The properties of a key enzyme of starch synthesis permit adjustment of this process under different day lengths.
Abstract
Arabidopsis (Arabidopsis thaliana) leaves synthesize starch faster in short days than in long days, but the mechanism that adjusts the rate of starch synthesis to daylength is unknown. To understand this mechanism, we first investigated whether adjustment occurs in mutants lacking components of the circadian clock or clock output pathways. Most mutants adjusted starch synthesis to daylength, but adjustment was compromised in plants lacking the GIGANTEA or FLAVIN-BINDING, KELCH REPEAT, F BOX1 components of the photoperiod-signaling pathway involved in flowering. We then examined whether the properties of the starch synthesis enzyme adenosine 5′-diphosphate-glucose pyrophosphorylase (AGPase) are important for adjustment of starch synthesis to daylength. Modulation of AGPase activity is known to bring about short-term adjustments of photosynthate partitioning between starch and sucrose (Suc) synthesis. We found that adjustment of starch synthesis to daylength was compromised in plants expressing a deregulated bacterial AGPase in place of the endogenous AGPase and in plants containing mutant forms of the endogenous AGPase with altered allosteric regulatory properties. We suggest that the rate of starch synthesis is in part determined by growth rate at the end of the preceding night. If growth at night is low, as in short days, there is a delay before growth recovers during the next day, leading to accumulation of Suc and stimulation of starch synthesis via activation of AGPase. If growth at night is fast, photosynthate is used for growth at the start of the day, Suc does not accumulate, and starch synthesis is not up-regulated.
Many plants store starch as a product of photosynthesis in leaves during day then degrade it to provide sugar for continued metabolism and growth at night. It is well established that rates of starch synthesis adjust to changes in daylength. Adjustments occur in plants acclimated to different daylengths: the shorter the day, the greater the proportion of photosynthate allocated to starch and hence available for metabolism at night (Chatterton and Silvius, 1979; Baysdorfer and Robinson, 1985; Hewitt et al., 1985; Lorenzen and Ewing, 1992; Lu et al., 2005; Gibon et al., 2009; Sulpice et al., 2014). In Arabidopsis (Arabidopsis thaliana), starch synthesis is also accelerated in the light period following a single, unexpected extension of the night (Gibon et al., 2004a). The mechanisms that adjust the rate of starch synthesis to daylength are not known. Neither the nature of the signals from daylength nor the manner in which they influence starch synthesis are understood, and it is not known whether the acclimation of the rate of starch synthesis to different daylengths involves the same mechanisms as its immediate adjustment following an unexpected extension of the night.
By contrast, the short-term control of starch synthesis is well understood. Partitioning of photosynthate into starch in the chloroplast is largely determined by the balance between the rate of photosynthetic carbon assimilation and the rate of Suc synthesis. Assimilate for Suc synthesis is exported from the chloroplast as triose phosphate, in exchange for inorganic phosphate (Pi) released during its cytosolic conversion to Suc. The rate of Suc synthesis is regulated via action of Fru 2,6-bisphosphate on the cytosolic Fru 1,6-bisphosphatase and allosteric and posttranslational regulation of Suc phosphate synthase (Stitt and Quick, 1989; Stitt et al., 2010). If the rate of Suc synthesis falls, for example in response to feedback from decreased utilization by sink organs, the amount of Pi available for exchange with triose phosphate is decreased, leading to retention of a larger proportion of the triose phosphate in the chloroplast and its use for starch synthesis. Conversely, if the rate of photosynthetic carbon assimilation falls, for example as a result of shading or stomatal closure, the concentration of triose phosphate in the chloroplast tends to fall, and the rate of starch synthesis decreases. Thus, a change in either the rate of photosynthetic carbon assimilation or the rate of Suc synthesis can rapidly bring about a change in the amount of photosynthate allocated to starch synthesis.
These short-term adjustments of starch synthesis are brought about by modulation of the activity of adenosine 5′-diphosphate-glucose pyrophosphorylase (AGPase). This enzyme catalyzes the first committed step in starch biosynthesis: the synthesis of ADP-Glc and pyrophosphate from ATP and Glc 1-P. It is composed of large and small subunits and is subject to several levels of posttranslational regulation. The allosteric properties of the enzyme potentially allow it to regulate flux into starch in response to the rate of cytosolic Suc synthesis. High levels of triose phosphate in the chloroplast result in high 3-phosphoglyceraldehyde (3-PGA) levels, which activate the enzyme. Low levels of triose phosphate result in low 3-PGA and high Pi levels, which inhibit the enzyme (Heldt et al., 1977; Stitt et al., 1987). AGPase is also activated in the light and by sugars through reduction via NADP-thioredoxin reductase C and the thioredoxin Trx f1, which causes the loss of a disulfide bond between the two small subunits. Redox activation greatly increases the sensitivity of the enzyme to allosteric regulation by 3-PGA and Pi (Preiss, 1988; Kavakli et al., 2002; Tiessen et al., 2002; Hendriks et al., 2003; Michalska et al., 2009; Thormählen et al., 2013). Both large and small subunits are reported to be phosphorylated (Reiland et al., 2009, 2011; Nakagami et al., 2010; Umezawa et al., 2013; Wu et al., 2013), but the functional significance of these modifications is not known. Flux control analysis using Arabidopsis mutants with reduced activities of starch biosynthetic enzymes shows that AGPase can exert strong control over the rate of starch synthesis during photosynthesis, although the degree of control varies with environmental conditions (Neuhaus and Stitt, 1990; Sun et al., 1999; Hädrich et al., 2011). These mechanisms enable coordinated control of the use of Calvin-Benson cycle metabolites for starch and Suc synthesis, preventing the depletion of cycle intermediates and the inhibition of photosynthesis.
It is not clear whether the mechanism that mediates short-term regulation of starch synthesis forms part of the unknown mechanism(s) by which starch synthesis is adjusted to daylength. One possibility is that a daylength-dependent signal sets an underlying default level of partitioning between Suc and starch or a target quantity of starch to be accumulated by the anticipated end of the day and that the short-term control described above operates on top of this default setting. We showed recently that the rate of starch degradation in leaves at night is controlled in part by the circadian clock (Graf et al., 2010; Scialdone et al., 2013): perhaps an analogous, clock-based mechanism ensures that sufficient starch is synthesized during the day to meet the carbon demands of the plant during the night. Daylength-dependent signals might affect partitioning through modulation of the abundance or properties of key enzymes of starch and/or Suc synthesis. Consistent with this idea, plants grown in short days have higher maximum catalytic activities of AGPase relative to Suc phosphate synthase than plants grown in long days (Gibon et al., 2009; Sulpice et al., 2014), due, in particular, to a rise of AGPase activity and protein before dawn in short but not in long days (Gibon et al., 2004b). Modulation of the redox sensitivity of AGPase can also affect partitioning: complementation of an AGPase-deficient Arabidopsis mutant with a mutagenized AGPase that is redox insensitive and mimics a constitutively redox-activated enzyme resulted in faster rates of starch accumulation in short days and low irradiance and the maintenance of slightly higher starch levels throughout the diel cycle in long day or high light regimes. However, these plants were still able to adjust the rate of starch synthesis in response to changes in daylength, indicating that redox modulation of activity is not essential for the response to photoperiod (Hädrich et al., 2012).
A second possibility is that the general relationship between starch synthesis and daylength is determined indirectly by daylength-dependent variation in the demand for photosynthate for growth. We showed recently that the temporal pattern of growth varies with daylength (Sulpice et al., 2014): our data are consistent with growth rates at night and at the start of the day being high in long days but lower in short days. Differences in growth rate impose different demands for Suc, hence different rates of Suc synthesis at the start of the day. Such differences are expected to influence the rate of starch synthesis via the AGPase-dependent mechanism described above.
To shed further light on mechanisms that gear the rate of starch synthesis to daylength in Arabidopsis, we first examined the importance for this relationship of components of the circadian clock and related photoperiod-signaling pathways. We then examined whether the properties of AGPase that are crucial for adjustment of starch synthesis over a single photoperiod are also important for the relationship between starch synthesis and the length of the photoperiod. This was achieved by replacing the endogenous Arabidopsis AGPase with a deregulated bacterial AGPase and examining the impact upon the response of starch synthesis to daylength and to an unexpected extension of the night.
RESULTS
Adjustment of Starch Synthesis to Daylength Requires Some Components of the Circadian Clock and Photoperiod-Signaling Pathways
We examined whether the adjustment of starch synthesis to daylength required either central components of the circadian clock or components of related photoperiod-signaling pathways. The rate of starch synthesis in the first 6 h of the light period was measured in plants grown in either 6-h light/18-h dark (6:18 plants) or 12-h light/12-h dark (12:12 plants). Wild-type plants were compared with plants lacking components of the circadian clock or related photoperiod-signaling pathways. The mutant early flowering3 (elf3) and the double mutant circadian clock associated1 × late elongated hypocotyl (cca1 lhy) lack core components of the circadian clock. ELF3 is a component of the evening complex of the clock (Dixon et al., 2011; Nusinow et al., 2011; Herrero et al., 2012). elf3 mutants lose circadian rhythmicity in continuous light and are defective in the circadian control of hypocotyl and root elongation (Covington et al., 2001; Nusinow et al., 2011; Yazdanbakhsh et al., 2011). CCA1 and LHY are partially redundant transcription factors that are part of the central clock oscillator; the double mutant has a free-running period of about 17 h (Alabadí et al., 2001; Mizoguchi et al., 2002). The gigantea (gi) mutant has a complex phenotype that reflects the multiple functions of the GI protein in both inputs to and outputs from the clock (Mizoguchi et al., 2005; Martin-Tryon et al., 2007; Sawa et al., 2007; Sawa and Kay, 2011). Among other functions, the protein is part of the photoperiod-signaling pathway that links the circadian clock to the triggering of flowering. The flavin-binding, kelch repeat, f box1 (fkf1) and quadruple cycling DNA-binding one finger (dof) factor (cdf) mutants lack proteins that interact with GI in the flowering-time pathway (an F-box protein and four mutually redundant DOF transcription factors, respectively), and the co mutant lacks the CONSTANS (CO) protein that regulates expression of FLOWERING LOCUS T, the signal that moves from leaves to the shoot apex to trigger flowering (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009; Song et al., 2012). GI and FKF form a complex in a photoperiod-dependent manner, which targets CDF proteins for degradation. CDF, GI, and FKF1 also bind directly to the CO promoter during the day. These mechanisms are key determinants of CO protein abundance and hence the timing of flowering in relation to daylength (Imaizumi et al., 2005; Sawa et al., 2007; Song et al., 2013). Finally, the phytochrome-interacting factor4 (pif4) mutant lacks a basic helix-loop-helix factor that acts downstream of light- and clock-signaling pathways to influence various aspects of development and growth (Huq and Quail, 2002; Nozue et al., 2007; de Lucas et al., 2008; Leivar and Quail, 2011).
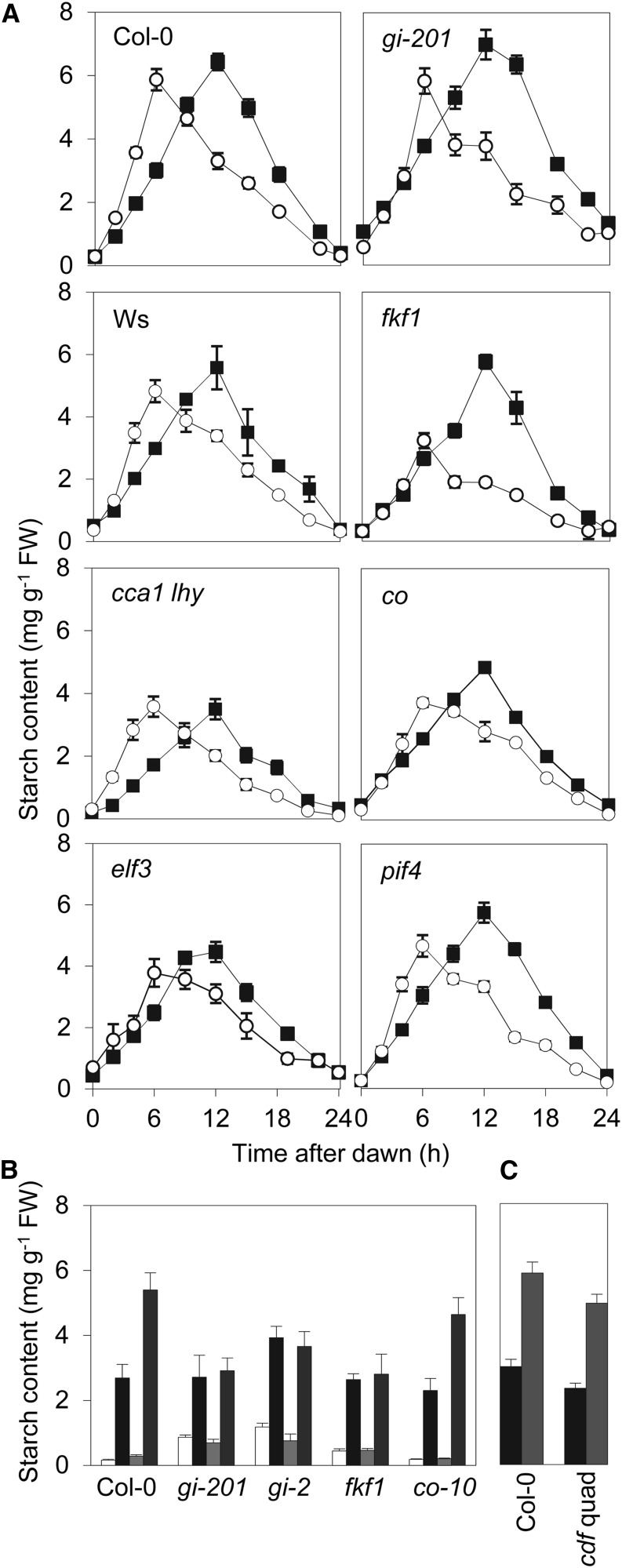
As expected from previous observations (Gibon et al., 2004a; Lu et al., 2005; Sulpice et al., 2014), the rate of starch synthesis in wild-type plants (ecotypes Columbia [Col-0] and Wassilewskija [Ws]) in 6:18 conditions was higher than in 12:12 conditions (Fig. 1A; mean rates and statistical significance of differences between photoperiods are given in Supplemental Table S1). This was also true for the clock mutants cca1 lhy and elf3 and for the co (Fig. 1, A and B; Supplemental Table S1), pif4 (Fig. 1A), and quadruple cdf (Fig. 1C) mutants lacking components of light- and photoperiod-signaling pathways. Although cca1 lhy and elf3 mutants accumulated less starch than the respective wild-type plants, a difference was still found between short and long photoperiods in each of these mutants. However, the fkf1 mutant had the same rate of starch synthesis in the two different daylengths (Fig. 1, A and B; Supplemental Table S1). Two gi mutants (carrying either the gi-2 or gi-201 alleles) showed a variable response. In four out of six experiments, these mutants behaved like the fkf1 mutant, in that the rate of starch synthesis was the same in 6:18 and 12:12 conditions (as in Fig. 1). In the remaining experiments, the rate of starch synthesis was higher in 6:18 than in 12:12 conditions. The reasons for this variation between experiments were not apparent.
Figure 1.
Response of starch synthesis to short days in clock and photoperiod mutants. For all experiments, values are means ± se of measurements made on six plants. A, Rosette starch content over 24 h in wild-type (Col-0 and Ws) and mutant Arabidopsis plants grown in 12:12 (white circle) or 6:18 (black square) conditions. The cca1 lhy double mutant is in the Ws background; other mutants are in the Col-0 background. See Supplemental Table S1 for calculated rates of starch synthesis. B, Starch contents at the end of the night (white and light gray) and after 6 h of the light period (black and dark gray) in wild-type (Col-0) and mutant plants grown in 12:12 (white and black) and 6:18 (light gray and dark gray) conditions. C, Starch contents after 6-h light in wild-type (Col-0) and cdf quadruple mutant plants grown in 12:12 (black) and 6:18 (gray) conditions. FW, Fresh weight.
Expression of AGPase Subunits Is Not Strongly Influenced by Daylength
To discover whether AGPase may be the target for mechanisms that adjust the rate of starch synthesis according to photoperiod, we first examined whether patterns of expression of its subunits differ between photoperiods. The plant enzyme is a heterotetramer of two large and two small subunits. Arabidopsis has one functional gene encoding the small subunit (AGPASE SMALL SUBUNIT1 [APS1], At5g48300) and four genes encoding large subunits (AGPASE LARGE SUBUNIT1 [APL1], At5g19220; APL2, At1g27680; APL3, At4g39210; and APL4, At2g21590). The large subunit genes are strongly differentially expressed, both developmentally and in response to environmental stimuli. Different large subunits are believed to confer different regulatory properties on the enzyme (Sokolov et al., 1998; Crevillén et al., 2003; 2005). Thus, differences in the relative levels of expression of the large subunits can potentially give rise to enzymes with different properties.
There was little difference between 6:18 and 12:12 conditions in the levels of transcripts for the AGPase subunits. In both conditions, transcripts for APS1 and APL1 were most abundant. Transcripts for APL2, APL3, and APL4 were much less abundant (Supplemental Fig. S1). Consistent with this result, we showed previously that APL1 accounted for 85% of peptides derived from large subunits in AGPase purified from wild-type rosettes (Hädrich et al., 2012). To check whether the subunit composition of the enzyme is affected by daylength, we used an antiserum to APS1 to immunoprecipitate proteins from crude extracts of leaves grown in 6:18 or 12:12 conditions. In two independent experiments, APS1 was the most abundant immunoprecipitated protein, as expected, and APL1 was also present in the immunoprecipitate (Supplemental Fig. S2). No other AGPase subunits were detected by immunological tests. Taken together, these results show that leaf AGPase consists largely of APS1 and APL1 under both 6:18 and 12:12 conditions.
A Deregulated Form of AGPase Complements an AGPase-Deficient Mutant But Fails to Restore Daylength-Dependent Control of Starch Synthesis
To discover whether the regulatory properties of AGPase activity are important for the response of starch synthesis to photoperiod, we replaced the Arabidopsis enzyme with a modified form of AGPase from Escherichia coli that lacks regulatory properties. The native E. coli enzyme is a monomeric protein that is not subject to redox activation and is activated by Fru 1,6-bisphosphate and inhibited by AMP and Pi. The modified form of the enzyme used in our experiments, Glycogen C Triple Mutant (GlgC-TM), contains three amino acid changes that together greatly reduce the sensitivity of its activity to allosteric regulation by these metabolites (Preiss and Romeo, 1994; Sakulsingharoj et al., 2004). The E. coli enzyme also lacks the Cys residues that are involved in the posttranslational redox regulation of higher plant AGPase.
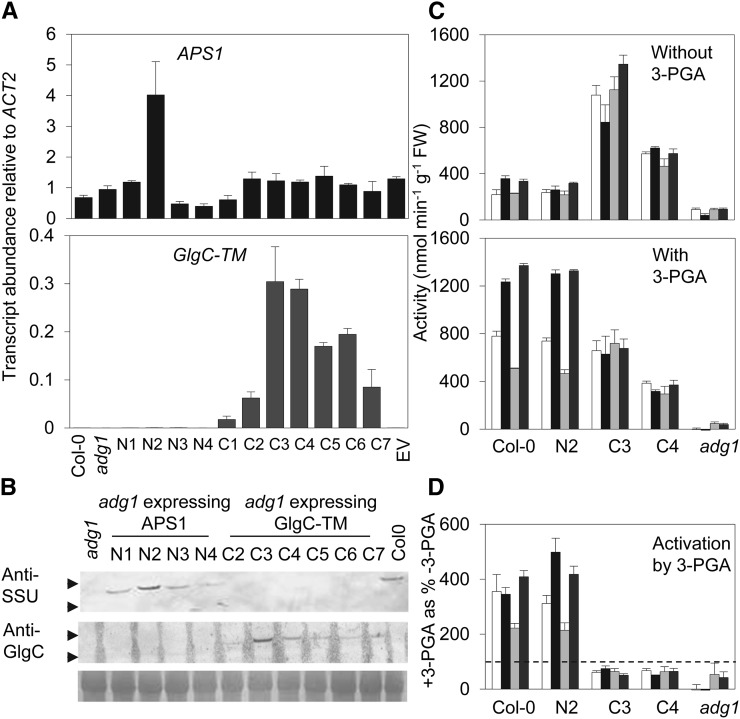
A complementary DNA (cDNA) encoding GlgC-TM was introduced into the AGPase-deficient adpglucose pyrophosphorylase1-1 (adg1-1) mutant under the control of the native Arabidopsis APS1 promoter and 5′and 3′ untranslated regions (UTRs). The protein was directed to the chloroplast by inclusion of the N-terminal chloroplast transit peptide from APS1, in frame with the GlgC-TM coding sequence. The adg1-1 mutant carries a mutation in the gene encoding APS1. Although APS1 transcript is still present, there is no detectable APS1 protein, very little AGPase activity or starch in leaves, and no accumulation of AGPase large subunits (Lin et al., 1988; Wang et al., 1998; Hädrich et al., 2012). As controls, adg1-1 was also transformed with the empty vector and with a cDNA encoding the native APS1, with the same promoter and UTR sequences. Immunoblotting with specific antisera confirmed the presence of APS1 protein in lines carrying the APS1 construct and of GlgC-TM protein in lines carrying the GlgC-TM construct. For both transgenes, there was a positive correlation across transgenic lines between levels of transcript and levels of the encoded proteins (Fig. 2, A and B). AGPase activity was also restored in both sorts of lines (Fig. 2C).
Figure 2.
AGPase transcript and protein levels and activities in adg1 transgenic plants expressing native APS1 or GlgC-TM. A, Transcript abundance of APS1 (top) and GlgC-TM (bottom) in wild-type (Col-0) plants, adg1 mutants, and transgenic lines of adg1 transformed with either APS1 (lines N1–N4) or GlgC-TM (lines C1–C7) on the APS1 promoter or the empty vector (EV). Measurements were made by qPCR on RNA from plants grown in 12:12 conditions and harvested after 6 h of light and were relative to ACTIN2 (ACT2). Values are means ± se of measurements made on four plants. B, Immunoblots showing APS1 (top) and GlgC-TM (middle) proteins in extracts of leaves grown and harvested as in A. Each lane contains proteins from 1.65 mg fresh weight (FW) of leaf tissue. For APS1, the blot was probed with an antiserum against the small subunit (SSU) of potato (Solanum tuberosum) AGPase (Tiessen et al., 2002). Note that APS1 protein is present in Col-0 plants and transgenic lines of adg1 transformed with APS1 (N1–N4) and absent from adg1 and lines expressing GlgC-TM (C2–C7). GlgC-TM protein is present only in transgenic lines of adg1 transformed with GlgC-TM. The positions of 50-kD (top arrow) and 40-kD (bottom arrow) molecular markers are indicated: The expected molecular masses of mature APS1 and GlgC-TM proteins are 49 and 48 kD, respectively. The bottom section shows, as a loading control, a blot of the same extracts stained with Ponceau S to reveal the Rubisco large subunit band. C, Activities of AGPase in leaf extracts, measured in the absence (top) or presence (bottom) of 5 mm 3-PGA in Col-0 plants, adg1 mutants, and adg1 expressing either APS1 (line N2) or GlgC-TM (lines C3 and C4) on the APS1 promoter. Extracts were of plants grown in 12:12 conditions and harvested at ZT3 (white) or ZT15 (black) or grown in 6:18 conditions and harvested at ZT3 (light gray) or ZT9 (dark gray). Values are means ± se of measurements made on three plants. D, Activation of AGPase by 3-PGA, calculated as (+3-PGA/–3-PGA) × 100, from data in C. The dotted line shows activity in the absence of 3-PGA (100%).
APS1 and GlgC-TM lines with high levels of expression of the respective transgenes were selected for more detailed characterization. When assayed in the absence of 3-PGA, AGPase activity in APS1 line N2 was similar to that of wild-type plants under both 12:12 and 6:18 growth conditions, whereas activity in GlgC-TM lines C3 and C4 was higher than that of wild-type plants under both photoperiods (P < 0.05, ANOVA; Fig. 2C). Addition of 3-PGA led to a 2- to 4-fold stimulation of activity in extracts of both wild-type and APS1 transgenic leaves (Fig. 2, C and D). As observed previously (Gibon et al., 2004b), activity at the end of the night tended to be higher than activity at the end of the day. By contrast, and as expected from the lack of regulatory properties of the GlgC-TM protein, addition of 3-PGA to assays of extracts of GlgC-TM leaves did not stimulate AGPase activity, in fact activity was somewhat lower with than without 3-PGA. This apparent inhibition is likely to reflect a minor interference of 3-PGA in the enzymatic cycling assay, rather than a direct effect on the bacterial AGPase. In the presence of 3-PGA, activity in the C3 line was comparable with that in wild-type and APS1 lines at the end of the day, and activity in the C4 line was about 50% lower. Activity in C3 and C4 lines was essentially the same at the end of the day and the end of the night (Fig. 2, C and D). When assays were performed in the presence of 3-PGA to allow full activity of the native enzyme, activity in GlcC-TM lines was similar to that in wild-type plants at the end of the day and lower at the end of the night.
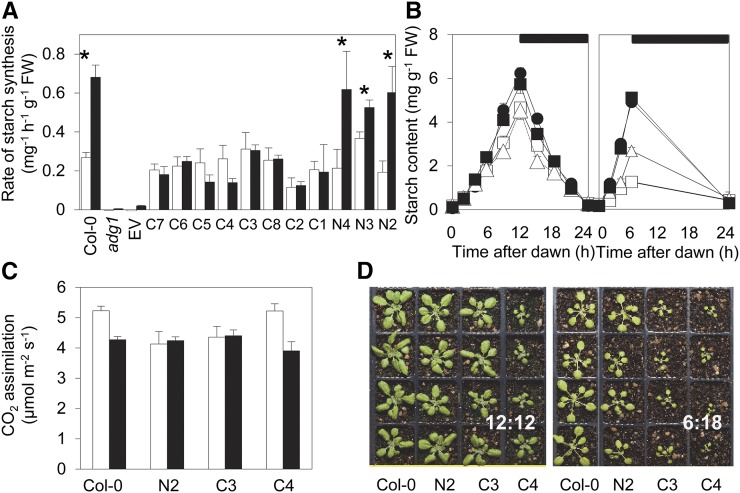
Expression of either APS1 or GlgC-TM in adg1-1 plants restored starch synthesis in leaves. In most GlgC-TM and APS1 lines, the rate of starch synthesis was comparable with the wild type in 12:12 conditions (Fig. 3A), and as a result, starch contents at the end of the day were close to those of wild-type plants (Fig. 3B). In independently grown batches of plants, end-of-day starch contents in several GlgC-TM lines were 60% to 100% of wild-type values. Transformation with the empty vector did not restore starch synthesis. The rates and patterns of starch accumulation and loss over 24 h in 12:12 conditions were similar in wild-type, APS1, and GlgC-TM plants (Fig. 3, A and B; Supplemental Fig. S3A; Supplemental Table S1). As expected, both wild-type and APS1 plants had higher rates of starch synthesis in 6:18 than in 12:12 conditions. These results are consistent with those of Hädrich et al. (2012), who found that both wild-type and APS1 plants had higher rates of starch synthesis in 8:16 than in 12:12 conditions. Importantly, starch synthesis in GlgC-TM plants was not accelerated in 6:18 conditions: the rates were not significantly different from those observed in 12:12 conditions (Fig. 3, A and B; Supplemental Fig. S3A; Supplemental Table S1). The difference in response of rates of starch synthesis to short days was not due to different responses of the rate of photosynthesis: rates of carbon assimilation were relatively unaffected by daylength and were not significantly different in wild-type and GlgC-TM plants in 12:12 and 6:18 conditions (Fig. 3C).
Figure 3.
Complementation of the adg1 phenotype by expression of GlgC-TM. A, Rates of starch synthesis in 12:12 (white) and 6:18 (black) conditions in wild-type (Col-0) plants, adg1 mutants, and transgenic lines of adg1 transformed with either APS1 (lines N2–N4) or GlgC-TM (lines C1–C8) on the APS1 promoter or the empty vector (EV). For each line and condition, measurements of starch contents were made on six rosettes at the beginning and end of the light period, and the rate of synthesis (± se) was calculated by linear regression. Asterisks indicate lines where rates of starch synthesis were statistically significantly different between the two daylengths (P < 0.05, multiple linear regression). B, Starch content over 24 h in plants grown in 12:12 (left, 21-d-old plants) or 6:18 (right, 28-d-old plants) conditions. Black circle indicates the wild type (Col-0), black square indicates APS1 control line N2, white triangle indicates GlgC-TM line C3, and white square indicates GlgC-TM line C4. Values are means ± se of measurements made on six plants. See Supplemental Table S1 for calculated rates of starch synthesis. C, Rates of CO2 assimilation. Measurements were made between 1 and 4 h into the photoperiod at 380 μL L−1 CO2, 210 μmol quanta m–2 s–1 light intensity, and 20°C. White bars indicate 12:12 conditions. Black bars indicate 6:18 conditions. Values are means ± se of measurements on five plants. There were no significant differences in the rate of CO2 assimilation between either transgenic line expressing GlgC-TM AGPase and the wild type or the APS1 control line (N2) in either photoperiod (Student’s t test, P > 0.05). The rate of assimilation was slightly but significantly lower in the N2 line relative to the wild type in the 12:12 photoperiod (P = 0.037) but not in 6:18. D, Plants grown in 12:12 (20 d old, left) or 6:18 (27 d old, right) conditions. Plants are the wild type (Col-0), APS1 control line N2, and GlgC-TM lines C3 and C4. FW, Fresh weight.
Growth rates of GlgC-TM plants were more affected by growth in short days than those of either wild-type or APS1 plants (Fig. 3D). For wild-type and APS1 control plants, the maximum relative rates of rosette expansion were comparable in 12:12 and 6:18 conditions (achieved in the 13- to 16-d time interval in 12:12 conditions and the 20- to 23-d time interval in 6:18 conditions). However, for lines C3 and C4, the relative rates of rosette expansion in these time intervals were 44% and 38% lower, respectively, in 6:18 than in 12:12 conditions (Supplemental Fig. S4B).
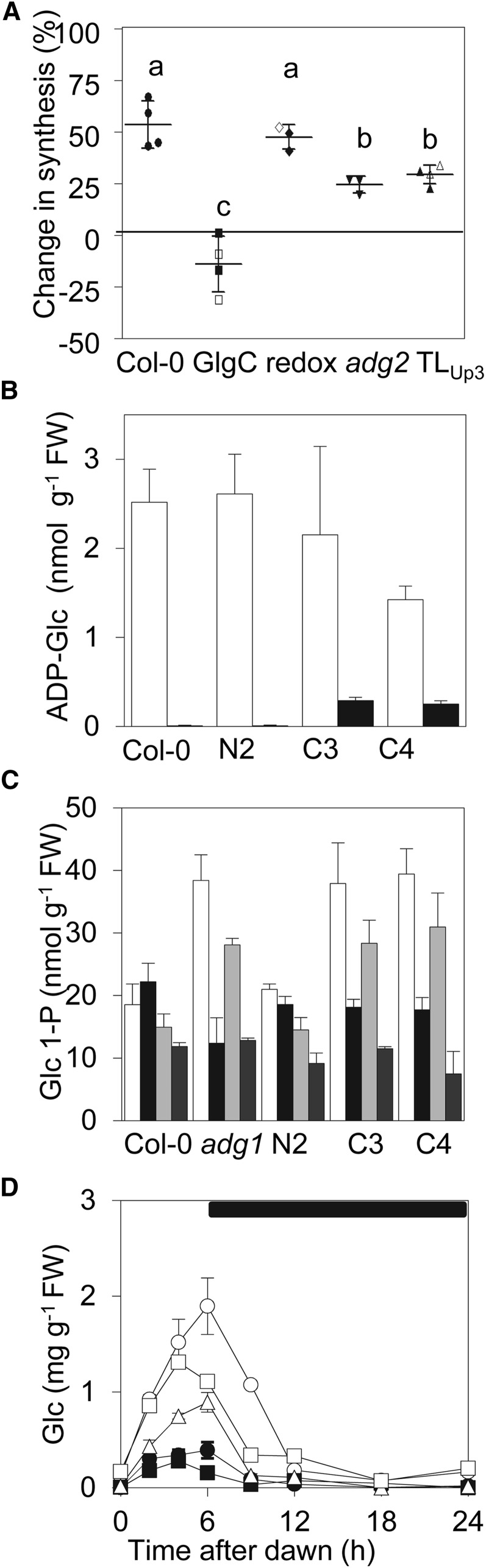
We checked whether GlgC-TM plants had also lost the capacity to accelerate the rate of starch synthesis following a single, unexpected extension of the night, in other words, whether they were deficient in acute as well as acclimatory responses to decreased daylength (Gibon et al., 2004a; Smith and Stitt, 2007). In four independent experiments, the rate of starch synthesis in wild-type plants following an unexpected 4-h extension of the night was faster than following a normal 12-h night. However, starch synthesis in the GlgC-TM line C3 was unaltered or lower following an unexpected extension of the night (Fig. 4A).
Figure 4.
Effects of an extended night on the rate of starch synthesis, and differences in primary metabolites between wild-type and GlgC-TM plants. A, Effect on the rate of starch synthesis of a single extended night. Rates were estimated from measurements (means of values from six plants) at the beginning and end of the day in plants in 12:12 conditions and plants grown in these conditions but subjected to a 4-h extension of the night prior to the day of measurement. At least three separate experiments were performed for each type of plant (independent symbols) with six biological replicates in each. Plants were the wild type (Col-0), GlgC-TM transgenic lines (GlgC), transgenic lines expressing a constitutively redox activated form of AGPase (redox), the adg2 mutant, and transgenic lines expressing a form of AGPase with altered sensitivity to allosteric modification (TLUp3). For GlgC-TM, constitutively redox activated, and TLUp3 plants, two independent lines were used (white and black symbols). Values are the percentage change in the rate of starch synthesis between the end of a normal night and the end of an extended night. For each genotype, the central bar is the mean and the top and bottom bars the sd of the means across all experiments. Means indicated with a different letter (a–c) are statistically significantly different (Newman-Keuls multiple comparison test, P < 0.05). The range of rates of starch synthesis (mg h–1 g–1 fresh weight [FW]) across all experiments following a normal night and an extended night were: Col-0, 0.51 to 0.81 and 0.74 to 1.29; GlgC-TM, 0.27 to 0.43 and 0.20 to 0.35; redox, 0.55 to 0.91 and 0.83 to 1.28; adg2, 0.59 to 0.64 and 0.69 to 0.74; and TLUp3, 0.88 to 1.08 and 1.07 to 1.42. B, ADP-Glc contents of wild-type (Col-0) plants, APS1 control line N2, and GlgC-TM lines C3 and C4. Plants were grown in 12:12 conditions and harvested at 3 h (white) and 15 h (black) after dawn. Values are means ± sd of measurements on six plants. C, Glc 1-P contents of plants as in C grown in 12:12 conditions and harvested at ZT3 (white) or ZT15 (black) or grown in 6:18 conditions and harvested at ZT3 (light gray) or ZT9 (dark gray). Values are means ± sd of measurements on five plants. D, Glc content over 24 h of plants grown in 6:18 conditions. Black circle indicates the wild type (Col-0), black rectangle indicates APS1 control line N2, white triangle indicates GlgC-TM line C3, white rectangle indicates GlgC-TM line C4, and white circle indicates adg1 mutant. The Glc content of transgenic lines C3 and C4 at 6 h after dawn was significantly higher than that of the wild type and the N2 control line but significantly lower than in the adg1 mutant in both 12:12 and 6:18 conditions (in 12:12: the wild type = 0.089 ± 0.03, N2 = 0.12 ± 0.04, C3 = 0.26 ± 0.03 [P < 0.05 versus the wild type or N2, Student’s t test], C4 = 0.44 ± 0.09 [P < 0.01], and adg1-1 = 1.30 ± 0.05 [P < 0.01 versus C3 or C4]; in 6:18: the wild type = 0.39 ± 0.08, N2 = 0.16 ± 0.02, C3 = 0.89 ± 0.10 [P < 0.01 versus the wild type or N2], C4 = 1.11 ± 0.05 [P < 0.01], and adg1-1 = 1.90 ± 0.29 [P < 0.05 versus C3 or C4]). Values are means ± se of measurements on six plants. Where not visible, error bars are smaller than symbols.
Expression of a Deregulated AGPase Perturbs ADP-Glc, Sugar-Phosphate, and Sugar Metabolism
To examine the metabolic consequences of expression of GlgC-TM, we compared levels of a range of primary metabolites in adg1-1, GlgC-TM, wild-type, and control APS1 plants in 12:12 and 6:18 conditions. Expression of native APS1 largely eliminated the metabolic differences between adg1-1 and wild-type plants under both 12:12 and 6:18 conditions, but in GlgC-TM plants, levels of several primary metabolites were different from those in wild-type plants. Levels of the AGPase product ADP-Glc in GlgC-TM plants were different from those in either wild-type or adg1-1 mutants. In wild-type plants, ADP-Glc levels were high during the day (at Zeitgeber Time 3 [ZT3]) and essentially undetectable at night (3 h after the onset of darkness) in 12:12 and 6:18 conditions. As reported previously (Ragel et al., 2013), ADP-Glc was almost undetectable in adg1-1 plants under all conditions (Fig. 4B; Supplemental Fig. S3B). Levels in GlgC-TM plants tended to be lower during the day (at ZT3) than those in wild-type plants. In a first batch of plants grown in 12:12 conditions, the level was not statistically significantly different in GlgC-TM line C3 and wild-type plants but was 50% lower in line C4 than in wild-type plants (Fig. 4B). In a second batch, the level in lines C3 and C4 was only about 35% or less of that in wild-type plants in both 12:12 and 6:18 conditions (Supplemental Fig. S3B). In both batches and both photoperiods, plants of GlgC-TM lines contained much more ADP-Glc at night than wild-type plants (Fig. 4B; Supplemental Fig. S3B).
Levels of Glc 6-P, Fru 6-P, and the AGPase substrate Glc 1-P were about 2-fold higher in GlgC-TM lines than in wild-type plants and similar to those in the adg1-1 mutant during the day in both 12:12 and 6:18 conditions (Fig. 4C; Supplemental Fig. S3B; Supplemental Table S2). For Glc, differences between GlgC-TM and wild-type plants were greater in 6:18 than in 12:12 conditions. Levels during the light period were much higher in lines C3 and C4 than in wild-type plants in 6:18 conditions (Fig. 4D; Supplemental Fig. S3C). Suc and Fru followed the same trend, but differences between GlgC-TM and wild-type plants were less marked than for Glc (Supplemental Fig. S3C). At night, there were no marked differences between sugar levels in the various lines. Levels of a range of other primary metabolites were also significantly different between GlgC-TM plants and wild-type plants (Suc 6-P, trehalose 6-P, glycerol 3-P, pyruvate, 2-oxoglutarate, succinate, malate, and fumarate; P < 0.05, ANOVA), but these differences were generally much less than those between adg1-1 and wild-type plants (Supplemental Table S2).
We examined whether the phenotype of GlgC-TM plants may be influenced by unexpected pleiotropic effects of the transgene on other aspects of the capacity for primary metabolism. To check for such effects, we measured activities of other enzymes of photosynthetic carbon assimilation, glycolysis, and nitrogen assimilation. The activity of UDPglucose pyrophosphorylase was 17% to 30% higher in GlgC-TM lines than in wild-type plants at ZT3 and 30% to 50% higher at ZT15. Activities of all of the other enzymes examined (phosphoglucomutase, phosphoglucoisomerase, Rubisco, glucokinase, fructokinase, triose phosphate isomerase, transketolase, Suc phosphate synthase, and nitrate reductase) were very similar in wild-type plants and GlgC-TM lines (Supplemental Table S3).
The Allosteric Properties of AGPase Are Important for the Adjustment of Starch Synthesis to Daylength
To discover which regulatory properties of AGPase are required for the adjustment of the rate of starch synthesis to different photoperiods, we considered two properties believed to be important for control of the rate of starch synthesis over a single light period: redox activation and allosteric regulation by 3-PGA and Pi. To test the importance of redox activation, we made use of adg1-1 plants complemented with a mutated form of APS1 in which formation of the disulfide bond between small subunits is prevented (expressing a pAPS1::APS1C81S construct; Hädrich et al., 2012). AGPase in these lines thus mimics a permanently redox-activated form of the enzyme. Like wild-type plants, both lines with constitutively activated AGPase had higher rates of starch synthesis in 6:18 than in 12:12 conditions (Fig. 5A). This result is consistent with our earlier observation that these lines have higher rates of starch synthesis in 8:16 conditions than in 12:12 conditions (Hädrich et al., 2012; Figs. 2 and 4 ). Both lines with constitutively activated AGPase also resembled wild-type plants in having higher rates of starch synthesis following an unexpected 4-h extension of the night than following a normal 12-h night (Fig. 4A).
Figure 5.
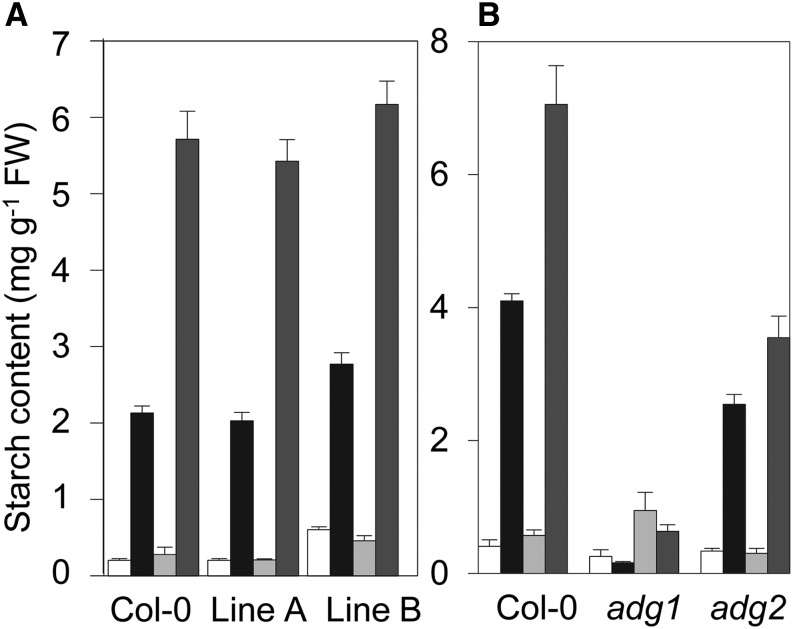
Starch contents during growth in 12:12 and 6:18 conditions in wild-type plants and plants expressing modified forms of AGPase. A, Comparison of the wild type (Col-0) and two transgenic lines expressing a constitutively redox activated form of AGPase (the adg1 mutant transformed with a pAPS1::APS1C81S construct). B, Comparison of the wild type (Col-0), the adg1 mutant lacking the small subunit of AGPase, and a mutant lacking the APL1 large subunit of AGPase (adg2). See Supplemental Table S1 for calculated rates of starch synthesis. White and black bars are starch contents at the end of the night and after 6 h of the light period, respectively, in 12:12 conditions. Light-gray and dark-gray bars are starch contents at the end of the night and after 6 h of the light period, respectively, in 6:18 conditions. Values are means ± se of measurements made on six rosettes. FW, Fresh weight.
To assess whether AGPase allosteric properties may be important in the adjustment of the rate of starch synthesis to different daylengths, we also examined adjustment in the adg2 mutant, which lacks the APL1 subunit of AGPase (Wang et al., 1997). Leaves of this mutant retain residual AGPase activity (about 5% of the wild type) due to either the formation of active APS1 homotetramers or the association of APS1 with one of the other three minor large subunits (APL2, APL3, and APL4). Purified AGPase from the adg2 mutant is less sensitive to activation by 3-PGA and more sensitive to inhibition by Pi than the enzyme from wild-type plants (Li and Preiss, 1992). In two experiments, starch synthesis was 76% and 105% greater in 6:18 than in 12:12 conditions for wild-type plants but only 45% and 17% greater for adg2 plants (Fig. 5B). Starch synthesis in adg2 plants also responded less strongly than in wild-type plants following a single, unexpected extension of the night. Whereas the rate for wild-type plants was on average about 50% higher after an extended night than after a normal night, the rate was only 23% higher in adg2 plants (Fig. 4A). Thus the allosteric properties of AGPase have an important influence on the capacity to adjust the rate of starch synthesis to altered daylengths.
To gain further information about the importance of the regulatory properties of AGPase, we investigated the effect of a single, unexpected extension of the night on the subsequent rate of starch synthesis in transgenic plants expressing an engineered form of AGPase with altered sensitivity to allosteric modulation (Fig. 4A). The TLUp3 transgenic lines lack wild-type APL1 but express a mutated form of this subunit (Up3) that renders AGPase more sensitive to activation by 3-PGA (2.5-fold lower A0.5 values than the wild-type enzyme) and less sensitive to inhibition by Pi in the presence of 3-PGA (a 3-fold higher Pi concentration required for inhibition of the Up3 than the wild-type enzyme at 1 mm 3-PGA; Obana et al., 2006). When two independent TLUp3 lines were subjected to an unexpected extension of the night, starch synthesis was less stimulated during the following day than in wild-type plants. Rates in wild-type plants were about 50% higher after an extended than a normal night; for the TLUp3 lines, rates were only about 30% higher (Fig. 4A).
DISCUSSION
Adjustment of Starch Synthesis to Photoperiod May Involve the FKF1-GI Complex
Our experiments show that the rate of starch synthesis is still accelerated in short-day conditions in plants lacking central components of the morning loop or the evening complex of the circadian clock (the cca1 lhy and elf3 mutants, respectively) or components of the photoperiod-signaling pathway that uses signals from the circadian clock to control flowering time (the quadruple cdf and co mutants) or the transcription factor PIF4, a signaling hub that integrates circadian clock, light, temperature, and hormone signals in the control of growth and morphogenesis. Even though cca1 lhy and elf3 mutants accumulated less starch than the respective wild types, they still showed a higher rate of starch accumulation in short photoperiods compared with long photoperiods. It remains possible that one or more of these proteins is involved in the fine-tuning of the rate of starch synthesis to daylength; however, none of them individually is required for the underlying mechanism by which adjustment occurs.
Two components of the photoperiod-signaling pathway that controls flowering time appeared to be required for normal adjustment of starch synthesis to daylength. Under our experimental conditions, this adjustment did not occur in mutants lacking FKF1 and occurred only in some batches of plants lacking GI. In long days, FKF1 forms a complex with GI and CDF proteins that activates expression of CO, a downstream component of the photoperiod pathway that controls flowering time (Sawa et al., 2007). Given that starch synthesis is not correctly adjusted to daylength in either fkf1 or gi mutants, it is tempting to speculate that this daylength-dependent FKF1-GI complex may be part of the mechanism by which adjustment occurs. The fact that CO and the CDF proteins are not required for adjustment of starch synthesis then implies that the photoperiod pathway controlling the rate of starch synthesis diverges from that controlling flowering time immediately downstream of the FKF1-GI complex. However, this interpretation must be treated with extreme caution because both FKF1 and GI have numerous other functions. FKF1 promotes degradation of the circadian clock components PSEUDO-RESPONSE REGULATOR5 and TIMING OF CAB EXPRESSION1 and interacts with the clock component ZEITLUPE in the control of expression of LHY (Baudry et al., 2010). The GI protein appears to act as a scaffold for numerous signaling functions. It mediates light inputs to the evening loop of the circadian clock (Pokhilko et al., 2012), and it has been implicated in, for example, drought, cold, salt, and oxidative stress responses, phytochrome signaling, stomatal opening, the response of the circadian clock to Suc, and the formation of phloem parenchyma cell wall ingrowths believed to be important for Suc export from leaves (Huq et al., 2000; Sothern et al., 2002; Mizoguchi et al., 2005; Cao et al., 2006; David et al., 2006; Edwards et al., 2010; Dalchau et al., 2011).
Adjustment of the Rate of Starch Synthesis to Daylength Does Not Involve Alterations in the Maximum Catalytic Activity or Subunit Composition of AGPase
We investigated whether AGPase is the target for the hypothetical mechanism that adjusts the rate of starch synthesis to daylength. As discussed above, redox and allosteric regulation of AGPase activity is central to the adjustment of partitioning of assimilate between Suc and starch during short-term regulation of starch synthesis. Initial investigation of the enzyme established that neither subunit composition nor maximum catalytic activity is substantially affected by daylength. Transcript levels of the APS1 and APL1 subunits were comparable in 6:18 and 12:12 conditions, and these were the only subunits detected in immunoprecipitation experiments in the two conditions. AGPase activity in leaf extracts was the same in the two conditions in the absence of 3-PGA and slightly lower during the day in 6:18 than in 12:12 conditions in the presence of 5 mm 3-PGA. Thus, the increased rate of starch accumulation in short photoperiods does not require either an adjustment of the subunit composition or an increased maximum catalytic activity of AGPase.
The Regulatory Properties of AGPase Are Crucial for Adjustment of the Rate of Starch Synthesis to Daylength
To investigate the importance of the regulatory properties of AGPase for adjustment of starch synthesis to daylength, we replaced the native enzyme with a mutant, deregulated form of the single subunit enzyme GlgC from E. coli (GlgC-TM). Transgenic plants selected for study had maximum catalytic activities of AGPase in both 12:12 and 6:18 conditions that were higher than those of wild-type plants. In the presence of the activator 3-PGA, AGPase activities were comparable in transgenic and wild-type plants. These AGPase activities (300 to 800 nmol min–1 g–1 fresh weight at the end of the day) are about 10-fold higher than the observed rates of starch accumulation (about 30 to 70 nmol min–1 g–1 fresh weight). Thus, the transgenic lines appear to be suitable material with which to investigate the importance of AGPase regulatory properties.
In 12:12 conditions, the phenotypes of plants expressing deregulated AGPase were largely comparable with those of wild-type plants. End-of-day starch contents were 60% to 100% of wild-type values, and growth rates were at least 70% of wild-type values. Similarly, Li et al. (2012) reported that plants in which the native AGPase was replaced by GlgC had essentially wild-type patterns of starch turnover in 18:6 conditions. Our GlgC-TM plants also had wild-type values for a range of enzyme activities and metabolite levels of central carbon metabolism.
Nevertheless, there were differences between GlgC-TM lines and wild-type plants in metabolite pools directly related to starch synthesis. Levels of hexose phosphates were higher during the day than in wild-type plants, and levels of ADP-Glc were lower during the day and substantially higher at night in GlcG-TM lines than in wild-type plants. These differences suggest that the allosteric properties of the native enzyme are important for maintaining an appropriate concentration of ADP-Glc for starch synthesis during the day and ensuring that ADP-Glc synthesis is minimal at night. The plant and E. coli enzymes have similar affinities for their substrates: reported KMs for the plant and bacterial enzymes, respectively, are 0.14 and 0.13 mm for Glc 1-P and 0.7 and 0.3 mm for ATP. However, allosteric activation of the native plant enzyme by 3-PGA increases its affinity for these substrates by 5- to 10-fold (Li and Preiss, 1992; Li et al., 2012). We propose that differences in ADP-Glc and hexose phosphate pools between wild-type plants and GlgC-TM lines can be explained by the activation and hence increased substrate affinity of the plant enzyme during the day and its inhibition by phosphate at night.
Transgenic plants in which the native enzyme was replaced by the deregulated GlgC-TM were unable to adjust the rate of starch synthesis in response to daylength. Whereas the rate of starch synthesis was doubled in short days in wild-type plants, the rate in GlgC-TM plants was unaffected or reduced. Failure to accelerate the rate of starch synthesis in short days was accompanied by high levels of Glc during the day and growth rates that were substantially reduced relative to those of wild-type plants. There was also no acceleration of starch synthesis in GlgC-TM plants following an extended night. One possible explanation for the failure of GlgC-TM plants to adjust the rate of synthesis might be that AGPase activity in these plants is not sufficient to permit rates higher than those observed in 12:12 conditions. The low ADP-Glc and high hexose phosphate levels in the light in GlgC-TM plants in 12:12 conditions could be interpreted in this way. However, as discussed above, AGPase activity in extracts of GlgC-TM plants is comparable with that in wild-type plants and greatly in excess of observed rates of starch synthesis, and substrate affinities of the plant and E. coli enzymes are comparable. It seems highly likely that differences between wild-type and GlgC-TM plants in hexose phosphate and ADP-Glc pool sizes in 12:12 conditions, and in the response of starch synthesis to 6:18 conditions, are attributable to the different kinetic properties of the native and GlgC-TM enzymes. Modulation of the substrate affinity of the wild-type enzyme through 3-PGA activation means that a high activity can be achieved at much lower substrate concentrations in vivo than is the case for the GlgC-TM enzyme. Thus, our data are consistent with the idea that regulatory properties of the native AGPase are essential for adjustment of the rate of starch synthesis to daylength.
We investigated two possible ways in which regulatory properties of the plant AGPase might permit adjustment of the rate of starch synthesis to different daylengths. First, we considered whether redox modulation of enzyme activity is essential for adjustment of starch synthesis to daylength. This is not the case: transgenic plants containing a redox-insensitive AGPase behave like wild-type plants in that starch synthesis is accelerated in response to growth in short days (Hädrich et al., 2012) and following an unexpected extension of the night to almost the same extent as in wild-type plants. While it remains possible that redox regulation is involved in adjusting the rate of starch synthesis in wild-type plants, our results show that it can be substituted by other mechanisms. Second, we investigated whether the allosteric properties of the enzyme may permit adjustment of the rate of starch synthesis to different daylengths. Our results are consistent with this possibility. We used two sets of plants containing AGPase with altered allosteric properties: the adg2 mutant in which AGPase is less sensitive to 3-PGA activation and more sensitive to inhibition by Pi (Li and Preiss, 1992) and transgenic lines expressing a modified large subunit that renders AGPase more sensitive to activation and less sensitive to inhibition (Obana et al., 2006). In both sets of plants, the acceleration of starch synthesis following an unexpected extension of the night was only about one-half that in wild-type plants. Considerable further work will be required to understand precisely what aspects of allosteric regulation are important, but this result shows that modification of allosteric properties can reduce the capacity for adjustment of starch synthesis in response to daylength.
These data suggest that the adjustment of starch synthesis to daylength is dependent upon posttranslational modulation of AGPase activity, most likely at the level of allosteric regulation. One possible explanation for this dependence is that changes in daylength bring about changes in the capacity for, or regulation of, the pathway of Suc synthesis. If, for example, the capacity for Suc synthesis is reduced relative to the capacity for starch synthesis in short days, this could lead to an increased allocation of newly assimilated carbon into starch. Consistent with this possibility, the ratio of activities of AGPase and Suc phosphate synthase is a little higher in short days than in 12:12 conditions (Gibon et al., 2009; Sulpice et al., 2014).
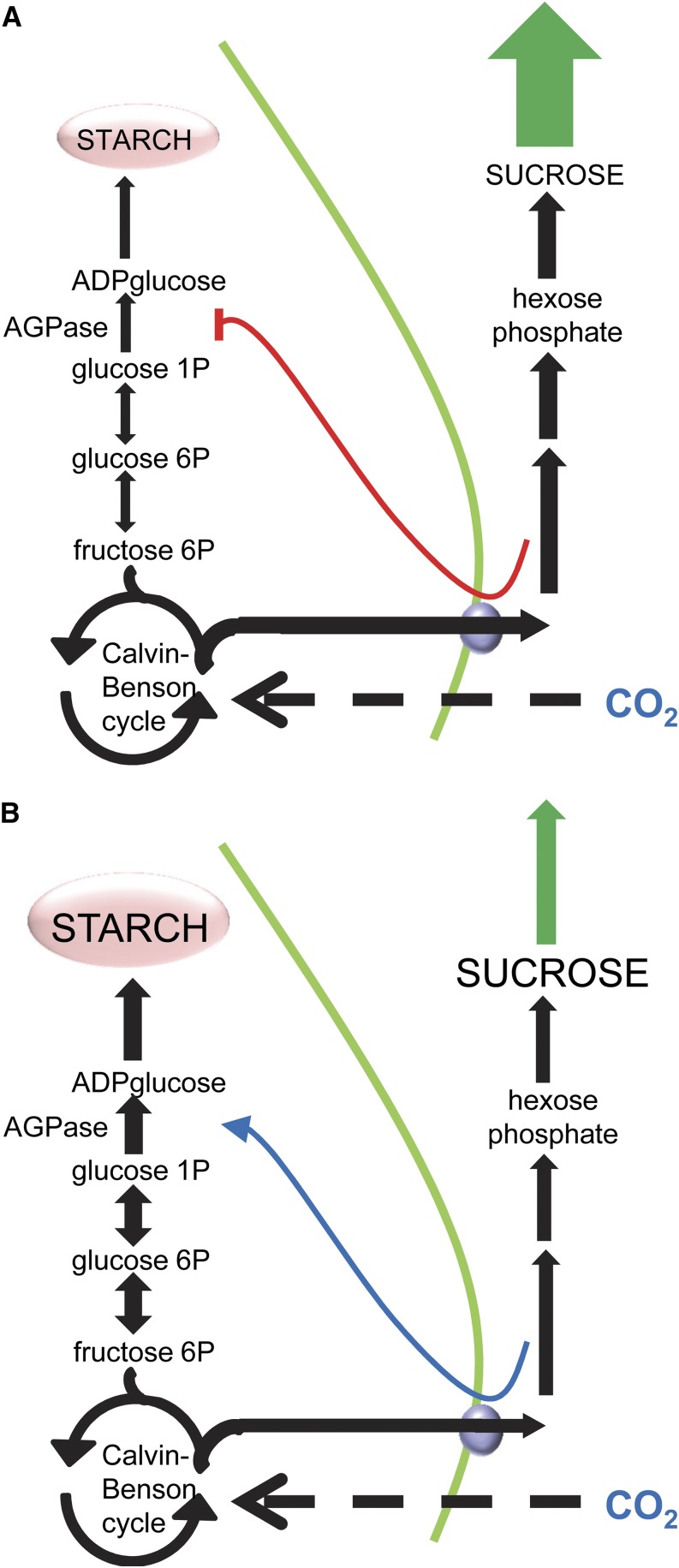
A second explanation is that the rate of starch synthesis is determined to a large extent by growth rate at the start of the day. Low growth rates will result in accumulation of Suc in the leaf, reduced rates of Suc synthesis, accumulation of phosphorylated intermediates, and hence acceleration of starch synthesis via allosteric activation of AGPase. High growth rates will deplete Suc pools, accelerate Suc synthesis, and thus result in low rates of starch synthesis via allosteric inhibition of AGPase (Fig. 6). There is considerable indirect evidence that this explanation can account for the daylength dependence of the rate of starch synthesis, as follows. Levels of Suc rise rapidly at the start of the light period when Arabidopsis plants are grown in short days but not when the light period is 12 h or longer (Sulpice et al., 2014). In plants grown in 12:12 conditions, levels of Suc rise more rapidly following an unexpected 4-h extension of the night than following a normal night (Gibon et al., 2004a). In both cases, the rapid rise in Suc has been proposed to reflect low rates of Suc utilization for growth. Growth at night (assessed as incorporation of carbon from starch and pools of primary metabolites into structure) decreases progressively as the photoperiod is shortened (Pilkington et al., 2014; Sulpice et al., 2014), so at the end of the night, growth rates will be lower in 6:18 conditions than in 12:12 conditions. Demand for newly assimilated carbon will therefore be lower at the start of a 6-h than a 12-h day. Similarly, growth rates (assessed as root extension) are much lower following an unexpected extension of the night than following a normal night (Yazdanbakhsh et al., 2011), hence demand for newly assimilated carbon will be lower following an extended night than a normal night.
Figure 6.
Proposed relationship between demand for carbon for growth and the rate of starch accumulation. A, At the start of the day in 12:12 conditions, demand for Suc for growth (green arrow) is relatively high. This stimulates flux of carbon from the Calvin-Benson cycle into the cytosol via the triose-P-Pi transporter (purple), creating conditions of relatively high Pi and low 3-PGA in the chloroplast and thus inhibiting AGPase (red line) and causing a relatively low rate of starch synthesis. B, At the start of the day in 6:18 conditions or following an unexpected extension of the night in 12:12 conditions, demand for Suc for growth is low. Suc and intermediates of the pathway build up in the cytosol, resulting in a relatively low flux of triose phosphates from the Calvin-Benson cycle into the cytosol, relatively high 3-PGA and low Pi in the chloroplast, and thus activation of AGPase and a relatively high rate of starch synthesis.
If the above explanation is correct, the adjustment of the rate of starch synthesis to daylength can be seen as an indirect response to the differing patterns of growth over the day-night cycle in different photoperiods. Any factor that influences the diel pattern of carbon demand for growth may thus indirectly influence the rate of starch accumulation during the day. Several mutants and transgenics defective in circadian clock function have altered diel patterns of growth and of starch turnover (Yazdanbakhsh et al., 2011; Ruts et al., 2012). It is tempting to speculate that the failure of fkf1 and gi mutants to adjust the rate of starch synthesis to daylength may reflect altered diel patterns of growth in these mutants. Further work will be required to discover whether this is the case.
Adjustment of Starch Metabolism to Daylength Is Essential for Normal Rates of Growth
In short days, plants expressing deregulated AGPase had both lower allocation of carbon into starch and greater reductions in growth rate than wild-type plants. It seems highly likely that the low growth rate of the transgenic plants is at least in part a direct consequence of low starch levels and hence reduced carbon availability at night. Mutants with reduced carbon availability at night (e.g. the glucan, water dikinase [gwd, starch excess1] mutant defective in starch degradation and the adg1 and phosphoglucomutase1 [pgm1] mutants defective in starch synthesis; Caspar et al., 1985, 1991; Lin et al., 1988) have wild-type growth rates in continuous light but reduced growth rates in day-night cycles. For gwd and pgm mutants, reduced overall growth appears to be attributable specifically to strong inhibition of growth at night (Wiese et al., 2007; Yazdanbakhsh et al., 2011).
The growth rates of plants expressing unregulated AGPase may also be adversely affected by ADP-Glc synthesis at night. In wild-type plants, the redox and allosteric regulatory properties of AGPase effectively prevent synthesis of ADP-Glc at night. By contrast, transgenic plants with unregulated AGPase contain levels of ADP-Glc at night that are up to 25% of those during the day. Thus, starch degradation in the GlgC-TM lines may be accompanied by resynthesis, reducing the amount of carbon available for growth at night and resulting in futile ATP consumption. Assuming that the level of ADP-Glc at night is a measure of the rate of starch synthesis, nighttime starch synthesis in GlgC-TM lines would be 25% to 50% of the net rate of starch mobilization in 12:12 conditions and 75% to 150% of the net rate of starch mobilization in 6:18 conditions, pointing to a wasteful cycle of starch synthesis and degradation that operates faster in shorter days. We observed previously that ADP-Glc was also elevated at night in lines containing a redox-insensitive AGPase (Hädrich et al., 2012), suggesting that redox inactivation of the enzyme also acts to prevent starch synthesis at night. However, nighttime ADP-Glc levels in these lines (approximately 0.05 nmol g–1 fresh weight) were 30-fold lower than those in GlgC-TM lines, suggesting that allosteric regulation makes a much larger contribution to prevention of starch synthesis at night than redox inactivation. Taken as a whole, our results emphasize the importance for plant productivity of effective regulatory networks that link growth to carbon allocation and availability (Sulpice et al., 2014).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutants cca1 lhy, elf3-1, fkf1, gi-201, and gi-2 were kindly provided by Andrew Millar (University of Edinburgh), co-10 by Caroline Dean (John Innes Centre), pif4-101 by Vinod Kumar (John Innes Centre), and the cdf quadruple mutant (cdf1-R/cdf2-1/cdf3-1/cdf5-1) by George Coupland (Max-Planck Institute for Molecular Plant Breeding). The APL1 TLUp3 transgenic lines were kindly provided by Tom Okita (Washington State University). Mutations were in the Col-0 background apart from cca1 lhy (Ws background).
Plants were grown in soil in growth cabinets at 20°C with a light intensity of 120 μmol quanta m–2 s–1. Unless otherwise stated, plants were harvested at stage 1.12 (Boyes et al., 2001). This stage was reached after 21 d in 12-h-light/12-h-dark cycles and after 28 d in 6-h-light/18-h-dark cycles. For measurement of AGPase transcripts in different photoperiods (Supplemental Fig. S1), plants were grown as described in Sulpice et al. (2014).
Generation of GlgC-TM Lines
A cDNA encoding GlgC-TM was kindly provided by Tom Okita (Washington State University). Using overlap PCR, this was cloned with the first 213 bp of the Arabidopsis APS1 open reading frame (encoding the chloroplast transit peptide). The upstream 655 bp of APS1 promoter sequence was fused to the 5′ end, and 302 bp of 3′ UTR downstream of the APS1 stop codon was fused to the 3′ end. A corresponding fragment with the endogenous APS1 gene flanked with the same promoter and terminator sequences was amplified by PCR. Both were cloned into the pWGB6 binary vector (Nakagawa et al., 2007). Mutant adg1-1 plants were grown in constant light and transformed with the constructs by Agrobacterium tumefaciens floral dip (Clough and Bent, 1998). Independent homozygous T2 lines that tested positive for restored starch synthesis by iodine staining were selected for further analysis.
Quantitative PCR Analysis of APL, APS1, and GlgC-TM Transcripts
Quantitative PCR (qPCR) analysis of APS1, APL1, APL2, APL3, and APL4 transcripts shown in Supplemental Figure S1 was performed with the high-throughput profiling method described by Czechowski et al. (2004). qPCR analysis of APS1 and GlgC-TM transcripts shown in Figure 2 was performed as described by Graf et al. (2010). Primers are shown in Supplemental Table S4.
Immunoprecipitation
Frozen, ground tissue was homogenized in extraction medium (50 mm Tris-HCl, pH 7.5, 10% [v/v] glycerol, 2 mm EDTA, 5 mm MgCl2, 150 mm NaCl, 1% [v/v] Triton X-100, 1× Sigma protease inhibitor cocktail, and 2 mm dithiothreitol [DTT]). Insoluble material was removed by centrifugation for 10 min at 12,000g. Ten microliters of antiserum raised against the potato (Solanum tuberosum) AGPase small subunit (Tiessen et al., 2002) was added to 1 mL of protein extract containing 1 mg of soluble protein and incubated with mixing at 4°C for 3 h. Three milligrams of protein-A beads (Sigma, reconstituted in extraction medium) was added and incubated with mixing at 4°C for 16 h. Samples were centrifuged at 12,000g for 10 min, and beads were washed once in 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 150 mm NaCl, and 1% (v/v) Triton X-100 and then twice in 50 mm Tris-HCl, pH 8. Beads were resuspended in Laemmli sample buffer containing 1% (v/v) β-mercaptoethanol and then denatured at 100°C for 10 min. Eluted proteins were separated by electrophoresis on a 12% (w/v) polyacyramide-SDS gel and stained with Coomassie Brilliant Blue. A gel slice containing proteins of 40 to 60 kD was incubated with trypsin, and peptides were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Immunoblotting
Tissue was extracted and extracts subjected to SDS-PAGE following denaturation in Laemmli sample buffer containing 1% (v/v) β-mercaptoethanol, as for immunoprecipitation above. Proteins were transferred onto nitrocellulose blots that were probed with antisera raised against the potato AGPase small subunit (Tiessen et al., 2002) to detect APS1 or with antisera raised against GlgC (kindly provided by Javier Pozueta-Romero [Instituto de Agrobiotecnología, Universidad Pública de Navarra]; Bahaji et al., 2011). SDS-PAGE and immunoblotting were as described by Barratt et al. (2001), using a secondary antibody conjugated to alkaline phosphatase and the SIGMAFAST BCIP/NBT reagent (Sigma-Aldrich).
Measurement of Photosynthesis
CO2 assimilation rates were measured between ZT1 and ZT4 on plants at ambient light intensity, temperature, and CO2 concentration, using a LI-6400 Infra-Red Gas Analyzer equipped with a LI-6400-17 whole-plant chamber and LI-6400-18 RGB light source (LI-COR Biosciences). Plants were allowed to acclimatize in the chamber for 10 min before measurements were made.
Metabolite Analysis and Enzyme Assays
Starch and soluble sugars were extracted and quantified as previously described (Critchley et al., 2001; Delatte et al., 2005). Rates of starch synthesis during the light period were calculated by linear regression (see Supplemental Table S1). Other metabolites were measured by high-performance anion-exchange chromatography coupled to tandem mass spectrometry (Lunn et al., 2006). Enzymes were extracted in media containing DTT and activities measured on a robotized platform according to Gibon et al. (2004b, 2009) and Sulpice et al. (2007). Significant changes in metabolite levels and enzyme activities were determined using a general linear model and ANOVA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Diel changes in levels of transcripts encoding AGPase large subunits.
Supplemental Figure S2. Immunoprecipitation with an antiserum to the AGPase small subunit.
Supplemental Figure S3. Metabolite levels in wild-type and GlgC-TM plants grown in different daylengths.
Supplemental Figure S4. Relative growth rates of wild-type and GlgC-TM plants grown in different daylengths.
Supplemental Table S1. Rates of starch synthesis in different photoperiods.
Supplemental Table S2. Metabolite contents of rosettes.
Supplemental Table S3. Activities of enzymes of primary metabolism.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Caroline Dean (John Innes Centre), Vinod Kumar (John Innes Centre), Andrew Millar (University of Edinburgh), Tom Okita (Washington State University), George Coupland (Max-Planck Institute for Molecular Plant Breeding), and Javier Pozueta-Romero (Instituto de Agrobiotecnología, Universidad Pública de Navarra) for kind gifts of materials used in this research and Zigmunds Orlovskis (John Innes Centre) and Carlos Caceres (John Innes Centre) for advice on statistical analyses.
Glossary
- AGPase
adenosine 5-diphosphate-glucose pyrophosphorylase
- Pi
inorganic phosphate
- 3-PGA
3-phosphoglyceraldehyde
- Col-0
ecotype Columbia
- Ws
ecotype Wassilewskija
- cDNA
complementary DNA
- UTR
untranslated region
- qPCR
quantitative PCR
- DTT
dithiothreitol
Footnotes
This work was supported by a grant from the European Commission FP7 collaborative project TiMet (contract no. 245142 to S.T.M., O.F., A.F., R.S., M.S., and A.M.S.) and the Max Planck Society (to N.K., B.E., R.F., J.E.L., and M.S.) and an Institute Strategic Programme Grant to the John Innes Centre from the Biotechnology and Biological Sciences Research Council (no. BB/J004561/1 to J.B. and A.M.S.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bahaji A, Li J, Ovecka M, Ezquer I, Muñoz FJ, Baroja-Fernández E, Romero JM, Almagro G, Montero M, Hidalgo M, et al. (2011) Arabidopsis thaliana mutants lacking ADP-glucose pyrophosphorylase accumulate starch and wild-type ADP-glucose content: further evidence for the occurrence of important sources, other than ADP-glucose pyrophosphorylase, of ADP-glucose linked to leaf starch biosynthesis. Plant Cell Physiol 52: 1162–1176 [DOI] [PubMed] [Google Scholar]
- Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127: 655–664 [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysdorfer C, Robinson JM (1985) Sucrose and starch synthesis in spinach plants grown under long and short photosynthetic periods. Plant Physiol 79: 838–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Jiang S, Zhang R (2006) The role of GIGANTEA gene in mediating the oxidative stress response and in Arabidopsis. Plant Growth Regul 48: 261–270 [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton NJ, Silvius JE (1979) Photosynthate partitioning into starch in soybean leaves: I. Effects of photoperiod versus photosynthetic period duration. Plant Physiol 64: 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Ballicora MA, Mérida A, Preiss J, (2003) The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme. J Biol Chem 278: 28508–28515 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Ventriglia T, Pinto F, Orea A, Mérida A, Romero JM (2005) Differential pattern of expression and sugar regulation of Arabidopsis thaliana ADP-glucose pyrophosphorylase-encoding genes. J Biol Chem 280: 8143–8149 [DOI] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan GB, Gonçalves JM, et al. (2011) The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA 108: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Armbruster U, Tama N, Putterill J (2006) Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 580: 1193–1197 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Martin AP, Andriunas F, Offler CE, Patrick JW, McCurdy DW (2010) GIGANTEA is a component of a regulatory pathway determining wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana. Plant J 63: 651–661 [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004b) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M (2004a) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hädrich N, Gibon Y, Schudoma C, Altmann T, Lunn JE, Stitt M (2011) Use of TILLING and robotised enzyme assays to generate an allelic series of Arabidopsis thaliana mutants with altered ADP-glucose pyrophosphorylase activity. J Plant Physiol 168: 1395–1405 [DOI] [PubMed] [Google Scholar]
- Hädrich N, Hendriks JHM, Kötting O, Arrivault S, Feil R, Zeeman SC, Gibon Y, Schulze WX, Stitt M, Lunn JE (2012) Mutagenesis of cysteine 81 prevents dimerization of the APS1 subunit of ADP-glucose pyrophosphorylase and alters diurnal starch turnover in Arabidopsis thaliana leaves. Plant J 70: 231–242 [DOI] [PubMed] [Google Scholar]
- Heldt HW, Chon CJ, Maronde D, Herold A, Stankovic ZS, Walker DA, Kraminer A, Kirk MR, Heber U (1977) Role of orthophosphate and other factors in the regulation of starch formation in leaves and isolated chloroplasts. Plant Physiol 59: 1146–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt JD, Casey LL, Zobel RW (1985) Effect of day length and night temperature on starch accumulation and degradation in soybean. Ann Bot (Lond) 56: 513–522 [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Kavakli IH, Kato C, Choi SB, Kim KH, Salamone PR, Ito H, Okita TW (2002) Generation, characterization, and heterologous expression of wild-type and up-regulated forms of Arabidopsis thaliana leaf ADP-glucose pyrophosphorylase. Planta 215: 430–439 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Almagro G, Muñoz FJ, Baroja-Fernández E, Bahaji A, Montero M, Hidalgo M, Sánchez-López AM, Ezquer I, Sesma MT, et al. (2012) Post-translational redox modification of ADP-glucose pyrophosphorylase in response to light is not a major determinant of fine regulation of transitory starch accumulation in Arabidopsis leaves. Plant Cell Physiol 53: 433–444 [DOI] [PubMed] [Google Scholar]
- Li L, Preiss J (1992) Characterization of ADPglucose pyrophosphorylase from a starch-deficient mutant of Arabidopsis thaliana (L.). Carbohydr Res 227: 227–2391291049 [Google Scholar]
- Lin TP, Caspar T, Somerville C, Preiss J (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh, lacking ADPglucose pyrophosphorylase activity. Plant Physiol 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen JH, Ewing EE (1992) Starch accumulation in leaves of potato (Solanum tuberosum L.) during the first 18 days of photoperiod treatment. Ann Bot (Lond) 69: 481–485 [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138: 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P (2009) NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Neuhaus H, Stitt M (1990) Control analysis of photosynthate partitioning: impact of reduced activity of ADP-glucose pyrophosphorylase or plastid phosphoglucomutase on the fluxes to starch and sucrose in Arabidopsis thaliana L. Heynh. Planta 182: 445–454 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana Y, Omoto D, Kato C, Matsumoto K, Nagai Y, Kavakli IH, Hamada S, Edwards GE, Okita TW, Matsui H, et al. (2006) Enhanced turnover of transitory starch by expression of up-regulated ADP-glucose pyrophosphorylases in Arabidopsis thaliana. Plant Sci 170: 1–11 [Google Scholar]
- Pilkington SM, Encke B, Krohn N, Höhne M, Stitt M, Pyl ET (2014) Relationship between starch degradation and carbon demand for maintenance and growth in Arabidopsis thaliana in different irradiance and temperature regimes. Plant Cell Environ (in press) [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. (1988) Biosynthesis of starch and its regulation. InPreiss J, ed, The Biochemistry of Plants, Vol 14 Academic Press, New York, pp 181–254 [Google Scholar]
- Preiss J, Romeo T (1994) Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. InCohn WE, Moldave K, eds, Progress in Nucleic Acid Research and Molecular Biology, Vol 47 Academic Press, San Diego, pp 299–329 [DOI] [PubMed] [Google Scholar]
- Ragel P, Streb S, Feil R, Sahrawy M, Annunziata MG, Lunn JE, Zeeman S, Mérida Á (2013) Loss of starch granule initiation has a deleterious effect on the growth of Arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol 163: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix JD, et al. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruts T, Matsubara S, Wiese-Klinkenberg A, Walter A (2012) Aberrant temporal growth pattern and morphology of root and shoot caused by a defective circadian clock in Arabidopsis thaliana. Plant J 72: 154–161 [DOI] [PubMed] [Google Scholar]
- Sakulsingharoj C, Choi SB, Hwang SK, Edwards GE, Bork J, Meyer CR, Preiss J, Okita TW (2004) Engineering starch biosynthesis for increasing rice seed weight: the role of the cytoplasmic ADP-glucose pyrophosphorylase. Plant Sci 167: 1323–1333 [Google Scholar]
- Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 11698–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Mugford ST, Feike D, Skeffington A, Borrill P, Graf A, Smith AM, Howard M (2013) Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife 2: e00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Déjardin A, Kleczkowski LA (1998) Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem J 336: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sothern RB, Tseng TS, Orcutt SL, Olszewski NE, Koukkari WL (2002) GIGANTEA and SPINDLY genes linked to the clock pathway that controls circadian characteristics of transpiration in Arabidopsis. Chronobiol Int 19: 1005–1022 [DOI] [PubMed] [Google Scholar]
- Stitt M, Huber S, Kerr P (1987) Control of photosynthetic sucrose synthesis. InHatch MD, Boardman NK, eds. The Biochemistry of Plants, Vol. 10 Academic Press, New York, pp 327–409 [Google Scholar]
- Stitt M, Lunn J, Usadel B (2010) Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Stitt M, Quick WP (1989) Photosynthetic carbon partitioning: its regulation and possibilities for manipulation. Physiol Plant 77: 633–641 [Google Scholar]
- Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, Stitt M (2014) Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant 7: 137–155 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Tschoep H, Von Korff M, Büssis D, Usadel B, Höhne M, Witucka-Wall H, Altmann T, Stitt M, Gibon Y (2007) Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ 30: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Sun J, Okita TW, Edwards GE (1999) Modification of carbon partitioning, photosynthetic capacity, and O2 sensitivity in Arabidopsis plants with low ADP-glucose pyrophosphorylase activity. Plant Physiol 119: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P (2013) Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Wang SM, Chu B, Lue WL, Yu TS, Eimert K, Chen J (1997) adg2-1 represents a missense mutation in the ADPG pyrophosphorylase large subunit gene of Arabidopsis thaliana. Plant J 11: 1121–1126 [DOI] [PubMed] [Google Scholar]
- Wang SM, Lue WL, Yu TS, Long JH, Wang CN, Eimert K, Chen J (1998) Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 13: 63–70 [DOI] [PubMed] [Google Scholar]
- Wiese A, Christ MM, Virnich O, Schurr U, Walter A (2007) Spatio-temporal leaf growth patterns of Arabidopsis thaliana and evidence for sugar control of the diel leaf growth cycle. New Phytol 174: 752–761 [DOI] [PubMed] [Google Scholar]
- Wu XN, Sanchez Rodriguez C, Pertl-Obermeyer H, Obermeyer G, Schulze WX (2013) Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol Cell Proteomics 12: 2856–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Sulpice R, Graf A, Stitt M, Fisahn J (2011) Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ 34: 877–894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.