A central component of the circadian clock suppresses variegation in chloroplast biogenesis through synergistic interactions between cytokinin and gibberellin signaling networks.
Abstract
The immutans (im) variegation mutant of Arabidopsis (Arabidopsis thaliana) is an ideal model to gain insight into factors that control chloroplast biogenesis. im defines the gene for PTOX, a plastoquinol terminal oxidase that participates in the control of thylakoid redox. Here, we report that the im defect can be suppressed during the late stages of plant development by gigantea (gi2), which defines the gene for GI, a central component of the circadian clock that plays a poorly understood role in diverse plant developmental processes. imgi2 mutants are late flowering and display other well-known phenotypes associated with gi2, such as starch accumulation and resistance to oxidative stress. We show that the restoration of chloroplast biogenesis in imgi2 is caused by a development-specific derepression of cytokinin signaling that involves cross talk with signaling pathways mediated by gibberellin (GA) and SPINDLY (SPY), a GA response inhibitor. Suppression of the plastid defect in imgi2 is likely caused by a relaxation of excitation pressures in developing plastids by factors contributed by gi2, including enhanced rates of photosynthesis and increased resistance to oxidative stress. Interestingly, the suppression phenotype of imgi can be mimicked by crossing im with the starch accumulation mutant, starch excess1 (sex1), perhaps because sex1 utilizes pathways similar to gi. We conclude that our studies provide a direct genetic linkage between GI and chloroplast biogenesis, and we construct a model of interactions between signaling pathways mediated by gi, GA, SPY, cytokinins, and sex1 that are required for chloroplast biogenesis.
The immutans (im) variegation mutant of Arabidopsis (Arabidopsis thaliana), first studied by Rédei and Röbbelen more than 50 years ago, is an ideal developmental system to gain insight into the poorly understood mechanisms of chloroplast biogenesis (Rédei, 1963, 1967; Röbbelen, 1968; Putarjunan et al., 2013). This process entails the differentiation of chloroplasts from proplastids in the leaf meristem or from etioplasts in illuminated dark-grown seedlings (Pogson and Albrecht, 2011). Green/white sectoring in all normally green organs of im is caused by a nuclear recessive gene, and cells in the mutant have a uniform im/im genotype (Rédei, 1963, 1967; Röbbelen 1968); all im alleles reported to date are null (Putarjunan et al., 2013). Whereas chloroplasts in the green sectors resemble those in the wild type with respect to pigment composition and morphology, plastids in the white sectors are vacuolated, lack lamellar structures, and accumulate phytoene, an early (colorless) C40 intermediate in the carotenoid biosynthetic pathway (Wetzel et al., 1994). Cloning of IM by map-based and transfer DNA-tagging methods revealed that the gene codes for a plastid membrane-localized plastoquinol oxidase (PTOX) that bears functional and structural similarity to the alternative oxidase class of mitochondrial inner membrane ubiquinol terminal oxidases (Carol et al., 1999; Wu et al., 1999).
It is now well established that PTOX functions as a terminal oxidase in the redox regulation of developing and mature thylakoids by transferring electrons from plastoquinol to molecular oxygen, forming water and plastoquinone (PQ; McDonald et al., 2011). In addition to linear electron transport, PTOX activity can be coupled to other processes that feed electrons to the PQ pool, including chlororespiration, cyclic electron flow around PSI, and the two desaturation steps of carotenoid biosynthesis catalyzed by phytoene desaturase and ζ-carotene desaturase (Albrecht et al., 1995; Peltier and Cournac, 2002; Okegawa et al., 2010; Putarjunan et al., 2013). It can also act as a safety valve to dissipate excess absorbed energy, thereby preventing the overreduction of photosynthetic electron carriers and protecting PSI and PSII from photoinhibition (McDonald et al., 2011; Foudree et al., 2012; Putarjunan et al., 2013). In the absence of PTOX, developing thylakoids become overreduced and phytoene accumulates, either because of a decreased supply of PQ available to phytoene desaturase and/or because electron transfer from phytoene to an overreduced PQ pool is not energetically favorable (Shahbazi et al., 2007; Rosso et al., 2009). This blockage prevents the accumulation of downstream (photoprotective) carotenoids, generating photooxidized (white) plastids. The extent of photooxidation, and white sector formation, is promoted by growth under restrictive conditions of high light or low temperature (Rosso et al., 2009).
Our current working hypothesis of the mechanism of im variegation is based on the premise that white sector formation is positively correlated with excitation pressures (EPs; Rosso et al., 2009); EP is a relative measure of the reduction state of the first stable electron acceptor of PSII (Dietz et al., 1985; Hüner et al., 1998). According to the threshold hypothesis, above-threshold EPs predispose developing im plastids to photooxidation, while plastids with below-threshold EPs develop into normal chloroplasts (Wetzel et al., 1994; Putarjunan et al., 2013). The observation that im contains heteroplastidic cells (with normal and abnormal plastids) argues that chloroplast biogenesis is also plastid autonomous (Wetzel et al., 1994; i.e. that EPs vary from plastid to plastid in the developing leaf primordium). This variation is promoted by the unique location of each plastid in the developing leaf, which dictates that each plastid has a unique biochemistry. For instance, plastids are subjected to widely varying concentrations of photosynthetic substrates such as light and CO2 that are present in steep gradients across the leaf lamina (Smith et al., 1997). A final assumption of our model is that the chaotic pattern of im variegation in the mature leaf is a reflection of the pattern of dicot leaf development: white plastids in cells of the leaf primordium give rise to clones of white plastids, cells, and sectors in the mature leaf, whereas chloroplasts in the developing primordium produce green cells and sectors.
We have turned to second-site suppressor analysis to identify genes that modify the im variegation phenotype as an approach to gain entrance into mechanisms of PTOX function, im variegation, and chloroplast biogenesis (Putarjunan et al., 2013). Our operating assumption is that suppressors can provide information about PTOX function and activity per se (e.g. interaction suppressors) as well as knowledge about factors and pathways that act early in chloroplast biogenesis but that might not be easily accessible by other means (bypass suppressors). In our search for im suppressors, we were intrigued by an observation of Rédei (1967) that vigorous vegetative growth of im could be obtained by crossing it with gigantea (gi), a late-flowering supervital mutant that had been recently isolated (Rédei, 1962). Unfortunately, Rédei (1967) did not describe the chloroplast phenotype of the imgi double mutants or their phenotype during development; the traits were assumed to act independently. However, it has since become apparent that supervitality is one of many strikingly diverse, and seemingly unrelated, phenotypes caused by GI deficiency. Others include alterations in period lengths of circadian rhythms (Park et al., 1999), resistance to paraquat (Kurepa et al., 1998a), impairment in phytochrome B signaling (Huq et al., 2000), enhanced accumulation of starch (Eimert et al., 1995) and anthocyanins (Kurepa et al., 1998b), elevated expression of detoxifying enzymes such as superoxide dismutase (SOD) and ascorbate peroxidase (APX) in the presence of paraquat (Cao et al., 2006), and increased sensitivity to freezing (Cao et al., 2007). GI was cloned nearly 40 years after Rédei’s initial description of gi (Fowler et al., 1999), and molecular analyses since that time have revealed that GI acts as a scaffold protein for the assembly of complexes that mediate protein-protein stability/activity via the 26S proteasome (Kim et al., 2007; Sawa et al., 2007; Yu et al., 2008b). It also interacts with a number of components of the circadian clock-associated flowering pathway in Arabidopsis and plays an important role in photoperiodic flowering (Fowler et al., 1999; Park et al., 1999; Sawa and Kay, 2011).
We were prompted by the observations of Rédei (1967) to revisit imgi as a tool to understand PTOX function and mechanisms of chloroplast biogenesis. In this article, we show that gi suppresses im variegation during the late stages of plant development by generating all-green versus variegated newly emerging leaves. We demonstrate that this suppression occurs via interactions between GI and cytokinin-mediated signaling pathways and that these paths are integrated by the GA response inhibitor SPINDLY (SPY). These interactions are described in a model that we test, showing that a reduction in GA signaling late in gi development derepresses cytokinin signaling, which reprograms processes required for chloroplast differentiation. Importantly, these processes include an enhancement of photosynthetic rates and an elevated reactive oxygen species (ROS) scavenging capacity. We show that when gi is combined with im in the double mutant, these processes act in concert to relax EPs, thereby promoting the formation of photosynthetically competent chloroplasts and suppression of the im defect.
RESULTS
Variegation Is Suppressed in imgi2 during Late Plant Development
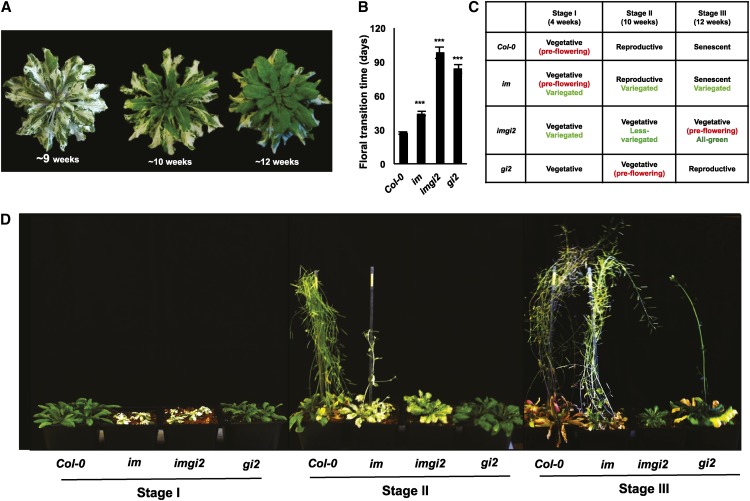
To examine interactions between im and gi, double mutants were generated by crossing null alleles of im (spotty) and gi (gi2; Park et al., 1999; Wu et al., 1999); the genotypes of the double mutants were identified in the F2 generation by PCR amplification. imgi2 plants have higher than normal leaf numbers and are late flowering (Fig. 1, A and B). This delay in the floral transition is similar to gi2 but unlike im and the wild type. Whereas imgi2 is variegated during early plant development, perhaps the most striking phenotype of imgi2 is a suppression of variegation that occurs during late development, commencing approximately 9 weeks after germination. After this time, newly emerging leaves are all green. This suggests that gi2 is able to suppress im variegation in a development-dependent manner.
Figure 1.
Growth stages of imgi2, gi2, im, and the wild type. A, imgi2 rosettes at 9, 10, and 12 weeks after germination. Plants were grown at 22°C under conditions of continuous illumination (100 μmol m−2 s−1). B, Time taken to achieve the floral transition in the wild type (Col-0), im, imgi2, and gi2. Values represent averages ± se of five independent replicates relative to Col-0 (***P < 0.001). C, Summary table of developmental stages associated with Col-0, im, imgi2, and gi2 at 4 weeks (stage I), 10 weeks (stage II), and 12 weeks (stage III) after germination. D, Representative Col-0, im, imgi2, and gi2 plants examined at the three developmental stages.
Because valid interpretations of the experiments in this report rely on the ability to compare wild-type and mutant plants at the same stage of development (versus the same chronological age), it was necessary to obtain a global assessment of the developmental profiles of imgi2, the wild type (Columbia-0 [Col-0]), im, and gi2. These data are shown in Figure 1D and summarized in Figure 1C. For these experiments, we chose to examine plants at three time points after germination, designated stage I (4 weeks), stage II (10 weeks), and stage III (12 weeks). These times were selected because they correlate with the timing of the dramatic phenotypic changes in imgi2 leaf coloration: emerging leaves are variegated during early vegetative development (stage I, until approximately 9 weeks), less variegated starting at about 10 weeks (stage II), and finally all green during late vegetative development (stage III, 12 weeks). Flowering commences 14 or so weeks after imgi2 germination (Fig. 1B). The development of gi2 proceeds in a manner similar to imgi2, with the exception that gi2 begins to flower earlier (during stage III). Finally, im and the wild type have similar growth profiles, with developmental phases that are reduced in duration compared with gi2 or imgi2: stage I corresponds to the vegetative phase, stage II to flowering, and stage III to senescence. It should be pointed out that the four genotypes in these experiments were grown under conditions of continuous illumination to eliminate circadian clock-mediated effects of gi2 (Park et al., 1999).
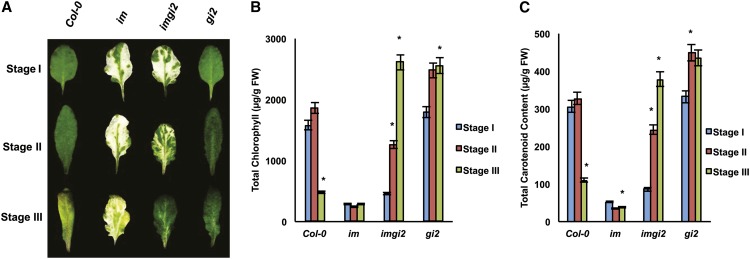
In Figure 2, we were interested in assessing the morphology and pigment contents of fully expanded leaves from the various plants at each developmental stage. Figure 2A shows that the four types of leaves are morphologically similar at stage I but that, by stage II, they have become more diverse in structure. At stage II, im and imgi2 leaves differ from one another and from wild-type and gi2 leaves; the latter two closely resemble one another. By stage III, imgi2 leaves have begun to look more like those of gi2.
Figure 2.
Leaf morphologies and pigment analyses. A, Leaf developmental stages of representative, fully expanded leaves of Col-0, im, imgi2, and gi2. B and C, Chlorophyll and carotenoid contents of the leaves in A. Values represent averages ± se of three independent replicates relative to stage I plants (*P < 0.05). FW, Fresh weight.
Figure 2, B and C, shows the pigment contents (on a fresh weight basis) of the leaves in Figure 2A. Not surprisingly, pigment accumulation in im and imgi2 leaves reflects the extent of their variegation, with imgi2 closely resembling im during early development and gi2 during late development. However, a striking feature of these analyses is that pigment amounts in gi2 are significantly higher than in the wild type at all developmental stages. This is also true for stage III (all-green) imgi2 leaves (i.e. the suppression phenotype is accompanied by high, gi2-like pigment levels). Because other imgi2 traits assessed in Figures 1 and 2 also resemble those found in gi2 more than in im, we conclude that gi2 is epistatic to im, or partially so, with respect to flowering time, leaf morphology, and pigment contents.
Cytokinins Play a Role in gi-Mediated Suppression of im Variegation
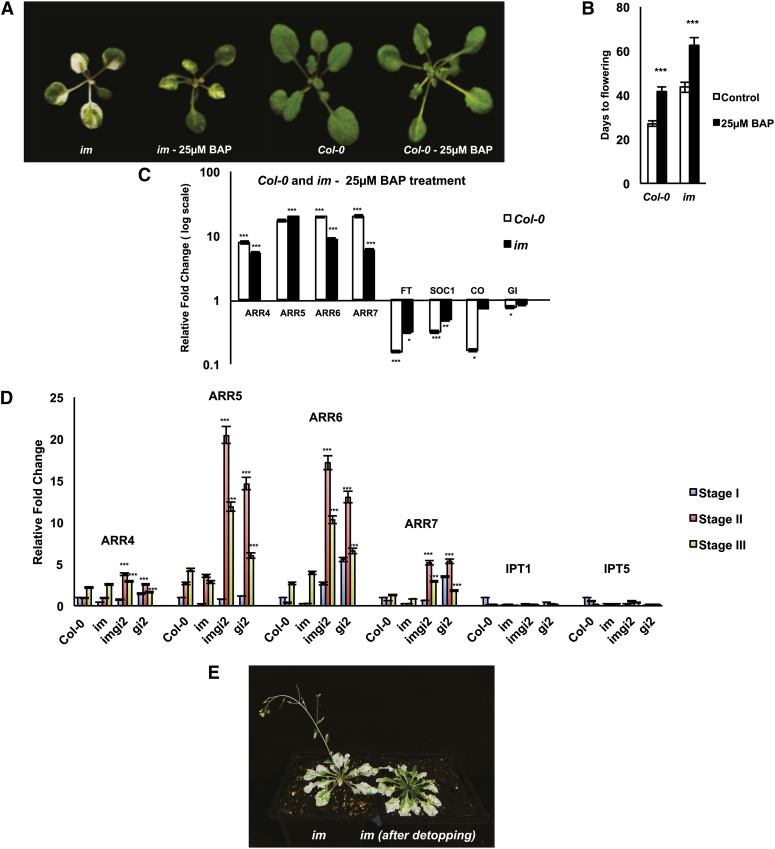
The suppression of variegation that occurs in imgi2 during stages II and III is striking and is reminiscent of the phenotype of im plants that have been detopped; subsequent leaves that emerge are all green (Fig. 3E). Although hormone levels do not necessarily correlate with signal strength, it has been proposed that cytokinins are involved in the detopping response in bean (Phaseolus vulgaris; Wang et al., 1977) and tobacco (Nicotiana tabacum; Zavaleta-Mancera et al., 1999). These observations prompted us to ask whether cytokinins play a role in the mechanism of variegation suppression in imgi2.
Figure 3.
Cytokinin responses. A, Three-week-old im and Col-0 plants were sprayed with 25 μm BAP every 3 d for a period of 2 weeks; shown are plants after treatment for 2 weeks. B, Time taken to achieve the floral transition in the plants in A. Values represent averages ± se of five independent replicates (***P < 0.001 relative to control plants). C, Total cell RNAs were isolated from im and Col-0 leaves that emerged after the 2-week BAP treatment, and real-time qRT-PCR analyses were performed to examine the expression of cytokinin response genes (ARR4–ARR7) and marker genes for the floral transition. Values are normalized to ACTIN and relative to control im and Col-0 and represent averages ± se of three technical and two biological replicates (***P < 0.001, **P < 0.01, *P < 0.05). D, Real-time qRT-PCR for cytokinin response genes (ARR4–ARR7) and cytokinin biosynthesis genes (IPT1 and IPT5) in Col-0, im, imgi2, and gi2 across all three developmental stages. The values are normalized to ACTIN and relative to Col-0 stage I plants for each gene and represent averages ± se of three technical and two biological replicates (***P < 0.001 and **P < 0.01 relative to Col-0 stage I plants). E, Representative im and detopped im plants approximately 7 weeks after germination; im was detopped at approximately 6 weeks.
As a first approach to address this question, we sprayed 3-week-old Col-0 and im for a period of 2 weeks with 6-benzylaminopurine (BAP), a widely used synthetic adenine-type cytokinin. As illustrated in Figure 3, A and C, variegation is suppressed in im leaves that emerge after BAP treatment, and this suppression is correlated with sharp, up-regulated expression of genes for type A Arabidopsis response regulators (ARR4–ARR7). These marker genes are rapidly induced at the transcript level in response to cytokinins in Arabidopsis (Hutchison and Kieber, 2002). BAP treatment also causes a delay in the transition to flowering in both Col-0 and im (Fig. 3B); as expected, this is accompanied by the down-regulated expression of marker genes for the floral transition, including CONSTANS, FLOWERING LOCUS T, and SUPPRESSOR OF CONSTANS1 (Koornneef et al., 1998; Levy and Dean, 1998; Fig. 3C).
In addition to an elevated expression in BAP-treated im (Fig. 3C), ARRs are dramatically up-regulated in stage II and III imgi2 leaves (Fig. 3D). However, this induction is not accompanied by enhanced expression of two representative cytokinin biosynthetic genes, IPT1 and IPT5 (Fig. 3D). These genes code for two isozymes of isopentenyltransferase, which catalyzes the first rate-limiting step in the biosynthesis of isoprene cytokinins via the methylerythritol phosphate pathway (Sakakibara, 2006). The lack of response of IPT1 and IPT5 might not be surprising, since IPTs are expressed primarily in root tips, immature seeds, young inflorescences, trichomes, and pollen but not in leaves (Miyawaki et al., 2004).
Although we were unable to detect dramatic changes at the transcript level for genes regulating cytokinin biosynthesis in imgi2 leaves, we considered the possibility of conducting hormone quantification assays to assess the contribution of hormone levels to the cytokinin-mediated events in Figure 3. However, we questioned our ability to identify and obtain enough of the appropriate tissues for analysis. The reason for this is that PTOX is required for chloroplast biogenesis (not for steady-state photosynthesis in the mature leaf; Rosso et al., 2006), and hence it is reasonable to assume that suppression of the im defect (by elevated cytokinin signaling) occurs sometime during the proplastid-to-chloroplast conversion in the leaf meristem and leaf primordium; this occurs sometime just prior to flowering. Knowing these times and being able to identify and collect enough cells that contain developing plastids in the meristem and primordium would be technically challenging. Therefore, we cannot rule out the possibility that hormone levels, as well as the hormone response, are altered in the developing leaf primordium of BAP-treated and imgi2 leaves.
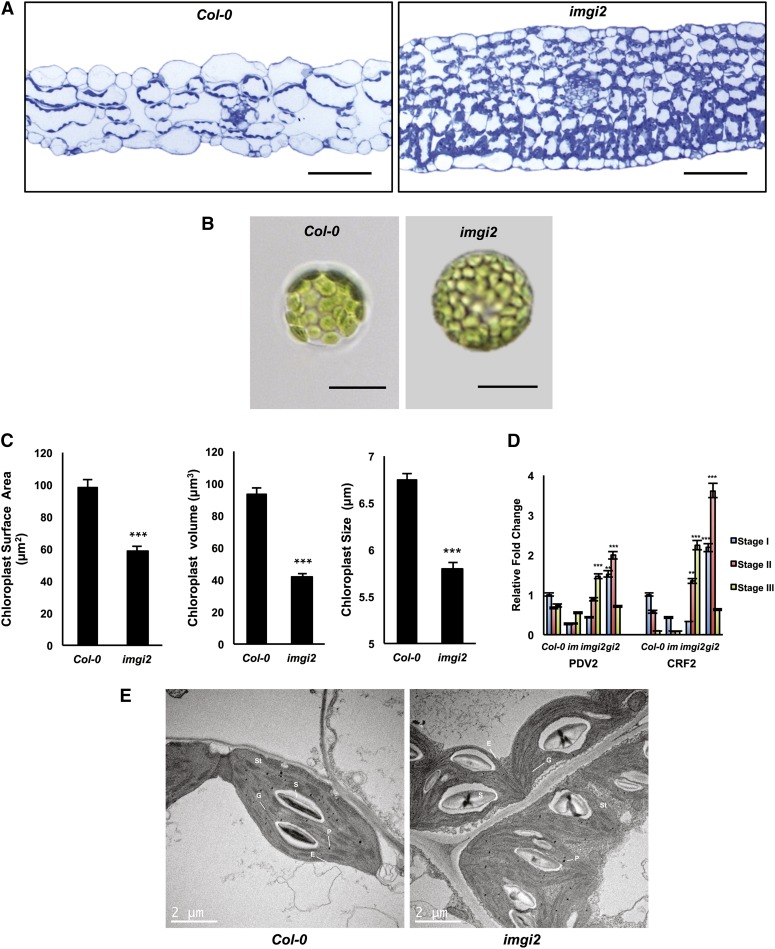
If gi-mediated alterations in cytokinin signaling play a major role in the suppression of im variegation, then we might expect to find other traits in imgi2 that are well-known characteristics of a cytokinin response, such as enhanced leaf blade thickness, starch content, cell number, and rates of chloroplast division (Lichtenthaler and Buschmann, 1978; Stoynova et al., 1996; Werner et al., 2003; Okazaki et al., 2009). In support of this hypothesis, light micrographs of leaf cross sections from the wild type and imgi2 at the same developmental stage (just prior to flowering, stage I versus stage III, respectively) show that imgi2 leaves are significantly thicker than normal, devoid of air spaces, and filled with many tightly packed cells, most of which appear to be smaller than normal (Fig. 4A). Figure 4B shows that imgi2 cells also have higher than normal chloroplast numbers, as illustrated by light microscopy of protoplasts from the wild type and imgi2 (again, isolated from leaves at the same developmental stages, as in Fig. 4A). The chloroplasts in imgi2 protoplasts also have significant reductions in surface area, volume, and diameter compared with the wild type (Fig. 4C). These alterations suggest that rates of chloroplast division are enhanced in imgi2 leaves.
Figure 4.
imgi2 leaf and chloroplast morphologies. A, Light micrographs of cross sections from fully expanded Col-0 and imgi2 leaves at the same developmental stage (just prior to flowering) stained with 1% (w/v) Toluidine Blue. Bars = 50 μm. B, Bright-field images of imgi2 and Col-0 protoplasts isolated from leaves (as in A). Bars = 20 μm. C, Chloroplast surface area, volume, and length in Col-0 and imgi2. Values represent averages ± se of 50 independent replicates. imgi2 plastids show a significant decrease in all three parameters in comparison with Col-0 (***P < 0.001). D, Expression analysis using real-time qRT-PCR for PDV2 and CRF2 in Col-0, im, imgi2, and gi2 across all three developmental stages. Values are normalized to ACTIN and relative to Col-0 stage I plants and represent averages ± se of three technical and two biological replicates (***P < 0.001 and **P < 0.01). E, Transmission electron micrographs of Col-0 and imgi2 chloroplasts from leaves just prior to flowering. E, Envelope; G, grana; P, plastoglobule; S, starch granule; St, stroma.
The analyses in Figure 4, B and D, were modeled after experiments in Arabidopsis from the Miyagishima group (Okazaki et al., 2009). They examined the role of cytokinins in chloroplast division in wild-type plants treated with exogenous cytokinins as well as in transgenic plants that overexpress CYTOKININ RESPONSE FACTOR2 (CRF2), a transcription factor induced by cytokinins. They observed that cells in both types of plants had higher chloroplast numbers and smaller chloroplast sizes than wild-type cells (similar to our findings), and they concluded that cytokinin treatment directly enhances chloroplast division. They attributed this enhancement to a specific up-regulation of two cytokinin-responsive genes that encode two essential components of the plastid division apparatus: PLASTID DIVISION1 (PDV1) and PDV2 (Miyagishima et al., 2006; Glynn et al., 2008). We suggest that a similar cytokinin-dependent mechanism is operative in imgi2, inasmuch as the alterations in chloroplast number and size in imgi2 are accompanied by a sharp up-regulation of CRF2 and PDV2 expression (Fig. 4D). Importantly, this up-regulation occurs late in development, coincident with the time when gi-mediated effects on cytokinin synthesis/signaling are proposed to be triggered and newly emerging im leaves become all green.
In summary, the experiments in Figures 1 to 4 are consistent with the hypothesis that cytokinins play a significant role in gi2-mediated suppression of im variegation and that the developmental dependence of the suppression phenotype is caused by an up-regulation of cytokinin signaling during stages II and III of plant development. We further propose that this up-regulation results in a reprogramming of plastid and leaf development. These hypotheses raise two important questions. (1) What is the linkage between cytokinin response and gi? (2) How does gi-mediated cytokinin signaling mechanistically suppress the defect in chloroplast biogenesis caused by the loss of PTOX in im?
Mechanistic Relationship between GI and Cytokinin-Mediated Signaling Pathways: A Testable Model
The data in Figures 1 to 4 provide support for the idea that interactions between signaling pathways mediated by GI and cytokinins play an important role in chloroplast biogenesis. To understand how gi and cytokinin-mediated signaling are integrated during chloroplast development, we focused on GA signaling as a possible coordinating factor. The reason for this is illustrated in Figure 5, which shows a comprehensive, testable model of how GA signaling might couple GI and cytokinin-mediated signaling pathways. We will then test elements of this model.
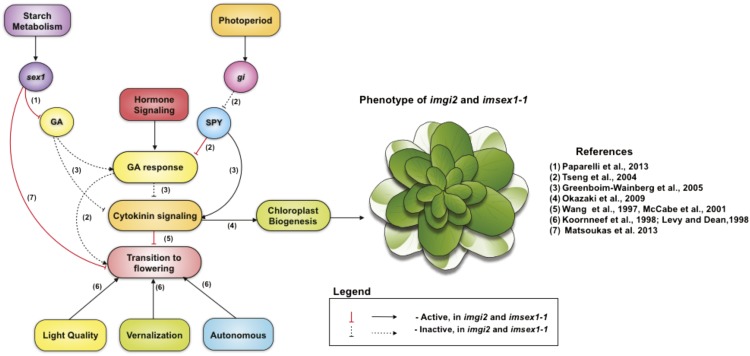
Figure 5.
Mechanism of variegation suppression in imgi2 and imsex1-1. Through its interaction with SPY and GA signaling, gi causes a derepression of cytokinin signaling that alters chloroplast biogenesis as well as the floral transition time in imgi2. imsex1-1 plants resemble the phenotype of imgi2 plants, possibly by utilizing the same downstream components (cytokinin-GA signaling pathways) as imgi2, causing the suppression of variegation late in development.
The model in Figure 5 is based on the following observations from our experiments and those of others. (1) The transition to flowering in Arabidopsis is controlled in a positive fashion by a number of factors, including photoperiod, light quality, vernalization, and the autonomous pathway, as well as by hormone signaling pathways (Koornneef et al., 1998; Levy and Dean, 1998). (2) Cytokinins delay the transition to flowering in our study (Fig. 3) and in other studies (Wang et al., 1997; McCabe et al., 2001). (3) Cytokinins positively influence chloroplast biogenesis, plastid gene expression, pigment contents, and rates of photosynthesis, although the molecular mechanisms are unclear (Figs. 3 and 4; Parthier, 1979; Lerbs et al., 1984; Chory et al., 1994; Kusnetsov et al., 1994; Yaronskaya et al., 2006, Okazaki et al., 2009). (4) Cytokinin-mediated responses in Arabidopsis are repressed by exogenous application of GA (i.e. up-regulation of GA/GA responses inhibits cytokinin signaling (Greenboim-Wainberg et al., 2005). (5) Arabidopsis spy mutants have up-regulated GA responses, and all GA-deficient phenotypes are suppressed in the mutant (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Swain et al., 2001). Cytokinin responses are also repressed in spy (Greenboim-Wainberg et al., 2005). It has been proposed that SPY represses GA signaling (Tseng et al., 2004) and promotes cytokinin responses either via GA or independently (Greenboim-Wainberg et al., 2005). (6) spy mutants suppress the late-flowering phenotype of gi (Tseng et al., 2004), indicating that SPY and GI act in concert to regulate the flowering time response in Arabidopsis. Consistent with this idea, SPY and GI interact with one another in yeast two-hybrid experiments (Tseng et al., 2004). (7) SPY controls the transition to flowering in two ways: by inhibiting GA/GA responses (Tseng et al., 2004) and by promoting cytokinin signaling directly (Greenboim-Wainberg et al., 2005; Fleishon et al., 2011). An important element in our proposed mechanism of variegation suppression relies on the observation that GA/GA responses are highly stimulated just prior to the floral transition (Koornneef et al., 1998; Levy and Dean, 1998; Tseng et al., 2004); that is, a reduced GA response at this time would delay flowering.
Given these considerations, we predict that an absence of GI should result in a derepression of SPY, and this in turn should trigger an inhibition of GA signaling (Fig. 5). Because elevated GA signaling is critical for the transition to flowering, a repression of GA signaling at this time would have two major consequences, both of which we have observed in imgi2 and both of which are hallmarks of the suppression phenotype: (1) rescue of the variegation phenotype caused by derepression of cytokinin signaling during stage II; and (2) delayed flowering.
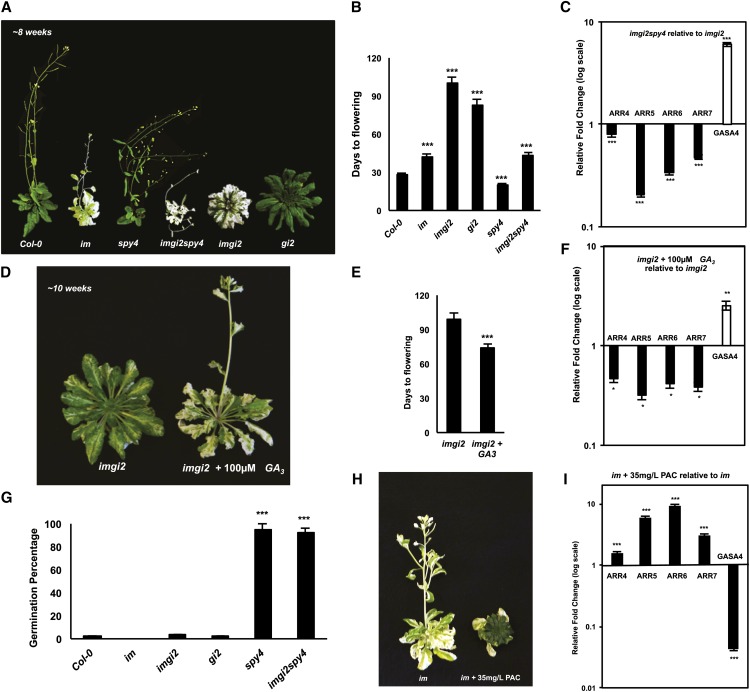
This model lends itself to a number of predictions. First, spy mutants should be able to reverse the effects of derepressed cytokinin signaling in stage II imgi2 plants (i.e. the variegation suppression/late-flowering phenotypes of imgi2 should be abrogated in triple mutants [imgi2spy]). To test this hypothesis, we crossed imgi2 with spy4 (a strong allele of spy; Jacobsen et al., 1996) and identified triple mutants in the segregating F2 population. Consistent with our model, imgi2spy4 mutants were variegated throughout their development (Fig. 6A), and they flowered after about 6 weeks, similar to wild-type and im plants (Fig. 6B). Also as predicted by the model, the alterations in the triple mutant were accompanied by a marked increase in the expression of GASA4, a representative GA response gene, and by a decrease in the expression of the cytokinin response genes ARR4 to ARR7 (i.e. GA signaling was up-regulated and cytokinin signaling was down-regulated). Taken together, these data indicate that SPY activity is essential to integrate GI and chloroplast biogenesis.
Figure 6.
Rescue of imgi2 by spy and GA. A, Representative Col-0, im, spy4, imgi2spy4, imgi2, and gi2 at approximately 8 weeks after germination. Plants were grown as in Figure 1A. B, Time taken to achieve the floral transition in Col-0, im, spy4, imgi2spy4, imgi2, and gi2. Values represent averages ± se of five independent replicates (***P < 0.001 relative to Col-0 plants). C, qRT-PCR analysis of cytokinin response genes (ARR4–ARR7) and a GA response gene, GASA4, in imgi2spy4 and imgi2 at the same developmental stage (just prior to flowering). Expression values were normalized to ACTIN and relative to imgi2 plants and represent averages ± se of three technical and two biological replicates (***P < 0.001). D, Four-week-old imgi2 plants were watered with or without 100 μm GA3 twice weekly for 6 weeks. E, Time taken to achieve the floral transition in the plants in D. Values represent averages ± se of five independent replicates (***P < 0.001 relative to imgi2 plants of the same age). F, qRT-PCR analysis of cytokinin response genes (ARR4–ARR7) and a GA response gene, GASA4, in the plants in D. Values were normalized to ACTIN and relative to nontreated imgi2 plants and represent averages ± se of three technical and two biological replicates (**P < 0.01 and *P < 0.05). G, Col-0, im, imgi2, gi2, spy4, and imgi2spy4 seeds were plated on MS medium supplemented with 35 mg L−1 PAC. The average germination percentage was determined after 10 d from 75 seeds sown on three independent plates (25 seeds per plate). Values represent averages ± se for percentage of seedlings that germinated on each of the plates (***P < 0.001 relative to Col-0). H, Representative im and PAC-treated im plants. Plants were grown as in Figure 1A and watered with or without PAC (35 mg L−1); PAC treatment commenced approximately 2 weeks after germination and continued for another 4 weeks. I, qRT-PCR analysis of cytokinin response genes (ARR4–ARR7) and the GA response gene, GASA4, in the plants in H (i.e. 6 weeks after germination). Expression values were normalized to ACTIN and relative to im control plants and represent averages ± se of three technical and two biological replicates (***P < 0.001).
A second prediction of our model is that an enhanced GA response should cause a repression of cytokinin signaling and, consequently, a reversal of the suppression phenotypes associated with imgi2 (all green, delayed flowering). To test this, we sprayed 4-week-old imgi2 plants with GA3 for 6 weeks. In accord with our model, the treated plants were variegated at all developmental stages and transitioned to flowering earlier than the nontreated imgi2 plants (Fig. 6, D and E). In addition, as predicted, these changes were accompanied by a marked down-regulation of the cytokinin response genes ARR4 to ARR7 and by an up-regulation of the GA response gene GASA4 (Fig. 6F). It might be noted that the application of GA3 to imgi2 was not as effective as spy (in the imgi2spy4 plants) in promoting time to flowering (Fig. 6, B versus E).
A third prediction of the model in Figure 5 is that the suite of phenotypes associated with imgi2 should be mimicked by inhibiting GA biosynthesis in im (i.e. inhibition of GA signaling [by inhibiting GA biosynthesis] should result in the derepression of cytokinin signaling). To test this hypothesis, we grew Col-0, im, imgi2, gi2, imgi2spy4, and spy4 seeds on Murashige and Skoog (MS) plates supplemented with paclobutrazol (PAC), an inhibitor of GA biosynthesis. Because GA is essential for germination, we anticipated that only seeds in the spy4 background should germinate; this was confirmed (Fig. 6G). To inhibit GA in a mutant lacking PTOX without inhibiting germination, we grew im on soil for 2 weeks and then watered the plants with PAC (35 mg L−1) for 4 weeks. Consistent with our hypothesis, PAC treatment suppressed the im variegation phenotype and newly emerging leaves were all green (Fig. 6H). As anticipated if PAC inhibits GA, this suppression was accompanied by a sharp decrease in the expression of the GA response gene GASA4 (Fig. 6I) as well as by a marked increase in expression of the cytokinin response genes ARR4 to ARR7. These findings are in accord with the model and indicate that an inhibition of GA results in a derepression of cytokinin signaling.
In summary, we conclude that there is a strong correlation between the greening/late-flowering phenotypes associated with imgi2 and the cytokinin response. We also conclude that GA signaling and SPY are essential components that integrate gi and cytokinin signaling to control chloroplast biogenesis.
How Does Cytokinin-Mediated gi Signaling Mechanistically Suppress im Variegation?
Although it is widely recognized that cytokinins modulate processes of chloroplast biogenesis, the mechanisms are poorly understood (Parthier, 1979; Lerbs et al., 1984; Chory et al., 1994; Kusnetsov et al., 1994; Yaronskaya et al., 2006). According to the threshold hypothesis of im variegation, developing plastids with lower than threshold EPs (i.e. relatively oxidized PQ pools) differentiate into normal chloroplasts and give rise to green sectors in the mature leaf, whereas developing plastids with higher than threshold EPs (i.e. overreduced PQ pools) accumulate phytoene and become blocked in the production of photoprotective, downstream carotenoids; these plastids are susceptible to photooxidation and give rise to white sectors in mature leaves (Rosso et al., 2009). To identify the mechanism(s) by which gi2 suppresses im variegation, we reasoned that the signaling pathways in Figure 5 likely result in the production of factors that cause a relaxation of EPs, such that more developing plastids have EPs within the range compatible with normal chloroplast biogenesis. Thus, our question became: how do cytokinins and GA control EPs?
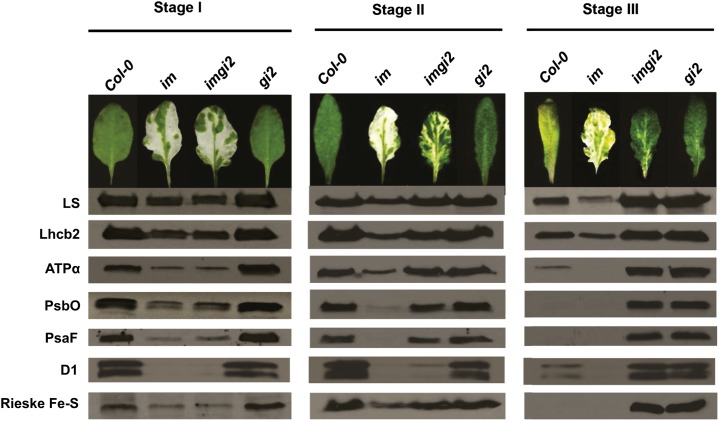
To address this question, we felt that it would be helpful to first obtain baseline information about the ultrastructure and protein composition of imgi2 chloroplasts. Figure 4E shows transmission electron micrographs of representative chloroplasts from fully expanded wild-type and imgi2 leaves just prior to flowering: imgi2 chloroplasts are smaller than normal (thus confirming the protoplast data in Fig. 4, B and C) but otherwise have normal-appearing internal membrane structures. We next conducted western immunoblot analyses to examine the accumulation of seven representative photosynthetic proteins from im, gi2, imgi2, and Col-0 leaves at developmental stages I to III; the proteins were loaded on the gels on an equal fresh weight basis. Figure 7 shows that, during stage I, each of the seven proteins accumulates to a similar extent in gi2 and the wild type and also to a similar, although reduced, extent in im and imgi2; the decreases in im and imgi2 are expected and caused by the presence of white tissues in the whole-leaf samples (Aluru et al., 2007). During stage II, the pattern of protein accumulation in imgi2 begins to resemble that of gi2 (a notable exception being D1), such that by stage III, all seven proteins have accumulated to high levels and the imgi2 and gi2 profiles are indistinguishable. Stage III wild-type and im leaves, on the other hand, are senescent and, not surprisingly, have much reduced amounts of all seven proteins.
Figure 7.
Photosynthetic protein accumulation. Total leaf proteins were isolated from the top two fully expanded rosette leaves of Col-0, im, imgi2, and gi2 at stages I to III. The proteins were loaded on an equal fresh weight basis for electrophoresis via 12% SDS-PAGE. Immunoblot analysis used polyclonal antibodies to the large subunit of Rubisco (LS), the light-harvesting complex protein (Lhcb2), the α-subunit of ATP synthase (ATPα), PSII subunit O (PsbO), PSI subunit F (PsaF), PSII reaction center protein (D1), and the Rieske Fe-S center of the cytochrome b6f complex (Rieske Fe-S).
Taken together, the data in Figures 4E and 7 indicate that gi2 is able to attenuate the impact of a loss of PTOX on chloroplast biogenesis late in plant development, such that all-green leaves are produced whose chloroplasts resemble those in gi2 with respect to ultrastructure and protein composition. This suggests that gi2 is epistatic to im with respect to these traits, commencing at stage II, thus expanding the list of traits enumerated earlier (i.e. leaf morphology, pigment composition, and flowering time; Figs. 1, A and B, and 2, A–C) and the expression of genes for the cytokinin response (Figs. 3D and 4D). Given the widespread number of gi traits that are epistatic to im, we asked whether the suppression of EPs in im could be caused by one or more gi phenotypes that have been documented over the years that could theoretically ameliorate EPs. These include an elevated expression of detoxifying enzymes such as SOD and APX in the presence of paraquat (Cao et al., 2006) and enhanced accumulation of starch (Eimert et al., 1995). Do either of these factors play a role in gi-mediated suppression of im variegation?
ROS Detoxification.
One of the most intriguing phenotypes of gi is its resistance to hydrogen peroxide (H2O2) and paraquat (Kurepa et al., 1998a). Paraquat is a superoxide (O2−) radical-generating herbicide that acts by intercepting the flow of electrons at PSI (Preston, 1994); O2− and H2O2 are produced by an interaction of paraquat radicals with molecular oxygen (Slade, 1966). The resistance of gi2 to paraquat is caused by an elevated expression of the chloroplast-detoxifying enzymes SOD and APX in the presence of the herbicide (Cao et al., 2006).
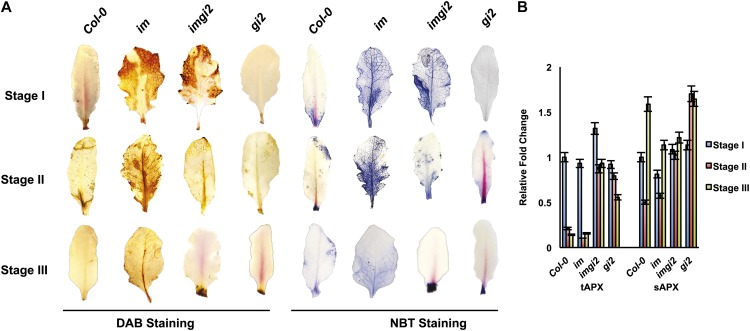
To obtain an overview of the role of ROS generation and quenching in the suppression of im variegation, we assayed the accumulation of O2− and H2O2 in im, gi2, imgi2, and Col-0 leaves by nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) staining, respectively. Figure 8A shows that ROS accumulation is very low in gi2 throughout development, consistent with previous observations (Kurepa et al., 1998a), but that O2− and H2O2 levels are high in im and imgi2, especially early in development. Strikingly, gi2 is able to sharply reduce the high amounts of ROS that are present in im. This suppression occurs during stages II and III of imgi2 development (when newly emerging leaves are becoming all green) and is accompanied by prolonged, high-level expression of stromal and thylakoid APXs (Shigeoka et al., 2002; Fig. 8B). Similar to imgi2, gi2 has high levels of APX expression at all three developmental stages; in contrast, expression levels fall markedly in the wild type and im after stage I (just prior to flowering) and either continue to fall (thylakoid APX) or increase dramatically (stromal APX) during senescence (stage III). Considered together, the data in Figure 8 suggest that gi2 is epistatic to im with respect to having an enhanced ROS scavenging capacity during late plant development. We conclude that this could be a contributing factor to reduce EPs in imgi2, thus suppressing variegation.
Figure 8.
ROS accumulation. A, DAB and NBT staining to detect H2O2 and O2−, respectively, in Col-0, im, imgi2, and gi2 across all three developmental stages. B, Expression analysis using real-time qRT-PCR for stromal APX (sAPX) and thylakoid APX (tAPX) genes in Col-0, im, imgi2, and gi2 at stages I to III. Expression values are normalized to ACTIN and relative to stage I Col-0 plants and represent averages ± se of three technical and two biological replicates.
Starch Accumulation.
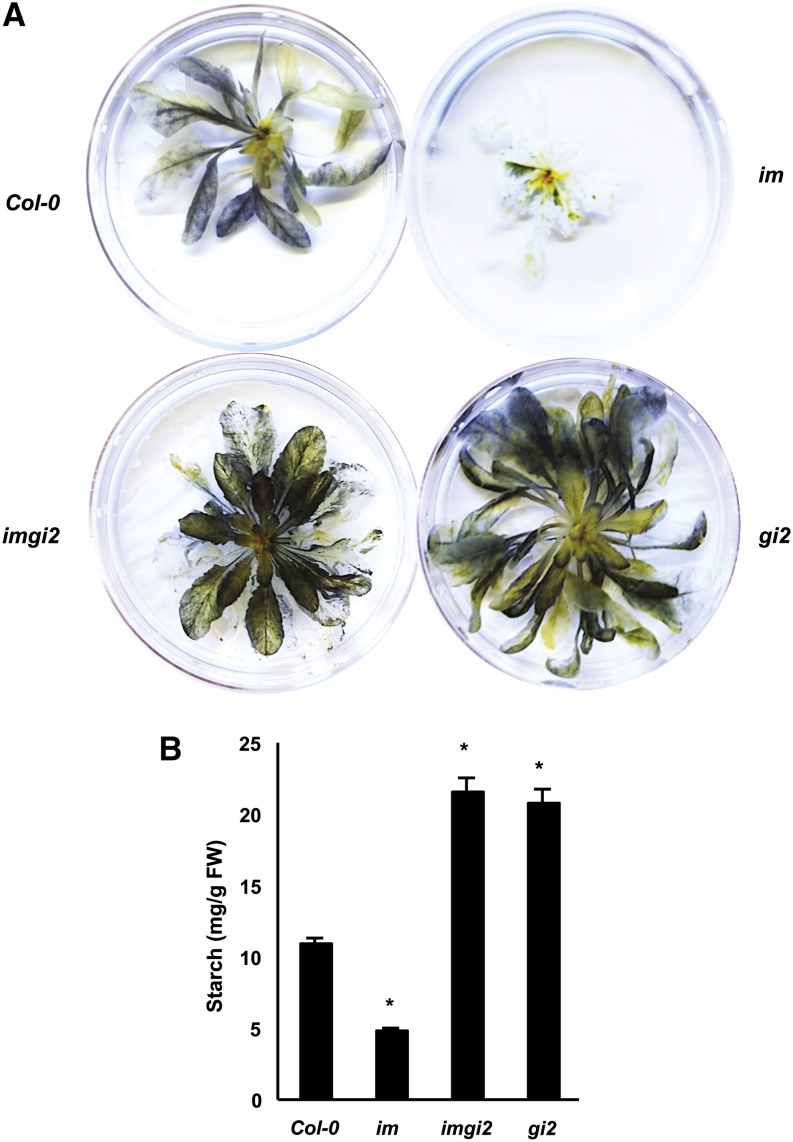
Another well-characterized phenotype of gi that has the potential to ameliorate EPs is starch accumulation (Eimert et al., 1995). To explore this question in imgi2, we monitored leaf starch content by staining intact plants (the wild type, im, imgi2, and gi2) with Lugol’s iodine solution; the plants were removed from the soil and stained when they were at the same developmental stage (just prior to flowering). Figure 9A shows that starch levels are significantly elevated in imgi2 and gi2 compared with im and the wild type. This conclusion was confirmed by quantitative measurements: vegetative leaves of prebolting imgi2 and gi2 accumulate about twice as much starch as the wild type on a fresh weight basis (Fig. 9B). The high levels of starch are also apparent from electron micrographs (Fig. 4).
Figure 9.
Starch accumulation. A, Qualitative analysis of starch accumulation in Col-0, im, imgi2, and gi2 at the same developmental stage (just prior to flowering). Plants were removed from the soil and stained with potassium iodide/iodine solution (Lugol’s iodine), destained in water for 1 h, and then photographed. B, Quantitative analysis of starch levels in Col-0, im, imgi2, and gi2 at the same developmental stage (prior to flowering). Values represent averages ± se of three independent replicates (*P < 0.05 relative to Col-0 plants). FW, Fresh weight.
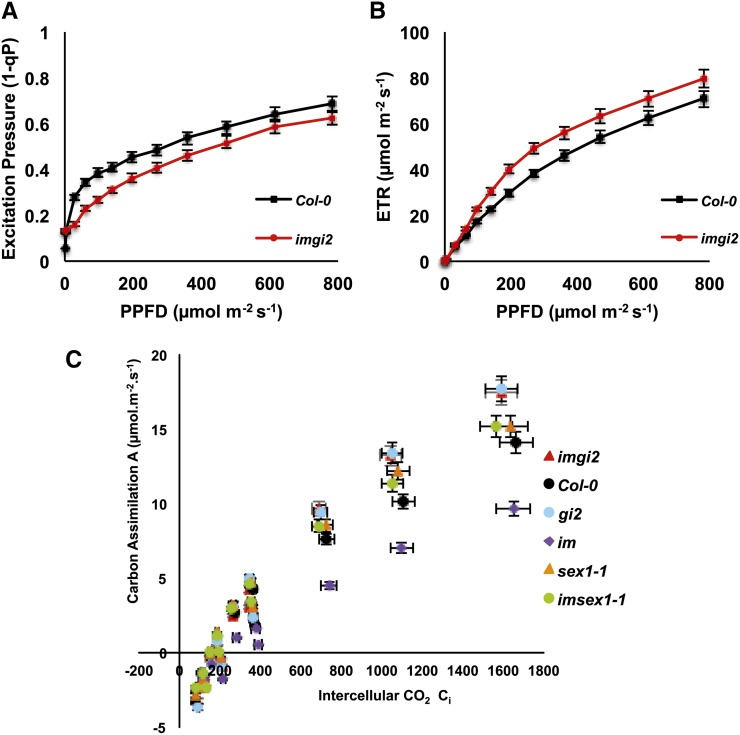
One explanation for these results is that imgi2 and gi2 have higher than normal rates of carbon assimilation. To test this hypothesis, we generated carbon assimilation rate-internal CO2 concentration curves at a constant photosynthetic photon flux density (200 μmol m−2 s−1) using fully expanded leaves from plants at the same stage of development (just prior to flowering). Figure 10C shows that carbon assimilation rates are significantly higher in the all-green leaves of imgi2 and gi2 versus the wild type; lower than normal values for im reflect the presence of contaminating white sectors in the leaf samples (Aluru et al., 2007). It is likely that the enhancement in assimilation in imgi2 is caused, in part, by increased photosynthetic electron transport capacity, inasmuch as chlorophyll fluorescence analyses revealed that imgi2 leaves have lower than normal EPs and higher than normal electron transport rates (Fig. 10, A and B). The latter experiments were conducted under a range of light intensities using leaves from imgi2 and the wild type at the same stage of development (just prior to flowering).
Figure 10.
Photosynthesis and carbon assimilation. A and B, Chlorophyll fluorescence parameters were measured on detached leaves from imgi2 and Col-0 under varying light intensities. Excitation pressure is a measure of the redox state of the first stable electron acceptor of PSII and is quantified using the parameter 1-qP (photochemical quenching), and ETR is the relative linear electron transport rate. C, CO2 assimilation rates (A) with respect to the internal CO2 concentrations (Ci) in Col-0, im, imgi2, gi2, imsex1-1, and sex1-1 plants at the same developmental stage (just prior to flowering). The A-Ci curves were plotted at a photosynthetic photon flux density of 200 μmol m−2 s−1. Values represent averages ± se of five biological replicates.
Regardless of the precise mechanism, the data in Figures 9 and 10 indicate that gi2 is epistatic to im with respect to carbon assimilation and starch accumulation. Enhanced rates of carbon assimilation and photosynthetic electron transport in imgi2 would be a powerful way to decrease EP thresholds and promote chloroplast biogenesis.
Can starch excess1 Rescue im?
Given that enhanced ROS scavenging and carbon assimilation are two mechanisms that could play a direct role in gi-mediated relaxation of EPs in im, we wanted to examine whether either of these is sufficient to suppress im variegation. Early attempts to rescue the im variegation phenotype by the overexpression of components of the ROS scavenging system (SOD and APX) were not successful, likely because these components are redundant and act in concert to scavenge ROS (Giacomelli et al., 2007). To examine whether starch accumulation is sufficient, we crossed im with starch excess1 (sex1), a well-characterized starch accumulation mutant that, interestingly, also displays a late-flowering phenotype (Matsoukas et al., 2013); we used null alleles of both im (spotty allele) and sex1 (sex1-1; Yu et al., 2001) for this cross. SEX1 is a starch-related α-glucan/water dikinase that controls the accessibility of degradative enzymes to the starch granule, and starch accumulates in sex1 mutants because it lacks this accessibility (Yu et al., 2001; Yano et al., 2005). Surprisingly, we found that imsex1-1 double mutants recapitulate the phenotype of imgi2 (i.e. they are late flowering [approximately 10 weeks to achieve the floral transition] and produce variegated leaves early in development but all-green leaves later in development; Fig. 11). imsex1-1 and sex1-1 plants also have enhanced rates of carbon assimilation compared with the wild type, similar to that observed for gi2 and imgi2 (Fig. 10C). Considered together, these data indicate, first, that sex1-mediated signaling is able to rescue im variegation in a development-specific manner, and second, that sex1 is epistatic to im with respect to flowering time, photosynthetic carbon assimilation, and chloroplast biogenesis, commencing late in plant development. This suggests that imsex1-1 and imgi2 resemble one another with respect to a number of traits.
Figure 11.
Suppression of variegation in imsex1-1 double mutants. Representative Col-0, im, imsex1-1, and sex1-1 plants are shown approximately 10 weeks after germination. The imsex1-1 plants are late flowering and produce variegated leaves early in development but all-green leaves later in development, similar to imgi2 plants.
DISCUSSION
Signaling Events That Control Chloroplast Biogenesis
For a number of years, we have used im as a tool to gain insight into mechanisms of photosynthetic electron transport and chloroplast development (Yu et al., 2007; Liu et al., 2010; Foudree et al., 2012; Putarjunan et al., 2013). These studies have included the characterization of second-site suppressors that rescue the variegation phenotype of im, producing all-green plants. These suppressors have allowed us to identify elements that are important for PTOX activity and, more broadly, elements that integrate pathways of chloroplast biogenesis (Fu et al., 2012; Putarjunan et al., 2013). Because PTOX plays a crucial role in maintaining the redox state of developing thylakoids, a unique feature of im bypass suppressors is that they provide a way to access factors that play a role in the early events of chloroplast biogenesis but that are difficult to identify using techniques of biochemistry and molecular biology because of their abundance and/or location in difficult-to-study meristematic and leaf primordial tissues. In this report, we found that the defect in im plastids can be suppressed by gi in developing chloroplasts of leaves that emerge late in plant development. Interestingly, Rédei (1962, 1963) crossed im with gi over 50 years ago as way to generate vigorous im plants for study but did not report on the late-greening phenotype of the double mutant; he also assumed that gi merely provided a growth phenotype and did not affect the im plastid phenotype per se.
Importantly, our analyses enabled us to develop a testable model of gi-mediated pathways that control chloroplast biogenesis using im as a reporter (Fig. 5). This model is based on published observations as well as on findings in this study. The robustness of the model is reflected in our ability to confirm elements of it experimentally. One of our central findings is that gi mediates chloroplast biogenesis (and flowering time) via an effect on GA and cytokinin signaling. Support for this hypothesis came from our observation that spy reverses the variegation suppression and late-flowering phenotypes of imgi2. This indicates that SPY activity plays an important role in chloroplast biogenesis and also highlights the significance of cross talk between the GA and cytokinin signaling pathways in the plastid development process. While our data suggest that the hormone response plays a dominant role in the suppression mechanism, we cannot rule out the possibility that hormone levels are also altered. Determining whether this is the case would be a challenging undertaking because it would require the ability to identify and obtain enough tissues in which the chloroplasts of the meristem and leaf primordium are still developing (i.e. the stage at which PTOX first functions and when the im defect is suppressed).
In this context, it is worth emphasizing that the uniqueness of imgi2 lies in its ability to manipulate hormone signaling pathways that are essential for the floral transition in such a manner that developmental reprogramming of imgi2 plastids occurs at this time. In other words, the defect in im plastids is suppressed late rather than early in development and is not expressed continuously throughout development, as might be expected with constitutive overexpression of cytokinin biosynthesis/response genes. Our model emphasizes GA, because GA is involved in stimulating the transition to flowering by transcriptionally activating genes directly controlling the floral transition; GA/GA responses also are expressed most highly just prior to this stage (Koornneef et al., 1998; Levy and Dean, 1998. Furthermore, transcriptomic analyses in gi mutants of rice (osgi-1) have revealed remarkable increases in the transcript accumulation of genes coding for GA 2-oxidase, which is involved in inactivating bioactive GAs (Itoh and Izawa, 2011).
The phenotypic similarity between imgi2 and imsex1-1 is striking and raises questions about the integration of sex1 into the model in Figure 5. Although speculative at this point, one possibility is that there is a convergence of gi- and sex1-mediated pathways at GA/GA signaling and the utilization of identical elements downstream from GA that involve derepression of cytokinin signaling. Support for this hypothesis comes from recent studies showing that GA biosynthesis is repressed in sex1-1 as a response to sugar starvation, which arises from the primary lesion in the mutant and its inability to break down starch (Paparelli et al., 2013). Interestingly, gi mutants display a similar sugar starvation phenotype (Cao et al., 2007). A detailed understanding of interactions between sex1 and GA awaits further investigation.
It should be noted that exogenously supplied cytokinins are able to rescue the phenotypes of other nuclear gene-induced variegations like pale cress (pac; Grevelding et al., 1996) and amidotransferase-deficient2 (atd2; van der Graaff et al., 2004), producing all-green plants. Although the function of the PAC gene is unknown, pac mutants have a light-conditional phenotype that results in photooxidized plastids early in development, perhaps because PAC affects the accumulation of carotenoids and abscisic acid (Grevelding et al., 1996; Meurer et al., 1998; Holding et al., 2000). On the other hand, atd2 (and other alleles of the same mutant gene, such as altered APX2 expression13 and chloroplast import apparatus1) defines the gene for ATASE2, the first enzyme of purine biosynthesis, and loss of this critical enzyme has pleiotropic effects, including a light-conditional phenotype and the accumulation of ROS (Sun et al., 2001; van der Graaff et al., 2004; Woo et al., 2011). Because pac and atd2 have different primary lesions in chloroplast development, we consider it likely that their rescued phenotypes, following cytokinin treatment, rely on the utilization of different batteries of cytokinin-regulated genes and perhaps different processes than those utilized in imgi2. In this context, it will be interesting to determine whether cytokinin-treated pac and atd2 are also late flowering, similar to BAP-treated im, and whether the pac and atd2 variegations can be suppressed by gi and sex1-1.
Molecular Mechanism(s) of im Suppression
Based on our current conception that EP thresholds govern im variegation (Rosso et al., 2009), the findings in this article support the idea that a lack of GI attenuates EP thresholds in imgi2 via an aberrant derepression of cytokinin signaling during late plant development. This causes suppression of the im defect, because EP thresholds are reduced to such an extent that all (or nearly all) developing plastids in the leaf primordium are able to differentiate into photosynthetically competent chloroplasts. Given that cytokinins play a pivotal role in photosynthesis and chloroplast biogenesis, it is perhaps surprising that the molecular mechanisms are so poorly understood. In perhaps the best-studied case, cytokinin control of chloroplast division has been traced to specific up-regulation of two nuclear genes whose products, PDV1 and PDV2, are imported into the plastid posttranslationally, where they serve as components of the plastid division machinery (Miyagishima et al., 2006; Glynn et al., 2008). By analogy, our question can be stated as: how does cytokinin signaling compensate for a loss of PTOX at the level of the photosynthetic membrane; that is, what are the changes in cytokinin-mediated gene expression that mitigate EPs during chloroplast biogenesis?
One widely used approach to address this type of question involves the identification of candidate genes via global expression analyses, followed by rescue of a mutant phenotype. This approach is fraught with difficulties (e.g. too many genes, and gene interactions, to sort through) and subject to a great degree of serendipity. We were able to avoid these problems by taking advantage of the epistatic interactions between gi2 and im, which allowed us to narrow down the possible factors that ameliorate EPs in imgi2 by asking whether any of the gi2 phenotypes characterized over the past 50 years of GI research could logically play a role in modulating EPs and, thereby, rescue the im defect.
The first gi phenotype we examined was ROS scavenging capacity, which is up-regulated in gi in the presence of paraquat, a ROS generator (Cao et al., 2006). Given that developing im membranes are overreduced, im also acts as a ROS generator (Ivanov et al., 2012); this was confirmed in Figure 8. We found that ROS accumulation is drastically reduced in imgi2 in late plant development to levels that are characteristic of gi2 (i.e. gi2 is epistatic to im with respect to this trait). This leads to the reasonable hypothesis that derepression of cytokinin signaling in late imgi2 development causes an up-regulation of gene expression for ROS detoxification enzymes, as illustrated for APX (Fig. 8B), and that this results in an enhanced ROS scavenging capacity in developing imgi2 plastids, a reduction of EPs, and suppression of the im defect. This proposal is consistent with the observations of others that cytokinin signaling and chloroplast ROS scavenging capacity are linked (Pogany et al., 2004; Cortleven et al., 2014) and also with the finding that plastid-localized ROS scavenging enzymes like SOD and APX are specifically increased in maize (Zea mays) seedlings treated with PAC (Pinhero et al., 1997). In accord with our model, the latter might be a cytokinin response, rather than a GA response, inasmuch as inhibition of GA (via PAC) would be anticipated to derepresses cytokinin signaling.
The second feature of gi2 that we investigated was its striking ability to accumulate starch. We discovered that this is accompanied by enhanced rates of photosynthetic carbon assimilation. Using biochemical and molecular techniques, we have previously shown that the green sectors of fully expanded im leaves behave as sources and the whites sectors as sinks. The green sectors also have enhanced photosynthetic capacity, as monitored by chlorophyll fluorescence, oxygen evolution, and carbon fixation rates (Aluru et al., 2001, 2007). Profiling experiments have revealed that this enhancement is likely caused by a reprogramming of photosynthetic gene expression in the green cells, perhaps elicited during leaf development in response to sink demand (i.e. elevated photosynthetic capacity is part of a growth strategy to compensate for total reductions in source tissue; Aluru et al., 2001, 2007). This explanation does not seem to apply to imgi2, however, because imgi2 plants are not variegated. Rather, we propose that gi2 is epistatic to im with respect to this trait and that when it is combined with im in the double mutant, one consequence is that EPs are attenuated to such an extent that the im defect is suppressed. Because we cannot rule out the possibility of interactions between im and gi2 that might modify the elevated photosynthesis trait, gi2 (versus imgi2) might be a cleaner system to assess mechanistically how photosynthetic rates are controlled by cytokinin. The answer to this question has obvious implications for practical and applied studies of mechanisms of carbon sequestration.
In this context, it should be pointed out that imsex1-1 suppressors resemble the phenotype of imgi2, perhaps because both SEX1 and GI utilize the same pathways downstream from GA (as discussed earlier). If this is the case, several predictions can be made. For example, sex1-1 should show the same suite of chloroplast phenotypes as gi2, including enhanced ROS scavenging capacity in response to paraquat. Predictions such as these can be tested in future investigations. Other mutants with a starch accumulation/late-flowering phenotype also might be found that rescue im, and if these mutants utilize the pathways in Figure 5, they could be used to investigate elements and interactions upstream of GA (like GI and SEX1) that regulate chloroplast biogenesis.
Given what we know about the primacy of redundancy and plasticity in signaling pathways in plants, it is perhaps naïve to expect that a single mechanism would be responsible for suppressing im variegation, especially in a genetic background that involves loss of a factor as critical for the cell as GI. As illustrated here, this loss leads to alterations in hormone signaling that could conceivably impact a number of processes that affect EPs. Two prime candidates identified in this article include enhanced photosynthesis and enhanced ROS scavenging, and we propose that these two constitute a prominent response pathway in chloroplasts. We conclude that our experiments show the utility and power of the suppressor approach for gaining insight into pathways and processes that govern chloroplast biogenesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants in this study were grown on soil at 22°C under continuous illumination (approximately 100 µmol m–2 s–1). Because im is light sensitive, it was germinated for 10 d under low illumination (15 μmol m−2 s−1) prior to transfer to normal light conditions. Mutant crosses in the im background were treated in a similar manner. gi2 and sex1-1 seeds were obtained from the Arabidopsis Biological Resource Center, and spy4 seeds were a generous gift from Neil Olszewski (University of Minnesota, St. Paul). Genotypes of double and triple mutant progeny were identified by PCR using the derived cleaved-amplified polymorphic sequence primers in Supplemental Table S1.
Pigment Analyses
The two top pairs of fully expanded rosette leaves from Col-0, im, imgi2, and gi2 at stages I to III of development were harvested and ground in liquid N2. Pigments were extracted in the dark at 4°C using 19:1 ethanol:H2O (v/v). Total chlorophyll and carotenoid contents were calculated as described (Lichtenthaler, 1987).
Hormone Response Assays
Three-week-old Col-0 and im plants were sprayed with 25 μm BAP (Sigma-Aldrich) once every 3 d for a period of 2 weeks. Leaves that emerged after treatment were used for experimentation. For treatment with GA3, imgi2 plants were sprayed with 100 μm GA3 (dissolved in water) at around 4 weeks after germination for a period of 6 weeks (two times per week). Endogenous GA was inhibited using PAC (Sigma-Aldrich) at a concentration of 35 mg L−1. For plate assays, MS plates were supplemented with 35 mg L−1 PAC, and for soil assays, 2-week-old im plants were watered with PAC for a period of 4 weeks (three times per week).
Light and Transmission Electron Microscopy
Fully expanded rosette leaves from Col-0 and imgi2 plants at the same developmental stage (stages I and III, respectively) were used for light and transmission electron micrograph imaging. Samples were fixed, stained, and examined as described by Horner and Wagner (1980).
Chloroplast Ultrastructure
Chloroplast surface area, volume, and size were calculated from light micrograph images of protoplasts from Col-0 and imgi2 as described previously (Li et al., 2013). Protoplasts were isolated as described by Yoo et al. (2007), and 50 independent protoplasts were examined for each parameter. Chloroplast size refers to maximal length, and chloroplast surface area and volume were calculated assuming an ellipsoidal shape using the Cesaro formula (Ivanova and P’yankov, 2002), where surface area = 4 × π × (a × b2)2/3 and volume = (4/3) × π × (a × b2), where a = chloroplast length/2 and b = chloroplast width/2.
Protein Manipulations
Total cell proteins were isolated from Col-0, im, imgi2, and gi2 leaves by previously described procedures (Yu et al., 2004) using leaf samples similar to those described above for pigment analyses. In brief, leaves were weighed, frozen in liquid N2, and homogenized in 2× SDS sample buffer. The homogenates were incubated at 65°C for 2 h, then centrifuged (14,000 rpm for 10 min), and the supernatants were collected and electrophoresed through 12% SDS polyacrylamide gels on an equal fresh weight basis. Western immunoblot analyses were carried out by established protocols using polyclonal antibodies to the large subunit of Rubisco, the light-harvesting complex protein, the α-subunit of ATP synthase, PSII subunit O, PSI subunit F, PSII reaction center protein, and the Rieske Fe-S center of the cytochrome b6f complex. All of these antibodies have been characterized previously (Yu et al., 2008a). The SuperSignal West Pico chemiluminescence kit (Pierce) was used for signal detection.
Transcript Analysis by Quantitative Reverse Transcription-PCR
Total cell RNAs were isolated from Arabidopsis leaf tissues using the Trizol RNA reagent (Invitrogen), then reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen). The reverse transcription reaction products were diluted 10-fold, and quantitative reverse transcription (qRT)-PCR was then carried out in a total volume of 25 µL using the Maxima SYBR Green qPCR Master Mix (Thermo Scientific). Expression values were normalized to ACTIN, and relative expression values were calculated using the equation 2dCT, where dCT (for cycle threshold) = CT (reference gene) – CT (target gene). Primers used in the expression analyses are listed in Supplemental Table S1.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence measurements were conducted as described (Baerr et al., 2005) on the top two pairs of rosette leaves from Col-0 and imgi2 at the same developmental stage (stages I and III, respectively) using a PAM-2500 chlorophyll fluorometer (Heinz Walz). The leaves were dark adapted (15 min) prior to measurement of steady-state light response curves for EP (the redox state of the first electron acceptor of PSII) and for relative linear electron transport rates.
ROS Staining Assays
Procedures to detect O2− using NBT (Sigma-Aldrich) have been described by Ramel et al. (2009). In brief, leaves of Col-0, im, imgi2, and gi2 were vacuum infiltrated with 3.5 mg mL−1 NBT in 10 mm potassium phosphate buffer containing 10 mm NaN3. After 2 h, the stained leaves were bleached by boiling for 5 min in an ethanol:glycerol:acetic acid solution (3:1:1, v/v/v). The stained leaves were stored in an ethanol:glycerol solution (4:1, v/v).
Procedures to detect H2O2 using DAB (Sigma-Aldrich) have been described by Clarke (2009). In brief, Col-0, im, imgi2, and gi2 leaves were vacuum infiltrated with a freshly prepared DAB solution (1 mg mL−1; pH 3.8), then immersed in the same solution for approximately 8 h. After staining, the leaves were bleached by boiling for 10 min in 19:1 ethanol:H2O (v/v) and stored in an ethanol:glycerol (4:1, v/v) solution.
Starch Staining and Quantification
To qualitatively assess starch contents, Col-0, im, imgi2, and gi2 plants were removed from soil just prior to bolting and bleached by boiling in 50 mL of 80% (v/v) ethanol for approximately 30 min. The plants were then stained for starch by immersion for 5 min in a solution of Lugol’s iodine (1% [w/v] potassium iodide and 0.1% [w/v] iodine). After staining, the plants were destained in distilled, deionized water for approximately 1 h; photographs were taken immediately.
For starch quantification, Col-0, im, imgi2, and gi2 plants were removed form the soil just prior to bolting, then weighed, boiled in 50 mL of 80% (v/v) ethanol, and ground to a slurry using a mortar and pestle. The slurries were centrifuged (8,500 rpm for 10 min), and the pellets were washed twice with 80% ethanol, resuspended in 15 mL of distilled, deionized water, and boiled for 30 min. Total starch content was quantified using a Glc assay kit (R-Biopharm) according to the manufacturer’s instructions.
CO2 Assimilation Measurements
A LI-COR 6400 (Li-Cor) was used to measure CO2 assimilation rates with respect to the internal CO2 concentration. The CO2 assimilation rate-internal CO2 concentration curves were plotted at a photosynthetic photon flux density of 200 μmol m−2 s−1, and the reference and sample infrared gas analyzers were matched automatically before taking each measurement.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g22260, NC_003075.7; At1g22770, NC_003070.9; At3g11540, NC_003074.8; and At1g10760, NC_003070.9.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Glossary
- PTOX
plastoquinol oxidase
- PQ
plastoquinone
- EP
excitation pressure
- SOD
superoxide dismutase
- APX
ascorbate peroxidase
- ROS
reactive oxygen species
- Col-0
Columbia-0
- BAP
6-benzylaminopurine
- MS
Murashige and Skoog
- PAC
paclobutrazol
- H2O2
hydrogen peroxide
- O2−
superoxide
- NBT
nitroblue tetrazolium
- DAB
3,3′-diaminobenzidine
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DEFG02–94ER20147 to S.R.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Albrecht M, Klein A, Hugueney P, Sandmann G, Kuntz M (1995) Molecular cloning and functional expression in E. coli of a novel plant enzyme mediating ζ-carotene desaturation. FEBS Lett 372: 199–202 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Stessman DJ, Spalding MH, Rodermel SR (2007) Alterations in photosynthesis in Arabidopsis lacking IMMUTANS, a chloroplast terminal oxidase. Photosynth Res 91: 11–23 [DOI] [PubMed] [Google Scholar]
- Baerr JN, Thomas JD, Taylor BG, Rodermel SR, Gray GR (2005) Differential photosynthetic compensatory mechanisms exist in the immutans mutant of Arabidopsis thaliana. Physiol Plant 124: 390–402 [Google Scholar]
- Cao S, Jiang S, Zhang R (2006) The role of GIGANTEA gene in mediating the oxidative stress response and in Arabidopsis. Plant Growth Regul 48: 261–270 [Google Scholar]
- Cao S, Song YQ, Su L (2007) Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biol Plant 51: 359–362 [Google Scholar]
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M (1999) Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis: det mutants have an altered response to cytokinins. Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD. (2009) Phenotypic analysis of Arabidopsis mutants: diaminobenzidine stain for hydrogen peroxide. Cold Spring Harb Protoc 6: pdb-prot4981. [DOI] [PubMed] [Google Scholar]
- Cortleven A, Nitschke S, Klaumünzer M, Abdelgawad H, Asard H, Grimm B, Riefler M, Schmülling T (2014) A novel protective function for cytokinin in the light stress response is mediated by the ARABIDOPSIS HISTIDINE KINASE2 and ARABIDOPSIS HISTIDINE KINASE3 receptors. Plant Physiol 164: 1470–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Schreiber U, Heber U (1985) The relationship between the redox state of QA and photosynthesis in leaves at various carbon-dioxide, oxygen and light regimes. Planta 166: 219–226 [DOI] [PubMed] [Google Scholar]
- Eimert K, Wang SM, Lue WI, Chen J (1995) Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishon S, Shani E, Ori N, Weiss D (2011) Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol 190: 609–617 [DOI] [PubMed] [Google Scholar]
- Foudree A, Putarjunan A, Kambakam S, Nolan T, Fussell J, Pogorelko G, Rodermel S (2012) The mechanism of variegation in immutans provides insight into chloroplast biogenesis. Front Plant Sci 3: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A, Liu H, Yu F, Kambakam S, Luan S, Rodermel S (2012) Alternative oxidases (AOX1a and AOX2) can functionally substitute for plastid terminal oxidase in Arabidopsis chloroplasts. Plant Cell 24: 1579–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli L, Masi A, Ripoll DR, Lee MJ, van Wijk KJ (2007) Arabidopsis thaliana deficient in two chloroplast ascorbate peroxidases shows accelerated light-induced necrosis when levels of cellular ascorbate are low. Plant Mol Biol 65: 627–644 [DOI] [PubMed] [Google Scholar]
- Glynn JM, Froehlich JE, Osteryoung KW (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevelding C, Suter-Crazzolara C, von Menges A, Kemper E, Masterson R, Schell J, Reiss B (1996) Characterisation of a new allele of pale cress and its role in greening in Arabidopsis thaliana. Mol Gen Genet 251: 532–541 [DOI] [PubMed] [Google Scholar]
- Holding DR, Springer PS, Coomber SA (2000) The chloroplast and leaf developmental mutant, pale cress, exhibits light-conditional severity and symptoms characteristic of its ABA deficiency. Ann Bot (Lond) 86: 953–962 [Google Scholar]
- Horner HT, Wagner BL (1980) The association of druse crystals with the developing stomium of Capsicum annum (Solanaceae) anthers. Am J Bot 67: 1347–1360 [Google Scholar]
- Hüner NPA, Oquist G, Sarhan F (1998) Energy balance and acclimation to light and cold. Trends Plant Sci 3: 224–230 [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Izawa T (2011) A study of phytohormone biosynthetic gene expression using a circadian clock-related mutant in rice. Plant Signal Behav 6: 1932–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AG, Rosso D, Savitch LV, Stachula P, Rosembert M, Oquist G, Hurry V, Hüner NPA (2012) Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynth Res 113: 191–206 [DOI] [PubMed] [Google Scholar]
- Ivanova LA, P’yankov VI (2002) Structural adaptation of the leaf mesophyll to shading. Russ J Plant Physiol 49: 419–431 [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ (1998) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Smalle J, Van Montagu M, Inzé D (1998a) Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J 14: 759–764 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle J, Van Montagu M, Inzé D (1998b) Effects of sucrose supply on growth and paraquat tolerance of the late-flowering gi-3 mutant. Plant Growth Regul 26: 91–96 [Google Scholar]
- Kusnetsov VV, Oelmüller R, Sarwat M, Porfirova SA, Cherepneva GN, Herrmann RG, Kulaeva ON (1994) Cytokinins, abscisic acid and light affect accumulation of chloroplast proteins in Lupinus luteus cotyledons, without notable effect on steady-state mRNA levels. Planta 194: 318–327 [Google Scholar]
- Lerbs S, Lerbs W, Klyachko NL, Romanko EG, Kulaeva ON, Wollgiehn R, Parthier B (1984) Gene expression in cytokinin- and light-mediated plastogenesis of Cucurbita cotyledons: ribulose-1,5-bisphosphate carboxylase/oxygenase. Planta 162: 289–298 [DOI] [PubMed] [Google Scholar]
- Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10: 1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ren B, Ding L, Shen Q, Peng S, Guo S (2013) Does chloroplast size influence photosynthetic nitrogen use efficiency? PLoS ONE 8: e62036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lichtenthaler HK, Buschmann C (1978) Control of chloroplast development by red light, blue light and phytohormones. InAkoyunoglou G, ed, Chloroplast Development. Elsevier, Amsterdam, pp 801–816 [Google Scholar]
- Liu X, Yu F, Rodermel S (2010) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Matsoukas IG, Massiah AJ, Thomas B (2013) Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ 36: 1802–1811 [DOI] [PubMed] [Google Scholar]
- McCabe MS, Garratt LC, Schepers F, Jordi WJRM, Stoopen GM, Davelaar E, van Rhijn JHA, Power JB, Davey MR (2001) Effects of P(SAG12)-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol 127: 505–516 [PMC free article] [PubMed] [Google Scholar]
- McDonald AE, Ivanov AG, Bode R, Maxwell DP, Rodermel SR, Hüner NPA (2011) Flexibility in photosynthetic electron transport: the physiological role of plastoquinol terminal oxidase (PTOX). Biochim Biophys Acta 1807: 954–967 [DOI] [PubMed] [Google Scholar]
- Meurer J, Grevelding C, Westhoff P, Reiss B (1998) The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol Gen Genet 258: 342–351 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY, Froehlich JE, Osteryoung KW (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y, Kobayashi Y, Shikanai T (2010) Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J 63: 458–468 [DOI] [PubMed] [Google Scholar]
- Paparelli E, Parlanti S, Gonzali S, Novi G, Mariotti L, Ceccarelli N, van Dongen JT, Kölling K, Zeeman SC, Perata P (2013) Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25: 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Parthier B. (1979) The role of phytohormones (cytokinins) in chloroplast development. Biochem Physiol Pflanz 174: 173–214 [Google Scholar]
- Peltier G, Cournac L (2002) Chlororespiration. Annu Rev Plant Biol 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Pinhero RG, Rao MV, Paliyath G, Murr DP, Fletcher RA (1997) Changes in activities of antioxidant enzymes and their relationship to genetic and paclobutrazol-induced chilling tolerance of maize seedlings. Plant Physiol 114: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany M, Koehl J, Heiser I, Elstner E, Barna B (2004) Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to Tobacco necrosis virus and confers tolerance to oxidative stress. Physiol Mol Plant Pathol 65: 39–47 [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. (1994) Mechanisms of resistance to herbicides interacting with photosystem I. InPowles SB, Holtum JAM, eds, Herbicide Resistance in Plants: Biology and Biochemistry. Lewis Publishers, Chelsea, MI, p 61 [Google Scholar]
- Putarjunan A, Liu X, Nolan T, Yu F, Rodermel S (2013) Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth Res 116: 437–453 [DOI] [PubMed] [Google Scholar]
- Ramel F, Sulmon C, Bogard M, Couée I, Gouesbet G (2009) Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei GP. (1962) Supervital mutants of Arabidopsis. Genetics 47: 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei GP. (1963) Somatic instability caused by a cysteine-sensitive gene in Arabidopsis. Science 139: 767–769 [DOI] [PubMed] [Google Scholar]
- Rédei GP. (1967) Biochemical aspects of a genetically determined variegation in Arabidopsis. Genetics 56: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röbbelen G. (1968) Genbedingte Rotlicht-Empfindlichkeit der Chloroplasten differenzierung bei Arabidopsis. Planta 80: 237–254 [Google Scholar]
- Rosso D, Bode R, Li W, Krol M, Saccon D, Wang S, Schillaci LA, Rodermel SR, Maxwell DP, Hüner NP (2009) Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D, Ivanov AG, Fu A, Geisler-Lee J, Hendrickson L, Geisler M, Stewart G, Krol M, Hurry V, Rodermel SR, et al. (2006) IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol 142: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 11698–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M, Gilbert M, Labouré AM, Kuntz M (2007) Dual role of the plastid terminal oxidase in tomato. Plant Physiol 145: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- Slade P. (1966) The fate of paraquat applied to plants. Weed Res 6: 158–167 [Google Scholar]
- Smith WK, Vogelmann TC, DeLucia EH, Bell DT, Shepherd KA (1997) Leaf form and photosynthesis: do leaf structure and orientation interact to regulate internal light and carbon dioxide? BioScience 47: 785–793 [Google Scholar]
- Stoynova EZ, Iliev LK, Georgiev GT (1996) Structural and functional alterations in radish plants induced by the phenylurea cytokinin 4-PU-30. Biol Plant 38: 237–244 [Google Scholar]
- Sun CW, Chen LJ, Lin LC, Li HM (2001) Leaf-specific upregulation of chloroplast translocon genes by a CCT motif-containing protein, CIA 2. Plant Cell 13: 2053–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 126: 1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Salomé PA, McClung CR, Olszewski NE (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Hooykaas P, Lein W, Lerchl J, Kunze G, Sonnewald U, Boldt R (2004) Molecular analysis of “de novo” purine biosynthesis in solanaceous species and in Arabidopsis thaliana. Front Biosci 9: 1803–1816 [DOI] [PubMed] [Google Scholar]
- Wang J, Letham DS, Cornish E, Wei K, Hocart CH, Michael M, Stevenson KR (1997) Studies of cytokinin action and metabolism using tobacco plants expressing either the ipt or the GUS gene controlled by a chalcone synthase promoter ipt and GUS gene expression, cytokinin levels and metabolism. Aust J Plant Physiol 24: 673–683 [Google Scholar]
- Wang TL, Thompson AG, Horgan R (1977) A cytokinin glucoside from the leaves of Phaseolus vulgaris L. Planta 135: 285–288 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CM, Jiang CZ, Meehan LJ, Voytas DF, Rodermel SR (1994) Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J 6: 161–175 [DOI] [PubMed] [Google Scholar]
- Woo NS, Gordon MJ, Graham SR, Rossel JB, Badger MR, Pogson BJ (2011) A mutation in the purine biosynthetic enzyme ATASE2 impacts high light signalling and acclimation response in green and chlorotic sectors of Arabidopsis leaves. Funct Plant Biol 38: 401–419 [DOI] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Nakamura M, Yoneyama T, Nishida I (2005) Starch-related α-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol 138: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya E, Vershilovskaya I, Poers Y, Alawady AE, Averina N, Grimm B (2006) Cytokinin effects on tetrapyrrole biosynthesis and photosynthetic activity in barley seedlings. Planta 224: 700–709 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 30: 350–365 [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S (2008a) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008b) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. (2001) The Arabidopsisb sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta-Mancera HA, Thomas BJ, Thomas H, Scott IM (1999) Regreening of senescent Nicotiana leaves. II. Redifferentiation of plastids. J Exp Bot 50: 1683–1689 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.