Abstract
Evolutionary theory predicts that genetic constraints should be widespread, but empirical support for their existence is surprisingly rare. Commonly applied univariate and bivariate approaches to detecting genetic constraints can underestimate their prevalence, with important aspects potentially tractable only within a multivariate framework. However, multivariate genetic analyses of data from natural populations are challenging because of modest sample sizes, incomplete pedigrees, and missing data. Here we present results from a study of a comprehensive set of life history traits (juvenile survival, age at first breeding, annual fecundity, and longevity) for both males and females in a wild, pedigreed, population of red deer (Cervus elaphus). We use factor analytic modeling of the genetic variance–covariance matrix (G) to reduce the dimensionality of the problem and take a multivariate approach to estimating genetic constraints. We consider a range of metrics designed to assess the effect of G on the deflection of a predicted response to selection away from the direction of fastest adaptation and on the evolvability of the traits. We found limited support for genetic constraint through genetic covariances between traits, both within sex and between sexes. We discuss these results with respect to other recent findings and to the problems of estimating these parameters for natural populations.
Keywords: genetic correlations, life history trade-off, heritability, sexual antagonism, selection
EVOLUTIONARY theory predicts low equilibrium genetic variation for fitness and fitness-related traits, because alleles that have negative effects on fitness should have been removed by selection, whereas those with positive effects should have reached fixation (Fisher 1958; Falconer 1981; Charlesworth 1987). The observation of strong selection on, and yet the persistence of genetic variation in, fitness-related traits when examined in isolation has led to the extension of this theory to multiple traits and the expectation of multivariate genetic constraints (Walsh and Blows 2009), where the majority of genetic variation segregating within a population should be the result of genes that have opposing effects on fitness through their effects on different traits (Falconer 1981; Lande 1982; Houle 1991; Roff 1996). Extension of theory to multiple traits has led to the prediction that, at equilibrium, further evolutionary change in traits under strong selection should be constrained or prohibited by genetic trade-offs—effectively, a lack of genetic variation in the direction of selection (Blows and Walsh 2009). However, despite their intuitive theoretical appeal, empirical support for these concepts is surprisingly scarce (Walsh and Blows 2009), especially for wild populations experiencing natural environments (Kruuk et al. 2008). Here, we use a multivariate framework to explore the role of genetic associations between life history traits in a wild population of red deer (Cervus elaphus) on the Isle of Rum, Northwest Scotland, and to consider in particular the relationship between traits expressed in either sex.
By far the most common approach to studying genetic trade-offs and genetic constraints is to estimate the bivariate genetic correlation between two traits (Blows and Hoffmann 2005; Walsh and Blows 2009). In particular, there has been a focus on the search for genetic correlations approaching −1 (or +1 if traits are selected in opposite directions) as these would represent an absolute constraint to bivariate trait evolution. However, given the inherently multivariate nature of selection and phenotypic variation, the focus on bivariate correlations may give a misleading impression of the extent of genetic constraints and in particular may lead to an underestimate of their importance (Dickerson 1955; Pease and Bull 1988). Indeed a mixture of positive and negative genetic correlations of intermediate magnitude can still result in limited or no genetic variation in the direction of selection when considering more than two traits (Walsh and Blows 2009). The focus on bivariate genetic correlations also ignores the importance of genetic variances as a potential source of genetic constraint (Agrawal and Stinchcombe 2009; Mcguigan and Blows 2010). Ultimately the degree to which multiple traits respond to selection is determined both by the distribution of genetic variances across those traits and by the genetic covariances among them, which jointly determine the amount of genetic variation that exists in the direction of selection (Lande 1979; Blows 2007). As such, it has been suggested that bivariate correlations are a poor indicator of genetic constraint and a more multivariate approach has been advocated (Dickerson 1955; Pease and Bull 1988; Walsh and Blows 2009; Houle et al. 2011).
As well as focusing on bivariate genetic correlations, the majority of previous studies have also focused on genetic correlations among traits expressed in the same sex as a potential cause of constraint (Roff 1996; Kruuk et al. 2008; Poissant et al. 2010). However, between-sex genetic correlations may also be important. For example, it has been hypothesized that because males and females often differ greatly in their reproductive roles and thus selective optima for different traits (Cox and Calsbeek 2009) and yet share the majority of their genome, there is the potential for sexually antagonistic genetic variation to exist, whereby genes that are beneficial to one sex are detrimental to the other (Lande 1980; Rice 1984; Bonduriansky and Chenoweth 2009). Evidence in support of sexually antagonistic genetic variation is accumulating from both laboratory (Chippindale et al. 2001; Fedorka and Mousseau 2004; Lewis et al. 2011) and natural populations (Brommer et al. 2007; Foerster et al. 2007; Mainguy et al. 2009; Cox and Calsbeek 2010).
Our aim in this study was to use multivariate techniques to assess the potential for genetic constraints to the evolution of four life history traits in a wild population of red deer (C. elaphus) on the Isle of Rum, Scotland. Previous studies in this population have shown genetic variation for numerous traits (Kruuk et al. 2000; Wilson et al. 2007; Nussey et al. 2008; Clements et al. 2011) and also, in line with theoretical predictions, that the heritability of traits decreases with increasing association with fitness [i.e., increasing strength of selection (Kruuk et al. 2000)]. Life history traits in the Rum red deer population have lower heritabilities than morphological traits, but this is largely due to an increase in environmental variance for these traits (Kruuk et al. 2000). There is also evidence of sexually antagonistic genetic variation in the population (Foerster et al. 2007), with negative genetic correlations between estimates of male and female fitness, but the strength of this evidence differs slightly, depending on the measure of fitness used (see Foerster et al. 2007 and the associated supplementary information). More recently, Morrissey et al. (2012b) used a multivariate method proposed by Agrawal and Stinchcombe (2009) to provide evidence for genetic constraint through antagonistic correlations between female adult survival and female reproductive traits.
Here we extend the multivariate analysis of genetic constraint in the Rum red deer population to include both males and females and to study the effect of both genetic variances and within- and between-sex genetic covariances in generating constraint. We consider four life history traits, which together form a comprehensive set of all life history traits that determine individual fitness: survival to breeding age, age at first reproduction, longevity, and annual reproductive success. Our aims were split into two parts: (1) to quantify the genetic variance–covariance matrix (G) for females, males, and both sexes, with particular focus on characterizing the major multivariate axes of variation; and (2) to assess the degree of constraint imposed by the structure of G relative to the direction of selection using, first, estimates of the angle between the vector of selection and the vector of the predicted response (Smith and Rausher 2008) (“deflection” of the predicted response, θ) and, second, the length that the vector of the predicted response travels in the direction of selection [“evolvability,” e(β) (Hansen and Houle 2008)]. Part 2 necessarily involves characterization of the phenotypic process of selection on the different life history traits, which we undertake using a regression-based approach to estimate selection gradients. Although such estimates may not lead to robust predictions of evolutionary responses in situations when other, excluded, traits contribute to associations between trait and fitness (Rausher 1992; Morrissey et al. 2010), in an analysis of selection of complete life histories, all pathways by which effects of multivariate phenotype influence fitness are represented, because life history completely determines fitness. Thus there is by definition no unaccounted-for trait–fitness covariance in an analysis of complete life histories (we return to this point in the Discussion). Here, in particular, we assess estimates of deflection (θ) and evolvability [e(β)] when genetic covariances are fixed to zero vs. not zero, to assess the importance of genetic variance vs. covariances in generating any constraint.
Materials and Methods
General information
Study population:
We used individual life history information from red deer born between 1971 and 2007 in the study population in the North Block of the Isle of Rum, Inner Hebrides, Scotland (57° 03′ N, 06° 21′ W) (Clutton-Brock 1982). Individuals are recognizable from natural markings or artificial tags and data on life history traits are collected in weekly censuses conducted throughout the year and more intensive daily surveys during calving (May to July, when ∼50% of females give birth to a single calf) and mating (September to November) seasons (for details on data collection see Clutton-Brock 1982; Kruuk et al. 2002). Since 1982, ∼70% of calves have been caught soon after birth and artificially marked and an ear punch taken for genetic analysis. Other individuals have been sampled postmortem, from cast antlers, or by immobilization. All sampled individuals are genotyped at up to 15 microsatellite loci (Walling et al. 2010).
Pedigree determination:
Maternity was assigned with certainty based on observed associations between mothers and calves (Pemberton et al. 1988; Walling et al. 2010). Paternity was inferred from a combination of phenotypic and behavioral data (male age and the length of time a male held a female in his harem during her estrus window) and genetic data, using the paternity inference programs MasterBayes (Hadfield et al. 2006) and COLONY2 (Wang 2004; Wang and Santure 2009). Individual paternity assignments were accepted when their individual-level confidence was ≥80% (giving an average confidence in assignments of >98%). For paternal links, “dummy” sires were created to represent half-sibships between groups of offspring identified by COLONY to have a common (unsampled) father. For full details on paternity inference see Walling et al. (2010).
Life history traits studied:
We analyzed within- and between-sex variances and covariances in four life history traits that represent the major components of fitness in this population, measured separately in each sex to give a total of eight traits. The traits were as follows:
Survival to breeding age (SBA): Defined as 0 for all animals known to have died before and 1 for all animals known to have survived to May 1 in the year in which they reached the age of 3 years. Individuals whose fate was unknown or who died as a result of being shot outside the study area were removed from all analyses, resulting in a sample size of 1126 females (born to 462 mothers) and 1114 males (born to 437 mothers).
Age at first reproduction (AFR): The age in years at which a female first produced a calf or at which a male was first assigned paternity (N = 519 females and 149 males; many more males than females fail to breed). Because first breeding at an early age necessarily has a positive direct effect on fitness, AFR was multiplied by −1 in all multivariate analyses so that trade-offs would be represented by negative covariances and correlations.
Adult longevity (L): For both sexes, the age in years at death for individuals that survived to at least 3 years of age (i.e., SBA = 1). Individuals that emigrated from the study area and thus for whom age at death was unknown were removed from the data set, as were individuals that died as a result of being shot outside the study area (121 females and 172 males in total). For both sexes, data were limited to individuals that were born before 1995 because for cohorts born from 1995 onward <80% of individuals were dead. This resulted in longevity values for 338 females and 245 males.
Annual breeding success (ABS): Defined as a repeated measure for all adults of breeding age. Females received a 0/1 score, with 1 representing years in which they produced a calf and 0 for years in which they did not. Females were included only if they were ≥3 years old and survived to at least 6 years of age; for each female ABS was recorded for each year until her death (or until 2008 if still living), so that several possible breeding attempts were included. This gave 3859 records from 439 individual females. For males, ABS was defined as the number of calves to which a male was assigned paternity for any given year in which males were ≥3 years old during the mating season (rut) and also were seen in the study area during the rut. Males were assigned an ABS score of 0 for a given year if they were seen in the study area during the corresponding rut, but not assigned paternity of any calves born the following year. Calves born in calendar year x are sired in the rut of calendar year x − 1. Thus male ABS is assigned to the year in which the calves were born rather than sired so that estimates of year-to-year variation in ABS in either sex correspond to the same calves. Paternity was assigned as described above and gave a total of 2004 records of ABS from 570 individual males.
For analyses of selection on these traits (see below), we also required estimates of individuals’ lifetime breeding success, defined as the total number of offspring produced across a lifetime, restricted to individuals known to have died a natural death.
To prevent differences in scale causing particular life history traits to dominate loadings in the factor analytic models described below, all traits were standardized to unit phenotypic variance by dividing by their standard deviation. In addition, AFR and male ABS were square-root transformed prior to standardization to better approximate normality.
A total of 1188 females were measured for at least one of the four life history traits under consideration in this study. The informative pedigree for these traits consisted of 1327 maternal links (including mothers that lacked phenotypic information but were informative for relatedness) and 893 paternal links. For males a total of 1369 individuals were measured for at least one trait and the informative pedigree consisted of 1586 maternal links and 1077 paternal links (of which 141 were dummy sires). When combining data on both sexes, 2557 individuals were scored for at least one trait, with the informative pedigree consisting of 2368 maternal links and 1573 paternal links. For all analyses, the pedigree had a maximum depth of eight generations with a mode of three generations of information for each individual.
Part 1: Variance decomposition: Estimating G
Univariate analysis:
For each trait, variance components and appropriate random effects structures were initially investigated using univariate animal models of the general form
| (1) |
where y is a vector of phenotypic observations, μ is the mean, b is a vector of fixed effects, u is a vector of random effects, X and Z are design matrices linking individual records to the appropriate fixed and random effects, and e is a vector of residual errors.
Animal models are a form of linear mixed-effect model that use pedigree information to decompose phenotypic variance into components due to additive genetic and other effects (Henderson 1976; Kruuk 2004). The random effects fitted (and thus variance components estimated) differed between traits: additive genetic (VA), year of birth (VBY), and residual (VR) effects were modeled for all traits; maternal effects [VM—the influence of a mother’s phenotype on that of her offspring, independent of additive genetic effects (Kruuk and Hadfield 2007)] were modeled for early life traits SBA and AFR; and permanent environment [VPE—constant environmental influences on an individual’s phenotype across repeated measures on that individual (Kruuk and Hadfield 2007)] and year of measure (VYR) effects for ABS were modeled because of its repeated measures on individuals across multiple years. Birth year and year of measurement were included to test for between-cohort and between-year environmental variation, respectively, such as that due to population density and climate variables (Kruuk et al. 2002). The statistical significance of random effects was tested by comparing full models to models excluding specific random effects, using likelihood-ratio tests, with twice the difference in log-likelihood being χ2 distributed with 1 d.f. for every additional parameter fitted. Nonsignificant random effects, apart from additive genetic effects, were removed from final models. Fixed effects previously shown to be important in this system were also included and are detailed in Supporting Information, File S1.
Multivariate analysis
Phenotypic covariation:
To estimate within-sex phenotypic covariances among traits, we ran multivariate equivalents of the models represented by Equation 1, where y now represents a matrix of phenotypic observations of all traits measured within each sex and μ is a vector of means for each phenotypic trait. SBA was not included in these models because only individuals that score 1 for SBA can have a phenotypic value for any other trait and thus the phenotypic covariance between SBA and other traits is undefined. These models contained fixed effects as described in File S1, but only a single, individual-level, random effect defining individual-level variance (VI, equivalent to individual repeatability) for all traits. By fixing the residual variance for single-measures traits (AFR and L) to zero, this model structure allows the residual (after correcting for fixed effects) variance for these traits to be represented by the individual-level variance, allowing estimation of the phenotypic covariance between AFR, L, and the individual-level repeatability of ABS (Morrissey et al. 2012a). This does not imply that we can estimate the repeatable component of variation for traits (AFR and L) that are measured only once; rather, the model structure allows estimation of the biologically interesting phenotypic relationship between AFR, L, and the repeatable component of ABS, which is the phenotypic covariance between these traits (Morrissey et al. 2012a). Between-sex phenotypic covariances are undefined and thus set to zero, as no sex ever expresses the phenotype of the opposite sex.
Genetic (co)variation:
We estimated G matrices for females (Gf) and males (Gm) separately and then for both sexes together (Gbs). To do this, we again ran full multivariate equivalents of the model represented in Equation 1 for each sex and then both sexes combined, but this time included the additive genetic and all significant random effects identified in the univariate models (above). Covariances were estimated between all variance components where they were definable. As in models of phenotypic covariances, residual variances for singly measured traits (AFR and L) were fixed to zero, allowing estimation of the covariance between residual and nongenetic permanent environment variances of single- and repeated-measures traits, respectively. Nongenetic random effects were modeled as variance-correlation matrices with correlations constrained to be positive definite (i.e., bounded by ±1) as these proved more stable and gave parameter estimates that were within the realms of possibility (Gilmour et al. 2009). The significance of correlations was assessed using likelihood-ratio tests as above, comparing models where correlations were estimated to those where the correlation was fixed at 0.
To overcome issues associated with the large number of parameters to be estimated in a multivariate G matrix (see File S1) we used factor analytic modeling (factor analysis, FA) techniques (Wright 1932). We discuss these methods in detail in File S1, but give a brief overview here. We first constructed sex-specific models of the four life history traits in either sex and then considered both sexes together. Specifically we modeled the genetic variance–covariance matrix (G) as a product of a number m of independent linear combinations of the original (p) traits,
| (2) |
where is a (potentially reduced-rank) estimate of G, Λ is a lower triangle matrix of constants that represent loadings of each trait on each factor, and T is the transpose of a matrix. FA can be performed in ASReml (Thompson et al. 2003; Gilmour et al. 2009) and the significance of additional factors can be assessed by comparing the log-likelihoods of models with sequentially more (or fewer) factors. Twice the difference between the log-likelihoods of successive models was assumed to be chi-square distributed with d.f. equal to the change in d.f. between models in statistical hypothesis tests. A full-rank FA model, with Λ representing a lower triangle of a matrix of dimension equal to the number of traits (for Equation 2), provides a multivariate estimate of G, with identical values and associated likelihood to an unconstrained estimate.
Although FA has been used to assess the rank of G (e.g., Mezey and Houle 2005; Hine and Blows 2006), doing so may result in an underestimate of the rank of G (Hill and Thompson 1978; Meyer and Kirkpatrick 2008; see File S1 for more details). Here, we took an alternative approach of “building up” an FA model, adding additional factors until either G was full rank {rank Λ = p [four (within-sex models) or eight (both-sex models) in this case]} or it was not possible to add additional factors to a model (due to failure of convergence). FA modeling allows estimation of (i.e., ΛΛT) that contains the maximum possible variance estimable given the data and thus provides the best possible estimate of G with which to subsequently assess its potential to generate evolutionary constraint (see below). Because the leading factors to be estimated are those that contain the most variance, any unestimable factors in our analysis should explain considerably less variance than those that have already been estimated and would thus contribute much less to a predicted response to selection than those that are included.
Part 2: Assessing the potential for genetic constraint to evolution
The majority of methods for estimating multivariate genetic constraint center around Lande’s (1979) formulation of the multivariate breeders’ equation
| (3) |
where is a vector of predicted changes in trait means over a single generation, G is the multivariate genetic variance–covariance matrix, and β is the vector of directional selection gradients (Lande 1979; Lande and Arnold 1983). When considering traits expressed in each sex, this becomes
| (4) |
(Lande 1980), where Gf is the additive genetic (co)variance matrix for females, Gm is the additive genetic (co)variance matrix for males, B is the genetic covariance matrix between the sexes, T is the transpose of a matrix, βm is the vector of selection gradients for male traits, and βf is the vector of selection gradients for female traits (Lande 1980). The factor 1/2 is required because male and female parents make equal autosomal contributions to the offspring of both sexes. Equation 4 demonstrates that the predicted response to selection () is scaled and deflected away from the direction of maximal adaptation (i.e., the direction in which population mean fitness increases most rapidly as a function of phenotype), as defined by the vector of selection gradients (β), by G. The degree of genetic constraint can therefore be summarized by comparing the vector of the predicted response to selection () to the vector of selection itself (β). Comparison of vectors can be done in two related ways (details below); doing so first required calculation of selection gradients for the traits under study.
Calculating selection gradients (β)
We initially calculated selection differentials (S) for all traits in this study from the covariance between the trait and relative fitness (Lande and Arnold 1983). Traits were standardized as for genetic analyses and the same fixed-effects structures were fitted to the regression models as to the animal models above. Because individuals have to survive to breeding age (i.e., score SBA = 1) to score for all other traits (AFR, L, and ABS), we analyzed selection as a two-step process, analyzing selection on SBA separately from selection on AFR, L, and ABS. Relative fitness for selection on SBA was defined as the ratio of lifetime breeding success (the total number of offspring produced) of an individual to the sex-specific population mean lifetime breeding success of individuals with known SBA. Relative fitness for selection on all other traits (AFR, L, and ABS) was defined as the ratio of lifetime breeding success of an individual to the sex-specific population mean lifetime breeding success of individuals that survived to breeding age (i.e., had an SBA score of 1) and had a known phenotype for at least one of AFR, L, and ABS.
To calculate S for each trait, bivariate models of the form in Equation 1 were fitted with both relative fitness and the trait of interest (i.e., female SBA, male SBA, female AFR, etc.) as dependent variables. For the single-measures traits (SBA, AFR, and L), no random effects (other than residual) were fitted and the selection differentials were estimated as the phenotypic covariance between the standardized trait and relative fitness. For the repeated-measures trait ABS, the relationship with fitness was calculated by fitting a bivariate model of ABS and fitness with an individual-level term for both traits, similar to the models used above. The residual variance for fitness was then fixed at zero, forcing the residual variance for fitness to be represented by the individual-level term and thus allowing the estimation of the phenotypic covariance between relative fitness and the individual repeatability of ABS (similar to that above; see also Morrissey et al. 2012a). Sample sizes for these models were 757 for female SBA, 262 for female AFR, 278 for female L, 254 individuals with 2479 observations for female ABS, 723 for male SBA, 81 for male AFR, 121 for male L, and 127 individuals with 849 measures of ABS.
Sex-specific selection gradients (β) were then calculated from the equation
| (5) |
where β is a vector of selection gradients, P is the phenotypic variance–covariance matrix for each sex (calculated as above, Table S2), and S is a vector of selection differentials (Lande and Arnold 1983). The phenotypic covariance between SBA and all other traits is undefined because only individuals that score 1 for SBA can score for any other trait. Thus standardized selection gradients for SBA are equal to the selection differential. Results from this approach are similar to results removing SBA from all analyses (data not shown), apart from the specific case of female evolvability (see Discussion in File S1). We calculated three vectors of selection gradients: a vector of selection gradients on female traits (βf); a vector of selection gradients on male traits (βm); and then, by combining βf and βm, a vector of selection gradients for both female and male traits (βbs). Standard errors for S and P are estimated within ASReml.
Metrics of constraint
Deflection (θ), the angle between the predicted response to selection () and the selection gradient (β):
To assess the strength of genetic constraints to evolution we calculated the angle (θ) between the vector of selection (β) and the predicted response to selection () (Smith and Rausher 2008) as
| (6) |
θ provides an estimate of the degree to which G deflects evolutionary trajectories away from the direction of selection and is thus a representation of the degree of genetic constraint. The use of angles between vectors can be extended (Agrawal and Stinchcombe 2009) to assess the influence of genetic covariances on the response to selection by calculating the predicted response to selection (nc) when all covariances within G are set to zero (Gnc, where nc = no covariances). Different angles can then be calculated to assess the effect of various aspects of G on the predicted response to selection (), with larger angles suggesting an increase in constraint. We calculated four angles for within- and both-sex analyses: θ1, the angle between β(f, m, or bs) and (f, m, or bs), the combined effect of unequal genetic variances and nonzero covariances on deflection; θ2, the angle between β(f, m, and bs) and (f, m, and bs)nc, the extent to which unequal variances cause deflection; θ3, the angle between (f, m, or bs) and (f, m, and bs)nc, the effect of nonzero covariances on the direction of the predicted response to selection; and θ4, the difference between θ1 and θ2 (θ1 − θ2), the amount that genetic covariances alter deflection (positive values indicate covariances increase constraint).
For the both-sex analysis, we calculated three additional angles: θ5_bs, the angle between βbs and when just the between-sex covariances are set to zero (i.e., using Gnbs in Equation 4 to calculate nbs, where nbs = no between-sex covariances), which represents the combined effect of unequal genetic variances and within-sex covariances on deflection; θ6_bs, the angle between bs and nbs, the effect of nonzero between-sex covariances on the direction of the predicted response to selection; and θ7_bs, the difference between θ1_bs and θ5_bs (θ1_bs − θ5_bs), the amount that between-sex covariances alter deflection (positive values indicate increased constraint, see Table 2).
Table 2. Estimates of deflection (θ) for females, males, and both sexes.
| Parameter | Description | Angle (°) | 95% CI |
|---|---|---|---|
| Females | |||
| θ1_f | Angle between f and βf, effect of unequal variances and nonzero within-sex covariances | 17.6 | 9.46–50.8 |
| θ2_f | Angle between fnc and βf, effect of unequal variances | 12.6 | 2.66–46.9 |
| θ3_f | Angle between f and fnc, effect of within-sex covariances on the direction of the response to selection | 11.9 | 5.36–41.6 |
| θ4_f | θ1 − θ2, effect of within-sex covariances on constrainta | 5.06 | −4.36–33.2 |
| Males | |||
| θ1_m | Angle between m and βm, effect of unequal variances and nonzero within-sex covariances | 52.2 | 26.2–72.9 |
| θ2_m | Angle between mnc and βm, effect of unequal variances | 49.6 | 15.0–74.6 |
| θ3_m | Angle between m and mnc, effect of within-sex covariances on the direction of the response to selection | 17.9 | 8.17–53.0 |
| θ4_m | θ1 − θ2, effect of within-sex covariances on constrainta | 2.57 | −13.8–31.2 |
| Both sexes | |||
| θ1_bs | Angle between bs and βbs, effect of unequal variances and nonzero within- and between-sex covariances | 34.9 | 24.5–62.5 |
| θ2_bs | Angle between bsnc and βbs, effect of unequal variances | 32.3 | 15.9–58.1 |
| θ3_bs | Angle between bs and bsnc, effect of within- and between-sex covariances on the direction of the response to selection | 24.9 | 16.3–47.5 |
| θ4_bs | θ1 − θ2, effect of nonzero within- and between-sex covariances on constrainta | 2.54 | −8.91–27.5 |
| θ5_bs | Angle between nbs and βbs, effect of unequal variances and nonzero within-sex covariances | 42.7 | 26.9–64.0 |
| θ6_bs | Angle between bs and nbs, effect of between-sex covariances on the direction of the response to selection | 14.6 | 8.43–33.3 |
| θ7_bs | θ1 – θ5, effect of nonzero between-sex covariances on constrainta | −7.80 | −13.0–9.73 |
Ninety-five percent credible intervals were calculated by simulation as described in Materials and Methods.
Positive values suggest covariances increase constraint.
Evolvability e(β):
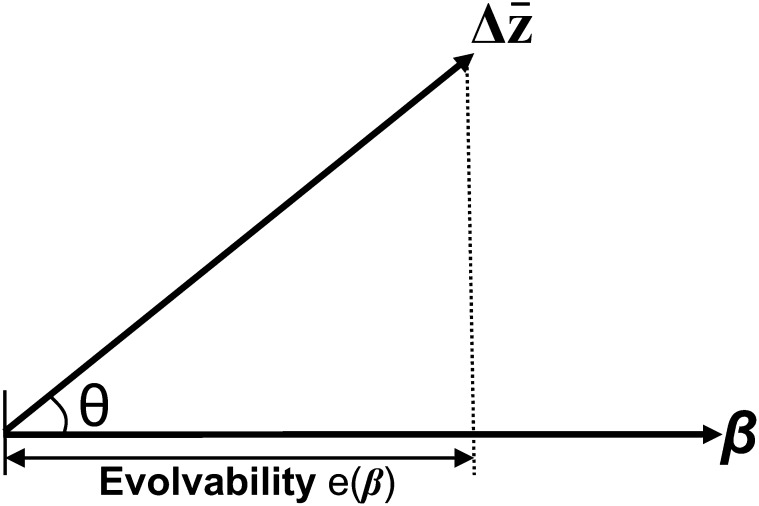
Although θ provides a good measure of the degree to which is deflected away from β by G, it does not take into account any effect of G on the magnitude of the responses. Thus another way of assessing constraint is to calculate Hansen and Houle’s (2008) evolvability metric [e(β)] (Figure 1) defined as
| (7) |
where β, G, and T are as defined above and || is the norm of the vector.
Figure 1.
Two-dimensional illustration of deflection (θ) and evolvability [e(β)], the measures of constraint. θ is the angle between the vector of selection (β) and the predicted response to selection (). Evolvability e(β) is the length of the projection of onto β, as a proportion of the length of β (Equation 7), and represents the magnitude of the predicted response to selection in the direction of selection; adapted from Hansen and Houle (2008).
Evolvability corresponds to the length of the projection of onto β and thus describes the length of the response in the direction of selection (Figure 1) and is standardized by the strength of selection (i.e., is given as a proportion of the length (norm) of the vector of selection (|β|2 in Equation 7). It summarizes the effect of G on in terms of both deflecting away from the direction of β and adjusting the magnitude of the response. For comparison with evolvability, a useful benchmark is the average evolvability (ē) over random selection gradients, defined as
| (8) |
where λi are eigenvalues of G and the sum is over all k eignevalues (Hansen and Houle 2008). This is equivalent to the average additive genetic variance of the traits and thus provides a measure of the evolutionary potential of G independent of the strength and direction of selection (Hansen and Houle 2008; Innocenti and Chenoweth 2013). Calculating this average evolvability over random selection gradients for females (f), males (m), and both sexes (bs) gives 0.111, 0.224, and 0.181, respectively.
As with deflection, we calculated a number of values of evolvability. For female, male, and both-sex models, we calculated the evolvability using G(f, m, and bs), where both genetic variances and covariances were estimated [evolvability e(βf), e(βm), and e(βbs)]. e(βf), e(βm), and e(βbs) provide an estimate of the effect of both genetic variances and covariances on evolvability. We also calculated evolvability fixing genetic covariances to 0 [i.e., using G(f, m and bs)nc], providing an estimate of the evolvability based on the genetic variances alone [evolvability e(βf)nc, e(βm)nc, and e(βbs)nc]. e(βx) and e(βx)nc can then be compared to provide an estimate of the degree to which genetic covariances alter evolvability,
| (9) |
where x = f, m, or bs (Morrissey et al. 2012b). Thus an Re_x value <1 suggests that genetic covariances reduce the predicted evolvability compared to the effect of genetic variances and increase constraint (Morrissey et al. 2012b).
For both-sex models we also calculated evolvability fixing between sex covariances to zero [G(bs)nbs giving evolvability e(βbs)nbs]. e(βbs)nbs allowed calculation of two additional values of Re: (1) Re_bs_nbs = e(βbs)/e(βbs)nbs and (2) Re_bs_nbs.nc = e(βbs)nbs /e(βbs)nc. Re_bs_nbs compares evolvability with and without between-sex genetic covariances fixed to 0, with a value of less than one indicating that between-sex genetic covariances reduce predicted evolvability and thus increase the constraint. Re_bs_nbs.nc compares evolvability with and without within-sex genetic covariances fixed to 0, with a value of less than one indicating that within-sex genetic covariances reduce the predicted evolvability and thus increase constraint.
All models were run in ASReml version 3.0 (Gilmour et al. 2009). The restricted maximum-likelihood procedures used here assume that residuals are normally distributed, having conditioned on the fixed and random effects structures, although they are likely to be fairly robust to departures from these assumptions (Lynch and Walsh 1998, p. 784). Even so, SBA and female ABS are binary traits. As such, estimates of the heritability of these traits based on observed data may be underestimates compared to estimates based on an underlying liability scale (Lynch and Walsh 1998; Roff 2001). However, because this underestimate of the variance also affects any estimate of the covariance, estimates of the genetic correlations should be unbiased (Brotherstone et al. 1990; Lynch and Walsh 1998; Roff 2001). Although methods exist that allow appropriate error structures for each trait to be fitted (Hadfield 2010), these methods do not currently allow FA models to be fitted and so could not be used here. In addition, a generalized model would generate estimates of (co)variance components on a latent scale that would not readily be combined with estimates of selection gradients to predict the response to selection (). As such, we present estimates from models assuming normally distributed residuals throughout. Simulation-based credible intervals (see below) and the comparison of vectors were performed in R version 2.12.0 (R Development Core Team 2010).
Estimates of β and G have associated error and thus so do values calculated from them [e.g., , θ, and e(β)]. Errors in these estimates were approximated using an MC simulation algorithm (see also Morrissey et al. 2012a). Briefly, we drew 100,000 multivariate random normal (MVN) values of S, P, and G, using the maximum-likelihood estimates of these parameters (from ASReml) as the mean and the variance covariance matrices of these parameter estimates as the variance (again these are given in ASReml). These 100,000 values were then combined as appropriate in Equations 5, 4, 6, 7, and 9 to produce 100,000 estimates of β, , θ, e(β), and Re. The 95% credible interval (CI) around these values was then calculated using the quantile function in R and used as an estimate of the 95% credible interval around each parameter estimate. It should be noted that this method assumes the sampling errors in the estimates of variances and covariances are multivariate normal. For angles θ1, θ2, θ3, θ5_bs, and θ6_bs (which are all defined as angles between two vectors) and for all values of evolvability, estimates cannot be negative and thus interpreting a lack of overlap of the 95% CI with zero as indicative of the value differing from zero is not valid. As such, statistical hypothesis tests have limited meaning and we therefore assessed statistical support for substantially nonzero values by examining the distribution of MC samples (see Figure 2, Figure 3, and Figure 4). In practice, this involves visual inspection of the distributions of estimates and, when the distribution is concentrated close to zero (i.e., is associated with left truncation and strong right skew), drawing conclusions equivalent to those associated with failure to reject a null hypothesis.
Figure 2.
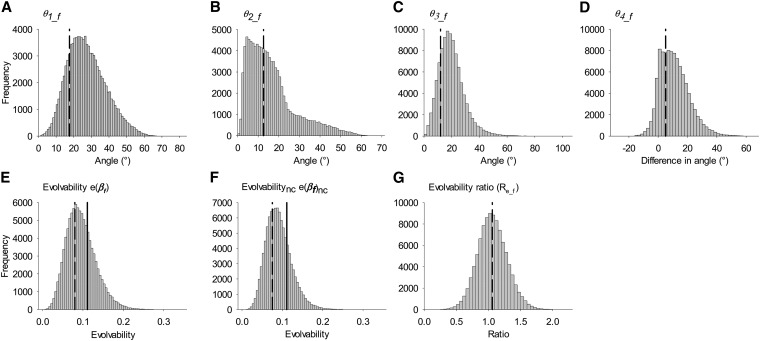
The simulated distribution of estimates of θ and e(β) for females. (A) θ1_f; (B) θ2_f; (C) θ3_f; (D) θ4_f; (E) e(βf); (F) e(βf)nc; (G) Re_f produced by carrying through the errors in the estimation of Gf and βf. Values <0 cannot exist except for θ4_f, and thus the distributions are presented to aid in interpretation of whether the simulated distributions are distinct from zero, i.e., have a normal distribution that is not highly concentrated near (ramped up against) zero. Dashed lines show the position of the “best estimate”, i.e., the estimate when using the maximum-likelihood estimate of the parameters of Gf and βf; this is the value given in Table 2 and Table 3. For E and F, solid lines show the position of the average evolvability over random selection gradients (ēf); see Materials and Methods for details.
Figure 3.
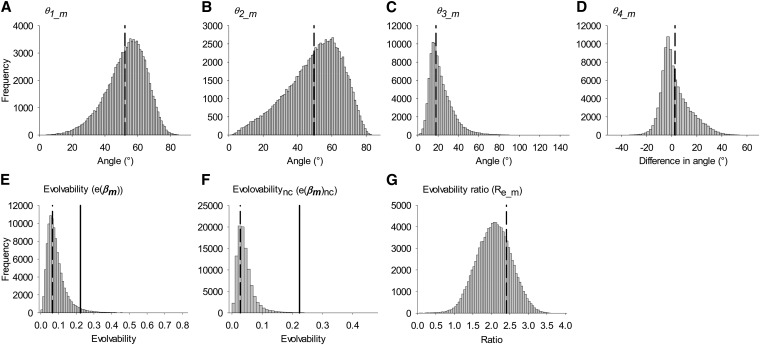
The simulated distribution of estimates of θ and e(β) for males. (A) θ1_m; (B) θ2_m; (C) θ3_m; (D) θ4_m; (E) e(βm); (F) e(βm)nc; (G) Re_m. Values <0 cannot exist except for θ4_m. Dashed lines show the position of the “best estimate,” Solid lines show the position of the average evolvability over random selection gradients (ēm); see Materials and Methods for details.
Figure 4.
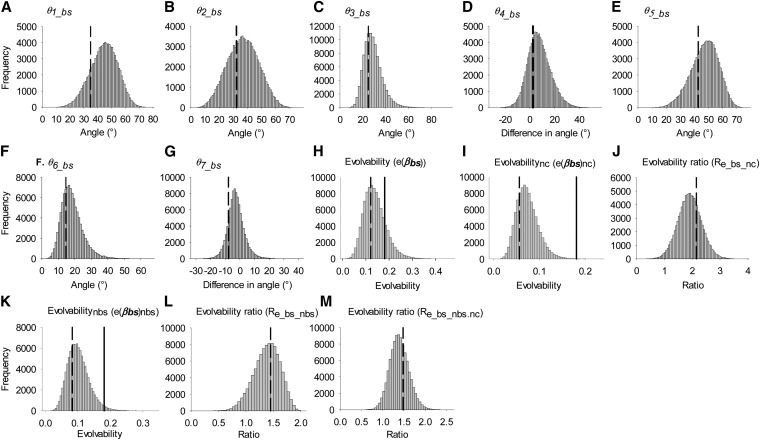
The simulated distribution of estimates of θ and e(β) for both-sex models. (A) θ1_bs; (B) θ2_bs; (C) θ3_bs; (D) θ4_bs; (E) θ5_bs; (F) θ6_bs; (G) θ7_bs; (H) e(βbs); (I) e(βbs)nc; (J) Re_bs_nc, the ratio e(βbs)/e(βbs)nc; (K) e(βbs)nbs; (L) Re_bs_nbs, the ratio e(βbs)/e(βbs)nbs; (M) Re_bs_nbs.nc, the ratio e(βbs)nbs/e(βbs)nc. Values <0 cannot exist except for θ4_bs and θ7_bs. Dashed lines show the position of the “best estimate.” Solid lines show the position of the average evolvability over random selection gradients (ēbs); see Materials and Methods for details.
Results
Part 1: Variance decomposition: Estimating G
Univariate analysis:
There was evidence of significant additive genetic variance for all female life history traits apart from female longevity and for male ABS but not other male life history traits (Table S1). Nongenetic random effects followed expected patterns with maternal and birth year effects being significant only for early life history traits although not all early life history traits in males (Table S1).
Multivariate analysis:
Multivariate models of phenotypic covariance within each sex indicated positive phenotypic correlations among all traits (Table S2), with all but one significantly greater than zero. In addition, all but one estimable nongenetic covariances among traits were positive (Table S3). Full-rank FA models of the genetic covariance matrix (G) converged for females, but for males and both sexes the maximal-rank models that would converge were 2 (of a maximum of 4) and 4 (of 8), respectively (Table S4). Although statistical comparison provided support only for lower-rank models for all G matrices (Table S4), we used the highest-rank model that converged in all subsequent analyses, for the reasons described in Materials and Methods and File S1. Genetic covariances between traits within and between the sexes were a mix of positive and negative values, although within-sex genetic covariances in males were all positive (Table S3, Table S5, and Table S6). A principal component analysis (PCA) of Gbs revealed a major axis of genetic variation that loaded positively on all male traits and female L, but negatively although weakly on female SBA, AFR, and ABS (Table S7C). A similar pattern was apparent when examining the sexes separately: negative associations between female longevity and the other female traits compared to positive associations among all male traits (Table S7, A and B).
Part 2: Assessing the potential for genetic constraint to evolution
All selection differentials and gradients were positive and strong, as life history traits must be by definition (Table 1). Given such strong positive selection, visual inspection of the estimated genetic parameters (Table S3, Table S5, Table S6, and Table S8) suggested an aspect of constraint for female traits in the general pattern of negative genetic covariance between survival and reproductive traits (Table S5 and Table S8). In males, an overall pattern of facilitation of adaptive evolution dominated as estimated genetic covariances were all positive (Table S6); however, it is unclear from the multiple imprecise estimates alone whether such an interpretation is really justified. Similarly, interpretation of the multiple modest between-sex genetic correlations (Table S3) is difficult from consideration of the estimates alone, necessitating consideration of metrics that integrate over the implications of all aspects of G and β.
Table 1. Selection differentials (±SE) and selection gradients (95% CI) for (standardized) male and female life history traits.
| Trait | Selection differential (S) | Selection gradient (β) |
|---|---|---|
| Female SBA | 1.25 ± 0.07 | NA |
| Female AFR | 0.186 ± 0.037 | 0.137 (0.108, 0.167) |
| Female L | 0.538 ± 0.046 | 0.531 (0.505, 0.558) |
| Female ABS | 0.140 ± 0.022 | 0.0776 (0.0603, 0.0936) |
| Male SBA | 1.71 ± 0.13 | NA |
| Male AFR | 0.413 ± 0.134 | 0.180 (−0.042, 0.412) |
| Male L | 0.708 ± 0.134 | 0.498 (0.324, 0.672) |
| Male ABS | 0.468 ± 0.063 | 0.419 (0.306, 0.515) |
Selection differentials and associated standard errors were calculated in ASReml; selection gradients were calculated using the formula for either sex. Because P was undefined between survival to breeding age (SBA) and all other traits, only selection differentials for this trait could be estimated. P for age at first reproduction (AFR), longevity (L), and annual breeding success (ABS) in both sexes is presented in Table S2. As before, note that AFR is premultiplied by −1 such that positive values indicate selection for earlier reproduction. Values in boldface type are significantly greater than zero based on either log-likelihood ratio tests comparing models with the parameter fixed to zero vs. estimated (selection differentials) or whether or not the 95% credible interval overlaps zero (selection gradients).
Metrics of constraint
Angle of deflection (θ) for females:
θ1_f was small [17.6°, less than midway between 0° (no constraint) and 90° (an absolute constraint)], but appeared greater than zero (the distribution is not highly concentrated near zero, Figure 2A), suggesting that unequal genetic variances and/or nonzero genetic covariances deflected the direction of the predicted response to selection away from the direction of the vector of selection, but by a small amount. θ2_f, the effect of unequal genetic variances alone, was also small (12.6°, Table 2, Figure 2B) but in this case the simulated distribution was highly concentrated near zero, suggesting little evidence that unequal genetic variances deflect from β. θ3_f, the effect of nonzero genetic covariances on the direction of the predicted response to selection, was 11.9° (Table 2) and distinct from zero (Figure 2C), suggesting genetic covariances have a mild effect on the predicted response to selection. Finally, θ4_f (the difference between θ1_f and θ2_f) was merely 5.06° (Table 2, Figure 2D) and the 95% credible interval overlapped zero.
Evolvability for females:
The evolvability of female traits when considering genetic variances and covariances [e(βf)] was 0.0801 (Table 3, Figure 2E). The distribution of e(βf) (Figure 2E) suggested this value was distinct from 0, but not from the average evolvability (f) of 0.111. The evolvability of female traits due to genetic variances alone [e(βf)nc] was 0.0753 (Table 3) and appeared distinct from 0, but not from f (Figure 2F). Re_f, the ratio of e(βf)/e(βf)nc, showed no evidence of differing from 1 (Re_f = 1.06; 95% CI = 0.63–1.53, Table 3, Figure 2G), implying little effect of genetic covariances between female traits on evolvability.
Table 3. Measures of evolvability [e(β)] and the ratio of evolvability calculated with and without genetic covariances (Re) for female (f), male (m), and both-sex (bs) models.
| Description | Estimate (95% CI) |
|---|---|
| Female | |
| Evolvability e(βf) | 0.0801 (0.0363, 0.177) |
| Evolvabilitync e(βf)nc | 0.0753 (0.0396, 0.162) |
| Evolvability ratio Re_f [e(βf)/e(βf)nc] | 1.06 (0.631, 1.53) |
| Male | |
| Evolvability e(βm) | 0.0659 (0.0218, 0.235) |
| Evolvabilitync e(βm)nc | 0.0274 (0.0106, 0.120) |
| Evolvability ratio Re_m [e(βm)/e(βm)nc] | 2.41 (1.18, 2.97) |
| Both sexes | |
| Evolvability e(βbs) | 0.121 (0.0626, 0.244) |
| Evolvabilitync e(βbs)nc | 0.0561 (0.0350, 0.128) |
| Evolvability ratio Re_bs_nc [e(βbs)/e(βbs)nc] | 2.15 (1.11, 2.75) |
| Evolvabilitynbs e(βbs)nbs | 0.0829 (0.0475, 0.176) |
| Evolvability ratio Re_bs_nbs [e(βbs)/e(βbs)nbs] | 1.45 (0.883, 1.79) |
| Evolvability ratio Re_bs_nbs.nc [e(βbs)nbs/e(βbs)nc] | 1.48 (0.981, 1.86) |
Evolvability and the 95% CIs are calculated as described in Materials and Methods. nc, all genetic correlations fixed to 0; nbs, between-sex genetic correlations fixed to 0.
Angle of deflection (θ) for males:
θ1_m was intermediate in magnitude (52.2°, Table 2, Figure 3A) and its distribution was distinct from zero (Figure 3A). The effect of unequal genetic variances alone was similar in magnitude (θ2_m = 49.6°, Table 2) and again appeared distinct from 0 (Figure 3B), suggesting significant deflection. The effect of genetic covariances on the direction of m (θ3_m) was small (17.9°, Table 2), but the simulated distribution (Figure 3C) suggested this value was distinct from zero and thus that genetic covariances altered the direction of the predicted response to selection in males. There was no evidence of genetic covariances increasing constraint: θ4_m (the difference between θ1_m and θ2_m) was only 2.57° (95% CI = −13.8–31.2, Table 2, Figure 3D).
Evolvability for males:
The evolvability of male traits when considering genetic variances and covariances [e(βm)] was 0.0659 (Table 3) and appeared distinct from 0 but not quite from m [Figure 3E, m = 0.224, upper 95% CI of e(βm) = 0.235]. The evolvability of male traits when considering only genetic variances [e(βm)nc] was 0.0274 (Table 3) and appeared distinct from 0 and also <m (Figure 3F). Re_m, the ratio e(βm)/e(βm)nc, was significantly >1 (2.41; 95% CI = 1.18–2.97, Table 3, Figure 3G,), suggesting that genetic covariances between male life history traits significantly increased evolvability and thus facilitated the predicted evolutionary response.
Angle of deflection (θ) combining both sexes:
Angles for the both-sex analysis are presented in Table 2 and Figure 4, A–G. The general pattern was one of the vector of the predicted response to selection being deflected away from the vector of selection by a moderate amount [e.g., θ1_bs = 34.9° (Table 2), and θ1_bs appeared distinct from zero (Figure 4A)], but that within- and between-sex genetic covariances did not increase deflection and thus constraint (θ4_bs = 2.54°; 95% CI = −8.91–27.5, Table 2, Figure 4D). Unequal genetic variances alone caused deflection of intermediate magnitude (θ2_bs = 32.3°, Table 2, Figure 4B). Although between-sex covariances appeared to alter the direction of the predicted response to selection (θ6_bs = 14.6°, Table 2, Figure 4F), there was little evidence that between-sex covariances increased constraint in terms of θ (θ7_bs = −7.80°, Table 2, Figure 4G).
Evolvability combining both sexes:
Predictions of evolvability are presented in Table 3 and Figure 4, H–M. In general, evolvability was >0, but lower than the average evolvability over random selection gradients (bs) (Figure 4, H, I, and K). However, there was no evidence that genetic covariances caused a reduction in evolvability (i.e., increase in constraint). Instead, evolvability was higher when including genetic covariances than when fixing them to zero (Figure 4, J, L, and M; Re_bs_nc = 2.15, 95% CI = 1.11–2.75; Re_bs_nbs = 1.45, 95% CI = 0.883–1.79; Re_bs_nbs.nc = 1.48, 95% CI = 0.981–1.86, although the 95% CIs of both Re_bs_nbs and Re_bs_nbs.nc overlapped 1; see Table 3). Thus the positive effects of covariances on evolvability appeared to be due to both between- and within-sex covariances.
Discussion
These results provide a detailed investigation of genetic constraints to life history evolution in a wild population of red deer. Studies that apply multiple measures of genetic constraint, particularly measures of evolvability, to data from a wild animal population are extremely rare. Teplitsky et al. (2014) assess multivariate evolvability, and also the effect of genetic correlations on the predicted rate of adaptation, in an analysis of morphological traits in 10 different bird populations and found general nonalignment between selection and genetic variance (see also Simonsen and Stinchcombe 2011 for a field study in plants and Morrissey et al. 2012b, but note Morrissey et al. do not present explicit estimates of evolvability). In contrast, in our analyses, although a substantial proportion of the estimates of individual genetic covariances were negative (11/28; Table S3), in general we found overall genetic constraints to be relatively mild and to result mainly from genetic variances rather than from genetic covariances among traits. In particular, we found little evidence that genetic covariances among traits in males or between the sexes generate constraint; rather, genetic covariances in males and between the sexes appear to facilitate the predicted response to selection in terms of evolvability and any constraint occurs primarily from the pattern of genetic variances.
Measures of constraint
In general, the deflection of the predicted response to selection away from the vector of selection (i.e., the direction of fastest adaptation) was small. This was particularly true in females (θ1_f and θ2_f < 20°), while in the male and both-sex models, deflection was of intermediate magnitude (θ1_m, θ1_bs, θ2_m, and θ2_bs >30° but <53°). In females the small deflection and thus constraint that was evident appeared to result from a combination of unequal genetic variances and nonzero genetic covariances, since neither one caused significant deflection alone. However, in males and both-sex models, genetic covariances caused limited deflection, while unequal genetic variances caused significant (albeit still not large) deflection and thus constraint.
Conclusions regarding the extent of constraint were slightly different when considering evolvability. For female and both-sex models, evolvability was similar to the average evolvability, a baseline that indicates the evolutionary potential of G independent of the direction of selection relative to G (Hansen and Houle 2008; Innocenti and Chenoweth 2013). Thus, the pattern of genetic variances and covariances relative to the direction of selection does not appear to greatly restrict the evolutionary potential of females or both sexes combined in comparison to the evolutionary potential of G [e(βf) was 72% of f, e(βbs)nc was 67% of bs]. However, for males, evolvability was low compared to the average evolvability particularly when genetic covariances were fixed to zero [e(βm) was 29% of m and e(βm)nc was 12% of m]. This suggests constraint in the pattern of genetic variances relative to the direction of selection in males when compared to the evolutionary potential of G. The effect of genetic covariances on the predicted evolvability differed slightly between female vs. male and both-sex models. For females, genetic covariances had very little effect on the predicted evolvability, as opposed to the slight increase in deflection and thus constraint as measured by deflection. In male and both-sex models evolvability increased as a result of genetic covariances and thus, while causing only a very slight increase in evolvability in absolute terms [i.e., small changes in e(β)], caused a large increase in evolvability in relative terms (Re_m and Re_bs_nc estimates were >2). This suggests that genetic covariances increased the predicted evolvability when considering males and both sexes, by increasing the magnitude of the predicted response in the direction of maximally increasing fitness.
Comparison with other results from the Rum red deer population
Detailed discussion of the comparison with previous results from the Rum red deer population is given in File S1. However, it should be noted here that despite patterns of genetic covariances between female traits being similar to those of a previous study (Morrissey et al. 2012b), the results presented in the present study provide weaker evidence for genetic constraint than that in two previous studies (Foerster et al. 2007; Morrissey et al. 2012b). These differences appear to be driven by the treatment of survival to breeding age, pointing to parent–offspring patterns/processes being a potential key area for future study of genetic constraints in this population (see File S1 for further discussion).
Comparison with results from other populations
Our results provide evidence for relatively modest genetic constraint to evolution, particularly as a result of genetic covariances between traits. Although rare, other estimates of multivariate genetic constraint in the literature have provided stronger evidence of constraint (Blows et al. 2004; Hine and Blows 2006; Smith and Rausher 2008; Lewis et al. 2011; Simonsen and Stinchcombe 2011; Gosden et al. 2012; Williams et al. 2012; Teplitsky et al. 2014). The reason for the difference in the magnitude of constraint between our results and those of previous studies is difficult to assess, given the multivariate nature of the techniques. Previous studies have tended to focus on combinations of traits, among which one might predict strong correlations. For example, a number of studies focus on Drosophila cuticular hydrocarbons (Blows et al. 2004; Gosden et al. 2012), many of which are built from the same amino acids and thus might be expected to share biosynthetic pathways (Blows et al. 2004); in their large-scale analyses of 10 bird populations, Teplitsky et al. (2014) considered four different morphological traits, all of which were positively correlated. Our study focuses on different components of fitness that might not be expected to be functionally related—at least not via immediate and simple biochemical relationships. Having said this, Lewis et al. (2011) analyze life history traits including development time and longevity in the Indian meal moth and also find evidence of strong constraint.
In addition, our results are from a wild rather than a laboratory population. While very useful, laboratory studies necessarily deal with populations that are experiencing selection pressures that are not “natural” selection and that they may have experienced for only a limited time period compared to wild populations; alternatively, selection pressures may have been very consistent compared to those experienced in wild populations. Thus differences in the patterns of selection between laboratory and natural populations may also contribute to differences in results. For example, a recent study of diet preferences in a population of Drosophila melanogaster that was not laboratory adapted found weak evidence for multivariate constraint (Reddiex et al. 2013). Teplitsky et al.’s (2014) analysis suggests multivariate constraints to the evolution of morphological traits in wild avian populations, but this appears to be due to antagonistic selection on positively genetically correlated traits. In one other study of a wild population, Coltman et al. (2005) analyze covariation in life history traits in bighorn sheep (Ovis canadensis) and find substantial angles between β and the first three principal components of G: 117°, 73°, and 103°. Applying the same technique to the results in our study (PC1–3 in Table S7C compared to the vector of selection gradients in Table 1) gives angles of 76°, 68°, and 121°, which would perhaps suggest a stronger genetic constraint. However, when considering the effect of G on deflection and evolvability, genetic constraint is less apparent. Our study therefore illustrates the difference in conclusions that may be drawn from alternative approaches to quantifying evolutionary constraints and argues for a range of different metrics to be explored (see, for example, Simonsen and Stinchcombe 2011). Given the overall paucity of data on this subject, and the potential tendency for significant rather than nonsignificant evidence of constraint to be published earlier, it will be interesting to see the patterns that emerge from future studies in wild populations.
The results of our study suggest a lack of genetic constraint to the evolution of life history traits in this population and hence to the maintenance of genetic variance in the direction of selection. As such, traits should have the potential to respond rapidly to natural selection and yet a lack of response to natural selection is commonly observed in wild populations (e.g., Kruuk et al. 2001; Merilä et al. 2001). Explaining the maintenance of genetic variance for quantitative traits is a core area of research in evolutionary biology and a number of potential explanations, other than multivariate genetic constraint, have been proposed (Via and Lande 1985; Gillespie and Turelli 1989; Charlesworth and Hughes 2000; Johnson and Barton 2005). Of particular relevance in natural populations may be the presence of environmental variation and thus the potential for genotype-by-environment interactions and variable selection to maintain genetic variation (Gillespie and Turelli 1989; Charmantier and Garant 2005; Bell 2010; Morrissey and Hadfield 2012). Certainly there is the potential for environmental variation both temporally and spatially within the red deer population studied here (Coulson et al. 1997; Stopher et al. 2012) and thus for genotype-by-environment interactions and variable selection to be important in the maintenance of genetic variation for quantitative traits. Given the current lack of data on microevolutionary parameters in natural populations and how they vary with environment (but see, for example, Charmantier and Garant 2005; Wilson et al. 2006; Robinson et al. 2009), further research into this area would be of interest. However, it should be noted that current evidence suggests patterns of directional selection are remarkably stable across study systems, although this pattern does not necessarily hold within any particular population (Siepielski et al. 2009; Morrissey and Hadfield 2012).
Finally, it is worth clarifying here that multiple regression-based selection analysis is expected to provide robust inference when complete life history data are analyzed. As we and others have discussed in the past, multiple regression-based selection gradient estimates may fail to represent the true direct effects of traits on fitness when unmeasured variables cause trait–fitness covariance (Robertson 1966; Rausher 1992; Kruuk et al. 2003; Morrissey et al. 2010, 2012a). Two alternative strategies are available to ensure that quantitative genetic predictions of evolutionary trajectories are robust. First, one might seek to estimate the additive genetic covariances of traits with relative fitness, which gives complete prediction of evolutionary change (Robertson 1966; reviewed in Morrissey et al. 2010). However, estimation of the genetic covariance of traits with relative fitness, despite being an application of the “secondary theorem of selection” (Robertson 1968), actually tells us very little about the phenotypic process of selection—a trait that covaries genetically with relative fitness may not itself be selected at all (for example, the trait may not causally influence fitness but may be genetically correlated with a trait that does; discussed in Morrissey et al. 2010, 2012a; Morrissey 2014). The second strategy is to include sufficient variables in multiple-regression analyses to account for all trait–fitness covariance. These variables may, for example, be features of the environment that ultimately cause trait–fitness covariance. Alternatively, the included traits may not necessarily be those that ultimately cause trait–fitness variation, but rather phenotypic traits by which the effects of the causal traits are mediated. For example, if an environmental variable E causes variation in survival S and reproduction R, regression of relative fitness w on S will give the wrong selection gradient for S. However, analyses regressing either w on S and E or w on S and R (even without the ultimate source of covariance, E) will give correct selection gradient estimates for S and in the latter case, the bonus of the correct selection gradient for R; in each case correct refers to the value of the selection gradient that provides the correct evolutionary predictions when used in the Lande equation (this may be initially unintuitive; for a simple demonstration, see R console procedure at end of paragraph). Thus, while it is typically impossible to be sure that all relevant traits are included, a comprehensive set of life history traits (e.g., survival, age at first reproduction, annual reproduction, in both sexes, as herein) is a special case where all pathways mediating any effects of traits or environmental variables on fitness are necessarily included, because life history completely determines fitness; there are no missing traits, in the sense that all effects of variables on fitness are mediated by life history.
Enter the following in an R console:
#simulate data according to hypothetical example in text, true LH betas = 1
n<-10000; E<-rnorm(n,0,1); S<-1*E+rnorm(n,0,1); R<-1*E+rnorm(n,0,1); w<-1*R+1*S+rnorm(n,1,1);
#missing variable problem
summary(lm(w∼S))
#solution 1: include ultimate effects
summary(lm(w∼S+E))
#solution 2: include mediating effects (complete life history)
summary(lm(w∼S+R)).
Conclusions
This study attempts to quantify the multivariate genetic constraint, considering both within- and between-sex genetic (co)variation, in a wild population experiencing a natural environment. Overall we found estimates of genetic constraint were mild and that patterns of genetic variances rather than genetic covariances were the main source of genetic constraint. There was little support for the contention that between-sex genetic covariances caused constraint, indicating that sexual antagonism may not be as strong as previous results from this population suggested. Finally, the degree of genetic constraint in this study was much lower than in previous studies based predominantly on laboratory populations. Given the lack of data on natural populations and the difficulty of estimating these parameters in such populations, we encourage further analysis of this type to assess whether the apparently lower level of genetic constraint in natural populations is a general pattern.
Supplementary Material
Acknowledgments
We thank Scottish Natural Heritage (SNH) for permission to work on the Isle of Rum and SNH staff on Rum for their support and assistance. We are also very grateful to the numerous field assistants and volunteers who have helped with data collection, in particular Fiona Guinness and, more recently, Ali Morris, Sean Morris and Martyn Baker. We also thank Jarrod Hadfield, Dan Nussey, Katie Stopher, Ian White, Alastair Wilson, the “Wild Evolution Group” at the University of Edinburgh, and two anonymous reviewers for very helpful discussions and comments on previous versions of this manuscript. The long-term study has been largely funded by the Natural Environment Research Council (NERC). This research was supported by a NERC grant (to L.E.B.K., T.H.C.-B., and J.M.P.), a NERC postdoctoral fellowship (to C.A.W.), a National Sciences and Engineering Research Council postdoctoral fellowship (to M.B.M.), and an Australian Research Council Future Fellowship (to L.E.B.K.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.164319/-/DC1.
Communicating editor: S. F. Chenoweth
Literature Cited
- Agrawal A. F., Stinchcombe J. R., 2009. How much do genetic covariances alter the rate of adaptation? Proc. R. Soc. Lond. B Biol. Sci. 276: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows M., Walsh B., 2009. Spherical cows grazing in flatland: constraints to selection and adaptation, pp. 83–101 in Adaptation and Fitness in Animal Populations - Evolutionary and Breeding Perspectives on Genetic Resource Management, edited by Van Der Werf J., Graser H. U., Frankham R., Gondro C. Springer-Verlag, Dordrecht, The Netherlands. [Google Scholar]
- Blows M. W., 2007. A tale of two matrices: multivariate approaches in evolutionary biology. J. Evol. Biol. 20: 1–8. [DOI] [PubMed] [Google Scholar]
- Blows M. W., Hoffmann A. A., 2005. A reassessment of genetic limits to evolutionary change. Ecology 86: 1371–1384. [Google Scholar]
- Blows M. W., Chenoweth S. F., Hine E., 2004. Orientation of the genetic variance-covariance matrix and the fitness surface for multiple male sexually selected traits. Am. Nat. 163: 329–340. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R., and S. F. Chenoweth, 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24: 280–288. [DOI] [PubMed] [Google Scholar]
- Brommer J. E., Kirkpatrick M., Qvarnström A., Gustafsson L., 2007. The intersexual genetic correlation for lifetime fitness in the wild and its implications for sexual selection. PLoS ONE 2: e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherstone S., Mcmanus C. M., Hill W. G., 1990. Estimation of genetic parameters for linear and miscellaneous type traits in Holstein-Friesian dairy cattle. Livest. Prod. Sci. 26: 177–192. [Google Scholar]
- Charlesworth B., 1987. The heritability of fitness, pp. 21–40 in Sexual Selection: Testing the Alternatives, edited by Bradbury J. W., Andersson M. B. John Wiley & Sons, New York. [Google Scholar]
- Charlesworth B., Hughes K. A., 2000. The maintenance of genetic variation in life history traits, pp. 369–392 in Evolutionary Genetics from Molecules to Morphology, edited by Singh R. S., Krimbas C. B. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Charmantier A., Garant D., 2005. Environmental quality and evolutionary potential: lessons from wild populations. Proc. Biol. Sci. 272: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale A. K., Gibson J. R., Rice W. R., 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M. N., Clutton-Brock T. H., Guinness F. E., Pemberton J. M., Kruuk L. E. B., 2011. Variances and covariances of phenological traits in a wild mammal population. Evolution 65: 788–801. [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T., 1982. The red deer of Rhum. Nat. Hist. 91: 42. [Google Scholar]
- Coltman D. W., O’donoghue P., Hogg J. T., Festa-Bianchet M., 2005. Selection and genetic (co)variance in bighorn sheep. Evolution 59: 1372–1382. [PubMed] [Google Scholar]
- Coulson T., Albon S., Guinness F., Pemberton J., Cluttonbrock T., 1997. Population substructure, local density, and calf winter survival in red deer (Cervus elaphus). Ecology 78: 852–863. [Google Scholar]
- Cox R. M., Calsbeek R., 2009. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am. Nat. 173: 176–187. [DOI] [PubMed] [Google Scholar]
- Cox R. M., Calsbeek R., 2010. Sex-specific selection and intraspecific variation in sexual size dimorphism. Evolution 64: 798–809. [DOI] [PubMed] [Google Scholar]
- Dickerson G. E., 1955. Genetic slippage in response to selection for multiple objectives. Cold Spring Harb. Symp. Quant. Biol. 20: 213–224. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., 1981. Introduction to Quantitative Genetics. Longman Group, Essex, UK. [Google Scholar]
- Fedorka K. M., Mousseau T. A., 2004. Female mating bias results in conflicting sex-specific offspring fitness. Nature 429: 65–67. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1958 The Genetical Theory of Natural Selection. Dover, New York. [Google Scholar]
- Foerster K., Coulson T., Sheldon B. C., Pemberton J. M., Clutton-Brock T. H., et al. , 2007. Sexually antagonistic genetic variation for fitness in red deer. Nature 447: 1107–1109. [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., Turelli M., 1989. Genotype-environment interactions and the maintenance of polygenic variation. Genetics 121: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, A. R., B. J. Gogel, B. R. Cullis, and R. Thompson, 2009 Asreml User Guide Release 3.0 VSN International, Hemel Hempstead, UK. Available at: www.vsni.co.uk.
- Gosden, T. P., K.-L. Shastri, P. Innocenti, and S. F. Chenoweth, 2012 The B-matrix harbors significant and sex-specific constraints on the evolution of multicharacter sexual dimorphism. Evolution 66: 2106–2116. [DOI] [PubMed]
- Hadfield J. D., 2010. MCMC methods for multi-response generalised linear mixed models: the mcmcglmm r package. J. Stat. Softw. 33: 1–22.20808728 [Google Scholar]
- Hadfield J. D., Richardson D. S., Burke T., 2006. Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol. Ecol. 15: 3715–3730. [DOI] [PubMed] [Google Scholar]
- Hansen T. F., Houle D., 2008. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 21: 1201–1219. [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1976. A simple method for computing the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32: 69–83. [Google Scholar]
- Hill W. G., Thompson R., 1978. Probabilities of non-positive definite between-group or genetic covariance matrices. Biometrics 34: 429–439. [Google Scholar]
- Hine E., Blows M. W., 2006. Determining the effective dimensionality of the genetic variance-covariance matrix. Genetics 173: 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., 1991. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution 45: 630–648. [DOI] [PubMed] [Google Scholar]
- Houle D., Pélabon C., Wagner G. P., Hansen T. F., 2011. Measurement and meaning in biology. Q. Rev. Biol. 86: 3–34. [DOI] [PubMed] [Google Scholar]
- Innocenti P., Chenoweth S. F., 2013. Interspecific divergence of transcription networks along lines of genetic variance in Drosophila: dimensionality, evolvability and constraint. Mol. Biol. Evol. 30: 1358–1367. [DOI] [PubMed] [Google Scholar]
- Johnson T., Barton N., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L. E. B., 2004. Estimating genetic parameters in natural populations using the “animal model”. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359: 873–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L. E. B., Hadfield J. D., 2007. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 20: 1890–1903. [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Clutton-Brock T. H., Slate J., Pemberton J. M., Brotherstone S., et al. , 2000. Heritability of fitness in a wild mammal population. Proc. Natl. Acad. Sci. USA 97: 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L. E. B., Merila J., Sheldon B. C., 2001. Phenotypic selection on a heritable size trait revisited. Am. Nat. 158: 557–571. [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Slate J., Pemberton J. M., Brotherstone S., Guinness F. E., et al. , 2002. Antler size in red deer: heritability and selection but no evolution. Evolution 56: 1683–1695. [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Merila J., Sheldon B. C., 2003. When environmental variation short-circuits natural selection. Trends Ecol. Evol. 18: 207–209. [Google Scholar]
- Kruuk L. E. B., Slate J., Wilson A. J., 2008. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 39: 525–548. [Google Scholar]
- Lande R., 1979. Quantitative genetic analysis of multivariate evolution, applied to brain: body size allometry. Evolution 33: 402–416. [DOI] [PubMed] [Google Scholar]
- Lande R., 1980. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34: 292–305. [DOI] [PubMed] [Google Scholar]
- Lande R., 1982. A quantitative genetic theory of life history evolution. Ecology 63: 607–615. [Google Scholar]
- Lande R., Arnold S. J., 1983. The measurement of selection on correlated characters. Evolution 37: 1210–1226. [DOI] [PubMed] [Google Scholar]
- Lewis Z., Wedell N., Hunt J., 2011. Evidence for strong intralocus sexual conflict in the Indian meal moth, Plodia interpunctella. Evolution 65: 2085–2097. [DOI] [PubMed] [Google Scholar]
- Lynch, A., and B. Walsh, 1998 Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Mainguy, J., S. D. Côté, M. Festa-Bianchet, and D. W. Coltman, 2009 Father-offspring phenotypic correlations suggest intralocus sexual conflict for a fitness-linked trait in a wild sexually dimorphic mammal. Proc. R. Soc. B Biol. Sci. 276: 4067–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcguigan K., Blows M. W., 2010. Evolvability of individual traits in a multivariate context: partitioning the additive genetic variance into common and specific components. Evolution 64: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Merilä J., Sheldon B. C., Kruuk L. E. B., 2001. Explaining stasis: microevolutionary studies in natural populations. Genetica 112–113: 199–222. [PubMed] [Google Scholar]
- Meyer K., Kirkpatrick M., 2008. Perils of parsimony: properties of reduced-rank estimates of genetic covariance matrices. Genetics 180: 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey J. G., Houle D., 2005. The dimensionality of genetic variation for wing shape in Drosophila melanogaster. Evolution 59: 1027–1038. [PubMed] [Google Scholar]
- Morrissey M. B., 2014. Selection and evolution of causally covarying traits. Evolution 68: 1748–1761. [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Hadfield J. D., 2012. Directional selection in temporally replicated studies is remarkably consistent. Evolution 66: 435–442. [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Kruuk L. E. B., Wilson A. J., 2010. The danger of applying the breeder’s equation in observational studies of natural populations. J. Evol. Biol. 23: 2277–2288. [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Parker D. J., Korsten P., Pemberton J. M., Kruuk L. E. B., et al. , 2012a The prediction of adaptive evolution: empirical application of the secondary theorem of selection and comparison to the breeder’s equation. Evolution 66: 2399–2410. [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Walling C. A., Wilson A. J., Pemberton J. M., Clutton-Brock T. H., et al. , 2012b Genetic analysis of life-history constraint and evolution in a wild ungulate population. Am. Nat. 179: E97–E114. [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Wilson A. J., Morris A., Pemberton J., Clutton-Brock T., et al. , 2008. Testing for genetic trade-offs between early- and late-life reproduction in a wild red deer population. Proc. R. Soc. Lond. B Biol. Sci. 275: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease C. M., Bull J. J., 1988. A critique of methods for measuring life history trade-offs. J. Evol. Biol. 1: 293–303. [Google Scholar]
- Pemberton J. M., Albon S. D., Guinness F. E., Clutton-Brock T. H., Berry R. J., 1988. Genetic variation and juvenile survival in red deer. Evolution 42: 921–934. [DOI] [PubMed] [Google Scholar]
- Poissant J., Wilson A. J., Coltman D. W., 2010. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution 64: 97–107. [DOI] [PubMed] [Google Scholar]
- R Development Core Team , 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rausher M. D., 1992. The measurement of selection on quantitative traits: biases due to environmental covariances between traits and fitness. Evolution 46: 616–626. [DOI] [PubMed] [Google Scholar]
- Reddiex A. J., Gosden T. P., Bonduriansky R., Chenoweth S. F., 2013. Sex-specific fitness consequences of nutrient intake and the evolvability of diet preferences. Am. Nat. 182: 91–102. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1984. Sexually chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Robertson A., 1966. A mathematical model of the culling process in dairy cattle. Anim. Prod. 8: 95–108. [Google Scholar]
- Robertson A., 1968. The spectrum of genetic variation, pp. 5–16 in Population Biology and Evolution, edited by Lewontin R. C. Syracuse University Press, New York. [Google Scholar]
- Robinson M. R., Wilson A. J., Pilkington J. G., Clutton-Brock T. H., Pemberton J. M., et al. , 2009. The impact of environmental heterogeneity on genetic architecture in a wild population of soay sheep. Genetics 181: 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. A., 1996. The evolution of genetic correlations: an analysis of patterns. Evolution 50: 1392–1403. [DOI] [PubMed] [Google Scholar]
- Roff D. A., 2001. The threshold model as a general purpose normalizing transformation. Heredity 86: 404–411. [DOI] [PubMed] [Google Scholar]
- Siepielski A. M., Dibattista J. D., Carlson S. M., 2009. It’s about time: the temporal dynamics of phenotypic selection in the wild. Ecol. Lett. 12: 1261–1276. [DOI] [PubMed] [Google Scholar]
- Simonsen A. K., Stinchcombe J. R., 2011. Quantifying evolutionary genetic constraints in the ivyleaf morning glory, Ipomoea hederacea. Int. J. Plant Sci. 171: 972–986. [Google Scholar]
- Smith R. A., Rausher M. D., 2008. Selection for character displacement is constrained by the genetic architecture of floral traits in the ivyleaf morning glory. Evolution 62: 2829–2841. [DOI] [PubMed] [Google Scholar]
- Stopher K. V., Walling C. A., Morris A., Guinness F. E., Clutton-Brock T. H., et al. , 2012. Shared spatial effects on quantitative genetic parameters: accounting for spatial autocorrelation and home range overlap reduces estimates of heritability in wild red deer. Evolution 66: 2411–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitsky C., Tarka M., Moller A. P., Nakagawa S., Balbontin J., et al. , 2014. Assessing multivariate constraints to evolution across ten long-term avian studies. PLoS ONE 9(3): e90444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Cullis B., Smith A., Gilmour A., 2003. A sparse implementation of the average information algorithm for factor analytic and reduced rank variance models. Aust. N. Z. J. Stat. 45: 445–459. [Google Scholar]
- Via S., Lande R., 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39: 505–522. [DOI] [PubMed] [Google Scholar]
- Walling C. A., Pemberton J. M., Hadfield J. D., Kruuk L. E. B., 2010. Comparing parentage inference software: re-analysis of a red deer pedigree. Mol. Ecol. 19: 1914–1928. [DOI] [PubMed] [Google Scholar]
- Walsh B., Blows M. W., 2009. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 40: 41–59. [Google Scholar]
- Wang J., 2004. Sibship reconstruction from genetic data with typing errors. Genetics 166: 1963–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Santure A. W., 2009. Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181: 1579–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. R., Van Heerwaarden B., Dowling D. K., Sgro C. M., 2012. A multivariate test of evolutionary constraints for thermal tolerance in Drosophila melanogaster. J. Evol. Biol. 25: 1415–1426. [DOI] [PubMed] [Google Scholar]
- Wilson A. J., Pemberton J. M., Pilkington J. G., Coltman D. W., Mifsud D. V., et al. , 2006. Environmental coupling of selection and heritability limits evolution. PLoS Biol. 4: 1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., Nussey D. H., Pemberton J. M., Pilkington J. G., Morris A., et al. , 2007. Evidence for a genetic basis of aging in two wild vertebrate populations. Curr. Biol. 17: 2136–2142. [DOI] [PubMed] [Google Scholar]
- Wright S., 1932. General, group and special size factors. Genetics 17: 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.