Two complementary inorganic carbon uptake systems show distinct functions in the CO2-concentrating mechanism and both are essential for limiting CO2 acclimation.
Abstract
The limiting-CO2 inducible CO2-concentrating mechanism (CCM) of microalgae represents an effective strategy to capture CO2 when its availability is limited. At least two limiting-CO2 acclimation states, termed low CO2 and very low CO2, have been demonstrated in the model microalga Chlamydomonas reinhardtii, and many questions still remain unanswered regarding both the regulation of these acclimation states and the molecular mechanism underlying operation of the CCM in these two states. This study examines the role of two proteins, Limiting CO2 Inducible A (LCIA; also named NAR1.2) and LCIB, in the CCM of C. reinhardtii. The identification of an LCIA-LCIB double mutant based on its inability to survive in very low CO2 suggests that both LCIA and LCIB are critical for survival in very low CO2. The contrasting effects of individual mutations in LCIB and LCIA compared with the effects of LCIB-LCIA double mutations on growth and inorganic carbon-dependent photosynthetic O2 evolution reveal distinct roles of LCIA and LCIB in the CCM. Although both LCIA and LCIB are essential for very low CO2 acclimation, LCIB appears to function in a CO2 uptake system, whereas LCIA appears to be associated with a HCO3− transport system. The contrasting and complementary roles of LCIA and LCIB in acclimation to low CO2 and very low CO2 suggest a possible mechanism of differential regulation of the CCM based on the inhibition of HCO3− transporters by moderate to high levels of CO2.
The CO2 concentration in Earth’s atmosphere has declined significantly since the origin of photosynthesis, and the current atmospheric CO2 level is a major limiting factor for optimal photosynthesis in many plant species. Rubisco, the central enzyme catalyzing CO2 assimilation, has a low affinity for CO2 and a slow catalytic turnover rate for the carboxylation reaction, possibly as an evolutionary relic, and also catalyzes the competing oxygenation reaction between O2 and ribulose-1,5-bisphosphate that releases fixed CO2 through photorespiration. As a result, several adaptive strategies have evolved to improve photosynthetic efficiency by raising the CO2 concentration at the site of Rubisco to increase the carboxylation rates and to suppress the wasteful photorespiration pathway. Among them, the cyanobacterial/microalgal CO2-concentrating mechanism (CCM) appears to be one of the most effective strategies for CO2 enrichment. These CCMs deploy diverse, active inorganic carbon (Ci) uptake systems, allowing cells to accumulate intracellular Ci up to 1,000-fold from low CO2 environments (Badger and Price, 2003; Moroney and Ynalvez, 2007; Price et al., 2008; Spalding, 2008). Because microalgae and cyanobacteria contribute a great portion of global CO2 sequestration through photosynthesis (Behrenfeld et al., 2001), and have shown enormous potential as an alternative future energy source (Sheehan et al., 1998; Wijffels and Barbosa, 2010), enhanced knowledge of the microalgal CCM will provide not only insights into the CCM’s influences on natural environments, but also guidance for the potential application of bioengineering approaches to enhance biomass productivity in economically important algae strains, or to improve photosynthetic carbon fixation in crop species that lack a CCM.
Although microalgal and cyanobacterial CCMs exhibit a very high level of diversity with regard to components and mechanisms (Badger and Spalding, 2000), these CCMs share some major molecular characteristics, including Rubisco sequestration in a specialized microcompartment, catalyzed interconversion of Ci species by carbonic anhydrase (CA), and energy-dependent, active Ci uptake systems. The eukaryotic CCM has been extensively studied in the model organism Chlamydomonas reinhardtii during the last 3 to 4 decades. Compared with the prokaryotic, cyanobacterial CCM, the eukaryotic, microalgal CCM appears complex because of the involvement of additional subcellular compartments and regulatory systems (Wang et al., 2011). Some areas of uncertainty regarding key features of the microalgal CCM remain to be addressed. First, Ci, including the charged species, bicarbonate (HCO3−), must cross both the plasma membrane and the chloroplast envelope to reach Rubisco, yet the roles, if any, of proposed and confirmed Ci transporters, including the plasma membrane proteins HLA3 and Limiting CO2 Inducible1 (LCI1) and the chloroplast envelope proteins LCIA (NAR1.2), CCP1, and CCP2 (Im and Grossman, 2002; Miura et al., 2004; Pollock et al., 2004; Mariscal et al., 2006; Duanmu et al., 2009a; Ohnishi et al., 2010), in acclimation and Ci uptake under different limiting CO2 conditions are not yet clearly defined. Second, previous physiological studies have demonstrated that active uptake of both CO2 and HCO3− occurs in C. reinhardtii (Moroney and Tolbert, 1985; Sültemeyer et al., 1989), but the molecular components and the underlying mechanism responsible for active CO2 uptake are still largely unknown. Third, the tight regulation of CCM functional components is still not well understood (Spalding et al., 2002). Physiological studies in wild-type C. reinhardtii cells have suggested that within the range of so-called limiting or low CO2 (the meaning of low CO2 is not universally defined in many previous publications, often ranging from air level of CO2 to nearly zero CO2), at least two distinct acclimation states, low CO2 (approximately 0.03%–0.5%) and very low CO2 (<0.02%), can be defined (Vance and Spalding, 2005). However, recent transcriptome studies (Brueggeman et al., 2012; Fang et al., 2012) failed to identify any changes that occur at the transcription abundance level between low CO2 and very low CO2 conditions, so it is still unclear how the CCM is differentially regulated between these two limiting CO2 acclimation states.
The first line of clear genetic evidence showing multiple, limiting-CO2 acclimation states came from identification of mutants with an air dier growth phenotype (Wang and Spalding, 2006); these mutants die in air level of CO2 (0.03%–0.05%) but can survive in either high CO2 (5%) or very low CO2 (<0.02%), and their ability to accumulate Ci is severely compromised in low CO2 but fairly normal in very low CO2. The mutation causing this air dier phenotype has been unequivocally linked to the LCIB gene (Wang and Spalding, 2006), which encodes a novel chloroplast protein that forms a heteromultimeric complex with its close homolog LCIC (Yamano et al., 2010; Wang and Spalding, 2014). It is unclear how LCIB is involved in Ci accumulation, largely because of its unknown biochemical characteristics and the lack of homologs of known function in other organisms. Genetic analysis of LCIB/CAH3 double mutants indicated that LCIB functions downstream of CAH3, a thylakoid lumen CA (Duanmu et al., 2009b). Because CAH3 catalyzes dehydration of accumulated HCO3− to provide CO2 for Rubisco inside the pyrenoid (Spalding et al., 1983b; Moroney et al., 2011), it has been hypothesized that LCIB captures CO2 leaked from the pyrenoid, possibly by unidirectionally hydrating CO2 back to HCO3− (Duanmu et al., 2009b). If this hypothesis is correct, it also implies that LCIB may also function in active Ci accumulation by unidirectionally hydrating externally diffused CO2 into the stromal HCO3− pool (Wang and Spalding, 2014). LCIB and the LCIB/LCIC complex change location within the chloroplast when cells are transferred from low CO2 (or high CO2) to very low CO2 conditions (Yamano et al., 2010; Wang and Spalding, 2014; Yamano et al., 2014), suggesting that a regulatory mechanism exists to regulate the functions of LCIB, possibly associated with low CO2 and very low CO2 acclimation states.
The growth phenotype and physiological characteristics of LCIB mutants clearly demonstrate that LCIB is indispensable for Ci acquisition in low CO2, but is dispensable in very low CO2 when other Ci uptake systems are still functional. It is also clear, however, that even though it is dispensable, LCIB contributes to viability in very low CO2 (Duanmu et al., 2009a), although neither its level of contribution to nor its precise role in Ci uptake under these conditions has been established. Two putative Ci transporters have been demonstrated to contribute LCIB-independent Ci uptake in C. reinhardtii. Knockdown of HLA3, a gene encoding an ATP-binding cassette transporter protein, causes growth defects in LCIB mutants in very low CO2 and decreased Ci accumulation especially at a high pH (Duanmu et al., 2009a). HLA3 is predicted to be localized on the plasma membrane, and therefore is very likely involved in HCO3− transport at this location. LCIA, another putative Ci transporter that might be involved in Ci uptake in very low CO2, belongs to a formate-nitrite transporter (FNT) family (Mariscal et al., 2006), and is predicted to be targeted to the chloroplast envelope (Miura et al., 2004). Although it shows significant identity to the C. reinhardtii nitrate assimilation-related (NAR) gene family and has also been named NAR1.2, unlike other NAR genes, LCIA expression is regulated by CO2 but not by nitrogen source (Miura et al., 2004; Mariscal et al., 2006), and introduction of an LCIA gene into Xenopus laevis oocytes reportedly increased HCO3− uptake (Mariscal et al., 2006). Although no LCIA single knockout or knockdown mutants have been reported, simultaneous co-knockdown of LCIA with HLA3 resulted in decreased Ci accumulation and decreased growth in very low CO2, especially when combined with LCIB mutations (Duanmu et al., 2009a). Although it appears that their contribution to Ci uptake may be important for the growth of C. reinhardtii under very low CO2, HLA3- or LCIA-associated Ci uptake in the absence of LCIB cannot support growth of LCIB mutants in air-level CO2, as evidenced by the air dier phenotype. This suggests that the primary role for these putative Ci transporters may be in very low CO2 conditions, but it is not clear whether, or how, HLA3 and LCIA are differentially regulated between very low CO2 and low CO2.
This study was undertaken primarily to dissect the molecular mechanisms underlying the functional acclimation to very low CO2 in C. reinhardtii. We identified an LCIA-LCIB double mutant that is unable to survive in very low CO2. The effects of the single LCIB mutation, the LCIA mutation, and the combined LCIA and LCIB mutations on growth and Ci-dependent photosynthetic O2 evolution reveal possible functions of LCIB and LCIA in the CCM, and demonstrate that both LCIA and LCIB contribute to Ci acquisition and acclimation under very low CO2 conditions but that they play distinct and complementary roles. Analyses of these data also revealed a possible mechanism to explain the curious air dier phenotype of LCIB mutants and the regulation between low CO2 and very low CO2 acclimation states.
RESULTS
Impact of LCIB Mutation on Ci-Dependent O2 Evolution
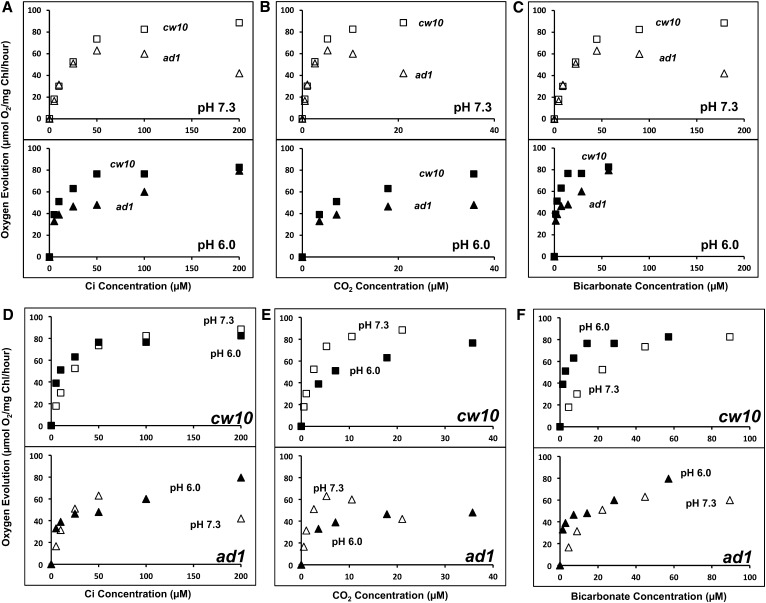
LCIB mutations cause severe inhibition of Ci uptake and photosynthesis in low CO2, but appear to have much less impact on photosynthesis in very low CO2 (Spalding et al., 1983a; Wang and Spalding, 2006; Duanmu et al., 2009a; Yamano et al., 2010; Duanmu and Spalding, 2011). To better understand this dichotomy, we more closely examined the impact of the LCIB mutation on very low CO2 acclimation and Ci species dependence of photosynthesis in the LCIB mutant by comparing photosynthetic O2 evolution in very low CO2 acclimated cells of the wild type and the LCIB mutant air dier1 (ad1; Wang and Spalding, 2006) at two pHs. As shown in Figure 1A, at an acidic pH (pH 6.0), where the relative abundance of CO2 is higher, O2 evolution in ad1 was severely inhibited in 10 to 100 µm Ci but regained wild type rates at 200 µm Ci. However, at pH 7.3, where HCO3− is much more abundant than CO2, O2 evolution rates in ad1 were very similar to those of wild-type cells at 0 to 50 µm Ci but were inhibited relative to the wild type in 50 to 200 µm Ci. Furthermore, photosynthetic rates in ad1 actually decreased as Ci concentration was increased between 50 µm and 200 µm. The different responses of ad1 at these two pHs suggest that the function of LCIB, which is absent in ad1, is Ci species dependent. When the photosynthetic O2 evolution rates are plotted as a function of the calculated CO2 or HCO3− concentrations (Fig. 1, B and C), the apparent inhibition of O2 evolution in ad1 relative to the wild type occurs in the same CO2 concentration range, but in different total Ci or HCO3− concentration ranges, at both pHs, and this CO2 range corresponds to CO2 concentrations associated with the low CO2 acclimation state (Vance and Spalding, 2005), even though the algae were acclimated to very low CO2. These results suggest that the LCIB mutant is compromised strictly in CO2 uptake and specifically in the low CO2 range. Furthermore, the different responses of ad1 at two pHs also suggest that another element, possibly a HCO3− transport pathway, is responsible for the apparently normal photosynthetic O2 evolution at lower Ci concentrations (<50 µm at pH 7.3; <10 µm at pH 6.0), equivalent to < 7 µm CO2 at either pH, which corresponds to the range of CO2 concentrations associated with the very low CO2 acclimation state (Vance and Spalding, 2005). The rates of photosynthetic O2 evolution in ad1 are very similar at either pH when plotted as a function of the HCO3− concentration, but not the total Ci or CO2 concentration (Fig. 1, D–F), which supports the suggestion that a HCO3− transport system is responsible for the growth and Ci uptake of the LCIB mutant in very low CO2.
Figure 1.
Photosynthetic activity in the wild type and the LCIB mutant ad1. A, Ci-dependent O2 evolution at pH 7.3 and 6.0. Strains cw10 (squares) and ad1 (triangles) were grown in high CO2 and then switched to very low CO2 for 24 h before the measurement. Two hundred units per milliliter of bovine CA was present to ensure the equilibrium of CO2 and HCO3−. B and C, The photosynthetic activities were plotted against CO2 (B) and HCO3− (C) concentrations calculated from total Ci at pH 6.0 (black squares for cw10 or black triangles for ad1) and pH 7.3 (white squares for cw10 or white triangles for ad1). D to F, Comparison of the photosynthetic activities of cw10 and ad1 at pH 7.3 and 6.0 plotted against total Ci (D) or CO2 (E) and HCO3− (F) concentrations calculated from total Ci. The experiments showed similar results with two independent cultures, although results from only one experiment are shown. Each data point represents an average of three technical replicates, and the coefficient of variation for each data point, the ratio of the sd (n = 3) to the average, is less than 10% for all data. The O2 evolution rates in cw10 and ad1 at 4,000 µm Ci are as follows: in pH 7.3, cw10 (101 ± 2) and ad1 (97 ± 5); and in pH 6.0, cw10 (93 ± 3) and ad1 (91 ± 6).
Generation of Additional Mutations in an LCIB Mutant Background
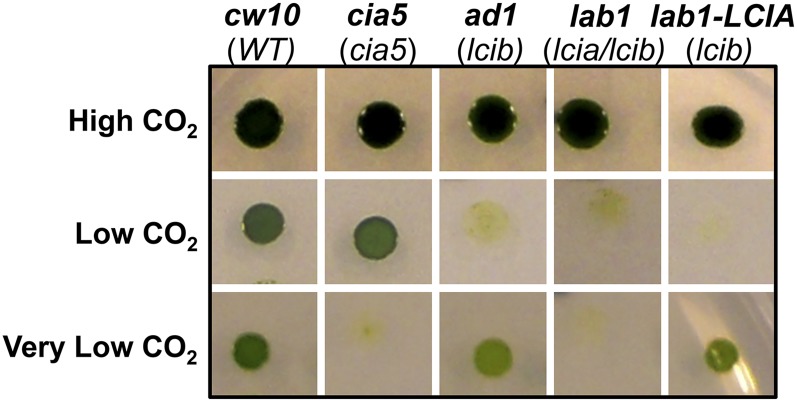
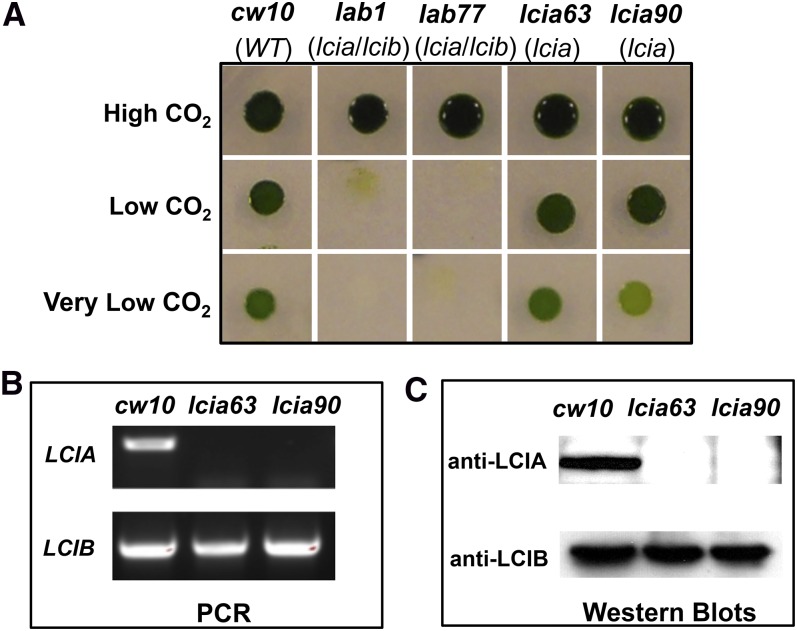
Although the LCIB-associated Ci uptake is missing in LCIB mutants, other Ci uptake systems appear still functional, as evidenced by the nearly normal photosynthetic rates of ad1 in the lowest Ci concentrations (<50 µm at pH 7.3; <10 µm at pH 6.0; Fig. 1A) and by its apparently normal growth in very low CO2 (Fig. 2). To facilitate identification of Ci uptake systems responsible in LCIB mutants for active Ci uptake and growth in very low CO2, ad1 was mutagenized by random insertion of an aminoglycoside 3-phosphotransferase (Aph8) gene-bearing plasmid (Sizova et al., 2001), selected for paromomycin resistance (paraR) and screened for inability to grow in very low CO2. Five mutants with the target phenotype were identified from about 2,000 screened paraR transformants. Whereas ad1 shows a typical air dier growth phenotype and thus can still grow in very low CO2, these new mutants were unable to grow in either very low CO2 or low CO2, but grew as well as the wild type in high CO2 (Fig. 2). The genomic DNA flanking the Aph8 insert from these new double mutants was isolated and sequenced, which confirmed that all were independent mutants. One mutant in which the insert was found to have disrupted the LCIA gene was named lab1 (LCIA-LCIB double mutant), and was further characterized.
Figure 2.
Identification of lab1, an LCIA-LCIB double mutant. A, Growth of the wild type (cw10) and mutant strains on minimal medium agar plates in different concentrations of CO2: high CO2 (5%), low CO2 (0.04%), and very low CO2 (0.01%). The mutant genes are shown in parentheses below the strain names. lab1-LCIA is a cell line generated from LCIA-complemented lab1. Strain cia5 is a classic mutant lacking a limiting CO2 acclimation response. WT, Wild type.
LCIA Mutation in lab1 Responsible for the Lethal Growth Phenotype in Very Low CO2
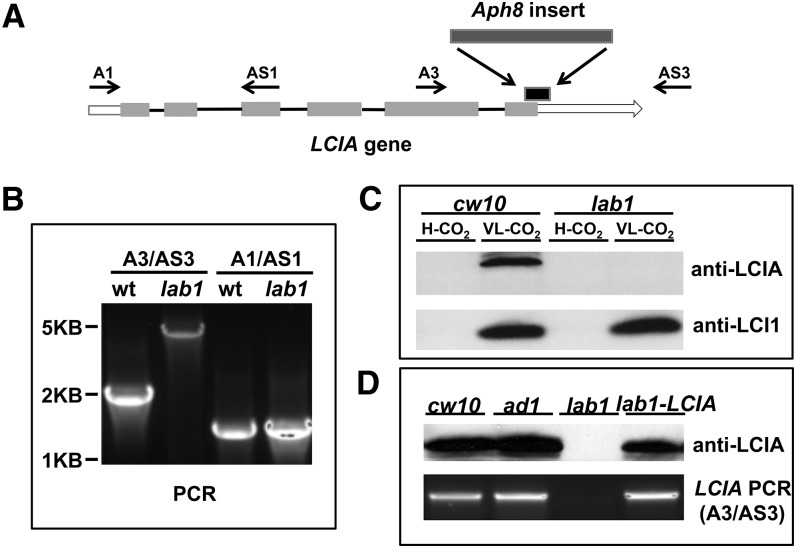
The plasmid bearing Aph8 in lab1 is inserted in the sixth exon of LCIA, causing a 243 base deletion, including 133 nucleotides in the coding sequence and 110 nucleotides in the 3′ untranslated region (UTR; Fig. 3A). The insertion in LCIA was confirmed by PCR with a pair of primers that amplify the genomic DNA in this region (Fig. 3B). In wild-type cells, expression of the LCIA protein is induced by limiting CO2. However, even though the insertion in lab1 is located near the 3′ end of LCIA and causes a deletion of only 44 C-terminal amino acids, western immunoblots were unable to detect any LCIA protein in lab1 (Fig. 3C), either because LCIA is not properly transcribed or not properly translated or because its transcripts or polypeptides are quickly degraded.
Figure 3.
Analysis of lab1, an LCIA-LCIB double mutant. A, Insertion of Aph8 in lab1. The region of deletion in LCIA caused by the Aph8 insertion is shown with a black box above the LCIA gene. B, Amplification of genomic DNA fragments in LCIA by PCR from cw10 and lab1. The locations of primers in LCIA are shown in A. The primers A1 and AS1 were used to amplify a region without insertion; A3 and AS3 were used to amplify a region disrupted by the Aph8 insert in lab1, and the larger size of PCR product in lab1 confirmed the insertion. C, Western-blot analysis of expression of LCIA and LCI1 in cells acclimated to high CO2 or very low CO2. LCI1 is a protein encoded by LCI1, a limiting CO2-induced gene, and was used as the control. No difference in LCI1 induction was observed between the wild type and lab1. D, Recovery of induced expression of LCIA in LCIA-complemented lab1. Wild-type cw10 and mutants ad1, lab1, and LCIA-complemented lab1 were grown in high CO2, and then shifted into very low CO2 for 12 h. LCIA protein was detected by western immunoblots with a specific LCIA antibody.
A genetic cross between lab1 and wild-type strain CC620 was performed to confirm whether the lethal growth phenotype in lab1 is caused by the mutation in LCIA. Because lab1 has both a bacterial phleomycin resistance (Ble) gene, (confers zeocin resistance [zeoR]) insert (causing the LCIB deletion) and an Aph8 insert (causing the LCIA truncation), progeny carrying both LCIB and LCIA mutations were selected for both zeoR and paraR. More than 50 random progeny were tested for growth in different CO2 concentrations, and all progeny carrying both mutations (confirmed by PCR) showed the same growth phenotype as lab1 (lethal in both low CO2 and very low CO2), indicating that the inability of lab1 to grow in very low CO2 is genetically linked to the insertion in LCIA. We further cloned a genomic DNA fragment containing the wild-type LCIA gene, including 1.1 kb of the upstream sequence (presumably including the LCIA promoter region) and the entire 3′ UTR, introduced this gene into the lab1 mutant, and found that this LCIA genomic clone complemented growth of lab1 in very low CO2. All complemented lines restored the air dier growth phenotype of the host strain ad1 (Fig. 2), and the presence of a wild-type LCIA gene and the expression of LCIA protein induced by limiting CO2 in these lines were confirmed by PCR and western immunoblots (Fig. 3D). These results demonstrate that the lethal growth phenotype in very low CO2 in the LCIA-LCIB double mutant is caused by the LCIA mutation.
Defective Photosynthesis in lab1 Double Mutants
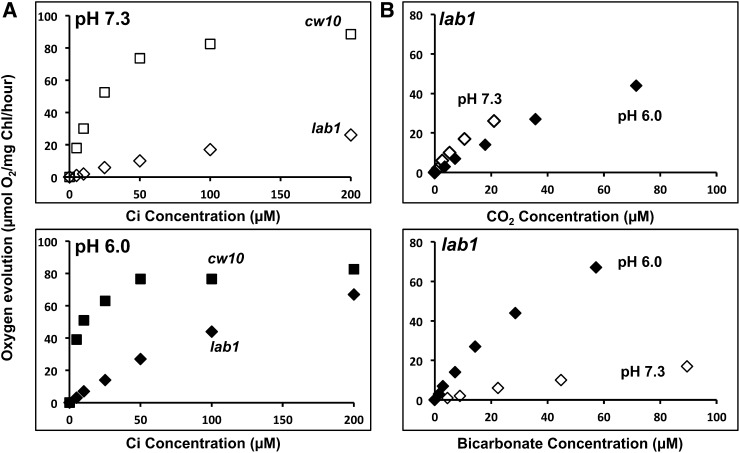
The inability of lab1 to grow in very low CO2 suggests that photosynthesis driven by active Ci uptake systems may be defective. Compared with that of the wild type, Ci-dependent O2 evolution in lab1 was severely inhibited in Ci concentrations ≤ 200 µm at pH 7.3 and markedly decreased in Ci concentrations ≤ 100 µm at pH 6.0 (Fig. 4A). Photosynthetic O2 evolution in lab1 was similar at either pH when plotted as a function of the calculated CO2 concentration (Fig. 4B), suggesting that photosynthetic CO2 assimilation remaining in the absence of both LCIB and LCIA is largely supported through Ci diffusion (Spalding and Ogren, 1983). Because photosynthesis in LCIB single mutants appears to be dependent on HCO3− uptake, as evidenced by the apparent dependence of photosynthetic O2 evolution in ad1 on the concentration of HCO3− (Fig. 1C), the highly reduced and apparently CO2-dependent photosynthesis in LCIA-LCIB double mutants implies that LCIA is required for much of the active HCO3− uptake in ad1.
Figure 4.
Ci-dependent photosynthetic oxygen evolution in cw10 and lab1. A, Ci-dependent O2 evolution at pH 7.3 and 6.0. Strains cw10 (squares) and lab1 (diamonds) were grown in high CO2 and then switched to very low CO2 for 24 h before the measurement. Two hundred units per milliliter of bovine CA was present to ensure the equilibrium of CO2 and HCO3−. B, The photosynthetic activities of lab1 measured at pH 7.3 (white diamonds) and pH 6.0 (black diamonds) were plotted against CO2 or HCO3− concentration calculated from total Ci. The experiments showed similar results with two independent cultures, although results from only one experiment are shown. Each data point represents an average of three technical replicates, and the coefficient of variation for each data point, the ratio of the sd (n = 3) to the average, is less than 10% for all data. The O2 evolution rates of lab1 at 4,000 µm Ci are as follows: (78 ± 3) in pH 7.3 and (79 ± 6) in pH 6.0. The rates of cw10 are the same in Figure 1.
Decreased HCO3−-Dependent Photosynthesis in LCIA Single Mutants
The impacts of an LCIA single mutation on growth and photosynthesis were evaluated in progeny bearing only the LCIA mutation. These were identified as paraR but zeocin-sensitive progeny from the genetic cross of lab1 with CC620 described above. PCR and western-blot analysis confirmed that these paraR but zeocin-sensitive progeny carry the LCIA mutation but retain the wild-type LCIB gene (Fig. 5, B and C). In contrast with lab1 or ad1, the LCIA single-gene mutants did not show any obvious growth defect in spot tests at either low CO2 or very low CO2 (Fig. 5A), indicating that loss of LCIA alone has no gross impact on growth and photosynthesis under these conditions.
Figure 5.
Generation and analysis of LCIA single mutants. A, Growth of the wild type, lab1 double mutant, and LCIA single mutants. Strains lab77 (an LCIA-LCIB double mutant progeny), lcia63, and lcia90 (LCIA single-mutant progeny) were derived from a genetic cross between lab1 and wild-type strain CC620. B, PCR with specific primers amplifying the disrupted regions in LCIA or LCIB. C, Western immunoblots with specific LCIA or LCIB antibody. Wild-type cells and mutants lcia-63 and lcia90 were grown in high CO2, and then shifted into very low CO2 for 12 h prior to analysis. WT, Wild type.
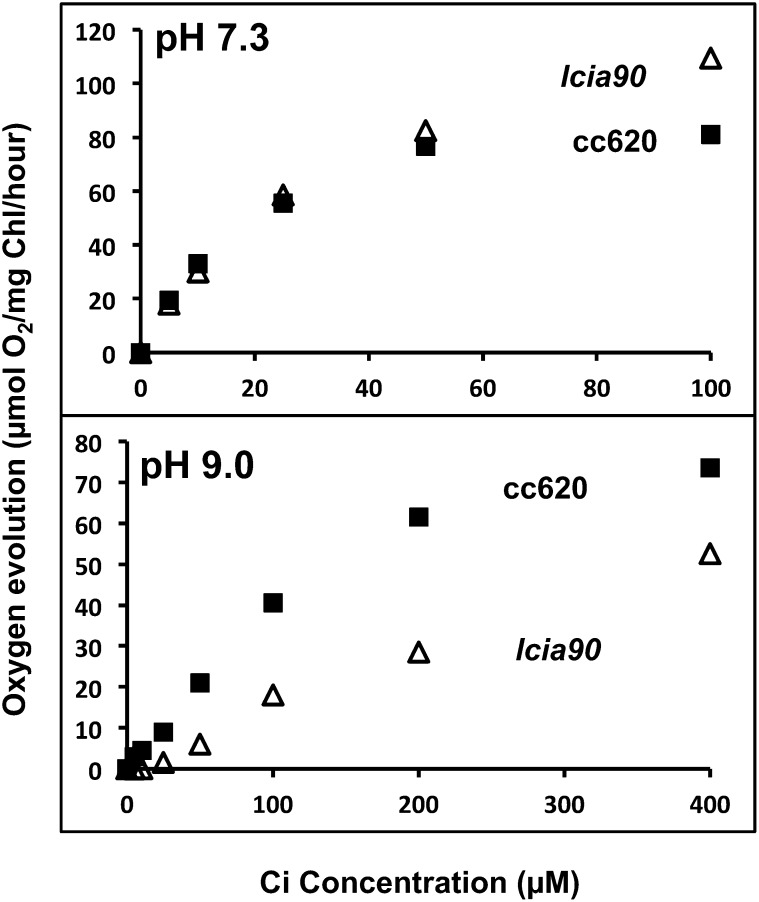
Ci-dependent photosynthetic O2 evolution in several LCIA mutant progeny was similar to that of wild-type cells at pH 7.3, as illustrated by the comparison between one LCIA mutant, lcia90, and its wild-type parent CC620 (Fig. 6). Although the Ci-dependent photosynthetic activities of lcia90 and the wild type are very similar at pH 7.3, photosynthesis was significantly inhibited in lcia90 relative to the wild type at pH 9.0. Because almost all Ci at pH 9 is in the form of HCO3−, decreased photosynthesis under these conditions is consistent with the mutation in LCIA causing a defect in active HCO3− uptake.
Figure 6.
Ci-dependent photosynthesis in the LCIA mutant. Ci-dependent photosynthetic O2 evolution measured at pH 7.3 and 9.0. Strains CC620 (black squares) and lcia90 (white triangles) were grown in high CO2 and then switched to very low CO2 for 24 h before the measurement. The experiments showed similar results with two independent cultures, although results from only one experiment are shown. Each data point represents an average of three technical replicates, and the coefficient of variation for each data point, the ratio of the sd (n = 3) to the average, is less than 10% for all data. The O2 evolution rates of CC620 and lcia90 at 4,000 µm Ci are as follows: in pH 7.3, CC620 (95 ± 2) and lcia90 (132 ± 4); and in pH 9.0, CC620 (87 ± 4) and lcia90 (112 ± 2).
DISCUSSION
We have demonstrated that two C. reinhardtii proteins, LCIA and LCIB, both contribute to CCM-mediated Ci assimilation in very low CO2, exemplified by growth and photosynthesis of mutants defective in LCIB and LCIA. Whereas LCIB or LCIA single-gene mutants show no obvious growth defects in very low CO2, combination of these mutations in the LCIA-LCIB double mutant results in a lethal phenotype under very low CO2 conditions. This demonstrates that LCIB- and LCIA-associated Ci uptake systems act either complementarily or synergistically in Ci uptake. The effects of LCIA or LCIB single mutations and LCIA-LCIB double mutations on Ci-dependent photosynthesis at different pHs reveal that LCIA and LCIB play distinct roles in different active Ci uptake processes. Whereas LCIB appears involved in active CO2 uptake, LCIA is more likely involved in a HCO3− transport pathway.
LCIB-Based CO2 Uptake System
The essentiality of LCIB in CCM-mediated Ci accumulation was demonstrated by a number of earlier studies with LCIB mutants, and LCIB was previously proposed to function in direct Ci transport, Ci transport regulation, recapture of CO2 escaping from the pyrenoid and/or the thylakoid lumen, or physically preventing CO2 escape from pyrenoids (Spalding et al., 1983a; Miura et al., 2004; Wang and Spalding, 2006, 2014; Duanmu et al., 2009b; Yamano et al., 2010). Here we demonstrate that the absence of LCIB causes photosynthetic defects under low Ci conditions that correlate with CO2 concentrations. At a slightly acidic pH (pH 6.0), where CO2 comprises more than one-half of the total Ci, Ci-dependent photosynthetic O2 evolution in the LCIB mutant ad1 was significantly inhibited at 10 to 100 µm Ci (approximately 7–70 µm CO2), but at pH 7.3, where HCO3− dominates, photosynthetic O2 evolution was not affected below 50 µm Ci (approximately 6 µm CO2), but showed progressively greater inhibition at higher Ci concentrations. Yamano et al. (2010) reported an inhibition of photosynthetic O2 evolution in the LCIB mutants above 200 µm Ci at pH 7.8 (approximately 7 µm CO2), but no obvious inhibition was observed below 100 µm Ci. It is clear that the lack of LCIB in ad1 results in an apparent inhibition of O2 evolution at similar CO2 concentrations regardless of the pH, and that the lower limit of those CO2 concentrations in all cases (6–7 µm or approximately 0.02%; Fig. 1C) is very similar to the gas-phase CO2 concentration of 0.02% that we previously defined as the upper limit of the very low CO2 acclimation state based on physiological characteristics in a wild-type strain and on the air dier phenotype (Vance and Spalding, 2005; Wang and Spalding, 2014). Below this CO2 concentration, LCIB mutants grow, and above this concentration, up to at least air CO2 level (0.03%–0.05% CO2), LCIB mutants cannot survive.
When CO2 significantly exceeds air CO2 level, CO2 diffusion alone may be enough to allow the mutants to survive. The CO2 concentration at which LCIB mutants regained wild-type O2 evolution rates at pH 6.0 (Fig. 1A; 200 µm Ci, corresponding to 140 µm or 0.42% CO2,) is also close to the Ci concentration at pH 7.8 where cells were reported by Yamano et al. (2010) to regain wild-type photosynthetic rates (4500 µm Ci at pH 7.8, corresponding to 170 µm or 0.5% CO2). This CO2 concentration is also close to the upper limit of the low CO2 acclimation state according to the previous physiological study with wild-type cells by Vance and Spalding (2005). On the basis of these data, LCIB likely plays a major role in the CCM via active CO2 uptake, especially in the range of low CO2 (0.02%–0.5%), although the underlying biochemical mechanism is not clear.
We recently proposed that LCIB may function similarly to the cyanobacterial ChpX/Y proteins in converting CO2 to HCO3− (Duanmu et al., 2009b; Wang and Spalding, 2014). ChpX/Y proteins in cyanobacteria are proposed to convert CO2 to HCO3− unidirectionally, in contrast with the bidirectional activity of CAs, via their linkage to NADH dehydrogenase complexes (Price et al., 2002). If LCIB does function in this manner, it should also accelerate HCO3− accumulation by hydrating externally diffused CO2 in the chloroplast stroma, which has an alkaline pH in light. Although this proposed active role of LCIB in CO2 uptake into the accumulated HCO3− pool is consistent with and can explain the observed physiological characteristics of LCIB mutants, LCIB/CAH3 double mutants (Duanmu et al., 2009b), and LCIB/LCIA double mutants, it is still speculative and needs to be rigorously tested at the biochemical level.
Function of LCIA in Ci Uptake
In contrast with the photosynthetic effect of LCIB mutations, the impact of an LCIA mutation, either alone or in combination with an LCIB mutation, on photosynthetic O2 evolution is more significant at higher pH, indicating that LCIA is likely involved in HCO3− uptake. LCIA has a predicted chloroplast envelope location (Miura et al., 2004), which we confirmed by immunolocalization (see Fig. 8A). Although active Ci uptake across the chloroplast envelope has been demonstrated (Sültemeyer et al., 1988; Moroney and Mason, 1991; Amoroso et al., 1998) and LCIA was previously identified as a possible chloroplast envelope Ci transporter (Miura et al., 2004; Mariscal et al., 2006; Duanmu et al., 2009a), no clear, direct evidence links LCIA to active, chloroplast envelope Ci uptake. In addition to the lack of direct evidence for Ci transport by LCIA, other potential Ci transporters, such as CCP1 and CCP2, are also present on the chloroplast envelope (Ramazanov et al., 1993). In addition, chloroplast Ci uptake also might be explained by the activity of LCIB.
Figure 8.
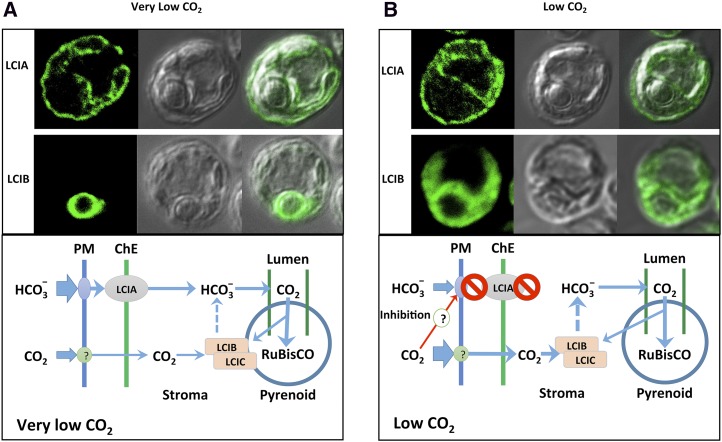
Models for acclimation to low and very low CO2 in C. reinhardtii. A and B, Immunolocalization of LCIA and LCIB and working models illustrating possible roles played by the LCIA-associated HCO3− uptake system and the LCIB-associated CO2 uptake system in very low CO2 grown cells (A) and low CO2 grown cells (B). No obvious changes of LCIA localization were observed between low and very low CO2 conditions. Dashed lines represent the hypothesized function of the LCIB-LCIC complex in conversion of CO2 to HCO3−, which is then delivered to CAH3 within the lumen of the pyrenoid-associated thylakoid tubules for dehydration to CO2. A detailed explanation is provided in “Discussion.” ChE, Chloroplast envelope; PM, plasma membrane.
LCIA belongs to an FNT family (Mariscal et al., 2006). Multiple recent studies have shown that the bacterial FNT proteins form a pentameric complex with a structure similar to aquaporin and behave more like channels than active transporters (Wang et al., 2009; Lü et al., 2012a, 2013). Nonselective permeability of FNT proteins to several structurally similar small, anionic molecules also has been demonstrated in artificial lipid membranes (Lü et al., 2012b). It seems improbable that LCIA would function as a HCO3− channel in the chloroplast envelope, because the membrane potential of the chloroplast envelope is inside negative (Demmig and Gimmler, 1983; Wu et al., 1991; Fuks and Homblé, 1999), meaning that HCO3− (or any anion) would have to move against an electrical gradient to enter the chloroplast. However, if LCIA does function as a channel to facilitate passive entry of HCO3− into the chloroplast, it would have to depend on the activity of plasma membrane Ci transporters (e.g. HLA3 and LCI1; Duanmu et al., 2009a; Ohnishi et al., 2010) to generate a sufficiently high HCO3− concentration in the relatively small volume between the plasma membrane and chloroplast envelope to establish a HCO3− concentration gradient sufficient to overcome the electrical gradient and push HCO3− across the inner envelope. An LCIA channel would also have to be highly regulated to prevent reversed flow when the HCO3− gradient was not favorable for uptake. Ci uptake studies with isolated chloroplasts from LCIA mutants and LCIA overexpression lines will help to clarify whether LCIA is directly responsible for HCO3− uptake into chloroplasts and, if so, whether it performs as an active Ci transporter or as an anion channel. Future biochemical, biophysical, and structural characterizations of LCIA will also be required to clarify the molecular mechanism underlying LCIA-associated Ci uptake.
CO2 Inhibition of LCIA-Mediated Ci Uptake
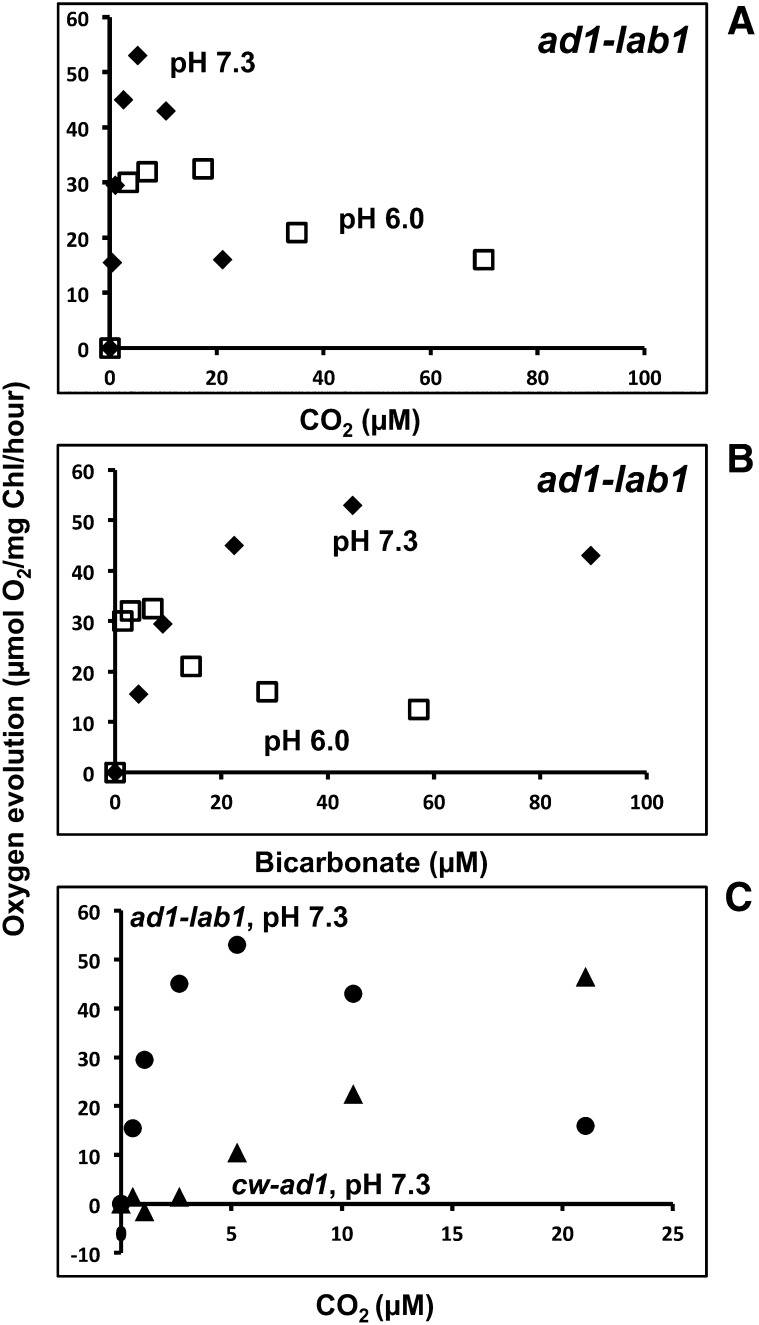
As shown in Figure 1B, the photosynthetic O2 evolution in an LCIB mutant progressively decreased as the Ci concentration increased beyond 50 µm at pH 7.3. The O2 evolution rate of ad1 at 200 µm Ci (approximately 22 µm CO2) was significantly reduced compared with that at 50 µm at this pH. Yamano et al. (2010) reported similar responses at a higher pH (pH 7.8) in LCIB mutants, in which the mutants showed O2 evolution rates similar to those of the wild type at 0 to 100 µm Ci, but reduced rates at higher Ci concentrations. The most significant inhibition occurred at 500 to 700 µm Ci (approximately 19–27 µm CO2). In these higher pHs (pH 7.3 or pH 7.8), the dominant Ci species is HCO3−, and the reduced activity at higher Ci concentrations cannot be simply explained by a defect in CO2 uptake caused by an LCIB mutation, because the Ci-dependent photosynthetic rates in these mutants should then at least remain constant, if not increase, with increased Ci concentrations. Because it seems clear that an LCIA-associated system contributes significant Ci uptake in cells lacking LCIB, this observed Ci inhibition of photosynthetic O2 evolution must represent inhibition of LCIA-associated Ci uptake activity.
We reasoned that if LCIA-mediated Ci uptake activity is inhibited by Ci, this should be evident in the calculated difference in photosynthetic activity between ad1and lab1, which should represent the contribution of LCIA to the photosynthetic performance of ad1. When we calculate this LCIA contribution from O2 evolution data obtained at pH 6.0 and pH 7.3 and plot it as a function of calculated CO2 and HCO3− concentrations (Fig. 7, A and B), it becomes evident that Ci inhibition of LCIA-associated Ci uptake activity responds to CO2 rather than to either HCO3− or total Ci concentrations. Furthermore, the apparent minimum inhibitory CO2 concentration occurs just above that defined as very low CO2 (approximately 7 µm CO2 at room temperature), and becomes more significant at a CO2 concentration range equivalent to air level (approximately 12 µm CO2 at room temperature) and above, implying that air level or low CO2 inhibits the LCIA-associated Ci uptake system.
Figure 7.
Contributions of LCIA and LCIB to O2 evolution and CO2 inhibition of LCIA-dependent photosynthesis. A and B, Contributions to O2 evolution activities of LCIA and LCIB. Contribution of LCIA to O2 evolution activities in ad1 as a function of CO2 (A) or HCO3− (B) concentration at pH 6.0 (white squares) and pH 7.3 (black diamonds). The LCIA contribution was calculated by subtracting photosynthetic O2 evolution activity in lab1 from that in ad1. C, Contributions of LCIA and LCIB to photosynthesis as a function of the CO2 concentration. The contribution of LCIA (black circles) was calculated as above, and the contribution of LCIB (black triangles) was calculated by subtracting photosynthetic O2 evolution activity in ad1 from that in wild-type (cw10) cells. In all cases, CO2 and HCO3− concentrations are calculated from total Ci concentration and pH, assuming that the addition of CA maintains CO2-HCO3− equilibrium.
The Ci response of the LCIB mutant at lower pH (pH 6.0) is different from that at higher pH, in that the photosynthetic O2 evolution rates of ad1 are inhibited relative to the wild type at pH 6.0 but plateau rather than decreasing with increasing CO2 concentration, as observed at pH 7.3 (Fig. 1). In this CO2 concentration range, the rate of O2 evolution is much higher at pH 7.3 than that at pH 6.0 (Fig. 1B), possibly because the HCO3− concentration is higher at pH 7.3 and can support a much higher rate of HCO3− transport. Therefore, the inhibition of this higher HCO3− transport rate at pH 7.3 has a greater impact on the overall rate of O2 evolution, resulting in a quantitative decrease in the O2 evolution rate. The observation that the rate of O2 evolution at about 20 µm CO2 is the same at pH 6.0 and pH 7.3 supports this interpretation.
The inhibition of LCIA activity by air level of CO2 could explain the puzzling air dier growth phenotype of LCIB mutants, because HCO3− uptake associated with LCIA (and other Ci transporters) should otherwise allow LCIB mutants to survive in low CO2 as it does in very low CO2. This CO2 inhibition appears to occur very quickly, because it can be seen almost immediately after Ci concentrations exceed the inhibitory CO2 concentration in photosynthetic O2 evolution measurements. If CO2 is responsible for the apparent inhibition of LCIA activity, this inhibition could be caused directly by CO2 itself or through an unidentified CO2 sensor protein, and this inhibition might extend to other target proteins, such as other Ci transporters. If this inhibition occurs and impacts survivability of LCIB mutants in air CO2 level (i.e. is responsible for the air dier phenotype), it must take place at the post-translational or allosteric level, because it occurs so rapidly, and because no significant changes beyond those detected in low CO2 conditions have been detected for LCIA or other potential Ci transporter genes at the transcriptional level under very low CO2 conditions, (Brueggeman et al., 2012; Fang et al., 2012).
We can use logic similar to that we used to calculate the contribution of LCIA to ad1, and plot the apparent contribution of LCIB to photosynthetic O2 evolution of the wild-type strain cw10 (i.e. subtract the O2 evolution rate of ad1 from that of cw10). If we then compare that calculated LCIB contribution with the apparent contribution of LCIA to ad1 (Fig. 7C), it becomes evident that the apparent LCIA contribution declines at CO2 concentrations where the apparent LCIB contribution becomes substantial. This relationship would explain why LCIA and LCIB both contribute to growth in very low CO2 but only LCIB appears essential for growth in low CO2 (e.g. air level of CO2).
Distinct Roles Played by LCIA and LCIB in Low CO2 and Very Low CO2 Acclimation States
Because of slow diffusion of CO2 in water, Ci concentration can often change dramatically on a daily basis in an aquatic environment, especially in the natural soil environment of C. reinhardtii when cell population increases and photosynthesis activities are high. The presence of multiple limiting CO2 acclimation states in microalgae may reflect an evolutionary adaptation to this aspect of their natural habitats. The apparent inhibition of LCIA activity by CO2 concentrations at and above air level, together with the subcellular localizations of and apparent complementary roles for LCIA and LCIB (Fig. 8), suggest a possible explanation for how and why C. reinhardtii acclimates to different limiting CO2 conditions. Figure 8 illustrates a proposed working model that could explain the functions of LCIA and LCIB in the C. reinhardtii CCM in these two acclimation states.
In this hypothetical model, at very low CO2 (Fig. 8A), both LCIB and LCIA contribute to Ci accumulation. An LCIA-associated HCO3− transport pathway is largely responsible for HCO3− uptake into the chloroplast, possibly in concert with other HCO3− transporter(s) on the plasma membrane and/or the chloroplast envelope. LCIB, located around the pyrenoid, traps CO2, either escaping from the pyrenoid or entering from outside the cell, into the stromal HCO3− pool. This stromal HCO3− pool then provides substrate to the pyrenoid thylakoid tubules for CAH3-catalyzed dehydration to CO2 for Rubisco, as described in previous CCM models (Spalding et al., 1983b; Mitra et al., 2005; Spalding, 2008; Moroney et al., 2011).
In low CO2 (Fig. 8B), the majority of LCIB protein complex in chloroplasts are located throughout the stromal space. In this condition, the hypothetical model predicts that LCIB-associated CO2 uptake into the stromal HCO3− pool plays a more dominant role, and Ci uptake associated with LCIA and other HCO3− transporters is inhibited by CO2 in the low CO2 concentration range. Again, the stromal HCO3− accumulated through LCIB activity provides substrate to the pyrenoid thylakoid tubules for CAH3-catalyzed dehydration to CO2 for Rubisco as described in previous CCM models.
Assuming that the LCIB-mediated CO2 uptake system provides an adequate supply of substrate CO2 for Rubisco in the low CO2 range and that simple CO2 diffusion can do so at higher CO2 concentrations, such CO2 inhibition of HCO3− transport at air concentrations and above may reflect a versatile regulatory mechanism present in eukaryotic algae for acclimating quickly to changes in CO2 availability that frequently occur in their natural environments. By switching rapidly from energy-intensive HCO3− transport systems to a CO2 uptake system (possibly energetically less costly) when CO2 becomes more abundant, algae cells can divert more solar energy for photosynthetic carbon fixation and other metabolic reactions to enable faster growth at a lower energy cost.
The well-documented regulation by transcript abundance should prevent even the synthesis of the HCO3− transporters under high CO2 conditions, in which the transporters clearly are not needed. However, in lower CO2 concentration ranges, such as 5 to 50 µm (0.015%–0.15%) CO2, it may be advantageous for the cells to have the capacity to respond quickly to either activate or inactivate HCO3− transport in response to CO2 availability. Fluctuations in CO2 concentration near the surface of the natural soil environment of C. reinhardtii may occur over time frames too short for transcriptional regulation to act effectively, but the post-translational or allosteric regulation proposed here would allow HCO3− transporters to shift rapidly between active and inactive states in response to CO2 concentration fluctuations. Further research will be required to determine whether this rapid regulation actually occurs via CO2 inhibition of the CCM HCO3− transporter pathway.
MATERIALS AND METHODS
Cultures and Growth Conditions
The wild-type strain cw10 (CC849) and CC620 and the mutant strains of Chlamydomonas reinhardtii were maintained and grown as previously described (Wang and Spalding, 2006). For inducing very low CO2 acclimation, liquid cell cultures were first grown in minimal medium (pH 7.3) to late log phase in high CO2 gas (5% CO2 [v/v] in air), and then switched to very low CO2 by bubbling very low CO2 gas though the liquid phase for 20 to 24 h. The very low CO2 (0.008%–0.01%) gas was obtained by passing normal air through a saturated sodium hydroxide solution and remixing with normal air to achieve the desired final concentration.
Isolation of Mutants, Identification of Mutation, and Genetic Analysis
Spot growth testing, mutant screening, and transformation were performed as previously described (Wang and Spalding, 2006). Strain ad1 was transformed with linearized pSI103d plasmid (Sizova et al., 2001). Transformed cells were kept in high CO2 and selected on minimal medium plates supplemented with 10 μg mL−1 paromomycin. Paromomycin-resistant transformants were transferred to duplicate plates for screening by growth spot tests in high CO2 and very low CO2.
The genomic DNA fragments flanking the insertion in mutants were identified by the restriction enzyme site-directed amplification PCR (González-Ballester et al., 2005). The primary PCR was carried out in a volume of 25 µL containing 2.5 µL of 10× PCR buffer (Invitrogen), 0.25 µL of Taq polymerase (Invitrogen), 1 µL of 25 mm MgCl2, 0.4 µL of 10 mm deoxynucleotides, 0.75 µL of dimethyl sulfoxide, 5 pmol of primer RB1, 15 pmol of degenerate primers (González-Ballester et al., 2005), and 50 ng of genomic DNA as the DNA template. The secondary PCR was carried out in a volume of 25 µL with the same reagents as the primary RCR except 5 pmol of Q0 primer was used to replace the degenerate primers and 1 µL of 25:1 diluted PCR product from the primary reaction was used as the DNA template. The parameters for the PCR cycle were set up according to Dent et al. (2005).
The genetic crosses between lab1 and CC620 were performed according to Harris (2009). The progeny bearing both Ble and Aph8 were selected on agar plates made from Tris-acetate-phosphate medium supplemented with 10 µg mL−1 paromomycin and 10 µg mL−1 zeocin. The progeny bearing only Aph8 was selected on agar medium supplemented with 10 µg mL−1 paromomycin, and then their zeoR was tested on plates supplemented with 10 µg mL−1 of zeocin. PCR amplification with primers specific for LCIB or LCIA was used to confirm the defects in LCIA or LCIB in these progeny. To complement lab1, a genomic DNA fragment including the LCIA coding sequence, putative promoter region, and 3′ UTR was cloned into pBluescript vector, and used to transform lab1. The successful transformants were directly selected in very low CO2 on minimal medium plates and untransformed lab1 was used as the control. The genomic DNA from the transformants was isolated and PCR amplified with specific primers in the region of LCIA where the insertion is located.
Measurements of Photosynthetic O2 Evolution
Photosynthetic O2 evolution was measured at 25°C with a Clark-type oxygen electrode controlled by an Oxy-Lab unit (Hansatech). Cells from liquid cultures were collected by centrifugation and suspended in 4 mL of N2-saturated buffers to a final chlorophyll concentration of 20 μg mL−1. The buffers used were MOPS-Tris (25 mm, pH 7.3), MES-KOH (25 mm, pH 6.0), or AMPSO (25 mm, pH 9.0). For each cell preparation, internal and external Ci were depleted under illumination (500 μmol photons m−2 s−1) as judged by cessation of O2 evolution before each measurement. The Ci-dependent O2 evolution was initiated by addition of various concentrations of NaHCO3. For most measurements, the subsequent addition of Ci was delayed until the previous O2 evolution rate approached zero. When O2 evolution rates were extremely slow, such as in high pH or in lab1 mutants, the subsequent Ci was added to a desired concentration after previous O2 evolution proceeded at least 1 min or longer; and the Ci concentration at each next addition was calculated based on the amount of Ci consumed from the previous Ci addition as reflected by the amount of O2 evolved. Because O2, when reaching high levels, appears to have negative influence on photosynthesis, the O2 level in cell suspension was kept low by flushing with N2 gas when it exceeded 200 nmol mL−1. Two hundred units per milliliter of bovine CA (C2624; Sigma) was present in the measurements at pH 6.0 and 7.3 to ensure the equilibrium of CO2 and HCO3−.
Western-Blot Analysis and Immunolocalization
For protein analyses, harvested cells were directly dissolved in a 1× SDS-PAGE buffer in the presence of 2-mercaptoethanol at 80°C for 5 min. The resulting lysates were passed through a 26-gauge needle to reduce viscosity and then separated on 12% (v/v) SDS-polyacrylamide gels. Immunoblotting with specific antibodies was performed to detect the specific proteins by chemiluminescence (SuperSignal Wester Pico; Thermo Scientific).
To generate LCIA-specific antibodies, five LCIA complementary DNA fragments encoding different predicted soluble portions in LCIA protein were PCR amplified from a complementary DNA library (Wang and Spalding, 2006) and combined by recombination PCR. The resulting mini-LCIA sequence was cloned into pET-28a vector, and transformed into BL21 (DE3) Escherichia coli cells for overexpression. The expressed mini-LCIA was purified from E. coli and used to raise LCIA polyclonal antiserum. An LCIA monoclonal antibody against a synthetic peptide ENAINVGAYK was also generated (Abmart). LCIB antiserum against purified LCIB was previously described (Duanmu et al., 2009b). LCI1 antiserum was a gift from James V. Moroney (Louisiana State University).
For immunolocalization, cell suspension was placed on precharged microscope slides (ProbeOn Plus; FisherBiotech) for 1 to 3 min, and then quickly fixed in −20°C methanol for 10 min. Immunofluorescence staining was performed according to Cole et al. (1998). Antiserum against LCIA or LCIB was used at a dilution of 1:1,000 as primary antibody, and fluorescein isothiocyanate-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) was used at a dilution of 1:150 as the secondary antibody for immunofluorescence. After final washing with phosphate-buffered saline, the slides were mounted using ProLong Gold reagents (Invitrogen), and digital images of stained cells were acquired with a Leica SP5 X MP confocal microscope.
Acknowledgments
We thank Dr. James V. Moroney (Department of Biological Sciences, Louisiana State University) for the kind gift of LCI1 antibody and Dr. Deqiang Duanmu (Department of Molecular Biosciences, University of California, Davis) for advice on the article.
Glossary
- CCM
CO2-concentrating mechanism
- Ci
inorganic carbon
- CA
carbonic anhydrase
- paraR
paromomycin resistance
- UTR
untranslated region
- zeoR
zeocin resistance
Footnotes
This work was supported by the U.S. Department of Energy, Office of Science (grant no. DEFG02–12ER16335 to Y.W. and M.H.S.) and the National Science Foundation, Directorate for Biological Sciences (grant no. MCB–0952323 to M.H.S.).
Articles can be viewed online without a subscription.
References
- Amoroso G, Sultemeyer D, Thyssen C, Fock HP (1998) Uptake of HCO3− and CO2 in cells and chloroplasts from the microalgae Chlamydomonas reinhardtii and Dunaliella tertiolecta. Plant Physiol 116: 193–201 [Google Scholar]
- Badger M, Spalding M (2000) CO2 Acquisition, Concentration and Fixation in Cyanobacteria and Algae. Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- Badger MR, Price GD (2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54: 609–622 [DOI] [PubMed] [Google Scholar]
- Behrenfeld MJ, Randerson JT, McClain CR, Feldman GC, Los SO, Tucker CJ, Falkowski PG, Field CB, Frouin R, Esaias WE, et al. (2001) Biospheric primary production during an ENSO transition. Science 291: 2594–2597 [DOI] [PubMed] [Google Scholar]
- Brueggeman AJ, Gangadharaiah DS, Cserhati MF, Casero D, Weeks DP, Ladunga I (2012) Activation of the carbon concentrating mechanism by CO2 deprivation coincides with massive transcriptional restructuring in Chlamydomonas reinhardtii. Plant Cell 24: 1860–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig B, Gimmler H (1983) Properties of the isolated intact chloroplast at cytoplasmic K+ concentrations: I. Light-induced cation uptake into intact chloroplasts is driven by an electrical potential difference. Plant Physiol 73: 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Miller AR, Horken KM, Weeks DP, Spalding MH (2009a) Knockdown of limiting-CO2-induced gene HLA3 decreases HCO3- transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 106: 5990–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Spalding MH (2011) Insertional suppressors of Chlamydomonas reinhardtii that restore growth of air-dier lcib mutants in low CO2. Photosynth Res 109: 123–132 [DOI] [PubMed] [Google Scholar]
- Duanmu D, Wang Y, Spalding MH (2009b) Thylakoid lumen carbonic anhydrase (CAH3) mutation suppresses air-dier phenotype of LCIB mutant in Chlamydomonas reinhardtii. Plant Physiol 149: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH (2012) Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24: 1876–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks B, Homblé F (1999) Passive anion transport through the chloroplast inner envelope membrane measured by osmotic swelling of intact chloroplasts. Biochim Biophys Acta 1416: 361–369 [DOI] [PubMed] [Google Scholar]
- González-Ballester D, de Montaigu A, Galván A, Fernández E (2005) Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal Biochem 340: 330–335 [DOI] [PubMed] [Google Scholar]
- Harris EH. (2009) The Chlamydomonas Sourcebook. Academic Press, Oxford [Google Scholar]
- Im CS, Grossman AR (2002) Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J 30: 301–313 [DOI] [PubMed] [Google Scholar]
- Lü W, Du J, Schwarzer NJ, Gerbig-Smentek E, Einsle O, Andrade SL (2012b) The formate channel FocA exports the products of mixed-acid fermentation. Proc Natl Acad Sci USA 109: 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü W, Du J, Schwarzer NJ, Wacker T, Andrade SLA, Einsle O (2013) The formate/nitrite transporter family of anion channels. Biol Chem 394: 715–727 [DOI] [PubMed] [Google Scholar]
- Lü W, Schwarzer NJ, Du J, Gerbig-Smentek E, Andrade SL, Einsle O (2012a) Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc Natl Acad Sci USA 109: 18395–18400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscal V, Moulin P, Orsel M, Miller AJ, Fernández E, Galván A (2006) Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 157: 421–433 [DOI] [PubMed] [Google Scholar]
- Mitra M, Mason CB, Xiao Y, Ynalvez RA, Lato SM, Moroney JV (2005) The carbonic anhydrase gene families of Chlamydomonas reinhardtii. Can J Bot 83: 780–795 [Google Scholar]
- Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, et al. (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B (2011) The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth Res 109: 133–149 [DOI] [PubMed] [Google Scholar]
- Moroney JV, Mason CB (1991) The Role of the chloroplast in inorganic carbon acquisition by Chlamydomonas reinhardtii. Can J Bot 69: 1017–1024 [Google Scholar]
- Moroney JV, Tolbert NE (1985) Inorganic carbon uptake by Chlamydomonas reinhardtii. Plant Physiol 77: 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ynalvez RA (2007) Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot Cell 6: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Mukherjee B, Tsujikawa T, Yanase M, Nakano H, Moroney JV, Fukuzawa H (2010) Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell 22: 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Prout DL, Godfrey AC, Lemaire SD, Moroney JV (2004) The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long-term growth, but are not necessary for efficient photosynthesis, in a low-CO2 environment. Plant Mol Biol 56: 125–132 [DOI] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Price GD, Maeda S, Omata T, Badger MR (2002) Modes of active inorganic carbon uptake in the cyanobacterium, Synechococcus sp. PCC7942. Funct Plant Biol 29: 131–149 [DOI] [PubMed] [Google Scholar]
- Ramazanov Z, Mason CB, Geraghty AM, Spalding MH, Moroney JV (1993) The low CO2-inducible 36-kilodalton protein is localized to the chloroplast envelope of Chlamydomonas reinhardtii. Plant Physiol 101: 1195–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan J, Dunahay T, Benemann J, Roessler P (1998) A Look Back at the U.S. Department of Energy’s Aquatic Species Program: Biodiesel from Algae. Report No. NREL/TP-580-24190. National Renewable Energy Laboratory, Golden, CO [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Spalding MH. (2008) Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. J Exp Bot 59: 1463–1473 [DOI] [PubMed] [Google Scholar]
- Spalding MH, Ogren WL (1983) Evidence for a saturable transport component in the inorganic carbon uptake of Chlamydomonas reinhardii. FEBS Lett 154: 335–338 [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL (1983a) Reduced inorganic carbon transport in a CO2-requiring mutant of Chlamydomonas reinhardii. Plant Physiol 73: 273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL (1983b) Carbonic anhydrase-deficient mutant of Chlamydomonas reinhardii requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol 73: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Van K, Wang Y, Nakamura Y (2002) Acclimation of Chlamydomonas to changing carbon availability. Funct Plant Biol 29: 221–230 [DOI] [PubMed] [Google Scholar]
- Sültemeyer DF, Klöck G, Kreuzberg K, Fock HP (1988) Photosynthesis and apparent affinity for dissolved inorganic carbon by cells and chloroplasts of Chlamydomonas reinhardtii grown at high and low CO2 concentrations. Planta 176: 256–260 [DOI] [PubMed] [Google Scholar]
- Sültemeyer DF, Miller AG, Espie GS, Fock HP, Canvin DT (1989) Active CO2 transport by the green-alga Chlamydomonas reinhardtii. Plant Physiol 89: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance P, Spalding MH (2005) Growth, photosynthesis and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Can J Bot 85: 796–805 [Google Scholar]
- Wang Y, Duanmu D, Spalding MH (2011) Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth Res 109: 115–122 [DOI] [PubMed] [Google Scholar]
- Wang Y, Huang Y, Wang J, Cheng C, Huang W, Lu P, Xu YN, Wang P, Yan N, Shi Y (2009) Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature 462: 467–472 [DOI] [PubMed] [Google Scholar]
- Wang Y, Spalding MH (2006) An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 10110–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spalding MH (2014) LCIB in the Chlamydomonas CO2-concentrating mechanism. Photosynth Res 121: 185–192 [DOI] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ (2010) An outlook on microalgal biofuels. Science 329: 796–799 [DOI] [PubMed] [Google Scholar]
- Wu W, Peters J, Berkowitz GA (1991) Surface charge-mediated effects of Mg2+ on K+ flux across the chloroplast envelope are associated with regulation of stromal pH and photosynthesis. Plant Physiol 97: 580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Asada A, Sato E, Fukuzawa H (2014) Isolation and characterization of mutants defective in the localization of LCIB, an essential factor for the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Photosynth Res 121: 193–200 [DOI] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H (2010) Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol 51: 1453–1468 [DOI] [PubMed] [Google Scholar]