ABA promotes arbuscular mycorrhizal colonization in Medicago truncatula via a Ser/Thr phosphatase.
Abstract
Legumes can establish intracellular interactions with symbiotic microbes to enhance their fitness, including the interaction with arbuscular mycorrhizal (AM) fungi. AM fungi colonize root epidermal cells to gain access to the root cortex, and this requires the recognition by the host plant of fungus-made mycorrhizal factors. Genetic dissection has revealed the symbiosis signaling pathway that allows the recognition of AM fungi, but the downstream processes that are required to promote fungal infection are poorly understood. Abscisic acid (ABA) has been shown to promote arbuscule formation in tomato (Solanum lycopersicum). Here, we show that ABA modulates the establishment of the AM symbiosis in Medicago truncatula by promoting fungal colonization at low concentrations and impairing it at high concentrations. We show that the positive regulation of AM colonization via ABA requires a PROTEIN PHOSPHATASE 2A (PP2A) holoenzyme subunit, PP2AB′1. Mutations in PP2AB′1 cause reduced levels of AM colonization that cannot be rescued with permissive ABA application. The action of PP2AB′1 in response to ABA is unlinked to the generation of calcium oscillations, as the pp2aB′1 mutant displays a normal calcium response. This contrasts with the application of high concentrations of ABA that impairs mycorrhizal factor-induced calcium oscillations, suggesting different modes of action of ABA on the AM symbiosis. Our work reveals that ABA functions at multiple levels to regulate the AM symbiosis and that a PP2A phosphatase is required for the ABA promotion of AM colonization.
Plants have evolved ingenious strategies to cope with environmental challenges, including partnerships with other organisms in mutualistic symbioses (Thompson, 2005). The arbuscular mycorrhizal (AM) symbiosis is one of the most abundant mutualistic interactions (Smith and Read, 2008) and is formed between more than 80% of terrestrial plant species and fungi in the phylum Glomeromycota (Schüßler et al., 2001; Smith and Read, 2008). This mutualistic symbiosis evolved over 450 million years ago (Remy et al., 1994), which attests to a remarkable selective advantage for both symbiotic partners. As obligate symbionts, AM fungi rely on carbon provided by their plant hosts to complete their life cycle. In exchange, the fungi not only improve the mineral nutrition (particularly phosphate) of the partner but also enhance protection against pathogens as well as drought tolerance (Boomsma and Vyn, 2008; Smith and Read, 2008; Hao et al., 2012; Jung et al., 2012).
Over the past decade, tremendous progress has been made in understanding how the AM symbiosis is established; it is initiated from phosphate-deprived plants that secrete strigolactones to stimulate AM metabolism, spore germination, and hyphae branching within the vicinity of the root (Akiyama et al., 2005; Besserer et al., 2006; Kretzschmar et al., 2012). The AM fungus releases mycorrhizal factors, including sulfated lipochitooligosaccharides and nonsulfated lipochitooligosaccharides (NS-LCOs; Maillet et al., 2011) and chitooligosaccharides (Genre et al., 2013), which are recognized by the plant host to activate the common symbiosis signaling pathway (Harrison, 2012), leading to calcium oscillations in the nuclear region (Kosuta et al., 2008; Chabaud et al., 2011; Sieberer et al., 2012). Following its recognition by the host plant, the AM fungus invades the root epidermal cells and grows through the root cortex, where it initiates highly branched hyphae, the so-called arbuscules in inner root cortical cells. Arbuscules are believed to be the main site for nutrient exchange between the two partners (Harrison, 2012), and this is supported by phosphate transporters such as PHOSPHATE TRANSPORTER4 (PT4) that are localized to the periarbuscular membrane (Javot et al., 2007; Pumplin et al., 2012). The arbuscule is a transient structure (Alexander et al., 1989; Javot et al., 2007), and mutation of PT4 leads to its premature senescence, indicating plant regulation of the process in response to the degree of phosphate released (Javot et al., 2007). Mycorrhization is also regulated via a microRNA, miR171h, and it is thought that this regulation may prevent overcolonization of the roots (Liu et al., 2011; Lauressergues et al., 2012).
Abscisic acid (ABA) is a key abiotic stress signal, and previous studies have demonstrated a role for ABA in the regulation of AM infection (Herrera-Medina et al., 2007). In an ABA biosynthetic mutant of tomato (Solanum lycopersicum), sitiens (Taylor et al., 1988; Harrison et al., 2011), AM colonization, arbuscule formation, and functionality are impaired (Herrera-Medina et al., 2007), despite a residual amount of ABA maintained in this mutant (Herde et al., 1999). These results highlight that ABA positively regulates processes associated with the arbuscular mycorrhiza (Herrera-Medina et al., 2007; Martín-Rodríguez et al., 2011). However, this effect was at least in part explained by an indirect effect of the sitiens mutation on ethylene signaling (Herrera-Medina et al., 2007; Martín-Rodríguez et al., 2011). In contrast to a positive role during AM colonization, ABA acts as a negative regulator during the root nodule symbiosis (Suzuki et al., 2004; Ding et al., 2008). At the root epidermis, ABA was shown to inhibit early signaling such as nodulation factor-induced calcium spiking, early gene expression, and bacterial infection (Ding et al., 2008). In the root cortex, ABA negatively regulates nodule initiation in response to cytokinin (Ding et al., 2008). In addition, it is believed that ABA also negatively regulates nitrogen fixation (Tominaga et al., 2009). Although AM and root nodule symbioses share a common signaling pathway, the opposite regulation by ABA implies different effects on symbiosis-specific signaling pathways and/or developmental processes. However, at this stage, we cannot discriminate whether this difference in symbiotic responses simply reflects differential effects for ABA in different plant species; for instance, in Medicago truncatula, ABA promotes lateral root emergence, while in Arabidopsis (Arabidopsis thaliana), it impairs lateral root emergence (Liang and Harris, 2005; De Smet et al., 2006; Laplaze et al., 2007).
Major progress over the last few years has unraveled the earliest events of ABA perception, involving the ABA receptors (PYRABACTIN RESISTANCE [PYR]/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR [RCAR]), type 2C protein phosphatases (PP2Cs), and SUCROSE NONFERMENTATION-RELATED PROTEIN KINASE2 (SnRK2; Cutler et al., 2010; Hauser et al., 2011; Santiago et al., 2012). The PYR/RCAR receptors bind ABA and inhibit PP2C activity, removing an inhibition of SnRK2 that phosphorylates numerous downstream targets such as ion channels, transcription factors, and NADPH oxidase (Kulik et al., 2011), and regulates the expression of ABA-responsive genes such as basic leucine zipper transcription factors or ABA-responsive element-binding factors (Finkelstein and Lynch, 2000; Hoth et al., 2002). Downstream of ABA perception, genetic studies have revealed additional protein phosphatases involved in ABA signaling in the PP2A class (Cutler et al., 2010). In contrast to PP2Cs, which are magnesium- and manganese-dependent monomeric enzymes, PP2As are holoenzymes composed of three subunits: a catalytic subunit, c, that complexes with two regulatory units, a and b (Farkas et al., 2007). Arabidopsis has five, three, and 17 PP2Ac, PP2Aa, and PP2Ab, respectively (Farkas et al., 2007). Among these, the Arabidopsis mutant rcn1 (for designated roots curl in naphthylphthalamic acid), impaired in a PP2Aa, is insensitive to ABA, suggesting that RCN positively regulates ABA signaling. This is in contrast to members of the PP2C class of protein phosphatases that are negative regulators of ABA signaling (Kwak et al., 2002; Pernas et al., 2007). So far, little is known about the function of the PP2Ab subunit, and nothing has been investigated in relation to its role in ABA signaling (Farkas et al., 2007; Leivar et al., 2011). The b subunit, which is further classified into B, B′, and B′′ subfamilies, is believed to determine the substrate specificity and the subcellular localization of the PP2A holoenzymes (Matre et al., 2009).
The previous work on ABA during AM colonization focused on a mutant defective in ABA biosynthesis, which had off-target effects on ethylene levels (Herrera-Medina et al., 2007; Martín-Rodríguez et al., 2011). In order to clarify the role of ABA during AM colonization, we targeted components of ABA signaling and show that at least some components of ABA signaling are involved in the promotion of AM colonization. Our work reveals that a PP2A protein complex is likely associated with the promotion of AM colonization and that at least one member of this complex is transcriptionally activated by ABA and during AM colonization.
RESULTS
ABA Modulates AM Colonization in M. truncatula
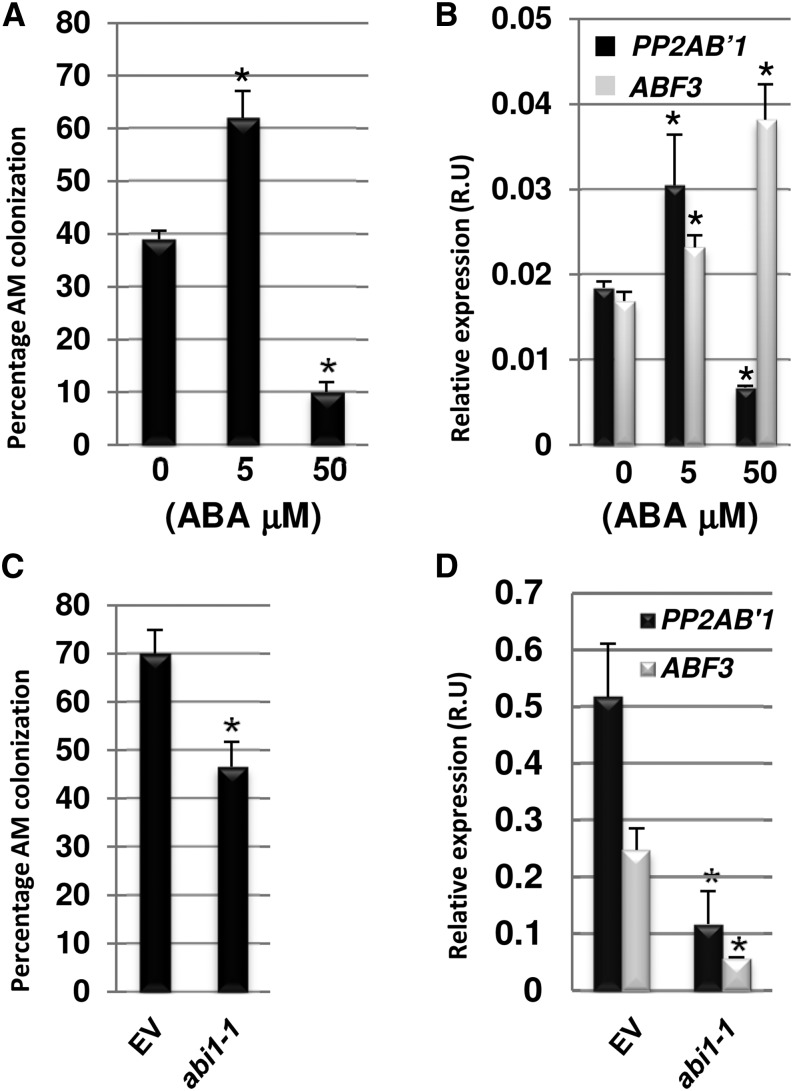
ABA has been shown to regulate AM colonization in tomato, and we tested the effect of ABA treatment on AM colonization in M. truncatula. External application of 5 μm ABA significantly enhanced AM colonization, while 50 μm ABA reduced AM colonization but did not impair arbuscule formation (Fig. 1A; Supplemental Figs. S1 and S2). The ABA marker gene ABA binding factor3 (ABF3; Hoth et al., 2002) was induced upon these ABA treatments, implying appropriate perception of ABA (Fig. 1B). These results suggest that ABA can modulate AM colonization in a concentration-dependent manner, where high concentrations of ABA impair AM colonization, in contrast to low concentrations, which promote the AM symbiosis. We previously reported that ABA modulates nodulation factor-induced calcium spiking (Ding et al., 2008). Therefore, we questioned whether the modulation of AM colonization in an ABA concentration-dependent manner occurred via regulation of the symbiotic signaling pathway leading to calcium oscillations. The role of ABA in mycorrhizal factor-induced calcium spiking was assessed using an M. truncatula line stably transformed with the calcium reporter cameleon YC3.6. The plants were grown on medium with 0, 5, or 50 μm ABA and analyzed for mycorrhizal factor (NS-LCO)-induced calcium spiking (Maillet et al., 2011). We found that calcium spiking was not altered in plants grown on 5 μm ABA (Supplemental Fig. S3), but at 50 μm ABA, only 50% of cells displayed calcium spiking, and this calcium spiking had a reduced frequency in comparison with plants treated with 5 μm ABA or untreated with ABA (Supplemental Fig. S3). These results show that permissive ABA concentrations do not influence NS-LCO-induced calcium spiking, in contrast to higher concentrations of ABA, which can impair NS-LCO activation of the symbiotic signaling pathway and AM colonization.
Figure 1.
ABA modulates AM colonization and PP2AB′1 expression. A, Quantification of AM colonization in the wild type after 4 weeks of inoculation with R. irregularis and external application of 0, 5, or 50 μm ABA. Values are means ± se (n = 16) from one biological replicate (for biological replicates 2 and 3, see Supplemental Fig. S1). Asterisks indicate significant differences between the ABA-treated roots (5 and 50 μm ABA) and the control (0 μm ABA; Student’s t test; P ≤ 0.01). B, Quantitative RT-PCR to monitor PP2AB′1 and ABF3 expression in AM colonized roots watered with 0, 5, or 50 μm ABA. Asterisks indicate statistical significance relative to the untreated control (Student’s t test; P ≤ 0.01). R.U, Relative units. C, Transformation of the Arabidopsis dominant-negative allele of abi1-1 in the wild type leads to a decrease of R. irregularis colonization at 7 weeks after inoculation. Values are means ± se (n = 10 [empty vector] and n = 14 [abi1-1]) from one biological replicate (for biological replicates 2 and 3, see Supplemental Fig. S4). The asterisk indicates a significant difference from the wild type transformed with the empty vector (EV; Student’s t test; P ≤ 0.01). D, Quantitative RT-PCR to monitor PP2AB′1 expression in roots transformed with abi1-1. ABF3, up-regulated in response to ABA, was used as an ABA marker. PP2AB′1 and ABF3 expression was significantly down-regulated in roots expressing abi1-1 in comparison with the empty vector. Asterisks indicate significant differences relative to the empty vector control (Student’s t test; P ≤ 0.01). Expression is normalized to EF-1α in B and D.
To further test the function of ABA during AM colonization of M. truncatula, we used a dominant negative allele of a PP2C from Arabidopsis. PP2Cs sit at the core of the ABA receptor complex, and the Arabidopsis abscisic acid insensitive1-1 (abi1-1) allele, mutated in a PP2C, dominantly suppresses ABA signaling in Arabidopsis as well as other plant species (Hagenbeek et al., 2000; Gampala et al., 2001; Wu et al., 2003; Ding et al., 2008). We overexpressed Arabidopsis abi1-1 in M. truncatula roots via Agrobacterium rhizogenes-mediated gene transfer (Boisson-Dernier et al., 2001) and assessed the effect on Rhizophagus irregularis colonization (Fig. 1C; Supplemental Fig. S4). Roots expressing abi1-1 showed a 30% reduction of AM colonization in comparison with roots expressing the empty vector (Fig. 1C; Supplemental Fig. S4). Arabidopsis abi1-1 transformation suppresses ABF3 expression (Fig. 1D), implying that abi1-1 does indeed block ABA signaling.
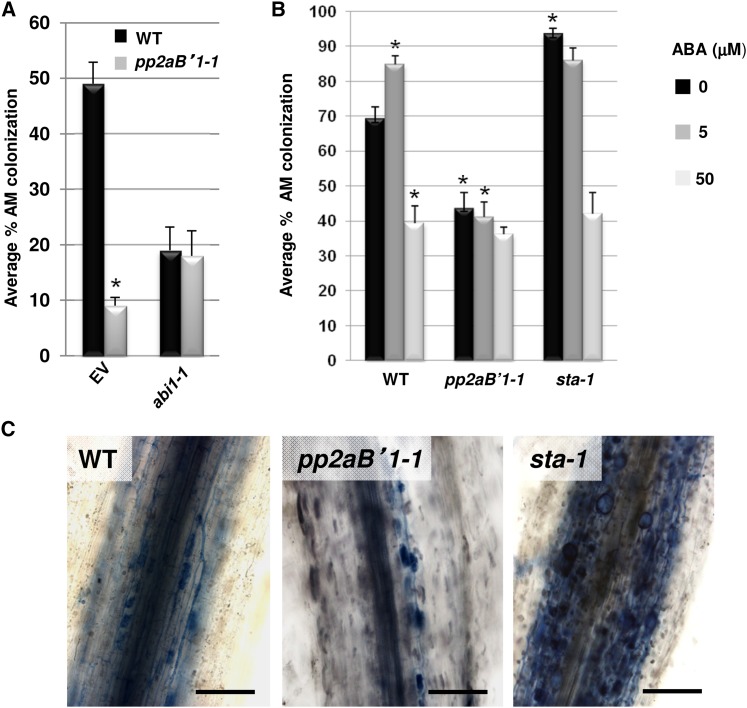
In a previous study, we identified the M. truncatula sensitivity to abscisic acid-1 (sta-1) mutant, which is impaired in ABA modulation of the root nodule symbiosis due to a hypersensitivity to ABA during nodule organogenesis in the root cortex (Ding et al., 2008). In the absence of ABA treatment, sta-1 was hypercolonized by AM fungi (Fig. 5, B and C), supporting the role of ABA in promoting AM colonization. Combining our results with external ABA application, abi1-1 transformation, and the effect of the sta-1 mutation on AM colonization, we conclude that ABA positively regulates AM colonization in M. truncatula, and this is consistent with previous work in tomato (Martin-Rodriguez et al., 2010; Martín-Rodríguez et al., 2011). However, high concentrations of ABA impair the symbiosis signaling pathway in the root epidermis in a manner analogous to its effect on nodulation factor signaling (Ding et al., 2008), and this is likely to lead to the impairment of AM colonization that we observed upon external application of high concentrations of ABA.
Figure 5.
PP2AB′1 acts downstream of ABA biosynthesis and perception during AM colonization. A, Quantification of AM colonization in the pp2aB′1-1 mutant transformed with abi1-1 or empty vector (EV) at 5 weeks after inoculation with R. irregularis. Expression of abi1-1 impaired AM colonization in the wild type (WT) similarly to pp2aB′1-1 expressing abi1-1. Values are means ± se (n = 10) from one biological replicate (for biological replicates 2 and 3, see Supplemental Fig. S17). The asterisk indicates significance in comparison with the wild type (Student’s t test; P ≤ 0.001). B, External application of ABA does not restore AM colonization in the pp2aB′1 mutant. AM colonization is shown in pp2aB′1-1 and sta-1 mutants watered with 0, 5, or 50 µm ABA for 4 weeks; the colonization is observed 4 weeks after R. irregularis inoculation. Asterisks indicate significant differences compared with the wild type watered with 0 µm ABA or with the wild type with the same ABA regime (Student’s t test; P < 0.001). Values are means ± se (n = 12). C, Bright-field images of wild-type as well as pp2aB′1-1 and sta-1 roots inoculated with R. irregularis for 4 weeks and watered with 0 µm ABA. The fungal structures are ink stained (blue coloration). Bars = 70 µm.
A PP2AB′1 Induced upon R. irregularis Infection and Regulated upon ABA Treatment
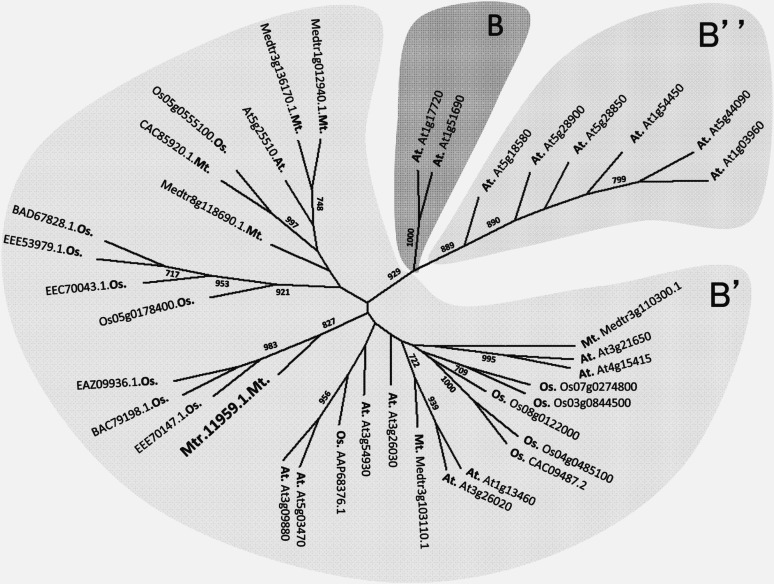
The finding that ABA promotes AM colonization led us to question whether there were ABA signaling components specific to the AM symbiosis. Interestingly, a component of the PP2A holoenzyme, the Medicago sativa PP2Ab expressed in roots, is induced by ABA (Tóth et al., 2000), suggesting that the PP2A holoenzymes might be involved in ABA signaling in Medicago spp. Since the PP2A subunits are induced by the conditions associated with their effect (Ariño et al., 1993; Casamayor et al., 1994), we explored the expression of the PP2Aa, PP2Ab, and PP2Ac gene families during mycorrhizal colonization using the M. truncatula gene expression atlas (Benedito et al., 2008). None of the PP2Aa and PP2Ac genes were strongly induced in mycorrhized roots, but one PP2Ab gene was highly induced during mycorrhizal colonization (Supplemental Fig. S5). This mycorrhiza-induced PP2Ab is part of the PP2AB′ subfamily (Fig. 2; Supplemental Fig. S6). This gene, whose probe set is Mtr11959.1, was not induced upon nodulation factor treatment or in mature nodules (Supplemental Fig. S7, A and C). This M. truncatula PP2AB′ has three rice (Oryza sativa) homologs but no closely related Arabidopsis homolog (Fig. 2). We chose to name it PP2AB′1.
Figure 2.
Mtr.11959.1 is a regulatory B′ subunit of the Ser/Thr phosphatase type 2A family. The unrooted phylogenetic tree was calculated from the alignment presented in Supplemental Figure S4, including the amino acid sequences from the B, B′, and B′′ regulatory subunits of the Ser/Thr phosphatases type 2A of Arabidopsis (At.), rice (Os.), and M. truncatula (Mt.). Branch labels indicate bootstrap values (n = 1,000) of the consensus tree. Low bootstrap values are not shown. Labels at the branch tips include unique identifiers (GenBank/Uniprot) followed by the species designation; for Mtr.11959.1, the GenBank identifier is KC859637.
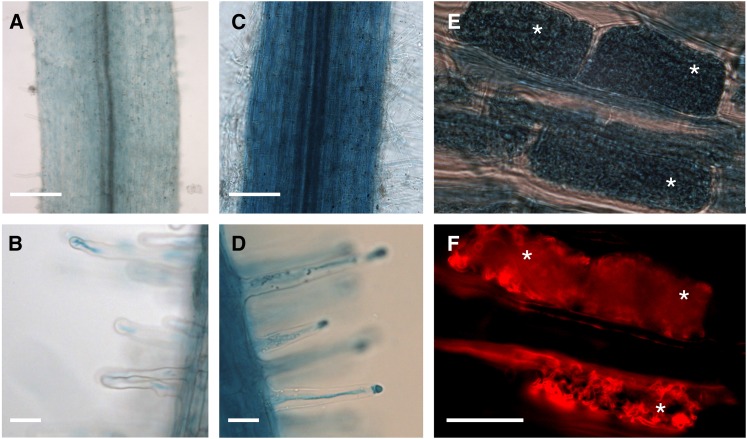
In order to confirm the gene expression atlas data, we checked the induction levels of PP2AB′1 using quantitative reverse transcriptase (qRT)-PCR (Supplemental Fig. S7B) and found induction upon AM colonization (Supplemental Figs. S7B). To further validate this, we monitored the expression of the PP2AB′1 promoter driving GUS (note that this promoter region was used in complementation studies; Fig. 4D; Supplemental Fig. S8). PP2AB′1 showed constitutive expression in all root cell layers (Fig. 3, A and B). In correlation with the quantitative RT-PCR analysis (Supplemental Fig. S7B), AM colonization led to stronger pPP2AB′1-GUS expression, and this enhancement occurred in all root cell layers (Fig. 3, C and D), including cortical cells containing arbuscules or not (Fig. 3, E and F).
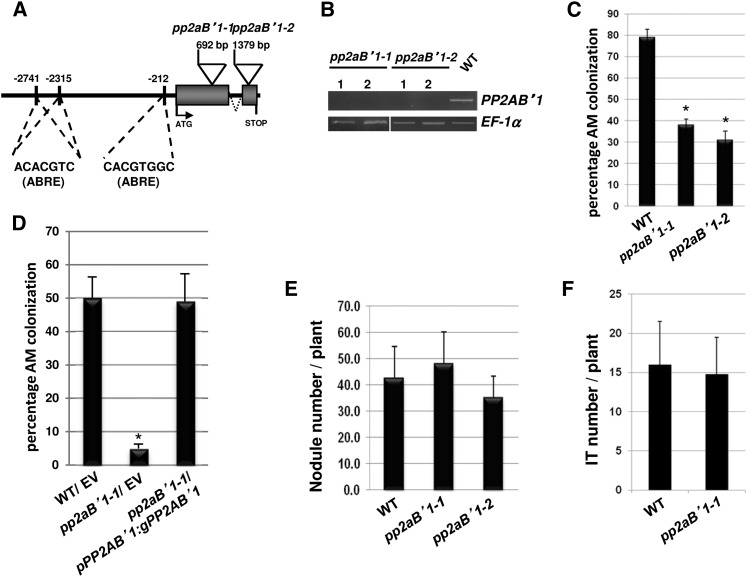
Figure 4.
pp2aB′1 mutants are impaired in AM colonization. A, Structure of the PP2AB′1 gene showing the positions of the retrotransposon Tnt1 insertions of the pp2aB′1-1 and pp2aB′1-2 mutant alleles. Three ABREs are predicted in the promoter of PP2AB′1 as indicated. B, RT-PCR to detect the accumulation of full-length PP2AB′1 transcript (1,290 bp) in mRNA from roots of two homozygous plants identified for pp2aB′1-1 and pp2aB′1-2 mutant alleles and in the wild type (WT). EF-1α expression was used as a constitutive control. C, AM colonization of wild-type and pp2aB′1 roots at 6 weeks after inoculation with R. irregularis. Values are means ± se (n = 10) from one biological replicate (for biological replicates 2 and 3, see Supplemental Fig. S13). The colonization between the wild type and pp2aB′1 mutant lines differs significantly, as indicated by asterisks (Student’s t test; P ≤ 0.0001). D, AM colonization of pp2aB′1-1 transformed roots complemented with the PP2AB′1 genomic sequence driven by its own promoter (pPP2AB′1:gPP2AB′1) or empty vector (EV) and wild-type transformed roots with the empty vector at 5 weeks after inoculation with R. irregularis. Values are means ± se (n = 10) from one biological replicate (for biological replicate 2, see Supplemental Fig. S8). The asterisk indicates statistical significance relative to the wild-type/empty vector control (Student’s t test; P ≤ 0.0001). E, Nodule quantification on the wild type and the two pp2aB′1 mutant alleles at 19 dpi with S. meliloti 2011 (optical density = 0.001). Values are means ± sd (n = 30) from three biological replicates. F, Infection thread (IT) number per wild type and pp2aB′1-1 mutant plant formed 7 dpi with an S. meliloti strain expressing LacZ. Values are means ± sd (n = 18) from two biological replicates.
Figure 3.
Expression analysis of the PP2AB′1 promoter in AM colonized roots. Promoter GUS activity in A. rhizogenes-transformed roots expressing the PP2AB′1 promoter-β-glucuronidase fusion was assessed 4 weeks after inoculation with R. irregularis in C to F and without R. irregularis in A and B. F shows a fluorescence microscopy image of the corresponding bright-field image in E showing R. irregularis stained with wheat germ agglutinin Alexa Fluor 594, which fluoresces red. White asterisks indicate cortical cells containing arbuscules. Bars = 250 µm (A and C), 50 µm (B and D), and 20 µm (F).
The presence in the putative promoter region of PP2AB′1 of three abscisic acid-responsive elements (ABREs; Fig. 4A) strongly suggests that PP2AB′1 expression is regulated via ABA signaling. To test whether ABA modulates PP2AB′1 expression at the concentrations promoting or impairing AM colonization, we applied 0, 5, and 50 μm ABA in the absence of AM colonization (Supplemental Fig. S9). PP2AB′1 expression was up-regulated by 5 μm ABA, but in contrast to ABF3 expression, which increased upon rising ABA concentrations, the expression of PP2AB′1 was down-regulated with 50 μm ABA treatment (Supplemental Fig. S9). PP2AB′1 expression was also down-regulated in roots transformed with abi1-1, which impairs ABA signaling (Fig. 1D). The modulation of PP2AB′1 expression by ABA was further confirmed in AM-inoculated roots, with an up-regulation upon 5 μm ABA and a down-regulation upon 50 μm ABA (Fig. 1B). The down-regulation of PP2AB′1 following 50 μm ABA application was further confirmed by analysis of the PP2AB′1 promoter driving GUS (Supplemental Fig. S10). Taken together, the similarity between the expression pattern of PP2AB′1 in response to ABA and R. irregularis infection and the AM colonization levels upon ABA treatments suggests that PP2AB′1 may be a new regulator of ABA signaling during the AM symbiosis.
PP2AB′1 Mutations Impair AM Colonization
To test for a role of PP2AB′1 during AM colonization, we generated an RNA interference (RNAi) construct targeting PP2AB′1. Transgenic roots of M. truncatula were obtained by A. rhizogenes-mediated gene transfer (Boisson-Dernier et al., 2001). Roots showing red fluorescence (the Discosoma spp. red fluorescent protein gene DsRed was included in the transfer DNA) were retained and inoculated with R. irregularis. After 5 weeks of inoculation, the AM colonization on the RNAi PP2AB′1 roots, whose PP2AB′1 expression was impaired (Supplemental Fig. S11A), was reduced significantly by 50%, in comparison with roots transformed with the empty vector (Supplemental Fig. S11, B and C). This observation suggests that PP2AB′1 positively regulates AM colonization.
To further validate the function of PP2AB′1, transposable element of Nicotiana tabacum cell type1 (Tnt1) mutagenized lines (Tadege et al., 2008) of M. truncatula were used for PCR-based reverse genetic screening (Cheng et al., 2011), and this identified two independent mutant lines, pp2aB′1-1 (NF3036) and pp2aB′1-2 (NF584), harboring Tnt1 insertions in PP2AB′1 exons at positions 692 and 1,379 bp, respectively (Fig. 4A). Both lines were predicted to be null mutants, as evidenced by the fact that no mRNA was detectable for PP2AB′1 in the mutant lines (Fig. 4B; Supplemental Fig. S12). Following R. irregularis inoculation, we observed a reduction of 50% AM root length colonization in both pp2aB′1-1 and pp2aB′1-2 compared with the wild type (Fig. 4C; Supplemental Fig. S13). However, fungal structures typical of the AM symbiosis, including hyphae, arbuscules, and vesicles, were all observed (Supplemental Figs. S14 and S15). This reduced colonization in pp2aB′1-1 was complemented by introducing the genomic sequence of PP2AB′1 driven by 3 kb of its own promoter region (Fig. 4D; Supplemental Fig. S8). Although PP2AB′1 expression was not enhanced upon nodulation factor or Sinorhizobium meliloti treatment (Supplemental Fig. S7, A and C), we investigated whether PP2AB′1 had a role in the root nodule symbiosis. After 7 d of inoculation with an S. meliloti strain expressing β-galactosidase (LacZ), infection threads formed on the mutant line pp2aB′1-1 at a frequency similar to that in the wild type (Fig. 4F). Moreover, 19 d post inoculation (dpi) with S. meliloti, nodules formed on pp2aB′1-1 at the same rate as on the wild type (Fig. 4E) and nitrogen fixation occurred at an equivalent level (Supplemental Fig. S16). Altogether, these results demonstrate that PP2AB′1 is required for appropriate AM colonization and is not required for nodulation in M. truncatula.
AM Colonization Mediated by ABA Is Dependent on PP2AB′1
We would predict that PP2AB′1 functions downstream of ABA perception by the ABA receptor complex. To test this, we assessed the effect of ABA treatment and abi1-1 on mycorrhizal colonization in pp2aB′1-1. Transformation with abi1-1 (Fig. 5A; Supplemental Fig. S17) and treatment with ABA (Fig. 5B) had no impact and did not rescue the AM colonization levels of pp2aB′1-1, indicating that ABA regulation of AM colonization is completely dependent upon PP2AB′1. We propose that the regulation of PP2AB′1 by ABA signaling allows ABA promotion of mycorrhizal colonization. The mechanism of ABA modulation may simply be through the promotion of PP2AB′1 expression, and notably, the induction of PP2AB′1 reflects ABA impact on AM colonization (compare Fig. 1, A and B).
PP2AB′1 Modulates ABA Sensitivity in Leaves and Seeds
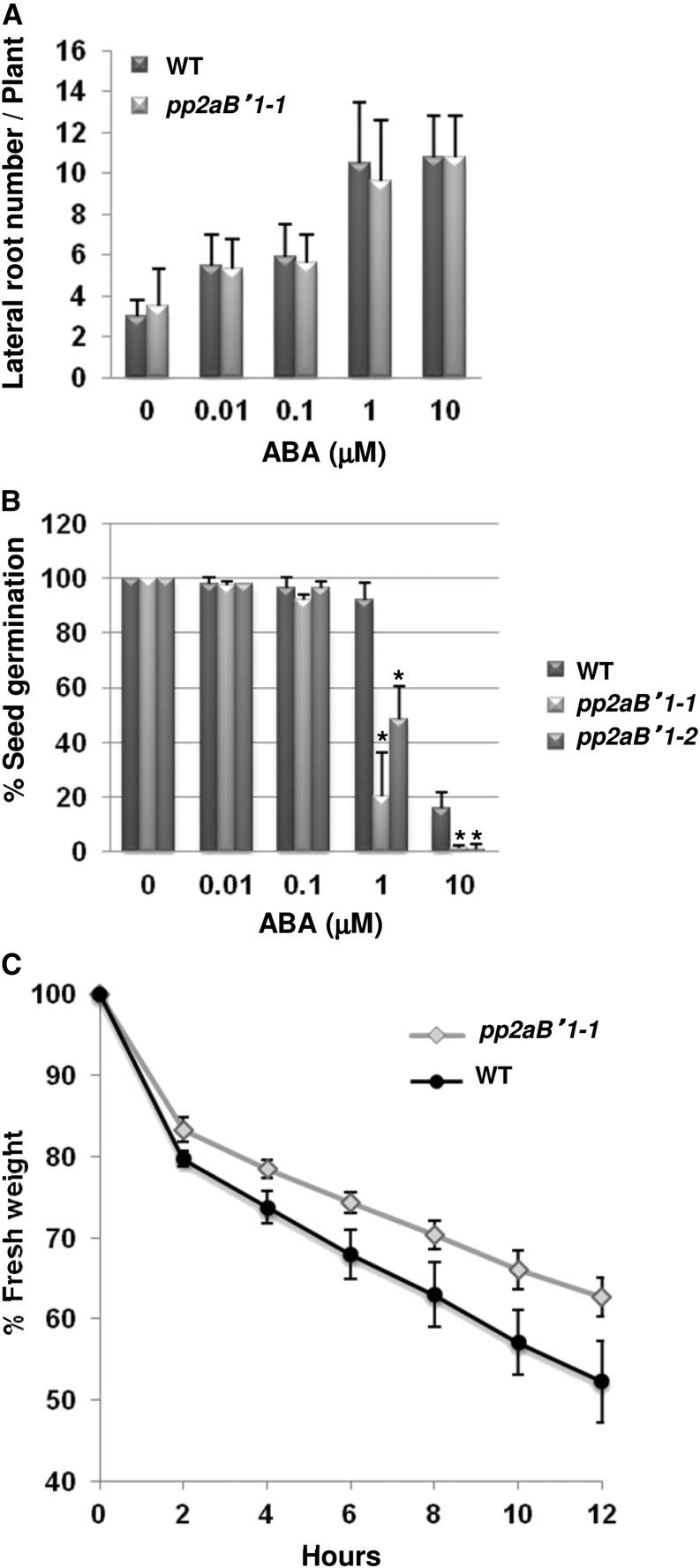
PP2Aa and PP2Ac subunits have been shown to promote ABA signaling in a variety of tissues (Kwak et al., 2002; Pernas et al., 2007). To assess whether PP2AB′1 had a specific AM function, we assessed the pp2aB′1 mutants for lateral root emergence, seed germination, and water loss from the leaf, all of which are controlled by ABA (Finkelstein et al., 2002; Liang et al., 2007). By growing the plants under different concentrations of ABA, we observed no significant difference between the wild type and pp2aB′1-1 for lateral root emergence (Fig. 6A), but pp2aB′1 mutants were hypersensitive to ABA during seed germination at higher ABA concentrations (Fig. 6B). Furthermore, leaves of pp2aB′1-1 are less susceptible to water loss when excised from the plant (Fig. 6C), suggesting either a reduced stomatal opening or increased protection of cells from dehydration; both processes are promoted by ABA (Yamaguchi-Shinozaki and Shinozaki, 2006; Okamoto et al., 2013). We conclude that PP2AB′1 functions in ABA promotion of seed germination, regulation of water loss from leaves, and ABA promotion of AM colonization but not in the ABA regulation of lateral root emergence or the root nodule symbiosis.
Figure 6.
PP2AB′1 regulates ABA responses in leaves and seeds. A, The number of lateral roots formed in pp2aB′1-1 is similar to that in the wild type (WT) in response to ABA treatment. Values are means ± sd from three biological replicates (n = 30). B, The germination of pp2aB′1 mutant seeds is hypersensitive to ABA. Values are means ± sd from three biological replicates. Asterisks indicate significant differences in comparison with the wild type (Student’s t test; P ≤ 0.01; n = 300). C, The fresh weight of detached leaves was used to measure relative water loss over time. pp2aB′1-1 shows slower water loss than the wild type. Values are means ± sd from three biological replicates.
Previous analyses suggest that the plant hormone ethylene increases AM epidermal infection in M. truncatula (Penmetsa et al., 2008) or fungal spread in tomato roots (Martín-Rodríguez et al., 2011) and that the sitiens mutant that affects ABA biosynthesis modifies AM colonization at least in part due to the effect of the mutation on ethylene levels (Martín-Rodríguez et al., 2011). One possible explanation for the mycorrhizal defects in the pp2aB′1 mutant is a role for this gene in ethylene responses. To test this, we assessed whether the pp2aB′1 mutant is perturbed in ethylene responses by measuring the hypocotyl and root length of wild-type and pp2aB′1 seedlings in the presence of various concentrations of 1-aminocyclopropane carboxylic acid (ACC). The pp2aB′1-1 mutant displays all aspects of the ethylene response, with reduction of root and hypocotyl length in a dose-dependent manner (Supplemental Fig. S18). This indicates that PP2AB′1 does not function in the plant’s response to ethylene.
pp2aB′1 Mutants Are Impaired in AM Propagation
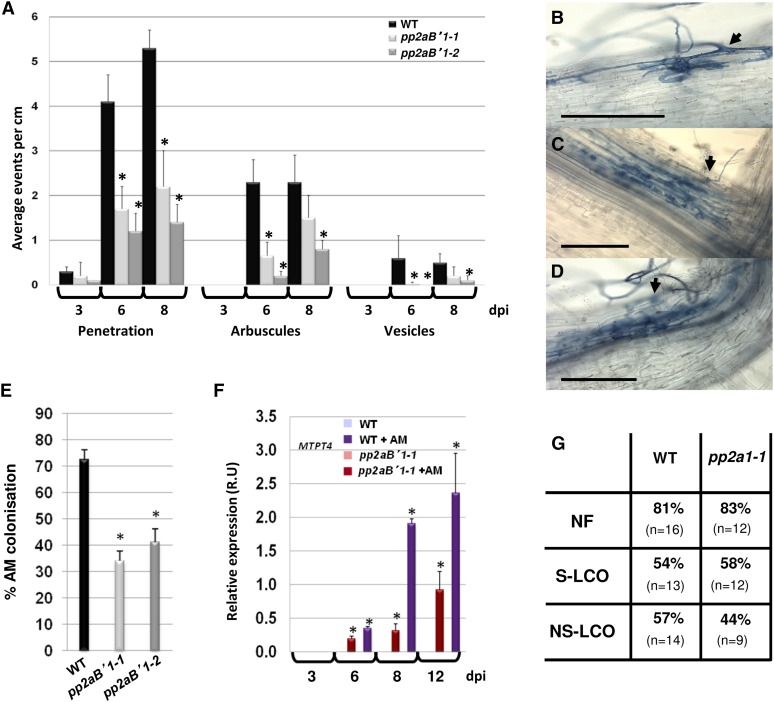
PP2AB′1 and ABA signaling could function at many different stages of the AM interaction, leading to promotion of the association. To follow the mycorrhizal fungal progression, we undertook a detailed analysis of pp2aB′1 mutants during colonization by R. irregularis (Fig. 7A). One-week-old seedlings were inoculated with germinated R. irregularis spores, and at 3, 6, and 8 dpi, plants were evaluated for the number of AM penetration events into epidermal cells (Fig. 7B) and AM penetration events into underlying cortical cells leading to arbuscules and vesicles (Fig. 7, C and D; Supplemental Figs. S14 and S15). The total root length colonization at 12 dpi was also scored (Fig. 7E). We monitored PP2AB′1 expression in the wild type over time (Supplemental Fig. S19) and found the induction of PP2AB′1 correlated with high AM colonization (Fig. 7E). At 3 dpi, the levels of fungal penetration were too low for a confident assessment of differences between the wild type and the mutants. However, penetration of AM into the mutant epidermal cells was observed, meaning that mutation in PP2AB′1 does not block epidermal entry. But at 6 and 8 dpi, the pp2aB′1 mutants showed clear reductions in the density of AM penetration of epidermal cells and reductions in the levels of arbuscules and vesicles (Fig. 7A). If we consider a ratio of epidermal penetration events to arbuscules (Supplemental Fig. S20), it is equivalent between the wild type and the pp2aB′1-1 mutant. This suggests that a primary defect of the fungal propagation is during the penetration of epidermal cells, and once the fungus has initiated infection, these infection events effectively progress to cortical colonization with resultant arbuscules and vesicles. To confirm the AM fungal colonization levels observed in the pp2aB′1 mutants, we checked the expression of the mycorrhizal marker gene PT4 (Fig. 7F). Consistent with the density of arbuscules formed, PT4 expression was impaired in the pp2aB′1-1 mutant (Fig. 7F). These results suggest that PP2AB′1 plays a role in AM propagation by promoting the infection of epidermal cells, but it also functions in growth of the fungus through the root cortex. We assessed whether PP2AB′1 regulates the early signaling pathway required for the generation of nucleus-localized calcium spiking in response to mycorrhizal factors. Both the sulfated and nonsulfated mycorrhizal factors (NS-LCO) induced calcium spiking in root hair cells in the pp2aB′1-1 mutant at a frequency similar to that in wild-type plants (Fig. 7G). These results demonstrate that PP2AB′1 is not required for early signaling but acts downstream of calcium spiking to promote AM penetration of the root epidermis and colonization of the root cortex.
Figure 7.
PP2AB′1 is required for AM penetration and propagation. A to E, AM propagation analysis 3, 6, 8, and 12 dpi with germinating R. irregularis spores infecting wild-type (WT) and pp2aB′1 mutant roots. A, The numbers of penetration events with intraradical mycelium (B), penetration events with cortical cells containing arbuscules (C), and penetration events with cortical cells containing arbuscules and vesicles (D; Supplemental Fig. S10) were quantified per cm of root. B to D, Bright-field images show the propagation of the fungus infecting the pp2aB′1-1 mutant 6 dpi. Black arrows show the fungal penetration. Bars = 200 μm. The AM fungus is ink stained. E, Percentage of AM root length colonization with R. irregularis germinating spores 12 dpi. F, Quantitative RT-PCR to monitor MtPT4 expression in wild-type M. truncatula roots infected with R. irregularis (WT + AM) and in pp2aB′1-1 during fungal propagation (A–E). The expression is normalized to EF-1α level. Values are means ± se from three biological replicates. Asterisks indicate significant differences from the wild type (A and E) or from the control noninoculated roots (the wild type and pp2aB′1-1) in F (Student’s t test; P ≤ 0.01; n = 24). G, Calcium spiking measurements in the wild type and pp2aB′1-1 mutants in response to 10−9 m nodulation factor (NF), 10−8 m sulfated lipochitooligosaccharides (S-LCO), and 10−6 m NS-LCO. The percentage of cells spiking is indicated, and n = number of cells recorded.
DISCUSSION
Phytohormones are known to mediate the AM symbiosis (Hause et al., 2007), with strigolactones involved in presymbiotic signaling (Akiyama et al., 2005; Besserer et al., 2006; Kretzschmar et al., 2012; Lauressergues et al., 2012) and ethylene postulated to increase AM infection but not cortical colonization of M. truncatula (Penmetsa et al., 2008). In tomato, ABA was shown to have a direct effect on arbuscule formation and functionality as well as an indirect effect on plant susceptibility to AM fungi by inhibition of ethylene (Herrera-Medina et al., 2007; Martin-Rodriguez et al., 2010; Martín-Rodríguez et al., 2011). In this study, we identify a new component of the ABA pathway, PP2AB′1, which is required for AM colonization and whose expression is controlled by ABA. The effect of external ABA application on the promotion of AM colonization was entirely dependent on the presence of PP2AB′1, implying that the positive action of ABA on AM colonization is predominantly via PP2AB′1. Moreover, in comparison with the tomato sitiens mutant (Herrera-Medina et al., 2007), pp2ab′1 mutants show neither an arbuscule defect nor impairment in the ethylene response. This observation implies that the regulation of AM colonization by ABA via PP2AB′1 occurs in a different pathway to that of SITIENS, which appears to be involved in the ABA regulation of arbuscule development. However, this difference may also reflect subtle species-specific differences in the function of ABA during the regulation of AM colonization.
Although the level of ABA in mycorrhized M. truncatula roots has not been investigated, previous reports suggest that AM fungi induce ABA accumulation in the colonized roots of soybean (Glycine max) and maize (Zea mays; Danneberg et al., 1992; Meixner et al., 2005). This accumulation could be due to endogenous ABA production but may also derive from the fungus (Esch et al., 1994). Therefore, in a hypothetical scenario, ABA accumulation in response to AM colonization leads to the induction of PP2AB′1 expression, which contains three ABREs within its promoter region. While the induction of PP2AB′1 is likely important for its function, there must be a basal level of this protein that is necessary for AM colonization and that may be induced at constitutive ABA levels, since we observed a phenotype in the pp2aB′1 mutants at time points prior to the induction of PP2AB′1. Notably, PP2AB′1 showed constitutive expression throughout the root prior to fungal colonization.
PP2As form heteromeric complexes; thus, we assume that PP2AB′1 functions in a complex with PP2Aa and PP2Ac subunits. The PP2Aa and PP2Ac components must be constitutively expressed, as we found no members of these gene families that were induced upon AM colonization. The mutant analysis indicates that PP2AB′1 predominantly functions at the root epidermis. Interestingly, a role for a PP2Ac at the root epidermis has been described in tomato, in which silencing of a PP2Ac leads to enhanced resistance to fungal pathogens (He et al., 2004). The PP2Ab family is believed to determine the substrate specificity of the PP2A holoenzymes, in contrast to the PP2Ac subunit, which is responsible for the catalytic activity. Thus, the PP2AB′ part of a holoenzyme complex might target a protein that is then dephosphorylated by a PP2Ac. The protein(s) targeted might be either required for or act as a negative regulator of AM colonization downstream of the common signaling pathway. However, it remains to be proven that PP2AB′1 does indeed act in a functional protein phosphatase complex.
ABA is also a regulator of the association with nitrogen-fixing rhizobial bacteria (Ding et al., 2008); however, this mode of action does not involve PP2AB′1. ABA modulates rhizobial colonization via suppression of the symbiotic signaling pathway but also via suppression of the cytokinin activation of nodule organogenesis (Ding et al., 2008). In common with the action of ABA on the rhizobial interaction, ABA suppressed the NS-LCO activation of calcium oscillations, and this probably explains the negative effect of higher concentrations of ABA on AM colonization. However, unlike the rhizobial interaction, lower concentrations of ABA positively regulate AM colonization. This opposing effect of different ABA concentrations on AM colonization may be explained if one considers a temporal process of fungal colonization. At the initial stages of the interaction, the presence of the fungus activates the symbiotic signaling pathway, which may lead to the promotion of ABA levels. These in turn induce PP2AB′1 expression, which facilitates AM colonization. Perhaps under prolonged exposure to AM fungi, sufficient levels of ABA accumulate that ultimately suppress the symbiotic signaling pathway, and this may be associated with down-regulating early signaling in cells already colonized by the fungus. Alternatively, stressful conditions that promote ABA production may block the symbiotic signaling pathway and thus inhibit the earliest stages of the AM interaction. Consistent with this, drought stress in M. truncatula suppresses PP2AB′1 expression (Supplemental Fig. S21), and this may be associated with the inhibition of AM colonization under certain plant stresses.
In Arabidopsis, ABA-dependent SnRK2 activity is controlled by a panel of positive and negative regulatory components (Fujita et al., 2011). As such, the SnRK2-interacting calcium sensor impairs SnRK2 activity in a calcium-dependent manner (Bucholc et al., 2011). The ABA regulation of cellular processes is intrinsically connected to calcium. Thereby, ABA can increase the cellular calcium sensitivity (Kim et al., 2010) as well as modulate the expression of similar genes containing ABA-calcium-responsive elements, notably ABREs (Whalley et al., 2011). In this regard, it is notable that symbiosis signaling involves calcium oscillations, and these may directly impact aspects of ABA signaling. Thus, the processes of symbiosis and ABA signaling may be more closely intertwined than we had imagined previously.
AM fungi activate the symbiotic signaling pathway of plants, and this is required to allow fungal colonization of the root. We propose that PP2AB′1 acts within a holoenzyme to dephosphorylate target protein(s) that are associated with mycorrhizal infection. As the absence of PP2AB′1 leads to a decrease in mycorrhizal infection and does not impair mycorrhizal lipochitooligosaccharide or nodulation factor-induced calcium spiking, possible targets are likely specific to AM colonization, functioning downstream of the common signaling pathway. ABA signaling appears to play an important positive role in AM colonization, and this function is predominantly via the promotion of the PP2A holoenzyme complex. The fact that ABA signaling promotes AM colonization but suppresses rhizobial colonization may be significant for discriminating between these different symbionts.
MATERIALS AND METHODS
Phylogenetic Analysis
Protein sequences similar to MtPP2AB′1 were retrieved by protein BLAST using the Medicago truncatula version 3.5 database, the Arabidopsis (Arabidopsis thaliana) database, and the rice (Oryza sativa) database. Multiple sequence alignments were performed using the ClustalW (version 1.83) program (Thompson et al., 1994) and edited using the BioEdit sequence alignment editor (Hall, 1999). The final alignment used for phylogenetic analysis is shown in Supplemental Figure S4. Phylogenetic analysis was performed with Phylip programs (version 3.69; Felsenstein, 2004). A distance matrix method employing the Jones-Taylor-Thornton model was used to compare the sequences, and a tree was built using the neighbor-joining clustering method (Saitou and Nei, 1987). One thousand bootstrapped data sets were used to indicate the confidence of each tree clade. The unrooted neighbor-joining tree was generated using PhyloDraw version 0.8 (Graphics Application Laboratory, Pusan National University).
Identification of PP2AB′1 Sequence
Full-length PP2AB′1 complementary DNA was obtained by RT-PCR using SuperScriptIII (Invitrogen) and 3′ and 5′ RACE methods using the GENE Racer Kit (Invitrogen) according to the manufacturer’s instructions. The primers were designed on the available Mtr11959.1 EST and are listed in Supplemental Table S1. PCR and nested PCR were performed with the DNA polymerase Phusion (New England Biolabs) using primers P1/GeneRacer 5′ primer and P2/GeneRacer 5′ nested primer for the 5′ RACE and P3/oligo(dT) primers and P3/GeneRacer 3′ primer for 3′ RACE. Fragments obtained were subsequently cloned into the pENTR Zero Blunt Topo vector (Invitrogen) and sequenced. Full-length PP2AB′1 complementary DNA or genomic sequences were amplified using primers P4/P5.
Gene Expression Analyses
To monitor gene expression, RNAs were extracted from root tissue with the Plant RNeasy Kit (Qiagen). RNAs were treated with TURBO DNA-free (Ambion), and 500 ng of RNA was retrotranscribed using SuperScriptII reverse transcriptase (Invitrogen). Gene expression was monitored by SYBR Green-based quantitative PCR on a Bio-Rad thermocycler using gene-specific primers. For the reactions, ELONGATION FACTOR-1α (EF-1α) or Ubiquitin expression was used for normalization. The primers used (P15–P28) are listed in Supplemental Table S1.
Identification of Homozygous pp2aB′1 Mutant Lines
DNA was extracted with the DNeasy 96 Plant Kit (Qiagen). Genotyping of segregating seedling populations was performed by PCR using the following primer combinations: Tnt1 insertions were identified with P6/P7 for PP2AB′1-1 and P6/P9 for PP2AB′1-2, and the wild type was identified with P6/P8. RT-PCR to amplify PP2AB′1 mRNA was performed using primers P4/P5. The primers used are listed in Supplemental Table S1.
Agrobacterium rhizogenes-Mediated Gene Transfer
The A. rhizogenes-mediated gene transfer was performed according to Boisson-Dernier et al. (2001) using A. rhizogenes strain AR1193 or ARquaI (Stougaard et al., 1987). The A. rhizogenes strain AR1193 was transformed with the constructs generated as follows. The RNAi PP2AB′1 construct was produced by amplification of a 105-bp fragment of PP2AB′1 EST with the primers P10/P11. The amplified product was subcloned into pENTR/D-TOPO (Invitrogen) and subsequently into the RNAi vector pK7GWIWG2D(II)R as described previously (Capoen et al., 2011) by gateway reaction using the LR clonase mix (Invitrogen). The construct pPP2AB′1:gPP2AB′1 used to complement the pp2aB′1 mutant was generated by amplification of the 3-kb promoter region of PP2AB′1 and its genomic sequence with the primers P12/P13. The amplified product was subcloned into pDONR207 (Invitrogen) by gateway reaction using the BP clonase mix (Invitrogen) and subsequently transferred into the vector pK7RD v/o 35S as described previously (Groth et al., 2010) by gateway reaction using the LR clonase mix (Invitrogen). The pPP2AB′1:GUS construct was generated by amplification of the 3-kb upstream region of gPP2AB′1 with the primers P12/P14, cloned into pDONR207 by gateway reaction using the BP clonase mix (Invitrogen) and subcloned into the vector pKGWFS7 (Karimi et al., 2002) by gateway reaction using the LR clonase mix (Invitrogen). The A. rhizogenes strain ARquaI was transformed with the construct containing ABI1-1 that was described previously (Ding et al., 2008). The primers are listed in Supplemental Table S1.
Plant Growth Conditions and AM and Nodulation Assays
Plants were grown in controlled-environment rooms at 22°C (80% humidity, 16-h photoperiod, and 300 μmol m−2 s−1). To monitor AM root length colonization, plants were grown in sterilized Terragreen/Sand (Oil-Dri) and inoculated with Rhizophagus irregularis (Endorize) to the ratio Terragreen:sand:spore-containing substrate 5:5:1. To monitor AM root length colonization in response to ABA or in overexpressing ABI1-1 transformed roots, plants were grown in sterilized Terragreen/Sand (Oil-Dri) to a ratio of 1:1 and inoculated with 500 R. irregularis spores (Symplanta). To reduce variation in AM colonization, biological replicates were initiated 4 d apart using the same fungal inoculum and grown under the same conditions. When treated this way, we found little variance between biological replicates. Another adaption that reduced variance between experiments was to increase the number of wild-type plants in each experiment. At different time points after inoculation, we screened colonization levels in wild-type plants and only initiated the full experiment when a sufficient level of colonization had occurred in wild-type plants. This reduced variance between different experiments, although we were not able to perform this prior control when performing experiments with A. rhizogenes-transformed roots. The fungal structures were stained in acidic ink as follows: roots were cleared in 10% (w/v) KOH for 15 min at 95°C, washed three times in water, and subsequently stained in acidic ink (5% [v/v] ink and 5% [v/v] acetic acid) for 4 min at 95°C. The AM root length colonization was quantified using the grid intersect method (Giovanetti and Mosse, 1980). To monitor the development of AM symbiosis on the pp2aB′1 mutant and wild-type ecotype R108, 7-d-old plants grown on buffered nodulation medium (BNM) plates (Ehrhardt et al., 1992) were planted in sterilized Terragreen/Sand (Oil-Dri) with 800 R. irregularis spores from soil substrate exposed to chive (Allium schoenoprasum) roots for 8 weeks. Plants were harvested at 3, 6, 8, and 12 dpi and acidic ink stained as described previously. For the 3-, 6-, and 8-d inoculations, the root length was measured and the infection progression was evaluated with the Leica DM6000 microscope. For nodulation assays, 1-week-old plants were grown in Terragreen/Sand (Oil-Dri) to a ratio of 1:1 and inoculated with approximately 4 mL of Sinorhizobium meliloti 2011 diluted with water to optical density at 600 nm = 0.001. Nodules were scored after 19 d.
GUS and Wheat Germ Agglutinin Staining
Transformed roots were stained for GUS activity with 5-bromo-4-chloro-3-indolyl-β-D-glucuronide for 3 h at 37°C in the dark according to Jefferson et al. (1987) and fixed. AM fungal structures were subsequently stained with 0.2 μm mL−1 wheat germ agglutinin Alexa Fluor 594 conjugate (Invitrogen) for 6 h (Ivashuta et al., 2005). The Leica DM6000 microscope was used for inspection and documentation.
Acetylene Reduction Assay
Nitrogenase activity was determined by assaying acetylene reduction. Acetylene reduction of nodulated root systems was measured in 2-mL sealed vials in the presence of 10% acetylene. After 2 h of incubation, 10 μL of acetylene was injected into a Photovac 10S Plus gas chromatograph to assess ethylene production. Samples were compared with a standard curve generated against a 5 µL L−1 ethylene standard, and nitrogenase activity was expressed as acetylene reduction assay units: ethylene products/(time [h] × nodule numbers).
ABA Activation of Lateral Root Formation
Wild-type ecotype R108 and mutant seeds were scarified, surface sterilized, and sown on 0.8% (w/v) water-agar medium. Seeds were cold treated at 4°C in the dark for 4 d and then moved to room temperature in the dark to germinate for 24 h. Germinated seeds were transferred to BNM (Ehrhardt et al., 1992) supplied with different concentrations of ABA (Sigma; A1049). The lateral roots that developed were scored after 3 weeks of growth in controlled-environment rooms at 23°C (16-h photoperiod and 300 μmol m−2 s−1).
ABA Inhibition of Seed Germination
Wild-type ecotype R108 and mutant seeds were scarified, surface sterilized, and sown on BNM supplemented with different concentrations of ABA (Sigma; A1049). Seeds were cold treated at 4°C in the dark for 4 d and then moved to room temperature in the dark. Germination was assessed after 24 h.
Water Loss Assay
Water loss was measured from detached leaves of wild-type ecotype R108 and mutants. For each biological replicate, 10 leaves from 4-week-old plants grown in controlled-environment rooms at 22°C (80% humidity, 16-h photoperiod, and 300 μmol m−2 s−1) were cut and weighed over time. The assay was performed at room temperature (approximately 23°C) under dim-light conditions with 70% relative humidity. The water loss was calculated as the percentage fresh weight at each time point.
Root Growth Response to ACC
Root growth response and hypocotyl growth were assessed in wild-type R108 and mutant pp2aB′1-1. For each biological replicate, 10 plants were grown on BNM (Ehrhardt et al., 1992) supplemented with 0, 5, 25, or 100 μm ACC (Sigma; A3903). The lengths of the roots and hypocotyls were measured after 5 d.
Calcium Spiking Analyses
For analysis of mutants, we microinjected root hair cells with calcium-responsive dyes as described (Wais et al., 2000). Micropipettes were pulled from filamented capillaries on a pipette puller (model 773; Campden Instruments). These were loaded with Oregon Green dextran (10,000 molecular weight; Molecular Probes) and Texas Red dextran (10,000 molecular weight; Molecular Probes). Injections were performed using iontophoresis with currents generated from a cell amplifier (model Intra 767; World Precision Instruments) and a stimulus generator made to our specifications (World Precision Instruments). Cells were analyzed on an inverted epifluorescence microscope (model TE2000; Nikon) using a monochrometer (model Optoscan; Cairns Research) to generate specific wavelengths of light. During image capture, the image was split using the Optosplit (Cairn Research), and each image passed through a filter for either Oregon Green or Texas Red emissions prior to exposure on the CCD chip (model ORCA-ER; Hamamatsu). Data were analyzed using Metaflor (Universal Imaging).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KC859637.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of AM colonization in wild type after 4 weeks of inoculation with R. irregularis and external application of 0, 5, or 50 μM ABA.
Supplemental Figure S2. Arbuscule formation in the wild type and pp2aB′1 is not altered by external application of 50 μM ABA.
Supplemental Figure S3. ABA (50 μM) impaired NS-LCO-induced calcium spiking.
Supplemental Figure S4. AM colonization of transformed roots expressing the Arabidopsis dominant-negative allele of abi1-1 in the wild type at 7 weeks post inoculation.
Supplemental Figure S5. Expression analyses of M. truncatula Ser/Thr protein phosphatase PP2A a, b, and c units.
Supplemental Figure S6. Alignment of PP2AB, B′, and B″ proteins from M. truncatula, Arabidopsis, and rice.
Supplemental Figure S7. Regulation of PP2AB′1 expression by AM.
Supplemental Figure S8. Complementation of pp2aB′1-1 mutant.
Supplemental Figure S9. PP2AB′1 expression is modulated by ABA in the absence of arbuscular mycorrhiza.
Supplemental Figure S10. ABA (50 μM) impaired PP2AB′1 expression in mycorrhized roots.
Supplemental Figure S11. Downregulation of PP2AB′1 impairs AM colonization.
Supplemental Figure S12. QRT-PCR analyses of PP2AB′1 expression in WT and mutant alleles pp2aB′1-1 and pp2aB′1-2.
Supplemental Figure S13. AM colonization of WT and pp2aB′1 roots at 6 weeks post inoculation with R. irregularis.
Supplemental Figure S14. AM fungus propagation in pp2aB′1-1 mutant roots after 6 dpi with R. irregularis spores.
Supplemental Figure S15. AM fungus propagation in pp2aB′1-1 mutant roots 6 dpi with R. irregularis spores.
Supplemental Figure S16. Quantification of nitrogenase activity responsible for N2 fixation using reduction of actelylene to ethylene.
Supplemental Figure S17. Quantification of AM colonization in pp2aB′1-1 mutant transformed with abi1-1 or empty vector.
Supplemental Figure S18. Sensitivity of wild-type and pp2aB′1-1 seedlings to exogenous ACC.
Supplemental Figure S19. Expression of PP2AB′1 during fungal propagation.
Supplemental Figure S20. Average ratio of arbuscules formed per penetration event at 6 and 8 dpi.
Supplemental Figure S21. Drought stress represses PP2AB′1 expression.
Supplementary Material
Acknowledgments
We thank Paul Bailey and Jeremy Murray for technical advice on the phylogenetic and database analyses, respectively. We also thank Donna Cousins for providing germinating R. irregularis spore cultures, Meritxell Antolin-Llovera and Martin Parniske for providing the vector pK7RD v/o 35S, and Allan Downie for critical reading of the article.
Glossary
- AM
arbuscular mycorrhizal
- ABA
abscisic acid
- NS-LCO
nonsulfated lipochitooligosaccharide
- ABREs
abscisic acid-responsive elements
- RNAi
RNA interference
- ACC
1-aminocyclopropane carboxylic acid
- RT
reverse transcription
- BNM
buffered nodulation medium
Footnotes
This work was supported by the European Research Council as SYMBIOSIS and by the Biotechnology and Biological Sciences Research Council (grant no. BB/J004553/1).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alexander T, Toth R, Meier R, Weber HC (1989) Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular-arbuscular mycorrhizae in grasses. Can J Bot 67: 2505–2521 [Google Scholar]
- Ariño J, Pérez-Callejón E, Cunillera N, Camps M, Posas F, Ferrer A (1993) Protein phosphatases in higher plants: multiplicity of type 2A phosphatases in Arabidopsis thaliana. Plant Mol Biol 21: 475–485 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boomsma CR, Vyn TJ (2008) Maize drought tolerance: potential improvements through arbuscular mycorrhizal symbiosis? Field Crops Res 108: 14–31 [Google Scholar]
- Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G (2011) SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem 286: 3429–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané JM, et al. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Pérez-Callejón E, Pujol G, Ariño J, Ferrer A (1994) Molecular characterization of a fourth isoform of the catalytic subunit of protein phosphatase 2A from Arabidopsis thaliana. Plant Mol Biol 26: 523–528 [DOI] [PubMed] [Google Scholar]
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P (2011) Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Cheng X, Wen J, Tadege M, Ratet P, Mysore KS (2011) Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Danneberg G, Latus C, Zimmer W, Hundeshagen B, Schneider-Poetsch H, Bothe H (1992) Influence of vesicular-arbuscular mycorrhiza on phytohormone balances in maize (Zea mays L.). Plant Physiol 141: 33–39 [Google Scholar]
- De Smet I, Zhang H, Inzé D, Beeckman T (2006) A novel role for abscisic acid emerges from underground. Trends Plant Sci 11: 434–439 [DOI] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GE (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Esch H, Hundeshagen B, Schneider-Poetsch H, Bothe H (1994) Demonstration of abscisic acid in spores and hyphae of the arbuscular-mycorrhizal fungus Glomus and in the N2-fixing cyanobacterium Anabaena variabilis. Plant Sci 99: 9–16 [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2004) PHYLIP (Phylogeny Inference Package), Version 3.6. Department of Genome Sciences, University of Washington, Seattle [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Gampala SS, Hagenbeek D, Rock CD (2001) Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J Biol Chem 276: 9855–9860 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, et al. (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202 [DOI] [PubMed] [Google Scholar]
- Giovanetti M, Mosse B (1980) An evaluation of techniques for measuring vesicular-arbuscule mycorrhizal infection in roots. New Phytol 84: 489–500 [Google Scholar]
- Groth M, Takeda N, Perry J, Uchida H, Dräxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, et al. (2010) NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbeek D, Quatrano RS, Rock CD (2000) Trivalent ions activate abscisic acid-inducible promoters through an ABI1-dependent pathway in rice protoplasts. Plant Physiol 123: 1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hao Z, Fayolle L, van Tuinen D, Chatagnier O, Li X, Gianinazzi S, Gianinazzi-Pearson V (2012) Local and systemic mycorrhiza-induced protection against the ectoparasitic nematode Xiphinema index involves priming of defence gene responses in grapevine. J Exp Bot 63: 3657–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E, Burbidge A, Okyere JP, Thompson AJ, Taylor IB (2011) Identification of the tomato ABA-deficient mutant sitiens as a member of the ABA-aldehyde oxidase gene family using genetic and genomic analysis. Plant Growth Regul 64: 301–309 [Google Scholar]
- Harrison MJ. (2012) Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 15: 691–698 [DOI] [PubMed] [Google Scholar]
- Hause B, Mrosk C, Isayenkov S, Strack D (2007) Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry 68: 101–110 [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Anderson JC, del Pozo O, Gu YQ, Tang X, Martin GB (2004) Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J 38: 563–577 [DOI] [PubMed] [Google Scholar]
- Herde O, Pena Cortes H, Wasternack C, Willmitzer L, Fisahn J (1999) Electric signaling and Pin2 gene expression on different abiotic stimuli depend on a distinct threshold level of endogenous abscisic acid in several abscisic acid-deficient tomato mutants. Plant Physiol 119: 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Medina MJ, Steinkellner S, Vierheilig H, Ocampo Bote JA, García Garrido JM (2007) Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol 175: 554–564 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, VandenBosch KA, Vance CP, Harrison MJ, Gantt JS (2005) RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17: 2911–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ (2012) Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol 38: 651–664 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GE (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases: key regulators of plant response to abiotic stresses. OMICS 15: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, Boronat A, Campos N (2011) Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Harris JM (2005) Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am J Bot 92: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304: 297–307 [DOI] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Martín-Rodríguez JÁ, León-Morcillo R, Vierheilig H, Ocampo JA, Ludwig-Müller J, García-Garrido JM (2011) Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol 190: 193–205 [DOI] [PubMed] [Google Scholar]
- Martin-Rodriguez JA, Morcillo RL, Vierheilig H, Ocampo JA, Ludwig-Müller J, Garrido JM (2010) Mycorrhization of the notabilis and sitiens tomato mutants in relation to abscisic acid and ethylene contents. J Plant Physiol 167: 606–613 [DOI] [PubMed] [Google Scholar]
- Matre P, Meyer C, Lillo C (2009) Diversity in subcellular targeting of the PP2A B’eta subfamily members. Planta 230: 935–945 [DOI] [PubMed] [Google Scholar]
- Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H (2005) Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 222: 709–715 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR (2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA 110: 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, Lichtenzveig J, Gish JC, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, et al. (2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J 55: 580–595 [DOI] [PubMed] [Google Scholar]
- Pernas M, García-Casado G, Rojo E, Solano R, Sánchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51: 763–778 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Zhang X, Noar RD, Harrison MJ (2012) Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci USA 109: E665–E672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Betz K, Antoni R, Gonzalez-Guzman M, Rodriguez L, Márquez JA, Rodriguez PL (2012) Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci 182: 3–11 [DOI] [PubMed] [Google Scholar]
- Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers AC, Barker DG (2012) A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J 69: 822–830 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (2008) Mycorrhizal Symbiosis, Ed 3. Academic Press and Elsevier, London. [Google Scholar]
- Stougaard J, Abildsten D, Marker K (1987) The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet 207: 251–255 [Google Scholar]
- Suzuki A, Akune M, Kogiso M, Imagama Y, Osuki K, Uchiumi T, Higashi S, Han SY, Yoshida S, Asami T, et al. (2004) Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol 45: 914–922 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Taylor IB, Linforth RST, Al-Naieb RJ, Bowman WR, Marples BA (1988) The wilty tomato mutants flacca and sitiens are impaired in the oxidation of ABA-aldehyde to ABA. Plant Cell Environ 11: 739–745 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. (2005) The Geographic Mosaic of Coevolution. University of Chicago Press, Chicago [Google Scholar]
- Tominaga A, Nagata M, Futsuki K, Abe H, Uchiumi T, Abe M, Kucho K, Hashiguchi M, Akashi R, Hirsch AM, et al. (2009) Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of Lotus japonicus. Plant Physiol 151: 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth EC, Vissi E, Kovács I, Szöke A, Ariño J, Gergely P, Dudits D, Dombrádi V (2000) Protein phosphatase 2A holoenzyme and its subunits from Medicago sativa. Plant Mol Biol 43: 527–536 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarié J, Long SR (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HJ, Sargeant AW, Steele JF, Lacoere T, Lamb R, Saunders NJ, Knight H, Knight MR (2011) Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 23: 4079–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sanchez JP, Lopez-Molina L, Himmelbach A, Grill E, Chua NH (2003) The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant J 34: 307–315 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.