The Australian Proteaceae Hakea prostrata transcriptionally regulates only a small number of genes to generate polar leaf lipid profiles associated with delayed greening and efficient phosphorus use.
Abstract
Hakea prostrata (Proteaceae) is adapted to severely phosphorus-impoverished soils and extensively replaces phospholipids during leaf development. We investigated how polar lipid profiles change during leaf development and in response to external phosphate supply. Leaf size was unaffected by a moderate increase in phosphate supply. However, leaf protein concentration increased by more than 2-fold in young and mature leaves, indicating that phosphate stimulates protein synthesis. Orthologs of known lipid-remodeling genes in Arabidopsis (Arabidopsis thaliana) were identified in the H. prostrata transcriptome. Their transcript profiles in young and mature leaves were analyzed in response to phosphate supply alongside changes in polar lipid fractions. In young leaves of phosphate-limited plants, phosphatidylcholine/phosphatidylethanolamine and associated transcript levels were higher, while phosphatidylglycerol and sulfolipid levels were lower than in mature leaves, consistent with low photosynthetic rates and delayed chloroplast development. Phosphate reduced galactolipid and increased phospholipid concentrations in mature leaves, with concomitant changes in the expression of only four H. prostrata genes, GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE1, N-METHYLTRANSFERASE2, NONSPECIFIC PHOSPHOLIPASE C4, and MONOGALACTOSYLDIACYLGLYCEROL3. Remarkably, phosphatidylglycerol levels decreased with increasing phosphate supply and were associated with lower photosynthetic rates. Levels of polar lipids with highly unsaturated 32:x (x = number of double bonds in hydrocarbon chain) and 34:x acyl chains increased. We conclude that a regulatory network with a small number of central hubs underpins extensive phospholipid replacement during leaf development in H. prostrata. This hard-wired regulatory framework allows increased photosynthetic phosphorus use efficiency and growth in a low-phosphate environment. This may have rendered H. prostrata lipid metabolism unable to adjust to higher internal phosphate concentrations.
Many Australian Proteaceae spp. are adapted to severely phosphorus (P)-impoverished soils and operate at very low leaf P status without compromising rates of photosynthesis (Denton et al., 2007; Lambers et al., 2012b; Sulpice et al., 2014). A large number of these species are endemic to the South West Australian Floristic Region, which features highly weathered, P-impoverished soils (Hopper, 2009; Laliberté et al., 2012; Hayes et al., 2014). Not only do southwestern Australian Proteaceae spp. use P-mining strategies to acquire inorganic phosphate (Pi; Shane et al., 2003; Shane and Lambers, 2005; Lambers et al., 2012a), but they also use leaf P very efficiently. They achieve this efficiency by allocating P to mesophyll cells rather than to epidermal cells (Shane et al., 2004; Lambers et al., 2015), as is common for other dicotyledons (Conn and Gilliham, 2010), by extensively replacing phospholipids by galactolipids and sulfolipids during leaf development (Lambers et al., 2012b) and by operating at very low ribosomal RNA (rRNA) levels in their leaves (Sulpice et al., 2014). The low abundance of rRNA economizes on P and also decreases the rate of protein synthesis and the resulting demand for P. In addition, many southwestern Australian Proteaceae spp. show delayed greening (Lambers et al., 2012b). Their young leaves have especially low levels of plastidic rRNA and a very low photosynthetic capacity. As the leaves develop, the levels of plastidic rRNA increase, concomitant with an increase in photosynthetic capacity, while their cytosolic rRNA levels decline (Sulpice et al., 2014).
The three major classes of polar membrane lipids in plants are phospholipids, glycoglycerolipids, and sphingolipids. During normal plant development, phospholipids are the dominant polar lipid fraction in plasma membranes, while the galactoglycerolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) are found exclusively in plastidic membranes (Moellering and Benning, 2011). Phospholipids are predominantly synthesized in the endoplasmic reticulum (ER; Nerlich et al., 2007; Gibellini and Smith, 2010) and constitute about one-third of the organic P pool (Bieleski, 1973; Poirier et al., 1991). Plants with low P status show lipid remodeling and replace some of their phospholipids with galactolipids and sulfolipids (Benning et al., 1995; Andersson et al., 2005; Hölzl and Dörmann, 2007; Tjellström et al., 2008; Shimojima and Ohta, 2011; Lambers et al., 2012b; Yuzawa et al., 2012; Shimojima et al., 2013). Genes involved in galactolipid and sulfolipid synthesis are strongly induced in response to low Pi supply in Arabidopsis (Arabidopsis thaliana), while those involved in de novo synthesis of phospholipids are repressed (Hammond et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007; Lan et al., 2012; Woo et al., 2012).
During P-limited growth, DGDG is found in extraplastidic membranes, including plasma membranes, mitochondrial membranes, and the tonoplast (Härtel et al., 2000; Jouhet et al., 2004; Andersson et al., 2005). However, this extraplastidic pool of DGDG differs from the large pool associated with the chloroplasts (Douce and Joyard, 1990). In Arabidopsis, the plastidic pool is synthesized by the sequential activities of the type A MGDG synthase MGD1, in the inner envelope membrane, and the DGDG synthase DGD1, in the outer envelope membrane (Dörmann et al., 1999; Dubots et al., 2010). The extraplastidic pool is synthesized via the inducible type B MGDG synthases MGD2 and MGD3 as well as DGD2, which are all found in the outer envelope membrane (Kelly and Dörmann, 2002; Kelly et al., 2003; Benning and Ohta, 2005; Kobayashi et al., 2009b). For this low-P-dependent lipid remodeling to occur, extraplastidic phospholipids are hydrolyzed by either PHOSPHOLIPASE C (PLC) or PLD to produce the substrate for MGDG synthases, diacylglycerol (DAG). In the case of PLD, this occurs via phosphatidic acid (PA) and its subsequent dephosphorylation by either LIPID PHOSPHATE PHOSPHATASE (LPP) or PHOSPHATIDIC ACID PHOSPHOHYDROLASE (PAH) enzymes (Pierrugues et al., 2001; Qin and Wang, 2002; Jouhet et al., 2003; Andersson et al., 2005; Nakamura et al., 2007, 2009; Pokotylo et al., 2013). PLC action produces DAG and P-containing lipid head groups. To release Pi for use in other metabolic pathways, the activity of a unique phosphoethanolamine/phosphocholine phosphatase of the haloacid dehalogenase family, PECP1, is required, which is strongly induced in P-limited Arabidopsis (Misson et al., 2005; May et al., 2012). Transcripts encoding Arabidopsis PLDς2, and to a lesser extent PLDς1, isoforms are induced by Pi deprivation, and both enzymes function redundantly in phosphatidylcholine (PC) hydrolysis, contributing to DGDG accumulation in roots of P-limited plants but not in their shoots (Cruz-Ramírez et al., 2006; Li et al., 2006). The Arabidopsis PLC isoforms that show transcriptional responses to the plant’s P status in roots and leaves are NONSPECIFIC PHOSPHOLIPASE C4 (NPC4) and NPC5, respectively (Misson et al., 2005; Nakamura et al., 2005; Morcuende et al., 2007; Gaude et al., 2008). Alternatively, phospholipids can be deacylated by either phospholipase A in combination with lysophospholipase, phospholipase B, or lipid acyl hydrolase activities (Matos and Pham-Thi, 2009). GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE (GDPD) subsequently hydrolyzes glycerophosphodiesters such as glycerophosphocholine into glycerol-3-phosphate and choline (Cheng et al., 2011).
In addition to neutral galactolipids, anionic sulfolipids may also replace phospholipids in plastids of P-limited plants (Essigmann et al., 1998). Transcripts encoding all three enzymes in the sulfolipid biosynthetic pathway identified so far, the plastidic UDP-GLUCOSE PYROPHOSPHORYLASE3 (UGP3), the UDP-sulfoquinovose synthase SQD1, and the sulfoquinovosyldiacylglycerol (SQDG) synthase SQD2, are coordinately induced in Arabidopsis plants with low P status (Essigmann et al., 1998; Yu et al., 2002; Hammond et al., 2003; Shimojima, 2011). SQDG species play a role under P-limiting conditions, as demonstrated by the reduced growth of the sqd2 mutant under Pi deficiency (Yu et al., 2002). Since SQDG is not able to fully compensate the photosynthetic impairment and growth inhibition in the Arabidopsis plastidic phosphatidylglycerolphosphate synthase1 mutant, which features a 30% reduction in phosphatidylglycerol (PG) levels, it is not clear how SQDG contributes to maintaining metabolic functions in plants with low P status (Xu et al., 2002; Frentzen, 2004).
While the ability to replace phospholipids with galactolipids and sulfolipids in response to low P availability is highly conserved in plants (Yuzawa et al., 2012), little is known about the importance of this trait for the functioning of species in severely P-impoverished environments or the molecular mechanisms that allow lipid remodeling in these species. Here, we address the question of how this trait may contribute to the high P-use efficiency of a southwestern Australian Proteaceae species, Hakea prostrata. This is one of six Proteaceae species we previously studied in their natural habitat (Lambers et al., 2012b; Sulpice et al., 2014). Here, we analyzed changes in lipid profiles and the levels of transcripts encoding lipid remodeling enzymes during leaf development in glasshouse-grown H. prostrata plants. In addition, we assessed how lipid and associated transcript profiles respond to Pi availability. Our aim was to address the following questions: How are lipid-remodeling pathways regulated during leaf development in H. prostrata? Do components of these pathways in H. prostrata respond to external Pi availability? and Do the observed metabolite and transcript profiles of this species suggest unique adaptive responses to a severely P-impoverished environment?
RESULTS
Plant Biomass Is Not Responsive to Short-Term Changes in External Pi Supply in H. prostrata
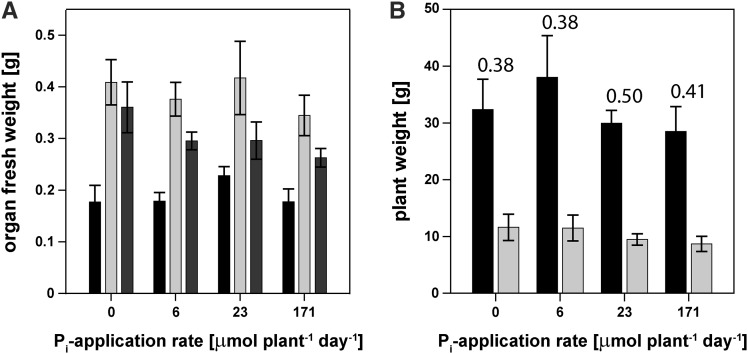
We first analyzed the impact of the external Pi supply on plant growth. Growth at a standard application rate of 6 µmol Pi plant−1 d−1 induced cluster-root production in all seedlings. When this application rate was continued, one-third of the plants produced new cluster roots during the 21-d treatment period. When Pi was withdrawn from the nutrient solution, plants had an average of 6 ± 2 cluster roots per plant at harvest. At the intermediate and high Pi supplies (23 and 171 µmol Pi plant−1 d−1, respectively), no cluster roots were produced, and newly formed white root tips were much thicker and shorter than at lower Pi supplies. Shoot-to-root dry weight ratios were similar across the four Pi treatments, ranging from 3.1 ± 0.5 for P-limited plants that received no Pi during the treatment period to 2.5 ± 0.2 for plants growing at the highest Pi supply. Interestingly, despite showing clear signs of P toxicity, plants at the highest Pi supply produced the same number of leaves as plants under the standard and intermediate application rates (9 ± 3 leaves plant−1), while plants without continuous Pi supply tended to produce fewer new leaves (5 ± 2 leaves plant−1). Young leaf fresh weight tended to be greater at the intermediate Pi supply, indicating faster growth rates of young leaves. However, there was no final trend in the weights of individual mature and senescing leaves across treatments (Fig. 1A). Total plant biomass was similar across treatments (Fig. 1B). Plants at the highest Pi supply showed 36% lower chlorophyll concentrations on a leaf area basis (Supplemental Fig. S1A). Irrespective of Pi supply, anthocyanins accumulated in young leaves and senescing leaves but not in mature leaves (Supplemental Fig. S1B).
Figure 1.
Biomass accumulation of H. prostrata plants grown at a range of Pi supplies. A, Biomass of young (black bars), mature (light gray bars), and senescing (dark gray bars) leaves. B, Total plant fresh weight (black bars) and dry weight (gray bars) after 28 weeks of treatment with the root-to-shoot dry weight ratios given above the columns. Four-month-old nursery-supplied seedlings were grown in nutrient solution for 12 weeks before being exposed to the indicated Pi supplies (experiment A). After 3 weeks of treatment, a destructive harvest was undertaken. Fresh weights are means ± se; n = 3 (young leaves) to 6. No significant differences were found between treatments.
Leaf Pi Accumulation and Inhibition of Its Conversion into Organic P Compounds
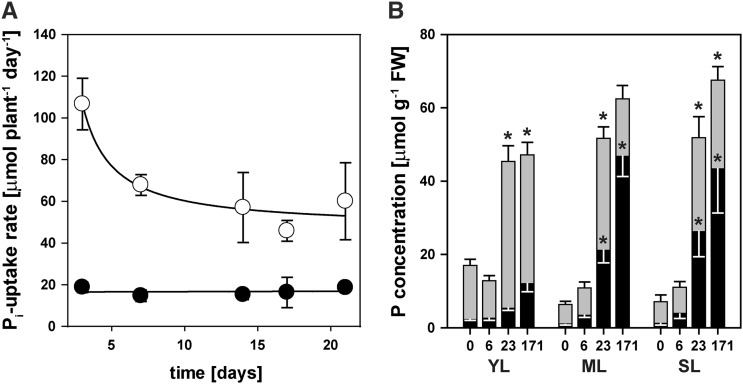
Plants under our standard, intermediate, and highest Pi treatments took up all, more than 82%, and 62% of the available Pi, respectively, at the first solution exchange (Fig. 2A). Plants without Pi supply did not release Pi into the growth medium. Net Pi uptake rates remained the same over the remaining 18 d for plants at the intermediate Pi supply. By contrast, plants at the highest Pi supply reduced their Pi uptake rate by a further 50% over the following 13 d, and between 16 to 22 d they took up only about 35% of the provided Pi. Nevertheless, their final uptake rate of 55 µmol Pi plant−1 d−1 was still three times greater than that of plants at intermediate Pi supply. The rapid Pi uptake at the highest Pi supply led to significant Pi accumulation of about 45 µmol Pi g−1 fresh weight in both mature and senescing leaves (Fig. 2B).
Figure 2.
P relations in H. prostrata plants grown at a range of Pi supplies. A, Whole-plant net Pi uptake rates for plants grown at a daily supply rate of 23 µmol Pi plant−1 (black circles) or 171 µmol plant−1 (white circles). Values shown are means ± se; n = 3. The curve was fitted using an exponential decay function [f = a × exp(b/(x + c)); r2 = 0.971]. B, Accumulation of Pi (black bars) or Po (gray bars; Po = total P – Pi) in young (YL), mature (ML), and senescing (SL) leaves of plants exposed to different Pi supplies (0, 6, 23, or 171 µmol plant−1 d−1) in experiment A. FW, Fresh weight. Values shown are means ± se; n = 4 (young leaves) to 6. Significant differences between treatments for each organ relative to P-limited plants are indicated with asterisks at P < 0.001.
Young leaves showed an increase in Pi concentration at higher external Pi supplies, but this increase was much smaller than that in mature or senescing leaves. This was most likely due to the greater ability of young leaves to convert Pi into organic phosphate (Po) compounds (Fig. 2B). In plants that did not receive any Pi during the treatment phase, the Po fraction in young leaves was three times greater (15 µmol P g−1 fresh weight) than in mature leaves (5 µmol P g−1 fresh weight, Fig. 2B; P < 0.001), indicating that H. prostrata reallocated resources to growing organs under these conditions (Lambers et al., 2012b). The Po concentration in young leaves increased by up to 3-fold in high external Pi treatments. This is similar to the increase in protein concentration in these leaves (see below). Mature and senescing leaves of P-limited H. prostrata plants had a free Pi concentration (1 µmol P g−1 fresh weight) that was approximately 50% less than that in young leaves (P < 0.001). While the free Pi pool in mature and senescing leaves increased more than 40-fold when high external Pi was supplied, their Po pool was much smaller than that in young leaves under these conditions.
The contribution of Po to the total P pool in young leaves was 85% across treatments, except at the highest Pi supply, where it was 74%. In mature and senescing leaves, the contribution of Po to the total P pool decreased with increasing Pi supply, from 84% to 25% and from 83% to 36%, respectively (Fig. 2B).
Leaf RNA, Protein, Photosynthetic Pigment, and Starch Concentrations
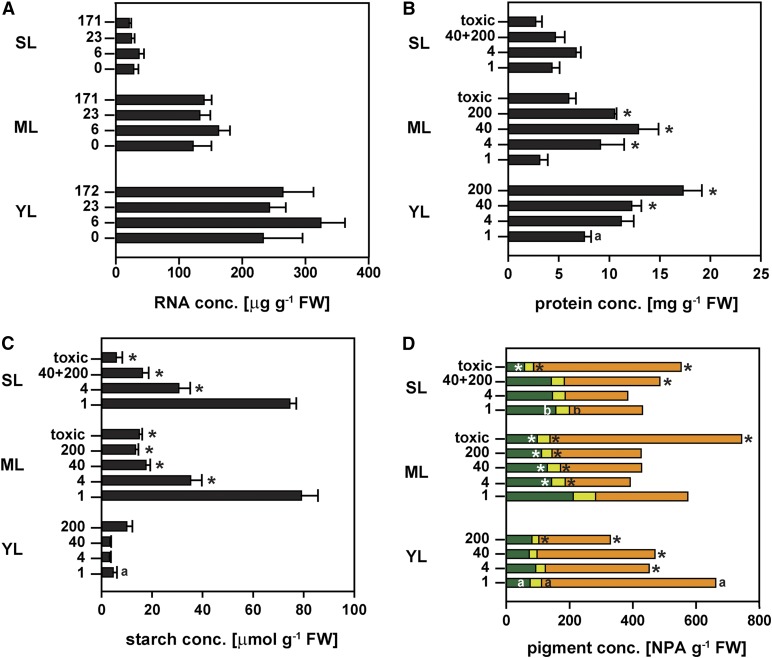
Consistent with higher Po concentrations, young leaves contained 285 ± 19 µg total RNA g−1 fresh weight, about twice as much as mature leaves (139 ± 9 µg g−1 fresh weight; Fig. 3A). RNA concentrations were extremely low in senescing leaves (Fig. 3A). The RNA level, however, was largely independent of the Pi supply in young, mature, and senescing leaves. Given that the majority of total RNA is rRNA, the change in RNA concentration during leaf development in these glasshouse-grown seedlings resembled the decrease in rRNA that was found between young and mature leaves of H. prostrata in the field (Sulpice et al., 2014).
Figure 3.
Dependence of key metabolite abundance on Pi supply during leaf development of H. prostrata. A, Total RNA concentration. B, Total protein concentration. C, Starch concentration. D, Concentrations of chlorophyll a (dark green bars), chlorophyll b (light green bars), and pheophytin (orange bars). RNA, protein, and starch concentrations (A–C) and normalized signal intensities (D; NPA, normalized peak area) g−1 fresh weight (FW) are plotted against the leaf developmental stage (YL, young leaf; ML, mature leaf; SL, senescing leaf) at the indicated Pi supply (0, 6, 23, or 171 µmol plant−1 d−1 for experiment A, in A; 1, 4, 40, or 200 µmol plant−1 d−1 for experiment B, in B–D). Data for healthy senescing leaves of plants grown at 40 and 200 µmol plant−1 d−1 were combined (40+200). The label toxic designates leaves with visible P-toxicity symptoms from plants at the two highest Pi application rates. Values shown are means ± se; n = 2 to 3 (A) or n = 3 to 6 (B–D). Significant leaf stage-specific differences relative to leaves of plants receiving 1 µmol plant−1 d−1 are shown (*P < 0.05). Metabolites with significant differences between young and mature leaves as well as between senescing and mature leaves of P-limited plants are also shown (a and b, respectively).
Total protein levels in P-limited plants were almost twice as high in young leaves as those at other leaf developmental stages (Fig. 3B). This resembled observations on H. prostrata plants in the field (Sulpice et al., 2014). The developmental changes in leaf protein concentrations closely tracked the changes in total RNA (see above). However, leaf protein concentration responded more strongly than leaf RNA concentration to the Pi supply. In young leaves, an increase in the Pi supply resulted in a 3-fold increase in total protein concentration. In healthy mature leaves, total protein concentrations were highest at 40 µmol Pi plant−1 d−1. Such large increases in leaf protein concentration with increasing Pi supply have not been reported in other plants, where changes in protein concentrations are relatively small (Lauer et al., 1989; Rao and Terry, 1989). It was also noteworthy that in H. prostrata, increased Pi supply did not lead to an increase in leaf fresh weight (Fig. 1A) but instead to an increase in protein concentration. Total protein concentration was very low in leaves showing P-toxicity symptoms (Fig. 3B). The protein concentration in senescing leaves was generally lower than that in the other leaves and did not change markedly with external Pi supply.
Levels of starch in young leaves at midday were generally low and did not respond to Pi supply (Fig. 3C). Starch concentrations were highest in mature and senescing leaves of P-limited plants and decreased with increasing Pi supply, confirming that the plants grown at the low Pi supply were indeed P limited (Rao and Terry, 1995; Ciereszko et al., 2005; Wissuwa et al., 2005). However, the marginal stimulation of leaf growth and overall plant biomass production by increased Pi supply in H. prostrata raises the question of why leaf starch concentrations decreased.
In young leaves, chlorophyll levels were lower than those in mature leaves and unresponsive to Pi supply (Fig. 3D). Surprisingly, the chlorophyll degradation intermediate pheophytin was 10-fold more abundant than chlorophylls in young leaves and showed a negative correlation with Pi availability (Fig. 3D). The opposite was true for mature leaves, where pheophytin levels were low and unresponsive to Pi supply, while chlorophyll concentrations were highest in mature leaves of P-limited plants and decreased with increasing Pi supply. In senescing leaves, chlorophylls and pheophytin were unresponsive to moderate increases in Pi supply. As an early indicator of P toxicity, chlorophyll levels were lowest in mature and senescing leaves of plants at the highest Pi supply, with a concomitant increase in the chlorophyll degradation intermediate pheophytin.
H. prostrata Lacks Orthologs of Arabidopsis Lipid-Remodeling Genes That Are Induced by P Limitation
In the H. prostrata transcriptome data set derived from leaves and roots of P-limited plants, transcripts for orthologs of P-responsive lipid-remodeling genes in Arabidopsis were largely absent, apart from those that are single-copy genes in Arabidopsis (Table I). However, a paralog encoding the strongly Pi starvation-inducible type B MGDG synthase MGD3 was present in the H. prostrata transcriptome assembly. From the selection of predicted H. prostrata complementary DNA (cDNA) contigs that encode putative orthologs of P-responsive lipid-remodeling genes in Arabidopsis (Table I), we chose a subset for relative transcript expression analysis (Supplemental Table S1). Observed differences in relative transcript abundance suggest that de novo phospholipid biosynthesis involving H. prostrata N-METHYLTRANSFERASE2 (HpNMT2) was suppressed in mature leaves compared with that in young leaves of P-limited plants (Fig. 4, column ML–no Pi; for individual transcript profiles, see Supplemental Table S2). At the same time, HpPECP1 transcripts were 5-fold more abundant, suggesting an increased release of Pi from phosphocholine and phosphoethanolamine, two phospholipid head groups. Consistent with the replacement of phospholipids by sulfolipids during leaf development, the abundance of H. prostrata SULFITE REDUCTASE (HpSIR) transcripts encoding plastid-localized sulfite reductase was 5-fold higher. This may indicate a higher demand for sulfate reduction, a rate-limiting step for sulfolipid biosynthesis in Arabidopsis (Khan et al., 2010; Shimojima, 2011). By contrast, the gene encoding the first committed step in the pathway, HpUGP3, showed a less than 2-fold induction, and the transcript abundance of HpSQD1 and HpSQD2, which were already very highly expressed in young leaves, did not change as the leaves matured. Galactolipid biosynthesis was probably more active in mature leaf chloroplasts, with a 2-fold increase in the transcript abundance for HpMGD1.
Table I. Key lipid-remodeling genes in Arabidopsis and their presence in the H. prostrata transcriptome.
Genes in regular lightface do not have an ortholog in the H. prostrata data set. Genes highlighted in boldface are those with H. prostrata orthologs that were selected for quantitative reverse transcription-PCR. For more information, see Supplemental Table S1. EM, Endomembranes; FPKM, fragments per kilobase of exon per million fragments mapped; IEM, chloroplast inner envelope membrane; N/A, not applicable; ND, not determined; OEM, chloroplast outer envelope membrane; PM, plasma membrane.
| Pathway | Arabidopsis Genome Initiative Code | Gene | Subcellular Localization (The Arabidopsis Information Resource; Joyard et al., 2010) | Reference | Fold Induction (Misson et al., 2005) | H. prostrata Ortholog? | FPKM |
|---|---|---|---|---|---|---|---|
| Galactolipid biosynthesis | At4g31780 | MGD1 | IEM | Benning and Ohta (2005) | ND | Yes | 13 |
| At5g20410 | MGD2 | OEM | Benning and Ohta (2005) | 24.8 | Yes | 14.1 | |
| At2g11810 | MGD3 | OEM | Benning and Ohta (2005) | 55.7 | Yes | 6.5 | |
| At3g11670 | DGD1 | OEM | Benning and Ohta (2005) | 3.6 | Yes | 29.1 | |
| At4g00550 | DGD2 | OEM | Benning and Ohta (2005) | ND | Yes | 18.6 | |
| Sulfolipid biosynthesis | At5g04590 | SIR | Stroma | Khan et al. (2010) | ND | Yes | 35.4 |
| At3g56040 | UGP3 | Stroma | Okazaki et al. (2009) | 10.2 | Yes | 11.3 | |
| At4g33030 | SQD1 | Stroma | Sanda et al. (2001) | 4.4 | Yes | 50.8 | |
| At5g01220 | SQD2 | IEM | Yu et al. (2002) | 14.6 | Yes | 73.5 | |
| Phospholipid biosynthesis | At3g18000 | NMT1 | Cytosol | Cruz-Ramirez et al. (2004) | 3.7 | ND | N/A |
| At1g48600 | NMT2 | Cytosol | BeGora et al. (2010) | ND | Yes | 17.2 | |
| At1g73600 | NMT3 | Cytosol | BeGora et al. (2010) | 0.09 | ND | N/A | |
| At2g32260 | CCT1 | Cytosol | Inatsugi et al. (2009) | 2.7 | Yes | 0.1 | |
| At4g15130 | CCT2 | Cytosol | Inatsugi et al. (2009) | ND | ND | N/A | |
| At1g13560 | AAPT1 | PM, Golgi | Goode and Dewey (1999) | ND | Yes | 28.3 | |
| At3g25585 | AAPT2 | PM | Goode and Dewey (1999) | ND | ND | N/A | |
| Phospholipid degradation phospholipase C | At1g07230 | NPC1 | PM, EM | Pokotylo et al. (2013) | ND | Yes | 74.9 |
| At2g26870 | NPC2 | PM, EM | Pokotylo et al. (2013) | ND | Yes | 40 | |
| At3g03520 | NPC3 | PM, tonoplast | Pokotylo et al. (2013) | ND | ND | N/A | |
| At3g03530 | NPC4 | PM | Nakamura et al. (2005) | ND | Yes | 4.6 | |
| At3g03540 | NPC5 | Cytosol | Gaude et al. (2008) | 241.6 | ND | N/A | |
| At3g48610 | NPC6 | PM (?) | Pokotylo et al. (2013) | ND | Yes | ||
| Phospholipase D | At3g15730 | PLDα1 | PM, EM | Li et al. (2008) | ND | Yes | 152.6 |
| At1g52570 | PLDα2 | OEM (?) | Eliáš et al. (2002) | ND | ND | 5.7 | |
| At5g25370 | PLDα3 | PM associated | Hong et al. (2008) | ND | ND | N/A | |
| At1g55180 | PLDα4/ε | PM associated | Hong et al. (2009) | ND | Yes | 3.3 | |
| At2g42010 | PLDβ1 | PM, nuclear (?) | Qin et al. (1997) | ND | Yes | 11.3 | |
| At4g00240 | PLDβ2 | PM, nuclear (?) | Qin et al. (1997) | ND | ND | N/A | |
| At4g11850 | PLDγ1 | EM associated | Hong et al. (2009) | ND | ND | N/A | |
| At4g11830 | PLDγ2 | OEM, PM (?) | Hong et al. (2009) | ND | ND | N/A | |
| At4g11840 | PLDγ3 | PM | Qin et al. (1997) | ND | ND | N/A | |
| At4g35790 | PLDδ | PM associated | Li et al. (2008) | ND | Yes | 12.5 | |
| At3g16785 | PLDζ1 | PM, cytosol | Li et al. (2006) | ND | Yes | 2 | |
| At3g05630 | PLDζ2 | Tonoplast | Li et al. (2006) | 54.1 | ND | N/A | |
| Glycerophosphodiester phosphodiesterase | At3g02040 | GDPD1 | Stroma | Cheng et al. (2011) | 14.2 | Yes | 7.2 |
| At5g41080 | GDPD2 | Cytosol | Cheng et al. (2011) | ND | ND | N/A | |
| At5g43300 | GDPD3 | Cytosol | Cheng et al. (2011) | ND | ND | N/A | |
| At1g71340 | GDPD4 | Mitochondrion | Cheng et al. (2011) | ND | Yes | 4.8 | |
| At1g74210 | GDPD5 | PM, tonoplast | Cheng et al. (2011) | 6.7 | Yes | 25.4 | |
| At5g08030 | GDPD6 | PM, ER | Cheng et al. (2011) | 156.1 | ND | N/A | |
| At1g66970 | GDPDL1 | IEM associated | Cheng et al. (2011) | ND | Yes | 12.7 | |
| At1g66980 | GDPDL2 | PM associated | Cheng et al. (2011) | ND | ND | N/A | |
| At4g26690 | GDPDL3 | PM associated | Cheng et al. (2011) | ND | ND | N/A | |
| At5g55480 | GDPDL4 | PM associated | Cheng et al. (2011) | ND | Yes | 12.7 | |
| At3g20520 | GDPDL5 | PM associated | Cheng et al. (2011) | ND | ND | N/A | |
| At5g58050 | GDPDL6 | PM associated | Cheng et al. (2011) | ND | ND | N/A | |
| At5g58170 | GDPDL7 | PM associated | Cheng et al. (2011) | ND | ND | N/A | |
| Phosphatidate phosphatase lipins (Mg2+ dependent) | At2g01180 | LPPα1 | PM | Pierrugues et al. (2001) | ND | ND | N/A |
| At1g15080 | LPPα2 | PM | Pierrugues et al. (2001) | ND | Yes | 23.3 | |
| At3g02600 | LPPα3 | PM | Pierrugues et al. (2001) | ND | Yes | 4 | |
| At3g18220 | LPPα4 | PM | Katagiri et al. (2005) | ND | ND | N/A | |
| At4g22550 | LPPβ | PM | Nakamura et al. (2007) | ND | Yes | 5.2 | |
| At5g03080 | LPPγ | IEM | Nakamura et al. (2007) | ND | Yes | 27.5 | |
| At3g58490 | LPPδ / SPP1 | ER | Nakagawa et al. (2012) | ND | Yes | 13.4 | |
| At3g50920 | LPPε1 | IEM | Nakamura et al. (2007) | ND | Yes | 11.1 | |
| At5g66450 | LPPε2 | IEM | Nakamura et al. (2007) | ND | Yes | 11.1 | |
| At3g09560 | PAH1 | ER associated | Nakamura et al. (2009) | ND | Yes | 5.6 | |
| At5g42870 | PAH2 | ER associated | Nakamura et al. (2009) | ND | Yes | 4.2 | |
| Phosphocholine/phosphoethanolamine phosphatase | At1g17710 | PECP1 | Cytosol | May et al. (2012) | 45.2 | Yes | 166.9 |
Figure 4.
Hierarchical cluster analysis of relative transcript abundance from lipid-remodeling genes as dependent on P status across H. prostrata leaf development. Mean log2 expression ratios (−ΔΔCt [difference in threshold cycle number relative to reference gene and control sample]) relative to the normalized expression in young leaves from P-limited plants with three biological replicates for each leaf stage are shown (experiment A). Data were normalized against the transcript abundance of HpACTIN7 and H. prostrata YELLOW-LEAF-SPECIFIC GENE8 (HpYLS8) reference genes. The clusters were generated using squared Euclidean distance and complete linkage (Dysvik and Jonassen, 2001). The linkage groups highlighted in red are described further in “Results.” Details on individual transcript expression patterns can be found in Supplemental Table S2. YL, Young leaf; ML, mature leaf.
The phospholipase NPC4 has been associated with P-dependent lipid remodeling in Arabidopsis roots (Nakamura et al., 2005). Here, phospholipases HpNPC2, HpNPC4, HpPLDα2, HpPLDα4, and HpLPPα2 were all suppressed in mature compared with young leaves of P-limited H. prostrata, indicating that they are not involved in the developmentally regulated lipid remodeling.
Transcript Abundances for Genes Involved in Lipid Remodeling Show Very Little Response to Pi Availability or Internal P Status in H. prostrata
Out of the 28 putative lipid-remodeling genes tested, very few showed P-dependent expression profiles in leaves similar to those described previously in Arabidopsis (Misson et al., 2005; Müller et al., 2007). In mature leaves, relative transcript abundance of HpNMT2 showed a positive correlation with Pi supply (Fig. 3). Consistent with this, HpGDPD1, HpMGD3, and HpSIR transcripts that were highly abundant in mature leaves of P-limited plants showed a lower abundance with increasing Pi availability. HpPECP1 transcript amounts remained high in mature leaves, irrespective of Pi supply. HpPLDα2, HpPLDα4, and HpLPPα2 transcripts showed an overall lower abundance in mature leaves, irrespective of the plant’s P status. HpNPC4 transcript abundance, which was already low in mature leaves of P-limited plants, decreased further with increasing Pi supply and was 4-fold lower in mature leaves of plants with P-toxicity symptoms. By contrast, HpMGD3, HpSIR, and HpLPPα2 had higher transcript abundance in mature leaves of plants at the highest Pi supply than in those of plants at intermediate Pi supply. In young leaves that otherwise showed even less P-dependent transcriptional regulation than mature leaves, HpMGD3, HpNPC4, HpGDPD1, and HpLPPα3 transcript amounts were lowest in plants given the highest Pi supply.
Polar Lipid Profiles Change during Leaf Development and in Response to Pi Supply
Given the relatively small changes in transcript abundance described above, we investigated how the lipid composition changed during leaf development and in response to Pi supply in H. prostrata. For these analyses, we used plants grown in experiment B (see “Materials and Methods”).
Free fatty acid levels were similar across leaf developmental stages. Mature leaves of P-limited plants had higher free fatty acid levels than leaves of P-sufficient plants. At the highest Pi supply, fatty acid levels were lower in young leaves (Supplemental Table S3). Mature and senescing leaves with P-toxicity symptoms had higher levels of free fatty acids. Lyso-PC levels were highest in young leaves and showed little response to external Pi availability, but these lipid species accumulated in mature and senescing leaves of plants showing P-toxicity symptoms (Supplemental Table S3).
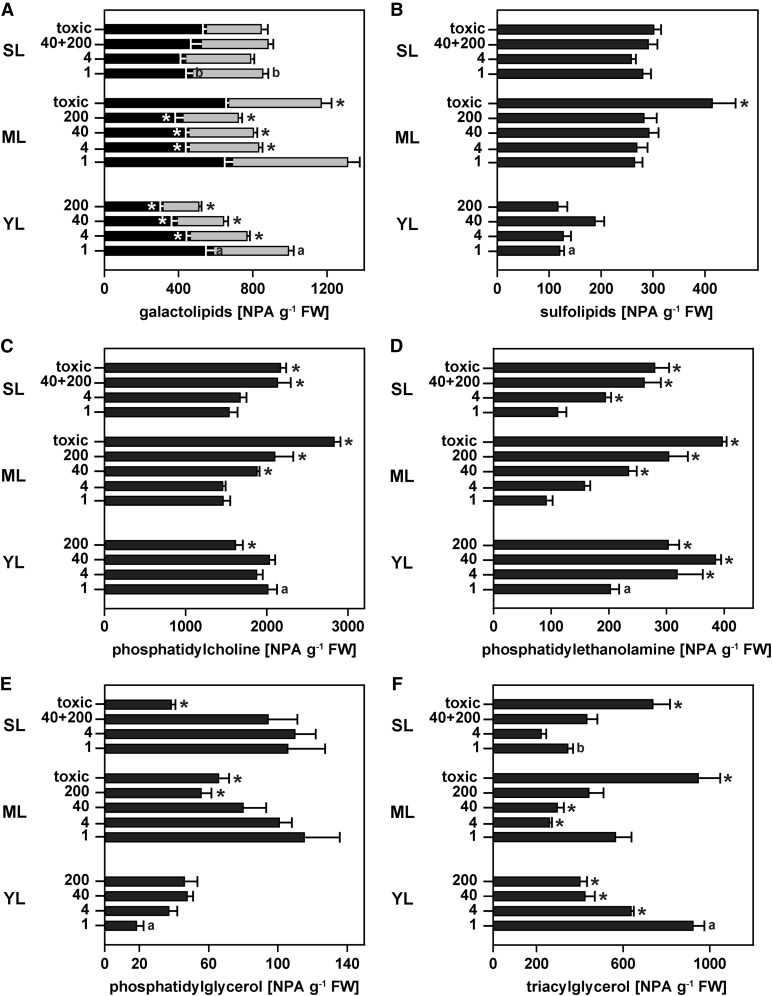
Concentrations of galactolipids decreased by as much as 50% in young and mature leaves with increasing Pi supply, while they did not respond to Pi availability in senescing leaves (Fig. 5A). MGDG and DGDG levels were higher in mature leaves compared with young and senescing leaves of P-limited plants. DGDG levels were also higher in mature leaves showing P-toxicity symptoms than in those without symptoms. The majority of galactolipids were found in group II (b) of the hierarchical cluster analysis (Supplemental Fig. S2). This group was most likely associated with lipid remodeling under P-limiting conditions, with a strong negative correlation to Pi supply.
Figure 5.
Lipid profiles across leaf developmental stages of H. prostrata plants grown at a range of Pi supplies. A, Galactolipids MGDG (black bars) and DGDG (gray bars). B, Sulfolipid SQDG. C, PC. D, PE. E, PG. F, TAG. Normalized signal intensities g−1 fresh weight (FW) are plotted against the leaf developmental stage (YL, young leaf; ML, mature leaf; SL, senescing leaf) at the indicated Pi application rates (1, 4, 40, and 200 µmol plant−1 d−1) in experiment B. Data for healthy senescing leaves of plants grown at 40 and 200 µmol plant−1 d−1 were combined (40+200). The label toxic designates leaves with visible P-toxicity symptoms from plants grown at the two highest Pi application rates. Values shown are means ± se; n = 3 to 6. Significant leaf stage-specific differences relative to leaves of plants receiving 1 µmol Pi plant−1 d−1 are indicated (*P < 0.05). Significant differences between young and mature leaves as well as between senescing and mature leaves of P-limited plants are also shown (a and b, respectively). Further details for individual lipid species are outlined in Supplemental Table S3.
Sulfolipid concentrations were more than 2-fold higher in mature and senescing leaves than in young leaves (Fig. 5B). Unlike galactolipids, concentrations of sulfolipids did not decrease with increasing Pi supply, but they were higher in mature leaves showing P-toxicity symptoms (Fig. 5B). While some of the most abundant sulfolipids were also found in the lipid remodeling group II (b), SQDG 34:1 and SQDG 36:3 associated with group II (a) alongside all three PG species detected in this study (Supplemental Fig. S2). The fact that these lipids are predominantly found in chloroplast membranes (Marechal et al., 1997) could associate group II (a) with delayed greening in H. prostrata.
The most abundant glycoglycerolipid species in leaves were MGDG 36:6, DGDG 36:6, and SQDG 36:6 (Supplemental Table S3). MGDG 34:3, DGDG 34:3, and SQDG 34:3 were also very strongly represented in all three leaf stages. MGDG 34:6 species were detected, while DGDG 34:6 and SQDG 34:6 were either not present or below the detection limit, consistent with the current model for the desaturation of acyl chains of thylakoid lipids in the chloroplast (Li-Beisson et al., 2010; Boudière et al., 2014).
Concentrations of the two main phospholipid classes in H. prostrata, PC and phosphatidylethanolamine (PE), were higher in young leaves of P-limited plants than in mature and senescing leaves (Fig. 5, C and D). This was mostly due to a decline in 36:6 PC species carrying 18-C acyl chains with a higher degree of unsaturation in older leaves (Supplemental Table S3). Consistent with the P-dependent lipid remodeling observed in other plants, PC species in mature and senescing leaves showed a positive correlation with Pi supply (group IV in Supplemental Fig. S2), but their levels were constant in young leaves (Fig. 5C). PE concentrations gradually increased with increasing Pi supply across all leaf developmental stages (Fig. 5D). At the highest Pi supply, however, both PC and PE levels dropped significantly in young leaves.
In contrast to the other phospholipids, concentrations of PG, the main phospholipid in the chloroplast inner envelope and thylakoid membranes (Marechal et al., 1997), were highest in mature and senescing leaves and lowest in young leaves of P-limited plants (Fig. 5E). All PG species clustered together in group II (a) and, therefore, may primarily be involved in chloroplast and more specifically thylakoid membrane development during delayed greening in H. prostrata (Supplemental Fig. S2). In young leaves, PG showed a positive correlation with Pi supply. However, in mature leaves, PG concentrations showed a negative correlation with Pi supply. PG levels were also more than 50% lower in senescent leaves that showed P-toxicity symptoms.
Consistent with a high demand for transient carbon storage during leaf development, triacylglycerol (TAG) levels were highest in young leaves of H. prostrata and tended to decline with increasing Pi availability (Fig. 5F; group I in Supplemental Fig. S2). TAG species with long acyl chains and a higher degree of unsaturation were almost exclusively found in young leaves (Supplemental Fig. S2). In mature and senescing leaves, normalized signal intensities of TAG were lowest at 4 μmol Pi plant−1 d−1, but TAG levels were considerably higher in leaves with toxicity symptoms. This profile, therefore, was very similar to the one observed for galactolipids. The lipid species that accumulated in leaves with P-toxicity symptoms differed from the ones found in young leaves or in mature leaves of P-limited plants and were mostly part of group III (Supplemental Fig. S2).
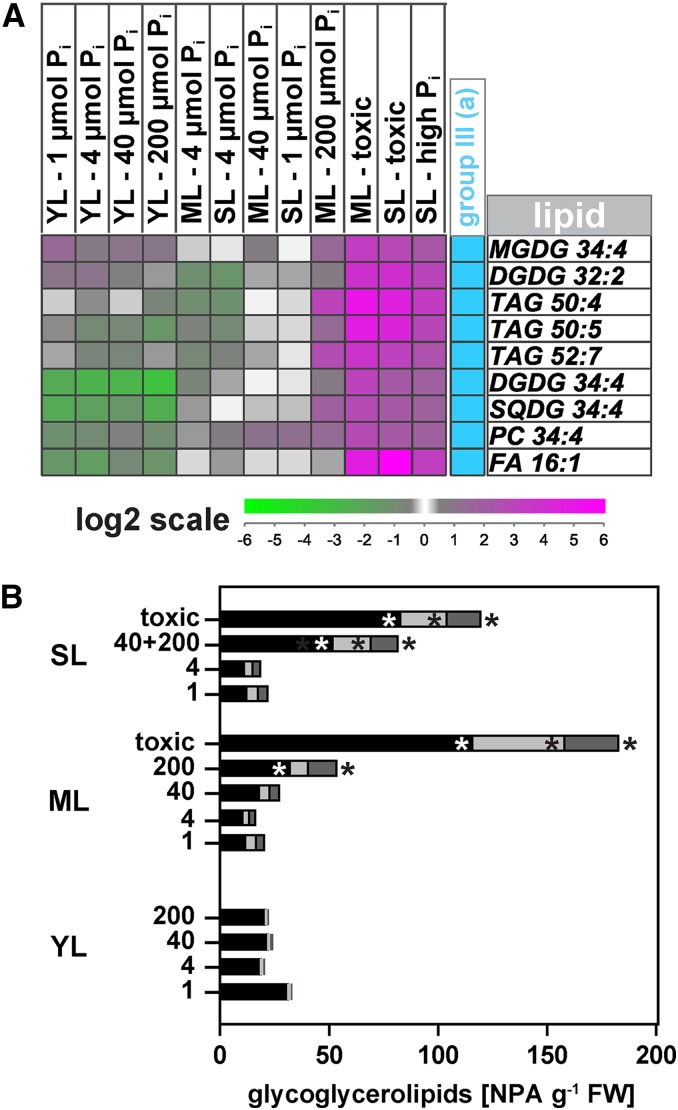
A Group of Polar Lipids Associated with the Chloroplast Is Highly Sensitive to Excess Pi Accumulation in Leaves
One group of lipids stood out from all others in the hierarchical cluster analysis. Group III was similar to the PG group II (a) (Fig. 6A; Supplemental Fig. S2). The 34:x (x = number of double bonds in hydrocarbon chain) sulfolipids (SQDG 34:1–34:4), 34:x PC species (34:1–34:4), PE 34:4, as well as the 34:x PG species (34:2–34:4) in group II (a) were all very low in abundance in young leaves compared with mature leaves (Supplemental Table S3). In a complete reversal of the normal Pi-dependent lipid-remodeling profile, PG levels decreased with increasing Pi supply in mature and senescing leaves, while at the same time, 34:x SQDG species increased. The 34:x PC species, on the other hand, showed the expected positive correlation with Pi supply. In contrast with group II (a), however, all of the group III lipids showed a very strong accumulation in leaves with P-toxicity symptoms; MGDG 34:4 and DGDG 34:4 were 8- to 15-fold more abundant than in leaves of P-limited plants (Fig. 6B). Similarly, sulfolipid SQDG 34:4 as well as its more saturated precursors SQDG 34:1, 34:2, and 34:3 in groups II (a) and III (b) increased 4- to 6-fold in both mature and senescing leaves at the two highest Pi supplies, regardless of whether the leaves were healthy or had P-toxicity symptoms (Fig. 6B; Supplemental Fig. S2). Interestingly, group III (a) also included free palmitoleic acid as well as a number of TAG species (TAG 50:4, 50:5, and 52:7) that showed the same profile as the select group of glycerolipids. They were more than 10-fold more abundant in leaves with P-toxicity symptoms than in comparable asymptomatic leaves (Fig. 6A; Supplemental Table S3).
Figure 6.
Profile of a subset of putative chloroplast-associated lipids that, unlike most glycoglycerolipids, are also highly abundant in leaves with P toxicity symptoms. A, Enlarged profile of group III (a) from the hierarchical cluster analysis shown in Supplemental Figure S2. Normalized mass spectral signal values are expressed relative to those in mature leaves of P-limited plants. B, Normalized signal intensities of glycoglycerolipids in group III (a) that are most likely synthesized via the prokaryotic pathway: MGDG 34:4 (black bars), DGDG 34:4 (light gray bars), and SQDG 34:4 (dark gray bars). Values are plotted against the leaf developmental stage (YL, young leaf; ML, mature leaf; SL, senescing leaf) at the indicated Pi application rates (1, 4, 40, and 200 µmol plant−1 d−1) in experiment B. Data for healthy senescing leaves of plants grown at 40 and 200 µmol plant−1 d−1 were combined (40+200). The label toxic designates leaves with visible P-toxicity symptoms from plants grown at the two highest Pi application rates. Values shown are means; n = 3 to 6. Significant leaf stage-specific differences relative to leaves of P-limited plants are indicated (*P < 0.05). Further details on individual lipid species are outlined in Supplemental Table S3. FW, Fresh weight.
While SQDG 34:1 to 34:3 profiles clustered in groups II (a) and III (b), the more saturated precursors of PE and galactolipids with 34:1 to 34:3 acyl chains were much more abundant in young than in mature leaves. These lipids showed the classic lipid-remodeling response, with MGDG 34:1 and DGDG 34:1 decreasing in abundance and PE 34:1 increasing in abundance with increasing Pi supply. Unlike PE 34:1, the 34:1 galactolipids were also quite abundant in mature leaves of P-limited plants and decreased with increasing Pi supply.
DISCUSSION
Increased Pi Supply Does Not Greatly Alter Leaf Growth Rates But Does Alter Leaf Metabolite Composition, Including a Large Increase in Protein Concentration
Proteaceae spp. like H. prostrata grow with a very low P availability in their natural habitat (Lambers et al., 2012a). The total leaf P concentrations of plants in the field (Lambers et al., 2012b) resemble those in plants with the lowest Pi supply in our experimental system. While higher root and shoot biomass production are observed in the longer term when Pi supply is increased (Shane et al., 2004), plant biomass production was not altered significantly by a moderate to large increase in Pi supply over the experimental period in this study and even decreased slightly at the highest Pi supply. However, Pi supply resulted in a slight increase in the production of new leaves and major changes in leaf composition. Young H. prostrata leaves showed a large increase in protein concentration, increased chlorophyll levels and chlorophyll a/b ratios, lower pheophytin and anthocyanin concentrations, as well as an altered lipid profile. Mature leaves showed positive responses to moderate increases in Pi supply, such as increased protein and phospholipid levels and reduced starch, galactolipid, and TAG accumulation (Fig. 7). Surprisingly, H. prostrata did not show the higher shoot-to-root ratio that typically accompanies a higher Pi supply in other plants (Marschner et al., 1996). Two factors may explain why growth did not immediately increase with an increase in Pi supply. First, growth costs are higher at moderate levels of Pi than at low Pi because of greater investment in protein per unit fresh weight in both young and mature H. prostrata leaves. The assimilation of inorganic nitrogen and sulfur, amino acid synthesis, and protein synthesis are far more costly than the synthesis of polysaccharide cell walls, lipids, or secondary metabolites (Penning de Vries et al., 1974; Warner, 1999; Amthor, 2010). Second, H. prostrata has previously been shown to have a lower photosynthetic performance at higher leaf P concentrations (Shane et al., 2004). In agreement, high Pi supply resulted in lower chlorophyll and PG levels in this study. A decrease in carbon gain may contribute to the lower than expected biomass gain at elevated Pi supply.
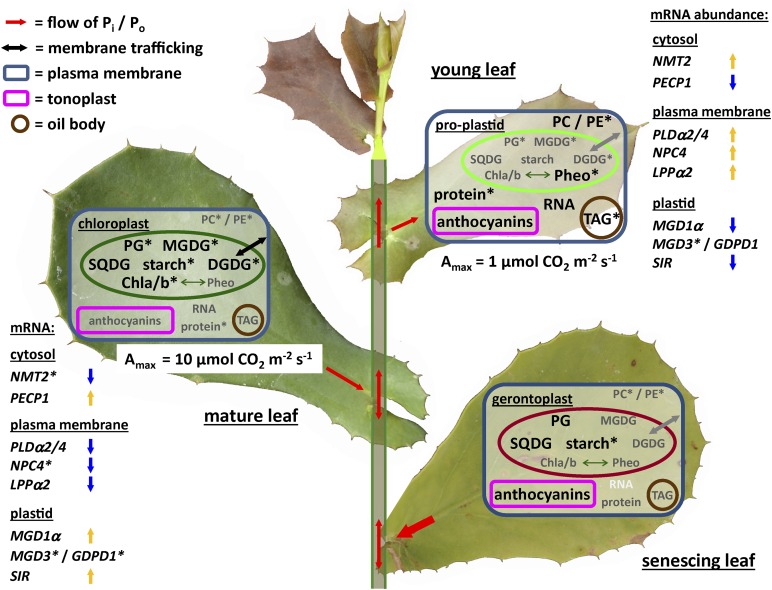
Figure 7.
Summary of the changes in metabolite and transcript abundance during leaf development in H. prostrata. Young leaves are able to develop on plants with very low P status due to delayed chloroplast development that allows them to allocate limited P resources to growth. As leaves mature, P reserves are mobilized by phospholipid degradation, and thylakoid membranes develop fully. Leaf senescence progresses slowly, beginning with the degradation of rRNA pools, while plasma and chloroplast membranes are maintained. Ultimately, P remobilization from fully senescent leaves can be as high as 80% (Denton et al., 2007). To reduce the complexity of the figure, protein and RNA pools are shown only in the cytosol, representing their overall concentration across compartments. In higher plants, PC and PE are mostly found in the plasma membrane and endomembranes, while PGs, MGDGs, and SQDGs are predominantly found in chloroplasts. DGDGs are also found in extraplastidic membranes of P-limited plants, but their distribution is unclear in H. prostrata. Metabolite levels are indicated (black type, high abundance; gray type, low abundance; white type, absent). Changes in transcript abundance between young and mature leaves are indicated by blue (down-regulated) and yellow (up-regulated) arrows. Genes are grouped according to the predicted subcellular localization of the encoded proteins. Asterisks indicate Pi-dependent changes in abundance. Amax, Photosynthetic capacity; Chla/b, chlorophylls; Pheo, pheophytin; Pi, phosphate; Po, phloem-mobile organophosphates (e.g. phosphocholine, sugar phosphates, ATP, etc.).
Young H. prostrata Leaves Are Shielded against the Consequences of Low Pi Availability
Organ growth depends heavily on P-containing organic compounds such as RNA, DNA, phospholipids, ATP, and phosphorylated sugars; P-efficient species and cultivars tend to allocate a greater proportion of their limiting P resources to growing and differentiating tissues (Kavanová et al., 2006; Aziz et al., 2014; Sulpice et al., 2014). Proteaceae spp. in their natural habitat have relatively high levels of phospholipids in their young expanding leaves compared with mature leaves (Lambers et al., 2012b), suggesting that phospholipids are indispensable, for example in processes such as cell division or expansion (Jackowski, 1996; Cruz-Ramírez et al., 2004; Gagne and Clark, 2010). Here, we show that young H. prostrata leaves also had a more than 3-fold larger organic P fraction than mature leaves. This is a much more significant difference than their 1.5-fold greater phospholipid fraction, given that H. prostrata also economizes on other organic P pools in mature leaves, especially the rRNA pool (Sulpice et al., 2014).
Interestingly, young leaves also appeared to be shielded from P toxicity, as they had much lower Pi concentrations than mature leaves under high-P conditions. Two factors might contribute to this. First, they may be able to utilize the P more rapidly for growth and are metabolically more active, as suggested by the high levels of Po. Second, as their gas-exchange activity is fairly low (Lambers et al., 2012b), the evaporative water loss via stomata would be less than in mature leaves, resulting in less import of Pi via the transpiration stream. The observation that total P per unit fresh weight was only slightly lower in young than in mature leaves of plants supplied with high Pi, however, indicates that the key factor may be their ability to utilize P.
Young H. prostrata Leaves Show Delayed Development of Functional Chloroplasts, Including Changes in Pigment and Lipid Metabolite Concentrations, as Well as Altered Profiles of Transcripts from Lipid-Remodeling Genes
Consistent with higher phospholipid levels, HpNMT2 transcripts encoding a putative phosphocholine-producing enzyme involved in de novo phospholipid biosynthesis (Bolognese and McGraw, 2000; Jost et al., 2009; BeGora et al., 2010) were more abundant in young than in mature leaves of P-limited H. prostrata. Interestingly, higher Po concentrations and NMT transcript abundance in young leaf tissues are also striking features of a highly P-efficient wheat (Triticum aestivum) cultivar (Aziz et al., 2014). Just as in H. prostrata, early wheat leaf developmental stages (e.g. leaf primordia, elongating blades, and mature leaf bases) are white and heterotrophic, as in many monocotyledons, while the activation of chloroplast transcription and translation occurs at later stages of development, in parallel with the greening of the leaf blade (Masle, 2000; Kusumi et al., 2010; Li et al., 2010). Lipid profiles in developing maize (Zea mays) leaves show some similarities to those in H. prostrata. Concentrations of MGDG species increased almost 4-fold, those of DGDG and SQDG species increased 3-fold, and those of PG species increased 2-fold, while concentrations of both PC and PE species decreased by more than 3-fold in sections farther up the maize leaf blade compared with the leaf base (Leech et al., 1973). These results in maize are consistent with chloroplast maturation and thylakoid development in mesophyll and bundle sheath cells in those segments. In P-limited H. prostrata plants, mature leaf lipid profiles were matched by the higher abundance of transcripts encoding the putative plastid-localized type A MGDG synthase HpMGD1 and the sulfite reductase HpSIR compared with young leaves. Transcripts for the putative phosphatase HpPECP1 showed a similar expression pattern. This phosphatase, therefore, could play a central role in the replacement of phospholipids in extraplastid membranes by galactolipids and the release of Pi (May et al., 2012) to support metabolic processes associated with chloroplast development.
The control of chloroplast RNA metabolism is critical for the efficient use of P in plants (Marchive et al., 2009). We reported earlier that delayed chloroplast maturation during leaf development in Proteaceae spp. may allow for sequential P allocation, first to cytosolic rRNA and later to plastidic rRNA pools, as a means of making the most of this sparingly available resource (Sulpice et al., 2014). Here, we present further evidence of this smart P-allocation strategy, which is summarized in Figure 7. In young H. prostrata leaves, pheophytin was highly abundant while chlorophylls only accumulated as the leaves matured, a situation similar to that found in maize, where pheophytin accumulates in etiolated leaves prior to light-regulated conversion into chlorophyll (Ignatov and Litvin, 1995). The chlorophyll a/b ratio was also very low in young H. prostrata leaves (Sulpice et al., 2014) and resembled ratios found in shade leaves, with 2-fold lower CO2 assimilation rates (Lichtenthaler et al., 2007). In young leaves of H. prostrata, the low total chlorophyll and low chlorophyll a/b ratio are accompanied by a 10-fold lower photosynthetic capacity compared with maximum Rubisco activity in mature leaves (Lambers et al., 2012b; Sulpice et al., 2014). Delayed greening, therefore, could involve the accumulation of chlorophyll intermediates, such as pheophytin or protochlorophyllide, in young leaf mesophyll cells (Matile et al., 1999).
Young H. prostrata leaves contain high anthocyanin concentrations. These may compensate for low levels of photosynthetic pigments and protect these leaves against photodamage and reactive oxygen species (Lee and Collins, 2001; Hatier and Gould, 2008) as well as provide protection against herbivory (Lev-Yadun et al., 2004). The pigment distribution in H. prostrata leaves is very different from that found in Arabidopsis, where anthocyanins accumulate in older leaves (Diaz et al., 2006) and chloroplast development takes place early during leaf development (Breeze et al., 2011), with high chlorophyll and Rubisco concentrations in young and expanding leaves. Another protective mechanism for young leaf plastids in H. prostrata could be the higher concentrations of more saturated 34:x PE and galactolipid species; a recent study in cotton (Gossypium hirsutum) found that young leaves have a higher proportion of saturated fatty acid side chains in chloroplast membrane lipids, and this is subject to seasonal variation and correlates with changes in their photosynthetic thermotolerance (Hall et al., 2014).
Further evidence for the delayed development of functional chloroplasts in H. prostrata includes (1) the sharp increase in starch accumulation, which can only occur in photosynthetically active chloroplasts, in mature leaves compared with young leaves of P-limited plants, and (2) the extremely low levels of SQDG and PG in young leaves, both of which are lipids essential for and found primarily in functional photosynthetic membranes (Marechal et al., 1997; Babiychuk et al., 2003; Shimojima et al., 2009). In Arabidopsis, lipid species found primarily in the chloroplast account for 75% of polar leaf lipids (Devaiah et al., 2006), while in mature H. prostrata leaves, this fraction was only about 37% (Supplemental Table S3). The smaller proportions of preferentially plastid-localized lipids suggest that H. prostrata leaves contain fewer or smaller chloroplasts on a fresh weight basis. This is especially true if we assume that some of the galactolipids are in fact localized in extraplastid membranes of mature leaves of P-limited plants. This assessment is consistent with the observations by Sulpice et al. (2014) and suggests a very high efficiency of the photosynthetic machinery, given that overall leaf area-based rates of photosynthesis in Proteaceae spp. leaves are similar to those of other plants (Denton et al., 2007; Lambers et al., 2012b).
Delayed Greening Is Only Partially Reversed by Increased Pi Supply
The delayed greening of young leaves is partially reversed at higher Pi supply. With increasing Pi availability, young leaves had higher chlorophyll a/b ratios, lower pheophytin, anthocyanin, and group II (a) and III sulfolipid levels (Supplemental Fig. S2), as well as higher RNA, protein, and in particular PG concentrations than analogous leaves from P-limited plants. However, the decrease in concentrations of pheophytin and anthocyanins was not matched by similar increases in chlorophyll a and b, which indicates that this reversal is incomplete.
In H. prostrata, transcripts encoding putative phospholipases HpNPC2, HpNPC4, HpPLDα2, HpPLDα4, and HpLPPα2 were more abundant in young leaves irrespective of Pi supply. This suggests that these enzymes are not involved in the release of Pi from membrane lipids but rather in developmentally controlled lipid remodeling. In Arabidopsis, PLDα activity is very prominent in metabolically active organs (Fan et al., 1999). These lipases, therefore, might be important for the turnover of phospholipids and associated signaling processes during organ growth (Dhonukshe et al., 2003; Gardiner et al., 2003; Potocký et al., 2003; Wang, 2005). Reduced PLDα activity in mature leaves, on the other hand, could delay leaf senescence (Fan et al., 1997; Lee et al., 2012), promoting leaf longevity, which is characteristic of Proteaceae spp. like H. prostrata (Wright et al., 2002; Denton et al., 2007). The facts that NPC4 is only involved in lipid remodeling in the roots of P-limited Arabidopsis (Nakamura et al., 2005) and that HpNPC4 transcripts are more abundant in young H. prostrata leaves without functional chloroplasts suggest that this phospholipase is important for lipid turnover in sink tissues. Consistent with extensive lipid-remodeling activity during H. prostrata leaf development, TAG levels were very high in young leaves of P-limited H. prostrata. It has been proposed that the transient accumulation of TAG could act as a buffer against the toxic effects of free fatty acids released from membranes (Fan et al., 2013; Troncoso-Ponce et al., 2013) and as a transient, readily accessible carbon storage pool (Zhang et al., 2009). High TAG accumulation, therefore, may partially compensate for the lower starch levels found in these organs.
H. prostrata Leaves Senesce Slowly with Early Remobilization of P from RNA
Southwestern Australian Proteaceae spp. remobilize scarce nutrients, such as P, very efficiently (Denton et al., 2007), perhaps through a process that extends cell viability (Hörtensteiner and Feller, 2002). Plants grown at the lowest Pi supply in this study showed similar leaf P concentrations compared with plants sampled in the field (Lambers et al., 2012b). In the oldest leaves of these P-limited plants, the photosynthetic pigment fraction declined by 30% compared with that in mature leaves. Anthocyanin concentrations also increased in these older leaves, which has been suggested to facilitate nutrient resorption by protecting against high radiation levels when chlorophyll levels decline (Hoch et al., 2003; Ougham et al., 2005; Zhang et al., 2013). Given that we were unable to isolate significant amounts of RNA from leaves in these very early stages of senescence, P resorption appears to start with the degradation of rRNA pools, which make up to about 50% of the total organic P fraction in photoautotrophic organisms (Veneklaas et al., 2012; Raven, 2013). In P-limited plants, protein concentrations in senescing leaves were similar to those in mature leaves, despite the large decline in the total RNA pool, indicating slow protein turnover. However, a clear decrease in protein concentration between mature and senescing leaves was seen for plants growing with an intermediate or high P supply. The overall normalized signal intensity for polar lipids in senescing leaves was about 80% of that at the other leaf developmental stages (Supplemental Table S3). Proportions of galactolipids and TAG, in particular, were much smaller than those in mature leaves. Interestingly, overall Pi and Po pools, and phospholipid levels in particular, did not decrease to the same extent during this early stage of leaf senescence as they do in other plants (Thompson et al., 1998).
The contrast between lower chlorophyll and galactolipid concentrations and relatively constant levels of PG and SQDG in senescing leaves suggests that the degradation of thylakoid membranes in the developing gerontoplast has not started (Koiwai et al., 1981; Matile et al., 1999; Kaup et al., 2002; Kolodziejek et al., 2003). A decline in phospholipid concentration during leaf senescence in other plant species increases membrane leakiness (Thompson et al., 1998). An extended lifetime for cellular membranes appears to be crucial for prolonging transport functions (Gniazdowska et al., 1999; Rilfors and Lindblom, 2002; Tjellström et al., 2010). This would give Proteaceae spp. like H. prostrata the time necessary to proficiently remobilize P from senescing leaves (Denton et al., 2007). Nutrient resorption in these long-lived leaves, therefore, seems to proceed relatively slowly, most likely leading to the transient accumulation of low-Mr Po catabolites prior to their export (Zhang et al., 2013).
The P Responsiveness of Regulatory Networks Is Reduced in H. prostrata Compared with That in Arabidopsis
Mature leaves of P-limited H. prostrata contained similar levels of starch, Pi, and Po as leaves of 3-week-old P-limited Arabidopsis seedlings (Nilsson et al., 2007). Lipid remodeling similar to that observed in P-limited Arabidopsis shoots (Kobayashi et al., 2009a) also occurred in H. prostrata leaves; galactolipids showed a negative correlation with Pi supply in all except senescing leaves, and phospholipids showed a positive correlation with Pi supply in all except young H. prostrata leaves. The decrease in galactolipid concentrations in young and mature leaves with Pi supply could be attributed to the P-dependent 4- to 5-fold decreases in transcripts encoding putative H. prostrata orthologs of the phosphodiesterase GDPD1 and the type B MGDG synthase MGD3 (Dubots et al., 2010; Cheng et al., 2011). The concomitant increase in phospholipids was accompanied by a modest 2-fold decrease in HpNPC4 and a 4-fold increase in HpNMT2 expression. While these changes in transcript abundance were moderate and affected very few of the lipid-remodeling genes examined in H. prostrata, leaves of P-limited Arabidopsis seedlings rapidly accumulated 4- to 60-fold more transcripts for a whole suite of genes encoding lipid-remodeling enzymes (Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007; Lan et al., 2012). The moderate transcriptional response in H. prostrata shows that while this species has not entirely lost the capacity to regulate P use for lipid metabolism through changes in gene expression, the magnitude of the response is greatly reduced compared with that in other plants. While profiles of Arabidopsis transcripts correlate well with changes in lipid profiles in response to environment (Szymanski et al., 2014), lipid metabolism in H. prostrata appears to be mostly regulated at the posttranscriptional level, perhaps through other mechanisms that are known to operate in other species like selective RNA translation and protein turnover (Galland et al., 2014) or allosteric enzyme regulation by PA and other lipid metabolites (Ohlrogge and Browse, 1995; Dubots et al., 2012). H. prostrata is restricted to low-P environments. It is thus likely that the P-starvation response that is well documented in other plants would never be triggered under natural low-P conditions in H. prostrata and that regulatory components found in plants such as Arabidopsis and rice (Oryza sativa; Rouached et al., 2010; Lin et al., 2014) have been attenuated or lost. This possibility is supported by the absence of orthologs in the H. prostrata transcriptome database for many lipid genes that are induced by the myeloblastosis transcription factor PHOSPHATE STARVATION RESPONSE1 in response to Pi limitation in Arabidopsis (Bustos et al., 2010; Acevedo-Hernández et al., 2012). Instead, transcript profiles suggested that rate-limiting steps in the pathway, such as increased flux through the sulfate assimilation pathway mediated by SIR (Khan et al., 2010), MGDG synthesis by the type A MGDG synthase MGD1 in the chloroplast (Dubots et al., 2010), and the release of Pi from the phosphocholine head group (May et al., 2012), are mostly developmentally controlled.
Chloroplast Lipid Metabolism Is Perturbed at Higher Leaf Pi Concentrations
The most remarkable and unusual response to increasing Pi supply was the decrease in PG levels in mature H. prostrata leaves. In higher plants, there are two pathways for glycoglycerolipid synthesis: the prokaryotic pathway of the chloroplast and the eukaryotic pathway that involves PA synthesis in the ER (Roughan and Slack, 1982; Ohlrogge and Browse, 1995). PG production via the prokaryotic pathway is essential for chloroplast function and growth in Arabidopsis (Babiychuk et al., 2003; Xu et al., 2006). Lower PG levels in mature H. prostrata leaves appear to be accompanied by a compensatory increase in sulfolipid species of the 34:x acyl chain configuration that were mainly found in groups II (a) and III of the hierarchical cluster analysis. This differs from the situation in Arabidopsis, where levels of all phospholipids show a positive correlation and all glycoglycerolipids show a negative correlation with Pi supply (Dörmann and Benning, 2002). By contrast, 36:x SQDG species, which are synthesized from the ER-derived secondary DAG pool in the chloroplast (Fritz et al., 2007; Li-Beisson et al., 2010; Boudière et al., 2014), clustered in group II (b) and showed the typically documented decrease with increasing Pi supply in H. prostrata. These contrasting responses resulted in an overall sulfolipid profile that responded very little to altered Pi availability. While the majority of galactolipids in mature leaves decreased with increasing Pi supply, some 34:x galactolipid species also showed an unexpected positive response to Pi supply. In Arabidopsis leaves, 34:x lipid species show mixed acyl chain configurations suggesting biosynthesis from both ER-derived and chloroplast DAG pools, making it hard to determine their precise origin. In Arabidopsis leaves, PG 34:4, SQDG 34:3/34:4, MGDG 34:4 to 34:6, and DGDG 34:1 species almost exclusively carried the C-16 fatty acid in the sn-2 position (DGDG 34:4 species were not detected; Devaiah et al., 2006; Maatta et al., 2012), which would suggest that these lipid species are synthesized from the primary DAG pool in the chloroplast inner envelope membrane via the so-called prokaryotic pathway (Fritz et al., 2007; Li-Beisson et al., 2010; Boudière et al., 2014). In Arabidopsis, both PG and sulfolipids are essential for chloroplast structure and function, but only about 30% of PG species are actually produced in the chloroplast (Xu et al., 2002; Yu and Benning, 2003). PG and SQDG production in the chloroplast are tightly regulated in an antagonist fashion (Essigmann et al., 1998). The fact that H. prostrata appears to produce most of its PG in the chloroplast might account for the fact that only the sulfolipid (and galactolipid) species connected to the primary DAG pool of the chloroplast show the typical compensatory response to lower PG levels in mature leaves (Fritz et al., 2007; Shimojima et al., 2009; Boudière et al., 2014), despite their overall higher P status. The inferred contribution of the prokaryotic pathway for MGDG synthesis rose to as much as 20% in leaves with toxicity symptoms compared with about 2% in leaves of P-limited plants (Supplemental Table S3). The glycoglycerolipids found in group II (b), on the other hand, seem to be synthesized from the secondary DAG pool derived from the ER. Their metabolite profiles suggest that they are more responsive to the plants’ P status and are involved in P-dependent lipid remodeling in extraplastid membranes. The disparity in the regulation of these two groups in response to Pi supply might provide an excellent tool to further dissect the regulation of prokaryotic and eukaryotic pathways (Roughan and Slack, 1982; Shimojima and Ohta, 2011) using H. prostrata. They also could be indicative of a strong adaptive mechanism in H. prostrata that enables it to maintain high PG levels in mature leaf chloroplasts under low Pi availability, ensuring optimal photosynthetic performance (Lambers et al., 2012b).
This specialized adaptation of lipid metabolism might provide a further reason why H. prostrata does not show a positive growth response with highest Pi supply: in mature and senescing leaves with P-toxicity symptoms, free fatty acid, lyso-PC, and TAG levels almost doubled. Most notably, the concentrations of PG species and starch were significantly lower than those in healthy leaves, while PC, PE, galactolipid, and sulfolipid concentrations were higher. These metabolite profiles would indicate a severe perturbation of chloroplast function. Given that HpNPC4 transcript abundance was lowest in mature leaves with P-toxicity symptoms, overall lipid turnover at the plasma membrane (Nakamura et al., 2005) also appears to be greatly reduced under conditions of excessive Pi accumulation in leaves. It will be interesting to further explore if the observed swelling of H. prostrata palisade mesophyll cells that accumulate the highest concentrations of Pi (Shane et al., 2004) is indicative of a loss of membrane integrity and function caused by the imbalance in lipid biosynthesis between the ER and chloroplast (Sakurai et al., 2003; Eastmond et al., 2010).
CONCLUSION
Phospholipids are the major lipid class in plant cell membranes and essential for cell division and expansion. Higher plants characterized to date exhibit some degree of lipid remodeling during P starvation. H. prostrata has adapted to its low-P environment partly through delayed greening and reduced chloroplast size or chloroplast numbers in mature leaves and extensive lipid remodeling during leaf development. This is accompanied by the developmental regulation of PG levels and key lipid-remodeling genes and their reduced P responsiveness. Other aspects of leaf metabolism, on the other hand, are highly responsive to Pi: protein synthesis in particular is tightly linked to Pi supply without being coupled to the developmental programs controlling leaf expansion and leaf size. Future work will further explore the metabolic adjustments, enzymatic properties, and regulatory networks that define H. prostrata’s high photosynthetic P-use efficiency but may have rendered its lipid metabolism and associated regulatory networks sensitive to high soil P.
MATERIALS AND METHODS
Plant Growth
Washed roots of 4-month-old soil-grown Hakea prostrata seedlings (Men of the Trees Nursery) were treated with 5% (v/v) sodium hypochlorite for 15 min to avoid fungal contamination. Individual seedlings were transferred to 5-L black plastic pots covered with black panels and gray foam plugs to support the shoots. The seedlings were grown in a temperature-controlled glasshouse at the University of Western Australia in cooling tanks that kept the root temperature at 19°C. Each pot contained 4 L of a continuously aerated reduced-strength Hoagland solution [40 μm Ca(NO3)2, 20 μm K2SO4, 10 μm KH2PO4, 11 μm MgSO4, 2 μm FeNaEDTA, 4 μm KCl, 0.05 μm MnCl2, 0.02 μm ZnSO4, 0.004 μm CuSO4, 0.5 μm H3BO3, 0.003 μm CoCl2, 0.01 μm (NH4)6Mo7O24, and 10 μm Na2O3Si, pH 5.8] that was changed weekly.
During experiment A, the average minimum and maximum temperatures were 14°C at night and 22°C during the day, and the average maximum light intensity between 12 noon and 2 pm was 820 µmol m−2 s−1 (70% transmission), with sunrise at 7.30 am and sunset at 5:30 pm. Relative humidity varied from 47% (day) to 66% (night). After 12 weeks, similarly sized seedlings were selected for growth in nutrient solution containing different Pi concentrations: 0, 5, 20, and 150 μm KH2PO4. The plants were randomly assigned to each treatment, and the nutrient solution was changed twice per week, resulting in average application rates of 0, 6, 23, and 171 µmol Pi plant−1 d−1. After 21 d of treatment, leaves (young, mature [i.e. the most recently fully expanded leaves on the main stem], and senescing [i.e. the oldest, chlorotic leaves on the main stem]) were harvested around midday, between 11 am and 4 pm, weighed, frozen in liquid N2, and stored at −80°C. Remaining roots, stems, and leaves were weighed separately and added to the mass of the collected samples to give the total fresh weight of the seedlings. This material was dried at 60°C for 4 d to determine plant dry weight.
For the polar lipid profiling work (experiment B), 6-month-old cuttings propagated from a mature H. prostrata plant (Nuts about Natives) were established as described above, but with a 4-week acclimation period and an experimental period of 12 weeks in the autumn/winter of 2011 (March to July 2011), with sunrise at 7:30 am and sunset at 5:30 pm, average temperatures of 27°C (day) and 17°C (night), average relative humidity of 49% (day) and 72% (night), and average midday maxima of around 810 µmol photons m−2 s−1, 37% relative humidity, and 35°C. The experimental period started with application rates of 0, 0.8, 40, and 100 μmol KH2PO4 plant−1 d−1 for 4 weeks. Due to signs of severe P limitation at the lowest Pi supply and the absence of P-toxicity symptoms at the highest Pi supply, application rates were adjusted to 0.8, 4, 40, and 200 μmol KH2PO4 plant−1 d−1 for the remaining 8 weeks. Plants were harvested as described above, except that mature leaves from plants exposed to the two highest Pi supplies that displayed signs of P toxicity such as chlorosis and blotchiness (most likely due to localized cell death) were harvested separately from leaves that showed no visible symptoms.
H. prostrata is an outcrossing species, and seed material used in nurseries is collected from the wild. To ensure that our chosen treatments triggered the expected Pi responses, phenotypic data were recorded for all plants. Plants that did not show the typical responses were excluded from the downstream analyses.
Phosphate and Total P Analyses
To determine Pi uptake rates, samples were taken from the spent nutrient solution from three pots per treatment when the solution was replaced. To determine the amount of Pi within plant organs, 50 mg of ground and frozen plant material was combined with 700 μL of 1% (v/v) acetic acid and homogenized (Precellys 24 Tissue Disruptor; Bertin Technologies) for three cycles of 30 s at 4,500 rpm. The cleared supernatant was stored at −20°C. For total P analysis, 50 mg of oven-dried leaf tissue was digested to convert esterified P to Pi (Zasoski and Burau, 1977), and the ash was resuspended in 5 mL of deionized water. The Pi concentration of all samples was determined using an ammonium molybdate/ascorbic acid-based assay (Ames, 1966).
Sequence Identification and Primer Design
Unique H. prostrata orthologs of 26 Arabidopsis (Arabidopsis thaliana) genes of interest associated with membrane-lipid remodeling were identified in a database (InParanoid; Remm et al., 2001) generated using open-reading frame translations of H. prostrata young leaf and root transcriptomes (http://www.onekp.com/). Unique primer pairs for the selected H. prostrata transcripts were designed in QuantPrime (Supplemental Table S1; Arvidsson et al., 2008). The specificity of the primer amplicons was confirmed by melting-curve analysis. Primer efficiencies were determined across cDNA samples in independent quantitative PCR assays using the LinReg algorithm (Ruijter et al., 2009).
RNA Extraction, Reverse Transcription, and Quantitative PCR
Ground frozen leaf powder (100 mg) was mixed with 700 µL of 100 mm Tris-HCl, pH 8, 500 mm LiCl, 10 mm EDTA, 1% (w/v) lithium dodecyl sulfate, 5 mm dithiothreitol, 0.5% (w/v) polyvinylpyrrolidone, and 0.5% (w/v) polyvinylpolypyrrolidone. Following the addition of 350 µL of Fruit-mate (Takara Biotechnology), samples were homogenized as described above. After several chloroform:isoamyl alcohol (24:1) extractions, RNA was concentrated by isopropanol precipitation, followed by another precipitation of the resuspended RNA in 2 m LiCl. The RNA pellet was washed with 2 m LiCl followed by two washes with 70% (v/v) ethanol, air dried, and resuspended in 20 µL of RNase-free water. After removal of contaminating DNA (RQ1 DNase; Promega), the RNA concentration was determined (NanoDrop 1000 Spectrophotometer; Thermo Scientific). The average residual genomic DNA contamination was less than 0.01%. About 1 µg of total RNA was reverse transcribed according to the manufacturer’s instructions (Tetro cDNA synthesis kit; Bioline). Quantitative PCR and threshold cycle determination were performed (7500 FAST Real-Time PCR System; Applied Biosystems) in reactions that contained 2.5 μL of cDNA (synthesized from approximately 25 ng of total RNA), 2.5 μL of gene-specific primer mix (1.2 μm each), and 5 μL of PCR master mix (Power SYBR Green; Applied Biosystems). The normalized threshold fluorescence level was set to 0.2. The relative expression levels of transcripts of interest were normalized to orthologs of the reference genes ACTIN7 (Hong et al., 2008) and YLS8 (Czechowski et al., 2005) after validation using the geNORM algorithm (Vandesompele et al., 2002).
Polar Lipid and Photosynthetic Pigment Profiling
Frozen leaf powder (50 mg) was extracted as described by Hummel et al. (2011). Pellets of the final organic phase were stored at −80°C prior to analysis by ultra-performance liquid chromatography (UPLC) quadrupole time-of-flight mass spectrometry (Burgos et al., 2011). The quantity of each lipid or pigment was derived as the normalized mass spectral signal relative to the internal standard 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (PC 34:0; 17:0/17:0). These values were then divided by the fresh weight of the extracted sample to give the normalized mass spectral signal per gram fresh weight. Alternatively, the values were divided by the total normalized signal to express them as percentages of the normalized mass spectral signal. All chemicals used were UPLC/mass spectrometry grade (BioSolve).
To account for possible chlorophyll degradation during extraction, chlorophyll a/b concentrations were also determined spectrophotometrically using methanol extraction of freshly harvested mature leaf discs (Supplemental Fig. S1). Profiles across treatments were found to be comparable. We were not able to confidently annotate phosphatidylserine, PA, or DAG-derived peaks in the mass spectra of any of our H. prostrata leaf samples. At this stage, it is unclear whether this is due to low levels of these lipids, the lack of the entire phosphatidylserine pathway, or a very rapid turnover of DAG. Our UPLC-mass spectrometry-based analytical approach did not allow the resolution of PA lipid species.
Starch and Total Protein Determination
The first pellet from the lipid extraction (see above) was resuspended in 400 µL of 0.1 m NaOH and heated for 30 min at 120°C with vigorous shaking. The cooled supernatant was assayed for protein at 595 nm against a bovine serum albumin standard (Bradford, 1976). The starch pellet was suspended in 80 µL of 0.5 m HCl and 0.1 m acetate, pH 4.9, buffer. The starch was degraded for 16 h at 37°C using an amyloglucosidase/α-amylase enzyme mixture, and the Glc end-product concentration was determined via NADPH production by Glc-6-P dehydrogenase (Hendriks et al., 2003).
Statistics
Statistical significance between treatments was determined using ANOVA in SigmaStat version 12.3 and defined as P ≤ 0.05. Unless stated otherwise, two-way ANOVA followed by Tukey’s posthoc test was used to separate means. Hierarchical clustering analysis was performed using J-Express 2012 (Norwegian Bioinformatics Platform and Norwegian Microarray Consortium [http://www.molmine.com]; Dysvik and Jonassen, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Accumulation of photosynthetic pigments and anthocyanins in leaves of H. prostrata plants grown at a range of Pi supplies.
Supplemental Figure S2. Hierarchical cluster analysis of changes in lipid profiles during leaf development and in response to Pi supply.
Supplemental Table S1. Sequence information on H. prostrata transcripts encoding orthologs of known lipid-remodeling genes in Arabidopsis, including quantitative reverse transcription-PCR primer sequences used in this study.
Supplemental Table S2. Detailed information on the relative abundance of individual transcripts in young and mature H. prostrata leaves in response to phosphate supply.
Supplemental Table S3. Detailed information on the relative abundance of individual lipid species along the leaf developmental gradient and in response to phosphate supply in H. prostrata.
Supplementary Material
Acknowledgments
We thank Robert D. Pontré for help with growing the first cohort of plants and Dr. Rebecca Ullmann for optimizing the RNA extraction from H. prostrata. We also thank Dana Schindelasch for preparing the extracts for the polar lipid profiling work, Melanie Höhne for conducting the total protein and starch assays, and the anonymous reviewers for the excellent comments that helped to greatly improve this article.
Glossary
- P
phosphorus
- Pi
inorganic phosphate
- rRNA
ribosomal RNA
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- ER
endoplasmic reticulum
- DAG
diacylglycerol
- PA
phosphatidic acid
- SQDG
sulfoquinovosyldiacylglycerol
- PG
phosphatidylglycerol
- PC
phosphatidylcholine
- Po
organic phosphate
- cDNA
complementary DNA
- PE
phosphatidylethanolamine
- TAG
triacylglycerol
- UPLC
ultra-performance liquid chromatography
Footnotes
This work was supported by the Australian Research Council (grant no. DP110101120 to H.L. and P.M.F. and grant no. DP140100148 to P.M.F. and P.G.), a 2009 Public Service Department Scholarship from Malaysia (to T.K.), a University of Western Australia Fay Gale Travel Fellowship (to R.J.), and the Max Planck Society.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Acevedo-Hernández G, Oropeza-Aburto A, Herrera-Estrella L (2012) A specific variant of the PHR1 binding site is highly enriched in the Arabidopsis phosphate-responsive phospholipase DZ2 coexpression network. Plant Signal Behav 7: 914–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8: 115–118 [Google Scholar]
- Amthor JS. (2010) From sunlight to phytomass: on the potential efficiency of converting solar radiation to phyto-energy. New Phytol 188: 939–959 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS (2005) Phosphate-limited oat: the plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Müller-Röber B (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz T, Finnegan PM, Lambers H, Jost R (2014) Organ-specific phosphorus-allocation patterns and transcript profiles linked to phosphorus efficiency in two contrasting wheat genotypes. Plant Cell Environ 37: 943–960 [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Müller F, Eubel H, Braun HP, Frentzen M, Kushnir S (2003) Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J 33: 899–909 [DOI] [PubMed] [Google Scholar]
- BeGora MD, Macleod MJ, McCarry BE, Summers PS, Weretilnyk EA (2010) Identification of phosphomethylethanolamine N-methyltransferase from Arabidopsis and its role in choline and phospholipid metabolism. J Biol Chem 285: 29147–29155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Huang ZH, Gage DA (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys 317: 103–111 [DOI] [PubMed] [Google Scholar]
- Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bieleski RL. (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol Plant Mol Biol 24: 225–252 [Google Scholar]
- Bolognese CP, McGraw P (2000) The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol 124: 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudière L, Michaud M, Petroutsos D, Rébeillé F, Falconet D, Bastien O, Roy S, Finazzi G, Rolland N, Jouhet J, et al. (2014) Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim Biophys Acta 1837: 470–480 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos A, Szymanski J, Seiwert B, Degenkolbe T, Hannah MA, Giavalisco P, Willmitzer L (2011) Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J 66: 656–668 [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA (2005) Interactive effects of phosphate deficiency, sucrose and light/dark conditions on gene expression of UDP-glucose pyrophosphorylase in Arabidopsis. J Plant Physiol 162: 343–353 [DOI] [PubMed] [Google Scholar]
- Conn S, Gilliham M (2010) Comparative physiology of elemental distributions in plants. Ann Bot (Lond) 105: 1081–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, González-Ortega E, Herrera-Estrella L (2004) The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L (2006) Phospholipase Dζ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton MD, Veneklaas EJ, Freimoser FM, Lambers H (2007) Banksia species (Proteaceae) from severely phosphorus-impoverished soils exhibit extreme efficiency in the use and re-mobilization of phosphorus. Plant Cell Environ 30: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TWJ, Munnik T (2003) Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell 15: 2666–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Balbo I, Benning C (1999) Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284: 2181–2184 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Benning C (2002) Galactolipids rule in seed plants. Trends Plant Sci 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Douce R, Joyard J (1990) Biochemistry and function of the plastid envelope. Annu Rev Cell Biol 6: 173–216 [DOI] [PubMed] [Google Scholar]
- Dubots E, Audry M, Yamaryo Y, Bastien O, Ohta H, Breton C, Maréchal E, Block MA (2010) Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J Biol Chem 285: 6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubots E, Botté C, Boudière L, Yamaryo-Botté Y, Jouhet J, Maréchal E, Block MA (2012) Role of phosphatidic acid in plant galactolipid synthesis. Biochimie 94: 86–93 [DOI] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I (2001) J-Express: exploring gene expression data using Java. Bioinformatics 17: 369–370 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Quettier AL, Kroon JTM, Craddock C, Adams N, Slabas AR (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22: 2796–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliáš M, Potocký M, Cvrčková F, Žárský V (2002) Molecular diversity of phospholipase D in angiosperms. BMC Genomics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yan C, Xu C (2013) Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis. Plant J 76: 930–942 [DOI] [PubMed] [Google Scholar]
- Fan L, Zheng S, Cui D, Wang X (1999) Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol 119: 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zheng S, Wang X (1997) Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen M. (2004) Phosphatidylglycerol and sulfoquinovosyldiacylglycerol: anionic membrane lipids and phosphate regulation. Curr Opin Plant Biol 7: 270–276 [DOI] [PubMed] [Google Scholar]
- Fritz M, Lokstein H, Hackenberg D, Welti R, Roth M, Zähringer U, Fulda M, Hellmeyer W, Ott C, Wolter FP, et al. (2007) Channeling of eukaryotic diacylglycerol into the biosynthesis of plastidial phosphatidylglycerol. J Biol Chem 282: 4613–4625 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Clark SE (2010) The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22: 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L (2014) Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol Cell Proteomics 13: 252–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J, Collings DA, Harper JDI, Marc J (2003) The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol 44: 687–696 [DOI] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Gibellini F, Smith TK (2010) The Kennedy pathway: de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62: 414–428 [DOI] [PubMed] [Google Scholar]
- Gniazdowska A, Szal B, Rychter AM (1999) The effect of phosphate deficiency on membrane phospholipid composition of bean (Phaseolus vulgaris L.) roots. Acta Physiol Plant 21: 263–269 [Google Scholar]
- Goode JH, Dewey RE (1999) Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol Biochem 37: 445–457 [Google Scholar]
- Hall TD, Chastain DR, Horn PJ, Chapman KD, Choinski JS Jr (2014) Changes during leaf expansion of ΦPSII temperature optima in Gossypium hirsutum are associated with the degree of fatty acid lipid saturation. J Plant Physiol 171: 411–420 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatier JHB, Gould KS (2008) Foliar anthocyanins as modulators of stress signals. J Theor Biol 253: 625–627 [DOI] [PubMed] [Google Scholar]
- Hayes P, Turner BL, Lambers H, Laliberté E (2014) Foliar nutrient concentrations and resorption efficiency in plants of contrasting nutrient-acquisition strategies along a 2-million-year dune chronosequence. J Ecol 102: 396–410 [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Singsaas EL, McCown BH (2003) Resorption protection: anthocyanins facilitate nutrient recovery in autumn by shielding leaves from potentially damaging light levels. Plant Physiol 133: 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzl G, Dörmann P (2007) Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res 46: 225–243 [DOI] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper SD. (2009) OCBIL theory: towards an integrated understanding of the evolution, ecology and conservation of biodiversity on old, climatically buffered, infertile landscapes. Plant Soil 322: 49–86 [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P (2011) Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov NV, Litvin FF (1995) Light-regulated pigment interconversion in pheophytin/chlorophyll-containing complexes formed during plant leaves greening. Photosynth Res 46: 445–453 [DOI] [PubMed] [Google Scholar]
- Inatsugi R, Kawai H, Yamaoka Y, Yu Y, Sekiguchi A, Nakamura M, Nishida I (2009) Isozyme-specific modes of activation of CTP:phosphorylcholine cytidylyltransferase in Arabidopsis thaliana at low temperature. Plant Cell Physiol 50: 1727–1735 [DOI] [PubMed] [Google Scholar]
- Jackowski S. (1996) Cell cycle regulation of membrane phospholipid metabolism. J Biol Chem 271: 20219–20222 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Shaw J, Masle J (2009) Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J Biol Chem 284: 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Bligny R, Joyard J, Block MA (2003) Transient increase of phosphatidylcholine in plant cells in response to phosphate deprivation. FEBS Lett 544: 63–68 [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N (2010) Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res 49: 128–158 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K (2005) An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J 43: 107–117 [DOI] [PubMed] [Google Scholar]
- Kaup MT, Froese CD, Thompson JE (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanová M, Lattanzi FA, Grimoldi AA, Schnyder H (2006) Phosphorus deficiency decreases cell division and elongation in grass leaves. Plant Physiol 141: 766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Dörmann P (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Froehlich JE, Dörmann P (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, et al. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009a) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakamura Y, Ohta H (2009b) Type A and type B monogalactosyldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol Biochem 47: 518–525 [DOI] [PubMed] [Google Scholar]
- Koiwai A, Matsuzaki T, Suzuki F, Kawashima N (1981) Changes in total and polar lipids and their fatty acid composition in tobacco leaves during growth and senescence. Plant Cell Physiol 22: 1059–1065 [Google Scholar]
- Kolodziejek I, Koziol J, Waleza M, Mostowska A (2003) Ultrastructure of mesophyll cells and pigment content in senescing leaves of maize and barley. J Plant Growth Regul 22: 217–227 [Google Scholar]
- Kusumi K, Chono Y, Shimada H, Gotoh E, Tsuyama M, Iba K (2010) Chloroplast biogenesis during the early stage of leaf development in rice. Plant Biotechnol 27: 85–90 [Google Scholar]
- Laliberté E, Turner BL, Costes T, Pearse SJ, Wyrwoll KH, Zemunik G, Lambers H (2012) Experimental assessment of nutrient limitation along a 2-million-year dune chronosequence in the south-western Australia biodiversity hotspot. J Ecol 100: 631–642 [Google Scholar]
- Lambers H, Bishop JG, Hopper SD, Laliberté E, Zúñiga-Feest A (2012a) Phosphorus-mobilization ecosystem engineering: the roles of cluster roots and carboxylate exudation in young P-limited ecosystems. Ann Bot (Lond) 110: 329–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Cawthray GR, Giavalisco P, Kuo J, Laliberté E, Pearse SJ, Scheible WR, Stitt M, Teste F, Turner BL (2012b) Proteaceae from severely phosphorus-impoverished soils extensively replace phospholipids with galactolipids and sulfolipids during leaf development to achieve a high photosynthetic phosphorus-use-efficiency. New Phytol 196: 1098–1108 [DOI] [PubMed] [Google Scholar]
- Lambers H, Clode P, Hawkins HJ, Laliberté E, Oliveira R, Reddell P, Shane MW, Stitt M, Weston P (2015) Metabolic adaptations of the non-mycotrophic Proteaceae to soil with a low phosphorus availability. InPlaxton WC, Lambers H, eds, Phosphorus Metabolism in Plants in the Post-genomic Era: From Gene to Ecosystem. Wiley-Blackwell, Oxford, UK (in press) [Google Scholar]
- Lan P, Li W, Schmidt W (2012) Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteomics 11: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer MJ, Pallardy SG, Blevins DG, Randall DD (1989) Whole leaf carbon exchange characteristics of phosphate deficient soybeans (Glycine max L.). Plant Physiol 91: 848–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Collins TM (2001) Phylogenetic and ontogenetic influences on the distribution of anthocyanins and betacyanins in leaves of tropical plants. Int J Plant Sci 162: 1141–1153 [Google Scholar]
- Lee J, Welti R, Roth M, Schapaugh WT, Li J, Trick HN (2012) Enhanced seed viability and lipid compositional changes during natural ageing by suppressing phospholipase Dα in soybean seed. Plant Biotechnol J 10: 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech RM, Rumsby MG, Thomson WW (1973) Plastid differentiation, acyl lipid, and fatty acid changes in developing green maize leaves. Plant Physiol 52: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Yadun S, Dafni A, Flaishman MA, Inbar M, Izhaki I, Katzir G, Ne’eman G (2004) Plant coloration undermines herbivorous insect camouflage. BioEssays 26: 1126–1130 [DOI] [PubMed] [Google Scholar]
- Li M, Welti R, Wang X (2006) Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation: roles of phospholipases D ζ1 and D ζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol 142: 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Li W, Wang R, Li M, Li L, Wang C, Welti R, Wang X (2008) Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J Biol Chem 283: 461–468 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, doi/10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Ac A, Marek MV, Kalina J, Urban O (2007) Differences in pigment composition, photosynthetic rates and chlorophyll fluorescence images of sun and shade leaves of four tree species. Plant Physiol Biochem 45: 577–588 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Leong SJ, Chiou TJ (2014) Long-distance call from phosphate: systemic regulation of phosphate starvation responses. J Exp Bot 65: 1817–1827 [DOI] [PubMed] [Google Scholar]
- Maatta S, Scheu B, Roth MR, Tamura P, Li M, Williams TD, Wang X, Welti R (2012) Levels of Arabidopsis thaliana leaf phosphatidic acids, phosphatidylserines, and most trienoate-containing polar lipid molecular species increase during the dark period of the diurnal cycle. Front Plant Sci 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, Wu H, Fernie AR, Fait A, Stern DB (2009) Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol 151: 905–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal E, Block MA, Dorne AJ, Douce R, Joyard J (1997) Lipid synthesis and metabolism in the plastid envelope. Physiol Plant 100: 65–77 [Google Scholar]
- Marschner H, Kirkby EA, Cakmak I (1996) Effect of mineral nutritional status on shoot-root partitioning of photoassimilates and cycling of mineral nutrients. J Exp Bot 47: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Masle J. (2000) The effects of elevated CO2 concentrations on cell division rates, growth patterns, and blade anatomy in young wheat plants are modulated by factors related to leaf position, vernalization, and genotype. Plant Physiol 122: 1399–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hortensteiner S, Thomas H (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50: 67–95 [DOI] [PubMed] [Google Scholar]
- Matos AR, Pham-Thi AT (2009) Lipid deacylating enzymes in plants: old activities, new genes. Plant Physiol Biochem 47: 491–503 [DOI] [PubMed] [Google Scholar]
- May A, Spinka M, Köck M (2012) Arabidopsis thaliana PECP1: enzymatic characterization and structural organization of the first plant phosphoethanolamine/phosphocholine phosphatase. Biochim Biophys Acta 1824: 319–325 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Kato M, Takahashi Y, Shimazaki K, Tamura K, Tokuji Y, Kihara A, Imai H (2012) Degradation of long-chain base 1-phosphate (LCBP) in Arabidopsis: functional characterization of LCBP phosphatase involved in the dehydration stress response. J Plant Res 125: 439–449 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci USA 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Tsuchiya M, Ohta H (2007) Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin. J Biol Chem 282: 29013–29021 [DOI] [PubMed] [Google Scholar]
- Nerlich A, von Orlow M, Rontein D, Hanson AD, Dörmann P (2007) Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol 144: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, Tanaka H, Matsuda F, Hirai A, Hirai MY, et al. (2009) A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21: 892–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougham HJ, Morris P, Thomas H (2005) The colors of autumn leaves as symptoms of cellular recycling and defenses against environmental stresses. Curr Top Dev Biol 66: 135–160 [DOI] [PubMed] [Google Scholar]
- Penning de Vries FWT, Brunsting AHM, van Laar HH (1974) Products, requirements and efficiency of biosynthesis: a quantitative approach. J Theor Biol 45: 339–377 [DOI] [PubMed] [Google Scholar]
- Pierrugues O, Brutesco C, Oshiro J, Gouy M, Deveaux Y, Carman GM, Thuriaux P, Kazmaier M (2001) Lipid phosphate phosphatases in Arabidopsis: regulation of the AtLPP1 gene in response to stress. J Biol Chem 276: 20300–20308 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokotylo I, Pejchar P, Potocký M, Kocourková D, Krčková Z, Ruelland E, Kravets V, Martinec J (2013) The plant non-specific phospholipase C gene family: novel competitors in lipid signalling. Prog Lipid Res 52: 62–79 [DOI] [PubMed] [Google Scholar]
- Potocký M, Eliás M, Profotová B, Novotná Z, Valentová O, Zárský V (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family: characterization of a calcium-independent and phosphatidylcholine-selective PLD ζ 1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin WS, Pappan K, Wang XM (1997) Molecular heterogeneity of phospholipase D (PLD): cloning of PLD gamma and regulation of plant PLD gamma, -beta, and -alpha by polyphosphoinositides and calcium. J Biol Chem 272: 28267–28273 [DOI] [PubMed] [Google Scholar]
- Rao IM, Terry N (1989) Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet: I. Changes in growth, gas exchange, and Calvin cycle enzymes. Plant Physiol 90: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao IM, Terry N (1995) Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet: IV. Changes with time following increased supply of phosphate to low-phosphate plants. Plant Physiol 107: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. (2013) RNA function and phosphorus use by photosynthetic organisms. Front Plant Sci 4: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Rilfors L, Lindblom G (2002) Regulation of lipid composition in biological membranes: biophysical studies of lipids and lipid synthesizing enzymes. Colloids Surf B Biointerfaces 26: 112–124 [Google Scholar]
- Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Roughan PG, Slack CR (1982) Cellular-organization of glycerolipid metabolism. Annu Rev Plant Physiol Plant Mol Biol 33: 97–132 [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Hagio M, Gombos Z, Tyystjarvi T, Paakkarinen V, Aro EM, Wada H (2003) Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiol 133: 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C (2001) Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J Biol Chem 276: 3941–3946 [DOI] [PubMed] [Google Scholar]
- Shane MW, de Vos M, de Roock S, Cawthray GR, Lambers H (2003) Effects of external phosphorus supply on internal phosphorus concentration and the initiation, growth and exudation of cluster roots in Hakea prostrata R. Br. Plant Soil 248: 209–219 [Google Scholar]
- Shane MW, Lambers H (2005) Cluster roots: a curiosity in context. Plant Soil 274: 101–125 [Google Scholar]
- Shane MW, McCully ME, Lambers H (2004) Tissue and cellular phosphorus storage during development of phosphorus toxicity in Hakea prostrata (Proteaceae). J Exp Bot 55: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Shimojima M. (2011) Biosynthesis and functions of the plant sulfolipid. Prog Lipid Res 50: 234–239 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Ohta H (2011) Critical regulation of galactolipid synthesis controls membrane differentiation and remodeling in distinct plant organs and following environmental changes. Prog Lipid Res 50: 258–266 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Ohta H, Nakamura Y (2009) Biosynthesis and function of chloroplast lipids. Lipids in Photosynthesis: Essential and Regulatory Functions 30: 35–55 [Google Scholar]
- Shimojima M, Watanabe T, Madoka Y, Koizumi R, Yamamoto MP, Masuda K, Yamada K, Masuda S, Ohta H (2013) Differential regulation of two types of monogalactosyldiacylglycerol synthase in membrane lipid remodeling under phosphate-limited conditions in sesame plants. Front Plant Sci 4: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Ishihara H, Schlereth A, Cawthray GR, Encke B, Giavalisco P, Ivakov A, Arrivault S, Jost R, Krohn N, et al. (2014) Low levels of ribosomal RNA partly account for the very high photosynthetic phosphorus-use efficiency of Proteaceae species. Plant Cell Environ 37: 1276–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski J, Brotman Y, Willmitzer L, Cuadros-Inostroza Á (2014) Linking gene expression and membrane lipid composition of Arabidopsis. Plant Cell 26: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Froese CD, Madey E, Smith MD, Hong Y (1998) Lipid metabolism during plant senescence. Prog Lipid Res 37: 119–141 [DOI] [PubMed] [Google Scholar]
- Tjellström H, Andersson MX, Larsson KE, Sandelius AS (2008) Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ 31: 1388–1398 [DOI] [PubMed] [Google Scholar]
- Tjellström H, Hellgren LI, Wieslander A, Sandelius AS (2010) Lipid asymmetry in plant plasma membranes: phosphate deficiency-induced phospholipid replacement is restricted to the cytosolic leaflet. FASEB J 24: 1128–1138 [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, Cao X, Yang Z, Ohlrogge JB (2013) Lipid turnover during senescence. Plant Sci 205-206: 13–19 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, et al. (2012) Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol 195: 306–320 [DOI] [PubMed] [Google Scholar]
- Wang X. (2005) Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol 139: 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Gamat G, Ismail AM (2005) Is root growth under phosphorus deficiency affected by source or sink limitations? J Exp Bot 56: 1943–1950 [DOI] [PubMed] [Google Scholar]
- Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH (2012) The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright IJ, Westoby M, Reich PB (2002) Convergence towards higher leaf mass per area in dry and nutrient-poor habitats has different consequences for leaf life span. J Ecol 90: 534–543 [Google Scholar]
- Xu C, Härtel H, Wada H, Hagio M, Yu B, Eakin C, Benning C (2002) The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol 129: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu B, Cornish AJ, Froehlich JE, Benning C (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Yu B, Benning C (2003) Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J 36: 762–770 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa Y, Nishihara H, Haraguchi T, Masuda S, Shimojima M, Shimoyama A, Yuasa H, Okada N, Ohta H (2012) Phylogeny of galactolipid synthase homologs together with their enzymatic analyses revealed a possible origin and divergence time for photosynthetic membrane biogenesis. DNA Res 19: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasoski RJ, Burau RG (1977) A rapid nitric‐perchloric acid digestion method for multi‐element tissue analysis. Commun Soil Sci Plant Anal 8: 425–436 [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Yang QY, Lee DW, Goldstein G, Cao KF (2013) Extended leaf senescence promotes carbon gain and nutrient resorption: importance of maintaining winter photosynthesis in subtropical forests. Oecologia 173: 721–730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.