Flowering time after vernalization and gene expression in flowering time pathways correlate with altitude, suggesting phenological adaptation.
Abstract
Steep environmental gradients provide ideal settings for studies of potentially adaptive phenotypic and genetic variation in plants. The accurate timing of flowering is crucial for reproductive success and is regulated by several pathways, including the vernalization pathway. Among the numerous genes known to enable flowering in response to vernalization, the most prominent is FLOWERING LOCUS C (FLC). FLC and other genes of the vernalization pathway vary extensively among natural populations and are thus candidates for the adaptation of flowering time to environmental gradients such as altitude. We used 15 natural Arabidopsis (Arabidopsis thaliana) genotypes originating from an altitudinal gradient (800–2,700 m above sea level) in the Swiss Alps to test whether flowering time correlated with altitude under different vernalization scenarios. Additionally, we measured the expression of 12 genes of the vernalization pathway and its downstream targets. Flowering time correlated with altitude in a nonlinear manner for vernalized plants. Flowering time could be explained by the expression and regulation of the vernalization pathway, most notably by AGAMOUS LIKE19 (AGL19), FLOWERING LOCUS T (FT), and FLC. The expression of AGL19, FT, and VERNALIZATION INSENSITIVE3 was associated with altitude, and the regulation of MADS AFFECTING FLOWERING2 (MAF2) and MAF3 differed between low- and high-altitude genotypes. In conclusion, we found clinal variation across an altitudinal gradient both in flowering time and the expression and regulation of genes in the flowering time control network, often independent of FLC, suggesting that the timing of flowering may contribute to altitudinal adaptation.
Environmental gradients, such as temperature or water availability, provide an ideal setting to study how species adapt to contrasting environmental scenarios (Reich et al., 2003; Keller et al., 2013). Many studies have shown that phenotypic plant traits such as leaf number, allocation to reproductive biomass, and height change along environmental gradients (Etterson, 2004; Leger and Rice, 2007; Fischer et al., 2011), and some studies could correlate environmental clines to changes in allelic frequencies at specific candidate genes (Manel et al., 2010; Poncet et al., 2010; Fischer et al., 2013).
Although allelic variation at genes with major effects may explain variation in some phenotypes, fine-tuning of other quantitative traits along an environmental gradient may require an adjustment of larger regulatory networks (Whitehead and Crawford, 2006; Hodgins-Davis and Townsend, 2009; Hodgins et al., 2013). In Arabidopsis (Arabidopsis thaliana), numerous genetic pathways have been studied extensively, mainly using laboratory accessions (Shinozaki and Yamaguchi-Shinozaki, 2007; Wellmer and Riechmann, 2010; Ó’Maoiléidigh et al., 2014). However, how consistently such pathways are expressed in natural populations, and how they respond to different environmental conditions, often remains unclear. Studying the expression of genetic pathways in natural genotypes originating from an environmental cline under a variety of climatic scenarios provides an ideal approach to understanding how plants can adapt to contrasting environments along a climatic gradient.
Across altitudes, environmental gradients are particularly steep: climatic conditions, including temperature, solar radiation, and precipitation, may change dramatically on a small geographic scale (Körner, 2007), while daylength and other factors remain constant. Many phenotypic traits, such as height, total seed weight, leaf size, and allocation to vegetative reproduction, have been found to change along altitudinal gradients in plants (Byars et al., 2007; Gonzalo-Turpin and Hazard, 2009; Fischer et al., 2011). Among these, the timing of flowering (i.e. the transition from vegetative growth to the reproductive phase) is a key developmental phase transition in seasonal alpine environments, as its accuracy is crucial for reproductive success: too-early flowering increases the risk of encountering detrimental frost (Kollas et al., 2013), whereas time for seed maturation may run out if flowering starts too late (Inouye and Wielgolaski, 2003; Chuine, 2010). These contrasting selective pressures may change along an altitudinal gradient, where the vegetation period becomes shorter with increasing altitude.
In Arabidopsis, an annual weed native to Eurasia and northern Africa, two different life cycles have been described (Koornneef et al., 2004; Alonso-Blanco et al., 2009): summer annuals germinate and flower within one growing season and do not require winter to initiate flowering; winter annuals germinate usually in autumn, overwinter as vegetative rosettes, and flower in the following spring. Accessions expressing a winter-annual life cycle need vernalization (a prolonged cold period) in order to initiate flowering; otherwise, they remain in a vegetative rosette stage for an extended period of time.
On the molecular level, the transition to flowering is among the best-studied processes in plants (Wellmer and Riechmann, 2010; Andrés and Coupland, 2012), and in Arabidopsis, several genetic pathways controlling flowering are known. Signals from the vernalization pathway, photoperiod pathway, autonomous pathway, GA pathway, and plant age all contribute to ensuring the correct timing of flowering (Ehrenreich et al., 2009; Wellmer and Riechmann, 2010; Srikanth and Schmid, 2011). Within the vernalization pathway, a number of key players have been identified (Andrés and Coupland, 2012; Schmitz and Amasino, 2012; Song et al., 2012; Zografos and Sung, 2012). In winter annuals, a functional FRIGIDA (FRI) allele is required to activate FLOWERING LOCUS C (FLC). FLC strongly suppresses the flowering promoters FLOWERING LOCUS T (FT) and AGAMOUS LIKE20 (AGL20; also referred to as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1) and thus inhibits flowering. During vernalization, VERNALIZATION INSENSITIVE3 (VIN3) represses FLC, and the repressed state is maintained in subsequent warm periods by epigenetic silencing (Crevillén and Dean, 2011; Zografos and Sung, 2012), allowing FT, AGL20, and, through positive feedback with AGL20, AGL24 (Liu et al., 2008) to initiate flowering. Many natural populations and most laboratory accessions, among them Columbia-0 (Col-0), carry nonfunctional FRI or FLC alleles and thus respond only weakly to vernalization, resulting in fast flowering, summer-annual life cycles (Johanson et al., 2000; Gazzani et al., 2003; Shindo et al., 2005).
In addition to the well-studied FLC branch of the vernalization pathway, FLC-independent components of the vernalization response have been identified. For example, AGL19 has been found to promote flowering following vernalization without interacting with FLC (Schönrock et al., 2006), and relatives of FLC, the MADS AFFECTING FLOWERING genes (MAF1–MAF5; MAF1 is also referred to as FLOWERING LOCUS M [FLM]; De Bodt et al., 2003), have been shown to inhibit flowering in a similar way to FLC (Ratcliffe et al., 2003; Scortecci et al., 2003; Werner et al., 2005; Sung et al., 2006; Gu et al., 2013). Genes MAF2 to MAF5 (Ratcliffe et al., 2003) are arranged in a tandem gene array and vary extensively among natural populations (Caicedo et al., 2009; Rosloski et al., 2010), and several recent studies have associated this polymorphic region with natural variation in flowering time (Salomé et al., 2011; Silady et al., 2011; Lasky et al., 2012; Fournier-Level et al., 2013; Grillo et al., 2013), making these genes interesting candidates for studying associations between flowering time and ecological parameters.
Associating genetic variation at a single gene with latitude or altitude has often proven to be difficult (Shindo et al., 2005; Stinchcombe et al., 2005; Méndez-Vigo et al., 2011), although Caicedo et al. (2004) found evidence for epistatic interactions between FRI and FLC alleles associated with latitude. Interestingly, some recent studies suggest that regulatory processes within the vernalization pathway may contribute to natural phenotypic variability (Shindo et al., 2006; Strange et al., 2011). Overall, the response to vernalization appears to be a complex process in natural populations, potentially involving epigenetic regulation of a number of genes. Therefore, to gain a better understanding of the involvement of this complex genetic network in the response to ecological parameters, it is essential to study multiple interacting genes of the vernalization pathway simultaneously.
Here, we used 15 natural Arabidopsis genotypes originating from an altitudinal cline (800–2,700 m) in the Swiss Alps to study the associations between vernalization, flowering initiation, gene expression and regulation, and altitude. Importantly, all genotypes originated from a restricted geographic range; thus, confounding effects such as differences in daylength, as found along latitudinal clines, can be excluded. We measured flowering time and the expression of 12 genes of the vernalization pathway under different vernalization scenarios to assess whether the response to vernalization is associated with altitude. In particular, we tested the hypotheses that (1) flowering time correlates with altitude; (2) genotypes from high altitudes need longer vernalization periods to initiate flowering reliably; (3) gene expression and regulation of the vernalization pathway can explain flowering time; (4) gene expression and regulation of the vernalization pathway is associated with altitude; and (5) FLC-independent branches of the vernalization pathway are important for initiating flowering and, thus, for altitudinal adaptation in natural populations.
RESULTS
Flowering Time
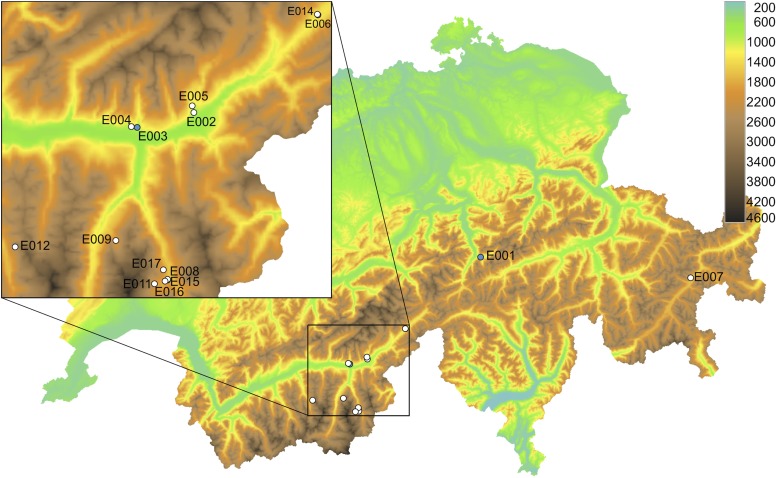
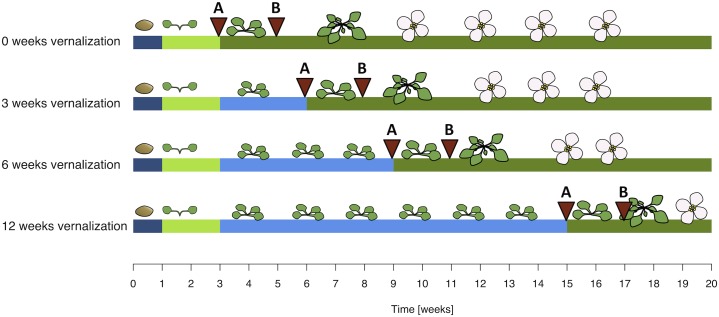
Fifteen genotypes of Arabidopsis originating from an altitudinal gradient in the Swiss Alps ranging from 800 to 2,700 m above sea level (m.a.s.l.; Table I; Fig. 1) as well as accessions Col-0 and Col-FRI (carrying a functional FRI allele in a Columbia background) were grown in climate chambers under four different vernalization scenarios: no vernalization and 3, 6, and 12 weeks of vernalization (Fig. 2). Two genotypes were excluded from most analyses due to potentially different habitat conditions and uncertain altitudinal origins (Supplemental Fig. S1); thus, most analyses were performed using the remaining 13 genotypes.
Table I. Natural genotypes collected in the Swiss Alps.
Fifteen genotypes of Arabidopsis originating from a range of altitudes in the Swiss Alps were used in this study; genotypes in italic were excluded from most analyses.
| Accession | Nearest Settlement | Latitude (N) | Longitude (E) | Altitude |
|---|---|---|---|---|
| m | ||||
| E001 | Bristen | 46°46′01.7′′ | 08°42′11.8′′ | 800 |
| E002 | Naters | 46°19′56.0′′ | 07°59′12.0′′ | 850 |
| E003 | Eggerberg | 46°18′47.1′′ | 07°5238.7′′ | 900 |
| E004 | Ausserberg | 46°15′50.4′′ | 07°52′00.9′′ | 1,000 |
| E005 | Geimen | 46°20′27.3′′ | 07°59′02.6′′ | 1,030 |
| E006 | Ritzingen | 46°27′37.3′′ | 08°13′33.7′′ | 1,360 |
| E007 | Brail | 46°39′20.7′′ | 10°01′07.6′′ | 2,040 |
| E008 | Saas Fee | 46°06′40.1′′ | 07°56′04.5′′ | 1,719 |
| E009 | Graechen | 46°09′47.8′′ | 07°50′12.3′′ | 1,850 |
| E011 | Saas Fee | 46°06′20.3′′ | 07°54′35.0′′ | 2,012 |
| E012 | Zinal | 46°09′20.4′′ | 07°38′40.0′′ | 2,700 |
| E014 | Ritzingen | 46°27′39.6′′ | 08°13′34.9′′ | 1,368 |
| E015 | Saas Fee | 46°06′38.6′′ | 07°56′03.8′′ | 1,730 |
| E016 | Saas Fee | 46°06′33.4′′ | 07°55′46.2′′ | 1,792 |
| E017 | Saas Fee | 46°07′26.3′′ | 07°55′36.9′′ | 1,949 |
Figure 1.
Origins of natural genotypes from the Swiss Alps collected from an altitudinal gradient (800–2,700 m). White circles indicate genotypes analyzed in this study, while blue circles indicate genotypes excluded from most analyses. Colors indicate altitude.
Figure 2.
Experimental design. Four different treatments were applied to 15 natural genotypes and two control genotypes of Arabidopsis: no vernalization and 3, 6, and 12 weeks of vernalization. Dark blue indicates stratification treatment to break seed dormancy (4°C, dark), light green indicates the first growing period under short-day conditions (22°C/20°C, 8-h/16-h day/night), light blue indicates vernalization treatment (4°C, 8-h/16-h day/night), while dark green indicates the second growth period under long-day conditions (22°C/20°C, 16-h/8-h day/night). Of each genotype, three plants each were collected at two time points per treatment: 1 d before long-day conditions started (i.e. at the last day of the vernalization treatment for vernalized plants; time A) and 10 d later during growth under long-day conditions (time B).
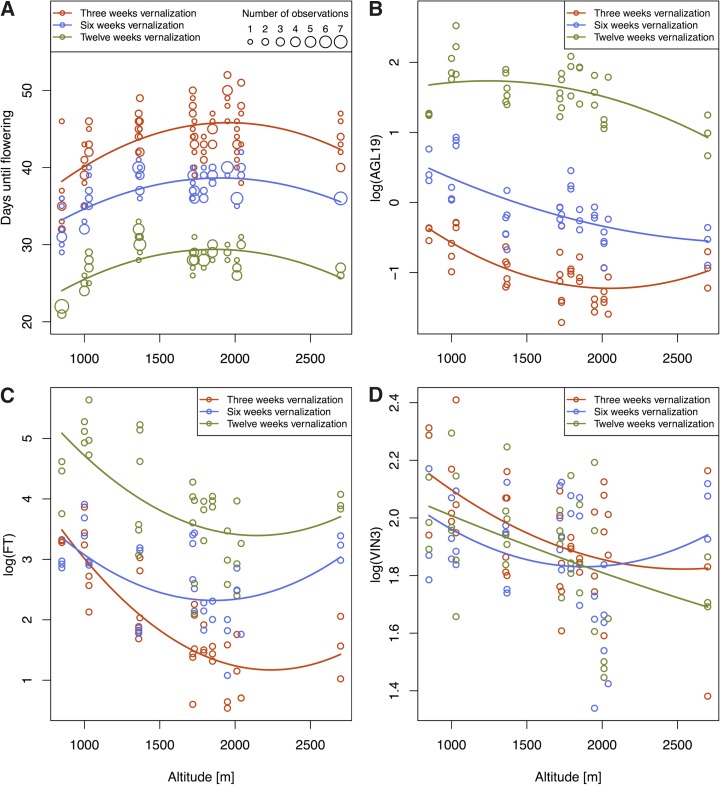
Altitude had a significant effect on flowering time when all treatments were analyzed together (Supplemental Table S1; Supplemental Fig. S2), even though the power of this statistical model was greatly reduced by the high variation in flowering time within genotypes of nonvernalized plants. When only vernalized plants were used, both linear and quadratic terms of altitude were highly significant (Table II; Fig. 3A). Lowland genotypes were under all vernalization treatments the first to flower, while the latest flowering genotypes originated from between 1,700 and 2,100 m.a.s.l. Vernalization treatment had a strong effect on flowering time, with flowering time decreasing with increasing length of vernalization. Interactions between altitude and vernalization treatment indicated a slight decrease of the linear component of altitude with increasing duration of vernalization (i.e. the difference between earliest and latest flowering genotypes became slightly smaller with increasing duration of vernalization). No interaction effect between the quadratic term of altitude and treatment was found.
Table II. Effects of altitude on flowering time of vernalized plants.
A linear mixed-effects model was used with altitude (linear and quadratic terms) and vernalization treatment including their interactions as fixed factors and genotypes as random factors. Heteroscedasticity between treatments was accounted for by using the weights argument in the lme function.
| Condition | Estimate | se | tdfa | P |
|---|---|---|---|---|
| Altitude, linear | 30.769 | 10.235 | 3.006292 | 0.003 |
| Altitude, quadratic | −33.856 | 10.202 | −3.319292 | 0.001 |
| 6 Weeks of vernalization | −6.538 | 0.304 | −21.512292 | <0.001 |
| 12 Weeks of vernalization | −15.789 | 0.290 | −54.442292 | <0.001 |
| Altitude (linear) × 6 weeks | −10.800 | 5.321 | −2.030292 | 0.043 |
| Altitude (quadratic) × 6 weeks | 7.340 | 5.297 | 1.386292 | 0.167 |
| Altitude (linear) × 12 weeks | −13.379 | 5.071 | −2.638292 | 0.009 |
| Altitude (quadratic) × 12 weeks | 5.463 | 5.056 | 1.081292 | 0.281 |
t value and degrees of freedom.
Figure 3.
Days until flowering and gene expression for 13 Arabidopsis genotypes originating from an altitudinal gradient in the Swiss Alps. Colors denote different vernalization treatments. Lines show the predicted regression slope for each vernalization treatment calculated from a linear mixed-effects model with altitude (linear and quadratic) and treatment as fixed and genotype as random factors. A, Days until flowering. Circle size indicates the number of plants flowering at a given day. B, Expression of AGL19 at early collection time. C, Expression of FT at late collection time. D, Expression of VIN3 at early collection time.
As genotype E012 originated from a considerably higher altitude than all other genotypes included in this study and thus may have a strong effect on the fit of the quadratic component, the analysis was repeated without E012. In this model, the linear term of altitude became even more significant, whereas the quadratic term remained significant, although less so than with E012 included (Supplemental Table S2; Supplemental Fig. S3).
When genotypes E001 and E003 (originally excluded because of their uncertain origin) were included in the model, the association of altitude with flowering time was no longer significant (Supplemental Table S3; Supplemental Fig. S1A).
Variation in days to flowering decreased with longer vernalization (i.e. flowering was more strongly synchronized within genotypes after longer vernalization). Nonvernalized plants consistently showed higher variation than vernalized plants (z = 3.668, adjusted P < 0.001 for all comparisons), 3 weeks of vernalization showed higher variation than 6 or 12 weeks (3 versus 6 weeks: z = 2.936, adjusted P = 0.004; 3 versus 12 weeks: z = 3.668, adjusted P < 0.001), whereas 6 and 12 weeks showed comparable variances in flowering time (z = 1.380, adjusted P = 0.168).
Because no direct measurements of climatic data were available from the original collection sites, we used extrapolated environmental data to test whether climatic factors correlated with flowering time. All temperature parameters (both linear and quadratic components) showed highly significant associations with flowering time (Supplemental Table S4), the quadratic component of precipitation showed a weakly significant association, whereas the other tested parameters (solar radiation, habitat exposition, and habitat slope) were nonsignificant (data not shown). However, extrapolated environmental parameters may not accurately reflect conditions at the exact collection sites, because microclimatic conditions can vary substantially on small spatial scales as a consequence of pronounced topographic variation in alpine habitats (Scherrer and Körner, 2011). Therefore, and as the strength of the temperature effect was similar to the effect of altitude, all further analyses were computed with altitude as a measure of the environmental gradient.
Gene Expression and Flowering Time
For each vernalization treatment, the expression of 12 genes of the vernalization pathway and its downstream targets (Supplemental Table S5) was measured at two time points: once at the last day of the vernalization treatment (or at the last day of the short-day treatment for nonvernalized plants) and once 10 d later (i.e. under warm and long-day conditions; Fig. 2).
We first analyzed whether gene expression could explain flowering time of the wild genotypes by computing linear mixed-effects models with all genes included as fixed factors and genotypes as random factors separately for early and late collection times. On the one hand, we randomly assigned each gene expression data a flowering date of a randomly chosen subset of the same treatment and genotype to include variance of both flowering time and gene expression per genotype; on the other hand, we calculated models using means of both flowering time and gene expression data (per treatment and genotype), which was statistically less powerful due to the reduced data set (Table III). Under both model scenarios, the expression of AGL19 and FLC had significant effects on flowering time both at early and late collection times, whereas FRI had significant effects at early collection time and FT at late collection time. MAF2 showed significant associations with flowering time both at early and late collection times in the model that incorporated variances, whereas these associations were lost when only mean values were used. Similarly, the effects of AGL20 and AGL24 at early collection time and of FRI at late collection time could only be observed in the more powerful models incorporating variances.
Table III. Effect of (logarithmic) gene expression on flowering time separately for early and late collection times.
Linear mixed-effects models were used with genes included as fixed factors and genotypes as random factors. Significant gene expression effects are shown in boldface.
| Gene | Early Collection |
Late Collection |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | se | tdfa | P | Pb | Estimate | se | tdfa | P | Pb | |
| AGL19 | −3.437 | 0.487 | −7.05890 | <0.001 | ** | −2.862 | 0.540 | −5.30384 | <0.001 | * |
| FLC | 2.582 | 0.534 | 4.83290 | <0.001 | * | 2.667 | 0.298 | 8.95784 | <0.001 | ** |
| MAF2 | 1.870 | 0.621 | 3.01190 | 0.003 | −1.745 | 0.707 | −2.46784 | 0.016 | ||
| FRI | −3.533 | 1.196 | −2.95590 | 0.004 | * | 3.070 | 1.392 | 2.20584 | 0.030 | |
| AGL20 | −1.152 | 0.410 | −2.81090 | 0.006 | −0.282 | 0.436 | −0.64784 | 0.519 | ||
| AGL24 | 1.669 | 0.816 | 2.04490 | 0.044 | −1.258 | 0.812 | −1.54984 | 0.125 | ||
| VIN3 | 1.351 | 0.954 | 1.41690 | 0.160 | −0.281 | 0.463 | −0.60684 | 0.546 | ||
| MAF5 | 2.575 | 2.009 | 1.28290 | 0.203 | −0.856 | 1.359 | −0.62984 | 0.531 | ||
| MAF3 | 0.169 | 0.425 | 0.39690 | 0.693 | −0.461 | 0.343 | −1.34384 | 0.183 | ||
| MAF4 | −1.267 | 3.397 | −0.37390 | 0.710 | 0.728 | 6.960 | 0.10584 | 0.917 | ||
| FT | −0.272 | 0.851 | −0.32090 | 0.750 | −1.047 | 0.367 | −2.85684 | 0.005 | * | |
t Value and degrees of freedom. bSignificant P values of linear mixed-effects models, where the mean gene expression of each gene (per genotype and treatment) was tested against the mean flowering time of each treatment, separately for the two collection times: *P < 0.05 and **P < 0.01.
When genotypes E001 and E003 were included in the models (Supplemental Table S6), the significant effects of AGL19 and FT remained similar, whereas the effects of FLC could only be observed in the models that incorporated variances within genotypes. Furthermore, MAF4 was now significantly associated with flowering time at late collection time for both model types, whereas at early collection time, the model calculated with mean flowering time and gene expression only showed a trend (P < 0.1).
When we looked at the regulation of the vernalization pathway using gene regulatory network (GRN) analyses with data at both collection times of vernalized plants, a number of edges (correlations between genes), most prominently AGL20-AGL24, VIN3-FLC, and FT-AGL20, significantly correlated with flowering time (Table IV). When genotypes E001 and E003 were included, the same edges showed significant associations with flowering time (Supplemental Table S7).
Table IV. Correlation coefficients between gene regulation of the vernalization pathway and flowering time.
GRN analysis revealed that several edges (Pearson correlations between gene pairs) of the vernalization pathway correlated significantly with flowering time in vernalized plants, indicating that flowering time can be explained by the regulation of genes analyzed in this study.
| Edge | Correlation | tdfa | P | Adjusted Pb |
|---|---|---|---|---|
| AGL20-AGL24 | −0.716 | −6.07435 | <0.001 | *** |
| VIN3-FLC | −0.625 | −4.74035 | <0.001 | *** |
| FT-AGL20 | −0.607 | −4.51835 | <0.001 | *** |
| FLC-FT | 0.519 | 3.59035 | 0.001 | ** |
| VIN3-AGL19 | −0.516 | −3.56635 | 0.001 | ** |
| MAF4-AGL20 | 0.382 | 2.44935 | 0.019 | ns |
| VIN3-MAF3 | 0.364 | 2.20932 | 0.034 | ns |
| MAF3-FT | −0.341 | −2.05332 | 0.048 | ns |
| VIN3-MAF4 | 0.325 | 2.03435 | 0.050 | ns |
| MAF2-AGL20 | −0.288 | −1.77735 | 0.084 | ns |
| MAF4-FT | −0.230 | −1.39935 | 0.170 | ns |
| MAF3-AGL20 | 0.222 | 1.28832 | 0.207 | ns |
| MAF5-FT | −0.107 | −0.63535 | 0.529 | ns |
| VIN3-AGL24 | −0.105 | −0.62435 | 0.537 | ns |
| MAF2-FT | 0.070 | 0.41635 | 0.680 | ns |
| MAF5-AGL20 | 0.061 | 0.36335 | 0.719 | ns |
| VIN3-MAF5 | 0.049 | 0.29135 | 0.773 | ns |
| FLC-AGL20 | −0.022 | −0.12835 | 0.899 | ns |
| FRI-FLC | −0.019 | −0.11135 | 0.912 | ns |
| VIN3-MAF2 | 0.000 | 0.00235 | 0.998 | ns |
t Value and degrees of freedom. bP values were adjusted following Benjamini and Hochberg (1995): ***adjusted P < 0.001; **adjusted P < 0.01; and ns, adjusted P > 0.05.
Gene Expression and Altitude
To quantify the effect of altitude on gene expression, linear mixed-effects models were calculated separately for each gene and collection time using vernalized plants (Supplemental Table S8). Several genes showed linear and quadratic associations with altitude, particularly AGL19 (early and late collection time), FT (late collection time), and VIN3 (early collection time; Fig. 3, B–D). The expression of many genes was strongly affected by vernalization treatments, but there were also exceptions (e.g. VIN3 at early collection time). Similar to the relationships between flowering time and altitude, quadratic relationships between altitude and gene expression were identified for a number of genes (e.g. FT, where genotypes with the lowest expression originated from altitudes between 1,700 and 2,100 m.a.s.l.; Fig. 3C).
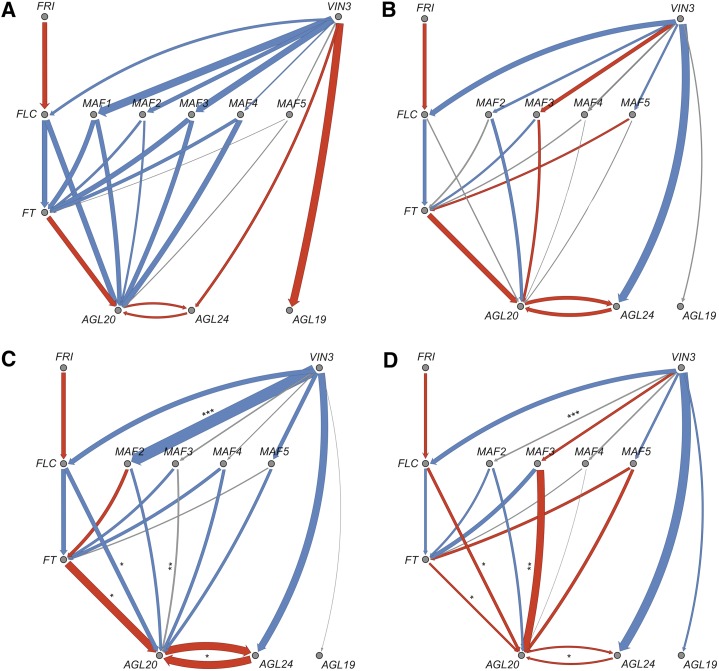
Regulatory processes of the vernalization pathway were examined by computing GRNs. A GRN computed for Col-0 and Col-FRI at late collection times demonstrated that the vernalization network in our study followed expectations from the literature (Fig. 4A). The same holds true for a GRN computed for all natural accessions of our study, with the exception of the VIN3-AGL24 edge, where our data showed negative correlations where positive correlations would be expected (Fig. 4B). However, when early expression data were correlated with late expression data, a positive relationship was found between VIN3 and AGL24 (Supplemental Fig. S4), suggesting that the earlier negative correlation may be an artifact of analyzing early and late data together. Further differences between natural genotypes and expectations from the literature based on laboratory accessions were edges connected to MAF3 (VIN3-MAF3 and MAF3-AGL20), which showed positive correlations in our analysis where negative correlations were expected (Gu et al., 2013; Kim and Sung, 2013). To test whether GRNs could be associated with altitude, GRNs were calculated separately for each genotype and the strength of each edge was then correlated with altitude of origin. The edge FT-AGL20 seemed to be significantly correlated with altitude (strong correlations for genotypes from low altitudes and no correlations at high altitudes); however, after correcting for multiple testing, the association of this edge with altitude was no longer significant (Supplemental Table S9). To increase statistical power, we computed GRNs for low-altitude and high-altitude genotypes separately and tested for differences between edges. Several edges differed significantly between the two groups, most prominently VIN3-MAF2 (strong negative correlation at low altitudes and no correlation at high altitudes) and MAF3-AGL20 (no correlation at low altitudes and strong positive correlation at high altitudes; Table V; Fig. 4, C and D).
Figure 4.
GRNs of the vernalization pathway with edges (correlation between two genes) as known from the literature. Edge width indicates the strength of correlation. Red edges indicate correlations with r > 0.1, and blue edges indicate correlations with r < −0.1. A, GRN for genotype Columbia (Col-0 and Col-FRI; expression data of FRI were set to 0 for Col-0, as FRI is nonfunctional in this genotype) at late collection time follows expectations from the literature. B, GRN for wild genotypes. Most edges follow expectations, except for the VIN3-AGL24 edge, where we find a negative correlation when a positive correlation would be expected (but compare with Supplemental Fig. S3). Another exception is MAF3, which interacts positively with both upstream VIN3 and downstream AGL20, when negative correlations would be expected for both. C, GRN for genotypes from low altitudes. Asterisks indicate that edges differed significantly from genotypes from high altitudes (*P < 0.05, **P < 0.01, and ***P < 0.001). D, GRN for genotypes from high altitudes. Asterisks indicate that edges differed significantly from genotypes from low altitudes.
Table V. Gene regulation differences between low- and high-altitude genotypes.
Shown are Pearson correlations between gene pairs (edges) of low- and high-altitude genotypes (Cor Low and Cor High, respectively; including the sample size n) and the differences between the edges of contrasting altitudes calculated with a z test.
| Edge | Cor Lown | Cor Highn | z | P | Adjusted Pa |
|---|---|---|---|---|---|
| VIN3-MAF2 | −0.71672 | 0.064109 | 6.232 | <0.001 | *** |
| MAF3-AGL20 | −0.08772 | 0.45985 | 3.568 | <0.001 | ** |
| FLC-AGL20 | −0.25172 | 0.170109 | 2.767 | 0.006 | * |
| AGL24-AGL20 | 0.48372 | 0.106109 | 2.718 | 0.007 | * |
| FT-AGL20 | 0.47572 | 0.116109 | 2.584 | 0.010 | * |
| MAF5-AGL20 | −0.17072 | 0.184109 | 2.314 | 0.021 | ns |
| MAF2-FT | 0.19272 | −0.117109 | 2.018 | 0.044 | ns |
| MAF4-AGL20 | −0.18372 | −0.002109 | 1.178 | 0.239 | ns |
| VIN3-AGL24 | −0.45172 | −0.558109 | 0.934 | 0.350 | ns |
| MAF4-FT | −0.18472 | −0.044109 | 0.917 | 0.359 | ns |
| FLC-FT | −0.27872 | −0.145109 | 0.897 | 0.370 | ns |
| VIN3-AGL19 | −0.00172 | −0.114109 | 0.738 | 0.461 | ns |
| VIN3-MAF5 | −0.23672 | −0.131109 | 0.703 | 0.482 | ns |
| MAF5-FT | 0.05472 | 0.154109 | 0.655 | 0.513 | ns |
| MAF3-FT | −0.15272 | −0.24585 | 0.592 | 0.554 | ns |
| FRI-FLC | 0.25972 | 0.172109 | 0.591 | 0.555 | ns |
| VIN3-MAF3 | 0.06972 | 0.13685 | 0.411 | 0.681 | ns |
| VIN3-MAF4 | −0.03272 | −0.073109 | 0.265 | 0.791 | ns |
| VIN3-FLC | −0.30572 | −0.273109 | 0.227 | 0.820 | ns |
| MAF2-AGL20 | −0.15672 | −0.139109 | 0.112 | 0.911 | ns |
P values were adjusted following Benjamini and Hochberg (1995): ***adjusted P < 0.001; **adjusted P < 0.01; *adjusted P < 0.05; and ns, adjusted P > 0.05.
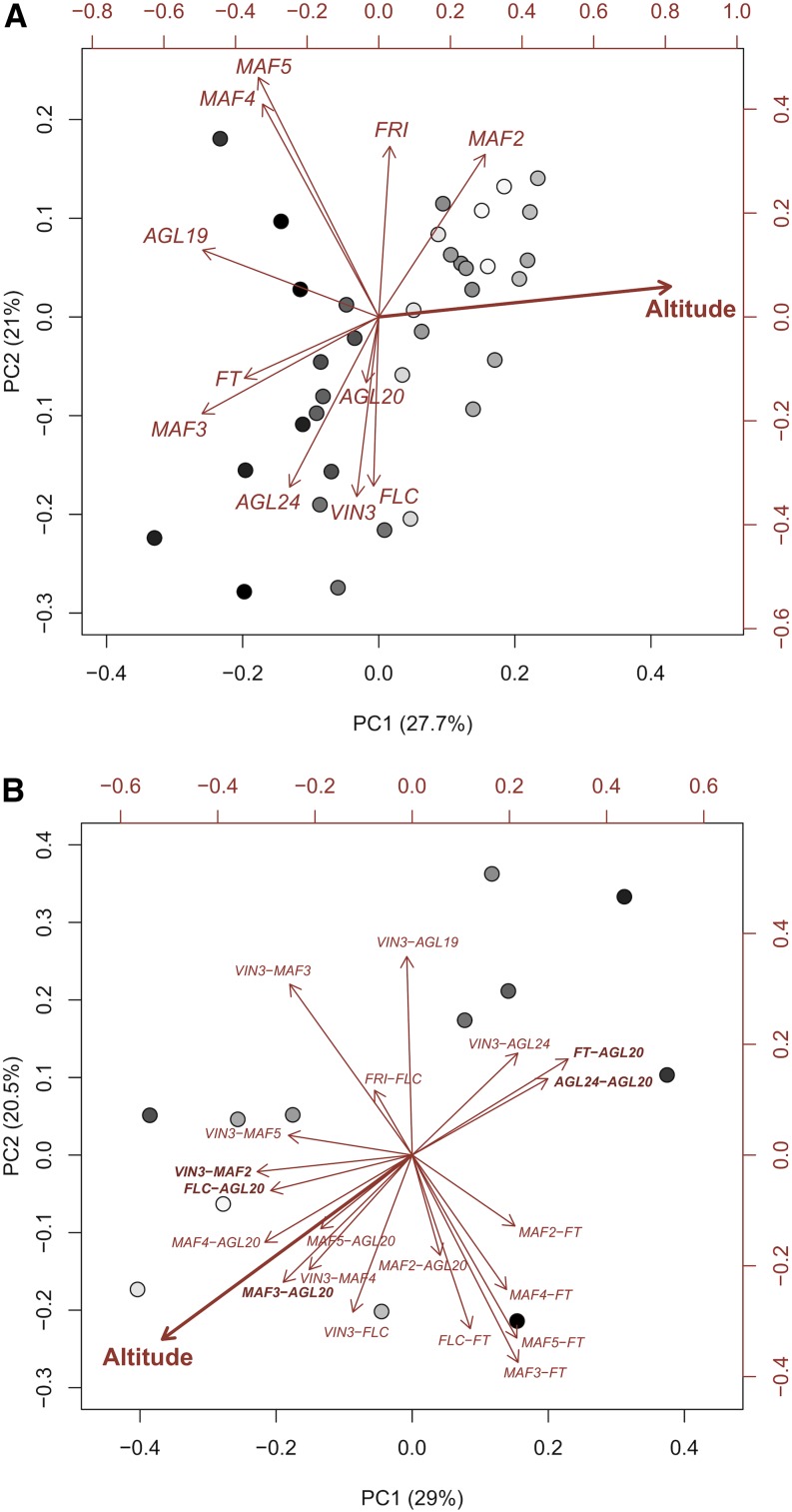
As several genes and edges of the vernalization pathway seemed to be involved in adaptation to altitude, we performed principal component analysis (PCA) to test whether the variance in the whole data set could be associated with altitude. PCAs including all genes were calculated separately for each vernalization treatment and collection time, and altitude of origin was then correlated with the first two principal components and added to the graphs. In all treatments, altitude correlated strongly with the first and/or second principal component, with the exceptions of the nonvernalized plants at early collection time (Fig. 5A; Supplemental Fig. S5). Similarly, temperature strongly correlated with the same first and/or second principal component as altitude, whereas the other environmental variables (precipitation, solar radiation, exposition, and slope) showed no consistent associations with either of the first principal components (an example is shown in Supplemental Fig. S6), corroborating that altitudinal effects are probably mainly due to the temperature cline. Similar to the phenotypic data, plants originating from the highest altitude (genotype E012) were not always the most extreme, sometimes grouping more closely to intermediate-altitude genotypes (e.g. after 3 weeks of vernalization, early collection time; Supplemental Fig. S5). Generally, high-altitude genotypes clustered more closely together, while low-altitude genotypes were more heterogenous. Of the FLC/MAF clade, only MAF4 and MAF5 consistently showed similar correlations with the first two principal components, indicating substantial differences in expression among genes in this gene family. A PCA was also calculated for the edges of the whole data set (Fig. 5B). Again, altitude strongly correlated with the first and second principal components, indicating that regulation of the vernalization pathway may be adjusted to altitude.
Figure 5.
A, PCA of the expression of 11 genes of the flowering pathway measured after 3 weeks of vernalization at late collection time. The gradation from light to dark indicates the altitudinal gradient from high to low. Altitude of origin strongly correlated with the first principal component (PC1; r = 0.814) and weakly with the second principal component (PC2; r = 0.059). B, PCA of 21 edges (correlation between gene pairs) of the vernalization pathway of all gene expression data combined (i.e. all vernalization treatments and collection times). The gradation from light to dark indicates the altitudinal gradient from high to low. Altitude of origin strongly correlates with the first principal component (PC1; r = −0.514) and the second principal component (PC2; r = −0.333).
DISCUSSION
Timing of Flowering
We found strong associations of flowering time with altitude of origin when 13 Arabidopsis genotypes from the Swiss Alps were vernalized, similar to recent studies conducted with populations from the Iberian peninsula (Méndez-Vigo et al., 2011) or the Pyrenees (Montesinos-Navarro et al., 2011). However, unlike the findings of Montesinos-Navarro et al. (2011), flowering time did not increase linearly with altitude in our study; instead, we observed a quadratic relationship: while there was a relatively strong increase in flowering time from low to intermediate altitudes, the genotypes from intermediate to high altitudes did not differ substantially. A quadratic relationship with altitude may reflect the action of two opposing selective pressures along an altitudinal gradient: the chances of encountering severe cold periods relatively late in the vegetation period increase with altitude, raising the risk of freezing during sensitive flowering stages (Kollas et al., 2013). As a consequence, natural selection is expected to favor later flowering genotypes at higher altitudes. However, the growing season becomes progressively shorter with increasing altitude. For annual plants in particular, this means that overly late flowering may not leave enough time to complete the life cycle before autumn frosts (Inouye and Wielgolaski, 2003; Chuine, 2010). Thus, selection may favor earlier flowering genotypes at high altitudes with strongly reduced vegetation periods. The results from our study suggest that the later flowering of intermediate-altitude genotypes relative to low-altitude genotypes may be an adaptation to the increasing risk of late freezing in spring, whereas the strongly reduced vegetation period at the highest collection sites may have reduced or even reversed the effect of delayed flowering for genotypes from the highest altitudes. It is important to note that our results do not contradict the linear correlations between flowering time and altitude reported by Montesinos-Navarro et al. (2011), as their populations originated from altitudes between 109 and 1,668 m, whereas the quadratic relationship in our experiment only became apparent when genotypes from altitudes between 1,500 and 2,700 m were included.
On a physiological level, the delayed flowering after vernalization observed in high-altitude genotypes may reflect either increased vernalization requirements (plants need longer vernalization periods to induce flowering efficiently) or, independent of vernalization duration, a slower induction of flowering after vernalization. The interactions between altitude and vernalization treatment observed in our experiment point toward an increased vernalization requirement for genotypes from high altitudes, because the longer the vernalization lasted, the smaller the differences in flowering time between the low- and high-altitude genotypes. Furthermore, synchronization of flowering time within genotypes increased with longer vernalization times, indicating that the short vernalization treatments (no vernalization and 3 weeks of vernalization) were insufficient to reliably induce flowering. However, despite a general acceleration of flowering with longer vernalization (compare with Shindo et al., 2006), overall, the effect of altitude on flowering time remained remarkably constant. Even after 12 weeks of vernalization, plants from low altitudes were clearly the earliest to flower. While vernalization duration may play some role in reliably inducing flowering in high-altitude genotypes, in general, these genotypes were always slower to induce flowering than low-altitude genotypes, suggesting an intrinsic delay in flowering after vernalization in comparison with low-altitude genotypes.
Without vernalization, variation in flowering time within genotypes was extensive, suggesting that an extended cold period is required in our genotypes to synchronize flowering. Furthermore, unlike the results of Méndez-Vigo et al. (2011), we found only a weak association of flowering time with altitude when nonvernalized plants were included in the analysis. However, we did not include genotypes from similarly low altitudes (0–600 m), while it was these populations that initiated flowering exceptionally fast in the study by Méndez-Vigo et al. (2011). In comparison, our lowland genotypes originated from 800 m and higher in the Swiss Alps, an altitude and latitude where winter is comparably harsh. Under natural conditions, all our genotypes experience vernalization, either as overwintering rosettes, as simulated in our experiment, or as imbibed seeds, which has been shown to have very similar effects to vernalization in the rosette stage (Lin et al., 2005); thus, conditions without vernalization are most likely not encountered naturally by our genotypes.
The effects of altitude on flowering time were very similar to the effects of average yearly temperature and other temperature parameters. While precipitation showed a weak quadratic association with altitude, we conclude that the effect of altitude can be explained to a large degree by decreasing temperature with increasing altitude. However, as the extrapolated environmental data might not reflect microclimatic conditions at collection sites (Scherrer and Körner, 2011), there may be other, unmeasured environmental factors contributing to adaptation to the altitudinal gradient.
The association of altitude with flowering time could not be observed when genotypes E001 and E003 were included. These genotypes had been excluded from the main data set due to different habitats and potentially uncertain altitudinal origins. Thus, altitudinal effects may be observable only when habitats along an altitudinal gradient are comparable and may be harder to detect if genotypes originate from a wider geographical range with more heterogenous habitats. Larger scale studies incorporating more genotypes from a wider geographical range, ideally from several local altitudinal gradients, would be required to disentangle habitat and altitude effects.
Gene Expression and Flowering Time
The expression and regulation of several genes belonging to the vernalization pathway were found to be associated with flowering time in the investigated natural genotypes. Both at early and late collection times, two genes were strongly associated with flowering time: FLC showed strong positive correlations, suggesting a delaying effect on flowering time, whereas AGL19 exhibited strong negative correlations, which may indicate an inducing effect on flowering. FLC is the usual suspect and has repeatedly been found to be an important regulator of flowering time (Caicedo et al., 2004; Shindo et al., 2006; Amasino, 2010), similar to its downstream targets FT and AGL20 (Liu et al., 2008; Schwartz et al., 2009). In our study, FT showed strong negative correlations with flowering time at late collection, suggesting a strong flowering-inducing effect, whereas a weaker negative association was observed for AGL20 at early collection time, with similar potential implications. In contrast, much less is known about the role of AGL19 in controlling flowering time. Interestingly, AGL19 has been shown to induce flowering upon vernalization independently of FLC (Schönrock et al., 2006; Alexandre and Hennig, 2008), and our findings indicate that it may indeed act as an important promoter of flowering in natural Arabidopsis populations. This suggests that both FLC-dependent and independent branches of the vernalization pathway act together to regulate flowering time in our wild genotypes.
Another FLC-independent gene associated with flowering time in our experiments was MAF2: at the early collection time, its expression correlated positively with flowering time, which would be expected from its described function as an inhibitor of flowering (Ratcliffe et al., 2003; Gu et al., 2013). On the other hand, at late collection time, there was a weak but significant negative correlation with flowering time, suggesting the promotion of flowering by MAF2. Posé et al. (2013) associated a splice variant of MAF1 (also known as FLM) that is expressed at low temperature with flowering inhibition, whereas another splice variant of MAF1 expressed at high temperature was associated with the activation of flowering. In MAF2, temperature-dependent splice variants have also been detected (Balasubramanian et al., 2006), and one may speculate whether the results reported here may reflect temperature-sensitive splice variants with opposing effects on flowering time. With our data, however, we cannot distinguish splice variants of MAF2; thus, testing this hypothesis would require further studies. Furthermore, the effect of MAF2 could be observed only in the models incorporating variances of flowering time and gene expression, and they disappeared when only mean values per genotype and treatment were used. Thus, further studies would first have to confirm that MAF2 has an effect on flowering time in the investigated genotypes.
At early collection time, FRI correlated negatively with flowering time, which is compatible with a role in the promotion of flowering time. This would contradict expectations from the literature (Johanson et al., 2000; Song et al., 2012). However, the expression of FRI generally may be difficult to interpret in our study, as our assay did not distinguish between functional and nonfunctional alleles. This is reflected in similar FRI expression levels in all genotypes, including Col-0, which is known to carry a nonfunctional FRI allele (Johanson et al., 2000).
When genotypes E001 and E003 were included in the analyses, the association of FLC with flowering time became less clear (the effect became nonsignificant when mean values of flowering time and gene expression were used in the model), whereas MAF4 newly showed positive correlations with flowering time, in particular at late collection times. This may indicate that, in addition to FLC, MAF4 also may contribute to flowering inhibition in these genotypes.
When analyzing the vernalization network using GRNs, several edges correlated significantly with flowering time. Among them, VIN3-FLC, FLC-FT, FT-AGL20, and AGL20-AGL24 all belonged to the well-established FLC-dependent branch of the vernalization pathway. Only the edge VIN3-AGL19 of the FLC-independent branch was significant, accentuating the importance of AGL19 in potentially promoting flowering in the investigated wild genotypes. Further studies will have to reveal whether AGL19 is indeed involved in the promotion of flowering in these genotypes and how widespread this function of AGL19 is in other natural populations.
Gene Expression and Altitude
Altitude of origin of our alpine genotypes was strongly associated with both gene expression and regulation of the vernalization pathway. Surprisingly, although FLC showed strong correlations with flowering time, we did not find consistent associations of FLC expression with altitude. Similarly, other studies that attempted to associate FLC with ecological and geographical parameters often failed to find simple patterns (Caicedo et al., 2004; Shindo et al., 2005; Méndez-Vigo et al., 2011). Although we found that AGL20 expression correlated negatively with FLC in genotypes from low but not from high altitudes, suggesting an altitude-dependent inhibition of AGL20 by FLC, our results indicate that, in addition, FLC-independent genes may be involved in the coordination of flowering time along our altitudinal cline.
AGL19 expression was strongly associated with altitude at both early and late collection times, with expression generally decreasing with increasing altitude and, thus, potentially inducing flowering more strongly in genotypes from low altitudes. Similarly, FT expression at late collection time was negatively associated with altitude, suggesting that both genes could be important not only in promoting flowering in these wild genotypes but also in fine-tuning flowering time along an altitudinal gradient. VIN3 could be a potential link between AGL19 and FT, as it is needed for AGL19 activation by vernalization (Schönrock et al., 2006) and indirectly activates FT by inhibiting FLC and the MAF genes. However, the VIN3-AGL19 edge was independent of altitude in our study; thus, either a so far unknown link between AGL19 and FT regulated the expression of both genes or two independent branches of the vernalization pathway contributed to the coordination of flowering time along the altitudinal gradient.
Interestingly, allelic variation in the promoter region of FT was recently found to affect FT expression and consequently flowering time (Schwartz et al., 2009; Strange et al., 2011). As FT is downstream of the vernalization pathway and further integrates signals from the photoperiod pathway and other environmental cues (Michaels, 2009), it was not possible with our data to disentangle whether FT itself was involved in adaptation to altitude. However, the expression of FT seemed to be carried farther downstream, as in low-altitude genotypes FT showed strong positive correlations with AGL20, which in turn positively interacted with AGL24, whereas in high-altitude genotypes, these interactions were much weaker. Thus, flowering may be initiated in a different way.
Although the VIN3-AGL19 edge was independent of altitude, the expression of VIN3 decreased with increasing altitude at early collection time. Furthermore, the correlation of VIN3 and MAF2 differed strongly between low- and high-altitude genotypes: at low altitudes, VIN3 showed strong negative correlations with MAF2, whereas no association between these genes was found at high altitudes. Ratcliffe et al. (2003) found that MAF2 was mainly involved in inhibiting flowering after comparably short periods of cold treatment. Low-altitude populations are at highest risk of warm periods during early winter, where initiation of flowering could prove fatal. Thus, MAF2 may be important in suppressing early flowering in low-altitude genotypes, whereas in high-altitude genotypes, this function may not be equally important.
Another edge involving a MAF gene, MAF3-AGL20, was also connected to altitude. However, contrary to expectations, in high-altitude genotypes, we found a strong positive correlation of MAF3 with the flowering promoter AGL20 and a weaker positive correlation with VIN3. This contradicts the emerging evidence that all MAF genes are involved in inhibiting flowering (Ratcliffe et al., 2003; Kim et al., 2009; Gu et al., 2013), as we would expect negative correlations of MAF3 both with VIN3 and with AGL20. In our study, the Columbia lines (Col-0 and Col-FRI) showed the expected negative correlations with VIN3 and AGL20, and only the natural genotypes deviated from expectations. A switch in MAF3 from flowering inhibitor to flowering promoter may seem at first unlikely. However, within the MAF clade, MAF2 and MAF3 have been found to be highly variable in Arabidopsis (Caicedo et al., 2009; Rosloski et al., 2010), which could facilitate a neofunctionalization of this potentially redundant gene clade (De Smet and Van de Peer, 2012; Wang et al., 2012). Interestingly, Rosloski et al. (2013) found that overexpression of a MAF2 variant interfered with endogenous FLC and MAF gene expression, leading to an acceleration of flowering instead of the expected inhibition. If high MAF3 expression could similarly interfere with the expression of FLC and other MAF genes in high-altitude genotypes, this could provide an explanation for the altered vernalization requirements of these genotypes. However, with our data, we cannot further explore this hypothesis, and additional molecular studies are needed to reveal the exact nature of the positive correlation of MAF3 with AGL20 and its role in regulating flowering time along an altitudinal gradient.
Taken together, low-altitude populations followed expectations from the literature much more consistently, with potential inhibition of MAF2 by VIN3 and AGL20 by FLC, strong positive interactions between FT-AGL20 and AGL20-AGL24, and strong induction of flowering by AGL19 and FT. At high altitudes, a thoroughly different way of activating AGL20 through MAF3 may lead to the required vernalization response, which may reflect the extreme environmental conditions in which these populations have to survive (Hardie and Hutchings, 2010). Most remarkable, though, seems the negligible role of FLC in the adjustment of flowering time along altitude. While MAF2 and MAF3 are known to vary extensively among natural populations (Caicedo et al., 2009; Rosloski et al., 2010) and have repeatedly been identified as important flowering time regulators in quantitative trait locus analyses using wild accessions (Salomé et al., 2011; Silady et al., 2011; Lasky et al., 2012; Fournier-Level et al., 2013; Grillo et al., 2013), the evolutionary properties of AGL19 and its distribution among natural populations will need further investigation.
Overall, the observed correlation of flowering time with altitude could not be attributed to a single gene or edge of the vernalization pathway. Rather, a complex, mainly FLC-independent interplay between gene expression and gene regulation seemed to underlie the observed adaptation to altitude. This was further supported by the PCA, which revealed that altitude was strongly associated with both the overall variance of gene expression and the variance of the edge strength in the flowering control GRN.
CONCLUSION
It has often been assumed that the various branches of the vernalization pathway are to some extent redundant and may provide robustness to the initiation of flowering (Alexandre and Hennig, 2008; Gu et al., 2013). We suggest that this redundancy enables natural populations to fine-tune their flowering time using the various branches of the vernalization pathway to respond and adapt to different environmental conditions. Thus, rather than the often expected antagonistic pleiotropy, where one allele is favorable in one environment and another allele of the same gene is beneficial in a different environment, we found a genetic phenomenon more similar to conditional neutrality, where one allele is beneficial in one environment and neutral in another and another allele of a different gene is beneficial in another environment (Colautti et al., 2012; Fournier-Level et al., 2013).
Previous studies have demonstrated that accurate timing of flowering can have direct fitness impacts (Pigliucci and Marlow, 2001; Volis et al., 2004; Ehrlén and Münzbergová, 2009; Chuine, 2010), suggesting an important role for flowering time in plant adaptation. Whether the observed association of altitude with flowering time, gene expression, and gene regulation truly reflects adaptation to altitude in our study remains to be tested further under natural conditions in the field, as climate chamber experiments cannot truly substitute for natural conditions (Mishra et al., 2012). However, the observed responses to the different vernalization scenarios revealed consistent results in terms of flowering time, and these could be corroborated in gene expression and gene regulation analyses. This study further revealed that studying natural accessions, in combination with standard laboratory lines, can provide novel insights into the genetic control of key life-history traits.
MATERIALS AND METHODS
Plant Material
We used second or third generation single-seed descendants of 15 natural Arabidopsis (Arabidopsis thaliana) genotypes originally collected in the Swiss Alps at altitudes ranging from 800 to 2,700 m (Fig. 1; Table I). Control genotypes were Col-0 and, as Col-0 carries a nonfunctional FRI allele and thus responds to vernalization only weakly (Johanson et al., 2000), Col-FRI, which carries a functional FRI from the San Feliu accession in a Columbia background (Lee et al., 1993; Lee and Amasino, 1995).
Experimental Design
Four different treatments were applied to test the effects of vernalization on the different genotypes: no vernalization and 3, 6, and 12 weeks of vernalization (Fig. 2). Plants were grown in individual 7- × 7- × 8-cm pots on Bio-Universalerde (Oekohum), an all-purpose soil without peat. On average, five seeds per pot were sown and stratified for 1 week at 4°C in the dark to break seed dormancy. Then, the pots were transferred to warm and short-day conditions (22°C/20°C, 8-h/16-h day/night) for a first growth period of 2 weeks. During this time, excessive seedlings were removed to leave one plant per pot. Plants of the three vernalization treatments were then transferred to cold and short-day conditions (4°C, 8-h/16-h day/night) to be vernalized for 3, 6, and 12 weeks, respectively. After the vernalization treatment (or after the short-day growth period for plants without vernalization), all plants were transferred to warm and long-day conditions (22°C/20°C, 16-h/8-h day/night) until all plants started to flower. The whole experiment was conducted in climate chambers, light conditions were 120 µmol s−1 m−2/0 µmol s−1 m−2 day/night, and relative humidity was 50%/60% day/night. Plants were watered once per week with tap water containing Solbac (Andermatt Biocontrol) diluted according to the manufacturer’s instructions to avoid insect infestation in the soil. Within each treatment, all pots were arranged randomly on 28-pot trays, and throughout the experiment, the positions of the trays were randomized three times per week to avoid position effects.
To measure flowering time, nine plants per treatment per genotype were grown (i.e. 612 plants in total). The first day a flower bud opened was recorded as the day of first flowering. To compare flowering times between treatments, vernalization time was subtracted from total days until flowering (i.e. flowering time was calculated as days each plant grew in warm conditions until first flowering). To collect plant material for molecular analyses, six plants per treatment and genotype were grown as described (i.e. an additional 408 plants). Three whole plants were collected at the last day of the vernalization treatment or at the last day of the short-day conditions for nonvernalized plants (time A in Fig. 2). Ten days later (i.e. in warm and long-day conditions), three more plants each were collected (time B in Fig. 2). All plants were collected shortly before end-of-day conditions, as at this time many genes of the flowering pathway are most active (Imaizumi and Kay, 2006). Only aboveground material was collected and immediately frozen in liquid nitrogen. Plant material was then stored at −80°C until RNA was extracted.
Molecular Analyses
RNA was extracted for each collected plant individually using the RNeasy Plant Mini Kit (Qiagen) coupled with on-column DNA digestion using the RNase-Free DNase Set (Qiagen). For a subset of 38 samples, RNA quantity and integrity were analyzed using the Agilent 2100 Bioanalyzer and RNA 6000 Nano Kit (Agilent). RNA was reverse transcribed to complementary DNA using Moloney murine leukemia virus reverse transcriptase (Promega). For all RNA extractions, control samples without reverse transcription were tested for genomic DNA contamination using the 7500 FAST Real Time PCR System (Applied Biosystems; software version 2.0.1), primers for ACTIN2 (Supplemental Table S1), and the Universal Probe Library (Roche; Supplemental Table S1). The expression of 12 genes of the flowering pathway plus four reference genes for expression normalization was analyzed for all samples (Supplemental Table S1) using the Universal Probe Library (Roche) and 10 48.48 Dynamic Array Integrated Fluidic Circuits (Fluidigm). We followed the manufacturer’s instructions with the exception of replacing TaqMan Universal PCR Master Mix with TaqMan Gene Expression Master Mix (Applied Biosystems). Each gene was replicated three times per 48.48 array, providing three technical replicates per gene and sample. Three samples were analyzed on each of the 10 48.48 arrays to allow for interrun calibrations. Additionally, two nontemplate controls were included on each array.
Data Normalization
Data were exported from Fluidigm Real-Time PCR Analysis version 3.1.3 with quality threshold set as 0.65, linear baseline correction, and the cycling threshold method as Auto (Global). Primer efficiency was estimated using LinReg version 12.16, importing the data as baseline corrected. Mean values of all three technical replicates of two 48.48 assays per gene were calculated (excluding nontemplate controls [i.e. the mean of 3 × 2 × 45 = 270 samples per gene]; Supplemental Table S1). Data were further analyzed using qbase PLUS version 2.3. Interrun calibration was calculated using three samples for nine arrays and two samples for one array due to missing data to take array effects into account. GeNorm implemented in qbase PLUS was used to identify the most stable reference genes, and consequently, S-ADENOSYLMETHIONINE SYNTHETASE2 was excluded from all further analyses. Gene expression intensity was normalized against the mean expression of Col-FRI over all treatments. Technical replicates were allowed to vary up to 1.5 cycling threshold values. If technical replicates were more diverse, the most aberrant values were manually excluded. Mean values of the three technical replicates of each sample were exported and analyzed using R (R Development Core Team, 2010).
Statistical Analysis
All statistical analyses were conduced using R (R Development Core Team, 2010). E001, the genotype from the lowest altitude, flowered consistently late in comparison with all other genotypes (Supplemental Fig. S1A) and also expressed genes differently when compared with other genotypes (Supplemental Fig. S1B). This genotype originated from a geographically very distinct area (north of the Alps versus central alpine valleys); thus, climatic conditions of the habitat differed strongly in comparison with other lowland genotypes (Supplemental Table S10). Furthermore, this genotype was collected in immediate proximity of an alpine river, which may indicate that seeds have been washed down from a higher altitude. Thus, we decided to exclude this genotype from most analyses. Similarly, E003, another lowland genotype, showed strongly delayed flowering and different gene expression after 3 weeks of vernalization in comparison with other genotypes (Supplemental Fig. S1). Although after 6 and 12 weeks of vernalization the flowering time and gene expression were similar to other genotypes, we decided to exclude E003 based on the habitat where it was originally collected: similar to E001, E003 was also collected in a river valley. Furthermore, the habitat was facing north; thus, solar radiation (Supplemental Table S10), snow cover, humidity, and temperature conditions were likely to be different from all other sites. Additionally, the proximity to an alpine river might again make it problematic to assess adaptation to altitude. Most analyses were thus computed using the 13 remaining genotypes.
To assess the effect of altitude on the time to flowering, a linear mixed-effects model was used with altitude (linear and quadratic terms) and treatment as well as their interactions as fixed and genotype as random factors (R function lme in package nlme). Heteroscedasticity between treatments was accounted for by using the weights argument. As synchronization of flowering time within genotypes was very low for nonvernalized plants, vernalized plants were subsequently analyzed separately. To test whether both linear and quadratic terms of altitude remained significant without the genotype from the highest altitude, E012, a model without E012 was calculated. To test the effect of altitude on all natural genotypes independent of habitat differences, the same model was calculated with E001 and E003 included.
To test whether environmental factors correlated with flowering time, interpolated geographic information system data (ArcMap 10 [Esri]) were used to extract climatic variables such as temperature (average, minimal, and maximal temperatures during winter, summer, and the whole year), yearly precipitation, yearly solar radiation, habitat slope, and exposition (Supplemental Table S10). These climatic factors have been collected over 30 years (1961–1990) at 25-m resolution (Zimmermann and Kienast, 2009). The variables were tested separately for effects on flowering time using linear mixed-effects models as described above.
To assess whether the synchronization in flowering time was altered with increasing vernalization time, the variance of flowering time for each genotype and treatment was calculated, then pairwise Wilcoxon signed rank tests were performed between treatments. P values were subsequently adjusted for multiple testing following Benjamini and Hochberg (1995).
Among the 12 studied genes of the flowering pathway, MAF1 was the only gene for which expression could be detected only in a subset of the 13 genotypes, namely in E002, E004, E016, and E017. Therefore, we excluded MAF1 from all further analyses. Similarly, detected MAF3 levels were consistently extremely low in genotype E017, making it unclear whether this gene was truly lowly expressed in E017 or whether there was a technical amplification problem for MAF3 in this genotype; thus, E017 was excluded from MAF3 analyses. When Col-0 was included in the analyses, expression values for FRI were set to 0, as this gene is nonfunctional in Col-0 (Johanson et al., 2000).
To test whether gene expression in our experiments followed expectations from the literature, a GRN analysis calculating edges (Pearson correlations between gene pairs) of the vernalization pathway known from the literature was computed using the Columbia genotypes (Col-0 and Col-FRI). The same analysis was performed using the chosen genotypes originating from the Swiss Alps. Furthermore, a GRN was calculated using upstream gene expression from the early collection time and downstream gene expression from the late collection time.
All analyses studying the effect of gene expression on flowering time were performed using vernalized plants only, as in nonvernalized plants, synchronization of flowering was very low. To incorporate variation in flowering date within genotypes and vernalization treatments, the up to nine flowering dates per genotype and treatment were randomly assigned to three groups, and the mean values of these groups were then randomly allocated to the respective three gene expression data set per collection time, genotype, and vernalization treatment. To test whether flowering time could be explained by the expression of genes of the vernalization pathway, a linear mixed-effects model with all genes as fixed and genotypes as random factors was calculated separately for early and late collection times. The same tests were performed using mean flowering times and mean gene expression values for each genotype and vernalization treatment. These tests were also performed using all natural genotypes, including E001 and E003. However, as the gene expression values for E003 after 3 weeks of vernalization were too different from all other values and, thus, could not be fitted in any model including all genotypes, these values again had to be excluded from the analyses.
Additionally, edges (Pearson correlations between gene pairs) of the vernalization pathway known from the literature were calculated separately for each genotype and vernalization treatment and then correlated with average flowering times to test whether gene regulation could explain flowering time. This was calculated both for the chosen genotypes as well as all genotypes (including E001 and E003).
The effect of altitude on gene expression was calculated exclusively for the chosen genotypes. Linear mixed-effects models were calculated separately for each gene and collection time with altitude (linear and quadratic terms), vernalization treatment (excluding nonvernalized plants), and their interactions as fixed and genotype as random factors. Additionally, known edges were calculated separately for each genotype and then correlated with altitude using the full data set (i.e. vernalized and nonvernalized plants). To increase statistical power, edges of low-altitude genotypes (altitudinal range from 850 to 1,030 m.a.s.l. [i.e. genotypes E002, E004, and E005]) were compared with edges of high-altitude genotypes (altitudinal range from 1,850 to 2,700 m.a.s.l. [i.e. E009, E017, E011, E007, and E012]) using a z-test (R package psych, function r.test).
To test whether altitude had an effect on gene expression of the whole vernalization network, PCAs were calculated for each treatment and collection time. Altitude of origin as well as five environmental variables (average yearly temperature, yearly precipitation, yearly solar radiation, exposition, and slope) were then correlated with the first two principal components and added to the graph. Similarly, a PCA was calculated for the effect of the edges of the vernalization pathway, calculating the edges separately for each genotype using the whole data set (i.e. all vernalization treatments and collection times together). Again, altitude of origin was then correlated with the first two principal components and added to the graph. When necessary, P values were adjusted for multiple testing following Benjamini and Hochberg (1995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Days until flowering and expression of FT at late collection plotted against altitude of origin including genotypes E001 and E003.
Supplemental Figure S2. Days until flowering plotted against altitude including nonvernalized plants.
Supplemental Figure S3. Days until flowering plotted against altitude of vernalized plants excluding the genotype from the highest altitude, E012.
Supplemental Figure S4. GRN of early expression data with late expression data.
Supplemental Figure S5. PCAs of the gene expression of the vernalization pathway separately for each treatment and collection time.
Supplemental Figure S6. PCA of the gene expression of the vernalization pathway including environmental variables.
Supplemental Table S1. Effect of altitude on flowering date, including nonvernalized plants.
Supplemental Table S2. Effect of altitude on flowering date of vernalized plants without the genotype from the highest altitude, E012.
Supplemental Table S3. Effect of altitude on flowering date of vernalized plants including genotypes E001 and E003.
Supplemental Table S4. Effect of yearly average temperature on flowering date of vernalized plants.
Supplemental Table S5. Genes, primers, and primer efficiency used in this study.
Supplemental Table S6. Effect of gene expression on flowering time including genotypes E001 and E003.
Supplemental Table S7. Effect of gene regulation on flowering time including genotypes E001 and E003.
Supplemental Table S8. Effect of altitude on gene expression of vernalized plants.
Supplemental Table S9. GRN analysis correlated with altitude of origin.
Supplemental Table S10. Environmental parameters for each original genotype.
Supplementary Material
Acknowledgments
We thank Maja Frei for taking care of the plants, Claudia Michel for technical support, Karsten Rohweder for help with Figure 1 and the extraction of the climatic data, Cristina Madeira Alexandre and Yvonne Steinbach for help with the Universal Probe Library and the primers, and the St. George’s University of Grenada for providing L.S. with a work space. We also thank Aria Minder, Tania Torossi, Stefan Zoller, Jean-Claude Walser, and the Eidgenössisch Technische Hochschule Zürich Genetic Diversity Center for technical support. Finally, we thank two anonymous reviewers for excellent comments on an earlier version of this article.
Glossary
- Col-0
Columbia-0
- Col-FRI
a functional FRIGIDA allele in a Columbia background
- GRN
gene regulatory network
- PCA
principal component analysis
Footnotes
This work was supported by the Competence Center Environment and Sustainability of the Eidgenössisch Technische Hochschule Domain in the framework of the GeneMig project.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alexandre CM, Hennig L (2008) FLC or not FLC: the other side of vernalization. J Exp Bot 59: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Byars SG, Papst W, Hoffmann AA (2007) Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution 61: 2925–2941 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Richards C, Ehrenreich IM, Purugganan MD (2009) Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2-5 flowering time gene cluster. Mol Biol Evol 26: 699–711 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004) Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I. (2010) Why does phenology drive species distribution? Philos Trans R Soc Lond B Biol Sci 365: 3149–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colautti RI, Lee CR, Mitchell-Olds T (2012) Origin, fate, and architecture of ecologically relevant genetic variation. Curr Opin Plant Biol 15: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Dean C (2011) Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol 14: 38–44 [DOI] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Van de Peer Y, Theissen G (2003) And then there were many: MADS goes genomic. Trends Plant Sci 8: 475–483 [DOI] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y (2012) Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15: 168–176 [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD (2009) Candidate gene association mapping of Arabidopsis flowering time. Genetics 183: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlén J, Münzbergová Z (2009) Timing of flowering: opposed selection on different fitness components and trait covariation. Am Nat 173: 819–830 [DOI] [PubMed] [Google Scholar]
- Etterson JR. (2004) Evolutionary potential of Chamaecrista fasciculata in relation to climate change. I. Clinal patterns of selection along an environmental gradient in the great plains. Evolution 58: 1446–1458 [DOI] [PubMed] [Google Scholar]
- Fischer M, Weyand A, Rudmann-Maurer K, Stoecklin J (2011) Adaptation of Poa alpina to altitude and land use in the Swiss Alps. Alp Bot 121: 91–105 [Google Scholar]
- Fischer MC, Rellstab C, Tedder A, Zoller S, Gugerli F, Shimizu KK, Holderegger R, Widmer A (2013) Population genomic footprints of selection and associations with climate in natural populations of Arabidopsis halleri from the Alps. Mol Ecol 22: 5594–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Wilczek AM, Cooper MD, Roe JL, Anderson J, Eaton D, Moyers BT, Petipas RH, Schaeffer RN, Pieper B, et al. (2013) Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol Ecol 22: 3552–3566 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo-Turpin H, Hazard L (2009) Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J Ecol 97: 742–751 [Google Scholar]
- Grillo MA, Li C, Hammond M, Wang L, Schemske DW (2013) Genetic architecture of flowering time differentiation between locally adapted populations of Arabidopsis thaliana. New Phytol 197: 1321–1331 [DOI] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DC, Hutchings JA (2010) Evolutionary ecology at the extremes of species’ ranges. Environ Rev 18: 1–20 [Google Scholar]
- Hodgins KA, Lai Z, Nurkowski K, Huang J, Rieseberg LH (2013) The molecular basis of invasiveness: differences in gene expression of native and introduced common ragweed (Ambrosia artemisiifolia) in stressful and benign environments. Mol Ecol 22: 2496–2510 [DOI] [PubMed] [Google Scholar]
- Hodgins-Davis A, Townsend JP (2009) Evolving gene expression: from G to E to GxE. Trends Ecol Evol 24: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Inouye DW, Wielgolaski FE (2003) High altitude climates. InSchwartz MD, ed, Phenology: An Integrative Environmental Science. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 195–214 [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Keller I, Alexander JM, Holderegger R, Edwards PJ (2013) Widespread phenotypic and genetic divergence along altitudinal gradients in animals. J Evol Biol 26: 2527–2543 [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollas C, Körner C, Randin CF (2013) Spring frost and growing season length co-control the cold range limits of broad-leaved trees. J Biogeogr 41: 773–783 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Körner C. (2007) The use of ‘altitude’ in ecological research. Trends Ecol Evol 22: 569–574 [DOI] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH (2012) Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol Ecol 21: 5512–5529 [DOI] [PubMed] [Google Scholar]
- Lee I, Amasino RM (1995) Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol 108: 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Bleecker A, Amasino R (1993) Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet 237: 171–176 [DOI] [PubMed] [Google Scholar]
- Leger EA, Rice KJ (2007) Assessing the speed and predictability of local adaptation in invasive California poppies (Eschscholzia californica). J Evol Biol 20: 1090–1103 [DOI] [PubMed] [Google Scholar]
- Lin SI, Wang JG, Poon SY, Su CL, Wang SS, Chiou TJ (2005) Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol 137: 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Manel S, Poncet BN, Legendre P, Gugerli F, Holderegger R (2010) Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Mol Ecol 19: 3824–3835 [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C (2011) Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol 157: 1942–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra Y, Jänkänpää HJ, Kiss AZ, Funk C, Schröder WP, Jansson S (2012) Arabidopsis plants grown in the field and climate chambers significantly differ in leaf morphology and photosystem components. BMC Plant Biol 12: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Pico FX, Tonsor SJ (2011) Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol 189: 282–294 [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Graciet E, Wellmer F (2014) Gene networks controlling Arabidopsis thaliana flower development. New Phytol 201: 16–30 [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Marlow ET (2001) Differentiation for flowering time and phenotypic integration in Arabidopsis thaliana in response to season length and vernalization. Oecologia 127: 501–508 [DOI] [PubMed] [Google Scholar]
- Poncet BN, Herrmann D, Gugerli F, Taberlet P, Holderegger R, Gielly L, Rioux D, Thuiller W, Aubert S, Manel S (2010) Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina. Mol Ecol 19: 2896–2907 [DOI] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB, Wright IJ, Cavender-Bares J, Craine JM, Oleksyn J, Westoby M, Walters MB (2003) The evolution of plant functional variation: traits, spectra, and strategies. Int J Plant Sci 164: S143–S164 [Google Scholar]
- Rosloski SM, Jali SS, Balasubramanian S, Weigel D, Grbic V (2010) Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics 186: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski SM, Singh A, Jali SS, Balasubramanian S, Weigel D, Grbic V (2013) Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Mol Biol 81: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RA, Yant L, Mott R, Weigel D (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D, Körner C (2011) Topographically controlled thermal-habitat differentiation buffers alpine plant diversity against climate warming. J Biogeogr 38: 406–416 [Google Scholar]
- Schmitz RJ, Amasino RM (2007) Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim Biophys Acta 1769: 269–275 [DOI] [PubMed] [Google Scholar]
- Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, Hennig L (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, Kobayashi Y, Maloof JN, Borevitz JO, Chory J, et al. (2009) Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183: 723–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K, Michaels SD, Amasino RM (2003) Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol 52: 915–922 [DOI] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Silady RA, Effgen S, Koornneef M, Reymond M (2011) Variation in seed dormancy quantitative trait loci in Arabidopsis thaliana originating from one site. PLoS ONE 6: e20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C (2012) Vernalization: a cold-induced epigenetic switch. J Cell Sci 125: 3723–3731 [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Caicedo AL, Hopkins R, Mays C, Boyd EW, Purugganan MD, Schmitt J (2005) Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am J Bot 92: 1701–1707 [DOI] [PubMed] [Google Scholar]
- Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C (2011) Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6: e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM (2006) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volis S, Verhoeven KJF, Mendlinger S, Ward D (2004) Phenotypic selection and regulation of reproduction in different environments in wild barley. J Evol Biol 17: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang X, Paterson AH (2012) Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci 1256: 1–14 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL (2010) Gene networks controlling the initiation of flower development. Trends Genet 26: 519–527 [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL (2006) Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA 103: 5425–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann NE, Kienast F (2009) Predictive mapping of alpine grasslands in Switzerland: species versus community approach. J Veg Sci 10: 469–482 [Google Scholar]
- Zografos BR, Sung S (2012) Vernalization-mediated chromatin changes. J Exp Bot 63: 4343–4348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.