Abstract
The application of quantitative genetics in plant and animal breeding has largely focused on additive models, which may also capture dominance and epistatic effects. Partitioning genetic variance into its additive and nonadditive components using pedigree-based models (P-genomic best linear unbiased predictor) (P-BLUP) is difficult with most commonly available family structures. However, the availability of dense panels of molecular markers makes possible the use of additive- and dominance-realized genomic relationships for the estimation of variance components and the prediction of genetic values (G-BLUP). We evaluated height data from a multifamily population of the tree species Pinus taeda with a systematic series of models accounting for additive, dominance, and first-order epistatic interactions (additive by additive, dominance by dominance, and additive by dominance), using either pedigree- or marker-based information. We show that, compared with the pedigree, use of realized genomic relationships in marker-based models yields a substantially more precise separation of additive and nonadditive components of genetic variance. We conclude that the marker-based relationship matrices in a model including additive and nonadditive effects performed better, improving breeding value prediction. Moreover, our results suggest that, for tree height in this population, the additive and nonadditive components of genetic variance are similar in magnitude. This novel result improves our current understanding of the genetic control and architecture of a quantitative trait and should be considered when developing breeding strategies.
Keywords: Genomic selection, G-BLUP, nonadditive, realized relationship matrices, dominance relationship matrix, GenPred, shared data resource
QUANTITATIVE genetics and its applications in plant and animal breeding have largely focused on additive models. Under idealized conditions, such as those described by Cockerham (1954) and Kempthorne (1954), genetic values due to additive and nonadditive effects are orthogonal. However, these conditions are often not met in breeding populations, with the consequence that genetic values due to additive and nonadditive effects may be confounded. Under these conditions, a large proportion of variance due to interactions of alleles (dominance and epistasis) can manifest as additive variance (Hill et al. 2008). For the same reason, with most commonly used family structures, it is difficult to dissect genetic variance into additive, dominance, and epistatic effects. With standard pedigree models, variance estimates of these elements are highly correlated, reflecting confounding effects (Lynch and Walsh 1998; Hill 2010). The proportion of additive variance attributable to interactions of alleles largely depends on the distribution of allele frequencies at causal loci (Lu et al. 1999; Hill et al. 2008; Zuk et al. 2012). This affects the estimation of variance components and breeding value (BV) predictions (Vanderwerf and Deboer 1989; Palucci et al. 2007), as well as the ability to dissect the genetic architecture of the trait at the causal level. Understanding the genetic architecture of a trait is also useful for defining breeding strategies and for maximizing genetic gains. For instance, individual genetic differences due to nonadditive effects can be exploited by designing mating schemes that maximize favorable allelic combinations, particularly if family or clonal propagation are possible in the breeding program.
Separation of additive and nonadditive genetic components with standard pedigree-based models requires specific family structures, which are commonly available in plant or animal breeding programs. In practice, estimation of variance due to dominance and additive effects involves mating designs with large numbers of close, typically full-sib relatives. Partitioning epistasis requires, in addition, either inbreds or vegetatively propagated (clonal) populations. In perennial plants, inbreds are not used because of their long generation time and because severe inbreeding depression often occurs. Thus, clonal populations are an alternative to explore the full genetic architecture in these species (Foster and Shaw 1988). Several studies aimed at partitioning genetic variance into its various components detected small dominance and negligible epistatic effects (Foster and Shaw 1988; Mullin et al. 1992; Wu 1996; Isik et al. 2003, 2005; Costa E Silva et al. 2004, 2009; Baltunis et al. 2007, 2008, 2009; Araujo et al. 2012). These results do not necessarily imply that such effects are not important. Instead, the contribution of nonadditive effects may be masked by effects due to the distribution of allele frequencies (e.g., Hill et al. 2008). These results may also reflect the limitations imposed by the data/family structure available or the genetic information used (pedigrees), which only allows for estimation of the expected degree of genetic similarity.
Genome-wide genotypic data can identify, with a high level of certainty, the actual fraction of allele sharing between pairs of individuals. In pedigree-based genetic relationships, each element in the numerator relationship matrix (A matrix) is defined as the expected fraction of shared alleles assuming an infinitesimal model. However, due to Mendelian sampling, the values of the realized genomic relationships (AG matrix), constructed from molecular marker information, deviate from their expected value (Vanraden 2008; Hill and Weir 2011). One way of incorporating molecular marker information for prediction of genetic values consists of replacing, in a genomic best linear unbiased predictor (BLUP) analysis, the pedigree-based relationship matrices (P-BLUP) with marker-based counterparts (or genetic, G-BLUP) (Vanraden 2008). Indeed, G-BLUP is one of the most frequently used methods that combine molecular information to predict BVs and has shown remarkably good predictive performance in animal and plant breeding populations (Hayes et al. 2009; Habier et al. 2010; Veerkamp et al. 2011; de los Campos et al. 2012; Heslot et al. 2012).
Genomic BLUP is a well-known and easily understood methodology. In the context of genome-wide selection (GWS), it is equivalent to ridge regression BLUP (RR-BLUP) (Vanraden 2008; de los Campos et al. 2012). Similar to P-BLUP, G-BLUP can be extended to account for nonadditive effects by replacing pedigree-based relationship matrices due to nonadditive effects (Mrode 2005), with their marker-based counterpart. This is because dominance and epistatic interaction (e.g., additive by additive, dominance by dominance, and additive by domiance) relationship matrices can also be constructed using molecular information, as is currently done with AG. Use of dominance and epistasis matrices of realized genetic relationships may increase the precision of estimates derived from data in poorly structured populations and may also increase the power to dissect genetic variance into components due to main and interaction effects.
Evidence indicates that G-BLUP based on AG yields more accurate predictions of breeding value and of future phenotypes than its pedigree-based counterpart (A) (Vanraden 2008; de los Campos et al. 2009; Hayes et al. 2009; Crossa et al. 2010; Heslot et al. 2012; Resende et al. 2012b; Muñoz et al. 2013). This suggests that use of the realized genomic similarity (AG) increases (relative to A) the ability of the model to uncover the genetic components of the phenotypic data. However, it is not clear whether the power to partition genetic variance into additive and nonadditive components can also be improved by the use of the realized genomic relationships. If so, this would lead to a finer dissection of the genetic architecture of complex traits that could have profound impacts on the future design and implementation of breeding strategies. The objective of this study is to assess the extent to which the use of marker-based additive and nonadditive relationship matrices improves the precision of partitioning genetic variance into its components. For this assessment, tree height from a clonal population of Pinus taeda L. was evaluated with a series of models that account for additive, dominance, and first-order epistatic interactions (additive by additive, dominance by dominance, and additive by dominance) implemented with either pedigree or molecular marker information.
Materials and Methods
Data
Field data from a single experimental trial from the CCLONES population (see Baltunis et al. 2007 and Resende et al. 2012a for details) was used in this study. The response variable total tree height (HT, m) was used. The population was generated by crossing 32 parents in a circular mating design with additional off-diagonal crosses, resulting in 70 full-sib families with an average of 13.5 individuals per family. Each individual was clonally replicated (ramet) and a clonal field trial was established using single-tree plots with eight replicates (one ramet per replicate), in a resolvable alpha-incomplete block design (Williams et al. 2002). Four of the replicates were grown under high intensity management while the rest were under an operational-like regime.
A subset of the CCLONES population, composed of 951 individuals from 61 families, was genotyped using the Illumina Infinium platform (Illumina, San Diego (Eckert et al. 2010) with 7216 SNPs, each representing a unique pine EST contig. A total of 4853 SNPs were polymorphic and were used for further analyses.
Relationship matrices
A marker-based additive relationship matrix (AG) was constructed following the method described by Yang et al. (2010). The pairwise relationship for individuals j and k was defined by
where m is the total number of markers, w is an indicator variable representing the number of copies of a given allele, and pi is the observed allele frequency of the ith SNP. To reduce the sampling variance of the entries of AG*, we expanded the formula proposed by Yang et al. (2010) and adjusted each value of the AG* matrix by shrinking it toward its expectation. The the adjusted AG was obtained as
where Cjk represents all values of AG* that belong to the same class in Ajk (e.g., full-sib individuals, where Ajk = 0.5). The resulting AG was used to correct the original pedigree as previously detailed (Muñoz et al. 2013), and it was verified that the estimated genomic coefficients and their standard deviations were within expectations according to Simeone et al. (2011).
In addition, a molecular marker-based dominance relationship matrix (DG) was constructed. To build a dominance relationship matrix, we created an incidence matrix (S) for effects due to dominance S = {sij}, where sij was parameterized to be coded 1 if the genotype was heterozygous and 0 if the marker genotype was homozygous for either class. The matrix S was further standardized to have mean zero by using: sij = 1 − 2pjqj if the individual is heterozygous, sij = 0 if the individual has missing data, and sij = 0 − 2pjqj otherwise.
Using the above, we expanded the theory for the AG matrix to construct DG as
where the denominator is the sum of the variances of sij under Hardy–Weinberg equilibrium, and the other terms were previously defined. This extension to construct DG was also used by Su et al. (2012). Another parameterization has been proposed for the dominance genomic relationship matrix (Vitezica et al. 2013), which generates a different partition of the genetic variance. We also evaluated this parameterization and included the results in the supporting material.
Pedigree-based relationship matrices for additive (A) and dominance (D) effects were computed using standard methods (Lynch and Walsh 1998; Mrode 2005). Following existing theory (Cockerham 1954; Kempthorne 1954; Henderson 1985; Gianola and de los Campos 2008), the covariance matrices due to first degree epistatic terms were computed using Hadamard products (i.e., cell-by-cell product denoted as #) of the following form: (i) additive-by-additive interactions (A#A or AG#AG); (ii) dominance-by-dominance interactions (D#D or DG#DG); and (iii) additive-by-dominance interactions (A#D or AG#DG) for pedigree and marker-based methods, respectively.
Genetic analyses
All analyses were carried out in the software ASReml v3.0 (Gilmour et al. 2009), which fits mixed models with complex datasets using sparse matrix methods. ASReml is equipped with the residual maximum likelihood (REML) for variance component estimation using the average information algorithm (Gilmour et al. 1995).
Five models were fit using the pedigree-based matrices (models 1–5) and five using the marker-based matrices (models 6–10). These models range from a simple additive model to a full model including additive, dominance, and epistatic effects. The full model (i.e., model 5 or 10) is described below:
where y is the phenotypic HT response, β is a vector of fixed effects (i.e., silvicultural treatment and replicate), i ∼ N(0, Iσ2i) is a vector of the random incomplete block effects within replication, a ∼ N(0, C1σ2a) is a vector of random additive effects of individuals and C1 is a relationship matrix due to additive effects either from pedigree (A) or markers (AG), t1 ∼ N(0, C1⊗Iσ2t1) is a vector of random additive by silviculture-type interactions, d ∼ N(0, C2σ2d) is a vector of random individual dominance effects and C2 is a relationship matrix due to dominance effects that was computed either from pedigree (D) or markers (DG), t2 ∼ N(0, C2⊗Iσ2t2) is a vector of random dominance by silviculture-type interactions, ∼ N(0, C1#C1σ2iaa) is either a vector of random additive-by-additive interaction, a vector of random dominance-by-dominance interactions ∼ N(0, C2#C2σ2idd), or a vector of random additive-by-dominance interactions ∼ N(0, C1#C2σ2iad), and e ∼ N(0, Iσ2e) is a vector of random residual effects. Above, matrices X and Z1–Z6, are incidence matrices for fixed and random effects, respectively, and I denotes an identity matrix, and ⊗ and # represent the Kronecker and Hadamard (cell by cell) product, respectively.
Under the above model, the narrow-sense heritability can be estimated as the dominance to total variance ratio as the epistatic to total variance ratio as and the broad-sense heritability as The is the estimated additive variance, is the estimated dominance variance, and , , and are the total phenotypic, epistatic, and total genetic variance, respectively, that changed accordingly to the model being fit (Table 1).
Table 1. Summary of models, fitted effects, and relationship matrices used in the study.
| Model |
Relationship matrix used (information used, A, D = pedigree, AG, DG = markers) |
|||
|---|---|---|---|---|
| Number | Code | Additive | Dominance | Epistasis |
| 1 | P_A | A | ||
| 6 | M_A | AG | ||
| 2 | P_AD | A | D | |
| 7 | M_AD | AG | DG | |
| 3 | P_A#A | A | D | A#A |
| 8 | M_A#A | AG | DG | AG#AG |
| 4 | P_D#D | A | D | D#D |
| 9 | M_D#D | AG | DG | DG#DG |
| 5 | P_A#D | A | D | A#D |
| 10 | M_A#D | AG | DG | AG#DG |
Model comparisons
Models were compared using the Akaike information criterion (AIC) (Akaike 1974). Precision of variance components estimates, and their dependency, was assessed using the asymptotic variance–covariance matrix of estimates of variance parameters (V). The asymptotic sampling correlation matrix of estimates (F) was computed as where L is a diagonal matrix containing the diagonal elements of V. Inspection of the off-diagonal elements of the F matrix allows assessing sampling correlation of variance estimates. To have an overall assessment of dependency between the estimates, eigenvalues of F were examined. The standard error of the prediction (SEP) was estimated for each model as the square root of the prediction error variance (PEV), which is obtained by extracting the elements of the diagonal of the generalized inverse of the coefficient matrix from the linear mixed model equations (left hand side), and scaled by the error variance. In short form, the PEVs correspond to the (Mrode, 2005, p. 51).
Predictive ability and stability of the models in estimating breeding and genetic values were evaluated. The predictive ability of a model’s breeding value was defined as the correlation between the estimated breeding value and the phenotypic average of all the ramets (clones). These values were calculated when all the data were used without cross-validation (BV-all). The predictive ability of a model’s total genetic value (sum of BV-all, dominance effect, and epistatic effect) was defined as the correlation between the predicted total genetic value and the phenotype average of all the clones using all the data without cross-validation (GV-all). Prediction models were assessed under cross-validation (Kohavi 1995) to obtain predicted breeding value (BV-cv) and predicted total genetic value (GV-cv) with a random sub-sampling partitioning, fixed for all models. The stability of the predictive models were evaluated as the correlation between the BV-all and BV-cv, and between GV-all and GV-cv, and was defined as a measure of the dependency of the predictive breeding value on the phenotype. The mean square error (MSE) was calculated between BV-all and BV-cv within each model using standard methods. Finally, the capacity of the model to predict ranking position of the top 10% of the individuals, simulating a selection scenario, was evaluated as the correlation between the ranking position using the BV-all and the ranking position using the BV-cv.
Results
The genetic parameters and goodness-of-fit statistics, estimated for each model, are summarized in Table 2. Both P_A and M_A models had narrow-sense heritability (h2) >0.30. After including the dominance effect in the pedigree-based model (P_AD), h2 decreased by ∼26% and the dominance ratio (d2) estimate was small (0.06) and nonsignificant (2 × SE(d2) > 0.06). When the dominance effect was included with the molecular marker-based model (M_AD), the h2 decreased 47%, to 0.20, and d2 increased to 0.12. With the M_AD model, the dominance variance represents 60% of the additive value and 39% of the total genetic variation. We further extended these models to include the additive-by-additive, dominance-by-dominance, and additive-by-dominance first-order epistatic interaction factors in three separate models. In pedigree-based models, P_A#A, P_D#D, and P_A#D, the estimations of variance components for additive and dominance varied only slightly from those of the P_AD model (Supporting Information, Table S1). Moreover, epistasis estimates were zero in all three models. When the additive-by-additive, dominance-by-dominance, and additive-by-dominance interactions were added (models M_A#A, M_D#D, and M_A#D), the narrow-sense heritability dropped by >30% and the dominance ratio by 80%, compared to the M_AD model. The epistatic ratio (i2) was estimated at 0.15, 0.12, and 0.14 for the M_A#A, M_D#D, and M_A#D models, respectively (Table 2). The alternative parameterization for the dominance genomic relationship matrix proposed by Vitezica et al. (2013) showed similar results regarding the partition of additive and nonadditive effects (Table S2).
Table 2. Estimates of genetic parameters (standard errors in parentheses) and goodness-of-fit measures.
| P_A | M_A | P_AD | M_AD | P_A#A | M_A#A | P_D#D | M_D#D | P_A#D | M_A#D | |
|---|---|---|---|---|---|---|---|---|---|---|
| h2 SE(h2) | 0.32 (0.017) | 0.347 (0.018) | 0.235 (0.047) | 0.199 (0.035) | 0.233 (0.047) | 0.088 (0.044) | 0.228 (0.046) | 0.139 (0.036) | 0.231 (0.047) | 0.1251 (0.038) |
| d2 SE(d2) | NA | NA | 0.056 (0.033) | 0.117 (0.029) | 0.055 (0.033) | 0.023 (0.034) | 0.058 (0.032) | 0.009 (0.033) | 0.056 (0.043) | 0.006 (0.035) |
| i2 SE(i2) | NA | NA | NA | NA | 0.000 (0.000) | 0.154 (0.038) | 0.000 (0.000) | 0.121 (0.028) | 0.000 (0.000) | 0.135 (0.031) |
| H2 SE(H2) | 0.32 (0.017) | 0.347 (0.018) | 0.29 (0.021) | 0.316 (0.018) | 0.288 (0.021) | 0.264 (0.019) | 0.286 (0.021) | 0.269 (0.018) | 0.288 (0.021) | 0.266 (0.018) |
| LogL | −1299.40 | −1323.73 | −1295.37 | −1307.63 | −1294.83 | −1293.53 | −1293.90 | −1292.19 | −1294.38 | −1292.54 |
| AIC | 2606.80 | 2655.46 | 2602.74 | 2627.26 | 2605.66 | 2603.06 | 2603.80 | 2600.38 | 2604.76 | 2601.08 |
Each column represents a different model. See Table 1 for matrices included in the model.
Goodness-of-fit statistics show that inclusion of nonadditive effects improved slightly the model fit for pedigree-based models and substantially for marker-based models (Table 2). The marker-based models M_A#D and M_D#D yielded the best fit of the data; however, fitting differences among the more complex models were small. Thus, the dependency of the random component estimates was evaluated to further differentiate the best model.
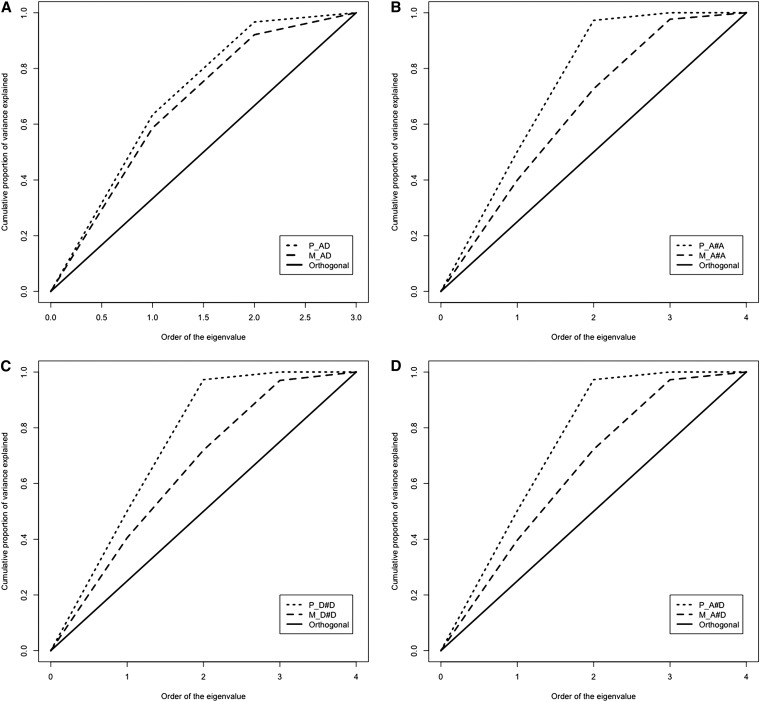
We studied the sampling correlation among the variance component estimates, to assess which of the nine models shows less dependency and thus partitioned the genetic variance better (Table S3). Figure 1 shows the cumulative proportion of variance explained by high order eigenvalues of the sampling variance–covariance matrix of estimates derived from models including additive-plus-dominance, additive-by-additive, dominance-by-dominance, and additive-by-dominance epistatic interactions, for pedigree- and marker-based models. As reference, the distribution of eigenvalues for a perfect orthogonal correlation matrix, representing the ideal model (all of the eigenvalues equal to 1) is included. In all cases, the marker-based cumulative distributions are closer to the orthogonal distribution, suggesting less dependency between estimates of variance components. Indeed, the sampling correlation between estimates of variance components due to additive and dominance effects decreases in absolute value from 0.90 with the P_AD to 0.70 with the M_AD model (Table S2). In general, all the marker-based models that include epistasis outperform their pedigree-based counterpart (Figure 1, B–D). Models M_D#D and M_A#D showed the smallest sampling correlations between additive and dominance/epistasis, with absolute correlation values <0.45 (Table S3).
Figure 1.
Cumulative proportion of variance explained by eigenvalues for models considering A plus D from pedigree (P_AD) vs. markers (M_AD) (A), A#A interaction from pedigree (P_A#A) vs. markers (M_A#A) (B), D#D interaction from pedigree (P_D#D) vs. markers (M_D#D) (C), and considering A#D interaction from pedigree (P_A#D) vs. markers (M_A#D) (D). The diagonal represents an orthogonal correlation matrix.
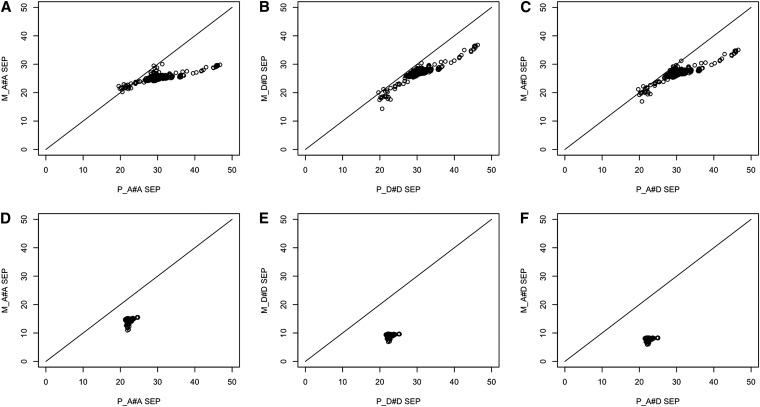
The standard error of the predictions (SEP) of BV and dominance value (DV) were compared for the pedigree and markers models including additive by additive, dominance by dominance, and additive by dominance (Figure 2). Values <45° reference line indicate that marker-based models have smaller SEPs. The SEPs for BVs from the marker-based models were smaller than the pedigree-based models in 99.2% of the cases (Figure 2, A–C). In the case of the SEPs of DVs, a clear advantage was observed for marker-based models (y-axis) over pedigree-based models (x-axis), with SEP on average 52% lower for the marker-based models (Figure 2, D–F).
Figure 2.
Standard error of the prediction (SEP) for pedigree-based (x-axis) against their counterpart marker-based models (y-axis). SEP for BV prediction model including A#A interaction (A), D#D interaction (B), and including A#D interaction (C). SEP for dominance value (DV) prediction model including A#A interaction (D), D#D interaction (E), and A#D interaction (F).
The predictive ability of breeding value and genetic value for the pedigree-based and marker-based models are shown in Table 3. The highest predictive ability for BV was obtained with the pedigree additive model (P_A). A slight decrease in the BV prediction ability was observed when nonadditive effects were included in the pedigree-based model (0.86), and a much larger decrease was observed for the marker-based models (0.76). All models evaluated reached similar GV predictive ability.
Table 3. Model of predictive ability and stability.
| Model | Predictive ability |
Predictive stability |
|||
|---|---|---|---|---|---|
| Breeding value* | Genetic value** | Breeding value | MSE (BV) | 10% rank cor (BV) | |
| P_A | 0.89 | — | 0.64 | 1335.67 | 0.17 |
| M_A | 0.87 | — | 0.66 | 1294.23 | 0.17 |
| P_AD | 0.86 | 0.89 | 0.72 | 681.53 | 0.12 |
| M_AD | 0.82 | 0.88 | 0.74 | 418.83 | 0.31 |
| P_A#A | 0.86 | 0.89 | 0.73 | 669.99 | 0.15 |
| M_A#A | 0.76 | 0.89 | 0.85 | 82.80 | 0.46 |
| P_D#D | 0.86 | 0.89 | 0.73 | 638.58 | 0.18 |
| M_D#D | 0.77 | 0.89 | 0.86 | 161.78 | 0.43 |
| P_A#D | 0.86 | 0.89 | 0.73 | 657.22 | 0.16 |
| M_A#D | 0.76 | 0.89 | 0.86 | 208.15 | 0.42 |
Correlation between the phenotypic average of all the ramets (phe) and BV-all, and
correlation between phe and total genetic value (GV-all = BV-all + DV + epistatic value). Stability in a cross-validation; correlation between BV-all and BV-cv, mean square error (MSE) and correlation of ranking positions for the top 10% individuals [10% rank cor(BV)].
Predictive stability can be viewed as a measure of how much the prediction of the breeding value and genetic value using all the data (BV-all and GV-all) depend on the individual phenotype (Table 3). Predictions based on models with markers are more stable than those derived from pedigree models (3% increase when comparing M_A to P_A). In the pedigree-based models, inclusion of nonadditive effects increased the stability to predict BV by 13, 14, and 14% for P_A#A, P_D#D, and P_A#D, respectively, while inclusion of nonadditive effects in the more complex marker-based models, increased the BV prediction stability by >29% when compared with the M_A and by >33% when compared to P_A. The mean square error (MSE) decreased by ∼50% from the additive models (P_A) to the more complex pedigree-based models. The addition of nonadditive effects to the marker-based models decreased the MSE even further by >68% and up to 94% decrease in the case of model P_A#A (Table 3).
In a breeding program, it is important to predict the trend and magnitude of the complete set of individuals in the population; however, it is often more important to predict the best performing individuals (potential selections). Here we ranked all individuals based on BV-all and BV-cv and evaluated the ranking correlation of the top 10%, emulating the selection of the top 10% of genotypes (Table 3). When the pedigree-based matrix was replaced by the marker-based matrix in the additive models (P_A and M_A), the capacity to predict the top 10% remained the same. However, this capacity increased substantially for the more complex marker-based models where the predictive stability of the top 10% of genotypes increased 82–170% (Table 3).
Discussion
Here we assessed the use of marker- and pedigree-based models to separate additive from nonadditive variances for height, in a structured population of loblolly pine. We showed that the two approaches are dramatically distinct in their capacity to properly partition the genetic variance into its various components, with marker-based models being significantly more effective in accounting for nonadditive variances. In the pedigree-based models, inclusion of nonadditive effects decreased the estimated narrow-sense heritability by 26%. This result is expected because depending on the distribution of allele frequencies, a sizable proportion of variance due to nonadditive effects can be manifest as additive variance (Lu et al. 1999; Zuk et al. 2012). In marker (pedigree) models 71% (57%) of the decrease in additive variance was captured by the dominance variance, suggesting that indeed, dominance is making a substantial contribution to the estimated additive variance obtained when dominance is ignored. This phenomena has been postulated theoretically (e.g., Falconer and Mackay 1996, p. 126) and observed in multiple studies (Wei and Van Der Werf 1993; Winkelman and Peterson 1994; Rodriguez-Almeida et al. 1995; Pante et al. 2002). In addition, when pedigree-based models included nonadditive effects, the conclusions were not different from what has been commonly observed that nonadditive effects represent a small fraction of the total genetic variation (Isik et al. 2003; Costa E Silva et al. 2004; Baltunis et al. 2007; Araujo et al. 2012). In contrast, marker-based models with additive and nonadditive effects yield a substantially different variance partitioning than their counterparts using the pedigree models. The additive variance decreased as dominance was included in the model and it further decreased when dominance and epistasis were considered. These models indicate that, for this population and trait, nonadditive effects are as important as additive effects, and dramatically larger than predicted by the pedigree-based matrices. These changes in the magnitude of variance components have already been observed when the relationship matrix derived from markers is used instead of the pedigree-based relationship matrix, in the context of additive genomic selection models (Lee et al. 2010).
The value for AIC varied modestly for the best models, with no clear advantage of one model relative to the others. This is not surprising; if additive effects capture part of the effects due to dominance and epistasis, the additive model should not suffer much if these components are omitted. However, these models varied considerably in the partitioning of the genetic variance components, thus changing not only the inference but also the potential decisions taken in the breeding strategy.
We also assessed the dependency of the random effects estimates to discriminate the best model, given the small differences for AIC in the different models. The level of confounding between components was very different in the pedigree- and marker-based models. The most unambiguous dissection of the genetic variance occurs when estimates of variance components are uncorrelated, i.e., the sampling correlation among the model effects will be closer to zero and all eigenvalues of this correlation matrix close to one (Hill 2010). In the models that included additive and dominance, and additive, dominance, and epistatic effects, the correlation matrices indicated that those derived from molecular markers partitioned the genetic effects more precisely, although the partition is still not fully orthogonal. The parameterizations of these paired models were identical, except for the origin of the relationship matrices (pedigree or marker based). The limited capacity of pedigree-based models to partition these components is not surprising, as all relationship matrices are derived from the pedigree additive relationship matrix (Mrode 2005) and, therefore, are strongly correlated (Visscher 2009). The models M_A#A, M_D#D, and M_A#D had the lowest correlation between additive and nonadditive, showing a partition substantially better than that of pedigree-based models (Table S3). These results support the finding that pedigree-based models are inadequate in separating the additive from nonadditive effects, as their results are comparable to those of additive models (Hill et al. 2008). On the other hand, the use of the matrix derived from markers has already been related to a better capacity to separate random effects in a model (Lee et al. 2010). Thus, we conclude that the use of the marker-based relationship matrices increase substantially the capacity to separate additive and nonadditive genetic effects.
Assessment of prediction accuracy further support the conjecture that in pedigree-based models, additive components can capture a large proportion of the variance due to interaction terms (Hill et al. 2008). Consequently, there is limited gain in this scenario, by including nonadditive effects. On the other hand, for marker-based models, the ability to predict the mean phenotype with the BV decreased when nonadditive effects were included in the model, and the maximum predictability (0.89) was only reached when additive and nonadditive values were considered together (GV). This indicates that pedigree-based models potentially overestimate the additive effects, which is likely to be due to an inflated additive variance estimate that also represents some of the nonadditive components. Inflation from epistasis, for example, falls apart as recombination breaks down favorable combinations of alleles. This is a problem for breeding programs because overestimates of BV inflates genetic gains, but the portion due to nonadditive effects is transient and cannot be captured if controlled sexual reproduction is used. Additionally, the genetic architecture of the trait will be predicted to be simpler than it actually is.
In breeding programs the true breeding value is never known. Thus, the prediction models including all the available data (BV-all) is usually used as the best BV estimation. We evaluated the stability of BV estimates by comparing the results obtained with all data, with the results from cross-validation for pedigree- and marker-based models. This is a measure of the influence of an individual’s phenotype on the predicted breeding value. We observed that models with nonadditive relationship matrices are more stable and produce estimates of breeding values in independent sets that are more similar to the BV-all. The inclusion of nonadditive relationship matrices yields models that predict BVs more stably than additive pedigree-based (animal model) and marker-based models (traditional G-BLUP). In addition, in this cross-validation scheme, the MSE of the model M_A#A decreased >15- and 8-fold when compared with both additive models and full pedigree-based models, respectively. These results indicate that, for this trait, a considerable increase in the stability cannot be reached simply by replacing the A matrix by the AG matrix but also needs to incorporate nonadditive effects in the model.
Overall, our study supports the hypothesis that additive effects can capture a large proportion of the genetic variance from dominance and epistasis. This is in part due to the fact that, in breeding populations, additive and nonadditive genetic components are not typically independent. However, we also show that with relationship matrices derived from markers, the genetic variances were partitioned more precisely than using only pedigree information. Moreover, our estimates suggest that in this population, for tree height, the additive and nonadditive components of the genetic variance are similar in magnitude. While further research is needed in other species, traits, and populations, we show that variance estimates can be inadequately estimated if only pedigree information is used. This study improves our current understanding of the genetic control and architecture of a quantitative trait and should be considered when developing effective breeding strategies.
Supplementary Material
Acknowledgments
The authors thank members of the Forest Biology Research Cooperative at the University of Florida for their support in establishing, maintaining, and measuring the field trial used in this study. The work was supported by the National Science Foundation Plant Genome Research Program (award no. 0501763), the Foundational Program (award no. 2013-67013-21159), the Department of Energy (award no. 2013-67009-21200), and the Plant Breeding and Education Program (award no. 2010-85117-20569) from the US Department of Agriculture, National Institute of Food and Agriculture.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171322/-/DC1.
Communicating editor: N. Yi
Literature Cited
- Akaike H., 1974. New look at statistical-model identification. Transactions on Automatic Control. AC19: 716–723. [Google Scholar]
- Araujo J. A., Borralho N. M. G., Dehon G., 2012. The importance and type of non-additive genetic effects for growth in Eucalyptus globulus. Tree Genet. Genomes 8: 327–337. [Google Scholar]
- Baltunis B. S., Huber D. A., White T. L., Goldfarb B., Stelzer H. E., 2007. Genetic gain from selection for rooting ability and early growth in vegetatively propagated clones of loblolly pine. Tree Genet. Genomes 3: 227–238. [Google Scholar]
- Baltunis B. S., Martin T. A., Huber D. A., Davis J. M., 2008. Inheritance of foliar stable carbon isotope discrimination and third-year height in Pinus taeda clones on contrasting sites in Florida and Georgia. Tree Genet. Genomes 4: 797–807. [Google Scholar]
- Baltunis B. S., Wu H. X., Dungey H. S., Mullin T. J. T., Brawner J. T., 2009. Comparisons of genetic parameters and clonal value predictions from clonal trials and seedling base population trials of radiata pine. Tree Genet. Genomes 5: 269–278. [Google Scholar]
- Cockerham C. C., 1954. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics 39: 859–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E Silva J., Borralho N. M. G., Potts B. M., 2004. Additive and non-additive genetic parameters from clonally replicated and seedling progenies of Eucalyptus globulus. Theor. Appl. Genet. 108: 1113–1119. [DOI] [PubMed] [Google Scholar]
- Costa E Silva J., Borralho N. M. G., Araujo J. A., Vaillancourt R. E., Potts B. M., 2009. Genetic parameters for growth, wood density and pulp yield in Eucalyptus globulus. Tree Genet. Genomes 5: 291–305. [Google Scholar]
- Crossa J., Campos G. D. L., Perez P., Gianola D., Burgueno J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Naya H., Gianola D., Crossa J., Legarra A., et al. , 2009. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2012. Whole genome regression and prediction methods applied to plant and animal breeding. Genetics 193(2): 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert A. J., van Heerwaarden J., Wegrzyn J. L., Nelson C. D., Ross-Ibarra J., et al. , 2010. Patterns of population structure and environmental associations to aridity across the range of Loblolly Pine (Pinus taeda L., Pinaceae). Genetics 185: 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Addison Wesley Longman, Essex, England. [Google Scholar]
- Foster G. S., Shaw D. V., 1988. Using clonal replicates to explore genetic-variation in a perennial plant-species. Theor. Appl. Genet. 76: 788–794. [DOI] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., 2008. Inferring genetic values for quantitative traits non-parametrically. Genet. Res. 90: 525–540. [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Thompson R., Cullis B. R., 1995. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51: 1440–1450. [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R., 2009. ASReml User Guide Release 3.0. VSN International, Hemel Hempstead, UK. [Google Scholar]
- Habier D., Tetens J., Seefried F.-R., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1985. Best linear unbiased prediction of nonadditive genetic merits in noninbred populations. J. Anim. Sci. 60: 111–117. [Google Scholar]
- Heslot N., Yang H.-P., Sorrells M. E., Jannink J.-L., 2012. Genomic selection in plant breeding: a comparison of models. Crop Sci. 52: 146–160. [Google Scholar]
- Hill W., 2010. Understanding and using quantitative genetic variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W., Goddard M., Visscher P., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4(2): e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isik F., Li B. L., Frampton J., 2003. Estimates of additive, dominance and epistatic genetic variances from a clonally replicated test of loblolly pine. For. Sci. 49: 77–88. [Google Scholar]
- Isik F., Goldfarb B., LeBude A., Li B. L., McKeand S., 2005. Predicted genetic gains and testing efficiency from two loblolly pine clonal trials. Can. J. Forest Research 35: 1754–1766. [Google Scholar]
- Kempthorne O., 1954. The correlation between relatives in a random mating population. Proc. R. Soc. Lond. B Biol. Sci. 143: 103–113. [PubMed] [Google Scholar]
- Kohavi, R., 1995 The power of decision tables. Machine Learning: ECML-95. pp. 174–189. [Google Scholar]
- Lee S. H., Goddard M. E., Visscher P. M., van der Werf J. H. J., 2010. Using the realized relationship matrix to disentangle confounding factors for the estimation of genetic variance components of complex traits. Genet. Sel. Evol. 42: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P. X., Huber D. A., White T. L., 1999. Potential biases of incomplete linear models in heritability estimation and breeding value prediction. Can. J. Forest Research 29: 724–736. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Mrode, R. A., 2005 Linear Models for the Prediction of Animal Breeding Values. CABI Publishing Series, Cambridge, UK. [Google Scholar]
- Mullin T. J., Morgenstern E. K., Park Y. S., Fowler D. P., 1992. Genetic-parameters from a clonally replicated test of black spruce (Picea mariana). Can. J. Forest Research 22: 24–36. [Google Scholar]
- Muñoz P., Resende M. F., Huber D. A., Quezada T., Resende M. D., et al. , 2013. Genomic relationship matrix for correcting pedigree errors in breeding populations: impact on genetic parameters and genomic selection accuracy. Crop Sci. 54: 1115–1123. [Google Scholar]
- Pante M. J., Gjerde B., McMillan I., Misztal I., 2002. Estimation of additive and dominance genetic variances for body weight at harvest in rainbow trout Oncorhynchus mykiss Oncorhynchus mykiss. Aquaculture 204: 383–392. [Google Scholar]
- Palucci V., Schaeffer L. R., Miglior F., Osborne V., 2007. Non-additive genetic effects for fertility traits in Canadian Holstein cattle. Genet. Sel. Evol. 39: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Visscher P. M., Goddard M. E., 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11: 800–805. [DOI] [PubMed] [Google Scholar]
- Resende M. F. R., Jr, Muñoz P., Acosta J. J., Peter G. F., Davis J. M., et al. , 2012a Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol. 193: 617–624. [DOI] [PubMed] [Google Scholar]
- Resende M. F., Jr, Muñoz P., Resende M. D., Garrick D. J., Fernando R. L., et al. , 2012b Accuracy of genomic selection methods in a standard data set of Loblolly Pine (Pinus taeda L.). Genetics 190: 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Almeida F. A., Van Vleck L. D., Wilham R. L., Northcutt S. L., 1995. Estimation of non-additive genetic variances in three synthetic lines of beef cattle using an animal model. J. Anim. Sci. 73: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Simeone R., Miztal I., Aguilar I., Legarra A., 2011. Evaluation of the utility of the diagonal elements of the genomic relationship matrix as a diagnostic tool to detect mislabelled genotyped animals in a broiler chicken population. J. Anim. Breed. Genet. 12895): 386–393. [DOI] [PubMed] [Google Scholar]
- Su G., Christensen O. F., Ostersen T., Henryon M., Lund M. S., 2012. Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS ONE 7(9): e45293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwerf J. H. J., Deboer W., 1989. Influence of nonadditive effects on estimation of genetic-parameters in dairy-cattle. J. Dairy Sci. 72: 2606–2614. [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Veerkamp R. F., Mulder H. A., Thompson R., Calus M. P. L., 2011. Genomic and pedigree-based genetic parameters for scarcely recorded traits when some animals are genotyped. J. Dairy Sci. 94: 4189–4197. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., 2009. Whole genome approaches to quantitative genetics. Genetica 136: 351–358. [DOI] [PubMed] [Google Scholar]
- Vitezica Z.G., Varona L., Legarra A. 2013. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195: 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., van der Werf J. H. J., 1993. Animal model estimation of additive and dominance variances in egg production traits of poultry. J. Anim. Sci. 71: 57–65. [DOI] [PubMed] [Google Scholar]
- Williams E. R., Matheson A. C., Harwood C. E., 2002. Experimental Design and Analysis for Tree Improvement, Ed. 2 Commonwealth Scientific and Industrial Research Organization, Melbourne. [Google Scholar]
- Winkelman A. M., Peterson R. G., 1994. Genetic parameters heritabilities, dominance ratios, and genetic correlations for body weight and length of chinook salmon after 9 and 22 months of saltwater rearing. Aquaculture 125: 30–36. [Google Scholar]
- Wu R. L., 1996. Detecting epistatic genetic variance with a clonally replicated design: models for low- vs. high-order non allelic interaction. Theor. Appl. Genet. 93: 102–109. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.