The sequenced genomes of three conifer species provide the first assemblies of gymnosperm genomes and perspectives on the largest plant genomes sequenced to date.
Abstract
Insights from sequenced genomes of major land plant lineages have advanced research in almost every aspect of plant biology. Until recently, however, assembled genome sequences of gymnosperms have been missing from this picture. Conifers of the pine family (Pinaceae) are a group of gymnosperms that dominate large parts of the world’s forests. Despite their ecological and economic importance, conifers seemed long out of reach for complete genome sequencing, due in part to their enormous genome size (20–30 Gb) and the highly repetitive nature of their genomes. Technological advances in genome sequencing and assembly enabled the recent publication of three conifer genomes: white spruce (Picea glauca), Norway spruce (Picea abies), and loblolly pine (Pinus taeda). These genome sequences revealed distinctive features compared with other plant genomes and may represent a window into the past of seed plant genomes. This Update highlights recent advances, remaining challenges, and opportunities in light of the publication of the first conifer and gymnosperm genomes.
Conifers are the most widely distributed group of gymnosperms, with 600 to 630 species in 69 genera, including 220 to 250 species of the Pinaceae family (Wang and Ran, 2014). Coniferous forests cover an estimated 39% of the world’s forests (Armenise et al., 2012). Conifers dominate many natural and planted forests in the northern hemisphere and are also planted as exotics for commercial forestry in the southern hemisphere. The importance of conifers for global ecosystem services, their value for forestry-dependent economies, and their contrasting biology with angiosperms are major drivers behind efforts to understand the complex structure, functions, and evolution of their genomes. However, owing to their nonmodel system attributes (i.e. slow-growing and long-lived life history traits), extremely large genome size (Fig. 1), and repeat-rich genome sequence with repeats mostly in the form of transposable elements, no reports of a conifer genome assembly, or any gymnosperm genome for that matter (Soltis and Soltis, 2013), were published until recently. Following early releases of the white spruce (Picea glauca) and loblolly pine (Pinus taeda) genome sequences in public databases (e.g. National Center for Biotechnology Information and http://dendrome.ucdavis.edu/treegenes/), a series of articles described the first conifer genome assemblies for Norway spruce (Picea abies; Nystedt et al., 2013) and interior white spruce, a genetic admix of white spruce (Birol et al., 2013) and loblolly pine (Neale et al., 2014; Zimin et al., 2014). Norway spruce is a prominent forest tree in northern Europe. White spruce is a dominant tree species across the large Canadian forest landscape. Loblolly pine dominates commercial forestry in the southeastern United States. White spruce, Norway spruce, and loblolly pine represent some of the most economically important conifers worldwide, and they are the subjects of important tree improvement/breeding programs (Mullin et al., 2011). This Update highlights significant insights obtained from these genomes as well as some ongoing challenges and recent developments in conifer genomics.
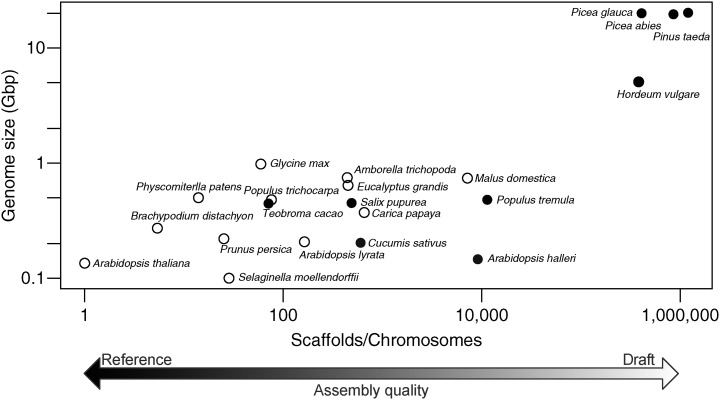
Figure 1.
Size and assembly of conifer genomes compared with other plant genomes. Genome size is plotted against the number of scaffolds divided by the haploid chromosome number for a range of plant species. As such, an assembly that reconstructs a genome with perfect contiguity will have a value of 1, and values greater than 1 represent increasing genome fragmentation. Genome assemblies that utilized Sanger sequencing either in full or in part are represented as white circles. Assemblies constructed using only next generation sequencing technologies are represented as black circles. Both axes are plotted on a log10 scale. With the exception of Populus tremula, Hordeum vulgare, and the three conifer genomes, all genomes were obtained from the Phytozome resource (version 10; http://phytozome.jgi.doe.gov/). The early release draft assembly of P. tremula was obtained from the PopGenIE.org FTP resource (ftp://popgenie.org/popgenie/UPSC_genomes/UPSC_Draft_Assemblies/Current/Genome/) and H. vulgare ‘Morex’ from the Munich Information Center for Protein Sequences barley genome database FTP resource (ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/sequences/). The conifer genomes are detailed by Birol et al. (2013), Nystedt et al. (2013), and Zimin et al. (2014).
CONIFER GENOME ASSEMBLIES
Characteristics of the Norway spruce, white spruce, and loblolly pine genomes and the corresponding sequence assemblies are summarized in Table I. The assembly statistics shown in Table I reflect sequences that are at least 500 bp and are calculated for the published assemblies using the fac utility within the ABySS package (Simpson et al., 2009).
Table I. Characteristics of the three conifer genomes and the corresponding sequence assemblies published for white spruce (Birol et al., 2013), Norway spruce (Nystedt et al., 2013), and loblolly pine (Zimin et al., 2014).
Contiguity statistics reflect sequences of length 500 bp or longer, and the total reconstruction figures represent the scaffold-level assemblies with undetermined bases (n) not contributing to the counts.
| Characteristic | Norway Spruce | White Spruce | Loblolly Pine |
|---|---|---|---|
| C value (1C pg)a | 20.01 | 16.15 | 22.10 |
| Genome size (Gb)b | 19.6 | 20.8 | 22 |
| Karyotype | 2n = 24 | 2n = 24 | 2n = 24 |
| Assembly | |||
| Contig N50 (kb) | 0.6 | 5.4 | 8.2 |
| Scaffold N50 (kb) | 0.7 | 22.9 | 66.9 |
| No. of scaffolds (million) | 10.3 | 7.1 | 14.4 |
| Total reconstruction (Gb) | 12.0 | 20.8 | 20.1 |
| Gene space | |||
| Predicted No. of genes | 70,968 | 56,064 | 50,172 |
| Reconstruction of 248 core eukaryotic genesc (complete/at least partial) | 124/189 | 95/184 | 185/203 |
Amount of DNA in picograms contained in a haploid cell as reported by Murray et al. (2012). bAs reported by Birol et al. (2013), Nystedt et al. (2013), and Zimin et al. (2014). cAs described by Parra et al. (2009).
The Norway spruce genome project (Nystedt et al., 2013) assembled a draft of the 19.6-Gb nuclear genome using a hierarchical sequencing strategy combining fosmid pools with both haploid and diploid whole-genome shotgun (WGS) and transcriptome (RNA sequencing [RNA-Seq]) sequence data. The resulting assembly (P. abies 1.0) included 4.3 Gb in more than 10-kb scaffolds, with this subset of the assembly covering an estimated 63% of all protein-coding genes. The total reconstruction reaches 10 Gb, when scaffolds of 500 bp or longer are considered. Transcript assemblies of the sequenced Norway spruce individual as well as full-length complementary DNA (cDNA) reference sequences from Sitka spruce (Picea sitchensis; Ralph et al., 2008) were used to assess gene space contiguity of the genome assembly.
The white spruce genome project (Birol et al., 2013) assembled a draft genome of 20.8 Gb consisting of 4.9 million scaffolds, with a N50 scaffold size of 20.4 kb. The sequenced individual (PG29) represents an elite parent in an advanced breeding program of the British Columbia Ministry of Forests. It originated from a western Canadian white spruce population near Prince George, British Columbia. This population was recently found to have ancient features of a genetic admix of white spruce with Engelmann spruce (Picea engelmannii) and Sitka spruce (De La Torre et al., 2014a; Hamilton et al., 2014). The white spruce genome was reconstructed entirely from WGS sequences of multiple libraries and assembled using the ABySS software (Simpson et al., 2009). Another novelty of the approach involved complementation of high-coverage short-read data from the HiSeq2000 platform with low coverage of longer reads from a modified MiSeq platform to support the assembly process. Increased read length together with paired-end reads from longer fragments had a major impact on assembly contiguity through their effect on the optimum k-mer length (the length cutoff for specific read-to-read alignments during assembly). The optimum k-mer length was higher (k = 109 bp) when using both short and long reads compared with using only short reads (k = 101 bp). The assembly quality was evaluated using white spruce bacterial artificial chromosome sequences (Hamberger et al., 2009; Keeling et al., 2010) and Sitka spruce and white spruce cDNAs (Ralph et al., 2008; Rigault et al., 2011).
The loblolly pine genome project (Neale et al., 2014; Wegrzyn et al., 2014; Zimin et al., 2014) completed a draft assembly of 17.6 Gb consisting of 2.1 million scaffolds and an N50 scaffold size of 72.9 kb. Most of the sequence data were from WGS sequencing of a single megagametophyte (haploid female gametophyte). Longer fragment mate-pair libraries were made with diploid DNA from needles of the maternal tree to produce long-range linking libraries. Two methods were used to link the assembly over large distances: mate-pair jumping libraries and fosmid ends or DiTags. The genome assembly was aided by condensing high numbers of paired-end reads into a smaller set of superreads, making the assembly computationally feasible, and by supporting the additional scaffolding by transcriptome assemblies.
A major focus when assembling the very large and repeat-rich conifer genomes using short-read sequencing technologies is to reconstruct the gene space as contiguously as possible. This has been particularly challenging because of the level of overall assembly fragmentation (e.g. approximately one-third of genes appeared to be fragmented across more than one scaffold in the Norway spruce genome assembly), combined with the challenge of distinguishing assembly fragments of functional genes from pseudogenes (Nystedt et al., 2013; Wegrzyn et al., 2014). To maximize gene space contiguity, the three conifer genome assemblies implemented scaffolding approaches using information from de novo transcript assemblies or unassembled RNA-Seq data.
The Norway spruce genome assembly (Nystedt et al., 2013) used a different strategy to utilize RNA-Seq data for scaffolding. Raw, unassembled paired-end RNA-Seq reads from a collection of 22 samples were first digitally normalized and subsequently stringently aligned to the assembly, which had already been scaffolded using paired-end and long-insert mate-pair libraries. To avoid introducing assembly errors, only uniquely mapped read pairs were used as scaffolding evidence, resulting in the formation of 11,528 new scaffolds and improving the representation of genes fully contained within a single assembly scaffold. Further improvements to transcriptome-based scaffolding are under development and evaluation. For the loblolly pine genome assembly, additional long-range paired reads, particularly DiTag pairs, were used to increase scaffold lengths and maximize gene space contiguity (Zimin et al., 2014). In addition, a large number of mapped single-nucleotide polymorphisms (SNPs) will allow the placement of loblolly pine genome sequence scaffolds onto the 12 chromosomes providing concrete physical anchors for the genome assembly.
GENE SPACE ANNOTATION
Despite limitations of gene space contiguity, gene space annotations using both automated and manual approaches have been developed for the three sequenced conifer species. In Norway spruce, automated annotation using a combination of AUGUSTUS (Stanke et al., 2004) and EUGENE (Schiex et al., 2001) identified a total of 70,968 genes. Of these, 28,354 were classified as high confidence based on EST or transcript support (Nystedt et al., 2013). In white spruce, automated annotation with MAKER (Holt and Yandell, 2011) identified 56,064 genes. Completion of these annotations, although automated, requires weeks to months of devoted computer cluster time. MAKER-P (Campbell et al., 2014) allows optimization and improvement of the automated annotation process. MAKER-P was used with the loblolly pine assembly to produce ab initio gene predictions together with SNAP (Korf, 2004) and AUGUSTUS. After applying multiexon and protein domain filters to remove abundant pseudogenes and fragmented genes, 50,172 genes were identified in loblolly pine. The majority of these (97%) align to known genes in other species, and 15,653 were reported as high confidence (Wegrzyn et al., 2014).
UNIQUE FEATURES OF CONIFER GENOMES
Gymnosperms and angiosperms differ in a number of features, of which contrasting reproductive biology and water-conducting systems are most prominent. The publication of three conifer genomes provided insights into the composition and structure of gymnosperm genomes and their differences from angiosperms. The genome of the basal angiosperm Amborella trichopoda, the sole living species of the sister lineage to all other extant flowering plants, provides a key reference for gymnosperm and angiosperm comparisons (Amborella Genome Project, 2013). In brief, the gymnosperm genomes as represented by conifers are extremely large, although the number of protein-coding sequences is not proportionally larger compared with sequenced angiosperm genomes. The conifer genomes contain very long introns and an abundance of repeat-rich content mostly in the form of transposable elements and a smaller contribution of tandem repeats. Transposable elements are mainly long terminal repeat-retrotransposons (LTR-RTs), with fewer non-long terminal repeat (LTR) retrotransposons, such as long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), and DNA transposons.
Accumulation of Transposable Elements Increased the Size of Gymnosperm Genomes
In contrast to angiosperms, in which genomes have been shaped by several whole-genome and smaller duplication events, there is no evidence of recent whole-genome duplication in gymnosperms and the basal angiosperm A. trichopoda (Kovach et al., 2010; Amborella Genome Project, 2013; Nystedt et al., 2013). Only one ancient whole-genome duplication event that predated the angiosperm-gymnosperm split (approximately 350 million years ago) is known to have occurred (Jiao et al., 2011; Amborella Genome Project, 2013). Instead, it appeared that gymnosperm genome size increased over millions of years as a result of a massive accumulation of LTR-RTs such as the Ty3/Gypsy, Ty1/Copia, and Gymny superfamilies, together with limited removal of transposable elements through unequal recombination (Nystedt et al., 2013). These findings support previous studies using bacterial artificial chromosome clones constructed for species such as white spruce, loblolly pine, maritime pine (Pinus pinaster), and bald cypress (Taxodium distichum), which found that abundant and diverse retrotransposons were the main components of the nongenic portion and accounted for the large genome size in pines (Kovach et al., 2010; Magbanua et al., 2011) and spruce (Hamberger et al., 2009). Homology analysis revealed that 62% of the loblolly pine genome is composed of retrotransposons, of which 70% are LTR-RTs, mainly Gypsy and Copia (Neale et al., 2014; Wegrzyn et al., 2014). Non-LTR retrotransposons including LINEs and SINEs are usually found at lower frequencies in conifers (Wegrzyn et al., 2014). Loblolly pine LINEs accounted for 2.35% of the genome, higher than previously reported for the same species (Wegrzyn et al., 2013) and for other conifers such as Scots pine (Pinus sylvestris; 0.52%) and Norway spruce (0.96%; Nystedt et al., 2013). Very low ratios of SINEs were found in gymnosperms and in some angiosperm species such as Tausch's goatgrass (Aegilops tauschii; Jia et al., 2013; Nystedt et al., 2013; Wegrzyn et al., 2013, 2014). As in the Norway spruce genome, the average age of LTR-RTs (Gypsy and Copia) in A. trichopoda was much older than in all other angiosperm species (the insertion time of LTRs in A. trichopoda was approximately 40 million years ago; Amborella Genome Project, 2013), possibly reflecting the ancient nature of this species. Comparison of Norway spruce and five additional gymnosperm genomes, of Scots pine, Siberian fir (Abies sibirica), common juniper (Juniperus communis), European yew (Taxus baccata), and gnetum (Gnetum gnemon), sequenced at low coverage, indicated that the diversity of transposable elements is shared among gymnosperms (Nystedt et al., 2013).
Besides transposable elements, tandem repeats (minisatellites, microsatellites, and satellites) also contribute to the highly repetitive content of conifer genomes; however, they represent less than 3% of these genomes (Wegrzyn et al., 2014). Main components of telomeres and centromeres, tandem repeats are involved in epigenetic responses on heterochromatin and gene expression regulation (Wegrzyn et al., 2014).
Long Introns
A notable characteristic of the predicted gene structures in the three sequenced conifer genomes is the abundance of long introns, with the largest observed intron lengths among the longest found in any plant species, only comparable with the repeat-rich genomes of A. trichopoda, grapevine (Vitis vinifera), and corn (Zea mays). Introns of more than 20 kb and up to 68 kb in length were found in the Norway spruce genome (Nystedt et al., 2013). Based on a high-confidence set of 15,653 transcripts, 48,720 introns, an average intron length of 2.4 kb, and a maximum length of 158 kb were found in the loblolly pine genome (Wegrzyn et al., 2014). Long introns appear largely conserved between pine and spruce (Sena et al., 2014). However, overall, the presence of very long introns does not appear to be a major contributor to the very large conifer genome size. In loblolly pine, introns contained 54.28% retrotransposons (34.57% of these were LTR-RTs) and 3.52% DNA transposons (Wegrzyn et al., 2014). Higher repetitive content was found in longer introns in loblolly pine, since intron expansion can be partially attributed to the generation of repeats (Wegrzyn et al., 2014).
Noncoding and Short RNAs
Small noncoding RNAs and short RNAs (sRNAs), which contribute to the epigenetic silencing of transposable elements, show differences in gymnosperms and angiosperms (Morin et al., 2008; Nystedt et al., 2013). RNA sequencing in Norway spruce revealed previously uncharacterized sRNAs (Nystedt et al., 2013). Although thought to be absent in gymnosperms (Morin et al., 2008; Yakovlev et al., 2010), the recent genome analysis found 24-nucleotide sRNAs in gymnosperms, but at lower abundance than in angiosperms (Nystedt et al., 2013). The presence of 24-nucleotide sRNAs was highly specific to reproductive tissues, largely associated with transposable elements. In contrast, the diversity of 21-nucleotide sRNAs was higher, with many repeat-associated sRNAs. Novel sRNAs of uncharacterized function were also reported in lodgepole pine (Pinus contorta; Morin et al., 2008) and in Norway spruce (Yakovlev et al., 2010; Nystedt et al., 2013).
Long noncoding RNAs involved in RNA processing or transcriptional and posttranscriptional gene regulation exhibited more sample-specific expression distributions than protein-coding transcripts. They were shorter and contained fewer exons, as also reported in humans (Nystedt et al., 2013). Analyses of the Norway spruce transcriptome data suggested that long noncoding RNAs are prevalent, yet largely uncharacterized, in conifers (Nystedt et al., 2013).
Reproductive Biology, Water-Conducting Systems, and Secondary Metabolism
Gymnosperms and angiosperms differ in their reproductive systems and water-conducting xylem tissues, and conifers also have characteristic secondary metabolism. Phylogenetic analysis identified gymnosperm-specific patterns of gene duplications and evolutionary trajectories for several different gene families (Nystedt et al., 2013; Zerbe et al., 2013; Neale et al., 2014). These include gene families related to cell wall and xylem formation, such as cellulose synthases and VASCULAR NAC DOMAIN, and gene families for transcription factors, such as KNOTTED-LIKE HOMEOBOX CLASS1. Examples of lineage-specific expansions of defense-related genes of secondary metabolism are gymnosperm-specific families of cytochrome P450s and terpene synthases (TPS-d). Gymnosperm-specific evolutionary trajectories were found for FLOWERING LOCUS T/TERMINAL FLOWER1-like genes, putative regulators of reproduction and seasonal growth cessation and bud set. The loblolly pine genome also revealed expansion of a particular class of Toll-interleukin receptor/nucleotide-binding/Leucine-rich repeat genes, potentially involved in pathogen resistance (Neale et al., 2014).
Conifer lignin biosynthesis is a feature highlighted by Neale et al. (2014) in the analysis of the loblolly pine genome assembly. The xylem of conifers differs from that of woody angiosperms in lignin composition (guaiacyl rich versus mixed syringil and guaiacyl), hemicellulose composition (mixed heteromannans versus xylan rich), and water-conducting cell types (tracheids versus vessels; Weng et al., 2010; Scheller and Ulvskov, 2010). For comparison, the basal angiosperm A. trichopoda has xylan-rich woody cell walls, typical of angiosperms, but lacks vessels; the relative abundance of syringil subunits in the mixed syringil and guaiacyl lignin is lower in A. trichopoda (13%) than in other angiosperm species (greater than 50%; Amborella Genome Project, 2013). These features of A. trichopoda cell walls may be representative of the biological transition between gymnosperms and angiosperms (Amborella Genome Project, 2013). A comprehensive set of expressed genes of lignin biosynthesis has been identified in the loblolly pine genome (Neale et al., 2014). The apparent lack of a gene encoding ferulate 5-hydroxylase, involved in the biosynthesis of syringil subunits, is in agreement with conifer lignin composition (Neale et al., 2014).
Although the origin of angiosperm flowers may be partially explained by the presence of novel gene lineages, the majority (70%) of floral genes, including genes involved in floral timing and initiation, meristem identity, and floral structure, were present in the most recent common ancestor of all extant seed plants (Amborella Genome Project, 2013). These include the MADS box genes, whose duplications and diversification predated the angiosperm-gymnosperm split (Amborella Genome Project, 2013). However, the number of MADS box genes appeared to be, in general, much higher in gymnosperms than in angiosperms (Gramzow et al., 2014). Most of these genes in gymnosperms were type II MADS box genes, while the number of type I MADS box genes was low (Nystedt et al., 2013; Gramzow et al., 2014).
Conifer-Specific Gene Families
Comparative genomics studies have begun to investigate conifer- or gymnosperm-specific gene families and their evolution (Nystedt et al., 2013; Wegrzyn et al., 2014). Pavy et al. (2013a) found a set of 1,911 (486 after e-value adjustment) apparently species-specific sequences in white spruce. These sequences were generally associated with high substitution rates. Nystedt et al. (2013) found 6,615 gene families in Norway spruce, of which 1,021 were apparently species specific and overrepresenting genes involved in DNA repair and methylation. In a broader gene family analysis, Wegrzyn et al. (2014) compared the genome of loblolly pine with 13 other plant species and identified 7,053 gene families in loblolly pine, of which 1,554 were conifer specific, containing at least one sequence in loblolly pine, Sitka spruce, and Norway spruce. These apparently conifer-specific gene families overrepresented genes generally annotated as involved in protein and nucleic acid binding and hydrolase activity, with an additional large contribution from small molecule binding (Wegrzyn et al., 2014).
FUNCTIONAL GENOMICS IN CONIFERS
For functional characterization of the gene-coding space of conifers, an abundance of ESTs and full-length cDNAs, as well as microarray tools and proteome sequences, and whole-transcriptome resources have been developed for several species to identify genes involved in processes such as wood formation, abiotic stress, somatic embryogenesis, and the defense and resistance against insects and pathogens (MacKay et al., 2012; Canales et al., 2014; Cañas et al., 2014). In the absence of mutant lines, functional genomics in conifers relies critically on approaches of metabolite profiling, protein and enzyme biochemistry, and transformation. Using these tools in conjunction with transcriptome and genome sequences, biochemical processes have been identified that feature prominently in conifers, such as the biosynthesis of oleoresin terpenoids and phenolics in conifer defense. For example, new functions of cytochrome P450 genes and enzymes in diterpene resin acid biosynthesis (Hamberger et al., 2011) and new routes in tetrahydroxystilbenes biosynthesis (Hammerbacher et al., 2011) were discovered based on biochemically exploring spruce transcriptomes. The transcriptome resources also enabled the functional characterization of several dozen spruce and pine terpene synthases of the TPS-c, TPS-e, and TPS-d gene families (Keeling et al., 2010, 2011b; Hall et al., 2013). TPS-c and TPS-e genes are essential for GA phytohormome biosynthesis. TPS-d genes are important for conifer resistance against pests and pathogens. The TPS genes represent the largest group of functionally characterized members of any conifer gene family (Zerbe and Bohlmann, 2014). Characterization of spruce TPS-d genes provides examples of combined genomics, transcriptomics, proteomics, and biochemical approaches for gene characterization in nonmodel systems (Hall et al., 2011), including the discovery of novel enzyme mechanisms (Keeling et al., 2011a) and genomic features of copy number variations in spruce defense genes (Hall et al., 2011). The recent publication of a new white spruce resistance gene, encoding a functional glycosylhydrolase specialized for the release of acetophenone metabolites that are active in the resistance of white spruce against spruce budworm, highlighted how transcriptomics and biochemical genomics approaches can be applied in native forest tree populations for the successful de novo discovery of conifer defense genes (Mageroy et al., 2014). Functional genomics approaches also enabled new progress in transcriptional networks involved in conifer secondary cell wall assembly, identifying a NAC gene involved in wood formation (Duval et al., 2014), and in secondary metabolism, showing evolutionary conservation and lineage-specific evolution and function of MYB regulators (Bomal et al., 2014).
SNP DISCOVERY
Sequencing of cDNAs, genomes, and transcriptomes accelerated the discovery of SNPs and the development of large SNP databases for several conifers (Howe et al., 2013; Pavy et al., 2013a; Canales et al., 2014; Pinosio et al., 2014). The landscape of nucleotide diversity showed that the frequency of nucleotide polymorphisms varied significantly across gene families as a function of genes that appear to be specific to conifers and the breadth of expression patterns (Pavy et al., 2013a). SNPs have been used to design high-throughput genotyping chips of broad coverage in species of spruce and pine as well as Douglas fir (Pseudotsuga menziesii) and other conifer species (Chancerel et al., 2011, 2013; Pavy et al., 2012, 2013b; Howe et al., 2013). They have been used to conduct various structural and population genome scans for genetic linkage mapping (see below) and for large-scale association studies with phenotypic and environmental variation in spruces, pines, and Douglas fir (Eckert et al., 2012; Prunier et al., 2013; De La Torre et al., 2014b). Present efforts aim at transcriptome-wide association studies and genome-wide association studies (GWAS) utilizing tens of thousands of genetic variants of diverse nature using genotyping chips or resequencing methods (Yeaman et al., 2014).
GENETIC LINKAGE MAPS
High-density genetic linkage maps have been developed for several conifer species, including white spruce (Pavy et al., 2012), loblolly pine (Martinez-Garcia et al., 2013; Neves et al., 2014), and maritime pine (Chancerel et al., 2011, 2013; Plomion et al., 2014). Three maritime pine linkage maps with 1,015 to 1,131 markers were produced using a 12,000-SNP genotyping array. These three maps were combined to a 1.712-centimorgan (cM) composite map of 1,838 SNP markers to estimate genome-wide levels of linkage disequilibrium and genetic diversity (Plomion et al., 2014). Linkage disequilibrium decayed rapidly and mostly extended over short physical distances, as in other outcrossing, long-lived species (Pavy et al., 2012; Plomion et al., 2014). The white spruce map comprised 1,801 genes distributed among the 12 linkage groups with a map length of 2.083 cM (Pavy et al., 2012). A loblolly pine map was constructed using exome sequence-capture genotyping and comprised 2,841 genes distributed among the 12 linkage groups with an average of one marker every 0.58 cM (Neves et al., 2014). Martinez-Garcia et al. (2013) used a combination of different markers and two reference three-generation outbred pedigrees to construct a high-density loblolly pine consensus map containing 2,466 markers with a map length of 1.476 cM and average marker density of 0.62 cM per marker. Comparative analyses indicated high levels of conservation of macrosynteny and colinearity among spruce and pine maps (Pavy et al., 2012). Given a divergence time of more than 100 million years between the spruce and pine lineages (Savard et al., 1994; Wang and Ran, 2014), this paralysis of genome macrostructure appeared unprecedented in seed plants.
APPLICATIONS OF CONIFER GENOMICS IN TREE BREEDING
The prospect of acquiring novel breeding tools and methods has been part of the rationale for sequencing genomes of economically and ecologically important conifers. Genomics-based breeding methods could shorten the long breeding cycle for conifers and improve the efficiency of genetic selection schemes (Burdon and Wilcox, 2011). The identification of genetic markers that explain a significant part of the variation in quantitative traits of interest for breeding has been pursued to unlock this potential by using approaches ranging from quantitative trait locus detection to GWAS (Burdon and Wilcox, 2011; de Miguel et al., 2014). In addition, genomic selection was proposed to afford a better fit with breeding objectives because of its ability to predict the genetic values of individuals (Jannink et al., 2010; Grattapaglia and Resende, 2011). The application and accuracy of genomic selection have been shown for Eucalyptus spp., loblolly pine, and white spruce (Resende et al., 2012; Zapata-Valenzuela et al., 2013; Beaulieu et al., 2014). These advances are significant considering the low linkage disequilibrium, the largely undomesticated status, and the wide genetic diversity of conifer breeding populations in addition to their large genome size. The availability of genome sequences and their polymorphisms will enable and accelerate the development of more efficient GWAS or genomic selection methods along with basic knowledge of the structure of conifer genomes. For example, work is under way aligning transcriptomics-discovered SNPs to the white spruce genome assembly to increase the repeatability of markers when transferred to DNA-based genotyping platforms. It is expected that, as conifer genome assemblies improve, and with more genotypes being sequenced, they will help to usher in more reliable and easily adaptable marker selection approaches. This could occur by better targeting SNPs in causative variants, with less reliance on linkage disequilibrium (Hayes et al., 2013), as well as by breeders developing a better understanding of SNP frequency and effects from historical population structures or hybridization events. Furthermore, with continued improvements and reductions in the costs of genotyping, such as large-scale genome coverage by genotyping-by-sequencing methods (Elshire et al., 2011), the cost effectiveness of using genomics-based breeding values is expected to offset some of the costs of phenotyping, particularly for expensive wood quality or complex pest resistance traits.
CONIFER GENOME DATABASES
The Dendrome database (https://dendrome.ucdavis.edu/) was the first to comprehensively host conifer genome data and a wealth of other important information for the tree genetics community, including updated versions of the loblolly pine genome assembly (Zimin et al., 2014; http://loblolly.ucdavis.edu). The Conifer Genome Integrative Explorer (ConGenIE; http://congenie.org) database was launched with the release of the Norway spruce genome sequence (Nystedt et al., 2013) and has been extended to include the white spruce and loblolly pine genome assemblies. The ConGenIE database design enables hosting genomes of multiple conifer species, and work is under way to cross-link between species under the umbrella PlantGenIE domain (http://plantgenie.org). This includes further development of the ComPlEx resource for investigating cross-species gene expression network conservation/divergence (Netotea et al., 2014; http://complex.plantgenie.org), where spruce RNA-Seq data have been integrated. In support of conifer genome annotation and comparative genomics, ORCAE (Sterck et al., 2012) and Genome Plaza (Van Bel et al., 2012), respectively, have been set up for conifer genomes at the University of Ghent in Belgium, modeled after existing platforms for other plant genomes, and will be integrated with the ConGenIE resource.
CONCLUSION
The genome sequences of conifers revealed distinctive features compared with other sequenced plant genomes. Structural features such as the large size of their genomes without recent whole-genome duplication, the very high occurrence of repeat elements, and the stability of macrostructure over long periods of time appear unique among known seed plant genomes. Although current conifer genome assemblies provide an important new resource for biological analyses, the road toward fully assembled and annotated conifer genomes still presents significant technological challenges. While much preliminary information about the gene space of conifer genomes can be gleaned from ab initio and computational comparisons with angiosperms, research in functional genomics beyond gene or protein profiling is critical to explore conifer genes and gene families, such as those of wood formation in gymnosperms, conifer-specific secondary metabolism, and defense against insect pests and disease.
Glossary
- WGS
whole-genome shotgun
- RNA-Seq
RNA sequencing
- cDNA
complementary DNA
- SNP
single-nucleotide polymorphism
- LTR-RTs
long terminal repeat-retrotransposons
- LTR
long terminal repeat
- LINEs
long interspersed nuclear elements
- SINEs
short interspersed nuclear elements
- sRNAs
short RNAs
- GWAS
genome-wide association studies
- cM
centimorgan
- ConGenIE
Conifer Genome Integrative Explorer
Footnotes
This work was supported by the European 7th Framework Program under the ProCoGen project (to A.R.D.L.T. and P.K.I.) and by Genome Canada, Genome British Columbia, and Genome Quebec through the Large-Scale Applied Research Project Program under the SMarTForests Project (to I.B., J.Bou., S.J.M.J., C.I.K., J.M.K., K.R., A.Y., P.Z., and J.Boh.).
References
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Armenise L, Simeone M, Piredda R, Schirone B (2012) Validation of DNA barcoding as an efficient tool for taxon identification and detection of species diversity in Italian conifers. Eur J For Res 131: 1337–1353 [Google Scholar]
- Beaulieu J, Doerksen TK, Clément S, Mackay J, Bousquet J (2014) Accuracy of genomic selection models in a large population of open-pollinated families in white spruce. Heredity 113: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MMS, Keeling CI, Brand D, Vandervalk BP, et al. (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Duval I, Giguère I, Fortin É, Caron S, Stewart D, Boyle B, Séguin A, MacKay JJ (2014) Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. J Exp Bot 65: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon RD, Wilcox PL (2011) Integration of molecular markers in breeding. InPlomion C, Bousquet J, Kole C, eds, Genetics, Genomics and Breeding of Conifers. CRC Press and Edenbridge Science Publishers, New York, pp 276–322 [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. (2014) MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol 164: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J, Bautista R, Label P, Gómez-Maldonado J, Lesur I, Fernández-Pozo N, Rueda-López M, Guerrero-Fernández D, Castro-Rodríguez V, Benzekri H, et al. (2014) De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology. Plant Biotechnol J 12: 286–299 [DOI] [PubMed] [Google Scholar]
- Cañas RA, Canales J, Gómez-Maldonado J, Avila C, Cánovas FM (January 3, 2014) Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol http://dx.doi.org/10.1093/treephys/tpt113 [DOI] [PubMed] [Google Scholar]
- Chancerel E, Lamy JB, Lesur I, Noirot C, Klopp C, Ehrenmann F, Boury C, Provost GL, Label P, Lalanne C, et al. (2013) High-density linkage mapping in a pine tree reveals a genomic region associated with inbreeding depression and provides clues to the extent and distribution of meiotic recombination. BMC Biol 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancerel E, Lepoittevin C, Le Provost G, Lin YC, Jaramillo-Correa JP, Eckert AJ, Wegrzyn JL, Zelenika D, Boland A, Frigerio JM, et al. (2011) Development and implementation of a highly-multiplexed SNP array for genetic mapping in maritime pine and comparative mapping with loblolly pine. BMC Genomics 12: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert AJ, Wegrzyn JL, Cumbie WP, Goldfarb B, Huber DA, Tolstikov V, Fiehn O, Neale DB (2012) Association genetics of the loblolly pine (Pinus taeda, Pinaceae) metabolome. New Phytol 193: 890–902 [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Roberts DR, Aitken SN (2014a) Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol Ecol 23: 2046–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, Wang T, Jaquish B, Aitken SN (2014b) Adaptation and exogenous selection in the Picea glauca × P. engelmannii hybrid zone and its implications for forest management under climate change. New Phytol 201: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miguel M, Cabezas JA, de María N, Sánchez-Gómez D, Guevara MA, Vélez MD, Sáez-Laguna E, Díaz LM, Mancha JA, Barbero MC, et al. (2014) Genetic control of functional traits related to photosynthesis and water use efficiency in Pinus pinaster Ait. drought response: integration of genome annotation, allele association and QTL detection for candidate gene identification. BMC Genomics 15: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval I, Lachance D, Giguère I, Bomal C, Morency MJ, Pelletier G, Boyle B, MacKay JJ, Séguin A (2014) Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J Exp Bot 65: 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Weilandt L, Theißen G (2014) MADS goes genomic in conifers: towards determining the ancestral set of MADS-box genes in seed plants. Ann Bot (Lond) 114: 1407–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genomes 7: 241–255 [Google Scholar]
- Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J (2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J 65: 936–948 [DOI] [PubMed] [Google Scholar]
- Hall DE, Zerbe P, Jancsik S, Quesada AL, Dullat H, Madilao LL, Yuen M, Bohlmann J (2013) Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol 161: 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Hall D, Yuen M, Oddy C, Hamberger B, Keeling CI, Ritland C, Ritland K, Bohlmann J (2009) Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Ohnishi T, Hamberger B, Séguin A, Bohlmann J (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol 157: 1677–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JA, De La Torre AR, Aitken SN (2014) Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genet Genomes (in press) [Google Scholar]
- Hammerbacher A, Ralph SG, Bohlmann J, Fenning TM, Gershenzon J, Schmidt A (2011) Biosynthesis of the major tetrahydroxystilbenes in spruce, astringin and isorhapontin, proceeds via resveratrol and is enhanced by fungal infection. Plant Physiol 157: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Lewin HA, Goddard ME (2013) The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet 29: 206–214 [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GT, Yu J, Knaus B, Cronn R, Kolpak S, Dolan P, Lorenz WW, Dean JF (2013) A SNP resource for Douglas-fir: de novo transcriptome assembly and SNP detection and validation. BMC Genomics 14: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink JL, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics 9: 166–177 [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J (2010) Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol 152: 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Madilao LL, Zerbe P, Dullat HK, Bohlmann J (2011a) The primary diterpene synthase products of Picea abies levopimaradiene/abietadiene synthase (PaLAS) are epimers of a thermally unstable diterpenol. J Biol Chem 286: 21145–21153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Ralph SG, Jancsik S, Hamberger B, Dullat HK, Bohlmann J (2011b) Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol 11: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. (2004) Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach A, Wegrzyn JL, Parra G, Holt C, Bruening GE, Loopstra CA, Hartigan J, Yandell M, Langley CH, Korf I, et al. (2010) The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics 11: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay J, Dean JFD, Plomion C, Peterson DG, Cánovas FM, Pavy N, Ingvarsson PK, Savolainen O, Guevara MA, Fluch S, et al. (2012) Towards decoding the conifer giga-genome. Plant Mol Biol 80: 555–569 [DOI] [PubMed] [Google Scholar]
- Magbanua ZV, Ozkan S, Bartlett BD, Chouvarine P, Saski CA, Liston A, Cronn RC, Nelson CD, Peterson DG (2011) Adventures in the enormous: a 1.8 million clone BAC library for the 21.7 Gb genome of loblolly pine. PLoS ONE 6: e16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mageroy MH, Parent GJ, Germanos G, Giguère I, Delvas N, Maaroufi H, Bauce É, Bohlmann J, Mackay JJ (November 6, 2014) Expression of the β-glucosidase gene Pgßglu-1 underpins natural resistance of white spruce against spruce budworm. Plant J DOI: 10.1111/tpj.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia PJ, Stevens KA, Wegrzyn JL, Liechty J, Crepeau M, Langley CH, Neale DB (2013) Combination of multipoint maximum likelihood (MML) and regression mapping algorithms to construct a high-density genetic linkage map for loblolly pine (Pinus taeda L.). Tree Genet Genomes 9: 1529–1535 [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin TJ, Andersson B, Bastien JC, Beaulieu J, Burdon RD, Dovrak WS, King JN, Kondo T, Krakowski J, Lee SJ, et al. (2011) Economic importance, breeding objectives and achievements. InPlomion C, Bousquet J, Kole C, eds, Genetics, Genomics and Breeding of Conifers. CRC Press and Edenbridge Science Publishers, New York, pp 40–127 [Google Scholar]
- Murray BG, Leitch IJ, Bennett MD (2012) Gymnosperm DNA C-values database (release 5.0, December 2012). http://www.kew.org/cvalues (accessed March 2014)
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netotea S, Sundell D, Street NR, Hvidsten TR (2014) ComPlEx: conservation and divergence of co-expression networks in A. thaliana, Populus and O. sativa. BMC Genomics 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves LG, Davis JM, Barbazuk WB, Kirst M (2014) A high-density gene map of loblolly pine (Pinus taeda L.) based on exome sequence capture genotyping. G3 (Bethesda) 4: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I (2009) Assessing the gene space in draft genomes. Nucleic Acids Res 37: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Deschênes A, Blais S, Lavigne P, Beaulieu J, Isabel N, Mackay J, Bousquet J (2013a) The landscape of nucleotide polymorphism among 13,500 genes of the conifer Picea glauca, relationships with functions, and comparison with Medicago truncatula. Genome Biol Evol 5: 1910–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Gagnon F, Rigault P, Blais S, Deschênes A, Boyle B, Pelgas B, Deslauriers M, Clément S, Lavigne P, et al. (2013b) Development of high-density SNP genotyping arrays for white spruce (Picea glauca) and transferability to subtropical and Nordic congeners. Mol Ecol Resour 13: 324–336 [DOI] [PubMed] [Google Scholar]
- Pavy N, Pelgas B, Laroche J, Rigault P, Isabel N, Bousquet J (2012) A spruce gene map infers ancient plant genome reshuffling and subsequent slow evolution in the gymnosperm lineage leading to extant conifers. BMC Biol 10: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinosio S, González-Martínez SC, Bagnoli F, Cattonaro F, Grivet D, Marroni F, Lorenzo Z, Pausas JG, Verdú M, Vendramin GG (2014) First insights into the transcriptome and development of new genomic tools of a widespread circum-Mediterranean tree species, Pinus halepensis Mill. Mol Ecol Resour 14: 846–856 [DOI] [PubMed] [Google Scholar]
- Plomion C, Chancerel E, Endelman J, Lamy JB, Mandrou E, Lesur I, Ehrenmann F, Isik F, Bink MC, van Heerwaarden J, et al. (2014) Genome-wide distribution of genetic diversity and linkage disequilibrium in a mass-selected population of maritime pine. BMC Genomics 15: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, Pelgas B, Gagnon F, Desponts M, Isabel N, Beaulieu J, Bousquet J (2013) The genomic architecture and association genetics of adaptive characters using a candidate SNP approach in boreal black spruce. BMC Genomics 14: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SG, Chun HJ, Kolosova N, Cooper D, Oddy C, Ritland CE, Kirkpatrick R, Moore R, Barber S, Holt RA, et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende MDV, Resende MF Jr, Sansaloni CP, Petroli CD, Missiaggia AA, Aguiar AM, Abad JM, Takahashi EK, Rosado AM, Faria DA, et al. (2012) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194: 116–128 [DOI] [PubMed] [Google Scholar]
- Rigault P, Boyle B, Lepage P, Cooke JE, Bousquet J, MacKay JJ (2011) A white spruce gene catalog for conifer genome analyses. Plant Physiol 157: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard L, Li P, Strauss SH, Chase MW, Michaud M, Bousquet J (1994) Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc Natl Acad Sci USA 91: 5163–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schiex T, Moisan A, Rouze P (2001) EUGENE: an eukaryotic gene finder that combines several sources of evidence. InGascual O, Sagot MF, eds, Computational Biology. Springer Verlag, Berlin, pp 118–133 [Google Scholar]
- Sena JS, Giguère I, Boyle B, Rigault P, Birol I, Zuccolo A, Ritland K, Ritland C, Bohlmann J, Jones S, et al. (2014) Evolution of gene structure in the conifer Picea glauca: a comparative analysis of the impact of intron size. BMC Plant Biol 14: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2013) A conifer genome spruces up plant phylogenomics. Genome Biol 14: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32: W309–W312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L, Billiau K, Abeel T, Rouzé P, Van de Peer Y (2012) ORCAE: online resource for community annotation of eukaryotes. Nat Methods 9: 1041. [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ran JH (2014) Evolution and biogeography of gymnosperms. Mol Phylogenet Evol 75: 24–40 [DOI] [PubMed] [Google Scholar]
- Wegrzyn JL, Liechty JD, Stevens KA, Wu LS, Loopstra CA, Vasquez-Gross HA, Dougherty WM, Lin BY, Zieve JJ, Martínez-García PJ, et al. (2014) Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196: 891–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Lin BY, Zieve JJ, Dougherty WM, Martínez-García PJ, Koriabine M, Holtz-Morris A, deJong P, Crepeau M, Langley CH, et al. (2013) Insights into the loblolly pine genome: characterization of BAC and fosmid sequences. PLoS ONE 8: e72439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev IA, Fossdal CG, Johnsen Ø (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol 187: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Yeaman S, Hodgins KA, Suren H, Nurkowski KA, Rieseberg LH, Holliday JA, Aitken SN (2014) Conservation and divergence of gene expression plasticity following c. 140 million years of evolution in lodgepole pine (Pinus contorta) and interior spruce (Picea glauca×Picea engelmannii). New Phytol 203: 578–591 [DOI] [PubMed] [Google Scholar]
- Zapata-Valenzuela J, Whetten RW, Neale D, McKeand S, Isik F (2013) Genomic estimated breeding values using genomic relationship matrices in a cloned population of loblolly pine. G3 (Bethesda) 3: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe P, Bohlmann J (2014) Bioproducts, biofuels, and perfumes: conifer terpene synthases and their potential for metabolic engineering. Recent Adv Phytochem 44: 85–107 [Google Scholar]
- Zerbe P, Hamberger B, Yuen MMS, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Bach SS, Bohlmann J (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol 162: 1073–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A, Stevens KA, Crepeau MW, Holtz-Morris A, Koriabine M, Marçais G, Puiu D, Roberts M, Wegrzyn JL, de Jong PJ, et al. (2014) Sequencing and assembly of the 22-gb loblolly pine genome. Genetics 196: 875–890 [DOI] [PMC free article] [PubMed] [Google Scholar]