Eliminating β-hexosaminidase3 activity and terminal sialylation yields a human protease inhibitor that resembles the serum-derived counterpart.
Abstract
Plants are increasingly being used as an expression system for complex recombinant proteins. However, our limited knowledge of the intrinsic factors that act along the secretory pathway, which may compromise product integrity, renders process design difficult in some cases. Here, we pursued the recombinant expression of the human protease inhibitor α1-antitrypsin (A1AT) in Nicotiana benthamiana. This serum protein undergoes intensive posttranslational modifications. Unusually high levels of recombinant A1AT were expressed in leaves (up to 6 mg g−1 of leaf material) in two forms: full-length A1AT located in the endoplasmic reticulum displaying inhibitory activity, and secreted A1AT processed in the reactive center loop, thus rendering it unable to interact with target proteinases. We found that the terminal protein processing is most likely a consequence of the intrinsic function of A1AT (i.e. its interaction with proteases [most likely serine proteases] along the secretory pathway). Secreted A1AT carried vacuolar-type paucimannosidic N-glycans generated by the activity of hexosaminidases located in the apoplast/plasma membrane. Notwithstanding, an intensive glycoengineering approach led to secreted A1AT carrying sialylated N-glycan structures largely resembling its serum-derived counterpart. In summary, we elucidate unique insights in plant glycosylation processes and show important aspects of postendoplasmic reticulum protein processing in plants.
Recombinant protein-based drugs are among the fastest growing areas of development in the pharmaceutical industry. Consequently, there is a demand for exploring new production systems. Plants are increasingly being used for the expression of recombinant proteins, primarily because of their remarkable production speed and yield (for review, see Gleba et al., 2014). The highly conserved secretory pathway between human and plant cells allows similar, if not identical, protein folding, assembly, and posttranslational modifications. Importantly, plants are able to synthesize complex N-glycans, a prerequisite for the in vivo activity of many therapeutically interesting proteins. Despite substantial differences in N-glycan diversity, we and others have shown that plants are highly amendable to glycan engineering and allow proteins with controlled human-type N-glycosylation profiles to be generated (Castilho and Steinkellner, 2012). Moreover, it has even been possible to reconstruct entire human glycosylation pathways, which was shown by the introduction of the human sialylation and O-glycosylation processes in Nicotiana benthamiana (Castilho et al., 2010, 2012). These accomplishments render plants suitable for the production of human proteins that require a complex glycosylation profile.
Notwithstanding, to use plants as a versatile expression host for complex human proteins, it is important to fully understand intracellular processes. Particularly detailed knowledge about constraints along the plant cell secretory pathway, including proteolytic processing, is required, because these constraints may compromise protein integrity and quality. Despite major achievements in controlling protein-bound oligosaccharide formation, some plant glycosylation peculiarities are not entirely understood. For example, plant cells synthesize so-called paucimannosidic N-glycans, a type of truncated glycans usually absent in mammals (Lerouge et al., 1998). The biosynthesis and physiological significance of this N-glycan formation has yet to be completely explained (Strasser et al., 2007; Liebminger et al., 2011). Another process not fully understood in plants is subcellular localization of proteins. Aberrant intracellular deposition and as a consequence, incorrect glycosylation of recombinant proteins are often reported. For example, recombinant proteins designed for secretion are frequently also located in the endoplasmic reticulum (ER) and as a consequence, carry oligomannosidic carbohydrates instead of the desired complex-type glycans (Loos et al., 2011; Schneider et al., 2014a). By contrast, KDEL-tagged proteins designed for ER retention are sometimes partially secreted (Van Droogenbroeck et al., 2007; Niemer et al., 2014). How and at which biosynthetic stage these plant-specific peculiarities arise are largely unpredictable, which makes controlled expression of recombinant proteins with features authentically to their natural counterparts a difficult task.
One human protein that is pharmaceutically interesting, and thus needed in large amounts at high quality, is α1-antitrypsin (A1AT). This highly glycosylated protease inhibitor from the serpin superfamily interacts with a wide variety of proteases (Gettins, 2002). Like other serpins, A1AT is characterized by an exposed and mobile reactive center loop (RCL) with a Met (358M) residue acting as bait for specific target proteinases (Travis and Salvesen, 1983). The main biological role of plasma A1AT is to prevent excessive action of leukocyte-derived Ser proteinases, especially neutrophil elastase, in the circulatory system (Blank and Brantly, 1994). Therapeutic A1AT used in augmentation therapies is currently purified from pooled human serum, and the treatment can cost up to $100,000 per year per patient (Alkins and O’Malley, 2000). Concerns over the supply and safety of the products have urged searches for alternative recombinant sources of A1AT. Recombinant A1AT has been produced in human and nonhuman cell production systems with limited success (Blanchard et al., 2011; Brinkman et al., 2012; Ross et al., 2012; Lee et al., 2013). The production suffers from two major drawbacks: low expression levels and/or incorrect glycosylation (Garver et al., 1987; Chang et al., 2003; McDonald et al., 2005; Hasannia et al., 2006; Karnaukhova et al., 2006; Plesha et al., 2007; Agarwal et al., 2008; Nadai et al., 2009; Huang et al., 2010; Arjmand et al., 2011; Jha et al., 2012). The mature plasma-derived 52-kD protein has three N-linked glycosylation sites that are mainly decorated with disialylated structures (Kolarich et al., 2006). Sialylated N-glycans are a well-known requisite for the plasma half-life of A1AT (Mast et al., 1991; Lindhout et al., 2011; Lusch et al., 2013); the difficulties associated with obtaining them hamper the generation of biologically active A1AT in many expression systems.
Here, we pursued the expression of recombinant human A1AT in glycoengineered N. benthamiana and investigated the system’s ability to generate active sialylated variants. Unusually high amounts of A1AT were obtained using a plant viral-based transient expression system. The inhibitor was efficiently secreted to the intercellular space (IF); however, peptide mapping showed that the secreted A1AT was truncated at both the N and C termini. Mass spectrometry (MS) -based N-glycan analysis of IF-derived A1AT showed that vacuolar typical paucimannosidic N-glycans were present. By expressing A1AT in Arabidopsis (Arabidopsis thaliana) knockout plants lacking β-N-acetylhexosaminidase (HEXO) activity (Liebminger et al., 2011), we showed that paucimannosidic structures are generated by the action of HEXO3 located at the plasma membrane.
Coexpression with the mammalian genes necessary for in planta sialylation allowed the synthesis of disialylated A1AT, and sialylation levels could be increased by the synthesis of multiantennary glycans. By contrast, full-length A1AT purified from total soluble extracts exhibited ER-typical oligomannosidic carbohydrates. Using live-cell imaging, a GFP-tagged A1AT fusion did, indeed, exhibit aberrant ER-associated deposition of full-length A1AT. Elastase inhibition assays showed that ER-retained A1AT exhibits inhibitory activity, whereas the IF-derived truncated form was rendered inactive by cleavage within its RCL.
RESULTS
A1AT Is Efficiently Expressed in N. benthamiana Leaves
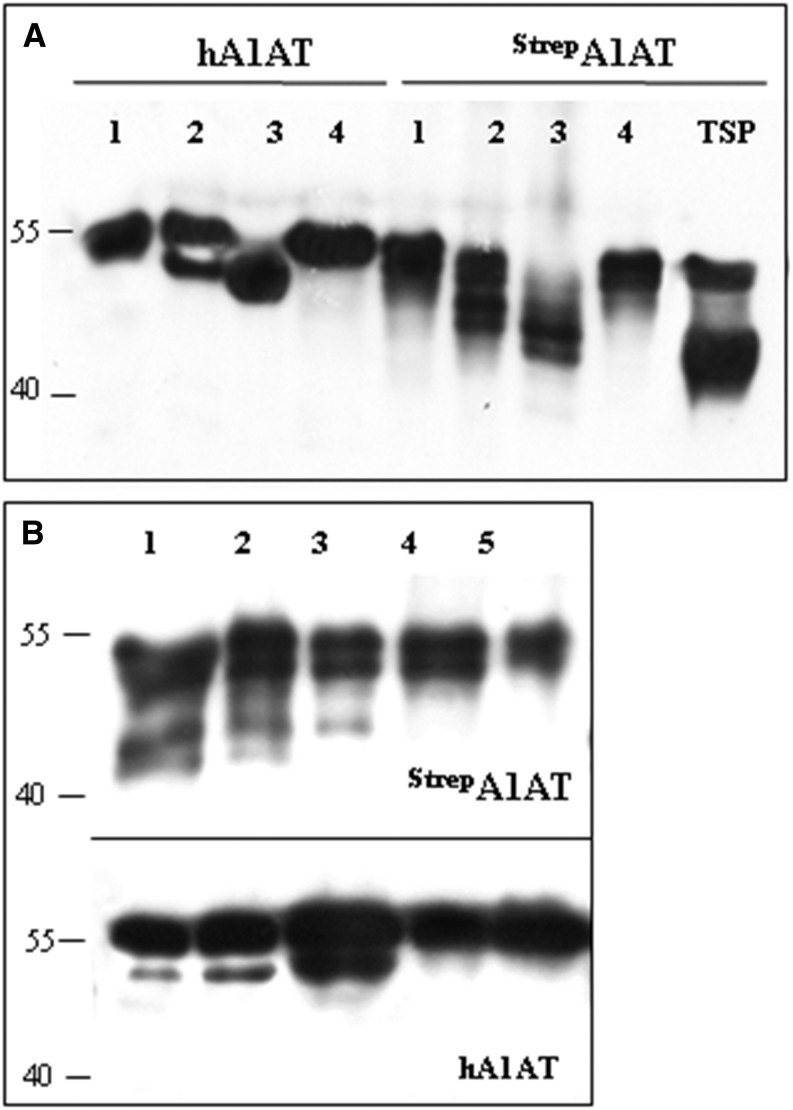
A plant viral-based transient expression system (Marillonnet et al., 2005) was used for recombinant expression of human A1AT. Complementary DNA (cDNA) of A1AT lacking the N-terminal signal peptide (Supplemental Fig. S1) and carrying either an N- or C-terminal Strep-II tag was cloned into the magnICON vector pICHα26211 (Schneider et al., 2014a). The recombinant protein was targeted for secretion using the barley (Hordeum vulgare) α-amylase signal peptide present in the vector. The resulting constructs (StrepA1AT and A1ATStrep; Fig. 1A) were transferred into Agrobacteria spp. and delivered to plant leaves by agroinfiltration. Expression of strep-tagged A1AT was monitored 5 d postinfiltration by western blotting using antibodies against the protein or the strep tag (Fig. 2A). Two signals of 40 and 52 kD were detected in total soluble protein (TSP) extracts, and the 52-kD band corresponded to the size of the full-length protein. A band corresponding to the full-length size was also present in StrepA1AT purified from TSP (Fig. 2A). Analysis of the intercellular fluid, representing the secretome of cells, using anti-A1AT antibodies showed the presence of only a 40-kD band, and no signal was detected using strep tag-specific antibodies. Similar results were obtained upon expression of A1ATStrep (data not shown). These results indicate the expression of full-length A1AT but also, an additional A1AT variant with truncations at both termini. The expression level of A1AT in TSP was up to 6 mg g−1 of leaf material, about 60% of which can be attributed to the 40-kD form (Supplemental Fig. S2).
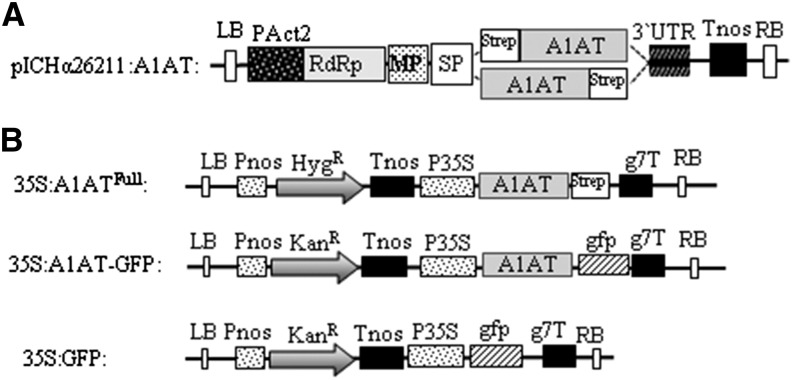
Figure 1.
Schematic representation of A1AT expression vectors. A, Tobacco mosaic virus-based magnICON vector (pICHα26211) carrying the barley α-amylase signal peptide (SP) was used for transient expression. A StrepII tag was N- or C-terminally fused to the A1AT cDNA gene lacking the endogenous signal peptide (StrepA1AT and A1ATStrep). B, Binary vectors used to express A1AT. For stable transformation in Arabidopsis, a C-terminal StrepII tag was fused to full-length A1AT (35S:A1ATFull; amino acids 1–418); for subcellular localization studies, A1AT was fused to GFP (35S:A1AT-GFP). A binary vector carrying GFP was used for control experiments (35S:GFP). g7T, Agrobacterium spp. gene 7 terminator; HygR, hygromycin B phosphotransferase gene; KanR, neomycin phosphotransferase 2 gene; LB, left border; MP, movement protein from the tobacco mosaic virus; PAct2, Arabidopsis actin2 promoter; Pnos, nopaline synthase gene promoter; P35S, cauliflower mosaic virus 35S gene promoter; RB, right border; RdRp, RNA-dependent RNA polymerase from turnip vein clearing virus; Tnos, nopaline synthase gene terminator; 3′UTR, 3′-untranslated region.
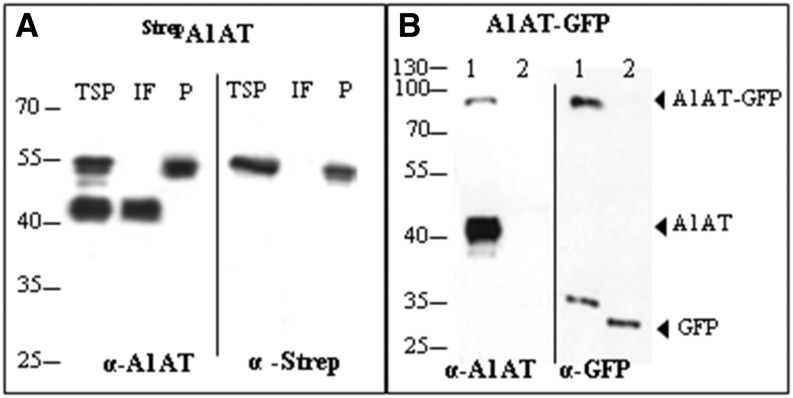
Figure 2.
A1AT is expressed as full-length and truncated protein versions. A, Immunoblot analysis of StrepA1AT expressed in N. benthamiana using anti-A1AT (α-A1AT) and anti-StrepII (α-Strep) antibodies. TSP extract, IF, or Strep tag-purified A1AT (P) was loaded on SDS-PAGE gels under reducing conditions. B, Immunoblot analysis of A1AT-GFP expression using α-A1AT and anti-GFP (α-GFP) antibodies. TSP extracts expressing the A1AT-GFP fusion protein (lane 1) or GFP alone (lane 2) were loaded on SDS-PAGE gels. Arrows indicate protein bands corresponding to the full-length fusion protein (A1AT-GFP), the 40-kD fragment (A1AT), and GFP. Protein sizes are indicated in kilodaltons.
A strong band at position 40 kD but not 52 kD was seen by SDS-PAGE and Coomassie Brilliant Blue staining of IF-derived proteins after infiltration with StrepA1AT (Fig. 3A), indicating that only the truncated version of A1AT is secreted. By contrast, three bands in the range of 50 to 55 kD that reacted with both anti-A1AT and antistrep antibodies were seen in StrepA1AT purified from TSP (Fig. 3B). The different sizes at close proximity may be attributable to differences in glycosylation of StrepA1AT.
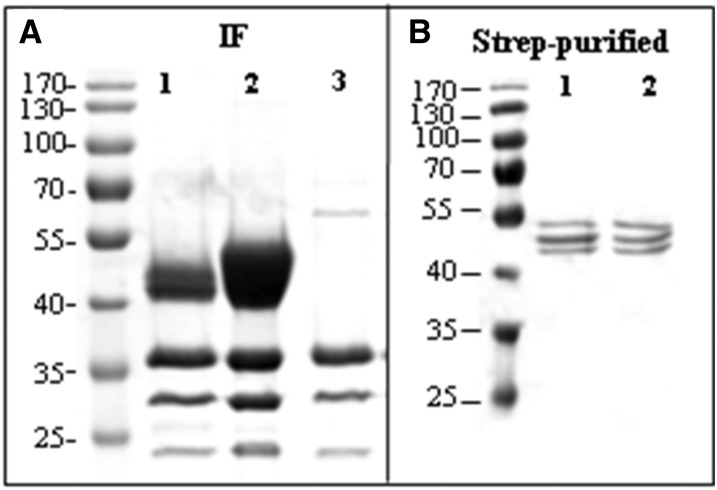
Figure 3.
Truncated A1AT is efficiently secreted to the intercellular fluid. A, Coomassie Brilliant Blue staining of proteins isolated from the IF of leaves infiltrated with StrepA1AT (lane 1), StrepA1AT coinfiltrated with mammalian genes necessary for protein trisialylation (StrepA1ATTriSia; lane 2), and Agrobacteria spp. carrying an empty plasmid (lane 3). A strong band corresponding to truncated A1AT is visible at the position slightly larger than 40 kD. Bands smaller than 40 kD are associated with Agrobacteria spp. infection. B, Coomassie Brilliant Blue staining of A1AT purified from TSP. Lanes 1 and 2 show purified StrepA1AT and StrepA1ATTriSia, respectively. Protein sizes are indicated in kilodaltons.
A1AT Exhibits Truncated N and C termini
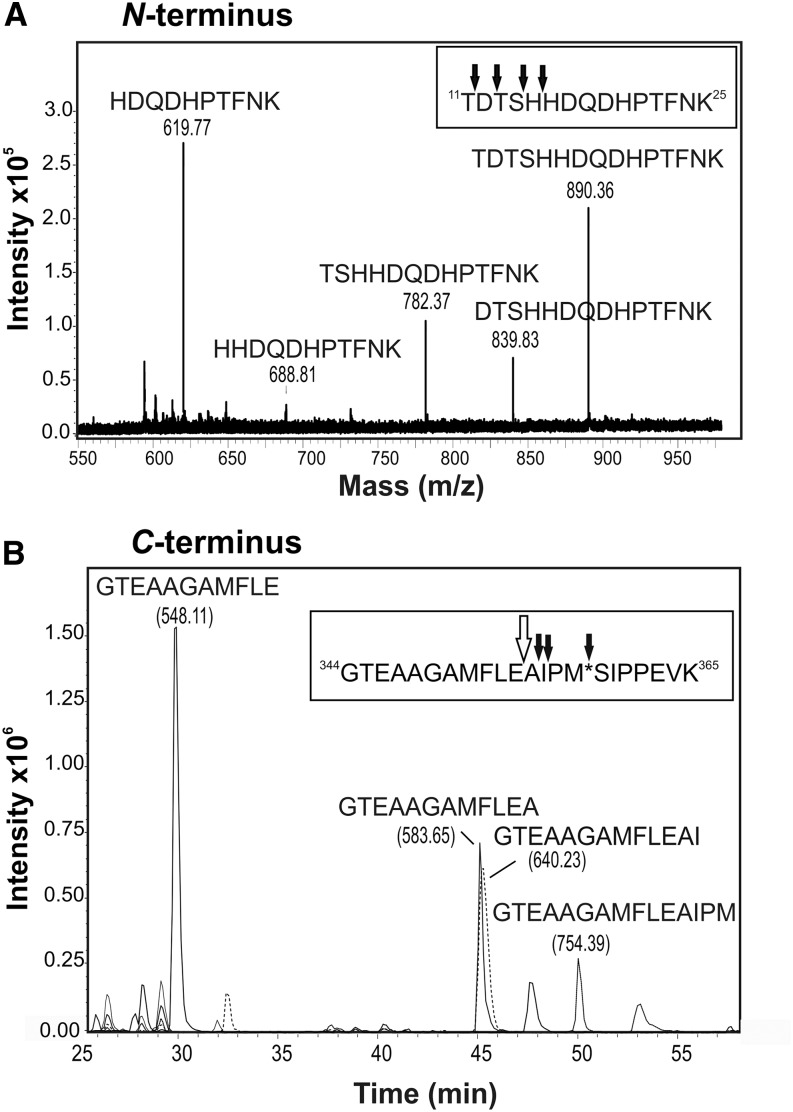
Liquid chromatography (LC)-electrospray ionization (ESI)-MS/MS was used to carry out peptide mapping of the IF-derived 40-kD A1AT and confirmed the presence of the A1AT protein backbone, but also, it revealed truncations at both ends (Fig. 4). The first peptide found corresponded to 11TDTSHHDQDHPTFNK25. One cleavage site in the mature protein sequence (Supplemental Fig. S1) was after 10K. This may have arisen from tryptic cleavage of the N-terminal sequence, but the expected N-terminal tryptic peptide (1EDPQGDAAQK10) was never detected. Also, several other peptides lacking up to 15 amino acids from the N terminus (16HDQDHPTFNK25) were detected. These fragments differing by 1 amino acid suggest additional trimming by amino-peptidase activity (Fig. 4A). C-terminal analysis of 40-kD A1AT revealed that the four most C-terminal tryptic peptides were missing. The full-length tryptic peptide carrying the RCL (344GTEAAGAMFLEAIPMSIPPEVK365) was also not detected, but several truncated forms of it could be found. The major cleavage occurred after 354E, thereby destroying the RCL (Fig. 4B; Supplemental Fig. S1). Other terminal fragments were found to a lesser extent (Fig. 4B).
Figure 4.
Secreted 40-kD A1AT exhibits N- and C-terminal truncations. A, Identification of the N terminus of secreted tryptic-digested A1AT by LC-ESI-MS/MS. All found N-terminal peptide variants eluted in a window of 3 min, allowing their presentation in the sum spectrum shown. Several peptides lacking up to 15 amino acids at the N terminus (16HDQDHPTFNK25) were detected. Black arrows indicate cleavage sites. B, Identification of the C terminus of secreted tryptic-digested A1AT by LC-ESI-MS/MS. Reversed-phase LC separated the differently truncated C-terminal peptides over the whole chromatogram; thus, extracted ion chromatograms for all variants detected are shown. The major peptide representing the C terminus was identified as 344GTEAAGAMFLE354. The peptide (344GTEAAGAMFLEAIPM*SIPPEVK365) carrying the center of the reactive loop (asterisk) was not detected. Black arrows indicate cleavage sites; the major cleavage site is indicated by the white arrow. All peptide variants were confirmed by MS/MS spectra, and their masses are indicated in parentheses. m/z, Mass-to-charge ratio.
To investigate which proteinases are involved in the conversion of A1AT from an active (native) to an inactive (truncated) state, we incubated commercially available human plasma α1-antitrypsin (hA1AT) and purified plant-derived A1AT (StrepA1AT) with N. benthamiana IF isolated in neutral and acidic pH buffers. Acidic pH mimics the conditions of IF in planta (Grignon and Sentenac, 1991; Gao et al., 2004), where several proteases are active (Kaschani et al., 2010). Incubation of hA1AT and purified StrepA1AT with phosphate-buffered saline (PBS) did not alter the protein sizes detected by western-blot analysis (Fig. 5A, lane 1). After incubation with IF at pH 7.0, both proteins were detected as full-length versions, but truncated fragments were also visible (Fig. 5A, lane 2). When acidic IF (pH 5.0) was used, only truncated A1AT was detected (Fig. 5A, lane 3). After incubation of A1AT with IF previously heated to 95°C to inactivate possible proteases present, only full-length A1AT was detected (Fig. 5A, lane 4).
Figure 5.
A1AT can be processed in vitro by interactions with endogenous plant proteases. A, Immunoblot analysis of plasma- and plant-derived purified A1AT (hA1AT and StrepA1AT) exposed to intercellular fluid. Although no major differences in protein size are visible after incubation with PBS (lane 1) and boiled IF (lane 4), significant alterations in A1AT size are visible postexposure to IF at pH 7.0 (lane 2) or 5.0 (lane 3). A TSP extract from leaves expressing StrepA1AT served as the control. B, Immunoblot analysis of plant- and plasma-derived A1AT (StrepA1AT and hA1AT) exposed to intercellular fluid (pH 5.0) pretreated with protease inhibitors. The following inhibitors were used at the indicated final concentrations: 3.3 mm CA-074 (lane 1), 4.6 mm E-64 (lane 2), 3.3 mm leupeptin (lane 3), 6.7 mm PMSF (lane 4), and 33× proteinase inhibitor cocktail (lane 5). For detection, antibodies against A1AT were used. Protein sizes are indicated in kilodaltons.
The 40-kD A1AT Is Generated by Interactions with Plant Proteases
Proteases are abundant in several subcellular compartments along the secretory pathway and in the apoplast. To assess which type of protease might be involved in A1AT conversion, we preincubated IF samples extracted at pH 5.0 with different protease inhibitors before addition of purified plant StrepA1AT or plasma-derived A1AT (Fig. 5B). A protease inhibitor cocktail designed to eliminate proteolysis in plant extracts completely abrogated A1AT processing. Similarly, the addition of the Ser protease inhibitor phenylmethylsulfonyl fluoride (PMSF), which also displays some reactivity with Cys proteases, completely prevented breakdown of hA1AT (Fig. 5B). Taken together, these results strongly indicate that plant-derived A1AT interacts with endogenous Ser proteases residing in secretory compartments or the apoplast, ultimately resulting in its cleavage and concomitant inactivation.
Plant-Derived A1AT Is Active in Vitro
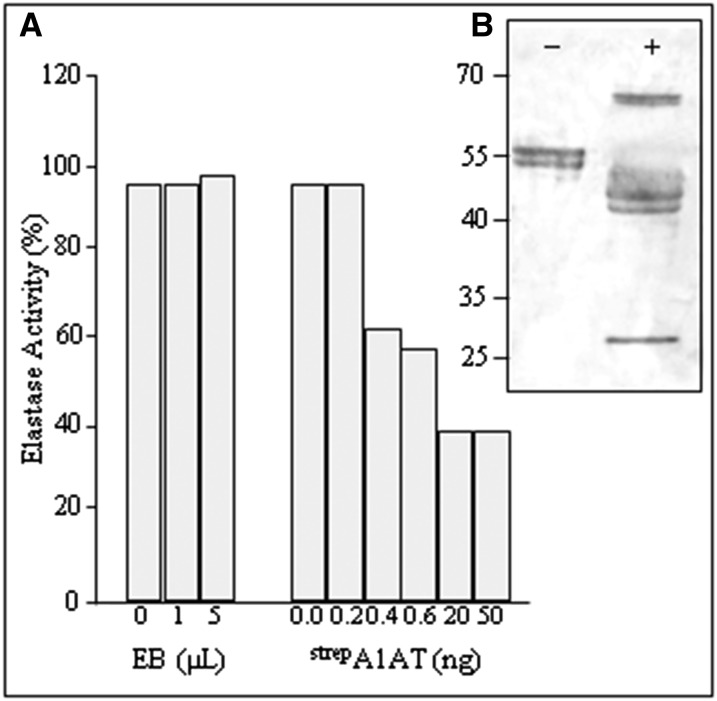
A1ATs, like other serpins, are molecular mouse traps, consisting of a bait and a swinging arm (Pike et al., 2002). Serpins are only partially stable in their active form, but they can snap into a far more stable form when they interact with a protease. When the protease binds to the bait (i.e. 358M), it forms a bond and begins to perform its normal cleavage reaction (Gettins, 2002). The flexible loop of A1AT, now cleaved and free to move, traps the protease. In the case of elastase, A1AT binds almost irreversibly to its active site, and the complex does not dissociate after cleavage. Here, we evaluated the biological activity of the purified plant-derived StrepA1AT as an inhibitor of porcine pancreatic elastase (PPE), a natural target protease. Incubation of 10 ng (or 0.38 µm) of elastase with 20 ng (approximately 0.38 µm) of purified StrepA1AT shows 40% of residual elastase activity. The incubation of stoichiometric amounts of the enzymes reveals that 60% of plant-derived purified A1AT is biologically active, and higher amounts of StrepA1AT (50 ng and more) do not result in more elastase inactivation (Fig. 6A).
Figure 6.
Plant-derived full-length A1AT is active in vitro. A, Inhibitory activity of StrepA1AT. SensoLyte Rh110 Elastase Assay Kit was used to estimate the potency of purified StrepA1AT to inhibit PPE using a flurogenic substrate. Elastase was incubated with different amounts of either elution buffer (EB) used for StrepA1AT purification (0–5 µL; control for autofluorescence) or purified StrepA1AT (0–50 ng). Histogram bars depict residual elastase activity (in percentages). B, Band shift assay of purified plant-derived StrepA1AT incubated with (+) or without (−) PPE, subjected to SDS-PAGE, and analyzed by Coomassie Brilliant Blue staining. The 65-kD band corresponds to the A1AT-PPE protein complex, whereas the approximately 40- and 25-kD bands represent the truncated A1AT and PPE, respectively, after complex dissociation. Protein sizes are indicated in kilodaltons.
The A1AT inhibition mechanism involves the formation of a 1:1 covalent complex between A1AT and PPE, which is also associated with some cleavage of the inhibitor by the protease. This unique property of the irreversible A1AT-PPE complex permits activity measurements of A1AT by SDS-PAGE band shift assays. Incubation of purified plant-derived A1AT with PPE resulted in the generation of a high Mr band (approximately 65) not present in the control sample. In addition, lower Mr bands represent truncated A1AT (approximately 40) and PPE (approximately 26) after complex dissociation (Fig. 6B). Peptide mapping revealed that PPE cleavage occurred after 358M, confirming that plant-derived A1AT interacts with target proteases in the same manner as its natural counterpart (Supplemental Fig. S3).
The 40-kD A1AT Exhibits Unusual N-Glycosylation
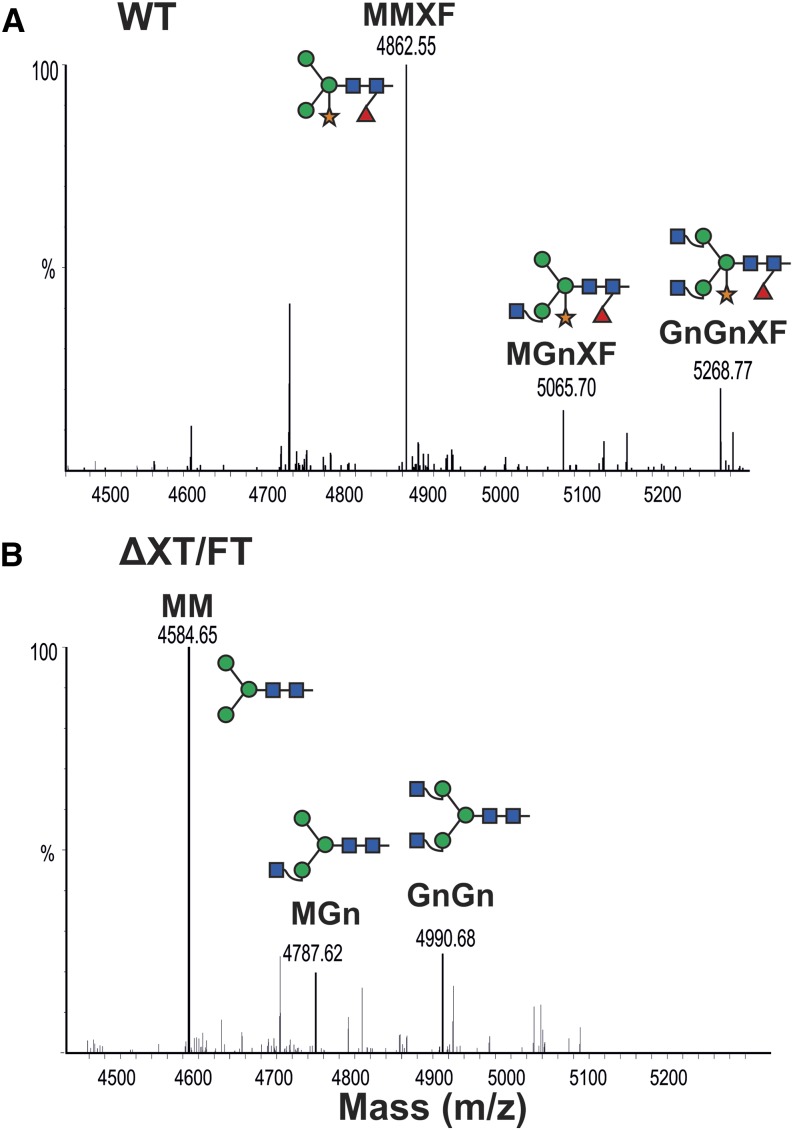
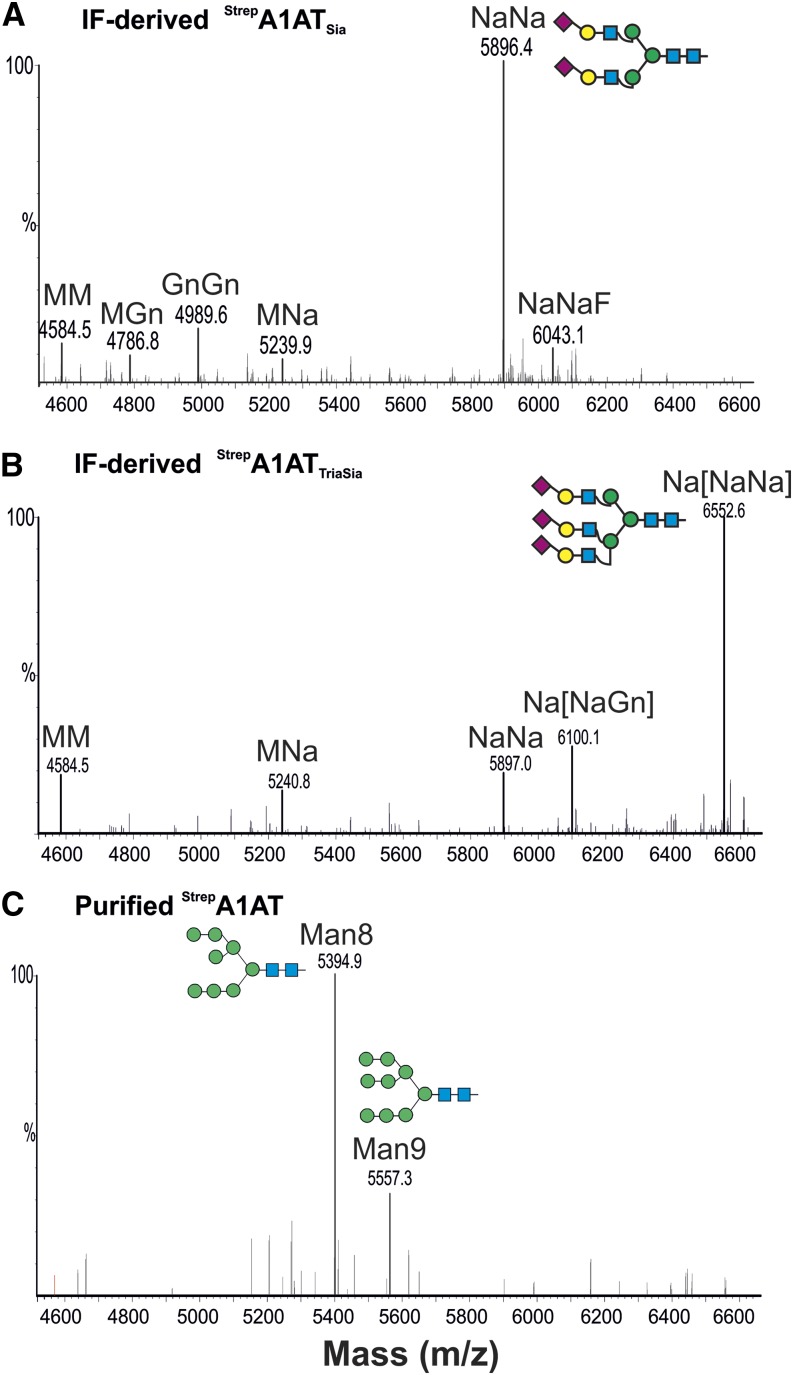
A1AT has three N-linked glycosylation sites (46Asn, 83Asn, and 247Asn), which are fully occupied in serum A1AT (Kolarich et al., 2006). We expressed StrepA1AT in the N. benthamiana wild type and the glycosylation mutant ∆XT/FT where the activities of the xylosyltransferase (XT) and core fucosyltransferase (FT) have been downregulated (Strasser et al., 2008), aiming to generate A1AT with a human-type glycosylation. This mutant lacks plant-specific Xyl and core Fuc residues and instead, synthesizes human-type complex structures with terminal GlcNAc residues (GlcNAc2Man3GlcNAc2 [GnGn] structures). We analyzed the glycosylation status of the plant-derived A1AT using LC-ESI-MS. Most IF-derived A1AT (40 kD) exhibited paucimannosidic structures (Man3(α1,3Fuc)(β1,2Xyl)GlcNAc2 [MMXF] or Man3GlcNAc2 [MM]; Fig. 7). These structures result from trimming of nonreducing terminal GlcNAc residues by β-hexosaminidases (Liebminger et al., 2011) and are considered typical for vacuolar plant glycoproteins (Gomord et al., 2010). Because we analyzed secreted A1AT, our data indicate that paucimannosidic N-glycans are also present on nonvacuolar glycoproteins.
Figure 7.
Secreted A1AT is decorated with paucimannosidic N-glycans. LC-ESI-MS of trypsin-digested A1AT collected from the IF. StrepA1AT was transiently expressed in N. benthamiana wild-type (WT; A) and glycosylation mutant (ΔXT/FT; B) plants. The N-glycosylation profile of glycopeptide2 (K/70ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR101) is shown. Peaks were labeled using the ProGlycAn system (www.proglycan.com). Adjacent illustrations display the respective N-glycans using standard symbols. m/z, Mass-to-charge ratio. [See online article for color version of this figure.]
HEXO3 Forms Paucimannosidic N-Glycans on Secreted A1AT
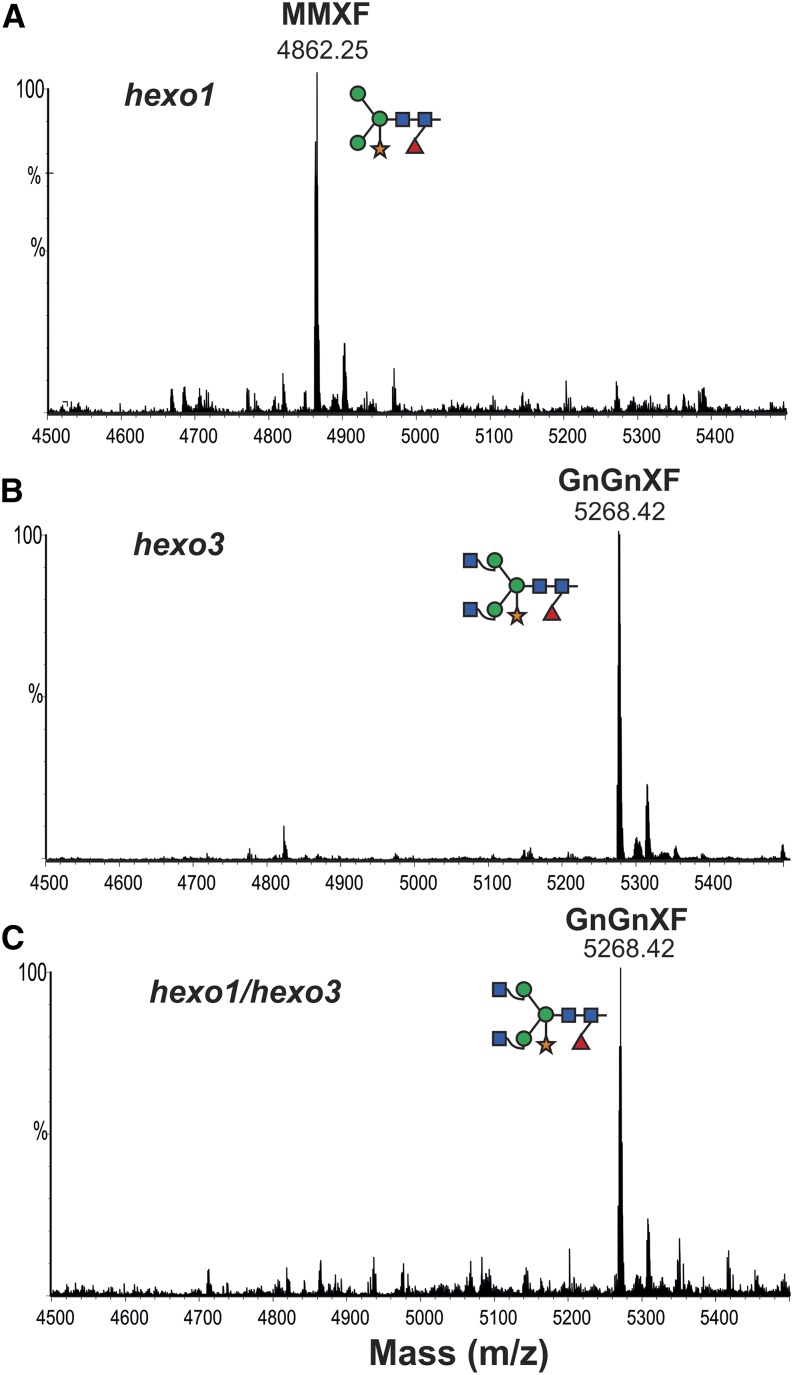
Two HEXOs (HEXO1 and HEXO3) responsible for the formation of paucimannosidic N-glycans have been identified in Arabidopsis (Liebminger et al., 2011). Here, we used Arabidopsis single-knockout (hexo1 and hexo3) and double-knockout plants (hexo1/hexo3) to investigate how these enzymes contribute to the generation of paucimannosidic structures on secreted A1AT. A binary vector encoding full-length A1AT, including the native signal peptide (1–418 amino acids; 35S:A1ATFull; Fig. 1B), was used to stably transform wild-type (Columbia-0 [Col-0]), hexo1, hexo3, and hexo1/hexo3 plants. Western-blot analysis of TSP extracted from transformed plants using anti-A1AT-specific antibodies showed two signals: one at position approximately 52 kD and the other at approximately 40 kD (similar to N. benthamiana; Supplemental Fig. S4). The 40-kD form was recovered from IF and subjected to N-glycan analysis using LC-ESI-MS. The glycosylation profile of A1AT derived from the Arabidopsis wild type and hexo1 lacking the vacuolar HEXO revealed the presence of mainly paucimannosidic structures (i.e. MMXF), as observed for A1AT expressed in N. benthamiana (Fig. 8A; Supplemental Fig. S4). By contrast, the glycosylation profile of A1AT expressed in hexo3 and hexo1/hexo3 mutants lacking the β-hexosaminidase associated with the plasma membrane exhibited primarily complex-type N-glycans (i.e. GnGnXF; Fig. 8, B and C). Only minor amounts of MMXF structures were detected on A1AT purified from IF of hexo3. Because both HEXO1 and HEXO3 are capable of converting GnGnXF into MMXF (Strasser et al., 2007), these results firmly establish that the paucimannosidic N-glycans decorating A1AT are generated within the secretory pathway and not caused by transient passage of the protein through vacuolar compartments.
Figure 8.
Arabidopsis HEXO3 generates paucimannosidic structures on secreted A1AT. LC-ESI-MS analysis of trypsin-digested A1ATFull expressed in Arabidopsis HEXO knockout lines (Liebminger et al., 2011). Glycan analysis was performed on the 40-kD A1AT collected from IF from hexo1 knockout (A), hexo3 knockout (B), and hexo1/hexo3 double knockout (C). Peaks were labeled using the ProGlycAn system (www.proglycan.com). Adjacent illustrations display the respective N-glycans using standard symbols. m/z, Mass-to-charge ratio. [See online article for color version of this figure.]
N-Glycans of Secreted A1AT Can Be Efficiently Decorated with Terminal Sialic Acid
The extent of sialylation is known to play a significant role in the serum half-life of A1AT (Mast et al., 1991). We set out to generate sialylated plant-derived A1AT that mimics the glycosylation profile of plasma-derived A1AT (Supplemental Fig. S5) by expressing StrepA1AT in N. benthamiana ∆XT/FT together with mammalian genes (in a total of six genes) necessary for in planta sialylation of N-glycans (Castilho et al., 2010). N-glycosylation profiling of IF-derived 40-kD A1AT (StrepA1ATSia) using LC-ESI-MS showed that almost exclusively complex-type sialylated glycans were present (up to 80%; Fig. 9A; Supplemental Table S1). This glycosylation profile highly mimics the plasma-derived counterpart. Because several studies have reported that raising the sialylation extent increases A1AT half-life (Lindhout et al., 2011; Lusch et al., 2013), we aimed to generate A1AT that carries additional sialic acid residues. One way of boosting sialic acid content is to increase the number of glycan antennae providing the appropriate acceptor substrates for terminal sialylation. Thus, we expressed the human enzymes needed for the formation of tri- and tetraantennary glycans (i.e. N-acetylglucosaminyltransferease IV [GnTIV] and GnTV; Castilho et al., 2011b, 2013). GnTIV and GnTV expressed individually with the genes for sialylation were able to efficiently branch the α1,3 and α1,6 arms, respectively (about 50%; StrepA1ATTriSia; Fig. 9B; Supplemental Fig. S5; Supplemental Table S1). Compared with StrepA1AT, StrepA1ATTriSia protein revealed a clear shift in size when isolated from the IF (Fig. 3A). In comparison, synthesis of tetraantennary glycans by simultaneous expression of GnTIV and GnTV was less effective (about 4%; Supplemental Table S1). Interestingly, it seems that protein sialylation is an effective way to protect the trimming of β-1,2-GlcNAc residues, because paucimannosidic (MM) structures were present only at very low levels (3%–5%; Fig. 9, A and B; Supplemental Table S1).
Figure 9.
A1AT N-glycan engineering allows the generation of highly sialylated structures. LC-ESI-MS profiles of trypsin-digested StrepA1AT (glycopeptide2) coexpressed with the genes necessary for in planta sialylation in ΔXT/FT. The 40-kD peptide collected from the IF was analyzed. A, StrepA1AT was coexpressed with the genes necessary for the synthesis of disialylated N-glycans (StrepA1ATSia). B, StrepA1AT was coexpressed with the genes necessary for the synthesis of multiantennary sialylated N-glycans (StrepA1ATTriSia). C, StrepA1AT purified from TSP extracts and coexpressed with sialylation genes. Oligomannosidic peaks corresponding to the detected structures are indicated. Peaks were labeled using the ProGlycAn system (www.proglycan.com). Adjacent illustrations display the respective N-glycans using standard symbols. m/z, Mass-to-charge ratio. [See online article for color version of this figure.]
Glycan analysis of A1AT purified from TSP (the wild type or ΔXT/FT) with and without coexpression of sialylation genes revealed the presence of peaks corresponding to oligomannosidic structures (Man8GlcNAc2 and Man9GlcNAc2; Fig. 9C; data not shown). The presence of such carbohydrates, typical for ER-resident proteins, and the absence of full-length A1AT in the IF support the idea that the truncation of A1AT largely occurs along the secretory pathway or in the apoplast.
A1AT-GFP Is Located in the ER
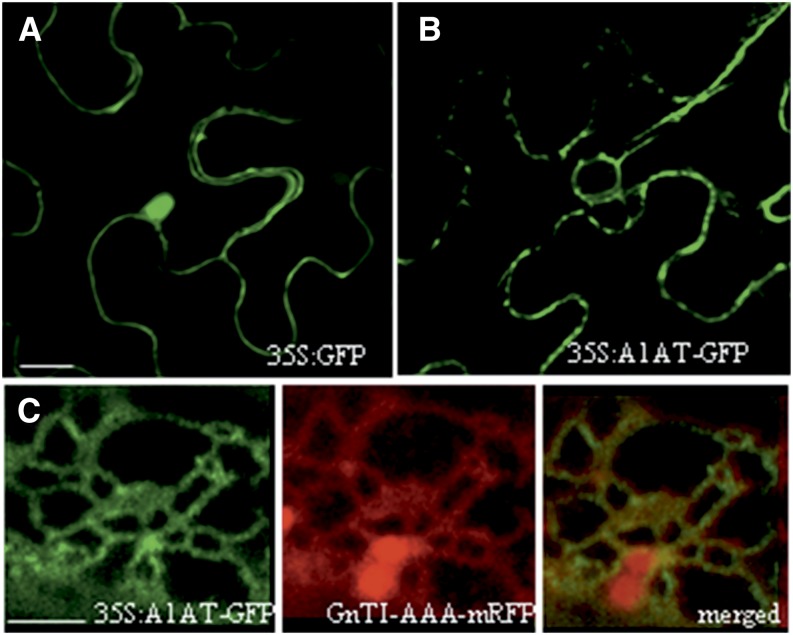
A binary vector containing a C-terminal GFP fusion was generated (35S:A1AT-GFP; Fig. 1B) to monitor subcellular localization of recombinant A1AT. The integrity of the expressed A1AT-GFP fusion protein was analyzed using western blots with anti-A1AT and anti-GFP antibodies (Fig. 2B). The results showed that the intact A1AT-GFP fusion protein (approximately 80 kD) was detected when either anti-A1AT or anti-GFP antibodies were used. In addition, a 40-kD protein band was detected with anti-A1AT antibodies but not anti-GFP antibodies. This indicates that the 40-kD protein is truncated at least on the C terminus. In addition, anti-GFP antibodies detected an approximately 30-kD band in the A1AT-GFP sample, which was slightly larger than the GFP used as the control (35S:GFP; Fig. 1B). This band probably represents GFP that still carries the A1AT fragment C terminus to the cleavage site in the RCL (approximately 4 kD; Fig. 2B).
As a control, the expression of GFP (35S:GFP) in N. benthamiana leaves was first monitored by live-cell confocal laser-scanning microscopy (CLSM). As expected, the small GFP protein was able to diffuse to the nucleus and localized mainly in the cytoplasm (Fig. 10A). By comparison, the CLSM image of cells expressing 35S:A1AT-GFP showed signals in primarily structures resembling the ER network and also, punctuate structures associated with the plasma membrane (Fig. 10B). No fluorescent staining was detected in the nucleus or the cytoplasm. To confirm ER retention of the intact A1AT fusion protein, we performed colocalization studies using the ER marker monomeric red fluorescent protein (mRFP) GnTI-CAAATS-mRFP (a fusion protein that is predominantly retained in the ER; Schoberer et al., 2009). The two fluorescent signals showed a significant overlap, suggesting at least partial ER retention of A1AT-GFP (Fig. 10C).
Figure 10.
Expression of A1AT-GFP shows ER localization. A, CLSM image of a representative leaf epidermal cell expressing GFP (35S:GFP). GFP is detected in the nucleus and the cytoplasm but not detected in the IF. B, CLSM image of a representative leaf epidermal cell expressing A1AT fused to GFP (35S:A1AT-GFP). The fluorescence signal displays a reticulate pattern resembling the ER. Red dots represent the Golgi apparatus. C, CLSM image from a cell coexpressing 35S:A1AT-GFP, and the ER marker GnTI-CAAATS-mRFP displays a significant colocalization of the two proteins (merged image). Red dots represent Golgi stacks. Bar = 20 μm in A and B. Bar = 2.5 μm in C.
The combined results of subcellular localization studies and glycan analysis of full-length and truncated A1AT strongly suggest that a fraction of the active protein is retained within the ER and thereby, escapes attack by nontarget proteinases. These data show that the A1AT-cleaving enzymes reside in a post-ER compartment.
DISCUSSION
In this study, we pursued the recombinant expression of human A1AT in N. benthamiana. This complex serum protein served as a model to monitor constraints along the plant cell secretory pathway compromising protein integrity. We obtained unusually high expression levels of StrepA1AT (estimated at 6 mg of recombinant protein per 1 g of leaf material). Such extraordinary amounts are usually only reported for expression of reporter proteins like GFP (Marillonnet et al., 2004, 2005), and would certainly meet the requirements for commercial production. Nevertheless, we were confronted with the fact that the 52-kD full-length StrepA1AT accounts for only about 40% of the recombinantly expressed protein. The majority of StrepA1AT exists as a truncated 40-kD polypeptide. The protein lacks up to 15 amino acids at the N terminus and up to 41 amino acids at the C terminus. Importantly, truncation at the C terminus destroys the RCL and thereby, abolishes the inhibitory activity of the protein. Similar hydrolysis at the C terminus (354E) has been observed when human A1AT is proteolytically inactivated by bacterial Cys proteinases (Nelson et al., 1999). Although target proteinases (e.g. elastase, trypsin, and α-chymotrypsin) cleave A1AT at 358M, other nontarget proteases have different cleavage sites on the RCL (Nelson et al., 1998). Cleavage of the RCL at 354E has been found to be typical of the lysosomal Cys proteinase cathepsin L (Johnson et al., 1986) and nontarget Ser proteases (Nelson et al., 1998). Considerable amounts of truncated protein were also obtained in previous attempts to express recombinant A1AT in rice (Oryza sativa) cells (Terashima et al., 1999; Huang et al., 2001;Trexler et al., 2002; McDonald et al., 2005), N. benthamiana (Sudarshana et al., 2006), and Nicotiana tabacum chloroplasts (Nadai et al., 2009). Nevertheless, A1AT produced in transgenic tomato (Solanum lycopersicum) plants does not show degradation (Jha et al., 2012).
The accumulation of the 40-kD polypeptide in the IF and the confinement of the residual full-length protein to the ER indicate that IF-resident proteases contribute to the conversion process. This is consistent with the notion that the plant apoplast is a proteinase-rich environment (Doran, 2006; van der Hoorn, 2008; Goulet et al., 2012). In support of this notion, in vitro assays showed that, when exposed to N. benthamiana IF, both plasma-derived and purified plant-derived A1AT are converted into lower Mr versions. This conversion was enhanced in vitro at acidic pH, mimicking the in planta conditions (Grignon and Sentenac, 1991; Gao et al., 2004), and blocked when proteinases were inactivated by high temperature. In addition, our results indicated that Ser proteases are the most likely contributors to the conversion process. Similar results have been reported for hA1AT when spiked in N. tabacum cell culture supernatant (Huang et al., 2009) and the IF-mediated fragmentation of proteolysis-prone monoclonal antibodies (Niemer et al., 2014).
The mode of action of A1AT is to trap target proteases that bind to its bait on the RCL and cleave it. The protease becomes destabilized and partially unfolded, changing the shape of its active site so that it is no longer active (Elliott et al., 1996). However, various proteinases can cleave A1AT within its RCL without being trapped. Previous studies have shown that the inhibitor can be inactivated by interaction with bacterial proteases, thereby causing the conversion of native A1AT into an inactive form of decreased Mr (Potempa et al., 1986; Nelson et al., 1998, 1999). This seems to have also been the case in our studies. Indeed, it seems likely that StrepA1AT accumulates first as a full-length active protein (native state), which acts as bait for host Ser proteases. However, in contrast to elastase (a target protease), the complex formed between A1AT and the nontarget plant proteases is reversible, with the 40-kD A1AT reflecting the converted state of the protein after complex dissociation. A1AT analyzed after incubation with target proteases (chymotrypsin, trypsin, and elastase) or nontarget proteases (papain and bacterial proteases; Johnson and Travis, 1978; Potempa et al., 1986) exhibited C-terminal truncations highly similar to those observed in our studies. Certainly, activity assays of N. benthamiana leaf extracts suggest an abundance of Ser-type proteases (i.e. trypsin- and chymotrypsin-like proteases) in the apoplast (Goulet et al., 2012).
Although C-terminal truncations of recombinant A1AT have been previously reported, N-terminal cleavage has so far not been investigated. However, analysis of the migration pattern of serum A1AT by isoelectric focusing suggested N-terminal truncations to be present (Kolarich et al., 2006). Interestingly, engineered A1AT with designed truncations at the N terminus showed higher inhibitory activity than the native protein (Pirooznia et al., 2013).
Another result of our studies, which we did not expect, was the presence of paucimannosidic N-glycans on IF-derived A1AT (MMXF or MM depending on whether the expression host was the wild type or ∆XT/FT). Although these truncated N-glycans are frequently found in plants, secreted recombinant glycoproteins usually carry complex N-glycans, like GnGnXF (Strasser et al., 2008; Castilho et al., 2011a, 2011b; Schneider et al., 2014a). Nevertheless, two studies have reported the presence of paucimannosidic oligosaccharides on recombinant proteins, maize (Zea mays)- and N. tabacum-derived human lactoferrin (Samyn-Petit et al., 2003), and bovine follicle-stimulating hormone expressed in N. benthamiana (Dirnberger et al., 2001). It is well known that the formation of this glycoform relies on the activity of HEXOs, which remove terminal GlcNAc residues from complex N-glycans (Strasser et al., 2007; Liebminger et al., 2011). Despite the presence of HEXO3 in the plasma membrane, it is commonly suggested that this glycan structure is generated in the vacuoles (Gomord et al., 2010). In this study, vacuolar HEXO1 does not seem to play a major role in the N-glycan processing of A1AT. Instead, our results indicate that complex N-glycans of A1AT are converted to paucimannosidic structures, most probably in the extracellular space by the action of HEXO3. This is a highly interesting finding, because most endogenous and recombinant proteins targeted to the apoplast are not processed by HEXO3. Why some proteins, like A1AT, are processed, whereas others, like human transferrin, are not processed by β-hexosaminidases when expressed and isolated in a similar way (Castilho et al., 2011b) is not known. Although it cannot be completely excluded that spatial separation is responsible for the observed differences, it seems more likely that the intrinsic properties of these proteins, such as their tertiary structures, are the major determinants. Recent advances in computer modeling of complex proteins and glycan structures designed to supplement experimental data (Nagae and Yamaguchi, 2012; Loos et al., 2014) should make it easier to explain this largely unexplored field.
Despite the unusual glycosylation profile of secreted A1AT, coexpression with genes of the sialylation pathway resulted in the generation of fully disialylated N-glycans similar to those in plasma A1AT. The formation of paucimannosidic structures could be largely inhibited. The results indicate that the presence of sialic acid residues protects terminal GlcNAc residues from being removed by HEXO3. In insect cells, the synthesis of paucimannosidic structures could be prevented by capping terminal GlcNAc residues with β1,4-Gal (Ailor et al., 2000). In contrast, our attempts to β1,4-galactosylate A1AT by the overexpression of the highly active human β1,4-galactosyltransferase (Strasser et al., 2009) proved unsuccessful (data not shown), probably because of the presence of potent β-galactosidases in the secretory pathway of plant cells. Hence, only capping of the newly synthesized β1,4-Gal with sialic acid residues prevented their removal as well as GlcNAc residues from complex N-glycan structures.
Purified full-length StrepA1AT was found to carry only ER-typical oligomannosidic structures. This finding complies with that of the subcellular localization studies, in which A1AT-GFP was found to exhibit an ER-associated staining pattern. Partial aberrant subcellular localization of mammalian recombinant proteins expressed in plants has been reported (Loos et al., 2011; Schneider et al., 2014b). It seems that the repertoire of chaperones and folding factors, which is mainly responsible for correct protein folding and ER protein export, differs to some extent between plant and mammalian cells (Gupta and Tuteja, 2011; Denecke et al., 2012). The molecular interactions that allow the ER to retain proteins with no defined ER retrieval or retention signal are not entirely understood, but it is possible that interactions with ER quality components retain incompletely folded or aberrant proteins in the ER. Notwithstanding, we observed that the purified A1AT displayed elastase inhibitory activity, which indicates correct folding.
Processed versions of A1AT were obtained in three different ways: from in planta IF deposition, purified StrepA1AT incubated with PPE, and plasma A1AT after exposure to IF. The three derivatives showed different migration patterns on SDS-PAGE, despite having virtually identical protein backbones, which was confirmed by peptide mapping. Differences in migration can be explained by distinct glycosylation profiles (i.e. paucimannosidic [IF-derived StrepA1AT], oligomannosidic [purified StrepA1AT], and sialylated glycans [plasma-derived A1AT]).
Here, we efficiently decorated plant-derived A1AT with sialylated structures. By coexpressing α2,6-sialyltransferase (ST6), secreted plant-derived A1AT carries exclusively α2,6-linked terminal sialic acid (Castilho et al., 2010) corresponding to the native therapeutic counterpart (Prolastin) and in sharp contrast to A1AT expressed in Chinese hamster ovary cells, where mainly α2,3-linked sialylation is present (Lee et al., 2013). Pharmacokinetic (PK) studies in rats have shown that the mean residence time of A1AT can be modified by varying the sialic acid linkage (Brinkman et al., 2012). Using the glycoengineering approach described here, it has been possible to generate recombinant proteins decorated with α2,6- or α2,3-sialic acid or a mixture of them, depending on the sialyltransferase used (ST6, ST3, or both; A. Castilho and H. Steinkellner, unpublished data). This procedure allows A1AT to be generated with tailored PK values. This means, for example, that the PK of A1AT can be artificially increased by α2,3-sialylation, which would reduce the dose needed per patient. However, using the α2,6-sialylated counterpart may be advantageous because of the more natural linkage. Apart from the sialic acid linkage, the overall sialylation content of A1AT influences PK values (Lindhout et al., 2011; Lusch et al., 2013). Here, we were able to increase the levels of A1AT sialylation in planta through the generation of triantennary structures, which indeed, showed a significant increase in the sialic acid content over its plasma-derived counterpart. This could contribute to improved PK profiles, which has also been shown for other therapeutic proteins (Elliott et al., 2003).
In summary, we provide unique insights into how complex human glycoproteins are generated and processed in plants. Although the plant-based expression of some therapeutically interesting proteins (e.g. antibodies and certain enzymes) seems to be a success story (Grabowski et al., 2014; Hiatt et al., 2014), our results highlight that it is hard to predict how complex human proteins actually perform when expressed in plants. Upon expression of human A1AT, we observed some unexpected phenomena, such as aberrant subcellular localization, post-ER processing at the N and C termini, and unusual glycosylation of the protein. Although we could only partially explain the first two phenomena, we could gain valuable unique insights into the proteolytic network of the secretory pathway, which might enable the design of strategies to overcome the unwanted processing of plant-produced A1AT. Importantly, we were able to uncover the process that underlies the synthesis of paucimannosidic structures on secretory proteins and provided two independent approaches (knockout of HEXO3 activities and sialylation) to direct their glycosylation pattern toward human-like structures. Overall, our results contribute to a better understanding of the secretory pathway in plant cells.
MATERIALS AND METHODS
A1AT Expression Vectors
Human A1AT cDNA (BC0156429) was purchased from the German Resource Centre for Genome Research (IRALp962E2033Q2). Two vector systems were used to express A1AT in plants: the plant viral-based magnICON vector pICH26211α carrying the barley α-amylase signal peptide for protein secretion (Fig. 1A; Schneider et al., 2014a) and standard binary vectors (Fig. 1B). A1AT cDNA (lacking the endogenous signal peptide) was appended with a strep tag on the N (StrepA1AT) and C (A1ATStrep) termini by means of PCR (Supplemental Table S2). Flanking BsaI restriction sites were included in the primer sequences to allow cloning of the PCR fragment into the pICH26211α vector. A1AT was also expressed in plants using standard binary vectors. This was done by amplifying the full-length A1AT sequence (amino acids 1–418, including the native signal peptide) as an XbaI-BamHI fragment and cloning it into the binary vector pPT2 (35S:A1ATFull) or p20 to generate a C-terminal GFP fusion (35S:A1AT-GFP’ Castilho et al., 2008).
All binary vectors were transferred into Agrobacterium tumefaciens strain UIA143, and magnICON constructs were transformed into strain GV3101 pMP90. Primers used in this investigation are listed in Supplemental Table S2.
Binary Vector for Modulation of A1AT N-Glycosylation
The binary vectors used to modulate the A1AT glycosylation pattern are described elsewhere. Briefly, biosynthesis of nucleotide-activated N-acetyl-neuraminic acid was achieved by the concomitant expression of UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase, N-acetylneuraminic acid phosphate synthase, and CMP-N-acetylneuraminic acid synthase (Castilho et al., 2008). β1,4-Galactosyltransferase was expressed as a fusion of the catalytic domain to the cytoplasmic tail, transmembrane domain, and stem region of ST6 (Strasser et al., 2009). The transport of sialic acid into the Golgi and its transfer to terminal Gal was achieved by the expression of the CMP-sialic acid transporter and ST6 as described previously (Castilho et al., 2010). For the synthesis of multiantennary glycans, we used binary vectors containing chimeric forms of GnTIV and GnTV targeted to medial Golgi compartments (Castilho et al., 2011b).
Plant Material and Plant Transformation
Nicotiana benthamiana wild-type, ∆XT/FT glycosylation mutants (Strasser et al., 2008), and Arabidopsis (Arabidopsis thaliana) plants (Col-0, hexo1, hexo3, and hexo1/hexo3 hexosaminidase single and double knockouts; Liebminger et al., 2011) were grown in a growth chamber at 22°C with a 16-h-light/8-h-dark photoperiod.
Agrobacterium spp. carrying the appropriate magnICON and binary vectors was used in agroinfiltration experiments to transiently express recombinant proteins (A1AT and proteins for glycan engineering) in N. benthamiana. magnICON and binary vectors were infiltrated at optical density at 600 nm (OD600) = 0.1 and OD600 = 0.05, respectively. For subcellular localization experiments, binary vectors were diluted to OD600 = 0.03, and samples were harvested at 3 d postinfiltration.
Floral dipping was used for stable expression of A1AT in Arabidopsis wild-type Col-0 and HEXO knockout lines (Liebminger et al., 2011) using 35S:A1ATFull.
Isolation of Intercellular Fluid and Purification of A1AT
IF was collected using fully expanded leaves as described previously (Castilho et al., 2011b). The pH of the IF isolation buffer (100 mm Tris-HCl, pH 7.5, 10 mm MgCl2, and 2 mm EDTA) was adjusted to 5.0 or 7.0 according to the experiment. One N. benthamiana leaf yields 500 µL of IF, whereas 200 µL of IF is recovered from 20 Arabidopsis leaves. For TSP extraction, infiltrated leaf material was ground in a swing mill (MM2000; Retsch) for 2 min at amplitude 70; then, 1 volume (v/w) of 1× PBS was added. After incubation on ice for 10 min, the extracts were centrifuged (10,000g for 5 min at 4°C). A1AT was purified from TSP extracts by affinity chromatography using Strep-Tactin MacroPrep (IBA2-1505) according to the manufacturer’s instructions.
Immunoblotting
Proteins were fractionated in 12% (w/v) SDS-PAGE under reducing conditions. Western blotting was done using A1AT-specific antibodies (1:2,000 rabbit anti-hA1AT; 49088; Abcam) or tag-specific antibodies (1:5,000 mouse anti-StrepII tag IBA2-1507 and 1:2,000 mouse anti-GFP; TP401; Amsbio). Detection was performed using horseradish peroxidase-conjugated secondary antibodies (anti-rabbit A0545 and anti-mouse IgG [both diluted 1:10,000]; A2554; Sigma Aldrich). Clarity Western enhanced chemiluminescence reagents from Bio-Rad Laboratories were used as substrates.
Quantification of A1AT expression was carried out by immunoblot analysis using A1AT-specific antibodies. TSPs were extracted from leaves expressing StrepA1AT and diluted (1:100; 1:200; 1:500, and 1:1,000); 5 µL was loaded on SDS-PAGE and analyzed by western blotting using antibodies against A1AT. Commercially available plasma-derived A1AT (A6150; Sigma) was used as a reference. Western-blot images captured with Gel Doc EQ were analyzed using Quantity One (Bio-Rad Laboratories) software to measure relative band intensities.
A1AT Incubation Studies
Purified plant-derived A1AT and plasma-derived A1AT (approximately 60 ng) were incubated for 1 h at 37°C with 20 µL of concentrated IF (pH 5.0 and 7.0) isolated from wild-type N. benthamiana; 10 µL of Ser and Cys proteases inhibitors (14 mm E-64, 10 mm leupeptin, and 20 mm PMSF; Sigma), cathepsin B inhibitor (10 mm CA-074; Sigma), and proteinase inhibitor cocktail (100×; Sigma) was added to 20 µL of concentrated IF (pH 5.0) and incubated overnight at 37°C. Afterward, 60 ng of hA1AT or purified plant-derived A1AT was added, and the reaction mixtures were incubated at 37°C for 1 h.
All samples were mixed with 3× Laemmli sample buffer, and proteins were denatured by incubation at 95°C for 5 min. Finally, samples were loaded on 12% (w/v) SDS-PAGE and analyzed by western blot.
Glycosylation Analysis and Peptide Mapping
A1AT was resolved by 12% (w/v) SDS-PAGE, S-alkylated, digested with trypsin, and analyzed by LC-ESI-MS (Stadlmann et al., 2008; Pabst et al., 2012). This digestion allows site-specific analysis of three N-glycosylation sites: Gp1, R/40QLAHQSNSTNIFFSPVSIATAFAMLSLGTK69; Gp2, K/70ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR101; and Gp3, K/243YLGNATAIFFLPDEGK259. Arabidopsis-derived A1AT was analyzed using a different nano-LC-ESI-MS setup to account for the small amounts of material available. Peptides were separated with a PepMap 100 C-18 column (150 × 0.075 mm, 3-µm particle size; LC packings) using a Dionex Ultimate 3000 nano-HPLC System with a flow rate of 0.4 µL per minute. The gradient applied was identical to the standard setup gradient, but the aqueous solvent was changed to 0.1% (v/v) formic acid. Peptides were ionized with a captive spray source and analyzed on a Bruker maXis 4G in the data-dependent acquisition mode using collision-induced (activated) dissociation. Glycopeptides were identified by their mass and the appearance of glycan fragment ions in MS2 spectra.
Identification of N and C termini was carried out by tryptic peptide mapping using LC-ESI-MS/MS (Pabst et al., 2012).
CLSM
Expression of A1AT C-terminally tagged with GFP (35S:A1AT-GFP) was monitored using an upright Leica TCS SP5 CLSM. The vector 35S:GFP was used to compare the subcellular localization of the A1AT fusion protein with that of GFP. GnTI-CAAATS-mRFP (Schoberer et al., 2009) was used as an ER marker in colocalization studies. Dual-color imaging of GFP- and mRFP-expressing cells was performed simultaneously using a 488-nm argon laser line and a 561-nm helium/neon laser line. The images obtained were processed in Adobe Photoshop CS4.
Estimation of Functional Activity of A1AT
The elastase inhibitory activity of the plant-derived strep tag-purified A1AT was assayed using the SensoLyte Rh110 Elastase Assay Kit (Fluorimetric; 72179; Anaspec). The inhibitory capacity of A1AT was calculated as the residual activity of elastase measured using a fluorogenic substrate, and the procedure followed the manufacturer’s instructions. In each reaction, 10 ng of elastase was incubated with plant-derived purified StrepA1AT at different concentrations ranging from 0 to 80 ng. Elution buffer used for StrepA1AT purification was used as a negative control to test sample autofluorescence.
A band shift assay was used to analyze A1AT and PPE complex formation. Briefly, 500 ng of plant-derived A1AT was incubated with 1,300 ng of PPE (E7885; Sigma) in 20 mm Tris-HCl buffer (pH 8.0) at 37°C for 20 min. All samples were analyzed by western blot using anti-A1AT antibodies or stained with Coomassie Brilliant Blue.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence of the mature hA1AT protein lacking the signal peptide (amino acids 1–394).
Supplemental Figure S2. Determination of StrepA1AT expression level in N. benthamiana.
Supplemental Figure S3. Identification of the amino- and carboxyl-terminal amino acid sequences of the StrepA1AT 40-kD band after interaction with elastase.
Supplemental Figure S4. Expression profiling and glycan analysis of A1AT produced in Arabidopsis Col-0.
Supplemental Figure S5. LC-ESI-MS-based N-glycosylation profiles of plasma- and plant-derived A1AT.
Supplemental Table S1. Relative abundance of major glycoforms detected on secreted StrepA1AT (given in percentage).
Supplemental Table S2. List of primers used in this investigation to express hA1AT in plants.
Supplementary Material
Acknowledgments
We thank Thomas Hackl (Department of Applied Genetics and Cell Biology, University of Natural Resources and Life Sciences, Vienna) for excellent technical support, Jennifer Schoberer (Department of Applied Genetics and Cell Biology, University of Natural Resources and Life Sciences) for supplying the GnTI-CAAATS-mRFP ER marker, and colleagues from Icon Genetics GmbH for providing access to the magnICON expression system.
Glossary
- cDNA
complementary DNA
- CLSM
confocal laser-scanning microscopy
- Col-0
Columbia-0
- ER
endoplasmic reticulum
- ESI
electrospray ionization
- IF
intercellular space
- LC
liquid chromatography
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- PK
pharmacokinetic
- PMSF
phenylmethylsulfonyl fluoride
- PPE
porcine pancreatic elastase
- RCL
reactive center loop
- TSP
total soluble protein
Footnotes
This work was supported by the Austrian Research Promotion Agency and Icon Genetics GmbH in the frame of Laura Bassi Centres of Expertise PlantBioP (grant no. 822757) and the Austrian Science Fund (grant no. L575–B13).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agarwal S, Singh R, Sanyal I, Amla DV (2008) Expression of modified gene encoding functional human alpha-1-antitrypsin protein in transgenic tomato plants. Transgenic Res 17: 881–896 [DOI] [PubMed] [Google Scholar]
- Ailor E, Takahashi N, Tsukamoto Y, Masuda K, Rahman BA, Jarvis DL, Lee YC, Betenbaugh MJ (2000) N-glycan patterns of human transferrin produced in Trichoplusia ni insect cells: effects of mammalian galactosyltransferase. Glycobiology 10: 837–847 [DOI] [PubMed] [Google Scholar]
- Alkins SA, O’Malley P (2000) Should health-care systems pay for replacement therapy in patients with alpha(1)-antitrypsin deficiency? A critical review and cost-effectiveness analysis. Chest 117: 875–880 [DOI] [PubMed] [Google Scholar]
- Arjmand S, Bidram E, Lotfi AS, Shamsara M, Mowla SJ (2011) Expression and purification of functionally active recombinant human alpha 1-antitrypsin in methylotrophic yeast Pichia pastoris. Avicenna J Med Biotechnol 3: 127–134 [PMC free article] [PubMed] [Google Scholar]
- Blanchard V, Liu X, Eigel S, Kaup M, Rieck S, Janciauskiene S, Sandig V, Marx U, Walden P, Tauber R, et al. (2011) N-glycosylation and biological activity of recombinant human alpha1-antitrypsin expressed in a novel human neuronal cell line. Biotechnol Bioeng 108: 2118–2128 [DOI] [PubMed] [Google Scholar]
- Blank CA, Brantly M (1994) Clinical features and molecular characteristics of alpha 1-antitrypsin deficiency. Ann Allergy 72: 105–120 [PubMed] [Google Scholar]
- Brinkman E, Hack C, Van den Nieuwenhof I, Inventors. January 22, 2013. Recombinant human alpha1-antitrypsin. Patent Publication No. 2012/0214747: A1 [Google Scholar]
- Castilho A, Bohorova N, Grass J, Bohorov O, Zeitlin L, Whaley K, Altmann F, Steinkellner H (2011a) Rapid high yield production of different glycoforms of Ebola virus monoclonal antibody. PLoS ONE 6: e26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Gattinger P, Grass J, Jez J, Pabst M, Altmann F, Gorfer M, Strasser R, Steinkellner H (2011b) N-glycosylation engineering of plants for the biosynthesis of glycoproteins with bisected and branched complex N-glycans. Glycobiology 21: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Neumann L, Daskalova S, Mason HS, Steinkellner H, Altmann F, Strasser R (2012) Engineering of sialylated mucin-type O-glycosylation in plants. J Biol Chem 287: 36518–36526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Neumann L, Gattinger P, Strasser R, Vorauer-Uhl K, Sterovsky T, Altmann F, Steinkellner H (2013) Generation of biologically active multi-sialylated recombinant human EPOFc in plants. PLoS ONE 8: e54836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Pabst M, Leonard R, Veit C, Altmann F, Mach L, Glössl J, Strasser R, Steinkellner H (2008) Construction of a functional CMP-sialic acid biosynthesis pathway in Arabidopsis. Plant Physiol 147: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Steinkellner H (2012) Glyco-engineering in plants to produce human-like N-glycan structures. Biotechnol J 7: 1088–1098 [DOI] [PubMed] [Google Scholar]
- Castilho A, Strasser R, Stadlmann J, Grass J, Jez J, Gattinger P, Kunert R, Quendler H, Pabst M, Leonard R, et al. (2010) In planta protein sialylation through overexpression of the respective mammalian pathway. J Biol Chem 285: 15923–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Choi JY, Jin BR, Roh JY, Olszewski JA, Seo SJ, O’Reilly DR, Je YH (2003) An improved baculovirus insecticide producing occlusion bodies that contain Bacillus thuringiensis insect toxin. J Invertebr Pathol 84: 30–37 [DOI] [PubMed] [Google Scholar]
- Denecke J, Aniento F, Frigerio L, Hawes C, Hwang I, Mathur J, Neuhaus JM, Robinson DG (2012) Secretory pathway research: the more experimental systems the better. Plant Cell 24: 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnberger D, Steinkellner H, Abdennebi L, Remy JJ, van de Wiel D (2001) Secretion of biologically active glycoforms of bovine follicle stimulating hormone in plants. Eur J Biochem 268: 4570–4579 [DOI] [PubMed] [Google Scholar]
- Doran PM. (2006) Foreign protein degradation and instability in plants and plant tissue cultures. Trends Biotechnol 24: 426–432 [DOI] [PubMed] [Google Scholar]
- Elliott PR, Lomas DA, Carrell RW, Abrahams JP (1996) Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nat Struct Biol 3: 676–681 [DOI] [PubMed] [Google Scholar]
- Elliott S, Lorenzini T, Asher S, Aoki K, Brankow D, Buck L, Busse L, Chang D, Fuller J, Grant J, et al. (2003) Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat Biotechnol 21: 414–421 [DOI] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver RI Jr, Chytil A, Karlsson S, Fells GA, Brantly ML, Courtney M, Kantoff PW, Nienhuis AW, Anderson WF, Crystal RG (1987) Production of glycosylated physiologically “normal” human alpha 1-antitrypsin by mouse fibroblasts modified by insertion of a human alpha 1-antitrypsin cDNA using a retroviral vector. Proc Natl Acad Sci USA 84: 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins PG. (2002) Serpin structure, mechanism, and function. Chem Rev 102: 4751–4804 [DOI] [PubMed] [Google Scholar]
- Gleba YY, Tusé D, Giritch A (2014) Plant viral vectors for delivery by Agrobacterium. Curr Top Microbiol Immunol 375: 155–192 [DOI] [PubMed] [Google Scholar]
- Gomord V, Fitchette AC, Menu-Bouaouiche L, Saint-Jore-Dupas C, Plasson C, Michaud D, Faye L (2010) Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol J 8: 564–587 [DOI] [PubMed] [Google Scholar]
- Goulet C, Khalf M, Sainsbury F, D’Aoust MA, Michaud D (2012) A protease activity-depleted environment for heterologous proteins migrating towards the leaf cell apoplast. Plant Biotechnol J 10: 83–94 [DOI] [PubMed] [Google Scholar]
- Grabowski GA, Golembo M, Shaaltiel Y (2014) Taliglucerase alfa: an enzyme replacement therapy using plant cell expression technology. Mol Genet Metab 112: 1–8 [DOI] [PubMed] [Google Scholar]
- Grignon C, Sentenac H (1991) Ph and ionic conditions in the apoplast. Annu Rev Plant Physiol Plant Mol Biol 42: 103–128 [Google Scholar]
- Gupta D, Tuteja N (2011) Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal Behav 6: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasannia S, Lotfi AS, Mahboudi F, Rezaii A, Rahbarizadeh F, Mohsenifar A (2006) Elevated expression of human alpha-1 antitrypsin mediated by yeast intron in Pichia pastoris. Biotechnol Lett 28: 1545–1550 [DOI] [PubMed] [Google Scholar]
- Hiatt A, Bohorova N, Bohorov O, Goodman C, Kim D, Pauly MH, Velasco J, Whaley KJ, Piedra PA, Gilbert BE, et al. (2014) Glycan variants of a respiratory syncytial virus antibody with enhanced effector function and in vivo efficacy. Proc Natl Acad Sci USA 111: 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sutliff TD, Wu L, Nandi S, Benge K, Terashima M, Ralston AH, Drohan W, Huang N, Rodriguez RL (2001) Expression and purification of functional human alpha-1-Antitrypsin from cultured plant cells. Biotechnol Prog 17: 126–133 [DOI] [PubMed] [Google Scholar]
- Huang TK, Plesha MA, Falk BW, Dandekar AM, McDonald KA (2009) Bioreactor strategies for improving production yield and functionality of a recombinant human protein in transgenic tobacco cell cultures. Biotechnol Bioeng 102: 508–520 [DOI] [PubMed] [Google Scholar]
- Huang TK, Plesha MA, McDonald KA (2010) Semicontinuous bioreactor production of a recombinant human therapeutic protein using a chemically inducible viral amplicon expression system in transgenic plant cell suspension cultures. Biotechnol Bioeng 106: 408–421 [DOI] [PubMed] [Google Scholar]
- Jha S, Agarwal S, Sanyal I, Jain GK, Amla DV (2012) Differential subcellular targeting of recombinant human α1-proteinase inhibitor influences yield, biological activity and in planta stability of the protein in transgenic tomato plants. Plant Sci 196: 53–66 [DOI] [PubMed] [Google Scholar]
- Johnson D, Travis J (1978) Structural evidence for methionine at the reactive site of human alpha-1-proteinase inhibitor. J Biol Chem 253: 7142–7144 [PubMed] [Google Scholar]
- Johnson DA, Barrett AJ, Mason RW (1986) Cathepsin L inactivates alpha 1-proteinase inhibitor by cleavage in the reactive site region. J Biol Chem 261: 14748–14751 [PubMed] [Google Scholar]
- Karnaukhova E, Ophir Y, Golding B (2006) Recombinant human alpha-1 proteinase inhibitor: towards therapeutic use. Amino Acids 30: 317–332 [DOI] [PubMed] [Google Scholar]
- Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, Ilyas M, Win J, Kamoun S, van der Hoorn RA (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol 154: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarich D, Weber A, Turecek PL, Schwarz HP, Altmann F (2006) Comprehensive glyco-proteomic analysis of human alpha1-antitrypsin and its charge isoforms. Proteomics 6: 3369–3380 [DOI] [PubMed] [Google Scholar]
- Lee KJ, Lee SM, Gil JY, Kwon O, Kim JY, Park SJ, Chung HS, Oh DB (2013) N-glycan analysis of human α1-antitrypsin produced in Chinese hamster ovary cells. Glycoconj J 30: 537–547 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé AC, Gomord V, Faye L (1998) N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38: 31–48 [PubMed] [Google Scholar]
- Liebminger E, Veit C, Pabst M, Batoux M, Zipfel C, Altmann F, Mach L, Strasser R (2011) Beta-N-acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. J Biol Chem 286: 10793–10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout T, Iqbal U, Willis LM, Reid AN, Li J, Liu X, Moreno M, Wakarchuk WW (2011) Site-specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc Natl Acad Sci USA 108: 7397–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos A, Gruber C, Altmann F, Mehofer U, Hensel F, Grandits M, Oostenbrink C, Stadlmayr G, Furtmüller PG, Steinkellner H (2014) Expression and glycoengineering of functionally active heteromultimeric IgM in plants. Proc Natl Acad Sci USA 111: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos A, Van Droogenbroeck B, Hillmer S, Grass J, Pabst M, Castilho A, Kunert R, Liang M, Arcalis E, Robinson DG, et al. (2011) Expression of antibody fragments with a controlled N-glycosylation pattern and induction of endoplasmic reticulum-derived vesicles in seeds of Arabidopsis. Plant Physiol 155: 2036–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusch A, Kaup M, Marx U, Tauber R, Blanchard V, Berger M (2013) Development and analysis of alpha 1-antitrypsin neoglycoproteins: the impact of additional N-glycosylation sites on serum half-life. Mol Pharm 10: 2616–2629 [DOI] [PubMed] [Google Scholar]
- Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y (2004) In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA 101: 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y (2005) Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 23: 718–723 [DOI] [PubMed] [Google Scholar]
- Mast AE, Enghild JJ, Pizzo SV, Salvesen G (1991) Analysis of the plasma elimination kinetics and conformational stabilities of native, proteinase-complexed, and reactive site cleaved serpins: comparison of alpha 1-proteinase inhibitor, alpha 1-antichymotrypsin, antithrombin III, alpha 2-antiplasmin, angiotensinogen, and ovalbumin. Biochemistry 30: 1723–1730 [DOI] [PubMed] [Google Scholar]
- McDonald KA, Hong LM, Trombly DM, Xie Q, Jackman AP (2005) Production of human alpha-1-antitrypsin from transgenic rice cell culture in a membrane bioreactor. Biotechnol Prog 21: 728–734 [DOI] [PubMed] [Google Scholar]
- Nadai M, Bally J, Vitel M, Job C, Tissot G, Botterman J, Dubald M (2009) High-level expression of active human alpha1-antitrypsin in transgenic tobacco chloroplasts. Transgenic Res 18: 173–183 [DOI] [PubMed] [Google Scholar]
- Nagae M, Yamaguchi Y (2012) Function and 3D structure of the N-glycans on glycoproteins. Int J Mol Sci 13: 8398–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Potempa J, Kordula T, Travis J (1999) Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem 274: 12245–12251 [DOI] [PubMed] [Google Scholar]
- Nelson D, Potempa J, Travis J (1998) Inactivation of alpha1-proteinase inhibitor as a broad screen for detecting proteolytic activities in unknown samples. Anal Biochem 260: 230–236 [DOI] [PubMed] [Google Scholar]
- Niemer M, Mehofer U, Torres Acosta JA, Verdianz M, Henkel T, Loos A, Strasser R, Maresch D, Rademacher T, Steinkellner H, et al. (2014) The human anti-HIV antibodies 2F5, 2G12, and PG9 differ in their susceptibility to proteolytic degradation: down-regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant-based expression platforms. Biotechnol J 9: 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Chang M, Stadlmann J, Altmann F (2012) Glycan profiles of the 27 N-glycosylation sites of the HIV envelope protein CN54gp140. Biol Chem 393: 719–730 [DOI] [PubMed] [Google Scholar]
- Pike RN, Bottomley SP, Irving JA, Bird PI, Whisstock JC (2002) Serpins: finely balanced conformational traps. IUBMB Life 54: 1–7 [DOI] [PubMed] [Google Scholar]
- Pirooznia N, Hasannia S, Arab SS, Lotfi AS, Ghanei M, Shali A (2013) The design of a new truncated and engineered alpha1-antitrypsin based on theoretical studies: an antiprotease therapeutics for pulmonary diseases. Theor Biol Med Model 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesha MA, Huang TK, Dandekar AM, Falk BW, McDonald KA (2007) High-level transient production of a heterologous protein in plants by optimizing induction of a chemically inducible viral amplicon expression system. Biotechnol Prog 23: 1277–1285 [DOI] [PubMed] [Google Scholar]
- Potempa J, Watorek W, Travis J (1986) The inactivation of human plasma alpha 1-proteinase inhibitor by proteinases from Staphylococcus aureus. J Biol Chem 261: 14330–14334 [PubMed] [Google Scholar]
- Ross D, Brown T, Harper R, Pamarthi M, Nixon J, Bromirski J, Li CM, Ghali R, Xie H, Medvedeff G, et al. (2012) Production and characterization of a novel human recombinant alpha-1-antitrypsin in PER.C6 cells. J Biotechnol 162: 262–273 [DOI] [PubMed] [Google Scholar]
- Samyn-Petit B, Wajda Dubos JP, Chirat F, Coddeville B, Demaizieres G, Farrer S, Slomianny MC, Theisen M, Delannoy P (2003) Comparative analysis of the site-specific N-glycosylation of human lactoferrin produced in maize and tobacco plants. Eur J Biochem 270: 3235–3242 [DOI] [PubMed] [Google Scholar]
- Schneider JD, Castilho A, Neumann L, Altmann F, Loos A, Kannan L, Mor TS, Steinkellner H (2014a) Expression of human butyrylcholinesterase with an engineered glycosylation profile resembling the plasma-derived orthologue. Biotechnol J 9: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JD, Marillonnet S, Castilho A, Gruber C, Werner S, Mach L, Klimyuk V, Mor TS, Steinkellner H (2014b) Oligomerization status influences subcellular deposition and glycosylation of recombinant butyrylcholinesterase in Nicotiana benthamiana. Plant Biotechnol J 12: 832–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Vavra U, Stadlmann J, Hawes C, Mach L, Steinkellner H, Strasser R (2009) Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F (2008) Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8: 2858–2871 [DOI] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Schoberer J, Svoboda B, Liebminger E, Glössl J, Altmann F, Steinkellner H, Mach L (2007) Enzymatic properties and subcellular localization of Arabidopsis beta-N-acetylhexosaminidases. Plant Physiol 145: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Castilho A, Stadlmann J, Kunert R, Quendler H, Gattinger P, Jez J, Rademacher T, Altmann F, Mach L, et al. (2009) Improved virus neutralization by plant-produced anti-HIV antibodies with a homogeneous beta1,4-galactosylated N-glycan profile. J Biol Chem 284: 20479–20485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Schähs M, Stiegler G, Quendler H, Mach L, Glössl J, Weterings K, Pabst M, Steinkellner H (2008) Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol J 6: 392–402 [DOI] [PubMed] [Google Scholar]
- Sudarshana MR, Plesha MA, Uratsu SL, Falk BW, Dandekar AM, Huang TK, McDonald KA (2006) A chemically inducible cucumber mosaic virus amplicon system for expression of heterologous proteins in plant tissues. Plant Biotechnol J 4: 551–559 [DOI] [PubMed] [Google Scholar]
- Terashima M, Murai Y, Kawamura M, Nakanishi S, Stoltz T, Chen L, Drohan W, Rodriguez RL, Katoh S (1999) Production of functional human alpha 1-antitrypsin by plant cell culture. Appl Microbiol Biotechnol 52: 516–523 [DOI] [PubMed] [Google Scholar]
- Travis J, Salvesen GS (1983) Human plasma proteinase inhibitors. Annu Rev Biochem 52: 655–709 [DOI] [PubMed] [Google Scholar]
- Trexler MM, McDonald KA, Jackman AP (2002) Bioreactor production of human alpha(1)-antitrypsin using metabolically regulated plant cell cultures. Biotechnol Prog 18: 501–508 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Van Droogenbroeck B, Cao J, Stadlmann J, Altmann F, Colanesi S, Hillmer S, Robinson DG, Van Lerberge E, Terryn N, Van Montagu M, et al. (2007) Aberrant localization and underglycosylation of highly accumulating single-chain Fv-Fc antibodies in transgenic Arabidopsis seeds. Proc Natl Acad Sci USA 104: 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.