Infection of barley with bacteria induces systemic resistance against a secondary Xanthomonas translucens infection and does not appear to be associated with salicylic acid.
Abstract
Leaf-to-leaf systemic immune signaling known as systemic acquired resistance is poorly understood in monocotyledonous plants. Here, we characterize systemic immunity in barley (Hordeum vulgare) triggered after primary leaf infection with either Pseudomonas syringae pathovar japonica (Psj) or Xanthomonas translucens pathovar cerealis (Xtc). Both pathogens induced resistance in systemic, uninfected leaves against a subsequent challenge infection with Xtc. In contrast to systemic acquired resistance in Arabidopsis (Arabidopsis thaliana), systemic immunity in barley was not associated with NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 or the local or systemic accumulation of salicylic acid. Instead, we documented a moderate local but not systemic induction of abscisic acid after infection of leaves with Psj. In contrast to salicylic acid or its functional analog benzothiadiazole, local applications of the jasmonic acid methyl ester or abscisic acid triggered systemic immunity to Xtc. RNA sequencing analysis of local and systemic transcript accumulation revealed unique gene expression changes in response to both Psj and Xtc and a clear separation of local from systemic responses. The systemic response appeared relatively modest, and quantitative reverse transcription-polymerase chain reaction associated systemic immunity with the local and systemic induction of two WRKY and two ETHYLENE RESPONSIVE FACTOR (ERF)-like transcription factors. Systemic immunity against Xtc was further associated with transcriptional changes after a secondary/systemic Xtc challenge infection; these changes were dependent on the primary treatment. Taken together, bacteria-induced systemic immunity in barley may be mediated in part by WRKY and ERF-like transcription factors, possibly facilitating transcriptional reprogramming to potentiate immunity.
To protect themselves from microbial pathogens, plants are equipped with an array of defense strategies, one of which is the ability to prime defense. Primed plants are in a state of heightened alert, allowing a faster and stronger reaction to pathogen attack, compared with naive, unprimed plants (Conrath et al., 2006; Conrath, 2011). A similar state of heightened alert is established in systemic, uninfected tissues of plants undergoing a primary infection in either aboveground or belowground tissues. Depending on the site of the primary infection and the virulence of the attacker, this form of induced resistance is often referred to as induced systemic resistance (ISR) or systemic acquired resistance (SAR). ISR is triggered on the colonization of plant roots by nonpathogenic soil microbes and protects aboveground tissues of both dicots and monocots from necrotrophic pathogens and pests (Pieterse et al., 2012; Balmer et al., 2013b; Walters et al., 2013). SAR, on the other hand, is induced in systemic, uninfected tissues of a plant on prior foliar pathogen challenge and is predominantly effective against biotrophic pathogens (for review, see Vlot et al., 2009; Fu and Dong, 2013). Although SAR in dicots is mainly studied as a leaf-to-leaf response, leaf-to-root SAR-like immune signaling was recently reported in the monocot banana (Musa spp.; Wu et al., 2013).
In dicots, including Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum), SAR is associated with salicylic acid (SA)-mediated immune signaling (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013) triggered during a primary infection of the plant upon recognition of either pathogen-associated molecular patterns or pathogen effectors (Cameron et al., 1994; Jones and Dangl, 2006; Mishina and Zeier, 2007; Liu et al., 2010; Spoel and Dong, 2012; Breitenbach et al., 2014). Pathogen-associated molecular patterns induce pathogen-associated molecular pattern-triggered immunity (PTI), which reduces the in planta propagation of virulent pathogens (Jones and Dangl, 2006; Henry et al., 2013). Effector-triggered immunity (ETI) in response to the direct or indirect recognition of pathogen effectors by canonical plant RESISTANCE proteins strongly impedes the growth of avirulent pathogens (Jones and Dangl, 2006; Bonardi and Dangl, 2012; Henry et al., 2013). ETI is a faster and stronger response that, in contrast to PTI, culminates in hypersensitive response-mediated death of the infected site and surrounding cells, restricting the pathogen to the site of infection (Jones and Dangl, 2006; Tsuda et al., 2009; Tsuda and Katagiri, 2010; Maekawa et al., 2011; Henry et al., 2013).
SAR in dicots is associated with several putative long-distance signals, some of which appear to be conditionally required for SAR (Attaran et al., 2009; Liu et al., 2011). These signals include the methylated derivative of SA methyl salicylate (Park et al., 2007), glycerol-3-phosphate (Chanda et al., 2011), the diterpenoid dehydroabietinal (Chaturvedi et al., 2012), the C9 dicarboxylic acid azelaic acid (Jung et al., 2009), and the lipid transfer proteins DEFECTIVE IN INDUCED RESISTANCE1 (DIR1; Maldonado et al., 2002), DIR1-like (Champigny et al., 2013), and AZELAIC ACID INDUCED1 (AZI1; Jung et al., 2009). Glycerol-3-phosphate might be part of a positive feedback loop, with DIR1 and AZI1 acting downstream of azelaic acid (Yu et al., 2013; Gao et al., 2014; Shah et al., 2014), which accumulates by autooxidation of C18 unsaturated fatty acids, possibly downstream of nitric oxide and reactive oxygen species, to promote SAR (Zoeller et al., 2012; Wang et al., 2014; Wittek et al., 2014). In addition, the nonprotein amino acid pipecolic acid is essential for SAR (Návarová et al., 2012; Shah and Zeier, 2013; Zeier, 2013). Signaling in the systemic SAR signal-perceiving tissue appears to converge on FLAVIN-DEPENDENT MONOOXYGENASE1 (Mishina and Zeier, 2006) and/or SA for the propagation or maintenance of immune signaling in the systemic tissue (Dempsey and Klessig, 2012; Shah and Zeier, 2013), leading to the expression of SAR marker genes, including PATHOGENESIS-RELATED1 (PR1; van Loon et al., 2006).
Local resistance, induced by either chemicals or pathogens, appears correlated with a function of SA in monocots. Application of SA or one of its functional analogs, 2,6-dichloroisonicotinic acid or S-methyl benzo-1,2,3-thiadiazole-7-carbothioate (BTH), induces local acquired resistance (LAR) in barley (Hordeum vulgare), wheat (Triticum aestivum), and maize (Zea mays; Kogel et al., 1994; Görlach et al., 1996; Morris et al., 1998; Makandar et al., 2012). Similarly, probenazole, an elicitor of SA-dependent immunity in Arabidopsis (Yoshioka et al., 2001), protects maize from corn leaf blight caused by Cochliobolus heterostrophus (Yang et al., 2011) and rice (Oryza sativa) from diseases including rice blast by a mechanism that is correlated with SA homeostasis (Umemura et al., 2009; Walters et al., 2013). The master regulator of SA signaling, NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), is conserved between dicots and monocots (Kogel and Langen, 2005; Balmer et al., 2013b), and overexpression of AtNPR1 either primes or enhances SA-associated disease resistance in wheat or rice, respectively (Makandar et al., 2006, 2012; Balmer et al., 2013b; Sharma et al., 2013). Irrespective of the importance of SA, signals contained in petiole (phloem) exudates from SAR signal-emitting Arabidopsis leaves effectively protect wheat from head blight caused by Fusarium graminearum (Chaturvedi et al., 2008). Furthermore, transcriptional changes associated with a LAR-like immune response in distal parts of barley leaves adjacent to sites inoculated with Pseudomonas syringae pv tomato carrying the effector locus AvrRpm1 revealed commonalities with Arabidopsis SAR (Colebrook et al., 2012). Taken together, signaling associated with induced resistance, including a role of SA, appears relatively conserved between dicots and monocots.
Early reports of biologically induced (SAR-like) systemic immunity in monocots include enhanced resistance against virulent Erisyphe graminis f. sp. hordei in systemic, uninfected leaves of barley preinfected with virulent or avirulent isolates of the same pathogen (Hwang and Heitefuss, 1982) and enhanced resistance against Pyricularia oryzae in systemic tissues of rice preinfected with P. syringae (Smith and Metraux, 1991). Systemic immunity protecting banana from Fusarium spp. wilt is induced by infection of a leaf with an avirulent isolate of Fusarium oxysporum and is accompanied by increased SA levels in the roots, the site of the secondary challenge inoculation (Wu et al., 2013). Similarly, infection of a maize leaf with Colletotrichum graminicola induces SA accumulation and resistance against a secondary C. graminicola challenge infection in systemic leaves (Balmer et al., 2013a). Xanthomonas oryzae pv oryzae causes bacterial blight in rice and induces systemic resistance against bacterial streak disease caused by X. oryzae pv oryzicola in transgenic rice plants with suppressed expression of MITOGEN-ACTIVATED PROTEIN (MAP) KINASE6 (Shen et al., 2010). Suppression of MAP KINASE6 is further associated with elevated SA levels, while the expression of PR1a is induced in systemic, uninfected leaves during the establishment of systemic immunity. Finally, systemic resistance in wheat against stripe rust (Puccinia striiformis f. sp. tritici) is induced by a local infection with an incompatible isolate of the same pathogen and is associated with glycerol-3-phosphate (Yang et al., 2013), suggesting that systemic immunity in wheat depends on similar mechanisms as SAR in Arabidopsis.
Although a growing body of evidence points to a role of SA in SAR-like immunity in monocots, there is little detailed knowledge on signaling cascades in monocot SAR. Here, we show that infection of a barley leaf with one of two bacterial pathogens induces an immune response in systemic, uninfected leaves that does not appear to be associated with SA. Instead, systemic immunity is associated with the accumulation in the systemic tissue of gene transcripts encoding transcription factors (TFs), including WRKY and ETHYLENE RESPONSIVE FACTOR (ERF)-like TFs. Transcriptional changes after the secondary infection suggest that these TFs potentiate immunity upon challenge infection of the systemic tissue.
RESULTS
Bacterial Growth Curves on Barley ‘Golden Promise’ and ‘Barke’
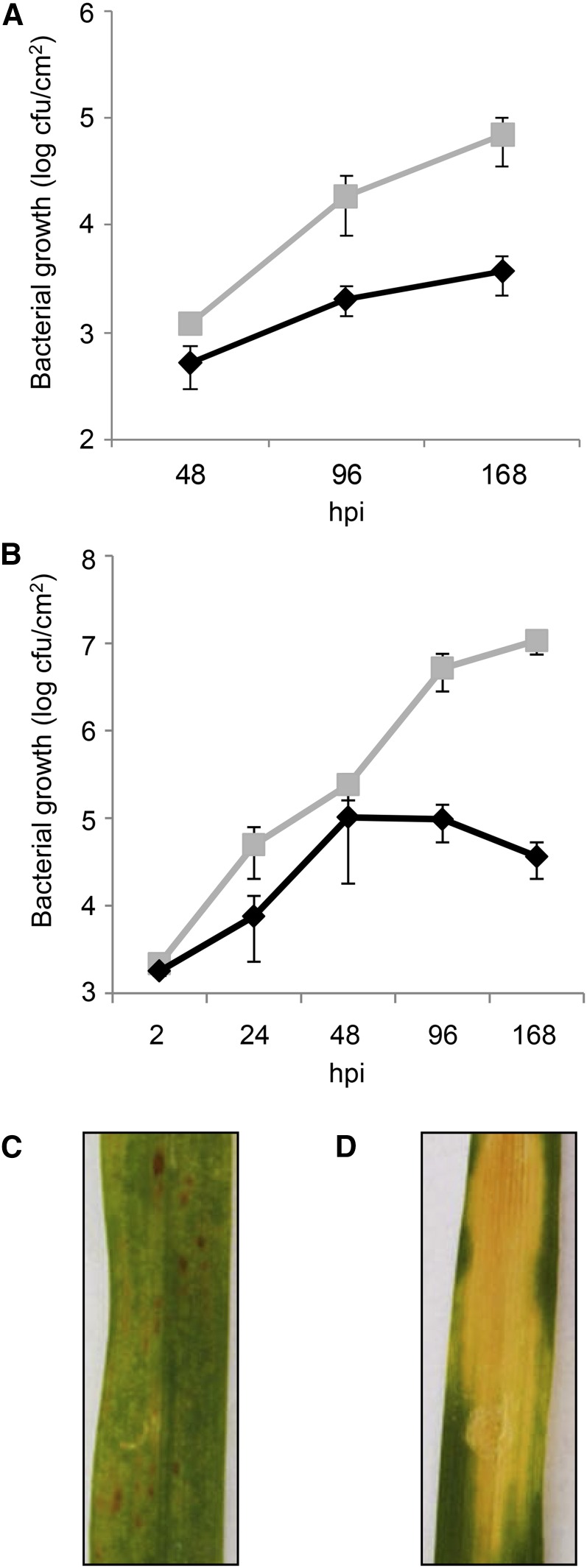
P. syringae pv japonica (Psj) and Xanthomonas translucens pv cerealis (Xtc) were selected from a panel of bacteria analyzed previously for their virulence on barley (Morrissey, 2007). Here, we infected leaves of 4-week-old barley ‘Golden Promise’ (GP) and ‘Barke’ plants by spray inoculation and syringe infiltration, respectively, and monitored the growth of the bacteria in the infected leaves (Fig. 1, A and B). After spray inoculation of GP plants, Psj displayed a slow but steady growth on barley leaves until at least 7 d post inoculation (dpi), whereas Xtc titers reached higher levels especially at 4 and 7 dpi (Fig. 1A). After syringe infiltration of ‘Barke’ plants, Psj displayed growth until 2 dpi, after which the bacterial density in the leaf remained the same or was reduced at 7 dpi (Fig. 1B). Because both spray and infiltration inoculations were followed by limited Psj propagation that was accompanied by the appearance of small brown spots reminiscent of hypersensitive response lesions (Fig. 1C; Supplemental Fig. S1A), Psj appeared to be avirulent on barley GP and ‘Barke.’ In line with this hypothesis, we did not detect Psj in leaves systemic to the site of inoculation, suggesting that the bacteria did not spread from infected to systemic sites (Supplemental Fig. S1C). By contrast, Xtc appeared virulent, growing to high titers after spray and infiltration inoculation of GP and ‘Barke’ plants, respectively (Fig. 1, A and B). Symptoms were hardly if at all visible at 7 d after spray inoculation of GP plants (Supplemental Fig. S1B) and included spreading yellowing lesions at 7 d after infiltration inoculation of ‘Barke’ plants (Fig. 1D), most likely due to the higher in planta Xtc titers reached after infiltration compared with spray inoculation (compare Fig. 1, B and A). At 7 and 9 d after infiltration inoculation of ‘Barke’ plants, Xtc titers in the range of 100 to 200 colony-forming units (cfu) cm−2 leaf were detected in leaves that were systemic to the site of inoculation (Supplemental Fig. S1C), suggesting that the virulent bacteria spread in barley plants.
Figure 1.
Primary infections. A and B, Titers of Psj (black diamonds) and Xtc (gray squares) in barley GP leaves at 48, 96, and 168 h after spray inoculation (A) and in barley ‘Barke’ from 2 to 168 h after infiltration inoculation (B). Values indicated are means of four replicates ± sd. C and D, Representative images of ‘Barke’ leaves 7 d after infiltration with Psj (C) or Xtc (D). These experiments were repeated two times with similar results and confirmed by at least five independent bacterial titer measurements at 120 h post (infiltration) inoculation (hpi [5 dpi]; Supplemental Fig. S9; data not shown). [See online article for color version of this figure.]
Psj and Xtc Trigger Systemic Resistance against Xtc
In order to investigate if barley plants are capable of mounting a systemic defense response, the first true leaf of 4-week-old ‘Barke’ plants was infected by infiltration inoculation with 106 cfu mL−1 either Psj or Xtc or treated with 10 mm MgCl2 as a mock control. Five days later, the next two leaves (leaves 2 and 3) were challenge infected by infiltration inoculation with 105 cfu mL−1 Xtc, and the resulting in planta Xtc titers were determined at 4 dpi. Because Xtc titers were significantly lower in challenge-infected leaves 2 and 3 of preinfected compared with mock-pretreated plants (Fig. 2A), the primary infections with Psj or Xtc had induced systemic resistance. Pilot experiments, in which the systemic Xtc challenge infection was performed at 3, 4, 5, 6, or 7 d after primary treatment, showed that systemic immunity was activated from 3 d after primary treatment (in one out of two experiments) until at least 5 d after primary treatment (Supplemental Fig. S2). Systemic resistance triggered by Psj or Xtc at 5 dpi was a robust response seen under greenhouse conditions in all tested barley cultivars, including ‘Barke’ (Fig. 2A), GP (Fig. 2E), and ‘Ingrid’ (Supplemental Fig. S3). Because Xtc was mobile in ‘Barke’ plants (Supplemental Fig. S1C), we cannot exclude that a subfraction of bacteria detected in the secondary infected tissue of Xtc-preinfected plants was derived from the primary inoculation. This subfraction could have caused LAR against the secondary infection. However, as Psj was immobile in ‘Barke,’ the immunity, detected systemically to this primary inoculum, was a phenomenon of true systemic immunity.
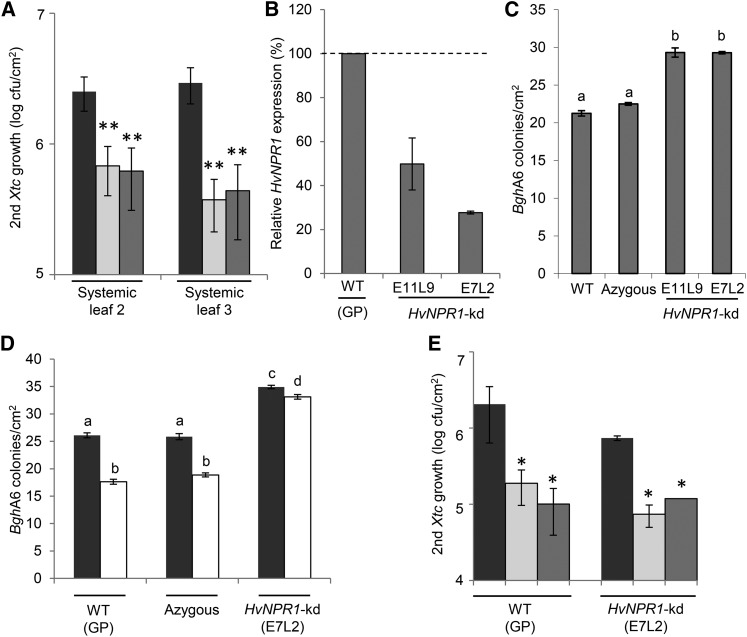
Figure 2.
Psj and Xtc induce systemic immunity in barley independently of HvNPR1. A, Barley ‘Barke’ plants were pretreated in leaf 1 with 10 mm MgCl2 (mock; black bars), Xtc (light gray bars), or Psj (dark gray bars). Five days later, (systemic) leaves 2 and 3 were infected with Xtc. Xtc titers in both systemic secondary (second) infected leaves are shown at 4 dpi. Values indicated are means of four replicates ± sd. B, HvNPR1 transcript accumulation was normalized to that of HvUBIQUITIN in GP wild-type (WT) plants and in T1 plants of HvNPR1-kd lines E11L9 and E7L2. The normalized HvNPR1 transcript accumulation in the wild type was set at 100%. Values indicated are means ± sd of two independent experiments consisting of three technical replicates each. C, First leaf segments of 7-d-old T1 seedlings of the genotypes indicated at bottom were infected with BghA6, and the resulting number of BghA6 colonies per cm2 was determined at 6 dpi. Values indicated are means of two experiments ± se, respectively, including 13 and 22 plants of each genotype. D, Five-day-old T1 seedlings of the genotypes indicated at bottom were treated by soil drench with water-solved wettable powder (black bars) or with BTH (white bars), and first leaf segments were infected with BghA6 2 d later. The resulting number of BghA6 colonies per cm2 was determined at 7 dpi. Values indicated are means ± se of 13 to 21 plants per genotype and pretreatment. E, Systemic immunity was induced and analyzed as in A in four plants of each genotype as indicated at bottom. In A and E, asterisks indicate statistically significant differences from the mock controls (*P < 0.05, **P < 0.01, Student’s t test). In C and D, results marked with different letters above the bars are statistically different (P < 0.01, Student’s t test [C] or one-way ANOVA [D]). These experiments were repeated two times (E) to at least three times (A–D) with similar results.
Bacteria-Triggered Systemic Immunity Is Independent of HvNPR1
Because SAR in Arabidopsis is dependent on the key SA signaling regulator NPR1 (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013; Fu and Dong, 2013), we investigated if systemic immunity in barley is associated with the barley NPR1 homolog HvNPR1 HOMOLOG1 (NH1; referred to as HvNPR1 throughout this work). HvNPR1 shared 78% identity at the amino acid level with both rice NH1 and maize NPR1 and 83% identity with Brachypodium distachyon NPR1-like (Supplemental Fig. S4). Here, we knocked down HvNPR1 transcript accumulation in transgenic barley GP plants by RNA interference (RNAi)-mediated gene silencing. Two independent HvNPR1 knockdown (kd) lines, E11L9 and E7L2, respectively accumulated approximately 45% and 30% of the wild-type HvNPR1 transcript level (Fig. 2B). Seven-day-old segregating T1 seedlings of both lines were infected with the virulent isolate A6 of the powdery mildew fungus Blumeria graminis f. sp. hordei (BghA6). Fungal growth was documented by monitoring the number of B. graminis f. sp. hordei (Bgh) pustules (colonies) per cm2 of leaf at 6 dpi (Fig. 2C). In parallel, genomic DNA of each plant was analyzed by PCR for the presence or absence of the RNAi construct, and the plants of each line that had lost the RNAi construct (by segregation) were pooled and considered as the azygous control group. Detached leaves of both E11L9 and E7L2 HvNPR1-kd plants displayed enhanced susceptibility to BghA6 compared with the GP wild-type and azygous controls (Fig. 2C), suggesting that resistance in barley against Bgh was at least in part mediated by HvNPR1.
Subsequently, we investigated if resistance induced by the SA functional analog BTH is associated with HvNPR1 in barley. For comparison, AtNPR1 is required for BTH-induced priming of defense-associated molecular events in Arabidopsis (Kohler et al., 2002). It was previously shown that soil-drench treatment of barley seedlings with BTH enhances resistance to Bgh in the leaves of treated plants (Beßer et al., 2000; Fig. 2D). Here, 5-d-old GP wild-type seedlings and segregating T1 seedlings of the transgenic line that accumulated the lowest level of HvNPR1 transcripts (E7L2; Fig. 2B) were treated with BTH by soil drench as described in “Materials and Methods.” Two days later, detached leaves of the treated plants were infected with BghA6, and the resulting BghA6 colonies were counted at 7 dpi. As expected (Beßer et al., 2000), BTH enhanced resistance to BghA6 in the GP wild-type plants and in the azygous control group (determined by PCR as described above; Fig. 2D). Also, a low and significant BTH-induced resistance to BghA6 was observed in the HvNPR1-kd plants, which we attributed to the remaining approximately 30% of HvNPR1 transcripts in the E7L2 compared with wild-type plants (Fig. 2B). BTH-induced resistance to BghA6 was compromised in the HvNPR1-kd plants compared with the wild-type and azygous control plants (Fig. 2D), suggesting that BTH acts through HvNPR1 in barley.
In contrast to basal and BTH-induced resistance to BghA6, which were compromised in the HvNPR1-kd plants, systemic immunity to Xtc induced by primary inoculations of homozygous (T4) plants of HvNPR1-kd line E7L2 with Psj or Xtc was unchanged compared with systemic immunity in GP wild-type plants (Fig. 2E). The HvNPR1 transcript levels in the E7L2 plants that were used in the experiment shown in Figure 2E were approximately 30% of the level in the GP wild-type plants from the same experiment, as determined by quantitative reverse transcription (qRT)-PCR (Supplemental Fig. S5A). Also, homozygous T4 E7L2 plants displayed compromised BTH-induced resistance to BghA6 compared with wild-type plants (Supplemental Fig. S5B). Taken together, the data suggest that HvNPR1 is required for full basal and BTH-induced immunity to Bgh but not for bacteria-triggered systemic immunity in barley.
Local and Systemic Accumulation of SA, Jasmonic Acid, and Abscisic Acid during Systemic Immune Signaling
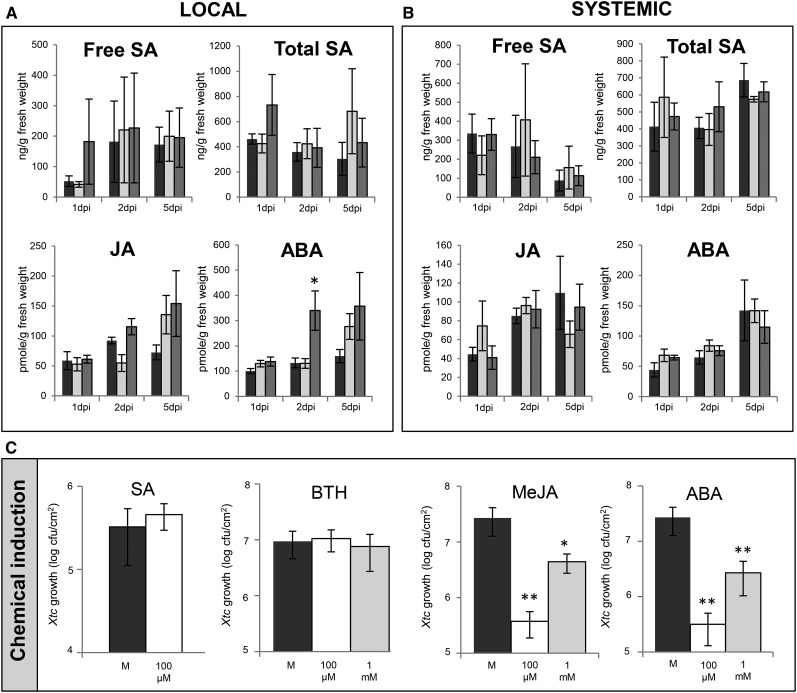
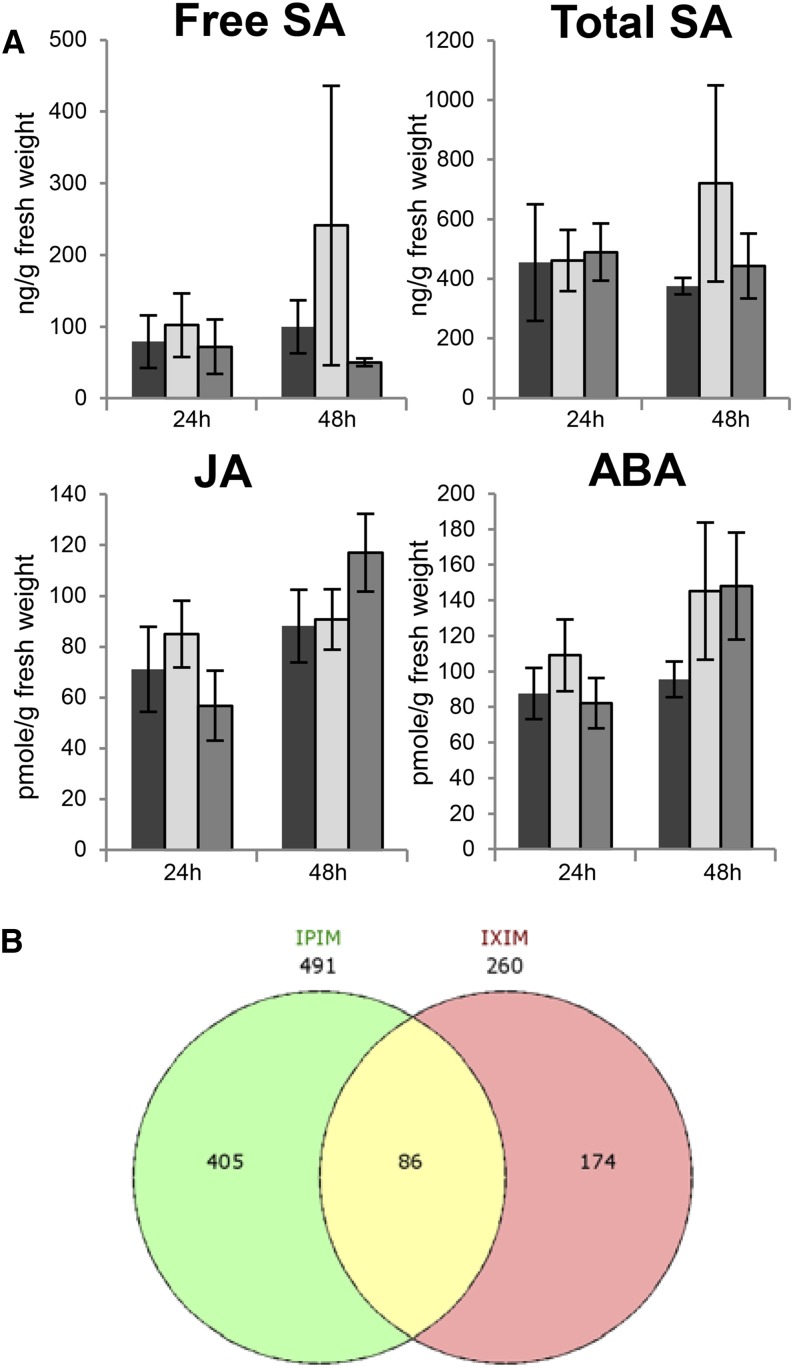
In dicots, SAR is strongly associated with SA (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013; Fu and Dong, 2013), whereas ISR is associated with jasmonic acid (JA)/ethylene (ET) and abscisic acid (ABA) signaling (Pieterse et al., 2012; Balmer et al., 2013b; Walters et al., 2013). In addition, induced resistance in maize leaves after infection of the roots with C. graminicola is associated with the induction of SA and ABA accumulation in the leaves (Balmer et al., 2013a). Here, we compared the levels of SA, JA, and ABA in local treated and systemic untreated leaves of mock-treated barley GP plants and GP plants that were infected with either Psj or Xtc. We investigated the SA, JA, and ABA accumulation relatively early after infection, at 1 and 2 dpi, arguing that essential signaling steps might occur prior to the establishment of systemic immunity. In addition, we measured the local and systemic SA, JA, and ABA levels at 5 dpi, the time point of the systemic challenge infection. First, we did not detect significant differences in the accumulation of free or total SA in the local treated or systemic tissue of infected compared with mock-treated plants (Fig. 3, A and B). At 1 dpi, the accumulation of free and total SA in leaves that had been infected with Psj was moderately elevated, but the difference from mock-treated plants was not significant. Similarly, we observed a slight increase in the level of JA in leaves that were infected with Psj or Xtc compared with that in mock-treated leaves at 5 dpi, but this was not significant (Fig. 3A). By contrast, a significant ABA induction was observed in Psj-infected compared with mock-treated leaves at 2 dpi (Fig. 3A). The Psj-infected leaves maintained a similar level of ABA until 5 dpi, whereas the ABA levels in mock-treated and Xtc-infected leaves continued to rise, reaching comparable levels to those in the Psj-infected tissue at 5 dpi. In the systemic, uninfected leaves, we did not observe differences in JA or ABA accumulation after a local infection with Psj or Xtc or after a local mock treatment (Fig. 3B). Together, bacteria-induced systemic immunity in barley does not appear to be associated with the local or systemic accumulation of SA but might be associated with a moderate local induction of JA and especially ABA. Because changes in the accumulation of JA and ABA in response to infection remained local and were small and mostly insignificant, we speculate that these phytohormones, similar to SA, might not play a prominent role in the establishment of systemic immunity in barley.
Figure 3.
Relationship between systemic immunity in barley and the phytohormones SA, JA, and ABA. A and B, Free SA, total SA, JA, and ABA levels in local barley GP leaves that were treated with 10 mm MgCl2 (mock; black bars) or infected with Xtc (light gray bars) or Psj (dark gray bars; A) and in systemic untreated leaves to the same treatments (B). Samples were taken at 1, 2, and 5 dpi. SA levels are means ± se of three biologically independent replicates (with the exception of two replicates for free SA on local Xtc infection at 1 dpi). JA and ABA levels are means ± se of at least five technical replicates from at least three biologically independent experiments. C, Systemic resistance induced by SA, the functional SA analog BTH, or by MeJA or ABA in barley GP. Plants were treated in leaf 1 with water (mock [M] for BTH) or 0.06% ethanol (mock for SA, MeJA, and ABA) or with 100 µm or 1 mm SA, BTH, MeJA, or ABA as indicated. Five days later, systemic leaf 2 was infected with Xtc. Xtc titers in the challenge-infected tissue are shown at 4 dpi as averages of five replicates ± sd. These experiments were repeated at least three times with similar results. In A and C, asterisks above the bars indicate statistically significant differences from mock treatment (*P < 0.05, **P < 0.01, Student’s t test).
Jasmonic Acid Methyl Ester and ABA Applications Trigger Systemic Resistance to Xtc
Although we found only moderate differences, if any, in the regulation of SA, JA, and ABA accumulation during local or systemic responses, we tested if these phytohormones trigger a systemic immune response against Xtc in barley. First, treatment of the first true leaf (leaf 1) of 4-week-old barley GP plants with two different concentrations of BTH induced the expression of the SA-responsive BARLEY CHEMICALLY INDUCED4 gene (Beßer et al., 2000) in the treated leaves (Supplemental Fig. S6A). However, growth of an Xtc inoculum in the systemic tissue was not restricted, indicating that BTH did not induce systemic immunity against Xtc (Fig. 3C). For comparison, a local application of 100 µm BTH induced SAR in Arabidopsis ecotype Columbia-0, reducing the growth of a systemic P. syringae pv tomato inoculum compared with that in negative control plants to similar levels as in the positive control, in which SAR was induced with avirulent P. syringae pv tomato delivering the effector AvrRpm1 (Supplemental Fig. S6B). Similar to BTH, a local application of 100 µm SA did not trigger systemic immunity against Xtc in barley GP plants. By contrast, treatment of leaf 1 of 4-week-old GP plants with 100 µm or 1 mm jasmonic acid methyl ester (MeJA) or ABA triggered resistance against Xtc in the systemic, untreated tissue, with the lower concentrations consistently inducing a stronger systemic resistance response than the higher concentrations (Fig. 3C). Although ET often signals together with JA (Pieterse et al., 2012), a local application of the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC) did not trigger systemic immunity against Xtc in GP plants (Supplemental Fig. S7A). Taken together, it seems unlikely that SA is involved in bacteria-induced systemic immunity in barley, whereas we cannot exclude a role of JA and ABA signaling in this response.
Local and Systemic Gene Regulation Associated with Systemic Immunity
To better understand the molecular basis of leaf-to-leaf systemic immunity triggered by Psj or Xtc, we generated RNA sequencing (RNA-seq) data of samples of (local) infected and systemic, uninfected leaves of ‘Barke’ plants that were infected with Psj or Xtc or given a mock treatment. Because systemic resistance was most reproducible if the systemic challenge infection was performed at 5 d after primary treatment (Fig. 2; Supplemental Fig. S2), transcript accumulation was analyzed at 5 dpi. We generated complementary DNA (cDNA) libraries from two biologically independent replicates of local (leaf 1) Xtc-infected tissue and three biologically independent replicates of each of the following conditions: local, mock-treated tissue and local, Psj-infected tissue as well as the systemic, untreated tissues (leaves 2 and 3) of mock-treated, Psj-infected, and Xtc-infected plants. At least 25 million reads were sequenced (100-bp read length) from each cDNA library and mapped on the barley ‘Morex’ genome (Mayer et al., 2012). The transcriptome mapping percentage was comparable to Mayer et al. (2012), with between 75% and 80% expressed genes out of a total of 24,440 annotated barley genes (Supplemental Fig. S8). As experimental controls, we monitored the local Psj and Xtc titers at 5 dpi as well as systemic immunity against Xtc in plants of two of the experiments that were included in the RNA-seq analysis (Supplemental Fig. S9, A and B).
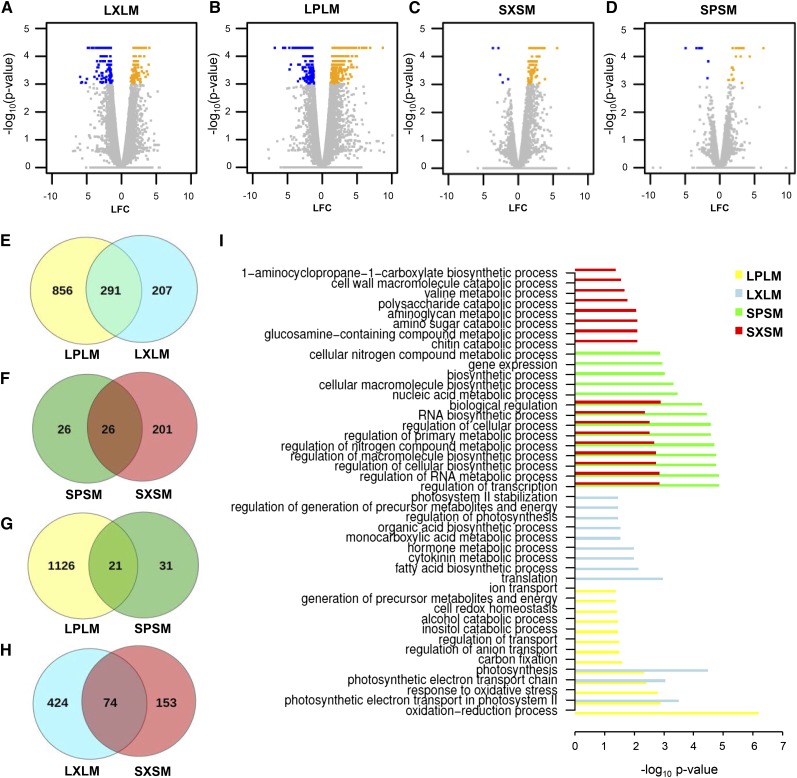
Transcript accumulation abundance was estimated using fragments per kilobase of transcript per million mapped reads as reported by Cufflinks (Trapnell et al., 2009), and differential expression between samples from infected and mock-treated plants was quantified using Cuffdiff (Trapnell et al., 2012). Subsequently, we selected sets of genes that were differentially expressed (false discovery rate [FDR]-adjusted P < 0.05) upon Psj or Xtc infection as compared with the mock treatment either locally or systemically (Fig. 4, A–D; Supplemental Table S1). In local infected leaves, the presumed incompatible interaction between ‘Barke’ plants and Psj induced changes in the accumulation of more than twice as many transcripts compared with the compatible interaction with Xtc, with limited overlap between transcripts regulated by Psj or Xtc (Fig. 4E). By contrast, fewer transcripts were regulated in the systemic, uninfected tissue of Psj-infected compared with Xtc-infected plants (Fig. 4F). Irrespective of the bacterial inducer, the expression of considerably fewer genes was regulated in the systemic, uninfected tissue compared with the infected tissue, with transcripts regulated in the systemic tissue hardly overlapping with their counterparts in the local, infected tissue (Fig. 4, G and H). Taken together, Psj and Xtc triggered partially distinct transcriptional profiles, with both systemic responses clearly differing from responses in the local, infected tissue.
Figure 4.
RNA-seq analysis of local and systemic transcript accumulation during the establishment of systemic immunity. A to D, Volcano plots summarizing transcript accumulation in local Xtc-infected (LX; A) or local Psj-infected (LP; B) tissue relative to local mock-treated (LM) tissue and transcript accumulation in systemic leaves of the same Xtc-infected (SX; C) or Psj-infected (SP; D) plants relative to mock-treated (SM) plants. The data represent means of two (LX) to three biologically independent replicates. The −log10 P value on the y axis represents an FDR-corrected P value, and transcripts with P < 0.05 are depicted in yellow for up-regulated transcripts and in blue for down-regulated transcripts. E to H, Venn diagrams comparing transcripts in data sets LPLM and LXLM (E), SPSM and SXSM (F), LPLM and SPSM (G), and LXLM and SXSM (H). I, Statistically significant GO terms (P < 0.05, conditional hypergeometric tests) of the domain Biological Process enriched in data sets LPLM (yellow), LXLM (blue), SPSM (green), and SXSM (red). LFC, Log fold change.
In support of the limited overlap in the genes that were regulated in the infected compared with the systemic, uninfected tissues (Fig. 4, G and H), the corresponding enriched Gene Ontology (GO) terms of the domain Biological Process did not overlap between the respective data sets (Fig. 4I). Also, limited overlap was observed in the enriched GO terms that were associated with responses to Psj and Xtc either locally or systemically (Fig. 4, E, F, and I). Since both pathogens induced systemic immunity, we reasoned that the genes and the associated GO terms that were regulated in response to both infections could be central to the mechanism of systemic immunity. Three GO terms were commonly enriched among transcripts regulated in local Psj- or Xtc-infected tissue (Fig. 4I). These were photosynthesis and two terms related to photosynthetic electron transport. Systemically, the GO terms that were enriched among transcripts that were regulated by both infections were mostly related to metabolic and biosynthetic processes of RNA and other macromolecules.
In the systemic, uninfected tissue, 26 transcripts were commonly regulated by Psj and Xtc, displaying the same direction and a similar extent of change irrespective of the pathogen used (Fig. 4F; Supplemental Table S1). Eleven of these transcripts appeared to code for TFs (Table I). Of these, four transcripts encoded zinc finger-containing proteins that might act as TFs (Gangappa and Botto, 2014) and one transcript encoded a protein with a plant TF-associated B3 domain (MLOC_71395; Swaminathan et al., 2008). Five additional transcripts encoded proteins that were annotated as WRKY TFs (MLOC_45055, HvWRKY22; MLOC_52504, HvWRKY28; and MLOC_60890, HvWRKY38/1; Table I) or ERFs (MLOC_73358, HvERF4; and MLOC_24530, HvERF-like; Table I). Finally, the protein encoded by MLOC_44411 displayed high similarity with ERF4 or ERF4-like proteins from B. distachyon, foxtail millet (Setaria italica), and maize and, similar to HvERF-like, contained the conserved APETALA2 (AP2) domain that is typical for AP2-ERFs and the ERF-associated amphiphilic repression motif that is typical for ERFs functioning as transcriptional repressors (Nakano et al., 2006; Licausi et al., 2013; Supplemental Fig. S10). Therefore, we propose that MLOC_44411 encodes an AP2-ERF that we refer to as HvERF44411 (Table I).
Table I. Putative transcription factors that were systemically regulated in response to local Psj and Xtc infections.
The relative transcript accumulation in the systemic tissue of Psj-infected and Xtc-infected compared with mock-treated plants is listed with the corresponding FDR-corrected P values derived from the RNA-seq analysis summarized in Supplemental Table S1. Gene annotations correspond to Mayer et al. (2012) or were derived from http://plants.ensembl.org. The transcript accumulation corresponding to the loci marked in boldface was analyzed by qRT-PCR (Figure 5).
| Locus |
Psj Induced |
Xtc Induced |
Annotation | ||
|---|---|---|---|---|---|
| Fold Change (log2) | P | Fold Change (log2) | P | ||
| MLOC_24530 HvERF-like | 3.21 | 5.00E-05 | 2.15 | 5.00E-05 | Ethylene-responsive factor-like transcription factor IPR016177 (DNA binding, integrase type) |
| MLOC_73358 HvERF4 | 2.31 | 5.00E-05 | 1.52 | 2.50E-04 | Ethylene-responsive transcription factor4 IPR016177 (DNA binding, integrase type) |
| MLOC_44411 HvERF_44411 | 3.05 | 5.00E-05 | 2.70 | 5.00E-05 | AP2-ERF transcription factora |
| MLOC_71395 | 6.27 | 5.00E-05 | 5.58 | 5.00E-05 | B3 domain-containing protein IPR015300 (DNA-binding pseudobarrel domain) |
| MLOC_45055 HvWRKY22 | 1.73 | 2.50E-04 | 2.81 | 5.00E-05 | WRKY TF 22 IPR003657 (DNA-binding WRKY) |
| MLOC_52504 HvWRKY28 | 1.98 | 1.25E-03 | 2.46 | 1.50E-04 | WRKY TF 28 IPR003657 (DNA-binding WRKY) |
| MLOC_60890 HvWRKY38/1b | 1.24 | 7.00E-04 | 1.84 | 5.00E-05 | WRKY TF 38 IPR003657 (DNA-binding WRKY) |
| MLOC_52308 | 1.40 | 1.05E-03 | 1.54 | 3.20E-03 | Ring finger protein, putative IPR013083 (zinc finger, RING/FYVE/PHD type) |
| MLOC_6731 | 2.35 | 5.00E-05 | 2.04 | 3.00E-04 | RING-H2 finger protein 2B (length = 147) IPR013083 (zinc finger, RING/FYVE/PHD type) |
| MLOC_65033 | 1.17 | 3.10E-03 | 1.43 | 6.00E-04 | Zinc finger protein1 IPR007087 (zinc finger, C2H2) |
| MLOC_63682 | 2.32 | 1.00E-04 | 1.61 | 2.55E-03 | CBF6-201, sequence-specific DNA-binding transcription factor activity |
Supplemental Figure S10. bHvWRKY38 (http://plants.ensembl.org; Marè et al., 2004) corresponds to HvWRKY1 (Eckey et al., 2004; Shen et al., 2007; Liu et al., 2014).
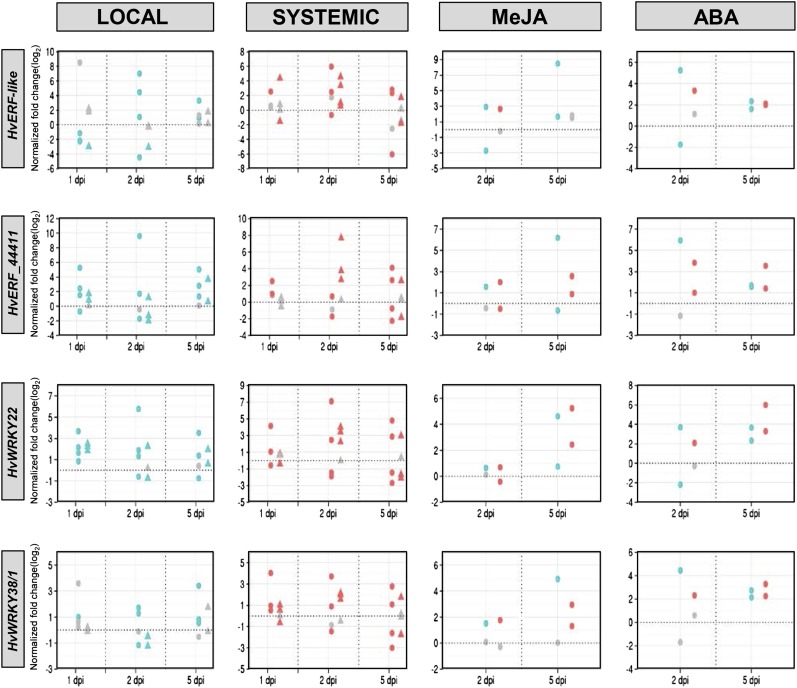
The local and systemic accumulation of four of the putative TF-encoding transcripts was studied by qRT-PCR in three to four biologically independent experiments (Fig. 5). To this end, the first true leaf of 4-week-old barley GP plants was infected with Psj or Xtc or treated with a mock solution, and the local treated leaves as well as the next systemic untreated leaf (leaf 2) were harvested at 1, 2, and 5 dpi. As experimental controls, we performed Xtc challenge infections of the systemic tissue at 5 dpi, confirming that systemic immunity was induced in each experiment (Supplemental Fig. S9, C–F). Transcript accumulation was determined for HvERF-like, HvERF44411, HvWRKY22, and HvWRKY38/1 (Table I; Fig. 5). Locally, Psj induced more pronounced transcriptional changes compared with Xtc, especially for HvERF-like, HvERF44411, and HvWRKY38/1 (Fig. 5). The induction of these transcripts by Psj was detected at 1, 2, and/or 5 dpi. HvWRKY22 transcript accumulation was locally induced by both Psj and Xtc at each of the tested time points, with the most consistent induction detected at 1 dpi (Fig. 5). Systemically, HvERF44411, HvWRKY22, and HvWRKY38/1 transcript accumulation was induced by Xtc mainly at 2 dpi and by Psj at 1 dpi and in approximately half of the experiments at 2 or 5 dpi (Fig. 5). The accumulation of HvERF-like transcripts was systemically induced by both Psj and Xtc, with the most consistent induction detected at 2 dpi (Fig. 5). Together, these data suggest that bacteria-induced systemic immunity in barley is associated with the local and/or systemic induction of HvERF-like, HvERF44411, HvWRKY22, and HvWRKY38/1 transcript accumulation.
Figure 5.
Transcript accumulation of HvERF-like, HvERF44411, HvWRKY22, and HvWRKY38/1 (as indicated at left) at 1, 2, and/or 5 dpi in local infected leaves (first or left-most column; blue), systemic uninfected leaves of locally infected plants (second column; red), and treated (blue) and systemic untreated (red) leaves of plants treated with MeJA (third column) or ABA (fourth or right-most column). Transcript accumulation was determined by qRT-PCR, and each data point represents the average of three technical replicates. Data points depicted in color represent significant differences from the respective mock controls (P < 0.05, unpaired Student’s t test); nonsignificant differences are depicted in gray. Local/systemic transcript accumulation was normalized to that of HvERF1α and is shown relative to the normalized transcript accumulation in the respective mock controls for local and systemic transcript accumulation upon Psj (circles) or Xtc (triangles) infection. The local and systemic transcript accumulation schemes include data from three and four biologically independent experiments, respectively; replicates are vertically aligned. MeJA/ABA transcript accumulation was normalized to that of HvERF1α, and the local (blue) and systemic (red) transcript accumulation in response to the application of 100 µm MeJA or ABA is shown relative to the normalized transcript accumulation in the respective mock controls. The schemes include data from two biologically independent experiments (vertically aligned).
As described above, local MeJA and ABA applications to leaf 1 of 4-week-old barley GP plants induced systemic immunity to Xtc in leaf 2 (Fig. 3C). Hence, we investigated the transcript accumulation of the four potentially systemic immunity-associated TF genes in the local treated and systemic untreated tissue at 2 and 5 d after treatment of barley GP plants with 100 µm MeJA or ABA (Fig. 5). MeJA application induced the most pronounced transcript accumulation changes at 5 dpi compared with 2 dpi. A moderate local or systemic induction of HvERF-like, HvERF44411, and HvWRKY38/1 was observed at 2 dpi in one out of two experiments. At 5 dpi, all of the studied transcripts were locally induced in at least one out of two replicate experiments and systemically in both experiments, with the exception of HvERF-like, which was not systemically regulated (Fig. 5). The transcript accumulation changes induced by ABA were evident at both of the time points tested, albeit more robustly detected at 5 dpi compared with 2 dpi (Fig. 5). At 5 dpi, all of the transcripts tested were locally and systemically induced, strengthening a potential association between the encoded TFs and systemic immunity in barley.
Primary Infection Affects Transcriptional Responses after Secondary Challenge Infection
Because the transcript accumulation of a number of TFs was systemically induced after a local infection of barley with Psj or Xtc, we investigated if these might potentiate or prime responses after the systemic challenge infection to promote systemic immunity. First, we investigated a possible potentiation of SA, JA, and ABA accumulation at 24 and 48 h after the systemic Xtc challenge infection (Fig. 6A). To this end, barley GP plants were first mock treated or infected with Psj or Xtc in leaf 1. Five days later, systemic untreated leaves were infected with Xtc and sampled at 24 and 48 h post infection (hpi). The free and total SA levels did not differ in Psj/Xtc-treated, Xtc/Xtc-treated, and mock/Xtc-treated plants (Fig. 6A), indicating that SA accumulation was not potentiated upon challenge infection of preinfected compared with mock-pretreated plants as measured at 24 or 48 hpi. Similarly, we did not detect a significant potentiation of JA or ABA accumulation, although ABA levels appeared to be slightly elevated at 48 h after Xtc challenge infection of preinfected compared with mock-pretreated plants (Fig. 6A). Taken together, bacteria-induced systemic immunity in barley did not appear to be associated with the potentiation of SA, JA, or ABA accumulation.
Figure 6.
SA, JA, ABA, and gene expression analysis after systemic challenge infection. Barley GP (A) or ‘Barke’ (B) plants were treated in leaf 1 with 10 mm MgCl2 (mock; black bars in A) or infected with Xtc (light gray bars in A) or Psj (dark gray bars in A). Five days later, systemic untreated leaves were infected with Xtc and harvested at 24 and/or 48 hpi. A, Free SA, total SA, JA, and ABA levels in the systemic Xtc challenge-infected tissue are shown as means ± se of at least two (free SA at 24 h post Xtc infection) to three biologically independent replicates for free and total SA and at least five technical replicates from at least three biologically independent experiments for JA and ABA. B, Venn diagram of microarray probes hybridizing to transcripts that differentially accumulated in the systemic Xtc challenge-infected tissue at 24 hpi of plants locally pretreated with either Xtc (IX) or Psj (IP) compared with mock-pretreated plants (IM). [See online article for color version of this figure.]
Subsequently, we investigated a possible potentiation of gene expression changes at 24 h after the systemic Xtc challenge infection. To this end, transcript accumulation was compared in samples from either Psj/Xtc-treated or Xtc/Xtc-treated plants with mock/Xtc-treated plants by using Agilent 44K barley microarrays. We analyzed data sets from three biologically independent replicate experiments using barley ‘Barke’ plants. Statistical analysis of the data revealed that transcripts hybridizing to 491 microarray probes were regulated on Psj/Xtc treatment compared with the mock/Xtc control (P < 0.01, greater than 1.5-fold change; Supplemental Table S2). Xtc/Xtc as compared with mock/Xtc treatment resulted in differential accumulation of transcripts hybridizing to 260 probes (Supplemental Table S2). Of these, transcripts hybridizing to 86 probes overlapped with transcripts regulated on Psj/Xtc treatment, with consistent up- or down-regulation on both Psj/Xtc and Xtc/Xtc treatments (Fig. 6B; Supplemental Table S2). It should be noted that we did not analyze Psj/mock or Xtc/mock compared with mock/mock treatments and cannot exclude that differences in transcript accumulation were independent of the nature of the secondary treatment. Nevertheless, because the secondary challenge inoculum was the same in all cases (Xtc), differential expression of genes was dependent on and thus possibly primed by the primary treatment, as illustrated in Table II for transcripts hybridizing to six different microarray probes. The 44K microarrays contained one probe that might hybridize to HvWRKY38/1 transcripts; this probe did not detect differences in transcript accumulation between mock/Xtc, Psj/Xtc, and Xtc/Xtc treatments, suggesting that HvWRKY38/1 transcript accumulation was not (further) potentiated upon Xtc infection of preinfected plants. The microarrays did not contain probes for the other three TF genes that were systemically induced prior to the systemic challenge infection.
Table II. Examples of putative priming or potentiation of systemic Xtc-induced gene expression changes by primary/local Psj and Xtc infections.
Plants were treated in (local) leaf 1 with 10 mm MgCl2 (mock), Psj, or Xtc. Five days later, all treated plants were inoculated in (systemic) leaves 2 and 3 with Xtc, and gene expression was analyzed in these leaves at 24 h after Xtc inoculation using 44K barley microarrays (Agilent). Putatively primed regulation of gene expression is summarized for three transcripts with elevated accumulation after the systemic Xtc challenge infection of locally preinfected plants compared with control-treated plants (in boldface) and three transcripts with reduced accumulation after the systemic Xtc challenge infection of locally preinfected plants compared with control-treated plants (in lightface).
| Agilent Probe Identifier | GenBank Accession No. | Description of Barley Sequence (Genespring GX11; Agilent) | Average Normalized Expression at 24 h after Xtc Infection Systemic to the Primary/Local Treatmentsa |
Putatively Primed Regulation of Systemic Gene Expression in Response to Xtc by the Primary/Local Psj and Xtc Treatments Compared with the Primary/Local Mock Treatment |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mock (IM) | Psj (IP) | Xtc (IX) | IPIMb | Pa | IXIMc | Pa | |||
| A_13_P062881 | AJ462829 | Subspecies vulgare, mRNA | 63.03 | 180.74 | 175.90 | +2.87 | 0.0042 | +2.79 | 6.71E-05 |
| A_13_P102730 | BF262337 | Seedling green leaf EST, mRNA | 20.17 | 57.11 | 51.63 | +2.83 | 0.0082 | +2.56 | 0.0009 |
| A_13_P044391 | AJ485291 | mRNA | 141.01 | 380.32 | 291.35 | +2.70 | 0.001 | +2.07 | 0.0059 |
| A_13_P061976 | AJ467036 | mRNA | 33.23 | 13.97 | 18.79 | −2.38 | 5.48E-05 | −1.77 | 0.0004 |
| A_13_P030016 | BF256646 | Seedling root EST, mRNA | 146.20 | 55.30 | 86.97 | −2.64 | 0.0003 | −1.68 | 0.0093 |
| A_13_P018166 | BM816774 | Subspecies vulgare, mRNA, similar to hydroxymethylglutaryl-CoA reductase (NADPH), 4-α-glucanotransferase, hydroxymethylglutaryl-CoA reductase | 480.82 | 223.39 | 291.0 | −2.15 | 0.0007 | −1.65 | 0.0016 |
Data extracted from Supplemental Table S2. 2IPIM, Induced gene expression in Psj-pretreated compared with mock-pretreated plants. cIXIM, Induced gene expression in Xtc-pretreated compared with mock-pretreated plants.
To obtain insight into significantly regulated biological processes upon secondary challenge infection of preinfected compared with mock-pretreated plants, we analyzed the full 44K array data sets in MapMan (Usadel et al., 2005) without statistical or fold-change cutoffs. As input, average fold changes per probe (of all three biologically independent replicates) were included comparing either Psj/Xtc or Xtc/Xtc treatment with mock/Xtc treatment. Subsequently, we analyzed both average data sets for pathways or processes (termed bins in MapMan) that were significantly regulated, with a Wilcoxon rank sum test and FDR-corrected P value of less than 0.05 (Supplemental Table S3). In addition, the same analysis was performed for each individual microarray replicate (Supplemental Table S3), and bins, representing individual biological processes, were selected from the average data sets as differentially regulated only if the bin or one or more associated subbins were significantly regulated in at least two out of three replicates (Supplemental Fig. S11). Three bins were commonly regulated upon challenge infection irrespective of the pathogen used for the primary infection (Supplemental Fig. S11). These included bin 10 (cell wall), bin 11 (lipid metabolism), and bin 19 (tetrapyrrole synthesis). In addition, several subbins were significantly regulated in Xtc/Xtc-treated plants that were included in significantly regulated bins in Psj/Xtc-treated plants, or vice versa (Supplemental Fig. S11). These included, for example, bins related to protein synthesis and signaling. The considerable overlap in bins that were potentially primed or potentiated by primary Psj or Xtc infections suggests that systemic immunity induced by Psj and Xtc converges on similar mechanisms.
DISCUSSION
The establishment of systemic immunity/SAR involves profound metabolic and transcriptional changes that are well documented in dicotyledonous plants and include the accumulation of SA and SA-associated gene transcripts in the systemic, uninfected tissue during the establishment of SAR (Vlot et al., 2009; Truman et al., 2010; Fu and Dong, 2013; Gruner et al., 2013). Here, we show that infection of barley with either Psj or Xtc induces systemic immunity against Xtc (Fig. 2) in a manner that does not appear to be associated with SA (Figs. 2, 3, and 6). Contrary to dicots studied so far (Vlot et al., 2009; Fu and Dong, 2013), systemic immunity triggered by primary Psj or Xtc infection of barley was not affected by RNAi-mediated silencing of HvNPR1 (Fig. 2) or associated with locally or systemically elevated levels of free or total SA (Fig. 3). Also, primary treatment of barley with SA or its functional analog BTH did not induce systemic immunity against Xtc (Fig. 3). Although local applications of MeJA or ABA induced systemic immunity to Xtc, bacteria-induced systemic immunity was associated with moderate but mostly insignificant increases in the local and not systemic JA and ABA levels (Fig. 3). Thus, SA, JA, and ABA might not play prominent roles during bacteria-induced systemic immunity in barley.
Similar to HvNPR1 promoting resistance to Bgh, rice NPR1 promotes resistance in rice against X. oryzae as well as BTH-induced resistance to the rice blast pathogen Magnaporthe grisea (Chern et al., 2005; Sugano et al., 2010). Also, overexpression of AtNPR1 in rice enhances resistance to bacterial and fungal pathogens and induces a BTH-triggered lesion-mimic phenotype that appears to be associated with SA signaling (Chern et al., 2001; Fitzgerald et al., 2004; Quilis et al., 2008). In wheat, both SA and AtNPR1 prime resistance against the head blight pathogen F. graminearum, with AtNPR1 enhancing the SA-primed immune response (Makandar et al., 2006, 2012). Infection of barley with Bgh is not associated with elevated SA levels (Hückelhoven et al., 1999). Nevertheless, barley GP plants with reduced HvNPR1 transcript accumulation displayed enhanced susceptibility to this pathogen (Fig. 2), suggesting that HvNPR1 is involved in defense against Bgh. Similar to AtNPR1 (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013; Fu and Dong, 2013), HvNPR1 might be important for signaling downstream of SA, which in turn might be relevant for defense in barley against the biotrophic powdery mildew fungus. In support of this hypothesis, the SA functional analog BTH induced resistance in barley against Bgh, and this was at least partially dependent on HvNPR1 (Fig. 2). It should be noted that Arabidopsis transcriptional responses to BTH only partially overlap with responses to SA and suggest that BTH induces a surplus of responses in addition to SA (Gruner et al., 2013). The BTH-induced transcriptional response is strikingly similar to the systemic transcriptional response in Arabidopsis during SAR (Gruner et al., 2013). Nevertheless, BTH did not induce systemic immunity in barley (Fig. 3). Also, bacteria-induced systemic immunity was not compromised in the HvNPR1-kd plants (Fig. 2). Taken together, HvNPR1 and possibly SA are required for full resistance in barley to Bgh, but HvNPR1 is not involved in bacteria-induced systemic immunity.
Local transcript accumulation changes triggered by Psj or Xtc infection appropriately reflected the apparent avirulent and virulent nature of the respective pathogens in barley, with Psj inducing more transcriptional changes than the more virulent pathogen Xtc. In Arabidopsis, avirulent, ETI-inducing and virulent, PTI-inducing pathogens are thought to trigger similar transcriptional responses, where ETI may have evolved as a fortification of PTI, inducing a stronger response that uses interactions within the signaling network differently compared with PTI (Tao et al., 2003; Tsuda et al., 2009; Tsuda and Katagiri, 2010). In barley, more than half of the transcripts locally regulated by Xtc were similarly regulated in Psj-infected leaves (Fig. 4). In line with a possibly fortified response elicited by Psj as compared with Xtc, Psj triggered the regulation of more genes compared with Xtc (Fig. 4). Systemically, Psj induced the more modest response, with fewer regulated transcripts compared with the response to Xtc (Fig. 4). In spite of limited overlap between locally and systemically regulated transcripts in response to Xtc, we cannot exclude that part of the systemically regulated transcripts were directly targeted by the Xtc infection traveling from the infected leaf to the systemic tissue. However, as most gene expression changes in response to either Psj or Xtc were unique either locally or systemically, we conclude that at least part of the systemic immune response induced by Xtc was a result of systemic signaling similar to the response induced by Psj.
By assuming that transcripts whose accumulation was regulated by both Psj and Xtc might be central to the mechanism of systemic immunity, we associated bacteria-induced systemic immunity in barley with the enhanced transcription in the systemic tissue of up to 11 putative TFs (Table I). Of these, two ERF and two WRKY TFs were locally and systemically induced at one or more time points after a local Psj or Xtc infection and after a local, systemic immunity-inducing MeJA or ABA application (Figs. 3 and 5). Because the four tested TFs were not equally strongly or consistently induced among the biologically independent replicate experiments, we speculate that there might be at least a partial redundancy between these and possibly other TFs acting together during the establishment of systemic immunity. HvERF-like and HvERF44411 belong to the AP2-ERF family of plant-specific TFs (Supplemental Fig. S10), with 122 predicted family members in the Arabidopsis genome and 139 in rice (Nakano et al., 2006). AP2-ERFs have been associated with developmental processes but also with responses to biotic and abiotic stresses that are associated with JA and/or ABA signaling (for review, see Licausi et al., 2013). AtERF1, for instance, preferentially binds to one of two different gene promoter cis-elements, depending on the type of stress encountered integrating JA, ABA, and ET signaling (Cheng et al., 2013). AtERF4 expression is induced by ABA, JA, ET, and abiotic stress, and overexpression of AtERF4 represses ABA responses, whereas it may enhance JA responsiveness (Yang et al., 2005; Memelink, 2009). Other plant AP2-ERFs, including OsERF922 and wheat PATHOGEN-INDUCED ERF1, also appear to regulate abiotic stress tolerance or disease resistance against necrotrophic pathogens mediated by JA, ABA, or ET (Pré et al., 2008; Liu et al., 2012; Moffat et al., 2012; Maruyama et al., 2013; Zhu et al., 2014). Notably, HvERF-like and HvERF44411 were systemically induced by MeJA and/or ABA application (Fig. 5), but HvERF-like transcript accumulation was repressed systemically to a local ET application to barley, whereas HvERF44411 was not systemically regulated by ET (Supplemental Fig. S7B). Similar to AP2-ERFs, WRKY TFs are often associated with stress tolerance, responding to biotic and abiotic triggers and to different phytohormones, including SA, JA, and ABA (Agarwal et al., 2011). An increasing body of evidence suggests that WRKY TFs function on the interface between SA, JA, and ABA signaling, balancing different stress tolerance pathways to optimize resistance to biotic and abiotic influences in both Arabidopsis and rice (Pandey and Somssich, 2009; Agarwal et al., 2011; Sharma et al., 2013). HvWRKY22, for example, is orthologous with AtWRKY30/41/53 (Mangelsen et al., 2008) that are associated with abiotic stress tolerance or the regulation of the cross talk between SA and JA (Miao and Zentgraf, 2007; Higashi et al., 2008; Scarpeci et al., 2013). HvWRKY38/1 is orthologous with AtWRKY18/40/60 (Shen et al., 2007; Mangelsen et al., 2008; Liu et al., 2014). The accumulation of the corresponding barley transcript is induced by Bgh and abiotic factors, including cold, drought, and wounding stress (Eckey et al., 2004; Marè et al., 2004; Liu et al., 2014). Notably, HvWRKY38/1 is a negative regulator of barley resistance to Bgh (Eckey et al., 2004; Shen et al., 2007). Taken together, the systemic immunity-associated TFs identified in this study or their homologous counterparts in other plant species are associated with stress tolerance responses that are related to JA and/or ABA signaling or might act on the interface between JA and SA signaling or abiotic and biotic stress tolerance.
In Arabidopsis, contrasting reports debate a possible role of JA in promoting SAR (Truman et al., 2007; Attaran et al., 2009; Shah and Zeier, 2013), while ABA is thought to repress SA signaling and SAR (Yasuda et al., 2008; Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). With JA also mostly antagonizing SA signaling in Arabidopsis, bacteria-induced systemic immunity in barley appears more closely related to Arabidopsis ISR, which shares similarity with abiotic rather than biotic stress responses and is associated with JA, ET, and ABA rather than SA signaling (Pieterse et al., 2012). Alternatively, responses to biotrophic and necrotrophic pathogens might mechanistically differ between barley and Arabidopsis. PR1 gene expression, for example, is induced in barley and wheat by (hemi)biotrophic pathogens but (unlike in Arabidopsis) not by SA or BTH (Molina et al., 1999; Jarosch et al., 2003; van Loon et al., 2006). Also, the barley MICRORCHIDIA (MORC) ATPases HvMORC1 and HvMORC2 negatively regulate resistance to Bgh, whereas AtMORC1 and AtMORC2 promote resistance in Arabidopsis to the (hemi)biotrophic bacterial pathogen P. syringae (Langen et al., 2014). In contrast to AtMORC1, ectopic expression of HvMORC1 does not complement the hypersusceptible phenotype of Arabidopsis Atmorc1/Atmorc2 double mutants to P. syringae, although AtMORC1 and HvMORC1 have similar enzymatic activities (Langen et al., 2014). The extent of possible mechanistic similarities and differences between barley and Arabidopsis immune-related responses requires further investigation.
The differential gene expression observed at 24 h after Xtc challenge infection of preinfected compared with mock-pretreated plants suggests that at least part of the gene expression changes occurring during the execution phase of systemic immunity in barley are primed or potentiated. Arabidopsis can be primed for defense by SAR and the defense-inducing compound β-aminobutyric acid (Ton et al., 2005; Conrath et al., 2006; Conrath, 2011; Návarová et al., 2012). In both cases, priming is mediated at least in part by the natural priming agent pipecolic acid that accumulates in a number of plant species, including rice (Návarová et al., 2012; Zeier, 2013). Primed plants or tissues respond faster and stronger to a subsequent stress than unprimed plants. In Arabidopsis SAR, SA and/or SA-associated gene transcripts accumulate faster and to a higher level after challenge inoculation of primed (or SAR-induced) tissues as compared with unprimed tissues (Beckers et al., 2009; Jung et al., 2009; Návarová et al., 2012). Such a mechanism of primed induced resistance is not unprecedented in barley. Local application of P. fluorescens primes barley head tissue for 88 gene expression changes upon a Fusarium culmorum challenge infection of the treated tissue (Petti et al., 2010). Moreover, systemic leaves of plants pretreated with the root-colonizing and plant growth-promoting fungus Piriformospora indica display enhanced resistance to Bgh (Waller et al., 2005). Similar to our leaf-to-leaf response, this root-to-leaf ISR response does not appear to be correlated with SA or JA signaling but with relatively few gene expression changes in the aboveground, systemic tissue (Waller et al., 2005, 2008) that might prime the leaves for both apoplast alkalinization and expression changes of a specific set of genes, including three PR genes, upon Bgh infection (Felle et al., 2009; Molitor et al., 2011). Alternatively, leaf-to-leaf and root-to-leaf systemic immune responses might be primed independently of the (limited) gene expression changes observed in the putatively primed tissue but instead might be regulated by, for example, posttranslational protein modifications (Beckers et al., 2009). Together, the data suggest that both pathogen-induced systemic immunity and ISR in barley are based on mechanisms that are reminiscent of priming.
In conclusion, infection of barley leaves with Psj or Xtc triggers systemic immunity to Xtc, possibly via one or more of up to 11 TFs, including two ERFs and two WRKY TFs, that might prime the systemic tissue for gene expression changes upon a challenge infection (Supplemental Fig. S12). Future research efforts will be concentrated on unraveling the mechanistic details of barley systemic immunity, including the specific roles of individual TFs and possibly MeJA or ABA. In addition, the spectrum of pathogens against which bacteria-induced systemic immunity in barley acts is under further investigation.
MATERIALS AND METHODS
Plants and Pathogens
Barley (Hordeum vulgare ‘Barke’ and GP) were used throughout this study. Prior to sowing, seeds were sterilized in 1% (v/v) sodium hypochlorite for 5 min, rinsed three times for 10 min with sterilized water, and then sown on fertilized soil (Einheitserde Classic of the Bayerische Gaertnereigenossenschaft). For bacterial infections and systemic immunity experiments, the plants were grown in the greenhouse with the additional lights HQI-TS 400W/D (Osram) in 12-h-light/12-h-dark cycles at 70% relative humidity and were kept at 24°C during the day and 20°C during the night. All experiments were carried out during the months of October through March of four consecutive winter seasons. Pseudomonas syringae pv japonica strain LMG5659 and Xanthomonas translucens pv cerealis strain LMG7393 were obtained from the Laboratory of Microbiology UGent (LMG) collection of the Belgian Coordinated Collections of Microorganisms. The bacteria were maintained on LMG Agar Medium containing 15 g L−1 trypton, 5 g L−1 soja-pepton, 5 g L−1 sodium chloride, and 18 g L−1 agar (pH adjusted to 7.3). For Bgh infection experiments, barley GP seeds were surface sterilized with a sodium hypochlorite solution containing 6% (v/v) active chlorine for 2 h and germinated for 3 d on filter paper. Seedlings were transferred to soil (Fruhstorfer Erde, Hawita Gruppe) and maintained in a climate chamber at 20°C/18°C (day/night) with 60% relative humidity and a photoperiod of 16 h (180 μmol m−2 s−1 photon flux density). The obligate biotrophic powdery mildew fungus Blumeria graminis f. sp. hordei, race A6 (Wiberg, 1974), was maintained on barley ‘Siri.’ For the experiment shown in Supplemental Figure S6B, Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 and P. syringae pv tomato with or without AvrRpm1 were maintained and used for the SAR assay as described (Breitenbach et al., 2014).
Generation of HvNPR1-kd Plants
To knock down the barley NPR1 gene (accession no. AM050559), sense and antisense fragments of HvNPR1 (corresponding to amino acids 204–333 of the protein) were amplified by PCR from leaf cDNA using primers SmaI-NPR1 and BamHI-NPR1 (sense fragment) and SpeI-NPR1 and SphI-NPR1 (antisense fragment; Supplemental Table S4). The HvNPR1 amplicons (401 bp) were ligated into pJP26 (Christensen et al., 2004) downstream of the cauliflower mosaic virus 35S promoter using SmaI and BamHI (sense fragment) as well as SpeI and SphI (antisense fragment) restriction sites, and the final construct was confirmed by sequencing. For the production of transgenic plants, the hairpin construct under the control of the 35S promoter was cloned into the binary vector pLH6000 (DNA Cloning Service), and the resulting construct (pLH6000-35S::NPR1-RNAi) was electroporated (Gene Pluser; Biometra) into Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) to transform spring barley GP as described by Imani et al. (2011). T0 regenerated and T1 segregating plants were tested for the presence of the HvNPR1-kd construct by PCR using genomic DNA and primers p-35S_F and BamHI-NPR1-R (Supplemental Table S4). HvNPR1 transcript levels were determined by qRT-PCR with primers HvNPR1_F and HvNPR1_R (Supplemental Table S4). The expression values were normalized to the housekeeping gene HvUBIQUITIN with the primers HvUBQ_F and HvUBQ_R (Supplemental Table S4).
Bacterial Inoculations and Systemic Resistance Assay
Bacteria were grown on LMG plates overnight at 28°C, and inoculi were resuspended from the plates in 10 mm MgCl2 and diluted to the appropriate concentration for infection by using the equation optical density at 600 nm = 0.2 = 108 cfu mL−1. For bacteria growth curve analyses, two fully expanded leaves of 4-week-old barley plants were sprayed with 108 cfu mL−1 Psj or Xtc in 0.01% (v/v) Tween 20 or infiltrated with 105 cfu mL−1 Psj or Xtc in 10 mm MgCl2. Bacterial titers in leaves were determined at different time points essentially as described (Vlot et al., 2008). Per biologically independent experiment, four to five technical replicates were harvested per time point and inoculum. Per technical replicate, three 6-mm leaf discs were taken spaced equally across the basal half of one infected leaf. Bacteria were extracted by gently shaking the leaf discs for 1 h in 10 mm MgCl2 containing 0.01% (v/v) Silwet L-77 (Lehle Seeds). Bacterial titers were determined by serial dilution of the extracted bacteria in 10 mm MgCl2 followed by plating of 20 µL per dilution on LMG plates. Colonies were grown for 2 d at 28°C, counted, and converted back to cfu cm−2 leaf.
For systemic resistance induction, the first true leaf (leaf 1; Supplemental Fig. S12) of 4-week-old barley plants was syringe infiltrated with either 10 mm MgCl2 (mock) or 106 cfu mL−1 of either Psj or Xtc. Five days after the primary treatment, the next two fully expanded leaves of the treated plants (leaves 2 and 3; Supplemental Fig. S12) were syringe infiltrated with 105 cfu mL−1 Xtc. Xtc titers in the secondary infected leaves were determined at 4 dpi. Four to five technical repetitions (as defined above) were included in each biologically independent replicate experiment.
Fungal Infection Assay
Seven-day-old seedlings were infected with BghA6 as follows. After complete emergence (at 7 d), first leaf segments were placed on 0.5% (w/v) agar in water containing 20 mg L−1 benzimidazole (Merck Schuchardt), inoculated with BghA6 spores (density, 5 conidia per mm2), and maintained in the climate chamber for 6 d. BghA6 colony numbers were counted on 2.5 cm2 of each leaf segment using a stereomicroscope and converted back to colonies cm−2.
Chemically Induced Resistance Assays
BTH purchased commercially under the trade name BION (Ciba Geigy) and ACC (Sigma-Aldrich) were dissolved in water to produce stock solutions of 100 mm. SA (Sigma-Aldrich), MeJA (95%; Sigma-Aldrich), and ±ABA (98%; ACROS Organics) were dissolved in ethanol at a concentration of 100 mm. For systemic resistance assays, all stock solutions were stored in the refrigerator for a maximum time of 1 month and diluted with sterile water to the required concentration prior to plant treatment. Ethanol diluted with sterile water to similar concentrations as included in the SA, MeJA, and ABA treatments was used as the respective control. For systemic resistance tests, the first true leaf of 4-week-old barley GP plants was syringe infiltrated with the appropriate concentration of the different chemicals. Five days later, the next fully expanded leaf was syringe infiltrated with 105 cfu mL−1 Xtc. Xtc titers were determined 4 d after the infection. Five technical repetitions (as defined above) were included in each biologically independent replicate experiment. For the analysis of BTH-induced resistance to Bgh, plants were treated with a BION (Novartis, Syngenta) soil drench. To this end, 2-d-old synchronized germinated seedlings were grown in 200-mL capacity pots in soil and kept in a climate chamber with the conditions described above. Three days later, 8 mg of BION (water-dispersible granular/wettable powder formulation with 50% active ingredient in water) was applied as a soil drench per 200 mL of soil volume. Control plants were treated similarly with the same volume of water-suspended wettable powder. Two days after the BTH treatment, first leaf segments were infected with BghA6 as described above. The resulting BghA6 colonies were counted as described above at 7 dpi.
RNA Isolation and qRT-PCR
RNA was isolated using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was synthesized from 1.5 µg of total RNA by reverse transcription using SuperScript II (Invitrogen, Life Technologies). Quantitative PCR was performed using the SensiMix SYBR Low ROX kit (Bioline) on a 7500 Fast qPCR system (Applied Biosystems) with the primers listed in Supplemental Table S4. Relative quantitation was performed using the Sequence Detection Software version 1.3.1 (Applied Biosystems). The significance of differences from mock treatment in Figure 5 was determined by unpaired Student’s t tests using the Graphpad Web interface (http://www.graphpad.com/quickcalcs/ttest1).
Hormone Measurements
Free and total SA was quantified as described (von Saint Paul et al., 2011; Breitenbach et al., 2014). JA and ABA were quantified simultaneously using a standardized ultra-performance liquid chromatography-tandem mass spectrometry-based method as described (Balcke et al., 2012). Of most biologically independent experiments, multiple technical repetitions were performed using separately harvested material from different plants within the same experiment.
Transcript Profiling and Bioinformatics Analyses
RNA Sample Definition and Preparation
Four-week-old barley ‘Barke’ plants were used for these experiments. For the RNA-seq analyses, the first true leaf (leaf 1) was used for the treatments, and leaves 2 and 3 were pooled and harvested as the systemic tissue. Transcript accumulation in the local, treated tissue was analyzed 5 d after treatment of leaf 1 with 10 mm MgCl2 (local mock), 106 cfu mL−1 Xtc (local Xtc), or 106 cfu mL−1 Psj (local Psj). Transcript accumulation in the systemic, untreated tissue was analyzed in leaves 2 and 3 at 5 d after treatment of leaf 1 with 10 mm MgCl2 (systemic mock), 106 cfu mL−1 Xtc (systemic Xtc), or 106 cfu mL−1 Psj (systemic Psj). For the microarray analysis of transcript accumulation in the systemic challenged tissue, RNA was isolated from leaves 2 and 3 at 24 h after a secondary infection of systemic mock, systemic Xtc, and systemic Psj leaves with 105 cfu mL−1 Xtc. For each biologically independent RNA sample, total RNA was isolated from a pool of three technical replicates using the NucleoSpin RNA Plant kit (Macherey-Nagel) according to the manufacturer’s instructions. The quantity and purity of the RNA was determined with a Nanodrop 2000 apparatus (Thermo Scientific). The integrity of the RNA was verified using the Agilent RNA 6000 Nano kit with the Agilent 2100 Bioanalyzer (Agilent Technologies).
RNA-seq
Library preparation and RNA-seq were carried out as described in the Illumina TruSeq RNA Sample Preparation Version 2 Guide, the Illumina HiSeq 1000 System User Guide (Illumina), and the KAPA Library Quantification Kit-Illumina/ABI Prism User Guide (Kapa Biosystems).
In brief, the poly(A)-containing mRNA was isolated from 1 µg of total RNA by using poly(T) oligonucleotide-attached magnetic beads. Following purification, the mRNA was fragmented to an average insert size of 100 to 200 bases using divalent cations under elevated temperature (94°C for 4 min). The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers, followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H. The resulting cDNA fragments then went through an end-repair process, including the addition of a single A base, the ligation of the adapters, and a purification step. Finally, cDNA libraries were created by PCR enrichment. The libraries were quantified using the KAPA SYBR FAST ABI Prism Library Quantification Kit. Equimolar amounts of each library were used for cluster generation on cBot (TruSeq PE Cluster Kit Version 3). The sequencing run was performed on a HiSeq 1000 instrument using the indexed, 2 × 100 cycles paired-end protocol and the TruSeq SBS Version 3 Kit. Image analysis and base calling resulted in .bcl files, which were converted into .fastq files by the CASAVA1.8.2 software.
RNA-seq Analysis
The sequence reads were mapped on the barley ‘Morex’ 50× whole-genome sequence (Mayer et al., 2012) using TopHat 2.0.8 (Trapnell et al., 2009) and Bowtie2 2.1.0 (Langmead et al., 2009) with default parameter settings and an expected mean insert size of 150 bp. Cufflinks 2.1.1 (Trapnell et al., 2010) was used to assemble the mapped RNA-seq reads into transcripts to quantify their relative abundance. Differentially expressed genes were selected using the Cuffdiff tool using default settings (Trapnell et al., 2010) and filtered for an FDR-adjusted P < 0.05. The GO terms overrepresented for the differentially expressed genes were reported using the GOstat package from R Bioconductor (Falcon and Gentleman, 2007). Statistically significant GO terms tested by conditional hypergeometric tests (P < 0.05) were considered to be enriched. After normalization, the RNA-seq data were interactively browsed and investigated using the RNASeqExpressionBrowser (Nussbaumer et al., 2014). The corresponding RNASeqExpressionBrowser can be accessed at http://mips.helmholtz-muenchen.de/plant/RNASeqExpressionBrowser/.
Microarrays and Data Analysis
The 4x44K custom barley arrays (Agilent Technologies) were processed for one-color microarray-based gene expression analysis (Low Input Quick Amp Labeling; Agilent Technologies). All procedures were performed strictly according to the manufacturer’s instructions.
The background subtracted raw data of all samples were extracted using Feature Extraction version 9 (Agilent Technologies). A composite file of all data sets was generated in Genespring GX version 11.5. Subsequently, multiple biologically independent data sets were normalized against each other using the quantile algorithm in CARMAweb 1.5 (Rainer et al., 2006). Differentially expressed genes were selected by using a moderated limma Student’s t test, filtering for P < 0.01 and a fold change greater than 1.5.
For functional categorization of the complete Agilent 44K probe set, MapMan version 3.6.ORC1 was used. The 44K Agilent probes were mapped against MapMan Hordeum vulgare Hvu1_AGILENT_DESIGNID_021623 (http://mapman.gabipd.org). As input, we used the log2 fold change per probe from the average of three biologically independent replicates or from each replicate individually. The Wilcoxon rank sum test with Benjamini-Hochberg FDR correction was performed to determine the overrepresentation of one bin over all other bins. We considered as significant only bins that were both significantly enriched (FDR-corrected P < 0.05) in the average data set and either themselves or in the form of one of their subbins significantly enriched (FDR-corrected P < 0.05) in at least two out of the three individual replicates.
The RNA-seq and array data from this work are deposited at the European Bioinformatics Institute (www.ebi.ac.uk) under accession numbers E-MTAB-3063 (RNA-seq) and E-MTAB-3056 (array data).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primary infections.
Supplemental Figure S2. Secondary challenge time-course experiments.
Supplemental Figure S3. Systemic resistance in barley ‘Ingrid.’
Supplemental Figure S4. HvNPR1 (CAJ19095).
Supplemental Figure S5. HvNPR1 transcript levels and BTH-induced resistance to BghA6 in homozygous T4 plants of HvNPR1-kd line E7L2.
Supplemental Figure S6. Barley and Arabidopsis responses to BTH application.
Supplemental Figure S7. ACC does not induce systemic immunity in barley to Xtc.
Supplemental Figure S8. Mapping statistics of RNA-seq data.
Supplemental Figure S9. Systemic immunity controls.
Supplemental Figure S10. Multiple sequence alignment including HvERF44411.
Supplemental Figure S11. MapMan output summary of putatively primed bins/biological processes.
Supplemental Figure S12. Working model of bacteria-induced systemic immunity in barley.
Supplemental Table S1. Summary of RNA-seq analysis.
Supplemental Table S2. Microarray probes detecting differentially regulated transcripts upon secondary Xtc infection.
Supplemental Table S3. MapMan analysis of enriched biological processes upon secondary Xtc infection.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Robert Dudler (Institute of Plant Biology, University of Zurich) for kindly providing the RNAi expression vector, Gerd Balcke (Leibniz Institute of Plant Biochemistry) for help with liquid chromatography-mass spectrometry measurements, Elisabeth Georgii (Helmholtz Zentrum Muenchen [HMGU]) for helpful discussions, Rebekka Kastner (HMGU) and Hagen Stellmach (Leibniz Institute of Plant Biochemistry) for excellent technical support, J. Barbro Winkler (HMGU) and Peter Kary (HMGU) for support in the greenhouse, and Joerg-Peter Schnitzler (HMGU) and Joerg Durner (HMGU) for critically reading the article.
Glossary
- ISR
induced systemic resistance
- SAR
systemic acquired resistance
- SA
salicylic acid
- PTI
pathogen-associated molecular pattern-triggered immunity
- ETI
effector-triggered immunity
- BTH
S-methyl benzo-1,2,3-thiadiazole-7-carbothioate
- LAR
local acquired resistance
- TF
transcription factor
- Psj
Pseudomonas syringae pv japonica
- Xtc
Xanthomonas translucens pv cerealis
- GP
Golden Promise
- dpi
days post inoculation
- cfu
colony-forming units
- RNAi
RNA interference
- BghA6
Blumeria graminis f. sp. hordei virulent isolate A6
- Bgh
Blumeria graminis f. sp. Hordei
- qRT
quantitative reverse transcription
- JA
jasmonic acid
- ET
ethylene
- ABA
abscisic acid
- MeJA
jasmonic acid methyl ester
- ACC
1-aminocyclopropane-1-carboxylic acid
- RNA-seq
RNA sequencing
- cDNA
complementary DNA
- PMID
PubMed Indexed for MEDLINE
- FDR
false discovery rate
- GO
Gene Ontology
- hpi
hours post infection
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SFB924 projects B6 to A.C.V. and Z02 to K.F.X.M.), by the Ministry of Science, Research, and Technology of Iran (to V.B.), and by the Helmholtz Society Portfolio Sustainable Bioeconomy (to R.H.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Agarwal P, Reddy MP, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38: 3883–3896 [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcke GU, Handrick V, Bergau N, Fichtner M, Henning A, Stellmach H, Tissier A, Hause B, Frolov A (2012) An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer D, de Papajewski DV, Planchamp C, Glauser G, Mauch-Mani B (2013a) Induced resistance in maize is based on organ-specific defence responses. Plant J 74: 213–225 [DOI] [PubMed] [Google Scholar]
- Balmer D, Planchamp C, Mauch-Mani B (2013b) On the move: induced resistance in monocots. J Exp Bot 64: 1249–1261 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beßer K, Jarosch B, Langen G, Kogel KH (2000) Expression analysis of genes induced in barley after chemical activation reveals distinct disease resistance pathways. Mol Plant Pathol 1: 277–286 [DOI] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K (2013) Salicylic acid: an old hormone up to new tricks. Mol Plant Pathol 14: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Dangl JL (2012) How complex are intracellular immune receptor signaling complexes? Front Plant Sci 3: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach HH, Wenig M, Wittek F, Jordá L, Maldonado-Alconada AM, Sarioglu H, Colby T, Knappe C, Bichlmeier M, Pabst E, et al. (2014) Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1, and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol 165: 791–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5: 715–725 [Google Scholar]
- Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK (2013) Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front Plant Sci 4: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, Yu K, Sekine KT, Gao QM, Selote D, Hu Y, Stromberg A, Navarre D, et al. (2011) Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat Genet 43: 421–427 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J (2008) Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Venables B, Petros RA, Nalam V, Li M, Wang X, Takemoto LJ, Shah J (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J 71: 161–172 [DOI] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162: 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact 18: 511–520 [DOI] [PubMed] [Google Scholar]
- Chern MS, Fitzgerald HA, Yadav RC, Canlas PE, Dong X, Ronald PC (2001) Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J 27: 101–113 [DOI] [PubMed] [Google Scholar]
- Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjaer MF, Dudler R, Schweizer P (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact 17: 109–117 [DOI] [PubMed] [Google Scholar]
- Colebrook EH, Creissen G, McGrann GRD, Dreos R, Lamb C, Boyd LA (2012) Broad-spectrum acquired resistance in barley induced by the Pseudomonas pathosystem shares transcriptional components with Arabidopsis systemic acquired resistance. Mol Plant Microbe Interact 25: 658–667 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, et al. (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF (2012) SOS: too many signals for systemic acquired resistance? Trends Plant Sci 17: 538–545 [DOI] [PubMed] [Google Scholar]
- Eckey C, Korell M, Leib K, Biedenkopf D, Jansen C, Langen G, Kogel KH (2004) Identification of powdery mildew-induced barley genes by cDNA-AFLP: functional assessment of an early expressed MAP kinase. Plant Mol Biol 55: 1–15 [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Felle HH, Waller F, Molitor A, Kogel KH (2009) The mycorrhiza fungus Piriformospora indica induces fast root-surface pH signaling and primes systemic alkalinization of the leaf apoplast upon powdery mildew infection. Mol Plant Microbe Interact 22: 1179–1185 [DOI] [PubMed] [Google Scholar]
- Fitzgerald HA, Chern MS, Navarre R, Ronald PC (2004) Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact 17: 140–151 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gao QM, Kachroo A, Kachroo P (2014) Chemical inducers of systemic immunity in plants. J Exp Bot 65: 1849–1855 [DOI] [PubMed] [Google Scholar]
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E, Yadeta KA, Coaker G (2013) Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol 199: 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2008) Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol Genet Genomics 279: 303–312 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Fodor J, Preis C, Kogel KH (1999) Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol 119: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BK, Heitefuss R (1982) Induced resistance of spring barley to Erysiphe graminis f. sp. hordei. Phytopathol Z 103: 41–47 [Google Scholar]
- Imani J, Li L, Schäfer P, Kogel KH (2011) STARTS: a stable root transformation system for rapid functional analyses of proteins of the monocot model plant barley. Plant J 67: 726–735 [DOI] [PubMed] [Google Scholar]
- Jarosch B, Jansen M, Schaffrath U (2003) Acquired resistance functions in mlo barley, which is hypersusceptible to Magnaporthe grisea. Mol Plant Microbe Interact 16: 107–114 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kogel KH, Beckhove U, Dreschers J, Münch S, Rommé Y (1994) Acquired resistance in barley: the resistance mechanism induced by 2,6-dichloroisonicotinic acid is a phenocopy of a genetically based mechanism governing race-specific powdery mildew resistance. Plant Physiol 106: 1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel KH, Langen G (2005) Induced disease resistance and gene expression in cereals. Cell Microbiol 7: 1555–1564 [DOI] [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen G, von Einem S, Koch A, Imani J, Pai SB, Manohar M, Ehlers K, Choi HW, Claar M, Schmidt R, et al. (2014) The compromised recognition of turnip crinkle virus1 subfamily of microrchidia ATPases regulates disease resistance in barley to biotrophic and necrotrophic pathogens. Plant Physiol 164: 866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Liu D, Chen X, Liu J, Ye J, Guo Z (2012) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 63: 3899–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Leib K, Zhao P, Kogel KH, Langen G (2014) Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and -2 as repressors of the pathogen-inducible gene HvGER4c. Mol Genet Genomics 289: 1331–1345 [DOI] [PubMed] [Google Scholar]
- Liu PP, Bhattacharjee S, Klessig DF, Moffett P (2010) Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Mol Plant Pathol 11: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, von Dahl CC, Klessig DF (2011) The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol 157: 2216–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact 19: 123–129 [DOI] [PubMed] [Google Scholar]
- Makandar R, Nalam VJ, Lee H, Trick HN, Dong Y, Shah J (2012) Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol Plant Microbe Interact 25: 431–439 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu ÜH, Harter K, Jansson C, Wanke D (2008) Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 55: 399–416 [DOI] [PubMed] [Google Scholar]
- Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K, Sato T, Yamaguchi J (2013) The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci 213: 79–87 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, Matsumoto T, Sato K, Schulman A, Muehlbauer GJ,et al. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–717 [DOI] [PubMed] [Google Scholar]
- Memelink J. (2009) Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A, Görlach J, Volrath S, Ryals J (1999) Wheat genes encoding two types of PR-1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol Plant Microbe Interact 12: 53–58 [DOI] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, Pons-K Hnemann J, Samans B, Kogel KH, Waller F (2011) Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Mol Plant Microbe Interact 24: 1427–1439 [DOI] [PubMed] [Google Scholar]
- Morris SW, Vernooij B, Titatarn S, Starrett M, Thomas S, Wiltse CC, Frederiksen RA, Bhandhufalck A, Hulbert S, Uknes S (1998) Induced resistance responses in maize. Mol Plant Microbe Interact 11: 643–658 [DOI] [PubMed] [Google Scholar]
- Morrissey KL (2007) Biological and chemical induction of systemic resistance in the barley powdery mildew pathosystem. PhD thesis. Forschungszentrum Juelich, Juelich, Germany [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer T, Kugler KG, Bader KC, Sharma S, Seidel M, Mayer KFX (2014) RNASeqExpressionBrowser: a web interface to browse and visualize high-throughput expression data. Bioinformatics 30: 2519–2520 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Petti C, Khan M, Doohan F (2010) Lipid transfer proteins and protease inhibitors as key factors in the priming of barley responses to Fusarium head blight disease by a biocontrol strain of Pseudomonas fluorescens. Funct Integr Genomics 10: 619–627 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J, Peñas G, Messeguer J, Brugidou C, San Segundo B (2008) The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact 21: 1215–1231 [DOI] [PubMed] [Google Scholar]
- Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z (2006) CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res 34: W498–W503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Mueller-Roeber B, Valle EM (2013) Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol 83: 265–277 [DOI] [PubMed] [Google Scholar]
- Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA (2014) Signaling by small metabolites in systemic acquired resistance. Plant J 79: 645–658 [DOI] [PubMed] [Google Scholar]
- Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC (2013) Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant 6: 250–260 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ülker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64: 86–99 [DOI] [PubMed] [Google Scholar]
- Smith JA, Metraux JP (1991) Pseudomonas syringae pv. syringae induces systemic resistance to Pyricularia oryzae in rice. Physiol Mol Plant Pathol 39: 451–461 [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Sugano S, Jiang CJ, Miyazawa S, Masumoto C, Yazawa K, Hayashi N, Shimono M, Nakayama A, Miyao M, Takatsuji H (2010) Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol Biol 74: 549–562 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T (2008) The plant B3 superfamily. Trends Plant Sci 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Métraux JP, Mauch-Mani B (2005) Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman WM, Bennett MH, Turnbull CG, Grant MR (2010) Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol 152: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K, Satou J, Iwata M, Uozumi N, Koga J, Kawano T, Koshiba T, Anzai H, Mitomi M (2009) Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J 57: 463–472 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu PP, Cameron RK, Park SW, Yang Y, Kumar D, Zhou F, Padukkavidana T, Gustafsson C, Pichersky E, et al. (2008) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J 56: 445–456 [DOI] [PubMed] [Google Scholar]
- von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Kessler T, Schmitt-Kopplin P, Schäffner AR (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23: 4124–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Hückelhoven R, Neumann C, von Wettstein D, et al. (2005) The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA 102: 13386–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Mukherjee K, Deshmukh SD, Achatz B, Sharma M, Schäfer P, Kogel KH (2008) Systemic and local modulation of plant responses by Piriformospora indica and related Sebacinales species. J Plant Physiol 165: 60–70 [DOI] [PubMed] [Google Scholar]
- Walters DR, Ratsep J, Havis ND (2013) Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot 64: 1263–1280 [DOI] [PubMed] [Google Scholar]
- Wang C, El-Shetehy M, Shine MB, Yu K, Navarre D, Wendehenne D, Kachroo A, Kachroo P (2014) Free radicals mediate systemic acquired resistance. Cell Reports 7: 348–355 [DOI] [PubMed] [Google Scholar]
- Wiberg A. (1974) Genetical studies of spontaneous sources of resistance to powdery mildew in barley. Hereditas 77: 89–148 [DOI] [PubMed] [Google Scholar]
- Wittek F, Hoffmann T, Kanawati B, Bichlmeier M, Knappe C, Wenig M, Schmitt-Kopplin P, Parker JE, Schwab W, Corina Vlot A (2014) Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. J Exp Bot 65: 5919–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yi G, Peng X, Huang B, Liu E, Zhang J (2013) Systemic acquired resistance in Cavendish banana induced by infection with an incompatible strain of Fusarium oxysporum f. sp. cubense. J Plant Physiol 170: 1039–1046 [DOI] [PubMed] [Google Scholar]
- Yang KH, Huang CJ, Liu YH, Chen CY (2011) Efficacy of probenazole for control of southern corn leaf blight. J Pestic Sci 36: 235–239 [Google Scholar]
- Yang Y, Zhao J, Liu P, Xing H, Li C, Wei G, Kang Z (2013) Glycerol-3-phosphate metabolism in wheat contributes to systemic acquired resistance against Puccinia striiformis f. sp. tritici. PLoS ONE 8: e81756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Nakashita H, Klessig DF, Yamaguchi I (2001) Probenazole induces systemic acquired resistance in Arabidopsis with a novel type of action. Plant J 25: 149–157 [DOI] [PubMed] [Google Scholar]
- Yu K, Soares JM, Mandal MK, Wang C, Chanda B, Gifford AN, Fowler JS, Navarre D, Kachroo A, Kachroo P (2013) A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Reports 3: 1266–1278 [DOI] [PubMed] [Google Scholar]
- Zeier J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36: 2085–2103 [DOI] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol 164: 1499–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller M, Stingl N, Krischke M, Fekete A, Waller F, Berger S, Mueller MJ (2012) Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol 160: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.