Mutation of a cytochrome P450 reductase affects lignin biosynthesis and improves cell wall breakdown, making the enzyme a potential target in cell wall engineering for biofuel production.
Abstract
ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE1 (ATR1) and ATR2 provide electrons from NADPH to a large number of CYTOCHROME P450 (CYP450) enzymes in Arabidopsis (Arabidopsis thaliana). Whereas ATR1 is constitutively expressed, the expression of ATR2 appears to be induced during lignin biosynthesis and upon stresses. Therefore, ATR2 was hypothesized to be preferentially involved in providing electrons to the three CYP450s involved in lignin biosynthesis: CINNAMATE 4-HYDROXYLASE (C4H), p-COUMARATE 3-HYDROXYLASE1 (C3H1), and FERULATE 5-HYDROXYLASE1 (F5H1). Here, we show that the atr2 mutation resulted in a 6% reduction in total lignin amount in the main inflorescence stem and a compositional shift of the remaining lignin to a 10-fold higher fraction of p-hydroxyphenyl units at the expense of syringyl units. Phenolic profiling revealed shifts in lignin-related phenolic metabolites, in particular with the substrates of C4H, C3H1 and F5H1 accumulating in atr2 mutants. Glucosinolate and flavonol glycoside biosynthesis, both of which also rely on CYP450 activities, appeared less affected. The cellulose in the atr2 inflorescence stems was more susceptible to enzymatic hydrolysis after alkaline pretreatment, making ATR2 a potential target for engineering plant cell walls for biofuel production.
Economic and environmental concerns have stimulated the production and use of renewable materials and fuels. Plant cell wall biomass has the potential to become a renewable feedstock for processing into chemicals and liquid fuels in the biorefinery (Vanholme et al., 2013b). Plant cell walls contain polysaccharides (mainly cellulose and hemicelluloses) that can be hydrolyzed into fermentable sugars via enzymatic saccharification. However, the presence of lignin in the cell wall severely hampers the saccharification process (Chen and Dixon, 2007; Van Acker et al., 2013). As a consequence, lignin must be removed, or at least partially cleaved, using biomass pretreatments that require chemicals and energy, prior to saccharification. These pretreatments contribute a significant cost to the production of cell wall-based biofuels in the biorefinery. Strategies to alter lignin content or composition are promising for reducing the need for and/or costs of pretreatments (Sticklen, 2006; Chen and Dixon, 2007; Weng and Chapple, 2010; Fu et al., 2011; Jung et al., 2013; Van Acker et al., 2013, 2014).
Lignin is a polymer derived from three differentially ring-methoxylated 4-hydroxycinnamyl alcohols called monolignols. The two major monolignols incorporated into the lignin polymer in angiosperms are coniferyl alcohol and sinapyl alcohol, which give rise to guaiacyl (G) and syringyl (S) units, respectively (Fig. 1). Lignin also incorporates small amounts of p-coumaryl alcohol, which gives rise to p-hydroxyphenyl (H) units. A number of other hydroxyphenylpropanoids, and other phenolic monomers, can be incorporated into the lignin polymer as well (Baucher et al., 1998; Boerjan et al., 2003; Ralph et al., 2004; Vanholme et al., 2012a). The structural genes involved in lignin biosynthesis have been rather well elucidated, at least in model species such as Arabidopsis (Arabidopsis thaliana), poplar (Populus spp.), and alfalfa (Medicago sativa; Bonawitz and Chapple, 2010; Vanholme et al., 2010a), although additional genes are still being discovered (Berthet et al., 2011; Vanholme et al., 2012b, 2013c; Zhao et al., 2013; Petrik et al., 2014).
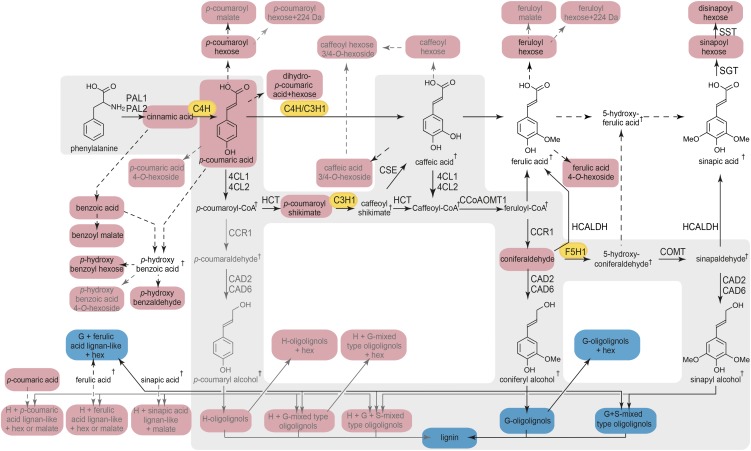
Figure 1.
General phenylpropanoid- and monolignol-specific pathways and metabolic map of the changes in phenolic metabolism in atr2 mutant stems. The main route toward lignin (i.e. the general phenylpropanoid- and monolignol-specific pathway) is marked with a gray background. Metabolites that accumulate in atr2 mutants are marked in red, and those that decrease in atr2 mutants are marked in blue. Metabolic routes that are active in stems of wild-type plants are given in black. Metabolic routes of which the metabolites are near or below the detection limit in wild-type plants but detected in atr2 mutants are given in gray. Metabolites that are below the detection limit in both wild-type and mutant plants are indicated with daggers. The three CYP450s are marked in orange. PAL, PHENYLALANINE AMMONIA LYASE; C4H, CINNAMATE 4-HYDROXYLASE; 4CL, 4-COUMARATE:COENZYME A LIGASE; HCT, p-HYDROXYCINNAMOYL-COENZYME A:QUINATE SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE; C3H, p-COUMARATE 3-HYDROXYLASE; CSE, CAFFEOYL SHIKIMATE ESTERASE, CCoAOMT:CAFFEOYL-COENZYME A O-METHYLTRANSFERASE; CCR, CINNAMOYL-COENZYME A REDUCTASE; F5H, FERULATE 5-HYDROXYLASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; CAD, CINNAMYL ALCOHOL DEHYDROGENASE; HCALDH, HYDROXYCINNAMALDEHYDE DEHYDROGENASE; SGT, SINAPATE 1-GLUCOSYLTRANSFERASE; SST, SINAPOYLGLUCOSE:SINAPOYLGLUCOSE SINAPOYLTRANSFERASE. For each enzymatic step of the general phenylpropanoid- and monolignol-specific pathways, specific gene family members are given that largely contribute to lignification in the inflorescence stem (Sibout et al., 2005; Vanholme et al., 2012b).
Starting with Phe, monolignols are synthesized via the general phenylpropanoid- and monolignol-specific pathways (Fig. 1). Up to three hydroxylation steps that are catalyzed by specific CYTOCHROME P450 (CYP450) oxidases occur on the aromatic ring; these are the 4-, 3-, and 5-hydroxylations catalyzed by C4H, C3H1, and F5H1 (Fig. 1; Meyer et al., 1996; Franke et al., 2002; Schilmiller et al., 2009). C4H, C3H1, and F5H1 belong to the class II type CYP450s. Members of this class rely on CYTOCHROME P450 REDUCTASE (CPR) activity to transfer electrons from NADPH either directly to the CYP450s or via CYTOCHROME B5s (CB5s; Ilan et al., 1981; Hannemann et al., 2007; Jensen and Møller, 2010; Im and Waskell, 2011). Animals and yeast carry only one CPR gene, whereas higher plants often have two or more (Ro et al., 2002), possibly because of the relatively higher number of CYP450 enzymes in plants. Both CYP450s and CPRs are generally accepted to be localized at the membrane of the endoplasmic reticulum (they occur in a 15:1 ratio in rat liver microsomal membranes; Shephard et al., 1983), although a number of CPR-requiring CYP450s have been predicted and/or shown to be localized in the chloroplast (Ro et al., 2002; Bak et al., 2011).
Arabidopsis has 244 CYP450 genes and 28 CYP450 pseudogenes (Bak et al., 2011). In contrast, only three CPR genes have been identified in the Arabidopsis genome. These CPRs are annotated as ARABIDOPSIS THALIANA P450 REDUCTASE1 (ATR1), ATR2, and ATR3 (Urban et al., 1997; Varadarajan et al., 2010). Of these three genes, ATR1 and ATR2 encode genuine CPR proteins that share 64% amino acid identity, according to The Arabidopsis Information Resource genome annotation version 10. ATR3 has been described as a putative CPR but shares only 25.7% and 24% amino acid identity with ATR1 and ATR2, respectively. Unlike ATR1 and ATR2, recombinant ATR3 was unable to support human CYP1A2, and there is no experimental evidence to date that it can support CYP450s (Varadarajan et al., 2010). In addition, Arabidopsis has two genes coding for CYTOCHROME B5 REDUCTASE (CBR) proteins. CBR1 is located at the membrane of the endoplasmic reticulum but is mainly involved in electron transport for the biosynthesis of polyunsaturated fatty acids, whereas CBR2 is in the mitochondrial membrane, and there is no evidence yet that it could take part in the endoplasmic reticulum electron transport system (Wayne et al., 2013).
ATR1 and ATR2 catalyze the same reaction and can reduce C4H in yeast and baculovirus expression systems with the same efficiency (Urban et al., 1997; Mizutani and Ohta, 1998). Despite this observation, there is clear evidence that ATR1 and ATR2 have distinct functions in the plant. For example, ATR1 is constitutively expressed, whereas ATR2 is induced by wounding and light (Mizutani and Ohta, 1998). In addition, the expression of ATR2, but not that of ATR1, is correlated with the expression of a number of genes from the phenylpropanoid pathway; microarray analysis of Arabidopsis rosette leaves subjected to cold treatment under high-light conditions displayed a coordinated expression of ATR2 with phenylpropanoid biosynthetic genes such as PAL2, CCR1, and CCoAOMT1 (Soitamo et al., 2008). Also, transcript profiling of Arabidopsis stems during fiber differentiation showed that expression of ATR2, in contrast to that of ATR1, is coregulated with a set of genes of the general phenylpropanoid- and monolignol-specific pathways, including the three CYP450 genes C4H, C3H1, and F5H1 (Ehlting et al., 2005).
Given that the ATR enzymes may need to serve up to 244 different CYP450 enzymes in Arabidopsis, it is surprising that one of them, ATR2, is coexpressed with the lignin biosynthetic genes. This coexpression suggests that ATR2 provides electrons preferentially to the three CYP450s of the phenylpropanoid and monolignol biosynthetic pathways, making ATR2 a good target for lignin modification. To test this hypothesis, we studied the consequences of the down-regulation of ATR2 in Arabidopsis on phenolic metabolism, lignin structure, and saccharification potential. As we will show, our data confirm that ATR2 has a role in the lignin biosynthesis pathway and that ATR2 mutation reduces the electron donor activity to the three CYP450s involved in lignin biosynthesis, as indicated by the shift in the phenolic profile and lignin composition. Additionally, the atr2 mutant biomass has improved saccharification efficiency.
RESULTS
ATR2 Is Coexpressed with Lignin Biosynthetic Genes
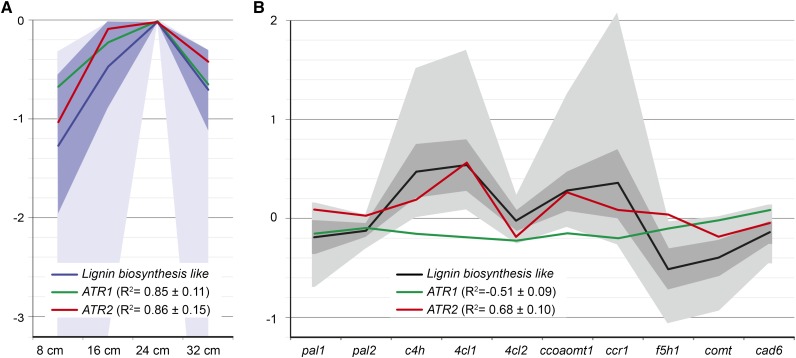
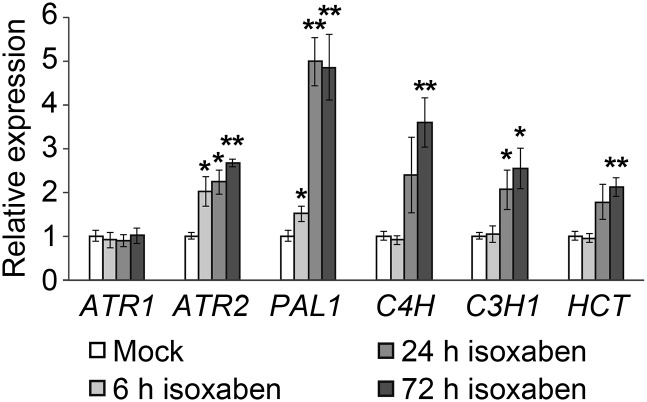
We have previously investigated the system-wide responses of lignin biosynthesis perturbation in Arabidopsis (Vanholme et al., 2012b). To this end, a total of 20 mutants, each mutated in a single lignin biosynthetic gene (two mutant alleles for 10 genes), were investigated with transcriptomics and metabolomics (Vanholme et al., 2012b). This enabled the identification of genes that were coexpressed with known lignin biosynthetic genes and, therefore, that are good candidates to be involved in lignin biosynthesis. Two of the candidates, TRANSALDOLASE2 and CSE, were pathway genes subsequently proven via reverse genetics to be involved in lignification (Vanholme et al., 2012b, 2013c). Reevaluation of the transcriptome data showed that both ATR1 and ATR2 followed a similar expression profile to those of lignin biosynthetic genes at various stages of wild-type Arabidopsis inflorescence stem development (Fig. 2). Moreover, it is apparent that ATR2, but not ATR1, is coexpressed with lignin biosynthetic genes in the different lignin biosynthesis mutants (Fig. 2). The potential coexpression of ATR2 and ATR1 with lignin biosynthetic genes was further investigated by quantitative reverse transcription (qRT)-PCR in Arabidopsis seedlings treated with isoxaben. This herbicide impairs cellulose biosynthesis, which in turn induces stress-related processes such as ectopic lignification (Caño-Delgado et al., 2003; Duval and Beaudoin, 2009). Wild-type seedlings were grown in liquid cultures for 3 d and subjected to treatment with isoxaben for 6, 24, and 72 h. qRT-PCR analysis showed that the expression levels of PAL1, C4H, C3H1, and HCT, which are genes involved in general phenylpropanoid biosynthesis, and ATR2 were significantly induced after 24 h of isoxaben treatment (Fig. 3). More specifically, a similar increase in expression was observed for PAL1 and ATR2 (i.e. the response was already significant after 6 h of treatment). In contrast, ATR1 expression was not affected by isoxaben treatment (Fig. 3). These data again show that ATR2 is coexpressed with phenylpropanoid biosynthetic genes.
Figure 2.
Expression of ATR2 follows that of lignin biosynthetic genes (evaluation of data from Vanholme et al. [2012b]). A, Both ATR1 and ATR2 are coexpressed with lignin biosynthetic genes during inflorescence stem development (at an inflorescence stem height of 8, 16, 24, and 32 cm). The blue line, dark-blue area, and light-blue area indicate average, sd, and spread of the expression, respectively, of the 1,760 probes with a lignin biosynthesis-like expression profile over development (Vanholme et al., 2012b, figure 3, cluster C). The green line indicates expression of ATR1, and the red line indicates expression of ATR2. R2 values are average correlation coefficients ± sd of ATR1 and ATR2 with each of the 1,760 probes with a lignin biosynthesis-like expression profile. B, ATR2, but not ATR1, is coexpressed with genes involved in lignin biosynthesis in lignin mutant backgrounds. The black line, dark gray area, and light gray area indicate average, sd, and spread of the expression, respectively, of the 62 genes with a lignin biosynthesis-like expression profile in the lignin mutants (Vanholme et al., 2012b, figure 9). The green line indicates expression of ATR1, and the red line indicates expression of ATR2. R2 values are average correlation coefficients ± sd of ATR1 and ATR2 with each of the 62 genes with a lignin biosynthesis-like expression profile (Vanholme et al. 2012b).
Figure 3.
qRT-PCR analysis of wild-type seedlings either untreated or treated with 80 nm isoxaben for 6, 24, and 72 h. Data represent mean values of four biological pools of 20 seedlings each, and error bars represent se. The relative expression in mock conditions was set to 1 for each gene tested. *0.05 > P > 0.01, **P < 0.01.
Growth and Development of atr2 Mutants
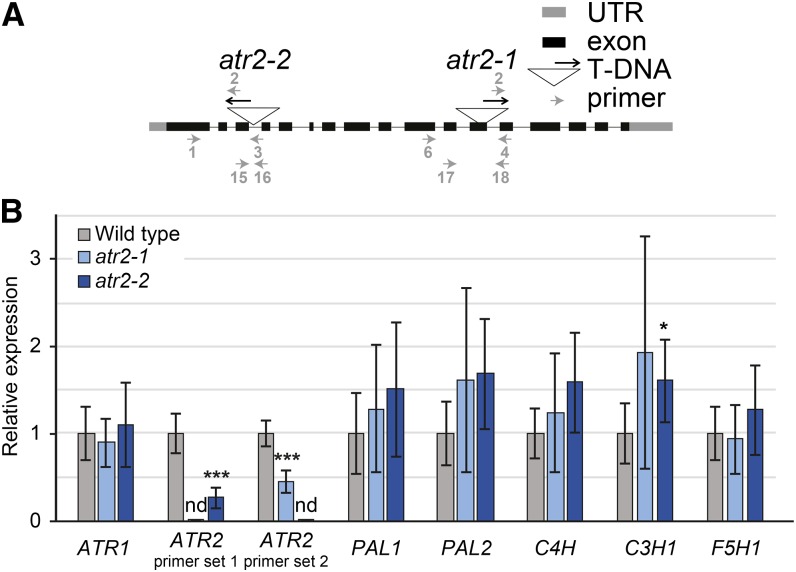
To further investigate the role of ATR2 during lignin biosynthesis, we isolated two insertion mutants from the SALK collection, SALK_152766 and SALK_026053 (Alonso et al., 2003). The transfer DNA (T-DNA) insertions in the ATR2 genes were validated by PCR, and the corresponding mutant alleles were designated atr2-1 and atr2-2, respectively (Fig. 4). Allele atr2-2 contains a T-DNA in the third intron of ATR2, whereas atr2-1 contains a T-DNA in the 12th exon (Fig. 4). Next, ATR2 expression levels in inflorescence stems were determined via qRT-PCR with two sets of primers. Primer set 1 spanned the atr2-1 T-DNA insertion position, and primer set 2 spanned the atr2-2 T-DNA insertion position. The ATR2 transcript levels in atr2-1 and atr2-2 mutants were below the detection limit when measured with the respective T-DNA-spanning primers, proving that the native ATR2 mRNA was absent or close to being absent (Fig. 4). However, residual ATR2 expression was detected in atr2-1 mutants using primer set 2 and in atr2-2 mutants using primer set 1, indicating that residual truncated mRNA fragments of ATR2 were present in the atr2 mutants. The expression levels of ATR1, the constitutive CPR, remained unaltered in both mutant lines, demonstrating that the reduction in ATR2 transcript levels was not compensated by an increased ATR1 expression (Fig. 4).
Figure 4.
Characterization of atr2 T-DNA insertion mutants. A, Representation of the ATR2 genomic structure. Exons are represented as black boxes and introns as gray lines. The T-DNA insertion sites for the two alleles atr2-1 and atr2-2 are indicated by triangles. The orientation of each T-DNA is indicated by a black arrow pointing in the direction of the left border. The primers used to confirm the mutants are represented as gray arrows. The numbers of the primers correspond to those given in “Materials and Methods.” UTR, Untranslated region. B, Expression analysis of ATR1, ATR2, PAL1, PAL2, C4H, C3H1, and F5H1 in atr2-1 and atr2-2 mutants and the wild type as determined via qRT-PCR. Two primer pairs were used to determine the expression of ATR2. Primer sets 1 (i.e. primers 17 and 18) and 2 (i.e. primers 15 and 16) spanned the atr2-1 and atr2-2 T-DNA insertion positions, respectively. For each gene tested, the relative expression was normalized to the one of the wild type. nd, Signal below detection limit. *0.05 > P > 0.01, ***P < 0.001.
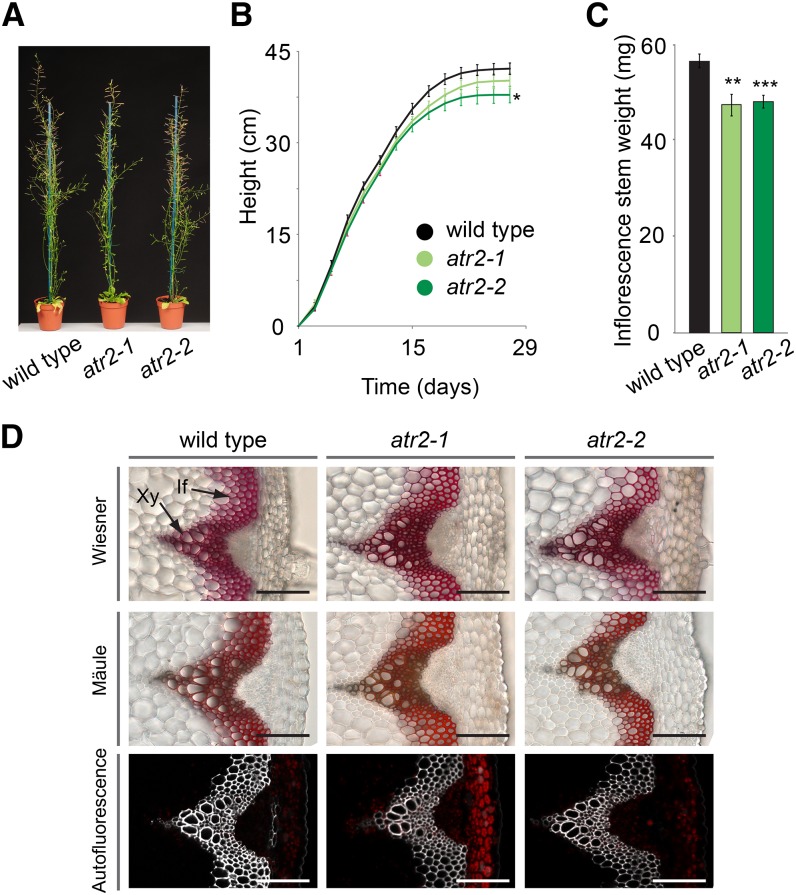
To investigate whether mutation of ATR2 had any effect on growth, the mutants were grown in soil under short-day conditions for 8 weeks and subsequently transferred to long-day conditions. Short-day conditions lead to the development of large vegetative rosettes, which allow the development of a large inflorescence stem upon a shift to long-day conditions. The final height of the atr2-1 mutant was not significantly different from that of the wild type, whereas atr2-2 was 10% smaller at final height (Fig. 5). Analysis of the final dry weight of the main stem revealed 19% and 17% reductions for atr2-1 and atr2-2, respectively, compared with the wild type (Fig. 5).
Figure 5.
Phenotypic characterization of atr2 mutants. A, Phenotype of fully grown plants after 8 weeks of short-day growth conditions followed by 5 weeks of long-day growth conditions. B, Growth curves. Height was monitored every 2 d. At the final stage, atr2-2 was significantly smaller compared with the wild type. C, The main inflorescence stem of atr2-1 and atr2-2 showed reduced weight compared with that of the wild type. For B and C, error bars represent se. *0.05 > P > 0.01, **0.01 > P > 0.001, ***P < 0.001. D, Transverse stem sections of atr2 mutants and the wild type. Mäule and Wiesner staining and lignin autofluorescence are shown. If, Interfascicular fibers; Xy, xylem. Bars = 100 µm.
In order to examine potential changes in the morphology of the vascular tissues caused by the mutated ATR2, transverse sections from the bottom part of stems from fully grown but not yet senesced plants were investigated via microscopy. The morphology of the vasculature as well as the lignin was visualized with Wiesner and Mäule staining and via autofluorescence. Neither collapsed xylem vessels, nor any obvious changes in the intensity of either Mäule or Wiesner staining or autofluorescence, was observed (Fig. 5).
atr2 Mutants Show Altered Levels of Phenylpropanoids, Benzenoids, Lignan-Like Molecules, and Oligolignols
To assess the consequences of the mutation of ATR2 at the metabolic level, phenolic profiling of methanol-soluble metabolites was performed via ultra-HPLC-mass spectrometry (MS) on inflorescence stems from plants that were around 24 cm tall. This procedure enables the detection of several classes of aromatic compounds, including metabolites of the lignin biosynthetic pathway and derivatives thereof, as well as glucosinolates and flavonols. A total of 1,067 peaks were integrated in the chromatograms of atr2-1, atr2-2, and wild-type samples (Supplemental Table S1). Principal component analysis showed that atr2 mutant samples separated from wild-type samples based on a combination of the first and second principal components (Supplemental Fig. S1), indicating metabolic differences between the atr2 mutants and wild-type plants. Univariate statistical analysis was applied to select peaks with significantly different abundances in the atr2 mutants as compared with their levels in wild-type plants. Using stringent filters (i.e. P < 0.01, a 2-fold difference in abundance between mutant and the wild type, and a peak intensity of greater than 500), 71 and 73 peaks were found to be significantly lower in abundance in atr2-1 and atr2-2, respectively, of which 61 were in common (Supplemental Fig. S1; Supplemental Table S2); 184 and 229 peaks were found to be significantly higher in abundance in atr2-1 and atr2-2, respectively, of which 177 were in common (Supplemental Fig. S1; Supplemental Table S2). The relatively high fraction of differential peaks that were shared by both mutants indicates that most of the detected differential peaks can be attributed to the defective ATR2 gene.
Further analysis showed that the 61 peaks that were significantly lower in abundance in both atr2 mutants could be assigned to 47 compounds (Supplemental Table S3), of which 30 could be structurally characterized based on their mass-to-charge ratio (m/z), retention time, and tandem mass spectrometry (MS/MS) fragmentation spectrum (Table I; Supplemental Fig. S2). Twenty-six of these 30 characterized compounds belonged to the classes of oligolignols, hexosylated oligolignols, or G(8-O-4/8-5)phenylpropenoic acids/esters. As all of these compounds had at least one G unit, their reduced abundance indicated that the flux toward coniferyl alcohol was reduced in the atr2 mutants. In addition, the abundances of two aliphatic and two indolic glucosinolates (3-methyl sulfinyl glucosinolate, 2-hydroxy-3-butenylglucosinolate, indol-3-yl methylglucosinolate, and 4-hydroxyindol-3-yl-methylglucosinolate) were lower in both atr2 mutants. Because several CYP450-dependent reactions are involved in the biosynthesis of these glucosinolates from their precursor amino acids (Supplemental Fig. S3), the reduced abundance in the atr2 mutants could be a direct result of the lost CYP450 activity. On the other hand, the biosynthesis of all other glucosinolates also relies on two or more CYP450-dependent reactions, but only the four glucosinolates mentioned above were retrieved in the list of compounds with reduced abundance. To investigate if the other glucosinolates could have been missed due to the stringent filters (P < 0.01 and 2-fold difference), a targeted search for aliphatic and indolic glucosinolates was performed (Supplemental Fig. S3). However, no general tendency for a reduced abundance of aliphatic or indolic glucosinolates was apparent, indicating that the effect of atr2 mutation on the CYP450s active in the glucosinolate pathways was moderate or nonexistent or, potentially, only secondary.
Table I. Metabolites with a reduced abundance in atr2 mutants.
An overview of the identified compounds of which the abundance was lower in the atr2-1 and atr2-2 mutants is given. For each compound, average peak areas of wild-type and atr2 mutant samples are given. For the structural elucidation of the compounds, see Supplemental Table S2 and Supplemental Figure S2.
| No. | Retention Time | m/z | Name | Wild Type | atr2-1 | Change in atr2-1:Wild-Type Ratio | atr2-2 | Change in atr2-2:Wild-Type Ratio | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | se | Mean | se | Mean | se | ||||||
| min | |||||||||||
| Glucosinolates | |||||||||||
| 1 | 1.28 | 422.025 | 3-Methylsulfinylpropyl glucosinolate | 176,502 | 7,833 | 48,551 | 4,389 | 0.28 | 46,856 | 2,768 | 0.27 |
| 2 | 3.82 | 388.077 | 2-Hydroxy-3-butenylglucosinolate | 2,701 | 293 | 526 | 129 | 0.19 | 386 | 51 | 0.14 |
| 3 | 3.61 | 895.117 | Indol-3-ylmethylglucosinolate (dimer) | 10,432 | 1,004 | 3,219 | 456 | 0.31 | 2,997 | 273 | 0.29 |
| 4 | 2.10 | 847.149 | 4-Hydroxyindol-3-ylmethylglucosinolate (dimer)a | 2,194 | 129 | 926 | 173 | 0.42 | 1,064 | 159 | 0.49 |
| Oligolignols | |||||||||||
| 5 | 8.84 | 375.145 | G(8-O-4)G | 274 | 58 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 6 | 9.14 | 375.146 | G(8-O-4)G | 222 | 57 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 7 | 12.97 | 339.125 | G(8-5)Ga | 2,063 | 288 | 33 | 17 | 0.02 | 33 | 12 | 0.02 |
| 8 | 15.45 | 337.109 | G(8-5)G'a | 877 | 79 | 13 | 9 | 0.02 | 81 | 27 | 0.09 |
| 9 | 12.80 | 369.136 | S(8-5)Ga | 208 | 41 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 10 | 13.12 | 553.208 | G(8-O-4)G(8-5)G | 356 | 71 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 11 | 13.42 | 553.209 | G(8-O-4)G(8-5)G | 574 | 65 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 12 | 14.71 | 583.218 | G(8-O-4)S(8-5)G | 7,656 | 817 | 78 | 44 | 0.01 | 51 | 25 | 0.01 |
| 13 | 15.52 | 583.219 | G(8-O-4)S(8-5)G | 2,158 | 301 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 14 | 16.25 | 581.203 | G(8-O-4)S(8-5)G' | 829 | 100 | 19 | 19 | 0.02 | 5 | 5 | 0.01 |
| 15 | 12.77 | 555.224 | G(8-O-4)G(red8-5)G | 456 | 80 | 0 | 0 | 0.00 | 5 | 5 | 0.01 |
| 16 | 16.66 | 613.232 | G(8-O-4)S(8-8)S | 37 | 15 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 17 | 16.96 | 583.219 | G(8-O-4)S/G(8-8)G/S | 1,260 | 233 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| 18 | 18.00 | 809.306 | G(8-O-4)G(8-O-4)S(8-8)S | 272 | 63 | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
| Hexosylated oligolignols | |||||||||||
| 19 | 6.06 | 583.203 | G 4-O-hexoside(8-O-4)Gb | 523 | 80 | 24 | 18 | 0.05 | 46 | 30 | 0.09 |
| 20 | 7.61 | 613.212 | G 4-O-hexoside(8-O-4)Sb | 778 | 71 | 46 | 19 | 0.06 | 48 | 21 | 0.06 |
| 21 | 8.20 | 521.203 | G(red8-8)G hexoside | 412 | 60 | 42 | 17 | 0.10 | 53 | 28 | 0.13 |
| 22 | 9.04 | 521.202 | G(red8-8)G hexoside | 882 | 54 | 365 | 73 | 0.41 | 407 | 77 | 0.46 |
| 23 | 9.00 | 565.193 | G 4-O-hexoside(8-5)Gb | 3,748 | 298 | 576 | 91 | 0.15 | 1,053 | 136 | 0.28 |
| 24 | 9.14 | 567.208 | G 4-O-hexoside(red8-5)Gb | 2,975 | 321 | 182 | 81 | 0.06 | 286 | 91 | 0.10 |
| 25 | 10.68 | 521.203 | G(red8-5)G hexoside | 525 | 65 | 159 | 36 | 0.30 | 149 | 23 | 0.28 |
| G(8-O-4/8-5)phenylpropenoic acid/ester | |||||||||||
| 26 | 5.98 | 551.177 | G(8-O-4)feruloyl hexose | 2,527 | 118 | 744 | 147 | 0.29 | 736 | 105 | 0.29 |
| 27 | 11.68 | 775.246 | G(8-O-4)feruloyl hexose + 224 D | 7,413 | 878 | 2,154 | 319 | 0.29 | 3,243 | 383 | 0.44 |
| 28 | 11.89 | 775.246 | G(8-O-4)feruloyl hexose + 224 D | 4,438 | 405 | 1,318 | 171 | 0.30 | 2,025 | 235 | 0.46 |
| 29 | 9.54 | 533.167 | G(8-5)feruloyl hexose | 492 | 39 | 0 | 0 | 0.00 | 16 | 10 | 0.03 |
| 30 | 10.08 | 533.166 | G(8-5)feruloyl hexose | 562 | 44 | 229 | 24 | 0.41 | 116 | 20 | 0.21 |
Compounds detected as fragments of the parent ion. bCompounds detected as formic acid adducts.
Similar to glucosinolate biosynthesis, two CYP450s (C4H and FLAVONOID-3'-HYDROXYLASE) are involved in the biosynthesis of flavonol glycosides (Supplemental Fig. S3), so a reduction in the abundance of flavonol glycosides could be anticipated in atr2 mutants. However, a targeted search showed that kaempferol glycosides were not differentially abundant between atr2 and the wild type (Supplemental Fig. S3). There was a tendency for lower quercetin and isorhamnetin glycoside levels (the levels were about 63%–85% in atr2 mutants as compared with those in the wild type), but these reductions were only significant (P < 0.05) in atr2-1 and not in atr2-2 (Supplemental Fig. S3).
The 177 peaks that were higher in abundance in both atr2 mutants as compared with wild-type plants could be assigned to 117 compounds (Supplemental Table S2), of which 63 could be structurally characterized (Table II; Supplemental Fig. S2). Many of these compounds were below the detection limit in wild-type samples and only observed in the atr2 mutants (Table II; Supplemental Table S1). All identified compounds could be classified into one of the following metabolic classes: phenylpropanoids, benzenoids, oligolignols, hexosylated oligolignols, or H(8-O-4/8-5)phenylpropenoic acids/esters. The majority of the identified phenylpropanoids were esters or hexosides of p-coumaric acid, ferulic acid, or caffeic acid. p-Coumaric acid and p-coumaroyl shikimate, the substrates of the C3H1/C4H complex and C3H1, respectively, were either not or barely detectable in the wild type but accumulated in the atr2 mutants. Similarly, cinnamic acid and coniferaldehyde, the in vivo substrates of C4H and F5H1, respectively, were only detected in atr2 mutants and not in the wild type. The benzenoids that accumulated in atr2 mutants all contained benzoic acid or p-hydroxybenzoic acid moieties and were most likely derived via chain shortening of cinnamic and p-coumaric acids (Hertweck et al., 2001; Boatright et al., 2004).
Table II. Metabolites with an increased abundance in atr2 mutants.
An overview of the identified compounds of which the abundance was higher in the atr2-1 and atr2-2 mutants is given. For each compound, average peak areas of wild-type and atr2 mutant samples are given. Infinity (inf) is given when a compound is below detection limit in the wild type. For the structural elucidation of the compounds, see Supplemental Table S2 and Supplemental Figure S2.
| No. | Retention Time | m/z | Name | Wild Type | atr2-1 | Change in atr2-1:Wild-Type Ratio | atr2-2 | Change in atr2-2:Wild Type Ratio | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | se | Mean | se | Mean | se | ||||||
| min | |||||||||||
| Phenylpropanoids | |||||||||||
| 31 | 13.73 | 147.045 | Cinnamic acid | 0 | 0 | 300 | 99 | inf | 235 | 41 | inf |
| 32 | 6.75 | 163.040 | p-Coumaric acid | 2 | 1 | 1,147 | 136 | 593 | 2,241 | 438 | 1,159 |
| 33 | 4.47 | 325.093 | p-Coumaroyl hexose | 330 | 21 | 10,773 | 1,695 | 33 | 22,378 | 4,991 | 68 |
| 34 | 5.05 | 325.093 | p-Coumaroyl hexose | 22 | 12 | 2,534 | 408 | 114 | 4,305 | 1,051 | 194 |
| 35 | 11.66 | 549.162 | p-Coumaroyl hexose + 224 D | 0 | 0 | 674 | 134 | inf | 1,279 | 245 | inf |
| 36 | 13.36 | 549.161 | p-Coumaroyl hexose + 224 D | 0 | 0 | 728 | 196 | inf | 1,441 | 331 | inf |
| 37 | 7.80 | 279.052 | p-Coumaroyl malate | 0 | 0 | 390 | 73 | inf | 358 | 70 | inf |
| 38 | 8.26 | 279.052 | p-Coumaroyl malate | 0 | 0 | 314 | 93 | inf | 260 | 80 | inf |
| 39 | 7.89 | 319.083 | p-Coumaroyl shikimate | 0 | 0 | 4,164 | 347 | inf | 4,808 | 540 | inf |
| 40 | 9.49 | 319.084 | p-Coumaroyl shikimate | 0 | 0 | 198 | 53 | inf | 254 | 44 | inf |
| 41 | 3.00 | 325.094 | p-Coumaric acid 4-O-hexoside | 0 | 0 | 791 | 77 | inf | 1,458 | 132 | inf |
| 42 | 3.71 | 327.110 | Dihydro-p-coumaric acid + hexose | 0 | 0 | 564 | 121 | inf | 1,173 | 185 | inf |
| 43 | 3.90 | 327.108 | Dihydro-p-coumaric acid + hexose | 5,099 | 215 | 12,814 | 973 | 3 | 16,108 | 572 | 3 |
| 44 | 3.31 | 341.089 | Caffeoyl hexose | 0 | 0 | 667 | 109 | inf | 1,075 | 187 | inf |
| 45 | 2.85 | 341.089 | Caffeic acid 3/4-O-hexoside | 66 | 26 | 2,391 | 255 | 36 | 3,528 | 264 | 54 |
| 46 | 3.97 | 341.089 | Caffeic acid 3/4-O-hexoside | 266 | 23 | 2,085 | 176 | 8 | 2,710 | 149 | 10 |
| 47 | 5.44 | 341.088 | Caffeic acid 3/4-O-hexoside | 0 | 0 | 637 | 102 | inf | 712 | 111 | inf |
| 48 | 2.97 | 503.141 | Caffeoyl hexose 3/4-O-hexoside | 0 | 0 | 1,053 | 130 | inf | 1,683 | 247 | inf |
| 49 | 5.28 | 355.104 | Feruloyl hexose | 377 | 25 | 2,362 | 261 | 6 | 3,852 | 597 | 10 |
| 50 | 5.80 | 355.105 | Feruloyl hexose | 0 | 0 | 187 | 43 | inf | 265 | 65 | inf |
| 51 | 12.31 | 217.051 | Feruloyl hexose + 224 Da | 7 | 5 | 203 | 43 | 29 | 397 | 67 | 58 |
| 52 | 13.55 | 355.104 | Feruloyl hexose + 224 Da | 0 | 0 | 119 | 41 | inf | 277 | 69 | inf |
| 53 | 8.77 | 309.062 | Feruloyl malate | 0 | 0 | 1,273 | 239 | inf | 577 | 116 | inf |
| 54 | 9.13 | 309.062 | Feruloyl malate | 0 | 0 | 1,013 | 161 | inf | 810 | 101 | inf |
| 55 | 3.69 | 355.105 | Ferulic acid 4-O-hexoside | 20 | 10 | 398 | 86 | 20 | 519 | 43 | 26 |
| 56 | 5.48 | 385.114 | Sinapoyl hexose | 8,150 | 713 | 23,447 | 2,222 | 3 | 27,655 | 2,809 | 3 |
| 57 | 6.06 | 205.051 | Sinapoyl hexosea | 23 | 10 | 185 | 17 | 8 | 275 | 43 | 12 |
| 58 | 13.22 | 591.171 | Disinapoyl Glc | 666 | 113 | 4,152 | 843 | 6 | 4,499 | 765 | 7 |
| 59 | 10.21 | 177.056 | Coniferaldehyde | 0 | 0 | 181 | 45 | inf | 392 | 35 | inf |
| Benzenoids | |||||||||||
| 60 | 9.16 | 237.041 | Benzoyl malate | 500 | 38 | 1,581 | 181 | 3 | 2,250 | 179 | 5 |
| 61 | 5.44 | 121.030 | p-Hydroxybenzaldehyde | 81 | 13 | 374 | 39 | 5 | 694 | 64 | 9 |
| 62 | 2.64 | 299.078 | p-Hydroxybenzoyl hexose | 0 | 0 | 320 | 100 | inf | 632 | 199 | inf |
| 63 | 1.85 | 299.078 | p-Hydroxybenzoic acid 4-O-hexoside | 1,738 | 101 | 5,093 | 398 | 3 | 6,300 | 292 | 4 |
| Oligolignols | |||||||||||
| 64 | 12.32 | 309.114 | H(8-5)Ga | 0 | 0 | 346 | 75 | inf | 439 | 52 | inf |
| 65 | 13.16 | 279.104 | H(8-5)Ha | 0 | 0 | 108 | 47 | inf | 170 | 42 | inf |
| 66 | 13.09 | 523.197 | H(8-O-4)G(8-5)G | 0 | 0 | 518 | 107 | 5,318 | 735 | 54 | 7,540 |
| 67 | 14.95 | 521.182 | H(8-O-4)G(8-5)G | 0 | 0 | 301 | 95 | inf | 674 | 56 | inf |
| 68 | 15.12 | 521.182 | H(8-O-4)G(8-5)G | 0 | 0 | 708 | 131 | inf | 1,386 | 117 | inf |
| 69 | 15.94 | 523.197 | H(8-O-4)G(8-5)G | 0 | 0 | 1,223 | 261 | inf | 1,913 | 98 | inf |
| 70 | 16.76 | 523.198 | H(8-O-4)G(8-8)G | 0 | 0 | 497 | 143 | inf | 870 | 77 | inf |
| 71 | 13.85 | 493.187 | H(8-O-4)H/G(8-5)G/H | 0 | 0 | 227 | 69 | inf | 478 | 41 | inf |
| 72 | 14.41 | 553.208 | H(8-O-4)S(8-5)G | 16 | 12 | 2,985 | 425 | 190 | 3,597 | 117 | 229 |
| 73 | 15.16 | 553.208 | H(8-O-4)S(8-5)G | 10 | 7 | 2,341 | 335 | 225 | 2,759 | 74 | 265 |
| 74 | 16.46 | 551.192 | H(8-O-4)S(8-5)G' | 0 | 0 | 2,457 | 464 | inf | 4,413 | 369 | inf |
| 75 | 17.18 | 551.192 | H(8-O-4)S(8-5)G' | 0 | 0 | 948 | 225 | inf | 1,726 | 161 | inf |
| Hexosylated oligolignols | |||||||||||
| 76 | 6.00 | 621.218 | H(8-8)H dihexoside | 119 | 22 | 11,070 | 1,461 | 93 | 17,861 | 2,405 | 150 |
| 77 | 9.00 | 459.166 | H(8-8)H O-4-hexoside | 0 | 0 | 1,952 | 261 | inf | 3,808 | 406 | inf |
| 78 | 9.60 | 489.177 | H(8-8)G hexoside | 0 | 0 | 187 | 50 | inf | 220 | 41 | inf |
| 79 | 6.59 | 651.230 | H(8-8)G dihexoside | 0 | 0 | 569 | 90 | inf | 775 | 111 | inf |
| 80 | 9.16 | 505.171 | H O-4-hexoside(8-5)Hb | 0 | 0 | 4,748 | 538 | 2,149 | 8,575 | 950 | 222,225 |
| 81 | 7.50 | 621.219 | H 8-O-hexoside (8-5)H 8-O-hexoside | 0 | 0 | 211 | 92 | inf | 591 | 194 | inf |
| 82 | 8.53 | 535.181 | H/G(8-5)G/H 4-O-hexosideb | 0 | 0 | 1,903 | 262 | inf | 3,375 | 319 | inf |
| 83 | 10.88 | 461.180 | H(red8-5)H hexoside | 0 | 0 | 129 | 44 | inf | 443 | 65 | inf |
| H(8-O-4/8-5)phenylpropenoic acids/esters | |||||||||||
| 84 | 11.20 | 715.225 | H(8-O-4)p-coumaroyl hexose + 224 D | 0 | 0 | 575 | 175 | inf | 1,357 | 207 | inf |
| 85 | 11.59 | 715.228 | H(8-O-4)p-coumaroyl hexose + 224 D | 0 | 0 | 187 | 65 | 2,149 | 543 | 106 | 6,255 |
| 86 | 10.80 | 473.145 | H 4/9-O hexoside(8-5)p-coumaric acid | 0 | 0 | 172 | 44 | inf | 703 | 187 | inf |
| 87 | 13.62 | 427.103 | H(8-5)p-coumaroyl malate | 0 | 0 | 462 | 159 | inf | 786 | 112 | inf |
| 88 | 13.91 | 427.104 | H(8-5)p-coumaroyl malate | 0 | 0 | 546 | 197 | inf | 831 | 131 | inf |
| 89 | 6.07 | 521.166 | H(8-O-4)ferulic acid hexoside | 0 | 0 | 228 | 46 | inf | 829 | 195 | inf |
| 90 | 11.52 | 745.235 | H(8-O-4)feruloyl hexose + 224 D | 4 | 4 | 1,569 | 397 | 445 | 3,297 | 511 | 936 |
| 91 | 9.11 | 475.125 | H(8-O-4)feruloyl malate | 9 | 5 | 3,252 | 631 | 348 | 4,165 | 474 | 445 |
| 92 | 9.95 | 475.126 | H(8-O-4)feruloyl malate | 3 | 2 | 932 | 277 | 364 | 1,107 | 168 | 433 |
| 93 | 13.16 | 457.114 | H(8-5)feruloyl malate | 0 | 0 | 904 | 184 | inf | 1,186 | 160 | inf |
| 94 | 10.84 | 389.127 | H(8-O-4)sinapoyl malatea | 0 | 0 | 292 | 102 | inf | 455 | 67 | inf |
Compounds detected as fragments of the parent ion. bCompounds detected as formic acid adducts.
All accumulating oligolignols, hexosylated oligolignols, and H(8-O-4/8-5)phenylpropenoic acids/esters contained at least one H unit. This observation indicates a severe increase in the flux toward p-coumaryl alcohol (i.e. the precursor of H units) in atr2 mutants. In some cases, the phenylpropenoic acid/ester moiety in the H(8-O-4/8-5)phenylpropenoic acids/esters class was derived from p-coumaric acid. This observation is in line with the increased amount of p-coumaric acid-containing phenylpropanoids in atr2 mutants and indicative of an increased flux toward p-coumaric acid or a reduced consumption of this metabolite.
atr2 Mutants Are Affected in Lignin Content and Composition
Fully senesced stems of wild-type plants and the atr2 mutants were evaluated for lignin content and composition. Soluble compounds were removed from these stems by applying a sequential extraction to produce cell wall residues (CWRs; Jarvis et al., 2000; Van Acker et al., 2013). For both atr2 mutants and the wild type, the proportion of CWR relative to the dry weight of the stem was similar at approximately 80% (Table III).
Table III. Cell wall characterization of atr2.
The CWR is the material remaining after sequential extraction of dry senesced stem biomass with hydrophobic and hydrophilic solvents. Lignin content was measured with the acetyl bromide assay. Cellulose content was measured with the Updegraff method. Monomer composition was determined by thioacidolysis. P values are from Student’s t test: *0.05 > P > 0.01, **0.01 > P > 0.001, and ***P < 0.001; n = 8. No diff, No difference.
| Parameter | Wild Type | atr2-1 | P | Change in atr2-1 versus the Wild Type | atr2-2 | P | Change in atr2-2 versus the Wild Type | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | se | Mean | se | Mean | se | |||||
| % | % | |||||||||
| CWR (%) | 77.8 | 0.6 | 76.2 | 0.6 | 0.08 | No diff | 77.0 | 1.1 | 0.3 | No diff |
| Lignin per CWR (%) | 20.8 | 0.4 | 19.6 | 0.2 | 0.03* | −6 | 19.6 | 0.2 | 0.03* | −6 |
| Cellulose per CWR (%) | 30.9 | 1.2 | 31.2 | 0.4 | 0.84 | No diff | 32.5 | 1.7 | 0.5 | No diff |
| H units per CWR (µmol mg−1) | 0.38 | 0.03 | 5.9 | 0.32 | 4.6E-07*** | +1,454 | 6.1 | 0.25 | 5.0E-08*** | +1,516 |
| G units per CWR (µmol mg−1) | 48.4 | 1.28 | 41.9 | 2.97 | 0.06 | No diff | 42.3 | 1.51 | 7.8E-03** | −13 |
| S units per CWR (µmol mg−1) | 23.0 | 1.23 | 14.8 | 1.01 | 1.5E-04*** | −35 | 13.6 | 0.50 | 5.9E-06*** | −41 |
| S/G ratio | 0.47 | 0.02 | 0.32 | 0.01 | 2.7E-04*** | −32 | 0.36 | 0.01 | 8.7E-05*** | −25 |
| (H+G+S) per Lignin (µmol mg−1) | 344.7 | 10.9 | 319.4 | 20.8 | 0.3 | No diff | 317.0 | 9.5 | 0.08 | No diff |
The total lignin amount present in these prepared CWRs was estimated by the spectrophotometric acetyl bromide assay. In this method, the cell wall polymers are derivatized by acetyl bromide and dissolved in acetic acid. A reduction in lignin content of 6% was detected in both atr2-1 and atr2-2 (Table III).
Senesced stem material of both mutant lines was analyzed further for potential changes in lignin composition. This was first done by thioacidolysis, a procedure that comprises cleavage of the β-O-4-ether bonds of the lignin polymer to release hydroxyphenyl-trithioethyl ether monomers, followed by derivatization for subsequent identification and quantification by gas chromatography coupled to MS. Thioacidolysis showed significant changes in lignin composition in both atr2 lines, with H units increasing up to 16-fold (Table III). Thioacidolysis-released S units decreased significantly by 35% and 41% in atr2-1 and atr2-2 (Table III). Levels of thioacidolysis-released G units were significantly reduced by 13% for the atr2-2 line compared with the wild type based on CWR (Table III). A similar decrease in thioacidolysis-released G units was also measured for atr2-1 but was not significant below the 0.05 P value threshold (P = 0.06). Consequently, reductions in the S/G ratio by 32% and 25% were observed for atr2-1 and atr2-2, respectively. The degree of lignin condensation inversely correlates with the sum of H, G, and S units released from the lignin by thioacidolysis. This sum was not significantly different from that of the wild type (Table III).
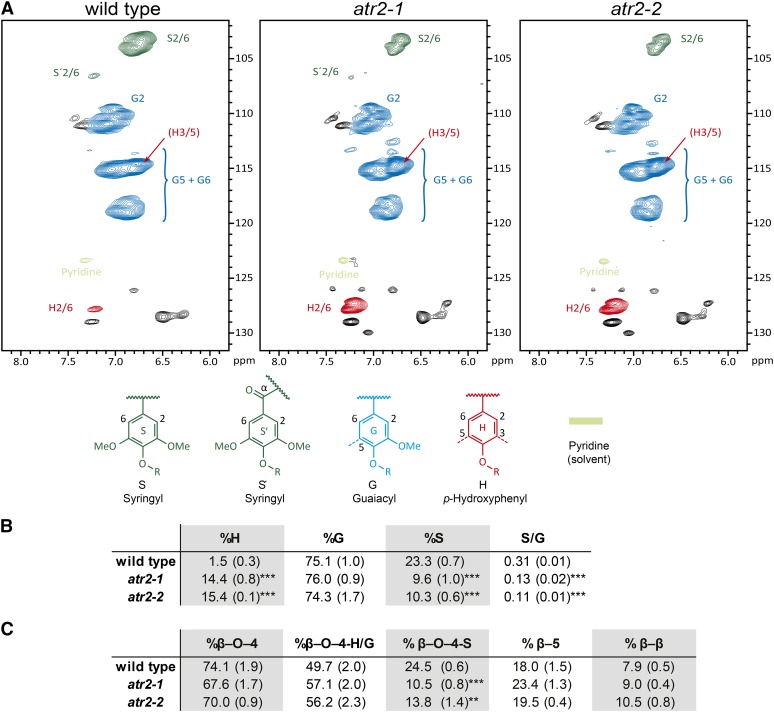
Changes in both lignin aromatic and side chain regions were further revealed by applying two-dimensional 1H-13C correlation heteronuclear single quantum coherence (HSQC) NMR on whole-plant cell wall samples of the wild type and the atr2 mutants prepared from senesced main stems (Fig. 6; Supplemental Fig. S4). In contrast to thioacidolysis, which determines the composition of units linked only by ether bonds, NMR analysis of the aromatic region allows the investigation of all monomers irrespective of their mode of linkage. In addition, interunit linkage types can be determined by use of the aliphatic region of the spectrum. Compared with the wild type, the fraction of H units in the lignin of atr2 mutants was increased about 10-fold (Fig. 6). The relative fraction of G units was not significantly different, but the relative fraction of S units was reduced in atr2 mutants by almost 60% compared with the wild type. Accordingly, the S/G ratio decreased significantly from 0.31 in the wild type to 0.13 and 0.11 in the atr2-1 and atr2-2 mutants (Fig. 6). The HSQC spectrum fully resolved correlations for the β-aryl ether (β-O-4), phenylcoumaran (β-5), and resinol (β-β) units. The β-syringyl ether linkages, (β-O-4)S, were reduced, reflecting the lower S unit content, whereas those linkages in which the aryl moiety is an H or G unit, (β-O-4)H/G, were seemingly not affected (Fig. 6).
Figure 6.
NMR lignin characterization. A, Two-dimensional HSQC NMR spectra of the lignin aromatic region of senesced stem material from the wild type, atr2-1, and atr2-2. The volume integrals of the S2/6, G2, and H2/6 contour peaks, when scaled (by 2 for G), represents the quantity of each monomer. B, Overview of the relative amounts of the different monolignol-derived units and the S/G ratio as determined from the HSQC aromatic region. C, Overview of the relative amount (as determined by uncorrected volume integrals) of interunit linkage types (β-O-4, β-5, and β-β) as determined from the HSQC side chain region (for the spectra of the side chain regions, see Supplemental Fig. S4). **0.01 > P > 0.001, ***P < 0.001; n = 3.
Transcriptional Feedback on CYP450s Involved in Lignin Biosynthesis
To investigate the possible feedback caused by ATR2 deficiency (and the concomitant shifts in phenolic metabolism) on the transcript levels of the lignin-associated CYP450s, the expression of C4H, C3H1, and F5H1 in inflorescence stems was investigated via qRT-PCR (Fig. 4B). In addition, expression levels of PAL1 and PAL2 were used as controls, as these are not CYP450-encoding genes but also involved in lignin biosynthesis. No significant differences were observed in the levels of the measured transcripts between the atr2-1 mutant and the wild type. In the atr2-2 mutant, C3H1 expression was significantly increased as compared with the wild type (P = 0.049), and the expression of PAL2 and C4H was on the border of being increased significantly, with P values of 0.059 and 0.064, respectively.
atr2 Mutant Biomass Is Easier to Saccharify
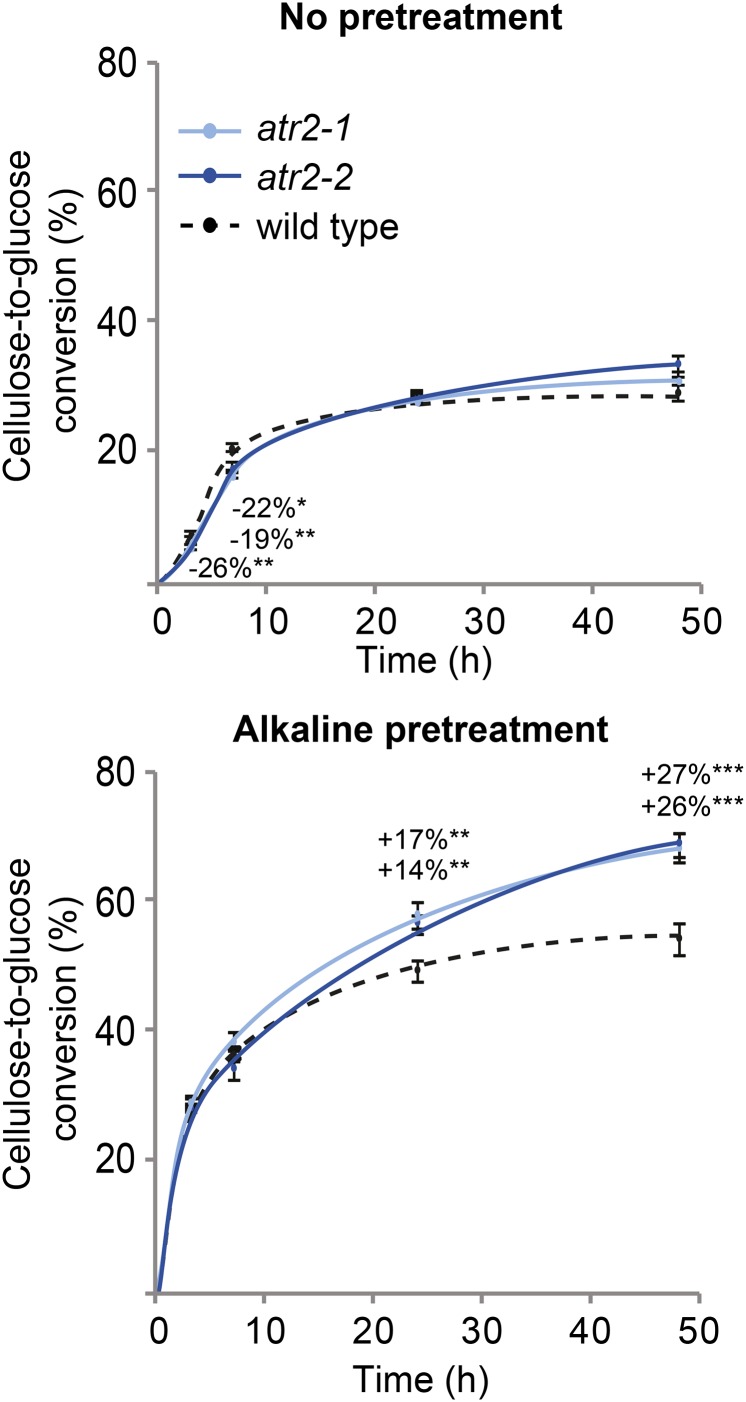
Lignin content and composition generally have an impact on cell wall digestibility. Because atr2 mutants have reduced lignin levels and shifts in lignin composition compared with the wild type, we investigated the saccharification of senesced atr2 mutant stems. First, the cellulose content of the prepared CWRs was determined by the Updegraff method. In this method, the crystalline cellulose polymer that remains after acetic and nitric acid treatment is broken down to Glc by sulfuric acid; the amount of released Glc is determined spectrophotometrically after derivatization with anthrone. No significant changes in crystalline cellulose levels were detected in the atr2 mutant lines when compared with the wild type (Table III). Saccharification measures the amount of Glc released from the enzymatic breakdown of cellulose present in the plant cell wall. The applied procedure included either no pretreatment or an alkaline (6.25 mm NaOH) pretreatment to loosen the cell wall components, thereby facilitating access of the enzymes to the cell wall polysaccharides. The released Glc was measured after 3, 7, 24, and 48 h. Without pretreatment, a significantly reduced Glc release was observed after 3 h of saccharification for both atr2 mutants and after 7 h for atr2-1 only. However, after 48 h of saccharification, atr2-2 released more Glc than the wild type. The cellulose-to-Glc conversion improved strikingly after alkaline pretreatment; after 24- and 48-h incubation times, 26% and 27% improvements were reached in atr2-1 and atr2-2, respectively, as compared with the wild type (Fig. 7).
Figure 7.
Saccharification data of the atr2 mutants. Samples were saccharified with either no pretreatment or an alkaline pretreatment (6.25 mm NaOH). The cellulose-to-Glc conversion was measured at 3, 7, 24, and 48 h (n = 8). Error bars represent se. *0.05 > P > 0.01, **0.01 > P > 0.001, ***P < 0.001; n = 8.
DISCUSSION
ATR2 Is Involved in Lignification
ATR2-deficient plants have 6% less lignin with a different composition compared with wild-type plants (Table III; Fig. 6). The composition of lignin has traditionally been an important clue in determining at which position in the pathway the gene in question is acting. A relative increase in H units has been reported in plants in which the flux from p-coumaroyl-CoA to caffeoyl-CoA and caffeic acid was disturbed (i.e. in plants with reduced HCT, C3H, and/or CSE activity; Abdulrazzak et al., 2006; Ralph et al., 2006, 2012; Coleman et al., 2008; Vanholme et al., 2013c; Bonawitz et al., 2014). The 10- to 15-fold increase in H units in atr2 mutants (Table III; Fig. 6), therefore, is likely a direct result of reduced C3H1 activity because of the compromised ATR electron transfer. In addition, the lignin in atr2 mutants has a reduced S/G ratio (Fig. 6). Commonly, plants with a lower flux toward coniferaldehyde (e.g. c4h, 4cl1, c3h1, and ccoaomt1) have lignin with higher S/G ratios, because the remaining flux through the pathway is preferentially used by the rate-limiting enzyme F5H1 and not by cinnamyl alcohol dehydrogenase (Meyer et al., 1996; Van Acker et al., 2013; Bonawitz et al., 2014; Wang et al., 2014). Thus, the reduced S/G ratio in atr2 mutants strongly suggests that the F5H1 activity is also negatively affected by reduced ATR2 activity (Fig. 6).
Mutation of ATR2 Causes Widespread Changes in Aromatic Metabolism
Phenolic profiling of the atr2 mutants revealed large differences in the metabolite pool compared with the wild type. Previously, it has been shown that lignin mutants display changes in their phenolic metabolome and that they commonly accumulate the substrate of the deficient enzyme or derivatives thereof (Vanholme et al., 2012b, 2013c). In particular, c4h Arabidopsis mutants accumulate cinnamic acid (Schilmiller et al., 2009), c3h1 Arabidopsis mutants and C3H down-regulated poplar accumulate p-coumaroyl shikimate (Coleman et al., 2008; Bonawitz et al., 2014), and mutants in F5H1 accumulate coniferaldehyde (Vanholme et al., 2012b). In the atr2 mutants, cinnamic acid, p-coumaroyl shikimate, and coniferaldehyde were all significantly higher in abundance compared with the wild type (Fig. 1; Table II). The accumulation of these compounds can be linked directly to the role of ATR2 in serving electrons to C4H, C3H1, and F5H1.
The accumulating phenylpropanoids and benzenoids in atr2 mutants were derivatives of p-coumaric, caffeic, ferulic, and sinapic acids (Table II). Most of them were detected in their hexosylated or malated form. Hexosylation occurred as 9-O-esters, as 3/4-O-hexosides, or both. Hexosylation and malation of accumulating phenylpropanoids have been suggested as either a strategy for the plant to detoxify potentially harmful metabolites or a passive process wherein accumulating phenylpropanoids serve as surrogates for the wild-type substrates (Vanholme et al., 2010b). Accumulating metabolites have been commonly detected when individual steps of the lignin biosynthetic pathway were perturbed. It has been shown that c4h mutants accumulate metabolites derived from cinnamic acid and ferulic acid (Vanholme et al., 2012b), f5h1 mutants accumulate metabolites derived from ferulic acid (Vanholme et al., 2012b), and plants with strongly reduced C3H1 activity have increased levels of p-coumaric acid-derived metabolites (Bonawitz et al., 2014). The accumulation of p-coumarates, caffeates, and ferulates and their benzenoid derivatives in atr2 mutants are a logical consequence of the reduced activity of C4H, C3H1, and F5H1.
Sinapate esters such as sinapoyl malate and sinapoyl hexose are known to protect wild-type Arabidopsis against harmful UV-B radiation and, therefore, are mainly localized in the epidermis (Chapple et al., 1992; Landry et al., 1995). Paradoxically, sinapate esters accumulated in the atr2 mutants (Table II), whereas f5h1, c3h1, and c4h mutants have been reported to have fewer sinapate esters (Franke et al., 2002; Vanholme et al., 2012b). This observation might logically be explained by the fact that ATR2 electron donation is important for lignifying cells, but epidermis cells that accumulate sinapate esters do not lignify. Therefore, it is possible that the mutation in ATR2 has no negative effect on the F5H1 activity in the epidermis; electrons might be supplied to C4H, C3H1, and F5H1 by the constitutive ATR1 in this cell type for the biosynthesis of sinapate esters.
In addition to phenylpropanoid and lignin biosynthesis, several other pathways rely on CYP450 enzymes. For instance, cyp79f1 mutants and Arabidopsis plants down-regulated for CYP79F1 have reduced levels of short-chain aliphatic glucosinolates (Hansen et al., 2001; Chen et al., 2003), cyp79f2 mutants were affected in the biosynthesis of long-chain aliphatic glucosinolates (Chen et al., 2003), and cyp83a1 mutants have reduced levels of both short- and long-chain aliphatic glucosinolates (Hemm et al., 2003). The cyp79b1, cyp79b2, cyp83b1, and cyp81f2 mutants and the cyp79b1 cyp79b2 double mutant have reduced levels of indolic glucosinolates (Zhao et al., 2002; Naur et al., 2003; Bednarek et al., 2009). In addition, c4h and f3′h mutants have less flavonol glycosides (Schoenbohm et al., 2000; Vanholme et al., 2012b). On the other hand, c3h1 mutants accumulate flavonoid glycosides, but this has been attributed to a stress-induced synthesis and not metabolic overflow (Abdulrazzak et al., 2006; Bonawitz et al., 2014). However, a targeted phenolic profiling revealed that a mutation of ATR2 had only moderate effects on the abundance of glucosinolates and flavonol glycosides (Supplemental Fig. S3). Again, this indicates that ATR2 has a preferential role in lignin biosynthesis in Arabidopsis. However, this does not rule out the possibility that ATR2 activity is needed for glucosinolate and flavonol synthesis in other cell types or during stress.
The differences in lignin composition and phenolic metabolite levels between the atr2 mutants and wild-type plants can logically be explained by lower C4H, C3H1, and F5H1 activities in the atr2 mutants. The ability of ATR2 to donate electrons to C4H was already proven in vitro in yeast and baculovirus expression systems (Urban et al., 1997; Mizutani and Ohta, 1998). Here, we provided in planta evidence for this interaction. To exclude that these reduced activities were due to a lower expression of the corresponding genes, we determined the expression levels of C4H, C3H1, and F5H1 in the inflorescence stem of atr2 mutants. The expression levels of these genes were not reduced in the atr2 mutants as compared with the wild type. In contrast, the expression of C3H1 was significantly higher in atr2-2 and the expression of C4H was on the border of being significantly higher (Fig. 4B). These data indicate that the reduced C4H, C3H1, and F5H1 activities are not the result of reduced gene expression. More likely, the reduced C4H, C3H1, and F5H1 activities are caused by the compromised electron-donating capacities of CPRs in the atr2 mutants. Notably, our results do not exclude the possibility that C4H, C3H1, and F5H1 activities were reduced via a posttranscriptional mechanism (e.g. on the protein activity level, where particular accumulating phenolics would act as inhibitors of the CYP450s).
Mutation of ATR2 Has Modest Effects on Plant Growth
Fully grown atr2-2 plants were only slightly smaller at final height than their wild-type counterparts, whereas atr2-1 was similar in size (Fig. 5). However, the reduction in dry weight of the main inflorescence stem in both atr2 mutants showed that normal plant development was affected (Fig. 5). A reduction in biomass has been observed previously in Arabidopsis plants with reduced lignin levels (e.g. in c4h, c3h1, cse, and ccr1 mutants; Jones et al., 2001; Franke et al., 2002; Do et al., 2007; Schilmiller et al., 2009; Van Acker et al., 2013; Vanholme et al., 2013c). Several hypotheses have been put forward to explain the growth penalty of low-lignin plants, of which some were proven (Bonawitz and Chapple, 2013). For example, a reduced amount of lignin results in weaker vessel cell walls and, in extreme cases, in vessel collapse, which in turn affects the transport of water and solutes (Turner and Somerville, 1997; Yang et al., 2013). Strengthening the cell walls of c4h mutants by vessel-specific expression of C4H could restore the growth phenotype, even though overall lignin levels were still reduced compared with the wild type (Yang et al., 2013). It was also recently discovered that the transcriptional coregulatory complex, Mediator, is a key component for mediating the plant’s response commonly associated with lignin biosynthesis reduction (stunted growth and less lignin). Mutation of genes encoding components of the Mediator complex resulted in a partial reversion of the growth penalty of c3h1 mutants (Bonawitz et al., 2014). We have not observed any deformation of cell walls in atr2 mutants (Fig. 5), but as ATR2 affects the functioning of C3H1, it is possible that the atr2 mutation triggers the Mediator-regulated transcriptional cascade, resulting in reduced biomass.
Because Arabidopsis has 244 genes encoding CYP450s, it is striking that atr2 mutants did not display stronger morphological defects. ATR1 and ATR2 are the only known genes in Arabidopsis for which the corresponding proteins have genuine CPR activity, meaning that they have the ability to transfer electrons from NADPH to CYP450s. It has been reported that ATR1 is constitutively expressed whereas ATR2 is mainly induced by wounding and light (Mizutani and Ohta, 1998). We further note that ATR2, but not ATR1, expression is inducible in seedlings treated with isoxaben, a cellulose inhibitor that induces several processes, including ectopic lignification (Fig. 3). The differences in ATR1 and ATR2 expression were also apparent after reevaluation of published data (Fig. 2). That ATR1 appears to be constitutively expressed suggests it to be partially redundant with ATR2 function, and this may explain the absence of strong morphological defects in atr2 mutants. However, we cannot rule out the possibility that the truncated ATR2 transcripts detected in both atr2 mutant alleles (Fig. 4) still provide leaky ATR2 activity and that a full knockout would have revealed stronger phenotypes, as was the case, for example, for the allelic c4h mutant series (Schilmiller et al., 2009). Taken together, our results support the hypothesis that ATR1 provides CPR activity constitutively, fulfilling the need for electrons in most cell types during development, and that ATR2 is induced in cases where additional CPR activity is needed that cannot be fulfilled by ATR1 alone (e.g. during lignification).
ATR2 Homologs as Targets to Improve Saccharification in Crops
The presence of lignin in the cell wall physically hinders the enzymatic hydrolysis of cellulose into Glc (Alvira et al., 2010). The saccharification efficiency can be improved by applying a pretreatment to render the cellulose more accessible to cellulases. Different pretreatments have specific effects on the cell wall material (Mosier et al., 2005; Carvalheiro et al., 2008; Alvira et al., 2010). We have tested alkaline (NaOH) pretreatment prior to the saccharification of cell wall material of wild-type plants and atr2 mutants and compared it with the saccharification efficiencies of cell walls without pretreatment. Although no improvement in saccharification efficiency was observed without pretreatment, a 26% to 27% increase in cellulose-to-Glc conversion was observed with alkaline-pretreated atr2 mutants stems (Fig. 7). The improved saccharification of atr2 mutant biomass may be explained by the reduced lignin content but also by the alterations of the lignin structure. Enrichment in p-coumaryl alcohol results in shorter lignin polymers because the H units tend to stop the polymerization process (Russell et al., 2006; Ziebell et al., 2010; Vanholme et al., 2012a, 2013a). This is supported by the identity of the oligolignols that accumulated in the atr2 mutant: they all had a terminal H unit, in most cases β-O-4 linked to a G unit (Table II). Shorter lignins will be more easily extracted during the alkaline pretreatment.
Besides ATR2, other members of the electron transport chain also might become targets for cell wall engineering. It was apparent from Vanholme et al. (2012b) that there is at least one CB5 coexpressed with lignin biosynthetic genes in a similar way to ATR2. Another CB5 has been shown by tandem affinity purification to interact with C4H in Arabidopsis cell cultures grown under standard conditions and under conditions inducing differentiation to tracheary elements (Bassard et al., 2012). Functional analysis of these CB5s will shed further light on their putative role as electron donors of the CYP450s involved in lignification.
In conclusion, ATR or other components of the CYP450 electron chain such as CB5s may become interesting candidates for the genetic engineering of lignocellulosic crops if the yield penalty can be overcome (e.g. by vessel-specific complementation or by engineering the Mediator complex; Yang et al., 2013; Bonawitz et al., 2014).
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines of ATR2 (At4g30210), SALK_152766 and SALK_026053, were ordered from the Nottingham Arabidopsis Stock Centre and called atr2-1 and atr2-2, respectively. Homozygous lines were screened by PCR using two primer combinations to distinguish heterozygous lines from homozygous ones: for the presence of the atr2-2 allele, 5′-CTCGTTTGGAGGAGATCCG-3′ (primer 1) and 5′-GCCCTTTGACGTTGGAGTC-3′ (primer 2), and for the absence of the atr2-2 allele, primer 1 and 5′-CATCTCCATATCTACATCCAG-3′ (primer 3); for the presence of the atr2-1 allele, primer 2 and 5′-GTGGAACACACTCCCTTATG-3′ (primer 4), and for the absence of the atr2-1 allele, primer 4 and 5′-CACACCAATCAGCAGCTCAC-3′ (primer 6).
Isoxaben Treatment
Seeds were germinated in 25 mL of liquid Murashige and Skoog medium (0.5× Murashige and Skoog medium + 1% [w/v] Glc, pH 5.7), and seedlings were grown in a 16-/8-h light/dark cycle at 21°C with gentle agitation (130 rpm). After 3 d, isoxaben dissolved in dimethyl sulfoxide (DMSO) was added (80 nm final concentration), and the same amount of DMSO was also added to the control. Twenty seedlings per repeat were harvested at 0, 6, 24, and 72 h after isoxaben addition.
qRT-PCR Analysis of Stems and Seedlings
To determine the transcript levels in the atr2 mutants, the basal 2 cm of the inflorescence stems was sampled from plants with a height of about 24 cm. Total RNA of seedlings and stem material was prepared using the RNeasy Plant Mini Kit (Qiagen). The RNA from the seedlings was treated with RQ1 RNase-Free DNase (Promega), and the RNA from stems was treated with DNA-free (Ambion) to remove traces of genomic DNA. Subsequent complementary DNA preparation was done using the iScript cDNA Synthesis Kit (Bio-Rad). Relative expression levels were determined with the LightCycler 480 Real-Time SYBR Green PCR System (Escoubas et al., 1995). For qRT-PCR of seedlings, CYCLIN-DEPENDENT KINASE A1 (At3g48750) was used as the reference gene, whereas for qRT-PCR of stem material, the gene encoding S-adenosyl-l-Met-dependent methyltransferases superfamily protein (At2g32170) was used. To measure ATR2 expression in the experiment performed on seedlings, primers 13 and 14 were combined, while for the experiment performed with stem material, primer combinations 15/16 (primer set 2) and 17/18 (primer set 1) were used. Primers are listed in Supplemental Table S3.
Plant Growth and Harvest
Plants were randomized and germinated in short-day conditions (21°C, 9 h of light, 120 µE s−1 m−2; 18°C, 15 h of darkness) on soil in pots 5.3 cm wide × 5.5 cm high. Plants were moved to long-day conditions (16-/8-h light/dark cycle) after 8 weeks to allow bolting. For metabolomics, plants were grown until the stem reached 22 to 26 cm in height. The bottom 10 cm of the main stem was then harvested, frozen in liquid nitrogen, and deprived of all side branches. For saccharification and cell wall compositional analyses, plants were grown until full senescence. The bottom 15 cm of the main stem was harvested and analyzed. For microscopy, stems of fully grown but not yet senesced plants were used.
Microscopy
The bottom 2 cm of inflorescence stems was embedded in 7% (w/v) agarose. Using a vibratome from Campden Instruments, slices 100 μm thick were made and stained as described by Rohde et al. (2004) with slight modifications: Wiesner staining was performed with a drop of a mixture of 1 g of phloroglucinol (Sigma-Aldrich) in 100 mL of 95% (v/v) ethanol and 16 mL of 37% (v/v) HCl. For Mäule staining, the samples were incubated for 5 min in 1% (w/v) KMnO4, and after rinsing with water, the samples were incubated in 37% (v/v) HCl, after which a drop of NH4OH was added to the sample. Images were taken with an Axioskop 2 (Zeiss) microscope. The photographs showing lignin and chlorophyll autofluorescence were taken with an LSM 710 (Zeiss) microscope. The fluorescence signal for lignin was 410 nm with filter 420 to 635 nm, and the fluorescence signal for chlorophyll was 470 nm with filter 650 to 755 nm.
Metabolomics
Approximately 50 mg of frozen ground stem tissue was extracted at room temperature by shaking with 500 µL of methanol. After centrifugation, the supernatant was transferred to new tubes and lyophilized. The pellet was resuspended in equal volumes of cyclohexane and ultrapure water (approximately 50 µL each) such that the portion of initially used stem tissue was 1 mg µL−1 aqueous phase. After vortexing, samples were centrifuged, and the aqueous phase was transferred to vials for injection of 15 µL onto the ultra-HPLC-MS system. n = 8 for the wild type and atr2-2, and n = 7 for atr2-1. Ultra-HPLC-MS analysis was performed as described by Vanholme et al. (2013c).
Metabolomic Data Processing
From the resulting chromatograms, 1,067 deisotoped peaks (compound ions) were integrated and aligned via Progenesis QI, each compound ion having an m/z and a retention time. Statistics (ANOVA with posthoc Student’s t test) and principal component analysis (Pareto scaling, with all 1,067 peaks included) was performed in Progenesis QI extension EZinfo. Statistical analyses were performed on arcsinh-transformed ion intensities. The following thresholds for significance were used: P value for ANOVA < 0.01, and differentially significant P < 0.01 in both atr2-1 and atr2-2.
Saccharification
Saccharification was performed according to Van Acker et al. (2013). For alkali pretreatment, 1 mL of NaOH (0.25%, w/v) was added to the dry CWR, and the samples were shaken at 70°C for 3 h. After centrifugation, the pellet was washed three times with ultrapure water, followed by an overnight extraction with 1 mL of 70% ethanol at 55°C and shaking at 750 rpm. After centrifugation, the pellet was washed an additional three times with 1 mL of 70% ethanol and once with 1 mL of acetone.
Lignin Amount via Acetyl Bromide
The determination of the lignin amount was done for eight biological replicates according to Van Acker et al. (2013).
Cellulose Amount
Cellulose was measured on 3 mg of dry stems with sulfuric acid and anthrone in accordance with Foster et al. (2010). Briefly, hemicellulose was removed by adding 1 mL of 2 m trifluoroacetic acid and followed by incubation for 1 h at 99°C while shaking at 750 rpm. After centrifugation at 10,000 rpm, the pellet was washed three times with 0.5 mL of water and twice with 0.5 mL of acetone before being dried and weighed. Next, 1 mL of Updegraff reagent was added to the pellet followed by incubation at 100°C for 30 min. After centrifugation at 10,000 rpm for 15 min, the supernatant was discarded. The pellet was washed three times with 1 mL of acetone. The pellet was left to air dry overnight, and the next day, 175 µL of 72% (v/v) H2SO4 was added. The samples were incubated for 30 min at room temperature, whereupon the samples were vortexed with an additional incubation time of 15 min at room temperature. A total of 825 µL of water was added, and the samples were vortexed again. The samples were centrifuged at 10,000 rpm for 10 min. Ten microliters was diluted with 90 µL of water on a 96-well plate. To each sample, 200 µL of freshly prepared anthrone reagent (2 mg anthrone mL−1 pure H2SO4) was added. After incubation at 80°C for 30 min, the samples were cooled on the bench and the absorption was measured at 625 nm.
Lignin Composition via Thioacidolysis
This was performed in accordance with Robinson and Mansfield (2009).
NMR Sample Preparation
The gel samples of whole-plant cell walls for NMR experiments were prepared as described previously (Kim and Ralph, 2010). The dried Arabidopsis stems were preground for 1 min in a Retsch MM400 mixer mill at 30 Hz using zirconium dioxide (ZrO2) vessels (10 mL) containing ZrO2 ball bearings (2 × 10 mm). The preground cell walls were extracted with distilled water (ultrasonication, 1 h, three times) and 80% ethanol (ultrasonication, 1 h, three times). Isolated cell walls were dried and ball milled using a Retsch PM100 planetary ball mill at 600 rpm using ZrO2 vessels (50 mL) containing ZrO2 ball bearings (10 × 10 mm). Each sample (100 mg) was ground for 25 min (interval, 5 min; break, 5 min). The cell walls were collected directly in the NMR tubes and formed gels in DMSO-d6:pyridine-d5 (4:1).
NMR Experiments
NMR experiments on the whole-plant cell wall gel samples were performed as described previously (Kim et al., 2008; Kim and Ralph, 2010). NMR spectra were acquired on a Bruker Biospin Avance 500-MHz spectrometer fitted with a cryogenically cooled 5-mm TCI (for triple resonance cryoprobe optimized for 1H and 13C observation) gradient probe with inverse geometry (proton coils closest to the sample). The central DMSO solvent peak was used as an internal reference (δC 39.5 ppm, δH 2.49 ppm). The 13C-1H correlation experiment was an adiabatic HSQC experiment (Bruker standard pulse sequence hsqcetgpsisp.2; phase-sensitive gradient-edited two-dimensional HSQC using adiabatic pulses for inversion and refocusing; Kupče and Freeman, 2007). HSQC experiments were carried out using the following parameters: acquired from 10 to 0 ppm in F2 (1H) with 1,000 data points (acquisition time, 100 ms) and from 200 to 0 ppm in F1 (13C) with 400 increments (F1 acquisition time, 8 ms) of 72 scans with a 500-ms interscan delay; the d24 (for maximum duration of the chemical shift selective filter with constant time) delay was set to 0.89 ms (1/8J, J: 145 Hz). The total acquisition time was 5 h. Processing used typical matched Gaussian apodization in F2 and squared cosine-bell and one level of linear prediction (32 coefficients) in F1. Volume integration of contours in HSQC plots from data that were not linear predicted used Bruker’s TopSpin 3.1 software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenolic profiling.
Supplemental Figure S2. MS/MS spectra and the reasoning for the tentative structural identification of 33 metabolites of which the MS/MS spectra were not published before.
Supplemental Figure S3. Pathways dependent on CYP450s with metabolites that were detected via ultra-HPLC-MS.
Supplemental Figure S4. Two-dimensional HSQC NMR spectra of the lignin side chain and polysaccharide region of atr2-1 and atr2-2 compared with the wild type.
Supplemental Table S1. The 1,067 peaks detected in wild-type plants and atr2-1 and atr2-2 mutants.
Supplemental Table S2. The 238 peaks significantly different in abundance in atr2-1 and atr2-2 mutants as compared with wild-type plants.
Supplemental Table S3. Overview of primers used for qRT-PCR
Supplementary Material
Glossary
- qRT
quantitative reverse transcription
- T-DNA
transfer DNA
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- MS/MS
tandem mass spectrometry
- CWR
cell wall residue
- HSQC
heteronuclear single quantum coherence
- G
guaiacyl
- S
syringyl
- H
p-hydroxyphenyl
- DMSO
dimethyl sulfoxide
- ZrO2
zirconium dioxide
Footnotes
This work was supported by the European Commission’s Directorate General for Research within the 7th Framework Program (grant nos. 211982 [RENEWALL] and 270089 [MULTIBIOPRO]), by Stanford University’s Global Climate and Energy Projects Towards New Degradable Lignin Types and Efficient Biomass Conversion: Delineating the Best Lignin Monomer-Substitutes, by the Department of Energy Great Lakes Bioenergy Research Center (grant no. DE–FC02–07ER64494), by Ghent University (Hercules program grant no. AUGE/014, Bijzondere Onderzoeksfonds-Zware Apparatuur grant no. 174PZA05, and Biotechnology for a Sustainable Economy grant no. 01MRB510W), by the Agency for Innovation by Science and Technology (predoctoral fellowship to L.S.), and by the Research Foundation-Flanders (postdoctoral fellowship to R.V.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, et al. (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140: 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alvira P, Tomás-Pejó E, Ballesteros M, Negro MJ (2010) Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour Technol 101: 4851–4861 [DOI] [PubMed] [Google Scholar]
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D (2011) Cytochromes p450. The Arabidopsis Book 9: doi/10.1199/tab.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W (1998) Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci 17: 125–197 [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J, Negre F, Chen X, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N (2004) Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Carvalheiro F, Duarte LC, Gírio FM (2008) Hemicellulose biorefineries: a review on biomass pretreatments. J Sci Ind Res (India) 67: 849–864 [Google Scholar]
- Chapple CCS, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen S, Glawischnig E, Jørgensen K, Naur P, Jørgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33: 923–937 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105: 4501–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L (2007) Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Duval I, Beaudoin N (2009) Transcriptional profiling in response to inhibition of cellulose synthesis by thaxtomin A and isoxaben in Arabidopsis thaliana suspension cells. Plant Cell Rep 28: 811–830 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp 37: 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, et al. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann F, Bichet A, Ewen KM, Bernhardt R (2007) Cytochrome P450 systems: biological variations of electron transport chains. Biochim Biophys Acta 1770: 330–344 [DOI] [PubMed] [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA (2001) Cytochrome p450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem 276: 11078–11085 [DOI] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck C, Jarvis AP, Xiang L, Moore BS, Oldham NJ (2001) A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid β-oxidation. ChemBioChem 2: 784–786 [DOI] [PubMed] [Google Scholar]
- Ilan Z, Ilan R, Cinti DL (1981) Evidence for a new physiological role of hepatic NADPH:ferricytochrome (P-450) oxidoreductase: direct electron input to the microsomal fatty acid chain elongation system. J Biol Chem 256: 10066–10072 [PubMed] [Google Scholar]
- Im SC, Waskell L (2011) The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b5. Arch Biochem Biophys 507: 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis AP, Schaaf O, Oldham NJ (2000) 3-Hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 212: 119–126 [DOI] [PubMed] [Google Scholar]
- Jensen K, Møller BL (2010) Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71: 132–141 [DOI] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Jung JH, Vermerris W, Gallo M, Fedenko JR, Erickson JE, Altpeter F (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J 11: 709–716 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Akiyama T (2008) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. BioEnergy Res 1: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupče E, Freeman R (2007) Compensated adiabatic inversion pulses: broadband INEPT and HSQC. J Magn Reson 187: 258–265 [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D (1998) Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana: gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol 116: 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96: 673–686 [DOI] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, et al. (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Akiyama T, Coleman HD, Mansfield SD (2012) Effects on lignin structure of coumarate 3-hydroxylase downregulation in poplar. BioEnergy Res 5: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Ro DK, Ehlting J, Douglas CJ (2002) Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol 130: 1837–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AR, Mansfield SD (2009) Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J 58: 706–714 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WR, Burkitt MJ, Scobbie L, Chesson A (2006) EPR investigation into the effects of substrate structure on peroxidase-catalyzed phenylpropanoid oxidation. Biomacromolecules 7: 268–273 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381: 749–753 [DOI] [PubMed] [Google Scholar]
- Shephard EA, Phillips IR, Bayney RM, Pike SF, Rabin BR (1983) Quantification of NADPH:cytochrome P-450 reductase in liver microsomes by a specific radioimmunoassay technique. Biochem J 211: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre EM, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro EM (2008) Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticklen M. (2006) Plant genetic engineering to improve biomass characteristics for biofuels. Curr Opin Biotechnol 17: 315–319 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Van Acker R, Leplé JC, Aerts D, Storme V, Goeminne G, Ivens B, Légée F, Lapierre C, Piens K, Van Montagu MCE, et al. (2014) Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 111: 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Cesarino I, Goeminne G, Kim H, Marroni F, Van Acker R, Vanholme R, Morreel K, Ivens B, Pinosio S, et al. (2013a) Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol 198: 765–776 [DOI] [PubMed] [Google Scholar]
- Vanholme B, Desmet T, Ronsse F, Rabaey K, Van Breusegem F, De Mey M, Soetaert W, Boerjan W (2013b) Towards a carbon-negative sustainable bio-based economy. Front Plant Sci 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013c) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010a) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012a) Metabolic engineering of novel lignin in biomass crops. New Phytol 196: 978–1000 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010b) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012b) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan J, Guilleminot J, Saint-Jore-Dupas C, Piégu B, Chabouté ME, Gomord V, Coolbaugh RC, Devic M, Delorme V (2010) ATR3 encodes a diflavin reductase essential for Arabidopsis embryo development. New Phytol 187: 67–82 [DOI] [PubMed] [Google Scholar]
- Wang JP, Naik PP, Chen HC, Shi R, Lin CY, Liu J, Shuford CM, Li Q, Sun YH, Tunlaya-Anukit S, et al. (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26: 894–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LL, Wallis JG, Kumar R, Markham JE, Browse J (2013) Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell 25: 3052–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS, Sun L, Zheng K, Tang K, Auer M, et al. (2013) Engineering secondary cell wall deposition in plants. Plant Biotechnol J 11: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285: 38961–38968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.