2002. Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 19: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Villa-Angulo R., Matukumalli L., Gill C., Choi J., Van Tassell C., et al. , 2009. High-resolution haplotype block structure in the cattle genome. BMC Genet. 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Adams A. M., Frisse L. A., Qian Y., Hudson R. R., et al. , 2005. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA 102: 18508–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N. R., 2005. Allele frequencies and the r2 measure of linkage disequilibrium: impact on design and interpretation of association studies. Twin Res. Hum. Genet. 8: 87–94. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scripts and FREGENE parameter files used to generate simulated sequence data for each of the four scenarios are given in File S1, File S2, File S3, and File S4. Due to the large size and number of data sets generated for this study, the data have not been deposited in a public repository, but the authors are willing to share data files on request.